- 1Institute of Animal Science, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Precision Livestock and Nutrition Unit, Gembloux Agro-Bio Tech, TERRA Teaching and Research Centre, University of Liège, Gembloux, Belgium
- 3State Key Laboratory of Animal Nutrition, Beijing, China
Salmonella enteritidis (SE) is a foodborne pathogen that negatively affects both animal and human health. Controlling poultry SE infection will have great practical significance for human public health, as poultry are considered to be important sources and carriers of the disease. In this study, the splenic transcriptomes of challenged-susceptible (S), challenged-resistant (R) and non-challenged (C) chicks (3-days old, specific-pathogen-free White Leghorn) were characterized in order to identify the immune-related gene markers and pathways. A total of 934 significant differentially expressed genes (DEGs) were identified in comparisons among the C, R and S birds. First reported here, the DEGs involved in the Forkhead box O (FoxO) signaling pathway, especially FoxO3, were identified as potential markers for host resistance to SE infection. The challenged-susceptible birds exhibited strong activation of the FoxO signaling pathway, which may be a major defect causing immune cell apoptosis as part of SE-induced pathology; these S birds also showed weak activation of mitogen-activated protein kinase (MAPK)-related genes, contrasting with strong splenic activation in the R birds. Interestingly, suppression of several pathways in the immune response against Salmonella, including cytokine-cytokine receptor interaction and Jak-STAT, was only found in S birds and there was evidence of cross-talk among these pathways, perhaps contributing to susceptibility to Salmonella infection. These findings will help facilitate understanding resistance and susceptibility to SE infection in the earliest phases of the host immune response through Salmonella-induced pathways, provide new approaches to develop strategies for SE prevention and treatment, and may enhance innate resistance by genetic selection in animals.
Background
Salmonella enteritidis (SE) is an enteric bacterium that can colonize chickens, contaminating meat and eggs; it does not cause production losses, but birds carry a bacterial burden with non-obvious symptoms, thereby constituting an insidious risk for public health (Barrow et al., 2012; Calenge and Beaumont, 2012; Kogut and Arsenault, 2017). SE is among the top ranking food-borne pathogens causing huge economic and human life losses. Poultry are considered to be important sources and carriers of the disease. Although use of appropriate control measures can reduce Salmonella contamination in poultry, Salmonella cases continue (Inns et al., 2015). Control of SE, therefore, is highly desirable from the perspective of both animal and human health. In recent years, genetic selection of birds is considered to be an efficient and permanent way to control Salmonella infection (Berthelot et al., 1998; Kaiser and Lamont, 2001; Gou et al., 2012; Kogut et al., 2012; Li et al., 2017b). A better understanding of host immunological response mechanisms should be given priority in achieving this goal.
The main route of SE infection is the oral intake of contaminated feed or water. From the intestinal tract, SE can quickly enter the bloodstream and colonize the internal organs including liver, spleen and heart (Chappell et al., 2009). The spleen plays a major role in detecting cell damage during bacterial infection and in the pathogenic mechanisms of bacterial clearance (Altamura et al., 2001). Increasing evidence suggests that the spleen plays a greater role in immune function in avian than in mammalian species, and is responsible for an immediate immune reaction after recognizing pathogens by filtering antigens from the blood (Smith and Hunt, 2004; Li et al., 2017a).
Although there have been several previous studies focusing on the splenic transcriptome following infection with Salmonella enterica (Zhou and Lamont, 2007; Matulova et al., 2012), avian pathogenic Escherichia coli (APEC) (Sandford et al., 2011; Nie et al., 2012) and virus (Wang et al., 2006; Haq et al., 2010; Smith et al., 2011), little is known about immune-related genes and pathways between resistant and susceptible birds during the course of SE pathogenesis. This paper identifies genes and pathways that are differently expressed in susceptible versus resistant chickens, after challenge with SE, to aid understanding of host immune resistance to SE infection; an earlier report (Li et al., 2017a) presented differences that were apparent at the miRNA level.
Materials and Methods
Ethics Statement
All animal care and experimental procedures were approved by the Institute of Animal Sciences, Chinese Academy of Agricultural Sciences (approval number: IASCAAS-AE20140615).
Animals and Sample Collection
Specific-pathogen-free chicks (White Leghorn) were obtained from the Beijing Laboratory Animal Research Center and were treated as described earlier (Gou et al., 2012; Li et al., 2017a). Groups of 30 SE-challenged chicks were initially screened at 0.5, 1, 2, 4, 6, and 8-days post infection at 3 days of age; 24-h post infection was found to be optimal for showing differences (clinical symptoms and bacterial burden) between the 3 groups to best expose potential differences in mRNA expression. The challenged-susceptible (S) chicks exhibited severe clinical symptoms (diarrhea, drooping wings and dying) and higher bacterial loads (>107 cfu/10 μL blood) compared with the others. Chicks with only slight clinical symptoms and lower bacterial loads (<105 cfu/10 μL blood) were identified as challenged-resistant (R) birds. Six challenged chickens conforming to the requirements (3 R and 3 S) were selected from the 30 chickens sampled at 24 h. No Salmonella were detected in the PBS-challenged chicks and 3 were randomly chosen from 15 chicks as controls (C) at same time-point. As shown in Supplementary Figure S1, the number of SE (log10 cfu) measured in blood or spleen were closely related (R2 = 0.892, n = 30). The bacterial burden in blood of S chicks exceeded that in R chicks (Supplementary Figure S2, P < 0.01).
RNA Extraction, cDNA Library Preparation, and RNA Sequencing
Total splenic RNA was extracted from each of the 9 birds, using RNeasy Plus Micro Kit (74034) (Qiagen, Hilden, Germany) following the manufacturer’s protocol. The total RNA quantity was evaluated using Bioanalyzer 2100 and RNA Integrity Number (RIN) scores exceeding 8.0. For each sample, approximately 3 μg of total RNA was depleted of ribosomal RNA (Epicentre Ribo-Zero Gold Kit, Illumina, San Diego, CA, United States). Following purification, the RNA fractions were broken into small pieces using divalent cations at high temperature. And the final cDNA library was generated using reverse transcription amplification of cleaved RNA fragments. Sample Preparation Kit (Illumina, San Diego, CA, United States), and paired-end sequencing was performed on an Illumina Hiseq2000 by LC-BIO (LC Sciences, Houston, TX, United States) and 100 bp paired-end reads were generated. Quality control of reads was determined by FastQC software (v0.10.1), details of which were described earlier (Li et al., 2017a). In brief, clear data were obtained from the raw reads, eliminating contamination with sequencing adapters or poly-N and low quality reads (Q values < 20), along with potential residual ribosome RNA. Clean reads were aligned to the reference genome (Gallus gallus 4.0) database using TopHat (Trapnell et al., 2009) software (v2.0.9) and Bowtie (Langmead and Salzberg, 2012) (v2.0.0), and the mapped transcripts were assembled de novo using Cufflinks (Trapnell et al., 2010). The RNA-seq data can be obtained from the BIG Data Center Members (2017) database with the accession number CRA000463.
Differentially Expressed Genes and Function Enrichment Analysis
Fragments per kilobase of exon per million mapped reads (FPKM) was used to quantify the abundance of mRNAs using the Cufflinks package (v2.1.1). Analysis of DEGs between the 3 groups of chickens was performed with a false discovery rate (FDR < 0.1), P < 0.05, and |fold change| >1.5. The DEGs were used to implement GO and Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis using KOBAS (Xie et al., 2011) (v3.0) and DAVID (Dennis et al., 2003) (v6.8). Volcano plots were performed using OmicShare tools1. The normalized read counts of some mRNAs were set to be 0.01 for further calculation if they had no reads in the library.
Validation of DE Genes by Quantitative Real-Time PCR (qPCR)
To validate the DEGs identified by RNA-seq, qPCR analyses were performed to measure transcript abundance of 16 selected genes (IL10RB, TNFSF10, LAMP1, ZNF207, CCND1, GJA1, FTH, HBBA, GAL1, CREBBP, BRI3BP, SOCS1, ICOS, CTLA4, AVD, and IL8) in an ABI 7500 Detection System (Applied Biosystems, Foster, CA, United States). The candidate genes were selected for their involvement in multiple immune response pathways and their levels of differential expression (high, mean FPKM > 1000; middle, 70 < mean FPKM < 200; low, mean FPKM < 50) in the RNA-seq analysis. cDNA was obtained from the same individual samples used in RNA-seq. The qPCR amplification was as follows: each qPCR reaction (20 μl), run in triplicate, consisted of either 1 μl of template cDNA, 10 μl of 2 × KAPA SYBR FAST qPCR Master Mix (Roche, Shanghai, PRC), 0.4 μl ROX Low, 0.5 μl of each primers, and 7.6 μl PCR-grade water. The qPCR program was performed following the instructions of ABI 7500 with default parameters. 2-ΔΔCt method (Livak and Schmittgen, 2001) was used to calculate the relative mRNA abundance. β-actin was used as the housekeeping gene and all primers of examined genes are described in Supplementary Table S5. Three independent replications were used for each assay and data are presented as means ± SD. Student’s t-test was used to compare the different expression of genes in each comparison and P < 0.05 was considered to be statistically significant.
Results
Sequencing of Splenic Transcriptomes
Next generation sequencing of splenic samples collected at 24-h post-infection produced minimum amount of 11G raw data for each of the 9 libraries. Around 95% of the clean reads had quality scores exceeding the Q 20 value. After removing the interference data, an average of 71.4% high quality reads was mapped to the chicken reference genome Gallus gallus 4 (see Supplementary Table S1). Among the total of 15,278 detected genes, 1,666 were novel and 11,169 genes were considered in further statistical analysis. Volcano plots, integrating both the P-value and fold-change of each transcript were constructed, to show the general scattering of the transcripts and filter the DEGs between the S vs. C, R vs. C and S vs. R, comparisons (Figures 1A–C).
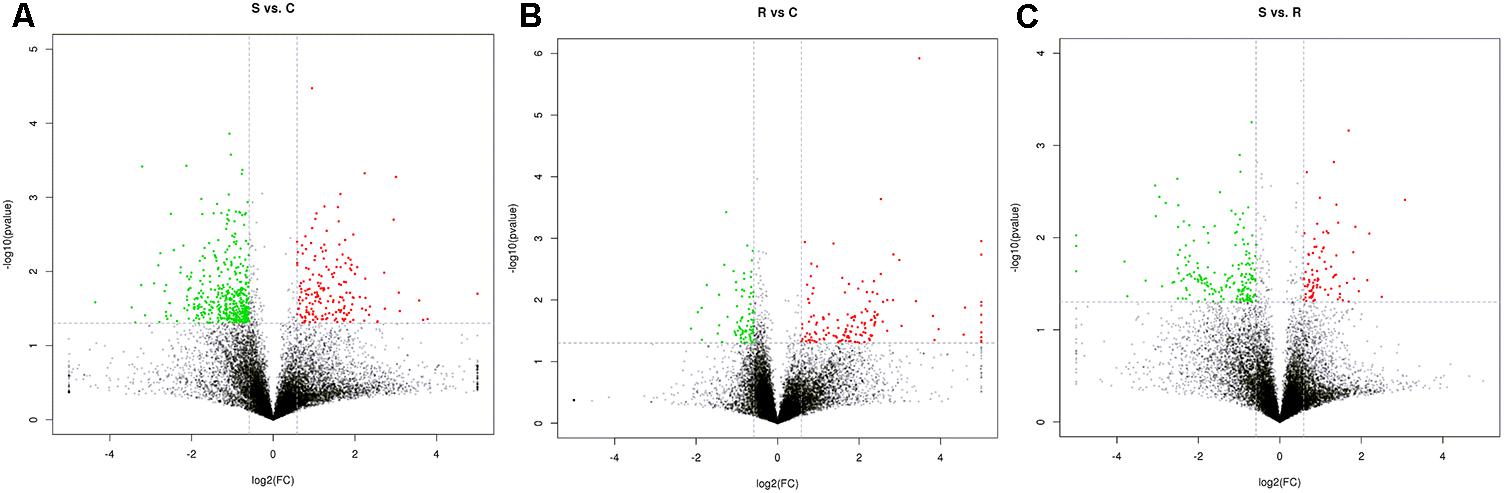
FIGURE 1. Identification of genes differentially expressed among S, R, and C chickens during SE infection. Volcano plot showing differentially expressed genes (DEGs) S vs. C (A), S vs. R (B), and R vs. C (C) after Salmonella infection. S, challenged-susceptible; R, challenged-resistant; C, non-challenged controls.
Differential Expression of mRNAs in Response to Salmonella Infection
The DEGs in spleens of the controls, resistant and susceptible birds were examined. A total of 934 significant DEGs were identified among the S, R and C chickens (Figure 2A). As shown in Figure 2, 588 genes differed between S and C (176 up- and 412 down-regulated); for R vs. C, 234 differed (145 up- and 89 down-regulated); and 265 genes (80 up- and 185 down-regulated) were DEGs between S and R birds (Supplementary Tables S2–S4). A total of 32 DEGs were shared in comparisons between S vs. C and R vs. C birds while 144 DEGs were uniquely expressed in the S vs. R comparison. Only 4 DEGs were co-expressed in the 3 contrasts. Hierarchical clustering analysis of the DEGs in each comparison demonstrated that it was appropriate to classify the challenged birds as being resistant or susceptible from their phenotypic evaluation (Figures 2B–D). Several immune-related genes such as SOCS1, CXCR4 and FOS were significantly up-regulated (P < 0.01) after challenge with Salmonella (log2 FC 3.06, 3.69, and 2.50, respectively).
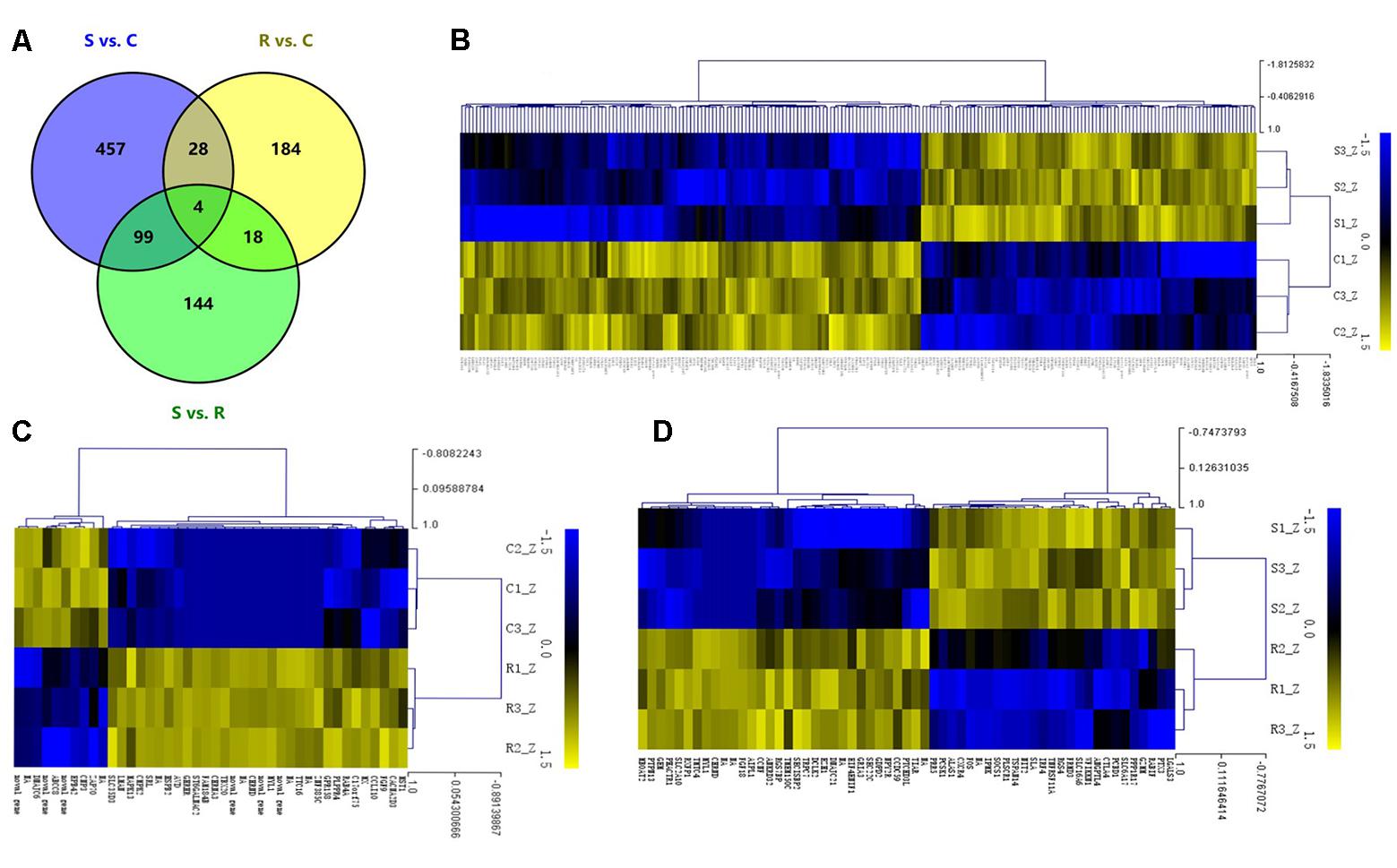
FIGURE 2. Different expression profiles of unique mRNAs in host immune response to SE infection. (A) Venn diagram shows the overlap of DEGs among the 3 groups; numbers are the DEGs in each comparison. (B–D) The heat map of unique DEGs in S vs. C, R vs. C, and S vs. R, respectively (FC > 2 and FDR < 0.05). S, challenged-susceptible; R, challenged-resistant; C, non-challenged controls.
Significant GO Terms and KEGG Analysis
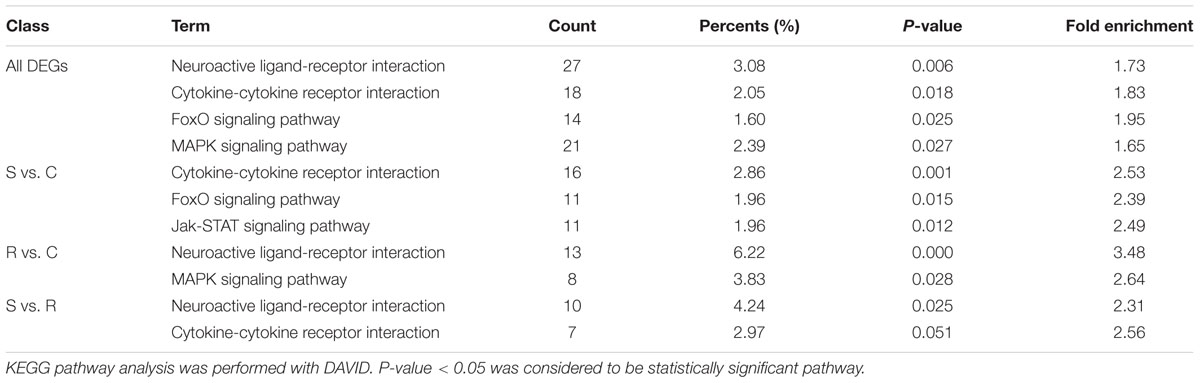
All the DEGs and unique DEGs in each comparison among S, R, and C birds were analyzed using GO and KEGG enrichment. In this study, potential function analysis of all 934 DEGs showed that some significantly enriched GO Terms were mainly involved in channel activity and transmembrane transport but several top immune-related terms were also enriched (P < 0.05), including regulation of toll-like receptor signaling pathway (GO:0034121), immune response-activating signal transduction (GO:0002757), B cell receptor signaling pathway (GO:0050853) and regulation of response to stimulus (GO:0048583) (Table 1). In addition, Neuroactive ligand-receptor interaction, Cytokine-cytokine receptor interaction and FoxO signaling pathway were significantly changed in response to SE infection (P < 0.03, Table 2).
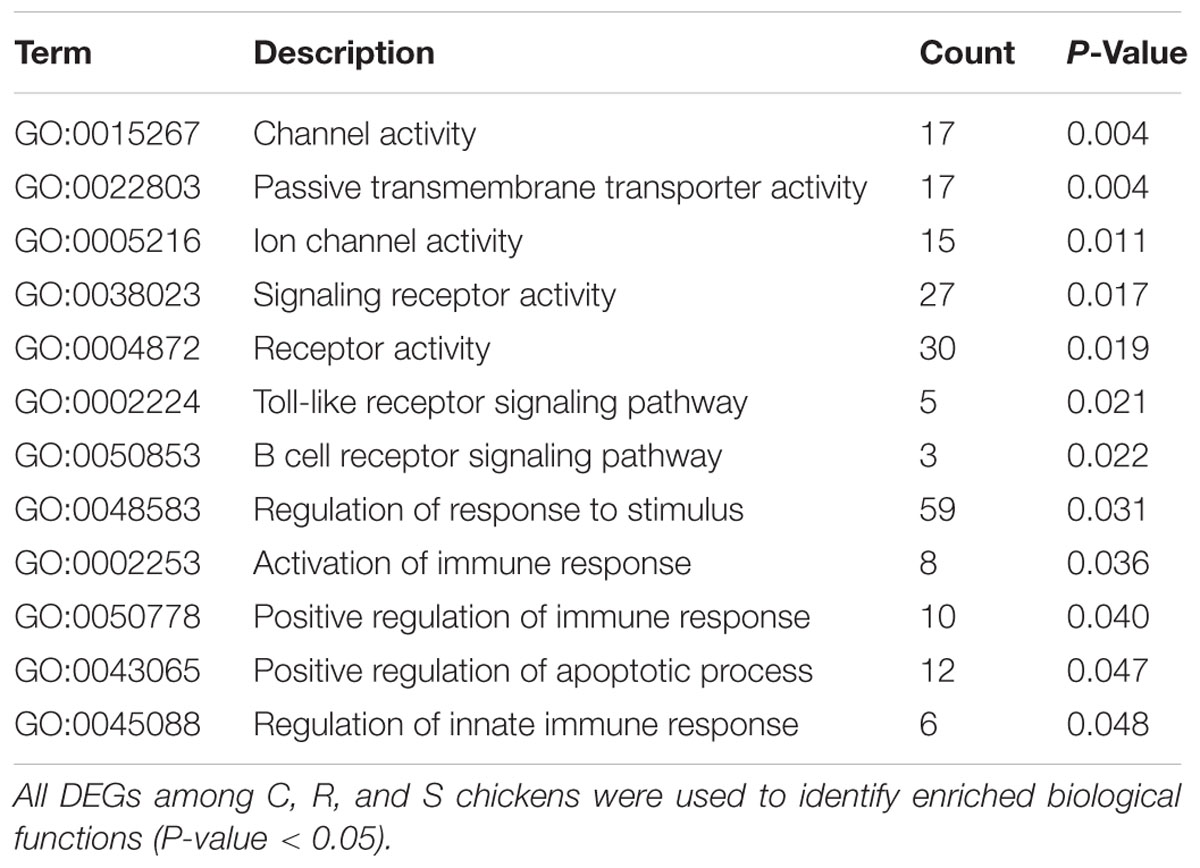
TABLE 1. Immune-related biological processes identified by gene ontology analysis of differentially expressed genes.
Potential functional analyses for host immune responses to SE infection between S and R chickens were further characterized. In the S vs. R comparisons, Neuroactive ligand-receptor interaction and Cytokine-cytokine receptor interaction pathway were enriched (P < 0.05) (Table 2); the top 3 enriched GO terms were intracellular ligand-gated ion channel activity, ligand-gated channel activity and ligand-gated ion channel activity (P < 0.01). For S vs. C, 3 pathways were enriched (P < 0.05), viz. Cytokine-cytokine receptor interaction, FoxO signaling pathway and Jak-STAT signaling pathway (Table 2); and several top GO terms of the immune response were enriched (P < 0.01), including B cell receptor signaling pathway (GO:0050853), regulation of response to stimulus (GO:0048583), and immune response-regulating signaling pathway (GO:0002764). In the R vs. C comparisons, Neuroactive ligand-receptor interaction and MAPK signaling pathway were enriched (P < 0.05) (Table 2); the enriched GO terms (P < 0.01) were mainly involved in channel activity, transmembrane transport, cardiac muscle cell proliferation and receptor activity, such as cation channel activity (GO:0005261), transmembrane transporter complex (GO:1902495), signaling receptor activity (GO:0038023) and oxygen transport (GO:0015671). These results indicate that compared to resistant birds, susceptible birds extensively initiate their pathways of immune response, signal transduction, and signal molecules and interaction, presumably in an attempt to resist SE infection.
Quantitative Real-Time PCR (qPCR) Validation
The qPCR assays were conducted to validate 16 selected DEGs from RNA-seq: IL10RB, TNFSF10, LAMP1, ZNF207, CCND1, GJA1, FTH, HBBA, GAL1, CREBBP, BRI3BP, SOCS1, ICOS, CTLA4, AVD, and IL8. Pearson’s correlation of the fold-changes between qPCR and RNA-seq was 0.92 (Figure 3). Overall, the RNA-seq results were considered to be reliable and appropriate for further analysis.
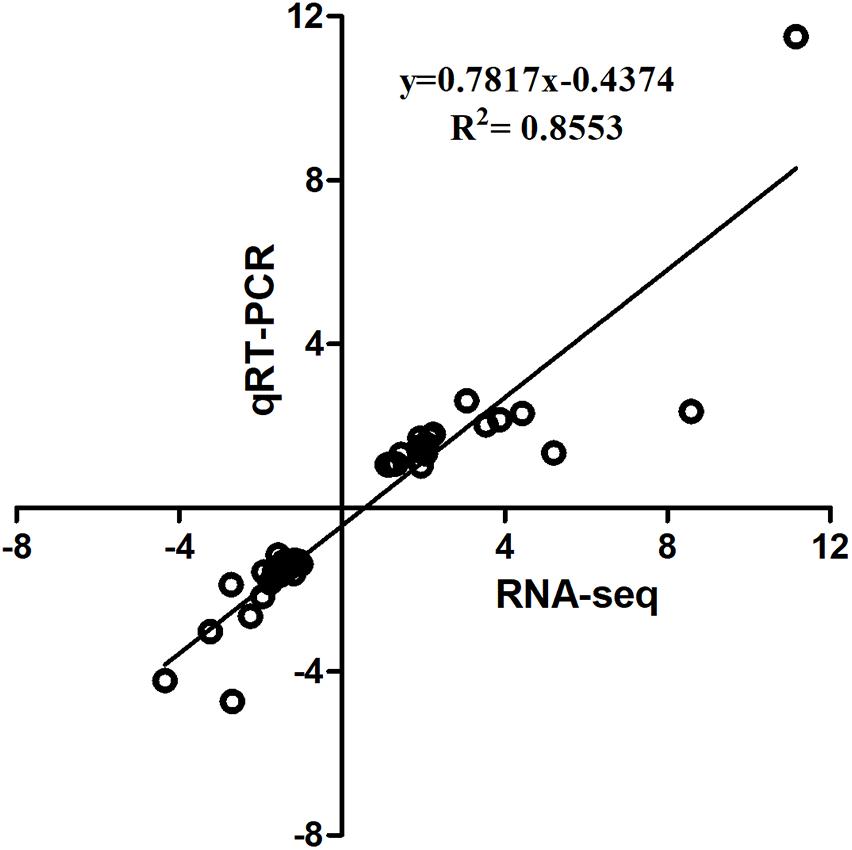
FIGURE 3. Linear regression fitted for Log2 Fold Change (FC) of selected genes determined via qPCR and RNA-seq. The selected genes in each comparison were used for linear regression analysis. Log2 FC in RNA-seq equals 2-ΔΔCt in qPCR for each comparison.
Discussion
Although there are previous studies focused on the chicken splenic transcriptome following Salmonella challenge, the novel experimental design of the current study enabled exposure of the resistance and susceptibility mechanisms of phenotypically different birds in host immune response to Salmonella enteritidis infection. In this research, both the extent of clinical symptoms and the bacterial load in blood were used to assess birds after infection to distinguish resistant from susceptible chickens. And RNA-seq was used here to identify differences splenic mRNAs expression profiles in chicks following SE infection. A total of 934 DEGs were identified among the C, R, and S birds. After SE infection, several up-regulated unique DEGs were mainly related to immune function, such as FOS, SOCS1, IL-18, IKBKB, CXCR4, CTAL4, IL10RA, IL10RB, IL1RAP, and AVD. These findings differ from the splenic results after Salmonella infection (Matulova et al., 2012), although they did identify AVD and immune responsive gene 1 (IRG1). The differences may have arisen from genetics or ages of the chickens used, or the bacterium used for challenge (Zekarias et al., 2002; Beaumont et al., 2009; Redmond et al., 2009). In generation of the heatmaps, genes included were largely driven by the S chicks. Comparing the change for each of the 3 groups, they clustered as expected based on earlier contrast comparisons (Figure 2).
It was clear, from the bacterial burden in blood, that septic infection occurred in the challenged birds. In response to systemic infection with S. typhimurium, pro-inflammatory cytokines that are host-produced are critical for the control of bacterial growth but bacterial clearance is dependent on the successful activation of CD4+ T cells, especially in peripheral immune organs (Talbot et al., 2009). Unfortunately, high doses of LPS or Salmonella can result in production of excess amounts of pro-inflammatory cytokines, or a “cytokine storm,” leading to endotoxin shock or sepsis-related deaths (Cohen, 2002; Clark and Vissel, 2017; Netea et al., 2017). Thus, the potential influence of over-expression of inflammatory cytokines due to hypersensitivity response to SE in susceptible birds was also considered in this study.
After challenge with SE, the significantly changed pathways included Cytokine-cytokine receptor interaction, FoxO signaling pathway, Neuroactive ligand-receptor interaction and MAPK signaling pathway. Consistent with previous studies (Chiang et al., 2008; Matulova et al., 2012; Li et al., 2017a), many immune-related pathways (Cytokine-cytokine receptor interaction, MAPK, and Jak-STAT signaling pathway) have been identified in susceptible chickens following Salmonella infection. Importantly, the FoxO signaling pathway is reported here for the first time.
The Forkhead box O (FOXO) is one subfamily of the fork head transcription factor family with important roles in cell fate decisions, including cellular differentiation, apoptosis, cell proliferation, DNA damage and repair and as mediators of oxidative stress (Vurusaner et al., 2012; Farhan et al., 2017). FoxO activity is mainly regulated by the PI3K (phosphoinositide 3-kinase) pathway, whereas FoxO function is negatively “fine-tuned” by protein kinase B (PKB; also known as AKT), casein kinase 1 (CK1) and IκB kinase (IKK) (Peng, 2008). Research shows that FoxO3 gene is strongly considered to regulate lymphoid homeostasis in host immune system (Lin et al., 2004; Lu et al., 2017). For instance, FoxO3a overexpression induces apoptosis in a human leukemia T cell line (Brunet et al., 1999), murine CTLL-2 T cell line (Stahl et al., 2002), murine pre-B cell line Ba/F3 (Dijkers et al., 2002), murine peritoneal macrophages (Senokuchi et al., 2008) and BCG-infected macrophages (Haoues et al., 2014). Deficiency of FoxO3a in mice leads to spontaneous, autoreactive helper T cell activation and Th1 and Th2 cytokine production (Peng, 2008), which is required for controlling bacterial growth and clearance following Salmonella infection (Chappell et al., 2009; Talbot et al., 2009). In addition, Foxo3a clearly plays critical roles in neutrophil survival, as demonstrated by Foxo3a-deficient mice which are resistant to both peritonitis and arthritis (Jonsson et al., 2005). In the current study, signaling adapter molecules EGFR, IRS1 and PIK3CD of the PI3K pathway, as well as IKBKB gene of IκB kinase, were all significantly reduced in S vs. C birds. Of particular note, the gene FoxO3 and apoptosis gene Bcl-6 were significantly up-regulated only in susceptible (S) birds (Figure 4). These results, together with what is known of the immunobiology of avian systemic salmonellosis, indicate that the FoxO signaling pathway plays an important role in response against SE infection. Based on the above analysis, it is hypothesized that the hyperactive FoxO3 in susceptible chickens might both enhance apoptosis of T, and B lymphocytes and macrophages in the spleen, and constrain production of Th1 and Th2 cytokine, all of which are necessary for immunological clearance during the early stage of Salmonella infection.
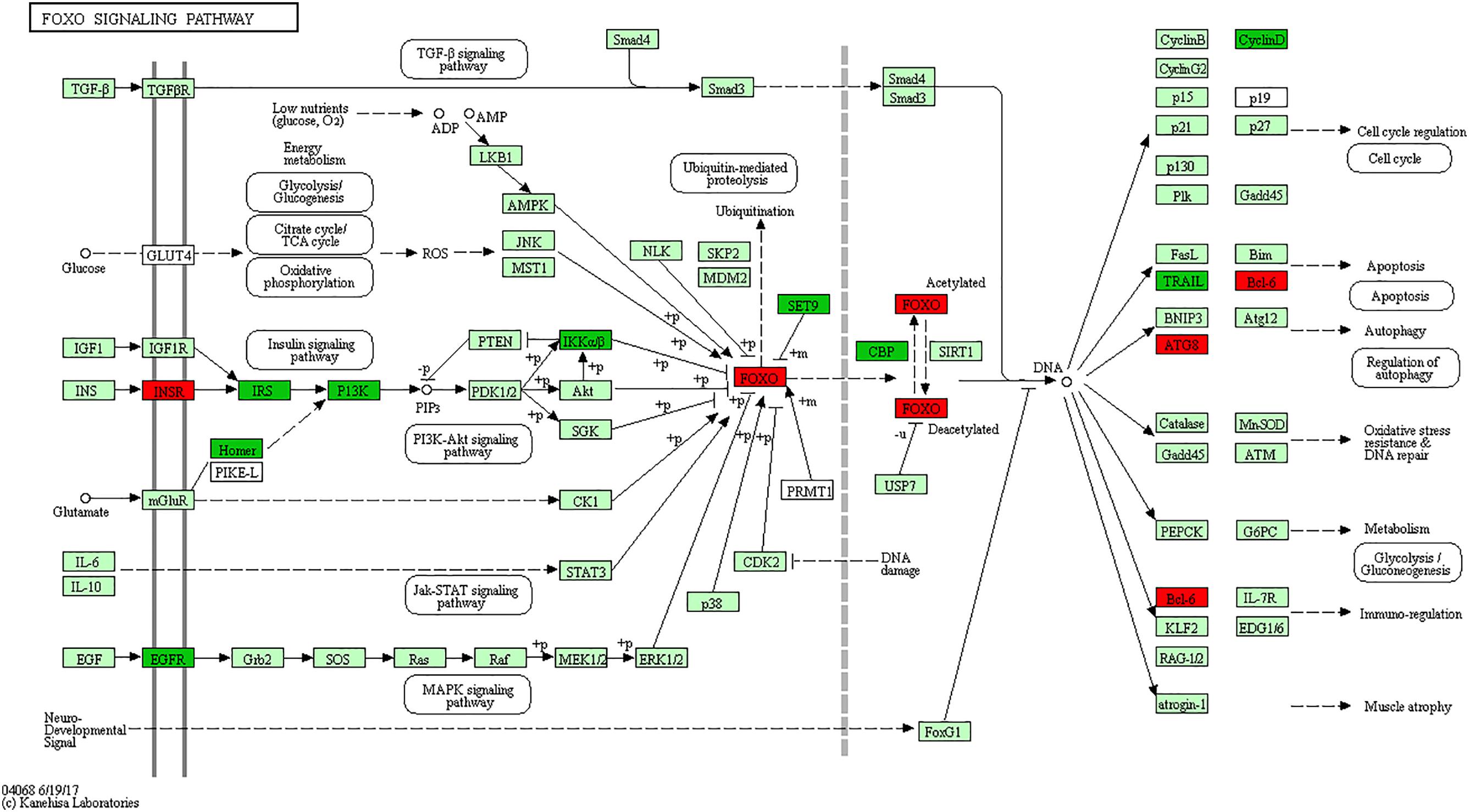
FIGURE 4. FoxO signaling pathway. Red, significantly up-regulated differentially expressed genes. Green, significantly down-regulated differentially expressed genes. Light green represents the genes involved in the pathway.
Many immune-related pathways were significantly induced in S birds (S vs. C or S vs. R comparisons), including cytokine-cytokine receptor interaction, Jak-STAT, MAPK signaling pathway and neuroactive ligand-receptor interaction (Table 2). Interestingly, cytokine-cytokine receptor interaction, MAPK, and Jak-STAT signaling had cross-talk with activating the FoxO signaling pathway (Figure 5). These results suggested that multiple signaling pathway cascades control Salmonella invasion and clearance. In addition, increased expression of many genes in these identified pathways, in response to APEC infection, have also been demonstrated (Li et al., 2011; Sandford et al., 2012; Sun et al., 2015, 2016).
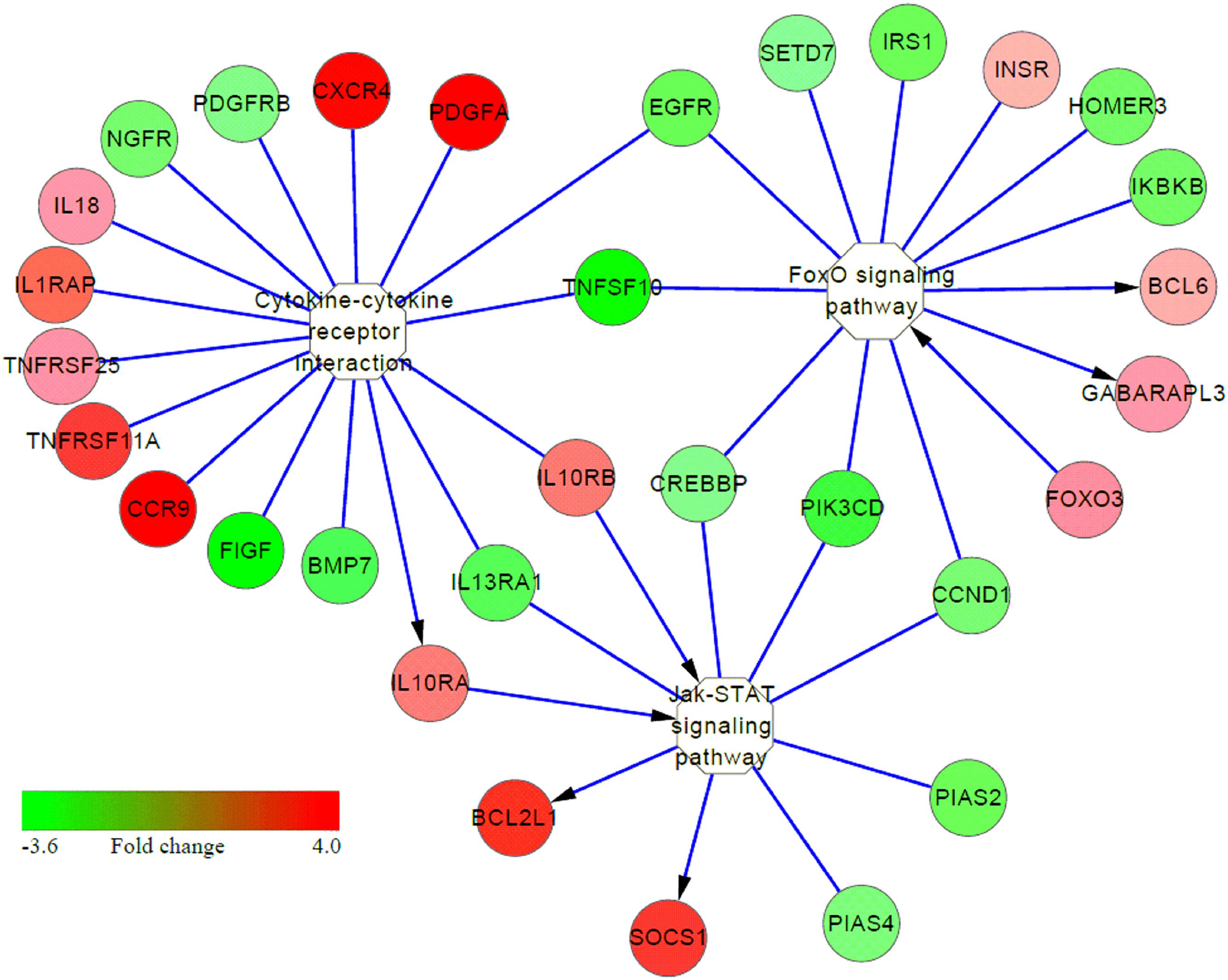
FIGURE 5. Interaction among several signaling pathways involved in susceptibility to Salmonella enteritidis infection. Color column from red to green represents the fold change (FC 4.0 to –3.6) of genes in S vs. C or S vs. R chickens. Red represents up-regulation and green represents down-regulation; yellow represents the pathway. The arrow represents direct activation.
CXCR4 is expressed in multifarious types of cancer. This cytokine receptor and its ligand are also involved in the recruitment of T-cells at the site of the immune or inflammatory reactions (Yusuf et al., 2005; Sallusto and Baggiolini, 2008). TNFRSF11A is a member of the tumor necrosis factor receptor (TNFR) molecular sub-family, also known as receptor activator of nuclear factor κB (RANK). Most commonly, over-expression of RANK alone is sufficient to activate the NF-κB pathway (Tracey et al., 2008). Interleukin 10 receptor A and B (IL10RA, and IL10RB) are expressed in most immune cells, and very low expression levels have been observed on a variety of non-hematopoietic cells (Moore et al., 2001; Yao et al., 2012). Differential expression of IL-10RA plays an important role in IL-10-mediated immune regulation, and activation of monocytes and neutrophils increases mRNA expression, whereas expression levels have been shown to decrease following stimulation of human T-cells, B-cells and NK cells (Carson et al., 1995; Jurlander et al., 1997; Denning et al., 2000; Yao et al., 2012). IL-1 receptor accessory protein (IL1RAP) mediates the response to IL-1, IL-33, and IL-36 and has been shown to regulate the inflammatory response, as well as activation of T lymphocytes and mast cells (Barreyro et al., 2012; Boraschi and Tagliabue, 2013; Dinarello, 2018). High IL1RAP expression is associated with poor overall survival in acute myeloid leukemia (AML) patients (Barreyro et al., 2012); although several receptors genes were increased here, only IL18 genes were significantly up-regulated. The expression of IL18 was lower in tumor-associated macrophages cultured with metastatic gastric cancer cell lines (Shen et al., 2012). In the current study, the key genes (CXCR4, TNFRSF11A, IL10RAP, and IL10RB) in the cytokine-cytokine receptor interaction pathway had increased expression, both in S vs. C and S vs. R comparisons. While the inflammatory cytokines response is critical for the control of bacterial growth (Cross et al., 1995), excessive cytokines production can lead to endotoxic shock or sepsis-related deaths (Cohen, 2002; Clark and Vissel, 2017; Netea et al., 2017). Overall, these results suggest that susceptible birds showed hypersensitivity to acute SE infections and that the cytokine-cytokine receptor interaction pathway is an important mediator in SE-induced pathogenesis.
The Jak-STAT pathway is needful to ensure T and B-cell development (Rawlings et al., 2004). In the present study, various genes involved in the Jak-STAT signaling pathway had increased expression in susceptible birds than in non-infected controls, including IL10RA, IL10RB, BCL2-like 1 (BCL2L1) and suppressor of cytokine signaling 1 (SOCS1). BCL2L1 is one the family of Bcl-2 proteins with important roles in the regulation of mitochondrial pathway of apoptosis (Cory and Adams, 2002) and SOCS1 is a negative regulator of LPS-induced macrophage activation (Kinjyo et al., 2002; Alvarez et al., 2017). Interestingly, three overlapping elements were found in Jak-STAT and cytokine-cytokine receptor interaction pathways in susceptible birds, both in S vs. C and S vs. R comparisons (Figure 5). These results indicate that susceptible birds extensively activate key pathways of immune response, signal transduction, and signal molecules and interaction in an attempt to resist SE infection, but fail to do so and succumb.
In addition, resistant chicks seem to activate the MAPK signaling in regulating the host response to SE infection. MAPK signaling was shown to be activated in chicks when pathogenic bacteria invaded (Withanage et al., 2004). It was reported that P38 MAPK is very important for B-cell development and is a survival mediator for T-cells in human inflamed tissues (Huang et al., 2009). In the current study, 5 up-regulated DE genes (MAPT, MAPK13, CACNA2D3, CACNG5 and FGF9) participated in activating MAPK signaling pathway in resistant birds (Supplementary Table S3). These results are consistent with SE-infected, resistant birds increasing proliferation of T- and B- lymphocytes in the spleen to achieve protection against Salmonella.
Conclusion
The current study is the first to characterize the splenic transcriptomes of 3 categories of chicks in response to infection with Salmonella enteritidis; challenged birds that were resistant (R), those that succumbed clinically (S) and non-challenged controls (C). A total of 934 DEGs were identified in comparisons between the C, R and S birds (R vs. C, S vs. C, and S vs. R). The DE genes involved in the FoxO signaling pathway, especially FoxO3, is reported here for the first time, and is identified as a potential marker of host resistance to SE infection. There was strong activation of the FoxO signaling pathway in S birds, which may be a major defect causing intracellular apoptosis of immune cells as part of SE-induced pathologenesis. Challenged-susceptible birds failed to activate components of the MAPK-related pathway in spleen whereas it was strongly activated in the R birds. Although cytokine-cytokine receptor interaction and Jak-STAT signaling pathway were activated extensively and there was cross-talk between them in challenged-susceptible birds, their role in negatively regulating the immune response against Salmonella likely aggravated the bacterial infections. However, the chicks used here had a consistent genetic background (White Leghorn), health status and management, a potential confounding genetic effect such as population stratification may be present, possibly affecting the results. This may influence the screening of candidate genes using randomly selected control chicks. Further analysis using birds where this possibility is eliminated is warranted. These findings will facilitate the understanding of resistance and susceptibility to SE infection in the earliest phases of the host immune response and provides new approaches to developing strategies for SE prevention and treatment.
Author Contributions
PL and WF performed the experiments and data analysis and draft writing. NE contributed to experimental design and revised the manuscript. RL, QL, MZ, and HC contributed to animal experiments and data analysis. GZ and JW helped design the study and supervised and coordinated the study. All authors reviewed the manuscript.
Funding
The research was supported by grants from National Natural Science Foundation of China (No. 31572393) and National Key Technology R&D Program (2015BAD03B03) and China Agricultural Science and Technology Innovation Project (ASTIP-IAS04), and the earmarked fund for Modern Agro-Industry Technology Research System (CARS-41).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer CH and handling Editor declared their shared affiliation.
Acknowledgments
The authors thank W. Bruce Currie, Emeritus Professor, Cornell University, for his contributions to presenting the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00256/full#supplementary-material
Abbreviations
AVD, avidin; BCL2L1, B-cell lymphoma-2 like 1; Bcl-6, B-cell CLL/lymphoma 6; BRI3BP, brain protein I3-binding protein; CACNA2D3, calcium voltage-gated channel auxiliary subunit alpha 2 delta 3; CACNG5, calcium voltage-gated channel auxiliary subunit gamma 5; CCND1, cyclin D1; CREB, cAMP responsive element-binding protein; CREBBP, CREB binding protein; CTLA4, cytotoxic T-lymphocyte associated protein 4; CXCR4, C-X-C motif chemokine receptor 4; DEGs, differentially expressed genes; EGFR, epidermal growth factor receptor; FDR, false discovery rate; FGF9, fibroblast growth factor 9; FOS, Fos proto-oncogene, AP-1 transcription factor subunit; FoxO, Forkhead box-O transcription factor; FTH, ferritin heavy chain 1; GAL1, Galactokinase 1, also known as AvBD1, avian beta-defensin 1; GJA1, gap junction protein alpha 1; GO, Gene ontology; HBBA, Hemoglobin beta, subunit A; ICOS, Inducible T-cell costimulator; IKBKB, Inhibitor of nuclear factor kappa B kinase subunit beta; IL, Interleukin; IL10RA/B, Interleukin 10 receptor subunit alpha/beta; IL1RAP, Interleukin 1 receptor accessory protein; IRS1, Insulin receptor substrate 1; Jak-STAT, Janus kinase-signal transducer and activator of transcription; KEGG, Kyoto encyclopedia of genes and genomes; LAMP1, lysosomal associated membrane protein 1; MAPK, mitogen-activated protein kinase; MAPT, Microtubule associated protein tau; PIK3CD, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta; SE, Salmonella enteritidis; SOCS1, Suppressor of cytokine signaling 1; Th, helper T cell; TNFRSF11A, TNF receptor superfamily member 11a; TNFSF10, tumor necrosis factor superfamily member 10; ZNF207, zinc finger protein 207.
Footnotes
References
Altamura, M., Altamura, M., Caradonna, L., Amati, L., Pellegrino, N., Urgesi, G., et al. (2001). Splenectomy and sepsis: the role of the spleen in the immune-mediated bacterial clearance. Immunopharmacol. Immunotoxicol. 23, 153–161. doi: 10.1081/IPH-100103856
Alvarez, A. P., Glosson-Byers, N., Brandt, S., Wang, S., Wong, H., Sturgeon, S., et al. (2017). SOCS1 is a negative regulator of metabolic reprogramming during sepsis. JCI Insight 2:e92530. doi: 10.1172/jci.insight.92530
Barreyro, L., Will, B., Bartholdy, B., Zhou, L., Todorova, T. I., Stanley, R. F., et al. (2012). Overexpression of IL-1 receptor accessory protein in stem and progenitor cells and outcome correlation in AML and MDS. Blood 120, 1290–1298. doi: 10.1182/blood-2012-01-404699
Barrow, P., Jones, M., Smith, A., and Wigley, P. (2012). The long view: Salmonella-the last forty years. Avian Pathol. 41, 413–420. doi: 10.1080/03079457.2012.718071
Beaumont, C., Chapuis, H., Protais, J., Sellier, N., Menanteau, P., Fravalo, P., et al. (2009). Resistance to Salmonella carrier state: selection may be efficient but response depends on animal’s age. Genet. Res. 91, 161–169. doi: 10.1017/S0016672309000135
Berthelot, F., Beaumont, C., Mompart, F., Girard-Santosuosso, O., Pardon, P., and Duchet-Suchaux, M. (1998). Estimated heritability of the resistance to cecal carrier state of Salmonella enteritidis in chickens. Poult. Sci. 77, 797–801. PMID: 9628525 doi: 10.1093/ps/77.6.797
BIG Data Center Members (2017). The BIG Data Center: from deposition to integration to translation. Nucleic Acids Res. 45, D18–D24. doi: 10.1093/nar/gkw1060
Boraschi, D., and Tagliabue, A. (2013). The interleukin-1 receptor family. Semin. Immunol. 25, 394–407. doi: 10.1016/j.smim.2013
Brunet, A., Bonni, A., Zigmond, M. J., Lin, M. Z., Juo, P., Hu, L. S., et al. (1999). Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96, 857–868. doi: 10.1016/S0092-8674(00)80595-4
Calenge, F., and Beaumont, C. (2012). Toward integrative genomics study of genetic resistance to Salmonella and Campylobacter intestinal colonization in fowl. Front. Genet. 3:261. doi: 10.3389/fgene.2012.00261
Carson, W. E., Lindemann, M. J., Baiocchi, R., Linett, M., Tan, J. C., Chou, C.-C., et al. (1995). The functional characterization of interleukin-10 receptor expression on human natural killer cells. Blood 85, 3577–3585. PMID: 7540068
Chappell, L., Kaiser, P., Barrow, P., Jones, M. A., Johnston, C., and Wigley, P. (2009). The immunobiology of avian systemic salmonellosis. Vet. Immunol. Immunopathol. 128, 53–59. doi: 10.1016/j.vetimm.2008.10.295
Chiang, H. I., Swaggerty, C. L., Kogut, M. H., Dowd, S. E., Li, X., Pevzner, I. Y., et al. (2008). Gene expression profiling in chicken heterophils with Salmonella enteritidis stimulation using a chicken 44 K Agilent microarray. BMC Genomics 9:526. doi: 10.1186/1471-2164-9-526
Clark, I. A., and Vissel, B. (2017). The meteorology of cytokine storms, and the clinical usefulness of this knowledge. Semin. Immunopathol. 39, 505–516. doi: 10.1007/s00281-017-0628-y
Cory, S., and Adams, J. M. (2002). The Bcl-2 family: regulators of the cellular life-or-death switch. Nat. Rev. Cancer 2, 647–656. doi: 10.1038/nrc883
Cross, A., Asher, L., Seguin, M., Yuan, L., Kelly, N., Hammack, C., et al. (1995). The importance of a lipopolysaccharide-initiated, cytokine-mediated host defense mechanism in mice against extraintestinally invasive Escherichia coli. J. Clin. Invest. 96, 676–686. doi: 10.1172/JCI118110
Denning, T. L., Campbell, N. A., Song, F., Garofalo, R. P., Klimpel, G. R., Reyes, V. E., et al. (2000). Expression of IL-10 receptors on epithelial cells from the murine small and large intestine. Int. Immunol. 12, 133–139. PMID: 10653848 doi: 10.1093/intimm/12.2.133
Dennis, G., Sherman, B. T., Hosack, D. A., Yang, J., Gao, W., Lane, H. C., et al. (2003). DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4:R60. PMID: 12734009
Dijkers, P. F., Birkenkamp, K. U., Lam, E. W. F., Thomas, N. S. B., Lammers, J.-W. J., Koenderman, L., et al. (2002). FKHR-L1 can act as a critical effector of cell death induced by cytokine withdrawal. J. Cell Biol. 156, 531–542. doi: 10.1083/jcb.200108084
Dinarello, C. A. (2018). Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol. Rev. 281, 8–27. doi: 10.1111/imr.12621
Farhan, M., Wang, H., Gaur, U., Little, P. J., Xu, J., and Zheng, W. (2017). FOXO signaling pathways as therapeutic targets in cancer. Int. J. Biol. Sci. 13, 815–827. doi: 10.7150/ijbs.20052
Gou, Z., Liu, R., Zhao, G., Zheng, M., Li, P., Wang, H., et al. (2012). Epigenetic modification of TLRs in leukocytes is associated with increased susceptibility to Salmonella enteritidis in chickens. PLoS One 7:e33627. doi: 10.1371/journal.pone.0033627
Haoues, M., Refai, A., Mallavialle, A., Barbouche, M. R., Laabidi, N., Deckert, M., et al. (2014). Forkhead box O3 (FOXO3) transcription factor mediates apoptosis in BCG-infected macrophages. Cell. Microbiol. 16, 1378–1390. doi: 10.1111/cmi.12298
Haq, K., Brisbin, J. T., Thanthrige-Don, N., Heidari, M., and Sharif, S. (2010). Transcriptome and proteome profiling of host responses to Marek’s disease virus in chickens. Vet. Immunol. Immunopathol. 138, 292–302. doi: 10.1016/j.vetimm.2010.10.007
Huang, G., Shi, L. Z., and Chi, H. (2009). Regulation of JNK and p38 MAPK in the immune system: signal integration, propagation and termination. Cytokine 48, 161–169. doi: 10.1016/j.cyto.2009.08.002
Inns, T., Lane, C., Peters, T., Dallman, T., Chatt, C., McFarland, N., et al. (2015). A multi-country Salmonella enteritidis phage type 14b outbreak associated with eggs from a German producer:‘near real-time’application of whole genome sequencing and food chain investigations, United Kingdom, May to September 2014. Euro Surveil. 20:21098. PMID: 25953273 doi: 10.2807/1560-7917.ES2015.20.16.21098
Jonsson, H., Allen, P., and Peng, S. L. (2005). Inflammatory arthritis requires Foxo3a to prevent Fas ligand–induced neutrophil apoptosis. Nat. Med. 11, 666–671. doi: 10.1038/nm1248
Jurlander, J., Lai, C.-F., Tan, J., Chou, C., Geisler, C. H., Schriber, J., et al. (1997). Characterization of interleukin-10 receptor expression on B-cell chronic lymphocytic leukemia cells. Blood 89, 4146–4152.
Kaiser, M., and Lamont, S. (2001). Genetic line differences in survival and pathogen load in young layer chicks after Salmonella enterica serovar Enteritidis exposure. Poult. Sci. 80, 1105–1108. doi: 10.1093/ps/80.8.1105
Kinjyo, I., Hanada, T., Inagaki-Ohara, K., Mori, H., Aki, D., Ohishi, M., et al. (2002). SOCS1/JAB is a negative regulator of LPS-induced macrophage activation. Immunity 17, 583–591. doi: 10.1016/S1074-7613(02)00446-6
Kogut, M. H., and Arsenault, R. J. (2017). Immunometabolic phenotype alterations associated with the induction of disease tolerance and persistent asymptomatic infection of Salmonella in the chicken intestine. Front. Immunol. 8:372. doi: 10.3389/fimmu.2017.00372
Kogut, M. H., Chiang, H. I., Swaggerty, C. L., Pevzner, I. Y., and Zhou, H. (2012). Gene expression analysis of Toll-like receptor pathways in heterophils from genetic chicken lines that differ in their susceptibility to Salmonella enteritidis. Front. Genet. 3:121. doi: 10.3389/fgene.2012.00121
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, G., Tivendale, K. A., Liu, P., Feng, Y., Wannemuehler, Y., Cai, W., et al. (2011). Transcriptome analysis of avian pathogenic Escherichia coli O1 in chicken serum reveals adaptive responses to systemic infection. Infect. Immun. 79, 1951–1960. doi: 10.1128/IAI.01230-10
Li, P., Fan, W., Li, Q., Wang, J., Liu, R., Everaert, N., et al. (2017a). Splenic microRNA expression profiles and integration analyses involved in host responses to Salmonella enteritidis infection in chickens. Front. Cell. Infect. Microbiol. 7:377. doi: 10.3389/fcimb.2017.00377
Li, P., Wang, H., Zhao, X., Gou, Z., Liu, R., Song, Y., et al. (2017b). Allelic variation in TLR4 is linked to resistance to Salmonella Enteritidis infection in chickens. Poult. Sci. 96, 2040–2048. doi: 10.3382/ps/pex010
Lin, L., Hron, J. D., and Peng, S. L. (2004). Regulation of NF-κB, Th activation, and autoinflammation by the forkhead transcription factor Foxo3a. Immunity 21, 203–213. doi: 10.1016/j.immuni.2004.06.016
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, L., Barbi, J., and Pan, F. (2017). The regulation of immune tolerance by FOXP3. Nat. Rev. Immunol. 17, 703–717. doi: 10.1038/nri.2017.75
Matulova, M., Rajova, J., Vlasatikova, L., Volf, J., Stepanova, H., Havlickova, H., et al. (2012). Characterization of chicken spleen transcriptome after infection with Salmonella enterica serovar Enteritidis. PLoS One 7:e48101. doi: 10.1371/journal.pone.0048101
Moore, K. W., de Waal Malefyt, R., Coffman, R. L., and O’Garra, A. (2001). Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 19, 683–765. doi: 10.1146/annurev.immunol.19.1.683
Netea, M. G., Balkwill, F., Chonchol, M., Cominelli, F., Donath, M. Y., Giamarellos-Bourboulis, E. J., et al. (2017). A guiding map for inflammation. Nat. Immunol. 18, 826–831. doi: 10.1038/ni.3790
Nie, Q., Sandford, E. E., Zhang, X., Nolan, L. K., and Lamont, S. J. (2012). Deep sequencing-based transcriptome analysis of chicken spleen in response to avian pathogenic Escherichia coli (APEC) infection. PLoS One 7:e41645. doi: 10.1371/journal.pone.0041645
Rawlings, J. S., Rosler, K. M., and Harrison, D. A. (2004). The JAK/STAT signaling pathway. J. Cell Sci. 117, 1281–1283. doi: 10.1242/jcs.00963
Redmond, S. B., Chuammitri, P., Andreasen, C. B., Palić, D., and Lamont, S. J. (2009). Chicken heterophils from commercially selected and non-selected genetic lines express cytokines differently after in vitro exposure to Salmonella enteritidis. Vet. Immunol. Immunopathol. 132, 129–134. doi: 10.1016/j.vetimm.2009.05.010
Sallusto, F., and Baggiolini, M. (2008). Chemokines and leukocyte traffic. Nat. Immunol. 9, 949–952. doi: 10.1038/ni.f.214
Sandford, E. E., Orr, M., Balfanz, E., Bowerman, N., Li, X., Zhou, H., et al. (2011). Spleen transcriptome response to infection with avian pathogenic Escherichia coli in broiler chickens. BMC Genomics 12:469. doi: 10.1186/1471-2164-12-469
Sandford, E. E., Orr, M., Shelby, M., Li, X., Zhou, H., Johnson, T. J., et al. (2012). Leukocyte transcriptome from chickens infected with avian pathogenic Escherichia coli identifies pathways associated with resistance. Results Immunol. 2, 44–53. doi: 10.1016/j.rinim.2012.02.003
Senokuchi, T., Liang, C.-P., Seimon, T. A., Han, S., Matsumoto, M., Banks, A. S., et al. (2008). Forkhead transcription factors (FoxOs) promote apoptosis of insulin-resistant macrophages during cholesterol-induced endoplasmic reticulum stress. Diabetes Metab. Res. Rev. 57, 2967–2976. doi: 10.2337/db08-0520
Shen, Z., Seppänen, H., Vainionpää, S., Ye, Y., Wang, S., Mustonen, H., et al. (2012). IL10, IL11, IL18 are differently expressed in CD14+ TAMs and play different role in regulating the invasion of gastric cancer cells under hypoxia. Cytokine 59, 352–357. doi: 10.1016/j.cyto.2012.04.033
Smith, J., Sadeyen, J.-R., Paton, I. R., Hocking, P. M., Salmon, N., Fife, M., et al. (2011). Systems analysis of immune responses in Marek’s disease virus-infected chickens identifies a gene involved in susceptibility and highlights a possible novel pathogenicity mechanism. J. Virol. 85, 11146–11158. doi: 10.1128/JVI.05499-11
Smith, K. G., and Hunt, J. L. (2004). On the use of spleen mass as a measure of avian immune system strength. Oecologia 138, 28–31. doi: 10.1007/s00442-003-1409-y
Stahl, M., Dijkers, P. F., Kops, G. J., Lens, S. M., Coffer, P. J., Burgering, B. M., et al. (2002). The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J. Immunol. 168, 5024–5031. doi: 10.4049/jimmunol.168.10.5024
Sun, H., Liu, P., Nolan, L. K., and Lamont, S. J. (2015). Novel pathways revealed in bursa of fabricius transcriptome in response to extraintestinal pathogenic Escherichia coli (ExPEC) infection. PLoS One 10:e0142570. doi: 10.1371/journal.pone.0142570
Sun, H., Liu, P., Nolan, L. K., and Lamont, S. J. (2016). Thymus transcriptome reveals novel pathways in response to avian pathogenic Escherichia coli infection. Poult. Sci. 95, 2803–2814. doi: 10.3382/ps/pew202
Talbot, S., Tötemeyer, S., Yamamoto, M., Akira, S., Hughes, K., Gray, D., et al. (2009). Toll-like receptor 4 signalling through MyD88 is essential to control Salmonella enterica serovar Typhimurium infection, but not for the initiation of bacterial clearance. Immunology 128, 472–483. doi: 10.1111/j.1365-2567.2009.03146.x
Tracey, D., Klareskog, L., Sasso, E. H., Salfeld, J. G., and Tak, P. P. (2008). Tumor necrosis factor antagonist mechanisms of action: a comprehensive review. Pharmacol. Ther. 117, 244–279. doi: 10.1016/j.pharmthera.2007.10.001
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., Van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Vurusaner, B., Poli, G., and Basaga, H. (2012). Tumor suppressor genes and ROS: complex networks of interactions. Free Radic. Biol. Med. 52, 7–18. doi: 10.1016/j.freeradbiomed.2011.09.035
Wang, X., Rosa, A. J., Oliverira, H. N., Rosa, G. J., Guo, X., Travnicek, M., et al. (2006). Transcriptome of local innate and adaptive immunity during early phase of infectious bronchitis viral infection. Viral Immunol. 19, 768–774. doi: 10.1089/vim.2006.19.768
Withanage, G., Kaiser, P., Wigley, P., Powers, C., Mastroeni, P., Brooks, H., et al. (2004). Rapid expression of chemokines and proinflammatory cytokines in newly hatched chickens infected with Salmonella enterica serovar typhimurium. Infect. Immun. 72, 2152–2159. doi: 10.1128/IAI.72.4.2152-2159.2004
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483
Yao, Q., Fischer, K. P., Tyrrell, D. L., and Gutfreund, K. S. (2012). cDNA cloning, genomic structure, molecular characterization and mRNA expression analysis of the Pekin duck interleukin-10 receptor 1. Int. J. Immunogenet. 39, 55–67. doi: 10.1111/j.1744-313X.2011.01058.x
Yusuf, F., Rehimi, R., Dai, F., and Brand-Saberi, B. (2005). Expression of chemokine receptor CXCR4 during chick embryo development. Anat. Embryol. 210, 35–41. doi: 10.1007/s00429-005-0013-9
Zekarias, B., Ter Huurne, A. A., Landman, W. J., Rebel, J. M., Pol, J. M., and Gruys, E. (2002). Immunological basis of differences in disease resistance in the chicken. Vet. Res. 33, 109–125. doi: 10.1051/vetres:2002001
Keywords: spleen transcriptome, Salmonella enteritidis, resistant and susceptible, immune-related genes and pathway, chicken
Citation: Li P, Fan W, Everaert N, Liu R, Li Q, Zheng M, Cui H, Zhao G and Wen J (2018) Messenger RNA Sequencing and Pathway Analysis Provide Novel Insights Into the Susceptibility to Salmonella enteritidis Infection in Chickens. Front. Genet. 9:256. doi: 10.3389/fgene.2018.00256
Received: 19 February 2018; Accepted: 26 June 2018;
Published: 13 July 2018.
Edited by:
Farai Catherine Muchadeyi, Agricultural Research Council of South Africa (ARC-SA), South AfricaReviewed by:
Charles Hefer, Agricultural Research Council of South Africa (ARC-SA), South AfricaEvangelina López De Maturana, Centro Nacional de Investigaciones Oncológicas CNIO – Fundación Cáncer FUCA, Spain
Huaijun Zhou, University of California, Davis, United States
Copyright © 2018 Li, Fan, Everaert, Liu, Li, Zheng, Cui, Zhao and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guiping Zhao, emhhb2d1aXBpbmdAY2Fhcy5jbg== Jie Wen, d2VuamllQGNhYXMuY24=
†These authors have contributed equally to this work.
 Peng Li
Peng Li Wenlei Fan1,3†
Wenlei Fan1,3† Nadia Everaert
Nadia Everaert