- 1Fish Genetics Unit, Institute of Genome Biology, Leibniz Institute for Farm Animal Biology (FBN), Dummerstorf, Germany
- 2Research Station Aquaculture Born, Institute of Fisheries, Mecklenburg-Vorpommern Research Centre for Agriculture and Fisheries (LFA MV), Born, Germany
The complex and still poorly understood nature of thermoregulation in various fish species complicates the determination of the physiological status on the basis of diagnostic marker genes and indicative molecular pathways. The present study aimed to compare the physiological impacts of both gradual and acute temperature rise from 18 to 24°C on maraena whitefish in aquaculture. Microarray-based transcriptome profiles in the liver, spleen and kidney of heat-stressed maraena whitefish revealed the modulation of a significantly higher number of genes in those groups exposed to gradually rising temperatures compared with the acutely stressed groups, which might reflect early adaptation mechanisms. Moreover, we suggest a common set of 11 differentially expressed genes that indicate thermal stress induced by gradual or acute temperature rise in the three selected tissues. Besides the two pathways regulated in both data sets unfolded protein response and aldosterone signaling in epithelial cells, we identified unique tissue- and stress type-specific pathways reflecting the crossroads between signal transduction, metabolic and immunologic pathways to cope with thermal stress. In addition, comparing lists of differentially regulated genes with meta-analyzed published data sets revealed that “acute temperature rise”-responding genes that encode members of the HSP70, HSP90, and HSP40 families; their functional homologs; co-chaperones and stress-signal transducers are well-conserved across different species, tissues and/or cell types and experimental approaches.
Introduction
The most economically significant fish species produced in the European Union and Norway are rainbow trout Oncorhynchus mykiss and Atlantic salmon Salmo salar. While these two salmonid species have been domesticated for decades (Tymchuk et al., 2009), the cultivation of the maraena whitefish Coregonus maraena, another salmonid fish, started only in the mid-1990s in Finland (Kause et al., 2011; Brietzke et al., 2016). Additionally, other European countries have established whitefish brood stocks for aquaculture industry (Arndt, 2001; Fopp-Bayat et al., 2015). Whitefish is a species-rich genus of mostly cold-water benthivores that grow optimally at water temperatures between 13 and 18°C (McCormick et al., 1971), while higher temperatures above 22°C impair growth (Jobling, 1981; Ficker et al., 2016) and temperatures above 26°C are lethal (Edsall et al., 1970; Edsall and Rottiers, 1976) for several coregonid species. According to a recently published mathematical model, increasing water temperatures due to global warming will reduce the biomass of wild whitefish populations by between 3 and 8% over a period of 50 years (Ficker et al., 2016).
Independent of global climate changes, farmed whitefish may occasionally or even persistently suffer from adverse housing conditions (Korytár et al., 2016). These include seasonally increased temperatures—closely linked to hypoxia—as whitefish have not yet or barely been selected for traits proven to be genetically correlated to stress resistance, as has been successfully carried out and documented for rainbow trout and Atlantic salmon (Pickering and Pottinger, 1989; Fevolden et al., 1991; Rexroad et al., 2013). Maraena whitefish is considered a highly stress-susceptible species (Arndt, 2001; Cingi et al., 2010; Korytár et al., 2016) and is thus a suitable model organism for studying physiological welfare aspects.
Alterations from thermal optima affect a wide range of molecular aspects: temperature-sensitive kinetics of enzymatic and non-enzymatic proteins are described, along with the temperature-dependent stability of nucleic acids and the saturation levels of either rigid or flexible cell membranes, as reviewed in Pörtner et al. (2007). These molecular-level perturbations are eventually decisive for the health and well-being of farmed fish, or may even make the difference between survival and death in aquaculture facilities lacking thermal refugia.
Heat-shock proteins (HSPs) are prominently investigated responders to different kinds of stress, primarily thermal stress. In recent years, however, research has not been limited to the physiological consequences of thermal stress to the response of HSPs and a pre-selected panel of related genes of interest. Instead, global approaches, such as microarray or RNAseq technologies, were utilized to study the mechanistic link between the environment and transcriptome and decipher comprehensive gene expression changes in salmonid fish species exposed to non-optimal or even lethal ambient temperatures. Such studies across different tissues and cell types proved on the one hand the common upregulation of apoptosis-related genes (Lewis et al., 2010; Rebl et al., 2013; Tomalty et al., 2015) and genes involved in innate immunity (Jeffries et al., 2012; Rebl et al., 2013; Tomalty et al., 2015; Verleih et al., 2015; Li et al., 2017), detoxification (Rebl et al., 2013; Li et al., 2017) and ion transport (Rebl et al., 2013; Tomalty et al., 2015) in addition to a diverse panel of induced stress-related genes (Vornanen et al., 2005; Lewis et al., 2010; Quinn et al., 2011a,b; Rebl et al., 2013; Anttila et al., 2014; Jeffries et al., 2014; Narum and Campbell, 2015; Tomalty et al., 2015; Verleih et al., 2015). On the other hand, genes involved in cold response (Jeffries et al., 2012; Rebl et al., 2013; Verleih et al., 2015) and mRNA translation (Jeffries et al., 2012) were found to be downregulated. Despite their differences, all these scientific reports document the conserved upregulation of particular genes, biofunctions and/or pathways in salmonid fish suffering from different kinds of thermal stress and contribute progressively to our understanding of thermoregulation and thus the well-being of farmed fish. Digital technologies will soon become prominent in farm animal production and high-throughput detection of biomarkers in farmed fish and ambient water are expected to regulate aquaculture husbandry conditions in optimal ranges in the near future.
The present study subjected the first generation of maraena whitefish under small-scale farming conditions to both gradual and acute rise in temperature from 18 to 24°C in order to record plasma parameters and differentially expressed (DE) genes to derive regulated biofunctions and pathways. These datasets were compared with meta-analyzed published datasets from different salmonid fish species to extract a common list of DE features with the potential to indicate temperature stress in salmonid aquaculture.
Materials and Methods
Fish and Experimental Design
Maraena whitefish were bred from wild-fish eggs at the Institute of Fisheries, LFA M-V (Born, Germany), where the temperature experiments described below were conducted. Whitefish at the age of 50 weeks (349 days) post-hatch were randomly transferred from the recirculation system into nine 300-L indoor tanks at a density of ~30 kg/m3 for 7 d. Each of the nine tanks was identical (0.74 m length × 0.58 m width × 0.72 m height) receiving brackish water from the Darss-Zingst Bodden chain with 2.5–6 practical salinity units. Water was pretreated by drum filter and moving bed biofilm reactors in complement with UV radiation and exchanged about 0.5 times per hour. Monitoring characteristic parameters such as the concentrations of dissolved O2, , , , or pH in all tanks throughout the trial ensured a consistently high water quality. Maraena whitefish were fed commercial dry pellets by automatic feeders that distributed the food during the light period 12 h/d.
Two of the nine experimental tanks (termed as A and B in Figure 1) were assigned as “temperature reference” tanks and kept at 18°C for the experimental period of 12 d. The water temperature in two other tanks (C and D), assigned as “gradual temperature rise” was elevated from 18 to 24°C by 0.5°C increments per day,. The remaining five tanks were used for “acute temperature rise.” Water temperature was kept constant at 18°C in two of these five tanks (tanks E and F) for 11 d, but on day 12, maraena whitefish were transferred from these tanks into 24°C-warm tanks (G and H), where the fish were kept for 1 h until sampling. To account for handling procedures during transfer, another fish group, assigned as “handling reference,” was transposed from an 18°C-tempered tank (F) to another 18°C tank (I), in parallel to those groups that were transferred to 24°C tanks (G and H). The handling and sampling procedures of maraena whitefish have been conducted in compliance with terms of the German Animal Welfare Act [§ 4(3) TierSchG] and approved by the institute's ethics commission. Throughout the experiment, no mortality or disease occurred among maraena whitefish and no technical problems were registered.
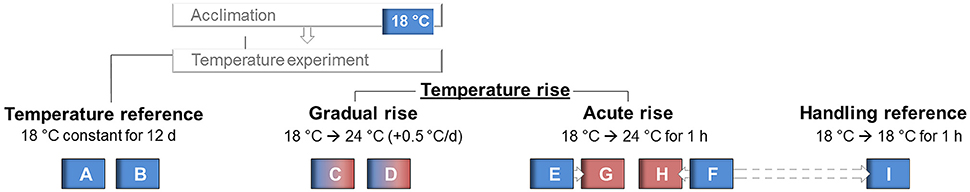
Figure 1. Experimental setup of the temperature experiment subjected to 50-week old maraena whitefish. Temperature in the “reference” tanks (labeled A, B, and I) was kept constant at 18°C throughout the experiment, while water temperature was increased up to 24°C in the “gradual temperature rise” tanks (labeled C, D). For the “acute temperature rise” experiments, maraena whitefish were transferred from 18°C tanks (labeled E, F) to 24°C tanks (labeled G, H).
Sampling and RNA Preparation
At the end of the experiment, fish averaged 30.3 cm ± 1.7 cm in length (mean ± STD), and 300.6 ± 68.5 g in weight. Blood was sampled from the caudal vein of each whitefish using 5-ml plastic syringes filled with 500 μl 0.5 M EDTA (pH 8.0) solution. Tissue samples (entire liver; head and trunk kidney; entire spleen) were taken from seven maraena whitefish from each group.
Tissue fragments of liver, kidney and spleen were snap-frozen in liquid N2 and stored at −80°C until separate extraction of individual RNA with TRIzol reagent (Invitrogen, Carlsbad, CA, US). RNeasy Mini Kit (Qiagen, Hilden, Germany) in complement with the RNase-free DNase Set (Qiagen) gave high-quality RNA yields suitable for microarray hybridizations and subsequent quantitative real-time PCR (qPCR) assays.
Nucleic acid concentrations were quantified by measuring the 260:280 nm absorbance ratio with the NanoDrop 1000 spectrophotometer (NanoDrop Technologies/Thermo Fisher Scientific, Darmstadt, Germany). RNA was quality-checked with the Agilent 2100 Bioanalyzer platform (Agilent Technologies, Waldbronn, Germany) revealing RNA integrity number values ranging from 9.2 to 9.8.
Plasma Cortisol and Glucose Analyses
Blood samples of seven fish from each tank (A, B, C, D, G, H, and I) were centrifuged (4°C, 1700 rcf) and the supernatant was kept on ice until measurement of plasma parameters. According to the manufacturer's instructions, we analyzed the samples using a Cortisol ELISA Assay (DRG Instruments GmbH, Marburg, Germany) and the Glucose Assay Kit II (BioVision, Milpitas, California, US) with the Beckman Coulter DTX 800/880 Series Multimode Detector (Beckman Coulter, Brea, CA, US) for measuring absorbance at 450 nm.
cRNA Synthesis, Array Hybridization, and Data Analysis
Seven individual RNA samples from the same tissue (liver, kidney, spleen) and treatment (reference; gradual temperature rise; acute temperature rise) for each experimental replicate were proportionally pooled and labeled according to the One-Color Microarray-Based Gene Expression Analysis protocol, version 6.6. Briefly, 100 ng of each total RNA sample was amplified and labeled with the fluorescent dye cyanine 3 (Cy3) using the Agilent Low Input Quick Amp Labeling Kit (Agilent Technologies). The dye-incorporation of Cy3-labeled cRNA was measured with the ND-1000 spectrophotometer (NanoDrop Technologies/Thermo Fisher Scientific) revealing a rate between 12 and 20 fmoL/ng. Following fragmentation, 600 ng of Cy3-labeled cRNA was mixed with hybridization buffer (Agilent Gene Expression Hybridization Kit; Agilent Technologies) and hybridized to 8 × 60 K Agilent-049158 Salmon Oligo Microarrays (Agilent Technologies; Gene Expression Omnibus platform: GPL21057) at 65°C for 17 h. Microarrays were then washed once with the Agilent Gene Expression Wash Buffer 1 for 1 min at room temperature followed by a second wash with preheated Agilent Gene Expression Wash Buffer 2 at 37°C for 1 min.
The hybridized Agilent microarrays were scanned immediately after drying at 3-μm resolution (microarray scanner system G2505C; Agilent Technologies). The Agilent Feature Extraction Software (FES) 10.7.3.1 was used to read out the microarray image files. Extracted data were further analyzed using the limma package of the R version 3.1.1/Bioconductor suite (Smyth, 2005). Only genes with a corrected p-value of < 0.05 according to Benjamini and Hochberg (1995), and an absolute fold change of >2.0 were deemed DE genes and considered for further evaluation. Principal component analysis was done using the Rosetta Resolver Data Reduction (Weng et al., 2006). DE genes were re-annotated using the Basic Local Alignment Search Tool BLASTx. We considered only transcripts with unique BLAST results (coverage and sequence identity of >80% and E-value < 1 × 10−4) and joined redundant probes to prevent repeated counting of the same gene in subsequent analyses, such as comparisons of gene expression across different treatments using Venn Diagrams (Hulsen et al., 2008).
Annotated genes and corresponding fold-change (FC) values were imported into the Ingenuity program (Ingenuity Pathway Analyses/Qiagen) to evaluate the canonical pathways of interacting genes and visualize these in global functional networks. Benjamini–Hochberg multiple-testing was used for cut-off criterion. The Ingenuity Knowledge Base represents however a collection of experimental observations in mammalian species; therefore, we excluded the canonical pathways of mammalian diseases or exclusively mammalian pathways. Pathway enrichment analyses were complemented and cross-checked with DAVID-generated lists of KEGG pathways (Huang et al., 2009). In the following, predicted pathways and biofunctions are indicated by italic font. Standard scores (z-scores) were used to evaluate whether a particular pathway was activated (z > 1) or repressed (z < 1); for pathways with z-scores around 0, no prediction could be made.
Confirmation of DE Genes by Fluidigm 48.48 Real-Time qPCR
Quantitative PCR was performed using the integrated microfluidic circuit technology of the Fluidigm Gene Expression chips (Fluidigm, Munich, Germany) and EvaGreen fluorescence dyes (Bio-Rad, Munich, Germany) to validate array-predicted mRNA abundances. The software Pyrosequencing Assay Design (version 1.0.6; Biotage, Uppsala, Sweden) suggested optimal whitefish-specific primers to amplify target fragments between 150 and 190 bp (Table 1). In addition, we used primer pairs that were originally designed for the amplification of gene fragments from trout and revealed a perfect conservation among rainbow trout and maraena whitefish (listed in Table 1). The resulting PCR products were ligated into a pGEM-T Easy Vector (Promega, Mannheim, Germany) and sequenced (Applied Biosystems 3130 Genetic Analyzer; Life Technologies) to verify the respective gene fragments.
RNA samples (used previously for microarray hybridization) were individually reverse-transcribed into cDNA using the Reverse Transcription Master Mix (Fluidigm), preamplified using the PreAmp Master Mix (Fluidigm) and then treated with exonuclease I (New England BioLabs, Frankfurt/Main, Germany). All three steps followed the manufacturers' instructions. In the meantime, the primer assays were pipetted in duplicate into the 48.48 microfluidic circuits. Subsequently, the chip was primed in the MX IFC Controller (Fluidigm). Afterwards, the preamplified cDNA samples (n = 7 per tissue and treatment) were separately loaded on the primed 48.48 chip and analyzed with the Biomark HD system using the manufacturer's thermal protocol “GE Fast 48 × 48 PCR+Melt v2.pcl” (application type: gene expression; passive reference: ROX; assay: single probe).
Standard curves were performed for each primer pair using the LightCycler 96 System (Roche, Mannheim, Germany) in complement with the 2 × SensiFAST SYBR No-ROX Kit (Bioline, Luckenwalde, Germany) (Table 1). The qBase+ software (Biogazelle, Ghent University, Belgium) evaluated the suitability of a range of putative, whitefish-specific reference genes (see Altmann et al., 2015) based on averaged GeNorm stability values of M < 0.37 and coefficients of variation (CV) between 0.12 and 0.15. RPL9, RPL32, and RPS20 were recorded as internal normalizer genes for liver samples, while EEF1A1b, RPL9, RPL32, and RPS20 were used as normalizer genes for kidney and spleen samples.
Quantitative PCR data were analyzed using the Fluidigm Real-Time PCR Analysis software v. 4.0.1 and evaluated based on a linear regression of standard curves for each individual amplicon. Pearson's correlation test (http://www.socscistatistics.com/tests/pearson/) was used to assess the concordance between the microarray and qPCR results.
The full complement of microarray data was deposited in the NIH/NLM Gene Expression Omnibus (GEO, accession code: GSE95422), contributing to the initiative “Functional Annotation of All Salmonid Genomes” (Macqueen et al., 2017).
Meta-Analysis
The GEO database was scanned for gene expression studies focusing on gradual and acute temperature stress in salmonid fish. The gene lists of eligible microarray studies were searched for expression values of the respective target genes. The raw data from RNA-seq runs were received from the European Bioinformatics Institute database (EMBL-EBI) and reads were subsequently assembled to the respective target genes by using the Unipro UGENE bioinformatics software (Golosova et al., 2014). Total reads were counted and normalized to read counts of the reference genes RPL9, RPL32, and RPS20.
Results
The present study compared the physiological impacts of gradual and acute temperature rise from 18to 24°C on maraena whitefish in aquaculture. Figure 1 provides an overview of the experimental setup. Although plasma cortisol (ranging from 7.0 to 273.7 ng/ml) and glucose levels (2.3–8.3 nmol/μl) did not show any significant differences between the analyzed groups due to high standard deviations (Figure 2), we recorded significant changes in gene expression in the challenged “gradual temperature rise” and “acute temperature rise” groups compared with the “temperature reference” group (Figure 3). The number of DE genes was 5-fold smaller in the “acute temperature rise” group compared with the number of DE genes in the “gradual rise” group (Supplementary Figures 1A,B). Moreover, the percentage contribution to the entirety of DE genes by the liver decreased in acute temperature rise (compared to gradual rise) on account of the spleen, while the percentage contribution by the kidney remained at a similar level.
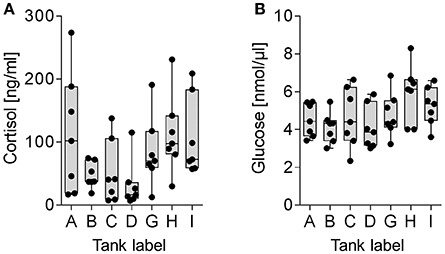
Figure 2. Plasma diagnostics of maraena whitefish kept in reference tanks (labeled A, B, I, abscissa), “gradual temperature rise” tanks (labeled C, D) and “acute temperature rise” tanks (labeled G, H). Box and whisker plots illustrate (A) cortisol levels (ng/ml) and (B) glucose levels (nmol/μl) in the plasma of seven individuals. Individual measuring points are indicated by black dots.
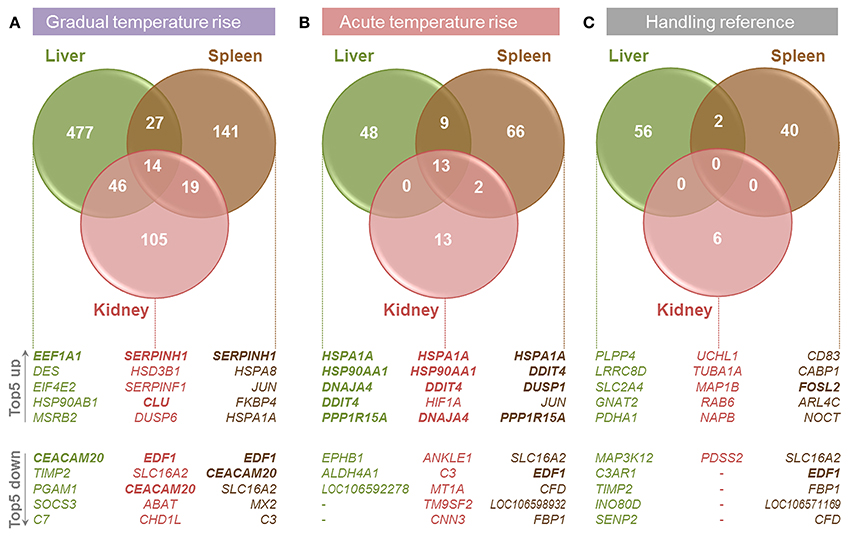
Figure 3. Overview of transcriptome analyses in the liver (green circles), spleen (brown), and kidney (red) of maraena whitefish exposed to (A) gradual temperature rise, (B) acute temperature rise and (C) handling relative to the data obtained for the “temperature reference” group. The number of DE genes is illustrated by Venn diagrams and the five most up- and down-regulated genes in the respective tissues are listed below. Commonly regulated features in all three tissues per treatment are printed in bold.
Tissue-Specific Responses to Gradual Temperature Rise
In the liver of maraena whitefish, gradual temperature rise provoked the highest number of DE genes (n = 564, relative to the “temperature reference” group) compared with the spleen (n = 201) and kidney (n = 184) (Figure 3A). Diverse hepatic signaling pathways were predominantly attributed to blood cells (B cell receptor, erythropoietin, GM-CSF signaling, and CD40 signaling) (Table 2). In addition, the analysis predicted pathways directly (ERK/MAPK signaling) or indirectly (ILK signaling, angiopoietin signaling) involved in the response to stress.
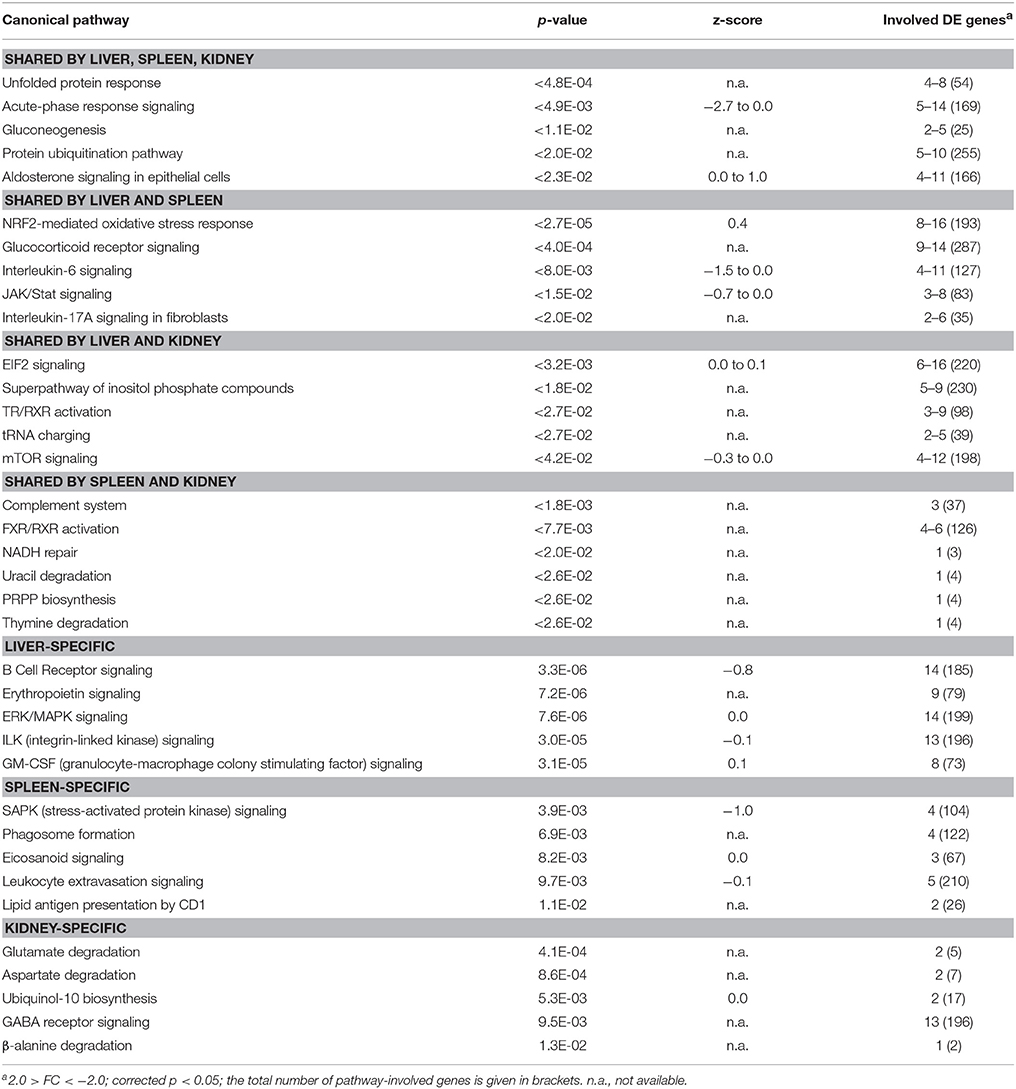
Table 2. TOP5 list of functional pathways regulated in maraena whitefish exposed to gradual heat stress (18–24°C).
In the spleen, gradual heat stress had an even larger impact on several immune pathways in leukocytes including lymphocytes and macrophages (phagosome formation, eicosanoid signaling, leukocyte extravasation signaling, lipid antigen presentation by CD1), whereas metabolic pathways were mainly found in the kidney (glutamate, aspartate, and β-alanine degradation, ubiquinol-10 biosynthesis) (Table 2).
Tissue-Specific Responses to Acute Temperature Rise
Acute temperature stress induced a significantly lower number of DE genes than did gradual temperature stress in the liver (n = 70, relative to “temperature reference”), spleen (n = 90), and kidney (n = 28) (Figure 3B). In the spleen, these regulated features were predicted to participate in the second messenger-triggered response to challenging stimuli (cAMP-mediated signaling, G-protein coupled receptor signaling, protein kinase A signaling, ceramide signaling) (Table 3). Also, in the liver and kidney, a general response to stress was reflected by indicative pathways, such as the stress-induced control of translation (EIF2 signaling, regulation of eIF4 and p70S6K signaling), stress signaling (PCP pathway, mTOR signaling), and metabolic responses (glycolysis) (Table 3).
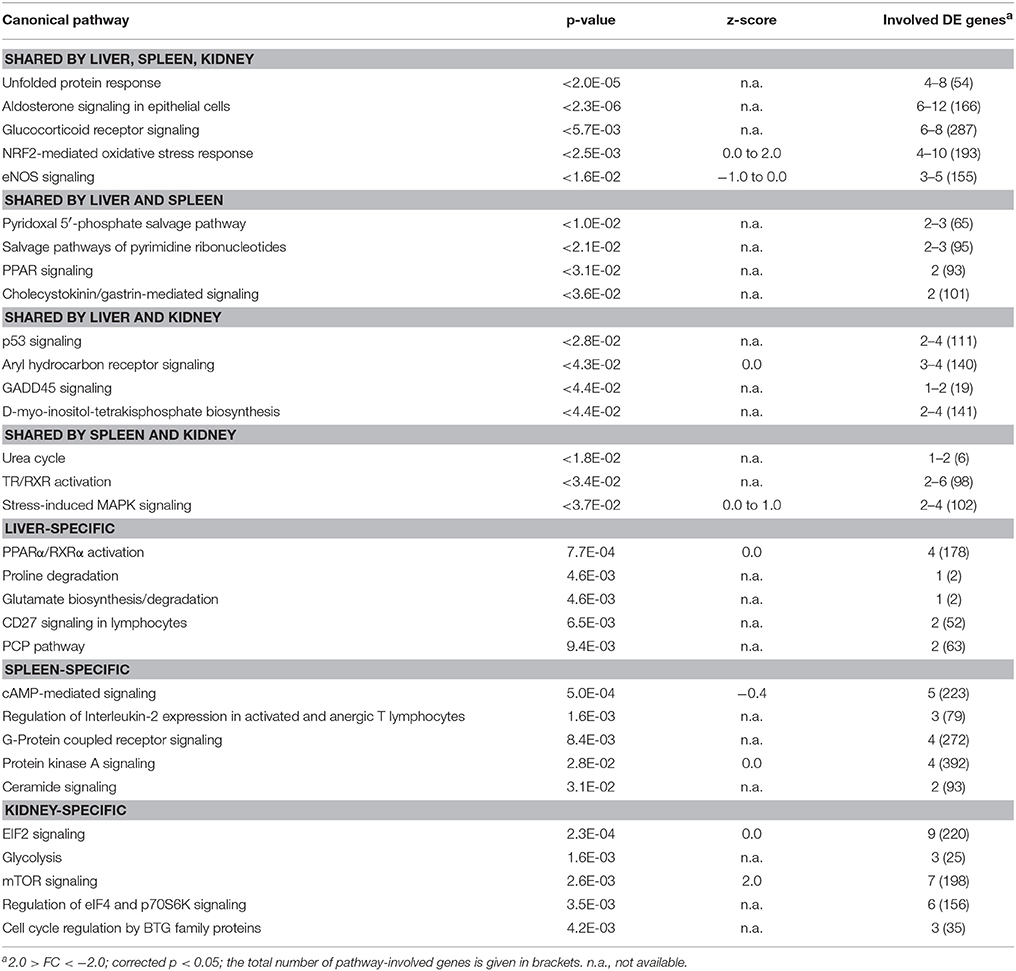
Table 3. TOP5 list of functional pathways regulated in maraena whitefish exposed to acute heat stress (18–24°C).
Tissue-Specific Responses to Handling Stress
As we expected that not only would the sudden temperature change affect gene expression in the “acute temperature rise” group, but also the transfer procedure itself, we analyzed the gene expression after transferring fish from one tank to another (Figure 3C). This handling stress had, as expected, a detectable impact on the mRNA levels of a particular number of genes in the liver (n = 58, relative to “temperature reference”), spleen (n = 42) and, to a lesser extent, kidney (n = 6). Of note, “gradual temperature rise” and “handling reference” shared 18 features in the liver and 22 features (including EDF1) in the spleen, while “acute temperature rise” and “handling reference” shared 15 features (including DDIT4) in the liver and 35 features (including EDF1, PPP1R15A, FOSL2) in the spleen. Regarding the kidney, there was no overlap between the DE gene lists among the respective groups.
The Responses to Gradual and Acute Temperature Rise Are Characterized by Distinct Gene-Expression Profiles
Pairwise comparisons revealed that a gradual temperature rise from 18 to 24°C induced the differential expression of a 2-fold (spleen) to 8-fold (liver) larger set of genes than did the acute temperature rise in the same range (Figures 3A,B and Supplementary Figure 1). In contrast, the lists of DE genes in fish transferred from the 18-°C tempered tanks to equally warm tanks (“handling reference”) or to tanks with 6°C warmer water (“acute temperature rise”) were similarly large (Figures 3B,C).
The comparison of DE gene sets in the liver, spleen, and kidney after gradual temperature rise pointed to 12 shared DE features (including duplicated EEF1A1 copies and an unknown feature; c.f. Figure 4A, upper panel). This gene set includes both upregulated (EEF1A1) and downregulated features (SPCS2, DDX5, EDF1) linked with the biofunctions regulation of transcription and/or translation. Furthermore, it contains upregulated genes involved in unfolded protein response (SERPINH1, CLU) and downregulated genes related to response to cold (YBX2a, CIRBP) and glucocorticoid receptor signaling (AGT, FKBP5). Ingenuity bioinformatic analyses predicted that upstream transcription factor 1 (USF1) and the interaction of the transcriptional activators heat shock factors 1 and 2 (HSF1,−2), bHLH transcription factor MYC, CCAAT/enhancer binding protein beta (CEBPB), and deactivated tumor protein p53 (TP53) control the expression of the majority of the above genes, at least in mammals (Figure 4B).
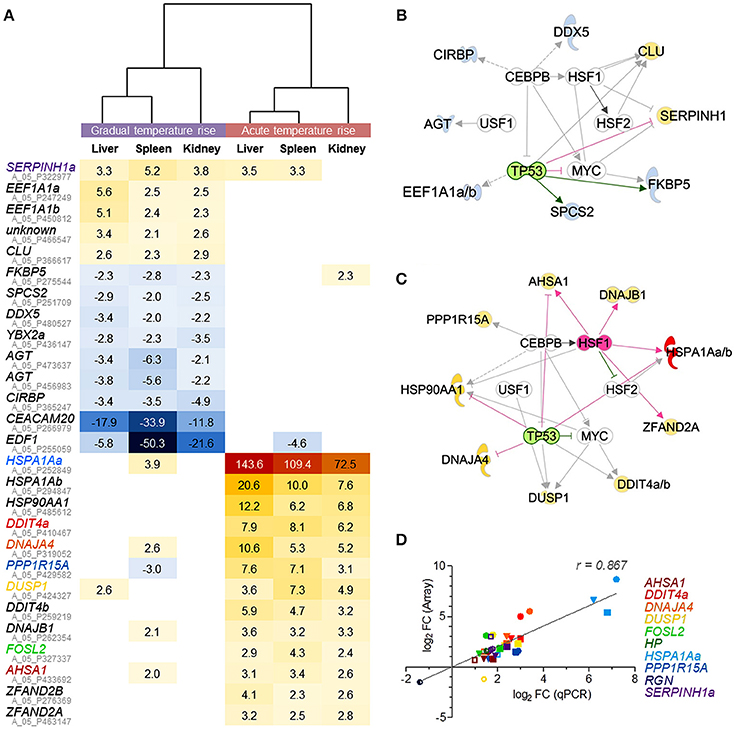
Figure 4. Microarray-based prediction of DE genes for gradual and acute temperature stress in maraena whitefish. (A) Hierarchical representation of DE features (p < 0.05; q < 0.05; −2.0 > FC < 2.0) shared by the liver, spleen, and kidney of maraena whitefish exposed to “gradual” (left panel) or “acute temperature rise” (right panel) as measured with microarray technology. The intensity and shade of the colored cells represent the normalized ratios, obtained after pairwise analysis with the “temperature reference group.” Gene symbols are listed with their respective Agilent IDs (in gray); the coloring of gene symbols (corresponding to the symbol colors in D) indicates that 10 selected FC values were validated using microfluidic qPCR. Interaction networks were generated using the IPA software to predict potential upstream regulators responsible for altered gene expression after (B) “gradual temperature rise” and (C) “acute temperature rise.” These regulators are arranged in the inner circle; predicted activation and inhibition are highlighted in pink and green, respectively. The affected genes are arranged in the outer circle; observed upregulation and downregulation are highlighted according to the colors of the heat map in (A). The relationships between regulators and genes are displayed by arrows indicating induction (pink), repression (green) or uncertainty due to lack of knowledge or the state of downstream factors (gray). (D) The linear correlation plot shows the concordance of array (ordinate) and qPCR (abscissa) results for selected genes. Normalized log2 FC values were plotted for “gradual temperature rise” vs. “temperature reference” (open symbols), “acute temperature rise” vs. “temperature reference” (filled symbols) and “gradual” vs. “acute temperature rise” (semi-open symbols) comparisons in in the liver (circle), spleen (square) or kidney (triangle). Pearson's correlation is indicated. Symbol colors correspond with the colors of the gene symbols, as listed on the right margin and in (A).
A different set of 13 genes (including duplicated HSPA1A and DDIT4 gene copies) was commonly upregulated across the liver, spleen and kidney after acute temperature rise (Figure 4A). Again, USF1, HSF2, MYC, CEBPB, and activated HSF1 with simultaneously inactivated TP53 were predicted to promote the expression of most of these 13 genes (Figure 4C) contributing to the pathways aldosterone signaling (HSPA1Aa, -b, HSP90AA1, DNAJB1, DUSP1) and unfolded protein response (HSPA1Aa, -b, PPP1R15A, DNAJA4), involving most likely co-chaperones (AHSA1) and factors with HSP-like functions (encoded by ZFAND2A, ZFAND2B), as well as factors regulating cell proliferation and cell death (FOSL2, DDIT) (Figure 4A, lower panel).
In conclusion, the upstream regulatory networks in Figures 4B,C suggest that an identical set of partially modulated transcriptional activators trigger very distinct responses to either gradual or acute temperature rise in the liver, spleen, and kidney. Although the two pathways unfolded protein response and aldosterone signaling in epithelial cells belong to the most-regulated pathways after gradual and acute temperature stress (Tables 2, 3), both are constituted by completely different gene sets. In addition, the two different treatment conditions activated several distinct stress type- and tissue-specific pathways.
Microfluidic qPCR Data Confirm the Microarray-Identified Expression Differences of Selected Genes
The microfluidic qPCR technology was used to verify the array-predicted expression differences of ten selected genes induced by either gradual or acute temperature rise and that are known to be involved in different mammalian cellular pathways (cf. Tables 2, 3), i.e., DDIT4a (mTOR signaling), DNAJA4 (NRF2-mediated oxidative stress response), DUSP1 (p38 MAPK signaling), HP (acute-phase response signaling), HSPA1Aa (aldosterone signaling in epithelial cell, unfolded protein response), PPP1R15A (EIF2 signaling), RGN (glucose/glucose-1-phosphate degradation) and SERPINH1a (collagen biosynthesis), as well as AHSA1 and FOSL2 (Figure 4D). Profiles across these ten target genes confirmed a high concordance (Pearson product-moment correlation coefficient, r = 0.9) between both technologies. Moreover, qPCR data confirmed the statistical significance of the large majority of microarray-predicted FC values of fish exposed to the “gradual temperature rise” or “acute temperature rise” relative to the “temperature reference” fish (Supplementary Table 1).
Meta-Analysis Endorses the Modulation of Particular Gene-Expression Profiles Upon Acute Temperature Stress in Different Salmonid Species
Our array analysis identified 22 genes (excluding duplicated copies) that were commonly regulated by gradual and acute temperature rise in the three investigated maraena whitefish tissues (cf. Figure 4A). Based on this list, we screened similar bioprojects having detected differentially regulated genes upon heat stress in salmonid fish species using microarray (Vornanen et al., 2005; Lewis et al., 2010; Quinn et al., 2011a,b; Rebl et al., 2013; Anttila et al., 2014; Jeffries et al., 2014; Verleih et al., 2015) or RNA-seq technique (Narum and Campbell, 2015; Tomalty et al., 2015). It should be noted that these investigations cover a broad temperature range (4–28°C) and also a broad range of tissues (including heart ventricle, kidney, gills, and blood cells), apart from different feeding regimes, sampling procedures and housing conditions.
The enquiry confirmed, however, that the maraena-whitefish genes induced by acute temperature rise in our study have the potential to reliably indicate acute heat stress in salmonid fish in general, as they were also significantly upregulated in at least three out of the seven acute-stress studies (Figure 5). The only exception is DNAJA4, which was found to be upregulated by a factor of three in one single array-based investigation (Verleih et al., 2015). In contrast to acute-stress-responding genes, the genes responding to gradual stress revealed a rather occasional accordance with our list of DE genes from maraena whitefish. Nevertheless, we found for SERPINH1, EEF1A1, and CIRBP one to three gradual-stress studies supporting our results.
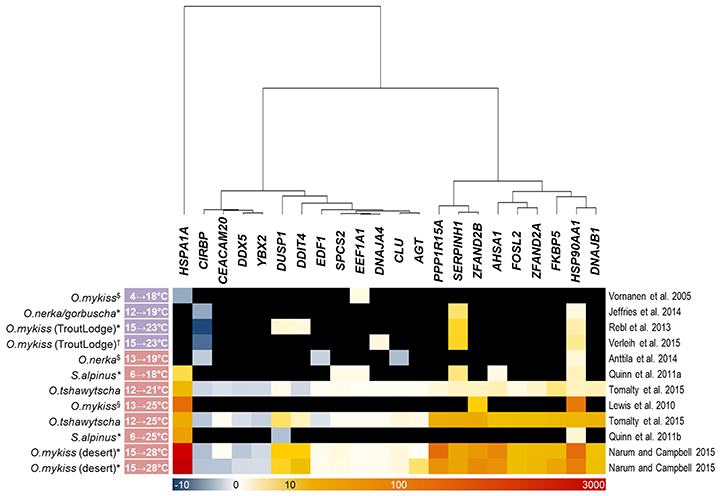
Figure 5. Thermoregulated genes in salmonid fish species after gradual and acute temperature rise (marked with a purple and red label, respectively, alongside the experimental temperature range given at the left-hand side) investigated in previous studies. The intensity and shade of the colored cells correspond to the color bar below the heat map and reflect FC expression values relative to the respective control groups; black fields denote missing information (only referring to microarray studies). Note that the included studies focused on different salmonid species (incl. strains) and—if applicable—tissues, marked with ($) for heart ventricle, (*) for gills, (†) for kidney, and (§) for red blood cells, as indicated. A literature reference is given at the right-hand side.
Discussion
Improper husbandry conditions generally equate either to the sudden and possibly repeated appearance of one or even a combination of distinct environmental stressors (acute stress) or the persistent presence of stressful stimuli (chronic stress). Fish are—like all vertebrates—able to buffer exogenously induced perturbations to their homeostasis, which is reflected by various physiological alterations, including modulated gene expression. Therefore, particular gene-expression profiles indicate the type of stress exposure. The fast and reliable detection of an informative set of animal-based marker genes indicating the type and the severity of stress exposure would clearly improve welfare concepts and hence the health of farmed fish in aquaculture facilities.
In the present study, maraena whitefish were subjected to gradual or acute heat stress from 18 to 24°C. As expected, the majority of the commonly regulated genes in the investigated tissues encode cytoprotectors and key responders to stress, as discussed below. Moreover, we recorded the levels of the “traditional” plasma parameters cortisol and glucose, but we did not detect any statistically significant differences across the investigated groups. Previous analyses in salmonids revealed that cortisol concentrations of about 100 mg/ml and higher reflect stressful conditions (Procarione et al., 1999; Gesto et al., 2008; Conde-Sieira et al., 2010; Tahmasebi-Kohyani et al., 2012). We detected similarly high cortisol concentrations with high inter-individual variance in all groups analyzed. Since the maraena whitefish used in the present study were the first generation maintained in an anthropogenic environment and therefore considered as sensitive as wild fish, the elevated cortisol concentrations suggestively indicate the immediate response to the final sampling procedure rather than to the experimental stressor (see also Ortuño et al., 2002; Gräns et al., 2016). Additionally, numerous researches have expressed doubt about the reliability and information value of both parameters under certain circumstances (Martinez-Porchas et al., 2009; Roberts et al., 2010; Aerts et al., 2015; Rebl et al., 2017), also because preformed cortisol can be rapidly released upon exposition to an acute stressor, while gene expression changes take significantly longer periods of time (Das et al., 2018) and should thus be better suited to assess fish's long-term response to stress.
Both, gradual and acute temperature stress activated the expression of distinct—almost non-overlapping—sets of genes reflecting specific strategies either to adapt to slightly elevating ambient temperatures or to survive a heat shock. The repeated appearance of a low-intensity stressor was proven to eventually lead to attenuated stress responses. This desensitization process represents an important step on the way to the adaptation to moderately stressful conditions, as reviewed in von Borell (2000), and is characterized by comprehensive alterations in gene expression, as observed in this study for maraena whitefish within only 12 days in a gradually increasing ambient temperature. In contrast, harmful stress stimuli are likely to intensify the physiological response, known as sensitization. Provided that our temperature trial evoked two different responses in the direction of desensitization or sensitization, both responses were necessarily expected to be characterized by a cluster of particular DE genes.
DE Genes and Specific Pathways Indicate the Response to Gradual Temperature Rise
The “gradual temperature stress” experiment was configured to mimic the seasonal elevation of water temperature in open aquaculture facilities. Surprisingly, we found downregulated features commonly present in liver, spleen, and kidney of gradually—but not acutely—stressed maraena whitefish. A conclusive explanation for this is that few of those genes are activated upon cooling, i.e., the cold-inducible RNA-binding protein (CIRBP), Y-box protein 2 (YBX2) and angiotensinogen (AGT).
CIRBP is probably the best characterized “cold-shock protein” in vertebrates (Phadtare et al., 1999). It is significantly upregulated after cold-shock (Gracey et al., 2004) and consequently down-regulated in fish during heat stress (Rebl et al., 2013; Verleih et al., 2015). Our research at gene-expression databases confirmed that reduced CIRBP levels are frequent indicators of gradual and also acute heat stress. For instance, in heat-stressed Chinook salmon Oncorhynchus tshawytscha, CIRBP has been downregulated together with YBX2 (Tomalty et al., 2015). YBX2 (alias MSY2 and CSDA3) also belongs to the “cold-shock proteins” containing a “cold-shock conserved site” mediating the survival of prokaryotes at temperatures below the optimal growth temperature (Phadtare et al., 1999).
Low temperatures activate the renin–angiotensin–aldosterone system, of which angiotensinogen (AGT, alias SERPINA8) is a principal effector elevating blood pressure, thus reducing blood flow and heat emission (Hiramatsu et al., 1984; Cassis et al., 1998). Vice versa, the reduced AGT levels observed in gradually heat-stressed maraena whitefish most likely indicate accelerated blood flow to allow for increased oxygen transport rate in order to counteract hypoxia. Angiotensinogen is considered an acute-phase protein (Soden et al., 1994) that is involved in acute-phase response signaling in maraena whitefish exposed to gradual, but not acute, temperature rise. Here, AGT was predicted to act in conjunction with members of the complement cascade (C3, CFB), mitogen-activated protein kinases (MAPK), haptoglobin (HP) and other acute-phase proteins. The regulation of the acute-phase response signaling together with complement system during gentle temperature stress over a longer period of time has been previously observed in rainbow trout (Rebl et al., 2013).
The heat-shock protein HSP47, encoded by the SERPINH1 gene, emerged in the present study as a clearly increased feature in different tissues of maraena whitefish challenged either by gradual or acute temperature rise. Previous studies into the thermal response of salmonids have demonstrated the significant potential of SERPINH1 as a biomarker for heat stress in various tissues of different salmonid species, i.e., sockeye salmon Oncorhynchus nerka (Jeffries et al., 2012), rainbow trout O. mykiss (Rebl et al., 2013; Verleih et al., 2015; Wang et al., 2016; Li et al., 2017), Pacific salmon O. gorbuscha and O. nerka (Jeffries et al., 2014), Chinook salmon O. tshawytscha (Tomalty et al., 2015) and lake whitefish Coregonus clupeaformis (Stefanovic et al., 2016). HSP47 localizes to the endoplasmic reticulum (ER) and mainly chaperones collagen (Nagata et al., 1988; Nakai et al., 1992), a major component of the extracellular matrix, in mammals and in fish (Bhadra and Iovine, 2015). In this respect, it is worth noting that the predominant secondary structures in collagen are concatenated (hydroxyl-)proline residues and the related pathways (hydroxyl-)proline degradation were overexpressed in the liver of maraena whitefish that were challenged by gradual and acute temperature rise. Besides SERPINH1, we detected more upregulated members of the HSP family in the liver (HSP90AB1, HSPB5, HSPA8), spleen (HSPA8, HSP1A1, HSP90AB1, DNAJA4, DNAJB1) and kidney (HSPA5) of gradually stressed maraena whitefish, but these were not uniformly induced in all three tissues. We refer in this context to FK506-binding protein 5 (FKBP5), which inhibits the activity of the heterocomplex of HSP90 and the glucocorticoid receptor (Nair et al., 1997; Wochnik et al., 2005). FKBP5 was downregulated in gradually stressed maraena whitefish indicating most likely a removal of barriers to enhance the responsiveness of the glucocorticoid receptor (Gillespie et al., 2009) reflected by the modulation of the glucocorticoid receptor signaling pathway in the liver and spleen. Interestingly, we and others (Narum and Campbell, 2015; Tomalty et al., 2015) found that acute stress provokes the upregulation of FKBP5, suggesting that the factor itself is tightly controlled dependent on the intensity, type and/or duration of stress.
Comparable to the SERPINH1 mRNA abundance, the level of mRNA coding for clusterin (CLU) and the eukaryotic translation elongation factor 1-alpha 1 (EEF1A1, represented by duplicated gene copies) were significantly increased after gradual stress in maraena whitefish. CLU (alias APOJ) is not an HSP, but is considered as an extra- and intra-cellular chaperone that binds to misfolded proteins and inhibits their aggregation (Wyatt et al., 2009). Reflecting these vital functions, CLU has been referred to a “sensitive biosensor of environmental insults, specifically oxidative stress” (Trougakos, 2013) and “a protector against stress” in mammals (Criswell and Boothman, 2005). EEF1A1 has been detected as an upregulated feature in heat-stressed rainbow trout (Vornanen et al., 2005). The encoded elongation factor supports the binding of heat-shock factor 1 to its response element in mammalian target promoters (Shamovsky et al., 2006) and ensures the fast export from the nucleus and the efficient translation of HSP1A1-encoding mRNAs (Vera et al., 2014). Moreover, EEF1A1 is inhibits the pro-apoptotic actions of TP53 (Blanch et al., 2013). This effect might have been enhanced by the down-regulation of DDX5, as observed in the present study in gradually stressed maraena whitefish. The DEAD-box helicase 5 is also termed p68 and since it acts as a co-activator of murine TP53 (Nicol et al., 2013) and is thus involved in multiple cellular decisions on survival and stress response in a context-dependent manner (reviewed in Shih and Lee, 2014).
Similarly, pleiotropic functions are attributed to the genes encoding endothelial differentiation-related factor 1 (EDF1), subunit 2 of the signal peptidase complex (SPCS2) and the carcinoembryonic antigen-related cell adhesion molecule 20 (CEACAM20), which were commonly repressed by gradual temperature rise in maraena whitefish. EDF1 (alias MBF1) interacts with calmodulin (Mariotti et al., 2004) and a member of the JUN family (Kabe et al., 1999) to suppress endothelial differentiation (Dragoni et al., 1998) and regulate a number of intracellular proteins, mainly signal transducers including those associated with the response to oxidative stress (Jindra et al., 2004). Members of the CEACAM family have manifold regulatory functions in epithelial, endothelial, and immune cells and it has been suggested that CEACAM20 transduces stimulatory signals (Zebhauser et al., 2005). The signal peptidase complex (SPC) is vital for the synthesis of all membrane and secretory proteins (Kalies and Hartmann, 1996). The downregulation of the subunit 2-encoding gene SPCS2 in stressed maraena whitefish could indicate a generally reduced protein synthesis to leave energy resources for the dominant stress coping mechanisms. In this respect, the strong dependency on available glucose has been attributed to SPC subunit 2 besides its particular role during the unfolded protein response in mammalian cells (Hassler et al., 2015). In line with that, the gluconeogenesis pathway has been modulated as a prominent pathway in the liver, spleen, and kidney of gradually stressed maraena whitefish, but only in the kidney of acutely stressed maraena whitefish. The “neuroendocrine response to stress is characterized by excessive gluconeogenesis” (Marik and Bellomo, 2013), and it can be assumed that the duration of stress conditions enhance the hyperglycemia phenotype.
Altogether, we measured the dynamic regulation of almost two-hundred to more than five-hundred genes in the liver, spleen and kidney of maraena whitefish exposed to gradually rising temperature. This could strongly indicate the body's efforts to adapt to a moderate stressor. Despite the high number of DE genes, we identified only 11 genes that were both, up- and down-regulated—most likely in a systemic manner.
DE Genes and Specific Pathways Indicate the Response to Acute Temperature Rise
Our “acute temperature rise” experiment exposed maraena whitefish to nearly critical temperatures, as proven for other coregonids (Edsall et al., 1970; Edsall and Rottiers, 1976) in terms of a “temperature shock.” Eleven genes showed pronounced inductions across the three investigated tissues of maraena whitefish. Our literature research confirmed that all these DE genes have been described earlier as highly regulated features after heat or hypoxic conditions.
The HSP70 family member HSPA1Aa was the most strongly upregulated gene of the present study and seems thus to be a biomarker candidate indicating a sudden “heat shock” in maraena whitefish. In fact, HSPA1A is the best investigated and most heat-inducible gene among vertebrates (Kampinga et al., 2009), including salmonid fish (Smith et al., 1999; Lewis et al., 2010; Quinn et al., 2011a; Narum and Campbell, 2015; Tomalty et al., 2015). A previous investigation on acute heat shock in Atlantic cod Gadus morhua (Hori et al., 2010) revealed that the mRNA abundance of HSPA1A increases by a level similar to that detected in the present study. The chaperone activity of HSPA proteins is generally regulated by DNAJ/HSP40 family members. In humans, the co-chaperone DNAJB1 is the most widely expressed and most heat-inducible member of the DNAJ family. In the three tissues investigated in maraena whitefish, DNAJB1 was significantly upregulated together with DNAJA4. The latter gene is upregulated as early as 2 h after exposure to heat stress in chickens (Slawinska et al., 2016), a species with limited thermoregulatory capacities. Furthermore, DNAJA4 is induced in the course of an unfolded protein response in Atlantic salmon (Bower and Johnston, 2010) and early after bacterial infection in channel catfish Ictalurus punctatus (Song et al., 2014). Though it has been documented that increased DNAJA4 levels elevate the synthesis of cholesterol, a precursor of the stress hormone cortisol (Robichon et al., 2006), we detected only a slight tendency of higher cortisol concentrations in the “acute temperature rise” group compared to the “gradual temperature rise” group.
A second chaperone/co-chaperone pair that emerged as a strongly regulated features in the present study is HSP90 and AHSA1 encoding the activator of heat-shock 90-kDa protein ATPase homolog 1 (alias AHA1). HSP90AA1 is already known as a universal biomarker indicating heat stress, in mammals and fish (Buckley et al., 2006; Quinn et al., 2011a,b; Anttila et al., 2014; Jeffries et al., 2014; Tomalty et al., 2015; Verleih et al., 2015). The ATPase activity of HSP90 is stimulated by AHSA1, which chaperones its own client proteins (Tripathi et al., 2014). In line with this, AHSA1 has been identified as heat-shock responsive in Chinook salmon (Tomalty et al., 2015) and channel catfish (Liu et al., 2013).
Besides the canonical members of the HSP family, the two zinc finger AN1 type-containing protein-encoding genes ZFAND2A and -2B were upregulated in acutely stressed maraena whitefish. ZFAND2A (alias AIRAP) has been assessed as functionally homologous to canonical HSPs, as its functionality in humans is induced by heat and depends strictly on HSF1 (Rossi et al., 2010). The paralog ZFAND2B (alias AIRAPL) interacts with the proteasome, thereby stimulating the degradation of misfolded proteins (Braunstein et al., 2015). Moreover, ZFAND2A has already been suggested as a genetic marker in Atlantic cod G. morhua correlating with habitat differences in salinity and oxygen (Berg et al., 2015).
Our pathway analysis identified the unfolded protein response (the major ER stress pathway) and the aldosterone signaling in epithelial cells (the major pathway of electrolyte homeostasis) as two characteristic pathways for both acute and gradual heat stress in maraena whitefish. In the “acute temperature rise” group, both pathways were predominantly constituted by the HSPA, HSPC/HSP90 and DNAJ members of the diverse HSP family and this gene set overlaps only marginally with the respective one of the “gradual temperature rise” group. In this respect, we refer to the gene coding for the regulatory subunit 15A of protein phosphatase 1 (PPP1R15A, alias GADD34) that was upregulated in acutely stressed trout (Narum and Campbell, 2015; Tomalty et al., 2015) and maraena whitefish in the current investigation. The stress-inducible PPP1R15A (Fornace et al., 1988) supports the cellular recovery from the unfolded protein response, as suggested for mouse (Kojima et al., 2003) and grouper Epinephelus spp. (Lu et al., 2012). The pathway analysis revealed additionally that NRF2 (Nuclear factor erythroid 2-related factor 2)-mediated oxidative stress response together with the eNOS signaling pathway was activated in all three investigated tissues of acutely stressed maraena whitefish, possibly mirroring the increased utilization of oxygen accompanied by elevated rates of reactive oxygen species in cold-water fish.
The immediate-early gene FOSL2 (alias FRA2) encodes the leucine zipper-containing FOS-like antigen 2 heterodimerizing with proteins of the JUN family, which are upregulated together with FOSL2 by “acute temperature rise” (cf. panel “TOP5 up” in Figures 3C,D). FOSL2 and JUN form the transcription factor AP-1, which in turn regulates cell proliferation, differentiation, and transformation. Cell proliferation is among the regulated pathways in brown trout Salmo trutta suffering from oxidative stress with FOSL2 as one of the most strongly upregulated features (Uren Webster and Santos, 2015). Stress experiments in mice have disclosed that mRNA levels of FOSL2 recover fast (Weinberg et al., 2007) suggesting that this gene might be indeed an informative indicator of acute stress responses. The experimental exposition of zebrafish to acute oxidative stress induced a series of genes involved in stress response, including FOS together with HSP1A1, HSP90AA1, and DNAJB1 (Hahn et al., 2014). In that study, DUSP1 (alias MKP1, PTPN10) also emerged as a fast responsive gene. The encoded dual specificity phosphatase 1 is a short-lived protein (Brondello et al., 1999) that dephosphorylates the mitogen-activated protein kinases ERK, JNK, and p38, and is thus intimately involved in stress signal transduction and, most likely, also in acutely stressed maraena whitefish.
The DNA damage-inducible transcript 4 DDIT4 (alias REDD-1 and Rtp801) is another factor involved in stress signal transduction. DDIT4 has been identified as a heat- and hypoxia-responsive target gene of the hypoxia-inducible factor 1 and controls the proapoptotic mTOR signaling (Shoshani et al., 2002; Brugarolas et al., 2004). Also in the present study, the mRNA levels of two DDIT4 isoforms increased in all investigated tissues, concurrently with modulated mTOR signaling. The modulation of the DDIT4 expression across several tissues results most likely from glucocorticoids circulating in stressed individuals (Wong et al., 2010).
In summary, our study demonstrates that an acute temperature rise induces a significantly smaller set of DE genes in the liver, spleen and kidney of maraena whitefish than does a gradual temperature rise. The temperature-responsive genes commonly regulated in the respective tissues of whitefish may represent interesting candidates for future biomarker studies, as supported by several scientific reports about their functional involvement in stress-coping mechanisms across different animal models and experimental challenges.
Conclusions
Stress has been defined as “a condition that disturbs the normal function of the biological system” (Sørensen et al., 2003). Vertebrates have developed different strategies to launch finely tuned responses for coping with different types of stressors. The present study investigated the influence of both acute and gradual change in ambient water temperature on maraena whitefish up to a critical temperature of 24°C. We clearly observed distinct transcriptomic responses to both kinds of thermal stress. In the three stress-relevant tissues (liver, spleen and kidney), 3 genes were commonly upregulated (SERPINH1, EEF1A1, CLU) and 8 genes were commonly downregulated (EDF1, CEACAM20, CIRBP, AGT, YBX2, DDX5, SPCS2, FKBP5) after gradual temperature rise. However, we noticed that the collection of genes responding to “gradual temperature rise” identified for other salmonid fish species so far is rather diverse and obviously strictly dependent on the duration of the experiment and the sampling time point, as well as on the chosen temperature range, the species and the tissues or cell types investigated. In contrast, the list of 11 genes responding to “acute temperature rise” (HSP1A1, HSP90AA1, DDIT4, DNAJA4, PPP1R15A, DUSP1, DNAJB1, FOSL2, AHSA1, ZFAND2A, and -B) that were exclusively upregulated after heat shock, is mostly in accordance with previous reports on acute-stress genes in vertebrates and specifically in salmonid fish, although rainbow trout and maraena whitefish, for instance, are at different stages of domestication and hence differentially adapted to anthropogenic environments. Subsequent analyses are required to evaluate if the identified list of DE genes constitutes a suitable set of biomarkers allowing for diagnostic predictions about the welfare status of whitefish in aquaculture facilities.
Author Contributions
TG, SA, and AR designed research. RB organized the production of fish and provided experimental facilities. MN, SA, MV, AR, and TG sampled fish and prepared samples for hybridization experiments. MN performed measurement of plasma parameters. AR and MN conducted RT-qPCR assays. AR and MV analyzed data. AR wrote the paper.
Funding
This project of the Campus bioFISCH M-V was financed by project VI-560/730-32614 (Maraena whithefish genomics) granted by the European Fisheries Fund (EFF) and the Ministry of Agriculture, the Environment and Consumer Protection Mecklenburg-Western Pomerania, Germany. The publication of this article was funded by the open access funds of the Leibniz Association and the Leibniz Institute for Farm Animal Biology (FBN).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer SFC and handling Editor declared their shared affiliation.
Acknowledgments
We thank Brigitte Schöpel, Ingrid Hennings and Luisa Falkenthal for their excellent technical assistance and Frieder Hadlich for help with data analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00241/full#supplementary-material
References
Aerts, J., Metz, J. R., Ampe, B., Decostere, A., Flik, G., and De Saeger, S. (2015). Scales tell a story on the stress history of fish. PLoS ONE 10:e0123411. doi: 10.1371/journal.pone.0123411
Altmann, S., Rebl, A., Kühn, C., and Goldammer, T. (2015). Identification and de novo sequencing of housekeeping genes appropriate for gene expression analyses in farmed maraena whitefish (Coregonus maraena) during crowding stress Identification and de novo sequencing of housekeeping genes appropriate maraena. Fish. Physiol. Biochem. 41, 397–412. doi: 10.1007/s10695-014-9991-y
Anttila, K., Eliason, E. J., Kaukinen, K. H., Miller, K. M., and Farrell, A. P. (2014). Facing warm temperatures during migration: cardiac mRNA responses of two adult Oncorhynchus Nerka populations to warming and swimming challenges. J. Fish Biol. 84, 1439–1456. doi: 10.1111/jfb.12367
Arndt, G.-M. (2001). Wiederansiedlung, Bestandserhöhung und Bestandsmanagement der Großen Maräne (Coregonus lavaretus) sowie Bestandseinschätzung und –stabilisierung der Kleinen Maräne (Coregonus albula) in Mecklenburg-Vorpommern. Jahresheft Fisch und Umwelt. 71–80. Available online at: https://www.yumpu.com/de/document/view/30661252/jahresheft-2001-fisch-und-umwelt-mecklenburg-vorpommern-ev
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300.
Berg, P. R., Jentoft, S., Star, B., Ring, K. H., Knutsen, H., Lien, S., et al. (2015). Adaptation to low salinity promotes genomic divergence in atlantic cod (Gadus morhua L.). Genome Biol. Evol. 7, 1644–1663. doi: 10.1093/gbe/evv093
Bhadra, J., and Iovine, M. K. (2015). Hsp47 mediates Cx43-dependent skeletal growth and patterning in the regenerating fin. Mech. Dev. 138, 364–374. doi: 10.1016/j.mod.2015.06.004
Blanch, A., Robinson, F., Watson, I. R., Cheng, L. S., and Irwin, M. S. (2013). Eukaryotic translation elongation factor 1-alpha 1 inhibits p53 and p73 dependent apoptosis and chemotherapy sensitivity. PLoS ONE 8:e66436. doi: 10.1371/journal.pone.0066436
Bower, N. I., and Johnston, I. A. (2010). Discovery and characterization of nutritionally regulated genes associated with muscle growth in Atlantic salmon. Physiol Genomics 42A, 114–130. doi: 10.1152/physiolgenomics.00065.2010
Braunstein, I., Zach, L., Allan, S., Kalies, K.-U., and Stanhill, A. (2015). Proteasomal degradation of preemptive quality control (pQC) substrates is mediated by an AIRAPL-p97 complex. Mol. Biol. Cell 26, 3719–3727. doi: 10.1091/mbc.E15-02-0085
Brietzke, A., Borchel, A., Altmann, S., Nipkow, M., Rebl, A., Brunner, R. M., et al. (2016). Transcriptome sequencing of maraena whitefish (Coregonus maraena). Mar. Genomics 29, 27–29. doi: 10.1016/j.margen.2016.05.006
Brondello, J. M., Pouysségur, J., and McKenzie, F. R. (1999). Reduced MAP kinase phosphatase-1 degradation after p42/p44MAPK-dependent phosphorylation. Science 286, 2514–2517.
Brugarolas, J., Lei, K., Hurley, R. L., Manning, B. D., Reiling, J. H., Hafen, E., et al. (2004). Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes. Dev. 18, 2893–2904. doi: 10.1101/gad.1256804
Buckley, B. A., Gracey, A. Y., and Somero, G. N. (2006). The cellular response to heat stress in the goby Gillichthys mirabilis: a cDNA microarray and protein-level analysis. J. Exp. Biol. 209, 2660–2677. doi: 10.1242/jeb.02292
Cassis, L., Laughter, A., Fettinger, M., Akers, S., Speth, R., Burke, G., et al. (1998). Cold exposure regulates the Renin-Angiotensin system. J. Pharmacol. Exp. Ther. 286, 718–726.
Cingi, S., Keinänen, M., and Vuorinen, P. J. (2010). Elevated water temperature impairs fertilization and embryonic development of whitefish Coregonus lavaretus. J. Fish Biol. 76, 502–521. doi: 10.1111/j.1095-8649.2009.02502.x
Conde-Sieira, M., Aguilar, A. J., López-Patiño, M. A., Míguez, J. M., and Soengas, J. L. (2010). Stress alters food intake and glucosensing response in hypothalamus, hindbrain, liver, and Brockmann bodies of rainbow trout. Physiol. Behav. 101, 483–493. doi: 10.1016/j.physbeh.2010.07.016
Criswell, T., and Boothman, D. A. (2005). Regulation and functions of clusterin: a protector against stress. Acta. Med. Nagasaki 50, S55–S62. doi: 10.11343/amn.50.s55
Das, C., Thraya, M., and Vijayan, M. M. (2018). Nongenomic cortisol signaling in fish. Gen. Comp. Endocrinol. doi: 10.1016/j.ygcen.2018.04.019. [Epub ahead of print].
Dragoni, I., Mariotti, M., Consalez, G. G., Soria, M. R., and Maier, J. A. (1998). EDF-1, a novel gene product down-regulated in human endothelial cell differentiation. J. Biol. Chem. 273, 31119–31124. doi: 10.1074/JBC.273.47.31119
Edsall, T. A., and Rottiers, D. V. (1976). Temperature tolerance of young-of-the-year lake whitefish, Coregonus clupeaformis. J. Fish Res. Board Can. 33, 177–180. doi: 10.1139/f76-021
Edsall, T. A., Rottiers, D. V., and Brown, E. H. (1970). Temperature tolerance of bloater (Coregonus Hoyi). J. Fish Res. Board Can. 27, 2047–2052. doi: 10.1139/f70-228
Fevolden, S. E., Refstie, T., and Røed, K. H. (1991). Selection for high and low cortisol stress response in Atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss). Aquaculture 95:53–65. doi: 10.1016/0044-8486(91)90072-F
Ficker, H., Mazzucco, R., Gassner, H., Wanzenböck, J., and Dieckmann, U. (2016). Stocking strategies for a pre-alpine whitefish population under temperature stress. Ecol. Modell. 320, 170–176. doi: 10.1016/j.ecolmodel.2015.10.002
Fopp-Bayat, D., Kaczmarczyk, D., and Szczepkowski, M. (2015). Genetic characteristics of Polish whitefish (Coregonus lavaretus maraena) broodstocks – recommendations for the conservation management. Czech. J. Anim. Sci. 60, 171–177. doi: 10.17221/8131-CJAS
Fornace, A. J., Alamo, I., and Hollander, M. C. (1988). DNA damage-inducible transcripts in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 85, 8800–8804.
Gesto, M., Soengas, J., and Míguez, J. (2008). Acute and prolonged stress responses of brain monoaminergic activity and plasma cortisol levels in rainbow trout are modified by PAHs (naphthalene, β-). Aquat. Toxicol. 86, 341–351. doi: 10.1016/j.aquatox.2007.11.014
Gillespie, C. F., Phifer, J., Bradley, B., and Ressler, K. J. (2009). Risk and resilience: genetic and environmental influences on development of the stress response. Depress. Anxiety 26, 984–992. doi: 10.1002/da.20605
Golosova, O., Henderson, R., Vaskin, Y., Gabrielian, A., Grekhov, G., Nagarajan, V., et al. (2014). Unipro UGENE NGS pipelines and components for variant calling, RNA-seq and ChIP-seq data analyses. PeerJ. 2:e644. doi: 10.7717/peerj.644
Gracey, A. Y., Fraser, E. J., Li, W., Fang, Y., Taylor, R. R., Rogers, J., et al. (2004). Coping with cold: an integrative, multitissue analysis of the transcriptome of a poikilothermic vertebrate. Proc. Natl. Acad. Sci. U.S.A. 101, 16970–16975. doi: 10.1073/pnas.0403627101
Gräns, A., Niklasson, L., Sandblom, E., Sundell, K., Algers, B., Berg, C., et al. (2016). Stunning fish with CO2 or electricity: contradictory results on behavioural and physiological stress responses. Animal 10, 294–301. doi: 10.1017/S1751731115000750
Hahn, M. E., McArthur, A. G., Karchner, S. I., Franks, D. G., Jenny, M. J., Timme-Laragy, A. R., et al. (2014). The transcriptional response to oxidative stress during vertebrate development: effects of tert-Butylhydroquinone and 2,3,7,8-Tetrachlorodibenzo-p-Dioxin. PLoS ONE 9:e113158. doi: 10.1371/journal.pone.0113158
Hassler, J. R., Scheuner, D. L., Wang, S., Han, J., Kodali, V. K., Li, P., et al. (2015). The IRE1α/XBP1s pathway is essential for the glucose response and protection of β cells. PLoS Biol. 13:e1002277. doi: 10.1371/journal.pbio.1002277
Hiramatsu, K., Yamada, T., and Katakura, M. (1984). Acute effects of cold on blood pressure, renin-angiotensinaldosterone system, catecholamines and adrenal steroids in man. Clin. Exp. Pharmacol. Physiol. 11, 171–179. doi: 10.1111/j.1440-1681.1984.tb00254.x
Hori, T. S., Gamperl, A. K., Afonso, L. O., Johnson, S. C., Hubert, S., Kimball, J., et al. (2010). Heat-shock responsive genes identified and validated in Atlantic cod (Gadus morhua) liver, head kidney and skeletal muscle using genomic techniques. BMC Genomics 11:72. doi: 10.1186/1471-2164-11-72
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Hulsen, T., de Vlieg, J., and Alkema, W. (2008). BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9:488. doi: 10.1186/1471-2164-9-488
Jeffries, K. M., Hinch, S. G., Sierocinski, T., Clark, T. D., Eliason, E. J., Donaldson, M. R., et al. (2012). Consequences of high temperatures and premature mortality on the transcriptome and blood physiology of wild adult sockeye salmon (Oncorhynchus nerka). Ecol. Evol. 2, 1747–1764. doi: 10.1002/ece3.274
Jeffries, K. M., Hinch, S. G., Sierocinski, T., Pavlidis, P., and Miller, K. M. (2014). Transcriptomic responses to high water temperature in two species of Pacific salmon. Evol. Appl. 7, 286–300. doi: 10.1111/eva.12119
Jindra, M., Gaziova, I., Uhlirova, M., Okabe, M., Hiromi, Y., and Hirose, S. (2004). Coactivator MBF1 preserves the redox-dependent AP-1 activity during oxidative stress in Drosophila. EMBO J. 23, 3538–3547. doi: 10.1038/sj.emboj.7600356
Jobling, M. (1981). Temperature tolerance and the final preferendum-rapid methods for the assessment of optimum growth temperatures. J. Fish Biol. 19, 439–455. doi: 10.1111/j.1095-8649.1981.tb05847.x
Kabe, Y., Goto, M., Shima, D., Imai, T., Wada, T., Morohashi, K., et al. (1999). The role of human MBF1 as a transcriptional coactivator. J. Biol. Chem. 274, 34196–34202.
Kalies, K. U., and Hartmann, E. (1996). Membrane topology of the 12- and the 25-kDa subunits of the mammalian signal peptidase complex. J. Biol. Chem. 271, 3925–3929.
Kampinga, H. H., Hageman, J., Vos, M. J., Kubota, H., Tanguay, R. M., Bruford, E. A., et al. (2009). Guidelines for the nomenclature of the human heat shock proteins. Cell. Stress Chaperones 14, 105–111. doi: 10.1007/s12192-008-0068-7
Kause, A., Quinton, C., Airaksinen, S., Ruohonen, K., and Koskela, J. (2011). Quality and production trait genetics of farmed European whitefish, Coregonus lavaretus. J. Anim. Sci. 89, 959–971. doi: 10.2527/jas.2010-2981
Kojima, E., Takeuchi, A., Haneda, M., Yagi, A., Hasegawa, T., Yamaki, K., et al. (2003). The function of GADD34 is a recovery from a shutoff of protein synthesis induced by ER stress: elucidation by GADD34-deficient mice. FASEB J. 17, 1573–1575. doi: 10.1096/fj.02-1184fje
Korytár, T., Nipkow, M., Altmann, S., Goldammer, T., Köllner, B., and Rebl, A. (2016). Adverse husbandry of maraena whitefish directs the immune system to increase mobilization of myeloid cells and pro-inflammatory responses. Front. Immunol. 7:631. doi: 10.3389/FIMMU.2016.00631
Lewis, J. M., Hori, T. S., Rise, M. L., Walsh, P. J., and Currie, S. (2010). Transcriptome responses to heat stress in the nucleated red blood cells of the rainbow trout (Oncorhynchus mykiss). Physiol. Genomics 42, 361–373. doi: 10.1152/physiolgenomics.00067.2010
Li, Y., Huang, J., Liu, Z., Zhou, Y., Xia, B., Wang, Y., et al. (2017). Transcriptome analysis provides insights into hepatic responses to moderate heat stress in the rainbow trout (Oncorhynchus mykiss). Gene 619, 1–9. doi: 10.1016/j.gene.2017.03.041
Liu, S., Wang, X., Sun, F., Zhang, J., Feng, J., Liu, H., et al. (2013). RNA-Seq reveals expression signatures of genes involved in oxygen transport, protein synthesis, folding, and degradation in response to heat stress in catfish. Physiol. Genomics 45, 462–476. doi: 10.1152/physiolgenomics.00026.2013
Lu, M.-W., Ngou, F.-H., Chao, Y.-M., Lai, Y.-S., Chen, N.-Y., Lee, F.-Y., et al. (2012). Transcriptome characterization and gene expression of Epinephelus spp in endoplasmic reticulum stress-related pathway during betanodavirus infection in vitro. BMC Genomics 13:651. doi: 10.1186/1471-2164-13-651
Macqueen, D. J., Primmer, C. R., Houston, R. D., Nowak, B. F., Bernatchez, L., Bergseth, S., et al. (2017). Functional Annotation of All Salmonid Genomes (FAASG): an international initiative supporting future salmonid research, conservation and aquaculture. BMC Genomics 18:484. doi: 10.1186/s12864-017-3862-8
Marik, P. E., and Bellomo, R. (2013). Stress hyperglycemia: an essential survival response! Crit. Care 17:305. doi: 10.1186/cc12514
Mariotti, M., De Benedictis, L., Maier, J. A. M., and Ballabio, E. (2004). The dual role of endothelial differentiation-related factor-1 in the cytosol and nucleus: modulation by protein kinase A. Cell. Mol. Life Sci. 61, 1069–1074. doi: 10.1007/s00018-004-4016-0
Martinez-Porchas, M., Martinez-Cordova, L. T., and Ramos-Enriquez, R. (2009). Cortisol and Glucose: reliable indicators of fish stress? J. Aquat. Sci. 4, 158–178.
McCormick, J. H., Jones, B. R., and Syrett, R. F. (1971). Temperature Requirements for Growth and Survival of Larval Ciscos (Coregonus artedii). J. Fish Res. Board Can. 28, 924–927. doi: 10.1139/f71-134
Nagata, K., Saga, S., and Yamada, K. M. (1988). Characterization of a novel transformation-sensitive heat-shock protein (HSP47) that binds to collagen. Biochem. Biophys. Res. Commun. 153, 428–434.
Nair, S. C., Rimerman, R. A., Toran, E. J., Chen, S., Prapapanich, V., Butts, R. N., et al. (1997). Molecular cloning of human FKBP51 and comparisons of immunophilin interactions with Hsp90 and progesterone receptor. Mol. Cell. Biol. 17, 594–603.
Nakai, A., Satoh, M., Hirayoshi, K., and Nagata, K. (1992). Involvement of the stress protein HSP47 in procollagen processing in the endoplasmic reticulum. J. Cell. Biol. 117, 903–914.
Narum, S. R., and Campbell, N. R. (2015). Transcriptomic response to heat stress among ecologically divergent populations of redband trout. BMC Genomics 16:103. doi: 10.1186/s12864-015-1246-5
Nicol, S. M., Bray, S. E., Black, H. D., Lorimore, S. A., Wright, E. G., Lane, D. P., et al. (2013). The RNA helicase p68 (DDX5) is selectively required for the induction of p53-dependent p21 expression and cell-cycle arrest after DNA damage. Oncogene 32, 3461–3469. doi: 10.1038/onc.2012.426
Ortuño, J., Esteban, M. A., and Meseguer, J. (2002). Effects of phenoxyethanol on the innate immune system of gilthead seabream (Sparus aurata L.) exposed to crowding stress. Vet. Immunol. Immunopathol. 89, 29–36. doi: 10.1016/S0165-2427(02)00183-6
Phadtare, S., Alsina, J., and Inouye, M. (1999). Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 2, 175–180. doi: 10.1016/S1369-5274(99)80031-9
Pickering, A. D., and Pottinger, T. G. (1989). Stress responses and disease resistance in salmonid fish: effects of chronic elevation of plasma cortisol. Fish. Physiol. Biochem. 7, 253–258. doi: 10.1007/BF00004714
Pörtner, H. O., Peck, L., and Somero, G. (2007). Thermal limits and adaptation in marine Antarctic ectotherms: an integrative view. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 362, 2233–2258. doi: 10.1098/rstb.2006.1947
Procarione, L., Barry, T., and Malison, J. (1999). Effects of high rearing densities and loading rates on the growth and stress responses of juvenile rainbow trout. North Am. J. Aquacult. 61, 91–96.
Quinn, N. L., McGowan, C. R., Cooper, G. A., Koop, B. F., and Davidson, W. S. (2011a). Identification of genes associated with heat tolerance in Arctic charr exposed to acute thermal stress. Physiol. Genomics 43, 685–696. doi: 10.1152/physiolgenomics.00008.2011
Quinn, N. L., McGowan, C. R., Cooper, G. A., Koop, B. F., and Davidson, W. S. (2011b). Ribosomal genes and heat shock proteins as putative markers for chronic, sublethal heat stress in Arctic charr: applications for aquaculture and wild fish. Physiol. Genomics 43, 1056–1064. doi: 10.1152/physiolgenomics.00090.2011
Rebl, A., Verleih, M., Köbis, J. M., Kühn, C., Wimmers, K., Köllner, B., et al. (2013). Transcriptome profiling of gill tissue in regionally bred and globally farmed rainbow trout strains reveals different strategies for coping with thermal stress. Mar. Biotechnol. 15, 445–460. doi: 10.1007/s10126-013-9501-8
Rebl, A., Zebunke, M., Borchel, A., Bochert, R., Verleih, M., and Goldammer, T. (2017). Microarray-predicted marker genes and molecular pathways indicating crowding stress in rainbow trout (Oncorhynchus mykiss). Aquaculture 473, 355–365. doi: 10.1016/j.aquaculture.2017.03.003
Rexroad, C. E., Vallejo, R. L., Liu, S., Palti, Y., and Weber, G. M. (2013). Quantitative trait loci affecting response to crowding stress in an f2 generation of rainbow trout produced through phenotypic selection. Mar. Biotechnol. 15, 613–627. doi: 10.1007/s10126-013-9512-5
Roberts, R. J., Agius, C., Saliba, C., Bossier, P., and Sung, Y. Y. (2010). Heat shock proteins (chaperones) in fish and shellfish and their potential role in relation to fish health: a review. J. Fish Dis. 33, 789–801. doi: 10.1111/j.1365-2761.2010.01183.x
Robichon, C., Varret, M., Le Liepvre, X., Lasnier, F., Hajduch, E., Ferré, P., et al. (2006). DnaJA4 is a SREBP-regulated chaperone involved in the cholesterol biosynthesis pathway. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1761, 1107–1113. doi: 10.1016/j.bbalip.2006.07.007
Rossi, A., Trotta, E., Brandi, R., Arisi, I., Coccia, M., and Gabriella Santoro, M. (2010). AIRAP, a new human heat shock gene regulated by heat shock factor 1. J. Biol. Chem. 285, 13607–13615. doi: 10.1074/jbc.M109.082693
Shamovsky, I., Ivannikov, M., Kandel, E. S., Gershon, D., and Nudler, E. (2006). RNA-mediated response to heat shock in mammalian cells. Nature 440, 556–560. doi: 10.1038/nature04518
Shih, J. W., and Lee, Y. H. W. (2014). Human DExD/H RNA helicases: emerging roles in stress survival regulation. Clin. Chim. Acta 436, 45–58. doi: 10.1016/j.cca.2014.05.003
Shoshani, T., Faerman, A., Mett, I., Zelin, E., Tenne, T., Gorodin, S., et al. (2002). Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol. Cell. Biol. 22, 2283–2293. doi: 10.1128/mcb.22.7.2283-2293.2002
Slawinska, A., Hsieh, J. C., Schmidt, C. J., and Lamont, S. J. (2016). Heat stress and lipopolysaccharide stimulation of chicken macrophage-like cell line activates expression of distinct sets of genes. PLoS ONE 11:e0164575. doi: 10.1371/journal.pone.0164575
Smith, T. R., Tremblay, G. C., and Bradley, T. M. (1999). Characterization of the heat shock protein response of Atlantic salmon (Salmo salar). Fish. Physiol. Biochem. 20, 279–292. doi: 10.1023/A:1007743329892
Smyth, G. K. (2005). “Limma: linear models for microarray data,” in Bioinformatics and Computational Biology Solutions Using R and Bioconductor, eds R. Gentleman, V. Carey, S. Dudoit, R. Irizarry, and W. Huber (New York, NY: Springer), 397–420.
Soden, M., Klett, C., Hasmann, T., and Hackenthal, E. (1994). Angiotensinogen: an acute-phase protein? Hypertension 23, I126–I130.
Song, L., Zhang, J., Li, C., Yao, J., Jiang, C., Li, Y., et al. (2014). Genome-wide identification of hsp40 genes in channel catfish and their regulated expression after bacterial infection. PLoS ONE 9:e115752. doi: 10.1371/journal.pone.0115752
Sørensen, J. G., Kristensen, T. N., and Loeschcke, V. (2003). The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 6, 1025–1037. doi: 10.1046/j.1461-0248.2003.00528.x
Stefanovic, D. I., Manzon, L. A., McDougall, C. S., Boreham, D. R., Somers, C. M., Wilson, J. Y., et al. (2016). Thermal stress and the heat shock response in embryonic and young of the year juvenile lake whitefish. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 193, 1–10. doi: 10.1016/j.cbpa.2015.12.001
Tahmasebi-Kohyani, A., Keyvanshokooh, S., Nematollahi, A., Mahmoudi, N., and Pasha-Zanoosi, H. (2012). Effects of dietary nucleotides supplementation on rainbow trout (Oncorhynchus mykiss) performance and acute stress response. Fish Physiol. Biochem. 38, 431–440. doi: 10.1007/s10695-011-9524-x
Tomalty, K. M. H., Meek, M. H., Stephens, M. R., Rincón, G., Fangue, N. A., May, B. P., et al. (2015). Transcriptional response to acute thermal exposure in juvenile chinook salmon determined by RNAseq. G3 5, 1335–1349. doi: 10.1534/g3.115.017699
Tripathi, V., Darnauer, S., Hartwig, N. R., and Obermann, W. M. J. (2014). Aha1 can act as an autonomous chaperone to prevent aggregation of stressed proteins. J. Biol. Chem. 289, 36220–36228. doi: 10.1074/jbc.M114.590141
Trougakos, I. P. (2013). The molecular chaperone apolipoprotein J/clusterin as a sensor of oxidative stress: implications in therapeutic approaches - a mini-review. Gerontology 59, 514–523. doi: 10.1159/000351207
Tymchuk, W., Sakhrani, D., and Devlin, R. (2009). Domestication causes large-scale effects on gene expression in rainbow trout: analysis of muscle, liver and brain transcriptomes. Gen. Comp. Endocrinol. 164, 175–183. doi: 10.1016/j.ygcen.2009.05.015
Uren Webster, T. M., and Santos, E. M. (2015). Global transcriptomic profiling demonstrates induction of oxidative stress and of compensatory cellular stress responses in brown trout exposed to glyphosate and Roundup. BMC Genomics 16:32. doi: 10.1186/s12864-015-1254-5
Vera, M., Pani, B., Griffiths, L. A., Muchardt, C., Abbott, C. M., Singer, R. H., et al. (2014). The translation elongation factor eEF1A1 couples transcription to translation during heat shock response. Elife 3:e03164. doi: 10.7554/eLife.03164
Verleih, M., Borchel, A., Krasnov, A., Rebl, A., Korytár, T., Kühn, C., et al. (2015). Impact of thermal stress on kidney-specific gene expression in farmed regional and imported rainbow trout. Mar. Biotechnol. 17, 576–592. doi: 10.1007/s10126-015-9640-1
von Borell, E. (2000). Stress and coping in farm animals. Arch. Fur. Tierzucht Archives Anim. Breed 43, 144–152.
Vornanen, M., Hassinen, M., Koskinen, H., and Krasnov, A. (2005). Steady-state effects of temperature acclimation on the transcriptome of the rainbow trout heart. Am. J. Physiol. Regul. Integr. Comp. Physiol. 289, R1177–R1184. doi: 10.1152/ajpregu.00157.2005
Wang, Y., Liu, Z., Li, Z., Shi, H., Kang, Y., Wang, J., et al. (2016). Effects of heat stress on respiratory burst, oxidative damage and SERPINH1 (HSP47) mRNA expression in rainbow trout Oncorhynchus mykiss. Fish Physiol. Biochem. 42, 701–710. doi: 10.1007/s10695-015-0170-6
Weinberg, M. S., Girotti, M., and Spencer, R. L. (2007). Restraint-induced fra-2 and c-fos expression in the rat forebrain: relationship to stress duration. Neuroscience 150, 478–486. doi: 10.1016/j.neuroscience.2007.09.013
Weng, L., Dai, H., Zhan, Y., He, Y., Stepaniants, S. B., and Bassett, D. E. (2006). Rosetta error model for gene expression analysis. Bioinformatics 22, 1111–1121. doi: 10.1093/bioinformatics/btl045
Wochnik, G. M., Regg, J., Abel, G. A., Schmidt, U., Holsboer, F., and Rein, T. (2005). FK506-binding Proteins 51 and 52 differentially regulate dynein interaction and nuclear translocation of the glucocorticoid receptor in mammalian cells. J. Biol. Chem. 280, 4609–4616. doi: 10.1074/jbc.M407498200
Wong, S., Tan, K., Carey, K. T., Fukushima, A., Tiganis, T., and Cole, T. J. (2010). Glucocorticoids stimulate hepatic and renal catecholamine inactivation by direct rapid induction of the dopamine sulfotransferase Sult1d1. Endocrinology 151, 185–194. doi: 10.1210/en.2009-0590
Wyatt, A., Yerbury, J., Poon, S., Dabbs, R., and Wilson, M. (2009). Chapter 6 The chaperone action of clusterin and its putative role in quality control of extracellular protein folding. Adv. Cancer Res. 104, 89–114. doi: 10.1016/S0065-230X(09)04006-8
Keywords: aquaculture, DE genes, salmonid fish, transcriptome, temperature challenge, welfare
Citation: Rebl A, Verleih M, Nipkow M, Altmann S, Bochert R and Goldammer T (2018) Gradual and Acute Temperature Rise Induces Crossing Endocrine, Metabolic, and Immunological Pathways in Maraena Whitefish (Coregonus maraena). Front. Genet. 9:241. doi: 10.3389/fgene.2018.00241
Received: 13 April 2018; Accepted: 19 June 2018;
Published: 19 July 2018.
Edited by:
Nguyen Hong Nguyen, University of the Sunshine Coast, AustraliaReviewed by:
Scott Francis Cummins, University of the Sunshine Coast, AustraliaBam Dev Paneru, Middle Tennessee State University, United States
Copyright © 2018 Rebl, Verleih, Nipkow, Altmann, Bochert and Goldammer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tom Goldammer, dG9tLmdvbGRhbW1lckB1bmktcm9zdG9jay5kZQ==
 Alexander Rebl
Alexander Rebl Marieke Verleih
Marieke Verleih Mareen Nipkow
Mareen Nipkow Simone Altmann1
Simone Altmann1 Tom Goldammer
Tom Goldammer