
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 29 May 2018
Sec. Evolutionary and Population Genetics
Volume 9 - 2018 | https://doi.org/10.3389/fgene.2018.00192
 Shangguo Feng1,2*
Shangguo Feng1,2* Yujia Zhu1
Yujia Zhu1 Chenliang Yu3
Chenliang Yu3 Kaili Jiao1
Kaili Jiao1 Mengying Jiang1
Mengying Jiang1 Jiangjie Lu1
Jiangjie Lu1 Chenjia Shen1
Chenjia Shen1 Qicai Ying1
Qicai Ying1 Huizhong Wang1*
Huizhong Wang1*Physalis is an important genus in the Solanaceae family. It includes many species of significant medicinal value, edible value, and ornamental value. However, many Physalis species are easily confused because of their similar morphological traits, which hinder the utilization and protection of Physalis resources. Therefore, it is necessary to create fast, sensitive, and reliable methods for the Physalis species authentication. Intended for that, in this study, species-specific sequence-characterized amplified region (SCAR) markers were developed for accurate identification of the closely related Physalis species P. angulata, P. minima, P. pubescens, and P. alkekengi var. franchetii, based on a simple and novel marker system, start codon targeted (SCoT) marker. A total of 34 selected SCoT primers yielded 289 reliable SCoT loci, of which 265 were polymorphic. Four species-specific SCoT fragments (SCoT3-1404, SCoT3-1589, SCoT5-550, and SCoT36-520) from Physalis species were successfully identified, cloned, and sequenced. Based on these selected specific DNA fragments, four SCAR primers pairs were developed and named ST3KZ, ST3MSJ, ST5SJ, and ST36XSJ. PCR analysis of each of these primer pairs clearly demonstrated a specific amplified band in all samples of the target Physalis species, but no amplification was observed in other Physalis species. Therefore, the species-specific SCAR primer pairs developed in this study could be used as powerful tools that can rapidly, effectively, and reliably identify and differentiate Physalis species.
The genus Physalis (Solanaceae family) consists of 75–120 species, which are mainly distributed in American tropical and temperate regions (Chinese Academy of Sciences, 1978; Whitson and Manos, 2005; Wei et al., 2012; Feng et al., 2016b). However, P. angulata, P. minima, P. pubescens, and P. alkekengi var. franchetii are mainly distributed in China (Chinese Academy of Sciences, 1978). They have a variety of pharmacological activities, such as anti-oxidant, anti-inflammatory, and anti-cancer effects, and most of these species have been used as Chinese medicinal herbs for the treatment of malaria, rheumatism, hepatitis, asthma, and cancer for a long time (Ji et al., 2013; Ding et al., 2014; Continuing Professional Education Committee, 2015; Xu et al., 2016). Furthermore, P. alkekengi var. franchetii, a standard Physalis medicinal plant, has been included in the Pharmacopoeia of China (Continuing Professional Education Committee, 2015). In addition, some Physalis species including P. philadelphica, P. peruviana, and P. pubescens have important ornamental value and edible value, and have been cultivated in many regions of the world, such as China and Europe. In recent years, increasing attention has been paid to the genus Physalis, particularly their phytochemical and pharmacology characteristics (Ji et al., 2013; Ding et al., 2014; Li et al., 2014; Yang et al., 2016; Zhang and Tong, 2016; Sun et al., 2017). However, the research on classification and identification of Physalis species is weak. The morphological characteristics of Physalis species are extremely similar and are easily influenced by their environment and the plant developmental stage, which makes differentiation very difficult and sometimes impossible using morphological methods (Wei et al., 2012; Feng et al., 2016b). In addition, owing to overexploitation and increased urbanization, the natural Physalis resources have become endangered, particularly in many areas of China (Feng et al., 2016b). These problems are inhibiting the use of Physalis and the development of the Physalis industry. Therefore, it is necessary to establish a quick and effective species identification method for the utilization and protection of Physalis resources.
In contrast to phenotypic characters, molecular markers are independent of environmental conditions and show higher levels of reliability. They have been widely used as important tools by modern taxonomists to improve phylogenetic analysis and to species authenticate many different plants (Mulpuri et al., 2013; Feng et al., 2015a,b). At present, only a few molecular markers have been applied to the genetic relationship and diversity study of Physalis species (Whitson and Manos, 2005; Vargas-Ponce et al., 2011; Wei et al., 2012; Labate and Robertson, 2015; Feng et al., 2016b).
As a simple and novel marker system, start codon targeted (SCoT) marker was developed based on the short conserved region flanking the start codon (ATG) in plant genes (Collard and Mackill, 2009). SCoT marker requires no sequence information and is correlated with functional genes and corresponding traits (Mulpuri et al., 2013). Compared with random amplified polymorphic DNA (RAPDs), inter-simple sequence repeats (ISSR), and simple sequence repeats (SSRs), SCoT employs longer primers (18 nucleotide bases 18-mer) that produce more polymorphisms. It has been extensively used in genetic diversity studies, phylogenetic analysis, and for the marker-assisted breeding of many plants (Collard and Mackill, 2009; Luo et al., 2010, 2011, 2012; Xiong et al., 2011; Guo et al., 2012; Mulpuri et al., 2013; Feng et al., 2015a, 2016a). However, because it is relatively sensitive to experimental conditions and the complexity of PCR products when used as RAPDs and ISSRs, SCoT markers are rarely used directly for species identification. In order to alleviate the problems that arise when using conventional molecular marker techniques, the sequence characterized amplified region (SCAR) marker system was developed (Paran and Michelmore, 1993). As one of the PCR-based genetic markers, SCARs could be derived from RAPD (Joseph et al., 2014), ISSR (Lee et al., 2011), SCoT (Mulpuri et al., 2013), inter-retrotransposon amplified polymorphism (IRAP) (Xiao et al., 2011; Mandoulakani et al., 2015), and intron length polymorphisms (ILP) (Shimada et al., 2009) markers. It represents a specific, defined genomic DNA fragment that is detected by PCR amplification using a pair of specific primers. Compared to conventional molecular markers, SCARs have proven to be simpler, more reproducible and more reliable when used for plant identification at the intra-and/or inter-specific level (Lee et al., 2011; Cirillo et al., 2012; Mulpuri et al., 2013; Marieschi et al., 2016).
The objective of this study was to develop species-specific SCAR markers for identifying four popular Physalis species (P. minima, P. angulata, P. alkekengi var. franchetii, and P. pubescens) based on SCoT analysis. Because SCoT is related to functional genes and corresponding traits, SCoT-SCAR has better stability and specificity.
A total of 20 individuals belonging to four Physalis species collected from their natural distribution areas in China were used to screen specific markers (Table 1). These individuals are popular medicinal plants in the genus Physalis, such as P. alkekengi var. franchetii, which is included in the Chinese Pharmacopoeia (Continuing Professional Education Committee, 2015). Further, 16 individuals of P. angulata, 10 individuals of P. pubescens, 10 individuals of P. alkekengi var. franchetii, and 10 individuals of P. minima were used to validate the developed SCAR markers (Supplementary Table S1). In order to identify and confirm the collected samples at the morphological level, the specimens stored in the herbarium at the Institute of Botany, Chinese Academy of Sciences, China1 were used to confirm the collected samples. Genomic DNA was isolated from the fresh, young leaf tissues of all the collected samples and the integrity and quality of the DNA was evaluated as previously described (Feng et al., 2016b).
Following a study by Collard and Mackill (2009), a total of 36 SCoT primers, synthesized by the Shanghai Sangon Biological Engineering Technology and Service Co., Ltd., China, were used for an initial primer screen, which utilized two samples of each tested species. These primers produced clearly separated bands and allowed stable and rich polymorphisms to be selected (Table 2). The PCR reactions were conducted in a total volume of 20 μl containing 1 × PCR buffer [200 mM Tris–HCl (pH 8.8), 100 mM KCl, 100 mM (NH4)2SO4, 20 mM MgCl2, 1% TritonX-100], 0.4 mM dNTPs, 0.5 μM each primer, 1 U Taq DNA polymerase (Beijing Dingguo Changsheng Biotechnology Co., Ltd., China), and genomic DNA template 50 ng. The amplification was performed using the following PCR program: 5 min at 94°C, followed by 35 cycles of 50 s at 94°C, 50 s at 50–60°C (depending on the annealing temperature of each primer), 1.5 min at 72°C, and a final extension at 72°C for 10 min. The PCR was performed in a gradient thermal cycler (A200) (Hangzhou Longgene Scientific Instruments Co., Ltd., Zhejiang, China). The PCR products were separated on 1.5% (W/V) agarose gel using Trans2K DNA markers (TransGen Biotech, Beijing, China) as size standards in Tris-acetate buffer stained with GelStain (TransGen Biotech) and photographed under UV light. The experiments were repeated at least two times.
The SCoT amplified bands were scored visually, but this was assisted by Quantity One software (Version 4.6.2, Bio-Rad Technical Service Department, United States). Only clear, unambiguous, and reproducible SCoT fragments were scored as present (1) or absent (0). The NTSYS-pc version 2.10e software was used to conduct the cluster analysis (Rohlf, 2000). The unweighted pair group method with arithmetic mean (UPGMA) was used to construct a dendrogram based on similarity matrices that had been calculated along with a simple matching (SM) coefficient (Nei and Li, 1979). In addition, a UPGMA analysis was performed based on the genetic distances using MEGA 6.0 software.
The SCoT profiles were compared to select amplicons present in a particular species and their absence in all the other species, which meant that they could be considered as species-specific markers. Several suitable marker bands were obtained using primers SCoT2, SCoT3, SCoT4, SCoT5, SCoT19, SCoT22, and SCoT36 (Table 3). The selected marker bands were excised from 1.5% agarose gel and were purified using a DNA Fragment Quick Purification/Recovery Kit (Beijing Dingguo Changsheng Biotechnology Co., Ltd.). The purified DNA fragments were cloned in a pUCm-T vector (Shanghai Sunny Biotechnology Co., Ltd.) and incorporated into ultra-competent Escherichia coli strain Trans5α cells. The transformed bacterial colonies were selected by colony PCR and clones with correctly sized inserts were sequenced using M13 forward and M13 reverse primers by the Shanghai Sunny Biotechnology Co., Ltd.
The sequence similarities of the obtained sequences were identified using the nucleotide databases of BLASTN2 program and the sequences were deposited in GenBank (GenBank accession numbers: MF566104, MF566105, MF566106, and MF566107) (Clark et al., 2016) (Table 4). The SCAR primers were designed using Primer Premier 5 software (Lalitha, 2000). Four SCAR primer pairs, named ST3KZ, ST3MSJ, ST5SJ, and ST36XSJ, were designed based on the sequenced SCoT fragments (Table 4). The designed primers were 19- to 22-mers with annealing temperatures (Tm) between 49.9 and 61.9°C. They were based on the obtained specific SCoT fragments and synthesized by Sangon Biotech (Shanghai) Co., Ltd. SCAR amplification was performed in 20 μl of reaction mixture containing 1 × PCR buffer [200 mM Tris–HCl (pH 8.8), 100 mM KCl, 100 mM (NH4)2SO4, 20 mM MgCl2, 1% TritonX-100], 0.4 mM of dNTPs, 0.5 μM of forward primer, 0.5 μM of reverse primer, 50 ng of DNA, and 1 U Taq DNA polymerase (Beijing Dingguo Changsheng Biotechnology Co., Ltd.). The SCAR-PCR amplifications were carried out with an initial denaturation at 94°C for 5 min, followed by 35 cycles at 94°C for 50 s, a working annealing temperature, which depended on each SCAR primer pair (Table 4), for 50 s, 72°C for 1.5 min, and with a final extension at 72°C for 10 min. The amplification products were resolved by electrophoresis on 1.5% agarose gel and detected by GelStain (TransGen Biotech) staining.
After the initial primer screening, 34 SCoT primers that produced clear and repeatable polymorphic patterns were chosen for further study (Table 2). These 34 primers produced 289 reliable SCoT bands, of which 265 bands were polymorphic. The polymorphic bands per primer ranged from 3 (SCoT9 and SCoT18) to 13 (SCoT22), with an average of 7.8. The percentage of polymorphic bands in each primer varied from 55.6 to 100.0%, with an average of 90.2%. Three representative profiles (SCoT2, SCoT3, and SCoT36) are shown in Figure 1.
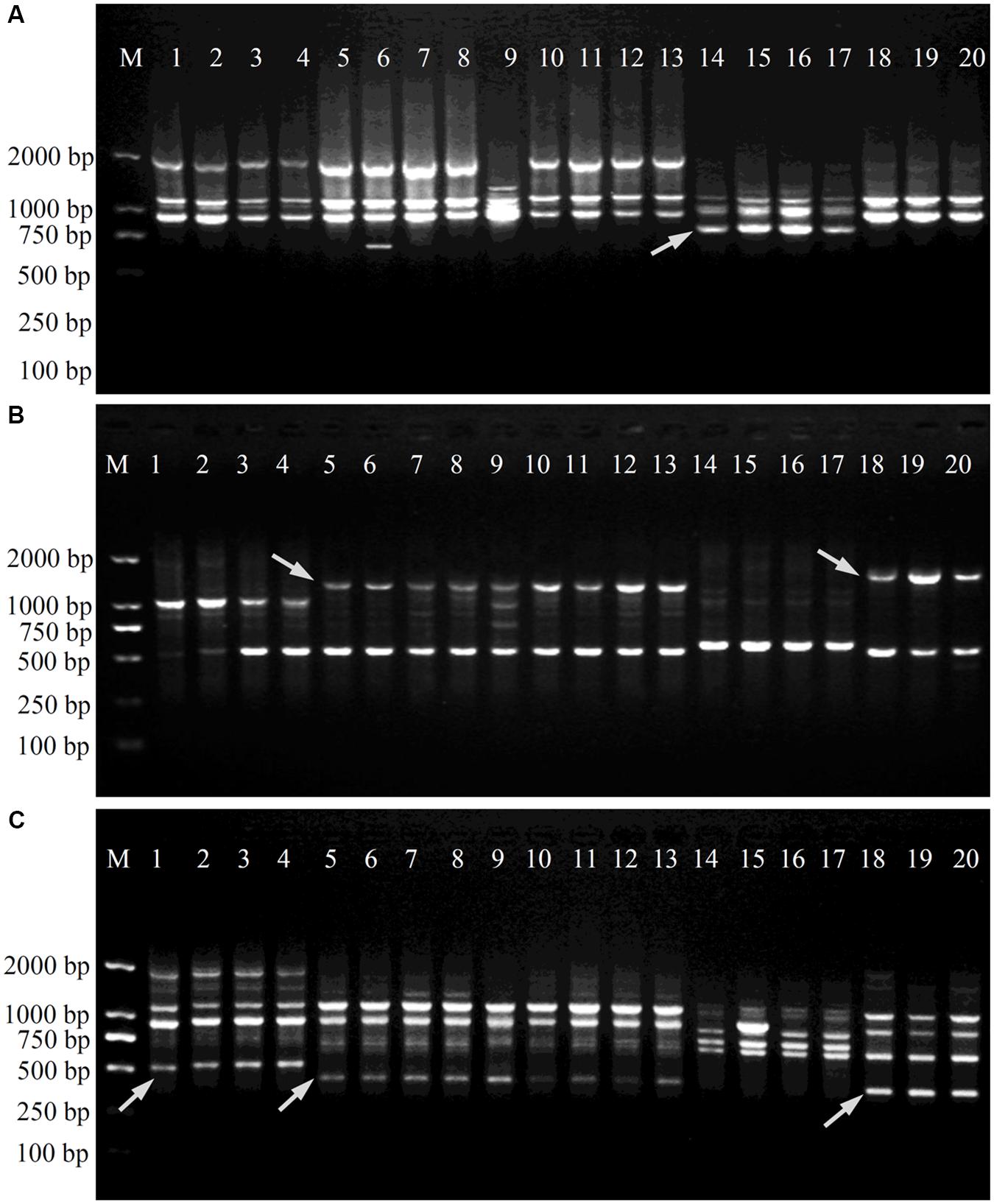
FIGURE 1. Amplification profiles of primers SCoT2 (A), SCoT3 (B) and SCoT36 (C). Lane M: Trans2K DNA Marker; Lanes 1–4: four individuals belonging to P. minima, Lanes 5–13: nine individuals belonging to P. angulata, Lanes 14–17: four individuals belonging to P. alkekengi var. franchetii, Lanes 18–20: three individuals belonging to P. pubescens. Details of Physalis individuals given in Table 1; Arrowheads represent specific amplified bands in all individuals of target Physalis species.
A total of 289 loci were accounted for the phylogenetic analysis. The genetic similarity among Physalis samples was estimated using the binary data matrices produced by SCoTs (Supplementary Table S2). The UPGMA dendrogram grouped Physalis samples into four main groups with a similarity of 0.622 (Figure 2). The UPGMA tree showed that all of the samples from the same species were grouped into one group and that an obvious boundary existed between species (group I contained the four samples from P. minima, group II contained the nine samples from P. angulata, group III contained the three samples from P. pubescens, while all the samples from P. alkekengi var. franchetii, which were far apart from any of the other Physalis species, were grouped into IV) (Figure 2). The similar results were obtained by UPGMA analysis based on the genetic distances using MEGA 6.0 software (Supplementary Figure S1).
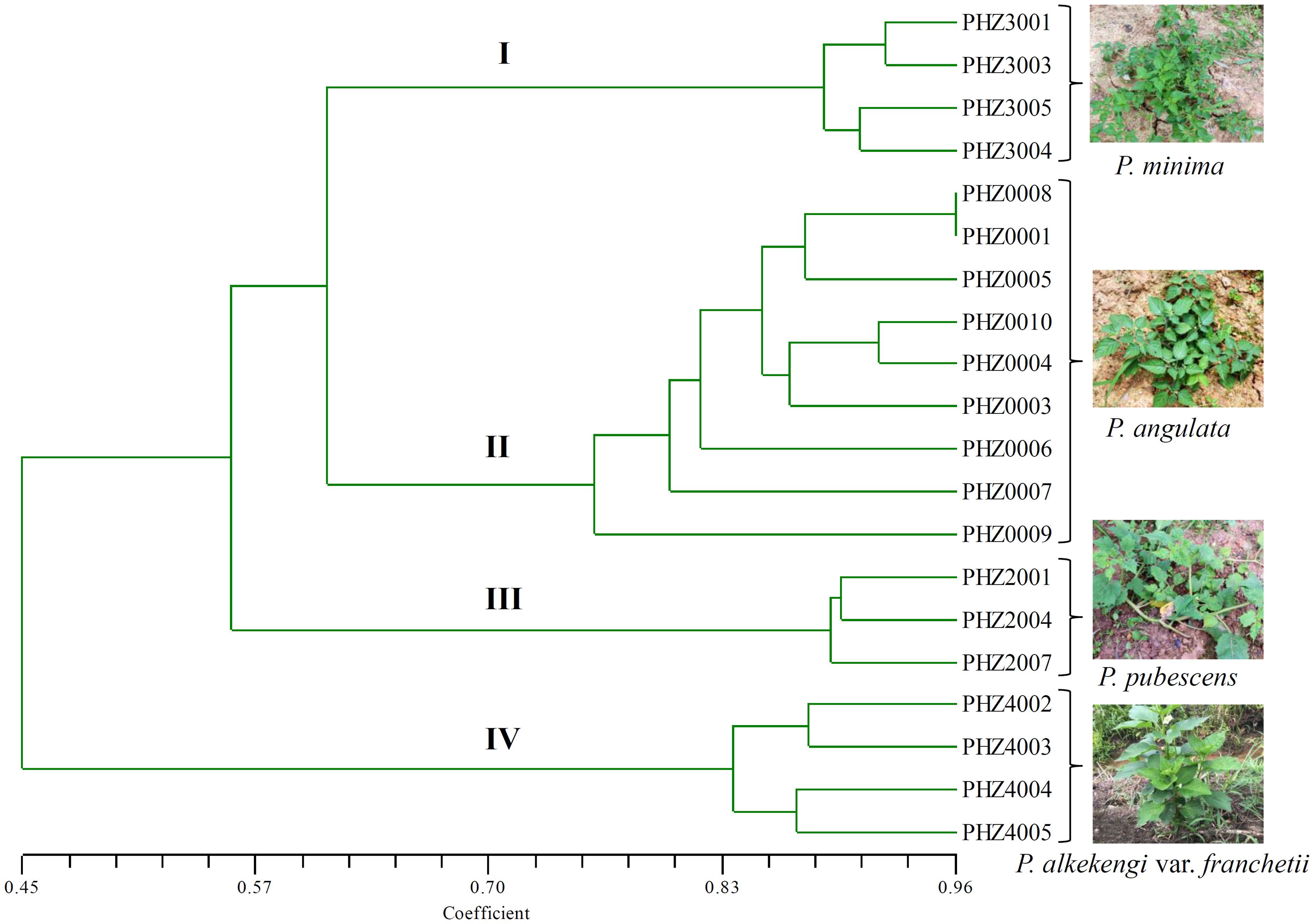
FIGURE 2. Dendrogram of Physalis species in this study using UPGMA cluster analysis based on genetic similarities of DNA fingerprinting patterns from the 34 SCoT primers. Numbers (I–IV) indicates Physalis samples were grouped into four groups with a similarity of 0.622.
A total of 11 SCoT fragments, each specific for a particular species and absent in the SCoT profiles of all the other species samples (SCoT2-730, SCoT5-550, and SCoT22-662 were specific for P. alkekengi var. franchetii; SCoT3-1404, SCoT19-590, and SCoT36-462 were specific for P. angulata; SCoT3-1589, SCoT22-537, and SCoT36-417 were specific for P. pubescens; and SCoT4-1412 and SCoT36-520 were specific for P. minima), were identified (Table 3). During cloning and sequencing, 7 of the 11 specific SCoT fragments were rejected due to technical problems (these fragments were not successfully cloned, leading to the failure of sequencing). The remaining four fragments, SCoT3-1404, SCoT3-1589, SCoT5-550, and SCoT36-520, were successfully cloned and sequenced. Their sequences were deposited in GenBank and their accession numbers are shown in Table 3. The blast results showed that most of the SCoT fragments had some similarities with other sequences deposited in GenBank. SCoT3-1404 had 80% identity with the Solanum lycopersicum chromosome ch10, a complete genome (GenBank accession: HG975522). SCoT3-1589 showed 80% identity with the RNA-directed DNA polymerase homolog mRNA of Daucus carota subsp. sativus (GenBank accession: XM_017389806), and SCoT36-520 had a high similarity (91%) with the corresponding region of mRNA from Brassica oleracea var. oleracea (GenBank accession: XM_013769290). However, SCoT5-550 showed no similarity with other known sequences at the sequence-similarity level.
In order to verify the specificity of the SCAR markers, each designed primer pair was tested after amplification using DNA extracted from 20 samples of four Physalis species (Table 1). After optimization of the PCR conditions, the optimum working annealing temperatures of these primer pairs (ST3KZ, ST3MSJ, ST5SJ, and ST36XSJ) were 65°C, 56°C, 61°C, and 60°C, respectively (Table 4). The amplification profiles of the SCAR primers are shown in Figure 3. The profile of the primer pair ST3KZF/R showed (Figure 3A) that a clear specific band representing 1102 bp was detected in all P. angulata samples, but no PCR band was found in the samples from P. minima, P. pubescens, and P. alkekengi var. franchetii. Similarly, all the tested individuals from the remaining three species contained a specific band detected by ST3MSJF/R (a band with 1479 bp for P. pubescens, Figure 3B), ST5SJF/R (a band with 463 bp for P. alkekengi var. franchetii, Figure 3C), and ST36XSJF/R (a band with 444 bp for P. minima, Figure 3D). The SCAR markers were further validated in 16 P. angulata, 10 P. pubescens, 10 P. alkekengi var. franchetii, and 10 P. minima individuals and the amplification of the markers at 1102, 1479, 463, and 444 bp ascertained the parentage of the target species (Figure 4).
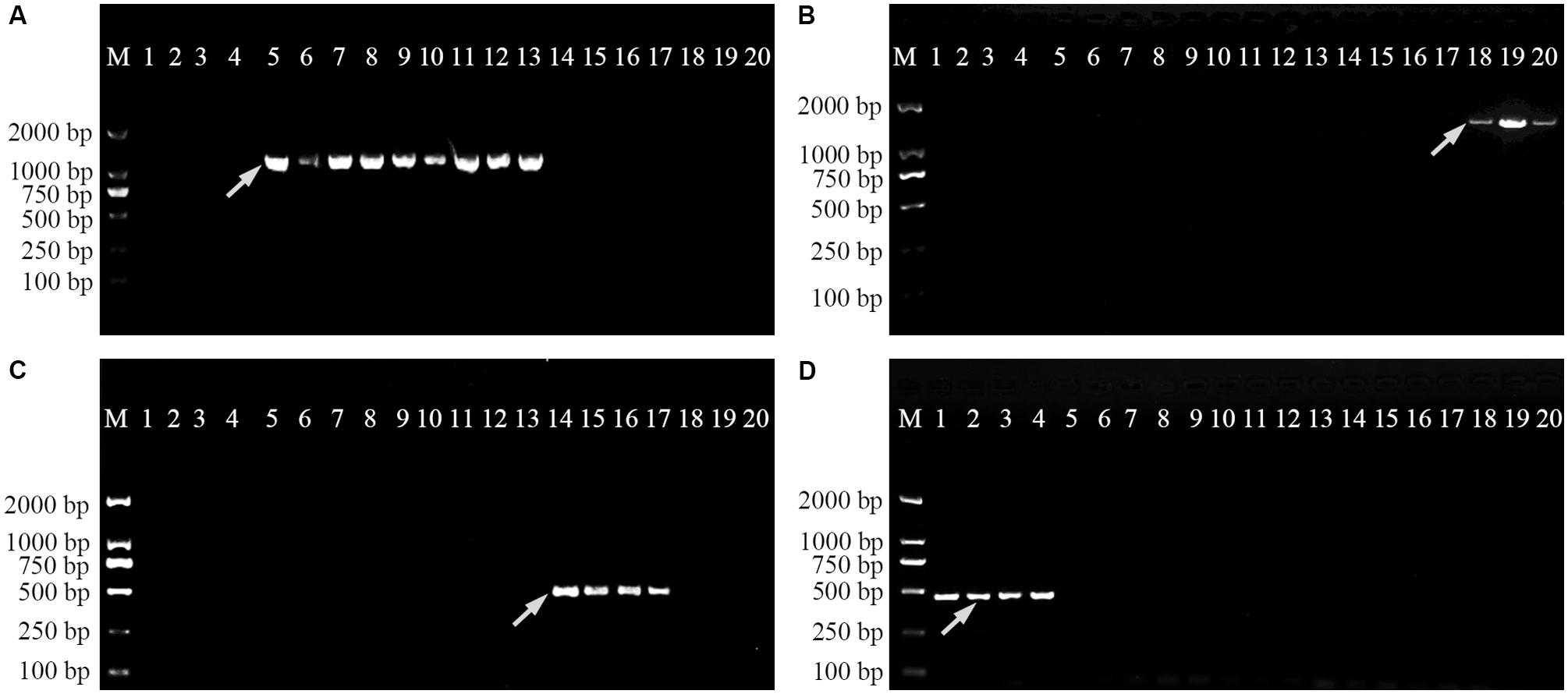
FIGURE 3. Amplification profiles of primer pairs ST3KZ (A), ST3MSJ (B), ST5SJ (C), and ST36XSJ (D) in 20 Physalis samples. Lane M: Trans2K DNA Marker; Lanes 1–4: four individuals belonging to P. minima, Lanes 5–13: nine individuals belonging to P. angulata, Lanes 14–17: four individuals belonging to P. alkekengi var. franchetii, Lanes 18–20: three individuals belonging to P. pubescens. Details of Physalis individuals given in Table 1; Arrowheads represent species-specific markers.
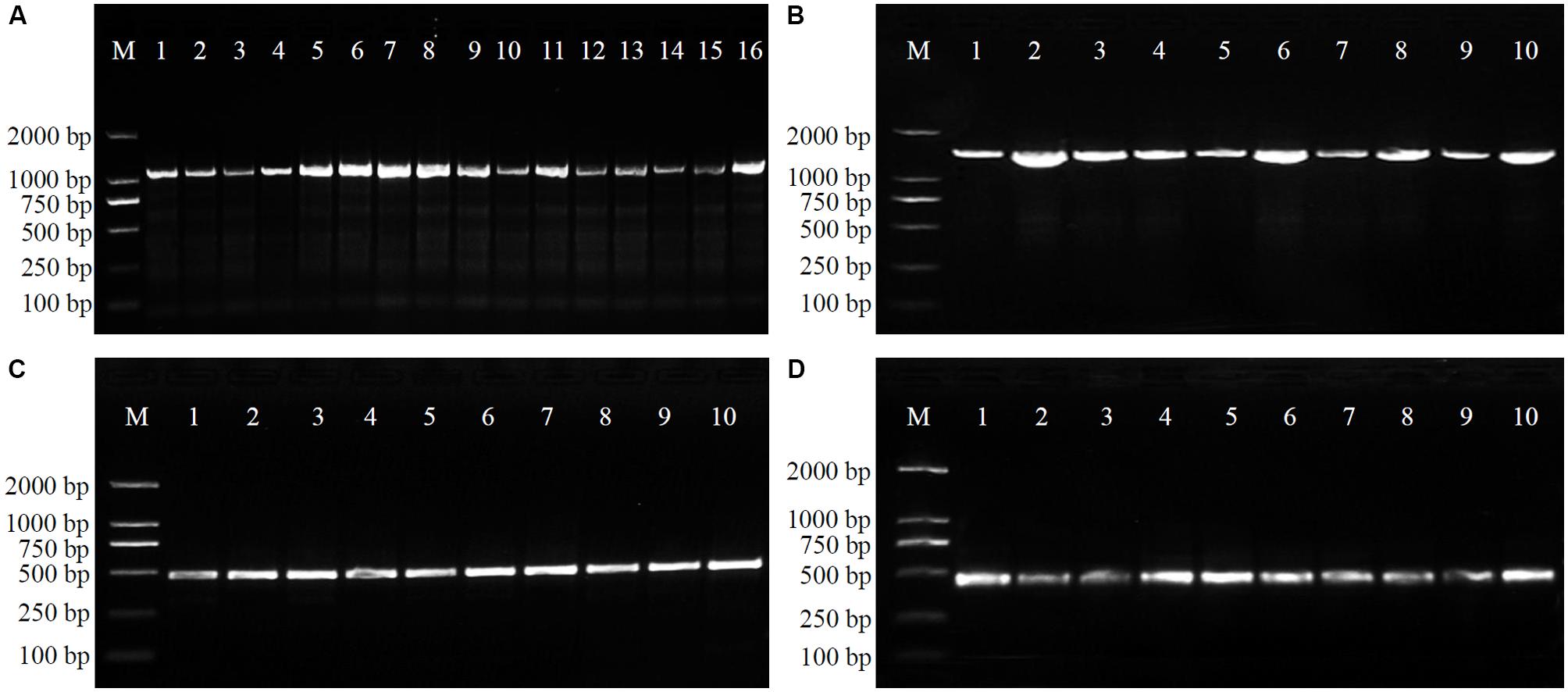
FIGURE 4. Amplification profiles of primer pairs ST3KZ in 16 P. angulata individuals (A), ST3MSJ in 10 P. pubescens individuals (B), ST5SJ in 10 P. alkekengi var. franchetii individuals (C), and ST36XSJ in 10 P. minima individuals (D). Details of Physalis individuals given in Supplementary Table S1.
In recent years, Physalis species have attracted the attention of many different scientists because of their significant nutritional value, edible fruit, and potential medicinal value. Many Physalis species have been used as raw materials to extract the active chemical constituents for new drug development (Fang et al., 2012; Ji et al., 2013; Ding et al., 2014; Li et al., 2014; Sang-Ngern et al., 2016; Zhang and Tong, 2016). Traditionally, Physalis identification was mainly dependent on morphological traits (Axelius, 1996; Martinez, 1998). However, the morphological features between Physalis species are extremely similar. In addition, morphological characteristics are often influenced by plant variation and growth habitats (Whitson and Manos, 2005). These problems have greatly hindered the utilization and protection of Physalis resources. DNA molecular marker technologies provide fast, effective, and accurate methods for identifying similar looking plants.
Several different molecular markers, including ISSR, SSR, InDel, SNP, ITS2, and psbA-trnH intergenic region, have been used for evaluating the genetic diversity and relationships of Physalis species (Vargas-Ponce et al., 2011; Wei et al., 2012; Garzon-Martinez et al., 2015; Zamora-Tavares et al., 2015; Feng et al., 2016b, 2018). This is the first study to identify Physalis species based on SCoT analysis. SCoT is a novel, simple and reliable gene-targeted marker technique, and has commonly been used to reveal genetic diversity and relationships in a wide range of plants, such as mango (Luo et al., 2010), grape (Guo et al., 2012), Mangifera indica (Luo et al., 2012), Dendrobium (Feng et al., 2015a), and Chrysanthemum morifolium (Feng et al., 2016a).
In this study, the amplified SCoT loci contained 265 polymorphic loci, which represented a polymorphism rate of 90.2%. This was higher than the polymorphic loci rate detected among mango cultivars (65.82%) (Luo et al., 2011), and M. indica (73.82%) (Luo et al., 2012), but similar to the proportion reported among C. morifolium, which was about 90% (Feng et al., 2016a). The high polymorphic rates detected in this study indicated that the SCoT technique has the potential ability to differentiate Physalis species. In recent years, the taxonomy of Physalis is regarded as one of the most intricate challenges in Solanaceae (Whitson and Manos, 2005; Olmstead et al., 2008; Feng et al., 2016b). In our previous studies, we found that several previously defined infrageneric taxa of Physalis are not monophyletic, and suggested that P. alkekengi var. franchetii should be recognized as a small genus (Feng et al., 2016b, 2018). In Whitson and Manos’ (2005) study, it was indicated that P. alkekengi, P. carpenter, and P. microphysa were atypical Physalis species and should be recognized as three small genera. In this study, the dendrogram constructed using the UPGMA method contained four groups, and indicated that different samples from the same Physalis species can be grouped on one branch (Figure 2). All samples from P. alkekengi var. franchetii constituted a separate group (IV), which was distant from any other Physalis species and this result confirmed with the findings of previous studies (Whitson and Manos, 2005; Feng et al., 2016b).
The SCoT markers could be used for species authentication by converting the species-specific SCAR markers for some other plant species. For example, Mulpuri et al. (2013) developed a SCAR marker based on SCoT analysis, which could be used to identify the toxic and non-toxic accessions of Jatropha curcas. A SCoT-derived SCAR marker was developed to distinguish tall/dwarf trait in arecanut (Rajesh et al., 2016). Hao et al. (2018) developed a SCoT-Based SCAR Marker for rapid authentication of Taxus Media among other related Taxus species (T. chinensis, T. cuspidate, and T. fauna) (Hao et al., 2018). In this study, SCoT markers generated 11 species-specific SCoT bands, four (SCoT3-1404, SCoT3-1589, SCoT5-550, and SCoT36-520) of which were successfully cloned, sequenced, and converted into SCAR markers. Usually, SCAR markers have been developed from traditional fingerprinting methods, such as RAPD (Joseph et al., 2014), ISSR (Lee et al., 2011), and AFLP amplicons (Choi et al., 2008).
Each of the SCAR markers (ST3KZ, ST3MSJ, ST5SJ, and ST36XSJ) developed in the study produced a specific amplicon of a certain length for each target Physalis species, but no amplicons were produced by other, non-target Physalis species (Figures 3, 4 and Table 4). These results revealed that SCAR markers might be used to identify and differentiate Physalis species. Of course, in the future, we will collect more Physalis species to further verify the reliability of our results. An increasing number of studies have reported that SCAR markers have been used to authenticate a variety of plant species, such as Panax japonicas (Choi et al., 2008), Casuarina equisetifolia (Ghosh et al., 2011), Jatropha curcas (Mulpuri et al., 2013), Rosa (Bashir et al., 2014), Commiphora (Sairkar et al., 2016), and Dasypyrum species (Hu et al., 2017).
Of course, in order to identify and classify Physalis species more accurately, it is best to conduct relevant studies from the genome level. At present, the complete chloroplast genome of P. peruviana have been sequenced (GenBank accession: NC_026570), which provides a new method for more accurate identification and phylogenetic reconstruction of Physalis species in the future.
A fast, sensitive, and reliable method for identifying Physalis species based on SCAR markers was developed from SCoT fragments that were specific for four target Physalis species. The specific primer pairs developed in this study clearly demonstrated that the specific amplicon was only present in the target Physalis species, while no amplification was observed in the other Physalis non-target species. The resulting specific SCAR primers were shown to be powerful tools that could be used to rapidly and reliably screen Physalis species.
SF and HW conceived and designed the study. HW, SF, and JL collected the plant samples. CY, KJ, YZ, MJ, and QY performed the experiments. SF, KJ, JL, and CS analyzed the data. SF and HW wrote the manuscript.
This study was supported in part by the National Natural Science Foundation of China (31470407), the Hangzhou Scientific and Technological Program (20150932H04, 20150932H03), and the Zhejiang Provincial Public Welfare Technology Applied Research Foundation of China (2014C32090).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00192/full#supplementary-material
FIGURE S1 | A UPGMA analysis for tested Physalis species in this study was performed based on the genetic distances using MEGA 6.0 software. Numbers (I–IV) indicates Physalis samples were grouped into four groups.
TABLE S1 | List of Physalis samples used for SCAR markers validation.
TABLE S2 | The pairwise similarity coefficient calculated by SCoT markers.
Axelius, B. (1996). The phylogenetic relationships of the physaloid genera (Solanaceae) based on morphological data. Am. J. Bot. 83, 118–124. doi: 10.1007/s13364-012-0075-z
Bashir, K. M., Awan, F. S., Khan, I. A., Khan, A. I., and Usman, M. (2014). Identification and authentication of Rosa species through development of species-specific SCAR marker(s). Genet. Mol. Res. 13, 4130–4139. doi: 10.4238/2014.May.30.8
Choi, Y. E., Ahn, C. H., Kim, B. B., and Yoon, E. S. (2008). Development of species specific AFLP-derived SCAR marker for authentication of Panax japonicus C. A. MEYER. Biol. Pharm. Bull. 31, 135–138.
Cirillo, A., Del Gaudio, S., Di Bernardo, G., Galano, G., Galderisi, U., and Cipollaro, M. (2012). A new SCAR marker potentially useful to distinguish Italian cattle breeds. Food Chem. 130, 172–176.
Clark, K., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., and Sayers, E. W. (2016). GenBank. Nucleic Acids Res. 44, D67–D72. doi: 10.1093/nar/gkv1276
Collard, B. C. Y., and Mackill, D. J. (2009). Start codon targeted (SCoT) polymorphism: a simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol. Biol. Rep. 27, 86–93. doi: 10.1007/s11105-008-0060-5
Continuing Professional Education Committee (2015). Pharmmacopoeia of the People’s Republic of China. Beijing: Chemical Industry Press, 360–361.
Ding, H., Hu, Z., Yu, L., Ma, Z., Ma, X., Chen, Z., et al. (2014). Induction of quinone reductase (QR) by withanolides isolated from Physalis angulata L. var. villosa Bonati (Solanaceae). Steroids 86, 32–38. doi: 10.1016/j.steroids.2014.04.015
Fang, S. T., Liu, J. K., and Li, B. (2012). Ten new withanolides from Physalis peruviana. Steroids 77, 36–44. doi: 10.1016/j.steroids.2011.09.011
Feng, S., He, R., Yang, S., Chen, Z., Jiang, M., Lu, J., et al. (2015a). Start codon targeted (SCoT) and target region amplification polymorphism (TRAP) for evaluating the genetic relationship of Dendrobium species. Gene 567, 182–188. doi: 10.1016/j.gene.2015.04.076
Feng, S., Jiang, Y., Wang, S., Jiang, M., Chen, Z., Ying, Q., et al. (2015b). Molecular Identification of Dendrobium Species (Orchidaceae) based on the DNA Barcode ITS2 region and its application for phylogenetic study. Int. J. Mol. Sci. 16, 21975–21988. doi: 10.3390/ijms160921975
Feng, S., Jiao, K., Zhu, Y., Wang, H., Jiang, M., and Wang, H. (2018). Molecular identification of species of Physalis (Solanaceae) using a candidate DNA barcode: the chloroplast psbA-trnH intergenic region. Genome 61, 15–20. doi: 10.1139/gen-2017-0115
Feng, S., He, R.-F., Jiang, M.-Y., Lu, J.-J., Shen, X.-X., Liu, J.-J., et al. (2016a). Genetic diversity and relationships of medicinal Chrysanthemum morifolium revealed by start codon targeted (SCoT) markers. Sci. Hortic. 201, 118–123.
Feng, S., Jiang, M., Shi, Y., Jiao, K., Shen, C., Lu, J., et al. (2016b). Application of the ribosomal DNA ITS2 region of Physalis (Solanaceae): DNA barcoding and phylogenetic study. Front. Plant Sci. 7:1047. doi: 10.3389/fpls.2016.01047
Garzon-Martinez, G. A., Osorio-Guarin, J. A., Delgadillo-Duran, P., Mayorga, F., Enciso-Rodriguez, F. E., Landsman, D., et al. (2015). Genetic diversity and population structure in Physalis peruviana and related taxa based on InDels and SNPs derived from COSII and IRG markers. Plant Gene 4, 29–37. doi: 10.1016/j.plgene.2015.09.003
Ghosh, M., Chezhian, P., Sumathi, R., and Yasodha, R. (2011). Development of SCAR marker in Casuarina equisetifolia for species authentication. Trees Struct. Funct. 25, 465–472.
Guo, D. L., Zhang, J. Y., and Liu, C. H. (2012). Genetic diversity in some grape varieties revealed by SCoT analyses. Mol. Biol. Rep. 39, 5307–5313. doi: 10.1007/s11033-011-1329-6
Hao, J., Jiao, K., Yu, C., Guo, H., Zhu, Y., Yang, X., et al. (2018). Development of SCoT-Based SCAR marker for rapid authentication of Taxus Media. Biochem. Genet. doi: 10.1007/s10528-018-9842-0 [Epub ahead of print].
Hu, M., Baum, B. R., and Johnson, D. A. (2017). SCAR markers that discriminate between Dasypyrum species and cytotypes. Genet. Resour. Crop Evol. 64, 505–514.
Ji, L., Yuan, Y. L., Ma, Z. J., Chen, Z., Gan, L. S., Ma, X. Q., et al. (2013). Induction of quinone reductase (QR) by withanolides isolated from Physalis pubescens L. (Solanaceae). Steroids 78, 860–865. doi: 10.1016/j.steroids.2013.05.008
Joseph, K. S., Murthy, H. N., and Ravishankar, K. V. (2014). Development of SCAR marker for sex identification in dioecious Garcinia gummi-gutta. Trees Struct. Funct. 28, 1645–1651.
Labate, J., and Robertson, L. (2015). Nucleotide diversity estimates of tomatillo (Physalis philadelphica) accessions including nine new inbred lines. Mol. Breed. 35:105. doi: 10.1007/s11032-015-0302-9
Lee, J. W., Kim, Y. C., Jo, I. H., Seo, A. Y., Lee, J. H., Kim, O. T., et al. (2011). Development of an ISSR-Derived SCAR Marker in Korean Ginseng Cultivars (Panax ginseng C. A. Meyer). J. Ginseng Res. 35, 52–59.
Li, X., Zhao, J. P., Yang, M., Liu, Y. L., Li, Z. C., Li, R. Y., et al. (2014). Physalins and withanolides from the fruits of Physalis alkekengi L. var. franchetii (Mast.) Makino and the inhibitory activities against human tumor cells. Phytochem. Lett. 10, 95–100.
Luo, C., He, X. H., Chen, H., Hu, Y., and Ou, S. J. (2012). Genetic relationship and diversity of Mangifera indica L.: revealed through SCoT analysis. Genet. Res. Crop Evol. 59, 1505–1515. doi: 10.1007/s10722-011-9779-1
Luo, C., He, X. H., Chen, H., Ou, S. J., and Gao, M. P. (2010). Analysis of diversity and relationships among mango cultivars using start codon targeted (SCoT) markers. Biochem. Syst. Ecol. 38, 1176–1184. doi: 10.1016/j.bse.2010.11.004
Luo, C., He, X. H., Chen, H., Ou, S. J., Gao, M. P., Brown, J. S., et al. (2011). Genetic diversity of mango cultivars estimated using SCoT and ISSR markers. Biochem. Syst. Ecol. 39, 676–684. doi: 10.1016/j.bse.2011.05.023
Mandoulakani, B. A., Yaniv, E., Kalendar, R., Raats, D., Bariana, H. S., Bihamta, M. R., et al. (2015). Development of IRAP- and REMAP-derived SCAR markers for marker-assisted selection of the stripe rust resistance gene Yr15 derived from wild emmer wheat. Theor. Appl. Genet. 128, 211–219. doi: 10.1007/s00122-014-2422-8
Marieschi, M., Torelli, A., Beghe, D., and Bruni, R. (2016). Authentication of Punica granatum L.: development of SCAR markers for the detection of 10 fruits potentially used in economically motivated adulteration. Food Chem. 202, 438–444. doi: 10.1016/j.foodchem.2016.02.011
Martinez, M. (1998). Revision of Physalis section Epeteiorhiza (Solanaceae). Ann. Ins. Biol. Bot. 69, 71–117. doi: 10.1002/ece3.102
Mulpuri, S., Muddanuru, T., and Francis, G. (2013). Start codon targeted (SCoT) polymorphism in toxic and non-toxic accessions of Jatropha curcas L. and development of a codominant SCAR marker. Plant Sci. 207, 117–127. doi: 10.1016/j.plantsci.2013.02.013
Nei, M., and Li, W. H. (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U.S.A. 76, 5269–5273.
Olmstead, R. G., Bohs, L., Migid, H. A., Santiago-Valentin, E., Garcia, V. F., and Collier, S. M. (2008). A molecular phylogeny of the Solanaceae. Taxon 57, 1159–1181.
Paran, I., and Michelmore, R. W. (1993). Development of reliable PCR-based markers linked to downy mildew resistance genes in lettuce. Theor. Appl. Genet. 85, 985–993. doi: 10.1007/BF00215038
Rajesh, M. K., Sabana, A. A., Rachana, K. E., Rahman, S., Ananda, K. S., and Karun, A. (2016). Development of a SCoT-derived SCAR marker associated with tall-type palm trait in arecanut and its utilization in hybrid (dwarf x tall) authentication. Indian J. Genet. Plant Breed. 76, 119–122.
Rohlf, F. J. (2000). NTSYS-PC: Numerical Taxonomy and Multivariate Analysis System, Version 2.00. Setauket, NY: Exeter Software.
Sairkar, P. K., Sharma, A., and Shukla, N. P. (2016). SCAR marker for identification and discrimination of Commiphora wightii and C. myrrha. Mol. Biol. Int. 2016:1482796. doi: 10.1155/2016/1482796
Sang-Ngern, M., Youn, U. J., Park, E. J., Kondratyuk, T. P., Simmons, C. J., Wall, M. M., et al. (2016). Withanolides derived from Physalis peruviana (Poha) with potential anti-inflammatory activity. Bioorg. Med. Chem. Lett. 26, 2755–2759. doi: 10.1016/j.bmcl.2016.04.077
Shimada, N., Nakatsuka, T., Nakano, Y., Kakizaki, Y., Abe, Y., Hikage, T., et al. (2009). Identification of gentian cultivars using SCAR markers based on intron-length polymorphisms of flavonoid biosynthetic genes. Sci. Hortic. 119, 292–296.
Sun, C. P., Nie, X. F., Kang, N., Zhao, F., Chen, L. X., and Qiu, F. (2017). A new phenol glycoside from Physalis angulata. Nat. Prod. Res. 31, 1059–1065. doi: 10.1080/14786419.2016.1269102
Vargas-Ponce, O., Perez-Alvarez, L. F., Zamora-Tavares, P., and Rodriguez, A. (2011). Assessing genetic diversity in mexican husk tomato species. Plant Mol. Biol. Rep. 29, 733–738.
Wei, J., Hu, X., Yang, J., and Yang, W. (2012). Identification of single-copy orthologous genes between Physalis and Solanum lycopersicum and analysis of genetic diversity in Physalis using molecular markers. PLoS One 7:e50164. doi: 10.1371/journal.pone.0050164
Whitson, M., and Manos, P. S. (2005). Untangling Physalis (Solanaceae) from the Physaloids: a two-gene phylogeny of the Physalinae. Syst. Bot. 30, 216–230.
Xiao, Y., Dai, Y. H., Lu, Y. Y., Liu, W., Wang, Z. R., and Bian, Y. B. (2011). Development of IRAP-SCAR marker for strain identification in Lentinula edodes. World J. Microbiol. Biotechnol. 27, 1731–1734.
Xiong, F. Q., Zhong, R. C., Han, Z. Q., Jiang, J., He, L. Q., Zhuang, W. J., et al. (2011). Start codon targeted polymorphism for evaluation of functional genetic variation and relationships in cultivated peanut (Arachis hypogaea L.) genotypes. Mol. Biol. Rep. 38, 3487–3494. doi: 10.1007/s11033-010-0459-6
Xu, X. M., Guan, Y. Z., Shan, S. M., Luo, J. G., and Kong, L. Y. (2016). Withaphysalin-type withanolides from Physalis minima. Phytochem. Lett. 15, 1–6. doi: 10.1007/s10753-016-0485-1
Yang, Y. K., Xie, S. D., Xu, W. X., Nian, Y., Liu, X. L., Peng, X. R., et al. (2016). Six new physalins from Physalis alkekengi var. franchetii and their cytotoxicity and antibacterial activity. Fitoterapia 112, 144–152. doi: 10.1016/j.fitote.2016.05.010
Zamora-Tavares, P., Vargas-Ponce, O., Sanchez-Martinez, J., and Cabrera-Toledo, D. (2015). Diversity and genetic structure of the husk tomato (Physalis philadelphica Lam.) in Western Mexico. Genet. Resour. Crop Evol. 62, 141–153.
Keywords: Physalis species, species-specific, SCoT markers, SCAR markers, marker development
Citation: Feng S, Zhu Y, Yu C, Jiao K, Jiang M, Lu J, Shen C, Ying Q and Wang H (2018) Development of Species-Specific SCAR Markers, Based on a SCoT Analysis, to Authenticate Physalis (Solanaceae) Species. Front. Genet. 9:192. doi: 10.3389/fgene.2018.00192
Received: 07 November 2017; Accepted: 11 May 2018;
Published: 29 May 2018.
Edited by:
Badri Padhukasahasram, Illumina (United States), United StatesReviewed by:
Leonardo Marino-Ramirez, National Institutes of Health (NIH), United StatesCopyright © 2018 Feng, Zhu, Yu, Jiao, Jiang, Lu, Shen, Ying and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shangguo Feng, c2hhbmdndW8wMDdAMTI2LmNvbQ== Huizhong Wang, d2h6NjJAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.