
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 13 December 2017
Sec. RNA
Volume 8 - 2017 | https://doi.org/10.3389/fgene.2017.00200
A correction has been applied to this article in:
Corrigendum: Novel Genomic and Evolutionary Perspective of Cyanobacterial tRNAs
Transfer RNA (tRNA) plays a central role in protein synthesis and acts as an adaptor molecule between an mRNA and an amino acid. A tRNA has an L-shaped clover leaf-like structure and contains an acceptor arm, D-arm, D-loop, anti-codon arm, anti-codon loop, variable loop, Ψ-arm and Ψ-loop. All of these arms and loops are important in protein translation. Here, we aimed to delineate the genomic architecture of these arms and loops in cyanobacterial tRNA. Studies from tRNA sequences from 61 cyanobacterial species showed that, except for few tRNAs (tRNAAsn, tRNALeu, tRNAGln, and tRNAMet), all contained a G nucleotide at the 1st position in the acceptor arm. tRNALeu and tRNAMet did not contain any conserved nucleotides at the 1st position whereas tRNAAsn and tRNAGln contained a conserved U1 nucleotide. In several tRNA families, the variable region also contained conserved nucleotides. Except for tRNAMet and tRNAGlu, all other tRNAs contained a conserved A nucleotide at the 1st position in the D-loop. The Ψ-loop contained a conserved U1-U2-C3-x-A5-x-U7 sequence, except for tRNAGly, tRNAAla, tRNAVal, tRNAPhe, tRNAThr, and tRNAGln in which the U7 nucleotide was not conserved. However, in tRNAAsp, the U7 nucleotide was substituted with a C7 nucleotide. Additionally, tRNAArg, tRNAGly, and tRNALys of cyanobacteria contained a group I intron within the anti-codon loop region. Maximum composite likelihood study on the transition/transversion of cyanobacterial tRNA revealed that the rate of transition was higher than the rate of transversion. An evolutionary tree was constructed to understand the evolution of cyanobacterial tRNA and analyses revealed that cyanobacterial tRNA may have evolved polyphyletically with high rate of gene loss.
Transfer RNAs (tRNA) are short non-coding RNAs comprised of 75–95 nucleotides, universally present in all organisms from the prokaryotes to the eukaryotes. The 75–95 residues polynucleotide sequences of tRNAs fold back upon themselves and form hydrogen-bonded clover leaf-like structures that fold into L-shaped tertiary structure (Holley et al., 1965; Wilusz, 2015). The tRNA is organized into a linear double stranded helix. Determination of the three-dimensional structure of a tRNA was a landmark discovery for modern molecular biology research. The major function of the tRNA is to bridge between an amino acid and an mRNA during protein synthesis where the tRNA transfers the cognate amino acid to the translating polypeptide chain. The clover leaf-like structure of the tRNA is comprised of an acceptor arm, D-arm, D-loop, anti-codon arm, anti-codon loop, variable loop, Ψ-arm, and Ψ-loop (Figure 1; Kirchner and Ignatova, 2014; Mohanta and Bae, 2017). The acceptor arm is 7 base pairs long, the D-arm is 3–4 base pairs, the D-loop is 4–12 nucleotides, the anti-codon arm is 5 base pairs, the anti-codon loop is 7 nucleotides, the variable loop is 4–23 nucleotides, the Ψ-arm is 5 base pairs and the Ψ-loop is 7 nucleotides long (Figure 1; Kirchner and Ignatova, 2014). The heterogeneity in the length of tRNAs is caused by variable number of bases in D-loop and the variable loop. The most important functional parts of a tRNA are: the anti-codon triplet, which reads the codons of a messenger RNA; a 3′-CCA tail where the cognate amino acid is attached and the Ψ-arm and Ψ-loop that hold the ribosome machinery.
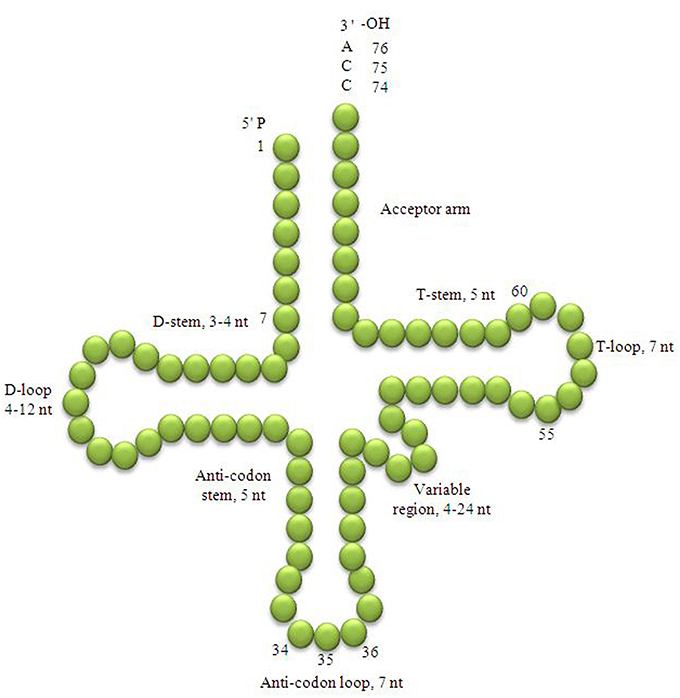
Figure 1. Clover leaf-like structure of tRNA. The tRNA possess the acceptor arm (7 nt), D-arm (3–4 nt), D-loop (4–12 nt), anti-codon arm (5 nt), anti-codon loop (7 nt), variable region (4–23 nt), Ψ-arm (5 nt) and Ψ-loop (7 nt). The D-arm, D-loop, and variable region possess variable number of nucleotides whereas the nucleotide number in the acceptor arm, anti-codon arm, anti-codon loop, Ψ-arm and Ψ-loop is always constant. The anti-codon nucleotides in the anti-codon loop is always numbered 34, 35, and 36 whereas, the nucleotides of C-C-A tail is always numbered with 74, 75, and 76.
There are 21 different types of iso-acceptor families for the 20 amino acids, one for each amino acid and one for selenocysteine. In prokaryotes, an isoacceptor family may have one (tRNATrp, and tRNAMet) to six tRNA members (tRNASer, tRNAArg, and tRNALeu). To date, four different types of tRNA genes have been reported so far including intron-containing, non-intronic, split and permutated types (Randau et al., 2005; Fujishima et al., 2009; Chan et al., 2011). The intron-containing and non-intronic tRNAs are encoded in a single gene, whereas the split tRNAs are encoded in two or more separate genes; in permutated tRNAs, the 3′ half of the tRNA is located up-stream to the 5′ half of the tRNA (Tocchini-Valentini et al., 2009; Maruyama et al., 2010; Soma, 2014). The intron-containing, permutated and split tRNA genes are called “disrupted tRNA genes” whereas the non-intronic tRNA genes are called “continuous tRNA genes” (Sugahara et al., 2009; Kanai, 2015). The disrupted tRNA genes may have evolved earlier than the continuous genes (Di Giulio, 2008a,b; Fujishima et al., 2009). The tRNA mini helix is the most ancient form of tRNA and forms the top half of the tRNA that contains the acceptor arm and the Ψ-arm (Weiner and Maizels, 1987, 1999; Shi et al., 1992, 1998; Sun and Caetano-Anollés, 2008). The tRNA mini-helix is a validated substrate for aminoacyl-tRNA synthetases and the 3′-CCA tail adding enzymes (Shi et al., 1992, 1998). The genomic tag hypothesis suggests that the upper half of the tRNA had evolved earlier than the lower half. Phylogenetic analysis also suggests that the top half of the tRNA is more ancient than the bottom half (Sun and Caetano-Anollés, 2008). The evolution of tRNA occurred with respect to its surrounding environment and is far more complex than that of proteins, because tRNA perform multiple roles in the cell (Navarre and Schneewind, 1999; Kirchner and Ignatova, 2014). tRNA interacts with numerous molecules; during protein synthesis, it interacts with amino-acyl tRNA synthetase, ribosome and messenger RNA, and a large number of transcription factors and other enzymes (Nirenberg and Leder, 1964). tRNAGly acts as a structural element of the peptidoglycan of the bacterial cell wall whereas tRNALys and tRNAAla are used to aminoacylate membrane lipid to permeate cationic antibiotics (Roy and Ibba, 2008). Cyanobacteria are the most ancient living organism whose existence dates back to 3.3–3.5 billion years (Schopf and Packer, 1987; Altermann et al., 2006). Because it is most likely that the tRNA genes date back to the age of cyanobacteria, and studying the genomic and evolutionary perspectives of cyanobacterial tRNA is very important. Given their high level of conservation it is especially important to study the genomics, origin and evolution of tRNAs. Here, a comprehensive study of cyanobacterial tRNA was conducted by analyzing the tRNA sequences of 61 cyanobacterial species.
The genomic tRNA sequences of cyanobacteria were searched from Joint Genome Portal (http://genome.jgi.doe.gov/) and cyanobacterial genome database of National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/). In total, tRNA sequences from 61 cyanobacterial species were retrieved and analyzed (Supplementary data 1). The downloaded genomic tRNA sequences of cyanobacterial species were analyzed using tRNAScan-SE software (Lowe and Eddy, 1997). All the sequences analyzed during this study were found to encode for tRNA. Different statistical parameters used to run the tRNAScan-SE software were as follows: sequence source, bacterial/mixed; search mode, default; and genetic code for tRNA isotype prediction, universal. The tRNAScan-SE software is the most accurate software and it identifies 99–100% tRNA genes from DNA sequences. It produces less than one false positive per 15 gigabases (Lowe and Eddy, 1997).
Multiple sequence alignment was conducted to identify the conserved consensus sequences of cyanobacterial tRNA. Multalin software was used to conduct the multiple sequence alignment of cyanobacterial tRNA using the default parameters as described previously (Mohanta et al., 2015a,b,c, 2016) with minor modification.
To analyze the transition and transversion rate of the cyanobacterial tRNA, a clustal file for each of the tRNA gene family was generated using the multiple sequence alignment program MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/). The generated clustal file was downloaded and converted to a MEGA file format. The generated MEGA file of the cyanobacterial tRNA was uploaded in MEGA6 software to analyze the transition/transversion rate (Tamura et al., 2013). Following statistical parameters were used to analyze the transition/transversion rate: analysis, estimate transition/transversion bias (MCL); scope, all selected taxa; statistical method, maximum composite likelihood; substitution type, nucleotide; model/method, Tamura-Nei model; and gaps/missing data treatment, pair wise deletion.
A phylogenetic tree was constructed to understand the evolutionary aspects of cyanobacterial tRNAs. All the genomic tRNA sequences of cyanobacterial tRNA were subjected to clustal omega server to construct a clustal file. Generated clustal file of the cyanobacterial tRNA was converted to MEGA file format using MEGA6 software. The MEGA files of the cyanobacterial tRNA were subjected to MEGA6 software to construct the phylogenetic trees (Tamura et al., 2013). Following statistical parameters were used to construct the phylogenetic tree: analysis, phylogeny reconstruction; scope, all selected taxa; statistical method, neighbor-joining; test of phylogeny, bootstrap method; no. of bootstrap replicate, 1,000; substitution type, nucleotide; model/method, maximum composite likelihood; substitution to include, d: transition+transversion; rates among sites, uniform rates; pattern among lineage, same (homogeneous); and gaps/missing data treatment, pair wise deletion. The pairwise distances and substitution model parameters were estimated by maximizing the composite likelihood because maximum likelihood approach can accurately estimate the parameters that drive the evolutionary process (Akaike, 1998; Varin et al., 2011; Xu and Reid, 2011; Zhao et al., 2015). Phylogenetic trees with low bootstrap values were collapsed with 50% cutoff values.
To understand the duplication and loss event of cyanobacterial tRNA, all the clustal files generated to study the transition/transversion rate of individual tRNA family were used to construct the phytlogenetic tree. Twenty phylogenetic trees were generated separately for each tRNA gene family using the similar statistical parameters as mentioned previously. A species tree of studied cyanobacterial species was downloaded from NCBI (https://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi). The phylogenetic tree of the cyanobacterial tRNA and species tree of cyanobacterial species were subjected to Notung2.6 software and further reconciliation of gene tree with the species tree led to finding of duplicated and loss genes of cyanobacterial tRNA (Chen et al., 2000; Vernot et al., 2008).
To understand the basic genomic and evolutionary aspects of cyanobacterial tRNAs, genomic tRNA sequences of cyanobacterial species were downloaded from the Joint Genome Institute (JGI) genome portal and National Center for Biotechnology Information (NCBI) (Table 1). In total, the tRNA sequences of 61 cyanobacterial species were downloaded and analyzed for their conserved genomic aspects. A conserved genomic sequence is required for the conserved clover leaf-like structure of a tRNA. Although significant conservation is present in tRNAs at the nucleotide level, occasionally the conserved structure of tRNAs varies at the family level.
In tRNAGly of the Pseudanabaena sp. PCC 7367 (gene id: 2504679288) was found to contain only a conserved C2 nucleotide instead of the conserved G1-C2-G3 (Table 2, Supplementary Figure 1). Similarly, the acceptor arm of Cylindrospermum stagnale PCC 7417 (gene id: 2509767604) was found to contain G1-G2-A3 nucleotides. The nucleotide U8 and A9 in the acceptor arm of tRNAGly was found to be conserved (Table 3). The D-arm was found to be unconserved whereas the D-loop has conserved A1-x2-G4-G5 nucleotides followed by the presence of conserved U2-x-C4-C5-A6 nucleotides in anti-codon arm (Table 2). The variable loop was found to be unconserved whereas the Ψ-arm has conserved G5 nucleotide and the Ψ-loop has conserved U1-U2-C3-x-A5 nucleotides (Table 2).
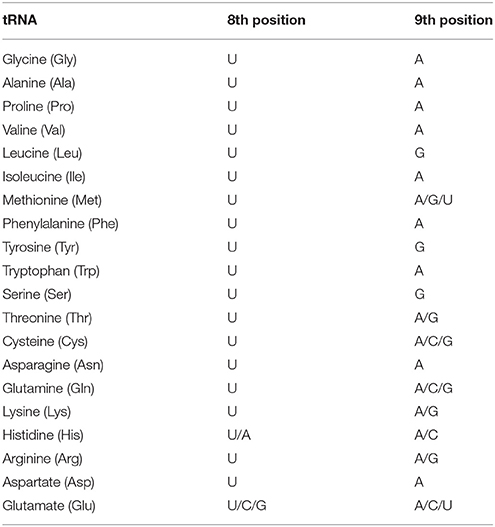
Table 3. Variations of nucleotide conservation at the 8th and 9th position of tRNAs in the 5′ end of the acceptor arm in cyanobacteria.
In tRNAAla, the acceptor arm was found to contain conserved G1-G2-G3 nucleotides at the 5′end (Table 2) whereas tRNAAla of Pseudanabaena sp. PCC 6802 (gene id: 2507088349) and Microchaete sp. PCC 7126 (gene id: 2509784201) was found to contain A1-G2-G3 nucleotides. The nucleotide U8 and A9 in the acceptor arm were found to be conserved (Table 3) followed by the presence of conserved G1-C2-U3-C4 nucleotides in the D-arm. The D-loop contained conserved A1-x2-U4-G5-G6-x-A8 nucleotides followed by the presence of conserved U2-x-G4-C5-A6 nucleotides in the anti-codon arm. A G5 nucleotide was found to be conserved in the variable region as well as in the Ψ-arm followed by the presence of conserved U1-U2-C3-x-A5 nucleotides in the Ψ- loop (Table 2).
The acceptor arm of tRNAPro was found to contain conserved C1-x-G3-G4-x2-G7 nucleotides (Table 2). The nucleotide U8 and A9 in the acceptor arm and C4 in the D-arm were found to be conserved followed by the presence of conserved A1-G2-x6-A9 nucleotides in the D-loop (Tables 2, 3). The anti-codon loop contained conserved U1-U2-x-G4-G5-G6 followed by the presence of conserved G3-x-C5 nucleotides in the variable region. The Ψ-arm contained conserved G1-x2-G4-G5 nucleotides followed by the presence of conserved U1-U2-C3-x-A5-A6-U7 nucleotides in the Ψ- loop (Table 2).
The acceptor arm of tRNAVal was found to contain conserved G1-G2-x-C4 nucleotides (Table 2) except for species Cyanobacterium sp. ESFC 1 (gene id: 2517646710), Microcoleus vaginatus FGP-2 (gene id: 2506348931), Nostoc sp. PCC 7107 (gene id: 2503741640), Kamptonema formosum PCC 6407 (gene id: 2508873173), Pseudanabaena sp. PCC 6802 (gene id: 2507089028), Oscillatoria acuminate PCC 6304 (gene id: 2509421383), Microchaete sp.PCC 7126 (gene id: 2509784199), and Microcoleus sp. PCC 7113 (gene id: 2509433894). The tRNAVal of Pleurocapsa sp. PCC 7319 (gene id: 2509708986). They were found to contain U1 nucleotide instead of the G1 or A1 nucleotides. The U8 and A9 nucleotides were conserved in tRNAVal (Table 3) followed by the presence of conserved C2-x-C4 nucleotides in the D-arm. The D-loop contained conserved A1-G2-x3-G6-x-U8-A9 nucleotides followed by the presence of conserved U1-U2-x-A4-C5-A6 nucleotides in the anti-codon arm (Table 2). The variable region contained conserved G3-U4-C5 nucleotides, followed by the presence of conserved G4-G5 nucleotides in the Ψ-arm and U1-U2-C3-x-A5 nucleotides in the Ψ- loop (Table 2).
The acceptor arm of tRNALeu contained variable nucleotides. However, more than half of the tRNALeu were contained G1-C2-G3 nucleotides (Table 2). The U8 and U9 nucleotide were conserved at the 8th and 9th position, respectively (Table 3), whereas the D-arm did not contain any conserved nucleotide sequences whereas the D-loop contained conserved A1-A2-x2-G5-G6-x-A8 nucleotides. The anti-codon loop contained conserved U2-x-A4 nucleotides followed by the presence of G5 nucleotide in the Ψ-arm and U1-U2-C3-x-A5-x-U7 nucleotides in the Ψ- loop (Table 2). No conserved nucleotides were found in the variable loop.
The acceptor arm of tRNAIle contained conserved G1-G2-G3-C4 nucleotides (Table 2). The nucleotides U8 and A9 were also found to be conserved in their respective position (Table 3) followed by the presence of conserved G1-C2-U3-C4 nucleotides in the D-arm and A1-x4-G6-x2-A9 nucleotides in the D-loop. The anti-codon loop contained conserved C1-U2-x-A4-U5-A6-A7 nucleotides followed by the presence of conserved G3-U4 nucleotides in the variable region, G4-G5 nucleotides in the Ψ-arm and U1-U2-C3-x-A5-x-U7 nucleotides in the Ψ-loop (Table 2).
In tRNAMet, no consensus conserved nucleotides were found at the 5′ end of the acceptor arm. A few tRNAs were contained G1-G2-C3 nucleotides whereas some other were contained C1-G2-C3, or C1-C2-A3 nucleotides (Table 2). The U nucleotide at the 8th position was conserved in the acceptor arm whereas the 9th position was occupied by either A9/G9 or U9 nucleotide. The D-arm and the D-loop was found to contain conserved G2 and G6 nucleotide, respectively. The anti-codon loop contained conserved C1-U2-C3-A4-U5-A6-A7 nucleotides followed by the presence of conserved G5 nucleotide in the Ψ-arm and U1-U2-C3-x-A5-x-U7 nucleotides in the Ψ-loop (Table 2).
The acceptor arm of tRNAPhe contained conserved C1/G1-C2-C3-x2-G6 nucleotides (Table 2). The nucleotides U8 and A9 in the acceptor arm were conserved (Table 3) followed by the presence of conserved G1-C2-U3-C4 nucleotides in the D-arm and A1-G2-U3-U4-G5-G6-U7 nucleotides in the D-loop, respectively. In the majority of the cases, the anti-codon arm contained conserved G4 nucleotide and the anti-codon loop contained conserved U2-G3-A4-A5-x-A7 nucleotides. The variable loop contained conserved G3-U4-C5 nucleotides followed by the presence of conserved G5 nucleotide the Ψ-arm and U1-U2-C3-x-A5 nucleotides in the Ψ-loop (Table 2).
In tRNATyr, the acceptor arm contained conserved G1-G2-G3-U4-C5 nucleotides (Table 2). The U8 and G9 nucleotides in the acceptor arm were conserved (Table 3) followed by the presence of conserved G1-x-C3-C4 nucleotides in the D-arm and A1-G2-U3-G4-G5-U6-U7-A8 nucleotides in the D-loop, respectively (Table 2). The anti-codon loop contained conserved U2-G3-U4-A5 nucleotides followed by the presence of conserved G4-G5 nucleotides in the Ψ-arm and U1-U2-C3-x-A5-x-U7 nucleotides in the Ψ-loop (Table 2).
In the majorities of cases, the acceptor arm of tRNATrp contained a conserved G1 nucleotide whereas few others were found to contain A1 nucleotide. The U8 nucleotide of the acceptor arm was conserved whereas A9 nucleotide was sometimes substituted with G9 nucleotide (Tables 2, 3). The D-arm contained conserved G1-U2 nucleotides followed by the presence of conserved A1-x3-G5 nucleotides in the D-loop. The anti-codon arm contained conserved G4-U5 nucleotides followed by the presence of conserved C1-U2-C3-C4-A5-A6-A7 nucleotides in the anti-codon loop, a G5 nucleotide in the Ψ-arm and U1-U2-C3-x-A5-x-U7 nucleotides in the Ψ-loop, respectively (Table 2).
In tRNASer, the acceptor arm contained conserved G1-G2-A3 nucleotides (Table 2). A few of the tRNASer were contained U1 or G1 nucleotides instead of G1-G2-A3 consensus nucleotide sequence. The U8 and G9 nucleotide were found to be conserved (Table 3) followed by the presence of conserved G1-C2 nucleotides in the D-arm and A2-x2-G5-G6 nucleotides in the D-loop, respectively. The anti-codon loop contained conserved U2-x3-A5-A6 nucleotides (Table 2) followed by the presence of conserved C21 nucleotide in variable region, A4/G4-G5 nucleotides in the Ψ-arm and U1-U2-C3-x-A5-x-U7 nucleotides in the Ψ-loop, respectively (Table 2).
The acceptor arm of tRNAThr contained conserved G1-C2 nucleotides (Table 2). The U8 nucleotide was found to be conserved whereas the 9th position was occupied by A9 or G9 nucleotide (Table 3). The D-arm contained conserved G1-C2-x-C4 nucleotides and A1-x1−4-G3-G4-x-A6 nucleotides in the D-loop, respectively. The anti-codon loop contained conserved U2-x-G4-U5-A6-A7 nucleotides followed by the presence of conserved G3 nucleotide in the variable loop, G5 nucleotide in the Ψ-arm and U1-U2-C3-x-A5 nucleotides in the Ψ-loop, respectively (Table 2). At the 7th position of the Ψ-loop, the U7 nucleotide was sometimes substituted with C7 nucleotide.
The acceptor arm of tRNACys contained a conserved G1 nucleotide. The U8 and A9 nucleotides were found to be conserved while in some cases, A9 nucleotide was substituted with either C9 or G9 nucleotides (Table 3). The D-arm contained conserved G1-C2-C3 nucleotides followed by the presence of conserved A1-A2-G3-x-G5-G6-U7 and C1-U2-G3-C4-A5-A6-A7 nucleotides in the D-loop and anti-codon loop, respectively. The C5 nucleotide was conserved in the variable region of the tRNACys followed by the presence of conserved G5 and U1-U2-C3-x-A5-x-U7 nucleotides in the Ψ-arm and Ψ-loop, respectively (Table 2).
The acceptor arm of tRNAAsn contained conserved U1-C2-C3-x-C5 nucleotides (Table 2) with a few exceptions. The U8 and A9 nucleotides of the acceptor arm were conserved (Table 3) followed by the presence of conserved G1-C2-U3 nucleotides in the D-arm and A1-x1−2-G3-G4 nucleotides in the D-loop, respectively (Table 2). The anti-codon loop contained conserved C1-U2-G3-U4-U5-A6-A7 nucleotides followed by the presence of conserved G3-U4 nucleotides in the variable region, G5 in the Ψ-arm and U1-U2-C3-x-A5-x-U7 nucleotides in the Ψ-loop, respectively (Table 2).
The acceptor arm of tRNAGln contained conserved U1-G2-x3−4-G6 nucleotides (Table 2). The U8 nucleotide of the acceptor arm was conserved whereas the 9th position was substituted with either A9/C9, or G9 nucleotides (Table 3). The D-loop contained conserved A1/U1-A2-x2-G4 nucleotides followed by the presence of conserved U1-U2-U3-U4-G5 nucleotides in the anti-codon loop, A3-G4-G5 nucleotides in the Ψ-arm and U1-U2-C3-x-A5 nucleotides in the Ψ-loop, respectively (Table 2).
The acceptor arm at tRNALys contained conserved G1-G2-G3 nucleotides (Table 2). In a few cases, the G1 nucleotide at the 1st position was substituted with either C1 or U1 nucleotide. The nucleotide U8 and A9 in the acceptor arm were conserved (Table 3) followed by the presence of conserved C2 nucleotide in the D-arm and A1-x2-G4-G5 nucleotides in the D-loop. The anti-codon loop contained conserved C1-U2-U3-U4-U5-A6-A7 nucleotides, followed by the presence of conserved G4-G5 sequence in the Ψ-arm and U1-U2-C3-x-A5-x-U7 nucleotides in the Ψ-loop (Table 2). In a few cases, the U7 nucleotide in the Ψ-loop was substituted with C7 nucleotide (Table 2).
In tRNAHis, the acceptor arm contained conserved G1-C2/U2-G3-x3-G7 nucleotides with a few exceptions (Table 2). The U8 and A9 nucleotides in the acceptor arm were conserved followed by the presence of conserved G1-C2-C3 nucleotides in the D-arm and A1-A2-G3-x-G5-G6-U7 nucleotides in the D-loop, respectively (Table 2). The anti-codon arm contained conserved G4-G5 nucleotides followed by the presence of conserved U1-U2-G3-U4-G5 nucleotides in the anti-codon loop, G1-x-G3-G4-G5 nucleotides in the Ψ-arm and U1-U2-C3-x-A5-x-U7nucleotides in the Ψ-loop. In a few cases, the U7 nucleotide in the Ψ-loop was substituted with C7 nucleotide (Table 2).
In tRNAArg, the acceptor arm at the 5′ end contained conserved G1-G2 nucleotides (Table 2). In a few cases, the G1 nucleotide at the 1st position substituted by A1 or C1 nucleotide. The U8 nucleotide in the acceptor arm was conserved whereas the 9th position was substitutes with A9/G9 or C9 nucleotides (Table 3). The D-arm was found to contain conserved G1 nucleotide, followed by the presence of conserved A1-x2−3-G4-x3-A8 nucleotide in the D-loop and C1/U1-U2-x-C4 nucleotides in the anti-codon loop (Table 2). The Ψ-arm and Ψ-loop was found to contain conserved G4-G5 and U1-U2-C3-x-A5-x-U7 nucleotides, respectively (Table 2).
The acceptor arm of tRNAAsp contained conserved G1-G2-G3 nucleotides (Table 2). A few species were found to contain C1 nucleotide instead of G1 nucleotide at the 1st position. The U8 and A9 nucleotides in the acceptor arm were conserved (Table 3), followed by the presence of conserved G1-x-U3-C4 and A1-x2−3-G4-G5-U6-x-A8 nucleotides in the D-arm and D-loop, respectively (Table 2). The anti-codon loop contained conserved C1-U2-G3-U4-C5-A6 nucleotides followed by the presence of conserved A1-x-G3-U4 nucleotides in the variable region, G1-x-G3-x-G5 nucleotides in the Ψ-arm and U1-U2-C3-x-A5-x-C7 nucleotides in the Ψ-loop, respectively (Table 2). The C7 nucleotide in the Ψ-loop sometimes substituted with U7 nucleotide.
In tRNAGlu, the majorities of species were contained conserved G1 nucleotide in the acceptor arm (Table 2). In a few species, G1 nucleotide was substituted by A1 nucleotide at the first position. The U8 nucleotide in the acceptor arm was conserved whereas the 9th position was substituted by A9/G9 or C9 nucleotides (Table 3). The D-arm was devoid of any conserved nucleotide sequence whereas the D-loop contained conserved G5 nucleotide. The anti-codon loop contained conserved U2-U3-U4-C5 nucleotides, followed by the presence of conserved G3-x-G5 and U1-U2-C3-x-A5-x-U7 nucleotides in the Ψ-arm and Ψ-loop, respectively. The U7 nucleotide in the Ψ-loop sometimes substituted with the C7 nucleotide.
In our study, cyanobacterial tRNA was found to contain group I introns. The intron was present in the anti-codon loop region of the tRNA in the Nostoc sp. PCC 7524 (gene id: 2509813156), Gloeocapsa sp. PCC 73106 (gene id: 2508643885), and Nostoc sp. PCC 7107 (gene id: 2503742551) (Figure 2). tRNAArg (ACG) of Nostoc sp. PCC 7524 contained an intron from nucleotide 39–60 whereas in tRNAGly (TCC) of the Nostoc sp. PCC 7107 the intron was found from nucleotide 39–87. One of the genes encoding tRNALys (CTT) of the Gloeocapsa sp. PCC 73106 was found to contain an intron, from nucleotide 38 to 74. Previous studies reported that an intron was present only in tRNALeu (UAA) and tRNAfMet (UAC) of cyanobacterial tRNA (Paquin et al., 1997; Rudi and Jakobsen, 1999). However in our study, we found that the cyanobacterial group I intron was also present in tRNAArg, tRNAGly, and tRNALys (Figure 2). Multiple sequence alignment has shown the presence of conserved T-x-G-x2-T and G-x-C motifs in the introns of cyanobacterial tRNA (Figure 3). Sequence alignment between the introns of Gloeocapsa sp. PCC 73106 (tRNALys) and Nostoc sp. PCC 7107 (tRNAGly) showed the presence of conserved T-T-x2-C, C-T-T-G-x-C-T, A-A-G-x-C, G-A-A-G-A-A-T, and G-G motifs. The length of the intron ranged between 22 and 49 nucleotides. The introns of the Nostoc sp. PCC 7524 (gene id: 2509813156) and Nostoc sp. PCC 7107 (gene id: 2503742551) were found to be 22 and 38 nucleotides, respectively, whereas that of the Gloeocapsa sp. PCC 73106 (gene id: 2508643885) was 49 nucleotides long. No introns were found in other parts of the tRNA except for the anti-codon loop.
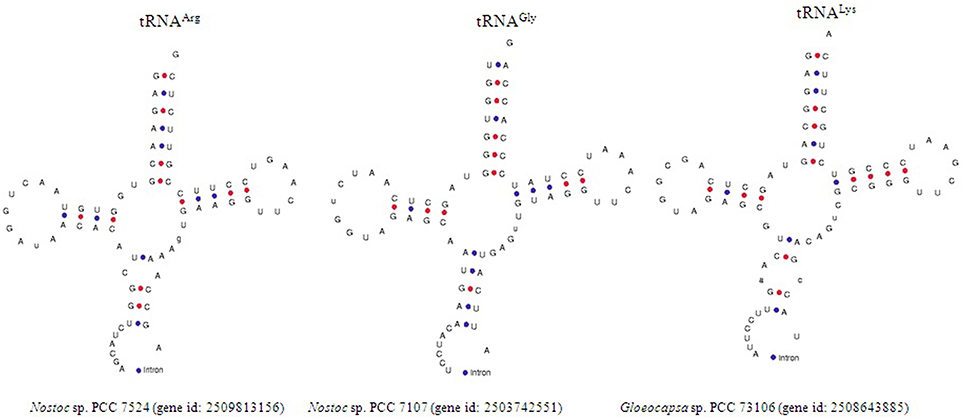
Figure 2. Figure representing the presence of group I intron in the cyanobacterial tRNA. Nostoc sp. PCC 7524 (gene id: 2509813156) was found to encode tRNAArg, and Nostoc sp. PCC 7107 (gene id: 2503742551) was found to encode tRNAGly whereas, Gleocapsa sp. PCC 73106 (gene id: 2508643885) was found to encode for tRNALys. The red and blue marks indicate G-C and A-U bonding, respectively.
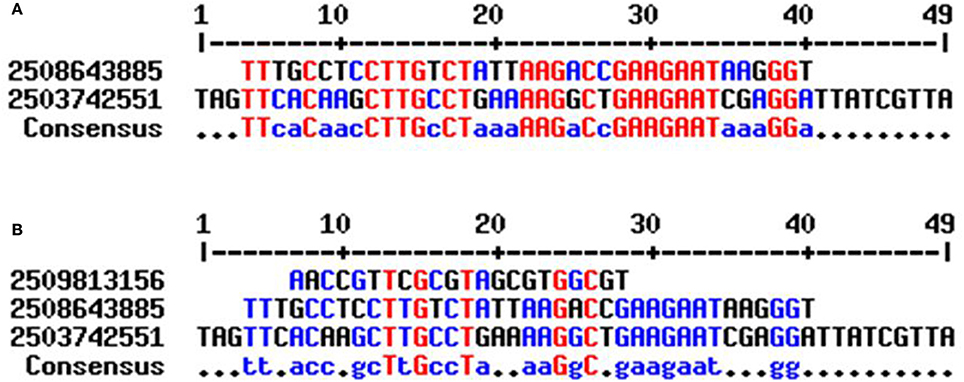
Figure 3. Sequence alignment of cyanobacterial group I intron. (A) Sequence alignment of cyanobacterial tRNA showed, the introns found in Nostoc sp. PCC 7107 (gene id: 2503742551) and Gleocapsa sp. PCC 73106 (gene id: 2508643885) share conserved consensus sequences (B) whereas the intron of Nostoc sp. PCC 7524 (gene id: 2509813156) did not share any conserved sequences with other introns. This showed that there are two different groups of cyanobacterial group I intron.
The genes of cyanobacterial tRNA are found to range from 55 to 96 nucleotides; a tRNA gene possessing only 55 nucleotides is the smallest gene reported thus far. Studying the substitution pattern of mutation is important for understanding the molecular basis of evolution. Therefore, we examined the substitutions (transition/transversion) of cyanobacterial tRNA. The mutation, which changes a purine (A<>G) nucleotide to another purine or a pyrimidine nucleotide to another pyrimidine (T<>C) is called transition, whereas a mutation that changes a purine nucleotide to a pyrimidine and vice versa, is called a transversion. In our study, we found that the rate of transition, in cyanobacterial tRNA, was higher than that of transversion (Table 4). The highest rate of transition was 42.05 in tRNATrp followed by 25.66 in tRNACys, where an adenine was substituted with a guanine (Table 4); the highest rate of transversion was 9.7 in tRNASer, where a uracil and a cytosine were substituted with a guanine (Table 4). The highest rate of transition from a guanine to an adenine was found in tRNATrp (28.89), whereas the highest rate of transition from a uracil to a cytosine was found in tRNATyr (Table 4). The highest rate of transition from a cytosine to a uracil was found in tRNATyr. The lowest rate of transition was found in tRNASer where a guanine was substituted with an adenine. Similarly, the lowest rate of transversion was found in tRNATrp where a uracil and a cytosine were substituted by an adenine (Table 4). The transition/transversion bias of tRNATrp was highest, followed by tRNAAsp and tRNATyr (Table 4).
In addition to substitution (transition/transversion), gene duplication and loss play crucial roles in the evolution of a gene. Duplication study of cyanobacterial tRNA showed that tRNA genes in all 20 tRNAs families, were duplicated by more than 50% (Table 5, Supplementary Figure 1). The highest duplication of 85% was observed in tRNAIle, whereas the lowest duplication was observed in tRNAHis (Table 5). The highest percentage of conditional duplication was found in tRNAAsn (20.75%), whereas the lowest percentage was found in tRNAIle (5.26%), followed by tRNAGln (5.68%) and tRNAAsp (5.84%) (Table 5). Unlike duplication and conditional duplication, the highest percentage of losses with respect to the species tree (Figure 4), was found in tRNAAsp (284.93%) followed by tRNAGlu (280.76%). The low level of loss was found in tRNAArg (221.45%) followed by tRNAAla (226.63%).
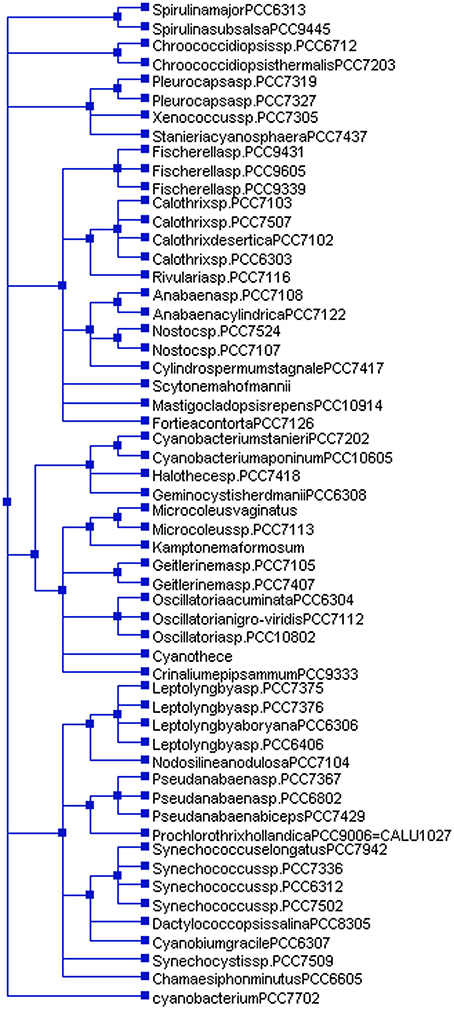
Figure 4. Species tree of cyanobacteria. The species tree was constructed using NCBI taxonomy browser (https://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi). Cyanobacterium sp. PCC 7702 in the species tree falls independently with regard to other cyanobacterial species. This show, cyanobacterial groups show polyphyletic origins.
Understanding the evolution of cyanobacterial tRNA is important for delineating the evolution of tRNA, its evolutionary lineages, and subsequent diversification. In this study, we constructed a phylogenetic tree by examining all the tRNAs of the studied species (Figure 5). The phylogenetic tree was divided into five distinct clusters (Supplementary Figure 1). Cluster I contained tRNAAla (red), tRNAVal (blue), tRNAIle (purple), tRNAArg (teal), tRNALys (lime), tRNAGlu (green), tRNAAsp (maroon), and tRNAGly (green). Cluster II was found to contain tRNAPro (red), tRNAThr (fuchsia), tRNAPhe (blue), tRNAMet (olive), tRNAAsn (navy), and tRNAPhe (blue) (Supplementary Figure 1). Cluster III contained tRNAArg (teal) and tRNATrp (fuchsia) whereas cluster IV had tRNALys (lime), tRNALeu (maroon), tRNAGln (aqua), tRNALeu (maroon), tRNAVal (blue), tRNAGlu (green), tRNAHis (red), tRNAAla (black), tRNALeu (maroon), tRNAGly (green), tRNATrp (fuschia), tRNAGln (aqua), tRNAHis (red), tRNACys (navy), tRNATyr (gray), tRNAMet (olive), and tRNASer (fuschia) (Supplementary Figure 1). Cluster V contained tRNAArg (teal), tRNAMet (olive), tRNAAla (red), tRNAMet (olive), and tRNAGln (aqua). In cluster I, tRNAAla and tRNAVal were present together with tRNAVal and tRNAIle. In cluster I tRNAArg was shared by tRNAAsp, and tRNAGly. Cluster II was shared by tRNAPro, tRNAThr, tRNAPhe, tRNAMet, and tRNAAsn (Supplementary Figure 1). Cluster III was shared by tRNAArg and tRNATrp. Cluster IV contained tRNALys, tRNALeu, tRNAGln, tRNAVal, tRNAHis, tRNAAla, tRNAGly, tRNATrp, tRNAGln, tRNACys, tRNATyr, tRNAMet, and tRNASer. Cluster V contained tRNAAla, tRNAMet, tRNAGln, and tRNAArg. tRNAAla was distributed in clusters I, V, and VI whereas tRNAGln was distributed in clusters IV, V, and VI. tRNAVal, tRNALys, and tRNAGly were distributed in clusters I and IV; tRNAAsn was distributed in cluster II and IV, and tRNATrp was distributed in cluster III and IV (Supplementary Figure 1). Due to low bootstrap values in few clades, it was difficult to infer the phylogenetic result properly. Therefore, we collapsed the clades with low bootstrap values (cut-off values for condensed tree was 50%) (Figure 5, Supplementary Figure 1). The overall tree architecture remained unchanged post-collapsing the clades with 50% cut-off values. Several potentially novel evolutionary trends were found in the phylogenetic analysis of cyanobacterial tRNA. In between tRNAMet and tRNAAsn of cluster II, several other tRNAs were also found. In cluster V, tRNAAsp and tRNAAsn were found in close to tRNAMet, suggesting their possible evolution from tRNAMet. Similarly, a few tRNAsVal (Microcoleus sp. PCC 7113, gene id: 2509433894; Leptolyngbya boryana PCC 6306, gene id: 2509801628) were present close to tRNAAla. This suggests that, tRNAVal has likely evolved from tRNAAla. In cluster V, tRNAGly (Microcoleus sp. PCC 7113, gene id: 2509433891) was clustered with tRNAMet, reflecting it's evolution from tRNAMet.
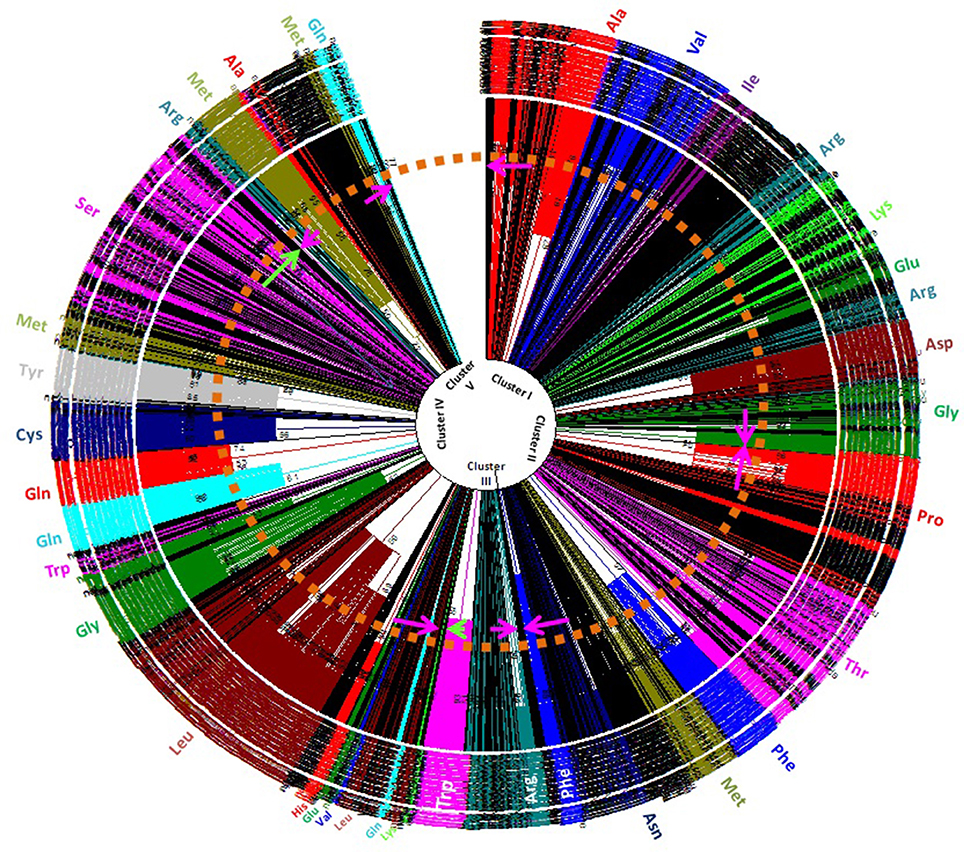
Figure 5. Phylogenetic tree of cyanobacterial tRNAs. Phylogenetic analysis revealed polyphyletic origin of cyanobacterial tRNA. Besides this, it also revealed that tRNAGln, tRNAMet, tRNAAla, and tRNAArg were most probably evolved earlier than others tRNAs. The other tRNAs mostly evolved from these tRNAs with subsequent modification (transition/transversion), duplication and eventual loss of old genes.
Nucleotide sequence conservation is an important phenomenon that demonstrates conserved functional role. Therefore, understanding the conserved nucleotide consensus sequences in cyanobacterial tRNA was very important. In this study we found that except for tRNAAsn, tRNALeu, tRNAGln, and tRNAMet, all the tRNAs were contained conserved G1 nucleotide at the 1st position in the acceptor arm. tRNALeu and tRNAMet did not contain any conserved nucleotides at the 1st position whereas tRNAAsn and tRNAGln contained a conserved U1 nucleotide at the 1st position. With a few exceptions, the G2 nucleotide was conserved at the 2nd position in tRNAAla, tRNAVal, tRNAIle, tRNAPhe, tRNATyr, tRNASer, tRNAGln, tRNALys, tRNAHis, tRNAArg, and tRNAAsp. The nucleotide C2 was conserved in tRNAGly and tRNAThr whereas tRNAHis contained either a C2 or a U2 nucleotide. At the 3rd position, the G3 nucleotide was conserved in tRNAGly, tRNAAla, tRNAPro, tRNAIle, tRNATyr, tRNALys, and tRNAAsp, whereas tRNAPhe contained a C3; tRNAHis contained C3/U3; whereas tRNASer contained an A3 nucleotide. At the 4th position of the acceptor arm, tRNAVal and tRNAIle contained the conserved C4 nucleotide whereas tRNATyr contained a U4 nucleotide and tRNAPro contained the G4 nucleotide. In tRNA, the C5 nucleotide was conserved, whereas tRNAPhe and tRNAGln contained the conserved G6 nucleotide. In all canonical tRNA, 1st, 2nd, and 3rd base of the acceptor arm pairs with the 72nd, 71st, and 70th base, respectively of the acceptor arm. Their conserved structure is highly important to have proper functional clover leaf-like structure of tRNA. At the 7th position, tRNAPro and tRNAHis contained conserved G7 nucleotide. In tRNATyr, all the nucleotides in the positions 1–5 were conserved across the species. In the 1st and 2nd positions of the acceptor arm of tRNA, the G1-G2 conserved consensus sequence present in tRNAAla, tRNAVal, tRNAIle, tRNATyr, tRNASer, tRNALys, tRNAArg, and tRNAAsp whereas the G1-C2 conserved consensus sequence present in tRNAGly, tRNAPhe, tRNAThr, and tRNALys. The nucleotide, at the 2nd position of the acceptor arm, was substituted with either G2 or C2, whereas only tRNAHis had a U2 at the 2nd position. Only tRNAAsn and tRNAGln contained a U1 nucleotide at the 1st position of the acceptor arm whereas tRNATyr contained conserved U4 nucleotide at the 4th position and tRNAHis contained the conserved U2 nucleotide at the 2nd position (Table 2). Except the above-mentioned four tRNAs, none of the other tRNAs were found to encode U nucleotide in the acceptor arm. Previously, the 8th position of tRNA between the acceptor arm and the D-arm was thought to contain a conserved U8 nucleotide (Table 3). In this study, the majority of tRNAs were found to contain a conserved U8 nucleotide. However, tRNAHis was found to contain either a U8 or an A8 whereas tRNAGlu contained a U8, C8 or a G8 nucleotide (Table 3). This showed that the 8th position of cyanobacterial tRNA can be substituted with any nucleotide. Similarly, at the 9th position, the majority of tRNAs were found to contain a conserved A9 nucleotide (Table 3). However, tRNALeu, tRNATyr and tRNASer were found to contain a G9 nucleotide whereas tRNAThr, tRNALys, and tRNAArg contained an A9 or a G9 nucleotide (Table 3). tRNAHis contained an A9 or a C9 nucleotide at the 9th position whereas tRNACys and tRNAGln contained either a A9, C9, or a G9 (Table 3). tRNAMet contained either an A9, G9 or a U9 nucleotide whereas tRNAGlu contained an A9, C9, or a U9 nucleotide at the 9th position. This negated the previous assumption about the conservation of the U8 nucleotide in cyanobacterial tRNA and showed that it can be prone to variations; the major variation were observed in tRNAGlu which was found to contain varied nucleotides at 8th and 9th position.
In the D-arm, the majority of tRNA contained a conserved G1 nucleotide at the 1st position except for tRNAGly, tRNAPro, tRNAVal, tRNAMet, tRNAGln, tRNALys, and tRNAGlu. In total of 20 tRNA families, 10 families were found to contain a conserved C2 nucleotide at the 2nd position (Table 2). These were tRNAAla, tRNAVal, tRNAIle, tRNAPhe, tRNASer, tRNAThr, tRNACys, tRNAAsn, tRNALys, and tRNAHis whereas tRNAMet was found to contain a G2 and tRNATrp contained a U2 nucleotide at the 2nd position (Table 2). With respect to the first two nucleotides in the D-arm, the G1-C2 conserved consensus sequence can be found in tRNAAla, tRNAIle, tRNAPhe, tRNASer, tRNAThr, tRNACys, tRNAAsn, and tRNAHis. At the 3rd position, the U3 nucleotide conserved in tRNAAla, tRNAIle, tRNAPhe, tRNAAsn, and tRNAAsp. At the 4th position of the D-arm, the C4 nucleotide was found to be conserved in tRNAAla, tRNAVal, tRNAIle, tRNAPhe, tRNATyr, tRNAThr, and tRNAAsp (Table 2). The D-stem expected to have rigid structure due to high G/C content, but due to the presence of more A-U and G-U base pairing, it seems rather weak (Hardt et al., 1993). Previous study reported that D-arm play important role in recognition of aminoacyl-tRNA synthetase and is highly variable and possess unusual conformation due to over-crowding of G residue (Smith and Yarus, 1989; Hardt et al., 1993). However, in our study we found that D-arm is also highly conserved, suggesting its common functional role.
Except for tRNAMet and tRNAGlu, 18 tRNA families were found to contain a conserved A1 nucleotide at the 1st position in the D-loop. At the 2nd position, the G2 nucleotide was conserved in tRNAPro, tRNAVal, tRNAPhe, and tRNATyr, whereas tRNAThr contained a U2 nucleotide at the 2nd position (Table 2). In the 3rd position, the U3 nucleotide was conserved in tRNAPhe, and tRNATyr and the G3 nucleotide was conserved in tRNACys, tRNAAsn, and tRNAHis. In the 4th position of the D-loop, the G4 nucleotide was conserved in tRNAGly, tRNATyr, tRNAThr, tRNAAsn, tRNAGln, tRNALys, tRNAArg, and tRNAAsp whereas at the same position tRNAPhe contained conserved U4 nucleotide (Table 2). At the 5th position, the G5 nucleotide was found to be conserved in tRNAGly, tRNAAla, tRNALeu, tRNAPhe, tRNATyr, tRNATrp, tRNASer, tRNACys, tRNALys, tRNAHis, tRNAAsp and tRNAGlu. The majority of tRNAs were found to contain a conserved G5 nucleotide whereas tRNAThr contain a U5 nucleotide. At the 6th position, tRNAAla, tRNAVal, tRNALeu, tRNAIle, tRNAMet, tRNAPhe, tRNASer, tRNACys, and tRNAHis contained the G6 nucleotide whereas tRNATyr, and tRNAAsp contained the U6 nucleotide. At the 7th position of the D-loop, the U7 nucleotide was found to be conserved in tRNAPhe, tRNATyr, tRNACys, and tRNAHis whereas the 8th position had a conserved A8 nucleotide in tRNAAla, tRNALeu, tRNATyr, tRNAArg and tRNAAsp. The A9 nucleotide was conserved in tRNAVal, and tRNAIle. This indicates that a great variation was present in the nucleotide composition of the D-loop. However, the A1 nucleotide was conserved in the majority of the cases, whereas conservation of nucleotides in the other parts of the D-loop was specific to an individual tRNA family. The D-loop interacts with the Ψ-loop via long range interactions where G18 and G19 interact with Ψ55 and C56, respectively (Hanawa-Suetsugu et al., 2001). Although the position is dynamic, except for tRNAGln, all tRNAs were contained at least one conserved G nucleotide in the D-loop, suggesting their common functional role.
The anti-codon arm of tRNA was less conserved than other parts. In tRNAPhe, tRNATrp and tRNAHis, the G4 nucleotide was found to be conserved, whereas tRNATrp had a conserved U5 nucleotide and in tRNAHis, the G5 nucleotide was conserved. The anti-codon loop, which reads the codon of an mRNA during protein translation, is the most important part of tRNA. The C1 nucleotide was found to be conserved at the 1st position of the anti-codon loop in tRNAIle, tRNAMet, tRNATrp, tRNACys, tRNAAsn, tRNALys, and tRNAAsp. Additionally, the U1 nucleotide at the 1st position of the anti-codon loop, was found to be conserved in tRNAPro, tRNAVal, tRNAGln, tRNAHis, and tRNAArg (Table 2). Occasionally, tRNAArg contained a C1 nucleotide in the anti-codon loop. All the tRNAs were found to contain a conserved U2 nucleotide at the 2nd position of the anti-codon loop. The A6 nucleotide, at the 6th position of the anti-codon loop, was conserved in tRNAGly, tRNAAla, tRNAVal, tRNAIle, tRNAMet, tRNATrp, tRNASer, tRNAThr, tRNACys, tRNAAsn, tRNALys, and tRNAAsp (Table 2). In the 7th position of the anti-codon loop, the A7 nucleotide was conserved in tRNAIle, tRNAMet, tRNATrp, tRNAThr, tRNACys, tRNAAsn, and tRNALys. The presence of a conserved A6 nucleotide at the 6th position suggests the possible entry site for group I intron in the anti-codon loop.
Previous study reported that the variable loop of tRNA is dynamic in nature and does not contain any conserved nucleotides. However, in our study we found that the variable loop contained conserved nucleotides at the specific positions at the level of individual tRNA family (Table 2). tRNAVal and tRNAPhe were found to contain the conserved G3-U4-C5 nucleotides at the 3rd, 4th, and 5th positions of the variable loop, whereas tRNAPro contained the conserved G3-x-C5 nucleotides at 3rd and 5th position (Table 2). In tRNAIle and tRNAAsn, the G3-U4 nucleotides were found to be conserved at 3rd and 4th positions. At the 5th position, the C5 nucleotide was conserved in tRNAPro, tRNAVal, tRNAPhe, and tRNACys (Table 2). This study indicates that the G3, U4, and C5 nucleotides at 3rd, 4th, and 5th position, respectively, were the most likely to be conserved in the variable loop of the cyanobacterial tRNA.
All the tRNAs were found to contain a conserved G5 nucleotide in the Ψ-arm. Several tRNAs contained a conserved G4 nucleotide at the 4th position whereas others was found to contain conserved G3 nucleotide. The conserved G nucleotide at the 4th and 5th position was detected as the G4-G5 configuration in tRNAPro, tRNAVal, tRNAIle, tRNATyr, tRNASer, tRNAGln, tRNALys, tRNAHis, and tRNAArg.
The Ψ-loop of all the tRNA contained the conserved U1-U2-C3-x-A5 nucleotides at the 1st, 2nd, 3rd, and 5th positions, respectively. Few other tRNAs were found to contain the extended conserved U1-U2-C3-x-A5-x-U7 sequence from the 1st to the 7th position of the Ψ-loop (Table 2). The tRNA containing the conserved U1-U2-C3-x-A5-x-U7 nucleotides were tRNALeu, tRNAIle, tRNAMet, tRNATyr, tRNATrp, tRNASer, tRNACys, tRNAAsn, tRNALys, tRNAHis, tRNAArg, and tRNAGlu (Table 2). tRNAPro was found to contain the conserved U1-U2-C3-x-A5-A6-U7 consensus sequence, where the A6 nucleotide was found to be conserved at the 6th position. In tRNAAsp, it was found to contain conserved U1-U2-C3-x-A5-x-C7 consensus sequence in which the U7 nucleotide substituted with a C7 nucleotide (Table 2). These analyses show that tRNAs were conserved in the acceptor arm, D-arm, D-loop, anti-codon arm, anti-codon loop, the variable loop and the Ψ-arm at the individual family level with majority of tRNAs showing greater conservation in the Ψ-loop.
In spite of the presence of highly conserved genetic architecture of cyaobacterial tRNA, they also found to possess introns within it. Previous studies reported that an intron was present only in tRNALeu (UAA) and tRNAfMet (UAC) of cyanobacterial tRNA (Paquin et al., 1997; Rudi and Jakobsen, 1999). However in our study, we found that the cyanobacterial group I intron was also present in tRNAArg, tRNAGly, and tRNALys (Figure 3). Because cyanobacteria are a prokaryotic organism, the intron, present in cyanobacterial tRNA can be compared with the group I intron in the plastid. Previous studies reported that the group I intron, found in the plastid, are immobile (Rudi and Jakobsen, 1999). The group I introns found in various other locations including the mitochondria, nucleus, bacteria, and bacteriophages share conserved sequence motifs and have evolved via lateral gene transfer (Rudi and Jakobsen, 1999). Cyanobacterial group I introns were also possessed conserved motifs. Rudi and Jakobsen (1999) have classified the group I intron as intron element P, Q, R, and S. Cluster analysis revealed that tRNAArg of the Nostoc sp. PCC 7524 were grouped in intron element P, whereas the introns of the Nostoc sp. PCC 7107 and Gloeocapsa sp. PCC 73106 were clustered independently. None of these were clustered with intron elements Q, R, or S. This suggests that there may be another conserved intron element in the cyanobacteria which is yet to be elucidated. The presence of an intron, in the anti-codon loop of cyanobacterial tRNA agrees with the exon theory of the evolution of tRNA genes where modern tRNA is reported to have evolved by duplication of one half of tRNA (Di Giulio, 1992). Report suggests that the introns of cyanobacterial tRNA moved via lateral gene transfer (Marck and Grosjean, 2002).
Self-splicing group I intron of cyanobacterial tRNAfMet in genera Dermocarpa, Scytonema, and Synechocystis possess open reading frame (ORF). The intron of Synechocystis possesses ORF of 150 codons. The introns in cyanobacteria distributed sporadically without any correlation between relatedness in intron sequence corroborates with the lateral transfer of intron (Dujon, 1989; Biniszkiewicz et al., 1994). Sporadic occurrence of these group I introns in homologous genes of closely related species show consistent similarity in the mobility. The group I introns are suggested to be either mobile and ancient in origin (Rudi and Jakobsen, 1999). Closely related group I intron are inserted in the UAA anit-codon of tRNALeu of cyanobacteria and chloroplast (Biniszkiewicz et al., 1994). Study of introns from 22 cyanobacterial strains by Rudi and Jakobsen (1999) demonstrated relatively recent gain and/or loss of intron in some cyanobacterial lineage (Rudi and Jakobsen, 1999). The presence of self-splicing group I intron in tRNALeu at the same position of the same gene in cyanobacteria and chloroplast indicated that this intron predates the invasion of eukaryotic cells by endosymbiosis (Reinhold-Hurek and Shub, 1992). The group I intron of tRNALeu have higher homology with the group I intron of tRNAIle (CAU) and tRNAArg (CCU) (Rudi and Jakobsen, 1997, 1999).
Evolution of a gene occurred through random mutation/substitution. The pattern and frequencies of nucleotide substitution is largely depends on the mutational events of the gene (Shimizu et al., 1989; Zhang and Gerstein, 2003; Arnheim and Calabrese, 2009). Although transitions are more frequent in coding genes because they are less likely to result in amino acid substitutions due to a wobble in the genetic code, the presence of a higher rate of transition in cyanobacterial tRNA is intriguing. A universal bias may favor transition over transversion and a diverse rate of transitions/transversions, combined with the bias may help to explain the multiple evolutionary rates and lineages. The rates of transition/transversion bias of tRNATrp, tRNAAsp, and tRNATyr were 8.409, 4.131, and 3.658, respectively; these were quite higher than the rates of other tRNAs (Table 6). At a low level of genetic divergence, the transition/transversions bias remains high, whereas at a high level of genetic divergence, the transition/transversion bias remains low (Yang and Yoder, 1999). This indicates that tRNATrp, tRNAAsp, and tRNATyr have a very low level of genetic divergence, whereas other tRNAs have a very high rate of genetic divergence. Therefore, cyanobacterial species have only a fewer number of tRNATrp, tRNAAsp, and tRNATyr genes in their genome compared to other tRNAs and rest of the tRNAs have a very high rate of divergence. Due to high genetic divergence, the frequency of other tRNAs are more abundant than tRNATrp, tRNAAsp, and tRNATyr. Silent mutation can have significant impact on exhibiting genetic divergence which accompany through selection/genetic drift or novel adaptation (Palumbi, 1994).
Understanding the process of evolution and function of new gene is important in genomics and evolutionary biology; gene duplication is a powerful process in generating novel gene function via sub-functionalization and neo-functionalization, whereas loss of a gene can greatly shape the gene family (Lynch and Conery, 2000; Niimura and Nei, 2007; Rasmussen and Kellis, 2012; Teufel et al., 2016). Gene duplication is a fundamental process in the evolution of novel species, especially in eukaryotes in which it plays major roles in creating novel gene functions (Cotton and Page, 2005; Blomme et al., 2006; Zhao et al., 2015). Similarly, the loss of a gene occurs via segmental deletion, and pseudogenization preserves a minimum number of functional gene copies (Ohno, 1970; Lynch and Conery, 2000; Ting et al., 2004; Cotton and Page, 2005; Blomme et al., 2006; Demuth et al., 2006). Due to the lowest percentage of duplication, the genome of the cyanobacteria encodes only one to two tRNAHis genes per genome, whereas the number of tRNAIle genes is more numerous due to highest percentage of duplication (Table 5). The low level of duplication and conditional duplication may responsible for the low frequency in the occurrence of tRNAHis, tRNAAsn, and tRNAAsp in the cyanobacterial genome. Coupled with the low percentage of duplication, conditional duplication and the high percentage of gene loss led to the low frequency of tRNAHis, tRNAAsn, tRNAGln, and tRNAAsp genes in the cyanobacterial genome. Similarly, low percentage of gene loss, coupled with the high percentage of gene duplication and conditional duplication in tRNAArg and tRNAAla, resulted multiple and variable copies of tRNAArg and tRNAAla in the cyanobacterial genome (Table 5). Previous studies showed that tRNAs are the classic example of gene families that were continuously duplicated and lost (Withers et al., 2006; Tang et al., 2009; Bermudez-Santana et al., 2010; Rogers et al., 2010). The operon or tRNA clusters are highly dynamic and unstable genomic region. Therefore, it became an easy target for duplication or loss event. However, in cyanobacteria, the loss event was predominant over the duplication and conditional duplication.
Numerous studies have been conducted on the evolution of tRNA. Reports suggest that tRNA hairpin structures were the precursors of tRNA, having evolved from the combination of different sub-structures, where the top half (acceptor arm) of the tRNA is considered older than the bottom half containing the anti-codon arm (Sun and Caetano-Anollés, 2008). According to the “genomic tag” hypothesis, the upper half of the tRNA harbors the ancestral genomic information and the bottom half provides the specificity for the genetic code (Weiner and Maizels, 1987). The hairpin structure having the anti-codon loop may have evolved as an intermediary in the process of protein synthesis. Introns in tRNA are usually present in the anti-codon loop, and the 5′ and 3′ halves of the tRNA were likely the mini-genes before their merger (Di Giulio, 1992, 2004). The presence of an intron in tRNA suggests that the two modules of tRNA were separated by introns; hence, tRNA is the exon part of the gene described by the “exonic gene theory” of the origin of tRNA (Di Giulio, 1995). The tRNA molecule is universally present in all cellular organisms and is transferred by “horizontal gene transfer.” It's ubiquitous presence in all cellular lineages strongly supports its early origin, explaining its presence in organisms of early origin. Fossil record suggests that cyanobacteria are older than 2 billion years and indirect evidence suggests that they may be older than 2.45 billion years (Amard and Bertrand-Sarfati, 1997; Bekker et al., 2004; Schirrmeister et al., 2011; Mohanta et al., 2017). The presence of tRNAArg and tRNAMet in clusters I, III, V, and VI in the phylogenetic tree reflected their ancient origin (Figure 5); hence, their co-distribution in different clusters may have been followed by subsequent evolution of tRNAGln and tRNAAla. The presence of tRNAArg and tRNAMet in multiple clusters vividly illustrates that they have undergone duplication and subsequently diverged into different species. The distribution of tRNAIle, tRNAGlu, tRNAAsp, tRNApro, tRNAThr, tRNAGly, tRNATrp, tRNAHis, tRNACys, tRNATyr, and tRNASer was confined to a single cluster only (Figure 5). This suggests that they have evolved recently, compared with tRNAMet, tRNAArg, tRNAAla, and tRNAGln (Figure 5). In cluster I, tRNAArg grouped twice and present close to tRNAAsp and tRNAGly, suggesting their evolution from tRNAArg (Figure 5). In cluster I, tRNAAla was present close to tRNAVal; this suggests that tRNAVal evolved from tRNAAla. In cluster II, tRNApro was grouped separately whereas tRNAThr, tRNAPhe, and tRNAAsn were grouped with tRNAMet, suggesting their evolution from tRNAAla (Figure 5). Cluster III contained tRNAArg and tRNATrp, suggesting possible evolution of tRNATrp from tRNAArg. In cluster IV, tRNAGln present close to tRNAHis, and tRNAMet present closer to tRNATyr and tRNACys. This suggests that tRNAHis may have evolved from tRNAGln whereas tRNATyr and tRNACys may have evolved from tRNAMet. tRNASer found in cluster IV shared a branch with tRNAMet, suggesting the evolution of tRNASer from tRNAMet. tRNAGln, tRNAMet, and tRNAAla were found together in cluster V, suggesting that these tRNAs may have coevolved together, and later diversified and evolved into other tRNAs. These tRNAs were grouped with several pseudo-tRNAs in the cluster of tRNAAsn (Figure 5, Supplementary Figure 1). This suggests that these tRNAs are likely in the verge of becoming pseudo-tRNAs and will be lost from the evolutionary lineage. tRNAPhe was present as sister branch to tRNAAsn in cluster III, suggesting their evolution from tRNAAsn. Several tRNAsLys, tRNAsHis, and tRNAsAla in cluster IV were grouped with the pseudo-tRNAs, suggesting their possible loss of function and, hence, extinction from the evolutionary lineage by pseudogenization. However, the evolutionary history of cyanobacterial tRNAs may be more complicated. The presence of common secondary and tertiary structures occurring with an invariant number of nucleotide residues, 20 invariant gene families, and similar functional roles during the translation is the phenomena that are complicated to delineate. It is assumed that for a total of 20 tRNA gene families, there would be 20 branches, corresponding to a group in each tRNA gene family, in the phylogenetic tree. However, this assumption is incorrect. The differences between tRNA families, with regard to their respective amino acid and tRNA specificity were found overlap significantly. The non-isoacceptor tRNAs shared the phylogenetic groups with the isoacceptors, suggesting their evolution from multiple lineages. This shows that tRNAs had evolved by multiple duplication with extensive genetic mutation and subsequent loss of old genes.
Sequence analysis revealed that cyanobacterial tRNAs were dynamic and transiently conserved within their respective tRNA families. However, the U1-U2-C3 conserved consensus sequence in the Ψ-loop was found in all of the studied tRNAs. Several cyanobacterial tRNAs contained a group I intron in the anti-codon region; further study is required to discover more introns and to understand their specific genomic and evolutionary aspects. The rate of mutational transition of cyanobacterial tRNAs was higher than that of transversion; cyanobacterial tRNA may have evolved polyphyletically by loss of tRNA genes.
TM: Conceived the idea, performed the study, analyzed data, drafted and revised the manuscript. AS: Revised the manuscript. FA: Revised the manuscript. HB: Revised the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was carried out with the support of the Next-Generation Biogreen 21 Program (PJ011113), Rural Development Administration, Korea. Authors also like to extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through research group No (RG-1438-078).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2017.00200/full#supplementary-material
Supplementary Figure 1. Phylogenetic tree of cyanobacterial tRNAs. The phylogenetic clades with low bootstrap replicates were collapsed with 50% cutoff values.
Supplementary data 1. Supplementary data showing genomic tRNA sequences of 61 cyanobacterial species.
Akaike, H. (1998). “Information theory and an extension of the maximum likelihood principle,” in Selected Papers of Hirotugu Akaike, eds E. Parzen, K. Tanabe, and G. Kitagawa (New York, NY: Springer), 199–213.
Altermann, W., Kazmierczak, J., Oren, A., and Wright, T. (2006). Cyanobacterial calcification and its rock-building potential during 3.5 billion years of Earth history. Geobiology 4, 147–166. doi: 10.1111/j.1472-4669.2006.00076.x
Amard, B., and Bertrand-Sarfati, J. (1997). Microfossils in 2000 Ma old cherty stromatolites of the Franceville Group, Gabon. Precambrian Res. 81, 197–221. doi: 10.1016/S0301-9268(96)00035-6
Arnheim, N., and Calabrese, P. (2009). Understanding what determines the frequency and pattern of human germline mutations. Nat. Rev. Genet. 10, 478–488. doi: 10.1038/nrg2529
Bekker, A., Holland, H. D., Wang, P. L., Rumble, D., Stein, H. J., Hannah, J. L., et al. (2004). Dating the rise of atmospheric oxygen. Nature 427, 117–120. doi: 10.1038/nature02260
Bermudez-Santana, C., Attolini, C. S., Kirsten, T., Engelhardt, J., Prohaska, S. J., Steigele, S., et al. (2010). Genomic organization of eukaryotic tRNAs. BMC Genomics 11:270. doi: 10.1186/1471-2164-11-270
Biniszkiewicz, D., Cesnaviciene, E., and Shub, D. A. (1994). Self-splicing group I intron in cyanobacterial initiator methionine tRNA: evidence for lateral transfer of introns in bacteria. EMBO J. 13, 4629–4635.
Blomme, T., Vandepoele, K., De Bodt, S., Simillion, C., Maere, S., and Van de Peer, Y. (2006). The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 7:R43. doi: 10.1186/gb-2006-7-5-r43
Chan, P. P., Cozen, A. E., and Lowe, T. M. (2011). Discovery of permuted and recently split transfer RNAs in Archaea. Genome Biol. 12, R38–R38. doi: 10.1186/gb-2011-12-4-r38
Chen, K., Durand, D., and Farach-Colton, M. (2000). NOTUNG: a program for dating gene duplications and optimizing gene family trees. J. Comput. Biol. 7, 429–447. doi: 10.1089/106652700750050871
Cotton, J. A., and Page, R. D. (2005). Rates and patterns of gene duplication and loss in the human genome. Proc. R. Soc. B. 272, 277–283. doi: 10.1098/rspb.2004.2969
Demuth, J. P., De Bie, T., Stajich, J. E., Cristianini, N., and Hahn, M. W. (2006). The evolution of mammalian gene families. PLoS ONE 1:e85. doi: 10.1371/journal.pone.0000085
Di Giulio, M. (1992). On the origin of the transfer RNA molecule. J. Theor. Biol. 159, 199–214. doi: 10.1016/S0022-5193(05)80702-7
Di Giulio, M. (1995). Was it an ancient gene codifying for a hairpin RNA that, by means of direct duplication, gave rise to the primitive tRNA molecule? J. Theor. Biol. 177, 95–101. doi: 10.1016/S0022-5193(05)80007-4
Di Giulio, M. (2004). The origin of the tRNA molecule: implications for the origin of protein synthesis. J. Theor. Biol. 226, 89–93. doi: 10.1016/j.jtbi.2003.07.001
Di Giulio, M. (2008a). The split genes of Nanoarchaeum equitans are an ancestral character. Gene 421, 20–26. doi: 10.1016/j.gene.2008.06.010
Di Giulio, M. (2008b). Transfer RNA genes in pieces are an ancestral character. EMBO Rep. 9, 820. doi: 10.1038/embor.2008.153
Dujon, B. (1989). Group I introns as mobile genetic elements: facts and mechanistic speculations—a review. Gene 82, 91–114. doi: 10.1016/0378-1119(89)90034-6
Fujishima, K., Sugahara, J., Kikuta, K., Hirano, R., Sato, A., Tomita, M., et al. (2009). Tri-split tRNA is a transfer RNA made from 3 transcripts that provides insight into the evolution of fragmented tRNAs in archaea. Proc. Natl. Acad. Sci. U.S.A. 106, 2683–2687. doi: 10.1073/pnas.0808246106
Hanawa-Suetsugu, K., Bordeau, V., Himeno, H., Muto, A., and Felden, B. (2001). Importance of the conserved nucleotides around the tRNA-like structure of Escherichia coli transfer-messenger RNA for protein tagging. Nucleic Acids Res. 29, 4663–4673. doi: 10.1093/nar/29.22.4663
Hardt, W. D., Schlegl, J., Erdmann, V. A., and Hartmann, R. K. (1993). Role of the D arm and the anticodon arm in tRNA recognition by eubacterial and eukaryotic RNase P enzymes. Biochemistry 32, 13046–13053. doi: 10.1021/bi00211a014
Holley, R. W., Apgar, J., Everett, G. A., Madison, J. T., Marquisee, M., Merrill, S. H., et al. (1965). Structure of a ribonucleic acid. Science (80-.). 147, 1462–1465. doi: 10.1126/science.147.3664.1462
Kanai, A. (2015). Disrupted tRNA genes and tRNA fragments: a perspective on tRNA gene evolution. Life 5, 321–331. doi: 10.3390/life5010321
Kirchner, S., and Ignatova, Z. (2014). Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 16, 98–112. doi: 10.1038/nrg3861
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.0955
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science (80-.). 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Marck, C., and Grosjean, H. (2002). tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 8, 1189–1232. doi: 10.1017/S1355838202022021
Maruyama, S., Sugahara, J., Kanai, A., and Nozaki, H. (2010). Permuted tRNA genes in the nuclear and nucleomorph genomes of photosynthetic eukaryotes. Mol. Biol. Evol. 27, 1070–1076. doi: 10.1093/molbev/msp313
Mohanta, T. K., Arora, P. K., Mohanta, N., Parida, P., and Bae, H. (2015a). Identification of new members of the MAPK gene family in plants shows diverse conserved domains and novel activation loop variants. BMC Genomics 16:58. doi: 10.1186/s12864-015-1244-7
Mohanta, T. K., and Bae, H. (2017). Analyses of genomic tRNA reveal presence of novel tRNAs in Oryza sativa. Front. Genet. 8:90. doi: 10.3389/fgene.2017.00090
Mohanta, T. K., Mohanta, N., Mohanta, Y. K., and Bae, H. (2015b). Genome-wide identification of calcium dependent protein kinase gene family in plant lineage shows presence of novel D-x-D and D-E-L motifs in EF-hand domain. Front. Plant Sci. 6:1146. doi: 10.3389/fpls.2015.01146
Mohanta, T. K., Mohanta, N., Mohanta, Y. K., Parida, P., and Bae, H. (2015c). Genome-wide identification of calcineurin B-like (CBL) gene family of plants reveals novel conserved motifs and evolutionary aspects in calcium signaling events. BMC Plant Biol. 15:189. doi: 10.1186/s12870-015-0543-0
Mohanta, T. K., Mohanta, N., Parida, P., Panda, S. K., Ponpandian, L. N., and Bae, H. (2016). Genome-wide identification of mitogen-activated protein kinase gene family across fungal lineage shows presence of novel and diverse activation loop motifs. PLoS ONE 11:e0149861. doi: 10.1371/journal.pone.0149861
Mohanta, T. K., Pudake, R. N., and Bae, H. (2017). Genome-wide identification of major protein families of cyanobacteria and genomic insight into the circadian rhythm. Eur. J. Phycol. 52, 1–17. doi: 10.1080/09670262.2016.1251619
Navarre, W. W., and Schneewind, O. (1999). Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63, 174–229.
Niimura, Y., and Nei, M. (2007). Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS ONE 2:e708. doi: 10.1371/journal.pone.0000708
Nirenberg, M., and Leder, P. (1964). RNA codewords and protein synthesis. Science (80-.). 145, 1399–1407.
Palumbi, S. (1994). Genetic divergence, reproductive isolation, speciation. Annu. Rev. Ecol. Syst. 25, 547–572. doi: 10.1146/annurev.es.25.110194.002555
Paquin, B., Kathe, S. D., Nierzwicki-bauer, S. A., and Shub, D. A. (1997). Origin and evolution of group I introns in cyanobacterial tRNA genes. Origin and Evolution of Group I Introns in Cyanobacterial tRNA Genes. J. Bacteriol. 179, 6798–6806. doi: 10.1128/jb.179.21.6798-6806.1997
Randau, L., Münch, R., Hohn, M. J., Jahn, D., and Söll, D. (2005). Nanoarchaeum equitans creates functional tRNAs from separate genes for their 5′- and 3′-halves. Nature 433, 537–541. doi: 10.1038/nature03233
Rasmussen, M. D., and Kellis, M. (2012). Unified modeling of gene duplication, loss, and coalescence using a locus tree. Genome Res. 22, 755–765. doi: 10.1101/gr.123901.111
Reinhold-Hurek, B., and Shub, D. (1992). Self-splicing introns in tRNA genes of widely divergent bacteria. Nature 357, 173–176. doi: 10.1038/357173a0
Rogers, H. H., Bergman, C. M., and Griffiths-Jones, S. (2010). The evolution of tRNA genes in Drosophila. Genome Biol. Evol. 2, 467–477. doi: 10.1093/gbe/evq034
Roy, H., and Ibba, M. (2008). RNA-dependent lipid remodeling by bacterial multiple peptide resistance factors. Proc. Natl. Acad. Sci. U.S.A. 105, 4667–4672. doi: 10.1073/pnas.0800006105
Rudi, K., and Jakobsen, K. S. (1997). Cyanobacterial tRNALeu(UAA) group I introns have polyphyletic origin. FEMS Microbiol. Lett. 156, 293–298. doi: 10.1111/j.1574-6968.1997.tb12743.x
Rudi, K., and Jakobsen, K. S. (1999). Complex evolutionary patterns of tRNA Leu(UAA) group I introns in the cyanobacterial radiation. J. Bacteriol. 181, 3445–3451.
Schirrmeister, B. E., Anisimova, M., Antonelli, A., and Bagheri, H. C. (2011). Evolution of cyanobacterial morphotypes: taxa required for improved phylogenomic approaches. Commun. Integr. Biol. 4, 424–427. doi: 10.4161/cib.16183
Schopf, J. W., and Packer, B. M. (1987). Early archean (3.3-billion to 3.5-billion-year-old) microfossils from Warrawoona Group, Australia. Science (80-.). 237, 70–73.
Shi, J. P., Martinis, S. A., and Schimmel, P. (1992). RNA tetraloops as minimalist substrates for aminoacylation. Biochemistry 31, 4931–4936. doi: 10.1021/bi00136a002
Shi, P. Y., Weiner, A. M., and Maizels, N. (1998). A top-half tDNA minihelix is a good substrate for the eubacterial CCA-adding enzyme. RNA 4, 276–284.
Shimizu, N., Okamoto, T., Moriyama, E. N., Takeuchi, Y., Gojobori, T., and Hoshino, H. (1989). Patterns of nucleotide substitutions and implications for the immunological diversity of human immunodeficiency virus. FEBS Lett. 250, 591–595. doi: 10.1016/0014-5793(89)80802-6
Smith, D., and Yarus, M. (1989). Transfer RNA structure and coding specificity. J. Mol. Biol. 206, 489–501. doi: 10.1016/0022-2836(89)90496-8
Soma, A. (2014). Circularly permuted tRNA genes: their expression and implications for their physiological relevance and development. Front. Genet. 5:63. doi: 10.3389/fgene.2014.00063
Sugahara, J., Fujishima, K., Morita, K., Tomita, M., and Kanai, A. (2009). Disrupted tRNA gene diversity and possible evolutionary scenarios. J. Mol. Evol. 69, 497. doi: 10.1007/s00239-009-9294-6
Sun, F. J., and Caetano-Anollés, G. (2008). The origin and evolution of tRNA inferred from phylogenetic analysis of structure. J. Mol. Evol. 66, 21–35. doi: 10.1007/s00239-007-9050-8
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tang, D. T., Glazov, E. A., McWilliam, S. M., Barris, W. C., and Dalrymple, B. P. (2009). Analysis of the complement and molecular evolution of tRNA genes in cow. BMC Genomics 10:188. doi: 10.1186/1471-2164-10-188
Teufel, A. I., Liu, L., and Liberles, D. A. (2016). Models for gene duplication when dosage balance works as a transition state to subsequent neo- or sub-functionalization. BMC Evol. Biol. 16:45. doi: 10.1186/s12862-016-0616-1
Ting, C. T., Tsaur, S. C., Sun, S., Browne, W. E., Chen, Y. C., Patel, N. H., et al. (2004). Gene duplication and speciation in Drosophila: evidence from the Odysseus locus. Proc. Natl. Acad. Sci. U.S.A. 101, 12232–12235. doi: 10.1073/pnas.0401975101
Tocchini-Valentini, G. D., Fruscoloni, P., and Tocchini-Valentini, G. P. (2009). Processing of multiple-intron-containing pretRNA. Proc. Natl. Acad. Sci. U.S.A. 106, 20246–20251. doi: 10.1073/pnas.0911658106
Varin, C., Reid, N., and Firth, D. (2011). An overview of composite likelihood methods. Stat. Sin. 21, 5–42.
Vernot, B., Stolzer, M., Goldman, A., and Durand, D. (2008). Reconciliation with non-binary species trees. J. Comput. Biol. 15, 981–1006. doi: 10.1089/cmb.2008.0092
Weiner, A. M., and Maizels, N. (1987). tRNA-like structures tag the 3′ ends of genomic RNA molecules for replication: implications for the origin of protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 84, 7383–7387. doi: 10.1073/pnas.84.21.7383
Weiner, A. M., and Maizels, N. (1999). The genomic tag hypothesis: modern viruses as molecular fossils of ancient strategies for genomic replication, and clues regarding the origin of protein synthesis. Biol. Bull. 196, 327–330. doi: 10.2307/1542962
Wilusz, J. E. (2015). Controlling translation via modulation of tRNA levels. Wiley Interdiscip. Rev. RNA 6, 453–470. doi: 10.1002/wrna.1287
Withers, M., Wernisch, L., and dos Reis, M. (2006). Archaeology and evolution of transfer RNA genes in the Escherichia coli genome. RNA 12, 933–942. doi: 10.1261/rna.2272306
Xu, X., and Reid, N. (2011). On the robustness of maximum composite likelihood estimate. J. Stat. Plan. Inference 141, 3047–3054. doi: 10.1016/j.jspi.2011.03.026
Yang, Z., and Yoder, A. D. (1999). Estimation of the transition/transversion rate bias and species sampling. J. Mol. Evol. 48, 274–283. doi: 10.1007/PL00006470
Zhang, Z., and Gerstein, M. (2003). Patterns of nucleotide substitution, insertion and deletion in the human genome inferred from pseudogenes. Nucleic Acids Res. 31, 5338–5348. doi: 10.1093/nar/gkg745
Keywords: cyanobacteria, tRNA, evolution, intron, transition, transversion
Citation: Mohanta TK, Syed AS, Ameen F and Bae H (2017) Novel Genomic and Evolutionary Perspective of Cyanobacterial tRNAs. Front. Genet. 8:200. doi: 10.3389/fgene.2017.00200
Received: 12 June 2017; Accepted: 21 November 2017;
Published: 13 December 2017.
Edited by:
Ashok Sharma, Augusta University, United StatesReviewed by:
Jian Lu, National Institutes of Health (NIH), United StatesCopyright © 2017 Mohanta, Syed, Ameen and Bae. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tapan K. Mohanta, bm9zdG9jLnRhcGFuQGdtYWlsLmNvbQ==
Hanhong Bae, aGFuaG9uZ2JhZUB5bnUuYWMua3I=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.