- Department of Microbiology and Molecular Medicine, Centre Médical Universitaire (CMU), Faculty of Medicine, University of Geneva, Geneva, Switzerland
The NOT genes encode subunits of the conserved Ccr4-Not complex, a global regulator of gene expression, and in particular of mRNA metabolism. They were originally identified in a selection for increased resistance to histidine starvation in the yeast S. cerevisiae. Recent work indicated that the Not5 subunit, ortholog of mammalian CNOT3, determines global translation levels by defining binding of the Ccr4-Not scaffold protein Not1 to ribosomal mRNAs during transcription. This is needed for optimal translation of ribosomal proteins. In this work we searched for mutations in budding yeast that were resistant to histidine starvation using the same selection that originally led to the isolation of the NOT genes. We thereby isolated mutations in ribosome-related genes. This common phenotype of ribosome mutants and not mutants is in good agreement with the positive role of the Not proteins for translation. In this regard, it is interesting that frequent mutations in RPL5 and RPL10 or in CNOT3 have been observed to accumulate in adult T-cell acute lymphoblastic leukemia (T-ALL). This suggests that in metazoans a common function implicating ribosome subunits and CNOT3 plays a role in the development of cancer. In this perspective we suggest that the Ccr4-Not complex, according to translation levels and fidelity, could itself be involved in the regulation of amino acid biosynthesis levels. We discuss how this could explain why mutations have been identified in many cancers.
Introduction
Cells use highly coordinated cascades of regulatory mechanisms to precisely define the production of specific gene products, which in turn determine development and differentiation, allow the cell to respond to the stressful environment or to adapt to new food sources. Sophisticated programs of gene expression integrate activities from multiple factors acting at different steps along the gene expression pathway. Each and every step of this pathway can be modulated, and many factors or protein complexes act at single steps, and in less frequent cases act at a couple of different steps. However, only one factor has been connected to most steps of the gene expression pathway, and this is the Ccr4-Not complex, which has recently been reviewed quite extensively (Collart and Timmers, 2004; Collart and Panasenko, 2012; Doidge et al., 2012; Miller and Reese, 2012; Collart, 2013; Collart et al., 2013; Panepinto et al., 2013; Reese, 2013; Wahle and Winkler, 2013; Winkler and Balacco, 2013; Chapat and Corbo, 2014; Inada and Makino, 2014; Panasenko, 2014; Shirai et al., 2014; Temme et al., 2014; Villanyi and Collart, 2015; Collart, 2016).
Ccr4-Not was first discovered in budding yeast (Denis, 1984; Collart and Struhl, 1993, 1994; Bai et al., 1999; Chen et al., 2001) where it is composed of 9 subunits, the Not1-Not5 proteins, Ccr4, and 3 Ccr4-associated factors, Caf1, Caf40, and Caf130. All of these subunits have orthologs in metazoans with the exception of Caf130. Metazoans have 2 orthologs for Caf1 called CNOT7 and CNOT8, 2 orthologs for Ccr4 called CNOT6 and CNOT6L and carry CNOT3 as a functional homolog of both Not5 and Not3, the products of a gene duplication event in budding yeast. Subunits with no yeast ortholog such as CNOT10 and CNOT11 are present in the metazoan Ccr4-Not complex (Albert et al., 2000). The metazoan ortholog of yeast Not4 is not a stable subunit of the metazoan Ccr4-Not complex but it does functionally complement the deletion of yeast Not4 (Albert et al., 2000; Bhaskar et al., 2015).
Multiple Activities of the Ccr4-Not Complex
Two different enzymatic activities are associated with the Ccr4-Not complex: deadenylation and ubiquitination. Ccr4 and Caf1 mediate deadenylation (Tucker et al., 2001, 2002), the first and rate-limiting step for mRNA degradation in eukaryotes, while Not4, a RING E3 ligase, mediates ubiquitination (Albert et al., 2002). It poly-ubiquitinates and destabilizes some substrates (Cooper et al., 2012; Gronholm et al., 2012; Gulshan et al., 2012) and mono-ubiquitinates others without any consequence for their stability (Panasenko et al., 2006; Panasenko and Collart, 2012). Other non-enzymatic activities of the Ccr4-Not complex have been reported. For instance Not2, Not3 and Not5 are thought to promote decapping by interaction with the Pat1 protein (Maillet and Collart, 2002; Chen et al., 2014; Mathys et al., 2014; Rouya et al., 2014; Nishimura et al., 2015; Ozgur et al., 2015; Alhusaini and Coller, 2016). In contrast to these negative functions in expression of the genome, the Ccr4-Not complex also plays positive roles. For instance yeast Not5 promotes co-translational assembly of RNA Polymerase II (RNAPII) and SAGA, a function that correlates with the association of Not1 to relevant mRNAs (Villanyi et al., 2014; Kassem et al., 2017). Moreover, the Ccr4-Not complex can bind transcription elongation complexes and promote elongation of a backtracked RNAPII (Kruk et al., 2011). Recently an essential positive role of Not5 for production of the translation machine was uncovered. It is mediated by binding of Not1 to ribosomal mRNAs during their production in the nucleus (imprinting) (Gupta et al., 2016). Indeed, ribosomal protein mRNAs are enriched within the pool of mRNAs that can be immunopecipitated with Not1. The binding of Not1 to this category of mRNAs requires Not5 and negatively correlates with the level of these mRNAs in total extracts, but positively with their level in polysome fractions, with production of new ribosomal proteins and with global cellular translation levels. The fact that Not5 is needed in the nucleus to promote this Not1 binding to ribosomal mRNAs, and that Not1 binds to intronic sequences, indicated that Not1 was binding to newly produced mRNAs, hence the term “imprinting” (Gupta et al., 2016).
In vivo the deadenylase is functional when tethered to target mRNAs with the Not1 scaffold (reviewed in Collart, 2016). In contrast ubiquitination by Not4 does not always require its association with Not1 (reviewed in Collart, 2013). Tethering of the Not1 scaffold to mRNAs can also repress translation in a manner that is independent of any enzymatic activity of the complex. This is thought to occur via the interaction of the complex with proteins such as the eIF4E-binding proteins or the DDX6 RNA helicase (called Dhh1 in budding yeast).
It is intriguing that tethering of the Ccr4-Not machinery to mRNAs can promote translation and co-translational events (Villanyi et al., 2014; Gupta et al., 2016; Kassem et al., 2017) or promote mRNA silencing and degradation (Finoux and Seraphin, 2006). These opposite outcomes might be determined by the cellular compartment, in which Not1 initially binds mRNAs. Indeed it could be that the global architecture of the Ccr4-Not complex in the nucleus and the cytoplasm is different. Tethering of Not5 out of the yeast nucleus does not lead to co-depletion of nuclear Not1, supporting the idea that different Not1 complexes exist (Gupta et al., 2016). However, this issue still needs to be clarified and the role of the Not subunits in particular, associated with both repression and activation of gene expression, needs to be better defined.
Ribosome Mutants and NOT Mutants are Similarly Resistant to Histidine Starvation
To consolidate our understanding of the functions mediated by the Not proteins we repeated the genetic selection in budding yeast that led to their isolation (Collart and Struhl, 1993). The idea was that we should isolate new mutations in the NOT genes, and potentially also additional mutations defining the cellular function affected by the Not proteins. We screened for new mutants that could grow on medium lacking histidine and containing 5 mM 3-aminotriazole (AT), a competitive inhibitor of the His3 enzyme (Collart and Struhl, 1993). His3 is necessary for yeast cells to produce histidine, and transcription of the HIS3 gene, like other amino acid biosynthetic genes, is under the control of the Gcn4 transcriptional activator. In wild type cells the translation of Gcn4 is regulated by the presence of upstream open reading frames (ORFs) and its production increases in response to starvation. For the selection, we used a starting strain (MATa ura3-52 trp1-Δ1 leu2-PET56 gal2 gcn4-Δ1; Hope and Struhl, 1986) with a deletion of the endogenous GCN4, carrying a plasmid expressing a mutant Gcn4 (YCp88-Sc4363) with a truncated activation domain (Hope and Struhl, 1986) and expressed from the constitutive DED1 promoter lacking uORFs (Collart and Struhl, 1993).
From the new recessive mutants isolated that were resistant to 5 mM AT in the growth medium, 9 strains carried mutations in NOT1, 2 in NOT2, 14 in NOT3, and 2 in NOT4. The mutations isolated in NOT5 have been described (Oberholzer and Collart, 1998). Fifteen other recessive mutants had slow growth or temperature sensitive phenotypes that co-segregated with AT-resistance. Surprisingly they defined 15 different complementation groups. We isolated clones complementing the mutations for 6 of these and sequenced the clone ends to identify the genomic fragments. Each clone carried either a ribosomal protein gene or a gene important for ribosome biogenesis. To determine whether these mutants were defective in ribosome biogenesis, we analyzed the polysome profiles of the 6 mutants by fractionation of total cellular extracts on a sucrose gradient. All 6 had defective polysome profiles, even at the permissive temperature (data not shown). In particular shoulders indicative of the presence of half-mers were visible in all mutant profiles (see below). This indicated that ribosomes were altered, and consistently, mutations were identified for each mutant in the ribosome-related gene. Sub-clones carrying these genes, but not sub-clones lacking these genes, complemented the mutant phenotypes (Table 1).
To confirm that the identified mutations were responsible for AT-resistance we focused on RPL10, integrated a URA3 marker gene at the RPL10 locus and confirmed by crosses and tetrad dissection that it co-segregated with the mutant phenotype. We also recovered the mutant gene on a plasmid and confirmed that it could not complement the temperature sensitive growth phenotype. Hence, mutations in genes that impair ribosome biogenesis and decrease global translation, like mutations in the NOT genes, lead to resistance to histidine starvation.
Specific Not1 Binding Responds to Decreased Protein Synthesis
HIS3 mRNA is increased in not mutants (Collart and Struhl, 1993, 1994; Oberholzer and Collart, 1998) and we observed a similar increase in the rpl10 mutant (data not shown). This most likely contributes to AT-resistance, possibly together with an increase in free amino acids due to reduced translation. These findings indicate that defective or reduced protein synthesis, as observed in not mutants or in ribosome-related mutants, is connected, possibly by the means of a cellular signal, to a relative increase in the transcript levels of an amino acid biosynthesis gene, namely HIS3. This raises the question of what the nature of the signal is. This signal cannot be the well-established eIF2α phosphorylation and translational up-regulation of Gcn4, known to respond to amino acid starvation, since the strain used in our selection expresses a Gcn4 derivative without uORFs.
We considered the possibility that the Not proteins themselves were part of the signaling pathway, since amino acid biosynthesis gene products are amongst the most up-regulated newly produced proteins in not5Δ (Gupta et al., 2016). Moreover, Not1 is significantly less associated with HIS3 mRNA in not5Δ, and HIS3 mRNA levels increase in total extracts and in polysomes (Gupta et al., 2016). To test this idea, we chose to compare 2 isogenic strains that had as only difference that they expressed a randomly chosen ribosomal protein gene at different levels. We prepared 2 rpl13bΔ stains containing plasmids expressing Rpl13b at different levels. One plasmid carried the endogenous RPL13B gene (promoter, intron, and terminator: PIT), and one plasmid contained the RPL13B promoter, open reading frame (ORF), and the heterologous CYC1 terminator (POC). It also lacked an intron (constructs depicted on Figure 1A). Cells expressing PIT had higher levels of polysomes compared to POC, but the polysomes had shoulders indicative of the presence of ribosome half-mers and ribosome biogenesis defects (Figure 1B). Indeed, in a normal polysome profile the identified peaks indicate one extra ribosome per mRNA (dimer, trimer, tetramer…). Half-mers instead indicate an unequal number of 40S and 60S ribosomal subunits on the mRNA. Expression of RPL13B from PIT was lower than from POC (Figure 1C, left panel). This different expression of Rpl13b induced differences in relative expression of other cellular mRNAs such as other ribosomal mRNAs (e.g., RPS22A) or HIS3 (Figure 1C, middle and right panels). This correlated with changes in relative binding of Not1 to those mRNAs in the 2 strains (Figure 1D) and in relative presence of those mRNAs in monosomes and polysomes (Figure 1E). NIP1 mRNA, whose association with Not1 does not change with different expression of ribosomal mRNAs (Gupta et al., 2016), was used for normalization.
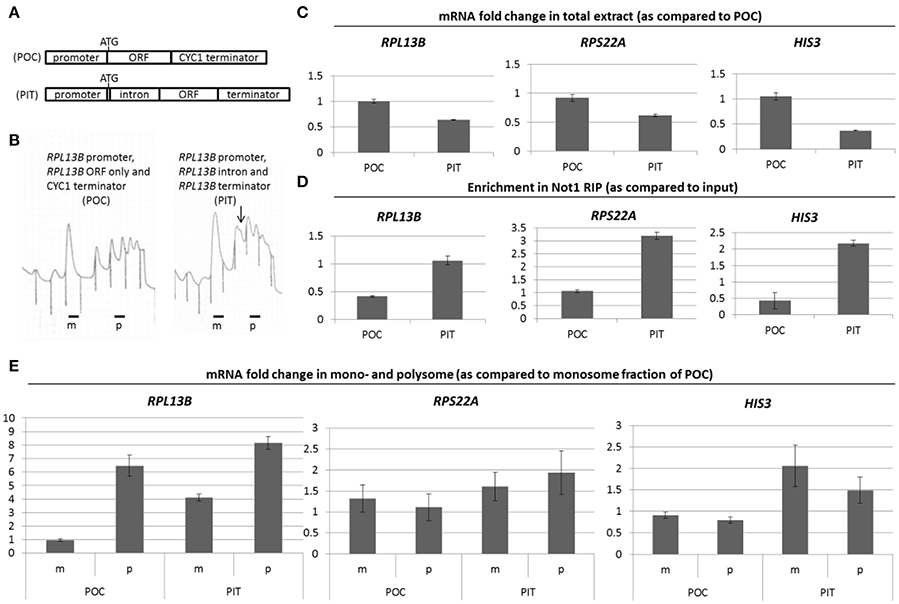
Figure 1. (A) RPL13B constructs. Similarly to the majority of yeast RP genes with introns, in the case of RPL13B the entire ORF is encoded in the 2nd exon except for a methionine which is encoded in the 1st exon. Construct “PIT” contains all the endogenous elements of the RPL13B gene. The intron is removed and the terminator is changed to a CYC1 terminator in “POC.” (B) Extracts from cells expressing PIT or POC were separated on a sucrose gradient to follow the polysome profiles. The arrow points to a shoulder on the disome peak. (C,D) Levels of the indicated mRNAs were measured in total extracts (C) or in Not1 immunoprecipitates (D). (E) Levels of the indicated mRNAs were measured in the monosome (m) and polysome (p) fractions as previously described (Gupta et al., 2016). The specific mRNAs measured are RPL13B, RPS22A, and HIS3 as indicated, and normalization was to the NIP1 mRNA levels.
Hence changes in ribosome production lead to modification of Not1 binding to HIS3 and ribosomal mRNAs, and to changes in relative translation of those mRNAs.
Resistance to Starvation Resulting from Ribosome or Not Protein Mutations Could Explain their Frequent Occurrence in Cancer
This study reveals that mutations in yeast ribosomal genes or ribosome biogenesis genes, like mutations in the NOT genes that affect global translation (Gupta et al., 2016), lead to increased resistance to histidine starvation. In metazoan cells, altering Not function or disrupting the translation machine, can also have similar phenotypes. Indeed, mutations in CNOT3, the ortholog of Not5, or in the RPL5 and RPL10 ribosomal proteins, are associated with adult T-cell acute lymphoblastic leukemia (T-ALL) (De Keersmaecker et al., 2013). T-ALL is an aggressive malignancy caused by the accumulation of genomic lesions leading to altered gene dosage. RPL5 and RPL10 occupy neighboring positions in the 60S subunit of the ribosomes, next to the central protuberance, which is in close vicinity to both the P, A, and E sites of the ribosomes. Around 8% of pediatric T-ALL patients harbor an Arg98Ser mutation in RPL10, which was shown to reduce translation fidelity (Sulima et al., 2014). RPL22 on the other hand forms the narrowest constriction of the ribosomal exit tunnel (Nakatogawa and Ito, 2002). Mutations in RPL22 are believed to alter protein synthesis efficacy. CNOT3 in T-ALL patients frequently carries missense mutations in Arg57 that affects splicing and reduces CNOT3 levels (De Keersmaecker et al., 2013). According to our work in yeast this should lead to reduced global translation and also to defects in co-translational complex assembly and consequently to elevated protein aggregation (Panasenko and Collart, 2012; Villanyi et al., 2014; Gupta et al., 2016; Kassem et al., 2017). Thus it appears that alteration of the process of translation in cancer cells can ensure their survival.
Numerous reports have described the presence of frequent mutations in ribosome subunits in tumor cells and different mechanisms have been considered to explain how RP mutations contribute to tumorigenesis or tumor maintenance (reviewed in Ruggero, 2013; Wang et al., 2015; Goudarzi and Lindstrom, 2016). Three main pathways have generally been considered: global suppression of protein synthesis, specific suppression of protein synthesis or finally extra-ribosomal functions. Our current observations suggest that relative up-regulation of amino acid biosynthesis genes resulting from global alteration of the translation process might contribute to sustain growth of the tumor cells. Since Not5 in particular appears to be involved in the regulatory loop linking translation to amino acid biosynthesis, the ortholog CNOT3 may have a tumor suppressor function. This is certainly compatible with the fact that mutations in CNOT3 have been identified in many tumors (Collart et al., 2013).
It is important to note that an impact of CNOT3 on amino acid biosynthesis is unlikely to be the only mechanism, by which a mutation of CNOT3 in tumor cells might contribute to tumor survival. This protein has very broad cellular functions and it has for instance been described as a modifier of gene expression that leads to incomplete penetrance of PRFP31 mutations in retinitis pigmentosa (Venturini et al., 2012). We have reported that in yeast Not1 and Not5 are important for gene expression homeostasis, buffering between transcription, translation and mRNA decay to maintain steady state protein levels (Villanyi and Collart, 2015). The deletion of Not5 leads to very slow growth of yeast cells, and the deletion of both alleles of CNOT3 is embryonic lethal in mice. Moreover, CNOT3+/− mice have many disturbed physiological functions (Morita et al., 2011; Shirai et al., 2014) indicating a very essential function of this protein (reviewed in Collart et al., 2013). It could be that the lower dose of CNOT3 in T-ALL cells does not disturb the major gene expression buffering function of CNOT3, but does have an impact on protein folding and complex assembly. The same could be true for the frequent RPL5, RPL10, and RPL22 mutations: affecting translation fidelity or efficacy has obvious dosage compensatory effects, as improperly assembled or folded proteins are likely to be non-functional, and prone to degradation or aggregation. In this context it is interesting to note that it has been reported that null mutants of specific Ccr4-Not subunits reduce the viability of aneuploid yeast cells (Tange et al., 2012). There may be a very fine balance between the impairment of Ccr4-Not function that can ensure survival of tumor cells with genomic lesions, or instead more severe ccr4-not mutations that will be toxic.
Conclusion
The Ccr4-Not complex acts at all stages of the gene expression pathway, and has both positive and negative effects on gene expression. Clarifying which contribution of Ccr4-Not regulation is important or lost in specific biological contexts is a real challenge. Work in vivo cannot easily distinguish direct from indirect effects, whereas studies in vitro define what the complex can do, but not how and when this is relevant in vivo. Here we point out that perturbation of translation is positively affecting amino acid biosynthesis that can facilitate cancer cell survival. We suggest that Not5 might not only regulate production of the translation machinery but also participate in the cross-talk between translation and amino acid production. Finally, we put forward the idea that impaired translation might contribute to dosage compensation via production of non-functional proteins. As the story of Ccr4-Not unfolds, it is clear that its understanding requires an open mind.
Author Contributions
MC contributed to experimental design, interpretation, and writing. SK contributed to interpretation and writing. ZV contributed to experimental work, interpretation, and writing.
Funding
This work was supported by grant [31003a_135794] from the Swiss National Science Foundation awarded to MC.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Ursula Oberholzer for the initial genetic analyses, Yassin Mouhajir, Janina Blattner, and Laetitia Maillard for their technical help, and finally Ravish Rashpa and Olesya Panasenko for a critical reading of the manuscript.
References
Albert, T. K., Hanzawa, H., Legtenberg, Y. I., de Ruwe, M. J., van den Heuvel, F. A., Collart, M. A., et al. (2002). Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 21, 355–364. doi: 10.1093/emboj/21.3.355
Albert, T. K., Lemaire, M., van Berkum, N. L., Gentz, R., Collart, M. A., and Timmers, H. T. (2000). Isolation and characterization of human orthologs of yeast CCR4-NOT complex subunits. Nucleic Acids Res. 28, 809–817. doi: 10.1093/nar/28.3.809
Alhusaini, N., and Coller, J. (2016). The deadenylase components Not2p, Not3p, and Not5p promote mRNA decapping. RNA 22, 709–721. doi: 10.1261/rna.054742.115
Bai, Y., Salvadore, C., Chiang, Y. C., Collart, M. A., Liu, H. Y., and Denis, C. L. (1999). The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol. Cell. Biol. 19, 6642–6651. doi: 10.1128/MCB.19.10.6642
Bhaskar, V., Basquin, J., and Conti, E. (2015). Architecture of the ubiquitylation module of the yeast Ccr4-Not complex. Structure 23, 921–928. doi: 10.1016/j.str.2015.03.011
Chapat, C., and Corbo, L. (2014). Novel roles of the CCR4-NOT complex. Wiley Interdiscip. Rev. RNA 5, 883–901. doi: 10.1002/wrna.1254
Chen, J., Rappsilber, J., Chiang, Y. C., Russell, P., Mann, M., and Denis, C. L. (2001). Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J. Mol. Biol. 314, 683–694. doi: 10.1006/jmbi.2001.5162
Chen, Y., Boland, A., Kuzuoğlu-Öztürk, D., Bawankar, P., Loh, B., Chang, C. T., et al. (2014). A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol. Cell 54, 737–750. doi: 10.1016/j.molcel.2014.03.034
Collart, M. A. (2013). The NOT4 RING E3 ligase: a relevant player in co-translational quality control. ISRN Mol. Biol. 2013:548359. doi: 10.1155/2013/548359
Collart, M. A. (2016). The Ccr4-Not complex is a key regulator of eukaryotic gene expression. Wiley Interdiscip. Rev. RNA, 7, 438–454. doi: 10.1002/wrna.1332
Collart, M. A., and Panasenko, O. O. (2012). The Ccr4–not complex. Gene 492, 42–53. doi: 10.1016/j.gene.2011.09.033
Collart, M. A., Panasenko, O. O., and Nikolaev, S. I. (2013). The Not3/5 subunit of the Ccr4-Not complex: a central regulator of gene expression that integrates signals between the cytoplasm and the nucleus in eukaryotic cells. Cell. Signal. 25, 743–751. doi: 10.1016/j.cellsig.2012.12.018
Collart, M. A., and Struhl, K. (1993). CDC39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible HIS3 promoters. EMBO J. 12, 177–186.
Collart, M. A., and Struhl, K. (1994). NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 8, 525–537. doi: 10.1101/gad.8.5.525
Collart, M. A., and Timmers, H. T. (2004). The eukaryotic Ccr4-not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog. Nucleic Acid Res. Mol. Biol. 77, 289–322. doi: 10.1016/S0079-6603(04)77008-7
Cooper, K. F., Scarnati, M. S., Krasley, E., Mallory, M. J., Jin, C., Law, M. J., et al. (2012). Oxidative-stress-induced nuclear to cytoplasmic relocalization is required for Not4-dependent cyclin C destruction. J. Cell Sci. 125, 1015–1026. doi: 10.1242/jcs.096479
De Keersmaecker, K., Atak, Z. K., Li, N., Vicente, C., Patchett, S., Girardi, T., et al. (2013). Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat. Genet. 45, 186–190. doi: 10.1038/ng.2508
Denis, C. L. (1984). Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics 108, 833–834.
Doidge, R., Mittal, S., Aslam, A., and Winkler, G. S. (2012). Deadenylation of cytoplasmic mRNA by the mammalian Ccr4-Not complex. Biochem. Soc. Trans. 40, 896–901. doi: 10.1042/BST20120074
Finoux, A. L., and Seraphin, B. (2006). In vivo targeting of the yeast Pop2 deadenylase subunit to reporter transcripts induces their rapid degradation and generates new decay intermediates. J. Biol. Chem. 281, 25940–25947. doi: 10.1074/jbc.M600132200
Goudarzi, K. M., and Lindstrom, M. S. (2016). Role of ribosomal protein mutations in tumor development (Review). Int. J. Oncol. 48, 1313–1324. doi: 10.3892/ijo.2016.3387
Gronholm, J., Kaustio, M., Myllymäki, H., Kallio, J., Saarikettu, J., Kronhamn, J., et al. (2012). Not4 enhances JAK/STAT pathway-dependent gene expression in Drosophila and in human cells. FASEB J. 26, 1239–1250. doi: 10.1096/fj.11-195875
Gulshan, K., Thommandru, B., and Moye-Rowley, W. S. (2012). Proteolytic degradation of the Yap1 transcription factor is regulated by subcellular localization and the E3 ubiquitin ligase Not4. J. Biol. Chem. 287, 26796–26805. doi: 10.1074/jbc.M112.384719
Gupta, I., Villanyi, Z., Kassem, S., Hughes, C., Panasenko, O. O., Steinmetz, L. M., et al. (2016). Translational capacity of a cell is determined during transcription elongation via the Ccr4-not complex. Cell Rep. 15, 1782–1794. doi: 10.1016/j.celrep.2016.04.055
Hope, I., and Struhl, K. (1986). Functional dissection of a eukaryotic transcriptional activator protein. Cell 46, 885–894. doi: 10.1016/0092-8674(86)90070-X
Inada, T., and Makino, S. (2014). Novel roles of the multi-functional CCR4-NOT complex in post-transcriptional regulation. Front. Genet. 5:135. doi: 10.3389/fgene.2014.00135
Kassem, S., Villanyi, Z., and Collart, M. A. (2017). Not5-dependent co-translational assembly of Ada2 and Spt20 is essential for functional integrity of SAGA. Nucleic Acids Res. 45, 1186–1199. doi: 10.1093/nar/gkw1059
Kruk, J. A., Dutta, A., Fu, J., Gilmour, D. S., and Reese, J. C. (2011). The multifunctional Ccr4-Not complex directly promotes transcription elongation. Genes Dev. 25, 581–593. doi: 10.1101/gad.2020911
Maillet, L., and Collart, M. A. (2002). Interaction between Not1p, a component of the Ccr4-not complex, a global regulator of transcription, and Dhh1p, a putative RNA helicase. J. Biol. Chem. 277, 2835–2842. doi: 10.1074/jbc.M107979200
Mathys, H., Basquin, J., Ozgur, S., Czarnocki-Cieciura, M., Bonneau, F., Aartse, A., et al. (2014). Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol. Cell 54, 751–765. doi: 10.1016/j.molcel.2014.03.036
Miller, J. E., and Reese, J. C. (2012). Ccr4-Not complex: the control freak of eukaryotic cells. Crit. Rev. Biochem. Mol. Biol. 47, 315–333. doi: 10.3109/10409238.2012.667214
Morita, M., Oike, Y., Nagashima, T., Kadomatsu, T., Tabata, M., Suzuki, T., et al. (2011). Obesity resistance and increased hepatic expression of catabolism-related mRNAs in Cnot3+/− mice. EMBO J. 30, 4678–4691. doi: 10.1038/emboj.2011.320
Nakatogawa, H., and Ito, K. (2002). The ribosomal exit tunnel functions as a discriminating gate. Cell 108, 629–636. doi: 10.1016/S0092-8674(02)00649-9
Nishimura, T., Padamsi, Z., Fakim, H., Milette, S., Dunham, W. H., Gingras, A. C., et al. (2015). The eIF4E-binding protein 4E-T Is a component of the mRNA decay machinery that bridges the 5' and 3' termini of target mRNAs. Cell Rep. 11, 1425–1436. doi: 10.1016/j.celrep.2015.04.065
Oberholzer, U., and Collart, M. A. (1998). Characterization of NOT5 that encodes a new component of the Not protein complex. Gene 207, 61–69. doi: 10.1016/S0378-1119(97)00605-7
Ozgur, S., Basquin, J., Kamenska, A., Filipowicz, W., Standart, N., and Conti, E. (2015). Structure of a human 4E-T/DDX6/CNOT1 complex reveals the different interplay of DDX6-binding proteins with the CCR4-NOT complex. Cell Rep. 13, 703–711. doi: 10.1016/j.celrep.2015.09.033
Panasenko, O. O. (2014). The role of the E3 ligase Not4 in cotranslational quality control. Front. Genet. 5:141. doi: 10.3389/fgene.2014.00141
Panasenko, O. O., and Collart, M. A. (2012). Presence of Not5 and ubiquitinated Rps7A in polysome fractions depends upon the Not4 E3 ligase. Mol. Microbiol. 83, 640–653. doi: 10.1111/j.1365-2958.2011.07957.x
Panasenko, O., Landrieux, E., Feuermann, M., Finka, A., Paquet, N., and Collart, M. A. (2006). The yeast Ccr4-Not complex controls ubiquitination of the nascent-associated polypeptide (NAC-EGD) complex. J. Biol. Chem. 281, 31389–31398. doi: 10.1074/jbc.M604986200
Panepinto, J. C., Heinz, E., and Traven, A. (2013). The cellular roles of Ccr4-NOT in model and pathogenic fungi-implications for fungal virulence. Front. Genet. 4:302. doi: 10.3389/fgene.2013.00302
Reese, J. C. (2013). The control of elongation by the yeast Ccr4-not complex. Biochim. Biophys. Acta 1829, 127–133. doi: 10.1016/j.bbagrm.2012.09.001
Rouya, C., Siddiqui, N., Morita, M., Duchaine, T. F., Fabian, M. R., and Sonenberg, N. (2014). Human DDX6 effects miRNA-mediated gene silencing via direct binding to CNOT1. RNA 20, 1398–1409. doi: 10.1261/rna.045302.114
Ruggero, D. (2013). Translational control in cancer etiology. Cold Spring Harb. Perspect. Biol. 5:a012336. doi: 10.1101/cshperspect.a012336
Shirai, Y. T., Suzuki, T., Morita, M., Takahashi, A., and Yamamoto, T. (2014). Multifunctional roles of the mammalian CCR4-NOT complex in physiological phenomena. Front. Genet. 5:286. doi: 10.3389/fgene.2014.00286
Sulima, S. O., Patchett, S., Advani, V. M., De Keersmaecker, K., Johnson, A. W., and Dinman, J. D. (2014). Bypass of the pre-60S ribosomal quality control as a pathway to oncogenesis. Proc. Natl. Acad. Sci. U.S.A. 111, 5640–5645. doi: 10.1073/pnas.1400247111
Tange, Y., Kurabayashi, A., Goto, B., Hoe, K. L., Kim, D. U., Park, H. O., et al. (2012). The CCR4-NOT complex is implicated in the viability of aneuploid yeasts. PLoS Genet. 8:e1002776. doi: 10.1371/journal.pgen.1002776
Temme, C., Simonelig, M., and Wahle, E. (2014). Deadenylation of mRNA by the CCR4-NOT complex in Drosophila: molecular and developmental aspects. Front. Genet. 5:143. doi: 10.3389/fgene.2014.00143
Tucker, M., Staples, R. R., Valencia-Sanchez, M. A., Muhlrad, D., and Parker, R. (2002). Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 21, 1427–1436. doi: 10.1093/emboj/21.6.1427
Tucker, M., Valencia-Sanchez, M. A., Staples, R. R., Chen, J., Denis, C. L., Parker, R., et al. (2001). The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104, 377–386. doi: 10.1016/S0092-8674(01)00225-2
Venturini, G., Rose, A. M., Shah, A. Z., Bhattacharya, S. S., and Rivolta, C. (2012). CNOT3 is a modifier of PRPF31 mutations in retinitis pigmentosa with incomplete penetrance. PLoS Genet. 8:e1003040. doi: 10.1371/journal.pgen.1003040
Villanyi, Z., and Collart, M. A. (2015). Ccr4-Not is at the core of the eukaryotic gene expression circuitry. Biochem. Soc. Trans. 43, 1253–1258. doi: 10.1042/BST20150167
Villanyi, Z., Ribaud, V., Kassem, S., Panasenko, O. O., Pahi, Z., Gupta, I., et al. (2014). The Not5 subunit of the ccr4-not complex connects transcription and translation. PLoS Genet. 10:e1004569. doi: 10.1371/journal.pgen.1004569
Wahle, E., and Winkler, G. S. (2013). RNA decay machines: deadenylation by the Ccr4-Not and Pan2-Pan3 complexes. Biochim. Biophys. Acta 1829, 561–570. doi: 10.1016/j.bbagrm.2013.01.003
Wang, W., Nag, S., Zhang, X., Wang, M. H., Wang, H., Zhou, J., et al. (2015). Ribosomal proteins and human diseases: pathogenesis, molecular mechanisms, and therapeutic implications. Med. Res. Rev. 35, 225–285. doi: 10.1002/med.21327
Keywords: Ccr4-Not complex, histidine starvation, CNOT3, ribosome, translation, T-ALL leukemia
Citation: Collart MA, Kassem S and Villanyi Z (2017) Mutations in the NOT Genes or in the Translation Machinery Similarly Display Increased Resistance to Histidine Starvation. Front. Genet. 8:61. doi: 10.3389/fgene.2017.00061
Received: 04 March 2017; Accepted: 04 May 2017;
Published: 22 May 2017.
Edited by:
Subbaya Subramanian, University of Minnesota, United StatesCopyright © 2017 Collart, Kassem and Villanyi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martine A. Collart, bWFydGluZS5jb2xsYXJ0QHVuaWdlLmNo
 Martine A. Collart
Martine A. Collart Sari Kassem
Sari Kassem Zoltan Villanyi
Zoltan Villanyi