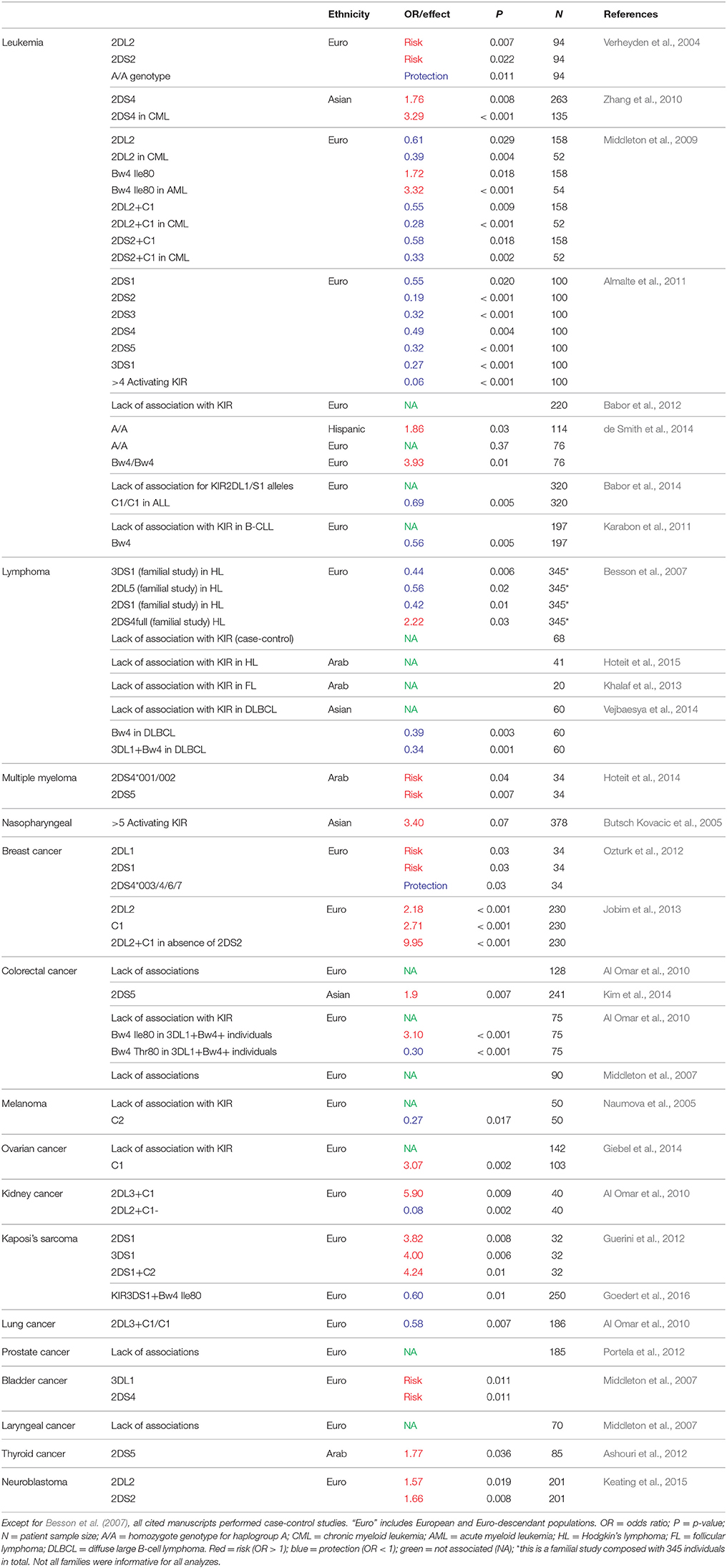
- Laboratório de Genética Molecular Humana, Departamento de Genética, Universidade Federal do Paraná, Curitiba, Brazil
The polymorphism of killer cell immunoglobulin-like receptors (KIR) has been associated with several diseases, including infection, autoimmunity and cancer. KIR molecules are a family of receptors expressed on the surface of natural killer cells (NK), frontline defense of innate immunity against microorganisms and neoplastic cells. Some studies have shown conflicting results concerning the role that KIR polymorphism plays in tumor susceptibility, particularly in leukemia and lymphoma. Interestingly, the presence of HLA ligands is sometimes strongly associated with several types of cancer and apparently is not related with their interaction with KIR. This manuscript briefly reviews the uncommon polymorphism of KIR and critically summarizes the recent findings with regards of the importance of KIR variation for cancer susceptibility.
Introduction
Natural killer (NK) cells were initially discovered because of their ability to kill virus-induced murine leukemic cells without prior sensibilization (Kiessling et al., 1975; Herberman and Ortaldo, 1981) and have been implicated in tumor surveillance and early recognition of microbial infections. NK cells present a variety of surface receptors that can either enhance or diminish their response against the target cell. That includes the killer cell immunoglobulin-like receptors (KIR), encoded by a region at chromosome 19 called leukocyte receptor complex (LRC; Wilson et al., 2000).
The KIR gene complex, located at 19q13.42 (Wende et al., 1999; Liu et al., 2000), consists of a cluster of homologous genes that has suffered extensive expansion and contraction (Wende et al., 1999; Martin et al., 2003). Fourteen KIR genes (KIR2DL1-5, KIR2DS1-5, KIR3DL1-3, KIR3DS1, and two pseudogenes (KIR2DP1 and KIR3DP1) have been described. Not all KIR genes are present in all individuals; this uncommon presence/absence polymorphism generates a broad variety of haplotypes that differ among individuals and populations. The diversity of haplotypes combined with increased number of alleles in each locus make nearly impossible two non-related individuals to carry the same KIR variants.
The KIR polymorphism has been studied in several populations across the globe (Norman et al., 2001; Augusto et al., 2012b, 2013, 2015a, 2016; Hollenbach et al., 2012, 2013) and more than 500 KIR gene-content genotypes have been described among over 200 worldwide populations (Gonzalez-Galarza et al., 2015). However, allelic diversity is still poorly known.
The nomenclature of KIR genes is based on the structure of the mature protein. KIR genes encode two or three (2D or 3D) extracellular immunoglobulin domains that may have short (S) or long (L) cytoplasmic tails (Colonna et al., 1996). Except by KIR2DL4 (Kikuchi-Maki et al., 2003), all molecules that present long cytoplasmic tails are inhibitory and all KIR that present short tails transduce activating signals. KIR haplotypes can be divided in two major groups: (1) haplogroup A, which classically consists of a fixed number of genes, mostly inhibitory; and (2) haplogroup B, composed by a large variation of gene-content combinations, characterized by the presence of more activating genes (Uhrberg et al., 2002).
HLA (human leukocyte antigens) class I are MHC (major histocompatibility complex) molecules that bind self and non-self peptides and display them on the cell surface for recognition by appropriate cells of the immune system. Additionally, HLA are known ligands for KIR. HLA-C2 allotypes are recognized by KIR2DL1 (Wagtmann et al., 1995; Fan et al., 1996; Winter and Long, 1997); KIR2DS1 also binds HLA-C2, but at lower affinity (Stewart et al., 2005). HLA-C1 and some C2 allotypes are bound by KIR2DL2/3 (Winter et al., 1998), and predicted to be ligand for KIR2DS2/3. HLA function is primarily related to presentation of antigens and regulation of immune responses. During the course of evolution, HLA-A and HLA-B apparently kept their main role as T cell receptor (TCR) ligands while HLA-C seems to have had evolved as primarily KIR ligands (Older Aguilar et al., 2010). Still, several HLA-A and -B molecules interact with KIR. HLA-Bw4, that comprises about 40% of all HLA-B molecules (Müller et al., 1989) plus a subset of HLA-A (A*23, A*24, and A*32; Kostyu et al., 1980), are recognized by KIR3DL1 (Cella et al., 1994; Stern et al., 2008). Despite the lack of direct evidence (Gillespie et al., 2007; O'Connor et al., 2007), the homology with KIR3DL1 and the numerous disease association studies suggest that KIR3DS1 also recognizes HLA-Bw4. KIR3DL2 recognizes HLA-A3/A11 (Döhring et al., 1996; Hansasuta et al., 2004), B27 (Shaw et al., 2014; Hatano et al., 2015) and HLA-F (Goodridge et al., 2013). As product of gene conversion with KIR3DL2, KIR2DS4 also binds HLA-A11 (Graef et al., 2009) and HLA-F (Goodridge et al., 2013). HLA-A11 is also a ligand for KIR2DS2 (Liu et al., 2014).
KIR polymorphism has been associated with infection, autoimmunity and cancer (van der Slik et al., 2003; Khakoo and Carrington, 2006; Yamada et al., 2007; Kulkarni et al., 2008; Augusto et al., 2012a, 2015b). The importance of KIR for reproduction is also well-documented (Hiby et al., 2004, 2014; Trowsdale and Moffett, 2008; Nakimuli et al., 2015). There is strong evidence that KIR and HLA are coevolving as an integrated system (Augusto and Petzl-Erler, 2015) and that KIR-driven pressures are balancing HLA haplotypes (Capittini et al., 2012; Fasano et al., 2014; Nemat-Gorgani et al., 2014; Augusto et al., 2015a).
As consequence of infection or malignancy, abnormal cells may exhibit reduced expression of self-MHC molecules. NK cells are able to recognize and to attack those cells with low expression of self-MHC molecules (Parham, 2004). Over the last two decades, KIR genes have been reported among those most strongly associated with disease susceptibility. Because the recognition of HLA by KIR modulates NK function, and also because these cells are important for attacking tumors, variation in KIR and HLA have been thought to intensely interfere in the risk of developing cancer. This hypothesis was corroborated by case control studies that showed association of KIR presence/absence and leukemia (Verheyden et al., 2004; Zhang et al., 2010). However, as we critically discuss in this review, as more case-control studies have further been performed, it now seems that the role of KIR presence/absence variation in cancer susceptibility may not be as strong as initially believed.
KIR Polymorphism in Leukemia
Acute lymphoblastic leukemia (ALL) is a malignancy in the bone marrow that leads to abnormal production and consequent excess of juvenile lymphocytes. ALL comprises ~75% of all cases of leukemia and normally occurs in children. ALL is a heterogeneous group of cancers that typically implicates B- or T-cell precursors, therefore subdivided in B-ALL or T-ALL. Differently, chronic lymphocytic leukemia (CLL) is a slow-growing tumor of lymphoid cells and usually occurs in individuals over 55 years of age. Myeloid leukemia causes rapid growth of myeloid cells; its acute form (AML) occurs either in children or in adults and its chronic form (CML) affects primarily adults.
KIR presence/absence in leukemia was initially explored by Verheyden et al., who reported KIR2DL2 and KIR2DS2 increased in patients (Verheyden et al., 2004). Both genes are present in haplogroup B; therefore, the authors could demonstrate that haplotype A was protective (Pc = 0.01). Primarily inhibitory genes compose haplotype A, suggesting that inhibitory KIR could protect against leukemia. Limitations of this study were the mixture of all four types of leukemia listed above within the patient group and the fact that the impact of KIR polymorphism in each form is not necessarily the same. Despite these limitations, the association of KIR polymorphism and leukemia appeared substantial. However, these results were not totally supported in a larger Chinese cohort (Zhang et al., 2010). Zhang and colleagues showed that KIR2DL2 was not significantly increased in patients (p = 0.10). The presence of KIR2DS4, however, was significantly increased in the total patients' sample (OR = 1.76, p = 0.008), but this effect seemed to be driven by CML subgroup (OR = 3.29, p < 0.001). Although Zhang's study did not analyze KIR haplotypes, activating KIR were slightly more frequent in patients (not significant), what partially corroborated Verheyden's findings. In opposition to all these previous results, however, Middleton et al. showed that KIR2DL2 was protecting against leukemia (Middleton et al., 2009).
In 2011, Almalte and colleagues analyzed a Canadian-French cohort composed by 145 B-ALL patients and 30 T-ALL and compared them to 245 controls (Almalte et al., 2011). In that study, the authors showed strong protective associations for the presence of all six activating KIR analyzed. They have not analyzed presence/absence of inhibitory genes, what challenge the interpretation those results due to the impossibility of analyzing the linkage disequilibrium between loci or verifying the KIR genotypes/haplotypes. Remarkably, an European cohort was further analyzed by Babor et al. and their results diverged from all former studies (Babor et al., 2012). Babor et al. reported no association between KIR presence/absence and leukemia, despite the fact that KIR frequencies in Babor's Canadian-French cohort did not differ from the cohorts from other studies. After that, another research group analyzed over 300 patients and performed another study (Oevermann et al., 2015). Applying careful and rigorous analyzes, Oevermann et al. corroborated Babor's results and reported absence of association of KIR presence/absence and leukemia. Lack of association was reported again in Thai patients (Vejbaesya et al., 2014).
Subsequent results brought some light to this discussion by showing the presence of homozygosity for haplotype A was associated with increased risk of developing leukemia in Hispanic, but not in Euro-descendants (de Smith et al., 2014). de Smith's explanation was that possibly the role played by KIR in leukemia may vary among ethnic groups.
Interestingly, three studies have shown stronger associations of leukemia with HLA: HLA-C2 (Babor et al., 2014) and specially HLA-Bw4 (Bw4/Bw4; OR = 3.9, p = 0.01; de Smith et al., 2014) and Bw4Ile80 (OR = 3.32, p = 0.0005). Although Bw4 and C2 are putative KIR ligands, due to conflicting results regarding KIR in leukemia, it is difficult to believe that these HLA associations are related to their interaction with KIR, but probably other HLA-related immune mechanisms.
Together, all these studies lead us to interpret that KIR genes probably don't play a major role in leukemia susceptibility, and this effect may vary in different ethnic groups. Additionally, HLA polymorphism has a stronger effect in leukemia susceptibility than KIR. This conclusion is also supported by another study, which showed only a trend of association for the presence of five or six activating KIR genes (p = 0.06), but a strong association for the presence of Bw4 (OR = 0.56; p = 0.005) in CLL patients (Karabon et al., 2011). Another interesting result from this same study is that, in general, the combinations KIR3DL1/S1+Bw4 presented similar odds ratios when compared to Bw4 alone. The association of the pair KIR3DS1+Bw4 (OR = 0.46; p = 0.003) being similar to Bw4 individually suggests that the effect appears to be driven mostly by Bw4. To explore KIR-HLA in the allelic level or expression studies like the one performed by Obama et al. (2007) could be a key to bring some light to this subject.
Lymphoma and Multiple Myeloma
The presence of large tumor cells derived from a germinal center B cell, known as Hodgkin and Reed-Sternberg, characterizes Hodgkin lymphoma (HL; Re et al., 2005). Epstein-Barr virus (EBV) is the major environmental factor associated with HL; approximately 40% of HL patients in the Western community tested positive for EBV (Küppers, 2009). Considering the importance of KIR for virus elimination, it is plausible to consider them as candidate genes for HL association studies. A familial study with 90 French families and 255 first-degree siblings was the first analysis of KIR polymorphism in HL (Besson et al., 2007). They reported negative association for the presence of KIR3S1, KIR2DL5, KIR2DS1 and KIR2DS5 (0.42 < OR < 0.56; 0.006 < P < 0.05). In that same study, they could not replicate their own results in a case-control study with 68 patients. Lack of association was also reported in a Lebanese case control study with 41 patients and 120 controls (Hoteit et al., 2015). It is important to notice that both case-control studies that reported lack of association were composed of small samples, what makes difficult to exclude the relevance of KIR polymorphism for HL pathogenesis. Furthermore, a familial study is more powerful than a case-control study, especially in the example above, in which Besson et al. performed deep analyzes, including EBV status of each HL patient.
KIR variation was also studied in non-Hodgkin lymphomas and multiple myeloma. Similarly from what was shown for ALL, no associations were seen for individual KIR genes in diffuse large B-cell lymphoma (DLBCL; Vejbaesya et al., 2014). Despite the lack of association with KIR variation in DLBCL, significant association was reported for the presence of HLA-Bw4 (OR = 0.39; p = 0.003). Presence of HLA-Bw4 alone showed similar odds ratio when comparing to the receptor/ligand pair KIR3DL1+HLA-Bw4 (OR = 0.34; p = 0.0006), what suggests that HLA alone was driving the effect. In multiple myeloma, KIR2DS5 and some alleles of KIR2DS4 were associated with increased risk (Hoteit et al., 2014), but again, the sample size was not large enough to allow more conclusive assumptions. Comprehensive studies with large and well-characterized cohorts need to be performed to verify the real impact of KIR-HLA in these diseases.
Breast Cancer
A pilot study analyzed the presence/absence of KIR in breast cancer (Ozturk et al., 2012). In that study, the authors analyzed 33 patients and 77 controls and reported borderline associations: KIR2DS1 associated with increased risk (p = 0.03) and KIR2DL1 increased in controls (p = 0.02). In addition, the authors performed allelic typing for KIR2DS4 and the alleles KIR2DS4*003/4/6/7 were overrepresented in controls (p = 0.03). Although the authors suggested that KIR variation might be involved in breast cancer pathogenesis, the small cohort and the borderline associations didn't provide conclusive results. These suggestions could not be corroborated by another study in a larger cohort of predominantly euro-descendants from Brazil (230 patients and 278 controls; Jobim et al., 2013). Jobim et al. reported a strong association for the presence of KIR2DL2 (OR = 2.7; p < 10−8) and for the presence of HLA-C1 (OR = 2.7; p < 10−7). The strong associations reported for breast cancer in the Brazilian cohort suggest that KIR2DL2 combined with its ligand C1 are, in fact, altering susceptibility to breast cancer. It is important to notice that the combination KIR2DL2+C1 in the absence of KIR2DS2 presented odds ratio as strong as 9.9 (Pc < 0.001).
Colon and Rectal Cancers
Absence of association of KIR with colorectal cancer was reported in Europeans (Middleton et al., 2007); different from Koreans, in which KIR2DS5 was increased in patients (OR = 1.9; p = 0.0007; Kim et al., 2014). Al Omar et al. also reported lack of association of KIR and colon cancer (Al Omar et al., 2010); interestingly, they showed a strong association of the presence of HLA-Bw4 (Bw4Ile80, OR = 3.1, p = 0.0001; Bw4The80, OR = 0.3, p = 0.0001 in individuals KIR3DL1+Bw4+). HLA-Bw4Ile80 is a stronger ligand for KIR (Cella et al., 1994); although this association suggests that KIR may interfere in the colon cancer susceptibility, it is important to note the lack of association for KIR+HLA pairs.
Other Cancers
Figure 1 and Table 1 summarize the associations and effect seen for KIR and ligands in several types of cancer. Nasopharyngeal cancer (NPC) is another example of neoplasm in which HLA polymorphism plays a major role in its susceptibility. In Chinese, HLA-B58 and HLA-A11 have been shown to confer risk and protection, respectively, for the development of NPC (Chan et al., 1983; Hildesheim et al., 2002; Lu et al., 2003). Even though HLA-A11 is a ligand for KIR3DL2 and KIR2DS4 (Döhring et al., 1996; Hansasuta et al., 2004; Graef et al., 2009), these relationships have not been investigated in NPC yet. The presence of five or more activating KIR conferred risk to EBV positive NPC patients; HLA-Cw4 was also reduced in NPC patients (Butsch Kovacic et al., 2005). HLA polymorphism seems to play a strong effect also in other cancers, such melanoma and ovarian, when comparing to a small or no effect of KIR for the susceptibility of those diseases (Naumova et al., 2005; Giebel et al., 2014).
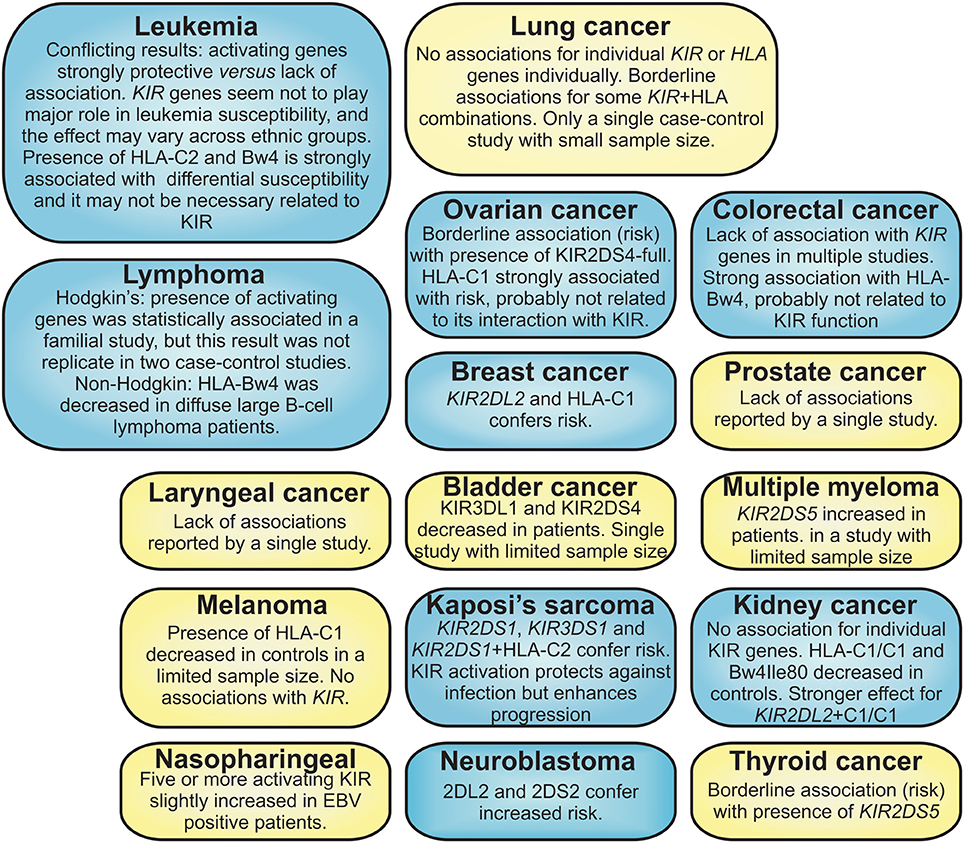
Figure 1. Summary of the cancers for which KIR polymorphism have been analyzed. Blue boxes = statistically significant association of cancer with KIR and/or HLA ligand; yellow boxes = borderline associations, lack of association or studies with reduced sample size.
The combination KIR2DL2+C1/C1 was strongly associated with protection in kidney patients (OR = 0.08; p = 0.002, n = 40 patients). Similarly from what was seen for breast cancer, the combination KIR-HLA showed stronger effect than either KIR or HLA isolated, suggesting the role of KIR-HLA combinations for the risk to develop this disease (Naumova et al., 2007; Al Omar et al., 2010; Giebel et al., 2014).
Activating KIR genes were associated with Kaposi's sarcoma (KS), a complication of KS-associated herpesvirus (KSHV) infection (Antman and Chang, 2000). Activating genes (KIR2DS1, KIR3DS1, and the combination KIR2DS1+HLA-C2) were significantly increased in individuals with classic KS (Guerini et al., 2012). Goedert et al. showed that KIR activation might decrease the risk of KSHV infection in an Italian cohort, while might enhance KSHV dissemination and progression to KS if infection occurs (Goedert et al., 2016).
Concluding Remarks
Despite the number of studies, it is still difficult to fully comprehend the role of KIR variation in cancer. One of the reasons is the reduced number of studies that analyzed large and well-characterized cohorts. Some studies have shown strong association with some types of cancer, but lack of association in several other studies and conflicting results suggest that the role of KIR presence/absence polymorphism may vary in different cancers. It is also clear that further studies with larger cohorts are needed.
Leukemia and lymphoma are examples of diseases for which mostly divergent results have been reported. It is interesting, however, that despite the conflict regarding KIR, the presence of HLA ligands has been consistently associated with different types of cancer. Even more interesting is the fact that many studies have shown that combinations KIR-HLA did not exhibit stronger effect than HLA alone. This suggests that the associations with HLA are possibly not related to KIR interaction. All these studies together suggest that KIR presence/absence polymorphism possibly does not play a major role in cancer. Considering the importance of NK for killing neoplastic cells, and the growing number of studies reporting KIR-HLA association with diseases, this conclusion can be quite intriguing.
It is important, however, to emphasize that lack of association with KIR presence/absence does not mean that KIR is not relevant for cancer. First, presence/absence polymorphism doesn't take the allelic polymorphism in consideration. KIR allelic variation is poorly known and rarely studied in diseases. Lack of genetic association does not discard the possibility of the cancer being associated with KIR differential expression levels, what confers another layer of complexity. Finally, the epigenetic mechanisms that regulate KIR-HLA should be studied especially in cancer, as it has been extensively demonstrated the importance of epigenetic regulation for tumor development. The comprehension of how KIR-HLA may be implicated in cancer is beyond presence/absence polymorphism, and perhaps beyond genetics. Different approaches have to be carefully considered, not only for KIR-HLA, but also for all genes that could impact cancer susceptibility.
Author Contributions
The author confirms being the sole contributor of this work and approved it for publication.
Funding
I thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Capes) and the Science without Borders Program for the research fellowship.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer AC and handling editor declared their shared affiliation, and the handling editor states that the process nevertheless met the standards of a fair and objective review
Acknowledgments
Thanks to Maria Dias da Silva and Danielle Malheiros for kindly reading this manuscript.
References
Almalte, Z., Samarani, S., Iannello, A., Debbeche, O., Duval, M., Infante-Rivard, C., et al. (2011). Novel associations between activating killer-cell immunoglobulin-like receptor genes and childhood leukemia. Blood 118, 1323–1328. doi: 10.1182/blood-2010-10-313791
Al Omar, S., Middleton, D., Marshall, E., Porter, D., Xinarianos, G., Raji, O., et al. (2010). Associations between genes for killer immunoglobulin-like receptors and their ligands in patients with solid tumors. Hum. Immunol. 71, 976–981. doi: 10.1016/j.humimm.2010.06.019
Antman, K., and Chang, Y. (2000). Kaposi's sarcoma. N. Engl. J. Med. 342, 1027–1038. doi: 10.1056/NEJM200004063421407
Ashouri, E., Dabbaghmanesh, M. H., Rowhanirad, S., Bakhshayeshkaram, M., Ranjbar Omrani, G., and Ghaderi, A. (2012). Activating KIR2DS5 receptor is a risk for thyroid cancer. Hum. Immunol. 73, 1017–1022. doi: 10.1016/j.humimm.2012.07.325
Augusto, D. G., Amorim, L. M., Farias, T. D. J., and Petzl-Erler, M. L. (2016). KIR and HLA genotyping of Japanese descendants from Curitiba, a city of predominantly European ancestry from Southern Brazil. Hum. Immunol. 77, 336–337. doi: 10.1016/j.humimm.2016.01.011
Augusto, D. G., Hollenbach, J. A., and Petzl-Erler, M. L. (2015a). A deep look at KIR-HLA in Amerindians: comprehensive meta-analysis reveals limited diversity of KIR haplotypes. Hum. Immunol. 76, 272–280. doi: 10.1016/j.humimm.2015.01.025
Augusto, D. G., Lobo-Alves, S. C., Melo, M. F., Pereira, N. F., and Petzl-Erler, M. L. (2012a). Activating KIR and HLA Bw4 ligands are associated to decreased susceptibility to pemphigus foliaceus, an autoimmune blistering skin disease. PLoS ONE 7:e39991. doi: 10.1371/journal.pone.0039991
Augusto, D. G., O'Connor, G. M., Lobo-Alves, S. C., Bass, S., Martin, M. P., Carrington, M., et al. (2015b). Pemphigus is associated with KIR3DL2 expression levels and provides evidence that KIR3DL2 may bind HLA-A3 and A11 in vivo. Eur. J. Immunol. 45, 2052–2060. doi: 10.1002/eji.201445324
Augusto, D. G., and Petzl-Erler, M. L. (2015). KIR and HLA under pressure: evidences of coevolution across worldwide populations. Hum. Genet. 134, 929–940. doi: 10.1007/s00439-015-1579-9
Augusto, D. G., Piovezan, B. Z., Tsuneto, L. T., Callegari-Jacques, S. M., and Petzl-Erler, M. L. (2013). KIR gene content in amerindians indicates influence of demographic factors. PLoS ONE 8:e56755. doi: 10.1371/journal.pone.0056755
Augusto, D. G., Zehnder-Alves, L., Pincerati, M. R., Martin, M. P., Carrington, M., and Petzl-Erler, M. L. (2012b). Diversity of the KIR gene cluster in an urban Brazilian population. Immunogenetics 64, 143–152. doi: 10.1007/s00251-011-0565-1
Babor, F., Manser, A. R., Fischer, J. C., Scherenschlich, N., Enczmann, J., Chazara, O., et al. (2014). KIR ligand C2 is associated with increased susceptibility to childhood ALL and confers an elevated risk for late relapse. Blood 124, 2248–2251. doi: 10.1182/blood-2014-05-572065
Babor, F., Manser, A., Schönberg, K., Enczmann, J., Borkhardt, A., Meisel, R., et al. (2012). Lack of association between KIR genes and acute lymphoblastic leukemia in children. Blood 120, 2770–2–author reply 2772. doi: 10.1182/blood-2012-07-440495
Besson, C., Roetynck, S., Williams, F., Orsi, L., Amiel, C., Lependeven, C., et al. (2007). Association of killer cell immunoglobulin-like receptor genes with Hodgkin's lymphoma in a familial study. PLoS ONE 2:e406. doi: 10.1371/journal.pone.0000406
Butsch Kovacic, M., Martin, M., Gao, X., Fuksenko, T., Chen, C.-J., Cheng, Y.-J., et al. (2005). Variation of the killer cell immunoglobulin-like receptors and HLA-C genes in nasopharyngeal carcinoma. Cancer Epidemiol. Biomarkers Prev. 14, 2673–2677. doi: 10.1158/1055-9965.EPI-05-0229
Capittini, C., Tinelli, C., Guarene, M., Pasi, A., Badulli, C., Sbarsi, I., et al. (2012). Possible KIR-driven genetic pressure on the genesis and maintenance of specific HLA-A,B haplotypes as functional genetic blocks. Genes Immun. 13, 452–457. doi: 10.1038/gene.2012.14
Cella, M., Strominger, J. L., Longo, A., Ferrara, G. B., and Colonna, M. (1994). NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J. Exp. Med. 180, 1235–1242. doi: 10.1084/jem.180.4.1235
Chan, S. H., Day, N. E., Kunaratnam, N., Chia, K. B., and Simons, M. J. (1983). HLA and nasopharyngeal carcinoma in Chinese–a further study. Int. J. Cancer 32, 171–176. doi: 10.1002/ijc.2910320206
Colonna, M., Lanier, L. L., and Long, E. O. (1996). Inhibitory MHC class I receptors on NK and T cells: a standard nomenclature. Immunol. Today 17:100. doi: 10.1016/0167-5699(96)80590-1
de Smith, A. J., Walsh, K. M., Ladner, M. B., Zhang, S., Xiao, C., Cohen, F., et al. (2014). The role of KIR genes and their cognate HLA class I ligands in childhood acute lymphoblastic leukemia. Blood 123, 2497–2503. doi: 10.1182/blood-2013-11-540625
Döhring, C., Scheidegger, D., Samaridis, J., Cella, M., and Colonna, M. (1996). A human killer inhibitory receptor specific for HLA-A1,2. J. Immunol. 156, 3098–3101.
Fan, Q. R., Garboczi, D. N., Winter, C. C., Wagtmann, N., Long, E. O., and Wiley, D. C. (1996). Direct binding of a soluble natural killer cell inhibitory receptor to a soluble human leukocyte antigen-Cw4 class I major histocompatibility complex molecule. Proc. Natl. Acad. Sci. U.S.A. 93, 7178–7183. doi: 10.1073/pnas.93.14.7178
Fasano, M. E., Rendine, S., Pasi, A., Bontadini, A., Cosentini, E., Carcassi, C., et al. (2014). The distribution of KIR-HLA functional blocks is different from north to south of Italy. Tissue Antigens 83, 168–173. doi: 10.1111/tan.12299
Giebel, S., Boratyn-Nowicka, A., Karabon, L., Jedynak, A., Pamula-Pilat, J., Tecza, K., et al. (2014). Associations between genes for killer immunoglobulin-like receptors and their ligands in patients with epithelial ovarian cancer. Hum. Immunol. 75, 508–513. doi: 10.1016/j.humimm.2014.04.002
Gillespie, G. M. A., Bashirova, A., Dong, T., McVicar, D. W., Rowland-Jones, S. L., and Carrington, M. (2007). Lack of KIR3DS1 binding to MHC class I Bw4 tetramers in complex with CD8+ T cell epitopes. AIDS Res. Hum. Retroviruses 23, 451–455. doi: 10.1089/aid.2006.0165
Goedert, J. J., Martin, M. P., Vitale, F., Lauria, C., Whitby, D., Qi, Y., et al. (2016). Risk of classic kaposi sarcoma with combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci: a population-based case-control study. J. Infect. Dis. 213, 432–438. doi: 10.1093/infdis/jiv413
González-Galarza, F. F., Takeshita, L. Y. C., Santos, E. J. M., Kempson, F., Maia, M. H. T., da Silva, A. L. S., et al. (2015). Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 43, D784–D788. doi: 10.1093/nar/gku1166
Goodridge, J. P., Burian, A., Lee, N., and Geraghty, D. E. (2013). HLA-F and MHC class I open conformers are ligands for NK cell Ig-like receptors. J. Immunol. 191, 3553–3562. doi: 10.4049/jimmunol.1300081
Graef, T., Moesta, A. K., Guethlein, L. A., Parham, P., Norman, P. J., Abi-Rached, L., et al. (2009). KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J. Exp. Med. 206, 2557–2572. doi: 10.1084/jem.20091010
Guerini, F. R., Mancuso, R., Agostini, S., Agliardi, C., Zanzottera, M., Hernis, A., et al. (2012). Activating KIR/HLA complexes in classic Kaposi's Sarcoma. Infect. Agents Cancer 7:9. doi: 10.1186/1750-9378-7-9
Hansasuta, P., Dong, T., Thananchai, H., Weekes, M., Willberg, C., Aldemir, H., et al. (2004). Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur. J. Immunol. 34, 1673–1679. doi: 10.1002/eji.200425089
Hatano, H., Shaw, J., Marquardt, K., Zhang, Z., Gauthier, L., Chanteux, S., et al. (2015). The D0 Ig-like domain plays a central role in the stronger binding of KIR3DL2 to B27 free H chain dimers. J. Immunol. 194, 1591–1601. doi: 10.4049/jimmunol.1402214
Herberman, R. B., and Ortaldo, J. R. (1981). Natural killer cells: their roles in defenses against disease. Science 214, 24–30. doi: 10.1126/science.7025208
Hiby, S. E., Apps, R., Chazara, O., Farrell, L. E., Magnus, P., Trogstad, L., et al. (2014). Maternal KIR in combination with paternal HLA-C2 regulate human birth weight. J. Immunol. 192, 5069–5073. doi: 10.4049/jimmunol.1400577
Hiby, S. E., Walker, J. J., O'shaughnessy, K. M., Redman, C. W. G., Carrington, M., Trowsdale, J., et al. (2004). Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J. Exp. Med. 200, 957–965. doi: 10.1084/jem.20041214
Hildesheim, A., Apple, R. J., Chen, C.-J., Wang, S. S., Cheng, Y.-J., Klitz, W., et al. (2002). Association of HLA class I and II alleles and extended haplotypes with nasopharyngeal carcinoma in Taiwan. J. Natl. Cancer Inst. 94, 1780–1789. doi: 10.1093/jnci/94.23.1780
Hollenbach, J. A., Augusto, D. G., Alaez, C., Bubnova, L., Fae, I., Fischer, G., et al. (2013). 16th IHIW: Population global distribution of Killer Immunoglobulin-like Receptor (KIR) and ligands. Int. J. Immunogenet. 40, 39–45. doi: 10.1111/iji.12028
Hollenbach, J. A., Nocedal, I., Ladner, M. B., Single, R. M., and Trachtenberg, E. A. (2012). Killer cell immunoglobulin-like receptor (KIR) gene content variation in the HGDP-CEPH populations. Immunogenetics 64, 719–737. doi: 10.1007/s00251-012-0629-x
Hoteit, R., Abboud, M., Bazarbachi, A., Salem, Z., Shammaa, D., Zaatari, G., et al. (2015). KIR genotype distribution among Lebanese patients with Hodgkin's lymphoma. Meta Gene 4, 57–63. doi: 10.1016/j.mgene.2015.02.004
Hoteit, R., Bazarbachi, A., Antar, A., Salem, Z., Shammaa, D., and Mahfouz, R. (2014). KIR genotype distribution among patients with multiple myeloma: higher prevalence of KIR 2DS4 and KIR 2DS5 genes. Meta Gene 2, 730–736. doi: 10.1016/j.mgene.2014.09.008
Jobim, M. R., Jobim, M., Salim, P. H., Portela, P., Jobim, L. F., Leistner-Segal, S., et al. (2013). Analysis of KIR gene frequencies and HLA class I genotypes in breast cancer and control group. Hum. Immunol. 74, 1130–1133. doi: 10.1016/j.humimm.2013.06.021
Karabon, L., Jedynak, A., Giebel, S., Wołowiec, D., Kielbinski, M., Woszczyk, D., et al. (2011). KIR/HLA gene combinations influence susceptibility to B-cell chronic lymphocytic leukemia and the clinical course of disease. Tissue Antigens 78, 129–138. doi: 10.1111/j.1399-0039.2011.01721.x
Keating, S. E., Ní Chorcora, C., Dring, M. M., Stallings, R. L., O'Meara, A., and Gardiner, C. M. (2015). Increased frequencies of the killer immunoglobulin-like receptor genes KIR2DL2 and KIR2DS2 are associated with neuroblastoma. Tissue Antigens 86, 172–177. doi: 10.1111/tan.12608
Khakoo, S. I., and Carrington, M. (2006). KIR and disease: a model system or system of models? Immunol. Rev. 214, 186–201. doi: 10.1111/j.1600-065x.2006.00459.x
Khalaf, R., Hoteit, R., Yazbek, S., El Hajj, N., Otrock, Z., Khansa, S., et al. (2013). Natural Killer Cell Immunoglobulin-like Receptor (KIR) genotypes in Follicular Lymphoma patients: results of a pilot study. Gene 525, 136–140. doi: 10.1016/j.gene.2013.03.144
Kiessling, R., Klein, E., Pross, H., and Wigzell, H. (1975). “Natural” killer cells in the mouse. II. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Characteristics of the killer cell. Eur. J. Immunol. 5, 117–121. doi: 10.1002/eji.1830050209
Kikuchi-Maki, A., Yusa, S.-I., Catina, T. L., and Campbell, K. S. (2003). KIR2DL4 is an IL-2-regulated NK cell receptor that exhibits limited expression in humans but triggers strong IFN-gamma production. J. Immunol. 171, 3415–3425. doi: 10.4049/jimmunol.171.7.3415
Kim, H.-J., Choi, H.-B., Jang, J.-P., Baek, I.-C., Choi, E.-J., Park, M., et al. (2014). HLA-Cw polypmorphism and killer cell immunoglobulin-like receptor (KIR) gene analysis in Korean colorectal cancer patients. Int. J. Surg. 12, 815–820. doi: 10.1016/j.ijsu.2014.06.012
Kostyu, D. D., Cresswell, P., and Amos, D. B. (1980). A public HLA antigen associated with HLA-A9, Aw32, and Bw4. Immunogenetics 10, 433–442. doi: 10.1007/BF01572579
Kulkarni, S., Martin, M. P., and Carrington, M. (2008). The Yin and Yang of HLA and KIR in human disease. Semin. Immunol. 20, 343–352. doi: 10.1016/j.smim.2008.06.003
Küppers, R. (2009). The biology of Hodgkin's lymphoma. Nat. Rev. Cancer 9, 15–27. doi: 10.1038/nrc2542
Liu, J., Xiao, Z., Ko, H. L., Shen, M., and Ren, E. C. (2014). Activating killer cell immunoglobulin-like receptor 2DS2 binds to HLA-A*11. Proc. Natl. Acad. Sci. U.S.A. 111, 2662–2667. doi: 10.1073/pnas.1322052111
Liu, W. R., Kim, J., Nwankwo, C., Ashworth, L. K., and Arm, J. P. (2000). Genomic organization of the human leukocyte immunoglobulin-like receptors within the leukocyte receptor complex on chromosome 19q13.4. Immunogenetics 51, 659–669. doi: 10.1007/s002510000183
Lu, C.-C., Chen, J.-C., Jin, Y.-T., Yang, H.-B., Chan, S.-H., and Tsai, S.-T. (2003). Genetic susceptibility to nasopharyngeal carcinoma within the HLA-A locus in Taiwanese. Int. J. Cancer 103, 745–751. doi: 10.1002/ijc.10861
Martin, M. P., Bashirova, A., Traherne, J., Trowsdale, J., and Carrington, M. (2003). Cutting edge: expansion of the KIR locus by unequal crossing over. J. Immunol. 171, 2192–2195. doi: 10.4049/jimmunol.171.5.2192
Middleton, D., Diler, A. S., Meenagh, A., Sleator, C., and Gourraud, P. A. (2009). Killer immunoglobulin-like receptors (KIR2DL2 and/or KIR2DS2) in presence of their ligand (HLA-C1 group) protect against chronic myeloid leukaemia. Tissue Antigens 73, 553–560. doi: 10.1111/j.1399-0039.2009.01235.x
Middleton, D., Vilchez, J. R., Cabrera, T., Meenagh, A., Williams, F., Halfpenny, I., et al. (2007). Analysis of KIR gene frequencies in HLA class I characterised bladder, colorectal and laryngeal tumours. Tissue Antigens 69, 220–226. doi: 10.1111/j.1399-0039.2006.00792.x
Müller, C. A., Engler-Blum, G., Gekeler, V., Steiert, I., Weiss, E., and Schmidt, H. (1989). Genetic and serological heterogeneity of the supertypic HLA-B locus specificities Bw4 and Bw6. Immunogenetics 30, 200–207. doi: 10.1007/BF02421207
Nakimuli, A., Chazara, O., Hiby, S. E., Farrell, L., Tukwasibwe, S., Jayaraman, J., et al. (2015). A KIR B centromeric region present in Africans but not Europeans protects pregnant women from pre-eclampsia. Proc. Natl. Acad. Sci. U.S.A. 112, 845–850. doi: 10.1073/pnas.1413453112
Naumova, E., Mihaylova, A., Ivanova, M., and Mihailova, S. (2007). Impact of KIR/HLA ligand combinations on immune responses in malignant melanoma. Cancer Immunol. Immunother. 56, 95–100. doi: 10.1007/s00262-006-0151-9
Naumova, E., Mihaylova, A., Stoitchkov, K., Ivanova, M., Quin, L., and Toneva, M. (2005). Genetic polymorphism of NK receptors and their ligands in melanoma patients: prevalence of inhibitory over activating signals. Cancer Immunol. Immunother. 54, 172–178. doi: 10.1007/s00262-004-0575-z
Nemat-Gorgani, N., Edinur, H. A., Hollenbach, J. A., Traherne, J. A., Dunn, P. P. J., Chambers, G. K., et al. (2014). KIR diversity in Māori and Polynesians: populations in which HLA-B is not a significant KIR ligand. Immunogenetics 66, 597–611. doi: 10.1007/s00251-014-0794-1
Norman, P. J., Stephens, H. A., Verity, D. H., Chandanayingyong, D., and Vaughan, R. W. (2001). Distribution of natural killer cell immunoglobulin-like receptor sequences in three ethnic groups. Immunogenetics 52, 195–205. doi: 10.1007/s002510000281
Obama, K., Kubota, R., Tara, M., Furukawa, Y., Osame, M., and Arimura, K. (2007). Killer cell immunoglobulin-like receptor/3DL2 expression in adult T-cell leukaemia. Br. J. Haematol. 138, 666–667. doi: 10.1111/j.1365-2141.2007.06704.x
O'Connor, G. M., Guinan, K. J., Cunningham, R. T., Middleton, D., Parham, P., and Gardiner, C. M. (2007). Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J. Immunol. 178, 235–241. doi: 10.4049/jimmunol.178.1.235
Oevermann, L., Firnkorn, M., Michaelis, S., Müller, S., Schaeffeler, E., Schrappe, M., et al. (2015). No association between the presence of killer-cell immunoglobulin-like receptor genes and susceptibility to childhood ALL. Blood 125, 3355–3357. doi: 10.1182/blood-2015-02-628339
Older Aguilar, A. M., Guethlein, L. A., Adams, E. J., Abi-Rached, L., Moesta, A. K., and Parham, P. (2010). Coevolution of killer cell Ig-like receptors with HLA-C to become the major variable regulators of human NK cells. J. Immunol. 185, 4238–4251. doi: 10.4049/jimmunol.1001494
Ozturk, O. G., Gun, F. D., and Polat, G. (2012). Killer cell immunoglobulin-like receptor genes in patients with breast cancer. Med. Oncol. 29, 511–515. doi: 10.1007/s12032-011-9932-x
Parham, P. (2004). Killer cell immunoglobulin-like receptor diversity: balancing signals in the natural killer cell response. Immunol. Lett. 92, 11–13. doi: 10.1016/j.imlet.2003.11.016
Portela, P., Jobim, L. F., Salim, P. H., Koff, W. J., Wilson, T. J., Jobim, M. R., et al. (2012). Analysis of KIR gene frequencies and HLA class I genotypes in prostate cancer and control group. Int. J. Immunogenet. 39, 423–428. doi: 10.1111/j.1744-313X.2012.01115.x
Re, D., Küppers, R., and Diehl, V. (2005). Molecular pathogenesis of Hodgkin's lymphoma. J. Clin. Oncol. 23, 6379–6386. doi: 10.1200/JCO.2005.55.013
Shaw, J., Hatano, H., and Kollnberger, S. (2014). The biochemistry and immunology of non-canonical forms of HLA-B27. Mol. Immunol. 57, 52–58. doi: 10.1016/j.molimm.2013.05.243
Stern, M., Ruggeri, L., Capanni, M., Mancusi, A., and Velardi, A. (2008). Human leukocyte antigens A23, A24, and A32 but not A25 are ligands for KIR3DL1. Blood 112, 708–710. doi: 10.1182/blood-2008-02-137521
Stewart, C. A., Laugier-Anfossi, F., Vély, F., Saulquin, X., Riedmuller, J., Tisserant, A., et al. (2005). Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc. Natl. Acad. Sci. U.S.A. 102, 13224–13229. doi: 10.1073/pnas.0503594102
Trowsdale, J., and Moffett, A. (2008). NK receptor interactions with MHC class I molecules in pregnancy. Semin. Immunol. 20, 317–320. doi: 10.1016/j.smim.2008.06.002
Uhrberg, M., Parham, P., and Wernet, P. (2002). Definition of gene content for nine common group B haplotypes of the Caucasoid population: KIR haplotypes contain between seven and eleven KIR genes. Immunogenetics 54, 221–229. doi: 10.1007/s00251-002-0463-7
van der Slik, A. R., Koeleman, B. P. C., Verduijn, W., Bruining, G. J., Roep, B. O., and Giphart, M. J. (2003). KIR in type 1 diabetes: disparate distribution of activating and inhibitory natural killer cell receptors in patients versus HLA-matched control subjects. Diabetes 52, 2639–2642. doi: 10.2337/diabetes.52.10.2639
Vejbaesya, S., Sae-Tam, P., Khuhapinant, A., and Srinak, D. (2014). Killer cell immunoglobulin-like receptors in Thai patients with leukemia and diffuse large B-cell lymphoma. Hum. Immunol. 75, 673–676. doi: 10.1016/j.humimm.2014.04.004
Verheyden, S., Bernier, M., and Demanet, C. (2004). Identification of natural killer cell receptor phenotypes associated with leukemia. Leukemia 18, 2002–2007. doi: 10.1038/sj.leu.2403525
Wagtmann, N., Rajagopalan, S., Winter, C. C., Peruzzi, M., and Long, E. O. (1995). Killer cell inhibitory receptors specific for HLA-C and HLA-B identified by direct binding and by functional transfer. Immunity 3, 801–809. doi: 10.1016/1074-7613(95)90069-1
Wende, H., Colonna, M., Ziegler, A., and Volz, A. (1999). Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm. Genome 10, 154–160. doi: 10.1007/s003359900961
Wilson, M. J., Torkar, M., Sheer, D., Haude, A., Milne, S., Jones, T., et al. (2000). Plasticity in the organization and sequences of human KIR/ILT gene families. Proc. Natl. Acad. Sci. U.S.A. 97, 4778–4783. doi: 10.1073/pnas.080588597
Winter, C. C., and Long, E. O. (1997). A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. J. Immunol. 158, 4026–4028.
Winter, C. C., Parham, P., Long, E. O., Gumperz, J. E., and Wagtmann, N. (1998). Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J. Immunol. 161, 571–577.
Yamada, E., Walker, B. D., Phair, J., Martin, M. P., Goedert, J. J., Martin, J. N., et al. (2007). Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 39, 733–740. doi: 10.1038/ng2035
Keywords: killer cell immunoglobulin-like receptors, HLA genes, cancer, association, susceptibility
Citation: Augusto DG (2016) The Impact of KIR Polymorphism on the Risk of Developing Cancer: Not as Strong as Imagined? Front. Genet. 7:121. doi: 10.3389/fgene.2016.00121
Received: 21 September 2015; Accepted: 14 June 2016;
Published: 28 June 2016.
Edited by:
Heather Cunliffe, University of Otago, New ZealandReviewed by:
Aniruddha Chatterjee, University of Otago, New ZealandShicheng Guo, University of California San Diego, USA
Copyright © 2016 Augusto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Danillo G. Augusto, ZGFuaWxsb0BhdWd1c3RvLmJpby5icg==
 Danillo G. Augusto
Danillo G. Augusto