
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet., 01 February 2016
Sec. Genetics of Common and Rare Diseases
Volume 7 - 2016 | https://doi.org/10.3389/fgene.2016.00003
 Changbing Shen1
Changbing Shen1 Jing Gao2
Jing Gao2 Yujun Sheng1
Yujun Sheng1 Jinfa Dou1
Jinfa Dou1 Fusheng Zhou1
Fusheng Zhou1 Xiaodong Zheng1
Xiaodong Zheng1 Randy Ko3
Randy Ko3 Xianfa Tang1
Xianfa Tang1 Caihong Zhu1
Caihong Zhu1 Xianyong Yin4
Xianyong Yin4 Liangdan Sun1
Liangdan Sun1 Yong Cui5
Yong Cui5 Xuejun Zhang1,2*
Xuejun Zhang1,2*Vitiligo is an autoimmune disease with a strong genetic component, characterized by areas of depigmented skin resulting from loss of epidermal melanocytes. Genetic factors are known to play key roles in vitiligo through discoveries in association studies and family studies. Previously, vitiligo susceptibility genes were mainly revealed through linkage analysis and candidate gene studies. Recently, our understanding of the genetic basis of vitiligo has been rapidly advancing through genome-wide association study (GWAS). More than 40 robust susceptible loci have been identified and confirmed to be associated with vitiligo by using GWAS. Most of these associated genes participate in important pathways involved in the pathogenesis of vitiligo. Many susceptible loci with unknown functions in the pathogenesis of vitiligo have also been identified, indicating that additional molecular mechanisms may contribute to the risk of developing vitiligo. In this review, we summarize the key loci that are of genome-wide significance, which have been shown to influence vitiligo risk. These genetic loci may help build the foundation for genetic diagnosis and personalize treatment for patients with vitiligo in the future. However, substantial additional studies, including gene-targeted and functional studies, are required to confirm the causality of the genetic variants and their biological relevance in the development of vitiligo.
Vitiligo is a relatively common skin disease, and is an acquired pigmentary disorder characterized by areas of depigmented skin resulting from loss of epidermal melanocytes. The prevalence of this disease varies from 0.2% to 1% in various global populations without sex predilection (Spritz, 2008). The pathogenesis of vitiligo remains elusive, although many theories such as autoimmune hypothesis, genetics theory, reactive oxygen species model, zinc-α2-glycoprotein deficiency hypothesis, viral theory, intrinsic theory and biochemical, molecular and cellular alterations accounting for loss of functioning melanocytes in vitiligo were elaborated to clarify vitiligo pathogenesis and showed that it was a multifactorial disease involving many different interactions (Mohammed et al., 2015). In this review, we mainly summarize the recent studies in the genetics of vitiligo through genome-wide association studies (GWASs), with a focus on the susceptibility genes or loci (Table 1) that have been identified to date, which implicate important pathways in the pathogenesis of vitiligo.
The earliest evidence relating to the genetic basis of vitiligo was a description provided by Addison in the year of 1855, Addison presented a patient with idiopathic adrenal insufficiency, generalized vitiligo (GV), and pernicious anemia (PA; Addison, 1855). In the 1950s, perhaps the first time to address the possible inheritance of GV, Stüttgen and Teindel described multigenerational families with multiple cases of GV and other autoimmune diseases, concluding the possibility that GV is likely an autosomal dominant inheritance disease (Teindel, 1950; Stüttgen, 1950). Many subsequent studies around the world confirmed frequent clustering of vitiligo cases within families (Alkhateeb et al., 2003; Zhang et al., 2004b; Sun et al., 2006; Zhang et al., 2009). The genetics of vitiligo cannot be explained by simple Mendelian genetics, and it is characterized by incomplete penetrance, multiple susceptibility loci, and genetic heterogeneity factors. Interestingly, many of the risk loci that have been identified are shared between vitiligo and other autoimmune diseases by GWASs (Table 2), implying that common molecular pathways exist among various autoimmune disorder processes.
Over the past several decades, a large number of genes and genomic regions involved in vitiligo susceptibility have been revealed through linkage analysis and candidate gene studies (Spritz, 2012). Candidate gene association studies are best suited to detect genetic signals that represent relatively common causal variants with modest effect sizes. Moreover, candidate gene association studies are relatively easy to carry out, usually involving a simple comparison of allele frequencies in cases and controls. At least 33 different candidate genes for vitiligo have been reported on the basis of such studies (Birlea et al., 2011). However, such studies are usually subject to false positive results because of the ethnic differences in case-control analyses, inadequate statistical power and statistical fluctuation, and inadequate correction for multiple testing both within and across studies.
Genome-wide linkage study is a method, when executed correctly, identifies genetic loci of vitiligo in multiplex families. These studies help to determine the position of the genetic marker inherited together with a specific disease. Genome-wide linkage studies in the Caucasian population multiplex vitiligo families identified additional linkage signals on chromosomes 7, 8, 9, 11, 13, 17, 19, and 22 (Fain et al., 2003; Spritz et al., 2004). In addition, this parallels with the genetic linkage studies of vitiligo in Chinese Han population which detected linkage signals on chromosomes 1, 4, 6, 14, and 22 (Chen et al., 2005; Liang et al., 2007). Normally, genetic loci discovered through genome-wide linkage studies encompass several megabases. The diversity of proposed regions has created a challenge in fine mapping.
Genome-wide association studies is a high through put technology, capable of “pin-pointing” disease-causing genes. Since 2005, GWAS has been proved to be the most powerful and efficient study design thus far in identifying genetic variants that are associated with complex diseases. More than 1000 types of complex diseases and traits have been investigated by this approach. Since 2010, several GWASs have been performed in patients with vitiligo in various ethnic populations (Birlea et al., 2010; Quan et al., 2010; Jin et al., 2011, 2010a, 2012a; Cheong et al., 2013), these studies have confirmed genetic associations of almost 40 genes and loci with vitiligo during the past 5 years (Table 1; Figure 1). Recently, Next-Generation DNA Re-Sequencing and exome sequencing analysis have also been used for identifying variants of genes for vitiligo (Jin et al., 2012b).
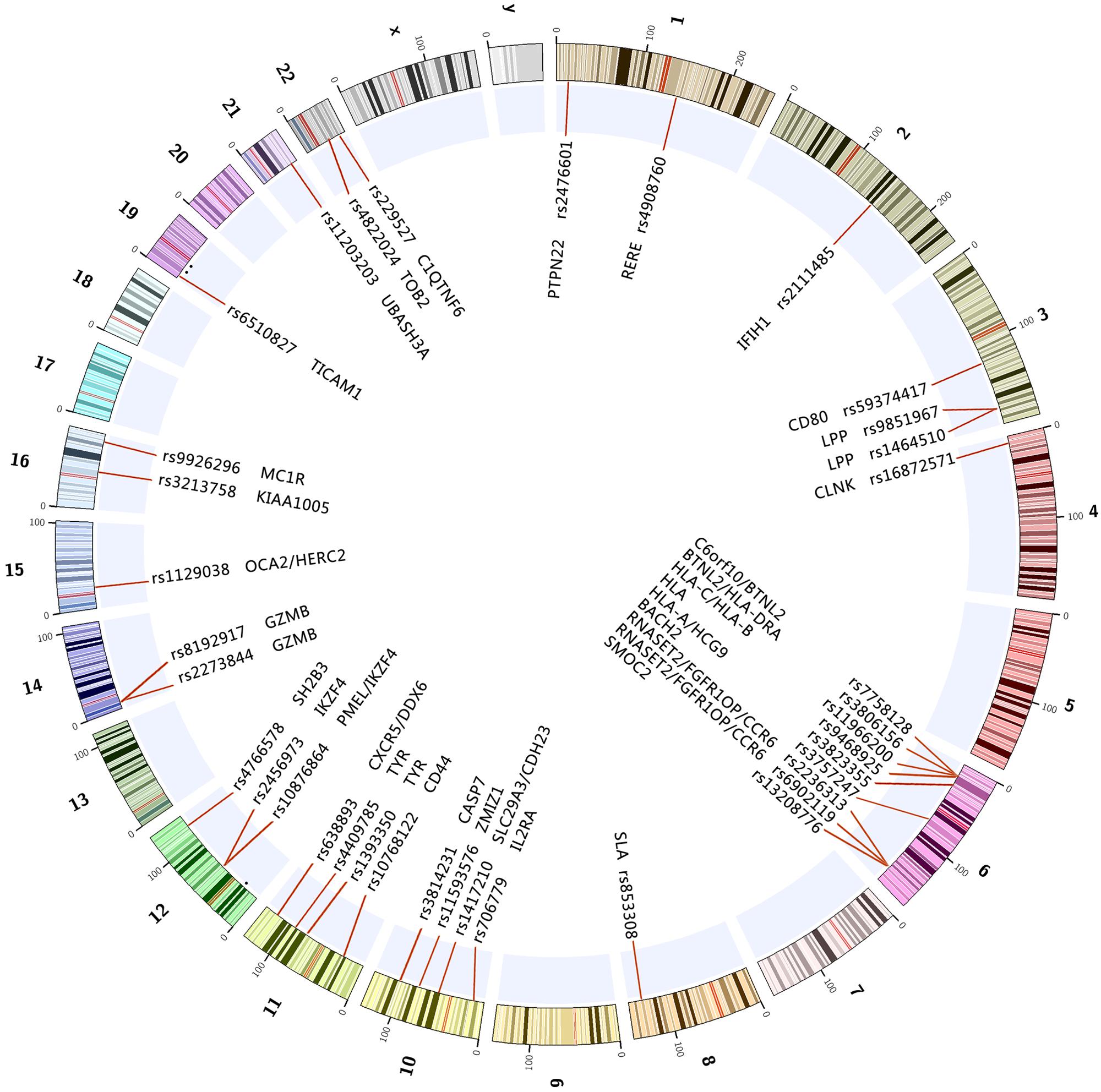
FIGURE 1. The reported genes and loci on the chromosomes associated with vitiligo through GWAS and GWAS meta-analysis up to 2015-10. All of the above marked regions and genes on the chromosomes achieved genome-wide significance (p < 5E-8) in at least one study.
The human leukocyte antigen (HLA) is the most gene-dense region of the genome, encoding more than 120 functional genes in humans which are distributed over a 3.6 Mbp region. Many previous studies have found genes in the HLA region associated with vitiligo, such as HLA-A∗02, HLA-A∗30, HLA-B∗13, HLA-C∗0602, HLA-DRB1∗04, HLA-DRB1∗07 and HLA-DQB1∗03 (Tastan et al., 2004; Zhang et al., 2004a; Liu et al., 2007). GWASs for vitiligo have detected major association signals in the MHC on chromosome 6p21.3 in Caucasian and Chinese populations, and there are some specific associations differed between the two populations. In Caucasians, two major association peaks in MHC region were detected, the peak in class I gene region, principally between HLA-A and HCG9, and in the class II region, mainly between HLA-DRB1 and HLA-DQA1, in linkage disequilibrium (LD) with HLA-DRB1∗04 (Jin et al., 2010a). In the Chinese population study, the major MHC association signal was in the class III gene region, though there was also some evidence for independent association in the class II region (Quan et al., 2010). In Chinese Han population, GWAS for vitiligo identified two independent association signals (rs11966200 and rs9468925) within the HLA region. Further analyses have suggested that the strong association at rs11966200 might reflect the reported association of the HLA-A∗3001, HLA-B∗1302, HLA-C∗0602 and HLA-DRB1∗0701 alleles and the association at rs9468925 might represent a previously unknown HLA susceptibility allele (HLA-C/HLA-B) (Quan et al., 2010). Additional studies found that rs9468925 is associated with clinical features of GV (Liu et al., 2012), and more importantly, rs9468925 in HLA-C/HLA-B is associated with both psoriasis (PS) and vitiligo (Zhu et al., 2011), providing first important evidence that two major skin diseases share a common genetic locus in the MHC, and revealing that this genetic locus may share the same molecular mechanism for PS and vitiligo. Genome-wide analysis has identified rs532098 in the vicinity of HLA-DRB1-DQA1, showing suggestive evidence of the age of onset for GV (Jin et al., 2011). All evidences shown here clearly suggest that HLA genes represent attractive therapeutic targets for vitiligo pathogenesis.
Vitiligo is a common autoimmune disease, around 20% of vitiligo patients manifest concomitant occurrence of other autoimmune diseases, particularly autoimmune thyroid disease (AMD), rheumatoid arthritis (RA), late-onset type I diabetes (T1D), PS, PA, systemic lupus erythematosus (SLE), and Addison’s disease (AD; Birlea et al., 2010). Outside MHC region, some susceptibility genes encode immunoregulatory proteins which involve in biological pathways that are most likely influencing the development of vitiligo.
CD44 encodes a cell surface glycoprotein with various functions, including a role in T cell development (Baaten et al., 2010), and is associated with lupus (Ramos et al., 2011). CD80 encodes a surface protein on activated B-cells, monocytes, and dendritic cells that co-stimulates T cell priming (Peach et al., 1995). Vitiligo is a CD8 T cell-mediated autoimmune disease and promotes the longevity of memory T cell responses to melanoma. Studies have found that HA-specific CD8 T cells are adoptively transferred into mice expressing HA as a self-antigen in the pancreas, and HA-specific T cells proliferate in draining lymph nodes and upregulated CD44 (Hernandez et al., 2001). GWAS meta analysis (GWAS-MA) of vitiligo has showed suggestive association of SNP rs4330287 and imputed SNP rs59374417 in CD80, which was confirmed by replication study and overall meta-analysis (Jin et al., 2012a). Flow cytometric analysis has found that the percentage of CD80+ monocytes are significantly increased in the vitiligo group compared with the controls (Basak et al., 2008), which may indicate alterations of monocyte function in the pathogenesis of vitiligo.
GWAS-MA has identified vitiligo-associated loci TG/SLA and BACH2 in European-derived white (CEU) population (Jin et al., 2012a). TG encodes thyroglobulin, SLA encodes Src-like adaptor protein, an inhibitor of T- and B-cell receptor signaling. It is not apparent what role thyroglobulin might play in vitiligo pathogenesis, suggesting association of vitiligo with the TG/SLA locus may derive from SLA, rather than TG (Jin et al., 2012a). Studies have suggested that SLA might likewise account for reported association with autoimmune thyroid disease (ATD; Tomer and Greenberg, 2004) and acute lymphoblastic leukemia (ALL; Mansha et al., 2010). The GWAS-MA for vitiligo showed suggestive association of SNPs (nt 90941239-91915693) spanning BACH2, particularly rs3757247, confirmed by the replication study and overall meta-analysis (Jin et al., 2012a). BACH2 encodes a transcriptional repressor of B cells (Sasaki et al., 2000), which is a key regulator of CD4(+) T-cell differentiation that prevents inflammatory disease by controlling the balance between tolerance and immunity. Genetic polymorphisms analysis shows that BACH2 is associated with asthma, Crohn’s disease (CD), multiple sclerosis (MS) and T1D (Roychoudhuri et al., 2013).
IFIH1 and TICAM1 are involved in the innate immune response system (Kato et al., 2006). GWAS-MA for vitiligo has shown genome-wide level association with SNP rs2111485 in IFIH1 (Jin et al., 2012a). IFIH1 encodes an interferon-induced RNA helicase involved in antiviral innate immune responses, associated with T1D (Smyth et al., 2006), Graves’ disease (GD) (Sutherland et al., 2007), MS (Martinez et al., 2008), PS (Li et al., 2010), and possibly lupus (Gateva et al., 2009). TICAM1, also known as TIR domain-containing adaptor-inducing IFN-b (TRIF), eventually activates transcription factors (TF), interferon regulatory factor-3 (IRF-3), NF-κB and AP-1, leading to the induction of type I interferons and inflammatory cytokines (Kumeta et al., 2014). TICAM1 encodes a toll-like receptor adaptor molecule 1, which mediates innate immune responses to viral pathogens (Seya et al., 2009). Viral factor has been implicated in the etiopathogenesis of vitiligo, we speculate that TICAM1 might act as a viral factor in the pathogenesis of vitiligo.
PTPN22, UBASH3A, and CLNK are the T-cell–receptor signaling pathway genes. PTPN22 620W allele plays a role in autoimmune disorders, and underscores the importance of a genetically mediated autoimmune mechanism in the pathogenesis of vitiligo. Evidence shown that the PTPN22 1858C/T variants contribute to risk of GV in European Caucasian and Mexican populations (Laberge et al., 2008; Garcia-Melendez et al., 2014), but it does not appear to play a similar role in the Jordanian population and Turkish generalized-vitiligo patients (Alkhateeb et al., 2013; Akbas et al., 2014). A meta-analysis was conducted of the association between PTPN22 1858 C/T polymorphisms and vitiligo and found PTPN22 C1858T polymorphism is associated with vitiligo susceptibility in European population (Song et al., 2013). Variants of PTPN22 also are associated with a number of different autoimmune diseases, including RA (Begovich et al., 2004) and SLE (Kyogoku et al., 2004). There was an association between GV and nine SNPs in the region spanning UBASH3A on chromosome 21q22.3; of these SNPs, rs2839511 showed genome wide significance (Jin et al., 2010a). Functional prediction of the variants in non-MHC vitiligo loci identified predicted deleterious variants at UBASH3A confer protection from vitiligo (Jin et al., 2012a). UBASH3A encodes a regulator of T-cell–receptor signaling and is associated with T1D (Concannon et al., 2008), the genotype distribution of rs2277798 is significantly associated with hematuria in SLE patients (Liu et al., 2015). At 4p16.1, the GWAS-MA for vitiligo showed suggestive association of SNPs (nt 10702156–10729386) upstream of CLNK, including rs16872571 and several imputed SNPs, particularly rs11940117, confirmed by the replication study and overall meta-analysis. CLNK encodes a mast cell immunoreceptor signal transducer, a positive regulator in immunoreceptor signaling (Wu and Koretzky, 2004). Haplotype analysis has shown that the TCATTCTGA haplotype of CLNK is more frequent among patients with gout (Jin et al., 2015).
IKZF4, IL2RA, and BTNL2 are involved in T-cell activation. Two GWASs have identified IKZF4 as one of the susceptible genes for GV (Jin et al., 2012a; Tang et al., 2013). IKZF4, is a critical mediator of Foxp3-dependent gene silencing in T cell reguration (Treg), interacts directly with Foxp3 and is necessary for gene silencing without affecting the expression of Foxp3 activated genes (Pan et al., 2009). IKZF4 maybe another biological candidate gene for vitiligo and influences the development of vitiligo. Further fine mapping and function analysis required to determine the causal variants within this locus for vitiligo. Besides, studies have also found IKZF4 is associated with T1D (Hakonarson et al., 2008) and alopecia areata (AA; Petukhova et al., 2010). There are 25 SNPs in the region of IL2RA (encoding the interleukin-2-receptor alpha chain) on chromosome 10p15.1, 8 of which showed genome wide significance, SNPs rs706779 and rs7090530 had the strongest association with GV (Jin et al., 2010a). Elevated serum interleukin-2-receptor levels indicate T-cell activation in GV (Honda et al., 1997; Wang et al., 2009). The variants of IL2RA have been shown to be associated with T1D (Vella et al., 2005), RA (Hinks et al., 2009), and SLE (Carr et al., 2009). Study identified a quantitative trait locus for vitiligo age of onset, located near c6orf10-BTNL2 (rs7758128), a region that is also associated with GV susceptibility (Jin et al., 2011). BTNL2 encodes an immunoglobulin superfamily membrane protein implicated in T-cell activation. Variants in BTNL2 may play a role which involved in vitiligo susceptibility versus vitiligo age of onset. The BTNL2 gene region has been associated with susceptibility to many other autoimmune diseases, such as T1D (He et al., 2009), RA (Cui et al., 2009), SLE (Orozco et al., 2005), PS (Feng et al., 2009), and GD (Simmonds et al., 2006).
CXCR5, CCR6, and SH2B3 encode chemokine or cytokine receptors. Association analyses identified that rs638893 at 11q23.3 is associated with vitiligo in the Chinese Han population, and rs638893 is located in an intergenic region between CXCR5 and DDX6 (Tang et al., 2013). CXCR5 encodes a multi-pass membrane protein that belongs to the CXC chemokine receptor family. This cytokine receptor binds to B-lymphocyte chemoattractant (BLC), and is involved in B-cell migration into B-cell follicles of spleen and Peyer patches. CXCR5 has also been shown to have an important role in the pathogenesis of colorectal cancer (CC) (Qi et al., 2014), SLE (Zhang et al., 2014), and MS (Lill et al., 2013). At 6q27, GWAS of GV in the Chinese populations identified rs6902119 in CCR6 with genome wide significance (Quan et al., 2010). Another study showed that the most significant association SNPs rs6902119 and rs2301436 in CCR6 were observed, SNPs rs6902119 and rs2301436 are in moderate LD, and logistic regression analysis indicated that association of GV with rs2301436 might be secondary to LD with rs6902119 (Jin et al., 2010b). CCR6 encodes chemokine receptor 6 and is favorably expressed by immature dendritic cells and memory T cells (Schutyser et al., 2003). When binding to its ligand (CCL20), CCR6 may result in the chemoattraction of immune cells, which might have a role in skin and mucosal surfaces under homeostatic and inflammatory conditions (Le Borgne et al., 2006). CCR6 is also associated with inflammatory bowel disease (IBD) (Barrett et al., 2008). The GWAS-MA showed association with SNPs (nt 111708458-112906415) within and near SH2B3, particularly rs3184504 and imputed SNP rs4766578, located downstream, within ATXN2 (Jin et al., 2012a). ATXN encodes Ataxin-2, and is causal for spinocerebellar ataxia type 2. SH2B3 encodes a member of the SH2B adaptor family of proteins, which are involved in a range of signaling activities by growth factor and cytokine receptors. SH2B3 thus seems more likely relevant to vitiligo susceptibility than ATXN2. SH2B3 is also involved in the development regulation of both B and T cells, and associated with some immune diseases, including T1D (Devalliere and Charreau, 2011), RA (Coenen et al., 2009), and lupus (Li et al., 2010).
Vitiligo is one of the most common pigment disorders of the skin and hair and results from a selective destruction of melanocytes. Vitiligo patients have a progressive loss of melanocytes, predominantly in areas of skin subject to physical abrasion or at pressure points, leading to white patches appearing on the body. In normal physiological circumstances, melanin pigment is generated by the melanocytes and transferred to the surrounding keratinocytes to produce skin complexion and hair coloration (Smith and Sturm, 2010). GWASs for vitiligo have also identified some susceptible genes which showed genome-wide significant association level with the related function that influence the activity of melanocyte.
ZMIZ1 locus at 10q22, encodes a protein related to protein inhibitor of activated STAT (PIAS). PIAS3, a related member of the PIAS protein family, can inhibit the transcriptional activity of microphthalmia transcription factor (MITF), which has been demonstrated to be a key regulator of melanocyte development, function and survival (Garraway et al., 2005). Another study confirmed ZMIZ1 as a novel susceptibility locus for vitiligo and further suggested rs1408944 to be the putative causal variant that potentially interrupts TF binding and thus the transcriptional regulation of ZMIZ1 (Sun et al., 2014). In addition, ZMIZ1 might be associated with type II diabetes (T2D) (Matsuba et al., 2015), CD (Yang et al., 2015), and IBD (Jakobsen et al., 2014).
PMEL encodes a melanocyte-specific type I transmembrane glycoprotein. The encoded protein is enriched in melanosomes, which are the melanin-producing organelles in melanocytes, and plays an essential role in the structural organization of premelanosomes (McGlinchey et al., 2009). Skin biopsy transcriptome analysis found that PMEL has a decreased expression in vitiligo lesional skin compared to vitiligo perilesional normal skin (Tang et al., 2013). In addition, the antigen-specific CD8+ T cells exhibit reactivity to modified PMEL peptide epitopes in HLA-A2-positive vitiligo patients (Mandelcorn-Monson et al., 2003), which also supports the notion that there is a cell-mediated immunopathologic mechanism in vitiligo.
GWAS identifies two SNPs (rs1847134 and rs1393350) in association with the TYR gene region which showed genome-wide significance with vitiligo, and haplotype analysis reveals a strong association with a block of six SNPs (rs1018528, rs10765198, rs1847134, rs1393350, rs1126809, and rs1806319) in tight LD (Jin et al., 2010a). TYR encodes tyrosinase, an enzyme of the melanocyte that catalyzes the rate-limiting steps of melanin biosynthesis and constitutes a major autoantigen in GV (Rezaei et al., 2007). Next-generation DNA re-sequencing identifies common variants of TYR and HLA-A that modulate the risk of GV via antigen presentation (Jin et al., 2012b). The biological interaction between HLA-A and TYR shows an apparent inverse relationship between susceptibility to GV versus malignant melanoma (Spritz, 2010), suggesting that GV may result from dysregulation of normal processes of immune surveillance against melanoma.
MC1R, encoding the receptor protein for melanocyte-stimulating hormone (MSH), is a regulator of melanogenesis and minor vitiligo autoantigen, associating with malignant melanoma and with skin and hair color (Dessinioti et al., 2011). Expression shows MC1R is marked significantly different between lesional and non-lesional vitiligo skin (Kingo et al., 2007). The frequency of five MC1R coding region SNPs: Arg67Gln (G200A), Val92Met (G274A), Ile120Thr (T359C), Arg160Arg (C478A), and Gln163Arg (A488G) in Korean vitiligo patients and normal controls did not reach statistical significance (Na et al., 2003). However, another study shows that the Arg160Trp allele of MC1R gene may be able to protect against vitiligo (Szell et al., 2008). Further study need to be conducted to confirm this conclusion between vitiligo and MC1R coding region SNPs.
In the Chinese Han population, GWAS for vitiligo identifies a risk locus at 6q27, which contains three genes: RNASET2, FGFR1OP and CCR6. RNASET2 is potentially involved in tumorigenesis and associated with human malignancies and chromosomal rearrangement. Overexpression of RNASET2 hypersensitizes cells to oxidative stress, thus promoting cell death during peroxide exposure and stationary-phase onset (Thompson and Parker, 2009). Therefore, RNASET2 regulates the oxidative stress and intervenes the initial pathogenic event in melanocyte destruction in vitiligo. FGFR1OP encodes a largely hydrophilic centrosomal protein that is required for anchoring microtubules to subcellular structures. Loss of FGFR1OP causes apoptosis in the G1 phase of the cell cycle, with accumulation of a 32-kDa p53 tumor suppressor isoform and NOXA and FAS transcripts, suggesting that FGFR1OP is necessary for cell-cycle progression and survival (Acquaviva et al., 2009). Mutations in this gene not only associate with vitiligo (Quan et al., 2010), but also are associated with CD (Yang et al., 2014), GD (Chu et al., 2011).
At 15q12–q13.1, GWAS-MA shows suggestive association of SNPs spanning OCA2 upstream to within HERC2, especially SNP rs12913832 and imputed SNP rs1129038 (Jin et al., 2012a). OCA2 encode melanocyte antigens presented by HLA-A∗0215, for vitiligo protection is associated with reduced functional protein, and for susceptibility to vitiligo and melanoma constitute genetic opposites, perhaps modulating immune surveillance for melanoma (Jin et al., 2012a). Mutations in OCA2 gene associate with oculocutaneous albinism type II (Rimoldi et al., 2014) and melanoma (Hawkes et al., 2013). HERC2 belongs to the HERC gene family that encodes a group of unusually large proteins, which contain multiple structural domains. Variants within HERC2 down-regulate transcription of the OCA2 allele in cis (Jin et al., 2012a). Mutations in HERC2 are associated with the development of angelman syndrome (AS; Harlalka et al., 2013), global developmental delay (GDD), and autismspectrum disorder (AD) (Puffenberger et al., 2012).
Vitiligo is an acquired and progressive hypomelanotic disease that manifests as circumscribed depigmented patches on the skin. The interactions between melanocytes and other typical skin cells, particularly keratinocytes, may be an interpretation for the cause of vitiligo. Some genes have the role in gene repression, apoptosis and cell survival, inflammation, and cytotoxic cells, its possible involvement in the progression vitiligo.
RERE is the locus on chromosome 1p36.23, which encodes the arginine–glutamic acid dipeptide repeats protein. The protein is a transcriptional corepressor that is highly expressed in lymphoid cells and is thought to regulate apoptosis (Wang and Tsai, 2008). There are 40 SNPs in RERE were identified as having an association with GV especially rs301819 showed genome-wide significance, the most strongly associated SNPs in the RERE region were in tight LD; no haplotype showed a significantly stronger association than any component SNP (Jin et al., 2012a). Another SNP rs4908760 in RERE is the strongest associated SNP with genome-wide significance in the GWAS study (Jin et al., 2010a).
At 10q25.3, GWAS-MA for vitiligo shows suggestive association of SNPs spanning CASP7, particularly rs3814231, which is confirmed by the replication and meta-analysis study (Jin et al., 2012a). CASP7 encodes a member of the cysteine-aspartic acid protease (caspase) family, and sequential activation of caspases plays a central role in the execution-phase of cell apoptosis and inflammation (Lamkanfi and Kanneganti, 2010). CASP7 also is associated with RA (Garcia-Lozano et al., 2007), and may be a candidate gene for T1D (Babu et al., 2003).
Genome-wide association studies in European-derived whites have demonstrated genetic association between vitiligo and GZMB (Jin et al., 2012a; Jin et al., 2010a). GZMB is a caspase much alike serine protease that mediates two processes: immune-induced target-cell apoptosis mediated by cytotoxic T cells (CTLs), and natural killer cells and activation-induced cell death or type 2 helper T cells, which terminates the immune response (Trapani and Sutton, 2003; Devadas et al., 2006). Next-generation DNA re-sequencing has identified a direct causal role for the GZMB rs8192917-C-rs11539752-C haplotype (55R-94A) in the pathogenesis of GV (Ferrara et al., 2013). GZMB is only genetically associated with juvenile idiopathic arthritis (JIA; Donn et al., 2008) and Behcet’s disease (BD; Kucuksezer et al., 2009), which suggests the possibility that GZMB may be relatively specific for melanocyte-directed autoimmune susceptibility.
There are some vitiligo susceptible genes identified by GWAS and reach at the genome-wide significance level, but the function of these genes in the pathogenesis and development of vitiligo is still unclear.
At 22q13.2, the GWAS-MA showed association with SNPs in a broad region (nt 41707054-42062822), particularly rs79008, upstream of TOB2, and several imputed SNPs, including rs4822024, between ZC3H7B and TEF (Jin et al., 2012a). TOB2 locus on 22q13.2, encodes a regulator of cell cycle progression involved in T cell tolerance (Jia and Meng, 2007). However, the assignment of TOB2 as causal remains uncertain in the pathogenesis of vitiligo.
Genome-wide association studies in an isolated European population identified SMOC2 as a risk locus for GV (Birlea et al., 2010). However, another study shows that the variant rs13208776 in SMOC2 gene does not play a major role in increasing the risk of vitiligo in Jordanian Arab patients (Alkhateeb et al., 2010), maybe due to the different genetic background in these two populations. In the skin, SMOC2 is mainly present in the basal levels of the epidermis, and SMOC2-stimulated attachment of primary keratinocytes in culture (Maier et al., 2008). The study has also found that SMOC2 may play a role in autoimmune thyroid disease (AITD) susceptibility as a dominant polymorphism (Alkhateeb et al., 2013).
KIAA1005, also known as retinitis pigmentosa GTPase regulator-interacting protein 1-like (RPGRIP1L) gene, encodes a protein that can localize to the basal body-centrosome complex or to primary cilia and centrosomes in ciliated cells. In KIAA1005, the genotype and allele frequencies of 3854 G > A (1264 Asp > Asn) in vitiligo patients are significantly different compared to healthy controls. The GG frequency is lower and AA frequency is higher in vitiligo, suggesting the A allele at the KIAA1005 G3854A may increase susceptibility to vitiligo (Cheong et al., 2013). Multiple variants of the KIAA1005 gene have also been associated with certain clinical manifestations, particularly ciliopathies as in DNAH5 with neurological, renal and ocular manifestations (Delous et al., 2007).
GWAS has revealed rs1417210 at 10q22.1 to have a strong association with vitiligo (Tang et al., 2013). This SNP is located in an LD block that contains SLC29A3 and CDH23. SLC29A3 encodes a nucleoside transporter. The encoded protein plays a role in cellular uptake of nucleosides, nucleobases, and their related correspondents. Mutations in this gene have been associated with H syndrome (Molho-Pessach et al., 2008). CDH23 is a member of the cadherin superfamily, whose genes encode calcium dependent cell-cell adhesion glycoproteins. The encoded protein is thought to be involved in stereocilia organization and hair bundle formation. Whole-exome sequencing (WES) has identified that CDH23 mutations cause hearing loss in Koreans families (Woo et al., 2014).
The LPP gene locus on 3q28, encodes a member of a subfamily of LIM domain proteins that are characterized by a N-terminal proline-rich region and three C-terminal LIM domains. LPP has also been associated with celiac disease and RA (Coenen et al., 2009). Rs638893 located in an intergenic region between DDX6 and CXCR5 is associated with vitiligo (Tang et al., 2013). DDX6 encodes a member of the DEAD box protein family, which is a RNA helicase found in P-bodies and stress granules, and functions in translation suppression and mRNA degradation (Weston and Sommerville, 2006). Rs229527 in C1QTNF6 has shown to have an association with GV (Jin et al., 2010a). C1QTNF6 denotes the C1q and tumor necrosis factor–related protein 6 gene. C1QTNF6-RAC2 at 22q12.3-13.1 has reached a genome-wide significant association is a novel susceptibility loci for GD (Zhao et al., 2013), and GWAS-MA also identifies C1QTNF6 as one of risk loci for T1D (Cooper et al., 2008).
In general, genes and locus only explain partial variation of heritability (Manolio et al., 2009), gene–gene (or genetic variants) interactions are strongly believed to contribute to the genetic risk of common diseases (Cordell, 2009). Transmission disequilibrium and family based association statistical tests found the SNP markers in regions 7p13, 7q11, and 9q22 were significantly associated with GV, tagging SNPs for these regions represented by rs6960920, rs734930, and rs4744411, respectively (Spritz et al., 2004). The investigators examined the potential genetic interactions for these independently identified loci using two-way tests (and three-way tests in the context of the previously identified NLRP1 gene tagged by the rs6502867). Notably, all three SNPs showed significant interaction with the NLRP1 gene in predicting the GV phenotype (Jin et al., 2010c). The pairwise interaction analysis between 6q27, 10q22 and the two MHC SNPs (rs11966200 and rs9468925) were performed in the Chinese Han population, but no significant genetic interaction (P > 0.05 after correction for multiple testing) was identified (Quan et al., 2010).
It is observed that the autoimmunity feature of vitiligo is supported through the significant linkage to the MHC region on 6p21-p22 and evidence provided for the association of HLA-DR with vitiligo. The epistatic interaction between rs2269577 (XBP1) and HLADRB1∗07 was tested by using logistic regression analysis and found that the full model with both the main and interactive effects was better than the model with only the main effect to fit the data (Ren et al., 2009). Stratified association analysis of rs2269577 by HLA-DRB1∗07 allele has shown that the association at rs2269577 is significant in both the patients carrying and not carrying the -DRB1∗07 allele. However, the association seems to be stronger in patients carrying the HLA-DRB1∗07 allele (Ren et al., 2009). XBP1 as one of biological candidate genes for vitiligo may be due to its plausible role in the development of the disease through its interaction with HLA-DR.
GWAS identifies RNASET2 as a susceptible gene to vitiligo in the Chinese Han population (Quan et al., 2010), but the function of RNASET2 in vitiligo pathogenesis or in melanocyte apoptosis is unknown. In vitro analyses have indicated that overexpression of RNASET2 is inducible in cultured primary human melanocytes and keratinocytes through stressful conditions, exposure to ultraviolet irradiation, hydrogen peroxide, and inflammatory factors, respectively, and lead to increased cell apoptosis via the tumor necrosis factor receptor-associated factor 2 (TRAF2)-caspases pathway through the physical interaction of RNASET2 with TRAF2 (Wang et al., 2014). Hence, RNASET2 may contribute to vitiligo pathogenesis by inhibiting TRAF2 expression.
In the past five years, GWASs have contributed tremendously to the identification of key loci that were associated with the risk of developing vitiligo. These genes may provide novel therapeutic and prophylactic targets for new interventional approaches to treat and prevent vitiligo. Developments in this area will be exciting and influence the therapeutic approaches for the suppression of vitiligo in the future. We summarized and evaluated the importance of these loci in their respective molecular signaling pathways, and suggested new etiologic clues to vitiligo development. Considering that most of these genetic associations are restricted to moderate effects, large sample size studies are required in future investigations in order for these subtle variations to be detected. With the increasing number of GWASs being conducted, it is desirable to combine these findings across these studies to improve the statistical power. Meta-analysis of multiple GWASs improves the power to detect more associations, and to investigate the heterogeneity or consistency of these associations across different datasets and study populations. Beyond gene-association approaches, functional and gene-targeted assays, whole exome sequencing are required to identify the causal variants and understand their biological function.
XZ is the principal investigator for this review article and has contributed to the concept and planning of the article, collection of data, and reporting of the work described. CS, JG, YS, FZ, XZ, RK, XT, CZ, XY, and JD contributed to the planning of the article, collection of data, and reporting of the work described. XY, LS, and YC are the other principal investigators for this review article and have contributed to the concept of the manuscript, planning of the article, collection of data, and reporting of the work described. All authors contributed to drafting the article or revising it critically for important intellectual content.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This review article was supported by grants from the General Program of National Natural Science Foundation of China (81402591, 1408085QH177, 81171506, 81273302, 81000691, 81072460, 8127330).
Acquaviva, C., Chevrier, V., Chauvin, J. P., Fournier, G., Birnbaum, D., and Rosnet, O. (2009). The centrosomal FOP protein is required for cell cycle progression and survival. Cell Cycle 8, 1217–1227. doi: 10.4161/cc.8.8.8248
Addison, T. (1855). On the constitutional and local effects of disease of the suprarenal capsules. Med. Classics 1937, 244–293.
Akbas, H., Dertlioglu, S. B., Dilmec, F., and Atay, A. E. (2014). Lack of association between PTPN22 gene +1858 C>T polymorphism and susceptibility to generalized vitiligo in a Turkish population. Ann. Dermatol. 26, 88–91. doi: 10.5021/ad.2014.26.1.88
Alkhateeb, A., Al-Dain Marzouka, N., and Qarqaz, F. (2010). SMOC2 gene variant and the risk of vitiligo in Jordanian Arabs. Eur. J. Dermatol. 20, 701–704.
Alkhateeb, A., Fain, P. R., Thody, A., Bennett, D. C., and Spritz, R. A. (2003). Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res. 16, 208–214. doi: 10.1034/j.1600-0749.2003.00032.x
Alkhateeb, A., Marzouka, N. A., and Tashtoush, R. (2013). Variants in PTPN22 and SMOC2 genes and the risk of thyroid disease in the Jordanian Arab population. Endocrine 44, 702–709. doi: 10.1007/s12020-013-9908-z
Baaten, B. J., Li, C. R., and Bradley, L. M. (2010). Multifaceted regulation of T cells by CD44. Commun. Integr. Biol. 3, 508–512. doi: 10.4161/cib.3.6.13495
Babu, S. R., Bao, F., Roberts, C. M., Martin, A. K., Gowan, K., Eisenbarth, G. S., et al. (2003). Caspase 7 is a positional candidate gene for IDDM 17 in a Bedouin Arab family. Ann. N. Y. Acad. Sci. 1005, 340–343. doi: 10.1196/annals.1288.054
Barrett, J. C., Hansoul, S., Nicolae, D. L., Cho, J. H., Duerr, R. H., Rioux, J. D., et al. (2008). Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 40, 955–962. doi: 10.1038/ng.175
Basak, P. Y., Adiloglu, A. K., Koc, I. G., Tas, T., and Akkaya, V. B. (2008). Evaluation of activatory and inhibitory natural killer cell receptors in non-segmental vitiligo: a flow cytometric study. J. Eur. Acad. Dermatol. Venereol. 22, 970–976. doi: 10.1111/j.1468-3083.2008.02681.x
Begovich, A. B., Carlton, V. E., Honigberg, L. A., Schrodi, S. J., Chokkalingam, A. P., Alexander, H. C., et al. (2004). A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am. J. Hum. Genet. 75, 330–337. doi: 10.1086/422827
Birlea, S. A., Gowan, K., Fain, P. R., and Spritz, R. A. (2010). Genome-wide association study of generalized vitiligo in an isolated European founder population identifies SMOC2, in close proximity to IDDM8. J. Invest. Dermatol. 130, 798–803. doi: 10.1038/jid.2009.347
Birlea, S. A., Jin, Y., Bennett, D. C., Herbstman, D. M., Wallace, M. R., McCormack, W. T., et al. (2011). Comprehensive association analysis of candidate genes for generalized vitiligo supports XBP1, FOXP3, and TSLP. J. Invest. Dermatol. 131, 371–381. doi: 10.1038/jid.2010.337
Carr, E. J., Clatworthy, M. R., Lowe, C. E., Todd, J. A., Wong, A., Vyse, T. J., et al. (2009). Contrasting genetic association of IL2RA with SLE and ANCA-associated vasculitis. BMC Med. Genet. 10:22. doi: 10.1186/1471-2350-10-22
Chen, J. J., Huang, W., Gui, J. P., Yang, S., Zhou, F. S., Xiong, Q. G., et al. (2005). A novel linkage to generalized vitiligo on 4q13-q21 identified in a genomewide linkage analysis of Chinese families. Am. J. Hum. Genet. 76, 1057–1065. doi: 10.1086/430279
Cheong, K. A., Kim, N. H., Noh, M., and Lee, A. Y. (2013). Three new single nucleotide polymorphisms identified by a genome-wide association study in Korean patients with vitiligo. J. Korean Med. Sci. 28, 775–779. doi: 10.3346/jkms.2013.28.5.775
Chu, X., Pan, C. M., Zhao, S. X., Liang, J., Gao, G. Q., Zhang, X. M., et al. (2011). A genome-wide association study identifies two new risk loci for Graves’ disease. Nat. Genet. 43, 897–901. doi: 10.1038/ng.898
Coenen, M. J., Trynka, G., Heskamp, S., Franke, B., van Diemen, C. C., Smolonska, J., et al. (2009). Common and different genetic background for rheumatoid arthritis and coeliac disease. Hum. Mol. Genet. 18, 4195–4203. doi: 10.1093/hmg/ddp365
Concannon, P., Onengut-Gumuscu, S., Todd, J. A., Smyth, D. J., Pociot, F., Bergholdt, R., et al. (2008). A human type 1 diabetes susceptibility locus maps to chromosome 21q22.3. Diabetes 57, 2858–2861. doi: 10.2337/db08-0753
Cooper, J. D., Smyth, D. J., Smiles, A. M., Plagnol, V., Walker, N. M., Allen, J. E., et al. (2008). Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat. Genet. 40, 1399–1401. doi: 10.1038/ng.249
Cordell, H. J. (2009). Detecting gene-gene interactions that underlie human diseases. Nat. Rev. Genet. 10, 392–404. doi: 10.1038/nrg2579
Cui, J., Taylor, K. E., Destefano, A. L., Criswell, L. A., Izmailova, E. S., Parker, A., et al. (2009). Genome-wide association study of determinants of anti-cyclic citrullinated peptide antibody titer in adults with rheumatoid arthritis. Mol. Med. 15, 136–143. doi: 10.2119/molmed.2009.00008
Delous, M., Baala, L., Salomon, R., Laclef, C., Vierkotten, J., Tory, K., et al. (2007). The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat. Genet. 39, 875–881. doi: 10.1038/ng2039
Dessinioti, C., Antoniou, C., Katsambas, A., and Stratigos, A. J. (2011). Melanocortin 1 receptor variants: functional role and pigmentary associations. Photochem. Photobiol. 87, 978–987. doi: 10.1111/j.1751-1097.2011.00970.x
Devadas, S., Das, J., Liu, C., Zhang, L., Roberts, A. I., Pan, Z., et al. (2006). Granzyme B is critical for T cell receptor-induced cell death of type 2 helper T cells. Immunity 25, 237–247. doi: 10.1016/j.immuni.2006.06.011
Devalliere, J., and Charreau, B. (2011). The adaptor Lnk (SH2B3): an emerging regulator in vascular cells and a link between immune and inflammatory signaling. Biochem. Pharmacol. 82, 1391–1402. doi: 10.1016/j.bcp.2011.06.023
Donn, R., Ellison, S., Lamb, R., Day, T., Baildam, E., and Ramanan, A. V. (2008). Genetic loci contributing to hemophagocytic lymphohistiocytosis do not confer susceptibility to systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. 58, 869–874. doi: 10.1002/art.23270
Fain, P. R., Gowan, K., LaBerge, G. S., Alkhateeb, A., Stetler, G. L., Talbert, J., et al. (2003). A genomewide screen for generalized vitiligo: confirmation of AIS1 on chromosome 1p31 and evidence for additional susceptibility loci. Am. J. Hum. Genet. 72, 1560–1564. doi: 10.1086/375451
Feng, B. J., Sun, L. D., Soltani-Arabshahi, R., Bowcock, A. M., Nair, R. P., Stuart, P., et al. (2009). Multiple Loci within the major histocompatibility complex confer risk of psoriasis. PLoS Genet 5:14. doi: 10.1371/journal.pgen.1000606
Ferrara, T. M., Jin, Y., Gowan, K., Fain, P. R., and Spritz, R. A. (2013). Risk of generalized vitiligo is associated with the common 55R-94A-247H variant haplotype of GZMB (encoding granzyme B). J. Invest. Dermatol. 2013 Jun;133(6):1677-9. doi: 10.1038/jid.2013.5.
Garcia-Lozano, J. R., Torres, B., Fernandez, O., Orozco, G., Alvarez-Marquez, A., Garcia, A., et al. (2007). Caspase 7 influences susceptibility to rheumatoid arthritis. Rheumatology 46, 1243–1247. doi: 10.1093/rheumatology/kem096
Garcia-Melendez, M. E., Salinas-Santander, M., Sanchez-Dominguez, C., Gonzalez-Cardenas, H., Cerda-Flores, R. M., Ocampo-Candiani, J., et al. (2014). Protein tyrosine phosphatase PTPN22 +1858C/T polymorphism is associated with active vitiligo. Exp. Ther. Med. 8, 1433–1437.
Garraway, L. A., Widlund, H. R., Rubin, M. A., Getz, G., Berger, A. J., Ramaswamy, S., et al. (2005). Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 436, 117–122. doi: 10.1038/nature03664
Gateva, V., Sandling, J. K., Hom, G., Taylor, K. E., Chung, S. A., Sun, X., et al. (2009). A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat. Genet. 41, 1228–1233. doi: 10.1038/ng.468
Hakonarson, H., Qu, H. Q., Bradfield, J. P., Marchand, L., Kim, C. E., Glessner, J. T., et al. (2008). A novel susceptibility locus for type 1 diabetes on Chr12q13 identified by a genome-wide association study. Diabetes Metab. Res. Rev. 57, 1143–1146. doi: 10.2337/db07-1305
Harlalka, G. V., Baple, E. L., Cross, H., Kuhnle, S., Cubillos-Rojas, M., Matentzoglu, K., et al. (2013). Mutation of HERC2 causes developmental delay with Angelman-like features. J. Med. Genet. 50, 65–73. doi: 10.1136/jmedgenet-2012-101367
Hawkes, J. E., Cassidy, P. B., Manga, P., Boissy, R. E., Goldgar, D., Cannon-Albright, L., et al. (2013). Report of a novel OCA2 gene mutation and an investigation of OCA2 variants on melanoma risk in a familial melanoma pedigree. J. Dermatol. Sci. 69, 30–37. doi: 10.1016/j.jdermsci.2012.09.016
He, C., Hamon, S., Li, D., Barral-Rodriguez, S., and Ott, J. (2009). MHC fine mapping of human type 1 diabetes using the T1DGC data. Diabetes. Obes. Metab. 1, 53–59. doi: 10.1111/j.1463-1326.2008.01003.x
Hernandez, J., Aung, S., Redmond, W. L., and Sherman, L. A. (2001). Phenotypic and functional analysis of CD8(+) T cells undergoing peripheral deletion in response to cross-presentation of self-antigen. J. Exp. Med. 194, 707–717. doi: 10.1084/jem.194.6.707
Hinks, A., Ke, X., Barton, A., Eyre, S., Bowes, J., Worthington, J., et al. (2009). Association of the IL2RA/CD25 gene with juvenile idiopathic arthritis. Arthritis Rheum. 60, 251–257. doi: 10.1002/art.24187
Honda, Y., Okubo, Y., and Koga, M. (1997). Relationship between levels of soluble interleukin-2 receptors and the types and activity of vitiligo. J. Dermatol. 24, 561–563. doi: 10.1111/j.1346-8138.1997.tb02292.x
Jakobsen, C., Cleynen, I., Andersen, P. S., Vermeire, S., Munkholm, P., Paerregaard, A., et al. (2014). Genetic susceptibility and genotype-phenotype association in 588 Danish children with inflammatory bowel disease. J. Crohns Colitis 8, 678–685. doi: 10.1016/j.crohns.2013.12.010
Jia, S., and Meng, A. (2007). Tob genes in development and homeostasis. Dev. Dyn. 236, 913–921. doi: 10.1002/dvdy.21092
Jin, T. B., Ren, Y., Shi, X., Jiri, M., He, N., Feng, T., et al. (2015). Genetic variations in the CLNK gene and ZNF518B gene are associated with gout in case-control sample sets. Rheumatol. Int. 35, 1141–1147. doi: 10.1007/s00296-015-3215-3
Jin, Y., Birlea, S. A., Fain, P. R., Ferrara, T. M., Ben, S., Riccardi, S. L., et al. (2012a). Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat. Genet. 44, 676–680. doi: 10.1038/ng.2272
Jin, Y., Ferrara, T., Gowan, K., Holcomb, C., Rastrou, M., Erlich, H. A., et al. (2012b). Next-generation DNA re-sequencing identifies common variants of TYR and HLA-A that modulate the risk of generalized vitiligo via antigen presentation. J. Invest. Dermatol. 132, 1730–1733. doi: 10.1038/jid.2012.37
Jin, Y., Birlea, S. A., Fain, P. R., Gowan, K., Riccardi, S. L., Holland, P. J., et al. (2010a). Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. N. Engl. J. Med. 362, 1686–1697. doi: 10.1056/NEJMoa0908547
Jin, Y., Birlea, S. A., Fain, P. R., Mailloux, C. M., Riccardi, S. L., Gowan, K., et al. (2010b). Common variants in FOXP1 are associated with generalized vitiligo. Nat. Genet. 42, 576–578. doi: 10.1038/ng.602
Jin, Y., Riccardi, S. L., Gowan, K., Fain, P. R., and Spritz, R. A. (2010c). Fine-mapping of vitiligo susceptibility loci on chromosomes 7 and 9 and interactions with NLRP1 (NALP1). J. Invest. Dermatol. 130, 774–783. doi: 10.1038/jid.2009.273
Jin, Y., Birlea, S. A., Fain, P. R., Gowan, K., Riccardi, S. L., Holland, P. J., et al. (2011). Genome-wide analysis identifies a quantitative trait locus in the MHC class II region associated with generalized vitiligo age of onset. J. Invest. Dermatol. 131, 1308–1312. doi: 10.1038/jid.2011.12
Kato, H., Takeuchi, O., Sato, S., Yoneyama, M., Yamamoto, M., Matsui, K., et al. (2006). Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105. doi: 10.1038/nature04734
Kingo, K., Aunin, E., Karelson, M., Philips, M. A., Ratsep, R., Silm, H., et al. (2007). Gene expression analysis of melanocortin system in vitiligo. J. Dermatol. Sci. 48, 113–122. doi: 10.1016/j.jdermsci.2007.06.004
Kucuksezer, U. C., Duymaz-Tozkir, J., Gul, A., and Saruhan-Direskeneli, G. (2009). No association of granzyme B gene polymorphism with Behcet’s disease. Clin. Exp. Rheumatol. 27 (2 Suppl 53), S102.
Kumeta, H., Sakakibara, H., Enokizono, Y., Ogura, K., Horiuchi, M., Matsumoto, M., et al. (2014). The N-terminal domain of TIR domain-containing adaptor molecule-1, TICAM-1. J. Biomol. NMR 58, 227–230. doi: 10.1007/s10858-014-9819-1
Kyogoku, C., Langefeld, C. D., Ortmann, W. A., Lee, A., Selby, S., Carlton, V. E., et al. (2004). Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am. J. Hum. Genet. 75, 504–507. doi: 10.1086/423790
Laberge, G. S., Birlea, S. A., Fain, P. R., and Spritz, R. A. (2008). The PTPN22-1858C>T (R620W) functional polymorphism is associated with generalized vitiligo in the Romanian population. Pigment Cell Melanoma Res. 21, 206–208. doi: 10.1111/j.1755-148X.2008.00443.x
Lamkanfi, M., and Kanneganti, T. D. (2010). Caspase-7: a protease involved in apoptosis and inflammation. Int. J. Biochem. Cell Biol. 42, 21–24. doi: 10.1016/j.biocel.2009.09.013
Le Borgne, M., Etchart, N., Goubier, A., Lira, S. A., Sirard, J. C., van Rooijen, N., et al. (2006). Dendritic cells rapidly recruited into epithelial tissues via CCR6/CCL20 are responsible for CD8+ T cell crosspriming in vivo. Immunity 24, 191–201. doi: 10.1016/j.immuni.2006.01.005
Li, Y., Liao, W., Cargill, M., Chang, M., Matsunami, N., Feng, B. J., et al. (2010). Carriers of rare missense variants in IFIH1 are protected from psoriasis. J. Invest. Dermatol. 130, 2768–2772. doi: 10.1038/jid.2010.214
Liang, Y., Yang, S., Zhou, Y., Gui, J., Ren, Y., Chen, J., et al. (2007). Evidence for two susceptibility loci on chromosomes 22q12 and 6p21-p22 in Chinese generalized vitiligo families. J. Invest. Dermatol. 127, 2552–2557. doi: 10.1038/sj.jid.5700904
Lill, C. M., Schjeide, B. M., Graetz, C., Ban, M., Alcina, A., Ortiz, M. A., et al. (2013). MANBA, CXCR5, SOX8, RPS6KB1 and ZBTB46 are genetic risk loci for multiple sclerosis. Brain 136, 1778–1782. doi: 10.1093/brain/awt101
Liu, J., Ni, J., Leng, R. X., Pan, H. F., and Ye, D. Q. (2015). Association of UBASH3A gene polymorphisms and systemic lupus erythematosus in a Chinese population. Gene 565, 116–121. doi: 10.1016/j.gene.2015.04.005
Liu, J., Tang, H., Zuo, X., Liang, B., Wang, P., Sun, L., et al. (2012). A single nucleotide polymorphism rs9468925 of MHC region is associated with clinical features of generalized vitiligo in Chinese Han population. J. Eur. Acad. Dermatol. Venereol. 26, 1137–1141. doi: 10.1111/j.1468-3083.2011.04259.x
Liu, J. B., Li, M., Chen, H., Zhong, S. Q., Yang, S., Du, W. D., et al. (2007). Association of vitiligo with HLA-A2: a meta-analysis. J. Eur. Acad. Dermatol. Venereol. 21, 205–213. doi: 10.1111/j.1468-3083.2006.01899.x
Maier, S., Paulsson, M., and Hartmann, U. (2008). The widely expressed extracellular matrix protein SMOC-2 promotes keratinocyte attachment and migration. Exp. Cell Res. 314, 2477–2487. doi: 10.1016/j.yexcr.2008.05.020
Mandelcorn-Monson, R. L., Shear, N. H., Yau, E., Sambhara, S., Barber, B. H., Spaner, D., et al. (2003). Cytotoxic T lymphocyte reactivity to gp100, MelanA/MART-1, and tyrosinase, in HLA-A2-positive vitiligo patients. J. Invest. Dermatol. 121, 550–556. doi: 10.1046/j.1523-1747.2003.12413.x
Manolio, T. A., Collins, F. S., Cox, N. J., Goldstein, D. B., Hindorff, L. A., Hunter, D. J., et al. (2009). Finding the missing heritability of complex diseases. Nature 461, 747–753. doi: 10.1038/nature08494
Mansha, M., Carlet, M., Ploner, C., Gruber, G., Wasim, M., Wiegers, G. J., et al. (2010). Functional analyses of Src-like adaptor (SLA), a glucocorticoid-regulated gene in acute lymphoblastic leukemia. Leuk. Res. 34, 529–534. doi: 10.1016/j.leukres.2009.06.029
Martinez, A., Santiago, J. L., Cenit, M. C., de Las Heras, V., de la Calle, H., Fernandez-Arquero, M., et al. (2008). IFIH1-GCA-KCNH7 locus: influence on multiple sclerosis risk. Eur. J. Hum. Genet. 16, 861–864. doi: 10.1038/ejhg.2008.16
Matsuba, R., Sakai, K., Imamura, M., Tanaka, Y., Iwata, M., Hirose, H., et al. (2015). Replication study in a japanese population to evaluate the association between 10 SNP loci, identified in european genome-wide association studies, and type 2 diabetes. PLoS ONE 10:e0126363. doi: 10.1371/journal.pone.0126363
McGlinchey, R. P., Shewmaker, F., McPhie, P., Monterroso, B., Thurber, K., and Wickner, R. B. (2009). The repeat domain of the melanosome fibril protein Pmel17 forms the amyloid core promoting melanin synthesis. Proc. Natl. Acad. Sci. U.S.A. 106, 13731–13736. doi: 10.1073/pnas.0906509106
Mohammed, G. F., Gomaa, A. H., and Al-Dhubaibi, M. S. (2015). Highlights in pathogenesis of vitiligo. World J. Clin. Cases 3, 221–230. doi: 10.12998/wjcc.v3.i3.221
Molho-Pessach, V., Lerer, I., Abeliovich, D., Agha, Z., Abu Libdeh, A., Broshtilova, V., et al. (2008). The H syndrome is caused by mutations in the nucleoside transporter hENT3. Am. J. Hum. Genet. 83, 529–534. doi: 10.1016/j.ajhg.2008.09.013
Na, G. Y., Lee, K. H., Kim, M. K., Lee, S. J., Kim, D. W., and Kim, J. C. (2003). Polymorphisms in the melanocortin-1 receptor (MC1R) and agouti signaling protein (ASIP) genes in Korean vitiligo patients. Pigment. Cell Res. 16, 383–387. doi: 10.1034/j.1600-0749.2003.00062.x
Orozco, G., Eerligh, P., Sanchez, E., Zhernakova, S., Roep, B. O., Gonzalez-Gay, M. A., et al. (2005). Analysis of a functional BTNL2 polymorphism in type 1 diabetes, rheumatoid arthritis, and systemic lupus erythematosus. Hum. Immunol. 66, 1235–1241. doi: 10.1016/j.humimm.2006.02.003
Pan, F., Yu, H., Dang, E. V., Barbi, J., Pan, X., Grosso, J. F., et al. (2009). Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science 325, 1142–1146. doi: 10.1126/science.1176077
Peach, R. J., Bajorath, J., Naemura, J., Leytze, G., Greene, J., Aruffo, A., et al. (1995). Both extracellular immunoglobin-like domains of CD80 contain residues critical for binding T cell surface receptors CTLA-4 and CD28. J. Biol. Chem. 270, 21181–21187. doi: 10.1074/jbc.270.36.21181
Petukhova, L., Duvic, M., Hordinsky, M., Norris, D., Price, V., Shimomura, Y., et al. (2010). Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature 466, 113–117. doi: 10.1038/nature09114
Puffenberger, E. G., Jinks, R. N., Wang, H., Xin, B., Fiorentini, C., Sherman, E. A., et al. (2012). A homozygous missense mutation in HERC2 associated with global developmental delay and autism spectrum disorder. Hum. Mutat. 33, 1639–1646. doi: 10.1002/humu.22237
Qi, X. W., Xia, S. H., Yin, Y., Jin, L. F., Pu, Y., Hua, D., et al. (2014). Expression features of CXCR5 and its ligand, CXCL13 associated with poor prognosis of advanced colorectal cancer. Eur. Rev. Med. Pharmacol. Sci. 18, 1916–1924.
Quan, C., Ren, Y. -Q., Xiang, L. -H., Sun, L. -D., Xu, A. -E., Gao, X. -H., et al. (2010). Genome-wide association study for vitiligo identifies susceptibility loci at 6q27 and the MHC. Nat. Genet. 42, 614–618. doi: 10.1038/ng.603
Ramos, P. S., Williams, A. H., Ziegler, J. T., Comeau, M. E., Guy, R. T., Lessard, C. J., et al. (2011). Genetic analyses of interferon pathway-related genes reveal multiple new loci associated with systemic lupus erythematosus. Arthritis Rheum. 63, 2049–2057. doi: 10.1002/art.30356
Ren, Y., Yang, S., Xu, S., Gao, M., Huang, W., Gao, T., et al. (2009). Genetic variation of promoter sequence modulates XBP1 expression and genetic risk for vitiligo. PLoS Genet 5:e1000523. doi: 10.1371/journal.pgen.100052319
Rezaei, N., Gavalas, N. G., Weetman, A. P., and Kemp, E. H. (2007). Autoimmunity as an aetiological factor in vitiligo. J. Eur. Acad. Dermatol. Venereol. 21, 865–876. doi: 10.1111/j.1468-3083.2007.02228.x
Rimoldi, V., Straniero, L., Asselta, R., Mauri, L., Manfredini, E., Penco, S., et al. (2014). Functional characterization of two novel splicing mutations in the OCA2 gene associated with oculocutaneous albinism type II. Gene 537, 79–84. doi: 10.1016/j.gene.2013.11.102
Roychoudhuri, R., Hirahara, K., Mousavi, K., Clever, D., Klebanoff, C. A., Bonelli, M., et al. (2013). BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature 498, 506–510. doi: 10.1038/nature12199
Sasaki, S., Ito, E., Toki, T., Maekawa, T., Kanezaki, R., Umenai, T., et al. (2000). Cloning and expression of human B cell-specific transcription factor BACH2 mapped to chromosome 6q15. Oncogene 19, 3739–3749. doi: 10.1038/sj.onc.1203716
Schutyser, E., Struyf, S., and Van Damme, J. (2003). The CC chemokine CCL20 and its receptor CCR6. Cytokine Growth Factor. Rev. 14, 409–426. doi: 10.1016/S1359-6101(03)00049-2
Seya, T., Matsumoto, M., Ebihara, T., and Oshiumi, H. (2009). Functional evolution of the TICAM-1 pathway for extrinsic RNA sensing. Immunol. Rev. 227, 44–53. doi: 10.1111/j.1600-065X.2008.00723.x
Simmonds, M. J., Heward, J. M., Barrett, J. C., Franklyn, J. A., and Gough, S. C. (2006). Association of the BTNL2 rs2076530 single nucleotide polymorphism with Graves’ disease appears to be secondary to DRB1 exon 2 position beta74. Clin. Endocrinol. 65, 429–432. doi: 10.1111/j.1365-2265.2006.02586.x
Smith, A. G., and Sturm, R. A. (2010). Multiple genes and locus interactions in susceptibility to vitiligo. J. Invest. Dermatol. 130, 643–645. doi: 10.1038/jid.2009.403
Smyth, D. J., Cooper, J. D., Bailey, R., Field, S., Burren, O., Smink, L. J., et al. (2006). A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat. Genet. 38, 617–619. doi: 10.1038/ng1800
Song, G. G., Kim, J. H., and Lee, Y. H. (2013). The CTLA-4 +49 A/G, CT60 A/G and PTPN22 1858 C/T polymorphisms and susceptibility to vitiligo: a meta-analysis. Mol. Biol. Rep. 40, 2985–2993. doi: 10.1007/s11033-012-2370-9
Spritz, R. A. (2008). The genetics of generalized vitiligo. Curr. Dir. Autoimmun. 10, 244–257. doi: 10.1159/000131501
Spritz, R. A. (2010). The genetics of generalized vitiligo: autoimmune pathways and an inverse relationship with malignant melanoma. Genome Med 2, 78. doi: 10.1186/gm199
Spritz, R. A. (2012). Six decades of vitiligo genetics: genome-wide studies provide insights into autoimmune pathogenesis. J. Invest. Dermatol. 132, 268–273. doi: 10.1038/jid.2011.321
Spritz, R. A., Gowan, K., Bennett, D. C., and Fain, P. R., (2004). Novel vitiligo susceptibility loci on chromosomes 7 (AIS2) and 8 (AIS3), confirmation of SLEV1 on chromosome 17, and their roles in an autoimmune diathesis. Am. J. Hum. Genet. 4, 188–191. doi: 10.1086/381134
Stüttgen, G. (1950). Die Vitilgio in erbbiologischer Betrachtung. Z. Haut. Geschlenkskr. 9, 451–456.
Sun, X., Xu, A., Wei, X., Ouyang, J., Lu, L., Chen, M., et al. (2006). Genetic epidemiology of vitiligo: a study of 815 probands and their families from south China. Int. J. Dermatol. 45, 1176–1181. doi: 10.1111/j.1365-4632.2006.02907.x
Sun, Y., Zuo, X., Zheng, X., Zhou, F., Liang, B., Liu, H., et al. (2014). A comprehensive association analysis confirms ZMIZ1 to be a susceptibility gene for vitiligo in Chinese population. J. Med. Genet. 51, 345–353. doi: 10.1136/jmedgenet-2013-102233
Sutherland, A., Davies, J., Owen, C. J., Vaikkakara, S., Walker, C., Cheetham, T. D., et al. (2007). Genomic polymorphism at the interferon-induced helicase (IFIH1) locus contributes to Graves’ disease susceptibility. J. Clin. Endocrinol. Metab. 92, 3338–3341. doi: 10.1210/jc.2007-0173
Szell, M., Baltas, E., Bodai, L., Bata-Csorgo, Z., Nagy, N., Dallos, A., et al. (2008). The Arg160Trp allele of melanocortin-1 receptor gene might protect against vitiligo. Photochem. Photobiol. 84, 565–571. doi: 10.1111/j.1751-1097.2008.00296.x
Tang, X. F., Zhang, Z., Hu, D. Y., Xu, A. E., Zhou, H. S., Sun, L. D., et al. (2013). Association analyses identify three susceptibility Loci for vitiligo in the Chinese Han population. J. Invest. Dermatol. 133, 403–410. doi: 10.1038/jid.2012.320
Tastan, H. B., Akar, A., Orkunoglu, F. E., Arca, E., and Inal, A. (2004). Association of HLA class I antigens and HLA class II alleles with vitiligo in a Turkish population. Pigment. Cell Res. 17, 181–184. doi: 10.1111/j.1600-0749.2004.00141.x
Thompson, D. M., and Parker, R. (2009). The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J. Cell Biol. 185, 43–50. doi: 10.1083/jcb.200811119
Tomer, Y., and Greenberg, D. (2004). The thyroglobulin gene as the first thyroid-specific susceptibility gene for autoimmune thyroid disease. Trends Mol. Med. 10, 306–308. doi: 10.1016/j.molmed.2004.05.002
Trapani, J. A., and Sutton, V. R. (2003). Granzyme B: pro-apoptotic, antiviral and antitumor functions. Curr. Opin. Immunol. 15, 533–543. doi: 10.1016/S0952-7915(03)00107-9
Vella, A., Cooper, J. D., Lowe, C. E., Walker, N., Nutland, S., Widmer, B., et al. (2005). Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. Am. J. Hum. Genet. 76, 773–779. doi: 10.1086/429843
Wang, J., Wicker, L. S., and Santamaria, P. (2009). IL-2 and its high-affinity receptor: genetic control of immunoregulation and autoimmunity. Semin. Immunol. 21, 363–371. doi: 10.1016/j.smim.2009.04.004
Wang, L., and Tsai, C. C. (2008). Atrophin proteins: an overview of a new class of nuclear receptor corepressors. Nucl. Recept. Signal. 6, 31.
Wang, Q., Jiang, M., Wu, J., Ma, Y., Li, T., Chen, Q., et al. (2014). Stress-induced RNASET2 overexpression mediates melanocyte apoptosis via the TRAF2 pathway in vitro. Cell Death Dis. 23, 539. doi: 10.1038/cddis.2013.539
Weston, A., and Sommerville, J. (2006). Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res. 34, 3082–3094. doi: 10.1093/nar/gkl409
Woo, H. M., Park, H. J., Park, M. H., Kim, B. Y., Shin, J. W., Yoo, W. G., et al. (2014). Identification of CDH23 mutations in Korean families with hearing loss by whole-exome sequencing. BMC Med. Genet. 15:46. doi: 10.1186/1471-2350-15-46
Wu, J. N., and Koretzky, G. A. (2004). The SLP-76 family of adapter proteins. Semin. Immunol. 16, 379–393. doi: 10.1016/j.smim.2004.08.018
Yang, S. K., Hong, M., Choi, H., Zhao, W., Jung, Y., Haritunians, T., et al. (2015). Immunochip analysis identification of 6 additional susceptibility loci for Crohn’s disease in Koreans. Inflamm. Bowel Dis. 21, 1–7. doi: 10.1097/MIB.0000000000000268
Yang, S. K., Hong, M., Zhao, W., Jung, Y., Baek, J., Tayebi, N., et al. (2014). Genome-wide association study of Crohn’s disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut 63, 80–87. doi: 10.1136/gutjnl-2013-305193
Zhang, J., Zhang, Y., Yang, J., Zhang, L., Sun, L., Pan, H. F., et al. (2014). Three SNPs in chromosome 11q23.3 are independently associated with systemic lupus erythematosus in Asians. Hum. Mol. Genet. 23, 524–533. doi: 10.1093/hmg/ddt424
Zhang, X. J., Liu, H. S., Liang, Y. H., Sun, L. D., Wang, J. Y., Yang, S., et al. (2004a). Association of HLA class I alleles with vitiligo in Chinese Hans. J. Dermatol. Sci. 35, 165–168. doi: 10.1016/j.jdermsci.2004.05.003
Zhang, X. J., Liu, J. B., Gui, J. P., Li, M., Xiong, Q. G., Wu, H. B., et al. (2004b). Characteristics of genetic epidemiology and genetic models for vitiligo. J. Am. Acad. Dermatol. 51, 383–390. doi: 10.1016/j.jaad.2003.12.044
Zhang, Z., Xu, S. X., Zhang, F. Y., Yin, X. Y., Yang, S., Xiao, F. L., et al. (2009). The analysis of genetics and associated autoimmune diseases in Chinese vitiligo patients. Arch. Dermatol. Res. 301, 167–173. doi: 10.1007/s00403-008-0900-z
Zhao, S. X., Xue, L. Q., Liu, W., Gu, Z. H., Pan, C. M., Yang, S. Y., et al. (2013). Robust evidence for five new Graves’ disease risk loci from a staged genome-wide association analysis. Hum. Mol. Genet. 22, 3347–3362. doi: 10.1093/hmg/ddt183
Keywords: Vitiligo, GWASs, genetic susceptibility, genes, loci
Citation: Shen C, Gao J, Sheng Y, Dou J, Zhou F, Zheng X, Ko R, Tang X, Zhu C, Yin X, Sun L, Cui Y and Zhang X (2016) Genetic Susceptibility to Vitiligo: GWAS Approaches for Identifying Vitiligo Susceptibility Genes and Loci. Front. Genet. 7:3. doi: 10.3389/fgene.2016.00003
Received: 14 October 2015; Accepted: 11 January 2016;
Published: 01 February 2016.
Edited by:
Elizabeth Helen Kemp, University of Sheffield, UKReviewed by:
Sulev Kõuks, University of Tartu, EstoniaCopyright © 2016 Shen, Gao, Sheng, Dou, Zhou, Zheng, Ko, Tang, Zhu, Yin, Sun, Cui and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuejun Zhang, YXl6eGpAdmlwLnNpbmEuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.