- 1Department of Immunology and Microbial Science, The Scripps Research Institute, La Jolla, CA, USA
- 2Kellogg School of Science and Technology, The Scripps Research Institute, La Jolla, CA, USA
MicroRNAs (miRNAs) are endogenously encoded single-stranded RNAs of about 22 nucleotides (nts) in length that play essential roles in a large variety of physiological processes in animals and plants (Ambros, 2004; Bushati and Cohen, 2007). Mature miRNAs are integrated into the RNA-induced silencing complex (RISC), whose core component is one of the Argonaute family proteins. MiRNAs then direct RISCs to target mRNAs, which are recognized through partial sequence complementarity. Bioinformatic prediction and experimental target gene identification have shown that a miRNA binds mRNAs of hundreds of protein coding genes, which often span a broad spectrum of functional categories (Bartel, 2009; Chi et al., 2009; Hafner et al., 2010). The functional consequence of miRNA-target mRNA interaction and the mechanism of miRNA action have been under intensive investigation and remain a matter of hot debate. It was initially thought that miRNAs repress the protein output of a small number of target genes without significantly affecting their mRNA levels in animals (Lee et al., 1993; Wightman et al., 1993). Subsequent genetic studies in C. elegans and zebrafish showed that miRNAs promote the degradation of their target mRNAs (Bagga et al., 2005; Giraldez et al., 2006). Later, a series of genome-wide studies of in vitro cultured mammalian cell lines transiently transfected with chemically synthesized miRNA mimics led to the conclusion that the predominant functional consequence of miRNA action is target mRNA degradation (Guo et al., 2010). A follow-up study employing temporal dissection of zebrafish development seems to reconcile these two opposite observations by revealing that translational repression precedes target mRNA decay, and suggesting that the immediate outcome of miRNA-target mRNA interaction is translation inhibition but mRNA degradation can follow (Bazzini et al., 2012). Similarly, re-analysis of the previous datasets from cultured cell lines transiently transfected with synthetic miRNA mimics also found that translation repression precedes mRNA degradation (Larsson and Nadon, 2013).
However, the model miRNA used in the aforementioned zebrafish study, miR-430, is unique in that its expression is rapidly induced and reaches millions of copies per cell in a few hours after fertilization. This expression level of miR-430 is at least 10 times more than all mature miRNAs combined in a mammalian cell, and serves the single purpose of degrading its target genes, maternal mRNAs, at the maternal-zygotic transition (Giraldez et al., 2006). Mammalian cells often express 100–200 different species of miRNAs (Kuchen et al., 2010), with a total amount of 1–2 × 105 copies of mature miRNAs in a cell (Calabrese et al., 2007; Janas et al., 2012). The most abundant miRNAs are often expressed at the level of ~2 × 104 copies per cell (Neilson et al., 2007; Kuchen et al., 2010). As an extreme example, miR-122 is expressed at the estimated level of 5 × 104 copies per cell in hepatocytes (Chang et al., 2004; Jopling et al., 2005). This is still about 20 times lower than the million-copy-per-cell expression level of miR-430 in zebrafish embryos. Considering that the estimated copy number of Argonaute proteins in a mammalian cell is of the same order of magnitude as the total amount of mature miRNAs (1.5 × 104–1.7 × 105; Janas et al., 2012; Wang et al., 2012), the million-copy-per-cell expression level of miR-430 is unlikely to be physiologically relevant in mammalian cells. Therefore, the in vivo mechanism of action of mammalian miRNAs remains to be a central question in the field of miRNA research.
In contrast to these desperate efforts to search for a unified model of miRNA mechanism of action, studies of individual functional targets in primary cells or tissues from miRNA mutant mice are painting a rather different picture. Depending on miRNAs, target genes, and cellular contexts, the outcome of miRNA-target mRNA interactions could be predominantly translation repression or mRNA degradation, or a mixture of both. This heterogeneity in miRNA mechanisms of action has been increasingly recognized as more and more miRNA mutant mice are generated and analyzed (Olive et al., 2015), but a comprehensive review of relevant literature is still missing.
Here we sought to summarize the relative contribution of translation repression and mRNA degradation to miRNA regulation of functional targets in miRNA mutant mice. We focused on miRNA target genes whose protein and mRNA levels were measured concurrently in primary cells or tissues from mutant mice with genetic ablation or transgenic expression of individual miRNA genes. This includes a total of 159 target genes from 77 miRNA mutant mice (Table S1; Zhao et al., 2005, 2007; Lu et al., 2007, 2009, 2014; van Rooij et al., 2007; Vigorito et al., 2007; Dorsett et al., 2008; Liu et al., 2008, 2011, 2012, 2014; Wang et al., 2008, 2013a,b, 2014, 2015a,b; Boettger et al., 2009; Callis et al., 2009; O'Connell et al., 2009, 2010; Poy et al., 2009; Shan et al., 2009; Williams et al., 2009; Xin et al., 2009; Miyaki et al., 2010; Patrick et al., 2010; Yu et al., 2010; Biton et al., 2011; Boldin et al., 2011; Dunand-Sauthier et al., 2011; Jiang et al., 2011; Jordan et al., 2011; Ma et al., 2011, 2013; Nakamura et al., 2011; Sanuki et al., 2011; Shibata et al., 2011; Aurora et al., 2012; Callegari et al., 2012; Caruso et al., 2012; Dong et al., 2012; Gurha et al., 2012; Horie et al., 2012, 2013; Hsu et al., 2012; Liang et al., 2012, 2015; Mori et al., 2012; Tsai et al., 2012; Ucar et al., 2012; Wei et al., 2012, 2014; Zhuang et al., 2012; Belkaya et al., 2013; Bian et al., 2013; Danielson et al., 2013; Dorhoi et al., 2013; Dudda et al., 2013; Gebeshuber et al., 2013; Guo et al., 2013; Hasuwa et al., 2013; Heidersbach et al., 2013; Henao-Mejia et al., 2013; Khan et al., 2013; Mok et al., 2013; Song et al., 2013, 2014; Stadthagen et al., 2013; Tan et al., 2013; Wystub et al., 2013; Agudo et al., 2014; Ahmed et al., 2014; Burger et al., 2014; Chapnik et al., 2014; Dahan et al., 2014; Escobar et al., 2014; Giusti et al., 2014; Hu et al., 2014; Krzeszinski et al., 2014; Latreille et al., 2014; Pan et al., 2014; Stickel et al., 2014; Cushing et al., 2015; Jin et al., 2015; Kosaka et al., 2015; Kramer et al., 2015; Li et al., 2015a,b,c; Parchem et al., 2015; Sullivan et al., 2015; Sun et al., 2015; Tung et al., 2015; Xu et al., 2015; Yan et al., 2015; Zhang et al., 2015). Our analysis showed that 48% target genes are predominantly regulated by translation repression (76/159), 29% are regulated mainly by mRNA degradation (46/159), and 23% are regulated by both (37/159) (Figure 1). It is still unclear what determines the dominant mode of miRNA mechanism of action. As most of these studies measured target gene mRNA and protein levels under steady-state conditions, we speculate that differences in miRNA mechanism of action are not solely determined by the expression kinetics of miRNA or target mRNAs (Bazzini et al., 2012; Béthune et al., 2012; Djuranovic et al., 2012), but are instead attributed to cell type-, target mRNA-, or even miRNA-specific factors. Interestingly, almost all target genes identified in developing cells or tissues are mainly regulated by mRNA degradation, such as day 0 or day 2.5 cardiac cells (Heidersbach et al., 2013; Wei et al., 2014), embryonic stem cell-derived neurons (Tung et al., 2015), thymocytes (Belkaya et al., 2013; Henao-Mejia et al., 2013; Burger et al., 2014), bone marrow cells (Song et al., 2013), embryonic heart (Wystub et al., 2013; Liang et al., 2015), embryonic yolk sac (Wang et al., 2008), embryonic and neonatal epithelium (Ahmed et al., 2014), and fetal liver (Patrick et al., 2010). This is in sharp contrast to target genes identified in terminally differentiated cells, which are predominantly regulated by translation repression. It is conceivable that mRNA degradation gets rid of target gene mRNAs in a non-reversible way and provides an efficient way for cell fate determination, while translation repression is immediate, transient, and reversible, which is more suitable for differentiated cells to respond to environmental stresses. Our analysis also suggests miRNA-specific functional consequences. Several groups independently observed that target genes of miR-17~92 (Lu et al., 2007; Shan et al., 2009; Jiang et al., 2011; Bian et al., 2013; Danielson et al., 2013; Jin et al., 2015), miR-214 (Aurora et al., 2012; Wang et al., 2013b; Li et al., 2015b), miR-143/145 (Boettger et al., 2009; Jordan et al., 2011; Caruso et al., 2012; Dahan et al., 2014), and miR-146 (Boldin et al., 2011; Guo et al., 2013; Stickel et al., 2014) tend to be regulated at the translational level, but target genes of miR-122 (Hsu et al., 2012; Tsai et al., 2012), miR-140 (Miyaki et al., 2010; Nakamura et al., 2011) and miR-142 (Chapnik et al., 2014; Kramer et al., 2015; Sun et al., 2015) are often regulated by mRNA degradation. Interestingly, among miR-155 target genes, some are predominantly regulated by translation repression, some are mainly regulated by mRNA degradation, while the others are regulated by both mechanisms (Vigorito et al., 2007; Dorsett et al., 2008; Lu et al., 2009, 2014; O'Connell et al., 2009, 2010; Dudda et al., 2013; Escobar et al., 2014; Hu et al., 2014; Jin et al., 2015; Wang et al., 2015a), suggesting that different target genes of the same miRNA can be regulated through different mechanisms even in the same cell. It is a tempting possibility that cis-elements in mature miRNAs and target mRNAs determine the mechanism of miRNA action. Future investigation is warranted to identify these cis-elements, if they exist at all.
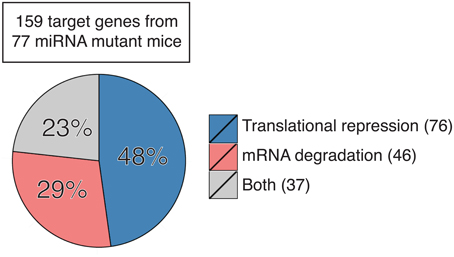
Figure 1. Contribution of translation repression and mRNA degradation to miRNA regulation of target gene expression in primary cells and tissues from miRNA mutant mice. Number in parentheses indicates the number of genes in each category. See also Table S1 for detailed information.
From a practical standpoint, measuring target gene protein levels is preferred to mRNA levels for the purpose of studying the effect of a miRNA on its target genes. Even for target genes predominantly regulated by mRNA degradation, the miRNA effect can still be captured by measuring their protein abundance. In the same vein, translatome analysis is more appropriate for measuring the global effect of a miRNA on its target genes, while transcriptome analysis often failed to identify any significant effect of miRNA deletion on its target genes, despite the obvious functional consequences in mutant mice (Matkovich et al., 2010; Boldin et al., 2011; Jiang et al., 2011; Agudo et al., 2014; Sullivan et al., 2015; Yuan et al., 2015). In the broader context of gene expression regulation, accumulating evidence shows that proteome and transcriptome are not sufficiently correlated to act as proxies for each other (Payne, 2015). miRNA-mediated translation regulation may play an important role in the de-coupling of translatome from transcriptome. We speculate that miRNAs emerged during evolution to increase the complexity of gene regulation, thereby contributing to the diversity of organisms.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Li-Fan Lu (UCSD) for discussion and Jovan Shepherd for critical reading of this manuscript. This study is supported by the PEW Charitable Trusts, Cancer Research Institute, Lupus Research Institute, and National Institute of Health (R01AI087634, R01AI089854, and R56 AI110403 to CX). CX is a Pew Scholar in Biomedical Sciences.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fgene.2015.00328
References
Agudo, J., Ruzo, A., Tung, N., Salmon, H., Leboeuf, M., Hashimoto, D., et al. (2014). The miR-126-VEGFR2 axis controls the innate response to pathogen-associated nucleic acids. Nat. Immunol. 15, 54–62. doi: 10.1038/ni.2767
Ahmed, M. I., Alam, M., Emelianov, V. U., Poterlowicz, K., Patel, A., Sharov, A. A., et al. (2014). MicroRNA-214 controls skin and hair follicle development by modulating the activity of the Wnt pathway. J. Cell Biol. 207, 549–567. doi: 10.1083/jcb.201404001
Aurora, A. B., Mahmoud, A. I., Luo, X., Johnson, B. A., van Rooij, E., Matsuzaki, S., et al. (2012). MicroRNA-214 protects the mouse heart from ischemic injury by controlling Ca(2)(+) overload and cell death. J. Clin. Invest. 122, 1222–1232. doi: 10.1172/JCI59327
Bagga, S., Bracht, J., Hunter, S., Massirer, K., Holtz, J., Eachus, R., et al. (2005). Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122, 553–563. doi: 10.1016/j.cell.2005.07.031
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Bazzini, A. A., Lee, M. T., and Giraldez, A. J. (2012). Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336, 233–237. doi: 10.1126/science.1215704
Belkaya, S., Murray, S. E., Eitson, J. L., de la Morena, M. T., Forman, J. A., and van Oers, N. S. (2013). Transgenic expression of microRNA-185 causes a developmental arrest of T cells by targeting multiple genes including Mzb1. J. Biol. Chem. 288, 30752–30762. doi: 10.1074/jbc.M113.503532
Béthune, J., Artus-Revel, C. G., and Filipowicz, W. (2012). Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 13, 716–723. doi: 10.1038/embor.2012.82
Bian, S., Hong, J., Li, Q., Schebelle, L., Pollock, A., Knauss, J. L., et al. (2013). MicroRNA cluster miR-17-92 regulates neural stem cell expansion and transition to intermediate progenitors in the developing mouse neocortex. Cell Rep. 3, 1398–1406. doi: 10.1016/j.celrep.2013.03.037
Biton, M., Levin, A., Slyper, M., Alkalay, I., Horwitz, E., Mor, H., et al. (2011). Epithelial microRNAs regulate gut mucosal immunity via epithelium-T cell crosstalk. Nat. Immunol. 12, 239–246. doi: 10.1038/ni.1994
Boettger, T., Beetz, N., Kostin, S., Schneider, J., Krüger, M., Hein, L., et al. (2009). Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J. Clin. Invest. 119, 2634–2647. doi: 10.1172/JCI38864
Boldin, M. P., Taganov, K. D., Rao, D. S., Yang, L., Zhao, J. L., Kalwani, M., et al. (2011). miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 208, 1189–1201. doi: 10.1084/jem.20101823
Burger, M. L., Xue, L., Sun, Y., Kang, C., and Winoto, A. (2014). Premalignant PTEN-deficient thymocytes activate microRNAs miR-146a and miR-146b as a cellular defense against malignant transformation. Blood 123, 4089–4100. doi: 10.1182/blood-2013-11-539411
Bushati, N., and Cohen, S. M. (2007). microRNA functions. Annu. Rev. Cell Dev. Biol. 23, 175–205. doi: 10.1146/annurev.cellbio.23.090506.123406
Calabrese, J. M., Seila, A. C., Yeo, G. W., and Sharp, P. A. (2007). RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 104, 18097–18102. doi: 10.1073/pnas.0709193104
Callegari, E., Elamin, B. K., Giannone, F., Milazzo, M., Altavilla, G., Fornari, F., et al. (2012). Liver tumorigenicity promoted by microRNA-221 in a mouse transgenic model. Hepatology 56, 1025–1033. doi: 10.1002/hep.25747
Callis, T. E., Pandya, K., Seok, H. Y., Tang, R. H., Tatsuguchi, M., Huang, Z. P., et al. (2009). MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J. Clin. Invest. 119, 2772–2786. doi: 10.1172/JCI36154
Caruso, P., Dempsie, Y., Stevens, H. C., McDonald, R. A., Long, L., Lu, R., et al. (2012). A role for miR-145 in pulmonary arterial hypertension: evidence from mouse models and patient samples. Circ. Res. 111, 290–300. doi: 10.1161/CIRCRESAHA.112.267591
Chang, J., Nicolas, E., Marks, D., Sander, C., Lerro, A., Buendia, M. A., et al. (2004). miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 1, 106–113. doi: 10.4161/rna.1.2.1066
Chapnik, E., Rivkin, N., Mildner, A., Beck, G., Pasvolsky, R., Metzl-Raz, E., et al. (2014). miR-142 orchestrates a network of actin cytoskeleton regulators during megakaryopoiesis. Elife 3:e01964. doi: 10.7554/eLife.01964
Chi, S. W., Zang, J. B., Mele, A., and Darnell, R. B. (2009). Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460, 479–486. doi: 10.1038/nature08170
Cushing, L., Costinean, S., Xu, W., Jiang, Z., Madden, L., Kuang, P., et al. (2015). Disruption of miR-29 leads to aberrant differentiation of smooth muscle cells selectively associated with distal lung vasculature. PLoS Genet. 11:e1005238. doi: 10.1371/journal.pgen.1005238
Dahan, D., Ekman, M., Larsson-Callerfelt, A. K., Turczynska, K., Boettger, T., Braun, T., et al. (2014). Induction of angiotensin-converting enzyme after miR-143/145 deletion is critical for impaired smooth muscle contractility. Am. J. Physiol. Cell Physiol. 307, C1093–C1101. doi: 10.1152/ajpcell.00250.2014
Danielson, L. S., Park, D. S., Rotllan, N., Chamorro-Jorganes, A., Guijarro, M. V., Fernandez-Hernando, C., et al. (2013). Cardiovascular dysregulation of miR-17-92 causes a lethal hypertrophic cardiomyopathy and arrhythmogenesis. FASEB J. 27, 1460–1467. doi: 10.1096/fj.12-221994
Djuranovic, S., Nahvi, A., and Green, R. (2012). miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336, 237–240. doi: 10.1126/science.1215691
Dong, C., Wang, H., Xue, L., Dong, Y., Yang, L., Fan, R., et al. (2012). Coat color determination by miR-137 mediated down-regulation of microphthalmia-associated transcription factor in a mouse model. RNA 18, 1679–1686. doi: 10.1261/rna.033977.112
Dorhoi, A., Iannaccone, M., Farinacci, M., Faé, K. C., Schreiber, J., Moura-Alves, P., et al. (2013). MicroRNA-223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. J. Clin. Invest. 123, 4836–4848. doi: 10.1172/JCI67604
Dorsett, Y., McBride, K. M., Jankovic, M., Gazumyan, A., Thai, T. H., Robbiani, D. F., et al. (2008). MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity 28, 630–638. doi: 10.1016/j.immuni.2008.04.002
Dudda, J. C., Salaun, B., Ji, Y., Palmer, D. C., Monnot, G. C., Merck, E., et al. (2013). MicroRNA-155 is required for effector CD8(+) T cell responses to virus infection and cancer. Immunity 38, 742–753. doi: 10.1016/j.immuni.2012.12.006
Dunand-Sauthier, I., Santiago-Raber, M. L., Capponi, L., Vejnar, C. E., Schaad, O., Irla, M., et al. (2011). Silencing of c-Fos expression by microRNA-155 is critical for dendritic cell maturation and function. Blood 117, 4490–4500. doi: 10.1182/blood-2010-09-308064
Escobar, T. M., Kanellopoulou, C., Kugler, D. G., Kilaru, G., Nguyen, C. K., Nagarajan, V., et al. (2014). miR-155 activates cytokine gene expression in Th17 cells by regulating the DNA-binding protein jarid2 to relieve polycomb-mediated repression. Immunity 40, 865–879. doi: 10.1016/j.immuni.2014.03.014
Gebeshuber, C. A., Kornauth, C., Dong, L., Sierig, R., Seibler, J., Reiss, M., et al. (2013). Focal segmental glomerulosclerosis is induced by microRNA-193a and its downregulation of WT1. Nat. Med. 19, 481–487. doi: 10.1038/nm.3142
Giraldez, A. J., Mishima, Y., Rihel, J., Grocock, R. J., Van Dongen, S., Inoue, K., et al. (2006). Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–79. doi: 10.1126/science.1122689
Giusti, S. A., Vogl, A. M., Brockmann, M. M., Vercelli, C. A., Rein, M. L., Trumbach, D., et al. (2014). MicroRNA-9 controls dendritic development by targeting REST. Elife 3:e02755. doi: 10.7554/eLife.02755
Guo, H., Ingolia, N. T., Weissman, J. S., and Bartel, D. P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840. doi: 10.1038/nature09267
Guo, Q., Zhang, J., Li, J., Zou, L., Zhang, J., Xie, Z., et al. (2013). Forced miR-146a expression causes autoimmune lymphoproliferative syndrome in mice via downregulation of fas in germinal center B cells. Blood 121, 4875–4883. doi: 10.1182/blood-2012-08-452425
Gurha, P., Abreu-Goodger, C., Wang, T., Ramirez, M. O., Drumond, A. L., van Dongen, S., et al. (2012). Targeted deletion of microRNA-22 promotes stress-induced cardiac dilation and contractile dysfunction. Circulation 125, 2751–2761. doi: 10.1161/CIRCULATIONAHA.111.044354
Hafner, M., Landthaler, M., Burger, L., Khorshid, M., Hausser, J., Berninger, P., et al. (2010). Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141. doi: 10.1016/j.cell.2010.03.009
Hasuwa, H., Ueda, J., Ikawa, M., and Okabe, M. (2013). MiR-200b and miR-429 function in mouse ovulation and are essential for female fertility. Science 341, 71–73. doi: 10.1126/science.1237999
Heidersbach, A., Saxby, C., Carver-Moore, K., Huang, Y., Ang, Y. S., de Jong, P. J., et al. (2013). microRNA-1 regulates sarcomere formation and suppresses smooth muscle gene expression in the mammalian heart. Elife 2:e01323. doi: 10.7554/eLife.01323
Henao-Mejia, J., Williams, A., Goff, L. A., Staron, M., Licona-Limón, P., Kaech, S. M., et al. (2013). The microRNA miR-181 is a critical cellular metabolic rheostat essential for NKT cell ontogenesis and lymphocyte development and homeostasis. Immunity 38, 984–997. doi: 10.1016/j.immuni.2013.02.021
Horie, T., Baba, O., Kuwabara, Y., Chujo, Y., Watanabe, S., Kinoshita, M., et al. (2012). MicroRNA-33 deficiency reduces the progression of atherosclerotic plaque in apoE(-/-) mice. J. Am. Heart Assoc. 1:e003376. doi: 10.1161/JAHA.112.003376
Horie, T., Nishino, T., Baba, O., Kuwabara, Y., Nakao, T., Nishiga, M., et al. (2013). MicroRNA-33 regulates sterol regulatory element-binding protein 1 expression in mice. Nat. Commun. 4, 2883. doi: 10.1038/ncomms3883
Hsu, S. H., Wang, B., Kota, J., Yu, J., Costinean, S., Kutay, H., et al. (2012). Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J. Clin. Invest. 122, 2871–2883. doi: 10.1172/JCI63539
Hu, R., Kagele, D. A., Huffaker, T. B., Runtsch, M. C., Alexander, M., Liu, J., et al. (2014). miR-155 promotes t follicular helper cell accumulation during chronic, low-grade inflammation. Immunity 41, 605–619. doi: 10.1016/j.immuni.2014.09.015
Janas, M. M., Wang, B., Harris, A. S., Aguiar, M., Shaffer, J. M., Subrahmanyam, Y. V., et al. (2012). Alternative RISC assembly: binding and repression of microRNA-mRNA duplexes by human Ago proteins. RNA 18, 2041–2055. doi: 10.1261/rna.035675.112
Jiang, S., Li, C., Olive, V., Lykken, E., Feng, F., Sevilla, J., et al. (2011). Molecular dissection of the miR-17-92 cluster's critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood 118, 5487–5497. doi: 10.1182/blood-2011-05-355644
Jin, H. Y., Gonzalez-Martin, A., Miletic, A., Lai, M., Knight, S., Sabouri-Ghomi, M., et al. (2015). Transfection of microRNA mimics should be used with caution. Front. Genet. 6:340. doi: 10.3389/fgene.2015.00340
Jopling, C. L., Yi, M., Lancaster, A. M., Lemon, S. M., and Sarnow, P. (2005). Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309, 1577–1581. doi: 10.1126/science.1113329
Jordan, S. D., Kruger, M., Willmes, D. M., Redemann, N., Wunderlich, F. T., Bronneke, H. S., et al. (2011). Obesity-induced over expression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat. Cell Biol. 13, 434. doi: 10.1038/ncb2211
Khan, A. A., Penny, L. A., Yuzefpolskiy, Y., Sarkar, S., and Kalia, V. (2013). MicroRNA-17~92 regulates effector and memory CD8 T-cell fates by modulating proliferation in response to infections. Blood 121, 4473–4483. doi: 10.1182/blood-2012-06-435412
Kosaka, A., Ohkuri, T., Ikeura, M., Kohanbash, G., and Okada, H. (2015). Transgene-derived overexpression of miR-17-92 in CD8(+) T-cells confers enhanced cytotoxic activity. Biochem. Biophys. Res. Commun. 458, 549–554. doi: 10.1016/j.bbrc.2015.02.003
Kramer, N. J., Wang, W. L., Reyes, E. Y., Kumar, B., Chen, C. C., Ramakrishna, C., et al. (2015). Altered lymphopoiesis and immunodeficiency in miR-142 null mice. Blood 125, 3720–3730. doi: 10.1182/blood-2014-10-603951
Krzeszinski, J. Y., Wei, W., Huynh, H., Jin, Z., Wang, X., Chang, T. C., et al. (2014). miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature 512, 431–U460. doi: 10.1038/nature13375
Kuchen, S., Resch, W., Yamane, A., Kuo, N., Li, Z., Chakraborty, T., et al. (2010). Regulation of microRNA expression and abundance during lymphopoiesis. Immunity 32, 828–839. doi: 10.1016/j.immuni.2010.05.009
Larsson, O., and Nadon, R. (2013). Re-analysis of genome wide data on mammalian microRNA-mediated suppression of gene expression. Translation 1:e24557. doi: 10.4161/trla.24557
Latreille, M., Hausser, J., Stützer, I., Zhang, Q., Hastoy, B., Gargani, S., et al. (2014). MicroRNA-7a regulates pancreatic beta cell function. J. Clin. Invest. 124, 2722–2735. doi: 10.1172/JCI73066
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. doi: 10.1016/0092-8674(93)90529-Y
Li, C. J., Cheng, P., Liang, M. K., Chen, Y. S., Lu, Q., Wang, J. Y., et al. (2015a). MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. J. Clin. Invest. 125, 1509–1522. doi: 10.1172/JCI77716
Li, K., Zhang, J., Yu, J., Liu, B., Guo, Y., Deng, J., et al. (2015b). MicroRNA-214 suppresses gluconeogenesis by targeting activating transcriptional factor 4. J. Biol. Chem. 290, 8185–8195. doi: 10.1074/jbc.M114.633990
Li, Z., Chen, P., Su, R., Li, Y., Hu, C., Wang, Y., et al. (2015c). Overexpression and knockout of miR-126 both promote leukemogenesis through targeting distinct gene signaling. Blood 126, 2005–2015. doi: 10.1182/blood-2015-04-639062
Liang, C., Zhu, H., Xu, Y., Huang, L., Ma, C., Deng, W., et al. (2012). MicroRNA-153 negatively regulates the expression of amyloid precursor protein and amyloid precursor-like protein 2. Brain Res. 1455, 103–113. doi: 10.1016/j.brainres.2011.10.051
Liang, D., Li, J., Wu, Y., Zhen, L., Li, C., Qi, M., et al. (2015). miRNA-204 drives cardiomyocyte proliferation via targeting Jarid2. Int. J. Cardiol. 201, 38–48. doi: 10.1016/j.ijcard.2015.06.163
Liu, N., Bezprozvannaya, S., Shelton, J. M., Frisard, M. I., Hulver, M. W., McMillan, R. P., et al. (2011). Mice lacking microRNA 133a develop dynamin 2-dependent centronuclear myopathy. J. Clin. Invest. 121, 3258–3268. doi: 10.1172/JCI46267
Liu, N., Bezprozvannaya, S., Williams, A. H., Qi, X., Richardson, J. A., Bassel-Duby, R., et al. (2008). microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 22, 3242–3254. doi: 10.1101/gad.1738708
Liu, N., Williams, A. H., Maxeiner, J. M., Bezprozvannaya, S., Shelton, J. M., Richardson, J. A., et al. (2012). microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J. Clin. Invest. 122, 2054–2065. doi: 10.1172/JCI62656
Liu, S. Q., Jiang, S., Li, C., Zhang, B., and Li, Q. J. (2014). miR-17-92 cluster targets phosphatase and tensin homology and ikaros family zinc finger 4 to promote TH17-mediated inflammation. J. Biol. Chem. 289, 12446–12456. doi: 10.1074/jbc.M114.550723
Lu, D., Nakagawa, R., Lazzaro, S., Staudacher, P., Abreu-Goodger, C., Henley, T., et al. (2014). The miR-155-PU.1 axis acts on Pax5 to enable efficient terminal B cell differentiation. J. Exp. Med. 211, 2183–2198. doi: 10.1084/jem.20140338
Lu, L. F., Thai, T. H., Calado, D. P., Chaudhry, A., Kubo, M., Tanaka, K., et al. (2009). Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity 30, 80–91. doi: 10.1016/j.immuni.2008.11.010
Lu, Y., Thomson, J. M., Wong, H. Y., Hammond, S. M., and Hogan, B. L. (2007). Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev. Biol. 310, 442–453. doi: 10.1016/j.ydbio.2007.08.007
Ma, W., Hu, S., Yao, G., Xie, S., Ni, M., Liu, Q., et al. (2013). An androgen receptor-microrna-29a regulatory circuitry in mouse epididymis. J. Biol. Chem. 288, 29369–29381. doi: 10.1074/jbc.M113.454066
Ma, X. D., Kumar, M., Choudhury, S. N., Becker Buscaglia, L. E., Barker, J. R., Kanakamedala, K., et al. (2011). Loss of the miR-21 allele elevates the expression of its target genes and reduces tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 10144–10149. doi: 10.1073/pnas.1103735108
Matkovich, S. J., Wang, W., Tu, Y., Eschenbacher, W. H., Dorn, L. E., Condorelli, G., et al. (2010). MicroRNA-133a protects against myocardial fibrosis and modulates electrical repolarization without affecting hypertrophy in pressure-overloaded adult hearts. Circ. Res. 106, 166–175. doi: 10.1161/CIRCRESAHA.109.202176
Miyaki, S., Sato, T., Inoue, A., Otsuki, S., Ito, Y., Yokoyama, S., et al. (2010). MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 24, 1173–1185. doi: 10.1101/gad.1915510
Mok, Y., Schwierzeck, V., Thomas, D. C., Vigorito, E., Rayner, T. F., Jarvis, L. B., et al. (2013). MiR-210 is induced by Oct-2, regulates B cells, and inhibits autoantibody production. J. Immunol. 191, 3037–3048. doi: 10.4049/jimmunol.1301289
Mori, M., Nakagami, H., Rodriguez-Araujo, G., Nimura, K., and Kaneda, Y. (2012). Essential role for miR-196a in brown adipogenesis of white fat progenitor cells. PLoS Biol. 10:e1001314. doi: 10.1371/journal.pbio.1001314
Nakamura, Y., Inloes, J. B., Katagiri, T., and Kobayashi, T. (2011). Chondrocyte-specific microRNA-140 regulates endochondral bone development and targets dnpep to modulate bone morphogenetic protein signaling. Mol. Cell. Biol. 31, 3019–3028. doi: 10.1128/MCB.05178-11
Neilson, J. R., Zheng, G. X. Y., Burge, C. B., and Sharp, P. A. (2007). Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 21, 578–589. doi: 10.1101/gad.1522907
O'Connell, R. M., Chaudhuri, A. A., Rao, D. S., and Baltimore, D. (2009). Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl. Acad. Sci. U.S.A. 106, 7113–7118. doi: 10.1073/pnas.0902636106
O'Connell, R. M., Kahn, D., Gibson, W. S., Round, J. L., Scholz, R. L., Chaudhuri, A. A., et al. (2010). MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 33, 607–619. doi: 10.1016/j.immuni.2010.09.009
Olive, V., Minella, A. C., and He, L. (2015). Outside the coding genome, mammalian microRNAs confer structural and functional complexity. Sci. Signal. 8, re2. doi: 10.1126/scisignal.2005813
Pan, D., Mao, C., Quattrochi, B., Friedline, R. H., Zhu, L. J., Jung, D. Y., et al. (2014). MicroRNA-378 controls classical brown fat expansion to counteract obesity. Nat. Commun. 5, 4725. doi: 10.1038/ncomms5725
Parchem, R. J., Moore, N., Fish, J. L., Parchem, J. G., Braga, T. T., Shenoy, A., et al. (2015). miR-302 is required for timing of neural differentiation, neural tube closure, and embryonic viability. Cell Rep. 12, 760–773. doi: 10.1016/j.celrep.2015.06.074
Patrick, D. M., Zhang, C. C., Tao, Y., Yao, H., Qi, X., Schwartz, R. J., et al. (2010). Defective erythroid differentiation in miR-451 mutant mice mediated by 14-3-3zeta. Genes Dev. 24, 1614–1619. doi: 10.1101/gad.1942810
Payne, S. H. (2015). The utility of protein and mRNA correlation. Trends Biochem. Sci. 40, 1–3. doi: 10.1016/j.tibs.2014.10.010
Poy, M. N., Hausser, J., Trajkovski, M., Braun, M., Collins, S., Rorsman, P., et al. (2009). miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc. Natl. Acad. Sci. U.S.A. 106, 5813–5818. doi: 10.1073/pnas.0810550106
Sanuki, R., Onishi, A., Koike, C., Muramatsu, R., Watanabe, S., Muranishi, Y., et al. (2011). miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat. Neurosci. 14, 1125–1134. doi: 10.1038/nn.2897
Shan, S. W., Lee, D. Y., Deng, Z., Shatseva, T., Jeyapalan, Z., Du, W. W., et al. (2009). MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat. Cell Biol. 11, 1031–1038. doi: 10.1038/ncb1917
Shibata, M., Nakao, H., Kiyonari, H., Abe, T., and Aizawa, S. (2011). MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J. Neurosci. 31, 3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011
Song, R., Walentek, P., Sponer, N., Klimke, A., Lee, J. S., Dixon, G., et al. (2014). miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature 510, 115. doi: 10.1038/nature13413
Song, S. J., Ito, K., Ala, U., Kats, L., Webster, K., Sun, S. M., et al. (2013). The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell 13, 87–101. doi: 10.1016/j.stem.2013.06.003
Stadthagen, G., Tehler, D., Høyland-Kroghsbo, N. M., Wen, J., Krogh, A., Jensen, K. T., et al. (2013). Loss of miR-10a activates lpo and collaborates with activated Wnt signaling in inducing intestinal neoplasia in female mice. PLoS Genet. 9:e1003913. doi: 10.1371/journal.pgen.1003913
Stickel, N., Prinz, G., Pfeifer, D., Hasselblatt, P., Schmitt-Graeff, A., Follo, M., et al. (2014). MiR-146a regulates the TRAF6/TNF-axis in donor T cells during GVHD. Blood 124, 2586–2595. doi: 10.1182/blood-2014-04-569046
Sullivan, R. P., Leong, J. W., Schneider, S. E., Ireland, A. R., Berrien-Elliott, M. M., Singh, A., et al. (2015). MicroRNA-15/16 antagonizes Myb to control NK cell maturation. J. Immunol. 195, 2806–2817. doi: 10.4049/jimmunol.1500949
Sun, Y., Oravecz-Wilson, K., Mathewson, N., Wang, Y., McEachin, R., Liu, C., et al. (2015). Mature T cell responses are controlled by microRNA-142. J. Clin. Invest. 125, 2825–2840. doi: 10.1172/JCI78753
Tan, C. L., Plotkin, J. L., Venø, M. T., Von Schimmelmann, M., Feinberg, P., Mann, S., et al. (2013). MicroRNA-128 governs neuronal excitability and motor behavior in mice. Science 342, 1254–1258. doi: 10.1126/science.1244193
Tsai, W. C., Hsu, S. D., Hsu, C. S., Lai, T. C., Chen, S. J., Shen, R., et al. (2012). MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J. Clin. Invest. 122, 2884–2897. doi: 10.1172/JCI63455
Tung, Y. T., Lu, Y. L., Peng, K. C., Yen, Y. P., Chang, M., Li, J., et al. (2015). Mir-17 ~ 92 governs motor neuron subtype survival by mediating nuclear pten. Cell Rep. 11, 1305–1318. doi: 10.1016/J.Celrep.2015.04.050
Ucar, A., Gupta, S. K., Fiedler, J., Erikci, E., Kardasinski, M., Batkai, S., et al. (2012). The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat. Commun. 3, 1078. doi: 10.1038/ncomms2090
van Rooij, E., Sutherland, L. B., Qi, X., Richardson, J. A., Hill, J., and Olson, E. N. (2007). Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316, 575–579. doi: 10.1126/science.1139089
Vigorito, E., Perks, K. L., Abreu-Goodger, C., Bunting, S., Xiang, Z., Kohlhaas, S., et al. (2007). microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 27, 847–859. doi: 10.1016/j.immuni.2007.10.009
Wang, D., Zhang, Z., O'Loughlin, E., Lee, T., Houel, S., O'Carroll, D., et al. (2012). Quantitative functions of Argonaute proteins in mammalian development. Genes Dev. 26, 693–704. doi: 10.1101/gad.182758.111
Wang, D., Zhang, Z., O'Loughlin, E., Wang, L., Fan, X., Lai, E. C., et al. (2013a). MicroRNA-205 controls neonatal expansion of skin stem cells by modulating the PI(3)K pathway. Nat. Cell Biol. 15, 1153–1163. doi: 10.1038/ncb2827
Wang, H. P., Flach, H., Onizawa, M., Wei, L., McManus, M. T., and Weiss, A. (2014). Negative regulation of hif1a expression and T(H)17 differentiation by the hypoxia-regulated microRNA miR-210. Nat. Immunol. 15, 393. doi: 10.1038/ni.2846
Wang, J., Yu, F., Jia, X., Iwanowycz, S., Wang, Y., Huang, S., et al. (2015a). MicroRNA-155 deficiency enhances the recruitment and functions of myeloid-derived suppressor cells in tumor microenvironment and promotes solid tumor growth. Int. J. Cancer 136, E602–E613. doi: 10.1002/ijc.29151
Wang, K., Liu, C. Y., Zhang, X. J., Feng, C., Zhou, L. Y., Zhao, Y., et al. (2015b). MiR-361-regulated prohibitin inhibits mitochondrial fission and apoptosis and protects heart from ischemia injury. Cell Death Differ. 22, 1058–1068. doi: 10.1038/cdd.2014.200
Wang, S. S., Aurora, A. B., Johnson, B. A., Qi, X., McAnally, J., Hill, J. A., et al. (2008). The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 15, 261–271. doi: 10.1016/j.devcel.2008.07.002
Wang, X., Guo, B., Li, Q., Peng, J., Yang, Z., Wang, A., et al. (2013b). MiR-214 targets ATF4 to inhibit bone formation. Nat. Med. 19, 93–100. doi: 10.1038/nm.3026
Wei, J., Shi, Y., Zheng, L., Zhou, B., Inose, H., Wang, J., et al. (2012). MiR-34s inhibit osteoblast proliferation and differentiation in the mouse by targeting SATB2. J. Cell Biol. 197, 509–521. doi: 10.1083/jcb.201201057
Wei, Y., Peng, S., Wu, M., Sachidanandam, R., Tu, Z., Zhang, S., et al. (2014). Multifaceted roles of miR-1s in repressing the fetal gene program in the heart. Cell Res. 24, 278–292. doi: 10.1038/cr.2014.12
Wightman, B., Ha, I., and Ruvkun, G. (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862. doi: 10.1016/0092-8674(93)90530-4
Williams, A. H., Valdez, G., Moresi, V., Qi, X., McAnally, J., Elliott, J. L., et al. (2009). MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 326, 1549–1554. doi: 10.1126/science.1181046
Wystub, K., Besser, J., Bachmann, A., Boettger, T., and Braun, T. (2013). miR-1/133a clusters cooperatively specify the cardiomyogenic lineage by adjustment of myocardin levels during embryonic heart development. PLoS Genet. 9:e1003793. doi: 10.1371/journal.pgen.1003793
Xin, M., Small, E. M., Sutherland, L. B., Qi, X., McAnally, J., Plato, C. F., et al. (2009). MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 23, 2166–2178. doi: 10.1101/gad.1842409
Xu, S., Ou, X., Huo, J., Lim, K., Huang, Y., Chee, S., et al. (2015). Mir-17-92 regulates bone marrow homing of plasma cells and production of immunoglobulin G2c. Nat. Commun. 6, 6764. doi: 10.1038/ncomms7764
Yan, S., Xu, Z., Lou, F., Zhang, L., Ke, F., Bai, J., et al. (2015). NF-kappaB-induced microRNA-31 promotes epidermal hyperplasia by repressing protein phosphatase 6 in psoriasis. Nat. Commun. 6, 7652. doi: 10.1038/ncomms8652
Yu, D., dos Santos, C. O., Zhao, G., Jiang, J., Amigo, J. D., Khandros, E., et al. (2010). miR-451 protects against erythroid oxidant stress by repressing 14-3-3zeta. Genes Dev. 24, 1620–1633. doi: 10.1101/gad.1942110
Yuan, S., Tang, C., Zhang, Y., Wu, J., Bao, J., Zheng, H., et al. (2015). Mir-34b/c and mir-449a/b/c are required for spermatogenesis, but not for the first cleavage division in mice. Biol. Open 4, 212–223. doi: 10.1242/bio.201410959
Zhang, L., Ke, F., Liu, Z., Bai, J., Liu, J., Yan, S., et al. (2015). MicroRNA-31 negatively regulates peripherally derived regulatory T-cell generation by repressing retinoic acid-inducible protein 3. Nat. Commun. 6, 7639. doi: 10.1038/ncomms8639
Zhao, Y., Ransom, J. F., Li, A., Vedantham, V., von Drehle, M., Muth, A. N., et al. (2007). Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 129, 303–317. doi: 10.1016/j.cell.2007.03.030
Zhao, Y., Samal, E., and Srivastava, D. (2005). Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436, 214–220. doi: 10.1038/nature03817
Keywords: microRNA mechanism of action, translation regulation, mRNA degradation, miR-430, transient transfection of miRNA mimics, miRNA mutant mice
Citation: Jin HY and Xiao C (2015) MicroRNA Mechanisms of Action: What have We Learned from Mice? Front. Genet. 6:328. doi: 10.3389/fgene.2015.00328
Received: 31 August 2015; Accepted: 22 October 2015;
Published: 16 November 2015.
Edited by:
Ghanshyam Upadhyay, City College of New York-CUNY, USAReviewed by:
Sandeep Kumar, State University of New York College of Optometry, USAIsrar Ahmad, University of Alabama at Birmingham, USA
Copyright © 2015 Jin and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changchun Xiao, Y3hpYW9Ac2NyaXBwcy5lZHU=
 Hyun Yong Jin
Hyun Yong Jin Changchun Xiao
Changchun Xiao