- Department of Biomedical Sciences and Pathobiology, Virginia–Maryland College of Veterinary Medicine, Virginia Polytechnic Institute and State University, Blacksburg, VA, USA
In recent years, IL-17A (IL-17), a pro-inflammatory cytokine, has received intense attention of researchers and clinicians alike with documented effects in inflammation and autoimmune diseases. IL-17 mobilizes, recruits and activates different cells to increase inflammation. Although protective in infections, overproduction of IL-17 promotes inflammation in autoimmune diseases such as multiple sclerosis, rheumatoid arthritis, psoriasis, among others. Regulating IL-17 levels or action by using IL-17-blocking antibodies or IL-17R antagonist has shown to attenuate experimental autoimmune diseases. It is now known that in addition to IL-17-specific transcription factor, RORγt, several other transcription factors and select microRNAs (miRNA) regulate IL-17. Given that miRNAs are dysregulated in autoimmune diseases, a better understanding of transcriptional factors and miRNA regulation of IL-17 expression and function will be essential for devising potential new therapies. In this review, we will overview IL-17 induction and function in relation to autoimmune diseases. In addition, current findings on transcriptional regulation of IL-17 induction and plausible interplay between IL-17 and miRNA in autoimmune diseases are highlighted.
Introduction
Interleukin 17 (IL-17) was initially termed in Rouvier et al. (1993) as cytotoxic T lymphocyte-associated antigen-8 (CTLA-8) when it was first cloned from a rodent cDNA sequence. Subsequently, IL-17 was also identified in humans (Yao et al., 1995). Among the members of IL-17 family, IL-17A (hence referred as IL-17) and IL-17F are known for their important functional and biological properties. IL-17A and IL-17F are 50% homologous and map to the same chromosomal loci. They exist either as homodimers or IL-17A/F heterodimers (Liang et al., 2007). IL-17A is known to be secreted by many cell types including: CD4+ (Th17), CD8+ (Tc17), γδ+ T cells, natural killer cells, mast cells, neutrophils among other cell types (Rachitskaya et al., 2008; Lin et al., 2011; Zhao et al., 2012; Gelderblom et al., 2014; Li et al., 2014a; Ravichandran et al., 2014).
Although IL-17 is known to have protective effects in infections, increased IL-17 and/or aberrant responses to IL-17 has been shown to aggravate disease conditions and contribute to tissue injury as observed in many autoimmune diseases (Weaver and Murphy, 2007). IL-17 stimulates the production of various inflammatory mediators such as IL-6, IL-8, CNTF, TGF-β2, IL-10, BMP6, IL-1α, TNF-α, CCL19, CCL4, and M-CSF, CXCL1, CXCL2, CCL2, CCL12, CCL20 (Kang et al., 2010; Nardinocchi et al., 2014; Rodgers et al., 2014); MCP-1, KP, macrophage inflammatory protein (MIP)-2, TIMP-1, granulocyte chemotactic protein-2 (GCP-2) and matrix metalloproteinases (MMPs) -3, 9, and 13 (Qiu et al., 2009) and nitric oxide, HGF, MCP-1, KC, MIP-2, PGE1, PGE2, and VGEF (Numasaki et al., 2004). Studies have demonstrated that IL-17 cosynergizes with different ligands and signaling pathways such as toll-like receptor (TLR) ligands, B cell-activating factor (BAFF), IFNγ, IL-1β, CD40-ligand and TNFα to fine-tune inflammatory responses (Woltman et al., 2000; Andoh et al., 2001; Liu et al., 2014a; Nardinocchi et al., 2014; Francois et al., 2015).
IL-17 and Autoimmune Diseases
In healthy homeostatic conditions, the levels of IL-17A in human sera are undetectable, however, the levels are markedly increased in inflammatory bowel disease, psoriasis, systemic lupus erythematosus (SLE), multiple sclerosis (MS), and rheumatoid arthritis (RA; Wang et al., 2012; Jiang et al., 2014; Babaloo et al., 2015; Mease, 2015; Talaat et al., 2015). IL-23 has been shown to be critical for expansion, and/or survival and stabilization of Th17 cells by activating STAT3 and partially STAT4 (Aggarwal et al., 2003; Harrington et al., 2005). Interaction of IL-23-producing APCs and Th17 cells has been shown to have a role in many autoimmune diseases. In support of this view, targeting IL-23 pathway, IL-17 production or action by using IL-17R antagonist and IL-17A-blocking antibodies have been shown to attenuate autoimmune diseases (Hueber et al., 2010; Yeilding et al., 2011; Leonardi et al., 2012; Papp et al., 2012; Sofen et al., 2014). In this regard, several clinical trials are underway to treat psoriasis. These include, ustekinumab anti-p40-IL-23 mAb and guselkumab, an anti-IL-23-specific mAb, ixekizumab and secukinumab (anti–IL-17A mAbs) and brodalumab (an anti-IL-17RA mAb) are currently under clinical trials. A recent study has demonstrated that a combination of inhibition of IL-23 and IL-17 is more efficacious in treating Th17-mediated autoimmunity in mouse models (Mangan et al., 2015). In addition, human recombinant IL-37 has also been shown to decrease IL-17 expression and Th17 cell frequency in PBMCs and CD4+ T cells from RA patients (Ye et al., 2015). Together these studies have shown promising results of targeting IL-17 induction and signaling pathways in the treatment of chronic autoimmune diseases.
Molecular Aspects of IL-17 Induction
Although initial reports showed that TGFβ1 inhibits IL-17A production in a dose-dependent manner in human naïve CD4+ T cells (Acosta-Rodriguez et al., 2007), other studies have shown that low concentrations of TGFβ1 in combination with either IL-21 (Yang et al., 2008a), or IL-1β and IL-23 (Manel et al., 2008; Duhen and Campbell, 2014) or IL-1β, IL-23, and IL-6 (Volpe et al., 2008) promote differentiation of human CD4+ T cells into Th17 cells. Interestingly, it was recently shown that TGFβ3-induced Th17 cells have pathogenic effector signature when compared to TGFβ1-induced Th17 cells (Lee et al., 2012). Studies have also shown that IL-17 expression may be transient and not a terminal/end-stage Th cell differentiation (Kurschus et al., 2010; Hirota et al., 2011). Further, there is dynamic plasticity among Th subsets and Th17 differentiation is highly dependent on the kind of stimuli (polarizing conditions) received from the local tissues.
Sentinel cells of innate immune system (neutrophils, γδ T, Lti, Paneth, and iNKT cells) also secrete IL-17 (Cua and Tato, 2010). These cells constitutively express transcriptional regulators for IL-17 induction (discussed in the next section), therefore, upon activation produce IL-17 within hours of stimuli (Sutton et al., 2009; Cua and Tato, 2010). The next section will focus on multiple transcription factors involved in the positive and negative regulation of IL-17 (Figure 1).
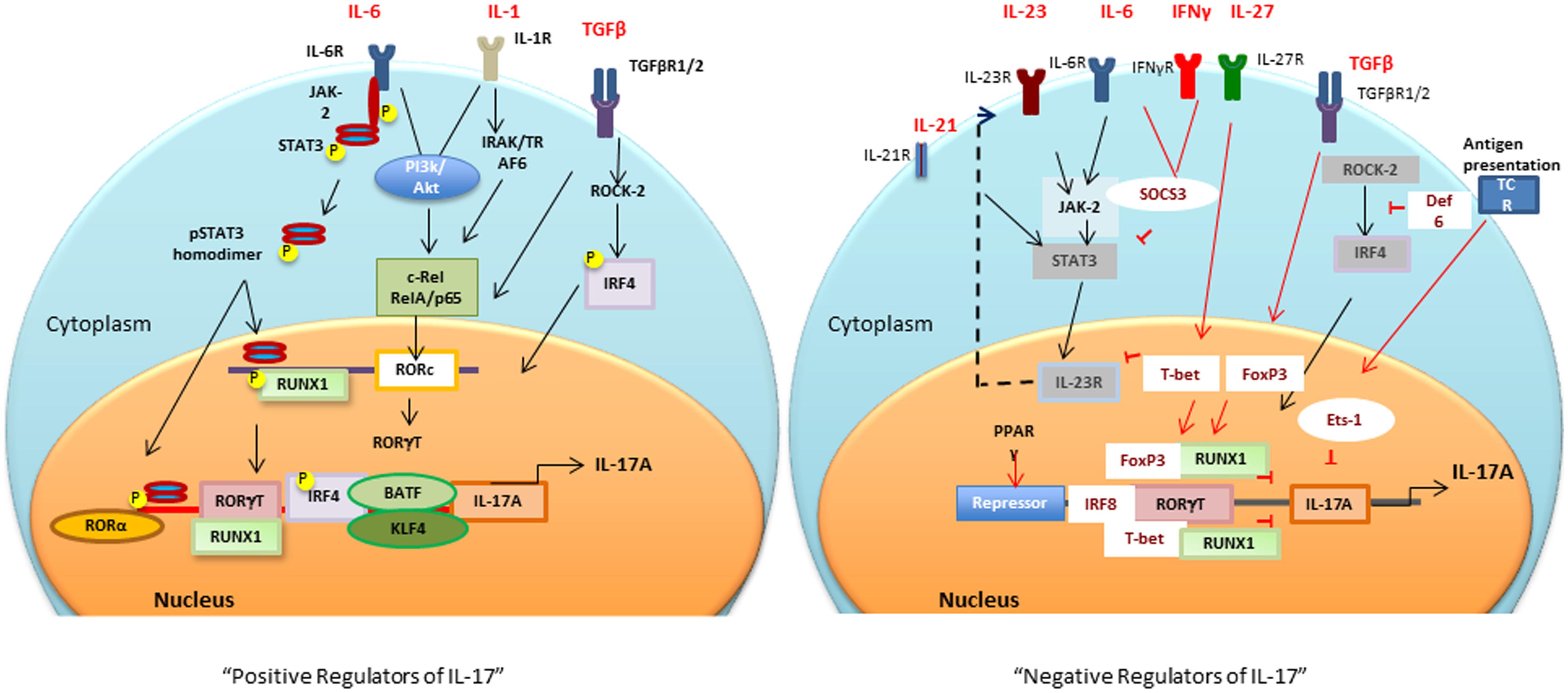
FIGURE 1. Positive and negative transcriptional tegulators of IL-17 induction: Different cytokines and antigen specific stimuli trigger (black arrows and lines) different signaling cascades for activation of RORc and consequently Il17 gene. Negative regulators (red arrows and lines), T-bet or FoxP3 interaction with RUNX1 prevents RORγt-RUNX1 interaction, which prevents RORγt-mediated IL-17 induction. Def6 binding to IRF4 prevents ROCK2-mediated IRF4 phosphorylation and subsequent IL-17 induction. PPARγ, peroxisome proliferator activated receptor γ; SOCS, suppressors of cytokine signaling; TCR, T cell receptor; BATF, B cell-activating transcription factor; IL, interleukin; TGFβ, transforming growth factor β; RORγt, retinoic acid-related orphan receptor γt; STAT, signal transducer and activator of transcription; IRF-4, interferon-inducible factor-4; RUNX1, Runt-related transcription factor 1; IRAK, IL-1 receptor-associated kinase; TRAF6, TNF receptor associated factor-6; ROCK, Rho-associated serine/threonine kinases.
Positive IL-17 Regulators
RORγt and RUNX1
It is now well established that for Th17 differentiation, it is critical to have TGFβ1 in the presence of IL-1, IL-6, or IL-21 to decrease suppressive FoxP3 and upregulate RORc gene encoded unique lineage-specific transcription factor, RORγt, an retinoic acid related-orphan nuclear receptor (Bettelli et al., 2006; Ivanov et al., 2006; McGeachy et al., 2007; Yang et al., 2008b; Biswas et al., 2010; Ikeda et al., 2014). Runt-related transcription factor 1 (Runx1) regulates Th17 differentiation by upregulating RORγt expression and by direct binding to RORγt (Zhang et al., 2008; Liu et al., 2015). Interestingly, a study has shown that IL-17-secreting Treg cells (Tr17) have co-expression of FoxP3, RORγt, Runx1, and Runx3 (Li et al., 2012). A recent study has shown that RORγt-transcriptional activity, and subsequent IL-17 induction is increased by Sirtuin 1 (SIRT1), a protein deacetylase. Inhibition of SIRT1 by chemical Ex-527 based inhibition or T cell specific deletion strongly suppressed the development of experimental autoimmune encephalitis (EAE) in mice (Lim et al., 2015). In addition, a selective RORγt inverse agonist (TMP778) has been shown to inhibit Th17 signature gene expression, and IL-17 production from Tc17 and γδ T cells indicating the therapeutic potential of targeting RORγt in inflammatory conditions (Skepner et al., 2014).
STAT3
Activation of IL-6R (ligand binding IL-6Rα and signal transducing gp130) by IL-6 results in activation of JAK-2/STAT3 pathway. Activated STAT3 binds to the promoter of IL-17A and IL-17F (Chen et al., 2006). Depletion of either STAT3 or gp130 in CD4+ T cells by utilizing Cre-loxP recombination results in decreased RORγt expression and Th17 differentiation, suggesting that IL-6-gp130-STAT3 regulate IL-17 induction at least in part by regulating RORγt levels (Nishihara et al., 2007). In vivo inhibition of JAK2-STAT3 pathway by AG490 was recently shown to decrease Th17 but increase Tregs in collagen-induced arthritis mice (Park et al., 2014).
NF-κB
It has recently been shown that members of NF-κB family RelA (p65) and c-Rel bind to promoters of RORγ and RORγt, respectively (Ruan et al., 2011). The positive role of NF-κB in IL-17 induction was further substantiated by the findings that activation of NF-κB increases secretion of IL-17 (Huang et al., 2008). c-Rel deficient mice have decreased EAE due to impaired activation of RORc gene and subsequently decreased Th17 development (Lazarevic et al., 2011). Peripheral blood mononuclear cells (PBMCs) from RA patients have increased IL-17 induction by activation of PI3K/Akt, which increases the DNA binding activity of NF-κB (Kim et al., 2005). In addition, in DC and CD4 T cells co-culture system, dendritic cells (DCs) that are deficient in RelB have decreased induction of IL-12p70, IL-23, and IL-6 when compared to control DCs, thereby resulting in decreased Th17- and Th1-related markers but increased Th2 and Treg markers (Yang et al., 2010).
Interferon Regulatory Factor 4
Interferon regulatory factor 4 (IRF4) is also critical for IL-17 and IL-21 induction (Ciofani et al., 2012; Huber and Lohoff, 2014). It has been shown that IRF4-deficient mice have decreased RORα and RORγt expression but increased FoxP3 levels (Brustle et al., 2007; Huber et al., 2008). TGFβ-mediated activation of Rho-ROCK pathway, promotes phosphorylation of IRF4 by ROCK kinase. Once phosphorylated, IRF4 translocates to nuclei and binds to IL-17 and IL-21 promoters (Biswas et al., 2010; Mudter et al., 2011). It is noteworthy that in autoimmune models such as MRL/lpr, there is enhanced ROCK2 activation concomitant with increased IRF4 function and IL-17 levels (Biswas et al., 2010).
Other IL-17 Promoting Transcription Factors
Kruppel-like factor (KLF) 4, has been shown to regulate Th17 development by binding to the IL-17 promoter directly without altering RORγt expression (Lebson et al., 2010; An et al., 2011). Basic leucine zipper transcription factor, ATF-like (BATF), synergizes with RORγt to induce IL-17 by direct interaction with conserved intergenic elements of Il17A/Il17F loci (Jordan-Williams et al., 2013). Additionally, other signaling pathway Rho-associated kinase, ROCK2 (Zanin-Zhorov and Waksal, 2015), Ets-family transcription factor (Etv5; Pham et al., 2014), Sphingosine 1-phosphate- /type 1 S1P receptors (S1P1s; Liao et al., 2007), RORα (Yang et al., 2008b) and aryl hydrocarbon receptor (Hayes et al., 2014) have also been shown to be important for the induction of IL-17.
Negative IL-17 Regulators
Both IFNγ and IL-4 markedly inhibit Th17 differentiation (Harrington et al., 2005; Yeh et al., 2014). In addition, IL-25 and IL-27 have been found to negatively regulate Th17 cells (Batten et al., 2006; Kleinschek et al., 2007; Tang et al., 2015). Suppressor of cytokine signaling (SOCS3) also negatively regulates IL-6-gp160 signal transduction resulting in decreased IL-17 (Babon et al., 2014; Vartoukian et al., 2014). FoxP3 interacts directly with RORγt through the exon 2 region and forkhead domain of FoxP3 and suppresses the activation of the IL-17 promoter (Ichiyama et al., 2008). Moreover, it has also been shown that Runx1 interacts with FoxP3 and negatively regulates Th17 differentiation (Zhang et al., 2008). T-bet, a Th1-lineage specific transcription factor, also suppresses Th17 development by binding to Runx1 via tyrosine 304 of T-bet. This T-bet-Runx1 binding has been shown to block the transactivation of RORc gene and therefore IL-17 induction (Lazarevic et al., 2011). These studies indicate that Runx1 associates either with RORγt to upregulate IL-17, or with T-bet /FoxP3 to suppress Th17 differentiation (Gocke et al., 2007). Interestingly, a recent report has shown that in EAE, pathogenic IFNγ-secreting Th17 cells have co-expression of T-bet, Runx-1 or -3 (Wang et al., 2014). These findings suggest a critical role of Runx1 in differentiation of Th cells into different specific Th cell lineages.
Another T-bet interacting transcription factor, v-ets erythroblastosis virus E26 oncogene homolog 1 (ETS-1), has been shown to inhibit Th17 differentiation. ETS-1-deficient mice have increased IL-17 levels (without affecting RORγt), suggesting that ETS-1 is a negative regulator of IL-17 (Moisan et al., 2007). Recently, IRF8, also known as ICSBP, has been shown to physically interact with RORγt and regulate IL-17 by silencing Th17 differentiation and downregulating Th17-associated genes (Qi et al., 2009; Ouyang et al., 2011). Peroxisome proliferator-activated receptor γ (PPARγ) also acts as an intrinsic suppressor of Th17. It prevents the removal of repressor complexes from RORγt promoter, resulting in decreased RORγt expression and RORγt-induced Th17 differentiation (Klotz et al., 2009). In addition, there are other compounds such as digoxin and signaling molecules [e.g., STAT1, STAT4, STAT5, STAT6, NFAT, SOCS1, early growth response gene 2 (Egr-2), IRF-4 binding protein Def6, epidermal fatty acid binding protein (E-FABP)] which negatively regulate IL-17 induction (Fujita-Sato et al., 2011; Huh et al., 2011; Grange et al., 2013; Miao et al., 2013). Overall these reports confirm that there are multiple transcription factors, which fine-tune and tightly regulate IL-17 induction. In the next section, we will focus on the miRNAs, which epigenetically either regulate IL-17 induction or response in different autoimmune and other disease conditions.
IL-17 and miRNA
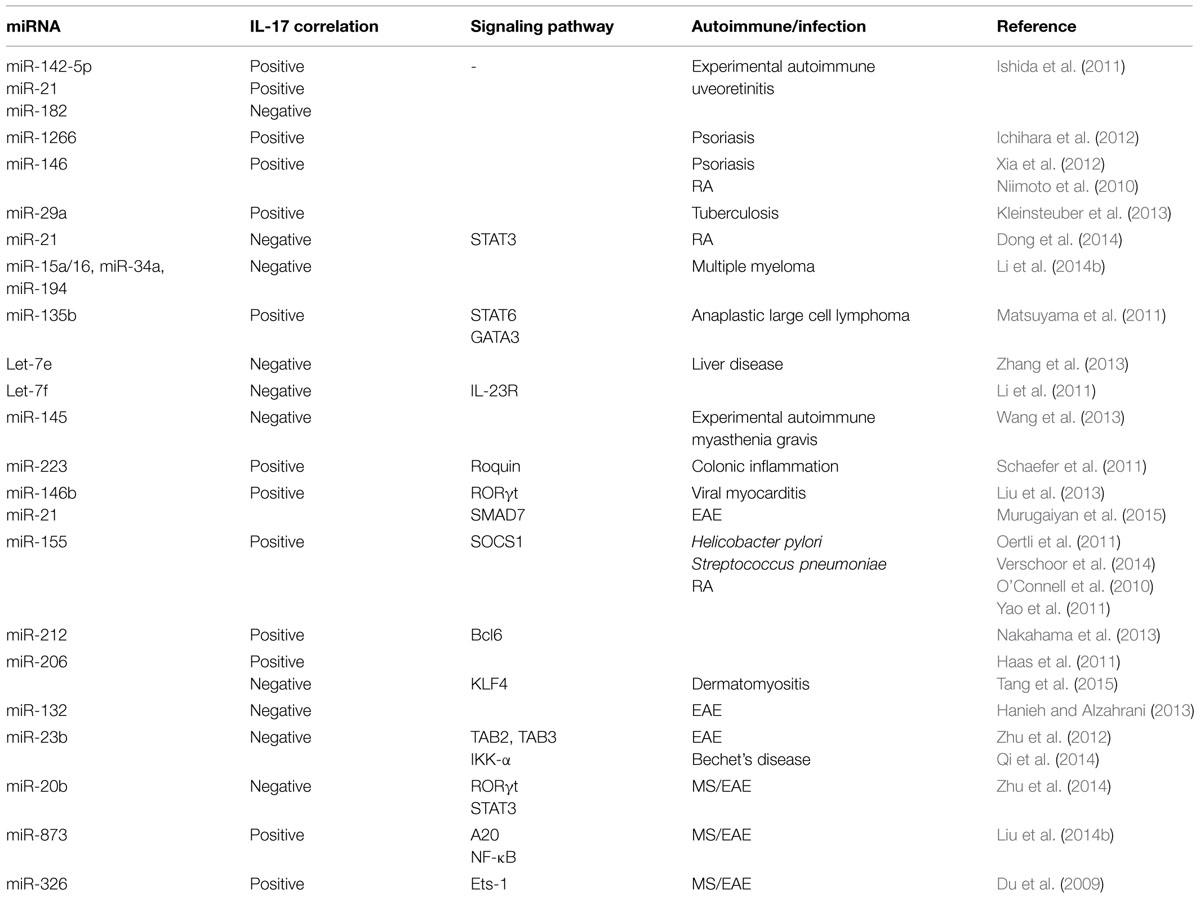
Positive correlations in miRNA expression and IL-17 levels have been observed in different studies (Table 1). In experimental autoimmune uveoretinitis (EAU), miR-142-5p and miR-21 were increased but miR-182 decreased in eye. The kinetic changes in these miRNA paralleled with increased IL-17 levels (Ishida et al., 2011). Given that IL-17 levels are increased in skin lesions and sera of psoriasis vulgaris patients, it is noteworthy that miR-1266 levels, a putative regulator of IL-17A, were also increased in the sera of these patients (Ichihara et al., 2012). miR-146 is also positively correlated with IL-17A levels in psoriasis (Xia et al., 2012) and in RA disease severity, and is co-expressed with IL-17A in the PBMC and synovium in RA patients (Niimoto et al., 2010). Inverse relationship between select miRNAs and IL-17 has also been observed in autoimmune conditions and other diseases (Table 1). For example, combined treatment with anti-TNFα/disease-modifying antirheumatic drugs (anti-TNFα/DMARDs) increased miR-16-5p, miR-23-3p, miR125b-5p, miR-126-3p, miRN-146a-5p, miR-223-3pmiRNA with a concomitant decrease in TNFα, interleukin (IL)-6, IL-17, rheumatoid factor (RF), and C-reactive protein (CRP; Castro-Villegas et al., 2015). Imbalance of Th17/Treg in RA patients has been associated with decreased miR-21 levels, increased STAT3 activation and decreased STAT5/pSTAT5 protein and FoxP3 mRNA levels (Dong et al., 2014). In experimental autoimmune myasthenia gravis (EAMG), there is marked downregulation of miR-145 expression. Administration of lentiviral-miR-145 decreased EAMG disease severity by decreased IL-17 production (Wang et al., 2013). miRNA dysregulation has been best studied in cancer and in several cancer studies miRNA regulation of IL-17 has been demonstrated, which may have implications for autoimmune diseases (Matsuyama et al., 2011; Arisawa et al., 2012; Zhang et al., 2013; Li et al., 2014b). A positive correlation in expession of miR-133b and miR-206 and IL-17 in both αβ and γδ T cells in human and inbred mouse strains have also been reported (Haas et al., 2011). It was found that these miRNAs were clustered nearly 45 kb upstream of Il17a/f locus (Haas et al., 2011). On the other hand, in dermatomyositis patients, there is downregulation of miR-206 and upregulation of miR-206 predicted target KLF4, a positive regulator of IL-17 and Th17 cells (Tang et al., 2015). It is likely that miR-206 regulation of IL-17 may be context dependent.
A recent report has shown that miR-21 increased Th17 differentiation by targeting and depleting a negative regulator of TGF-β signaling SMAD-7 (Murugaiyan et al., 2015). Treatment of wild type mice with anti miR-21 oligonucleotide decreased EAE clinical severity along with decreased Th17 cells (Murugaiyan et al., 2015). In MS patients, there is downregulation of miR-20b. In EAE, miR-20b overexpression decreased disease severity by decreasing Th17 differentiation by targeting RORγt and STAT3 (Zhu et al., 2014). There is upregulation of miR-873 in brain tissue of EAE mice and in IL-17 activated mouse primary astrocytes (Liu et al., 2014b). In EAE model, miR-873 induced by IL-17 stimulation aggravated disease severity and increased inflammation by targeting A20/NF-κ (Liu et al., 2014b). Importantly, Du et al. (2009) reported that miR-326 expression correlated with MS disease severity in human patients. Further in EAE mice, miR-326 played an important role in pathogenesis by regulating Th-17 cell differentiation through translational inhibition of Ets-1, a negative regulator of Th17 differentiation (). In MS patients there is decreased expression of an IL-6-related miRNA, miR-26a (Zhang et al., 2015). In vivo silencing of miR-26a increased Th17-related cytokines and EAE severity (Zhang et al., 2015).
miR-155 deficiency results in decreased severity of different autoimmune diseases such as EAE, collagen induced arthritis (CIA) by impairment of Th17 polarization and decreased IL-17 levels (O’Connell et al., 2010; Bluml et al., 2011; Murugaiyan et al., 2011). The above studies indicate a strong correlation between miR-155 expression and Th17 differentiation, which is potentially mediated by miR-155 targeting of signaling molecule, SOCS1 (Yao et al., 2011, 2012).
Interleukin 17 has been shown to down regulate miR-23b expression in human fibroblast-like synoviocytes, mouse primary kidney cells and astrocytes. miR-23b suppresses IL-17-mediated autoimmune inflammation by targeting TNF-α- or IL-1β-induced NF-κB activation by targeting TGF-β-activated kinase 1/MAP3K7 binding protein 2 (TAB2), TAB3 and inhibitor of NF-κB kinase subunit α (IKK-α; Zhu et al., 2012). Behcet’s disease (BD) patients have increased activation of Notch pathway and Th17 response but decreased miR-23b (Qi et al., 2014). These studies indicate the potential of miR-23b as a therapeutic target for IL-17-related autoimmune conditions. miR-21 levels are also increased in BD patients and decrease in miR-21 in herpes simplex virus (HSV)-induced BD mouse model decreased serum IL-6 and IL-17 levels and improved disease condition (Choi et al., 2015).
AHR activation results in upregulation of miR-132/212 cluster under Th17 inducing conditions (Nakahama et al., 2013). However, overexpression of miR-132 in CD4 T cells from EAE mice decreased IL-17 and IFNγ and T cell proliferation (Hanieh and Alzahrani, 2013). Interestingly, miR-212 targeted B-cell lymphoma 6, a negative regulator of Th17 differentiation (Nakahama et al., 2013). These findings suggest that miRNA regulation and correlation with IL-17 is dependent on disease model.
Taken together, these studies indicate that IL-17 is regulated by interplay of multiple transcription factors and miRNAs and vary with different disease condition and cell type studied. It is recognized that in addition to miRNAs, other epigenetic mechanisms such as histone modifications and DNA methylation also contribute to autoimmune diseases. However, this topic is beyond the scope of this concise focused review.
Concluding Comments
Overall, IL-17 is regarded as a potent proinflammatory cytokine that is essential for defense against pathogens. However, dysregulated IL-17 production or response has been associated with tissue damage in various inflammatory diseases. Given that IL-17 has now been associated with many inflammatory and autoimmune diseases, a better understanding of IL-17 induction and regulation is imperative to institute novel effective targeted therapeutic strategies. While RORγt, is considered as a IL-17-specific transcription factor, recent collective data clearly show that induction of IL-17 is regulated by multiple transcription factors. Transcription factors that positively and negatively regulate IL-17 have now been identified. In addition, post-transcriptional regulation of IL-17 by specific miRNAs is now increasingly appreciated. Aberrant miRNA expression is reported in several human autoimmune diseases (Dai and Ahmed, 2011; Dai et al., 2013; Khan et al., 2015). Dysregulated miRNA expression profiles have the potential to serve as good diagnostic and prognostic marker and/or therapeutic targets. Signature miRNA profile can be potentially used as novel biomarkers for Th17-mediated immune reactions. However, more in-depth and mechanistic studies are required to further define the role of miRNAs in IL-17 induction and interplay of miRNA with IL-17-related transcription factors and signaling pathways. Since blocking a major cytokine such as IL-17 may have unintended consequences, more refined (tissue-dependent) manipulation of select IL-17-regulating-miRNAs may be a viable therapeutic option in some diseases.
Author Contributions
DK and SA designed the work, drafted and revised the work and finally approved the version to be published and agree to be accountable for all aspects of the work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Studies were supported by American Autoimmune Related Diseases Association (AARDA), NIH T35 grant, 9T350DO1187-06, Lupus Foundation of America (LFA) Novel Pilot Project (208-11-110B-033-918-1), Alliance for Lupus Research (ALR, Award No: 219636), and Virginia-Maryland Regional College of Veterinary Medicine (VMRCVM) Intramural Research Competition Grant (IRC 17385).
References
Acosta-Rodriguez, E. V., Napolitani, G., Lanzavecchia, A., and Sallusto, F. (2007). Interleukins 1beta and 6 but not transforming growth factor-beta are essential for the differentiation of interleukin 17-producing human T helper cells. Nat. Immunol. 8, 942–949. doi: 10.1038/ni1496
Aggarwal, S., Ghilardi, N., Xie, M. H., de Sauvage, F. J., and Gurney, A. L. (2003). Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J. Biol. Chem. 278, 1910–1914. doi: 10.1074/jbc.M207577200
An, J., Golech, S., Klaewsongkram, J., Zhang, Y., Subedi, K., Huston, G. E., et al. (2011). Kruppel-like factor 4 (KLF4) directly regulates proliferation in thymocyte development and IL-17 expression during Th17 differentiation. FASEB J. 25, 3634–3645. doi: 10.1096/fj.11-186924
Andoh, A., Takaya, H., Makino, J., Sato, H., Bamba, S., Araki, Y., et al. (2001). Cooperation of interleukin-17 and interferon-gamma on chemokine secretion in human fetal intestinal epithelial cells. Clin. Exp. Immunol. 125, 56–63. doi: 10.1046/j.1365-2249.2001.01588.x
Arisawa, T., Tahara, T., Shiroeda, H., Matsue, Y., Minato, T., Nomura, T., et al. (2012). Genetic polymorphisms of IL17A and pri-microRNA-938, targeting IL17A 3′-UTR, influence susceptibility to gastric cancer. Hum. Immunol. 73, 747–752. doi: 10.1016/j.humimm.2012.04.011
Babaloo, Z., Aliparasti, M., Babaie, F., Almasi, S., Baradaran, B., and Farhoodi, M. (2015). The role of Th17 cells in patients with relapsing-remitting multiple sclerosis: interleukin-17A and interleukin-17F serum levels. Immunol. Lett. 164, 76–80. doi: 10.1016/j.imlet.2015.01.001
Babon, J. J., Varghese, L. N., and Nicola, N. A. (2014). Inhibition of IL-6 family cytokines by SOCS3. Semin. Immunol. 26, 13–19. doi: 10.1016/j.smim.2013.12.004
Batten, M., Li, J., Yi, S., Kljavin, N. M., Danilenko, D. M., Lucas, S., et al. (2006). Interleukin 27 limits autoimmune encephalomyelitis by suppressing the development of interleukin 17-producing T cells. Nat. Immunol. 7, 929–936. doi: 10.1038/ni1375
Bettelli, E., Carrier, Y., Gao, W., Korn, T., Strom, T. B., Oukka, M., et al. (2006). Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238. doi: 10.1038/nature04753
Biswas, P. S., Gupta, S., Chang, E., Song, L., Stirzaker, R. A., Liao, J. K., et al. (2010). Phosphorylation of IRF4 by ROCK2 regulates IL-17 and IL-21 production and the development of autoimmunity in mice. J. Clin. Invest. 120, 3280–3295. doi: 10.1172/JCI42856
Bluml, S., Bonelli, M., Niederreiter, B., Puchner, A., Mayr, G., Hayer, S., et al. (2011). Essential role of microRNA-155 in the pathogenesis of autoimmune arthritis in mice. Arthritis Rheum. 63, 1281–1288. doi: 10.1002/art.30281
Brustle, A., Heink, S., Huber, M., Rosenplanter, C., Stadelmann, C., Yu, P., et al. (2007). The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat. Immunol. 8, 958–966. doi: 10.1038/ni1500
Castro-Villegas, C., Perez-Sanchez, C., Escudero, A., Filipescu, I., Verdu, M., Ruiz-Limon, P., et al. (2015). Circulating miRNAs as potential biomarkers of therapy effectiveness in rheumatoid arthritis patients treated with anti-TNFalpha. Arthritis Res. Ther. 17:49. doi: 10.1186/s13075-015-0555-z
Chen, Z., Laurence, A., Kanno, Y., Pacher-Zavisin, M., Zhu, B. M., Tato, C., et al. (2006). Selective regulatory function of Socs3 in the formation of IL-17-secreting T cells. Proc. Natl. Acad. Sci. U.S.A. 103, 8137–8142. doi: 10.1073/pnas.0600666103
Choi, B., Kim, H. A., Suh, C. H., Byun, H. O., Jung, J. Y., and Sohn, S. (2015). The relevance of miRNA-21 in HSV-induced inflammation in a mouse model. Int. J. Mol. Sci. 16, 7413–7427. doi: 10.3390/ijms16047413
Ciofani, M., Madar, A., Galan, C., Sellars, M., Mace, K., Pauli, F., et al. (2012). A validated regulatory network for Th17 cell specification. Cell 151, 289–303. doi: 10.1016/j.cell.2012.09.016
Cua, D. J., and Tato, C. M. (2010). Innate IL-17-producing cells: the sentinels of the immune system. Nat. Rev. Immunol. 10, 479–489. doi: 10.1038/nri2800
Dai, R., and Ahmed, S. A. (2011). MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl. Res. 157, 163–179. doi: 10.1016/j.trsl.2011.01.007
Dai, R., McReynolds, S., Leroith, T., Heid, B., Liang, Z., and Ahmed, S. A. (2013). Sex differences in the expression of lupus-associated miRNAs in splenocytes from lupus-prone NZB/WF1 mice. Biol. Sex. Differ. 4:19. doi: 10.1186/2042-6410-4-19
Dong, L., Wang, X., Tan, J., Li, H., Qian, W., Chen, J., et al. (2014). Decreased expression of microRNA-21 correlates with the imbalance of Th17 and Treg cells in patients with rheumatoid arthritis. J. Cell. Mol. Med. 18, 2213–2224. doi: 10.1111/jcmm.12353
Du, C., Liu, C., Kang, J., Zhao, G., Ye, Z., Huang, S., et al. (2009). MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat. Immunol. 10, 1252–1259. doi: 10.1038/ni.1798
Duhen, T., and Campbell, D. J. (2014). IL-1beta promotes the differentiation of polyfunctional human CCR6+CXCR3+ Th1/17 cells that are specific for pathogenic and commensal microbes. J. Immunol. 193, 120–129. doi: 10.4049/jimmunol.1302734
Francois, A., Gombault, A., Villeret, B., Alsaleh, G., Fanny, M., Gasse, P., et al. (2015). B cell activating factor is central to bleomycin- and IL-17-mediated experimental pulmonary fibrosis. J. Autoimmun. 56, 1–11. doi: 10.1016/j.jaut.2014.08.003
Fujita-Sato, S., Ito, S., Isobe, T., Ohyama, T., Wakabayashi, K., Morishita, K., et al. (2011). Structural basis of digoxin that antagonizes RORgamma t receptor activity and suppresses Th17 cell differentiation and interleukin (IL)-17 production. J. Biol. Chem. 286, 31409–31417. doi: 10.1074/jbc.M111.254003
Gelderblom, M., Arunachalam, P., and Magnus, T. (2014). gammadelta T cells as early sensors of tissue damage and mediators of secondary neurodegeneration. Front. Cell. Neurosci. 8:368. doi: 10.3389/fncel.2014.00368
Gocke, A. R., Cravens, P. D., Ben, L. H., Hussain, R. Z., Northrop, S. C., Racke, M. K., et al. (2007). T-bet regulates the fate of Th1 and Th17 lymphocytes in autoimmunity. J. Immunol. 178, 1341–1348. doi: 10.4049/jimmunol.178.3.1341
Grange, M., Verdeil, G., Arnoux, F., Griffon, A., Spicuglia, S., Maurizio, J., et al. (2013). Active STAT5 regulates T-bet and eomesodermin expression in CD8 T cells and imprints a T-bet-dependent Tc1 program with repressed IL-6/TGF-beta1 signaling. J. Immunol. 191, 3712–3724. doi: 10.4049/jimmunol.1300319
Haas, J. D., Nistala, K., Petermann, F., Saran, N., Chennupati, V., Schmitz, S., et al. (2011). Expression of miRNAs miR-133b and miR-206 in the Il17a/f locus is co-regulated with IL-17 production in alphabeta and gammadelta T cells. PLoS ONE 6:e20171. doi: 10.1371/journal.pone.0020171
Hanieh, H., and Alzahrani, A. (2013). MicroRNA-132 suppresses autoimmune encephalomyelitis by inducing cholinergic anti-inflammation: a new Ahr-based exploration. Eur. J. Immunol. 43, 2771–2782. doi: 10.1002/eji.201343486
Harrington, L. E., Hatton, R. D., Mangan, P. R., Turner, H., Murphy, T. L., Murphy, K. M., et al. (2005). Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 6, 1123–1132. doi: 10.1038/ni1254
Hayes, M. D., Ovcinnikovs, V., Smith, A. G., Kimber, I., and Dearman, R. J. (2014). The aryl hydrocarbon receptor: differential contribution to T helper 17 and T cytotoxic 17 cell development. PLoS ONE 9:e106955. doi: 10.1371/journal.pone.0106955
Hirota, K., Duarte, J. H., Veldhoen, M., Hornsby, E., Li, Y., Cua, D. J., et al. (2011). Fate mapping of IL-17-producing T cells in inflammatory responses. Nat. Immunol. 12, 255–263. doi: 10.1038/ni.1993
Huang, M. C., Liao, J. J., Bonasera, S., Longo, D. L., and Goetzl, E. J. (2008). Nuclear factor-kappaB-dependent reversal of aging-induced alterations in T cell cytokines. FASEB J. 22, 2142–2150. doi: 10.1096/fj.07-103721
Huber, M., Brustle, A., Reinhard, K., Guralnik, A., Walter, G., Mahiny, A., et al. (2008). IRF4 is essential for IL-21-mediated induction, amplification, and stabilization of the Th17 phenotype. Proc. Natl. Acad. Sci. U.S.A. 105, 20846–20851. doi: 10.1073/pnas.0809077106
Huber, M., and Lohoff, M. (2014). IRF4 at the crossroads of effector T-cell fate decision. Eur. J. Immunol. 44, 1886–1895. doi: 10.1002/eji.201344279
Hueber, A. J., Asquith, D. L., Miller, A. M., Reilly, J., Kerr, S., Leipe, J., et al. (2010). Mast cells express IL-17A in rheumatoid arthritis synovium. J. Immunol. 184, 3336–3340. doi: 10.4049/jimmunol.0903566
Huh, J. R., Leung, M. W., Huang, P., Ryan, D. A., Krout, M. R., Malapaka, R. R., et al. (2011). Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORgammat activity. Nature 472, 486–490. doi: 10.1038/nature09978
Ichihara, A., Jinnin, M., Oyama, R., Yamane, K., Fujisawa, A., Sakai, K., et al. (2012). Increased serum levels of miR-1266 in patients with psoriasis vulgaris. Eur. J. Dermatol. 22, 68–71.
Ichiyama, K., Yoshida, H., Wakabayashi, Y., Chinen, T., Saeki, K., Nakaya, M., et al. (2008). Foxp3 inhibits RORgammat-mediated IL-17A mRNA transcription through direct interaction with RORgammat. J. Biol. Chem. 283, 17003–17008. doi: 10.1074/jbc.M801286200
Ikeda, S., Saijo, S., Murayama, M. A., Shimizu, K., Akitsu, A., and Iwakura, Y. (2014). Excess IL-1 signaling enhances the development of Th17 cells by downregulating TGF-beta-induced Foxp3 expression. J. Immunol. 192, 1449–1458. doi: 10.4049/jimmunol.1300387
Ishida, W., Fukuda, K., Sakamoto, S., Koyama, N., Koyanagi, A., Yagita, H., et al. (2011). Regulation of experimental autoimmune uveoretinitis by anti-delta-like ligand 4 monoclonal antibody. Invest. Ophthalmol. Vis. Sci. 52, 8224–8230. doi: 10.1167/iovs.11-7756
Ivanov, II, McKenzie, B. S., Zhou, L., Tadokoro, C. E., Lepelley, A., Lafaille, J. J., et al. (2006). The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133. doi: 10.1016/j.cell.2006.07.035
Jiang, W., Su, J., Zhang, X., Cheng, X., Zhou, J., Shi, R., et al. (2014). Elevated levels of Th17 cells and Th17-related cytokines are associated with disease activity in patients with inflammatory bowel disease. Inflamm. Res. 63, 943–950. doi: 10.1007/s00011-014-0768-7
Jordan-Williams, K. L., Poston, S., and Taparowsky, E. J. (2013). BATF regulates the development and function of IL-17 producing iNKT cells. BMC Immunol. 14:16. doi: 10.1186/1471-2172-14-16
Kang, Z., Altuntas, C. Z., Gulen, M. F., Liu, C., Giltiay, N., Qin, H., et al. (2010). Astrocyte-restricted ablation of interleukin-17-induced Act1-mediated signaling ameliorates autoimmune encephalomyelitis. Immunity 32, 414–425. doi: 10.1016/j.immuni.2010.03.004
Khan, D., Dai, R., and Ansar Ahmed, S. (2015). Sex differences and estrogen regulation of miRNAs in lupus, a prototypical autoimmune disease. Cell. Immunol. 294, 70–79. doi: 10.1016/j.cellimm.2015.01.004
Kim, K. W., Cho, M. L., Park, M. K., Yoon, C. H., Park, S. H., Lee, S. H., et al. (2005). Increased interleukin-17 production via a phosphoinositide 3-kinase/Akt and nuclear factor kappaB-dependent pathway in patients with rheumatoid arthritis. Arthritis Res. Ther. 7, R139–R148. doi: 10.1186/ar1470
Kleinschek, M. A., Owyang, A. M., Joyce-Shaikh, B., Langrish, C. L., Chen, Y., Gorman, D. M., et al. (2007). IL-25 regulates Th17 function in autoimmune inflammation. J. Exp. Med. 204, 161–170. doi: 10.1084/jem.20061738
Kleinsteuber, K., Heesch, K., Schattling, S., Kohns, M., Sander-Jülch, C., Walzl, G., et al. (2013). Decreased expression of miR-21, miR-26a, miR-29a, and miR-142-3p in CD4+ T cells and peripheral blood from tuberculosis patients. PLoS ONE 8:e61609. doi: 10.1371/journal.pone.0061609
Klotz, L., Burgdorf, S., Dani, I., Saijo, K., Flossdorf, J., Hucke, S., et al. (2009). The nuclear receptor PPAR gamma selectively inhibits Th17 differentiation in a T cell-intrinsic fashion and suppresses CNS autoimmunity. J. Exp. Med. 206, 2079–2089. doi: 10.1084/jem.20082771
Kurschus, F. C., Croxford, A. L., Heinen, A. P., Wortge, S., Ielo, D., and Waisman, A. (2010). Genetic proof for the transient nature of the Th17 phenotype. Eur. J. Immunol. 40, 3336–3346. doi: 10.1002/eji.201040755
Lazarevic, V., Chen, X., Shim, J. H., Hwang, E. S., Jang, E., Bolm, A. N., et al. (2011). T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat. Immunol. 12, 96–104. doi: 10.1038/ni.1969
Lebson, L., Gocke, A., Rosenzweig, J., Alder, J., Civin, C., Calabresi, P. A., et al. (2010). Cutting edge: the transcription factor Kruppel-like factor 4 regulates the differentiation of Th17 cells independently of RORgammat. J. Immunol. 185, 7161–7164. doi: 10.4049/jimmunol.1002750
Lee, Y., Awasthi, A., Yosef, N., Quintana, F. J., Xiao, S., Peters, A., et al. (2012). Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 13, 991–999. doi: 10.1038/ni.2416
Leonardi, C., Matheson, R., Zachariae, C., Cameron, G., Li, L., Edson-Heredia, E., et al. (2012). Anti–interleukin-17 monoclonal antibody ixekizumab in chronic plaque psoriasis. N. Engl. J. Med. 366, 1190–1199. doi: 10.1056/NEJMoa1109997
Li, L., Patsoukis, N., Petkova, V., and Boussiotis, V. A. (2012). Runx1 and Runx3 are involved in the generation and function of highly suppressive IL-17-producing T regulatory cells. PLoS ONE 7:e45115. doi: 10.1371/journal.pone.0045115
Li, S., Joseph, C., Becourt, C., Klibi, J., Luce, S., Dubois-Laforgue, D., et al. (2014a). Potential role of IL-17-producing iNKT cells in type 1 diabetes. PLoS ONE 9:e96151. doi: 10.1371/journal.pone.0096151
Li, Y., Li, D., Yan, Z., Qi, K., Chen, L., Zhang, Z., et al. (2014b). Potential relationship and clinical significance of miRNAs and Th17 cytokines in patients with multiple myeloma. Leuk. Res. 38, 1130–1135. doi: 10.1016/j.leukres.2014.07.005
Li, Z., Wu, F., Brant, S. R., and Kwon, J. H. (2011). IL-23 receptor regulation by Let-7f in human CD4+ memory T cells. J. Immunol. 186, 6182–6190. doi: 10.4049/jimmunol.1000917
Liang, S. C., Long, A. J., Bennett, F., Whitters, M. J., Karim, R., Collins, M., et al. (2007). An IL-17F/A heterodimer protein is produced by mouse Th17 cells and induces airway neutrophil recruitment. J. Immunol. 179, 7791–7799. doi: 10.4049/jimmunol.179.11.7791
Liao, J. J., Huang, M. C., and Goetzl, E. J. (2007). Cutting edge: alternative signaling of Th17 cell development by sphingosine 1-phosphate. J. Immunol. 178, 5425–5428. doi: 10.4049/jimmunol.178.9.5425
Lim, H. W., Kang, S. G., Ryu, J. K., Schilling, B., Fei, M., Lee, I. S., et al. (2015). SIRT1 deacetylates RORgammat and enhances Th17 cell generation. J. Exp. Med. 212, 607–617. doi: 10.1084/jem.20132378
Lin, A. M., Rubin, C. J., Khandpur, R., Wang, J. Y., Riblett, M., Yalavarthi, S., et al. (2011). Mast cells and neutrophils release IL-17 through extracellular trap formation in psoriasis. J. Immunol. 187, 490–500. doi: 10.4049/jimmunol.1100123
Liu, G., Guo, J., Liu, J., Wang, Z., and Liang, D. (2014a). Toll-like receptor signaling directly increases functional IL-17RA expression in neuroglial cells. Clin. Immunol. 154, 127–140. doi: 10.1016/j.clim.2014.07.006
Liu, X., He, F., Pang, R., Zhao, D., Qiu, W., Shan, K., et al. (2014b). Interleukin-17 (IL-17)-induced microRNA 873 (miR-873) contributes to the pathogenesis of experimental autoimmune encephalomyelitis by targeting A20 ubiquitin-editing enzyme. J. Biol. Chem. 289, 28971–28986. doi: 10.1074/jbc.M114.577429
Liu, H. P., Cao, A. T., Feng, T., Li, Q., Zhang, W., Yao, S., et al. (2015). TGF-beta converts Th1 cells into Th17 cells through stimulation of Runx1 expression. Eur. J. Immunol. 45, 1010–1018. doi: 10.1002/eji.201444726
Liu, Y. L., Wu, W., Xue, Y., Gao, M., Yan, Y., Kong, Q., et al. (2013). MicroRNA-21 and -146b are involved in the pathogenesis of murine viral myocarditis by regulating TH-17 differentiation. Arch. Virol. 158, 1953–1963.
Manel, N., Unutmaz, D., and Littman, D. R. (2008). The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nat. Immunol. 9, 641–649. doi: 10.1038/ni.1610
Mangan, P. R., Su, L. J., Jenny, V., Tatum, A. L., Picarillo, C., Skala, S., et al. (2015). Dual inhibition of IL-23 and IL-17 offers superior efficacy in mouse models of autoimmunity. J. Pharmacol. Exp. Ther. doi: 10.1124/jpet.115.224246 [Epub ahead of print].
Matsuyama, H., Suzuki, H. I., Nishimori, H., Noguchi, M., Yao, T., Komatsu, N., et al. (2011). miR-135b mediates NPM-ALK-driven oncogenicity and renders IL-17-producing immunophenotype to anaplastic large cell lymphoma. Blood 118, 6881–6892. doi: 10.1182/blood-2011-05-354654
McGeachy, M. J., Bak-Jensen, K. S., Chen, Y., Tato, C. M., Blumenschein, W., McClanahan, T., et al. (2007). TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat. Immunol. 8, 1390–1397. doi: 10.1038/ni1539
Mease, P. J. (2015). Inhibition of interleukin-17, interleukin-23 and the TH17 cell pathway in the treatment of psoriatic arthritis and psoriasis. Curr. Opin. Rheumatol. 27, 127–133. doi: 10.1097/BOR.0000000000000147
Miao, T., Raymond, M., Bhullar, P., Ghaffari, E., Symonds, A. L., Meier, U. C., et al. (2013). Early growth response gene-2 controls IL-17 expression and Th17 differentiation by negatively regulating Batf. J. Immunol. 190, 58–65. doi: 10.4049/jimmunol.1200868
Moisan, J., Grenningloh, R., Bettelli, E., Oukka, M., and Ho, I. C. (2007). Ets-1 is a negative regulator of Th17 differentiation. J. Exp. Med. 204, 2825–2835. doi: 10.1084/jem.20070994
Mudter, J., Yu, J., Zufferey, C., Brustle, A., Wirtz, S., Weigmann, B., et al. (2011). IRF4 regulates IL-17A promoter activity and controls RORgammat-dependent Th17 colitis in vivo. Inflamm. Bowel Dis. 17, 1343–1358. doi: 10.1002/ibd.21476
Murugaiyan, G., Beynon, V., Mittal, A., Joller, N., and Weiner, H. L. (2011). Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J. Immunol. 187, 2213–2221. doi: 10.4049/jimmunol.1003952
Murugaiyan, G., da Cunha, A. P., Ajay, A. K., Joller, N., Garo, L. P., Kumaradevan, S., et al. (2015). MicroRNA-21 promotes Th17 differentiation and mediates experimental autoimmune encephalomyelitis. J. Clin. Invest. 125, 1069–1080. doi: 10.1172/JCI74347
Nakahama, T., Hanieh, H., Nguyen, N. T., Chinen, I., Ripley, B., Millrine, D., et al. (2013). Aryl hydrocarbon receptor-mediated induction of the microRNA-132/212 cluster promotes interleukin-17-producing T-helper cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 110, 11964–11969. doi: 10.1073/pnas.1311087110
Nardinocchi, L., Sonego, G., Passarelli, F., Avitabile, S., Scarponi, C., Failla, C. M., et al. (2014). Interleukin-17 and interleukin-22 promote tumor progression in human nonmelanoma skin cancer. Eur. J. Immunol. 45, 922–931. doi: 10.1002/eji.201445052
Niimoto, T., Nakasa, T., Ishikawa, M., Okuhara, A., Izumi, B., Deie, M., et al. (2010). MicroRNA-146a expresses in interleukin-17 producing T cells in rheumatoid arthritis patients. BMC Musculoskelet. Disord. 11:209. doi: 10.1186/1471-2474-11-209
Nishihara, M., Ogura, H., Ueda, N., Tsuruoka, M., Kitabayashi, C., Tsuji, F., et al. (2007). IL-6-gp130-STAT3 in T cells directs the development of IL-17+ Th with a minimum effect on that of Treg in the steady state. Int. Immunol. 19, 695–702. doi: 10.1093/intimm/dxm045
Numasaki, M., Lotze, M. T., and Sasaki, H. (2004). Interleukin-17 augments tumor necrosis factor-alpha-induced elaboration of proangiogenic factors from fibroblasts. Immunol. Lett. 93, 39–43. doi: 10.1016/j.imlet.2004.01.014
O’Connell, R. M., Kahn, D., Gibson, W. S., Round, J. L., Scholz, R. L., Chaudhuri, A. A., et al. (2010). MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 33, 607–619. doi: 10.1016/j.immuni.2010.09.009
Oertli, M., Engler, D. B., Kohler, E., Koch, M., Meyer, T. F., and Müller, A. (2011). MicroRNA-155 is essential for the T cell-mediated control of Helicobacter pylori infection and for the induction of chronic Gastritis and Colitis. J. Immunol. 187, 3578–3586. doi: 10.4049/jimmunol.1101772
Ouyang, X., Zhang, R., Yang, J., Li, Q., Qin, L., Zhu, C., et al. (2011). Transcription factor IRF8 directs a silencing programme for TH17 cell differentiation. Nat. Commun. 2:314. doi: 10.1038/ncomms1311
Papp, K., Cather, J. C., Rosoph, L., Sofen, H., Langley, R. G., Matheson, R. T., et al. (2012). Efficacy of apremilast in the treatment of moderate to severe psoriasis: a randomised controlled trial. Lancet 380, 738–746. doi: 10.1016/S0140-6736(12)60642-4
Park, J. S., Lee, J., Lim, M. A., Kim, E. K., Kim, S. M., Ryu, J. G., et al. (2014). JAK2-STAT3 blockade by AG490 suppresses autoimmune arthritis in mice via reciprocal regulation of regulatory T cells and Th17 cells. J. Immunol. 192, 4417–4424. doi: 10.4049/jimmunol.1300514
Pham, D., Sehra, S., Sun, X., and Kaplan, M. H. (2014). The transcription factor Etv5 controls TH17 cell development and allergic airway inflammation. J. Allergy Clin. Immunol. 134, 204–214. doi: 10.1016/j.jaci.2013.12.021
Qi, C. F., Li, Z., Raffeld, M., Wang, H., Kovalchuk, A. L., and Morse, H. C. III (2009). Differential expression of IRF8 in subsets of macrophages and dendritic cells and effects of IRF8 deficiency on splenic B cell and macrophage compartments. Immunol. Res. 45, 62–74. doi: 10.1007/s12026-008-8032-2
Qi, J., Yang, Y., Hou, S., Qiao, Y., Wang, Q., Yu, H., et al. (2014). Increased Notch pathway activation in Behcet’s disease. Rheumatology (Oxford) 53, 810–820. doi: 10.1093/rheumatology/ket438
Qiu, Z., Dillen, C., Hu, J., Verbeke, H., Struyf, S., Van Damme, J., et al. (2009). Interleukin-17 regulates chemokine and gelatinase B expression in fibroblasts to recruit both neutrophils and monocytes. Immunobiology 214, 835–842. doi: 10.1016/j.imbio.2009.06.007
Rachitskaya, A. V., Hansen, A. M., Horai, R., Li, Z., Villasmil, R., Luger, D., et al. (2008). Cutting edge: NKT cells constitutively express IL-23 receptor and RORgammat and rapidly produce IL-17 upon receptor ligation in an IL-6-independent fashion. J. Immunol. 180, 5167–5171. doi: 10.4049/jimmunol.180.8.5167
Ravichandran, J., Jackson, R. J., Trivedi, S., and Ranasinghe, C. (2014). IL-17A Expression in HIV-specific CD8 T cells is regulated by IL-4/IL-13 following HIV-1 prime-boost immunization. J. Interferon Cytokine Res. 35, 176–185. doi: 10.1089/jir.2014.0078
Rodgers, J. M., Robinson, A. P., Rosler, E. S., Lariosa-Willingham, K., Persons, R. E., Dugas, J. C., et al. (2014). IL-17A activates ERK1/2 and enhances differentiation of oligodendrocyte progenitor cells. Glia 63, 768–779. doi: 10.1002/glia.22783
Rouvier, E., Luciani, M. F., Mattéi, M. G., Denizot, F., and Golstein, P. (1993). CTLA-8, cloned from an activated T cell, bearing AU-rich messenger RNA instability sequences, and homologous to a herpesvirus saimiri gene. J. Immunol. 150, 5445–5456.
Ruan, Q., Kameswaran, V., Zhang, Y., Zheng, S., Sun, J., Wang, J., et al. (2011). The Th17 immune response is controlled by the Rel-RORgamma-RORgamma T transcriptional axis. J. Exp. Med. 208, 2321–2333. doi: 10.1084/jem.20110462
Schaefer, J. S., Montufar-Solis, D., Vigneswaran, N., and Klein, J. R. (2011). Selective upregulation of microRNA expression in peripheral blood leukocytes in IL-10-/- mice precedes expression in the colon. J. Immunol. 187, 5834–5841. doi: 10.4049/jimmunol.1100922
Skepner, J., Ramesh, R., Trocha, M., Schmidt, D., Baloglu, E., Lobera, M., et al. (2014). Pharmacologic inhibition of RORgammat regulates Th17 signature gene expression and suppresses cutaneous inflammation in vivo. J. Immunol. 192, 2564–2575. doi: 10.4049/jimmunol.1302190
Sofen, H., Smith, S., Matheson, R. T., Leonardi, C. L., Calderon, C., Brodmerkel, C., et al. (2014). Guselkumab (an IL-23-specific mAb) demonstrates clinical and molecular response in patients with moderate-to-severe psoriasis. J. Allergy Clin. Immunol. 133, 1032–1040. doi: 10.1016/j.jaci.2014.01.025
Sutton, C. E., Lalor, S. J., Sweeney, C. M., Brereton, C. F., Lavelle, E. C., and Mills, K. H. (2009). Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity 31, 331–341. doi: 10.1016/j.immuni.2009.08.001
Talaat, R. M., Mohamed, S. F., Bassyouni, I. H., and Raouf, A. A. (2015). Th1/Th2/Th17/Treg cytokine imbalance in systemic lupus erythematosus (SLE) patients: correlation with disease activity. Cytokine 72, 146–153. doi: 10.1016/j.cyto.2014.12.027
Tang, S. C., Fan, X. H., Pan, Q. M., Sun, Q. S., and Liu, Y. (2015). Decreased expression of IL-27 and its correlation with Th1 and Th17 cells in progressive multiple sclerosis. J. Neurol. Sci. 348, 174–180. doi: 10.1016/j.jns.2014.11.035
Vartoukian, S. R., Tilakaratne, W. M., Seoudi, N., Bombardieri, M., Bergmeier, L., Tappuni, A. R., et al. (2014). Dysregulation of the suppressor of cytokine signalling 3-signal transducer and activator of transcription-3 pathway in the aetiopathogenesis of Sjogren’s syndrome. Clin. Exp. Immunol. 177, 618–629. doi: 10.1111/cei.12377
Verschoor, C. P., Dorrington, M. G., Novakowski, K. E., Kaiser, J., Radford, K., Nair, P., et al. (2014). MicroRNA-155 is required for clearance of Streptococcus pneumoniae from the nasopharynx. Infect. Immun. 82, 4824–4833. doi: 10.1128/IAI.02251-14
Viladomiu, M., Hontecillas, R., Pedragosa, M., Carbo, A., Hoops, S., Michalak, P., et al. (2012). Modeling the role of peroxisome proliferator-activated receptor gamma and microRNA-146 in mucosal immune responses to Clostridium difficile. PLoS ONE 7:e47525. doi: 10.1371/journal.pone.0047525
Volpe, E., Servant, N., Zollinger, R., Bogiatzi, S. I., Hupe, P., Barillot, E., et al. (2008). A critical function for transforming growth factor-beta, interleukin 23 and proinflammatory cytokines in driving and modulating human T(H)-17 responses. Nat. Immunol. 9, 650–657. doi: 10.1038/ni.1613
Wang, J., Zheng, S., Xin, N., Dou, C., Fu, L., Zhang, X., et al. (2013). Identification of novel MicroRNA signatures linked to experimental autoimmune myasthenia gravis pathogenesis: down-regulated miR-145 promotes pathogenetic Th17 cell response. J. Neuroimmune Pharmacol. 8, 1287–1302. doi: 10.1007/s11481-013-9498-9
Wang, W., Shao, S., Jiao, Z., Guo, M., Xu, H., and Wang, S. (2012). The Th17/Treg imbalance and cytokine environment in peripheral blood of patients with rheumatoid arthritis. Rheumatol. Int. 32, 887–893. doi: 10.1007/s00296-010-1710-0
Wang, Y., Godec, J., Ben-Aissa, K., Cui, K., Zhao, K., Pucsek, A. B., et al. (2014). The transcription factors T-bet and Runx are required for the ontogeny of pathogenic interferon-gamma-producing T helper 17 cells. Immunity 40, 355–366. doi: 10.1016/j.immuni.2014.01.002
Weaver, C. T., and Murphy, K. M. (2007). The central role of the Th17 lineage in regulating the inflammatory/autoimmune axis. Semin. Immunol. 19, 351–352. doi: 10.1016/j.smim.2008.01.001
Woltman, A. M., de Haij, S., Boonstra, J. G., Gobin, S. J., Daha, M. R., and van Kooten, C. (2000). Interleukin-17 and CD40-ligand synergistically enhance cytokine and chemokine production by renal epithelial cells. J. Am. Soc. Nephrol. 11, 2044–2055.
Xia, P., Fang, X., Zhang, Z. H., Huang, Q., Yan, K. X., Kang, K. F., et al. (2012). Dysregulation of miRNA146a versus IRAK1 induces IL-17 persistence in the psoriatic skin lesions. Immunol. Lett. 148, 151–162. doi: 10.1016/j.imlet.2012.09.004
Yang, H., Zhang, Y., Wu, M., Li, J., Zhou, W., Li, G., et al. (2010). Suppression of ongoing experimental autoimmune myasthenia gravis by transfer of RelB-silenced bone marrow dentritic cells is associated with a change from a T helper Th17/Th1 to a Th2 and FoxP3+ regulatory T-cell profile. Inflamm. Res. 59, 197–205. doi: 10.1007/s00011-009-0087-6
Yang, L., Anderson, D. E., Baecher-Allan, C., Hastings, W. D., Bettelli, E., Oukka, M., et al. (2008a). IL-21 and TGF-beta are required for differentiation of human T(H)17 cells. Nature 454, 350–352. doi: 10.1038/nature07021
Yang, X. O., Pappu, B. P., Nurieva, R., Akimzhanov, A., Kang, H. S., Chung, Y., et al. (2008b). T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity 28, 29–39. doi: 10.1016/j.immuni.2007.11.016
Yao, R., Ma, Y., Du, Y., Liao, M., Li, H., Liang, W., et al. (2011). The altered expression of inflammation-related microRNAs with microRNA-155 expression correlates with Th17 differentiation in patients with acute coronary syndrome. Cell. Mol. Immunol. 8, 486–495. doi: 10.1038/cmi.2011.22
Yao, R., Ma, Y. L., Liang, W., Li, H. H., Ma, Z. J., Yu, X., et al. (2012). MicroRNA-155 modulates Treg and Th17 cells differentiation and Th17 cell function by targeting SOCS1. PLoS ONE 7:e46082. doi: 10.1371/journal.pone.0046082
Yao, Z., Painter, S. L., Fanslow, W. C., Ulrich, D., Macduff, B. M., Spriggs, M. K., et al. (1995). Human IL-17: a novel cytokine derived from T cells. J. Immunol. 155, 5483–5486.
Ye, L., Jiang, B., Deng, J., Du, J., Xiong, W., Guan, Y., et al. (2015). IL-37 alleviates rheumatoid arthritis by suppressing IL-17 and IL-17-triggering cytokine production and limiting Th17 cell proliferation. J. Immunol. 194, 5110–5119. doi: 10.4049/jimmunol.1401810
Yeh, W. I., McWilliams, I. L., and Harrington, L. E. (2014). IFNgamma inhibits Th17 differentiation and function via Tbet-dependent and Tbet-independent mechanisms. J. Neuroimmunol. 267, 20–27. doi: 10.1016/j.jneuroim.2013.12.001
Yeilding, N., Szapary, P., Brodmerkel, C., Benson, J., Plotnick, M., Zhou, H., et al. (2011). Development of the IL-12/23 antagonist ustekinumab in psoriasis: past, present, and future perspectives. Ann. N. Y. Acad. Sci. 1222, 30–39. doi: 10.1111/j.1749-6632.2011.05963.x
Zanin-Zhorov, A., and Waksal, S. D. (2015). ROCKing cytokine secretion balance in human T cells. Cytokine 72, 224–225. doi: 10.1016/j.cyto.2014.12.025
Zhang, F., Meng, G., and Strober, W. (2008). Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat. Immunol. 9, 1297–1306. doi: 10.1038/ni.1663
Zhang, R., Tian, A., Wang, J., Shen, X., Qi, G., and Tang, Y. (2015). miR26a modulates Th17/T reg balance in the EAE model of multiple sclerosis by targeting IL6. Neuromol. Med. 17, 24–34. doi: 10.1007/s12017-014-8335-5
Zhang, S., Liang, R., Luo, W., Liu, C., Wu, X., Gao, Y., et al. (2013). High susceptibility to liver injury in IL-27 p28 conditional knockout mice involves intrinsic interferon-gamma dysregulation of CD4+ T cells. Hepatology 57, 1620–1631. doi: 10.1002/hep.26166
Zhao, F., Hoechst, B., Gamrekelashvili, J., Ormandy, L. A., Voigtlander, T., Wedemeyer, H., et al. (2012). Human CCR4+ CCR6+ Th17 cells suppress autologous CD8+ T cell responses. J. Immunol. 188, 6055–6062. doi: 10.4049/jimmunol.1102918
Zhu, E., Wang, X., Zheng, B., Wang, Q., Hao, J., Chen, S., et al. (2014). miR-20b suppresses Th17 differentiation and the pathogenesis of experimental autoimmune encephalomyelitis by targeting RORgammat and STAT3. J. Immunol. 192, 5599–5609. doi: 10.4049/jimmunol.1303488
Keywords: interleukin 17, autoimmune, transcription, microRNA, inflammation
Citation: Khan D and Ansar Ahmed S (2015) Regulation of IL-17 in autoimmune diseases by transcriptional factors and microRNAs. Front. Genet. 6:236. doi: 10.3389/fgene.2015.00236
Received: 16 February 2015; Accepted: 22 June 2015;
Published: 14 July 2015.
Edited by:
Sundararajan Jayaraman, University of Illinois College of Medicine, USAReviewed by:
Beisi Xu, St. Jude Children’s Research Hospital, USASuhasni Gopalakrishnan, University of Southern California, USA
Sundararajan Jayaraman, University of Illinois College of Medicine, USA
Copyright © 2015 Khan and Ansar Ahmed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: S. Ansar Ahmed, Department of Biomedical Sciences and Pathobiology, Virginia–Maryland College of Veterinary Medicine, Virginia Polytechnic Institute and State University, Phase II, Duck Pond Drive, Blacksburg, VA 24060, USA,YW5zcmFobWRAdnQuZWR1
†Present addres: Deena Khan, Cincinnati Children’s Hospital and Medical Center, Cincinnati, OH, USA
 Deena Khan†
Deena Khan† S. Ansar Ahmed
S. Ansar Ahmed