
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 02 July 2015
Sec. Evolutionary and Population Genetics
Volume 6 - 2015 | https://doi.org/10.3389/fgene.2015.00228
This article is part of the Research Topic Evolution of organismal form: from regulatory interactions to developmental processes and biological patterns View all 11 articles
Paired box transcription factors play important role in development and tissue morphogenesis. The number of Pax homologs varies among species studied so far, due to genome and gene duplications that have affected PAX family to a great extent. Based on sequence similarity and functional domains, four Pax classes have been identified in chordates, namely Pax1/9, Pax2/5/8, Pax3/7, and Pax4/6. Numerous splicing events have been reported mainly for Pax2/5/8 and Pax6 genes. Of significant interest are those events that lead to Pax proteins with presumed novel properties, such as altered DNA-binding or transcriptional activity. In the current study, a thorough analysis of Pax2/5/8 splicing events from cephalochordates and vertebrates was performed. We focused more on Pax2/5/8 and Pax6 splicing events in which the paired domain is involved. Three new splicing events were identified in Oryzias latipes, one of which seems to be conserved in Acanthomorphata. Using representatives from deuterostome and protostome phyla, a comparative analysis of the Pax6 exon-intron structure of the paired domain was performed, during an attempt to estimate the time of appearance of the Pax6(5a) mRNA isoform. As shown in our analysis, this splicing event is characteristic of Gnathostomata and is absent in the other chordate subphyla. Moreover, expression pattern of alternative spliced variants was compared between cephalochordates and fish species. In summary, our data indicate expansion of alternative mRNA variants in paired box region of Pax2/5/8 and Pax6 genes during the course of vertebrate evolution.
Transcription factors encoded by genes of the paired box (PAX) family are highly conserved throughout metazoan phyla and hold a vital role in embryonic development. The association of different PAX subfamilies with organ and tissue morphogenesis, such as the thymus, central nervous system (CNS), enteric nervous system, kidneys, ear, thyroid, neural crest, vertebrae, and midbrain-hindbrain boundary (MHB) formation has been the object of various studies (summarized in Noll, 1993; Chi and Epstein, 2002; Paixao-Cortes et al., 2013; Blake and Ziman, 2014). Certain members of the PAX family are characterized as the master control genes for eye morphogenesis (Gehring, 1996, 2002, 2012; Kozmik, 2008; Klimova and Kozmik, 2014) and genetic defects are linked to the onset of eye-related diseases, e.g., small eye in mouse or aniridia in human. Mutations in Pax genes are also correlated with diseases of the kidney and the CNS, but various types of cancer as well (see Chi and Epstein, 2002; Lang et al., 2007; Paixao-Cortes et al., 2013; Blake and Ziman, 2014 and references therein).
Gene and genome duplications, followed by gene losses, helped shape PAX gene family, leading to a varying number of Pax homologs in metazoan phyla studied so far (reviewed in Noll, 1993; Breitling and Gerber, 2000; Hoshiyama et al., 2007; Paixao-Cortes et al., 2013). Different subfamilies have been identified and are classified according to the similarity in sequence, functional domains they possess as well as their expression patterns (see Stuart et al., 1994; Blake and Ziman, 2014 and references therein). PaxB is apparently the oldest member and is present in sponges and cnidarians (Kozmik et al., 2003; Hoshiyama et al., 2007; Hill et al., 2010). Pox neuro and single genes from Pax1/9, Pax2/5/8, Pax4/6, and Pax3/7 classes are present in cephalochordates (Short and Holland, 2008; Takatori et al., 2008). Pox neuro is present in Drosophila (Bopp et al., 1989), but is lost in the lineages of tunicates and vertebrates. Two Pax258 genes are present in urochordates, due to a duplication prior to ascidian and larvacean diversification (Wada et al., 2003; Canestro et al., 2005). As a result of two rounds of whole-genome duplication (Escriva et al., 2002; Putnam et al., 2008) and subsequent gene losses, the coelacanth Latimeria chalumnae possesses nine Pax genes, i.e., discrete gene copies from each of the Pax1/9, Pax2/5/8, Pax3/7, and Pax4/6 classes, suggesting that all members of the PAX family were present in the ancestor that gave rise to the tetrapod lineage (Paixao-Cortes et al., 2013). More than nine genes are present in teleost fishes (reviewed in Ravi et al., 2013), due to the so-called third round of genome duplication (Jaillon et al., 2004; Van De Peer, 2004).
All Pax proteins contain a DNA-binding domain in their N-terminus, known as the paired domain (PD), as well as transactivation and inhibitory domains in their C-terminus. The PD consists of 128 aminoacids and is made up by two helix-turn-helix (HTH) subdomains, known as PAI and RED or N-terminal and C-terminal respectively, joined through a linker (Czerny et al., 1993; Xu et al., 1999). The Pax4/6 class contains an additional DNA-binding homeodomain, while the Pax2/5/8 possesses a partial homeodomain and an octapeptide motif, the latter known to interact with members of the Groucho family of co-repressors (Eberhard et al., 2000; Kreslova et al., 2002). Classes Pax1/9 and Pax3/7 both contain the octapeptide motif, yet the former lacks the homeodomain (Chi and Epstein, 2002). Pax loci which encode for proteins with truncated paired domain have been identified in Drosophila, C. elegans, as well as representatives of Hemichordata and Echinodermata (Chisholm and Horvitz, 1995; Cinar and Chisholm, 2004; Howard-Ashby et al., 2006; Friedrich and Caravas, 2011; Ravi et al., 2013).
Gene/genome duplication is a driving force for evolution (Bergthorsson et al., 2007; Maere and Van De Peer, 2010) and this could be nicely exemplified by the well-studied PAX gene family, where many duplicates were preserved in the genome and obtained new functions and new domains of expression (neofunctionalization), or original gene functions were partitioned (subfunctionalization) between duplicates (Pfeffer et al., 1998; Bassham et al., 2008; Kleinjan et al., 2008, reviewed in Holland and Short, 2010).
At posttranscriptional level, alternative splicing is also known to promote evolution, protein diversity and development of novel functions in eukaryotic genomes (reviewed in Nilsen and Graveley, 2010; Chen et al., 2012; Kelemen et al., 2013). In fact, it has been suggested that in the case of Pax genes the impact of alternative splicing on functional motifs is more intense than gene duplication and subsequent divergence of the duplicated genes (Short and Holland, 2008). It has been shown that alternative spicing usually takes place in a tissue or developmental stage-specific manner (Wang et al., 2008a; Kelemen et al., 2013). Depending on which exonic segments are cut-out and whether intronic regions are retained in the transcripts, splicing events can be clustered into four major groups, namely (1) exon skipping, (2) alternative 3′-, (3) alternative 5′-splice sites, and (4) intron-inclusion (Koralewski and Krutovsky, 2011). These four types of events can occur independently or in combination with other incidents, such as mutually exclusive exons, alternative initiation and alternative polyadenylation (Wang et al., 2008a; Koralewski and Krutovsky, 2011; Kelemen et al., 2013).
Alternative splicing of Pax genes has been observed in various species, from protostomes (Fu and Noll, 1997; Cinar and Chisholm, 2004) to cephalochordates (Glardon et al., 1998; Kozmik et al., 1999; Short and Holland, 2008; Holland and Short, 2010; Short et al., 2012) and vertebrates (Kozmik et al., 1993, 1997; Poleev et al., 1995; Heller and Brandli, 1997, 1999; Lun and Brand, 1998; Short et al., 2012), where multiple incidents from all major groups of splicing events were present. In principle, splice forms seem to have diverged between lineages, some of them are species- or genus-specific (Heller and Brandli, 1999; Short et al., 2012), nevertheless several splice isoforms seem to be evolutionary conserved (Kwak et al., 2006; Short and Holland, 2008; Short et al., 2012; Ravi et al., 2013). The majority of reported splice events regards the transactivation and inhibitory domain in the C-terminal part of the Pax proteins (Kozmik et al., 1993; Ward et al., 1994; Nornes et al., 1996; Tavassoli et al., 1997; Kreslova et al., 2002; Robichaud et al., 2004), still there is an increasing number of events affecting paired domain and consequently DNA binding capacity (Kozmik et al., 1993, 1997; Zwollo et al., 1997; Short and Holland, 2008; Short et al., 2012). One such example is the Pax6(5a) isoform, where inclusion of exon 5a (Walther and Gruss, 1991; Glaser et al., 1992; Puschel et al., 1992) leads to a protein with interrupted paired domain that recognizes an altered DNA binding sequence (Epstein et al., 1994). Apparently, this event is quite conserved among vertebrate lineages, with differences in size and peptide sequence of exon 5a between fish and tetrapods (Ravi et al., 2013). In some cases, alternatively spliced Pax isoforms exhibit temporally and spatially differentiated expression patterns (Kozmik et al., 1993, 1997; Heller and Brandli, 1997; Short and Holland, 2008) and have been associated with cancer and genetic disorders (reviewed in Wang et al., 2008b; Holland and Short, 2010).
Previous studies have shown that any insertion in the conserved paired domain, no matter if it is a single-aminoacid extension or a whole exon cassette, modifies DNA binding capacity and attributes differentiated functions to the isoforms bearing the insertion (Kozmik et al., 1997; Azuma et al., 2005).
In the present study, we sought to identify splicing events in Pax2/5/8 and Pax6 classes that affect the paired domain and study the expression patterns of these alternative spliced transcripts. We identified three new splicing events in Oryzias latipes Pax2 genes, one of which seems to be highly conserved in Acanthomorphata. We detected a re-occurring splicing event in O. latipes and Danio rerio Pax6 genes generating the exon 5a insertion. Using our data set we tried to elucidate the time point at which the exon 5a-insertion appeared and the extent of its conservation in various phylogenetic groups.
Nucleotide and aminoacid sequences for annotated Pax2/5/8 and Pax6 genes were obtained using proper keywords, through NCBI GenBank (Benson et al., 2013), ENSEMBL release 78 (Cunningham et al., 2015), the UCSC Genome Browser database (Karolchik et al., 2014), the SpBase (Sea Urchin Genome Database, Cameron et al., 2009) and the JGI (Grigoriev et al., 2012). The retrieved Pax genes were crosschecked using GENSCAN (Burge and Karlin, 1997), BLASTx and version 0.9 of the NNSPLICE splice predictor (Reese et al., 1997).
For various taxonomic groups (e.g., Chondricthyes:Holopocephali) there are available genomes, but no Pax genes are annotated in public databases. In order to include representatives from these groups, we conducted BLAST searches against the NCBI GenBank and wgs subdivision (Trace archive), using known homologs from Deuterostomia species. Where required, small contigs or scaffolds were fused using Merger of the EMBOSS software suite (Rice et al., 2000) and gene structure was defined using GENSCAN (Burge and Karlin, 1997), BLASTx and splice predictor (Reese et al., 1997). ScanProsite (De Castro et al., 2006) was used to detect conserved functional domains in newly identified genes. In addition, adjacent genes of the de novo predicted Pax genes were also predicted/annotated and gene order was compared to known Pax syntenic regions through the Genomicus website v78.01 (Louis et al., 2013). In all cases, the NNSPLICE was used for the prediction of possible alternative acceptor and donator sites.
PipMaker (Schwartz et al., 2000) was used along with BLAST, in order to locate putative sequence conservation among species. Alignment of Pax6 paired domains from various species was performed using the MUSCLE algorithm (Edgar, 2004) included in Mega version 5.0 (Tamura et al., 2011).
In order to validate already annotated or newly predicted Pax homologs, BLAST searches were performed against the ESTs subdivision. Collected ESTs were aligned with predicted coding sequences from genome analyses and mRNA sequences—if available—in order to detect putative splicing events not recognized so far.
Specimens of Branchiostoma floridae were collected from Old Tampa Bay, Florida, USA. Gametes were obtained and embryos were raised, as previously described (Holland and Yu, 2004). B. lanceolatum adults were collected in Banyuls-sur-Mer, France, prior to summer breeding season and raised in the lab until spawning. The spawning of males and females was induced by temperature shift (Fuentes et al., 2007). B. lanceolatum and B. floridae embryos were developed at 16°C and 26°C, respectively. Embryos of inbred strains of Oryzias latipes (Cab) and Danio rerio (AB) were used for all experiments. O. latipes and D. rerio embryonic stages were determined according to Iwamatsu (2004) and Kimmel et al. (1995). Housing of animals and in vivo experiments were performed after approval by the Animal Care Committee of the Institute of Molecular Genetics (study ID#36/2007) and in compliance with national and institutional guidelines (ID#12135/2010-17210).
Total RNA was isolated from embryos using the Trizol reagent (Ambion). Random-primed cDNA was prepared in a 20 μl reaction from 500 ng of total RNA using SuperScript VILO cDNA Synthesis kit (Invitrogen).
cDNA was subjected to PCR using DreamTaq polymerase (Thermo Scientific) for 30 cycles under the following conditions: 1 min at 98°C, 30 s at 60°C, 30 s at 72°C. Primers for this analysis are provided in Table 1. PCR products were analyzed on 2.5% agarose gel and bands of interest were eluted, cloned to pCR-Blunt II (Invitrogen) and sequenced (GATC Biotech sequencing service, Germany).
Exhaustive search through databases and literature, in combination with de novo analysis of available ESTs and mRNA sequences (Table S1), revealed numerous splicing events in chordate members of the Pax2/5/8 class (Figure 1). Some of these events seem to characterize specific orthologs and are present in cephalochordates, fish and mammals (e.g., exon 2 of Pax5 gene), while others are much less conserved (e.g., exon 3a of mouse Pax5). Branchiostoma floridae appears to experience the largest number of splicing events, however no event of insertion in the paired domain has been reported so far and no such event could be predicted using splice prediction software or available ESTs/mRNA sequences.
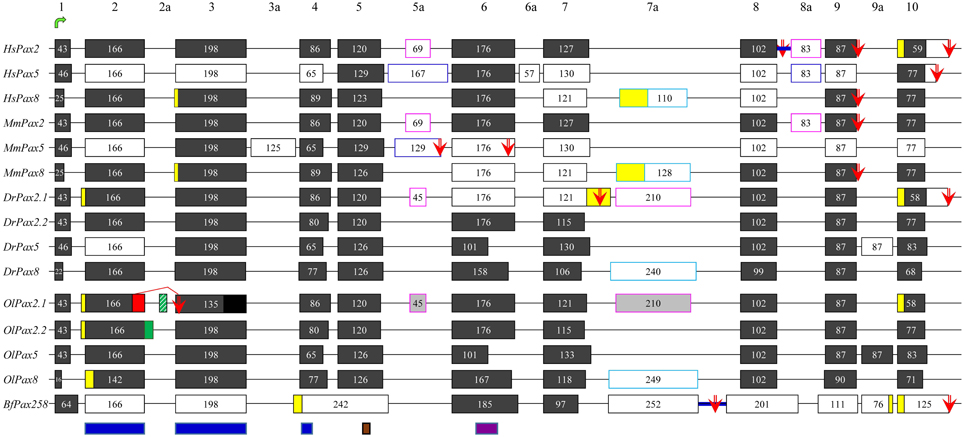
Figure 1. Exon-intron organization and alternative splicing events of Pax2/5/8 representatives from vertebrates and cephalochordates. Dark gray and white boxes represent constitutive and alternatively spliced exons, respectively. Yellow boxes denote alternatively spliced parts of exons, due to different 5′ or 3′ splicing donors/acceptors. The suffix “a” is used for non-canonical exons, characteristic of the different Pax2/5/8 genes (different colors of outline is used for different orthologs). For O. latipes, light gray boxes were predicted due to high similarity to respective exons in D. rerio, whereas black box shows the non-sequenced part of exon 3. Lines represent introns (not drawn to scale). Blue thick lines represent intron retention events. Green arrow points to exon containing translation initiation, while red arrows stand for alternative stop codons. Blue, brown, and purple boxes define borders of paired domain, the octapeptide, and partial homeodomain, respectively. Alternative splicing events detected in the present study are indicated by red and green boxes (alternative 5′ splice donors) and green stripped box (exon cassette). Previously published data are included (Dressler et al., 1990; Krauss et al., 1991; Kozmik et al., 1993, 1997, 1999; Ward et al., 1994; Poleev et al., 1995; Zwollo et al., 1997; Lun and Brand, 1998; Pfeffer et al., 1998; Borson et al., 2002; Robichaud et al., 2004; Kwak et al., 2006; Short and Holland, 2008; Arseneau et al., 2009; Busse et al., 2009).
A single Oryzias latipes Pax2 gene, namely OlPax2.2, located on chromosome 19 (NC_019877), has been used in previous studies (Paixao-Cortes et al., 2013). Our search through NCBI revealed an annotated Pax-2a-like gene on chromosome 15 (NW_004088010.1). It must be noted that the aminoacid sequence encoded by the first half of this gene exhibits no similarity to the paired domain of other Pax genes and it is not supported by ESTs, a fact that indicates an erroneous gene prediction, caused by a non-sequenced area in this genomic region. We assume that the first coding exon as well as the two exons coding for the paired domain of OlPax2.1 are located in this non-sequenced region. The record of an unplaced scaffold (NW_004093539, Table S1) was retrieved through BLAST. It apparently corresponds partly to the non-sequenced region of chromosome 15 and contains the 5′ UTR, the first exon of OlPax2.1 gene and part of the first intron.
In order to retrieve more information on OlPax2.1 and to elucidate how many OlPax2 (either Pax2.1 or Pax2.2) transcripts exist, we searched for different ESTs and mRNA sequences using OlPax2.2 and Danio rerio Pax2.1 as queries. There are only two ESTs (AM320053 and AM321390) for OlPax2.1 that contain the first coding exon, as well as the complete exon 2 and part of exon 3, encoding for N- and C- paired subdomains, respectively. Using the genomic scaffolds and available ESTs collectively (Table S1), an almost complete OlPax2.1 gene was reconstructed (Figure 1). For OlPax2.2 gene, one cDNA sequence and four ESTs were retrieved (Table S1), comparison of which revealed both 5′ and 3′ alternatively spliced parts of exon 2 encoding for the N-terminal of paired domain (see Figure 1).
Exon-to-exon comparison of Pax2/5/8 genes between O. latipes and D. rerio, shows that in principle there is conservation in the sequence, number, size, and borders of exons and some indication for alternatively spliced exons in OlPax2 genes (light gray boxes in Figure 1), which are not suggested by the available ESTs. Retrieved ESTs encoding both OlPax2.1 and OlPax2.2 support a 5′ alternative splicing donor site in exon 2 (Figure 1, Table S1); the insertion is 12 bp long, and results to four additional aminoacids, exactly at the beginning of the paired domain. The same insertion has been reported for D. rerio Pax2.1 (Lun and Brand, 1998), as well.
In the present study we identified two splicing events that lead to insertion of extra aminoacids in the paired domain of OlPax2 genes. More specifically, an alternatively spliced 21-bp exon was detected between exons 2 and 3 of fish Pax2.1 genes, which is annotated in some species (e.g., Poecilia reticulata and Maylandia zebra). This exon could not be detected in silico in the genome of O. latipes, due to the fact that the intron between exons 2 and 3 is not sequenced. Through BLAST searches and de novo analysis of Pax2.1 genes we spotted this exon in numerous representatives from different orders of Acanthomorphata (Table S2), while PipMaker alignment reveals a high degree of sequence conservation among compared species (Figure 2). A putative exon with proper splice sites has been identified in the respective genomic region of three Cyprinidae species (D. rerio, Pimephales promelas, and Cyprinus carpio). Even though this exon is highly conserved in these species, the encoded aminoacids are quite dissimilar from those of the Acanthomorphata 21-bp exon (Figure 2). In both cases, inclusion of this exon leads to an alternative transcript, which incorporates seven extra aminoacids toward the end of the a3 helix of the PAI subdomain (Figure 2).
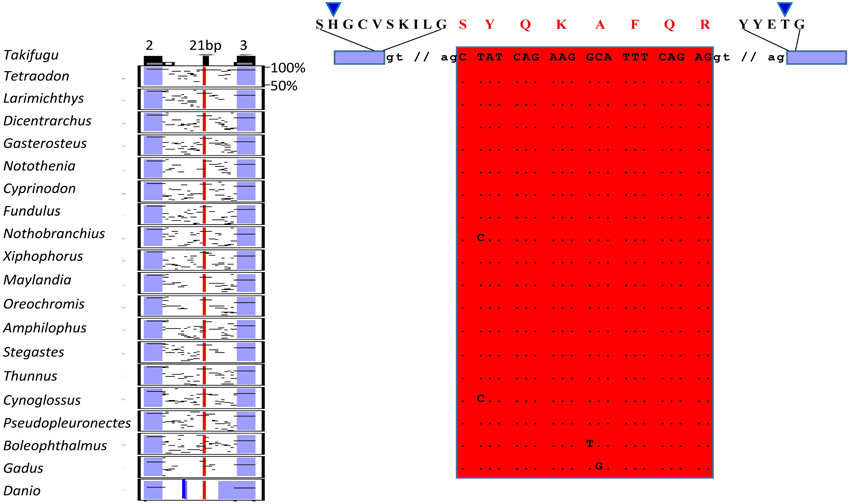
Figure 2. Alternatively spliced isoform OlPax2.1(2a+). Pip diagram of paired-encoding (2 and 3, blue boxes) and alternatively spliced exon (21 bp, red box) in Acanthomorphata species. Blue thick line represents alternatively spliced exon predicted in three Cyprinidae species. Dots stand for conserved residues, small case letters correspond to intronic nucleotides. Blue arrowheads point to PAI a3 helix.
In regard to OlPax2.2, the available mRNA sequence in GenBank and our analysis revealed a 24-bp in-frame extension at the 3′ end of exon 2 (Figures 1, 3, Table S1), which does not alter the downstream translation (Figure 3). This isoform, to which we will refer as OlPax2.2(ext24+), is due to an alternative splicing donor downstream the canonical one (Figure 3). In D. rerio, a similar isoform is neither supported by splicing prediction software nor by available mRNA sequences.
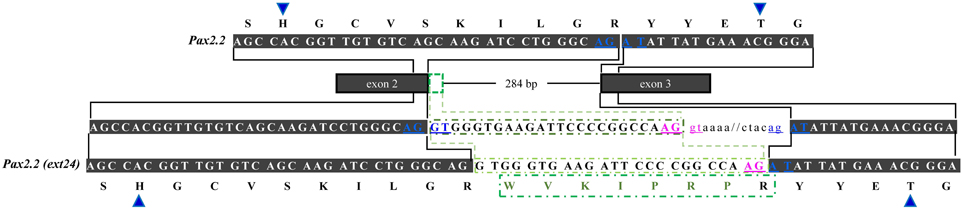
Figure 3. Alternatively spliced isoform OlPax2.2(ext24+). Dark gray boxes represent constitutive exons, green-outlined box shows part of intron 2 (24 bp) retained in OlPax2.2. Thin line represents intron 2 and intronic sequences are in small case letters. Canonical splice sites are in blue letters. Blue arrowheads point to PAI a3 helix. Green letters represent the extra aminoacids incorporated in OlPax2.1(ext24+), when the downstream alternative splicing donor (pink letters) is used.
It should be noted that splicing prediction analysis of OlPax2.1 gene revealed the presence of an alternative donor site in exon 2, upstream the canonical one. Deletion of 35 bp at the 3′ end of exon 2 causes a frameshift and leads consequently to a premature stop codon at the beginning of exon 3 (Figure S1). A similar donor site was not in silico identified in D. rerio (data not shown).
In teleosts and tetrapods studied so far, the major part of Pax6 paired domain is encoded by two exons, responsible for the N- and C-subdomains, with a size of 131 and 216 bp, respectively. In Tetrapoda and in the Pax6.1 copy of teleosts, a small exon of varying size (36–42 bp), namely 5a, has been shown to be included in alternative transcripts, causing an in-frame insertion in the paired domain (Ravi et al., 2013). We wanted to identify at which point of evolutionary history this exon appeared and investigate a putative correlation of the appearance of this exon with the exon/intron organization of Pax6 homologs. For this reason, already annotated Pax6 homologs were collected from public databases and available genomes and EST sequences from non-jawed vertebrates, cephalochordates, tunicates, hemichordates, echinoderms, as well as Drosophila and C. elegans were analyzed (Table S3).
Analysis of a genomic scaffold from Leucoraja erinacea, containing the Pax6 ortholog, provides evidence that besides Holocephali (Ravi et al., 2013), the 5a exon is also present in Elasmonbranchii, the second subclass of Chondricthyes. Unfortunately, no genomes from hagfishes are publicly available, yet the two Pax6 mRNA sequences that were retrieved from Eptatretus bergeri (Table S3), do not provide any indication of an exon5a-like insertion in the paired domain.
In regard to Hyperoartia, genomic scaffolds containing parts of the Pax6 genes from Petromyzon marinus and Lethenteron japonicum were retrieved and analyzed, along with two Pax6 mRNA from the species L. japonicum and Lampetra fluviatilis. Apparently, there are more than one Pax6 genes in the L. japonicum genome, yet the low genome coverage for both P. marinus and L. japonicum (5× and 20×, respectively) does not allow for safer conclusions. In all cases, there is no evidence for the existence of the exon 5a in Hyperoartia.
Existing models for Stongylocentrotus purpuratus predict two truncated neighboring Pax6 proteins that contain either the paired or the homeobox domain. Taking into account the provided information in SpBase mentioning that the gene models are incomplete and an intervening sequence appears to be missing in a scaffold gap in the Spur3.1 assembly (Howard-Ashby et al., 2006; Cameron et al., 2009), we re-evaluated the prediction and tried to re-construct the SpPax6 homolog.
Our focus was on the exon-intron structure in the region of paired domain (Figure 4). It is apparent, from our analysis, that the size and borders of the two exons encoding the main part of paired domain underwent various changes in different taxonomic groups (Figure 4). More specifically, in basal deuterostomes, such as Echinodermata and Hemichordata, there is one large exon with a size of 347 bp encoding for the first 115 aminoacids of the paired domain. Therefore, there is no intervening non-coding region in the respective position to vertebrate's intron, or in other words “space” for insertion of an alternatively spliced exon in the paired domain. In cephalochordates and tunicates, this large exon has split into two exons, first of which has a size of 166 bp, still larger than the N-subdomain encoding exon in vertebrates, and the second one is 181 bp long, slightly smaller than the respective exon in vertebrates. The 166 bp exon encodes a peptide including the first four aminoacids of a3 helix of the N-subdomain (Xu et al., 1995) and ends shortly after the position where the exon 5a is inserted, i.e., downstream of a3 helix. Thorough in silico search in the intronic sequence flanked by the paired-encoding exons in tunicates and cephalochordates did not reveal a putative alternatively spliced exon cassette similar to the exon 5a (Figure 4, Figure S2). It seems that paired-encoding exons obtained fixed borders and size of 131 and 216 bp for PAI and RED, respectively before diversification of cyclostomes and preserved them throughout vertebrates, nonetheless “birth” of exon 5a probably appeared in Gnathostomata (Figure 4, Figure S2). It should be noted that the major re-arrangements concern the genomic region encoding for the PAI a3 helix, in contrast to the high conservation observed toward the C-subdomain.
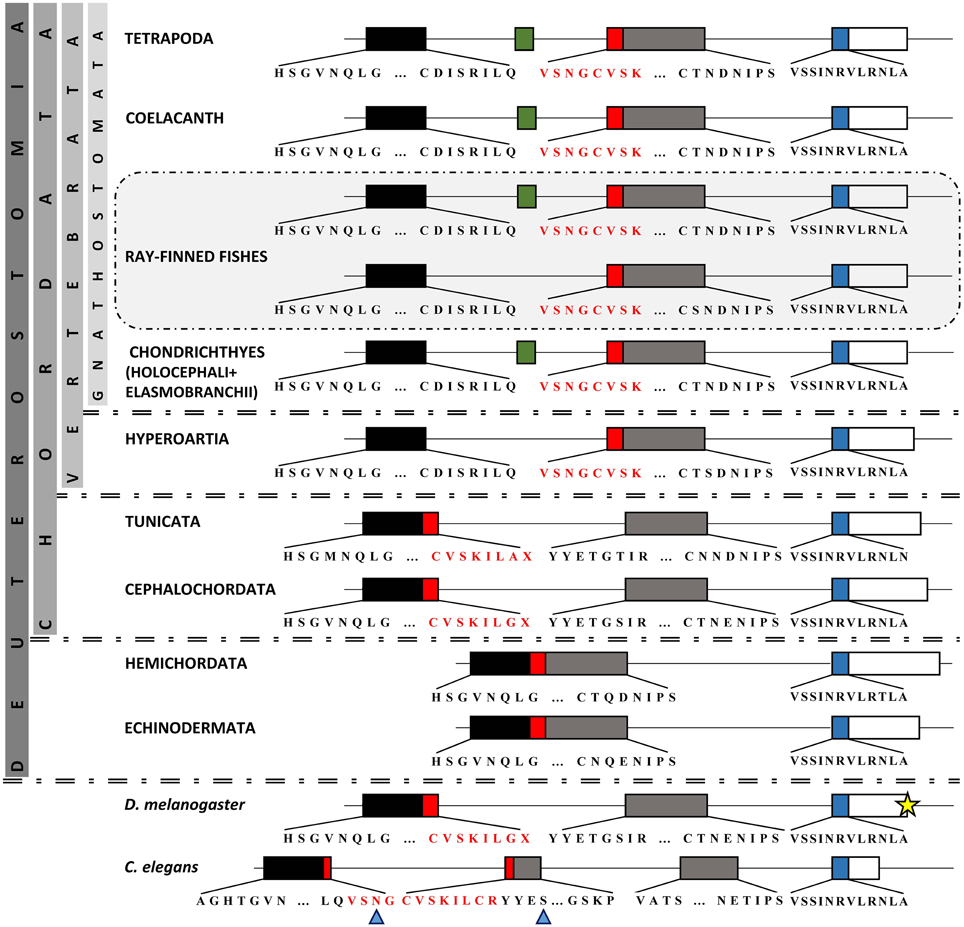
Figure 4. Comparison of Pax6 paired domain among representatives from Protostomia and Deuterostomia. Genomic organization of paired domain-encoding exons. Boxes represent exons, thin lines represent introns (the latter are not drawn to scale). Different colors of exons correspond to different subdomains. Green box represents the previously reported exon 5a (Walther and Gruss, 1991). Red box represents the section of paired domain that underwent re-arrangement throughout evolution. For practical reasons, the third exon (665 bp) in Drosophila is not depicted to its full length (yellow star). The residue X reveals an asymmetrical exon. Blue arrowheads point to PAI a3 helix.
To verify in silico predicted alternative splice isoforms (Figures 2–4, Figure S1) and compare their expression across various developmental stages, we performed RT-PCR using RNA from different embryonic stages of Branchiostoma lanceolatum, B. floridae, Oryzias latipes, and Danio rerio (Figure 5).
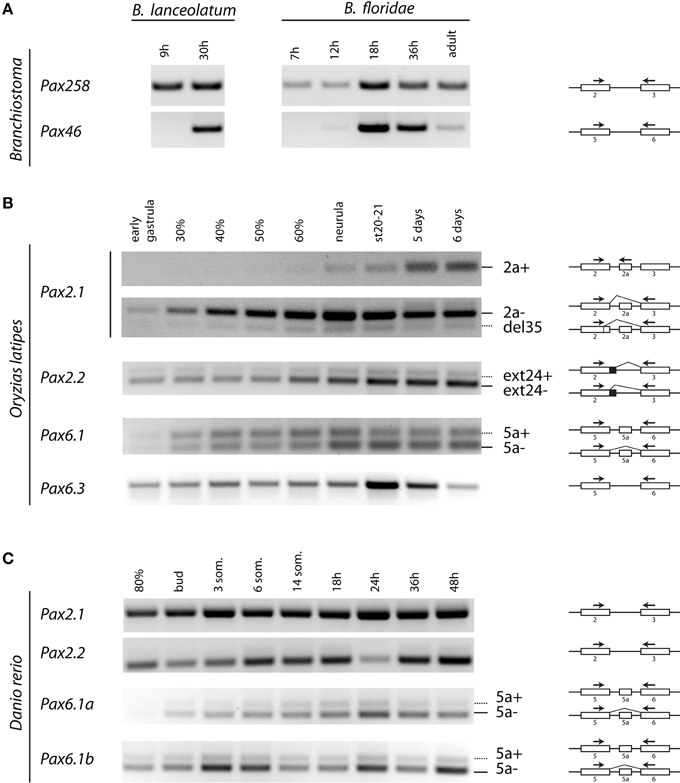
Figure 5. Temporal expression of Pax2 and Pax6 genes during development of Branchiostoma lanceolatum, B. floridae, Oryzias latipes, and Danio rerio embryogenesis. cDNA sequences were amplified across the paired encoding exons, as indicated by arrows. (A) For B. lanceolatum and B. floridae, single isoforms were detected for Pax258 and Pax46 genes. (B) For O. latipes Pax2.1, an isoform containing exon 2a was detectable from neurula stage. Pax2.1(del35) and Pax2.2(ext24+) as well as Pax6.1(5a) isoforms are expressed across all developmental stages. Pax6.3 gene does not possess exon 5a. (C) No alternatively spliced isoforms were detected in the case of D. rerio Pax2.1 and Pax2.2 genes. Isoforms including exon 5a are present for both Pax6.1a and Pax6.1b genes.
In agreement with our in silico analysis, RT-PCR (Figure 5) and DNA sequencing (data not shown) using primers located in exons encoding PAI and RED domains revealed that B. lanceolatum and B. floridae express single Pax258 and Pax46 isoforms.
Three splice isoforms of OlPax2 genes have been in silico predicted. Lack of information concerning intron 2 of OlPax2.1 prohibited the in silico detection of the 21-bp exon, characteristic of Acanthomorphata. However, the presence of 21-bp exon in the O. latipes genome was experimentally verified, using a proper set of primers (Table 1), one of which was specifically designed on the 21-bp exon characteristic of Acanthomorphata (Figure 2). Expression of the alternatively spliced OlPax2.1(2a+) isoform was detectable from neurula and later developmental stages (Figure 5). In contrast, the truncated isoform OlPax2.1(del35), which results from a deletion in exon 2 and a premature stop codon in the 3rd exon (Figure S1), was present across all developmental stages (Figure 5). OlPax2.1(del35) was expressed at much lower level than dominant OlPax2.1(2a-) isoform. Sequencing of this isoform revealed that it makes use of the alternative donor site in exon 2, as predicted by in silico analysis (Figure S1). The extended isoform OlPax2.2(ext24+) (Figure 3) was present at detectable level throughout the examined stages (Figure 5). In the case of D. rerio Pax2.1 and Pax2.2 genes, no alternative splice variants were detected, in agreement with splicing prediction analysis.
The alternatively spliced isoform OlPax6.1(5a-) was expressed approximately at the same level as isoform OlPax6.1(5a+) (Figure 5). In agreement with previous studies (Ravi et al., 2013), O. latipes Pax6.3 gene does not possess the equivalent of exon 5a. Variants bearing exon 5a were observed for both D. rerio Pax6.1a and Pax6.1b genes, and in both cases, expression level of isoform 5a- was relatively higher than isoform 5a+.
In silico and experimental data, collectively, demonstrate increased complexity of splicing events in vertebrate paired domain of Pax genes in comparison to cephalochordates.
Paired box (Pax) genes encode for transcription factors that are considered key players in organogenesis and embryonic development. The presence of Pax genes in a variety of organisms and the evolution of the PAX family has been the object of various studies (Hill et al., 2010; Paixao-Cortes et al., 2013; Ravi et al., 2013). Whole-genome duplications as well as lineage-specific gene duplications provide additional possibilities for diversified evolution and/or speciation (Bergthorsson et al., 2007; Maere and Van De Peer, 2010). These processes are considered to have played important role in shaping the number of Pax homologs in various taxonomic groups (see Paixao-Cortes et al., 2013; Ravi et al., 2013 and references therein), but same applies for alternative splicing, a posttranslational mechanism that also promoted evolution and complexity of Pax proteins (Glardon et al., 1998; Short et al., 2012).
In the present study, we wanted to evaluate the degree of alternative splicing taking place in various lineages, as well as to identify splicing events that are either evolutionary conserved or characteristic of cephalochordates and not vertebrates or vice versa. For this purpose, we collected annotated homologs from Pax2/5/8 and Pax4/6 classes from public databases (NCBI, Ensembl, UCSC, JGI and SpBase). Furthermore, we analyzed de novo genomes, ESTs and mRNA sequences from different species, in order to enrich our dataset with taxonomic groups not present in previous studies. Our second focus was Pax isoforms from cephalochordates and vertebrates, that differ in the paired domain, and their expression patterns across different developmental stages.
Apart from partially reconstructing the Oryzias latipes Pax2.1, using available scaffolds and EST sequences, we identified three new splicing events in the Pax2 genes of O. latipes. The OlPax2.1(2a+) isoform is reminiscent of the 5a isoform found in Pax6 homologs (Ravi et al., 2013, this study), as it incorporates a 21-bp in-frame-exon located in the intron between the two exons encoding for the paired domain. Our analysis showed that this exon is present in numerous species from various orders of Acanthomorphata and exhibits a high degree of conservation among compared species (Figure 2). We presume that sequence conservation of this mRNA splice form over a wide phylogenetic distance also implies conservation of this isoform's function. A similar exon in terms of location, yet quite divergent in terms of sequence, was in silico predicted only in three Cyprinidae species (D. rerio, Pimephales promelas, and Cyprinus carpio).
The second alternatively spliced isoform, namely OlPax2.2(ext24+), results from the use of an alternative splice donor downstream the canonical one at the end of OlPax2.2 exon 2. In this case, extra aminoacids are incorporated in the middle of a3 helix of the PAI subdomain, with no influence on the downstream sequence. A similar isoform could not be detected in D. rerio, neither experimentally nor in silico. The sequences surrounding the normal splice junctions of exon 2-intron 2 are highly conserved between D. rerio and O. latipes Pax2.2 genes, yet there is no proper donor-acceptor site in the region of D. rerio (AG-AG) that corresponds to the alternative splice site of O. latipes.
Both OlPax2.1(2a+) and OlPax2.2(ext24+) transcripts bear an insertion in the recognition a3 helix of PAI subdomain. Previous studies on insertions in the paired domain of Pax genes have proven that, regardless of the number of the inserted aminoacids, disruption of this helix, which is responsible for all major groove DNA contacts of the N-terminal subdomain (Xu et al., 1995, 1999) is expected to inactivate the DNA-binding function of the N-terminal HTH motif, which subsequently leads to severe restriction in the DNA-binding sequence specificity of the paired domain (Kozmik et al., 1997).
The importance of alternative splicing as a mechanism for divergent evolution is established. In the case of Pax genes, the fact that insertions in the paired domain may preferentially guide Pax proteins, namely Pax6(5a) and Pax8(S), to the control region of genes containing a modified binding site (5aCON-like sequence, Kozmik et al., 1997), in other words insertions add new target-genes in the repertoire of genes controlled by Pax genes, may be indicative of a mechanism through which alternative splicing contributes to the increase of complexity at the level of protein function.
The isoform OlPax2.1(del35) makes use of an alternative 5′ splicing donor, upstream of the normal splicing site in exon 2 (N-terminal of paired domain). As mentioned before, the exact junction sequence between exon 2 and intron 2 is not known, nevertheless, the sequence at the normal end of exon 2 (CAG) is in agreement with the optimal consensus for 5′ splice sites (Stephens and Schneider, 1992), in contrast to the sequences at the alternative upstream 5′ splice donor (CGG/GT, Figure S1). As it has been observed before for the Pax8 gene (Kozmik et al., 1997), there is a higher abundance and constitutive splicing of the Pax2 mRNA relative to the alternative transcript (Figure 5), a fact that could be attributed to different affinities by which the spliceosomes may recognize the two 5′ donor sites. A similar truncated isoform could not be detected neither during Pax2.1 transcript analysis of D. rerio, nor by in silico analysis.
The alternative isoform OlPax2.1(del35), lacks the greater part of a3 helix of PAI domain and ends at a premature stop codon exactly at the beginning of exon 3. Truncated isoforms are not a rare phenomenon, given the fact that approximately 35% of alternatively spliced human transcripts have been found to contain a premature termination codon, rendering them as candidates for non-sense- mediated decay (Green et al., 2003; Lewis et al., 2003). It has been proposed that most low copy number alternative isoforms produced in human cells are likely to be non-functional, therefore we assume that this is also the case for OlPax2.1(del35). Deletion of a3 helix has been observed in one of the Pax6 isoforms in B. floridae (Glardon et al., 1998), yet this deletion does not influence downstream translation and hence its functionality. A 32 bp deletion in mouse is responsible for splotch phenotype in mouse (Epstein et al., 1991). In addition, there are accumulating reports about heterozygous deletions of parts of PAI subdomain in general or a3 helix in specific, most of which cause a frame shift and a premature stop codon (Schimmenti et al., 1997; Fletcher et al., 2005) and are correlated with diseases in human (e.g., renal-coloboma syndrome, oligomeganephronia).
In regard to the Pax6 class and the alternative splice isoform Pax6(5a), our analysis showed that an important re-arrangement of coding and non-coding sequences in the region of paired domain took place during evolution. Although conservation of the position of introns has been noted between highly divergent eukaryotes, the number and placement of the majority of introns are dynamically fluctuating during evolution (Hartung et al., 2002; Rogozin et al., 2003). In Hemichordata and Echinodermata, the exon-intron organization does not allow for any type of insertions in the paired domain. In other lineages compared, paired domain is encoded by exons disrupted by one or more introns. Incidents of intron gain and loss as well as intron sliding have been reported for various genes (Hartung et al., 2002), whereas the intron density, i.e., the average number of intron per gene does not necessarily coincide with the position of the genome on the evolutionary tree (Jeffares et al., 2006). We assume that the 5a insertion is characteristic of Gnathostomata. Introns are required for alternative splicing and alternative splicing increases the size of the proteome, thus increasing the level of complexity in higher eukaryotes. Moreover, introns have been found to harbor many conserved non-coding elements, necessary for gene regulation (Irvine et al., 2008; Bhatia et al., 2014).
Unique isoforms were detected during expression pattern study of Branchiostoma Pax258 and Pax46 genes. This is in agreement with in silico analysis, during which no splicing events involving the paired domain were predicted. In contrast, new alternative spliced variants were identified for fish species. Previous studies have shown that there is no developmental regulation of paired domain alternative splice forms of Pax6 and Pax8, as opposed to splicing events affecting the C-terminal sequences of Pax8 protein (Kozmik et al., 1993, 1997). In principle, non-constitutive OlPax2 isoforms are expressed at low levels, therefore at this stage, it is not easy to conclude as to the regulation of these isoforms. Nonetheless there is an indication of a temporal regulation of OlPax2.1(2a+) isoform, which requires further investigation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was supported by grant LH12047 from the Ministry of Education, Youth and Sports of the Czech Republic and the project “BIOCEV—Biotechnology and Biomedicine Centre of the Academy of Sciences and Charles University” (CZ.1.05/1.1.00/02.0109). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fgene.2015.00228
Arseneau, J. R., Laflamme, M., Lewis, S. M., Maïcas, E., and Ouellette, R. J. (2009). Multiple isoforms of PAX5 are expressed in both lymphomas and normal B-cells. Br. J. Haematol. 147, 328–338. doi: 10.1111/j.1365-2141.2009.07859.x
Azuma, N., Tadokoro, K., Asaka, A., Yamada, M., Yamaguchi, Y., Handa, H., et al. (2005). The Pax6 isoform bearing an alternative spliced exon promotes the development of the neural retinal structure. Hum. Mol. Genet. 14, 735–745. doi: 10.1093/hmg/ddi069
Bassham, S., Canestro, C., and Postlethwait, J. (2008). Evolution of developmental roles of Pax2/5/8 paralogs after independent duplication in urochordate and vertebrate lineages. BMC Biol. 6:35. doi: 10.1186/1741-7007-6-35
Benson, D. A., Cavanaugh, M., Clark, K., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., et al. (2013). GenBank. Nucleic Acids Res. 41, D36–D42. doi: 10.1093/nar/gks1195
Bergthorsson, U., Andersson, D. I., and Roth, J. R. (2007). Ohno's dilemma: evolution of new genes under continuous selection. Proc. Natl. Acad. Sci. U.S.A. 104, 17004–17009. doi: 10.1073/pnas.0707158104
Bhatia, S., Monahan, J., Ravi, V., Gautier, P., Murdoch, E., Brenner, S., et al. (2014). A survey of ancient conserved non-coding elements in the PAX6 locus reveals a landscape of interdigitated cis-regulatory archipelagos. Dev. Biol. 387, 214–228. doi: 10.1016/j.ydbio.2014.01.007
Blake, J. A., and Ziman, M. R. (2014). Pax genes: regulators of lineage specification and progenitor cell maintenance. Development 141, 737–751. doi: 10.1242/dev.091785
Bopp, D., Jamet, E., Baumgartner, S., Burri, M., and Noll, M. (1989). Isolation of two tissue-specific Drosophila paired box genes, Pox meso and Pox neuro. EMBO J. 8, 3447–3457.
Borson, N. D., Lacy, M. Q., and Wettstein, P. J. (2002). Altered mRNA expression of Pax5 and Blimp-1 in B cells in multiple myeloma. Blood 100, 4629–4639. doi: 10.1182/blood.V100.13.4629
Breitling, R., and Gerber, J. K. (2000). Origin of the paired domain. Dev. Genes Evol. 210, 644–650. doi: 10.1007/s004270000106
Burge, C., and Karlin, S. (1997). Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94. doi: 10.1006/jmbi.1997.0951
Busse, A., Rietz, A., Schwartz, S., Thiel, E., and Keilholz, U. (2009). An intron 9 containing splice variant of PAX2. J. Transl. Med. 7:36. doi: 10.1186/1479-5876-7-36
Cameron, R. A., Samanta, M., Yuan, A., He, D., and Davidson, E. (2009). SpBase: the sea urchin genome database and web site. Nucleic Acids Res. 37, D750–D754. doi: 10.1093/nar/gkn887
Canestro, C., Bassham, S., and Postlethwait, J. (2005). Development of the central nervous system in the larvacean Oikopleura dioica and the evolution of the chordate brain. Dev. Biol. 285, 298–315. doi: 10.1016/j.ydbio.2005.06.039
Chen, L., Tovar-Corona, J. M., and Urrutia, A. O. (2012). Alternative splicing: a potential source of functional innovation in the eukaryotic genome. Int. J. Evol. Biol. 2012:596274. doi: 10.1155/2012/596274
Chi, N., and Epstein, J. A. (2002). Getting your pax straight: pax proteins in development and disease. Trends Genet. 18, 41–47. doi: 10.1016/S0168-9525(01)02594-X
Chisholm, A. D., and Horvitz, H. R. (1995). Patterning of the Caenorhabditis elegans head region by the Pax-6 family member vab-3. Nature 377, 52–55. doi: 10.1038/377052a0
Cinar, H. N., and Chisholm, A. D. (2004). Genetic Analysis of the Caenorhabditis elegans pax-6 Locus: roles of paired domain-containing and nonpaired domain-containing isoforms. Genetics 168, 1307–1322. doi: 10.1534/genetics.104.031724
Cunningham, F., Amode, M. R., Barrell, D., Beal, K., Billis, K., Brent, S., et al. (2015). Ensembl 2015. Nucleic Acids Res. 43, D662–D669. doi: 10.1093/nar/gku1010
Czerny, T., Schaffner, G., and Busslinger, M. (1993). DNA sequence recognition by pax proteins: bipartite structure of the paired domain and its binding site. Genes Dev. 7, 2048–2061. doi: 10.1101/gad.7.10.2048
De Castro, E., Sigrist, C. J. A., Gattiker, A., Bulliard, V., Langendijk-Genevaux, P. S., Gasteiger, E., et al. (2006). ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34, W362–W365. doi: 10.1093/nar/gkl124
Dressler, G. R., Deutsch, U., Chowdhury, K., Nornes, H. O., and Gruss, P. (1990). Pax2, a new murine paired-box-containing gene and its expression in the developing excretory system. Development 109, 787–795.
Eberhard, D., Jimenez, G., Heavey, B., and Busslinger, M. (2000). Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 19, 2292–2303. doi: 10.1093/emboj/19.10.2292
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Epstein, D. J., Vekemans, M., and Gros, P. (1991). Splotch (sp2H), a mutation affecting development of the mouse neural tube, shows a deletion within the paired homeodomain of Pax-3. Cell 67, 767–774. doi: 10.1016/0092-8674(91)90071-6
Epstein, J. A., Glaser, T., Cai, J., Jepeal, L., Walton, D. S., and Maas, R. L. (1994). Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 8, 2022–2034. doi: 10.1101/gad.8.17.2022
Escriva, H., Manzon, L., Youson, J., and Laudet, V. (2002). Analysis of lamprey and hagfish genes reveals a complex history of gene duplications during early vertebrate evolution. Mol. Biol. Evol. 19, 1440–1450. doi: 10.1093/oxfordjournals.molbev.a004207
Fletcher, J., Hu, M., Berman, Y., Collins, F., Grigg, J., Mciver, M., et al. (2005). Multicystic Dysplastic kidney and variable phenotype in a family with a novel deletion mutation of PAX2. J. Am. Soc. Nephrol. 16, 2754–2761. doi: 10.1681/ASN.2005030239
Friedrich, M., and Caravas, J. (2011). New insights from hemichordate genomes: prebilaterian origin and parallel modifications in the paired domain of the pax gene eyegone. J. Exp. Zool. B Mol. Dev. Evol. 316, 387–392. doi: 10.1002/jez.b.21412
Fu, W., and Noll, M. (1997). The Pax2 homolog sparkling is required for development of cone and pigment cells in the Drosophila eye. Genes Dev. 11, 2066–2078. doi: 10.1101/gad.11.16.2066
Fuentes, M., Benito, E., Bertrand, S., Paris, M., Mignardot, A., Godoy, L., et al. (2007). Insights into spawning behavior and development of the European amphioxus (Branchiostoma lanceolatum). J. Exp. Zool. B Mol. Dev. Evol. 308, 484–493. doi: 10.1002/jez.b.21179
Gehring, W. J. (1996). The master control gene for morphogenesis and evolution of the eye. Genes Cells 1, 11–15. doi: 10.1046/j.1365-2443.1996.11011.x
Gehring, W. J. (2002). The genetic control of eye development and its implications for the evolution of the various eye-types. Int. J. Dev. Biol. 46, 65–73.
Gehring, W. J. (2012). The evolution of vision. Wiley Interdiscip. Rev. Dev. Biol. 3, 1–40. doi: 10.1002/wdev.96
Glardon, S., Holland, L. Z., Gehring, W. J., and Holland, N. D. (1998). Isolation and developmental expression of the amphioxus Pax-6 gene (AmphiPax-6): insights into eye and photoreceptor evolution. Development 125, 2701–2710.
Glaser, T., Walton, D. S., and Maas, R. L. (1992). Genomic structure, evolutionary conservation and aniridia mutations in the human PAX6 gene. Nat. Genet. 2, 232–239. doi: 10.1038/ng1192-232
Green, R. E., Lewis, B. P., Hillman, R. T., Blanchette, M., Lareau, L. F., Garnett, A. T., et al. (2003). Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics 19(Suppl. 1), i118–i121. doi: 10.1093/bioinformatics/btg1015
Grigoriev, I. V., Nordberg, H., Shabalov, I., Aerts, A., Cantor, M., Goodstein, D., et al. (2012). The genome portal of the department of energy joint Genome Institute. Nucleic Acids Res. 40, D26–D32. doi: 10.1093/nar/gkr947
Hartung, F., Blattner, F. R., and Puchta, H. (2002). Intron gain and loss in the evolution of the conserved eukaryotic recombination machinery. Nucleic Acids Res. 30, 5175–5181. doi: 10.1093/nar/gkf649
Heller, N., and Brandli, A. W. (1997). Xenopus Pax-2 displays multiple splice forms during embryogenesis and pronephric kidney development. Mech. Dev. 69, 83–104. doi: 10.1016/S0925-4773(97)00158-5
Heller, N., and Brandli, A. W. (1999). Xenopus Pax-2/5/8 orthologues: novel insights into Pax gene evolution and identification of Pax-8 as the earliest marker for otic and pronephric cell lineages. Dev. Genet. 24, 208–219. doi: 10.1002/(sici)1520-6408(1999)24:3/4<208::aid-dvg4>3.0.co;2-j
Hill, A., Boll, W., Ries, C., Warner, L., Osswalt, M., Hill, M., et al. (2010). Origin of Pax and Six gene families in sponges: single PaxB and Six1/2 orthologs in Chalinula loosanoffi. Dev. Biol. 343, 106–123. doi: 10.1016/j.ydbio.2010.03.010
Holland, L. Z., and Short, S. (2010). Alternative splicing in development and function of chordate endocrine systems: a focus on Pax genes. Integr. Comp. Biol. 50, 22–34. doi: 10.1093/icb/icq048
Holland, L. Z., and Yu, J. K. (2004). Cephalochordate (amphioxus) embryos: procurement, culture, and basic methods. Methods Cell. Biol. 74, 195–215. doi: 10.1016/S0091-679X(04)74009-1
Hoshiyama, D., Iwabe, N., and Miyata, T. (2007). Evolution of the gene families forming the Pax/Six regulatory network: isolation of genes from primitive animals and molecular phylogenetic analyses. FEBS Lett. 581, 1639–1643. doi: 10.1016/j.febslet.2007.03.027
Howard-Ashby, M., Materna, S. C., Brown, C. T., Chen, L., Cameron, R. A., and Davidson, E. H. (2006). Identification and characterization of homeobox transcription factor genes in Strongylocentrotus purpuratus, and their expression in embryonic development. Dev. Biol. 300, 74–89. doi: 10.1016/j.ydbio.2006.08.039
Irvine, S. Q., Fonseca, V. C., Zompa, M. A., and Antony, R. (2008). Cis-regulatory organization of the Pax6 gene in the ascidian Ciona intestinalis. Dev. Biol. 317, 649–659. doi: 10.1016/j.ydbio.2008.01.036
Iwamatsu, T. (2004). Stages of normal development in the medaka Oryzias latipes. Mech. Dev. 121, 605–618. doi: 10.1016/j.mod.2004.03.012
Jaillon, O., Aury, J. M., Brunet, F., Petit, J. L., Stange-Thomann, N., Mauceli, E., et al. (2004). Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431, 946–957. doi: 10.1038/nature03025
Jeffares, D. C., Mourier, T., and Penny, D. (2006). The biology of intron gain and loss. Trends Genet. 22, 16–22. doi: 10.1016/j.tig.2005.10.006
Karolchik, D., Barber, G. P., Casper, J., Clawson, H., Cline, M. S., Diekhans, M., et al. (2014). The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 42, D764–D770. doi: 10.1093/nar/gkt1168
Kelemen, O., Convertini, P., Zhang, Z., Wen, Y., Shen, M., Falaleeva, M., et al. (2013). Function of alternative splicing. Gene 514, 1–30. doi: 10.1016/j.gene.2012.07.083
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. doi: 10.1002/aja.1002030302
Kleinjan, D. A., Bancewicz, R. M., Gautier, P., Dahm, R., Schonthaler, H. B., Damante, G., et al. (2008). Subfunctionalization of duplicated zebrafish pax6 genes by cis-regulatory divergence. PLoS Genet. 4:e29. doi: 10.1371/journal.pgen.0040029
Klimova, L., and Kozmik, Z. (2014). Stage-dependent requirement of neuroretinal Pax6 for lens and retina development. Development 141, 1292–1302. doi: 10.1242/dev.098822
Koralewski, T. E., and Krutovsky, K. V. (2011). Evolution of exon-intron structure and alternative splicing. PLoS ONE 6:e18055. doi: 10.1371/journal.pone.0018055
Kozmik, Z., Czerny, T., and Busslinger, M. (1997). Alternatively spliced insertions in the paired domain restrict the DNA sequence specificity of Pax6 and Pax8. EMBO J. 16, 6793–6803. doi: 10.1093/emboj/16.22.6793
Kozmik, Z., Daube, M., Frei, E., Norman, B., Kos, L., Dishaw, L. J., et al. (2003). Role of Pax genes in eye evolution: a cnidarian PaxB gene uniting Pax2 and Pax6 functions. Dev. Cell. 5, 773–785. doi: 10.1016/S1534-5807(03)00325-3
Kozmik, Z., Holland, N. D., Kalousova, A., Paces, J., Schubert, M., and Holland, L. Z. (1999). Characterization of an amphioxus paired box gene, AmphiPax2/5/8: developmental expression patterns in optic support cells, nephridium, thyroid-like structures and pharyngeal gill slits, but not in the midbrain-hindbrain boundary region. Development 126, 1295–1304.
Kozmik, Z., Kurzbauer, R., Dorfler, P., and Busslinger, M. (1993). Alternative splicing of Pax-8 gene transcripts is developmentally regulated and generates isoforms with different transactivation properties. Mol. Cell. Biol. 13, 6024–6035. doi: 10.1128/MCB.13.10.6024
Kozmik, Z. (2008). The role of Pax genes in eye evolution. Brain Res. Bull. 75, 335–339. doi: 10.1016/j.brainresbull.2007.10.046
Krauss, S., Johansen, T., Korzh, V., and Fjose, A. (1991). Expression of the zebrafish paired box gene pax[zf-b] during early neurogenesis. Development 113, 1193–1206.
Kreslova, J., Holland, L. Z., Schubert, M., Burgtorf, C., Benes, V., and Kozmik, Z. (2002). Functional equivalency of amphioxus and vertebrate Pax258 transcription factors suggests that the activation of mid-hindbrain specific genes in vertebrates occurs via the recruitment of Pax regulatory elements. Gene 282, 143–150. doi: 10.1016/S0378-1119(01)00840-X
Kwak, S. J., Vemaraju, S., Moorman, S. J., Zeddies, D., Popper, A. N., and Riley, B. B. (2006). Zebrafish pax5 regulates development of the utricular macula and vestibular function. Dev. Dyn. 235, 3026–3038. doi: 10.1002/dvdy.20961
Lang, D., Powell, S. K., Plummer, R. S., Young, K. P., and Ruggeri, B. A. (2007). PAX genes: roles in development, pathophysiology, and cancer. Biochem. Pharmacol. 73, 1–14. doi: 10.1016/j.bcp.2006.06.024
Lewis, B. P., Green, R. E., and Brenner, S. E. (2003). Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. U.S.A 100, 189–192. doi: 10.1073/pnas.0136770100
Louis, A., Muffato, M., and Roest Crollius, H. (2013). Genomicus: five genome browsers for comparative genomics in eukaryota. Nucleic Acids Res. 41, D700–D705. doi: 10.1093/nar/gks1156
Lun, K., and Brand, M. (1998). A series of no isthmus (noi) alleles of the zebrafish pax2.1 gene reveals multiple signaling events in development of the midbrain-hindbrain boundary. Development 125, 3049–3062.
Maere, S., and Van De Peer, Y. (2010). “Duplicate retention after small- and large-scale duplications,” in Evolution after Gene Duplication, eds K. Dittmar and D. Liberles (Hoboken, NJ: John Wiley & Sons, Inc.), 31–56.
Nilsen, T. W., and Graveley, B. R. (2010). Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463. doi: 10.1038/nature08909
Noll, M. (1993). Evolution and role of Pax genes. Curr. Opin. Genet. Dev. 3, 595–605. doi: 10.1016/0959-437X(93)90095-7
Nornes, S., Mikkola, I., Krauss, S., Delghandi, M., Perander, M., and Johansen, T. (1996). Zebrafish Pax9 encodes two proteins with distinct C-terminal transactivating domains of different potency negatively regulated by adjacent N-terminal sequences. J. Biol. Chem. 271, 26914–26923. doi: 10.1074/jbc.271.43.26914
Paixao-Cortes, V. R., Salzano, F. M., and Bortolini, M. C. (2013). Evolutionary history of chordate PAX genes: dynamics of change in a complex gene family. PLoS ONE 8:e73560. doi: 10.1371/journal.pone.0073560
Pfeffer, P. L., Gerster, T., Lun, K., Brand, M., and Busslinger, M. (1998). Characterization of three novel members of the zebrafish Pax2/5/8 family: dependency of Pax5 and Pax8 expression on the Pax2.1 (noi) function. Development 125, 3063–3074.
Poleev, A., Wendler, F., Fickenscher, H., Zannini, M. S., Yaginuma, K., Abbott, C., et al. (1995). Distinct functional properties of three human paired-box-protein, PAX8, isoforms generated by alternative splicing in thyroid, kidney and Wilms' tumors. Eur. J. Biochem. 228, 899–911. doi: 10.1111/j.1432-1033.1995.0899m.x
Puschel, A. W., Gruss, P., and Westerfield, M. (1992). Sequence and expression pattern of pax-6 are highly conserved between zebrafish and mice. Development 114, 643–651.
Putnam, N. H., Butts, T., Ferrier, D. E., Furlong, R. F., Hellsten, U., Kawashima, T., et al. (2008). The amphioxus genome and the evolution of the chordate karyotype. Nature 453, 1064–1071. doi: 10.1038/nature06967
Ravi, V., Bhatia, S., Gautier, P., Loosli, F., Tay, B. H., Tay, A., et al. (2013). Sequencing of Pax6 loci from the elephant shark reveals a family of Pax6 genes in vertebrate genomes, forged by ancient duplications and divergences. PLoS Genet. 9:e1003177. doi: 10.1371/journal.pgen.1003177
Reese, M. G., Eeckman, F. H., Kulp, D., and Haussler, D. (1997). Improved splice site detection in Genie. J. Comput. Biol. 4, 311–323. doi: 10.1089/cmb.1997.4.311
Rice, P., Longden, I., and Bleasby, A. (2000). EMBOSS: the European molecular biology open software suite. Trends Genet. 16, 276–277. doi: 10.1016/S0168-9525(00)02024-2
Robichaud, G. A., Nardini, M., Laflamme, M., Cuperlovic-Culf, M., and Ouellette, R. J. (2004). Human Pax-5 C-terminal isoforms possess distinct transactivation properties and are differentially modulated in normal and malignant B cells. J. Biol. Chem. 279, 49956–49963. doi: 10.1074/jbc.M407171200
Rogozin, I. B., Wolf, Y. I., Sorokin, A. V., Mirkin, B. G., and Koonin, E. V. (2003). Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr. Biol. 13, 1512–1517. doi: 10.1016/S0960-9822(03)00558-X
Schimmenti, L. A., Cunliffe, H. E., Mcnoe, L. A., Ward, T. A., French, M. C., Shim, H. H., et al. (1997). Further delineation of renal-coloboma syndrome in patients with extreme variability of phenotype and identical PAX2 mutations. Am. J. Hum. Genet. 60, 869–878.
Schwartz, S., Zhang, Z., Frazer, K. A., Smit, A., Riemer, C., Bouck, J., et al. (2000). PipMaker–a web server for aligning two genomic DNA sequences. Genome Res. 10, 577–586. doi: 10.1101/gr.10.4.577
Short, S., and Holland, L. Z. (2008). The evolution of alternative splicing in the Pax family: the view from the Basal chordate amphioxus. J. Mol. Evol. 66, 605–620. doi: 10.1007/s00239-008-9113-5
Short, S., Kozmik, Z., and Holland, L. Z. (2012). The function and developmental expression of alternatively spliced isoforms of amphioxus and Xenopus laevis Pax2/5/8 genes: revealing divergence at the invertebrate to vertebrate transition. J. Exp. Zool. B Mol. Dev. Evol. 318, 555–571. doi: 10.1002/jez.b.22460
Stephens, R. M., and Schneider, T. D. (1992). Features of spliceosome evolution and function inferred from an analysis of the information at human splice sites. J. Mol. Biol. 228, 1124–1136. doi: 10.1016/0022-2836(92)90320-J
Stuart, E. T., Kioussi, C., and Gruss, P. (1994). Mammalian Pax genes. Annu. Rev. Genet. 28, 219–236. doi: 10.1146/annurev.ge.28.120194.001251
Takatori, N., Butts, T., Candiani, S., Pestarino, M., Ferrier, D. K., Saiga, H., et al. (2008). Comprehensive survey and classification of homeobox genes in the genome of amphioxus, Branchiostoma floridae. Dev. Genes Evol 218, 579–590. doi: 10.1007/s00427-008-0245-9
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tavassoli, K., Ruger, W., and Horst, J. (1997). Alternative splicing in PAX2 generates a new reading frame and an extended conserved coding region at the carboxy terminus. Hum. Genet. 101, 371–375. doi: 10.1007/s004390050644
Van De Peer, Y. (2004). Tetraodon genome confirms Takifugu findings: most fish are ancient polyploids. Genome Biol. 5:250. doi: 10.1186/gb-2004-5-12-250
Wada, S., Tokuoka, M., Shoguchi, E., Kobayashi, K., Di Gregorio, A., Spagnuolo, A., et al. (2003). A genomewide survey of developmentally relevant genes in Ciona intestinalis. II. Genes for homeobox transcription factors. Dev. Genes Evol. 213, 222–234. doi: 10.1007/s00427-003-0321-0
Walther, C., and Gruss, P. (1991). Pax-6, a murine paired box gene, is expressed in the developing CNS. Development 113, 1435–1449.
Wang, E. T., Sandberg, R., Luo, S., Khrebtukova, I., Zhang, L., Mayr, C., et al. (2008a). Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476. doi: 10.1038/nature07509
Wang, Q., Fang, W. H., Krupinski, J., Kumar, S., Slevin, M., and Kumar, P. (2008b). Pax genes in embryogenesis and oncogenesis. J. Cell. Mol. Med. 12, 2281–2294. doi: 10.1111/j.1582-4934.2008.00427.x
Ward, T. A., Nebel, A., Reeve, A. E., and Eccles, M. R. (1994). Alternative messenger RNA forms and open reading frames within an additional conserved region of the human PAX-2 gene. Cell Growth Differ. 5, 1015–1021.
Xu, H. E., Rould, M. A., Xu, W., Epstein, J. A., Maas, R. L., and Pabo, C. O. (1999). Crystal structure of the human Pax6 paired domain–DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding. Genes Dev 13, 1263–1275. doi: 10.1101/gad.13.10.1263
Xu, W., Rould, M. A., Jun, S., Desplan, C., and Pabo, C. O. (1995). Crystal structure of a paired domain-DNA complex at 2.5 A resolution reveals structural basis for Pax developmental mutations. Cell 80, 639–650. doi: 10.1016/0092-8674(95)90518-9
Keywords: Pax258, Pax6, alternative splicing, paired domain, splice variants
Citation: Fabian P, Kozmikova I, Kozmik Z and Pantzartzi CN (2015) Pax2/5/8 and Pax6 alternative splicing events in basal chordates and vertebrates: a focus on paired box domain. Front. Genet. 6:228. doi: 10.3389/fgene.2015.00228
Received: 28 May 2015; Accepted: 15 June 2015;
Published: 02 July 2015.
Edited by:
Hector Escriva, Centre National de la Recherche Scientifique, FranceReviewed by:
Manuel Irimia, Centre for Genomic Regulation, SpainCopyright © 2015 Fabian, Kozmikova, Kozmik and Pantzartzi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chrysoula N. Pantzartzi, Department of Transcriptional Regulation, Institute of Molecular Genetics, Videnska 1083, Prague 14220, Czech Republic,Y2hyeXNvdWxhLnBhbnR6YXJ0emlAaW1nLmNhcy5jeg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.