
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 20 April 2012
Sec. RNA
Volume 3 - 2012 | https://doi.org/10.3389/fgene.2012.00056
MicroRNAs (miRNAs), small non-coding regulatory RNAs that regulate gene expression at the post-transcriptional level, are master regulators of a wide array of cellular processes. Altered miRNA expression could be a determinant of disease development and/or progression and manipulation of miRNA expression represents a potential avenue of therapy. Exosomes are cell-derived extracellular vesicles that promote cell–cell communication and immunoregulatory functions. These “bioactive vesicles” shuttle various molecules, including miRNAs, to recipient cells. Inappropriate release of miRNAs from exosomes may cause significant alterations in biological pathways that affect disease development, supporting the concept that miRNA-containing exosomes could serve as targeted therapies for particular diseases. This review briefly summarizes recent advances in the biology, function, and therapeutic potential of exosomal miRNAs.
Exosomes are small (30–90 nm) extracellular vesicles derived from the multivesicular body (MVB) sorting pathway. These vesicles are produced by a wide variety of cell types including reticulocytes, epithelial cells, neurons, and tumor cells (Thery et al., 2009). Exosomes have been isolated from bronchoalveolar lavage, urine, serum, bile, and breast milk (Thery, 2011). Although previously considered to be cellular waste products (Thery, 2011), recent studies have demonstrated that exosomes are “bioactive vesicles” that promote intercellular communication and immunoregulatory processes by shuttling molecules between cells (Thery et al., 2009; Pegtel et al., 2010).
One of the most recent exciting findings is that microRNAs (miRNAs) exist in exosomes and these exosomal miRNAs can be functionally delivered to target cells (Montecalvo et al., 2012). miRNAs are small non-coding regulatory RNAs that bind to complementary sequences in the 3′-untranslated region (3′UTR) of target mRNAs, leading to either translational repression or target degradation of the specific mRNA (Bartel, 2004, 2009). miRNAs play a pivotal regulatory role in diverse biological processes including but not limited to, development and cell signaling, proliferation and differentiation (Bartel, 2004, 2009). While miRNA expression patterns are incompletely defined in most cell types under normal conditions, aberrant expression of certain miRNAs has been described in diverse human diseases, suggesting that miRNAs can be used as biomarkers for disease development and as a potential target of therapy (Jiang et al., 2009). Several publicly available databases (e.g., miR2Disease) have been established which document the relationships between the expression of specific miRNAs and the development and/or progression of specific human diseases (Jiang et al., 2009). Identification of miRNAs key to disease pathogenesis provides new insights into the development of therapeutic targets in the treatment of human diseases, particularly for some forms of cancer. Currently, several approaches to manipulate miRNA function are being studied in clinical trials (Kasinski and Slack, 2011). Because inappropriately released miRNA-containing exosomes have been shown to significantly alter biological pathways integral to disease development (Record et al., 2011), it is postulated that exosomes could be used to deliver therapeutic molecules in a variety of human diseases. Here, we briefly summarize recent advances in the biology, function and therapeutic potential of exosomal miRNAs.
Exosomes are the newest family member of “bioactive vesicles.” These bioactive vesicles were originally described during maturation of reticulocytes into erythrocytes. These small vesicles contained the transferrin receptor which is downregulated in mature erythrocytes (Pan and Johnstone, 1983). Exosomes are also secreted by a variety of cell types including mast cells (Laulagnier et al., 2004), B lymphocyte cell lines, dendritic cells (Laulagnier et al., 2004), platelets, astrocytes, neurons, and epithelial cells (Hogan et al., 2009; Zhou et al., 2011). Exosomes enter the circulatory system and bodily fluids and have been detected in plasma, bronchoalveolar lavage, blood, urine, bile (Masyuk et al., 2010), ascites (Rupp et al., 2011), breast milk, and cerebrospinal fluid. Tumor-derived exosomes have been identified in the plasma of patients with lung adenocarcinoma, glioblastoma multiforme, malignant glioma, and ovarian carcinoma ascites (Skog et al., 2008; Rabinowits et al., 2009; Rupp et al., 2011).
The formation and release of exosomes are tightly regulated processes governed by multiple signaling mechanisms. Various stimuli influence exosome secretion including activation of a variety of receptors including the P2X receptor by ATP on monocytes and neutrophils, the thrombin receptor on platelets, and toll-like receptor-4 (TLR-4) by bacterial lipopolysaccharide (LPS) on dendritic cells (Bhatnagar and Schorey, 2007). Morphological studies suggest that exosomes are derived from the MVB sorting pathway. The MVBs fuse with the plasma membrane and release the vesicles into the extracellular milieu as exosomes. The exosomes consist of a lipid bilayer membrane surrounding a small cytosol that lacks cellular organelles such as mitochondria, lysosomes, endoplasmic reticulum, nucleus, and Golgi apparatus.
The formation and release of exosomes involves contractile machinery that draws opposing membranes together before pinching off the membrane connection and releasing the vesicle into the extracellular space (Cocucci et al., 2009). SNARE (soluble NSF attachment protein receptor) complexes are proposed to be involved in bringing the membranes together in opposition, as there is an energy requirement for the membrane fusion process (Sudhof and Rothman, 2009). SNAREs can be divided into vesicle (v-SNAREs) and target SNARES (t-SNAREs; Fukuda et al., 2000). The complex begins assembling at the N-terminal region of the SNARE motifs and proceeds toward the C-termini which anchors the interacting proteins in the membranes (Sutton et al., 1998). An intermediate complex composed of SNAP-25 and syntaxin-1, which interacts with synaptobrevin-2 then forms. Recent studies from Thery’s group demonstrated that several Rab family proteins, including Rab27a and Rab27b, are key regulators of the exosome secretion pathway (Ostrowski et al., 2010). Several lines of evidence support a role for Rab27a and Rab27b in MVB docking at the plasma membrane. Rab27a silencing increases the size of MVBs; alternatively, Rab27b silencing results in redistribution of the MVBs to the perinuclear region (Ostrowski et al., 2010). Another Rab family member protein, Rab35, is a target of TBC1D10A-C, which regulates exosome secretion and inhibiting Rab35 activity induces intracellular accumulation of endosomal vesicles and impaired exosome secretion (Hsu et al., 2010). Recent studies revealed that exosomes can be released into the extracellular environment by the outward budding and fission of the plasma membrane, a pathway called the exosome and microvesicle (EMV) budding pathway. The progression of the EMV pathway is regulated by multiple processes including the elevation of intracellular calcium and reorganization of the cell’s structural scaffolding. The current model of exosome formation and release of exosomes is diagrammed in Figure 1.
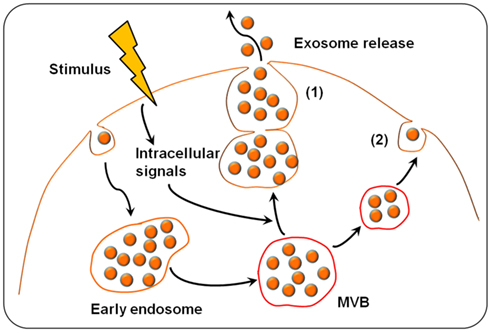
Figure 1. Exosome biogenesis and release. Exosomes are derived from the multivesicular bodies (MVBs) which are known as late endosomes. Extracellular stimuli (e.g., microbial infection) can induce exosome formation and trigger exosome release. The exosomes can be released into the extracellular environment by either fusion of MVBs with the cell surface (1) or budding pathway (2). Fusion of MVBs with the plasma membrane surface involves the SNARE protein complex that brings the two membranes together. Budding exosomes are released from cytoplasm by budding of cell plasma membrane in response to cell stimulation.
Exosomes released from donor cells can be taken up by recipient cells, where an array of biological processes can be impacted in the recipient cells, including cell proliferation and differentiation, apoptosis, and the immune response (Abusamra et al., 2005; Yu et al., 2007). Exosomes have been implicated as mediators in the communication between T cells and antigen-presenting cells (APCs) at the immune synapse. Professional APCs secrete major histocompatibility complex class II (MHC II) carrying exosomes (Buschow et al., 2010), and these exosomes can stimulate antigen-specific MHC class II-restricted T cell responses (Raposo et al., 1996). Intestinal epithelial cell-derived exosomes containing αvβ6 integrin and food antigen induced dendritic cell tolerance and tolerogenic DCs in a model of tolerance induction. In addition, the presence of these intestinal epithelial cell-derived exosomes alsoimpacted the development of antigen-specific T regulatory cells (Chen et al., 2011). Additional normal cellular processes influenced by exosomal contents include myelin membrane biogenesis and neuronal plasticity (Bakhti et al., 2011) in the central nervous system (CNS) and decreasing cholangiocyte proliferation via interactions with cholangiocyte cilia (Masyuk et al., 2010).
Exosomes may also be involved in pathogenic processes. Extracellular amyoid fibril formation, a key feature of Alzheimer’s disease, occurs at an accelerated rate in the presence of GM1 ganglioside-containing exosomes (Yuyama et al., 2008). These data suggest that targeting GM1 in exosomes may provide a mechanism to slow the rate of plaque formation. Tumor-derived exosomes may also significantly influence the development and effectiveness of anti-tumor immune responses (Rupp et al., 2011). Exposure of peripherial blood mononuclear cells (PBMCs) to exosomes isolated from ascites of ovarian cancer patients impaired the cytotoxic activity of the PBMCs in the presence of dendritic cells. In addition to containing tumor antigens, exosomes secreted from cancer cells also have been shown to contain two molecules critical to immunoregulation, PD-L1 and FasL. The presence of FasL and TRAIL in exosomes may contribute to apoptosis of cells of the immune response, thereby serving as a mechanism of tumor escape from immunosurveillance (Peng et al., 2011). For example, exosomes from plasma of cancer patients have been shown to induce T cell apoptosis (Ichim et al., 2008), reminiscent of Fas-FasL counterattack that has been described in some cancers (Alderson et al., 1995; O’Connell et al., 1998). Intercellular communication via exosomes is not limited to communications between mammalian cells. Interestingly, exosomes have been isolated from the protozoan parasite, Leishmania, and these exosomes deliver protozoan proteins to macrophages in the vertebrate host. Similar to what is observed with mammalian-derived exosomes, protozoan exosomes may influence the host immune response and regulate cell signaling pathways (Silverman et al., 2010). Exosomes can also transport pathogen or virus proteins to neighboring and/or distant cells and enhance disease spread (Schorey and Bhatnagar, 2008). The infectious form of the prion protein, PrPsc, has been found in exosomes. Exosomally derived PrPsc is infectious to mice (Vella et al., 2007). This finding suggests that in humans or wildlife, exosomally derived PrPSc could serve as a source of infectious material in the host which accelerates the spread of the misfolded protein through the body. Also, because exosomes are in mucus, urine, and blood, disease spread between hosts may be enhanced by the presence of PrPSc-containing exosomes.
Exosomes are considered extracellular organelles which can affect neighboring cells following delivery of their contents into target cells (Silverman and Reiner, 2011). It has been speculated that the composition of exosomes is quite heterogenic, depending on the cellular origin of the exosome. Large-scale proteomics and phosphoproteomics studies of exosomes derived from various cell types suggest that the exosomes shuttle wide array of biologically relevant molecules, including lipids, carbohydrates, RNAs, and proteins (Schorey and Bhatnagar, 2008), as outlined in Figure 2. Detailed information regarding the contents of exosomes is available (http://dir.nhlbi.nih.gov/papers/lkem/exosome). The contents of urinary exosomes have been characterized by Knepper’s group. This study identified 1132 proteins contained within the exosomes. In addition, phosphoproteomic studies identified several unique phosphorylation sites (Gonzales et al., 2009). This study, along with others, support the potential for exosomes obtained from bodily fluids to serve as biomarkers of disease development and/or progression.
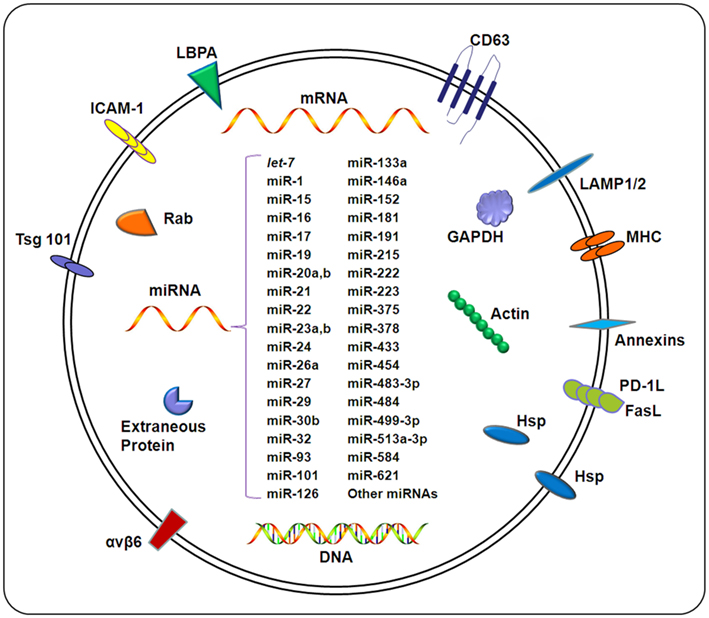
Figure 2. Composition of exosomes. Exosomes carry a wide array of molecules including proteins, DNAs, mRNAs, and miRNAs, depending on a variety of factors including the cell type from which the exosome originates, the state of health of the host, and extracellular stimuli. The contents of exosomes can be transferred from origin cells to target cells, resulting in an elaborate intercellular communication network. Exosomal miRNAs confirmed by different methods (e.g., microarray and qPCR) under different pathological conditions are listed in this figure. For details, also see Table 1.
To date, over 1000 human miRNAs have been identified and it is predicted that miRNAs control the expression of approximately 60% of all human genes (Bentwich et al., 2005; Krol et al., 2010). Accumulating data indicate that miRNAs comprise an essential component in the complex regulatory networks controlling a wide array of cellular processes, including the timing of developmental processes, cell proliferation/differentiation, apoptosis, organ development (Tiscornia and Izpisua Belmonte, 2010), and immune regulation (O’Neill et al., 2011; Zhou et al., 2011). Several excellent reviews have been recently published on these topics (O’Neill et al., 2011; Zhou et al., 2011).
Aberrant miRNA expression has been linked to the initiation and progression of certain cancers (Mencia et al., 2009). An example of a miRNA associated with cancer development (that is, an oncogenic miRNA or oncomiR) is miR-21. miRNA-21 targets tumor suppressor genes and upregulation of miR-21 is associated with a wide variety of cancers including breast, ovaries, cervix, colon, lung, liver, brain, esophagus, prostate, pancreas, and thyroid cancers (Volinia et al., 2006; Lui et al., 2007). miRNAs are also involved in developmental processes in vertebrates. Studies utilizing genetically modified mice have demonstrated the critical requirement for miRNAs in cardiac development, as mice deficient in Dicer experience cardiac failure (Chen et al., 2008). In the CNS, the role of miRNAs in regulating neuronal development, plasticity and differentiation is under exploration (Guo et al., 2002; Gao et al., 2010). In Parkinson’s disease, miR-34b and miR-34c are decreased and this decrease in expression is accompanied by a decrease in cell viability of dopaminergic neurons (Minones-Moyano et al., 2011). In Huntington’s disease, both murine models of the disease as well as human tissue samples have demonstrated that RE1 silencing transcription (REST) factor, a transcriptional repressor, is not appropriately expressed, and can be found in the nucleus of neurons rather than the cytoplasm. REST dysregulation results in a wide array of miRNAs are repressed, thereby resulting in an overexpression of mRNAs in the CNS of individuals with Huntington’s disease (Johnson et al., 2008; Buckley et al., 2010).
The small size (23–24 nucleotides in length) of mature miRNA molecules makes them very attractive targets for drug development. It may be possible to restore normal levels of mature forms of dysregulated miRNAs in disease states and provide therapeutic benefits to the host by appropriately regulating the level of expression of their target genes. A variety of delivery approaches have been used to deliver miRNA precursors into cells to overexpress miRNAs of interest in vitro (Hu et al., 2009; Kota et al., 2009). In vivo studies have been performed in mice to test the therapeutic potential of miRNAs. Intranasal administration of cholesterol-conjugated anti-miR-126 to the asthmatic mice resulted in reduced airway disease characterized by decreased T-helper-2 cytokine production, decreased airway hyperresponsiveness and muscus hypersecretion, and a reduction in airway-infiltrating eosinophils and neutrophils compared to control-treated asthmatic mice (Mattes et al., 2009). Other means of delivery of antisense miRNAs has also been successful. Inhibition of miR-122 by systemic administration of a miR-122 antisense oligonucleotide reduced plasma cholesterol levels and decreased hepatic fatty acid and cholesterol synthesis rates in normal mice. Therapeutic administration of anti-miR-122 also reduced levels of triglycerides and improved hepatic steatosis in diet-induced obese mice (Esau et al., 2006). Mice appeared healthy after one month of antisense therapy. The lack of obvious toxicity further cements the potential of miRNAs as therapeutic molecules.
The potential for use of miRNAs against cancer has been extensively studied. A recent study using plasmid-expressed miR-155 in nasopharyngeal carcinoma cells demonstrated decreased expression of manganese superoxide dismutase in miR-155 overexpressing CNE1 cells and reduced resistance to ionizing radiation (Du et al., 2011). In vivo administration of a lentivirus-expressing miR-26a resulted in inhibited tumorigenicity of nasopharyngeal cells in nude mice (Lu et al., 2011). Systemic delivery of synthetic miR-16 reduced the growth of metastatic prostate tumors by downregulating multiple cell-cycle genes in a prostate cancer xenograft model (Takeshita et al., 2010). Let-7 miRNAs are proposed to function as tumor suppressors by negatively regulating multiple oncogenes (RAS, MYC, HMGA2) and cell-cycle promoters (CDC25A, CDK6, CCND2; Johnson et al., 2005; Mayr et al., 2007). Administration of let-7 miRNA prevented tumor formation in a mouse model of non-small cell lung cancer (Kumar et al., 2008). Intratumoral injection of synthetic let-7b miRNA significantly reduced tumor growth, induced necrotic cores in the tumors, and reduced NRAS and CDK6 expression in tumors in a mouse model of lung cancer (Trang et al., 2010). Such approaches to manipulate the expression of miRNA targets in the context of disease are currently being explored in clinical trials (Kasinski and Slack, 2011).
A recent breakthrough in exosome biology is that RNA molecules, in particular, miRNAs, are present in these vesicles. To date, studies have demonstrated that miRNAs in exosomes can influence target cell function (Kosaka et al., 2010; Mittelbrunn et al., 2011). Hunter et al. (2008) identified miRNAs expressed in circulating plasma microvesicles from normal subjects, providing the basis for future work examining the predictive role of peripheral blood miRNA signatures in human disease, as well as defining the biological processes regulated by particular miRNAs. Koh et al. demonstrated that miRNAs are also found in the extracellular environment of human embryonic stem cell-derived mesenchymal stem cells (hES-MSC). Significant differences in expression profile of miRNAs in the intracellular and extracellular environment of hES-MSC cultures were identified. Interestingly, the let-7 miRNA family is highly expressed in both intra- and extra-cellular samples of hES-MSC (Koh et al., 2010). Advances in miRNA array techniques have provided significant advances in miRNA profiling of exosomes. Exosomes contain a substantial amount of small RNAs, but little or no ribosomal RNA compared to the levels observed in the donor cells (Valadi et al., 2007). Moreover, selective packaging of miRNAs into exosomes appears to occur, as the miRNA profiles in exosomes do not reflect the miRNA profiles observed in the parental cells (Valadi et al., 2007; Skog et al., 2008; Taylor and Gercel-Taylor, 2008; Rabinowits et al., 2009; Mittelbrunn et al., 2011). Exosomes isolated from T cells, B cells, and dendritic cells had miRNA expression profiles unique from their parent cells. The authors also demonstrated that there was antigen-driven unidirectional transfer of miRNAs from the T cell to the APC mediated by CD63+ exosomes (Mittelbrunn et al., 2011). The mechanism for the selective packaging of RNAs into exosomes is unclear. Compared to cellular RNAs, exosomal RNAs are more stable (Keller et al., 2011), and are reportedly resistant to degradation during prolonged storage and freeze/thaw cycles (Reid et al., 2011). The stability of the contents of these membrane-bound structures adds to the attractiveness of exosomes as biomarkers of cancer and other diseases.
Exosomal miRNAs have been implicated in many of exosome-mediated biological functions (Wang et al., 2010). Exosomes derived from a human (HMC-1) and mouse (MC/9) mast cell lines transport RNA to neighboring mast cells, impacting the function of the recipient mast cells (Valadi et al., 2007; Schorey and Bhatnagar, 2008). Impairment of exosome production reduces the transfer of miRNAs to APCs. The miRNAs transferred at the immune synapse are functional and can alter gene expression in the recipient APCs (Mittelbrunn et al., 2011). Together, the current data make a strong case for role of exosomally-derived miRNAs as mediators of cell function.
Aberrant expression of miRNAs has been identified in numerous diseases (Table 1). In addition, exosomal miRNAs are considered as biomarkers for many pathological states (Michael et al., 2010; Ciesla et al., 2011). Exosomes from diseased individuals contain miRNAs not found in normal, healthy subjects (Taylor and Gercel-Taylor, 2008; Rabinowits et al., 2009). Many miRNAs, including virus miRNAs, are in exosomes. Nevertheless, it is unknown how exosomal miRNAs function in the recipient cells. Exosomes released from Epstein–Barr virus (EBV)-infected B95-8 LCL cells were identified in monocyte-derived dendritic cells, resulting in gene silencing in recipient cells (Pegtel et al., 2010). Furthermore, exosomal-loaded viral miRNAs suggest that exosome may serve as a vesicle to contribute to disease spread or pathogenesis (Meckes et al., 2010; Pegtel et al., 2011). Interestingly, macrophages have been shown to influence the invasiveness of breast cancer cells through exosome-mediated delivery of oncogenic miRNAs from the macrophages to the cancerous cells (Yang et al., 2011). Exosomes released from dendritic cells can fuse with the target dendritic cells and release their contents into the target cells, resulting in mRNA silencing (Montecalvo et al., 2012). A recent report from the Lim group demonstrated that miRNA-containing stroma-derived exosomes contribute to B cell quiescence. The transfer of miRNAs from bone marrow stroma to B cells may participate in dormancy of bone marrow metastases (Lim et al., 2011).
The levels of specific miRNAs in certain disease states may provide insight into disease pathogenesis and/or potential mechanisms of repair. miR-133a levels are elevated in circulating exosomes derived from injured myocardium in patients with cardiovascular diseases (Kuwabara et al., 2011). In addition, miR-133a is implicated to regulate NFATc4, a protein that contributes to cardiac hypertrophy (Li et al., 2010). Treatment with miR-133a inhibitors reduced the level of cardiac hypertrophy (Li et al., 2010).
Exosomal miRNA profiles of circulating tumor exosomes are unique from those in normal controls (Taylor and Gercel-Taylor, 2008). Because of this unique exosomal miRNA signature, screening tests utilizing circulating exosomal miRNAs as markers of disease might be used to identify tumor development. Unique miRNA profiles have been described in exosomes from patients with lung cancer, glioblastoma, and hepatocellular carcinoma (Rabinowits et al., 2009). In cancer, a variety of processes are impacted by exosomal miRNAs. While in cases such as lung adenocarcinoma, the miRNA signatures are different between cancer patients and normal controls, the exosome-loaded miRNAs are similar in nature to the miRNAs isolated from the tumors. In the case of hepatocellular carcinoma, some of the miRNAs transferred by exosomes can downregulate transforming growth factor-β activated kinase-1 (TAK1) pathway in hepatocarcinogenesis. This finding suggests that miRNAs from the exosomes may be involved in the local spread, intrahepatic metastases, or multifocal growth in hepatocellular carcinoma (Kogure et al., 2011). Liu et al. (2010) has demonstrated that tumor-derived exosomes are pivotal in promoting tumor metastasis via a proinflammatory cytokine-driven expansion of myeloid-derived suppressor cells via the MyD88 pathway. Taken together, current exosomal miRNA studies have opened a new window for the development of biomarkers for important human diseases.
Due to their ability to efficiently shuttle small molecules between cells, exosomes are an extremely promising therapeutic tool for numerous diseases. Alternatively, because of the pathogenic significance of exosomal miRNAs in an array of diseases, drug development focusing on targeting the release of exosomal miRNA contents has begun. The use of exosomes as therapeutic delivery vehicles covers a wide array of diseases, including but not limited to cancer, virus-induced diseases, and parasitic diseases. Of particular note, the Wood laboratory successfully expressed Lamp2b, an exosomal membrane protein, fused to a neuron-penetrating RVG peptide in the dendritic cells isolated from mice. Exosomes released from these cells were loaded with exogenous siRNA to GAPDH by electroporation. Intravenously injected RVG-targeted exosomes delivered GAPDH siRNA specifically to neurons, microglia, oligodendrocytes in the brain, resulting in a specific gene knockdown (Alvarez-Erviti et al., 2011). Akao et al. reported that chemically modified miRNAs (i.e., miR-143BPs) are detectable in exosomes secreted from THP-1 macrophages following miR-143BP transfection. More importantly, increased miR-143BP levels were detected in the serum, tumor, and kidney of the host animals when the mice were intravenously injected with the THP-1 macrophages transfected with miR-143BP (Akao et al., 2011). These data suggest that manipulating exosomal miRNAs ex vivo may be an efficient tool to deliver miRNAs to target specific organs. Exosomes have also been utilized successfully to deliver siRNAs to specific cell types in vivo in mice (van den Boorn et al., 2011). Thus far, studies on manipulation of exosomal miRNAs are primarily in animal models and the clinical application of this work is unclear, although it is obvious that, (1) exosomes could serve as targets of therapy and (2), delivery miRNAs to target cells through exosomes is feasible. Therefore, functional manipulation of miRNAs in exosomes and in vivo delivery of exosomal miRNAs would be novel targets for therapeutic intervention for a variety of human diseases. With the advance of technology that allows us to manipulate the miRNA contents in exosomes as well as exosomal membrane proteins, the future studies on exosomal miRNAs will benefit all patients by providing efficient diagnosis and disease-specific therapy.
Currently, ExoCarta (Version 3.1) contains information on 11,261 protein entries, 2375 mRNA entries, and 764 miRNA entries that were obtained from 134 exosomal studies (Mathivanan et al., 2011). There are several advantages for the use of exosomes/microvesicles to deliver information to a target cell. Exosomes can shuttle mRNAs and miRNAs directly to recipient cells which increases the likelihood that this information will alter the function of target cells. The release of exosomes allows cell–cell communication regardless of the distance between the cells. Exosomes have been identified in body fluids and blood, indicating that this exchange of information between organs may occur via exosomes (Qazi et al., 2010). Additionally, exosomes may contain cell-specific factors that allow the exosomes to target specific cells for delivery of the exosomal contents (proteins, mRNAs, miRNAs, and DNAs) to recipient cells with a high degree of efficiency. Exosomal miRNAs may reflect disease specific-aberrations. Emerging evidence supports the hypothesis that disease diagnosis can occur by examining levels of miRNA in exosomes isolated from body fluids. Finally, manipulation of the miRNAs within exosomes may provide an effective tool to target therapy to specific cells and organs.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was partially supported by National Natural Science Foundation of China No. 31101809 (to Guoku Hu), the National Institute of Health AI095532, and Tobacco Settlement Foundation of Nebraska LB506 and CU LB595 (to Xian-Ming Chen).
Abusamra, A. J., Zhong, Z., Zheng, X., Li, M., Ichim, T. E., Chin, J. L., and Min, W. P. (2005). Tumor exosomes expressing Fas ligand mediate CD8+ T-cell apoptosis. Blood Cells Mol. Dis. 35, 169–173.
Akao, Y., Iio, A., Itoh, T., Noguchi, S., Itoh, Y., Ohtsuki, Y., and Naoe, T. (2011). Microvesicle-mediated RNA molecule delivery system using monocytes/macrophages. Mol. Ther. 19, 395–399.
Alderson, M. R., Tough, T. W., Davis-Smith, T., Braddy, S., Falk, B., Schooley, K. A., Goodwin, R. G., Smith, C. A., Ramsdell, F., and Lynch, D. H. (1995). Fas ligand mediates activation-induced cell death in human T lymphocytes. J. Exp. Med. 181, 71–77.
Alvarez-Erviti, L., Seow, Y., Yin, H., Betts, C., Lakhal, S., and Wood, M. J. (2011). Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 29, 341–345.
Bakhti, M., Winter, C., and Simons, M. (2011). Inhibition of myelin membrane sheath formation by oligodendrocyte-derived exosome-like vesicles. J. Biol. Chem. 286, 787–796.
Bentwich, I., Avniel, A., Karov, Y., Aharonov, R., Gilad, S., Barad, O., Barzilai, A., Einat, P., Einav, U., Meiri, E., Sharon, E., Spector, Y., and Bentwich, Z. (2005). Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 37, 766–770.
Bhatnagar, S., and Schorey, J. S. (2007). Exosomes released from infected macrophages contain Mycobacterium avium glycopeptidolipids and are proinflammatory. J. Biol. Chem. 282, 25779–25789.
Buckley, N. J., Johnson, R., Zuccato, C., Bithell, A., and Cattaneo, E. (2010). The role of REST in transcriptional and epigenetic dysregulation in Huntington’s disease. Neurobiol. Dis. 39, 28–39.
Buschow, S. I., Van Balkom, B. W., Aalberts, M., Heck, A. J., Wauben, M., and Stoorvogel, W. (2010). MHC class II-associated proteins in B-cell exosomes and potential functional implications for exosome biogenesis. Immunol. Cell Biol. 88, 851–856.
Chen, J. F., Murchison, E. P., Tang, R., Callis, T. E., Tatsuguchi, M., Deng, Z., Rojas, M., Hammond, S. M., Schneider, M. D., Selzman, C. H., Meissner, G., Patterson, C., Hannon, G. J., and Wang, D. Z. (2008). Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc. Natl. Acad. Sci. U.S.A. 105, 2111–2116.
Chen, X., Song, C. H., Feng, B. S., Li, T. L., Li, P., Zheng, P. Y., Chen, X. M., Xing, Z., and Yang, P. C. (2011). Intestinal epithelial cell-derived integrin {alpha}{beta}6 plays an important role in the induction of regulatory T cells and inhibits an antigen-specific Th2 response. J. Leukoc. Biol. 90, 751–759.
Ciesla, M., Skrzypek, K., Kozakowska, M., Loboda, A., Jozkowicz, A., and Dulak, J. (2011). MicroRNAs as biomarkers of disease onset. Anal. Bioanal. Chem. 401, 2051–2061.
Cocucci, E., Racchetti, G., and Meldolesi, J. (2009). Shedding microvesicles: artefacts no more. Trends Cell Biol. 19, 43–51.
da Silveira, J. C., Veeramachaneni, D. N., Winger, Q. A., Carnevale, E. M., and Bouma, G. J. (2012). Cell-secreted vesicles in equine ovarian follicular fluid contain miRNAs and proteins: a possible new form of cell communication within the ovarian follicle. Biol. Reprod. 86, 71.
Du, Z. M., Hu, L. F., Wang, H. Y., Yan, L. X., Zeng, Y. X., Shao, J. Y., and Ernberg, I. (2011). Upregulation of MiR-155 in nasopharyngeal carcinoma is partly driven by LMP1 and LMP2A and downregulates a negative prognostic marker JMJD1A. PLoS ONE 6:e19137. doi:10.1371/journal.pone.0019137
Esau, C., Davis, S., Murray, S. F., Yu, X. X., Pandey, S. K., Pear, M., Watts, L., Booten, S. L., Graham, M., Mckay, R., Subramaniam, A., Propp, S., Lollo, B. A., Freier, S., Bennett, C. F., Bhanot, S., and Monia, B. P. (2006). miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3, 87–98.
Fukuda, R., Mcnew, J. A., Weber, T., Parlati, F., Engel, T., Nickel, W., Rothman, J. E., and Sollner, T. H. (2000). Functional architecture of an intracellular membrane t-SNARE. Nature 407, 198–202.
Gao, J., Wang, W. Y., Mao, Y. W., Graff, J., Guan, J. S., Pan, L., Mak, G., Kim, D., Su, S. C., and Tsai, L. H. (2010). A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466, 1105–1109.
Gonzales, P. A., Pisitkun, T., Hoffert, J. D., Tchapyjnikov, D., Star, R. A., Kleta, R., Wang, N. S., and Knepper, M. A. (2009). Large-scale proteomics and phosphoproteomics of urinary exosomes. J. Am. Soc. Nephrol. 20, 363–379.
Grange, C., Tapparo, M., Collino, F., Vitillo, L., Damasco, C., Deregibus, M. C., Tetta, C., Bussolati, B., and Camussi, G. (2011). Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res. 71, 5346–5356.
Guo, C. J., Li, Y., Tian, S., Wang, X., Douglas, S. D., and Ho, W. Z. (2002). Morphine enhances HIV infection of human blood mononuclear phagocytes through modulation of beta-chemokines and CCR5 receptor. J. Investig. Med. 50, 435–442.
Hogan, M. C., Manganelli, L., Woollard, J. R., Masyuk, A. I., Masyuk, T. V., Tammachote, R., Huang, B. Q., Leontovich, A. A., Beito, T. G., Madden, B. J., Charlesworth, M. C., Torres, V. E., Larusso, N. F., Harris, P. C., and Ward, C. J. (2009). Characterization of PKD protein-positive exosome-like vesicles. J. Am. Soc. Nephrol. 20, 278–288.
Hsu, C., Morohashi, Y., Yoshimura, S., Manrique-Hoyos, N., Jung, S., Lauterbach, M. A., Bakhti, M., Gronborg, M., Mobius, W., Rhee, J., Barr, F. A., and Simons, M. (2010). Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J. Cell Biol. 189, 223–232.
Hu, G., Zhou, R., Liu, J., Gong, A. Y., Eischeid, A. N., Dittman, J. W., and Chen, X. M. (2009). MicroRNA-98 and let-7 confer cholangiocyte expression of cytokine-inducible Src homology 2-containing protein in response to microbial challenge. J. Immunol. 183, 1617–1624.
Hunter, M. P., Ismail, N., Zhang, X., Aguda, B. D., Lee, E. J., Yu, L., Xiao, T., Schafer, J., Lee, M. L., Schmittgen, T. D., Nana-Sinkam, S. P., Jarjoura, D., and Marsh, C. B. (2008). Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE 3:e3694. doi:10.1371/journal.pone.0003694
Ichim, T. E., Zhong, Z., Kaushal, S., Zheng, X., Ren, X., Hao, X., Joyce, J. A., Hanley, H. H., Riordan, N. H., Koropatnick, J., Bogin, V., Minev, B. R., Min, W. P., and Tullis, R. H. (2008). Exosomes as a tumor immune escape mechanism: possible therapeutic implications. J. Transl. Med. 6, 37.
Jiang, Q., Wang, Y., Hao, Y., Juan, L., Teng, M., Zhang, X., Li, M., Wang, G., and Liu, Y. (2009). miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 37, D98–D104.
Johnson, R., Zuccato, C., Belyaev, N. D., Guest, D. J., Cattaneo, E., and Buckley, N. J. (2008). A microRNA-based gene dysregulation pathway in Huntington’s disease. Neurobiol. Dis. 29, 438–445.
Johnson, S. M., Grosshans, H., Shingara, J., Byrom, M., Jarvis, R., Cheng, A., Labourier, E., Reinert, K. L., Brown, D., and Slack, F. J. (2005). RAS is regulated by the let-7 microRNA family. Cell 120, 635–647.
Kasinski, A. L., and Slack, F. J. (2011). MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat. Rev. Cancer 11, 849–864.
Keller, S., Ridinger, J., Rupp, A. K., Janssen, J. W., and Altevogt, P. (2011). Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 9, 86.
Kogure, T., Lin, W. L., Yan, I. K., Braconi, C., and Patel, T. (2011). Intercellular nanovesicle-mediated microRNA transfer: A mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology 54, 1237–1248.
Koh, W., Sheng, C. T., Tan, B., Lee, Q. Y., Kuznetsov, V., Kiang, L. S., and Tanavde, V. (2010). Analysis of deep sequencing microRNA expression profile from human embryonic stem cells derived mesenchymal stem cells reveals possible role of let-7 microRNA family in downstream targeting of hepatic nuclear factor 4 alpha. BMC Genomics 11(Suppl. 1), S6. doi:10.1186/1471-2164-11-S1-S6
Kosaka, N., Iguchi, H., Yoshioka, Y., Takeshita, F., Matsuki, Y., and Ochiya, T. (2010). Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 285, 17442–17452.
Kota, J., Chivukula, R. R., O’donnell, K. A., Wentzel, E. A., Montgomery, C. L., Hwang, H. W., Chang, T. C., Vivekanandan, P., Torbenson, M., Clark, K. R., Mendell, J. R., and Mendell, J. T. (2009). Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137, 1005–1017.
Krol, J., Loedige, I., and Filipowicz, W. (2010). The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610.
Kumar, M. S., Erkeland, S. J., Pester, R. E., Chen, C. Y., Ebert, M. S., Sharp, P. A., and Jacks, T. (2008). Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. U.S.A. 105, 3903–3908.
Kuwabara, Y., Ono, K., Horie, T., Nishi, H., Nagao, K., Kinoshita, M., Watanabe, S., Baba, O., Kojima, Y., Shizuta, S., Imai, M., Tamura, T., Kita, T., and Kimura, T. (2011). Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ. Cardiovasc. Genet. 4, 446–454.
Laulagnier, K., Motta, C., Hamdi, S., Roy, S., Fauvelle, F., Pageaux, J. F., Kobayashi, T., Salles, J. P., Perret, B., Bonnerot, C., and Record, M. (2004). Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem. J. 380, 161–171.
Li, Q., Lin, X., Yang, X., and Chang, J. (2010). NFATc4 is negatively regulated in miR-133a-mediated cardiomyocyte hypertrophic repression. Am. J. Physiol. Heart Circ. Physiol. 298, H1340–H1347.
Lim, P. K., Bliss, S. A., Patel, S. A., Taborga, M., Dave, M. A., Gregory, L. A., Greco, S. J., Bryan, M., Patel, P. S., and Rameshwar, P. (2011). Gap junction-mediated import of microRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells. Cancer Res. 71, 1550–1560.
Liu, Y., Xiang, X., Zhuang, X., Zhang, S., Liu, C., Cheng, Z., Michalek, S., Grizzle, W., and Zhang, H. G. (2010). Contribution of MyD88 to the tumor exosome-mediated induction of myeloid derived suppressor cells. Am. J. Pathol. 176, 2490–2499.
Lu, J., He, M. L., Wang, L., Chen, Y., Liu, X., Dong, Q., Chen, Y. C., Peng, Y., Yao, K. T., Kung, H. F., and Li, X. P. (2011). MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 71, 225–233.
Lui, W. O., Pourmand, N., Patterson, B. K., and Fire, A. (2007). Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 67, 6031–6043.
Luo, S. S., Ishibashi, O., Ishikawa, G., Ishikawa, T., Katayama, A., Mishima, T., Takizawa, T., Shigihara, T., Goto, T., Izumi, A., Ohkuchi, A., Matsubara, S., and Takeshita, T. (2009). Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol. Reprod. 81, 717–729.
Masyuk, A. I., Huang, B. Q., Ward, C. J., Gradilone, S. A., Banales, J. M., Masyuk, T. V., Radtke, B., Splinter, P. L., and Larusso, N. F. (2010). Biliary exosomes influence cholangiocyte regulatory mechanisms and proliferation through interaction with primary cilia. Am. J. Physiol. Gastrointest. Liver Physiol. 299, G990–G999.
Mathivanan, S., Fahner, C. J., Reid, G. E., and Simpson, R. J. (2011). ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 40, D1241–D1244.
Mattes, J., Collison, A., Plank, M., Phipps, S., and Foster, P. S. (2009). Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc. Natl. Acad. Sci. U.S.A. 106, 18704–18709.
Mayr, C., Hemann, M. T., and Bartel, D. P. (2007). Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315, 1576–1579.
Meckes, D. G. Jr., Shair, K. H., Marquitz, A. R., Kung, C. P., Edwards, R. H., and Raab-Traub, N. (2010). Human tumor virus utilizes exosomes for intercellular communication. Proc. Natl. Acad. Sci. U.S.A. 107, 20370–20375.
Mencia, A., Modamio-Hoybjor, S., Redshaw, N., Morin, M., Mayo-Merino, F., Olavarrieta, L., Aguirre, L. A., Del Castillo, I., Steel, K. P., Dalmay, T., Moreno, F., and Moreno-Pelayo, M. A. (2009). Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 41, 609–613.
Michael, A., Bajracharya, S. D., Yuen, P. S., Zhou, H., Star, R. A., Illei, G. G., and Alevizos, I. (2010). Exosomes from human saliva as a source of microRNA biomarkers. Oral. Dis. 16, 34–38.
Minones-Moyano, E., Porta, S., Escaramis, G., Rabionet, R., Iraola, S., Kagerbauer, B., Espinosa-Parrilla, Y., Ferrer, I., Estivill, X., and Marti, E. (2011). MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum. Mol. Genet. 20, 3067–3078.
Mittelbrunn, M., Gutierrez-Vazquez, C., Villarroya-Beltri, C., Gonzalez, S., Sanchez-Cabo, F., Gonzalez, M. A., Bernad, A., and Sanchez-Madrid, F. (2011). Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2, 282.
Montecalvo, A., Larregina, A. T., Shufesky, W. J., Beer Stolz, D., Sullivan, M. L., Karlsson, J. M., Baty, C. J., Gibson, G. A., Erdos, G., Wang, Z., Milosevic, J., Tkacheva, O. A., Divito, S. J., Jordan, R., Lyons-Weiler, J., Watkins, S. C., and Morelli, A. E. (2012). Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 119, 756–766.
Müller, G., Schneider, M., Biemer-Daub, G., and Wied, S. (2011). Microvesicles released from rat adipocytes and harboring glycosylphosphatidylinositol-anchored proteins transfer RNA stimulating lipid synthesis. Cell. Signal. 23, 1207–1223.
O’Connell, J., Bennett, M. W., O’sullivan, G. C., Roche, D., Kelly, J., Collins, J. K., and Shanahan, F. (1998). Fas ligand expression in primary colon adenocarcinomas: evidence that the Fas counterattack is a prevalent mechanism of immune evasion in human colon cancer. J. Pathol. 186, 240–246.
O’Neill, L. A., Sheedy, F. J., and Mccoy, C. E. (2011). MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat. Rev. Immunol. 11, 163–175.
Ohshima, K., Inoue, K., Fujiwara, A., Hatakeyama, K., Kanto, K., Watanabe, Y., Muramatsu, K., Fukuda, Y., Ogura, S., Yamaguchi, K., and Mochizuki, T. (2010). Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PloS ONE 5, e13247. doi:10.1371/journal.pone.0013247
Ostrowski, M., Carmo, N. B., Krumeich, S., Fanget, I., Raposo, G., Savina, A., Moita, C. F., Schauer, K., Hume, A. N., Freitas, R. P., Goud, B., Benaroch, P., Hacohen, N., Fukuda, M., Desnos, C., Seabra, M. C., Darchen, F., Amigorena, S., Moita, L. F., and Thery, C. (2010). Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12, 19–30; sup pp. 11–13.
Pan, B. T., and Johnstone, R. M. (1983). Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell 33, 967–978.
Pegtel, D. M., Cosmopoulos, K., Thorley-Lawson, D. A., Van Eijndhoven, M. A., Hopmans, E. S., Lindenberg, J. L., De Gruijl, T. D., Wurdinger, T., and Middeldorp, J. M. (2010). Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. U.S.A. 107, 6328–6333.
Pegtel, D. M., Van De Garde, M. D., and Middeldorp, J. M. (2011). Viral miRNAs exploiting the endosomal-exosomal pathway for intercellular cross-talk and immune evasion. Biochim. Biophys. Acta 1809, 715–721.
Peng, P., Yan, Y., and Keng, S. (2011). Exosomes in the ascites of ovarian cancer patients: origin and effects on anti-tumor immunity. Oncol. Rep. 25, 749–762.
Qazi, K. R., Torregrosa Paredes, P., Dahlberg, B., Grunewald, J., Eklund, A., and Gabrielsson, S. (2010). Proinflammatory exosomes in bronchoalveolar lavage fluid of patients with sarcoidosis. Thorax 65, 1016–1024.
Rabinowits, G., Gercel-Taylor, C., Day, J. M., Taylor, D. D., and Kloecker, G. H. (2009). Exosomal microRNA: a diagnostic marker for lung cancer. Clin. Lung Cancer 10, 42–46.
Raposo, G., Nijman, H. W., Stoorvogel, W., Liejendekker, R., Harding, C. V., Melief, C. J., and Geuze, H. J. (1996). B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 183, 1161–1172.
Record, M., Subra, C., Silvente-Poirot, S., and Poirot, M. (2011). Exosomes as intercellular signalosomes and pharmacological effectors. Biochem. Pharmacol. 81, 1171–1182.
Reid, G., Kirschner, M. B., and Van Zandwijk, N. (2011). Circulating microRNAs: Association with disease and potential use as biomarkers. Crit. Rev. Oncol. Hematol. 80, 193–208.
Rupp, A. K., Rupp, C., Keller, S., Brase, J. C., Ehehalt, R., Fogel, M., Moldenhauer, G., Marme, F., Sultmann, H., and Altevogt, P. (2011). Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol. Oncol. 122, 437–446.
Schorey, J. S., and Bhatnagar, S. (2008). Exosome function: from tumor immunology to pathogen biology. Traffic 9, 871–881.
Silverman, J. M., Clos, J., De’oliveira, C. C., Shirvani, O., Fang, Y., Wang, C., Foster, L. J., and Reiner, N. E. (2010). An exosome-based secretion pathway is responsible for protein export from Leishmania and communication with macrophages. J. Cell. Sci. 123, 842–852.
Silverman, J. M., and Reiner, N. E. (2011). Exosomes and other microvesicles in infection biology: organelles with unanticipated phenotypes. Cell. Microbiol. 13, 1–9.
Skog, J., Wurdinger, T., Van Rijn, S., Meijer, D. H., Gainche, L., Sena-Esteves, M., Curry, W. T. Jr., Carter, B. S., Krichevsky, A. M., and Breakefield, X. O. (2008). Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476.
Sudhof, T. C., and Rothman, J. E. (2009). Membrane fusion: grappling with SNARE and SM proteins. Science 323, 474–477.
Sutton, R. B., Fasshauer, D., Jahn, R., and Brunger, A. T. (1998). Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395, 347–353.
Takeshita, F., Patrawala, L., Osaki, M., Takahashi, R. U., Yamamoto, Y., Kosaka, N., Kawamata, M., Kelnar, K., Bader, A. G., Brown, D., and Ochiya, T. (2010). Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol. Ther. 18, 181–187.
Taylor, D. D., and Gercel-Taylor, C. (2008). MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110, 13–21.
Thery, C. (2011). Exosomes: secreted vesicles and intercellular communications. F1000 Biol. Rep. 3, 15.
Thery, C., Ostrowski, M., and Segura, E. (2009). Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593.
Tiscornia, G., and Izpisua Belmonte, J. C. (2010). MicroRNAs in embryonic stem cell function and fate. Genes Dev. 24, 2732–2741.
Trang, P., Medina, P. P., Wiggins, J. F., Ruffino, L., Kelnar, K., Omotola, M., Homer, R., Brown, D., Bader, A. G., Weidhaas, J. B., and Slack, F. J. (2010). Regression of murine lung tumors by the let-7 microRNA. Oncogene 29, 1580–1587.
Valadi, H., Ekstrom, K., Bossios, A., Sjostrand, M., Lee, J. J., and Lotvall, J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659.
van den Boorn, J. G., Schlee, M., Coch, C., and Hartmann, G. (2011). SiRNA delivery with exosome nanoparticles. Nat. Biotechnol. 29, 325–326.
Vella, L. J., Sharples, R. A., Lawson, V. A., Masters, C. L., Cappai, R., and Hill, A. F. (2007). Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J. Pathol. 211, 582–590.
Volinia, S., Calin, G. A., Liu, C. G., Ambs, S., Cimmino, A., Petrocca, F., Visone, R., Iorio, M., Roldo, C., Ferracin, M., Prueitt, R. L., Yanaihara, N., Lanza, G., Scarpa, A., Vecchione, A., Negrini, M., Harris, C. C., and Croce, C. M. (2006). A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261.
Wang, K., Zhang, S., Weber, J., Baxter, D., and Galas, D. J. (2010). Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 38, 7248–7259.
Yang, M., Chen, J., Su, F., Yu, B., Lin, L., Liu, Y., Huang, J. D., and Song, E. (2011). Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol. Cancer 10, 117.
Yu, S., Liu, C., Su, K., Wang, J., Liu, Y., Zhang, L., Li, C., Cong, Y., Kimberly, R., Grizzle, W. E., Falkson, C., and Zhang, H. G. (2007). Tumor exosomes inhibit differentiation of bone marrow dendritic cells. J. Immunol. 178, 6867–6875.
Yuyama, K., Yamamoto, N., and Yanagisawa, K. (2008). Accelerated release of exosome-associated GM1 ganglioside (GM1) by endocytic pathway abnormality: another putative pathway for GM1-induced amyloid fibril formation. J. Neurochem. 105, 217–224.
Keywords: microRNAs, exosomes, post-transcriptional regulation, cancer, therapy
Citation: Hu G, Drescher KM and Chen X-M (2012) Exosomal miRNAs: biological properties and therapeutic potential. Front. Gene. 3:56. doi: 10.3389/fgene.2012.00056
Received: 10 January 2012; Accepted: 27 March 2012;
Published online: 20 April 2012.
Edited by:
Michael Rossbach, Genome Institute of Singapore, SingaporeReviewed by:
Chris Sullivan, University of Texas at Austin, USACopyright: © 2012 Hu, Drescher and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Xian-Ming Chen, Department of Medical Microbiology and Immunology, Creighton University, Criss II, Room 529, 2500 California Plaza, Omaha, NE 68178, USA. e-mail:eGlhbm1pbmdjaGVuQGNyZWlnaHRvbi5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.