
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 12 July 2011
Sec. Nutrigenomics
volume 2 - 2011 | https://doi.org/10.3389/fgene.2011.00045
Dietary supplementation with ω-3 polyunsaturated fatty acids (ω-3 PUFAs), specifically the fatty acids docosahexaenoic acid (DHA; 22:6 ω-3) and eicosapentaenoic acid (EPA; 20:5 ω-3), is known to have beneficial health effects including improvements in glucose and lipid homeostasis and modulation of inflammation. To evaluate the efficacy of two different sources of ω-3 PUFAs, we performed gene expression profiling in the liver of mice fed diets supplemented with either fish oil (FO) or krill oil (KO). We found that ω-3 PUFA supplements derived from a phospholipid krill fraction (KO) downregulated the activity of pathways involved in hepatic glucose production as well as lipid and cholesterol synthesis. The data also suggested that KO-supplementation increases the activity of the mitochondrial respiratory chain. Surprisingly, an equimolar dose of EPA and DHA derived from FO modulated fewer pathways than a KO-supplemented diet and did not modulate key metabolic pathways regulated by KO, including glucose metabolism, lipid metabolism and the mitochondrial respiratory chain. Moreover, FO upregulated the cholesterol synthesis pathway, which was the opposite effect of krill-supplementation. Neither diet elicited changes in plasma levels of lipids, glucose, or insulin, probably because the mice used in this study were young and were fed a low-fat diet. Further studies of KO-supplementation using animal models of metabolic disorders and/or diets with a higher level of fat may be required to observe these effects.
The liver plays a major role in controlling whole body carbohydrate and lipid metabolism by using dietary ω-6 and ω-3 fatty acids and their respective products as sensors for nutritional state. These fatty acids have the ability to target specific transcription factors, e.g., peroxisome proliferator-activated receptor α (PPARα), sterol regulator element binding protein-1c (SREBF1) and the heterodimer from MLX interacting protein-like (MLXIPL or ChREBP) and Max-like protein X (MLX; Jump et al., 2008a). Consequently, fatty acids have the ability to control transcription factor activity, which in turn regulates gene expression and metabolic processes such as glycolysis, lipid biosynthesis, fatty acid elongation, desaturation, and oxidation. Furthermore, ω-3 fatty acids have the ability to influence the abundance and activity of nuclear receptors by affecting their proteolytic processing and phosphorylation states (Jump, 2004; Dentin et al., 2005a; Xu et al., 2006). However, these specific regulatory effects are not shared by all fatty acids. Two ω-3 polyunsaturated fatty acids (ω-3 PUFAs) that can regulate gene expression, and are linked to many beneficial effects in human health and disease, are eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA; Riediger et al., 2009). These long-chain fatty acids are enriched in krill and fish, though the EPA and DHA found in krill oil (KO) and fish oil (FO) differ in their structural form: EPA and DHA in FO are in the form of triglycerides, whereas KO contains the majority of the EPA and DHA esterified into phospholipids (Winther et al., 2011). In addition to a high level of phospholipids, KO contains the antioxidant astaxanthin that protects the unsaturated bonds from oxidative damage (Tou et al., 2007). The different structural fractions of fatty acids might affect incorporation into cells, susceptibility to peroxidation, and bioavailability resulting in different molecular effects or different tissue specificities (Wijendran et al., 2002; Batetta et al., 2009; Maki et al., 2009; Graf et al., 2010; Ulven et al., 2011).
Despite the known differences in fatty acid and micronutrient compositions in FO and KOs, it is unknown if there are differential effects of the two compounds in vivo. Therefore, we performed a study to determine if consumption of a diet supplemented with either FO or KO altered gene expression profiles of liver tissue of mice fed diets varying in their lipid source (i.e., FO and KO) but containing equal levels of of EPA and DHA. Our analysis revealed that ω-3 PUFAs of different origin have mostly non-overlapping effects at the gene expression level, such that markedly fewer genes and metabolic pathways were regulated by a FO (vs. KO) supplemented diet.
Male CBA/J mice were purchased from Jackson Laboratory at 6 weeks of age and were individually housed and fed 84 kcal/week of a diet based on the AIN93M formulation (Reeves, 1997). Starting at 2 months of age, mice were maintained on the Control diet (containing 4% lipid from soybean oil) or were fed one of the following two test diets (n = 10 mice per group): supplemented with FO (FO: 18/12 cholesterol-reduced FO, GC Rieber Oil AS, Kristiansund, Norway), or supplemented with Superba™ KO (KO, Aker BioMarine ASA, Norway). Both test diets were based on modifications of the Control diet and were formulated by substituting an equivalent amount of the soybean oil with either 1.1% FO or 1.5% KO. Fish and krill oil diets did not differ considerably in the total amount of ω-3 PUFAs as the content of EPA and DHA in the diets were 0.31% (FO) and 0.29% (KO). Fatty acid composition of the diets are shown in Table 1. All diets were formulated by Bio-Serv (Frenchtown, NJ, USA).
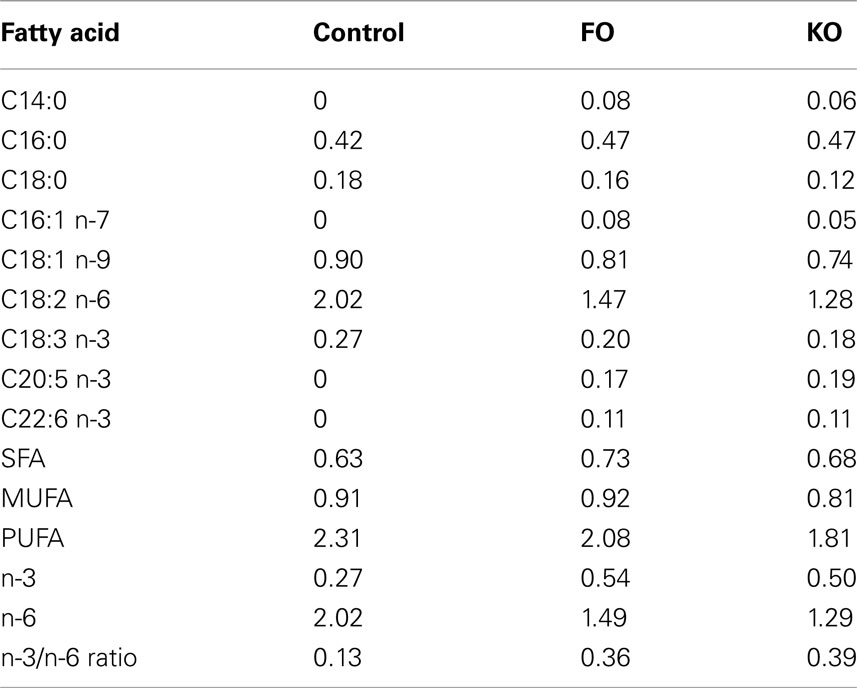
Table 1. Fatty acid composition (grams per 100 g of diet) of the three diets as determined from reference values for the different lipid sources.
Body weight was measured once every 2 weeks. At 5 months of age, mice were euthanized by cervical dislocation after an overnight fast, blood was collected from the body cavity, and tissues were rapidly dissected, flash frozen in liquid nitrogen, and stored at −80°C. All experimental protocols received approval from the Animal Care and Use Committee at the William S. Middleton Memorial Veterans Hospital (Madison, WI, USA), which is fully compliant with the American Physiological Society’s “Guiding Principles in the Care and Use of Animals” (American Physiological Society, 2002).
Gene expression profiling was performed as described previously (Barger et al., 2008). Briefly, total RNA was extracted from liver tissue of seven mice per group and was processed according to standard protocols described by Affymetrix. Samples were hybridized on the Affymetrix Mouse Genome 430 2.0 array which represents >20,000 known genes. To determine the effect of a test diet on the expression of a gene, the average signal intensity for the treated group was compared to the average signal intensity for that gene in the control group. Comparisons between groups were made using two-tailed t-tests (experimental vs. Control); a gene was considered to be significantly changed by treatment at p < 0.01.
To identify functional classes of genes changed by treatment, we performed parametric analysis of gene set enrichment (PAGE). This technique allows for an unbiased and highly sensitive method of detecting classes of genes that are modulated by treatment (Kim and Volsky, 2005). In addition, PAGE generates a z-score indicating if a gene class was activated (z-score > 0) or repressed (z-score < 0) by treatment. We grouped genes into functional classes using the Reactome hierarchy (http://www.reactome.org; Vastrik et al., 2007) and only considered Reactome terms that were annotated with at least 10 but not more than 1000 genes per term. Gene functional classes were considered to be significantly altered by treatment at p < 0.001. Raw microarray data have been archived at NCBI-GEO at can be accessed at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE29572.
Lipids and glucose were measured enzymatically on a Hitachi 917 system (Roche Diagnostics GmbH, Mannheim, Germany) using the triglyceride (GPO–PAP) and cholesterol (CHOD–PAP) kits from Roche Diagnostics (Mannheim, Germany) and the phospholipid kit from DiaSys Diagnostic Systems GmbH (Holzheim, Germany). Plasma insulin was measured using an ELISA kit from Millipore (Billerica, MA, USA).
Individually housed male, 2-month-old CBA/J mice were fed one of three low-fat diets that were isocaloric, but differed in their source of lipids: a control diet containing soybean oil (Control), or a control diet either supplemented with FO or KO and balanced for EPA and DHA concentration. Biweekly bodyweight did not differ among the groups at any time point (data not shown). At 5 months of age the mice were euthanized and tissues were collected for gene expression analysis by microarray and plasma was collected for lipid measurements.
To determine if different sources of ω-3 PUFAs have differential effects at the molecular level, we performed gene expression profiling in liver and compared the effect of FO and KO diets to the Control diet. Of the 20,118 genes represented on the array, 192 genes were significantly changed in expression by FO, whereas 4,892 genes were significantly changed in expression by KO (p < 0.01 treated vs. control). Of the 51 genes that were significantly changed by both diets, 44/51 genes were changed in the same direction. Similarly, pathway analysis using PAGE revealed a more robust effect of KO vs FO. Specifically, 52 pathways were significantly modulated by KO, whereas only four pathways were influenced by FO in the diet (p < 0.001; Figure 1). All four pathways modulated by FO were also modulated by KO; however, one pathway (“Cholesterol biosynthesis”) was differentially regulated between the two diets such that FO increased the activity of this pathway, whereas KO decreased the activity. Because the two experimental diets contained identical concentrations of EPA and DHA (0.3%), these data clearly show that KO is more bioactive than FO on gene expression in the liver.

Figure 1. Metabolic pathways regulated by either fish oil (FO) or krill oil (KO) in mouse liver. Pathway analysis (PAGE, see Materials and Methods) was used to identify functional classes of genes from the Reactome database that were differentially regulated in supplemented diets compared to the Control diet at p < 0.001. Red or blue fills represent the magnitude of the z-score and indicate if a pathway was significantly increased or decreased in activity (respectively) by treatment. Each row represents an individual Reactome pathway and each column represents the effect of either FO or KO on that pathway. Only those Reactome IDs represented by at least 10, but not more than 1000, genes are shown. Pathways indicated by bold are discussed in detail in the manuscript.
Pathway analysis of the hepatic transcriptome revealed that there was a decrease in the activity of the pathway “Glucose metabolism” (Reactome ID 969399) for both the KO and FO diets compared to controls (Figure 2). Although only one gene in this pathway (Pklr, pyruvate kinase, liver and red blood cell) was significantly changed in expression by FO, PAGE revealed that the pathway overall was modulated because there were modest, yet consistent effects of FO on genes in this functional category (data not shown). The ability of ω-3 PUFAs to decrease glucose metabolism, compared to controls with the same energy intake, may be regulated at the early stages of glycolysis by decreasing glucose uptake through downregulation of the liver-specific glucose transporter (Slc2a2). Indeed, KO showed a trend for 25% decreased Slc2a2 expression (−1.3-fold change; p = 0.037).

Figure 2. Gene expression heatmap for genes involved in glucose metabolism. PAGE analysis showed a statistically significant effect of fish oil (FO) and krill oil (KO) on the Reactome pathway “Glucose metabolism.” All genes annotated in this pathway and changed in expression by at least one treatment are shown. Two-tailed t-tests were used to determine significant changes at p < 0.01 between treatment and the Control group.
Parametric analysis of gene set enrichment also revealed a significant downregulation (p < 0.001) in the pathway “Gluconeogenesis” (Reactome ID 969398) only in the KO group (Figure 3A). In addition, several genes known to regulate hepatic glucose production (but not yet annotated by the Reactome consortium) were decreased in expression in the KO group, providing further evidence for decreased hepatic gluconeogenesis. Two of these genes, peroxisome proliferator-activated receptor γ coactivator 1α (Ppargc1a or Pgc-1α) and hepatocyte nuclear factor 4α (Hnf4a), are master regulators of metabolic gene transcription and have potent physiological effects on hepatic gluconeogenesis (Yoon et al., 2001). Both Ppargc1a (Figure 3B) and Hnf4a (Figure 3C) expression were significantly decreased by the KO diet (−1.5-fold change, p = 0.01 and −3.6-fold change, p < 0.001, respectively), as was a downstream target of these transcriptional regulators, phosphoenolpyruvate carboxykinase 1 (Pck1). The Pck1 gene encodes an enzyme that exerts control over hepatic glucose production, and Pck1 was significantly decreased in expression only by KO (−2.6-fold change, p = 0.0002, Figure 3D). Despite the decreased expression of genes involved in gluconeogenesis, we did not observe significant alterations in the levels of plasma glucose in either the FO or KO groups compared to the Control group (Table 2).
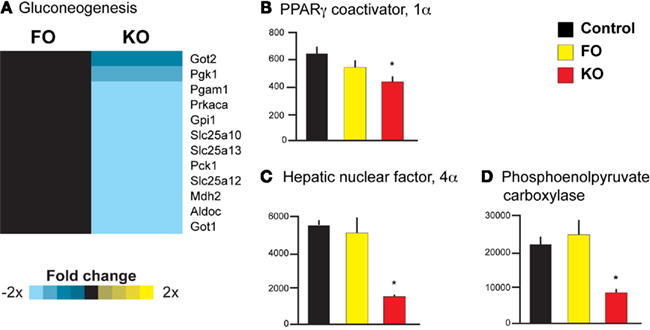
Figure 3. Gene expression heatmap and transcriptional regulators of gluconeogenesis. Pathway analysis revealed a significant downregulation of the Reactome pathway “Gluconeogenesis” by KO (A). In addition, only KO significantly decreased the expression of genes regulating hepatic gluconeogenesis, including Ppargc1a(B), Hnf4a (C), and Pck1 (D).
Gene expression profiling and pathway analysis also suggested that FO and KO differ in their ability to regulate hepatic lipid metabolism. Specifically, KO significantly downregulated the activity of the pathway “Metabolism of lipids and lipoproteins” (Reactome ID 969435), whereas FO had no effect on this pathway (Figure 4A). Two key regulators of the hepatic lipid metabolism are the transcription cofactors SREBF1 and MLXIPL. Both were significantly decreased in expression in the KO group (−2.4-fold change, p = 0.002 for Srebf1 and −5.8-fold change, p < 0.001 for Mlxipl), whereas FO had no effect on the expression of these genes (Figures 4B,C). In addition to the decreased expression of these transcriptional cofactors, three downstream targets of these regulatory genes (Dentin et al., 2005b) were significantly decreased after supplementing KO to the diet (p < 0.001): ATP citrate lyase (Acly) catalyzes the conversion of citrate to acetyl CoA for synthesis of fatty acids and showed a −2.0-fold change with KO (Figure 4A); fatty acid synthase (Fasn, Figure 4A) and acetyl CoA carboxylase alpha (Acaca, Figure 4D), catalyze two of the initial steps of fatty acid synthesis and both were decreased with KO supplementation (−3.3-fold and −2.7-fold change, respectively).
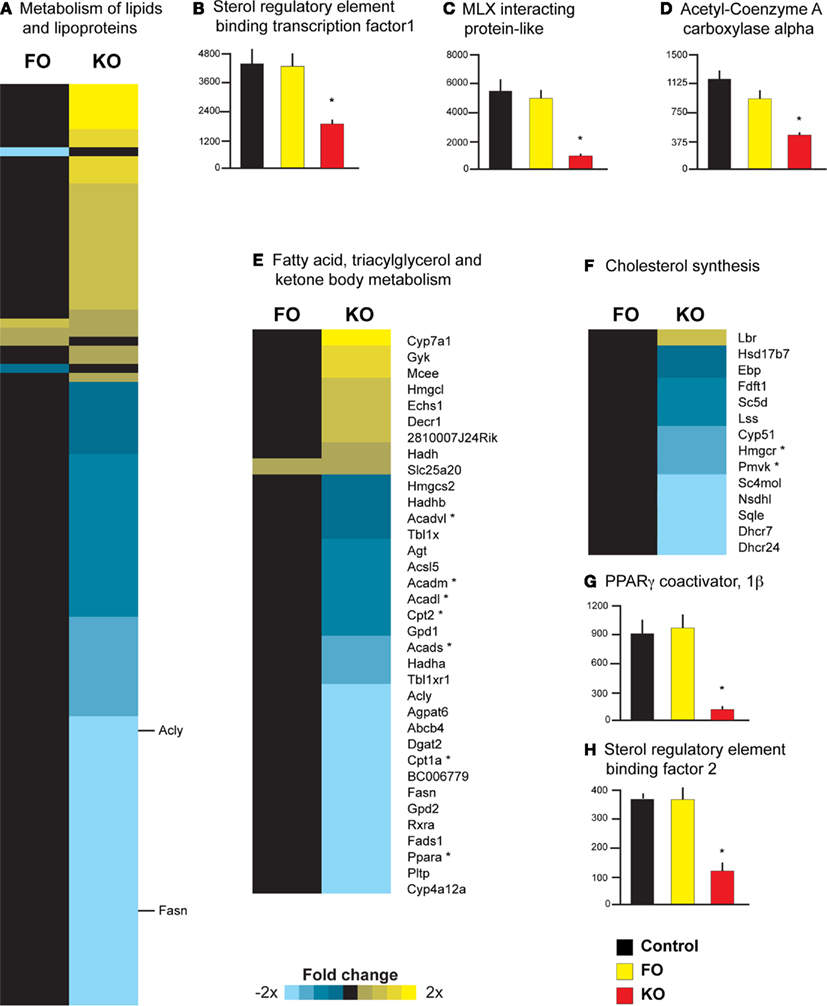
Figure 4. Diverse effects of krill oil on genes regulating lipid metabolism in mouse liver. Pathway analysis revealed a significant downregulation by KO on the Reactome pathway “Metabolism of lipids and lipoproteins” (A) as well as three key transcriptional regulators of lipid metabolism, including Srebf1(B), Mlxipl (C), and Acaca (D). Individual gene symbols could not be represented for all genes in this pathway; two genes described in the Results section are highlighted with their respective gene symbols. In addition, KO downregulated the activity of the Reactome pathway “Fatty acid, triacylglycerol and ketone body metabolism” (E); genes highlighted in the Results section are indicated with an asterisk. Pathway analysis also revealed that FO upregulated, whereas KO downregulated, the Reactome pathway “Cholesterol metabolism” (F). Two regulatory genes in cholesterol synthesis are highlighted in the Results and indicated with an asterisk. Two transcriptional cofactors that are upstream regulators of cholesterol synthesis [Ppargc1b, (G) and Srebf2, (H)] were also decreased in expression only by KO.
Krill oil, but not FO, supplementation also was associated with a significant downregulation of the pathway “Fatty acid, triacylglycerol and ketone body metabolism” (Reactome ID 969434; Figure 4E). Significant decreases in the expression of key genes involved in mitochondrial β-oxidation were observed only in the KO group, including the transcription factor PPARα (−5.1-fold change) and the liver isoform of the rate-limiting enzymes carnitine palmitoyltransferase 1a and 2 (Cpt1a, −2.4-fold change and Cpt2, −1.6-fold change; Figure 4D). Moreover, four genes encoding enzymes involved in mitochondrial fatty acid β-oxidation (Acads, Acadm, Acadl, and Acadvl) were all significantly decreased in expression in the KO group (−1.4 to −1.8-fold change, see Figure 4D). FO however, did not affect the expression of any of the above genes.
Parametric analysis of gene set enrichment analysis further revealed that FO significantly upregulated the Reactome pathway “Cholesterol biosynthesis” (Reactome ID 969930), while a KO diet downregulated this pathway (Figure 4F). Specifically, KO resulted in a significant decrease in the expression of two key genes in the pathway of cholesterol metabolism: 3-hydroxy-3-methylglutaryl-coenzyme A reductase (Hmgcr, the gene encoding the rate-limiting enzyme for cholesterol synthesis, −1.9-fold change) and phosphomevalonate kinase (Pmvk, the gene encoding the protein that catalyzes the fifth condensation reaction in cholesterol synthesis, −2.0-fold change; see Figure 4F). Other studies suggest that the hyperlipidemic effects of dietary saturated fats are mediated through increased activity of Ppargc1b and Srebf2 (Lin et al., 2005; Lelliott et al., 2007), and we observed a significant (p < 0.001) downregulation of these genes in the KO group (−7.9-fold change for Ppargc1b, Figure 4G and −3.2-fold change for Srebf2, Figure 4H). Thus, these data support previous studies demonstrating that the liver is sensitive to the saturation of dietary fatty acids, and that PPARGC1B and SREBF2 activity may be the important regulators of cholesterol synthesis in response to dietary fatty acid saturation.
Plasma analysis showed no significant changes in triglyceride, total cholesterol, free fatty acids, phospholipids, or insulin levels in either treatment groups (Table 2).
Parametric analysis of gene set enrichment revealed that only feeding of a KO and not FO diet to mice resulted in a significant activation of the Reactome pathway “Respiratory electron transport” (ID 969800, Figure 5A). This response was largely driven by an increased expression of genes encoding subunits of Complex I (NADH dehydrogenase). Noteworthy is also the increased expression of sirtuin 3 (Sirt3) only in the KO treated animals (1.7-fold change, Figure 5B). Sirt3 is a regulator of mitochondrial energy metabolism, particularly Complex I (Ahn et al., 2008).
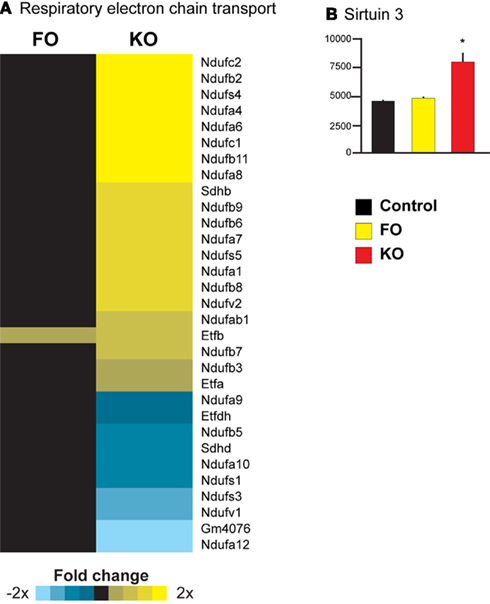
Figure 5. Upregulation of mitochondrial energy metabolism in liver of mice consuming KO. Pathway analysis revealed a significant upregulation of the pathway “Respiratory electron transport” in the KO group (A). A transcriptional regulator of nuclear genes regulating mitochondrial energy metabolism, Sirt3, was also upregulated by KO (B).
Gene expression profiling and pathway analysis demonstrated that krill-supplementation regulates genes with diverse aspects of hepatic energy metabolism, including glucose metabolism, lipid biosynthesis, fatty acid metabolism, and cholesterol biosynthesis. Importantly, these transcriptional effects are in strong agreement with published reports showing beneficial effects of krill-supplementation on circulating levels of glucose and cholesterol (Batetta et al., 2009; Tandy et al., 2009). Although we did not observe changes in plasma glucose or lipids in this study, this is likely because our study involved young, healthy mice fed a low-fat diet. For example, two recent studies demonstrate that KO supplementation improves parameters of hepatic lipid metabolism in genetic and diet-induced animal models of obesity (Batetta et al., 2009; Tandy et al., 2009). Because hepatic lipid accumulation (steatosis) is associated with insulin resistance and the metabolic syndrome in humans (Parekh and Anania, 2007), the transcriptomic findings from this study provide evidence that KO may have beneficial effects on hepatic lipid metabolism associated with metabolic disease in humans (Bunea et al., 2004).
Elevated hepatic glucose production is associated with metabolic disease in humans (Natali and Ferrannini, 2006) and we observed that KO supplementation favorably altered the expression of genes regulating hepatic glucose metabolism. For example, the expression of Ppargc1a and Hnf4a is increased in the liver of humans with type 2 diabetes and in mouse models of diabetes (Herzig et al., 2001; Puigserver, 2005; Liang et al., 2009), and we observed that expression of these genes was decreased after KO treatment (Figures 3A,B). Suppression of gluconeogenesis and hepatic glucose output via modulation of PGC-1α activity has been proposed as a therapeutic target for the treatment of type 2 diabetes, provided that the intervention does not oppose the beneficial effects of Ppargc1a expression in other tissues. One study has shown that KO lowers circulating glucose in humans (Bunea et al., 2004), suggesting that the krill supplements used in this study may functionally decrease hepatic glucose production and increase glucose uptake in tissues other than liver. Transcriptomic analysis in other tissues of krill-treated mice would reveal if PGC-1α activity on regulation of metabolic genes is seen in other peripheral tissues.
Interestingly, FO had rather modest effects on metabolic pathways regulated by the KO-supplemented diet (Figure 1). For example the pathway “Fatty acid, triacylglycerol and ketone body metabolism” was not modulated by a FO-supplemented diet (Figure 4E). The reason for the discrepancy between the effect of FO in this study and previous reports (Price et al., 2000; Clarke, 2004; Jump, 2008b) showing that ω-3 PUFAs increase fatty acid oxidation is not clear, but might be related to the relatively low dose of EPA and DHA used in the current study. In our study, the content of EPA and DHA was approximately 0.3%, whereas FO-supplemented diets usually contain much higher doses, with concentrations around 5% (Mori et al., 2007; Wergedahl et al., 2009; Tsuduki et al., 2011). Nonetheless, we observed marked transcriptional effects of KO at a low dose, suggesting that krill-derived ω-3 PUFAs are bioactive, when present in the diet at a low concentration. One characteristic of KO is the predominant presence of EPA and DHA in the phospholipid form. The amount of phospholipids in KO is around 40–45%. It is a complex mixture of 69 different choline-containing phospholipid molecules (Le Grandois et al., 2009; Winther et al., 2011). The majority of ω-3 fatty acids are EPA and DHA, but also stearidonic acid and docosapentaenoic acid are present in smaller quantities. FO contains ω-3 fatty acids in triglyceride form. This might explain the different bioavailabilities and tissue distributions observed for EPA and DHA in phospholipid versus triglyceride form (Wijendran et al., 2002; Maki et al., 2009; Graf et al., 2010; Ulven et al., 2011). The exact mechanisms behind are not known, but it has been proposed that the different molecular forms could determine in which blood pool the fatty acids end up, which might influence tissue distribution (Amate et al., 2001). Different hepatic levels of ω-3 PUFAs might in turn explain the observed differential gene expression profiles between the KO and FO groups.
In addition to showing that KO affects multiple metabolic pathways, our data also suggests a molecular mechanism by which krill-derived ω-3 PUFAs alter the expression of key transcriptional cofactors that have downstream effects on expression of metabolic genes. For example, expression and activity of the genes encoding the transcriptional cofactors Srebf1 and Mlxipl are increased by insulin, resulting in a stimulation of glycolysis and hepatic lipogenesis leading to lipid accumulation and insulin resistance in the liver (Dentin et al., 2005b; Iizuka and Horikawa, 2008). Conversely, inhibition or deficiency of these proteins ameliorates metabolic abnormalities in mouse models of the metabolic syndrome (Iizuka et al., 2004; Dentin et al., 2006a). We found that KO robustly decreased the expression of these genes (Figures 4B,C) which is in agreement with other studies showing that ω-3 PUFAs decrease the activity of these proteins (Xu et al., 2002; Sekiya et al., 2003; Dentin et al., 2006b). The observation that KO reduced Fasn and Acaca also supports the finding in a study in rat, which showed that KO significantly reduced the activity of three key enzymes in lipogenesis in liver (Ferramosca et al., 2011).
The current study also provides evidence that krill-supplementation may stimulate mitochondrial respiratory activity, a pathway which is repressed in humans with type 2 diabetes (Mootha et al., 2003). We found a coordinated upregulation of nuclear-encoded genes regulating mitochondrial electron transport (Figure 5A), and this agrees with a large number of studies showing that increases in the PUFA content of the diet are associated with changes in metabolic rate (Hulbert et al., 2005). In addition to an increase of genes coding for subunits of complex I of the electron transport chain, a change in the expression of Sirt3 was observed with KO treatment. Sirtuins are a family of genes associated with metabolic control, and in humans elevated Sirt3 expression is associated with increased lifespan (Rose et al., 2003; Bellizzi et al., 2005; Lanza et al., 2008). It is therefore interesting to note that KO, but not FO significantly upregulated Sirt3 in this study. SIRT3 deacetylates several targets, including proteins of complex I, which is important for maintaining basal ATP levels (Ahn et al., 2008). A KO-induced stimulation of ATP production could be of interest in combating aging-associated decline of ATP levels.
Our data demonstrate a marked effect of KO on the regulation of genes and pathways involved in hepatic energy metabolism. Specifically, the effects of KO were the opposite of the metabolic dysregulation seen in obesity and type 2 diabetes. Dietary supplementation with KO elicited a transcriptional profile that suggests improved glucose, lipid, and cholesterol metabolism and increased mitochondrial respiration. Furthermore, our data also suggest a mechanism whereby krill-derived ω-3 PUFAs may influence metabolism through upstream transcriptional activation of PGC-1α, PGC-1β, SREBF1, and MLXIPL. The differential expression of key transcriptional regulators likely has multiplicative effects on the expression of downstream genes, and this may explain why a large number of genes were changed in expression in response to KO. Surprisingly, these effects were not observed in mice receiving a diet supplemented with FO containing an equivalent level of PUFAs, emphasizing the influence that the structural form of ω-3 PUFAs (esterified to either phospholipids or triglycerides) has on exerting a biological response. Overall, the data provide support for a role of KO in ameliorating abnormalities seen with obesity and insulin resistance; further studies in other animal models will be needed to confirm if the gene expression patterns observed in this study have effects on metabolic disease.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Tsuyoshi Kayo, Liv Kristine Øysæd and Eili Tranheim Kase for excellent technical assistance. This work was supported by a grant from NordForsk, Grant number 070010, MitoHealth (to Lena Burri and Rolf K. Berge) and funding from Aker BioMarine ASA, Oslo, Norway.
Ahn, B. H., Kim, H. S., Song, S., Lee, I. H., Liu, J., Vassilopoulos, A., Deng, C. X., and Finkel, T. (2008). A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc. Natl. Acad. Sci. U.S.A. 105, 14447–14452.
Amate, L., Gil, A., and Ramirez, M. (2001). Feeding infant piglets formula with long-chain polyunsaturated fatty acids as triacylglycerols or phospholipids influences the distribution of these fatty acids in plasma lipoprotein fractions. J. Nutr. 131, 1250–1255.
American Physiological Society. (2002). Guiding principles for research involving animals and human beings. Am. J. Physiol. Regul. Integr. Comp. Physiol. 283, R281–R283.
Barger, J. L., Kayo, T., Vann, J. M., Arias, E. B., Wang, J., Hacker, T. A., Wang, Y., Raederstorff, D., Morrow, J. D., Leeuwenburgh, C., Allison, D. B., Saupe, K. W., Cartee, G. D., Weindruch, R., and Prolla, T. A. (2008). A low dose of dietary resveratrol partially mimics caloric restriction and retards aging parameters in mice. PLoS ONE 3, e2264. doi: 10.1371/journal.pone.0002264
Batetta, B., Griinari, M., Carta, G., Murru, E., Ligresti, A., Cordeddu, L., Giordano, E., Sanna, F., Bisogno, T., Uda, S., Collu, M., Bruheim, I., Di Marzo, V., and Banni, S. (2009). Endocannabinoids may mediate the ability of (n-3) fatty acids to reduce ectopic fat and inflammatory mediators in obese Zucker rats. J. Nutr. 139, 1495–1501.
Bellizzi, D., Rose, G., Cavalcante, P., Covello, G., Dato, S., De Rango, F., Greco, V., Maggiolini, M., Feraco, E., Mari, V., Franceschi, C., Passarino, G., and De Benedictis, G. (2005). A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics 85, 258–263.
Bunea, R., El Farrah, K., and Deutsch, L. (2004). Evaluation of the effects of Neptune Krill Oil on the clinical course of hyperlipidemia. Altern. Med. Rev. 9, 420–428.
Clarke, S. D. (2004). The multi-dimensional regulation of gene expression by fatty acids: polyunsaturated fats as nutrient sensors. Curr. Opin. Lipidol. 15, 13–18.
Dentin, R., Benhamed, F., Hainault, I., Fauveau, V., Foufelle, F., Dyck, J. R., Girard, J., and Postic, C. (2006a). Liver-specific inhibition of ChREBP improves hepatic steatosis and insulin resistance in ob/ob mice. Diabetes 55, 2159–2170.
Dentin, R., Denechaud, P. D., Benhamed, F., Girard, J., and Postic, C. (2006b). Hepatic gene regulation by glucose and polyunsaturated fatty acids: a role for ChREBP. J. Nutr. 136, 1145–1149.
Dentin, R., Benhamed, F., Pegorier, J. P., Foufelle, F., Viollet, B., Vaulont, S., Girard, J., and Postic, C. (2005a). Polyunsaturated fatty acids suppress glycolytic and lipogenic genes through the inhibition of ChREBP nuclear protein translocation. J. Clin. Invest. 115, 2843–2854.
Dentin, R., Girard, J., and Postic, C. (2005b). Carbohydrate responsive element binding protein (ChREBP) and sterol regulatory element binding protein-1c (SREBP-1c): two key regulators of glucose metabolism and lipid synthesis in liver. Biochimie 87, 81–86.
Ferramosca, A., Conte, L., and Zara, V. (2011). A krill oil supplement diet reduces the activities of the mitochondrial tricarboxylate carrier and of the cytosolic lipogenic enzymes in rats. J. Anim. Physiol. Anim. Nutr. doi: 10.1111/j.1439-0396.2011.01135.x. [Epub ahead of print].
Graf, B. A., Duchateau, G. S., Patterson, A. B., Mitchell, E. S., Van Bruggen, P., Koek, J. H., Melville, S., and Verkade, H. J. (2010). Age dependent incorporation of 14C-DHA into rat brain and body tissues after dosing various 14C-DHA-esters. Prostaglandins Leukot. Essent. Fatty Acids 83, 89–96.
Herzig, S., Long, F., Jhala, U. S., Hedrick, S., Quinn, R., Bauer, A., Rudolph, D., Schutz, G., Yoon, C., Puigserver, P., Spiegelman, B., and Montminy, M. (2001). CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413, 179–183.
Hulbert, A. J., Turner, N., Storlien, L. H., and Else, P. L. (2005). Dietary fats and membrane function: implications for metabolism and disease. Biol. Rev. Camb. Philos. Soc. 80, 155–169.
Iizuka, K., Bruick, R. K., Liang, G., Horton, J. D., and Uyeda, K. (2004). Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc. Natl. Acad. Sci. U.S.A. 101, 7281–7286.
Iizuka, K., and Horikawa, Y. (2008). ChREBP: a glucose-activated transcription factor involved in the development of metabolic syndrome. Endocr. J. 55, 617–624.
Jump, D. B. (2004). Fatty acid regulation of gene transcription. Crit. Rev. Clin. Lab. Sci. 41, 41–78.
Jump, D. B., Botolin, D., Wang, Y., Xu, J., Demeure, O., and Christian, B. (2008a). Docosahexaenoic acid (DHA) and hepatic gene transcription. Chem. Phys. Lipids 153, 3–13.
Jump, D. B. (2008b). N-3 polyunsaturated fatty acid regulation of hepatic gene transcription. Curr. Opin. Lipidol. 19, 242–247.
Kim, S. Y., and Volsky, D. J. (2005). PAGE: parametric analysis of gene set enrichment. BMC Bioinform. 6, 144. doi: 10.1186/1471-2105-6-144
Lanza, I. R., Short, D. K., Short, K. R., Raghavakaimal, S., Basu, R., Joyner, M. J., Mcconnell, J. P., and Nair, K. S. (2008). Endurance exercise as a countermeasure for aging. Diabetes 57, 2933–2942.
Le Grandois, J., Marchioni, E., Zhao, M., Giuffrida, F., Ennahar, S., and Bindler, F. (2009). Investigation of natural phosphatidylcholine sources: separation and identification by liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS2) of molecular species. J. Agric. Food Chem. 57, 6014–6020.
Lelliott, C. J., Ljungberg, A., Ahnmark, A., William-Olsson, L., Ekroos, K., Elmgren, A., Arnerup, G., Shoulders, C. C., Oscarsson, J., and Linden, D. (2007). Hepatic PGC-1beta overexpression induces combined hyperlipidemia and modulates the response to PPARalpha activation. Arterioscler. Thromb. Vasc. Biol. 27, 2707–2713.
Liang, H., Balas, B., Tantiwong, P., Dube, J., Goodpaster, B. H., O’Doherty, R. M., Defronzo, R. A., Richardson, A., Musi, N., and Ward, W. F. (2009). Whole body overexpression of PGC-1alpha has opposite effects on hepatic and muscle insulin sensitivity. Am. J. Physiol. Endocrinol. Metab. 296, E945–E954.
Lin, J., Yang, R., Tarr, P. T., Wu, P. H., Handschin, C., Li, S., Yang, W., Pei, L., Uldry, M., Tontonoz, P., Newgard, C. B., and Spiegelman, B. M. (2005). Hyperlipidemic effects of dietary saturated fats mediated through PGC-1beta coactivation of SREBP. Cell 120, 261–273.
Maki, K. C., Reeves, M. S., Farmer, M., Griinari, M., Berge, K., Vik, H., Hubacher, R., and Rains, T. M. (2009). Krill oil supplementation increases plasma concentrations of eicosapentaenoic and docosahexaenoic acids in overweight and obese men and women. Nutr. Res. 29, 609–615.
Mootha, V. K., Lindgren, C. M., Eriksson, K. F., Subramanian, A., Sihag, S., Lehar, J., Puigserver, P., Carlsson, E., Ridderstrale, M., Laurila, E., Houstis, N., Daly, M. J., Patterson, N., Mesirov, J. P., Golub, T. R., Tamayo, P., Spiegelman, B., Lander, E. S., Hirschhorn, J. N., Altshuler, D., and Groop, L. C. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273.
Mori, T., Kondo, H., Hase, T., Tokimitsu, I., and Murase, T. (2007). Dietary fish oil upregulates intestinal lipid metabolism and reduces body weight gain in C57BL/6J mice. J. Nutr. 137, 2629–2634.
Natali, A., and Ferrannini, E. (2006). Effects of metformin and thiazolidinediones on suppression of hepatic glucose production and stimulation of glucose uptake in type 2 diabetes: a systematic review. Diabetologia 49, 434–441.
Parekh, S., and Anania, F. A. (2007). Abnormal lipid and glucose metabolism in obesity: implications for nonalcoholic fatty liver disease. Gastroenterology 132, 2191–2207.
Price, P. T., Nelson, C. M., and Clarke, S. D. (2000). Omega-3 polyunsaturated fatty acid regulation of gene expression. Curr. Opin. Lipidol. 11, 3–7.
Puigserver, P. (2005). Tissue-specific regulation of metabolic pathways through the transcriptional coactivator PGC1-alpha. Int. J. Obes. (Lond.) 29(Suppl. 1), S5–S9.
Reeves, P. G. (1997). Components of the AIN-93 diets as improvements in the AIN-76A diet. J. Nutr. 127, 838S–841S.
Riediger, N. D., Othman, R. A., Suh, M., and Moghadasian, M. H. (2009). A systemic review of the roles of n-3 fatty acids in health and disease. J. Am. Diet. Assoc. 109, 668–679.
Rose, G., Dato, S., Altomare, K., Bellizzi, D., Garasto, S., Greco, V., Passarino, G., Feraco, E., Mari, V., Barbi, C., Bonafe, M., Franceschi, C., Tan, Q., Boiko, S., Yashin, A. I., and De Benedictis, G. (2003). Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp. Gerontol. 38, 1065–1070.
Sekiya, M., Yahagi, N., Matsuzaka, T., Najima, Y., Nakakuki, M., Nagai, R., Ishibashi, S., Osuga, J., Yamada, N., and Shimano, H. (2003). Polyunsaturated fatty acids ameliorate hepatic steatosis in obese mice by SREBP-1 suppression. Hepatology 38, 1529–1539.
Tandy, S., Chung, R. W., Wat, E., Kamili, A., Berge, K., Griinari, M., and Cohn, J. S. (2009). Dietary krill oil supplementation reduces hepatic steatosis, glycemia, and hypercholesterolemia in high-fat-fed mice. J. Agric. Food Chem. 57, 9339–9345.
Tou, J. C., Jaczynski, J., and Chen, Y. C. (2007). Krill for human consumption: nutritional value and potential health benefits. Nutr. Rev. 65, 63–77.
Tsuduki, T., Honma, T., Nakagawa, K., Ikeda, I., and Miyazawa, T. (2011). Long-term intake of fish oil increases oxidative stress and decreases lifespan in senescence-accelerated mice. Nutrition 27, 334–337.
Ulven, S. M., Kirkhus, B., Lamglait, A., Basu, S., Elind, E., Haider, T., Berge, K., Vik, H., and Pedersen, J. I. (2011). Metabolic effects of krill oil are essentially similar to those of fish oil but at lower dose of EPA and DHA, in healthy volunteers. Lipids 46, 37–46.
Vastrik, I., D’eustachio, P., Schmidt, E., Gopinath, G., Croft, D., De Bono, B., Gillespie, M., Jassal, B., Lewis, S., Matthews, L., Wu, G., Birney, E., and Stein, L. (2007). Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 8, R39.
Wergedahl, H., Gudbrandsen, O. A., Rost, T. H., and Berge, R. K. (2009). Combination of fish oil and fish protein hydrolysate reduces the plasma cholesterol level with a concurrent increase in hepatic cholesterol level in high-fat-fed Wistar rats. Nutrition 25, 98–104.
Wijendran, V., Huang, M. C., Diau, G. Y., Boehm, G., Nathanielsz, P. W., and Brenna, J. T. (2002). Efficacy of dietary arachidonic acid provided as triglyceride or phospholipid as substrates for brain arachidonic acid accretion in baboon neonates. Pediatr. Res. 51, 265–272.
Winther, B., Hoem, N., Berge, K., and Reubsaet, L. (2011). Elucidation of phosphatidylcholine composition in krill oil extracted from Euphausia superba. Lipids 46, 25–36.
Xu, J., Cho, H., O’Malley, S., Park, J. H., and Clarke, S. D. (2002). Dietary polyunsaturated fats regulate rat liver sterol regulatory element binding proteins-1 and -2 in three distinct stages and by different mechanisms. J. Nutr. 132, 3333–3339.
Xu, J., Christian, B., and Jump, D. B. (2006). Regulation of rat hepatic L-pyruvate kinase promoter composition and activity by glucose, n-3 polyunsaturated fatty acids, and peroxisome proliferator-activated receptor-alpha agonist. J. Biol. Chem. 281, 18351–18362.
Keywords: Krill oil, fish oil, polyunsaturated fatty acids, gene transcription, liver, metabolism, EPA, DHA
Citation: Burri L, Berge K, Wibrand K, Berge RK and Barger JL (2011) Differential effects of krill oil and fish oil on the hepatic transcriptome in mice. Front. Gene. 2:45. doi: 10.3389/fgene.2011.00045
Received: 22 March 2011;
Paper pending published: 11 April 2011;
Accepted: 25 June 2011;
Published online: 12 July 2011.
Edited by:
Lynnette Robin Ferguson, The University of Auckland, New ZealandReviewed by:
Sander Kersten, Wageningen University, NetherlandsCopyright: © 2011 Burri, Berge, Wibrand, Berge and Barger. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Jamie L. Barger, LifeGen Technologies, LLC, 510 Charmany Drive, Suite 263, Madison, WI 53719, USA. e-mail:amFtaWUubC5iYXJnZXJAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.