- 1 Animal and Bioscience Research Department, Animal and Grassland Research and Innovation Centre, Teagasc, Athenry, County Galway, Ireland
- 2 Animal and Bioscience Research Department, Animal and Grassland Research and Innovation Centre, Teagasc, Fermoy, County Cork, Ireland
- 3 Moorepark Food Research Centre, Teagasc, Fermoy, County Cork, Ireland
- 4 Animal and Bioscience Research Department, Animal and Grassland Research and Innovation Centre, Teagasc, Grange, Dunsany, County Meath, Ireland
- 5 Animal Genomics Laboratory, UCD School of Agriculture, Food Science and Veterinary Medicine, University College Dublin, Belfield, Dublin, Ireland
Insulin-like growth factor 1 (IGF-1) has been shown to be associated with fertility, growth, and development in cattle. The aim of this study was to (1) identify novel single nucleotide polymorphisms (SNPs) in the bovine IGF-1 gene and alongside previously identified SNPs (2) determine their association with traits of economic importance in Holstein-Friesian dairy cattle. Nine novel SNPs were identified across a panel of 22 beef and dairy cattle by sequence analysis of the 5′ promoter, intronic, and 3′ regulatory regions, encompassing ∼5 kb of IGF-1. Genotyping and associations with daughter performance for milk production, fertility, survival, and measures of body size were undertaken on 848 Holstein-Friesian AI sires. Using multiple regression analysis nominal associations (P < 0.05) were identified between six SNPs (four novel and two previously identified) and milk composition, survival, body condition score, and body size. The C allele of AF017143 a previously published SNP (C-512T) in the promoter region of IGF-1 predicted to introduce binding sites for transcription factors HSF1 and ZNF217 was associated (P < 0.05) with increased cow carcass weight (i.e., an indicator of mature cow size). Novel SNPs were identified in the 3′ region of IGF-1 were associated (P < 0.05) with functional survival and chest width. The remaining four SNPs, all located within introns of IGF-1 were associated (P < 0.05) with milk protein yield, milk fat yield, milk fat concentration, somatic cell score, carcass conformation, and carcass fat. Results of this study further demonstrate the multifaceted influences of IGF-1 on milk production and growth related traits in cattle.
Introduction
Insulin-like growth factor 1 (IGF-1) plays an important physiological role in growth regulation, development, metabolism, and lactation in cattle (Daughaday and Rotwein, 1989; Hossner et al., 1997; Breier, 1999; Lucy, 2008). It regulates differentiation including the maintenance of differentiated function in numerous tissues and in specific cell types (Werner et al., 1994). IGF-1 also stimulates the anabolic and mitogenic activity of growth hormone in various tissues (Laron, 2001). The primary source of circulatory IGF-1 is the liver however it is also produced locally in a tissue specific manner (Miller et al., 1981; Schwander et al., 1983; Thissen et al., 1994; Etherton, 2004).
Systemic IGF-1 has been associated with a variety of fertility and reproductive performance measurements in cattle including age at first calving, post-partum resumption of ovarian cyclicity, conception rate to first service, twin ovulation rate, and preimplantation embryo development (Echternkamp et al., 2004; Velazquez et al., 2005; Yilmaz et al., 2006; Patton et al., 2007; Wathes et al., 2007; Velazquez, 2008). Serum IGF-1 has been shown to be highly heritable, with repeatability estimates to the order of 0.48 during the post-weaning period in Angus cattle (Davis and Simmen, 1997). This suggests a strong additive genetic control of IGF-1, a gene which is regulated at both a transcriptional and translational level (Wang et al., 2003). IGF-1 has been shown to contribute to genetic variation in traits such as carcass fatness, live and carcass weight, average daily gain, as well as body size, food conversion efficiency, milk production, and fat deposition (Davis et al., 1995; Davis and Simmen, 2000; Johnston et al., 2001).
Quantitative trait loci (QTL) overlapping the IGF-1 region on BTA5 have been reported (Allan et al., 2009; Kim et al., 2009; Sahana et al., 2010). However despite the physiological importance of IGF-1 there are few published studies analyzing the relationship between IGF-1 variants and performance. Some studies have analyzed a single nucleotide polymorphism (SNP) in the promoter region (C-512T) of IGF-1, AF017143, and a SNP located between exon 3 and 4, rs29012855. AF017143 has been shown to be positively associated with growth traits in Angus cattle (Ge et al., 2001). More recently, Islam et al. (2009), genotyped two purebred cattle populations including 204 Angus and 186 Charolais steers and a hybrid population of 455 cattle for AF017143 and rs29012855. Both SNPs associated with back fat thickness, average back fat, and lean meat yield in the Angus population although only rs29012855 associated with slaughter and carcass weight in the Angus steers and rib eye area in the Charolais cattle (Islam et al., 2009).
The objective of the present study was to identify novel SNPs in IGF-1 and together with AF017143 and rs29012855 determine their association with a number of performance traits in Holstein-Friesian cattle.
Materials and Methods
IGF-1 SNP Discovery
DNA extraction
Blood samples were obtained from 22 cattle of differing breeds including four Belgian Blue – Holstein-Friesian crossbreds, four Aberdeen Angus – Holstein-Friesian crossbreds as well as four, four and six purebred Charolais, Simmental and Holstein-Friesian cattle respectively. DNA was extracted from blood using a proteinase K/salting out/ethanol precipitation extraction method (adapted from Montgomery and Sise, 1990). DNA was also extracted from thawed–frozen semen from Holstein-Friesian AI bulls (n = 914) using a Maxwell® 16 instrument and Maxwell® 16 Tissue DNA Purification Kits from Promega (Southampton, UK). Prior to DNA extraction washed semen was incubated overnight in lysis buffer (Heyen et al., 1997). Following extraction, the quality and quantity of DNA derived from both blood and semen were assessed using a Nanodrop® spectrophotometer (Thermo Scientific, USA) and agarose gel electrophoresis.
Re-sequencing and SNP discovery
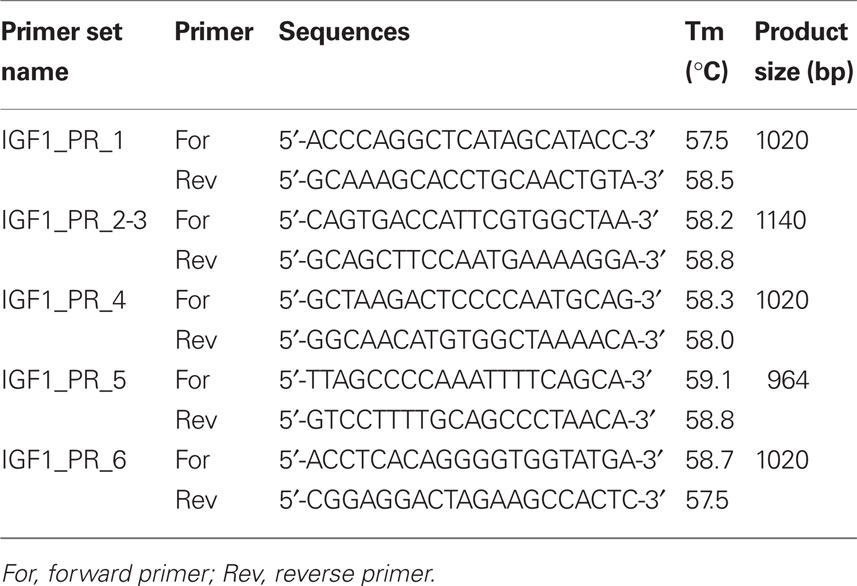
Putative SNPs in IGF-1 (including 5 kb upstream and downstream of the gene to encompass promoter and regulatory regions) were identified using the ENSEMBL database1 and the Btau_4.0 (October, 2007) assembly (accession no. ENSBTAG00000011082). Bovine specific PCR primers were designed using the Primer3 program (Rozen and Skaletsky, 2000) and the NCBI primer BLAST tool2 to amplify approximately 1000 bp fragments of the gene flanking the SNPs identified in ENSEMBL (Table 1). Cycling conditions were as follows: initial denaturation at 94°C for 4 min followed by 30 cycles 94°C for 30 s, 56°C for 30 s, 72°C for 2 min, and a final extension step of 72°C for 5 min. Each PCR reaction was carried out in 50 μl final volume and contained 20 ng genomic DNA, 1 μM primers, 2 mM MgCl2, 1 U Platinum® Taq polymerase, and 1× PCR buffer (Invitrogen Life Sciences, Dublin, Ireland). PCR products were purified and sequenced commercially by Eurofins MWG Operon (Martinsried, Germany). Sequence data were manually checked for quality including background noise, peak intensity, and accuracy via the chromatograms using Chromas Lite v2.013 and analyzed using the BLAST tool from NCBI to confirm their identity and position on bovine chromosome 54.
Sequence alignments and identification of SNPs including comparison with the reference Ensembl sequence and published literature were performed using Clustal W (Larkin et al., 2007) and Chromas Lite v2.01 programs5. This allowed validation of published SNPs and the identification of putative novel SNPs within and flanking IGF-1.
Association Study
Genotyping
All SNPs identified during the re-sequencing process (n = 10) alongside additional published SNPs (n = 6; AF017143, rs43434843, rs43434842, rs43434841, rs43434839, and rs43434840) were genotyped across 914 Holstein-Friesian sires. The SNP genotyping was carried out commercially using the Sequenom MassArray® iPLEX Gold assay (Sequenom, San Diego, CA, USA). As a quality control measure, 25 animals, originating from 25 different samples of extracted genomic DNA were genotyped twice for all SNPs. Mean concordance across all IGF-1 SNPs and all duplicates was 99.8%. Where discordance existed, the SNP genotype for the sample in question was discarded.
Data editing and phenotypes
An iterative algorithm was used to simultaneously discard both SNPs and individuals with poor genotype call rates (Waters et al., 2011). This resulted in genotypes of 66 individuals being discarded with all IGF-1 SNPs remaining in the study. A measure of linkage disequilibrium (r2; Hill and Robertson, 1968) was calculated between each pair-wise combination of the segregating SNPs (Emigh, 1980).
Daughter yield deviations (DYD; expressed on the scale of predicted transmitting ability; PTA) and PTAs, as well as associated data reliabilities, for a range of performance traits evaluated by the Irish Cattle Breeding Federation in the January 2009 domestic genetic evaluations were available for inclusion in the analysis. Models used in genetic evaluations in Ireland, as well as variance components, are summarized in detail by Berry et al. (2007). DYD for 305 day milk, fat, and protein yield as well as geometric mean somatic cell score (SCS; loge somatic cell count) are estimated in Ireland using a repeatability animal model across the first five lactations. PTA for calving interval and survival are estimated using a multi-trait animal model, including data from the first three lactations. PTA for milk yield are used to adjust survival for differences in genetic merit of milk yield; hence, this survival trait is functional survival. PTA for cow carcass weight, progeny carcass weight, progeny carcass fat score, and progeny carcass conformation score, measured at slaughter, are estimated in a multi-trait animal model that includes weaning weight, live-weight of the animal between 300 and 600 days of age, feed intake, and skeletal and muscular linear traits. Cows slaughtered between 875 and 4000 days of age were included in the evaluation of cow carcass weight while male progeny slaughtered between 300 and 1200 days of age and female progeny slaughtered between 300 and 875 days of age were included in the evaluation of the remaining three carcass traits. Genetic evaluations for linear type traits are undertaken as part of a joint evaluation in the UK and Ireland. The estimated breeding values (EBVs) were standardized to the mean and standard deviation of the base population. PTAs were de-regressed using the procedure outlined by Berry et al. (2009):
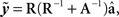
Where 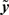 is the de-regressed PTA,
is the de-regressed PTA, 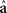 is a avector of PTAs from the genetic evaluations, R is a diagonal matrix where each element is
is a avector of PTAs from the genetic evaluations, R is a diagonal matrix where each element is 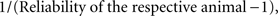 and A is the numerator relationship matrix.
and A is the numerator relationship matrix.
Parental contribution to the reliability of each DYD or PTA was removed using the approach of Harris and Johnson (1998):
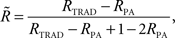
Where 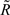 is the reliability less the parental contribution, RTRAD is the reliability from the traditional genetic evaluation (i.e., includes information from all relatives), RPA is parental average reliability.
is the reliability less the parental contribution, RTRAD is the reliability from the traditional genetic evaluation (i.e., includes information from all relatives), RPA is parental average reliability.
Only sires with a reliability (less parental contribution) of >60% for the trait under investigation were retained for inclusion in the association analysis. A total of 742 sires fulfilled these criteria for inclusion in the analysis of milk, fat, and protein yield as well as milk fat and protein concentration; the number of sires included in the association analysis with SCS, calving interval, and survival was 703, 501, and, 477, respectively. The number of sires with a reliability of >60% for the carcass traits was 446 and the number of sires with a reliability of >60% for the linear type traits varied from 484 to 551.
Statistical analysis
The association between each SNP and performance was quantified using weighted mixed models in ASREML (Gilmour et al., 2009) with genotyped individual included as a random effect and average expected relationships amongst individuals accounted for through the numerator relationship matrix. Year of birth (divided into five yearly intervals) and percent Holstein of the individual sire were included as fixed effects in the model. In all instances the dependent variable was DYD for milk, yield, fat yield, protein yield, and SCS and de-regressed PTA for the remaining traits, weighted by their respective reliability less the parental contribution.
Genotype was included in the analysis as a continuous variable coded as the number of copies of a given allele. Bonferroni correction of the significance levels was undertaken to account for multiple testing across all SNPs by all traits. Additionally, a multiple regression model was progressively built for each trait where at least one SNP was associated (P < 0.05) with the trait in the univariate analyses.
Transcription Factor and microRNA Binding Site Analysis
Bioinformatic analysis was performed on SNPs in the promoter regions of the IGF-1 gene to examine the effects of allele substitution on predicted transcription factor binding sites using MatInspector software package (Quandt et al., 1995) and microRNA binding sites using MicroInspector software (Rusinov et al., 2005).
Results
Re-Sequencing and SNP Discovery
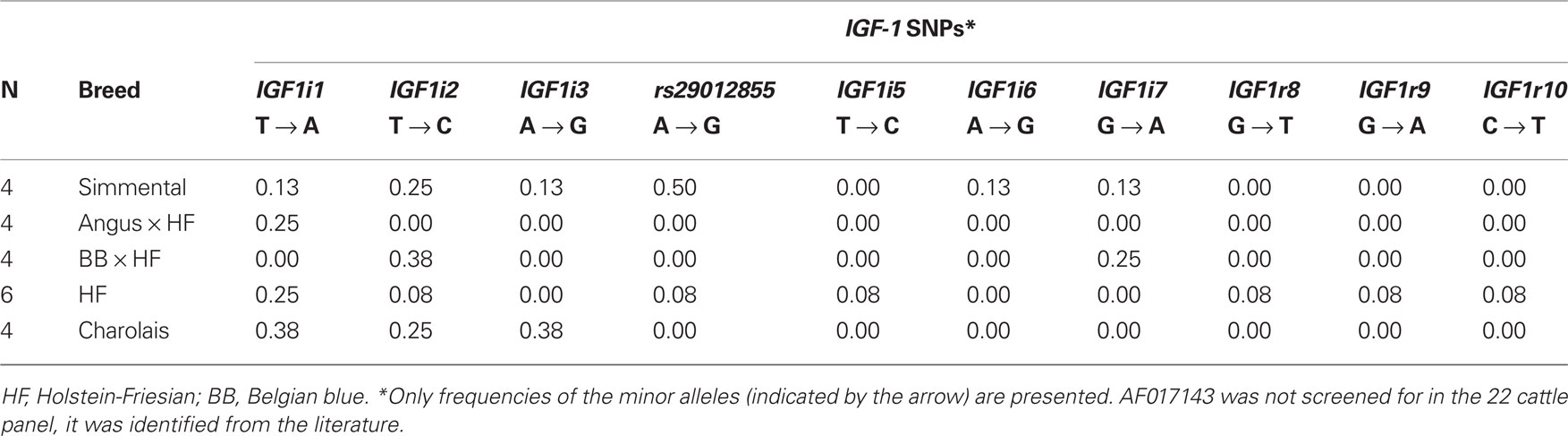
Ten SNPs were identified via sequencing in the 22 cattle, including nine which were putatively novel and one SNP which was previously published, rs29012855. The frequency of these 10 SNPs in the 22 cattle SNP discovery panel is shown in Table 2. In addition to these detected SNPs, one SNP, AF017143 previously discovered 512 bp upstream from the IGF-1 start codon (Ge et al., 1997, 2001) and the five published SNPs originally identified in Ensembl were also included in the genotyping process.
The seven previously published SNPs were all located within introns of the gene, with the exception of AF017143 and rs43434840. Of the nine putative novel SNPs, three (IGF1r8, IGF1r9, and IGF1r10) were identified <2 kb downstream of IGF-1, while six SNPs (IGF1i1, IGF1i2, IGF1i3, IGF1i5, IGF1i6, IGF1i7) were all located within introns of IGF-1. To the knowledge of the authors, the nine putative novel SNPs discovered in the current study have not been described previously in the literature or in dbSNP and have been submitted to dbSNP6.
Association Study
Summary statistics
Of the total 16 SNPs examined, five previously identified SNPs (rs43434843, rs43434842, rs43434841, rs43434839, and rs43434840) were monomorphic in this population of Holstein-Friesian sires and were therefore excluded from the association analyses. Details of the 11 segregating SNPs including location on BTA5 relative to IGF-1 is illustrated in Figure 1, while the nomenclature, flanking sequences including summary statistics of allele frequencies are described in Table 3. The minor allele frequency (MAF) for the 11 segregating SNPs ranged from 0.03 (IGF1i7 and IGF1r9) to 0.43 (AF017143). Eight of the 11 SNPs (IGF1i3, rs29012855, IGF1i5 – r10) had MAF between 0.03 and 0.05 with minor alleles present in the heterozygous genotype only for IGF1r8 and IGF1r10 and heterozygote frequencies between 5 and 10%. The remaining three SNPs (AF017143, IGF1i1, and IGF1i2) had MAF between 0.32 and 0.43 and heterozygote frequencies of 46–47%. None of the SNPs deviated from Hardy–Weinberg equilibrium.
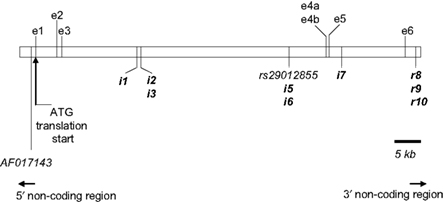
Figure 1. Annotation of IGF-1 (white box) on BTA5 as per build 4. 0 of the Bos taurus genome sequence. The physical location of each of the 11 segregating SNPs used in this study is indicated in relation to contig BTA5. The putative nine novel SNPs are highlighted in bold (with the IGF1 prefix omitted for clarity). The two previously reported SNPs are listed as per their dbSNP accession number (http://www.ncbi.nlm.nih.gov/projects/SNP) or GenBank reference (http://www.ncbi.nlm.nih.gov/genbank/). Three of the reported alternatively spliced IGF-1 transcripts [ENSBTAT00000014713 (IGF-1); ENSBTAT00000057516 (Q27962_BOVIN) and ENSBTAT00000043595 (Q7YS34_BOVIN)] are shown here. Exon 4a and 4b share overlapping sequences. The IGF-1 transcript is transcribed by exons 1, 3, 4b, and 6; Q27962_BOVIN is transcribed from exons 4a and 5 and Q7YS34_BOVIN is transcribed from exons 2, 3, 4a, and 6.
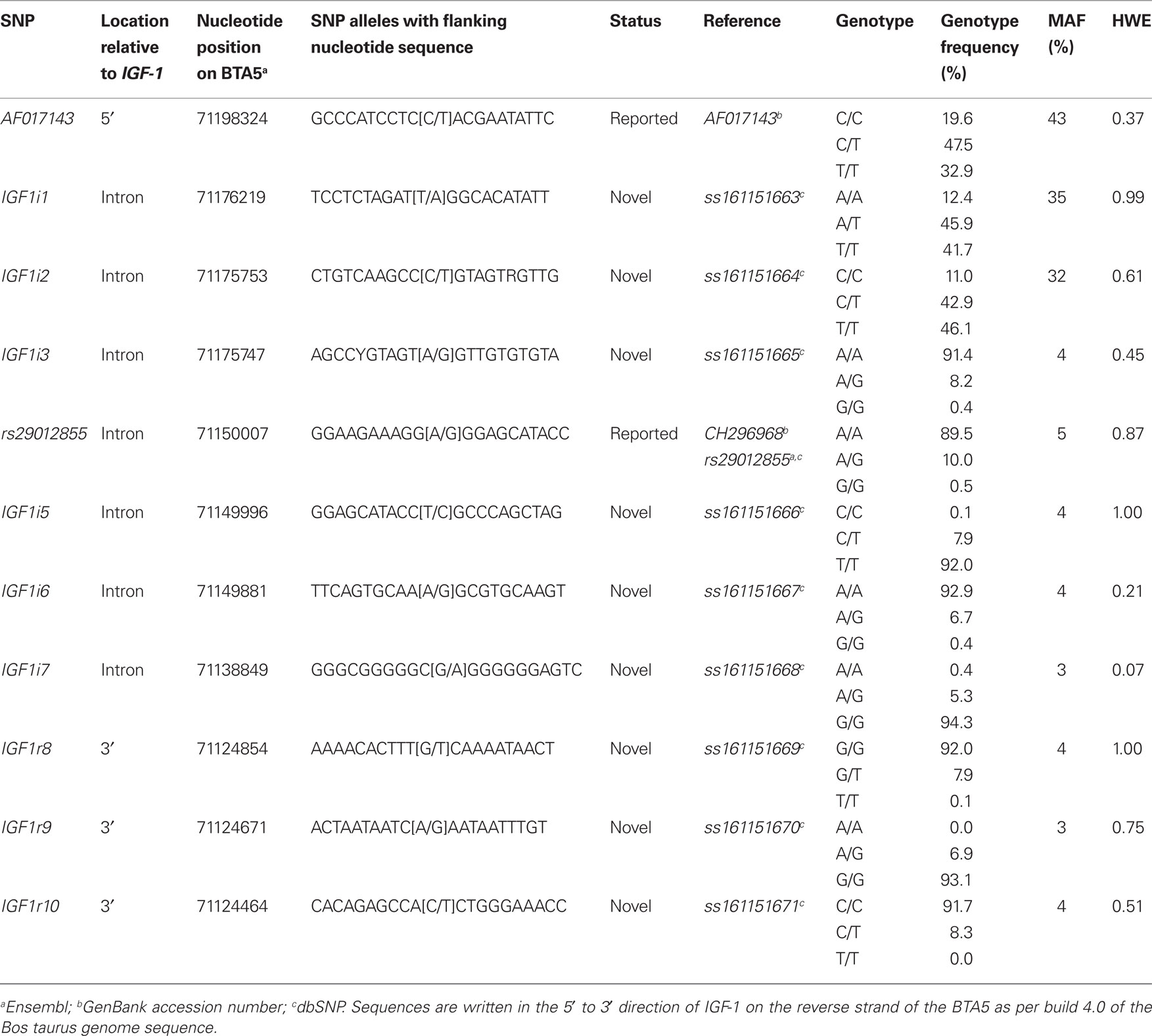
Table 3. Location, nomenclature, genotype frequency, minor allele frequency (MAF), and significance of deviation from Hardy–Weinberg equilibrium (HWE) for the 11 segregating SNPs in the 848 Holstein-Friesian sires.
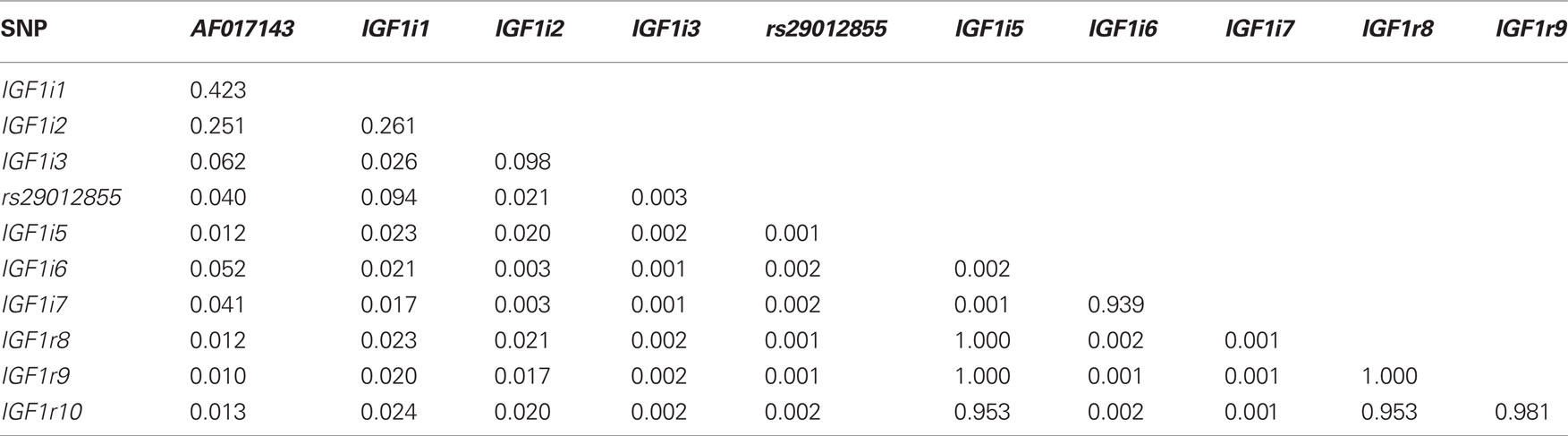
Table 4 summarizes the extent of linkage disequilibrium among the 11 segregating SNPs in the Holstein-Friesian sample population. Strong LD existed between IGF1r8, IGF1r9, and IGF1r10, all of which were within 390 base pairs. IGF1i5, IGF1r8, and IGF1r9 were also in complete LD and as expected IGF1i5 was in strong LD with IGF1r10. With the exception of IGF1i6 and IGF1i7, the LD between all remaining SNPs was weak. Since IGF1i5 and IGF1r9 were in complete LD with IGF1r8 and the latter SNP had the greatest call rate, IGF1i5 and IGF1r9 were omitted from further analysis.
Association analysis
The significant allelic substitution effects of the 11 SNPs on milk yield, milk composition, survival, carcass, and body size traits, when included individually in the model, are listed in Table 5. However following adjustment for multiple testing no significant association remained. Significant associations, when estimated using a multiple regression model, are presented in Table 6 and only these are discussed herein and the significance levels are nominal values and not adjusted for multiple testing. Due to the relatively low LD and low MAF for some of the SNPs haplotype analysis did not provide any further insight into the associations and are therefore not presented.
Table 5. Allelic substitution effect (standard error in parenthesis) between the 11 segregating SNPs and milk production, somatic cell score (SCS), survival, carcass, and body size traits when included individually in the statistical model.
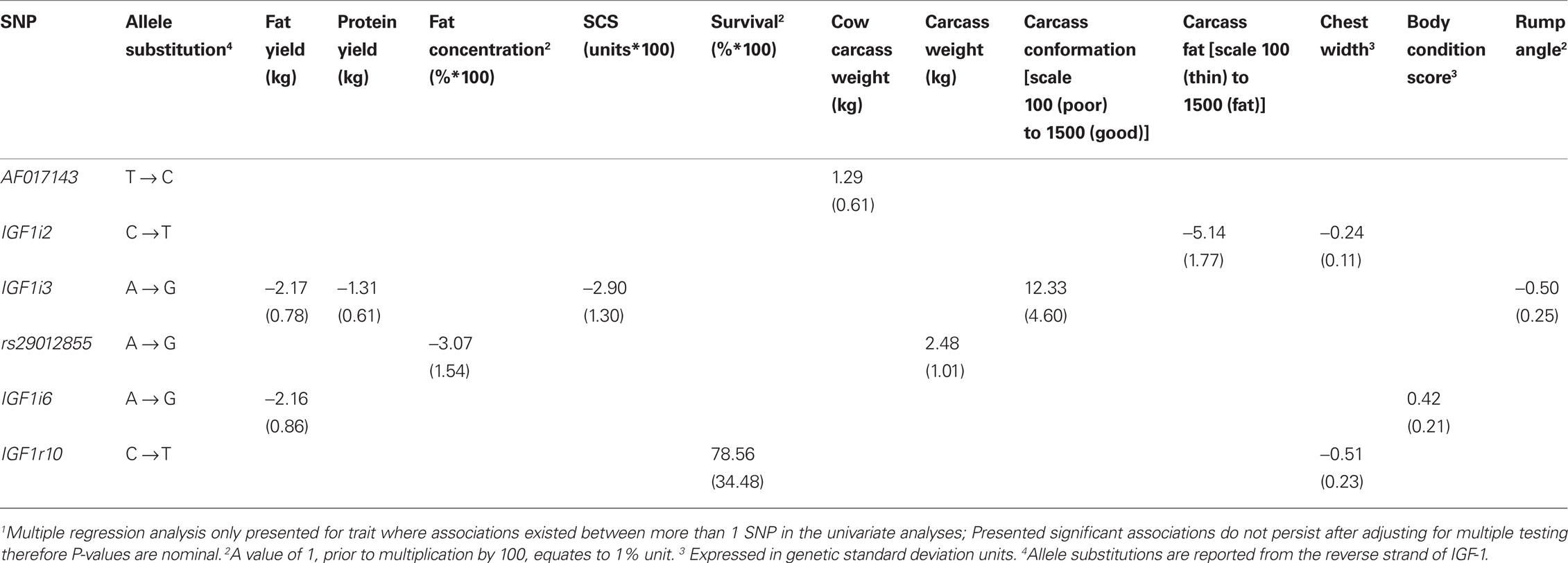
Table 6 Associations (P < 0.05) between IGF-1 SNPs and milk composition, survival, body condition score, and carcass traits when estimated using multiple regression models1.
Milk production, somatic cell count, calving interval, and survival
No significant association between any SNPs and either milk yield, milk protein concentration, or calving interval was evident. The G allele of rs29012855 was associated (P < 0.05) with reduced milk fat percentage. Similarly, the G allele of IGF1i6 and the G allele of IGF1i3 were associated independently (P < 0.05) with reduced fat yields. Also, IGFi3 was the only SNP associated with protein yield with the G allele associated with decreased yield. Only one SNP, the G allele of IGF1i3 was associated (P < 0.05) with decreased somatic cell count. The T alleles in both IGF1r8 and IGF1r10 SNPs associated (P < 0.05) with improved survival. However, when either IGF1r8 or IGF1r10 were included in the model, the association between survival and IGF1r8 or IGF1r10 was no longer significant (Table 6), attributable to the strong LD between both SNPs.
Carcass and body size related traits
In the multiple regression analysis the G allele of rs29012855 was associated (P < 0.05) with decreased carcass weight while the G allele of AF017143 was associated (P < 0.05) with increased cow carcass weight. IGF1i3 was associated with carcass conformation; the G allele associated with improved conformation and reduced rump angle although no homozygous GG animals were present in this sample population. IGF1i2 (T allele) was associated (P < 0.05) with decreased carcass fat.
No SNP was significantly associated (P > 0.05) with stature, angularity, or rump width (data not shown). However, IGF1i2 (T allele) and IGF1r10 (T allele) were all negatively associated (P < 0.05) with chest width. IGF1i6 (G allele) was positively associated (P < 0.05) with body condition score.
Discussion
There is a dearth of information available on polymorphisms in bovine IGF-1 and their effects on economically important traits in cattle. The present study describes the identification of previously unreported SNPs in the intronic and in the 3′ non-coding regions of IGF-1 which associated with milk composition, somatic cell count, survival, cow carcass weight, carcass fat, body condition score, and body size. Following adjustment for multiple testing no SNP remained significantly associated with performance. However, identification of patterns/clusters of associations between adjacent SNPs, as well as results from multiple regression analysis, can provide useful information into regions of the genes that may be associated with performance.
Although all 11 SNPs were in Hardy–Weinberg equilibrium, eight were segregating with low MAFs of ≤5% and two SNPs were present in the heterozygote form only. Other studies from our group have shown almost identical MAF for 10 of these SNPs (excluding AF017143) in a separate population of 610 commercial Holstein-Friesian dairy cows (Lynch et al., 2010). This provides supporting evidence of their segregation at low MAF within Holstein-Friesian cattle. Low frequency variants have being speculated to contribute to the missing heritability described from large scale genome wide association studies of complex traits in many species including humans (Maher, 2008; Manolio et al., 2009). Although substantially larger sample sizes are required to establish with confidence the effects of low frequency or rare variants on complex traits, the SNPs identified herein may represent such variants. This study therefore highlights the benefits of re-sequencing in addition to SNP chip based genome wide association analyses to facilitate detection of uncommon or rare variants affecting complex phenotypes.
The C allele of AF017143 has previously been documented to be positively associated with higher IGF-1 gene and protein expression levels in Polish Holstein-Friesian cattle (Maj et al., 2008). Ge et al. (2001) also reported a positive association between the C allele and post-weaning weight gain, ultrasound back fat thickness, and average carcass back fat. However, Islam et al. (2009) reported a negative association with meat yield in Angus beef cattle. In our study we failed to observe any association between the AF017143 SNP in Holstein-Friesian sires and carcass fat scores in their progeny. The inconsistency between studies may be a result of breed differences with Angus beef cattle maturing earlier and having greater carcass fat at slaughter compared with Holstein-Friesian cattle. It may also be due to different LD patterns within breeds which may account for the absence of an association of this SNP in the Charolais cattle analyzed by Islam et al. (2009). However, in the present study, the C allele of AF017143 was associated with heavier cow carcasses but not associated with progeny carcass weight, suggesting that AF017143 is associated with mature cow size rather than size at a younger age. Siadkowska et al. (2006) reported AF017143 positively associated with live body weight at slaughter and cold carcass weight in a population of 131 fifteen-month-old Holstein-Friesian bulls. These observations are consistent with the role of the somatotropic axis through the action of IGF-1. It plays a key role in the regulation of the metabolism and physiology of mammalian growth and is the main regulator of post-natal somatic growth, stimulating anabolic processes such as cell division, skeletal growth, and protein synthesis (Curi et al., 2005).
Potential mechanisms for an association between AF017143 and carcass weight may lie in its location in the promoter region of IGF-1 and the possible influence on transcriptional factor binding sites (TFBS). Analysis of TFBS in silico predicted that the C allele of AF017143 introduced two new TFBS that are abrogated by the T allele (data not shown). One of which is for Heat Shock Factor 1 (HSF1), a known transcriptional repressor (Xie et al., 2002) which may act to repress IGF-1 expression. Similarly the C allele introduces a predicted TFBS for Zinc finger protein 217 from the zinc finger family of transcription factors. Although less is known about ZNF217, research in other species suggests that it too acts as a transcriptional repressor (Banck et al., 2009).
It is well established that gene transcription is extensively and co-ordinately regulated. While introns are known to carry regulatory sequences, they may not have a direct involvement in the regulation of transcription of highly expressed genes. Systematic differences in motif distributions do suggest that introns play a role in the rate of transcription (Zhang et al., 2008). Of the seven SNPs located within introns of the IGF-1 gene, six including five novel (IGF1i2, IGF1i3, IGF1i5, IGF1i6, IGF1i7) and one previously reported (rs29012855) were associated with at least two performance traits, emphasizing the integral role of IGF-1 in milk production, growth, and development in cattle.
In the current study, IGF1i2 associated with carcass fat and chest width, while other studies from this group (Lynch et al., 2010) reported associations between IGF1i2 and body condition score at calving in a cohort of 241 dairy cows. Body condition score in the current study was associated with IGF1i6 (despite its low MAF). Both IGF1i2 and IGF1i6 are located between exons 3 and 4 of the IGF-1 gene and it is possible a causative polymorphism affecting body condition score is in LD with this region of IGF-1 exhibiting different LD patterns between the cattle populations. Indeed, previous studies have described a relationship between systemic IGF-1 and carcass fat; Davis and Simmen (2000) reported that Angus bulls with lower plasma IGF-1 concentrations had higher marbling scores and back fat thickness. Similarly, circulating IGF-1 was found to be negatively correlated with carcass fat percentage, fat accretion rate, and fat thickness in Simmental crossbred bulls (Anderson et al., 1988). Interestingly, the stage of differentiation of precursor cells into mature fat cells is accompanied by enhanced expression of IGF-1 in transgenic mice indicating a role for IGF-1 in fat cell developmental processes (Rajkumar et al., 1999).
Systemic IGF-1 has been shown to be positively correlated with milk yield and milk fat concentrations (Moyes, 2004; Rose et al., 2005). However there are no reports in the literature of associations between IGF-1 SNPs and milk production traits. Furthermore none of the SNPs analyzed in this study displayed an association with milk yield. However the G alleles of two novel SNPs, IGF1i3, and IGF1i6, were associated with decreased milk fat and protein (IGFi3 only) yield. Consistent with this, IGF-1 is known to play an important role in mammary gland growth and function by regulating several cellular processes (Akers, 2006) including the stimulation of protein synthesis in the epithelial cells of the mammary gland (Burgos and Cant, 2010). Additionally, significant associations (IGF1i3) were observed with somatic cell count (SCC). Liebe and Schams (1998) reported that the concentration profile of IGF-1 in milk corresponded well with SCC and concluded that this was possibly an important measurement in monitoring the state of udder health. Ruffer (2003) also found that IGF-1 concentrations increased significantly in milk collected from healthy udder quarters that were adjacent to quarters showing SCC of more than 100,000/ml and mastitis.
To the knowledge of the authors, associations between rs29012855 and performance have only been previously reported by Islam (2009) in 455 hybrid, 206 Angus, and 186 Charolais beef cattle. Islam (2009) reported positive associations between the G allele and lean meat yield, slaughter, and carcass weight in the Angus steers. Islam (2009) also reported negative associations between the G allele and back fat thickness in the Angus steers and rib eye area in the Charolais cattle. This is in agreement with our findings where the G allele associated with heavier carcasses but also decreased milk fat percentage. This is the first reporting of rs29012855 being associated with milk production variables. A MAF of 1.9, 5.1, and 3.5% was observed for rs29012855 in the Angus, Charolais, and hybrid populations respectively, providing further evidence that rs29012855 is segregating at low MAF in cattle (Islam, 2009). The low MAF of rs29012855 and other SNPs in this study suggests a potential antagonistic relationship with other variants under positive selection. The minor alleles of IGF1i3, rs29012855, IGF1i6, and IGF1i7 were all significantly negatively associated with milk production variables and their low frequency in Holstein-Friesian cattle may be due to the intense selection for milk production over the last four decades in dairy cattle (Diskin et al., 2006).
The importance of mutations located in the 3′UTR was evident in a study by Clop et al. (2006), where a G to an A transition in the 3′UTR of the myostatin gene introduced illegitimate miRNA target sites resulting in muscular hypertrophy in Texel sheep. None of the three 3′UTR novel SNPs in this study (IGF1r8, IGF1r9, and IGF1r10) occur in any region of the IGF-1 where miRNA are predicted to bind. However these three mutations are in high LD, within 390 bp, and significantly associated with functional survival. Although none of the SNPs in this study associated with reproductive performance measured as calving interval, reproductive performance, particularly in grass based production systems, is a major determinant of cow survival (Diskin et al., 2006). Therefore this may indicate an indirect effect on calving interval perhaps through fitness per se.
Other studies from our group using this population of sires have identified SNPs in other candidate genes including other members of the somatotropic axis, i.e., Growth Hormone (Mullen et al., 2010) and Growth Hormone Receptor (Waters et al., 2011). The possible effects of these and IGF-1 variants on epistasis especially in established pathways such as the somatotropic axis is unknown. Although untested, the multiple associations within traits and pleiotropic effects observed with IGF-1 and the other genes analyzed in this population suggests independent effects. This may contribute to the multifactorial nature of complex phenotypes. Whether the effects observed herein are due to these IGF-1 SNPs directly, or with LD with causative variants, affecting systemic IGF-1 levels remains to be ascertained. Future work could include targeted enrichment encompassing entire genes and regulatory regions or whole genome sequencing approaches in an attempt to identify causative polymorphisms. If such studies were augmented with functional genomic analyses and validated in independent populations, greater insight into the genetics underpinning performance would be gained.
Conclusion
The effects of IGF-1 on mammalian post-natal growth and developmental processes including metabolism and nutrient partitioning are well established. To what degree polymorphisms in IGF-1 contribute to these effects is unknown. However this study indicates, polymorphisms including low frequency variants located within IGF-1 or nearby regions of the genome are affecting performance in cattle.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge the Irish Cattle Breeding Federation (ICBF) for access to estimates of sire merit. Research funding was provided by the Irish Dairy Research Trust and the Research Stimulus Fund (RSF-06-0353; RSF-06-0409). Michael Paul Mullen’s contribution was funded by a Science Foundation Ireland grant (07/SRC/B1156).
Footnotes
- ^http://www.ensembl.org accessed April 2008.
- ^http://www.ncbi.nlm.nih.gov/tools/primer-blast/
- ^http://www.technelysium.com.au/chromas_lite.htm, manual inspection of sequence data, accessed 14th October 2008.
- ^http://www.ncbi.nlm.nih.gov/genome/seq/BlastGen/BlastGen.cgi?taxid = 9913, confirmation of sequence data with bovine database, accessed 14th October 2008.
- ^http://www.technelysium.com.au/chromas_lite.htm, manual inspection of sequence data, accessed 14th October 2008.
- ^http://www.ncbi.nlm.nih.gov/projects/SNP, registered SNP data with dbSNP 11th September 2009.
References
Akers, R. M. (2006). Major advances associated with hormone and growth factor regulation of mammary growth and lactation in dairy cows. J. Dairy Sci. 89, 1222–1234.
Allan, M. F., Kuehn, L. A., Cushman, R. A., Snelling, W. M., Echternkamp, S. E., and Thallman, R. M. (2009). Confirmation of quantitative trait loci using a low-density single nucleotide polymorphism map for twinning and ovulation rate on bovine chromosome 5. J. Anim. Sci. 87, 46–56.
Anderson, P. T., Bergen, W. G., Merkel, R. A., Enright, W. J., Zinn, S. A., Refsal, K. R., and Hawkins, D. R. (1988). The relationship between composition of gain and circulating hormones in growing beef bulls fed three dietary crude protein levels. J. Anim. Sci. 66, 3059–3067.
Banck, M. S., Li, S., Nishio, H., Wang, C., Beutler, A. S., and Walsh, M. J. (2009). The ZNF217 oncogene is a candidate organizer of repressive histone modifiers Epigenetics 4, 100–106.
Berry, D., Kearney, F., and Harris, B. (2009). “Genomic selection in Ireland,” in Proceedings of the Interbull International workshop, Uppsala.
Berry, D. P., Shalloo, L., Cromie, A. R., Veerkamp, R. F., Dillion, P., Amer P. R., Kearney, J. F., Evans, R. D., and Wickham, B. (2007). The Economic Breeding Index: A Generation On. Technical report to the Irish Cattle Breeding Federation, 29–34.
Breier, B. H. (1999). Regulation of protein and energy metabolism by the somatotropic axis. Domest. Anim. Endocrinol. 17, 209–218.
Burgos, S. A., and Cant, J. P. (2010). IGF-1 stimulates protein synthesis by enhanced signaling through mTORC1 in bovine mammary epithelial cells. Domest. Anim. Endocrinol. 38, 211–221.
Clop, A., Marcq, F., Takeda, H., Pirottin, D., Tordoir, X., Bibe, B., Bouix, J., Caiment, F., Elsen, J. M., Eychenne, F., Larzul, C., Laville, E., Meish, F., Milenkovic, D., Tobin, J., Charlier, C., and Georges, M. (2006). A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat. Genet. 38, 813–818.
Curi, R. A., Oliveira, H. N., Silveira, A. C., and Lopes, C. R. (2005). Effects of polymorphic microsatellites in the regulatory region of IGF1 and GHR on growth and carcass traits in beef cattle. Anim. Genet. 36, 58–62.
Daughaday, W. H., and Rotwein, P. (1989). Insulin-like growth factors I and II. Peptide, messenger ribonucleic acid and gene structures, serum, and tissue concentrations. Endocr. Rev. 10, 68–91.
Davis, M. E., Bishop, M. D., Park, N. H., and Simmen, R. C. (1995). Divergent selection for blood serum insulin-like growth factor I concentration in beef cattle. J. Anim. Sci. 73, 1927–1932.
Davis, M. E., and Simmen, R. C. (1997). Genetic parameter estimates for serum insulin-like growth factor I concentration and performance traits in Angus beef cattle J. Anim. Sci. 75, 317–324.
Davis, M. E., and Simmen, R. C. (2000). Genetic parameter estimates for serum insulin-like growth factor-I concentration and carcass traits in Angus beef cattle. J. Anim. Sci. 78, 2305–2313.
Diskin, M. G., Murphy, J. J., and Sreenan, J. M. (2006). Embryo survival in dairy cows managed under pastoral conditions. Anim. Reprod. Sci. 96, 297–311.
Echternkamp, S. E., Roberts, A. J., Lunstra, D. D., Wise, T., and Spicer, L. J. (2004). Ovarian follicular development in cattle selected for twin ovulations and births. J. Anim. Sci. 82, 459–471.
Etherton, T. D. (2004). Somatotropic function: the somatomedin hypothesis revisited. J. Anim. Sci. 82, E239–E244.
Ge, W., Davis, M. E., and Hines, H. C. (1997). Two SSCP alleles detected in the 5’-flanking region of bovine IGF1 gene. Anim. Genet. 28, 155–156.
Ge, W., Davis, M. E., Hines, H. C., Irvin, K. M., and Simmen, R. C. (2001). Association of a genetic marker with blood serum insulin-like growth factor-I concentration and growth traits in Angus cattle. J. Anim. Sci. 79, 1757–1762.
Gilmour, A. R., Cullis, B. R., Welham, S. J., and Thompson, R. (2009). ASREML Reference Manual. Orange, NSW: New South Wales Agriculture, Orange Agricultural Institute.
Harris, B., and Johnson, D. (1998). Approximate reliability of genetic evaluations under an animal model. J. Dairy Sci. 81, 2723–2728.
Heyen, D. W., Beever, J. E., Da, Y., Evert, R. E., Green, C., Bates, S. E. R., Ziegle, J. S., and Lewin, H. A. (1997). Exclusion probabilities of 22 bovine microsatellite markers in fluorescent multiplexes for semiautomated parentage testing. Anim. Genet. 28, 21–27.
Hill, W. G., and Robertson, A. (1968). Linkage disequilibrium in finite populations. Theor. Appl. Genet. 38, 226–231.
Hossner, K. L., McCusker, R. H., and Dodson, M. V. (1997). Insulin-like growth factors and their binding proteins in domestic animals. Anim. Sci. 64, 1–15.
Islam, K. K. (2009). Association Analyses of SNPs in Candidate Genes with Body Fat Deposition and Carcass Merit Traits in Beef Cattle. M.Sc., University of Alberta, Edmonton, AB.
Islam, K. K., Vinsky, M., Crews, R. E., Okine, E., Moore, S. S., Crews, D. H., and Li, C. (2009). Association analyses of a SNP in the promoter of IGF1 with fat deposition and carcass merit traits in hybrid, Angus and Charolais beef cattle. Anim. Genet. 40, 766–769.
Johnston, D. J., Herd, R. M., Reverter, A., and Oddy, V. H. (2001). Heritability of IGF-I in beef cattle and its association with growth and carcase traits. Proc. Assoc. Adv. Anim. Breed. Genet. 14, 163–166.
Kim, E. S., Shi, X., Cobanoglu, O., Weigel, K., Berger, P. J., and Kirkpatrick, B. W. (2009). Refined mapping of twinning-rate quantitative trait loci on bovine chromosome 5 and analysis of insulin-like growth factor-1 as a positional candidate gene. J. Anim. Sci. 87, 835–843.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948.
Liebe, A., and Schams, D. (1998). Growth factors in milk: interrelationships with somatic cell count. J. Dairy Res. 65, 93–100.
Lucy, M. C. (2008). Functional differences in the growth hormone and insulin-like growth factor axis in cattle and pigs: implications for post-partum nutrition and reproduction. Reprod. Domest. Anim. 43(Suppl. 2), 31–39.
Lynch, C. O., Mullen, M. P., Waters, S. M., Howard, D. J., O’Boyle, P., Berry, D. P., Kenny, D. A., Buckley, F., Horan, B., and Diskin, M. G. (2010). “Single nucleotide polymorphisms in the growth hormone and insulin-like growth factor 1 genes are associated with production and fertility traits in dairy cows,” in BSAS Advances in Animal Biosciences (BSAS & IGAPA 12th-14th April 2010, Whitla Hall, Queen’s University, Belfast, Northern Ireland), (Belfast: Cambridge University Press), 314.
Maj, A., Snochowski, M., Siadkowska, E., Rowinska, B., Lisowski, P., Robakowska-Hyzorek, D., Oprzadek, J., Grochowska, R., Kochman, K., and Zwierzchowski, L. (2008). Polymorphism in genes of growth hormone receptor (GHR) and insulin-like growth factor-1 (IGF1) and its association with both the IGF1 expression in liver and its level in blood in Polish Holstein-Friesian cattle. Neuro Endocrinol. Lett. 29, 981–989.
Manolio, T. A., Collins, F. S., Cox, N. J., Goldstein, D. B., Hindorff, L. A., Hunter, D. J., McCarthy, M. I., Ramos, E. M., Cardon, L. R., Chakravarti, A., Cho, J. H., Guttmacher, A. E., Kong, A., Kruglyak, L., Mardis, E., Rotimi, C. N., Slatkin, M., Valle, D., Whittemore, A. S., Boehnke, M., Clark, A. G., Eichler, E. E., Gibson, G., Haines, J. L., Mackay, T. F. C., McCarroll, S. A., and Visscher, P. M. (2009). Finding the missing heritability of complex diseases. Nature 461, 747–753.
Miller, L. L., Schalch, D. S., and Draznin, B. (1981). Role of the liver in regulating somatomedin activity: effects of streptozotocin diabetes and starvation on the synthesis and release of insulin-like growth factor and its carrier protein by the isolated perfused rat liver. Endocrinology 108, 1265–1271.
Montgomery, G. W., and Sise, J. A. (1990). Extraction of DNA from sheep white blood cells. N. Z. J. Agric. Res. 33, 437–441.
Moyes, T. E. (2004). Variation in Concentrations of Insulin-like Growth Factor-1 (IGF-1) in Pasture-fed Holstein-Friesian Cows. Ph.D. thesis, University of Melbourne, Melbourne, Australia.
Mullen, M. P., Berry, D. P., Howard, D. J., Diskin, M. G., Lynch, C. O., Berkowicz, E. W., Magee, D. A., MacHugh, D. E., and Waters., S. M. (2010). Associations between novel SNPs in the Bos taurus growth hormone gene and performance traits in Holstein-Friesian dairy cattle. J. Dairy Sci. 93, 5959–5969.
Patton, J., Kenny, D. A., McNamara, S., Mee, J. F., O’Mara, F. P., Diskin, M. G., and Murphy, J. J. (2007). Relationships among milk production, energy balance, plasma analytes, and reproduction in Holstein-Friesian cows. J. Dairy Sci. 90, 649–658.
Quandt, K., Frech, K., Karas, H., Wingender, E., and Werner, T. (1995). MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23, 4878–4884.
Rajkumar, K., Modric, T., and Murphy, L. J. (1999). Impaired adipogenesis in insulin-like growth factor binding protein-1 transgenic mice. J. Endocrinol. 162, 457–465.
Rose, M. T., Weekes, T. E. C., and Rowlinson, P. (2005). Correlation of blood and milk components with the milk yield response to bovine somatotropin in dairy cows. Domest. Anim. Endocrinol. 28, 296–307.
Rozen, S., and Skaletsky, H. J. (2000). “Primer3 on the WWW for general users and for biologist programmers,” in Bioinformatics Methods and Protocols: Methods in Molecular Biology, eds S. Misener and S. A. Krawetz (Totowa, NJ: Humana Press), 365–386.
Ruffer, U. (2003). “Influences on the concentrations of IGF-1 in bovine milk – a field study,” in Schriftenreihe des Instituts für Tierzucht und Tierhaltung der Christian-Albrechts-Universität zu Kiel, Universitätsbibliothek, Gießen, H. 139.
Rusinov, V., Baev, V., Minkov, I. N., and Tabler, M. (2005). MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res. 33, W696–W700.
Sahana, G., Guldbrandtsen, B., Bendixen, C., and Lund, M. S. (2010). Genome-wide association mapping for female fertility traits in Danish and Swedish Holstein cattle. Anim. Genet. 41, 579–588.
Schwander, J. C., Hauri, C., Zapf, J., and Froesch, E. R. (1983). Synthesis and secretion of insulin-like growth factor and its binding protein by the perfused rat liver: dependence on growth hormone status. Endocrinology 113, 297–305.
Siadkowska, E., Zwierzchowski, L., Oprza?dek, J., StrzaLkowska, N., Bagnicka, E., and Krzylewski, J. (2006). Effect of polymorphism in IGF-1 gene on production traits in Polish Holstein-Friesian cattle. Anim. Sci. Pap. Rep. 24, 225–237.
Thissen, J.-P., Ketelslegers, J.-M., and Underwood, L. E. (1994). Nutritional regulation of the insulin-like growth factors. Endocr. Rev. 15, 80–101.
Velazquez, M. (2008). Assisted reproductive technologies in cattle: applications in livestock production, biomedical research and conservation biology. Ann. Rev. Biomed. Sci. 10, 36–62.
Velazquez, M. A., Newman, M., Christie, M. F., Cripps, P. J., Crowe, M. A., Smith, R. F., and Dobson, H. (2005). The usefulness of a single measurement of insulin-like growth factor-1 as a predictor of embryo yield and pregnancy rates in a bovine MOET program. Theriogenology 64, 1977–1994.
Wang, Y., Price, S. E., and Jiang, H. (2003). Cloning and characterization of the bovine class 1 and class 2 insulin-like growth factor-I mRNAs. Domest. Anim. Endocrinol. 25, 315–328.
Waters, S. M., McCabe, M. S., Howard, D. J., Giblin, L., Magee, D. A., MacHugh, D. E., and Berry, D. P. (2011). Associations between newly discovered polymorphisms in the Bos taurus growth hormone receptor gene and performance traits in Holstein-Friesian dairy cattle. Anim. Genet. 42, 39–49.
Wathes, D. C., Bourne, N., Cheng, Z., Mann, G. E., Taylor, V. J., and Coffey, M. P. (2007). Multiple correlation analyses of metabolic and endocrine profiles with fertility in primiparous and multiparous cows. J. Dairy Sci. 90, 1310–1325.
Werner, H., Adamo, M., Roberts, C. T. Jr., Eroith, D. L., and Gerald, L. (1994) “Molecular and cellular aspects of insulin-like growth factor action,” in Vitamins and Hormones, Vol. 48, ed. G. Litwack (Elsevier, Oxford), 1–58.
Xie, Y., Chen, C., Stevenson, M. A., Auron, P. E., and Calderwood, S. K. (2002). Heat shock factor 1 represses transcription of the IL-1beta gene through physical interaction with the nuclear factor of interleukin 6. J. Biol. Chem. 277, 11802–11810.
Yilmaz, A., Davis, M. E., and Simmen, R. C. M. (2006). Analysis of female reproductive traits in Angus beef cattle divergently selected for blood serum insulin-like growth factor I concentration. Theriogenology 65, 1180–1190.
Keywords: single nucleotide polymorphism, performance, Holstein-Friesian dairy cattle
Citation: Mullen MP, Berry DP, Howard DJ, Diskin MG, Lynch CO, Giblin L, Kenny DA, Magee DA, Meade KG and Waters SM (2011) Single nucleotide polymorphisms in the insulin-like growth factor 1 (IGF-1) gene are associated with performance in Holstein-Friesian dairy cattle. Front. Gene. 2:3. doi: 10.3389/fgene.2011.00003
Received: 22 November 2010;
Accepted: 02 February 2011;
Published online: 16 February 2011.
Edited by:
Steve Bishop, University of Edinburgh, UKReviewed by:
Dirk-Jan De Koning, Swedish University of Agricultural Sciences, SwedenGeorgia Hadjipavlou, Agricultural Research Institute, Cyprus
Kathryn Kemper, University of Melbourne, Australia
Copyright: © 2011 Mullen, Berry, Howard, Diskin, Lynch, Giblin, Kenny, Magee, Meade and Waters. This is an open-access article subject to an exclusive license agreement between the authors and Frontiers Media SA, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Michael Paul Mullen, Animal and Bioscience Research Department, Animal and Grassland Research and Innovation Centre, Teagasc, Mellows Campus, Athenry, County Galway, Ireland. e-mail:bWljaGFlbC5tdWxsZW5AdGVhZ2FzYy5pZQ==


 Dawn J. Howard1
Dawn J. Howard1