
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Genome Ed. , 10 February 2025
Sec. Genome Editing in Plants
Volume 7 - 2025 | https://doi.org/10.3389/fgeed.2025.1537148
This article is part of the Research Topic Genome Editing for Plant Immunity Research: From Understanding Multidimensional Regulation of Immune Responses Towards Breeding Disease-Resistant Crop Plants Resilient to Climate Changes View all articles
Various pathogens severely threaten tomato yield and quality. Advances in understanding plant-pathogen interactions have revealed the intricate roles of resistance (R) and susceptibility (S) genes in determining plant immunity. While R genes provide targeted pathogen resistance, they are often vulnerable to pathogen evolution. Conversely, S genes offer a promising avenue for developing broad-spectrum and durable resistance through targeted gene editing. Recent breakthroughs in CRISPR/Cas-based technologies have revolutionized the manipulation of plant genomes, enabling precise modification of S genes to enhance disease resistance in tomato without compromising growth or quality. However, the utilization of the full potential of this technique is challenging due to the complex plant-pathogen interactions and current technological limitations. This review highlights key advances in using gene editing tools to dissect and engineer tomato S genes for improved immunity. We discuss how S genes influence pathogen entry, immune suppression, and nutrient acquisition, and how their targeted editing has conferred resistance to bacterial, fungal, and viral pathogens. Furthermore, we address the challenges associated with growth-defense trade-offs and propose strategies, such as hormonal pathway modulation and precise regulatory edits, to overcome these limitations. This review underscores the potential of CRISPR-based approaches to transform tomato breeding, paving the way for sustainable production of disease-resistant cultivars amidst escalating global food security challenges.
Tomato (Solanum lycopersicum L.) is one of the most economically important horticulture crops, representing one of the top produced and consumed vegetables worldwide. Tomatoes are popular as both fresh and processed vegetable that benefit human health with bioactive compounds like flavonoids, lycopene, and ascorbic acid (Borguini and Ferraz Da Silva Torres, 2009). In the context of rapid climate change, adverse environmental factors limit tomato growth and result in substantial yield loss and fruit quality deterioration (Bacelar et al., 2024). Along with these limiting factors, infectious diseases caused by a wide range of pathogens pose significant threats to crop yield and quality (Potnis et al., 2015; Zhang et al., 2022).
Co-evolution of plants and their surrounding pathogenic microorganisms has enabled plants to evolve a two-tiered immune system to combat pathogen infections (Jones and Dangl, 2006). The first line of plant immunity, known as pattern-triggered immunity (PTI), offers broad-spectrum defense response, which is triggered when plant cell surface receptors sense pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs). Many pathogens can secrete virulent effectors into host cells, facilitating infection by breaching the first line of defense. Thus, plants employed a second layer of defense, known as effector-triggered immunity (ETI) to counter pathogens. The ETI is a more specialized and robust defense that is triggered when plant intracellular immune receptors, mostly the nucleotide-binding/leucine-rich-repeat receptors (NB-LRRs or NLRs) class, detect and recognize pathogen effector proteins, often leading to hypersensitive response (HR), localized cell death and induction of systemic acquired resistance (Jones et al., 2024). PTI and ETI act synergistically and mutually potentiate each other to trigger a robust defense against pathogen (Ngou et al., 2021; Yuan et al., 2021). Numerous transcriptional factors and hormones highly regulate the interplay between PTI and ETI.
The competition of resistance (R) genes and susceptibility (S) genes during plant-pathogen interactions determines if the plant will resist or be affected by a disease during interactions with pathogens. R genes are broadly grouped into two classes: typical R genes, which include NLRs and membrane-localized receptor-like kinases or proteins (RLKs/RLPs); and atypical R genes, which possess diverse architectures and functions associated with transcriptional regulation, kinases, translocation of substrates and hormone signaling (Sun et al., 2024). Typical R genes in plants mainly recognize specific pathogenic proteins or effectors. Gaining disease resistance by incorporating typical R genes can be challenging because these genes constantly evolve under strong positive selection due to the rapid evolution of pathogenic effectors. In contrast, many atypical R genes exhibit broad-spectrum and durable resistance and thus can be promising candidates for the resistant crop breeding (Sun et al., 2024).
S genes are defined as any plant genes that allow compatibility with pathogens and facilitate infection, and they can be further categorized into three subclasses depending on diverse functions (van Schie and Takken, 2014). The first type of S genes helps pathogens enter the host plant by regulating cell wall structure, cuticle properties, and stomata opening. The second type suppresses the immune response of plants, especially through transcriptional and hormonal regulation. The third type allows pathogens to access nutrients and grow by controlling sugar transport and metabolite production (van Schie and Takken, 2014; Koseoglou et al., 2022).
Pesticides have been used worldwide to control plant disease for over half a century. However, their overuse presents a substantial risk to the environment, ecological stability, and human health (Kimm et al., 2017). Breeding disease-resistant crops using new breeding technologies including gene editing is an effective and sustainable strategy for plant protection (Manzoor et al., 2024). Natural genetic variation and artificially induced genetic diversity, combined with modern molecular and genetic tools, including deep sequencing technologies and gene editing, have accelerated the discovery of genomic regions or alleles associated with immunity in tomato. So far, dozens of genes related to plant immunity, including R genes and S genes in tomato have been mapped and cloned (Rothan et al., 2019), and the role of these genes in regulating disease resistance was characterized in CRISPR edited tomato plants (Table 1).
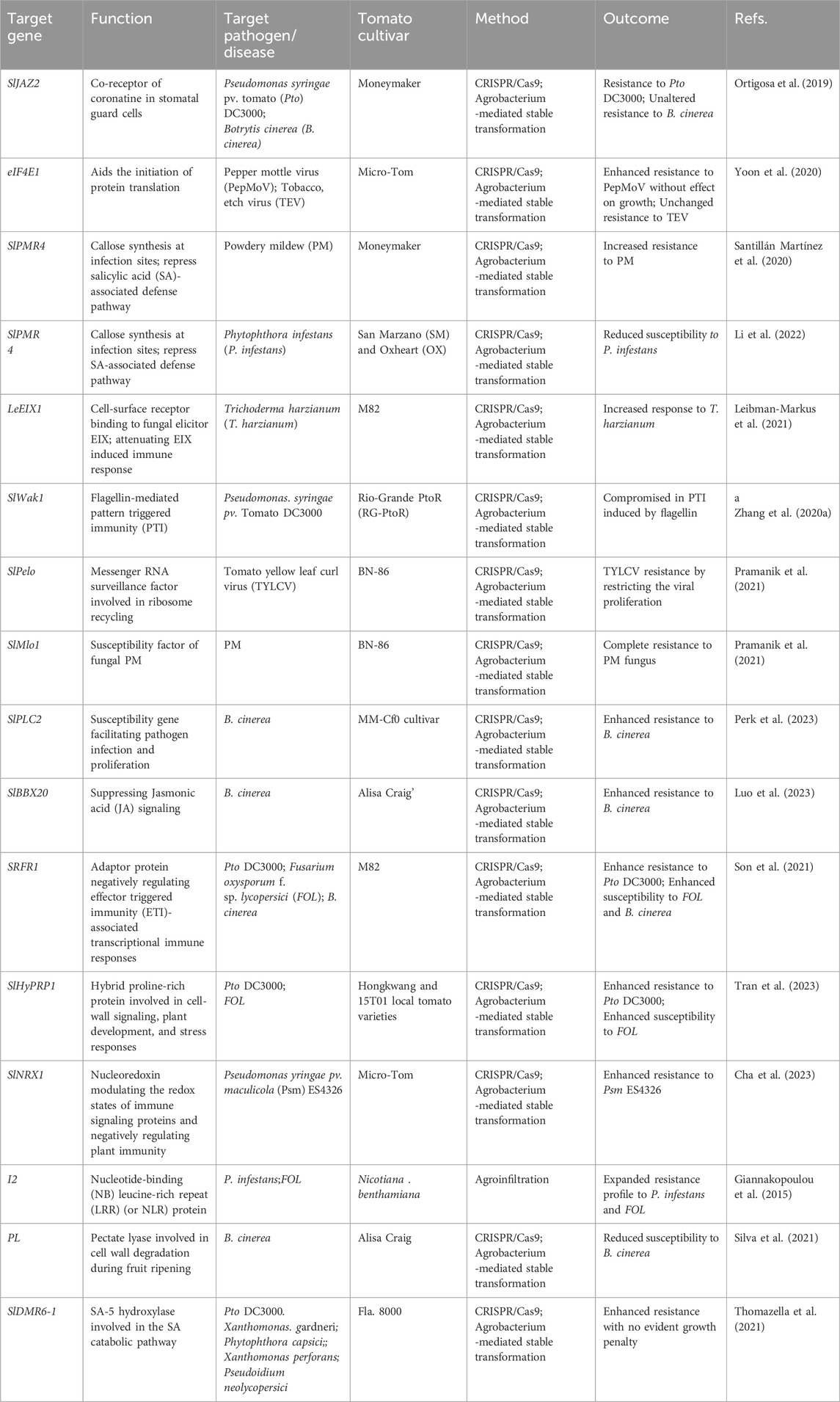
Table 1. Application of CRISPR/Cas-based gene editing tool for engineering resistance against pathogens in tomato plants.
Disease resistance can be ensured via the incorporation and introgression of desired R genes into crops through different breeding approaches, including conventional breeding and transgenic technology (Derbyshire et al., 2024). However, R gene associated resistance is mostly disrupted because their targeted effectors are generally under strong negative selection (Sacristán et al., 2021; Bent and Mackey, 2007). Additionally, the incorporation of R genes into host genome via conventional breeding can be time-consuming or results in genetically modified organisms (GMOs) by means of genetic engineering techniques, which will provoke public concern (Rozas et al., 2022). A better strategy is using mutated S genes for sustainable and broad-spectrum resistance. Various gene-editing technologies, particularly CRISPR/Cas-based gene editing tools, have opened up new avenues to precisely engineer S genes for resistance breeding. CRISPR/Cas9 and its derivatives have been developed for a wide variety of applications, including gene knockout, gene knock-in, gene regulation, and epigenetic editing (Altpeter et al., 2016; Čermák et al., 2015; Tyumentseva et al., 2023). Recently, gene editing has greatly expedited our understanding of plant-pathogen interaction and the development of host resistance against various biotic stresses in many crops (Zhao et al., 2022). This mini-view summarizes the recent applications of genome editing technology in developing disease-resistant tomato cultivars, discusses the current obstacles that may restrict the use of CRISPR in breeding disease-resistant crops, and proposes some viewpoints that may help overcome these challenges.
Pathogens usually need to break through the plant cell wall to infect a plant. This process activates various cell wall structure-related genes. Recent studies showed that knocking out a cell wall structure-related gene pectate lyase (PL) in tomato cultivar Ailsa Craig (AC) using CRISPR/Cas9 resulted in significantly reduced degradation of pectin, increased fruit firmness and resistance against fungal disease (Wang et al., 2018; 2019; Silva et al., 2021; Ortega-Salazar et al., 2024), suggesting a strong link between pectin and cell wall-mediated plant immunity.
Instead of breaching the cell wall, some pathogens enter the host apoplast via entry portals like stomata with the help of S genes. SlJAZ2 which encodes a major co-receptor of coronatine (COR) in tomato stomatal guard cells could facilitate Pseudomonas syringae pv. tomato (Pto) DC3000 colonization by stimulating stomata opening. Knocking out SlJAZ2 in the tomato cultivar Moneymaker using CRISPR/Cas9 resulted in enhanced resistance to (Pto) DC3000 while did not affect resistance to the necrotrophic fungal pathogen Botrytis cinerea (B. cinerea) that does not rely on stomata for penetration (Ortigosa et al., 2019). The tomato S gene Phospholipase C2 (SlPLC2), which can be induced by fungal elicitor xylanase, was required for B. cinerea proliferation. Knock-downing the expression of SlPLC2 by virus-induced gene silencing and knocking out SlPLC2 by CRISPR/Cas9 in the tomato cultivar Moneymaker without Cf resistance genes (MM-Cf0) resulted in enhanced resistance to B. cinerea, accompanied by decreased reactive oxygen species (ROS) production and altered salicylic acid (SA) and jasmonic acid (JA) signaling pathways (Gonorazky et al., 2016; Perk et al., 2023). Cell surface localized receptors can bind microbial elicitors and mediate plant defense response. In tomato plants, cell-surface decoy receptor LeEIX1 has the ability to bind ethylene-inducing xylanase (EIX), a fungal elicitor secreted by Trichoderma spp, and attenuate EIX-induced signaling and host defense (Ron and Avni, 2004). Knocking out LeEIX1 in tomato M82 cultivar using CRISPR/Cas9 led to stronger host immune activation and enhanced disease resistance against Trichoderma in LeEIX1-edited lines compared with wild-type (WT) control plants (Leibman-Markus et al., 2021).
Some S genes facilitate pathogen survival by providing nutrients or supporting microbial metabolism. One of the best characterized S gene family is mildew resistance locus o (Mlo) which encodes membrane-associated proteins and was reported to confer susceptibility to powdery mildew (PM) disease in many plant species (Acevedo-Garcia et al., 2014). Targeted mutagenesis of SlMlo1, the major contributor to PM susceptibility, in both tomato BN-86 and Moneymaker cultivars using CRISPR/Cas9 resulted in fully resistant plants to the PM fungus without compromising plant growth and fruit development (Nekrasov et al., 2017; Pramanik et al., 2021). SlPelo was previously discovered to encode a messenger RNA surveillance factor and render susceptibility to Tomato yellow leaf curl virus (TYLCV) (Lapidot et al., 2015). More recently, the role of SlPelo in regulating TYLCV infection in the elite tomato cultivar BN-86was validated in SlPelo-edited mutants generated by CRISPR/Cas9, and the results indicated that SlPelo was a susceptibility factor of yellow leaf curl disease caused by TYLCV (Pramanik et al., 2021). Another well-studied S gene related to PM is POWDERY MILDEW RESISTANT 4 (PMR4). PMR4 was reported to inhibit SA defense signaling pathway (Nishimura et al., 2003). CRISPR/Cas-9 mediated mutagenesis of SlPMR4 in susceptible tomato cultivar moneymaker resulted in reduced susceptibility to PM, accompanied by a higher occurrence of hypersensitive response-like cell death at infection sites in the CRISPR edited line compared with a control plant, which was likely to be induced by SA signaling pathway (Santillán Martínez et al., 2020). More recently, knocking out SlPMR4 in two widely grown Italian tomato cultivars including San Marzano (SM) and Oxheart (OX) by CRISPR/Cas9, indicated that SlPMR4 conferred susceptibility to Late Blight (LB), a fungal disease caused by Phytophthora infestans (Li et al., 2022). Genes required for viruses to maintain their lifecycle can also be regarded as S genes. Yoon et al. (2020) generated CRISPR/Cas9-derived mutations in the eukaryotic translation initiation factor 4E (eIF4E) in the tomato cultivar Micro-Tom, and evaluated the role of eIF4E in Potyvirus resistance. Results demonstrated that eIF4E was a susceptible factor which is necessary for pepper mottle virus (PepMoV) infection (Yoon et al., 2020).
The third class of S genes act as negative regulators of host innate immune response, many of which regulate hormone signaling pathways. Disabling such S genes is likely to be a promising strategy to obtain durable and broad-spectrum disease resistance. Inactivating a tomato DOWNY MILDEW RESISTANCE gene (SlDMR6-1) in the tomato Fla. 8000 variety using CRISPR/Cas9 enhanced resistance to bacterial, oomycete, and fungal pathogens, correlating with increased SA (Thomazella et al., 2021). Moreover, SlDMR6-1 displayed SA-5 hydroxylase activity, which could explain the increased SA level in the SlDMR6-1 homozygous mutants (Thomazella et al., 2021). More recently, CRISPR/Cas9 was employed to edit two tomato NUCLEOREDOXIN (SlNRX) genes including SlNRX1 and SlNRX2 in the Mocro-Tom cultivar, and characterization of the CRISPR edited plants unraveled the negative role of SlNRX1, a member of the nucleocytoplasmic THIOREDOXIN subfamily, in modulating SA-dependent immune response to both bacterial and fungal infection in tomato (Cha et al., 2023).
Another important plant hormone JA regulates various biological processes, including plant immunity, through complex modules involving multiple transcription factors (TFs) or regulators. Manipulating a number of MYC2-TARGETED BHLH (MTB) genes which negatively regulate JA-mediated defense response in the tomato AC cultivar by CRISPR/Cas9 resulted in stronger resistance to herbivore attack compared to WT plants, without compromising normal plant growth (Liu et al., 2019). By integrating CRISPR/Cas9 with RNA sequencing, SlBBX20, a gene belonging to the B-box (BBX) family in tomato, was knocked out in the tomato AC cultivar and found to negatively regulate resistance to B. cinerea in tomato plants by attenuating JA signaling (Luo et al., 2023).
The contrasting effect of host genes in regulating resistance to different pathogens was observed in many plants. A functional study of tomato SRFR1 using CRISPR-based gene editing tool revealed its negative role in immune response to Pto DC3000 via regulating SA-pathway defense genes, while it functioned as a positive regulator to necrotrophic pathogens Fusarium oxysporum f. sp. lycopersici (FOL) by modulating JA/ethylene genes (Son et al., 2021). Similarly, a tomato gene SlHyPRP1encoding a proline-rich protein involved in cell wall signaling was reported as a negative regulator of defense to Pto DC3000 but a positive regulator of immunity to FOL (Tran et al., 2023). It has been suggested that defense against biotrophic pathogens is largely regulated by SA signaling pathway while JA/ethylene mainly dominates in facilitating host defense response against necrotrophic pathogens (Glazebrook, 2005). In addition to JA and SA, other hormones including auxins, cytokinins (CK), abscisic acid, gibberellins (GA), brassinosteroids, strigolactones, as well as nitric oxide, are likely to act antagonistically in the regulation of plant-pathogen interactions (Huang et al., 2020). The regulation of plant defense is largely dependent on multiple hormone pathways often interconnected by complex transcription module. The antagonistic role of host genes in regulating defense against different types of pathogens might be explained by different attack strategies of pathogens and complex strategies of host plants to counteract the invasion of pathogens.
Gene editing technology has been used to elucidate the molecular mechanisms of R genes in regulating immune response in tomato. For instance, the role of a cell wall-associated kinase gene named SlWak1 was characterized in CRISPR mutant lines generated in the Grande PtoR (RG-PtoR) genetic background, suggesting SlWak1, acting in a complex with Fls2 and Fls3, positively regulate immune signaling at later stages of PTI in the apoplast upon the inoculation of P. syringae pv. tomato (Pst) (Zhang et al., 2020a). So far, more than 150 genes in tomato associated with host immunity to various pathogens have been edited by gene editing technologies, and detailed information can be found in Plant Genome Editing Database (PGED) (Zheng et al., 2019). Among them, 63 candidate genes were further molecularly characterized in CRISPR edited lines for their gRNA efficiency, specificity of modifications and heritability of the mutations (Zhang et al., 2020b). Many of the selected genes encode cell surface localized pattern recognition receptors (PRRs), kinases, transporters and TFs, and their role in plant immunity requires further investigation.
Incorporating R genes from natural germplasm resources into cultivated species by classical or transgenic breeding has been used for enhancing disease resistance in many crops including tomato. However, this is a lengthy and laborious process. R genes usually defend against specific pathogens, and their effectiveness may not last long because they often fail to recognize frequently mutated pathogen effectors of newly evolved pathogens. Engineering R genes using CRISPR-based technology to achieve broad-spectrum resistance might be a promising solution to this challenge. Giannakopoulou et al. (2015) engineered a tomato NLR gene named I2 and investigated if the mutation in I2 could alter plant response to the pathogen effector AVR3a in Nicotiana benthamiana by agroinfiltration assay. It was shown that mutation of I2 resulted in markedly increased response to AVR3a, suggesting altered resistance to pathogens could be achieved by engineering synthetic immune receptors (Giannakopoulou et al., 2015).
Another approach to achieve resistance is manipulating S genes, a class of plant genes that facilitate pathogen penetration and proliferation or supports compatibility with pathogens. Enhanced disease resistance can be achieved by manipulating S genes using gene editing. Several strategies have been developed to generate transgene-free CRISPR edited crops which are defined as non- GMO; these include elimination of transgenic sequences via genetic segregation, and transient expression of CRISPR/Cas9 editor via ribonucleoprotein (RNP)-mediated CRISPR genome editing method (Gu et al., 2021; Ahmad et al., 2023). The options for utilizing both R and S genes to develop disease-resistant genotypes are summarized in Figure 1. However, the defense-growth trade-off or fitness cost of silencing S genes is common in plants, which are species and condition-specific. Modifying the regulatory element precisely might be a solution. Plant growth-defense trade-off has remained a challenge when engineering disease resistance. Numerous studies have suggested enhanced defense usually results in inhibited growth and development, and this process is modulated by complex network involving interactions between multiple regulators and hormones (Giolai and Laine, 2024; He et al., 2022). In tomato, several modules were reported to fine-turn trade-off of plant growth-defense. For example, using a systematical approach including CRISPR/Cas9 and transcriptional regulation methods, the RALF2-FER-MYB63 module was found to fine-tune root growth and resistance against FOL through regulating the deposition of lignin in tomato cultivar Condine Red (Fan et al., 2024). JA was shown to involve in maintaining a balance between lateral root (LR) development and root-knot nematode (RKN) susceptibility via SlMYB-mediated transcriptional regulation in tomato (Zhao et al., 2023). A more recent study using three different immunity elicitors to investigate the effect of systemic acquired resistance (SAR) and induced systemic resistance (ISR) pathways on tomato development indicated that growth and defense could be positively correlated through alterations to the CK/GA balance (Leibman-Markus et al., 2023). This study challenges the classic model of the growth-defense trade-off, suggesting that growth promotion and induced resistance can be co-dependent. It further shows that defense priming can occur within a specific developmental window and that growth-defense trade-off can be uncoupled through the modulation of certain hormonal pathways. Therefore, uncoupling the antagonism between hormonal pathways opens new avenues for applying gene editing tools to develop crops with enhanced biotic stress resistance. So far, only a limited number of S genes have been identified and manipulated in tomato for conferring resistance to pathogens. A wealth of available tomato genetic resources and multi-omics dataset, along with newly developed web-based platforms, including the Tomato multi-omics data Analysis Platform (TomAP) (http://bioinformatics.cau.edu.cn/TomAP/) (Cao et al., 2024) and Solanaceae Information Resource (SoIR) (https://soir.bio2db.com) (Liu et al., 2024), will facilitate the identification of key genes or modules linked to immune response in tomato. Future research should also focus on developing novel CRISPR/Cas-based toolbox and using high-throughput genetic screens to improve the editing of disease resistance genes. The present review proposes that critical challenges in sustainable agriculture, such as dependency on pesticides and drawbacks of conventional methods, can be addressed by leveraging gene editing technology. Additionally, it proposes multi-omics approaches coupled with advanced gene editing tools provide an effective way for precisely engineering disease resistance for crop improvement.
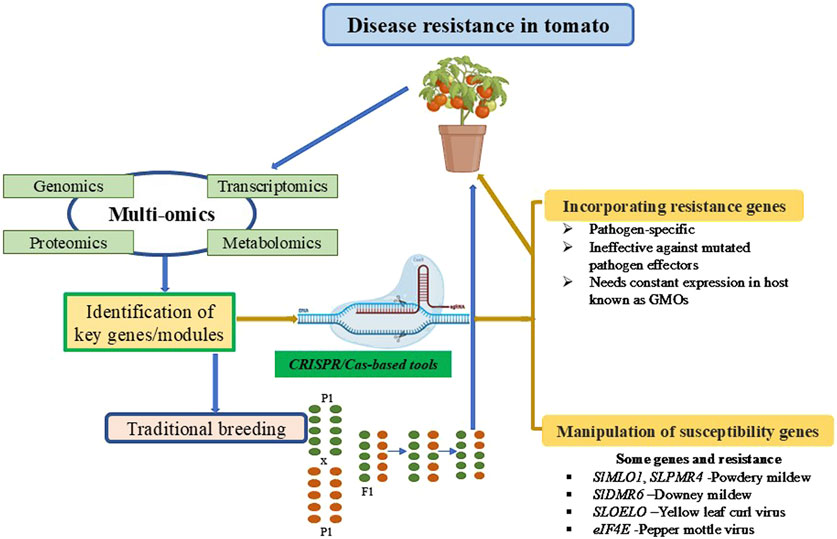
Figure 1. An integrated framework for improving disease resistance in tomatoes through multi-omics approaches and genome editing technologies. Genomics, transcriptomics, proteomics, and metabolomics are utilized to identify key genes or modules associated with disease resistance. These insights are applied through two complementary strategies: traditional breeding methods and CRISPR/Cas-based genome editing tools. CRISPR/Cas technology facilitates the incorporation of resistance (R) genes and the manipulation of susceptibility (S) genes, enabling precise genetic modifications to enhance tomato disease resistance. This systematic approach combines modern molecular tools with conventional practices to develop robust, disease-resistant tomato cultivars.
DW: Writing–original draft, Writing–review and editing. PM: Writing–review and editing. MR: Writing–review and editing. LY: Writing–review and editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acevedo-Garcia, J., Kusch, S., and Panstruga, R. (2014). Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol. 204 (2), 273–281. doi:10.1111/nph.12889
Ahmad, A., Jamil, A., and Munawar, N. (2023). GMOs or non-GMOs? The CRISPR conundrum. Front. Plant Sci. 14, 1232938. doi:10.3389/fpls.2023.1232938
Altpeter, F., Springer, N. M., Bartley, L. E., Blechl, A. E., Brutnell, T. P., Citovsky, V., et al. (2016). Advancing crop transformation in the era of genome editing. Plant Cell 28 (7), 1510–1520. doi:10.1105/tpc.16.00196
Bacelar, E., Pinto, T., Anjos, R., Morais, M. C., Oliveira, I., Vilela, A., et al. (2024). Impacts of climate change and mitigation strategies for some abiotic and biotic constraints influencing fruit growth and quality. Plants 13 (14), 1942. doi:10.3390/plants13141942
Bent, A. F., and Mackey, D. (2007). Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45, 399–436. doi:10.1146/annurev.phyto.45.062806.094427
Borguini, R. G., and Ferraz Da Silva Torres, E. A. (2009). Tomatoes and tomato products as dietary sources of antioxidants. Food Rev. Int. 25 (4), 313–325. doi:10.1080/87559120903155859
Cao, Y., She, J., Li, Z., Liu, Y., Tian, T., You, Q., et al. (2024). TomAP: a multi-omics data analysis platform for advancing functional genomics research in tomatoes. New Crop. 1, 100002. doi:10.1016/j.ncrops.2023.10.001
Čermák, T., Baltes, N. J., Čegan, R., Zhang, Y., and Voytas, D. F. (2015). High-frequency, precise modification of the tomato genome. Genome Biol. 16, 232. doi:10.1186/s13059-015-0796-9
Cha, J. Y., Uddin, S., Macoy, D. M., Shin, G.-I., Jeong, S. Y., Ali, I., et al. (2023). Nucleoredoxin gene SINRX1 negatively regulates tomato immunity by activating SA signaling pathway. Plant Physiol. biochem. 200, 107804. doi:10.1016/j.plaphy.2023.107804
Derbyshire, M. C., Newman, T. E., Thomas, W. J. W., Batley, J., and Edwards, D. (2024). The complex relationship between disease resistance and yield in crops. Plant Biotechnol. J. 22 (9), 2612–2623. doi:10.1111/pbi.14373
Fan, Y., Bai, J., Wu, S., Zhang, M., Li, J., Lin, R., et al. (2024). The RALF2-FERONIA-MYB63 module orchestrates growth and defense in tomato roots. New Phytol. 243 (3), 1123–1136. doi:10.1111/nph.19865
Giannakopoulou, A., Steele, J. F. C., Segretin, M. E., Bozkurt, T. O., Zhou, J., Robatzek, S., et al. (2015). Tomato I2 immune receptor can be engineered to confer partial resistance to the oomycete Phytophthora infestans in addition to the fungus Fusarium oxysporum. Mol. Plant-Microbe Interact. 28 (12), 1316–1329. doi:10.1094/MPMI-07-15-0147-R
Giolai, M., and Laine, A. L. (2024). A trade-off between investment in molecular defense repertoires and growth in plants. Science 386 (6722), 677–680. doi:10.1126/science.adn2779
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43 (1), 205–227. doi:10.1146/annurev.phyto.43.040204.135923
Gonorazky, G., Guzzo, M. C., Abd-El-Haliem, A. M., Joosten, M. H. A. J., and Laxalt, A. M. (2016). Silencing of the tomato phosphatidylinositol-phospholipase C2 (SlPLC2) reduces plant susceptibility to Botrytis cinerea. Mol. Plant Pathol. 17 (9), 1354–1363. doi:10.1111/mpp.12365
Gu, X., Liu, L., and Zhang, H. (2021). Transgene-free genome editing in plants. Front. Genome 3, 805317. doi:10.3389/fgeed.2021.805317
He, Z., Webster, S., and He, S. Y. (2022). Growth-defense trade-offs in plants. Curr. Biol. 32 (12), R634–R639. doi:10.1016/j.cub.2022.04.070
Huang, S., Zhang, X., and Fernando, W. G. D. (2020). Directing trophic divergence in plant-pathogen interactions: antagonistic phytohormones with no doubt? Front. Plant Sci. 11, 600063. doi:10.3389/fpls.2020.600063
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444 (7117), 323–329. doi:10.1038/nature05286
Jones, J. D. G., Staskawicz, B. J., and Dangl, J. L. (2024). The plant immune system: from discovery to deployment. Cell 187 (9), 2095–2116. doi:10.1016/j.cell.2024.03.045
Kimm, K. H., Kabir, E., and Jahan, S. A. (2017). Exposure to pesticides and the associated human health effects. Sci. Total Environ. 1 (575), 525–535. doi:10.1016/j.scitotenv.2016.09.009
Koseoglou, E., van der Wolf, J. M., Visser, R. G. F., and Bai, Y. (2022). Susceptibility reversed: modified plant susceptibility genes for resistance to bacteria. Trends Plant Sci. 27 (1), 69–79. doi:10.1016/j.tplants.2021.07.018
Lapidot, M., Karniel, U., Gelbart, D., Fogel, D., Evenor, D., Kutsher, Y., et al. (2015). A novel route controlling begomovirus resistance by the messenger RNA surveillance factor pelota. PLOS Genet. 11 (10), e1005538. doi:10.1371/journal.pgen.1005538
Leibman-Markus, M., Gupta, R., Pizarro, L., Gershony, O., Rav-David, D., Elad, Y., et al. (2021). Gene editing of the decoy receptor LeEIX1 increases host receptivity to Trichoderma bio-control. Front. Fungal Biol. 2, 678840. doi:10.3389/ffunb.2021.678840
Leibman-Markus, M., Schneider, A., Gupta, R., Marash, I., Rav-David, D., Carmeli-Weissberg, M., et al. (2023). Immunity priming uncouples the growth–defense trade-off in tomato. Development 150 (21), dev201158. doi:10.1242/dev.201158
Li, R., Maioli, A., Yan, Z., Bai, Y., Valentino, D., Milani, A. M., et al. (2022). CRISPR/Cas9-based knock-out of the PMR4 gene reduces susceptibility to late blight in two tomato cultivars. Int. J. Mol. Sci. 23 (23), 14542. doi:10.3390/ijms232314542
Liu, Y., Du, M., Deng, L., Shen, J., Fang, M., Chen, Q., et al. (2019). MYC2 regulates the termination of jasmonate signaling via an autoregulatory negative feedback loop. Plant Cell 31 (1), 106–127. doi:10.1105/tpc.18.00405
Liu, Z., Shen, S., Li, C., Zhang, C., Chen, X., Fu, Y., et al. (2024). SoIR: a comprehensive Solanaceae information resource for comparative and functional genomic study. Nucleic Acids Res. 53, D1623–D1632. doi:10.1093/nar/gkae1040
Luo, D., Sun, W., Cai, J., Hu, G., Zhang, D., Zhang, X., et al. (2023). SlBBX20 attenuates JA signalling and regulates resistance to Botrytis cinerea by inhibiting SlMED25 in tomato. Plant Biotechnol. J. 21 (4), 792–805. doi:10.1111/pbi.13997
Manzoor, S., Nabi, S. U., Rather, T. R., Gani, G., Mir, Z. A., Wani, A. W., et al. (2024). Advancing crop disease resistance through genome editing: a promising approach for enhancing agricultural production. Front. Genome 6, 1399051. doi:10.3389/fgeed.2024.1399051
Nekrasov, V., Wang, C., Win, J., Lanz, C., Weigel, D., and Kamoun, S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7 (1), 482. doi:10.1038/s41598-017-00578-x
Ngou, B. P. M., Ahn, H.-K., Ding, P., and Jones, J. D. G. (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592 (7852), 110–115. doi:10.1038/s41586-021-03315-7
Nishimura, M. T., Stein, M., Hou, B.-H., Vogel, J. P., Edwards, H., and Somerville, S. C. (2003). Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301 (5635), 969–972. doi:10.1126/science.1086716
Ortega-Salazar, I., Crum, D., Sbodio, A. O., Sugiyama, Y., Adaskaveg, A., Wang, D., et al. (2024). Double CRISPR knockout of pectin degrading enzymes improves tomato shelf-life while ensuring fruit quality. Plants, People, Planet 6 (2), 330–340. doi:10.1002/ppp3.10445
Ortigosa, A., Gimenez-Ibanez, S., Leonhardt, N., and Solano, R. (2019). Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol. J. 17 (3), 665–673. doi:10.1111/pbi.13006
Perk, E. A., Arruebarrena Di Palma, A., Colman, S., Mariani, O., Cerrudo, I., D’Ambrosio, J. M., et al. (2023). CRISPR/Cas9-mediated phospholipase C 2 knock-out tomato plants are more resistant to Botrytis cinerea. Planta 257 (6), 117. doi:10.1007/s00425-023-04147-7
Potnis, N., Timilsina, S., Strayer, A., Shantharaj, D., Barak, J. D., Paret, M. L., et al. (2015). Bacterial spot of tomato and pepper: diverse Xanthomonas species with a wide variety of virulence factors posing a worldwide challenge. Mol. Plant Pathol. 16 (9), 907–920. doi:10.1111/mpp.12244
Pramanik, D., Shelake, R. M., Park, J., Kim, M. J., Hwang, I., Park, Y., et al. (2021). CRISPR/Cas9-mediated generation of pathogen-resistant tomato against tomato yellow leaf curl virus and powdery mildew. Int. J. Mol. Sci. 22 (4), 1878. doi:10.3390/ijms22041878
Ron, M., and Avni, A. (2004). The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16 (6), 1604–1615. doi:10.1105/tpc.022475
Rothan, C., Diouf, I., and Causse, M. (2019). Trait discovery and editing in tomato. Plant J. 97 (1), 73–90. doi:10.1111/tpj.14152
Rozas, P., Kessi-Pérez, E. I., and Martínez, C. (2022). Genetically modified organisms: adapting regulatory frameworks for evolving genome editing technologies. Biol. Res. 55 (1), 31. doi:10.1186/s40659-022-00399-x
Sacristán, S., Goss, E. M., and Eves-van den Akker, S. (2021). How do pathogens evolve novel virulence activities? Mol. Plant Microbe Interact. 34 (6), 576–586. doi:10.1094/MPMI-09-20-0258-IA
Santillán Martínez, M. I., Bracuto, V., Koseoglou, E., Appiano, M., Jacobsen, E., Visser, R. G. F., et al. (2020). CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew. BMC Plant Biol. 20 (1), 284. doi:10.1186/s12870-020-02497-y
Silva, C. J., van den Abeele, C., Ortega-Salazar, I., Papin, V., Adaskaveg, J. A., Wang, D., et al. (2021). Host susceptibility factors render ripe tomato fruit vulnerable to fungal disease despite active immune responses. J. Exp. Bot. 72 (7), 2696–2709. doi:10.1093/jxb/eraa601
Son, G. H., Moon, J., Shelake, R. M., Vuong, U. T., Ingle, R. A., Gassmann, W., et al. (2021). Conserved opposite functions in plant resistance to biotrophic and necrotrophic pathogens of the immune regulator SRFR1. Int. J. Mol. Sci. 22 (12), 6427. doi:10.3390/ijms22126427
Sun, P., Han, X., Milne, R. J., and Li, G. (2024). Trans-crop applications of atypical R genes for multipathogen resistance. Trends Plant Sci. 29 (10), 1103–1112. doi:10.1016/j.tplants.2024.05.004
Thomazella, D. P. de T., Seong, K., Mackelprang, R., Dahlbeck, D., Geng, Y., Gill, U. S., et al. (2021). Loss of function of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. 118 (27), e2026152118. doi:10.1073/pnas.2026152118
Tran, M. T., Son, G. H., Song, Y. J., Nguyen, N. T., Park, S., Thach, T. V., et al. (2023). CRISPR-Cas9-based precise engineering of SlHyPRP1 protein towards multi-stress tolerance in tomato. Front. Plant Sci. 14, 1186932. doi:10.3389/fpls.2023.1186932
Tyumentseva, M., Tyumentsev, A., and Akimkin, V. (2023). CRISPR/Cas9 landscape: current state and future perspectives. Int. J. Mol. Sci. 24 (22), 16077. doi:10.3390/ijms242216077
van Schie, C. C. N., and Takken, F. L. W. (2014). Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 52 (1), 551–581. doi:10.1146/annurev-phyto-102313-045854
Wang, D., Samsulrizal, N., Yan, C., Allcock, N. S., Craigon, J., Blanco-Ulate, B., et al. (2018). Characterisation of CRISPR mutants targeting genes modulating pectin degradation in ripening tomato. Plant Physiol. 179 (2), 01187–02018. doi:10.1104/pp.18.01187
Wang, D., Yeats, T. H., Uluisik, S., Rose, J. K. C., and Seymour, G. B. (2019). Fruit softening: revisiting the role of pectin. Trends Plant Sci. 23 (4), 302–310. doi:10.1016/j.tplants.2018.01.006
Yoon, Y. J., Venkatesh, J., Lee, J. H., Kim, J., Lee, H. E., Kim, D. S., et al. (2020). Genome editing of eIF4E1 in tomato confers resistance to pepper mottle virus. Front. Plant Sci. 11, 1098. doi:10.3389/fpls.2020.01098
Yuan, M., Jiang, Z., Bi, G., Nomura, K., Liu, M., Wang, Y., et al. (2021). Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592 (7852), 105–109. doi:10.1038/s41586-021-03316-6
Zhang, N., Pombo, M. A., Rosli, H. G., and Martin, G. B. (2020a). Tomato wall-associated kinase SlWak1 depends on Fls2/Fls3 to promote apoplastic immune responses to Pseudomonas syringae. Plant Physiol. 183 (4), 1869–1882. doi:10.1104/pp.20.00144
Zhang, N., Roberts, H. M., Van Eck, J., and Martin, G. B. (2020b). Generation and molecular characterization of CRISPR/Cas9-induced mutations in 63 immunity-associated genes in tomato reveals specificity and a range of gene modifications. Front. Plant Sci. 11, 10. doi:10.3389/fpls.2020.00010
Zhang, S., Griffiths, J. S., Marchand, G., Bernards, M. A., and Wang, A. (2022). Tomato brown rugose fruit virus: an emerging and rapidly spreading plant RNA virus that threatens tomato production worldwide. Mol. Plant Pathol. 23 (9), 1262–1277. doi:10.1111/mpp.13229
Zhao, W., Liang, J., Huang, H., Yang, J., Feng, J., Sun, L., et al. (2023). Tomato defence against Meloidogyne incognita by jasmonic acid-mediated fine-tuning of kaempferol homeostasis. New Phytol. 238 (4), 1651–1670. doi:10.1111/nph.18837
Zhao, Y., Zhu, X., Chen, X., and Zhou, J. M. (2022). From plant immunity to crop disease resistance. J. Genet. Genomics 49 (8), 693–703. doi:10.1016/j.jgg.2022.06.003
Keywords: CRISPR, crop improvement, immunity, susceptibility gene, tomato
Citation: Wang D, Mandal P, Rahman MS and Yang L (2025) Engineering tomato disease resistance by manipulating susceptibility genes. Front. Genome Ed. 7:1537148. doi: 10.3389/fgeed.2025.1537148
Received: 30 November 2024; Accepted: 21 January 2025;
Published: 10 February 2025.
Edited by:
Prasenjit Saha, Planet 13 Holdings, Inc., United StatesReviewed by:
Elida R. Robinson, Cibus, United StatesCopyright © 2025 Wang, Mandal, Rahman and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Duoduo Wang, ZHVvZHVvLndhbmdAdW5oLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.