- 1Vilasrao Deshmukh College of Agricultural Biotechnology, Vasantrao Naik Marathwada Krishi Vidyapeeth, Latur, India
- 2Division of Applied Life Science (BK21 Four), Division of Life Science, Plant Molecular Biology and Biotechnology Research Centre (PMBBRC), Gyeongsang National University, Jinju, Republic of Korea
Climate change threatens global crop yield and food security due to rising temperatures, erratic rainfall, and increased abiotic stresses like drought, heat, and salinity. Gene editing technologies, including CRISPR/Cas9, base editors, and prime editors, offer precise tools for enhancing crop resilience. This review explores the mechanisms of these technologies and their applications in developing climate-resilient crops to address future challenges. While CRISPR/enables targeted modifications of plant DNA, the base editors allow for direct base conversion without inducing double-stranded breaks, and the prime editors enable precise insertions, deletions, and substitutions. By understanding and manipulating key regulator genes involved in stress responses, such as DREB, HSP, SOS, ERECTA, HsfA1, and NHX; crop tolerance can be enhanced against drought, heat, and salt stress. Gene editing can improve traits related to root development, water use efficiency, stress response pathways, heat shock response, photosynthesis, membrane stability, ion homeostasis, osmotic adjustment, and oxidative stress response. Advancements in gene editing technologies, integration with genomics, phenomics, artificial intelligence (AI)/machine learning (ML) hold great promise. However, challenges such as off-target effects, delivery methods, and regulatory barriers must be addressed. This review highlights the potential of gene editing to develop climate-resilient crops, contributing to food security and sustainable agriculture.
Introduction
The increase in global population and severe climate change are primary challenges to food security. Higher greenhouse gas emissions lead to increased atmospheric temperatures. It is projected that an average increase of 2°C by the year 2100 (Scafetta, 2024). This will cause substantial economic losses in agriculture and food production (Abdelrahman et al., 2021). Climate change presents abiotic stresses via salinity, drought, and temperature stress that affect crop physiology, reduce productivity, and threaten global food security (Abdelrahman et al., 2021) Moreover, climate change negatively impacts soil microbial populations and their enzymatic functions, particularly in arid regions (Cooper et al., 2018). It alters plants’ physiological and metabolic processes, affecting growth, pest dynamics, and agricultural productivity. These factors raise the risk of pest invasions and plant diseases, exacerbating fragile food production (Abdelrahman et al., 2021).
Rapid and unpredictable changes in climatic conditions command novel solutions to develop resilient crops that can withstand the challenges of extreme drought, heat, and salinity. Gene editing technologies like CRISPR/Cas9, transcription activator-like effector nucleases (TALENs), zinc finger nucleases (ZFNs), and base editors provide a significant opportunity for crop improvement (Biswas et al., 2021). These methods allow for precise, targeted modifications in a plant’s genetic makeup, facilitating the development of crops with enhanced traits, including increased yield, pest and disease resistance, and improved resilience to environmental stresses (Ahmad A. et al., 2021) Gene editing holds immense potential for addressig global agricultural challenges, including improved climate resilience, enhanced nutritional content, increased yield and productivity, pest and disease resistance, and faster breeding cycles (Biswas et al., 2021; Kumar et al., 2017).
Gene editing technologies
CRISPR/Cas9
CRISPR/Cas9 was initially discovered in Escherichia coli (Ishino et al., 1987). CRISPR/Cas functions as an adaptive immune defense in bacteria, safeguarding their genomic DNA from viral (phage) attacks by degrading the DNA with the help of RNA guidance and the Cas9 protein (Mallapaty, 2019; Li et al., 2020a). CRISPR/Cas systems are classified into two main classes (Jinek et al., 2012). Class 1 systems require multiple effector proteins, whereas Class 2 systems use a single protein. Class 1 is further divided into types I, III, and IV, while Class 2 includes types II, V, and VI (Makarova et al., 2011; 2015). Type II systemsprimarily use Cas9 and are the most widely applied (Wiedenheft et al., 2012; Sorek et al., 2013). The CRISPR/Cas9 mechanism incorporates fragments of foreign DNA into the CRISPR locus, which are later transcribed into CRISPR RNA (crRNA). This crRNA binds with trans-activating crRNA (tracrRNA), enabling Cas9 to identify and cleave the target foreign DNA sequence (Jinek et al., 2012).
Cas9 requires the presence of a conserved protospacer-adjacent motif (PAM) sequence upstream of the crRNA binding region to accurately identify the target sequence (Jinek et al., 2012). The CRISPR/Cas9 complex is composed of the Cas9 endonuclease, trans-activating crRNA (tracrRNA), CRISPR RNA (crRNA), and RNase III (El-Mounadi et al., 2020; Mohanta et al., 2017; Jansing et al., 2019). The tracrRNA and crRNA can be combined into a single-guide RNA (sgRNA), which incorporates elements of both (Kumar et al., 2019). Cas9, which cleaves double-stranded DNA (dsDNA), has two active domains: The His-Asn-His (HNH) and RuvC-like domains. These domains cut the dsDNA three base pairs upstream of the PAM sequence (5′-NGG or 5′-NAG) (Jiang and Doudna, 2017; Hille and Charpentier, 2016; Manghwar et al., 2019). The HNH domain cleaves the strand complementary to the crRNA, while the RuvC-like domain cleaves the opposite strand, creating double-stranded breaks (DSBs) that are repaired via non-homologous end joining (NHEJ) or homology-directed repair (HDR) (Kumar et al., 2019; Jiang and Doudna, 2017).
The sgRNA is approximately 100 nucleotides long and includes a 20-nucleotide guide sequence at the 5′end, which directs the complex to the target site, followed by the PAM sequence. The 3′end of the sgRNA forms a loop structure that aids in binding to the target DNA. Together, the sgRNA and Cas9 create a ribonucleoprotein (RNP) complex capable of cleaving the target DNA. The crRNA plays a key role in recognizing the target DNA and assists the RNP complex in binding by forming an R-loop-like structure (Manghwar et al., 2019). The loop structure formation activates both active domains of the Cas9 endonuclease, resulting in the cleavage of double-stranded DNA (dsDNA) and the production of blunt ends (Hille and Charpentier, 2016). For Streptococcus pyogenes Cas9, the recognized PAM sequence is typically 5′-NGG-3′, although 5′-NAG-3′ is sometimes tolerated (Hsu et al., 2013; Jiang et al., 2013; Mali et al., 2013).
Research has shown that the guide RNA (gRNA) forms a heteroduplex with the complementary DNA strand within a positively charged groove situated between the HNH and RuvC-like domains of Cas9 (Lu et al., 2022). An arginine-rich motif in Cas9 is essential for PAM recognition (Anders et al., 2014). It is found that the displacement of the DNA strand triggers a conformational shift in Cas9, positioning the non-target DNA strand within the RuvC domain and moving the HNH domain closer to the target strand, thus facilitating the cleavage of both strands (Jiang et al., 2016). This adaptability allows CRISPR/Cas systems to create unidirectional double-stranded breaks in DNA efficiently. Such breaks activate cellular DNA repair pathways, introducing precise mutations at the target site.
This method is widely used for gene knockout by inducing insertions and deletions (INDELs) through the nonhomologous end joining (NHEJ) repair pathway. Alternatively, when a donor template similar to the target DNA is available, genes can be integrated or corrected via the homology-directed repair (HDR) pathway (Gaj et al., 2016). Since the gRNA facilitates target recognition, CRISPR/Cas9 serves as a powerful genome editing tool, eliminating the need to design custom proteins for each target site. Its ease of programming, precise cutting capability, and versatile system variants have driven significant progress in the field (Figure 1). This cost-effective and user-friendly technology enables precise targeting, editing, modification, regulation, and labeling of genomic loci across various cell types and organisms (Doudna and Charpentier, 2014). CRISPR/Cas9 has yielded remarkable achievements, including improvements in nutritional content (Li A. et al., 2018), the creation of male-sterile maize (Li et al., 2017) and wheat (Okada et al., 2019), the development of disease-resistant crops (Zhang et al., 2017), and the production of herbicide-resistant plants (Sun et al., 2016). Notably, in 2021, Japan introduced the world’s first CRISPR/Cas9-edited tomato, i.e., Sicilian Rouge High GABA tomato (Waltz, 2022), engineered to have increased levels of γ-aminobutyric acid (GABA) (Ezura, 2022).
CRISPR/Cas9 technology holds a significant edge over zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs). The CRISPR/Cas9 system employs a programmable single-guide RNA (sgRNA) for sequence-specific DNA targeting, enabling unprecedented precision in genome editing through its complementarity-driven recognition mechanism, which significantly reduces molecular complexity compared to traditional gene-editing approaches (Doggalli et al., 2024). This has accelerated advancements in plant breeding and molecular research. A key advantage of CRISPR/Cas9 is its multiplexing capability, allowing simultaneous targeting of multiple genes and facilitating the rapid development of complex trait combinations (Figure 2). This feature is especially valuable for engineering disease resistance and studying gene interactions (Goberna et al., 2022). Additionally, its high efficiency and potential for creating transgene-free crop varieties have made it an indispensable tool for developing plants with enhanced traits, such as higher yields, improved quality, and increased disease resistance (Zhang and Qi, 2019). CRISPR/Cas9 also has limitations, notably the risk of off-target effects, where the Cas9 enzyme may cleave similar but unintended DNA sequences. This can result in unexpected gene structure and function changes, potentially leading to undesirable phenotypes. To mitigate these risks, researchers have developed strategies such as improving sgRNA design, using truncated or modified sgRNAs, exploring Cas9 mutants and orthologues, and employing the “double nicking” strategy. Applying CRISPR/Cas9 technology to crop improvement faces several challenges, including delivery issues and regulatory hurdles. Addressing these challenges is crucial for advancing CRISPR/Cas9 technology in plant biotechnology and fully realizing its potential in crop improvement.
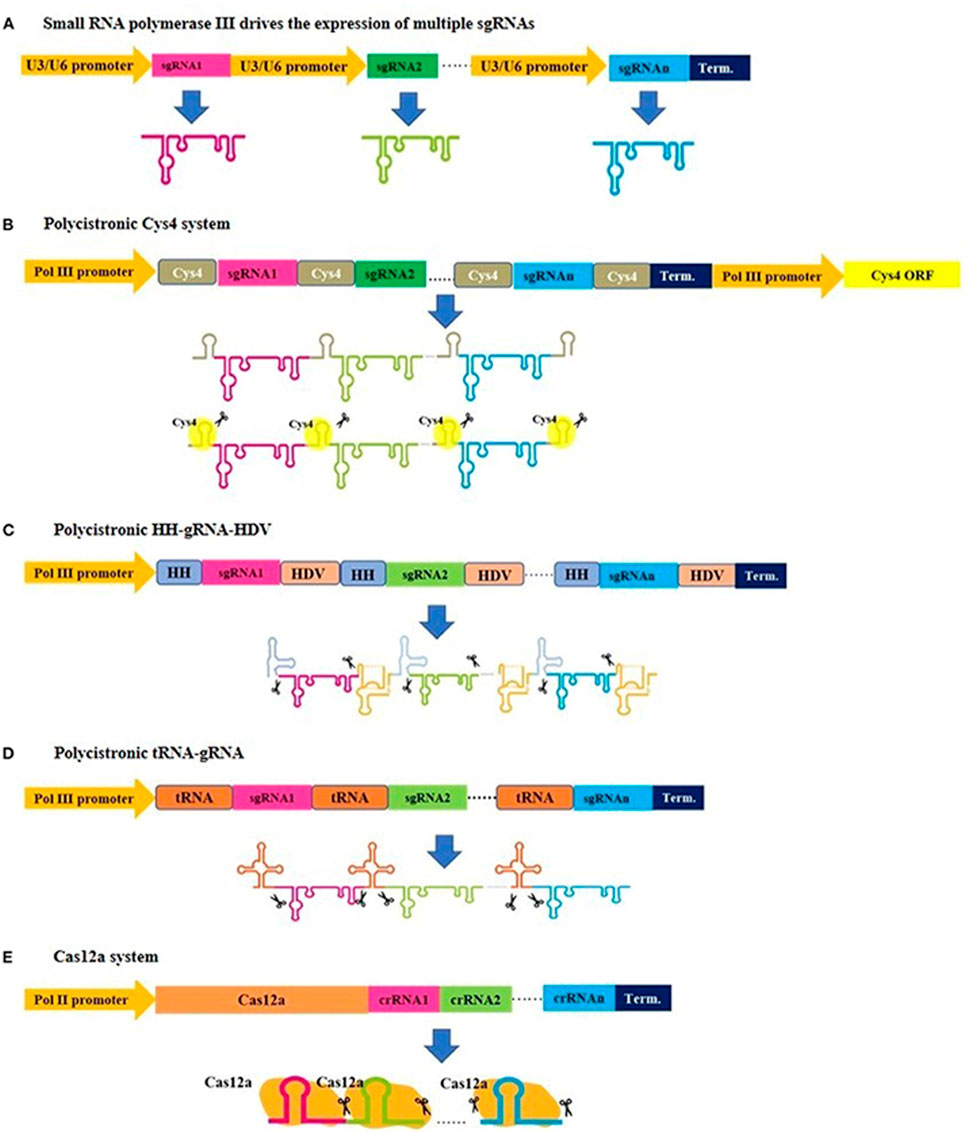
Figure 2. An illustration demonstrating various strategies for expressing multiplex gRNA cassettes in plants. (A) Small gRNAs are cloned after U3 or U6 promoters and derived by small RNA polymerase III to generate individual gRNAs. (B–D) Small gRNAs are cloned to be transcribed into a single transcript, and subsequent posttranscriptional processing is needed for gRNA separation, where Csy4, tRNAs, and hammerhead ribozyme regulate this separation. Similarly, a single transcript is generated in the (E) Cas12a system, but this system has a gRNA self-cleaving feature and does not require additional elements for posttranscriptional processing. (Figure from Abdelrahman et al., 2021; Copyright: CC BY License).
Base editors
Base editing technology is an advanced genome editing method derived from the CRISPR/Cas9 system. It empowers precise single-base substitutions without inducing double-strand breaks or requiring donor DNA templates (Molla et al., 2021; Negishi et al., 2019). This method employs engineered deaminases to convert specific nucleotides, such as cytosine to thymine (C-to-T) or adenine to guanine (A-to-G), enabling targeted genetic modifications with high efficiency and specificity (Huang et al., 2023). Unlike traditional CRISPR/Cas9, which often results in insertions or deletions (indels) due to DSB repair, base editing yields more predictable outcomes, making it particularly beneficial for plant genome engineering (Li et al., 2023). The mechanism involves a modified CRISPR/Cas9 system, where a catalytically inactive Cas9 protein is fused to an active deaminase enzyme. The process begins with a single-guide RNA (sgRNA) directing the Cas9 protein to a specific genomic location (Yang X. et al., 2024). Once bound, the Cas9 creates a single-strand break, allowing the deaminase to access the target base. Depending on the base editor used—either a cytidine base editor (CBE) or an adenine base editor (ABE)—the deaminase converts cytidine (C) to uracil (U) or adenine (A) to inosine (I). During DNA replication, these modifications result in the desired C ⋅ G to T ⋅ A or A ⋅ T to G ⋅ C substitutions (Azameti and Dauda, 2021).
Base editing systems consist of molecular components that function together effectively. Cytosine base editors (CBEs) typically include a deaminase (variants of DddA or APOBEC) fused to TALE arrays for targeted binding, enabling C-to-T conversions (Wang X. et al., 2024; Zhang D. et al., 2024). Recent findings like the TadA-8e-derived CBEs and mTCBE variants have achieved editing efficiencies of up to 81% in rice (Wang Y.-H. et al., 2024; Fan, 2024). The A3A-CBE variant offers a broad editing range, effectively targeting multiple sites simultaneously (Luo et al., 2023). Adenine base editors (ABEs) are dCas9 linked to an adenine deaminase, facilitating A-to-G conversions. For example, the ABE8e variant has demonstrated enhanced editing capabilities in various crops (Fan, 2024; Arantes et al., 2024). Additionally, dual base editors have been developed, combining both CBE and ABE functionalities to enable simultaneous editing of cytosines and adenines, thereby expanding the potential for genetic modifications in plants (Fan et al., 2024; Wang X. et al., 2024).
The alteration of GmAITR genes, leading togmaitr36 double and gmaitr23456 quintuple mutants in soybean using CRISPR/Cas9, has shown enhanced salinity tolerance, highlighting base editing’s potential to improve abiotic stress responses (Wang J. et al., 2021). Moreover, base editing technologies have demonstrated high efficiency and specificity in various crops, including rice and Arabidopsis, allowing for the precise development of desirable traits compared to conventional breeding techniques (Ren et al., 2021; Lyzenga et al., 2021). Recent studies have successfully introduced herbicide resistance and created beneficial mutations in crops, showcasing the practical applications of these systems in agricultural biotechnology (Fan et al., 2024; Zhong et al., 2024). Such precision accelerates the breeding process and helps address challenges related to genetic diversity and linkage drag common in traditional methods. Additionally, the capability for simultaneous multiple edits enhances its utility in crop improvement, enabling the development of varieties with desirable traits such as disease resistance and improved nutritional content (Azameti and Dauda, 2021). Despite its advantages, challenges persist in optimizing editing ranges and enhancing the efficiency of specific base conversions, especially in diverse plant species (Zhong et al., 2024). Additionally, the potential for off-target effects requires further optimizing these systems (Zhang W. et al., 2024). Overall, base editing marks a significant advancement in the toolkit for crop improvement and functional genomic research (Sun et al., 2024; Li et al., 2023).
Adenine base editors (ABEs) are engineered tools comprising nCas9 (D10A) fused with an artificially evolved adenosine deaminase, which catalyzes the conversion of adenine (A) to inosine (I), subsequently leading to A:T to G:C base substitutions during DNA repair and replication (Gaudelli et al., 2017). The initial ABE7.10 construct was developed by fusing nCas9 (D10A) with a heterodimer of wild-type adenine deaminase TadA and an evolved variant TadA7.10, enabling an editing window spanning positions 4–8 nt in the protospacer region, with the PAM located at positions 21–23 nt. Enhancements in editing efficiency were achieved by optimizing the codon usage and incorporating an additional nuclear localization sequence (NLS) for use in mammalian cells (Koblan et al., 2018). To further improve performance, ABEmax was developed by adding NLS motifs at both termini of ABE7.10, resulting in editing efficiencies below 50% at most target loci (Hua et al., 2018; Li C. et al., 2018; Yan et al., 2018). ABEmax facilitated A: T to G:C conversions at loci such as OsACC, OsMPK6, OsSERK2, and OsWRKY45 in rice with editing frequencies of 17.6%, 32.1%, and 62.3%, respectively (Yan et al., 2018). Additionally, a simplified ABE variant, ABE-P1S (TadA7.10-nCas9 D10A), demonstrated superior editing efficiency in rice compared to the commonly used TadATadA7.10-nCas9 D10A fusion (Hua et al., 2020).
Subsequently, ABE8e was created by incorporating TadA8e, a more efficient adenine deaminase variant that evolved from TadA7.10 (Gaudelli et al., 2020; Richter et al., 2020). ABE8e exhibited a significantly higher deamination rate, enhancing A-to-G conversion efficiency (Richter et al., 2020). A V106W mutation in TadA8e was introduced to minimize off-target effects (Richter et al., 2020). A rice-optimized version, rABE8e, was later developed by combining codon-optimized monomeric TadA8e with bis-bpNLS (dual NLSs at both termini), leading to markedly improved editing efficiencies on NG-PAM and NGG-PAM target sequences compared to ABEmax in rice. rABE8e achieved near-complete editing efficiencies and a higher homozygous substitution ratio within the editing window, particularly at positions A5 and A6 (Wei et al., 2021).
Recently, the ABE toolbox was further advanced with the development of PhieABE, which integrates hyTadA8e (TadA8e with a single-stranded DNA-binding domain) to achieve significantly enhanced base editing activity and expanded editing windows compared to standard ABE8e systems (Tan et al., 2022). Finally, TadA9, an optimized adenosine deaminase harboring V82S and Q154R mutations, was developed for rice (Yan et al., 2021). TadA9 is compatible with multiple Cas9 variants, including nSpCas9, nSpCas9-NG, nScCas9, and SpRY, with near-PAM-less capability. Compared to TadA8e, TadA9 extends the editing window, enabling efficient editing of previously challenging endogenous target sites and exhibiting robust editing efficiency in commercial rice cultivars (Yan et al., 2021).
Cytosine base editor (CBE), termed Td-CBEs or TadCBEs, was recently developed through two primary methodological approaches: strategic re-engineering and phage-assisted continuous evolution of the adenine deaminase TadA-8e, targeting efficient and specific CRISPR-based cytosine base editing (Neugebauer et al., 2022; Chen et al., 2022). The introduction of an N46L mutation in TadA-8e strategically ablated its inherent adenine deaminase activity. By systematically fusing a diverse array of TadA-8e mutants with uracil glycosylase inhibitors (UGIs), researchers generated multiple Td-CBEs demonstrating either comparable activity to CBE4max or enhanced accuracy for C: G to T: A base editing (Chen et al., 2022).
Concurrently, through phage-assisted continuous evolution, an optimized TadA8e capable of cytidine deamination was successfully engineered (Neugebauer et al., 2022). This evolved TadA cytidine deaminase variant incorporates mutations within DNA-binding residues, substantially modifying enzyme selectivity to preferentially catalyze deoxycytidine deamination over deoxyadenosine. Relative to conventional CBEs, TadA-derived cytosine base editors (TadCBEs) demonstrate comparable or superior on-target activity, a more compact molecular architecture, and significantly reduced Cas-independent DNA and RNA off-target interactions (Neugebauer et al., 2022).
Prime editors
Prime editing (PE) provides significant improvements in genome editing by enabling precise DNA modifications—including targeted insertions, deletions, and substitutions—without causing double-strand breaks in plants (Kim et al., 2023; Perroud et al., 2023; Xu Y. et al., 2023). The PE system consists of two main components: a prime editor that combines a catalytically impaired Cas9 nickase with a reverse transcriptase and a prime editing guide RNA (pegRNA) that directs the editing to a specific genomic location while encoding the desired genetic modification (Mikhaylova et al., 2024; Volodina et al., 2024; Zhang W. et al., 2024; Liu et al., 2023). This unique combination facilitates the installation of all 12 types of point mutations and small indels without the need for donor DNA templates (Liu J. et al., 2022).
PE offers several advantages over CRISPR/Cas9 and other genome editing techniques, including higher precision, reduced off-target effects, and independence from the cell’s repair mechanisms, which minimizes unintended mutations (Volodina et al., 2024; Zeng et al., 2024; Xu Z. et al., 2023). Recent studies have demonstrated a significant increase in PE efficiency, with variants like PE6c achieving over threefold increases in editing efficiency in rice (Cao et al., 2024). Additionally, incorporating T5 exonuclease into the PE system has resulted in up to 2.9-fold increases in editing efficiency across various plant cells (Liang et al., 2023), while integrating cellular factors, such as the small RNA-binding protein La, has further enhanced efficiency (Yan et al., 2024). Engineered pegRNAs (epegRNAs) have also shown improved activity and precision in editing essential plant genes (Salem et al., 2023). Despite its promise, several challenges remain in developing and applying PE technology, including the delivery of large PE components, the need for further optimization across diverse organisms, and improving efficiency and specificity in various plant species (Hosseini et al., 2024; Zeng et al., 2024; Huang and Liu, 2023).
PE empowers the rapid development of novel traits in agriculturally important crops by enabling precise genome modifications without donor DNA (Mikhaylova et al., 2024). It holds promise for enhancing traits such as yield, stress resistance, and nutritional content, which are critical for food security amid climate change (et al., 2024). Successful applications include rice, where conditional knockdown of OsMLH1 has improved PE systems while maintaining fertility (Liu X. et al., 2024). In wheat, the development of ePPEplus has significantly boosted editing efficiency by 33-fold, allowing multiplex editing of up to eight genes (Ni et al., 2023). Moreover, an enhanced prime editing methodology in Physcomitrium patens has enabled routine editing, showcasing the potential for gene modification through direct selection (Perroud et al., 2023).
Ongoing research and development are essential to address these challenges and fully realize the potential of prime editing in crop improvement and genetic research (Li J. et al., 2022; Huang and Liu, 2023). As scientists continue to refine and expand PE capabilities, it is set to play an increasingly important role in advancing agricultural biotechnology and tackling global food security challenges (Ma et al., 2018).
New emerging gene editing technologies
Gene editing technologies have revolutionized the field of biotechnology, offering precise tools for modifying genetic material. Among these technologies, Fanzor represents a novel class of RNA-guided DNA endonucleases that have been identified in eukaryotes and their viruses. Fanzors are homologous to the prokaryotic TnpB proteins and have been detected in various eukaryotic genomes, suggesting a widespread presence beyond prokaryotic systems (Jiang et al., 2023). These enzymes are characterized by their ability to be programmed by RNA to target specific DNA sequences, making them a promising tool for genome editing applications in eukaryotic cells (Jiang et al., 2023). Fanzors function as RNA-programmable DNA endonucleases, similar to the well-known CRISPR/Cas systems. They possess a rearranged catalytic site within the RuvC domain, which is crucial for their endonuclease activity. Unlike some other nucleases, Fanzors lack collateral cleavage activity, which can be advantageous for precise genome editing (Jiang et al., 2023). The evolutionary analysis of Fanzors indicates that they have adapted extensively to function in eukaryotic cells, acquiring features such as introns and nuclear localization signals (Jiang et al., 2023). This adaptation suggests a long-term evolutionary process that has enabled Fanzors to integrate effectively into eukaryotic cellular machinery. Fanzors are derived from a unique lineage of bacterial enzymes, specifically the IS607 TnpBs, which have evolved into two distinct types in eukaryotes: Fanzor1s and Fanzor2s (Yoon et al., 2023; Marraffini and Sontheimer, 2010). This evolutionary pathway highlights the transition from prokaryotic to eukaryotic systems, with Fanzors co-evolving alongside their associated transposases (Yoon et al., 2023). The ability of Fanzors to be harnessed for genome editing in human cells underscores their potential as a versatile tool in biotechnology, offering new possibilities for genetic research and therapeutic applications (Jiang et al., 2023). Fanzor represents a significant advancement in gene editing technology, with its unique RNA-guided mechanism and evolutionary adaptation making it a promising candidate for future biotechnological innovations.
Gene editing using TnpB, a transposon-associated protein, represents a novel approach in the field of genome editing. TnpB is an RNA-guided DNA endonuclease that has been identified as a potential precursor to the well-known CRISPR/Cas systems, specifically Cas12 nucleases. This discovery has opened new avenues for biotechnological applications and genome editing techniques. RNA-Guided DNA Cleavage. TnpB functions as an RNA-guided endonuclease, similar to CRISPR/Cas systems. It utilizes a guide RNA, derived from its own mRNA, to direct the cleavage of DNA at specific sites. This RNA, known as omegaRNA (ωRNA), is processed by TnpB itself, enabling it to target and cleave DNA in a sequence-specific manner (Karvelis et al., 2021; Nety et al., 2023). Transposon-Associated Motif (TAM): The DNA cleavage by TnpB occurs adjacent to a specific sequence known as the transposon-associated motif (TAM). For instance, TnpB from Deinococcus radiodurans targets the 5′TTGAT motif, while TnpB from Sulfolobus islandicus targets the 5′TA motif. This specificity allows TnpB to generate double-stranded DNA breaks, which are crucial for genome editing applications (Karvelis et al., 2021; Xu Y. et al., 2023). Reprogrammability and Evolutionary Significance: TnpB can be reprogrammed to target different DNA sequences, making it a versatile tool for genome editing. This reprogrammability is akin to the flexibility seen in CRISPR systems, and it highlights the evolutionary link between TnpB and CRISPR/Cas nucleases. TnpB’s evolutionary journey from a transposon-encoded protein to a potential genome editing tool underscores its functional and evolutionary flexibility (Altae-Tran et al., 2023; Altae-Tran et al., 2021). Biotechnological Potential: The discovery of TnpB’s RNA-guided nuclease activity expands the toolkit available for genome editing, particularly in organisms where traditional CRISPR systems may not be as effective. TnpB’s ability to function across a range of temperatures and its presence in diverse organisms, including archaea, further enhance its potential for biotechnological applications (Xu Z. et al., 2023; Altae-Tran et al., 2021). TnpB represents a promising new system for genome editing, with its RNA-guided DNA cleavage mechanism offering a novel approach that complements existing CRISPR technologies. Its evolutionary connection to CRISPR/Cas systems and its re-programmability make it a valuable addition to the field of genetic engineering.
CRISPR-associated transposases (CASTs) represent a novel approach in genome engineering, leveraging the RNA-guided DNA binding capabilities of CRISPR systems to facilitate the insertion of large genetic payloads without the need for DNA double-strand breaks. This method offers a promising alternative to traditional genome editing techniques, such as nuclease-based and prime editing approaches, due to its potential for high efficiency and programmability (George et al., 2023a; Walker et al., 2023a; George et al., 2023a). CASTs utilize nuclease-deficient CRISPR effectors to direct the integration of DNA at specific target sites. This process is primarily guided by RNA, which ensures the precise insertion of genetic material. The Type V-K CAST system, for instance, employs Cas12k to achieve accurate target selection, thereby facilitating RNA-dependent transposition (George et al., 2023a; George et al., 2023b). In addition to the RNA-guided mechanism, Type V-K CASTs also exhibit an RNA-independent transposition pathway. This untargeted integration is primarily driven by the availability of TnsC filaments, which preferentially bind to AT-rich sites. The TnsB transposase further refines the specificity of the insertion site by recognizing specific sequence motifs (George et al., 2023a; George et al., 2023b). The transposition process involves complex protein-protein and protein-DNA interactions. Key components such as TnsB, TnsC, and TniQ form a transpososome complex that facilitates the integration of transposons. The Type I-F Vibrio cholerae CAST system, for example, requires the integration host factor (IHF) for efficient transposition, highlighting the importance of cellular factors in the assembly of the transpososome (Walker et al., 2023a; Walker et al., 2023b). CRISPR-guided transposons offer a versatile and efficient tool for genome engineering, with the ability to insert large genetic payloads accurately. The dual pathways of RNA-guided and RNA-independent transposition provide flexibility in target site selection, while the understanding of protein interactions and sequence requirements enhances the precision of these systems. As research progresses, CAST systems hold the potential to revolutionize genome editing applications across various fields (Guan et al., 2013).
Key negative regulators in climate resilience
Negative regulator genes help plants cope with abiotic stresses like drought and salinity, hence playing a crucial part in stress responses and climate resilience. Somenegative regulators often encode transcription factors that inhibit stress-responsive pathways, fine-tuning physiological responses (Yang Z. et al., 2024; Sun et al., 2008). For example, ARR1, ARR10, and ARR12act as negative regulators and are critical in modulating drought responses, suggesting that targeting these genes through gene editing could enhance climate resilience (Nguyen et al., 2017).
In rice, OsWRKY12 functions as a negative regulator by repressing genes involved in abscisic acid (ABA) signaling and secondary cell wall biosynthesis, thereby decreasing drought tolerance (Jia et al., 2024). Similarly, PgRAV-04 in pearl millet negatively impacts drought tolerance by increasing sensitivity in transgenic plants (Wang Y.-H. et al., 2024). In soybean, GmPRR3b suppresses the expression of GmABF3, a key player in the ABA signaling pathway, affecting drought response (Li et al., 2024). While studies on TaWRKY genes in wheat suggest their involvement in drought stress response, specific negative regulators remain to be conclusively identified (Shakam et al., 2024). In cotton, GhVIM28 acts as a negative regulator under salt stress, indicating a potential role in drought tolerance as well (Yang X. et al., 2024). Additionally, GhDi19-3 and GhDi19-4 help reduce sensitivity to salt stress by regulating reactive oxygen species (ROS) levels and are involved in calcium and ABA signaling pathways (Zhao et al., 2022a). The GhRR7 gene negatively regulates drought stress responses through its role in reactive oxygen removal systems (Zhao et al., 2022b). The miR394 pathway, targeting F-Box proteins ZmLCR1 and ZmLCR2, is linked to drought tolerance, as mutants in these genes show improved drought survival, indicating their role as negative regulators (Miskevish et al., 2023). Furthermore, drought stress represses miR166, leading to the upregulation of its target gene, ATHB14-LIKE, which enhances drought tolerance. This feedback mechanism highlights miR166’s role as a negative regulator in drought response (Ni et al., 2023). The ZmGA20ox3 gene demonstrated as prominent negative regulator for enhancing drought tolerance in maize seedling, reduces Anthesis-Silking Interval (ASI) delay and decreasing the yield loss significantly in the field under drought conditions (Liu Y. et al., 2024).
These findings illustrate the complex interplay of negative regulators in enhancing drought tolerance across different crops. However, the potential negative impact of these genes on yield under non-stress conditions warrants further investigation.
Various negative regulator genes modulating stress responses influence horticultural crops’ drought tolerance. For example, the NtAITR family of ABA-induced transcription repressors in tobacco negatively regulates drought tolerance. CRISPR/Cas9 editing of NtAITRs has enhanced drought tolerance, suggesting their role in repressing ABA signaling pathways (Li M. et al., 2022). In tomato, SlWRKY6, while primarily a positive regulator, can interact with other WRKY proteins to exhibit negative regulation under certain conditions, affecting drought response mechanisms (Chen et al., 2024). In rose, RcPP2C24 has been identified as a negative regulator that reduces drought tolerance by promoting stomatal opening, leading to increased water loss during drought conditions (Shen et al., 2024).
While bZIP transcription factors are generally associated with positive regulation, some bZIP factors can act as negative regulators under specific conditions, influencing drought stress responses through complex interactions with other signaling pathways (Tao et al., 2022). These findings underscore the intricate balance of gene regulation in plant responses to drought, where negative regulators are crucial for modulating stress tolerance mechanisms. However, targeting these genes for crop improvement poses a complex challenge, as their functions can vary significantly across different species and environmental contexts.
Key positive regulators in climate resilience
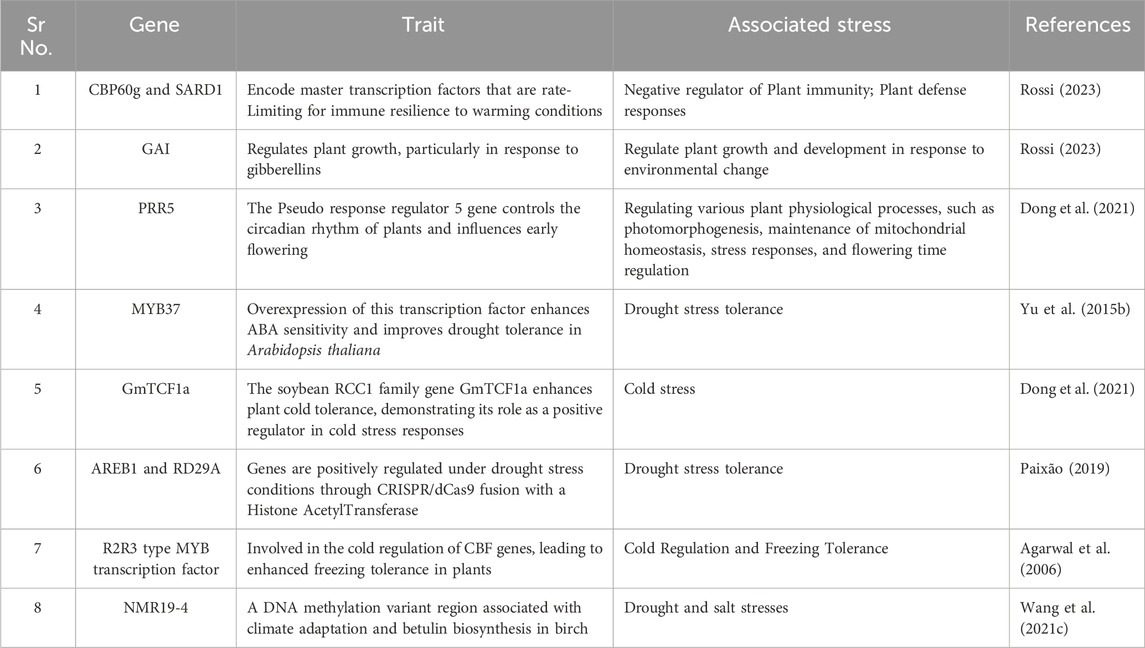
Positive regulator genes are essential for orchestrating plant responses to environmental stressors, significantly enhancing resilience and phenotypic plasticity (Hu et al., 2023). These genes modulate various physiological processes, including stress responses, growth regulation, and defense mechanisms (Table 1). A notable example is the ERECTA gene family, which plays a critical role in drought tolerance by influencing root system architecture and transpiration efficiency in both Arabidopsis thaliana and economically important crops like Oryza sativa (Kulkarni et al., 2017; Wen J. et al., 2019). Additionally, transcription factors such as MYB37 and CaAIEF1 have been shown to enhance ABA sensitivity and drought tolerance in Arabidopsis and Capsicum annuum, respectively, highlighting their importance in stress response pathways (Yu C. et al., 2015; Hong et al., 2017; Cheng et al., 2022). Plant glutathione S-transferases (GSTs), glycinebetaine aldehyde dehydrogenase (BADH), choline monooxygenase (CMO) and flavanone-3-hydroxylase (F3H) were involved in the protecting plants against diverse abiotic and biotic stresses (Fan, 2024). The potential target genes like Na+/H+ antiporter viz., RtNHX1, potassium transporter gene viz., RtHKT1 and Group II WRKY transcription factor viz., RtWRKY23 from recretohalophyte R. trigyna demonstrated prominent sources for abiotic stress tolerance in Arabidopsis (Fan, 2024).
In the context of thermotolerance, the Heat Shock Factor A1 (HsfA1) family proteins serve as master regulators of the heat stress response (Liu Y. et al., 2022), orchestrating a complex transcriptional cascade that enhances plant resilience to elevated temperatures (Mishra et al., 2002). Research in Solanum lycopersicum and Zea mays has highlighted the critical roles of HsfA1 and related genes (ZmHsf05, ZmHsf12) in activating heat stress-inducible genes and heat shock proteins, thereby improving thermotolerance (Guo et al., 2020). Molecular mechanisms underlying salt tolerance involve a diverse array of genes, including ion transporters (OsCIPK9, TaHKT9), transcription factors (WRKY75, BnaABF2), and genes involved in reactive oxygen species (ROS) scavenging (PtGSTF1), all contributing to salinity stress adaptation (Zhou et al., 2023; Du et al., 2023; Ishka and Julkowska, 2023; Ishka et al., 2024; Zhao et al., 2016; Gao et al., 2022; Li J. et al., 2022). Additionally, genes such as AtHDA19 and ATILL6 regulate hormonal signaling cascades and defense mechanisms, enhancing plant immunity against pathogens and abiotic stresses (Wang L. et al., 2020; Wang Z. et al., 2020; Khan et al., 2021).
An enhancement of drought tolerance is reported through CRISPR/Cas9 editing of the TaDREB3 gene in Triticum aestivum (Kim et al., 2018) and knocking out the OsbZIP46 gene in Oryza sativa (Tang J. et al., 2019). Improved thermotolerance has been achieved by modifying the HsfA1 gene in S. lycopersicum, which upregulates heat stress-inducible genes and heat shock proteins (Mishra et al., 2002). Additionally, editing genes involved in ion homeostasis and ROS scavenging has significantly improved salt tolerance in various crops (Leawtrakun et al., 2024; Wani et al., 2020). Targeted modification of the ERECTA gene in A. thaliana has also enhanced drought tolerance by optimizing root architecture and transpiration efficiency (Wen W. et al., 2019; Lim et al., 2018). ABA-stimulated Calcium-dependent protein kinases (CDPKs) from grape berry, i.e., ACPK1 is involved in ABA signal transduction as a positive regulator (Yu et al., 2007).
Epigenetic mechanisms are crucial in regulating gene expression and stress responses in plants. Modifications like DNA methylation and histone changes significantly enhance stress responses in leguminous crops, such as Cicer arietinum (Chandana et al., 2022). The chromatin’s plasticity during environmental changes suggests that chromatin regulators and associated enzymes could be key targets for epigenetic engineering to improve climate resilience (Kumar, 2017; Kumar, 2018). Recent studies have also underscored the importance of small RNAs and the plant microbiome in boosting climate resilience. Non-coding RNAs (ncRNAs) are vital for regulating gene expression and stress responses, contributing to sustainable yields under climate change (Yadav et al., 2024). Additionally, the plant-associated microbiome enhances growth, fitness, and resistance to climate-induced stresses, highlighting the role of microbial interactions in strengthening plant resilience (Wahdan et al., 2021).
Novel trait development for climate resilience in crops
Recent studies highlight the potential of gene editing techniques, such as CRISPR/Cas9, to modify key genetic traits associated with drought resistance in various crops. One promising application is the manipulation of gibberellin synthesis pathways, which are crucial for plant growth during drought conditions. Liu Y. et al. (2024) demonstrated that editing the ZmGA20ox3 gene in maize improved plant architecture and enhanced drought tolerance. This suggests that targeted modifications in hormone regulation can significantly bolster a plant’s ability to withstand water scarcity. Furthermore, advancements in genomics-assisted breeding have enabled researchers to identify drought-related genes in crop wild relatives. These genes can be incorporated into modern cultivars to improve their drought resistance (Kapoor et al., 2021). The integration of stay-green traits, which prolong photosynthetic activity during drought, is another critical area of focus. Research has identified potential targets within the PIN-FORMED gene family that could be manipulated to develop stay-green cereal varieties, ensuring yield stability under drought conditions (Wong et al., 2023). Research on chickpeas has demonstrated that targeting specific genes can improve drought tolerance, contributing to developing more resilient leguminous crops (Roy and Sandhu, 2024). Similarly, studies on sorghum have highlighted genome editing’s potential to enhance stress tolerance, which is vital for sustaining food security amid climate change (Parikh et al., 2021).
Moreover, a comprehensive approach that integrates multi-omics strategies—combining genomics, proteomics, and phenomics—has been emphasized to improve the efficiency of breeding programs aimed at generating climate-resilient crops. This integration allows for a better understanding of plant responses to abiotic stresses and facilitates the identification of novel genetic targets for editing (Ndlovu, 2020; Shahzad et al., 2021). Such advanced biotechnological interventions are increasingly essential for developing sustainable agricultural practices that can mitigate drought stress (Şimşek et al., 2024).
Gene editing for drought and heat tolerance
Water use efficiency (WUE) is another trait that can help design resilient crops. For instance, studies have shown that manipulating genes involved in root growth, such as TaDREB2 and TaERF3 in wheat, can significantly improve drought resilience (Kim et al., 2017). In grapevines, regulating stomatal density by editing genes like VvEPFL9-1 has resulted in improved water conservation and higher WUE under drought conditions (Clemens et al., 2022). Additionally, Song et al. (2023) proposed a model for the evolution of stomatal regulators in C3 and C4 crops, highlighting opportunities for enhancing drought tolerance through genetic modification. Manipulating stress response pathways has also shown great promise. For example, knocking out the AITR gene in Arabidopsis enhanced both drought and salinity tolerance (Chen et al., 2021). Targeting ABA signaling genes, such as PYL receptors, has improved drought tolerance across various species (Kumar, 2023). Multi-omics approaches have identified targets like OsBADH2 and OsMPK2 in rice that could enhance drought resilience (Kumar, 2023). In leguminous crops, successful editing of the 4CL and RVE7 genes in chickpea protoplasts has demonstrated potential for improving drought tolerance (Badhan et al., 2021). Additionally, engineering trehalose metabolism in Arabidopsis, where a CRISPR-edited line mimicked the substrate binding site of the trehalase enzyme, resulted in increased drought tolerance (Mahalingam et al., 2022). This case illustrates the potential of gene editing to modify metabolic pathways, enhancing stress tolerance and opening avenues for similar approaches in other cultivated crops. In addition to metabolic engineering, manipulating root architecture has emerged as a key focus in drought tolerance research. For instance, Nascimento et al. (2023) explored genes that promote root development, enhanced root growth, and improved water uptake, demonstrating their potential as targets for CRISPR/Cas9 gene editing. Their findings highlighted how knocking out certain genes negatively impacts root development under drought conditions, ultimately enhancing overall crop resilience to water scarcity.
Manipulating genes involved in the ABA signaling pathway through gene editing has shown promising results in enhancing drought tolerance. Furthermore, the application of gene editing techniques extends beyond model plants; for example, targeting the GhCLA1 gene in cotton has been shown to improve drought tolerance (Gao et al., 2017). This study not only confirmed the efficacy of gene editing in a major crop but also paved the way for future applications aimed at enhancing drought resilience in economically important species across various crops.
Heat stress poses a significant threat to agricultural productivity; here, we focuson case studies in heat shock response, photosynthesis, and membrane stability. Heat shock proteins (HSPs) protect cellular functions under thermal stress. Several HSP-encoding genes, including HSP70, have been identified as crucial for heat tolerance in various cereal crops (Baldoni et al., 2021). The expression of these genes has been enhanced through gene editing techniques, successfully improving survival rates under high-temperature stress in crops like wheat and maize. Additionally, the transcription factor HsfA2 has been discovered; its overexpression promotes the expression of downstream heat-responsive genes, making it essential for developing heat stress tolerance (Xie et al., 2023; Xie et al., 2021).
Photosynthesis, another critical physiological process affected by heat stress, has also been the focus of research. One study demonstrated that grafting cucumber onto heat-tolerant rootstocks significantly reduced heat-induced photosynthesis (Xu et al., 2018). Proteomic analysis revealed that key enzymes involved in photosynthesis were upregulated in the grafted plants, showcasing the potential for gene editing to enhance photosynthetic efficiency and productivity under elevated temperatures.
Membrane stability is crucial for maintaining cellular integrity during heat stress. Research has highlighted the role of ethylene-responsive transcription factors in regulating membrane stability in cotton under high-temperature stress (Liang et al., 2021). Moreover, Zha et al. (2023) described how overexpressing genes such as VvDREB2c, HSP70, and HsfA2 improve heat tolerance in Arabidopsis. The role of salicylic acid in heat tolerance has also been explored through gene editing. Researchers demonstrated that exogenous application of salicylic acid could enhance heat tolerance in waxy maize by modulating the expression of heat shock response genes (Guo et al., 2022).
Another example isthat engineering SlHyPRP1 protein domains in tomatoes has improved multi-stress tolerance, including heat stress. Genome-edited tomato lines have demonstrated better germination and vegetative growth under heat-stress conditions, showcasing the potential of these edited genes to enhance heat resilience (Tran et al., 2023). Moreover, a notable case involves the overexpression of the heat shock factor TaHsfA6f in wheat, which resulted in increased ABA levels, thereby improving tolerance to multiple abiotic stresses, including heat (Bi et al., 2020). In rice, researchers have exploited quantitative trait loci (QTLs) associated with heat stress tolerance, targeting candidate genes related to heat shock proteins (HSPs) and calmodulin-binding proteins to enhance heat tolerance (Kilasi et al., 2018). Moreover, in Maize, the Complex Trait Loci (CTL, Figure 3) have been used for novel trait development. The role of ABA in mediating heat stress tolerance has also been explored. ABA is critical in preventing pollen abortion under high-temperature stress in rice spikelets, and manipulating ABA signaling pathways through gene editing could enhance heat tolerance during critical reproductive stages (Rezaul et al., 2018). Additionally, research has highlighted the proactive role of heat shock factor C2a in wheat, which operates through an ABA-mediated regulatory pathway to protect developing grains from heat stress (Hu et al., 2017). By enhancing the expression of this transcription factor using CRISPR/Cas9, researchers can potentially improve heat tolerance in wheat during crucial growth stages, thus safeguarding yields in hot tropical climates.
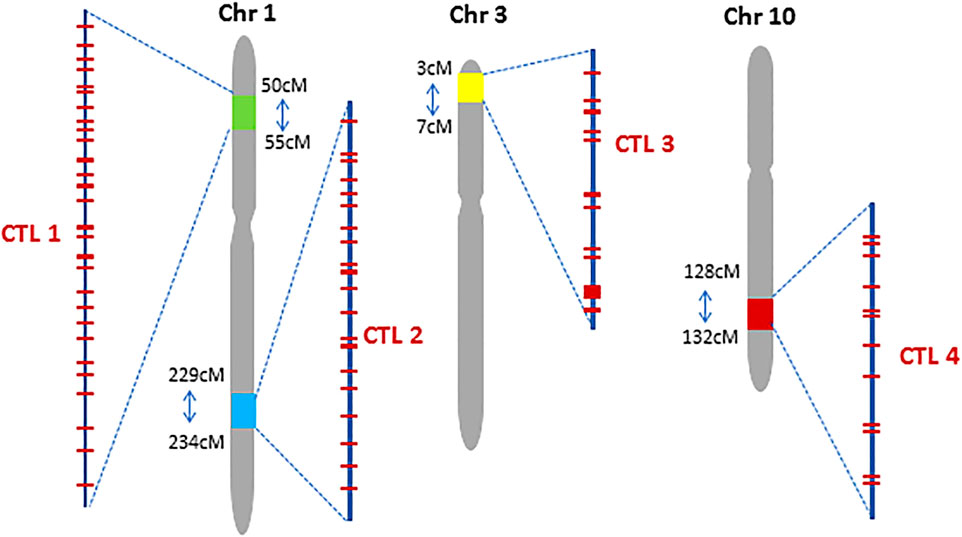
Figure 3. Chromosomal location of four Complex Trait Loci (CTL) in the maize genome. Red bars within each CTL represent preselected CRISPR targeting sites (Figure from Gao et al., 2022; Copyright: CC BY License).
Gene editing for salt tolerance
By stacking multiple edited genes, researchers can greatly improve crop resilience to various stressors, such as drought, salinity, and heat. This approach synthesizes findings from numerous studies that focus on integrating multiple traits for robust climate resilience. The combined editing of genes to enhance stress tolerance offers a promising strategy for developing crops that can withstand challenging environmental conditions.
Salt stress is a major abiotic challenge that significantly impacts agricultural productivity, making the enhancement of salt tolerance in crops essential for ensuring food security in saline areas. Plants manage salt stress through mechanisms that regulate ion homeostasis, particularly involving sodium (Na+) and potassium (K+) transporters. One study highlighted that using CRISPR/Cas9 to mutate GmAIT genes in soybeans improved salinity tolerance (Wang T. et al., 2021). The AITR transcription factors play a key role in controlling ion transport and homeostasis. By knocking out these factors, researchers observed improved K+/Na+ ratios in soybean plants, which are crucial for maintaining cellular function during salt stress. This example demonstrates how precise gene editing can directly modify ion transport pathways to enhance salt tolerance in crops.
In rice, the OsCIPK9 gene has been identified as a crucial regulator of sodium ion homeostasis. It interacts with OsSOS3, affecting salt-related transport and improving salt tolerance (Zhou et al., 2023). Editing the genomic region of OsCIPK9 could create new alleles that enhance the plant’s ability to manage sodium levels during salt stress. Osmotic adjustments are another critical aspect of salt tolerance. The CRISPR/Cas9, gene editing approach, can enhance the expression of specific transporters involved in osmotic regulation, thereby improving the plant’s ability to maintain turgor pressure and cellular integrity under saline conditions. This strategy is particularly relevant for crops grown in saline soils, where osmotic stress can severely limit growth and yield. Furthermore, the integration of multi-omics approaches, such as comparative transcriptomic analyses, has helped identify essential genes associated with salt stress response in sorghum (Jeon et al., 2023). These insights can be exploited to edit targets that enhance salt tolerance through various mechanisms, including ion homeostasis, osmotic adjustments, and oxidative stress management (Yuan et al., 2024).
Oxidative stress response is also vital for plant adaptation to salt stress. MYB transcription factors regulate ion homeostasis and control reactive oxygen species (ROS) levels during salt stress (Ahmad et al., 2023). Editing these transcription factors with CRISPR/Cas9 can accelerate a plant’s ability to manage oxidative stress, thereby improving overall salt tolerance. Additionally, a study reviewed various transgenic approaches to improve crop salt tolerance, emphasizing the importance of pyramiding multiple genes involved in salt stress responses (Hsu et al., 2020; Kotula et al., 2020; Liu et al., 2021). By combining gene editing strategies with traditional breeding methods, researchers can develop crops with enhanced resilience to salinity, ultimately leading to improved agricultural productivity in saline environments. Understanding the genetic basis of salt tolerance in halophytes is also necessary. Researchers identified some target genes in glycophytes (Grigore and Vicente, 2023) and could exploit opportunities through gene editing to confer resilience to salinity.
One prominent example is the study focusing on the GhA08G1120 (GH3.5) gene in cotton. Researchers demonstrated that suppressing this gene resulted in enhanced drought and salt stress tolerance (Kirungu et al., 2019). The GH3.5 gene is involved in auxin metabolism, and its manipulation led to increased proline accumulation, which plays a crucial role in osmotic adjustment and the detoxification of reactive oxygen species (ROS). The OsMYB6 gene has been identified as a key regulator of abiotic stress responses in rice. Overexpressing OsMYB6 in transgenic rice improved resistance to drought and salinity stress (Tang Y. et al., 2019). This study highlighted several abiotic stress-related genes, including OsLEA3 and OsDREB2A, known to enhance stress tolerance. These examples underscore the potential of targeting transcription factors through gene editing to improve salt tolerance in crops.
The GmST1 gene in soybeans has also shown promise; it reduces ROS production and enhances sensitivity to ABA during salt stress. Overexpressing GmST1 in Arabidopsis improved drought tolerance and reduced water loss from leaves (Ren et al., 2016; Ma et al., 2021). Additionally, the role of auxin in salt tolerance has been explored through the CqEXPA50 gene identified in quinoa. Manipulating this gene, which is involved in auxin-mediated responses, enhances salt tolerance by improving cell expansion and osmotic adjustment (Sun et al., 2022; Zhang et al., 2021).
Targeting multiple genes simultaneously facilitates the pyramiding of traits, including enhanced root architecture, improved water use efficiency, and increased resistance to pests and diseases. This approach accelerates breeding and increases the likelihood of developing crops that can thrive under changing climatic conditions. The importance of root traits in enhancing crop resilience has also gained attention. Modulating root hair development can optimize nutrient and water uptake, thereby improving crop yield and resilience (Tsang et al., 2023). By targeting key genes that control root hair development through gene editing, breeders can enhance crops’ ability to access water and nutrients in challenging environments, further contributing to climate resilience. In addition to root traits, stacking traits related to osmotic adjustment and oxidative stress response are essential for developing resilient crops. Research is exploring the potential of multiplex-CRISPR gene editing constructs to accelerate genetic gains in underutilized crops, focusing on traits that enhance tolerance to abiotic stresses (Sharma et al., 2022).
Recent advancements in genome editing
The CRISPR variants Cas12 and Cas13 offered improvements in specificity and reduced off-target effects compared to Cas9 and proved to be a highly efficient gene editing tool with complementary properties and functionality to Cas9 (Zetsche et al., 2017; Zhang et al., 2017). The Cas12a does not require a tracrRNA for activation and is potentially exploited for multiplexed editing (Martin et al., 2024). Additionally, Cas12a′s trans-nuclease activity is utilized to detect sequence-specific nucleic acid (Chen et al., 2018). In contrast, Cas13 is an RNA-targeting enzyme useful for modifying gene expression at the transcript level.
The first-generation CRISPR-based gene editing tools face several challenges, including specificity, targeting scope, dependency on endogenous DSB repair mechanisms, absence of efficient delivery methods, lack of effective vectors and rigidity of cell or organisms, off-target activity, etc. (Kadam et al., 2018; Martin et al., 2024). Also, the requirement of a specific PAM Sequence for targeted genome editing is crucial, and even though it reduces off-target activity, it often results in suboptimal DNA cleavage, editing efficiencies (Ran et al., 2015; Edraki et al., 2019) and restricting the scope of targeted edition. Recent advancements in gene-editing technologies have led to the development of artificial Cas9 variants with relaxed PAM specificities, which improve efficiency but are associated with an increase in off-target effects and a decrease in target specificity (Ran et al., 2015). The control as well as stimulating, as well as enhancing efficiency of HDR within a coRanntrolled editing system ruling out adverse editing outcomes, especially large deletions, chromosomal rearrangements, and even chromosome loss (Kosicki et al., 2018; Cullot et al., 2019; Leibowitz et al., 2021).,Enhancement of HDR methods pertains touse of asymmetric ssODN templates (Richardson et al., 2016), Silent mutations introduction to obstruct recurrent cleavage at the target site (Paquet et al., 2016), tethering of the repair donor template to the break site (Carlson-Stevermer et al., 2017),and manipulation of the cell cycle combined with the delivery of pre-assembled Cas9-ribonucleoparticles (Lin et al., 2014).
The delivery of existing Cas9 or Cas12a enzymes and their guide RNAs are available in different methods, among which electroporation (nucleofection) or liposome-mediated transfection remain the methods of choice, and it is delivered in the form of RNA, plasmid DNA, or in ribonucleoprotein (RNP)complexes. The delivery of the enzymes expressed the limitations of less efficiency, tissue-specific, immunogenicity, and species-specificity, etc. (Chen et al., 2020; Cheng et al., 2023). The limitations of first-generation gene editing techniques have been enhanced by the latest evolved versatile new tools, including precision editing, safety concerns, and minimal unintended editing consequences. The currently available technologies provide amuch more custom-made approach to genome editing, with specificity for certain types of edits or delivery methods.
High-fidelity Cas9 variants
The variant SpCas9 has been developed to resolve the issue of off-target activity and improve specificity using two complementary approaches. Several other variants of Cas9 and Cas12a enzymes have been developed with improved specificity and efficiency, but their efficiency and specificity may vary depending on the different target DNA and utility (Kim et al., 2022). Nevertheless, each of the high-fidelity variants is not universally acceptable and possesses certain limitations (Martin et al., 2024).
Guide RNA modifications
Modifications to the guide RNA (gRNA) have been developed to reduce off-target effects, though these changes often come at the cost of editing efficiency. Various approaches have been explored, including truncated gRNA, 5′end modifications, hybrid RNA-DNA guides, and nucleotide substitutions within the gRNA sequence. Truncating the gRNA from 20 nucleotides to 17–18 nucleotides has been shown to reduce off-target activity (Fu et al., 2014), though it can also decrease editing efficiency (Sai, 2021). Modifications at the 5′end of the gRNA, such as the addition of secondary structures (Kocak et al., 2019) or unpaired nucleotides (Kulcsár et al., 2020), have also been found to lower off-target activity. The use of “hybrid” RNA-DNA guides has demonstrated significant improvements in gene editing specificity (Donohoue et al., 2021). Additionally, chemical modifications involving 2′-O-methyl or 2′-fluoro nucleotides and phosphorothioate linkages in the gRNA have proven to be effective strategies for enhancing both the specificity of Cas9 and the stability of the gRNA (Martin et al., 2024).
Alternative PAM genome editors
The requirement of aspecific PAM recognition site for CAS nucleases is becoming the major constraint in genome editing in several species. In this concern, to get rid of the availability of specific PAM sites, the modified variants of SpCas9 and Cas12a have been developed. The modified variants of SpCas9 and Cas12a bypass the need for specific PAM sites. Efforts to expand the PAM targeting range of SpCas9 have included structure-based rational engineering and amino acid substitutions in its PAM-interacting domain. These modifications have led to the creation of SpCas9 variants such as VQR, EQR, and VRER, which can recognize broader PAM sequences like NGAN, NGNG, and NGCG, respectively (Kleinstiver et al., 2015; Hirano et al., 2016).
Recent advancements have also focused on engineering SpCas9 variants that can target NRN PAM sequences, significantly broadening their application potential. Efforts are ongoing to develop PAM-free Cas9 molecules to eliminate PAM site constraints altogether, thereby increasing the flexibility and effectiveness of genome editingthrough structure-based rational engineering or by altering amino acid substitutions in the PAM-interacting domain (Walton et al., 2020). This could result in the development of wider-ranged VQR, EQR, and VRER SpCas9 variants, enabling the targeting of NGAN, NGNG, and NGCG PAMs, respectively (Kleinstiver et al., 2015; Hirano et al., 2016).
Base editing
The discovery of spontaneous hydrolytic deamination of cytosine converting C-G base pairs to T-A in humans highlighted the potential of base editing for direct base pair conversion without requiring double-strand breaks or homology repair templates (Komor, et al., 2016). Base editors consist of an inactive CRISPR/Cas9 (dCas9/Cas9 nickase) and a deaminase (cytosine or adenosine). They are classified into DNA and RNA-based editors. Current DNA base editors include cytosine base editors (CBEs) and adenine base editors (ABEs) for C-T and A-G conversions. First-generation base editors had limitations such as low efficiency and base excision repair (BER) activity that reversed edits (Komor et al., 2016). Second-generation editors, such as APOBEC-XTEN-dCas9-UGI, added an uracil DNA glycosylase inhibitor (UGI) to improve C-to-T conversion (Komor et al., 2016) but still had <0.1% indel formation, limiting precision (Azameti and Dauda, 2021).
Third-generation base editors, with rAPOBEC1 fused to the N-terminus and UGI at the C-terminus of nickase Cas9 D10A, offered higher editing efficiency and reduced off-target effects but required an NGG PAM sequence, limiting their scope (Kim et al., 2017). Fourth-generation editors (SpBE4, SaBE4) improved upon this by adding two UGI molecules to the C-terminus. The BE4-GAM variant further enhanced efficiency by fusing the Gam protein (a DNA end-binding protein) to the Cas9 nickase N-terminus.
The adenine base editor (ABE) comprises three main components: a mutant transfer RNA adenosine deaminase (TadA), sgRNA, and Cas9 nickase, facilitating A-to-G conversions (Gaudelli et al., 2017). To enhance editing efficiency and minimize off-target effects, approximately eight ABE variants have been developed. The ABE-Plant version 1 Simplified (ABE-P1S) demonstrated higher editing efficiency in rice compared to the commonly used ecTadA-ecTadA*7.10-nSpCas9 (D10A) fusion (Hua et al., 2020). In developing herbicide-resistant commercial rice, TadA9 showed compatibility with multiple nickase systems, including CRISPR/SpCas9, CRISPR/SpCas9-NG, CRISPR/SpRY, and CRISPR/ScCas9, achieving high editing efficiency across four herbicide target genes.
A dual-base editing system was created by fusing both cytidine and adenosine deaminases to Cas9, allowing simultaneous C→T and A→G substitutions using a single sgRNA within one target site (Li et al., 2020b). Addressing the limitations of ABEs and CBEs, which can only achieve up to 33% of possible base substitutions, researchers developed Transversion Base Editors to expand editing capabilities. Notably, Zhao et al. (2022a) introduced glycosylase base editors (GBEs), which enable C-to-A and C-to-G conversions.
PAM-less base editing
PAM-less base editing has expanded the possibilities in genome editing by providing access to previously unreachable PAM sequences. Normally, SpCas9 recognizes the ‘NGG’ PAM sequence; however, researchers have developed variants that reduce this specificity, enabling recognition of a single guanine (G) nucleotide. These include variants like xCas9-3.7, SpCas9-NG, and ScCas9 (Azameti and Dauda, 2021). Building on this, Walton et al. (2020) introduced the SpG variant, which can recognize a broader range of NGN PAMs. Further optimization led to SpRY, a variant capable of targeting nearly any PAM. Although this technology significantly broadens the targetable genome space, it has limitations. For example, SpCas9-NG shows reduced editing efficiency on 5′-NGC-3′ PAM targets and tends to increase off-target activity (Nishimasu et al., 2014).
Multiplex base editing systems
It provides a multifunctional CRISPR system that performs dual and tri-functional base editing but suffers from the requirement of each Cas protein specific to its own PAM sequence (Lian et al., 2017). The primer base editors CBE and ABE have been improved a lot through several modifications with increased specificity and reduced deaminase—induced off-target activity (Doman et al., 2020). Further base editing option has been expanded beyond ABE and CBE and covers A-to-C, A-to-Y and C-to-G (Martin et al., 2024) edits. Being an alternative to CRISPR/Cas nine and efficient system (Li M. et al., 2022; Veillet, et al., 2019), base editing catches the universal attraction of the scientist for functional genomics studies and in the quest of mining the important traits governing SNPs in various crop plants (Azameti and Dauda, 2021), improved crop varieties could be developed by the programmed and precise conversion of targeted single bases in the genomes of plants. It has been utilized for several applications, including single point mutations and targeted mutation induction for therapeutic corrections of diseases (Musunuru et al., 2021), mutational screening, and gene knockout among the genomes (Hanna et al., 2021; Que et al., 2010). Besides controlled editing, BEs are facing several limitations, including low efficiency, large editing windows, and off-target activity (Martin et al., 2024). Also, they possessundesired genotoxic effects by generating DSBs, deletions, and translocations at the on-target locus.
Synthetic gene activators
Synthetic gene activators are also a promising strategy that activates genes by tethering an autonomous transcription activation domain (TAD) to the gene promoter via a programmable DNA-binding module. Various Synthetic gene activators are available, including dCas9-TADs, zincfinger protein–TADs, and transcription activator-like effector (TALE)–TADs. Among which, the nuclease-dead S. pyogenes Cas9 (dCas9) protein provides simplicity and multiplexity (Didovyk et al., 2016). Further advancements in genome editing technologies have developed deactivated Cas9 (dCas9) based transcriptional activation systems (Li et al., 2017). In animal cells, the dCas9-based transcriptional activation systems, like VPR, SAM, and SunTag, are being utilized, while in plant cells, dCas9–TV system offers stronger transcriptional activation of single or multiple genes (Li et al., 2017). This method is particularly useful for correcting point mutations that cause genetic diseases or undesirable traits. For instance, base editing has improved disease resistance and enhanced nutritional profiles in crops like tomatoes and potatoes.
Prime editing
Prime editing is a more recent development that enables precise insertion, deletion, or replacement of DNA sequences with high accuracy. It uses a “prime editor” complex that includes a modified CRISPR protein and an engineered reverse transcriptase. This technology holds promise for fixing a broader range of genetic mutations with fewer unintended effects, potentially improving traits such as yield, pest resistance, and environmental resilience (Liu, et al., 2023). The prime editor system contains a fusion of nCas9 (H840A) and reverse transcriptase enzyme derived from Moloney murine leukemia virus (M-MLV RT), prime editing guide RNA (pegRNA)/sgRNA consisting of a reverse transcriptase template, and a primer-binding site at the 3' end of the sgRNA. It produces indels and base replacements without the limitations of specific PAM. Prime editing has been extensively utilized in rice, wheat, and maize and induced point mutations, deletions, and insertions (Lin et al., 2020; Xu et al., 2020b).
Epigenome editing
Epigenome editing involves modifying the epigenetic marks on DNA, such as DNA methylation or histone modification, to influence gene expression without altering the underlying DNA sequence (Kumar and Mohapatra, 2021). This approach can be used to fine-tune gene expression levels, which may help in optimizing crop traits like flowering time and stress responses. This can be achieved through fusion between nuclease-dead Cas9 with DNA and histone-modifying enzymes, which restructure the chromatin at precise loci of the genome and enable induction or repression of expression of the target gene (Nuñez et al., 2021). It can be achieved through CRISPR off as well as CRISPR on mechanism enabling silencing as well as reactivation of gene expression respectively (Nunezet al., 2021). It suffers from the challenges of binding Cas9 to off-target sites and influencing histone and chromatin modifiers, which inadvertently affect the transcription of off-target genes (Kuscu et al., 2014).
Bio-mimicking via promoter, allele, or gene replacement
The introduction of foreign DNA/genes into crops is treated as transgenic and often faces the challenges of lengthy regulatory concerns and consumer refusal. Such constraints can be overcome by CRISPR technology working on the principle of bio-mimicking. It offers the introduction of mutation instead of the whole gene using CRISPR, wherein the sequence of the target gene is converted into a desirable gene sequence that has a specific trait. It is being achieved through gene silencing or gene knockout via induced mutation within gene sequence and its replacement within the cultivated species, induction of mutations within allele as well as promoter region of the gene (Tan et al., 2022).
Emerging methods for efficient CRISPR delivery in plant systems
The advancement of plant genome editing through CRISPR technologies hinges critically on developing efficient delivery mechanisms that can overcome the inherent challenges posed by plant cellular structures. Researchers are actively exploring diverse strategies to successfully introduce CRISPR components into plant cells, addressing the complexities introduced by rigid cell walls and intricate genomic landscapes.
Non-viral delivery methods have emerged as a promising avenue for CRISPR component transmission. Innovative nanotechnology-based approaches, including inorganic nanoparticles, carbon nanotubes, liposomes, and protein- and peptide-based nanoparticles, offer significant advantages over traditional delivery techniques. These vectors demonstrate reduced immunogenic responses and lower cytotoxicity, making them particularly attractive for genetic modification strategies (Alghuthaymi et al., 2021).
Viral vector delivery, specifically virus-induced genome editing (VIGE), represents another sophisticated approach to CRISPR component introduction. By utilizing plant RNA viruses as transient delivery vectors, this method enables high-efficiency editing and facilitates the generation of DNA-free gene-edited plants. VIGE is especially valuable for achieving tissue-culture-free editing and enhancing plants’ biotic resistance mechanisms (Uranga and Daròs, 2022; Zhang C. et al., 2022).
Ribonucleoprotein (RNP) delivery has gained significant attention as a refined method for CRISPR component transmission. This approach minimizes potential risks associated with transgene integration and off-target effects, providing a safer alternative to conventional plasmid-based methodologies. Researchers have demonstrated impressive editing efficiencies using RNPs across various plant species, including rice and citrus, underscoring the method’s potential for generating transgene-free genome-edited plants (Zhang Y. et al., 2022; Zhang et al., 2019).
Traditional transformation techniques, such as Agrobacterium-mediated transformation and particle bombardment, continue to play a crucial role in CRISPR delivery. While these methods have historically been successful in a limited number of plant species and often require extensive tissue culture and regeneration procedures, ongoing research aims to enhance their capabilities. Current developments focus on achieving genotype-independent delivery and implementing DNA-free editing protocols (Laforest and Nadakuduti, 2022; Ghogare et al., 2021; González et al., 2021).
Despite considerable progress in developing novel delivery methods, significant challenges persist in establishing universally efficient and scalable CRISPR delivery mechanisms across diverse plant species. Key research priorities include improving plant regeneration from edited protoplasts and developing genotype-independent delivery technologies. Future research directions will concentrate on optimizing existing methodologies and exploring innovative approaches to enhance the precision and efficiency of CRISPR-based plant genome editing (Laforest and Nadakuduti, 2022; Nadakuduti and Enciso-Rodríguez, 2021; Erdoğan et al., 2023).
This multifaceted approach to CRISPR delivery reflects the dynamic and evolving landscape of plant genome editing, promising transformative potential for agricultural innovation and crop improvement.
Off-target effects in CRISPR/Cas technology and their mitigation strategies
Off-target effects are unintended genomic modifications occur when guide RNA (gRNA) binds to sequences similar to, but not identical with, the intended target, potentially resulting in undesirable DNA cleavage and mutations (Guo et al., 2023; Lopes and Prasad, 2024; Garrigues et al., 2023).
The fundamental mechanism of off-target effects stems from the gRNA’s ability to interact with non-target genomic sites sharing sequence homology. Consequentially, these interactions can generate diverse genetic alterations, including small insertions or deletions (indels), structural variations like translocations, inversions, and extensive deletions. Such modifications pose substantial risks, particularly in therapeutic applications (Lopes and Prasad, 2024; Mengstie et al., 2024). In bacterial systems, these off-target interactions can further precipitate gene silencing and cellular toxicity, underscoring the critical importance of precise molecular targeting (Rostain et al., 2023).
Researchers have recognized these challenges and developed sophisticated mitigation strategies to enhance CRISPR/Cas9’s safety and efficacy. Bioinformatics tools have been instrumental in this endeavor, employing computational approaches to optimize gRNA design, ensuring high specificity and minimizing off-target activity. These computational strategies assist researchers in strategically selecting target sites and validating experimental outcomes (Naeem and Alkhnbashi, 2023; Zhang et al., 2023).
Innovative technological approaches have further expanded the toolkit for managing off-target effects. Researchers have explored strategies such as utilizing Non-Homologous End-Joining (NHEJ)-deficient strains in organisms like filamentous fungi, which have demonstrated significant reductions in off-target mutations and enhanced genomic stability (Garrigues et al., 2023). Machine learning and deep learning models have emerged as powerful predictive tools, enabling researchers to anticipate and comprehend potential off-target activities with unprecedented precision (Vora et al., 2023).
Perhaps most promising are cutting-edge technological interventions like G-quadruplex-based CRISPR photoswitches. These innovative mechanisms provide spatiotemporal control over CRISPR activity, allowing researchers to precisely activate and deactivate the CRISPR system. Such controlled approaches substantially mitigate off-target risks by enabling more targeted genetic interventions (Deng et al., 2023).
Continuous technological advancements have been paramount in addressing these challenges. The development of high-fidelity Cas9 variants and strategically modified gRNAs represents a significant stride toward minimizing off-target effects, progressively refining the precision of gene editing technologies (Mengstie et al., 2024).
While off-target effects remain a substantial concern in CRISPR/Cas9 applications, particularly in therapeutic contexts, the multifaceted approach to mitigation demonstrates remarkable scientific ingenuity. Researchers are systematically addressing the technology’s inherent challenges by integrating computational prediction, technological innovation, and sophisticated molecular control mechanisms. These advancements not only enhance the safety and reliability of CRISPR/Cas9 but also expand its potential applications across diverse scientific and medical domains.
Application of artificial intelligence and phenomics in genomic editing
Artificial intelligence (AI) constitutes machine-learning algorithms, for example, deep neural network (DNN), artificial neural network (ANN), random forest (RF), support vector machine (SVM), and advanced hi-tech equipment like the internet of things (IoT) (Baduge et al., 2022). It fascinates a hi-tech system that is capable of handling big data in a short time and making judgments rapidly and more accurately than humans (Xu Y. et al., 2023). It saves breeders time in data identification and processing. It accelerates advanced breeding through the use of high-throughput genomics and phenomics, leading to the breeding of next-generation resilient crops (Khan et al., 2021). Moreover, machine learning (ML) tools help in genomic prediction, genomic selection, deep learning, and predictive analysis help to increase the planning, learning, reasoning, thinking, and action-taking abilities, enabling digital breeding and developing next-generation crops (Shaw et al., 2022).
Machine learning (ML) tools arebeing proposed to predict off-target placement throughout crop genomes. It also enables the training and test the possible regression points that predict off-target or on-target specificities (Kaul et al., 2020). AI is helping to overcome phenomics bottlenecks; it manages phenotypic data through algorithms and programs that convert sensory data into phenotypic information. It helps in model development and enables the understanding ofgenotype-phenotype relationships with interaction with different environmental conditions (Nabwire et al., 2021). AI assists in plant phenomics systems by identifying plant developmental stages, plant images, categorization of the crops and weeds, scoring and images of the diseases of crops, etc. (Khan et al., 2021). Moreover, AI has been used in gene function analysis. The available AI algorithms may be utilized to predict the cleavage ability of theCRISPR system. AI tools are extensively utilized in genome exploration, including the identification of protein-coding genes, regulatory elements, cell-to-cell gene expression and location, protein-protein interaction networks, and metabolic pathways. Prediction and analysis of the genetic features of an individual are becoming possible through the DeepSEA and DeepBind models (Zhou et al., 2023). The various laboratory data point features such as GC content of gRNA, the secondary structure of sgRNA, CpG island, chromatic structure, ORF sequence, the gene expression profile of various developmental stages, available sgRNA and their distributions, sequence and protein databases with detailed descriptions can be linked to formulating AI models in gene editing.
Thus, integrated use of Artificial intelligence (AI) technology assisted with phenomics and genomics tools combined with genome editing tools offers projective modeling in precision editing, reduced off-target activity, minimizing unintended genetic alterations, and increased editing efficiency with promising outcomes (Xiang et al., 2021). It is being used in the coming future to predict and mine more powerful and efficient specific nucleases through data analysis, modeling, and computational biology approach. However, the technology has come up with its own challenges, like the availability of qualitative and variable data, suitable predictions, interpretations, efficient decision-making systems, etc.
Prospects of multiple traits development in climate resilience
Stacking multiple traits in crops through gene editing technologies presents a promising avenue for developing comprehensive climate resilience. However, this approach also faces several challenges and considerations that must be addressed to ensure successful implementation. This response synthesizes insights from various studies on the complexities of pyramiding multiple edited genes for combined stress tolerance.
One primary challenge in stacking traits is the potential for negative interactions between different genetic modifications (Zonneveld et al., 2020). When multiple traits are introduced through gene editing, the interactions between these traits could lead to unforeseen physiological responses, potentially diminishing the overall effectiveness of the stacked traits (Malenica et al., 2021). Additionally, there is a risk of increased susceptibility to certain diseases or reduced fitness under specific environmental conditions, which poses a significant challenge in developing multi-trait crops (Panda et al., 2024; Dormatey et al., 2020).
A significant technical hurdle is the high-throughput mutant genotyping required to identify and confirm the presence of multiple edited genes in a single plant (Karunarathne et al., 2023). The complexity of plant genomes and gene interactions complicates the breeding process, as one gene’s expression can influence another’s expression, leading to unintended phenotypic outcomes (Kumar, 2023). Thus, there is a pressing need to understand the interactions between different stress response pathways to mitigate potential risks.
Another layer of complexity is the intricate nature of plant responses to environmental stresses. Plant responses to climate change can often contradict empirical expectations, making it challenging to predict how stacked traits will perform in real-world conditions (Putra et al., 2022). This complexity necessitates comprehensive modeling approaches and field trials to assess the effectiveness of stacked traits under varying environmental conditions. Additionally, the genetic background of the crop plays a critical role in the success of trait stacking, highlighting the importance of understanding the genetic architecture of the crop for successful trait pyramiding.
Considering the potential unintended consequences of a multiplex gene editing approach requires careful planning and robust solutions. While significant advances have been made in developing varieties tolerant to specific stresses, most efforts have focused on monogenic or oligogenic traits (Prohens et al., 2017). Furthermore, the socio-economic context surrounding the adoption of stacked trait varieties must be addressed, emphasizing effective communication and education about the benefits of these traits among end-user farmers.
Gene pyramiding has been effectively demonstrated in several crops, including rice, maize, and barley. Using CRISPR/Cas9 gene editing technologies, barley abiotic stress tolerance has been enhanced by targeting specific genes associated with drought and salinity resistance (Karunarathne et al., 2023; Yadav et al., 2023). Similarly, in maize and sorghum, a multi-trait improvement approach with resilience to acidic soils is ongoing (Guimarães and Magalhães, 2021). Regulatory considerations also play a critical role in deploying gene pyramiding strategies. The approval processes for genetically modified organisms (GMOs) vary significantly across countries, and the introduction of multi-gene constructs may face additional scrutiny due to concerns over environmental impact and food safety (Fan, 2024; Suza et al., 2018). This regulatory landscape can hinder the rapid adoption of innovative breeding techniques, delaying the availability of improved crop varieties to farmers.
Despite these challenges, the potential benefits of gene pyramiding for climate resilience are substantial. The pyramiding of genes associated with drought tolerance, salinity resistance, and nutrient acquisition has enhanced the overall resilience of crops like rice and maize (Pang et al., 2017; Shailani et al., 2020). Combining multiple stress tolerance traits allows a researcher to develop crop varieties that survive and thrive under adverse conditions. This comprehensive approach improves individual plant performance and contributes to greater agricultural sustainability by reducing the need for chemical inputs and enhancing soil health.
Multiplex genome editing and stacking multiple alleles with CRISPR/Cas can accelerate plant genetic improvement. However, the emergence of CRISPR crops has sparked international debates on regulation, risks, and differences between CRISPR-edited crops and GMOs. CRISPR has significantly reduced the cost of producing genome-edited crops, reshaping agriculture with precisely edited varieties (Bartkowski et al., 2018). Despite the economic benefits, concerns remain about unintended genome modifications and regulatory oversight due to the non-specific binding of sgRNA. The debate over legal, ethical, and policy issues surrounding CRISPR crops continues, with differing regulatory frameworks in different countries (Kondo and Taguchi, 2022). The international community is grappling with questions about regulatory oversight, safety data requirements, and the distinction between CRISPR-edited crops and GMOs (Zannoni, 2019). The regulatory landscape for CRISPR-edited crops is complex, with varying approaches to assessing and regulating these products. Developers and investors must navigate regulatory requirements before investing in CRISPR-edited crops. The choice of reagents and delivery methods for CRISPR/Cas systems can impact the final products and regulatory requirements. Understanding these factors is crucial for ensuring the acceptance and regulation of CRISPR-edited crops in the market (Ahmad M. et al., 2021).
Regulatory aspects and biosafety concerns
Conventional mutagenesis and genetic engineering involve random genetic modifications that can lead to unintended changes in the genome, potentially disrupting genes or regulatory elements. This often requires extensive screening and regulatory approval (Monarkh, 2020). New genome editing (GE) technologies possessing site-directed nucleases (SDNs) offer precise control over random genome modifications and mutations. Genome editing techniques, such as ZFNs, TALENs, and CRISPR/Cas, allow for targeted changes in the genome, resulting in cost-effective and efficient trait introduction without the need for exogenous genetic material.
Genome-edited mutants are classified into three types—SDN1, SDN2, and SDN3 (Figure 4), based on the nature of genome modifications and double-strand break (DSB) repair outcomes (Zannoni, 2019) SDN1 mutants involve precise point mutations that disrupt or silence genes without requiring a repair template. Although off-target effects in SDN1 mutants are debated, the resulting changes are similar to natural mutations (Zannoni, 2019; Sprink et al., 2016). SDN2 mutants, on the other hand, utilize a repair template to modify one or a few bases at a DSB site, yielding outcomes that mimic natural mutations and can be achieved with traditional breeding. However, SDN2 is limited for larger insertions due to its reliance on microhomology-mediated end joining (Verma et al., 2020). SDN3 mutants use a repair template with homologous regions of at least 500 base pairs flanking the DSB, enabling the insertion of new sequences, similar to transgenic or cisgenic approaches. Nonetheless, SDN3 applications in plants face challenges due to the low efficiency of homologous recombination (HDR), making them complex and less efficient (Verma et al., 2020; Podevin et al., 2013; Van de Wiel et al., 2017).
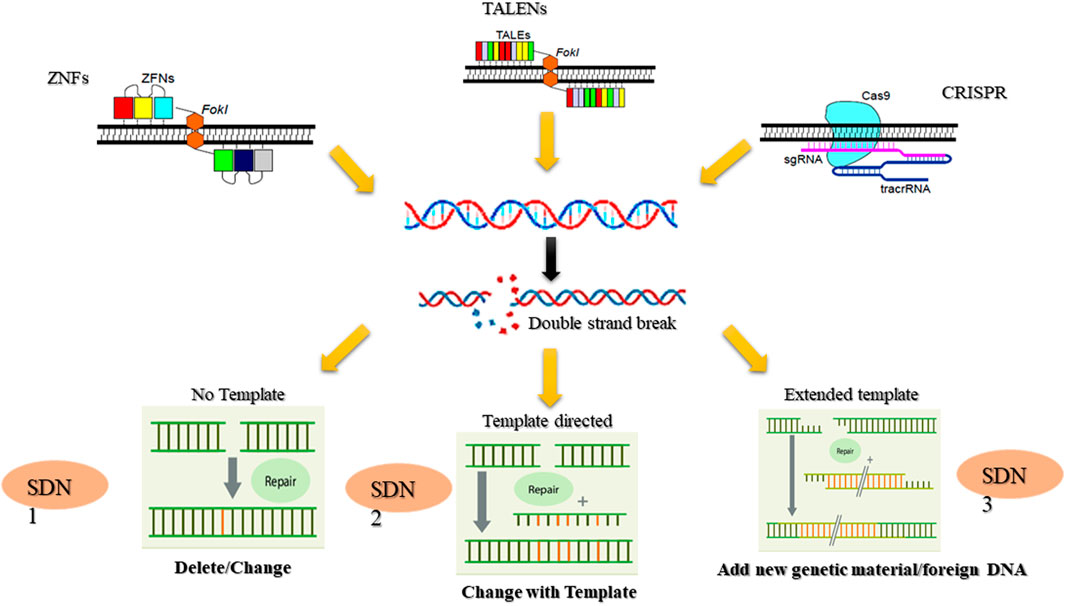
Figure 4. Genome editing and mechanisms using different tools, showing the occurrence of SDN1, SDN2, and SDN3 events.
The commercial use of genome editing technologies is prejudiced by national legislation in genetic engineering and biotechnology, as well as facing the obstacle of consumer acceptance. Genome editing techniques in plant breeding have evolved, with CRISPR/Cas systems being widely adopted. The rapid development of genome editing poses challenges to existing regulations regimes worldwide. Current regulations may differentiate between genome editing techniques (SDN-1, SDN-2, SDN-3) and the use of foreign DNA or sequence templates in the editing process (Kulcsár et al., 2020).
Public concerns about environmental and health risks drive the need for consistent global regulations. Public acceptance of genetically modified crops can be improved through education, transparent regulatory processes, and ongoing risk assessments (Eckerstorfer et al., 2019). The CRISPR/Cas9 system offers a promising approach to developing improved plant varieties with minimal concerns. Transgene-free crops produced by CRISPR/Cas9 may bypass GMO regulations and do not require isolated field tests or labeling in some countries. Regulatory approaches for genetically engineered crops should balance safety, legal definitions, and public acceptance (Hessels et al., 2009). Decision-makers should consider the economic impact and social perceptions of handling genome-edited products under different regulatory scenarios to promote innovation in agriculture while ensuring sustainability. Regulatory policies (Table 2) need to prioritize improving food security and dietary health without compromising environmental or ethical values (Friedman et al., 2023).
Increasing public acceptance of transgene-free crops can be achieved by raising awareness about CRISPR-based crops, building trust in safety regulations and developers, and clearly comparing risks and benefits (Ishii and Araki, 2016). The CRISPR/Cas9 system is considered the most effective method for developing improved plant varieties with minimal concerns. To address off-target issues, using the CRISPR system in the form of a rRNP and utilizing high-fidelity CRISPR variants can help minimize off-target effects (Kleinstiver et al., 2016). Modern technologies like whole genome sequencing can be used to assess potential off-target effects before creating genome-edited organisms. Transgene-free plants produced through CRISPR/Cas9 do not contain foreign DNA in the final product, potentially allowing them to bypass GMO regulations (Zetsche et al., 2017; Kim et al., 2017). This eliminates concerns about transgene flow to non-target species isolated field tests and GMO labeling requirements. However, in some countries, labelling of GM ingredients is mandatory, which can both build public trust and create resistance among certain consumer groups (Ishii and Araki, 2016; Voytas and Gao, 2014). For instance, a field trial of GM grapevine grafting in France faced disruptions from activists despite legal approval. The European Plant Sciences Organization has called for separating safety assessments and environmental risks from the labeling of genome-edited organisms to address these challenges (Zetsche et al., 2017).
DNA tagging in the genome of crops for the cultivation and marketing of genetically engineered organisms (GEOs) has been proposed (Ishii and Araki, 2016). However, this process involves additional gene modification steps and new GMO regulations, increasing costs for developers and companies (Duensing et al., 2018). Establishing appropriate regulatory guidelines is crucial to ensure the safety and legal definitions of genetically engineered crops. Clear regulatory rules can enhance public acceptance of GEOs, promote innovation in agriculture, and facilitate international trade (Whelan and Lema, 2015). Decision-makers should consider the economic impact of different regulatory scenarios for genome-edited products to anticipate social perceptions (Duensing et al., 2018). Political decisions should align with scientific recommendations to prevent excessive regulation that could hinder agricultural innovation and sustainability (Whelan and Lema, 2015). Regulatory policies for GEOs should emphasize the goal of enhancing food security and promoting a healthier diet while respecting environmental, religious, and ethical considerations.
Potential impact on food security and sustainable agriculture
Agriculture today faces newer challenges exacerbated by genetic erosion, the narrow genetic base of commercial crops and environmental degradation. There is an urgentneed to make agriculture more resilient and sustainable while still continuing to develop. The Agriculture sector is facing newer challenges like narrow gene pool and genetic erosion among commercial crops and environmental challenges, etc. This would generate the need to make agriculture more resilient and sustainable by developing high-yielding, stress-tolerant, and climate-smart varieties. The CRISPR gene editing technologies are being harnessed and improved avariety of cereals, legumes, fruits, andunderutilized dryland crops. The CRISPR technologyhas the potential to enhance crop productivity, climate resilience, and nutritional value, and address the global challenges related to food security and sustainable agriculture.
The prospects of Biodiversity are potentially utilizedto improve food and agriculture and mitigate the crisis of hunger and malnutrition. A diverse gene pool in crops can provide valuable traits for improving crop resilience, yield, and nutritional quality. Besides, it allows natural systems to better withstand and recover from environmental stresses, such as diseases, pests, and climate change. However, the decline of natural resources, including crop diversity, alarming the loss of gene pools and may lose the future climate-smart breeding prospects of crop varieties (Lenka et al., 2020). Considering the declining biodiversity resources, serious attention has been paid to food security and the nutrition status of available diets. To ensure the demand of healthy food, there is an urgent need to of biofortification of fruits, vegetables, and cereals with enriched nutritional compounds such as vitamins, amino acids, antioxidants, proteins, minerals and fatty acids, etc.
Current strategies for the improvement of crop varieties for changing climatic conditions include traditional and molecular breeding methods, speed breeding, nanotechnology, mineral fertilization, and transgenic technologies. Recent modern gene editing technologies, such as CRISPR/Cas9, offer highly precise genetic modifications by targeting specific genes for alteration or replacement. This allows for the development of crops with improved traits without introducing foreign genes. CRISPR technology is utilized potentially in crops and is being utilized to improve crop varieties withrespect to nutritional quality, yield, disease and pest resistance, consumer acceptance, economic concerns, and environmental suitability, etc. Novel ARGOS8 edited variants of maize generated through CRISPR/Cas9 showed higher grain yield under drought stress conditions (Shi et al., 2017a; and b). The gene and base-editing strategies has precisely edited granule bound starch synthase gene (StGBSSI) gene in the tetraploid potato and with impaired amylose biosynthesis (Veillet et al., 2019).
Future challenges and perspectives
With the assistance of AI and molecular engineering tools, CRISPR-based gene editing is gradually turning into a third-generation CRISPR-edited crop. The technology is showing tremendous potential in medical, agricultural, and environmental sciences. Particularly in agriculture, it has laid its footprint by generating broad-spectrum resistance against multiple pathogens, nutritional enhancement, etc. The false flax (C. sativa) exhibited enhanced omega-3 oil and was commercially released in the USA, showingrecord-breaking market space (Waltz, 2018). CRISPR modification has been extensively used in crop improvement by de novo meristem induction, targeting and editing efficiency can be enhanced by use of temperature-tolerant CRISPR/LbCas12a, largeerDNAinsertions with precision,use of heat-inducible CRISPR system in maize and breaking of genetic linkage via somatic chromosome engineering, etc., (Zaidi et al., 2020). This is an indicative of CRISPR based gene editing crop that has future potential.
Moreover, the other side of this technology has shown several concerns, particularly less specificity, genome complexity, editing efficiency, off-target effects, regulatory concerns, absence of testing methods, and lack of efficient delivery vectors and methods. In the absence of a transgene/foreign gene, the European Union (EU) is still treating the genome-edited crop as transgenic (Custers et al., 2019). The regulatory landscape for gene-edited crops varies globally. Some regions have adopted more permissive policies for gene-edited crops compared to genetically modified organisms (GMOs), reflecting evolving attitudes toward these technologies. Thusabsence of universal regulatory framework is also a concern affecting the research, cultivation, and marketing of gene-editedproducts. It warrants the need forcomprehensive plans for gene-edited crops from scientists and policymakers.
Apart from this, technological bottleneck, pertaining less to HDR efficiency affects gene replacement or deletions and large chromosomal segments. Third-generation AI-assisted gene editing methods would enable theprediction of on- and off-target edits, designing more powerful genome editors with increased editing efficiency, andaccelerating the pace of implementing safe agricultural use. Even third-generation CRISPR strategies are being introduced to engineer next-generation CRISPR crops, theystill facing the challenges of off-target edits, regulatory concerns, requirement of efficient delivery methods, recalcitrancy during transformation, requirement of extensive field trial for testing the performance.
Conclusion
In conclusion, genome editing technologies, particularly CRISPR/Cas9, present powerful tools for the modulation of positive regulator genes to enhance climate resilience in crops. By targeting key regulatory genes and signaling pathways involved in stress tolerance mechanisms, researchers can develop crop varieties better equipped to withstand the multifaceted challenges posed by climate change. However, a holistic approach that integrates epigenetic regulation, small RNA-mediated processes, and plant-microbiome interactions is crucial for maximizing the potential of these technologies in ensuring global food security in the face of rapidly changing environmental conditions.
Author contributions
RC: Conceptualization, Methodology, Writing–original draft, Writing–review and editing, Supervision. SJ: Writing–original draft, Writing–review and editing, Conceptualization, Methodology, Visualization. VH: Methodology, Writing–original draft, Writing–review and editing, Validation. AD: Writing–original draft, Writing–review and editing, Investigation, Resources. US: Methodology, Validation, Writing–original draft, Writing–review and editing, Formal Analysis. PJ: Methodology, Validation, Writing–original draft, Writing–review and editing. UK: Resources, Writing–original draft, Writing–review and editing, Conceptualization, Formal Analysis, Funding acquisition, Methodology. JH: Writing–original draft, Writing–review and editing, Funding acquisition, Project administration, Resources, Supervision.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by funding from the Establishment of Modern Biotechnology Laboratory for Sustainable Agricultural Development Project through the Chief Minister’s Fund from the Government of Maharashtra (GR No. MKV-1222/Sr. No.164/7A--Budget Allotment Letter # CFB/1774/2023 and Budget Allotment Letter # CFB/316/2024); and funded by the Basic Science Research Program and LAMP program through the National Research Foundation of Korea (NRF) funded by the Korean Government (MSIT) (RS-2022 NR074941 to USK; RS-2020-NR049590, RS-2024-00336624, and RS-2023-00301974 to JCH).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelrahman, M., Wei, Z., Rohila, J. S., and Zhao, K. (2021). Multiplex genome-editing technologies for revolutionizing plant biology and crop improvement. Front. Plant Sci. 12, 721203. doi:10.3389/fpls.2021.721203
Agarwal, M., Hao, Y., Kapoor, A., Dong, C., Fujii, H., Zheng, X., et al. (2006). A r2r3 type myb transcription factor is involved in the cold regulation of cbf genes and in acquired freezing tolerance. J. Biol. Chem. 281 (49), 37636–37645. doi:10.1074/jbc.m605895200
Ahmad, A., Ghouri, M. Z., Munawar, N., Ismail, M., Ashraf, S., and Aftab, S. O. (2021a). “Regulatory, ethical, and social aspects of CRISPR crops,” in CRISPR crops. Editors A. Ahmad, S. H. Khan, and Z. Khan (Singapore: Springer). doi:10.1007/978-981-15-7142-8_9
Ahmad, I., Zhu, G., Zhou, G., Younas, M., Suliman, M., Liu, J., et al. (2023). Integrated approaches for increasing plant yield under salt stress. Front. Plant Sci. 14, 1215343. doi:10.3389/fpls.2023.1215343
Ahmad, M., Ali, Q., Hafeez, M., and Malik, A. (2021b). Improvement for biotic and abiotic stress tolerance in crop plants. Biol. Clin. Sci. Res. J. 2021 (1). doi:10.54112/bcsrj.v2021i1.50
Alghuthaymi, M., Ahmad, A., Khan, Z., Khan, S., Ahmed, F., Faiz, S., et al. (2021). Exosome/Liposome-like nanoparticles: new carriers for CRISPR genome editing in plants. Int. J. Mol. Sci. 22, 7456. doi:10.3390/ijms22147456
Altae-Tran, H., Kannan, S., Demircioglu, F., Oshiro, R., Nety, S., McKay, L., et al. (2021). The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science 374, 57–65. doi:10.1126/science.abj6856
Altae-Tran, H., Shmakov, S., Makarova, K., Wolf, Y., Kannan, S., Zhang, F., et al. (2023). Diversity, evolution, and classification of the RNA-guided nucleases TnpB and Cas12. Proc. Natl. Acad. Sci. U. S. A. 120, e2308224120. doi:10.1073/pnas.2308224120
Anders, C., Niewoehner, O., Duerst, A., and Jinek, M. (2014). Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513 (7519), 569–573. doi:10.1038/nature13579
Arantes, P. R., Sinha, S., Chen, X., Saha, A., Patel, A. C., Sample, M., et al. (2024). Biophysical origin of adenine base editors’ improved editing efficiency. Biophysical J. 123 (3), 340a. doi:10.1016/j.bpj.2023.11.2067
Azameti, M. K., and Dauda, W. P. (2021). Base editing in plants: applications, challenges, and future prospects. Front. plant Sci. 12, 664997. doi:10.3389/fpls.2021.664997
Badhan, S., Ball, A., and Mantri, N. (2021). First report of CRISPR/cas9 mediated dna-free editing of 4cl and rve7 genes in chickpea protoplasts. Int. J. Mol. Sci. 22 (1), 396. doi:10.3390/ijms22010396
Baduge, S. K., Thilakarathna, S., Perera, J. S., Arashpour, M., Sharafi, P., Teodosio, B., et al. (2022). Artificial intelligence and smart vision for building and construction 4.0: machine and deep learning methods and applications. Autom. Constr. 141, 104440. doi:10.1016/j.autcon.2022.104440
Baldoni, E., Frugis, G., Martinelli, F., Benny, J., Paffetti, D., and Buti, M. (2021). A comparative transcriptomic meta-analysis revealed conserved key genes and regulatory networks involved in drought tolerance in cereal crops. Int. J. Mol. Sci. 22 (23), 13062. doi:10.3390/ijms222313062
Bartkowski, B., Theesfeld, I., Pirscher, F., and Timaeus, J. (2018). Snipping around for food: economic, ethical and policy implications of CRISPR/Cas genome editing. Geoforum 96, 172–180. doi:10.1016/j.geoforum.2018.07.017
Bi, H., Zhao, Y., Li, H., and Liu, W. (2020). Wheat heat shock factor tahsfa6f increases aba levels and enhances tolerance to multiple abiotic stresses in transgenic plants. Int. J. Mol. Sci. 21 (9), 3121. doi:10.3390/ijms21093121
Biswas, S., Zhang, D., and Shi, J. (2021). CRISPR/Cas systems: opportunities and challenges for crop breeding. Plant Cell Rep. 40, 979–998. doi:10.1007/s00299-021-02708-2
Bratlie, S., Halvorsen, K., Myskja, B. K., Mellegård, H., Bjorvatn, C., Frost, P., et al. (2019). A novel governance framework for GMO: a tiered, more flexible regulation for GMO s would help to stimulate innovation and public debate. EMBO Rep. 20 (5), e47812. doi:10.15252/embr.201947812
Cao, Z., Sun, W., Qiao, D., Wang, J., Li, S. N., Liu, X.-L., et al. (2024). PE6c greatly enhances prime editing in transgenic rice plants. J. Integr. Plant Biol. 66, 1864–1870. doi:10.1111/jipb.13738
Carlson-Stevermer, J., Abdeen, A. A., Kohlenberg, L., Goedland, M., Molugu, K., Lou, M., et al. (2017). Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA aptamer for precise gene editing. Nat. Commun. 8 (1), 1711. doi:10.1038/s41467-017-01875-9
Chandana, B., Mahto, R., Singh, R., Ford, R., Vaghefi, N., Gupta, S., et al. (2022). Epigenomics as potential tools for enhancing magnitude of breeding approaches for developing climate resilient chickpea. Front. Genet. 13, 900253. doi:10.3389/fgene.2022.900253
Chen, F., Alphonse, M., and Liu, Q. (2020). Strategies for nonviral nano particle-based delivery of CRISPR/Cas9 therapeutics. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 12, e1609. doi:10.1002/wnan.1609
Chen, H., Yu, S., Lu, A., Xiaohui, Y., Jie, L., Zemin, D., et al. (2024). Overexpression of SlWRKY6 enhances drought tolerance by strengthening antioxidant defense and stomatal closure via ABA signaling in Solanum lycopersicum L. Plant Physiology Biochem. 213, 108855. doi:10.1016/j.plaphy.2024.108855
Chen, J. S., Ma, E., Harrington, L. B., Da Costa, M. D., Tian, X., Palefsky, J. M., et al. (2018). CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439. doi:10.1126/science.aar6245
Chen, L., Zhu, B., Ru, G., Meng, H., Yan, Y., Hong, M., et al. (2022). Re-engineering the adenine deaminase TadA-8e for efficient and specific CRISPR-based cytosine base editing. Nat. Biotechnol. 41, 663–672. doi:10.1038/s415870-22-01532-7
Chen, S., Zhang, N., Zhou, G., Hussain, S., Ahmed, S., Tian, H., et al. (2021). Knockout of the entire family of AITR genes in arabidopsis leads to enhanced drought and salinity tolerance without fitness costs. BMC Plant Biol. 21 (1), 137. doi:10.1186/s12870-021-02907-9
Cheng, Y., Ma, Y., Zhang, N., Lin, R., Yuan, Y., Tian, H., et al. (2022). The R2R3 MYB transcription factor MYB71 regulates abscisic acid response in arabidopsis. Plants 11 (10), 1369. doi:10.3390/plants11101369
Cheng, Y., Zhang, Y., Li, G., Fang, H., Sretenovic, S., Fan, A., et al. (2023). CRISPR–Cas12a base editors confer efficient multiplexed genome editing in rice. Plant Commun. 4, 100601. doi:10.1016/j.xplc.2023.100601
Clemens, M., Faralli, M., Lagreze, J., Bontempo, L., Piazza, S., Varotto, C., et al. (2022). VvEPFL9-1 knock-out via CRISPR/Cas9 reduces stomatal density in grapevine. Front. Plant Sci. 13, 878001. doi:10.3389/fpls.2022.878001
Cooper, C. L., Li, Y., Yang, L. Q., and Brough, P. (2018). Well-being-oriented human resource management practices and employee performance: the role of resilience. J. Occup. Health Psychol. 23 (2), 197–207. doi:10.1002/hrm.21934
Cullot, G., Boutin, J., Toutain, J., Prat, F., Pennamen, P., Rooryck, C., et al. (2019). CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat. Commun. 10 (1), 1136. doi:10.1038/s41467-019-09006-2
Custers, R., Casacuberta, J. M., Eriksson, D., Sági, L., and Schiemann, J. (2019). Genetic alterations that do or do not occur naturally; consequences for genome edited organisms in the context of regulatory oversight. Front. Bioeng. Biotechnol. 6, 213. doi:10.3389/fbioe.2018.00213
Deng, H., Xu, H., Wang, Y., Jia, R. X., Feng, Y., Chen, H., et al. (2023). G-quadruplex-based CRISPR photoswitch for spatiotemporal control of genomic modulation. Nucleic Acids Res. 51, 4064–4077. doi:10.1093/nar/gkad178
Didovyk, A., Borek, B., Tsimring, L., and Hasty, J. (2016). Transcriptional regulation with CRISPR-Cas9: principles, advances, and applications. Curr. Opin. Biotechnol. 40, 177–184. doi:10.1016/j.copbio.2016.06.003
Dima, O., and Inzé, D. (2021). The role of scientists in policy making for more sustainable agriculture. Curr. Biol. 31 (5), R218–R220. doi:10.1016/j.cub.2021.01.090
Doggalli, G., Kavya, K., Singh, O. B., G, M. H., Srivastava, M., B, C. G., et al. (2024). Transformative gene editing methods: precision in genetically modified crops through trait modification. J. Sci. Res. Rep. 30, 615–628. doi:10.9734/jsrr/2024/v30i82283
Doman, J. L., Raguram, A., Newby, G. A., and Liu, D. R. (2020). Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 38 (5), 620–628. doi:10.1038/s41587-020-0414-6
Dong, H., Huang, Y., and Wang, K. (2021). The development of herbicide resistance crop plants using CRISPR/Cas9-mediated gene editing. Genes 12 (6), 912. doi:10.3390/genes12060912
Donohoue, P. D., Pacesa, M., Lau, E., Vidal, B., Irby, M. J., Nyer, D. B., et al. (2021). Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells. Mol. Cell 81, 3637–3649.e5. doi:10.1016/j.molcel.2021.07.035
Dormatey, R., Sun, C., Ali, K., Coulter, J., Bi, Z., and Bai, J. (2020). Gene pyramiding for sustainable crop improvement against biotic and abiotic stresses. Agronomy 10 (9), 1255. doi:10.3390/agronomy10091255
Doudna, J. A., and Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR/Cas9. Sci. (New York, N.Y.) 346 (6213), 1258096. doi:10.1126/science.1258096
Du, H., Zaman, S., Hu, S., and Che, S. (2023). Transcriptome profiling of unigenes participating in salt tolerance in purslane (Portulaca oleracea) under salinity stress. Acta Physiol. Plant. 45 (6), 76. doi:10.1007/s11738-023-03535-6
Duensing, N., Sprink, T., Parrott, W. A., Fedorova, M., Lema, M. A., Wolt, J. D., et al. (2018). Novel features and considerations for ERA and regulation of crops produced by genome editing. Front. Bioeng. Biotechnol. 6, 79. doi:10.3389/fbioe.2018.00079
Eckerstorfer, M. F., Engelhard, M., Heissenberger, A., Simon, S., and Teichmann, H. (2019). Plants developed by new genetic modification techniques comparison of existing regulatory frameworks in the EU and Non-EU countries. Front. Bioeng. Biotechnol. 7, 26. doi:10.3389/fbioe.2019.00026
Edraki, A., Mir, A., Ibraheim, R., Gainetdinov, I., Yoon, Y., Song, C. Q., et al. (2019). A compact, high-accuracy Cas9 with a dinucleotide PAM for in vivo genome editing. Mol. cell 73 (4), 714–726. doi:10.1016/j.molcel.2018.12.003
El-Mounadi, K., Morales-Floriano, M. L., and Garcia-Ruiz, H. (2020). Principles, applications, and biosafety of plant genome editing using CRISPR/Cas9. Front. Plant Sci. 11, 56. PMID: 32117392; PMCID: PMC7031443. doi:10.3389/fpls.2020.00056
Erdoğan, İ., Cevher-Keskin, B., Bilir, Ö., Hong, Y., and Tör, M. (2023). Recent developments in CRISPR/Cas9 genome-editing technology related to plant disease resistance and abiotic stress tolerance. Biology 12, 1037. doi:10.3390/biology12071037
Ezura, H. (2022). Letter to the editor: the world's first CRISPR tomato launched to a Japanese market: the social-economic impact of its implementation on crop genome editing. Plant cell physiology 63 (6), 731–733. doi:10.1093/pcp/pcac048
Fan, K. (2024). Precision pathway engineering in plants: enhancing crop resilience and productivity for sustainable agriculture. doi:10.32942/x2z624
Fan, T., Cheng, Y., Wu, Y., Liu, S., Tang, X., He, Y., et al. (2024). High performance TadA-8e derived cytosine and dual base editors with undetectable off-target effects in plants. Nat. Commun. 15 (1), 5103. doi:10.1038/s41467-024-49473-w
Friedman, C. E., Fayer, S., Pendyala, S., Chien, W. M., Loiben, A., Tran, L., et al. (2023). CRaTER enrichment for on-target gene editing enables generation of variant libraries in hiPSCs. J. Mol. Cell. Cardiol. 179, 60–71. doi:10.1016/j.yjmcc.2023.03.017
Fu, Y., Sander, J. D., Reyon, D., Cascio, V. M., and Joung, J. K. (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32 (3), 279–284. doi:10.1038/nbt.2808
Gaj, T., Sirk, S. J., Shui, S. L., and Liu, J. (2016). Genome-editing technologies: principles and applications. Cold Spring Harb. Perspect. Biol. 8 (12), a023754. doi:10.1101/cshperspect.a023754
Gao, H., Yu, C., Liu, R., Li, X., Huang, H., Wang, X., et al. (2022). The glutathione s-transferase ptgstf1 improves biomass production and salt tolerance through regulating xylem cell proliferation, ion homeostasis and reactive oxygen species scavenging in poplar. Int. J. Mol. Sci. 23 (19), 11288. doi:10.3390/ijms231911288
Gao, W., Long, L., Tian, X., Xu, F., Liu, J., Singh, P., et al. (2017). Genome editing in cotton with the CRISPR/cas9 system. Front. Plant Sci. 8, 1364. doi:10.3389/fpls.2017.01364
Garrigues, S., Peng, M., Kun, R., and De Vries, R. (2023). Non-homologous end-joining-deficient filamentous fungal strains mitigate the impact of off-target mutations during the application of CRISPR/Cas9. mBio 14, e0066823. doi:10.1128/mbio.00668-23
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471. doi:10.1038/nature24644
Gaudelli, N. M., Lam, D. K., Rees, H. A., Sola-Esteves, N. M., Barrera, L. A., Born, D. A., et al. (2020). Directed evolution of adenine base editors with increased activity and therapeutic application. Nat. Biotechnol. 38, 892–900. doi:10.1038/s41587-020-0491-6
George, J., Acree, C., Park, J., Kong, M., Wiegand, T., Pignot, Y., et al. (2023a). Mechanism of target site selection by type V-K CRISPR-associated transposases. Science 382, eadj8543. doi:10.1126/science.adj8543
George, J., Acree, C., Park, J., Kong, M., Wiegand, T., Pignot, Y., et al. (2023b). Mechanism of target site selection by type V-K CRISPR-associated transposases. bioRxiv. doi:10.1101/2023.07.14.548620
Ghogare, R., Ludwig, Y., Bueno, G., Slamet-Loedin, I., and Dhingra, A. (2021). Genome editing reagent delivery in plants. Transgenic Res. 30, 321–335. doi:10.1007/s11248-021-00239-w
Goberna, M. F., Whelan, A. I., Godoy, P., and Lewi, D. M. (2022). Genomic editing: the evolution in regulatory management accompanying scientific progress. Front. Bioeng. Biotechnol. 10, 835378. doi:10.3389/fbioe.2022.835378
González, M., Massa, G., Andersson, M., Oneto, C., Turesson, H., Storani, L., et al. (2021). Comparative potato genome editing: Agrobacterium tumefaciens-mediated transformation and protoplasts transfection delivery of CRISPR/Cas9 components directed to StPPO2 gene. Plant Cell, Tissue Organ Cult. (PCTOC) 145, 291–305. doi:10.1007/s11240-020-02008-9
Grigore, M., and Vicente, Ó. (2023). Wild halophytes: tools for understanding salt tolerance mechanisms of plants and for adapting agriculture to climate change. Plants 12 (2), 221. doi:10.3390/plants12020221
Guan, Q., Wu, J., Yue, X., Zhang, Y., and Zhu, J. (2013). A nuclear calcium-sensing pathway is critical for gene regulation and salt stress tolerance in arabidopsis. Plos Genet. 9 (8), e1003755. doi:10.1371/journal.pgen.1003755
Guimarães, C., and Magalhães, J. (2021). Recent molecular breeding advances for improving aluminium tolerance in maize and sorghum, 318–324. doi:10.1079/9781789245431.0018
Guo, C. X., Gao, F., and Guo, Y. (2023). Off-target effects in CRISPR/Cas9 gene editing. Front. Bioeng. Biotechnol. 11, 1143157. doi:10.3389/fbioe.2023.1143157
Guo, J., Wang, Z., Qu, L., Hu, Y., and Lu, D. (2022). Transcriptomic and alternative splicing analyses provide insights into the roles of exogenous salicylic acid ameliorating waxy maize seedling growth under heat stress. BMC Plant Biol. 22 (1), 432. doi:10.1186/s12870-022-03822-3
Guo, X., Liu, D., Tang, S., Zhang, L., Liu, S., and Du, Y. (2020). The maize heat shock factors ZmHsf05 and ZmHsf12 play critical roles in thermotolerance and heat stress response. Plant Physiology Biochem. 155, 289–298. doi:10.1186/s40779-020-00240-0
Hanna, R. E., Hegde, M., Fagre, C. R., DeWeirdt, P. C., Sangree, A. K., Szegletes, Z., et al. (2021). Massively parallel assessment of human variants with base editor screens. Cell 184 (4), 1064–1080.e20. doi:10.1016/j.cell.2021.01.012
Haque, E., Taniguchi, H., Hassan, M. M., Bhowmik, P., Karim, M. R., Śmiech, M., et al. (2018). Application of CRISPR/Cas9 genome editing technology for the improvement of crops cultivated in tropical climates: recent progress, prospects, and challenges. Front. plant Sci. 9, 617. doi:10.3389/fpls.2018.00617
Hessels, L. K., Van Lente, H., and Smits, R. (2009). In search of relevance: the changing contract between science and society. Sci. Public Policy 36 (5), 387–401. doi:10.3152/030234209x442034
Hille, F., and Charpentier, E. (2016). CRISPR-Cas: biology, mechanisms and relevance. Philosophical Trans. R. Soc. Lond. Ser. B, Biol. Sci. 371 (1707), 20150496. doi:10.1098/rstb.2015.0496
Hirano, S., Nishimasu, H., Ishitani, R., and Nureki, O. (2016). Structural basis for the altered PAM specificities of engineered CRISPR-cas9. Mol. Cell 61, 886–894. doi:10.1016/j.molcel.2016.02.018
Hong, E., Lim, C., Han, S., and Lee, S. (2017). Functional analysis of the pepper ethylene-responsive transcription factor, caaief1, in enhanced aba sensitivity and drought tolerance. Front. Plant Sci. 8, 1407. doi:10.3389/fpls.2017.01407
Hosseini, S. Y., Mallick, R., Mäkinen, P., and Ylä-Herttuala, S. (2024). Insights into prime editing technology: a deep dive into fundamentals, potentials and challenges. Hum. Gene Ther. 35, 649–668. doi:10.1089/hum.2024.043
Hsu, P. D., Scott, D. A., Weinstein, J. A., Ran, F. A., Konermann, S., Agarwala, V., et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31 (9), 827–832. doi:10.1038/nbt.2647
Hsu, Y., Chiu, C., Yap, R., Tseng, Y., and Wu, Y. (2020). Pyramiding bacterial blight resistance genes in tainung82 for broad-spectrum resistance using marker-assisted selection. Int. J. Mol. Sci. 21 (4), 1281. doi:10.3390/ijms21041281
Hu, X., Chen, D., Mclntyre, C., Dreccer, M., Zhengbin, Z., Drenth, J., et al. (2017). Heat shock factor c2a serves as a proactive mechanism for heat protection in developing grains in wheat via an aba-mediated regulatory pathway. Plant Cell Environ. 41 (1), 79–98. doi:10.1111/pce.12957
Hu, Y., Sperotto, R., Koubouris, G., Stojnić, S., and Bellaloui, N. (2023). Tree ecophysiology in the context of climate change. J. For. Res. 34 (1), 1–5. doi:10.1007/s11676-023-01596-4
Hua, K., Tao, X., Liang, W., Zhang, Z., Gou, R., and Zhu, J.-K. (2020). Simplified adenine base editors improve adenine base editing efficiency in rice. Plant Biotechnol. J. 18, 770–778. doi:10.1111/pbi.13244
Hua, K., Tao, X., Yuan, F., Wang, D., and Zhu, J. K. (2018). Precise A·T to G·C base editing in the rice genome. Mol. Plant 11, 627–630. doi:10.1016/j.molp.2018.02.007
Huang, L., Yang, C., Chen, Y., Deng, H., Liao, Z., and Xiao, H. (2023). CRISPR-mediated base editing: promises and challenges for a viable oncotherapy strategy. Hum. gene Ther. 34 (15-16), 669–681. doi:10.1089/hum.2023.045
Huang, Z., and Liu, G. (2023). Current advancement in the application of prime editing. Front. Bioeng. Biotechnol. 11, 1039315. doi:10.3389/fbioe.2023.1039315
Ishii, T., and Araki, M. (2016). Consumer acceptance of food crops developed by genome editing. Plant cell Rep. 35, 1507–1518. doi:10.1007/s00299-016-1974-2
Ishino, Y., Shinagawa, H., Makino, K., Amemura, M., and Nakata, A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169 (12), 5429–5433. PMID: 3316184; PMCID: PMC213968. doi:10.1128/jb.169.12.5429-5433.1987
Ishka, M., and Julkowska, M. (2023). Tapping into the plasticity of plant architecture for increased stress resilience. F1000research 12, 1257. doi:10.12688/f1000research.140649.1
Ishka, M., Sussman, H., Hu, Y., Alqahtani, M. D., Craft, E., Sicat, R., et al. (2024). Natural variation in salt-induced changes in root:shoot ratio revealssr3gas a negative regulator of root suberization and salt resilience in arabidopsis. doi:10.1101/2024.04.09.588564
Jansing, J., Schiermeyer, A., Schillberg, S., Fischer, R., and Bortesi, L. (2019). Genome editing in agriculture: technical and practical considerations. Int. J. Mol. Sci. 20 (12), 2888. PMID: 31200517; PMCID: PMC6627516. doi:10.3390/ijms20122888
Jeon, D., Kim, J., Kang, B., and Kim, C. (2023). Deciphering the genetic mechanisms of salt tolerance in sorghum bicolor l.: key genes and snp associations from comparative transcriptomic analyses. Plants 12 (14), 2639. doi:10.3390/plants12142639
Jia, S., Wang, C., Sun, W., Yan, X., Wang, W., Xu, B., et al. (2024). OsWRKY12 negatively regulates the drought-stress tolerance and secondary cell wall biosynthesis by targeting different downstream transcription factor genes in rice. Plant Physiology Biochem. 212, 108794. doi:10.1016/j.plaphy.2024.108794
Jiang, F., and Doudna, J. A. (2017). CRISPR/Cas9 structures and mechanisms. Annu. Rev. Biophys. 46, 505–529. Epub 2017 Mar 30. PMID: 28375731. doi:10.1146/annurev-biophys-062215-010822
Jiang, F., Taylor, D. W., Chen, J. S., Kornfeld, J. E., Zhou, K., Thompson, A. J., et al. (2016). Structures of a CRISPR/Cas9 R-loop complex primed for DNA cleavage. Sci. (New York, N.Y.) 351 (6275), 867–871. doi:10.1126/science.aad8282
Jiang, K., Lim, J., Sgrizzi, S., Trinh, M., Kayabolen, A., Yutin, N., et al. (2023). Programmable RNA-guided DNA endonucleases are widespread in eukaryotes and their viruses. Sci. Adv. 9, eadk0171. doi:10.1126/sciadv.adk0171
Jiang, W., Bikard, D., Cox, D., Zhang, F., and Marraffini, L. A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31 (3), 233–239. doi:10.1038/nbt.2508
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337 (6096), 816–821. Epub 2012 Jun 28. PMID: 22745249; PMCID: PMC6286148. doi:10.1126/science.1225829
Kadam, U. S., Shelake, R. M., Chavhan, R. L., and Suprasanna, P. (2018). Concerns regarding ‘off-target’activity of genome editing endonucleases. Plant Physiology Biochem. 131, 22–30. doi:10.1016/j.plaphy.2018.03.027
Kapoor, R., Datta, A., and Thomson, M. (2021). Fused graphical lasso recovers flowering time mutation genes in arabidopsis thaliana. Inventions 6 (3), 52. doi:10.3390/inventions6030052
Karunarathne, S., Walker, E., Sharma, D., Li, C., and Han, Y. (2023). Genetic resources and precise gene editing for targeted improvement of barley abiotic stress tolerance. J. Zhejiang Univ. Sci. B 24 (12), 1069–1092. doi:10.1631/jzus.b2200552
Karvelis, T., Druteika, G., Bigelyte, G., Budre, K., Žedaveinytė, R., Silanskas, A., et al. (2021). Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease. Nature 599, 692–696. doi:10.1038/s41586-021-04058-1
Kaul, T., Sony, S. K., Verma, R., Motelb, K. F. A., Prakash, A. T., Eswaran, M., et al. (2020). Revisiting CRISPR/Cas-mediated crop improvement: special focus on nutrition. J. Biosci. 45, 137–37. doi:10.1007/s12038-020-00094-7
Khan, M., Azawi, T., Pande, A., Mun, B., Lee, D., Hussain, A., et al. (2021). The role of nitric oxide-induced atill6 in growth and disease resistance in arabidopsis thaliana. Front. Plant Sci. 12, 685156. doi:10.3389/fpls.2021.685156
Kilasi, N., Singh, J., Vallejos, C., Ye, C., Jagadish, S., Kusolwa, P., et al. (2018). Heat stress tolerance in rice (oryza sativa l.): identification of quantitative trait loci and candidate genes for seedling growth under heat stress. Front. Plant Sci. 9, 1578. doi:10.3389/fpls.2018.01578
Kim, D., Alptekin, B., and Budak, H. (2017). CRISPR/cas9 genome editing in wheat. Funct. Integr. Genomics 18 (1), 31–41. doi:10.1007/s10142-017-0572-x
Kim, D., Alptekin, B., Budak, H., and Akpinar, B. A. (2018). CRISPR/Cas9-mediated targeting of TaDREB3 improves drought tolerance in wheat. Plant Biotechnol. J. 16 (7), 1380–1389.
Kim, J., Castroverde, C., Huang, S., Li, C., Hilleary, R., Seroka, A., et al. (2022). Increasing the resilience of plant immunity to a warming climate. Nature 607 (7918), 339–344. doi:10.1038/s41586-022-04902-y
Kim, S., Woods, W. S., Perez-Pinera, P., Song, J. S., and Song, J. S. (2023). Chromatin structure and context-dependent sequence features control prime editing efficiency. Front. Genet. 14, 1222112. doi:10.3389/fgene.2023.1222112
Kirungu, J., Magwanga, R., Pu, L., Cai, X., Wang, X., Peng, R., et al. (2019). Functional characterization of gh_a08g1120 (gh3.5) gene reveal their significant role in enhancing drought and salt stress tolerance in cotton. BMC Genet. 20 (1), 62. doi:10.1186/s12863-019-0756-6
Kleinstiver, B. P., Pattanayak, V., Prew, M. S., Tsai, S. Q., Nguyen, N. T., Zheng, Z., et al. (2016). High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529 (7587), 490–495. doi:10.1038/nature16526
Kleinstiver, B. P., Prew, M. S., Tsai, S. Q., Topkar, V. V., Nguyen, N. T., Zheng, Z., et al. (2015). Engineered CRISPR-Cas9 nucleases with altered PAM specific ities. Nature 523, 481–485. doi:10.1038/nature14592
Koblan, L. W., Doman, J. L., Wilson, C., Levy, J. M., Tay, T., Newby, G. A., et al. (2018). Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 36, 843–846. doi:10.1038/nbt.4172
Kocak, D. D., Josephs, E. A., Bhandarkar, V., Adkar, S. S., Kwon, J. B., and Gersbach, C. A. (2019). Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat. Biotechnol. 37 (6), 657–666. doi:10.1038/s41587-019-0095-1
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., and Liu, D. R. (2016). Programmable editing of a target base in genomic DNA without dou ble-stranded DNA cleavage. Nature 533, 420–424. doi:10.1038/nature17946
Kondo, K., and Taguchi, C. (2022). Japanese regulatory framework and approach for genome-edited foods based on latest scientific findings. Food Saf. 10 (3), 113–128. doi:10.14252/foodsafetyfscj.D-21-00016
Kosicki, M., Tomberg, K., and Bradley, A. (2018). Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36 (8), 765–771. doi:10.1038/nbt.4192
Kotula, L., Caparrós, P., Zörb, C., Colmer, T., and Flowers, T. (2020). Improving crop salt tolerance using transgenic approaches: an update and physiological analysis. Plant Cell Environ. 43 (12), 2932–2956. doi:10.1111/pce.13865
Kulcsár, P. I., Tálas, A., Tóth, E., Nyeste, A., Ligeti, Z., Welker, Z., et al. (2020). Blackjack mutations improve the on-target activities of increased fidelity variants of SpCas9 with 5′ G-extended sgRNAs. Nat. Commun. 11 (1), 1223. doi:10.1038/s41467-020-15021-5
Kulkarni, M., Soolanayakanahally, R., Ogawa, S., Uga, Y., Selvaraj, M., and andKagale, S. (2017). Drought response in wheat: key genes and regulatory mechanisms controlling root system architecture and transpiration efficiency. Front. Chem. 5, 106. doi:10.3389/fchem.2017.00106
Kumar, R., Kaur, A., Pandey, A., Mamrutha, H. M., and Singh, G. P. (2019). CRISPR-based genome editing in wheat: a comprehensive review and future prospects. Mol. Biol. Rep. 46 (3), 3557–3569. doi:10.1007/s11033-019-04761-3
Kumar, S. (2017). Epigenomics of plantandamp; rsquo; s responses to environmental stress. doi:10.20944/preprints201710.0185.v2
Kumar, S. (2018). Epigenomics of plant responses to environmental stress. Epigenomes 2 (1), 6. doi:10.3390/epigenomes2010006
Kumar, S. (2023). Climate-resilient agricultural practices for sustainable food production. J. Agron. Crop Sci. 209 (2), 168–182. doi:10.1007/s12652-021-03612-z
Kumar, S., Chen, W., and Novak, S. (2017). Trait stacking in modern agriculture: application of genome editing tools. Emerg. Top. Life Sci. 1 (2), 151–160. doi:10.1042/etls20170012
Kumar, S., and Mohapatra, T. (2021). Dynamics of dna methylation and its functions in plant growth and development. Front. Plant Sci. 12, 596236. doi:10.3389/fpls.2021.596236
Kuscu, C., Arslan, S., Singh, R., Thorpe, J., and Adli, M. (2014). Genome wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 32, 677–683. doi:10.1038/nbt.2916
Laforest, L., and Nadakuduti, S. (2022). Advances in delivery mechanisms of CRISPR gene-editing reagents in plants. Front. Genome Ed. 4, 830178. doi:10.3389/fgeed.2022.830178
Leawtrakun, J., Aesomnuk, W., Khanthong, S., Dumhai, R., Songtoasesakul, D., Phosuwan, S., et al. (2024). Identification of candidate genes for salt tolerance at seedling stage in rice using qtl-seq and chromosome segment substitution line-derived population. Agronomy 14 (5), 929. doi:10.3390/agronomy14050929
Leibowitz, M. L., Papathanasiou, S., Doerfler, P. A., Blaine, L. J., Sun, L., Yao, Y., et al. (2021). Chromothripsis as an on-target consequence of CRISPR–Cas9 genome editing. Nat. Genet. 53 (6), 895–905. doi:10.1038/s41588-021-00838-7
Lenka, B., Kulkarni, G., Moharana, A., Singh, A., Pradhan, G., and Muduli, L. (2020). Millets: promising crops for climate-smart agriculture. Int. J. Curr. Microbiol. Appl. Sci. 9 (11), 656–668. doi:10.20546/ijcmas.2020.911.081
Li, A., Jia, S., Yobi, A., Ge, Z., Sato, S. J., Zhang, C., et al. (2018a). Editing of an alpha-kafirin gene family increases, digestibility and protein quality in sorghum. Plant physiol. 177 (4), 1425–1438. doi:10.1104/pp.18.00200
Li, A., Tanner, M. R., Lee, C. M., Hurley, A. E., De Giorgi, M., Jarrett, K. E., et al. (2020a). AAV-CRISPR gene editing is negated by pre-existing immunity to Cas9. Mol. Ther. 28 (6), 1432–1441. Epub 2020 Apr 19. PMID: 32348718; PMCID: PMC7264438. doi:10.1016/j.ymthe.2020.04.017
Li, C., Chen, Y., Hu, Q., Yang, X., Zhao, Y., Lin, Y., et al. (2024). PSEUDO-RESPONSE REGULATOR 3b and transcription factor ABF3 modulate abscisic acid-dependent drought stress response in soybean. Plant Physiol. 195, 3053–3071. doi:10.1093/plphys/kiae269
Li, C., Zong, Y., Wang, Y., Jin, S., Zhang, D., Song, Q., et al. (2018b). Expanded base editing in rice and wheat using a Cas9- adenosine deaminase fusion. Genome Biol. 19, 59. doi:10.1186/s13059-018-1443-z
Li, J., Pan, Y., Cheng, Y., Tang, X., Zhao, Y., and Li, J. (2020b). The transcription factor OsbHLH034 regulates heat tolerance by modulating auxin and reactive oxygen species levels in rice. Plant Physiol. 183 (2), 730–748. doi:10.48550/arXiv.2010.08895
Li, J., Zhang, C., He, Y., Li, S., Yan, L., Li, Y., et al. (2022a). Plant base editing and prime editing: the current status and future perspectives. J. Integr. Plant Biol. 65, 444–467. doi:10.1111/jipb.13425
Li, J., Zhang, H., Si, X., Tian, Y., Chen, K., Liu, J., et al. (2017). Generation of thermosensitive male-sterile maize by targeted knockout of the ZmTMS5 gene. J. Genet. genomics = Yi chuan xue bao 44 (9), 465–468. doi:10.1016/j.jgg.2017.02.002
Li, M., Xu, L., Zhang, L., Li, X., Cao, C., Chen, L., et al. (2022b). Overexpression of mtr-mir319a contributes to leaf curl and salt stress adaptation in arabidopsis thaliana and medicagotruncatula. Int. J. Mol. Sci. 24 (1), 429. doi:10.3390/ijms24010429
Li, Y., Liang, J., Deng, B., Jiang, Y., Zhu, J., Chen, L., et al. (2023). Applications and prospects of CRISPR/Cas9-Mediated base editing in plant breeding. Curr. issues Mol. Biol. 45 (2), 918–935. doi:10.3390/cimb45020059
Lian, J., HamediRad, M., Hu, S., and Zhao, H. (2017). Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system. Nat. Commun. 8 (1), 1688. doi:10.1038/s41467-017-01695-x
Liang, Y., Gong, Z., Wang, J., Zheng, J., Ma, Y., Ling, M., et al. (2021). Nanopore-based comparative transcriptome analysis reveals the potential mechanism of high-temperature tolerance in cotton (gossypiumhirsutum l.). Plants 10 (11), 2517. doi:10.3390/plants10112517
Liang, Z., Wu, Y., Guo, Y., and Wei, S. (2023). Addition of the T5 exonuclease increases the prime editing efficiency in plants. J. Genet. Genomics 50, 582–588. doi:10.1016/j.jgg.2023.03.008
Lim, C. J., Hwang, J. E., Park, Y. J., Kim, J. K., Kim, J. A., and Kim, W. T. (2018). The pepper gene CaDIL1 positively regulates drought tolerance and ABA signaling. Plant Mol. Biol. 96 (3), 291–302. doi:10.3389/fpls.2018.01301
Lin, Q., Zong, Y., Xue, C., Wang, S., Jin, S., Zhu, Z., et al. (2020). Prime genome editing in rice and wheat. Nat. Biotechnol. 38, 582–585. doi:10.1038/.s41587-020-0455-x
Lin, Y., Cradick, T. J., Brown, M. T., Deshmukh, H., Ranjan, P., Sarode, N., et al. (2014). CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic acids Res. 42 (11), 7473–7485. doi:10.1093/nar/gku402
Liu, J., Novero, M., Scheler, B., Wang, W., and Sanchez-Baracaldo, P. (2021). COLD1 interacts with multiple genes to regulate temperature stress in rice. J. Exp. Bot. 72 (5), 1561–1573. doi:10.1016/j.cell.2015.01.046
Liu, J., Yao, Y., Xin, M., Peng, H., Ni, Z., and Sun, Q. (2022a). Shaping Polyploid wheat for success: origins, domestication, and the genetic improvement of agronomic traits. J. Integr. Plant Biol. 64 (2), 536–563. doi:10.1111/jipb.13210
Liu, T., Zou, J., Xi, Y., Wang, K., Rao, Y., and Chun, W. (2023). Development and application of prime editing in plants. Rice Sci. 30, 509–522. doi:10.1016/j.rsci.2023.07.005
Liu, X., Gu, D., Zhang, Y., Jiang, Y., Qin, R. Y., Li, J., et al. (2024a). Conditional knockdown of OsMLH1 to improve plant prime editing systems without disturbing fertility in rice. Genome Biol. 25, 131. doi:10.1186/s13059-024-03282-y
Liu, Y., Chen, Z., Zhang, C., Guo, J., Liu, Q., Yin, Y., et al. (2024b). Gene editing of ZmGA20ox3 improves plant architecture and drought tolerance in maize. Plant Cell Rep. 43 (1), 18. doi:10.1007/s00299-023-03090-x
Liu, Y., Zhou, X.-H., Huang, S., and Wang, X.-L. (2022b). Prime editing: a search and replace tool with versatile base changes. Yi Chuan 44, 993–1008. doi:10.16288/j.yczz.22-156
Lombardo, L., and Grando, M. S. (2020). Genetically modified plants for nutritionally improved food: a promise kept? Food Rev. Int. 36 (1), 58–76. doi:10.1080/87559129.2019.1613664
Lopes, R., and Prasad, M. (2024). Beyond the promise: evaluating and mitigating off-target effects in CRISPR gene editing for safer therapeutics. Front. Bioeng. Biotechnol. 11, 1339189. doi:10.3389/fbioe.2023.1339189
Lu, L., Wu, X., Wang, P., Zhu, L., Liu, Y., Tang, Y., et al. (2022). Halophyte nitrariabillardieri CIPK25 mitigates salinity-induced cell damage by alleviating H2O2 accumulation. Front. Plant Sci. 13, 961651. doi:10.3389/fpls.2022.961651
Luo, M., Pan, Y., He, Y., Ruhan, A., Wu, C., Huang, B., et al. (2023). Detecting SARS-CoV-2 BA. 2, BA. 4, and BA. 5 variants utilizing a robust RT-RPA-CRISPR/Cas12a-Based method—China, 2023. China CDC Wkly. 5 (26), 584–591. doi:10.46234/ccdcw2023.113
Lyzenga, W. J., Pozniak, C. J., and Kagale, S. (2021). Advanced domestication: harnessing the precision of gene editing in crop breeding. Plant Biotechnol. J. 19 (4), 660–670. doi:10.1111/pbi.13576
Ma, D., Gao, Z., Jiang, T., Chen, X., Ma, X., Shi, H., et al. (2021). CaCIPK3, a CBL-interacting protein kinase, is a positive regulator of salt and drought tolerance in Capsicum annuum L. Plant Physiology Biochem. 166, 218–229. doi:10.1038/s41438-021-00651-7
Ma, X., Mau, M., and Sharbel, T. F. (2018). Genome editing for global food security. Trends Biotechnol. 36 (2), 123–127. doi:10.1016/j.tibtech.2017.08.004
Mahalingam, R., Duhan, N., Kaundal, R., Smertenko, A., Nazarov, T., and andBregitzer, P. (2022). Heat and drought induced transcriptomic changes in barley varieties with contrasting stress response phenotypes. Front. Plant Sci. 13, 1066421. doi:10.3389/fpls.2022.1066421
Makarova, K., Wolf, Y., Alkhnbashi, O., Costa, F., Shah, S. A., Saunders, S. J., et al. (2015). An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 13, 722–736. doi:10.1038/nrmicro3569
Makarova, K. S., Aravind, L., Wolf, Y. I., and Koonin, E. V. (2011). Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol. Direct 6, 38. doi:10.1186/1745-6150-6-38
Malenica, N., Dunić, J., Vukadinović, L., Cesar, V., and Šimić, D. (2021). Genetic approaches to enhance multiple stress tolerance in maize. Genes 12 (11), 1760. doi:10.3390/genes12111760
Mali, P., Aach, J., Stranges, P. B., Esvelt, K. M., Moosburner, M., Kosuri, S., et al. (2013). CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31 (9), 833–838. doi:10.1038/nbt.2675
Mallapaty, S. (2019). Australian gene-editing rules adopt ‘middle ground. Nature 23. doi:10.1038/d41586-019-01282-8
Manghwar, H., Lindsey, K., Zhang, X., and Jin, S. (2019). CRISPR/Cas system: recent advances and future prospects for genome editing. Trends Plant Sci. 24 (12), 1102–1125. Epub 2019 Nov 11. PMID: 31727474. doi:10.1016/j.tplants.2019.09.006
Marraffini, L. A., and Sontheimer, E. J. (2010). CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11 (3), 181–190. PMID: 20125085; PMCID: PMC2928866. doi:10.1038/nrg2749
Martin, P., Oana, P., and Martin, J. (2024). Past, present, and future of CRISPR genome editing technologies. Cell 187, 1076–1100. doi:10.1016/j.cell.2024.01.042
Mengstie, M., Azezew, M., Dejenie, T., Teshome, A., Admasu, F., Teklemariam, A., et al. (2024). Recent advancements in reducing the off-target effect of CRISPR/Cas9 genome editing. Biol. Targets and Ther. 18, 21–28. doi:10.2147/BTT.S429411
Mikhaylova, E. V., Kuluev, B. R., Gerashchenkov, G. A., Chemeris, D. A., Garafutdinov, R. R., Kuluev, A. R., et al. (2024). Prime-editing methods and pegRNA design programs. Mol. Biol. 58 (1), 17–32. doi:10.1134/s0026893324010084
Mishra, S. K., Tripp, J., Winkelhaus, S., Tschiersch, B., Theres, K., Nover, L., et al. (2002). In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 16 (12), 1555–1567. doi:10.1101/gad.228802
Miskevish, F., Lodeyro, A., Ponso, M. A., Bouzo, C., Meeley, R., Timmermans, M. C., et al. (2023). Maize mutants in miR394-regulated genes show improved drought tolerance. bioRxiv 12. doi:10.1101/2023.12.05.570184
Mohanta, T. K., Bashir, T., Hashem, A., Abd Allah, E. F., and Bae, H. (2017). Genome editing tools in plants. Genes (Basel) 8 (12), 399. doi:10.3390/genes8120399
Molla, K. A., Sretenovic, S., Bansal, K. C., and Qi, Y. (2021). Precise plant genome editing using base editors and prime editors. Nat. plants 7 (9), 1166–1187. doi:10.1038/s41477-021-00991-1
Monarkh, V. (2020). GMO and health risks: selected issues. Agric. For. 14 (2), 245–254. doi:10.17707/AgricultForest.14.2.08
Musunuru, K., Chadwick, A. C., Mizoguchi, T., Garcia, S. P., DeNizio, J. E., Reiss, C. W., et al. (2021). In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature 593 (7859), 429–434. doi:10.1038/s41586-021-03534-y
Nabwire, S., Suh, H. K., Kim, M. S., Baek, I., and Cho, B. K. (2021). Review: application of artificial intelligence in phenomics. Sensors 21 (13), 4363. doi:10.3390/s21134363
Nadakuduti, S., and Enciso-Rodríguez, F. (2021). Advances in genome editing with CRISPR systems and transformation technologies for plant DNA manipulation. Front. Plant Sci. 11, 637159. doi:10.3389/fpls.2020.637159
Naeem, M., and Alkhnbashi, O. (2023). Current bioinformatics tools to optimize CRISPR/Cas9 experiments to reduce off-target effects. Int. J. Mol. Sci. 24, 6261. doi:10.3390/ijms24076261
Nascimento, F., Rocha, A., Soares, J., Mascarenhas, M., Ferreira, M., Lino, L., et al. (2023). Gene editing for plant resistance to abiotic factors: a systematic review. Plants 12 (2), 305. doi:10.3390/plants12020305
Ndlovu, N. (2020). Application of genomics and phenomics in plant breeding for climate resilience. Asian Plant Res. J., 53–66. doi:10.9734/aprj/2020/v6i430137
Negishi, K., Kaya, H., Abe, K., Hara, N., Saika, H., and Toki, S. (2019). An adenine base editor with expanded targeting scope using SpCas9-NGv1 in rice. Plant Biotechnol. J. 17 (8), 1476–1478. doi:10.1111/pbi.13120
Nety, S., Altae-Tran, H., Kannan, S., Demircioglu, F., Faure, G., Hirano, S., et al. (2023). The transposon-encoded protein TnpB processes its own mRNA into ωRNA for guided nuclease activity. CRISPR J. 6, 232–242. doi:10.1089/crispr.2023.0015
Neugebauer, M. E., Hsu, A., Arbab, M., Krasnow, N. A., McElroy, A. N., Pandey, S., et al. (2022). Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 38, 620–628. doi:10.1038/s41587-020-0414-6
Nguyen, N., Quail, M. M., and Hernday, A. D. (2017). An efficient, rapid, and recyclable system for CRISPR-mediated genome editing in Candida albicans. MSphere 2 (2), e00149–e01128. doi:10.1128/mSphereDirect.00149-17
Ni, P., Zhao, Y., Zhou, X., Liu, Z., Huang, Z., Ni, Z., et al. (2023). Efficient and versatile multiplex prime editing in hexaploid wheat. Genome Biol. 24 (1), 156. doi:10.1186/s13059-023-02990-1
Nishimasu, H., Ran, F. A., Hsu, P. D., Konermann, S., Shehata, S. I., Dohmae, N., et al. (2014). Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156 (5), 935–949. doi:10.1016/j.cell.2014.02.001
Nuñez, J. K., Chen, J., Pommier, G. C., Cogan, J. Z., Replogle, J. M., Adriaens, C., et al. (2021). Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell 184 (9), 2503–2519.e17. doi:10.1016/j.cell.2021.03.025
Okada, A., Arndell, T., Borisjuk, N., Sharma, N., Watson-Haigh, N. S., Tucker, E. J., et al. (2019). CRISPR/Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol. J. 17 (10), 1905–1913. doi:10.1111/pbi.13106
Panda, D., Baig, M. J., and Molla, K. A. (2024). “Genome editing and opportunities for trait improvement in pearl millet,” in Pearl millet in the 21st century: food-nutrition-climate resilience-improved livelihoods (Singapore: Springer Nature Singapore), 163–178.
Pang, Y., Chen, K., Wang, X., Wang, W., Xu, J., Ali, J., et al. (2017). Simultaneous improvement and genetic dissection of salt tolerance of rice (oryza sativa l.) by designed qtl pyramiding. Front. Plant Sci. 8, 1275. doi:10.3389/fpls.2017.01275
Paquet, D., Kwart, D., Chen, A., Sproul, A., Jacob, S., Teo, S., et al. (2016). Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533 (7601), 125–129. doi:10.1038/nature17664
Parikh, A., Brant, E., Baloğlu, M., and Altpeter, F. (2021). CRISPR/cas-mediated genome editing in sorghum — recent progress, challenges and prospects. Vitro Cell. Dev. Biol. - Plant 57 (4), 720–730. doi:10.1007/s11627-021-10215-y
Perroud, P.-F., Guyon-Debast, A., Casacuberta, J. M., Paul, W., Pichon, J.-P., Comeau, D., et al. (2023). Improved Prime Editing allows for routine predictable gene editing in Physcomitrium patens. J. Exp. Bot. 74, 6176–6187. doi:10.1093/jxb/erad189
Podevin, N., Davies, H. V., Hartung, F., Nogué, F., and andCasacuberta, J. M. (2013). Site-directed nucleases: a paradigm shift in predictable, knowledge-based plant breeding. Trends Biotechnol. 31 (6), 375–383. doi:10.1016/j.tibtech.2013.03.004
Prohens, J., Gramazio, P., Plazas, M., Dempewolf, H., Kilian, B., Díez, M., et al. (2017). Introgressiomics: a new approach for using crop wild relatives in breeding for adaptation to climate change. Euphytica 213 (7), 158. doi:10.1007/s10681-017-1938-9
Putra, A., Yen, J., and Fournier-Level, A. (2022). Forecasting trait responses in novel environments to aid seed provenancing under climate change. Mol. Ecol. Resour. 23 (3), 565–580. doi:10.1111/1755-0998.13728
Que, Q., Chilton, M., Fontes, C., He, C., Nuccio, M., Zhu, T., et al. (2010). Trait stacking in transgenic crops: challenges and opportunities. Gm. Crops 1 (4), 220–229. doi:10.4161/gmcr.1.4.13439
Ran, F. A., Cong, L., Yan, W. X., Scott, D. A., Gootenberg, J. S., Kriz, A. J., et al. (2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature 520 (7546), 186–191. doi:10.1038/nature14299
Ren, Q., Sretenovic, S., Liu, G., Zhong, Z., Wang, J., Huang, L., et al. (2021). Improved plant cytosine base editors with high editing activity, purity, and specificity. Plant Biotechnol. J. 19 (10), 2052–2068. doi:10.1111/pbi.13635
Ren, S., Lyle, C., Jiang, G., and andPenumala, A. (2016). Soybean salt tolerance 1 (gmst1) reduces ROS production, enhances aba sensitivity, and abiotic stress tolerance in arabidopsis thaliana. Front. Plant Sci. 7, 445. doi:10.3389/fpls.2016.00445
Rezaul, I., Feng, B., Chen, T., Fu, W., Zhang, C., Tao, L., et al. (2018). Abscisic acid prevents pollen abortion under high-temperature stress by mediating sugar metabolism in rice spikelets. Physiol. Plant. 165 (3), 644–663. doi:10.1111/ppl.12759
Richardson, C. D., Ray, G. J., DeWitt, M. A., Curie, G. L., and Corn, J. E. (2016). Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 34 (3), 339–344. doi:10.1038/nbt.3481
Richter, M. F., Zhao, K. T., Eton, E., Lapinaite, A., Newby, G. A., Thuronyi, B. W., et al. (2020). Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 38, 883–891. doi:10.1038/s41587-020-0453-z
Rossi, M. (2023). Advancements and challenges in gene therapy ap-proaches for sickle cell disease: a comprehensive.
Rostain, W., Grébert, T., Vyhovskyi, D., Pizarro, P., Bellingen, G., Cui, L., et al. (2023). Cas9 off-target binding to the promoter of bacterial genes leads to silencing and toxicity. Nucleic Acids Res. 51, 3485–3496. doi:10.1093/nar/gkad170
Roy, A., and Sandhu, R. (2024). Advancements in genetic enhancement: CRISPR/cas-mediated genome editing in leguminous crops. J. Adv. Biol. Biotechnol. 27 (6), 670–681. doi:10.9734/jabb/2024/v27i6927
Sai, Y. R. K. M. (2021). Cytosine extensions optimize case activity for telomere length regulation: implications for CRISPR-based therapies–A short communication. Ann. Mol. Genet. Med. 5 (1), 001–003. doi:10.17352/amgm.000009
Salem, A. R., Bryant, W. B., Doja, J., Griffin, S. H., Shi, X., Han, W., et al. (2023). Prime editing in mice with an engineered pegRNA. Vasc. Pharmacol. 154, 107269. doi:10.1016/j.vph.2023.107269
Scafetta, N. (2024). Impacts and risks of “realistic” global warming projections for the 21st century. Geosci. Front. 15 (2), 101774. doi:10.1016/j.gsf.2023.101774
Shahzad, A. N., Ahmed, M., Yasin, M. U., Rizwan, M., Imran, M., and Rajwana, I. A. (2021). Role of transcription factors in developing climate-resilient crops: future prospects and challenges. Plant Growth Regul. 95 (1), 15–32. doi:10.1016/j.sjbs.2021.01.028
Shailani, A., Joshi, R., Singla-Pareek, S., and Pareek, A. (2020). Stacking for future: pyramiding genes to improve drought and salinity tolerance in rice. Physiol. Plant. 172 (2), 1352–1362. doi:10.1111/ppl.13270
Shakam, H. M., Ebaid, M., and Hamza, D. A. (2024). Polymorphism detection of some transcription factors genes conferring drought tolerance in diverse wheat (Triticum aestivum L.) genotypes. Genotypes. Alexandria Sci. Exch. 45, 307–319. doi:10.21608/asejaiqjsae.2024.365960
Sharma, K., Palakolanu, S., Bhattacharya, J., Shankhapal, A., and Bhatnagar-Mathur, P. (2022). CRISPR for accelerating genetic gains in under-utilized crops of the drylands: progress and prospects. Front. Genet. 13, 999207. doi:10.3389/fgene.2022.999207
Shaw, R., Lokshin, A. E., Miller, M. C., Messerlian-Lambert, G., and Moore, R. G. (2022). Stacking machine learning algorithms for biomarker-based preoperative diagnosis of a pelvic mass. Cancers 14 (5), 1291. doi:10.3390/cancers14051291
Shen, Q., Ruan, H., Zhang, H., Wu, T., Zhu, K., Han, W., et al. (2024). Utilization of CRISPR-Cas genome editing technology in filamentous fungi: function and advancement potentiality. Front. Microbiol. 15, 1375120. doi:10.3389/fmicb.2024.1375120
Shi, J., Gao, H., Wang, H., Laftte, H. R., Archibald, R. L., Yang, M., et al. (2017a). ARGOS8 variants generated by CRISPRCas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15, 207–216. doi:10.1111/pbi.12603
Shi, T. Q., Liu, G. N., Ji, R. Y., Shi, K., Song, P., Ren, L. J., et al. (2017b). CRISPR/Cas9-based genome editing of the filamentous fungi: the state of the art. Appl. Microbiol. Biotechnol. 101, 7435–7443. doi:10.1007/s00253-017-8497-9
Shukla, M., Al-Busaidi, K. T., Trivedi, M., and Tiwari, R. K. (2018). Status of research, regulations and challenges for genetically modified crops in India. GM crops and food 9 (4), 173–188. doi:10.1080/21645698.2018.1529518
Şimşek, Ö., Isak, M. A., Dönmez, D., Dalda Şekerci, A., İzgü, T., and Kaçar, Y. A. (2024). Advanced biotechnological interventions in mitigating drought stress in plants. Plants 13 (5), 717. doi:10.3390/plants13050717
Song, Z., Wang, L., Lee, M., and Yue, G. (2023). The evolution and expression of stomatal regulators in c3 and c4 crops: implications on the divergent drought tolerance. Front. Plant Sci. 14, 1100838. doi:10.3389/fpls.2023.1100838
Sorek, R., Lawrence, C. M., and Wiedenheft, B. (2013). CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 82, 237–266. Epub 2013 Mar 11. PMID: 23495939. doi:10.1146/annurev-biochem-072911-172315
Sprink, T., Eriksson, D., Schiemann, J., and Hartung, F. (2016). Regulatory hurdles for genome editing: process-vs. product-based approaches in different regulatory contexts. Plant Cell Rep. 35 (7), 1493–1506. doi:10.1007/s00299-016-1990-2
Sun, S., Yu, J., Chen, F., Zhao, T., Fang, X., Li, Y., et al. (2008). Tiny, a dehydration-responsive element (dre)-binding protein-like transcription factor connecting the dre- and ethylene-responsive element-mediated signaling pathways in arabidopsis. J. Biol. Chem. 283 (10), 6261–6271. doi:10.1074/jbc.m706800200
Sun, W., Yao, M., Wang, Z., Chen, Y., Zhan, J., Yan, J., et al. (2022). Involvement of auxin-mediated cqexpa50 contributes to salt tolerance in quinoa (chenopodium quinoa) by interaction with auxin pathway genes. Int. J. Mol. Sci. 23 (15), 8480. doi:10.3390/ijms23158480
Sun, X., Zhang, H., Jia, Y., Li, J., and Jia, M. (2024). CRISPR/Cas9-based genome-editing technologies in engineering bacteria for the production of plant-derived terpenoids. Eng. Microbiol. 4, 100154. doi:10.1016/j.engmic.2024.100154
Sun, Y., Zhang, X., Wu, C., He, Y., Ma, Y., Hou, H., et al. (2016). Engineering herbicide-resistant rice plants through CRISPR/Cas9-Mediated homologous recombination of acetolactate synthase. Mol. plant 9 (4), 628–631. doi:10.1016/j.molp.2016.01.001
Suza, W., Kena, A., Akromah, R., Mugwanya, N., and Zeller, M. (2018). Fear holds back gene-edited crops — educate the public. Nature 563 (7733), 626. doi:10.1038/d41586-018-07547-y
Tan, J., Zeng, D., Zhao, Y., Wang, Y., Liu, T., Li, S., et al. (2022). PhieABEs: a PAM-less/free high-efficiency adenine base editor toolbox with wide target scope in plants. Plant Biotechnol. J. 20, 934–943. doi:10.1111/pbi.13774
Tang, J., Chu, C., and Feng, Q. (2019a). CRISPR/Cas9-mediated knockout of the rice transcription factor OsbZIP46 confers enhanced drought tolerance and ABA sensitivity. Plant Sci. 287, 110188. doi:10.3389/fpls.2019.00168
Tang, Y., Bao, X., Yu-ling, Z., Wu, Q., Guo, Y., XuHui, Y., et al. (2019b). Overexpression of a MYB family gene, OsMYB6, increases drought and salinity stress tolerance in transgenic rice. Front. Plant Sci. 10, 168. doi:10.3389/fpls.2019.00168
Tao, S., Chen, H., Li, N., and Liang, W. (2022). The application of the CRISPR-Cas system in antibiotic resistance. Infect. drug Resist. 15, 4155–4168. doi:10.2147/IDR.S370869
Tran, M., Son, G., Song, Y., Nguyen, N., Park, S., Thach, T., et al. (2023). CRISPR/Cas9-based precise engineering of slhyprp1 protein towards multi-stress tolerance in tomato. Front. Plant Sci. 14, 1186932. doi:10.3389/fpls.2023.1186932
Tsang, I., Atkinson, J. A., Rawsthorne, S., Cockram, J., and Leigh, F. (2023). Root hairs: an under-explored target for sustainable cereal crop production. doi:10.31219/osf.io/fh2u5
Uranga, M., and Daròs, J. (2022). Tools and targets: the dual role of plant viruses in CRISPR–Cas genome editing. Plant Genome 16, e20220. doi:10.1002/tpg2.20220
Van de Wiel, C., Schaart, J., Lotz, L., and Smulders, M. (2017). New traits in crops produced by genome editing techniques based on deletions. Plant Biotechnol. Rep. 11 (1), 1–8. doi:10.1007/s11816-017-0425-z
Veillet, F., Chauvin, L., Kermarrec, M. P., Sevestre, F., Merrer, M., Terret, Z., et al. (2019). The Solanum tuberosum GBSSI gene: a target for assessing gene and base editing in tetraploid potato. Plant cell Rep. 38 (9), 1065–1080. doi:10.1007/s00299-019-02426-w
Verma, P., Tandon, R., Yadav, G., and Gaur, V. (2020). Structural aspects of DNA repair and recombination in crop improvement. Front. Genet. 11, 574549. doi:10.3389/fgene.2020.574549
Volodina, O. V., Fabrichnikova, A. R., Anuchina, A. A., Mishina, O. S., Lavrov, A., and Smirnikhina, S. A. (2024). Evolution of prime editing systems: move forward to the treatment of hereditary diseases. Curr. Gene Ther. 25, 46–61. doi:10.2174/0115665232295117240405070809
Vora, D., Yadav, S., and Sundar, D. (2023). Hybrid multitask learning reveals sequence features driving specificity in the CRISPR/Cas9 system. Biomolecules 13, 641. doi:10.3390/biom13040641
Voytas, D. F., and Gao, C. (2014). Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biol. 12 (6), e1001877. doi:10.1371/journal.pbio.1001877
Wahdan, S., Tanunchai, B., Wu, Y., Sansupa, C., Schädler, M., Dawoud, T., et al. (2021). Deciphering trifolium pratense l. holobiont reveals a microbiome resilient to future climate changes. Microbiologyopen 10 (4), e1217. doi:10.1002/mbo3.1217
Walker, M., Klompe, S., Zhang, D., and Sternberg, S. (2023a). Novel molecular requirements for CRISPR RNA-guided transposition. Nucleic Acids Res. 51, 4519–4535. doi:10.1093/nar/gkad270
Walker, M., Klompe, S., Zhang, D., and Sternberg, S. (2023b). Transposon mutagenesis libraries reveal novel molecular requirements during CRISPR RNA-guided DNA integration. bioRxiv. doi:10.1101/2023.01.19.524723
Walton, R. T., Christie, K. A., Whittaker, M. N., and Kleinstiver, B. P. (2020). Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368 (6488), 290–296. doi:10.1126/science.aba8853
Waltz, E. (2018). With a free pass, CRISPR-edited plants reach market in record time. Nat. Biotechnol. 36 (1), 6–7. doi:10.1038/nbt0118-6b
Waltz, E. (2022). GABA-enriched tomato is first CRISPR-edited food to enter market. Nat. Biotechnol. 40 (1), 9–11. doi:10.1038/d41587-021-00026-2
Wang, J., Chen, B. W., Ali, S., Zhang, T. X., Wang, Y., Zhang, H., et al. (2021a). Epigenetic modification associated with climate regulates betulin biosynthesis in birch. J. For. Res. 34, 21–35. doi:10.1007/s11676-021-01424-7
Wang, L., Ahmad, B., Chen, L., Shi, X., Sun, R., Zhang, S., et al. (2020a). “Bioinformatics and expression analysis of histone modification genes in grapevine predict their involvement in seed development,” in Powdery mildew resistance, and hormonal signaling. doi:10.21203/rs.3.rs-27843/v3
Wang, T., Xun, H., Wang, W., Ding, X., Tian, H., Hussain, S., et al. (2021b). Mutation of GmAITR genes by CRISPR/Cas9 genome editing results in enhanced salinity stress tolerance in soybean. Front. plant Sci. 12, 779598. doi:10.3389/fpls.2021.779598
Wang, X., Fang, T., Lu, J., Tripathi, L., and Qi, Y. (2024a). Broad range plastid genome editing with monomeric TALE-linked cytosine and dual base editors. Plant Biotechnol. J. 22 (9), 2441–2443. doi:10.1111/pbi.14358
Wang, X., Guo, X., Ma, X., Luo, L., Fang, Y., Zhao, N., et al. (2021c). Development of new rice (oryza. sativa l.) breeding lines through marker-assisted introgression and pyramiding of brown planthopper, blast, bacterial leaf blight resistance, and aroma genes. Agronomy 11 (12), 2525. doi:10.3390/agronomy11122525
Wang, Y.-H., Ye, X., Zhao, B., Wang, W.-J., Zhou, Z.-F., Zhang, X.-Q., et al. (2024b). Comprehensive analysis of B3 family genes in pearl millet (Pennisetum glaucum) and the negative regulator role of PgRAV-04 in drought tolerance. Front. Plant Sci. 15, 1400301. doi:10.3389/fpls.2024.1400301
Wang, Z., Hong, Y., Zhu, G., Li, Y., Niu, Q., Yao, J., et al. (2020b). Loss of salt tolerance during tomato domestication conferred by variation in a Na+/K+ transporter. Embo J. 39 (10), e103256. doi:10.15252/embj.2019103256
Wani, S., Kumar, V., Khare, T., Rajasheker, G., Parveda, M., Solymosi, K., et al. (2020). Engineering salinity tolerance in plants: progress and prospects. Planta 251 (4), 76. doi:10.1007/s00425-020-03366-6
Wei, C., Wang, C., Jia, M., Guo, H. X., Luo, P. Y., Wang, M. G., et al. (2021). Efficient generation of homozygous substitutions in rice in one generation utilizing an rABE8e base editor. J. Integr. Plant Biol. 63, 1595–1599. doi:10.1111/jipb.13089
Wen, J., Jiang, F., Weng, Y., Sun, M., Shi, X., Zhou, Y., et al. (2019a). Identification of heat-tolerance qtls and high-temperature stress-responsive genes through conventional qtl mapping, qtl-seq and rna-seq in tomato. BMC Plant Biol. 19 (1), 398. doi:10.1186/s12870-019-2008-3
Wen, W., Li, D., Shen, Y., Li, W., Chen, H., and Li, X. (2019b). CRISPR/Cas9-mediated ERECTA gene editing enhances drought tolerance and water use efficiency in Arabidopsis. Plant Sci. 285, 38–45. doi:10.1186/s12870-019-2008-3
Whelan, A. I., and Lema, M. A. (2015). Regulatory framework for gene editing and other new breeding techniques (NBTs) in Argentina. GM crops and food 6 (4), 253–265. doi:10.1080/21645698.2015.1114698
Wiedenheft, B., Sternberg, S. H., and Doudna, J. A. (2012). RNA-guided genetic silencing systems in bacteria and archaea. Nature 482 (7385), 331–338. PMID: 22337052. doi:10.1038/nature10886
Wong, A., Oosterom, E., Godwin, I., and Borrell, A. (2023). Integrating stay-green and pin-formed genes: pin-formed genes as potential targets for designing climate-resilient cereal ideotypes. Aob Plants 15 (4), plad040. doi:10.1093/aobpla/plad040
Xiang, X., Corsi, G. I., Anthon, C., Qu, K., Pan, X., Liang, X., et al. (2021). Enhancing CRISPR-Cas9 gRNA efficiency prediction by data integration and deep learning. Nat. Commun. 12 (1), 3238. doi:10.1038/s41467-021-23576-0
Xie, H., Zhang, P., Jiang, C., Wang, Q., Guo, Y., Huang, T., et al. (2023). Combined transcriptomic and metabolomic analyses of high temperature stress response of quinoa seedlings. BMC Plant Biol. 23 (1), 292. doi:10.1186/s12870-023-04310-y
Xie, Y., Mao, Y., Xu, H., Liu, S., and Zhang, X. (2021). The Arabidopsis thaliana CBF1 transcription factor is a positive regulator of ABA signaling. J. Integr. Plant Biol. 63 (8), 1422–1435. doi:10.1111/jipb.13161
Xu, R., Li, J., Liu, X., Shan, T., Qin, R., and Wei, P. (2020b). Development of plant prime-editing systems for precise genome editing. Plant Commun. 1, 100043. doi:10.1016/j.xplc.2020.100043
Xu, Y., Liu, T., Wang, J., Xiong, B., Liu, L., and Peng, N. (2023a). Reprogramming an RNA-guided archaeal TnpB endonuclease for genome editing. Cell Discov. 9, 112. doi:10.1038/s41421-023-00615-2
Xu, Y., Yuan, Y., Du, N., Yu, W., Shu, S., Sun, J., et al. (2018). Proteomic analysis of heat stress resistance of cucumber leaves when grafted onto momordica rootstock. Hortic. Res. 5 (1), 53. doi:10.1038/s41438-018-0060-z
Xu, Z., Ma, D., Jia, X., Li, Y., Lu, Y., Xie, Z., et al. (2023b). Explore the dominant factor in prime editing via a view of DNA processing. Synthetic Syst. Biotechnol. 8, 371–377. doi:10.1016/j.synbio.2023.05.007
Yadav, A., Mathan, J., Dubey, A. K., and Singh, A. (2024). The emerging role of non-coding rnas (ncrnas) in plant growth, development, and stress response signaling. Non-Coding Rna 10 (1), 13. doi:10.3390/ncrna10010013
Yadav, R., Tripathi, M., Tiwari, S., Tripathi, N., Asati, R., Chauhan, S., et al. (2023). Genome editing and improvement of abiotic stress tolerance in crop plants. Life 13 (7), 1456. doi:10.3390/life13071456
Yan, D., Ren, B., Liu, L., Yan, F., Li, S., Wang, G., et al. (2021). High-efficiency and multiplex adenine base editing in plants using new TadA variants. Mol. Plant 14, 722–731. doi:10.1016/j.molp.2021.02.007
Yan, F., Kuang, Y., Ren, B., Wang, J., Zhang, D., Lin, H., et al. (2018). Highly efficient A·T to G·C base editing by cas9n-guided tRNA adenosine deaminase in rice. Mol. Plant 11, 631–634. doi:10.1016/j.molp.2018.02.008
Yan, J., Oyler-Castrillo, P., Ravisankar, P., Ward, C. C., Levesque, S., Jing, Y., et al. (2024). Improving prime editing with an endogenous small RNA-binding protein. Nature 628, 639–647. doi:10.1038/s41586-024-07259-6
Yang, X., Zhu, P., and Gui, J. (2024a). Advancements of CRISPR-mediated base editing in crops and potential applications in populus. Int. J. Mol. Sci. 25 (15), 8314. doi:10.3390/ijms25158314
Yang, Z., Lu, X., Wang, N., Mei, Z., Fan, Y., Zhang, M., et al. (2024b). GhVIM28, a negative regulator identified from VIM family genes, positively responds to salt stress in cotton. BMC Plant Biol. 24, 432. doi:10.1186/s12870-024-05156-8
Yoon, P., Skopintsev, P., Shi, H., Chen, L., Adler, B., Al-Shimary, M., et al. (2023). Eukaryotic RNA-guided endonucleases evolved from a unique clade of bacterial enzymes. Nucleic Acids Res. 51, 12414–12427. doi:10.1093/nar/gkad1053
Yu, C., Liu, Y., Ma, T., Liu, K., Xu, S., Zhang, Y., et al. (2015a). Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell stem cell 16 (2), 142–147. doi:10.1016/j.stem.2015.01.003
Yu, X. C., Zhu, S. Y., Gao, G. F., Wang, X. J., Zhao, R., Zou, K. Q., et al. (2007). Expression of a grape calcium-dependent protein kinase ACPK1 in Arabidopsis thaliana promotes plant growth and confers abscisic acid-hypersensitivity in germination, postgermination growth, and stomatal movement. Plant Mol. Biol. 64, 531–538. doi:10.1007/s11103-007-9172-9
Yu, Y., Wu, Z., Lü, K., Bi, C., Liang, S., Wang, X., et al. (2015b). Overexpression of the MYB37 transcription factor enhances abscisic acid sensitivity, and improves both drought tolerance and seed productivity in arabidopsis thaliana. Plant Mol. Biol. 90 (3), 267–279. doi:10.1007/s11103-015-0411-1
Yuan, H., Cheng, M., Wang, R., Wang, Z., Fan, F., Wang, W., et al. (2024). mir396b/grf6 module contributes to salt tolerance in rice. Plant Biotechnol. J. 22 (8), 2079–2092. doi:10.1111/pbi.14326
Zaidi, S. S. E. A., Mahas, A., Vanderschuren, H., and Mahfouz, M. M. (2020). Engineering crops of the future: CRISPR approaches to develop climate-resilient and disease-resistant plants. Genome Biol. 21 (1), 289. doi:10.1186/s13059-020-02204-y
Zannoni, L. (2019). Evolving regulatory landscape for genome-edited plants. CRISPR J. 2 (1), 3–8. doi:10.1089/crispr.2018.0016
Zeng, H., Daniel, T. C., Lingineni, A., Chee, K., Talloo, K., and Gao, X. (2024). Recent advances in prime editing technologies and their promises for therapeutic applications. Curr. Opin. Biotechnol. 86, 103071. doi:10.1016/j.copbio.2024.103071
Zetsche, B., Heidenreich, M., Mohanraju, P., Fedorova, I., Kneppers, J., DeGennaro, E. M., et al. (2017). Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nat. Biotechnol. 35 (1), 31–34. doi:10.1038/nbt.3737
Zha, Q., Yin, X., Xi, X., and Jiang, A. (2023). Heterologous vvdreb2c expression improves heat tolerance in arabidopsis by inducing photoprotective responses. Int. J. Mol. Sci. 24 (6), 5989. doi:10.3390/ijms24065989
Zhang, C., Liu, S., Li, X., Zhang, R., and Li, J. (2022a). Virus-induced gene editing and its applications in plants. Int. J. Mol. Sci. 23, 10202. doi:10.3390/ijms231810202
Zhang, D., Pries, V., and Boch, J. (2024a). Targeted C ⋅ G-to-T ⋅ A base editing with TALE-cytosine deaminases in plants. BMC Biol. 22, 99. doi:10.1186/s12915-024-01895-0
Zhang, G., Luo, Y., Dai, X., and Dai, Z. (2023). Benchmarking deep learning methods for predicting CRISPR/Cas9 sgRNA on- and off-target activities. Briefings Bioinforma. 24, bbad333. doi:10.1093/bib/bbad333
Zhang, H., Xu, W., Chen, H., Chen, J., Liu, X., Chen, X., et al. (2021). Transcriptomic analysis of salt tolerance-associated genes and diversity analysis using indel markers in yardlong bean (vigna unguiculata ssp. sesquipedialis). BMC Genomic Data 22 (1), 34. doi:10.1186/s12863-021-00989-w
Zhang, W., Petri, K., Ma, J., Lee, H., Tsai, C., Joung, J., et al. (2024b). Enhancing CRISPR prime editing by reducing misfolded pegRNA interactions. eLife 12. doi:10.7554/elife.90948.2
Zhang, Y., Bai, Y., Wu, G., Zou, S., Chen, Y., Gao, C., et al. (2017). Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. cell Mol. Biol. 91 (4), 714–724. doi:10.1111/tpj.13599
Zhang, Y., Cheng, Y., Fang, H., Roberts, N., Zhang, L., Vakulskas, C., et al. (2022b). Highly efficient genome editing in plant protoplasts by ribonucleoprotein delivery of CRISPR-cas12a nucleases. Front. Genome Ed. 4, 780238. doi:10.3389/fgeed.2022.780238
Zhang, Y., Guo, H., Zhou, L., Shi, Y., Wang, Y., and Yan, K. (2019). A positive regulator of fungal disease resistance, GhBOP1, is involved in lignin accumulation and activates defense-related genes in cotton. Plant Physiology Biochem. 141, 233–243. doi:10.3389/fpls.2021.743795
Zhang, Y., and Qi, Y. (2019). CRISPR enables directed evolution in plants. Genome Biol. 20 (1), 83–84. doi:10.1186/s13059-019-1693-4
Zhao, B., Hu, Y., Li, J., Yao, X., and Liu, K. (2016). Bnaabf2, a bzip transcription factor from rapeseed (brassica napus l.), enhances drought and salt tolerance in transgenic arabidopsis. Bot. Stud. 57 (1), 12. doi:10.1186/s40529-016-0127-9
Zhao, L., Guo, L., Lu, X., Malik, W. A., Zhang, Y.-L., Wang, J., et al. (2022a). Structure and character analysis of cotton response regulator genes family reveals that GhRR7 responses to draught stress. Biol. Res. 55, 27. doi:10.1186/s40659-022-00394-2
Zhao, L., Li, Y., Li, Y., Chen, W. F., Yao, J., Fang, S., et al. (2022b). Systematical characterization of the cotton Di19 gene family and the role of GhDi19-3 and GhDi19-4 as two negative regulators in response to salt stress. Antioxidants 11, 2225. doi:10.3390/antiox11112225
Zhong, D., Pan, H., Li, K., Zhou, Y., Zhao, F., Ye, L., et al. (2024). Targeted A-to-T and A-to-C base replacement in maize using an optimized adenine base editor. Plant Biotechnol. J. 22 (3), 541–543. doi:10.1111/pbi.14256
Zhou, Z., Tang, W., Sun, Z., Li, J., Yang, B., Liu, Y., et al. (2023). Oscipk9 interacts with ossos3 and affects salt-related transport to improve salt tolerance. Plants 12 (21), 3723. doi:10.3390/plants12213723
Keywords: genome editing, climate-resilience crops, abiotic and biotic stress, climate change, CRISPR-cas (clustered regularly interspaced short palindromic Repeats-CRISPRassociated) system, base editing (BE)
Citation: Chavhan RL, Jaybhaye SG, Hinge VR, Deshmukh AS, Shaikh US, Jadhav PK, Kadam US and Hong JC (2025) Emerging applications of gene editing technologies for the development of climate-resilient crops. Front. Genome Ed. 7:1524767. doi: 10.3389/fgeed.2025.1524767
Received: 08 November 2024; Accepted: 07 January 2025;
Published: 10 March 2025.
Edited by:
Amolkumar U. Solanke, Indian Council of Agricultural Research, IndiaReviewed by:
Mawuli K. Azameti, Council for Scientific and Industrial Research (CSIR), GhanaThant Zin Maung, Mahidol University, Thailand
Sang-Ho Chu, Kongju National University, Republic of Korea
Copyright © 2025 Chavhan, Jaybhaye, Hinge, Deshmukh, Shaikh, Jadhav, Kadam and Hong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: U. S. Kadam, dWxoYXNza2FkYW1AZ21haWwuY29t; J. C. Hong, amNob25nQGdudS5hYy5rcg==
 R. L. Chavhan
R. L. Chavhan S. G. Jaybhaye
S. G. Jaybhaye V. R. Hinge
V. R. Hinge A. S. Deshmukh
A. S. Deshmukh U. S. Shaikh
U. S. Shaikh P. K. Jadhav
P. K. Jadhav U. S. Kadam
U. S. Kadam J. C. Hong
J. C. Hong