
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genome Ed. , 26 June 2024
Sec. Genome Editing in Plants
Volume 6 - 2024 | https://doi.org/10.3389/fgeed.2024.1399051
This article is part of the Research Topic Women in Genome Editing View all 3 articles
 Subaya Manzoor1*
Subaya Manzoor1* Sajad Un Nabi2
Sajad Un Nabi2 Tariq Rasool Rather3
Tariq Rasool Rather3 Gousia Gani4
Gousia Gani4 Zahoor Ahmad Mir5
Zahoor Ahmad Mir5 Ab Waheed Wani6
Ab Waheed Wani6 Sajad Ali7*
Sajad Ali7* Anshika Tyagi7*
Anshika Tyagi7* Nazia Manzar8
Nazia Manzar8Modern agriculture has encountered several challenges in achieving constant yield stability especially due to disease outbreaks and lack of long-term disease-resistant crop cultivars. In the past, disease outbreaks in economically important crops had a major impact on food security and the economy. On the other hand climate-driven emergence of new pathovars or changes in their host specificity further poses a serious threat to sustainable agriculture. At present, chemical-based control strategies are frequently used to control microbial pathogens and pests, but they have detrimental impact on the environment and also resulted in the development of resistant phyto-pathogens. As a replacement, cultivating engineered disease-resistant crops can help to minimize the negative impact of regular pesticides on agriculture and the environment. Although traditional breeding and genetic engineering have been instrumental in crop disease improvement but they have certain limitations such as labour intensity, time consumption, and low efficiency. In this regard, genome editing has emerged as one of the potential tools for improving disease resistance in crops by targeting multiple traits with more accuracy and efficiency. For instance, genome editing techniques, such as CRISPR/Cas9, CRISPR/Cas13, base editing, TALENs, ZFNs, and meganucleases, have proved successful in improving disease resistance in crops through targeted mutagenesis, gene knockouts, knockdowns, modifications, and activation of target genes. CRISPR/Cas9 is unique among these techniques because of its remarkable efficacy, low risk of off-target repercussions, and ease of use. Some primary targets for developing CRISPR-mediated disease-resistant crops are host-susceptibility genes (the S gene method), resistance genes (R genes) and pathogen genetic material that prevents their development, broad-spectrum disease resistance. The use of genome editing methods has the potential to notably ameliorate crop disease resistance and transform agricultural practices in the future. This review highlights the impact of phyto-pathogens on agricultural productivity. Next, we discussed the tools for improving disease resistance while focusing on genome editing. We provided an update on the accomplishments of genome editing, and its potential to improve crop disease resistance against bacterial, fungal and viral pathogens in different crop systems. Finally, we highlighted the future challenges of genome editing in different crop systems for enhancing disease resistance.
Microbial pathogens are a constant threat to agricultural productivity, economy and food security (Manzoor et al., 2023). Furthermore, climate change not only affects the plant defence system but also increases the rate of plant disease outbreaks, their distribution and host specificity, thereby putting the global food supply and ecological biodiversity at risk (Bebber, 2015) primary goal of contemporary agriculture is to secure long-term food security while also preserving environmental sustainability. As the world’s population has continually increased in recent years, food security has emerged as a serious global concern. It has become increasingly important to consider the problem of feeding the world’s expanding population of billions while preserving the environment’s balance (Pandit et al., 2022). However, phytopathogens, which represent a danger to the sustainability of food globally, limit the yield of crops. Plant pests and pathogens lead to yield losses globally in rice up-to 30.3%, wheat (30.3%), maize (22.6%) and potato (17.2%) (Savary et al., 2012). Fungal diseases alone in cereals result in yield losses of about (15%–2%) and in extreme cases upto 60% (Rozewicz et al., 2021). Phoma blight in soybeans leads to a yield loss of 51.72% (Manzoor et al., 2023). Fusarium root rot in field peas leads to a yield loss of about 60% (Gossen et al., 2016). The agriculture sector mainly relies on chemical control methods to battle infections in the absence of genetic resistance. However, the use of pesticides and other chemical agents presents issues with the direct or indirect safety of other living things (Damalas et al., 2011). Repeated use of chemicals has polluted the water, air and soil ecosystems, thereby bioaccumulating at higher tropic levels. About 0.1% of chemicals reach the target pathogens, rest accumulate in the surrounding environment (Gill and Garg, 2014). Considering their extensive and sporadic application, phytopathogens have still grown more resilient to several chemical treatments (Ahmad et al., 2020). Colletotrichum truncatum which causes soybean anthracnose, has developed resistance against fungicides such as carbendazim, difenoconozole, azoxystrobin and penthiopyrad. Pathogens have undergone mutations in the beta–tubulin gene, the cytochrome b gene, and the gene encoding the succinate dehydrogenase subunit, which were used as target sites for fungicides (Poti et al., 2023). Reducing the reliance on chemical control in food production has become a critical goal to address the considerable impact of global climate change (Pachauri & Reisinger, A, 2007) and minimize the detrimental environmental effects connected with existing practices (Tilman et al., 2002). Crops with better qualities and performance have been made possible by the processes of plant domestication and breeding. The production of resistant plant types has the most promise for the sustainable control of plant diseases, providing advantages on the social, economic, and environmental fronts. To supply the nutritional needs of human populations, genetic resources that provide resistance to pests and diseases are crucial resources for crop improvement (Mundt, 1994). Traditional breeding techniques have historically been used in conventional resistance breeding to introduce naturally occurring or artificially generated mutant alleles into desired genotypes. Although these genetic methods of disease control have shown significant results over many years, they also have several drawbacks. Notably, such techniques can only be used on plants with adequate genetic diversity and crossbreeding propensity. Additionally, because these methods might transfer several characteristics, covering broad genomic areas rather than only the intended resistant trait (single gene insertion), they may be inaccurate and ambiguous. Additionally, the selection of progeny and the process of genetic crossover can be labour- and time-intensive. Also, most contemporary cultivars that have been chosen to increase yield values are comparatively more vulnerable to infections. Conventional practices must alter to handle the problems caused by the dynamic global food demand landscape, developing diseases, and the period of climate change. The availability of genome and transcriptome sequences requires the adaptation of standard methods (Gao, 2018). The following disadvantages of conventional resistance breeding are present: The first is how time- and labour-intensive it is to produce resistant cultivars. The second phenomenon is linkage drag, wherein top cultivars unintentionally acquire unfavourable traits along with the resistant gene. Third, traditional breeding is only applicable to crossable genotypes. An alternate strategy is mutation breeding, which involves inducing changes in the plant’s DNA using chemical or physical mutagenesis. This technique produces mutants, which are then subjected to stringent selection to assess their desirable traits. This approach also helps with the discovery and mapping of new genes in the genome. However, mutant breeding is a laborious and time-consuming procedure that normally takes 6–7 years to produce results. The existence of random mutations in the genome, which can occasionally be hard to recognize and anticipate, is a significant drawback of this method (Ahmad et al., 2020). Transgenic technology, in particular genetically modified (GM) crops, has emerged as a viable remedy in light of the difficulties faced by agricultural scientists and farmers. Transgenic technology is a flexible method that allows the inclusion of genes from many sources and is not restricted to crossable genotypes. Utilizing this technique, scientists have created crops with much higher yields, increased resilience to biotic and abiotic stressors, herbicide resistance, and greater nutritional value. Regulatory committees have highlighted considerable acceptability problems for transgenic crops, notwithstanding these benefits (Gao, 2021).
In recent years, the advancement in genome editing approach has revolutionized the plant biology and made it possible to precisely modify crops’ genetic makeup to produce desired features. The term “genome editing” refers to a range of technologies that enable the insertion, deletion, or modification of genetic information at certain sites within the genome of a living creature. The term “new breeding techniques” (NBT) refers to modern, precise molecular methods for focusing on one or more genes. The mechanism of DNA double-strand break (DSB) repair, in which sequence-particular nucleases recognize certain DNA sequences, is the basis for the core processing in genome editing. Nonhomologous end joining (NHEJ) and homologous recombination (HR) are the two main mechanisms by which DSBs are repaired (Voytas and Gao, 2014). Using NHEJ or HR to precisely alter genes, new plant types with favourable agronomic features may be created. While HR uses homologous sequences as templates to recreate the lacking DNA sequences at the breakpoint, NHEJ uses distinct enzymes for DSB repair. Even though NHEJ is error-prone and can result in insertion or deletion alterations, the majority of cells frequently utilize it for DSB repair. Although when a donor DNA template is present, the HR route predominates, leading to precise and targeted alterations. Genome editing uses a variety of sequence-specific nucleases, including base editing, meganucleases (MNs), transcription activator-like effector nucleases (TALENs), zinc finger nucleases (ZFNs), clustered regularly interspaced short palindromic repeats (CRISPR), and CRISPR-associated protein (Cas9). These genome editing tools provide democratic approaches and are distinguished by their quick and low-cost manufacture, making them available to public-private partnerships with nonprofit objectives in addition to private businesses and multinational corporations (Ricroch, 2019). They are already used in many public laboratories. Gene editing tools like CRISPR/Cas9, which are especially efficient and practical, function without the use of proteins or engineering processes. Cas9 nucleases target DNA with single-guide RNA (sgRNA), which is formed from the duplex-RNA structure incorporated with CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA). The method effectively targets certain genomic regions by changing the dual crRNA-tracrRNA structure into a single-guide RNA (Ahmad et al., 2020). The range of its uses has also been broadened by recent developments in CRISPR-Cas13 technology, base editors, and prime editors (Molla et al., 2020).
The growing abundance of recent studies on plant genome editing through GE technology suggests its practicality, owing to its heightened success rate and user-friendly nature. Beyond plant applications, CRISPR/Cas9 has been employed to target genes encoding proteins involved in interactions between host plants and fungal or oomycete pathogens. This facilitates the exploration of the molecular mechanisms underlying host-pathogen recognition and enables the development of screening systems for disease resistance (Fang and Tyler, 2016). In this review, we highlighted an overview of genome editing technologies and their applications in bolstering plant disease resistance. We delve into the mechanisms underlying genome editing and explore recent advancements in CRISPR/Cas-based tools, including base editing and prime editing. Furthermore, we investigate the potential impact of genome editing on sustainable farming practices, global food security and shaping the future of agriculture. Finally, we highlight the challenges and ethical concerns associated with the widespread adoption of genome-edited crops and propose potential strategies to address these issues. By highlighting recent developments and prospects in genome editing for plant disease resistance, this review aims to contribute to a deeper understanding of this innovative approach and its implications for agriculture and society.
Phytopathogens pose a significant threat to the agricultural economy, causing up to a 40% yearly production loss in economically important crops (Baldi and La Porta, 2020). The financial impact of these losses is substantial, amounting to an estimated $220 billion each year (FAO, 2019). The issue is intensified by the surge in global commerce, which facilitates the fast spread of invasive pathogens, resulting in significant crop damage and reduced yields (Baldi and La Porta, 2020). For instance, numerous soil-borne diseases, including plant-parasitic nematodes, Fusarium species, Rhizoctonia species, and Pythium species, can cause major production losses in legume crops, whether they occur alone or in combination (Panth et al., 2020). Similar to rice, Bipolaris oryzae, a dematiaceous hyphomycete, is responsible for the devastating brown spot diseases that affect rice. This contributed to the 1943 Bengal famine in India. The same fungus is still present in important rice-growing regions of the world, despite the fact that it has not recently produced any significant outbreaks (Sobanbabu et al., 2018). Worldwide crop losses and starvation have been attributed to graminicolous hyphomycetes, which are linked to cereal crops and their wild cousins. These phytopathogens have been shown to be highly destructive. Southern corn leaf blight, which is caused by Bipolaris maydis, has been linked to significant crop losses worldwide (Manzar et al., 2022) and Exserohilum turcicum, which causes northern corn leaf blight. Another typical leaf spot pathogen in barley and wheat is Bipolaris sorokiniana (Kashyap et al., 2023). Pyricularia oryzae, also known as Magnaporthe oryzae, is a fungal disease that causes frequent outbreaks and is thought to be the most devastating pathogen in rice cultivars. Up to 30% of the world’s rice harvest is lost to this disease every year, making it difficult to control and potentially causing humanitarian and economic problems, especially in Asia (Savary et al., 2019). Many studies on the molecular causes of illnesses and host-pathogen interactions use P. oryzae. Another invasive disease that started in the United Kingdom in 1994 is boxwood blight, which is now found in Europe, Asia, New Zealand, and North America (LeBlanc et al., 2018). Boxwood blight, as its name implies, is a disease that affects boxwood (Buxus spp.) and results in latent dieback of the leaves as well as fast defoliation (LeBlanc et al., 2018). Similarly, the growth and yield of boxwood in nurseries and landscape plantings are at risk due to recent outbreaks of boxwood blight disease, which is brought on by the fungus Calonectria pseudonaviculata. This poses a serious danger to the ornamental plant sector (LeBlanc et al., 2018). After contracting C. pseudonaviculata, plants become debilitated, which can lead to plant stress and secondary invader colonization, which frequently results in plant mortality. A wide variety of crops and non-crop plants are affected by foliar anthracnose, which is typically found in tropical and subtropical regions (LeBlanc et al., 2018). As a result, foliar Colletotrichum species are regarded as a major source of pre- and post-harvest loss of a wide range of high-value crops, and they are also regularly discovered in plant biosecurity interceptions. Apple mosaic virus (ApMV) and a recently reported novel virus, apple necrotic mosaic virus (ApNMV), linked to mosaic disease in apples, represent a significant economic threat to the apple industry (Nabi et al., 2020). Viral infections affect the entire plant system, are systemic, and are capable of affecting the overall health of orchards (Manzoor et al., 2023; Nabi et al., 2023). The Geminiviridae single-stranded DNA viruses are represented by two viruses: tomato yellow leaf curl virus (TYLCV) and african cassava mosaic virus (ACMV), both of which have enormous economic importance and cause huge economic losses, much of which is exacerbated by efficient transmission via whitefly vectors. The yearly losses from ACMV (and related species) are currently estimated to be between $1.9 and $2.7 billion, with the cassava disease epidemic in East and Central Africa generating significant misery and concerns.
The yield loss estimated due to various diseases is illustrated in (Table 1). Addressing these challenges is critical, and one potential approach is the use of genome editing technology in disease management. Genome editing has emerged as a cutting-edge technique with the potential to revolutionize the way we combat plant diseases. Researchers can increase plant resilience to pathogens by accurately changing their genetic composition, lowering susceptibility and output losses. This novel methodology offers a focused and effective method for creating crops with better disease resistance. Scientists may use genome editing tools like CRISPR-Cas9 to make precise changes to particular genes linked with disease susceptibility, therefore improving the plant’s natural defensive systems (Bi et al., 2024). The use of genome editing in agriculture holds promise for developing resilient crops that can survive the encounters posed by emerging diseases. As we strive to ensure global food production and mitigate the economic effects of plant diseases, genome editing emerges as a potent tool in the goal of sustainable and resilient agriculture. By harnessing the potential of genetic modification, we may aim to reduce the destructive consequences of plant diseases while also ensuring a more secure and productive future for agriculture.
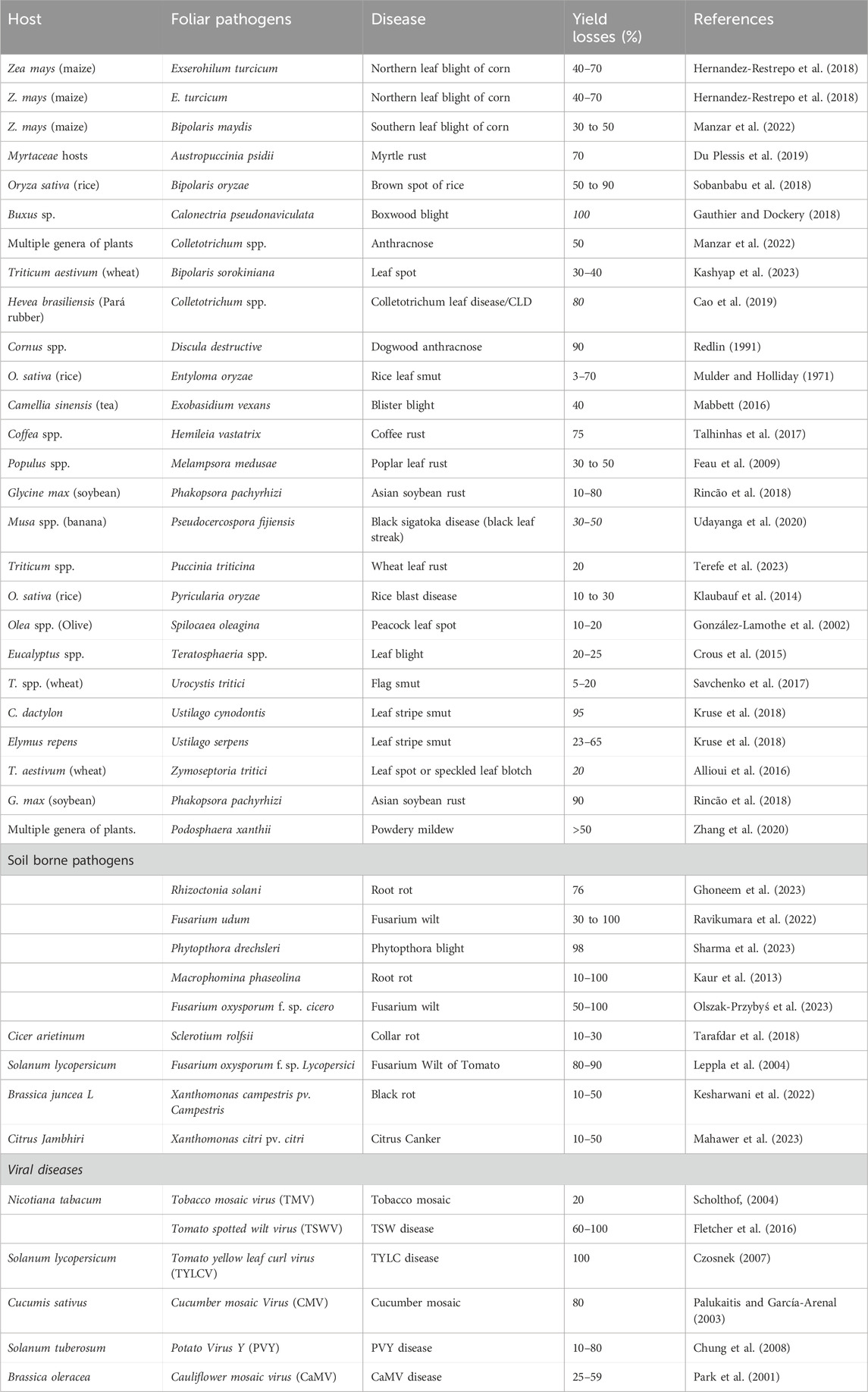
Table 1. Impact of foliar, soil borne pathogenic fungal, bacterial and viral diseases on crop productivity.
Plant disease control is critical for avoiding production losses in diverse crops. The majority of management techniques fall into three categories: physical, chemical, and biological. Plant disease control relies heavily on the indiscriminate use of chemical pesticides such as fungicides, bactericides, and insecticides that are detrimental to plant pathogens or vectors (Singh et al., 2020). However, the adverse effects of these pesticides and their breakdown products may represent a risk to the environment and human health prompting researchers and producers to investigate alternative and eco-friendly methods of disease management (Ali et al., 2018; 2023). Also, heavy reliance on chemical pesticides may foster the development of pesticide-resistant pathogens. Furthermore, frequent use can disrupt natural ecosystems and beneficial organisms (Pathak et al., 2022). There is a growing emphasis in research emphasize on the development of safer and more efficient fungicides and pesticides, including novel formulations and application techniques. On the other hand, biological control methods harness beneficial organisms to suppress plant pathogens. Using natural enemies, such as predators, parasitoids, and microorganisms, suppresses plant pathogens. It is eco-friendly and sustainable, minimizing the use of chemical pesticides and reducing the risk of pesticide resistance. However, it takes time for populations of natural enemies to establish and control the pathogen, which can pose unintended ecological impacts on non-target species (Kohl et al., 2019). Another important disease controlling tactics in agriculture is implementing cultural practices mitigating plant disease incidence and severity. Practices such as crop rotation, sanitation, and proper irrigation management contribute to disrupting pathogen populations and interrupting their disease cycles. However, cultural practices require significant labour or changes in farming practices, which could increase production costs. Additionally, cultural control alone may not be sufficient to manage severe pathogen infections (Singh et al., 2012). Developing crop varieties with genetic resistance to specific pathogens can offer long-term and sustainable protection. This approach reduces the dependence on chemical pesticides and keeps pathogens at bay (Pathania et al., 2021). However, developing resistant varieties through breeding or genetically engineered can be time-consuming. Additionally, pathogens may evolve to overcome plant resistance, rendering resistance ineffective over time (Miedaner, 2016). However, genome editing (GE) methods enable quick trait alteration in agricultural plants and has emerged promising tool for developing disease resistant crops.
One of the primary goals of plant pathologists is the identification and exploitation of potential targets that have evolved in disease resistance mechanisms that can be utilized for generating future disease resistance cultivars in sustainable agriculture. This requires an in-depth understanding of the molecular mechanisms of host-pathogen interactions and disease resistance. Plants evolved multi-layered defensive systems against microbial infections over time. Pre-formed physiological barriers play the first line of defense against microbial pathogens; however, plants may also generate adequate defensive responses after pathogen perception, which is mediated by cell wall plasma membrane-bound and intracellular immunological receptors that identify pathogen-derived molecules or by indirectly modifying host targets. With the advent of high-throughput tools, the identification and functional validation of genes related to host immunity, such as receptors, pathogen Avr proteins and effectors, pathogen-associated molecular patterns (PAMPs), channels, and signaling molecules, has provided a snapshot of plant immune systems and their signaling cascades in response to different pathogens. Now it is well documented that there are two tiers of plant immune systems: effector-triggered immunity (ETI) and PAMP-triggered immunity (PTI) (Jacob et al., 2013). Modifying or engineering genetic resistance traits is the most cost-effective strategy to improve disease resistance in crops. However, this requires vast knowledge about the molecular dynamics of host-pathogen interactions and plant immune responses. For developing long-term disease-resistant cultivars, there is a need to understand host-pathogen interaction using different tactics. For example, an in-depth understanding of disease epidemics and “hot spots” to screen disease-resistant or susceptible genotypes in the field during natural disease epidemics is important in order to choose resistant genotypes. After being cultivated in hotspot areas, the test genotypes can be examined for their disease resistance or susceptibility (Ye et al., 2002). Major obstacles that are often faced in the natural screening method could be avoided by screening disease-resistant genotypes under artificial circumstances (Ye et al., 2002). Genotypes can be screened in a glasshouse with controlled environmental conditions to monitor disease resistance using new tools. In the controlled glasshouse or greenhouse, the ideal photoperiod, relative humidity, and temperature may be readily changed to meet the needs for the advancement of the disease. Certain laboratory-based screening techniques, such as the whole plant infection, detached leaf test, purified phytotoxins or culture filtrates, and the cut-twig method, are commonly used to study host pathogen interactions and disease progression (Sharma et al., 1995).
In the past, traditional breeding and transgenic breeding have played key roles in improving disease resistance in various economically important crops. Like selecting resistant plants from commercial varieties, the utility of this method is limited. Introduction of resistant varieties into new areas; however, this method possesses the limitation of variety not performing well in new environments or becoming susceptible to specific diseases in new areas (Russell, 2013). By mating a superior recipient parent line with a donor line that possesses advantageous qualities, conventional plant breeding, predominantly cross-breeding, has long been used to enhance plant properties, including disease resistance. However, this whole method is labor-intensive, time-consuming, ineffective in transferring undesired genes to desired genes and rife with drawbacks (Gao, 2021). Another mainstay of disease resistance breeding programs is the establishment of appropriate and efficient screening methods. To achieve desired results, a thorough understanding of pathogenicity, virulence pattern, resistance type, and an efficient breeding strategy is needed. Effective strategies for screening disease resistance to multiple pathogens have been developed, and enough research has been conducted in the past 10 years to define the nature and durability of resistance (Podder et al., 2013). The development of molecular markers and next-generation sequencing techniques made it simple to pinpoint the genes and QTLs linked to particular disease resistance. Recent genomic techniques, such as bi-parental QTL mapping (Collard et al., 2005), association mapping and QTL Seq (Takagi et al., 2013), can be used to correctly quantify the connection between genes and molecular markers. Creating a mapping population, screening for polymorphic markers, phenotyping the population, genotyping the population using polymorphic markers, creating a linkage map, performing QTL analysis, and validating linked markers are all steps in the development of molecular markers. To screen a candidate, a molecular marker closely associated with the QTL or gene of interest may be used.
Under incompatible host-pathogen interactions, the host displays complete disease resistance against the pathogen when both the pathogen’s homologous avr and the host’s R gene are present (Flor, 1971). In the early 1900s, British scientist Rowland Biffen showed that R gene-mediated resistance might be beneficial in wheat (Triticum sp.) breeding (Biffen, 1905). Since then, a large number of R genes have been cloned and inserted into other species (De Wit et al., 1985), genera (Tai et al., 1999), and species borders (Song et al., 1995). For instance, in laboratory settings, the transfer of the R gene Rxo1 from maize (Zea mays) into rice (Oryza sativa) resulted in resistance to the bacterial streak disease (Zhao et al., 2005). Multiyear field trials conducted in tomatoes in commercial-type growth settings have shown that strong resistance to Xanthomonas sp. producing bacterial spot disease is conferred by tomatoes expressing the pepper gene (Kunwar et al., 2018). In the field, wheat transgenic for different alleles of the wheat resistance locus Pm3 showed racialized resistance to powdery mildew (Brunner et al., 2012). Strong field resistance to the causative agent of potato late blight, is exhibited by transgenic potatoes expressing the wild potato R gene RB or Rpi-vnt1.1 (Jones et al., 2014). Notably, the only genetically modified crop with increased resistance to a nonviral disease that has been licensed for commercial use to date is the transgenic potato expressing created by J.R. Simplot.
As pathogens can escape detection of their gene by the R gene, the disease resistance conferred by a single R gene is therefore frequently not durable in the field. Multiple R genes are sometimes inserted simultaneously, a practice known as stacking, to increase durability and widen the resistance spectrum (Mundt, 2018). Since the emergence of a pathogen strain that may overcome resistance supplied by several genes at once is a rare occurrence, resistance conferred by stacked R genes is projected to be long-lasting. Cross-breeding pre-existing R loci is one method of stacking R genes. Breeders can use marker-assisted selection to identify offspring with the desired R gene makeup (Das and Rao, 2015). For instance, three R genes—Xa21, Xa5, and Xa13 give resistance to X. oryzae pv. oryzae (Xoo) in the deep-water rice cultivar Jalmagna rice by cross-breeding and marker-assisted selection (Pradhan et al., 2015). Eight Xoo isolates were investigated, and the line that resulted with the stacked R genes displayed a high degree of disease resistance under field conditions (Pradhan et al., 2015). Even though marker-assisted selection has significantly improved selection efficiency, integrating several loci using this technique may still take a long time. Only a few quantitative traits controlling a remarkable variation in population can be selected by this method (Lamichhane and Thapa, 2022).
In chemical or physical mutagenesis, seeds are exposed to various mutagens to cause mutations in the plant’s DNA. This process is known as mutation breeding. The process’s mutants go through rigorous selection to assess desired traits and find new genes in the genome. However, mutant breeding is a laborious and drawn-out procedure that frequently takes 6–7 years to achieve the desired outcomes. The randomness of mutations, which makes their detection and prediction difficult, is a key methodology drawback. Several crops have effectively enhanced a variety of qualities through transgenic breeding, which requires moving a desired gene from one genome to another; examples include Bt cotton, Bt maize, transgenic rice resistant to blast, sheath blight, and false smut (Shen et al., 2023). To improve desired features, the gene of interest is carefully introduced into the host variety’s genome. However, the introduction of foreign DNA causes transgenic crops to be genetically changed, which raises concerns among the general public and the scientific community about their adoption (Ahmad et al., 2020).
Genome editing is one of the new genetic modification approaches that is sometimes attributed to “new breeding techniques,” and it offers encouraging availabilities for the sustainable development of crops in the future. With the use of gRNA and guided target sites, genome editing enables the exact targeting and destruction of certain negative regulators or genes, as well as the reorganization of chromosomes in elite variety genomes. The utilization of genome editing approaches, such as the CRISPR/Cas9 system, offers benefits including efficacy, robustness, cost-effectiveness, and the lack of foreign DNA, potentially enabling these crops to circumvent GMO restrictions and be marketed as non-GMOs (Ahmad et al., 2020).
Plant pathogens demonstrate diverse strategies, ranging from biotrophic to necrotrophic lifestyles, to efficiently colonize plant hosts (Andolfo and Ercolano, 2015). Biotrophs rely on live host cells, either completely or partially, for their life cycle completion, causing minimal damage to host cell walls and maintaining host viability. Examples include obligate biotrophic fungi like Puccinia graminis, which depend on living host cells for sustenance. In contrast, necrotrophs such as Botrytis cinerea kill host tissues during infection, using cell wall-degrading enzymes and toxins to obtain nutrients. These pathogens deploy various virulence factors, including effectors, toxins, and cell wall-degrading enzymes, to manipulate host physiology and evade immune recognition. Nevertheless, plants have developed sophisticated defense mechanisms to counter pathogen attacks. The plant immune system utilizes pattern recognition receptors (PRRs) to detect conserved microbial signatures, initiating immune responses mediated by defense hormones like salicylic acid, jasmonic acid, and ethylene. Additionally, plants harness resistance (R) genes, encoding intracellular receptors that specifically recognize pathogen effectors, leading to a localized hypersensitive response (HR) and systemic acquired resistance (SAR). Understanding this intricate interplay between pathogens and plant immunity is pivotal for comprehending disease dynamics and devising crop protection strategies (Jones and Dangl, 2006; Andolfo and Ercolano, 2015).
Plants lack an adaptive immune system, unlike animals (Kumar et al., 2011). Consequently, they rely on a complex array of defences to fend off pathogens. However, pathogens have evolved strategies to bypass or suppress these plant immunity mechanisms. Here’s how they accomplish this: The primary defense line for plants is the cell wall, a robust barrier primarily composed of cellulose and other constituents. Pathogens employ various tactics to subdue this first line of defense, known as pattern-triggered immunity (PTI). Some produce enzymes that degrade these components, facilitating entry. For example, fungal pathogens release enzymes like cutinases, cellulases, and pectinases to breach the plant cell wall. An instance is seen in Colletotrichum gloeosporioides, the pathogen responsible for oil tea anthracnose, which secretes the cutinase CglCUT1 to degrade the cuticle during infection (Wang et al., 2017). Others release molecules that mimic plant hormones, manipulating plant growth and impeding defense responses. Pseudomonas syringae, for instance, produces effectors that manipulate stomata, aiding pathogen entry (Zhang et al., 2022). Pathogen effectors can also target plasmodesmata (PD), enlarging pore size to control cytoplasmic continuity. During Fusarium oxysporum infection, effectors like Avr2 and Six5 enlarge PD pore size (Cao et al., 2018). Xanthomonas oleifera T3E XopR hijacks the Arabidopsis actin cytoskeleton during infection, disrupting host actin assembly (Sun et al., 2021). In a study by Jin et al. (2023), the bacterial pathogen P. syringae’s T3SS protein HrpP was found to facilitate effector translocation and manipulate plant immunity to promote bacterial infection. This underscores the ongoing arms race between plants and pathogens, where both constantly evolve strategies to outmaneuver each other. Pathogens often target receptor-like kinases to avoid recognition by the plant’s immune system. Effector proteins like CoNIS1 and MoNIS1 inhibit kinase activity, thereby suppressing immune activation (Irieda et al., 2019). Pathogens also act through effectors to disrupt phytohormone-dependent plant defenses. For instance, oomycete pathogens secrete isochorismatases that hinder salicylic acid (SA) biosynthesis, diminishing the plant’s ability to mount an effective defense response (Han et al., 2019). By employing these tactics, pathogens establish themselves within the plant and cause disease. Understanding these mechanisms is vital for developing strategies to protect crops and enhance plant disease resistance.
Genome editing refers to the exact alterations made to the genomic DNA of cells or animals using site-specific genome targeting methods, similar to targeted mutagenesis or site-directed insertion, deletion, or replacement (Ahmad et al., 2020). These cutting-edge methods have a great deal of promise to be effective tools for understanding gene function and promoting crop development. Sequence-specific nucleases are used in the procedure to recognize and target certain DNA sequences, resulting in the creation of double-stranded breaks (DSBs) at predetermined places. Diverse mechanisms, including non-homologous end-joining (NHEJ), micro-homology-mediated end-joining (MMEJ), and homology-directed repair (HDR) pathways, are used to restore DSBs (Figure 1) (Molla and Yang, 2020). Although NHEJ is the main DSB repair pathway, it frequently introduces mistakes that lead to insertion or deletion changes at the DSB sites. Conversely, while it does not happen often in plant systems, repair via the HDR route is error-free. CRISPR/Cas9 and other gene editing technologies, in particular, have a dramatic impact on fundamental research in plants and crop enhancement. Notably, one of the key benefits of these methods is the simplicity with which the transgenes originally used for genetic alteration may be removed by genetic segregation. As a result, gene-edited variants become identical to those created utilizing traditional breeding techniques (Zhang et al., 2017). By focusing on illness-related genes, genome editing has been used to ameliorate disease resistance (Zhang et al., 2017). Additionally, genome editing has been used to build broad-spectrum disease resistance in crops. This has been done either by using particular host-vulnerability genes (the S gene method) or by cleaving the genetic material of phytopathogens to prevent their growth (Zaidi et al., 2018).
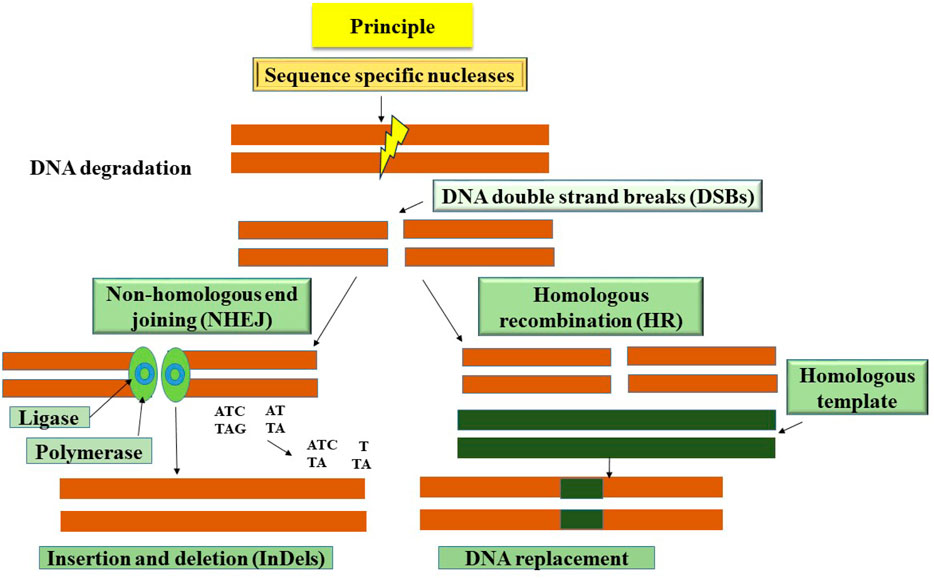
Figure 1. A schematic representation shows the basic principle of genome editing techniques to cleave double-stranded DNA at a targeted position on the genome. Various methods such as insertions, deletions or substitutions or gene insertion are used to disrupt the function of the gene using genome editing.
Single gene knockout and multiplex gene knockout stand out as two particularly influential uses of genome editing in modern plant science. Individual gene knockout: currently, precise deletion of target genes is the remarkably common and generally used use of genome editing. This method enables thorough investigations into gene activities and their consequences in diverse biological processes by selectively disrupting particular genes. Multiple genes may be targeted concurrently in different plant species via genome editing, a technique known as multiplex gene knockout. Researchers may explore complicated gene connections and regulatory networks using this multiplex gene knockout technology, providing greater insights into the intricacies of plant biology (Mushtaq et al., 2019). Large-scale deletions: When two double-stranded breaks (DSBs) are established on the same chromosome at a specific distance, the two points can be connected via the non-homologous end-joining (NHEJ) repair pathway, which results in the deletion of the intervening sequence. For particular research projects and crop development initiatives, similarly to the investigation of gene clusters and noncoding RNAs, this method of generating rather significant deletions has proven useful (Ordon et al., 2017). Gene knockout in polyploid plants: In polyploid crops lacking sufficient mutant resources, genome editing methods have shown outstanding value. Notably, genome editing tools have accomplished gene knockout in triploid plants, with citrus and apples serving as examples (Zhang et al., 2017). This has permitted precise genetic alterations in complicated polyploid genomes. Gene targeting is the deliberate use of genetic engineering techniques to substitute a DNA fragment exactly for another (gene replacement) or to precisely introduce a new sequence into a particular genomic location (gene knock-in) (Zhang et al., 2017). This focused strategy has enormous potential for specialized genetic alterations and might revolutionize crop improvement and other genetic treatments. In the past, the application of genome editing has been carried out in both model species Arabidopsis thaliana (Li et al., 2013) and Nicotiana benthamiana (Gao et al., 2015), as well as different crop systems such as rice, wheat, sorghum, soybean, corn, and potato, to improve diverse traits related to stress tolerance, growth, and yield (Rani et al., 2016). Similarly, by altering disease-related genes, genome editing has shown promise for improving disease resistance and providing a way to strengthen crop defences against pathogens (Mishra et al., 2021).
Plants defend against pathogen attacks by two-tiered immune systems, namely, PTI and ETI, that are triggered by cell surface-localized PRRs and intracellular nucleotide-binding domain leucine-rich repeat-containing receptors (NLRs) (Ali et al., 2018). Both PTI and ETI immune receptor classes eventually converge into many identical downstream responses, even though with differing amplitudes and dynamics; this is despite the fact that they involve separate activation processes and appear to require different early signaling components. Different tactics are employed by plants to fend against pathogen attacks. Immune receptors such as extracellular pattern-recognition receptors (PRRs) and nucleotide-binding leucine-rich repeat (NLR) receptors recognize pathogens or their derived molecules or effectors, which triggers a variety of defense signaling pathways to thwart pathogen attack (Figure 2). Activation of PTI and ETI lead a series biochemical events like cytosolic calcium burst, ROS formation and hormonal reprogramming that regulates plant immune responses. Genome editing can be used to manipulate several potential targets, of host immune system and pathogens including effector binding targets, S and R genes, hormonal pathways, and pathogen virulence factors, in order to improve disease resistance. Phytopathogens usually target susceptibility (S) genes in plants in order to multiply and cause disease progression more easily. Therefore, by altering these S genes, it may be possible to create long-lasting, broad-spectrum disease resistance by interfering with the host-pathogen relationship. Targeting S genes using genome editing is the most viable and transgenic-free approach to developing disease-resistant cultivars in sustainable agriculture. For instance, editing susceptibility genes like SWEETs (Sugars Will Eventually Be Exported Transporters) or receptor-like kinases (RLKs) can reduce pathogen susceptibility (Gupta et al., 2021). Additionally, manipulating defense-related genes like transcription factors or genes involved in hormone signaling pathways can bolster plant immunity (Kourelis and Van der Hoorn, 2018). On the pathogen side, editing virulence factors such as effectors or genes involved in pathogen recognition can attenuate pathogenicity. For example, CRISPR-mediated editing of effector genes in pathogens like Phytophthora infestans can impair their ability to cause disease (Fang and Tyler, 2016). Furthermore, targeting essential genes in pathogens through gene editing can render them non-viable or less virulent, thereby reducing disease severity (Nekrasov et al., 2017). Integrating these approaches offers a promising avenue for developing durable disease resistance in crops.
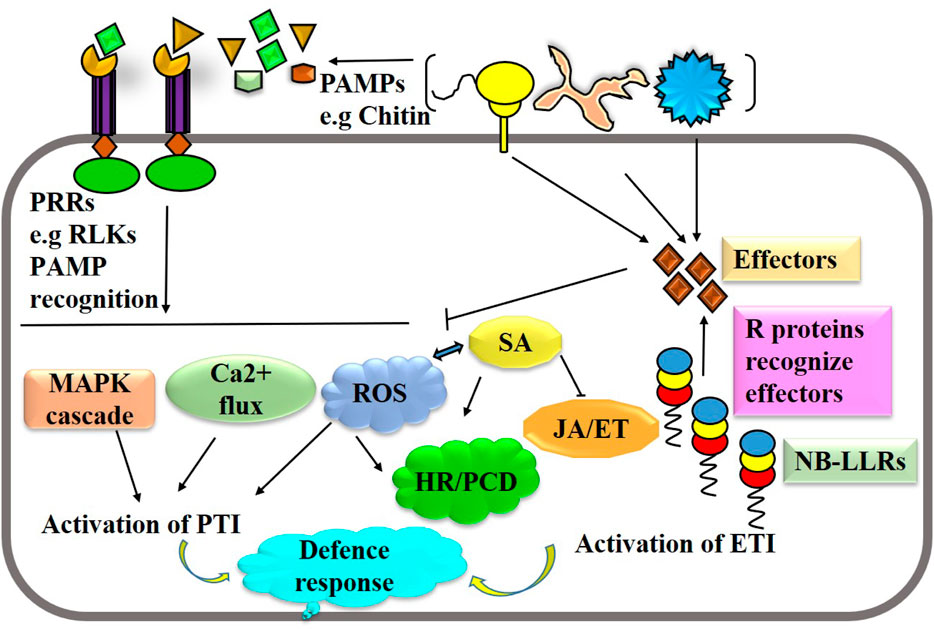
Figure 2. A simplified schematic illustration of the immune system in plants. Recognition of pathogen or derived elicitors by pattern-recognition receptors and effectors by nucleotide-binding leucine-rich repeat leads the activation of PTI and ETI. Activation of PTI and ETI lead a series biochemical events like cytosolic calcium burst, ROS formation and hormonal reprogramming that regulates plant immune responses.
Modern genome editing (GE) technologies have been instrumental in crop improvement, especially in terms of boosting disease resistance. The CRISPR/Cas9 system, CRISPR/Cas13, base editing, transcription activator-like effector nucleases (TALENs), zinc-finger nucleases (ZFNs), and meganucleases are some of these cutting-edge techniques which have been utilized from crop improvement (Karmakar et al., 2022). With the use of these versatile tools, crop genomes may be precisely and specifically modified, opening up the enormous potential to engineer disease resistance in crops by changing certain genes involved in the plant immune system. They are useful tools to improve crop food sustainability and security throughout the world because of their efficacy and adaptability. In this review, we discussed the application and limitations of different genome editing tools available for crop improvement.
One of the earliest sequence-specific nucleases used for targeted double-stranded DNA breaks was the meganuclease, also known as the homing endonuclease. Prokaryotes, archaea, and unicellular eukaryotes all include them naturally (Silva et al., 2011). These amazing enzymes are highly effective at cleaving double-stranded DNA at predetermined recognition sites, which generally include between 14 and 40 base pairs. Their recognition sites are larger than those of type II restriction enzymes frequently used in recombinant DNA technology, making them the most popular naturally occurring restriction enzymes (Carroll et al., 2017). Meganucleases may change their recognition sequence through protein engineering, allowing them to replace, remove, or change any sequence of interest in an extremely focused and effective manner. They are involved in the process of homing, which permits them to start recombination, and are encoded by mobile genetic elements. These genes can transmit genes horizontally because they are located in introns or intergenic regions (Stoddard, 2005). Meganucleases, which include DNA binding and DNA cleavage domains, cause double-stranded breaks at their recognition sites. Homologous recombination (HR) is then used to repair the lesions. By inserting the meganuclease coding sequence into the target gene, targeted genome engineering with designed meganucleases is made possible (Hafez and Hausner, 2012). Meganucleases have been used to precisely alter plant genomes since the 1990s and to learn more about the underlying principles controlling the integrity of plant genomes. However, their application in genome engineering is not as general as that of zinc-finger nucleases (ZFNs) or transcription activator-like effector nucleases (TALENs) due to certain limitations. Firstly, the DNA binding and cleavage domains in meganucleases overlap, which can compromise their catalytic activity (Stoddard, 2011). Secondly, meganucleases lack the modular DNA-binding domain architecture present in ZFNs and TALENs. Lastly, there are instances where meganucleases may exhibit sequence degeneracy, increasing the likelihood of off-target binding (Argast et al., 1998). Despite these limitations, meganucleases remain a promising avenue for targeted genome engineering and persist as a subject of research and development in the field of genetic modification.
The first sequence-specific nucleases previously used for genome editing in eukaryotes were zinc finger nucleases (ZFNs). ZFNs were used for the first time to modify the plant genome in 2005 (Lloyd et al., 2005). One of the most frequent DNA-binding domains in eukaryotes is the zinc finger domain, which was first determined in 1985 as a component of transcription factor IIIa in Xenopus oocytes. The zinc finger (ZF) domain is made up of a variety of Cys2His2 zinc fingers, each of which has around 30 amino acids and may bind to homologous triplets of nucleotides. The ability of ZFs to recognize all 64 potential nucleotide combinations makes it possible to develop ZFNs that can specifically target any 3–6 nucleotide triplet. There are two distinct functional groups in the ZFN monomer. A synthetic ZF domain at the N-terminus attaches to the target DNA, and a DNA cleavage domain called FokI is located at the C-terminus, where it causes double-strand breaks (DSBs) in the target genomic region. The FokI nuclease must dimerize to break double-stranded DNA. To create a DSB at the targeted DNA site, a set of ZFNs is necessary. Two distinct ZFNs must bind to the forward and reverse strands independently for FokI dimerization to occur (Zhang et al., 2017). Additionally, the forward and reverse target sequences should be separated by a 5–7 bp spacer region. ZFNs have been commonly employed for genome editing in both plant and animal systems for more than 10 years (Mushtaq et al., 2019). ZFNs have been used to change the endogenous gene ABA-INSENSITIVE-4 (ABI4) in Arabidopsis, for example, (Osakabe et al., 2010). ZFNs were employed in the deletion of a 4.3 kb integrated GUS gene sequence in tobacco that was surrounded by ZFN cleavage sites (Petolino et al., 2010). Additionally, ZFNs were used to target three homologous copies of the aceto-hydroxy acid synthase (AHAS) gene in hexaploid wheat (T. aestivum) to simultaneously knock down several genes (Ran et al., 2018). ZFNs have been successful in genome editing, but their design and aggregation are technically difficult, and it can be expensive to outsource the production of ZFN modules to commercial vendors (Ramirez et al., 2008). Despite these obstacles, ZFNs have significantly contributed to the evolution of genome editing and have been used in several ground-breaking investigations involving a variety of taxa.
When compared to ZFNs, TALENs, which are protein-based DNA-targeting enzymes, have been shown to be more efficient and selective. Like ZFNs, TALENs are made up of two domains: a target-specific DNA-binding domain that may be customized and a non-specific DNA cleavage domain (Fok1). The TAL effector (TALE) is the name of the DNA-binding domain of TALENs. When Xanthomonas bacteria infect their host plants, they release TAL effector proteins that bind to the plant’s DNA via a domain that contains some tandem 34–35 amino acid repeats (Malzahn et al., 2017). An N-terminal translocation signal, an acidic transcription-activation domain, and a central DNA-binding domain are all common features of TALE proteins. The repeat variable di-residues (RVDs), which are located next to each other at positions 12 and 13, make up the almost identical 33–35 amino acid repeats that make up the DNA-binding domain. Two different teams independently cracked the DNA binding specificity of TALE in 2009 (Moscou and Bogdanove, 2009). The DNA-binding domain’s repeat units each bind to a single nucleotide, and the RVDs inside the repeats have a significant connection with one particular nucleotide (A, G, C, or T). By choosing a certain aggregation of the four conspicuous repetitions to accommodate the proper RVDs, it is possible to create particular DNA-binding proteins thanks to the clear link between protein sequence and DNA recognition. Since TALENs are methylation-sensitive, they may be created and come together to target any DNA sequence (Kaya et al., 2017b). Similar to ZFNs, a dimerized functional FokI requires two Talen monomers to form. Since TALENs are simpler to assemble than ZFNs, more plants are using this editing technique (Zhang et al., 2017). Designing a sizable number of identical repeat sequences for TALENs, however, is one of the main technological hurdles addressed by scientists (Li et al., 2020). By accurately altering the Lipoxygenase (Lox3) gene, TALEN-based targeted mutagenesis has been effectively used to improve storage tolerance in rice (Ma et al., 2015).
Early 1987 witnessed the discovery of CRISPR, which has subsequently been found in numerous more bacterial species. However, CRISPR was first used to alter plant genes in 2013, and it has since taken over as the most extensively used gene editing approach (Nekrasov et al., 2017). Depending on how the Cas genes are arranged and their structural variation, the CRISPR-Cas system may be divided into two main groups. While Class 2 CRISPR-Cas systems are characterized by a single effector complex, Class 1 CRISPR-Cas systems use several protein effector complexes. Based on recent studies, there are two classes, six kinds, and more than 30 subtypes of CRISPR-Cas systems (Koonin and Makarova, 2022). Although there are many other CRISPR-Cas systems in nature, the Type II CRISPR-Cas9 system from Streptococcus pyogenes is the most straightforward. It uses two short RNAs for target identification and a single Cas9 endonuclease to cause double-strand breaks (DSB) in DNA (Liu et al., 2015). Prokaryotes are naturally home to CRISPR- and CRISPR-Cas systems, which act as a defense mechanism against mobile genetic elements like plasmids and phages (Figure 3). There are two primary parts to these systems. The first is a genomic region known as CRISPR, which has a collection of brief foreign-origin sequences known as spacers. These spacers allow the organism to recognize certain mobile genetic elements (MGEs) that it has previously encountered (Vercoe et al., 2013). Repeats, which are repeating regulatory sequences, separate the spacers. A key component of the CRISPR-Cas system is the CRISPR array, which comprises the leader sequence, spacers, and repeats (Yosef et al., 2012). The Cas proteins, which are often present close to the CRISPR array, are the second component of CRISPR-Cas systems. They are encoded by Cas genes. Effectors are components of the CRISPR-Cas system. Three stages make up the mechanism of CRISPR-Cas systems: adaptation, maturity, and interference. New spacers are added to the CRISPR array’s leader end during adaptation. The CRISPR array is translated through the maturation phase, producing pre-CRISPR RNA (pre-crRNA). The pre-crRNA undergoes further processing to become CRISPR RNA (crRNA) molecules, each of which has a single spacer and fragments of a repeat sequence on each side (Burmistrz et al., 2017). RNP complexes are created when these crRNAs join forces with Cas proteins. According to Semenova et al. (2011), the RNP complexes search nucleic acids for a sequence that matches the one encoded by the crRNA. The interference phase begins when the target sequence is recognized, which causes the recognized nucleic acid to degrade. In almost half of the known bacterial and archaeal species, CRISPR-Cas systems are present in the prokaryotic world (Van der Oost et al., 2014). Depending on the design of the effector complex, they are now divided into two groups (Makarova et al., 2018). While class 2 systems use single-subunit RNP complexes, class 1 RNPs have many subunits. Based on particular Cas proteins and the design of the CRISPR loci, each class is further classified into kinds and subtypes. Modern molecular biology has been changed by the discovery of CRISPR-Cas systems, which provide several desirable characteristics for future tools in this field. Due to the modular design of the CRISPR locus, it displays excellent specificity when recognizing target sequences, which can be readily changed by changing the crRNA sequence. The breadth of uses for CRISPR-Cas systems is also increased by altering the Cas proteins themselves (Brezgin et al., 2019). The CRISPR-Cas9 system stands out as the most practical foundation for the enlargement of new tools in research and biotechnology, even though the majority of the documented CRISPR-Cas systems target DNA sequences (Khosravi, 2021). However, the recent identification of RNA-targeting CRISPR-Cas systems offers fresh opportunities for the creation of novel research and biotechnology instruments.
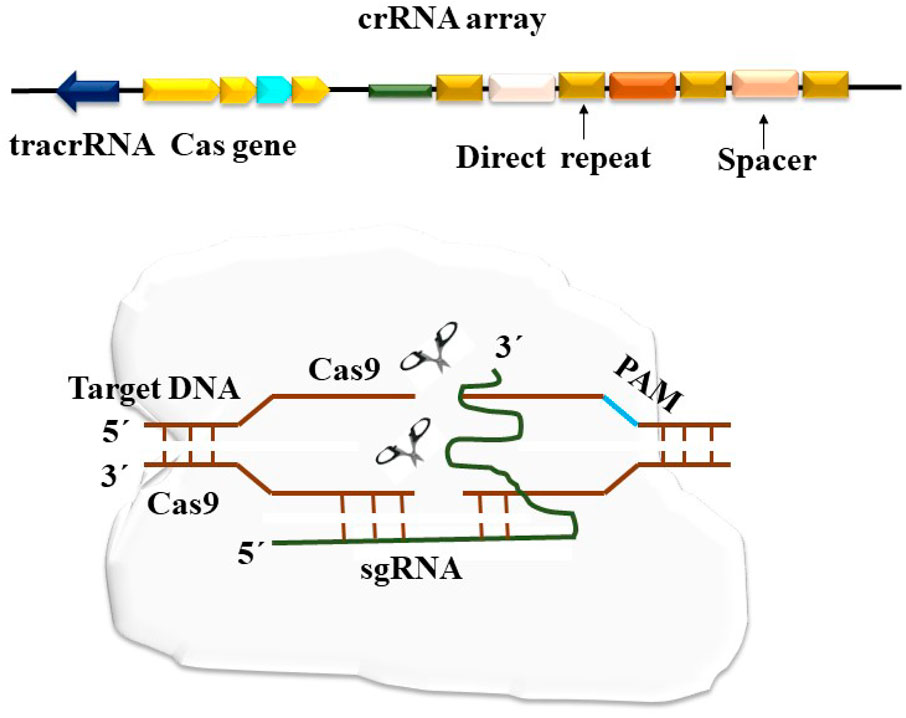
Figure 3. The schematic representation of the CRISPR/Cas9 system depicting a cluster of regularly spaced short palindrome repeats (CRISPR) and the CRISPR-associated nuclease 9 (Cas9). This system operates by inducing double-strand breaks (DSB) in the DNA strand. The Cas9 protein is directed by CRISPR RNA (crRNA) to perform its molecular function.
The following procedures are frequently used to modify the plant’s genome: First, guide RNAs (sgRNAs) are generated manually or with the use of online tools. They have a 20-bp target sequence at the 5′ end. Second, the kind of PAM sequence depicted in the target genomic area is used to determine which Cas variant is most appropriate (Molla et al., 2020). Thirdly, although this step is optional to reduce the possibility of failure, the effectiveness of the guide RNAs can be evaluated in vitro or using a transient protoplast transfection technique. Fourthly, a binary vector made up of one or more sgRNA expression cassettes and a Cas9 expression cassette is created. With this binary vector, biolistic transformation or agrobacterium-mediated transformation can be employed. Fifthly, either the agrobacterium or biolistic method are used to carry out genetic transformation. Sixth, the potential genome-edited plants are regenerated once the changed cells have been suitably chosen. Seventh, to verify effective genome editing in the target region, the putative plants are genotyped using a variety of techniques. The genome-edited plants can be rendered transgene-free by undergoing sexual segregation with the genetically altered plants when effective genome editing has been identified. To assist in the creation of guide RNAs, the choice of suitable editing tools, and the analysis of experimental data, several online applications have been developed (Figure 4). Precision genome and protein engineering is now possible thanks to the considerable advancement of CRISPR-Cas9-mediated gene editing brought about by the growth of bioinformatics-based design tools. Novel applications such as genomic disruption, transcriptional regulation, base editing, epigenetic modification, and prime editing have emerged as a result of this advancement (Molla et al., 2020). Base editing and prime editing technologies are two of the most promising toolkits for precise genome change, having the ability to create desired features in agricultural plants among the sophisticated genome editing methods (Molla et al., 2021). These methods open up a new prospect for precise genetic alterations in plants by overcoming the drawbacks of traditional CRISPR-Cas tools and low-efficiency HDR-mediated genome editing.
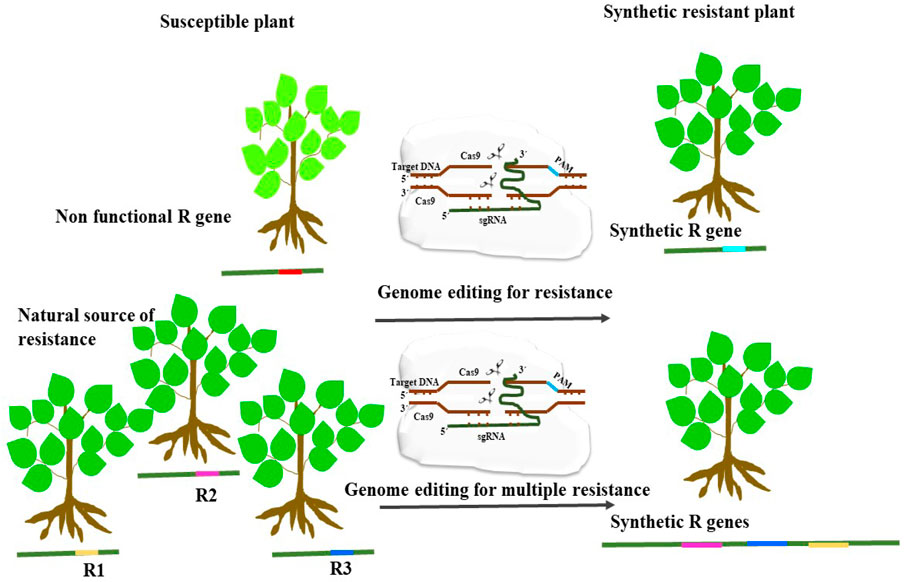
Figure 4. An illustration showing the application of genome editing for enhancing disease resistance by modifying non-functional pathogen recognition sites to synthesize synthetic functional R-gene. Another strategy is to engineer R gene for detecting multiple pathogen recognition sites which could provide resistance to several pathogens.
Cas13 enzymes have emerged as significant participants in the CRISPR field. Just a year after Cas13a (C2c2) was identified as an RNA-targeting CRISPR enzyme by Feng Zhang’s lab (Abudayyeh et al., 2019), they further advanced the technology by adapting Cas13b for exact RNA editing (Cox et al., 2017). The first RNA editing CRISPR tool was this ground-breaking technology, named REPAIR (RNA editing for programmed A to I (G) replacement). The group then unveiled RESCUE, an RNA editor that permits C-to-U changes, ushering in yet another development in the area. Compared to conventional DNA editing methods, RNA editing has many benefits. Since RNA editing does not need homology-directed repair (HDR) machinery, it may be used in cells that are not dividing. In addition, Cas13 enzymes are more adaptable than Cas9 and Cpf1 enzymes because they do not need a protospacer adjacent motif (PAM) sequence at the target locus. Some Cas13 enzymes favor targets with particular single-base proto-spacer flanking site (PFS) sequences, but LwaCas13a does not. Notably, direct genome editing is impossible because Cas13 enzymes lack the DNA-cleaving RuvC and HNH domains. As a result, Cas13-based RNA editing systems can prevent genomic off-targets or indels brought on by non-homologous end joining (NHEJ) and are likely to be reversible. According to Abudayyeh et al. (2019), CRISPR-Cas13a, also known as C2c2, is a single-effector Class II, Type VI RNA-guided RNA editing system. It has been demonstrated to successfully cleave molecules of single-stranded RNA (ssRNA). Cas13a’s adaptability is further increased by its ideal proto-spacer length (crRNA) and particular PFS preferences. Another notable aspect of the CRISPR-Cas13a system is multiplex editing. It can take a CRISPR array with several guides and process and produce mature crRNA from it, enabling the simultaneous targeting of many sequences from various genes. Both the CRISPR-Cas9 and CRISPR-Cas13a systems in plants have successfully undergone multiplex editing (Aman et al., 2018). Additionally, Cas13a shows promise in the sensitive detection of genetic molecules, which might be extremely helpful in the early identification of epidemics and the diagnosis of diseases. Cas13a may be reprogrammed to allow target RNA to start a collateral action, enabling it to identify particular RNA molecules in vivo and in vitro with great sensitivity and specificity. It is a possible candidate for the development of a lateral flow assay for the early observation of RNA viruses because of its superiority over presently used techniques like RT-PCR and qPCR in the capacity to identify a single copy of RNA. In conclusion, Cas13 enzymes have brought about a new era of RNA editing and detection, providing special benefits and fascinating opportunities for study in a variety of domains, including plant genetic engineering. These advancements have the potential to completely alter scientific research and open the door to discoveries.
Base editors are a ground-breaking addition to the toolkit for genome engineering, providing a quick and accurate way to substitute nucleotides without the necessity for donor templates or double-stranded breaks (DSBs) (Tiwari and Ghag, 2024). Cytidine and adenine deaminase-based base editors have been effectively created and applied for base editing in both plants and animals during the past few years. Constant improvements have increased the capabilities of CBEs and ABEs, making them more appropriate for agricultural plants. Examples of these improvements include reducing the catalytic window and using enhanced Cas9 variations. Base editors are chimeric proteins that demethylate cytosine or adenine bases in the genome; they consist of a DNA targeting module and a catalytic domain (Komor et al., 2016; Gaudelli et al., 2017). Besides, a catalytically inactive Cas9 endonuclease (dCas9) or a Cas9 nickase directed by a single-guide RNA (sgRNA) molecule can be used as the DNA targeting module. When it comes to dCas9, particular mutations (Asp10Ala and His840Ala) render its nuclease action inactive but leave it still able to bind DNA. An “R-loop,” or short length of unpaired DNA, is produced when dCas9-sgRNA binds to the target DNA. The tethered deaminase can change cytosines or adenines inside this tiny single-stranded area of roughly 5-8 nucleotides. Base editors can reduce the number of indels by introducing single-base alterations or replacements without triggering DSBs. Cytosine base editors (CBEs) and adenine base editors (ABEs) are the two primary categories of DNA base editors. While ABEs help with the translation of A-to-G or T-to-C, CBEs can convert C to T or G to A. Future crop development might be sped up with the use of these exact base editing tools, which have enormous promise for precision breeding in model plants and crops (Mishra et al., 2020). Despite the bright future, base editing technology is still in its inception. To increase efficiency and specificity, additional development and an extension of its editing scope are required. It could be conceivable to continually improve base editors and look at novel mutant forms of cytidine deaminase to boost DNA selectivity and lessen off-target effects. With these developments, the area of plant genetic engineering will undergo a revolution as extremely accurate base editors are extensively used in crop enhancement initiatives.
CBEs are effective tools that catalyse the transformation of cytosines into thymines. The cytidine deaminase enzyme, which separates an amino group from cytosine and produces uracil as a consequence, is responsible for this conversion. Following the correction of the uracil-guanine (U-G) mismatch via DNA repair pathways, uracil-adenine (U-A) base pairs are created. As a result, thymine is added to the freshly synthesized DNA strand, changing C-G into T-A. David Liu and his group at Harvard University in the United States created the first-generation CBE, sometimes referred to as BE1, in 2016. It had a 16-amino acid XTEN linker that connected the rat-derived APOBEC1 cytidine deaminase enzyme to a catalytically inactive Cas9 (dCas9) (Komor et al., 2016). A balance between the two proteins is maintained by the XTEN linker. The APOBEC family of cytidine deaminases, which function on single-stranded DNA and RNA as substrates, are naturally occurring enzymes that protect vertebrates against viral infections. The recurrent removal of uracil by uracil DNA glycosylase (UDG), which led to poor editing ability, was one of BE1’s key drawbacks. The second-generation base editor BE2 (APOBEC-XTEN-dCas9-UGI) was created to get around this restriction. It entailed incorporating a uracil DNA glycosylase inhibitor (UGI) into the DNA targeting module’s C-terminus, which increased editing effectiveness (Komor et al., 2016). Multiple cytosines (Cs) present inside the catalytic window provide a problem for CBEs, potentially leading to off-target action and the changing of non-target cytosines to uracil. Several BE3 variations were created using various Cas9 variants and non-canonical PAM sequences to overcome this problem. For instance, SpCas9 variants that target the NGAN, NGAG, NGCG, and NRT PAMs, respectively, such as VQR-BE3, EQR-BE3, VRER-BE3, and SaKKH-BE3, have demonstrated a 2.5-fold improvement in editing ability (Kim et al., 2017). The accuracy and efficacy of base editing have considerably enlarged thanks to these developments in CBE technology, creating new opportunities for precise genome engineering in a variety of taxa, along with plants and mammals.
Adenine bases may now be precisely modified in a programmed manner thanks to ABEs, which is an exciting development in genome editing technology (Gaudeli et al., 2017). Adenine DNA deaminases are not found in nature, in contrast to cytidine deaminases, which are naturally occurring enzymes. David Liu and his colleagues used Escherichia coli TadA (E. coli TadA) and carried out substantial protein engineering and guided advancement to create ABEs. With similarity to the APOBEC enzyme family, E. coli TadA is a tRNA adenine deaminase that transforms adenine to inosine in the single-stranded anticodon loop of tRNA Arg. By combining E. coli TadA with a catalytically imperfect CRISPR/Cas9 mutant, the first-generation adenine base editors, or ABEs, were produced (Gaudeli et al., 2017). ABE7.7, ABE7.8, and ABE7.9 from the newly created family of ABEs showed significant activity as adenine base editors and compatibility with a wider variety of target sequences. The seventh generation of ABE, also known as ABE7.10, was evolved later and greatly enhanced the effectiveness and product purity for converting A.T. to G.C. in a variety of target sequences. By permitting all four transitions—C to T, A to G, T to C, and G to A—in a programmable way, ABEs have considerably increased the range of base editing. Recently, base editor rBE5 was used in rice to target Pi-d2gene for modulating defense against rice blast fungus (Ran et al., 2018). ABEs are excellent instruments for precisely introducing point mutations due to their enhanced adaptability and efficiency. The use of ABEs for targeted base editing creates new avenues for precisely targeting certain genetic mutations and creating treatments.
To get over the constraints of HDR-mediated editing, prime editing represents a substantial leap in genome editing technology (Tiwari and Ghag, 2024). It can carry out a variety of modifications, including 1–80 bp deletions, 1–44 bp insertions, and all base substitutions in human cells (Anzalone et al., 2019). The primary editing method is built on a special guide RNA known as peg RNA that has a reverse transcriptase (RT) template with the needed alteration and a primer binding site (PBS). On the nicked strand of genomic DNA, the PBS sequence hybridizes with the complementary target sequence. The target DNA sequence is subsequently precisely edited by the RT, which uses this hybridized template to facilitate the synthesis of the necessary modifications. The RESCUE (RNA Editing for Specific C-to-U Exchange) system for RNA editing is another cutting-edge technique. The RESCUE system was created to address some inherent shortcomings of natural cytidine deaminases for RNA editing, and it has shown up to 42% editing efficiency in endogenous transcripts. But around the targeted nucleotide, RESCUE showed off-target edits, including C-to-U and A-to-I. Guanine mismatches were added to the guide RNA across from the off-target edits to lessen these off-target effects. This change enabled Rescue to make fewer local, off-target C-to-U edits. In addition, rational mutagenesis was carried out, resulting in the discovery of the RESCUE-S variant (Abudayyeh et al., 2019). This variant significantly reduced off-target C-to-U edits by about 45% and off-target A-to-I edits by about 94%, similar to the original RESCUE system. These developments in prime editing and the RESCUE system have enormous potential to broaden the application of precise genome editing and solve the problems brought on by off-target effects. These technologies can change the field of genetic engineering and open the door to numerous uses in research, biotechnology, and therapeutic interventions with additional optimization and refinement.
Feng Zhang and his team were the ones who first created RNA base editors, also known as ADAR (adenosine deaminase acting on RNA) base editors. Adenosine-to-inosine transformation in mammalian cells was made possible by the use of the naturally occurring ADAR enzyme and a catalytically inactive form of the Cas13 protein (dCas13) (Cox et al., 2017). Cas13 is a type VI RNA-guided RNase that is associated with CRISPR and can bind to RNA molecules. The Prevotella sp. Cas13b ortholog, one of the tested Cas13 enzymes, showed greater effectiveness and specificity in RNA binding and knockout operations. By catalysing the hydrolytic deamination of adenosine to inosine and thereby enabling precise base editing in RNA, the ADAR family of enzymes is in charge of the endogenous editing of transcripts (Nishikura, 2010). Targeted RNA editing of transcripts is possible with the help of this system, noted as RNA Editing for Programmable A to I Replacement (REPAIR). With even greater specificity compared to earlier RNA editing platforms, REPAIRv2—an improved version of the original REPAIR—was later produced (Stafforst et al., 2012). According to Ferreira et al. (2010), the REPAIR system has proven to be very successful at simulating protective alleles that protect against a variety of autoimmune diseases. It provides a favourable RNA editing platform with numerous uses in biotechnology, therapeutic interventions, and research. With the ability to precisely edit RNA, new avenues for treating genetic disorders and improving our knowledge of how genes work and are regulated are now possible. RNA-base editing holds great potential for a variety of applications as the technology develops and improves.
The use of disease-resistant cultivars is the most efficient and ecologically friendly method of controlling plant diseases. Most plant pathogens are known to make use of dominantly inherited genes, so-called S genes, in order to successfully develop disease and proliferate in their host. However, genetic disruption of these genes has proven to be one of the most effective methods for mitigating plant diseases and inducing long-term disease resistance. Interestingly, novel genome-editing methods provide prospects for controlling bacterial, viral, and fungal infections, as well as implementing pathogen resistance in plants (Karmakar et al., 2022). Among genome editing tools, CRISPR/Cas9 has been regarded as a successful technique due to its versatility, lower cost, ease of design and implementation, and increased success rate (Saraswat et al., 2024).
The rapid rate of plant pathogenic bacteria’s multiplication makes it difficult to control infestations of these diseases using conventional methods. However, specific genomic modifications have been successfully introduced into the sensitivity genes of host crop plants using genome editing technology. For instance, CRISPR-Cas9 technology was used to make resistance against the pathogen in the case of citrus canker, a serious bacterial disease caused by Xanthomonas citri sp. Citri (Table 2). It was discovered that the vulnerability gene Lateral Organ Boundaries 1 (CsLOB1) is essential for the pathogen’s expansion and the emergence of symptoms. The genome-edited citrus plants displayed protection from citrus canker disease without interfering with the gene’s typical developmental functions by targeting the effector binding element (EBEPthA4) of the CsLOB1 promoter, which is bound by the transcription activator-like effectors (TALEs) (Peng et al., 2017). Similar to bacterial blight, which has a devastating effect on rice production in Asia and Africa, X. oryzae pv. oryzae is the pathogen responsible. The pathogen secretes TALEs that bind to particular promoter sequences and cause the expression of the genes SWEET11, SWEET13, and SWEET14, sucrose-specific transporter genes that are crucial for disease susceptibility. Rice plants with disease resistance were produced by using TALEN-mediated genome editing to reduce the effector-induced transcriptional activation of the susceptibility gene OsSWEET14 (Li et al., 2012). CRISPR/Cas 9 was used to target the gene OsSWEET11 in rice to produce a bacterial leaf blight-resistant line (Roviqowati et al., 2024). It has been demonstrated that the CRISPR-Cas9 genome editing technology is a useful tool for causing mutations in particular genes to confer disease resistance in crops against dissimilar viral pathogens.
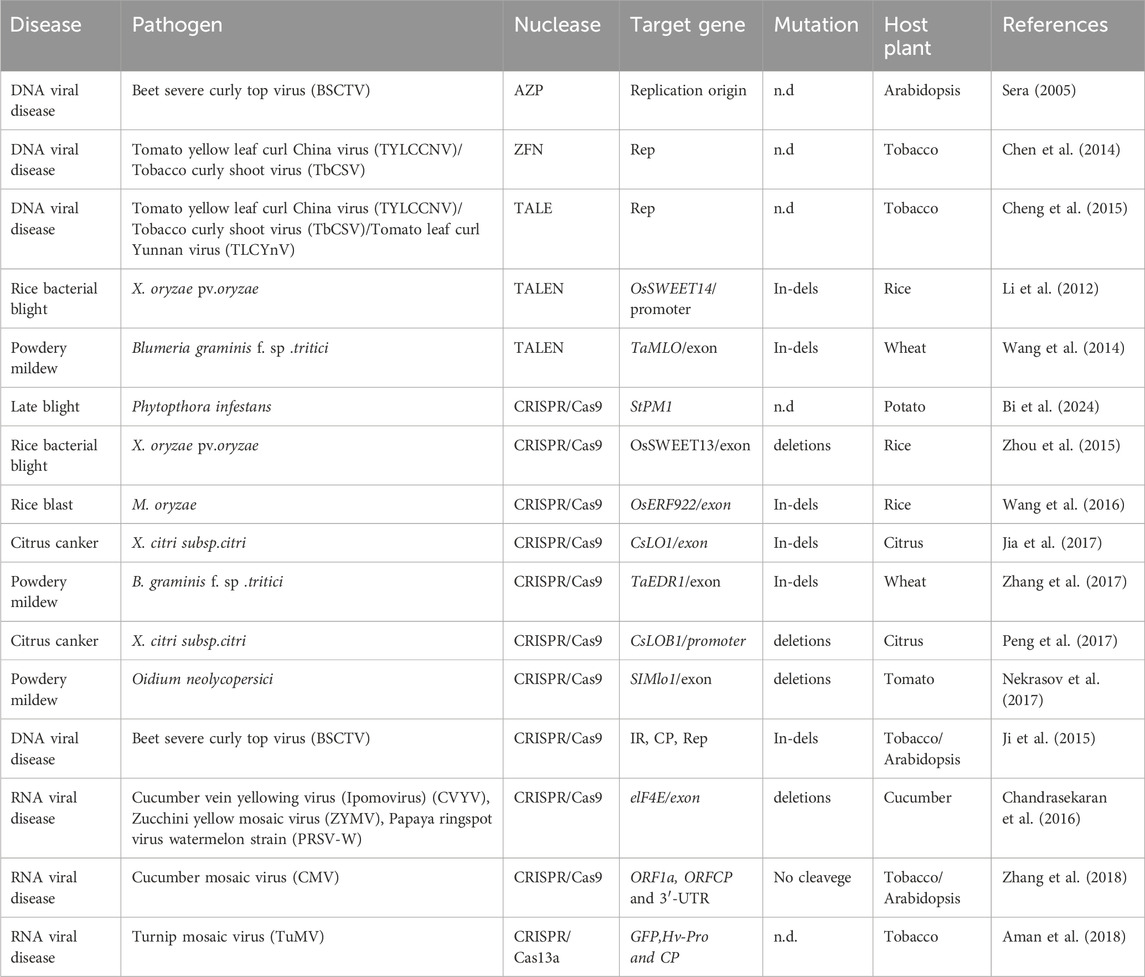
Table 2. Application of genome editing for improving disease resistance in plants by targeting different genes.
CRISPR-Cas technology is used in two main ways to build virus resistance in crops. In the first strategy, viral disease resistance is attained by carefully destroying the viral pathogen’s genome. For instance, the foundation of the sgRNA-Cas9 construct targeting the viral genome followed the development of plants resistant to the Geminivirus in Arabidopsis and N. benthamiana (Ji et al., 2015). Alike findings were obtained in N. benthamiana by designing sgRNAs against the coding and non-coding sequences of the tomato yellow leaf curl virus (TYLCV), which resulted in transgenic plants with decreased viral DNA accumulation and TYLCV resistance (Ali et al., 2015). The second strategy entails editing host genes that encode susceptibility factors (S genes) to make crops that are resistant to viruses. Genome editing was used to target the eIF4E gene in the cucumber (L.), which is a susceptibility factor. As a result, transformed plants have resistance to various phytoviruses and ipomoviruses (Chandrasekaran et al., 2016). Adopting the CRISPR-Cas9 system, the wheat dwarf virus (WDV), a member of the Geminiviridae family, was targeted at four sites in its genome to create stable WDV-resistant wheat lines with few off-target effects (Kis et al., 2019). Additionally, the CRISPR-Cas9 innovation has been profitably used to combat other viruses like the tomato yellow leaf curl virus (TYLCV), bean yellow dwarf virus, and beet severe curly top virus (BSCTV) (Ali et al., 2015; Baltes et al., 2015; Ji et al., 2015). Along with these methods, genome editing innovations have made it possible to introduce genetic variations that are naturally present in wild cultivars, particularly recessive traits that are primarily governed by genes like the translation initiation factor 4 gamma gene (eIF4G), which confers disease resistance. To confer protection against the Rice tungro spherical virus (RTSV), Macovei et al. (2018) modified the eIF4G gene of the IR64 rice variety using the CRISPR-Cas9 tool. For breeding and spreading hybrids of Musa species, the presence of endogenous banana streak virus (eBSV) in the B genome poses a significant barrier. To inactivate eBSV, CRISPR-Cas9 gene editing was used. This resulted in targeted mutations in all three open reading frames (ORFs) and the successful removal of integrated dsDNA from BSV from the banana genome. This innovation has made it easier to improve the Plantain B genome germplasm, according to Tripathi et al. (2019). The CRISPR-Cas9 system has also been profitably used to confer resistance against RNA viruses like the cucumber mosaic virus (CMV) and tobacco mosaic virus (TMV), in addition to DNA viruses. Reduced viral infection symptoms and viral RNA accumulation in the genome-edited plants were the results of targeting CMV and TMV with Francisella novicida Cas9 (FnCas9) in combination with particular sgRNAs (Zhang et al., 2018). CRISPR-Cas9 was used to target single-stranded DNA-A of the AV1 and AC1 genes in the Mung bean yellow mosaic virus to enhance resistance in tobacco against this virus (Talakayala et al., 2024). RNA viruses that harm plant cells can be targeted using the CRISPR-Cas13 system in addition to the CRISPR-Cas9 system. When different Cas13 systems were characterized by Mahas et al. (2019), they discovered that the LwaCas13a, PspCas13b and CasRx variants had the best defense against RNA viruses. CasRx is a highly targeted and programmable tool for transcriptome engineering because it has demonstrated high specificity for its target RNA viruses and minimal off-target effects in plant cells.
Plants that have been genetically changed modified the CRISPR-Cas9 system are significantly more resistant to fungal diseases. To increase protection against the fungus Magnaporthe oryzae, Wang et al. (2016) targeted ethylene-responsive factors (ERF) in rice. Similar to this, in hexaploid wheat, heritable mutations in the MILDEW-RESISTANCE LOCUS (MLO) genes were introduced using TALEN and CRISPR-Cas9 technologies, providing notable protection against powdery mildew disease (Wang et al., 2014). Additionally, employing genome deletion using CRISPR-Cas9 technology, Nekrasov et al. (2017) created the tomato variety “Tomelo,” which is resistant to the powdery mildew fungus. This variety was created quickly, and it is non-transgenic and free of foreign DNA sequences. To create a cultivar of potatoes that is resistant to late blight disease, Kieu et al. (2021) discovered potential S-genes and underwent genome editing. These susceptibility genes’ (S-genes) absence results in increased resistance to the late blight pathogen. Knocking out RECEPTOR-LIKE KINASE 902 (RLK902) by CRISPR-Cas9 in Brassica napus displayed a higher level of resistance against Sclerotinia sclerotiorum and B. cinerea (Zhao et al., 2024). Overall, the use of CRISPR-Cas technologies to build disease resistance in different crops has notable potential to increase agricultural output and fight plant diseases. These examples demonstrate how the evolution of disease-resistant crop plants is now possible thanks to genome editing technology that targets both host genes and pathogenic organisms. Genome editing’s capacity to make precise and targeted changes holds great potential for reducing the financial losses brought on by plant diseases and raising agricultural production worldwide.
The success rate of genome editing in conferring disease resistance against pathogens in plants varies depending on several factors, including the target crop, the specific pathogen, the genetic basis of resistance, and the efficiency of the editing technique employed. Some crops and pathogens have been more amenable to successful genome editing-mediated disease resistance than others. For example, in crops like rice, wheat, and tomato, researchers have achieved significant success in enhancing resistance against various pathogens such as rice blast fungus, wheat powdery mildew, and bacterial wilt in tomatoes through genome editing (Wang et al., 2014; Nekrasov et al., 2017; Li et al., 2018). However, success rates can vary when targeting different pathogens within the same crop species due to variations in the genetic basis of resistance and the complexity of the pathogen’s interaction with the plant. The choice of genome editing technique can also influence the success rate. CRISPR/Cas-based systems have emerged as powerful tools for precise genome editing in plants due to their efficiency, versatility, and relatively low cost compared to older techniques like zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) (Wang et al., 2014). Continuous improvements in CRISPR/Cas systems, such as the development of high-fidelity Cas variants and delivery methods, have further enhanced the success rates of genome editing-mediated disease resistance. The success of genome editing-mediated disease resistance depends on identifying and targeting genes or genetic elements that confer resistance to the pathogen. In some cases, single genes may confer strong resistance, making them ideal targets for editing. However, in other instances, resistance may be governed by multiple genes or quantitative trait loci (QTLs), requiring more complex editing strategies such as gene stacking or modification of regulatory elements (Zaidi et al., 2018). Efficient delivery of genome editing tools into plant cells remains a technical challenge, particularly for crops with recalcitrant or complex genomes, limiting the practical application of these techniques (Mushtaq et al., 2019).
Genome editing holds promise for bolstering plant disease resistance, but several limitations and barriers hinder its widespread application such as off-target effects, regulatory hurdles, Complex genetic interactions and ethical concerns. For example, genome editing techniques such as CRISPR/Cas9 may inadvertently incorporate mutations in unintended regions of the genome, potentially leading to unforeseen consequences (Chen et al., 2022). The regulatory frameworks governing genome-edited crops vary globally and can pose significant barriers to the commercialization and adoption of disease-resistant plants (Entine et al., 2021). The genetic basis of disease resistance in plants is often multifaceted, involving numerous genes and regulatory elements, making it challenging to engineer robust resistance using genome editing alone (Mushtaq et al., 2019). Manipulating the genomes of plants raises ethical questions related to environmental impact, biodiversity, and unintended consequences, necessitating careful consideration and public engagement (Chen et al., 2022).
The growing global demand for food, coupled with the challenges posed by climate change, has necessitated advancements in crop resilience and yield improvement strategies. Conventional plant breeding methods have been relied upon for crop amelioration, but the development of genome editing has provided a more precise and targeted approach to modifying crop plants and overcomes the limitations of conventional resistance breeding. Genome editing techniques, such as ZFNs, TALENs, and CRISPR-Cas9, have revolutionized molecular biology and crop sciences, enabling functional genomics and the enhancement of agricultural traits. The application of genome editing tools has demonstrated remarkable efficacy in enhancing crop immunity against various phytopathogens by modifying host disease susceptibility genes. The CRISPR-Cas system, in particular, is well-suited for multiplexing, allowing the targeting of multiple genes to evolve broad-spectrum resistance. Moreover, genome editing offers valuable insights into the molecular mechanisms of pathogenesis and host resistance, opening new avenues for disease diagnosis in plants. Genome editing methods are unparalleled in precision, allowing targeted changes at the gene, regulatory, and nucleotide levels. Genome editing technology can be used to leverage pathogen effector target sequences to develop novel sources of resistance in key agricultural crops, thereby reducing overreliance on pesticide applications and minimizing the necessity for genetically modified (GM) crops. Crops derived from genome editing with minor genomic modifications are virtually indistinguishable from their traditional counterparts, posing negligible risks to the environment, society, economy, and human health. Many countries, as well as India, have de-regulated genome-edited crop varieties that lack foreign DNA, fostering their adoption in agriculture. To further advance genome editing technology, research is needed to explore PAM-independent CRISPR applications and develop strategies to avoid and detect off-target effects more effectively. Additionally, efforts to develop smaller editing tools will enhance their delivery and editing efficiency in plant cells. As we move forward, the advantage of genome editing technologies will be noticed through the commercialization of disease-resistant, climate-resilient, and superior-quality crops. These advancements will not only strengthen global food security but also foster sustainable agricultural practices in the face of evolving environmental challenges.
Despite the challenges and variability in success rates, the future outlook for genome editing-mediated disease resistance in plants is promising. Continued advancements in genome editing technologies, coupled with increasing knowledge of plant-pathogen interactions and genetic mechanisms underlying resistance, are expected to improve success rates and expand the range of target crops and pathogens. Furthermore, ongoing research efforts are focused on developing innovative strategies to enhance the efficacy and durability of edited resistance traits, such as combining genome editing with other breeding approaches like marker-assisted selection and RNA interference (RNAi), as well as leveraging synthetic biology tools to engineer novel resistance mechanisms. Overall, genome editing holds immense potential for revolutionizing plant disease resistance breeding by providing precise and sustainable solutions to combat pathogens, thereby contributing to global food security and agricultural sustainability.
SM: Writing–review and editing, Writing–original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal Analysis, Data curation, Conceptualization. SN: Writing–review and editing, Writing–original draft. TR: Writing–review and editing, Writing–original draft. GG: Writing–review and editing, Writing–original draft. ZM: Writing–review and editing, Writing–original draft. AW: Writing–review and editing, Writing–original draft. SA: Writing–review and editing, Writing–original draft. AT: Writing–review and editing, Writing–original draft. NM: Writing–review and editing, Writing–original draft.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abudayyeh, O. O., Gootenberg, J. S., Franklin, B., Koob, J., Kellner, M. J., Ladha, A., et al. (2019). A cytosine deaminase for programmable single-base RNA editing. Science 365 (6451), 382–386. doi:10.1126/science.aax7063
Ahmad, N., Rahman, M. U., Mukhtar, Z., Zafar, Y., and Zhang, B. (2020a). A critical look on CRISPR-based genome editing in plants. J. Cell. Physiology 235 (2), 666–682. doi:10.1002/jcp.29052
Ahmad, S., Wei, X., Sheng, Z., Hu, P., and Tang, S. (2020b). CRISPR/Cas9 for development of disease resistance in plants: recent progress, limitations and future prospects. Briefings Funct. Genomics 19 (1), 26–39. doi:10.1093/bfgp/elz041
Ali, S., Ganai, B. A., Kamili, A. N., Bhat, A. A., Mir, Z. A., Bhat, J. A., et al. (2018). Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 212, 29–37. doi:10.1016/j.micres.2018.04.008
Ali, S., Tyagi, A., and Bae, H. (2023). Plant microbiome: an ocean of possibilities for improving disease resistance in plants. Microorganisms 11 (2), 392. doi:10.3390/microorganisms11020392
Ali, Z., Abulfaraj, A., Idris, A., Ali, S., Tashkandi, M., and Mahfouz, M. M. (2015). CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 16, 238–311. doi:10.1186/s13059-015-0799-6
Allioui, N., Siah, A., Brinis, L., and Reignault, P. (2016). Identification of QoI fungicide-resistant genotypes of the wheat pathogen Zymoseptoria tritici in Algeria. Phytopathol. Mediterr. 55 (1), 89–97. doi:10.14601/Phytopathol_Mediterr-16235
Aman, R., Ali, Z., Butt, H., Mahas, A., Aljedaani, F., Khan, M. Z., et al. (2018). RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 19, 1–9. doi:10.1186/s13059-017-1381-1
Andolfo, G., and Ercolano, M. R. (2015). Plant innate immunity multicomponent model. Front. plant Sci. 6, 987. doi:10.3389/fpls.2015.00987
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576 (7785), 149–157. doi:10.1038/s41586-019-1711-4
Argast, G. M., Stephens, K. M., Emond, M. J., and Monnat Jr, R. J. (1998). I-PpoI and I-CreI homing site sequence degeneracy determined by random mutagenesis and sequential in vitro enrichment. J. Mol. Biol. 280 (3), 345–353. doi:10.1006/jmbi.1998.1886
Baldi, P., and La Porta, N. (2020). Molecular approaches for low-cost point-of-care pathogen detection in agriculture and forestry. Front. Plant Sci. 11, 570862. doi:10.3389/fpls.2020.570862
Baltes, N. J., Hummel, A. W., Konecna, E., Cegan, R., Bruns, A. N., Bisaro, D. M., et al. (2015). Conferring resistance to geminiviruses with the CRISPR–Cas prokaryotic immune system. Nat. Plants 1 (10), 15145–15154. doi:10.1038/nplants.2015.145
Bebber, D. P. (2015). Range-expanding pests and pathogens in a warming world. Annu. Rev. Phytopathology 53, 335–356. doi:10.1146/annurev-phyto-080614-120207
Bi, W., Liu, J., Li, Y., He, Z., Chen, Y., Zhao, T., et al. (2024). CRISPR/Cas9-guided editing of a novel susceptibility gene in potato improves Phytophthora resistance without growth penalty. Plant Biotechnol. J. 22 (1), 4–6. doi:10.1111/pbi.14175
Biffen, R. H. (1905). Mendel’s laws of inheritance and wheat breeding. J. Agric. Sci. 1, 4–48. doi:10.1017/s0021859600000137
Brezgin, S., Kostyusheva, A., Kostyushev, D., and Chulanov, V. (2019). Dead Cas systems: types, principles, and applications. Int. J. Mol. Sci. 20 (23), 6041. doi:10.3390/ijms20236041
Brunner, S., Stirnweis, D., Diaz Quijano, C., Buesing, G., Herren, G., Parlange, F., et al. (2012). Transgenic Pm3 multilines of wheat show increased powdery mildew resistance in the field. Plant Biotechnol. J. 10, 398–409. doi:10.1111/j.1467-7652.2011.00670.x
Burmistrz, M., Rodriguez Martinez, J. I., Krochmal, D., Staniec, D., and Pyrc, K. (2017). Clustered regularly interspaced short palindromic repeat (CRISPR) RNAs in the Porphyromonas gingivalis CRISPR-Cas IC system. J. Bacteriol. 199 (23), e00275-17–e01128. doi:10.1128/JB.00275-17
Cao, L., Blekemolen, M. C., Tintor, N., Cornelissen, B. J. C., and Takken, F. L. W. (2018). The Fusarium oxysporum avr2-six5 effector pair alters plasmodesmatal exclusion selectivity to facilitate cell-to-cell movement of Avr2. Mol. Plant 11, 691–705. doi:10.1016/j.molp.2018.02.011
Cao, X., Xu, X., Che, H., West, J. S., and Luo, D. (2019). Three Colletotrichum species, including a new species, are associated to leaf anthracnose of rubber tree in Hainan, China. Plant Dis. 103 (1), 117–124. doi:10.1094/PDIS-02-18-0374-RE
Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., Pearlsman, M., et al. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. plant Pathol. 17 (7), 1140–1153. doi:10.1111/mpp.12375
Chen, J., Zhang, X., Rathjen, J. P., and Dodds, P. N. (2022). Direct recognition of pathogen effectors by plant NLR immune receptors and downstream signalling. Essays Biochem. 66 (5), 471–483. doi:10.1042/EBC20210072
Chen, W., Qian, Y., Wu, X., Sun, Y., Wu, X., and Cheng, X. (2014). Inhibiting replication of begomoviruses using artificial zinc finger nucleases that target viral-conserved nucleotide motif. Virus Genes. 48, 494–501. doi:10.1007/s11262-014-1041-4
Cheng, X., Li, F., Cai, J., Chen, W., Zhao, N., Sun, Y., et al. (2015). Artificial TALE as a convenient protein platform for engineering broad-spectrum resistance to begomoviruses. Viruses 7 (8), 4772–4782. doi:10.3390/v7082843
Chung, B. Y. W., Miller, W. A., Atkins, J. F., and Firth, A. E. (2008). An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. U. S. A. 105, 5897–5902. doi:10.1073/pnas.0800468105
Collard, B. C., Jahufer, M. Z. Z., Brouwer, J. B., and Pang, E. C. K. (2005). An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica 142, 169–196. doi:10.1007/s10681-005-1681-5
Cox, D. B., Gootenberg, J. S., Abudayyeh, O. O., Franklin, B., Kellner, M. J., Joung, J., et al. (2017). RNA editing with CRISPR-Cas13. Science 358 (6366), 1019–1027. doi:10.1126/science.aaq0180
Crous, P. W., Hawksworth, D. L., and Wingfield, M. J. (2015). Identifying and naming plant-pathogenic fungi: past, present, and future. Annu. Rev. Phytopathology 53, 247–267. doi:10.1146/annurevphyto-080614-120245
Czosnek, H. (2007). Tomato yellow leaf curl virus disease: management, molecular biology, breeding for resistance. Dordrecht, The Netherlands: Springer.
Damalas, C. A., and Eleftherohorinos, I. G. (2011). Pesticide exposure, safety issues, and risk assessment indicators. Int. J. Environ. Res. public health 8 (5), 1402–1419. doi:10.3390/ijerph8051402
Das, G., and Rao, G. J. N. (2015). Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front. Plant Sci. 6, 698. doi:10.3389/fpls.2015.00698
De Wit, P. J., Hofman, A. E., Velthuis, G. C., and Kuć, J. A. (1985). Isolation and characterization of an elicitor of necrosis isolated from intercellular fluids of compatible interactions of Cladosporium fulvum (syn. Fulvia fulva) and tomato. Plant Physiol. 77, 642–647. doi:10.1104/pp.77.3.642
Du Plessis, E., Granados, G. M., Barnes, I., Ho, W. H., Alexander, B. J. R., Roux, J., et al. (2019). The pandemic strain of Austropuccinia psidii causes myrtle rust in New Zealand and Singapore. Australas. Plant Pathol. 48 (3), 253–256. doi:10.1007/s13313-019-0624-x
Entine, J., Felipe, M. S. S., Groenewald, J. H., Kershen, D. L., Lema, M., McHughen, A., et al. (2021). Regulatory approaches for genome edited agricultural plants in select countries and jurisdictions around the world. Transgenic Res. 30 (4), 551–584. doi:10.1007/s11248-021-00257-8
Fang, Y., and Tyler, B. M. (2016). Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Mol. Plant Pathol. 17 (1), 127–139. doi:10.1111/mpp.12318
FAO (2019). New standards to curb the global spread of plant pests and diseases. Available at: https://www.fao.org/news/story/en/item/1187738/icode/.
Feau, N., Vialle, A., Allaire, M., Tanguay, P., Joly, D. L., Frey, P., et al. (2009). Fungal pathogen (mis-) identifications: a case study with DNA barcodes on Melampsora rusts of aspen and white poplar. Mycol. Res. 113, 713–724. doi:10.1016/j.mycres.2009.02.007
Ferreira, R. C., Pan-Hammarström, Q., Graham, R. R., Gateva, V., Fontán, G., Lee, A. T., et al. (2010). Association of IFIH1 and other autoimmunity risk alleles with selective IgA deficiency. Nat. Genet. 42 (9), 777–780. doi:10.1038/ng.644
Fletcher, S. J., Shrestha, A., Peters, J. R., Carroll, B. J., Srinivasan, R., Pappu, H. R., et al. (2016). The tomato spotted wilt virus genome is processed differentially in its plant host Arachis hypogaea and its thrips vector Frankliniella fusca. Front. Plant Sci. 7, 1349. doi:10.3389/fpls.2016.01349
Flor, H. H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. doi:10.1146/annurev.py.09.090171.001423
Gao, C. (2018). The future of CRISPR technologies in agriculture. Nat. Rev. Mol. Cell. Biol. 19 (5), 275–276. doi:10.1038/nrm.2018.2
Gao, C. (2021). Genome engineering for crop improvement and future agriculture. Cell. 184 (6), 1621–1635. doi:10.1016/j.cell.2021.01.005
Gao, J., Wang, G., Ma, S., Xie, X., Wu, X., Zhang, X., et al. (2015). CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol. Biol. 87, 99–110. doi:10.1007/s11103-014-0263-0
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable base editing of A· T to G· C in genomic DNA without DNA cleavage. Nature 551 (7681), 464–471. doi:10.1038/nature24644
Gauthier, N., and Dockery, J. (2018). “Boxwood blight,” in Plant pathology fact sheets:PPFS-or-W-20. Available at: http://plantpathology.ca.uky.edu/files/ppfs-or-w-20.pdf.
Ghoneem, K. M., El-Wakil, D. A., Ahmed, M. I., Kamel, H. M., Rashad, E. M., Al-Askar, A. A., et al. (2023). Biodiversity of Rhizoctonia solani in Phaseolus vulgaris seeds in East delta of Egypt. Agronomy 13 (5), 1317. doi:10.3390/agronomy13051317
Gill, H. K., and Garg, H. (2014). Pesticide: environmental impacts and management strategies. Pesticides-toxic Asp. 8 (187), 10–5772.
Gonzalez-Lamothe, R., Segura, R., Trapero, A., Baldoni, L., Botella, M. A., and Valpuesta, V. (2002). Phylogeny of the fungus Spilocaea oleagina, the causal agent of peacock leaf spot in olive. FEMS Microbiol. Lett. 210 (1), 149–155. doi:10.1111/j.1574-6968.2002.tb11174.x
Gossen, B. D., Conner, R. L., Chang, K. F., Pasche, J. S., McLaren, D. L., Henriquez, M. A., et al. (2016). Identifying and managing root rot of pulses on the northern great plains. Plant Dis. 100 (10), 1965–1978. doi:10.1094/PDIS-02-16-0184-FE
Gupta, P. K., Balyan, H. S., and Gautam, T. (2021). SWEET genes and TAL effectors for disease resistance in plants: present status and future prospects. Mol. Plant Pathol. 22 (8), 1014–1026. doi:10.1111/mpp.13075
Hafez, M., and Hausner, G. (2012). Homing endonucleases: DNA scissors on a mission. Genome 55 (8), 553–569. doi:10.1139/g2012-049
Han, X., and Kahmann, R. (2019). Manipulation of phytohormone pathways by effectors of filamentous plant pathogens. Front. Plant Sci. 10, 822. doi:10.3389/fpls.2019.00822
Hernandez-Restrepo, M., Madrid, H., Tan, Y. P., Da Cunha, K. C., Gene, J., Guarro, J., et al. (2018). Multi-locus phylogeny and taxonomy of Exserohilum. Persoonia Mol. Phylogeny Evol. Fungi 41, 71–108. doi:10.3767/persoonia.2018.41.05persoonia.41.05
Irieda, H., Inoue, Y., Mori, M., Yamada, K., Oshikawa, Y., Saitoh, H., et al. (2019). Conserved fungal effector suppresses PAMP-triggered immunity by targeting plant immune kinases. Proc. Natl. Acad. Sci. 116 (2), 496–505. doi:10.1073/pnas.1807297116
Jacob, F., Vernaldi, S., and Markawa, T. (2013). Evolution and conservation of plant NLR functions. Front. Immunol. 4, 297. doi:10.3389/fimmu.2013.00297
Ji, X., Zhang, H., Zhang, Y., Wang, Y., and Gao, C. (2015). Establishing a CRISPR–Cas-like immune system conferring DNA virus resistance in plants. Nat. Plants 1 (10), 15144–4. doi:10.1038/nplants.2015.144
Jia, H., Zhang, Y., Orbović, V., Xu, J., White, F. F., Jones, J. B., et al. (2017). Genome editing of the disease susceptibility gene Cs LOB 1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 15 (7), 817–823. doi:10.1111/pbi.12677
Jin, Y., Zhang, W., Cong, S., Zhuang, Q. G., Gu, Y. L., Ma, Y. N., et al. (2023). Pseudomonas syringae type III secretion protein HrpP manipulates plant immunity to promote infection. Microbiol. Spectr. 11 (3), e0514822–22. doi:10.1128/spectrum.05148-22
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444 (7117), 323–329. doi:10.1038/nature05286
Jones, J. D., Witek, K., Verweij, W., Jupe, F., Cooke, D., Dorling, S., et al. (2014). Elevating crop disease resistance with cloned genes. Philos. Trans. R. Soc. Lond B Biol. Sci. 369, 20130087. doi:10.1098/rstb.2013.0087
Karmakar, A., Taufiqa, S., Baig, M. J., and Molla, K. A. (2022). Increasing disease resistance in host plants through genome editing. Proc. Indian Natl. Sci. Acad. 88 (3), 417–429. doi:10.1007/s43538-022-00100-6
Kashyap, A. S., Manzar, N., Maurya, A., Mishra, D. D., Singh, R. P., and Sharma, P. K. (2023). Development of diagnostic markers and applied for genetic diversity study and population structure of Bipolaris sorokiniana associated with leaf blight complex of wheat. J. Fungi 9 (2), 153. doi:10.3390/jof9020153
Kaur, S., Dhillon, G. S., and Chauhan, V. B. (2013). Morphological and pathogenic variability in Macrophomina phaseolina isolates of pigeonpea (Cajanus cajan L.) and their relatedness using principle component analysis. Archives Phytopathology Plant Prot. 46 (19), 2281–2293. doi:10.1080/03235408.2013.792538
Kesharwani, A. K., Singh, D., Kulshreshtha, A., Kashyap, A. S., Avasthi, A. S., and Geat, N. (2022). Black rot disease incited by Indian race 1 of Xanthomonas campestris pv. campestris in Brassica juncea ‘Pusa Bold’ in India. Plant Dis. 107, 212. doi:10.1094/PDIS-04-22-0738-PDN
Khosravi, S. (2021). Development of a CRISPR-imaging toolset for imaging of genomic loci in living plants.
Kieu, N. P., Lenman, M., Wang, E. S., Petersen, B. L., and Andreasson, E. (2021). Mutations introduced in susceptibility genes through CRISPR/Cas9 genome editing confer increased late blight resistance in potatoes. Sci. Rep. 11 (1), 4487. doi:10.1038/s41598-021-83972-w
Kim, Y. B., Komor, A. C., Levy, J. M., Packer, M. S., Zhao, K. T., and Liu, D. R. (2017). Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 35 (4), 371–376. doi:10.1038/nbt.3803
Kis, A., Hamar, É., Tholt, G., Bán, R., and Havelda, Z. (2019). Creating highly efficient resistance against wheat dwarf virus in barley by employing CRISPR/Cas9 system. Plant Biotechnol. J. 17 (6), 1004–1006. doi:10.1111/pbi.13077
Klaubauf, S., Tharreau, D., Fournier, E., Groenewald, J. Z., Crous, P. W., de Vries, R. P., et al. (2014). Resolving the polyphyletic nature of Pyricularia (pyriculariaceae). Stud. Mycol. 79, 85–120. doi:10.1016/j.simyco.2014.09.004
Köhl, J., Kolnaar, R., and Ravensberg, W. J. (2019). Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy. Front. plant Sci. 10, 845. doi:10.3389/fpls.2019.00845
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., and Liu, D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533 (7603), 420–424. doi:10.1038/nature17946
Koonin, E. V., and Makarova, K. S. (2022). Evolutionary plasticity and functional versatility of CRISPR systems. PLoS Biol. 20 (1), e3001481. doi:10.1371/journal.pbio.3001481
Kourelis, J., and van der Hoorn, R. A. L. (2018). Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell. 30 (2), 285–299. doi:10.1105/tpc.17.00579
Kruse, J. M. (2018). The phylogeny of smut fungi (ustilaginomycotina) (PhD dissertation). Frankfurt am Main, Germany: Johann Wolfgang Goethe University.
Kumar, H., Kawai, T., and Akira, S. (2011). Pathogen recognition by the innate immune system. Int. Rev. Immunol. 30 (1), 16–34. doi:10.3109/08830185.2010.529976
Kunwar, S., Iriarte, F., Fan, Q., Evaristo da Silva, E., Ritchie, L., Nguyen, N. S., et al. (2018). Transgenic expression of EFR and Bs2 genes for field management of bacterial wilt and bacterial spot of tomato. Phytopathology 108, 1402–1411. doi:10.1094/PHYTO-12-17-0424-R
Lamichhane, S., and Thapa, S. (2022). Advances from conventional to modern plant breeding methodologies. Plant Breed. Biotechnol. 10 (1), 1–14. doi:10.9787/pbb.2022.10.1.1
LeBlanc, N., Salgado-Salazar, C., and Crouch, J. A. (2018). Boxwood blight: an ongoing threat to ornamental and native boxwood. Appl. Microbiol. Biotechnol. 102, 4371–4380. doi:10.1007/s00253-018-8936-2
Leppla, N., Momol, T., Nesheim, N., and Dusky, J. (2004). Plant, animal and human protection, FAS 2 Focus Area. United States: University of Florida/IFAS.
Li, C., Zong, Y., Wang, Y., Jin, S., Zhang, D., Song, Q., et al. (2018). Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 19, 59–9. doi:10.1186/s13059-018-1443-z
Li, H., Yang, Y., Hong, W., Huang, M., Wu, M., and Zhao, X. (2020). Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct. Target. Ther. 5 (1), 1. doi:10.1038/s41392-019-0089-y
Li, J. F., Norville, J. E., Aach, J., McCormack, M., Zhang, D., Bush, J., et al. (2013). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691. doi:10.1038/nbt.2654
Li, T., Liu, B., Spalding, M. H., Weeks, D. P., and Yang, B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30 (5), 390–392. doi:10.1038/nbt.2199
Liu, W., Zhu, X., Lei, M., Xia, Q., Botella, J. R., Zhu, J. K., et al. (2015). A detailed procedure for CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana. Sci. Bull. 60, 1332–1347. doi:10.1007/s11434-015-0848-2
Lloyd, A., Plaisier, C. L., Carroll, D., and Drews, G. N. (2005). Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc. Natl. Acad. Sci. 102 (6), 2232–2237. doi:10.1073/pnas.0409339102
Ma, L., Zhu, F., Li, Z., Zhang, J., Li, X., Dong, J., et al. (2015). TALEN-based mutagenesis of lipoxygenase LOX3 enhances the storage tolerance of rice (Oryza sativa) seeds. PLoS One 10 (12), e0143877. doi:10.1371/journal.pone.0143877
Mabbett, T. (2016). Epidemiology and management of Exobasidium vexans (blister blight disease) on Camellia sinensis (tea). Int. Pest Control 58 (2), 114.
Macovei, A., Sevilla, N. R., Cantos, C., Jonson, G. B., Slamet-Loedin, I., Čermák, T., et al. (2018). Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol. J. 16 (11), 1918–1927. doi:10.1111/pbi.12927
Mahas, A., Aman, R., and Mahfouz, M. (2019). CRISPR-Cas13d mediates robust RNA virus interference in plants. Genome Biol. 20, 263–316. doi:10.1186/s13059-019-1881-2
Mahawer, A. K., Dubey, A. K., Awasthi, O. P., Singh, D., Dahuja, A., Sevanthi, A. M., et al. (2023). Elucidation of physio-biochemical changes in citrus spp. Incited by Xanthomonas citri pv. Citri. Hortic. 9, 324. doi:10.3390/horticulturae9030324
Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2018). Classification and nomenclature of CRISPR-Cas systems: where from here? CRISPR J. 1 (5), 325–336. doi:10.1089/crispr.2018.0033
Malzahn, A., Lowder, L., and Qi, Y. (2017). Plant genome editing with TALEN and CRISPR. Cell. Biosci. 7 (1), 21–18. doi:10.1186/s13578-017-0148-4
Manzar, N., Kashyap, A. S., Maurya, A., Rajawat, M. V. S., Sharma, P. K., Srivastava, A. K., et al. (2022). Multi-gene phylogenetic approach for identification and diversity analysis of Bipolaris maydis and Curvularia lunata isolates causing foliar blight of Zea mays. J. Fungi 8, 802. doi:10.3390/jof8080802
Manzoor, S., Bhat, F. A., Baba, Z. A., Rather, T. R., Nisa, R. T., Parveen, S., et al. (2023). Phoma blight of soybean in Kashmir: etiology, relative yield losses and critical stage of management intervention. Legume Res. 46 (12), 1674–1679.
Manzoor, S., Mir, Z. A., Wani, T. A., Gulzar, H., Nabi, Z., Parveen, S., et al. (2023). Recent advances in diagnostic approaches for plant pathogen detection. Futur. Trends Biotechnol. IIP Ser. ISBN, 3.
Manzoor, S., Nabi, S. U., Baranwal, V. K., Verma, M. K., Parveen, S., Rather, T. R., et al. (2023). Overview on century progress in research on mosaic disease of apple (Malus domestica Borkh) incited by apple mosaic virus/apple necrotic mosaic virus. Virology 587, 109846. doi:10.1016/j.virol.2023.109846
Miedaner, T. (2016). “Breeding strategies for improving plant resistance to diseases,” in Advances in plant breeding strategies: agronomic, abiotic and biotic stress traits, 561–599.
Mishra, R., Joshi, R. K., and Zhao, K. (2020). Base editing in crops: current advances, limitations and future implications. Plant Biotechnol. J. 18 (1), 20–31. doi:10.1111/pbi.13225
Mishra, R., Zheng, W., Joshi, R. K., and Kaijun, Z. (2021). Genome editing strategies towards enhancement of rice disease resistance. Rice Sci. 28 (2), 133–145. doi:10.1016/j.rsci.2021.01.003
Molla, K. A., Karmakar, S., and Islam, M. T. (2020). Wide horizons of CRISPR-Cas-derived technologies for basic biology, agriculture, and medicine. CRISPR-Cas methods, 1–23. doi:10.1007/978-1-0716-0616-2_1
Molla, K. A., Sretenovic, S., Bansal, K. C., and Qi, Y. (2021). Precise plant genome editing using base editors and prime editors. Nat. Plants 7 (9), 1166–1187. doi:10.1038/s41477-021-00991-1
Molla, K. A., and Yang, Y. (2020). Predicting CRISPR/Cas9-induced mutations for precise genome editing. Trends Biotechnol. 38 (2), 136–141. doi:10.1016/j.tibtech.2019.08.002
Moscou, M. J., and Bogdanove, A. J. (2009). A simple cipher governs DNA recognition by TAL effectors. Science 326 (5959), 1501. doi:10.1126/science.1178817
Mulder, J. L., and Holliday, P. (1971). Entyloma oryzae. C.M.I. Descriptions of pathological fungi and bacteria 2.
Mundt, C. C. (1994). Use of host genetic diversity to control cereal diseases: implications of rice blast. Rice blast Dis.
Mundt, C. C. (2018). Pyramiding for resistance durability: theory and practice. Phytopathology 108, 792–802. doi:10.1094/PHYTO-12-17-0426-RVW
Mushtaq, M., Sakina, A., Wani, S. H., Shikari, A. B., Tripathi, P., Zaid, A., et al. (2019). Harnessing genome editing techniques to engineer disease resistance in plants. Front. plant Sci. 10, 550. doi:10.3389/fpls.2019.00550
Nabi, S. U., Baranwal, V. K., Yadav, M. K., and Rao, G. P. (2020). Association of Apple necrotic mosaic virus (ApNMV) with mosaic disease in commercially grown cultivars of apple (Malus domestica Borkh) in India. 3 Biotech. 10, 122–129. doi:10.1007/s13205-020-2117-6
Nabi, S. U., Mir, J. I., Yasmin, S., Din, A., Raja, W. H., Madhu, G. S., et al. (2023). Tissue and time optimization for real-time detection of apple mosaic virus and apple necrotic mosaic virus associated with mosaic disease of apple (Malus domestica). Viruses 15 (3), 795. doi:10.3390/v15030795
Nekrasov, V., Wang, C., Win, J., Lanz, C., Weigel, D., and Kamoun, S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7 (1), 482. doi:10.1038/s41598-017-00578-x
Nishikura, K. (2010). Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79, 321–349. doi:10.1146/annurev-biochem-060208-105251
Olszak-Przybyś, H., Korbecka-Glinka, G., and Patkowska, E. (2023). Identification and pathogenicity of Fusarium isolated from soybean in Poland. Pathogens 12, 1162. doi:10.3390/pathogens12091162
Ordon, J., Gantner, J., Kemna, J., Schwalgun, L., Reschke, M., Streubel, J., et al. (2017). Generation of chromosomal deletions in dicotyledonous plants employing a user-friendly genome editing toolkit. Plant J. 89 (1), 155–168. doi:10.1111/tpj.13319
Osakabe, K., Osakabe, Y., and Toki, S. (2010). Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proc. Natl. Acad. Sci. 107 (26), 12034–12039. doi:10.1073/pnas.1000234107
Pachauri, R. K., and Reisinger, A. (2007). Climate change 2007: synthesis report. Contribution of working groups I, II and III to the fourth assessment report of the Intergovernmental Panel on Climate Change. Geneva: IPCC.
Palukaitis, P., Roossinck, M. J., Dietzgen, R. G., and Francki, R. I. B. (1992). Cucumber mosaic virus. Adv. Virus Res. 41, 281–348. doi:10.1016/s0065-3527(08)60039-1
Pandit, M. A., Kumar, J., Gulati, S., Bhandari, N., Mehta, P., Katyal, R., et al. (2022). Major biological control strategies for plant pathogens. Pathogens 11 (2), 273. doi:10.3390/pathogens11020273
Panth, M., Hassler, S. C., and Baysal-Gurel, F. (2020). Methods for management of soilborne diseases in crop production. Agriculture 10 (1), 16. doi:10.3390/agriculture10010016
Park, H.-S., Hohn, T., Ryabova, L. A., and Ryabova, L. A. (2001). A plant viral "reinitiation" factor interacts with the host translational machinery. Cell. 106, 723–733. doi:10.1016/s0092-8674(01)00487-1
Pathak, V. M., Verma, V. K., Sharma, A., Dewali, S., Kumari, R., Mohapatra, A., et al. (2022). Current status of pesticide effects on environment, human health and it’s eco-friendly management as bioremediation: a comprehensive review. Front. Microbiol. 13, 962619. doi:10.3389/fmicb.2022.962619
Pathania, A., Singh, L., and Sharma, P. N. (2021). “Host plant resistance: an eco-friendly approach for crop disease management,” in Microbial biotechnology in crop protection (Singapore: Springer Singapore), 395–449.
Peng, A., Chen, S., Lei, T., Xu, L., He, Y., Wu, L., et al. (2017). Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene Cs LOB 1 promoter in citrus. Plant Biotechnol. J. 15 (12), 1509–1519. doi:10.1111/pbi.12733
Petolino, J. F., Worden, A., Curlee, K., Connell, J., StrangeMoynahan, T. L., Larsen, C., et al. (2010). Zinc finger nuclease-mediated transgene deletion. Plant Mol. Biol. 73, 617–628. doi:10.1007/s11103-010-9641-4
Podder, R., Banniza, S., and Vandenberg, A. (2013). Screening of wild and cultivated lentil germplasm for resistance to stemphylium blight. Plant Genet. Resour. 11, 26–35. doi:10.1017/S1479262112000329
Poti, T., Thitla, T., Imaiam, N., Arunothayanan, H., Doungsa-Ard, C., Kongtragoul, P., et al. (2023). Isolates of Colletotrichum truncatum with resistance to multiple fungicides from soybean in Northern Thailand. Plant Dis. 107 (9), 2736–2750. doi:10.1094/PDIS-08-22-1882-RE
Pradhan, S. K., Nayak, D. K., Mohanty, S., Behera, L., Barik, S. R., Pandit, E., et al. (2015). Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety, Jalmagna. Jalmagna. Rice (N Y) 8, 51. doi:10.1186/s12284-015-0051-8
Ramirez, C. L., Foley, J. E., Wright, D. A., Müller-Lerch, F., Rahman, S. H., Cornu, T. I., et al. (2008). Unexpected failure rates for modular assembly of engineered zinc fingers. Nat. methods 5 (5), 374–375. doi:10.1038/nmeth0508-374
Ran, Y., Patron, N., Kay, P., Wong, D., Buchanan, M., Cao, Y. Y., et al. (2018). Zinc finger nuclease-mediated precision genome editing of an endogenous gene in hexaploid bread wheat (Triticum aestivum) using a DNA repair template. Plant Biotechnol. J. 16 (12), 2088–2101. doi:10.1111/pbi.12941
Rana, D. S., Dass, A., Rajanna, G. A., and Kaur, R. (2016). Biotic and abiotic stress management in pulses. Indian J. Agron. 61, 238–248.
Rani, R., Yadav, P., Barbadikar, K. M., Baliyan, N., Malhotra, E. V., Singh, B. K., et al. (2016). CRISPR/Cas9: a promising way to exploit genetic variation in plants. Biotechnol. Lett. 38, 1991–2006. doi:10.1007/s10529-016-2195-z
Ravikumara, B. M., Naik, M. K., Telangre, R., and Sharma, M. (2022). Distribution and pathogenic diversity in Fusarium udum Butler isolates: the causal agent of pigeonpea Fusarium wilt. BMC Plant Biol. 22 (1), 147. doi:10.1186/s12870-022-03526-8
Redlin, S. (1991). Discula destructiva sp. nov., cause of dogwood anthracnose. Mycologia 83 (5), 633–642. doi:10.1080/00275514.1991.12026062
Ren, B., Yan, F., Kuang, Y., Li, N., Zhang, D., Zhou, X., et al. (2018). Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID mutant. Mol. plant 11 (4), 623–626. doi:10.1016/j.molp.2018.01.005
Ricroch, A. (2019). Global developments of genome editing in agriculture. Transgenic Res. 28, 45–52. doi:10.1007/s11248-019-00133-6
Rincao, M. P., Carvalho, M. C. D. C. G. D., Nascimento, L. C., Lopes-Caitar, V. S., Carvalho, K. D., Darben, L. M., et al. (2018). New insights into Phakopsora pachyrhizi infection based on transcriptome analysis in planta. Genet. Mol. Biol. 41 (3), 671–691. doi:10.1590/1678-4685-gmb-2017-0161
Rott, M. E., Kesanakurti, P., Berwarth, C., Rast, H., Boyes, I., Phelan, J., et al. (2018). Discovery of negative-sense RNA viruses in trees infected with apple rubbery wood disease by next-generation sequencing. Plant Dis. 102 (7), 1254–1263. doi:10.1094/PDIS-06-17-0851-RE
Roviqowati, F., Samanhudi, S. T., Purwanto, E., Sisharmini, A., Apriana, A., and Yunus, A. (2024). Introduction of CRISPR/Cas9 with the target genes to improve agronomic traits and leaf blight resistance in rice. SABRAO J. Breed. Genet. 56 (1), 29–44. doi:10.54910/sabrao2024.56.1.3
Różewicz, M., Wyzińska, M., and Grabiński, J. (2021). The most important fungal diseases of cereals—problems and possible solutions. Agronomy 11 (4), 714. doi:10.3390/agronomy11040714
Russell, G. E. (2013). Plant breeding for pest and disease resistance: studies in the agricultural and food sciences.
Saraswat, P., Singh, D., Singh, A., Mathur, S., Waswani, H., and Ranjan, R. (2024). “CRISPR/Cas techniques used in plant disease management,” in Biocontrol agents for improved agriculture (Academic Press), 331–351.
Savary, S., Ficke, A., Aubertot, J. N., and Hollier, C. (2012). Crop losses due to diseases and their implications for global food production losses and food security. Food Secur. 4 (4), 519–537. doi:10.1007/s12571-012-0200-5
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., McRoberts, N., and Nelson, A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3 (3), 430–439. doi:10.1038/s41559-018-0793-y
Savchenko, K. G., Carris, L. M., Demers, J., Manamgoda, D. S., and Castlebury, L. A. (2017). What causes flag smut of wheat? Plant Pathol. 66 (7), 1139–1148. doi:10.1111/ppa.12657
Scholthof, K. B. (2004). Tobacco mosaic virus: a model system for plant biology. Annu. Rev. Phytopathol. 42, 13–34. doi:10.1146/annurev.phyto.42.040803.140322
Semenova, E., Jore, M. M., Datsenko, K. A., Semenova, A., Westra, E. R., Wanner, B., et al. (2011). Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. 108 (25), 10098–10103. doi:10.1073/pnas.1104144108
Sera, T. (2005). Inhibition of virus DNA replication by artificial zinc finger proteins. J. virology 79 (4), 2614–2619. doi:10.1128/JVI.79.4.2614-2619.2005
Sharma, M., and Gaviyappanavar, R.Tarafdar (2023). Evaluation of fungicides and fungicide application methods to manage Phytophthora blight of pigeonpea. Agriculture 13, 633. doi:10.3390/agriculture13030633
Sharma, Y. R., Singh, G., and Kaur, L. (1995). A rapid technique for ascochyta blight resistance in chickpea. Int. Chickpea Pigeonpea Newsl. 2, 34–35.
Shen, E., Wang, X., Lu, Z., Zhou, F., Ma, W., Cui, Z., et al. (2023). Overexpression of a beta-1, 6-glucanase gene GluM in transgenic rice confers high resistance to rice blast, sheath blight and false smut. Pest Manag. Sci. 79 (6), 2152–2162. doi:10.1002/ps.7394
Silva, G., Poirot, L., Galetto, R., Smith, J., Montoya, G., Duchateau, P., et al. (2011). Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr. Gene Ther. 11 (1), 11–27. doi:10.2174/156652311794520111
Singh, A., Dhiman, N., Kar, A. K., Singh, D., Purohit, M. P., Ghosh, D., et al. (2020). Advances in controlled release pesticide formulations: prospects to safer integrated pest management and sustainable agriculture. J. Hazard. Mater. 385, 121525. doi:10.1016/j.jhazmat.2019.121525
Singh, V. K., and Chawla, S. (2012). Cultural practices: an eco-friendly innovative approach in plant disease management. India: International Book Publishers and Distributers. Dehradun–248, 1, 01-20.
Sobanbabu, G., Sabarinathan, K. G., Parthiban, V. K., and Ramamoorthy, V. (2018). Isolation, screening and identification of virulent isolates of Bipolaris oryzae causing rice Brown spot and Sarocladium oryzae causing sheath rot disease. Int. J. Curr. Microbiol. Appl. Sci. 7 (9), 930–939. doi:10.20546/ijcmas.2018.709.112
Song, W. Y., Wang, G. L., Chen, L. L., Kim, H. S., Pi, L. Y., Holsten, T., et al. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806. doi:10.1126/science.270.5243.1804
Stafforst, T., and Schneider, M. F. (2012). An RNA–deaminase conjugate selectively repairs point mutations. Angew. Chem. Int. Ed. 51 (44), 11166–11169. doi:10.1002/anie.201206489
Stoddard, B. L. (2005). Homing endonuclease structure and function. Q. Rev. Biophysics 38 (1), 49–95. doi:10.1017/S0033583505004063
Stoddard, B. L. (2011). Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure 19 (1), 7–15. doi:10.1016/j.str.2010.12.003
Sun, H., Zhu, X., Li, C., Ma, Z., Han, X., Luo, Y., et al. (2021). Xanthomonas effector XopR hijacks host actin cytoskeleton via complex coacervation. Nat. Commun. 12, 4064. doi:10.1038/s41467-021-24375-3
Tai, T. H., Dahlbeck, D., Clark, E. T., Gajiwala, P., Pasion, R., Whalen, M. C., et al. (1999). Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc. Natl. Acad. Sci. U. S. A. 96, 14153–14158. doi:10.1073/pnas.96.24.14153
Takagi, H., Abe, A., Yoshida, K., Kosugi, S., Natsume, S., Mitsuoka, C., et al. (2013). QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 74, 174–183. doi:10.1111/tpj.12105
Talakayala, A., Ankanagari, S., and Mallikarjuna, G. (2024). Augmenting the CRISPR/Cas9 System to Edit Mungbean yellow mosaic viral genome for enhanced disease resistance in Nicotiana Benthamiana.
Talhinhas, P., Batista, D., Diniz, I., Vieira, A., Silva, D. N., Loureiro, A., et al. (2017). The coffee leaf rust pathogen Hemileia vastatrix: one and a half centuries around the tropics. Mol. Plant Pathol. 18 (8), 1039–1051. doi:10.1111/mpp.12512
Tarafdar, A., Rani, T. S., Chandran, U. S. S., Ghosh, R., Chobe, D. R., and Sharma, M. (2018). Exploring combined effect of abiotic (soil moisture) and biotic (Sclerotium rolfsii sacc.) stress on collar rot development in chickpea. Front. Plant Sci. 9, 1154. doi:10.3389/fpls.2018.01154
Terefe, T. G., Visser, B., Pretorius, Z. A., and Boshoff, W. H. P. (2023). Physiologic races of Puccinia triticina detected on wheat in South Africa from 2017 to 2020. Eur. J. Plant Pathology 165 (1), 1–15. doi:10.1007/s10658-022-02583-x
Tilman, D., Cassman, K. G., Matson, P. A., Naylor, R., and Polasky, S. (2002). Agricultural sustainability and intensive production practices. Nature 418, 671–677. doi:10.1038/nature01014
Tiwari, A., and Ghag, S. B. (2024). Base editing and prime editing in horticulture crops: potential applications, challenges, and prospects. CRISPRized Horticulture Crops, 105–126.
Tripathi, J. N., Ntui, V. O., Ron, M., Muiruri, S. K., Britt, A., and Tripathi, L. (2019). CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun. Biol. 2 (1), 46. doi:10.1038/s42003-019-0288-7
Udayanga, D., Miriyagalla, S. D., Herath, I. S., Castlebury, L. A., Ferdinandez, H. S., and Manamgoda, D. S. (2020). Foliar pathogenic fungi: growing threats to global food security and ecosystem health.
Van Der Oost, J., Westra, E. R., Jackson, R. N., and Wiedenheft, B. (2014). Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nat. Rev. Microbiol. 12 (7), 479–492. doi:10.1038/nrmicro3279
Vercoe, R. B., Chang, J. T., Dy, R. L., Taylor, C., Gristwood, T., Clulow, J. S., et al. (2013). Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 9 (4), 1003454. doi:10.1371/journal.pgen.1003454
Voytas, D. F., and Gao, C. (2014). Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biol. 12 (6), 1001877. doi:10.1371/journal.pbio.1001877
Wang, F., Wang, C., Liu, P., Lei, C., Hao, W., Gao, Y., et al. (2016). Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PloS One 11 (4), e0154027. doi:10.1371/journal.pone.0154027
Wang, Y., Chen, J., Li, D.-W., Zheng, L., and Huang, J. (2017). CglCUT1 gene required for cutinase activity and pathogenicity of Colletotrichum gloeosporioides causing anthracnose of camellia oleifera. Eur. J. Plant Pathol. 147, 103–114. doi:10.1007/s10658-016-0983-x
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32 (9), 947–951. doi:10.1038/nbt.2969
Ye, G., McNell, D. L., and Hill, G. D. (2002). Breeding for resistance to lentil ascochyta blight. Plant Breed. 121, 185–191. doi:10.1046/j.1439-0523.2002.00705.x
Yosef, I., Goren, M. G., and Qimron, U. (2012). Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 40 (12), 5569–5576. doi:10.1093/nar/gks216
Zaidi, S. S. E. A., Mukhtar, M. S., and Mansoor, S. (2018). Genome editing: targeting susceptibility genes for plant disease resistance. Trends Biotechnol. 36, 898–906. doi:10.1016/j.tibtech.2018.04.005
Zhang, H., Si, X., Ji, X., Fan, R., Liu, J., Chen, K., et al. (2018). Genome editing of upstream open reading frames enables translational control in plants. Nat. Biotechnol. 36 (9), 894–898. doi:10.1038/nbt.4202
Zhang, H., Zhang, J., Lang, Z., Botella, J. R., and Zhu, J. K. (2017). Genome editing—principles and applications for functional genomics research and crop improvement. Crit. Rev. Plant Sci. 36 (4), 291–309. doi:10.1080/07352689.2017.1402989
Zhang, S., Li, C., Si, J., Han, Z., and Chen, D. (2022). Action mechanisms of effectors in plant-pathogen interaction. Int. J. Mol. Sci. 23 (12), 6758. doi:10.3390/ijms23126758
Zhang, S., Xu, B., and Liu, J. (2020). Mechanisms of different cultivars of Cucurbita pepo in resistance to Podosphaera xanthii infection through improvement of antioxidative defense system and gene expression.
Zhang, T., Zheng, Q., Yi, X., An, H., Zhao, Y., Ma, S., et al. (2018). Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol. J. 16 (8), 1415–1423. doi:10.1111/pbi.12881
Zhao, B., Lin, X., Poland, J., Trick, H., Leach, J., and Hulbert, S. (2005). A maize resistance gene functions against bacterial streak disease in rice. Proc. Natl. Acad. Sci. U. S. A. 102, 15383–15388. doi:10.1073/pnas.0503023102
Zhao, C., Zhang, Y., Gao, L., Xie, M., Zhang, X., Zeng, L., et al. (2024). Genome editing of RECEPTOR-LIKE KINASE 902 confers resistance to necrotrophic fungal pathogens in Brassica napus without growth penalties. Plant Biotechnol. J. 22 (3), 538–540. doi:10.1111/pbi.14253
Keywords: pathogens, disease resistance, genome editing crispr, meganucleases, TALEN, ZFN
Citation: Manzoor S, Nabi SU, Rather TR, Gani G, Mir ZA, Wani AW, Ali S, Tyagi A and Manzar N (2024) Advancing crop disease resistance through genome editing: a promising approach for enhancing agricultural production. Front. Genome Ed. 6:1399051. doi: 10.3389/fgeed.2024.1399051
Received: 11 March 2024; Accepted: 22 April 2024;
Published: 26 June 2024.
Edited by:
Humira Sonah, Central University of Haryana, IndiaReviewed by:
Mohammad Aslam, Donald Danforth Plant Science Center, United StatesCopyright © 2024 Manzoor, Nabi, Rather, Gani, Mir, Wani, Ali, Tyagi and Manzar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Subaya Manzoor, c3ViYXlhbWFuem9vcjk2QGdtYWlsLmNvbQ==; Sajad Ali, c2FqYWRhbGltaWNyb0B5dS5hYy5rcg==; Anshika Tyagi, dHlhZ2kuYW5zaGlrYTlAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.