- Department of Plant Science, Central University of Kerala, Kasaragod, Kerala, India
Plant genome editing, a recently discovered method for targeted mutagenesis, has emerged as a promising tool for crop improvement and gene function research. Many genome-edited plants, such as rice, wheat, and tomato, have emerged over the last decade. As the preliminary steps in the procedure for genome editing involve genetic transformation, amenability to genome editing depends on the efficiency of genetic engineering. Hence, there are numerous reports on the aforementioned crops because they are transformed with relative ease. Legume crops are rich in protein and, thus, are a favored source of plant proteins for the human diet in most countries. However, legume cultivation often succumbs to various biotic/abiotic threats, thereby leading to high yield loss. Furthermore, certain legumes like peanuts possess allergens, and these need to be eliminated as these deprive many people from gaining the benefits of such crops. Further genetic variations are limited in certain legumes. Genome editing has the potential to offer solutions to not only combat biotic/abiotic stress but also generate desirable knock-outs and genetic variants. However, excluding soybean, alfalfa, and Lotus japonicus, reports obtained on genome editing of other legume crops are less. This is because, excluding the aforementioned three legume crops, the transformation efficiency of most legumes is found to be very low. Obtaining a higher number of genome-edited events is desirable as it offers the option to genotypically/phenotypically select the best candidate, without the baggage of off-target mutations. Eliminating the barriers to genetic engineering would directly help in increasing genome-editing rates. Thus, this review aims to compare various legumes for their transformation, editing, and regeneration efficiencies and discusses various solutions available for increasing transformation and genome-editing rates in legumes.
1 Introduction
Proteins are an integral component of almost every part of our body. The recommended quantity of protein for individuals with minimal to intense physical activity ranges from 1 to 1.6 g per kg body weight per day (Wu, 2016). Legumes contain approximately 13–36 g of proteins per 100 g (Singh et al., 2022; Affrifah et al., 2023). Legumes are also rich in minerals, fibers, and bioactive compounds (Margier et al., 2018). The commonly cultivated grain legumes, also known as pulses, include soybeans, mung beans, field peas, cowpeas, pigeon peas, chickpeas, common beans, and lentils. Legumes are desirable for agriculture as well as they increase the yield of other crops (Zhao et al., 2022) by enhancing soil fertility and nitrogen content. However, legume cultivation suffers an average loss of 31.9%–69.6% due to abiotic (drought) and biotic (insects, diseases, and weeds) reasons (Sharma et al., 2016). Genome editing has recently revolutionized research in crop development as it offers a non-transgenic method of generating targeted mutants with desirable agronomic traits (Bhowmik et al., 2021; Jha et al., 2023; Singh et al., 2023).
2 Genome editing
Genome editing is the mutagenesis of desired portions of a gene or genome. Of the various methods used for genome editing, which are based on zinc-finger nuclease (ZFN), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) (Amritha and Shah, 2021), CRISPR/Cas9 has proven to be most effective for targeted genome editing in plants (Yin et al., 2017; Mao et al., 2019). CRISPR/Cas9-based genome editing involves site-specific cutting using the Cas9 endonuclease, guided by RNA (Ran et al., 2022). Recently, various versions of Cas (natural and synthetic) and similar nucleases have been reported (reviewed in Aksoy et al., 2022; Liu et al., 2019). Furthermore, various plant promoters for expressing the guided RNAs have previously been reported (reviewed by Kor et al., 2023). This method can be tailored to perform the insertion, deletion, or substitution of nucleotide(s) in the target site (Das et al., 2022; Xie et al., 2022). More details on the process and advances of genome editing have been elaborated in many previous reviews (Aksoy et al., 2022; Ran et al., 2022; Verma et al., 2023b). The mutants of approximately 28 crops including rice, tomato, wheat, and soybean, exhibiting economically important traits such as biotic/abiotic stress resistance and enhanced nutritional value, have been generated in the past few years (reviewed by Verma et al., 2023b; Ukhatovaa et al., 2023). Since transgene integration is not required in the mutants, this method has become the most widely used targeted transgene-free method (Verma et al., 2023b).
Apart from providing abiotic/biotic stress tolerance, genome editing has the potential to eliminate allergy-causing/antinutrient factors from legume crops, such as peanuts (Biswas et al., 2022) and grass peas (Xu et al., 2018; Verma et al., 2023a). Genome editing of legumes can also aid in the functional analysis of genes involved in symbiotic nitrogen fixation (Wang et al., 2017; Wang et al., 2019). Since limited genetic variants are available for cultivated legume crops, genome editing offers an excellent and efficient method for generating favorable mutants. However, there are few reports on legume genome editing due to their transformation recalcitrance.
3 Transformation as a prerequisite for genome editing
Since the genome-edited plants are non-transgenic, they are generated via steps that are common for making transgenic plants (Figure 1). Like genetic engineering, genome editing reagents are delivered into the plant cell using Agrobacterium-mediated or direct gene transfer methods, followed by antibiotic selection and regeneration. Once integrated into the plant genome, the genome editing construct expresses the reagents required for editing and completes the job. Thus, the T0 plants are hemizygous for two loci-—the transformed (harboring the genome editing construct) and the edited loci. Since the segregation of T1 plants would generate the desired homozygous genome-edited candidate, it has to be identified using detailed molecular analysis involving sequencing. The transformed non-edited T1 individuals are eliminated at this stage. The homozygosity of the edited plants is assured by selfing and generating T2 plants. Thus, although genome editing generates a non-transgenic mutant, it can be achieved only if the transformation procedure is followed.
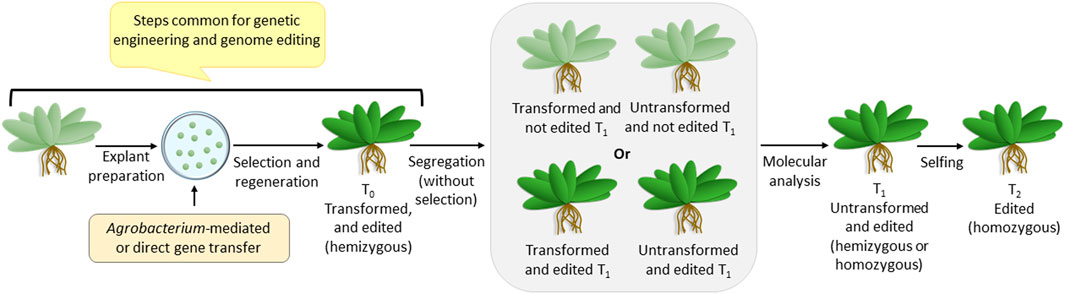
FIGURE 1. Overview of steps for transformation (also common for genome editing), followed by segregation, leading to a non-transgenic genome-edited plant.
4 Transformation recalcitrance of legumes
The main challenge faced during genome editing of legumes is their transformation recalcitrance. Reports on genome editing of plants such as rice and tomato are high (Jaganathan et al., 2018; Ukhatovaa et al., 2023) because they are transformation-amenable. Several crops are susceptible to transformation in comparison to the transformation of recalcitrant crops (Table 1). Table 1 shows that most legumes, excluding alfalfa and Lotus japonicus, have lower transformation efficiency. Even soybean, whose genetically modified versions are commercially cultivated in some countries, is known for its recalcitrance to transformation like other legumes (Xu et al., 2022). Only certain cultivars of soybean have generated an appreciable transformation efficiency. Susceptibility to transformation is desirable since it not only reduces the labor of handling more explants but also generates more edited individuals, thereby increasing the probability of obtaining desirable and clean individuals without off-target mutations. For the commercial release of an edited mutant, it is desirable that the best out of many is chosen after thorough genotype/phenotype screening.
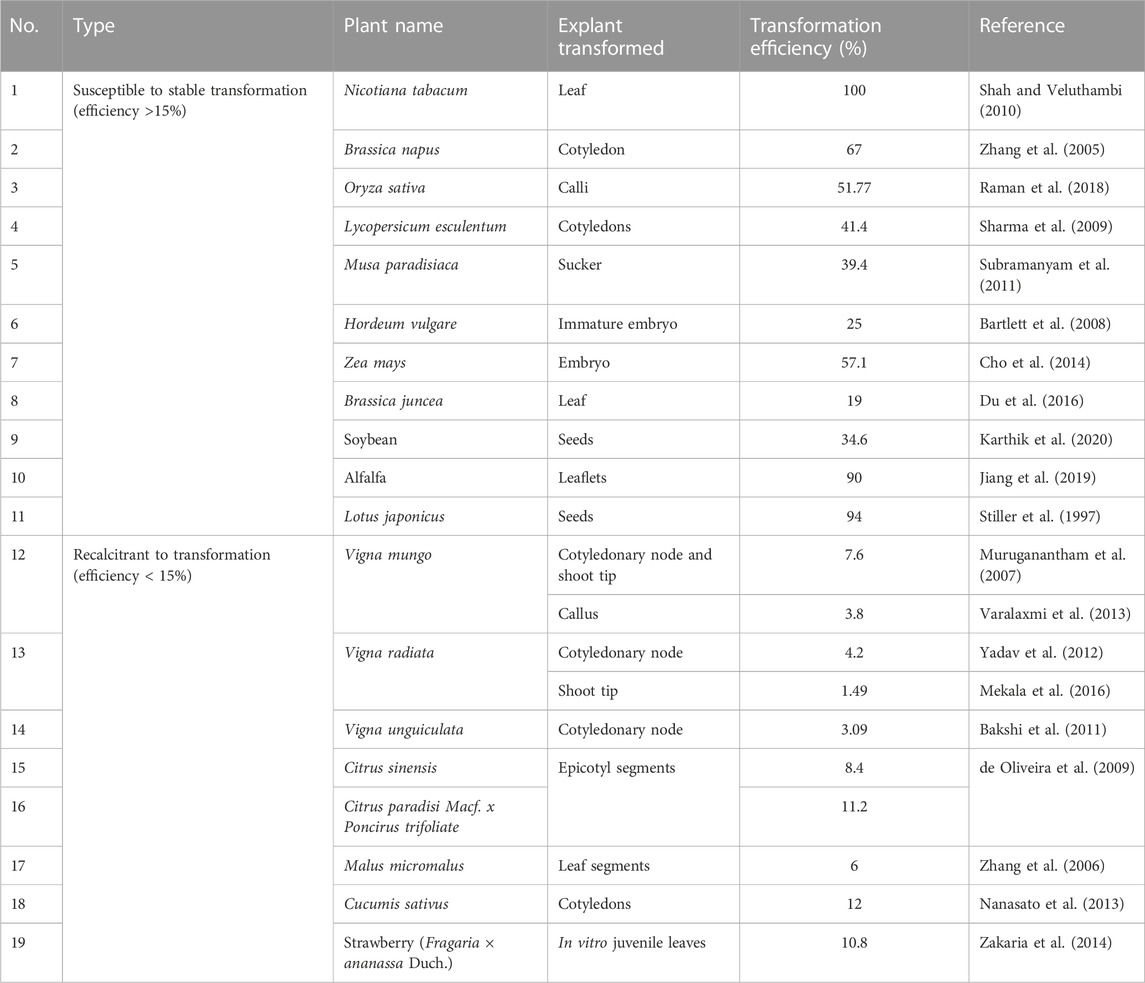
TABLE 1. Transformation efficiencies of some plants that are susceptible or recalcitrant to transformation.
4.1 Legume transformation
Previously, various explants and regeneration protocols have been attempted in different legume crops (Table 2). As evident in most cases, although the transformation efficiency seemed to be very high when transformed calli were counted based on the expression of reporter genes GUS/GFP/YFP, the number of transgenic plants drastically reduced after the antibiotic-containing media selection. It should also be noted that the true transformation efficiency can be calculated after thorough molecular screening. Although PCR confirms the transgenic nature of plants, the occurrence of clones due to the same transgenic events can be identified after junction fragment analysis by Southern hybridization (Shah and Veluthambi, 2010). Not all previous reports have characterized the junction fragment analysis by Southern hybridization (Table 2). Furthermore, PCR cannot rule out the possibility of transgene amplification due to Agrobacterium contamination. Most previous reports do not mention the stable inheritance of the transgene in the subsequent T1 generation. It could be possible that the T0 plants were chimeras and the transgene was lost in the subsequent generation, probably due to an insufficient number of transformed cells per plant.
5 Genome editing of legumes
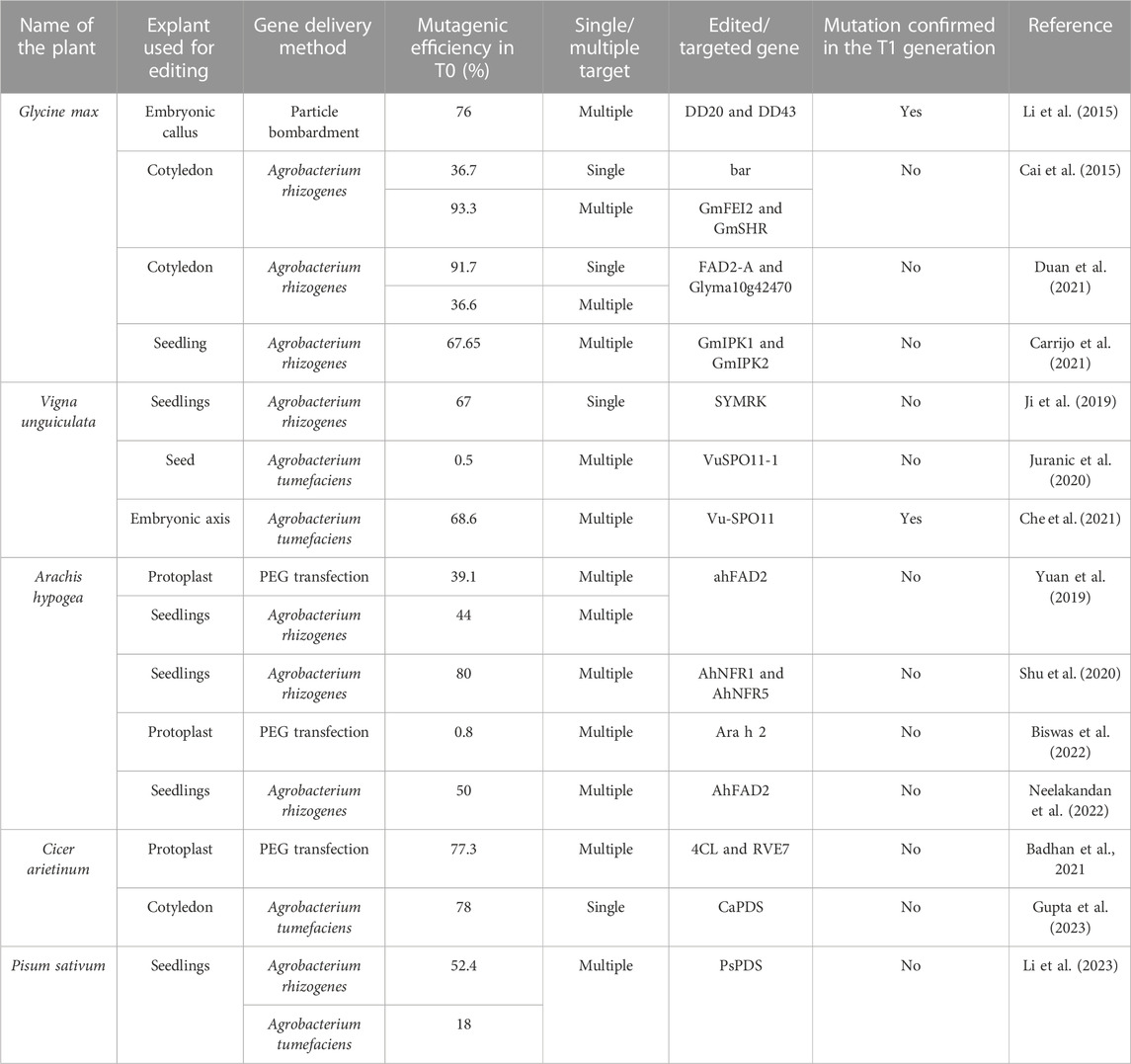
The availability of deeper information on the whole genome sequences and functional characterization of various genes in many legumes, including soybean, pigeon pea, chickpea, groundnut, common bean, mung bean, and cowpea (Shunmugam et al., 2018; Varshney et al., 2018) has opened vistas for crop improvement via genome editing (Kingsley et al., 2022). Examples of genome-edited legume crops are limited, and these include soybean (Sun et al., 2015; Lu and Tian, 2022), Lotus japonicus (Wang et al., 2017), Medicago truncatula (Meng et al., 2017; Jaudal et al., 2022), cowpea (Ji et al., 2019; Bridgeland et al., 2023), peanut (Yuan et al., 2019), and chickpea (Badhan et al., 2021; Gupta et al., 2023). Details on the traits of genome-edited legumes are elaborated in previous reviews (Bhowmik et al., 2021; Baloglu et al., 2022; Rasheed et al., 2022). The compilation of the previous reports (Table 4) indicates that although the genome editing efficiency in most cases was appreciably high in the T0 generation, most of these reports do not mention the inheritance of the edited trait in the T1 generation. Furthermore, it is to be noted that the editing efficiencies mentioned in most of these reports were based on the molecular characterization of the callus tissue and not the number of genome-edited T0 individuals. Most of these reports have not mentioned the exact number of edited T0 individuals obtained. This situation is comparable to the genetic transformation of legumes (Table 2), where the efficiency of obtaining GUS/GFP-positive calli was very high but not the transgenic individuals, and importantly, not the transgenic T1 individuals.
6 Causes for transformation recalcitrance
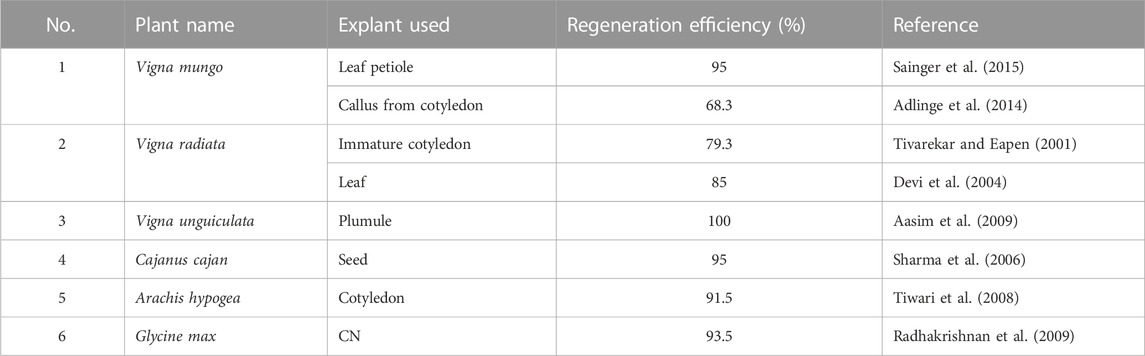
In order to identify the cause for transformation recalcitrance, it is important to recall the major steps in transformation (common for the direct or Agrobacterium-mediated method). Three major turning points are crucial for successful transformation (Figure 2). The first one is the effective entry of foreign DNA into the plant cell and its nucleus. The second is transgene integration, during which the cell is transformed. Editing can also take place in this step if the construct for the same is carried out. The third is the regeneration of transformants under selection pressure. The efficiency of the reporter (GUS/GFP/YFP) expression is a reflection of the efficiency of the first two steps. Thus, a high percentage of reporter expression observed in previous data (Table 2) indicates that the first two steps are successfully achieved in legumes. The transformation efficiency based on the T0 regeneration was extremely low in most of the previous cases. This indicates that the problem could be in the third step, which is regeneration under selection pressure. It appears that regeneration from the transformed cells does not occur frequently. Hence, although the high transformation of calli/explant indicates good transformation susceptibility in most cases, the failure of regeneration of transformed cells decreases the overall transformation efficiency. Nevertheless, the in vitro regeneration efficiency of most legumes is quite high (Table 3), when not subjected to transformation. This is an indication that the selection pressure during transformation adversely influences the regeneration efficiency in legumes. The reason for this remains unknown. Much standardization can make the recalcitrant legume crops more amenable to transformation (Bekalu et al., 2023) like their transformation-friendly cousins, alfalfa and Lotus japonicus (Table 1).
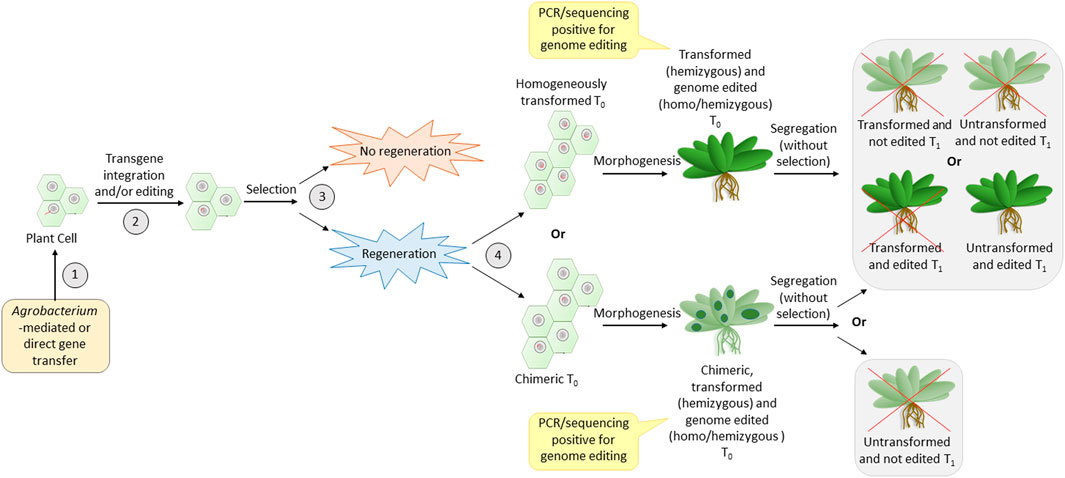
FIGURE 2. Most crucial steps of transformation that have an impact on the efficiency of genome editing. Numbers 1, 2, 3, and 4 are the steps—entry of DNA, integration accompanied by editing, selection, and regeneration of transformed calli, respectively. The red line entering into the nucleus and present inside the nucleus is the T-DNA. Red crosses indicate undesirable events.
Another observation with legumes is that the majority of reports do not show the inheritance of the transgenic (Table 2) or the edited loci (Table 4) to the T1 generation. This reduced heritability of the transgene/edited loci could probably be associated with the fourth step (Figure 2), where the regenerated plants could either be homogenously transformed or chimeras made of transformed and non-transformed tissues. The problem with chimeric plants is the unassured transfer of desired loci to the gametes. This is because the development of gametes with the desired loci depends on the development of floral meristem from transformed somatic cells, which, in turn, is proportional to the number of transformed cells in the regenerated plant. Hence, even if the first three steps are crossed, the fourth step may be a challenge in most legumes. To overcome this problem, it is better to generate plants via somatic embryogenesis and not via direct/indirect regeneration from calli/explant (Chandra and Pental, 2003). This is because somatic embryogenesis generates true-to-type clones (Gaj, 2004; Krishna et al., 2016). This can be achieved by standardization of the tissue culture medium and careful microscopic observation of the regenerating tissue to ensure the selection of somatic embryogenesis (Chandra and Pental, 2003; Pratap et al., 2018).
7 Strategies to enhance transformation
Many strategies have been previously discussed to improvise crop transformation to aid genome editing (Altpeter et al., 2016). When it comes to legumes, the transformation efficiency is directly proportional to regeneration efficiency. It is known that regeneration depends on multiple factors, such as culture media, hormone concentration, explant, and other supplements. Table 2 shows that many explants have been used in most legumes. Toward this point, soybean sets an excellent example of an explant-dependent enhancement of transformation. Although soybean is considered a crop recalcitrant to transformation (Xu et al., 2022), there are reports where this legume was transformed with an appreciable efficiency. The use of imbibed split seed with an attached partial embryonic axis resulted in 18.7% transformation, in comparison to the average efficiency of 8.7% (Pareddy et al., 2020). Various additives to the cocultivation/selection/regeneration media have been previously used. Another example of success is soybean, where the addition of sodium nitroprusside aided in uplifting the transformation rates up to 34.6% (Karthik et al., 2020). It should also be noted that the cultivar they used was Pusa 9712, which provided an appreciable efficiency of 23% even without adding sodium nitroprusside. On the other hand, Pareddy et al. (2020) used the cultivars, namely, Maverick and 20 proprietary elite, which provided a lesser average transformation efficiency (18.7%). Since the susceptibility to transformation is cultivar-dependent and the choice of cultivar depends on various agronomic conditions, more efforts are needed to standardize cultivar-specific transformation conditions. Although introgression has been used for the inter-cultivar transfer of transgenes, it is laborious and time-consuming. Pareddy et al. (2020) also reported that the A. tumefaciens strain EHA105 provided a better result (up to 23.5%) than EHA101 (up to 15.5%). EHA101 was previously reported to transform multiple soybean cultivars, such as Thorne, Williams, Williams 79, and Williams 82 (Paz et al., 2006). In chickpea, the A. tumefaciens strain GV3101 resulted in a better transformation efficiency of 17.56%, in comparison with two other strains EHA105 and LBA4404 with 8.54% and 5.43% efficiencies, respectively (Gupta et al., 2023). Agrobacterium rhizogenes also resulted in transformation in legumes such as common bean (Li et al., 2022), Robinia pseudoacacia L. (Han et al., 1993), and fenugreek (Garagounis et al., 2020).
The overexpression of morphogenetic regulator genes, such as BABY BOOM and WUSCHEL, increased the percentage of Agrobacterium-mediated transformation in monocots, like maize, sugarcane, rice, and sorghum (Lowe et al., 2016). This approach enhanced the regeneration efficiency of genome-edited crops as well (Debernardi et al., 2020; Chen et al., 2022). The transformation efficiency is proportional to the regeneration efficiency. The overexpression of maize GOLDEN2, a GARP transcription factor superfamily member that regulates several biological processes and phytohormone signaling pathways in plants, enhanced the regeneration of rice and maize calli by activating chloroplast development (Luo et al., 2023). Similarly, the homologs of GOLDEN2 from legumes (Wang et al., 2013) could help enhance regeneration in legumes as well. Incorporating such growth-promoting factors during transformation for gene editing may resolve the problem of regeneration of transformed cells in legumes.
Legumes serve as a poor host to most strains of Agrobacterium; bacteria alternative to Agrobacterium can offer a promising solution to enhance transformation events in legumes. Cho et al. (2022) reported the highest transformation efficiency (35%) for soybean using the novel bacteria Ochrobactrum haywardense H1, in comparison to two Agrobacterium strains AGL1 and LBA4404, with 26% and 12%, respectively. Other non-Agrobacterium natural genetic engineers, such as Ensifer adhaerens and Rhizobium etli (Rathore and Mullins, 2018), should also be explored for legume transformation.
8 Transformation-based strategies to facilitate genome editing
Most previously reported strategies on improvising plant genome editing are based on transformation, where transgene is initially integrated and then segregated out (He et al., 2022; Rasheed et al., 2022; Son and Park, 2022). Since most legumes fail to regenerate efficiently under antibiotic selection pressure, the alternative strategy based on reporter expression could be promising. Gao et al. (2016) introduced the novel strategy of expressing the fluorescent reporter mCherry along with the Cas9 construct in Arabidopsis thaliana plants. Although their transformation did not involve antibiotic selection, they could visually identify the transformed T1 plants by screening under UV. Similarly, He et al. (2022) proposed the use of the pigment-based RUBY reporter for genome editing as it gives a reddish coloration to the plants. Unlike the popularly known GUS reporter, RUBY does not require additional substrate/chemicals and can be used for live-screening of plants; unlike mCherry/GFP/other fluorescent reporters, its screening does not require UV (He et al., 2020). Thus, during legume transformation, if we use the gene coding for RUBY instead of the usual antibiotic marker gene, we may obtain more transformants due to the absence of antibiotic selection pressure. The non-transformed T1 plants can easily be segregated based on visual selection and subjected to molecular confirmation to identify the edited candidates.
9 Strategies for genome editing bypassing transformation
Deconstructed viral vectors have successfully been used for gene function analysis by silencing in legumes (Constantin et al., 2004; Zhang and Ghabrial, 2006) and other plants (Peyret and Lomonossoff, 2015). However, virus-mediated transformation is not a desirable method for stable transformation as it does not generate transformants that can inherit the transgene. Ironically, the lack of transgene integration has appeared to be a desirable feature for genome editing in plants. There are many recent reports on virus-mediated genome editing in Nicotiana benthamiana and a few other plants (Varanda et al., 2021; Zhang et al., 2022). There are no such reports on any legumes except for soybeans (Luo et al., 2021). However, although successful editing was demonstrated in this report, the edited plants were not generated. The major drawback observed in most of the previous reports utilizing the virus-mediated method, including the one on soybean, is that they deployed using the Agrobacterium-mediated method to generate Cas9-expression. This is because most viruses fail to cargo the Cas9 construct due to its large size for coding approximately 1,368 amino acids (aa). To tackle this situation, two solutions have been proposed in previous reviews (He et al., 2022; Zhang et al., 2022): one is to use nucleases with small coding regions such as CasΦ U (786 aa), Cas12f1 variants (400–600 aa), TnpB (400 aa), and IscBs (approximately 400 aa). The other is to use viruses that can carry longer constructs such as the Potato virus X, Barley yellow striate mosaic virus, and Sonchus yellow net virus (He et al., 2022; Zhang et al., 2022). Since legumes are hosts to a large number of viruses (Chatzivassiliou, 2021; Jha et al., 2023), their deployability, as carriers of genome-editing reagents, needs to be assessed. Previously, the pea early-browning virus of the pea plant was successfully used for heterologous genome editing via the CRISPR/Cas9 system in Nicotiana and Arabidopsis plants (Ali et al., 2018). Furthermore, the viral vectors previously used for gene silencing in legumes can be modified for carrying the constructs for genome editing. Some more lengths can be reduced by using a bidirectional promoter that would express both Cas9 and sgRNA (Ren et al., 2019).
Complementing the virus-based genome editing, we use mobile sgRNAs that can move to the apical meristem because these are augmented with sequences promoting cell-to-cell mobility (Ellison et al., 2020). Editing in the apical meristem is desirable because it gives rise to the floral meristem, thereby enhancing the chance of heritability of the edited loci. Furthermore, the genome editing efficiency of legumes is increased by enhancing the expression of the editing reagents by codon optimization and the use of efficient promoters, preferably from the same or related species (Baloglu et al., 2022). Many examples of these have been previously reviewed (Gu et al., 2021; Das et al., 2022; Verma et al., 2023a). When it comes to legumes, a higher genome editing efficiency of chickpea was achieved by using chickpea codon-optimized Cas9 and sgRNA driven by the M. truncatula U6.1 promoter (Gupta et al., 2023). Although most reports of plant genome editing use CRISPR derived from Prevotella and Francisella1 (Cpf1), Kim and Choi (2021) and Duan et al. (2021) demonstrated an efficient performance of CRISPR tools from other species, such as Acidaminococcus sp. and Lachnospiraceae bacterium in the legume crop, soybean.
Combining the previously reported usage of the innate visible marker such as phytoene desaturase (PDS) (Lu and Tian, 2022), herbicide resistance using acetohydroxy acid synthase (AHAS) (Wei et al., 2023), and viruses for genome editing, we proposed two “dual-editing” strategies (Figure 3) that can generate genome-edited legumes, bypassing the transformation step. Here, the viral genome will carry either one of the two constructs (Figures 3A, B). Apart from harboring the expression cassette of a desirable endonuclease (such as Cas9) and the sgRNA for a gene of interest (GOI), the viral genome will carry an additional sgRNA targeting either PDS (Figure 3A) or AHAS (Figure 3B), or genes with similar functions. Thus, the cells harboring the viral replicons can possibly be mutated for both loci. Although the infected T0 plants are chimeric, the flowers emerging from the double-mutant meristematic tissue can set double-mutant T1 seeds. Dual mutation would facilitate the visible selection of plants, thereby reducing the laborious screening through PCR. Thus, in the case where PDS is targeted, T1 will have phenotypes with reduced chlorophyll, which can be selected only by visual screening. Similarly, in the case where AHAS is targeted, the T1 phenotype can be selected after the elimination of unwanted individuals by herbicide application. As various plants have multiple homologs of PDS, the mutant phenotype may vary on a case-to-case basis. For example, two PDS homologs, known as LEAFY (LFY) and KORRIGAN1 (KOR1), are described in pea plants (Constantin et al., 2004). Although the former mutant had bleached leaves and distorted flowers, the inhibition of KOR1 expression significantly reduced shoot/root growth and did not affect flower development (Constantin et al., 2004). In such a situation, it would be desirable to choose the KOR1 homolog of PDS for the dual-editing strategy. Considering the high efficiency of viral-mediated genome editing (Gentzel et al., 2022; Zhang et al., 2022), not only will the possibility of inheritance of the mutant loci increase but there could also be chances of obtaining biallelic mutants for both the loci. The desirable homozygous single mutants for GOI can be segregated by selfing the T1 plants. Although this method can generate the desirable homozygous mutants mostly in the T2 generation, considering the annual life cycle of most legume crops, T2 plants can be obtained in a few months. Importantly, since most legume crops are not susceptible to transgene integration (via biolistic or Agrobacterium-mediated), this method could be effective as it does not involve Cas9 integration, as required in most previous reports of virus-mediated editing methods (Zhang et al., 2022). Furthermore, this type of transient expression of Cas9 is desirable because it reduces the off-target mutations due to the reduced time availability in the plant cells. Since the homologs of PDS, AHAS, or genes with similar functions are available in most plants (Hussain et al., 2021), this method of dual editing can have a broad-spectrum application. We believe that similar to this proposed visual screening, it may be possible to utilize other genes with mutants with evident phenotypes, such as the HYPERNODULATION ABERRANT ROOT FORMATION (Har1) (Wopereis et al., 2000), and the temperature-sensitive gene, such as brush mutant (Maekawa-Yoshikawa et al., 2009), of L. japonicus.
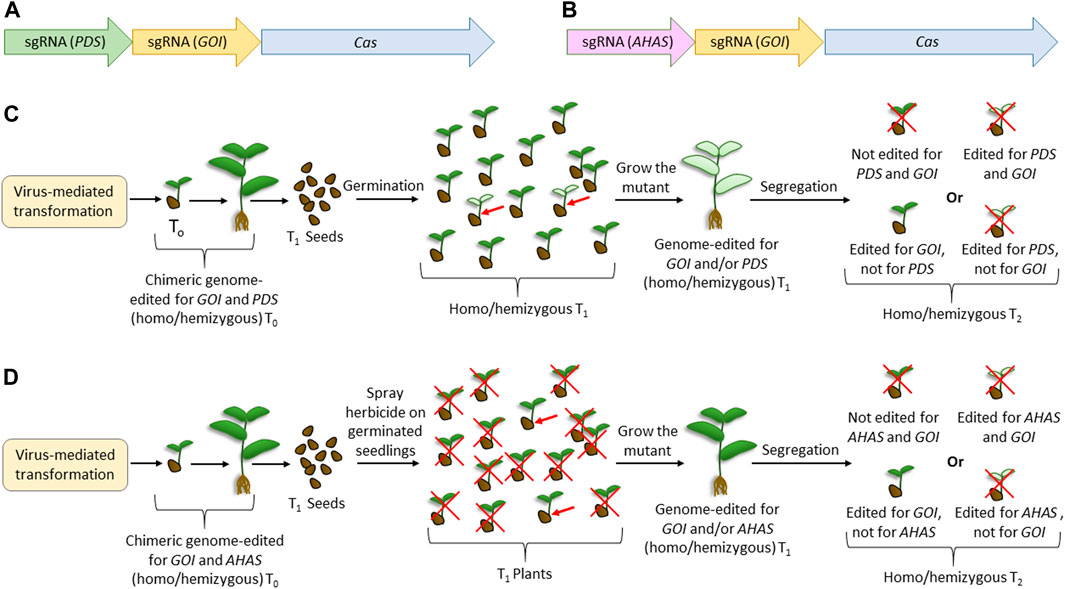
FIGURE 3. Proposed dual-editing models using the virus-mediated genome-editing method. (A) Editing cassette for expressing the sgRNAs of any gene of interest and phytoene desaturase, along with the construct for expressing Cas. (B) Editing cassette for expressing the sgRNAs of any gene of interest and acetohydroxy acid synthase, along with the construct for expressing Cas. (C) Steps for obtaining plants edited for GOI using the construct expressing sgRNA for PDS. (D) Steps for obtaining plants edited for GOI using the construct expressing sgRNA for AHAS. Cas is the gene encoding any efficient endonuclease desirable for genome editing. Red crosses indicate undesirable events. Red arrows indicate the expected genome-edited candidate in the experiments.
Previously, Lu et al. (2017) reported a genome editing strategy in rice using CYP81A6, which was analogous to our strategy of using AHAS. CYP81A6 encodes a cytochrome P450 protein, and its silencing renders the plants susceptible to the herbicide bentazon. Thus, Lu et al. (2017) developed a T-DNA-based genome editing vector harboring the Cas9 cassette, sgRNA for a desirable gene, and an RNA silencing construct for CYP81A6. Thus, the transgene-containing T1 plants were visually segregated after the application of bentazon. The two main differences between this strategy and the proposed strategy were that the former involves T-DNA integration and RNA silencing, whereas transient expression and genome editing were involved in the latter. Another similar strategy to the proposed dual-editing is the “dual gRNA” strategy, where one gRNA is aimed for the desirable editing of GOI, and the other is aimed for a large deletion of GOI so that this deletion can facilitate easy screening using PCR (Gao et al., 2016). Here, the homozygous mutants cannot be obtained in T1 since the same loci are differently edited in the same cell. In any case, both analogous methods are not recommended for legumes because they are based on transformation and involve transgene integration, which is not easily accepted by most legumes.
Since Cas9 protein and sgRNA are required for introducing desirable editing into the genome, the integration of the construct expressing these editing reagents is not a prerequisite. Woo et al. (2015) demonstrated the successful genome editing of A. thaliana, tobacco, lettuce, and rice by delivering the ribonucleoprotein complex comprising Cas9 protein and sgRNA (not the DNA expressing these) into the protoplast using the polyethylene glycol-mediated method. Similarly, Svitashev et al. (2016) and Zhang et al. (2016) edited the genomes of maize and wheat, respectively, by delivering the editing reagents into the protoplast using the biolistic method. Genome editing of the legume crop, soybean, using the ribonucleoprotein transfer via the protoplast culture was previously achieved by Kim and Choi (2021) and Seol et al. (2022). Recently, there have been recommendations for the use of nanoparticles, such as carbon nanotubes, carbon dots, magnetic nanoparticles, and mesoporous silicon nanoparticles, to deliver the ribonucleoprotein complex for editing, by traversing the cell wall (Naik et al., 2022; Verma et al., 2023b). Due to the lack of transgene integration, there is no antibiotic selection in this strategy involving the transport of the Cas9-containing ribonucleoprotein complex. Hence, the major drawback is the laborious PCR-based screening involved since the editing efficiency is quite low and the non-edited individuals outnumbered the edited individuals. We suggest that our “dual-editing” approach can ease the screening procedure. Instead of transporting sgRNAs through the virus (Figure 3), they can directly be introduced into the protoplasts. Since many legumes are amenable to protoplast culture and regeneration (Wiszniewska and Pindel, 2020), it is possible to attain genome editing by directly introducing the editing reagents into the protoplast.
10 Conclusion
Extensive standardization of transformation protocols has made soybean, alfalfa, and L. japonicus amenable to efficient genome editing (Baloglu et al., 2022; Bekalu et al., 2023). Similarly, standardization on other pulse crops should be encouraged. The in-depth research on plant–Agrobacterium interaction, regeneration, and development has aided in increasing the transformation in model plants like Arabidopsis and Nicotiana. It is possible that similar studies in transformation-recalcitrant legumes will aid in rectifying the post-transformation regeneration procedures. Using reporters like RUBY, instead of antibiotic markers, may reduce the adverse effect of selection pressure on regeneration. Methods bypassing transformation, such as the virus-mediated genome editing, could be more promising for legumes. Hence, legume viruses must be analyzed for their capacity to carry the cargo of genome editing reagents. Successful genome editing will aid in incorporating agronomically favorable traits in the legume crops, which serve as an alternative source of protein diet.
Author contributions
VN: writing and preparing tables. JS: concept design, writing, preparing figures, preparing tables, and corrections. All authors contributed to the article and approved the submitted version.
Acknowledgments
VN acknowledges DST INSPIRE for their fellowship.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aasim, M., Khawar, K. M., and Özcan, S. (2009). In vitro micropropagation from plumular apices of Turkish cowpea (Vigna unguiculata L.) cultivar Akkiz. Sci. Hortic. 122 (3), 468–471.
Adlinge, P. M., Samal, K. C., Kumara Swamy, R. V., and Rout, G. R. (2014). Rapid in vitro plant regeneration of black gram (Vigna mungo L. Hepper) var. sarala, an important legume crop. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 84, 823–827.
Affrifah, N. S., Uebersax, M. A., and Amin, S. (2023). Nutritional significance, value-added applications, and consumer perceptions of food legumes: A review, Legume Science, 2023, e192.
Aftabi, M., Teressa Negawo, A., and Hassan, F. (2018). Improved protocol for Agrobacterium-mediated transformation of pea (Pisum sativum). Mol. Biol. 7 (1), 1–6. doi:10.4172/2168-9547.1000202
Aksoy, E., Yildirim, K., Kavas, M., Kayihan, C., Yerlikaya, B. A., Çalik, I., et al. (2022). General guidelines for CRISPR/Cas-based genome editing in plants. Mol. Biol. Rep. 49, 12151–12164. doi:10.1007/s11033-022-07773-8
Ali, Z., Eid, A., Ali, A., and Mahfouz, M. M. (2018). Pea early-browning virus-mediated genome editing via the CRISPR/Cas9 system in Nicotiana benthamiana and Arabidopsis. Virus Res. 244, 333–337. doi:10.1016/j.virusres.2017.10.009
Altpeter, F., Springer, N. M., Bartley, L. E., Blechl, A. E., Brutnell, T. P., Citovsky, V., et al. (2016). Advancing crop transformation in the era of genome editing. Plant Cell. 28 (7), 1510–1520. doi:10.1105/tpc.16.00196
Amritha, P. P., and Shah, J. M. (2021). Can genetic engineering-based methods for gene function identification be eclipsed by genome editing in plants? A comparison of methodologies. Mol. Genet. Genomics 296 (3), 485–500. doi:10.1007/s00438-021-01769-y
Badhan, S., Ball, A. S., and Mantri, N. (2021). First report of CRISPR/Cas9 mediated DNA-free editing of 4CL and RVE7 genes in chickpea protoplasts. Int. J. Mol. Sci. 22 (1), 396. doi:10.3390/ijms22010396
Bakshi, S., Sadhukhan, A., Mishra, S., and Sahoo, L. (2011). Improved Agrobacterium-mediated transformation of cowpea via sonication and vacuum infiltration. Plant Cell. Rep. 30, 2281–2292. doi:10.1007/s00299-011-1133-8
Baloglu, M. C., Altunoglu, C. Y., Baloglu, P., Yildiz, A. B., Türkölmez, N., and Çiftçi, O. Y. (2022). Gene-editing technologies and applications in legumes: progress, evolution, and future prospects. Front. Genet. 13, 859437. doi:10.3389/fgene.2022.859437
Barik, D. P., Mohapatra, U., and Chand, P. L. (2005). Transgenic grasspea (lathyrus sativus L.): factors influencing agrobacterium-mediated transformation and regeneration. Plant Cell. Rep. 24, 523–531. doi:10.1007/s00299-005-0957-5
Bartlett, J. G., Alves, S. C., Smedley, M., Snape, J. W., and Harwood, W. A. (2008). High-throughput Agrobacterium-mediated barley transformation. Plant Methods 4 (1), 22–12. doi:10.1186/1746-4811-4-22
Bekalu, Z. E., Panting, M., Bæksted Holme, I., and Brinch-Pedersen, H. (2023). Opportunities and challenges of in vitro tissue culture systems in the era of crop genome editing. Int. J. Mol. Sci. 24, 11920. doi:10.3390/ijms241511920
Bhomkar, P., Upadhyay, C. P., Saxena, M., Muthusamy, A., Shiva Prakash, N., Pooggin, M., et al. (2008). Salt stress alleviation in transgenic Vigna mungo L. Hepper (blackgram) by overexpression of the glyoxalase I gene using a novel Cestrum yellow leaf curling virus (CmYLCV) promoter. Mol. Breed. 22, 183–184. doi:10.1007/s11032-008-9181-7
Bhowmik, P., Konkin, D., Polowick, P., Hodgins, C. L., Subedi, M., Xiang, D., et al. (2021). CRISPR/Cas9 gene editing in legume crops: opportunities and challenges. Legume Sci. 3, e96. doi:10.1002/leg3.96
Biswas, S., Wahl, N. J., Thomson, M. J., Cason, J. M., McCutchen, B. F., and Septiningsih, E. M. (2022). Optimization of protoplast isolation and transformation for a pilot study of genome editing in peanut by targeting the allergen gene Arah2. Int. J. Mol. Sci. 23 (2), 837. doi:10.3390/ijms23020837
Bridgeland, A., Biswas, S., Tsakirpaloglou, N., Thomson, M. J., and Septiningsih, E. M. (2023). Optimization of gene editing in cowpea through protoplast transformation and agroinfiltration by targeting the phytoene desaturase gene. Plos One 18 (4), e0283837. doi:10.1371/journal.pone.0283837
Cai, Y., Chen, L., Liu, X., Sun, S., Wu, C., Jiang, B., et al. (2015). CRISPR/Cas9-mediated genome editing in soybean hairy roots. PLoS One 10 (8), e0136064. doi:10.1371/journal.pone.0136064
Carrijo, J., Illa-Berenguer, E., LaFayette, P., Torres, N., Aragão, F. J., Parrott, W., et al. (2021). Two efficient CRISPR/Cas9 systems for gene editing in soybean. Transgenic Res. 30 (3), 239–249. doi:10.1007/s11248-021-00246-x
Celikkol Akcay, U., Mahmoudian, M., Kamci, H., Yucel, M., and Oktem, H. A. (2009). Agrobacterium tumefaciens-mediated genetic transformation of a recalcitrant grain legume, lentil (Lens culinaris Medik). Plant Cell. Rep. 28, 407–417. doi:10.1007/s00299-008-0652-4
Chabaud, M., de Carvalho-Niebel, F., and Barker, D. G. (2003). Efficient transformation of Medicago truncatula cv. Jemalong using the hypervirulent Agrobacterium tumefaciens strain AGL1. Plant Cell. Rep. 22, 46–51. doi:10.1007/s00299-003-0649-y
Chandra, A., and Pental, D. (2003). Regeneration and genetic transformation of grain legumes: an overview. Curr. Sci. 84 (3), 381–387. https://www.currentscience.ac.in/Volumes/84/03/0381.pdf.
Chatzivassiliou, E. K. (2021). An annotated list of legume-infecting viruses in the light of metagenomics. Plants (Basel) 10 (7), 1413. doi:10.3390/plants10071413
Chaudhury, D., Madanpotra, S., Jaiwal, R., Saini, R., Kumar, P. A., and Jaiwal, P. K. (2007). Agrobacterium tumefaciens-mediated high frequency genetic transformation of an Indian cowpea (Vigna unguiculata L. Walp.) cultivar and transmission of transgenes into progeny. Plant Sci. 172 (4), 692–700. doi:10.1016/j.plantsci.2006.11.009
Che, P., Chang, S., Simon, M. K., Zhang, Z., Shaharyar, A., Ourada, J., et al. (2021). Developing a rapid and highly efficient cowpea regeneration, transformation and genome editing system using embryonic axis explants. Plant J. 106 (3), 817–830. doi:10.1111/tpj.15202
Chen, Z., Debernardi, J. M., Dubcovsky, J., and Gallavotti, A. (2022). The combination of morphogenic regulators BABY BOOM and GRF-GIF improves maize transformation efficiency. bioRxiv. doi:10.1101/2022.09.02.506370
Cho, H. J., Moy, Y., Rudnick, N. A., Klein, T. M., Yin, J., Bolar, J., et al. (2022). Development of an efficient marker-free soybean transformation method using the novel bacterium Ochrobactrum haywardense H1. Plant Biotechnol. J. 20 (5), 977–990. doi:10.1111/pbi.13777
Cho, M. J., Wu, E., Kwan, J., Yu, M., Banh, J., Linn, W., et al. (2014). Agrobacterium-mediated high-frequency transformation of an elite commercial maize (Zea mays L.) inbred line. Plant Cell. Rep. 33, 1767–1777. doi:10.1007/s00299-014-1656-x
Constantin, G. D., Krath, B. N., MacFarlane, S. A., Nicolaisen, M., Elisabeth Johansen, I., and Lund, O. S. (2004). Virus-induced gene silencing as a tool for functional genomics in a legume species. Plant J. 40, 622–631. doi:10.1111/j.1365-313X.2004.02233.x
Crane, C., Wright, E., Dixon, R. A., and Wang, Z-Y. (2006). Transgenic Medicago truncatula plants obtained from Agrobacterium tumefaciens-transformed roots and Agrobacterium rhizogenes-transformed hairy roots. Planta 223, 1344–1354. doi:10.1007/s00425-006-0268-2
Das, D., Singha, D. L., Paswan, R. R., Chowdhury, N., Sharma, M., Sudhakar Reddy, P., et al. (2022). Recent advancements in CRISPR/Cas technology for accelerated crop improvement. Planta 255, 109. doi:10.1007/s00425-022-03894-3
de Oliveira, M. L. P., Febres, V. J., Costa, M. G. C., Moore, G. A., and Otoni, W. C. (2009). High-efficiency Agrobacterium-mediated transformation of citrus via sonication and vacuum infiltration. Plant Cell. Rep. 28, 387–395. doi:10.1007/s00299-008-0646-2
Debernardi, J. M., Tricoli, D. M., Ercoli, M. F., Hayta, S., Ronald, P., Palatnik, J. F., et al. (2020). A GRF–GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 38 (11), 1274–1279. doi:10.1038/s41587-020-0703-0
Devi, P., Radha, P., Sitamahalakshmi, L., Syamala, D., and Kumar, S. M. (2004). Plant regeneration via somatic embryogenesis in mung bean [Vigna radiata (L.) Wilczek]. Sci. Hortic. 99 (1), 1–8.
Du, H., Zeng, X., Zhao, M., Cui, X., Wang, Q., Yang, H., et al. (2016). Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J. Biotechnol. 217 (10), 90–97. doi:10.1016/j.jbiotec.2015.11.005
Duan, K., Cheng, Y., Ji, J., Wang, C., Wei, Y., and Wang, Y. (2021). Large chromosomal segment deletions by CRISPR/LbCpf1-mediated multiplex gene editing in soybean. J. Integr. Plant Biol. 63 (9), 1620–1631. doi:10.1111/jipb.13158
Egnin, M., Mora, A., and Prakash, C. S. (1998). Factors enhancing Agrobacterium tumefaciens-mediated gene transfer in peanut (Arachis hypogaea L.). Vitro Cell. Dev. Biol. - Plant 34, 310–318. doi:10.1007/BF02822740
Ellison, E. E., Nagalakshmi, U., Gamo, E. M., Huang, P. j., Dinesh-Kumar, S., and Voytas, D. F. (2020). Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nat. Plants 6, 620–624. doi:10.1038/s41477-020-0670-y
Gaj, M. D. (2004). Factors influencing somatic embryogenesis induction and plant regeneration with particular reference to Arabidopsis thaliana (L.) heynh. Heynh. Plant Growth Regul. 43, 27–47. doi:10.1023/b:grow.0000038275.29262.fb
Gao, X., Chen, J., Dai, X., Zhang, D., and Zhao, Y. (2016). An effective strategy for reliably isolating heritable and Cas9-free Arabidopsis mutants generated by CRISPR/Cas9-mediated genome editing. Plant Physiol. 171, 1794–1800. doi:10.1104/pp.16.00663
Garagounis, C., Georgopoulou, M. E., Beritza, K., and Papadopoulou, K. K. (2020). An Agrobacterium rhizogenes mediated hairy root transformation protocol for fenugreek. MethodsX 7, 101098. doi:10.1016/j.mex.2020.101098
Geetha, N., Venkatachalam, P., and Sita, G. L. (1999). Agrobacterium-mediated genetic transformation of pigeonpea (Cajanus cajan L.) and development of transgenic plants via direct organoenesis. Plant Biotechnol. 16 (3), 213–218. doi:10.5511/plantbiotechnology.16.213
Gentzel, I. N., Ohlson, E. W., Redinbaugh, M. G., and Wang, G. L. (2022). Vige: virus-induced genome editing for improving abiotic and biotic stress traits in plants. Stress Biol. 2, 2. doi:10.1007/s44154-021-00026-x
Gorji, A. H., Zolnori, Z., Jamasbi, A., and Khodadadzadeh, Y. (2014). Transformation fava beans by Agrobacterium using Chitinase, Glucanase and CryIA (b) genes. Int. J. Adv. Sci. Eng. Inf. Technol. 4 (2), 50–17. doi:10.18517/ijaseit.4.2.366
Gu, X., Liu, L., and Zhang, H. (2021). Transgene-free genome editing in plants. Front. Genome Ed. 3, 805317. doi:10.3389/fgeed.2021.805317
Gupta, S. K., Vishwakarma, N. K., Malakar, P., Vanspati, P., Sharma, N. K., and Chattopadhyay, D. (2023). Development of an Agrobacterium-delivered codon-optimized CRISPR/Cas9 system for chickpea genome editing. Protoplasma 260, 1437–1451. doi:10.1007/s00709-023-01856-4
Han, K. H., Keathley, D. E., Davis, J. M., and Gordon, M. P. (1993). Regeneration of a transgenic woody legume (Robinia pseudoacacia L., black locust) and morphological alterations induced by Agrobacterium rhizogenes-mediated transformation. Plant Sci. 88 (2), 149–157. doi:10.1016/0168-9452(93)90086-f
Hanafy, M., Pickardt, T., Kiesecker, H., and Jacobsen, H-J. (2005). Agrobacterium-mediated transformation of faba bean (Vicia faba L.) using embryo axes. Euphytica 142, 227–236. doi:10.1007/s10681-005-1690-4
Hanafy, M. S., El-Banna, A., Schumacher, H. M., Jacobsen, H.-J., and Hassan, F. S. (2013). Enhanced tolerance to drought and salt stresses in transgenic faba bean (Vicia faba L.) plants by heterologous expression of the PR10a gene from potato. Plant Cell. Rep. 32, 663–674. doi:10.1007/s00299-013-1401-x
He, Y., Mudgett, M., and Zhao, L. (2022). Advances in gene editing without residual transgenes in plants. Plant Physiol. 188, 1757–1768. doi:10.1093/plphys/kiab574
He, Y., Zhang, T., Sun, H., Zhan, H., and Zhao, Y. (2020). A reporter for noninvasively monitoring gene expression and plant transformation. Hortic. Res. 7, 152. doi:10.1038/s41438-020-00390-1
Hsieh, Y. F., Jain, M., Wang, J., and Gallo, M. (2017). Direct organogenesis from cotyledonary node explants suitable for Agrobacterium-mediated transformation in peanut (Arachis hypogaea L.). Plant Cell., Tissue Organ Cult. 128, 161–175. doi:10.1007/s11240-016-1095-1
Hussain, A., Ding, X., Alariqi, M., Manghwar, H., Hui, F., Li, Y., et al. (2021). Herbicide resistance: another hot agronomic trait for plant genome editing. Plants 10, 621. doi:10.3390/plants10040621
Ignacimuthu, S. (2000). Agrobacterium mediated transformation of Vigna sesquipedalis Koern (asparagus bean).
Ismael, K. A., and Antar, E.-B. N. (2014). Establishment of high-efficiency Agrobacterium-mediated transformation conditions of soybean callus. Indian J. Biotechnol. 13 (4), 459–463. http://nopr.niscpr.res.in/handle/123456789/30472.
Indurker, S., Misra, H. S., and Eapen, S. (2010). Agrobacterium-mediated transformation in chickpea (cicer arietinum L.) with an insecticidal protein gene: optimisation of different factors. Physiology Mol. Biol. Plants 16, 273–284. doi:10.1007/s12298-010-0030-x
Jaganathan, D., Ramasamy, K., Sellamuthu, G., Jayabalan, S., and Venkataraman, G. (2018). CRISPR for crop improvement: an update review. Front. Plant Sci. 9, 985. doi:10.3389/fpls.2018.00985
Jain, D., Jones, L., and Roy, S. (2023). Gene editing to improve legume-rhizobia symbiosis in a changing climate. Curr. Opin. Plant Biol. 71, 102324. doi:10.1016/j.pbi.2022.102324
Jaiwal, P. K., Kumari, R., Ignacimuthu, S., Potrykus, I., and Sautter, C. (2001). Agrobacterium tumefaciens-mediated genetic transformation of mungbean (vigna radiata L. Wilczek)—a recalcitrant grain legume. Plant Sci. 161 (2), 239–247. doi:10.1016/s0168-9452(01)00352-1
Jaudal, M., Mayo-Smith, M., Poulet, A., Whibley, A., Peng, Y., Zhang, L., et al. (2022). MtING2 encodes an ING domain PHD finger protein which affects Medicago growth, flowering, global patterns of H3K4me3, and gene expression. Plant J. 112 (4), 1029–1050. doi:10.1111/tpj.15994
Jha, U. C., Nayyar, H., Chattopadhyay, A., Beena, R., Lone, A. A., Naik, Y. D., et al. (2023). Major viral diseases in grain legumes: designing disease resistant legumes from plant breeding and OMICS integration. Front. Plant Sci. 14, 1183505. doi:10.3389/fpls.2023.1183505
Ji, J., Zhang, C., Sun, Z., Wang, L., Duanmu, D., and Fan, Q. (2019). Genome editing in cowpea Vigna unguiculata using CRISPR-Cas9. Int. J. Mol. Sci. 20 (10), 2471. doi:10.3390/ijms20102471
Jia, Y., Yao, X., Zhao, M., Zhao, Q., Du, Y., Yu, C., et al. (2015). Comparison of soybean transformation efficiency and plant factors affecting transformation during the Agrobacterium infection process. Int. J. Mol. Sci. 16 (8), 18522–18543. doi:10.3390/ijms160818522
Jian, B., Hou, W., Wu, C., Liu, B., Liu, W., Song, S., et al. (2009). Agrobacterium rhizogenes-mediated transformation of superroot-derived Lotus corniculatus plants: A valuable tool for functional genomics. BMC Plant Biol. 9, 78. doi:10.1186/1471-2229-9-78
Jiang, Q., Fu, C., and Wang, Z. Y. (2019). A unified Agrobacterium-mediated transformation protocol for alfalfa (Medicago sativa L.) and Medicago truncatula. Transgenic Plants Methods Protoc. 1864, 153–163. doi:10.1007/978-1-4939-8778-8_11
Juranić, M., Nagahatenna, D. S., Salinas-Gamboa, R., Hand, M. L., Sánchez-León, N., Leong, W. H., et al. (2020). A detached leaf assay for testing transient gene expression and gene editing in cowpea (Vigna unguiculata [L.] Walp.). Plant Methods 16 (1), 88–17. doi:10.1186/s13007-020-00630-4
Kapildev, G., Chinnathambi, A., Sivanandhan, G., Rajesh, M., Vasudevan, V., Mayavan, S., et al. (2016). High-efficient Agrobacterium-mediated in planta transformation in black gram (Vigna mungo (L.) Hepper). Acta Physiol. Plant. 38, 205–213. doi:10.1007/s11738-016-2215-6
Karmakar, S., Molla, K. A., Gayen, D., Karmakar, A., Das, K., Sarkar, S. N., et al. (2019). Development of a rapid and highly efficient Agrobacterium-mediated transformation system for pigeon pea [Cajanus cajan (L.) Millsp]. GM Crops Food 10, 115–138. doi:10.1080/21645698.2019.1625653
Karthik, S., Pavan, G., and Manickavasagam, M. (2020). Nitric oxide donor regulates Agrobacterium-mediated genetic transformation efficiency in soybean [Glycine max (L.) Merrill]. Plant Cell., Tissue Organ Cult. 141, 655–660. doi:10.1007/s11240-020-01808-3
Karthik, S., Pavan, G., Sathish, S., Siva, R., Kumar, P. S., and Manickavasagam, M. (2018). Genotype-independent and enhanced in planta Agrobacterium tumefaciens-mediated genetic transformation of peanut [Arachis hypogaea (L.)]. 3 Biotech. 8, 202. doi:10.1007/s13205-018-1231-1
Kim, H., and Choi, J. (2021). A robust and practical CRISPR/crRNA screening system for soybean cultivar editing using LbCpf1 ribonucleoproteins. Plant Cell. Rep. 40, 1059–1070. doi:10.1007/s00299-020-02597-x
Kimura, M., Cutler, S., and Isobe, S. (2015). A novel phenolic compound, chloroxynil, improves Agrobacterium-mediated transient transformation in Lotus japonicus. PLoS ONE 10 (7), e0131626. doi:10.1371/journal.pone.0131626
Kingsley, O., Lili, Y., Bo-hong, S., Ming-ming, Z., Zhang-Xiong, L., Hua-wei, G., et al. (2022). “Genetic improvement of minor crop legumes: prospects of de novo domestication,” in In legumes research - volume 1. Editors J. C. Jimenez-Lopez., and A. Clemente (UK: IntechOpen).
Kor, S. D., Chowdhury, N., Keot, A. K., Yogendra, K., Chikkaputtaiah, C., and Sudhakar Reddy, P. (2023). RNA pol III promoters—Key players in precisely targeted plant genome editing. Front. Genet. 13, 989199. doi:10.3389/fgene.2022.989199
Krishna, G., Reddy, P. S., Ramteke, P. W., Rambabu, P., Tawar, K. B., and Bhattacharya, P. (2011). Agrobacterium-mediated genetic transformation of pigeon pea [Cajanus cajan (L.) Millsp.] for resistance to legume pod borer Helicoverpa armigera. J. Crop Sci. Biotechnol. 14, 197–204. doi:10.1007/s12892-010-0063-2
Krishna, H., Alizadeh, M., Singh, D., Singh, U., Chauhan, N., Eftekhari, M., et al. (2016). Somaclonal variations and their applications in horticultural crops improvement. 3 Biotech. 6, 54. doi:10.1007/s13205-016-0389-7
Krishnamurthy, K. V., Suhasini, K., Sagare, A. P., Meixner, M., De Kathen, A., Pickardt, T., et al. (2000). Agrobacterium mediated transformation of chickpea (Cicer arietinum L.) embryo axes. Plant Cell. Rep. 19, 235–240. doi:10.1007/s002990050005
Kumar, A., Jaiwal, R., Sreevathsa, R., Chaudhary, and D., and Jaiwal, P. K. (2021a). Transgenic cowpea plants expressing Bacillus thuringiensis Cry2Aa insecticidal protein imparts resistance to Maruca vitrata legume pod borer. Plant Cell. Rep. 40 (3), 583–594. doi:10.1007/s00299-020-02657-2
Kumar, A., Sainger, M., Jaiwal, R., Chaudhary, D., and Jaiwal, P. K. (2021b). Tissue culture- and selection-independent Agrobacterium tumefaciens-mediated transformation of a recalcitrant grain legume, Cowpea (Vigna unguiculata L. Walp). Mol. Biotechnol. 63 (8), 710–718. doi:10.1007/s12033-021-00333-8
Liu, R. M., Liang, L. L., Freed, E., Chang, H., Oh, E., Liu, Z. Y., et al. (2019). Synthetic chimeric nucleases function for efficient genome editing. Nat. Commun. 10, 5524. doi:10.1038/s41467-019-13500-y
Li, G., Liu, R., Xu, R., Varshney, R. K., Ding, H., Li, M., et al. (2023). Photonic elementary cellular automata for simulation of complex phenomena. Crop J. 11 (1), 132–139. doi:10.1038/s41377-023-01180-9
Li, H. L., Fang, P. P., Hu, Y. N., Li, X. F., Xia, W. J., and Xu, P. (2022). An Agrobacterium rhizogenes strain R1000-mediated efficient hairy root transformation protocol for common bean. Legume Research-An Int. J. 45 (10), 1247–1251. doi:10.18805/lrf-680
Li, S., Cong, Y., Liu, Y., Wang, T., Shuai, Q., Chen, N., et al. (2017). Optimization of Agrobacterium-mediated transformation in soybean. Front. plant Sci. 8, 246. doi:10.3389/fpls.2017.00246
Li, Z., Liu, Z. B., Xing, A., Moon, B. P., Koellhoffer, J. P., Huang, L., et al. (2015). Cas9-guide RNA directed genome editing in soybean. Plant Physiol. 169 (2), 960–970. doi:10.1104/pp.15.00783
Lowe, K., Wu, E., Wang, N., Hoerster, G., Hastings, C., Cho, M. J., et al. (2016). Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell. 28 (9), 1998–2015. doi:10.1105/tpc.16.00124
Lu, H.-P., Liu, S.-M., Xu, S.-L., Chen, W.-Y., Zhou, X., Tan, Y.-Y., et al. (2017). CRISPR-S: an active interference element for a rapid and inexpensive selection of genome-edited, transgene-free rice plants. Plant Biotechnol. J. 15, 1371–1373. doi:10.1111/pbi.12788
Lu, Q. S. M., and Tian, L. (2022). An efficient and specific CRISPR-Cas9 genome editing system targeting soybean phytoene desaturase genes. BMC Biotechnol. 22 (1), 7. doi:10.1186/s12896-022-00737-7
Luo, W., Tan, J., Li, T., Feng, Z., Ding, Z., Xie, X., et al. (2023). Overexpression of maize GOLDEN2 in rice and maize calli improves regeneration by activating chloroplast development. Sci. China Life Sci. 66, 340–349. doi:10.1007/s11427-022-2149-2
Luo, Y., Na, R., Nowak, J. S., Qiu, Y., Lu, Q. S., Yang, C., et al. (2021). Development of a Csy4-processed guide RNA delivery system with soybean-infecting virus ALSV for genome editing. BMC Plant Biol. 21, 419. doi:10.1186/s12870-021-03138-8
Lurquin, P. F., Cai, Z., Stiff, C. M., and Fuerst, E. P. (1998). Half-embryo cocultivation technique for estimating the susceptibility of pea (Pisum sativum L.) and lentil (Lens culinaris Medik.) cultivars to Agrobacterium tumefaciens. Mol. Biotechnol. 9, 175–179. doi:10.1007/BF02760819
Maekawa-Yoshikawa, M., Müller, J., Takeda, N., Maekawa, T., Sato, S., Tabata, S., et al. (2009). The temperature-sensitive brush mutant of the legume Lotus japonicus reveals a link between root development and nodule infection by rhizobia. Plant Physiol. 149 (4), 1785–1796. doi:10.1104/pp.108.135160
Mao, Y., Botella, J. R., Liu, Y., and Zhu, J. K. (2019). Gene editing in plants: progress and challenges. Natl. Sci. Rev. 6 (3), 421–437. doi:10.1093/nsr/nwz005
Margier, M., Georgé, S., Hafnaoui, N., Remond, D., Nowicki, M., Du Chaffaut, L., et al. (2018). Nutritional composition and bioactive content of legumes: characterization of pulses frequently consumed in France and effect of the cooking method. Nutrients 10 (11), 1668. doi:10.3390/nu10111668
Mekala, G. K., Juturu, V. N., Mallikarjuna, G., Kirti, P. B., and Yadav, S. K. (2016). Optimization of Agrobacterium-mediated genetic transformation of shoot tip explants of green gram (Vigna radiata (L.) Wilczek). Plant Cell., Tissue Organ Cult. 127, 651–663. doi:10.1007/s11240-016-1085-3
Meng, Y., Hou, Y., Wang, H., Ji, R., Liu, B., and Wen, J. (2017). Targeted mutagenesis by CRISPR/Cas9 system in the model legume Medicago truncatula. Plant Cell Rep. 36 (2), 371–374.
Muruganantham, M., Amutha, S., Selvaraj, N., Vengadesan, G., and Ganapathi, A. (2007). Efficient Agrobacterium-mediated transformation of Vigna mungo using immature cotyledonary-node explants and phosphinothricin as the selection agent. Vitro Cell. Dev. Biology-Plant 43, 550–557. doi:10.1007/s11627-007-9060-7
Naik, B. J., Shimoga, G., Kim, S.-C., Manjulatha, M., Subramanyam Reddy, C., Palem, R. R., et al. (2022). CRISPR/Cas9 and nanotechnology pertinence in agricultural crop refinement. Front. Plant Sci. 13, 843575. doi:10.3389/fpls.2022.843575
Nanasato, Y., Konagaya, K. I., Okuzaki, A., Tsuda, M., and Tabei, Y. (2013). Improvement of Agrobacterium-mediated transformation of cucumber (Cucumis sativus L.) by combination of vacuum infiltration and co-cultivation on filter paper wicks. Plant Biotechnol. Rep. 7, 267–276. doi:10.1007/s11816-012-0260-1
Neelakandan, A. K., Wright, D. A., Traore, S. M., Ma, X., Subedi, B., Veeramasu, S., et al. (2022). Application of CRISPR/Cas9 system for efficient gene editing in peanut. Plants 11 (10), 1361. doi:10.3390/plants11101361
Nifantova, S. N., Komarnickiy, I. K., and Kuchuk, N. V. (2011). Obtaining of transgenic French bean plants (Phaseolus vulgaris L.) resistant to the herbicide pursuit by Agrobacterium-mediated transformation. Cytol. Genet. 45, 97–100. doi:10.3103/s0095452711020113
Pareddy, D., Chennareddy, S., Anthony, G., Sardesai, N., Mall, T., Minnicks, T., et al. (2020). Improved soybean transformation for efficient and high throughput transgenic production. Transgenic Res. 29, 267–281. doi:10.1007/s11248-020-00198-8
Pathak, M. R., and Hamzah, R. Y. (2008). An effective method of sonication-assisted Agrobacterium-mediated transformation of chickpeas. Plant Cell., Tissue Organ Cult. 93, 65–71. doi:10.1007/s11240-008-9344-6
Paz, M. M., Martinez, J. C., Kalvig, A. B., Fonger, T. M., and Wang, K. (2006). Improved cotyledonary node method using an alternative explant derived from mature seed for efficient Agrobacterium-mediated soybean transformation. Plant Cell. Rep. 25, 206–213. doi:10.1007/s00299-005-0048-7
Peyret, H., and Lomonossoff, G. P. (2015). When plant virology met Agrobacterium: the rise of the deconstructed clones. Plant Biotechnol. J. 13 (8), 1121–1135. doi:10.1111/pbi.12412
Polowick, P. L., Baliski, D. S., and Mahon, J. D. (2004). Agrobacterium tumefaciens-mediated transformation of chickpea (cicer arietinum L.): gene integration, expression and inheritance. Plant Cell. Rep. 23, 485–491. doi:10.1007/s00299-004-0857-0
Popelka, J. C., Gollasch, S., Moore, A., Molvig, L., and Higgins, T. J. (2006). Genetic transformation of cowpea (Vigna unguiculata L.) and stable transmission of the transgenes to progeny. Plant Cell. Rep. 25, 304–312. doi:10.1007/s00299-005-0053-x
Pratap, A., Prajapati, U., Singh, C. M., Gupta, S., Rathore, M., Malviya, N., et al. (2018). Potential, constraints and applications of in vitro methods in improving grain legumes. Plant Breed. 137, 235–249. doi:10.1111/pbr.12590
Radhakrishnan, R., Ramachandran, A., and Ranjitha Kumari, B. D. (2009). Rooting and shooting: dual function of thidiazuron in in vitro regeneration of soybean (Glycine max. L). Acta Physiol. Plant 31, 1213–1217.
Raji, A. A., Oriero, E., Odeseye, B., Odunlami, T., and Ingelbrecht, I. L. (2008). Plant regeneration and Agrobacterium-mediated transformation of African cowpea [Vigna unguiculata (L.) Walp] genotypes using embryonic axis explants. J. Food, Agric. Environ. 6 (3), 350–356. https://hdl.handle.net/10568/90876.
Raman, K. V., Aggarwal, D., Rao, S. R., Sreevathsa, R., Singh, A. K., Abdin, M. Z., et al. (2018). Rapid and efficient Agrobacterium mediated transformation of early scutellum derived calli of Indica rice. Indian J. Exp. Biol. 56, 20–28. http://nopr.niscpr.res.in/handle/123456789/43283.
Ramírez Rivera, N. G., García-Salinas, C., Aragão, F. J. L., and Díaz de la Garza, R. I. (2016). Metabolic engineering of folate and its precursors in Mexican common bean (Phaseolus vulgaris L.). Plant Biotechnol J. 14, 2021–2032. doi:10.1111/pbi.12561
Ran, F., Hsu, P., Wright, J., Agarwala, V., Scott, D. A., and Zhang, F. (2022). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308. doi:10.1038/nprot.2013.143
Rasheed, A., Barqawi, A. A., Mahmood, A., Nawaz, M., Shah, A. N., Bay, D. H., et al. (2022). CRISPR/Cas9 is a powerful tool for precise genome editing of legume crops: A review. Mol. Biol. Rep. 49, 5595–5609. doi:10.1007/s11033-022-07529-4
Rathore, D. S., and Mullins, E. (2018). Alternative Non-Agrobacterium based methods for plant transformation. Annu. Plant Rev. online, 891–908. doi:10.1002/9781119312994.apr0659
Raveendar, S., and Ignacimuthu, S. (2010). Improved Agrobacterium mediated transformation in cowpea vigna unguiculata L. Walp. Asian J. Plant Sci. 9 (5), 256–263. doi:10.3923/ajps.2010.256.263
Ren, Q., Zhong, Z., Wang, Y., You, Q., Li, Q., Yuan, M., et al. (2019). Bidirectional promoter-based CRISPR-Cas9 systems for plant genome editing. Front. Plant Sci. 10, 1173. doi:10.3389/fpls.2019.01173
Sağlam Yılmaz, S., Khawar, K. M., and Çiftçi, C. Y. (2022). Genetic transformation of common beans (Phaseolus vulgaris L.) through Agrobacterium tumefaciens carrying Cry1Ab gene. Mol. Biol. Rep. 49, 7195–7203. doi:10.1007/s11033-022-07637-1
Sainger, M., Chaudhary, D., Dahiya, S., Jaiwal, R., and Jaiwal, P. K. (2015). Development of an efficient in vitro plant regeneration system amenable to Agrobacterium-mediated transformation of a recalcitrant grain legume blackgram (Vigna mungo L. Hepper). Physiology Mol. Biol. Plants 21, 505–517. doi:10.1007/s12298-015-0315-1
Saini, R., and Jaiwal, P. K. (2007). Agrobacterium tumefaciens-mediated transformation of blackgram: an assessment of factors influencing the efficiency of uidA gene transfer. Biol. Plant. 51, 69–74. doi:10.1007/s10535-007-0014-z
Saini, R., and Jaiwal, P. K. (2005). Transformation of a recalcitrant grain legume, Vigna mungo L. Hepper, using Agrobacterium tumefaciens-mediated gene transfer to shoot apical meristem cultures. Plant Cell. Rep. 24, 164–171. doi:10.1007/s00299-005-0934-z
Saini, R., Jaiwal, S., and Jaiwal, P. K. (2003). Stable genetic transformation of Vigna mungo L. Hepper via Agrobacterium tumefaciens. Plant Cell. Rep. 21, 851–859. doi:10.1007/s00299-003-0574-0
Seol, M. A., Cho, S., and Jung, J. Y. (2022). Efficiency verification of CRISPR–Cas9-mediated mutagenesis of target gene sgRNA using soybean protoplasts. Plant Biotechnol. Rep. 16, 599–611.
Shah, J. M., and Veluthambi, K. (2010). DIANTHIN, a negative selection marker in tobacco, is non-toxic in transgenic rice and confers sheath blight resistance. Biol. Plant. 54, 443–450. doi:10.1007/s10535-010-0080-5
Sharma, H. C., Manuele, T., Bouhssini, M. E., and Ranga Rao, G. V. (2016). “Pest management in grain legumes: potential and limitations,” in Integrated pest management in the tropics. Editor D. P. Abrol (India: New India Publishing Agency), 275–292.
Sharma, K. K., Lavanya, M., and Anjaiah, V. (2006). Agrobacterium-mediated production of transgenic pigeonpea (Cajanus cajanL. Millsp.) expressing the synthetic Bt cry1Ab gene. Vitro Cell. Dev. Biology-Plant 42, 165–173. doi:10.1079/ivp2005730
Sharma, M. K., Solanke, A. U., Jani, D., Singh, Y., and Sharma, A. K. (2009). A simple and efficient Agrobacterium-mediated procedure for transformation of tomato. J. Biosci. 34 (3), 423–433. doi:10.1007/s12038-009-0049-8
Shu, H., Luo, Z., Peng, Z., and Wang, J. (2020). The application of CRISPR/Cas9 in hairy roots to explore the functions of AhNFR1 and AhNFR5 genes during peanut nodulation. BMC plant Biol. 20 (1), 417–515. doi:10.1186/s12870-020-02614-x
Shunmugam, A. S., Kannan, U., Jiang, Y., Daba, K. A., and Gorim, L. Y. (2018). Physiology based approaches for breeding of next-generation food legumes. Plants 7 (3), 72. doi:10.3390/plants7030072
Singh, C., Kumar, R., Sehgal, H., Bhati, S., Singhal, T., Nimmy, M. S., et al. (2023). Unclasping potentials of genomics and gene editing in chickpea to fight climate change and global hunger threat. Front. Genet. 14, 1085024. doi:10.3389/fgene.2023.1085024
Singh, N., Jain, P., Ujinwal, M., and Langyan, S. (2022). Escalate protein plates from legumes for sustainable human nutrition. Front. Nutr. 9, 977986. doi:10.3389/fnut.2022.977986
Singsit, C., Adang, M. J., Lynch, R. E., Anderson, W. F., Wang, A., Cardineau, G., et al. (1997). Expression of a Bacillus thuringiensis cryIA(c) gene in transgenic peanut plants and its efficacy against lesser cornstalk borer. Transgenic Res. 6, 169–176. doi:10.1023/a:1018481805928
Son, S., and Park, S. R. (2022). Challenges facing CRISPR/Cas9-based genome editing in plants. Front. Plant Sci. 13, 902413. doi:10.3389/fpls.2022.902413
Song, G-q., Han, X., Wiersma, A. T., Zong, X., Awale, H. E., and Kelly, J. D. (2020). Induction of competent cells for Agrobacterium tumefaciens-mediated stable transformation of common bean (Phaseolus vulgaris L.). PLoS ONE 15 (3), e0229909. doi:10.1371/journal.pone.0229909
Sonia, , Saini, R., Singh, R. P., and Jaiwal, P. K. (2007). Agrobacterium tumefaciens mediated transfer of Phaseolus vulgaris alpha-amylase inhibitor-1 gene into mungbean Vigna radiata (L.) Wilczek using bar as selectable marker. Plant Cell. Rep. 26, 187–198. doi:10.1007/s00299-006-0224-4
Stiller, J., Martirani, L., Tuppale, S., Chian, R. J., Chiurazzi, M., and Gresshoff, P. M. (1997). High frequency transformation and regeneration of transgenic plants in the model legume Lotus japonicus. J. Exp. Bot. 48 (7), 1357–1365. doi:10.1093/jxb/48.7.1357
Subramanyam, K., Subramanyam, K., Sailaja, K. V., Srinivasulu, M., and Lakshmidevi, K. (2011). Highly efficient Agrobacterium-mediated transformation of banana cv. Rasthali (AAB) via sonication and vacuum infiltration. Plant Cell. Rep. 30, 425–436. doi:10.1007/s00299-010-0996-4
Sun, X., Hu, Z., Chen, R., Jiang, Q., Song, G., Zhang, H., et al. (2015). Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci. Rep. 5, 10342.
Suraninpong, P., Chanprame, S., Cho, H. J., Widholm, J. M., and Waranyuwat, A. (2004). Agrobacterium-mediated transformation of mungbean [Vigna radiata (L.) Wilczek]. Walailak J. Sci. Technol. 1 (2), 38–48. https://wjst.wu.ac.th/index.php/wjst/article/view/184.
Surekha, C. H., Beena, M. R., Arundhati, A., Singh, P. K., Tuli, R., Dutta-Gupta, A., et al. (2005). Agrobacterium-mediated genetic transformation of pigeon pea [Cajanus cajan (L.) Millsp.] using embryonal segments and development of transgenic plants for resistance against Spodoptera. Plant Sci. 169 (6), 1074–1080. doi:10.1016/j.plantsci.2005.07.011
Svitashev, S., Schwartz, C., Lenderts, B., Young, J. K., and Cigan, A. M. (2016). Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat. Commun. 7, 13274. doi:10.1038/ncomms13274
Tazeen, S., and Mirza, B. (2004). Factors affecting Agrobacterium tumefaciens mediated genetic transformation of Vigna radiata (L.) Wilczek. Pak. J. Bot. 36 (4), 887–896. https://www.pakbs.org/pjbot/PDFs/36(4)/PJB36(4)887.pdf.
Tivarekar, S., and Eapen, S. (2001). High frequency plant regeneration from immature cotyledons of mungbean. Plant Cell Tissue Organ Cult. 66, 227–230.
Tiwari, S., Mishra, D. K., Singh, A., Singh, P. K., and Tuli, R. (2008). Expression of a synthetic cry1EC gene for resistance against Spodoptera litura in transgenic peanut (Arachis hypogaea L.). Plant Cell. Rep. 27 (6), 1017–1025. doi:10.1007/s00299-008-0525-x
Trieu, A. T., Burleigh, S. H., Kardailsky, I. V., Maldonado-Mendoza, I. E., Versaw, W. K., Blaylock, L. A., et al. (2000). Transformation of Medicago truncatula via infiltration of seedlings or flowering plants with Agrobacterium. Plant J. 22 (6), 531–541. doi:10.1046/j.1365-313x.2000.00757.x
Ukhatovaa, Y. V., Erastenkovaa, M. V., Korshikovaa, E. S., Krylovaa, E. A., Mikhailovaa, A. S., Semileta, T. V., et al. (2023). Improvement of crops using the CRISPR/Cas system: new target genes. Mol. Biol. 57, 375–397. doi:10.1134/s0026893323030135
Varalaxmi, Y., Prasanna, A., Yadav, S. K., Venkateswarlu, B., and Maheswari, M. (2013). Optimization of parameters for Agrobacterium mediated transformation of black gram (Vigna mungo L. Hepper) using cotyledon explants. Afr. J. Biotechnol. 12, 1209–1215. https://www.ajol.info/index.php/ajb/article/view/130949.
Varanda, C. M. R., Félix, M. d. R., Campos, M. D., Patanita, M., and Materatski, P. (2021). Plant viruses: from targets to tools for CRISPR. Viruses 13, 141. doi:10.3390/v13010141
Varshney, R. K., Thudi, M., Pandey, M. K., Tardieu, F., Ojiewo, C., Vadez, V., et al. (2018). Accelerating genetic gains in legumes for the development of prosperous smallholder agriculture: integrating genomics, phenotyping, systems modelling and agronomy. J. Exp. Bot. 69 (13), 3293–3312.
Verma, A., Kaur, L., Kaur, N., Bhardwaj, A., Pandey, A. K., and Kandoth, P. K. (2023a). An efficient hairy root system for genome editing of a β-ODAP pathway gene in Lathyrus sativus. bioRxiv. doi:10.1101/2023.04.03.535460
Verma, V., Kumar, A., Partap, M., Thakur, M., and Bhargava, B. (2023b). CRISPR-cas: A robust technology for enhancing consumer-preferred commercial traits in crops. Front. Plant Sci. 14, 1122940. doi:10.3389/fpls.2023.1122940
Wang, L., Rubio, M. C., Xin, X., Zhang, B., Fan, Q., Wang, Q., et al. (2019). CRISPR/Cas9 knockout of leghemoglobin genes in Lotus japonicus uncovers their synergistic roles in symbiotic nitrogen fixation. New Phytol. 224 (2), 818–832. doi:10.1111/nph.16077
Wang, L., Wang, L., Zhou, Y., and Duanmu, D. (2017). Use of CRISPR/Cas9 for symbiotic nitrogen fixation research in legumes. Prog. Mol. Biol. Transl. Sci. 149, 187–213. doi:10.1016/bs.pmbts.2017.03.010
Wang, P., Fouracre, J., Kelly, S., Karki, S., Gowik, U., Aubry, S., et al. (2013). Evolution of GOLDEN2-LIKE gene function in C3 and C4 plants. Planta 237, 481–495. doi:10.1007/s00425-012-1754-3
Wang, Y., Li, Z., Chen, X., Gu, Y., Zhang, L., and Qiu, L. (2022). An efficient soybean transformation protocol for use with elite lines. Plant Cell. Tissue Organ Cult. 151, 457–466. doi:10.1007/s11240-022-02312-6
Wei, T., Jiang, L., You, X., Ma, P., Xi, Z., and Wang, N. N. (2023). Generation of herbicide-resistant soybean by base editing. Biology 12, 741. doi:10.3390/biology12050741
Wiszniewska, A., and Pindel, A. (2010). Protoplast culture utilization in studies on legume crops. Acta Agric. Scand. Sect. B – Soil Plant Sci. 60 (5), 389–399. doi:10.1080/09064710903179545
Woo, J. W., Kim, J., Kwon, S. I., Corvalán, C., Cho, S. W., Kim, H., et al. (2015). DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. doi:10.1038/nbt.3389
Wopereis, J., Pajuelo, E., Dazzo, F. B., Jiang, Q., Gresshoff, P. M., De Bruijn, F. J., et al. (2000). Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. Plant J. 23 (1), 97–114. doi:10.1046/j.1365-313x.2000.00799.x
Wu, G. (2016). Dietary protein intake and human health. Food Funct. 7 (3), 1251–1265. doi:10.1039/c5fo01530h
Xie, Y., Haq, S. I. U., Jiang, X., Zheng, D., Feng, N., Wang, W., et al. (2022). Plant genome editing: CRISPR, base editing, prime editing, and beyond. Grassl. Res. 1, 234–243. doi:10.1002/glr2.12034
Xu, H., Guo, Y., Qiu, L., and Ran, Y. (2022). Progress in soybean genetic transformation over the last decade. Front. Plant Sci. 13, 900318. doi:10.3389/fpls.2022.900318
Xu, Q., Song, B., Liu, F., Song, Y., Chen, P., Liu, S., et al. (2018). Identification and characterization of β-Lathyrin, an abundant glycoprotein of grass pea (Lathyrus sativus L.), as a potential allergen. J. Agric. food Chem. 66 (32), 8496–8503. doi:10.1021/acs.jafc.8b02314
Yamada, T., Teraishi, M., Hattori, K., and Ishimoto, M. (2001). Transformation of azuki bean by Agrobacterium tumefaciens. Plant Cell., Tissue Organ Cult. 64, 47–54. doi:10.1023/a:1010635832468
Yadav, S. K., Katikala, S., Yellisetty, V., Kannepalle, A., Narayana, J. L., Maddi, V., et al. (2012). Optimization of Agrobacterium mediated genetic transformation of cotyledonary node explants of Vigna radiata. SpringerPlus 1 (1), 59–68. doi:10.1186/2193-1801-1-59
Yan, B., Srinivasa Reddy, M. S., Collins, G. B., and Dinkins, R. D. (2000). Agrobacterium tumefaciens-mediated transformation of soybean [Glycine max (L.) Merrill.] using immature zygotic cotyledon explants. Plant Cell. Rep. 19 (11), 1090–1097. doi:10.1007/s002990000236
Yang, S., Hu, Y., Cheng, Z., Rice, J. H., Miao, L., Ma, J., et al. (2019). An efficient Agrobacterium-mediated soybean transformation method using green fluorescent protein as a selectable marker. Plant Signal. Behav. 14 (7), 1612682. doi:10.1080/15592324.2019.1612682
Yin, K., Gao, C., and Qiu, J. L. (2017). Progress and prospects in plant genome editing. Nat. Plants 3 (8), 17107–17116. doi:10.1038/nplants.2017.107
Yuan, M., Zhu, J., Gong, L., He, L., Lee, C., Han, S., et al. (2019). Mutagenesis of FAD2 genes in peanut with CRISPR/Cas9 based gene editing. BMC Biotechnol. 19, 24–27. doi:10.1186/s12896-019-0516-8
Zakaria, H., Hussein, G. M., Abdel-Hadi, A. H. A., and Abdallah, N. A. (2014). Improved regeneration and transformation protocols for three strawberry cultivars. GM crops food 5 (1), 27–35. doi:10.4161/gmcr.27229
Zambre, M., Goossens, A., Cardona, C., Van Montagu, M., Terryn, N., and Angenon, G. (2005). A reproducible genetic transformation system for cultivated Phaseolus acutifolius (tepary bean) and its use to assess the role of arcelins in resistance to the Mexican bean weevil. Theor. Appl. Genet. 110, 914–924. doi:10.1007/s00122-004-1910-7
Zhang, C., and Ghabrial, S. A. (2006). Development of Bean pod mottle virus-based vectors for stable protein expression and sequence-specific virus-induced gene silencing in soybean. Virology 344 (2), 401–411. doi:10.1016/j.virol.2005.08.046
Zhang, C., Liu, S., Li, X., Zhang, R., and Li, J. (2022). Virus-induced gene editing and its applications in plants. Int. J. Mol. Sci. 23, 10202. doi:10.3390/ijms231810202
Zhang, Y., Liang, Z., Zong, Y., Wang, Y., Liu, J., Chen, K., et al. (2016). Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617. doi:10.1038/ncomms12617
Zhang, Y., Singh, M. B., Swoboda, I., and Bhalla, P. L. (2005). Agrobacterium-mediated transformation and generation of male sterile lines of Australian canola. Aust. J. Agric. Res. 56 (4), 353–361. doi:10.1071/ar04175
Zhang, Z., Sun, A., Cong, Y., Sheng, B., Yao, Q., and Cheng, Z. M. (2006). Agrobacterium-mediated transformation of the apple rootstock Malus micromalus Makino with the Rol C gene. Vitro Cell. Dev. Biology-Plant 42, 491–497. doi:10.1079/ivp2006812
Keywords: legume, pulses, transformation recalcitrance, genome editing, genetic engineering, CRISPR/Cas9, plant, virus-mediated genome editing
Citation: Nivya VM and Shah JM (2023) Recalcitrance to transformation, a hindrance for genome editing of legumes. Front. Genome Ed. 5:1247815. doi: 10.3389/fgeed.2023.1247815
Received: 26 June 2023; Accepted: 06 September 2023;
Published: 21 September 2023.
Edited by:
Qinlong Zhu, South China Agricultural University, ChinaReviewed by:
Naglaa A. Abdallah, Cairo University, EgyptTan Jiantao, Guangdong Academy of Agricultural Sciences (GDAAS), China
Copyright © 2023 Nivya and Shah. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jasmine M. Shah, amFzbWluZUBjdWtlcmFsYS5hYy5pbg==
 V. M. Nivya
V. M. Nivya Jasmine M. Shah
Jasmine M. Shah