
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genome Ed. , 24 February 2023
Sec. Genome Editing in Plants
Volume 5 - 2023 | https://doi.org/10.3389/fgeed.2023.1094965
Agricultural production relies on horticultural crops, including vegetables, fruits, and ornamental plants, which sustain human life. With an alarming increase in human population and the consequential need for more food, it has become necessary for increased production to maintain food security. Conventional breeding has subsidized the development of improved verities but to enhance crop production, new breeding techniques need to be acquired. CRISPR-Cas9 system is a unique and powerful genome manipulation tool that can change the DNA in a precise way. Based on the bacterial adaptive immune system, this technique uses an endonuclease that creates double-stranded breaks (DSBs) at the target loci under the guidance of a single guide RNA. These DSBs can be repaired by a cellular repair mechanism that installs small insertion and deletion (indels) at the cut sites. When equated to alternate editing tools like ZFN, TALENs, and meganucleases, CRISPR- The cas-based editing tool has quickly gained fast-forward for its simplicity, ease to use, and low off-target effect. In numerous horticultural and industrial crops, the CRISPR technology has been successfully used to enhance stress tolerance, self-life, nutritional improvements, flavor, and metabolites. The CRISPR-based tool is the most appropriate one with the prospective goal of generating non-transgenic yields and avoiding the regulatory hurdles to release the modified crops into the market. Although several challenges for editing horticultural, industrial, and ornamental crops remain, this new novel nuclease, with its crop-specific application, makes it a dynamic tool for crop improvement.
In the present world, chronic malnourishment engulfs at least one billion population with the loss of biodiversity, climate change, and continuously degrading agriculture systems (Foley et al., 2011). Contemporary agriculture systems will face an enormous problem as the world population increases day by day with an estimated 9 billion by 2050 (Gomiero et al., 2011). The yield enhancement spawned during the green revolution is now steady diwindling because of rapid climate change which limits crop production and needs plants that can resist adverse environments with a higher yield. Conventional breeding is a dilatory and laborious process, a more efficient and systematized method is much required (Ashkani et al., 2015). With the availability of genomic sequences for multifarious plants, genome editing furnishes with an opportunity to edit crop varieties with greater precision. This strategy includes sequence-specific nucleases which can induce DNA double-stranded break (DSB) at the specific target sites. Following DSBs, repairs are made through donor template-dependent homology dependent repair (HDR) or error-prone non-homologous end joining pathways that introduce tiny insertion and deletion (indels) events at the target sites (Schmidt et al., 2019).
CRISPR (Clustered regularly interspaced short palindromic repeat) -Cas (CRISPR-associated protein) is an adaptive immune system discovered in bacteria and archaea, relying on RNA-DNA interaction and nuclease for targeted cleavage. With minimal effort and cost, DSBs may be readily induced at any desired target genomic locations using CRISPR-Cas and their orthologs (Kumlehn et al., 2018).
Genome editing technology has better efficiency to change the genome architecture at precise locations, with appropriate accuracy. The generation of new varieties of plants producing fecund yields with environmental stress resistance has been possible using this genome editing approach. It is more difficult to modify all of the plant genes using a specific genome editing technology because of the complicated architecture of the plants. However, several genome editing technologies have been created that have improved genome editing in plants to overcome this type of problem. Some of the genome editing technology employed to edit genomes of the plant is CRISPR/Cas9, ZFNs (zinc finger nucleases), HR (homologous recombination), TALENs (transcription activator-like effector nucleases). Additionally, oligonucleotide-directed mutagenesis and editing of the site-directed sequence have the efficiency of genome editing at the single-nucleotide level. Lately, the ABEs (adenine base editors) has been developed to mutate A-T base pairs to G-C base pairs. The field of genome-based breeding has gained new opportunities because of genome editing technology. Earlier genome editing tools such as meganuclease (Silva et al., 2011), zinc finger nucleases (ZFNs) (Carroll, 2011), and transcription activator-like effector nucleases (TALENs) (Sun and Zhao, 2013) have shown effectiveness for PGE (plant genome editing), but require complex protein engineering for each target site. CRISPR-Cas has been used for a large number of crops to modify traits with great economic value (Li et al., 2020). CRISPR-Cas mediated genome editting offers fewer risks as compared to GM crops as the majority of edits involve few nucleotides change (Ahmad et al., 2021). As the editing reagents are segregated out in the subsequent generation there is no distinction between a natural mutant and a mutant caused by genome edit. Thus, the incorporation of genome editing in normal breeding practices should enhance the speed of precision crop breeding and improve modern agriculture (Miki et al., 2018). Fruits and vegetables are more nutrient-dense and include more bioactive phytochemicals, which are essential for the global population (Alfa and Arroo, 2019; Fraga et al., 2019; Liskova et al., 2019; Saiwal et al., 2019). Plants experience several environmental factors like dehydration, heat, cold, pathogen, and many other harmful abiotic and biotic stress. Around 25 to 40 percent of the fruits and vegetables grown worldwide are never consumed after being harvested (Gustavsson et al., 2011). Fruit, vegetable, and ornamental plant losses are estimated to be around 75% (Kitinoja and Tokala, 2018; Porat et al., 2018). Production of fruit, vegetable, and ornamental plant in developing countries is already insufficient to meet people’s nutritional needs (Kc et al., 2018). Worldwide efforts have been initiated to reduce the anticipated loss of fruits, vegetables, and ornamental plants. However, the causes of postharvest waste and loss are very complex. Technology-based breeding for new and improved fruits, vegetables, and ornamental plants provides better quality which is a crucial component of the long-term solution. The most significant advancement in plant breeding since the green revolution is gene editing. It has already been used to uncover new information about plants, and it also holds enormous promise for creating new crops with various desired traits (Wolter et al., 2019). Genome editing technology is used in plants to produce novel phenotypes that increases agricultural productivity.
In several commercial crops, successful application of the CRISPR-based genome editing tools have been utilized. It is for improving resistance against diverse biotic and abiotic stresses in association with enhanced quality of fruits and grain architectures (Parmar et al., 2017). In this comprehensive study, we explain the potential use of genome editing tools that can create a robust and measurable platform to improve traits related to nutritional quality, post-harvest yield losses, and resistance against biotic and abiotic stresses. The gene and genome editing tools viz, ZFN, TALEN, CRISPR/Cas9, and also base editing, prime editing, and epigenome editing tools has been discussed in a concise way. Finally, we discuss gene editing technologies and improvement strategies, which will provide a resource for new researchers for crop improvement.
A slight modification or epigenetic change in the genome structure or gene structure of any organism can tune up or bring about desired changes, such as better yield, high tolerance, and high storage susceptibility. Modern technology has allowed scientists to perform controlled alterations in the gene structure within plant genomes which can be widely classified as Gene Editing or Gene targeting. Any technique which can perform permanent modification at a specific site is called genome targeting and is widely used for the improvement of the plant. The cellular DNA repair mechanisms are triggered when a Double-Stranded Break (DSBs) occurs. Either the homology-directed repair (HDR) mechanism or the non-homologous end joining (NHEJ) technique mechanism repairs the double-stranded breaks. The first one being sensitive to error, causes many additions or deletions, thus causing deterioration of the gene or its by-product. The Second one although error-prone yet causes small changes like addition of transgene. This allows for the targeted mutagenesis that results from engineering the target locus.
In plant species, the NHEJ repair mechanism is more prevalent than HDR because the latter depends on both structure of donor DNA and the repair machinery of the target cell. It has proven to be a tough assignment; hence NHEJ is chosen as a better and more relevant approach. The discovery of innovative specific and rare-cutting restriction enzymes that can induce double-stranded breaks at specific genomic sites is essential for the development of DSB-dependent Genome Editing technologies in plant cells. Nucleases recognize long nucleotide sequences and bring upon double-stranded breaks at their target sites. These meganucleases can be engineered as per the target sequence to be mutated. The gene editing technologies uses different engineered nucleases such as meganucleases, zinc finger nucleases (ZFNs), Transcription activator-like effector nucleases (TALENs), and newly added CRISPR-Cas endonuclease (Mao et al., 2019). All these designed nucleases share the trait of coupling a programmable DNA binding activity with a sequence-independent endonuclease activity that is sufficiently selective to target a particular genomic location.
ZFN is an artificially engineered endonuclease system consisting of Zinc Finger Protein (ZFP) fused with the Fok1 restriction enzyme. Zinc Finger Protein consists of approximately 30 conserved amino acids in ββα configuration, which are classified as Type II endonuclease (Gaj et al., 2013). ZFN comprised two domains responsible for DNA cleavage; a synthetically designed Cys-Cys-His-His Zinc finger domain at the N-terminal and the non-specific DNA cleavage Domain of the Fok1 endonuclease at the C-terminal. ZFN domain has three to four zinc fingers, each of which can recognize three base pairs long stretch of the target DNA sequence and hence allows proper tethering with the target DNA (Petolino, 2015). The Fok1 cleavage domain dimerization is essential to cut the target DNA sequence. Two ZFN monomers orient themselves in opposite directions to the target DNA site (Figure 1). They are designed such that there is a flank of 5-6 bps DNA sequence between the two monomers, which becomes the restriction site of dimerized Fok1 endonuclease (Weinthal et al., 2010). Once the target site is cut, the cellular repair mechanism of the Non-Homologous End Joining mechanism repairs it with some errors like Insertions and deletions. ZFNs have been employed in model and crop plant species as site-specific mutagens to facilitate the integration of the targeted transgene into donor genome sequences, to promote the repair of defective transgenes, to replace donor DNA sequences with foreign DNA molecules, and to ease the integration of the targeted transgene into donor genome sequences.
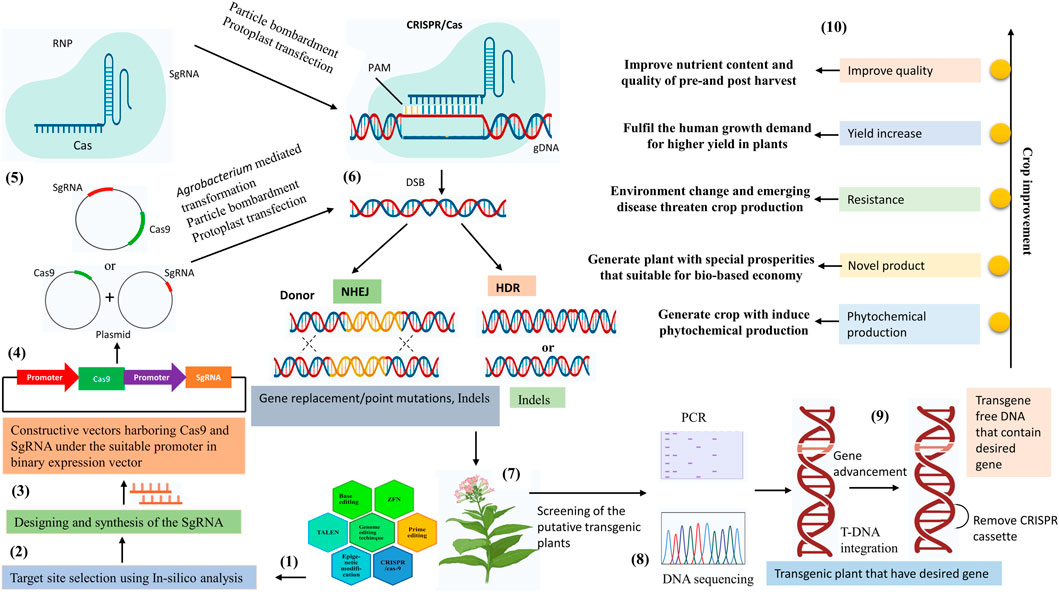
FIGURE 1. Overview of the CRISPR/Cas9 mediated genome editing in the plant. (1) Genome editing techniques. (2) Selection of the genomic target using In-silico analysis. (3) Design the sgRNA complementary to the target sequence. (4) Cloning of the designed Cas9 and sgRNA under suitable promoter in the binary expression vector. (5) The components of CRISPR/Cas9 tools construct transfer into plant cells, via Agrobacterium-mediated transformation, protoplast transfection, and particle bombardment. (6) CRISPR/Cas9 tools mediated genome editing in plants depend on the two main DSB (double-strand break) pathways. Gene replacement/point mutations, and indels are outcomes of the NHEJ (non-homologous end joining) pathway. Gene indels are outcomes of the HDR (homology-directed repair) pathway. (7) Development of the transgenic plant. (8) Identify the mutations in transgenic plants using DNA sequencing and PCR restriction enzyme assays and genotyping the transgenic plant with the desired mutation. (9) Removal of the CRISPR/Cas9 cassette (10) Crop improvement by the CRISPR/cas9 technology.
TALENs were developed as the fusion of Transcription Activator Like Effector with the catalytic domain of Fok1 endonuclease. TALEs proteins are found naturally in Xanthomonas proteobacterium and are involved in plant infection (Malzahn et al., 2017). During infection, they transfer the TALE proteins to the plant’s cell’s nucleus, bind to the promoter region of the target gene, and activate transcription (Figure 1). The structure of TALENs consists of three different domains: the N terminal contains a type III secretion system (T3SS), non-canonical repeats (NCR), and the C terminal has a transcription factor binding site, Nuclear Localization Signal (NLS), and an activation domain (Becker and Boch, 2021). Between these two domains, there exists a stretch of conserved 34 amino-acid long tandem repeats. Among them, the amino acid at the 12th and 13th positions are the ones responsible for the specificity of the TALENs. As in ZFN, TALENs also function as dimers, operating in opposite directions. When these two opposite TALENs fuse on the target gene, their Fok1 domain is responsible for mediating double-strand break. With the cell’s repair property, the DSB is repaired either by NHEJ or HDR mechanism resulting in insertion, deletion, or substitution and forming a mutant or knock-out variety.
CRISPR/Cas system was found to naturally exist as an RNA-guided DNA endonuclease system in the prokaryotic organism mostly by archaea and eubacteria. It helped to develop a target-specific immune response against foreign invaders, primarily viruses and plasmids, with the aid of the Cas protein, which cleaves the target sequence specifically and creates DSBs in the target DNA of the invading organism and hence destroys them. CRISPR stands for Clustered Regularly Interspaced Short Palindromic Repeats, and Cas are the CRISPR-associated genes. CRISPR consists of highly conserved short tandem repeats separated by short spacer sequences. Spacer sequences are unique sequences homologous with exogenous viral or plasmid DNA sequences (Bhowmik et al., 2021). These sequences are further used as recognition elements of foreign viral or plasmids and help bacteria to eliminate them. This is defined as the defense mechanism adopted by archaea and eubacteria. This technology is widely accepted and recognized for serving as machinery for genome editing and gene therapy for any organism/living system. It is the most precise, accurate, simple, and site-specific method of genetic engineering in plant systems. This technology has an immense capability to produce plants with the knock-out of undesirable characteristics.
The three stages of the CRISPR general mechanism are adaptation, expression, and interference. In the process of adapting, the bacteria take on the brief and distinctive protospacer sequences of the invaders and integrate them across nearby CRISPR loci in their genome. These sequences are recognized as novel CRISPR spacer sequences, which are added to the CRISPR array to create a memory of the invading organism. The next step is an expression in which the CRISPR locus is translated into pre-crRNA before being further processed into mature crRNAs, once the protospacer from the invasive organism is integrated into the CRISPR loci. In the last step, crRNA forms a complex with Cas protein, and the crRNA-Cas complex makes a complementary base pairing with the protospacer of the invader invading for the second time. Finally, crRNA-directed cleavage of the DNA follows with the aid of the Cas endonuclease protein. The system relies on two key molecules, Cas9 endonuclease, and a guide RNA, targeting a particular gene. gRNA (Spacer sequence) entails two RNA molecules, i.e., crRNA and tracrRNA, the former having the sequence complementary to the target DNA molecule and the latter acting as a binding partner for the Caspase 9 endonuclease enzyme. During exposure to invading virus or plasmid, short fragments of foreign DNA get integrated into CRISPR repeat-spacer array within host chromosomes as spacer DNA. They act as a marker of invading organisms. The transcription of this spacer DNA and enzymatic cleavage produces short CRISPR RNA (crRNA). During subsequent infection caused by the virus, hybridization occurs between crRNA and complementary foreign target sequence, marking it for sequence-specific cleavage by Cas9 endonuclease. The target site is recognized by a specific short conserved sequence motif, the PAM (Protospacer-adjacent motif) sequence, flanking between the target site of foreign invaders (Figure 1).
Makarova and her groups have immensely put their efforts into providing the evolutionary classification of the CRISPR-Cas system. In 2015 (Makarova and Koonin, 2015), the group emphasized 2 classes, 5 types, and 16 subtypes of the CRISPR system until recently, in 2020, where they provided an updated evolutionary classification including 2 classes, 6 subtypes, and 33 subtypes, based on the various effector modules and Cas protein compositions. The Class I CRISPR-Cas system comprises modules of several Cas proteins that bind and process the target as part of a complex called the cr-RNA-binding complex. A single, multidomain cr-RNA binding protein in the class II CRISPR-Cas system functions similarly to the entire class I system’s complex (Kato et al., 2022) Types 1, III, and IV are included in Class I, whereas Types II, V, and VI are included in Class II (Makarova et al., 2020).
The efficient delivery of the CRISPR/Cas9 complex to the target cell is an essential point to consider. It can be delivered in various forms, such as plasmid DNA, messenger RNA, or Ribonucleoproteins. Ribonucleoprotein (RNP), another emerging approach for genome editing, comprises of a Cas9 protein and a gRNA. Some of the critical points of RNP-based genome editing are its DNA/transgene-free genome editing approach which also ensures minimal off-target effects and being DNA-free, it offers reduced toxicity (Zhang et al., 2021).
The CRISPR-Cas9 system offers significant opportunities for genomic and epigenetic regulation in addition to its editing capabilities. The components of the CRISPR/Cas9 system Cas9 and sgRNA complex can serve as a scaffold to drive various effectors or markers to particular DNA sites. This feature of CRISPR-Cas9 has been implemented to modify the transcriptional level of gene expression, either to activate genes (CRISPRa) or to repress genes (CRISPRi) (La Russa and Qi, 2015). Even though being an important approach for genome editing, Double-Stranded Breaks open the door to off-target gene editing, hence unintended mutations. Therefore, other approaches, not including DSBs have been developed to escape these demerits. One such approach is using the dead Cas9 variant (dCas9) wherein the two catalytic domains are inactivated by point mutation and are inactive. They do not cause double-stranded breaks in the genome but still have strong binding efficiency to the target site in the genome. For this nature of dCas9, they are fused with other specific enzymes that can modify or bring the required changes in the genome. The two systems, CRISPRa/CRISPRi, also utilize nuclease-deactivated Cas9, whose catalytic domains are made disabled, and then fused to transcriptional modulators (Jensen et al., 2021).
Base editing, as the name suggests, is the tool whereby nucleotide base is either edited or modified with the guidance of sgRNA and the help of dCas9 protein fused with Activation-Induced Deaminases (AID) such as Cytidine deaminase or Adenine deaminase–based editors. Base editing is more effective because it does not call for double-strand breaks. Targeted editing for plants has utilized base editing.
Base editors- CBEs cause cytosine deamination to uracil, which then converts to thymidine by DNA replication or repair mechanism. CBEs have been immensely utilized for plant genome editing. As reported by Chen et al., CRISPR/CAS9-mediated base editing was performed for gain-of-functions mutation in Arabidopsis (Chen et al., 2017). Using Cytidine deaminase fused with Cas9 protein, Shimatani, and the group (Shimatani et al., 2017) were able to perform targeted base editing in rice and tomato for crop improvement.
Similarly, Adenine Deaminase (ABEs) fused with dCas9 causes base mutation from adenine to inosine, which can be further base paired with Cytosine. This cytosine pairs up with guanine in the newly synthesized strand, causing alteration in the dictating DNA. ABEs have already been used for genome editing, as reported in Arabidopsis, Rice, Wheat, etc.
Gene expression can be changed by epigenetic alterations like DNA acetylation, methylation, or Histone modification. These epigenetic modifiers fused with the dCAS9 protein have been established as a better tool for generating epigenetic alterations (Pan et al., 2021). These modifications can alter the expression level of genes without changing the parental DNA sequence. Author Kang and his group (Kang et al., 2019) demonstrated the use of CRISPR/Cas9 to generate methylation at specific CpG sites and successfully targeted the Oct4 gene. Gallego-Bartolome with his group (Gallego-Bartolomé et al., 2018), altered the CRISPR/dCas9 SunTag system to target DNA demethylation in plants. In plants, the loss of 5 mC (5-methyl Cytosine) at the FLOWERING WAGENINGEN (FWA) promoter stimulates FWA expression, which is accountable for late-flowering. They achieved this by introducing the human TET1 (TEN-ELEVENTRANSLOCATION1) catalytic domain fused with CRISPR/dCas9.
Prime editing has been developed as a new tool kit for CRISPR-mediated genome editing by Anzalone et al. (2019). Like base editing, prime-editing does not rely either on DSBs or donor templates. Prime-editing involves three main components: Cas9 nickase, which is fused with Reverse Transcriptase enzyme, and a prime-editing guide RNA (pegRNA). pegRNA differs from other sgRNA in terms of its features: a sequence complementary to the target location of the template DNA at the 5′ end, Primer Binding Site (PBS) at the 3′ end, and the sequence carrying desired changes next to PBS. The Cas9 is drafted to the target location of DNA by the complementary guide sequence (5′ end) of the pegRNA and creates nicks at PAM-containing DNA strand. The RT template acts as a template for the synthesis of altered genetic information on the exposed 3′-OH group of target DNA as a result of the development of a nick, which causes the 3′ nicked end of the template strand to hybridize with the PBS of pegRNA. Hybridization of target DNA and RT produces either a 3′ flap having desired sequence or a 5′ flap having the original sequence (Wada et al., 2020). Endonucleases preferentially cleave the 5′ flaps, forming a DNA duplex with one edited strand with the desired alterations and another strand that is the original strand. This mismatch is either stably integrated by cellular replication or is corrected by a mismatch repair mechanism, both opening a gate for stable integration of the desired sequence into the genome (Kantor et al., 2020).
Agricultural modifications have shifted the paradigm of consumer acceptance. Traditional extrinsic visual-quality attribute awareness has moved to intrinsic attributes. Nutritional, functional, and physio-chemical factors of fruits and vegetables have been magnified in public awareness. These factors comprise the essential vitamins, minerals, phytochemicals, and antioxidant content traits introgressed. The dynamic genotypic and environmental factors determine the above attribute along with handling post-harvest. Post-harvest loss has been a major bottleneck, and a struggle to diminish the yield gap by improving the nutritional toolbox for genome editing to feed the growing population. The economic and social burden of the loss prompts the potential of the new-editing tools. It must be exploited alongside the conventional ones that have been limiting success. Due to their effectiveness and specificity of cleavage recognition sites, either by RNA-directed SDNs or protein-directed SDNs, site-directed nucleases (SDNs) utilized in plant editing technologies are frequently used to modify genomes. It includes meganucleases, ZFNs, TALENs, and CRISPR/Cas used to achieve random, predicted, or precise insertion. These all accomplish precise genetic alterations by causing deliberate DNA double-strand breaks (DSBs) (Tripathi et al., 2021). Additionally, CRISPR can avoid other obstacles, including sterility, self-incompatibility, high heterozygosity, a low frequency of recovering desirable alleles and features, and long life cycles that make conventional breeding operations more difficult or impossible to complete (Shipman et al., 2021). With the easy elimination of undesirable traits, the in-depth investigation of non-interference about the constitutive genome editing on other cellular functionality has increased. To avoid any pleiotropic changes on regulatory genes, Somatic mutations can be generated by special promoters in specific cells, tissues, and organs of the plant genome using the CRISPR- TSKO (tissue-specific knockouts), or TSGE (tissue specific gene editing) were preferred. It generated more accurate KO (knock-out) variants in plants. DJ3S, p54/1.0—Cassava promoters targeted into root of Carrot and Arabidopsis (Singha et al., 2022). Similar to this, another gene-editing method regulates the production of the Cas protein in fruits using an inducible chimeric transcription factor (XVE). The activator is an assembly of bacterial repressor LexA(X), activating domain VP16(V), and regulatory region of human estrogen receptor (E) (Brand et al., 2006). The estradiol-induced XVE is an inducible expression system used in transgenic apples, tobacco, Arabidopsis (Zuo et al., 2001), and soybeans for reporter gene expression and overcoming certain limitations in other expression systems (Corrado and Karali, 2009). The outcomes of repair mechanisms chosen for genome editing applications that include site-directed nuclease-1 (SDN1) in kiwi-fruit (ACC oxidase) produce non-homologous end products after the cleavage of host DNA by CRISPR/Cas9. Non-homologous end joining (Brand et al., 2006). The outcomes of repair mechanisms for genome editing include site-directed nuclease-1 (SDN1), producing non-homologous end products after the cleavage of host DNA by CRISPR/Cas9. NHEJ causes indel mutations by gene silencing, gene inactivation, and gene knockout. SDN2 utilizes the plant Homology-Directed Repair (HDR) pathway to modify the gene’s activity by altering the sequence. The SDN2 techinique uses template DNA to produce an intended sequence modification at the DSB site. SDN3 technique uses a DNA insertion or substitution at a specific site in the DNA. Base and prime editing techniques are a type of SDN1 type. They can generate change in nucleotide without deploying a DSB or template DNA insertion in the locus targeted (Lin et al., 2020). The modifications of base and prime editing are comparable to the SDN2 technology. Oligo-directed mutagenesis (ODM) alters the genomic locus by targeted mutations. Successful application of ODM has been done on maize (Zhu et al., 2000), rice (Okuzaki and Toriyama, 2004), and oilseed rape (Gocal et al., 2015). The approach of SDN1, SDN2, and ODM targets mutation without the inclusion of exogenous DNA. But the SDN3 aims to target exogenous DNA insertion of various lengths (Modrzejewski et al., 2019). The delivery system of the SDNs includes PEG-fusion, electroporation, and biolistics, whereas, without the aid of any expression system. The chemically synthesized oligonucleotide used in ODM is supplied directly to the plant cell (Sandhya et al., 2020). However, the most reliable transformation for gene-editing is the Agrobacterium-mediated delivery in plants. Another approach to altering gene expression through DNA methylation has also been approached in orange and bell pepper (Cheng et al., 2018). Alongside genome editing, the combination of big data modeling and artificial intelligence (AI) monitoring has changed the course of field challenges. Most of the gene-editing tools has been researched on tomato and then progressed to achieve in several other fleshy fruits discovering numerous candidate gene targets (Rehman et al., 2022). Browning reactions in fruits post-harvest has also been triggering loss to the food industry. Potential research on natural bioactive compounds with anti-browning extracts (mangrove, green tea, thyme, pineapple) has been achieved to replace chemical additives by genome editing. An industrial approach for regulating the challenge has been worked through the activity control of the Polyphenol Oxidase (PPO) enzyme (Moon et al., 2020). Innovations targeting manipulation in the genome of plants have been extensively investigated and achieved at a spellbinding pace. ONM (Oligonucleotide directed mutagenesis), and ENs (Engineered nucleus) have been traditionally practiced and have been revolutionized with ZFNs, TALENs, EMNs, mi-RNA, and CRISPR. Modulation in the PPO and POD genes with miRNA, and CRISPR knock-out mutation reduced the browning activity in eggplant berries by 52% on the SmelPPO4-5-6 gene (Maioli et al., 2020). Verification has been done by HRFA (High-Resolution fragment analysis) on the T1 phenotypic lines generated (Hamdan et al., 2022). The post-harvest storage also critically implicates the quality and shelf-life of freshly harvested fruits. The pathogenic microbes and pests in favorable growth conditions cause two types of deterioration: 1) physiological-softening, ripening, and senescence and 2) microbial deterioration by fungal or bacterial growth. Therefore, optimized storage conditions with CA (controlled atmosphere), DCA (dynamic controlled atmosphere), or ULO (Ultra-low oxygen) have been evaluated as chemical-free storage. Apart from the storage conditions, chemical treatments of pesticides have been used against the microbial activity. The implications of the hazardous by-products and residues have reduced their demand for usage. Comprehensive research has developed alternatives to chemical treatments using the physical-natural blend. Led light treatment, essential oil application, and edible coating (EC) are a few examples (Sánchez-González et al., 2011). EC is a semipermeable film over the fresh stock of perishable produce that has been catching attention for extending post-harvest life (Fawole et al., 2020). EC preserved fruit quality of ripening, softening, colour transformation, formation of sugar, and loss of organic acid by lowering the exchange of gaseous metabolic process in plants (Ziv and Fallik, 2021). Exploration of enzymatic inhibitors, nano and microencapsulation of the bioactive compounds is the need of the hour for implementing techniques to improve and utilize the natural extracts. Extensive broad-spectrum research on protein and gene expression profiles would also determine its biochemical, physiological, and receptor signaling activity pathways. This will help generate the required information about the natural extract’s target and through efficient statistical approaches, it can be effectively used on a large scale. Genome editing on fruits has dissected an imminent network of signaling and biological pathways that is anticipated to aid in investigating hitherto undiscovered genes inducing positive post-harvest phenotypes (Table 1).
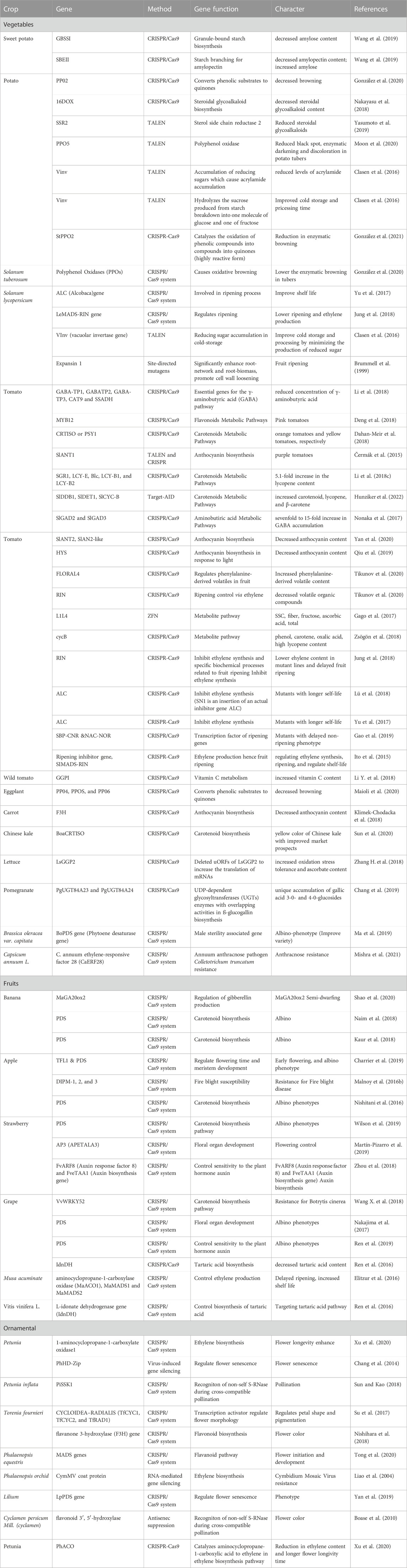
TABLE 1. List of the targeted genes that are modified by the CRISPR/Cas9 tool in fruits, vegetables, and ornamental plants.
There is currently a great deal of attention on the health advantages of consuming vegetables of the expansive array of nutrients found in them, such as vitamin supplements, minerals, antioxidants, dietary fiber, and phytochemical compounds (Septembre-Malaterre et al., 2018). Vegetables, whether consumed fresh or in processed form, defend against a wide range of non-transmitted diseases and assist in the reduction of such food insecurity in underdeveloped countries (Desjardins, 2014). Food losses and waste reduction are crucial to a country’s food security as intensive and extensive farming. Losses reduce the availability of food, contributing to food insecurity and waste food generating unnecessary emissions of CO2 as well as a failure of the food’s economic value. As a result, limiting post-harvest losses in agricultural products is critical for food availability and security in developing countries. Post-harvest losses are losses in the nutritional value, seed viability, and selling price of food that occur along the entire food supply chain, from harvest to consumption (Ghosh et al., 2011). Genome editing allows for the acquisition of multiple homozygous mutations in a single generation without foreign DNA, opening up new avenues for genomics and the breeding of crops and quality traits in vegetables. The tomato and potato have attracted the most focus on genome editing research and implementation.
The ability to alter important characteristics in cucumber, watermelon, lettuce, and broccoli has been demonstrated (Cardi et al., 2017). In tomato seeds, gene LEAFY-COTYLEDON1-LYKE4, that encodes the β -subunit of the transcription factor Y, is damaged by the transiently expressed zinc finger nucleases (ZFNs). Plants with mutations displayed a wide range of morphological changes as well as the expression of genes involved in hormone metabolism, especially the ethylene biosynthetic pathway. No off-target changes were found in the closely linked L1L2 and L1L3 gene sequences (Hilioti et al., 2016). GABA (Y-aminobutyric acid) is a chemical that promotes health and has received a lot of attention in traditional tomato breeding research. Using genome editing, multiple GABA pathway genes were modified, resulting in a 19-fold increase in GABA content in Solanum lycopersicum. Resistance to diseases through genetic modification is another new area of study in vegetables rapidly expanding as the CRISPR/Cas system as a genome editing tool. Genetically modified by deletion of 48 base pair in homozygous layout in MLO1 locus, tomato plants generated which showed resistance to powdery mildew disease affected by Oidium neolycopersici (Erpen-Dalla Corte et al., 2019). Editing the SIDMR6-1 gene also produced bacterial disease resistance (downy mildew resistance 6 gene) (Tripathi et al., 2020). The shift in the recent paradigm of abiotic stress tolerance research includes more horticulture crops for genetic modification. Significant advances have been made in the improvement of biotic stress tolerance (fungal infection and virus resistance) as in tomatoes and cucumbers (Jain, 2015; Shi et al., 2017). Multiple research studies have pivoted the potential functions in plant protection against abiotic and biotic stresses, as well as improving fruit quality, plant architectural features, and storage stability (Kulus, 2018). The N′ and C′ ends of eIF4E-were targeted by CRISPR/Cas in cucumbers by agrobacterium-mediated transformation, leading to resistance to cucumber vein yellow virus, pumpkin mosaic virus, and papaya ring spot mosaic virus (PRSV-W) (Chandrasekaran et al., 2016). Numerous diseases are brought on by fungi, which can significantly reduce crop output and quality. For example, downy and powdery mildew significantly reduce tomato production (Borrelli et al., 2018). CRISPR/Cas9 knockout of DMR6 homologous genes in tomatoes demonstrated resistance to Pseudomonas syringae, Phytophthora, and Xanthomonas spp (de Toledo Thomazella et al., 2016). Climate change is making crops more susceptible to abiotic stress. The invention of CRISPR/Cas9 has accelerated the production of new varieties. Several developmental pathways that are mediated by brassinosteroids (BR) involve the brassinazole-resistant 1 gene (BZR1). The agrobacterium-mediated CRISPR-mediated mutation in BZR1 (engaged in a variety of brassinosteroid (BR)-mediated developmental processes) inhibited the induction of RESPIRATORY BURST OXIDASE HOMOLOG1 (RBOH1) and the production of H2O2. Exogenous H2O2 restored heat tolerance in tomato bzr1 mutants (Yin et al., 2018). To develop larger fruit size than the WT fruits, Cold Spring Harbor Laboratory edited the tomato CLAVATA-WUSCHEL (CLV-WUS) stem cell gene CLV3’s promoter region using the CRISPR/Cas9 technique (Li T. et al., 2018). High concentrations of steroidal glycoalkaloids (SGAs) give potato tubers a foul flavor and make them dangerous to consume. St16DOX (steroid 16-hydroxylase) was removed from the potato SGA biosynthetic pathway using CRISPR/Cas9, producing SGA-free potato lines (Nakayasu et al., 2018).
Ornamental plant production is linked with various features and markets like cut flowers and decorative vegetation, interior and exterior houseplants, window ledges, bulbous and nursery plants. In the scenario of ever-increasing demand, the floriculture industries require advancements and more different kinds with privileged qualities. The major objectives of ornamental breeding programs include establishing uniqueness in a blooming plant, color diversity, and perfumes, as well as inflorescence development by enhancing the flower count, altering the duration and longevity of flowering. Flower colors are the most impactful for their economic interest of any characteristic. Yellow cyclamens, red iris, blue chrysanthemums and roses, for example, are not found in nature due to the absence of pigment pathways (Tanaka et al., 2009). New decorative plant cultivars have been produced using a variety of breeding techniques. Apart from enhancing plant architecture and disease resistance, traditional plant breeding practices like crossbreeding and mutation breeding have been employed to produce various patterns and colours in plants. The majority of present methods for ornamental plants are extremely heterozygous, which produces a multifaceted transmission of genetic traits and polyploidy with several limits and drawbacks (Azadi et al., 2016; Sharma and Messar, 2017). Only a few countries allowed genetically modified (GM) ornamental plants commercially. For instance, the blue rose “ApplauseTM” and the violet carnation “MoondustTM,” both of which are genetically modified (GM) to hoard delphinidin-based anthocyanins, were developed and commercialized in the global flower market by the Japanese company SUNTORY and the Australian biotechnology company Florigene. However, consumers prefer genetically modified crops to genetically engineered ornamental plants (Nishihara and Nakatsuka, 2011). The first genome of an ornamental plant to undergo CRISPR/Cas9 modification was Petunia hybrida (Subburaj et al., 2016). The NITRATE REDUCTASE (PhNR) gene exhibited maximal targeted mutagenesis by direct administration of designed RNA-guided endonuclease (RGEN) ribonucleoproteins (RNPs) induced in Petunia protoplast cells. This has been made possible by the quick advancement of genome editing tools like ZFN, TALENs, and CRISPR/Cas9. These methods also enable a comprehensive understanding of the metabolic activity of annotated genes in newly sequenced and assembled genomes. Flower colour modification is now widely used in many plant species through genome-editing tools. Mutagenesis in the dihydroflavonol-4-reductase (DFR) gene of Ipomoea nilotica using CRISPR/Cas9 resulted in the first floral colour alteration in higher plants (Watanabe et al., 2017). Watanabe modified the carotenoid cleavage dioxygenase 4 (CCD4) gene using CRISPR/Cas9 to produce mutant Ipomoea plant. CRISPR/Cas9 approaches are used to target multiple MADS genes involved in floral organ development in the Phalaenopsis orchid (Tong et al., 2020). The chrysanthemum is one of the most well-known and important floricultural crops. Despite a complete genome sequence, due to the enormous genomic size and ploidy stages with a higher fraction of repetitive sequences, CRISPR/Cas systems will probably be challenging to implement. The CRISPR/Cas9 system functions in this plant, according to just one study (Kishi-Kaboshi et al., 2017) (Table.1).
The gradual increase in customer demand for extrinsic quality characteristics in fruits and vegetables, such as size, color, texture, aroma, storage life, etc., has been considered crucial. With increased public awareness, nutrition enhancement became economically significant. The commercialization of genetically engineered approved horticulture crops included virus-resistant papaya, squash variety, non-browning apple, fleshy pineapple, insect tolerant eggplant (BT brinjal), Brassica oleracea-(BolC.GA4. a gene), Tomato-(PSY1,Mlo,GABA-TP1,TP2, SIAGL6 gene), Potato- (StMB44, StALS1 gene) and Lactuca sativa-(BIN2 gene). The demand for parthenocarpic fruits and their socio-economic impacts accelerated the potential for developing genome editing. High-quality cultivars have been developed by silencing genes involved in signaling, knockout technique, reverse genetics, transgenesis, etc. The notable accomplishments through CRISPR/Cas9 techniques have brought about successful trait modification through mutation for enhancing nutritional gains. Mutations included loss of function in tomato (SIALC, SIFUL1, Pectate lyase), banana (MaLCY epsilon, MaPDS), apple (MdPDS, MdDIMP4), grapes, kiwi fruit (AcPDS, AcCENs), soybean (GmPRR3b, 37, GmFT2a), potato (StPPO), strawberry (FaTM6), gene or promoter insertion, cis-regulatory alleles in tomato (SICLV3, lycopene beta cyclase), citrus (CsLOBs) frame-shift mutation, gene replacement in tomato (SIALC), etc. The modifications in the aforesaid horticultural crops included increased flower and fruit size, fruit ripening, inflorescence branching, enhanced ascorbic acid synthesis, fortification of beta-carotene (banana), the transformation of perennial to annual (kiwi fruit), decreased tuber-browning and enhanced quality of berry (strawberry) (Tiwari et al., 2021). In an elegant research study, it has been published that the flavor of fruit directly modulates certain organic acids that influence organoleptic characteristics. Hence improving or minimizing them has a direct impact on the general character of a fruit (Sweetman et al., 2009). Acidity, which is quite a significant trait of the fruit harvest, has a direct impact on fruit quality. But it is especially critical for later processing. Hence, early-ripening apples often have high levels of acidity and low levels of sugar, which lowers the market demand for fresh consumption and leads to a low proportion of total organic acids to total carbohydrates. Numerous recent research in the fields of omics and quantitative trait loci (QTLs) (Xu et al., 2012) revealed a significant cellular synchronization between nutrition, post-harvest fruit quality, and shelf-life variation (Lin et al., 2016). The effectiveness of UV-C usage to combat the post-harvest impacts on early-ripened fruits produced at room temperature include ripening delay, senescence, preserving the significant ratio of fruit firmness, biosynthesis of flavonoids, phenolic content, enhanced antioxidant, and defense responsive molecules (Artés et al., 2009). The different doses for the UV irradiations have a marked positive impact by lowering the acidity-to-sugar ratio of various fruits and vegetables to preserve them post-harvest. UV-C treatment has been well-established on apples, mango, strawberry, peach, tomato, etc., promoting the flavour. The irradiations generate malate degradation-improving the quality of fruit, plant defense response mechanism against microbes, activating the gene regulation for disease resistance, enhancing salubrious phytochemical content, and regulating the proportions of antioxidant ratio on exposure to the shorter wavelength of UV-C. For successful implementation of post-harvest storage conditions to enhance nutritional benefits, the various horticultural produce effect is being noted. It includes the specific plant parts, developmental stages targeted for treatment, doses used, cultivar specificity, and harvesting time of the fruits and vegetables.
Postharvest loss and waste are becoming increasingly unsustainable as global horticulture crop production is inadequate for meeting human nutritional needs. Postharvest loss is unintentional. It outlines the sporadic losses that farmers occasionally sustain at the hands of consumer, including physical harm, internal bleeding, premature spoilage, and bug damage, among other things. Ornamental popularity has exploded recently, with an average worth of $16 billion in 2015 (Kader, 2004; Porat et al., 2018). Ornamental crops have a high moisture content, and the cold-chain process results in a loss of up to 50% of farm value (Pranuthi et al., 2018). Value decreases by 15% for each additional day spent on transit. Furthermore, the vase-life of ornaments is usually just 10–12 days after consumer purchase (Mamias, 2018). So, it is important to move things along a cold chain quickly. Reduced respiration rate is achieved by using cold temperatures, which also increases shelf life. As vegetables and fruits travel from farmer to customer, various factors (processing, storage, and transportation conditions) contribute to such a deteriorative process. Understanding where food supply chain losses occur is critical for determining possible causes and improving best post-harvest practices (Gustavsson et al., 2011). Temperature, moisture content, level of ethylene hormone, and the storage proportion of O2 to CO2 must be managed to ensure yield shelf-life and quality (Kader, 2004). Infectious agent’s invasion attempts on harvested products are likely to caramelise the fruit ingredients, resulting in diseased or spore-covered fruits and their metabolic by-products. The products are unaesthetic and non-edible due to the acidic, bitter, foul-smelling, and toxicants produced (Encinas-Basurto et al., 2017).
Fruits that are overripe or underripe are more vulnerable to physiological disorders. Sometimes entire products are discarded as they are unsafe to eat (ömer AZABAĞAOĞLU, 2018). Incorrect cultural practises such as cold snaps, weather, water stress, heavy rainfalls, pathogens, physiological disorders, plant health, safeguards, water management, fertilization, and cutting possibly cause fruit and vegetable damage during the preharvest period. Harvesting period loss (4%–12%) is due to inaccurate harvesting time estimation, harvesting at the wrong time, inappropriate harvesting procedures implementation, and not applying pre-cooling to fruits such as cherries during the harvesting process (Özdemir et al., 2003). According to the International Refrigeration Institute (IIR), 23% of food waste in developing countries is generated when cooling systems are not used [(Dupont et al., 2009), 2009]. Constant cold storage ensures that the product reaches the consumer in pristine condition. The lack of adequate storage facilities is the leading cause of the degradation of both qualitative and quantitative factors of food production from food harvest to consumption level in underdeveloped countries (Tatlıdil et al., 2013). Fresh produce changes cannot be avoided, but they can be reduced with preventative measures, including cold temperature, relative humidity, appropriate transportation and packaging, and others (Ahmad and Siddiqui, 2015). The most popular ornamental plants, such as roses, liliums, lisianthus, chrysanthemums, and carnations, demonstrated varying levels of ethylene sensitivity and flower longevity. The use of breeding methods in conjunction with ethylene screening was only partially successful. It is possible to expand the life of ornamental plants by using molecular techniques (Netam, 2018). Gene-editing of 1-aminocyclopropane-1-carboxylate oxidase1 (PhACO1) gene which encodes for ethylene producing enzyme via CRISPR/Cas9 method resulted in decreased ethylene and delayed petal senescence in the petunia cultivar “Mirage Rose” (Xu et al., 2020). Mutations in the EPHEMERAL1 (EPH1) gene, which encodes the major regulator for petal senescence as a NAC transcription factor, resulted in delayed petal senescence in Japanese morning glory (Ipomoea nil “Violet”) plants (Shibuya et al., 2018). MaACO1 mutated through CRISPR/Cas9 system produced less ethylene and had a longer shelf life in the ripening stage (Hu et al., 2021). To date, the CRISPR/Cas9 gene-editing approach has already been effectively implemented in many fruit crop species, such as climacteric ripening species, like apples (Malnoy et al., 2016b; Nishitani et al., 2016), bananas (Kaur et al., 2018), kiwifruit (Wang Z. et al., 2018)), and non-climacteric ripening species, such as sweet orange (Jia and Wang, 2014), Duncan grapefruit (Jia et al., 2016), grapevine (Malnoy et al., 2016b), watermelon (Tian et al., 2017), cucumber (Chandrasekaran et al., 2016), and cultivated strawberries (Martín-Pizarro et al., 2019).
Abiotic stress, which includes dehydration, soil salinity, and high heat, poses serious risks to crop development, yield, and quality (Sharma and Gayen, 2021). The effect of adverse conditions on the plant is more common in tropical regions than temperate regions. High temperature creates more threat in tropical countries. Drought stress is another abiotic stress limiting crop development and production, which is continuously increasing due to global warming. Crop yields are becoming more and more susceptible due to unfavorable climate changes, deteriorating soil health, and declining air quality. Researchers are attempting to create a transgenic crop that can produce more and easily tolerate severe and variable conditions. CRISPR/Cas9 technology has been implemented to increase crop yield. Abiotic stress, such as drought, soil salinity, and high temperature, create major problems for the growth and development of the crop, drastically lowering agricultural production and quality. Resistance to the abiotic stress created by the many genes is a complex trait. With the use of nucleases that cause double-strand breaks by providing a new method for editing genes in molecular biology, which transformed the field of genome editing. ZFNs are credited as the 1st genome editing technique that used programmable nucleases to make a significant advancement in the field of genome engineering (Chandrasegaran and Carroll, 2016). After a while, TALENs (Transcription activator-like effector nucleases), which are entirely dependent on TALEs of the bacteria, additionally broaden genome engineering potential. TALENs were quickly adapted to approximately 40 different types of species for genome editing (Chandrasegaran and Carroll, 2016). Scientists worldwide have turned their attention to the CRISPR/Cas9 technologies since their discovery because of their numerous advantages over TALENs and ZFN (Mao et al., 2013). Unlike the previous technology, TALENs and ZFN, which use a protein motif to identify the target, CRISPR/Cas9 is based on RNA-DNA recognition to generate the double-strand break. Apart from this, there are many benefits of the CRISPR/Cas9 technology over the TALENs, and ZFN 1) Ease of the target design 2) ability to directly inject RNAs encoding the Cas9 protein and gRNA (guide RNA) to cause mutation, and 3) create targeted mutations in numerous genes in a single event because to the simplicity of multiplexing (Ma et al., 2015; Malzahn et al., 2017). A new genetic engineering tool is precise genome editing for crop improvement. Multiple techniques such as TALENs (Christian et al., 2010; Li et al., 2011; Miller et al., 2011), ZFNs (Maeder et al., 2008; Sander et al., 2011), RGENs (RNA-guided nucleases), and CRISPR/Cas9 (Jinek et al., 2012; Cong et al., 2013; Mali et al., 2013), all these types of technology developed for the targeted genome editing. CRISPR/Cas9 technology-based genome editing in the crop can be used to modify almost any sequence to show its function in the organism’s genome. CRISPR/Cas9 generated SIMAPK3 participates in tomato dehydration resistance by preventing oxidative damage to the cell wall generated by stress, further adjusting the transcription of genes linked to drought stress (Wang et al., 2017). Recently, CRISPR/Cas9-generated genome engineering for high-temperature resistance has been achieved by focusing on an S gene, SIAGL6 (SIGAMOUS-LIKE 6), in S. lycopersicum, which has a significant role in the improvement in fruit quality against high-temperature stress (Klap et al., 2017). In tomatoes involvement of the auxin in salinity stress and osmotic stress resistance and the role of the ARF4 (auxin response factor 4) providing the resistance and confirm the use of the CRISPR/Cas9 tools to develop tolerant plants (Bouzroud et al., 2020). In tomato genome editing by the CRISPR/Cas9 tools under the abiotic stress in various genes such as SIMAPK3 (Kong et al., 2012), SILOX, SIGST, SIDREB (Wang et al., 2017), SILBD40 (Liu et al., 2020), SINPR1 (Li et al., 2019), and SIMAPK6 (Li et al., 2021), under the abiotic stress condition. CRISPR/Cas tools were used to generate mutation in lettuce plants to understand better the mechanism of NCED4 (Bertier et al., 2018). During heat stress, CRISPR/Cas9 technology was used for silencing SIMAPK3 in tomatoes (Yu et al., 2019). This technique is used to increase the expression level of the ARGOS8 gene (a negative regulator of the ethylene response) to develop drought resistance maize crops, and the ARGOS8 gene promoter changed into GOS2. These traits had a major role in the enhancement of grain yields during drought stress conditions (Hirai et al., 2007). Overexpression of the biosynthesis of the melatonin gene in plants was identified as an abiotic stress resistance (Byeon and Back, 2016; Antoniou et al., 2017). Currently, the CRISPR/Cas9 genome engineering technique further expands the application of these tools to genome-wide screening for the improvement of the desired trait (Rodríguez-Leal et al., 2017; Mahas and Mahfouz, 2018). Gain- or loss-of-function mutants can result from precise base alterations made by the CRISPR/Cas9 system at desired gene locations. There is no doubt that CRISPR/Cas technology might replace conventional crop breeding methods. It depends on locating plant populations with genetic differences to produce desired features in crop cultivars.
The gRNA sequencing method can be used to identify novel allelic variations that match a specific desirable trait that could be introduced into plant populations by this base editing technique (Eid et al., 2018).
Biotic stress, such as insects, viruses, fungi, and bacteria, can attack plants and cause severe damage (Langner et al., 2018). CRISPR/Cas9 tools have been developed to obtain disease-tolerant plants (Arora and Narula, 2017). CRISPR/Cas technology is a beneficial method for creating stable transgenic lines. Following the development of CRISPR/Cas technology against bacterial, viral, and fungal infections, which result in significant losses in plants. Despite the information of technological advances being already reported in CRISPR/Cas9 technology for the fruits, vegetables, and ornamental plant improvement, not much more progress has been made towards the uses of CRISPR/Cas9 genome engineering tool for biotic stress resistance in fruits, vegetables, and ornamental crops. The response of the plants toward biotic stress is more complicated; during the biotic stress circumstances, several crucial genes are overexpressed and down-expressed.
For the inactivation of the DMR6 ortholog in Solanum Lycopersicum, researchers used the CRISPR/Cas9 system. They discovered that mutations in the dmr6 gene exhibit disease tolerance against various pathogens, including species Xanthomonas, Phytophthora capsica, and P. syringae, without causing any discernible adverse effects (de Toledo Thomazella et al., 2016). It has been demonstrated that tomato MAPK3 (mitogen-activated protein kinase-3) gains the potential to Botrytis cinerea (Zhang S. et al., 2018). Scientists used CRISPR/Cas9 to develop Solanum Lycopersicum JAZ2 (Jasmonatezim domain protein 2) repressors lacking C-terminal jasmonate associated domain (JAZ2Δjas). These repressors provide tolerance against the P. syringae, indicating CRISPR/Cas9 technology for the development of resistance fruit and its implementation in agriculture (Ortigosa et al., 2019). In Vitis vinifera, knockout of the WRKY5 by the CRISPR/Cas9 technology encoding a TF that has a significant role in biotic stress response, WRKY5 exhibits disease tolerance against B. cinerea (Wang X. et al., 2018). The CRISPR/Cas9 technology is crucial for the fruits such as apples (Malus domestica) and grapes (Vitis vinifera), which have long life cycles and generation times. Numerous papers have described the use of CRISPR/Cas9 so far with fruits like Citrus reticulate (Jia and Wang, 2014), grapes (Malnoy et al., 2016a; Ren et al., 2016; Wang et al., 2016), apples (Malnoy et al., 2016a; Nishitani et al., 2016), Solanum tuberosum (potato) (Andersson et al., 2017), banana (Musa balbisiana) (Tripathi et al., 2019), cucumber (Chandrasekaran et al., 2016), papaya (Gumtow et al., 2018), citrus (Jia et al., 2016; Peng et al., 2017). The fruits, vegetables, and ornamental plant production and growth are exposed to continuously increased risks of biotic stress. The CRISPR/Cas9 technology is used in applications to obtain new germplasm resources rapidly. As fruits, vegetables, and ornamental plants are sensitive to biotic stress, it can make achieving optimal yield difficult, emphasizing the significance of creating stress-resistant crops.
The application of CRISPR/Cas9-based genome editing technology in fruits, vegetables, and ornamental plants has specific significant methodological challenges that need to be addressed to appreciate the uniqueness of this technique fully. The following are some main problems with the CRISPR/Cas9 genome editing technique: 1) The first prerequisite for beginning genome editing work is the species’ whole genome information. Although notably in many crops, it is not available, which is more complicated and severely restricts the applicability of CRISPR/Cas9. 2) The reproducible and efficient gene transfer methodology containing particle bombardment, in vitro generation protocol, Agrobacterium-mediated transformation, due to their resistance to plant tissue culture, many fruits, vegetables, and ornamental plants have not been investigated for PEG-mediated transformation, etc. Another factor to consider is that the CRISPR/Cas9 technique involves a laborious and drawn-out process of desirable transformed/mutated selection and regeneration. 3) The fact that some crops have a complicated genetic structure makes them difficult to study from a genomic perspective. Because of their quantitative nature, the genes relating to post-harvest quality traits in many crops are still mostly unstudied. The CRISPR/Cas9 technique has the ability to insert modifications crop genomes. The use of CRISPR/Cas has been constrained in many countries caused of the ambiguity of the current biosafety regimes (SDN-1, SDN-2, and SDN-3) (Eckerstorfer et al., 2019). Additionally, the proponents of the CRISPR/Cas9 tool are more concerned about this “overregulation” according to the GMO laws, not because there is not a specific or unique regulatory framework. Hence, fruits, vegetables, and ornamental crops must be allowed to use the CRISPR/Cas9 tool. Delivery protocols and easy transformation must be developed for the modification of the desired plant generation in the case of recalcitrant crops. Additionally, in the circumstances of the apple and grape, adopting DNA-free genome editing technology based on the RNP (ribonucleoprotein) complex yields a better outcome for successfully creating mutant lines (Malnoy et al., 2016a), and potatoes (Andersson et al., 2018) by biolistic gun and PEG-mediated transformation. Many nations have already accepted and developed genome editing technologies and successfully commercialized genetically altered crops. Most countries decide the regulatory status of genome-edited organisms on a case-by-case basis and can use the current regulatory framework for GMOs (Eckerstorfer et al., 2019). The worldwide regulatory environment for genome-edited species is far more heterogeneous than conventional GMOs. The regulatory triggers for biosafety laws vary across nations. Although International bodies, such as the OECD, fully acknowledge this fact yet are still striving to harmonize the regulatory control of biotechnology. It is difficult to see how this cooperation among experts and comprehension of all these technologies will successfully address this issue. Overall, the adoption and formulation of suitable regulations requirements to be increased effectively to apply the capability of this technology in the better crop improvement to require high-quality nutrient-rich food with accessibility to the continuously growing world population.
Traditional breeding merely depends on sufficient variability that exists among the plant populations that have made a great contribution to modern agricultural practices. However, variations generated either from spontaneous natural mutations or developed using chemical or physical mutagens, are random and in low-frequency events. Moreover, desired variations among the elite cultivars may not arise at the same time, and it is a delitory process. Conventional breeding fails to produce such traits in a limited period of time. In contrast, genome editing serves as a modern molecular biology tool that can produce error-free targeted modifications.
It is urgently necessary to provide this constantly expanding population with enough food that has better nutritional value on a worldwide scale. This could only be achieved in horticultural, and fruit crops with the acquisition of the latest genome editing technologies. However, there is a need for a transparent and smooth regulatory system, which can provide brooder applicability for the rapid delivery of genome-edited crops to the consumer market. In this present review, we have made an effort to deliver the great potentiality and versatility of genome editing to enhance the quality of traditional vegetables, fruits, and ornamental plants in a very concise way. In many underdeveloped countries, the significant causes of vegetable and fruit loss are because of inadequate infrastructure. In contrast, in developed countries, the losses occur at the consumer level. The use of convenient technology to control post-harvest yield loss is a demanding process. Genome editing is a promising method for rapid trait improvement, considering ornamental plants. But three possible drawbacks exist, which need to be addressed. Firstly, there is a need for suitable transformation methods to deliver genome editing reagents in the target tissue. Secondly, proper identification of genes related to a specific trait with agronomic importance is still required. Recent advancement in genome sequencing technology has made it easy to sequence many diploid ornamental plants, but it remains a challenge to identify genes specific to the particular trait of interest. In the case of polyploid flowers like Chrysanthemums, and roses, the WGS is not available. The proteome and transcriptome of these ornamental plants can pave the way forward for the identification of specific genes of interest at a particular time or environmental condition. Regardless of these limitations, genome editing serves as an extraordinary and outstanding approach to modifying horticultural crops for improving required traits and expanding the availability of ornamental plants for benefit of the society.
DG conceived the idea and designed the concept. The manuscript was prepared with the potential contribution of PS, AP, RM, SD, SK and DG. DG corrected and approved the manuscript.
This work was supported by the SERB-DST grant (SRG/2020/001024), Central University of Rajasthan research support. DG is gratefully acknowledging the SERB-DST grant (SRG/2020/001024), UGC (BSR), and Central University of Rajasthan for research support. We thank the Council of Scientific and Industrial Research (CSIR), India for the Fellowship to RM and SERB for providing fellowship to SD, PS, and AP is gratefully acknowledging the Central University of Rajasthan for financial support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahmad, M. S., and Siddiqui, M. W. (2015). “Factors affecting postharvest quality of fresh fruits,” in Postharvest quality assurance of fruits (Springer), 7–32. doi:10.1007/978-3-319-21197-8_2
Ahmad, A., Munawar, N., Khan, Z., Qusmani, A. T., Khan, S. H., Jamil, A., et al. (2021). An outlook on global regulatory landscape for genome-edited crops. Int. J. Mol. Sci. 22, 11753. doi:10.3390/ijms222111753
Alfa, H. H., and Arroo, R. R. J. (2019). Over 3 decades of research on dietary flavonoid antioxidants and cancer prevention: What have we achieved? Phytochem. Rev. 18, 989–1004. doi:10.1007/s11101-019-09632-0
Andersson, M., Turesson, H., Nicolia, A., Fält, A.-S., Samuelsson, M., and Hofvander, P. (2017). Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 36, 117–128. doi:10.1007/s00299-016-2062-3
Andersson, M., Turesson, H., Olsson, N., Fält, A. S., Ohlsson, P., Gonzalez, M. N., et al. (2018). Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol. Plant. 164, 378–384. doi:10.1111/ppl.12731
Antoniou, C., Chatzimichail, G., Xenofontos, R., Pavlou, J. J., Panagiotou, E., Christou, A., et al. (2017). Melatonin systemically ameliorates drought stress-induced damage in M edicago sativa plants by modulating nitro-oxidative homeostasis and proline metabolism. J. Pineal Res. 62, e12401. doi:10.1111/jpi.12401
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. doi:10.1038/s41586-019-1711-4
Arora, L., and Narula, A. (2017). Gene editing and crop improvement using CRISPR-cas9 system. Front. Plant Sci. 8, 1932. doi:10.3389/fpls.2017.01932
Artés, F., Gómez, P., Aguayo, E., Escalona, V., and Artés-Hernández, F. (2009). Sustainable sanitation techniques for keeping quality and safety of fresh-cut plant commodities. Postharvest Biol. Technol. 51, 287–296. doi:10.1016/j.postharvbio.2008.10.003
Ashkani, S., Rafii, M. Y., Shabanimofrad, M., Miah, G., Sahebi, M., Azizi, P., et al. (2015). Molecular breeding strategy and challenges towards improvement of blast disease resistance in rice crop. Front. Plant Sci. 6, 886. doi:10.3389/fpls.2015.00886
Azadi, P., Bagheri, H., Nalousi, A. M., Nazari, F., and Chandler, S. F. (2016). Current status and biotechnological advances in genetic engineering of ornamental plants. Biotechnol. Adv. 34, 1073–1090. doi:10.1016/j.biotechadv.2016.06.006
Becker, S., and Boch, J. (2021). TALE and TALEN genome editing technologies. Gene Genome Ed. 2, 100007. doi:10.1016/j.ggedit.2021.100007
Bertier, L. D., Ron, M., Huo, H., Bradford, K. J., Britt, A. B., and Michelmore, R. W. (2018). High-resolution analysis of the efficiency, heritability, and editing outcomes of CRISPR/Cas9-induced modifications of NCED4 in lettuce (Lactuca sativa). G3 Genes, Genomes, Genet. 8, 1513–1521. doi:10.1534/g3.117.300396
Bhowmik, P., Konkin, D., Polowick, P., Hodgins, C. L., Subedi, M., Xiang, D., et al. (2021). CRISPR/Cas9 gene editing in legume crops: Opportunities and challenges. Legum. Sci. 3, e96. doi:10.1002/leg3.96
Boase, M. R., Lewis, D. H., Davies, K. M., Marshall, G. B., Patel, D., Schwinn, K. E., et al. (2010). Isolation and antisense suppression of flavonoid 3’, 5'-hydroxylasemodifies flower pigments and colour in cyclamen. BMC Plant Biol. 10, 107–112. doi:10.1186/1471-2229-10-107
Borrelli, V. M. G., Brambilla, V., Rogowsky, P., Marocco, A., and Lanubile, A. (2018). The enhancement of plant disease resistance using CRISPR/Cas9 technology. Front. Plant Sci. 9, 1245. doi:10.3389/fpls.2018.01245
Bouzroud, S., Gasparini, K., Hu, G., Barbosa, M. A. M., Rosa, B. L., Fahr, M., et al. (2020). Down regulation and loss of auxin response factor 4 function using CRISPR/Cas9 alters plant growth, stomatal function and improves tomato tolerance to salinity and osmotic stress. GenesGenes (Basel). 11, 272. doi:10.3390/genes11030272
Brand, L., Hörler, M., Nuesch, E., Vassalli, S., Barrell, P., Yang, W., et al. (2006). A versatile and reliable two-component system for tissue-specific gene induction in Arabidopsis. Plant Physiol. 141, 1194–1204. doi:10.1104/pp.106.081299106.081299
Brummell, D. A., Harpster, M. H., Civello, P. M., Palys, J. M., Bennett, A. B., and Dunsmuir, P. (1999). Modification of expansin protein abundance in tomato fruit alters softening and cell wall polymer metabolism during ripening. Plant Cell 11, 2203–2216. doi:10.1105/tpc.11.11.2203
Byeon, Y., and Back, K. (2016). Low melatonin production by suppression of either serotonin N-acetyltransferase or N-acetylserotonin methyltransferase in rice causes seedling growth retardation with yield penalty, abiotic stress susceptibility, and enhanced coleoptile growth under anoxic conditions. J. Pineal Res. 60, 348–359. doi:10.1111/jpi.12317
Cardi, T., Batelli, G., and Nicolia, A. (2017). Opportunities for genome editing in vegetable crops. Emerg. Top. Life Sci. 1, 193–207. doi:10.1042/ETLS20170033
Carroll, D. (2011). Genome engineering with zinc-finger nucleases. Genetics 188, 773–782. doi:10.1534/genetics.111.131433
Čermák, T., Baltes, N. J., Čegan, R., Zhang, Y., and Voytas, D. F. (2015). High-frequency, precise modification of the tomato genome. Genome Biol. 16, 232–315. doi:10.1186/s13059-015-0796-9
Chandrasegaran, S., and Carroll, D. (2016). Origins of programmable nucleases for genome engineering. J. Mol. Biol. 428, 963–989. doi:10.1016/j.jmb.2015.10.014
Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., Pearlsman, M., et al. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153. doi:10.1111/mpp.12375
Chang, X., Donnelly, L., Sun, D., Rao, J., Reid, M. S., and Jiang, C.-Z. (2014). A petunia homeodomain-leucine zipper protein, PhHD-Zip, plays an important role in flower senescence. PLoS One 9, e88320. doi:10.1371/journal.pone.0088320
Chang, L., Wu, S., and Tian, L. (2019). Effective genome editing and identification of a regiospecific gallic acid 4-O-glycosyltransferase in pomegranate (Punica granatum L.). Hortic. Res. 6, 123. doi:10.1038/s41438-019-0206-7
Charrier, A., Vergne, E., Dousset, N., Richer, A., Petiteau, A., and Chevreau, E. (2019). Efficient targeted mutagenesis in apple and first time edition of pear using the CRISPR-Cas9 system. Front. Plant Sci. 10, 40. doi:10.3389/fpls.2019.00040
Chen, Y., Wang, Z., Ni, H., Xu, Y., Chen, Q., and Jiang, L. (2017). CRISPR/Cas9-mediated base-editing system efficiently generates gain-of-function mutations in Arabidopsis. Sci. China Life Sci. 60, 520–523. doi:10.1007/s11427-017-9021-5
Cheng, J., Niu, Q., Zhang, B., Chen, K., Yang, R., Zhu, J.-K., et al. (2018). Downregulation of RdDM during strawberry fruit ripening. Genome Biol. 19, 212–214. doi:10.1186/s13059-018-1587-x
Christian, M., Cermak, T., Doyle, E. L., Schmidt, C., Zhang, F., Hummel, A., et al. (2010). Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186, 757–761. doi:10.1534/genetics.110.120717
Clasen, B. M., Stoddard, T. J., Luo, S., Demorest, Z. L., Li, J., Cedrone, F., et al. (2016). Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol. J. 14, 169–176. doi:10.1111/pbi.12370
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Sci. (80-. ) 339, 819–823. doi:10.1126/science.1231143
Corrado, G., and Karali, M. (2009). Inducible gene expression systems and plant biotechnology. Biotechnol. Adv. 27, 733–743. doi:10.1016/j.biotechadv.2009.05.006
Dahan-Meir, T., Filler-Hayut, S., Melamed-Bessudo, C., Bocobza, S., Czosnek, H., Aharoni, A., et al. (2018). Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 95, 5–16. doi:10.1111/tpj.13932
de Toledo Thomazella, D. P., Brail, Q., Dahlbeck, D., and Staskawicz, B. (2016). CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. bioRxiv [Preprint]. doi:10.1101/064824
Deng, L., Wang, H., Sun, C., Li, Q., Jiang, H., Du, M., et al. (2018). Efficient generation of pink-fruited tomatoes using CRISPR/Cas9 system. J. Genet. Genomics= Yi chuan xue bao 45, 51–54. doi:10.1016/j.jgg.2017.10.002
Desjardins, Y. (2014). Fruit and vegetables and health: An overview. Hortic. Plants People Places 3, 965–1000. doi:10.1007/978-94-017-8560-0_2
Dupont, J. L., El Ahmar, A., and Guilpart, J. (2009). The role of refrigeration in worldwide nutrition. International Institute of Refrigeration, 5th Informatory Note on Refrigeration and Food. 18, 2019. Available at: http://www.iifiir.org.
Eckerstorfer, M. F., Engelhard, M., Heissenberger, A., Simon, S., and Teichmann, H. (2019). Plants developed by new genetic modification techniques-Comparison of existing regulatory frameworks in the EU and Non-EU countries. Front. Bioeng. Biotechnol. 7, 26. doi:10.3389/fbioe.2019.00026
Eid, A., Alshareef, S., and Mahfouz, M. M. (2018). CRISPR base editors: Genome editing without double-stranded breaks. Biochem. J. 475, 1955–1964. doi:10.1042/BCJ20170793
Elitzur, T., Yakir, E., Quansah, L., Zhangjun, F., Vrebalov, J., Khayat, E., et al. (2016). Banana MaMADS transcription factors are necessary for fruit ripening and molecular tools to promote shelf-life and food security. Plant Physiol. 171, 380–391. doi:10.1104/pp.15.0186615.01866
Encinas-Basurto, D., Valenzuela-Quintanar, M. I., Sánchez-Estrada, A., Tiznado-Hernández, M. E., Rodríguez-Félix, A., and Troncoso-Rojas, R. (2017). Alterations in volatile metabolites profile of fresh tomatoes in response to Alternaria alternata (Fr.) Keissl. 1912 infection. Chil. J. Agric. Res. 77, 194–201. doi:10.4067/S0718-58392017000300194
Erpen-Dalla Corte, L., Mahmoud, L. M., Moraes, T. S., Mou, Z., Grosser, W. J., and Dutt, M. (2019). Development of improved fruit, vegetable, and ornamental crops using the CRISPR/Cas9 genome editing technique. Plants 8, 601. doi:10.3390/plants8120601
Fawole, O. A., Riva, S. C., and Opara, U. L. (2020). Efficacy of edible coatings in alleviating shrivel and maintaining quality of Japanese plum (Prunus salicina Lindl.) during export and shelf life conditions. Agronomy 10, 1023. doi:10.3390/agronomy10071023
Foley, J. A., Ramankutty, N., Brauman, K. A., Cassidy, E. S., Gerber, J. S., Johnston, M., et al. (2011). Solutions for a cultivated planet. Nature 478, 337–342. doi:10.1038/nature10452
Fraga, C. G., Croft, K. D., Kennedy, D. O., and Tomás-Barberán, F. A. (2019). The effects of polyphenols and other bioactives on human health. Food Funct. 10, 514–528. doi:10.1039/c8fo01997e
Gago, C., Drosou, V., Paschalidis, K., Guerreiro, A., Miguel, G., Antunes, D., et al. (2017). Targeted gene disruption coupled with metabolic screen approach to uncover the LEAFY COTYLEDON1-LIKE4 (L1L4) function in tomato fruit metabolism. Plant Cell Rep. 36, 1065–1082. doi:10.1007/s00299-017-2137-9
Gaj, T., Gersbach, C. a., and Barbas, C. F. (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405. doi:10.1016/j.tibtech.2013.04.004
Gallego-Bartolomé, J., Gardiner, J., Liu, W., Papikian, A., Ghoshal, B., Kuo, H. Y., et al. (2018). Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc. Natl. Acad. Sci. 115, E2125–E2134. doi:10.1073/pnas.1716945115
Gao, Y., Zhu, N., Zhu, X., Wu, M., Jiang, C.-Z., Grierson, D., et al. (2019). Diversity and redundancy of the ripening regulatory networks revealed by the fruitENCODE and the new CRISPR/Cas9 CNR and NOR mutants. Hortic. Res. 6, 39. doi:10.1038/s41438-019-0122-x
Ghosh, S., Meli, V. S., Kumar, A., Thakur, A., Chakraborty, N., Chakraborty, S., et al. (2011). The N-glycan processing enzymes α-mannosidase and β-D-N-acetylhexosaminidase are involved in ripening-associated softening in the non-climacteric fruits of capsicum. J. Exp. Bot. 62, 571–582. doi:10.1093/jxb/erq289
Gocal, G. F. W., Schöpke, C., and Beetham, P. R. (2015). “Oligo-mediated targeted gene editing,” in Advances in new technology for targeted modification of plant genomes (Springer), 73–89. doi:10.1007/978-1-4939-2556-8_5
Gomiero, T., Pimentel, D., and Paoletti, M. G. (2011). Is there a need for a more sustainable agriculture? CRC. Crit. Rev. Plant Sci. 30, 6–23. doi:10.1080/07352689.2011.553515
González, M. N., Massa, G. A., Andersson, M., Turesson, H., Olsson, N., Fält, A.-S., et al. (2020). Reduced enzymatic browning in potato tubers by specific editing of a polyphenol oxidase gene via ribonucleoprotein complexes delivery of the CRISPR/Cas9 system. Front. Plant Sci. 10, 1649. doi:10.3389/fpls.2019.01649
González, M. N., Massa, G. A., Andersson, M., Décima Oneto, C. A., Turesson, H., Storani, L., et al. (2021). Comparative potato genome editing: Agrobacterium tumefaciens-mediated transformation and protoplasts transfection delivery of CRISPR/Cas9 components directed to StPPO2 gene. Plant Cell, Tissue Organ Cult. 145, 291–305. doi:10.1007/s11240-020-02008-9
Gumtow, R., Wu, D., Uchida, J., and Tian, M. (2018). A Phytophthora palmivora extracellular cystatin-like protease inhibitor targets papain to contribute to virulence on papaya. Mol. Plant-Microbe Interact. 31, 363–373. doi:10.1094/MPMI-06-17-0131-FI
Gustavsson, J., Cederberg, C., Sonesson, U., van Otterdijk, R., and Meybeck, A. (2011). Global food losses and food waste: Extent, causes and prevention. FAO.
Hamdan, N., Lee, C. H., Wong, S. L., Fauzi, C. E. N. C. A., Zamri, N. M. A., and Lee, T. H. (2022). Prevention of enzymatic browning by natural extracts and genome-editing: A review on recent progress. Molecules 27, 1101. doi:10.3390/molecules27031101
Hilioti, Z., Ganopoulos, I., Ajith, S., Bossis, I., and Tsaftaris, A. (2016). A novel arrangement of zinc finger nuclease system for in vivo targeted genome engineering: The tomato LEC1-LIKE4 gene case. Plant Cell Rep. 35, 2241–2255. doi:10.1007/s00299-016-2031-x
Hirai, M. Y., Sugiyama, K., Sawada, Y., Tohge, T., Obayashi, T., Suzuki, A., et al. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. 104, 6478–6483. doi:10.1073/pnas.0611629104
Hu, C., Sheng, O., Deng, G., He, W., Dong, T., Yang, Q., et al. (2021). CRISPR/Cas9-mediated genome editing of MaACO1 (aminocyclopropane-1-carboxylate oxidase 1) promotes the shelf life of banana fruit. Plant Biotechnol. J. 19, 654–656. doi:10.1111/pbi.13534
Hunziker, J., Nishida, K., Kondo, A., Ariizumi, T., and Ezura, H. (2022). Phenotypic characterization of high carotenoid tomato mutants generated by the target-AID base-editing technology. Front. Plant Sci. 13, 848560. doi:10.3389/fpls.2022.848560
Ito, Y., Nishizawa-Yokoi, A., Endo, M., Mikami, M., and Toki, S. (2015). CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem. Biophys. Res. Commun. 467, 76–82. doi:10.1016/j.bbrc.2015.09.117
Jain, M. (2015). Function genomics of abiotic stress tolerance in plants: A CRISPR approach. Front. Plant Sci. 6, 375. doi:10.3389/fpls.2015.00375
Jensen, T. I., Mikkelsen, N. S., Gao, Z., Foßelteder, J., Pabst, G., Axelgaard, E., et al. (2021). Targeted regulation of transcription in primary cells using CRISPRa and CRISPRi. Genome Res. 31, 2120–2130. doi:10.1101/gr.275607.121
Jia, H., and Wang, N. (2014). Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS One 9, e93806. doi:10.1371/journal.pone.0093806
Jia, H., Orbovic, V., Jones, J. B., and Wang, N. (2016). Modification of the PthA4 effector binding elements in type I CsLOB1 promoter using cas9/sgRNA to produce transgenic duncan grapefruit alleviating XccΔpthA4: DCsLOB1.3 infection. Plant Biotechnol. J. 14, 1291–1301. doi:10.1111/pbi.12495
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi:10.1126/science.1225829
Jung, Y. J., Lee, G. J., Bae, S., and Kang, K. K. (2018). Reduced ethylene production in tomato fruits upon CRSPR/Cas9-mediated LeMADS-RIN mutagenesis. Hortic. Sci. Technol. 36 (3), 396–405. doi:10.12972/kjhst.20180039
Kader, A. A. (2004). Increasing food availability by reducing postharvest losses of fresh produce. V. Int. Postharvest Symp. 682, 2169–2176. doi:10.17660/ActaHortic.2005.682.296
Kang, J. G., Park, J. S., Ko, J.-H., and Kim, Y.-S. (2019). Regulation of gene expression by altered promoter methylation using a CRISPR/Cas9-mediated epigenetic editing system. Sci. Rep. 9, 11960–12012. doi:10.1038/s41598-019-48130-3
Kantor, A., McClements, M. E., and MacLaren, R. E. (2020). CRISPR-Cas9 DNA base-editing and prime-editing. Int. J. Mol. Sci. 21, 6240. doi:10.3390/ijms21176240
Kato, K., Okazaki, S., Schmitt-Ulms, C., Jiang, K., Zhou, W., Ishikawa, J., et al. (2022). RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease. Sciece (80-. ). 378, 882–889. doi:10.1126/science.add7347
Kaur, N., Alok, A., Shivani, , Kaur, N., Pandey, P., Awasthi, P., et al. (2018). CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome. Funct. Integr. Genomics 18, 89–99. doi:10.1007/s10142-017-0577-5
Kc, K. B., Dias, G. M., Veeramani, A., Swanton, C. J., Fraser, D., Steinke, D., et al. (2018). When too much isn’t enough: Does current food production meet global nutritional needs? PLoS One 13, e0205683. doi:10.1371/journal.pone.0205683
Kishi-Kaboshi, M., Aida, R., and Sasaki, K. (2017). Generation of gene-edited Chrysanthemum morifolium using multicopy transgenes as targets and markers. Plant Cell Physiol. 58, 216–226. doi:10.1093/pcp/pcw222
Kitinoja, L., and Tokala, V. Y. (2018). Brondy. AA review of global postharvest loss assessments in plant-based food crops: Recent findings and measurement gaps. J. Postharvest Technol. 6, 1–15.
Klap, C., Yeshayahou, E., Bolger, A. M., Arazi, T., Gupta, S. K., Shabtai, S., et al. (2017). Tomato facultative parthenocarpy results from Sl AGAMOUS-LIKE 6 loss of function. Plant Biotechnol. J. 15, 634–647. doi:10.1111/pbi.12662
Klimek-Chodacka, M., Oleszkiewicz, T., Lowder, L. G., Qi, Y., and Baranski, R. (2018). Efficient CRISPR/Cas9-based genome editing in carrot cells. Plant Cell Rep. 37, 575–586. doi:10.1007/s00299-018-2252-2
Kong, F., Wang, J., Cheng, L., Liu, S., Wu, J., Peng, Z., et al. (2012). Genome-wide analysis of the mitogen-activated protein kinase gene family in Solanum lycopersicum. Gene 499, 108–120. doi:10.1016/j.gene.2012.01.048
Kulus, D. (2018). Genetic resources and selected conservation methods of tomato. J. Appl. Bot. Food Qual. 91, 135–144. doi:10.5073/JABFQ.2018.091.019
Kumlehn, J., Pietralla, J., Hensel, G., Pacher, M., and Puchta, H. (2018). The CRISPR/cas revolution continues: From efficient gene editing for crop breeding to plant synthetic biology. J. Integr. Plant Biol. 60, 1127–1153. doi:10.1111/jipb.12734
La Russa, M. F., and Qi, L. S. (2015). The new state of the art: Cas9 for gene activation and repression. Mol. Cell. Biol. 35, 3800–3809. doi:10.1128/MCB.00512-15
Langner, T., Kamoun, S., and Belhaj, K. (2018). CRISPR crops: Plant genome editing toward disease resistance. Annu. Rev. Phytopathol. 56, 479–512. doi:10.1146/annurev-phyto-080417-050158
Li, T., Huang, S., Jiang, W. Z., Wright, D., Spalding, M. H., Weeks, D. P., et al. (2011). TAL nucleases (TALNs): Hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 39, 359–372. doi:10.1093/nar/gkq704
Li, R., Li, R., Li, X., Fu, D., Zhu, B., Tian, H., et al. (2018). Multiplexed CRISPR/Cas9-mediated metabolic engineering of γ-aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnol. J. 16, 415–427. doi:10.1111/pbi.12781
Li, T., Yang, X., Yu, Y., Si, X., Zhai, X., Zhang, H., et al. (2018). Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 36, 1160–1163. doi:10.1038/nbt.4273
Li, X., Wang, Y., Chen, S., Tian, H., Fu, D., Zhu, B., et al. (2018). Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front. Plant Sci. 9, 559. doi:10.3389/fpls.2018.00559
Li, Y., Wang, H., Zhang, Y., and Martin, C. (2018). Can the world’s favorite fruit, tomato, provide an effective biosynthetic chassis for high-value metabolites? Plant Cell Rep. 37, 1443–1450. doi:10.1007/s00299-018-2283-8
Li, R., Liu, C., Zhao, R., Wang, L., Chen, L., Yu, W., et al. (2019). CRISPR/Cas9-Mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol. 19, 38–13. doi:10.1186/s12870-018-1627-4
Li, Q., Sapkota, M., and van der Knaap, E. (2020). Perspectives of CRISPR/Cas-mediated cis-engineering in horticulture: Unlocking the neglected potential for crop improvement. Hortic. Res. 7, 36. doi:10.1038/s41438-020-0258-8
Li, Y., Yue, N., Basit, A., Li, Y., Zhang, D., Qin, L., et al. (2021). Targeted editing of SlMAPK6 using CRISPR/Cas9 technology to promote the development of axillary buds in tomato plants. J. Agric. Sci. 1. doi:10.5539/jas.v13n2p11
Liao, L.-J., Pan, I., Chan, Y.-L., Hsu, Y.-H., Chen, W.-H., and Chan, M.-T. (2004). Transgene silencing in Phalaenopsis expressing the coat protein of Cymbidium Mosaic Virus is a manifestation of RNA-mediated resistance. Mol. Breed. 13, 229–242. doi:10.1023/b:molb.0000022527.68551.30
Lin, Q., Qian, J., Zhao, C., Wang, D., Liu, C., Wang, Z., et al. (2016). Low temperature induced changes in citrate metabolism in ponkan (Citrus reticulata Blanco cv. Ponkan) fruit during maturation. PLoS One 11, e0156703. doi:10.1371/journal.pone.0156703
Lin, Q., Zong, Y., Xue, C., Wang, S., Jin, S., Zhu, Z., et al. (2020). Prime genome editing in rice and wheat. Nat. Biotechnol. 38, 582–585. doi:10.1038/s41587-020-0455-x
Liskova, A., Kubatka, P., Samec, M., Zubor, P., Mlyncek, M., Bielik, T., et al. (2019). Dietary phytochemicals targeting cancer stem cells. Molecules 24, 899. doi:10.3390/molecules24050899
Liu, L., Zhang, J., Xu, J., Li, Y., Guo, L., Wang, Z., et al. (2020). CRISPR/Cas9 targeted mutagenesis of SlLBD40, a lateral organ boundaries domain transcription factor, enhances drought tolerance in tomato. Plant Sci. 301, 110683. doi:10.1016/j.plantsci.2020.110683
Lü, P., Yu, S., Zhu, N., Chen, Y.-R., Zhou, B., Pan, Y., et al. (2018). Genome encode analyses reveal the basis of convergent evolution of fleshy fruit ripening. Nat. plants 4, 784–791. doi:10.1038/s41477-018-0249-z
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi:10.1016/j.molp.2015.04.007
Ma, C., Liu, M., Li, Q., Si, J., Ren, X., and Song, H. (2019). Efficient BoPDS gene editing in cabbage by the CRISPR/Cas9 system. Hortic. Plant J. 5, 164–169. doi:10.1016/j.hpj.2019.04.001
Maeder, M. L., Thibodeau-Beganny, S., Osiak, A., Wright, D. A., Anthony, R. M., Eichtinger, M., et al. (2008). Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell 31, 294–301. doi:10.1016/j.molcel.2008.06.016
Mahas, A., and Mahfouz, M. (2018). Engineering virus resistance via CRISPR–Cas systems. Curr. Opin. Virol. 32, 1–8. doi:10.1016/j.coviro.2018.06.002
Maioli, A., Gianoglio, S., Moglia, A., Acquadro, A., Valentino, D., Milani, A. M., et al. (2020). Simultaneous CRISPR/Cas9 editing of three PPO genes reduces fruit flesh browning in Solanum melongena L. Front. Plant Sci. 11, 607161. doi:10.3389/fpls.2020.607161
Makarova, K. S., and Koonin, E. V. (2015). Annotation and classification of CRISPR-Cas systems. CRISPR, 1311, 47–75. doi:10.1007/978-1-4939-2687-9_4
Makarova, K. S., Wolf, Y. I., Iranzo, J., Shmakov, S. A., Alkhnbashi, O. S., Brouns, S. J. J., et al. (2020). Evolutionary classification of CRISPR–cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83. doi:10.1038/s41579-019-0299-x
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Sci. (80-. ) 339, 823–826. doi:10.1126/science.1232033
Malnoy, M., Viola, R., Jung, M. H., Koo, O. J., Kim, S., Kim, J. S., et al. (2016a). DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 7, 1904. doi:10.3389/fpls.2016.01904
Malnoy, M., Viola, R., Jung, M. H., Koo, O. J., Kim, S., Kim, J. S., et al. (2016b). Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 6, 1–8. doi:10.3389/fpls.2016.01904
Malzahn, A., Lowder, L., and Qi, Y. (2017). Plant genome editing with TALEN and CRISPR. Cell Biosci. 7, 21. doi:10.1186/s13578-017-0148-4
Mamias, S. (2018). “The floriculture supply chain: Characteristics and prospects,” in Lecture presented at the seminar. Supply-chains in the agri-food sector as the UK leaves the EU (Amsterdam, The Netherlands. “
Mao, Y., Zhang, H., Xu, N., Zhang, B., Gou, F., and Zhu, J. K. (2013). Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol. Plant 6, 2008–2011. doi:10.1093/mp/sst121
Mao, Y., Botella, J. R., Liu, Y., and Zhu, J. K. (2019). Gene editing in plants: Progress and challenges. Natl. Sci. Rev. 6, 421–437. doi:10.1093/nsr/nwz005
Martín-Pizarro, C., Triviño, J. C., and Posé, D. (2019). Functional analysis of the TM6 MADS-box gene in the octoploid strawberry by CRISPR/Cas9-directed mutagenesis. J. Exp. Bot. 70, 885–895. doi:10.1093/jxb/ery400
Miki, D., Zhang, W., Zeng, W., Feng, Z., and Zhu, J.-K. (2018). CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation. Nat. Commun. 9, 1967–1969. doi:10.1038/s41467-018-04416-0
Miller, J. C., Tan, S., Qiao, G., Barlow, K. A., Wang, J., Xia, D. F., et al. (2011). A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148. doi:10.1038/nbt.1755
Mishra, R., Mohanty, J. N., Mahanty, B., and Joshi, R. K. (2021). A single transcript CRISPR/Cas9 mediated mutagenesis of CaERF28 confers anthracnose resistance in chilli pepper (Capsicum annuum L.). Planta 254, 5–17. doi:10.1007/s00425-021-03660-x
Modrzejewski, D., Hartung, F., Sprink, T., Krause, D., Kohl, C., and Wilhelm, R. (2019). What is the available evidence for the range of applications of genome-editing as a new tool for plant trait modification and the potential occurrence of associated off-target effects: A systematic map. Environ. Evid. 8, 27–33. doi:10.1186/s13750-019-0171-5
Moon, K. M., Kwon, E. B., Lee, B., and Kim, C. Y. (2020). Recent trends in controlling the enzymatic browning of fruit and vegetable products. Molecules 25, 2754. doi:10.3390/molecules25122754
Naim, F., Dugdale, B., Kleidon, J., Brinin, A., Shand, K., Waterhouse, P., et al. (2018). Gene editing the phytoene desaturase alleles of Cavendish banana using CRISPR/Cas9. Transgenic Res. 27, 451–460. doi:10.1007/s11248-018-0083-0
Nakajima, I., Ban, Y., Azuma, A., Onoue, N., Moriguchi, T., Yamamoto, T., et al. (2017). CRISPR/Cas9-mediated targeted mutagenesis in grape. PLoS One 12, e0177966. doi:10.1371/journal.pone.0177966
Nakayasu, M., Akiyama, R., Lee, H. J., Osakabe, K., Osakabe, Y., Watanabe, B., et al. (2018). Generation of α-solanine-free hairy roots of potato by CRISPR/Cas9 mediated genome editing of the St16DOX gene. Plant Physiol. biochem. 131, 70–77. doi:10.1016/j.plaphy.2018.04.026
Netam, N. (2018). Improving ornamental’s vase life through molecular approaches: A review. J. Pharmacogn. Phytochem. 7, 1687–1691.
Nishihara, M., and Nakatsuka, T. (2011). Genetic engineering of flavonoid pigments to modify flower color in floricultural plants. Biotechnol. Lett. 33, 433–441. doi:10.1007/s10529-010-0461-z
Nishihara, M., Higuchi, A., Watanabe, A., and Tasaki, K. (2018). Application of the CRISPR/Cas9 system for modification of flower color in Torenia fournieri. BMC Plant Biol. 18, 331–339. doi:10.1186/s12870-018-1539-3
Nishitani, C., Hirai, N., Komori, S., Wada, M., Okada, K., Osakabe, K., et al. (2016). Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 6, 31481–31488. doi:10.1038/srep31481
Nonaka, S., Arai, C., Takayama, M., Matsukura, C., and Ezura, H. (2017). Efficient increase of ɣ-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci. Rep. 7, 1–14. doi:10.1038/s41598-017-06400-y
Okuzaki, A., and Toriyama, K. (2004). Chimeric RNA/DNA oligonucleotide-directed gene targeting in rice. Plant Cell Rep. 22, 509–512. doi:10.1007/s00299-003-0698-2
ömerAZABAĞAOĞLU, M. (2018). Investigating fresh fruit and vegetables losses at contemporary food retailers. Sos. Bilim. Araştırma Derg. 7, 55–62.
Ortigosa, A., Gimenez-Ibanez, S., Leonhardt, N., and Solano, R. (2019). Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of Sl JAZ 2. Plant Biotechnol. J. 17, 665–673. doi:10.1111/pbi.13006
Özdemir, A. E., Dündar, Ö., Ertürk, E., and Dilbaz, R. (2003). Bazı yörelerimizde yetiştirilen starking delicious elmalarında derim öncesi ve derim sırasında görülen kayıpların belirlenmesi. IV. Ulus. Bahçe Bitk. Kongresi 8, 166–168.
Pan, C., Sretenovic, S., and Qi, Y. (2021). CRISPR/dCas-mediated transcriptional and epigenetic regulation in plants. Curr. Opin. Plant Biol. 60, 101980. doi:10.1016/j.pbi.2020.101980
Parmar, N., Singh, K. H., Sharma, D., Singh, L., Kumar, P., Nanjundan, J., et al. (2017). Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: A comprehensive review. 3 Biotech. 7, 1–35. doi:10.1007/s13205-017-0870-y
Peng, A., Chen, S., Lei, T., Xu, L., He, Y., Wu, L., et al. (2017). Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 15, 1509–1519. doi:10.1111/pbi.12733
Petolino, J. F. (2015). Genome editing in plants via designed zinc finger nucleases. Vitr. Cell. Dev. Biol. - Plant 51, 1–8. doi:10.1007/s11627-015-9663-3
Porat, R., Lichter, A., Terry, L. A., Harker, R., and Buzby, J. (2018). Postharvest losses of fruit and vegetables during retail and in consumers’ homes: Quantifications, causes, and means of prevention. Postharvest Biol. Technol. 139, 135–149. doi:10.1016/j.postharvbio.2017.11.019
Pranuthi, P., Suseela, T., Swami, D. V., Suneetha, D. R. S., and Vani, V. S. (2018). Effect of packing and storage conditions on physiological loss in weight, diameter of the flower, electrolyte leakage in extending the vase life of carnation cv. Kiro. Int. J. Curr. Microbiol. Appl. Sci. 7, 1278–1287. doi:10.20546/ijcmas.2018.712.158
Qiu, Z., Wang, H., Li, D., Yu, B., Hui, Q., Yan, S., et al. (2019). Identification of candidate HY5-dependent and-independent regulators of anthocyanin biosynthesis in tomato. Plant Cell Physiol. 60, 643–656. doi:10.1093/pcp/pcy236
Rehman, F., Gong, H., Bao, Y., Zeng, S., Huang, H., and Wang, Y. (2022). CRISPR gene editing of major domestication traits accelerating breeding for Solanaceae crops improvement. Plant Mol. Biol. 108, 157–173. doi:10.1007/s11103-021-01229-6
Ren, C., Liu, X., Zhang, Z., Wang, Y., Duan, W., Li, S., et al. (2016). CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay (Vitis vinifera L.). Sci. Rep. 6, 32289. doi:10.1038/srep32289
Ren, F., Ren, C., Zhang, Z., Duan, W., Lecourieux, D., Li, S., et al. (2019). Efficiency optimization of CRISPR/CAS9-mediated targeted mutagenesis in grape. Front. Plant Sci. 10, 612. doi:10.3389/fpls.2019.00612
Rodríguez-Leal, D., Lemmon, Z. H., Man, J., Bartlett, M. E., and Lippman, Z. B. (2017). Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470–480.e8. doi:10.1016/j.cell.2017.08.030
Saiwal, N., Dahiya, M., and Dureja, H. (2019). Nutraceutical insight into vegetables and their potential for nutrition mediated healthcare. Curr. Nutr. Food Sci. 15, 441–453. doi:10.2174/1573401314666180115151107
Sánchez-González, L., Vargas, M., González-Martínez, C., Chiralt, A., and Chafer, M. (2011). Use of essential oils in bioactive edible coatings: A review. Food Eng. Rev. 3, 1–16. doi:10.1007/s12393-010-9031-3
Sander, J. D., Dahlborg, E. J., Goodwin, M. J., Cade, L., Zhang, F., Cifuentes, D., et al. (2011). Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat. Methods 8, 67–69. doi:10.1038/nmeth.1542
Sandhya, D., Jogam, P., Allini, V. R., Abbagani, S., and Alok, A. (2020). The present and potential future methods for delivering CRISPR/Cas9 components in plants. J. Genet. Eng. Biotechnol. 18, 25–11. doi:10.1186/s43141-020-00036-8
Schmidt, C., Pacher, M., and Puchta, H. (2019). “DNA break repair in plants and its application for genome engineering,” in Transgenic plants (Springer), 237–266. doi:10.1007/978-1-4939-8778-8_17
Septembre-Malaterre, A., Remize, F., and Poucheret, P. (2018). Fruits and vegetables, as a source of nutritional compounds and phytochemicals: Changes in bioactive compounds during lactic fermentation. Food Res. Int. 104, 86–99. doi:10.1016/j.foodres.2017.09.031
Shao, X., Wu, S., Dou, T., Zhu, H., Hu, C., Huo, H., et al. (2020). Using CRISPR/Cas9 genome editing system to create MaGA20ox2 gene-modified semi-dwarf banana. Plant Biotechnol. J. 18, 17–19. doi:10.1111/pbi.13216
Sharma, P., and Gayen, D. (2021). Plant protease as regulator and signaling molecule for enhancing environmental stress-tolerance. Plant Cell Rep. 40, 2081–2095. doi:10.1007/s00299-021-02739-9
Sharma, R., and Messar, Y. (2017). Transgenics in ornamental crops: Creating novelties in economically important cut flowers. Curr. Sci. 113, 43–52. doi:10.18520/cs/v113/i01/43-52
Shi, J., Gao, H., Wang, H., Lafitte, H. R., Archibald, R. L., Yang, M., et al. (2017). ARGOS 8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15, 207–216. doi:10.1111/pbi.12603
Shibuya, K., Watanabe, K., and Ono, M. (2018). CRISPR/Cas9-mediated mutagenesis of the EPHEMERAL1 locus that regulates petal senescence in Japanese morning glory. Plant Physiol. biochem. 131, 53–57. doi:10.1016/j.plaphy.2018.04.036
Shimatani, Z., Kashojiya, S., Takayama, M., Terada, R., Arazoe, T., Ishii, H., et al. (2017). Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 35, 441–443. doi:10.1038/nbt.3833
Shipman, E. N., Yu, J., Zhou, J., Albornoz, K., and Beckles, D. M. (2021). Can gene editing reduce postharvest waste and loss of fruit, vegetables, and ornamentals? Hortic. Res. 8, 1. doi:10.1038/s41438-020-00428-4
Silva, G., Poirot, L., Galetto, R., Smith, J., Montoya, G., Duchateau, P., et al. (2011). Meganucleases and other tools for targeted genome engineering: Perspectives and challenges for gene therapy. Curr. Gene Ther. 11, 11–27. doi:10.2174/156652311794520111
Singha, D. L., Das, D., Sarki, Y. N., Chowdhury, N., Sharma, M., Maharana, J., et al. (2022). Harnessing tissue-specific genome editing in plants through CRISPR/Cas system: Current state and future prospects. Planta 255, 28–17. doi:10.1007/s00425-021-03811-0
Su, S., Xiao, W., Guo, W., Yao, X., Xiao, J., Ye, Z., et al. (2017). The CYCLOIDEA–RADIALIS module regulates petal shape and pigmentation, leading to bilateral corolla symmetry in Torenia fournieri (Linderniaceae). New Phytol. 215, 1582–1593. doi:10.1111/nph.14673
Subburaj, S., Chung, S. J., Lee, C., Ryu, S.-M., Kim, D. H., Kim, J.-S., et al. (2016). Site-directed mutagenesis in Petunia× hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep. 35, 1535–1544. doi:10.1007/s00299-016-1937-7
Sun, L., and Kao, T. (2018). CRISPR/Cas9-mediated knockout of PiSSK1 reveals essential role of S-locus F-box protein-containing SCF complexes in recognition of non-self S-RNases during cross-compatible pollination in self-incompatible Petunia inflata. Plant Reprod. 31, 129–143. doi:10.1007/s00497-017-0314-1
Sun, N., and Zhao, H. (2013). Transcription activator-like effector nucleases (TALENs): A highly efficient and versatile tool for genome editing. Biotechnol. Bioeng. 110, 1811–1821. doi:10.1002/bit.24890
Sun, B., Jiang, M., Zheng, H., Jian, Y., Huang, W.-L., Yuan, Q., et al. (2020). Color-related chlorophyll and carotenoid concentrations of Chinese kale can be altered through CRISPR/Cas9 targeted editing of the carotenoid isomerase gene BoaCRTISO. Hortic. Res. 7, 161. doi:10.1038/s41438-020-00379-w
Sweetman, C., Deluc, L. G., Cramer, G. R., Ford, C. M., and Soole, K. L. (2009). Regulation of malate metabolism in grape berry and other developing fruits. Phytochemistry 70, 1329–1344. doi:10.1016/j.phytochem.2009.08.006
Tanaka, Y., Brugliera, F., and Chandler, S. (2009). Recent progress of flower colour modification by biotechnology. Int. J. Mol. Sci. 10, 5350–5369. doi:10.3390/ijms10125350
Tatlıdil, F. F., Dellal, İ., and Bayramoğlu, Z. (2013). Food losses and waste in Turkey. Ctry. Rep. Prep. Natl. Adapt. Plans.
Tian, S., Jiang, L., Gao, Q., Zhang, J., Zong, M., Zhang, H., et al. (2017). Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep. 36, 399–406. doi:10.1007/s00299-016-2089-5
Tikunov, Y. M., Roohanitaziani, R., Meijer-Dekens, F., Molthoff, J., Paulo, J., Finkers, R., et al. (2020). The genetic and functional analysis of flavor in commercial tomato: The FLORAL4 gene underlies a QTL for floral aroma volatiles in tomato fruit. Plant J. 103, 1189–1204. doi:10.1111/tpj.14795
Tiwari, M., Kumar Trivedi, P., and Pandey, A. (2021). Emerging tools and paradigm shift of gene editing in cereals, fruits, and horticultural crops for enhancing nutritional value and food security. Food Energy Secur 10, e258. doi:10.1002/fes3.258
Tong, C., Wu, F., Yuan, Y., Chen, Y., and Lin, C. (2020). High-efficiency CRISPR/Cas-based editing of Phalaenopsis orchid MADS genes. Plant Biotechnol. J. 18, 889–891. doi:10.1111/pbi.13264,
Tripathi, J. N., Ntui, V. O., Ron, M., Muiruri, S. K., Britt, A., and Tripathi, L. (2019). CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun. Biol. 2, 46. doi:10.1038/s42003-019-0288-7
Tripathi, L., Ntui, V. O., and Tripathi, J. N. (2020). CRISPR/Cas9-based genome editing of banana for disease resistance. Curr. Opin. Plant Biol. 56, 118–126. doi:10.1016/j.pbi.2020.05.003
Tripathi, L., Ntui, V. O., Tripathi, J. N., and Kumar, P. L. (2021). Application of CRISPR/Cas for diagnosis and management of viral diseases of banana. Front. Microbiol. 11, 609784. doi:10.3389/fmicb.2020.609784
Wada, N., Ueta, R., Osakabe, Y., and Osakabe, K. (2020). Precision genome editing in plants: State-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol. 20, 1–12. doi:10.1186/s12870-020-02385-5
Wang, Y., Liu, X., Ren, C., Zhong, G.-Y., Yang, L., Li, S., et al. (2016). Identification of genomic sites for CRISPR/Cas9-based genome editing in the Vitis vinifera genome. BMC Plant Biol. 16, 96–97. doi:10.1186/s12870-016-0787-3
Wang, L., Chen, L., Li, R., Zhao, R., Yang, M., Sheng, J., et al. (2017). Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J. Agric. Food Chem. 65, 8674–8682. doi:10.1021/acs.jafc.7b02745
Wang, X., Tu, M., Wang, D., Liu, J., Li, Y., Li, Z., et al. (2018). CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol. J. 16, 844–855. doi:10.1111/pbi.12832
Wang, Z., Wang, S., Li, D., Zhang, Q., Li, L., Zhong, C., et al. (2018). Optimized paired-sgRNA/Cas9 cloning and expression cassette triggers high-efficiency multiplex genome editing in kiwifruit. Plant Biotechnol. J. 16, 1424–1433. doi:10.1111/pbi.12884
Wang, H., Wu, Y., Zhang, Y., Yang, J., Fan, W., Zhang, H., et al. (2019). CRISPR/Cas9-based mutagenesis of starch biosynthetic genes in sweet potato (Ipomoea Batatas) for the improvement of starch quality. Int. J. Mol. Sci. 20, 4702. doi:10.3390/ijms20194702
Watanabe, K., Kobayashi, A., Endo, M., Sage-Ono, K., Toki, S., and Ono, M. (2017). CRISPR/Cas9-mediated mutagenesis of the dihydroflavonol-4-reductase-B (DFR-B) locus in the Japanese morning glory Ipomoea (Pharbitis) nil. Sci. Rep. 7, 10028–10029. doi:10.1038/s41598-017-10715-1
Weinthal, D., Tovkach, A., Zeevi, V., and Tzfira, T. (2010). Genome editing in plant cells by zinc finger nucleases. Trends Plant Sci. 15, 308–321. doi:10.1016/j.tplants.2010.03.001
Wilson, F. M., Harrison, K., Armitage, A. D., Simkin, A. J., and Harrison, R. J. (2019). CRISPR/Cas9-mediated mutagenesis of phytoene desaturase in diploid and octoploid strawberry. Plant Methods 15, 45–13. doi:10.1186/s13007-019-0428-6
Wolter, F., Schindele, P., and Puchta, H. (2019). Plant breeding at the speed of light: The power of CRISPR/cas to generate directed genetic diversity at multiple sites. BMC Plant Biol. 19, 176–178. doi:10.1186/s12870-019-1775-1
Xu, K., Wang, A., and Brown, S. (2012). Genetic characterization of the Ma locus with pH and titratable acidity in apple. Mol. Breed. 30, 899–912. doi:10.1007/s11032-011-9674-7
Xu, J., Kang, B., Naing, A. H., Bae, S., Kim, J., Kim, H., et al. (2020). CRISPR/Cas9-mediated editing of 1-aminocyclopropane-1-carboxylate oxidase1 enhances Petunia flower longevity. Plant Biotechnol. J. 18, 287–297. doi:10.1111/pbi.13197
Yan, R., Wang, Z., Ren, Y., Li, H., Liu, N., and Sun, H. (2019). Establishment of efficient genetic transformation systems and application of CRISPR/Cas9 genome editing technology in Lilium pumilum DC. Fisch. and Lilium longiflorum White Heaven. Int. J. Mol. Sci. 20, 2920. doi:10.3390/ijms20122920
Yan, S., Chen, N., Huang, Z., Li, D., Zhi, J., Yu, B., et al. (2020). Anthocyanin Fruit encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription of SlMYBATV to fine-tune anthocyanin content in tomato fruit. New Phytol. 225, 2048–2063. doi:10.1111/nph.16272
Yasumoto, S., Umemoto, N., Lee, H. J., Nakayasu, M., Sawai, S., Sakuma, T., et al. (2019). Efficient genome engineering using Platinum TALEN in potato. Plant Biotechnol. 36, 167–173. doi:10.5511/plantbiotechnology.19.0805a
Yin, Y., Qin, K., Song, X., Zhang, Q., Zhou, Y., Xia, X., et al. (2018). BZR1 transcription factor regulates heat stress tolerance through FERONIA receptor-like kinase-mediated reactive oxygen species signaling in tomato. Plant Cell Physiol. 59, 2239–2254. doi:10.1093/pcp/pcy146
Yu, Q., Wang, B., Li, N., Tang, Y., Yang, S., Yang, T., et al. (2017). CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Sci. Rep. 7, 11874–11879. doi:10.1038/s41598-017-12262-1
Yu, W., Wang, L., Zhao, R., Sheng, J., Zhang, S., Li, R., et al. (2019). Knockout of SlMAPK3 enhances tolerance to heat stress involving ROS homeostasis in tomato plants. BMC Plant Biol. 19, 1–13. doi:10.1186/s12870-019-1939-z
Zhang, H., Si, X., Ji, X., Fan, R., Liu, J., Chen, K., et al. (2018). Genome editing of upstream open reading frames enables translational control in plants. Nat. Biotechnol. 36, 894–898. doi:10.1038/nbt.4202
Zhang, S., Wang, L., Zhao, R., Yu, W., Li, R., Li, Y., et al. (2018). Knockout of SlMAPK3 reduced disease resistance to Botrytis cinerea in tomato plants. J. Agric. Food Chem. 66, 8949–8956. doi:10.1021/acs.jafc.8b02191
Zhang, Y., Iaffaldano, B., and Qi, Y. (2021). CRISPR ribonucleoprotein-mediated genetic engineering in plants. Plant Commun. 2, 100168. doi:10.1016/j.xplc.2021.100168
Zhou, J., Wang, G., and Liu, Z. (2018). Efficient genome editing of wild strawberry genes, vector development and validation. Plant Biotechnol. J. 16, 1868–1877. doi:10.1111/pbi.12922
Zhu, T., Mettenburg, K., Peterson, D. J., Tagliani, L., and Baszczynski, C. L. (2000). Engineering herbicide-resistant maize using chimeric RNA/DNA oligonucleotides. Nat. Biotechnol. 18, 555–558. doi:10.1038/75435
Ziv, C., and Fallik, E. (2021). Postharvest storage techniques and quality evaluation of fruits and vegetables for reducing food loss. Agronomy 11, 1133. doi:10.3390/agronomy11061133
Zsögön, A., Čermák, T., Naves, E. R., Notini, M. M., Edel, K. H., Weinl, S., et al. (2018). De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 36, 1211–1216. doi:10.1038/nbt.4272
Keywords: CRISPR/Cas9, postharvest loss, fruits, vegetables, gene editing
Citation: Sharma P, Pandey A, Malviya R, Dey S, Karmakar S and Gayen D (2023) Genome editing for improving nutritional quality, post-harvest shelf life and stress tolerance of fruits, vegetables, and ornamentals. Front. Genome Ed. 5:1094965. doi: 10.3389/fgeed.2023.1094965
Received: 10 November 2022; Accepted: 03 February 2023;
Published: 24 February 2023.
Edited by:
Santosh Kumar Gupta, National Institute of Plant Genome Research (NIPGR), IndiaReviewed by:
Aftab Ahmad, University of Agriculture, Faisalabad, PakistanCopyright © 2023 Sharma, Pandey, Malviya, Dey, Karmakar and Gayen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dipak Gayen, ZGlwYWsuZ2F5ZW5AY3VyYWouYWMuaW4=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.