- 1Institute for Zoology, University of Cologne, Cologne, NRW, Germany
- 2Worm∼lab, Institute for Zoology, University of Cologne, Cologne, NRW, Germany
The phylum Nematoda harbors a huge diversity of species in a broad range of ecosystems and habitats. Nematodes share a largely conserved Bauplan but major differences have been found in early developmental processes. The development of the nematode model organism Caenorhabditis elegans has been studied in great detail for decades. These efforts have provided the community with a large number of protocols and methods. Unfortunately, many of these tools are not easily applicable in non-Caenorhabditis nematodes. In recent years it has become clear that many crucial genes in the C. elegans developmental toolkit are absent in other nematode species. It is thus necessary to study the developmental program of other nematode species in detail to understand evolutionary conservation and novelty in the phylum. Panagrolaimus sp. PS1159 is a non-parasitic nematode exhibiting parthenogenetic reproduction and we are establishing the species to comparatively study evolution, biodiversity, and alternative reproduction and survival strategies. Here, we demonstrate the first successful application of the CRISPR/Cas9 system for genome editing in Panagrolaimus sp. PS1159 and the closely related hermaphroditic species Propanagrolaimus sp. JU765 applying the non-homologous end joining and the homology-directed repair (HDR) mechanisms. Using microinjections and modifying published protocols from C. elegans and P. pacificus we induced mutations in the orthologue of unc-22. This resulted in a visible uncoordinated twitching phenotype. We also compared the HDR efficiency following the delivery of different single-stranded oligodeoxynucleotides (ssODNs). Our work will expand the applicability for a wide range of non-model nematodes from across the tree and facilitate functional analysis into the evolution of parthenogenesis, changes in the developmental program of Nematoda, and cryptobiosis.
1 Introduction
Understanding the molecular machinery controlling cellular fate during development of an organism is a major goal in developmental biology. Functional analysis of genes and how they are regulated aids our understanding of how, for example, the body shape is encoded in genomes (Levine and Davidson, 2005).
The phylum Nematoda comprises an estimated 1 million species (Lambshead and Boucher, 2003) occupying a broad range of habitats including terrestrial, marine, and aquatic environments (Holterman et al., 2019). Many species have adopted a parasitic lifestyle, or have evolved survival strategies to deal with extreme cold and desiccation (Shannon et al., 2005; Blaxter and Koutsovoulos, 2015). Throughout the Nematoda, different modes of reproduction are realized. For example, parthenogenesis, a form of asexual reproduction, occurs frequently. Despite their different biotopes and lifestyles, all nematodes share a morphologically similarly structured and largely conserved Bauplan (Poinar, 2011). In contrast to this conserved outcome of development, variations are found in early development on the cellular level with divergent cleavage patterns (Dolinski et al., 2001; Schulze and Schierenberg, 2009). This variability of embryogenesis shows that the patterns and structures observed in a single species, such as the derived nematode model Caenorhabditis elegans, do not allow a generalization across the phylum.
On the genetic level, many aspects of the molecular machinery for development have been studied and are understood in the nematode model system. However, it has recently become clear that many developmentally crucial C. elegans genes are absent in many other nematode species. This includes for example sex determination, axis formation and endo-mesoderm specific genes (Schiffer et al., 2013; Kraus et al., 2017; Schiffer et al., 2018). It is thus necessary to study the functions of genes and their regulation throughout the phylum. Some nematode species have been studied to greater detail, for example Pristionchus pacificus, a species from the same clade as C. elegans (Figure 1), but we lack approachable and informative systems from across the diversity of the tree. We are particularly interested in parthenogenetic nematodes, which initiate development without male input (as is present in C. elegans where sperm entry defines embryonic axis-formation (Goldstein and Hird, 1996)). To understand parthenogenesis as a trait it is necessary to establish new model systems. It is, for example, not present in any of the more than 50 Caenorhabditis species named to date, nor in species close to Pristionchus.
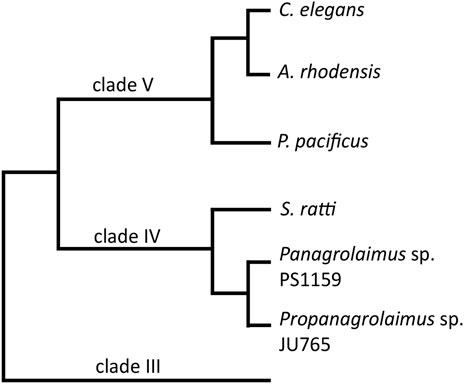
FIGURE 1. A cladogram showing the position of the two main species in this study, Panagrolaimus sp. PS1159 and Propanagrolaimus sp. JU765, in relation to the nematode model organisms C. elegans and P. pacificus, as well as A. rhodensis and S. ratti, species with reportedly successful use of the CRISPR system (modified from Blaxter and Koutsovoulos, 2015; Tandonnet et al., 2018).
In nematodes many species can easily be cultured in the laboratory, and the availability of more high-quality genomes through recent advances in sequencing technology is enabling us to define precise targets for gene knock-out studies. Unfortunately, many methods and protocols established in the model organisms do not work out-of-the-box in non-Caenorhabditis nematodes and organisms with less background on the genetic level. For example, RNAi has been used in a range of nematodes (Shannon et al., 2008; Ratnappan et al., 2016; Adams et al., 2019; Dulovic and Streit, 2019), but very often shows issues in efficiency and delivery (Félix, 2008; Nuez and Félix, 2012). Nematodes are often insensitive to this method and important genes of the pathway are missing (Koutsovoulos, personal communication). In our hands, RNAi by feeding did not result in reliable phenotypes in different parthenogenetic roundworms, e.g., Diploscapter coronatus, Acrobeloides nanus and Panagrolaimus sp. PS1159. The CRISPR technology promises to be more reliable, delivering consistent results across a broader range of species and thus opening new opportunities for functional analysis in many nematodes.
CRISPR/Cas9 is a powerful experimental tool for gene-editing (Wang et al., 2016). The simple design, its high efficiency, and low cost have made it the go-to-method in non-model organisms in recent years (Dickinson et al., 2020). The most important and widely adapted CRISPR/Cas application for gene editing is the type II system, which relies on one protein only (Makarova et al., 2011). The Streptococcus pyogenes Cas9 requires two small RNAs to build a ribonucleoprotein complex (RNP): the CRISPR RNA (crRNA) with a 20 bp guide sequence and its downstream protospacer-adjacent motif (PAM; NGG) that determines the target, and the universal trans-activating CRISPR RNA (tracrRNA). The crRNA binds to the DNA target by base-pair binding. Cas9 recognizes the PAM sequence, leading to cleavage of both strands 3 nucleotides away from the PAM (Doudna and Charpentier, 2014), followed by cellular repair mechanisms. The Non-homologous End Joining (NHEJ) pathway repairs the double-strand break (DSB) by ligating the ends. This mechanism sometimes results in the loss or gain of nucleotides (Davis and Chen, 2013), a frame shift, premature stop and therefore creating knock-out mutations. The homology-directed repair (HDR) is a more accurate system, in which user-provided DNA templates with homology arms serve as a guide to precisely repair the DSB (Mali et al., 2013; Paix et al., 2017) generating knock-in mutations. Cas9 can be delivered to the target DNA in various ways, for example via expression plasmids, or as a purified protein (Lino et al., 2018). It was shown that direct injection of the RNP complex into the adult gonad is more efficient and less time consuming than injection of plasmids (Paix et al., 2015). Over the past years, CRISPR/Cas9 was successfully applied in C. elegans. But the system has been proven to be useful also in non-model nematodes, such as C. briggsae (Culp et al., 2020), P. pacificus (Witte et al., 2015), the human- and rat-parasitic worms S. stercoralis and S. ratti (Gang et al., 2017), Auanema (Adams et al., 2019) and O. tipulae (Vargas-Velazquez et al., 2019).
Here we report the successful and reproducible use of microinjection and CRISPR/Cas9 in two new nematode strains from clade IV (Figure 1), the parthenogenetic strain Panagrolaimus sp. PS1159 and the hermaphroditic species Propanagrolaimus sp. JU765. We also provide an optimization of previously published protocols using modified microinjection needles, different target regions as well as temperature and concentration adjustment during the RNP complex assembly. Additionally, we compare the HDR efficiency following the delivery of different ssODNs.
2 Results
2.1 Microinjection in Panagrolaimus sp. PS1159
In nematodes CRISPR/Cas9 components are delivered through injection into the adult gonad. We found the cuticle in Panagrolaimus to be more robust and harder to penetrate than in C. elegans. At the same time the gonad appeared to be less tolerant to the injury resulting by the needles used for C. elegans. Thus, for successful microinjection in Panagrolaimus sp. PS1159 we adapted the C. elegans microinjection protocol (Evans, 2006) accordingly (Supplementary Video SV1).
Panagrolaimus worms were also less likely to stick on the 2% agarose pads. We therefore used higher concentrations of 2.5% agarose pads for good adhesion of the worms. To prepare injection needles using a puller (Sutter Instruments, P-2000), we use a 1-lined so called “bee stinger” program (Oesterle, 2018) as a basis for a 3-lined program (Table 1), producing sharper needles with a short taper. With these modifications we were able to easily inject C. elegans, P. pacificus, PS1159, and JU765 with a high survival rate.

TABLE 1. Needle puller P-2000 (A) One-line program “bee stinger needles” B) newly written 3-line program for our microinjection needles.
2.2 Mutation of unc-22 in PS1159
We targeted the PS1159 orthologue of the C. elegans unc-22 gene, which was previously successfully mutagenized by CRISPR/Cas9 in C. elegans (Kim et al., 2014), and the non-model nematodes S. stercoralis, and S. ratti (Gang et al., 2017). unc-22 encodes Twitchin, a large protein, which is involved in the regulation of muscle contraction (Moerman et al., 1988). Mutation of unc-22 in C. elegans results in motility defects and a twitching phenotype, which is easily detectable under the dissecting microscope.
2.2.1 Identification of the PS1159 unc-22 orthologue
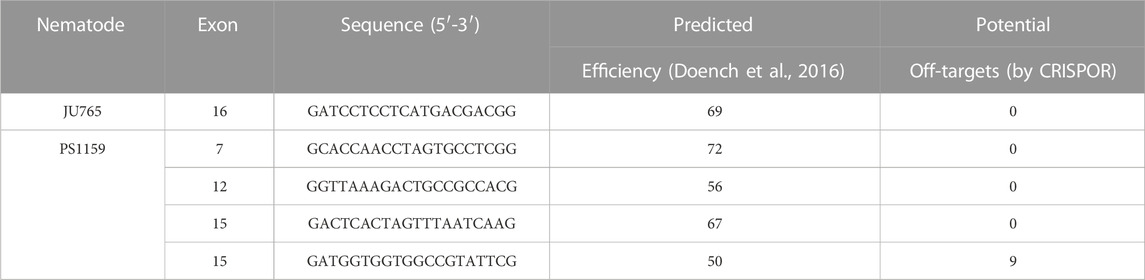
The Twitchin protein (UNC-22, isoform a) of C. elegans was downloaded from www.wormbase.org (WormBase ID CE33017) and blasted against the PS1159 genome (PRJEB32708) using the TBLASTN tool. A potential orthologue with 62.9% sequence similarity was selected (“PS1159_v2.g4256”). Reciprocal BLASTN against the C. elegans genome confirmed the C. elegans unc-22 gene as the best hit indicating the gene found is an unc-22 orthologue. Target guide RNA sequences, preferably following the form G(N)G(NGG) were searched for in the exonic regions of the unc-22 orthologue in our PS1159 genome assembly (Schiffer et al., 2019) using CRISPOR (Figure 2A; Concordet and Haeussler, 2018). Guide sequences with a GG motif at the 3′ end have been observed to have a higher cleavage efficiency in C. elegans (Farboud and Meyer, 2015). BLASTN of the chosen guide sequence revealed no significant off-target sequences in the entire genome. Two pairs of repair templates with homology arms of either 36 nt on both sides or 96 and 92 nt flanking the predicted Cas9 cleavage site with a built-in mutation and restriction enzyme XbaI site were produced (Figure 2A).
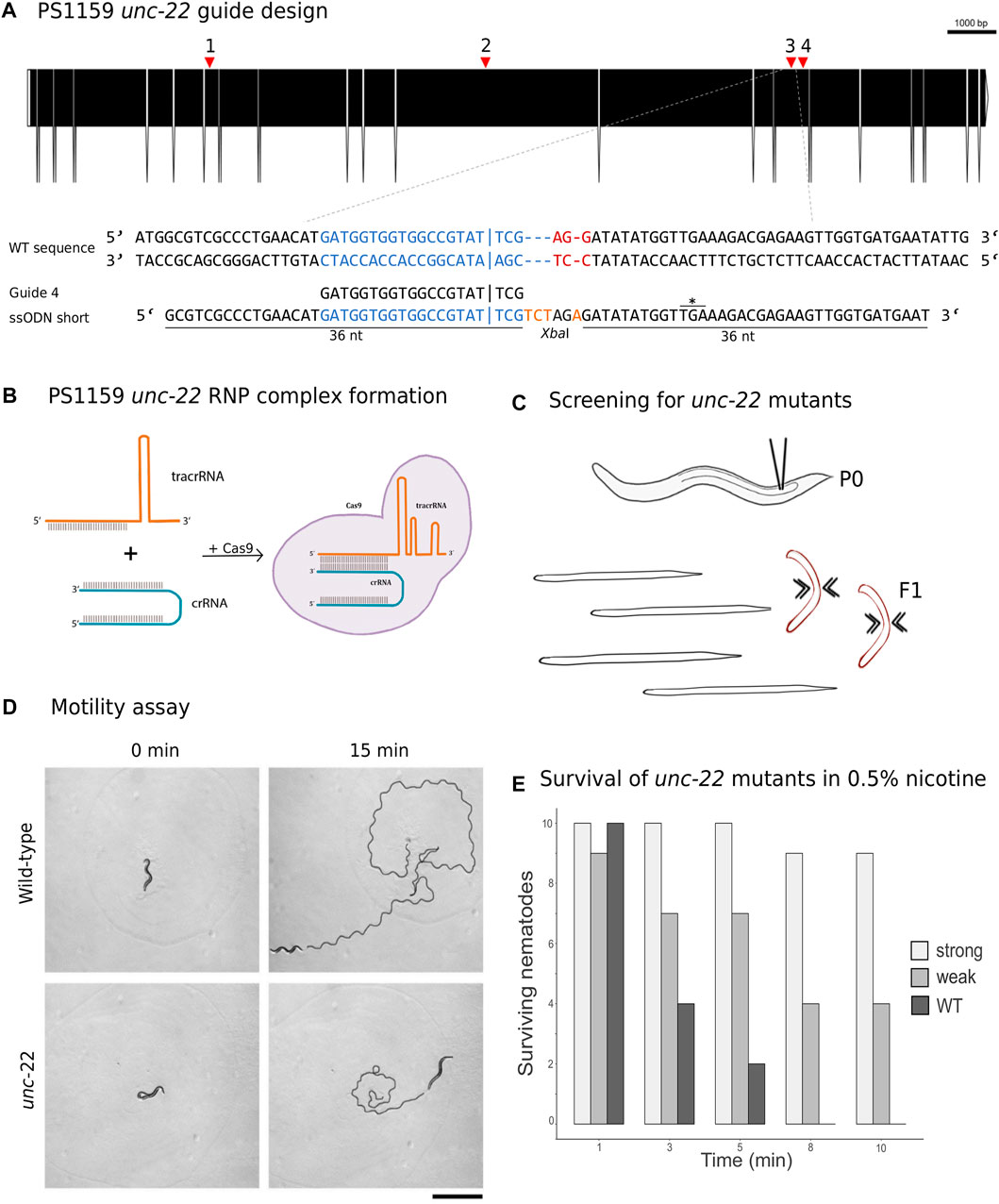
FIGURE 2. (A) CRISPR target site for Panagrolaimus sp. PS1159 unc-22. Predicted gene structure. Black boxes = exons. Red arrows = CRISPR target regions. Zoom into part of exon 15 coding strand with target region (blue) and PAM site (red). Vertical bar indicates the expected Cas9 cleavage site. Example of guide sequence and single-stranded oligodeoxynucleotide (ssODN) homology arms (36 nt) flanking the cleavage site on both sides and insertion of a XbaI cleavage site (orange). Asterisk shows in frame stop codon after successful incorporation of desired bases. Gene figure was designed by using the Exon-Intron Graphic Maker (version 4, http://wormweb.org/exonintron). (B) Ribonucleoprotein complex assembly. Cas9 protein, crRNA and tracrRNA are mixed and incubated. (C) RNP complexes are introduced into the syncytial part of the gonad of an adult worm (P0) by microinjection. Screen for twitching phenotype in F1 generation. (D) Motility assay: Observation of motility of wild-type worms vs strong twitching F1 in the time course of 15 min (n = 10); black line shows movement; scale bar = 1 mm. (E) Survival assay: Wild type worms and F1 progeny after injection with a phenotype (strong and weak) are screened in 0.5% nicotine for a total of 10 min for survival (n = 10 for each).
2.2.2 CRISPR-Cas9 targeting of PS1159 unc-22 causes a twitching phenotype
CRISPR/Cas9 components were assembled (Figure 2B) and introduced into the syncytial part of the gonad of PS1159 following the protocol of Paix et al. (2017). After injections, all worms were recovered on a single agar plate. After 12 days (generation time is 8–10 days in Panagrolaimus; Schiffer et al., 2019), 16 F1 animals showing reduced crawling activity and a twitching phenotype in water were transferred into single drops of Plectus nematode growth medium (PNGG; Supplementary Table S1). We were able to isolate four different twitchers with varying intensity of their twitching behavior, most likely indicating weak (C-shaped bending to the left and right side with twitching observable in head and tail region in liquid) and strong (no bending to both sides, strong twitching through whole body) phenotypes (Supplementary Video SV1). This phenotype was visible and consistent in future generations of the isolated worms. We did not observe any non-twitching phenotypes in the population of these generations. The motility on agar plates of strong twitchers was monitored and compared to wild-type worms. The movement of twitching worms appeared uncoordinated. In a comparative assay of 10 wild type worms versus 10 strong twitchers, wild type nematodes appeared to have moved a longer distance after 15 min, indicating reduced motility in mutants (Figure 2D).
In C. elegans, wild-type worms stop moving and eventually die under exposure of nicotinic acetylcholine receptor agonists, such as levamisole or nicotine. C. elegans unc-22 mutants are resistant to these poisons and twitching is enhanced by exposure (Lewis et al., 1980; Moerman et al., 1988). We tested exposure of PS1159 wild-type and putative CRISPR-induced unc-22 mutants to both, levamisole (1 mM, not shown) and 0.5% nicotine. 10 wild-type animals and 10 of each twitching phenotypes were analyzed (Figures 2C, E; Supplementary Video SV1). After 10 min all worms were placed in M9 buffer. No wild-type animals survived the procedure. By contrast, 9/10 strong twitchers, and 5/10 weak twitchers survived, suggesting that like in C. elegans wild-type PS1159 are not resistant to this poison and that the resistance of worms with different twitching phenotypes seems unequal. For all future experiments the progeny of injected worms were screened in water or, if confirmation was needed, additionally in 0.5% nicotine for 2–3 min, ensuring not to lose possible mutants. We also tested wild-type PS1159 worms from a mixed population plate (no differentiation between P0s or F generations) for spontaneous twitching behavior in water and 1% nicotine for 10 min to observe if this phenotype might occur in wild-type animals. 0/1300 of wild-type worms displayed a twitching phenotype. Observing 3 P0s (not injected) and their 135 F1 progeny revealed no uncoordinated phenotype. When performing the injection experiment without crRNA (11 P0s and 109 F1 animals) or without Cas9 (10 P0s and 297 F1s) no twitching animals were detected (Supplementary Table S4). Based on these results we did not expect spontaneous twitching or twitching caused by the injection with missing components of the complex.
2.3 Development of an optimal cultivation method after injections for PS1159
The transfer of single worms onto an agar plate, as routinely done for C. elegans and P. pacificus, was not optimal for Panagrolaimus sp. PS1159, because the worms frequently crawled out of the agar to the walls of the plates and dried out. To find the optimal way of cultivating the worms after microinjections, different approaches were taken. Injected worms were transferred either 1) into single PNGG drops on 12-well plates in a moist chamber individually, 2) all individuals onto a single agar plate or 3) onto single plates with each worm covered by a slice of agar. Although surviving the transfer into single PNGG drops, worms could not be screened properly because of strong bacterial growth in the drops. When worms were placed on a plate altogether, they remained on the plate but analysis of F1s from individual P0s was impossible. In the latter approach, almost all worms remained under the agar pad and laid eggs. Occasionally some worms still left and dried out, but the advantage of this method outweighed the loss of single individuals. However, since the reproduction cycle of PS1159 is much longer than in C. elegans and P. pacificus, mothers were kept at 25°C for at least 5–7 days on the plates, allowing them to lay more eggs.
2.4 Modification of the CRISPR mix preparation increases efficiency
During the first experiments, we were only able to isolate a few worms with a twitching phenotype and injection results were thus unsatisfactory and not consistent. After the development of the optimal cultivation strategy and the possibility to screen progeny of individual injected P0s, we aimed to adapt the protocol of the CRISPR injection mix to yield a higher efficiency.
2.4.1 Using a modified crRNA dramatically increases efficiency
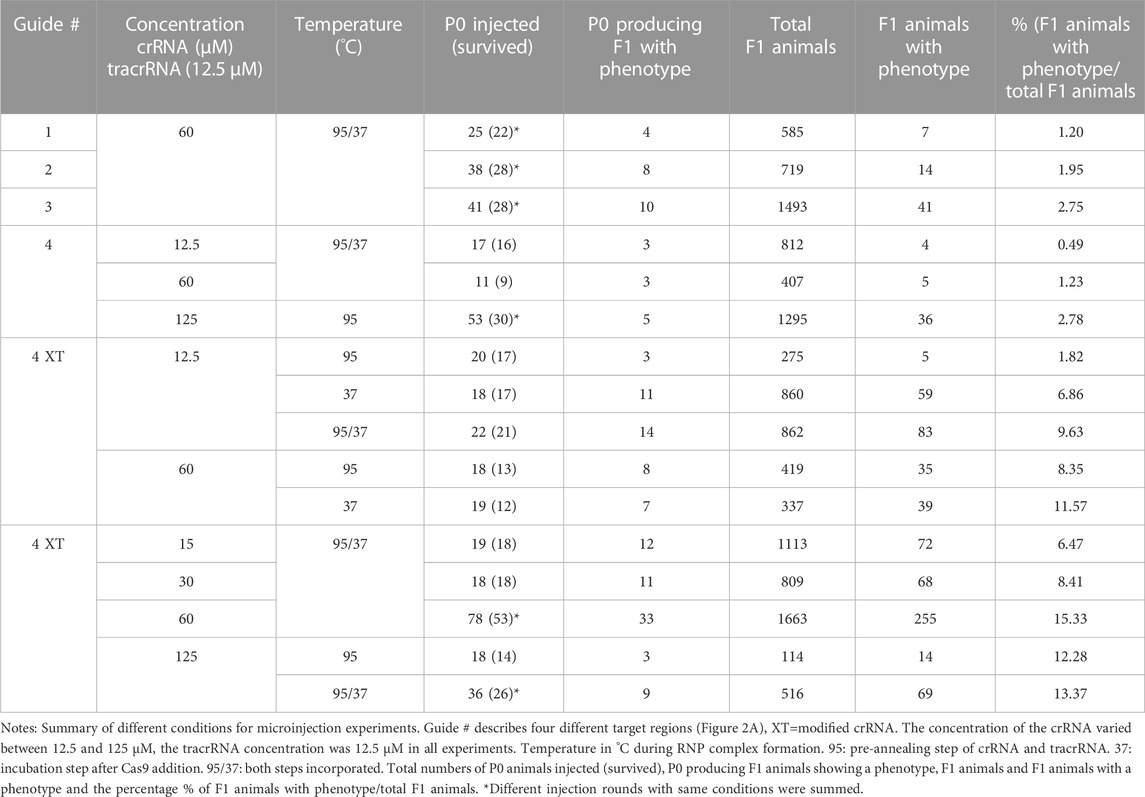
To examine if the genomic region of the double strand break affects the editing success, we used four guides (60 µM) targeting different regions in the gene (Figure 2A; Table 2). We injected guide 1 for the first target into a total of 25 P0s. 22 worms survived and produced a total of 585 F1 progeny. 7 F1s showed a twitching phenotype (1.2%, Table 3). Guide 2 was used in a total of 38 injected (28 survived) P0s producing 14/719 F1 animals with a phenotype (1.95%, Table 3). Using guide 3 in a total of 41 (28 survived) P0s led to 2.75% (41/1493 F1s) twitching animals. 11 P0s were injected using guide 4, resulting in 5/407 (1.23%) twitching F1s. Only minor variations were found using different target regions. But when using a modified crRNA from IDT (Alt-R CRISPR-Cas9 crRNA XT) for guide 4 (4 XT), which contains additional chemical modifications and providing an increased stability the efficiency rose dramatically to 15.33% (Table 3) with a total of 78 injected (53 survived) P0s and 255/1663 F1s showing an uncoordinated phenotype.
2.4.2 Changes of temperature optimizes complex formation
In Pristionchus pacificus the CRISPR mix protocol differs from the one used in C. elegans (Paix et al., 2017) by an additional pre-annealing step of the tracrRNA and crRNA (James W. Lightfoot, personal communication). We tested if the incubation temperature has an impact on complex formation in PS1159. Pre-annealing the crRNA (60 µM) and tracrRNA at 95°C for 5 min (Hiraga et al., 2021) and injecting 18 P0s resulted in 35/419 F1s (8.25%) of the progeny with a twitching phenotype. By contrast, when we replaced the 95°C annealing step with a 37°C incubation step for 10 min after adding the Cas9 (19 P0s), we obtained a twitching phenotype in 11.57% of the offspring (39/337 F1s). When we incorporated both the 95°C annealing step and the 37°C incubation step, the percentage of F1s with the phenotype increased to a mean of 15.33% (255/1663 F1s, Table 3).
2.4.3 Guide RNA concentration determines effect-size of gene editing
We then asked how the concentration of the crRNA affected the mutation frequency. We therefore injected the modified guide 4 XT (with the pre-annealing and incubation steps during complex assembly) and used concentrations of 12.5 µM resulting in 83/862 twitching F1s (9.63%), 15 µM (72/1113 F1s, 6.47%), 30 µM (68/809 F1s, 8.41%), 60 µM (255/1663 F1s, 15.33%), and 125 µM (69/516 F1s, 12.93%, Table 3), and discovered that increasing the crRNA concentration up to 60 µM improved the efficiency rate, but a concentration of 125 µM resulted in no further improvements (Table 3). After these findings we decided that our future experiments would be performed with that optimal concentration of 60 µM.
2.4.4 The optimal time-window for screening is during the first 48 h after injection
After establishing the enhanced protocol for the injection mix, we further investigated the optimal time window for observing twitching phenotypes putatively induced by CRISPR knock-outs in PS1159. After microinjections, 15 P0s were isolated onto single agar plates with a slice of agar allowing them to lay eggs and transferred them every 24 h for a total of 6 days (Figure 3A). The F1 progeny on every plate was screened after 5–7 days (adult stage) for a twitching phenotype. We found that in eggs laid in the first 24 h the average percentage of phenotypic adult animals was 25% and then dropped to 8.5% in the progeny laid between 24 and 48 h after injection (Figure 3B). After 72 h, there was still an average of 7.5% and then 0%–1.2% in the following 3 days. Based on these results we concluded that the preferred time-window to screen for phenotypes is 48 h post injection after considering the feasibility and simplicity of maintenance of the progeny and achieving mutations arising from a higher number of P0s. We thus discarded the injected P0 worms after that time.
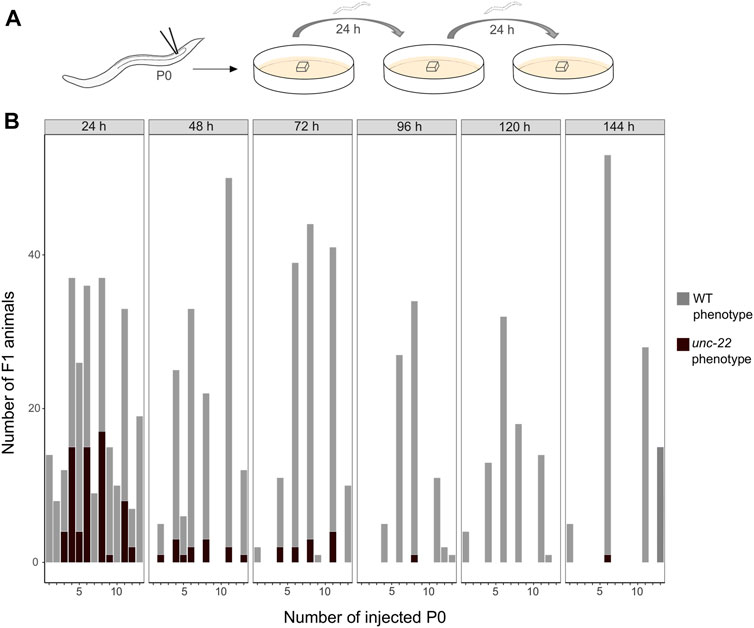
FIGURE 3. (A) Scheme of experimental workflow. Microinjection of adult P0 into gonad. Every 24 h individual worms are transferred onto fresh agar plates covered by a slice of agar. (B) Number of F1 animals with wild-type (light grey) and twitching (dark grey) phenotype per injected P0 (15 worms were injected in total) in time intervals of 24 h for a total of 6 days.
2.5 Sequencing verifies unc-22 mutation after double strand break within the target site
We aimed to molecularly characterize any mutations produced by gRNA/Cas9 injection and therefore amplified and sequenced the target region of 35 twitching and 15 non-twitching F1 animals using primers located 328 bp upstream and 454 bp downstream of the PAM (guide 4). Sanger sequencing of the PCR product confirmed different types of mutations near the cleavage site in 27 twitching individuals (Figure 4A for an example). To quantify and identify the genotypes from the trace data, we used the ICE tool from Synthego (Conant et al., 2022; Figure 4B). Panagrolaimus species are triploid parthenogenetic hybrids and the genomic implications of this mode of reproduction are not fully understood (Schiffer et al., 2019). Currently, we assume that offspring are clones, and all mutations are propagated in all subsequent generations (apart from random losses). Triploidy in Panagrolaimus likely arose from a hybridization event (Schiffer et al., 2019). Thus, the terms homo- and heterozygosity are not appropriate. We therefore will use the term “homeolog” to denote the different copies of the same gene in the triploid framework (Glover et al., 2016). To determine the exact location of the mutations, we cloned and subsequent sequenced each homeolog of one representative. The results were in accordance with the prediction from the Synthego Analysis tool (Figure 4C).
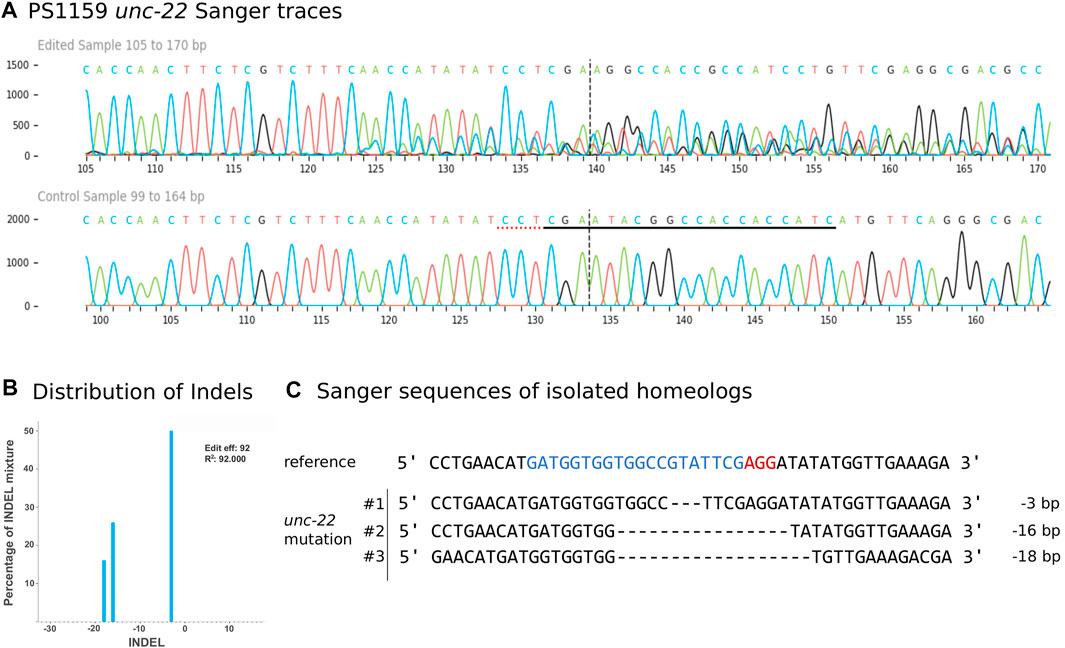
FIGURE 4. (A) Panagrolaimus sp. PS1159 unc-22 Sanger Sequencing traces after PCR of edited and wild-type (control) in the region around the guide sequence using the Synthego Performance Analysis, ICE Analysis. 2019. V3.0. Synthego. Black underlined region shows the guide sequence. Red underline represents the PAM site. Vertical dotted line shows expected cut site. (B) Predicted distribution and percentage of Indels in the entire population of genomes using ICE. (C) Sanger sequencing reveals deletions in isolated homeologs #1-#3 after DNA cloning. Target region (blue) and PAM site (red).
In 8 cases, after amplification only the WT sequences were observed even though a clear phenotype was visible, suggesting a deletion of at least one primer site. We attempted to localize the exact site of the deletion by increasing the PCR amplification region for four animals to up to 6,000 bp and used different primer combinations. For one animal (T4), we recovered both a ∼6,000 bp product and a ∼3,000 bp product (Supplementary Figure S1A). When we performed PCR with the reverse primer only, we also recovered a ∼3,000 bp product. We sequenced this product and found a deletion incorporating both the target and forward primer binding site. Furthermore, we found an inverted copy of the reverse primer site (Supplementary Figure S1C), explaining why we obtained a PCR product using reverse primer alone (Supplementary Figure S1A). In total, of 50 worms genotyped, 28/35 worms with the twitching phenotype had a corresponding mutation in the unc-22 gene. No mutations were detected in animals with a wild-type phenotype (0/15 worms).
2.6 Longer homology arms with modification increase efficiency
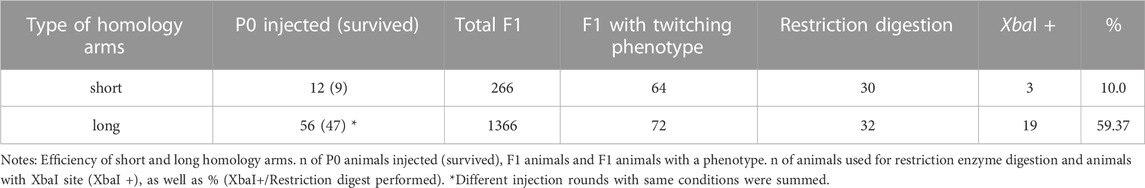
To simplify the detection of Cas9-induced mutations, we used homology arms with a built-in insert containing a recognition site for the restriction enzyme XbaI, which additionally delivers an in frame stop, thus allowing us to easily check for gene editing in the progeny with a phenotype via restriction digestion (Figure 5A). Restriction digestion was successful and Sanger sequencing after DNA cloning confirmed the desired insertion in at least one homeolog (Figure 5B for one representative). We designed longer homology arms (97/92 nt) with an additional incorporated phosphorothioate (PS) bond modification and observed the effect of knock-in efficiency when compared to the results of injections with 36 nt arms flanking the target site. We performed PCR and restriction digestion on 30/64 F1s that showed the twitching phenotype after injection of short (36 nt) homology arms and on 32/72 twitching animals with long (97/92 nt) homology arms. 10% of the twitching animals had the desired knock-in after injection of short arms (3 out of 30 worms), whereas 59.37% of the twitching animals had the desired knock-in after injection of the longer arms (19 of a total of 32 worms, Table 4).
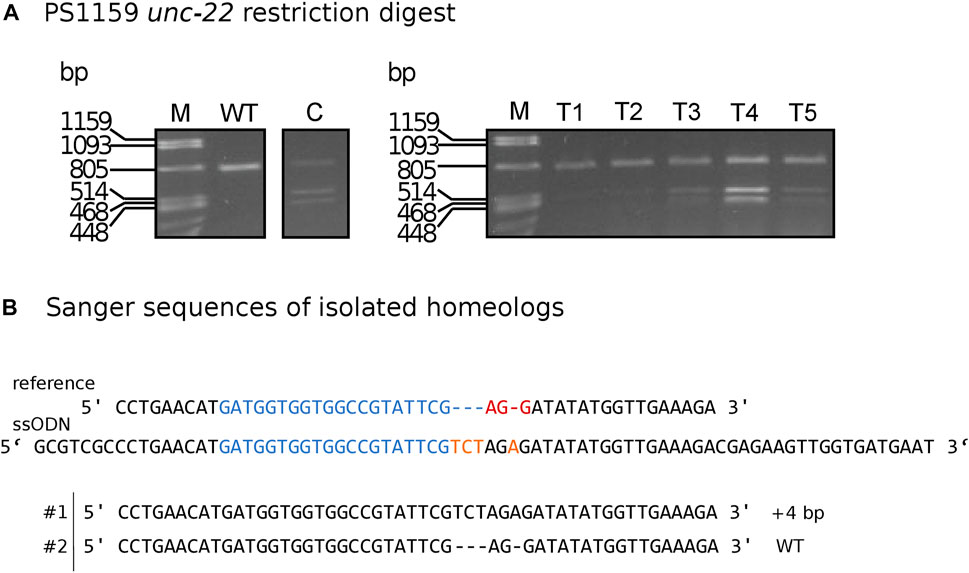
FIGURE 5. (A) Panagrolaimus sp. PS1159 unc-22: Representative examples of agarose gel electrophoresis after restriction digest with XbaI of knock-in experiments. Multiple band patterns of mutants with desired insertion. M = Marker Lambda PstI digest. C = positive control of approved knock-in positive F1. T1 - T5 = twitchers 1-5. (B) Sanger sequencing revealing insertion in one homeolog (#1) after DNA cloning of mutant after positive restriction digest. Wild-type sequence in homeolog #2. Target region (blue) and PAM site (red). XbaI cleavage site in dark yellow.
2.7 The same protocol can be applied in Propanagrolaimus sp. JU765
Following our improved protocol for PS1159, we asked whether we could transfer the injection mixture assembly and microinjection strategy to modify the genome of other non-model nematodes. We hence aimed to induce mutations in the Propanagrolaimus sp. JU765 orthologue of unc-22 (Figure 6A). After successful injections of 13 individuals, we screened the F1 progeny after 7 days and recovered 30.7% with a twitching phenotype (total F1 = 501) which could be observed in the future generations as well. To verify that the phenotype arose from a CRISPR induced knock-out, we amplified the target region of 19 animals (out of 154 twitching worms) with primers 437 bp upstream and 416 bp downstream of the PAM and performed a T7EI assay. The T7EI results indicate that 3 of the worms harbored a mutation within the region (Figure 6B). We identified the predicted position of indels of these 3 representatives with Sanger Sequencing and ICE analysis tool (Figure 6C). This result supports that we provide an effective protocol that might be useful in a variety of non-model species in the phylum Nematoda.
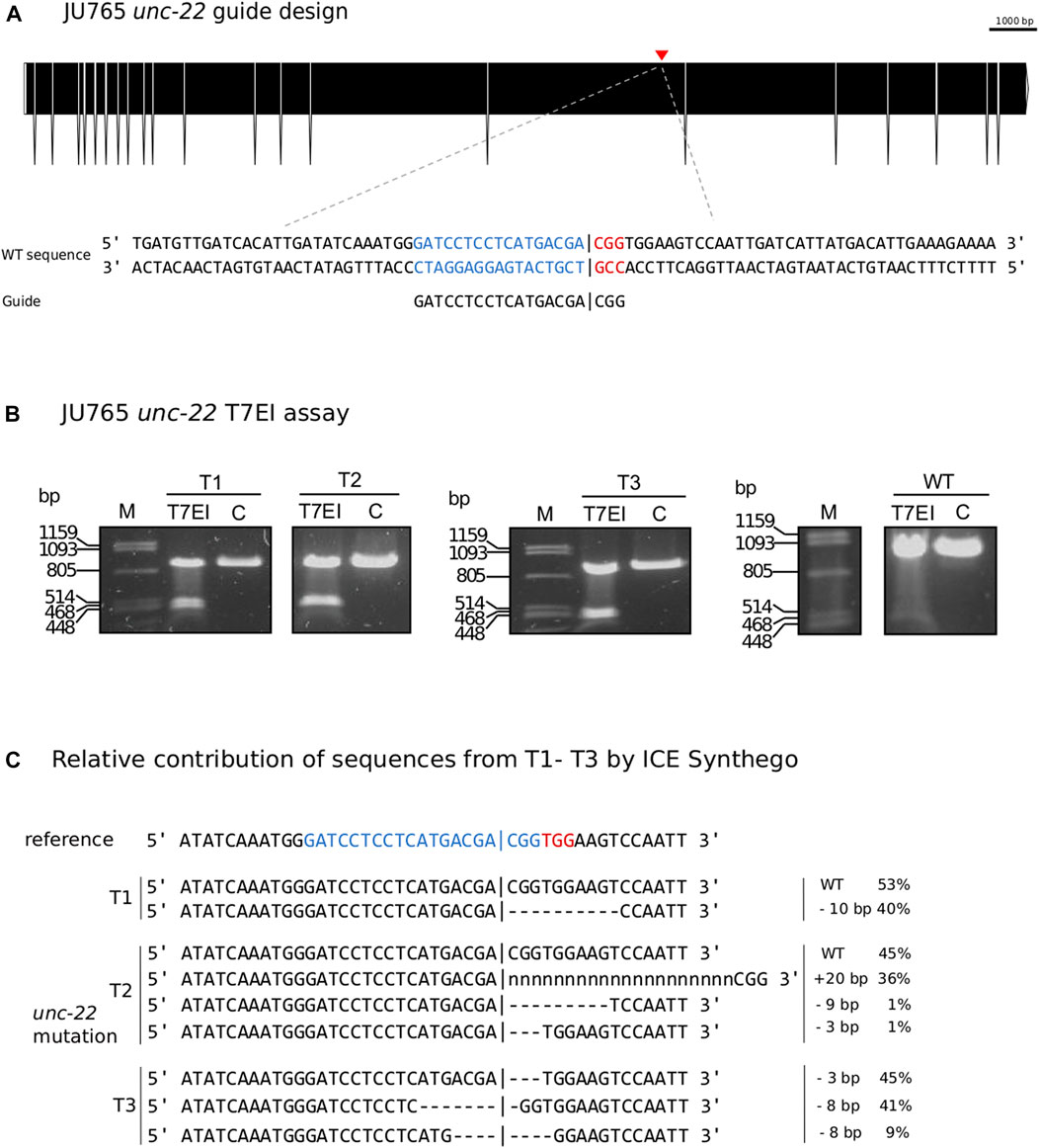
FIGURE 6. (A) CRISPR target site for Propanagrolaimus sp. JU765 unc-22. Predicted gene structure. Black boxes = exons. Red arrow = CRISPR target region. Zoom into part of exon 16 coding strand with target region (blue) and PAM site (red). Vertical bar indicates the expected Cas9 cleavage site. (B) Representative examples of 3 individuals T1-T3. Agarose gel electrophoresis of T7EI assay after knock-out experiments. Multiple band patterns detect indels. M = Marker Lambda PstI digest. C = negative control. T1 - T3 = twitchers 1-3. (C) Relative contribution of inferred sanger sequences in edited T1-T3 (PCR product) confirms indels in the target region. Target region (blue) and complementary PAM site (red). Vertical bar indicates the expected (actual) Cas9 cleavage site. Dashes indicate deletions and ‘n’ indicates insertions. The number of inserted (+) or deleted (−) nucleotides are indicated on the right with the proportion of that sequence inferred in the pool. Prediction produced using the Synthego Performance Analysis, ICE Analysis. 2019. v3.0. Synthego.
3 Discussion
CRISPR/Cas9 has become an indispensable tool for precise gene editing in different eukaryotes, and has revolutionized genetics in the past years in many model organisms (Chang et al., 2013; Cho et al., 2013; Cong et al., 2013; Miki et al., 2021). In this study, we demonstrate the successful and efficient application of the CRISPR/Cas9 system in the nematodes Panagrolaimus sp. PS1159 and Propanagrolaimus sp. JU765 by generating heritable unc-22 mutations. We further optimized the protocol for a higher cleavage efficiency in our nematode species.
Microinjection is a useful tool to introduce genetic material into an adult worm. In C. elegans this method has long been used for different approaches, including RNAi and CRISPR/Cas9 (Mello and Fire, 1995; Cinkornpumin and Hong, 2011; Conte et al., 2015; Ghanta et al., 2021) and has successfully been used in some non-model nematodes as well (Ratnappan et al., 2016; Adams et al., 2019). Because of differences in the cuticle and gonad, the published protocols for C. elegans did not yield results in Panagrolaimus sp. PS1159. We therefore adapted existing protocols for the microinjection pads and needles. With these changes it was possible to inject not only PS1159, but also C. elegans, P. pacificus, and Propanagrolaimus sp. JU765 with high survival rates.
We generated knock-out mutations in the PS1159 and JU765 orthologues of the C. elegans unc-22 gene, which resulted in a visible uncoordinated twitching phenotype with reduced motility increased by nicotine exposure. This phenotype is consistent with what has been observed in C. elegans and S. stercoralis. To determine if the phenotype was caused by mutations in the desired target, we recovered and isolated single worms with a phenotype. We amplified, cloned, and sequenced the regions spanning the predicted sites. We were able to identify indels showing that the genotypes were consistent with the observed phenotypes. However, in some cases after amplification we only observed wild-type sequences, which was in line with previous results from knock-out experiments in the nematodes Strongyloides and Pristionchus (Witte et al., 2015; Gang et al., 2017). We took a closer look and found a deletion of the guide sequence and an inversion of the reverse primer site, removing the forward primer site in one individual. It has been demonstrated in different organisms that repair of cleavage induced by Cas9 can lead to large deletions and genomic rearrangements using one or two single guide RNAs (Shin et al., 2017; Kosicki et al., 2018) and should therefore be considered as a possible outcome. To create knock-in mutations of small insertions we provided single-stranded homology arms on both sites. We were able to achieve even higher knock-in efficiency when using longer homology arms with a protection of the donor DNA with phosphorothioate modification. This result is in accordance with previous findings regarding optimized HDR donors (Liang et al., 2017; Schubert et al., 2021).
We found that best results can be obtained when using a more stable, modified version of a crRNA with an additional pre-annealing step with the tracrRNA before incubation with the Cas9 protein. It has previously been reported that certain chemical alterations on terminal ends can lead to increased metabolic stability of guide RNA and subsequent higher CRISPR/Cas9 efficiency (Hendel et al., 2015; Allen et al., 2021). Increasing the concentration of the modified crRNA resulted in an increase from ∼1-2% to up to 15%. Moreover, we observed the editing efficiency in the timeframe of 1 week and counted the frequency of mutations every 24 h, and our analysis indicates that the CRISPR effect in PS1159 performs at its best in eggs laid within the first 48 h post injection, leading to even up to 25% cleavage efficiency. In Pristionchus pacificus it has been reported that most mutations arose in eggs laid in the first 9 h after injections (Witte et al., 2015).
In summary, testing and adapting experimental conditions, such as needle size, temperature, and concentration of the RNP complex components during preparation as well as using modified reagents is necessary for successfully applying microinjections and CRISPR/Cas9 in divergent nematode species. Our work will facilitate functional analysis into the evolution of parthenogenesis, changes in the developmental program of Nematoda, and cryptobiosis. The unc-22 target might be a useful and easily identifiable co-marker for future functional analysis, in particular for large insertions of fluorescent markers in Panagrolaimidae, as it has been used in C. elegans (Arribere et al., 2014; Kim et al., 2014) routinely.
4 Materials and methods
4.1 Nematodes and maintenance
All experiments have been performed using the parthenogenetic Panagrolaimus sp. PS1159 and the hermaphroditic Propanagrolaimus sp. JU765, cultured on 55 mm diameter petri dishes at 20°C with low-salt agar (Lahl et al., 2003) seeded with Escherichia coli strain OP50 as a food source (Brenner, 1974). After injections worms were kept at 25°C (20°C for experiments with older protocol) either on single drops of 15 µL Plectus nematode growth medium (PNGG, Supplementary Table S1; 30 µL OP50 in 1,000 µL PNGG) on 12-well plates in a moist chamber, or on single agar plates covered with a slice of agar.
4.2 Selection of guide RNA targets
The C. elegans sequences of UNC-22 were downloaded from Wormbase (http://www.wormbase.org/db/get?name=CE33017;class=PROTEIN). TBLASTN and BLASTN tools (Altschul et al., 1990) were used to identify orthologues for PS1159 (PRJEB32708, Schiffer et al., 2019) and JU765 (PRJEB32708, Schiffer et al., 2019) in WormBase ParaSite (https://parasite.wormbase.org). For Propanagrolaimus sp. JU765 a potential orthologue with 63.5% similarity was selected (JU765_v2.g7216) We searched for potential guide RNAs and analyzed the putative off-targets using a web-based software (CRISPOR, http://crispor.tefor.net; Concordet and Haeussler, 2018). A list of guide RNAs and ssODNs can be found on Tables 2, 5.
4.3 Preparation of HDR donor
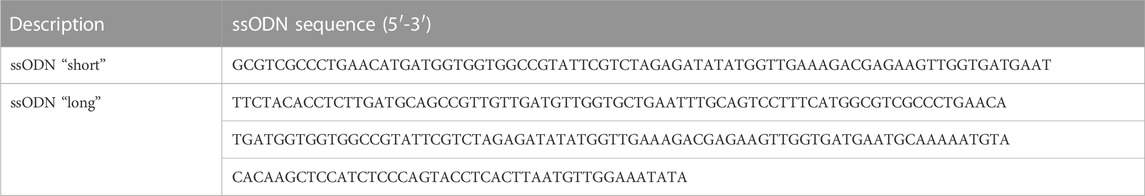
For knock-in of small insertions using the homology-directed repair (HDR) ssODN (single-stranded oligodeoxynucleotides) repair templates with 37 nt (“short”) and 97/92 nt (“long”) homology arms flanking the cleavage site with an inserted XbaI site followed by a stop codon were designed using Geneious Pro 5.5.6 (Kearse et al., 2012; Figures 2A, 6A).
4.4 Preparation of injection mix
1.25 µL tracrRNA (100 μM, catalog# 1073191, IDT) and different amounts of crRNA according to the tested concentration of 12.5 µM, 30 μM, 60 μM and 125 µM (Alt-R® CRISPR-Cas9 crRNA (XT), 10 nmol, IDT) were mixed and incubated at 95°C for 5 min following incubation at RT for 5 min 0.5 µL Cas9 (10 mg/mL, catalog# 10811059, IDT) was added and incubated at 37°C for 10 min for RNP (ribonucleoprotein) complex formation. 1 µL ssODN repair template (from 25 µM stock) was added and the mixture was filled up with TE buffer to 10 μL, centrifuged at 13.000 rpm for 5 min and kept on ice for injections.
4.5 Preparation of injection needles, agar pads and microinjection
2.5% agarose pads were produced, and microinjections performed as described in Evans (2006). PS1159 and JU765 are monodelphic and the syncytial region of the gonadal arm looks similar as in C. elegans. Borosilicate glass capillary (GB120F-10, 0.69 mm × 1.20 mm × 100 mm, Science products GmbH) were pulled in a needle puller (P-2000, Sutter instruments) and loaded with the final injection mixture. The needle was opened by gently tapping the tip against the edge of a broken piece of coverslip placed on an agar pad and covered with oil (Halocarbon oil 700, Sigma-Aldrich). An Inverted DIC microscope (Zeiss IM 35) equipped with standard Nomarski objectives (non-oil immersion, 6.3x and 40x) was used. A micromanipulator (Bachofer) with a needle holder with a fine mobility in the x, y and z axes (Piezo manipulator, PM10, Bachofer) was attached to the microscope and a pressure regulator (Pneumatic PicoPump PV 820) with a foot petal connection to a vacuum pump (Vacuubrand ME2C). Pressure was adjusted between 15 and 20 psi for injections. Worms were recovered in M9 buffer (Supplementary Table S3), transferred to a single agar plate, and covered with a small piece of agar at 25°C (video of microinjection steps in Supplementary Video SV1). Phenol red dye was used for the purpose of the Supplementary Video SV1. No dye was used for microinjection experiments.
4.6 Screening for knock-out mutations
4.6.1 Single worm PCR and Sanger Sequencing
Individual worms were picked and transferred into tubes with 10 µL H2O or M9 buffer. Tubes were placed at −80°C for at least 1 h 10 µL 2x worm lysis buffer (Supplementary Table S2) was added, incubated at 55°C for 3 h and heated at 95°C for 10 min. We designed primers to amplify the genomic DNA fragment containing the cut site. PCR products were resolved on a 0.8% agarose gel. Sanger sequencing was performed through Eurofins Scientific SE. Sequences were analyzed using the ICE CRISPR analysis tool v3 (https://www.synthego.com; Conant et al., 2022) and Geneious Pro 5.5.6 (Kearse et al., 2012). List of primers used for this study can be found on Supplementary Table S5.
4.6.2 Motility assay
Observation of motility of wild-type worms vs. strong twitching F1 in the time course of 15 min (n = 10); Single worms were transferred onto a drop of water in an agar plate. After the drop evaporated, the worms were allowed to acclimate for 1 min and the sinusoidal waves of the movement were observed (Figure 2D).
4.6.3 Survival assay
10 wild-type worms and 10 F1 progeny after injection with a phenotype (strong and weak) were screened in 0.5% nicotine for a total of 10 min for survival. Individual worms were first transferred into water and then nicotine. After 10 min worms were rescued and observed for survival in M9 buffer.
4.6.4 T7 Endonuclease I assay
10 µL single worm PCR product was used for T7 Endonuclease I assay according to the protocol from New England Biolabs Inc. Samples were loaded onto a 1.4% agarose gel.
4.7 Screening for knock-ins/HDR
4.7.1 Restriction digest
10 µL single worm PCR products of F2 individuals were mixed with 1 µL 10x Tango buffer, 1 µL XbaI (10 U/μL) enzyme and 13 µL H2O and incubated at 37°C for 3 h. Samples were loaded onto a 1.4% agarose gel.
4.8 Data analysis and editing
Gene structure diagrams were generated with the Exon-Intron Graphic Maker (Version 4, www.wormweb.org/exonintron). Plots were generated with R package ggplot2 (Wickham, 2016). Images and Videos were edited with InkScape 1.2, Adobe Illustrator CS6 v. 16.0.0 and Avidemux 2.8.0.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
VH conducted all major experiments, was involved in planning the experiments, analyzed the data, wrote the manuscript, designed figures. DC conducted some experiments and contributed to figure design. JS conducted some experiments. FI contributed to one experiment. PS conceived the study, wrote the manuscript, and supervised VH and DC. MK conceived the study, planned the experiments, analyzed data, edited the manuscript, supervised VH, JS, and FI and co-supervised DC.
Funding
VH is funded by a DFG ENP grant to PS (grant number: 434028868).
Acknowledgments
We thank James W. Lightfoot for sharing RNP complex components and protocols for pilot experiments. This manuscript is dedicated to the memory of Einhard Schierenberg.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgeed.2023.1078359/full#supplementary-material
References
Adams, S., Pathak, P., Shao, H., Lok, J. B., and Pires-daSilva, A. (2019). Liposome-based transfection enhances RNAi and CRISPR-mediated mutagenesis in non-model nematode systems. Sci. Rep. 9, 483. doi:10.1038/s41598-018-37036-1
Allen, D., Rosenberg, M., and Hendel, A. (2021). Using synthetically engineered guide RNAs to enhance CRISPR genome editing systems in mammalian cells. Front. Genome Ed. 2, 617910. doi:10.3389/fgeed.2020.617910
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi:10.1016/S0022-2836(05)80360-2
Arribere, J. A., Bell, R. T., Fu, B. X. H., Artiles, K. L., Hartman, P. S., and Fire, A. Z. (2014). Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846. doi:10.1534/genetics.114.169730
Blaxter, M., and Koutsovoulos, G. (2015). The evolution of parasitism in Nematoda. Parasitology 142, S26–S39. doi:10.1017/S0031182014000791
Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. doi:10.1093/genetics/77.1.71
Chang, N., Sun, C., Gao, L., Zhu, D., Xu, X., Zhu, X., et al. (2013). Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 23, 465–472. doi:10.1038/cr.2013.45
Cho, S. W., Lee, J., Carroll, D., Kim, J.-S., and Lee, J. (2013). Heritable gene knockout in Caenorhabditis elegans by direct injection of cas9–sgRNA ribonucleoproteins. Genetics 195, 1177–1180. doi:10.1534/genetics.113.155853
Cinkornpumin, J. K., and Hong, R. L. (2011). RNAi mediated gene knockdown and transgenesis by microinjection in the necromenic Nematode Pristionchus pacificus. J. Vis. Exp., e3270. doi:10.3791/3270
Conant, D., Hsiau, T., Rossi, N., Oki, J., Maures, T., Waite, K., et al. (2022). Inference of CRISPR edits from sanger trace data. CRISPR J. 5, 123–130. doi:10.1089/crispr.2021.0113
Concordet, J.-P., and Haeussler, M. (2018). CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245. doi:10.1093/nar/gky354
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi:10.1126/science.1231143
Conte, D., MacNeil, L. T., Walhout, A. J. M., and Mello, C. C. (2015). RNA interference in Caenorhabditis elegans. Curr. Protoc. Mol. Biol. 109, 261–326. doi:10.1002/0471142727.mb2603s109.3.30
Culp, E., Richman, C., Sharanya, D., Jhaveri, N., Van Den Berg, W., and Gupta, B. P. (2020). Genome editing in the nematode Caenorhabditis briggsae using the CRISPR/Cas9 system. Biol. Methods Protoc. 5, bpaa003. doi:10.1093/BIOMETHODS/BPAA003
Davis, A. J., and Chen, D. J. (2013). DNA double strand break repair via non-homologous end-joining. Transl. cancer Res. 2, 130–143. doi:10.3978/J.ISSN.2218-676X.2013.04.02
Dickinson, M. H., Vosshall, L. B., and Dow, J. A. T. (2020). Genome editing in non-model organisms opens new horizons for comparative physiology. J. Exp. Biol. 223, jeb221119. doi:10.1242/jeb.221119
Doench, J. G., Fusi, N., Sullender, M., Hegde, M., Vaimberg, E. W., Donovan, K. F., et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191. doi:10.1038/nbt.3437
Dolinski, C., Baldwin, J. G., and Thomas, W. K. (2001). Comparative survey of early embryogenesis of secernentea (nematoda), with phylogenetic implications. Can. J. Zoology 79, 82–94. doi:10.1139/z00-179
Doudna, J. A., and Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096. doi:10.1126/science.1258096
Dulovic, A., and Streit, A. (2019). RNAi-mediated knockdown of daf-12 in the model parasitic nematode Strongyloides ratti. PLOS Pathog. 15, e1007705. doi:10.1371/journal.ppat.1007705
Evans, T. C. (2006). Transformation and microinjection, WormBook, The C. elegans research community, WormBook. doi:10.1895/wormbook.1.108.1
Farboud, B., and Meyer, B. J. (2015). Dramatic enhancement of genome editing by CRISPR/Cas9 through improved guide RNA design. Genetics 199, 959–971. doi:10.1534/genetics.115.175166
Félix, M.-A. (2008). RNA interference in nematodes and the chance that favored Sydney Brenner. J. Biol. 7, 34. doi:10.1186/jbiol97
Gang, S. S., Castelletto, M. L., Bryant, A. S., Yang, E., Mancuso, N., Lopez, J. B., et al. (2017). Targeted mutagenesis in a human-parasitic nematode. PLoS Pathog. 13, e1006675. doi:10.1371/journal.ppat.1006675
Ghanta, K. S., Ishidate, T., and Mello, C. C. (2021). Microinjection for precision genome editing in Caenorhabditis elegans. Star. Protoc. 2, 100748. doi:10.1016/j.xpro.2021.100748
Glover, N. M., Redestig, H., and Dessimoz, C. (2016). Homoeologs: What are they and how do we infer them? Trends Plant Sci. 21, 609–621. doi:10.1016/j.tplants.2016.02.005
Goldstein, B., and Hird, S. N. (1996). Specification of the anteroposterior axis in Caenorhabditis elegans. Development 122, 1467–1474. doi:10.1242/dev.122.5.1467
Hendel, A., Bak, R. O., Clark, J. T., Kennedy, A. B., Ryan, D. E., Roy, S., et al. (2015). Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 33, 985–989. doi:10.1038/nbt.3290
Hiraga, H., Ishita, Y., Chihara, T., and Okumura, M. (2021). Efficient visual screening of CRISPR/Cas9 genome editing in the nematode Pristionchus pacificus. Dev. Growth & Differ. 63, 488–500. doi:10.1111/dgd.12761
Holterman, M., Schratzberger, M., and Helder, J. (2019). Nematodes as evolutionary commuters between marine, freshwater and terrestrial habitats. Biol. J. Linn. Soc. 128, 756–767. doi:10.1093/biolinnean/blz107
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi:10.1093/bioinformatics/bts199
Kim, H., Ishidate, T., Ghanta, K. S., Seth, M., Conte, D., Shirayama, M., et al. (2014). A Co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics 197, 1069–1080. doi:10.1534/genetics.114.166389
Kosicki, M., Tomberg, K., and Bradley, A. (2018). Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36, 765–771. doi:10.1038/nbt.4192
Kraus, C., Schiffer, P. H., Kagoshima, H., Hiraki, H., Vogt, T., Kroiher, M., et al. (2017). Differences in the genetic control of early egg development and reproduction between C. elegans and its parthenogenetic relative D. coronatus. EvoDevo 8, 16. doi:10.1186/s13227-017-0081-y
Lahl, V., Halama, C., and Schierenberg, E. (2003). Comparative and experimental embryogenesis of plectidae (nematoda). Dev. Genes Evol. 213, 18–27. doi:10.1007/s00427-002-0289-1
Lambshead, P. J. D., and Boucher, G. (2003). Marine nematode deep-sea biodiversity-hyperdiverse or hype? J. Biogeogr. 30, 475–485. doi:10.1046/j.1365-2699.2003.00843.x
Levine, M., and Davidson, E. H. (2005). Gene regulatory networks for development. Proc. Natl. Acad. Sci. 102, 4936–4942. doi:10.1073/pnas.0408031102
Lewis, J. A., Wu, C. H., Berg, H., and Levine, J. H. (1980). The genetics of levamisole resistance in the nematode Caenorhabditis elegans. Genetics 95, 905–928. doi:10.1093/genetics/95.4.905
Liang, X., Potter, J., Kumar, S., Ravinder, N., and Chesnut, J. D. (2017). Enhanced CRISPR/Cas9-mediated precise genome editing by improved design and delivery of gRNA, Cas9 nuclease, and donor DNA. J. Biotechnol. 241, 136–146. doi:10.1016/j.jbiotec.2016.11.011
Lino, C. A., Harper, J. C., Carney, J. P., and Timlin, J. A. (2018). Delivering CRISPR: A review of the challenges and approaches. Drug Deliv. 25, 1234–1257. doi:10.1080/10717544.2018.1474964
Makarova, K. S., Haft, D. H., Barrangou, R., J Brouns, S. J., Charpentier, E., Horvath, P., et al. (2011). Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Microbiol. 9, 467. doi:10.1038/nrmicro2577
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Sci. (New York, N.Y.) 339, 823–826. doi:10.1126/SCIENCE.1232033
Mello, C., and Fire, A. (1995). “Chapter 19 DNA transformation,” in Methods in cell biology, Caenorhabditis elegans: Modern biological analysis of an organism. Editors H. F. Epstein, and D. C. Shakes (Cambridge: Academic Press), 451–482. doi:10.1016/S0091-679X(08)61399-0
Miki, D., Zinta, G., Zhang, W., Peng, F., Feng, Z., and Zhu, J.-K. (2021). “CRISPR/Cas9-Based genome editing toolbox for Arabidopsis thaliana,” in Arabidopsis protocols, methods in molecular biology. Editors J. J. Sanchez-Serrano, and J. Salinas (New York, NY: Springer US), 121–146. doi:10.1007/978-1-0716-0880-7_5
Moerman, D. G., Benian, G. M., Barstead, R. J., Schriefer, L. A., and Waterston, R. H. (1988). Identification and intracellular localization of the unc-22 gene product of Caenorhabditis elegans. Genes & Dev. 2, 93–105. doi:10.1101/GAD.2.1.93
Nuez, I., and Félix, M.-A. (2012). Evolution of susceptibility to ingested double-stranded RNAs in Caenorhabditis nematodes. PLoS One 7, e29811. doi:10.1371/journal.pone.0029811
Oesterle, A. (2018). Pipette cookbook 2018. P-97 & P-1000 micropipette pullers. [online]. Available at: https://www.sutter.com/PDFs/pipette_cookbook.pdf.
Paix, A., Folkmann, A., Rasoloson, D., and Seydoux, G. (2015). High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9ribonucleoprotein complexes. Genetics 201, 47–54. doi:10.1534/genetics.115.179382
Paix, A., Folkmann, A., and Seydoux, G. (2017). Precision genome editing using CRISPR-Cas9 and linear repair templates in C. elegans. Methods (San Diego, Calif.) 121–122, 86–93. doi:10.1016/J.YMETH.2017.03.023
Poinar, G. O. (2011). “The evolutionary history of nematodes: As revealed in stone, amber and mummies,” in Nematology monographs and perspectives (Leiden: Brill).
Ratnappan, R., Vadnal, J., Keaney, M., Eleftherianos, I., O’Halloran, D., and Hawdon, J. M. (2016). RNAi-mediated gene knockdown by microinjection in the model entomopathogenic nematode Heterorhabditis bacteriophora. Parasit. Vectors 9, 160. doi:10.1186/s13071-016-1442-4
Schiffer, P. H., Danchin, E. G. J., Burnell, A. M., Creevey, C. J., Wong, S., Dix, I., et al. (2019). Signatures of the evolution of parthenogenesis and cryptobiosis in the genomes of panagrolaimid nematodes. iScience 21, 587–602. doi:10.1016/j.isci.2019.10.039
Schiffer, P. H., Kroiher, M., Kraus, C., Koutsovoulos, G. D., Kumar, S., Camps, J. I. R., et al. (2013). The genome of romanomermis culicivorax: Revealing fundamental changes in the core developmental genetic toolkit in nematoda. BMC Genomics 14, 923. doi:10.1186/1471-2164-14-923
Schiffer, P. H., Polsky, A. L., Cole, A. G., Camps, J. I. R., Kroiher, M., Silver, D. H., et al. (2018). The gene regulatory program of Acrobeloides nanus reveals conservation of phylum-specific expression. Proc. Natl. Acad. Sci. U. S. A. 115, 4459–4464. doi:10.1073/pnas.1720817115
Schubert, M. S., Thommandru, B., Woodley, J., Turk, R., Yan, S., Kurgan, G., et al. (2021). Optimized design parameters for CRISPR Cas9 and Cas12a homology-directed repair. Sci. Rep. 11, 19482. doi:10.1038/s41598-021-98965-y
Schulze, J., and Schierenberg, E. (2009). Embryogenesis of Romanomermis culicivorax: An alternative way to construct a nematode. Dev. Biol. 334, 10–21. doi:10.1016/j.ydbio.2009.06.009
Shannon, A. J., Browne, J. A., Boyd, J., Fitzpatrick, D. A., and Burnell, A. M. (2005). The anhydrobiotic potential and molecular phylogenetics of species and strains of Panagrolaimus (Nematoda, Panagrolaimidae). J. Exp. Biol. 208, 2433–2445. doi:10.1242/jeb.01629
Shannon, A. J., Tyson, T., Dix, I., Boyd, J., and Burnell, A. M. (2008). Systemic RNAi mediated gene silencing in the anhydrobiotic nematode Panagrolaimus superbus. BMC Mol. Biol. 9, 58. doi:10.1186/1471-2199-9-58
Shin, H. Y., Wang, C., Lee, H. K., Yoo, K. H., Zeng, X., Kuhns, T., et al. (2017). CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat. Commun. 8, 15464. doi:10.1038/ncomms15464
Tandonnet, S., Koutsovoulos, G., Adams, S., Cloarec, D., Parihar, M., Blaxter, M., et al. (2018). Chromosome-wide evolution and sex determination in a nematode with three sexes. doi:10.1101/466961
Vargas-Velazquez, A. M., Besnard, F., and Félix, M.-A. (2019). Necessity and contingency in developmental genetic screens: EGF, wnt, and semaphorin pathways in vulval induction of the nematode Oscheius tipulae. Genetics 211, 1315–1330. doi:10.1534/genetics.119.301970
Wang, H., La Russa, M., and Qi, L. S. (2016). CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 85, 227–264. doi:10.1146/annurev-biochem-060815-014607
Keywords: CRISPR, microinjection, nematodes, Panagrolaimus, C. elegans, unc-22
Citation: Hellekes V, Claus D, Seiler J, Illner F, Schiffer PH and Kroiher M (2023) CRISPR/Cas9 mediated gene editing in non-model nematode Panagrolaimus sp. PS1159. Front. Genome Ed. 5:1078359. doi: 10.3389/fgeed.2023.1078359
Received: 24 October 2022; Accepted: 23 January 2023;
Published: 03 February 2023.
Edited by:
Tessa Montague, Columbia University, United StatesReviewed by:
Sounak Sahu, National Cancer Institute at Frederick (NIH), United StatesHillel Schwartz, California Institute of Technology, United States
Copyright © 2023 Hellekes, Claus, Seiler, Illner, Schiffer and Kroiher. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Viktoria Hellekes, di5oZWxsZWtlc0B1bmkta29lbG4uZGU=
 Viktoria Hellekes
Viktoria Hellekes Denise Claus2
Denise Claus2