- 1CHU de Québec Research Center, Laval University, Quebec City, QC, Canada
- 2Department of Molecular Medicine, Laval University, Quebec City, QC, Canada
Advancements in genome editing make possible to exploit the functions of enzymes for efficient DNA modifications with tremendous potential to treat human genetic diseases. Several nuclease genome editing strategies including Meganucleases (MNs), Zinc Finger Nucleases (ZFNs), Transcription Activator-like Effector Nucleases (TALENs) and Clustered Regularly Interspaced Short Palindromic Repeats-CRISPR associated proteins (CRISPR-Cas) have been developed for the correction of genetic mutations. CRISPR-Cas has further been engineered to create nickase genome editing tools including Base editors and Prime editors with much precision and efficacy. In this review, we summarized recent improvements in nuclease and nickase genome editing approaches for the treatment of genetic diseases. We also highlighted some limitations for the translation of these approaches into clinical applications.
1 Introduction
Mutations of a single or several nucleotides in human genome are responsible for major hereditary health problems (Benusiglio et al., 2021; Samuelson et al., 2021; Xiao et al., 2021; Chen et al., 2022; Cobo et al., 2022). To date, about 7,000 hereditary diseases are estimated to be caused by monogenic mutations (Claussnitzer et al., 2020). Homologous recombination (HR) has long been proposed as an avenue to treat human genetic diseases and its efficiency can be increased by inducing the DNA double-strand breaks (DSBs) (Mao et al., 2008). A significant step forward in gene therapy has been the discovery of enzymes known as nucleases. These enzymes enable gene editing technologies to modify a specific DNA sequence within the natural cell environment for the correction of hereditary diseases (Durai, 2005; Cermak et al., 2011; Hwang et al., 2013).
Over the past decades, significant technology development have empowered bio-engineers with tools such as nuclease-mediated Meganucleases (MNs) (Epinat et al., 2003), Zinc Finger Nucleases (ZFNs) (Porteus and Baltimore, 2003; Miller et al., 2007), Transcription Activator-Like Effector Nucleases (TALENs) (Method of the Year 2011, 2012) and Clustered Regularly Interspaced Short Palindromic Repeats-CRISPR associated protein 9 (CRISPR-Cas9) (Jinek et al., 2012a; Ran et al., 2013b; Shalem et al., 2014b). These technologies grant us access to the genome for an accurate base-to-base modification without DSBs by deploying techniques such as Base editing and Prime editing (Gaudelli, 2017; Komor et al., 2017; Anzalone et al., 2019). Theoretically, these gene editing technology enable the replacement of single or multiple bases in any gene of interest at any given location. However, the gene editing effectiveness are influenced by at least three factors: (i) The type of gene editing (e.g., DNA base pair conversion, deletion, insertion, or a combination of the three above changes), (ii) the availability of a gene editing technology to achieve its desired outcome, (iii) the efficiency of the gene editing process. More importantly, the main bottleneck, the acquisition of tissue-specific edits and unwanted genome modification events, is still remains.
To explore how to edit the genome, in this review, we particularly focused on the mechanisms, the limitations and optimizations of six high-profile gene editing technologies as well as the recent progress of various types of genome editing tools used in clinical or preclinical research.
2 Nuclease-based genome engineering technologies
In the past decade, various nuclease gene editing technologies have been developed and widely used. These technologies empowered scientists to modify specific sequences in the genome of diverse organisms (Tzfira et al., 2003; Urnov et al., 2010). The most common nucleases-mediated gene editing technologies (Table 1) are MNs (Figure 1A), ZFNs (Figure 1B), TALENs (Figure 1C), and CRISPR-Cas9 (Figure 1D). These technologies combined specific DNA target recognition sequences and programmable endonucleases to induce the desired genomic DNA sequence alterations by introduction of DNA DSB resulting in insertions, deletions, gene replacements and nucleotide substitutions (Lo et al., 2013; Miyaoka et al., 2016). In eukaryotic cells, double-strand DNA cleavage by nucleases triggers two major DNA repair mechanisms including: (i) The non-homologous end joining (NHEJ) and microhomology-mediated end joining through re-ligation of the ends pathways, and (ii) the homology-directed repair (HDR) generated by repairing through a separate donor DNA template (Ranjha et al., 2018). Genome editing takes advantage of these DNA repair processes to produce desired genomic alteration in cell cultures and organisms. (Valerie and Povirk, 2003; Iliakis et al., 2004). However, the DNA DSBs can cause undesired outcomes such as insertions and deletions (Indels) as well as p53 activation (Naeem et al., 2020; Ihry et al., 2018).
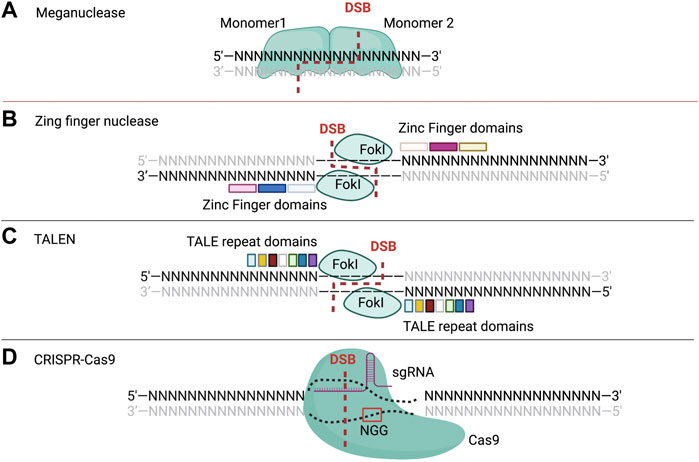
FIGURE 1. The (A) is depicting the two monomer domains (monomer 1 and monomer 2) of mega nuclease which cleave at the binding sites, resulting in the DNA double strand break. The (B) shows the two zinc finger domains of ZFNs made of three ZF motifs distinct binding sites (sequence of 3 nucleotides) and cleaving domains (FokI). The (C) represents TALE repeat domains (shown in colored squares) pair with distinct binding DNA nucleotides and cleaving domains (FokI). The (D) shows the single guide RNA complexed with Cas9 to open and cleave the DNA sequence, trough the recognition of the PAM sequence (by Cas9) which is NGG for SpCas9, and the target sequence (by sgRNA, single guide RNA). N represents any nucleotide amongst A, T, C, and G nucleotide.
2.1 Meganucleases
MNs also referred to as homing endonucleases recognize a large DNA stretches (12–40 base pairs) to facilitate cleavage in the most genomes (Rouet et al., 1994b). MNs are generally encoded by introns or inteins to promote homing of their respective genetic elements into intron or intein-free homologous allelic sites (Epinat et al., 2003). One of the distinctive features of these MNs is their high specificity, due to the tight coupling of their binding site. This tight coupling recognizes a single locus within the yeast nuclear or mitochondrial genome (Paques and Duchateau, 2007). The LAGLIDADG proteins are the most well-studied homing endonuclease. They interact with their targets by nonspecific interactions between the ß strands and the backbone of the target DNA through the recognition of a sequence of 2–4 bp region. Consequently, several new engineered endonuclease variations derived from the following homing endonuclease: (i) I-CreI which was discovered in the chloroplast genome of Chlamydomonas reinhardtii, and (ii) I-SceI which is present in the mitochondria of Saccharomyces cerevisiae (Prieto et al., 2007; Gao et al., 2010; Zekonyte et al., 2021; Lee et al., 2022). These engineered endonucleases enable in vivo and in vitro genetic modifications. Due to the tiny molecular weight of modified MNs makes the in vivo delivery possible. However, the editing efficiency of this strategy is low compared to later developed nuclease-mediated technologies (Aubert et al., 2016; Zhu et al., 2017). Furthermore, reengineering MNs to expand the spectrum of DNA target sequences is complex and laborious and therefore vastly limit its application (Gouble et al., 2006). Recently, Zekonyte et al. (2021) used the engineered I-CreI meganuclease administered via intravenous (IV) injection of AAV9 into mice for the correction of m.5024C>T mutation in the mt-tRNAAla gene, as a curative method for disorders caused by heteroplasmic mitochondrial DNA mutation. This resulted to the elimination of mutant mitochondrial DNA followed by the restoration of mt-tRNAAla level. Moreover, this approach has also been used in pig model of autosomal dominant Retinitis Pigmentosa (adRP) for the correction of P23H mutation in the rhodopsin (RHO) gene (Jalligampala et al., 2021) and in non-human primates for the modification of the proprotein convertase subtilisin/kexin type 9 (PCSK9) gene responsible for hypercholesterolemia (Wang et al., 2018a; Wang et al., 2021).
2.1.1 Meganuclease related limitations and perspectives
Since the introduction of MNs application, unexpected drawbacks are constantly being discovered. Some of these challenges are: (i) The targeted locus must contain a specific MN cleavage site for each endonuclease whereas the microbial self-splicing intervening sequence could specifically duplicate into recipient alleles of their host gene lacking such sequence (Rouet et al., 1994a; Stoddard, 2011), (ii) low efficacy (Chapdelaine et al., 2010), and (iii) potential genotoxicity (Suzuki et al., 2020). The natural repertory of homing endonucleases is limited to a finite number of proteins, most of them still being hypothetical or uncharacterized. Thus, for other protein, the cleavage site for each meganuclease has to be inserted into the target genome. Because of this flaw, the application of this technology is significantly limited. However, some engineered enzymes originated from meganuclease I-CreI (Arnould et al., 2011), I-SceI (Siegl et al., 2010) and I-DmoI (a monomeric meganuclease from the hyperthermophilic archaeon Desulfurococcus mobilis) (Molina et al., 2016) capable of cleaving DNA in specific genomic sites have been generated. I-CreI plays a critical role in the localization and occupancy of the catalytic metal ions, which is crucial for the DNA cleavage (Prieto et al., 2018). Wang et al. (2022) developed a transgenic Xenopus tropicalis line which is used for evaluating the potential effects of I-SceI mediated transgenesis and further understanding its mechanisms. The fusion of transcription activator-like effector (TALE) DNA-binding domains to MNs dramatically increases the efficiency by 35-fold compared to standalone MNs to modify T-cells receptor alpha (Boissel et al., 2014a). Furthermore, due to specific sites targeted by high cleavage specificity and the long length of the sequences (Petersen and Niemann, 2015b; Izmiryan et al., 2016), low off-target effects were detected in MNs because of the structure of meganucleases and the delivery methods. Some strategies could reduce the off-target by combining meganuclease with TALarray or I-TevI (a GIY-YIG enzyme) (Boissel et al., 2014b; Wang et al., 2018b).
2.2 Zinc finger nucleases
Zinc Finger Proteins (ZFPs) are artificially synthetic engineered hybrid heterodimeric proteins for site specific genome editing. ZFPs include a sequence of 3–6 peptides (called Zinc Finger (ZF) domains), each binding to a specific sequence of 3–6 base pairs for a specific attachment to a gene sequence (Durai et al., 2005). Two ZFPs are required to fuse with Flavobacterium okeanokoites endonuclease I (FokI) to induce a DSB at a specific genomic site. ZFNs are frequently used for gene silencing and knockout (Santiago et al., 2008; Gutschner et al., 2011; Gaj et al., 2012; Sun et al., 2018). Therefore, ZFNs have emerged as a versatile tool for gene targeting in various mammalian cells and organisms for the treatment of hereditary diseases (Almeida and Matos, 2019) and creation of animal models for diseases (Petersen and Niemann, 2015a).
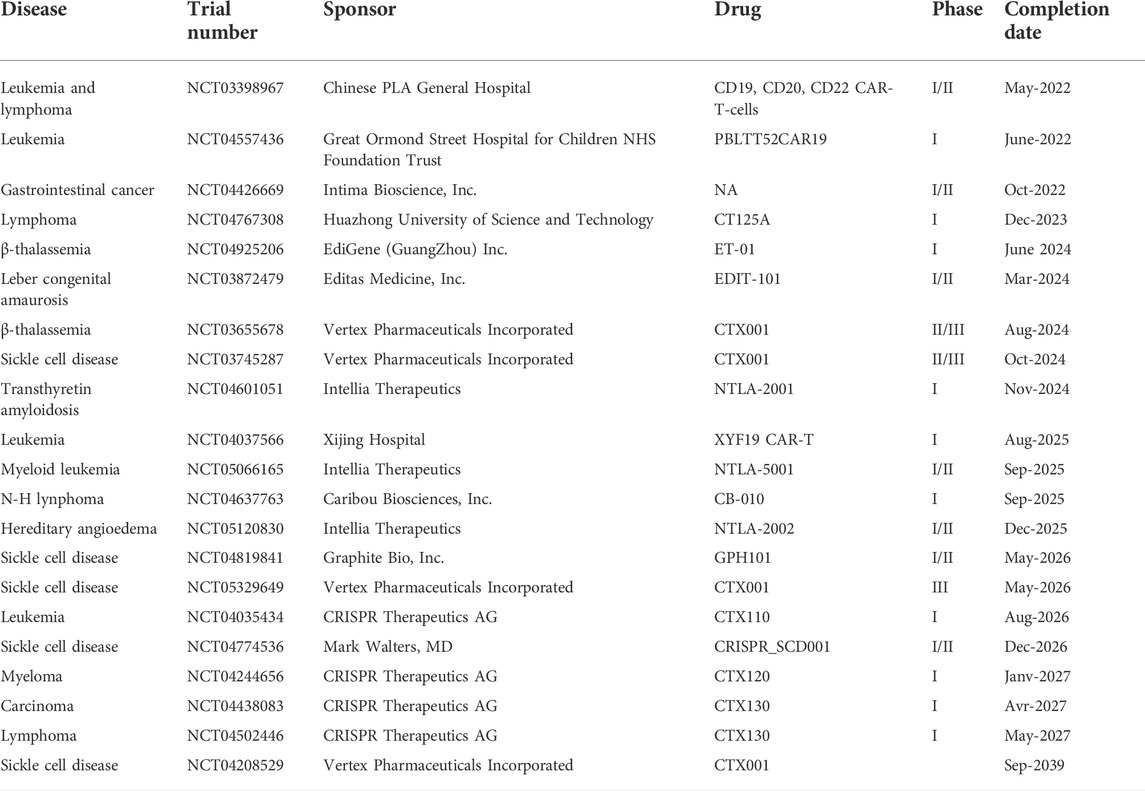
ZFNs represent the first gene editing method applied for clinical treatment of diseases. ZFNs have been used to modify autologous CD4+ T-cells to inhibit the function of the human chemokine (C-C motif) receptor 5 gene (CCR5) receptor and reduce the infection of these cells by HIV (Tebas et al., 2014). The results showed that infusion of genetically modified CD4+ T-cells was well tolerated, and the HIV viral load has been decreased in blood level of most patients. Another experiment also showed that the HIV-specific CD8+ T-cell responses are substantially restored (Tebas et al., 2021). The ZFN approach was also successfully used for the treatment of ß-hemoglobinopathies (Hoban et al., 2015; Chang et al., 2017; Psatha et al., 2018; Smith et al., 2019). Indeed, the phase I/II clinical trial (NCT03432364) sponsored by Sangamo Therapeutics Inc., aimed to assess the safety, tolerability and efficacy of ST-400 for the treatment of transfusion-dependent beta-thalassemia. The ST-400 are patient’s hematopoietic stem cells genetically modified by ZFNs to disrupt a specific and precise sequence of the enhancer of the BCL11A gene in order to boost the expression of fetal hemoglobin (HbF) (Bauer and Orkin, 2015; Masuda et al., 2016). A cohort of six participants will be completed by November 2022. In other studies involving two patients manifesting different genotypic profiles showed a prompt hematopoietic reconstitution with long term increased HbF levels; however, serious adverse events (e.g., Hypersensitivity) have been recorded with one patient as a result of reengineered ST-400 (Smith et al., 2019). Scientists at Bioverative Inc., a Sanofi company, are conducting a phase I/II clinical trials (NCT03653247) in a cohort of eight patients to evaluate the safety, tolerability and efficacy of autologous hematopoietic stem cell transplantation using BIVV003 for the treatment of severe Sickle Cell Disease (SCD) in adults. This trial was supported by encouraging preclinical results that showed a robust long-term engraftment of ex-vivo modified hematopoietic stem and progenitor cells (HSPC) from patients. In other phase I/II clinical trials conducted by Sangamo Therapeutics Inc., the UCSF Benioff Children’s Hospital delivered SB-318 (NCT02702115) and SB-913 (NCT03041324) into participants to insert the corrected copy of α-L-iduronidase (IDUA) and iduronate-2-sulfatase (IDS) transgenes respectively into the albumin locus to provide permanent liver specific expression of iduronidase. Preliminary results showed the evidence of albumin-IDS mRNA transcripts in liver and the hepatocytes are able to generate active IDS enzyme (Muenzer et al., 2019). The summary of ongoing clinical trials is presented in Table 2.
2.2.1 ZFN related limitations and perspectives
Although ZFNs exhibited their ability to modify a specific gene in mammalian cells, the strategy faces three major limitations. 1) Cannot cut arbitrary gene sequences (Durai, 2005). 2) A ZFN coding gene must be engineered for each specific target site (Porter et al., 2019). 3) The likelihood of off-target gene editing is another drawback of the ZFN technology (Pattanayak et al., 2011). At the beginning, the targeted sequence should contain 5′-GNN, 5′-ANN, 5′-CNN or 5′TNN (Dreier et al., 2001, 2005). Therefore, this technology is costly, laborious, time consuming and requires highly trained researchers for protein engineering (Porter et al., 2019). Recently, the liaison between the ZFP and FokI cleavage domain have been substituted to increase the number of distinct zinc-finger arrays enabling cleavage at a target genomic site (Paschon et al., 2019). In comparison to classical ZFNs, this technique reduced the off-target effect, boosted modification activities, and is more precise. More significantly, it can target and cleave at any intended base (Paschon et al., 2019). In addition, different strategies to engineer the ZFNs and reduce the off-target mutations have also been developed (Ji et al., 2018; Miller et al., 2019a). The assembly of a high specific ZFNs system is highly complex. Researchers isolated naturally occurring ZF modules with different sequence specificities to engineer ZF modules with altered DNA binding specificities (Xiong et al., 2013). Furthermore, several methods are available to increase the specificity and reduced the cellular toxicity of this system by improving the ZFN architecture to develop FokI nuclease domain variants, which could result in a 3,000-fold reduction in off-target indels. (Miller et al., 2007; Miller et al., 2019b).
2.3 Transcription activator-like effector nucleases
A class of naturally occurring DNA binding proteins called the Transcription Activator-Like Effector (TALE) has been identified in the plant pathogen Xanthomonas. These TALEs regulate the transcription of several host target genes (Sanjana et al., 2012). TALENs are artificial engineered proteins combining the DNA-binding properties of a TALE protein and the DNA cleavage of the FokI endonuclease (Moscou and Bogdanove, 2009). The central region of TALEs is composed of 34 amino acid repeats amongst which 32 are constant and 2 are variable and recognized as repeat variable diresidues (RVDs) (Scholze and Boch, 2011). RVDs are involved in the DNA target recognition (Boch et al., 2009). Two TALENs target binding sequences are required to form a FokI dimer that induces a DSB (Feng et al., 2014).
TALENs have successfully been used for the modification of T-cell receptors for the treatment of leukemia (Qasim et al., 2017; Benjamin et al., 2020). UCART19, a CAR-T-cell product engineered with TALENs, was tested in children and adults in phase I clinical trials (NCT02808442 and NCT02746952) to cure advanced lymphoid malignancies and refractory B-cell acute lymphoblastic leukemia (B-ALL). These trials demonstrated the potential of UCART19 in patients with aggressive leukemia, but significant adverse events such as cytokine release syndrome, acute graft-versus-host of the skin and infectious complications have also been observed (Benjamin et al., 2020). There are many phase I trials sponsored by Cellectis Inc., using programmed allogenic engineered T-cells expressing different CARs such as UCART123 (NCT04106076, NCT03203369), UCARTCS1A (NCT04142619) and UCART22 (NCT04150497) respectively to treat acute myeloid leukemia, blastic plasmacytoid dendritic cell neoplasm (BPDCN), multiple myeloma and CD22+ B cell acute lymphoblastic leukemia. Ongoing clinical trials using this approach are summarized in Table 3.
2.3.1 TALENs related limitations and perspectives
Compared to MNs and ZFNs, TALENs exhibits high efficiency, low off-target effects and are proven to target the mitochondrial DNA (Mussolino et al., 2014). Nonetheless, there are some constraints that prevent a more widespread deployment. 1) The repetitive sequences of TALEs make them difficult to construct using polymerase chain reaction (PCR) (Cermak et al., 2011). 2) TALENs are unable to target a methylated DNA, because the methylation of cytosine can potentially abrogate TALE binding and alter recognition by its normal RVD (Deng et al., 2019). Different approaches have been proposed to bypass the challenges associated with the TALEs repetitive sequences are the following: (i) design the ligation-independent cloning techniques (Reyon et al., 2012), (ii) the high-throughput solid-phase assembly (Schmid-Burgk et al., 2013), (iii) the Golden gate cloning (Cermak et al., 2015), and (iv) alternative one-day TALE assembly (Zhang et al., 2020). To improve the efficiency of gene editing, a new bicistronic TALEN termed T2A using classical TALEN coding sequences linked to different reporter molecules by 2A “self-cleaving peptide” has been developed. This improvement could help each TALEN monomer to transcribe from the same reading frame in order to increase the gene editing efficacy. (Mariano et al., 2014; Martín-Fernández et al., 2020). Additionally, Zhang et al. (2017) used deciphered TALEs for 5-hydroxymethylcytosine and 5-methylcytosine to achieve methylation-dependent genome editing and gene activation in vivo.
2.4 Clustered regularly interspaced short palindromic repeats—CRISPR associated protein 9
CRISPR-Cas9 system is a sophisticated gene editing tool that revolutionized the genome engineering field and generated excitement for the potential of novel therapeutic approaches to treat human diseases. The CRISPR-Cas system is divided into class I and class II (Makarova et al., 2015). The class I uses multi-protein complexes for nucleic acid cleavage and is subdivided into CRISPR-Cas types I, III, and IV. The class II uses a single protein effector domain for the cleavage and is subdivided in CRISPR-Cas type II, V, and VI. The CRISPR-Cas9 belongs to type II system, which is simple to use and thus become the most widely utilized tool for biological research and translational applications (Tang and Fu, 2018).
Over the past decade, CRISPR-Cas9 system has been modified and adapted to become a versatile tool for genome editing in eukaryotes (Ran et al., 2013a; Perez-Pinera et al., 2013; Shalem et al., 2014a; Gao et al., 2019; Casas-Mollano et al., 2020). The system requires a Cas9 nuclease and a single guide RNA (sgRNA) adapted from CRISPR RNA (crRNA), which specifies the target site (the spacer sequence), fused with a trans-activating RNA (tracrRNA), which forms a complex with the crRNA (the scaffold sequence) (Jinek et al., 2012a). The sgRNA forms a stable ribonucleoprotein complex with Cas9 nuclease which initially attaches to a protospacer adjacent motif (PAM) to initiate the first conformational changes of the protein. The targeting activity is driven by 20 nucleotides of RNA-DNA base-pairing between the target DNA strand protospacer and the complementary RNA strand and through interactions between the non-target DNA strand PAM. Subsequently, mediates the second conformational change of the protein which then becomes active. Once activated, the HNH domain of Cas9 cleaves the DNA strand to which the sgRNA is attached and the RuvC domain cleaves at the PAM strand (Jinek et al., 2012b). One of the advantages of CRISPR/Cas9 is that it only requires a sgRNA to specify the DNA sequence where a DSB needs to be generated.
There are many clinical trials based on CRISPR-Cas9 and half of these trials (Phase I and Phase II) have already been successfully completed, the ongoing clinical trials are summarized in Table 4. In a phase I trial (NCT03399448), Pennsylvania University used a multiplex CRISPR-Cas9 to knockout the TCRα, TCRβ, PD-1 genes to treat various malignancies (Stadtmauer et al., 2020). The results showed that the modified T cells engrafted in patients at stable levels for at least 9 months and were barely immunogenic, indicating the feasibility of CRISPR-Cas9 gene editing for cancer immunotherapy (Stadtmauer et al., 2020). In addition to that, some clinical trials moved the technology from ZFNs or TALENs to CRISPR-Cas9 (Yi and Li, 2016; Lu et al., 2020). This was done for example to knockout PD-1 and CD52 for the different types of cancer by electroporating Cas9 and a sgRNA to edit the cells ex vivo. The results indicated that the clinical applications of CRISPR-Cas9 gene-edited T-cells are generally safe and feasible (Lu et al., 2020; Frangoul et al., 2021; Modarai et al., 2021). The disruption of erythroid enhancer of the BCL11A gene by CRISPR-Cas9 for the treatment of ß-thalassemia has been observed with serious adverse events. The reported serious adverse events are sepsis and pneumonia in the presence of neutropenia, vaso-occlusive liver disease, abdominal pain and cholelithiasis after CTX001 (autologous CRISPR-Cas9–edited CD34+ HSPCs) intravenous (IV) administration, although the substantially raised of hemoglobin levels in fetal blood cells (Frangoul et al., 2021). Two clinical trials are currently conducted to evaluate the safety and efficacy of CRISPR-Cas9 strategies either to restore ß-globin expression in ß-thalassemic patients harboring the IVS-2-654C>T mutation (NCT04205435) or to de-repress γ-globin in transfusion dependent thalassemia (TDT) patients (NCT04211480), but no results are yet available. Recently, another study prepared NTLA-2001, an in vivo gene-editing therapeutic agent, made by lipid nanoparticles encapsulating messenger RNA of Cas9 protein and single guide RNA targeting misfolding transthyretin (TTR) responsible of transthyretin amyloidosis. The result showed that the TTR is durably knockout after a single dose of NTLA-2001 (Gillmore et al., 2021; Philippidis, 2021).
2.4.1 Clustered regularly interspaced short palindromic repeats—CRISPR associated protein 9 limitations and perspectives
The CRISPR-Cas9 technology is associated with some limitation: (i) There are some off-target effects, (ii) the PAM requirement initially limits the editing scope to only sequence near an NGG for canonical SpCas9, and (iii) a more efficient method of delivering the CRISPR/Cas9 system for in vivo applications, which includes a protein of about 160 kDa, is required (Woo et al., 2015). Fortunately, the impact of these shortcomings is reduced with the continuous improvement of this technology. Amongst these improvements, Cas9 variants (Kleinstiver et al., 2016a; Slaymaker et al., 2016; Chen et al., 2017; Liu et al., 2020) and engineered sgRNA (Doench et al., 2014; Kocak et al., 2019; Nelson et al., 2022a) have been created to improve specificity and efficiency. Subsequently, the Cas9 variants requiring different PAM sequences which contributes to solve the PAM restriction challenge has also been developed. These variants are described as follows: (i) VQR (D1135V/R1335Q/T1337R) variant which recognizes NGA sequence and (ii) EQR (D1135E/R1335Q/T1337R) variant which recognizes NGA PAM sequence instead of an NGG sequence (Kleinstiver et al., 2015), (iii) SpCas9-NG variant which enables the recognition of NG PAM sequence (Nishimasu et al., 2018), (iv) xCas9 binding to NG, GAT as well as GAA (Hu et al., 2018) and the newest variant (v) SpRY which recognizes the NYN PAM and nearly eliminates the PAM restriction (Walton et al., 2020). These PAM flexibilities significantly increased the genome accessibility. For the delivery part, some CRISPR/Cas9 tools were successfully used to edit cells in vitro. This ex vivo approach may have certain safety benefits especially regarding off-target gene editing. However, some versions of CRISPR-Cas9 with a sgRNA cannot be efficiently delivered in vivo due to their size. Split viruses have been developed to resolve this problem but with reduced expression of the fusion protein. The limitations and optimizations of CRISPR/Cas9 and other limitations such as immunotoxicity, DNA-damage toxicity, etc. have also been detailed. (Tang and Gu, 2020; Uddin et al., 2020; Liu et al., 2021a; Wang et al., 2021b; Khatibi et al., 2021; Maximiano et al., 2021).
3 Nickase-based genome engineering technologies
DSBs at targeted genomic loci could be associated with serious undesirable effects, including p53 activation (Haapaniemi et al., 2018), translocations (Ghosh et al., 2018), off-target mutation (Vakulskas and Behlke, 2019) and complex undesired products (Paquet et al., 2016). Furthermore, half of all known disease associated gene variants are point mutations (Landrum et al., 2016). Therefore, the Cas9 nickases emerged as useful tools with a targetable property (Cong et al., 2013). The Cas9n D10A and Cas9n H840A are Cas9 variants mediating the cleavage of a single strand of the DNA respectively in the gRNA complementary or non-complementary DNA strand. These Cas9 nickases have been fused with various enzymes, to develop new gene editing technologies, including Base editors [Cytidine base editor (CBE)], Adenosine base editor (ABE) and Prime editor (PE) (Figure 2).
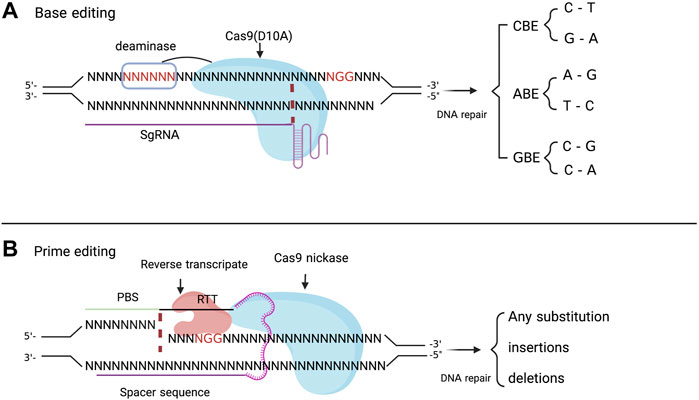
FIGURE 2. (A) represents a basic principle of base editor which is made of sgRNA that target a specific DNA sequence, the Cas9 nickase (D10A) which in interaction with sgRNA binds his recognition domain (PAM sequence) and cleaves the non-PAM DNA strand. The Cas9 is linked to a deaminase which modifies the targeted nucleotide in a window of 5 nucleotides (shown in red color) in the spacer sequence through CBE, ABE, and GBE. The (B) depicts the prime editing principle makes of pegRNA which include a spacer sequence, a primer binding site (PBS) and reverse transcript template (RTT), and Cas9 nickase fused with a reverse transcriptase. The pegRNA recognizes the target sequence and provide the desired sequence for modification. Once the Cas9n cleaves the DNA sequence, the reverse transcriptase uses the RTT as template for the synthesis of a new sequence containing the desired edit (gene substitution, insertion and deletion).
3.1 Base editor
Base editors are mainly used to target point mutations that may result in an altered DNA sequence with novel or enhanced functions and gene inactivation (Gaudelli, 2017; Gapinske et al., 2018; Zhang et al., 2019). Briefly DNA base editing requires two main components: a Cas9n fused with a deaminase and a sgRNA which binds to a specific DNA sequence (Komor et al., 2016). Three (3) types of base editors have been created including the Cytidine Base Editor (CBE) (Komor et al., 2016), Adenosine Base Editor (ABE) (Gaudelli, 2017) and Glycosylase Base Editors (GBE) (Zhao et al., 2021a) (Figure 2A).
Theoretically, base editors can function in both dividing and non-dividing cells to address several of single-nucleotide polymorphisms (SNPs) associated with human diseases.
Base editing is being used to study and treat hereditary diseases in a variety of cell types (Zeng et al., 2020; Tekel et al., 2021) and organisms (Kim et al., 2021c), including animal models of human hereditary diseases (Liu et al., 2018; Caso and Davies, 2022). Alternatively, some preclinical experiments demonstrated the prospects of using the base editing technologies to treat diseases. Due to the base editor, hA3A-BE3 creates C-to-T conversion at NGN PAM sites efficiently. Wang et al. (2020) showed that the binding site of BCL11A (TGACCA: −114 to −119) represents an ideal target site for binding with hA3A-BE3 to induce a mutation and raise the level of fetal γ-globin expression to ameliorate the ß-hemoglobinopathies. Furthermore, a transformer BE (tBE) system to eliminate unintended mutations was developed and delivered by AAV into mice creating a premature stop codon in proprotein convertase subtilisin/kexin type 9 (PCSK9) gene which significantly reduced the serum PCSK9 and cholesterol (Wang et al., 2021a). Newby et al. (2021) used mRNA encoding the Base editor to treat hematopoietic stem and progenitor cells (HSPCs) from patient with SCD generating high percentage conversion of the SCD allele (HBBS) into Makassar ß-globin (HBBG), which is a non-pathogenic variant. It has been shown that base edited CAR T-cells for combinational therapy against T-cell malignancies permitted to enhance molecular remission prior to allo-HSCT for T-cell malignancies (Georgiadis et al., 2021).
3.1.1 BE system limitations and optimizations
The BE system is mainly affected by three aspects: (i) the product purity, (ii) the off-target mutation and 3) the editing window of adjacent sites. Product purity refers to the percentage of edited sequencing reads [reads in which the targeted C has been converted into T but also C to R (G or A)] (Zhang et al., 2018). Furthermore, the product purity is also associated with the frequencies of Indels which are formed during the gene modifying process. Other studies have shown that over-expression of Uracil DNA glycosylase inhibitor (UGI) increased the purity of BE products in human cells (Komor et al., 2017; Jiang et al., 2018; Jang et al., 2021) In addition to UGI, Gehrke et al. (2018) used an engineered human enzyme known as APOBEC3A (eA3A) to develop the eA3A-BE3 base editor which improved target accuracy and reduced bystander mutations. To reduce the Indel formations, the BE4-Gam and CBEmax base editors have been developed (Komor et al., 2017; Huang et al., 2019). The genome targeting function of the base editor caused substantial off-target editing in genomic DNA and RNA (Gaudelli, 2017). Different bioinformatic tools have been developed to overcome the off-target problems including the CRISPR-Cas9/Cpf1 (Kim et al., 2020; Lee et al., 2020), BE-Designer, BE-Analyzer (Hwang and Bae. 2021) and Digenome-seq (Kim et al., 2021b). Furthermore, the engineered deaminases (Rees et al., 2017; Zhou et al., 2019) and gRNA sequences (Kleinstiver et al., 2016b; Hu et al., 2021) have also been built to significantly reduce the off target in the BE system. The range of the editing window for base editing varies according to different application (Kim et al., 2017; Wang et al., 2018b, Wang et al., 2020a; Huang et al., 2019; Tan et al., 2019; Dang et al., 2021; Fu et al., 2021). There are two solutions to set the best range for editing window. 1) When only one specific base pair is required to be changed accurately, the editing window should be minimized to increase the target base accuracy. 2) When CBE system is used to introduce premature stop codons, to produce large-scale saturation mutations, to screen gene function, to locate key amino acid positions in protein domains, etc., a large editing activity window is more advantageous.
3.2 Prime editing
Genome editing with base editors effectively induced C→T, G→A, A→G, T→C, C→G, and C→A base substitutions without inducing DSB (Gaudelli, 2017; Zhao et al., 2021b; Kurt et al., 2021). However, they are unable to correct variants beyond these six transition mutations, or other modifications like insertions and deletions of DNA fragments which are successfully achieved by Prime editing (PE). PE uses an engineered Cas9 nickase fused to a reverse transcriptase (RT) enzyme and a modified sgRNA known as prime editing guide RNA (pegRNA) (Figure 2B) (Anzalone et al., 2019). Recent efforts stepwise improved the efficiency of PE system to PEmax system (Chen et al., 2021) and engineer pegRNA known as epegRNA (Nelson et al., 2022b). PE makes possible the accurate insertion up to 1 kb (Wang et al., 2022a) and the deletion of up to 10 kb (Choi et al., 2022) DNA fragment.
As base editing, prime editing has not yet entered clinical trials due to its immature development. However, the potential of PE has so far been demonstrated during the past 2 years in vitro (Surun et al., 2020; Habib et al., 2022; Happi Mbakam et al., 2022; Petri et al., 2022; Tremblay et al., 2022) and in animal models (Liu et al., 2021b; Jang et al., 2022; Zheng et al., 2022; Zhi et al., 2022). Unfortunately, the gene modification efficiency is very low in some models. This may be due to the use of the split viruses to overcome the size of PE, which cannot be packed into a single viral delivery vector. Prime editing has the potential to increase the safety and expand the scope of genome-editing in T-cells showing that method is adaptable to enhance the efficiency of CAR T-cell therapy by concurrently introducing additional complex gene edits into T-cells (Petri et al., 2022).
3.2.1 Prime editing limitations and optimizations
The defects of Prime editing are somehow similar to classical CRISPR/Cas9 and base editing as described above, such as the PAM restriction, off-targets and large molecular weight delivery hindering. However, some new progresses have been obtained. Kweon et al. (2021) developed PE2 variants by using various SpCas9 variants named PE2-VQR, PE2-VRQR, PE2-NG, PE2-SpG, and PE2-SpRY. The PE2-SpRY enables targeting 94.4% of pathogenic variants. Furthermore, some studies demonstrated that the PE is not always efficient due to some unknown factors (Li et al., 2020; Lin et al., 2020; Xu et al., 2020). Recently, further research showed that the design of the pegRNA strongly affects the efficiency of prime editors. Researchers demonstrated that optimization of pegRNA sequence widely improved PE efficiency in different cell lines and the efficiency of installing or correcting disease-associated mutations (Jiang et al., 2020, 2022; Kim et al., 2021b; Lin et al., 2021; Nelson et al., 2021).
4 Conclusion
During the past decade, gene editing technologies got tremendous improvement in optimizations and applications. With the continuous optimizations of these technologies ZFNs, TALENs and CRISPR-Cas9 have already entered human clinical trials. To date, the majority of clinical applications of these technologies are focused on ex vivo gene editing therapeutics. Ex vivo editing is highly effective for many medical conditions, such as sickle cell disease, but genome editing should ideally be used for diseases that require in vivo cell modification. Having said that, the in vivo applications of CRISPR technologies is challenged by issues such as off-target editing, inefficiency, and the stimulation of counterproductive immune responses. Current research addressing these issues may launch new avenues for clinical applications of those nuclease-mediated technologies. Moreover, the novel innovations such as Base editing and Prime editing are still at the pre-clinical stage. All these approaches require DNA strand break that evokes DNA damage responses. Therefore, more efforts are needed to address these limitations including wide off-target events, genome stability, transcription-activation systems and cell proliferation to accelerate the treatment of genetic and infectious diseases.
Author contributions
YL and CHM conceived the manuscript, YL, CHM, EB, BS, and J-PT wrote the manuscript, J-PT supervised the work.
Funding
The work is supported by the grants from the Canadian Institute of Health Research and from the Foundation for Cell and Gene Therapy. YL is supported by the China Scholarship Council (CSC) Grant.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Almeida, M. J., and Matos, A. (2019). Designer nucleases: Gene-editing therapies using CCR5 as an emerging target in HIV. Curr. HIV Res. 17, 306–323. doi:10.2174/1570162X17666191025112918
Anzalone, A. v., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. doi:10.1038/s41586-019-1711-4
Arnould, S., Delenda, C., Grizot, S., Desseaux, C., Paques, F., Silva, G. H., et al. (2011). The I-CreI meganuclease and its engineered derivatives: Applications from cell modification to gene therapy. Protein Eng. Des. Sel. 24, 27–31. doi:10.1093/protein/gzq083
Aubert, M., Madden, E. A., Loprieno, M., Feelixge, H. S. D., Stensland, L., Huang, M.-L., et al. (2016). In vivo disruption of latent HSV by designer endonuclease therapy. JCI Insight 1, e88468. doi:10.1172/jci.insight.88468
Bauer, D. E., and Orkin, S. H. (2015). Hemoglobin switching’s surprise: The versatile transcription factor BCL11A is a master repressor of fetal hemoglobin. Curr. Opin. Genet. Dev. 33, 62–70. doi:10.1016/j.gde.2015.08.001
Benjamin, R., Graham, C., Yallop, D., Jozwik, A., Mirci-Danicar, O. C., Lucchini, G., et al. (2020). Genome-edited, donor-derived allogeneic anti-CD19 chimeric antigen receptor T cells in paediatric and adult B-cell acute lymphoblastic leukaemia: Results of two phase 1 studies. Lancet 396, 1885–1894. doi:10.1016/S0140-6736(20)32334-5
Benusiglio, P. R., Fallet, V., Sanchis-Borja, M., Coulet, F., and Cadranel, J. (2021). Lung cancer is also a hereditary disease. Eur. Respir. Rev. 30, 210045. doi:10.1183/16000617.0045-2021
Boch, J., Scholze, H., Schornack, S., Landgraf, A., Hahn, S., Kay, S., et al. (2009). Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512. doi:10.1126/science.1178811
Boissel, S., Jarjour, J., Astrakhan, A., Adey, A., Gouble, A., Duchateau, P., et al. (2014a). megaTALs: a rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res. 42, 2591–2601. doi:10.1093/nar/gkt1224
Boissel, S., Jarjour, J., Astrakhan, A., Adey, A., Gouble, A., Duchateau, P., et al. (2014b). megaTALs: a rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res. 42, 2591–2601. doi:10.1093/nar/gkt1224
Casas-Mollano, J. A., Zinselmeier, M. H., Erickson, S. E., and Smanski, M. J. (2020). CRISPR-cas activators for engineering gene expression in higher eukaryotes. CRISPR J. 3, 350–364. doi:10.1089/crispr.2020.0064
Caso, F., and Davies, B. (2022). Base editing and prime editing in laboratory animals. Lab. Anim. 56, 35–49. doi:10.1177/0023677221993895
Cermak, T., Doyle, E. L., Christian, M., Wang, L., Zhang, Y., Schmidt, C., et al. (2011). Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82. doi:10.1093/nar/gkr218
Cermak, T., Starker, C. G., and Voytas, D. F. (2015). Efficient design and assembly of custom TALENs using the Golden Gate platform. Methods Mol. Biol. 1239, 133–159. doi:10.1007/978-1-4939-1862-1_7
Chang, K.-H., Smith, S. E., Sullivan, T., Chen, K., Zhou, Q., West, J. A., et al. (2017). Long-term engraftment and fetal globin induction upon BCL11A gene editing in bone-marrow-derived CD34 + hematopoietic stem and progenitor cells. Mol. Ther. Methods Clin. Dev. 4, 137–148. doi:10.1016/j.omtm.2016.12.009
Chapdelaine, P., Pichavant, C., Rousseau, J., Pâques, F., and Tremblay, J. P. (2010). Meganucleases can restore the reading frame of a mutated dystrophin. Gene Ther. 17, 846–858. doi:10.1038/gt.2010.26
Chen, J. S., Dagdas, Y. S., Kleinstiver, B. P., Welch, M. M., Sousa, A. A., Harrington, L. B., et al. (2017). Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature 550, 407–410. doi:10.1038/nature24268
Chen, P. J., Hussmann, J. A., Yan, J., Knipping, F., Ravisankar, P., Chen, P.-F., et al. (2021). Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell. 184, 5635–5652.e29. e29. doi:10.1016/j.cell.2021.09.018
Chen, W., Dai, G., Qian, Y., Wen, L., He, X., Liu, H., et al. (2022). PIK3CA mutation affects the proliferation of colorectal cancer cells through the PI3K-MEK/PDK1-GPT2 pathway. Oncol. Rep. 47, 11. doi:10.3892/or.2021.8222
Choi, J., Chen, W., Suiter, C. C., Lee, C., Chardon, F. M., Yang, W., et al. (2022). Precise genomic deletions using paired prime editing. Nat. Biotechnol. 40, 218–226. doi:10.1038/s41587-021-01025-z
Claussnitzer, M., Cho, J. H., Collins, R., Cox, N. J., Dermitzakis, E. T., Hurles, M. E., et al. (2020). A brief history of human disease genetics. Nature 577, 179–189. doi:10.1038/s41586-019-1879-7
Cobo, I., Tanaka, T., Glass, C. K., and Yeang, C. (2022). Clonal hematopoiesis driven by DNMT3A and TET2 mutations: Role in monocyte and macrophage biology and atherosclerotic cardiovascular disease. Curr. Opin. Hematol. 29, 1–7. doi:10.1097/MOH.0000000000000688
Cong, L., Ran, F. A., Cox, D., Lin, S. L., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi:10.1126/science.1231143
Dang, L., Zhou, X., Zhong, X., Yu, W., Huang, S., Liu, H., et al. (2021). Correction of the pathogenic mutation in TGM1 gene by adenine base editing in mutant embryos. Mol. Ther. 30, 175–183. doi:10.1016/j.ymthe.2021.05.007
Deng, P., Carter, S., and Fink, K. (2019). Design, construction, and application of transcription activation-like effectors. Methods Mol. Biol., 47–58. doi:10.1007/978-1-4939-9065-8_3
Doench, J. G., Hartenian, E., Graham, D. B., Tothova, Z., Hegde, M., Smith, I., et al. (2014). Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267. doi:10.1038/nbt.3026
Dreier, B., Beerli, R. R., Segal, D. J., Flippin, J. D., and Barbas, C. F. (2001). Development of zinc finger domains for recognition of the 5′-ANN-3′ family of DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 276, 29466–29478. doi:10.1074/jbc.M102604200
Dreier, B., Fuller, R. P., Segal, D. J., Lund, C. v., Blancafort, P., Huber, A., et al. (2005). Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 280, 35588–35597. doi:10.1074/jbc.M506654200
Durai, S., Mani, M., Kandavelou, K., Wu, J., Porteus, M. H., Chandrasegaran, S., et al. (2005). Zinc finger nucleases: Custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 33, 5978–5990. doi:10.1093/nar/gki912
Durai, S., Mani, M., Kandavelou, K., Wu, J., Porteus, M. H., and Chandrasegaran, S. (2005). Zinc finger nucleases: Custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 33, 5978–5990. doi:10.1093/nar/gki912
Epinat, J. C., Arnould, S., Chames, P., Rochaix, P., Desfontaines, D., Puzin, C., et al. (2003). A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 31, 2952–2962. doi:10.1093/nar/gkg375
Feng, Y., Zhang, S., and Huang, X. (2014). A robust TALENs system for highly efficient mammalian genome editing. Sci. Rep. 4, 3632. doi:10.1038/srep03632
Frangoul, H., Altshuler, D., Cappellini, M. D., Chen, Y.-S., Domm, J., Eustace, B. K., et al. (2021). CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N. Engl. J. Med. 384, 252–260. doi:10.1056/NEJMoa2031054
Fu, J., Li, Q., Liu, X., Tu, T., Lv, X., Yin, X., et al. (2021). Human cell based directed evolution of adenine base editors with improved efficiency. Nat. Commun. 12, 5897. doi:10.1038/s41467-021-26211-0
Gaj, T., Guo, J., Kato, Y., Sirk, S. J., and Barbas, C. F. (2012). Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat. Methods 9, 805–807. doi:10.1038/nmeth.2030
Gao, H., Smith, J., Yang, M., Jones, S., Djukanovic, V., Nicholson, M. G., et al. (2010). Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 61, 176–187. doi:10.1111/j.1365-313X.2009.04041.x
Gao, Z., Herrera-Carrillo, E., and Berkhout, B. (2019). A single H1 promoter can drive both guide RNA and endonuclease expression in the CRISPR-cas9 system. Mol. Ther. Nucleic Acids 14, 32–40. doi:10.1016/j.omtn.2018.10.016
Gapinske, M., Luu, A., Winter, J., Woods, W. S., Kostan, K. A., Shiva, N., et al. (2018). CRISPR-SKIP: Programmable gene splicing with single base editors. Genome Biol. 19, 107. doi:10.1186/s13059-018-1482-5
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471. doi:10.1038/nature24644
Gehrke, J. M., Cervantes, O., Clement, M. K., Wu, Y., Zeng, J., Bauer, D. E., et al. (2018). An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat. Biotechnol. 36, 977–982. doi:10.1038/nbt.4199
Georgiadis, C., Rasaiyaah, J., Gkazi, S. A., Preece, R., Etuk, A., Christi, A., et al. (2021). Base-edited CAR T cells for combinational therapy against T cell malignancies. Leukemia 35, 3466–3481. doi:10.1038/s41375-021-01282-6
Ghosh, R., Das, D., and Franco, S. (2018). “The role for the DSB response pathway in regulating chromosome translocations65–87. doi:10.1007/978-981-13-0593-1_6
Gillmore, J. D., Gane, E., Taubel, J., Kao, J., Fontana, M., Maitland, M. L., et al. (2021). CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 385, 493–502. doi:10.1056/NEJMoa2107454
Gouble, A., Smith, J., Bruneau, S., Perez, C., Guyot, V., Cabaniols, J. P., et al. (2006). Efficient in toto targeted recombination in mouse liver by meganuclease-induced double-strand break. J. Gene Med. 8, 616–622. doi:10.1002/jgm.879
Gutschner, T., Baas, M., and Diederichs, S. (2011). Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res. 21, 1944–1954. doi:10.1101/gr.122358.111
Haapaniemi, E., Botla, S., Persson, J., Schmierer, B., and Taipale, J. (2018). CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 24, 927–930. doi:10.1038/s41591-018-0049-z
Habib, O., Habib, G., Hwang, G. H., and Bae, S. (2022). Comprehensive analysis of prime editing outcomes in human embryonic stem cells. Nucleic Acids Res. 50, 1187–1197. doi:10.1093/nar/gkab1295
Happi Mbakam, C., Rousseau, J., Tremblay, G., Yameogo, P., and Tremblay, J. P. (2022). Prime editing permits the introduction of specific mutations in the gene responsible for duchenne muscular dystrophy. Int. J. Mol. Sci. 23, 6160. doi:10.3390/ijms23116160
Hoban, M. D., Cost, G. J., Mendel, M. C., Romero, Z., Kaufman, M. L., Joglekar, A. v., et al. (2015). Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood 125, 2597–2604. doi:10.1182/blood-2014-12-615948
Hu, J. H., Miller, S. M., Geurts, M. H., Tang, W., Chen, L., Sun, N., et al. (2018). Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63. doi:10.1038/nature26155
Hu, Z., Wang, Y., Liu, Q., Qiu, Y., Zhong, Z., Li, K., et al. (2021). Improving the precision of base editing by bubble hairpin single guide RNA. MBio 12, e00342-21. doi:10.1128/mBio.00342-21
Huang, T. P., Zhao, K. T., Miller, S. M., Gaudelli, N. M., Oakes, B. L., Fellmann, C., et al. (2019). Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors. Nat. Biotechnol. 37, 626–631. doi:10.1038/s41587-019-0134-y
Hwang, G. H., and Bae, S. (2021). Web-based base editing toolkits: BE-Designer and BE-analyzer. Methods Mol. Biol. 2189, 81–88. doi:10.1007/978-1-0716-0822-7_7
Hwang, W. Y., Fu, Y., Reyon, D., Maeder, M. L., Tsai, S. Q., Sander, J. D., et al. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227–229. doi:10.1038/nbt.2501
Ihry, R. J., Worringer, K. A., Salick, M. R., Frias, E., Ho, D., Theriault, K., et al. (2018). p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. 24, 939–946. doi:10.1038/s41591-018-0050-6
Iliakis, G., Wang, H., Perrault, A. R., Boecker, W., Rosidi, B., Windhofer, F., et al. (2004). Mechanisms of DNA double strand break repair and chromosome aberration formation. Cytogenet. Genome Res. 104, 14–20. doi:10.1159/000077461
Izmiryan, A., Danos, O., and Hovnanian, A. (2016). Meganuclease-mediated COL7A1 gene correction for recessive dystrophic epidermolysis bullosa. J. Investig. Dermatol. 136, 872–875. doi:10.1016/j.jid.2015.11.028
Jalligampala, archana, Noel, Jennifer, JamesFransen, W., Wang, Wei, Jabbar, Maha, Hasan, Nazarul, et al. (2021). Rho 1-2 meganuclease, an allele-specific gene-editing therapy, rejuvenates rod photoreceptor structure and function in a pig model of autosomal dominant Retinitis Pigmentosa (adRP). Investigative Ophthalmology & Visual Science, 1478.
Jang, H., Jo, D. H., Cho, C. S., Shin, J. H., Seo, J. H., Yu, G., et al. (2022). Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases. Nat. Biomed. Eng. 6, 181–194. doi:10.1038/s41551-021-00788-9
Jang, H. K., Jo, D. H., Lee, S. N., Cho, C. S., Jeong, Y. K., Jung, Y., et al. (2021). High-purity production and precise editing of DNA base editing ribonucleoproteins. Sci. Adv. 7, eabg2661. doi:10.1126/sciadv.abg2661
Ji, H., Lu, P., Liu, B., Qu, X., Wang, Y., Jiang, Z., et al. (2018). Zinc-finger nucleases induced by HIV-1 tat excise HIV-1 from the host genome in infected and latently infected cells. Mol. Ther. Nucleic Acids 12, 67–74. doi:10.1016/j.omtn.2018.04.014
Jiang, T., Zhang, X. O., Weng, Z., and Xue, W. (2022). Deletion and replacement of long genomic sequences using prime editing. Nat. Biotechnol. 40, 227–234. doi:10.1038/s41587-021-01026-y
Jiang, W., Feng, S., Huang, S., Yu, W., Li, G., Yang, G., et al. (2018). BE-PLUS: A new base editing tool with broadened editing window and enhanced fidelity. Cell. Res. 28, 855–861. doi:10.1038/s41422-018-0052-4
Jiang, Y. Y., Chai, Y. P., Lu, M. H., Han, X. L., Lin, Q., Zhang, Y., et al. (2020). Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol. 21, 257. doi:10.1186/s13059-020-02170-5
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., Charpentier, E., et al. (2012a1979). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi:10.1126/science.1225829
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., Charpentier, E., et al. (2012b1979). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi:10.1126/science.1225829
Khatibi, S., Sahebkar, A., and Aghaee-Bakhtiari, S. H. (2021). CRISPR genome editing technology and its application in genetic diseases: A review. Curr. Pharm. Biotechnol. 22, 468–479. doi:10.2174/1389201021666200621161610
Kim, D., Kang, B. C., and Kim, J. S. (2021a). Identifying genome-wide off-target sites of CRISPR RNA-guided nucleases and deaminases with Digenome-seq. Nat. Protoc. 16, 1170–1192. doi:10.1038/s41596-020-00453-6
Kim, H. J., Oh, S. Y., and Lee, S. J. (2020). Single-base genome editing in corynebacterium glutamicum with the help of negative selection by target-mismatched CRISPR/Cpf1. J. Microbiol. Biotechnol. 30, 1583–1591. doi:10.4014/jmb.2006.06036
Kim, H. K., Yu, G., Park, J., Min, S., Lee, S., Yoon, S., et al. (2021b). Predicting the efficiency of prime editing guide RNAs in human cells. Nat. Biotechnol. 39, 198–206. doi:10.1038/s41587-020-0677-y
Kim, Y. B., Komor, A. C., Levy, J. M., Packer, M. S., Zhao, K. T., Liu, D. R., et al. (2017). Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 35, 371–376. doi:10.1038/nbt.3803
Kim, Y., Hong, S.-A., Yu, J., Eom, J., Jang, K., Yoon, S., et al. (2021c). Adenine base editing and prime editing of chemically derived hepatic progenitors rescue genetic liver disease. Cell. Stem Cell. 28, 1614–1624.e5. e5. doi:10.1016/j.stem.2021.04.010
Kleinstiver, B. P., Pattanayak, V., Prew, M. S., Tsai, S. Q., Nguyen, N. T., Zheng, Z., et al. (2016b). High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495. doi:10.1038/nature16526
Kleinstiver, B. P., Pattanayak, V., Prew, M. S., Tsai, S. Q., Nguyen, N. T., Zheng, Z., et al. (2016a). High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495. doi:10.1038/nature16526
Kleinstiver, B. P., Prew, M. S., Tsai, S. Q., Topkar, V. v., Nguyen, N. T., Zheng, Z., et al. (2015). Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485. doi:10.1038/nature14592
Kocak, D. D., Josephs, E. A., Bhandarkar, V., Adkar, S. S., Kwon, J. B., Gersbach, C. A., et al. (2019). Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat. Biotechnol. 37, 657–666. doi:10.1038/s41587-019-0095-1
Komor, A. C. E., Zhao, K. T., Packer, M. S., Gaudelli, N. M., Waterbury, A. L., Koblan, L. W., et al. (2017). Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci. Adv. 3, eaao4774. doi:10.1126/sciadv.aao4774
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., and Liu, D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424. doi:10.1038/nature17946
Kurt, I. C., Zhou, R., Iyer, S., Garcia, S. P., Miller, B. R., Langner, L. M., et al. (2021). CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. 39, 41–46. doi:10.1038/s41587-020-0609-x
Kweon, J., Yoon, J. K., Jang, A. H., Shin, H. R., See, J. E., Jang, G., et al. (2021). Engineered prime editors with PAM flexibility. Mol. Ther. 29, 2001–2007. doi:10.1016/j.ymthe.2021.02.022
Landrum, M. J., Lee, J. M., Benson, M., Brown, G., Chao, C., Chitipiralla, S., et al. (2016). ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 44, D862–D868. doi:10.1093/nar/gkv1222
Lee, H. J., Kim, H. J., and Lee, S. J. (2020). CRISPR-Cas9-mediated pinpoint microbial genome editing aided by target-mismatched sgRNAs. Genome Res. 30, 768–775. doi:10.1101/gr.257493.119
Lee, K. Z., Mechikoff, M. A., Parasa, M. K., Rankin, T. J., Pandolfi, P., Fitzgerald, K. S., et al. (2022). Repurposing the homing endonuclease I-SceI for positive selection and development of gene-editing technologies. ACS Synth. Biol. 11, 53–60. doi:10.1021/acssynbio.1c00340
Li, H., Li, J., Chen, J., Yan, L., and Xia, L. (2020). Precise modifications of both exogenous and endogenous genes in rice by prime editing. Mol. Plant 13, 671–674. doi:10.1016/j.molp.2020.03.011
Lin, Q., Jin, S., Zong, Y., Yu, H., Zhu, Z., Liu, G., et al. (2021). High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat. Biotechnol. 39, 923–927. doi:10.1038/s41587-021-00868-w
Lin, Q., Zong, Y., Xue, C., Wang, S., Jin, S., Zhu, Z., et al. (2020). Prime genome editing in rice and wheat. Nat. Biotechnol. 38, 582–585. doi:10.1038/s41587-020-0455-x
Liu, G., Lin, Q., Jin, S., and Gao, C. (2021a). The CRISPR-Cas toolbox and gene editing technologies. Mol. Cell. 82, 333–347. doi:10.1016/j.molcel.2021.12.002
Liu, M.-S., Gong, S., Yu, H.-H., Jung, K., Johnson, K. A., Taylor, D. W., et al. (2020). Engineered CRISPR/Cas9 enzymes improve discrimination by slowing DNA cleavage to allow release of off-target DNA. Nat. Commun. 11, 3576. doi:10.1038/s41467-020-17411-1
Liu, P., Liang, S. Q., Zheng, C., Mintzer, E., Zhao, Y. G., Ponnienselvan, K., et al. (2021b). Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice. Nat. Commun. 12, 2121. doi:10.1038/s41467-021-22295-w
Liu, Z., Chen, M., Chen, S., Deng, J., Song, Y., Lai, L., et al. (2018). Highly efficient RNA-guided base editing in rabbit. Nat. Commun. 9, 2717. doi:10.1038/s41467-018-05232-2
Lo, T. W., Pickle, C. S., Lin, S., Ralston, E. J., Gurling, M., Schartner, C. M., et al. (2013). Precise and heritable genome editing in evolutionarily diverse nematodes using TALENs and CRISPR/Cas9 to engineer insertions and deletions. Genetics 195, 331–348. -+. doi:10.1534/genetics.113.155382
Lu, Y., Xue, J., Deng, T., Zhou, X., Yu, K., Deng, L., et al. (2020). Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat. Med. 26, 732–740. doi:10.1038/s41591-020-0840-5
Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S., Costa, F., Shah, S. A., Saunders, S. J., et al. (2015). An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 13, 722–736. doi:10.1038/nrmicro3569
Mao, Z., Bozzella, M., Seluanov, A., and Gorbunova, V. (2008). DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell. Cycle 7, 2902–2906. doi:10.4161/cc.7.18.6679
Mariano, A., Xu, L., and Han, R. (2014). Highly efficient genome editing via 2A-coupled co-expression of two TALEN monomers. BMC Res. Notes 7, 628. doi:10.1186/1756-0500-7-628
Martín‐Fernández, J. M., Fleischer, A., Vallejo‐Diez, S., Palomino, E., Sánchez‐Gilabert, A., Ruiz, R., et al. (2020). New bicistronic TALENs greatly improve genome editing. Curr. Protoc. Stem Cell. Biol. 52, e104. doi:10.1002/cpsc.104
Masuda, T., Wang, X., Maeda, M., Canver, M. C., Sher, F., Funnell, A. P. W., et al. (2016). Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science 351, 285–289. doi:10.1126/science.aad3312
Maximiano, M. R., Tavora, F., Prado, G. S., Dias, S. C., Mehta, A., Franco, O. L., et al. (2021). CRISPR genome editing technology: A powerful tool applied to developing agribusiness. J. Agric. Food Chem. 69, 6379–6395. doi:10.1021/acs.jafc.1c01062
Miller, J. C., Holmes, M. C., Wang, J., Guschin, D. Y., Lee, Y. L., Rupniewski, I., et al. (2007). An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 25, 778–785. doi:10.1038/nbt1319
Miller, J. C., Patil, D. P., Xia, D. F., Paine, C. B., Fauser, F., Richards, H. W., et al. (2019a). Enhancing gene editing specificity by attenuating DNA cleavage kinetics. Nat. Biotechnol. 37, 945–952. doi:10.1038/s41587-019-0186-z
Miller, J. C., Patil, D. P., Xia, D. F., Paine, C. B., Fauser, F., Richards, H. W., et al. (2019b). Enhancing gene editing specificity by attenuating DNA cleavage kinetics. Nat. Biotechnol. 37, 945–952. doi:10.1038/s41587-019-0186-z
Miyaoka, Y., Chan, A. H., and Conklin, B. R. (2016). Detecting single-nucleotide substitutions induced by genome editing. Cold Spring Harb. Protoc., pdb.top090845. doi:10.1101/pdb.top090845
Modarai, S. R., Kanda, S., Bloh, K., Opdenaker, L. M., and Kmiec, E. B. (2021). Precise and error-prone CRISPR-directed gene editing activity in human CD34+ cells varies widely among patient samples. Gene Ther. 28, 105–113. doi:10.1038/s41434-020-00192-z
Molina, R., Besker, N., Marcaida, M. J., Montoya, G., Prieto, J., D’Abramo, M., et al. (2016). Key players in I-DmoI endonuclease catalysis revealed from structure and dynamics. ACS Chem. Biol. 11, 1401–1407. doi:10.1021/acschembio.5b00730
Moscou, M. J., and Bogdanove, A. J. (2009). A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501. doi:10.1126/science.1178817
Muenzer, J., Prada, C. E., Burton, B., Lau, H. A., Ficicioglu, C., Foo, C. W. P., et al. (2019). Champions: A phase 1/2 clinical trial with dose escalation of SB-913 ZFN-mediated in vivo human genome editing for treatment of mps II (hunter syndrome). Mol. Genet. Metabolism 126, S104. doi:10.1016/j.ymgme.2018.12.263
Mussolino, C., Alzubi, J., Fine, E. J., Morbitzer, R., Cradick, T. J., Lahaye, T., et al. (2014). TALENs facilitate targeted genome editing in human cells with high specificity and low cytotoxicity. Nucleic Acids Res. 42, 6762–6773. doi:10.1093/nar/gku305
Nelson, J. W., Randolph, P. B., Shen, S. P., Everette, K. A., Chen, P. J., Anzalone, A. v., et al. (2021). Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. 40, 402–410. doi:10.1038/s41587-021-01039-7
Nelson, J. W., Randolph, P. B., Shen, S. P., Everette, K. A., Chen, P. J., Anzalone, A. v., et al. (2022a). Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. 40, 402–410. doi:10.1038/s41587-021-01039-7
Nelson, J. W., Randolph, P. B., Shen, S. P., Everette, K. A., Chen, P. J., Anzalone, A. v., et al. (2022b). Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. 40, 402–410. doi:10.1038/s41587-021-01039-7
Newby, G. A., Yen, J. S., Woodard, K. J., Mayuranathan, T., Lazzarotto, C. R., Li, Y., et al. (2021). Base editing of haematopoietic stem cells rescues sickle cell disease in mice. Nature 595, 295–302. doi:10.1038/s41586-021-03609-w
Nishimasu, H., Shi, X., Ishiguro, S., Gao, L., Hirano, S., Okazaki, S., et al. (2018). Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361, 1259–1262. doi:10.1126/science.aas9129
Paques, F., and Duchateau, P. (2007). Meganucleases and DNA double-strand break-induced recombination: Perspectives for gene therapy. Curr. Gene Ther. 7, 49–66. doi:10.2174/156652307779940216
Paquet, D., Kwart, D., Chen, A., Sproul, A., Jacob, S., Teo, S., et al. (2016). Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125-129doi:10.1038/nature17664
Paschon, D. E., Lussier, S., Wangzor, T., Xia, D. F., Li, P. W., Hinkley, S. J., et al. (2019). Diversifying the structure of zinc finger nucleases for high-precision genome editing. Nat. Commun. 10, 1133. doi:10.1038/s41467-019-08867-x
Pattanayak, V., Ramirez, C. L., Joung, J. K., and Liu, D. R. (2011). Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat. Methods 8, 765–770. doi:10.1038/nmeth.1670
Perez-Pinera, P., Kocak, D. D., Vockley, C. M., Adler, A. F., Kabadi, A. M., Polstein, L. R., et al. (2013). RNA-guided gene activation by CRISPR-Cas9–based transcription factors. Nat. Methods 10, 973–976. doi:10.1038/nmeth.2600
Petersen, B., and Niemann, H. (2015a). Advances in genetic modification of farm animals using zinc-finger nucleases (ZFN). Chromosome Res. 23, 7–15. doi:10.1007/s10577-014-9451-7
Petersen, B., and Niemann, H. (2015b). Molecular scissors and their application in genetically modified farm animals. Transgenic Res. 24, 381–396. doi:10.1007/s11248-015-9862-z
Petri, K., Zhang, W., Ma, J., Schmidts, A., Lee, H., Horng, J. E., et al. (2022). CRISPR prime editing with ribonucleoprotein complexes in zebrafish and primary human cells. Nat. Biotechnol. 40, 189–193. doi:10.1038/s41587-021-00901-y
Philippidis, A. (2021). First positive clinical data for in vivo genome editing in humans opens “new era of medicine. Hum. Gene Ther. 32, 783–786. doi:10.1089/hum.2021.29170.bfs
Porter, S. N., Levine, R. M., and Pruett-Miller, S. M. (2019). A practical guide to genome editing using targeted nuclease technologies. Compr. Physiol. 9, 665–714. doi:10.1002/cphy.c180022
Porteus, M. H., and Baltimore, D. (2003). Chimeric nucleases stimulate gene targeting in human cells. Science 300, 763. doi:10.1126/science.1078395
Prieto, J., Redondo, P., Lopez-Mendez, B., D’Abramo, M., Merino, N., Blanco, F. J., et al. (2018). Understanding the indirect DNA read-out specificity of I-CreI Meganuclease. Sci. Rep. 8, 10286. doi:10.1038/s41598-018-28599-0
Prieto, J., Redondo, P., Padró, D., Arnould, S., Epinat, J.-C., Pâques, F., et al. (2007). The C-terminal loop of the homing endonuclease I-CreI is essential for site recognition, DNA binding and cleavage. Nucleic Acids Res. 35, 3262–3271. doi:10.1093/nar/gkm183
Psatha, N., Reik, A., Phelps, S., Zhou, Y., Dalas, D., Yannaki, E., et al. (2018). Disruption of the BCL11A erythroid enhancer reactivates fetal hemoglobin in erythroid cells of patients with β-thalassemia major. Mol. Ther. Methods Clin. Dev. 10, 313–326. doi:10.1016/j.omtm.2018.08.003
Qasim, W., Zhan, H., Samarasinghe, S., Adams, S., Amrolia, P., Stafford, S., et al. (2017). Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci. Transl. Med. 9, eaaj2013. doi:10.1126/scitranslmed.aaj2013
Ran, F. A., Hsu, P. D., Lin, C.-Y., Gootenberg, J. S., Konermann, S., Trevino, A. E., et al. (2013a). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 154, 1380–1389. doi:10.1016/j.cell.2013.08.021
Ran, F. A., Hsu, P. D., Wright, J., Agarwala, V., Scott, D. A., Zhang, F., et al. (2013b). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308. doi:10.1038/nprot.2013.143
Ranjha, L., Howard, S. M., and Cejka, P. (2018). Main steps in DNA double-strand break repair: An introduction to homologous recombination and related processes. Chromosoma 127, 187–214. doi:10.1007/s00412-017-0658-1
Rees, H. A., Komor, A. C., Yeh, W. H., Caetano-Lopes, J., Warman, M., Edge, A. S. B., et al. (2017). Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 8, 15790. doi:10.1038/ncomms15790
Reyon, D., Khayter, C., Regan, M. R., Joung, J. K., and Sander, J. D. (2012). Engineering designer transcription activator‐‐like effector nucleases (TALENs) by REAL or REAL‐fast assembly. Curr. Protoc. Mol. Biol. 100, Unit 12.15. doi:10.1002/0471142727.mb1215s100
Rouet, P., Smih, F., and Jasin, M. (1994a). Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 91, 6064–6068. doi:10.1073/pnas.91.13.6064
Rouet, P., Smih, F., and Jasin, M. (1994b). Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 14, 8096–8106. doi:10.1128/Mcb.14.12.8096
Samuelson, C., Radtke, S., Zhu, H., Llewellyn, M., Fields, E., Cook, S., et al. (2021). Multiplex CRISPR/Cas9 genome editing in hematopoietic stem cells for fetal hemoglobin reinduction generates chromosomal translocations. Mol. Ther. Methods Clin. Dev. 23, 507–523. doi:10.1016/j.omtm.2021.10.008
Sanjana, N. E., Cong, L., Zhou, Y., Cunniff, M. M., Feng, G., Zhang, F., et al. (2012). A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 7, 171–192. doi:10.1038/nprot.2011.431
Santiago, Y., Chan, E., Liu, P.-Q., Orlando, S., Zhang, L., Urnov, F. D., et al. (2008). Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc. Natl. Acad. Sci. U. S. A. 105, 5809–5814. doi:10.1073/pnas.0800940105
Scholze, H., and Boch, J. (2011). TAL effectors are remote controls for gene activation. Curr. Opin. Microbiol. 14, 47–53. doi:10.1016/j.mib.2010.12.001
Shalem, O., Sanjana, N. E., Hartenian, E., Shi, X., Scott, D. A., Mikkelsen, T. S., et al. (2014a1979). Genome-scale CRISPR-cas9 knockout screening in human cells. Science 343, 84–87. doi:10.1126/science.1247005
Shalem, O., Sanjana, N. E., Hartenian, E., Shi, X., Scott, D. A., Mikkelson, T., et al. (2014b1979). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. doi:10.1126/science.1247005
Siegl, T., Petzke, L., Welle, E., and Luzhetskyy, A. (2010). I-SceI endonuclease: A new tool for DNA repair studies and genetic manipulations in streptomyces. Appl. Microbiol. Biotechnol. 87, 1525–1532. doi:10.1007/s00253-010-2643-y
Slaymaker, I. M., Gao, L., Zetsche, B., Scott, D. A., Yan, W. X., Zhang, F., et al. (2016). Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88. doi:10.1126/science.aad5227
Smith, A. R., Schiller, G. J., Vercellotti, G. M., Kwiatkowski, J. L., Krishnamurti, L., Esrick, E. B., et al. (2019). Preliminary results of a phase 1/2 clinical study of zinc finger nuclease-mediated editing of BCL11A in autologous hematopoietic stem cells for transfusion-dependent beta thalassemia. Blood 134, 3544. doi:10.1182/blood-2019-125743
Stadtmauer, E. A., Fraietta, J. A., Davis, M. M., Cohen, A. D., Weber, K. L., Lancaster, E., et al. (2020). CRISPR-engineered T cells in patients with refractory cancer. Science, eaba7365. doi:10.1126/science.aba7365
Stoddard, B. L. (2011). Homing endonucleases: From microbial genetic invaders to reagents for targeted DNA modification. Structure 19, 7–15. doi:10.1016/j.str.2010.12.003
Sun, Z., Wang, M., Han, S., Ma, S., Zou, Z., Ding, F., et al. (2018). Production of hypoallergenic milk from DNA-free beta-lactoglobulin (BLG) gene knockout cow using zinc-finger nucleases mRNA. Sci. Rep. 8, 15430. doi:10.1038/s41598-018-32024-x
Surun, D., Schneider, A., Mircetic, J., Neumann, K., Lansing, F., Paszkowski-Rogacz, M., et al. (2020). Efficient Generation and Correction of Mutations in Human iPS Cells Utilizing mRNAs of CRISPR Base Editors and Prime, 11. doi:10.3390/genes11050511Genes. (Basel)
Suzuki, S., Ohta, K., Nakajima, Y., Shigeto, H., Abe, H., Kawai, A., et al. (2020). Meganuclease-based artificial transcription factors. ACS Synth. Biol. 9, 2679–2691. doi:10.1021/acssynbio.0c00083
Tan, J., Zhang, F., Karcher, D., and Bock, R. (2019). Engineering of high-precision base editors for site-specific single nucleotide replacement. Nat. Commun. 10, 439. doi:10.1038/s41467-018-08034-8
Tang, L. C., and Gu, F. (2020). Next-generation CRISPR-cas for genome editing: Focusing on the Cas protein and PAM. Yi Chuan 42, 236–249. doi:10.16288/j.yczz.19-297
Tang, Y., and Fu, Y. (2018). Class 2 CRISPR/Cas: An expanding biotechnology toolbox for and beyond genome editing. Cell. Biosci. 8, 59. doi:10.1186/s13578-018-0255-x
Tebas, P., Jadlowsky, J. K., Shaw, P. A., Tian, L., Esparza, E., Brennan, A. L., et al. (2021). CCR5-edited CD4+ T cells augment HIV-specific immunity to enable post-rebound control of HIV replication. J. Clin. Investig. 131, 144486. doi:10.1172/JCI144486
Tebas, P., Stein, D., Tang, W. W., Frank, I., Wang, S. Q., Lee, G., et al. (2014). Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 370, 901–910. doi:10.1056/NEJMoa1300662
Tekel, S. J., Brookhouser, N., Standage-Beier, K., Wang, X., and Brafman, D. A. (2021). Cytosine and adenosine base editing in human pluripotent stem cells using transient reporters for editing enrichment. Nat. Protoc. 16, 3596–3624. doi:10.1038/s41596-021-00552-y
Tremblay, G., Rousseau, J., Mbakam, C. H., and Tremblay, J. P. (2022). Insertion of the Icelandic mutation (A673T) by prime editing: A potential preventive treatment for familial and sporadic alzheimer’s disease. CRISPR J. 5, 109–122. doi:10.1089/crispr.2021.0085
Tzfira, T., Frankman, L. R., Vaidya, M., and Citovsky, V. (2003). Site-specific integration of Agrobacterium tumefaciens T-DNA via double-stranded intermediates. Plant Physiol. 133, 1011–1023. doi:10.1104/pp.103.032128
Uddin, F., Rudin, C. M., and Sen, T. (2020). CRISPR gene therapy: Applications, limitations, and implications for the future. Front. Oncol. 10, 1387. doi:10.3389/fonc.2020.01387
Urnov, F. D., Rebar, E. J., Holmes, M. C., Zhang, H. S., and Gregory, P. D. (2010). Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11, 636–646. doi:10.1038/nrg2842
Vakulskas, C. A., and Behlke, M. A. (2019). Evaluation and reduction of CRISPR off-target cleavage events. Nucleic Acid. Ther. 29, 167–174. doi:10.1089/nat.2019.0790
Valerie, K., and Povirk, L. F. (2003). Regulation and mechanisms of mammalian double-strand break repair. Oncogene 22, 5792–5812. doi:10.1038/sj.onc.1206679
Walton, R. T., Christie, K. A., Whittaker, M. N., and Kleinstiver, B. P. (2020). Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368, 290–296. doi:10.1126/science.aba8853
Wang, F., Zeng, Y., Wang, Y., and Niu, Y. (2020a). The development and application of a base editor in biomedicine. Biomed. Res. Int. 2020, 2907623. doi:10.1155/2020/2907623
Wang, J., He, Z., Wang, G., Zhang, R., Duan, J., Gao, P., et al. (2022a). Efficient targeted insertion of large DNA fragments without DNA donors. Nat. Methods 19, 331–340. doi:10.1038/s41592-022-01399-1
Wang, J., Lu, C., and Wei, S. (2022b). Whole-genome sequencing identifies I-SceI-mediated transgene integration sites in Xenopus tropicalis snai2: eGFP line. G3 (Bethesda). doi:10.1093/g3journal/jkac037
Wang, L., Breton, C., Warzecha, C. C., Bell, P., Yan, H., He, Z., et al. (2021a). Long-term stable reduction of low-density lipoprotein in nonhuman primates following in vivo genome editing of PCSK9. Mol. Ther. 29, 2019–2029. doi:10.1016/j.ymthe.2021.02.020
Wang, L. R., Li, L. X., Ma, Y. L., Hu, H. D., Li, Q., Yang, Y., et al. (2020b). Reactivation of gamma-globin expression through Cas9 or base editor to treat beta-hemoglobinopathies. Cell. Res. 30, 276–278. doi:10.1038/s41422-019-0267-z
Wang, L., Smith, J., Breton, C., Clark, P., Zhang, J., Ying, L., et al. (2018a). Meganuclease targeting of PCSK9 in macaque liver leads to stable reduction in serum cholesterol. Nat. Biotechnol. 36, 717–725. doi:10.1038/nbt.4182
Wang, X., Li, J., Wang, Y., Yang, B., Wei, J., Wu, J., et al. (2018b). Efficient base editing in methylated regions with a human APOBEC3A-Cas9 fusion. Nat. Biotechnol. 36, 946–949. doi:10.1038/nbt.4198
Wang, Y., Wang, H., Jian, Y., Luo, Z., Shao, H., and Zhang, W. (2021b). Strategies for optimization of the CRISPR-based genome editing system for enhanced editing specificity. Hum. Gene Ther. doi:10.1089/hum.2021.283
Woo, J. W., Kim, J., Kwon, S. il, Corvalán, C., Cho, S. W., Kim, H., et al. (2015). DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. doi:10.1038/nbt.3389
Xiao, T., Wu, B., Cao, Y., Liu, R., Cheng, G., Wang, L., et al. (2021). Genetic identification of pathogenic variations of the DMD gene: A retrospective study from 10, 481 neonatal patients based on next-generation sequencing data. Ann. Transl. Med. 9, 766. doi:10.21037/atm-20-7102
Xiong, K., Li, S., Zhang, H., Cui, Y., Yu, D., Li, Y., et al. (2013). Targeted editing of goat genome with modular-assembly zinc finger nucleases based on activity prediction by computational molecular modeling. Mol. Biol. Rep. 40, 4251–4256. doi:10.1007/s11033-013-2507-5
Xu, W., Zhang, C., Yang, Y., Zhao, S., Kang, G., He, X., et al. (2020). Versatile nucleotides substitution in plant using an improved prime editing system. Mol. Plant 13, 675–678. doi:10.1016/j.molp.2020.03.012
Yi, L., and Li, J. (2016). CRISPR-Cas9 therapeutics in cancer: Promising strategies and present challenges. Biochim. Biophys. Acta 1866, 197–207. doi:10.1016/j.bbcan.2016.09.002
Zekonyte, U., Bacman, S. R., Smith, J., Shoop, W., Pereira, C. v., Tomberlin, G., et al. (2021). Mitochondrial targeted meganuclease as a platform to eliminate mutant mtDNA in vivo. Nat. Commun. 12, 3210. doi:10.1038/s41467-021-23561-7
Zeng, J., Wu, Y., Ren, C., Bonanno, J., Shen, A. H., Shea, D., et al. (2020). Therapeutic base editing of human hematopoietic stem cells. Nat. Med. 26, 535–541. doi:10.1038/s41591-020-0790-y
Zhang, H., Pan, H., Zhou, C., Wei, Y., Ying, W., Li, S., et al. (2018). Simultaneous zygotic inactivation of multiple genes in mouse through CRISPR/Cas9-mediated base editing. Development 145, dev168906. doi:10.1242/dev.168906
Zhang, M., Zhou, C., Wei, Y., Xu, C., Pan, H., Ying, W., et al. (2019). Human cleaving embryos enable robust homozygotic nucleotide substitutions by base editors. Genome Biol. 20, 101. doi:10.1186/s13059-019-1703-6
Zhang, S., Wang, J., and Wang, J. (2020). One-day TALEN assembly protocol and a dual-tagging system for genome editing. ACS Omega 5, 19702–19714. doi:10.1021/acsomega.0c02396
Zhang, Y., Liu, L., Guo, S., Song, J., Zhu, C., Yue, Z., et al. (2017). Deciphering TAL effectors for 5-methylcytosine and 5-hydroxymethylcytosine recognition. Nat. Commun. 8, 901. doi:10.1038/s41467-017-00860-6
Zhao, D., Li, J., Li, S., Xin, X., Hu, M., Price, M. A., et al. (2021a). Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. 39, 35–40. doi:10.1038/s41587-020-0592-2
Zhao, D., Li, J., Li, S., Xin, X., Hu, M., Price, M. A., et al. (2021b). Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. 39, 35–40. doi:10.1038/s41587-020-0592-2
Zheng, C. W., Liang, S. Q., Liu, B., Liu, P. P., Kwan, S. Y., Wolfe, S. A., et al. (2022). A flexible split prime editor using truncated reverse transcriptase improves dual-AAV delivery in mouse liver. Mol. Ther. 30, 1343–1351. doi:10.1016/j.ymthe.2022.01.005
Zhi, S., Chen, Y., Wu, G., Wen, J., Wu, J., Liu, Q., et al. (2022). Dual-AAV delivering split prime editor system for in vivo genome editing. Mol. Ther. 30, 283–294. doi:10.1016/j.ymthe.2021.07.011
Zhou, C., Sun, Y., Yan, R., Liu, Y., Zuo, E., Gu, C., et al. (2019). Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature 571, 275–278. doi:10.1038/s41586-019-1314-0
Keywords: gene editing, ZFN, TALEN, CRISPR-cas, Cytidine Base Editor (CBE), Adenosine Base Editor (ABE), prime editing
Citation: Lu Y, Happi Mbakam C, Song B, Bendavid E and Tremblay J-P (2022) Improvements of nuclease and nickase gene modification techniques for the treatment of genetic diseases. Front. Genome Ed. 4:892769. doi: 10.3389/fgeed.2022.892769
Received: 09 March 2022; Accepted: 08 July 2022;
Published: 26 July 2022.
Edited by:
Ayal Hendel, Bar-Ilan University, IsraelReviewed by:
Beeke Wienert, Graphite Bio, Inc., United StatesKarim Benabdellah, Andalusian Autonomous Government of Genomics and Oncological Research (GENYO), Spain
Mollie Schubert, Integrated DNA Technologies, United States
Copyright © 2022 Lu, Happi Mbakam, Song, Bendavid and Tremblay. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jacques-P. Tremblay, amFjcXVlcy1wLnRyZW1ibGF5QGNyY2h1ZGVxdWViZWMudWxhdmFsLmNh
 Yaoyao Lu
Yaoyao Lu Cedric Happi Mbakam
Cedric Happi Mbakam Bo Song
Bo Song Eli Bendavid
Eli Bendavid Jacques-P. Tremblay
Jacques-P. Tremblay