
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genome Ed., 28 February 2022
Sec. Genome Editing in Plants
Volume 4 - 2022 | https://doi.org/10.3389/fgeed.2022.852867
Base editors, such as adenine base editors (ABE) and cytosine base editors (CBE), provide alternatives for precise genome editing without generating double-strand breaks (DSBs), thus avoiding the risk of genome instability and unpredictable outcomes caused by DNA repair. Precise gene editing mediated by base editors in citrus has not been reported. Here, we have successfully adapted the ABE to edit the TATA box in the promoter region of the canker susceptibility gene LOB1 from TATA to CACA in grapefruit (Citrus paradise) and sweet orange (Citrus sinensis). TATA-edited plants are resistant to the canker pathogen Xanthomonas citri subsp. citri (Xcc). In addition, CBE was successfully used to edit the acetolactate synthase (ALS) gene in citrus. ALS-edited plants were resistant to the herbicide chlorsulfuron. Two ALS-edited plants did not show green fluorescence although the starting construct for transformation contains a GFP expression cassette. The Cas9 gene was undetectable in the herbicide-resistant citrus plants. This indicates that the ALS edited plants are transgene-free, representing the first transgene-free gene-edited citrus using the CRISPR technology. In summary, we have successfully adapted the base editors for precise citrus gene editing. The CBE base editor has been used to generate transgene-free citrus via transient expression.
Unlike classical CRISPR systems that use Cas proteins, such as Cas9 and Cas12, nickase Cas9 (nCas9) derived base editors do not create double-strand breaks (DSBs). DSBs introduced by Cas proteins may pose the risk of genome instability and unpredictable outcomes caused by Non-homologous end joining (NHEJ) DNA repair mechanisms. Base editors provide alternative tools for precise genome editing without generating DSBs. Base editors are derived by tethering deoxynucleoside deaminase to a nCas9–gRNA complex that induces efficient and direct base substitutions in the genomic sequence (Rees and Liu, 2018). Among the available base editors, cytosine base editors (Komor et al., 2016; Nishida et al., 2016) and adenine base editors (Gaudelli et al., 2017) enable highly efficient and precise base substitutions in a narrow window of gRNA-targeting sites. Specifically, adenine base editors mediate the conversion of A⋅T to G⋅C, whereas cytosine base editors enable the conversion of C⋅G to T⋅A in genomic DNA. It is well known that base editors can introduce specific amino acid changes in a protein, thus can be used for site-specific mutagenesis. They can also be deployed to disrupt gene functions by altering splicing sites (splice donor, splice acceptor, and branch point). CBEs can introduce premature stop codons to knock out genes. Both ABEs and CBEs can modify cis-regulatory elements to fine-tune gene functions. They can also be utilized to mutate start codon ATG to interrupt protein translation (Kluesner et al., 2021; Molla et al., 2021).
The development of base editing stemmed from seminal studies in 2016 and 2017 (Komor et al., 2016; Nishida et al., 2016; Gaudelli et al., 2017). Since then, base editing has been applied to different fields of life science including plants (Molla et al., 2021). Base editors have been adopted in different plant species, including Arabidopsis (Kang et al., 2018; Xue et al., 2018; Bastet et al., 2019; Zhenxiang Li et al., 2019), rice (Li et al., 2017; Lu and Zhu, 2017; Shimatani et al., 2017; Hua et al., 2018; Li et al., 2018; Xue et al., 2018; Yan et al., 2018; Zong et al., 2018; Hao Li et al., 2019; Hua et al., 2019; Juan Li et al., 2020; Zeng et al., 2020; Ren et al., 2021), maize (Zong et al., 2017), wheat (Zong et al., 2017; Li et al., 2018; Zhang et al., 2019), tomato (Shimatani et al., 2017; Veillet et al., 2019a; Hunziker et al., 2020; Veillet et al., 2020), potato (Zong et al., 2018; Veillet et al., 2019a; Veillet et al., 2019b; Veillet et al., 2020), Nicotiana benthamiana (Wang et al., 2021), soybean (Cai et al., 2020), rapeseed (Kang et al., 2018; Wu et al., 2020; Cheng et al., 2021), cotton (Qin et al., 2020), watermelon (Tian et al., 2018), strawberry (Xing et al., 2020), apple (Malabarba et al., 2021), pear (Malabarba et al., 2021), and poplar tree (Gen Li et al., 2021). Base editors have not been reported in citrus. Previously, CRISPR/Cas has been successfully used in genome editing of citrus (Jia et al., 2017; Zhang F. et al., 2017; Jia et al., 2019b; Zhu et al., 2019; Huang et al., 2020; Jia and Wang, 2020; Huang et al., 2022) despite the challenges in citrus transformation owing to its recalcitrant nature. Importantly, we have developed a very efficient, improved CRISPR/Cas9 system for citrus genome editing (Huang et al., 2022), of which we took advantage for the base editors in citrus in this study.
Citrus is one of the most important fruit crops in the world and faces many disease challenges including citrus Huanglongbing and citrus canker (Gochez et al., 2018; Wang, 2019). Citrus canker is caused by Xanthomonas citri subsp. citri (Xcc). Most commercial citrus varieties, including grapefruit and sweet orange varieties, are susceptible to canker disease. Xcc causes the characteristic hypertrophy and hyperplasia symptoms on citrus tissues via secretion of PthA4, a transcriptional activator-like (TAL) effector, through the type III secretion system (Swarup et al., 1992; Hu et al., 2014). PthA4 enters the nucleus and activates the expression of the canker susceptibility (S) gene LATERAL ORGAN BOUNDARIES 1 (LOB1) via binding to the effector binding elements (EBE) in the promoter region (Hu et al., 2014). In previous studies, canker-resistant citrus plants were generated by editing the EBE or the coding region of LOB1 (Jia et al., 2019b; Jia and Wang, 2020; Jia et al., 2022; Huang et al., 2022). Intriguingly, in most cases, Xanthomonas TAL EBE in the promoter of S genes overlaps with or locates immediately downstream of the TATA box of the S genes (Huang et al., 2017). TATA box is a core promoter element conserved both in plants and animals, with the consensus sequence TATA (A/T)A (A/T). The TATA box is pivotal in transcriptional activation. The TATA box of the CsLOB1 promoter is overlapped with the EBE region. In this study, we aimed to test if the TATA box can be edited with ABE8e (Richter et al., 2020). We reasoned that editing of the EBE-associated TATA box may abolish or reduce the induction of S genes by Xanthomonas TAL effectors to generate Xanthomonas-resistant crops.
Unlike most economically important crops, citrus species reproduce through apomixis. Apomixis is a way of asexual reproduction with offspring genetically identical to the mother plant (Xia Wang et al., 2017). Apomixis facilitates fixing desired traits, hybrid vigor and heterozygosity. However, one of the disadvantages of apomixis is the lack of sexual crosses, hence the lack of genetic segregation in the next generation of citrus. Therefore, it is challenging to obtain transgene-free, gene-edited citrus through genetic segregation. Transgene-free gene-edited crops such as rice, maize, wheat, are usually obtained through genetic segregation in the next generation (Zong et al., 2018; Molla et al., 2020). In addition, fruit trees such as citrus, have a long juvenile period (5–10 years). Thus, it is crucial to generate transgene-free gene-edited citrus in the T0 generation. In this study, we explored the possibility to obtain transgene-free, gene-edited citrus through base editors, such as CBE.
In this study, we successfully employed base editor ABE8e to edit the TATA box of the LOB1 promoter in citrus and the edited plants were resistant to canker disease. By using CBE, we edited the citrus ALS gene and obtained herbicide-resistant, transgene-free citrus.
The binary vector backbone PC-35S was first modified to contain a CsU6-tRNA-gRNA scaffold cassette (Huang et al., 2020) with two AarI sites for the gRNA insertion. The vector also contains a unique XbaI site downstream of the 35S promoter and a unique EcoRI site right upstream of the HSP terminator. Dicot plant codon-optimized Cas9 gene from the pXH1 vector (Huang et al., 2020) was first mutated at D10 to A to make Cas9D10A nickase or nCas9. The evolved TadA8e (Richter et al., 2020) was PCR amplified using the ABE8e plasmid (Addgene) as template and primers ABE8-F1/R1; nCas9 was PCR amplified using primers ABE8-F2/R2 and pXH1 as template. The modified PC-35S was digested with EcoRI + XbaI. Ligation of TadA8e, nCas9, and EcoRI/XbaI-digested vector was performed using the in-fusion cloning method (Takara Bio) to make vector PC-ABE8e. The CmYLCV promoter (Huang et al., 2022), which confers high gene editing efficiency in citrus, was PCR amplified with primers CmY-F2/CmY-R2. The 35S promoter in PC-ABE8e was replaced with the CmYLCV promoter to make the final vector PC-CmYLCV-ABE8e. Primers LOBBE-F1/LOBBE-R1 (for gRNA GTTTATATAGAGAAAGGAAA) were annealed and cloned into AarI-digested PC-CmYLCV-ABE8e. All constructs were verified with Sanger sequencing.
The PC-ABE8e vector described above was digested with SbfI + BspEI to remove TadA8e. The CmYLCV promoter (Huang et al., 2022) was PCR amplified using primers CmY-F1/R3. The fragment A3A-RAD51DBD (Supplementary Information S1) was synthesized by Integrated DNA Technologies, Inc. (Coralville, IA, United States). Ligation of the CmYLCV promoter, A3A-RAD51DBD, and SbfI/BspEI-digested vector was performed using the in-fusion cloning method (Takara Bio) to make an intermediate vector CmYLCV-A3A-RAD51-nCas9. CmYLCV-A3A-RAD51-nCas9 was further digested with EcoRI. The UGI was PCR amplified using primers UGI-F1/R1 and cloned into EcoRI site of the CmYLCV-A3A-RAD51-nCas9 vector via the in-fusion cloning method (Takara Bio) to construct the final CBE vector. Two gRNAs for two different alleles of the citrus ALS gene were designed. Primers ALS-F/ALS-R were used to amplify gRNA scaffold-tRNA unit using the plasmid pXH1 (Huang et al., 2020) as template. The amplicon was digested with BsaI and cloned into the AarI-digested CBE to make the CBE-2xALS construct. All constructs were verified by Sanger sequencing.
The constructs were transformed into Agrobacterium strain EHA105. Agrobacterium-mediated transformation of citrus epicotyl was performed as described previously (Jia et al., 2019a; Huang et al., 2020; Huang et al., 2022). Shoots of GFP positive, TATA-edited citrus were micro-grated onto Carrizo rootstock seedlings. Survived plants were transplanted in soil after establishment in a glasshouse. For the selection of herbicide chlorsulfuron resistant citrus, citrus epicotyl segments were cultured on kanamycin-containing selection media (100 mg/L) for 1 week under dark at 30°C. After 1 week, the citrus epicotyl segments were transferred to chlorsulfuron (Fisher Scientific, Catalog No.50-255-082) containing media (150 nM) without kanamycin under light at room temperature. Every 3 weeks, the citrus epicotyl segments were transferred to new chlorsulfuron-containing media to select chlorsulfuron-resistant shoots. After three rounds of subculture with chlorsulfuron selection, chlorsulfuron-resistant shoots were visible on the media.
We performed genotyping of citrus transformants as described previously (Huang et al., 2022). Citrus genomic DNA was extracted using the CTAB (cetyltrimethylammonium bromide) method. Detection of editing in the target genes was performed via amplifying the target regions (primers in Supplementary Table S1) with high fidelity DNA polymerase Q5 (New England Biolabs, Ipswich, MA, United States), followed by cloning of PCR products and sequencing. Primers LOBpro-F1/LOBpro-R1 were used for the LOB1 promoter genotyping. Primers CsALSgt-F2/CsALSgt-R2 were used for ALS genotyping. Primers Cas9gt-F1/Cas9gt-R1 were used for Cas9 genotyping. Primers CsALSgt-F1/CsALSgt-R1 were used for the PCR detection of ALS.
Xanthomonas citri subsp. citri (Xcc) wild type strain 306 and dLOB2 containing Xcc pthA4:Tn5 (Hu et al., 2014; Zhang J. et al., 2017; Teper et al., 2020; Huang et al., 2022) were suspended in 20 mM MgCl2 at 108 CFU/ml. The bacterial suspensions were syringe-infiltrated into fully expanded young leaves of wild type grapefruit, sweet orange Hamlin plants, or edited lines. Three different leaves from each genotype were included for the Xcc inoculation assays. Inoculated plants were kept in a temperature-controlled (28°C) glasshouse with high humidity. Pictures were taken 8 days post inoculation for disease resistance evaluation.
Reverse Transcription-Quantitative (RT-qPCR) was performed essentially as described previously (Huang et al., 2022). Leaves of wild-type and a representative grapefruit TATA-edited plant with and without Xcc inoculation were sampled at 48 h post inoculation. The citrus house-keeping gene GAPDH was used as an endogenous control. The primers QLOB1-F1/QLOB1/R1 and GAPDH-F1/R1 for qPCR are listed in Supplementary Table S1.
To analyze potential off-targets, we analyzed the putative off-targets using a web-based software (http://crispr.hzau.edu.cn/cgi-bin/CRISPR2/CRISPR). Genomic DNA from transgenic lines was used as template, and the primers listed in Supplementary Table S1 were used to amplify the fragments spanning the off-targets. Finally, the PCR products were subjected to Sanger sequencing.
To test if precise gene editing works in citrus, we first tested adenine base editors (ABE), ABE8e which mediates A⋅T-to-G⋅C base changes (Richter et al., 2020). The citrus optimized ABE8e vector was constructed (Figure 1A). TadA8e (Richter et al., 2020) was N-terminally fused with Cas9D10A nickase. There is a nuclear localization signal (NLS) at each end of TadA8e- Cas9D10A to increase its nucleus transportation. The CsU6-tRNA-gRNA-scaffold unit for multiplex editing in this ABE system was described previously (Huang et al., 2020). We sought to edit the TATA box (Figures 1B,D) located upstream of the EBE of the promoter of the citrus canker susceptibility (S) gene LOB1 (locus ID: Cs7g27640, C. sinensis v2.0 genome) with ABE8e. A nearby NGG PAM site enables the TATA box within the editing window of ABE8e. The EBE region of the LOB1 promoter in citrus is responsible for binding by the TAL effector PthA4 of Xcc (Hu et al., 2014) to activate its expression. The TATA box of the LOB1 promoter overlaps with the EBE region (Figures 1B,D). Hu et al. (2014) previously showed that mutation of TATA box abolishes LOB1 induction by PthA4 in the transient assay. We first investigated if the ABE construct targeting the TATA box can edit the target as expected through Xcc-facilitated agroinfiltration of citrus leaves (Jia and Wang, 2014). The transient assay (Figure 1C) showed that the construct edited the TATA box as anticipated (Figures 1D,E). The ABE mutated TATA to CACA (from TATA to TGTG for the complementary strand). Test on another gene CsTub (Cs1g21050) through transient assay also demonstrated precise editing (Figures 1F,G).
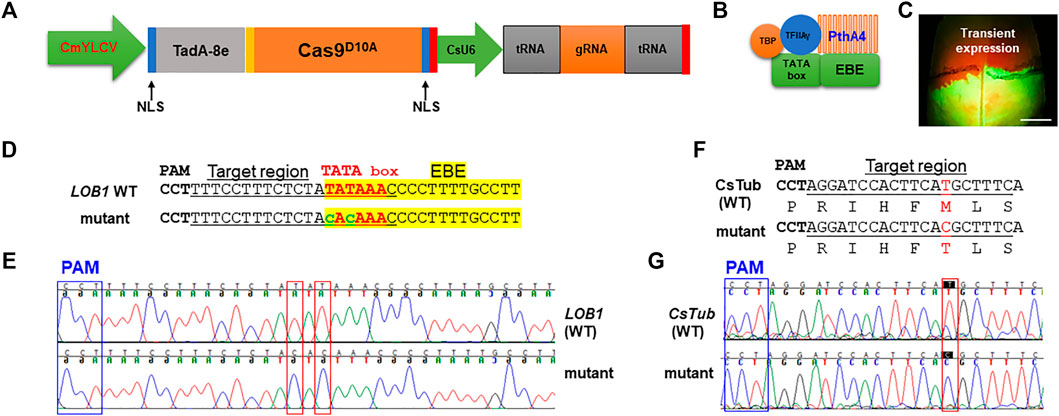
FIGURE 1. Precise gene editing in citrus with adenine base editor ABE8e via transient expression. (A) Illustration of ABE8e adenine base editing (ABE) system for citrus. CmYLCV (Huang et al., 2022), Cestrum yellow leaf curling virus promoter; TadA-8e (Richter et al., 2020), evolved Escherichia coli tRNA adenosine deaminase; Cas9D10A, Cas9 nickase; NLS, nuclear localization signal; CsU6, citrus U6 promoter. (B) TATA box located upstream of the LOB1 EBE region is associated with general transcription factors and the TAL effector PthA4 of Xanthomonas citri subsp. citri (Xcc). (C) Xcc-facilitated transient expression of the PC-CmYLCV-ABE8e-LOB1 construct in citrus leaf. The construct carries a GFP expression cassette. Scale bar, 1 cm. (D) Editing of TATA in the TATA box of the LOB1 promoter into CACA in transient assay. Underlined nucleotides were selected for gRNA design (gRNA: GTTTATATAGAGAAAGGAAA); EBE, Xanthomonas TAL effector binding element, highlighted with yellow; TATA box, in red font; edited sequences, in lower case with green font. (E) Chromatograms for (D), LOB1 WT (upper), and mutant (lower). Mutation sites are indicated within red rectangles. (F) Editing of the CsTub gene (Cs1g21050) in the transient expression assay. The amino acids are aligned under the corresponding DNA sequences. The mutation of T to C changes the corresponding amino acid from methionine (M) to threonine (T). (G) Chromatograms for (E), CsTub WT (upper) and mutant (lower). Mutation sites are indicated within red rectangles.
Biallelic editing of CsLOB1 coding region results in Xcc resistance while maintaining normal plant development and growth (data not shown). We expected that editing of the TATA box of CsLOB1 would not affect plant development and growth either. Agrobacterium-mediated stable transformation of grapefruit (Citrus paradise) and sweet orange (Citrus sinensis) Hamlin epicotyl tissues was conducted to edit the TATA box in the CsLOB1 promoter of both varieties. Two transgenic grapefruit plants and one transgenic sweet orange were obtained. Genotyping showed that in the grapefruit line #2 and Hamlin sweet orange mutant the TATA box was successfully edited (Figures 2A,B). The TATA box in the LOB1 promoter in grapefruit line #2 and Hamlin sweet orange mutant was 100% edited into CACA. The purity of editing was confirmed through direct sequencing of PCR products and colony sequencing of cloned PCR products. In grapefruit line #1, the first T in TATA box was 100% mutated to C, while 56% of the second T in the TATA box was mutated into C (9 clones out of 16). Another T to C editing was observed immediately upstream of TATA box in the grapefruit line #1 (31.2%, 5 clones out of 16). The bystander editing or proximal base editing has been observed in other studies too (Molla et al., 2020). Using the ABE system, we achieved high rate (66.7%) of biallelic/homozygous editing of the TATA box in the LOB1 promoter. After micro-grafting, one transgenic grapefruit plant and one transgenic sweet orange plant survived. Inoculation of the TATA-edited plants with Xcc demonstrated that the TATA-edited plants were resistant to Xcc (Figure 2C). On the contrary, all plants were susceptible to Xcc pthA4:Tn5 dLOB2, which carries a designer TAL dLOB2 for LOB2 induction to cause canker disease in citrus plants. LOB2, a LOB1 homolog, can cause canker symptoms when artificially induced, such as in the presence of designer TAL dLOB2 (Zhang F. et al., 2017; Teper et al., 2020; Huang et al., 2022). Xcc pthA4:Tn5 dLOB2 was included in the inoculation assay as a control. The onset of canker disease by Xcc pthA4:Tn5 dLOB2 on the same leaves excluded the possibility that lack of canker disease on the TATA-edited mutant plants is due to leaf age. To explore the outcome of TATA box editing on LOB1 induction by Xcc, we performed RT-qPCR for both wild type and TATA-edited grapefruit plant. The results showed that at 48 h after Xcc inoculation, LOB1 was induced about 64 folds in wild type citrus, while LOB1 was only induced 6.8 folds in TATA-edited citrus (Figure 2D). This result provides evidence that editing of TATA box of LOB1 dramatically compromises its inducibility by Xcc. The basal expression level of LOB1 is not significantly different between WT and TATA-edited citrus. These results demonstrated that base editor ABE8e can efficiently and precisely edit the citrus genome (both grapefruit and sweet orange). These results also showed a promising strategy by editing the EBE-associated TATA box of S genes to breed Xanthomonas-resistant crops through base editor ABE8e.
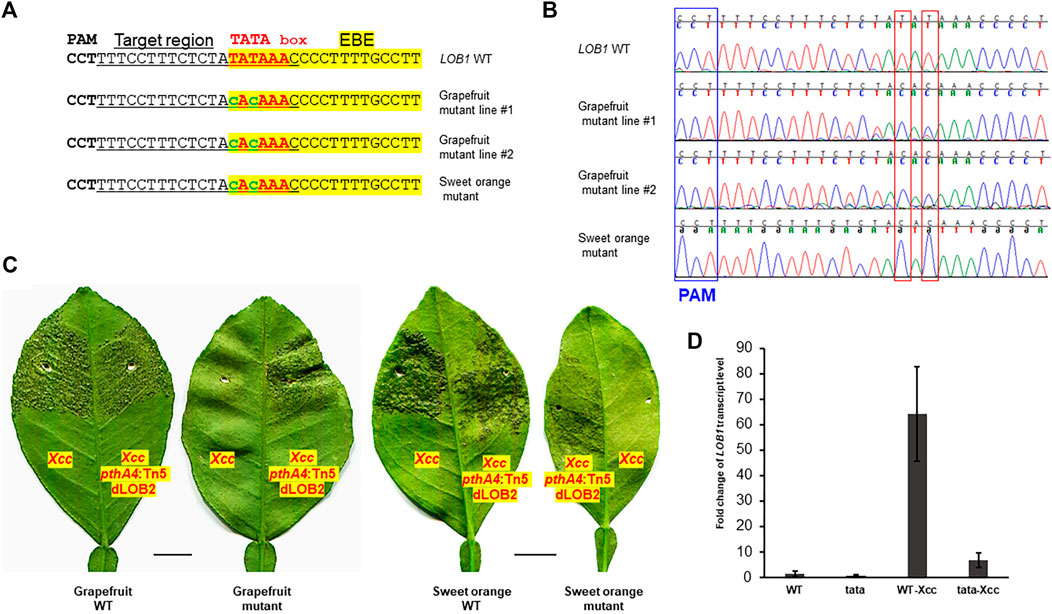
FIGURE 2. TATA-edited grapefruit (Citrus paradise) and sweet orange (Citrus sinensis) are resistant to the canker pathogen Xanthomonas citri subsp. citri (Xcc). (A) Edition of the TATA box into CACA in stable transgenic grapefruit and sweet orange. Underlined nucleotides were selected for gRNA design (gRNA: GTTTATATAGAGAAAGGAAA); EBE, Xanthomonas TAL effector PthA4 binding element, highlighted with yellow; TATA box, in red font; edited sequences, in lower case with green font. (B) Chromatograms for (A). Mutation sites are indicated within red rectangles. (C) Inoculation of WT and TATA-edited mutant plants with Xcc or Xcc pthA4:Tn5 dLOB2 in the indicated areas of leaves. Xcc pthA4:Tn5 dLOB2, an Xcc pthA4 mutant strain carrying a designer TAL effector dLOB2 for the induction of citrus LOB2 expression to cause canker symptoms. Xcc pthA4:Tn5 dLOB2 was used as control. Scale bar, 1 cm. (D) RT-qPCR analyses of CsLOB1 relative expression in leaf samples collected at 48 h post inoculation with Xcc (108 CFU/ml). Each treatment has three biological replicates. All expression levels were normalized to the WT. The GAPDH gene was used as an endogenous control. WT: wild type grapefruit plant. tata: TATA-edited grapefruit plant.
Next, we tested if cytosine base editors (CBE), which can mediate C⋅G-to-T⋅A base changes, work in citrus. A citrus optimized CBE vector was constructed (Figure 3A). We chooseAPOBEC3A (A3A) deaminase for citrus CBE considering its wide deamination window and high editing efficiency (Zong et al., 2018; Zhang et al., 2020). An inhibitor of uracil DNA glycosylase (UGI) was fused immediately downstream of nCas9 to increase efficacy of C to T editing. A single-stranded DNA-binding domain from Rad51 protein (RAD51-DBD) was inserted between the deaminases and nCas9, which was reported to substantially increase activity and an expand editing window of CBE (Zhang et al., 2020). Tan et al. (2022) recently reported that RAD51-DBD can also dramatically increase editing efficiency of ABE. This CBE system contains a CsU6-tRNA-gRNA-scaffold unit for multiplex editing. The final CBE construct also carries a GFP expression cassette for direct visualization of transgene integration. We first determined whether the citrus acetolactate synthase (ALS) gene (locus ID: Cs7g22130, C. sinensis v2.0 genome) can be edited with this CBE. Most citrus varieties, owing to their hybrid nature, have two different alleles of the ALS gene responsible for herbicide resistance. We designed two gRNAs targeting both ALS alleles. The transient expression result showed that the CBE construct edited both alleles of the ALS gene as expected (Figures 3B,C).
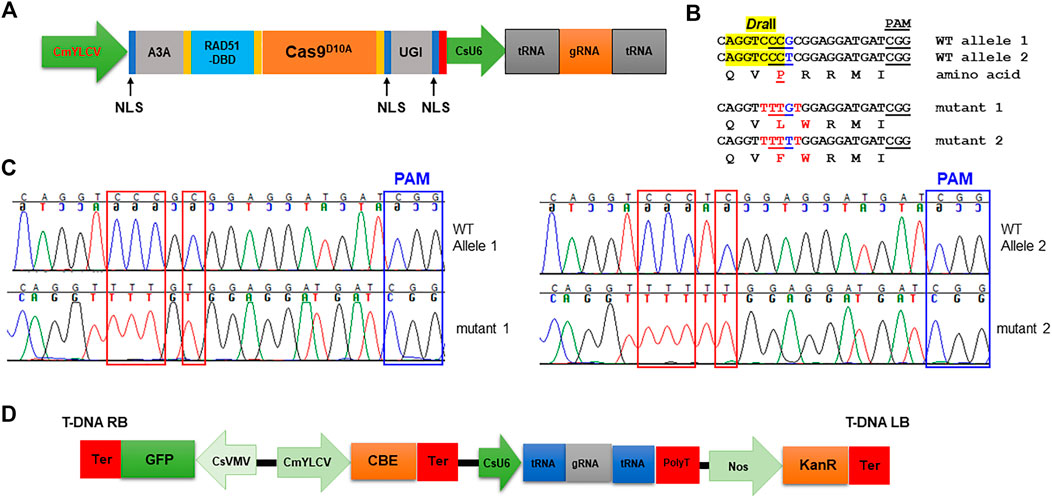
FIGURE 3. Precise gene editing in citrus with cytosine base editor A3A-RAD51-DBD via transient expression. (A) Illustration of cytosine base editing system (CBE). A3A, human APOBEC3A cytidine deaminase; RAD51-DBD, RAD51 DNA-binding domain; UGI, uracil glycosylase inhibitor; Cas9D10A, Cas9 nickase; NLS, nuclear localization signal; CsU6, citrus U6 promoter. (B) CBE base editing of the ALS gene via transient expression assay. There are two ALS alleles in citrus. Two gRNAs (gRNA1: CAGGTCCCGCGGAGGATGAT and gRNA2: CAGGTCCCTCGGAGGATGAT) were designed to edit both ALS alleles using the multiplex CBE construct. The amino acids are aligned under the corresponding DNA sequences. The restriction enzyme DraII-resistant PCR amplicon was subject to cloning and sequencing. The restriction enzyme DraII recognition site, highlighted in yellow. Edited sites, in red font. (C) Chromatograms for (B). Mutation sites are indicated within red rectangles. (D) T-DNA part of the CBE construct.
Acetolactate synthase (ALS) is an enzyme required for the biosynthesis of multiple branched-chain amino acids, such as valine, leucine, and isoleucine. Chlorsulfuron is a known ALS inhibitor that kills plants, thus being used as a herbicide. A single amino acid mutation in ALS genes in various plants confers resistance to chlorsulfuron (Kuang et al., 2020; Malabarba et al., 2021). For example, when the amino acid P in the “QVPRRMI” amino stretch of ALS protein is mutated to S or F (Figure 3B, Figure 4C), the plant becomes resistant to chlorsulfuron. We first tested the sensitivity of citrus to chlorsulfuron. The growth of citrus Carrizo citrange, a hybrid of Citrus sinensis “Washington” sweet orange X Poncirus trifoliata, was completely inhibited by chlorsulfuron (Figure 4A). Agrobacterium-mediated stable transformation of Carrizo epicotyl tissues was performed with chlorsulfuron selection in the culture media after 1 week of culture. Three chlorsulfuron-resistant plants grew on the chlorsulfuron-containing medium while most of the transformed epicotyl tissues did not grow (Figure 4B). One of these three plants showed green fluorescence, indicating transgene integration. Genotyping of the other two chlorsulfuron-resistant plants showed that both alleles of the ALS gene were edited in the two edited plants (Figures 4C,D). Only C-to-T substitutions, but no other by-product substitutions, were observed in the edited plants. The editing rendered the “QVPRRMI” amino stretch of ALS protein to “QVSRRMI” in one allele, and to “QVFWRMI” in another allele. Both mutant plants have same genotypes in ALS locus. Intriguingly, these two plants did not show green fluorescence although the construct used for transformation contains a GFP expression cassette (Figure 3D). To further confirm the absence of T-DNA integration, we performed PCR amplification of the Cas9 gene in the mutant plants and the Cas9 gene was undetectable in the herbicide-resistant citrus plants (Figure 4E). This indicates that the ALS-edited plants are transgene-free, which likely resulted from the transient expression of the CBE construct, representing the first transgene-free gene-edited citrus using the CRISPR technology.
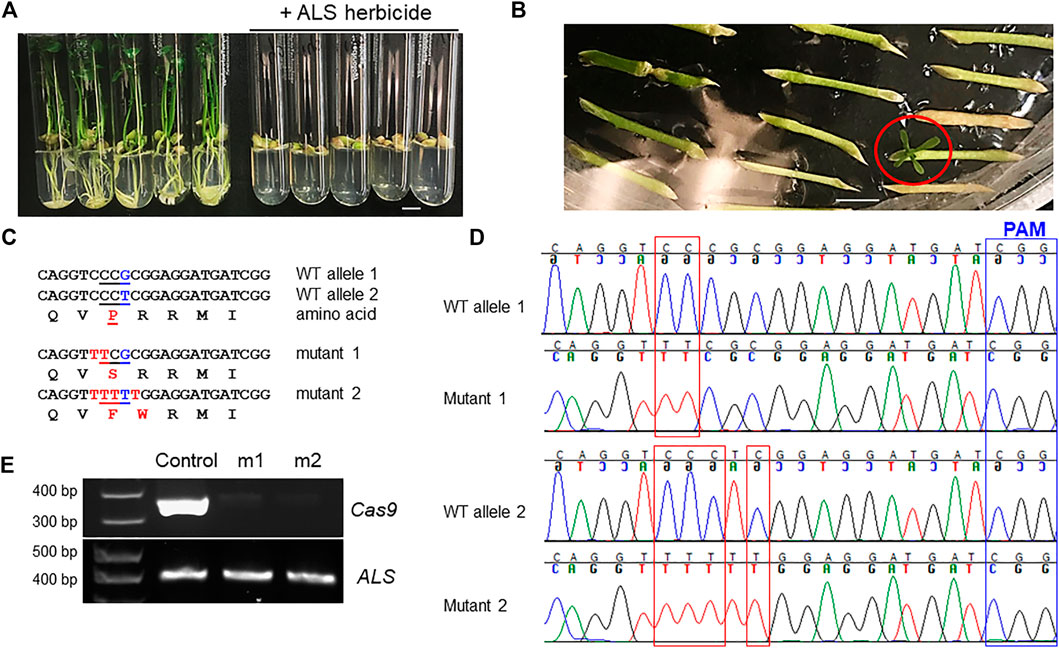
FIGURE 4. Transgene-free editing of the ALS gene in citrus confers herbicide resistance. (A) The growth of citrus seedlings (Carrizo citrange) was inhibited by the herbicide chlorsulfuron (300 nM). Scale bar, 1 cm. (B) Selection of herbicide-resistant Carrizo citrange on chlorsulfuron-containing media (150 nM). The chlorsulfuron-resistant regenerated plant was indicated by a red circle. Scale bar, 1 cm. (C) Sequencing results from chlorsulfuron-resistant mutant plants. The amino acids are aligned under the corresponding DNA sequences. Edited sites, in red font. (D) Chromatograms for (C). Mutation sites are indicated within red rectangles. Sanger sequencing results of the PCR amplicons that were cloned for colony sequencing. For each mutant plant, 14 clones were subjected to Sanger sequencing. (E) PCR of Cas9 and ALS for control plant (GFP positive) and herbicide chlorsulfuron-resistant citrus plants.
To investigate whether base editors can introduce off-target mutations, we amplified and sequenced top 12 potential off-target sites in both Grapefruit and sweet orange Hamlin TATA-edited mutant lines. The top potential off-targets all carry 4 or more mismatches. Sequencing results showed that no edits were detected at potential off-target sites (Supplementary Table S2). For ALS-edited plants, only 1 potential off-target exists with four or fewer mismatches within the protospacers. Sequencing results showed that no edits were detected at the potential off-target site.
TATA box plays an important role in recruiting the basal transcription factors for assembly into transcription machinery. The TATA box is a key determinant of promoter strength (Jores et al., 2021). Previously, Hu et al. (2014) showed that mutation of TATA box in the LOB1 promoter abolished the LOB1 induction by TAL effector PthA4 in transient assay. In this study, we successfully used an ABE [ABE8e variant (Richter et al., 2020)] to edit the TATA box of LOB1 in citrus. Mutation of TATA in the TATA box to CACA confers resistance to citrus canker disease caused by Xcc. To our knowledge, this is the first report that editing the TATA box in the promoter with an ABE can confer disease resistance in plants. The TATA box in the promoter region of overexpressed PMP22 was targeted by CRISPR/Cas9 to knock down its expression level to cure the disease Charcot-Marie-Tooth 1A (CMT1A) in mice model (Lee et al., 2020). Xanthomonas species can infect a wide range of crops, such as rice, wheat, citrus, tomato, pepper, cabbage, cassava, banana, mango, sugarcane, cotton, bean, strawberry, and lettuce. TAL effectors secreted by Xanthomonas species via type III secretion system can induce S genes to cause diseases. By editing the TATA box in the S genes with an ABE, our strategy may be applied to other crops for breeding of Xanthomonas-resistant crops.
ABE has been artificially evolved from ABE7.10 to ABE8e, which dramatically increases deamination activity (Richter et al., 2020). ABE8e has been adapted for base editing in plants, such as in rice (Juan Li et al., 2021; Ren et al., 2021; Wei et al., 2021; Xu et al., 2021; Yan et al., 2021). The editing efficiency varied from 33% to more than 93%. A recent report demonstrated that ABE8e achieved 30–60% editing efficiency in Nicotiana benthamiana (Wang et al., 2021). In our current study, we adapted ABE8e to edit the TATA box of LOB1 promoter in citrus and achieved high editing efficiency. We speculate that the highly efficient base editing in citrus can be attributed to both the highly efficient ABE8e and highly efficient improved CRISPR/Cas9 system that we developed (Huang et al., 2022). However, owing to the recalcitrant nature of citrus to genetic transformation, gene editing of citrus remains challenging despite the high efficacy of CRISPR/Cas9-mediated gene editing.
For many economically important crops, such as rice and maize, it is easy to obtain transgene-free gene-edited crops by simply choosing the segregating progenies that do not contain the CRISPR construct (Zong et al., 2018; Molla et al., 2020). For fruit trees such as citrus, it has been extremely difficult to obtain transgene-free gene-edited plants, which is because of the long juvenile period and reproduction of citrus through apomixis (Xia Wang et al., 2017). Due to the apomixis reproduction nature, citrus lacks genetic segregation in the next generation. Therefore, the CRISPR constructs in transgenic citrus cannot be segregated out like rice and maize. In this study, we successfully generated gene-edited, transgene-free citrus with a CBE. We obtained transgene-free, ALS-edited citrus by the selection of citrus plants on herbicide chlorsulfuron-containing media. The selection pressure exerted by herbicide chlorsulfuron facilitated the selection of ALS-edited plants, among which some are transgene-free through transient expression of CBE construct. A similar strategy has been reported previously in other crops (Veillet et al., 2019a). Chen et al. (2018) transiently expressed CRISPR/Cas9 construct through A. tumefaciens-mediated transformation and obtained transgene-free, gene-edited plants. These results in addition to ours show that it is possible to obtain transgene-free, gene-edited plants through A. tumefaciens-mediated transient expression of CRISPR/Cas constructs.
In our current study, we used A3A-based CBE (Zong et al., 2018) to edit the citrus ALS gene to confer herbicide resistance. It was reported that A3A-based CBE outperformed rAPOBEC1-BE3, hAID-BE3, and PmCDA1-BE3 in base editing efficiency (Zong et al., 2018; Cheng et al., 2021; Gen Li et al., 2021; Randall et al., 2021). Off-target mutations caused by CBEs have been reported in tomato, rice, and mouse (Shimatani et al., 2017; Jin et al., 2019; Zuo et al., 2019; Randall et al., 2021). Although we detected no off-target mutations at predicted off-target sites, we cannot rule out that there are gRNA-independent off-target mutations. Randall et al. (2021) reported low level of sgRNA-independent off-target mutations in tomato mediated by A3A-based CBE, though the difference is not statistically significant. They proposed that reduced expression level and/or duration of CBE can further reduce off-target mutations. Transient expression of a CBE in tomato to edit SlALS1 greatly reduced the risk of sgRNA-dependent off-target editing at the SlALS2 locus, compared to the constitutive expression of the CBE through stable transformation (Veillet et al., 2019a). In our study, we achieved ALS-edited citrus with CBE through transient expression. Therefore, according to the study by Randall et al. (2021), the ALS-edited citrus might have minimum off-target mutations. In contrast to CBE, ABEs produce much cleaner edits and do not generate genome-wide gRNA independent off-target mutations (Jin et al., 2019; Zuo et al., 2019). Noticeably, the rate of unwanted SNPs caused by ABEs is comparable to spontaneous mutations (Jin et al., 2019). Consistently, in our study we detected no off-target mutations.
It is probable to generate non-transgenic citrus varieties by simultaneously editing ALS and genes of interest by using our citrus optimized multiplex CBE and selecting the regenerated plants on herbicide chlorsulfuron-containing media. In this way, we can select transgene-free citrus with desired agronomic traits. For example, we may take advantage of ABE-CBE dual-editor (Chao Li et al., 2020) to simultaneously edit CsLOB1 EBE and CsALS to select transgene-free, canker-resistant citrus. It is challenging for many vegetatively propagated crops and hybrid crops to obtain transgene-free gene-edited plants. The strategy that we discussed here may also be applicable to other vegetatively propagated crops and hybrid crops to obtain transgene-free plants with desired traits.
In summary, we have successfully adapted base editors for citrus gene editing and have generated transgene-free gene-edited citrus plants. Such tools will be useful to tackle the challenges the citrus industry is facing, such as Huanglongbing (HLB) (Nian Wang et al., 2017; Wang, 2019).
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
XH designed and performed the experiments. YW performed micro-grafting and off-target experiments. XH and NW wrote the paper. NW supervised the study.
The research has been supported by USDA National Institute of Food and Agriculture grants # 2018-70016-27412, #2016-70016-24833, and #2019-70016-29796, USDA-NIFA Plant Biotic Interactions Program 2017-67013-26527, Florida Citrus Initiative, and Florida Citrus Research and Development Foundation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgeed.2022.852867/full#supplementary-material
ABE, adenine base editor; CBE, cytosine base editor; CRISPR, clustered regularly interspaced short palindromic repeats; EBE, TAL effector-binding element; gRNA, guide RNA; Xcc, Xanthomonas citri subsp. citri.
Bastet, A., Zafirov, D., Giovinazzo, N., Guyon-Debast, A., Nogué, F., Robaglia, C., et al. (2019). Mimicking Natural Polymorphism in eIF4E by CRISPR-Cas9 Base Editing Is Associated with Resistance to Potyviruses. Plant Biotechnol. J. 17, 1736–1750. doi:10.1111/pbi.13096
Cai, Y., Chen, L., Zhang, Y., Yuan, S., Su, Q., Sun, S., et al. (2020). Target Base Editing in Soybean Using a Modified CRISPR/Cas9 System. Plant Biotechnol. J. 18, 1996–1998. doi:10.1111/pbi.13386
Chao Li, C., Zhang, R., Meng, X., Chen, S., Zong, Y., Lu, C., et al. (2020). Targeted, Random Mutagenesis of Plant Genes with Dual Cytosine and Adenine Base Editors. Nat. Biotechnol. 38, 875–882. doi:10.1038/s41587-019-0393-7
Chen, L., Li, W., Katin-Grazzini, L., Ding, J., Gu, X., Li, Y., et al. (2018). A Method for the Production and Expedient Screening of CRISPR/Cas9-mediated Non-transgenic Mutant Plants. Hortic. Res. 5, 13. doi:10.1038/s41438-018-0023-4
Cheng, H., Hao, M., Ding, B., Mei, D., Wang, W., Wang, H., et al. (2021). Base Editing with High Efficiency in Allotetraploid Oilseed Rape by A3A‐PBE System. Plant Biotechnol. J. 19, 87–97. doi:10.1111/pbi.13444
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable Base Editing of AT to GC in Genomic DNA without DNA Cleavage. Nature 551, 464–471. doi:10.1038/nature24644
Gochez, A. M., Huguet-Tapia, J. C., Minsavage, G. V., Shantaraj, D., Jalan, N., Strauss, A., et al. (2018). Pacbio Sequencing of Copper-Tolerant Xanthomonas Citri Reveals Presence of a Chimeric Plasmid Structure and Provides Insights into Reassortment and Shuffling of Transcription Activator-like Effectors Among X. Citri Strains. Bmc Genomics 19, 16. doi:10.1186/s12864-017-4408-9
Hu, Y., Zhang, J., Jia, H., Sosso, D., Li, T., Frommer, W. B., et al. (2014). Lateral Organ Boundaries 1 Is a Disease Susceptibility Gene for Citrus Bacterial Canker Disease. Proc. Natl. Acad. Sci. 111, E521–E529. doi:10.1073/pnas.1313271111
Hua, K., Tao, X., Yuan, F., Wang, D., and Zhu, J.-K. (2018). Precise A·T to G·C Base Editing in the Rice Genome. Mol. Plant 11, 627–630. doi:10.1016/j.molp.2018.02.007
Hua, K., Tao, X., and Zhu, J.-K. (2019). Expanding the Base Editing Scope in rice by Using Cas9 Variants. Plant Biotechnol. J. 17, 499–504. doi:10.1111/pbi.12993
Huang, R., Hui, S., Zhang, M., Li, P., Xiao, J., Li, X., et al. (2017). A Conserved Basal Transcription Factor Is Required for the Function of Diverse TAL Effectors in Multiple Plant Hosts. Front. Plant Sci. 8, 1919. doi:10.3389/fpls.2017.01919
Huang, X., Wang, Y., Xu, J., and Wang, N. (2020). Development of Multiplex Genome Editing Toolkits for Citrus with High Efficacy in Biallelic and Homozygous Mutations. Plant Mol. Biol. 104, 297–307. doi:10.1007/s11103-020-01043-6
Huang, X., Wang, Y., and Wang, N. (2022). Highly Efficient Generation of Canker-Resistant Sweet Orange Enabled by an Improved CRISPR/Cas9 System. Front. Plant Sci. 12, 769907. doi:10.3389/fpls.2021.769907
Hunziker, J., Nishida, K., Kondo, A., Kishimoto, S., Ariizumi, T., and Ezura, H. (2020). Multiple Gene Substitution by Target-AID Base-Editing Technology in Tomato. Sci. Rep. 10, 20471. doi:10.1038/s41598-020-77379-2
Jia, H., and Wang, N. (2014). Xcc-facilitated Agroinfiltration of Citrus Leaves: a Tool for Rapid Functional Analysis of Transgenes in Citrus Leaves. Plant Cel. Rep. 33, 1993–2001. doi:10.1007/s00299-014-1673-9
Jia, H., and Wang, N. (2020). Generation of Homozygous Canker‐resistant Citrus in the T0 Generation Using CRISPR‐SpCas9p. Plant Biotechnol. Journal) 18, 1990–1992. doi:10.1111/pbi.13375
Jia, H., Wang, Y., Su, H., Huang, X., and Wang, N. (2022). LbCas12a-D156R Efficiently Edits LOB1 Effector Binding Elements to Generate Canker-Resistant Citrus Plants. Cells 11 (3), 315. doi:10.3390/cells11030315
Jia, H., Zhang, Y., Orbović, V., Xu, J., White, F. F., Jones, J. B., et al. (2017). Genome Editing of the Disease Susceptibility Gene CsLOB1 in Citrus Confers Resistance to Citrus Canker. Plant Biotechnol. J. 15, 817–823. doi:10.1111/pbi.12677
Jia, H., Zou, X., Orbovic, V., and Wang, N. (2019a). Genome Editing in Citrus Tree with CRISPR/Cas9. Methods Mol. Biol., 1917, 235–241. (Plant Genome Editing with CRISPR Systems). doi:10.1007/978-1-4939-8991-1_17
Jia, H., Orbović, V., and Wang, N. (2019b). CRISPR ‐LbCas12a‐mediated Modification of Citrus. Plant Biotechnol. J. 17, 1928–1937. doi:10.1111/pbi.13109
Jin, S., Zong, Y., Gao, Q., Zhu, Z., Wang, Y., Qin, P., et al. (2019). Cytosine, but Not Adenine, Base Editors Induce Genome-wide Off-Target Mutations in rice. Science 364, 292–295. doi:10.1126/science.aaw7166
Jores, T., Tonnies, J., Wrightsman, T., Buckler, E. S., Cuperus, J. T., Fields, S., et al. (2021). Synthetic Promoter Designs Enabled by a Comprehensive Analysis of Plant Core Promoters. Nat. Plants 7, 842–855. doi:10.1038/s41477-021-00932-y
Li, J., Qin, R., Zhang, Y., Xu, S., Liu, X., Yang, J., et al. (2020). Optimizing Plant Adenine Base Editor Systems by Modifying the Transgene Selection System. Plant Biotechnol. J. 18, 1495–1497. doi:10.1111/pbi.13304
Li, J., Xu, R., Qin, R., Liu, X., Kong, F., and Wei, P. (2021). Genome Editing Mediated by SpCas9 Variants with Broad Non-canonical PAM Compatibility in Plants. Mol. Plant 14, 352–360. doi:10.1016/j.molp.2020.12.017
Kang, B.-C., Yun, J.-Y., Kim, S.-T., Shin, Y., Ryu, J., Choi, M., et al. (2018). Precision Genome Engineering through Adenine Base Editing in Plants. Nat. Plants 4, 427–431. doi:10.1038/s41477-018-0178-x
Kluesner, M. G., Lahr, W. S., Lonetree, C. L., Smeester, B. A., Qiu, X., Slipek, N. J., et al. (2021). CRISPR-Cas9 Cytidine and Adenosine Base Editing of Splice-Sites Mediates Highly-Efficient Disruption of Proteins in Primary and Immortalized Cells. Nat. Commun. 12, 2437. doi:10.1038/s41467-021-22009-2
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., and Liu, D. R. (2016). Programmable Editing of a Target Base in Genomic DNA without Double-Stranded DNA Cleavage. Nature 533, 420–424. doi:10.1038/nature17946
Kuang, Y., Li, S., Ren, B., Yan, F., Spetz, C., Li, X., et al. (2020). Base-Editing-Mediated Artificial Evolution of OsALS1 in Planta to Develop Novel Herbicide-Tolerant Rice Germplasms. Mol. Plant 13, 565–572. doi:10.1016/j.molp.2020.01.010
Lee, J. S., Lee, J. Y., Song, D. W., Bae, H. S., Doo, H. M., Yu, H. S., et al. (2020). Targeted PMP22 TATA-Box Editing by CRISPR/Cas9 Reduces Demyelinating Neuropathy of Charcot-Marie-Tooth Disease Type 1A in Mice. Nucleic Acids Res. 48, 130–140. doi:10.1093/nar/gkz1070
Li, J., Sun, Y., Du, J., Zhao, Y., and Xia, L. (2017). Generation of Targeted Point Mutations in Rice by a Modified CRISPR/Cas9 System. Mol. Plant 10, 526–529. doi:10.1016/j.molp.2016.12.001
Li, C., Zong, Y., Wang, Y., Jin, S., Zhang, D., Song, Q., et al. (2018). Expanded Base Editing in rice and Wheat Using a Cas9-Adenosine Deaminase Fusion. Genome Biol. 19, 59. doi:10.1186/s13059-018-1443-z
Li, G., Sretenovic, S., Eisenstein, E., Coleman, G., and Qi, Y. (2021). Highly Efficient C‐to‐T and A‐to‐G Base Editing in a Populus Hybrid. Plant Biotechnol. J. 19, 1086–1088. doi:10.1111/pbi.13581
Li, H., Qin, R. Y., Liu, X. S., Liao, S. X., Xu, R. F., Yang, J. B., et al. (2019). CRISPR/Cas9-Mediated Adenine Base Editing in Rice Genome. Rice Sci. 26, 125–128. doi:10.1016/j.rsci.2018.07.002
Lu, Y., and Zhu, J.-K. (2017). Precise Editing of a Target Base in the Rice Genome Using a Modified CRISPR/Cas9 System. Mol. Plant 10, 523–525. doi:10.1016/j.molp.2016.11.013
Malabarba, J., Chevreau, E., Dousset, N., Veillet, F., Moizan, J., and Vergne, E. (2021). New Strategies to Overcome Present CRISPR/Cas9 Limitations in Apple and Pear: Efficient Dechimerization and Base Editing. Int. J. Mol. Sci. 22, 319. doi:10.3390/ijms22010319
Molla, K. A., Shih, J., and Yang, Y. (2020). Single-nucleotide Editing for Zebra3 and Wsl5 Phenotypes in Rice Using CRISPR/Cas9-mediated Adenine Base Editors. aBiotech 1, 106–118. doi:10.1007/s42994-020-00018-x
Molla, K. A., Sretenovic, S., Bansal, K. C., and Qi, Y. (2021). Precise Plant Genome Editing Using Base Editors and Prime Editors. Nat. Plants 7, 1166–1187. doi:10.1038/s41477-021-00991-1
Wang, N., Pierson, E. A., Setubal, J. C., Xu, J., Levy, J. G., Zhang, Y., et al. (2017). The Candidatus Liberibacter-Host Interface: Insights into Pathogenesis Mechanisms and Disease Control. Annu. Rev. Phytopathol. 55, 451–482. doi:10.1146/annurev-phyto-080516-035513
Nishida, K., Arazoe, T., Yachie, N., Banno, S., Kakimoto, M., Tabata, M., et al. (2016). Targeted Nucleotide Editing Using Hybrid Prokaryotic and Vertebrate Adaptive Immune Systems. Science 353, aaf8729. doi:10.1126/science.aaf8729
Qin, L., Li, J., Wang, Q., Xu, Z., Sun, L., Alariqi, M., et al. (2020). High‐Efficient and Precise Base Editing of CG to TA in the Allotetraploid Cotton ( Gossypium Hirsutum ) Genome Using a Modified CRISPR/Cas9 System. Plant Biotechnol. J. 18, 45–56. doi:10.1111/pbi.13168
Randall, L. B., Sretenovic, S., Wu, Y., Yin, D., Zhang, T., Eck, J. V., et al. (2021). Genome- and Transcriptome-wide Off-Target Analyses of an Improved Cytosine Base Editor. Plant Physiol. 187, 73–87. doi:10.1093/plphys/kiab264
Rees, H. A., and Liu, D. R. (2018). Base Editing: Precision Chemistry on the Genome and Transcriptome of Living Cells. Nat. Rev. Genet. 19, 770–788. doi:10.1038/s41576-018-0059-1
Ren, Q., Sretenovic, S., Liu, S., Tang, X., Huang, L., He, Y., et al. (2021). PAM-less Plant Genome Editing Using a CRISPR-SpRY Toolbox. Nat. Plants 7, 25–33. doi:10.1038/s41477-020-00827-4
Richter, M. F., Zhao, K. T., Eton, E., Lapinaite, A., Newby, G. A., Thuronyi, B. W., et al. (2020). Phage-assisted Evolution of an Adenine Base Editor with Improved Cas Domain Compatibility and Activity. Nat. Biotechnol. 38, 883–891. doi:10.1038/s41587-020-0453-z
Shimatani, Z., Kashojiya, S., Takayama, M., Terada, R., Arazoe, T., Ishii, H., et al. (2017). Targeted Base Editing in rice and Tomato Using a CRISPR-Cas9 Cytidine Deaminase Fusion. Nat. Biotechnol. 35, 441, 443-+.doi:10.1038/nbt.3833
Swarup, S., Yang, Y. N., Kingsley, M. T., and Gabriel, D. W. (1992). AnXanthomonas citriPathogenicity Gene,pthA,Pleiotropically Encodes Gratuitous Avirulence on Nonhosts. Mpmi 5, 204–213. doi:10.1094/mpmi-5-204
Tan, J., Zeng, D., Zhao, Y., and Zhu, Q. (2022). PhieABEs: a PAM-Less/free High-Efficiency Adenine Base Editor Toolbox with Wide Target Scope in Plants. Plant Biotechnol. J. doi:10.1111/pbi.13774
Teper, D., Xu, J., Li, J., and Wang, N. (2020). The Immunity of Meiwa Kumquat against Xanthomonas Citri Is Associated with a Known Susceptibility Gene Induced by a Transcription Activator-like Effector. Plos Pathog. 16, e1008886. doi:10.1371/journal.ppat.1008886
Tian, S., Jiang, L., Cui, X., Zhang, J., Guo, S., Li, M., et al. (2018). Engineering Herbicide-Resistant Watermelon Variety through CRISPR/Cas9-mediated Base-Editing. Plant Cel. Rep. 37, 1353–1356. doi:10.1007/s00299-018-2299-0
Veillet, F., Perrot, L., Chauvin, L., Kermarrec, M. P., Guyon-Debast, A., Chauvin, J. E., et al. (2019a). Transgene-Free Genome Editing in Tomato and Potato Plants Using Agrobacterium-Mediated Delivery of a CRISPR/Cas9 Cytidine Base Editor. Int. J. Mol. Sci. 20, 2. doi:10.3390/ijms20020402
Veillet, F., Chauvin, L., Kermarrec, M.-P., Sevestre, F., Merrer, M., Terret, Z., et al. (2019b). The Solanum tuberosum GBSSI Gene: a Target for Assessing Gene and Base Editing in Tetraploid Potato. Plant Cel. Rep. 38, 1065–1080. doi:10.1007/s00299-019-02426-w
Veillet, F., Perrot, L., Guyon-Debast, A., Kermarrec, M. P., Chauvin, L., Chauvin, J. E., et al. (2020). Expanding the CRISPR Toolbox in P. patens Using SpCas9-NG Variant and Application for Gene and Base Editing in Solanaceae Crops. Int. J. Mol. Sci. 21, 3. doi:10.3390/ijms21031024
Wang, Z. P., Liu, X. Y., Xie, X. D., Deng, L., Zheng, H., Pan, H., et al. (2021). ABE8e with Polycistronic tRNA-gRNA Expression Cassette Sig-Nificantly Improves Adenine Base Editing Efficiency in Nicotiana benthamiana. Int. J. Mol. Sci. 22, 11. doi:10.3390/ijms22115663
Wang, N. (2019). The Citrus Huanglongbing Crisis and Potential Solutions. Mol. Plant 12, 607–609. doi:10.1016/j.molp.2019.03.008
Wei, C., Wang, C., Jia, M., Guo, H. X., Luo, P. Y., Wang, M. G., et al. (2021). Efficient generation of homozygous substitutions in rice in one generation utilizing an rABE8e base editor. J. Integr. Plant Biol. 63, 1595–1599. doi:10.1111/jipb.13089
Wu, J., Chen, C., Xian, G., Liu, D., Lin, L., Yin, S., et al. (2020). Engineering Herbicide‐resistant Oilseed Rape by CRISPR/Cas9‐mediated Cytosine Base‐editing. Plant Biotechnol. J. 18, 1857–1859. doi:10.1111/pbi.13368
Wang, X., Xu, Y., Zhang, S., Cao, L., Huang, Y., Cheng, J., et al. (2017). Genomic Analyses of Primitive, Wild and Cultivated Citrus Provide Insights into Asexual Reproduction. Nat. Genet. 49, 765–772. doi:10.1038/ng.3839
Xing, S., Chen, K., Zhu, H., Zhang, R., Zhang, H., Li, B., et al. (2020). Fine-tuning Sugar Content in Strawberry. Genome Biol. 21, 230. doi:10.1186/s13059-020-02146-5
Xu, Z., Kuang, Y., Ren, B., Yan, D., Yan, F., Spetz, C., et al. (2021). SpRY Greatly Expands the Genome Editing Scope in rice with Highly Flexible PAM Recognition. Genome Biol. 22, 6. doi:10.1186/s13059-020-02231-9
Xue, C., Zhang, H., Lin, Q., Fan, R., and Gao, C. (2018). Manipulating mRNA Splicing by Base Editing in Plants. Sci. China Life Sci. 61, 1293–1300. doi:10.1007/s11427-018-9392-7
Yan, F., Kuang, Y., Ren, B., Wang, J., Zhang, D., Lin, H., et al. (2018). Highly Efficient A.T to G.C Base Editing by Cas9n-Guided tRNA Adenosine Deaminase in Rice. Mol. Plant 11, 631–634. doi:10.1016/j.molp.2018.02.008
Yan, D., Ren, B., Liu, L., Yan, F., Li, S., Wang, G., et al. (2021). High-efficiency and Multiplex Adenine Base Editing in Plants Using New TadA Variants. Mol. Plant 14 (5), 722–731. doi:10.1016/j.molp.2021.02.007
Zeng, D., Liu, T., Tan, J., Zhang, Y., Zheng, Z., Wang, B., et al. (2020). PhieCBEs: Plant High-Efficiency Cytidine Base Editors with Expanded Target Range. Mol. Plant 13, 1666–1669. doi:10.1016/j.molp.2020.11.001
Zhang, F., LeBlanc, C., Irish, V. F., and Jacob, Y. (2017). Rapid and Efficient CRISPR/Cas9 Gene Editing in Citrus Using the YAO Promoter. Plant Cel. Rep. 36, 1883–1887. doi:10.1007/s00299-017-2202-4
Zhang, J., Huguet ‐Tapia, J. C., Hu, Y., Jones, J., Wang, N., Liu, S., et al. (2017). Homologues of CsLOB1 in Citrus Function as Disease Susceptibility Genes in Citrus Canker. Mol. Plant Pathol. 18, 798–810. doi:10.1111/mpp.12441
Zhang, R., Liu, J., Chai, Z., Chen, S., Bai, Y., Zong, Y., et al. (2019). Generation of Herbicide Tolerance Traits and a New Selectable Marker in Wheat Using Base Editing. Nat. Plants 5, 480–485. doi:10.1038/s41477-019-0405-0
Zhang, X., Chen, L., Zhu, B., Wang, L., Chen, C., Hong, M., et al. (2020). Increasing the Efficiency and Targeting Range of Cytidine Base Editors through Fusion of a Single-Stranded DNA-Binding Protein Domain. Nat. Cel. Biol. 22, 740–750. doi:10.1038/s41556-020-0518-8
Li, Z., Xiong, X., Wang, F., Liang, J., and Li, J. F. (2019). Gene Disruption through Base Editing‐induced Messenger RNA Missplicing in Plants. New Phytol. 222, 1139–1148. doi:10.1111/nph.15647
Zhu, C., Zheng, X., Huang, Y., Ye, J., Chen, P., Zhang, C., et al. (2019). Genome Sequencing and CRISPR/Cas9 Gene Editing of an Early Flowering Mini‐Citrus ( Fortunella Hindsii ). Plant Biotechnol. J. 17, 2199–2210. doi:10.1111/pbi.13132
Zong, Y., Wang, Y., Li, C., Zhang, R., Chen, K., Ran, Y., et al. (2017). Precise Base Editing in rice, Wheat and maize with a Cas9-Cytidine Deaminase Fusion. Nat. Biotechnol. 35, 438–440. doi:10.1038/nbt.3811
Zong, Y., Song, Q., Li, C., Jin, S., Zhang, D., Wang, Y., et al. (2018). Efficient C-To-T Base Editing in Plants Using a Fusion of nCas9 and Human APOBEC3A. Nat. Biotechnol. 36, 950–953. doi:10.1038/nbt.4261
Keywords: citrus, base editing, adenine base editor, cytosine base editor, xanthomonas, LOB1, ALS, transgene-free
Citation: Huang X, Wang Y and Wang N (2022) Base Editors for Citrus Gene Editing. Front. Genome Ed. 4:852867. doi: 10.3389/fgeed.2022.852867
Received: 11 January 2022; Accepted: 10 February 2022;
Published: 28 February 2022.
Edited by:
Yiping Qi, University of Maryland, United StatesReviewed by:
Qinlong Zhu, South China Agricultural University, ChinaCopyright © 2022 Huang, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nian Wang, bmlhbndhbmdAdWZsLmVkdQ==
†ORCID: Xiaoen Huang, orcid.org/0000-0002-7514-1663; Yuanchun Wang, orcid.org/0000-0002-2165-2716; Nian Wang, orcid.org/0000-0001-7743-0728
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.