- Department of Biological Sciences, The George Washington University, Science and Engineering Hall, Washington, DC, United States
While piggyBac transposon-based transgenesis is widely used in various emerging model organisms, its relatively low transposition rate in butterflies and moths has hindered its use for routine genetic transformation in Lepidoptera. Here, we tested the suitability of a codon-optimized hyperactive piggyBac transposase (hyPBase) in mRNA form to deliver and integrate transgenic cassettes into the genome of the pantry moth Plodia interpunctella. Co-injection of hyPBase mRNA with donor plasmids successfully integrated 1.5–4.4 kb expression cassettes driving the fluorescent markers EGFP, DsRed, or EYFP in eyes and glia with the 3xP3 promoter. Somatic integration and expression of the transgene in the G0 injected generation was detectable from 72-h embryos and onward in larvae, pupae and adults carrying a recessive white-eyed mutation. Overall, 2.5% of injected eggs survived into transgene-bearing adults with mosaic fluorescence. Subsequent outcrossing of fluorescent G0 founders transmitted single-insertion copies of 3xP3::EGFP and 3xP3::EYFP and generated stable isogenic lines. Random in-crossing of a small cohort of G0 founders expressing 3xP3::DsRed yielded a stable transgenic line segregating for more than one transgene insertion site. We discuss how hyPBase can be used to generate stable transgenic resources in Plodia and other moths.
Introduction
Lepidoptera is a large insect order that comprises 160,000 species (Kristensen et al., 2007; Roskov et al., 2013), including a wide range of agricultural pests and ecosystem service providers, as well as important model systems for research in conservation biology, ecology, and evolutionary biology. In order to foster the potential of lepidopteran insects for functional genetics beyond the silkworm flagship system, for which transgenic resources already exist, we are developing the pantry moth Plodia interpunctella (hereafter Plodia; abbr. Pi), or Indianmeal moth, as an alternative laboratory organism amenable to routine genome editing and transgenesis. Plodia is a worldwide pest of stored food products, and exhibits convenient laboratory features that make it a promising system for the long-term maintenance of isogenic lines. In addition to its relatively short life cycle (25 days at 28°C) and ease of culture on a low-cost diet (Silhacek and Miller, 1972), Plodia cultures are resilient to inbreeding (Bartlett et al., 2018). Mass egg-laying can be stimulated by exposing their highly fecund females (Mbata, 1985) to CO2 gas, a property that allows the collection of synchronized embryos within the time frame of the first cell divisions, thus facilitating genetic transformation by microinjection (Dyby and Silhacek, 1997; Bossin et al., 2007). Finally, several genome assemblies and several transcriptomic resources have been published in this species (Harrison et al., 2012; Tang et al., 2017; Roberts et al., 2020; Heryanto et al., 2022; Kawahara et al., 2022).
Transgenesis techniques based on the piggyBac transposase (PBase) have been successfully implemented in a wide variety of insect model organisms and beyond (Handler, 2002; Gregory et al., 2016; Laptev et al., 2017). Butterflies and moths were shown to have transposition rates an order of magnitude lower than in beetles, mosquitoes and flies (Gregory et al., 2016), making routine transgenesis more challenging in the Lepidoptera order. A modified version of the transposase dubbed hyperactive piggyBac (hyPBase) was isolated from a mutant screen in 2011 (Yusa et al., 2011). HyPBase was later shown to dramatically increase transformation rates in flies and honeybees compared to its native version (Eckermann et al., 2018; Otte et al., 2018), and was also shown to provide practical transformation rates in Spodoptera noctuid moths (Chen and Palli, 2021).
Previously, delivery of the original pBase as a helper plasmid into Plodia syncytial embryos resulted in somatic transformation of fluorescent markers, but its efficiency for germline transformation was not reported (Bossin et al., 2007). Here, we extend the assessment of hyPBase transgenesis in Lepidoptera with a focus on the pyralid moth P. interpunctella, a pest of stored foods that is amenable to genome editing and genetic transformation (Bossin et al., 2007; Heryanto et al., 2022). In the current study, we injected an insect codon-optimized hyPBase as an mRNA (Otte et al., 2018) and monitored both the somatic and germline transformation rates of fluorescent markers driven by the 3xP3 promoter, a canonical promoter with strong activity in the ocular and glial tissues in Lepidoptera and other insects (Berghammer et al., 1999; Horn et al., 2002; Thomas et al., 2002). This approach robustly generated transgenic lines carrying various fluorescent protein markers, illustrating the suitability of hyPBase for routine genetic transformation in Plodia pantry moths. We discuss future strategies for establishing transgenic lines in emerging laboratory systems for lepidopteran functional genomics.
Results
We tested the suitability of hyPBase for transgenesis, using three donor plasmids that drive the expression of the fluorescent markers EGFP, DsRed, and EYFP. For each experiment, we report the levels of somatic transformation observed in the G0 injected generation, as well as our observations about the transmission of transgenes into further G1-3 generations.
hyPBase delivery of a 4.4 kb insert expressing 3xP3::EGFP
A practical transgenesis method must allow the delivery of relatively large cargos of several kilobases. To test the efficiency of hyPBase, we generated a piggyBac donor plasmid with a 4.4 kb insert with both a transgene and a transgenesis marker (Figure 1B). The cassette consisted of the mScarlet red fluorescent protein flanked by promoter and 3′UTR regions of the nanos-O gene Plodia homolog (nos-O), a germline determinant selected on its apparent specificity to gonadic tissues (Nakao and Takasu, 2019; Xu et al., 2022). The nos-O_prom::mScarlet component was an attempt to drive a fluorescent marker into the germline, as an exploratory experiment for future driving of Cas9 in germ cells to facilitate genome editing (Xu et al., 2022)— this did not yield positive results for this current study (see Discussion). As a transgenesis marker, we used a 3xP3::EGFP marker that labeled ocular tissues during previous somatic piggyBac transformation attempts in Plodia (Bossin et al., 2007). First, we injected this plasmid without hyPBase mRNA to control for episomal expression of the 3xP3::EGFP driver. These injections showed strong EGFP expression in large internal cells 48 h post-injection, suggesting episomal expression from the embryo vitellophages (Figure 2A). However, this signal was lost in 72-h old embryos, which only showed background levels of fluorescence or external autofluorescence artifacts at injection sites (Figure 2B). Thus, episomal expression of injected plasmids dissipates by 72 h of embryonic development and should not interfere with the screening of successful integration events at this stage and onwards.
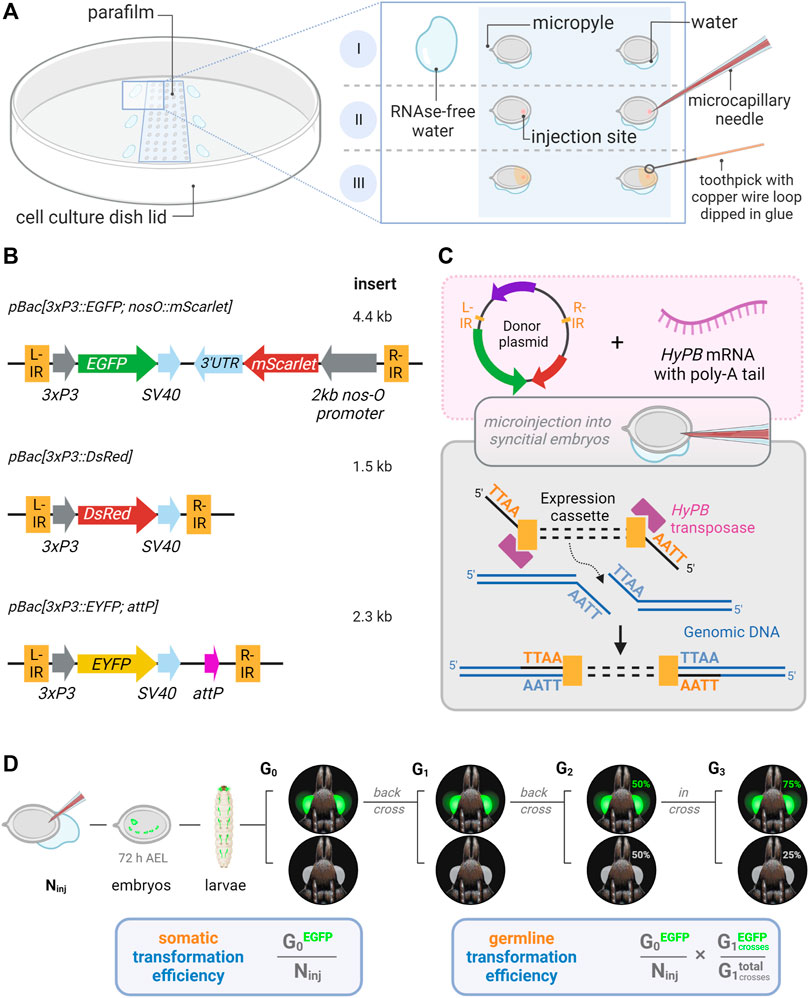
FIGURE 1. Microinjection procedure and transgenic constructs for the testing of hyperactive piggyBac transformation in Plodia. (A) Microinjection of P. interpunctella syncytial embryos. Gravid females oviposit en masse after CO2 narcosis, and eggs are collected and oriented on a parafilm strip in a tissue culture dish. A wet brush is used to position eggs, with water contact helping firm adhesion to the parafilm (I). Microinjection is performed on the side opposite to the micropyle (II). Peripheral droplets of water are used to periodically flush the injection capillary of yolk. Eggs are sealed with glue following injection (III). (B) Expression cassettes of donor plasmids carrying 3xP3 eye and glia fluorescent markers. IR = piggyBac internal repeats (L, left; R, right). (C) Transposon-mediated random integration following the injection of donor plasmid and hyPBase mRNA. (D) Somatic transformation efficiency (%) is equivalent to the number of potential G0 founders obtained out of 100 injected eggs. Germline transformation efficiency (%) factors proportion of transgenic G1 broods obtained from G0 outcrosses. Ninj = number of injected eggs. Made with Biorender.
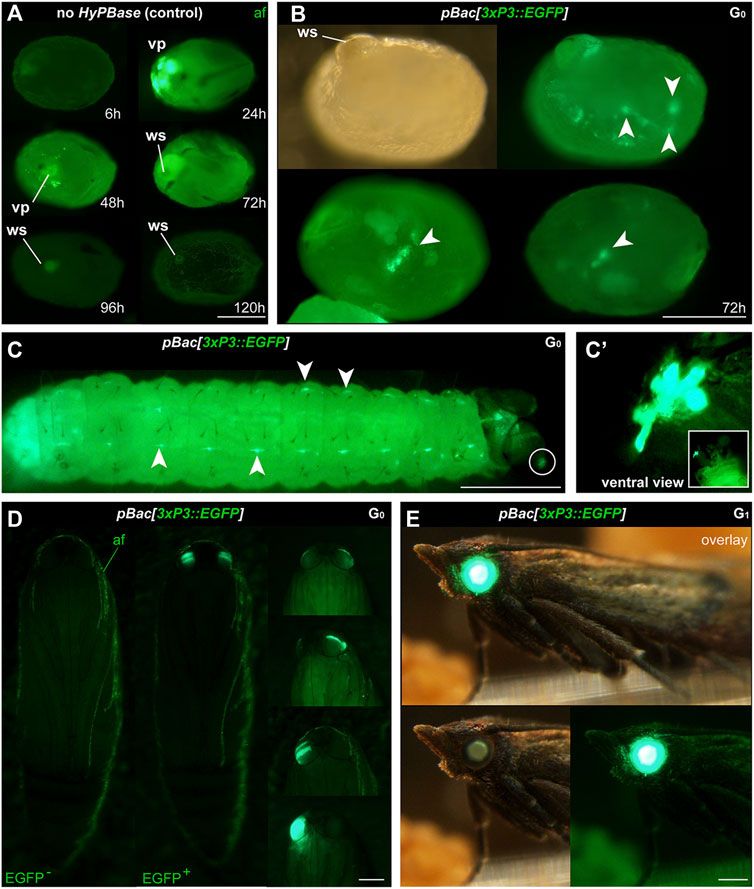
FIGURE 2. Phenotype of transgenic Plodia expressing EGFP in eyes and putative glia. (A) Control injections of pBac[3xP3::EGFP] show variable levels of green autofluorescence (af), most markedly at the injection wound site (ws). Episomal expression of EGFP in vitellophages (vp) is intense 24 h post-injection, reduced to background level after 48 h. (B) Donor pBac[3xP3::EGFP] + hyPBase mRNA injections resulted in 3xP3::EGFP expression, emerging as nervous system markings around 72 h post-injection (arrowheads). 23.7% of injected G0 eggs (262/1,104) showed a similar fluorescence during screening. (C) 3xP3::EGFP expression in a first instar larva, in ganglia of the Central Nervous System (consistent with an expected glial reporter activity of 3xP3), and in ocellar stemmata (circled, magnified in C′). (D) G0 mosaics of 3xP3::EGFP expression in pupal eyes. An EGFP-negative pupa is shown on the left for reference. (E) 3xP3::EGFP expression in a G1 Plodia adult with non-mosaic expression of EGFP in the eye (bottom right). EGFP is also visible in the brightfield (bottom right), with a green tint of the compound eyes in the Pi_wFog recessive white-eyed strain. Scale bars: A-C’ = 200 μm; D-E = 500 µm.
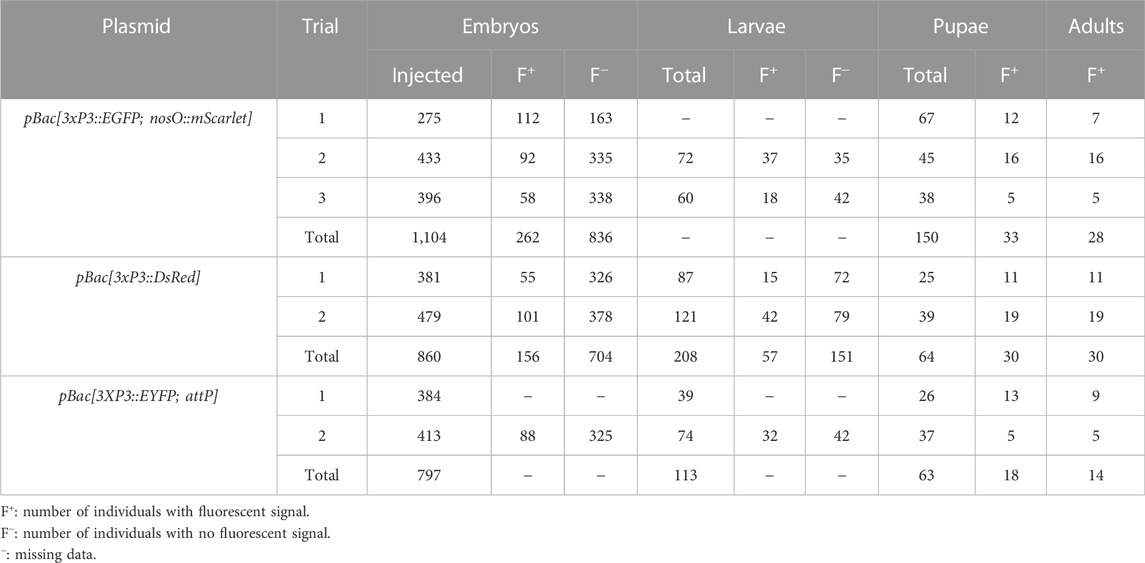
We then co-injected the donor plasmid pBac[3xP3::EGFP; nosO::mScarlet] with a hyPBase mRNA and monitored somatic transformation efficiencies throughout the G0 generation. In order to facilitate the screening of fluorescence, all experiments were performed in the Pi_wFog white-eyed strain that is devoid of screening pigments in eye tissues and also shows increased larval translucency (Heryanto et al., 2022). Transformed embryos and first instar hatchlings showed ocellar and glial EGFP fluorescence (Figures 2C, D), with 23.7% of injected eggs showing EGFP in 72-h embryos (Table 1). Injections produced viable larvae with persistent ocellar fluorescence, as well as eye fluorescence in pupae and adults (Figures 2E, F). Over several replicated experiments, we found that 16% of injected eggs resulted in pupae, of which 18.6% were EGFP+. Taking into account occasional pupal failure observed in normal rearing conditions, we determined that 2.5–3% of injected eggs become viable and fertile G0 somatic transformants.
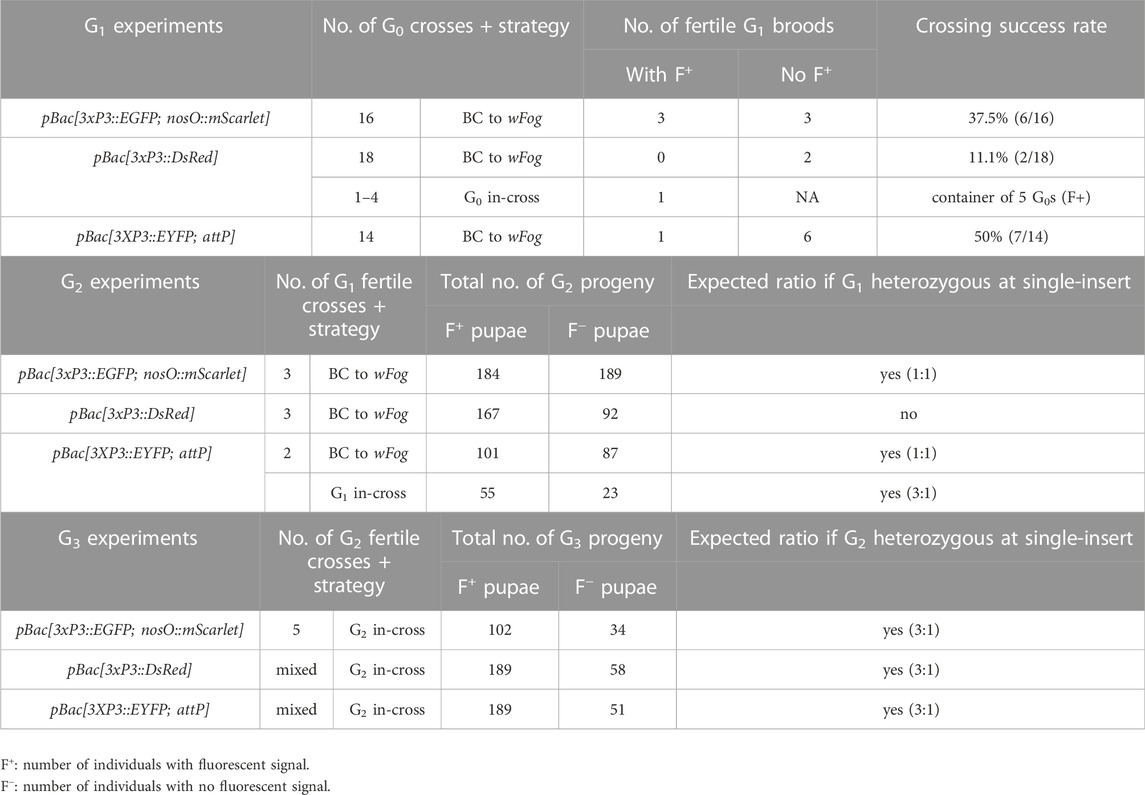
Next, we tested germline transmission by back-crossing G0 EGFP+ individuals to uninjected stock (Table 2). Out of six fertile pairs, 50% yielded EGFP+ G1 progeny, suggesting a practical level of germline mobilization among G0 founders. This result is mitigated by the fact that only six out a total of 16 single-pair matings (37.5%) were fertile and generated offspring in our conditions, suggesting that single-pair matings have limited success in our conditions (see Discussion). This establishes a germline efficiency rate of 0.94% (Figure 1D, GTE = 6/16 × 2.5% G0 founders), meaning that for 1000 G0 embryos injected, 9.4 embryos will survive as fertile founders passing the transgene to the G1 generation.
As we wanted to assess whether hyPBase would allow the rapid isolation of single-insertion lines, we needed to test if transgenes were integrated into multiple copies per G0 gamete, or if they could cause sterility. EGFP+ G1 individuals (N = 3) were back-crossed (Table 2) and produced a mean of 61 EGFP+ adults out of 124 emerged G2 per cross (49.3%), showing no statistical difference from an expected 50% ratio of a single insertion event (0.06< X2 < 0.46; df = 1; 0.10 < p < 0.80). Likewise, a total of five subsequent in-crosses (G2 EGFP+ x G2 EGFP+) each resulted in positive offspring ratios close to the expected 75% (0.06< X2 < 0.44; df = 1; 0.507 < p < 0.80). Of note, the Plodia-nosO:mScarlet transgene failed to drive red fluorescent signals detectable by epifluorescent and confocal microscopy in dissected ovaries, and we will explore the activity of alternative germline-driving promoters in the future (Nakao and Takasu, 2019; Xu et al., 2022). Overall, these data demonstrate that hyPBase provides practical transformation rates for a relatively large cargo insert, with at least 2.5% of injected zygotes yielding potential founders ready for isogenic line establishment after only one or two generations of backcrossing.
Evaluation of a 3xP3::DsRed donor vector
To expand the toolkit of transgenesis markers, we sought to test the activity of a pBac[3xP3::DsRed] donor vector for the screening of red eye fluorescence. We used the pHD-DsRed plasmid available through Addgene (Gratz et al., 2014, 2015), which carries a 1,146 bp 3xP3::DsRed-SV40 cassette tightly flanked by piggyBac internal repeats. Control injections without hyPBase mRNA revealed weak episomal expression in vitellophages and red background fluorescence (Figure 3A). Injection sites, which show non-specific autofluorescence under EGFP filter sets, do not fluoresce in the red channel. HyPBase-mediated insertion of pBac[3xP3::DsRed] resulted in glial signals in 14.4% of injected 72-h AEL (after egg laying) embryos, but intriguingly, no signal in the head region. Likewise, larval transformants showed sporadic signals in abdominal regions, seemingly nervous ganglia, but these patterns were always mosaic (Figure 3C). About 25/30 G0 DsRed+ pupae (83%) exhibited DsRed expression in the body (Figure 3D, G0 DsRedbody). DsRed fluorescence in the head region was observed in only five G0 pupae (Figure 3D, G0 DsRedeye), but its expression failed to reproduce the 3xP3::EGFP signal pattern in ocelli and eye tissues (Figure 2D).
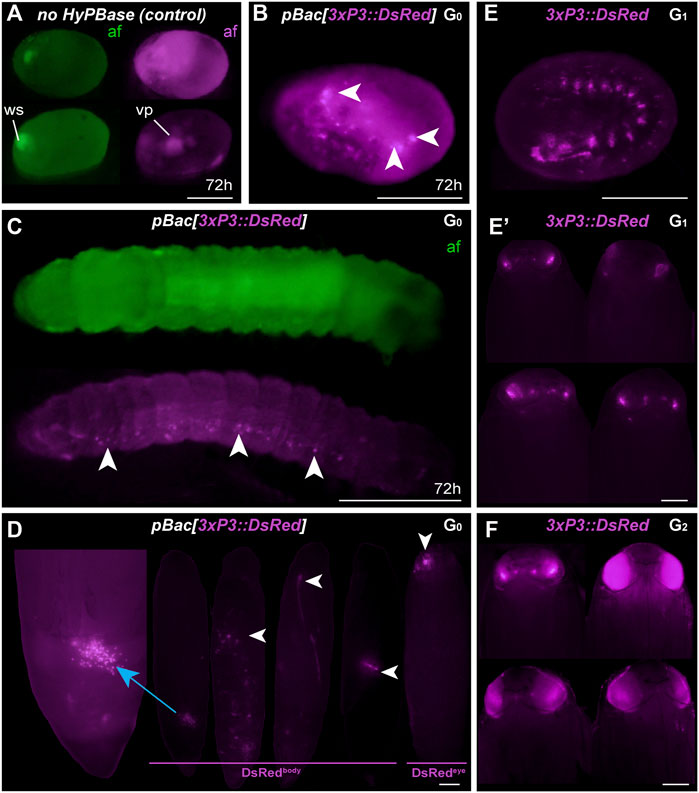
FIGURE 3. Somatic and germline transgenesis of 3xP3:DsRed. (A) Two control eggs (top and bottom rows) injected with only pBac[3xP3::DsRed] show background autofluorescence levels in the DsRed channel (af, magenta) at 72 h post-injection, including residual signal in vitellophages (vp). Wound site (ws) autofluorescence is limited to the EGFP channel (af, green). (B–C) HyPBase mRNA and pBac[3xP3::DsRed] result in glial expression of 3xP3::DsRed (magenta) in injected embryos. Ocellar expression was not observed in these experiments at the G0 phase. (D) G0 pupae showing various fluorescent signals in the abdomen (DsRedbody), a phenomenon not observed with other constructs. Expression in the head (DsRedeye) was occasionally seen at the G0 phase. (E) G1 transgenic embryo with non-mosaic expression of 3xP3::DsRed. (E′) G1 pupae showing weak eye fluorescent signals. These signals did not expand to the entire eye as the pupae developed, suggesting possible epigenetic effects. (F) G2 Plodia transgenic pupae obtained from G1 outcrosses resulted in pupae with bright 3xP3 fluorescence patterns that expanded throughout development. Variable intensity may be due to transgene copy number variation in this line. Scale bars: A-C, E = 200 μm; D, E′, F = 500 µm.
The presence of DsRed in abdominal regions suggested successful integration of the donor plasmid including in tissues close to the germline. To evaluate the germline transmission in G0 DsRedeye individuals, we backcrossed DsRedbody individuals to the uninjected stock. Only two out of 14 G0 DsRedeye backcrossed pairs gave G1 progeny (Table 2), and none inherited any DsRed fluorescence expression. In contrast, we recovered eggs from five G0 DsRedbody individuals that were incrossed liberally in a container, and showed full embryonic 3xP3::DsRed signals (Figure 3E). This salvaged stock resulted in six G1 pupae with DsRed expression in the eyes (Figure 3E) out of 52 isolated G1 pupae (11.5%), with no body phenotype observed. These six G1 DsRed+ Plodia were then individually crossed with Pi_wFog, three of which generated 83%, 70%, and 67% G2 DsRed+ G2 progeny (Figure 3F). As these ratios deviate from the 1:1 ratio expected in these crosses, we conclude that more than one insert occurred in the parental G0 founder germline. Finally, we used Splinkerette PCR (Potter and Luo, 2010; Shao and Lok, 2014) to map the piggyBac insertions in the G3 generation and found a single insertion site in the genome (Supplementary Table S1). We infer that a second insertion was either eliminated by chance when establishing the G3 line, or undetected due to the set of restriction enzymes used for digesting the genome.
The 3xP3 activity in this DsRed donor plasmid showed inconsistent results not seen with the EGFP donor, including absence of activity in G0 eye tissues, unusual abdominal fluorescent patches in G0 pupae, and reduced activity in G1 eyes. Intriguingly, full 3xP3::DsRed activity was recovered in G2-3 pupae, suggesting possible epigenetic effects of transient nature in earlier generations. This unusual behavior may be due to minor differences in the cassette proximal promoter (Supplementary Figure S1), to the compact design of this cassette (Figure 1B), or to other sequence features making the insert prone to abnormal expression.
Generation of 3xP3::EYFP transgenic lines carrying an attP docking site
We co-injected the pBac[3XP3::EYFP; attP] plasmid (Stern et al., 2017) with hyPBase mRNA into Pi_wFog. This donor includes an attP docking site (Figure 1B), a feature that may facilitate genetic engineering using site-specific recombination, if successfully integrated into the Plodia genome. Control injections show little background autofluorescence and vitellophage signals under the EYFP filter set (Figure 4A). Transgenic G0 embryos and larvae showed strong somatic 3xP3 activity consistent with ocular and glial expression (Figures 4B, C), with expected mosaic variations such as unilateral expression in one side of ocellus glia and ocelli-only expression. We recovered 14 pupae with mosaic G0 EYFP expression (Figure 2D) from a total of 63 surviving pupae, out of 797 embryos injected over two trials (Table 1).
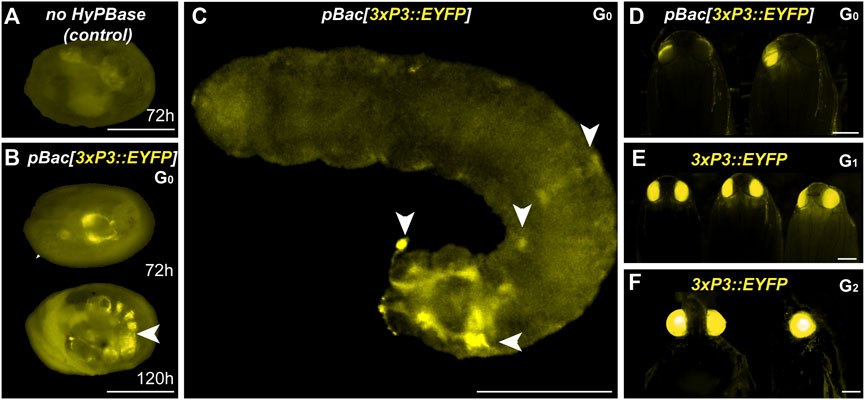
FIGURE 4. Somatic and germline transgenesis of 3xP3::EYFP in Plodia. (A) Weak background autofluorescence in the EYFP observation channel following control injection of the donor plasmid only. (B) Somatic activity of 3xP3::EYFP transgenes at 72 h and 120 h post injection in the late egg stage (C) Mosaic G0 3xP3::EYFP expression in a first instar larva, marking glia and ocellar stemmata (arrowheads). (D) Mosaic G0 3xP3::EYFP expression in pupal eyes. (E) 3xP3::EYFP expression in G1 Plodia pupae. (F) Ventral (left) and lateral views (right) of 3xP3::EYFP expression in G2 Plodia adults. Scale bars: A-C = 200 μm; D-F = 500 µm.
To estimate the efficiency of germline integration from these mosaic founders, we individually backcrossed the 14 EYFP+ G0 adults to single Pi_wFog individuals (Figure 4E; Table 2). Seven pairs gave progeny, among which only one cross generated progeny with 22 G1 EYFP+ pupal phenotypes out of 37 total isolated pupae, a ratio statistically close to the 50% proportion expected from a germline tissue heterozygous for a single insertion in the G0 founder (X2 = 1.32; df = 1, p = 0.25). To test if positive G1 individuals were heterozygous carriers for a single insertion, we simultaneously backcrossed 12 G1 EYFP+ to Pi_wFog and in-crossed five pairs of G1 EYFP+. Three of these crosses resulted in G2 EYFP+ progenies, with 59% and 48% positive ratios matching the 50% expected from backcrossing (0.18< X2 < 3.17; df = 1; 0.07 < p < 0.67), and a 71% positive ratio matching the expected 75% in the in-cross (X2 = 0.84, df = 1 p = 0.36). In summary, injection of pBac[3XP3::EYFP; attP] had a somatic transformation efficiency of 1.8%. The high level of mosaicism in G0 resulted in only one out of 14 successful backcrosses, resulting in a germline transformation efficiency of 0.13% (Figure 1D, GTE = 7.1.% x 1.8% G0 founders), but this event was successfully carried into a stable transgenic line. Similar to 3xP3::DsRed, Splinkerette PCR revealed a single piggyBac insertion into the Plodia genome in the G3 generation (Supplementary Table S1).
Discussion
Transformation efficiency rates of hyPBase in Plodia
In this study, we carried out somatic and stable germline transformation in Plodia interpunctella using the hyperactive piggyBac transposase (Yusa et al., 2011), and achieved high rates of somatic transformation with three independent piggyBac donor plasmids. We injected the transposase as a mRNA and used hyPBapis, a version of hyPBase codon-optimized for honeybees (Otte et al., 2018). Because Apis and Plodia both have an average GC content of around 35% (Jørgensen et al., 2007; Kawahara et al., 2022), we can reasonably expect compatibility in their codon usage biases.
Our study is the second to use an hyPBase mRNA as a transposase for transgenesis in a lepidopteran insect after the fall armyworm Spodoptera frugiperda (Chen and Palli, 2021). Plodia injections generated 15–40% of G0 somatic transformants when observed in 72-h embryos (mean of 22%), suggesting highly efficient integration. Importantly, while we expect a higher efficiency of hyPBase based on a previous report in Diptera (Eckermann et al., 2018), we have not directly compared the efficiency of hyPBase compared to pBase using our injection set-up and conditions here.
Across different trials, a mean of 2.5% of injected eggs expressed the transgene marker as adults, representing 30% of surviving adults. However, somatic fluorescence in the injected generation does not guarantee that the transgene has transposed into the germline, or that transgenic gametes are fertile. To assess transgene inheritability into the G1 generation, we backcrossed G0 fluorescent founders to non-transgenic individuals. We obtained three independent G1 lines expressing 3xP3::EGFP out of six fertile G0 crosses, and one line expressing 3xP3::EYFP out of seven fertile G0 crosses. Founders expressing 3xP3::DsRed showed unusual patterns of G0 mosaicism, possibly due to epigenetic regulatory effects (see Results section), and failed to propagate the transgene when mated in single outcrossing pairs (N = 14), but we recovered a stable insertion from G1 eggs that had been laid in a container where five G0 founders had been left to mate randomly, meaning that one out of 19 G0 transmitted 3xP3::DsRed.
In summary, our hyPBase mRNA-based injections in Plodia resulted in germline transformation efficiency rates of 0.18% (DsRed), 0.25% (EYFP), and 0.94% (EGFP). For comparison, Plutella transgenic experiments using pBase have efficiency rates of 0.43–0.65% (Gregory et al., 2016). Our Plodia injection protocol has a median pharate survival of 9%, much lower than the published Plutella adult survival rate of 27.8% (Gregory et al., 2016). Indeed, our injection methods favor speed and quantity over precision, using relatively wide-open needle bores that avoid clogging during injections, as well as a rapid but aggressive glue-based egg sealing procedure (Heryanto et al., 2022). Only 10–25% of eggs injected with piggyBac reagents hatched across trials in our conditions—as opposed to 21–60% in a previous Plodia microinjection report conducted by another group (Bossin et al., 2007) — but this is balanced by the fact that a single experimenter can inject about 400 pre-blastoderm embryos in a 2 h session with our procedure. Overall, the germline efficiency rates reported here mean that one fertile G0 founder was obtained for every 106 (EGFP), 555 (DsRed), and 777 (EYFP) injected embryos, making a 2–4 h injection effort (400–800 eggs) reasonably well suited for initiating each transgenic line attempt. Ultimately, practicality boils down to a trade-off between the number of injected embryos and their survival, and our data suggest that the high efficiency of hyPBase (Yusa et al., 2011; Eckermann et al., 2018) can make transgenesis feasible if one of these two factors is not optimal.
Other practical considerations for transgenesis in Lepidoptera
Mendelian segregation patterns observed at the G2 generations indicate that all four out of five stable lines originated as single-insertion events, with G0 founders likely carrying a single copy (Table 2). This feature can be used by experimenters to use various crossing strategies in the future, but we must highlight that single-mating strategies and crossing conditions resulted in few successful pairings in our initial attempts (e.g. 11–50% of G0 crosses, Table 2). This artificially lowered germline transmission rates, likely due to founders failing to mate in small containers in suboptimal conditions. As we gained experience with Plodia husbandry during these experiments, we increased mating success rates to 66–78% in subsequent generations (see Methods for the optimized procedure). Furthermore we recommend to mix one transgene carrier with two to three wild-type unmated adults of the opposite sex instead of one, as this maximizes the likelihood of successful mating in this system (Brower, 1975; Huang and Subramanyam, 2003).
Each of the three constructs we tested provided complementary information. The EGFP construct was the largest and delivered the highest germline transformation rate. Of note, the compact DsRed construct resulted in unusual G0 fluorescent patterns. While circumstantial, these observations bode well for using large inserts, and we caution that the pBac[3xP3::DsRed] (pHD-DsRed, Addgene #64703) has a more compact minimal promoter that might also explain its weaker expression (Supplementary Figure S1). The candidate germline driver of mScarlet, consisting of the proximal promoter and 3′UTR of nanos-O (Nakao and Takasu, 2019; Xu et al., 2019) cloned from the Plodia genome, failed to drive detectable fluorescence in ovarian tissues. We will investigate alternative germline promoters in the future (Xu et al., 2022), for instance by testing the PhiC31 site-specific integrase at the attP docking site from our new EYFP transgenic line (Yonemura et al., 2013; Haghighat-Khah et al., 2015; Stern et al., 2017; Stern, 2022). Both EGFP and EYFP showed robust and strong 3xP3-driven expression at all generations, without noticeable decrease over time in adult eyes (Das Gupta et al., 2015), with EYFP benefiting from lower autofluorescence effects than EGFP at various stages. We strategically used a white mutant strain deficient for eye-screening pigment, as routinely done in other insects to facilitate the screening of 3xP3-driven fluorescence (Stern et al., 2017; Klingler and Bucher, 2022), and this mutation also increases the translucency of Plodia larvae (Shirk, 2021; Heryanto et al., 2022). Of note, white mutations can be recessive-lethal in some lepidopteran species (Khan et al., 2017). Until alternative ways to generate depigmented eyes are found, this may limit the usefulness of 3xP3 drivers, especially in species where eggs and larvae are opaque and where screening becomes limited to narrow developmental windows (Das Gupta et al., 2015; Özsu et al., 2017). In such species, we suggest that stronger, more ubiquitous promoters of viral origin such as Op-ie2 and Hr5-ie1 may be more practical for transgenic screening (Martins et al., 2012; Xu et al., 2019, 2022). In future hyPBase transgenesis experiments, we intend to test the activity of viral promoters, potential ubiquitous promoters, and putative tissue-specific drivers for marking tissues such as the nervous system, wing epithelia, silk glands, and hemocytes. With these tools in hand, Plodia is well positioned to complement other organisms with functional genomics capacity like Bombyx and Plutella, due to its suitability for mass rearing, synchronized egg collection, and long-term maintenance of inbred lines.
Materials and methods
Plodia strains and rearing
The Pi_wFog strain (Heryanto et al., 2022) consists of an introgression of the recessive w-mutation (Shirk, 2021) from the Pi w- strain (origin: USA, kind gift of Paul Shirk), into the genetic background of the “Dundee” strain (origin: United Kingdom, kind gift of Mike Boots). Genome assemblies of both Pi w- and Pi_Dundee parental strains are available (Roberts et al., 2020; Kawahara et al., 2022). The resulting hybrid Pi_wFog strain has been maintained in inbred state for 3 years and used throughout this study. All rearing used previously published methods (Heryanto et al., 2022), using special containers and a wheat bran-sucrose-glycerol diet (Silhacek and Miller, 1972). A rearing temperature of 28°C resulted in a generation time of 28 days.
Plasmid constructs
The pBac[3xP3::EGFP; Tc’hsp5′-Gal4Delta-3′UTR] (Addgene plasmid # 86449) was used as a donor plasmid with piggyBac insertion repeats and the 3xP3::EGFP reporter (Schinko et al., 2010). To generate pBac[3xP3::EGFP; nosO_prom::mScarlet-nosO_3′UTR], an mScarlet cassette preceded by 2 kb of promoter sequence immediately upstream of the Plodia nanos-O start codon, was synthesized in the pUC-GW-Amp backbone by Genewiz and sub-cloned into the FseI and AscI restriction sites of pBac[3xP3::EGFP; Tc’hsp5′-Gal4Delta-3′UTR]. The pBac[3xP3::DsRed] (pHD-DsRed) and pBac[3XP3::EYFP; attP] plasmids were obtained from Addgene (#64703, and #86860) and used without modification (Gratz et al., 2014, 2015; Stern et al., 2017). All the 3xP3-driven fluorophore genes included an SV40 termination sequence.
Transposase mRNA and injection mixes
The pGEM-T_hyPBapis plasmid encodes a hyPBase that was codon-optimized for honeybees (Otte et al., 2018). The source plasmid was purified using the QIAprep Spin Miniprep Kit (Qiagen), linearized with NcoI-HF (New England Biolabs) and concentrated using acetate/ethanol precipitation. Around 500 ng of linearized template were transcribed using the mMESSAGE mMACHINE™ T7 ULTRA Transcription Kit (Invitrogen) and purified using the MEGAclear Transcription Clean-Up Kit (Invitrogen). After quantification with Nanodrop (Thermofisher), the solution was divided into 1,050 ng/μL one-time use aliquots and stored at −80°C.
Microinjections
Injection mixes consisted of 400 ng/μL hyPBase mRNA, 200 ng/μL donor plasmid, and 0.05% cell-culture grade Phenol Red (Sigma-Aldrich). Donor plasmids without hyPBase mRNA were injected in separate experiments as controls. Microinjection procedures (Figure 1A) followed a previously described procedure (Heryanto et al., 2022), with all embryo injections performed within 40 min after egg laying (AEL). Injected embryos were counted and kept in a rearing container with a small damp Kimwipe at 28°C. For the first 72 h, the container vent was covered with tape in order to maintain humidity saturation, a parameter that prevents egg desiccation. After 72 h, the vent was opened and the Kimwipe removed, and about five flakes of Plodia food added next to the eggs, in order to keep the emerging larvae within the injection dish. Mean emergence time of the Pi_wFog strain is 83 h AEL at 28°C for uninjected eggs, and is delayed by injection stress to 100–115 h AEL. Because of this variability, we report times of observation after injection in hours rather than in relative percentages.
Fluorescent microscopy
Larvae and adult Plodia were anesthetized in tissue culture dishes positioned over a cold metal block during microscopy observation. All pictures were taken under the Olympus SZX16 stereomicroscope equipped with a Lumencor SOLA Light Engine SM 5-LCR-VA lightsource or standard stereomicroscope brightfield lamp, and with a trinocular tube connected to an Olympus DP73 digital color camera. Separation of fluorescent channels was performed using Chroma Technology filter sets ET-EGFP 470/40 × 510/20 m, ET-EYFP 500/20 × 535/30 m, and AT-TRICT-REDSHFT 540/25x, 620/60 m.
Survival and G0 somatic transformation rates
Embryonic survival rates (“egg hatching” rates) were determined by the ratio of hatched eggs at 120 h AEL over the number of injected eggs (Ninj). Empty egg shells were counted for this purpose instead of first-instar hatchlings, which are difficult to count accurately in the presence of food. Pharate survival rates were determined by the ratio of pupae obtained from a given injection experiment, divided by Ninj, and thus accounts for mortality occurring at embryonic and larval stages. Pupal mortality was negligible, making pharate survival rates a reasonable proxy for overall adult survival, and is more convenient to couple to fluorescent screening than in mobile adults. G0 transformation rates were independently measured in embryos and in pupae. For embryos, eggs with bright, internal fluorescent signals consistent with an ocellar or glial expression were counted as positive (fluorescent, F+ in Table 1) around 72 h AEL, and non-fluorescent eggs were counted as negative (F‒). To isolate individual pupae, cardboard strips that are preferentially used as pupation sites (“hotels”) were added into containers containing fifth instar larvae, allowing a convenient isolation of individual Plodia pupae. Pupae were then extracted from these lodges and aligned on double-sided tape for fluorescence screening. Pupae with any glial or eye signal were counted as positive, while others were counted as negative. G0 somatic transformation efficiency rate was determined as the number of healthy adult individuals emerged from fluorescent pupae, and normalized by Ninj.
Controlled crosses for germline transmission
Germline transformation efficiency rates factored the somatic transformation efficiency rate by the proportion of attempted G0 backcrosses yielding transgenic offspring. G0 transgenic adults or late pupae exhibiting positive fluorescent signals (G0 F+) were crossed to a single unmated Pi_wFog adult of the opposite sex, by mixing in a 1.25 oz Plastic Souffle Cup (Solo) containing ∼0.2 g of diet and ∼1 cm2 of paper towel. Pi_wFog outcrossing mates were replaced if found dead before any visible egg laying. These cups were monitored for up to 2 weeks for any larval emergence, after which they were transferred to a vented rearing container with a bed of Plodia food (modified LocknLock containers described in Heryanto et al., 2022; 177 ml and 350 ml formats). At the wandering L5 stage, cardboard “hotels” were added into the containers for pupal isolation. G1 pupae with positive fluorescent signal were counted and backcrossed to an unmated Pi_wFog with the same procedure stated above. The resulting G2 pupae were in-crossed as sib-matings and maintained as isogenic stock in the G3 generations and henceforth.
Mapping of piggyBac insertions with splinkerette PCR
The Genomic DNA of 3 separate transgenic G3 Plodia adults carrying [3xP3::DsRed] or [3XP3::EYFP; attP] was isolated using the Quick-DNA Tissue/Insect Kits (Zymo Research) in 20 µL DNA Elution Buffer, RNAse-treated, and quantified by fluorimetry. Following the Splinkerette PCR protocols (Potter and Luo, 2010; Shao and Lok, 2014), 100 ng of the isolated DNA was digested with BfuCI (New Englands Biolabs). Two rounds of Splinkerette PCR were done using Q5 High-Fidelity DNA Polymerase (New England Biolabs) with the following cycle parameters: 98°C for 1 min; 30 cycles of 98°C for 20 s, 67°C (Tm of the primers targeting the insertion 5’ end) for 20 s, and 72°C for 2 min; and 72°C for 10 min. The PCR products were purified using the PureLink PCR Purification Kit (Thermofisher) prior to Sanger sequencing (Supplementary Table S1).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
CH and AM designed the study and wrote the manuscript. AM-V and AM advised on the methodology. CH performed the experiments and analyzed the data.
Funding
This work was supported by National Science Foundation Grant under NSF/IOS grant IOS-1923147.
Acknowledgments
We thank Paul Shirk for invaluable advice on the Plodia system, Patricia Hernandez for providing access to the fluorescent microscope used in this manuscript, Ioannis Eleftherianos for advice and laboratory access, and the members of the Martin lab for comments on the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgeed.2022.1074888/full#supplementary-material
References
Bartlett, L. J., Wilfert, L., and Boots, M. (2018). A genotypic trade-off between constitutive resistance to viral infection and host growth rate. Evolution 72, 2749–2757. doi:10.1111/evo.13623
Berghammer, A. J., Klingler, M., and A Wimmer, E. (1999). A universal marker for transgenic insects. Nature 402, 370–371. doi:10.1038/46463
Bossin, H., Furlong, R. B., Gillett, J. L., Bergoin, M., and Shirk, P. D. (2007). Somatic transformation efficiencies and expression patterns using the JcDNV and piggyBac transposon gene vectors in insects. Insect Mol. Biol. 16, 37–47. doi:10.1111/j.1365-2583.2006.00693.x
Brower, J. H. (1975). Plodia interpunctella: Effect of sex ratio on reproductivity. Ann. Entomol. Soc. Am. 68, 847–851. doi:10.1093/aesa/68.5.847
Chen, X., and Palli, S. R. (2021). Hyperactive piggyBac: Transposase-mediated Germline Transformation in the Fall Armyworm, Spodoptera frugiperda. J. Vis. Exp., e62714. doi:10.3791/62714
Das Gupta, M., Chan, S. K. S., and Monteiro, A. (2015). Natural loss of eyeless/Pax6 expression in eyes of Bicyclus anynana adult butterflies likely leads to exponential decrease of eye fluorescence in transgenics. PloS One 10, e0132882. doi:10.1371/journal.pone.0132882
Dyby, S., and Silhacek, D. L. (1997). Juvenile hormone agonists cause abnormal midgut closure and other defects in the moth, Plodia interpunctella (Lepidoptera: Pyralidae). Invertebr. Reprod. Dev. 32, 231–244. doi:10.1080/07924259.1997.9672629
Eckermann, K. N., Ahmed, H. M. M., KaramiNejadRanjbar, M., Dippel, S., Ogaugwu, C. E., Kitzmann, P., et al. (2018). Hyperactive piggyBac transposase improves transformation efficiency in diverse insect species. Insect biochem. Mol. Biol. 98, 16–24. doi:10.1016/j.ibmb.2018.04.001
Gratz, S. J., Rubinstein, C. D., Harrison, M. M., Wildonger, J., and O’Connor-Giles, K. M. (2015). CRISPR-Cas9 genome editing in Drosophila. Curr. Protoc. Mol. Biol. 111, 31. doi:10.1002/0471142727.mb3102s111
Gratz, S. J., Ukken, F. P., Rubinstein, C. D., Thiede, G., Donohue, L. K., Cummings, A. M., et al. (2014). Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971. doi:10.1534/genetics.113.160713
Gregory, M., Alphey, L., Morrison, N. I., and Shimeld, S. M. (2016). Insect transformation with piggyBac: Getting the number of injections just right. Insect Mol. Biol. 25, 259–271. doi:10.1111/imb.12220
Haghighat-Khah, R. E., Scaife, S., Martins, S., St John, O., Matzen, K. J., Morrison, N., et al. (2015). Site-specific cassette exchange systems in the Aedes aegypti mosquito and the Plutella xylostella moth. PLOS ONE 10, e0121097. doi:10.1371/journal.pone.0121097
Handler, A. M. (2002). Use of the piggyBac transposon for germ-line transformation of insects. Insect biochem. Mol. Biol. 32, 1211–1220. doi:10.1016/s0965-1748(02)00084-x
Harrison, P. W., Mank, J. E., and Wedell, N. (2012). Incomplete sex chromosome dosage compensation in the Indian meal moth, Plodia interpunctella, based on de novo transcriptome assembly. Genome Biol. Evol. 4, 1118–1126. doi:10.1093/gbe/evs086
Heryanto, C., Hanly, J. J., Mazo-Vargas, A., Tendolkar, A., and Martin, A. (2022). Mapping and CRISPR homology-directed repair of a recessive white eye mutation in Plodia moths. iScience 25, 103885. doi:10.1016/j.isci.2022.103885
Horn, C., Schmid, B. G. M., Pogoda, F. S., and Wimmer, E. A. (2002). Fluorescent transformation markers for insect transgenesis. Insect biochem. Mol. Biol. 32, 1221–1235. doi:10.1016/S0965-1748(02)00085-1
Huang, F., and Subramanyam, B. (2003). Effects of delayed mating on reproductive performance of Plodia interpunctella (hübner)(Lepidoptera: Pyralidae). J. Stored Prod. Res. 39, 53–63. doi:10.1016/s0022-474x(02)00018-8
Jørgensen, F. G., Schierup, M. H., and Clark, A. G. (2007). Heterogeneity in regional GC content and differential usage of codons and amino acids in GC-poor and GC-rich regions of the genome of Apis mellifera. Mol. Biol. Evol. 24, 611–619. doi:10.1093/molbev/msl190
Kawahara, A. Y., Storer, C. G., Markee, A., Heckenhauer, J., Powell, A., Plotkin, D., et al. (2022). Long-read HiFi sequencing correctly assembles repetitive heavy fibroin silk genes in new moth and caddisfly genomes. Gigabyte 2022, 1–14. doi:10.46471/gigabyte.64
Khan, S. A., Reichelt, M., and Heckel, D. G. (2017). Functional analysis of the ABCs of eye color in Helicoverpa armigera with CRISPR/Cas9-induced mutations. Sci. Rep. 7, 1–14. doi:10.1038/srep40025
Klingler, M., and Bucher, G. (2022). The red flour beetle T. castaneum: Elaborate genetic toolkit and unbiased large scale RNAi screening to study insect biology and evolution. EvoDevo 13, 14–11. doi:10.1186/s13227-022-00201-9
Kristensen, N. P., Scoble, M. J., and Karsholt, O. L. E. (2007). <p><strong>Lepidoptera phylogeny and systematics: The state of inventorying moth and butterfly diversity</strong></p>. Zootaxa 1668, 699–747. doi:10.11646/zootaxa.1668.1.30
Laptev, I. A., Raevskaya, N. M., Filimonova, N. A., and Sineoky, S. P. (2017). The piggyBac transposon as a tool in genetic engineering. Appl. Biochem. Microbiol. 53, 874–881. doi:10.1134/s000368381709006x
Martins, S., Naish, N., Walker, A. S., Morrison, N. I., Scaife, S., Fu, G., et al. (2012). Germline transformation of the diamondback moth, Plutella xylostella L., using the piggyBac transposable element: Germline transformation of diamondback moth. Insect Mol. Biol. 21, 414–421. doi:10.1111/j.1365-2583.2012.01146.x
Mbata, G. N. (1985). Some physical and biological factors affecting oviposition by Plodia interpunctella (Hubner) (Lepidoptera: Phycitidae). Int. J. Trop. Insect Sci. 6, 597–604. doi:10.1017/S1742758400009176
Nakao, H., and Takasu, Y. (2019). Complexities in Bombyx germ cell formation process revealed by Bm-nosO (a Bombyx homolog of nanos) knockout. Dev. Biol. 445, 29–36. doi:10.1016/j.ydbio.2018.10.012
Otte, M., Netschitailo, O., Kaftanoglu, O., Wang, Y., Jr, R. E. P., and Beye, M. (2018). Improving genetic transformation rates in honeybees. Sci. Rep. 8, 16534. doi:10.1038/s41598-018-34724-w
Özsu, N., Chan, Q. Y., Chen, B., Gupta, M. D., and Monteiro, A. (2017). Wingless is a positive regulator of eyespot color patterns in Bicyclus anynana butterflies. Dev. Biol. 429, 177–185. doi:10.1016/j.ydbio.2017.06.030
Potter, C. J., and Luo, L. (2010). Splinkerette PCR for mapping transposable elements in Drosophila. PloS One 5, e10168. doi:10.1371/journal.pone.0010168
Roberts, K. E., Meaden, S., Sharpe, S., Kay, S., Doyle, T., Wilson, D., et al. (2020). Resource quality determines the evolution of resistance and its genetic basis. Mol. Ecol. 29, 4128–4142. doi:10.1111/mec.15621
Roskov, Y., Kunze, T., Paglinawan, L., Orrell, T., Nicolson, D., Culham, A., et al. (2013). Species 2000 & ITIS catalogue of life, 2013 annual checklist.
Schinko, J. B., Weber, M., Viktorinova, I., Kiupakis, A., Averof, M., Klingler, M., et al. (2010). Functionality of the GAL4/UAS system in Tribolium requires the use of endogenous core promoters. BMC Dev. Biol. 10, 53–12. doi:10.1186/1471-213X-10-53
Shao, H., and Lok, J. B. (2014). Detection of piggyBac-mediated transposition by splinkerette PCR in transgenic lines of Strongyloides ratti. Bio. Protoc. 4, e1015. doi:10.21769/BioProtoc.1015
Shirk, B. D. (2021). Gene editing of the ABC transporter/white locus using CRISPR/Cas9 mutagenesis in the Indian meal moth (Plodia interpunctella).
Silhacek, D. L., and Miller, G. L. (1972). Growth and development of the Indian meal moth, Plodia interpunctella (Lepidoptera: Phycitidae), under laboratory mass-rearing Conditions1. Ann. Entomol. Soc. Am. 65, 1084–1087. doi:10.1093/aesa/65.5.1084
Stern, D. L., Crocker, J., Ding, Y., Frankel, N., Kappes, G., Kim, E., et al. (2017). Genetic and transgenic reagents for Drosophila simulans, D. mauritiana, D. yakuba, D. santomea, and D. virilis. G3 7, 1339–1347. doi:10.1534/g3.116.038885
Stern, D. L. (2022). Transgenic tools for targeted chromosome rearrangements allow construction of balancer chromosomes in non-melanogaster Drosophila species. G3 12, jkac030. doi:10.1093/g3journal/jkac030
Tang, P.-A., Wu, H.-J., Xue, H., Ju, X.-R., Song, W., Zhang, Q.-L., et al. (2017). Characterization of transcriptome in the Indian meal moth Plodia interpunctella (Lepidoptera: Pyralidae) and gene expression analysis during developmental stages. Gene 622, 29–41. doi:10.1016/j.gene.2017.04.018
Thomas, J. L., Da Rocha, M., Besse, A., Mauchamp, B., and Chavancy, G. (2002). 3xP3-EGFP marker facilitates screening for transgenic silkworm Bombyx mori L. from the embryonic stage onwards. Insect biochem. Mol. Biol. 32, 247–253. doi:10.1016/s0965-1748(01)00150-3
Xu, J., Chen, R., Chen, S., Chen, K., Tang, L., Yang, D., et al. (2019). Identification of a germline-expression promoter for genome editing in Bombyx mori. Insect Sci. 26, 991–999. doi:10.1111/1744-7917.12657
Xu, X., Harvey-Samuel, T., Siddiqui, H. A., Ang, J. X. D., Anderson, M. E., Reitmayer, C. M., et al. (2022). Toward a CRISPR-Cas9-based gene drive in the diamondback moth Plutella xylostella. CRISPR J. 5, 224–236. doi:10.1089/crispr.2021.0129
Yonemura, N., Tamura, T., Uchino, K., Kobayashi, I., Tatematsu, K., Iizuka, T., et al. (2013). phiC31-integrase-mediated, site-specific integration of transgenes in the silkworm, Bombyx mori (Lepidoptera: Bombycidae). Appl. Entomol. Zool. 48, 265–273. doi:10.1007/s13355-013-0182-6
Keywords: Plodia, Lepidoptera, transgenesis, piggyBac, microinjection, germline transformation, transposon
Citation: Heryanto C, Mazo-Vargas A and Martin A (2022) Efficient hyperactive piggyBac transgenesis in Plodia pantry moths. Front. Genome Ed. 4:1074888. doi: 10.3389/fgeed.2022.1074888
Received: 20 October 2022; Accepted: 05 December 2022;
Published: 23 December 2022.
Edited by:
Trevor Sorrells, Yale University, United StatesReviewed by:
Michael Perry, University of California, San Diego, United StatesSounak Sahu, National Cancer Institute at Frederick (NIH), United States
Copyright © 2022 Heryanto, Mazo-Vargas and Martin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arnaud Martin, arnaud@gwu.edu
†ORCID: Christa Heryanto, https://orcid.org/0000-0002-9917-5710; Anyi Mazo-Vargas, https://orcid.org/0000-0001-9644-2871; Arnaud Martin, https://orcid.org/0000-0002-5980-2249
 Christa Heryanto
Christa Heryanto