
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genome Ed., 13 January 2022
Sec. Genome Engineering and Neurologic Disorders
Volume 3 - 2021 | https://doi.org/10.3389/fgeed.2021.799722
This article is part of the Research TopicMethods, Applications and Developments of Genome Editing ToolsView all 5 articles
The replication-defective, non-pathogenic, nearly ubiquitous single-stranded adeno-associated viruses (AAVs) have gained importance since their discovery about 50 years ago. Their unique life cycle and virus-cell interactions have led to the development of recombinant AAVs as ideal genetic medicine tools that have evolved into effective commercialized gene therapies. A distinctive property of AAVs is their ability to edit the genome precisely. In contrast to all current genome editing platforms, AAV exclusively utilizes the high-fidelity homologous recombination (HR) pathway and does not require exogenous nucleases for prior cleavage of genomic DNA. Together, this leads to a highly precise editing outcome that preserves genomic integrity without incorporation of indel mutations or viral sequences at the target site while also obviating the possibility of off-target genotoxicity. The stem cell-derived AAV (AAVHSCs) were found to mediate precise and efficient HR with high on-target accuracy and at high efficiencies. AAVHSC editing occurs efficiently in post-mitotic cells and tissues in vivo. Additionally, AAV also has the advantage of an intrinsic delivery mechanism. Thus, this distinctive genome editing platform holds tremendous promise for the correction of disease-associated mutations without adding to the mutational burden. This review will focus on the unique properties of direct AAV-mediated genome editing and their potential mechanisms of action.
The completion of the sequencing of the human genome marked the start of the race to create targeted modifications for the study of gene function, generation of disease models, and therapeutic applications (Li et al., 2020; Galichet and Lovell-Badge, 2021). Subsequent extensive genome-wide association studies linked defined genetic mutations in the population with known diseases. However, despite intensive efforts in these areas, few curative genetic therapies have emerged to date. It is estimated that currently, less than 5% of rare genetic diseases have effective treatments (Brooks et al., 2020). Protein replacement therapies have been developed for many inherited diseases. However, the availability, cost, and quality of life issues pose significant barriers. Moreover, the inability of many recombinant proteins to cross the blood-brain barrier and their immunogenicity in patients with null mutations pose additional therapeutic challenges. Thus, the ability to precisely and permanently correct pathogenic mutations at the level of the genome without adding to the mutational burden has the potential to be transformative for genetic therapies of inherited and acquired diseases. Here, we will review the unique contributions of adeno-associated virus (AAV) vectors to the field of genome editing in the larger context of genetic medicine.
AAVs have emerged as efficient genetic modification vehicles due to efficient in vivo infectivity, non-pathogenicity, widespread tissue tropism, rare genomic integration, and their ability to infect and persist in non-dividing cells (Gaj et al., 2016; Epstein and Schaffer, 2017). AAVs are comprised of a family of natural human non-pathogenic, single-stranded, replication-defective parvoviruses (Berns and Linden, 1995; Srivastava, 2016). The single-stranded AAV genomes are bounded at either end by palindromic G-C-rich inverted terminal repeats (ITRs), which self-base-pair to form unique structures. AAV infection of target cells in the absence of helper virus coinfection results in latency which is the basis for the use of AAV as delivery vehicles for genetic material. AAV infection of target cells is initiated by binding to a receptor and/or a co-receptor (Figure 1). AAV virions are then internalized and translocated to the nucleus via an endosomal route and enter the nuclear pore complex (Junod et al., 2021). AAV virions then undergo uncoating in the nucleus, and the single-stranded genomes are released. AAV genomes localize to the periphery of the nuclei and colocalize with euchromatin, where active transcription and DNA repair are known to occur. For wild-type AAV, if a helper virus is present, replication ensues. AAV replication utilizes the AAV encoded Rep proteins, helper virus-encoded functions, and the cellular replication proteins RPA, RFC, PCNA, and DNA polymerase delta (Ni et al., 1998; Nash et al., 2007). Recombinant AAV vectors are doubly replication-deficient in the absence of AAV Rep/Cap genes and helper virus functions and do not undergo replication. After uncoating, the single-stranded genomes are converted to double-stranded multimeric circular concatemeric episomal forms that persist long-term in post-mitotic cells (Figure 2A) (Ferrari et al., 1996; Wang et al., 2007).
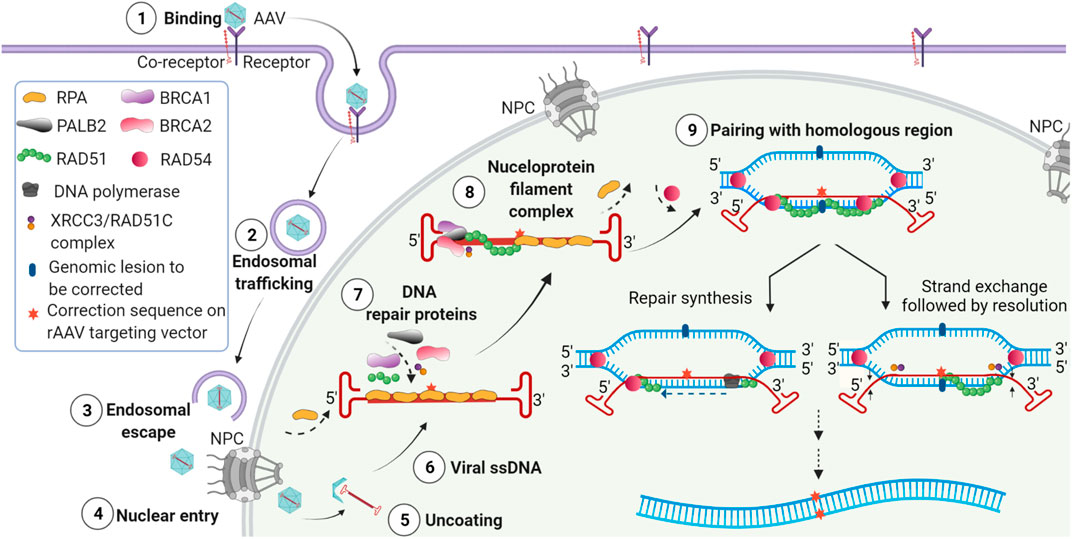
FIGURE 1. Model of events following transduction by AAV editing vectors leading to genome editing. (1) Binding of AAV virions to cell surface receptor and co-receptors and initiation of receptor-mediated endocytosis. (2) Endosomal entry and trafficking of the AAV virions. (3) Release of AAVs from endosomes. (4) Entry of AAV through the nuclear pore complex (NPC). (5) Uncoating of AAV virions and release of the single-stranded vector genome in the nucleus. (6) The released single stranded AAV vector genome with homology region and ITRs. (7) Recruitment of DNA repair proteins to the AAV editing genome containing the correction sequence. (8) Assembly of the HR complex on the AAV editing genome and formation of nucleoprotein filament complex. (9) Homology search between editing vector genome and chromosomal DNA leading to the pairing of homologous sequences and initiation of editing by repair synthesis (left) or strand exchange (right).
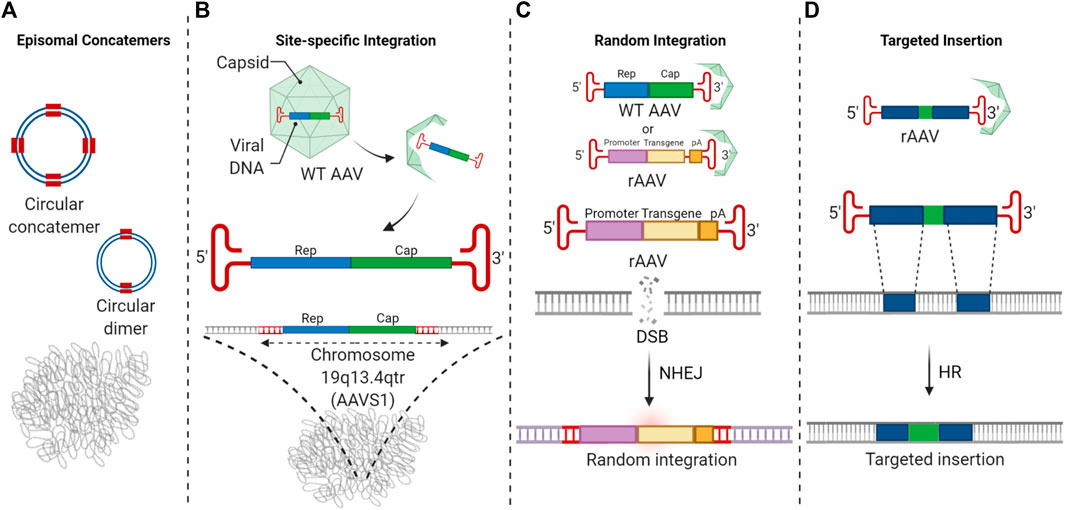
FIGURE 2. The fate of AAV vector genomes in the nucleus. (A) AAV vector genomes predominantly survive long-term as episomal concatemers. (B) Wild-type AAV and Rep 68/78 containing AAV vectors can undergo site-specific integration at the AAVS1. (C) A small fraction of AAV vector genomes undergo random integration at very low frequencies. This event involves the AAV ITRs. (D) HR-mediated AAV editing results in targeted insertion at chromosomal locations specified by the homology arms.
Recombinant AAV vectors have proven to be safe, well-tolerated, and effective gene therapy vectors for treating genetic diseases. Over 3,300 individuals have been treated with AAV vectors, and there are over 130 AAV trials registered on clinicaltrials.gov (Kuzmin et al., 2021). Two AAV vectors have received USFDA approval, Luxturna for a rare inherited retinal dystrophy and Zolgensma for spinal muscular atrophy. AAV vectors are primarily safe, with a few exceptions resulting from very high-dose treatments in specific disease settings (Kuzmin et al., 2021). It is expected that with improvements in vector design, production, and purification methods, the toxicities associated with high-dose treatment will be controlled. Thus, AAV vectors are well on their way to becoming established genetic therapies.
However, despite the tremendous promise of gene therapy to cure genetic diseases, several limitations remain. First, since AAV vector genomes primarily exist as nuclear episomes, the long-term durability of treatment is a concern. It is unclear whether the episomal AAV vector genomes will persist for the life of the patient. Since most cells in the adults are post-mitotic, it is likely that gene therapy will last for several years, as has been documented in many trials (Niemeyer et al., 2009; Nathwani et al., 2011; Nathwani et al., 2014). However, the consequences of cellular turnover in adult tissues on the persistence of episomal AAV are yet to be defined. Similarly, the long-term fate of AAV gene therapy in infants and children whose tissues are actively undergoing growth and expansion will become clear over time.
Secondly, while gene therapy is ideally suited for the treatment of autosomal and X-linked recessive diseases, where expression of a transgene that encodes a missing protein is sufficient to overcome the deficiency associated with the disease, treatment of autosomal dominant disorders is more challenging. For example, treatment with an AAV vector encoding the clotting factor VIII overcomes the deficiency in the X-linked recessive disease, hemophilia A. However, for autosomal dominant disorders, expression of mutant proteins may have a dominant-negative effect or cause toxicity, such as in Huntington’s disease. In such cases, strategies for allele-specific silencing of expression are necessitated but challenging to achieve with gene therapy.
Lastly, while gene therapy in its current iteration readily addresses diseases where the expression level is not critical, it is much harder to use to treat conditions where the tolerated window of transgene expression is narrow. In these cases, often, either too little or too much transgene expression leads to toxicity. An example of this is Rett syndrome, caused by mutations in the MECP2 gene (Amir et al., 1999). MECP2 expression is highly regulated in vivo, and either too much or too little expression leads to toxicity (Montgomery et al., 2018; D'Mello, 2021). Most gene therapy vectors utilize heterologous promoters since the natural chromosomal promoters of most disease-associated genes exceed the coding capacity of AAV vectors. Additionally, regulatory sequences for many genes have yet to be identified. Even when identified, the addition of regulatory sequences to AAV gene therapy vectors may again be limited by the coding capacity of AAV. Thus, achieving physiologic regulation of transgene expression from a gene therapy vector is challenging. Hence, despite the tremendous achievement of AAV gene therapy in curing previously incurable genetic diseases, other approaches are necessary to address these recognized limitations.
One solution is to repair pathogenic mutations precisely and accurately at the level of the genome such that the correction would last for life and natural physiologic gene regulation would be maintained. This is enabled by genome editing technologies.
A key breakthrough in genome editing was the observation that editing could be enhanced by homologous recombination (HR) (Capecchi, 1989) in human cells and in vivo (Smithies et al., 1985; Thomas and Capecchi, 1987). However, the frequency of HR usually is very low, ∼1 event in 106–109 cells, rendering it challenging for therapeutic applications (Capecchi, 1989). A key advance in genome modification was the observation that the creation of double-stranded DNA breaks (DSBs) at targeted sites could enhance editing through the use of HR (Rudin et al., 1989; Plessis et al., 1992; Choulika et al., 1995; Bibikova et al., 2001; Bibikova et al., 2003). This spurred the development of editing platforms based upon programmable nucleases to induce targeted DSBs. In the presence of correction DNA templates bearing homology to the target genomic sequence, a fraction of DSBs were found to undergo HR-based editing (Vasileva et al., 2006; Wang et al., 2016; Smith et al., 2018), resulting in a recent proliferation of programmable nuclease-based editing platforms. These include meganucleases (Stoddard, 2005; Smith et al., 2006), zinc finger nuclease (ZFNs) (Porteus and Baltimore, 2003; Urnov et al., 2005; Wood et al., 2011), TALENs (Boch et al., 2009; Christian et al., 2010; Wood et al., 2011; Zhang et al., 2011) and CRISPR/Cas9 (Horvath and Barrangou, 2010; Gasiunas et al., 2012; Jinek et al., 2012; Cong et al., 2013; Mali et al., 2013b). The CRISPR/Cas9 platform has recently been further refined to replace DSBs with single-stranded nicks resulting in the base editing and prime editing platforms (Anzalone et al., 2019; Matsoukas, 2020). However, it was shown that in addition to the high-fidelity homology dependent repair (HDR), DSBs could also be repaired by the Non-Homologous End Joining (NHEJ) pathway (Chapman et al., 2012). The NHEJ pathway, which is thought to have developed as a stop-gap repair mechanism to patch DNA breaks in the mid-cell cycle, is error-prone as it is carried out without a homologous repair template. DNA repair by NHEJ is often associated with insertion/deletion (indels) mutations. While HR, which primarily occurs during mitosis and utilizes sister chromatids as repair templates, is error-free and precise and geared towards maintaining genomic integrity. These DNA repair pathways are being leveraged for gene editing to enable the versatile and accurate correction of pathogenic mutations. The major genome editing platforms based upon programmable nucleases are briefly reviewed below.
Meganucleases are sequence-specific endonucleases that recognize 15–30 bp cleavage sites. Meganucleases such as homing endonucleases I-SceI and I-CreI (Hoess et al., 1982; Monteilhet et al., 1990; Hasan et al., 1994; Stoddard, 2005; Smith et al., 2006) have relatively high specificity and precision. Because of their relatively long recognition sequence, target sites are rare in any genome. Strategies have been developed to retarget them to novel sequences, thus expanding their use for genome editing (Smith et al., 2006; Silva et al., 2011). However, tailoring meganucleases is laborious and requires significant protein engineering (Smith et al., 2006). In addition, the lack of defined DNA binding and cleavage domains further hampers protein engineering efforts, rendering large-scale use challenging.
ZFNs are artificially engineered fusion proteins that consist of zinc finger DNA-binding domains and a DNA-cleavage domain (Bibikova et al., 2003; Porteus and Baltimore, 2003; Porteus and Carroll, 2005; Urnov et al., 2005; Miller et al., 2007; Wood et al., 2011). Each zinc finger is relatively small at about 30 amino acids and can bind a three base pair DNA sequence. Tandem zinc fingers are required to create sequence specificity to target a given locus. Complex and expensive protein engineering is necessary to achieve sequence specificity. However, high levels of affinity and specificity of the system are difficult to achieve, resulting in a high frequency of off-target cleavage.
TALENs are also engineered fusion proteins comprised of tandem arrays of 10–30 DNA recognition repeats and the FokI endonuclease (Boch et al., 2009; Moscou and Bogdanove, 2009; Christian et al., 2010; Cermak et al., 2011; Mahfouz et al., 2011; Miller et al., 2011; Wood et al., 2011; Zhang et al., 2011; Reyon et al., 2012). The repeats are derived from transcription activator-like effectors (TALEs) that contain 33–35 amino acids with two adjacent amino acids termed the repeat-variable di-residue (RVD). The RVDs confer binding specificity to one of the four DNA base pairs. Thus, engineering DNA binding domains to specific sequences can be achieved by stitching different repeats together. Although the process is straightforward, the construction of each TALEN array is time-consuming and labor-intensive, limiting their use in high-throughput applications.
The CRISPR/Cas9 system is one of the later additions to the toolbox of programmable nuclease-based genome editing (Haurwitz et al., 2010; Deltcheva et al., 2011; Gasiunas et al., 2012; Jinek et al., 2012; Cong et al., 2013; Mali et al., 2013b). It is built upon the adaptive immunity of bacteria and archaea, whereby the foreign DNA sequences (protospacers) of bacteriophage and plasmids are integrated into copies of repeat sequences named CRISPR (Jansen et al., 2002), which serve as marks of memory of previous attackers and act as a surveillance defense mechanism. Short guide RNAs transcribed from the protospacers form complexes with Cas nucleases to search and destroy incoming matching foreign DNA (Haft et al., 2005; Makarova et al., 2006). The more straightforward class II CRISPR system that consists of a single guide RNA (sgRNA) and Cas9 protein has been widely adapted for gene manipulations. The sgRNA consists of a scaffold sequence for binding to Cas9 and a 20 base pair spacer for sequence recognition that guides Cas9 to the cleavage target (Sapranauskas et al., 2011; Chylinski et al., 2013). The Cas9 nuclease recognizes a protospacer-adjacent motif (PAM) (Mojica et al., 2009) of 3′-NGG and cleaves 3-4 base pairs upstream of the PAM sequence. Since NGG motifs are abundant in most genomes, the CRISPR/Cas9 system offers flexible targeted cleavages at numerous genomic loci. By simply changing the protospacer sequence, any sequence in the genome could be potentially targeted and manipulated, making it ideal for high-throughput applications. These properties also allow easy adaptation for multiplexing by simply introducing multiple guide RNAs into the same cell to achieve multiple manipulations simultaneously (Cong et al., 2013). In addition, engineered Cas9 and the discovery of other Cas proteins that recognize different PAM sites further expand the flexibility of this platform. To avoid detrimental mutations associated with DSBs, new functions other than DNA cleavage have been engineered into Cas9 to mediate processes such as nicking (Mali et al., 2013a; Ran et al., 2013), gene activation (Cheng et al., 2013; Perez-Pinera et al., 2013; Qi et al., 2013; Konermann et al., 2015), gene suppression, (Bikard et al., 2013) directed base editing, and prime editing (Komor et al., 2016; Nishida et al., 2016). The latter two strategies that allow genome editing without donor DNA are discussed further below.
Directed base editing and prime editing allow genome editing without the requirement for DSBs or a donor template. Base editing utilizes a nuclease defective Cas9 mutant (dCas9) (Jinek et al., 2012) fused to a cytidine deaminase. The fusion protein then targets a genomic locus where the cytidine deaminase converts any C to U within a five base-pair window, thereby directly mediating a C to T (G to A on opposite strand) conversion (Komor et al., 2016). To broaden the scope of this platform, an engineered RNA deaminase was developed to convert the A-T base pair to a G-C base pair (Gaudelli et al., 2017). To further improve versatility and specificity, extensive engineering was also carried out on dCas9 to recognize different PAM sites and on the cytidine deaminase to narrow the targeting window from approximately 5 to 1-2 base pairs (Kim et al., 2017). However, the direct base editing strategy is limited to converting single bases but not for correction of insertion or deletion mutants.
The prime editing platform may address this limitation of base editing (Anzalone et al., 2019; Anzalone et al., 2020). The key components of prime editing are the prime editing guide RNA (pegRNA) and the Cas9 nickase-reverse transcriptase (RT) fusion protein. pegRNAs contain extra sequences at the 3′ end of the sgRNA that act as a priming site for the nicked DNA and serve as a template for reverse transcription. Target search is mediated by the spacer sequence of the pegRNA. Upon binding to the target strand, the displaced DNA strand, R-loop DNA, is nicked by Cas9 nickase (Ran et al., 2013). The 3′ end of the nicked DNA can then anneal to the prime RNA sequence and extend to copy the correction sequences by RT. The corrected ssDNA flap can then anneal back to the target strand, replace the 5′ flap and result in a heteroduplex with one copy of the corrected sequence. Further nicking of the unedited strand can result in the correction of both strands. Both direct editing and prime editing avoid double-strand DNA breaks, therefore, dramatically lowering DSB-induced on-target indels and, more critically, extensive scale gene rearrangement. However, the high frequency of off-target recognition remains the primary concern for therapeutic applications (Pattanayak et al., 2013; Tsai et al., 2015). Among the 20 base pair spacer sequences that confer specificity, only the 8–10 base seed sequence (Semenova et al., 2011; Wiedenheft et al., 2011) at the 3′ end is most stringent while the rest allows a certain degree of mismatch, resulting in off-target recognition.
All programmable nuclease-based platforms require the generation of DSBs or single-stranded nicks designed to enhance HR. However, each platform has certain limitations. Nuclease-mediated genomic DSBs undergo repair mostly via NHEJ, which occurs more frequently than HDR. Since NHEJ is an error-prone repair pathway, indel mutations are often observed at the target sites. These mutations span 1 to 10 base pairs and may result in frameshifts, leading to nonfunctional or mutated proteins. Indel mutations can be especially deleterious in essential genes or genes involved in tumor suppression. Evaluation of the mutation spectra associated with nuclease-based editing platforms shows that deletions are common with the TALENs platform while larger deletions are associated with ZFNs and CRISPR/Cas9 (Gabriel et al., 2011; Pattanayak et al., 2011). Occasional substitutions, inversions, duplications, and longer insertions or deletions of up to several hundred bases have also been observed, albeit at low frequencies (Fu et al., 2013; Kuscu et al., 2014; Tsai et al., 2015). Even in the presence of donor constructs bearing homology to the target sites, repair by NHEJ was observed more frequently than the higher fidelity HDR, likely because the NHEJ pathway was dominant and active at all stages of the cell cycle (Mukherjee et al., 2019). However, HDR could exhibit higher frequency than NHEJ in the presence of very high copy number of donor DNA or when NHEJ pathway genes were knocked down (Certo et al., 2011). Multiple efforts are underway to increase the frequency of HDR after DSB creation, including the use of small-molecule inhibitors of the NHEJ pathway.
Off-target cleavage is likely the most significant concern for programmable nuclease-based editing platforms since the DNA recognition specificity of the nucleases is not precise. This poses a considerable challenge due to the risk of mutagenesis throughout the genome. Promiscuous cutting by programmable nucleases at off-target sites in the genome has been extensively documented (Gabriel et al., 2011; Pattanayak et al., 2011; Fu et al., 2013; Pattanayak et al., 2013; Duan et al., 2014; Kuscu et al., 2014; Lin et al., 2014; Wang X. et al., 2015). DSBs created by the off-target action of programmable nucleases are also repaired via the error-prone NHEJ pathway, adding to the genomic mutational burden. Excessive generation of off-target DSBs has been shown to be cytotoxic (Porteus, 2006; Miller et al., 2007; Szczepek et al., 2007; Guo et al., 2010; Doyon et al., 2011). Improvements by in silico design have led to a reduction but not prevention of off-target cleavage. Significant off-target cleavage has been reported in these platforms, including CRISPR/Cas9. Off-target cleavage by Cas9 with up to 5 base pair mismatches between the target DNA and the guide RNA has been documented. Previous studies have shown that the different structures of the guide RNA can influence the cleavage of on-target and off-target sites, thereby raising concerns over their use.
Most nuclease-based editing platforms comprise of multiple components. The precise delivery of each element to the nuclei of target cells is critical for genome editing. While in vitro or ex vivo applications can be achieved by viral vector-mediated delivery or direct transfection of plasmid DNA or synthetic mRNA, in vivo delivery to target tissues remains challenging. Recently, AAVs have become the vector of choice for delivering programmable nucleases and the donor template for HDR or both (Wang et al., 2015; Sather et al., 2015; Dever et al., 2016; Wang et al., 2016). However, the limited coding capacity (∼4.8 kb) of AAV vectors remains a hurdle for delivering nuclease-based editing components.
All AAVs possess unique inherent genome editing properties that set them apart from all other editing platforms. They utilize a high-fidelity repair pathway that does not introduce additional mutations during the editing process and do not require the use of exogenous nucleases to create DSBs before editing. AAVs edit the cellular genome independently of the cell cycle stage and equally well in vitro and in vivo. Here, we will focus on the intrinsic and distinctive genome editing properties of AAV vectors and the potential mechanisms by which they function. It has long been known that AAV vectors mediate targeted insertion in the cellular genome (Russell and Hirata, 1998; Inoue et al., 1999; Miller et al., 2003; Porteus et al., 2003; Vasileva et al., 2006; Khan et al., 2011; Barzel et al., 2015; Smith et al., 2018). Early studies showed that gene targeting efficiency with AAV2 was approximately 2-3 logs higher than other methods (Russell and Hirata, 1998). Gene targeting in human cell lines was shown to occur at frequencies of approximately 0.1–1% (Russell and Hirata, 1998; Porteus et al., 2003). AAV gene targeting has been demonstrated at multiple genomic locations, including genes such as hypoxanthine phosphoribosyltransferase (HPRT), Type I collagen (COL1A1) (Hirata et al., 2002; Chamberlain et al., 2008), human and murine IL2RG (Hiramoto et al., 2018; Smith et al., 2018), the AAVS1 safe harbor locus (Smith et al., 2018), human phenylalanine hydroxylase (PAH) (Chen et al., 2020), and the murine ROSA26 locus (Miller et al., 2006; Smith et al., 2018). Recombinant AAV was used to site-specifically target therapeutic transgenes to the 3′ untranslated region of the albumin gene, such that expression was driven by the albumin promoter. Transgenes were coexpressed with a short hairpin RNA. Coupling this approach with targeted expression from the albumin promoter led to supraphysiologic levels of Factor IX expression (Barzel et al., 2015; Nygaard et al., 2016). Using a similar approach, expression of methylmalonyl-CoA mutase from the albumin promoter was shown to have efficacy in a murine model of methylmalonic acidemia (Chandler et al., 2021). In addition to targeted genomic insertions at specified locations, AAV vectors have been shown to mediate nucleotide substitutions (Smith et al., 2018) and small deletions.
While intracellular AAV vector genomes predominantly survive as episomes long-term (Figure 2A), in some instances, chromosomal integration of AAVs has been observed. There are two specific forms of AAV integration that are distinct from AAV-mediated genome editing. To distinguish between AAV integration and AAV editing, we will briefly review the forms of AAV integration.
In the absence of helper virus infection, preferential integration of wild-type (WT) AAV genomes was shown to occur at a specific genomic site on chromosome 19q13.2-13.4qtr, also known as the AAVS1 locus (Figure 2B) (Kotin and Berns, 1989; Kotin et al., 1990; Kotin et al., 1991; Samulski et al., 1991; Kotin et al., 1992). Thirty to seventy percent of integrated WT AAV is found at the AAVS1 locus. Site-specific integration by WT AAV requires the presence of the AAV-encoded Rep 68/78 proteins which bind to both the Rep binding site (RBS) element on AAVS1 and on the AAV genome (Linden et al., 1996; Hamilton et al., 2004). Following the formation of a double-stranded intermediate of the WT AAV genome, the AAV p5 promoter is activated to express the Rep proteins. Rep68/78 proteins then mediate complex formation between the AAV genome and AAVS1 locus and generate a nick at the terminal resolution site (TRS) in AAVS1, resulting in the integration of the AAV genome at the AAVS1 locus via a strand switch mechanism. The p5 Integration efficiency element (p5IEE) in the p5 promoter was shown to be required in cis to mediate integration (Philpott et al., 2002a; Philpott et al., 2002b). This element has been reported to also act as an origin of replication (Wang and Srivastava, 1997) which might recruit the replication machinery for strand switching. However, little is known about the host cell factors that are involved in Rep-mediated site-specific integration. Since site-specific integration by AAV at the AAVS1 locus is not associated with pathogenicity or toxicity, it is widely used as a safe harbor locus to insert reporter or therapeutic sequences (Dreyer et al., 2015; Stellon et al., 2021).
In addition to site-specific integration by Rep-containing AAV, Rep-free AAV vectors have been found to undergo random integration at very low frequencies (Figure 2C) (McCarty et al., 2004). Random integration of recombinant AAV in the host genome has been shown to involve the ITRs and potentially regions of microhomology with the genome, possibly indicating a role for the hairpin structure of ITR in NHEJ-mediated integration (Rutledge and Russell, 1997; Yang et al., 1997; Nakai et al., 1999). In these integration events, complete, partial, or rearranged ITR sequences are found at the junctions between chromosomal sequences and AAV vector genomes. Induction of DSBs was shown to result in a higher frequency of integrations, confirming the hypothesis that random integration of AAV is mediated via NHEJ in the presence of DSBs. However, in the absence of DSBs, AAV vectors integrate at very low frequencies, possibly at sites of spontaneous DNA breaks (Miller et al., 2004). AAV vectors have been shown to integrate at regions of genomic instability, for instance, satellite DNA sequences, palindromic sequences, rRNA encoding DNA repeats, and CpG islands. These regions may be more prone to spontaneous breaks resulting in deletions, insertions, and translocations (Nakai et al., 2003; Miller et al., 2005; Nakai et al., 2005; Inagaki et al., 2007; Nguyen et al., 2021). Random integration of AAV vectors into the cellular genome is a rare event that has not been associated with pathology in clinical trials to date, and AAV vectors are considered safe (Gaudet et al., 2013; Nathwani et al., 2014).
Two studies have extensively evaluated the sequence of edited chromosomal loci after AAV-mediated HR (Smith et al., 2018; Chen et al., 2020). Evaluation of numerous edited chromosomal sequences by both Sanger and NGS sequencing showed a clear absence of ITR insertions in both cases, indicating that ITRs are not inserted during AAV HR. The substrate for AAV-mediated HR is the single stranded AAV genome. Double-stranded AAV genomes do not undergo HR (Vasileva et al., 2006). In addition, long regions of homology are required for successful alignment of the editing vector and the target chromosomal locus. The presence of homology arms is critical for alignment with the chromosomal genomic sequence followed by crossover and resolution of the crossover junction within the homology regions at the 5′ and 3′ ends. Since the crossovers occur within the homology arms, the ITRs are excluded and not inserted into the chromosomal locus following AAV-mediated HR.
However, in the absence of homology arms, any double-stranded or self-complementary AAV DNA in the nuclei appear to be dropped into sites of the DSB regardless of whether they are created by nucleases associated with editing platforms or created by environmental conditions such as irradiation and chemicals. For random integration of AAV and in most nuclease-based platforms, the insertion of double-stranded AAV genomes into the sites of chromosomal DSBs results in the insertion of ITRs, often in rearranged forms.
AAV vectors specifically designed for genome editing in the absence of programmable nucleases function in a distinctly different manner from either WT AAV or Rep-free AAV gene transfer vectors. Infection of cells with single-stranded AAVs initiates a cellular DNA damage response (DDR), although no accompanying actual cellular DNA damage has been identified (Jurvansuu et al., 2005; Choi et al., 2006; Jurvansuu et al., 2007; Cervelli et al., 2008; Ingemarsdotter et al., 2010; Hirsch, 2015). The initiation of DDR in target cells likely provides an ideal environment for genome editing by AAV. It is possible that the single-stranded AAV genome with the structured ITRs resembles a stalled replication fork and therefore elicits a DDR in infected cells. AAV infection has been shown to cause cell cycle arrest, primarily in the G1 phase of the cell cycle associated with activation of checkpoint proteins ATR, Chk1, and H2AX (Jurvansuu et al., 2007; Fragkos et al., 2009). In cells bearing mutations of certain DNA repair proteins or p53, this AAV-induced DDR leads to catastrophic mitosis and cell death. In normal cells, however, no deleterious effects have been observed. The mechanism by which the cell cycle arrest is relieved in normal cells remains unknown. DDR activation in AAV infected cells likely contributes to the onset of HR if other conditions are met.
Increased targeted integration by AAVs was originally attributed to the increased availability of vector genomes in the nucleus, increased stability of AAV vector genomes due to the structured ITRs, and the potential of single-stranded genomes to participate in HR (Figure 2D). The hairpin structures formed by ITRs at the ends of the AAV genome likely prevent degradation of the single-stranded AAV genomes by nucleases, increasing their stability and availability for genome targeting in the presence of flanking homology arms. Similar findings have been reported in yeast and fungal species where it was observed that the linear plasmids with telomeres or terminal palindromic repeats were more stable in episomal configurations compared to those without telomeric ends. These were additionally found to be capable of integration if they contained regions of homology to the genomic sequence (Bijlani et al., 2019). It is likely that the structure of ITR and the junction with the single-stranded AAV genome is identified as stalled replication fork and initiates DDR resulting in the recruitment of DNA repair proteins (Shechter et al., 2004; Smith et al., 2018). The efficiency of HR-based gene targeting by AAVs may depend upon the design of the editing vectors and transduction conditions. While not absolute, improved editing efficiency was associated with an increase in multiplicity of infection (MOI), the use of longer homology arms, and central positioning of the insert sequences. However, in some studies, the use of asymmetric homology arms appeared to be more efficient in editing certain genes (Russell and Hirata, 1998; Hirata and Russell, 2000; Gaj et al., 2016; Smith et al., 2018). In some, but not all cases, it was found that editing increased over time, possibly due to delayed transduction of target cells or slow nuclear accumulation or uncoating of AAV, possibly correlated with cell cycle progression (Russell et al., 1994) and AAV capsid serotype (Smith et al., 2018). It has been reported that transcriptionally active sites and targeting loci transcribed in the opposite direction of the gene-targeting event may be easier to edit (Deyle et al., 2014; Spector et al., 2021). Genomic sequence and potential secondary structure of the target sites have also been shown to play important roles in guiding editing efficiencies. In general, however, it has been difficult to define a set of universal rules that would uniformly apply to AAV editing at all genomic loci. As of now, optimal editing constructs must be empirically defined for different genomic target loci. A broad range of genome modifications have been achieved using AAV-based genome engineering. These include introduction of nucleotide substitutions, deletions, and insertions at multiple loci in the genome.
Notably, genome editing by AAV does not require the addition of exogenous nucleases and likely utilizes a highly precise and probably non-canonical HR pathway (Figures 1 and 2D). However, there are specific critical requirements for AAV-based editing vectors. AAV editing vectors must include: 1) the presence of ITR sequences, 2) a single-stranded AAV vector genome, and 3) the presence of homology arms complementary to the target genomic sequences. Self-complementary and dimeric AAV genomes do not edit, and thus, the single-stranded configuration of AAV vectors is essential for gene editing (Hirata and Russell, 2000).
We have recently shown that AAV editing vectors belonging to clade F mediate exclusively HR-based, highly accurate and seamless, and efficient genome editing in human cells and mice without the requirement for exogenous nucleases (Figure 2D) (Smith et al., 2018). These vectors include a family of novel, naturally occurring AAVs isolated from healthy human hematopoietic stem cells (HSCs), termed AAVHSC (Smith et al., 2014; Chatterjee et al., 2020). AAVHSC editing was found to be precise and efficient, with no on-target insertion/deletion mutations and no evidence of genomic scarring or incorporation of residual viral sequences, including the ITRs. A variety of human cell types, including human CD34+ hematopoietic stem cells, liver sections, hepatic sinusoidal endothelial cells, myoblasts, and immortalized human B lymphoblastoid cell lines, were successfully edited using AAVHSCs. We reported a median editing efficiency of 24.2% for clade F AAVs, with some serotypes showing up to 50% efficiency. In comparison, the editing efficiency of clade B AAV was 2.12%, and clade E AAV was 1.7%. The reasons underlying the increased efficiency of certain AAV serotypes remain unknown.
Interestingly, we observed efficient editing of post-mitotic tissues in vivo. Sequence analyses again revealed high on-target precision with no indels or ITR insertions observed. In vivo targeted insertion of the promoterless luciferase open reading frame into intron one of the Rosa26 locus resulted in long-term luciferase expression, indicating that AAVHSCs were capable of efficient and stable in vivo genome editing. In addition to targeted insertion, AAVHSC editing vectors also mediated nucleotide substitutions with high accuracy. In another study, an AAVHSC15 editing vector was used to edit the PAH gene on chromosome 12, mutations of which result in phenylketonuria. The editing vector targeted the PAH cDNA to intron one of the PAH gene. In vivo editing in a chimeric human liver xenograft model resulted in editing 6% of human hepatocytes. No on-target mutations were found following NGS sequencing spanning both homology arms (Chen et al., 2020). Thus, AAVHSCs represent a promising platform for seamless, high-efficiency gene editing and corroborate the use of AAVs as a genome engineering tool.
Although the mechanism of AAV gene targeting has yet to be definitively delineated, we have recently shown that AAV editing requires homology to targeted chromosomal sequence, single-stranded AAV editing genomes, and the presence of BRCA2 in target cells (Smith et al., 2018) (Figure 1). In the absence of BRCA2, no editing was observed by any AAV serotype. Based upon the requirements for homology arms and BRCA2, our results strongly suggest that AAVs utilize the HR pathway to carry out editing (Vasileva and Jessberger, 2005; Vasileva et al., 2006; Smith et al., 2018). AAV is the only currently used genome editing platform that does not require the prior creation of double-stranded DNA breaks by exogenous nucleases and appears to exclusively use the high-fidelity HR pathway to mediate genome editing.
Only single-stranded AAV genomes can mediate editing, and self-complementary or double-stranded AAV genomes do not function in this capacity (Hirata and Russell, 2000). This requirement for single-stranded AAV genomes most likely reflects their role in annealing to homologous genomic sequences. We examined the genome forms of the editing vectors in the murine liver 6 months after intravenous injection of editing vectors (Smith et al., 2018). Notably, we did not detect either free monomeric or circular concatemeric vector genomes in the liver, suggesting that the editing vector genomes may not have initiated second-strand synthesis or did not form double-stranded concatamers commonly observed following AAV transduction (Smith et al., 2018). Upon nuclear entry and uncoating, single-stranded AAV genomes undergo second-strand synthesis and form circular concatamers (Figure 2A). It is possible that a prolonged single-stranded phase of editing vector genomes may enhance editing efficiencies. Thus, AAV serotypes that promote extended survival of single-stranded genomes may be more proficient at mediating HR.
Based on our observation that editing by every AAV serotype was abolished in the absence of BRCA2 (Smith et al., 2018), we concluded that BRCA2 plays a critical role in AAV editing and that there is no redundancy provided by any other alternate repair protein. We hypothesized that BRCA2 recognizes the double-strand to single strand junction at the ends of the ITRs, then recruits RAD51, displacing RPA from the coated single-stranded AAV genome and forms the nucleofilament necessary to initiate HR (Figure 1). This likely leads to the assembly of the HR complex, including BRCA1, PALB2, and Rad51 (Figure 1).
We previously reported that a reduction in editing efficiencies was observed with all AAV serotypes in cell lines bearing homozygous or compound heterozygous mutations in BLM helicase, FANCA, FANCC, FANCD2, and FANCF, indicating their possible involvement in AAV-mediated HR (Smith et al., 2018). However, another study showed that knockout of FANCM when combined with a knockout of BLM or RMI1 increased AAV HR (de Alencastro et al., 2021). Whether these differences in the HR-related outcomes were a result of the interaction of these proteins or were unique to the limited number of clones studied is unclear. ATR may also play a role in DNA repair upon recognition of the single-stranded AAV genome structure resembling a stalled replication fork. ATR in association with ATR interacting protein (ATRIP) is known to signal cell cycle checkpoint activation in the presence of stalled replication forks and single-stranded DNA gaps (Shechter et al., 2004). It is likely that some redundancy in the proteins involved accounted for the persistence of some editing activity in these cells. This is supported by reports of ATR colocalizing with the AAV genome in nuclear foci in the absence of ATM and NBS1 (Jurvansuu et al., 2005). Redundancy is known to exist for the proteins of the HR complex and is thought to be evolutionarily important for the maintenance of genomic integrity.
We observed little or no change in HR in cells bearing homozygous or compound heterozygous mutations in FANCB, ATM, NBS1, ERCC4/XPF, and RAG1. This was in contrast to the complete abolition of AAV HR in cell lines with compound heterozygous mutations in BRCA2 and a reduction of HR in cells with mutations in BLM helicase, FANCA, FANCC, FANCD2, and FANCF (Smith et al., 2018). Evaluation of the roles of these proteins in AAV editing will further clarify the mechanisms involved in AAV editing and any built-in redundancies in the involved pathways.
In addition to HR, AAVs may additionally use a non-canonical HR pathway. It has been shown that silencing of the NHEJ protein DNA-PK did not affect gene targeting efficiency; however, silencing of HR proteins, RAD54L or RAD54B, and partial silencing of the RAD51 paralogue XRCC3 reduced or abolished AAV gene targeting (Vasileva et al., 2006). RAD54L and RAD54B belong to the RAD52 epistasis group, and RAD54L facilitates strand exchange by RAD51 (Kanaar et al., 1996; Hiramoto et al., 1999; Sigurdsson et al., 2002). RAD51 replaces RPA on single-stranded DNA, forming a nucleoprotein filament (Jensen et al., 2010). XRCC3 binds to ssDNA along with RAD51C and slows down the replication fork progression upon DNA damage (Masson et al., 2001; Henry-Mowatt et al., 2003). Recently XRCC3 was shown to bind to DNA directly, promoting nucleoprotein formation by RAD51 (Forget et al., 2004). Thus, the involvement of HR proteins RAD54, RAD51, and BRCA2 suggests that the single-stranded AAV genome structure may indeed be recognized as a stalled replication fork, which sends a signal to BRCA2 (Figure 1). BRCA2 then recruits RAD51 and its paralogues to the site and stimulates RAD51-mediated nucleoprotein filament formation resulting in HR-mediated gene targeting.
The MRN (MRE11, RAD50, NBS1) complex associates with AAV vector genomes and limits the conversion of the single-stranded AAV genome to the double-stranded form (Sanlioglu et al., 2000; Cervelli et al., 2008). For AAV gene transfer vectors, MRN and ATM reduce transduction efficiency by preventing second strand synthesis resulting in reduced transgene transcription. However, for gene editing, recognition of the structured AAV genome and the subsequent binding by MRN may stimulate HR by inducing ATM in the presence of homologous sequences. This further underscores the lack of editing by self-complementary AAV genomes as single and double-stranded junctions are not available for assembly of the HR complex. The association of RAD52 and the MRN complex with AAV ITRs has also been reported (Zentilin et al., 2001; Schwartz et al., 2007). It was hypothesized that RAD52 might be involved in the replication of AAV via break-induced replication (BIR), where replication proceeds unidirectionally, resulting in a formation of double-stranded intermediate (Hendrickson, 2020). It is possible that in the case of AAV vectors, the absence of AAV Rep proteins and limited double-strand synthesis by the MRN complex and binding to Rad52/MRN complex may further protect against nuclease-mediated degradation of the single-stranded AAV editing genome. Rad52 also mediates homology search (Rothenberg et al., 2008; Jalan et al., 2019). Thus, upon binding to the AAV editing genome, it may be instrumental in guiding the editing genome to the homologous region in the cellular genome.
The HR mechanisms discussed above illustrate how AAVs may mediate gene targeting in dividing cells where HR occurs mainly during the S/G2 phases of the cell cycle. However, recent observations suggest that AAV also mediates genome editing in non-dividing HSCs, cardiomyocytes, and adult post-mitotic tissues in vivo, indicating that non-canonical HR pathways may be involved (Smith et al., 2018; Kohama et al., 2020). While little is known about how AAVs mediate HR in non-dividing cells, some common requirements appear to be critical. First, a high MOI of single-stranded AAV is essential to trigger HR in vitro. Furthermore, targeted insertion into cardiomyocytes was shown to utilize the Fanconi anemia pathway involved in single-strand break repair. Interestingly, an independent study uncovered a non-canonical HR pathway that used single-stranded donor templates to repair artificially generated single-stranded nicks at the target locus (Davis and Maizels, 2014). They further showed that this mechanism exhibited a much higher frequency of gene editing than the canonical HR, which required DSBs (Davis and Maizels, 2011). Whether AAVs interact with this pathway to mediate gene editing remains to be elucidated. If indeed they do, one critical missing link is the mechanism required to generate single-stranded breaks (SSBs). Do AAVs utilize random SSBs that occur at the target sites, or is it possible that SSBs are actively created at target sites? The probability of random SSBs occurring at a 2–4 kb homologous target site in the haploid genome is approximately one in a million. This is consistent with spontaneous targeting frequencies observed for plasmid DNAs (Porteus et al., 2003) but does not account for the significantly higher targeting frequency observed with AAV vectors. Therefore, it is likely that AAV editing may result in SSB generation at the target site. However, neither the AAV genome nor the capsid proteins have intrinsic nicking activity. It is possible that the AAV editing machinery recruits cellular nickases using the homology arms as a guide to direct the cleavage to a targeted genomic locus. Mre11 is a cellular nuclease that interacts with the AAV genome (Cervelli et al., 2008). Mre11 of the MRN complex plays a critical role in DNA repair by binding to DNA break sites and initiating resection through its exonuclease and endonuclease activities. As described above, several studies have shown that MRE11 mainly hinders AAV transduction by inhibiting second strand synthesis (Schwartz et al., 2007; Lentz and Samulski, 2015). It will be interesting to determine whether high MOIs of AAV editing vectors can alter MRE11 functions that may lead to the nicking of genomic DNA at target sites. Another possibility is that at high MOIs, single-stranded AAV genomes may hybridize to the homologous genomic DNA and activate DDR resulting in noncanonical HR. While there is little direct evidence supporting this model, recent studies may provide some hints.
Another noncanonical HR pathway that is active in non-dividing cells is transcription-coupled HR (TC-HR), reported in yeast and mammalian cells (Keskin et al., 2014; Wei et al., 2015). Here, RNA-templated recombination repair mechanism was identified in the G0/G1 cell cycle phase, mediated by the Cockayne syndrome B protein (CSB), RPA, RAD51, RAD51C, and RAD52 at the site of DNA breaks (Wei et al., 2015). This process was shown to be initiated by CSB, which further recruited the HR proteins to the site. TC-HR also contributed to DNA repair in post-mitotic neurons (Welty et al., 2018), with the recruitment of RAD52 to the site of DNA breaks. A gene targeting study conducted with AAVs concluded that transcriptionally active sites exhibited improved AAV-mediated HR (Spector et al., 2021). A study conducted in yeast reported the formation of Rad51 foci on DSBs in the G1 phase of the cell cycle in the absence of Rad52 foci, which is required for Rad51 recruitment (Smith et al., 2019). It was hypothesized that Rad52 may be present in low abundance, albeit at sufficient levels to recruit Rad51, but not enough to form Rad52 foci. Other factors may regulate HR during the G1 phase of the cell cycle. For instance, some histone modifications associated with transcription are also known to influence the recruitment of DNA repair proteins. For example, H3K36me3, a histone marker of transcription elongation, is known to recruit CtIP and RAD51 at transcriptionally active sites (Aymard et al., 2014), and TIP60-dependent H4 acetylation recruits BRCA1 to the sites of DSBs (Tang et al., 2013), which suggests possible regulation of HR during transcription. Histone marks H4K20me1 and H4K20me2 present in euchromatin regions that undergo active transcription recruit repair factor p53 binding protein1 (53BP1) to DSBs that plays a role in NHEJ (Botuyan et al., 2006). Thus, there is a balance between the HR and NHEJ repair pathways, and one potential regulator of the choice of the pathway appears to be histone modifications during transcription. This mechanism of TC-HR may also explain AAV-mediated HR in non-dividing cells where spontaneous breaks in the genome may occur during transcription rendering the genes accessible for editing.
It is well established that highly expressed genes commonly associate with R-loop formation where the RNA transcripts hybridize to the transcribed DNA strand resulting in the non-transcribed strand being exposed as single-stranded DNA (R-loop). This exposed section of single-stranded DNA is vulnerable to lesion formation that can result in DNA breaks (Crossley et al., 2019; Rinaldi et al., 2020). Since the R-loop structures formed during transcription are prone to damage (Aguilera and Garcia-Muse, 2012), it is possible that the damaged single-stranded genomic DNA in the R-loop structures hybridize to the single-stranded AAV genome and repair by HR ensues using AAV as the template. It has been shown that RAD52 binds to R-loop structures (Welty et al., 2018), which functions in homology search for repair. In such cases, the presence of a homologous sequence in the AAV editing vector genome in the vicinity of actively transcribing DNA could mediate targeted genome editing. Most lesions on one strand of DNA in the R-loop can be repaired by the classical nucleotide excision repair pathway (NER), which is not strictly dependent on cell division (Sollier et al., 2014; Marnef et al., 2017). Recently, an alternate repair mechanism was described that uses RNA transcripts instead of sister chromatids as repair templates and is therefore independent of cell division (Keskin et al., 2014; Meers et al., 2016; Welty et al., 2018; Ouyang et al., 2021). Both pathways are associated with transcription and are independent of the cell cycle. It would be interesting to investigate whether AAV vectors utilize these pathways to mediate gene editing.
Although the path from the bench to the clinic is long and arduous, clinical trials to test the safety and efficacy of therapeutic genome editing have been initiated. Therapeutic gene editing has focused upon genetic diseases and immuno-oncology (Mullard, 2020). Here we will focus primarily on gene editing for genetic diseases. Among the first editing platforms to enter the clinic were zinc finger nucleases and CRISPR/Cas9. ZFN and CRISPR/Cas9 have been used ex vivo to downregulate expression of CCR5, a receptor for HIV-1. Patients were infused with ex vivo edited CD4 cells. Ex vivo editing followed by infusion was found to be safe, although the level of edited cells was low in both studies (Tebas et al., 2014; Xu et al., 2019). One clinical trial tested the efficacy of in vivo systemic administration of ZFNs to treat mucopolysaccharidosis II (MPS II) (ClinicalTrials.gov Identifier: NCT03041324). AAV vectors encoding ZFNs targeted to the mutant iduronidase gene and a donor sequence encoding the wild type version were infused intravenously. Some modest benefits were noted, although they could not be definitively linked to the gene editing treatment due to limited sensitivity of the assays.
Hemoglobinopathies represent attractive targets for genome editing. Upregulation of fetal hemoglobin achieved by inactivation of BCL11A is an effective treatment for diseases caused by mutations in the beta globin gene. Two patients, one with transfusion dependent thalassemia and the other with sickle cell anemia were treated with autologous ex vivo CRISPR/Cas9 edited CD34+ hematopoietic stem cells. In both cases the BCL11A gene which suppresses fetal hemoglobin production was inactivated with CRISPR/Cas9. Following infusion of ex vivo edited cells, both patients expressed fetal hemoglobin. This resulted in the elimination of vaso-occlusive events and transfusion independence (Frangoul et al., 2021). Thus, while unanswered questions still remain, early results are encouraging.
In the first systemic in vivo trial of CRISPR/Cas9, lipid nanoparticles were used to deliver the mRNA for Cas9 and a guide RNA targeting transthyretin (TTR) (Gillmore et al., 2021). In this approach, the TTR gene was disabled by the introduction of a Cas9-mediated double stranded break in hepatocytes, resulting in a dose-dependent reduction in TTR expression. However, this approach targeted both the mutant and wild type TTR alleles and notably would be ineffective at reducing previously accumulated amyloid deposits. Nevertheless, promising early indications of efficacy were noted.
A clinical trial of CRISPR/Cas9 is underway to test subretinal delivery of an AAV5 vector encoding two guide RNAs and Cas9 designed to delete a mutation in the CEP290 gene associated with Leber’s Congenital Amaurosis (LCA), a common cause of childhood blindness (ClinicalTrials.gov Identifier: NCT03872479), (Maeder et al., 2019). Some efficacy was noted at the mid-dose levels, although adverse events including retinal tears, hemorrhage and inflammation were observed. (https://editasmedicine.gcs-web.com/static-files/22b32d3d-e38f-4e90-899c-a2e701872745), (Ledford, 2020).
Preclinical and investigational new drug (IND)-enabling studies in AAV-mediated therapeutic gene editing are being conducted by several groups. Notably, one study recently received IND clearance from the US Food and Drug Administration (FDA) for the evaluation of AAV-mediated gene editing for the treatment of phenylketonuria (PKU), an inborn error of metabolism caused by mutations in the phenylalanine hydroxylase gene, resulting in an inability to metabolize phenylalanine (https://www.homologymedicines.com/news-story/homology-medicines-announces-worlds-first-gene-editing-clinical-trial-for-pku). This trial will evaluate HMI-103, an AAVHSC15-based gene editing vector to edit the PAH gene and restore expression in PKU patients. While this is the first AAV genome editing study to enter the clinic, it will likely be followed by others to treat a spectrum of genetic diseases.
Although clinical gene editing is still in early stages, pioneering trials have generally yielded promising results. Long-term follow up of ex vivo, systemic and locally administered gene editing strategies using the diverse available platforms and delivery systems will be important to assess the overall safety and therapeutic efficacy and importantly, the potential impact of nuclease-mediated off target DNA breaks and the creation of indel mutations.
AAV editing vectors represent an orthogonal editing platform that differs significantly from all current nuclease-based gene editing tools. It is the only genome editing platform that exclusively utilizes the high-fidelity cellular HR repair pathway and does not require exogenous nucleases for the a priori creation of DSBs. Both of these properties together lead to a highly precise editing outcome that preserves genomic integrity with no incorporation of indel mutations or viral ITR at the target site while also removing the possibility of genotoxicity associated with the creation of off-target DNA breaks and subsequent mutagenesis following error-prone repair.
All AAV serotypes possess the inherent ability to mediate accurate editing of cellular genomes in vitro and in vivo. AAV-based editing incorporates the key hallmarks of HR, including the absolute requirement for BRCA2 and homology between the repair template and the genomic target. Notably, however, unlike classical HR, AAV editing is not restricted to dividing cells, and post-mitotic cells in vivo undergo efficient editing by AAVs. While all AAV serotypes mediate genome editing, specific serotypes display increased editing efficiency, offering the prospect of effective in vivo editing. AAV editing vectors also serve as their own in vivo delivery vehicles that have been proven safe in numerous gene therapy trials. Some AAV vectors also possess the ability to cross the blood-brain barrier and transduce cells of the central nervous system (CNS), thus potentially enabling the much-needed ability to repair mutations in the CNS. Further insights into the mechanism of how AAV co-opts the cellular HR pathway to carry out high-fidelity editing and how some AAV serotypes amplify the efficiency of HR will be necessary for the further evolution of genome editing for the permanent correction of inherited and acquired genetic diseases.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This work was supported in part by a grant from the W. M. Keck Foundation and by grant DISC2-10524 from California Institute for Regenerative Medicine. Research reported in this publication included work performed in City of Hope Cores supported by the National Cancer Institute of the National Institutes of Health under award number P30CA033572.
SC is an advisor to and holds equity in Homology Medicines Inc. AS is currently an employee of Allergan Aesthetics (an AbbVie Company).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors wish to thank Supriya Deshpande for the figures and manuscript preparation and Lakshmi Bugga, Colin Cook, and Yuman Fong for their support and discussions. The authors wish to thank the W. M. Keck Foundation and the California Institute for Regenerative Medicine for their generous support. Based in Los Angeles, the W. M. Keck Foundation was established in 1954 by the late W. M. Keck, founder of the Superior Oil Company. The Foundation’s grant making is focused primarily on pioneering efforts in the areas of medical research and science and engineering. The Foundation also supports undergraduate education and maintains a Southern California Grant Program that provides support for the Los Angeles community, with a special emphasis on children and youth. For more information, visit www.wmkeck.org. Figures in this manuscript were created using BioRender.
Aguilera, A., and García-Muse, T. (2012). R Loops: from Transcription Byproducts to Threats to Genome Stability. Mol. Cel. 46, 115–124. doi:10.1016/j.molcel.2012.04.009
Amir, R. E., Van Den Veyver, I. B., Wan, M., Tran, C. Q., Francke, U., and Zoghbi, H. Y. (1999). Rett Syndrome Is Caused by Mutations in X-Linked MECP2, Encoding Methyl-CpG-Binding Protein 2. Nat. Genet. 23, 185–188. doi:10.1038/13810
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search-and-Replace Genome Editing without Double-Strand Breaks or Donor DNA. Nature 576, 149–157. doi:10.1038/s41586-019-1711-4
Anzalone, A. V., Koblan, L. W., and Liu, D. R. (2020). Genome Editing with CRISPR-Cas Nucleases, Base Editors, Transposases and Prime Editors. Nat. Biotechnol. 38, 824–844. doi:10.1038/s41587-020-0561-9
Aymard, F., Bugler, B., Schmidt, C. K., Guillou, E., Caron, P., Briois, S., et al. (2014). Transcriptionally Active Chromatin Recruits Homologous Recombination at DNA Double-Strand Breaks. Nat. Struct. Mol. Biol. 21, 366–374. doi:10.1038/nsmb.2796
Barzel, A., Paulk, N. K., Shi, Y., Huang, Y., Chu, K., Zhang, F., et al. (2015). Promoterless Gene Targeting without Nucleases Ameliorates Haemophilia B in Mice. Nature 517, 360–364. doi:10.1038/nature13864
Berns, K. I., and Linden, R. M. (1995). The Cryptic Life Style of Adenoassociated Virus. Bioessays 17, 237–245. doi:10.1002/bies.950170310
Bibikova, M., Carroll, D., Segal, D. J., Trautman, J. K., Smith, J., Kim, Y.-G., et al. (2001). Stimulation of Homologous Recombination through Targeted Cleavage by Chimeric Nucleases. Mol. Cel. Biol. 21, 289–297. doi:10.1128/MCB.21.1.289-297.2001
Bibikova, M., Beumer, K., Trautman, J. K., and Carroll, D. (2003). Enhancing Gene Targeting with Designed Zinc finger Nucleases. Science 300, 764. doi:10.1126/science.1079512
Bijlani, S., Thevandavakkam, M. A., Tsai, H.-J., and Berman, J. (2019). Autonomously Replicating Linear Plasmids that Facilitate the Analysis of Replication Origin Function in Candida Albicans. mSphere 4, e00103–19. doi:10.1128/mSphere.00103-19
Bikard, D., Jiang, W., Samai, P., Hochschild, A., Zhang, F., and Marraffini, L. A. (2013). Programmable Repression and Activation of Bacterial Gene Expression Using an Engineered CRISPR-Cas System. Nucleic Acids Res. 41, 7429–7437. doi:10.1093/nar/gkt520
Boch, J., Scholze, H., Schornack, S., Landgraf, A., Hahn, S., Kay, S., et al. (2009). Breaking the Code of DNA Binding Specificity of TAL-type III Effectors. Science 326, 1509–1512. doi:10.1126/science.1178811
Botuyan, M. V., Lee, J., Ward, I. M., Kim, J.-E., Thompson, J. R., Chen, J., et al. (2006). Structural Basis for the Methylation State-specific Recognition of Histone H4-K20 by 53BP1 and Crb2 in DNA Repair. Cell 127, 1361–1373. doi:10.1016/j.cell.2006.10.043
Brooks, P. J., Ottinger, E. A., Portero, D., Lomash, R. M., Alimardanov, A., Terse, P., et al. (2020). The Platform Vector Gene Therapies Project: Increasing the Efficiency of Adeno-Associated Virus Gene Therapy Clinical Trial Startup. Hum. Gene Ther. 31, 1034–1042. doi:10.1089/hum.2020.259
Capecchi, M. R. (1989). Altering the Genome by Homologous Recombination. Science 244, 1288–1292. doi:10.1126/science.2660260
Cermak, T., Doyle, E. L., Christian, M., Wang, L., Zhang, Y., Schmidt, C., et al. (2011). Efficient Design and Assembly of Custom TALEN and Other TAL Effector-Based Constructs for DNA Targeting. Nucleic Acids Res. 39, e82. doi:10.1093/nar/gkr218
Certo, M. T., Ryu, B. Y., Annis, J. E., Garibov, M., Jarjour, J., Rawlings, D. J., et al. (2011). Tracking Genome Engineering Outcome at Individual DNA Breakpoints. Nat. Methods 8, 671–676. doi:10.1038/nmeth.1648
Cervelli, T., Palacios, J. A., Zentilin, L., Mano, M., Schwartz, R. A., Weitzman, M. D., et al. (2008). Processing of Recombinant AAV Genomes Occurs in Specific Nuclear Structures that Overlap with Foci of DNA-Damage-Response Proteins. J. Cel. Sci. 121, 349–357. doi:10.1242/jcs.003632
Chamberlain, J. R., Deyle, D. R., Schwarze, U., Wang, P., Hirata, R. K., Li, Y., et al. (2008). Gene Targeting of Mutant COL1A2 Alleles in Mesenchymal Stem Cells from Individuals with Osteogenesis Imperfecta. Mol. Ther. 16, 187–193. doi:10.1038/sj.mt.6300339
Chandler, R. J., Venturoni, L. E., Liao, J., Hubbard, B. T., Schneller, J. L., Hoffmann, V., et al. (2021). Promoterless, Nuclease‐Free Genome Editing Confers a Growth Advantage for Corrected Hepatocytes in Mice with Methylmalonic Acidemia. Hepatology 73, 2223–2237. doi:10.1002/hep.31570
Chapman, J. R., Taylor, M. R. G., and Boulton, S. J. (2012). Playing the End Game: DNA Double-Strand Break Repair Pathway Choice. Mol. Cel. 47, 497–510. doi:10.1016/j.molcel.2012.07.029
Chatterjee, S., Sivanandam, V., and Wong, K. K.-M. (2020). Adeno-Associated Virus and Hematopoietic Stem Cells: The Potential of Adeno-Associated Virus Hematopoietic Stem Cells in Genetic Medicines. Hum. Gene Ther. 31, 542–552. doi:10.1089/hum.2020.049
Chen, H.-M., Resendes, R., Ghodssi, A., Sookiasian, D., Tian, M., Dollive, S., et al. (2020). Molecular Characterization of Precise In Vivo Targeted Gene Integration in Human Cells Using AAVHSC15. PLoS One 15, e0233373. doi:10.1371/journal.pone.0233373
Cheng, A. W., Wang, H., Yang, H., Shi, L., Katz, Y., Theunissen, T. W., et al. (2013). Multiplexed Activation of Endogenous Genes by CRISPR-On, an RNA-Guided Transcriptional Activator System. Cel. Res. 23, 1163–1171. doi:10.1038/cr.2013.122
Choi, V. W., Mccarty, D. M., and Samulski, R. J. (2006). Host Cell DNA Repair Pathways in Adeno-Associated Viral Genome Processing. J. Virol. 80, 10346–10356. doi:10.1128/JVI.00841-06
Choulika, A., Perrin, A., Dujon, B., and Nicolas, J. F. (1995). Induction of Homologous Recombination in Mammalian Chromosomes by Using the I-SceI System of Saccharomyces cerevisiae. Mol. Cel. Biol. 15, 1968–1973. doi:10.1128/MCB.15.4.1968
Christian, M., Cermak, T., Doyle, E. L., Schmidt, C., Zhang, F., Hummel, A., et al. (2010). Targeting DNA Double-Strand Breaks with TAL Effector Nucleases. Genetics 186, 757–761. doi:10.1534/genetics.110.120717
Chylinski, K., Le Rhun, A., and Charpentier, E. (2013). The tracrRNA and Cas9 Families of Type II CRISPR-Cas Immunity Systems. RNA Biol. 10, 726–737. doi:10.4161/rna.24321
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339, 819–823. doi:10.1126/science.1231143
Crossley, M. P., Bocek, M., and Cimprich, K. A. (2019). R-Loops as Cellular Regulators and Genomic Threats. Mol. Cel. 73, 398–411. doi:10.1016/j.molcel.2019.01.024
D’Mello, S. R. (2021). MECP2 and the Biology of MECP2 Duplication Syndrome. J. Neurochem. 159, 29–60. doi:10.1111/jnc.15331
Davis, L., and Maizels, N. (2011). DNA Nicks Promote Efficient and Safe Targeted Gene Correction. PLoS One 6, e23981. doi:10.1371/journal.pone.0023981
Davis, L., and Maizels, N. (2014). Homology-Directed Repair of DNA Nicks via Pathways Distinct from Canonical Double-Strand Break Repair. Proc. Natl. Acad. Sci. USA 111, E924–E932. doi:10.1073/pnas.1400236111
de Alencastro, G., Puzzo, F., Pavel-Dinu, M., Zhang, F., Pillay, S., Majzoub, K., et al. (2021). Improved Genome Editing through Inhibition of FANCM and Members of the BTR Dissolvase Complex. Mol. Ther. 29, 1016–1027. doi:10.1016/j.ymthe.2020.10.020
Deltcheva, E., Chylinski, K., Sharma, C. M., Gonzales, K., Chao, Y., Pirzada, Z. A., et al. (2011). CRISPR RNA Maturation by Trans-Encoded Small RNA and Host Factor RNase III. Nature 471, 602–607. doi:10.1038/nature09886
Dever, D. P., Bak, R. O., Reinisch, A., Camarena, J., Washington, G., Nicolas, C. E., et al. (2016). CRISPR/Cas9 β-Globin Gene Targeting in Human Haematopoietic Stem Cells. Nature 539, 384–389. doi:10.1038/nature20134
Deyle, D. R., Hansen, R. S., Cornea, A. M., Li, L. B., Burt, A. A., Alexander, I. E., et al. (2014). A Genome-wide Map of Adeno-Associated Virus-Mediated Human Gene Targeting. Nat. Struct. Mol. Biol. 21, 969–975. doi:10.1038/nsmb.2895
Doyon, Y., Vo, T. D., Mendel, M. C., Greenberg, S. G., Wang, J., Xia, D. F., et al. (2011). Enhancing zinc-finger-nuclease Activity with Improved Obligate Heterodimeric Architectures. Nat. Methods 8, 74–79. doi:10.1038/nmeth.1539
Dreyer, A.-K., Hoffmann, D., Lachmann, N., Ackermann, M., Steinemann, D., Timm, B., et al. (2015). TALEN-Mediated Functional Correction of X-Linked Chronic Granulomatous Disease in Patient-Derived Induced Pluripotent Stem Cells. Biomaterials 69, 191–200. doi:10.1016/j.biomaterials.2015.07.057
Duan, J., Lu, G., Xie, Z., Lou, M., Luo, J., Guo, L., et al. (2014). Genome-wide Identification of CRISPR/Cas9 Off-Targets in Human Genome. Cel. Res. 24, 1009–1012. doi:10.1038/cr.2014.87
Epstein, B. E., and Schaffer, D. V. (2017). Combining Engineered Nucleases with Adeno-Associated Viral Vectors for Therapeutic Gene Editing. Adv. Exp. Med. Biol. 1016, 29–42. doi:10.1007/978-3-319-63904-8_2
Ferrari, F. K., Samulski, T., Shenk, T., and Samulski, R. J. (1996). Second-strand Synthesis Is a Rate-Limiting Step for Efficient Transduction by Recombinant Adeno-Associated Virus Vectors. J. Virol. 70, 3227–3234. doi:10.1128/JVI.70.5.3227-3234.1996
Forget, A. L., Bennett, B. T., and Knight, K. L. (2004). Xrcc3 Is Recruited to DNA Double Strand Breaks Early and Independent of Rad51. J. Cel. Biochem. 93, 429–436. doi:10.1002/jcb.20232
Fragkos, M., Jurvansuu, J., and Beard, P. (2009). H2AX Is Required for Cell Cycle Arrest via the P53/p21 Pathway. Mol. Cel. Biol. 29, 2828–2840. doi:10.1128/MCB.01830-08
Frangoul, H., Altshuler, D., Cappellini, M. D., Chen, Y.-S., Domm, J., Eustace, B. K., et al. (2021). CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia. N. Engl. J. Med. 384, 252–260. doi:10.1056/NEJMoa2031054
Fu, Y., Foden, J. A., Khayter, C., Maeder, M. L., Reyon, D., Joung, J. K., et al. (2013). High-frequency Off-Target Mutagenesis Induced by CRISPR-Cas Nucleases in Human Cells. Nat. Biotechnol. 31, 822–826. doi:10.1038/nbt.2623
Gabriel, R., Lombardo, A., Arens, A., Miller, J. C., Genovese, P., Kaeppel, C., et al. (2011). An Unbiased Genome-wide Analysis of Zinc-finger Nuclease Specificity. Nat. Biotechnol. 29, 816–823. doi:10.1038/nbt.1948
Gaj, T., Epstein, B. E., and Schaffer, D. V. (2016). Genome Engineering Using Adeno-Associated Virus: Basic and Clinical Research Applications. Mol. Ther. 24, 458–464. doi:10.1038/mt.2015.151
Galichet, C., and Lovell-Badge, R. (2021). Applications of Genome Editing on Laboratory Animals. Lab. Anim., 002367722199314. doi:10.1177/0023677221993141
Gasiunas, G., Barrangou, R., Horvath, P., and Siksnys, V. (2012). Cas9-crRNA Ribonucleoprotein Complex Mediates Specific DNA Cleavage for Adaptive Immunity in Bacteria. Proc. Natl. Acad. Sci. 109, E2579–E2586. doi:10.1073/pnas.1208507109
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable Base Editing of A*T to G*C in Genomic DNA without DNA Cleavage. Nature 551, 464–471. doi:10.1038/nature24644
Gaudet, D., Méthot, J., Déry, S., Brisson, D., Essiembre, C., Tremblay, G., et al. (2013). Efficacy and Long-Term Safety of Alipogene Tiparvovec (AAV1-LPLS447X) Gene Therapy for Lipoprotein Lipase Deficiency: an Open-Label Trial. Gene Ther. 20, 361–369. doi:10.1038/gt.2012.43
Gillmore, J. D., Gane, E., Taubel, J., Kao, J., Fontana, M., Maitland, M. L., et al. (2021). CRISPR-Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis. N. Engl. J. Med. 385, 493–502. doi:10.1056/NEJMoa2107454
Guo, J., Gaj, T., and Barbas, C. F. (2010). Directed Evolution of an Enhanced and Highly Efficient FokI Cleavage Domain for Zinc finger Nucleases. J. Mol. Biol. 400, 96–107. doi:10.1016/j.jmb.2010.04.060
Haft, D. H., Selengut, J., Mongodin, E. F., and Nelson, K. E. (2005). A Guild of 45 CRISPR-Associated (Cas) Protein Families and Multiple CRISPR/Cas Subtypes Exist in Prokaryotic Genomes. Plos Comput. Biol. 1, e60. doi:10.1371/journal.pcbi.0010060
Hamilton, H., Gomos, J., Berns, K. I., and Falck-Pedersen, E. (2004). Adeno-associated Virus Site-specific Integration and AAVS1 Disruption. J. Virol. 78, 7874–7882. doi:10.1128/JVI.78.15.7874-7882.2004
Hasan, N., Koob, M., and Szybalsk, W. (1994). Escherichia coli Genome Targeting I. Cre-Zox-Mediated In Vitro Generation of Ori− Plasmids and Their In Vivo Chromosomal Integration and Retrieval. Gene 150, 51–56. doi:10.1016/0378-1119(94)90856-7
Haurwitz, R. E., Jinek, M., Wiedenheft, B., Zhou, K., and Doudna, J. A. (2010). Sequence- and Structure-Specific RNA Processing by a CRISPR Endonuclease. Science 329, 1355–1358. doi:10.1126/science.1192272
Henry-Mowatt, J., Jackson, D., Masson, J.-Y., Johnson, P. A., Clements, P. M., Benson, F. E., et al. (2003). XRCC3 and Rad51 Modulate Replication fork Progression on Damaged Vertebrate Chromosomes. Mol. Cel. 11, 1109–1117. doi:10.1016/s1097-2765(03)00132-1
Hiramoto, T., Nakanishi, T., Sumiyoshi, T., Fukuda, T., Matsuura, S., Tauchi, H., et al. (1999). Mutations of a Novel Human RAD54 Homologue, RAD54B, in Primary Cancer. Oncogene 18, 3422–3426. doi:10.1038/sj.onc.1202691
Hiramoto, T., Li, L. B., Funk, S. E., Hirata, R. K., and Russell, D. W. (2018). Nuclease-free Adeno-Associated Virus-Mediated Il2rg Gene Editing in X-SCID Mice. Mol. Ther. 26, 1255–1265. doi:10.1016/j.ymthe.2018.02.028
Hirata, R. K., and Russell, D. W. (2000). Design and Packaging of Adeno-Associated Virus Gene Targeting Vectors. J. Virol. 74, 4612–4620. doi:10.1128/jvi.74.10.4612-4620.2000
Hirata, R., Chamberlain, J., Dong, R., and Russell, D. W. (2002). Targeted Transgene Insertion into Human Chromosomes by Adeno-Associated Virus Vectors. Nat. Biotechnol. 20, 735–738. doi:10.1038/nbt0702-735
Hirsch, M. L. (2015). Adeno-associated Virus Inverted Terminal Repeats Stimulate Gene Editing. Gene Ther. 22, 190–195. doi:10.1038/gt.2014.109
Hoess, R. H., Ziese, M., and Sternberg, N. (1982). P1 Site-specific Recombination: Nucleotide Sequence of the Recombining Sites. Proc. Natl. Acad. Sci. 79, 3398–3402. doi:10.1073/pnas.79.11.3398
Horvath, P., and Barrangou, R. (2010). CRISPR/Cas, the Immune System of Bacteria and Archaea. Science 327, 167–170. doi:10.1126/science.1179555
Inagaki, K., Lewis, S. M., Wu, X., Ma, C., Munroe, D. J., Fuess, S., et al. (2007). DNA Palindromes with a Modest Arm Length of ≳20 Base Pairs Are a Significant Target for Recombinant Adeno-Associated Virus Vector Integration in the Liver, Muscles, and Heart in Mice. J. Virol. 81, 11290–11303. doi:10.1128/JVI.00963-07
Ingemarsdotter, C., Keller, D., and Beard, P. (2010). The DNA Damage Response to Non-Replicating Adeno-Associated Virus: Centriole Overduplication and Mitotic Catastrophe Independent of the Spindle Checkpoint. Virology 400, 271–286. doi:10.1016/j.virol.2010.02.003
Inoue, N., Hirata, R. K., and Russell, D. W. (1999). High-fidelity Correction of Mutations at Multiple Chromosomal Positions by Adeno-Associated Virus Vectors. J. Virol. 73, 7376–7380. doi:10.1128/JVI.73.9.7376-7380.1999
Jalan, M., Olsen, K. S., and Powell, S. N. (2019). Emerging Roles of RAD52 in Genome Maintenance. Cancers 11, 1038. doi:10.3390/cancers11071038
Jansen, R., Embden, J. D. A. v., Gaastra, W., and Schouls, L. M. (2002). Identification of Genes that Are Associated with DNA Repeats in Prokaryotes. Mol. Microbiol. 43, 1565–1575. doi:10.1046/j.1365-2958.2002.02839.x
Jensen, R. B., Carreira, A., and Kowalczykowski, S. C. (2010). Purified Human BRCA2 Stimulates RAD51-Mediated Recombination. Nature 467, 678–683. doi:10.1038/nature09399
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337, 816–821. doi:10.1126/science.1225829
Junod, S. L., Saredy, J., and Yang, W. (2021). Nuclear Import of Adeno-Associated Viruses Imaged by High-Speed Single-Molecule Microscopy. Viruses 13, 167. doi:10.3390/v13020167
Jurvansuu, J., Raj, K., Stasiak, A., and Beard, P. (2005). Viral Transport of DNA Damage that Mimics a Stalled Replication fork. J. Virol. 79, 569–580. doi:10.1128/JVI.79.1.569-580.2005
Jurvansuu, J., Fragkos, M., Ingemarsdotter, C., and Beard, P. (2007). Chk1 Instability Is Coupled to Mitotic Cell Death of P53-Deficient Cells in Response to Virus-Induced DNA Damage Signaling. J. Mol. Biol. 372, 397–406. doi:10.1016/j.jmb.2007.06.077
Kanaar, R., Troelstra, C., Swagemakers, S. M. A., Essers, J., Smit, B., Franssen, J.-H., et al. (1996). Human and Mouse Homologs of the Saccharomyces cerevisiaeRAD54 DNA Repair Gene: Evidence for Functional Conservation. Curr. Biol. 6, 828–838. doi:10.1016/s0960-9822(02)00606-1
Keskin, H., Shen, Y., Huang, F., Patel, M., Yang, T., Ashley, K., et al. (2014). Transcript-RNA-templated DNA Recombination and Repair. Nature 515, 436–439. doi:10.1038/nature13682
Khan, I. F., Hirata, R. K., and Russell, D. W. (2011). AAV-mediated Gene Targeting Methods for Human Cells. Nat. Protoc. 6, 482–501. doi:10.1038/nprot.2011.301
Kim, Y. B., Komor, A. C., Levy, J. M., Packer, M. S., Zhao, K. T., and Liu, D. R. (2017). Increasing the Genome-Targeting Scope and Precision of Base Editing with Engineered Cas9-Cytidine Deaminase Fusions. Nat. Biotechnol. 35, 371–376. doi:10.1038/nbt.3803
Kohama, Y., Higo, S., Masumura, Y., Shiba, M., Kondo, T., Ishizu, T., et al. (2020). Adeno-associated Virus-Mediated Gene Delivery Promotes S-phase Entry-independent Precise Targeted Integration in Cardiomyocytes. Sci. Rep. 10, 15348. doi:10.1038/s41598-020-72216-y
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., and Liu, D. R. (2016). Programmable Editing of a Target Base in Genomic DNA without Double-Stranded DNA Cleavage. Nature 533, 420–424. doi:10.1038/nature17946
Konermann, S., Brigham, M. D., Trevino, A. E., Joung, J., Abudayyeh, O. O., Barcena, C., et al. (2015). Genome-scale Transcriptional Activation by an Engineered CRISPR-Cas9 Complex. Nature 517, 583–588. doi:10.1038/nature14136
Kotin, R. M., and Berns, K. I. (1989). Organization of Adeno-Associated Virus DNA in Latently Infected Detroit 6 Cells. Virology 170, 460–467. doi:10.1016/0042-6822(89)90437-6
Kotin, R. M., Siniscalco, M., Samulski, R. J., Zhu, X. D., Hunter, L., Laughlin, C. A., et al. (1990). Site-Specific Integration by Adeno-Associated Virus. Proc. Natl. Acad. Sci. 87, 2211–2215. doi:10.1073/pnas.87.6.2211
Kotin, R. M., Menninger, J. C., Ward, D. C., and Berns, K. I. (1991). Mapping and Direct Visualization of a Region-specific Viral DNA Integration Site on Chromosome 19q13-Qter. Genomics 10, 831–834. doi:10.1016/0888-7543(91)90470-y
Kotin, R. M., Linden, R. M., and Berns, K. I. (1992). Characterization of a Preferred Site on Human Chromosome 19q for Integration of Adeno-Associated Virus DNA by Non-homologous Recombination. EMBO J. 11, 5071–5078. doi:10.1002/j.1460-2075.1992.tb05614.x
Kuscu, C., Arslan, S., Singh, R., Thorpe, J., and Adli, M. (2014). Genome-wide Analysis Reveals Characteristics of Off-Target Sites Bound by the Cas9 Endonuclease. Nat. Biotechnol. 32, 677–683. doi:10.1038/nbt.2916
Kuzmin, D. A., Shutova, M. V., Johnston, N. R., Smith, O. P., Fedorin, V. V., Kukushkin, Y. S., et al. (2021). The Clinical Landscape for AAV Gene Therapies. Nat. Rev. Drug Discov. 20, 173–174. doi:10.1038/d41573-021-00017-7
Ledford, H. (2020). CRISPR Treatment Inserted Directly into the Body for First Time. Nature 579, 185. doi:10.1038/d41586-020-00655-8
Lentz, T. B., and Samulski, R. J. (2015). Insight into the Mechanism of Inhibition of Adeno-Associated Virus by the Mre11/Rad50/Nbs1 Complex. J. Virol. 89, 181–194. doi:10.1128/JVI.01990-14
Li, H., Yang, Y., Hong, W., Huang, M., Wu, M., and Zhao, X. (2020). Applications of Genome Editing Technology in the Targeted Therapy of Human Diseases: Mechanisms, Advances and Prospects. Sig Transduct Target. Ther. 5, 1. doi:10.1038/s41392-019-0089-y
Lin, Y., Cradick, T. J., Brown, M. T., Deshmukh, H., Ranjan, P., Sarode, N., et al. (2014). CRISPR/Cas9 Systems Have Off-Target Activity with Insertions or Deletions between Target DNA and Guide RNA Sequences. Nucleic Acids Res. 42, 7473–7485. doi:10.1093/nar/gku402
Linden, R. M., Ward, P., Giraud, C., Winocour, E., and Berns, K. I. (1996). Site-specific Integration by Adeno-Associated Virus. Proc. Natl. Acad. Sci. 93, 11288–11294. doi:10.1073/pnas.93.21.11288
Maeder, M. L., Stefanidakis, M., Wilson, C. J., Baral, R., Barrera, L. A., Bounoutas, G. S., et al. (2019). Development of a Gene-Editing Approach to Restore Vision Loss in Leber Congenital Amaurosis Type 10. Nat. Med. 25, 229–233. doi:10.1038/s41591-018-0327-9
Mahfouz, M. M., Li, L., Shamimuzzaman, M., Wibowo, A., Fang, X., and Zhu, J.-K. (2011). De Novo-engineered Transcription Activator-like Effector (TALE) Hybrid Nuclease with Novel DNA Binding Specificity Creates Double-Strand Breaks. Proc. Natl. Acad. Sci. 108, 2623–2628. doi:10.1073/pnas.1019533108
Makarova, K. S., Grishin, N. V., Shabalina, S. A., Wolf, Y. I., and Koonin, E. V. (2006). A Putative RNA-Interference-Based Immune System in Prokaryotes: Computational Analysis of the Predicted Enzymatic Machinery, Functional Analogies with Eukaryotic RNAi, and Hypothetical Mechanisms of Action. Biol. Direct 1, 7. doi:10.1186/1745-6150-1-7
Mali, P., Aach, J., Stranges, P. B., Esvelt, K. M., Moosburner, M., Kosuri, S., et al. (2013a). CAS9 Transcriptional Activators for Target Specificity Screening and Paired Nickases for Cooperative Genome Engineering. Nat. Biotechnol. 31, 833–838. doi:10.1038/nbt.2675
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., Dicarlo, J. E., et al. (2013b). RNA-Guided Human Genome Engineering via Cas9. Science 339, 823–826. doi:10.1126/science.1232033
Marnef, A., Cohen, S., and Legube, G. (2017). Transcription-Coupled DNA Double-Strand Break Repair: Active Genes Need Special Care. J. Mol. Biol. 429, 1277–1288. doi:10.1016/j.jmb.2017.03.024
Masson, J.-Y., Tarsounas, M. C., Stasiak, A. Z., Stasiak, A., Shah, R., Mcilwraith, M. J., et al. (2001). Identification and Purification of Two Distinct Complexes Containing the Five RAD51 Paralogs. Genes Dev. 15, 3296–3307. doi:10.1101/gad.947001
Matsoukas, I. G. (2020). Prime Editing: Genome Editing for Rare Genetic Diseases without Double-Strand Breaks or Donor DNA. Front. Genet. 11, 528. doi:10.3389/fgene.2020.00528
McCarty, D. M., Young, S. M., and Samulski, R. J. (2004). Integration of Adeno-Associated Virus (AAV) and Recombinant AAV Vectors. Annu. Rev. Genet. 38, 819–845. doi:10.1146/annurev.genet.37.110801.143717
Meers, C., Keskin, H., and Storici, F. (2016). DNA Repair by RNA: Templated, or Not Templated, that Is the Question. DNA Repair 44, 17–21. doi:10.1016/j.dnarep.2016.05.002
Miller, D. G., Petek, L. M., and Russell, D. W. (2003). Human Gene Targeting by Adeno-Associated Virus Vectors Is Enhanced by DNA Double-Strand Breaks. Mol. Cel. Biol. 23, 3550–3557. doi:10.1128/MCB.23.10.3550-3557.2003
Miller, D. G., Petek, L. M., and Russell, D. W. (2004). Adeno-associated Virus Vectors Integrate at Chromosome Breakage Sites. Nat. Genet. 36, 767–773. doi:10.1038/ng1380
Miller, D. G., Trobridge, G. D., Petek, L. M., Jacobs, M. A., Kaul, R., and Russell, D. W. (2005). Large-scale Analysis of Adeno-Associated Virus Vector Integration Sites in normal Human Cells. J. Virol. 79, 11434–11442. doi:10.1128/JVI.79.17.11434-11442.2005
Miller, D. G., Wang, P.-R., Petek, L. M., Hirata, R. K., Sands, M. S., and Russell, D. W. (2006). Gene Targeting In Vivo by Adeno-Associated Virus Vectors. Nat. Biotechnol. 24, 1022–1026. doi:10.1038/nbt1231
Miller, J. C., Holmes, M. C., Wang, J., Guschin, D. Y., Lee, Y.-L., Rupniewski, I., et al. (2007). An Improved Zinc-Finger Nuclease Architecture for Highly Specific Genome Editing. Nat. Biotechnol. 25, 778–785. doi:10.1038/nbt1319
Miller, J. C., Tan, S., Qiao, G., Barlow, K. A., Wang, J., Xia, D. F., et al. (2011). A TALE Nuclease Architecture for Efficient Genome Editing. Nat. Biotechnol. 29, 143–148. doi:10.1038/nbt.1755
Mojica, F. J. M., Díez-Villaseñor, C., García-Martínez, J., and Almendros, C. (2009). Short Motif Sequences Determine the Targets of the Prokaryotic CRISPR Defence System. Microbiology (Reading) 155, 733–740. doi:10.1099/mic.0.023960-0
Monteilhet, C., Perrin, A., Thierry, A., Colleaux, L., and Dujon, B. (1990). Purification and Characterization of Thein Vitroactivity of I-SceI, a Novel and Highly Specific Endonuclease Encoded by a Group I Intron. Nucl. Acids Res. 18, 1407–1413. doi:10.1093/nar/18.6.1407
Montgomery, K. R., Louis Sam Titus, A. S. C., Wang, L., and D’Mello, S. R. (2018). Elevated MeCP2 in Mice Causes Neurodegeneration Involving Tau Dysregulation and Excitotoxicity: Implications for the Understanding and Treatment of MeCP2 Triplication Syndrome. Mol. Neurobiol. 55, 9057–9074. doi:10.1007/s12035-018-1046-4
Moscou, M. J., and Bogdanove, A. J. (2009). A Simple Cipher Governs DNA Recognition by TAL Effectors. Science 326, 1501. doi:10.1126/science.1178817
Mukherjee, S., Abdisalaam, S., Bhattacharya, S., Srinivasan, K., Sinha, D., and Asaithamby, A. (2019). Mechanistic Link between DNA Damage Sensing, Repairing and Signaling Factors and Immune Signaling. Adv. Protein Chem. Struct. Biol. 115, 297–324. doi:10.1016/bs.apcsb.2018.11.004
Mullard, A. (2020). Gene-Editing Pipeline Takes off. Nat. Rev. Drug Discov. 19, 367–372. doi:10.1038/d41573-020-00096-y
Nakai, H., Iwaki, Y., Kay, M. A., and Couto, L. B. (1999). Isolation of Recombinant Adeno-Associated Virus Vector-Cellular DNA Junctions from Mouse Liver. J. Virol. 73, 5438–5447. doi:10.1128/JVI.73.7.5438-5447.1999
Nakai, H., Montini, E., Fuess, S., Storm, T. A., Grompe, M., and Kay, M. A. (2003). AAV Serotype 2 Vectors Preferentially Integrate into Active Genes in Mice. Nat. Genet. 34, 297–302. doi:10.1038/ng1179
Nakai, H., Wu, X., Fuess, S., Storm, T. A., Munroe, D., Montini, E., et al. (2005). Large-Scale Molecular Characterization of Adeno-Associated Virus Vector Integration in Mouse Liver. J. Virol. 79, 3606–3614. doi:10.1128/JVI.79.6.3606-3614.2005
Nash, K., Chen, W., Mcdonald, W. F., Zhou, X., and Muzyczka, N. (2007). Purification of Host Cell Enzymes Involved in Adeno-Associated Virus DNA Replication. J. Virol. 81, 5777–5787. doi:10.1128/JVI.02651-06
Nathwani, A. C., Rosales, C., Mcintosh, J., Rastegarlari, G., Nathwani, D., Raj, D., et al. (2011). Long-term Safety and Efficacy Following Systemic Administration of a Self-Complementary AAV Vector Encoding Human FIX Pseudotyped with Serotype 5 and 8 Capsid Proteins. Mol. Ther. 19, 876–885. doi:10.1038/mt.2010.274
Nathwani, A. C., Reiss, U. M., Tuddenham, E. G. D., Rosales, C., Chowdary, P., Mcintosh, J., et al. (2014). Long-term Safety and Efficacy of Factor IX Gene Therapy in Hemophilia B. N. Engl. J. Med. 371, 1994–2004. doi:10.1056/NEJMoa1407309
Nguyen, G. N., Everett, J. K., Kafle, S., Roche, A. M., Raymond, H. E., Leiby, J., et al. (2021). A Long-Term Study of AAV Gene Therapy in Dogs with Hemophilia A Identifies Clonal Expansions of Transduced Liver Cells. Nat. Biotechnol. 39, 47–55. doi:10.1038/s41587-020-0741-7
Ni, T.-H., Mcdonald, W. F., Zolotukhin, I., Melendy, T., Waga, S., Stillman, B., et al. (1998). Cellular Proteins Required for Adeno-Associated Virus DNA Replication in the Absence of Adenovirus Coinfection. J. Virol. 72, 2777–2787. doi:10.1128/JVI.72.4.2777-2787.1998
Niemeyer, G. P., Herzog, R. W., Mount, J., Arruda, V. R., Tillson, D. M., Hathcock, J., et al. (2009). Long-term Correction of Inhibitor-Prone Hemophilia B Dogs Treated with Liver-Directed AAV2-Mediated Factor IX Gene Therapy. Blood 113, 797–806. doi:10.1182/blood-2008-10-181479
Nishida, K., Arazoe, T., Yachie, N., Banno, S., Kakimoto, M., Tabata, M., et al. (2016). Targeted Nucleotide Editing Using Hybrid Prokaryotic and Vertebrate Adaptive Immune Systems. Science 353, aaf8729. doi:10.1126/science.aaf8729
Nygaard, S., Barzel, A., Haft, A., Major, A., Finegold, M., Kay, M. A., et al. (2016). A Universal System to Select Gene-Modified Hepatocytes In Vivo. Sci. Transl. Med. 8, 342ra379. doi:10.1126/scitranslmed.aad8166
Ouyang, J., Yadav, T., Zhang, J.-M., Yang, H., Rheinbay, E., Guo, H., et al. (2021). RNA Transcripts Stimulate Homologous Recombination by Forming DR-Loops. Nature 594, 283–288. doi:10.1038/s41586-021-03538-8
Pattanayak, V., Ramirez, C. L., Joung, J. K., and Liu, D. R. (2011). Revealing Off-Target Cleavage Specificities of Zinc-finger Nucleases by In Vitro Selection. Nat. Methods 8, 765–770. doi:10.1038/nmeth.1670
Pattanayak, V., Lin, S., Guilinger, J. P., Ma, E., Doudna, J. A., and Liu, D. R. (2013). High-Throughput Profiling of Off-Target DNA Cleavage Reveals RNA-Programmed Cas9 Nuclease Specificity. Nat. Biotechnol. 31, 839–843. doi:10.1038/nbt.2673
Perez-Pinera, P., Kocak, D. D., Vockley, C. M., Adler, A. F., Kabadi, A. M., Polstein, L. R., et al. (2013). RNA-Guided Gene Activation by CRISPR-Cas9-Based Transcription Factors. Nat. Methods 10, 973–976. doi:10.1038/nmeth.2600
Philpott, N. J., Giraud-Wali, C., Dupuis, C., Gomos, J., Hamilton, H., Berns, K. I., et al. (2002a). Efficient Integration of Recombinant Adeno-Associated Virus DNA Vectors Requires a P5- Rep Sequence in Cis. J. Virol. 76, 5411–5421. doi:10.1128/jvi.76.11.5411-5421.2002
Philpott, N. J., Gomos, J., Berns, K. I., and Falck-Pedersen, E. (2002b). A P5 Integration Efficiency Element Mediates Rep-dependent Integration into AAVS1 at Chromosome 19. Proc. Natl. Acad. Sci. 99, 12381–12385. doi:10.1073/pnas.182430299
Plessis, A., Perrin, A., Haber, J. E., and Dujon, B. (1992). Site-specific Recombination Determined by I-SceI, a Mitochondrial Group I Intron-Encoded Endonuclease Expressed in the Yeast Nucleus. Genetics 130, 451–460. doi:10.1093/genetics/130.3.451
Porteus, M. H., and Baltimore, D. (2003). Chimeric Nucleases Stimulate Gene Targeting in Human Cells. Science 300, 763. doi:10.1126/science.1078395
Porteus, M. H., and Carroll, D. (2005). Gene Targeting Using Zinc finger Nucleases. Nat. Biotechnol. 23, 967–973. doi:10.1038/nbt1125
Porteus, M. H., Cathomen, T., Weitzman, M. D., and Baltimore, D. (2003). Efficient Gene Targeting Mediated by Adeno-Associated Virus and DNA Double-Strand Breaks. Mol. Cel. Biol. 23, 3558–3565. doi:10.1128/MCB.23.10.3558-3565.2003
Porteus, M. H. (2006). Mammalian Gene Targeting with Designed Zinc finger Nucleases. Mol. Ther. 13, 438–446. doi:10.1016/j.ymthe.2005.08.003
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., et al. (2013). Repurposing CRISPR as an RNA-Guided Platform for Sequence-specific Control of Gene Expression. Cell 152, 1173. doi:10.1016/j.cell.2013.02.022
Ran, F. A., Hsu, P. D., Lin, C.-Y., Gootenberg, J. S., Konermann, S., Trevino, A. E., et al. (2013). Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell 154, 1380–1389. doi:10.1016/j.cell.2013.08.021
Reyon, D., Tsai, S. Q., Khayter, C., Foden, J. A., Sander, J. D., and Joung, J. K. (2012). FLASH Assembly of TALENs for High-Throughput Genome Editing. Nat. Biotechnol. 30, 460–465. doi:10.1038/nbt.2170
Rinaldi, C., Pizzul, P., Longhese, M. P., and Bonetti, D. (2020). Sensing R-Loop-Associated DNA Damage to Safeguard Genome Stability. Front. Cel. Dev. Biol. 8, 618157. doi:10.3389/fcell.2020.618157
Rothenberg, E., Grimme, J. M., Spies, M., and Ha, T. (2008). Human Rad52-Mediated Homology Search and Annealing Occurs by Continuous Interactions between Overlapping Nucleoprotein Complexes. Proc. Natl. Acad. Sci. 105, 20274–20279. doi:10.1073/pnas.0810317106
Rudin, N., Sugarman, E., and Haber, J. E. (1989). Genetic and Physical Analysis of Double-Strand Break Repair and Recombination in Saccharomyces cerevisiae. Genetics 122, 519–534. doi:10.1093/genetics/122.3.519
Russell, D. W., and Hirata, R. K. (1998). Human Gene Targeting by Viral Vectors. Nat. Genet. 18, 325–330. doi:10.1038/ng0498-325
Russell, D. W., Miller, A. D., and Alexander, I. E. (1994). Adeno-associated Virus Vectors Preferentially Transduce Cells in S Phase. Proc. Natl. Acad. Sci. 91, 8915–8919. doi:10.1073/pnas.91.19.8915
Rutledge, E. A., and Russell, D. W. (1997). Adeno-associated Virus Vector Integration Junctions. J. Virol. 71, 8429–8436. doi:10.1128/JVI.71.11.8429-8436.1997
Samulski, R. J., Zhu, X., Xiao, X., Brook, J. D., Housman, D. E., Epstein, N., et al. (1991). Targeted Integration of Adeno-Associated Virus (AAV) into Human Chromosome 19. EMBO J. 10, 3941–3950. doi:10.1002/j.1460-2075.1991.tb04964.x
Sanlioglu, S., Benson, P., and Engelhardt, J. F. (2000). Loss of ATM Function Enhances Recombinant Adeno-Associated Virus Transduction and Integration through Pathways Similar to UV Irradiation. Virology 268, 68–78. doi:10.1006/viro.1999.0137
Sapranauskas, R., Gasiunas, G., Fremaux, C., Barrangou, R., Horvath, P., and Siksnys, V. (2011). The Streptococcus Thermophilus CRISPR/Cas System Provides Immunity in Escherichia coli. Nucleic Acids Res. 39, 9275–9282. doi:10.1093/nar/gkr606
Sather, B. D., Romano Ibarra, G. S., Sommer, K., Curinga, G., Hale, M., Khan, I. F., et al. (2015). Efficient Modification of CCR5 in Primary Human Hematopoietic Cells Using a megaTAL Nuclease and AAV Donor Template. Sci. Transl. Med. 7, 307ra156. doi:10.1126/scitranslmed.aac5530
Schwartz, R. A., Palacios, J. A., Cassell, G. D., Adam, S., Giacca, M., and Weitzman, M. D. (2007). The Mre11/Rad50/Nbs1 Complex Limits Adeno-Associated Virus Transduction and Replication. J. Virol. 81, 12936–12945. doi:10.1128/JVI.01523-07
Semenova, E., Jore, M. M., Datsenko, K. A., Semenova, A., Westra, E. R., Wanner, B., et al. (2011). Interference by Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) RNA Is Governed by a Seed Sequence. Proc. Natl. Acad. Sci. 108, 10098–10103. doi:10.1073/pnas.1104144108
Shechter, D., Costanzo, V., and Gautier, J. (2004). Regulation of DNA Replication by ATR: Signaling in Response to DNA Intermediates. DNA Repair 3, 901–908. doi:10.1016/j.dnarep.2004.03.020
Sigurdsson, S., Van Komen, S., Petukhova, G., and Sung, P. (2002). Homologous DNA Pairing by Human Recombination Factors Rad51 and Rad54. J. Biol. Chem. 277, 42790–42794. doi:10.1074/jbc.M208004200
Silva, G., Poirot, L., Galetto, R., Smith, J., Montoya, G., Duchateau, P., et al. (2011). Meganucleases and Other Tools for Targeted Genome Engineering: Perspectives and Challenges for Gene Therapy. Cgt 11, 11–27. doi:10.2174/156652311794520111
Smith, J., Grizot, S., Arnould, S., Duclert, A., Epinat, J.-C., Chames, P., et al. (2006). A Combinatorial Approach to Create Artificial Homing Endonucleases Cleaving Chosen Sequences. Nucleic Acids Res. 34, e149. doi:10.1093/nar/gkl720
Smith, L. J., Ul-Hasan, T., Carvaines, S. K., Van Vliet, K., Yang, E., Wong, K. K., et al. (2014). Gene Transfer Properties and Structural Modeling of Human Stem Cell-Derived AAV. Mol. Ther. 22, 1625–1634. doi:10.1038/mt.2014.107
Smith, L. J., Wright, J., Clark, G., Ul-Hasan, T., Jin, X., Fong, A., et al. (2018). Stem Cell-Derived Clade F AAVs Mediate High-Efficiency Homologous Recombination-Based Genome Editing. Proc. Natl. Acad. Sci. USA 115, E7379–E7388. doi:10.1073/pnas.1802343115
Smith, M. J., Bryant, E. E., Joseph, F. J., and Rothstein, R. (2019). DNA Damage Triggers Increased Mobility of Chromosomes in G1-phase Cells. MBoC 30, 2620–2625. doi:10.1091/mbc.E19-08-0469
Smithies, O., Gregg, R. G., Boggs, S. S., Koralewski, M. A., and Kucherlapati, R. S. (1985). Insertion of DNA Sequences into the Human Chromosomal β-globin Locus by Homologous Recombination. Nature 317, 230–234. doi:10.1038/317230a0
Sollier, J., Stork, C. T., García-Rubio, M. L., Paulsen, R. D., Aguilera, A., and Cimprich, K. A. (2014). Transcription-coupled Nucleotide Excision Repair Factors Promote R-Loop-Induced Genome Instability. Mol. Cel. 56, 777–785. doi:10.1016/j.molcel.2014.10.020
Spector, L. P., Tiffany, M., Ferraro, N. M., Abell, N. S., Montgomery, S. B., and Kay, M. A. (2021). Evaluating the Genomic Parameters Governing rAAV-Mediated Homologous Recombination. Mol. Ther. 29, 1028–1046. doi:10.1016/j.ymthe.2020.11.025
Srivastava, A. (2016). In Vivo tissue-tropism of Adeno-Associated Viral Vectors. Curr. Opin. Virol. 21, 75–80. doi:10.1016/j.coviro.2016.08.003
Stellon, D., Tran, M. T. N., Talbot, J., Chear, S., Khalid, M., Pebay, A., et al. (2021). CRISPR/Cas-Mediated Knock-In of Genetically Encoded Fluorescent Biosensors into the AAVS1 Locus of Human-Induced Pluripotent Stem Cells. Methods Mol. Biol. doi:10.1007/7651_2021_422
Stoddard, B. L. (2005). Homing Endonuclease Structure and Function. Quart. Rev. Biophys. 38, 49–95. doi:10.1017/S0033583505004063
Szczepek, M., Brondani, V., Büchel, J., Serrano, L., Segal, D. J., and Cathomen, T. (2007). Structure-based Redesign of the Dimerization Interface Reduces the Toxicity of Zinc-finger Nucleases. Nat. Biotechnol. 25, 786–793. doi:10.1038/nbt1317
Tang, J., Cho, N. W., Cui, G., Manion, E. M., Shanbhag, N. M., Botuyan, M. V., et al. (2013). Acetylation Limits 53BP1 Association with Damaged Chromatin to Promote Homologous Recombination. Nat. Struct. Mol. Biol. 20, 317–325. doi:10.1038/nsmb.2499
Tebas, P., Stein, D., Tang, W. W., Frank, I., Wang, S. Q., Lee, G., et al. (2014). Gene Editing ofCCR5in Autologous CD4 T Cells of Persons Infected with HIV. N. Engl. J. Med. 370, 901–910. doi:10.1056/NEJMoa1300662
Thomas, K. R., and Capecchi, M. R. (1987). Site-directed Mutagenesis by Gene Targeting in Mouse Embryo-Derived Stem Cells. Cell 51, 503–512. doi:10.1016/0092-8674(87)90646-5
Tsai, S. Q., Zheng, Z., Nguyen, N. T., Liebers, M., Topkar, V. V., Thapar, V., et al. (2015). GUIDE-seq Enables Genome-wide Profiling of Off-Target Cleavage by CRISPR-Cas Nucleases. Nat. Biotechnol. 33, 187–197. doi:10.1038/nbt.3117
Urnov, F. D., Miller, J. C., Lee, Y.-L., Beausejour, C. M., Rock, J. M., Augustus, S., et al. (2005). Highly Efficient Endogenous Human Gene Correction Using Designed Zinc-finger Nucleases. Nature 435, 646–651. doi:10.1038/nature03556
Vasileva, A., and Jessberger, R. (2005). Precise Hit: Adeno-Associated Virus in Gene Targeting. Nat. Rev. Microbiol. 3, 837–847. doi:10.1038/nrmicro1266
Vasileva, A., Linden, R. M., and Jessberger, R. (2006). Homologous Recombination Is Required for AAV-Mediated Gene Targeting. Nucleic Acids Res. 34, 3345–3360. doi:10.1093/nar/gkl455
Wang, X. S., and Srivastava, A. (1997). A Novel Terminal Resolution-like Site in the Adeno-Associated Virus Type 2 Genome. J. Virol. 71, 1140–1146. doi:10.1128/JVI.71.2.1140-1146.1997
Wang, J., Xie, J., Lu, H., Chen, L., Hauck, B., Samulski, R. J., et al. (2007). Existence of Transient Functional Double-Stranded DNA Intermediates during Recombinant AAV Transduction. Proc. Natl. Acad. Sci. 104, 13104–13109. doi:10.1073/pnas.0702778104
Wang, J., Exline, C. M., Declercq, J. J., Llewellyn, G. N., Hayward, S. B., Li, P. W.-L., et al. (2015a). Homology-driven Genome Editing in Hematopoietic Stem and Progenitor Cells Using ZFN mRNA and AAV6 Donors. Nat. Biotechnol. 33, 1256–1263. doi:10.1038/nbt.3408
Wang, X., Wang, Y., Wu, X., Wang, J., Wang, Y., Qiu, Z., et al. (2015b). Unbiased Detection of Off-Target Cleavage by CRISPR-Cas9 and TALENs Using Integrase-Defective Lentiviral Vectors. Nat. Biotechnol. 33, 175–178. doi:10.1038/nbt.3127
Wang, J., Declercq, J. J., Hayward, S. B., Li, P. W.-L., Shivak, D. A., Gregory, P. D., et al. (2016). Highly Efficient Homology-Driven Genome Editing in Human T Cells by Combining Zinc-finger Nuclease mRNA and AAV6 Donor Delivery. Nucleic Acids Res. 44, e30. doi:10.1093/nar/gkv1121
Wei, L., Nakajima, S., Böhm, S., Bernstein, K. A., Shen, Z., Tsang, M., et al. (2015). DNA Damage during the G0/G1 Phase Triggers RNA-Templated, Cockayne Syndrome B-dependent Homologous Recombination. Proc. Natl. Acad. Sci. USA 112, E3495–E3504. doi:10.1073/pnas.1507105112
Welty, S., Teng, Y., Liang, Z., Zhao, W., Sanders, L. H., Greenamyre, J. T., et al. (2018). RAD52 Is Required for RNA-Templated Recombination Repair in post-mitotic Neurons. J. Biol. Chem. 293, 1353–1362. doi:10.1074/jbc.M117.808402
Wiedenheft, B., Van Duijn, E., Bultema, J. B., Waghmare, S. P., Zhou, K., Barendregt, A., et al. (2011). RNA-guided Complex from a Bacterial Immune System Enhances Target Recognition through Seed Sequence Interactions. Proc. Natl. Acad. Sci. 108, 10092–10097. doi:10.1073/pnas.1102716108
Wood, A. J., Lo, T.-W., Zeitler, B., Pickle, C. S., Ralston, E. J., Lee, A. H., et al. (2011). Targeted Genome Editing across Species Using ZFNs and TALENs. Science 333, 307. doi:10.1126/science.1207773
Xu, L., Wang, J., Liu, Y., Xie, L., Su, B., Mou, D., et al. (2019). CRISPR-Edited Stem Cells in a Patient with HIV and Acute Lymphocytic Leukemia. N. Engl. J. Med. 381, 1240–1247. doi:10.1056/NEJMoa1817426
Yang, C. C., Xiao, X., Zhu, X., Ansardi, D. C., Epstein, N. D., Frey, M. R., et al. (1997). Cellular Recombination Pathways and Viral Terminal Repeat Hairpin Structures Are Sufficient for Adeno-Associated Virus Integration In Vivo and In Vitro. J. Virol. 71, 9231–9247. doi:10.1128/JVI.71.12.9231-9247.1997
Zentilin, L., Marcello, A., and Giacca, M. (2001). Involvement of Cellular Double-Stranded DNA Break Binding Proteins in Processing of the Recombinant Adeno-Associated Virus Genome. J. Virol. 75, 12279–12287. doi:10.1128/JVI.75.24.12279-12287.2001
Keywords: AAV, genome editing, homologous recombination, gene therapy, genetic diseases, rare diseases
Citation: Bijlani S, Pang KM, Sivanandam V, Singh A and Chatterjee S (2022) The Role of Recombinant AAV in Precise Genome Editing. Front. Genome Ed. 3:799722. doi: 10.3389/fgeed.2021.799722
Received: 22 October 2021; Accepted: 14 December 2021;
Published: 13 January 2022.
Edited by:
Dirk Grimm, Heidelberg University, GermanyReviewed by:
Eli J. Fine, National Resilience, United StatesCopyright © 2022 Bijlani, Pang, Sivanandam, Singh and Chatterjee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saswati Chatterjee, c2NoYXR0ZXJqZWVAY29oLm9yZw==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.