A Corrigendum on
Targeted Gene Delivery: Where to Land
by Pavani, G., and Amendola, M. (2021). Front. Genome Ed. 2:609650. doi: 10.3389/fgeed.2020.609650
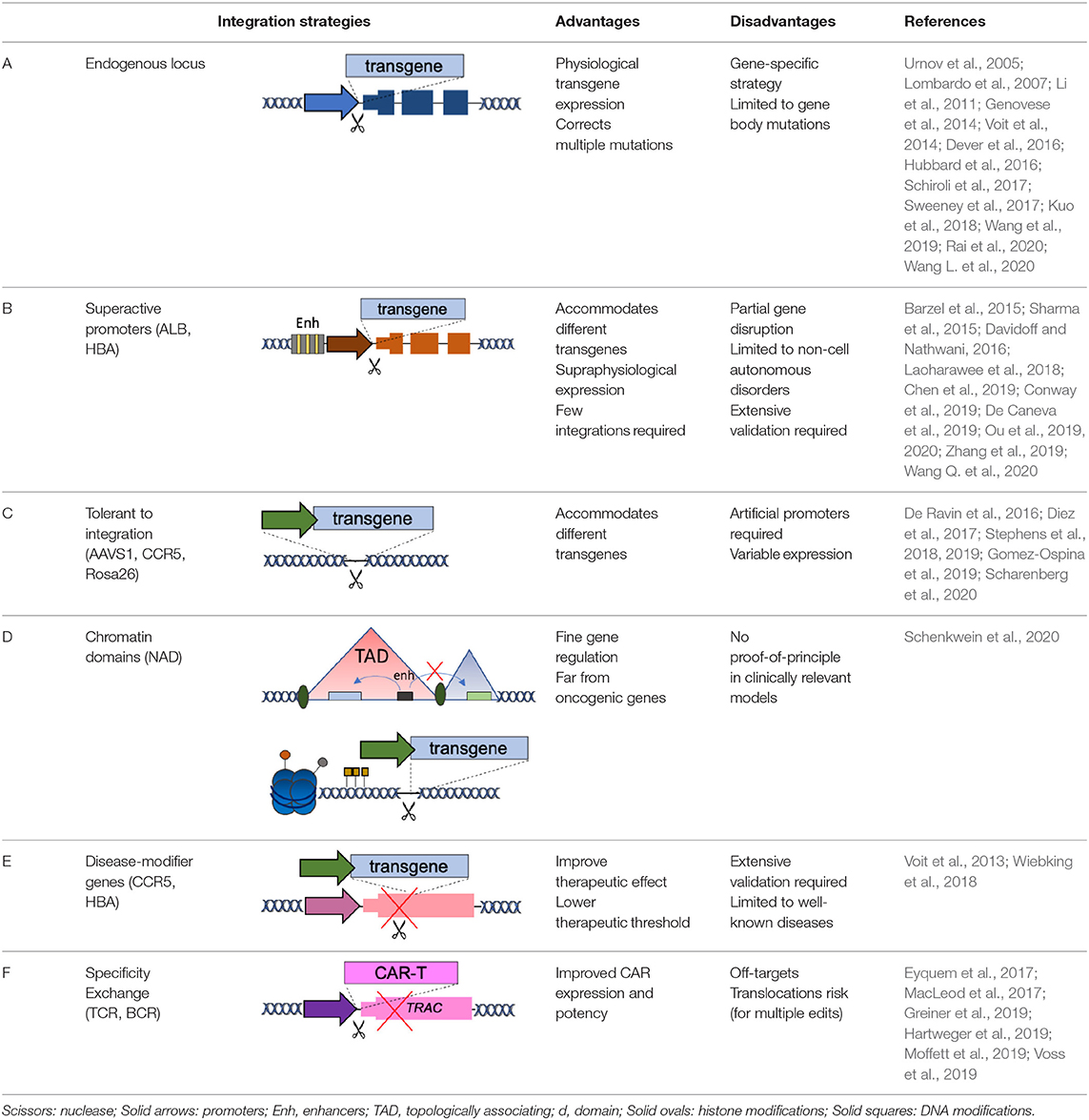
In the original article, there was a mistake in Table 1 as published. The references indicated in row B are wrong. The corrected Table 1 appears in the attached below.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
References
Barzel, A., Paulk, N. K., Shi, Y., Huang, Y., Chu, K., Zhang, F., et al. (2015). Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature 517, 360–364. doi: 10.1038/nature13864
Chen, H., Shi, M., Gilam, A., Zheng, Q., Zhang, Y., Afrikanova, I., et al. (2019). Hemophilia A ameliorated in mice by CRISPR-based in vivo genome editing of human factor. Sci. Rep. 9:16838. doi: 10.1038/s41598-019-53198-y
Conway, A., Mendel, M., Kim, K., McGovern, K., Boyko, A., Zhang, L., et al. (2019). Non-viral delivery of zinc finger nuclease mRNA enables highly efficient in vivo genome editing of multiple therapeutic gene targets. Mol. Ther. 27, 866–877. doi: 10.1016/j.ymthe.2019.03.003
Davidoff, A. M., and Nathwani, A. C. (2016). Genetic targeting of the albumin locus to treat Hemophilia. N. Engl. J. Med. 374, 1288–1290. doi: 10.1056/NEJMcibr1600347
De Caneva, A., Porro, F., Bortolussi, G., Sola, R., Lisjak, M., Barzel, A., et al. (2019). Coupling AAV-mediated promoterless gene targeting to SaCas9 nuclease to efficiently correct liver metabolic diseases. JCI Insight. 5:128863. doi: 10.1172/jci.insight.128863
De Ravin, S. S., Reik, A., Liu, P. Q., Li, L., Wu, X., Su, L., et al. (2016). Targeted gene addition in human CD34(+) hematopoietic cells for correction of X-linked chronic granulomatous disease. Nat. Biotechnol. 34, 424–429. doi: 10.1038/nbt.3513
Dever, D. P., Bak, R. O., Reinisch, A., Camarena, J., Washington, G., Nicolas, C. E., et al. (2016). CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature 539, 384–389. doi: 10.1038/nature20134
Diez, B., Genovese, P., Roman-Rodriguez, F. J., Alvarez, L., Schiroli, G., Ugalde, L., et al. (2017). Therapeutic gene editing in CD34(+) hematopoietic progenitors from Fanconi anemia patients. EMBO Mol. Med. 9, 1574–1588. doi: 10.15252/emmm.201707540
Eyquem, J., Mansilla-Soto, J., Giavridis, T., van der Stegen, S. J., Hamieh, M., Cunanan, K. M., et al. (2017). Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117. doi: 10.1038/nature21405
Genovese, P., Schiroli, G., Escobar, G., Tomaso, T. D., Firrito, C., Calabria, A., et al. (2014). Targeted genome editing in human repopulating haematopoietic stem cells. Nature 510, 235–240. doi: 10.1038/nature13420
Gomez-Ospina, N., Scharenberg, S. G., Mostrel, N., Bak, R. O., Mantri, S., Quadros, R. M., et al. (2019). Human genome-edited hematopoietic stem cells phenotypically correct Mucopolysaccharidosis type. Nat. Commun. 10:4045. doi: 10.1038/s41467-019-11962-8
Greiner, V., Bou Puerto, R., Liu, S., Herbel, C., Carmona, E. M., and Goldberg, M. S. (2019). CRISPR-mediated editing of the B cell receptor in primary human B cells. iScience 12, 369–378. doi: 10.1016/j.isci.2019.01.032
Hartweger, H., McGuire, A. T., Horning, M., Taylor, J. J., Dosenovic, P., Yost, D., et al. (2019). HIV-specific humoral immune responses by CRISPR/Cas9-edited B cells. J. Exp. Med. 216, 1301–1310. doi: 10.1084/jem.20190287
Hubbard, N., Hagin, D., Sommer, K., Song, Y., Khan, I., Clough, C., et al. (2016). Targeted gene editing restores regulated CD40L function in X-linked hyper-IgM syndrome. Blood 127, 2513–2522. doi: 10.1182/blood-2015-11-683235
Kuo, C. Y., Long, J. D., Campo-Fernandez, B., de Oliveira, S., Cooper, A. R., Romero, Z., et al. (2018). Site-specific gene editing of human hematopoietic stem cells for X-linked hyper-IgM syndrome. Cell Rep. 23, 2606–2616. doi: 10.1016/j.celrep.2018.04.103
Laoharawee, K., DeKelver, R. C., Podetz-Pedersen, K. M., Rohde, M., Sproul, S., Nguyen, H. O., et al. (2018). Dose-dependent prevention of metabolic and neurologic disease in murine MPS II by ZFN-mediated in vivo genome editing. Mol. Ther. 26, 1127–1136. doi: 10.1016/j.ymthe.2018.03.002
Li, H., Haurigot, V., Doyon, Y., Li, T., Wong, S. Y., Bhagwat, A. S., et al. (2011). In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 475, 217–221. doi: 10.1038/nature10177
Lombardo, A., Genovese, P., Beausejour, C. M., Colleoni, S., Lee, Y. L., Kim, K. A., et al. (2007). Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 25, 1298–1306. doi: 10.1038/nbt1353
MacLeod, D. T., Antony, J., Martin, A. J., Moser, R. J., Hekele, A., Wetzel, K. J., et al. (2017). Integration of a CD19 CAR into the TCR alpha chain locus streamlines production of allogeneic gene-edited CAR T cells. Mol. Ther. 25, 949–961. doi: 10.1016/j.ymthe.2017.02.005
Moffett, H. F., Harms, C. K., Fitzpatrick, K. S., Tooley, M. R., Boonyaratanakornkit, J., and Taylor, J. J. (2019). B cells engineered to express pathogen-specific antibodies protect against infection. Sci. Immunol. 4:aax0644. doi: 10.1126/sciimmunol.aax0644
Ou, L., DeKelver, R. C., Rohde, M., Tom, S., Radeke, R., St Martin, S. J., et al. (2019). ZFN-mediated in vivo genome editing corrects murine hurler syndrome. Mol. Ther. 27, 178–187. doi: 10.1016/j.ymthe.2018.10.018
Ou, L., Przybilla, M. J., Ahlat, O., Kim, S., Overn, P., Jarnes, J., et al. (2020). A highly efficacious PS gene editing system corrects metabolic and neurological complications of Mucopolysaccharidosis type I. Mol. Ther. 28, 1442–1454. doi: 10.1016/j.ymthe.2020.03.018
Rai, R., Romito, M., Rivers, E., Turchiano, G., Blattner, G., Vetharoy, W., et al. (2020). Targeted gene correction of human hematopoietic stem cells for the treatment of Wiskott - Aldrich Syndrome. Nat. Commun. 11:4034. doi: 10.1038/s41467-020-17626-2
Scharenberg, S. G., Poletto, E., Lucot, K. L., Colella, P., Sheikali, A., Montine, T. J., et al. (2020). Engineering monocyte/macrophage-specific glucocerebrosidase expression in human hematopoietic stem cells using genome editing. Nat. Commun. 11:3327. doi: 10.1038/s41467-020-17148-x
Schenkwein, D., Afzal, S., Nousiainen, A., Schmidt, M., and Yla-Herttuala, S. (2020). Efficient nuclease-directed integration of lentivirus vectors into the human ribosomal DNA locus. Mol. Ther. 28, 1858–1875. doi: 10.1016/j.ymthe.2020.05.019
Schiroli, G., Ferrari, S., Conway, A., Jacob, A., Capo, V., Albano, L., et al. (2017). Preclinical modeling highlights the therapeutic potential of hematopoietic stem cell gene editing for correction of SCID-X1. Sci. Transl. Med. 9:aan0820. doi: 10.1126/scitranslmed.aan0820
Sharma, R., Anguela, X. M., Doyon, Y., Wechsler, T., DeKelver, R. C., Sproul, S., et al. (2015). In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood 126, 1777–1784. doi: 10.1182/blood-2014-12-615492
Stephens, C. J., Kashentseva, E., Everett, W., Kaliberova, L., and Curiel, D. T. (2018). Targeted in vivo knock-in of human alpha-1-antitrypsin cDNA using adenoviral delivery of CRISPR/Cas9. Gene Ther. 25, 139–156. doi: 10.1038/s41434-018-0003-1
Stephens, C. J., Lauron, E. J., Kashentseva, E., Lu, Z. H., Yokoyama, W. M., and Curiel, D. T. (2019). Long-term correction of hemophilia B using adenoviral delivery of CRISPR/Cas9. J. Contr. Release. 298, 128–141. doi: 10.1016/j.jconrel.2019.02.009
Sweeney, C. L., Zou, J., Choi, U., Merling, R. K., Liu, A., Bodansky, A., et al. (2017). Targeted repair of CYBB in X-CGD iPSCs requires retention of intronic sequences for expression and functional correction. Mol. Ther. 25, 321–330. doi: 10.1016/j.ymthe.2016.11.012
Urnov, F. D., Miller, J. C., Lee, Y. L., Beausejour, C. M., Rock, J. M., Augustus, S., et al. (2005). Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435, 646–651. doi: 10.1038/nature03556
Voit, R. A., Hendel, A., Pruett-Miller, S. M., and Porteus, M. H. (2014). Nuclease-mediated gene editing by homologous recombination of the human globin locus. Nucl. Acids Res. 42, 1365–1378. doi: 10.1093/nar/gkt947
Voit, R. A., McMahon, M. A., Sawyer, S. L., and Porteus, M. H. (2013). Generation of an HIV resistant T-cell line by targeted “stacking” of restriction factors. Mol. Ther. 21, 786–795. doi: 10.1038/mt.2012.284
Voss, J. E., Gonzalez-Martin, A., Andrabi, R., Fuller, R. P., Murrell, B., McCoy, L. E., et al. (2019). Reprogramming the antigen specificity of B cells using genome-editing technologies. Elife 8:42995. doi: 10.7554/eLife.42995
Wang, L., Yang, Y., Breton, C., Bell, P., Li, M., Zhang, J., et al. (2020). A mutation-independent CRISPR-Cas9-mediated gene targeting approach to treat a murine model of ornithine transcarbamylase deficiency. Sci. Adv. 6:eaax5701. doi: 10.1126/sciadv.aax5701
Wang, L., Yang, Y., Breton, C. A., White, J., Zhang, J., Che, Y., et al. (2019). CRISPR/Cas9-mediated in vivo gene targeting corrects hemostasis in newborn and adult factor IX-knockout mice. Blood 133, 2745–2752. doi: 10.1182/blood.2019000790
Wang, Q., Zhong, X., Li, Q., Su, J., Liu, Y., Mo, L., et al. (2020). CRISPR-Cas9-mediated in vivo gene integration at the albumin locus recovers hemostasis in neonatal and adult hemophilia B mice. Mol. Ther. Methods Clin. Dev. 18, 520–531. doi: 10.1016/j.omtm.2020.06.025
Wiebking, V., Patterson, J. O., Martin, R., Chanda, M. K., Lee, C. M., Srifa, W., et al. (2018). Metabolic engineering generates a transgene-free safety switch for cell therapy. Nat. Biotechnol 2020:6. doi: 10.1038/s41587-020-0580-6
Keywords: genome editing, gene therapy, nuclease, CRISPR, targeted integration (TI), knock-in, safe harbor, homologous recombination (HR)
Citation: Pavani G and Amendola M (2021) Corrigendum: Targeted Gene Delivery: Where to Land. Front. Genome Ed. 3:682171. doi: 10.3389/fgeed.2021.682171
Received: 17 March 2021; Accepted: 06 April 2021;
Published: 17 May 2021.
Edited and reviewed by: Pietro Genovese, Boston Children's Hospital and Harvard Medical School, United States
Copyright © 2021 Pavani and Amendola. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mario Amendola, bWFtZW5kb2xhQGdlbmV0aG9uLmZy
†Present address: Giulia Pavani, The Children's Hospital of Philadelphia, Raymond G. Perelman Center for Cellular and Molecular Therapeutics, Philadelphia, PA, United States
 Giulia Pavani†
Giulia Pavani† Mario Amendola
Mario Amendola