
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genome Ed. , 25 February 2021
Sec. Genome Editing in Plants
Volume 3 - 2021 | https://doi.org/10.3389/fgeed.2021.632136
This article is part of the Research Topic Targeted Genome Editing in Crops View all 12 articles
Rice (Oryza sativa) production is damaged to a great extent by sheath blight disease (ShB). However, the defense mechanism in rice against this disease is largely unknown. Previous transcriptome analysis identified a significantly induced eukaryotic protein phosphatase 2A catalytic subunit 1 (PP2A-1) after the inoculation of Rhizoctonia solani. Five genes encoding PP2A exist in rice genome, and these five genes are ubiquitously expressed in different tissues and stages. Inoculation of R. solani showed that the genome edited pp2a-1 mutants using the CRISPR/Cas9 were more susceptible to ShB than the wild-type control, but other PP2A gene mutants exhibited similar response to ShB compared to wild-type plants. In parallel, PP2A-1 expression level was higher in the activation tagging line, and PP2A-1 overexpression inhibited plant height and promoted the resistance to ShB. PP2A-1-GFP was localized in the cytoplasm and nucleus. In addition, R. solani-dependent induction kinetics of pathogen-related genes PBZ1 and PR1b was lower in pp2a-1 mutants but higher in PP2A-1 activation line compared to those in the wild-type. In conclusion, our analysis shows that PP2A-1 is a member of protein phosphatase, which regulates rice resistance to ShB. This result broadens the understanding of the defense mechanism against ShB and provides a potential target for rice breeding for disease resistance.
R. solani is the causative agent of ShB in rice (Savary et al., 1995; Suryadi et al., 2013), and which damages rice during the entire growth period, and predominantly targets the leaves, sheaths, and panicles, eventually resulting in the withering and lodging of the entire plant. A severe form of ShB can lower the rice produce by ~50% (Savary et al., 2000). The rapid variation, wide host range, and high survival ability of the pathogen can make the disease control more challenging (Taheri and Tarighi, 2011; Yellareddygari et al., 2014; Singh et al., 2019). Currently, there is a dearth of ShB-resistant cultivars, therefore, the strategies to quell ShB involve the use of fungicides (Savary et al., 2000). However, fungicides directly affect the living environment of other microorganisms and increase the cost of cultivation. Thus, to develop ShB resistance in rice, it is necessary to isolate resistant cultivars and understand their underlying defense mechanisms against ShB.
Extensive studies have been performed to investigate the mechanism of rice defense against ShB. Overexpression of chitinase, β-1,3-glucanase, or OsPGIP1 (polygalacturonase-inhibiting protein) (Shah et al., 2009; Mao et al., 2014; Zhu et al., 2019), OsACS2 (key enzyme in ethylene synthesis) (Helliwell et al., 2013), OsGSTU5 (tau class glutathione-S-transferase 5) (Tiwari et al., 2020), and Os2H16 (Li et al., 2013, 2018) were found to promote rice resistance to ShB. In addition, BSR2 (broad-spectrum resistance 2) (Maeda et al., 2019) or a transcription factor complex including LPA1 (indeterminate domain 14, IDD14) and IDD13 (Sun et al., 2019, 2020) were reported to positively regulate rice resistance to ShB while SWEET11 (sugar will eventually be exported transporter 11) (Gao et al., 2018) exhibited a negative regulation. The transcription factor OsWRKYs also plays an important role in resistance to sheath blight (Peng et al., 2012, 2016; Wang et al., 2015; Jimmy and Babu, 2019; Yuan et al., 2020). In addition, salicylic acid-dependent immunity showed a positive regulation in ShB resistance in rice and Brachypodium distachyon (Kouzai et al., 2018).
Protein phosphatase also plays an important role in plant defense response. The protein phosphatases (PPs) with a vast array of structures and functions are mainly categorized as serine/threonine (Ser/Thr) PPs and protein tyrosine phosphatases (PTPs). PP1, PP2A, PP2B, and PP2C account for the sub-divisions of the protein tyrosine phosphatase group. The PP2A complex comprises three subunits: A, B, and C with scaffolding, regulatory, and catalytic roles, respectively (Yu et al., 2005; Durian et al., 2016). The role of PP2A protein in plant abiotic stress signal transduction has been confirmed. For instance, drought and elevated salinity induce high levels of OsPP2A-1 and OsPP2A-3, the closely associated genes coding for the C-subunit of PP2A (Yu et al., 2003). In Arabidopsis, the growth of roots and shoots is augmented by PP2A-C5 overexpression in the presence of several salts indicating the vital function of protein in growth to combat salinity (Hu et al., 2017). AtPP2A is involved in acclimation to light as well as when responding to pathogens, both based on the regulation of ROS (Rahikainen et al., 2016; Máthé et al., 2019). Exposure of wheat to R. cerealis or hydrogen peroxide showed elevated TaPP2Ac-4B and TaPP2Ac-4D RNA levels revealing the involvement of PP2A in the biotic stress response. Silencing of TaPP2A in wheat boosted the expression of ROS-scavenging and pathogenesis-related (PR) RNA molecules (Zhu et al., 2018). Resistance to Botrytis cinerea and leaf senescence in Arabidopsis involves the role of PP2A-B′γ. The swift induction of the gene coding for the heterotrimeric PP2A catalytic subunit, LePP2A-1 was observed when resistant tomato plants were challenged with Pseudomonas syringae pv. tomato (a virulent strain) (He et al., 2004). A mutation which was isolated from rice blast fungus was inserted into the promoter region of MoPPG1, a ser/thr-PP2A catalytic subunit (PP2Ac) gene, which made the mutant defective in the growth of vegetative mycelium and could not cause disease (Du et al., 2013). Fusarium graminearum contains three kinds of PP2A (FgPp2A, FgSit4, and FgPg1), which play a key role in the growth, development, and pathogenicity of fungi (Liu et al., 2018). Our recent transcriptomic study showed the sensitivity of PP2A-1 expression to R. solani infection (Yuan et al., 2020). However, PP2A function in rice defense to ShB is unknown.
In this study, PP2A-1 was significantly induced following R. solani inoculation. Further bioinformatics, genetic, and molecular analyses were performed to identify the function of PP2A family members in rice defense to ShB. Our results broaden the knowledge of the underlying ShB defense mechanisms and provide a potential target for resistant breeding in rice.
Four rice lines/cultivars, including Japonica rice cultivar Dongjin (DJ), Zhonghua11 (ZH11), pp2a-1 CRISPR/Cas-9 genome editing mutants in ZH11 background, and PP2A-1 activation tagging line (PP2A-1 OX) in DJ background were used in this study. All the rice lines used in this study were grown in a greenhouse in natural light. The type strain used in this study was R. solani AG1-IA.
The human codon-optimized hSpCas9 (Cong et al., 2013) was linked to the maize ubiquitin promoter (UBI) in an intermediate plasmid followed by its insertion into a binary pCAMBIA1300 vector (Cambia, Australia) harboring the HPT (hygromycin B phosphotransferase) gene. A point mutation kit (Transgen, China) was used to eliminate the original BsaI site in the backbone of pCAMBIA1300. A OsU6 promoter fragment (Feng et al., 2013), ccdB, a gene for negative selection flanked by two BsaI sites, and a pX260- derived sgRNA (Cong et al., 2013) were inserted employing an In-Fusion cloning kit (Takara, Japan) into this vector to produce the CRISPR/Cas9 binary vector pBGK032 (Figure 1). The vector was maintained in Escherichia coli strain DB3.1.
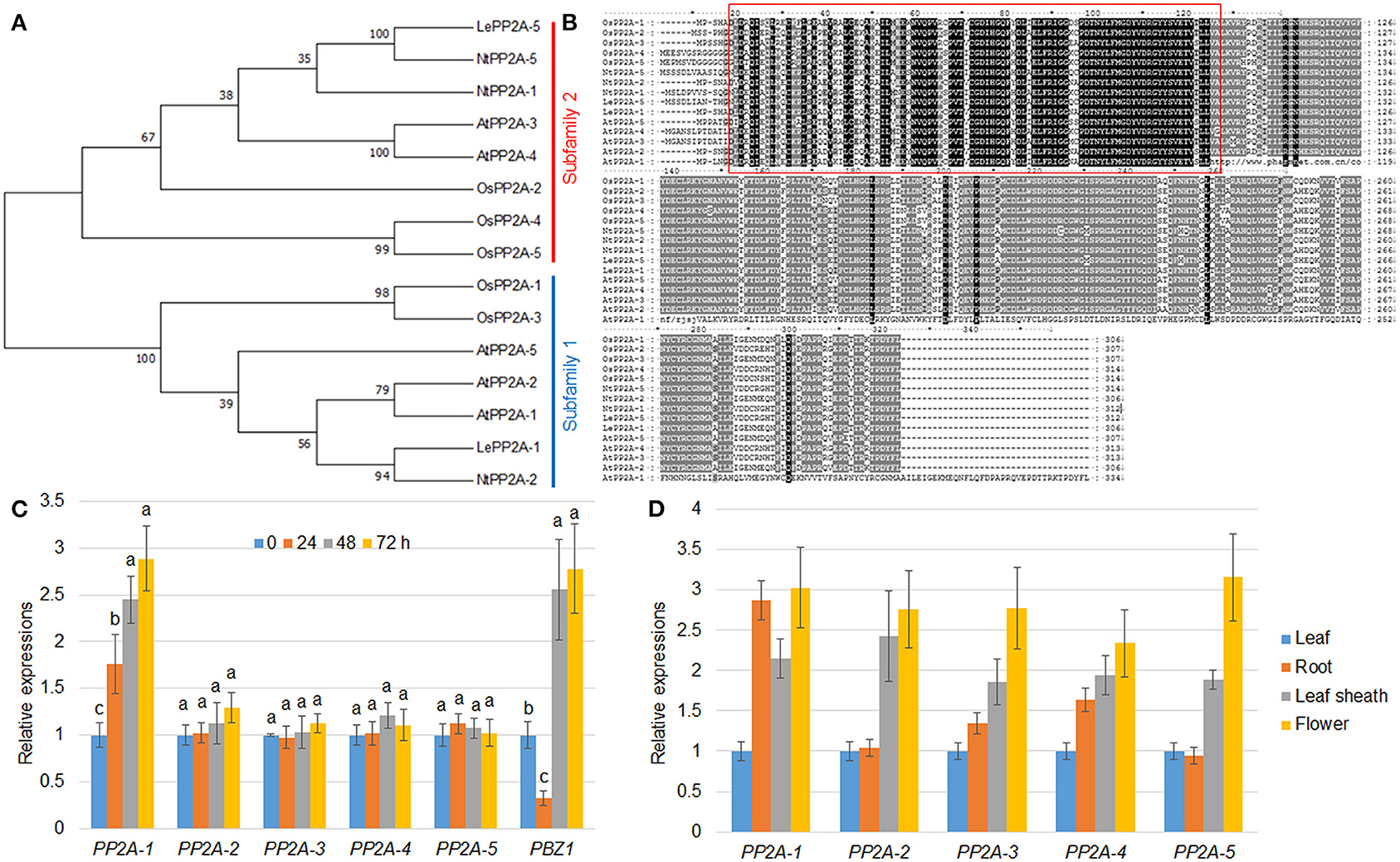
Figure 1. Phylogenetic and expression pattern analysis. (A) The maximum likelihood was utilized for the phylogenetic tree construction. The phylogenetic tree was generated by MEGA 7.0 on the basis of PP2A amino acid sequences from Oryza sativa (OsPP2As), Arabidopsis thaliana (AtPP2As), Solanum lycopersicum (LePP2As), and Nicotiana tabacum (NtPP2As). The PP2As were classified into subfamily 1 and 2 that were marked with blue and red, respectively. (B) The protein sequence identity of the PP2As listed in (A) was analyzed by the Clustal W program. The red box indicates a highly conserved region between PP2As. (C) R. solani inoculation-mediated PP2A expression. The five PP2A and PBZ1 gene expression patterns were evaluated after 0, 24, 48, and 72 h following R. solani inoculation. Normalization of expression was done with reference to the internal control ubiquitin. The experiments were done in triplicate. Different letters indicate significant differences at P < 0.05. (D) Tissue-specific expression patterns of five PP2A genes were analyzed using the RNA extracted from the leaf, root, leaf sheath, and flower. Ubiquitin was used as the internal control to normalize expression levels. The experiments were done in triplicate.
The targeting specificity and the selection of the 23-bp targeting sequences (including PAM) was done employing a BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) against the rice genome (Hsu et al., 2013). The designed targeting sequences were annealed to synthesize the oligo adaptors. The vector pBGK032 was restriction digested using BsaI and purified employing a DNA purification kit (Tiangen, China). This was ligated with 0.05 mM of oligo adaptor (10 μL) resulting in CRISPR/Cas9 plasmids, which were directly transformed in competent E. coli cells.
Agrobacterium tumefaciens strain EHA105 was transformed with the CRISPR/Cas9 plasmids followed by rice transformation following an earlier published protocol (Nishimura et al., 2006). The genomic DNA from these transformants was extracted, and PCR was conducted employing primer pairs flanking the designed target site. The Degenerate Sequence Decoding approach was applied to directly sequence and identified the PCR products (300–500 bp) method (Ma et al., 2015).
The total cellular RNA was extracted from the 1-month-old plant sheath, roots, leaves, or 3-month-old flower tissues. For analysis of R. solani-mediated gene expression, Trizol reagent (Invitrogen, China) was used to isolate the total RNA from 0.1 g of rice leaves, roots, leaf sheath, and flowers. Then the product was purified using the RNeasy mini kit (Promega, China) and RNase-Free DNase I (Promega, China) following manufacturer's instructions. The cDNAs were synthesized with M-MLV Reverse Transcriptase (Promega) kit following manufacturer's instructions.
The PP2A amino acid sequences were isolated from Arabidopsis, rice, tobacco, and tomato to perform phylogenetic analysis. MEGA7 software was used for multiple sequence alignment of the original sequences. The comparison results were edited by GeneDoc to export the multi-sequence alignment results graph. MEGA7 software was used for phylogenetic tree construction using the nearest neighbor-joining method (Kumar et al., 2016).
Reverse transcription using 2 μg of each purified RNA sample was done using a Prime Script TMRT Reagent Kit with gDNA Eraser (TaKaRa, China) in accordance with the provided instructions. qRT-PCR was performed on the ABI 7500 RT-PCR system (Applied Biosystems, United States). The composition of the mix was: 10 μL 2× SYBR Premix Ex Taq, 0.4 μL 50× ROX Reference Dye II, 0.4 μM of each primer, and 5 μL of the cDNA template (50-fold dilution) in a net volume of 20 μL. The conditions were: 95°C for 30 s; 95°C for 5 s, 58°C for 15 s, and 72°C for 34 s for 40 cycles. The 2[−DeltaDeltaC(T)] approach was employed to estimate the expression levels of target gene(s) relative (Livak and Schmittgen, 2001). Ubiquitin was used as an internal reference. Table 1 presents the primers in this study.
Rice plants were grown in the glasshouse for 1 month prior to inoculation with the pathogen R. solani AG1-IA. The second leaf of the main tiller was cut into 10-cm slices, placed on wet filter paper and stored in a culture dish (36 × 36 × 2.5 cm). In a completely randomized design, five leaves were placed in each plate, with a total of three replicate plates for each treatment. The fungal plug (7 mm in diameter) was cut from the Potato Dextrose Agar (PDA) plate with R. solani and placed on the back of the leaf. The leaves were cultured for 72 h at 25°C under continuous light, and the moisture of the filter paper was maintained with sterile water (Gao et al., 2018). Measurement from 0 (no lesion) to 9 (lesions occupying 90–100% of the leaf surface) was done after visual observation. Scores from one to eight represented 10–80% diseased leaf area (Prasad and Eizenga, 2008).
Rice plants that were cultured in a greenhouse for 1 month prior to the tillering stage were used for inoculation. The sheath of the first leaf of the main stem was inoculated with R. solani AG1-IA. The PDA fungal plug was inoculated into rice leaf, sprayed with sterile water, and the severity of the disease was determined after 24, 48, and 72 h.
PP2A-1 ORF region was amplified by PCR and moved into pCAMBIA1302 vector to create PP2A-1-GFP plasmid. The Agrobacterium-mediated transient expression approach was followed to introduce the fusion proteins into Nicotiana benthamiana (Kim et al., 2009). The location of the protein was monitored via GFP fluorescence with a confocal microscope (SP5; Leica, Solms, Germany).
The significant differences between different groups were analyzed using Microsoft Excel to compute the mean, standard deviation, and the Student's t-test. Dunnett's test was done employing the SPSS 19.0 statistical software.
Our previous transcriptome analysis identified that PP2A-1 expression was induced by the inoculation of R. solani AG1-IA (Yuan et al., 2020). Rice genome harbors five PP2A isoforms, and a phylogenetic analysis of PP2A proteins from Arabidopsis, rice, tobacco, and tomato revealed that OsPP2A-1 clustered with OsPP2A-3, NtPP2A-2, AtPP2A-1, AtPP2A-2, AtPP2A-5, and LePP2A-1, all of which belong to Subfamily II, while OsPP2A-5, OsPP2A-4, OsPP2A-2, AtPP2A-3, AtPP2A-4, NtPP2A-1, NtPP2A-5, and LePP2A-5 belong to Subfamily I (Figure 1A). The homologous sequence alignment of PP2As showed that OsPP2A-1 and OsPP2A-3 shared 98% similarity. The red box indicates a highly conserved region between the five PP2As (Figure 1B). qRT-PCR of the 5 PP2A genes was done for the verification of the transcriptome data post-inoculation with R. solani after 0, 24, 48, and 72 h. The results indicated that only PP2A-1 expression was induced by R. solani infection, and PP2A-1 expression was the highest 72 h after inoculation, while the other four PP2A genes did not respond to R. solani. PBZ1, a marker gene was used for evaluating pathogen infection, its expression was down-regulated at 24 h after inoculation, while it was up-regulated after 48 h and 72 h of inoculation (Figure 1C; Table 2). In addition, tissue-specific expression of PP2As was examined by qRT-PCR. All PP2As were expressed in root, leaf sheath, leaf, and flower tissues, while PP2A-1 was expressed highly in root and flower, indicating that PP2As were ubiquitously expressed in different tissues and developmental stages (Figure 1D).
To analyze the function of PP2A genes in rice defense to ShB, Crispr/Cas9 induced genome editing mutants for PP2As were generated. The PP2A genes consist of multiple exons and introns in the genome (Figure 2A). The sequencing of PP2A genome editing mutants revealed that pp2a-1 mutants have a genomic lesion in the first exon with 1 or 2-bp insertions (pp2a-1-1, pp2a-1-2) and the pp2a-2 mutant has a 1-bp deletion in the 11th exon. The pp2a-3, pp2a-4, and pp2a-5 mutants contained edited sequenced in the first exon with a 1-bp insertion, 2-bp insertion, and 1-bp deletion, respectively (Figure 2A). After inoculation with R. solani AG1-IA, pp2a-1 genome editing mutants in ZH11 background were more susceptible than ZH11 plants, showing obvious chlorosis (Figure 2B), while other pp2a genome editing mutants in DJ background had no obvious disease grade differences compared with that in wild-type plants (DJ) (Figure 2B). The lesion coverage on leaves of ZH11 (wild-type), pp2a-1-1, and pp2a-1-2 were 34.1, 60.8, and 59.2%, respectively. However, the lesion area on DJ, pp2a-2, pp2a-3, pp2a-4, and pp2a-5 were 48.2, 49.3, 50.1, 49.5, and 48.4%, respectively (Figure 2C).
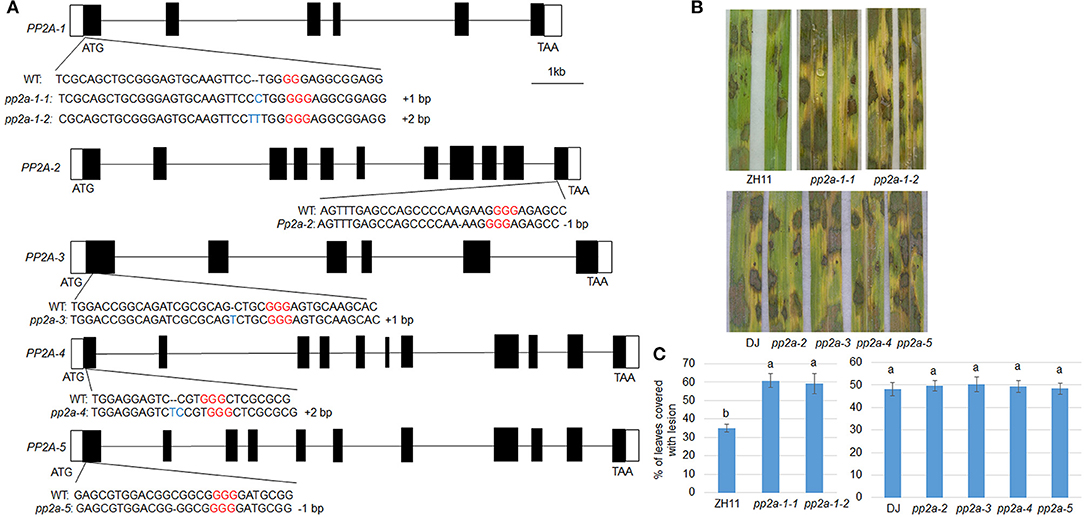
Figure 2. The genomic structure and defense response of PP2As. (A) Black and white boxes indicate the exon and UTR regions, respectively, while the lines indicate introns. The wild-type (WT) and CRISPR/Cas9-induced edited sequences are shown below the exon. (B) The leaves from the ZH11, pp2a-1 (#1, #2), DJ, pp2a-2, pp2a-3, pp2a-4, and pp2a-5 were inoculated with R. solani AG1-IA. (C) Shows the leaf surface lesion areas. Data show the average ± SE (n > 10). Different letters above the bars denote statistically significant differences (P < 0.05).
Since pp2a-1 mutants were susceptible to ShB, the PP2A-1 overexpression plants were further examined in response to ShB. We isolated a PP2A-1 activation tagging line (PP2A-1 OX) in a T-DNA insertional library (Jeong et al., 2002). In the activation tagging line, T-DNA was inserted in the promoter region in which four copies of the 35S promoter activated the PP2A-1 expression (Figure 3A). The qRT-PCR results indicated that PP2A-1 expression was significantly higher in hetero- and homozygous PP2A-1 activation tagging plants than wild type and the PP2A-1 expression level was significantly higher in homozygous compared to heterozygous plants (Figure 3B). PP2A-1 OX lines displayed a semi-dwarf phenotype, with homozygous plants significantly shorter than heterozygous and wild-type plants (Figure 3C). Next, we selected the homozygous PP2A-1 OX to inoculate R. solani AG1-IA. The lesion area of PP2A-1 homozygous overexpression plants was smaller than that of DJ (wild-type) after 48 h of inoculation (Figure 3D). The lesion coverage on leaves was 48.1 and 30.8%, respectively (Figure 3E), indicating that overexpression of PP2A-1 enhanced rice resistance to ShB. In addition, PP2A-1-GFP and free GFP were expressed in tobacco leaves, and PP2A-1-GFP signal was detected in the cytoplasm and nucleus (Figure 3F).
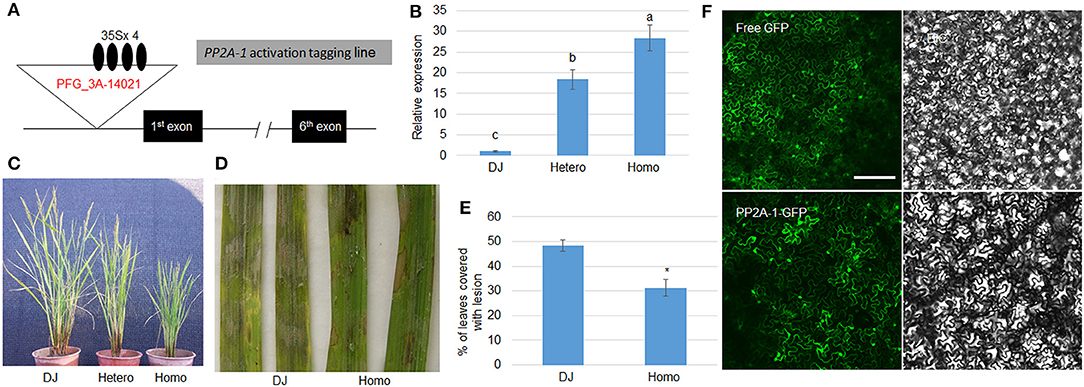
Figure 3. Genomic structure and defense response of PP2A-1 overexpressor and PP2A-1 subcellular localization. (A) Genomic structure of PP2A-1 activation tagging (PP2A-1 OX) line. Black boxes are demonstrative of exons and lines for introns. The T-DNA insertion site is shown as the white triangle and the black oval represents the 35S promoter. PFG_3A-14021 is the ID number of the mutant in the SALK database. (B) PP2A-1 expression levels in DJ (wild-type), PP2A-1 heterozygote, and homozygote plants. Normalization of expression was done with reference to the internal control ubiquitin. The experiments were done in triplicate. Significant changes are illustrated by various letters (P < 0.05). (C) Three-month-old DJ, PP2A-1 heterozygous, and homozygous plants. (D) Leaves from the DJ and PP2A-1 homozygous plants were inoculated with R. solani AG1-IA. (E) The lesion areas on the leaf surfaces are shown in (D). Data show the average ± SE (n > 10). Different letters above the bars denote statistically significant differences (P < 0.05). (F) Free GFP and PP2A-1-GFP were expressed in tobacco leaves. GFP and bright-field channels Scale bar = 20 μm.
PP2A-1 OX plants were less susceptible while pp2a-1 mutants were more susceptible to ShB compared to the wild-type control. The expression patterns of defense genes PBZ1 and PR1b in wild-type, pp2a-1, and PP2A-1 OX plants were examined following R. solani inoculation. qPCR results showed that there was no significant difference in the expression levels of PBZ1 and PR1b among wild-type, genome editing mutants, and overexpression lines with no R. solani inoculation. However, the expression level of PBZ1 in pp2a-1 mutants was significantly lower than that in control ZH11, while it was higher in PP2A-1 OX plants than that in wild-type DJ after 48 h of inoculation (Figure 4A). The expression pattern of PR1b was similar to that of PBZ1 at 48 h of inoculation, which showed lower and higher induction kinetics in pp2a-1 mutants and PP2A-1 OX compared to that in wild-type plants, respectively (Figure 4B).
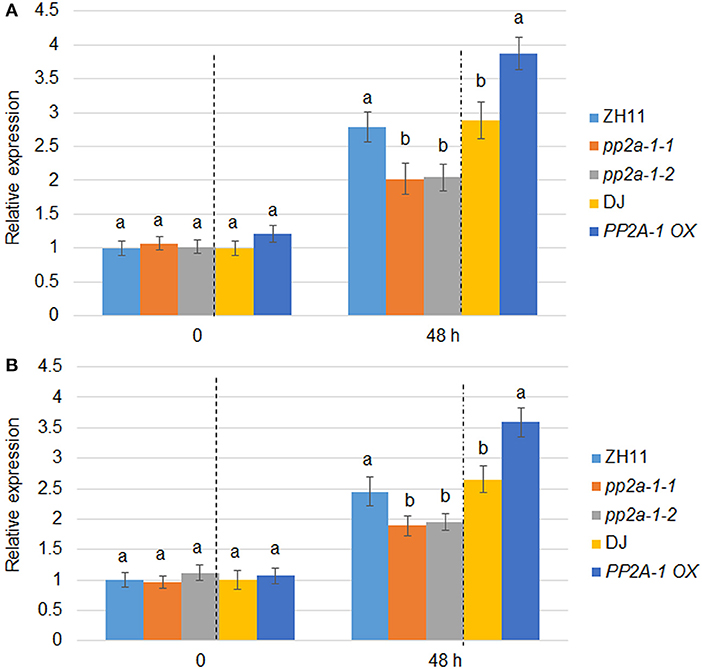
Figure 4. PBZ1 and PR1b expression patterns in wild-type, pp2a-1, and PP2A-1 overexpression plants. (A) PBZ1 and (B) PR1b expression after R. solani inoculation. The PBZ1 and PR1b gene expression patterns were evaluated after 0 and 48 h following R. solani inoculation. ZH11 is the control of pp2a-1-1 and pp2a-1-2, while DJ is the control of PP2A-1 OX plants. Normalization of expression was done with reference to the internal control ubiquitin. The experiments were done in triplicate. Significant changes are illustrated by various letters (P < 0.05).
Rice ShB caused by R. solani, bacterial leaf blight caused by Xanthomonas oryzae, and rice blast caused by Magnaporthe oryzae are three major diseases that significantly affect rice yield in China (Suryadi et al., 2013). The use of resistant varieties has been the primary means of disease control. However, due to the lack of resistant varieties and that sclerotium has a strong reproductive capacity, ShB control is challenging. Therefore, it is necessary to screen disease resistance genes and understand their resistance mechanism. Previous studies showed that PP2A regulates the development of lateral and primary roots, seed germination, and abiotic stress response against high concentration of sugar, salt, and drought (Yu et al., 2003; País et al., 2009; Liu et al., 2014; Hu et al., 2017). PP2A also plays important roles in biotic stress resistance. For example, AtPP2A is involved in regulation of PTI (pathogen-associated molecular pattern-triggered immunity) upon response to P. syringae pv. tomato (Pto) DC3000 infection; LePP2A gene was rapidly induced by inoculating with the model strain P. syringae pv. tomato; TaPP2A-4B and TaPP2A-4D may negatively regulate wheat defense response to R. cerealis infection by triggering the generation of ROS and PTI-mediated induction of PR genes (He et al., 2004; Segonzac et al., 2014; Durian et al., 2016; Zhu et al., 2018), suggesting that PP2A may be a key regulator of PAMP induced immunity. In rice, the induction of okadaic acid-dependent AMY3 and RCht2 (rice chitinase) transcription are regulated via the PP2A signal transduction pathway (Luan et al., 1993; Kim et al., 1998). However, the role of PP2A in rice disease resistance still remains unclear.
Our results indicate that the protein phosphatase 2A catalytic subunit OsPP2A-1 enhances resistance to sheath blight disease in rice. In our previous transcriptome analysis, OsPP2A-1 was significantly induced by R. solani inoculation (Yuan et al., 2020). The CRISPR/Cas9-mediated genome editing lines revealed higher susceptibility of pp2a-1 mutants to ShB, compared to wild-type control and other PP2A mutants (Figure 2). While PP2A-1 OX lines displayed a semi-dwarf phenotype, homozygous plants were significantly shorter than heterozygous and wild-type plants (Figure 3C). Inoculation of R. solani AG1-IA demonstrated that the PP2A-1 OX lines were less susceptible to ShB. The PP2As are ubiquitously expressed in different stages and tissues, and PP2A-1-GFP was localized at the cytosol and nucleus in tobacco leaves. The PP2A-1 expression level was significantly higher in the activation tagging line, with higher PP2A-1 expression inhibiting plant height while promoting ShB resistance.
The vital involvement of PP2A in responding to pathogens by plants has been demonstrated in recent studies (He et al., 2004; Zhu et al., 2018). For example, in A. thaliana, RLKs FLS2 (flagellin sensing receptor2) recognizes and EFR (EF-Tu receptor) is capable of recognizing the EF- Tu (elongation factor), both are PAMPs (pathogen-associated molecular pattern) of bacterial pathogens. The autophosphorylation and functioning of BAK1 (BRI1-associated kinase 1) is limited by PP2A-holoenzyme (Segonzac et al., 2014). PP2A-c4 and PP2A-a1 gene knockout mutants display a stronger resistance to virulent P. syringae pv. tomato DC3000 (Segonzac et al., 2014). The BSMV-VIGS (barley stripe mosaic virus–induced gene silencing) approach was applied to augment R. cerealis resistance in wheat attributed to TaPP2Ac-4B and TaPP2Ac-4D knock-down, suggesting the negative regulation of TaPP2A to wheat sharp eyespot. In many species, PP2A appears to function as a negative regulator, while OsPP2A-1 was found to positively regulate resistance to R. solani in this study. It seemed that the same gene may play different functions in response to infection of different types of pathogens. For example, WRKY transcription factors were more resistant to the hemibiotrophic bacterial pathogen P. syringae, but more susceptible to necrotrophic fungal pathogen B. cinerea in Arabidopsis (Xu et al., 2006), implying that PP2A-1 might play diverse functions when experiencing different stimuli. In our study, we confirmed that the CRISPR/Cas9-induced pp2a-1 genome editing mutants more susceptible to R. solani, while the other pp2as were similar to wild-type plants in response to R. solani infection. It may be valuable to dissect the associated molecular mechanism in the future research.
As mentioned earlier, PP2As comprises three subunits (A, B, and C). In the A. thaliana genome, these subunits are encoded by five genes of subunit C, three genes of subunit A, and 17 genes of subunit B (Farkas et al., 2007) to establish a minimum of 255 novel forms of the molecule. Immunity in plants is influenced by subunits A and B (with scaffolding and regulatory functioning, respectively). Resistance to P. syringae pv. tomato was augmented due to a subunit B-B′θ deficient mutation (Kataya et al., 2015). AtPP2A-B′γ enhances the negatively regulated defense against Myzus persicae (green peach aphid) and B. cinerea (a necrotrophic fungus) (Trotta et al., 2011; Rasool et al., 2014). PR protein phosphorylation (PR1, PR2–PR5) is augmented by mutations in subunit AtPP2A-B′γ (Trotta et al., 2011). The constitutive expression of PR1a, PR1b, and PR5 was induced by NbNPP4-1 and NbNPP4-2 silencing in N. benthamiana (He et al., 2004). PR2 levels were up-regulated by TaPP2A silencing (Zhu et al., 2018). PBZ1, a PR10 family protein accumulates in rice tissues which are in the process of cell mortality (Huang et al., 2016; Moselhy et al., 2016). In this study, PBZ1 and PR1b genes were up-regulated by R. solani infection, suggesting that that PBZ1 and PR1 play a role in ShB resistance in rice. The results indicated that R. solani-induced PBZ1 and PR1b expressions are under control of PP2A-1, suggesting that the expression of PP2A-1 might be through the activation of PR genes to promote rice defense.
Taken together, our findings suggest that the protein phosphatase 2A catalytic subunit, PP2A-1, regulates the defense response in rice to R. solani infection. This study revealed a new function of the rice PP2A in immune response, which provided a potential target for breeding ShB-resistant lines.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
QL, QM, and YX conceived and designed the studies and wrote the manuscript. QL, JC, VK, ZL, and DY collected and analyzed the data. All authors have read and approved the final version of the manuscript.
This work was supported by the Support Program for Science and Technology Innovation Talents of Shenyang (RC190489), and the Natural Science Foundation of Liaoning Province (2020-YQ-05).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Du, Y., Shi, Y., Yang, J., Chen, X., Xue, M., Zhou, W., et al. (2013). A serine/threonine-protein phosphatase PP2A catalytic subunit is essential for asexual development and plant infection in Magnaporthe oryzae. Curr. Genet. 59, 33–41. doi: 10.1007/s00294-012-0385-3
Durian, G., Rahikainen, M., Alegre, S., Brosche, M., and Kangasjarvi, S. (2016). Protein phosphatase 2A in the regulatory network underlying biotic stress resistance in plants. Front. Plant Sci. 7:812. doi: 10.3389/fpls.2016.00812
Farkas, I., Dombradi, V., Miskei, M., Szabados, L., and Koncz, C. (2007). Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci. 12, 169–176. doi: 10.1016/j.tplants.2007.03.003
Feng, Z., Zhang, B., Ding, W., Liu, X., Yang, D. L., Wei, P., et al. (2013). Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23, 1229–1232. doi: 10.1038/cr.2013.114
Gao, Y., Zhang, C., Han, X., Wang, Z. Y., Ma, L., Yuan, D. P., et al. (2018). Inhibition of OsSWEET11 function in mesophyll cells improves resistance of rice to sheath blight disease. Mol. Plant Pathol. 19, 2149–2161. doi: 10.1111/mpp.12689
He, X., Anderson, J. C., Pozo, O.d., Gu, Y.-Q., Tang, X., and Martin, G. B. (2004). Silencing of subfamily I of protein phosphatase 2a catalytic subunits results in activation of plant defenseresponses and localized cell death. Plant J. 38, 563–577. doi: 10.1111/j.1365-313X.2004.02073.x
Helliwell, E. E., Wang, Q., and Yang, Y. (2013). Transgenic rice with inducible ethylene production exhibits broad-spectrum disease resistance to the fungal pathogens Magnaporthe oryzae and Rhizoctonia solani. Plant Biotechnol. J. 11, 33–42. doi: 10.1111/pbi.12004
Hsu, P. D., Scott, D. A., Weinstein, J. A., Ran, F. A., Konermann, S., Agarwala, V., et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832. doi: 10.1038/nbt.2647
Hu, R., Zhu, Y., Wei, J., Chen, J., Shi, H., Shen, G., et al. (2017). Overexpression of PP2A-C5 that encodes the catalytic subunit 5 of protein phosphatase 2A in Arabidopsis confers better root and shoot development under salt conditions. Plant Cell Environ. 40, 150–164. doi: 10.1111/pce.12837
Huang, L. F., Lin, K. H., He, S. L., Chen, J. L., Jiang, J. Z., Chen, B. H., et al. (2016). Multiple patterns of regulation and overexpression of a ribonuclease-like pathogenesis-related protein gene, OsPR10a, conferring disease resistance in rice and Arabidopsis. PLoS ONE 11:e0156414. doi: 10.1371/journal.pone.0156414
Jeong, D. H., An, S., Kang, H. G., Moon, S., Han, J. J., Park, S., et al. (2002). T-DNA insertional mutagenesis for activation tagging in rice. Plant physiology 130, 1636–1644. doi: 10.1104/pp.014357
Jimmy, J. L., and Babu, S. (2019). Gene network mediated by WRKY13 to regulate resistance against sheath infecting fungi in rice (Oryza Sativa L.). Plant Sci. 280, 269–282. doi: 10.1016/j.plantsci.2018.12.017
Kataya, A. R. A, Heidari, B., and Lillo, C. (2015). Protein phosphatase 2A regulatory subunits affecting plant innate immunity, energy metabolism, and flowering time—joint functions among B'η subfamily members. Plant Signal. Behav. 10:e1026024. doi: 10.1080/15592324.2015.1026024
Kim, C. Y., Gal, S. W., Choe, M. S., Jeong, S. Y., Lee, S. I., Cheong, Y. H., et al. (1998). A new class II rice chitinase, Rcht2, whose induction by fungal elicitor is abolished by protein phosphatase 1 and 2A inhibitor. Plant Mol. Biol. 37, 523–534. doi: 10.1023/A:1005960313459
Kim, J.-G., Li, X., Roden, J. A., Taylor, K. W., Aakre, C. D., Su, B., et al. (2009). Xanthomonas T3S effector XopN suppresses PAMP-triggered immunity and interacts with a tomato atypical receptor-like kinase and TFT1. Plant Cell 21, 1305–1323. doi: 10.1105/tpc.108.063123
Kouzai, Y., Kimura, M., Watanabe, M., Kusunoki, K., Osaka, D., Suzuki, T., et al. (2018). Salicylic acid-dependent immunity contributes to resistance against Rhizoctonia solani, a necrotrophic fungal agent of sheath blight, in rice and Brachypodium distachyon. New Phytol. 217, 771–783. doi: 10.1111/nph.14849
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for Bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, N., Kong, L., Zhou, W., Zhang, X., Wei, S., Ding, X., et al. (2013). Overexpression of Os2H16 enhances resistance to phytopathogens and tolerance to drought stress in rice. Plant Cell Tissue Organ Cult. 115, 429–441. doi: 10.1007/s11240-013-0374-3
Li, N., Wei, S., Chen, J., Yang, F., Kong, L., Chen, C., et al. (2018). OsASR2 regulates the expression of a defence-related gene, Os2H16, by targeting the GT-1 cis-element. Plant Biotechnol. J. 16, 771–783. doi: 10.1111/pbi.12827
Liu, D., Li, A., Mao, X., and Jing, R. (2014). Cloning and characterization of TaPP2Ab B”-Alpha, a member of the PP2A regulatory subunit in wheat. PLoS ONE 9:e94430. doi: 10.1371/journal.pone.0094430
Liu, Z., Liu, N., Jiang, H., Yan, L., Ma, Z., and Yin, Y. (2018). The activators of type 2A phosphatases (PP2A) regulate multiple cellular processes via PP2A-dependent and -independent mechanisms in Fusarium graminearum. Mol. Plant Microbe Interact. 31, 1121–1133. doi: 10.1094/MPMI-03-18-0056-R
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luan, S., Li, W., Rusnak, F., Assmann, S. M., and Schreiber, S. L. (1993). Immunosuppressants implicate protein phosphatase regulation of K+ channels in guard cells. Proc. Natl. Acad. Sci. U.S.A. 90, 2202–2206. doi: 10.1073/pnas.90.6.2202
Ma, X., Chen, L., Zhu, Q., Chen, Y., and Liu, Y. G. (2015). Rapid decoding of sequence-specific nuclease-induced heterozygous and biallelic mutations by direct sequencing of PCR products. Mol. Plant 8, 1285–1287. doi: 10.1016/j.molp.2015.02.012
Maeda, S., Dubouzet, J. G., Kondou, Y., Jikumaru, Y., Seo, S., Oda, K., et al. (2019). The rice CYP78A gene BSR2 confers resistance to Rhizoctonia solani and affects seed size and growth in Arabidopsis and Rice. Sci. Rep. 9:587. doi: 10.1038/s41598-018-37365-1
Mao, B., Liu, X., Hu, D., and Li, D. (2014). Co-expression of RCH10 and AGLU1 confers rice resistance to fungal sheath blight Rhizoctonia solani and blast Magnorpathe oryzae and reveals impact on seed germination. World J. Microbiol. Biotechnol. 30, 1229–1238. doi: 10.1007/s11274-013-1546-3
Máthé, C., Garda, T., Freytag, C., and Hamvas, M. M. (2019). The role of serine-threonine protein phosphatase PP2A in plant oxidative stress signaling—facts and hypotheses. Int. J. Mol. Sci. 20:3028. doi: 10.3390/ijms20123028
Moselhy, S. S., Asami, T., Abualnaja, K. O., Al-Malki, A. L., Yamano, H., Akiyama, T., et al. (2016). Spermidine, a polyamine, confers resistance to rice blast. J. Pestic. Sci. 41, 79–82. doi: 10.1584/jpestics.D16-008
Nishimura, A., Aichi, I., and Matsuoka, M. (2006). A protocol for agrobacterium-mediated transformation in rice. Nat. Protoc. 1, 2796–2802. doi: 10.1038/nprot.2006.469
País, S. M., González, M. A., Téllez-Iñón, M. T., and Capiati, D. A. (2009). Characterization of potato (Solanum tuberosum) and tosdmato (Solanum lycopersicum) protein phosphatases type 2A catalytic subunits and their involvement in stress responses. Planta 230, 13–25. doi: 10.1007/s00425-009-0923-5
Peng, X., Hu, Y., Tang, X., Zhou, P., Deng, X., Wang, H., et al. (2012). Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 236, 1485–1498. doi: 10.1007/s00425-012-1698-7
Peng, X., Wang, H., J.-,Jang, C., Xiao, T., He, H., Jiang, D., et al. (2016). OsWRKY80-OsWRKY4 module as a positive regulatory circuit in rice resistance against Rhizoctonia solani. Rice 9:63. doi: 10.1186/s12284-016-0137-y
Prasad, B., and Eizenga, G. C. (2008). Rice sheath blight disease resistance identified in Oryza spp. Accessions. Plant Dis. 92, 1503–1509. doi: 10.1094/PDIS-92-11-1503
Rahikainen, M., Pascual, J., Alegre, S., Durian, G., and Kangasjärvi, S. (2016). PP2A phosphatase as a regulator of ROS signaling in plants. Antioxidants 5:8. doi: 10.3390/antiox5010008
Rasool, B., Karpinska, B., Konert, G., Durian, G., Denessiouk, K., Kangasjarvi, S., et al. (2014). Effects of light and the regulatory B-subunit composition of protein phosphatase 2A on the susceptibility of Arabidopsis thaliana to aphid (Myzus persicae) infestation. Front. Plant Sci. 5:405. doi: 10.3389/fpls.2014.00405
Savary, S., Castilla, N. P., Elazegui, F. A., McLaren, C. G., Ynalvez, M. A., and Teng, P. S. (1995). Direct and indirect effects of nitrogen supply and disease source structure on rice sheath blight spread. Phytopathology 85, 959–965. doi: 10.1094/Phyto-85-959
Savary, S., Willocquet, L., Elazegui, F. A., Castilla, N. P., and Teng, P. S. (2000). Rice pest constraints in tropical asia: quantification of yield losses due to rice pests in a range of production situations. Plant Dis. 84, 357–369. doi: 10.1094/PDIS.2000.84.3.357
Segonzac, C., Macho, A. P., Sanmartin, M., Ntoukakis, V., Sanchez-Serrano, J. J., and Zipfel, C. (2014). Negative control of BAK1 by protein phosphatase 2a during plant innate immunity. EMBO J. 33, 2069–2079. doi: 10.15252/embj.201488698
Shah, J. M., Raghupathy, V., and Veluthambi, K. (2009). Enhanced sheath blight resistance in transgenic rice expressing an endochitinase gene from Trichoderma virens. Biotechnol. Lett. 31, 239–244. doi: 10.1007/s10529-008-9856-5
Singh, P., Mazumdar, P., Harikrishna, J. A., and Babu, S. (2019). Sheath blight of rice: a review and identification of priorities for future research. Planta 250, 1387–1407. doi: 10.1007/s00425-019-03246-8
Sun, Q., Li, D. D., Chu, J., Yuan, D. P., Li, S., Zhong, L. J., et al. (2020). Indeterminate domain proteins regulate rice defense to sheath blight disease. Rice 13:15. doi: 10.1186/s12284-020-0371-1
Sun, Q., Li, T. Y., Li, D. D., Wang, Z. Y., Li, S., Li, D. P., et al. (2019). Overexpression of loose plant architecture 1 increases planting density and resistance to sheath blight disease via activation of PIN-FORMED 1a in rice. Plant Biotechnol. J. 17, 855–857. doi: 10.1111/pbi.13072
Suryadi, Y., Susilowati, D. N., Kadir, T. S., Zaffan, Z. R., and Hikmawati, N. Mubarik, N. R. (2013). Bioformulation of antagonistic bacterial consortium for controlling blast, sheath blight and bacterial blight diseases on rice. Asian J. Plant Pathol. 7, 92–108. doi: 10.3923/ajppaj.2013.92.108
Taheri, P., and Tarighi, S. (2011). Cytomolecular aspects of rice sheath blight caused by Rhizoctonia solani. Eur. J. Plant Pathol. 129, 511–528. doi: 10.1007/s10658-010-9725-7
Tiwari, M., Srivastava, S., Singh, P. C., Mishra, A. K., and Chakrabarty, D. (2020). Functional characterization of tau class glutathione-S-transferase in rice to provide tolerance against sheath blight disease. 3 Biotech 10:84. doi: 10.1007/s13205-020-2071-3
Trotta, A., Wrzaczek, M., Scharte, J., Tikkanen, M., Konert, G., Rahikainen, M., et al. (2011). Regulatory subunit B'gamma of protein phosphatase 2A prevents unnecessary defense reactions under low light in Arabidopsis. Plant physiol. 156, 1464–1480. doi: 10.1104/pp.111.178442
Wang, H., Meng, J., Peng, X., Tang, X., Zhou, P., Xiang, J., et al. (2015). Rice WRKY4 acts as a transcriptional activator mediating defense responses toward Rhizoctonia solani, the causing agent of rice sheath blight. Plant Mol. Biol. 89, 157–171. doi: 10.1007/s11103-015-0360-8
Xu, X., Chen, C., Fan, B., and Chen, Z. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant cell 18, 1310–1326. doi: 10.1105/tpc.105.037523
Yellareddygari, S, Reddy, M. S., Kloepper, J. W., Lawrence, K. S., and Fadamiro, H. (2014). Rice sheath blight: a review of disease and pathogen management approaches. J. Plant Pathol. Microbiol. 05:1000241. doi: 10.4172/2157-7471.1000241
Yu, R. M., Wong, M. M., Jack, R. W., and Kong, R. Y. (2005). Structure, evolution, and expression of a second subfamily of protein phosphatase 2A catalytic subunit genes in the rice plant (Oryza sativa L.). Planta 222, 757–768. doi: 10.1007/s00425-005-0018-x
Yu, R. M. K., Zhou, Y., Xu, Z.-F., Chye, M.-L., and Kong, R. Y. C. (2003). Two genes encoding protein phosphatase 2A catalytic subunits are differentially expressed in rice. Plant Mol. Biol. 51, 295–311. doi: 10.1023/a:1022006023273
Yuan, D. P., Xu, X. F., Hong, W.-J., Wang, S. T., Jia, X. T., Liu, Y., et al. (2020). Transcriptome analysis of rice leaves in response to Rhizoctonia solani infection and reveals a novel regulatory mechanism. Plant Biotechnol. Rep. 14, 559–573. doi: 10.1007/s11816-020-00630-9
Zhu, G., Liang, E., Lan, X., Li, Q., Qian, J., Tao, H., et al. (2019). ZmPGIP3 gene encodes a polygalacturonase-inhibiting protein that enhances resistance to sheath blight in rice. Phytopathology 109, 1732–1740. doi: 10.1094/PHYTO-01-19-0008-R
Keywords: PP2A-1, sheath blight, resistance, enhance, rice
Citation: Lin QJ, Chu J, Kumar V, Yuan DP, Li ZM, Mei Q and Xuan YH (2021) Protein Phosphatase 2A Catalytic Subunit PP2A-1 Enhances Rice Resistance to Sheath Blight Disease. Front. Genome Ed. 3:632136. doi: 10.3389/fgeed.2021.632136
Received: 22 November 2020; Accepted: 08 February 2021;
Published: 25 February 2021.
Edited by:
Huanbin Zhou, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Chuanxiao Xie, Chinese Academy of Agricultural Sciences, ChinaCopyright © 2021 Lin, Chu, Kumar, Yuan, Li, Mei and Xuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiong Mei, bWVpcWlvbmdAc3lhdS5lZHUuY24=; Yuan Hu Xuan, eHVhbnl1YW5odTExNUBzeWF1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.