
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genome Ed. , 21 January 2021
Sec. Genome Editing in Plants
Volume 2 - 2020 | https://doi.org/10.3389/fgeed.2020.627803
This article is part of the Research Topic Targeted Genome Editing in Crops View all 12 articles
Biotic diseases cause substantial agricultural losses annually, spurring research into plant pathogens and strategies to mitigate them. Nicotiana benthamiana is a commonly used model plant for studying plant–pathogen interactions because it is host to numerous plant pathogens and because many research tools are available for this species. The clustered regularly interspaced short palindromic repeats (CRISPR) system is one of several powerful tools available for targeted gene editing, a crucial strategy for analyzing gene function. Here, we demonstrate the use of various CRISPR-associated (Cas) proteins for gene editing of N. benthamiana protoplasts, including Staphylococcus aureus Cas9 (SaCas9), Streptococcus pyogenes Cas9 (SpCas9), Francisella novicida Cas12a (FnCas12a), and nCas9-activation-induced cytidine deaminase (nCas9-Target-AID). We successfully mutated Phytoene Desaturase (PDS) and Ethylene Receptor 1 (ETR1) and the disease-associated genes RNA-Dependent RNA Polymerase 6 (RDR6), and Suppressor of Gene Silencing 3 (SGS3), and confirmed that the mutated alleles were transmitted to progeny. sgs3 mutants showed the expected phenotype, including absence of trans-acting siRNA3 (TAS3) siRNA and abundant expression of the GFP reporter. Progeny of both sgs3 and rdr6 null mutants were sterile. Our analysis of the phenotypes of the regenerated progeny indicated that except for the predicted phenotypes, they grew normally, with no unexpected traits. These results confirmed the utility of gene editing followed by protoplast regeneration in N. benthamiana. We also developed a method for in vitro flowering and seed production in N. benthamiana, allowing the regenerants to produce progeny in vitro without environmental constraints.
Nicotiana benthamiana is a host to many plant pathogens, especially viruses, and is widely used to study plant–pathogen interactions (Goodin et al., 2008). Many tools for functional genomics are available for this species, including viral vectors, RNA interference (RNAi), ethyl methanesulfonate mutagenesis, agroinfiltration, protoplast transfection, and Agrobacterium-mediated stable transformation. These tools are useful for research in genomics, biochemistry, metabolomics, cell biology, and pathology, as well as other topics in agriculture (Derevnina et al., 2019).
Notwithstanding its many advantages, the fact that N. benthamiana is allotetraploid, with a very large genome (3.1 Gb) (Bombarely et al., 2012), makes it difficult to edit the genome of this plant and to obtain mutants for plant biological and gene functional studies. We chose to address this problem by using the powerful genome-editing tool CRISPR-Cas (clustered regularly interspaced short palindromic repeats-CRISPR-associated protein). CRISPR-Cas core technology involves programmable DNA cleavage by the Cas protein at DNA sites specified by the targeting sequence in a guide RNA (gRNA; review by Yue et al., 2020). The use of CRISPR-Cas has greatly accelerated plant research and crop breeding in recent years (Li et al., 2013; Nekrasov et al., 2013; Shan et al., 2013; Li and Xia, 2020; Yue et al., 2020).
Most genome editing studies in plants, including N. benthamiana, have involved Agrobacterium-mediated stable transformation to deliver DNA into target cells in order to express Cas protein and gRNA. However, mutant plants derived from Agrobacterium-mediated transformation could be considered genetically modified organisms (GMOs), especially for vegetatively propagated crops in which the transgenes cannot be removed from the genome by crossing. In dicots, however, most transformants are chimeric, and the edited allele cannot be transmitted to the progeny when the edited cells exist only in vegetative organs. Thus, as a less controversial alternative, plasmids encoding the Cas and gRNA sequences or pre-assembled Cas:gRNA ribonucleoprotein complexes (RNPs) can be delivered directly into protoplasts using transient transfection. Because the protoplast is a single cell, once gene editing has been performed, the entire regenerant derived from this edited protoplast will contain the same edited gene (Woo et al., 2015; Lin et al., 2018; Hsu et al., 2019). Although a similar type of delivery can also be achieved by particle bombardment, polyethylene glycol (PEG)-mediated protoplast transfection offers high transfection efficiency and high viability for robust gene editing while generating recombinant-DNA-free plants to circumvent GMO issues (Woo et al., 2015; Andersson et al., 2018; Lin et al., 2018).
The main bottleneck of this strategy, however, is protoplast regeneration. We previously established a protoplast regeneration system and a CRISPR-Cas gene editing system for polyploid tobacco (N. tabacum) (Lin et al., 2018; Hsu et al., 2019). Here we report a simple, highly robust protocol for streamlined CRISPR-mediated genome editing in N. benthamiana. This protocol, together with CRISPR genome editing and improved genomics resources, ushers in a new era of forward and reverse genetic analyses of this valuable model plant system.
Sterile N. benthamiana plantlets were propagated by cutting and grown in half-strength Murashige and Skoog (1/2 MS) medium supplemented with 30 mg/L sucrose and 1% agar, pH 5.7. These plantlets were incubated in a 26°C culture room (12 h light /12 h dark cycle) with a light density of 75 μmol m−2 s−1. The plantlets were subcutlured into fresh medium every month. For comparison with the seedlings derived from protoplasts and seed propagation, seeds were sown in 3-inch pots with peat moss, vermiculite and perlite in a ratio of 5:1:1. Each treatment had five repeats.
The protoplast isolation and transfection followed our previously published method with minor modification (Hsu et al., 2019). The protoplasts were isolated from the mature leaves of in vitro plantlets. Five to seven leaves (about 0.2–0.25 g) were used for 106 protoplast isolation. These leaves were put into a 6-cm glass petri dish with 10 ml digestion solution (1/4 MS liquid medium containing 1% cellulose, 0.5% Macerozyme, 3% sucrose and 0.4 M mannitol, pH 5.7) and cut into 0.5 cm-wide strips. The solution was incubated at room temperature in the dark overnight. The digested solution was diluted with 10 ml W5 (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES, and 5 mM glucose) solution and filtered by 40 μm nylon mesh. The solution was centrifuged at low-speed (200 × g) for 3 min to collect the protoplasts. The protoplasts were purified with 20% sucrose solution and washed in W5 solution three times. The protoplasts were transferred to a transfection buffer (1/2 MS solution supplemented with 3% sucrose, 0.4 M mannitol, 1 mg/L naphthaleneacetic acid (NAA), and 0.3 mg/L kinetin, 5 mM MES, pH 5.7) and the cell concentration was adjusted to 3 × 105/mL.
The protoplasts were transfected with plasmids by PEG-mediated transfection (Woo et al., 2015; Lin et al., 2018). CRISPR reagent DNA (40 μg in 40 μl) was added to 400 μl (1.2 × 105 protoplasts) and mixed carefully. Then the same volume of PEG solution was added and mixed, then left to stand for 30 min. To end the reaction, 3 ml of W5 was added and mixed well. The transfected protoplasts were collected by centrifugation at 200 × g for 3 min. The protoplasts were washed in 3 ml of W5 by centrifugation at 200 × g for 3 min.
Several target sites in N. benthamiana whose editing efficiencies have been confirmed in N. tabacum (Hsu et al., 2019) as well as new constructions were used in this study. The following Cas proteins and target genes were tested in N. benthamiana:
1. SaCas9: The binary plasmid (gPDA_Sa) was published by Kaya et al. (2016). The target gene is Phytoene Desaturase 1 (NbPDS-1), the target site is TTGCGATGCCTAACAAGCCAG.
2. FnCas12a: The binary plasmids (crNtPDS-1 and crNtPDS-2) were published by Endo et al. (2016). Target genes are NbPDS-1 and NbPDS-2, and the target sites are TCATCCAGTCCTTAACACTTAAAC(crNtPDS-1), and ACATGGCAATGAACACCTCATCTG (crNtPDS-1).
3. nCas9-Target-AID: The plasmid (pDicAID_nCas9-PmCDA-2A-NptII_ETR) is published in Shimatani et al. (2017) (Addgene ID: 91695). The target genes are NbETR1-1 and NbETR1-2. The target site is TGCACAAGAACCCATCTATA.
4. SpCas9: the vector commonly used for dicot transformation (pYLCRISPR/Cas9P35S-N) is used (Ma et al., 2015). The target genes were RNA-dependent RNA Polymerase 6 (NbRDR6-1 and NbRDR6-2), and Suppressor of Gene Silencing 3 (NbSGS3-1 and NbSGS3-2). For convenience, to validate the presence and efficiency of the mutations, double sgRNAs were present in a single construct for each gene (NbSGS3: AAGCAGTGCTGGGAAGCAAT, CTCATGCCACGATGGCCTTG; NbRDR6: GCCATGGCCTTCTCAAAGCT, GCAGTTCTATAGAAAACCAA). The sgRNAs were cloned into vectors.
Pooled protoplast DNA was used as a template to amplify the target genes for validation by sequencing. The putatively edited protoplasts were transferred to 5 cm diameter Petri dishes containing 3 ml 1/2 MS liquid medium supplemented with 3% sucrose, 0.4 M mannitol, 1 mg/L NAA, and 0.3 mg/L kinetin (1N0.3K) for plant regeneration. Callus formation occurred using protoplasts after 1 month of incubation in the dark. The calluses were subcultured in 9 cm diameter Petri dishes containing fresh medium with 1 mg/L 6-benzylaminopurine (1B) for 3–4-weeks in the light. Calluses that had turned green were then transferred to solid medium containing the same plant growth regulators. The explants were subcultured every 4 weeks until shoots formed after several subcultures. The shoots were subcultured in solid root medium (HB1: 3 g/L Hyponex No. 1, 2 g/L tryptone, 20 g/L sucrose, 1 g/L activated charcoal, 10 g/L Agar, pH 5.2). Adventitious roots formed at the bottoms of the containers (Figure 1).

Figure 1. Protoplast regeneration in N. benthamiana. The gray background indicates that the explants were incubated in the dark. All liquid media were supplemented with 0.4 M mannitol. The abbreviated names of the media are shown in the top left corner of the images. 1N0.3K: 1/2 MS basal medium supplemented with 1 mg/L NAA and 0.3 mg/L kinetin. 1B: ½ MS basal medium supplemented with 1 mg/L BA and 20 g/L sucrose. All solid media were solidified with 7 g/L Phytagel. 2B: 1/2 MS basal medium supplemented with 2 mg/L BA. HB1: 3 g/L Hyponex No. 1 (N:P:K = 7:6:19), 2 g/L tryptone, 20 g/L sucrose, and 1 g/L activated charcoal. (a) Transfected mesophyll protoplasts. Bar = 200 μm. (b) Microcallus formation after 1 month of incubation in 1N0.3K. Bar = 1 cm. (c) Microcallus amplification. (d) Microcalli transferred to 1B medium and incubated in the light. Bar = 1 cm. (e) Callus amplification in 2B. Bar = 1 cm. (f) Shoot formation in 2B. Bar = 1 cm. (g) Shoots incubated in HB1 for root formation. (h) In vitro seed formation. Bar = 5 mm. (i) Plantlet incubated in a growth chamber. Bar = 1 cm.
Two pairs of primers were designed to amplify the sgRNA-targeted DNA region for each target gene. PCR conditions were 94°C for 5 min, 35 cycles of denaturing (94°C for 30 s), annealing (55°C for 30 s), and polymerization (72°C for 30 s), followed by an extension reaction at 72°C for 5 min. The PCR product was sequenced by the Sanger method to determine the mutagenesis. The multiple sequences derived from mutated regenerated plants were separated using Poly Peak Parser (http://yosttools.genetics.utah.edu/PolyPeakParser/; Hill et al., 2014) or further confirmed by sequential T/A cloning and sequencing.
For protoplast regeneration, we placed protoplasts isolated from the leaves of in vitro-grown shoots (Figure 1a) in 1N0.3K liquid medium, incubated them in the dark for 1 month (Figure 1b), and then transferred them to fresh 1N0.3K medium and incubated them in the dark for another month (Figure 1c). Unlike in our previous method described for tobacco (Lin et al., 2018), we incubated N. benthamiana calluses directly in liquid 1B medium in light without embedding (Figure 1d). This step avoids the difficulty associated with embedding; however, it has the disadvantage that the calluses stick together and sometimes cannot be distinguished. After 1 month, we transferred the calluses larger than 3 mm to 1B solid medium and incubated them in light (Figure 1e). After several subcultures, shoots formed on the surface of the calluses (Figure 1f); this took more time for N. benthamiana than it does for N. tabacum (Lin et al., 2018). We then subcultured the shoots in solid HB1 medium and observed that adventitious roots formed without the need for plant growth regulators (Figure 1g). These plants could be further incubated successfully in test tubes, where they flowered and produced seeds (Figure 1h), or were transferred to a growth chamber for further growth (Figure 1i). The time required from protoplast isolation to regeneration was ~4–6 months.
To look for unexpected phenotypes in the regenerants, we randomly selected three regenerated plants (protoplasts #1, #2, and #3) and harvested their seeds. We grew the progeny in a growth chamber for 40 days and compared them to seedlings derived from seed propagation (seed #1, #2, and #3). We observed no significant differences in plant height between the regenerants and seed-derived plants (Figure 2). All plants flowered and produced seeds normally.
To demonstrate that this protocol can be used for CRISPR-mediated gene editing, we performed protoplast transfections using plasmids previously shown to be effective in N. tabacum (Hsu et al., 2019). We successfully used SaCas9, FnCas12a, and nCas9-Target-AID to obtain regenerated plants for target gene editing using this protocol. The efficiency of N. benthamiana transformation was similar to that of N. tabacum (Figure 3, Hsu et al., 2019). As in N. tabacum, three different Cas proteins were successfully used to edit different target genes simultaneously in a single N. benthamiana protoplast. The target site of SaCas9 has a mismatch in NbPDS-2, and there was 10.0% off-target editing of NbPDS-2 (Figure 3). In nCas9-Target-AID, we only observed mutation and no C to T editing regenerant was obtained. We designed NbSGS3 and NbRDR6 sgRNAs that can be used in both N. tabacum and N. benthamiana and introduced them into the SpCas9 plasmid. The regeneration results indicated that, except for sgRNA 2 in RDR6, these sgRNAs had target mutagenesis efficiencies in N. benthamiana (Figure 3) and N. tabacum (data not shown).
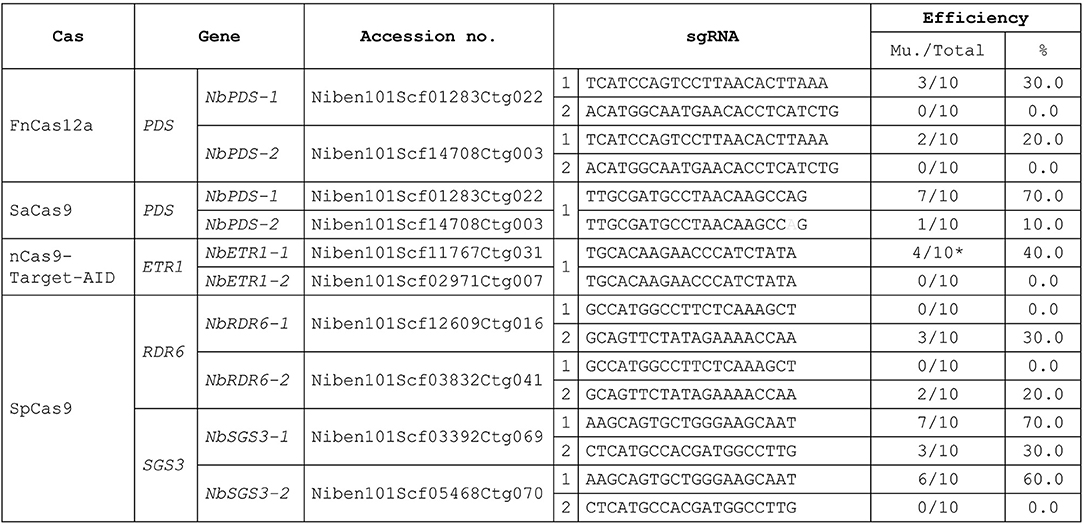
Figure 3. Target mutagenesis efficiencies. Mu, mutants. Gray: sgRNA mismatch. *: mutation. There were 10 regenerated plants (Total) analysis in each transfection. The mutants were confirmed by Sanger sequencing. % = No. of mutants/total no. of regenerated plants analyzed) X 100.
For the PDS gene study, we used SaCas9 and FnCas12a for targeted mutagenesis. When we used SaCas9 alone, since there was a mismatch in NbPDS-2 (Niben101Scf14708Ctg003), we obtained no NbPDS-1 and NbPDS-2 double knock out mutants, and found nbpds-1 null mutants with wild-type or heterozygous NbPDS-2. These mutants did not appear albino, the usual phenotype for this mutant, because NbPDS-2 was still functional, unlike our previous findings with N. tabacum in which double knock out mutants were obtained using the same plasmid (Lin et al., 2018). Although the mutagenesis efficiency was low, because the target sequences of NbPDS-1 and NbPDS-2 are identical, we still obtained albino nbpds-1 nbpds-1/nbpds-2 nbpds-2 double null mutants in regenerants derived from FnCas12a-mediated transfection. We also obtained NbPDS-1 nbpds-1/nbpds-2 nbpds-2 heterozygous T0 plants when the protoplasts were transfected with three plasmids (SaCas9, FnCas12a, and Target-AID). Albino mutants were detected in the T1 offspring, and their proportions and genotypes were as expected.
The homozygous nbrdr6-1 nbrdr6-2 double mutant derived from protoplast regeneration (nbrdr6#C13) was sterile, as are genome-edited mutants obtained via Agrobacterium-mediated transformation (Ludman and Fátyol, 2019; Matsuo and Atsumi, 2019), because they fail to produce seeds. Interestingly, two N. benthamiana sgs3-1 sgs3-2 biallelic mutants (nbsgs3-14 and nbsgs3-16) both produced seeds. We identified four editing “scars” in nbsgs3-14 (Figure 4A): a 1-bp substitution (E) and a 1-bp insertion (a) in NbSGS3-1, and a 5-bp deletion (d) and a 1-bp insertion in NbSGS3-2 (Figure 4B). Progeny with the EE/aa genotype could produce seeds, but dd/aa plants bore abnormal flowers that failed to produce fertile seeds (Figure 4C), as did the nbsgs3-14 aa/at progeny. All nbsgs3-14 progeny, regardless of their genotype, exhibited lower expression of the trans-acting secondary siRNA TAS3 than the wild type (Figure 4D). In sterile progeny (dd/aa in nbsgs3-14, aa/at in nbsgs3-16), no TAS3 siRNA was detected. These results indicate that RNA silencing was aberrant in the nbsgs3 mutants. RDR6 and SGS3 function in RNA silencing by reducing foreign gene expression. Similar to the enhanced transgene expression observed in Agrobacterium-infiltrated N. benthamiana rdr6 mutants (Ludman and Fátyol, 2019; Matsuo and Atsumi, 2019), GFP accumulated to higher levels in nbsgs3-16 than in wild type (Figure 4B).
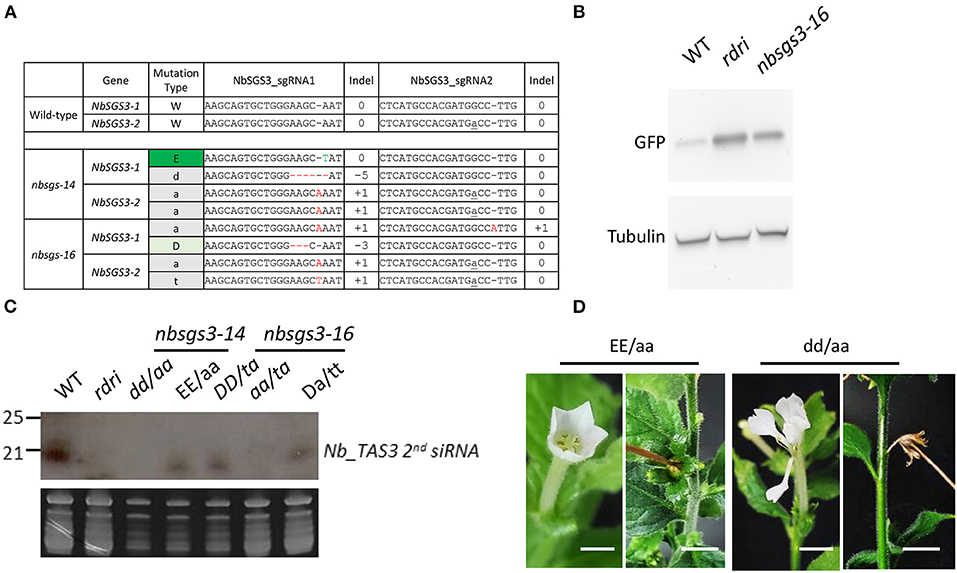
Figure 4. Mutants regenerated from CRISPR-Cas-edited Nicotiana benthamiana protoplasts. (A) Genotypes of F1 N. benthamiana sgs3 mutants used for small RNA analysis. NbSGS3-1: Niben101Scf03392Ctg069; NbSGS3-2: Niben101Scf05468Ctg070. Red -: deleted nucleotide. Letter in gray: inserted nucleotide. Letter in green: edited nucleotide. (B) Five-week-old N. benthamiana plants subjected to Agrobacterium-mediated transient infiltration with 1 OD Agrobacterium cultures (OD600 = 1) harboring binary vector with Green fluorescence protein (GFP) driven by the cauliflower mosaic virus 35S promoter. Leaves were harvested 3 d after infiltration. GFP and tubulin levels were analyzed by immunoblot analysis. (C) RNA gel blot analysis of the progeny of the Nbsgs mutants. WT, wild type; rdri, RNAi line of NbRDR6; D, 3-bp deletion. T insertion. (D) The progeny of Nbsgs3-14. Uppercase letters: in frame; lowercase letters: out of frame. E: base editing. a: A insertion. d: 5-bp deletion. Bar = 5 mm.
We incubated N. benthamiana plants regenerated from protoplasts in the same medium used for in vitro flowering of the orchid Erycina pusilla (Chiu et al., 2011). The N. benthamiana regenerants were able to flower (Figure 5a) and produce seeds (Figure 5b) in vitro. The seeds matured normally (Figure 5c) and germinated (Figure 5d). To investigate whether this medium can be widely used, we also incubated plants regenerated from protoplasts of other species in the same medium, including protoplasts from tobacco, broccoli, cauliflower, Arabidopsis, and rapid cycling Brassica oleracea. The tobacco, broccoli, and cauliflower plants did not flower in vitro, whereas the Arabidopsis and rapid cycling Brassica oleracea plants flowered but failed to produce seeds.

Figure 5. In vitro flowering and seed production in N. benthamiana. (a) In vitro flower. an: anther. Bar = 5 mm. (b) Mature fruit. Bar = 0.5 mm. (c) Open capsule. Bar = 0.5 mm. (d) Seedlings derived from in vitro seeds. Bar = 1 cm. (e) Bottle lip sealed with our newly developed sealing material. Bar = 1 cm. (f) Subculture in HB1 medium. Bar = 1 cm.
To increase seed production, we attempted to reduce the humidity inside the flask. For this purpose, we designed a new sealing material. A small piece of paper larger than the diameter of the bottle lip was placed inside two pieces of plastic film (Figure 5e), sealed over the lip, tied with a rubber band (Figure 5f), and sterilized by autoclaving. N. benthamiana produced seeds normally under these conditions, whereas the other species were still unable to produce seeds.
During this study, we determined that protoplasts derived from N. tabacum are easier to regenerate than protoplasts from N. benthamiana. The bottleneck in regenerating N. benthamiana plants occurs during the step from callus to shoot formation: whereas most calluses derived from N. tabacum can be regenerated into shoots after a single subculture, N. benthamiana requires two or three successive subcultures before shoots form, and the proportion of shoots that form in each subculture step is unpredictable. Nonetheless, N. benthamiana still has many advantages. Compared to N. tabacum, N. benthamiana requires less space, and it can flower all-year round, whereas in subtropical regions, N. tabacum cannot grow and produce flowers in the greenhouse in summer.
Protoplast regeneration has been used since 2016 for transcription activator-like effector nucleases (TALEN)-mediated gene editing (Li et al., 2016). Although this strategy has many advantages with regard to gene editing procedures, it is often avoided. Instead, protoplasts have mainly been used for protoplast fusion and mutation. Furthermore, protoplast regeneration is thought to result in many unanticipated mutations. Indeed, a study involving whole-genome sequencing of potato plants regenerated from protoplasts suggested that protoplast regeneration can cause numerous mutations and even chromosome rearrangements (Fossi et al., 2019). Because the tobacco genome is so large, we have not yet sequenced the entire genomes of gene-edited N. benthamiana regenerants, and thus we have not directly investigated their levels of mutations. However, our comparison of regenerated plants with seed-propagated offspring indicated that this protocol does not produce plants with phenotypic differences from the wild type.
In fact, unexpected mutations can occur in any tissue culture process and even under natural conditions (Lin and Chang, 1998; Yue et al., 2020). In crop breeding, even if mutations occur, desired offspring can be identified through selection from a wide range of gene-edited regenerated plants, without the ethical problems associated with the human application of genome editing (Tang et al., 2019). In plant research, the problem of unexpected mutations could be resolved by generating multiple mutations of the same gene, such as in Arabidopsis and rice knockout lines, or by transferring the edited gene to a wild-type plant by crossing.
Given concerns about the use of genetically modified crops, it is important to be able to produce genetically-edited crops without introducing foreign genes. In particular, although transgenes introduced via stable transformation can be removed from many plants through crossing, this is not the case for the many important crops that are propagated asexually, such as potato. The delivery of CRISPR reagents into cells by transient transfection, however, is widely regarded as transgene-free gene editing. Using protoplast regeneration, RNPs (Woo et al., 2015) or plasmids (Lin et al., 2018; Hsu et al., 2019) can be used as CRISPR reagents for transgene-free gene editing; this is the main reason that we use protoplast regeneration for gene editing of crops. In our experience, both RNPs and plasmids are effective for gene editing. When choosing gene-editing reagents, if no documented gene target sites are available to confirm editing efficiency, we use plasmids, which allow us to use multiple targets at once and are relatively cost-effective. For target insertion, we strongly recommend using RNPs, as plasmids may act as donor DNAs.
In addition, RNPs can be used to validate novel Cas proteins when the expected results are not obtained using plasmids. Since our Cas protein was translated and RNP-confirmed in vitro, we directly introduced these RNPs into protoplasts to validate that this protein functions in the species of interest. We used RNPs to monitor the efficiency of Cas12a proteins in N. benthamiana. Cas12a has a high target mutagenesis efficiency in Poaceae (Li et al., 2019). In dicots, LbCas12a has a higher editing efficiency than AsCas12a in soybean and tobacco protoplasts when delivered as RNP molecules (Kim et al., 2017). Similarly, when tested in rice, Arabidopsis, and maize, LbCas12a but not AsCas12a successfully edited target genes when these nuclease plasmids were delivered into protoplasts (Kim et al., 2017; Malzahn et al., 2019).
More importantly, the efficiency of Cas12a proteins is temperature dependent (Malzahn et al., 2019). These enzymes have high activity at 37°C, the temperature used for human cell culture, whereas plant transformation is performed at ~28°C, a temperature at which LbCas12a activity is reduced (Moreno-Mateos et al., 2017; Malzahn et al., 2019). Hence, it is likely that the absence of edited plants was due to the lower temperature along with the lower overall activity of AsCas12a. In both N. tabacum and N. benthamiana, the efficiencies of FnCas12a when using plasmids are <10%. When FnCas12a and AsCas12a RNPs (which were confirmed to have cleavage activity in vitro) were used, target mutagenesis did not occur, even when we increased the amount of RNP, raised the temperature, or changed the medium composition. Therefore, we suggest that the low target mutagenesis efficiency of Cas12a is due not to the low expression of this protein but to intracellular conditions unsuitable for its activity.
Because transient transfection can deliver multiple plasmids into the same protoplast at one time, Cas protein and sgRNAs do not necessarily need to be encoded by the same vector. The Agrobacterium transformation vector we used in the current study is a low copy number vector, which makes plasmid DNA extraction more difficult. Because the DNA does not need to be inserted into the chromosome, there is no need to clone these genes into the T-DNA vector. In addition, protoplast transfection is highly efficient and does not require a selectable marker for screening. Thus, to refine our method, we could simplify the vector structure and use a high copy number vector for CRISPR-mediated protoplast transfection. Alternatively, we could co-transfect in vitro transcribed sgRNA with the overexpression Cas protein vector to reduce the labor involved in construction (Zhang et al., 2016). When designing sgRNAs, we will not only use software to predict the efficiencies of the sgRNAs, but also select the relative positions of sgRNAs that have been successful for other species. For example, sgRNAs that were designed by this strategy based on the RDR6 and SGS3 sgRNAs that are effective in N. benthamiana were also effective in Solanum peruvianum. If there are many effective sgRNA candidates, we will choose the one that can be used in the largest number of species to increase usage. ETR1 sgRNA, which we used successfully in N. benthamiana and N. tabacum, is derived from tomato (Shimatani et al., 2017). Applying such a principle for sgRNA design to simultaneously induce mutations in multiple genes or gene families in heterozygous or polyploid plants is difficult because of mismatches. Because this is not a problem for off-target crops, it can instead be exploited for this type of multiplex gene mutation (Endo et al., 2015).
Most explants grow to the vegetative stage in vitro. By manipulating the medium and culture conditions, however, many plants can also be induced to flower in vitro. For example, bamboo has a juvenile period of several decades in the natural environment but can flower within 1 year in medium containing cytokinin (Lin and Chang, 1998). Plant species that can flower and be successfully pollinated, form fruit, and complete all growth stages in vitro are potentially good model plants for further study. For example, E. pusilla, which has these capabilities, serves as a model plant of the orchid family (Chiu et al., 2011). Here we demonstrated that N. benthamiana can bear fruit and produce seeds in vitro and that the use of HB1 medium and sealing film that we developed can increase fruiting and seed production. Although a speed breeding method has been developed to accelerate a plant's growth cycle and achieve year-round production (Ghosh et al., 2018), this method is quite expensive. It is important to develop an economical, space-saving method that can be used by all laboratories. The in vitro method developed in the study represents an alternative strategy for achieving this goal. However, this method cannot be used for all crops, an issue that will need to be addressed.
Although various protocols have been published for Agrobacterium-mediated stable transformation or DNA-free plant genome editing of N. benthamiana using virally delivered CRISPR-Cas (Ma et al., 2020), these techniques pose several problems, including issues related to the regulation of transgenic crops and the production of genetic chimeras. Protoplast regeneration represents an alternative approach for high-efficiency gene editing that avoids these complications. With this method, no foreign DNA is integrated into the chromosomes, the regenerated plants are derived from single cells, and all of the edited alleles can be passed on to the offspring. We also used this procedure to transfer large amounts of donor DNA to increase the efficiency of target DNA insertion. We believe that this system and the resulting mutants represent excellent tools for researchers using N. benthamiana for crop pathogen-related research.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
C-SL conceived and designed the experiments. C-TH, and Y-HY performed the CRISPR-Cas9 experiments. C-TH, Y-HY, and C-SL conducted the protoplast regeneration. C-TH, W-CL, Y-HY, and F-HW performed the molecular biology experiments and targeted mutagenesis analysis. W-CL performed small RNA Northern analysis and Western analysis. C-SL wrote the manuscript with input from all co-authors. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Qiao-Wei Cheng for help with tissue culture and protoplast transfection, Prof. Ho-Ming Chen for small RNA analysis, and Profs. Jen Sheen and Ming-Che Shih for discussion. This research was supported by Academia Sinica Innovative Translational Agricultural Research Administrative Office (AS-KPQ-107-ITAR-10; AS-KPQ-108-ITAR-10; AS-KPQ-109-ITAR-10), and the Ministry of Science and Technology (105-2313-B-001-007-MY3; 108-2313-B-001−011-; 109-2313-B-001−011-), Taiwan.
Andersson, M., Turesson, H., Olsson, N., Fält, A. S., Ohlsson, P., Gonzalez, M. N., et al. (2018). Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol. Plant. 164, 378–384. doi: 10.1111/ppl.12731
Bombarely, A., Rosli, H. G., Vrebalov, J., Moffett, P., Mueller, L. A., and Martin, G. B. (2012). A draft genome sequence of Nicotiana benthamiana to enhance molecular plant microbe biology research. Mol. Plant Microbe Interact. 25, 1523–1530. doi: 10.1094/MPMI-06-12-0148-TA
Chiu, Y. T., Lin, C. S., and Chang, C. (2011). In vitro fruiting and seed production in Erycina pusilla (L.) NH Williams and MW Chase. Propag. Ornam. Plants 11, 131–136.
Derevnina, L., Kamoun, S., and Wu, C. h. (2019). Dude, where is my mutant? Nicotiana benthamiana meets forward genetics. New Phytol. 221, 607–610. doi: 10.1111/nph.15521
Endo, A., Masafumi, M., Kaya, H., and Toki, S. (2016). Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci. Rep. 6:38169. doi: 10.1038/srep38169
Endo, M., Mikami, M., and Toki, S. (2015). Multi-gene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol. 56,41–47. doi: 10.1093/pcp/pcu154
Fossi, M., Amundson, K., Kuppu, S., Britt, A., and Comai, L. (2019). Regeneration of Solanum tuberosum plants from protoplasts induces widespread genome instability. Plant Physiol. 180, 78–86. doi: 10.1104/pp.18.00906
Ghosh, S., Watson, A., Gonzalez-Navarro, O. E., Ramirez-Gonzalez, R. H., Yanes, L., Mendoza-Suárez, M., et al. (2018). Speed breeding in growth chambers and glasshouses for crop breeding and model plant research. Nat. Protoc. 13, 2944–2963. doi: 10.1038/s41596-018-0072-z
Goodin, M. M., Zaitlin, D., Naidu, R. A., and Lommel, S. A. (2008). Nicotiana benthamiana: Its history and future as a model for plant-pathogen interactions. Mol. Plant Microbe Interact. 21:1015–1026. doi: 10.1094/MPMI-21-8-1015
Hill, J. T., Demarest, B. L., Bisgrove, B. W., Su, Y. C., Smith, M., and Yost, H. J. (2014). Poly peak parser: method and software for identification of unknown indels using Sanger sequencing of PCR products. Dev. Dyn. 243, 1632–1636. doi: 10.1002/dvdy.24183
Hsu, C. T., Cheng, Y. J., Yuan, Y. H., Hung, W. F., Cheng, Q. W., Wu, F. H., et al. (2019). Application of Cas12a and nCas9-activation-induced cytidine deaminase for genome editing and as a non-sexual strategy to generate homozygous/multiplex edited plants in the allotetraploid genome of tobacco. Plant Mol. Biol. 101, 355–371. doi: 10.1007/s11103-019-00907-w
Kaya, H., Mikami, M., Endo, A., Endo, M., and Toki, S. (2016). Highly specific targeted mutagenesis in plants using Staphylococcus aureus Cas9. Sci. Rep. 6:26871. doi: 10.1038/srep26871
Kim, H., Kim, S., Ryu, J., Kang, B. C., Kim, J. S., and Kim, S. G. (2017). CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 8:14406. doi: 10.1038/ncomms14406
Li, J., Stoddard, T. J., Demorest, Z. L., Lavoie, P. O., Luo, S., Clasen, B. M., et al. (2016). Multiplexed, targeted gene editing in N. benthamiana for glyco-engineering and monoclonal antibody production. Plant Biotechnol. J. 14, 533–542. doi: 10.1111/pbi.12403
Li, J. F., Norville, J. E., Aach, J., McCormack, M., Zhang, D. D., Bush, J., et al. (2013). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691. doi: 10.1038/nbt.2654
Li, S., and Xia, L. (2020). Precise gene replacement in plants through CRISPR/Cas genome editing technology: current status and future perspectives. aBIOTECH 1, 58–73. doi: 10.1007/s42994-019-00009-7
Li, S., Zhang, Y., Xia, L., and Qi, Y. (2019). CRISPR-Cas12a enables efficient biallelic gene targeting in rice. Plant Biotechnol. J. 18, 1351–1353. doi: 10.1111/pbi.13295
Lin, C.-S., Hsu, C.-T., Yang, L.-H., Lee, L.-Y., Fu, J.-Y., Cheng, Q.-W., et al. (2018). Application of protoplast technology to CRISPR/Cas9 mutagenesis: from single-cell mutation detection to mutant plant regeneration. Plant Biotechnol. J. 16,1295–1310. doi: 10.1111/pbi.12870
Lin, C. S., and Chang, W. C. (1998). Micropropagation of Bambusa edulis through nodal explants of field-grown culms and flowering of regenerated plantlets. Plant Cell Rep. 17, 617–620. doi: 10.1007/s002990050453
Ludman, M., and F?tyol, K. (2019). The virological model plant, Nicotiana benthamiana expresses a single functional RDR6 homeolog. Virology. 537, 143–148. doi: 10.1016/j.virol.2019.08.017
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant. 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Ma, X., Zhang, X., Liu, H., and Li, Z. (2020). Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9. Nat. Plants, 6, 773–779. doi: 10.1038/s41477-020-0704-5
Malzahn, A. A., Tang, X., Lee, K., Ren, Q., Sretenovic, S., Zhang, Y., et al. (2019). Application of CRISPR-Cas12a temperature sensitivity for improved genome editing in rice, maize, and Arabidopsis. BMC Biol. 17:9. doi: 10.1186/s12915-019-0629-5
Matsuo, K., and Atsumi, G. (2019). CRISPR/Cas9-mediated knockout of the RDR6 gene in Nicotiana benthamiana for efficient transient expression of recombinant proteins. Planta. 250, 463–473. doi: 10.1007/s00425-019-03180-9
Moreno-Mateos, M. A., Fernandez, J. P., Rouet, R., Vejnar, C. E., Lane, M. A., Mis, E., et al. (2017). CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat. Commun. 8:2024. doi: 10.1038/s41467-017-01836-2
Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J. D. G., and Kamoun, S. (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 691–693. doi: 10.1038/nbt.2655
Shan, Q. W., Wang, Y. P., Li, J., Zhang, Y., Chen, K. L., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686–688. doi: 10.1038/nbt.2650
Shimatani, Z., Kashojiya, S., Takayama, M., Terada, R., Arazoe, T., Ishii, H., et al. (2017). Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 35,441–443. doi: 10.1038/nbt.3833
Tang, J., Chen, L., and Liu, Y. G. (2019). Off-target effects and the solution. Nat. Plants 5:341–342. doi: 10.1038/s41477-019-0406-z
Woo, J. W., Kim, J., Kwon, S. I., Corvalan, C., Cho, S. W., Kim, H., et al. (2015). DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. doi: 10.1038/nbt.3389
Yue, J. J., Hong, C. Y., Wei, P., Tsai, Y. C., and Lin, C. S. (2020). How to start your monocot CRISPR/Cas project: plasmid design, efficiency detection, and offspring analysis. Rice. 13:9. doi: 10.1186/s12284-019-0354-2
Keywords: FnCas12a, nCas9-Target-AID, RDR6, SaCas9, SGS3, SpCas9
Citation: Hsu C-T, Lee W-C, Cheng Y-J, Yuan Y-H, Wu F-H and Lin C-S (2021) Genome Editing and Protoplast Regeneration to Study Plant–Pathogen Interactions in the Model Plant Nicotiana benthamiana. Front. Genome Ed. 2:627803. doi: 10.3389/fgeed.2020.627803
Received: 10 November 2020; Accepted: 28 December 2020;
Published: 21 January 2021.
Edited by:
Lanqin Xia, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Anshu Alok, Panjab University, IndiaCopyright © 2021 Hsu, Lee, Cheng, Yuan, Wu and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Choun-Sea Lin, Y3NsaW45OUBnYXRlLnNpbmljYS5lZHUudHc=; orcid.org/0000-0001-9566-2952
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.