- 1State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing, China
- 2National Key Laboratory of Crop Genetic Improvement and National Center of Plant Gene Research (Wuhan), Huazhong Agricultural University, Wuhan, China
- 3Guangdong Laboratory for Lingnan Modern Agricultural Science and Technology, Guangdong University of Petrochemical Technology, Maoming, China
- 4Key Laboratory of Plant Resource Conservation and Sustainable Utilization, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou, China
- 5Section of Cell and Developmental Biology, University of California, San Diego, La Jolla, CA, United States
CRISPR/Cas9 gene editing technology has been very effective in editing genes in many plant species including rice. Here we further improve the current CRISPR/Cas9 gene editing technology in both efficiency and time needed for isolation of transgene-free and target gene-edited plants. We coupled the CRISPR/Cas9 cassette with a unit that activates anthocyanin biosynthesis, providing a visible marker for detecting the presence of transgenes. The anthocyanin-marker assisted CRISPR (AAC) technology enables us to identify transgenic events even at calli stage, to select transformants with elevated Cas9 expression, and to identify transgene-free plants in the field. We used the AAC technology to edit LAZY1 and G1 and successfully generated many transgene-free and target gene-edited plants at T1 generation. The AAC technology greatly reduced the labor, time, and costs needed for editing target genes in rice.
Introduction
CRISPR/Cas9 genome editing technology has been widely used to generate targeted modifications of genes in many plant species (Feng et al., 2013; Li et al., 2013a; Mao et al., 2013; Miao et al., 2013; Nekrasov et al., 2013; Shan et al., 2013; Xie and Yang, 2013; Gao et al., 2015, 2016). Gene editing efficiency correlates with the expression level of Cas9 and higher expression of Cas9 usually leads to an increase in editing efficiency. Selecting transgenic plants with elevated Cas9 expression by analyzing Cas9 protein concentrations is laborious and time-consuming, Therefore, Cas9 expression levels are often monitored indirectly. Coupling a fluorescence marker with the Cas9 expression unit provides an effective approach for identify plants with elevated Cas9 concentrations (Gao et al., 2016; Wang and Chen, 2020). Another creative approach was to couple Cas9 expression with a guide RNA that can lead to a visible phenotype when the target gene is edited. Genes involved in trichome development have been targeted to visibly monitor gene editing efficiency (Wang et al., 2015; Miki et al., 2018). Whereas, the aforementioned methods have been effective, fluorescence markers need special equipment and are not suitable for field conditions. Targeting trichomes is very useful for Arabidopsis, but may not be effective for monocots such as rice. Additional markers that can be indicative of the levels of transgene expression will be very useful in conducting gene editing experiments.
In addition to improving gene editing efficiency, a main emphasis in editing plants/crops is to generate edited plants without any transgene residuals. Crops with any CRISPR/Cas9 component residual will unlikely receive approval for commercial applications from government regulatory agencies. The continuous presence of gene editing elements in plants may cause genetic instability and off-target events. Several strategies have been reported for isolating transgene-free and target gene-edited plants. Transient expression of Cas9 and gRNA genome editing complex through Agrobacterium-mediated infiltration has led to the identification of target gene-edited plants without any transgene residues in tobacco (Chen et al., 2018). Cas9 and gRNA can also be assembled into ribonucleoprotein (RNP) complexes and then delivered into plant cells using nano particles (Doyle et al., 2019; Landry and Mitter, 2019) or bombardment (Svitashev et al., 2016; Zhang et al., 2016). Because RNP contains no DNA, any edited plants are considered transgene-free, However, RNP method is extremely inefficient because the majority of the regenerated plants are not transformed due to a lack of selection pressure. Identification of transgene-free, target gene-edited plants can also be assisted by fluorescence markers (Gao et al., 2016; He et al., 2017a; Yu and Zhao, 2019; Ouyang et al., 2020) and positive or negative selection against chemicals (Lu et al., 2017; Wu et al., 2019; Li et al., 2020), Moreover, we developed a CRISPR/Cas9 gene editing system in which the transgenes undergo automatic self-elimination after the target gene has been edited (transgene killer CRISPR (TKC), greatly improving the efficiency for isolating the desired plants (He et al., 2018; 2019). Whereas, TKC system reduces labor and time needed for conducting gene editing in rice, it does not have a proxy for indicating Cas9 expression levels.
Anthocyanins are a large class of secondary metabolites (Tanaka et al., 2008), which are widely distributed in various tissues including flowers, stems, leaves, and fruits. Anthocyanins are water-soluble polyphenol pigments with vivid red, blue, purple, and other colors, Therefore, anthocyanins potentially can be used as a visible marker for visualizing transgenic events or the presence of transgenes, However, anthocyanin biosynthetic pathway is very complex and consists of at least eight genes (Zhu et al., 2017; Zheng et al., 2019). It is not realistic to couple the entire anthocyanin biosynthetic pathway with Cas9/gRNA units in a plasmid because the resulting plasmid would be too big for efficient transformation or cloning. Fortunately, anthocyanin biosynthesis pathway is under the control of key transcription factors including MYB, bHLH, and WD40 family genes (Zhang et al., 2014; Liu et al., 2015; Xu et al., 2015). Almost all of the anthocyanin biosynthesis structural genes are activated (or enhanced) by the MYB–bHLH–WD40 (MBW) complex (Xu et al., 2015; Zhu et al., 2017; Zheng et al., 2019), and the MYB protein is believed to be the key component in the allocation of specific gene expression patterns (Jaakola, 2013; Xu et al., 2015). Based on the number of imperfect repeats (R) domain(s), MYB genes are divided into four major groups: 1R (R1/2, R3-MYB), 2R (R2R3-MYB), 3R (R1R2R3-MYB), and 4R (four R1/R2-like repeats) (Liu et al., 2015). The R2R3-MYB is an activator for the synthesis of anthocyanins (Borevitz et al., 2000; Liu et al., 2015).
The ZmC1 gene, encoding an R2R3 MYB, regulates the synthesis of anthocyanins in the corn aleurone layer (Cone et al., 1986; Paz-Ares et al., 1986). Its ortholog, OsC1 gene controls the color change in leaf sheath and stigma in rice. Ectopic expression of OsC1 in different rice varieties results in accumulation of anthocyanins in various tissues and organs (Gao et al., 2011; Chin et al., 2016; Zhao et al., 2016), Therefore, ectopic expression of OsC1 may be used to activate anthocyanin biosynthesis, thus providing a visible marker in rice. Here, we show that the expression of OsC1 under the control of the rice ACTIN promoter leads to the accumulation of anthocyanin in calli, young seedlings, leaf vascular tissue, and grains, providing a visible marker for selecting transgenic plants. When the OsC1 unit is coupled with CRISPR/Cas9 units, target genes in almost all of the anthocyanin positive plants have been edited, suggesting that the threshold of visible anthocyanin accumulation is a good indication of high level Cas9 expression, Moreover, anthocyanin accumulation allows a visual differentiation between transgenic and non-transgenic plants at T1 generation without the need of conducting PCR and other molecular analysis, greatly accelerating the isolation of transgene free and target-gene edited plants.
Results
Activation of Anthocyanin Biosynthesis Serves as a Marker for the Presence of Transgenes
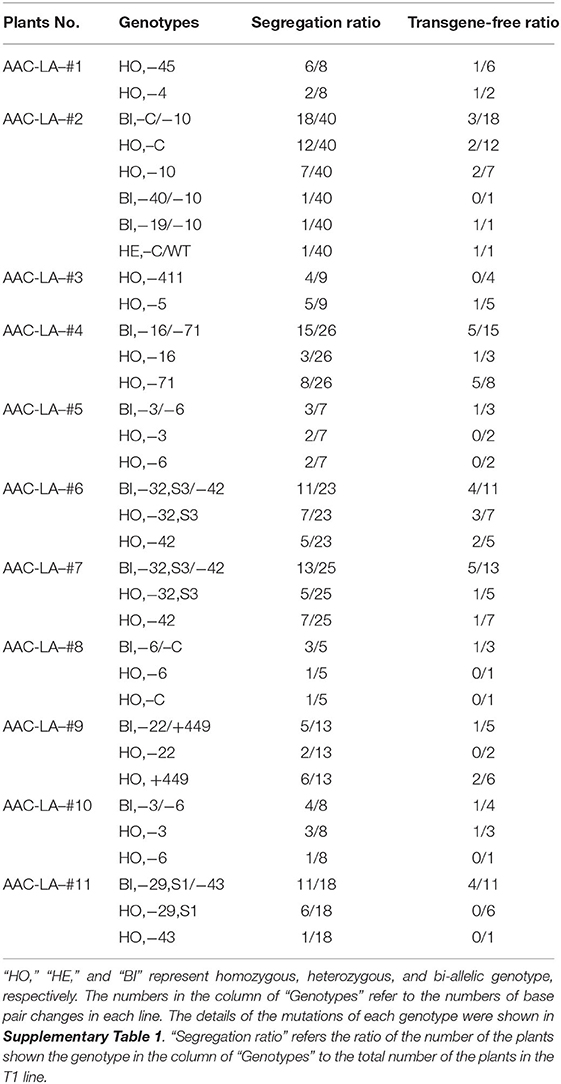
Ectopic expression of the R2R3-MYB gene in rice and other plants activated anthocyanin biosynthesis (Borevitz et al., 2000; Shin et al., 2006; Han et al., 2009; Li et al., 2017; Zhang et al., 2019). We chose the OsC1 (GenBank ID: MK636605), an R2R3-MYB gene, from the rice cultivar Heishuai (Zheng et al., 2019), which displays obvious purple color in leaves, stems, and grains. We placed OsC1 cDNA under the control of the rice ACTIN1 promoter, a constitutively activated promoter (Figure 1A). When plasmids containing the pACTIN-OsC1 unit were transformed into rice, we observed that anthocyanin accumulated in calli, leaves, and the tip of grains (Figure 1B), demonstrating that the pACTIN-OsC1 unit can be used to produce a visible color, which can facilitate the identification of transgenic events in calli and transgenic plants.

Figure 1. The pACTIN-OsC1 activates anthocyanin biosynthesis in rice. (A) A schematic description of the pACTIN-OsC1 unit in the vector used for rice transformation. The anthocyanin regulation gene OsC1 is under the control of the rice OsACTIN1 promoter. NOS T refers to the terminator of the nopaline synthase gene from Agrobacterium tumefaciens. (B) The AAC plasmid induced obvious accumulation of anthocyanin (purple color) in rice calli, vascular tissue in stems (transverse section), leaves, and the tip of grains. Scale bars 1 cm in tissue culture images.
Coupling Anthocyanin Biosynthesis With Genome Editing Units
We placed the pACTIN-OsC1 unit in adjacent to the cassettes for Cas9 expression and gRNA production, resulting in the AAC plasmid (Anthocyanin-marker Assisted CRISPR) (Figure 2A). We first tested whether our AAC plasmids can achieve efficient editing of the LAZY1 gene, which leads to agravitropic growth when compromised (Li et al., 2007), providing a visible phenotype for gene editing events. We cloned the LAZY1 gRNA unit into the AAC plasmid to generate the AAC-LA plasmid, which was subsequently transformed into rice through Agrobacterium-mediated transformation. We found that almost 80% of the T0 plants regenerated from the plate with two rounds hygromycin selection (10 days in a round) had purple color tissues (Figure 2B). Furthermore, we noticed that transferring the calli to antibiotic-free media after 7 days hygromycin selection (only one round), resulted in about 40% regenerated plants with purple color tissues (Figure 2B), suggesting that activation of anthocyanin can be used for selecting transgenic events with a decreased usage of antibiotic. Whereas, antibiotic or herbicide resistant markers are potentially detrimental to the environment, the native anthocyanin biosynthesis pathway offers a good alternative for selecting transformants.
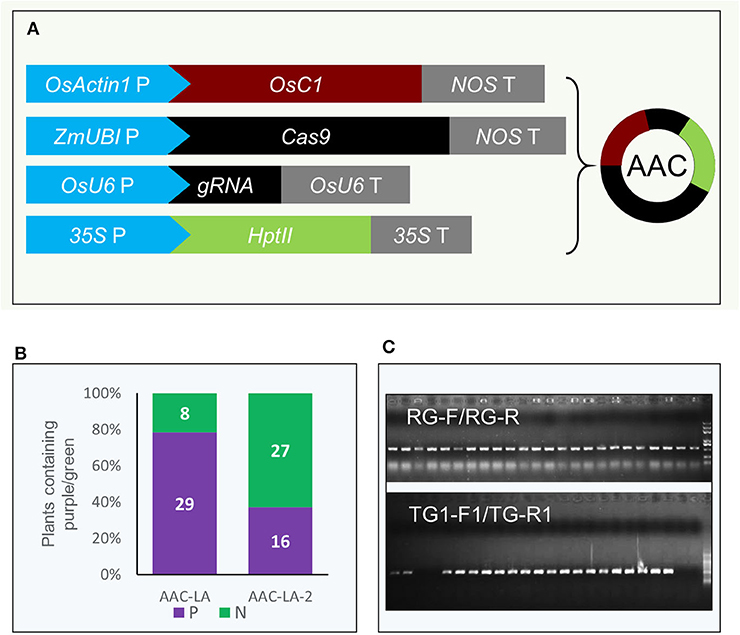
Figure 2. Anthocyanin accumulation as a marker for the presence of transgenes in gene editing experiments. (A) Coupling the pACTIN-OsC1 unit with the CRISPR/Cas9 genome editing units in the AAC (Anthocyanin-marker Assisted CRISPR) plasmid. The anthocyanin regulatory gene OsC1 is under the control of the rice OsACTIN1 promoter. NOS T refers to the terminator of the nopaline synthase gene from Agrobacterium tumefaciens. The rice codon-optimized Cas9 was placed under the control of the maize UBIQUITIN promoter. OsU6 P and OsU6 T represent rice U6 promoter and terminator, respectively. gRNA refers to guide RNA. The transcription of hygromycin phosphotransferase II gene HptII is under the control of the CaMV 35S promoter and the transcription was terminated with a CaMV 35S terminator. (B) Not all antibiotic resistant plants generated purple color in T0. AAC-LA was transgenic event screening from the plate with two rounds hygromycin selection (10 days in a round). AAC-LA-2 was regenerated from the calli on normal plate after 7 days hygromycin selection (only one round). “P” and “N” represent purple and normal color plants, respectively. (C) Verification of the transgenes in the rice plants by PCR. “RG-F/RG-R” and “TG-F1/TG-R1” refer to the primer pairs used for detection the presence of rice genomic DNA and the T-DNA of AAC plasmid, respectively. The primer information was described in Supplementary Table 3.
We observed that all of the T0 plants that displayed purple color contained the OsC1 expression cassette revealed by our PCR analyses (Figure 2C). We also found that some of the hygromycin-resistant plants without the purple color also contained the transgene, suggesting that a threshold level expression of the OsC1 is required in order to produce a visible color. The results suggest that we might be able to use the accumulation of anthocyanin as a proxy for monitoring Cas9 expression levels.
Facilitating the Isolation of Transgene-Free and Target Gene-Edited T1 Plants Using the Anthocyanin-Marker
We hypothesized that we might be able to visually identify transgene free T1 plants generated from the T0 plants that had obvious anthocyanin accumulation. We also used PCR-based assays to further confirm the absence of transgenes. We used two pairs of primers, TG-F1/TG-R1 and TG-F2/TG-R2 to amply specific DNA fragments from the AAC plasmid (Figure 3A). We found that the PCR results matched perfectly with the color based visual screen. For example, among 40 and 28 T1 plants from the AAC-LA-#2 and AAC-LA-#4 T0 plants, 9 and 12 plants did not display obvious anthocyanin accumulation, respectively (Figure 3B). Our PCR results identified the same plants as transgene-free (Figure 3A). Further analyses of the T1 plants from the AAC-LA–#1 to # 11 T0 plants demonstrated that the transgene-free T1 plants identified on basis of anthocyanin accumulation matched with those transgene-free plants identified by PCR assays (Figure 3B). Our results demonstrated that the anthocyanin-based visual screen for transgene-free T1 plants was effective and accurate. Such a visual assay greatly reduced labor and time for identifying transgene-free plants.
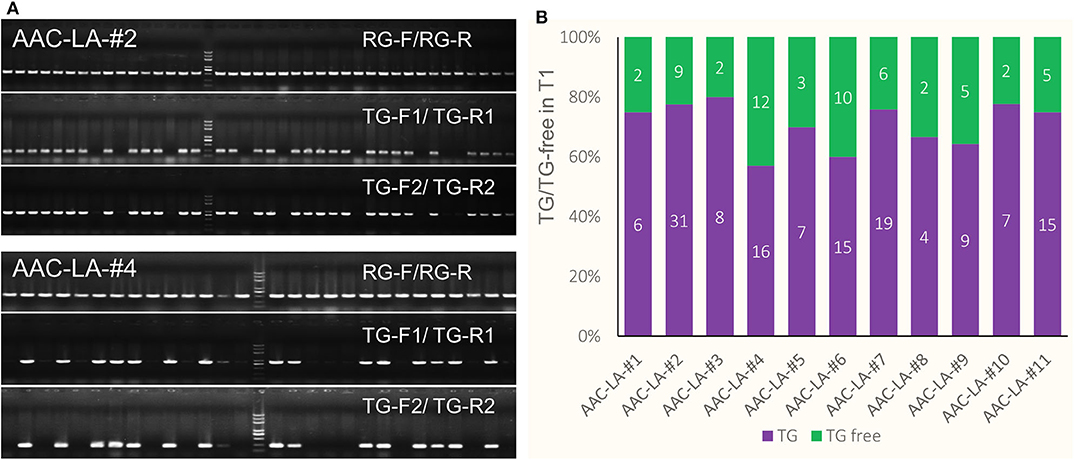
Figure 3. Isolation of transgene-free T1 plants by anthocyanin-marker assisted strategy. (A) The transgene-free plants from the progeny of the T0 plants that had purple color were identified by PCR and visual screen. AAC-LA–#2 and AAC-LA-#4 represent two AAC-LA T0 plants with purple color, respectively. (B) The segregation of the transgene (TG) and transgene-free (TG-free) plants from 11 AAC-LA T0 lines. The numbers on purple and green columns represent the numbers of purple and normal colored plants at T1 generation. The results were consistent with the results identified by PCR. “RG-F/RG-R” refers to primer pair used for checking the quality of rice genome DNA. “TG-F1/TG-R1,” and “TG-F2/TG-R2” represent two primer pairs used for detection the presence of T-DNA, respectively. The primers information was described in Supplementary Table 3.
Molecular Characterization of the Transgene-Free and Gene-Edited Plants
To analyze the molecular lesions in the LAZY1 gene in the T1 plants generated from the AAC-LA T0 plants that had displayed anthothyanin accumulation, we directly sequenced the LAZY1 fragment amplified using the primers of LAZY1-GTF/ LAZY1-GTR (Figure 4A). We analyzed T1 plants from 11 independent T0 plants, which had obvious anthocyanin accumulation (Table 1, Supplementary Table 1). Every single plant contained mutations at the target site except that a few plants failed to generate quality sequencing data. The T1 plants analyzed were either homozygous or bi-allelic except one T1 plant from AAC-LA-#2, which was heterozygous, demonstrating the power of AAC technology in gene editing (Table 1, Supplementary Table 1). For comparison, our previously experiments using our regular CRISPR vectors usually had the editing efficiency <80% (He et al., 2017b).
Figure 4. Identification of transgene-free and target gene-edited lazy1 mutants. (A) Target site of LAZY1 used in AAC-LA. A target sequence in the LAZY1 gene includes the PAM site GGG (marked green) is shown. LAZY1-GTF and LAZY1-GTR were the genotyping primer pair of LAZY1 CRISPR plants. Primers were listed in Supplementary Table 3. (B) Loss-of-function lazy1 mutants displayed an obvious tiller angle phenotype. WT, lazy1 and lazy1-3x refer to wild type, loss-of-function lazy1 mutant, and in-frame (multiple of three base pairs deletion) lazy1 mutant plants, respectively. (C) The mutation forms of the transgene-free and CRISPR-edited lazy1 mutants. The PAM site “GGG” in green required for Cas9 cleavage is marked in green. “WT” refers to the wild type plants. “HO,” “HE,” and “BI” represent homozygous, heterozygous, and bi-allelic genotypes, respectively. “-” refers to a deletion of one base pair. “S3” and “S1” means three and one base pair(s) substitution, respectively.
Further analysis of the mutations revealed some interesting patterns. We observed four lazy alleles in the progeny of AAC-LA−2 T0 plant: a “C” deletion, a 10-base pair deletion, a 40-base pair deletion, and a 19-base pair deletion. The lazy alleles resulted in six combinations of genotypes: eighteen “–C/−10” biallelic mutants, twelve “–C” homozygous mutants, seven “-10” homozygous mutants, one “−40/−19” biallelic, one “−19/−10” biallelic mutant, and a wild type plant in the T1 plants (Table 1, Supplementary Table 1). The first three genotypes were predominant, accounting for 92.5% (37/40) of the T1 plants. Because of the complex genotype (likely mosaic) in T0 plants, it is extremely important to analyzed T1 plants that no longer have the Cas9 transgenes.
Using anthocyanin as a visible marker for transgenes (Figure 3) of the T1 plants, we easily identified transgene-free lazy1 mutants of at the T1 generation from multiple independent T0 plants (Figures 4B,C). Some of T0 plants did not show the lazy mutant phenotype, but they harbored mutations with 3x of base pair deletion, such as the lines from AAC-LA–#1, 5, 6, 7, 8, 10 (Figure 4C, Table 1, Supplementary Table 1). Such 3x mutations did not cause frameshift and likely produced functional protein.
Testing the AAC Gene Editing System With Another Target
We assembled another construct AAC-G1 to target the G1 gene, which takes part in suppressing the development of the sterile lemma. When G1 is disrupted, the length of the sterile lemma increased dramatically, providing an easily scorable phenotype (Yoshida et al., 2009). We analyzed 88 T1 plants from 9 AAC-G1 T0 plants that displayed anthocyanin accumulation. The results of our visual screen for transgene-free plants (Figure 5A) were consistent with the results identified by PCR using primer pairs TG-F1/TG-R1 and TG-F2/TG-R2 (Figure 5B). For example, there were 5 and 2 transgene-free plants in the progeny of AAC-G1–#5 and AAC-G1–#6, respectively (Figures 5A,B). We also identified multiple alleles of g1 (Figures 5C,D, Supplementary Table 2). It was clear that all of the T1 plants sequenced were either homozygous or bi-allelic (Supplementary Table 2). Our results demonstrated that the anthocyanin-based screening for transgene-free and edited plants were very effective (Figures 5D,E).
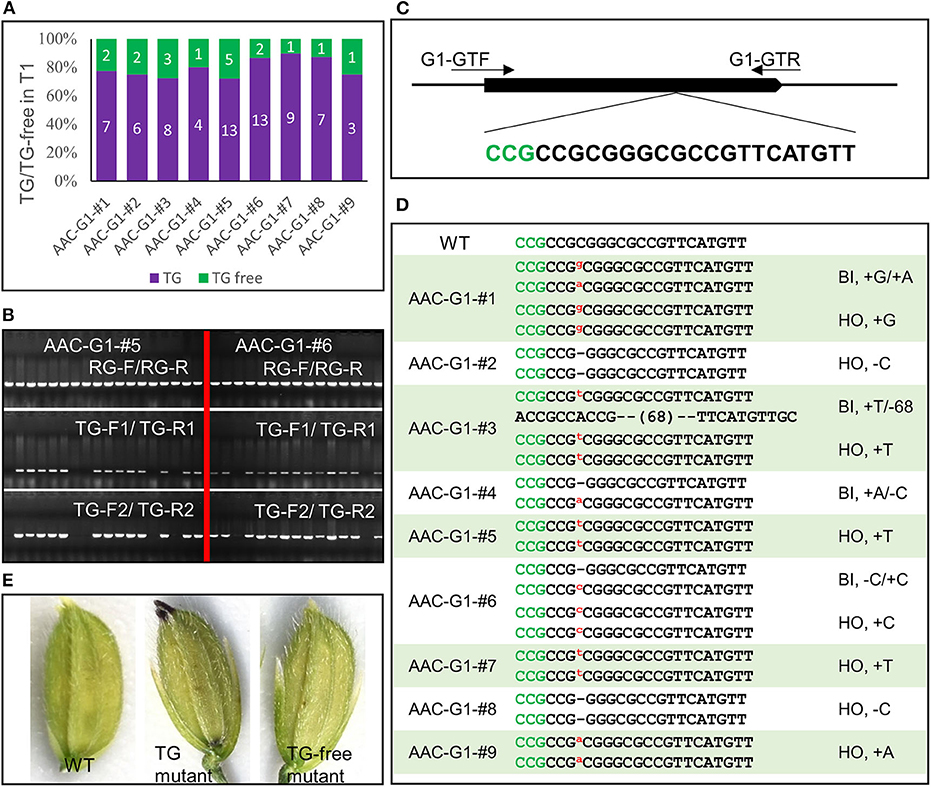
Figure 5. Identification transgene-free and gene-edited g1 mutants. (A) The segregation of the transgene (TG) and transgene-free (TG-free) plants from 9 AAC-G1 T1 lines. The numbers on purple and green columns represent the numbers of purple and normal color plants in T1 lines which were consistent with the results identified by PCR, respectively. (B) The transgene-free plants from the progeny of the purple-colored T0 plants were identified by PCR. AAC-G1–#5 and AAC-G1-# 6 refer to two AAC-G1 T0 plants containing the purple color, respectively. “RG-F/RG-R” refers to primer pair used for checking the quality of rice genome DNA. “TG-F1/TG-R1” and “TG-F2/TG-R2” represent two primer pairs used for detection the presence of T-DNA, respectively. The primers information was described in Supplementary Table 3. (C) Target site of G1 used in AAC-G1 plasmid. A target sequence including the PAM site CCG (marked green) was chosen. G1-GTF and G1-GTR are the genotyping primer pair of G1 CRISPR plants. Primers were listed in Supplementary Table 3. (D) The mutation forms of the transgene-free and CRISPR-edited g1 mutants. The PAM site “CCG” required for Cas9 cleavage is marked in green. “WT” refers to the wild-type plants. “HO” and “BI” represent homozygous and bi-allelic genotypes, respectively. “-” refers to a deletion of one base pair. “a,” “g,” “c” and “t” in red and superscript refers to an insertion of an “A,” “G,” “C,” and “T,” respectively. (E) The g1 mutant florets with (TG) or without transgene (TG-free) generated by AAC-G1.
Discussion
Plant transformation through tissue culture usually takes weeks before a positive result is obtained. The ability to identify transgenic events at the earliest stages of plant transformation offers advantages. We showed that anthocyanin biosynthesis can be activated at calli stage, providing a visual and functional assay for positive transformants very early on (Figure 6). Anthocyanin biosynthesis does not require exogenous substrates. It allows continuous monitoring transgene activities under sterile conditions, which is very useful in tissue culture. Because anthocyanin is a plant pigment existent in many plants including some rice varieties, it is not toxic to plants and causes fewer environmental impacts compared to antibiotic and herbicide-resistance screening. In addition, anthocyanins can absorb excessive ultraviolet lights and visible lights and remove free radicals to protect plants from ultraviolet rays, thereby providing protection for plants (Guo et al., 2008). Anthocyanins can also enhance plant resistance to drought and low temperature, and strengthen the ability to resist pathogenic bacteria, thereby protecting plants from biotic and abiotic stresses (Ahmed et al., 2014), Moreover, anthocyanins can also respond to external trauma by preventing oxidation (Gould et al., 2002), Therefore, the T0 plants generated using our AAC system might have better chance to survive after transferring them from the sterile environment to natural field.
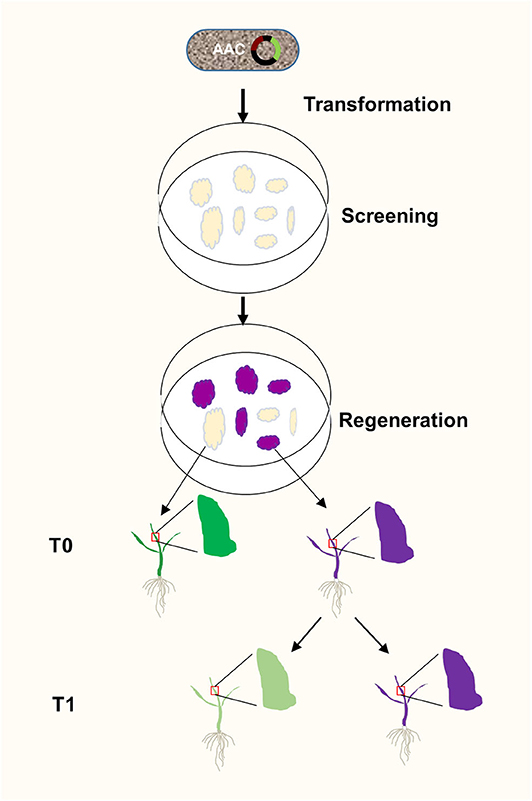
Figure 6. A flow chart of AAC-mediated isolation of transgene-free and target gene-edited rice plants. The AAC plasmid was transformed into rice calli through Agrobacterium-mediated transformation. At the calli stage, the OsC1 gene was expressed to generate purple callus, and the target gene is presumably being edited by Cas9/sgRNA complex. Consequently, at T1 stage, the transgene free plants without purple were selected from the progeny of the purple T0 plants.
We used the rice ACTIN promoter, which is a strong and ubiquitously active promoter, to drive OsC1 expression. Interestingly, anthocyanin accumulation was not ubiquitously distributed (Figure 1). We observed obvious purple color in calli, the vascular tissue of the stems and leaves, and the grains. But some other tissues did not display the purple color (Figure 1B). Previous studies showed that the anthocyanin biosynthesis pathway is regulated by the MYB-bHLH-WD40 (MBW) complex (Xu et al., 2015; Zhu et al., 2017; Zheng et al., 2019), and MYB protein is the key component (Jaakola, 2013; Xu et al., 2015). Our activation of anthocyanin biosynthesis by expressing the transcription factor OsC1 only works in plants that have all functional anthocyanin biosynthetic genes and sufficient amount of bHLH and WD40. Many popular white rice varieties have defects in some of the anthocyanin genes (Zheng et al., 2019), rendering them not suitable for our AAC technology. Recently, we developed a novel color reporter RUBY (He et al., 2020), which can generate red color in all eukaryotic cells include rice. For those rice cultivars that AAC is not suitable, RUBY can be used for color-assisted CRISPR technology. We introduced the pACTIN-OsC1 cassette to the rice variety Chao2-10 (Li et al., 2013b), which was previously shown to have all functional anthocyanin biosynthetic genes (Li et al., 2013b, 2017; Zheng et al., 2019), but lacked the MYB gene in the MBW complex. From a practical point view, the non-ubiquitous accumulation of anthocyanin actually is better because of its minimal impact on plant growth and development.
In our previous studies, we used antibiotics to screen the candidate gene-edited plants and found that some plants regenerated from antibiotic-resistance calli did not have target site mutations (He et al., 2017b). In this study, we found that all of the purple color plants were transgenic plants (Figures 2, 3, 5A,B) and all of the purple color plants had mutation at the target sites (Table 1, Supplementary Tables 1, 2). Our interpretation is that anthocyanin accumulation may be indicative of elevated Cas9 expression, which directly determines gene editing efficiency.
In summary, we have repurposed the anthocyanin biosynthesis pathway for serving as a visible marker for selecting plant transformation events and as a proxy for the presence of transgenes (Figure 6). Our system offers an alternative to fluorescence markers and antibiotic selections. It is especially powerful when used in tissue culture and in combination with gene editing machinery. The extend of anthocyanin accumulation might be indicative of the expression levels of other linked transgenes such as Cas9, thus providing a robust and speedy method for identifying plants with elevated gene editing efficiency.
Materials and Methods
The AAC Plasmid Construction
Primers used in this study were listed in the Supplementary Table 3. Our AAC plasmid contained two main expression cassettes: OsACTIN1 promoter-OsC1 and UBIQUITIN promoter-Cas9. OsACTIN1 promoter was amplified using primers of pCXUN-Act1PF and pCXUN-Act1PR from the plasmid pCB2006, which was kindly provided by Professor Lizhong Xiong (Xiao et al., 2009). The PCR product was cloned into pCXUN-Cas9 (He et al., 2017b) at the Kpn I site. The OsC1 gene was amplified from the cDNA plasmid provided by Dr. Hao Chen and Dr. Jie Zheng (Huazhong Agricultural University, Wuhan, China) by using primers of C1-Act1P-PCA9F and C1-Act1P-PCA9R, and the PCR fragment was inserted into the Kpn I site behind the OsACTIN1 promoter to complete the AAC (Anthocyanin-marker Assisted CRISPR) plasmid construction. Guide RNA production cassette was inserted into the Pme I site of the AAC plasmid. We generated the plasmid AAC-LA and AAC-G1, which produce guide RNAs from the rice U6 and U3 promoter to target the rice LAZY1 and G1 gene by overlapping PCR, respectively (He et al., 2019). The correct clones were confirmed by sequencing with primer AAC-PmeI-seqF.
Plant Transformation
The AAC-LA and AAC-G1 plasmids were transformed into Chao2-10 through Agrobacterium-mediated plant transformation following a protocol that was previously described (Hiei et al., 1994). T0 plants were visually scored for color phenotype at calli culture stage and at different growing stages in natural field. Seeds from each individual T0 plants containing purple color in vascular tissue of stems and leaves were harvested separately.
Characterization of T1 Plants
We randomly selected the T1 progenies of 11 T0 plants of AAC-LA and 9 independent AAC-G1 T0 plants to determine the efficiency of AAC system in editing the target genes. We used two primer pairs to amplify the specific regions of the AAC plasmids. TG-F1/TG-R1 was used to detect the presence of the OsC1 expression cassette, and TG-F2/TG-R2 was used to amply part of the CAS9 expression cassette. To check the quality of our genomic DNA samples, we used RG-F/RG-R for PCR reactions.
We used primer pairs LAZY1-GTF/LAZY1-GTF and G1-GTF/G1-GTR to amplify part of the LAZY1 and G1 genes from the AAC-LA and AAC-G1 T1 plants, respectively. We also directly sequenced the PCR products by using primers LAZY1-seq and G1-seq, respectively. For heterozygous or bi-allelic plants, the overlapping peaks were resolved using the publicly available Dsdecode site (http://skl.scau.edu.cn/dsdecode/) (Xie et al., 2017).
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
YH and YZ conceived the idea and wrote the first draft of the manuscript. YH, MZ, and JW conducted most of the experiments. MZ, JW, LO, RW, HS, LY, LW, MX, and HZ contributed to manuscript revision. All authors contributed to the article and approved the submitted version.
Funding
This work was partially supported by grants from the National Transgenic Science and Technology Program (2019ZX08010-001 and 2019ZX08010-003) to YH and HZ, the Fundamental Research Funds for the Central Universities (JCQY201903) and a startup fund from the Nanjing Agricultural University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor declared a past co-authorship with several of the authors YH, MX, and YZ.
Acknowledgments
We thank Professor Lizhong Xiong for providing pCB2006 plasmid. We are grateful to Dr. Hao Chen and Dr. Jie Zheng for kindly providing the cDNA of OsC1 and the seeds of Chao2-10.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgeed.2020.607982/full#supplementary-material
References
Ahmed, N. U., Park, J. I., Jung, H. J., Yang, T. J., Hur, Y., and Nou, I. S. (2014). Characterization of dihydroflavonol 4-reductase (DFR) genes and their association with cold and freezing stress in Brassica rapa. Gene 550, 46–55. doi: 10.1016/j.gene.2014.08.013
Borevitz, J. O., Xia, Y., Blount, J., Dixon, R. A., and Lamb, C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2394. doi: 10.1105/tpc.12.12.2383
Chen, L., Li, W., Katin-Grazzini, L., Ding, J., Gu, X., Li, Y., et al. (2018). A method for the production and expedient screening of CRISPR/Cas9-mediated non-transgenic mutant plants. Hortic Res. 5:13. doi: 10.1038/s41438-018-0023-4
Chin, H. S., Wu, Y. P., Hour, A. L., Hong, C. Y., and Lin, Y. R. (2016). Genetic and evolutionary analysis of purple leaf sheath in Rice. Rice 9:8. doi: 10.1186/s12284-016-0080-y
Cone, K. C., Burr, F. A., and Burr, B. (1986). Molecular analysis of the maize anthocyanin regulatory locus C1. Proc. Natl. Acad. Sci. U.S.A. 83, 9631–9635. doi: 10.1073/pnas.83.24.9631
Doyle, C., Higginbottom, K., Swift, T. A., Winfield, M., Bellas, C., Benito-Alifonso, D., et al. (2019). A simple method for spray-on gene editing in planta. bioRxiv [Prprint]. doi: 10.1101/805036
Feng, Z., Zhang, B., Ding, W., Liu, X., Yang, D. L., Wei, P., et al. (2013). Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23, 1229–1232. doi: 10.1038/cr.2013.114
Gao, D., He, B., Zhou, Y., and Sun, L. (2011). Genetic and molecular analysis of a purple sheath somaclonal mutant in japonica rice. Plant Cell Rep. 30, 901–911. doi: 10.1007/s00299-011-1004-3
Gao, X., Chen, J., Dai, X., Zhang, D., and Zhao, Y. (2016). An effective strategy for reliably isolating heritable and Cas9-free arabidopsis mutants generated by CRISPR/Cas9-mediated genome editing. Plant Physiol. 171, 1794–1800. doi: 10.1104/pp.16.00663
Gao, Y., Zhang, Y., Zhang, D., Dai, X., Estelle, M., and Zhao, Y. (2015). Auxin binding protein 1 (ABP1) is not required for either auxin signaling or arabidopsis development. Proc. Natl. Acad. Sci. U.S.A. 112, 2275–2280. doi: 10.1073/pnas.1500365112
Gould, K. S., McKelvie, J., and Markham, K. R. (2002). Do anthocyanins function as antioxidants in leaves? Imaging of H2O2 in red and green leaves after mechanical injury. Plant Cell Environ. 25, 1261–1269. doi: 10.1046/j.1365-3040.2002.00905.x
Guo, J., Han, W., and Wang, M. (2008). Ultraviolet and environmental stresses involved in the induction and regulation of anthocyanin biosynthesis: a review. Afr. J. Biotechnol. 7, 4966–4972. Available online at: https://www.ajol.info/index.php/ajb/article/view/59709
Han, Y. J., Kim, Y. M., Lee, J. Y., Kim, S. J., Cho, K. C., Chandrasekhar, T., et al. (2009). Production of purple-colored creeping bentgrass using maize transcription factor genes Pl and Lc through Agrobacterium-mediated transformation. Plant Cell Rep. 28, 397–406. doi: 10.1007/s00299-008-0648-0
He, Y., Wang, R., Dai, X., and Zhao, Y. (2017a). On Improving CRISPR for editing plant genes: ribozyme-Mediated Guide RNA production and fluorescence-based technology for isolating transgene-free mutants generated by CRISPR. Prog. Mol. Biol. Transl. Sci. 149, 151–166. doi: 10.1016/bs.pmbts.2017.03.012
He, Y., Zhang, T., Sun, H., Zhan, H., and Zhao, Y. (2020). A reporter for non-invasively monitoring gene expression and plant transformation. Hortic Res. 7:152. doi: 10.1038/s41438-020-00390-1
He, Y., Zhang, T., Yang, N., Xu, M., Yan, L., Wang, L., et al. (2017b). Self-cleaving ribozymes enable the production of guide RNAs from unlimited choices of promoters for CRISPR/Cas9 mediated genome editing. J. Genet. Genomics 44, 469–472. doi: 10.1016/j.jgg.2017.08.003
He, Y., Zhu, M., Wang, L., Wu, J., Wang, Q., Wang, R., et al. (2018). Programmed self-elimination of the CRISPR/Cas9 construct greatly accelerates the isolation of edited and transgene-free rice plants. Mol. Plant 11, 1210–1213. doi: 10.1016/j.molp.2018.05.005
He, Y., Zhu, M., Wang, L., Wu, J., Wang, Q., Wang, R., et al. (2019). Improvements of TKC technology accelerate isolation of transgene-free CRISPR/Cas9-edited rice plants. Rice Sci. 26, 109–117. doi: 10.1016/j.rsci.2018.11.001
Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313X.1994.6020271.x
Jaakola, L. (2013). New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 18, 477–483. doi: 10.1016/j.tplants.2013.06.003
Landry, M. P., and Mitter, N. (2019). How nanocarriers delivering cargos in plants can change the GMO landscape. Nat. Nanotechnol. 14, 512–514. doi: 10.1038/s41565-019-0463-5
Li, J., Qin, R., Zhang, Y., Xu, S., Liu, X., Yang, J., et al. (2020). Optimizing plant adenine base editor systems by modifying the transgene selection system. Plant Biotechnol. J. 18, 1495–1497. doi: 10.1111/pbi.13304
Li, J. F., Norville, J. E., Aach, J., McCormack, M., Zhang, D., Bush, J., et al. (2013a). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 31, 688–691. doi: 10.1038/nbt.2654
Li, P., Wang, Y., Qian, Q., Fu, Z., Wang, M., Zeng, D., et al. (2007). LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 17, 402–410. doi: 10.1038/cr.2007.38
Li, Y., Lin, Y., and Chen, H. (2017). Function of transcription factor OsC1 and OsPAC1 regulating biosynthesis of rice anthocyanin. J. Huazhong Agric. Univ. 1, 1–9. doi: 10.13300/j.cnki.hnlkxb.2017.01.001
Li, Y., Zhang, T., Shen, Z. W., Xu, Y., and Li, J. Y. (2013b). Overexpression of maize anthocyanin regulatory gene Lc affects rice fertility. Biotechnol Lett. 35, 115–119. doi: 10.1007/s10529-012-1046-9
Liu, J., Osbourn, A., and Ma, P. (2015). MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8, 689–708. doi: 10.1016/j.molp.2015.03.012
Lu, H. P., Liu, S. M., Xu, S. L., Chen, W. Y., Zhou, X., Tan, Y. Y., et al. (2017). CRISPR-S: an active interference element for a rapid and inexpensive selection of genome-edited, transgene-free rice plants. Plant Biotechnol. J. 15, 1371–1373. doi: 10.1111/pbi.12788
Mao, Y., Zhang, H., Xu, N., Zhang, B., Gou, F., and Zhu, J. K. (2013). Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol. Plant 6, 2008–2011. doi: 10.1093/mp/sst121
Miao, J., Guo, D., Zhang, J., Huang, Q., Qin, G., Zhang, X., et al. (2013). Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23, 1233–1236. doi: 10.1038/cr.2013.123
Miki, D., Zhang, W., Zeng, W., Feng, Z., and Zhu, J. K. (2018). CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation. Nat. Commun. 9:1967. doi: 10.1038/s41467-018-04416-0
Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J. D., and Kamoun, S. (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 691–693. doi: 10.1038/nbt.2655
Ouyang, L., Ma, M., and Li, L. (2020). An efficient transgene-free DNA-editing system for Arabidopsis using a fluorescent marker. Biotechnol. Lett. 42, 313–318. doi: 10.1007/s10529-019-02778-z
Paz-Ares, J., Wienand, U., Peterson, P. A., and Saedler, H. (1986). Molecular cloning of the c locus of Zea mays: a locus regulating the anthocyanin pathway. Embo J. 5, 829–833. doi: 10.1002/j.1460-2075.1986.tb04291.x
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686–688. doi: 10.1038/nbt.2650
Shin, Y. M., Park, H. J., Yim, S. D., Baek, N. I., Lee, C. H., An, G., et al. (2006). Transgenic rice lines expressing maize C1 and R-S regulatory genes produce various flavonoids in the endosperm. Plant Biotechnol. J. 4, 303–315. doi: 10.1111/j.1467-7652.2006.00182.x
Svitashev, S., Schwartz, C., Lenderts, B., Young, J. K., and Mark Cigan, A. (2016). Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat. Commun. 7:13274. doi: 10.1038/ncomms13274
Tanaka, Y., Sasaki, N., and Ohmiya, A. (2008). Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 54, 733–749. doi: 10.1111/j.1365-313X.2008.03447.x
Wang, J., and Chen, H. (2020). A novel CRISPR/Cas9 system for efficiently generating Cas9-free multiplex mutants in Arabidopsis. aBIOTECH. 1, 6–14. doi: 10.1007/s42994-019-00011-z
Wang, Z. P., Xing, H. L., Dong, L., Zhang, H. Y., Han, C. Y., Wang, X. C., et al. (2015). Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16:144. doi: 10.1186/s13059-015-0715-0
Wu, T. M., Huang, J. Z., Oung, H. M., Hsu, Y. T., Tsai, Y. C., and Hong, C. Y. (2019). H(2)O(2)-based method for rapid detection of transgene-free rice plants from segregating CRISPR/Cas9 genome-edited progenies. Int. J. Mol. Sci. 20:3885. doi: 10.3390/ijms20163885
Xiao, B. Z., Chen, X., Xiang, C. B., Tang, N., Zhang, Q. F., and Xiong, L. Z. (2009). Evaluation of seven function-known candidate genes for their effects on improving drought resistance of transgenic rice under field conditions. Mol. Plant. 2, 73–83. doi: 10.1093/mp/ssn068
Xie, K., and Yang, Y. (2013). RNA-guided genome editing in plants using a CRISPR-Cas system. Mol. Plant 6, 1975–1983. doi: 10.1093/mp/sst119
Xie, X., Ma, X., Zhu, Q., Zeng, D., Li, G., and Liu, Y. G. (2017). CRISPR-GE: a convenient software toolkit for CRISPR-based genome editing. Mol. Plant 10, 1246–1249. doi: 10.1016/j.molp.2017.06.004
Xu, W., Dubos, C., and Lepiniec, L. (2015). Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 20, 176–185. doi: 10.1016/j.tplants.2014.12.001
Yoshida, A., Suzaki, T., Tanaka, W., and Hirano, H. Y. (2009). The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc. Natl. Acad. Sci. U.S.A. 106, 20103–20108. doi: 10.1073/pnas.0907896106
Yu, H., and Zhao, Y. (2019). Fluorescence marker-assisted isolation of Cas9-free and CRISPR-edited Arabidopsis plants. Methods Mol. Biol. 1917, 147–154. doi: 10.1007/978-1-4939-8991-1_11
Zhang, R., Liu, J., Chai, Z., Chen, S., Bai, Y., Zong, Y., et al. (2019). Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat. Plants 5, 480–485. doi: 10.1038/s41477-019-0405-0
Zhang, Y., Butelli, E., and Martin, C. (2014). Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 19, 81–90. doi: 10.1016/j.pbi.2014.05.011
Zhang, Y., Liang, Z., Zong, Y., Wang, Y., Liu, J., Chen, K., et al. (2016). Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7:12617. doi: 10.1038/ncomms12617
Zhao, S., Wang, C., Ma, J., Wang, S., Tian, P., Wang, J., et al. (2016). Map-based cloning and functional analysis of the chromogen gene C in rice (Oryza sativa L.). J. Plant Biol. 59, 496–505. doi: 10.1007/s12374-016-0227-9
Zheng, J., Wu, H., Zhu, H., Huang, C., Liu, C., Chang, Y., et al. (2019). Determining factors, regulation system, and domestication of anthocyanin biosynthesis in rice leaves. New Phytol. 223, 705–721. doi: 10.1111/nph.15807
Keywords: CRISPR, transgene-free, anthocyanin, rice, AAC
Citation: He Y, Zhu M, Wu J, Ouyang L, Wang R, Sun H, Yan L, Wang L, Xu M, Zhan H and Zhao Y (2020) Repurposing of Anthocyanin Biosynthesis for Plant Transformation and Genome Editing. Front. Genome Ed. 2:607982. doi: 10.3389/fgeed.2020.607982
Received: 18 September 2020; Accepted: 09 November 2020;
Published: 03 December 2020.
Edited by:
Lanqin Xia, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Yiping Qi, University of Maryland, United StatesKan Wang, Iowa State University, United States
Copyright © 2020 He, Zhu, Wu, Ouyang, Wang, Sun, Yan, Wang, Xu, Zhan and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yubing He, eXViaW5naGVAbmphdS5lZHUuY24=
†These authors have contributed equally to this work
 Yubing He
Yubing He Min Zhu
Min Zhu Junhua Wu
Junhua Wu Lejun Ouyang3
Lejun Ouyang3 Rongchen Wang
Rongchen Wang Yunde Zhao
Yunde Zhao