
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Fungal Biol. , 13 November 2023
Sec. Fungi-Plant Interactions
Volume 4 - 2023 | https://doi.org/10.3389/ffunb.2023.1278525
In this study aiming to investigate potential fungal biocontrol agents for Fusarium culmorum, several isolates of Trichoderma spp. were evaluated for their antagonistic effects by means of transcriptional analyses. At first, 21 monosporic Trichoderma spp. isolates were obtained from natural wood debris and wood area soils in Manisa, Turkey. Trichoderma spp. Isolates were identified as belonging to four different species (T. atroviride, T. harzianum, T. koningii, and T. brevicompactum) by tef1-α sequencing. Then, the linear growth rate (LGR) of each species was calculated and determined to be in a range between 13.22 ± 0.71 mm/day (T. atroviride TR2) and 25.06 ± 1.45 mm/day (T. harzianum K30). Inter-simple sequence repeat (ISSR) genotyping validated the tef1-α sequencing results by presenting two sub-clusters in the dendrogram. We determined the genetically most similar (TR1 & TR2; 97.77%) and dissimilar (K9 & K17; 40.40%) individuals belonging to the same and different species, respectively. Dual sandwich culture tests (which are useful for antagonism studies) revealed that T. harzianum K21 (the least suppressive) and T. brevicompactum K26 (the most suppressive) isolates suppressed F. culmorum with growth rates of 3% and 46%, respectively. Expressions of genes previously associated with mycoparasitism-plant protection-secondary metabolism (nag1, tgf-1, and tmk-1) were tested by quantitative real-time polymerase chain reaction (qRT-PCR) in both those isolates. While there were no significant differences (p>0.05) in expression that were present in the K21 isolate, those three genes were upregulated with fold change values of 2.69 ± 0.26 (p<0.001), 2.23 ± 0.16 (p<0.001), and 5.38 ± 2.01 (p<0.05) in K26, meaning that the presence of significant alteration in the physiological processes of the fungus. Also, its mycoparasitism potential was tested on Triticum aestivum L. cv Basribey in planta, which was infected with the F. culmorum FcUK99 strain. Results of the trials, including specific plant growth parameters (weight or length of plantlets), confirmed the mycoparasitic potential of the isolate. It can be concluded that (i) nag1, tgf-1, and tmk-1 genes could be approved as reliable markers for evaluation of BCA capacities of Trichoderma spp. and (ii) the T. brevicompactum K26 strain can be suggested as a promising candidate for combating in F. culmorum diseases following the necessary procedures to ensure it is non-hazardous and safe.
Trichoderma is a fungal genus with more than 340 members, which are mainly found in soil and plants (Sariah et al., 2005; Amin and Razdan, 2010; Strauch et al., 2011). Trichoderma spp. is of great importance among filamentous fungi in terms of adapting to different ecological conditions and lifestyles and providing benefits to plant nutrition and immune defence. Trichoderma spp., which is saprophytic, contributes to recycling in nature by breaking down dead plant/organic residues and supporting essential nutrients for plant growth (Zeilinger et al., 2016; Singh et al., 2019). Thus, increasing the yield of the plant also prevents plant diseases caused by pathogens by attaching to plant roots and inhibiting the growth (Amin and Razdan, 2010; Mukherjee et al., 2012). Some fungal plant diseases could be controlled by this manner using Trichoderma instead of synthetic pesticides. With all these beneficial features and abilities, Trichoderma species can be mostly and widely used for biocontrol (Rojo et al., 2007; Zapparata et al., 2021; Yörük et al., 2022).
Microbial biocontrol agents have been preferred in recent years for the fight against the growth of pathogenic microorganisms (Mohamed and Haggag, 2006; Rojo et al., 2007; Zapparata et al., 2021; Yörük et al., 2022). In this context, Trichoderma spp. has been found suitable in controlling phytopathogenic fungi such as Fusarium sp., Rhizoctonia sp., and Sclerotium sp. (Elad et al., 1980; Matarese et al., 2012; Abbas et al., 2017; Rai et al., 2019). The most common biocontrol fungi are T. harzanium, T. afroharzanium, T. viride, T. virens, and T. atroviride (Mohamed and Haggag, 2006; Dubey et al., 2007; Amin and Razdan, 2010; Strauch et al., 2011). As a result, Trichoderma species provide a good opportunity to determine a useful strategy for both human/animal health and protecting agricultural products in an eco-friendly manner. In addition, some Trichoderma species are also known to show potential for industrial uses such as biodegradation of cellulose and biofuel production (Reithner et al., 2011; Druzhinina and Kubicek, 2017).
In literature, the valuable scientific studies and data on the active or secondary metabolites and volatile compounds produced by Trichoderma species are abundant in the study of the gene/gene clusters responsible for this metabolite production; its related activity is rare. Therefore, the molecular basis for the opportunistic success of Trichoderma is not yet fully understood, and access to detailed genetic characterization of genes encoding proteins of key importance to biological control strategies in Trichoderma spp. isolates is still limited (Küçük and Kivanc, 2005; Küçük et al., 2007; Reithner et al., 2011; Singh et al., 2019; Abbas et al., 2022; Gazdík et al., 2023). There is still limited knowledge of the linkage between genetic characteristics and biocontrol capacities of Trichoderma spp. isolates worldwide. Because of this gap in knowledge, studies on gene pathways related to mycoparasitism will gain growing momentum in coming years. Performing expression analyses in different Trichoderma spp. strains showing control activity under experimental/environmental conditions will also contribute to the determination of different genome features in terms of functional adaptation.
In recent years, Trichoderma sp. has been used as an effective biological control agent in the fight against Fusarium had blight (FHB) and root rot (RR) diseases caused by Fusarium spp (Matarese et al., 2012; Zapparata et al., 2021; Yörük et al., 2022; Gazdík et al., 2023; Risoli et al., 2023). FHB and RR diseases have been seen in plenty of plant species and have been often caused by F. graminearum and F. culmorum species worldwide (Matny, 2015; Yörük and Yli-Mattila, 2019; Mielniczuk and Skwaryło-Bednarz, 2020). These diseases inhibit plant growth, lead to yield loss, and harm the overall health of plants. Not only plant health but also animal and human health are threatened by FHB and RR epidemics. Besides, mycotoxins such as deoxynivalenol and zearalenone are accumulated on small grain cereals and their related products (Goswami and Kistler, 2004; Desjardins and Proctor, 2007; Matny, 2015; Miedaner et al., 2017; Yörük and Yli-Mattila, 2019; Mielniczuk and Skwaryło-Bednarz, 2020). Trichoderma species could act by different mechanisms in the control of FHB and RR disease. These mechanisms could be grouped into antagonism, competition, root protection, and stimulation of the plant immune system (Amin and Razdan, 2010; Hermosa et al., 2012; Matarese et al., 2012). In order to use the beneficial effects of Trichoderma species, factors such as selection of the appropriate Trichoderma species, application dosage, and timing of application must be considered. Also, detailed morphological, transcriptomic, and metabolomic analyses of Trichoderma species are needed to create an effective defence strategy.
Morphological and molecular identification of Trichoderma species, phylogenetic analysis, determination of mycoparasitic effects, observation of the effects of volatile compound production (on phytopathogenic fungus), detection of the presence of genes related to mycoparasitism and secondary metabolite production, and investigation of the expression levels of these target genes provide important information in terms of their potential use as biocontrol agents (BCA). Obtaining this information would provide an advantage in the use of Trichoderma spp. with its antagonistic effects, such as genetically engineered BCAs producing effective metabolites against phytopathogens. However, these positive effects should be confirmed by different morphological, physiological, and genetic methods. This way, within the scope of the current study, genetic identification was performed at the species level after the detailed morphological characterization of Trichoderma spp. isolates originating from Turkey. The presence and the expression of genes related to secondary metabolites production by these isolates and their effects as a biocontrol agent on F. culmorum, an important plant pathogen, were investigated. Thus, for the first time, identification, detailed genetic characterization, biocontrol potential, and comparative gene expression analysis in Trichoderma spp. from Turkey were carried out.
Trichoderma spp. isolates, which were previously obtained from natural wood debris and wood area soils from Manisa, Turkey, were used in this study (Table 1). Prior to in vitro cultivation assays, morphological observations, including branching type, phialide length, and conidia shape, were carried out following the protocols provided by Hoyos-Carvajal et al., 2009. Protocols provided by Irzykowska et al. (2013) and Yörük (2018) for solid and liquid cultures were used with slight modifications. After checking morphological characteristics carefully, each isolate was cultured on potato dextrose agar (PDA) plates for 7 days at 26°C ± 2°C for linear growth rate determination. Carboxymethylcellulose (CMC) medium, inducer medium for spore production, was used in disease severity determination analysis. Up to seven slices obtained from PDA cultures were added to CMC medium, and isolates were incubated at 23°C ± 2°C by shaking 150 rpm.
For the analysis of the linear growth rates of the 21 isolates obtained, the growth diameters (on PDA media) were measured) on days 4 and 7. The mean values as mm/day were recorded and statistical analyses were performed using descriptive statistics, Tukey’s post-test, and one-way analysis of variance (ANOVA) by Graphpad Prism 9.0. (Dotmatics, U.S.A.).
In species-specific identification of Trichoderma spp. isolates, gDNAs were isolated from single-spore isolates. SDS-based gDNA isolation was performed from 7-day-old cultures as reported by Niu et al. (2008) and Yörük et al. (2016). Fresh mycelium of 50 mg was homogenized using a microtube homogenizer (Merck-BeadBugTM, Germany) accompanied with 2 mg glass beads by vortexing samples at 3.000 rpm for 30s twice. The homogenized samples were then subjected to digestion, ligation, washing, and elution steps in gDNA isolation. The obtained gDNA molecules were analyzed qualitatively by 1% agarose gel electrophoresis (70 V for 45 min), and quantitatively by spectrophotometer (Thermo, U.S.A.) by measuring the absorbance values at 260 and 280 nm wavelengths for being used in PCR reaction. Quantitative analysis was used to adjust amounts of gDNA for PCRs.
The tef1-α gene, which is widely used for species-level diagnosis of fungi, due to it being highly conserved among fungal species, was selected for species-specific identification (Cai and Druzhinina, 2021; Xue et al., 2021; please see https://trichoderma.info/). The final concentrations of the components of PCRs were as follows: 50 ng gDNA, 1X PCR buffer, 2.5 mM MgCl2, 0.2 mM dNTP mixture, 10 pmol each primer (O’Donnell et al., 1998), and 0.04 U/μL Taq DNA polymerase (Episozyme, Turkey). Following pre-denaturation at 94 °C for 5 min in a thermal cycler, PCR was completed with 35 cycles and a final extension (5 minutes at 72 °C). A total of 35 cycles were performed at 94 °C for 30 s, 57 °C for 30 s, and 72 °C for 45 s. The presence of PCR bands was visualized by 1% agarose gel electrophoresis in the presence of a UV transilluminator at a 70 V constant current for 1 hour. PCR products were purified following the protocol (of the commercial kit) recommended by the manufacturer (Macherey-Nagel, Germany). Sanger sequencing process was performed using a BigDyeTM Terminator kit (Thermo, USA) with DNA sequencing system (Abi Prism-Thermo, U.S.A.). The sequencing reaction was carried out in a reaction volume of 20µL, including a 20ng PCR template, 3 pmol of each primer, and 1X Reaction Buffer by following manufacturer recommendation protocols. The chromatograms were visualized and analyzed with Chromas Pro software (Technelysium, Australia). The sequences were compared for the sequence similarities in the NCBI database by BlastN analysis. E value < 0.05, bit score >50, and query coverage >90% sequences were considered scientifically significant (Altschul et al., 1990). Multiple alignment of the obtained TEF1-α sequences was performed with ClustalW online software (please see https://www.genome.jp/tools-bin/clustalw). Aligned sequences were subjected to pairwise distance and Neighbor-Joining topology analysis via Mega 11.0 software and the Bootstrap support value was calculated for the phylogenetic tree with at least 1000 replicates in order to eliminate potential technical errors in phylogenetic analysis (Tamura et al., 2021). Principal component analysis for TEF1-α was carried out using R/R-Studio software. ‘.aln’ files obtained from multiple alignments were converted into ‘.phy’ files and then used in PCA analysis by use of R with ‘readxl’, ‘ape’, ‘phangorn’, and ‘factoextra’ packages.
Genotyping of Trichoderma spp. isolates were performed based on ISSR markers. These markers are particularly useful in genotyping and population genetic studies because they can reveal genetic diversity and relationships among individuals or isolates intra- or inter-specific levels. In total, 10 primers containing di-, tri-, and tetra-nucleotide motifs were used (Table 2). In ISSR-PCR, the components were combined in a final concentration consisting of 100 ng gDNA, 1X buffer, 3 mM MgCl2, 0.4 mM dNTP mixture, 10 pmol primer, 0.04 U/μL Taq DNA polymerase enzyme. The PCR reaction was performed in two-step loops to ensure strong binding of primers to the gDNA template. Following pre-denaturation at 94 °C for 5 min, PCR was performed in two steps. In the first stage, 7 cycles were cycled at 94°C for 1 min, 40°C for 1 min, and 72°C for 3 min. The next 35 cycles were performed at 94 °C for 1 min, 48 °C for 1 min, and 72 °C for 2 min. The final elongation step was completed at 72 °C for 10 min. PCR products were run on 2% agarose gel electrophoresis and visualized under a UV transilluminator. Genetic diversity was analyzed according to the Nei and Li coefficient (Nei and Li, 1979) by scoring the presence of bands with ‘1’ and absence with ‘0’ and using MVSP 3.2.1 (multi-variate statistical package-Kovach). 2D principal component analysis (PCA) graphics and dendrograms were also produced by MVSP software. Polymorphism informative content (PIC) and resolution power (RP) values were also evaluated according to previous publications (Roldan-Ruiz et al., 2000; Prevost and Wilkinson, 1999; Alkan et al., 2019).
In the study, the F. culmorum FcUK99 reference strain was used to demonstrate the mycoparasitic effects and BCA capacity of Trichoderma spp. isolates on it. The mycoparasitic effect and inhibition potential of both Trichoderma and FcUK99 isolates [the FcUK99 genome project was made, and this isolate has been the subject of many different morphological, physiological, and chemotype analyses and genetic studies (Urban et al., 2016)] were analyzed by subculturing 5-day-old cultures. The inhibition caused by Trichoderma species producing volatile compounds was determined by sandwich dual culture technique as reported by Yörük et al. (2022). Sandwich cultures were established in PDA by using a 0.25 cm2 agar plug of FcUK99 and Trichoderma isolates placed onto the PDA at the same time and conditions as a single culture. For this purpose, sandwich cultures were incubated for 5 days 26°C ± 2°C. The suppression observed in F. culmorum was evaluated according to common formula % inhibition = [(mycelial diameter control - mycelial diameter experiment)/mycelial diameter control] x 100 and the isolates were characterized according to the following value ranges (Popiel et al., 2008; Błaszczyk et al., 2017):
- Very active, > 19 mm suppression zone
- Effective, 13-19 mm suppression zone
- Partially active, 10-12.99 mm suppression zone
- No effect, <10 mm suppression zone
As target genes, nag1 (N-acetylglucosaminidase) and tgf-1 (histone acetyltransferase), previously found to be associated with secondary metabolism and mycoparasitism, and tmk1 (mitogen-activated protein kinase), which is associated with plant protection-secondary metabolism production (Brunner et al., 2003; Reithner et al., 2007; Gómez-Rodriguez et al., 2018), were selected to be used in qRT-PCR assays due to their potential for being a marker gene for mycoparasitism. Total RNA isolation from day 7 mycelium of Trichoderma spp. Mono and sandwich dual cultures was performed using Trizol-based monophasic compound (due to universal usage of this agent and its non-tissue/sample specificity). Mycelium (50-200 mg of wet weight) was homogenized with 1 mL of monophasic compound in a digestion device. The nucleoprotein complex was separated with chloroform, and total RNA molecules were precipitated with isoamyl alcohol and solubilized with RNase-free distilled water. After DNAseI treatment (Zymo Research, U.S.A.), Qualitative and quantitative analyses of total RNA were performed by 1% agarose gel electrophoresis and spectrophotometric measurements. From the quantified total RNAs, cDNA translation was performed using a commercial kit (Takara, Japan). cDNA translation followed the protocol recommended by the kit.
Differences in gene expression levels in dual cultures were investigated by qRT-PCR. The α-actin gene encodes a structural cytoskeletal protein and is known to be highly stable. This gene was used as a housekeeping gene, while genes detected by standard PCR were used as target genes. The qRT-PCR primers were designed with Primer3 software by accessing the accessions in the NCBI database (Supplementary File 1). qRT-PCR assays were performed based on “SYBR Green I” fluorophore. The reaction was performed in a total volume of 12µL. qR-TPCR Components 50 ng cDNA, 5 pmol forward and reverse primers, and1X SYBR Green Mix (Episozyme, Turkey) were combined in a qRT-PCR 96-wellplate. Samples were kept at 95°C for 2 minutes for pre-denaturation. Replication was performed in 45 cycles of 10 seconds at 95°C, 15 seconds at 60°C, and 20 seconds at 72°C. This was followed by a waiting phase of 30 seconds at 40°C and a melting curve temperature scan. The results were analyzed after three biological and three technical replicates. Standard plots were performed in 4 logarithmic phases at 1/4 dilution ratios. Gene expression profiles will be interpreted according to 2-ΔΔCT normalization values (Livak and Schmittgen, 2001).
Two genotypes showing different biocontrol activity for Trichoderma spp. as a high suppressor (K26) and low suppressor (K21) for F. culmorum FcUK99 strains were selected for in planta tests for evaluating their BCA potential on F. culmorum. A common wheat cultivar, Triticum aestivum L. cv. Basribey, was used as a plant host. The inoculum source was obtained from F. culmorum FcUK99 strain grown on carboxymethylcellulose (CMC) medium at 23 ± 2°C by shaking cultures of 50 mL with 150 rpm rotary shaker speed for 5 days. 1x106 macroconidium (filtered by 2X gauze) was added to the autoclaved soil with the ratio of 5:95/W:V/macroconidium:soil (Moya-Elizondo and Jacobsen, 2016; Moradi et al., 2017). After three days of inoculation, wheat seeds (surface sterilized as reported by Gargouri-Kammoun et al., 2009) and 1x107 spores of Trichoderma spp. (grown on potato dextrose broth [PDB] for 5 days at 23 ± 2°C by 150 rpm; including 20 µL Tween20) were incubated for 2 hours 23 ± 2°C by orbital shaker, and then seeds were left to be dried out in laminar flow. After 3 days of soil inoculation with F. culmorum, wheat seeds (treated with Trichoderma spp.) were planted onto plastic pots of 13 cm diameter and left for plant growth at 16/8 light and dark conditions at 26°C with 1500 lux for 3 weeks. Fresh weight (FW for gram), dry weight (DW for gram), and seedling length (SL for cm) of plantlets were recorded after 3 weeks. Statistical analysis was carried out as reported before using Graphpad Prism 9.0.
2D and 3D analysis were carried out for evaluating different technical protocol sets in a single way. For this purpose, firstly LGR values and mycoparasitism test (BCA) data were co-evaluated by PCA test by R/R-Studio using ‘readxl’, ‘devtools’, and ‘ggbiplot’ packages. Variation values for the dimensions and plots were produced by R. Similarly, by using R/R-Studio, each distinct technical procedure (LGR, BCA, gene expressions, DW, FW, and SL data) was co-evaluated in a single way for K21 and K26.
A total of 21 isolates of Trichoderma sp. were obtained by single spore isolation protocol from infected plant/soil sources taken from different regions. Four different species were detected via morphological identification criteria. The growth of the isolates (on PDA) was measured on petri on days 4 and 7, and LGRs (mm/day) were calculated. The highest linear growth rate was measured at 25.06 ± 1.45mm/day in the K30 isolate, while the lowest linear growth rate was calculated at 13.22 ± 1.78mm/day in the TR2 isolate (Figure 1).
gDNA was successfully isolated from all 21 isolates. TEF1-α was amplified ~0.8 kb in all isolates (Supplementary File 2), PCR products were purified from agarose gels and then sequenced. After BLASTN analysis, E values were recorded with the range from %91 to %99, respectively (Table 1). With the concordance of morphological characteristics of the isolates, 12 isolates belonged to T. harzianum, 7 isolates were characterized as T. atroviride, 1 isolate as T. koningii, and 1 isolate as T. brevicompactum. NJ topology analysis easily distinguished all isolates according to their origin of the species (Figure 2A). Similarly, PCA assays revealed two major groups with PCA1 and PCA2 scores of percentages of 89.2% and 5.6%, respectively (Figure 2B).
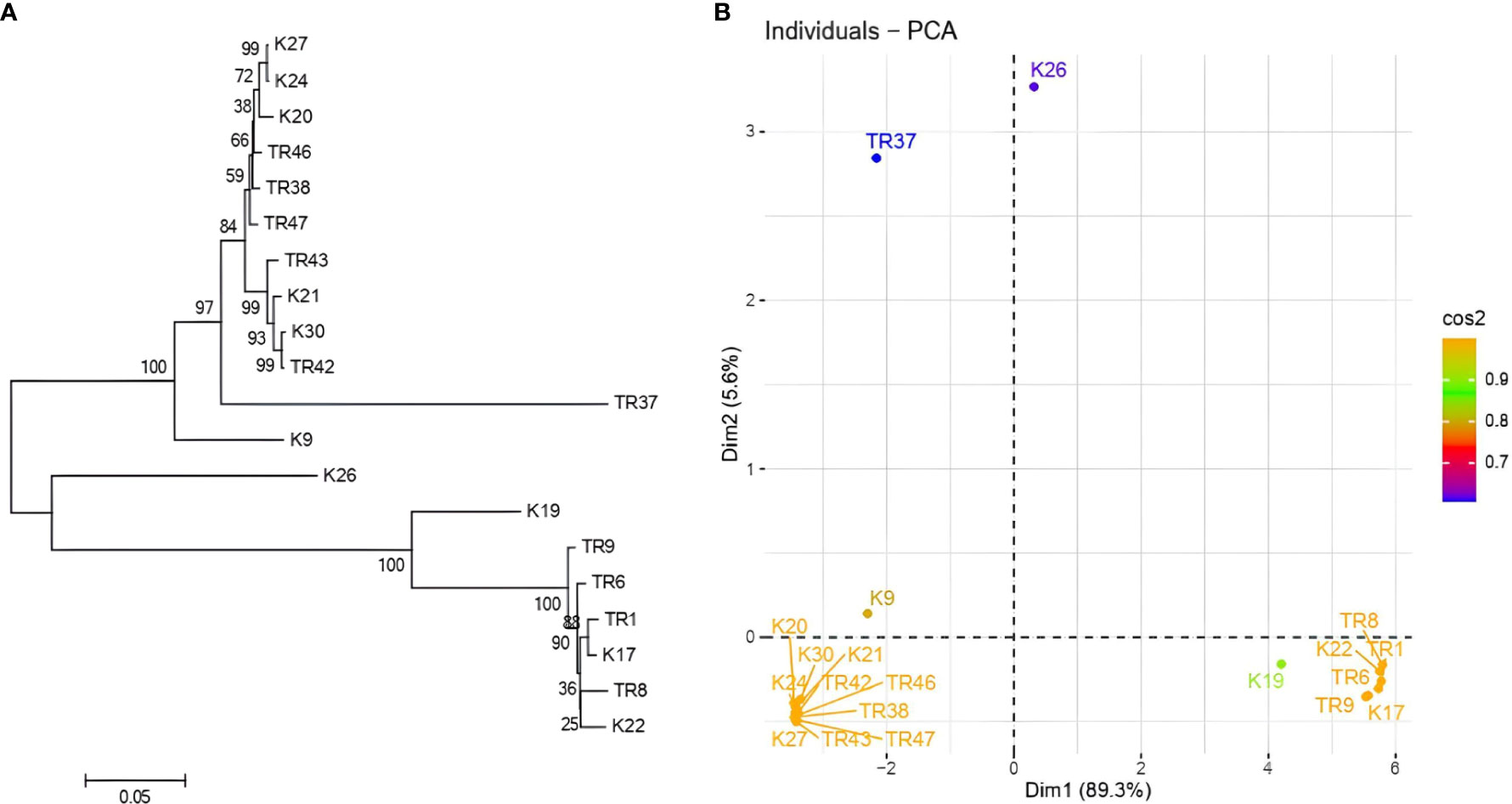
Figure 2 (A) NJ topology dendrogram from MEGA 11. Software, and (B) PCA graphic from R for tef1-α sequencing results in Trichoderma spp.
In total, 10 ISSR primers were used in the ISSR PCRs, and all of them yielded PCR products (Table 2). It was observed that 93 ISSR bands with 10 ISSR primers were generated in 21 Trichoderma isolates, and 88 bands were recorded as polymorphic (94.62%). The maximum number of ISSR bands (13 bands) was observed with the UBC866 and AYS10 primers while the minimum number of bands (3 bands) was recorded with the UBC872 primer. Figure 3A presents USSR profiling in the UBC828 primer. According to the ISSR analysis, a similarity matrix revealed that maximum similarity was present between TR1 and TR2 T. atroviride isolates with 97% similarity. Minimum similarity was detected as 30.6% between TR46 (T. harzianum) and TR8 (T. atroviride). The dendrogram (Figure 3B) and PCA biplot (Figure 3C) showed phylogenetic branching through two main branches. The members of T. harzianum and T. atroviride species showed co-clustering. Among the 21 isolates, 2 isolates with different species (T. koningii and T. brevicompactum) and showed a distant branching to T. atroviride and T. harzianum species.
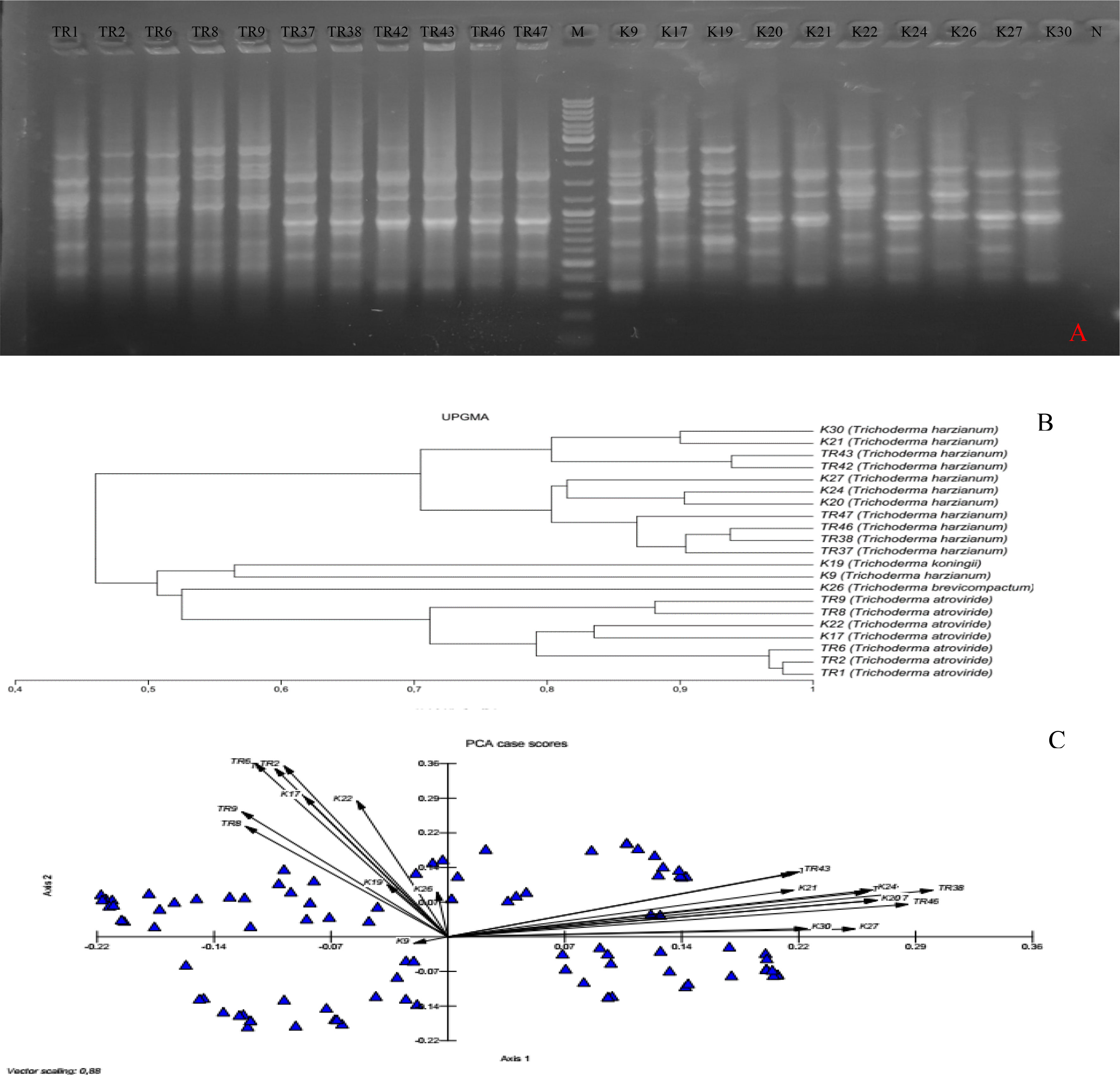
Figure 3 (A) UBC828 agarose gel profiling, (B) UPGMA dendrogram, (C) and PCA plot data obtained from MVSP 3.21 software.
LGR for FcUK99 treated with Trichoderma spp. isolates via sandwich cultures were recorded at the days 3, 4, and 5 of incubation. According to the measurement results obtained, among 21 different Trichoderma spp. isolates, the highest mycoparasitic effect on FcUK99 was found to be isolate K26 with 43% suppression, while the lowest mycoparasitic effect was observed in isolate K21 with 3% suppression (Figure 4). The overall mycoparasitic effect of all isolates was measured mean percentages of 20.77 ± 2.49. K21 and K26 isolates were selected for further molecular analysis.
In the study, K21 and K26 isolates were subjected to gene expression analysis. qRT-PCRs were used in expression analysis of the tgf-1, nag1, and tmk1 genes. Target gene expressions were normalized according to the α-actin gene. Total RNAs with high quality (Δ;260/280 = 1.9-2.0) and quantity (1-2 µg/µl) were obtained. The fold changes in expression of the tgf-1, nag1, and tmk1 genes were calculated as 1.15 ± 0.51, 1.16 ± 0.62, and 1.32 ± 0.35, respectively. No significant differences (p>0,05, ns) were present in the K21 isolate (Figure 5A). However, fold changes in gene expression levels of the tgf-1, nag1, and tmk1 genes in the K26 isolate were determined as 2.69 ± 0.26 (p<0.001, ****), 2.23 ± 0.16 (p<0.001, ****), and 5.38 ± 2.01 (p<0.05, *) respectively (Figure 5B). The expression of tgf-1, nag1, and tmk1 genes was found to be significantly altered in K26.
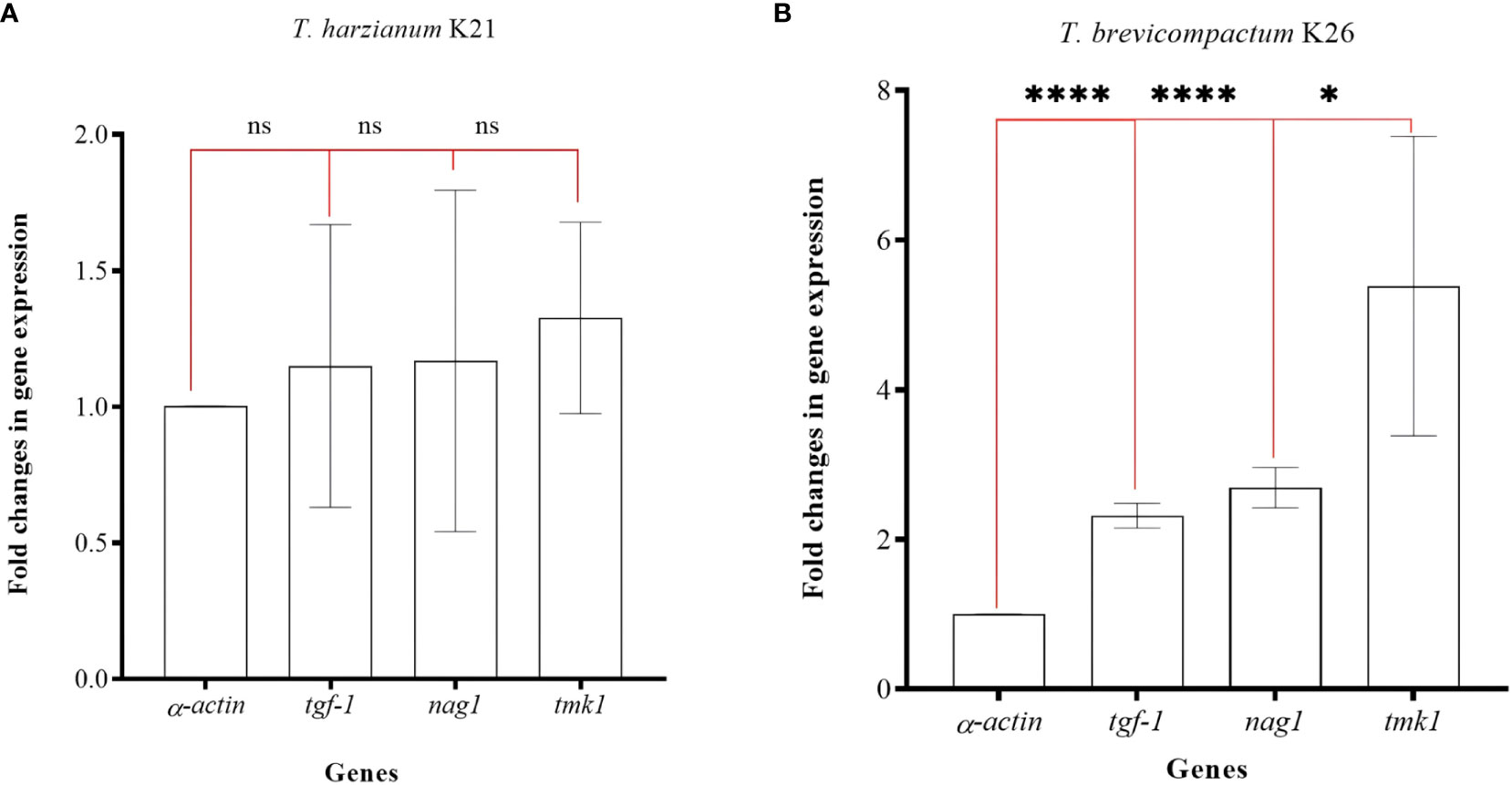
Figure 5 Fold changes in gene expression values for three genes in K21 (A) and K26 (B) isolates. ‘ns’ means no significant changes, ‘*’ means p>0.05, and ‘****’ means p<0.001.
After conducting the connection platform for phytopathogen, BCA, and plant host, physiological changes in plantlets were recorded. This way, the mycoparastism potential of selected Trichoderma isolates was tested on Triticum aestivum L. cv Basribey infected with F. culmorum FcUK99 strain. FW values were recorded as 0.18 ± 0.01 gr, 0.06 ± 0.008 gr (p<0.001, ****), 0.15 ± 0.02 gr (p>0.05, ns), and 0.15 ± 0.02 gr (p>0.05, ns) for the control plant (non-infected and non-BCA treated), positive control (only phytopathogen infected plant), experiment set for K21 (FcUK99 infected and K21), and experiment set for K26 (FcUK99 infected and K26 treated plant), respectively (Figure 5). In comparison to the control set, both K21 and K26 reduced the adverse effects of FcUK99 on wheat. DW values were calculated as 0.018 ± 0.001 gr, 0.009 ± 0.0009 gr (p<0.01, ***), 0.013 ± 0.001 gr (p>0.05, ns), and 0.015 ± 0.001 gr (p>0.05, ns) for the control plant (non-infected and non-BCA treated), positive control (only phytopathogen infected plant), experiment set for K21 (FcUK99 infected and K21), and experiment set for K26 (FcUK99 infected and K26 treated plant), respectively (Figure 5). Like DW results, both Trichoderma isolates reduced the adverse effects of FcUK99 on wheat. SL values were recorded as 25.58 ± 1.06 cm, 20.12 ± 2.07 cm (p<0.05, *), 24.62 ± 1.99 cm (p>0.05, ns), and 26.78 ± 0.52 cm (p>0.05, ns) for the control plant (non-infected and non-BCA treated), positive control (only phytopathogen infected plant), experiment set for K21 (FcUK99 infected and K21), and experiment set for K26 (FcUK99 infected and K26 treated plant), respectively (Figures 6A–D). Like FW and DW data, Trichoderma isolates reduced the potential growth of pathogen in wheat. The decreased level of disruption in F. culmorum-infected plantlets was clearly related to Trichoderma treatment.

Figure 6 3-week-old plantlet of T. aestivum L. cv. Basribey. (A) Control plant (non-infected and non-BCA treated), (B) positive control (only phytopathogen infected plant), (C) experiment set for K21 (FcUK99 infected and K21), and (D) experiment set for K26 (FcUK99 infected and K26 treated plant).
For LGR and BCA analysis, a PCA test was carried out using R. According to the PCA plot, data obtained for each Trichoderma isolate co-clustered in regard with species origin. Especially, T. atroviride isolates were distributed within the same ellipses for LGR data (Figure 7). Even if T. koningii K19 isolates were co-clustered in T. harzanium isolates and it showed similar pattern for LGR and BCA data, a clear distinction was present for T. brevicompactum K26 isolates for PCA biplot (Figure 7A). Percentages for axis 1 and axis 2 were 58.6% and 41.4% (showing a high level of variation for two different sets of test data), respectively. A PCA graphic was also obtained by way of R for a detailed comparison of tests carried out for the BCA capacity of K21 and K26 isolates (Figure 7B). A great degree of variation (with PC1 of 36.5% and PC2 of 27.2%) was detected for the characterization of K21 and K26 isolates by PCA test. Nearly every experimental procedure but LGR clearly distinguished between the two contrasting Trichoderma genotypes.
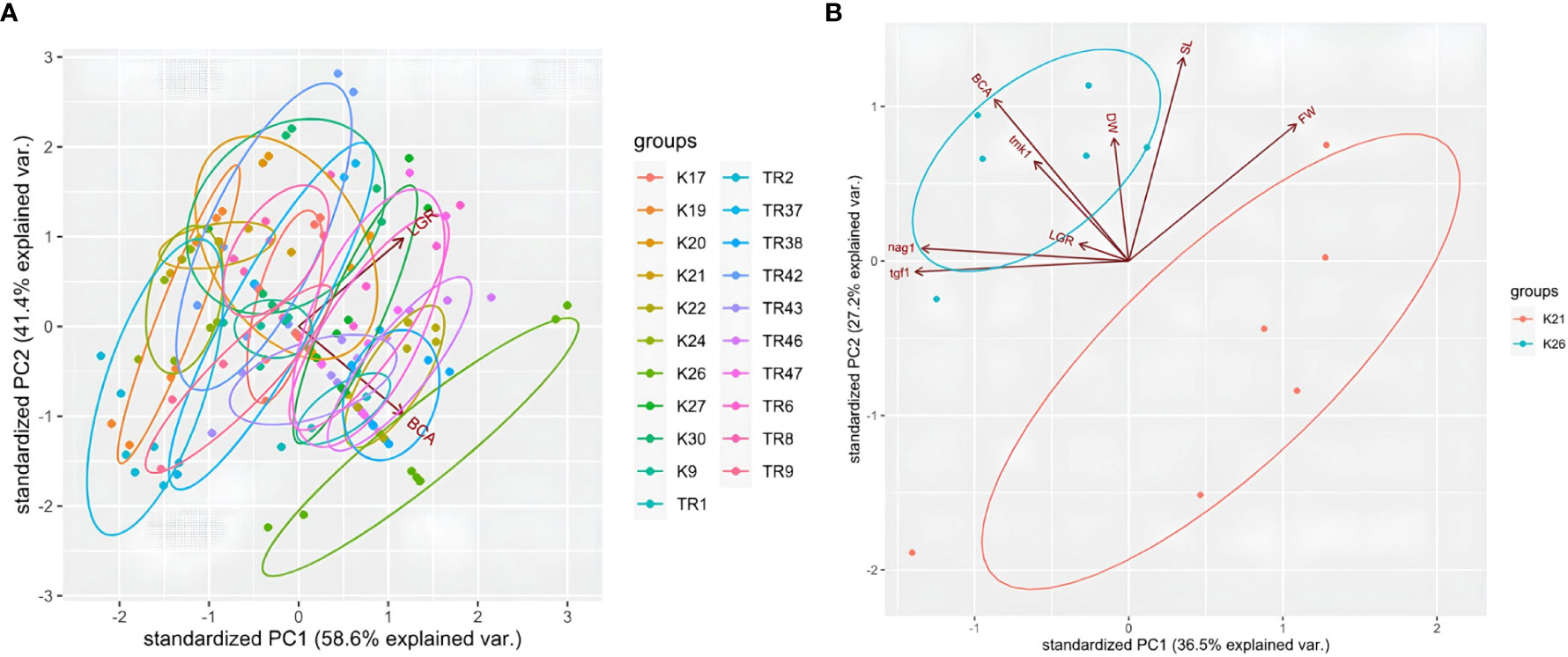
Figure 7 PCA plot for LGR and BCA analysis in 21 Trichoderma spp. isolates (A), and plot for K21 and K26 with LGR, BCA, tgf1, nag1, tmk1, DW, FW, and SL data (B).
The stress induced by treating plants with synthetic antifungal compounds promoted the resistance development in phytopathogenic fungi severely. The development of resistance in FHB and RR pathogens to common fungicides is estimated to be a highly severe problem in the next years (Chung et al., 2008; Yin et al., 2009; Zheng et al., 2015; Perlin et al., 2017). More effective disease management strategies (besides phytopathogen identification, crop rotations, novel compound usage, etc.) are needed to combat FHB and RR. In recent years, plant-derived secondary metabolites (PDSM) have started being used for in vitro tests. However, the amount of the PDSM needed for in vitro tests have been reported to be relatively higher in comparison to synthetic antifungal compounds (Elgat et al., 2020; Kong et al., 2022; Albayrak et al., 2023). Moreover, the production of PDSM (essentially essential oil derivatives) by biotechnological companies and/or obtaining these compounds could be non-cost-effective, laborious, and time-consuming. Also, in planta tests for PDSM effectiveness on plant protection are missing from the literature for nearly all publications. Thus, other strategies such as BCA usage could be more useful in FHB and RR disease management and control.
There is an extensive history of the use of Trichoderma spp. as a BCA against Fusarium oxysporum and therefore T. harzianum is the most widely known biocontrol species against phytopathogenic fungi (Mohamed and Haggag, 2006; Amin and Razdan, 2010; Yang et al., 2011; Hermosa et al., 2012; Rawat et al., 2013). T. asperellum, T. hamatum, and their products (culture filtrates or enzymes) have also been reported to be useful for use as biocontrol agents (Solanki et al., 2011; Kumar et al., 2012; Rai et al., 2019). However, in recent studies T. atroviride and other Trichoderma sp. strains have been focused on for controlling FHB, RR, and other Fusarium diseases (Matarese et al., 2012; Gómez-Rodriguez et al., 2018; Bouanaka et al., 2021; Zapparata et al., 2021; Özkale et al., 2023). In this study, members of four different Trichoderma species have been evaluated in terms of their mycoparasitisc capacities due to expressional analyses. In comparison to the results of previous studies, the BCA capacities of Trichoderma species used in this study against F. culmorum are of the acceptably effective and highly effective levels (Popiel et al., 2008; Błaszczyk et al., 2017). In particular, data obtained from LGR, BCA, and in planta tests revealed that the T. brevicompactum K26 isolate is found to be a strong suppressor of the growth of F. culmorum with BCA, LGR, DW, FW, and SL values of 43.99 ± 2.31, 22.5 ± 2.65, 0.016 ± 0.01, 0.15 ± 0.022, and 26.98 ± 0.47 respectively. The determination of the disease (FHB or RR) severity changes in Trichoderma spp.-treated plantlets could be investigated in further studies. Similarly, determination of Fusarium spp. infection levels in plant tissues after Trichoderma spp. application can also be investigated by qRT-PCR. The use of a qRT-PCR-based strategy for evaluating the potential BCA capacity of T. brevicompactum for Fusarium-infected plantlets is still missing from the literature. Besides, testing this activity of the isolate against the other Fusarium species related to FHB and RR diseases, such as F. graminearum, F. pseudograminearum, and F. avenaceum, in further studies would be very fruitful.
Phylogenetic analysis obtained from TEF1-α and ISSR fingerprinting revealed that two main sub-divisions, containing T. harzianum and T. atroviride isolates separately, were present in dendrograms and PCA plots. Moreover, both tef1-α and ISSR dendograms and TEF1-α and ISSR PCA plots presented T. brevicompactum and T. koningii isolates at the edge and extremes of the clusters. Similar results were obtained from ISSR and ITS sequencing data in Trichoderma harzianum and Trichoderma viride isolates from Delhi-India (Kumar and Sharma, 2011). However, they reported that genetic polymorphism analysis that they used did not establish a relationship between Trichoderma spp. isolates and their antagonism potential. In this study, similar patterns were detected between genetic diversity values and in vitro growth and the BCA capacity of the isolates. Moreover, contrast genotypes, K21 and K26, presented different patterns for their genetic similarity, mycoparasitism potential, and transcriptional activity for mycoparasitism-related genes.
Previous studies have not revealed a clear relationship between the mycoparasitism capacity, in vitro growth potential, mycoparasitism gene expressions, and plant protective effects of Trichoderma isolates (Ghutukade et al., 2015; Mazrou et al., 2020). In particular, correlation between mycoparasitism-related gene expression and physiological and genetic characteristics of Trichoderma isolates is missing from the literature. In addition, PIC and RP values showed that ISSR primers selected in this study presented high level of polymorphism and detailed genetic characterization for Trichoderma isolates. Mean PIC (0.47) and RP (8.67) values showed high levels of polymorphism in comparison to previous polymorphism studies (Jingade et al., 2018; Alkan et al., 2019; Aulakh et al., 2021; Yörük et al., 2022). Findings obtained from this study revealed that UBC866, UBC867, and AYS10 primers could provide good opportunity to distinguish Trichoderma isolates via ISSR fingerprinting assays.
Several similar studies on the relatedness between Trichoderma spp. genes and its mycoparasitic potential (Brunner et al., 2003; Reithner et al., 2007; Mukherjee et al., 2012; Sharma et al., 2013; Gómez-Rodriguez et al., 2018; Abbas et al., 2022) are found in literature. However, limited research on BCA-related genes and their potential use in characterizing Trichoderma isolates for their biocontrol capacities have been carried out. Three different genes and their relationships with mycoparasitism have already been confirmed previously (Brunner et al., 2003; Reithner et al., 2007; Gómez-Rodriguez et al., 2018), and they were used in gene expression analyses in this study. Each gene was significantly upregulated in a sandwich dual culture of K26 isolate in comparison to monoculturing of the strain. On the other hand, there were no significant changes for those three genes in K21 isolate. Our findings revealed that there were two contrast expression profiles in two contrast Trichoderma genotypes. K26, (the most. suppressive of Fc), showed significantly increased expressions of mycoparasitism-related genes when it was treated with F. culmorum in sandwich cultures. But no alteration in the expressions was quantified for K21 (relatively low level suppressive of Fc) in dual culture conditions. These findings concluded that (i) the genes selected in this study can be used as reliable marker genes for discriminating Trichoderma individuals for their biocontrol capacities, and (ii) changes in the expression of these genes may also be correlated with other physiological, genetic, and mycoparasitic characteristics of Trichoderma isolates. Therefore, further studies should be carried out by whole exome sequencing and/or ChiP-Seq experiments to provide more detailed information on Trichoderma isolates and their different mycoparasitic capacities.
The data presented in the study are deposited in the GenBank repository, accession numbers OR487466-OR487486 with release date 26 September, 2024.
ÖS: Conceptualization, Data curation, Methodology, Writing – original draft. EsÖ: Methodology, Writing – original draft, Investigation. EY: Methodology, Writing – original draft, Data curation, Formal Analysis. EvÖ: Methodology, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by the Research Fund of the Istanbul Yeni Yuzyil University under the project number İYYÜ-BAP-AP-2023-16.
The authors are grateful to Dr. Pierre Hellin from Walloon Agricultural Research Centre for providing the F. culmorum FcUK99 strain.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffunb.2023.1278525/full#supplementary-material
Abbas A., Jiang D., Fu Y. (2017). Trichoderma spp. as antagonist of Rhizoctonia solani. J. Plant Pathol Microbiol. 8 (3), 1–9. doi: 10.4172/2157-7471.10000402
Abbas A., Mubeen M., Zheng H., Sohail M. A., Shakeel Q., Solanki M. K., et al. (2022). Trichoderma spp. genes involved in the biocontrol activity against Rhizoctonia solani. Front. Microbiol. 13, 884469. doi: 10.3389/fmicb.2022.884469
Albayrak G., Yörük E., Teker T., Sefer Ö. (2023). Investigation of antifungal activities of myrcene on Fusarium reference strains. Arc. Microbiol. 205 (3), 82. doi: 10.1007/s00203-023-03420-3
Alkan M., Göre M. E., Bayraktar H., Özer G. (2019). Genetic variation of Fusarium spp. isolates associated with root and crown rot of winter wheat using retrotransposon-based iPBS assays. Int. J. Agri Wildlife Sci. 5 (2), 250–259. doi: 10.24180/ijaws.537423
Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 (3), 403–410. doi: 10.1016/S0022-2836(05)80360-2
Amin F., Razdan V. K. (2010). Potential of Trichoderma species as biocontrol agents of soil borne fungal propagules. J. Phytol. 2 (10), 38–41.
Aulakh S. K., Singh N., Buttar D. S., Choudhary A. K. (2021). Antagonistic potential and molecular characterization of Trichoderma asperellum against Rhizoctonia solani AG-3 causing black scurf of potato. Agric. Res. J. 58 (2), 216–223.
Błaszczyk L., Basińska-Barczak A., Ćwiek-Kupczyńska H., Gromadzka K., Popiel D., Stępień Ł. (2017). Suppressive effect of Trichoderma spp. on toxigenic Fusarium species. Pol. J. Microbiol. 66 (1), 85. doi: 10.5604/17331331.1234996
Bouanaka H., Bellil I., Harrat W., Boussaha S., Benbelkacem A., Khelifi D. (2021). On the biocontrol by Trichoderma afroharzianum against Fusarium culmorum responsible of Fusarium head blight and crown rot of wheat in Algeria. Egyp. J. Biol. Pest Cont. 31, 1–13. doi: 10.1186/s41938-021-00416-3
Brunner K., Peterbauer C. K., Mach R. L., Lorito M., Zeilinger S., Kubicek C. P. (2003). The Nag1 N-acetylglucosaminidase of Trichoderma atroviride is essential for chitinase induction by chitin and of major relevance to biocontrol. Curr. Genet. 43, 289–295. doi: 10.1007/s00294-003-0399-y
Cai F., Druzhinina I. S. (2021). In honor of John Bissett: Authoritative guidelines on molecular identification of Trichoderma. Fungal Diversity 107, 1–69. doi: 10.1007/s13225-020-00464-4
Chung W. H., Ishii H. H., Nishimura K., Ohshima M., Iwama T., Yoshimatsu H. (2008). Genetic analysis and PCR-based identification of major Fusarium species causing head blight on wheat in Japan. J. Gen. Plant Patho. 74, 364–374. doi: 10.1007/s10327-008-0110-8
Desjardins A. E., Proctor R. H. (2007). Molecular biology of Fusarium mycotoxins. Int. J. Food Microbiol. 119 (1-2), 47–50. doi: 10.1016/j.ijfoodmicro.2007.07.024
Druzhinina I. S., Kubicek C. P. (2017). Genetic engineering of Trichoderma reesei cellulases and their production. Micro. Biotechn. 10 (6), 1485–1499. doi: 10.1111/1751-7915.12726
Dubey S. C., Suresh M., Singh B. (2007). Evaluation of Trichoderma species against Fusarium oxysporum f. sp. ciceris for integrated management of chickpea wilt. Biol. Cont. 40 (1), 118–127.
Elad Y., Chet I., Katan J. (1980). Trichoderma harzianum: A biocontrol agent effective against Sclerotium rolfsii and Rhizoctonia solani. Phytopathol 70 (2), 119–121. doi: 10.1094/Phyto-70-119
Elgat W. A., Kordy A. M., Böhm M., Abdel-Megeed A., Salem M. Z. (2020). Eucalyptus camaldulensis, Citrus aurantium, and Citrus sinensis essential oils as antifungal activity against Aspergillus flavus, Aspergillus Niger, Aspergillus terreus, and Fusarium culmorum. Processes 8 (8), 1003. doi: 10.3390/pr8081003
Gargouri-Kammoun L., Gargouri S., Rezgui S., Trifi M., Bahri N., Hajlaoui M. R. (2009). Pathogenicity and aggressiveness of Fusarium and Microdochium on wheat seedlings under controlled conditions. Tunis. J. Plant Prot. 4, 135–144.
Gazdík Z., Vymětal T., Koprna R., Lojková L., Cerkal R. (2023). Control of malting barley Fusarium head blight by bioagents. Kvasny Prumysl. 69 (3), 747–754. doi: 10.18832/kp2023.69.747
Ghutukade K. S., Deokar C. D., Gore N. S., Chimote V. P., Kamble S. G. (2015). Molecular characterization of Trichoderma isolates by ISSR Marker. J. Agric. Vet. Sci. 8 (7), 1–5. doi: 10.9790/2380-08730105
Gómez-Rodriguez E. Y., Uresti-Rivera E. E., Patrón-Soberano O. A., Islas-Osuna M. A., Flores-Martínez A., Riego-Ruiz L., et al. (2018). Histone acetyltransferase TGF-1 regulates Trichoderma atroviride secondary metabolism and mycoparasitism. PloS One 13 (4), e0193872. doi: 10.1371/journal.pone.0193872
Goswami R. S., Kistler H. C. (2004). Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5 (6), 515–525. doi: 10.1111/j.1364-3703.2004.00252.x
Hermosa R., Viterbo A., Chet I., Monte E. (2012). Plant-beneficial effects of Trichoderma and of its genes. Microbiol 158 (1), 17–25. doi: 10.1099/mic.0.052274-0
Hoyos-Carvajal L., Orduz S., Bissett J. (2009). Genetic and metabolic biodiversity of Trichoderma from Colombia and adjacent neotropic regions. Fung. Genet. Biol 46 (9), 615–631. doi: 10.1016/j.fgb.2009.04.006
Irzykowska L., Bocianowski J., Baturo-Cieśniewska A. (2013). Association of mating-type with mycelium growth rate and genetic variability of Fusarium culmorum. Open Life Sci. 8 (7), 701–711. doi: 10.2478/s11535-013-0176-3
Jingade P., Sannasi S., Jha C. S., Mishra M. K. (2018). Molecular characterisation of Trichoderma species using SRAP markers. Arch. Phytopathol. Plant Prot. 51 (3-4), 128–138. doi: 10.1080/03235408.2018.1439871
Kong W., Huo H., Gu Y., Cao Y., Wang J., Liang J., et al. (2022). Antifungal activity of camphor against four phytopathogens of Fusarium. S. Afr. J. Bot. 148, 437–445. doi: 10.1016/j.sajb.2022.05.019
Küçük Ç., Kivanc M. (2005). Effect of formulation on the viabilıty of biocontrol agent, Trıchoderma harzianum conidia. Afr. J. Biotechn. 4 (6), 483–486.
Küçük Ç., Kivanç M., Kinaci E., Kinaci G. (2007). Biological efficacy of Trichoderma harzianum isolate to control some fungal pathogens of wheat (Triticum aestivum) in Turkey. Biologia 62, 283–286. doi: 10.2478/s11756-007-0049-9
Kumar A. M., Sharma P. (2011). A study on corroboration between DNA markers (RAPD, ISSR, ITS) and bio-control efficacy of Trichoderma species. Fungal Genom. Biol. 1 (103), 1–6.
Kumar D. P., Singh R. K., Anupama P. D., Solanki M. K., Kumar S., Srivastava A. K., et al. (2012). Studies on exo-chitinase production from Trichoderma asperellum UTP-16 and its characterization. Indian J. Microbiol. 52, 388–395. doi: 10.1007/s12088-011-0237-8
Livak J. K., Schmittgen T. D. (2001). Analysis of relative gene expression data using real time quantitative PCR and the 2-Δ;Δ;CT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Matarese F., Sarrocco S., Gruber S., Seidl-Seiboth V., Vannacci G. (2012). Biocontrol of Fusarium head blight: interactions between Trichoderma and mycotoxigenic Fusarium. Microbiol 158 (1), 98–106. doi: 10.1099/mic.0.052639-0
Matny O. N. (2015). Fusarium head blight and crown rot on wheat and barley: losses and health risks. Adv. Plants Agric. Res. 2 (1), 00039. doi: 10.15406/apar.2015.02.00039
Mazrou Y. S., Makhlouf A. H., Elseehy M. M., Awad M. F., Hassan M. M. (2020). Antagonistic activity and molecular characterization of biological control agent Trichoderma harzianum from Saudi Arabia. Egyp. J. Biol. Pest Cont. 30, 1–8. doi: 10.1186/s41938-020-0207-8
Miedaner T., Gwiazdowska D., Waśkiewicz A. (2017). Management of Fusarium species and their mycotoxins in cereal food and feed. Front. Microbiol. 8, 1543. doi: 10.3389/fmicb.2017.01543
Mielniczuk E., Skwaryło-Bednarz B. (2020). Fusarium head blight, mycotoxins and strategies for their reduction. Agronomy 10 (4), 509. doi: 10.3390/agronomy10040509
Mohamed H. A. L. A., Haggag W. M. (2006). Biocontrol potential of salinity tolerant mutants of Trichoderma harzianum against Fusarium oxysporum. Braz. J. Microbiol. 37, 181–191. doi: 10.1590/S1517-83822006000200016
Moradi M., Dehne H. W., Steiner U., Oerke E. C. (2017). Improved procedure for mass inoculum production of Fusarium species in a short period of time. Appl. Entomol. Phytopathol. 84 (2), 21–31.
Moya-Elizondo E. A., Jacobsen B. J. (2016). Integrated management of Fusarium crown rot of wheat using fungicide seed treatment, cultivar resistance, and induction of systemic acquired resistance (SAR). Biol. Cont. 92, 153–163. doi: 10.1016/j.biocontrol.2015.10.006
Mukherjee M., Mukherjee P. K., Horwitz B. A., Zachow C., Berg G., Zeilinger S. (2012). Trichoderma–plant–pathogen interactions: advances in genetics of biological control. Indian J. Microbiol. 52, 522–529. doi: 10.1007/s12088-012-0308-5
Nei M., Li W. H. (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. PNAS 76 (10), 5269–5273. doi: 10.1073/pnas.76.10.5269
Niu C., Kebede H., Auld D. L., Woodward J. E., Burow G., Wright R. J. (2008). A safe inexpensive method to isolate high quality plant and fungal DNA in an open laboratory environment. Afr. J. Biotechnol. 7 (16), 2818–2822.
O’Donnell K., Kistler H. C., Cigelnik E., Ploetz R. C. (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. PNAS 95 (5), 2044–2049. doi: 10.1073/pnas.95.5.2044
Özkale E., Yörük E., Budak M., Korkmaz E. M. (2023). Trichoderma atroviride suppresses Fusarium graminearum by altering primary and secondary metabolite biosynthesis profiling. Plant Pathol. 00, 1–14. doi: 10.1111/ppa.13768
Perlin D. S., Rautemaa-Richardson R., Alastruey-Izquierdo A. (2017). The global problem of antifungal resistance: prevalence, mechanisms, and management. Lancet Infect. Dis. 17 (12), e383–e392. doi: 10.1016/S1473-3099(17)30316-X
Popiel D., Kwasna A., Chelkowski J., Stepien L., Laskowska M. (2008). Impact of selected antagonistic fungi on Fusarium species-toxigenic cereal pathogens. Acta Mycologica 43 (1), 29–40.
Prevost A., Wilkinson M. J. (1999). A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor. Appl. Genet. 98, 107–112. doi: 10.1007/s001220051046
Rai S., Solanki M. K., Solanki A. C., Surapathrudu K. (2019). “Biocontrol potential of Trichoderma spp.: current understandings and future outlooks on molecular techniques,” in Plant Health Under Biotic Stress: Volume 2: Microbial Interactions. Singapore: Springer. 129–160. doi: 10.1007/978-981-13-6040-4_7
Rawat L., Singh Y., Shukla N., Kumar J. (2013). Salinity tolerant Trichoderma harzianum reinforces NaCl tolerance and reduces population dynamics of Fusarium oxysporum f. sp. ciceri in chickpea (Cicer arietinum L.) under salt stress conditions. Arch. Phytopathol. Plant Prot. 46 (12), 1442–1467.
Reithner B., Ibarra-Laclette E., Mach R. L., Herrera-Estrella A. (2011). Identification of mycoparasitism-related genes in Trichoderma atroviride. Appl. Environ. Microbiol. 77 (13), 4361–4370. doi: 10.1128/AEM.00129-11
Reithner B., Schuhmacher R., Stoppacher N., Pucher M., Brunner K., Zeilinger S. (2007). Signaling via the Trichoderma atroviride mitogen-activated protein kinase Tmk1 differentially affects mycoparasitism and plant protection. Fungal Genet. Biol. 44 (11), 1123–1133. doi: 10.1016/j.fgb.2007.04.001
Risoli S., Petrucci A., Vicente I., Sarrocco S. (2023). Trichoderma gamsii T6085, a biocontrol agent of Fusarium head blight, modulates biocontrol-relevant defence gene expression in wheat. Plant Pathol. 72, 1442–1452. doi: 10.1111/ppa.13773
Rojo F. G., Reynoso M. M., Ferez M., Chulze S. N., Torres A. M. (2007). Biological control by Trichoderma species of Fusarium solani causing peanut brown root rot under field conditions. Crop Prot. 26 (4), 549–555. doi: 10.1016/j.cropro.2006.05.006
Roldan-Ruiz I., Dendauw J., Van Bockstaele E., Depicker A., De Loose M. A. F. L. P. (2000). AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol. Breed. 6, 125–134. doi: 10.1023/A:1009680614564
Sariah M., Choo C. W., Zakaria H., Norihan M. S. (2005). Quantification and characterisation of Trichoderma spp. from different ecosystems. Mycopathol 159 (1), 113–117. doi: 10.1007/s11046-004-4432-6
Sharma V., Bhandari P., Singh B., Bhatacharya A., Shanmugam V. (2013). Chitinase expression due to reduction in fusaric acid level in an antagonistic Trichoderma harzianum S17TH. Ind. J. Microbiol. 53, 214–220. doi: 10.1007/s12088-012-0335-2
Singh V., Keswani C., Ray S., Upadhyay R. S., Singh D. P., Prabha R., et al. (2019). Isolation and screening of high salinity tolerant Trichoderma spp. with plant growth property and antagonistic activity against various soilborne phytopathogens. Arch. Phytopathol. Plant Prot. 52 (7-8), 667–680.
Solanki M. K., Singh N., Singh R. K., Singh P., Srivastava A. K., Kumar S., et al. (2011). Plant defense activation and management of tomato root rot by a chitin-fortified Trichoderma/Hypocrea formulation. Phytoparasitica 39, 471–481. doi: 10.1007/s12600-011-0188-y
Strauch O., Strasser H., Hauschild R., Ehlers R. U. (2011). “Proposals for bacterial and fungal biocontrol agents,” in Regulation of biological control agents. Ed. Ehlers R. U. (Dordrecht: Springer), 267–288.
Tamura K., Stecher G., Kumar S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi: 10.1093/molbev/msab120
Urban M., King R., Andongabo A., Maheswari U., Pedro H., Kersey P., et al. (2016). First draft genome sequence of a UK strain (UK99) of Fusarium culmorum. Genome Announc. 4 (5), 10–1128. doi: 10.1128/genomeA.00771-16
Xue M., Wang R., Zhang C., Wang W., Zhang F., Chen D., et al. (2021). Screening and identification of Trichoderma strains isolated from natural habitats in China with potential agricultural applications. BioMed. Res. Int. 2021, 7913950. doi: 10.1155/2021/7913950
Yang X., Chen L., Yong X., Shen Q. (2011). Formulations can affect rhizosphere colonization and biocontrol efficiency of Trichoderma harzianum SQR-T037 against Fusarium wilt of cucumbers. Biol. Fert. Soils. 47, 239–248. doi: 10.1007/s00374-010-0527-z
Yin Y., Liu X., Li B., Ma Z. (2009). Characterization of sterol demethylation inhibitor-resistant isolates of Fusarium asiaticum and F. graminearum collected from wheat in China. Phytopathology 99 (5), 487–497.
Yörük E. (2018). Tetraconazole leads to alterations in Fusarium graminearum at different molecular levels. Appl. Ecol. Environ. Res. 16 (5), 6155–6167.
Yörük E., Özkale E., Sefer Ö., Özsoy E. (2022). Trichoderma atroviride triggers alterations at epigenetics, transcripts, oxidative stress, and apoptosis levels on Fusarium graminearum. J. Plant Pathol. 104 (3), 1039–1047.
Yörük E., Tunali B., Kansu B., Ölmez F., Uz G., Zümrüt I. M., et al. (2016). Characterization of high-level deoxynivalenol producer Fusarium graminearum and F. culmorum isolates caused head blight and crown rot diseases in Turkey. J. Plant Dis. Prot. 123, 177–186. doi: 10.1007/s41348-016-0027-y
Yörük E., Yli-Mattila T. (2019). “Class B-trichothecene profiles of Fusarium species as causal agents of head blight,” Advancing Frontiers in Mycology and Mycotechnology: Basic and Applied Aspects of Fungi (Springer: Singapore), 347–376.
Zapparata A., Baroncelli R., Durling M. B., Kubicek C. P., Karlsson M., Vannacci G., et al. (2021). Fungal cross-talk: an integrated approach to study distance communication. Fungal Genet. Biol. 148, 103518. doi: 10.1016/j.fgb.2021.103518
Zeilinger S., Gruber S., Bansal R., Mukherjee P. K. (2016). Secondary metabolism in Trichoderma–chemistry meets genomics. Fungal Biol. Rev. 30 (2), 74–90. doi: 10.1016/j.fbr.2016.05.001
Keywords: biocontrol agent, Fusarium culmorum, Fusarium head blight (FHB), gene expression analysis, Trichoderma spp., root rot (RR)
Citation: Sefer Ö, Özsoy E, Yörük E and Özkale E (2023) Determining the biocontrol capacities of Trichoderma spp. originating from Turkey on Fusarium culmorum by transcriptional and antagonistic analyses. Front. Fungal Biol. 4:1278525. doi: 10.3389/ffunb.2023.1278525
Received: 16 August 2023; Accepted: 11 October 2023;
Published: 13 November 2023.
Edited by:
Lukasz Stepien, Polish Academy of Sciences, PolandReviewed by:
Manoj Kumar Solanki, University of Silesia in Katowice, PolandCopyright © 2023 Sefer, Özsoy, Yörük and Özkale. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Evrim Özkale, ZXZyaW1taWtvQGdtYWlsLmNvbQ==; ZXZyaW0udGFza2luQGJheWFyLmVkdS50cg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.