
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Fungal Biol. , 20 January 2023
Sec. Fungal Physiology and Metabolism
Volume 3 - 2022 | https://doi.org/10.3389/ffunb.2022.1081179
This article is part of the Research Topic The use of metabolic engineering techniques to increase the productivity of primary and secondary metabolites within filamentous fungi View all 5 articles
 Shigenari Yamaguchi1
Shigenari Yamaguchi1 Tomonori Fujioka1
Tomonori Fujioka1 Akira Yoshimi2,3
Akira Yoshimi2,3 Toshitaka Kumagai4
Toshitaka Kumagai4 Maiko Umemura5
Maiko Umemura5 Keietsu Abe2,6
Keietsu Abe2,6 Masayuki Machida5,7
Masayuki Machida5,7 Kiyoshi Kawai1*
Kiyoshi Kawai1*KK-1, a cyclic depsipeptide with 10 residues produced by a filamentous fungus Curvularia clavata BAUA-2787, is a promising pesticide active compound with high activity against many plant pathogens, especially Botrytis cinerea. As a first step toward the future mass production of KK-1 through synthetic biological approaches, we aimed to identify the genes responsible for the KK-1 biosynthesis. To achieve this, we conducted whole genome sequencing and transcriptome analysis of C. clavata BAUA-2787 to predict the KK-1 biosynthetic gene cluster. We then generated the overexpression and deletion mutants for each cluster gene using our originally developed transformation system for this fungus, and analyzed the KK-1 production and the cluster gene expression levels to confirm their involvement in KK-1 biosynthesis. As a result of these, a region of approximately 71 kb was found, containing 10 open reading frames, which were co-induced during KK-1 production, as a biosynthetic gene cluster. These include kk1B, which encodes nonribosomal peptide synthetase with a domain structure that is consistent with the structural features of KK-1, and kk1F, which encodes a transcription factor. The overexpression of kk1F increased the expression of the entire cluster genes and, consequently, improved KK-1 production, whereas its deletion decreased the expression of the entire cluster genes and almost eliminated KK-1 production, demonstrating that the protein encoded by kk1F regulates the expressions of the other nine cluster genes cooperatively as the pathway-specific transcription factor. Furthermore, the deletion of each cluster gene caused a reduction in KK-1 productivity, indicating that each gene is involved in KK-1 production. The genes kk1A, kk1D, kk1H, and kk1I, which showed a significant decrease in KK-1 productivity due to deletion, were presumed to be directly involved in KK-1 structure formation, including the biosynthesis of the constituent residues. kk1C, kk1E, kk1G, and kk1J, which maintained a certain level of KK-1 productivity despite deletion, were possibly involved in promoting or assisting KK-1 production, such as extracellular transportation and the removal of aberrant units incorporated into the peptide chain.
With growing global interest in the realization of a sustainable society, the reduction of environmental impacts in food systems is recognized as one of the most important issues worldwide (Brzozowski and Mazourek, 2018; Meemken and Qaim, 2018; Lazaroiu et al., 2019). Particularly in Europe, there has been strong concern about the impact on environment and biodiversity due to excessive dependence on chemical pesticides in agriculture. This has led the European Commission to establish concrete numerical targets to be achieved by 2030 for reducing the use of chemical pesticides and promoting organic agriculture in its “Farm to Fork Strategy,” which was released in May 2020 (https://food.ec.europa.eu/system/files/2020-05/f2f_action-plan_2020_strategy-info_en.pdf). This has been followed by a worldwide movement to tighten regulations on chemical pesticides, such as Japan’s strategy for sustainable food systems, called MeaDRI (https://www.maff.go.jp/e/policies/env/env_policy/meadri.html). Accordingly, the demand for new pesticides of natural origin, which are considered to have a relatively low impact on the environment and living organisms, is expected to increase further in the future.
The secondary metabolites produced by microorganisms such as fungi, actinomycetes, and bacteria, are diverse and include many compounds that exhibit remarkable biological activity (Pham et al., 2019; Jakubczyk and Dussart, 2020; De Simeis and Serra, 2021). Many of them have complex chemical structures based on carbon skeletons that result from characteristic biosynthetic mechanisms, including polyketide synthases, non-ribosomal peptide synthetases and terpene synthases (Pham et al., 2019; Jakubczyk and Dussart, 2020; De Simeis and Serra, 2021). When natural compounds with such complex structures have pesticidal activity, they are expected to act on different targets from existing chemical pesticides, leading to the development of pesticides with less environmental impacts, and providing a solution to important problems in modern agriculture, such as pesticide resistance in pests and weeds.
Some of the secondary metabolites produced by microorganisms have been commercialized as pesticides (Cantrell et al., 2012). For example, polyoxins are nucleoside antibiotics that are isolated from Streptomyces cacaoi and are mainly used for the control of plant pathogens (Isono et al., 1965; Isono et al., 1967). Polyoxins are a mixture of structurally similar compounds whose basic structure is composed of three moieties including the nucleoside skeleton, polyoxamic acid, and carbamoylpolyoxamic acid (Isono et al., 1969; Uramoto et al., 1981); and they are the only pesticides that specifically inhibit the biosynthesis of chitin (Endo and Misato, 1969; Endo et al., 1970), a major component of the fungal cell wall. Spinosyns are isolated from Saccharopolyspora spinosa and are macrolide compounds with insecticidal activity (Mertz and Yao, 1990; Kirst et al., 1992). The main products in the fermentation cultures are spinosyn A and spinosyn D (Kirst, 2010), and a mixture of these two has been commercialized as Spinosad. The basic structure consists of a tetracycle containing a 12-membered macrocyclic lactone attached to a natural sugar, rhamnose, and an amino sugar, forosamine (Kirst et al., 1992). Spinosyns exhibit insecticidal activity by acting on the nicotinic acetylcholine receptors in the insect neurotransmitter system (Orr et al., 2009; Crossthwaite et al., 2017). In addition to the metabolites that are specified above, many other secondary metabolites that exhibit pesticidal activity have been isolated; however, in many cases commercialization is difficult even if promising activity is confirmed. One of the major causes of this is the cost. When considering the industrial production of secondary metabolites with complex structures, fermentation methods with a microorganism, such as polyoxins and spinosyns, are a realistic production method, since synthetic organic chemistry methods are not profitable. However, in general, most of these secondary metabolites are produced in small quantities and often only under specific conditions. This makes it very difficult to ensure cost-effective and stable production as pesticides, which is a barrier to their practical application. To date, some progress has been made in improving productivity, especially in the field of pharmaceuticals, through the optimization of culture conditions and breeding by introducing random mutations (Parekh et al., 2000; Lee et al., 2002; Korbekandi et al., 2010; Siddique et al., 2014). However, it is difficult to achieve commercialization using only these methods because it is likely to take a long time to obtain high-producing strains, and agrochemicals need to be mass-produced at a lower cost than pharmaceuticals.
One solution to efficiently produce such valuable secondary metabolites is a synthetic biology approach that has recently attracted attention (Katz et al., 2018; Choi et al., 2019). With the recent development of next generation sequencing and bioinformatics, it has become easier to analyze genome information and the gene functions of microorganisms (Zhang et al., 2017; Hu et al., 2021). This has also facilitated the identification of genes that are related to the biosynthesis of certain secondary metabolites and the elucidation of biosynthetic pathways (Zhang et al., 2017). The cloning of these biosynthetic genes and their introduction into appropriate hosts, and the construction of systems that are specialized for the production of target compounds in the host by modifying these genes, i.e., the synthetic biological approach (Siddiqui et al., 2012; Jensen and Keasling, 2015; Lee et al., 2019), is expected to realize a large supply of natural products with useful activities. These approaches for the efficient production of useful compounds have made rapid progress in recent years (Cravens et al., 2019; Yang et al., 2020; Zhang et al., 2021). For example, in the pharmaceutical field, the productivity of the precursor of artemisinin, a terpenoid with antimalarial activity that was extracted from Artemisia annua, a kind of herb, was dramatically improved (Westfall et al., 2012; Paddon et al., 2013; Kung et al., 2018). Therefore, these approaches expand the possibilities for the efficient mass production and stable supply of natural compounds with potential agrochemical activity, which have previously been difficult to put into practical use due to their low productivity.
Here, we focused on an antifungal compound (CAS No. 143380-71-6), hereinafter called KK-1, which is produced by a filamentous fungus, Curvularia clavata (BAUA-2787 strain). KK-1 is probably identical to BK202, which was isolated and purified from Curvularia sp. by Novo Nordisk A/S (Bagsværd, Denmark) as described in patent WO1992005191A1 (Nielsen et al., 1992) and is a cyclic depsipeptide composed of one glycine, eight L-amino acids (four valines and one pipecolic acid, aspartic acid, isoleucine, and tyrosine each), and one D-lactic acid. The nitrogen atoms on the amino groups of five of these residues are methylated, and the hydroxy group on the phenyl of the tyrosine residue is also methylated [D-Lac/L-Pip/N-methyl-L-Val/L-Val/N-methyl-L-Asp/N-methyl-L-Val/N-methyl-Ile/Gly/N-methyl-L-Val/O-methyl-L-Tyr] (Figure 1, also refer to Yoshimi et al., 2018). KK-1 is a promising pesticide active compound with high activity against many plant pathogens, especially Botrytis cinerea, a pathogen that causes grey mold disease in a variety of agricultural and horticultural crops. However, a breakthrough is needed to enable a stable mass supply for practical use. Therefore, we aimed to identify the biosynthetic gene cluster as a first step to the mass production of KK-1 in the future through synthetic biological approaches. The genus Curvularia, as well as the genus Bipolaris, is a genus of filamentous ascomycetous fungi whose anamorph is Cochliobolus (Manamgoda et al., 2011), some of which are known as plant pathogens (Manamgoda et al., 2011), and several bioactive secondary metabolites have been isolated from fungi that belong to these genera (Khiralla et al., 2019; Elkhateeb et al., 2021). However, to date, there have been no reports published on Curvularia clavata regarding its ability to produce bioactive compounds or any enzymes related to secondary metabolic pathways, and its genome information has also not been sequenced.
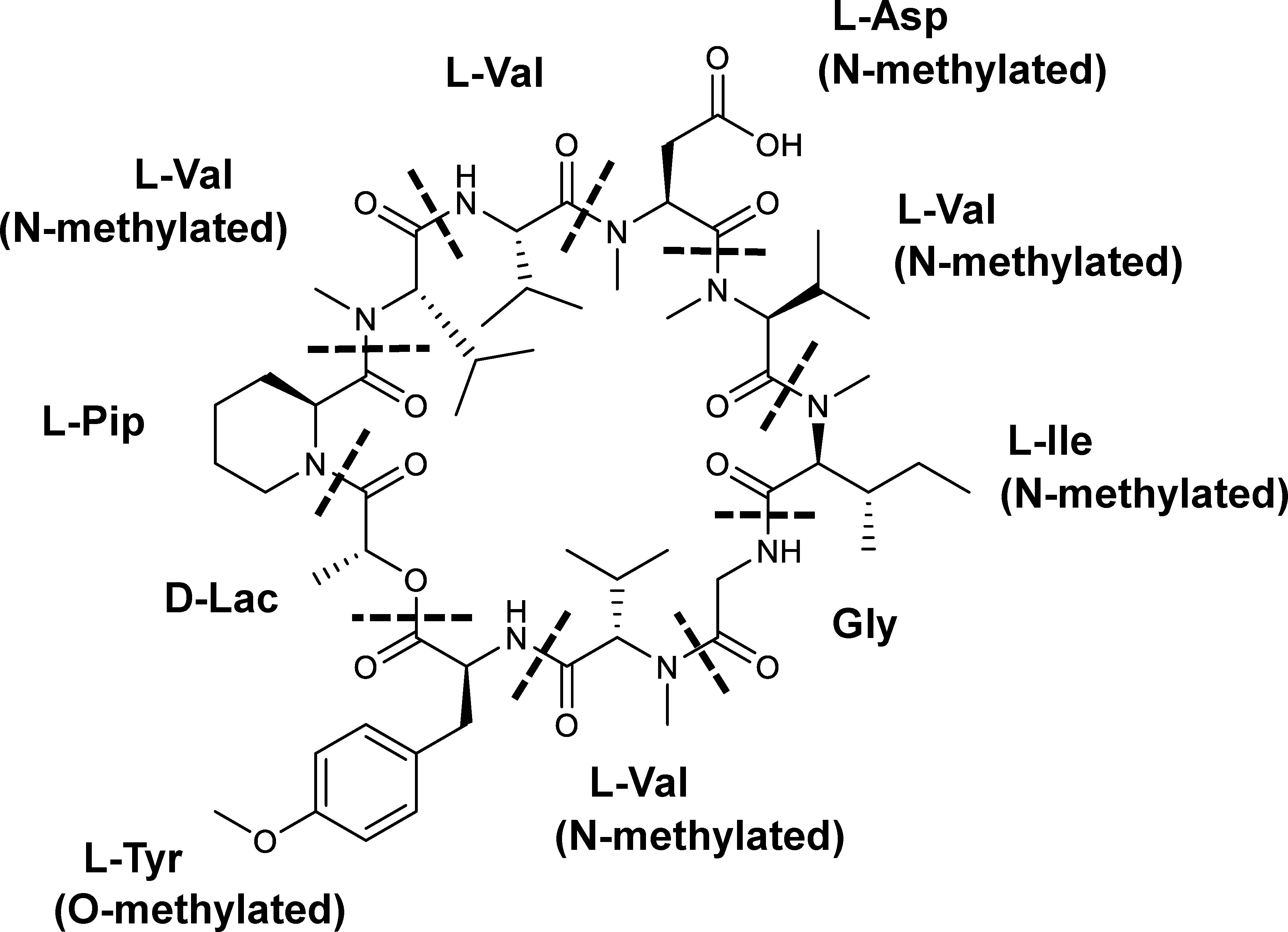
Figure 1 The chemical structure of KK-1. The constituent residues are separated by dotted lines: Lac, lactic acid residue; Pip, pipecolic acid residue; Val, valine residue; Asp, aspartic acid residue; Ile, isoleucine residue; Gly, glycine residue; Tyr, tyrosine residue.
Consequently, in this report, we have conducted whole genome sequencing of C. clavata BAUA-2787 as well as transcriptome analysis. Using these data, a gene cluster was identified that is involved in the biosynthesis of KK-1. Then, overexpression or deletion mutants of each cluster gene were generated. We also analyzed the KK-1 production of these mutants and the expression of the cluster genes to demonstrate that these genes are involved in the biosynthesis of KK-1.
Curvularia clavata BAUA-2787 was donated by Akita Konno Co., Ltd., Akita, Japan and registered at the National Institute of Technology and Evaluation (No. NITE BP-02399). The conidial suspension of C. clavata BAUA-2787 strain was prepared by adding sterile water to the colonies that were grown on CMA medium (Tanaka et al., 1991) [0.15% Ca(NO3)2·4H2O, 0.05% MgSO4·7H2O, 0.05% KCl, 0.04% KH2PO4, 0.003% K2HPO4, 0.1% tryptone, 0.1% yeast extract, 1% glucose, and 1.5% agar], scraping them with a sterile spatula, and filtering them through a Miracloth (Calbiochem, San Diego, CA, USA) to remove the mycelium. They were stored in 16% v/v glycerol at -80°C. All C. clavata wild-type and mutant strains that were derived from BAUA-2787 were cultured in CM (CMA minus agar) medium that was prepared in Erlenmeyer flasks, and they were inoculated with the conidial suspensions at a concentration of 103 conidia/mL.
The genomic DNA of C. clavata BAUA-2787 was extracted using a DNeasy Plant Mini Kit (QIAGEN, Manchester, UK) after 2 days of cultivation in 100 mL CM medium at 26°C and 130 rpm. The DNA libraries were prepared with a 5500 SOLiD Mate-Paired Library Kit and sequenced by a 5500xl SOLiD system (Life Technologies, Carlsbad, CA, USA). A Nextera DNA Sample Prep Kit was also used for the library preparation, and the libraries were sequenced using the MiSeq platform (Illumina, San Diego, CA, USA). The hybrid de novo genome assembly (Ikegami et al., 2015) was performed using the MiSeq and SOLiD short read data. Additionally, the protein-coding genes in the draft genome were predicted by a combination of the gene prediction programs, ALN (Gotoh, 2000) and GlimmerHMM (Majoros et al., 2004). NCBI BLAST (http://blast.ncbi.nlm.nih.gov/) and the Pfam protein family database (http://pfam.xfam.org/) were used for sequence homology and conserved domain searches, as appropriate.
A BlastP search was performed against the database of the amino acid sequences of the predicted proteins that was constructed from the genome information of C. clavata BAUA-2787. The sequences of NPS1-NPS12 were used as a query, which are non-ribosomal peptide synthetases (NRPSs) that were found in Cochliobolus heterostrophus, and some highly homologous genes were extracted. To predict more accurate coding sequences (CDSs) of these genes, a BlastX search was performed against the GenBank database using nucleic acid sequences from 3,000 bp upstream of the start codon to 3,000 bp downstream of the termination codon of these genes as queries. Then, the domain structures of the proteins that are encoded by these genes were predicted using InterProScan (http://www.ebi.ac.uk/InterProScan/) and antiSMASH (http://antismash.secondarymetabolites.org) to search for the NRPSs with a modular structure that is consistent with the structural features of KK-1.
Wild-type strains of C. clavata BAUA-2787 were cultured in 100 mL of CM medium that was prepared in a 300 mL Erlenmeyer flask at 130 rpm for 48 hours at two temperature conditions (26°C and 37°C). The total RNA of these samples was extracted with ISOGEN (Nippon Gene, Toyama, Japan) and purified using the RNeasy spin column (QIAGEN). Then, the quantity and purity of the extracted RNA were determined with an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). The RNA sequencing (RNA-Seq) libraries were prepared with a Truseq RNA Sample Prep Kit v2 (Illumina) and RNA-Seq was performed on a Miseq system (paired-end 2 × 75 bases). The obtained reads were mapped onto the draft genome of C. clavata BAUA-2787 using the TopHat program (Trapnell et al., 2009), and the reads per kilobase of exon per million mapped reads (RPKM) values were calculated from the raw read count data for every gene on the genome. The novel gene cluster mining tool, motif-independent de novo detection algorithm for secondary metabolite gene clusters (MIDDAS-M), which predicts co-regulated gene clusters using genome and transcriptomic data (Umemura et al., 2013), was then used to detect the significantly expressed gene clusters at 26°C when compared to 37°C.
We prepared 5 µL of the protoplast suspension of C. clavata BAUA-2787 wild-type strain according to the transformation method as described below. It was spotted in the center of CM-sucrose agar medium (CM medium containing 1.5% agar and 1.2M sucrose) supplemented with six concentrations (1, 2.5, 5, 10, 15, and 20 µg/mL) of aureobasidin A (AbA; Takara Bio, Shiga, Japan) or six concentrations (12.5, 25, 50, 100, 200, and 400 µg/mL) of hygromycin (Hyg; Fujifilm Wako Pure Chemical, Osaka, Japan). After 6 days of incubation at 26 ˚C, the mycelial growth inhibition was evaluated.
Wild-type strains of C. clavata BAUA-2787 were cultured in 100 mL of CM medium, which was prepared in a 300 mL Erlenmeyer flask at 30 ˚C and 130 rpm. After 40 hours, the mycelia were collected by filtration using a Miracloth, washed with sterilized water, and pressed with a spatula to lightly remove the water. The washed mycelia were suspended in 10 mL of protoplast forming solution [3 mg/mL Yatalase (Takara Bio), 0.3 mg/mL Lysing Enzymes from Trichoderma harziaum (Sigma-Aldrich, Steinheim am Albuch, Germany), 0.8 mol/L NaCl, and 10 mM sodium phosphate buffer (pH6.0)] and gently shaken at 30 ˚C and 80 rpm. After 3 hours, the cell wall debris was removed by filtration using a Miracloth, and the protoplasts were collected by centrifuging the flow-through. Next, the protoplast pellet was washed with 0.8 mol/L NaCl twice, suspended in Solution 1 [0.8 mol/L NaCl, 10 mM CaCl2, and 10 mM Tris-HCl buffer (pH8.0)] to 2 × 108 protoplasts/mL, and 1/5 volume of Solution 2 [40% (w/v) PEG4000, 50 mM CaCl2, and 50 mM Tris-HCl buffer (pH8.0)] was added with gentle mixing. Approximately 10 µg of pAUR316 (Takara Bio) was added to a 0.2 mL aliquot of the protoplast suspension, and this was incubated on ice for 10 min. Subsequently, 1 mL of Solution 2 was added to this suspension and incubated at room temperature for 15 min. Afterward, 0.2 mL of the suspension was mixed with 7 mL CM-sucrose top agar medium (CM medium containing 1% agar and 1.2M sucrose) with 10 μg/mL of AbA, and it was overlaid on the surface of the CM-sucrose agar medium (CM medium containing 1.5% agar and 1.2M sucrose) with 10 μg/mL of AbA prepared in petri dishes. These were incubated at 26°C until the colonies of the transformants appeared. Also, direct polymerase chain reaction (PCR) using a KAPA3G Plant PCR Kit (Kapa Biosystems, Wilmington, MA, USA) was performed on the growing colonies to confirm if they harbored pAUR316. The primer information is shown in Supplementary Table S1.
A Blastp search was performed using the amino acid sequence of the protein that is encoded by pyrG of Aspergillus nidulans (accession: AAB66359) as the query, and a gene that was found to be a homolog in C. clavata BAUA-2787 was designated as CcpyrG. The region from 2,005 bp upstream of the start codon to 1,261 bp downstream of the stop codon of CcpyrG was amplified from the genomic DNA of C. clavata BAUA-2787 using a primer set, CcPyrG-del_FW3/CcPyrG-del_RV3, and Phusion Hot Start II High-Fidelity DNA Polymerase (Thermo Fisher Scientific, Waltham, MA, USA). This fragment was phosphorylated by T4 polynucleotide kinase (Toyobo, Osaka, Japan) and ligated using T4 DNA ligase (Nippon Gene) in the pUC18 plasmid that was digested by SmaI (Takara Bio) and dephosphorylated by E. coli Alkaline Phosphatase (Toyobo) to construct the plasmid pUC18-CcpyrG. Subsequently, an inverse PCR using a primer set, CcPyrG-del_FW2/CcPyrG-del_RV2, and Phusion Hot Start II High-Fidelity DNA Polymerase was performed to amplify the fragment from pUC18-CcpyrG that was lacking the sequence from the CcpyrG promoter region to the CcpyrG terminator region. The fragment was phosphorylated by T4 polynucleotide kinase and self-ligated by T4 DNA ligase to construct the CcpyrG deletion plasmid. The DNA cassette, equivalent to the fragment consisting of the 1,150 bp fragment corresponding to the region from 2,005 bp to 856 bp upstream of the start codon of CcpyrG and the 1,079 bp fragment corresponding to the region from 183 bp to 1,261 bp downstream of the stop codon, which were directly connected to each other as homology arms for homologous recombination, was amplified from the CcpyrG deletion plasmid, using the primer set, CcPyrG-del_FW3/CcPyrG-del_RV3, and Phusion Hot Start II High-Fidelity DNA Polymerase. This DNA cassette was used for the generation of the CcpyrG deletion strain. The scheme described above is shown in Supplementary Figure S1 and the primer information is shown in Supplementary Table S1
Transformation was basically performed according to the method of introducing pAUR316 which was described above, but 0.2% (w/v) of uridine, 0.02% (w/v) of uracil, and 1 mg/mL of 5-fluoroorotic acid (Fujifilm Wako Pure Chemical) were added to the CM-sucrose top agar medium and CM-sucrose agar medium, respectively, instead of AbA. Then, a direct PCR using a KAPA3G Plant PCR Kit was conducted on the growing colonies to confirm that the target region containing CcpyrG was deleted by homologous recombination. The primer information is shown in Supplementary Table S1.
The plasmid for the overexpression of kk1F gene was constructed by inserting the four DNA fragments in the following order: the 1,000 bp region upstream of the start codon of Ccnmt1, which encodes a homolog of Nmt1 from Schizosaccharomyces pombe, as its promoter, the cDNA sequence of kk1F, the 355 bp region downstream of the stop codon of Ccnmt1 as its terminator, and aurAr, an AbA resistance gene that was isolated from Aspergillus nidulans, as a selectable marker in the pUC19 plasmid, using an In-Fusion HD Cloning Kit (Clontech, Mountain View, CA, USA). The promoter and terminator were amplified from the genomic DNA of C. clavata BAUA-2787, kk1F was amplified from the cDNA library of C. clavata BAUA-2787, and aurAr was amplified from a pAUR316 plasmid. The PCR was performed using Phusion Hot Start II High-Fidelity DNA Polymerase. The primer information is shown in Supplementary Table S1.
Transformation was basically performed according to the method of introducing pAUR316 as described above. A direct PCR using the KAPA3G Plant PCR Kit (Kapa Biosystems) was performed on the growing colonies to confirm that the target sequence was introduced into the chromosome. The scheme described above is shown in Supplementary Figure S2, and the primer information is shown in Supplementary Table S1.
The plasmid for each cluster gene deletion except kk1B gene was constructed by inserting the three DNA fragments in the following order: an approximately 1 kbp fragment that was homologous to the upstream region of the target gene as the homology left-arm, a 2,231 bp fragment corresponding to the region from 853 bp upstream of the CcpyrG start codon to 181 bp downstream of its stop codon as the CcpyrG selectable marker, and an approximately 1 kbp fragment homologous to the downstream region of the target gene as the homology right-arm in the pUC19 plasmid, using an In-Fusion HD Cloning Kit. The plasmid for kk1B deletion was also basically constructed using the same scheme as above but for the homology right-arm, the region from 4,482 bp to 5,463 bp downstream of the start codon of kk1B, corresponding to the inside of the open reading frame (ORF), was used. These DNA fragments were amplified from the genomic DNA of C. clavata BAUA-2787 using Phusion Hot Start II High-Fidelity DNA Polymerase. Each gene deletion plasmid was linearized by the appropriate restriction enzyme digestion and used for transformation. The primer information is shown in Supplementary Table S1, and the region of homology arms that was included in each deletion plasmid and the restriction enzymes that were used for the linearization of each plasmid are listed in Supplementary Table S2.
Transformation was basically performed according to the method of introducing pAUR316 as described above but the CcpyrG deletion strain was used as the host, the CM medium containing 0.2% (w/v) uridine and 0.02% (w/v) uracil was used during the initial culture process, and the CM-sucrose top agar medium and CM-sucrose agar medium without any addition were used for the transformant selection process. A direct PCR using the KAPA3G Plant PCR Kit was performed on the growing colonies to confirm that each cluster gene was deleted by homologous recombination and the transformants were obtained as homokaryons. The scheme described above is shown in Supplementary Figures S3, S4, and the primer information is shown in Supplementary Table S1.
A small amount of the culture supernatant was sterilized by filtration through a 0.22 µm membrane filter (Merck KGaA, Darmstadt, Germany), and paper disks with a diameter of 6 mm (Advantec, Tokyo, Japan, ø6 mm) were soaked in the supernatant. This paper disk was placed near the center of a potato dextrose agar plate in a 90 mm petri dish and B. cinerea was inoculated onto its opposite side. After 3 days of incubation at 26 ˚C, the KK-1 productivity was evaluated based on its effect on the growth of B. cinerea. The culture supernatant of the wild-type strain was used as a positive control, and the CM medium was used as a negative control.
C. clavata BAUA-2787 strains were grown on CM medium prepared in baffled Erlenmeyer flasks, the same volume of acetone was added, and it was sonicated. The sample was then extracted twice with the same volume of ethyl acetate. Then, the ethyl acetate layer was collected, dehydrated with anhydrous sodium sulfate, and evaporated. The extracts were dissolved in acetonitrile and analyzed by ultra-performance liquid chromatography (UPLC), high-performance liquid chromatography (HPLC), or HPLC-mass spectrometry (HPLC-MS). KK-1 was detected at a wavelength of 195 nm. The production of KK-1 was quantified by calculating the peak area of each UV chromatogram. The KK-1 that was isolated and purified from the culture medium of the wild-type strain was used for external calibration.
First, the UPLC measurements were carried out on an ACQUITY UPLC I-Class system (Waters, Milford, MA, USA) and performed using an ACQUITY UPLC BEH C18, 130 Å, 1.7 µm, 2.1 mm x 100 mm column (Waters) at a flow rate of 0.6 mL/min and using a gradient from 50% to 98% acetonitrile in the water containing 0.1% formic acid for 3 min. Second, the HPLC measurements were carried out on a Nexera-i LC-2040C HPLC system (Shimadzu, Kyoto, Japan) and performed using a CAPCELL PAK C18, SG120 Å, 5 µm, 4.6 mm x 250 mm column (Osaka Soda, Osaka, Japan) at a flow rate of 1.0 mL/min and using a gradient from 50% to 98% acetonitrile in water containing 0.1% formic acid for 15 min. Third, the HPLC-MS measurements were carried out on an ExionLC system (AB Sciex, Tokyo, Japan) coupled with a Triple QUAD 4500 system (AB Sciex). The HPLC was performed using a Kinetex 2.6 µm C18, 100 Å, LC column (150 x 4.6 mm; Phenomenex, Torrance, CA, USA) at a flow rate of 0.4 mL/min and using a gradient from 50% to 98% acetonitrile in water containing 0.1% formic acid for 8 min. The MS conditions were as follows: ionization mode, ESI; polarity, positive; turbo gas temperature, 300 ˚C; ion spray voltage, 5.5 kV; and scan range, m/z 500 to 1200.
The kk1F overexpression strain, its deletion strain, and the wild-type strain were cultured on 30 mL of CM medium, which was prepared in a 100 mL Erlenmeyer flask at 26 ˚C and 130 rpm, and RNA-Seq was performed as described above.
In many peptide secondary metabolites, the peptide backbone is known to be synthesized by non-ribosomal peptide synthetase (NRPS). NRPSs are large enzyme complexes that have modular structures, each containing multiple functional domains with specific activities and being capable of catalyzing the synthesis of peptide chains without depending on ribosomal machinery (Hur et al., 2012; Duban et al., 2022). Each module basically contains an adenylation domain (A domain), peptidyl carrier protein domain (PCP domain), and condensation domain (C domain), which are essential for the synthesis of the peptide backbone, and a thioesterase domain (TE domain) that is generally located at the C-terminus in the final module of NRPS and is responsible for the cyclization of the peptide chain (Hur et al., 2012; Duban et al., 2022). The other domains have the function of modifying the peptide backbone that is synthesized by these essential domains (Hur et al., 2012; Duban et al., 2022). Since the order and number of the module structures and the characteristics of their constituent domains are consistent with those of amino acid residues of the corresponding peptide, it is possible to presume that NRPS is involved in the biosynthesis of peptide secondary metabolites based on this information. Therefore, we searched for the NRPS gene that is responsible for the biosynthesis of the peptide backbone of KK-1 as a first step to identify the biosynthetic gene cluster in C. clavata genome.
We initially performed draft genome sequencing of C. clavata BAUA-2787 and obtained 142 sequences with an N50 value of 1,472,240 bp for a total of 30.6 Mbp at 50.3% GC content, as a result of the hybrid assembly combining the SOLiD and Illumina data. A total of 10,367 ORFs were predicted from this draft genome sequence. Subsequently, we searched the draft sequence for the genes encoding NRPS. In Cochliobolus heterostrophus, which is closely related to C. clavata, 12 NRPS genes (NPS1–NPS12) have been found (Lee et al., 2005). A BlastP search against the sequence information of the predicted proteins found in C. clavata BAUA-2787 was carried out using the sequences of NPS1–NPS12 as queries, resulting in 24 ORFs as candidates of the NRPS gene that is responsible for KK-1 synthesis, which were designated as Ccnps1–Ccnps24. The more accurate sequences of each of these ORFs were predicted by a homology search against the GenBank database, and it was shown that Ccnps4, Ccnps3, and Ccnps2 were one gene connected in this order. We called the gene Ccnps4-3-2. To find the NRPS gene that was consistent with the structural features of the cyclic peptide that is composed of 10 residues of KK-1, the domain structures of the proteins that were encoded by the above ORFs were subsequently examined using antiSMASH (Figure 2). Consequently, only Ccnps4-3-2 with a total length of 39,125 bp had 10 modules, each of which contained A, PCP, and C domains, suggesting that it is the NRPS gene that is involved in the synthesis of KK-1. In addition, the NRPS that is encoded by Ccnps4-3-2 has five N-methyltransferase domains (nMT domains), responsible for N-methylation of the amino acids that are incorporated into the same module and are located in the third, fifth, sixth, seventh, and ninth modules from the N-terminus, respectively. If the first module of the NRPS that is encoded by Ccnps4-3-2 is involved in the introduction of the L-lactic acid residue of KK-1, the order of the modules that contained the nMT domains perfectly matched that of the N-methylated amino acid residue in KK-1 (Figure 2). This also strongly suggests that Ccnps4-3-2 is probably the NRPS that is responsible for the synthesis of the basic peptide backbone of KK-1.
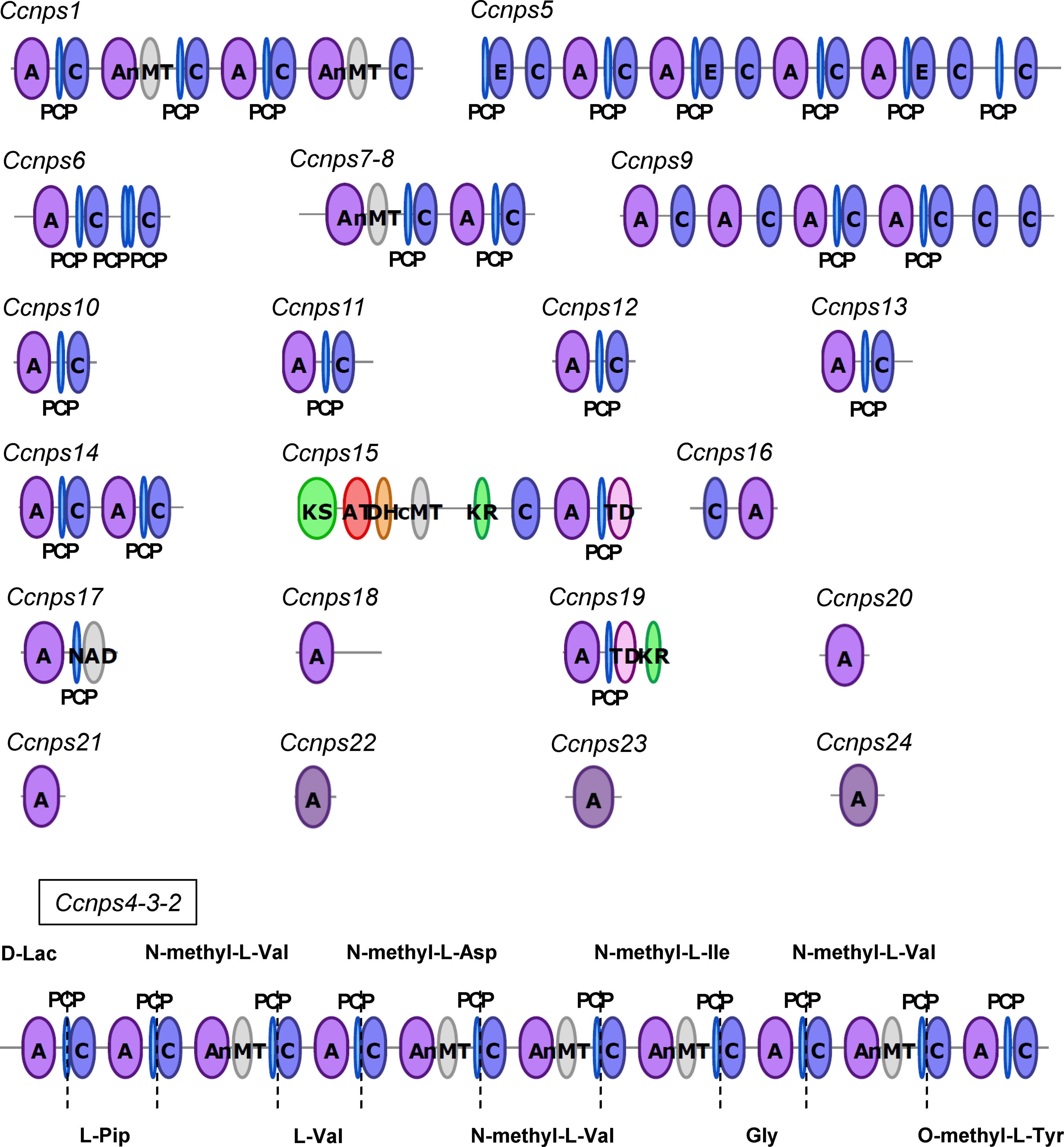
Figure 2 The domain structure of each non-ribosomal peptide synthetase that is possessed by Curvularia clavata BAUA-2787, which was deduced by antiSMASH: C, condensation domain; A, adenylation domain; PCP, peptidyl carrier protein domain; nMT, nitrogen methyl transferase; E, epimerization domain; KS, ketosynthase domain; AT, acyltransferase domain; DH, dehydratase domain; cMT, carbon methyltransferase; KR, ketoreductase domain; TD, terminal reductase domain; NAD, male sterility protein. Only the modular structure of NRPS that is encoded by Ccnps4-3-2 is consistent with the structural characteristics of KK-1.
In general, genes that are involved in the biosynthesis of secondary metabolites exist as clusters. For cyclosporine, which is used as an immunosuppressant agent, a series of genes including simA, which encodes the NRPS responsible for the synthesis of the basic backbone, has been identified as a putative biosynthetic gene cluster in an approximately 95 kb region of the Tolypocladium inflatum genome (Bushley et al., 2013). For daptomycin, a series of genes including three NRPS genes (dptA, dptBC, and dptD), which are essential for the synthesis of the basic backbone, are located at the same locus on the genome of Streptomyces roseosporus, and these genes are known to be involved in the biosynthesis of this antibiotic (Miao et al., 2005). Based on these cases, a group of KK-1 biosynthetic genes could also be present as a gene cluster containing Ccnps4-3-2. Therefore, for the 14 ORFs that were found upstream of Ccnps4-3-2 (designated orf(-14)–orf(-1)) and the other 14 ORFs found downstream (designated orf(+1)–orf(+14)), the function of the protein that is encoded by each gene was predicted (Table 1), and the region of approximately 71 kb containing a series of 10 genes from orf(-1) to orf(+8) that are likely to be involved in secondary metabolism was predicted as the KK-1 biosynthetic gene cluster (Figure 3A). These 10 genes are hereinafter referred to as kk1A–kk1J, and the NRPS gene, Ccnps4-3-2, corresponds to kk1B. About half of the region of the cluster was occupied by kk1B (Figure 3B; Table 2). The kk1F gene was predicted to encode a protein with a basic-region leucine zipper (bZIP) motif. This is the only putative transcription factor gene that was found in the cluster, and, therefore, was thought to be important in regulating the expression of the other cluster genes. The accuracy of the KK-1 cluster region assembly was later confirmed by performing long read sequencing with PacBio RS-II (data not shown). The 75 kb sequence that encompasses the above cluster region has been deposited in GenBank (accession: LC371755).
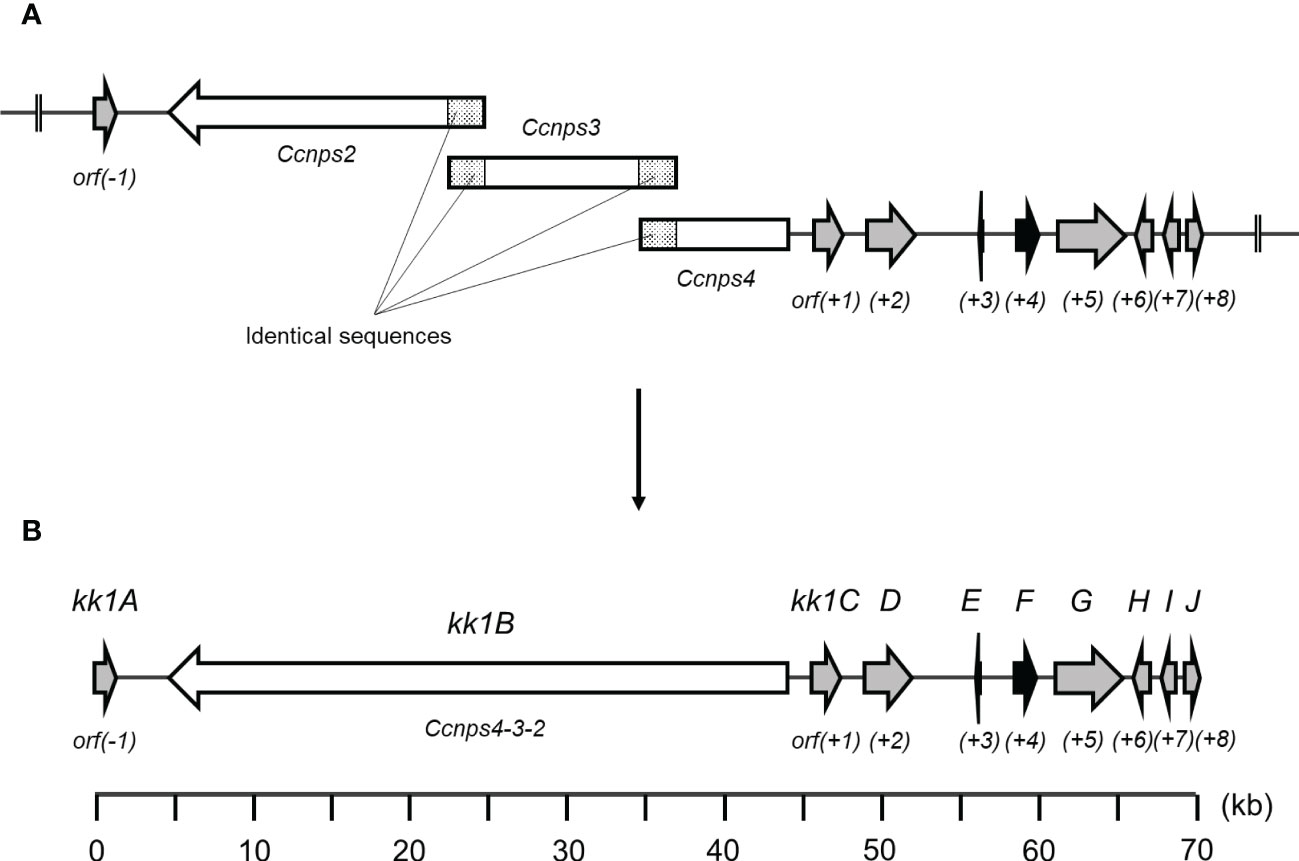
Figure 3 (A) A fragmentation image of Ccnps4-3-2, namely kk1B, into three ORFs (Ccnps2, Ccnps3, and Ccnps4) using the obtained sequencing data of Curvularia clavata BAUA-2787. (B) The genetic map and predicted gene arrangement of the KK-1 biosynthetic gene cluster (see Table 2). The gene kk1B, encoding non-ribosomal peptide synthetase (NRPS), is shown as a white arrow, and kk1F, encoding the transcription factor, is shown as a black arrow.
Cluster prediction was also performed using MIDDAS-M (Umemura et al., 2013), a sequence motif-independent detection method of secondary metabolism biosynthesis gene clusters. It can sensitively detect the gene clusters in which the genes are cooperatively regulated at transcriptional levels using genome sequence information that is combined with transcriptomic data that are obtained under compound-producing and non-producing conditions. C. clavata BAUA-2787 produced KK-1 well on the CM medium at 26 ˚C but less was produced at 37°C. Therefore, we set 26°C as the KK-1 production condition and 37°C as the KK-1 non-production condition, and RNA-Seq data were obtained under both conditions. As a result of cluster identification using MIDDAS-M and these data, a series of genes, including the 10 ORFs that were referred to above, were significantly detected (Figure 4), indicating that the expression of these genes was co-induced during KK-1 production. This result supports that this gene cluster is involved in the biosynthesis of KK-1.
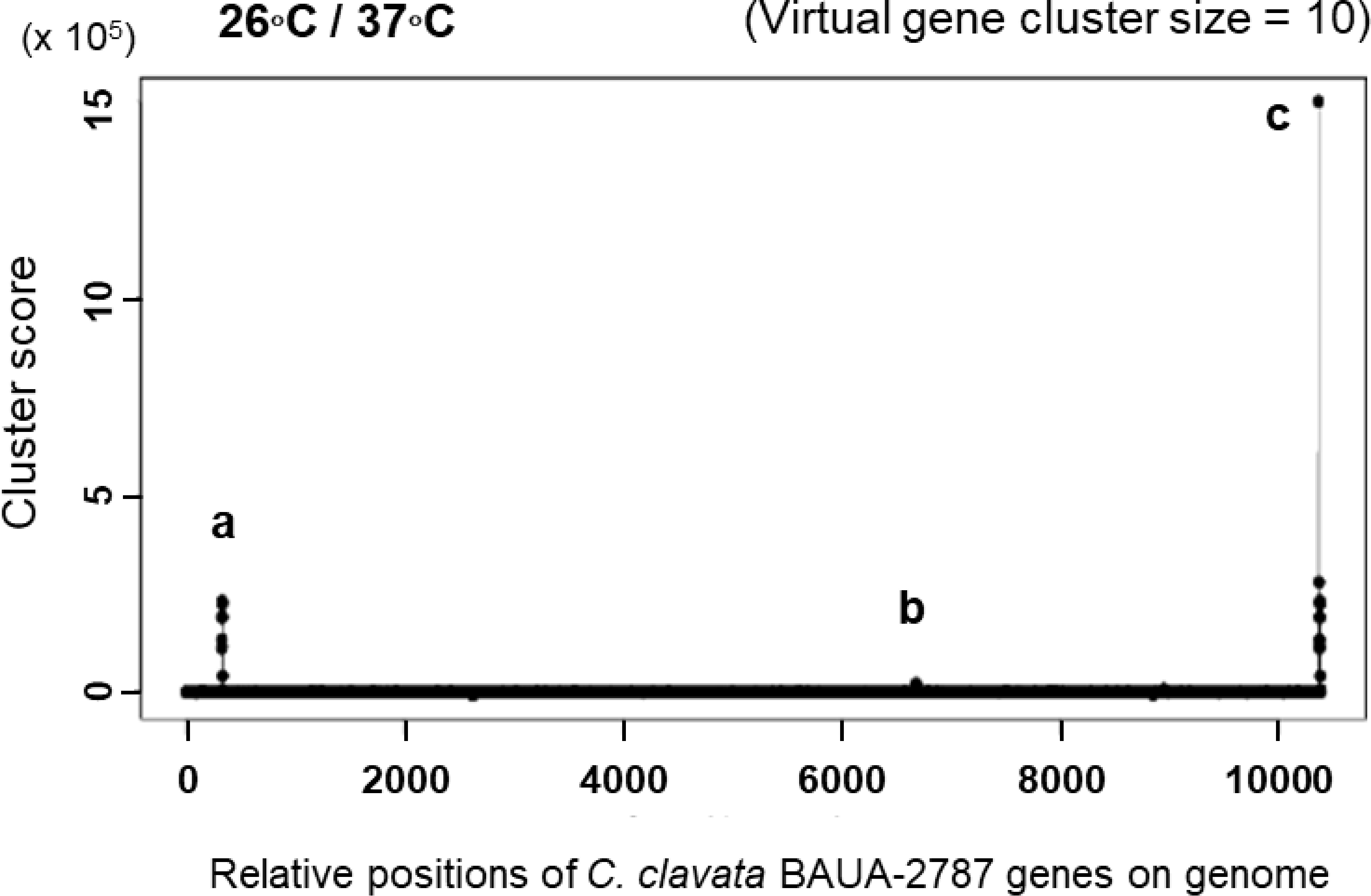
Figure 4 KK-1 cluster detection by MIDDAS-M. The gene clusters whose expression levels increased at 26 ˚C, the KK-1-producing culture temperature condition, when compared to 37˚C, the non-producing culture temperature condition. The analysis was performed on the data that consisted of the manually connected sequence of the putative KK-1 cluster, which was presumed to have been fragmented into three sequences (see Figure 3) and is at the end of all the obtained gene data at the right end of the horizontal axis. The three detected peaks are indicated by (A–C, (A), a fragmented KK-1 cluster containing Ccnps4; (B) a fragmented KK-1 cluster containing Ccnps2; and (C) a manually connected KK-1 cluster of the three fragments.
To analyze the function of each ORF, we started to develop a transformation system for C. clavata, which had not been previously established. Since the protoplast-PEG method had been well known as a transformation method for filamentous fungi, we first examined the conditions for the preparation of protoplasts of C. clavata BAUA-2787. As a result, we found that the protoplasts can be efficiently obtained by adding an enzyme solution that contains Yatalase and Lysing Enzymes from Trichoderma harziaum to culture mycelia on a 100 mL CM medium that is prepared in a 300 mL Erlenmeyer flask at 30°C and 130 rpm for 40 hours.
Next, the susceptibility of C. clavata BAUA-2787 protoplasts to AbA and Hyg was examined on the CM-sucrose agar medium to find drug-selectable markers for transformation. The results showed that AbA and Hyg almost completely inhibited mycelial growth from the protoplasts at concentrations of 10 μg/mL or higher and 200 μg/mL or higher, respectively (Figure 5), suggesting that these concentrations can be used to select transformants that confer resistance to the corresponding drugs.
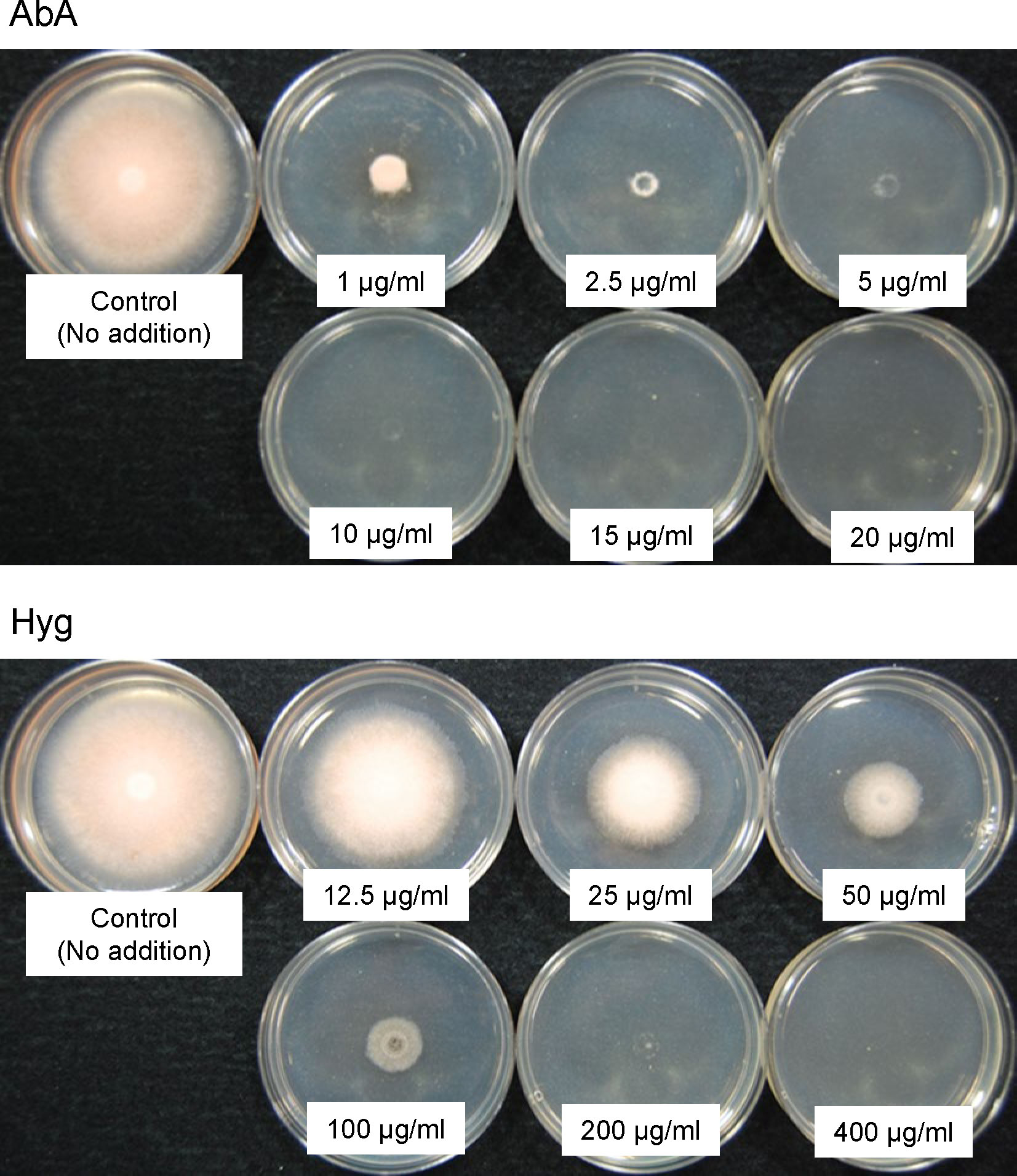
Figure 5 The susceptibility of the Curvularia clavata BAUA-2787 protoplast to aureobasidin A (AbA) and hygromycin (Hyg). The concentration of each agent in the medium is shown in the pictures.
With the necessary conditions in place, as described above, we attempted to transform C. clavata BAUA-2787 using the protoplast-PEG method and the pAUR316 plasmid, which is known as a shuttle vector for Aspergillus and contains the AbA resistance gene aurAr from Aspergillus nidulans as a selectable marker. Consequently, strains that could grow on the CM-sucrose agar medium containing 10 μg/mL AbA were obtained, and these strains also showed good growth when transferred to the CMA medium containing 2.5 μg/mL AbA. Furthermore, a PCR to amplify a partial sequence of pAUR316 revealed that these strains harbor pAUR316 (Figure 6), indicating that aurAr carried functions also in C. clavata BAUA2787, and the transformation system using AbA was successfully developed.
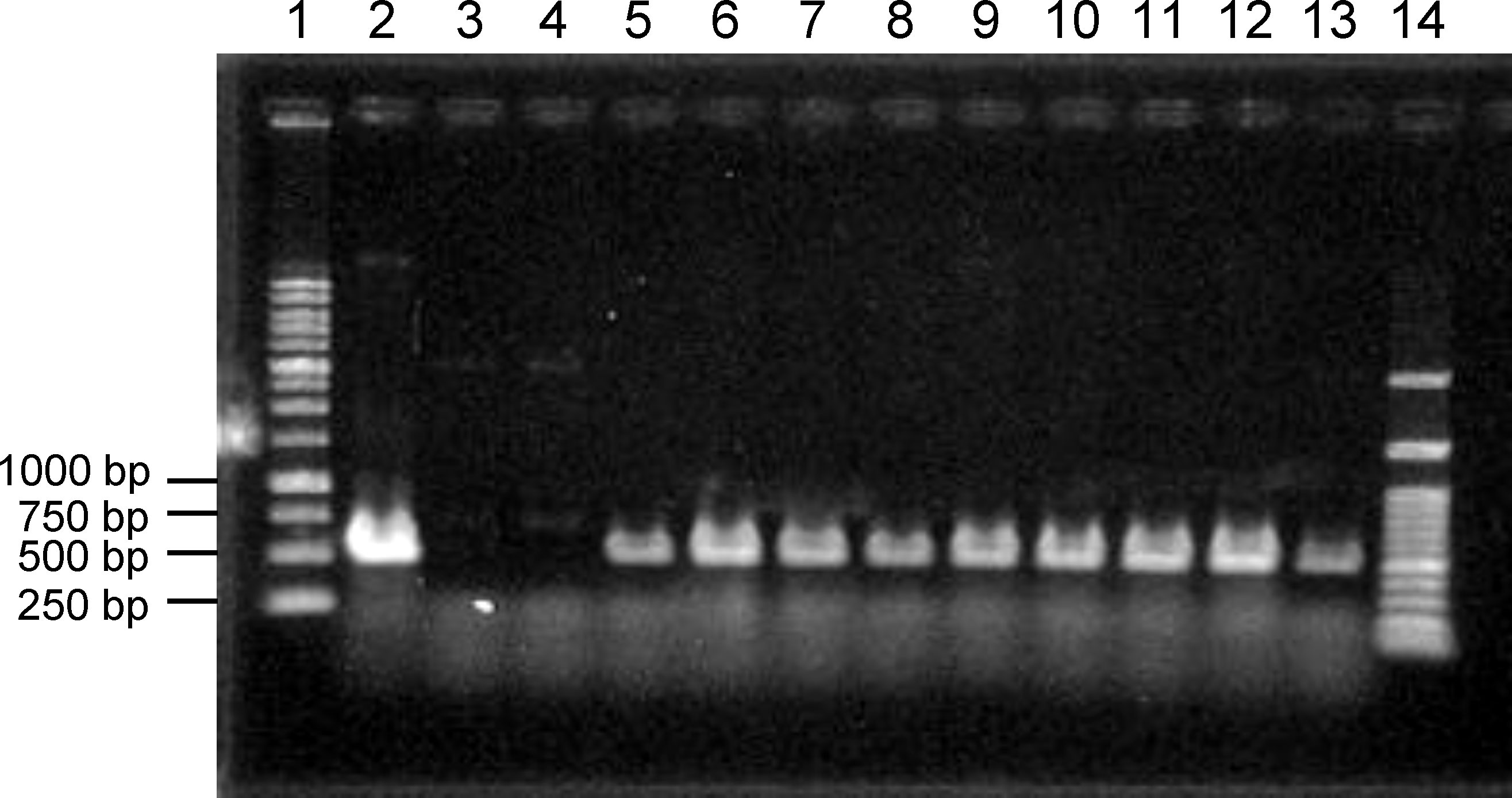
Figure 6 Agarose gel electrophoresis to confirm the introduction of pAUR316 into Curvularia clavata BAUA-2787. Lane 1, 1 kbp DNA ladder; lane 2, pAUR316 plasmid; lane 3, wild-type strain as the negative control; lane 4, non-transformant (negative result); lane 5-13, pAUR316 transformants; and lane 14, 100 bp DNA ladder.
In filamentous fungi, the pyrG gene, which is homologous to the Saccharomyces cerevisiae URA3 gene, is important for uridine synthesis and has often been used as a selectable auxotrophic marker for transformation (Van Hartingsveldt et al., 1987; Ling et al., 2013). In this study, the use of the pyrG homolog as an alternative selectable marker to the drug-resistant markers in C. clavata BAUA-2787 was examined. The pyrG gene encodes orotidine-5’-phosphate decarboxylase, which is responsible for the conversion of orotidine monophosphate to uridine monophosphate in the pyrimidine biosynthetic pathway, and the mutants lacking this function are auxotrophic for uridine and uracil and are also resistant to 5-fluoroorotic acid (5-FOA). A BlastP search revealed the gene which shows 55% similarity to the protein sequence that is encoded by pyrG of Aspergillus nidulans, and this gene was hereafter referred to as CcpyrG. Subsequently, to generate a CcpyrG deletion strain for use as a host, a CcpyrG deletion DNA cassette was prepared and used to transform the wild-type strain of C. clavata BAUA-2787 (Supplementary Figure S1). Therefore, we succeeded in obtaining the CcpyrG deletion strain, in which the DNA region containing CcpyrG was deleted by homologous recombination, and it became auxotrophic for uridine and uracil and 5-FOA-resistant.
Since kk1F, found in the putative biosynthetic gene cluster of KK-1, was predicted to be a transcription factor and was considered to regulate the expression of the other genes in the cluster, kk1F mutants were generated to analyze the function of this gene. Initially, the overexpression strain of kk1F was generated. As a promoter for overexpression, we focused on the gene which encodes a homolog of Nmt1 from Schizosaccharomyces pombe and Thi5p from Saccharomyces cerevisiae, and the gene shows 74% similarity to the conserved region (NMT1/THI5 like domain: Pfam 09084) of Nmt1 (accession: AAA35318) and was designated as Ccnmt1. Since the nmt1 gene has been known to have a strong promoter that can be regulated by thiamine in S. pombe (Maundrell, 1990) and Ccnmt1 is also highly expressed in a constant manner in C. clavata BAUA-2787 based on the RNA-Seq data, we decided to overexpress kk1F by using the Ccnmt-1 promoter sequence. Specifically, we constructed a plasmid with a DNA cassette in which kk1F was connected to the region that probably contained the Ccnmt1 promoter, and the wild-type strain of C. clavata BAUA-2787 was transformed using the plasmid (Supplementary Figure S2). Then, the transformants that were confirmed to have the target sequence introduced onto the chromosome were the kk1F overexpression strains.
KK-1 production of these strains was quantified on day 3 and day 7 of the cultures by LC-MS, and it was found to be increased when compared to the wild-type strain production on both days, reaching approximately double on day 7 (Figure 7A). Furthermore, the RNA-Seq analysis for day 2 and day 4 of the cultures revealed that the expression levels of the entire putative cluster of the kk1F overexpression strains were clearly higher than those of the wild-type strains, and the difference was greater on day 4 than on day 2 (Figure 7B). These results strongly suggested that stronger expression of kk1F increased the expression of the cluster genes, resulting in increased KK-1 productivity.
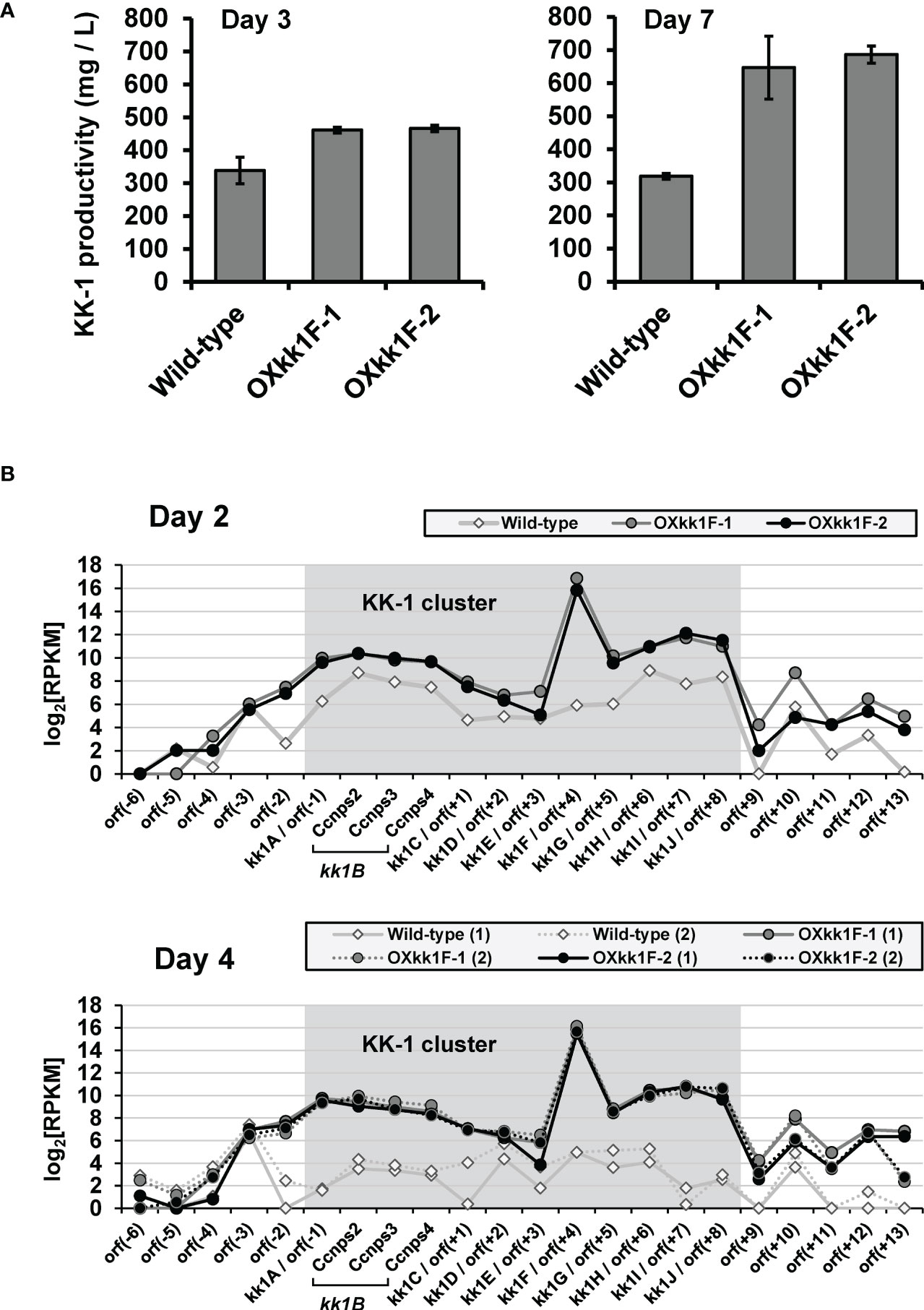
Figure 7 Analysis of kk1F overexpression strain. (A) The KK-1 productivity of the wild-type and two kk1F overexpression strains (OXkk1F-1, OXkk1F-2) on day 3 and day 7 of the culture. The error bars represent standard deviations (three biological replicates). (B) The reads per kilobase of exon per million mapped reads (RPKM) expression values in log2 scale for the putative KK-1 biosynthetic gene cluster corresponding to the gene IDs from kk1A to kk1J that are shown as shading and the surrounding genes in the above three strains on day 2 and day 4 of the culture. The numbers (1) and (2) indicate biological replicates. The region containing Ccnps2, Ccnps3, and Ccnps4 corresponds to kk1B, which encodes non-ribosomal peptide synthetase.
It should be noted that the compound corresponding to the peak used as an indicator for the quantification of KK-1 above was isolated and analyzed by infrared (IR) and proton nuclear magnetic resonance (1H-NMR) spectroscopy (Supplementary Figures S5, S6), and the respective spectra obtained certainly matched those of BK202, which is probably the same compound as KK-1, described in the patent WO1992005191A1 (Nielsen et al., 1992).
A kk1F deletion strain was also generated to examine KK-1 production and the cluster gene expression levels. A plasmid was constructed with sequences upstream and downstream of the kk1F ORF as homology arms for the deletion of this ORF by homologous recombination, and CcpyrG, including its putative promoter and terminator regions, was located between them as a selectable marker. Then, this plasmid, linearized with a restriction enzyme, was used to transform the C. clavata BAUA-2787 CcpyrG deletion strain (Supplementary Figure S3). Accordingly, the transformants that were confirmed to be homokaryons with kk1F deleted were the kk1F deletion strains.
Quantification of KK-1 on day 7 of the culture showed that the kk1F deletion strains hardly produced KK-1, and the antifungal activity of their culture supernatant against B. cinerea was also removed (Figure 8A). Furthermore, the deletion strains showed reduced transcriptional expression levels for the entire putative cluster when compared to those of the wild-type strains (Figure 8B). These results suggested that the deletion of kk1F led to reduced expression of the cluster genes, resulting in the loss of KK-1 productivity.
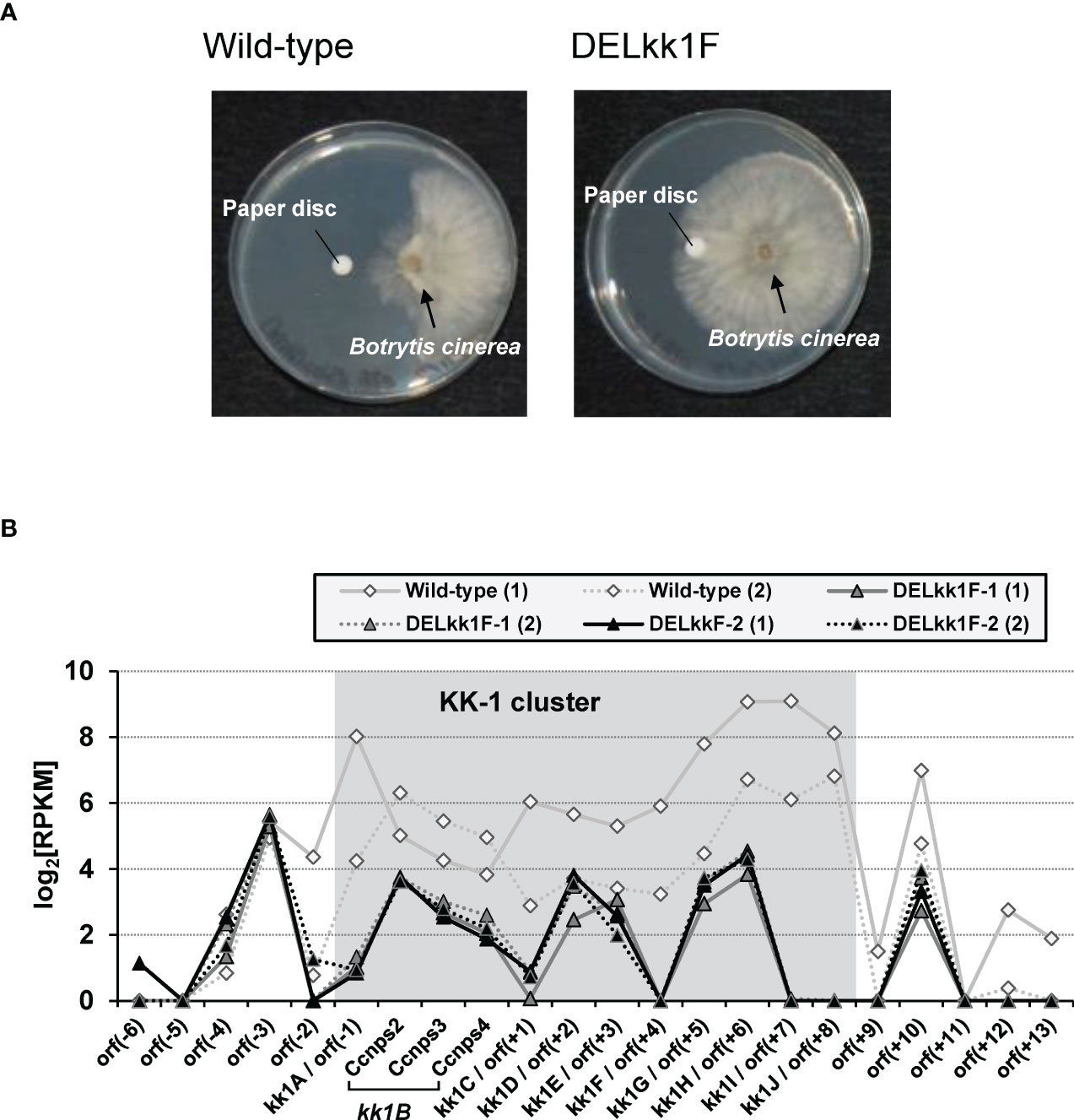
Figure 8 Analysis of the kk1F deletion strain. (A) Biological assays of the antifungal activity in the culture supernatants of the wild-type and kk1F deletion strains (DELkk1F). The pictures show the Botrytis cinerea colonies grown against paper discs soaked with the culture supernatant of each strain. (B) The reads per kilobase of exon per million mapped reads (RPKM) expression values in log2 scale for the putative KK-1 biosynthetic gene cluster corresponding to the gene IDs from kk1A to kk1J that are shown as shading and the surrounding genes of the wild-type and two kk1F deletion strains (DELkk1F-1, DELkk1F-2) on day 3 of the culture. The numbers (1) and (2) indicate biological replicates. The region containing Ccnps2, Ccnps3, and Ccnps4 corresponds to kk1B, which encodes non-ribosomal peptide synthetase.
To investigate the involvement of the putative cluster genes other than kk1F in KK-1 biosynthesis, the deletion mutants of each gene were generated and KK-1 productivity was evaluated. For each gene, a plasmid of almost the same design as that which was used to delete kk1F was constructed, and each of these plasmids was linearized by restriction enzymes and transformed into the C. clavata BAUA-2787 CcpyrG deletion strain (Supplementary Figures S3, S4). Thus, the transformants that were confirmed to be homokaryons with a deleted target gene were obtained for each gene deletion strain.
The KK-1 production of each mutant was then measured using day 7 of the culture (Figure 9). KK-1 production was completely lost in the kk1B deletion strain, and in four deletion strains (kk1A, kk1D, kk1H, and kk1I) it was markedly reduced to less than 10% when compared to that of the wild-type strain, suggesting that these genes are likely to be directly involved in the biosynthesis of KK-1. Furthermore, KK-1 production in the other four deletion strains (kk1C, kk1E, kk1G, and kk1J) was also lower than that in the wild-type strain, although the extent of the reduction was smaller than that of the above six mutants. This suggests that these genes are not essential for KK-1 synthesis but are involved in its production in some way by promoting or assisting productivity.
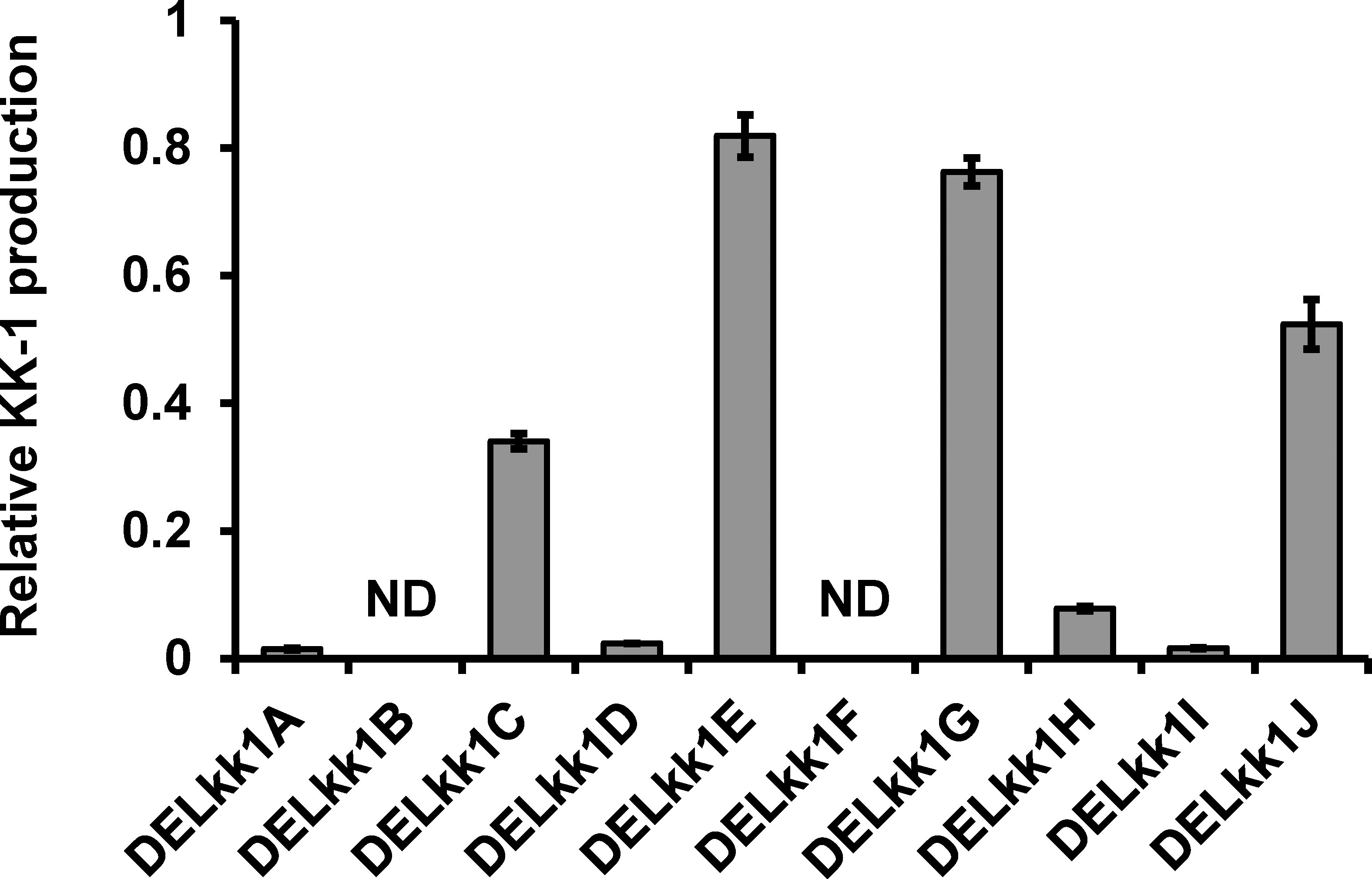
Figure 9 The relative KK-1 production of each cluster gene deletion strain when compared to that of the wild-type strain on day 7 of the culture. The abbreviations for the mutants are shown on the horizontal axis: kk1A deletion strain, DELkk1A; kk1B deletion strain, DELkk1B; kk1C deletion strain, DELkk1C; kk1D deletion strain, DELkk1D; kk1E deletion strain, DELkk1E; kk1F deletion strain, DELkk1F; kk1G deletion strain, DELkk1G; kk1H deletion strain, DELkk1H; kk1I deletion strain, DELkk1I; kk1J deletion strain, DELkk1J. The error bars represent standard deviations (three biological replicates). ND, not detected.
To determine whether kk1A, predicted to encode an S-adenosyl-L-methionine-dependent methyltransferase, is involved in the synthesis of O-methyltyrosine residues of KK-1, the metabolites in the culture medium of the kk1A deletion strain were analyzed and compared with those of the wild-type strain and the other cluster gene deletion strains using LC-MS (Figure 10). In addition to the peak of KK-1 (m/z = 1113 [M+H]+), five compounds (a, b, c, d, and e) with molecular weights corresponding to the mono-demethylated metabolites of KK-1 (m/z = 1099 [M+H]+) were detected in the wild-type strain (Figure 10A), all of which were more hydrophilic than KK-1. The production of all of these five compounds was ≤ 1/20 of that of KK-1, and each gene deletion strain basically produced them in proportion to the amount of KK-1 produced (Figure 10B). However, even though KK-1 production in the kk1A deletion strain was only about 1/100 of that of the wild-type strain, the production of compound c was maintained at the same level as that of the wild-type strain (Figure 10C), reversing the abundance ratio of KK-1 to compound c.
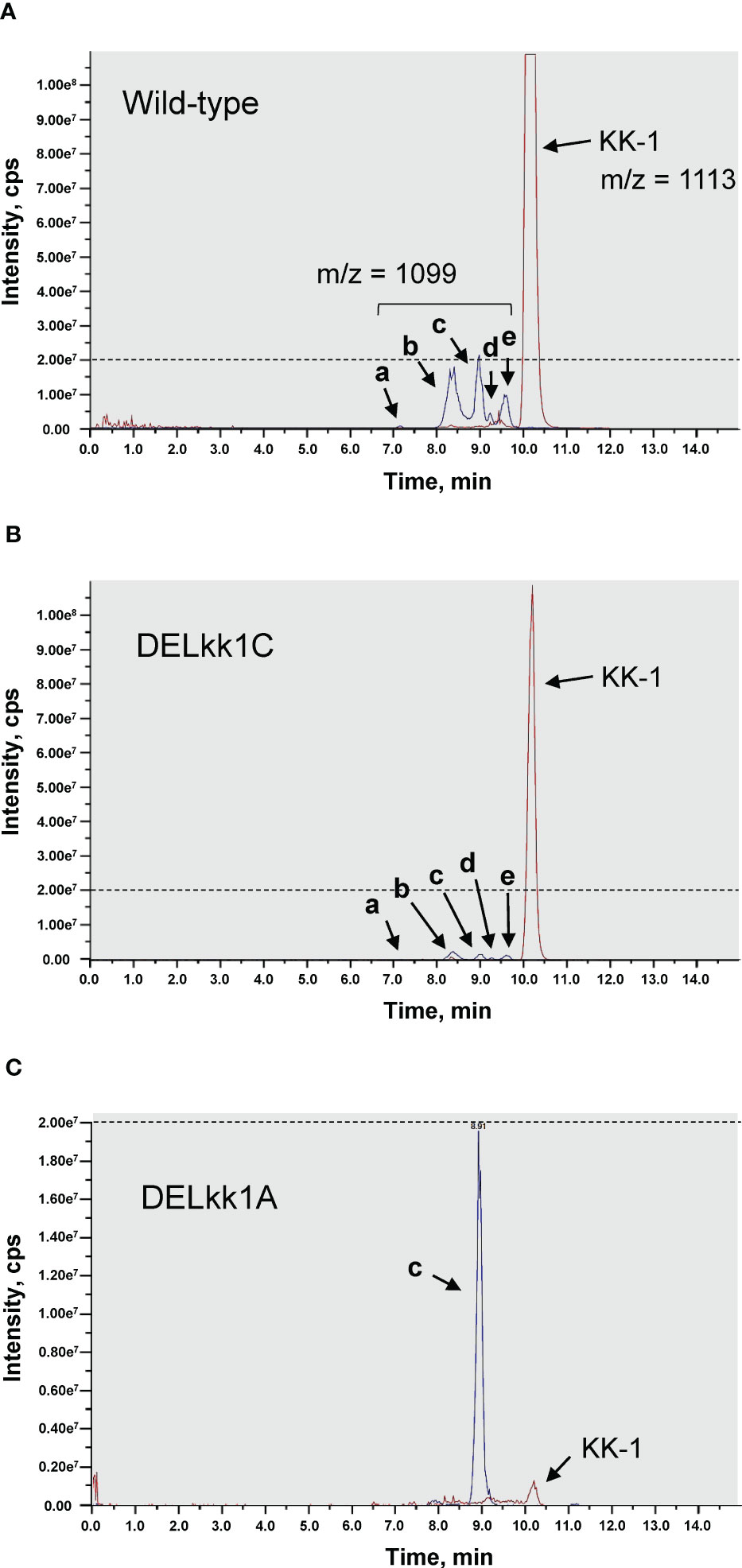
Figure 10 The HPLC-MS analysis of the culture extracts of the Curvularia clavata BAUA-2787 strains; (A) wild-type strain; (B) kk1C deletion strain (DELkk1C) as a representative of the KK-1 cluster gene deletion mutants except kk1A; and (C) kk1A deletion stain (DELkk1A). The extracted ion chromatograms are shown for KK-1 ([M+H+] = 1113) and its putative mono-demethylated metabolites (a, b, c, d and e, [M+H+] = 1099). The dotted lines are drawn at the peak intensity of 2.00e7 cps.
In this study, we sequenced a draft genome of C. clavata BAUA-2787 and identified a 71 kb region as the KK-1 biosynthetic gene cluster, which consists of 10 ORFs including kk1B. The kk1B gene encodes an NRPS with a modular structure that is consistent with the structural features of KK-1. Furthermore, for the first time, we reported on the development of a transformation system for C. clavata using the AbA resistant gene aurAr that is carried on the pAUR316 plasmid as a drug-resistant marker or the homologous pyrG gene on C. clavata BAUA-2787 as an auxotrophic marker. It was also shown that it is possible to modify the target region of the genome by homologous recombination. Consequently, the transformation systems that were developed in this work were used to confirm experimentally that six of the 10 ORFs are essential for KK-1 biosynthesis and the others play important roles in its enhancement.
In general, especially for bacterial NRPSs, the TE domain that is present at the C-terminus in the final module of NRPS is known to catalyze hydrolysis or macrocyclization to release the peptide (Hur et al., 2012; Duban et al., 2022) but the C-terminus of the NRPS that is encoded by kk1B lacks a TE domain and instead contains a C domain. It has been revealed that many of the NRPSs that are found in fungal genomes contain a condensation-like domain (CT domain) instead of a TE domain at the C-terminus, and that this domain is responsible for peptide macrocyclization (Gao et al., 2012). From these reports, we can speculate that in the NRPS that is encoded by kk1B, its C domain at the C-terminus in the final module is also responsible for releasing the peptide chain through macrocyclization.
The protein encoded by kk1F was predicted to have a bZIP domain and was the only gene encoding a transcription factor that was found within the putative KK-1 biosynthetic gene cluster. It is known that secondary metabolite clusters often contain a pathway specific transcription factor, which commonly regulate the expression of other cluster genes (Keller, 2019). A Zn(II)2Cys6-type transcription factor is well known as a pathway-specific transcription factor in fungi, such as AflR in the biosynthetic cluster of aflatoxin and its precursor sterigmatocystin in Aspergillus species (Yu et al., 1996; Flaherty and Payne, 1997; Ehrlich et al., 1999). Nevertheless, there have also been reported cases involving bZip-type transcription factors (Wang et al., 2008; Gao et al., 2011), suggesting that the protein encoded by kk1F likely regulates the expression of other genes in the KK-1 cluster. Therefore, kk1F overexpression and deletion strains were generated in this study, and we attempted to analyze the function of kk1F using these mutants. The results showed that the expression of the nine genes, except for kk1F, which constitutes the putative KK-1 cluster, was cooperatively regulated by the protein encoded by kk1F (Figures 7B, 8B), strongly supporting that these are the biosynthetic cluster genes of KK-1. Additionally, orf(-2) and orf(+10), located in the outer neighborhood of the cluster, also appeared to be co-expressed with the above nine genes. The protein encoded by orf(-2) shows similarity to the uncharacterized proteins of several plant pathogens, and that which is encoded by orf(+10) shows high similarity to known GDP-mannose transporters (Table 1). Although their involvement in KK-1 biosynthesis is not clear from this information alone and it has not been experimentally confirmed in this report, these two genes that are located on both ends of the KK-1 cluster may be strictly involved in KK-1 biosynthesis.
The four ORFs that were found in the KK-1 cluster, kk1A, kk1D, kk1H, and kk1I, probably contain the genes that are functionally required for KK-1 structure formation, because the KK-1 productivity of each deletion mutant was significantly reduced when compared to that of the wild-type strain (Figure 9). In particular, since three of the constituent residues of KK-1, D-lactic acid, L-pipecolic acid, and O-methyl-L-tyrosine, could not be synthesized by the enzyme domain of the NRPS encoded by kk1B alone, it was presumed that some of the above four genes were responsible for the biosynthesis of these residues.
In terms of the NRPSs that synthesize cyclic depsipeptides, there are known cases in which a certain A domain recognizes and incorporates an α-hydroxy carboxylic acid that has been previously synthesized by some enzyme instead of an amino acid. In this case, a cyclic depsipeptide, with an ester bond, is released through the cyclization of the peptide chain with the hydroxy group at the α-position of this residue as a nucleophile in the terminal module of the NRPS. For the biosynthesis of beauvericin, a cyclic depsipeptide that is produced by the entomopathogenic fungus Beauveria bassana, the A domain of the first module of NRPS that is encoded by the bbBeas gene was presumed to recognize and activate D-α-hydroxyisovaleric acid. This D-α-hydroxyisovaleric acid is synthesized from valine catabolism or pyruvate metabolism by NADPH-dependent α-ketoisovalerate reductase, which is encoded by the kivr gene, which is located upstream and adjacent to bbBeas (Xu et al., 2008). This has been experimentally confirmed using the kivr knockout strain (Xu et al., 2008). The cyclic depsipeptide antibiotic valinomycin, which is produced by Streptomyces tsusimaensis ATCC 15141 contains D-α-hydroxyisovaleric acid and L-lactic acid residues, both of which are presumably activated by the A domain within the corresponding module of the NRPSs that are encoded by vlm1 and vlm2 (Cheng, 2006). The synthesis of these residues is probably not derived from stand-alone enzymes but it is speculated to involve unusual enzyme domains that occur in the NRPSs (Cheng, 2006). The protein encoded by kk1H in the KK-1 cluster contains the catalytic domain of a D-isomer specific 2-hydroxyacid dehydrogenase and has high homology to known D-lactate dehydrogenase (Table 1). This enzyme functions to catalyze the reversible conversion of pyruvate into lactic acid, and it is likely to be involved in the synthesis of D-lactic acid, a constituent residue of KK-1 (Figure 11B). During the biosynthesis of KK-1, it is considered that the A domain (in the first module of NRPS that is encoded by kk1B) first activates free D-lactic acid, which is synthesized from pyruvic acid by the D-lactate dehydrogenase which is encoded by kk1H, and then it is incorporated into the PCP domain of the same module (Figure 11A). After a series of peptide elongation reactions to synthesize the precursor peptide chain, cyclization with the hydroxy group of the D-lactic acid as a nucleophile is presumed to occur in the final module of NRPS, resulting in the formation of the depsipeptide ester bond.
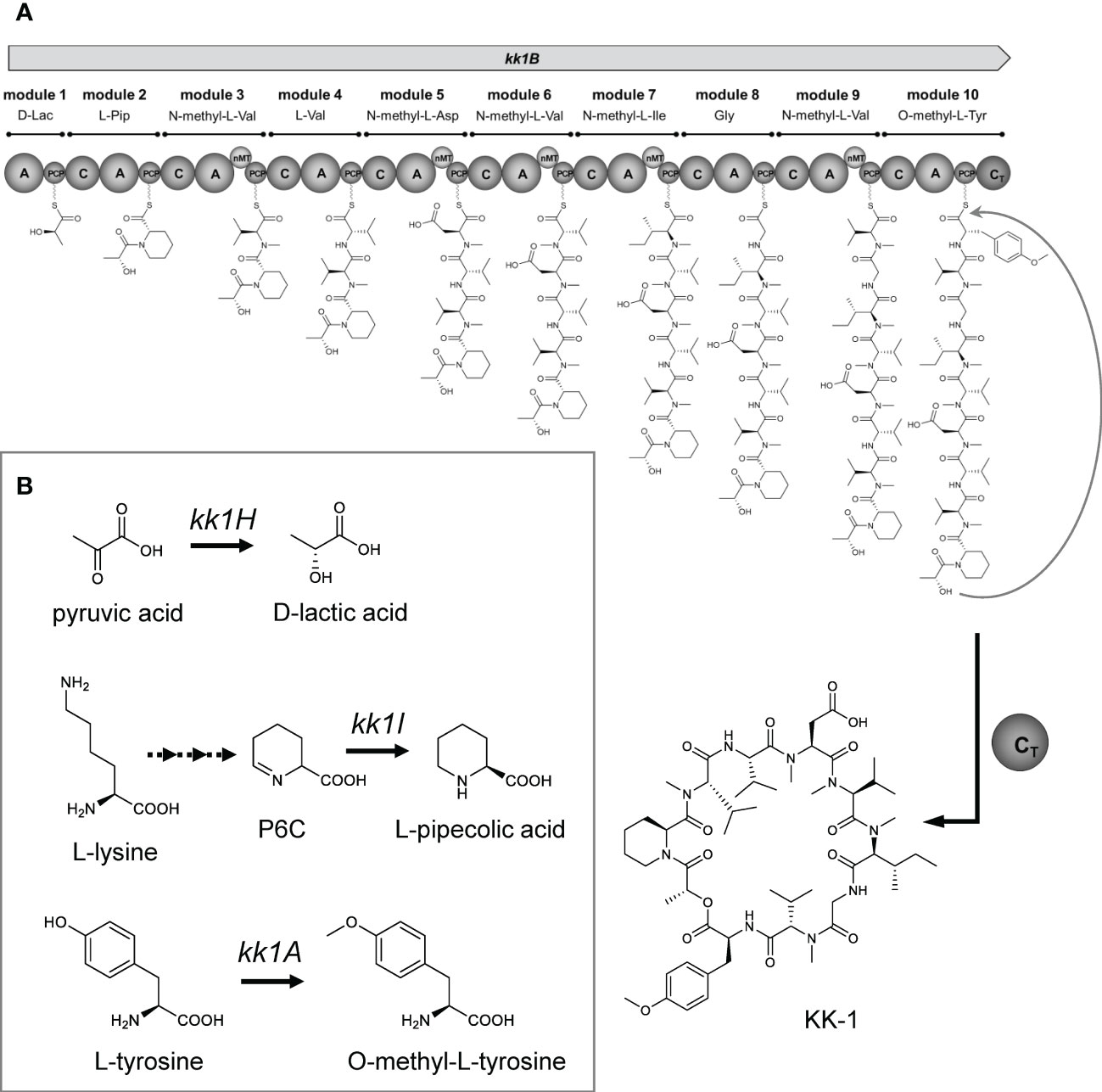
Figure 11 The proposed biosynthetic pathway for KK-1. (A) The proposed model for the KK-1 non-ribosomal peptide synthetase assembly line: C, condensation domain; A, adenylation domain; PCP, peptidyl carrier protein domain; nMT, nitrogen methyl transferase; CT, condensation-like domain. (B) The proposed pathway for the biosynthesis of non-amino acids or non-proteinogenic amino acids building blocks.
L-pipecolic acid, a constituent residue of KK-1, is biosynthesized from L-lysine, and several biosynthetic pathways have been identified in microorganisms (He, 2006). One is the pathway that directly biosynthesizes L-pipecolic acid from L-lysine by the catalysis of lysine cyclodeaminase. The proteins encoded by rapL found in the biosynthetic gene cluster of rapamycin (Gatto et al., 2006) and encoded by fkbL found in the biosynthetic gene cluster of tacrolimus (Motamedi and Shafiee, 1998) were known to be involved in the synthesis of pipecolic acid residues as a lysine cyclodeaminase. The following other pathways are also known: the spontaneous cyclization of aldehydes generated by the deamination of the α-position of lysine or the elimination of the ϵ-amino group via saccharopine as a metabolic intermediate in accordance with the lysine metabolic pathway, resulting in P2C (Δ1-piperideine-2-carboxylate) or P6C (Δ1-piperideine-6-carboxylate), respectively. These are further reduced by specific enzymes to generate L-pipecolic acid (He, 2006). For the P6C pathway, there are also known cases in which P6C is generated directly from lysine by ϵ-aminotransferase (He, 2006). It has been shown that the reduction of P6C to L-pipecolic acid in microorganisms is at least partially catalyzed by pyrroline-5-carboxylate reductase (Fujii et al., 2002), which is known to be universally present in almost all organisms and is responsible for the conversion of Δ1-pyrroline-5-carboxylate to L-proline as the final reaction in L-proline biosynthesis (Delauney and Verma, 1993; Csonka and Leisinger, 2007). For the cyclic tetrapeptide apicidin F, a known histone deacetylase inhibitor that is produced by Fusarium fujikuroi, the biosynthesis of its pipecolic acid residue has been experimentally confirmed to involve the APF3 gene that encodes pyrroline-5-carboxylate reductase, which is found in the biosynthetic gene cluster (Niehaus et al., 2014). The protein encoded by kk1I in the KK-1 biosynthetic cluster showed high sequence homology to known pyrroline-5-carboxylate reductase (Table 1), suggesting that this is an important gene for the synthesis of the L-pipecolic acid residue of KK-1 (Figure 11B). Thus, it is presumed that P6C, which is generated as a metabolic intermediate of lysine, is converted to L-pipecolic acid by the catalysis of pyrroline-5-carboxylate reductase, which is encoded by kk1I, and then it is recognized and incorporated by the A domain of the second module of NRPS, which is encoded by kk1B (Figure 11A).
The kk1A gene was predicted to encode a S-adenosyl-L-methionine-dependent methyltransferase based on sequence information and to have a conserved domain of O-methyltransferase (Table 1). S-adenosyl-L-methionine-dependent methyltransferases can transfer a methyl group from S-adenosyl-L-methionine, a universal methyl group donor in cells, to a nucleophilic acceptor such as C, O, N, or S, and they are involved in methylation in the biosynthesis of secondary metabolites (Roje, 2006). For the biosynthesis of skyllamycin, a cyclic depsipeptide that is produced by Streptomyces sp. Acta 2897 and has an O-methyltyrosine residue, it has been shown that the biosynthetic cluster gene sky37 (encoding S-adenosyl-L-methionine-dependent methyltransferase) is responsible for the O-methylation of tyrosine as a free amino acid before its activation as an aminoacyl-AMP by the corresponding A domain of NRPS (Pohle et al., 2011). Similarly, in KK-1, we hypothesized that kk1A is responsible for the synthesis of the O-methyl-L-tyrosine residues. To demonstrate this, the culture metabolites of the wild-type strain and each strain with cluster gene deletion were compared (Figure 10). The results showed that the culture extracts of each strain contained at least five metabolites (compound a, b, c, d, and e) that were probably mono-demethylated forms of KK-1, all of which were basically produced at less than 1/20th of the amount of KK-1. In contrast, for the kk1A deletion strain, only the production of compound c was unchanged from that of the wild-type strain, although the productivity of KK-1 was significantly reduced when compared to the wild-type strain. KK-1 contains six methylated amino acid residues: three N-methyl-L-Val and one of N-methyl-L-Asp, N-methyl-L-Ile, and O-methyl-L-Tyr each, of which the five N-methylated amino acids are synthesized by the function of the corresponding nMT domains of NRPS. If the mono-demethylated metabolites that were detected in the culture extracts of each C. clavata BAUA-2787 strain are by-products that were synthesized due to a low rate of recognition and incorporation of the unmethylated amino acids by each A domain, which should activate the methylated amino acids, compound c would be the demethylated form at the O-methyltyrosine residue of KK-1. In other words, while the deletion of kk1A prevented the conversion of free tyrosine to O-methyltyrosine and, consequently, the synthesis of KK-1, it did not affect the incorporation pattern of unmethylated normal tyrosine by the A domain of the 10th module of NRPS, which should correspond to the O-methyltyrosine residue. Thus, the productivity of compound c was presumably unchanged from that of the wild-type strain. These results strongly suggest that intracellular tyrosine is pre-converted to O-methyl-L-tyrosine by the function of O-methyltransferase, which is encoded by kk1A (Figure 11B), and is recognized and incorporated by the A domain of the 10th module of NRPS, which is encoded by kk1B (Figure 11A).
The four ORFs that were found in the KK-1 cluster, kk1C, kk1E, kk1G, and kk1J, were considered to be involved in KK-1 production in some way by promoting or assisting productivity because their KK-1 productivity was maintained at a certain level even though it was lower than that of the wild-type strain (Figure 9). The kk1E gene was a putative gene of 234 bp, which encoded a protein with a small molecular weight of 8.1 kDa and showed high homology to the N-terminal sequence of known GDP-mannose transporters (Table 1). The RNA-Seq data of each strain showed that although transcription at the kk1E locus was observed, the sequencing reads were only mapped to the region from 5’UTR to approximately 160 bp downstream of the start codon (data not shown), suggesting that its 3’ terminus was not transcribed at all. It is possible that kk1E is not a functional gene and that the transcript from this locus regulates the expression of some gene that is involved in the production of KK-1, such as non-coding RNAs. The protein encoded by kk1G is predicted to be an ATP-binding cassette (ABC) transporter (Table 1), comprising two transmembrane domains and two nucleotide-binding domains. Since this gene is regulated with other cluster genes by kk1F encoding the transcription factor, it probably functions as a KK-1-specific transporter and is responsible for the extracellular efflux of the produced KK-1. The function of this transporter was presumed to confer self-tolerance against KK-1 but kk1G deletion had no effect on the fungal growth (data not shown). It was also assumed that the accumulation of KK-1 in the cells due to a lack of the efflux system would cause the suppression of cluster gene expression but the final production of KK-1 in the kk1G deletion strain was only 20–30% lower than that in the wild-type strain (Figure 9). These findings suggest that the protein encoded by kk1G plays a role in promoting KK-1 efflux. This is similar to the function of the ABC transporter gene, sirA, in the biosynthetic gene cluster of the fungal toxin sirodesmin, which is produced by the plant pathogen Leptosphaeria maculans (Gardiner et al., 2005). It is also possible that there is another efflux system outside the KK-1 cluster, such as atrD, which encodes an ABC transporter that functions as a multiple drug resistance factor in Aspergillu nidulans (Andrade et al., 2000). The protein encoded by kk1J is predicted to belong to the α/β-hydrolase superfamily, and the Pfam search revealed that it contains the conserved domain of thioesterase (Table 1). Such thioesterases that are presented alone in the biosynthetic gene cluster of secondary metabolites are called type II thioesterases (TEIIs; Kotowska and Pawlik, 2014) and are distinguished from type I thioesterase, which is integrated into enzyme complexes such as NRPS and polyketide synthase (PKS) to release the synthesized products (Hur et al., 2012; Duban et al., 2022). Although, in most cases, TEIIs are not essential for the biosynthesis of the corresponding secondary metabolites, they are known to be important for efficient production because of their role in correcting products that are synthesized by NRPS and PKS (Schneider and Marahiel, 1998; Schwarzer et al., 2002; Kotowska and Pawlik, 2014). It is known that acyl residues that are incorrectly loaded into PCP domains block subsequent peptide chain elongation and cause the disruption of the module function of NRPS but TEII can remove them by hydrolysis to restore normal module function (Schwarzer et al., 2002; Yeh et al., 2004). Thus, in the biosynthesis of KK-1, TEII encoded by kk1J is likely to be involved in the corrective mechanism of peptide chain synthesis.
Currently, it is unclear how kk1C and kk1D are involved in the production of KK-1. In particular, the function of the protein encoded by kk1D is completely unknown because there is no similar protein with a known function but its role is very interesting because it is considered essential for KK-1 biosynthesis based on KK-1 productivity in the deletion strain (Figure 9).
Although further studies of the culture metabolites in each gene deletion or overexpression mutant are needed to more precisely understand the functions of the 10 ORFs that were found in the KK-1 cluster, it was revealed that they are at least involved in KK-1 production in some way. Yoshimi et al. (2018) succeeded in the heterologous production of KK-1 in Aspergillus oryzae in which eight of these 10 ORFs (except for kk1F, which encodes the specific transcription factor for the KK-1 pathway, and kk1E, which is probably not a gene) were all introduced under the control of the maltose-inducible amyB promoter. This provides further support that these genes are required for KK-1 biosynthesis. These results indicate that the efficient production of KK-1 may be achieved by studying the appropriate heterologous hosts and by using synthetic biology approaches to build metabolic pathways that are suitable for KK-1 production within those hosts. In this study, the C. clavata BAUA-2787 mutant in which kk1F was overexpressed under the control of the Ccnmt1 promoter showed approximately twice as much KK-1 production as the wild-type strain after 7 days of cultivation. Therefore, this suggests that further optimization of the promoter to express kk1F could lead to a drastic improvement in KK-1 productivity. Thus, the successful identification of the KK-1 biosynthetic gene cluster has enabled diverse approaches for the future realization of the mass production of KK-1.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, LC371755, https://www.ncbi.nlm.nih.gov/, BBC83956.1, https://www.ncbi.nlm.nih.gov/, BBC83957.1, https://www.ncbi.nlm.nih.gov/, BBC83958.1, https://www.ncbi.nlm.nih.gov/, BBC83959.1, https://www.ncbi.nlm.nih.gov/, BBC83960.1, https://www.ncbi.nlm.nih.gov/, BBC83961.1, https://www.ncbi.nlm.nih.gov/, BBC83962.1, https://www.ncbi.nlm.nih.gov/, BBC83963.1, https://www.ncbi.nlm.nih.gov/, BBC83964.1, https://www.ncbi.nlm.nih.gov/, BBC83965.1.
SY, TF, KA, MM and KK conceived and designed the experiments. SY, TF and TK carried out the genome sequence of the fungus. SY and TF performed the construction of the fungal strains. SY performed the essential experiments and analyzed the data. MU analyzed the data. SY, AY, KA, and KK wrote the paper. All authors contributed to the article and approved the submitted version.
This study was supported by the Commission for Development of Artificial Gene Synthesis Technology for Creating Innovative Biomaterial, the Ministry of Economy, Trade and Industry of Japan.
We thank Dr. Goro Terai for technical support in gene finding and Akita Konno Co., Ltd. for providing the fungal strain.
Author SY, TF, and KK are employed by Kumiai Chemical Industry Co., Ltd. and TK is employed by Fermlab Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffunb.2022.1081179/full#supplementary-material
Andrade A., Van Nistelrooy J., Peery R., Skatrud P., De Waard M. (2000). The role of ABC transporters from aspergillus nidulans in protection against cytotoxic agents and in antibiotic production. Mol. Gen. Genet. 263, 966–977. doi: 10.1007/PL00008697
Brzozowski L., Mazourek M. (2018). A sustainable agricultural future relies on the transition to organic agroecological pest management. Sustainability. 10:2023. doi: 10.3390/su10062023
Bushley K. E., Raja R., Jaiswal P., Cumbie J. S., Nonogaki M., Boyd A. E., et al. (2013). The genome of tolypocladium inflatum: Evolution, organization, and expression of the cyclosporin biosynthetic gene cluster. PloS Genet. 9:e1003496. doi: 10.1371/journal.pgen.1003496
Cantrell C. L., Dayan F. E., Duke S. O. (2012). Natural products as sources for new pesticides. J. Nat. Prod. 75, 1231–1242. doi: 10.1021/np300024u
Cheng Y. Q. (2006). Deciphering the biosynthetic codes for the potent anti-SARS-CoV cyclodepsipeptide valinomycin in streptomyces tsusimaensis ATCC 15141. Chembiochem. 7, 471–477. doi: 10.1002/cbic.200500425
Choi K. R., Jang W. D., Yang D., Cho J. S., Park D., Lee S. Y. (2019). Systems metabolic engineering strategies: Integrating systems and synthetic biology with metabolic engineering. Trends Biotechnol. 37, 817–837. doi: 10.1016/j.tibtech.2019.01.003
Cravens A., Payne J., Smolke C. D. (2019). Synthetic biology strategies for microbial biosynthesis of plant natural products. Nat. Commun. 10:2142. doi: 10.1038/s41467-019-09848-w
Crossthwaite A. J., Bigot A., Camblin P., Goodchild J., Lind R. J., Slater R., et al. (2017). The invertebrate pharmacology of insecticides acting at nicotinic acetylcholine receptors. J. Pestic Sci. 42, 67–83. doi: 10.1584/jpestics.D17-019
Csonka L. N., Leisinger T. (2007). Biosynthesis of proline. EcoSal Plus. 2. doi: 10.1128/ecosalplus.3.6.1.4
Delauney A. J., Verma D. P. S. (1993). Proline biosynthesis and osmoregulation in plants. Plant J. 4, 215–223. doi: 10.1046/j.1365-313X.1993.04020215.x
De Simeis D., Serra S. (2021). Actinomycetes: A never-ending source of bioactive compounds-an overview on antibiotics production. Antibiotics (Basel). 10:483. doi: 10.3390/antibiotics10050483
Duban M., Cociancich S., Leclere V. (2022). Nonribosomal peptide synthesis definitely working out of the rules. Microorganisms 10:577. doi: 10.3390/microorganisms10030577
Ehrlich K., Montalbano B., Cary J. (1999). Binding of the C6-zinc cluster protein, AFLR, to the promoters of aflatoxin pathway biosynthesis genes in aspergillus parasiticus. Gene. 230, 249–257. doi: 10.1016/S0378-1119(99)00075-X
Elkhateeb W. A., Kolaibe A. G. A., Daba G. M. (2021). Cochliobolus, drechslera, bipolaris, curvularia different nomenclature for one potent fungus. J. Pharm. Pharmacol. 4, 01–06. doi: 10.31579/2693-7247/031
Endo A., Kakiki K., Misato T. (1970). Mechanism of action of the antifugal agent polyoxin d. J. Bacteriol. 104, 189–196. doi: 10.1128/jb.104.1.189-196.1970
Endo A., Misato T. (1969). Polyoxin d, a competitive inhibitor of UDP-n-acetylglucosamine: Chitin n-acetylglucosaminyltransferase in neurospora crassa. Biochem. Biophys. Res. Commun. 37, 718–722. doi: 10.1016/0006-291X(69)90870-5
Flaherty J. E., Payne G. (1997). Overexpression of aflR leads to upregulation of pathway gene transcription and increased aflatoxin production in aspergillus flavus. Appl. Environ. Microbiol. 63, 3995–4000. doi: 10.1128/aem.63.10.3995-4000.1997
Fujii T., Mukaihara M., Agematu H., Tsunekawa H. (2002). Biotransformation of l-lysine to l-pipecolic acid catalyzed by l-lysine 6-aminotransferase and pyrroline-5-carboxylate reductase. Biosci. Biotechnol. Biochem. 66, 622–627. doi: 10.1271/bbb.66.622
Gao X., Chooi Y. H., Ames B. D., Wang P., Walsh C. T., Tang Y. (2011). Fungal indole alkaloid biosynthesis: Genetic and biochemical investigation of the tryptoquialanine pathway in penicillium aethiopicum. J. Am. Chem. Soc. 133, 2729–2741. doi: 10.1021/ja1101085
Gao X., Haynes S. W., Ames B. D., Wang P., Vien L. P., Walsh C. T., et al. (2012). Cyclization of fungal nonribosomal peptides by a terminal condensation-like domain. Nat. Chem. Biol. 8, 823–830. doi: 10.1038/nchembio.1047
Gardiner D. M., Jarvis R. S., Howlett B. J. (2005). The ABC transporter gene in the sirodesmin biosynthetic gene cluster of leptosphaeria maculans is not essential for sirodesmin production but facilitates self-protection. Fungal Genet. Biol. 42, 257–263. doi: 10.1016/j.fgb.2004.12.001
Gatto G. J., Boyne M. T., Kelleher N. L., Walsh C. T. (2006). Biosynthesis of pipecolic acid by RapL, a lysine cyclodeaminase encoded in the rapamycin gene cluster. J. Am. Chem. Soc. 128, 3838–3847. doi: 10.1021/ja0587603
Gotoh O. (2000). Homology-based gene structure prediction: Simplified matching algorithm using a translated codon (tron) and improved accuracy by allowing for long gaps. Bioinformatics. 16, 190–202. doi: 10.1093/bioinformatics/16.3.190
He M. (2006). Pipecolic acid in microbes: biosynthetic routes and enzymes. J. Ind. Microbiol. Biotechnol. 33, 401–407. doi: 10.1007/s10295-006-0078-3
Hu T., Chitnis N., Monos D., Dinh A. (2021). Next-generation sequencing technologies: An overview. Hum. Immunol. 82, 801–811. doi: 10.1016/j.humimm.2021.02.012
Hur G. H., Vickery C. R., Burkart M. D. (2012). Explorations of catalytic domains in non-ribosomal peptide synthetase enzymology. Nat. Prod Rep. 29, 1074–1098. doi: 10.1039/c2np20025b
Ikegami T., Inatsugi T., Kojima I., Umemura M., Hagiwara H., Machida M., et al. (2015). Hybrid De novo genome assembly using MiSeq and SOLiD short read data. PloS One. 10:e0126289. doi: 10.1371/journal.pone.0126289
Isono K., Asahi K., Suzuki S. (1969). Polyoxins, antifungal antibiotics. XIII. structure of polyoxins. J. Am. Chem. Soc. 91, 7490–7505. doi: 10.1021/ja01054a045
Isono K., Nagatsu J., Kawashima Y., Suzuki S. (1965). Studies on polyoxins, antifungal antibiotics: Part i. isolation and characterization of polyoxins a and b. Agric. Biol. Chem. 29, 848–854. doi: 10.1080/00021369.1965.10858475
Isono K., Nagatsu J., Kobinata K., Sasaki K., Suzuki S. (1967). Studies on polyoxins, antifungal antibiotics: Part v. isolation and characterization of polyoxins c, d, e, f, g, h and i. Agric. Biol. Chem. 31, 190–199. doi: 10.1080/00021369.1967.10858788
Jakubczyk D., Dussart F. (2020). Selected fungal natural products with antimicrobial properties. Molecules. 25:911. doi: 10.3390/molecules25040911
Jensen M. K., Keasling J. D. (2015). Recent applications of synthetic biology tools for yeast metabolic engineering. FEMS Yeast Res. 15, 1–10. doi: 10.1111/1567-1364.12185
Katz L., Chen Y. Y., Gonzalez R., Peterson T. C., Zhao H., Baltz R. H. (2018). Synthetic biology advances and applications in the biotechnology industry: A perspective. J. Ind. Microbiol. Biotechnol. 45, 449–461. doi: 10.1007/s10295-018-2056-y
Keller N. P. (2019). Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 17, 167–180. doi: 10.1038/s41579-018-0121-1
Khiralla A., Spina R., Saliba S., Laurain-Mattar D. (2019). Diversity of natural products of the genera curvularia and bipolaris. Fungal Biol. Rev. 33, 101–122. doi: 10.1016/j.fbr.2018.09.002
Kirst H. A. (2010). The spinosyn family of insecticides: Realizing the potential of natural products research. J. Antibiot (Tokyo). 63, 101–111. doi: 10.1038/ja.2010.5
Kirst H. A., Michel K. H., Mynderase J. S., Chio E. H., Yao R. C., Nakasukasa W. M., et al. (1992). Discovery, isolation, and structure elucidation of a family of structurally unique, fermentation-derived tetracyclic macrolides. Baker D. R., Fenyes J. G., Steffens J. J. (Eds.). Synthesis and chemistry of agrochemicals III. American Chemical Society, Washington, DC, pp. 214–225.
Korbekandi H., Darkhal P., Hojati Z., Abedi D., Hamedi J., Pourhosein M. (2010). Overproduction of clavulanic acid by UV mutagenesis of streptomyces clavuligerus. Iran J. Pharm. Res. 9, 177–181. doi: 10.1016/j.jbiotec.2007.07.931
Kotowska M., Pawlik K. (2014). Roles of type II thioesterases and their application for secondary metabolite yield improvement. Appl. Microbiol. Biotechnol. 98, 7735–7746. doi: 10.1007/s00253-014-5952-8
Kung S. H., Lund S., Murarka A., Mcphee D., Paddon C. J. (2018). Approaches and recent developments for the commercial production of semi-synthetic artemisinin. Front. Plant Sci. 9:87. doi: 10.3389/fpls.2018.00087
Lazaroiu G., Andronie M., Uta C., Hurloiu I. (2019). Trust management in organic agriculture: Sustainable consumption behavior, environmentally conscious purchase intention, and healthy food choices. Front. Public Health. 7:340. doi: 10.3389/fpubh.2019.00340
Lee N., Hwang S., Lee Y., Cho S., Palsson B., Cho B. K. (2019). Synthetic biology tools for novel secondary metabolite discovery in streptomyces. J. Microbiol. Biotechnol. 29, 667–686. doi: 10.4014/jmb.1904.04015
Lee B. N., Kroken S., Chou D. Y., Robbertse B., Yoder O. C., Turgeon B. G. (2005). Functional analysis of all nonribosomal peptide synthetases in cochliobolus heterostrophus reveals a factor, NPS6, involved in virulence and resistance to oxidative stress. Eukaryot Cell. 4, 545–555. doi: 10.1128/EC.4.3.545-555.2005
Lee S. D., Park S. W., Oh K. K., Hong S. I., Kim S. W. (2002). Improvement for the production of clavulanic acid by mutant streptomyces clavuligerus. Lett. Appl. Microbiol. 34, 370–375. doi: 10.1046/j.1472-765X.2002.01102.x
Ling S. O., Storms R., Zheng Y., Rodzi M. R., Mahadi N. M., Illias R. M., et al. (2013). Development of a pyrG mutant of aspergillus oryzae strain S1 as a host for the production of heterologous proteins. ScientificWorldJournal. 2013, 634317. doi: 10.1155/2013/634317
Majoros W. H., Pertea M., Salzberg S. L. (2004). TigrScan and GlimmerHMM: Two open source ab initio eukaryotic gene-finders. Bioinformatics. 20, 2878–2879. doi: 10.1093/bioinformatics/bth315
Manamgoda D. S., Cai L., Bahkali A. H., Chukeatirote E., Hyde K. D. (2011). Cochliobolus: An overview and current status of species. Fungal Divers. 51, 3–42. doi: 10.1007/s13225-011-0139-4
Maundrell K. (1990). nmt1 of fission yeast. a highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 265, 10857–10864. doi: 10.1016/S0021-9258(19)38525-4
Meemken E.-M., Qaim M. (2018). Organic agriculture, food security, and the environment. Annu. Rev. Resour Economics. 10, 39–63. doi: 10.1146/annurev-resource-100517-023252
Mertz F. P., Yao R. C. (1990). Saccharopolyspora spinosa sp. nov. isolated from soil collected in a sugar mill rum still. Int. J. Syst. Evol. Microbiol. 40, 34–39. doi: 10.1099/00207713-40-1-34
Miao V., Coeffet-Legal M. F., Brian P., Brost R., Penn J., Whiting A., et al. (2005). Daptomycin biosynthesis in streptomyces roseosporus: cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiol. (Reading). 151, 1507–1523. doi: 10.1099/mic.0.27757-0
Motamedi H., Shafiee A. (1998). The biosynthetic gene cluster for the macrolactone ring of the immunosuppressant FK506. Eur. J. Biochem. 256, 528–534. doi: 10.1046/j.1432-1327.1998.2560528.x
Niehaus E. M., Janevska S., Von Bargen K. W., Sieber C. M., Harrer H., Humpf H. U., et al. (2014). Apicidin f: Characterization and genetic manipulation of a new secondary metabolite gene cluster in the rice pathogen fusarium fujikuroi. PloS One. 9:e103336. doi: 10.1371/journal.pone.0103336
Nielsen R. I., Breinholt J., Jensen G. W. (1992). Fungicidally active compounds. WO1992005191A1. (Geneva, Switzerland: World Intellectual Property Organization)
Orr N., Shaffner A. J., Richey K., Crouse G. D. (2009). Novel mode of action of spinosad: Receptor binding studies demonstrating lack of interaction with known insecticidal target sites. Pestic Biochem. Physiol. 95, 1–5. doi: 10.1016/j.pestbp.2009.04.009
Paddon C. J., Westfall P. J., Pitera D. J., Benjamin K., Fisher K., Mcphee D., et al. (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 496, 528–532. doi: 10.1038/nature12051
Parekh S., Vinci V. A., Strobel R. J. (2000). Improvement of microbial strains and fermentation processes. Appl. Microbiol. Biotechnol. 54, 287–301. doi: 10.1007/s002530000403
Pham J. V., Yilma M. A., Feliz A., Majid M. T., Maffetone N., Walker J. R., et al. (2019). A review of the microbial production of bioactive natural products and biologics. Front. Microbiol. 10:1404. doi: 10.3389/fmicb.2019.01404
Pohle S., Appelt C., Roux M., Fiedler H. P., Sussmuth R. D. (2011). Biosynthetic gene cluster of the non-ribosomally synthesized cyclodepsipeptide skyllamycin: deciphering unprecedented ways of unusual hydroxylation reactions. J. Am. Chem. Soc. 133, 6194–6205. doi: 10.1021/ja108971p
Roje S. (2006). S-Adenosyl-L-methionine: beyond the universal methyl group donor. Phytochemistry. 67, 1686–1698. doi: 10.1016/j.phytochem.2006.04.019
Schneider A., Marahiel M. A. (1998). Genetic evidence for a role of thioesterase domains, integrated in or associated with peptide synthetases, in non-ribosomal peptide biosynthesis in bacillus subtilis. Arch. Microbiol. 169, 404–410. doi: 10.1007/s002030050590
Schwarzer D., Mootz H. D., Linne U., Marahiel M. A. (2002). Regeneration of misprimed nonribosomal peptide synthetases by type II thioesterases. Proc. Natl. Acad. Sci. U.S.A. 99, 14083–14088. doi: 10.1073/pnas.212382199
Siddique S., Syed Q., Adnan A., Qureshi F. A. (2014). Production and screening of high yield avermectin B1b mutant of streptomyces avermitilis 41445 through mutagenesis. Jundishapur J. Microbiol. 7:e8626. doi: 10.5812/jjm.8626
Siddiqui M. S., Thodey K., Trenchard I., Smolke C. D. (2012). Advancing secondary metabolite biosynthesis in yeast with synthetic biology tools. FEMS Yeast Res. 12, 144–170. doi: 10.1111/j.1567-1364.2011.00774.x
Tanaka C., Kubo Y., Tsuda M. (1991). Genetic analysis and characterization of cochliobolus heterostrophus colour mutants. Mycol Res. 95, 49–56. doi: 10.1016/S0953-7562(09)81360-9
Trapnell C., Pachter L., Salzberg S. L. (2009). TopHat: discovering splice junctions with RNA-seq. Bioinformatics. 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Umemura M., Koike H., Nagano N., Ishii T., Kawano J., Yamane N., et al. (2013). MIDDAS-m: motif-independent de novo detection of secondary metabolite gene clusters through the integration of genome sequencing and transcriptome data. PloS One. 8:e84028. doi: 10.1371/journal.pone.0084028
Uramoto M., Matsuoka M., Liehr J., Mccloskey J. A., Isono K. (1981). Polyoxin n, a new member of the polyoxin family. Agric. Biol. Chem. 45, 1901–1902. doi: 10.1080/00021369.1981.10864808
Van Hartingsveldt W., Mattern I. E., Van Zeijl C. M., Pouwels P. H., Van Den Hondel C. A. (1987). Development of a homologous transformation system for aspergillus niger based on the pyrG gene. Mol. Gen. Genet. 206, 71–75. doi: 10.1007/BF00326538
Wang S., Xu Y., Maine E. A., Wijeratne E. M., Espinosa-Artiles P., Gunatilaka A. A., et al. (2008). Functional characterization of the biosynthesis of radicicol, an Hsp90 inhibitor resorcylic acid lactone from chaetomium chiversii. Chem. Biol. 15, 1328–1338. doi: 10.1016/j.chembiol.2008.10.006
Westfall P. J., Pitera D. J., Lenihan J. R., Eng D., Woolard F. X., Regentin R., et al. (2012). Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc. Natl. Acad. Sci. U.S.A. 109, E111–E118. doi: 10.1073/pnas.1110740109
Xu Y., Orozco R., Wijeratne E. M., Gunatilaka A. A., Stock S. P., Molnar I. (2008). Biosynthesis of the cyclooligomer depsipeptide beauvericin, a virulence factor of the entomopathogenic fungus beauveria bassiana. Chem. Biol. 15, 898–907. doi: 10.1016/j.chembiol.2008.07.011
Yang D., Park S. Y., Park Y. S., Eun H., Lee S. Y. (2020). Metabolic engineering of escherichia coli for natural product biosynthesis. Trends Biotechnol. 38, 745–765. doi: 10.1016/j.tibtech.2019.11.007
Yeh E., Kohli R. M., Bruner S. D., Walsh C. T. (2004). Type II thioesterase restores activity of a NRPS module stalled with an aminoacyl-s-enzyme that cannot be elongated. Chembiochem. 5, 1290–1293. doi: 10.1002/cbic.200400077
Yoshimi A., Yamaguchi S., Fujioka T., Kawai K., Gomi K., Machida M., et al. (2018). Heterologous production of a novel cyclic peptide compound, KK-1, in aspergillus oryzae. Front. Microbiol. 9:690. doi: 10.3389/fmicb.2018.00690
Yu J., Butchko R. A., Fernandes M., Keller N. P., Leonard T. J., Adams T. H. (1996). Conservation of structure and function of the aflatoxin regulatory geneaflR fromAspergillus nidulans andA. flavus. Curr. Genet. 29, 549–555. doi: 10.1007/BF02426959
Zhang M. M., Qiao Y., Ang E. L., Zhao H. (2017). Using natural products for drug discovery: the impact of the genomics era. Expert Opin. Drug Discovery. 12, 475–487. doi: 10.1080/17460441.2017.1303478
Keywords: non-ribosomal peptide, biosynthetic gene cluster, antifungal agent, genome sequence, Curvularia clavata, natural fungicide, pesticide, grey mold
Citation: Yamaguchi S, Fujioka T, Yoshimi A, Kumagai T, Umemura M, Abe K, Machida M and Kawai K (2023) Discovery of a gene cluster for the biosynthesis of novel cyclic peptide compound, KK-1, in Curvularia clavata. Front. Fungal Biol. 3:1081179. doi: 10.3389/ffunb.2022.1081179
Received: 27 October 2022; Accepted: 15 December 2022;
Published: 20 January 2023.
Edited by:
István Pócsi, University of Debrecen, HungaryReviewed by:
Philipp Wiemann, Solugen, Inc., United StatesCopyright © 2023 Yamaguchi, Fujioka, Yoshimi, Kumagai, Umemura, Abe, Machida and Kawai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kiyoshi Kawai, ay1rYXdhaUBrdW1pYWktY2hlbS5jby5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.