- 1Center for Health and Bioresources, AIT Austrian Institute of Technology GmbH, Tulln, Austria
- 2Department of Microbiology and Ecosystem Science, Division of Terrestrial Ecosystem Research, University of Vienna, Vienna, Austria
The genus Trichoderma is among the best studied groups of filamentous fungi, largely because of its high relevance in applications from agriculture to enzyme biosynthesis to biofuel production. However, the physiological competences of these fungi, that led to these beneficial applications are intriguing also from a scientific and ecological point of view. This review therefore summarizes recent developments in studies of fungal genomes, updates on previously started genome annotation efforts and novel discoveries as well as efforts towards bioprospecting for enzymes and bioactive compounds such as cellulases, enzymes degrading xenobiotics and metabolites with potential pharmaceutical value. Thereby insights are provided into genomes, mitochondrial genomes and genomes of mycoviruses of Trichoderma strains relevant for enzyme production, biocontrol and mycoremediation. In several cases, production of bioactive compounds could be associated with responsible genes or clusters and bioremediation capabilities could be supported or predicted using genome information. Insights into evolution of the genus Trichoderma revealed large scale horizontal gene transfer, predominantly of CAZyme genes, but also secondary metabolite clusters. Investigation of sexual development showed that Trichoderma species are competent of repeat induced point mutation (RIP) and in some cases, segmental aneuploidy was observed. Some random mutants finally gave away their crucial mutations like T. reesei QM9978 and QM9136 and the fertility defect of QM6a was traced back to its gene defect. The Trichoderma core genome was narrowed down to 7000 genes and gene clustering was investigated in the genomes of multiple species. Finally, recent developments in application of CRISPR/Cas9 in Trichoderma, cloning and expression strategies for the workhorse T. reesei as well as the use genome mining tools for bioprospecting Trichoderma are highlighted. The intriguing new findings on evolution, genomics and physiology highlight emerging trends and illustrate worthwhile perspectives in diverse fields of research with Trichoderma.
Introduction
The genus Trichoderma belongs to the most beneficial group of fungi for humanity, which explains the extensive research efforts dedicated to biology and biotechnology with these fungi (Schuster and Schmoll, 2010; Druzhinina et al., 2011; Bischof et al., 2016; Guzman-Guzman et al., 2019). While industrial application of Trichoderma for protein production is limited to the descendants of a single species (Bischof et al., 2016; Paloheimo et al., 2016; Arnau et al., 2020), applications in agriculture for biocontrol and plant protection involve numerous species and strains (Kashyap et al., 2017; Lahlali et al., 2022; Tyskiewicz et al., 2022) and their high performance made Trichoderma the biocontrol agent with the highest market performance in terms of value, even higher than bacterial biocontrol agents together1 2. Due to their versatility, Trichoderma species serve as models for such important topics like mechanisms regulating plant cell wall degradation (Glass et al., 2013; Bischof et al., 2016), biocontrol (Harman et al., 2004a; Guzman-Guzman et al., 2019; Harman et al., 2021), effector like molecules (Ramirez-Valdespino et al., 2019) and light response (Schmoll et al., 2010; Carreras-Villaseñor et al., 2012; Schmoll, 2018a; 2018b). Additionally, Trichoderma spp. are a valuable source for natural products leveraged by screening genomes with constantly enhanced software tools (Rush et al., 2021). Recently, even a connection between the innate immune system of animals and Trichoderma was drawn (Medina-Castellanos et al., 2018). Interestingly, Trichoderma can initiate heritable plant priming responses, which are attributed to epigenetic regulation (Moran-Diez et al., 2021). Not only the active fungi themselves, but also extracts of Trichoderma spp. can inhibit growth and/or production of mycotoxins by pathogens (Stracquadanio et al., 2021). However, only a few years ago also a potential downside of the distribution of Trichoderma in nature became obvious with the detection of T. afroharzianum causing maize ear rot disease (Pfordt et al., 2020; Sanna et al., 2022). Consequently, species identification and genomic competences of these fungi deserve particular attention. Trichoderma spp. show an exceptional versatility in their preferred habitats and substrates with lifestyles ranging from mycoparasitism to plant saprotrophy and accordingly life in habitats characterized by feeding on fungi or decaying plant material, in soil or even as endophytes in living plant tissue, which changed during evolution several times (Chaverri and Samuels, 2013)
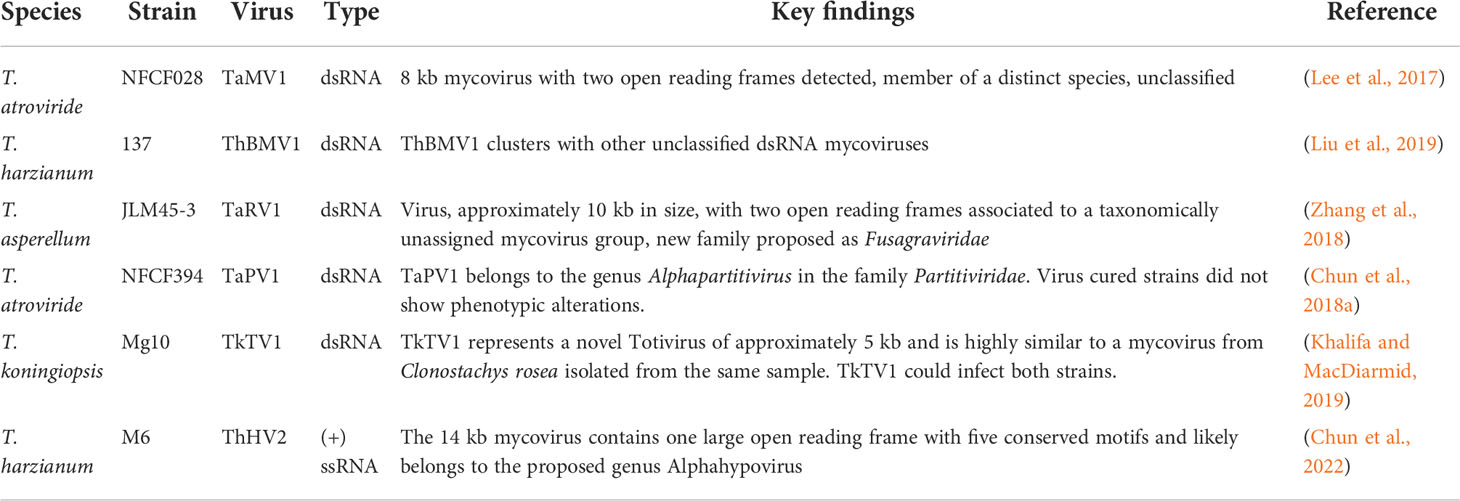
Starting from the genome sequence of T. reesei, which was published in 2008 (Martinez et al., 2008), the genomes of numerous Trichoderma model strains for enzyme production and biocontrol followed (Figure 1). The availability of the genome sequences of major model fungi of Trichoderma considerably contributed to detailed investigation of mechanisms of action and regulation of pathways (Sood et al., 2020), which is focused on systemic resistance of plants (Shoresh et al., 2010), colonization (Hinterdobler et al., 2021b; Taylor et al., 2021; Hafiz et al., 2022; Taylor et al., 2022), effector molecules (Ramirez-Valdespino et al., 2019) and plant-fungus-pathogen interactions (Mendoza-Mendoza et al., 2018; Macias-Rodriguez et al., 2020; Alfiky and Weisskopf, 2021) among others.
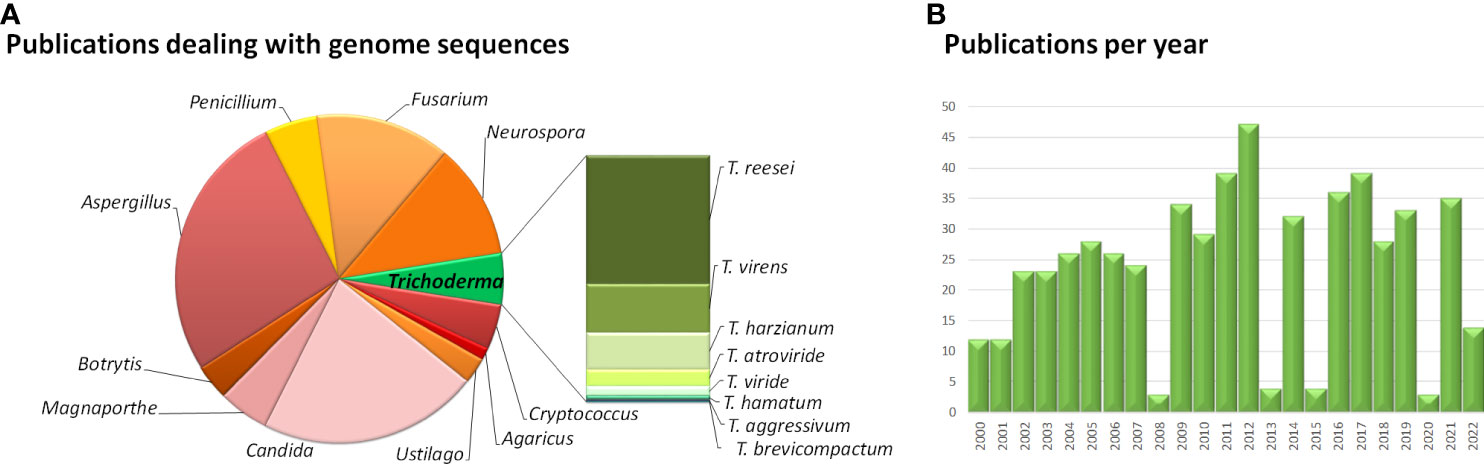
Figure 1 Overview of studies dealing with fungal genomes. (A) Results of a Pubmed search for "genome sequences” of the respective fungi. (B) Number of studies dealing with „Trichoderma genome sequences” per year published in Pubmed. The website pubmed.ncbi.nlm.nih.gov/was accessed in August 2022.
One crucial task in the future will be to seriously investigate both competences of Trichoderma strains that can be applied for human benefit and to balance these benefits with potential threats by strains with harmful characteristics for human and plant health. The imminent climate crisis and the aim of more sustainable and safe agriculture require increased research efforts to safely develop biocontrol applications and support of plant health by microbes without risking ecological or human adversities.
The NCBI taxonomy browser (https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/; accessed on July 11, 2022) lists more than 400 Trichoderma species and additionally over 1800 unclassified Trichoderma species. The ecophysiology and evolution of a first batch of more than 30 Trichoderma species is currently subject to a large scale sequencing effort with the Joint Genome Institute (JGI community sequencing project CSP-503464), which will be followed by analyses of another several hundred species to be analyzed.
Large scale research efforts for many species of Trichoderma revealed the evolutionary basis of the versatility of the fungi in this genus and provided important insights into their evolution. The genus evolved about 66 million years ago with later formation of the sections Longibrachiatum and Trichoderma and the clade Harzianum/Virens (Kubicek et al., 2019). Evolution of the genus was dominated by considerable gene gain (predominantly encoding ankyrins, HET domain proteins and transcription factors) and loss events around a core genome of exactly 7000 genes, which formed the basis for the diverse competences of Trichoderma (Kubicek et al., 2019). This core genome is dominated by genes involved in metabolism, with an important share of genes assigned to the KOG families “posttranslational modification, protein turnover and chaperones”, “transcription” and “carbohydrate transport and metabolism”, but also high numbers of glycoside hydrolases and fungal specific transcription factors (Kubicek et al., 2019). The constant adaptation and optimization of Trichoderma genomes is reflected in the finding that of the 105 orphan genes found in the core Trichoderma genome by comparing fungi out of this genus, most are under strong purifying selection (Kubicek et al., 2019).
Trichoderma – fungal pirates with a taste for plants
Being among the most important genera for industry and agriculture, the evolution of Trichoderma and their metabolic consequences is highly relevant for research. In recent years, enabled by sequencing efforts for several important Trichoderma species, intriguing new aspects on evolution in the genus Trichoderma were revealed, which in part explain their special characteristics. Horizontal or lateral gene transfer (HGT/LGT) has long been known to play a role in evolution, also in fungi, although it was considered to be more important in bacteria at first (Wang et al., 2015b). However, discrepancies of gene content or presence of whole clusters in closely related species can also be due to selective pressure and loss of genes, or a combination of selection and HGT (Hou et al., 2022).
An intriguing example for the latter phenomenon was found with the sorbicillin secondary metabolite gene cluster in T. reesei (Figure 2) (Druzhinina et al., 2016). Sorbicillinoids are yellow to orange secondary metabolites (Derntl et al., 2016; Salo et al., 2016; Guzman-Chavez et al., 2017), with weak antagonistic effects against bacteria (Duan et al., 2022; Hou et al., 2022) and pharmacological activity (Meng et al., 2016). The respective gene cluster contains two polyketide synthases, an FAD dependent monooxygenase, an MSF transporter and two transcription factors as most important components (Druzhinina et al., 2016; Monroy et al., 2017). The cluster is transcriptionally regulated by light and in response to different carbon sources (Monroy et al., 2017; Stappler et al., 2017) as well as by the two transcription factors in the cluster, YPR1 (Derntl et al., 2016; Derntl et al., 2017) and YPR2 (Hitzenhammer et al., 2019), by the carbon catabolite repressor CRE1 (Monroy et al., 2017) and by LAE1 (Karimi Aghcheh et al., 2013).
Figure 2 Schematic representation of selected secondary metabolite clusters in Trichoderma. (A) The SOR cluster in T. reesei along with characterized flanking genes. Functions and JGI protein IDs (T. reesei v2.0) are listed below the scheme. (B) The genomic locus of tri5 in T. arundinaceum and T. brevicompactum (Gutierrez et al., 2021). (C) The tri cluster in T. arundinaceum (Cardoza et al., 2011). Encoded proteins of the tri cluster and the tri5 genomic locus are given below the scheme.
The genes of this cluster are only in part present in other Trichoderma spp, but closely related to clusters in Penicillium notatum and other Eurotiomycetes as well as a few Sordariomycetes (Maskey et al., 2005; Harned and Volp, 2011) and their phylogeny is not in accordance with the Ascomycota phylogeny, which hints at HGT events from different ancestors. Investigation of the evolution of the SOR cluster revealed several HGT events between Acremonium chrysogenum, Penicillium rubens, Ustilaginoidea virens, Colletotrichum graminicola and Trichoderma, in which Trichoderma spp. appears to only have received SOR2 (Druzhinina et al., 2016). To explain the evolution of the remaining genes of the cluster, selection and gene loss were analyzed, which showed that the cluster arose in early Hypocreales, was complemented by HGT and is under strong purifying selection (Druzhinina et al., 2016). In T. reesei two additional genes with clear relations to the cluster and located close to it in the genome were detected. The gene encoding protein kinase USK1 (unique SOR cluster kinase) is located only 2.3 kb upstream of sor1 and is not syntenic in other fungi, not even in Trichoderma spp. (Beier et al., 2020). USK1 regulates VEL1 which is crucial for coordination of secondary metabolism with development (Bayram and Braus, 2012; Bazafkan et al., 2015) and several genes of the SOR cluster. Accordingly, lack of USK1 decreases production of sorbicillins considerably, but is also required for normal levels of alamethicine and paracelsins (Beier et al., 2020).
In a distance of about 60 kb upstream of the SOR cluster, the G-protein coupled receptor GPR8 (Hinterdobler et al., 2020a) is located, which is largely co-regulated with the SOR cluster under several conditions (Stappler et al., 2017). Although homologues of GPR8 are encoded in other Trichoderma spp. and related fungi, their locus in the genome is not syntenic and the homologous genes are mostly located on different scaffolds or chromosomes (Hinterdobler et al., 2020a). GPR8 is the only member of class VII (secretin like) G-protein coupled receptors in the currently investigated Trichoderma spp. (Gruber et al., 2013; Schmoll et al., 2016). While the ligand of GPR8 is not yet known, transcriptome analysis showed a considerable impact on gene regulation predominantly in darkness, which not only impacts secondary metabolism, but also carbon metabolism (Hinterdobler et al., 2020a). In accordance with its impact on the SOR cluster genes, the regulatory targets of GPR8 overlap also with those of the SOR cluster transcription factor YPR2 (Hitzenhammer et al., 2019) and production of sorbicillinoids, alamethicine, orsellinic acid and paracelsins is strongly decreased (Hinterdobler et al., 2020a). Despite their clear relations to the SOR cluster, the phylogenetic characteristics of both GPR8 and USK1 do not align with the phylogeny of Trichoderma within the ascomycetes, which hints at an involvement of either HGT or selection pressure on these genes during evolution as shown for the SOR cluster genes, which remains to be investigated.
In summary, the question remains, why this cluster was retained and even complemented during evolution in T. reesei, but not in closely related species. Since sorbicillins only have a relatively weak antagonistic function towards other microbes (Derntl et al., 2017), it is unlikely that their major relevance lies in competition and defense. Interestingly, transcriptome analyses revealed that the core SOR cluster genes are among those most strongly transcribed under conditions of sexual development in T. reesei (Dattenböck et al., 2018). Additionally, mutual transcriptional regulation of sor1, sor2 and sor5 establishes a pattern reminiscent of a feedback cycle, which acts positively in light and negatively in darkness (Monroy et al., 2017). Since deletion of gpr8 resulted in altered regulation of several sensing and signaling genes including eight G-protein coupled receptor genes, the function of sorbicillinoids as signaling molecules and of the SOR cluster as a tool for communication warrants further investigations.
A history of gene loss and potential re-acquisition via HGT was also proposed for a gene involved in trichothecene production. Trichothecenes are harmful mycotoxins, which are produced by several genera of the fungal order Hypocreales (Cardoza et al., 2011). The terpene synthase gene tri5 (Malmierca et al., 2013), which catalyzes formation of trichodiene is present in a part of Trichoderma species, but not in all cases with a functional copy (Vicente et al., 2020) (Gutierrez et al., 2021). In contrast to other fungi, the gene in Trichoderma is located outside of the trichothecene biosynthesis cluster (Figure 2) (Lindo et al., 2018; Proctor et al., 2018). This interesting evolution of gain and loss of tri5, sometimes even if the associated trichothecene biosynthesis cluster was not present in the genome, suggests a competitive advantage of trichodiene production, justifying the acquisition of the whole and parts of the cluster (Gutierrez et al., 2021).
A further case for the occurrence of LGT is the group of ceratoplatanins with three members in the core genome of Trichoderma (some Trichoderma species have also 4 ceratoplatanin genes) of which only one is efficiently expressed (Gaderer et al., 2014; Gaderer et al., 2015; Gao et al., 2020). Ceratoplatanins of Trichoderma are involved in the activation of plant defences and induce expression of peroxidase and dioxygenase encoding genes (Salas-Marina et al., 2015), reduce surface hydrophobicity and negatively affect root colonization (Gao et al., 2020). Analysis of evolution of EPL1-like ceratoplatanins in 37 Trichoderma genomes and numerous other genomes revealed their likely origin in Basidiomycota with a distribution to Ascomycota by several ancient events of LGT (also within Ascomycota) and subsequent diversification by birth-and-death evolution. Moreover, all four Trichoderma ceratoplatanins were found to be under purifying selection pressure (Gao et al., 2020).
For the evolution of hydrophobins, which form hydrophobic layers that cover fungal bodies and spores (Bayry et al., 2012; Guzman-Guzman et al., 2017) and broadly affect fungal physiology (Cai et al., 2021) a differential impact on fungal fitness was proposed (Cai et al., 2020). Especially T. harzianum HFB4 was shown to evolve under strong positive selective pressure, while most other hydrophobins were subject to purifying selection. Consequently, evolution of hydrophobins is suggested as an example of fitness tradeoffs during evolution (Kubicek et al., 2008; Cai et al., 2020).
However, the genomes of Trichoderma hold remnants of even more intriguing events, which shaped their physiological characteristics. Trichoderma species are known for their exceptional abilities to feed on a broad range of substrates from plant debris to dead fungi to parasitism on living pathogens and diverse degradation products related to these materials (Druzhinina et al., 2011; Druzhinina et al., 2018). Although at first different species were considered specialists for either degradation of cellulosic plant material or fungal biomass (Kubicek 2011), later studies showed that these metabolic competences are present in all Trichoderma species (Druzhinina and Kubicek, 2013). In fact, Trichoderma shares a last common ancestor with entomoparasitic fungi and the phylogenetic branch leading to the plant associated Nectriaceae diverged earlier in evolution (Druzhinina et al., 2018). Intriguingly, in depth analysis of nine Trichoderma genomes revealed that more than 40% of the plant cell wall degrading genes encoding CAZymes (Carbohydrate Active enZymes) of Trichoderma was gathered from other plant associated filamentous Ascomycota by LGT. The auxiliary protein swollenin (Saloheimo et al., 2002; Brotman et al., 2008) was even likely obtained from green plants by LGT (Druzhinina et al., 2018), which is supported by the finding that Trichoderma spp. are frequently found as members of endophytic fungal communities (Gazis and Chaverri, 2010; Yadav et al., 2022) and generally plant symbionts living in their rhizosphere (Harman et al., 2004a). In contrast to numerous genes encoding enzymes, their major regulators XYR1, ACE2 and ACE3 (Benocci et al., 2017) evolved by vertical gene transfer, but not by LGT (Druzhinina et al., 2018). The results of this study further suggest that Trichoderma spp. originally represented potent mycoparasites and that this very lifestyle allowed them to broaden their metabolic competences by collecting, combining and optimizing novel genes they encountered when feeding on fungal prey. Hence the plant cell wall degradation related CAZome of Trichoderma is distinct from other hypocrealean fungi. In addition to genes extending the nutrient versatility of Trichoderma, 123 further genes were detected, which were putatively obtained by other fungi. They include genes encoding four GPCRs of the PTH11 type, two of which were tested for a relevance in cellulase regulation previously (Stappler et al., 2017), the small unique protein OOC1 (Schmoll and Kubicek, 2005; Pang et al., 2021), the dehydrogenase GRD1, which impacts cellulase regulation in a light dependent manner (Schuster et al., 2011), the high affinity nitrate transporter NIT10, CIP1, which was identified as one of the VIP genes limiting hydrolysis performance of cellulase mixtures (Lehmann et al., 2016) and CON-13, which is putatively involved in asexual reproduction (Hager and Yanofsky, 1990).
With respect to CAZymes and polysaccharide degradation, the description of a novel glucuronan lyase system in T. parareesei (Druzhinina et al., 2010) adds a further highlight to the versatility of the genus (Pilgaard et al., 2022). Detailed analyses of activity and degradation products confirmed that this degradation system enables complete degradation of glucoronan, which is found in fungi like Agaricus bisporus – a possible prey of the mycoparasite T. parareesei, to easily absorbable dimers and monomers (Pilgaard et al., 2022).
A further interesting aspect of evolution was reported for the processivity of cellulases with T. reesei CEL6A and CEL7a as examples. While in fungi, family 7 glycoside hydrolases are highly processive, the same characteristic is adopted by bacteria for family 6 cellulases. In both cases, the presence of highly processive and less processive cellulases avoids “traffic jams” on the cellulose fibers, which would decrease efficiency. Consequently, the high relevance of efficient cellulose degradation for competitive success in nature obviously led to convergent evolution of cellulases in which the structural features of cellulases was shaped to optimize processivity and interplay of the cellulose degrading machinery in bacteria and fungi (Uchiyama et al., 2020).
Recombination and mating as tools of evolution - repeat induced point mutation and segmental aneuploidy in Trichoderma
Sexual development is crucial for propagation and evolution of a species (Bennett and Turgeon, 2016; Wallen and Perlin, 2018), especially if environmental conditions deteriorate (Debuchy et al., 2010; McDonald et al., 2016). In Trichoderma, numerous strains of diverse species were isolated from fruiting bodies (Jaklitsch and Voglmayr, 2015) – indicating that mating happens in nature – but only for T. reesei induction of sexual development was achieved under laboratory conditions (Seidl et al., 2009; Hinterdobler et al., 2020b). Although this novel tool for strain improvement opens up intriguing perspectives for industry, in that strains can be bred for enhanced performance, also drawbacks became obvious. Besides the female sterility of QM6a, which is due to a defect in the scaffolding protein HAM5 (Seidl et al., 2009; Linke et al., 2015; Tisch et al., 2017), the process of repeat induced point mutation may hamper optimization of genetically already modified production strains.
RIP was first discovered in Neurospora (Selker et al., 1987) as a mechanism for protection of the genome from transposable elements and mobile DNA (Gladyshev, 2017) and is operative in most Ascomycota (van Wyk et al., 2020). In case of a functional RIP mechanism, repetitive DNA sequences >400 bp cause C-5 cytosine methylation and deamination by the methyltransferases rid1 and dim2 prior to meiosis (Kouzminova and Selker, 2001; Li et al., 2018). Nevertheless, RIP was also observed for smaller duplicated regions (Gladyshev and Kleckner, 2014).
Genomes of organisms in which RIP is operative show a low number of repetitive sequences or transposon remnants (Gladyshev, 2017) as for example N. crassa (Krumlauf and Marzluf, 1980; Borkovich et al., 2004), which is also true for the genome of T. reesei (Martinez et al., 2008) although RIP was not unequivocally detected at first (Schuster et al., 2012). Nevertheless, operation of RIP is different in T. reesei compared to N. crassa (Li et al., 2018) with different dinucleotide preference and requirement of the methyltransferase genes rid1 and dim2 for normal sexual development in T. reesei but not N. crassa. Interestingly, T. reesei QM6a was found to have comparatively few transposon sequences, which are mostly located in AT-rich regions (Li et al., 2017).
Consequently, this mechanism is of utmost importance for evolution, especially considering the history of lateral gene transfer as well as gain and loss of genes as seen in Trichoderma. Additionally, evolution of the genome by gene duplication would be counteracted in a sexually propagating population of Trichoderma.
Besides RIP, another phenomenon decreased the enthusiasm after the achievement of sexual development under laboratory conditions. Already in the first crosses of QM6a with the fertile CBS999.97 (MAT1-1) strain, the diverse phenotypes of the progeny were obvious (M. Schmoll, unpublished). Detailed analysis then showed that of the 16 ascospores generated by meiosis and two rounds of postmeiotic mitosis four to eight were inviable and segmentally aneuploid ascospores were found (Chuang et al., 2015; Schmoll and Wang, 2016). The segmental duplication and deletion in the respective genome area caused loss of the polyketide synthase pks4, which is responsible for spore coloration (Atanasova et al., 2013) and loss of a xylanase and hence lignocellulosic biomass degradation efficiency increased in these progeny (Chuang et al., 2015). However, this process is not easily predictable in crosses between other nature isolates of T. reesei or among production strains.
High quality genome sequences of important plant symbionts
Besides strains of T. reesei, also strains of prototypical biocontrol agents were recently re-sequenced to get high quality genome sequences for reliable analysis of genome synteny and evolutionary events (Li et al., 2021b). This analysis revealed that the telomeric sequences are conserved between T. reesei CBS999.97, T. reesei QM6a, T. virens Gv29-8, T. virens FT-333, T. atroviride P1 and T. asperellum FT-101. Third generation sequencing now also enabled analysis of AT-rich blocks, which can hardly be aligned based on the short NGS-sequence reads. Interestingly, the T. reesei strains have a lower percentage of AT-rich blocks than the four other strains (7-8% versus 11-13%), which in part explains their larger genome sizes (Li et al., 2021b). Generally, the genome contents of the four Trichoderma species are highly divergent, with 2000 – 3000 species specific genes each and obviously all seven tested high quality Trichoderma genomes underwent extensive transposon invasions followed by RIP mutations. Additionally, evidence for considerable chromosome shuffling in Trichoderma was found (Li et al., 2021b). In addition to the previously reported 7000 core Trichoderma genes (Druzhinina et al., 2018), the four biocontrol agents possess 800 more conserved genes (Li et al., 2021b). An important divergence in gene content was observed for transcription factor genes, with variations in the subfamilies of fungal zinc-binuclear cluster domains and fungal specific transcription factor domains, while the gene numbers of the other transcription factor families were identical among the seven genomes. Moreover, the investigated high quality genome sequences support the hypothesis that CAZyme genes form physically linked CAZyme gene clusters in polysaccharide utilization loci. Besides several species specific clusters, T. reesei and T. virens were found to share 14 common CAZyme gene clusters whereas T. atroviride and T. asperellum share 18 (Li et al., 2021b). As for secondary metabolite gene clusters, T. reesei and T. virens share 27 common clusters and T. atroviride and T. asperellum have 37 clusters in common (Li et al., 2021b).
The frequent presence of genes associated with secondary metabolism in such clusters was already noted with the first genome analysis of T. reesei and also specific secondary metabolite clusters were detected (Martinez et al., 2008). Investigation of the distribution and evolution of Cytochrome P450 genes in several Trichoderma spp. now allowed for additional insights into clustering of secondary metabolite genes complemented by Cytochrome P450s. In the Trichoderma Cypome (the genome content of Cytochrome P450 encoding genes), 12 specific families of this functional group were detected (Chadha et al., 2018).
Intriguing news about well-known model Trichoderma species
In recent years, improvement of the quality of genome sequencing was subject to intensive research. The genome of T. reesei QM6a was subjected to proximity ligation scaffolding (Jourdier et al., 2017) using the GRAAL software tool (Marie-Nelly et al., 2014), which already yielded an improvement compared to the initial assembly (Martinez et al., 2008). Approximately at the same time, long read sequencing using the PacBio or Nanopore (ONT; flowcell 10.4) technologies (supported by Illumina short read sequencing to enhance quality) was increasingly used to improve the quality of genome sequences and so called “gold-standard” genomes became available for many organisms. For T. reesei QM6a (Li et al., 2017), this high quality sequence revealed numerous sequencing errors, indels and inversions in older genome assemblies (Martinez et al., 2008; Marie-Nelly et al., 2014; Jourdier et al., 2017) and yielded more than 1000 previously undetected genes. Telomer-to-telomer sequences became available for all seven chromosomes along with the sequence of the mitochondrial genome which is identical with that published previously (Chambergo et al., 2002), confirming the high quality of the sequence. Additionally, a comparison to the RutC30 genome indicates that the number of translocation in this strain is lower than reported earlier (Seidl et al., 2008; Peterson and Nevalainen, 2012). The long reads obtained with third generation sequencing even allowed for correct sequencing of the highly AT-rich centromere region, in which several genes were detected (Li et al., 2017).
Since the achievement of sexual development under laboratory conditions, this process is subject to detailed investigation for important determinants as well as its consequences in the Trichoderma genomes (Schmoll, 2013; Hinterdobler et al., 2020b). In the course of investigation of recombination during meiosis, high quality genome sequences of CBS999.97(MAT1-1) and CBS999.97 (MAT1-2) (Lieckfeldt et al., 2000; Seidl et al., 2009) were prepared by third generation sequencing (Li et al., 2021a). Like QM6a, they also have seven chromosomes. As RIP (Gladyshev, 2017) is operative in Trichoderma, the CBS999.97 genomes only contain 62 transposable elements (Li et al., 2021a). Phenotypes of QM6a and CBS999.97 strains show considerable differences and their progeny are unexpectedly diverse (Seidl et al., 2009; Li et al., 2017). Accordingly, a very high number of SNPs and indels (ca. 1 Mio) were detected between QM6a and the CBS999.97 strain, but only around 2700 between CBS999.97(MAT1-1) and CBS999.97(MAT1-2). However, during analysis, problems with “difficult to align” sequences were observed and therefore the specifically developed software tool TSETA (TGS to Enable Tetrad Analysis; (Liu et al., 2021)) was used, which then identified around 6 fold more alterations between QM6a and CBS999.97 with sizes of up to 61 kb (Li et al., 2021a). Interestingly, for QM6a more than 3000 nucleotides were subject to RIP, while in CBS999.97 (MAT1-1) this was the case for only 92 nucleotides (Li et al., 2021a).
Although QM6a represents a nature isolate, it still carries an important mutation in its genome: sexual development was hampered by female fertility (Seidl et al., 2009). Ten rounds of backcrossing of QM6a with the fertile CBS999.97 allowed for narrowing down the genomic locus which contains the mutation (Schmoll et al., 2013). Subsequently it could be confirmed that the MAPKinase scaffold protein HAM5, which is encoded by a gene in this very locus, shows several mutations causing frame shifts and lead to an unfunctional gene, which renders QM6a female sterile (Linke et al., 2015; Tisch et al., 2017). This knowledge now enables a complementation of the defect to facilitate strain improvement by breeding. However, QM6a and recombinant strains derived from it can now also be used as a female sterile test strains to evaluate male and female fertility and knowledge-based crossings to remove the HAM5 defect serve to investigate mating in homozygous crosses (Hinterdobler et al., 2020b).
Delving into the past – elucidation of the genetic basis of early and later random mutants
Since the advent of genome sequencing, the quest to elucidate the characteristics of high performance mutant strains of Trichoderma continues (Seidl and Seiboth, 2010). The number of mutations in these strains was greatly underestimated prior to knowledge on the genomes and hence, although many crucial functions of the mutated genes are already known (Seidl and Seiboth, 2010; Peterson and Nevalainen, 2012; Bischof et al., 2016), there are still numerous genes left, which are altered, but without deeper knowledge on their function or contribution to the phenotype of the respective mutant. Nevertheless, there was quite some progress in recent years.
One of the most important topics upon investigation of random mutants is gaining insight into the mechanism of cellulase regulation, both because of their relevance as homologously produced enzymes in Trichoderma and due to the efficiency of their promotors for heterologous protein production (Gudynaite-Savitch and White, 2016; Paloheimo et al., 2016; Arbige et al., 2019).
The random mutant strain QM9978 produces cellulases at a very low basic level, but cannot be induced under usual cellulase inducing conditions (Torigoi et al., 1996) and hence was used for comparative studies for decades (for example (Zeilinger et al., 2003; Schmoll et al., 2004)). Finally, in 2017, the defect of QM9978 was identified due to genome sequencing (Ivanova et al., 2017). Comparison with QM6a revealed 43 mutations, among them a translocation between chromosomes V and VII upstream of the transcription factor gene vib1, which caused the lack of cellulase induction in QM9978 (Ivanova et al., 2017). The homologue of VIB1 was previously reported as a link between glucose signaling and carbon catabolite repression and to be involved in regulation of plant cell wall degrading enzymes in N. crassa (Xiong et al., 2014). While overexpression of VIB1 did not increase cellulase gene expression in T. reesei (Ivanova et al., 2017), the same strategy led to significantly increased secreted cellulase activity in T. orientalis EU7-22 (Han et al., 2020).
The early random mutant T. reesei RUT C30 (Peterson and Nevalainen, 2012) is the most extensively studied mutant strain, as it serves as parental strain for many industrial production strains. Moreover, the considerable genomic rearrangements in this strain (Seidl et al., 2008) provided numerous hypothesis for strain improvement to be tested. Although quite some details on the basis for enhanced cellulase production of this strain are already known (Peterson and Nevalainen, 2012), it is still subject to research with the most recent finding, that the truncation of CRE1 present in RutC30 turns the repressor into an activator (Rassinger et al., 2018). Re-assembly and re-annotation of its genome showed diverse chromosomal rearrangements (Jourdier et al., 2017). The industrial application of derivatives of RUT C30 is also the reason for this strain to be used as parental strain for gene regulation studies. However, in this case, it has to be kept in mind that the numerous mutations of RUT C30, which also cause a considerably altered physiology including decreased growth and sporulation as well as weakened cell wall stability, do not allow for a reliable generalization of functional characteristics of individual genes to Trichoderma as a whole.
A less well known cellulase negative strain is T. reesei QM9136, which cannot grow on cellulose or form cellulases, but otherwise has a normal phenotype (Mandels et al., 1971; Nevalainen and Palva, 1978). In this strain, a frameshift mutation in the crucial cellulase transcription factor XYR1, which causes a truncation by 140 amino acids is responsible for the defect in cellulase production (Lichius et al., 2015). Additionally, 14 mutations have been detected which are likely irrelevant for cellulase production.
A descendant of the cellulase overproducer QM9414, which was subjected to several more mutagenic rounds is T. reesei PC-3-7 (Kawamori et al., 1985). Sequencing of this strain and comparison with the QM6a genome sequence yielded 260 SNPs in PC-3-7, of which most were located in non-coding regions. However, also in the important cellulase regulator genes ace1 and cre1 SNPs were detected. The SNP in CRE1 at amino acid 78 indeed caused a decrease in binding affinity of CRE1 to the cbh1 promotor, which in turn negatively affected cellulase production (Porciuncula Jde et al., 2013).
Bioprospecting in genomes of nature isolates
Investigation of the genome of novel biocontrol agents or biofertilizers is becoming increasingly interesting. On the one hand, these organisms are being distributed in nature in considerably larger amounts than would occur naturally. On the other hand, they can be sources for novel bioactive molecules (Karwehl and Stadler, 2016). In both cases the metabolic competences of a microbe can be a benefit or a threat to humanity. Especially the widespread application of biocontrol agents in the form of spores in low income countries raised questions as to the safety for farm workers there, but also the burden on consumers as well as nature in general (Konstantinovas et al., 2017; Hatvani et al., 2019). Clinically relevant Trichoderma strains are assumed to be limited to certain species (Kredics et al., 2003; Sandoval-Denis et al., 2014), but detailed knowledge on relevant secondary metabolite gene clusters is important to rule out selection of a potential harmful isolate for commercial application in agriculture. However, fungi including Trichoderma represent a potential source for valuable compounds to fight cancer or the counteract microbial resistance against common antibiotics (Viglas et al., 2021).
In all these cases detailed knowledge on the gene content of a given strain is crucial for knowledge based decisions on application (for an overview on recent genome level analyses of Trichoderma spp. aimed at biocontrol issues see Table 1). Additionally, a higher number of available genome sequences will enhance specificity of developed methods to detect biocontrol agents in agriculture (Kabani et al., 2002; Kredics et al., 2018), especially if identification of individual strains of a species is required – for example for following up distribution and habitat colonization of a biocontrol agent in the field. Unfortunately, genome level studies on health issues due to distribution of Trichoderma as biocontrol agents or of clinical isolates are still very rare.
Besides biocontrol and health related targets, also novel enzymes still remain an important commercial topic for which Trichoderma spp. are valuable (for an overview on recent genome level analyses of Trichoderma spp. aimed at bioprospecting see Table 2). Consequently, bioprospecting for novel, more efficient enzymes under diverse conditions – as required for various applications in industry – are still actively sought (enzymes reviews (Wilson, 2009; Singh et al., 2015; Arnau et al., 2020)).
Table 2 Recent genome level analyses of Trichoderma spp. targeted at bioprospecting of enzymes and bioactive compounds.
Importantly, numerous additional newly sequenced genomes of the genus Trichoderma became available in JGI mycocosm (https://mycocosm.jgi.doe.gov/mycocosm/home) recently, which are not specifically described here.
Mitochondrial genomes of Trichoderma
Mitochondria represent the powerhouse of every eukaryotic cell and are responsible for the production of ATP through the oxidative phosphorylation pathway and the aerobic citric acid (TCA) cycle and biosynthesis of metabolites like amino acids (Medina et al., 2020). Moreover, besides respiratory metabolism and energy production, mitochondria are also relevant for senescence during the cell cycle and maintenance of ion homeostasis (Basse, 2010; Chatre and Ricchetti, 2013). The mitogenome was shown to play a role in fungal virulence and emerged as an important target of fungicides (Medina et al., 2020).
In contrast to other cell organelles, mitochondria have their own genome, which can be present in multiple copies, also depending on growth conditions and is capable of independent replication and inheritance (Burger et al., 2003). Mitochondrial genomes have their own codon usage (Medina et al., 2020) and are highly variable, also with the extent of introns and their different sizes (Burger et al., 2003; Aguileta et al., 2014). Both high conservation of intronic sequences and their location within genes and species-specific introns were detected in Hypocreales, hence indicating an origin from a common ancestor as well as an alternative mechanism for intron evolution/transfer (Fonseca et al., 2020). Comparative analyses further showed a correlation between mitogenome length and the number and size of non-coding sequences in the mitochondrial genome of Hypocreales (Fonseca et al., 2020). Detailed sequence analysis revealed several cases of HGT of bacterial genera to fungi (Megarioti and Kouvelis, 2020). The different distribution of heavier and lighter nucleotides in the two strands of the mitochondrial genome enables their isolation by differential centrifugation (Garber and Yoder, 1983; Chambergo et al., 2002). The introns can contain so called homing endonucleases responsible for intron splicing (Megarioti and Kouvelis, 2020), of which the classes of LAGLIDADG and GIY-YIG are present also in fungal mtDNAs (Belfort et al., 2002; Stoddard, 2014). Such a gene conversion can happen through intron invasion and leads to altered size and composition of the mitochondrial genome (Wu and Hao, 2019; Medina et al., 2020). In fungi, mitochondrial genomes can be circular or linear with sizes from little more than 10 kb up to 200 kb (Pramateftaki et al., 2006; Losada et al., 2014; Liu et al., 2020), although the typical mitochondrial genome is much smaller. As for evolution of mitochondrial genomes, they were found to evolve more slowly than their nuclear genomes (Clark-Walker, 1991; Gaillardin et al., 2012), despite the high variability of non-coding regions.
In some cases, transfer of mitochondrial genes from the mitochondrial genome to the nuclear genome was observed and concerns genes like nad5, while for other genes no transfer events were detected (Fonseca et al., 2020; Medina et al., 2020). Over evolutionary time, a considerable number of the initially endosymbiotic genes were transferred to the nuclear genome and mitochondrial proteins are encoded by the nuclear genome and transported to the mitochondria (Burger et al., 2003). In the order Hypocreales, also duplication of mitochondrial genes is a common phenomenon as among the 17 core genes, only atp8, atp9 and cox3 were not detected in the respective nuclear genome (Fonseca et al., 2020).
The interest on research towards mitochondrial genomes remained very low with until recently just very few mitochondrial genomes published for the genus Trichoderma (Kwak, 2020; Kwak, 2021). In the last few years, with the advent of efficient and affordable genome sequencing methods, especially third generation sequencing, the numbers increased. Mitochondrial genomes in the Hypocreales are circular and range from about 24 kb to more than 100 kb in size (Fonseca et al., 2020). Thereby, the size of introns contributes to the variance of genome sizes (Medina et al., 2020).
Already in the 1990s, the relevance of mitochondrial functions for physiology of Trichoderma was investigated, which revealed a significant impact on cellulase production, in that a low oxygen tension negatively influenced their biosynthesis (Abrahao-Neto et al., 1995). This sensitivity of T. reesei to the functional state of mitochondria was subsequently associated with the 5’ region of the major cellulase gene, cbh1 (Carraro et al., 1998). A few years thereafter, the mitochondrial genome of T. reesei (Figure 3) was published (Chambergo et al., 2002). In T. reesei a role in oxidative stress resistance was proposed for mitochondria as well (Wang et al., 2015a). Recently, third generation sequencing yielded an update of mitochondrial genomes in T. reesei. The female sterile MAT1-2 strain QM6a (Li et al., 2017) was sequenced along with both mating types of T. reesei CBS999.97 (Seidl et al., 2009), which showed strikingly different sizes of the mitochondrial genomes yet a similar set of genes essential for mitochondrial functions (Li et al., 2021a). The mitochondrial genome of QM6a has 42 kb, while that of CBS999.97 (MAT1-1) has only 39 kb and only shares 75% of nucleotide sequence identity with that of QM6a. In contrast, the mitochondrial genomes of CBS999.97 (MAT1-1) and CBS999.97 (MAT1-2), which arose from the same crossing event (Seidl et al., 2009), are identical except for only six SNPs, hence reflecting maternal inheritance (Li et al., 2021a). Moreover, the mitochondrial genome sequences of T. atroviride P1, T. asperellum FT-101, T. virens Gv29-8 and Tvirens FT-333 vary considerably in size and analysis of high quality genomes of these strains indicates that mobile genetic elements played key roles in shaping the mitochondrial genomes in Trichoderma (Li et al., 2021b). Also three nuclear encoded mitochondrial sequences (NUMTs) were detected in Trichoderma, which are all located within an AT-rich block, which suggests that filamentous fungi and mammalian cells may have an evolutionarily conserved origin of NUMTs (Tsuji et al., 2012; Li et al., 2021b).
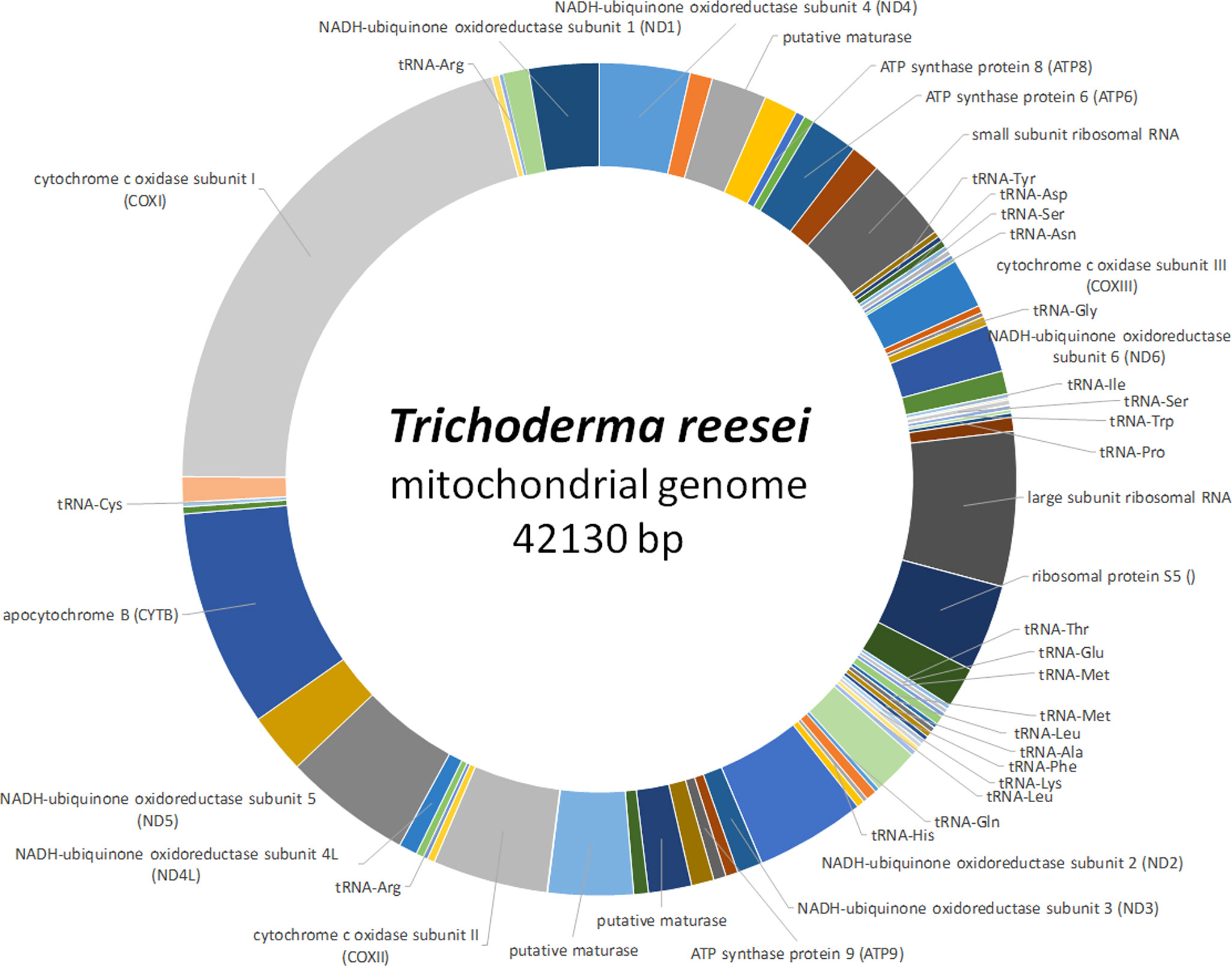
Figure 3 Schematic representation of the mitochondrial genome of T. reesei. The complete mitochondrial genome of T. reesei (GenBank accession number NC_003388) is shown along with encoded genes (Chambergo et al., 2002). Areas without label represent intergenic regions.
The mitochondrial genome of T. harzianum HB324 is circular and has a size of 32kb, with a GC content of 28% (Fonseca et al., 2020), which is in accordance with a generally high AT content in fungal mitogenomes (Medina et al., 2020). Fourteen genes associated with oxidative phosphorylation, 28 tRNA encoding genes as well as two ribosomal RNAs were detected in addition to a few hypothetical genes (Fonseca et al., 2020). As in many ascomycetes (Medina et al., 2020) all the genes were encoded on the same DNA strand in T. harzianum HB324. A comparison with other Trichoderma mitochondrial genomes showed a considerable variation in their structure and size even within the genus (Fonseca et al., 2020).
Another T. harzianum strain, CBS226.95, has a considerably smaller circular mitochondrial genome with only around 28 kb (Kwak, 2021). This mitogenome was further predicted to have evolved earlier than other Trichoderma species’ mitogenomes. It was further proposed that the evolution of Trichoderma mitochondria is influenced by their adaptive diversification depending on the diverse habitats from which Trichoderma strains were isolated concerning oxygen availability like soil, wood or living plants and fungi (Kwak, 2021).
Trichoderma atroviride ATCC26799 has a mitochondrial genome of 33 kb comprising the usual content of mitochondrial genes in the genus (Kwak, 2020). Gene order in the mitochondrial genomes of five Trichoderma spp. (T. reesei QM9414 (GenBank accession number AF447590), T. asperellum B05 (NC_037075), T. hamatum (MF287973), and T. gamsii KUC1747 (KU687109) is highly conserved, while intergenic regions, nucleotide composition bias, number of protein coding sequences and size of mitochondrial genomes was more variable (Kwak, 2020). Recently, also a mitochondrial genome of T. simonsii was reported (Chung et al., 2022), which clusters with several other Trichoderma mitogenomes including T. cornu-damae (Genbank accession number MW525445), T. lixii (NC_052832) and T. hamatum (NC_036144), most of which are not yet described in detail.
Mycoviruses of Trichoderma
Viruses are able to infect living organisms from bacteria to humans and are present in fungi as well (Myers and James, 2022). Viruses of fungi have been known for more than 5 decades (Ghabrial et al., 2015), but the detection of them in Trichoderma spp. happened quite recently. In most cases, infection of a fungus with a virus does not change the phenotype (Ghabrial and Suzuki, 2009; Son et al., 2015), which is a likely reason that viruses in Trichoderma did not receive much attention so far. Nevertheless, there are some intriguing examples of mycoviruses considerably altering physiology and organismic interactions: If Sclerotinia sclerotiorum is infected with the small DNA mycovirus SsHADV-1, it turns from a pathogen of Brassica spp. into a beneficial endophyte due to downregulation of major pathogenicity factor genes (Zhang et al., 2020). Even more fascinating is the three-way symbiosis of the endophytic fungus Curvularia protuberata, which allows its host plant Dichanthelium lanuginosum to grow at high temperatures only if it is infected by the mycovirus CThTV (Marquez et al., 2007). Although it was initially thought that mycoviruses have a relatively narrow host range, detection of a virus first described in Botrytis porri in B. squalosa and Sclerotinia sclerotiorum suggests that this is not the case and that mycoviruses can be transmitted between species (Wu et al., 2012; Nerva and Chitarra, 2021). Intriguingly, also mycoviruses from endophytes were found to replicate in a plant, although this phenomenon was so far only shown in vitro, not in nature (Nerva et al., 2017).
The model fungus for studying the interaction of mycoviruses with their hosts is Cryphonectria parasitica, the chestnut blight pathogen, in which infection by a so called hypovirus decreases virulence (Dawe and Nuss, 2013; Eusebio-Cope et al., 2015). Similar effects were shown in other plant pathogens, hence making mycoviruses an interesting subject to research towards biocontrol applications (Milgroom and Cortesi, 2004). In this respect, treatment of chestnut blight with mycoviruses exemplified the problem with such an application. No natural vectors are known for mycoviruses (Ghabrial and Suzuki, 2009; Son et al., 2015). They spread predominantly vertically by propagation via conidiospores, less efficiently via ascospores in ascomycetes (Ghabrial et al., 2015). Alternatively, infection occurs by hyphal anastomosis, which is limited by vegetative incompatibility, leading to programmed cell death if two incompatible fungi fuse (Daskalov et al., 2017). In case of multiple incompatibility groups in fungal population, spread of the viruses is very inefficient as is biocontrol in such a case (Xie and Jiang, 2014). The limited success of treatment of C. parasitica in the US compared to the imported strains in Europe is attributed to this problem (Myers and James, 2022). In Trichoderma, vegetative incompatibility was shown previously (Gomez et al., 1997), but is not yet sufficiently investigated to draw any conclusions as to the impact on biocontrol enhancements by mycovirus applications.
Another natural defense mechanism that limits infection of pathogens by mycoviruses is RNAi, meant to destroy intruding foreign nucleic acids (Segers et al., 2007). Interestingly, although mycoviruses often only have two genes encoded, they can counteract programmed cell death and vegetative incompatibility as well as RNAi (Hammond et al., 2008; Daskalov et al., 2017), which tips the balance again towards their benefit. Often, the presence of a mycovirus in a fungus causes lower growth rates which is interpreted as a lack of fitness (Ghabrial and Suzuki, 2009). Nevertheless, also co-evolution of mycoviruses with their hosts has been observed, although this observation cannot be generalized (Nerva and Chitarra, 2021).
In Trichoderma, the first mycovirus was described in 2009 (Jom-in and Akarapisan, 2009) and only a few followed thereafter. However, a study of more than 300 fungal isolates causing green mold disease revealed evidence for potential dsRNA mycoviruses of diverse groups in 32 isolates, indicating that mycoviruses are not uncommon in Trichoderma (Yun et al., 2016). The genomes of these mycoviruses are relatively small and they mostly encode only two proteins, a coat protein and an RNA dependent RNA polymerase (RdRp) (Figure 4). As reported from other fungi, mycoviruses often do not influence the phenotypes of their hosts or just have a minor impact on pyhsiology, which was also observed for several isolates from Trichoderma (Table 3).
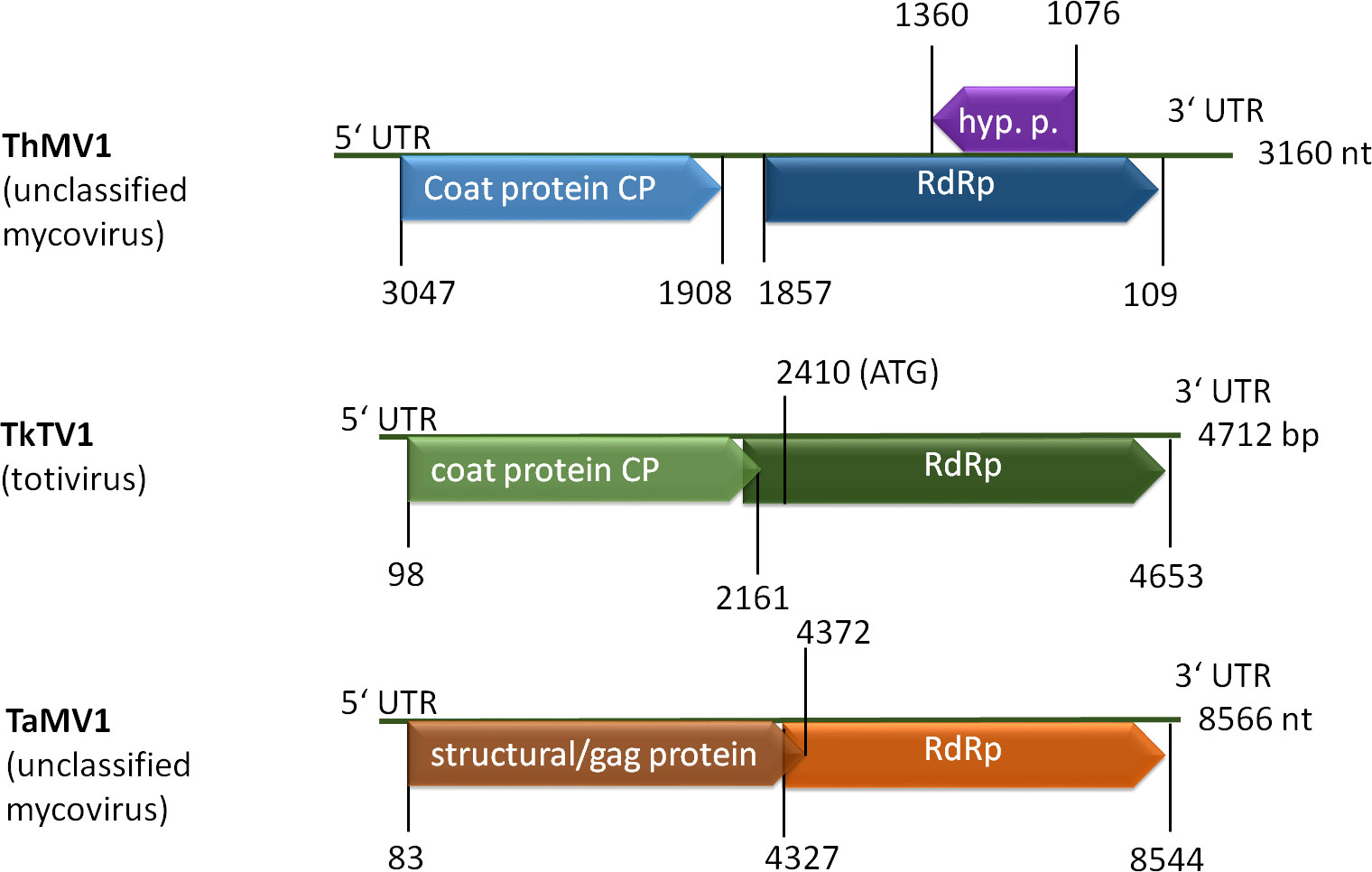
Figure 4 Schematic representation of selected mycovirus genomes. ThMV1 (Liu et al., 2019), TkTV1 (Khalifa and MacDiarmid, 2019) and TaMV1 (Lee et al., 2017) genomes comprise two ORFs coding for a coat protein (CP or GAG) and an RNA dependent RNA polymerase (RdRp). ThMV1 additionally comprises a small ORF encoding a hypothetical protein of unknown function.
In several cases however, presence of the mycovirus in the Trichoderma strain did cause phenotypic alterations, mostly enhanced mycoparasitic abilities:
A mycovirus in T. harzianum (ThMV1) was investigated in more detail and caused a decrease in biomass production, a slightly positive effect on plant health if T. harzianum was applied alone and somewhat better biocontrol activity against Fusarium oxysporum f. sp. cucumerinum in cucumber (Liu et al., 2019). The T. atroviride mycovirus TaMV1, a member of the proposed family Fusagraviridae does not impact conidiation or growth, but supported enhanced antifungal activity against the plant pathogen Rhizoctonia solani (Chun et al., 2020). For the T. harzianum mycovirus ThHV1, which shares similarity with betahypoviruses, an influence on mycoparasitism was shown in case of the presence of a defective genome. This mycovirus could be transmitted vertically to conidia, but could also infect another T. harzianum isolate as well as T. koningiopsis (You et al., 2019). The T. harzianum partitivirus 1 (ThPV1) enhances growth inhibition of co-cultured plant pathogens as well as glucanase activity (Chun et al., 2018b).
The still few reports on mycoviruses in Trichoderma indicate that investigation of mycoviruses and their interaction with the host as well as their influence on biocontrol of plant pathogens are emerging as an important new topic of research. Since it was shown that mycoviruses can have a broad host spectrum, transmission of a mycovirus from a pathogen to a mycoparasite like Trichoderma or vice versa is likely to be possible, yet the consequences for plant health, population structure and ecology are currently completely unknown. Additionally, the effects of mycovirus infections in Trichoderma are hardly predictable and loss of a mycovirus can easily happen during industrial cultivation, potentially leading to altered characteristics of the fungus. Consequently, it will be crucial to be aware of a potential mycovirus in a fungus if it is part of a commercial product in order to guarantee a stable, efficient product and knowledge on susceptibility of the fungus for mycovisuses abundant in the target pathogen will be beneficial.
In this respect, the increasing performance of next generation sequencing of transcriptomes (revealing RNA viruses) comes in handy, as a previous screening of available GenBank data shows (Gilbert et al., 2019). Detection of mycoviruses as a by-product of gene regulation studies is a worthwhile effort to gain important information on model organisms as well as production strains and biocontrol agents.
Genomic aspects of mycoremediation with Trichoderma
As the awareness of the community towards a healthy environment increases, contaminated sites, which could previously hardly be treated efficiently, comes into focus (Febbraio, 2017; Ford et al., 2022a; Sharma et al., 2022). The genomes of Trichoderma spp., originating from diverse habitats represent a treasure trove for the quest for enzymes and metabolic competences to detoxify or even mineralize dangerous chemicals (Tripathi et al., 2013; Zafra and Cortes-Espinosa, 2015; Repas et al., 2017; Mishra et al., 2021) and Trichoderma species are well known for their efficiency in remediation of soil and water pollution (Harman et al., 2004b). Fungi are generally very potent organisms for bioremediation (Kour et al., 2021), even outperforming bacteria (Dell'Anno et al., 2022). The decades-long application of Trichoderma spp. in biocontrol and in industry led to an in depth knowledge of their environmental safety (Nevalainen et al., 1994; Frisvad et al., 2018; Shenouda and Cox, 2021), which makes fungi of this genus prime candidates for application at natural contaminated sites with limited negative effects on the surrounding flora and mostly without the need for genetic modification. The chemical composition of pollutants is diverse, representing a considerable challenge for strain selection. In case of plastic degradation however, suitable enzyme classes for degradation of the respective structures can be defined beforehand (Verschoor et al., 2022).
Genome mining and omics analyses allow for delineating regulation pathways of suitable target enzymes, which is important for performance of a given strain, as not only the presence of degradative enzymes, but also their appropriate regulation under commercially viable conditions is crucial for applicability. This strategy is not yet routinely applied in Trichoderma, but in recent years, several interesting examples of detailed investigation of genomes, enzymes targeting certain pollutants and delineation of degradation pathways were reported.
Trichoderma lixii MUT3171 was isolated from a highly polluted environment and comparison of its genome was used to gain insight into potential degradation pathways of polycyclic aromatic hydrocarbons (PAHs) (Venice et al., 2020). Orthologues of genes encoding oxidoreductases, CAZymes and proteins responsible for biosurfactant biosynthesis were screened in 14 Trichoderma species. Additionally, also unique genes of MUT3171 including a quinoprotein alcohol dehydrogenase and a specialized mechanism of DNA repair were determined which may contribute to the ability of T. lixii to survive in this habitat (Venice et al., 2020). The dichlorodiphenyltrichloroethane (DDT) resistant T. hamatum strain FBL587 was investigated for its transcriptomic reaction to the pollutant (Davolos et al., 2021). This strain could degrade DDT and enhance degradation by Cucurbita phytoremediation. Especially Cytochrome P450 enzymes encoding genes were upregulated in the presence of DDT, but also transforming enzymes such as epoxide hydrolases, FAD-dependent monooxygenases, glycosyl and glutathione transferases as well as transporters (Davolos et al., 2021). The example of this detailed analysis shows how crucial genes for xenobiotic remediation can be narrowed down, which can represent an important step for strain screening and selection for future bioremediation purposes by fungal cultures or their isolated enzymes.
Currently, approaches to realize circular economy by valorization are in development – for example with Trichoderma spp. degrading waste biomass for production of metabolites (Shenouda and Cox, 2021; Shenouda et al., 2022), which should be extended to degradation of more problematic waste materials (Verschoor et al., 2022) like plastics or composites. In this respect the ability of adaptation of fungal strains to the substrate by crossing (Ashton and Dyer, 2016) can be a valuable tool for strain improvement towards plastics degradation and detoxification of pollutants. So far, only for T. reesei crossing was achieved under laboratory conditions (Seidl et al., 2009) and the method has successfully been applied to increase cellulase production by at least 10fold compared to RutC30 (M. Schmoll and S. Basyouni-Khamis, unpublished results). The availability of European nature isolates of T. reesei, which are sexually fertile (Hinterdobler et al., 2021a) is an important prerequisite to test a crossing approach for using plastics and/or their degradation products as carbon source to produce higher value chemicals and enzymes. Such a scheme of circular economy could serve as a blueprint for avoiding pollution and creating high value products with minimal pollution in the future, which makes further genome mining and efforts to achieve crossing with other Trichoderma spp. as well a worthwhile effort.
Tools for genome screening and manipulation
Fungal genomes are constantly subject to manipulation – mostly of course in nature in order to achieve optimal adaptation to the habitat or just to enable survival if environmental conditions deteriorate. Sexual development represents the major strategy of nature to modify the genome, but also a valuable tool for strain improvement in the lab (see above). However, also artificial methods for genome manipulation of fungi were developed further in recent years.
Functional analysis of genes and whole pathways are especially important to understand the physiology of Trichoderma and consequently, strategies for increasing the ease and efficiency of genome manipulation are constantly optimized (Guangtao et al., 2009; Schuster et al., 2012; Chum et al., 2017). A more recent addition is the TrichoGate cloning strategy which is mainly an adaptation of the Golden Gate cloning system to Trichoderma (Nogueira-Lopez et al., 2019). Using this system, vectors for different promotors, deletions, protein localization studies and overexpressions are introduced along with a vector for Agrobacterium mediated transformation.
Also the SES (synthetic expression system), which was previously established for Saccharomyces cerevisiae (Rantasalo et al., 2016; Rantasalo et al., 2019) represents an interesting addition to the toolset for genome manipulation in Trichoderma, which is focused on protein expression (Rantasalo et al., 2018). In this system induction of protein synthesis involves two cassettes for low and constitutive production of a synthetic transcription factor, which activates a promotor in a second cassette with strong and tunable expression (Rantasalo et al., 2018).
CRISPR – adaptation to Trichoderma and optimization
Traditionally, genome manipulation in Trichoderma spp. was done using protoplast transformation, electroporation, agrobacterium mediated transformation or biolistic transformation (Rodriguez-Iglesias and Schmoll, 2015; Schmoll and Zeilinger, 2021). However, in recent years, the versatile method of clustered regularly interspaced short palindromic repeat (CRISPR)-associated Cas9 (Hoof et al., 2018; Wang and Coleman, 2019; Rozhkova and Kislitsin, 2021) was also established for Trichoderma, particularly T. reesei (Liu et al., 2015; Zou and Zhou, 2021) and T. harzianum (Vieira et al., 2021) and subsequently optimized. Usually, the specific gRNA and the Cas9 protein are introduced as DNA into the host organisms which leads to constitutive exposure of the fungus to active Cas9. This can cause decreased viability and genome integrity of the host, but most importantly also unintended genome modifications. It was shown that in vitro assembly of Cas9 and gRNA prior to transformation of the nucleoprotein complex with a marker plasmid or the donor DNA was less prone to off target gene disruption than intracellular expression of Cas9 (Hao and Su, 2019; Rantasalo et al., 2019). Later on, the 5S rRNA promotor of A. niger was suggested for expression of the guide RNA, which enabled gene deletion using a donor DNA carrying only a 40 bp homology sequence and no selectable marker gene (Wang et al., 2021). Similarly, the promotors of two RNA polymerase III U6 snRNA genes were confirmed to be suitable for gRNA expression in T. reesei (Wu et al., 2020). Although the method of genome editing has become quite popular for modifications in fungi, the method via Cas9-CRISPR gRNA ribonucleoprotein complexes assembled in vitro is relatively low. Addition of chemicals like Triton X-100, inositol or benomyl led to increased efficiency in transformation as well as homologous integration (Zou et al., 2021). Nevertheless, the method is still relatively young and more improvements – for example as developed for Saccharomyces cerevisiae (Antony et al., 2022) – await adaptation and establishment in Trichoderma. Moreover, application CRISPR has not been established in several other Trichoderma species, which may be due to the capability of Trichoderma to clear their genomes of foreign DNA elements and hence further efforts are required.
Tools for genome mining
Secondary metabolites of fungi are generally an important resource for novel pharmaceuticals and antibiotics, but can also represent a threat to human health (Karwehl and Stadler, 2016; Bhattarai et al., 2021; Juraschek et al., 2022). Therefore it is very important to tackle new ways of genome mining and investigation, as was done for Ribosomally synthesized and posttranslationally modified peptides, RiPPs, in Trichoderma (Vignolle et al., 2020). These compounds add a further way of biosynthesis to polyketides and non-ribosomal peptides in that they are encoded within a precursor and subsequently processed after posttranslational modification. However, evaluation of the biological function of RiPPs is still at its beginnings and roles in defense, deterring mycophagy, support of nutrient acquisition, competition, but also in symbioses (Ford et al., 2022b). Despite the relevance of for example alpha-amanitin or phallacidin, which are formed by Amanita spp., current screening software is focused on bacterial sequences and reports on biosynthesis in fungi is scarce (Kessler and Chooi, 2022). The multistep approach now presented (Vignolle et al., 2020) and including manual inspection yielded a wide range of RiPP candidates for Trichoderma spp., from 6 in T. reesei to 222 in T. harzianum, which indicates a considerable relevance of these compounds for physiology of Trichoderma. Together with the potential importance of novel RiPPs as bioactive substances, their biological relevance to fungi and especially Trichoderma warrants further investigation.
A novel tool for screening for essential biosynthetic genes was developed to enable determination of biosynthetically relevant genes in clusters versus those which are not needed for secondary metabolite production within the cluster – the so-called gap genes (Vignolle et al., 2021). The tool “FunOrder” (Vignolle et al., 2021) was also tested for Trichoderma and applies computational molecular co-evolution to distinguish between biosynthetic genes and gap genes. Thereby FunOrder facilitates efficient heterologous expression of biosynthetic gene clusters as well as functional analysis of the underlying biochemical pathways.
Generally, for screening for secondary metabolite clusters responsible for novel bioactive compounds in Trichoderma several software packages are useful (Rush et al., 2021). Besides the well-known tool antiSMASH (Blin et al., 2019), which detects biosynthetic clusters for secondary metabolites, the tool amPEPpy (Lawrence et al., 2021) can be used for screening for antimicrobial peptides.
Author contributions
MoS drafted the manuscript, MiS edited the manuscript and all authors agreed on the final version of the manuscript.
Funding
Work of MoS and MiS was supported by the Austrian Research Fund (FWF), grant P31464 to MoS.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
- ^ https://www.marketsandmarkets.com/Market-Reports/biofungicide-market-8734417.html
- ^ https://www.mordorintelligence.com/industry-reports/global-fungi-based-agricultural-microbial-market
References
Abrahao-Neto J., Rossini C. H., El-Gogary S., Henrique-Silva F., Crivellaro O., El-Dorry H. (1995). Mitochondrial functions mediate cellulase gene expression in Trichoderma reesei. Biochemistry 34, 10456–10462. doi: 10.1021/bi00033a018
Aguileta G., De Vienne D. M., Ross O. N., Hood M. E., Giraud T., Petit E., et al. (2014). High variability of mitochondrial gene order among fungi. Genome Biol. Evol. 6, 451–465. doi: 10.1093/gbe/evu028
Alfiky A., Weisskopf L. (2021). Deciphering Trichoderma-plant-pathogen interactions for better development of biocontrol applications. J. Fungi (Basel) 7(1):61. doi: 10.3390/jof7010061
Antony J. S., Hinz J. M., Wyrick J. J. (2022). Tips, tricks, and potential pitfalls of CRISPR genome editing in Saccharomyces cerevisiae. Front. Bioeng Biotechnol. 10, 924914. doi: 10.3389/fbioe.2022.924914
Arbige M. V., Shetty J. K., Chotani G. K. (2019). Industrial enzymology: the next chapter. Trends Biotechnol. 37, 1355–1366. doi: 10.1016/j.tibtech.2019.09.010
Arnau J., Yaver D., Hjort M. C. (2020). “Strategies and challenges for the development of industrial enzymes using fungal cell factories,” in Grand challenges in fungal biotechnology. Ed. Nevalainen H. (Switzerland: Springer Nature Switzerland AG), 179–210.
Ashton G. D., Dyer P. (2016). “Sexual development in fungi and its uses in gene expression systems,” in Gene expression systems in fungi: Advancements and applications. Eds. Schmoll M., Dattenböck C. (Switzerland: Springer International Publishing), 335–350.
Atanasova L., Knox B. P., Kubicek C. P., Druzhinina I. S., Baker S. E. (2013). The polyketide synthase gene pks4 of Trichoderma reesei provides pigmentation and stress resistance. Eukaryot Cell 12, 1499–1508. doi: 10.1128/EC.00103-13
antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47 (W1), W81–W87. doi: 10.1093/nar/gkz310
Basse C. W. (2010). Mitochondrial inheritance in fungi. Curr. Opin. Microbiol. 13, 712–719. doi: 10.1016/j.mib.2010.09.003
Bayram O., Braus G. H. (2012). Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol. Rev. 36, 1–24. doi: 10.1111/j.1574-6976.2011.00285.x
Bayry J., Aimanianda V., Guijarro J. I., Sunde M., Latge J. P. (2012). Hydrophobins–unique fungal proteins. PloS Pathog. 8, e1002700. doi: 10.1371/journal.ppat.1002700
Bazafkan H., Dattenböck C., Böhmdorfer S., Tisch D., Stappler E., Schmoll M. (2015). Mating type dependent partner sensing as mediated by VEL1 in Trichoderma reesei. Mol. Microbiol. 96, 1103–1118. doi: 10.1111/mmi.12993
Beier S., Hinterdobler W., Monroy A. A., Bazafkan H., Schmoll M. (2020). The kinase USK1 regulates cellulase gene expression and secondary metabolite biosynthesis in Trichoderma reesei. Front. Microbiol. 11, 974. doi: 10.3389/fmicb.2020.00974
Belfort M., Derbyshire V., Parker M. M., Cousineau B., Lambowitz A. (2002). “Mobile introns: pathways and proteins,” in Mobile DNA II. Eds. Craig N. L., Craigie R., Gellert M., Lambowitz A. M. (Washington, DC: ASM Press), 761–783.
Bennett R. J., Turgeon B. G. (2016). Fungal sex: The ascomycota. Microbiol. Spectr. 4(5). doi: 10.1128/microbiolspec.FUNK-0005-2016
Benocci T., Aguilar-Pontes M. V., Zhou M., Seiboth B., De Vries R. P. (2017). Regulators of plant biomass degradation in ascomycetous fungi. Biotechnol. Biofuels 10, 152. doi: 10.1186/s13068-017-0841-x
Bhattarai K., Bhattarai K., Kabir M. E., Bastola R., Baral B. (2021). Fungal natural products galaxy: Biochemistry and molecular genetics toward blockbuster drugs discovery. Adv. Genet. 107, 193–284. doi: 10.1016/bs.adgen.2020.11.006
Bischof R. H., Ramoni J., Seiboth B. (2016). Cellulases and beyond: the first 70 years of the enzyme producer Trichoderma reesei. Microb. Cell Fact 15, 106. doi: 10.1186/s12934-016-0507-6
Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S. Y., et al. (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47(W1), W81–W87.
Borkovich K. A., Alex L. A., Yarden O., Freitag M., Turner G. E., Read N. D., et al. (2004). Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68, 1–108. doi: 10.1128/MMBR.68.1.1-108.2004
Brotman Y., Briff E., Viterbo A., Chet I. (2008). Role of swollenin, an expansin-like protein from Trichoderma, in plant root colonization. Plant Physiol. 147, 779–789. doi: 10.1104/pp.108.116293
Burger G., Gray M. W., Lang B. F. (2003). Mitochondrial genomes: anything goes. Trends Genet. 19, 709–716. doi: 10.1016/j.tig.2003.10.012
Cai F., Gao R., Zhao Z., Ding M., Jiang S., Yagtu C., et al. (2020). Evolutionary compromises in fungal fitness: hydrophobins can hinder the adverse dispersal of conidiospores and challenge their survival. ISME J. 14, 2610–2624. doi: 10.1038/s41396-020-0709-0
Cai F., Zhao Z., Gao R., Chen P., Ding M., Jiang S., et al. (2021). The pleiotropic functions of intracellular hydrophobins in aerial hyphae and fungal spores. PloS Genet. 17, e1009924. doi: 10.1371/journal.pgen.1009924
Canseco-Perez M. A., Castillo-Avila G. M., Chi-Manzanero B., Islas-Flores I., Apolinar-Hernandez M. M., Rivera-Munoz G., et al. (2018). Fungal screening on olive oil for extracellular triacylglycerol lipases: Selection of a Trichoderma harzianum strain and genome wide search for the genes. Genes (Basel) 9(2):62. doi: 10.3390/genes9020062
Cardoza R. E., Malmierca M. G., Hermosa M. R., Alexander N. J., McCormick S. P., Proctor R. H., et al. Identification of loci and functional characterization of trichothecene biosynthesis genes in filamentous fungi of the genus Trichoderma. Appl Environ Microbiol. (2011) 77(14):4867–77. doi: 10.1128/AEM.00595-11
Carraro D. M., Ferreira Junior J. R., Schumacher R., Pereira G. G., Hollenberg C. P., El-Dorry H. (1998). A region of the cellobiohydrolase I promoter from the filamentous fungus Trichoderma reesei mediates glucose repression in Saccharomyces cerevisiae, dependent on mitochondrial activity. Biochem. Biophys. Res. Commun. 253, 407–414. doi: 10.1006/bbrc.1998.9758
Carreras-Villaseñor N., Sanchez-Arreguin J. A., Herrera-Estrella A. H. (2012). Trichoderma: sensing the environment for survival and dispersal. Microbiology 158, 3–16. doi: 10.1099/mic.0.052688-0
Castrillo M. L., Bich G. A., Modenutti C., Turjanski A., Zapata P. D., Villalba L. L. (2017). First whole-genome shotgun sequence of a promising cellulase secretor, Trichoderma koningiopsis strain POS7. Genome Announc 5(37):e00823-17. doi: 10.1128/genomeA.00823-17
Chadha S., Mehetre S. T., Bansal R., Kuo A., Aerts A., Grigoriev I. V., et al. (2018). Genome-wide analysis of cytochrome P450s of Trichoderma spp.: annotation and evolutionary relationships. Fungal Biol. Biotechnol. 5, 12. doi: 10.1186/s40694-018-0056-3
Chambergo F. S., Bonaccorsi E. D., Ferreira A. J., Ramos A. S., Ferreira J.R.J.J.R., Abrahao-Neto J., et al. (2002). Elucidation of the metabolic fate of glucose in the filamentous fungus trichoderma reesei using expressed sequence tag (EST) analysis and cDNA microarrays. J. Biol. Chem. 277, 13983–13988. doi: 10.1074/jbc.M107651200
Chatre L., Ricchetti M. (2013). Prevalent coordination of mitochondrial DNA transcription and initiation of replication with the cell cycle. Nucleic Acids Res. 41, 3068–3078. doi: 10.1093/nar/gkt015
Chaverri P., Samuels G. J. (2013). Evolution of habitat preference and nutrition mode in a cosmopolitan fungal genus with evidence of interkingdom host jumps and major shifts in ecology. Evolution 67, 2823–2837. doi: 10.1111/evo.12169
Cheng P., Liu B., Su Y., Hu Y., Hong Y., Yi X., et al. (2017). Genomics insights into different cellobiose hydrolysis activities in two Trichoderma hamatum strains. Microb. Cell Fact 16, 63. doi: 10.1186/s12934-017-0680-2
Chuang Y. C., Li W. C., Chen C. L., Hsu P. W., Tung S. Y., Kuo H. C., et al. (2015). Trichoderma reesei meiosis generates segmentally aneuploid progeny with higher xylanase-producing capability. Biotechnol. Biofuels 8, 30. doi: 10.1186/s13068-015-0202-6
Chum P. Y., Schmidt G., Saloheimo M., Landowski C. P. (2017). Transient silencing of DNA repair genes improves targeted gene integration in the filamentous fungus Trichoderma reesei. Appl. Environ. Microbiol. 83, e00535–e00517. doi: 10.1128/AEM.00535-17
Chung D., Kwon Y. M., Yang Y. (2021). Telomere-to-telomere genome assembly of asparaginase-producing Trichoderma simmonsii. BMC Genomics 22, 830. doi: 10.1186/s12864-021-08162-4
Chung D., Kwon Y. M., Yang Y. (2022). The complete mitochondrial genome of Trichoderma simmonsii (Hypocreales: Hypocreaceae) from the southern coast of Korea. Mitochondrial DNA B Resour 7, 640–641. doi: 10.1080/23802359.2022.2060766
Chun J., Na B., Kim D. H. (2020). Characterization of a novel dsRNA mycovirus of Trichoderma atroviride NFCF377 reveals a member of "Fusagraviridae" with changes in antifungal activity of the host fungus. J. Microbiol. 58, 1046–1053. doi: 10.1007/s12275-020-0380-1
Chun J., So K. K., Ko Y. H., Kim D. H. (2022). Molecular characteristics of a novel hypovirus from Trichoderma harzianum. Arch. Virol. 167, 233–238. doi: 10.1007/s00705-021-05253-1
Chun J., Yang H. E., Kim D. H. (2018a). Identification and molecular characterization of a novel partitivirus from Trichoderma atroviride NFCF394. Viruses 10(11):578. doi: 10.3390/v10110578
Chun J., Yang H. E., Kim D. H. (2018b). Identification of a novel partitivirus of Trichoderma harzianum NFCF319 and evidence for the related antifungal activity. Front. Plant Sci. 9, 1699. doi: 10.3389/fpls.2018.01699
Clark-Walker G. D. (1991). Contrasting mutation rates in mitochondrial and nuclear genes of yeasts versus mammals. Curr. Genet. 20, 195–198. doi: 10.1007/BF00326232
Compant S., Gerbore J., Antonielli L., Brutel A., Schmoll M. (2017). Draft genome sequence of the root-colonizing fungus Trichoderma harzianum B97. Genome Announc 5(13):e00137-17. doi: 10.1128/genomeA.00137-17
Crucello A., Sforca D. A., Horta M. A., Dos Santos C. A., Viana A. J., Beloti L. L., et al. (2015). Analysis of genomic regions of Trichoderma harzianum IOC-3844 related to biomass degradation. PloS One 10, e0122122. doi: 10.1371/journal.pone.0122122
Daskalov A., Heller J., Herzog S., Fleissner A., Glass N. L. (2017). Molecular mechanisms regulating cell fusion and heterokaryon formation in filamentous fungi. Microbiol. Spectr. 5(2). doi: 10.1128/9781555819583.ch10
Dattenböck C., Tisch D., Schuster A., Monroy A. A., Hinterdobler W., Schmoll M. (2018). Gene regulation associated with sexual development and female fertility in different isolates of Trichoderma reesei. Fungal Biol. Biotechnol. 5, 9. doi: 10.1186/s40694-018-0055-4
Davolos D., Russo F., Canfora L., Malusa E., Tartanus M., Furmanczyk E. M., et al. (2021). A genomic and transcriptomic study on the DDT-resistant Trichoderma hamatum FBL 587: first genetic data into mycoremediation strategies for DDT-polluted sites. Microorganisms 9(8):1680. doi: 10.3390/microorganisms9081680
Dawe A. L., Nuss D. L. (2013). Hypovirus molecular biology: from koch's postulates to host self-recognition genes that restrict virus transmission. Adv. Virus Res. 86, 109–147. doi: 10.1016/B978-0-12-394315-6.00005-2
Debuchy R., Berteaux-Lecellier V., Silar P. (2010). “Mating systems and sexual morphogenesis in ascomycetes,” in Cellular and molecular biology of filamentous fungi. Eds. Borkovich K. A., Ebbole D. J. (Washington, DC: ASM Press), 501–535.
Dell'Anno F., Rastelli E., Buschi E., Barone G., Beolchini F., Dell'anno A. (2022). Fungi can be more effective than bacteria for the bioremediation of marine sediments highly contaminated with heavy metals. Microorganisms 10(5):993. doi: 10.3390/microorganisms10050993
Derntl C., Guzman-Chavez F., Mello-De-Sousa T. M., Busse H. J., Driessen A. J. M., Mach R. L., et al. (2017). In vivo study of the sorbicillinoid gene cluster in Trichoderma reesei. Front. Microbiol. 8, 2037. doi: 10.3389/fmicb.2017.02037
Derntl C., Rassinger A., Srebotnik E., Mach R. L., Mach-Aigner A. R. (2016). Identification of the main regulator responsible for synthesis of the typical yellow pigment produced by Trichoderma reesei. Appl. Environ. Microbiol. 82, 6247–6257. doi: 10.1128/AEM.01408-16
Druzhinina I. S., Chenthamara K., Zhang J., Atanasova L., Yang D., Miao Y., et al. (2018). Massive lateral transfer of genes encoding plant cell wall-degrading enzymes to the mycoparasitic fungus Trichoderma from its plant-associated hosts. PloS Genet. 14, e1007322. doi: 10.1371/journal.pgen.1007322
Druzhinina I. S., Komon-Zelazowska M., Atanasova L., Seidl V., Kubicek C. P. (2010). Evolution and ecophysiology of the industrial producer Hypocrea jecorina (anamorph Trichoderma reesei) and a new sympatric agamospecies related to it. PloS One 5, e9191. doi: 10.1371/journal.pone.0009191
Druzhinina I., Kubicek C. (2013). “Ecological genomics of trichoderma,” in The ecological genomics of fungi Hoboken, New Jersey, USA. Ed. Martin F. (John Wiley & Sons Inc), 89–116.
Druzhinina I. S., Kubicek E. M., Kubicek C. P. (2016). Several steps of lateral gene transfer followed by events of 'birth-and-death' evolution shaped a fungal sorbicillinoid biosynthetic gene cluster. BMC Evol. Biol. 16, 269. doi: 10.1186/s12862-016-0834-6
Druzhinina I. S., Seidl-Seiboth V., Herrera-Estrella A., Horwitz B. A., Kenerley C. M., Monte E., et al. (2011). Trichoderma: the genomics of opportunistic success. Nat. Rev. Microbiol. 9, 749–759. doi: 10.1038/nrmicro2637
Duan C., Wang S., Huo R., Li E., Wang M., Ren J., et al. (2022). Sorbicillinoid derivatives with the radical scavenging activities from the marine-derived fungus Acremonium chrysogenum C10. J. Fungi (Basel) 8(5):530. doi: 10.3390/jof8050530
Eusebio-Cope A., Sun L., Tanaka T., Chiba S., Kasahara S., Suzuki N. (2015). The chestnut blight fungus for studies on virus/host and virus/virus interactions: from a natural to a model host. Virology 477, 164–175. doi: 10.1016/j.virol.2014.09.024
Fanelli F., Liuzzi V. C., Logrieco A. F., Altomare C. (2018). Genomic characterization of Trichoderma atrobrunneum (T. harzianum species complex) ITEM 908: insight into the genetic endowment of a multi-target biocontrol strain. BMC Genomics 19, 662. doi: 10.1186/s12864-018-5049-3
Febbraio F. (2017). Biochemical strategies for the detection and detoxification of toxic chemicals in the environment. World J. Biol. Chem. 8, 13–20. doi: 10.4331/wjbc.v8.i1.13
Ferreira Filho J. A., Horta M., Beloti L. L., Dos Santos C. A., De Souza A. P. (2017). Carbohydrate-active enzymes in Trichoderma harzianum: a bioinformatic analysis bioprospecting for key enzymes for the biofuels industry. BMC Genomics 18, 779. doi: 10.1186/s12864-017-4181-9
Ferreira Filho J. A., Horta M., Dos Santos C. A., Almeida D. A., Murad N. F., Mendes J. S., et al. (2020). Integrative genomic analysis of the bioprospection of regulators and accessory enzymes associated with cellulose degradation in a filamentous fungus (Trichoderma harzianum). BMC Genomics 21, 757. doi: 10.1186/s12864-020-07158-w
Fonseca P. L. C., Badotti F., De-Paula R. B., Araujo D. S., Bortolini D. E., Del-Bem L. E., et al. (2020). Exploring the relationship among divergence time and coding and non-coding elements in the shaping of fungal mitochondrial genomes. Front. Microbiol. 11, 765. doi: 10.3389/fmicb.2020.00765
Ford R. E., Foster G. D., Bailey A. M. (2022b). Exploring fungal RiPPs from the perspective of chemical ecology. Fungal Biol. Biotechnol. 9, 12. doi: 10.1186/s40694-022-00144-9
Ford H. V., Jones N. H., Davies A. J., Godley B. J., Jambeck J. R., Napper I. E., et al. (2022a). The fundamental links between climate change and marine plastic pollution. Sci. Total Environ. 806, 150392. doi: 10.1016/j.scitotenv.2021.150392
Frisvad J. C., Moller L. L. H., Larsen T. O., Kumar R., Arnau J. (2018). Safety of the fungal workhorses of industrial biotechnology: update on the mycotoxin and secondary metabolite potential of Aspergillus niger, Aspergillus oryzae, and Trichoderma reesei. Appl. Microbiol. Biotechnol. 102, 9481–9515. doi: 10.1007/s00253-018-9354-1
Gaderer R., Bonazza K., Seidl-Seiboth V. (2014). Cerato-platanins: a fungal protein family with intriguing properties and application potential. Appl. Microbiol. Biotechnol. 98, 4795–4803. doi: 10.1007/s00253-014-5690-y
Gaderer R., Lamdan N. L., Frischmann A., Sulyok M., Krska R., Horwitz B. A., et al. (2015). Sm2, a paralog of the Trichoderma cerato-platanin elicitor Sm1, is also highly important for plant protection conferred by the fungal-root interaction of Trichoderma with maize. BMC Microbiol. 15, 2. doi: 10.1186/s12866-014-0333-0
Gaillardin C., Neuveglise C., Kerscher S., Nicaud J. M. (2012). Mitochondrial genomes of yeasts of the Yarrowia clade. FEMS Yeast Res. 12, 317–331. doi: 10.1111/j.1567-1364.2011.00782.x
Gao R., Ding M., Jiang S., Zhao Z., Chenthamara K., Shen Q., et al. (2020). The evolutionary and functional paradox of cerato-platanins in fungi. Appl. Environ. Microbiol. 86(13):e00696-20. doi: 10.1128/AEM.00696-20
Garber R. C., Yoder O. C. (1983). Isolation of DNA from filamentous fungi and separation into nuclear, mitochondrial, ribosomal, and plasmid components. Anal. Biochem. 135, 416–422. doi: 10.1016/0003-2697(83)90704-2
Gazis R., Chaverri P. (2010). Diversity of fungal endophytes in leaves and stems of wild rubber trees (Hevea brasiliensis) in Peru. Fungal Ecol. 3, 240–254. doi: 10.1016/j.funeco.2009.12.001
Ghabrial S. A., Caston J. R., Jiang D., Nibert M. L., Suzuki N. (2015). 50-plus years of fungal viruses. Virology 479-480, 356–368. doi: 10.1016/j.virol.2015.02.034
Ghabrial S. A., Suzuki N. (2009). Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 47, 353–384. doi: 10.1146/annurev-phyto-080508-081932
Gilbert K. B., Holcomb E. E., Allscheid R. L., Carrington J. C. (2019). Hiding in plain sight: New virus genomes discovered via a systematic analysis of fungal public transcriptomes. PloS One 14, e0219207. doi: 10.1371/journal.pone.0219207
Gladyshev E. (2017). Repeat-induced point mutation and other genome defense mechanisms in fungi. Microbiol. Spectr. 5(4), 10.1128/microbiolspec.FUNK-0042-2017. doi: 10.1128/9781555819583.ch33
Gladyshev E., Kleckner N. (2014). Direct recognition of homology between double helices of DNA in Neurospora crassa. Nat. Commun. 5, 3509. doi: 10.1038/ncomms4509
Glass N. L., Schmoll M., Cate J. H., Coradetti S. (2013). Plant cell wall deconstruction by ascomycete fungi. Annu. Rev. Microbiol. 67, 477–498. doi: 10.1146/annurev-micro-092611-150044
Gomez I., Chet I., Herrera-Estrella A. (1997). Genetic diversity and vegetative compatibility among Trichoderma harzianum isolates. Mol. Gen. Genet. 256, 127–135. doi: 10.1007/s004380050554
Gortikov M., Wang Z., Steindorff A. S., Grigoriev I. V., Druzhinina I. S., Townsend J. P., et al. (2022). Sequencing and analysis of the entire genome of the mycoparasitic bioeffector fungus Trichoderma asperelloides strain T 203 (Hypocreales). Microbiol. Resour Announc 11, e0099521. doi: 10.1128/mra.00995-21
Gruber S., Omann M., Zeilinger S. (2013). Comparative analysis of the repertoire of G protein-coupled receptors of three species of the fungal genus Trichoderma. BMC Microbiol. 13, 108. doi: 10.1186/1471-2180-13-108
Guangtao Z., Hartl L., Schuster A., Polak S., Schmoll M., Wang T., et al. (2009). Gene targeting in a nonhomologous end joining deficient Hypocrea jecorina. J. Biotechnol. 139, 146–151. doi: 10.1016/j.jbiotec.2008.10.007
Gudynaite-Savitch L., White T. C. (2016). “Fungal biotechnology for industrial enzyme production: Focus on (hemi)cellulase production strategies, advances and challenges,” in Gene expression systems in fungi: Advancements and applications. Eds. Schmoll M., Dattenböck C. (Switzerland: Springer International Publishing Switzerland), 395–435.
Gutierrez S., Mccormick S. P., Cardoza R. E., Kim H. S., Yugueros L. L., Vaughan M. M., et al. (2021). Distribution, function, and evolution of a gene essential for trichothecene toxin biosynthesis in Trichoderma. Front. Microbiol. 12, 791641. doi: 10.3389/fmicb.2021.791641
Guzman-Chavez F., Salo O., Nygard Y., Lankhorst P. P., Bovenberg R., Driessen A. J. M. (2017). Mechanism and regulation of sorbicillin biosynthesis by Penicillium chrysogenum. Microb. Biotechnol. 10, 958–968. doi: 10.1111/1751-7915.12736
Guzman-Guzman P., Aleman-Duarte M. I., Delaye L., Herrera-Estrella A., Olmedo-Monfil V. (2017). Identification of effector-like proteins in Trichoderma spp. and role of a hydrophobin in the plant-fungus interaction and mycoparasitism. BMC Genet. 18, 16. doi: 10.1186/s12863-017-0481-y
Guzman-Guzman P., Porras-Troncoso M. D., Olmedo-Monfil V., Herrera-Estrella A. (2019). Trichoderma species: Versatile plant symbionts. Phytopathology 109, 6–16. doi: 10.1094/PHYTO-07-18-0218-RVW
Hafiz F. B., Moradtalab N., Goertz S., Rietz S., Dietel K., Rozhon W., et al. (2022). Synergistic effects of a root-endophytic Trichoderma fungus and Bacillus on early root colonization and defense activation against Verticillium longisporum in rapeseed. Mol. Plant Microbe Interact. 35, 380–392. doi: 10.1094/MPMI-11-21-0274-R
Hager K. M., Yanofsky C. (1990). Genes expressed during conidiation in Neurospora crassa: molecular characterization of con-13. Gene 96, 153–159. doi: 10.1016/0378-1119(90)90247-O
Hammond T. M., Andrewski M. D., Roossinck M. J., Keller N. P. (2008). Aspergillus mycoviruses are targets and suppressors of RNA silencing. Eukaryot Cell 7, 350–357. doi: 10.1128/EC.00356-07
Han J., Xue Y., Li M., Li Y., Liu J., Gan L., et al. (2020). Effect of VIB gene on cellulase production of Trichoderma orientalis EU7-22. Appl. Biochem. Biotechnol. 191, 1444–1455. doi: 10.1007/s12010-020-03260-7
Hao Z., Su X. (2019). Fast gene disruption in Trichoderma reesei using in vitro assembled Cas9/gRNA complex. BMC Biotechnol. 19, 2. doi: 10.1186/s12896-018-0498-y
Harman G. E., Doni F., Khadka R. B., Uphoff N. (2021). Endophytic strains of trichoderma increase plants' photosynthetic capability. J. Appl. Microbiol. 130, 529–546. doi: 10.1111/jam.14368
Harman G. E., Howell C. R., Viterbo A., Chet I., Lorito M. (2004a). Trichoderma species–opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56. doi: 10.1038/nrmicro797
Harman G. E., Lorito M., Lynch J. M. (2004b). Uses of Trichoderma spp. to alleviate or remediate soil and water pollution. Adv. Appl. Microbiol. 56, 313–330. doi: 10.1016/S0065-2164(04)56010-0
Harned A. M., Volp K. A. (2011). The sorbicillinoid family of natural products: isolation, biosynthesis, and synthetic studies. Nat. Prod Rep. 28, 1790–1810. doi: 10.1039/c1np00039j
Hatvani L., Homa M., Chenthamara K., Cai F., Kocsube S., Atanasova L., et al. (2019). Agricultural systems as potential sources of emerging human mycoses caused by Trichoderma: a successful, common phylotype of Trichoderma longibrachiatum in the frontline. FEMS Microbiol. Lett. 366(21):fnz246. doi: 10.1093/femsle/fnz246
Hinterdobler W., Beier S., Monroy A. A., Berger H., Dattenbock C., Schmoll M. (2020a). The G-protein coupled receptor GPR8 regulates secondary metabolism in Trichoderma reesei. Front. Bioeng Biotechnol. 8, 558996. doi: 10.3389/fbioe.2020.558996
Hinterdobler W., Beier S., S. K., Schmoll M. (2020b). “Sexual development, its determinants and regulation in trichoderma reesei,” in Recent developments in trichoderma research. Eds. Zeilinger S., Druzhinina I., Singh H. B., Gupta V. K. (Amsterdam, The Netherlands: Elsevier), 185–206.
Hinterdobler W., Li G., Spiegel K., Basyouni-Khamis S., Gorfer M., Schmoll M. (2021a). Trichoderma reesei isolated from Austrian soil with high potential for biotechnological application. Front. Microbiol. 12, 552301. doi: 10.3389/fmicb.2021.552301
Hinterdobler W., Li G., Turra D., Schalamun M., Kindel S., Sauer U., et al. (2021b). Integration of chemosensing and carbon catabolite repression impacts fungal enzyme regulation and plant associations. bioRxiv. doi: 10.1101/2021.1105.1106.442915
Hitzenhammer E., Büschl C., Sulyok M., Schuhmacher R., Kluger B., Wischnitzki E., et al. (2019). YPR2 is a regulator of light modulated carbon and secondary metabolism in Trichoderma reesei. BMC Genomics 20, 211. doi: 10.1186/s12864-019-5574-8
Hoof J. B., Nodvig C. S., Mortensen U. H. (2018). Genome editing: CRISPR-Cas9. Methods Mol. Biol. 1775, 119–132. doi: 10.1007/978-1-4939-7804-5_11
Hou X., Zhang X., Xue M., Zhao Z., Zhang H., Xu D., et al. (2022). Recent advances in sorbicillinoids from fungi and their bioactivities (Covering 2016-2021). J. Fungi (Basel) 8(1):62. doi: 10.3390/jof8010062
Ivanova C., Ramoni J., Aouam T., Frischmann A., Seiboth B., Baker S. E., et al. (2017). Genome sequencing and transcriptome analysis of Trichoderma reesei QM9978 strain reveals a distal chromosome translocation to be responsible for loss of vib1 expression and loss of cellulase induction. Biotechnol. Biofuels 10, 209. doi: 10.1186/s13068-017-0897-7
Jaklitsch W. M., Voglmayr H. (2015). Biodiversity of Trichoderma (Hypocreaceae) in southern Europe and macaronesia. Stud. Mycol 80, 1–87. doi: 10.1016/j.simyco.2014.11.001
Jom-in S., Akarapisan A. (2009). Characterization of double stranded RNA in Trichoderma spp. isolates in Chiang mai province. J. Agric. Technol. 5, 261–270.
Jourdier E., Baudry L., Poggi-Parodi D., Vicq Y., Koszul R., Margeot A., et al. (2017). Proximity ligation scaffolding and comparison of two Trichoderma reesei strains genomes. Biotechnol. Biofuels 10, 151. doi: 10.1186/s13068-017-0837-6
Juraschek L. M., Kappenberg A., Amelung W. (2022). Mycotoxins in soil and environment. Sci. Total Environ. 814, 152425. doi: 10.1016/j.scitotenv.2021.152425
Kabani M., Beckerich J.-M., Brodsky J. L. (2002). Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol. Cell. Biol. 22, 4677–4689. doi: 10.1128/MCB.22.13.4677-4689.2002
Kannan C., Divya M., Rekha G., Barbadikar K. M., Maruthi P., Hajira S. K., et al. (2022). Whole genome sequencing data of native isolates of Bacillus and Trichoderma having potential biocontrol and plant growth promotion activities in rice. Data Brief 41, 107923. doi: 10.1016/j.dib.2022.107923
Karimi Aghcheh R., Druzhinina I. S., Kubicek C. P. (2013). The putative protein methyltransferase LAE1 of Trichoderma atroviride is a key regulator of asexual development and mycoparasitism. PloS One 8, e67144. doi: 10.1371/journal.pone.0067144
Karwehl S., Stadler M. (2016). Exploitation of fungal biodiversity for discovery of novel antibiotics. Curr. Top. Microbiol. Immunol. 398, 303–338. doi: 10.1007/82_2016_496
Kashyap P. L., Rai P., Srivastava A. K., Kumar S. (2017). Trichoderma for climate resilient agriculture. World J. Microbiol. Biotechnol. 33, 155. doi: 10.1007/s11274-017-2319-1
Kawamori M., Morikawa Y., Takasawa S. (1985). Inductive formation of celluases by l-sorbose in Trichoderma reesei. Appl. Microbiol. Biotechnol. 22, 235–236. doi: 10.1007/BF00253616
Kessler S. C., Chooi Y. H. (2022). Out for a RiPP: challenges and advances in genome mining of ribosomal peptides from fungi. Nat. Prod Rep. 39, 222–230. doi: 10.1039/D1NP00048A
Khalifa M. E., MacDiarmid R. M. (2019). A novel totivirus naturally occurring in two different fungal genera. Front. Microbiol. 10, 2318. doi: 10.3389/fmicb.2019.02318
Konstantinovas C., De Oliveira Mendes T. A., Vannier-Santos M. A., Lima-Santos J. (2017). Modulation of human immune response by fungal biocontrol agents. Front. Microbiol. 8, 39. doi: 10.3389/fmicb.2017.00039
Kour D., Kaur T., Devi R., Yadav A., Singh M., Joshi D., et al. (2021). Beneficial microbiomes for bioremediation of diverse contaminated environments for environmental sustainability: present status and future challenges. Environ. Sci. pollut. Res. Int. 28, 24917–24939. doi: 10.1007/s11356-021-13252-7
Kouzminova E., Selker E. U. (2001). dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO J. 20, 4309–4323. doi: 10.1093/emboj/20.15.4309
Kredics L., Antal Z., Doczi I., Manczinger L., Kevei F., Nagy E. (2003). Clinical importance of the genus trichoderma. a review. Acta Microbiol. Immunol. Hung 50, 105–117. doi: 10.1556/AMicr.50.2003.2-3.1
Kredics L., Chen L., Kedves O., Buchner R., Hatvani L., Allaga H., et al. (2018). Molecular tools for monitoring Trichoderma in agricultural environments. Front. Microbiol. 9, 1599. doi: 10.3389/fmicb.2018.01599
Krumlauf R., Marzluf G. A. (1980). Genome organization and characterization of the repetitive and inverted repeat DNA sequences in Neurospora crassa. J. Biol. Chem. 255, 1138–1145. doi: 10.1016/S0021-9258(19)86153-7
Kubicek C. P., Baker S., Gamauf C., Kenerley C. M., Druzhinina I. S. (2008). Purifying selection and birth-and-death evolution in the class II hydrophobin gene families of the ascomycete Trichoderma/Hypocrea. BMC Evol. Biol. 8, 4. doi: 10.1186/1471-2148-8-4
Kubicek C. P., Herrera-Estrella A., Seidl-Seiboth V., Martinez D. A., Druzhinina I. S., Thon M., et al. (2011). Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 12(4), R40. doi: 10.1186/gb-2011-12-4-r40
Kubicek C. P., Steindorff A. S., Chenthamara K., Manganiello G., Henrissat B., Zhang J., et al. (2019). Evolution and comparative genomics of the most common Trichoderma species. BMC Genomics 20, 485. doi: 10.1186/s12864-019-5680-7
Kuo H. C., Wang T. Y., Chen P. P., Chen R. S., Chen T. Y. (2015). Genome sequence of Trichoderma virens FT-333 from tropical marine climate. FEMS Microbiol. Lett. 362(7):fnv036. doi: 10.1093/femsle/fnv036
Kwak Y. (2020). Complete mitochondrial genome of the fungal biocontrol agent Trichoderma atroviride: genomic features, comparative analysis and insight into the mitochondrial evolution in Trichoderma. Front. Microbiol. 11, 785. doi: 10.3389/fmicb.2020.00785
Kwak Y. (2021). An update on Trichoderma mitogenomes: complete de novo mitochondrial genome of the fungal biocontrol agent Trichoderma harzianum (Hypocreales, sordariomycetes), an ex-neotype strain CBS 226.95, and tracing the evolutionary divergences of mitogenomes in Trichoderma. Microorganisms 9(8), 1564. doi: 10.3390/microorganisms9081564
Lahlali R., Ezrari S., Radouane N., Kenfaoui J., Esmaeel Q., El Hamss H., et al. (2022). Biological control of plant pathogens: a global perspective. Microorganisms 10(3), 596. doi: 10.3390/microorganisms10030596
Landeis A., Schmidt-Heydt M. (2021). Sequencing and analysis of the entire genome of the mycoparasitic fungus Trichoderma afroharzianum. Microbiol. Resour Announc 10 (15), e00211–e00221. doi: 10.1128/MRA.00211-21
Lawrence T. J., Carper D. L., Spangler M. K., Carrell A. A., Rush T. A., Minter S. J., et al. (2021). amPEPpy 1.0: a portable and accurate antimicrobial peptide prediction tool. Bioinformatics. 37 (14), 2058–2060. doi: 10.1093/bioinformatics/btaa917
Lee S. H., Yun S. H., Chun J., Kim D. H. (2017). Characterization of a novel dsRNA mycovirus of Trichoderma atroviride NFCF028. Arch. Virol. 162, 1073–1077. doi: 10.1007/s00705-016-3214-z
Lehmann L., Ronnest N. P., Jorgensen C. I., Olsson L., Stocks S. M., Jorgensen H. S., et al. (2016). Linking hydrolysis performance to Trichoderma reesei cellulolytic enzyme profile. Biotechnol. Bioeng 113, 1001–1010. doi: 10.1002/bit.25871
Li W. C., Chen C. L., Wang T. F. (2018). Repeat-induced point (RIP) mutation in the industrial workhorse fungus Trichoderma reesei. Appl. Microbiol. Biotechnol. 102, 1567–1574. doi: 10.1007/s00253-017-8731-5
Lichius A., Bidard F., Buchholz F., Le Crom S., Martin J., Schackwitz W., et al. (2015). Genome sequencing of the Trichoderma reesei QM9136 mutant identifies a truncation of the transcriptional regulator XYR1 as the cause for its cellulase-negative phenotype. BMC Genomics 16, 326. doi: 10.1186/s12864-015-1526-0
Lieckfeldt E., Kullnig C. M., Samuels G. J., Kubicek C. P. (2000). Sexually competent, sucrose- and nitrate-assimilating strains of Hypocrea jecorina (Trichoderma reesei) from south American soils. Mycologia 92, 374–380. doi: 10.1080/00275514.2000.12061170
Li W. C., Huang C. H., Chen C. L., Chuang Y. C., Tung S. Y., Wang T. F. (2017). Trichoderma reesei complete genome sequence, repeat-induced point mutation, and partitioning of CAZyme gene clusters. Biotechnol. Biofuels 10, 170. doi: 10.1186/s13068-017-0825-x
Li W. C., Lee C. Y., Lan W. H., Woo T. T., Liu H. C., Yeh H. Y., et al. (2021a). Trichoderma reesei Rad51 tolerates mismatches in hybrid meiosis with diverse genome sequences. Proc. Natl. Acad. Sci. U.S.A. 118(8):e2007192118. doi: 10.1073/pnas.2007192118
Li W. C., Lin T. C., Chen C. L., Liu H. C., Lin H. N., Chao J. L., et al. (2021b). Complete genome sequences and genome-wide characterization of Trichoderma biocontrol agents provide new insights into their evolution and variation in genome organization, sexual development, and fungal-plant interactions. Microbiol. Spectr. 9, e0066321. doi: 10.1128/Spectrum.00663-21
Li Z., Liu T. (2022). High-quality genome sequence data of Trichoderma gracile HK011-1, a fungal antagonistic agent against plant pathogens. Plant Dis. 106, 1035–1038. doi: 10.1094/PDIS-09-21-2006-A
Lindo L., Mccormick S. P., Cardoza R. E., Brown D. W., Kim H. S., Alexander N. J., et al. (2018). Effect of deletion of a trichothecene toxin regulatory gene on the secondary metabolism transcriptome of the saprotrophic fungus Trichoderma arundinaceum. Fungal Genet. Biol. 119, 29–46. doi: 10.1016/j.fgb.2018.08.002
Linke R., Thallinger G. G., Haarmann T., Eidner J., Schreiter M., Lorenz P., et al. (2015). Restoration of female fertility in Trichoderma reesei QM6a provides the basis for inbreeding in this industrial cellulase producing fungus. Biotechnol. Biofuels 8, 155. doi: 10.1186/s13068-015-0311-2
Liu W., Cai Y., Zhang Q., Chen L., Shu F., Ma X., et al. (2020). The mitochondrial genome of Morchella importuna (272.2 kb) is the largest among fungi and contains numerous introns, mitochondrial non-conserved open reading frames and repetitive sequences. Int. J. Biol. Macromol 143, 373–381. doi: 10.1016/j.ijbiomac.2019.12.056
Liu R., Chen L., Jiang Y., Zhou Z., Zou G. (2015). Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discovery 1, 15007. doi: 10.1038/celldisc.2015.7
Liu C., Li M., Redda E. T., Mei J., Zhang J., Wu B., et al. (2019). A novel double-stranded RNA mycovirus isolated from Trichoderma harzianum. Virol. J. 16, 113. doi: 10.1186/s12985-019-1213-x
Liu H. C., Li W. C., Wang T. F. (2021). TSETA: A third-generation sequencing-based computational tool for mapping and visualization of SNPs, meiotic recombination products, and RIP mutations. Methods Mol. Biol. 2234, 331–361. doi: 10.1007/978-1-0716-1048-0_22
Losada L., Pakala S. B., Fedorova N. D., Joardar V., Shabalina S. A., Hostetler J., et al. (2014). Mobile elements and mitochondrial genome expansion in the soil fungus and potato pathogen Rhizoctonia solani AG-3. FEMS Microbiol. Lett. 352, 165–173. doi: 10.1111/1574-6968.12387
Macias-Rodriguez L., Contreras-Cornejo H. A., Adame-Garnica S. G., Del-Val E., Larsen J. (2020). The interactions of Trichoderma at multiple trophic levels: inter-kingdom communication. Microbiol. Res. 240, 126552. doi: 10.1016/j.micres.2020.126552
Malmierca M. G., Cardoza R. E., Alexander N. J., McCormick S. P., Collado I. G., Hermosa R., Gutiérrez S. (2013). Relevance of trichothecenes in fungal physiology: disruption of tri5 in Trichoderma arundinaceum. Fungal Genet. Biol. 53, 22–33. doi: 10.1016/j.fgb.2013.02.001.
Mandels M., Weber J., Parizek R. (1971). Enhanced cellulase production by a mutant of Trichoderma viride. Appl. Microbiol. 21, 152–154. doi: 10.1128/am.21.1.152-154.1971
Marie-Nelly H., Marbouty M., Cournac A., Flot J. F., Liti G., Parodi D. P., et al. (2014). High-quality genome (re)assembly using chromosomal contact data. Nat. Commun. 5, 5695. doi: 10.1038/ncomms6695
Marquez L. M., Redman R. S., Rodriguez R. J., Roossinck M. J. (2007). A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science 315, 513–515. doi: 10.1126/science.1136237
Martinez D., Berka R. M., Henrissat B., Saloheimo M., Arvas M., Baker S. E., et al. (2008). Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 26, 553–560. doi: 10.1038/nbt1403
Maskey R. P., Grun-Wollny I., Laatsch H. (2005). Sorbicillin analogues and related dimeric compounds from Penicillium notatum. J. Nat. Prod 68, 865–870. doi: 10.1021/np040137t
McDonald M. J., Rice D. P., Desai M. M. (2016). Sex speeds adaptation by altering the dynamics of molecular evolution. Nature 531, 233–236. doi: 10.1038/nature17143
Medina-Castellanos E., Villalobos-Escobedo J. M., Riquelme M., Read N. D., Abreu-Goodger C., Herrera-Estrella A. (2018). Danger signals activate a putative innate immune system during regeneration in a filamentous fungus. PloS Genet. 14, e1007390. doi: 10.1371/journal.pgen.1007390
Medina R., Franco M. E. E., Bartel L. C., Martinez Alcantara V., Saparrat M. C. N., Balatti P. A. (2020). Fungal mitogenomes: relevant features to planning plant disease management. Front. Microbiol. 11, 978. doi: 10.3389/fmicb.2020.00978
Megarioti A. H., Kouvelis V. N. (2020). The coevolution of fungal mitochondrial introns and their homing endonucleases (GIY-YIG and LAGLIDADG). Genome Biol. Evol. 12, 1337–1354. doi: 10.1093/gbe/evaa126
Mendoza-Mendoza A., Zaid R., Lawry R., Hermosa R., Monte E., Horwitz B., et al. (2018). Molecular dialogues between Trichoderma and roots: role of the fungal secretome. Fungal Biol. Rev. 32, 62–85. doi: 10.1016/j.fbr.2017.12.001
Meng J., Wang X., Xu D., Fu X., Zhang X., Lai D., et al. (2016). Sorbicillinoids from fungi and their bioactivities. Molecules 21(6), 715. doi: 10.3390/molecules21060715
Milgroom M. G., Cortesi P. (2004). Biological control of chestnut blight with hypovirulence: a critical analysis. Annu. Rev. Phytopathol. 42, 311–338. doi: 10.1146/annurev.phyto.42.040803.140325
Mishra S., Lin Z., Pang S., Zhang W., Bhatt P., Chen S. (2021). Recent advanced technologies for the characterization of xenobiotic-degrading microorganisms and microbial communities. Front. Bioeng Biotechnol. 9, 632059. doi: 10.3389/fbioe.2021.632059
Monroy A. A., Stappler E., Schuster A., Sulyok M., Schmoll M. (2017). A CRE1- regulated cluster is responsible for light dependent production of dihydrotrichotetronin in Trichoderma reesei. PloS One, 12 (8), e0182530. doi: 10.1371/journal.pone.0182530
Moran-Diez M. E., Martinez De Alba A. E., Rubio M. B., Hermosa R., Monte E. (2021). Trichoderma and the plant heritable priming responses. J. Fungi (Basel) 7(4), 318. doi: 10.3390/jof7040318
Myers J. M., James T. Y. (2022). Mycoviruses. Curr. Biol. 32, R150–R155. doi: 10.1016/j.cub.2022.01.049
Nerva L., Chitarra W. (2021). “Mycoviruses: A hidden world within fungi,” in Encyclopedia of mycology. Eds. Zaragoza Ó., Casadevall A. (Oxford: Elsevier), 134–141.
Nerva L., Varese G. C., Falk B. W., Turina M. (2017). Mycoviruses of an endophytic fungus can replicate in plant cells: evolutionary implications. Sci. Rep. 7, 1908. doi: 10.1038/s41598-017-02017-3
Nevalainen K. M., Palva E. T. (1978). Production of extracellular enzymes in mutants isolated from Trichoderma viride unable to hydrolyze cellulose. Appl. Environ. Microbiol. 35, 11–16. doi: 10.1128/aem.35.1.11-16.1978
Nevalainen H., Suominen P., Taimisto K. (1994). On the safety of Trichoderma reesei. J. Biotechnol. 37, 193–200. doi: 10.1016/0168-1656(94)90126-0
Nogueira-Lopez G., Padilla-Arizmendi F., Inwood S., Lyne S., Steyaert J. M., Nieto-Jacobo M. F., et al. (2019). TrichoGate: An improved vector system for a large scale of functional analysis of Trichoderma genes. Front. Microbiol. 10, 2794. doi: 10.3389/fmicb.2019.02794
Pachauri S., Sherkhane P. D., Kumar V., Mukherjee P. K. (2020). Whole genome sequencing reveals major deletions in the genome of M7, a gamma ray-induced mutant of Trichoderma virens that is repressed in conidiation, secondary metabolism, and mycoparasitism. Front. Microbiol. 11, 1030. doi: 10.3389/fmicb.2020.01030
Paloheimo M., Haarmann T., Mäkinen S., Vehmaanperä J. (2016). “Production of industrial enzymes in trichoderma reesei,” in Gene expression systems in fungi: Advancements and applications. Eds. Schmoll M., Dattenböck C. (Heidelberg: Springer International), 23–58.
Pang A. P., Zhang F., Hu X., Luo Y., Wang H., Durrani S., et al. (2021). Glutamine involvement in nitrogen regulation of cellulase production in fungi. Biotechnol. Biofuels 14, 199. doi: 10.1186/s13068-021-02046-1
Peterson R., Nevalainen H. (2012). Trichoderma reesei RUT-C30 - thirty years of strain improvement. Microbiology 158, 58–68. doi: 10.1099/mic.0.054031-0
Pfordt A., Schiwek S., Karlovsky P., Von Tiedemann A. (2020). Trichoderma afroharzianum ear rot–a new disease on maize in Europe. Front. Agron. 2. doi: 10.3389/fagro.2020.547758
Pilgaard B., Vuillemin M., Munk L., Holck J., Meier S., Wilkens C., et al. (2022). Discovery of a novel glucuronan lyase system in Trichoderma parareesei. Appl. Environ. Microbiol. 88, e0181921. doi: 10.1128/AEM.01819-21
Porciuncula Jde O., Furukawa T., Mori K., Shida Y., Hirakawa H., Tashiro K., et al. (2013). Single nucleotide polymorphism analysis of a Trichoderma reesei hyper-cellulolytic mutant developed in Japan. Biosci. Biotechnol. Biochem. 77, 534–543. doi: 10.1271/bbb.120794
Pramateftaki P. V., Kouvelis V. N., Lanaridis P., Typas M. A. (2006). The mitochondrial genome of the wine yeast Hanseniaspora uvarum: a unique genome organization among yeast/fungal counterparts. FEMS Yeast Res. 6, 77–90. doi: 10.1111/j.1567-1364.2005.00018.x
Proctor R. H., Mccormick S. P., Kim H. S., Cardoza R. E., Stanley A. M., Lindo L., et al. (2018). Evolution of structural diversity of trichothecenes, a family of toxins produced by plant pathogenic and entomopathogenic fungi. PloS Pathog. 14, e1006946. doi: 10.1371/journal.ppat.1006946
Ramirez-Valdespino C. A., Casas-Flores S., Olmedo-Monfil V. (2019). Trichoderma as a model to study effector-like molecules. Front. Microbiol. 10, 1030. doi: 10.3389/fmicb.2019.01030
Rantasalo A., Czeizler E., Virtanen R., Rousu J., Lahdesmaki H., Penttila M., et al. (2016). Synthetic transcription amplifier system for orthogonal control of gene expression in Saccharomyces cerevisiae. PloS One 11, e0148320. doi: 10.1371/journal.pone.0148320
Rantasalo A., Landowski C. P., Kuivanen J., Korppoo A., Reuter L., Koivistoinen O., et al. (2018). A universal gene expression system for fungi. Nucleic Acids Res. 46, e111. doi: 10.1093/nar/gky558
Rantasalo A., Vitikainen M., Paasikallio T., Jantti J., Landowski C. P., Mojzita D. (2019). Novel genetic tools that enable highly pure protein production in Trichoderma reesei. Sci. Rep. 9, 5032. doi: 10.1038/s41598-019-41573-8
Rassinger A., Gacek-Matthews A., Strauss J., Mach R. L., Mach-Aigner A. R. (2018). Truncation of the transcriptional repressor protein Cre1 in Trichoderma reesei rut-C30 turns it into an activator. Fungal Biol. Biotechnol. 5, 15. doi: 10.1186/s40694-018-0059-0
Repas T. S., Gillis D. M., Boubakir Z., Bao X., Samuels G. J., Kaminskyj S. G. W. (2017). Growing plants on oily, nutrient-poor soil using a native symbiotic fungus. PloS One 12, e0186704. doi: 10.1371/journal.pone.0186704
Rodriguez-Iglesias A., Schmoll M. (2015). “Protoplast transformation for genome manipulation in fungi,” in Genetic transformation systems in fungi. Eds. Van Den Berg M. A., Maruthachalam K. (Switzerland: Springer International Publishing), 21–40.
Rozhkova A. M., Kislitsin V. Y. (2021). CRISPR/Cas genome editing in filamentous fungi. Biochem. (Mosc) 86, S120–S139. doi: 10.1134/S0006297921140091
Rush T. A., Shrestha H. K., Gopalakrishnan Meena M., Spangler M. K., Ellis J. C., Labbé J. L., et al. (2021). Bioprospecting Trichoderma: A systematic roadmap to screen genomes and natural products for biocontrol applications. Front. Fungal Biol. 2. doi: 10.3389/ffunb.2021.716511
Salas-Marina M. A., Isordia-Jasso M. I., Islas-Osuna M. A., Delgado-Sánchez P., Jiménez-Bremont J. F., Rodríguez-Kessler M., et al. (2015). The Epl1 and Sm1 proteins from Trichoderma atroviride and Trichoderma virens differentially modulate systemic disease resistance against different life style pathogens in Solanum lycopersicum. Front. Plant Sci. 6, 77. doi: 10.3389/fpls.2015.00077
Salo O., Guzman-Chavez F., Ries M. I., Lankhorst P. P., Bovenberg R. A., Vreeken R. J., et al. (2016). Identification of a polyketide synthase involved in sorbicillin biosynthesis by Penicillium chrysogenum. Appl. Environ. Microbiol. 82, 3971–3978. doi: 10.1128/AEM.00350-16
Saloheimo M., Paloheimo M., Hakola S., Pere J., Swanson B., Nyyssonen E., et al. (2002). Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur. J. Biochem. 269, 4202–4211. doi: 10.1046/j.1432-1033.2002.03095.x
Sandoval-Denis M., Sutton D. A., Cano-Lira J. F., Gene J., Fothergill A. W., Wiederhold N. P., et al. (2014). Phylogeny of the clinically relevant species of the emerging fungus Trichoderma and their antifungal susceptibilities. J. Clin. Microbiol. 52, 2112–2125. doi: 10.1128/JCM.00429-14
Sanna M., Pugliese M., Gullino M. L., Mezzalama M. (2022). First report of Trichoderma afroharzianum causing seed rot on maize in Italy. Plant Dis. doi: 10.1094/PDIS-12-21-2697-PDN
Schmoll M. (2013). “Sexual development in trichoderma - scrutinizing the aspired phenomenon,” in Trichoderma: biology and applications. Eds. Mukherjee P. K., Horwitz B. A., Singh U. S., Mukherjee M., Schmoll M. (Oxfordshire, UK: CAB International), 67–86.
Schmoll M. (2018a). Light, stress, sex and carbon - the photoreceptor ENVOY as a central checkpoint in the physiology of Trichoderma reesei. Fungal Biol. 122, 479–486. doi: 10.1016/j.funbio.2017.10.007
Schmoll M. (2018b). Regulation of plant cell wall degradation by light in Trichoderma. Fungal Biol. Biotechnol. 5, 10. doi: 10.1186/s40694-018-0052-7
Schmoll M., Dattenböck C., Carreras-Villasenor N., Mendoza-Mendoza A., Tisch D., Aleman M. I., et al. (2016). The genomes of three uneven siblings: footprints of the lifestyles of three Trichoderma species. Microbiol. Mol. Biol. Rev. 80, 205–327. doi: 10.1128/MMBR.00040-15
Schmoll M., Esquivel-Naranjo E. U., Herrera-Estrella A. (2010). Trichoderma in the light of day - physiology and development. Fungal Genet. Biol. 47, 909–916. doi: 10.1016/j.fgb.2010.04.010
Schmoll M., Kubicek C. P. (2005). ooc1, a unique gene expressed only during growth of Hypocrea jecorina (anamorph: Trichoderma reesei) on cellulose. Curr. Genet. 48, 126–133. doi: 10.1007/s00294-005-0585-1
Schmoll M., Tisch D., Schuster A., Freitag M., Pomraning K. R., Wang T. F. (2013). Introducing or inactivating female fertility in filamentous fungal cells. CAB International: Oxfordshire, UK.
Schmoll M., Wang T. F. (2016). “Sexual development in trichoderma,” in The mycota (Vol. i): Growth, differentiation and sexuality. Ed. Wendland J. (Switzerland: Springer International Publishing), 457–474.
Schmoll M., Zeilinger S. (2021). Resistance marker- and gene gun-mediated transformation of Trichoderma reesei. Methods Mol. Biol. 2234, 55–62. doi: 10.1007/978-1-0716-1048-0_4
Schmoll M., Zeilinger S., Mach R. L., Kubicek C. P. (2004). Cloning of genes expressed early during cellulase induction in Hypocrea jecorina by a rapid subtraction hybridization approach. Fungal Genet. Biol. 41, 877–887. doi: 10.1016/j.fgb.2004.06.002
Schuster A., Bruno K. S., Collett J. R., Baker S. E., Seiboth B., Kubicek C. P., et al. (2012). A versatile toolkit for high throughput functional genomics with Trichoderma reesei. Biotechnol. Biofuels 5, 1. doi: 10.1186/1754-6834-5-1
Schuster A., Kubicek C. P., Schmoll M. (2011). Dehydrogenase GRD1 represents a novel component of the cellulase regulon in Trichoderma reesei (Hypocrea jecorina). Appl. Environ. Microbiol. 77, 4553–4563. doi: 10.1128/AEM.00513-11
Schuster A., Schmoll M. (2010). Biology and biotechnology of Trichoderma. Appl. Microbiol. Biotechnol. 87, 787–799. doi: 10.1007/s00253-010-2632-1
Segers G. C., Zhang X., Deng F., Sun Q., Nuss D. L. (2007). Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. U.S.A. 104, 12902–12906. doi: 10.1073/pnas.0702500104
Seidl V., Gamauf C., Druzhinina I. S., Seiboth B., Hartl L., Kubicek C. P. (2008). The Hypocrea jecorina (Trichoderma reesei) hypercellulolytic mutant RUT C30 lacks a 85 kb (29 gene-encoding) region of the wild-type genome. BMC Genomics 9, 327. doi: 10.1186/1471-2164-9-327
Seidl V., Seibel C., Kubicek C. P., Schmoll M. (2009). Sexual development in the industrial workhorse Trichoderma reesei. Proc. Natl. Acad. Sci. U.S.A. 106, 13909–13914. doi: 10.1073/pnas.0904936106
Seidl V., Seiboth B. (2010). Trichoderma reesei: genetic approaches to improving strain efficiency. Biofuels 1, 343–354. doi: 10.4155/bfs.10.1
Selker E. U., Cambareri E. B., Jensen B. C., Haack K. R. (1987). Rearrangement of duplicated DNA in specialized cells of Neurospora. Cell 51, 741–752. doi: 10.1016/0092-8674(87)90097-3
Sharma P., Parakh S. K., Singh S. P., Parra-Saldivar R., Kim S. H., Varjani S., et al. (2022). A critical review on microbes-based treatment strategies for mitigation of toxic pollutants. Sci. Total Environ. 834, 155444. doi: 10.1016/j.scitotenv.2022.155444
Shenouda M. L., Ambilika M., Skellam E., Cox R. J. (2022). Heterologous expression of secondary metabolite genes in Trichoderma reesei for waste valorization. J. Fungi (Basel) 8(4):355. doi: 10.3390/jof8040355
Shenouda M. L., Cox R. J. (2021). Molecular methods unravel the biosynthetic potential of Trichoderma species. RSC Adv. 11, 3622–3635. doi: 10.1039/D0RA09627J
Shoresh M., Harman G. E., Mastouri F. (2010). Induced systemic resistance and plant responses to fungal biocontrol agents. Annu. Rev. Phytopathol. 48, 21–43. doi: 10.1146/annurev-phyto-073009-114450
Singh A., Taylor L. E. 2nd, Vander Wall T. A., Linger J., Himmel M. E., Podkaminer K., et al. (2015). Heterologous protein expression in Hypocrea jecorina: a historical perspective and new developments. Biotechnol. Adv. 33, 142–154. doi: 10.1016/j.biotechadv.2014.11.009
Son M., Yu J., Kim K. H. (2015). Five questions about mycoviruses. PloS Pathog. 11, e1005172. doi: 10.1371/journal.ppat.1005172
Sood M., Kapoor D., Kumar V., Sheteiwy M. S., Ramakrishnan M., Landi M., et al. (2020). Trichoderma: the "secrets" of a multitalented biocontrol agent. Plants (Basel) 9(6):762. doi: 10.3390/plants9060762
Stappler E., Dattenböck C., Tisch D., Schmoll M. (2017). Analysis of light- and carbon-specific transcriptomes implicates a class of G-protein-coupled receptors in cellulose sensing. mSphere 2, e00089–e00017. doi: 10.1128/mSphere.00089-17
Stoddard B. L. (2014). Homing endonucleases from mobile group I introns: discovery to genome engineering. Mob DNA 5, 7. doi: 10.1186/1759-8753-5-7
Stracquadanio C., Luz C., La Spada F., Meca G., Cacciola S. O. (2021). Inhibition of mycotoxigenic fungi in different vegetable matrices by extracts of Trichoderma species. J. Fungi (Basel) 7(6), 445. doi: 10.3390/jof7060445
Sun X., Cai Y. S., Yuan Y., Bian G., Ye Z., Deng Z., et al. (2019). Genome mining in Trichoderma viride J1-030: discovery and identification of novel sesquiterpene synthase and its products. Beilstein J. Org Chem. 15, 2052–2058. doi: 10.3762/bjoc.15.202
Tamizi A. A., Mat-Amin N., Weaver J. A., Olumakaiye R. T., Akbar M. A., Jin S., et al. (2022). Genome sequencing and analysis of Trichoderma (Hypocreaceae) isolates exhibiting antagonistic activity against the papaya dieback pathogen, Erwinia mallotivora. J. Fungi (Basel) 8(3), 246. doi: 10.3390/jof8030246
Taylor J. T., Harting R., Shalaby S., Kenerley C. M., Braus G. H., Horwitz B. A. (2022). Adhesion as a focus in Trichoderma-root interactions. J. Fungi (Basel) 8(4), 372. doi: 10.3390/jof8040372
Taylor J. T., Wang K.-D., Horwitz B., Kolomiets M., Kenerley C. (2021). Early transcriptome response of Trichoderma virens to colonization of maize roots. Front. Fungal Biol. 2. doi: 10.3389/ffunb.2021.718557
Tisch D., Pomraning K. R., Collett J. R., Freitag M., Baker S. E., Chen C. L., et al. (2017). Omics analyses of Trichoderma reesei CBS999.97 and QM6a indicate the relevance of female fertility to carbohydrate-active enzyme and transporter levels. Appl. Environ. Microbiol. 83(22), e01578-17. doi: 10.1128/AEM.01578-17
Torigoi E., Henrique-Silva F., Escobar-Vera J., Carle-Urioste J. C., Crivellaro O., El-Dorry H., et al. (1996). Mutants of Trichoderma reesei are defective in cellulose induction, but not basal expression of cellulase-encoding genes. Gene 173, 199–203. doi: 10.1016/0378-1119(96)00219-3
Tripathi P., Singh P. C., Mishra A., Chaudhry V., Mishra S., Tripathi R. D., et al. (2013). Trichoderma inoculation ameliorates arsenic induced phytotoxic changes in gene expression and stem anatomy of chickpea (Cicer arietinum). Ecotoxicol Environ. Saf. 89, 8–14. doi: 10.1016/j.ecoenv.2012.10.017
Tsuji J., Frith M. C., Tomii K., Horton P. (2012). Mammalian NUMT insertion is non-random. Nucleic Acids Res. 40, 9073–9088. doi: 10.1093/nar/gks424
Tyskiewicz R., Nowak A., Ozimek E., Jaroszuk-Scisel J. (2022). Trichoderma: The current status of its application in agriculture for the biocontrol of fungal phytopathogens and stimulation of plant growth. Int. J. Mol. Sci. 23(4), 2329. doi: 10.3390/ijms23042329
Uchiyama T., Uchihashi T., Nakamura A., Watanabe H., Kaneko S., Samejima M., et al. (2020). Convergent evolution of processivity in bacterial and fungal cellulases. Proc. Natl. Acad. Sci. U.S.A. 117, 19896–19903. doi: 10.1073/pnas.2011366117
van Bohemen A. I., Ruiz N., Zalouk-Vergnoux A., Michaud A., Robiou du Pont T., Druzhinina I., et al. (2021). Pentadecaibins I-V: 15-residue peptaibols produced by a marine-derived Trichoderma sp. of the Harzianum clade. J. Nat. Prod. 84 (4), 1271–1282. doi: 10.1021/acs.jnatprod.0c01355
van Wyk S., Wingfield B. D., De Vos L., van der Merwe N. A., Steenkamp E. T. (2020). Genome-wide analyses of repeat-induced point mutations in the ascomycota. Front. Microbiol. 11, 622368. doi: 10.3389/fmicb.2020.622368
Venice F., Davolos D., Spina F., Poli A., Prigione V. P., Varese G. C., et al. (2020). Genome sequence of Trichoderma lixii MUT3171, a promising strain for mycoremediation of PAH-contaminated sites. Microorganisms 8(9), 1258. doi: 10.3390/microorganisms8091258
Verschoor J. A., Kusumawardhani H., Ram A. F. J., De Winde J. H. (2022). Toward microbial recycling and upcycling of plastics: prospects and challenges. Front. Microbiol. 13, 821629. doi: 10.3389/fmicb.2022.821629
Vicente I., Baroncelli R., Morán-Diez M. E., Bernardi R., Puntoni G., Hermosa R., et al. (2020). Combined comparative genomics and gene expression analyses provide insights into the terpene synthases inventory in Trichoderma. Microorganisms 8 (10), 1603. doi: 10.3390/microorganisms8101603
Vieira A. A., Vianna G. R., Carrijo J., Aragão F. J. L., Vieira P. M. (2021). Generation of Trichoderma harzianum with pyr4 auxotrophic marker by using the CRISPR/Cas9 system. Sci. Rep. 11 (1), 1085. doi: 10.1038/s41598-020-80186-4
VÍglaš J., Dobiasova S., Viktorova J., Ruml T., Repiska V., Olejnikova P., et al. (2021). Peptaibol-containing extracts of Trichoderma atroviride and the fight against resistant microorganisms and cancer cells. Molecules 26(19), 6025. doi: 10.3390/molecules26196025
Vignolle G. A., Mach R. L., Mach-Aigner A. R., Derntl C. (2020). Novel approach in whole genome mining and transcriptome analysis reveal conserved RiPPs in Trichoderma spp. BMC Genomics 21, 258. doi: 10.1186/s12864-020-6653-6
Vignolle G. A., Schaffer D., Zehetner L., Mach R. L., Mach-Aigner A. R., Derntl C. (2021). FunOrder: A robust and semi-automated method for the identification of essential biosynthetic genes through computational molecular co-evolution. PloS Comput. Biol. 17, e1009372. doi: 10.1371/journal.pcbi.1009372
Wallen R. M., Perlin M. H. (2018). An overview of the function and maintenance of sexual reproduction in dikaryotic fungi. Front. Microbiol. 9, 503. doi: 10.3389/fmicb.2018.00503
Wang Q., Coleman J. J. (2019). Progress and challenges: Development and implementation of CRISPR/Cas9 technology in filamentous fungi. Comput. Struct. Biotechnol. J. 17, 761–769. doi: 10.1016/j.csbj.2019.06.007
Wang H., Sivonen K., Fewer D. P. (2015b). Genomic insights into the distribution, genetic diversity and evolution of polyketide synthases and nonribosomal peptide synthetases. Curr. Opin. Genet. Dev. 35, 79–85. doi: 10.1016/j.gde.2015.10.004
Wang G., Wang H., Xiong X., Chen S., Zhang D. (2015a). Mitochondria thioredoxin's backup role in oxidative stress resistance in Trichoderma reesei. Microbiol. Res. 171, 32–38. doi: 10.1016/j.micres.2015.01.005
Wang Q., Zhao Q., Liu Q., He X., Zhong Y., Qin Y., et al. (2021). CRISPR/Cas9-mediated genome editing in Penicillium oxalicum and Trichoderma reesei using 5S rRNA promoter-driven guide RNAs. Biotechnol. Lett. 43, 495–502. doi: 10.1007/s10529-020-03024-7
Wilson D. B. (2009). Cellulases and biofuels. Curr. Opin. Biotechnol. 20, 295–299. doi: 10.1016/j.copbio.2009.05.007
Wu C., Chen Y., Qiu Y., Niu X., Zhu N., Chen J., et al. (2020). A simple approach to mediate genome editing in the filamentous fungus Trichoderma reesei by CRISPR/Cas9-coupled in vivo gRNA transcription. Biotechnol. Lett. 42, 1203–1210. doi: 10.1007/s10529-020-02887-0
Wu B., Hao W. (2019). Mitochondrial-encoded endonucleases drive recombination of protein-coding genes in yeast. Environ. Microbiol. 21, 4233–4240. doi: 10.1111/1462-2920.14783
Wu M., Jin F., Zhang J., Yang L., Jiang D., Li G. (2012). Characterization of a novel bipartite double-stranded RNA mycovirus conferring hypovirulence in the phytopathogenic fungus Botrytis porri. J. Virol. 86, 6605–6619. doi: 10.1128/JVI.00292-12
Xie J., Jiang D. (2014). New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu. Rev. Phytopathol. 52, 45–68. doi: 10.1146/annurev-phyto-102313-050222
Xiong Y., Sun J., Glass N. L. (2014). VIB1, a link between glucose signaling and carbon catabolite repression, is essential for plant cell wall degradation by Neurospora crassa. PloS Genet. 10, e1004500. doi: 10.1371/journal.pgen.1004500
Yadav A. N., Kour D., Kaur T., Devi R., Yadav A. (2022). Endophytic fungal communities and their biotechnological implications for agro-environmental sustainability. Folia Microbiol. (Praha) 67, 203–232. doi: 10.1007/s12223-021-00939-0
You J., Zhou K., Liu X., Wu M., Yang L., Zhang J., et al. (2019). Defective RNA of a novel mycovirus with high transmissibility detrimental to biocontrol properties of Trichoderma spp. Microorganisms 7(11), 507. doi: 10.3390/microorganisms7110507
Yun S. H., Lee S. H., So K. K., Kim J. M., Kim D. H. (2016). Incidence of diverse dsRNA mycoviruses in Trichoderma spp. causing green mold disease of shiitake Lentinula edodes. FEMS Microbiol. Lett. 363(19), fnw220. doi: 10.1093/femsle/fnw220
Zafra G., Cortes-Espinosa D. V. (2015). Biodegradation of polycyclic aromatic hydrocarbons by Trichoderma species: a mini review. Environ. Sci. pollut. Res. Int. 22, 19426–19433. doi: 10.1007/s11356-015-5602-4
Zeilinger S., Schmoll M., Pail M., Mach R. L., Kubicek C. P. (2003). Nucleosome transactions on the Hypocrea jecorina ( Trichoderma reesei) cellulase promoter cbh2 associated with cellulase induction. Mol. Genet. Genomics 270, 46–55. doi: 10.1007/s00438-003-0895-2
Zhang H., Xie J., Fu Y., Cheng J., Qu Z., Zhao Z., et al. (2020). A 2-kb mycovirus converts a pathogenic fungus into a beneficial endophyte for Brassica protection and yield enhancement. Mol. Plant 13, 1420–1433. doi: 10.1016/j.molp.2020.08.016
Zhang T., Zeng X., Cai X., Liu H., Zeng Z. (2018). Molecular characterization of a novel double-stranded RNA mycovirus of Trichoderma asperellum strain JLM45-3. Arch. Virol. 163, 3433–3437. doi: 10.1007/s00705-018-3988-2
Zhou Y., Wang Y., Chen K., Wu Y., Hu J., Wei Y., et al. (2020). Near-complete genomes of two Trichoderma species: A resource for biological control of plant pathogens. Mol. Plant Microbe Interact. 33, 1036–1039. doi: 10.1094/MPMI-03-20-0076-A
Zou G., Xiao M., Chai S., Zhu Z., Wang Y., Zhou Z. (2021). Efficient genome editing in filamentous fungi via an improved CRISPR-Cas9 ribonucleoprotein method facilitated by chemical reagents. Microb. Biotechnol. 14, 2343–2355. doi: 10.1111/1751-7915.13652
Keywords: Trichoderma, Hypocrea, evolution, horizontal gene transfer, repeat induced point mutation, mycovirus, bioremediation, biocontrol
Citation: Schalamun M and Schmoll M (2022) Trichoderma – genomes and genomics as treasure troves for research towards biology, biotechnology and agriculture. Front. Fungal Biol. 3:1002161. doi: 10.3389/ffunb.2022.1002161
Received: 24 July 2022; Accepted: 25 August 2022;
Published: 14 September 2022.
Edited by:
Laszlo G. Nagy, Hungarian Academy of Sciences (MTA), HungaryReviewed by:
Enrique Monte, University of Salamanca, SpainFeng Marc Cai, Sun Yat-sen University, China
Copyright © 2022 Schalamun and Schmoll. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Monika Schmoll, bW9uaWthLnNjaG1vbGxAdW5pdmllLmFjLmF0
 Miriam Schalamun1
Miriam Schalamun1 Monika Schmoll
Monika Schmoll