- 1Laboratory of Applied Microbiology, Department of Microbial Biotechnology, Graduate School of Agricultural Science, Tohoku University, Sendai, Japan
- 2Laboratory of Filamentous Mycoses, Department of Fungal Infection, National Institute of Infectious Diseases, Tokyo, Japan
- 3Laboratory of Environmental Interface Technology of Filamentous Fungi, Graduate School of Agriculture, Kyoto University, Kyoto, Japan
- 4ABE-Project, New Industry Creation Hatchery Center, Tohoku University, Sendai, Japan
- 5Faculty of Food and Agricultural Sciences, Fukushima University, Fukushima, Japan
- 6Department of Biochemical Engineering, Graduate School of Engineering, Yamagata University, Yonezawa, Japan
- 7Food Microbiology Unit, School of Food and Agricultural Sciences, Miyagi University, Sendai, Japan
- 8Genome Biotechnology Laboratory, Kanazawa Institute of Technology, Hakusan, Japan
- 9Department of Applied Life Science, The United Graduate School of Agricultural Science, Tokyo University of Agriculture and Technology, Fuchu, Japan
- 10Department of Microbial Resources, Graduate School of Agricultural Science, Tohoku University, Sendai, Japan
α-1,3-Glucan is one of the main polysaccharides in the cell wall of Aspergillus nidulans. We previously revealed that it plays a role in hyphal aggregation in liquid culture, and that its molecular mass (MM) in an agsA-overexpressing (agsAOE) strain was larger than that in an agsB-overexpressing (agsBOE) strain. The mechanism that regulates its MM is poorly understood. Although the gene amyD, which encodes glycosylphosphatidylinositol (GPI)-anchored α-amylase (AmyD), is involved in the biosynthesis of α-1,3-glucan in A. nidulans, how it regulates this biosynthesis remains unclear. Here we constructed strains with disrupted amyD (ΔamyD) or overexpressed amyD (amyDOE) in the genetic background of the ABPU1 (wild-type), agsAOE, or agsBOE strain, and characterized the chemical structure of α-1,3-glucans in the cell wall of each strain, focusing on their MM. The MM of α-1,3-glucan from the agsBOE amyDOE strain was smaller than that in the parental agsBOE strain. In addition, the MM of α-1,3-glucan from the agsAOE ΔamyD strain was greater than that in the agsAOE strain. These results suggest that AmyD is involved in decreasing the MM of α-1,3-glucan. We also found that the C-terminal GPI-anchoring region is important for these functions.
Introduction
The fungal cell wall, composed mainly of polysaccharides, is essential for the survival of the fungus (Latgè et al., 2017). It has recently been understood that the cell wall is a highly dynamic structure; cell-wall components are synthesized by synthases and then reconstructed by glycosyltransferases to form a proper cell-wall architecture (Latgè and Beauvais, 2014; Latgè et al., 2017). The cell wall of filamentous fungi contains α-glucans, β-glucans, chitin, and galactomannan. Some fungi form an extracellular matrix, which includes secretory polysaccharides such as galactosaminogalactan (Sheppard and Howell, 2016; Yoshimi et al., 2016; Miyazawa et al., 2019). Cell-wall polysaccharides of some Aspergillus species can be fractionated into alkali-soluble and alkali-insoluble fractions (Fontaine et al., 2000; Yoshimi et al., 2013; Dichtl et al., 2015; Zhang et al., 2017b). The alkali-soluble fraction contains mainly α-1,3-glucan with interconnecting α-1,4-linkage and some galactomannan (Bernard and Latge, 2001; Latgè, 2010). The alkali-insoluble fraction is composed of chitin, β-1,6-branched β-1,3-glucan, and galactomannan (Fontaine et al., 2000; Bernard and Latge, 2001). Recently, Kang et al. (2018) investigated the molecular architecture of the cell wall in Aspergillus fumigatus using solid-state NMR and suggested that the structure of the cell wall consists of a rigid inner domain and a highly mobile outer shell. Even more recently, Chakraborty et al. (2021) reported that α-1,3-glucan of A. fumigatus is present in both alkali-soluble and -insoluble fractions.
In the human pathogenic dimorphic yeast Histoplasma capsulatum and the rice blast fungus Magnaporthe grisea, α-1,3-glucan functions as a stealth factor that prevents host immune recognition and consequently contributes to the establishment of invasion or infection (Rappleye et al., 2004, 2007; Fujikawa et al., 2009, 2012). In addition, the pathogenesis of an α-1,3-glucan-deficient strain is decreased in murine models infected with A. fumigatus (Henry et al., 2012; Beauvais et al., 2013). Recently, α-1,3-glucan was reported to stimulate the polarization of regulatory T-cells by inducing programmed death-ligand 1 expression on human dendritic cells (Stephen-Victor et al., 2017). Fontaine et al. (2010) revealed that α-1,3-glucan has adhesivity when the conidia of A. fumigatus germinate.
Grün et al. (2005) analyzed the detailed chemical structure of α-glucan in the cell wall of the fission yeast Schizosaccharomyces pombe and found that its molecular mass (MM) is 42,600 ± 5,200, which is equivalent to a degree of polymerization of 263 ± 32 (Grün et al., 2005). The α-glucans derived from S. pombe are composed of two chains of ≈120 residues of 1,3-linked α-glucose with 12 residues of 1,4-linked α-glucose at the reducing ends (Grün et al., 2005). In Aspergillus wentii, the water-insoluble (alkali-soluble) glucan has a MM of ≈850,000 and consists of 25 subunits (200 residues each) of α-1,3-glucan separated by short spacers composed of 1,4-linked α-glucan (Choma et al., 2013).
Aspergillus species have several α-1,3-glucan synthase genes: two in Aspergillus nidulans (agsA and agsB), three in A. fumigatus (AGS1–3) and Aspergillus oryzae (agsA–C), and five in Aspergillus niger (agsA–E). Disruptants of A. fumigatus that lack a single gene or all three genes have been constructed (Beauvais et al., 2005; Maubon et al., 2006; Henry et al., 2012); these strains lack α-1,3-glucan in the cell wall and are less pathogenic (Beauvais et al., 2013). In A. oryzae, agsB (orthologous to A. nidulans agsB) is the primary α-1,3-glucan synthase gene (Zhang et al., 2017b). An A. oryzae disruptant lacking all three genes loses its cell-wall α-1,3-glucan and forms small hyphal pellets under liquid culture conditions (Miyazawa et al., 2016). In A. niger, the expression of agsA (orthologous to A. fumigatus AGS3; no ortholog in A. nidulans) and agsE (orthologous to A. nidulans agsB) is upregulated in the presence of stress-inducing compounds in the cell wall (Damveld et al., 2005). In the kuro (black) koji mold Aspergillus luchuensis, disruption of agsE (orthologous to A. nidulans agsB) improves the protoplast formation (Tokashiki et al., 2019). Recently Uechi et al. revealed that A. luchuensis agsB (no ortholog in A. nidulans) plays a role in nigeran synthesis (Uechi et al., 2021). In A. nidulans, α-1,3-glucan in vegetative hyphae is synthesized mainly by AgsB (Yoshimi et al., 2013; He et al., 2014). The hyphae of a mutant deficient in α-1,3-glucan became fully dispersed, showing that α-1,3-glucan is a hyphal aggregation factor (Yoshimi et al., 2013; He et al., 2014). We recently constructed strains overexpressing agsA (agsAOE) and agsB (agsBOE) in the genetic background of, respectively, agsB and agsA disruptants. The peak MM of alkali-soluble glucan from agsAOE was 1,480,000 ± 80,000, which was four times that from the agsBOE (MM, 372,000 ± 47,000) (Miyazawa et al., 2018). The alkali-soluble glucan derived from these strains contains several 1,4-linked spacer structures interlinking the α-1,3-glucan subunits, which each contain 200 glucose residues (Miyazawa et al., 2018).
Outside of A. fumigatus, A. nidulans agsB and its orthologs are clustered with two α-amylase-encoding genes (amyD and amyG in A. nidulans) (He et al., 2014; Yoshimi et al., 2017; Miyazawa et al., 2020). The amyG gene encodes an intracellular α-amylase and is crucial for α-1,3-glucan synthesis (He et al., 2014). The amyD gene in A. nidulans encodes glycosylphosphatidylinositol (GPI)-anchored α-amylase. He et al. (2014) reported that α-1,3-glucan contents increased by 50% in an amyD-disrupted (ΔamyD) strain and halved in an amyD-overexpressing (actA(p)-amyD) strain, suggesting that amyD has a repressive effect on α-1,3-glucan synthesis. In addition, He et al. (2017) analyzed the chronological changes of α-1,3-glucan contents under liquid culture conditions. Whereas, the amount of α-1,3-glucan in strains that overexpressed the α-1,3-glucanase-encoding gene (mutA or agnB) was decreased after 20 h from inoculation, the amount of α-1,3-glucan in the cell wall of the amyDOE strain was half that of the wild-type strain from the initial stage of cultivation (He et al., 2017). He et al. (2017) suggested that AmyD decreased the amount of α-1,3-glucan in the cell wall by a mechanism independent of the effect of α-1,3-glucanase. The enzymatic characteristics of A. niger AgtA, which is encoded by an ortholog of A. nidulans amyD, have been reported (Van Der Kaaij et al., 2007). Although AgtA in A. niger barely hydrolyzed α-1,3-glucan, it had relatively high transglycosylation activity on donor substrates with maltooligosaccharides (Van Der Kaaij et al., 2007). Overall, AmyD seems to indirectly decrease the amount of α-1,3-glucan in the cell wall, but the detailed mechanism is still unknown.
Here, in a study of the function of amyD in α-1,3-glucan biosynthesis in A. nidulans, we constructed strains with overexpression or disruption of amyD in the genetic backgrounds of the wild-type, agsAOE, and agsBOE. We performed several chemical analyses of α-1,3-glucan derived from the strains, looking in particular at its MM, and examined the role of amyD in controlling the MM of α-1,3-glucan in the cell wall.
Materials and Methods
Strains and Growth Media
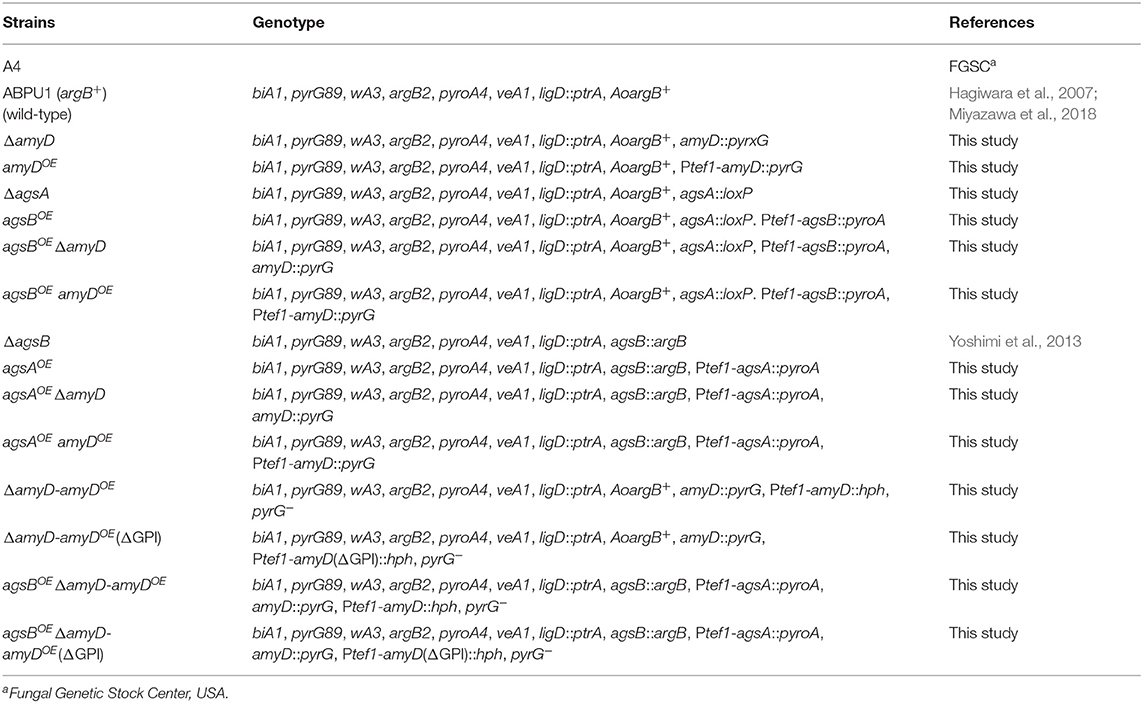
Strains are listed in Table 1. Czapek-Dox (CD) medium was used as the standard culture, as described previously (Fujioka et al., 2007; Miyazawa et al., 2018).
Construction of the agsA- and agsB-Overexpressing Strains
We newly constructed agsAOE and agsBOE strains for this study. To generate agsAOE, pAPyT-agsA plasmids (Miyazawa et al., 2018) were digested with NotI and transformed into a disrupted agsB (ΔagsB) strain (Supplementary Figure 1A). Correct integration of agsA overexpression cassettes was confirmed by PCR (Supplementary Figure 1B). To generate agsBOE, the disrupted agsA (ΔagsA) strain was first generated using the Cre/loxP marker recycling system (Zhang et al., 2017a). The pAPG-cre/DagsA plasmid (Miyazawa et al., 2018) was digested with EcoRI and transformed into the ABPU1 (argB+) strain. Candidate strains were selected on CD medium without uridine and uracil, and then cultured on CD medium with uridine and uracil and 1% xylose to induce Cre expression (Supplementary Figure 1C). Strains that required uridine and uracil were isolated, and then replacement of the agsA gene was confirmed by PCR (Supplementary Figure 1D). The pAPyT-agsB plasmid was digested with NotI and transformed into the ΔagsA strain (Supplementary Figure 1E). Correct integration of agsB overexpression cassettes was confirmed by PCR (Supplementary Figure 1B).
Construction of the amyDOE Strain
The amyDOE strain was constructed by replacing the native promoter with the constitutive tef1 promoter. The sequences of the primers are listed in Supplementary Table 1. To generate amyDOE, the plasmid pAPT-amyD was constructed (Supplementary Figure 2A). The 5′-non-coding region (amplicon 1) and the coding region (amplicon 2) of amyD were amplified from A. nidulans ABPU1 genomic DNA. The pyrG marker (amplicon 3) was amplified from the pAPG-cre/DagsA plasmid. The tef1 promoter (amplicon 4) was amplified from the pAPyT-agsB plasmid. The four amplicons and a SacI-digested pUC19 vector were fused using an In-Fusion HD Cloning Kit (Clontech Laboratories, Inc., Mountain View, CA, USA). The resulting plasmid was digested with SacI, and transformed into the ABPU1 (argB+), agsAOE, and agsBOE strains (Supplementary Figure 2B). Correct integration of the cassette was confirmed by PCR (Supplementary Figure 2C).
Disruption of the amyD Gene
In the first round of PCR, gene fragments containing the 5′-non-coding region (amplicon 1) and the coding region (amplicon 2) of amyD were amplified from ABPU1 genomic DNA, and the pyrG gene (amplicon 3) was amplified from A. oryzae genomic DNA (Supplementary Figure 2D). The three resulting fragments were gel-purified and fused into a disruption cassette in the second round of PCR. The resulting PCR product was gel-purified and transformed into the ABPU1 (argB+), agsAOE, and agsBOE strains (Supplementary Figure 2E). Replacement of the amyD gene was confirmed by PCR (Supplementary Figure 2F).
Expression of Complementary amyD Genes
The sequences of the primers are listed in Supplementary Table 1. A GPI-anchor modification site, the ω-site, was predicted with the GPI Prediction Server v. 3.0 (https://mendel.imp.ac.at/gpi/gpi_server.html), and the best score for the ω-site was Asn535 of AmyD. To remove the GPI anchor of AmyD, 54 nucleotides corresponding to the 18 amino acid residues from Asn535 in AmyD were deleted from the authentic amyD gene (Supplementary Figure 3A). To create complementary genes that have full-length open reading frames of either amyD or the gene without the GPI anchor–coding region, the plasmids pAHT-amyD, pAHdPT-amyD, pAHT-amyD(ΔGPI), and pAHdPT-amyD(ΔGPI) were first constructed (Supplementary Figure 3A). To construct pAHT-amyD, primers IF-Ptef1-hph-Fw and IF-amyD-up-hph-Rv were amplified by PCR using pAPT-amyD as a template (amplicon 1). The hygromycin-resistance gene hph (amplicon 2) was amplified with primers 397–5 and 397–3 from pSK397 (Krappmann et al., 2006). The two amplicons were fused using a NEBuilder HiFi DNA Assembly kit (New England Biolabs, Ipswich, MA, USA) according to the manufacturer's instructions. Then, to delete the GPI anchor–encoding region of amyD, PCR amplification was performed with primers ANamyD-dGPI-Fw and ANamyD-dGPI-Rv from the resulting pAHT-amyD plasmid with PrimeSTAR Max DNA Polymerase (Takara Bio Inc., Kusatsu, Japan). The amplified fragment was transformed into DH5α competent cells, and the pAHT-amyD(ΔGPI) plasmid was obtained (Supplementary Figure 3A). To construct pAHdPT-amyD, the first half (amplicon 1) and the second half (amplicon 2) of pyrG were amplified from A. oryzae genomic DNA. The fragment containing hph, tef1 promoter, and amyD (amplicon 3) was amplified from pAHT-amyD. The three amplicons were fused using a NEBuilder kit. For pAHdPT-amyD(ΔGPI) construction, the fragment containing hph, tef1 promoter, and amyD lacking its GPI anchor–coding region (amplicon 3′) was amplified from pAHT-amyD(ΔGPI). The three amplicons and the SacI-digested pUC19 vector were fused using an In-Fusion HD Cloning Kit (Supplementary Figure 3B). The pAHdPT-amyD and pAHdPT-amyD(ΔGPI) plasmids were digested with SacI and transformed into the ΔamyD and agsBOE ΔamyD strains (Supplementary Figure 3C). Correct integration of the cassettes was confirmed by PCR (Supplementary Figure 3D).
RNA Extraction and Quantitative Real-Time PCR
Mycelial cells cultured in CD liquid medium for 24 h were collected, and total RNA was extracted from the cells by using Sepasol-RNA I Super G (Nakalai Tesque, Kyoto, Japan) in accordance with the manufacturer's instruction. The total RNA (2.5 μg) was reverse-transcribed by using a SuperScript IV VILO Master Mix with ezDNase Enzyme (Invitrogen, Carlsbad, CA, United States). Quantitative real-time PCR was performed with a Mx3000P (Agilent Technologies, Santa Clara, CA, United States) with SYBR Green detection. For reaction mixture preparation, Thunderbird Next SYBR qPCR Mix (Toyobo Co., Ltd., Osaka, Japan) was used. Primers used for quantitative PCR are listed in Supplementary Table 1. An equivalent amount of cDNA, obtained from reverse transcription reactions using an equivalent amount of total RNA, was applied to each reaction mixture. The gene encoding histone H2B was used as a normalization reference (an internal control) for determining the target gene expression ratios.
Delipidization and Fractionation of Mycelial Cells
Cell walls were fractionated as previously described with some modification (Miyazawa et al., 2018). Mycelia cultured for 24 h in CD medium were collected by filtering through Miracloth (Merck Millipore, Darmstadt, Germany), washed with water, and freeze-dried. The mycelia were then pulverized in a MM400 bench-top mixer mill (Retch, Haan, Germany). The powder (1 g) was suspended in 25 mL of chloroform–methanol (3:1 vol/vol) and stirred at room temperature for 12 h to remove the total polar lipid content of the mycelial cells. The mixture was centrifuged (10,000× g, 10 min). The residue was suspended in chloroform–methanol, and the delipidizing procedure was repeated. Then the de-polar lipid residue was suspended in 40 mL of 0.1 M Na phosphate buffer (pH 7.0), and cell-wall components were fractionated by hot-water and alkali treatments, as described previously (Miyazawa et al., 2018). Hot-water–soluble, alkali-soluble, and alkali-insoluble fractions were obtained from this fractionation, and the alkali-soluble fraction was further separated into a fraction soluble in water at neutral pH (AS1) and an insoluble fraction (AS2). The monosaccharide composition of AS2 fractions was quantified according to Miyazawa et al. (2018).
To obtain mycelia cultured for 16 h, conidia (final conc. 5.0 × 105/mL) were inoculated into 200 mL CD medium and rotated at 160 rpm at 37°C. The mycelia were collected and fractionated as described above.
13C NMR Analysis
The AS2 fraction of each strain (50 mg) was suspended in 1 mL of 1 M NaOH/D2O and dissolved by vortexing. One drop of DMSO-d6 (deuterated dimethyl sulfoxide) was then added to each fraction and the solutions were centrifuged (3,000 × g, 5 min) to remove insoluble debris. 13C NMR spectra of the supernatants were obtained using a JNM-ECX400P spectrometer (JEOL, Tokyo, Japan) at 400 MHz at 35°C (72,000 scans). Chemical shifts were recorded relative to the resonance of DMSO-d6.
Determination of the Average Molecular Mass of Alkali-Soluble Glucan
The MM of alkali-soluble glucan was determined by gel permeation chromatography (GPC) according to the methods of Puanglek et al. (2016), with some modification. A GPC-101 system (Showa Denko Co. Ltd., Tokyo, Japan) with an ERC-3125S degasser (Showa Denko) and an RI-71S refractive index detector (Showa Denko) was used for the measurement. It was fitted with a GPC KD-G 4A guard column (Showa Denko) and a GPC KD-805 column (8.0 × 300 mm; Showa Denko). The eluent was 20 mM LiCl in N, N-dimethylacetamide (DMAc), and the flow rate was 0.6 mL/min at 40°C. The detector was normalized with polystyrene standards (SM-105; Showa Denko). With SmartChrom software (Jasco, Tokyo, Japan), the GPC profile was divided into virtual time slices (ni) with the height of each virtual slice from the base line (Hi) corresponding to a certain MM (Mi) obtained by calibrating the column. From these values, the number-average MM (Mn) and weight-average MM (Mw) were calculated as:
Polydispersity was calculated as Mw/Mn.
Smith Degradation
Smith degradation of the alkali-soluble glucan was performed as described (Miyazawa et al., 2018). In brief, the AS2 fraction (20 mg) was suspended in 0.1 M acetate buffer (pH 3.9), oxidized with sodium periodate, reduced with sodium borohydride, hydrolyzed with trifluoroacetic acid, and freeze-dried. These procedures resulted in selective hydrolyzing of the 1,4-linked glucose residues, which contain vicinal hydroxyl groups, but not the 1,3-glucose residues in alkali-soluble glucan. The Smith-degraded sample was dissolved in DMAc containing LiCl for GPC analysis.
Fluorescent Labeling of Cell-Wall Polysaccharides
Mycelial cells cultured for 16 h in CD liquid medium were dropped on a glass slide and dried at 55°C for 15 min. The cells were fixed and labeled with α-1,3-glucan-binding domain-fused green fluorescent protein (AGBD-GFP) (Suyotha et al., 2013) for α-1,3-glucan, fluorophore-labeled antibody for β-1,3-glucan, and fluorophore-labeled lectin for chitin. The cells were then imaged by confocal scanning microscopy as described (Miyazawa et al., 2018). Enzymatic digestion of β-1,3-glucan in the hyphal cells was performed as described (Miyazawa et al., 2018).
Western Blotting
Culture broth in CD liquid medium incubated for 24 h was filtered through Miracloth. Proteins in the supernatant were precipitated with trichloroacetic acid, separated by SDS-PAGE, and then transferred onto a polyvinylidene difluoride membrane. The membrane was blocked with the polyvinylidene difluoride blocking reagent for Can Get Signal (Toyobo). Antibodies of rabbit IgG against AmyD were developed with synthesized peptides (NH2-C+SGERAGELDVPMSK-COOH) (Eurofins Genomics, Tokyo, Japan) and used as the primary antibody, diluted with Can Get Signal (Toyobo). Antibody binding was visualized using a horseradish peroxidase–conjugated goat anti-rabbit IgG secondary antibody (Pierce Biotechnology, Rockford, IL, USA) and an ImmunoStar LD chemiluminescent substrate (Fujifilm Wako Pure Chemical Corp., Osaka, Japan).
Assay for Glucosyltransferase Activity
Supernatant obtained from CD liquid medium cultured for 24 h was concentrated 167 times and buffer-exchanged to 10 mM Tris·HCl (pH 8.0) in Nanosep Centrifugal Devices 10K (Pall, Port Washington, NY, USA). A 20-μL mixture containing 1 mM para-nitrophenyl (pNP)-α-maltopentaoside and 1 mM acarbose (Fujifilm Wako Pure Chemical Corp.), and 5 μL of the concentrated culture supernatant in 50 mM acetic acid/sodium acetate buffer (pH 5.5) was incubated for 20 min at 40°C. Samples (3 μL) were withdrawn from the reaction mixture and immediately inactivated by adding 40 μL of methanol. pNP-α-Maltopentaoside was prepared by following the method of Usui and Murata (1988). Then, 157 μL of water was added to each sample solution, which was analyzed by HPLC with a Jasco Intelligent System liquid chromatograph (Jasco). The bound material was eluted with 20% methanol at a flow rate of 1.0 mL/min at 40°C. The elution profiles were detected at 300 nm with a Unison UK C-18 column (4.6 × 250 mm, Imtakt, Kyoto, Japan).
Results
Characterization of Strains With Disrupted or Overexpressed amyD
We constructed amyDOE and ΔamyD strains by introducing the amyD cassettes for overexpression and disruption into the wild-type, agsAOE, and agsBOE strains (Supplementary Figure 2). The expression level of amyD in each strain was quantified in hyphal cells. Whereas, each disrupted strain (ΔamyD, agsAOE ΔamyD, and agsBOE ΔamyD) showed scarce amyD expression, each overexpressing strain (amyDOE, agsAOE amyDOE, and agsBOE amyDOE) showed significantly higher amyD expression than their parental strain (Figure 1).
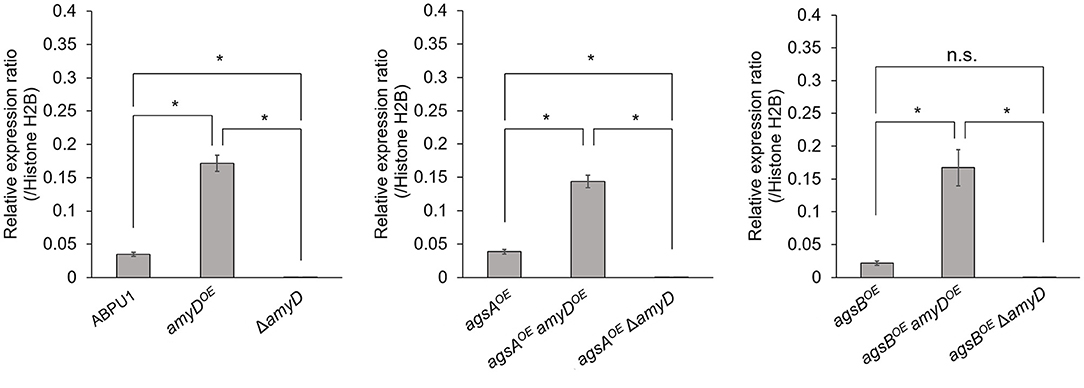
Figure 1. Transcript levels of amyD determined by quantitative PCR. Gene-specific primers are listed in Supplementary Table 1. Error bars represent the SEM calculated from three replicates. *Significant differences by Tukey's test (P < 0.05); n.s., not significant.
There was no significant difference in radial growth among the strains grown on agar plates for 5 days (Supplementary Figure 4). In liquid culture, the wild-type and ΔamyD strains formed tightly aggregated hyphal pellets; however, the hyphae of the amyDOE strain were almost fully dispersed (Figure 2). He et al. reported that the phenotype of their amyDOE strain resembles that of the ΔagsB strain in A. nidulans (He et al., 2014), which is consistent with our results (Figure 2). In agreement with our previous results (Miyazawa et al., 2018), the agsAOE and agsBOE strains formed, respectively, loosely and tightly aggregated pellets (Figure 2). Disruption of amyD did not affect the phenotypes of the agsAOE and agsBOE strains (Figure 2). Also, overexpression of amyD scarcely affected the phenotypes of the agsAOE and agsBOE strains (Figure 2).
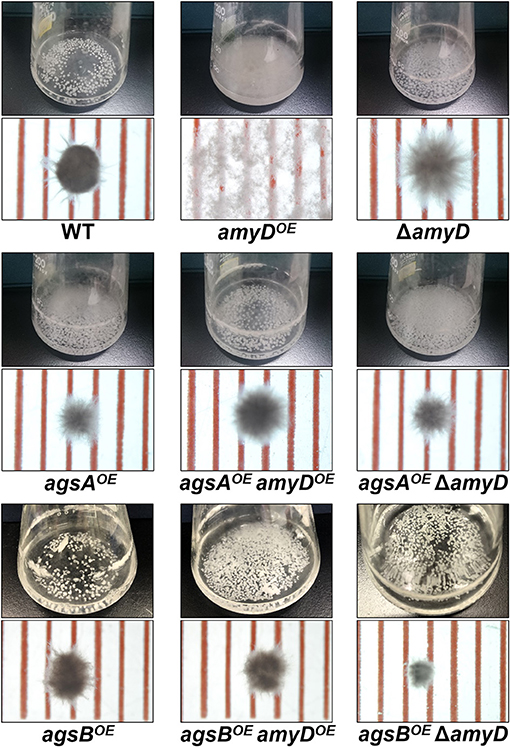
Figure 2. Growth characteristics of amyDOE and ΔamyD strains in liquid culture. Upper images, cultures in Erlenmeyer flasks; lower images, representative hyphal pellets of each strain under a stereomicroscope. Scale intervals are 1 mm.
Overexpression of amyD Resulted in a Decrease in Cell-Wall Alkali-Soluble Glucan
Cell-wall components of each strain were fractionated by a hot water–alkali treatment method, each fraction was weighed, and the monosaccharide composition of the AS2 fraction was quantified. The amount of glucose in the AS2 fraction was significantly lower in the amyDOE strain than in the wild-type strain (Figure 3; P < 0.05). That in the ΔamyD strain was similar to that in the wild-type strain (Figure 3). Those in the agsAOE amyDOE and agsAOE ΔamyD strains, which were constructed from the parental strain agsAOE, were almost the same (Figure 3). It was significantly lower in the agsBOE amyDOE strain than in the agsBOE and agsBOE ΔamyD strains (Figure 3; P < 0.05). These results indicate that AmyD acts to decrease the amount of alkali-soluble glucan in the wild-type and agsBOE strains, but not in the agsAOE strain, even when amyD is overexpressed.
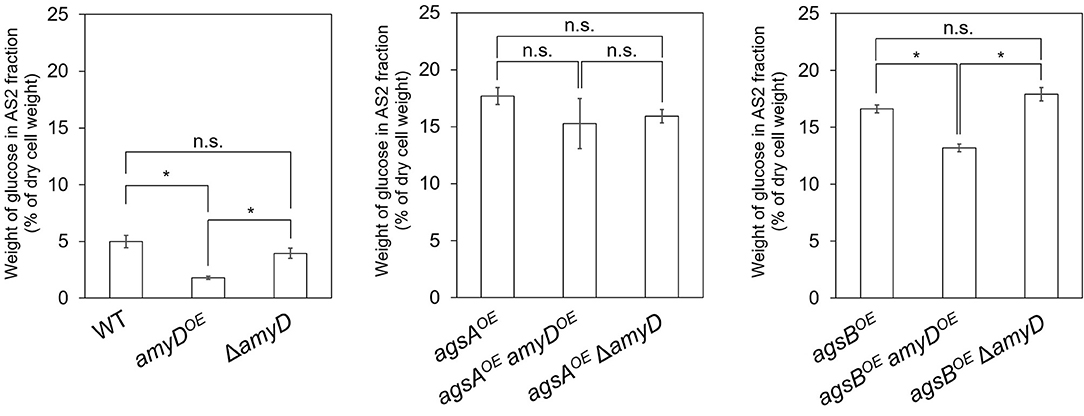
Figure 3. Amount of glucose in AS2 fractions. Conidia (5.0 × 105/mL) of each strain were inoculated into CD medium and rotated at 160 rpm at 37°C for 24 h. Values show glucose content of the AS2 fraction as a percentage of the total cell-wall weight. Error bars represent SEM calculated from three replicates. *Significant difference by Tukey's test (P < 0.05); n.s., not significant.
Overexpression of the amyD Gene Decreases the Molecular Mass of Alkali-Soluble Glucan
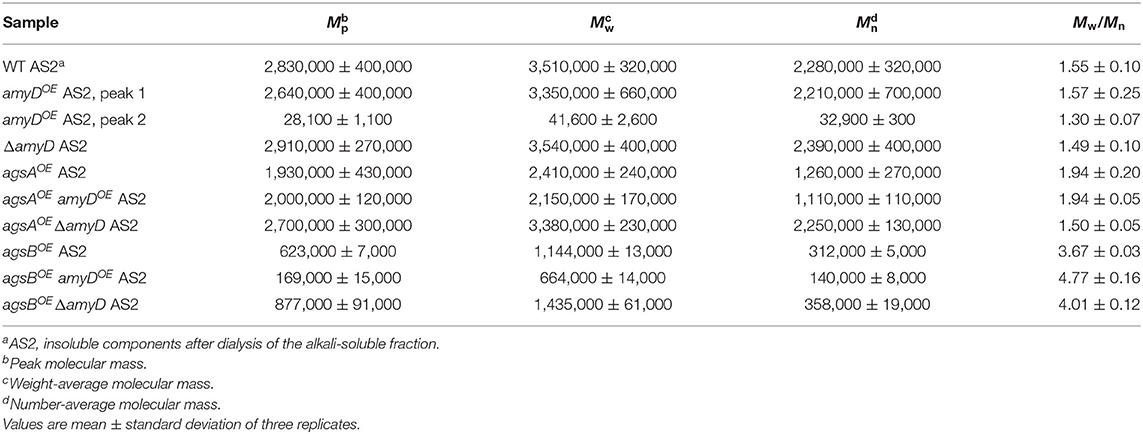
By 13C NMR analysis, the primary component in the AS2 fraction of the wild-type, amyDOE, and ΔamyD strains was found to be α-1,3-glucan, suggesting that amyD did not affect the primary components of alkali-soluble glucan (Supplementary Figure 5). To reveal whether the MM of alkali-soluble glucan was affected by disruption or overexpression of amyD, we determined the MM of alkali-soluble glucan in each strain by GPC analysis. Polystyrene (MM, 13,900–3,850,000) was used as a standard molecule to calibrate the column for size exclusion analysis. Although Mw is used to assess the physical properties of a polymer, the calculation of Mw favors molecules with a larger MM. Since Mn is the average of the MM values of the individual macromolecules, here we use Mn as the MM of alkali-soluble glucan. The Mn of the alkali-soluble glucan was 1,260,000 ± 270,000 in the agsAOE strain and 312,000 ± 5,000 in agsBOE strain (Figures 4A,B; Table 2), consistent with our previous results (Miyazawa et al., 2018). Although the Mn of alkali-soluble glucan in the agsAOE amyDOE strain (1,110,000 ± 110,000) was similar to that in the parental (agsAOE) strain, that of agsAOE ΔamyD was significantly greater (2,250,000 ± 130,000) than that of agsAOE (Figure 4A; Table 2; P < 0.05). In addition, the Mn of agsBOE amyDOE (140,000 ± 8,000) was significantly less than that of the parental (agsBOE) strain (Figure 4B; Table 2; P < 0.05). The Mn of alkali-soluble glucan in agsBOE ΔamyD (358,000 ± 19,000) was similar to that in agsBOE (Figure 4B; Table 2). Lastly, the Mn of alkali-soluble glucan in the wild-type (2,280,000 ± 320,000) and ΔamyD (2,390,000 ± 400,000) was larger than that in agsBOE (312,000 ± 5,000; Figure 4C; Table 2). The amyDOE strain had a primary peak at around 17 min (, 32,900 ± 300) and a secondary peak at 11 min (, 2,210,000 ± 700,000). These results suggest that AmyD degraded the alkali-soluble glucan eluted around 11 min to produce alkali-soluble glucan with a smaller MM (Figure 4C; Table 2).
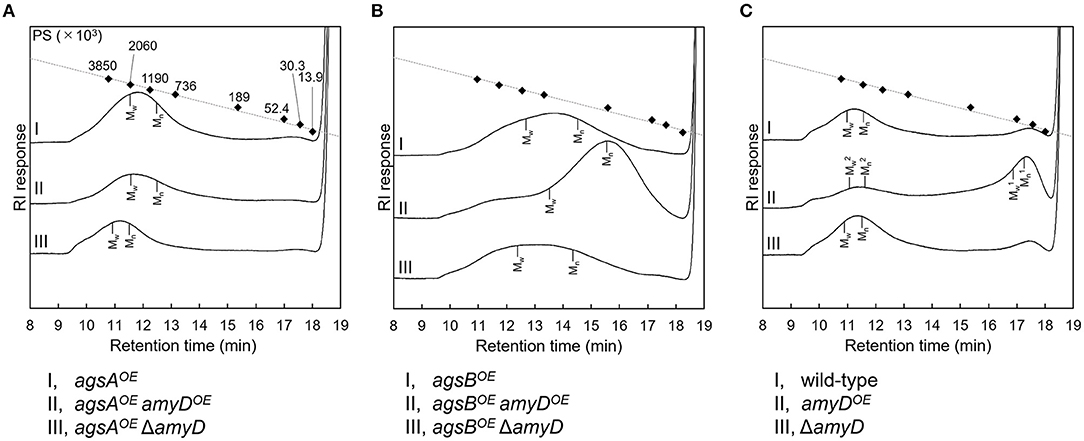
Figure 4. GPC elution profile of the AS2 fraction from the series of (A) agsAOE strains, (B) agsBOE strains, and (C) wild-type The AS2 fraction from 24-h-cultured mycelia of each strain was dissolved in 20 mM LiCl/DMAc. The elution profile was monitored by a refractive index detector. Molecular mass (MM) of the glucan peaks was determined from a calibration curve of polystyrene (PS) standards (♦). Mw, weight-average MM; Mn, number-average MM.
Although the alkali-soluble glucan in the wild-type strains was synthesized mainly by AgsB, its MM was larger than that in the agsBOE strain (Table 2). Additionally, when agsB was overexpressed, the amount of α-1,3-glucan was three times that in the wild-type (Figure 3). We supposed that some unknown glycosyl modification enzymes may contribute to the increase in MM of α-1,3-glucan in the wild-type, and that because the agsBOE strain produces more α-1,3-glucan, there is little modification by the unknown enzymes. Therefore, we determined the MM of alkali-soluble glucan extracted from 16-h cultured mycelia, which should be less affected by the modification enzyme than the 24-h cultured mycelia (He et al., 2017). Unexpectedly, the Mn of the alkali-soluble glucan in the mycelia cultured for 16 h was 1,980,000 ± 320,000, which was similar to that in the mycelia cultured for 24 h (1,930,000 ± 280,000; Supplementary Figure 6; Supplementary Table 2). We then evaluated the MM of alkali-soluble glucan in A4, which is the Glasgow wild-type of A. nidulans (Table 1), and found it had Mn = 2,224,000 ± 390,000 (Supplementary Table 3), which is similar to that in the wild-type strain.
To validate whether the degree of polymerization of α-1,3-glucan subunits in the alkali-soluble glucan was altered when the MM was changed by amyD disruption or overexpression, we applied Smith degradation to the alkali-soluble glucan from each strain to selectively cleave 1,4-linked glucan, and then determined the MM by GPC. One subunit of α-1,3-glucan in the alkali-soluble glucan is composed of ≈200 glucose residues (Choma et al., 2013; Miyazawa et al., 2018). The Smith-degraded alkali-soluble glucan in each strain had almost the same MM, equivalent to 300–400 glucose residues (Supplementary Figure 7; Supplementary Table 4), which suggests that AmyD activity does not decrease the degree of polymerization of the glucose residues in each α-1,3-glucan subunit.
Spatial Localization of α-1,3-Glucan in the Cell Wall Is Not Affected by amyD Disruption or Overexpression
We previously revealed that spatial localization of α-1,3-glucan in the cell wall changes according to its MM (Miyazawa et al., 2018); α-1,3-glucans in agsBOE cells are localized in the outer layer in the cell wall, whereas most of those in the agsAOE cells are masked by a β-1,3-glucan layer. In this study, disruption or overexpression of amyD altered the MM of alkali-soluble glucan (Figure 4; Table 2); therefore, we analyzed whether this alteration affected the spatial localization of α-1,3-glucan in the cell wall. In agreement with previous results (Miyazawa et al., 2018), the α-1,3-glucans with AGBD-GFP labeling showed clearly in the wild-type and agsBOE cells, but only weakly in agsAOE cells (Figure 5). The ΔamyD and amyDOE cells were also labeled with AGBD-GFP (Figure 5); fluorescent intensity in amyDOE was relatively low, which might be caused by a decrease in the amount of alkali-soluble glucan in the cell wall of amyDOE cells. The labeling with AGBD-GFP in agsAOE amyDOE and agsAOE ΔamyD cells was weak, as was that in the cells of the parental agsAOE strain (Figure 5). The agsBOE ΔamyD cells were clearly labeled with AGBD-GFP, as in the parental agsBOE (Figure 5). The AGBD-GFP labeling was slightly weaker in agsBOE amyDOE than in agsBOE, which might be attributable to a decrease in the amount of α-1,3-glucan. After treatment with β-1,3-glucanase, α-1,3-glucans of the hyphal cells in agsAOE, agsAOE amyDOE, and agsAOE ΔamyD cells were clearly labeled with AGBD-GFP (Supplementary Figure 8), suggesting that these strains have α-1,3-glucan in the inner layer of the cell wall in their hyphal cells. Taken together, these findings indicate that disruption or overexpression of amyD gene scarcely affected the spatial localization of α-1,3-glucan in the cell wall.
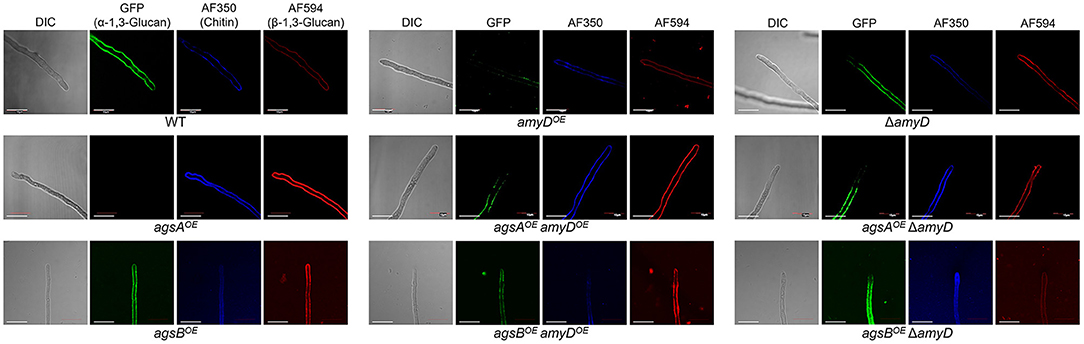
Figure 5. Localization of cell-wall polysaccharides of vegetative hyphae. Hyphae cultured for 16 h were fixed and stained with AGBD-GFP for α-1,3-glucan, fluorophore-labeled antibody for β-1,3-glucan, and fluorophore-labeled lectin for chitin. Scale bars are 10 μm.
The GPI Anchor Is Essential for the Effect of AmyD on Both the Amount and Molecular Mass of Alkali-Soluble Glucan
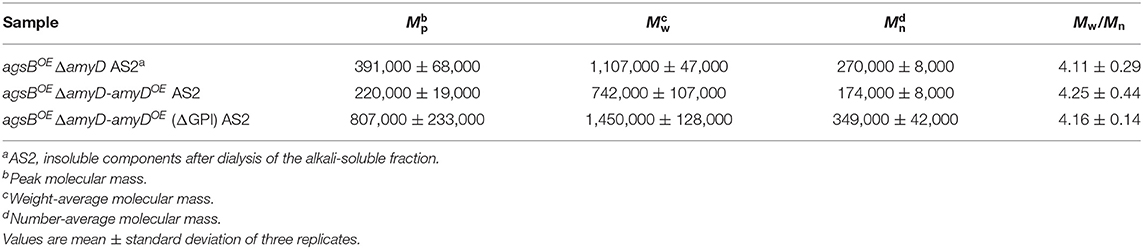
AmyD is thought to contain a GPI anchor at the C-terminal region. Fungal GPI anchor–type proteins are transferred from the plasma membrane to the cell wall by the activity of the GH76 family (Vogt et al., 2020). We speculated that localization in the cell wall would be essential for AmyD to reach the substrate, alkali-soluble glucan, so we constructed overexpression strains of amyD with and without the GPI-anchor site. Because we noticed that overexpression of amyD alters the phenotype or the alkali-soluble glucan, we used ΔamyD and agsBOE ΔamyD strains as hosts for the amyDOE strains. The hyphae of ΔamyD formed pellets in shake-flask culture (Figure 6). Those of ΔamyD-amyDOE were dispersed, as in amyDOE (Figure 6). Those of ΔamyD-amyDOE(ΔGPI) formed pellets, although the form was slightly different from that in the parental strain (Figure 6). Those of agsBOE ΔamyD, agsBOE ΔamyD-amyDOE, and agsBOE ΔamyD-amyDOE(ΔGPI) formed similar pellets (Figure 6). Although the ΔamyD-amyDOE hyphae had less AS2-Glc (1.13% ± 0.21%) than ΔamyD (5.68% ± 0.25%), the amount was restored in ΔamyD-amyDOE(ΔGPI) hyphae (5.17% ± 0.46%; Figure 7A). These results suggest that the GPI anchor of AmyD has an important negative effect on α-1,3-glucan biosynthesis. The hyphae of agsBOE ΔamyD-amyDOE had marginally less AS2-Glc (16.2% ± 0.6%) than those of agsBOE ΔamyD (17.6% ± 0.3%) and agsBOE ΔamyD-amyDOE(ΔGPI) (16.7% ± 0.5%; Figure 7B). We then evaluated the MM of alkali-soluble glucan in the cells of agsBOE ΔamyD, agsBOE ΔamyD-amyDOE, and agsBOE ΔamyD-amyDOE(ΔGPI). The Mn of the alkali-soluble glucan in agsBOE ΔamyD-amyDOE cells (174,000 ± 8,000) was smaller than that in agsBOE ΔamyD (270,000 ± 8,000; Figure 7C; Table 3; P < 0.05). The Mn of alkali-soluble glucan in agsBOE ΔamyD-amyDOE(ΔGPI) cells (349,000 ± 42,000) was similar to that in agsBOE ΔamyD (Figure 7C; Table 3). These results suggest that the GPI anchor of AmyD is also important for regulating the MM of alkali-soluble glucan.
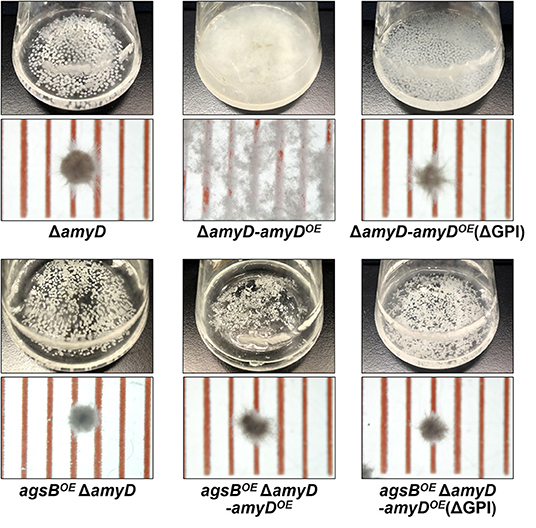
Figure 6. Growth characteristics of ΔamyD-amyDOE strains in liquid culture. Upper images, cultures in Erlenmeyer flasks; lower images, representative hyphal pellets of each strain under a stereomicroscope. Scale intervals are 1 mm.
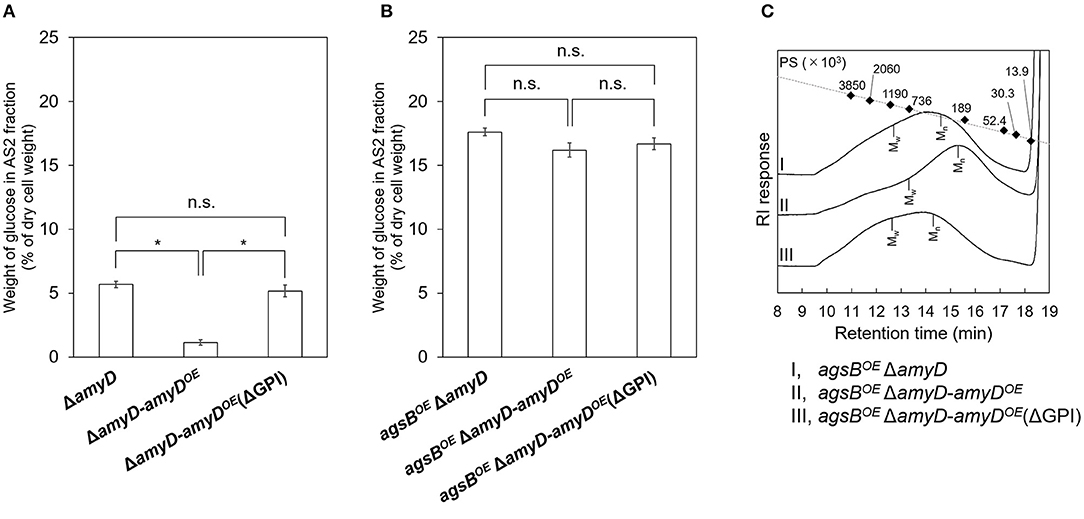
Figure 7. (A,B) Amounts of glucose and (C) GPC elution profiles of the AS2 fraction in ΔamyD-amyDOE strains. (A,B) Conidia (5.0 × 105/mL) of each strain were inoculated into CD medium and rotated at 160 rpm at 37°C for 24 h. Values show glucose content of AS2 fraction as a percentage of the total cell-wall weight. Error bars represent SEM calculated from three replicates. *Significant difference by Tukey's test (*P < 0.05); n.s., not significant. (C) The AS2 fraction from 24-h-cultured mycelia of each strain was dissolved in 20 mM LiCl/DMAc. The elution profile was monitored by a refractive index detector. Molecular mass (MM) of the glucan peaks was determined from a calibration curve of polystyrene (PS) standards (♦). Mw, weight-average MM; Mn, number-average MM.
Western blotting showed that secretion of AmyD in the culture supernatant could be detected only in the ΔamyD-amyDOE (ΔGPI) strain (Supplementary Figure 9A). Because AgtA in A. niger has relatively high transglycosylation activity on donor substrates with maltooligosaccharides (Van Der Kaaij et al., 2007), we evaluated the α-amylase activity in concentrated culture supernatants with pNP-α-maltopentaoside. Although various products possibly produced by coexisting α-glucosidase were detected in ΔamyD and both amyD-overexpressing strains, pNP-α-maltooctaoside (probably a transglycolylation product of AmyD) was detected only in the ΔamyD-amyDOE (ΔGPI) strain (data not shown). A histidine residue in region I, which is a highly conserved region of α-amylases, is substituted to the asparagine residue in AmyD and proteins encoded by orthologs of amyD (Van Der Kaaij et al., 2007). Substitution of the histidine residues in α-amylase increases the inhibitory constant (Ki) of a representative α-glucosidase inhibitor, acarbose (Svensson, 1994). Therefore, we evaluated the α-amylase activity in concentrated culture supernatants in the presence of acarbose. As expected, the hydrolysis product of pNP-α-maltopentaoside was hardly detected in ΔamyD (Supplementary Figure 9B). pNP-α-maltoside and pNP-α-maltooctaoside were clearly detected in the supernatant of ΔamyD-amyDOE (ΔGPI), but scarcely in that of ΔamyD-amyDOE (Supplementary Figure 9B). These results suggest that although enzymatically active AmyD is secreted into the culture supernatant, it cannot decrease the MM of α-1,3-glucan, leading us to suppose that active AmyD needs to be localized on the plasma membrane or in the cell wall space to regulate the MM of α-1,3-glucan.
Discussion
Although the GPI-anchored α-amylase AmyD is known to be involved in the biosynthesis of α-1,3-glucan in A. nidulans (He et al., 2014, 2017), the detailed mechanism remains unclear. Here, we looked at strains with disrupted or overexpressed amyD to analyze how AmyD affects the chemical properties of alkali-soluble glucan. The results reveal that overexpression of amyD not only decreased the MM of α-1,3-glucan, but also decreased the amount of α-1,3-glucan in the cell wall. The GPI anchor of AmyD was essential in both actions.
Overexpression of amyD affected the amount and MM of α-1,3-glucan in the wild-type and agsBOE strains, but not in the agsAOE strain (Figures 3, 4; Table 2). We previously reported that the MM of α-1,3-glucan controls where α-1,3-glucan is localized in the cell wall of A. nidulans; namely, that the α-1,3-glucan with a larger MM that is synthesized by AgsA is localized in the inner layer of the cell wall, and the smaller one that is synthesized by AgsB is localized in the outer layer (Miyazawa et al., 2018). To explain the effect of AmyD on the amount and MM of α-1,3-glucan, we formed the following two hypotheses from the results of this study. (1) Given that fungal GPI-anchored proteins are transferred from the plasma membrane to the cell wall (Orlean, 2012; Gow et al., 2017), our findings suggest that AmyD decreased the MM of α-1,3-glucan localized at the outer layer of the cell wall. The increased MM of alkali-soluble glucan in the agsAOE ΔamyD strain can be explained by its GPC elution profiles, which suggest that the MM of the polysaccharides was broadly distributed (Figure 4A); in other words, agsAOE ΔamyD had mainly α-1,3-glucan with larger MM [>623,000 (Mp of alkali-soluble glucan from agsBOE), 97.5%], but also had a small amount of α-1,3-glucan with small MM (<623,000, 2.5%). We speculate that this small amount of α-1,3-glucan with a smaller MM may be localized in the outer layer of the cell wall of agsAOE, where it is accessible to AmyD, which results in the small amount of α-1,3-glucan with a smaller MM. (2) Generally, as the degree of polymerization increases, the solubility of polysaccharides in water decreases (Guo et al., 2017). We have previously explained that after the biosynthesis of α-1,3-glucan on the plasma membrane, sugar chains are released to the outside of the membrane, where they are gradually insolubilized and immobilized to become a part of the cell wall (Miyazawa et al., 2018). Because α-1,3-glucan molecules with a larger MM might be more quickly insolubilized than those with a smaller MM, they are consequently localized in the inner layer of the cell wall, whereas α-1,3-glucan with a smaller MM might more likely be distributed toward the outer layer of the cell wall. α-1,3-Glucan synthesized with smaller MM in the agsBOE strain, which takes a relatively long time to become insoluble, seems to be more catalytically accessible by AmyD than α-1,3-glucan synthesized with larger MM in the agsAOE strain. To understand the relationship between the spatial localization of AmyD and α-1,3-glucan in the cell wall, immunoelectron microscopic and glycochemical analyses are necessary and are our future work.
AmyD of A. nidulans is considered to be a GPI-anchored protein (De Groot et al., 2009; He et al., 2014). It is well-known that many fungal GPI-anchored proteins are related to remodeling of the cell wall (Samalova et al., 2020). Proteins in the “defective in filamentous growth” (DFG) family recognize the GPI core glycan and then transfer to the β-1,3- or β-1,6-glucan (Muszkieta et al., 2019; Vogt et al., 2020), which allows GPI-anchored proteins to react with their substrates in the cell wall. Although there is no direct evidence that DFG family proteins contribute to transglycosylation in Aspergillus species, their role in cell-wall integrity in A. fumigatus was recently reported (Li et al., 2018; Muszkieta et al., 2019), which implies that DFG family proteins are important for transferring the GPI-core glycan to β-glucan in Aspergillus species. To reveal the importance of the GPI anchor in the function of AmyD, we evaluated the MM and amount of α-1,3-glucan in amyD-overexpressing strains with or without the GPI-anchoring site. Interestingly, decreases in the MM and the amount of α-1,3-glucan were not observed when the C-terminal GPI-anchoring site was deleted (Figure 7; Table 3); ΔamyD-amyDOE (ΔGPI) formed slightly altered pellets (Figure 6), suggesting that AmyD expressed without its GPI anchor has only partial functions. Furthermore, western blotting detected the secretion of AmyD in the culture supernatant from the ΔamyD-amyDOE (ΔGPI) strain (Supplementary Figure 9A). AgtA in A. niger has relatively high glucosyltransferase activity toward donor substrates with maltooligosaccharides (Van Der Kaaij et al., 2007). In a separate study, we expressed and purified A. oryzae AgtA (homologous to A. nidulans AmyD) in Pichia pastoris, and the purified A. oryzae AgtA showed α-amylase (hydrolysis and transferase) activity toward pNP-α-maltopentaoside (Koizumi et al., unpublished). Therefore, we evaluated the α-amylase activity in culture supernatants with pNP-α-maltopentaoside. Whereas, the hydrolysis product of pNP-α-maltopentaoside was hardly detected in ΔamyD (Supplementary Figure 9B), the supernatant from ΔamyD-amyDOE (ΔGPI) produced pNP-α-maltoside and pNP-α-maltooctaoside at an early stage of the reaction (Supplementary Figure 9B). These results suggest that enzymatically active AmyD that is secreted into the culture supernatant cannot decrease the amount and MM of α-1,3-glucan. Taken together, the results show that expression of AmyD with a GPI anchor is important for reaching the substrate, α-1,3-glucan, in the space of the cell wall.
Cell-wall polysaccharides are thought to be synthesized on the plasma membrane after the secretory vesicles containing polysaccharide synthases have been exported to the hyphal tip (Riquelme, 2013). On the basis of our previous findings (Miyazawa et al., 2020), we hypothesize the process of alkali-soluble glucan biosynthesis of A. nidulans to be as follows: (1) the intracellular domain of α-1,3-glucan synthase polymerizes 1,3-linked α-glucan chains from UDP-glucose as a substrate from the primers, which are maltooligosaccharides produced by intracellular α-amylase AmyG; (2) the elongated glucan chain is exported to the extracellular space through the multitransmembrane domain of α-1,3-glucan synthase; (3) the extracellular domain of α-1,3-glucan connects several chains of the elongated glucan to form mature alkali-soluble glucan. The mechanism underlying the distribution of mature alkali-soluble glucan to the cell-wall network is still unknown. However, the water solubility of newly synthesized glucan might be related to the spatial distribution of α-1,3-glucan in the cell wall, because localization of α-1,3-glucan varies according to the difference in MM (Miyazawa et al., 2018). Aspergillus niger AgtA (encoded by an ortholog of A. nidulans amyD) scarcely hydrolyzes α-1,3-glucan and shows weak hydrolytic activity to starch (Van Der Kaaij et al., 2007). Therefore, decrease of the MM of alkali-soluble glucan in the amyDOE strain could be caused by hydrolysis of the primer/spacer residues (1,4-linked α-glucan) rather than of the 1,3-linked α-glucan region. The mechanism underlying the decrease in the amount of α-1,3-glucan by AmyD is also unknown. He et al. (2017) reported that AmyD seems to directly repress α-1,3-glucan synthesis. We suspect that AmyD with a GPI anchor on the plasma membrane binds to the spacer residues of a glucan chain that is being just synthesized by α-1,3-glucan synthase, and competitively inhibits transglycosylation by the extracellular domain of α-1,3-glucan synthase to decrease the amount of alkali-soluble glucan in the cell wall.
The Mn of the alkali-soluble glucan from the wild-type strain was larger than that from the agsBOE, although the alkali-soluble glucan from both strains seemed to be synthesized mainly by AgsB (Figure 4; Table 2). The Mn of the alkali-soluble glucan in the 16-h-cultured mycelia from the wild-type was similar to that from the 24-h-cultured mycelia (Supplementary Figure 6; Supplementary Table 2). α-1,3-Glucan was clearly labeled with AGBD-GFP in the wild-type strain (Figure 5). These results suggest that α-1,3-glucan was located in the outer layer of the cell wall in the wild-type strain, consistent with the localization of α-1,3-glucan synthesized by AgsB. These results imply the existence of some factor that increases the MM of α-1,3-glucan. We surmise that once a matured α-1,3-glucan molecule synthesized by AgsB is localized in the outer layer of the cell wall, macromolecules are formed by interconnecting α-1,3-glucan or connecting α-1,3-glucan to other polysaccharides, resulting in a chemically stable complex. Although the difference was not significant, the MM of Smith-degraded alkali-soluble glucan in the wild-type strain was slightly higher (Supplementary Table 4) and its GPC profile had a broader distribution (Supplementary Figure 7) than those in the agsAOE and agsBOE strains, implying the existence of non-Smith-degradable glycosidic bonds (i.e., β-1,3-glycosidic bond) in the alkali-soluble fraction in the wild-type strain. It is well-known that β-glucan, chitin, and galactomannan are continuously modified by hydrolase or glycosyltransferase in the cell wall (Aimanianda et al., 2017; Henry et al., 2019; Muszkieta et al., 2019). However, an enzyme that modifies α-1,3-glucan has not been reported. The recent report by Kang et al. (2018) on the cell wall architecture of A. fumigatus suggested the presence of a covalent bond of α-1,3-glucan to β-1,3- and β-1,4-glucan. The report by Chakraborty et al. (2021) on cell wall organization by whole-cell NMR showed that α-1,3-glucan fractionated into both alkali-soluble and -insoluble fractions for the rigid and mobile portions. An enzyme that has a role in modifying α-1,3-glucan to allow its transition into the different portions needs to be identified in the near future.
Here, we revealed that AmyD in A. nidulans decreased the MM of the alkali-soluble glucan composed mainly of α-1,3-glucan in the cell wall and also the amount of alkali-soluble glucan. However, a complete picture of the biosynthesis of α-1,3-glucan has yet to be described, because the substrates or proteins associated with α-1,3-glucan synthesis have not been directly demonstrated. To unveil the true nature of the biosynthesis, further biochemical analysis of the α-1,3-glucan synthase is essential.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
KM, AY, TN, and KA conceived and designed the experiment. KM, TY, and AT performed most experiments and analyzed the data. KM, YK, and YT performed microscopic observation. KM, AY, MS, and YY constructed fungal mutants. SK performed 13C NMR. AK and SY produced AGBD-GFP. AK and MO performed the enzymatic assay. KM, AY, and KA wrote the paper. KA supervised this research and acquired funding. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Japan Society for Promotion of Science (JSPS) KAKENHI Grant Nos. 26292037 (KA), 18K05384 (KA), 20H02895 (KA) and 20K22773 (KM), and a Grant-in-Aid for JSPS Fellows Grant No. 18J11870 (KM). This work was also supported by the Institute for Fermentation, Osaka (Grant No. L-2018–2–014) (KA) and by the project JPNP20011 (KA), which is commissioned by the New Energy and Industrial Technology Development Organization (NEDO).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Professor Toshikazu Komoda (Miyagi University) for the NMR.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffunb.2021.821946/full#supplementary-material
References
Aimanianda, V., Simenel, C., Garnaud, C., Clavaud, C., Tada, R., Barbin, L., et al. (2017). The dual activity responsible for the elongation and branching of β-(1,3)-glucan in the fungal cell wall. mBio 8, e00619–e00617. doi: 10.1128/mBio.00619-17
Beauvais, A., Bozza, S., Kniemeyer, O., Formosa, C., Balloy, V., Henry, C., et al. (2013). Deletion of the α-(1,3)-glucan synthase genes induces a restructuring of the conidial cell wall responsible for the avirulence of Aspergillus fumigatus. PLoS Pathog. 9:e1003716. doi: 10.1371/journal.ppat.1003716
Beauvais, A., Maubon, D., Park, S., Morelle, W., Tanguy, M., Huerre, M., et al. (2005). Two α(1-3) glucan synthases with different functions in Aspergillus fumigatus. Appl. Environ. Microbiol. 71, 1531–1538. doi: 10.1128/AEM.71.3.1531-1538.2005
Bernard, M., and Latge, J. P. (2001). Aspergillus fumigatus cell wall: composition and biosynthesis. Med. Mycol. 39, 9–17. doi: 10.1080/mmy.39.1.9.17
Chakraborty, A., Fernando, L. D., Fang, W., Dickwella Widanage, M. C., Wei, P., Jin, C., et al. (2021). A molecular vision of fungal cell wall organization by functional genomics and solid-state NMR. Nat. Commun. 12:6346. doi: 10.1038/s41467-021-26749-z
Choma, A., Wiater, A., Komaniecka, I., Paduch, R., Pleszczynska, M., and Szczodrak, J. (2013). Chemical characterization of a water insoluble (1–>3)-α-D-glucan from an alkaline extract of Aspergillus wentii. Carbohydr. Polym. 91, 603–608. doi: 10.1016/j.carbpol.2012.08.060
Damveld, R. A., Vankuyk, P. A., Arentshorst, M., Klis, F. M., Van Den Hondel, C. A., and Ram, A. F. (2005). Expression of agsA, one of five 1,3-α-D-glucan synthase-encoding genes in Aspergillus niger, is induced in response to cell wall stress. Fungal Genet. Biol. 42, 165–177. doi: 10.1016/j.fgb.2004.11.006
De Groot, P. W. J., Brandt, B. W., Horiuchi, H., Ram, A. F. J., De Koster, C. G., and Klis, F. M. (2009). Comprehensive genomic analysis of cell wall genes in Aspergillus nidulans. Fungal Genet. Biol. 46, S72–S81. doi: 10.1016/j.fgb.2008.07.022
Dichtl, K., Samantaray, S., Aimanianda, V., Zhu, Z., Prevost, M. C., Latg,è, J. P., et al. (2015). Aspergillus fumigatus devoid of cell wall β-1,3-glucan is viable, massively sheds galactomannan and is killed by septum formation inhibitors. Mol. Microbiol. 95, 458–471. doi: 10.1111/mmi.12877
Fontaine, T., Beauvais, A., Loussert, C., Thevenard, B., Fulgsang, C. C., Ohno, N., et al. (2010). Cell wall α1-3glucans induce the aggregation of germinating conidia of Aspergillus fumigatus. Fungal Genet. Biol. 47, 707–712. doi: 10.1016/j.fgb.2010.04.006
Fontaine, T., Simenel, C., Dubreucq, G., Adam, O., Delepierre, M., Lemoine, J., et al. (2000). Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J. Biol. Chem. 275, 27594–27607. doi: 10.1074/jbc.M909975199
Fujikawa, T., Kuga, Y., Yano, S., Yoshimi, A., Tachiki, T., Abe, K., et al. (2009). Dynamics of cell wall components of Magnaporthe grisea during infectious structure development. Mol. Microbiol. 73, 553–570. doi: 10.1111/j.1365-2958.2009.06786.x
Fujikawa, T., Sakaguchi, A., Nishizawa, Y., Kouzai, Y., Minami, E., Yano, S., et al. (2012). Surface α-1,3-glucan facilitates fungal stealth infection by interfering with innate immunity in plants. PLoS Pathog. 8:e1002882. doi: 10.1371/journal.ppat.1002882
Fujioka, T., Mizutani, O., Furukawa, K., Sato, N., Yoshimi, A., Yamagata, Y., et al. (2007). MpkA-dependent and -independent cell wall integrity signaling in Aspergillus nidulans. Eukaryot. Cell 6, 1497–1510. doi: 10.1128/EC.00281-06
Gow, N. a,.R, Latge, J. P., and Munro, C.A. (2017). The fungal cell wall: structure, biosynthesis, and function. Microbiol. Spectr. 5:FUNK-0035-2016. doi: 10.1128/9781555819583.ch12
Grün, C. H., Hochstenbach, F., Humbel, B. M., Verkleij, A. J., Sietsma, J. H., Klis, F. M., et al. (2005). The structure of cell wall α-glucan from fission yeast. Glycobiology 15, 245–257. doi: 10.1093/glycob/cwi002
Guo, M. Q., Hu, X., Wang, C., and Ai, L. (2017). “Polysaccharides: structure and solubility,” in Solubility of Polysaccharides, ed Z. Xu. (London: IntechOpen), 7–21.
Hagiwara, D., Asano, Y., Marui, J., Furukawa, K., Kanamaru, K., Kato, M., et al. (2007). The SskA and SrrA response regulators are implicated in oxidative stress responses of hyphae and asexual spores in the phosphorelay signaling network of Aspergillus nidulans. Biosci. Biotechnol. Biochem. 71, 1003–1014. doi: 10.1271/bbb.60665
He, X., Li, S., and Kaminskyj, S. (2017). An amylase-like protein, AmyD, is the major negative regulator for α-glucan synthesis in Aspergillus nidulans during the asexual life cycle. Int. J. Mol. Sci. 18:695. doi: 10.3390/ijms18040695
He, X. X., Li, S. N., and Kaminskyj, S. G. W. (2014). Characterization of Aspergillus nidulans α-glucan synthesis: roles for two synthases and two amylases. Mol. Microbiol. 91, 579–595. doi: 10.1111/mmi.12480
Henry, C., Latg,è, J. P., and Beauvais, A. (2012). α1,3 Glucans are dispensable in Aspergillus fumigatus. Eukaryot. Cell 11, 26–29. doi: 10.1128/EC.05270-11
Henry, C., Li, J., Danion, F., Alcazar-Fuoli, L., Mellado, E., Beau, R., et al. (2019). Two KTR mannosyltransferases are responsible for the biosynthesis of cell wall mannans and control polarized growth in Aspergillus fumigatus. mBio 10, e02647–e02618. doi: 10.1128/mBio.02647-18
Kang, X., Kirui, A., Muszynski, A., Widanage, M. C. D., Chen, A., Azadi, P., et al. (2018). Molecular architecture of fungal cell walls revealed by solid-state NMR. Nat. Commun. 9:2747. doi: 10.1038/s41467-018-05199-0
Krappmann, S., Sasse, C., and Braus, G. H. (2006). Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end- joining-deficient genetic background. Eukaryot. Cell 5, 212–215. doi: 10.1128/EC.5.1.212-215.2006
Latgè, J. P.. (2010). Tasting the fungal cell wall. Cell. Microbiol. 12, 863–872. doi: 10.1111/j.1462-5822.2010.01474.x
Latgè, J. P., and Beauvais, A. (2014). Functional duality of the cell wall. Curr. Opin. Microbiol. 20, 111–117. doi: 10.1016/j.mib.2014.05.009
Latgè, J. P., Beauvais, A., and Chamilos, G. (2017). The cell wall of the human fungal pathogen Aspergillus fumigatus: biosynthesis, organization, immune response, and virulence. Annu. Rev. Microbiol. 71, 99–116. doi: 10.1146/annurev-micro-030117-020406
Li, J., Mouyna, I., Henry, C., Moyrand, F., Malosse, C., Chamot-Rooke, J., et al. (2018). Glycosylphosphatidylinositol anchors from galactomannan and GPI-anchored protein are synthesized by distinct pathways in Aspergillus fumigatus. J. Fungi 4:19. doi: 10.3390/jof4010019
Maubon, D., Park, S., Tanguy, M., Huerre, M., Schmitt, C., Prevost, M. C., et al. (2006). AGS3, an α(1-3)glucan synthase gene family member of Aspergillus fumigatus, modulates mycelium growth in the lung of experimentally infected mice. Fungal Genet. Biol. 43, 366–375. doi: 10.1016/j.fgb.2006.01.006
Miyazawa, K., Yoshimi, A., and Abe, K. (2020). The mechanisms of hyphal pellet formation mediated by polysaccharides, α-1,3-glucan and galactosaminogalactan, in Aspergillus species. Fungal Biol. Biotechnol. 7:10. doi: 10.1186/s40694-020-00101-4
Miyazawa, K., Yoshimi, A., Kasahara, S., Sugahara, A., Koizumi, A., Yano, S., et al. (2018). Molecular mass and localization of α-1,3-glucan in cell wall control the degree of hyphal aggregation in liquid culture of Aspergillus nidulans. Front. Microbiol. 9:2623. doi: 10.3389/fmicb.2018.02623
Miyazawa, K., Yoshimi, A., Sano, M., Tabata, F., Sugahara, A., Kasahara, S., et al. (2019). Both galactosaminogalactan and α-1,3-glucan contribute to aggregation of Aspergillus oryzae hyphae in liquid culture. Front. Microbiol. 10:2090. doi: 10.3389/fmicb.2019.02090
Miyazawa, K., Yoshimi, A., Zhang, S., Sano, M., Nakayama, M., Gomi, K., et al. (2016). Increased enzyme production under liquid culture conditions in the industrial fungus Aspergillus oryzae by disruption of the genes encoding cell wall α-1,3-glucan synthase. Biosci. Biotechnol. Biochem. 80, 1853–1863. doi: 10.1080/09168451.2016.1209968
Muszkieta, L., Fontaine, T., Beau, R., Mouyna, I., Vogt, M. S., Trow, J., et al. (2019). The glycosylphosphatidylinositol-anchored DFG family is essential for the insertion of galactomannan into the β-(1,3)-glucan–chitin core of the cell wall of Aspergillus fumigatus. mSphere 4, e00397–e00319. doi: 10.1128/mSphere.00397-19
Orlean, P.. (2012). Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 192, 775–818. doi: 10.1534/genetics.112.144485
Puanglek, S., Kimura, S., Enomoto-Rogers, Y., Kabe, T., Yoshida, M., Wada, M., et al. (2016). In vitro synthesis of linear α-1,3-glucan and chemical modification to ester derivatives exhibiting outstanding thermal properties. Sci. Rep. 6:30479. doi: 10.1038/srep30479
Rappleye, C. A., Eissenberg, L. G., and Goldman, W. E. (2007). Histoplasma capsulatum α-(1,3)-glucan blocks innate immune recognition by the β-glucan receptor. Proc. Natl. Acad. Sci. U. S. A. 104, 1366–1370. doi: 10.1073/pnas.0609848104
Rappleye, C. A., Engle, J. T., and Goldman, W. E. (2004). RNA interference in Histoplasma capsulatum demonstrates a role for α-(1,3)-glucan in virulence. Mol. Microbiol. 53, 153–165. doi: 10.1111/j.1365-2958.2004.04131.x
Riquelme, M.. (2013). Tip growth in filamentous fungi: a road trip to the apex. Annu. Rev. Microbiol. 67, 587–609. doi: 10.1146/annurev-micro-092412-155652
Samalova, M., Carr, P., Bromley, M., Blatzer, M., Moya-Nilges, M., Latg,é, J. P., et al. (2020). GPI anchored proteins in Aspergillus fumigatus and cell wall morphogenesis. Curr. Top. Microbiol. Immunol. 425, 167–186. doi: 10.1007/82_2020_207
Sheppard, D. C., and Howell, P. L. (2016). Biofilm exopolysaccharides of pathogenic fungi: lessons from bacteria. J. Biol. Chem. 291, 12529–12537. doi: 10.1074/jbc.R116.720995
Stephen-Victor, E., Karnam, A., Fontaine, T., Beauvais, A., Das, M., Hegde, P., et al. (2017). Aspergillus fumigatus cell wall α-(1,3)-glucan stimulates regulatory T-cell polarization by inducing PD-L1 expression on human dendritic cells. J. Infect. Dis. 216, 1281–1294. doi: 10.1093/infdis/jix469
Suyotha, W., Yano, S., Takagi, K., Rattanakit-Chandet, N., Tachiki, T., and Wakayama, M. (2013). Domain structure and function of α-1,3-glucanase from Bacillus circulans KA-304, an enzyme essential for degrading basidiomycete cell walls. Biosci. Biotechnol. Biochem. 77, 639–647. doi: 10.1271/bbb.120900
Svensson, B.. (1994). Protein engineering in the α-amylase family: catalytic mechanism, substrate specificity, and stability. Plant Mol. Biol. 25, 141–157. doi: 10.1007/BF00023233
Tokashiki, J., Hayashi, R., Yano, S., Watanabe, T., Yamada, O., Toyama, H., et al. (2019). Influence of α-1,3-glucan synthase gene agsE on protoplast formation for transformation of Aspergillus luchuensis. J. Biosci. Bioeng. 128, 129–134. doi: 10.1016/j.jbiosc.2019.01.018
Uechi, K., Yaguchi, H., Tokashiki, J., Taira, T., and Mizutani, O. (2021). Identification of genes involved in the synthesis of fungal cell wall component nigeran and regulation of its polymerization in Aspergillus luchuensis. Appl. Environ. Microbiol. 87:e0114421. doi: 10.1128/AEM.01144-21
Usui, T., and Murata, T. (1988). Enzymatic synthesis of p-nitrophenyl α-maltopentaoside in an aqueous-methanol solvent system by maltotetraose-forming amylase: a substrate for human amylase in serum. J. Biochem. 103, 969–972. doi: 10.1093/oxfordjournals.jbchem.a122395
Van Der Kaaij, R. M., Yuan, X. L., Franken, A., Ram, A. F., Punt, P. J., Van Der Maarel, M. J., et al. (2007). Two novel, putatively cell wall-associated and glycosylphosphatidylinositol-anchored α-glucanotransferase enzymes of Aspergillus niger. Eukaryot. Cell 6, 1178–1188. doi: 10.1128/EC.00354-06
Vogt, M. S., Schmitz, G. F., Varón Silva, D., Mösch, H. U., and Essen, L. O. (2020). Structural base for the transfer of GPI-anchored glycoproteins into fungal cell walls. Proc. Natl. Acad. Sci. U. S. A. 117, 22061–22067. doi: 10.1073/pnas.2010661117
Yoshimi, A., Miyazawa, K., and Abe, K. (2016). Cell wall structure and biogenesis in Aspergillus species. Biosci. Biotechnol. Biochem. 80, 1700–1711. doi: 10.1080/09168451.2016.1177446
Yoshimi, A., Miyazawa, K., and Abe, K. (2017). Function and biosynthesis of cell wall α-1,3-glucan in fungi. J. Fungi 3:63. doi: 10.3390/jof3040063
Yoshimi, A., Sano, M., Inaba, A., Kokubun, Y., Fujioka, T., Mizutani, O., et al. (2013). Functional analysis of the α-1,3-glucan synthase genes agsA and agsB in Aspergillus nidulans: AgsB is the major α-1,3-glucan synthase in this fungus. PLoS ONE 8:e54893. doi: 10.1371/journal.pone.0054893
Zhang, S., Ban, A., Ebara, N., Mizutani, O., Tanaka, M., Shintani, T., et al. (2017a). Self-excising Cre/mutant lox marker recycling system for multiple gene integrations and consecutive gene deletions in Aspergillus oryzae. J. Biosci. Bioeng. 123, 403–411. doi: 10.1016/j.jbiosc.2016.11.001
Keywords: cell wall, filamentous fungi, Aspergillus nidulans, glycosylphosphatidylinositol-anchored protein, α-amylase, α-1, 3-glucan
Citation: Miyazawa K, Yamashita T, Takeuchi A, Kamachi Y, Yoshimi A, Tashiro Y, Koizumi A, Ogata M, Yano S, Kasahara S, Sano M, Yamagata Y, Nakajima T and Abe K (2022) A Glycosylphosphatidylinositol-Anchored α-Amylase Encoded by amyD Contributes to a Decrease in the Molecular Mass of Cell Wall α-1,3-Glucan in Aspergillus nidulans. Front. Fungal Biol. 2:821946. doi: 10.3389/ffunb.2021.821946
Received: 25 November 2021; Accepted: 22 December 2021;
Published: 28 January 2022.
Edited by:
Katherine A. Borkovich, University of California, Riverside, United StatesReviewed by:
Tuo Wang, Louisiana State University, United StatesStephen J. Free, University at Buffalo, United States
Gerardo Díaz-Godínez, Autonomous University of Tlaxcala, Mexico
Copyright © 2022 Miyazawa, Yamashita, Takeuchi, Kamachi, Yoshimi, Tashiro, Koizumi, Ogata, Yano, Kasahara, Sano, Yamagata, Nakajima and Abe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keietsu Abe, a2VpZXRzdS5hYmUuYjVAdG9ob2t1LmFjLmpw
 Ken Miyazawa
Ken Miyazawa Takaaki Yamashita
Takaaki Yamashita Ayumu Takeuchi1
Ayumu Takeuchi1 Akira Yoshimi
Akira Yoshimi Motoaki Sano
Motoaki Sano Youhei Yamagata
Youhei Yamagata Keietsu Abe
Keietsu Abe