
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Fungal Biol. , 09 February 2021
Sec. Fungal Secondary Metabolites and Mycotoxins
Volume 2 - 2021 | https://doi.org/10.3389/ffunb.2021.632542
This article is part of the Research Topic The use of metabolic engineering techniques to increase the productivity of primary and secondary metabolites within filamentous fungi View all 5 articles
 Trong T. Dao1†
Trong T. Dao1† Kate M. J. de Mattos-Shipley2†
Kate M. J. de Mattos-Shipley2† Ian M. Prosser2†
Ian M. Prosser2† Katherine Williams2†
Katherine Williams2† Marija K. Zacharova1
Marija K. Zacharova1 Colin M. Lazarus2
Colin M. Lazarus2 Christine L. Willis1
Christine L. Willis1 Andrew M. Bailey2*
Andrew M. Bailey2*The use of filamentous fungi as cellular factories, where natural product pathways can be refactored and expressed in a host strain, continues to aid the field of natural product discovery. Much work has been done to develop host strains which are genetically tractable, and for which there are multiple selectable markers and controllable expression systems. To fully exploit these strains, it is beneficial to understand their natural metabolic capabilities, as such knowledge can rule out host metabolites from analysis of transgenic lines and highlight any potential interplay between endogenous and exogenous pathways. Additionally, once identified, the deletion of secondary metabolite pathways from host strains can simplify the detection and purification of heterologous compounds. To this end, secondary metabolite production in Aspergillus oryzae strain NSAR1 has been investigated via the deletion of the newly discovered negative regulator of secondary metabolism, mcrA (multicluster regulator A). In all ascomycetes previously studied mcrA deletion led to an increase in secondary metabolite production. Surprisingly, the only detectable phenotypic change in NSAR1 was a doubling in the yields of kojic acid, with no novel secondary metabolites produced. This supports the previous claim that secondary metabolite production has been repressed in A. oryzae and demonstrates that such repression is not McrA-mediated. Strain NSAR1 was then modified by employing CRISPR-Cas9 technology to disrupt the production of kojic acid, generating the novel strain NSARΔK, which combines the various beneficial traits of NSAR1 with a uniquely clean secondary metabolite background.
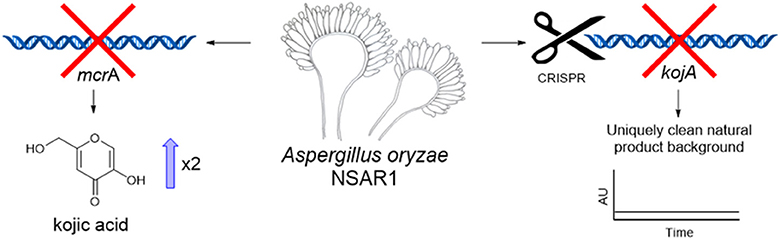
Graphical Abstract. Deletion of mcrA led to an increase in kojic acid yields in A. oryzae. Disruption of kojic acid production using CRISPR-Cas9 generated a novel NSAR1-based strain with a uniquely clean background for the heterologous production of natural products.
Natural products are hugely valuable small molecules, often of microbial origin, which are used in many industries including food and cosmetics, and particularly in medicine due to their varied and potent bioactivities. A pressing need for new lead compounds, especially antibiotics (Theuretzbacher et al., 2019), is driving ongoing natural product discovery, especially in relatively under-exploited groups of microorganisms such as filamentous fungi. An essential approach in natural product research is the heterologous expression of secondary metabolite (SM) pathways. This technique can be employed as a tool to investigate SM biosynthesis, as a way of discovering novel compounds, and as a means of production. The filamentous ascomycete Aspergillus oryzae has been used extensively as a heterologous host for the production of secondary metabolites (Watanabe et al., 1998; Heneghan et al., 2010; Chiba et al., 2013; Williams et al., 2016). Various factors make this fungus a suitable platform for such work: it has GRAS (“Generally Recognized As Safe”) status due to its long history of use in food and drink production, is generally atoxic (Barbesgaard et al., 1992), and importantly, produces few natural products. Thus, A. oryzae provides a relatively clean background for the production of heterologous compounds.
Strain engineering to improve A. oryzae as a heterologous host has included the development of the multiply auxotrophic strain NSAR1, which can be transformed with up to four separate integrative plasmids using different selectable complementation markers (Jin et al., 2004). This greatly improves the capacity of A. oryzae to express complex natural products, which often require the expression of multiple pathway genes. This capacity has been further expanded by construction of a set of corresponding plasmids, each containing four different expression cassettes that provide strong levels of expression in A. oryzae. Complementation alone therefore allows for the co-expression of up to 16 different genes in the host organism, while deployment of similar plasmids based on dominant selectable markers (the bar gene for resistance to glufosinate ammonium and ble gene for resistance to phleomycin) could provide a theoretical total of 24 expressible transgenes (Pahirulzaman et al., 2012; Lazarus et al., 2014).
A. oryzae has been successfully used as a platform for the expression of fungal natural product pathways using the system described above (Wasil et al., 2013; Lazarus et al., 2014; Bailey et al., 2016; Williams et al., 2016), as well as in similar independently developed vector systems (Fujii et al., 2011; Tagami et al., 2013). Such work has allowed complex biosynthetic pathways to be studied in detail, contributing significantly to the understanding of fungal SM biosynthesis and uncovering novel chemistry. Engineered A. oryzae strains can also be used to generate novel and potentially valuable structural diversity, through pathway engineering and biotransformations. Alberti et al. (2017) recently demonstrated the value of such an approach, establishing A. oryzae as a platform for the production of semi-synthetic derivatives of the valuable antibiotic pleuromutilin, one of which demonstrated enhanced antibiotic activity. This was achieved through the bio-conversion of chemically modified intermediates that were fed to a transgenic A. oryzae strain.
To fully exploit and further optimize NSAR1 as a “cellular factory,” it is important to characterize the natural metabolic capabilities of this strain. This would allow the exclusion of A. oryzae compounds when analyzing transformants and provide a better understanding of any potential interplay between endogenous A. oryzae metabolic pathways and the introduced natural product pathways. The main metabolite for which A. oryzae is known is kojic acid 1 (Figure 1), which is a sought-after product, believed to improve health and beauty, and used as a chelating agent in various industries including food and cosmetics (Bentley, 2006). Various strains of A. oryzae have been shown to produce additional secondary metabolites as detailed in a recent review by Frisvad et al. (2018), but in some cases these strains have been misidentified, and in general A. oryzae is known to be a poor producer of SMs.
The relatively low abundance of metabolites reported for A. oryzae strains is a characteristic that appears to have evolved since its divergence from the closely related A. flavus. A. oryzae is thought to be a “domesticated” relative of A. flavus (Barbesgaard et al., 1992); a hypothesis which is supported by genome analysis demonstrating that these species are 99.5% identical (Rokas et al., 2007). Comparison of gene expression profiles from SM biosynthetic gene clusters (BGCs) between A. oryzae and A. flavus shows that although numerous SM gene clusters are present in the genome of A. oryzae, they have been downregulated and, in many cases, appear totally silent under the tested conditions (Gibbons et al., 2012).
Recently, a novel negative regulator of secondary metabolism, McrA (multicluster regulator A), has been discovered in A. nidulans, where it has been shown to regulate at least 10 SM BGCs (Oakley et al., 2017). Deletion of mcrA was shown to stimulate SM production, even in strains carrying a deletion of the global SM regulator laeA (Bok and Keller, 2004). McrA appears to be conserved across many ascomycetes, with deletion of mcrA homologs in A. terreus and Penicillium canescens also resulting in increased SM production (Oakley et al., 2017).
The initial aim of this research was to identify and delete the mcrA homolog in A. oryzae in an attempt to induce SM production and thus better understand the metabolic capabilities of A. oryzae strain NSAR1. It was reasoned that the identification of functional biosynthetic pathways in A. oryzae could also direct strain improvements by targeting such pathways for deletion, thus creating an even cleaner background for future work employing A. oryzae as a cellular factory.
The A. oryzae mcrA homolog was identified by searching for any A. oryzae protein sequences on the NCBI non-redundant protein database using the A. nidulans McrA protein sequence as the query [accession number AN8694 (Oakley et al., 2017)]. Three homologous putative proteins were identified from three separate A. oryzae strains; A. oryzae 3.042 (Zhao et al., 2012), A. oryzae BCC7051 (Thammarongtham et al., 2018), and A. oryzae RIB40 (Machida et al., 2005) (accession numbers: EIT82532.1, OOO05597.1 and XP_023092425.1, respectively). These three predicted proteins were all different lengths (366, 318, and 234 amino acids, respectively). The latter two appear to be truncated at the N-terminus and have an additional 22 amino acid sequence internally (Supplementary Figure 1). Interrogating the genome sequences of each strain and re-annotating each gene manually determined that these apparent differences were due to incorrect annotation. The full protein sequence previously annotated in the genome of strain 3.042 (Zhao et al., 2012) is present and conserved with 100% identity in every strain. This protein sequence shares 62% identity with the A. nidulans McrA with 98% query coverage (Supplementary Figure 2). This is equivalent to that found and reported for other Aspergillus strains (Oakley et al., 2017), suggesting that this protein is also conserved in A. oryzae. A GAL4-like Zn2Cys6 binuclear cluster DNA-binding domain (accession number cd00067) was identified from residues 118–157, supporting the previous assertion that McrA is likely to function as a transcription factor, or have a similar DNA binding function.
To disrupt Ao_mcrA, a bipartite strategy (Nielsen et al., 2006) was used. A. oryzae NSAR1 was transformed with the relevant bi-partite knock-out fragments, containing homology to the mcrA flanking regions and the argB cassette for selection, and after multiple rounds of subculturing on selective media, nine of the resulting transformants were analyzed by PCR to evaluate integration of the targeting cassette and subsequent gene disruption (Supplementary Figure 3). Transformant AoΔmcrA-7 passed all diagnostic tests, with PCRs confirming correct integration of both bipartite fragments, and with the transformant being shown to be genetically pure by the absence of any wild-type mcrA (Supplementary Figure 3).
Metabolite production in strain AoΔmcrA-7 was assessed on a range of media; namely CMP, MEB, PDP, GN, and static rice, and compared to the parental strain; A. oryzae NSAR1, to identify any changes in the metabolic profile as a result of the gene disruption. On all media, the metabolic profiles were found to be similar, with no novel metabolites observed. CMP cultures were then used for further analysis, including purification and quantification, as this is the medium routinely employed for heterologous production of compounds in A. oryzae, and supports high yields of crude extract. Both NSAR1 and AoΔmcrA-7 were fermented in CMP media for 1 week in triplicate. Quantitative analysis by LCMS again demonstrated that there were no dramatic changes in the metabolic profile of AoΔmcrA-7 (Figure 2), with the only evident change being an increase in the production of kojic acid from 1.23 to 2.52 g/L (Supplementary Table 1).
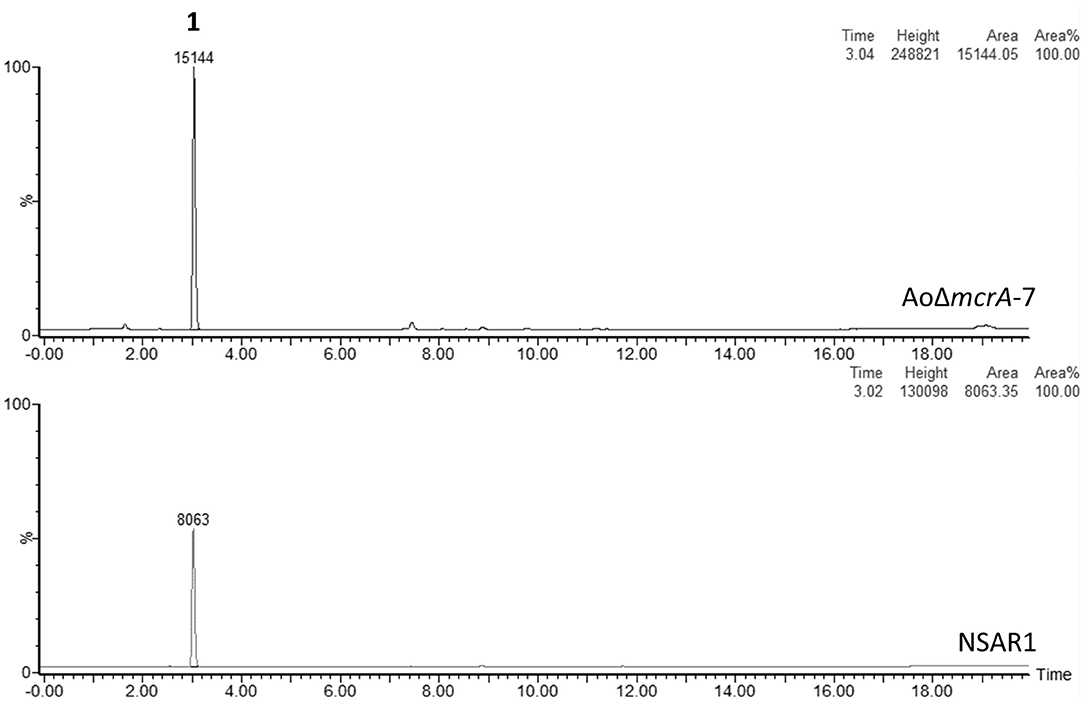
Figure 2. LCMS traces (ELSD) comparing the crude extracts of A. oryzae strain NSAR1 (bottom trace) and the mcrA deletion strain AoΔmcrA-7 (top trace). The only observable difference between the strains is an increase in kojic acid production (eluting at 3 min) in strain AoΔmcrA-7. The yields were quantified at 1.23 and 2.52 g/L for NSAR1 and AoΔmcrA-7, respectively.
Fermentation of ΔmcrA-7 was scaled up to purify minor compounds for NMR analysis and subsequent structural elucidation (Supplementary Figures 4–42). In addition to kojic acid 1, a kojic acid dimer 2 was also shown to be present in the extracts. Five diketopiperazines, 3–7, were also isolated and characterized as the known cyclo-[(D)Pro-(L)Tyr] (3), cyclo-[(D)Pro-(L)Val] (4), cyclo-[(D)Pro-(L)Ile] (5), cyclo-[(D)Pro-(L)Leu] (6), and cyclo-[(D)Pro-(L)Phe] (7) (Figure 3). Some of these compounds have previously been reported as being isolated from Aspergillus species (Dong et al., 2014; Shaaban et al., 2014; Uchoa et al., 2017). However, a comparison to extractions from CMP medium containing no A. oryzae demonstrated that in this case, these compounds were likely derived from the growth medium and not A. oryzae (Supplementary Figure 43).
To disrupt kojic acid production in strain NSAR1 kojA was mutated using CRISPR-Cas9 technology. KojA encodes an FAD-dependent oxidoreductase shown previously to be necessary for kojic acid production (Terabayashi et al., 2010). A plasmid named pAYargCas9-kojA2 was constructed, containing Cas9 under the control of the inducible PamyB promoter and a guide RNA cassette consisting of the Cas9-binding scaffold sequence and target-specific guide RNA, placed between the hammerhead (HH) and hepatitis delta virus (HDV) ribozymes. This design was based on that of Nødvig et al. (2015) and the use of ribozymes removes the requirement for an RNA polymerase III promoter. The targeting guide RNA (sequence GTCGCTCGAACAAAGAAACTCGG; complementary to nucleotides 90–109 of kojA) was designed using the online tool WU-CRISPR (Wong et al., 2015), and chosen based on its high predicted potency and 5′ location. An Interpro analysis (Mitchell et al., 2019) of KojA identified an FAD dependent oxidoreductase domain (IPR006076) located at residues 9–319, placing the target sequence toward the N-terminus of this domain. The approach used here is known to generate double-strand breaks (DSBs) at the target locus, with repair via the NHEJ pathway likely to introduce small indel (insertion-deletion) mutations (Nødvig et al., 2015). It was reasoned that any resulting indel mutations at this position, and particularly any out-of-frame mutations, would likely lead to a loss of function. pAYargCas9-kojA2 was also designed to contain the AMA1 autonomously replicating sequence, to allow for the subsequent loss of the plasmid and recycling of the selectable marker.
Plasmid pAYargCas9-kojA2 was used to transform A. oryzae strain NSAR1, generating 42 transformants, which were transferred onto secondary plates lacking arginine. After two rounds of subculturing under such selection, transformants were transferred to KA-Fe medium. This medium contains iron trichloride (FeCl3), which reacts with kojic acid to produce a distinct red color, thus acting as an initial screen of kojic acid production. One transformant, Ao-ko18, failed to trigger such a color change and was taken forward for further analysis. A ~1 kb region covering the targeted kojA site was amplified from both the original NSAR1 strain and Ao-ko18 and sequenced using primer pair kojA-F/kojA-R. An alignment of the sequences identified an 8 bp deletion in the mutant strain precisely at the target site (Figure 4). This out-of-frame deletion is sufficient for a loss of function for kojA.
To fully confirm a loss of kojic acid production a chemical extraction was performed for Ao-ko18 fermentation cultures and the metabolic profile of the resulting crude extract was analyzed by both TLC and HPLC analysis (Supplementary Figures 44, 45). This confirmed Ao-ko18 as an essentially “empty” strain, producing no major compounds under our laboratory production conditions.
Ao-ko18 was then subcultured multiple times on non-selective medium (MEA), to allow for the loss of the autonomously replicating plasmid pAYargCas9-kojA2. This loss was confirmed by restoration of arginine auxotrophy, and the strain renamed A. oryzae NSARΔK.
NSARΔK, and the original NSAR1 strain were both transformed with the pretenellin A 8 producing plasmid pTYGS-arg-TenC+TenS (Yang et al., 2019), to demonstrate the potential of using NSARΔK as a platform for the heterologous production of secondary metabolites. Two resulting strains, ΔK-Ten120 and N-Ten402, were fermented in production medium to induce the expression of the tenellin genes TenS and TenC. Both strains were confirmed as producing pretenellin A 8 in similar yields. In the case of ΔK-Ten120, the absence of kojic acid meant that the crude extract was almost pure pretenellin A 8 (Figure 5). The presence of kojic acid in large quantities can hinder the purification and downstream use of heterologously produced metabolites, making NSARΔK a valuable platform for the heterologous production of secondary metabolites.
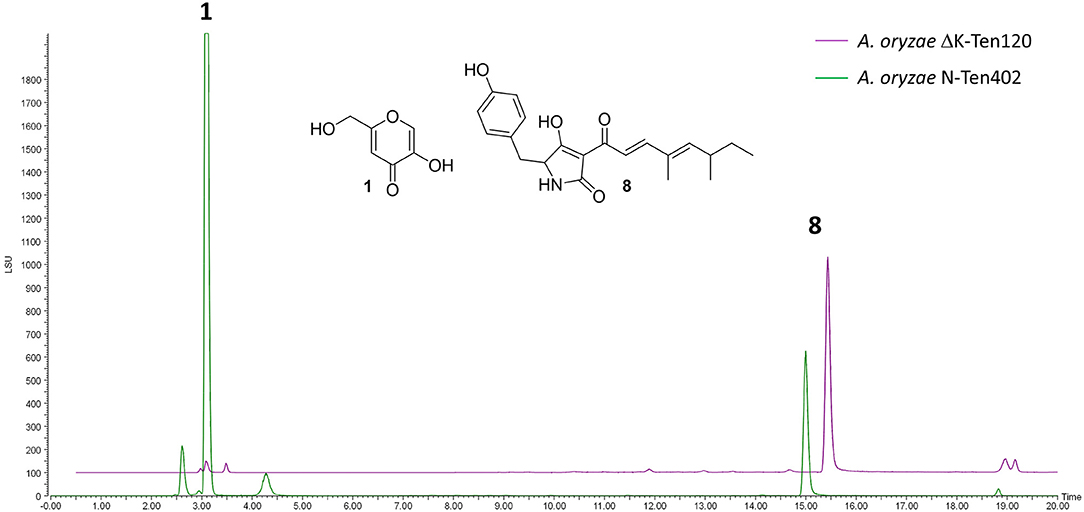
Figure 5. LCMS traces (ELSD) comparing CMP-agar crude extracts of A. oryzae strains heterologously producing pretenellin A 8 (eluting at 14.94 min). Strain N-Ten402 was generated by transforming NSAR1 and is therefore producing kojic acid 1 in addition to pretenellin A 8. Strain ΔK-Ten120 was generated using NSARΔK and is producing almost exclusively pretenellin A 8.
A 366 amino acid protein – named Ao_mcrA – was identified as being encoded in the genomes of Aspergillus oryzae strains 3.042, BCC7051, and RIB40. This protein shares 62% protein identity with the characterized McrA of Aspergillus nidulans (Oakley et al., 2017), demonstrating a similar level of homology to that found and reported for McrA proteins from other Aspergillus species (Oakley et al., 2017). The conserved GAL4-like Zn2Cys6 binuclear cluster DNA-binding domain could also be detected within the predicted protein sequence of Ao_mcrA.
Unlike in filamentous fungi previously investigated, disruption of Ao_mcrA was shown to have little impact on secondary metabolite (SM) production in A. oryzae strain NSAR1, with the only observable change being an increase in the already major metabolite, kojic acid, from 1.23 to 2.52 g/L. As kojic acid is a valuable and commercially produced compound, a similar deletion of McrA could be conducted in any producing strains as a potentially straightforward way of increasing yields. The lack of impact the Ao_mcrA deletion had on the NSAR1 metabolome provides further evidence that SM production has been downregulated in A. oryzae when compared to ancestral Aspergillus species, and that this downregulation has occurred through means other than McrA-mediated repression. Previous studies have shown that both A. oryzae and the closely related A. flavus are actually enriched in genes for secondary metabolism when compared to other Aspergillus species (Payne et al., 2006), suggesting that A. oryzae does have the genetic potential for SM production, but where A. flavus is a prolific producer of secondary metabolites, gene clusters in A. oryzae are downregulated (Gibbons et al., 2012). One interesting avenue for further research would be to determine whether McrA impacts the transcriptome of A. oryzae and thus assess whether the expression of gene clusters has been decoupled from McrA regulation, or whether a factor other than transcription is impacting the SM profile of A. oryzae. It would also be intriguing to determine whether McrA-mediated repression of secondary metabolism functions as a key regulatory factor in A. flavus, and thus whether changes in the role of McrA have occurred during the recent domestication of A. oryzae.
Although kojic acid 1 is a commercially valuable metabolite, its production can be problematic in the context of using A. oryzae as a heterologous host, making chemical extractions and purification of metabolites more time-consuming and costly. Therefore, CRISPR-Cas9 technology was harnessed to halt kojic acid 1 production in an NSAR1 derived strain, through targeted mutation of kojA, an FAD-dependent oxidoreductase shown previously to be necessary for kojic acid production (Terabayashi et al., 2010). The plasmid construction strategy was intentionally designed so that the addition of the target-specific sgRNA cassette was the final step. This means that the intermediate plasmid, pAYargCas9, which contains both the Cas9 and AMA1 cassettes but not the sgRNA cassette, could easily be adapted to target other loci in strain NSAR1. Targeting kojA led to the generation of a strain containing an 8 bp deletion at the target site, with a resulting loss of function and subsequent inability to produce kojic acid. The kojA mutant strain, named NSARΔK, was then used as a host for the production of pretenellin A 8, demonstrating the potential value of this strain as a platform for the production of commercially important heterologous compounds.
Slightly surprisingly, the CRISPR-Cas9 approach used here was found to be relatively inefficient, with only one out of 42 A. oryzae transformants demonstrating a loss of function for KojA. The low rate of disruption was not especially problematic in this instance, as rapid screening of kojic acid production is possible, but may be a hindrance in cases where disruption does not lead to a clear mutant phenotype. It is possible that the efficiency of targeting could be improved by the addition of a template for homology directed repair. Another consideration worth noting is the possibility of off-target mutations, which are known to occur with CRISPR-Cas9 editing (Zhang et al., 2015, Zischewski et al., 2017). Although multiple A. oryzae genome sequences are available, there is not currently a publicly available genome sequence for A. oryzae NSAR1. Sequencing the genomes of both NSAR1 and the novel strain NSARΔK would allow for an assessment of off-target mutations in strain NSARΔK and would provide genome data to support any further applications of these useful strains.
Finally, the isolation of apparent minor metabolites from crude extracts, which were later identified as diketopiperazines originating in the culturing medium, serves as a useful warning to the natural product community to consider all possible sources of small molecules and to document their presence for exclusion from future analyses.
Together these findings advanced our understanding of A. oryzae NSAR1 and led to the development of a new strain for heterologous production, namely strain NSARΔK. As a filamentous factory, A. oryzae NSARΔK offers a uniquely clean background.
Escherichia coli strain TOP10 (Invitrogen) was used as a host for plasmids and was maintained according to standard procedures (Sambrook and Russell, 2001). Saccharomyces cerevisiae strain YPH499 (Stratagene) was used for plasmid assembly by homologous recombination and was maintained on YPAD medium (1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) glucose, 0.004% (w/v) adenine sulfate, 1.5% (w/v) agar) at 28°C. A. oryzae NSAR1 was obtained as a gift from the Kitamoto group and was maintained on MEA medium (Sigma) at 28°C (Jin et al., 2004).
Transformation of A. oryzae was performed according to the protocol outlined by Williams et al. (2016).
A. oryzae strains were cultured in 10 ml GN medium [2% (w/v) D(+)-glucose monohydrate, 1 % (w/v) Nutrient broth No. 2 (Oxoid, UK)] at 28°C with shaking at 200 rpm, then pelleted, lyophilised and ground under liquid nitrogen. Genomic DNA was prepared using the GenElute Plant Genomic DNA Miniprep kit (Sigma).
Ao_mcrA was deleted from A. oryzae strain NSAR1 using the bipartite method (Nielsen et al., 2006). This method relies on the splitting of the selection marker (with a ~500 bp overlap), which leads to the requirement for homologous recombination between the two selection marker halves for selection to occur. This requirement for homologous recombination leads to higher gene disruption levels.
A plasmid was constructed using homologous recombination in S. cerevisiae to provide a template for the bi-partite fragment amplification (Figure 6). Flanking regions either side of Ao_mcrA were amplified using the proof-reading polymerase KAPA HiFi HotStart (Roche), with the primer pairs AO-mcrAKO-P1F/AO-mcrAKO-P2R and AO-mcrAKO-P3F/AO-mcrAKOP4R. The argB cassette which provides selection in A. oryzae NSAR1 was amplified using primer pair PargB-P5F/TargB-P8R using the plasmid pTYarg as the template (Lazarus et al., 2014). All three fragments were recombined into the E. coli/yeast shuttle plasmid pE-YA (Pahirulzaman et al., 2012), which had been linearised with NotI. The resulting recombinant plasmid, pE-YA-mcrAKO, was used as a template for PCR amplification of the bi-partite fragments using primers AO-mcrAKO-P1F with argB-P6R, and argB-P7F with AO-mcrAKOP4R. Primers used are shown in Table 1.
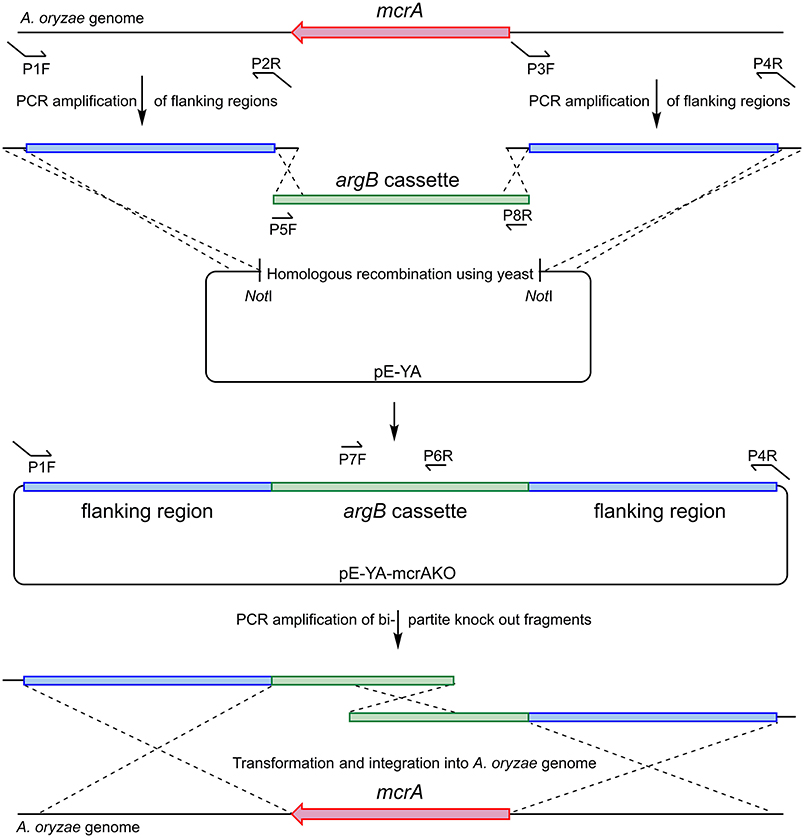
Figure 6. Construction of bi-partite fragments used for the deletion of Ao_mcrA and recombination into the A. oryzae genome.

Table 1. Primers used in this work. Compound primers, with tails (underlined) were designed to allow for yeast recombination.
A kojA mutant strain of NSAR1 was created using CRISPR-Cas9 technology, through the construction and application of plasmid pAYargCas9-kojA2.
Plasmid pAYargCas9-kojA2 was built from the A. oryzae expression plasmid pTYGS-arg (Lazarus et al., 2014) in multiple steps (Figure 7). Firstly, cas9 was recombined into pTYGS-arg by yeast recombination. Cas9 was amplified from plasmid pKWGF2::hCas9 (Nekrasov et al., 2013) in two overlapping fragments, to allow the removal of an internal AscI site, using the primer pairs PamyB-Cas9-F/Cas9-Asc-R and Cas9-Asc-F/Cas9-TamyB-R. These fragments were combined with pTYGS-arg digested with NotI, placing cas9 within the amyB cassette and creating plasmid pTYargCas9. pTYargCas9 was then adapted to contain the AMA1 autonomously replicating sequence to allow for extrachromosomal replication rather than integration, and the subsequent loss of the plasmid to recycle the selectable marker. This was achieved by digesting pTYargCas9 with NheI and inserting AMA1 into this break in two overlapping fragments which were amplified from pFC330 (Nødvig et al., 2015) using the primer pairs AMA1-1F/AMA1-2R and AMA1-2F/AMA1-1R. Again, this was done via yeast recombination, yielding the plasmid pAYargCas9. Finally, the single chimeric guide RNA (sgRNA) was constructed by recombining two PCR products into pAYargCas9 to place them between the gpdA promoter and the enolase terminator. This utilized the design of Nødvig and co-workers (Nødvig et al., 2015) where the sgRNA is embedded within a larger transcript from which it is released by the action of two ribozyme sequences; the 5′-end hammerhead (HH) and 3′-end hepatitis delta virus (HDV), which flank the sgRNA. This allows for the use of standard RNA polymerase II promoters, removing the requirement for an RNA polymerase III promoter. Portions of the sgRNA cassette were amplified from plasmid pFC334 (Nødvig et al., 2015) using the primer pairs Pgpd-kojA2-HH-F/Guide-kojA2-R and HH-kojA2-Guide-F/Teno-HDV-R. These primers introduced the necessary kojA-specific sequences including the protospacer, the sgRNA backbone and the variable HH sequence. The two resulting PCR products were combined with pAYargCas9 that had been digested with AscI as well as a short PCR product (amplified using primer pair ExAdhSeq.F/ExSeqAdh.R) to repair an AscI break site in another cassette and yeast recombination was conducted (Figure 7). This produced the final plasmid; pAYargCas9-kojA2, which was used to transform A. oryzae NSAR1. Primers used in construction of this plasmid are shown in Table 1.
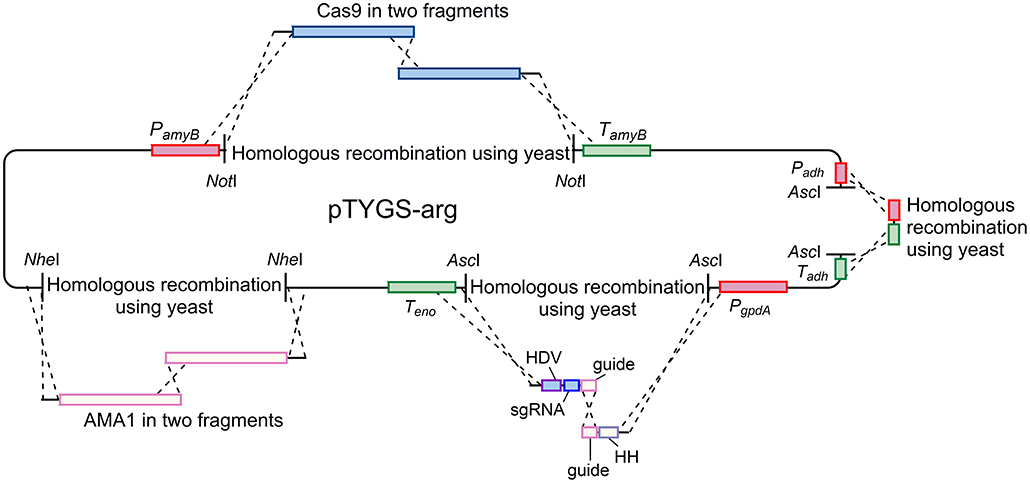
Figure 7. Diagram demonstrating the strategy used to construct the pAYargCas9-kojA2 plasmid. Cas9, AMA1, and the guide RNA were each introduced via consecutive yeast recombinations, as described in the text.
Putative ΔkojA transformants were screened by culturing on KA-Fe solid medium (5% (w/v) glucose, 0.25% (w/v) yeast extract, 0.1% (w/v) K2HPO4, 0.05% (w/v) MgSO4.7H2O, 1 mM FeCl3, 2% (w/v) agar, pH 5) (Terabayashi et al., 2010). Kojic acid reacts with the ferrous chloride in the medium and causes a clear color change to red. An absence of color change therefore indicates a loss of kojic acid production.
gDNA from nine putative ΔmcrA A. oryzae strains was analyzed using 4 sets of primers (Table 1). The first pair tested for integration of the “left hand” flanking region (primer pair AO-mcrA-test-F/pArgB-Bg-R). These primers should only amplify a product if integration has occurred correctly. The second pair (TargB-Nd-F/AO-mcrA-test-R) tested for correct integration of the “right-hand” flanking region. The third pair (AO-mcrA-F/AO-mcrA-R) tested for the presence of the wild-type Ao_mcrA gene. If the transformants are genetically pure, then no product should be amplified. As final confirmation, primers AO-mcrA-test-F/AO-mcrA-test-R were used to amplify over the whole disrupted region. Primers used are shown in Table 1. Analytical PCR was performed using Biomix Red (Bioline).
gDNA from A. oryzae NSAR1 and the putative ΔkojA transformant was analyzed using the primer pair kojA-F/kojA-R (Table 1) which amplified a ~1 kb product. This was subsequently sequenced to identify the 8 bp deletion in the ΔkojA transformant. Amplification of fragments for sequencing was performed using the high-fidelity KAPA HiFi HotStart (Roche).
Sample solutions (20 μl) were analyzed using a Waters LCMS system comprising of a Phenomenex KINETEX column (2.6 μ, C18, 100 Å, 4.6 × 100 mm) equipped with a Phenomenex Security Guard precolumn (Luna C5 300 Å) eluted at 1 ml/min. Detection was by Waters 2998 Diode Array detector between 200 and 600 nm; Waters 2424 ELSD and Waters SQD-2 mass detector operating simultaneously in ES+ and ES− modes between 100 and 800 m/z. Solvent A was HPLC grade water containing 0.05% formic acid, and solvent B was HPLC grade acetonitrile containing 0.05% formic acid. Gradient program was as follows: 5–95% of B over 15 min. 0 min, 5% B; 1 min, 5% B; 2 min, 40% B; 15 min, 95% B; 17 min, 95% B; 18 min, 5% B; 20 min, 5% B; flow rate 1 ml/min.
Instruments used; Varian 400-MR (400 MHz), Varian VNMRS500 (500 MHz), Bruker 500 Cryo (500 MHz), or Varian VNMRS600 Cryo (600 MHz). Chemical shifts (δ) quoted in parts per million (ppm) and coupling constants (J) in Hertz (Hz), rounded to 0.5 Hz intervals. Two-dimensional NMR techniques (HSQC, COZY, HMBC) were used routinely for the assignment of structures.
Initial screens of strain AoΔmcrA-7 were conducted in CMP (3.5% (w/v) Czapek Dox broth, 2% (w/v) maltose, 1% (w/v) peptone), MEB (1.7% (w/v) malt extract, 0.3% (w/v) mycological peptone), PDB (2.4% (w/v) potato dextrose broth), GN [2% (w/v) D(+)-glucose monohydrate, 1% (w/v) Nutrient broth No. 2 (Oxoid, UK)] and rice (50 g of rice, autoclaved in 50 ml of distilled H2O). Cultures were grown for 1 week at 28°C with shaking at 180 rpm, with the exception of the rice culture which is a static culture (28°C). Cultures were extracted with an equal volume of ethyl acetate and analyzed by LCMS.
For purification of compounds, strain AoΔmcrA-7 was cultured for 3 days in CMP medium at 28°C. This seed culture was then used to inoculate 2 l of CMP. The flasks were shaken at 180 rpm at 28°C for 1 week. The broth was filtered by Buchner filtration and extracted with ethyl acetate (1.5 l × 3). The organic layers were concentrated under reduced pressure to give 7.4 g of crude extract. The major product 1 (3.6 g) was crystallized out from ethyl acetate. The mother liquor was then chromatographed by reversed-phase silica gel column chromatography eluted with acetonitrile/water (v/v, from 1:3 to 1:0) to yield 3 fractions (F1–F3). From fraction F2 (320 mg), 2 (7.4 mg), a mixture of 3 and 4 (13.5 mg), 5 (3.8 mg), 6 (2.5 mg), and 7 (3.5 mg) were afforded by preparative LCMS (Luna 5μ C18 100 Å, 250 × 10 mm, 8 ml/min) using a 20 min 5–60% acetonitrile in water (+ 0.1% formic acid).
Kojic acid (1), white crystals, UV λmax 216, 268 nm, ESI-MS m/z 143 [M + H]+. 1H NMR (500 MHz, CD3OD, δH): 4.41 (2H, s, H-6), 6.50 (1H, s, H-4), 7.96 (1H, s, H-1); δC (125 MHz, CD3OD) 61.4 (C-6), 110.9 (C-4), 141.2 (C-1), 147.6 (C-2), 170.6 (C-5), 177.1 (C-3). These data were in agreement with the reported literature (Kingsbury et al., 1976).
Kojic acid dimer (2), white amorphous powder, UV λmax 216, 273 nm, ESI-MS m/z 283 [M + H]+. 1H NMR (500 MHz, CD3OD, δH): 4.35 (1H, d, J = 12.0 Hz, H-12a), 4.41 (1H, d, J = 12.0 Hz, H-12b), 5.94 (1H, s, H-6), 6.49 (1H, s, H-10), 6.73 (1H, s, H-4), 7.94 (1H, s, H-1); δC (125 MHz, CD3OD) 61.1 (C-12), 65.9 (C-6), 110.1 (C-10), 111.7 (C-4), 141.2 (C-1), 144.5 (C-8), 147.6 (C-2), 147.9 (C-7), 168.1 (C-5), 170.1 (C-11), 176.6 (C-9), 176.7 (C-3). These data were in agreement with the reported literature (Nakagawa et al., 1993).
A mixture (1:1) of compounds 3 and 4 was obtained as a white amorphous powder. Cyclo-[(D)Pro-(L)Tyr] (3), ESI-MS m/z 261 [M + H]+. 1H NMR (500 MHz, CD3OD, δH): 1.95 (2H, m, H-3a and 4a), 2.04 (1H, m, H-4b), 2.33 (1H, m, H-3b), 3.07 (2H, m, H-8), 3.57 (2H, m, H-5), 4.22 (1H, m, H-2), 4.38 (1H, m, H-7), 6.72 (2H, d, J = 8.5 Hz, H-11 and H-13), 7.04 (2H, d, J = 8.5 Hz, H-10 and H-14); δC (125 MHz, CD3OD) 23.3 (C-4), 29.5 (C-3), 37.7 (C-8), 46.2 (C-5), 57.9 (C-7), 60.1 (C-2), 116.2 (C-11 and C-13), 127.6 (C-9), 132.1 (C-10), 157.7 (C-12), 166.9 (C-6), 172.5 (C-1). These data were in agreement with the reported literature (Kumar et al., 2012).
Cyclo-[(D)Pro-(L)Val] (4), ESI-MS m/z 197 [M + H]+. 1H NMR (500 MHz, CD3OD, δH): 0.95 (3H, d, J = 7.5 Hz, H-9), 1.11 (3H, d, J = 7.5 Hz, H-10), 1.24 (1H, m, H-3a), 1.81 (2H, m, H-4), 2.11 (1H, m, H-3b), 2.51 (1H, m, H-8), 3.36 (1H, m, H-5a), 3.52 (1H, m, H-5b), 4.05 (1H, m, H-7), 4.07 (1H, m, H-2); δC (125 MHz, CD3OD) 16.7 (C-9), 18.4 (C-10), 22.7 (C-4), 29.4 (C-3), 29.9 (C-8), 45.9 (C-5), 60.1 (C-2), 61.5 (C-7), 167.6 (C-6), 170.7 (C-1). These data were in agreement with the reported literature (Kwon et al., 2001).
Cyclo-[(D)Pro-(L)Ile] (5), white amorphous powder, [α] + 72° (c 0.01, MeOH), ESI-MS m/z 211 [M + H]+. 1H NMR (500 MHz, CD3OD, δH): 0.96 (3H, d, J = 7.5 Hz, H-10), 1.08 (3H, d, J = 7.5 Hz, H-11), 1.34 (1H, m, H-9a), 1.47 (1H, m, H-9b), 1.95 (2H, m, H-3a and H-4a), 2.04 (1H, m, H-4b), 2.34 (1H, m, H-3b), 3.07 (1H, m, H-8), 3.50-3.58 (2H, m, H-5), 4.09 (1H, m, H-7), 4.21 (1H, m, H-2); δC (125 MHz, CD3OD) 12.6 (C-10), 15.5 (C-11), 23.2 (C-4), 25.4 (C-9), 29.6 (C-3), 37.1 (C-8), 60.0 (C-2), 61.3 (C-7), 167.5 (C-6), 172.4 (C-1). These data were in agreement with the reported literature (Bull et al., 1998).
Cyclo-[(D)Pro-(L)Leu] (6), white amorphous powder, [α] – 142° (c 0.01, MeOH), UV λmax 210 nm, ESI-MS m/z 211 [M + H]+. 1H NMR (500 MHz, CD3OD, δH): 0.97 (6H, m, H-10 and H-11), 1.53 (1H, m, H-8a), 1.91 (2H, m, H-4a and H-9), 1.95 (1H, m, H-9b), 2.03 (1H, m, H-4b), 2.31 (1H, m, H-3b), 3.53 (1H, m, H-5), 4.14 (1H, m, H-7), 4.27 (1H, m, H-2); δC (125 MHz, CD3OD) 20.8 (C-10), 21.9 (C-11), 22.2 (C-4), 24.4 (C-9), 27.7 (C-3), 37.9 (C-8), 45.0 (C-5), 53.2 (C-7), 58.9 (C-2), 167.5 (C-6), 171.3 (C-1). These data were in agreement with the reported literature (Kumar et al., 2012).
Cyclo-[(D)Pro-(L)Phe] (7), white amorphous powder, [α] + 51° (c 0.01, MeOH), UV λmax 210 nm, ESI-MS m/z 245 [M + H]+. 1H NMR (500 MHz, CD3OD, δH): 1.24 (1H, m, H-3a), 1.81 (2H, m, H-4), 2.11 (1H, m, H-3a), 3.19 (2H, m, H-8), 3.40 (1H, m, H-5a), 3.54 (1H, m, H-5b), 4.10 (1H, m, H-2), 4.45 (1H, m, H-7), 7.23 – 7.31 (5H, m, H-10, 11, 12, 13, 14); δC (125 MHz, CD3OD) 22.8 (C-4), 29.4 (C-3), 38.2 (C-8), 45.9 (C-5), 57.7 (C-7), 60.1 (C-2), 128.1 (C-12), 129.4 (C-11 and C-13), 131.0 (C-10 and C-14), 137.3 (C-9), 166.8 (C-6), 170.9 (C-1). These data were in agreement with the reported literature (Kumar et al., 2013).
Strains ΔK-Ten120 and N-Ten402 were cultured on solid CMP medium (3.5% (w/v) Czapek Dox broth, 2% (w/v) maltose, 1% (w/v) peptone, 2% (w/v) agar) for 5 days at 28°C. Subsequently, 1 cm2 plugs of agar from the rim and the center of growing colonies were extracted in 2 ml of a solution of ethyl acetate, dichloromethane and methanol (3:2:1) with 1% acetic acid. After 1 h in an ultrasonic bath, the liquid was removed and dried. The crude extract was then dissolved in 200 μl acetonitrile and analyzed by HPLC-MS/UV/ELSD.
Acetonitrile solutions containing specific kojic acid concentrations were prepared (0.25–1.5 mg/ml) and injected into our HPLC system in triplicate. The calibration curve was created by regression analysis plotting the kojic acid concentrations (mg/ml) with the area of the peak obtained from the ELSD chromatogram. The relationship was linear within the 0.25–1.50 mg/ml concentration range and the lower limit of detection was 0.20 mg/ml (Supplementary Figure 46). The obtained linear regression equation, y = 18784x – 5527.7 (with the coefficient of correlation r2 = 0.98), was then applied to quantify kojic acid in the crude extracts. All analyses were performed in triplicate, and the statistical analysis of the data was achieved by using Microsoft Office Excel 2016.
Strains A. oryzae NSAR1 and AoΔmcrA-7 were cultured for 2 days in CMP media at 28°C. From this seeding culture, 200 μl was used to inoculate conical flasks containing 50 ml of CMP. The flasks were then shaken at 180 rpm at 28°C for 1 week. The broths were separated from the mycelium, and a 10 ml portion of each culture was extracted with ethyl acetate (10 ml) by shaking for 3 min. A 5 ml portion of the organic layer was then concentrated under nitrogen gas to give a crude extract. The crude extract was then dissolved in acetonitrile at the concentration of 1 mg/ml for HPLC injection. Each extraction was performed in triplicate.
The original contributions generated for the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
AB and CL contributed to the design of the research. CL contributed to plasmid design and construction. TD, KdM-S, IP, KW, and MZ performed the experimental work. TD, KdM-S, IP, KW, and CW were involved in interpretation of data. KdM-S and KW drafted the manuscript. CL, CW, and AB had supervisory roles. All authors contributed to the article and approved the submitted version.
TD, KdM-S, IP, KW, and MZ are funded by the MRC (MR/N029909/1).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffunb.2021.632542/full#supplementary-material
SM, Secondary Metabolite; GRAS, Generally Recognized As Safe; mcrA, multicluster regulator A; BGC, Biosynthetic Gene Cluster; PCR, Polymerase Chain Reaction; LCMS, Liquid Chromatography Mass Spectrometry; ELSD, Evaporative Light Scattering Detector; HPLC, High Pressure Liquid Chromatography; TLC, Thin-Layer Chromatography; CRISPR, Clustered, Regularly Interspaced Short Palindromic Repeats; Cas, CRISPR-associated proteins; FAD, Flavin adenine dinucleotide; UV, Ultraviolet; ESI-MS, Electrospray ionization - Mass Spectrometry; NMR, Nuclear magnetic resonance; PamyB, promoter for A. oryzae α-amylase; Padh, promoter for A. oryzae alcohol dehydrogenase; PgpdA, promoter for Aspergillus nidulans glyceraldehyde-3-phosphate dehydrogenase; Peno, promoter for A. oryzae enolase; argB, Aspergillus nidulans ornithine carbamoyltransferase gene; AMA1, autonomously maintained in Aspergillus; RNA, Ribonucleic acid; sgRNA, single chimeric guide RNA; HH, 5′-end hammerhead ribozyme sequence; HDV, 3′-end hepatitis delta virus ribozyme sequence; rpm, revolutions per minute.
Alberti, F., Khairudin, K., Venegas, E. R., Davies, J. A., Hayes, P. M., Willis, C. L., et al. (2017). Heterologous expression reveals the biosynthesis of the antibiotic pleuromutilin and generates bioactive semi-synthetic derivatives. Nat. Commun. 8:1831. doi: 10.1038/s41467-017-01659-1
Bailey, A. M., Alberti, F., Kilaru, S., Collins, C. M., de Mattos-Shipley, K., Hartley, A. J., et al. (2016). Identification and manipulation of the pleuromutilin gene cluster from Clitopilus passeckerianus for increased rapid antibiotic production. Sci. Rep. 6:25202. doi: 10.1038/srep25202
Barbesgaard, P., Heldt-Hansen, H. P., and Diderichsen, B. (1992). On the safety of Aspergillus oryzae: a review. Appl. Microbiol. Biotechnol. 36, 569–572. doi: 10.1007/BF00183230
Bentley, R. (2006). From miso, sake and shoyu to cosmetics: a century of science for kojic acid. Nat. Prod. Rep. 23, 1046–1062. doi: 10.1039/b603758p
Bok, J. W., and Keller, N. P. (2004). LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryotic Cell 3, 527–535. doi: 10.1128/EC.3.2.527-535.2004
Bull, S. D., Davies, S. G., Parkin, R. M., and Sanchez-Sancho, F. (1998). The biosynthetic origin of diketopiperazines derived from D-proline. J. Chem. Soc. Perkin. Trans. 1, 2313–2320. doi: 10.1039/a802702a
Chiba, R., Minami, A., Gomi, K., and Oikawa, H. (2013). Identification of ophiobolin F synthase by a genome mining approach: a sesterterpene synthase from Aspergillus clavatus. Org. Lett. 15, 594–597. doi: 10.1021/ol303408a
Dong, Y., Cui, C. B., Li, C. W., Hua, W., Wu, C. J., Zhu, T. J., et al. (2014). Activation of dormant secondary metabolite production by introducing neomycin resistance into the deep-sea fungus, Aspergillus versicolor ZBY-3. Mar. Drugs 12, 4326–4352. doi: 10.3390/md12084326
Frisvad, J. C., Moller, L. L. H., Larsen, T. O., Kumar, R., and Arnau, J. (2018). Safety of the fungal workhorses of industrial biotechnology: update on the mycotoxin and secondary metabolite potential of Aspergillus niger, Aspergillus oryzae, and Trichoderma reesei. Appl. Microbiol. Biotechnol. 102, 9481–9515. doi: 10.1007/s00253-018-9354-1
Fujii, R., Minami, A., Tsukagoshi, T., Sato, N., Sahara, T., Ohgiya, S., et al. (2011). Total biosynthesis of diterpene aphidicolin, a specific inhibitor of DNA polymerase alpha: heterologous expression of four biosynthetic genes in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 75, 1813–1817. doi: 10.1271/bbb.110366
Gibbons, J. G., Salichos, L., Slot, J. C., Rinker, D. C., McGary, K. L., King, J. G., et al. (2012). The evolutionary imprint of domestication on genome variation and function of the filamentous fungus Aspergillus oryzae. Curr. Biol. 22, 1403–1409. doi: 10.1016/j.cub.2012.05.033
Heneghan, M. N., Yakasai, A. A., Halo, L. M., Song, Z., Bailey, A. M., Simpson, T. J., et al. (2010). First heterologous reconstruction of a complete functional fungal biosynthetic multigene cluster. Chembiochem 11, 1508–1512. doi: 10.1002/cbic.201000259
Jin, F. H., Maruyama, J.-I., Juvvadi, P. R., Arioka, M., and Kitamoto, K. (2004). Development of a novel quadruple auxotrophic host transformation system by argB gene disruption using adeA gene and exploiting adenine auxotrophy in Aspergillus oryzae. FEMS Microbiol. Lett. 239, 79–85. doi: 10.1016/j.femsle.2004.08.025
Kingsbury, C. A., Cliffton, M., and Looker, J. H. (1976). C-13 nuclear magnetic-resonance spectra of kojic acid and other 4-pyrone derivatives. J. Org. Chem. 41, 2777–2780. doi: 10.1021/jo00878a033
Kumar, S. N., Mohandas, C., Nambisan, B., Kumar, D. R. S., and Lankalapalli, R. S. (2013). Isolation of proline-based cyclic dipeptides from Bacillus sp N strain associated with rhabitid entomopathogenic nematode and its antimicrobial properties. World J. Microbiol. Biotechnol. 29, 355–364. doi: 10.1007/s11274-012-1189-9
Kumar, S. N., Mohandas, C., Siji, J. V., Rajasekharan, K. N., and Nambisan, B. (2012). Identification of antimicrobial compound, diketopiperazines, from a Bacillus sp N strain associated with a rhabditid entomopathogenic nematode against major plant pathogenic fungi. J. Appl. Microbiol. 113, 914–924. doi: 10.1111/j.1365-2672.2012.05385.x
Kwon, O. S., Park, S. H., Yun, B. S., Pyun, Y. R., and Kim, C. J. (2001). Cyclo(D-Pro-L-Val), a specific ss-glucosidase inhibitor produced by Aspergillus sp F70609. J Antibiot. 54, 179–181. doi: 10.7164/antibiotics.54.179
Lazarus, C. M., Williams, K., and Bailey, A. M. (2014). Reconstructing fungal natural product biosynthetic pathways. Nat. Prod. Rep. 31, 1339–1347. doi: 10.1039/C4NP00084F
Machida, M., Asai, K., Sano, M., Tanaka, T., Kumagai, T., Terai, G., et al. (2005). Genome sequencing and analysis of Aspergillus oryzae. Nature 438, 1157–1161. doi: 10.1038/nature04300
Mitchell, A. L., Attwood, T. K., Babbitt, P. C., Blum, M., Bork, P., Bridge, A., et al. (2019). InterPro in 2019, improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47, D351–D360. doi: 10.1093/nar/gky1100
Nakagawa, S., Nakanishi, O., Furuya, T., Okumya, T., and Sugihara, R. (1993). Physiologically active kojic acid dimer, its manufacture with Pseudonocardia species, and skin-lightening preparations containing the dimer. Japanese Patent No. JPH05310727A. JPO (Japan patent Office).
Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J. D., and Kamoun, S. (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 691–693. doi: 10.1038/nbt.2655
Nielsen, M. L., Albertsen, L., Lettier, G., Nielsen, J. B., and Mortensen, U. H. (2006). Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal Genet. Biol. 43, 54–64. doi: 10.1016/j.fgb.2005.09.005
Nødvig, C. S., Nielsen, J. B., Kogle, M. E., and Mortensen, U. H. (2015). A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS ONE 10:e0133085. doi: 10.1371/journal.pone.0133085
Oakley, C. E., Ahuja, M., Sun, W.-W., Entwistle, R., Akashi, T., Yaegashi, J., et al. (2017). Discovery of McrA, a master regulator of Aspergillus secondary metabolism. Mol. Microbiol. 103, 347–365. doi: 10.1111/mmi.13562
Pahirulzaman, K. A. K., Williams, K., and Lazarus, C. M. (2012). A toolkit for heterologous expression of metabolic pathways in Aspergillus oryzae. Meth. Enzymol. 517, 241–260. doi: 10.1016/B978-0-12-404634-4.00012-7
Payne, G. A., Nierman, W. C., Wortman, J. R., Pritchard, B. L., Brown, D., Dean, R. A., et al. (2006). Whole genome comparison of Aspergillus flavus and A. oryzae. Med. Mycol. 44, S9–11. doi: 10.1080/13693780600835716
Rokas, A., Payne, G., Fedorova, N. D., Baker, S. E., Machida, M., Yu, J., et al. (2007). What can comparative genomics tell us about species concepts in the genus Aspergillus? Stud. Mycol. 59, 11–17. doi: 10.3114/sim.2007.59.02
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual, 3rd Edn. New York, NY: Cold Spring Harbor Laboratory Press.
Shaaban, M., El-Metwally, M. M., and Nasr, H. (2014). A new diketopiperazine alkaloid from Aspergillus oryzae. Nat. Prod. Res. 28, 86–94. doi: 10.1080/14786419.2013.841687
Tagami, K., Liu, C. W., Minami, A., Noike, M., Isaka, T., Fueki, S., et al. (2013). Reconstitution of biosynthetic machinery for indole-diterpene paxilline in Aspergillus oryzae. J. Am. Chem. Soc. 135, 1260–1263. doi: 10.1021/ja3116636
Terabayashi, Y., Sano, M., Yamane, N., Marui, J., Tamano, K., Sagara, J., et al. (2010). Identification and characterization of genes responsible for biosynthesis of kojic acid, an industrially important compound from Aspergillus oryzae. Fungal Genet. Biol. 47, 953–961. doi: 10.1016/j.fgb.2010.08.014
Thammarongtham, C., Nookaew, I., Vorapreeda, T., Srisuk, T., Land, M. L., Jeennor, S., et al. (2018). Genome characterization of oleaginous Aspergillus oryzae BCC7051, a potential fungal-based platform for lipid production. Curr. Microbiol. 75, 57–70. doi: 10.1007/s00284-017-1350-7
Theuretzbacher, U., Outterson, K., Engel, A., and Karlen, A. (2019). The global preclinical antibacterial pipeline. Nat. Rev. Microbiol. 18, 275–285. doi: 10.1038/s41579-019-0288-0
Uchoa, P. K. S., Pimenta, A. T. A., Braz-Filho, R., de Oliveira, Md,CF, Saraiva, N. N., Rodrigues, B. S. F., et al. (2017). New cytotoxic furan from the marine sediment-derived fungi Aspergillus niger. Nat. Prod. Res. 31, 2599–2603. doi: 10.1080/14786419.2017.1283499
Wasil, Z., Pahirulzaman, K. A. K., Butts, C., Simpson, T. J., Lazarus, C. M., and Cox, R. J. (2013). One pathway, many compounds: heterologous expression of a fungal biosynthetic pathway reveals its intrinsic potential for diversity. Chem. Sci. 4, 3845–3856. doi: 10.1039/c3sc51785c
Watanabe, A., Ono, Y., Fujii, I., Sankawa, U., Mayorga, M. E., Timberlake, W. E., et al. (1998). Product identification of polyketide synthase coded by Aspergillus nidulans wA gene. Tetrahedron Lett. 39, 7733–7736. doi: 10.1016/S0040-4039(98)01685-2
Williams, K., Szwalbe, A. J., Mulholland, N. P., Vincent, J. L., Bailey, A. M., Willis, C. L., et al. (2016). Heterologous production of fungal maleidrides reveals the cryptic cyclization involved in their biosynthesis. Angew Chem. Int. Ed. 55, 6783–6787. doi: 10.1002/anie.201511882
Wong, N., Liu, W., and Wang, X. (2015). WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 16:218. doi: 10.1186/s13059-015-0784-0
Yang, X. L., Friedrich, S., Yin, S., Piech, O., Williams, K., Simpson, T. J., et al. (2019). Molecular basis of methylation and chain-length programming in a fungal iterative highly reducing polyketide synthase. Chem. Sci. 10, 8478–8489. doi: 10.1039/C9SC03173A
Zhang, X. H., Tee, L. Y., Wang, X. G., Huang, Q. S., and Yang, S. H. (2015). Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids 4:e264. doi: 10.1038/mtna.2015.37
Zhao, G., Yao, Y., Qi, W., Wang, C., Hou, L., Zeng, B., et al. (2012). Draft genome sequence of Aspergillus oryzae strain 3.042. Eukaryotic Cell 11, 1178–1178. doi: 10.1128/EC.00160-12
Keywords: heterologous host, Aspergillus, natural products, mcrA, secondary metabolism, kojic acid, NSAR1, NSARΔK
Citation: Dao TT, de Mattos-Shipley KMJ, Prosser IM, Williams K, Zacharova MK, Lazarus CM, Willis CL and Bailey AM (2021) Cleaning the Cellular Factory–Deletion of McrA in Aspergillus oryzae NSAR1 and the Generation of a Novel Kojic Acid Deficient Strain for Cleaner Heterologous Production of Secondary Metabolites. Front. Fungal Biol. 2:632542. doi: 10.3389/ffunb.2021.632542
Received: 23 November 2020; Accepted: 18 January 2021;
Published: 09 February 2021.
Edited by:
Nancy Keller, University of Wisconsin-Madison, United StatesReviewed by:
Berl Oakley, University of Kansas, United StatesCopyright © 2021 Dao, de Mattos-Shipley, Prosser, Williams, Zacharova, Lazarus, Willis and Bailey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrew M. Bailey, YW5keS5iYWlsZXlAYnJpc3RvbC5hYy51aw==
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.