- 1Department of Plant Molecular Biology, University of Delhi South Campus, New Delhi, India
- 2Faculty of Forestry and Wood Sciences, Czech University of Life Sciences Prague, Prague, Czechia
- 3Molecular Biology Research Lab, Department of Zoology, Deshbandhu College, University of Delhi, New Delhi, India
- 4Delhi School of Climate Change and Sustainability, Institution of Eminence, Maharishi Kannad Bhawan, University of Delhi, New Delhi, India
Forests are potential habitats for immense terrestrial ecosystems and aquatic biodiversity, performing an essential role in ecological preservation and regulation of climate. The anthropogenic pressures on the forests lead to forest loss, fragmentation and degradation. Requirements for sustainable methodologies for forest protection are of utmost priority under the climate change regime. Among forest trees, poplar trees (Populus L.) have attracted attention in global forestry as a promising material for improving the quality and quantity of urban landscapes. These plants provide wood, which can be utilized as raw resources for the paper industry and as a potential source of biofuel. However, several biotic stresses, such as attacks by pests and pathogens, severely affect poplar production and productivity. The improvement of Populus trees through conventional tree breeding methods is restricted due to their long-life cycles and the lack of suitable donors with resistance genes. Populus has been utilized as a model plant for studying gene functions due to its highly efficient genetic transformation capabilities. The present review will provide a comprehensive overview of pest and pathogen attacks on poplar, focusing on their infection mechanisms, transmission routes, and control strategies. Additionally, it will examine the most widely used genetic transformation methods (gene gun-mediated, Agrobacterium tumefaciens-mediated, protoplast transformation, micro-RNA mediated and micro-RNA clustered regularly interspaced short palindromic repeats (CRISPR)-associated (CRISPR-Cas) systems methods and RNA interference) for improving tolerance in poplar trees against pest and pathogens attack. Furthermore, it will delve into prospects, challenges, and recent advances in molecular biology tools and their safe application for genetic transformation to improve insect and pest resistance in poplar trees. Finally, the regeneration of transgenic poplar trees with enhanced resistance, developed through various genetic engineering techniques, is discussed.
1 Introduction
Forest trees have several pivotal roles, such as maintaining ecology, climate regulation, providing raw materials for the construction of buildings, greening roads, and being an energy source (Trumbore et al., 2015). Among these forest trees, poplar (Populus spp.) (known as ‘the people’s tree’) is one of the most widespread trees in the world (Xi et al., 2021; Yevtushenko and Misra, 2019). The poplars are essential for maintaining the world’s ecological balance and socio-economic wellbeing (Häggman et al., 2013). The first use of the poplar cultivar started in 1700–1720 when Populus nigra ‘Italica’ (P. nigra, Lombardy nigra) was used in Italy, Europe and North America. There was a rapidly increased demand for poplar plants after World War II when Europe was devastated by the lack of readily available wood for construction and fuel. Consequently, the domestication and cultivation of the genus poplar started in Europe by introducing eastern cottonwood (Populous deltoides) and followed by hybrids black poplar (P. nigra) (a hybrid of P. ×canadensis) for fulfilling the increased demands for woods (Stanton et al., 2009).
Poplar trees (family Salicaceae) are tall, deciduous, dioecious, paleopolyploids, or ancient polyploids, naturally diverse, fast-growing and widely distributed globally (Lubrano, 1992), especially in temperate, sub-temperate and sub-tropical regions of Northern hemisphere and in tropical Africa also (Tuskan et al., 2006; Guleria et al., 2022). These poplars, except aspens and Asian mountain balsam poplars, grow widely in several regions like hot-arid and desert-like regions of central Asia and Africa, alpine forests in Europe and North America, as well as riparian zones like river banks and flood plains (Guleria et al., 2022). Poplars are the dominant species in these habitats for tolerating and sustaining in the complete flood. China, Turkey, France, India and the Po River plain of Italy are the largest poplar farming areas for worldwide wood supply, whereas Italy, Spain, France and Hungary provide a landscape of 0.5 out of 0.61 million hectares for poplar farming only. Poplars shape global forests and woodlands in their natural habitats. Besides being domesticated as an agroforestry tree, they provide timber, fuel wood, plywood, industrial roundwood, sports materials, pallets, paper pulp for the paper industry and fodder (Kollert et al., 2014). In addition, Populus species are cultivated as energy crops/biofuel (ethanol) in Europe (England and Italy) for carbon sequestration and sustainable bioenergy production in USA because of its high biomass production in a relatively short time (Dou et al., 2017). They are also used in the phytoremediation of toxins, e.g., heavy metals (Cd, Pb, As, and Hg) from contaminated soils, indicating ozone pollution as a bio-indicator, rehabilitation of fragile ecosystems and restoration of forest landscapes (Alahabadi et al., 2017).
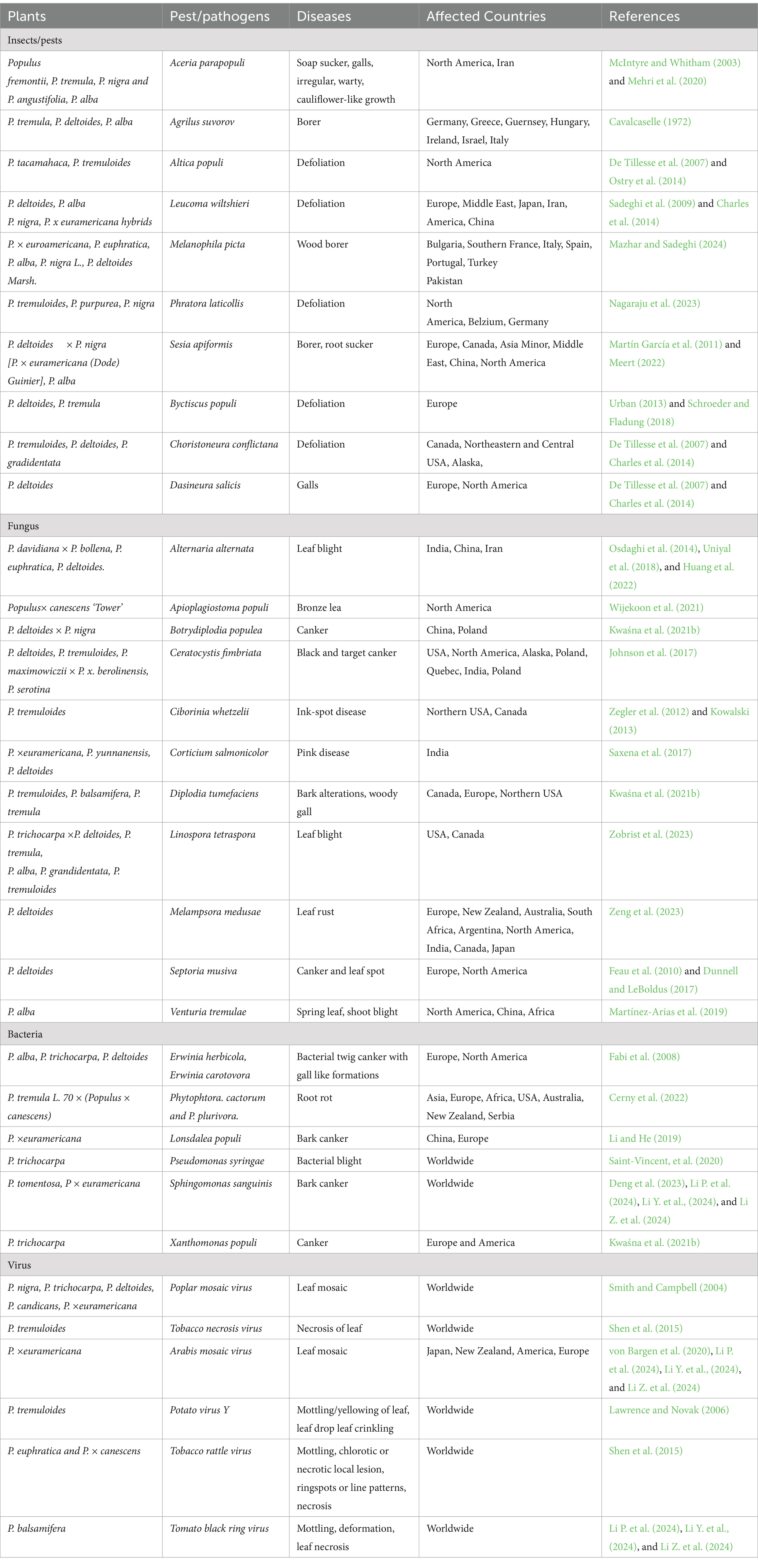
Poplar (Genus: Populus) comprises 32–40 species based on taxonomic and morphological traits (Cronk, 2005; Douglas, 2017). There is a record of a total 582 Populus species, with more than 100 species names recognized worldwide. The higher number of species is due to the presence of naturally occurring hybrids (The Plant List, 2013). According to Eckenwalder (1996), the genus Populus is classified into six groups including the cottonwoods (Aigeiros), aspens (Populus), balsam poplars (Tacamahaca), large-leaf or swamp poplars (Leucoides) and (Abaso) and Afro-Asian poplars (Turanga), and but the Flora of China recognized 71 species from five sections (except Abaso) (Park et al., 2004; Table 1). Species boundaries among poplars are sometimes variable as intrasectional and intersectional hybridization occurs among them. However, this has not been supported by molecular evidence. Hence relationships between these sections are reported to be controversial (Wang et al., 2014; Liu et al., 2017; Zhang et al., 2018). During 1950s, poplar was introduced in India from the United States of America. Since then, P. deltioides, have been cultivated in India and cover an area of 270,000 ha in India, according to the report of the Indian Council of Forestry Research and Education (ICFRE, 2016; Eqbal and Ansari, 2024). Several researches have proved that the genus populus is a rich source of active metabolites, like phenolic compounds, terpenoids, and flavonoids in different parts like stems, buds, leaves and bark (Guleria et al., 2022). These poplar trees have been used to cure various ailments and have several pharmacological properties such as antioxidants, antimicrobials, anticancer, and anti-inflammatory (Guleria et al., 2022).
Poplar was the first woody perennial tree used as a model/experimental tree species among forest trees worldwide for understanding several aspects such as taxonomy, genetics, evolution and the genomics of wood formation for decades because of their small genome size, clonal propagation, fast growth, easy transformation, and long-life cycle (Taylor, 2002). In addition, poplar genome has been completely sequenced after Arabidopsis and rice. Moreover, it is the most advanced genomics resource of any forest tree, having reference genomes for several tree species. The black cottonwood (P. trichocarpa) was the first forest tree whose genome was sequenced entirely, and currently, there are massive genomic resources available for other poplar species. Interestingly, P. tomentosa and P. euphratica genomes have been extensively studied, while others have been ignored. All Populus species contain 19 haploid genomes (Shi et al., 2024). However, massive rearrangement and diploidization of the whole genome of poplar have been reported. The arabidopsis-poplar genome comparative model approach has been used efficiently in many cases (Rottmann et al., 2000; Jansson and Douglas, 2007). The genus Populus is an excellent model for studying the molecular genetic mechanisms involved in pathogen defense responses in several forest trees. The availability of Populus genome sequences enhances the efficiency of the substantial molecular tool kit that already exists for Populus species, including expressed sequence tags (ESTs) collection and microarrays for transcriptome. In addition, these tools can be applied with valuable pedigrees and genetic maps developed for Populus breeding for decades (Sterky et al., 1998; Frewen et al., 2000; Hertzberg et al., 2001; Cervera et al., 2001; Bhalerao, 2003; Andersson Gunnerås et al., 2006). Such pedigrees have been proven useful and efficient in revealing Populus loci responsible for conferring resistance to fungal pathogens attacks (Goué-Mourier et al., 1996). Poplar cultivation is severely affected by several ranges of pest insects and pathogens such as fungi, bacteria, and viruses, resulting in reduced growth and quantity and quality of wood. These biotic stresses comprise the complex interactions between hosts, pests, pathogens, and environmental factors, negatively affecting the poplar population (Seserman, 2018). The efficiency of traditional methods for protecting poplar farming from attack of pests and pathogens is hindered by factors like climatic instability, global warming, flood, drought, high temperatures and humidity. Consequently, new biotechnological approaches like genetic transformation and genome editing are required to overcome these limitations of traditional breeding. These tools are utilized to increase the quality and yield of wood and improve pest and pathogen resistance in forest trees, including poplar (Li P. et al., 2024; Li Y. et al., 2024; Li Z. et al., 2024). This paper primarily summarized the details of pest and pathogen attacks that cause diseases in poplar trees. We also examined the mechanism of infection, transmission route, the role of lignin in protecting poplar against pests and pathogens and the mode of control of diseases in poplar. Here, we also emphasized conventional breeding and its limitation for poplar plantations on a large scale. The application of different tools of genetic transformation and genome editing for developing transgenic poplar resistant to pests and pathogens was discussed in detail. Finally, we summarize the regeneration of transgenic poplar trees that have been modified by incorporating defense genes, such as those conferring pest and pathogen resistance.
2 Poplar susceptibility to biotic stresses
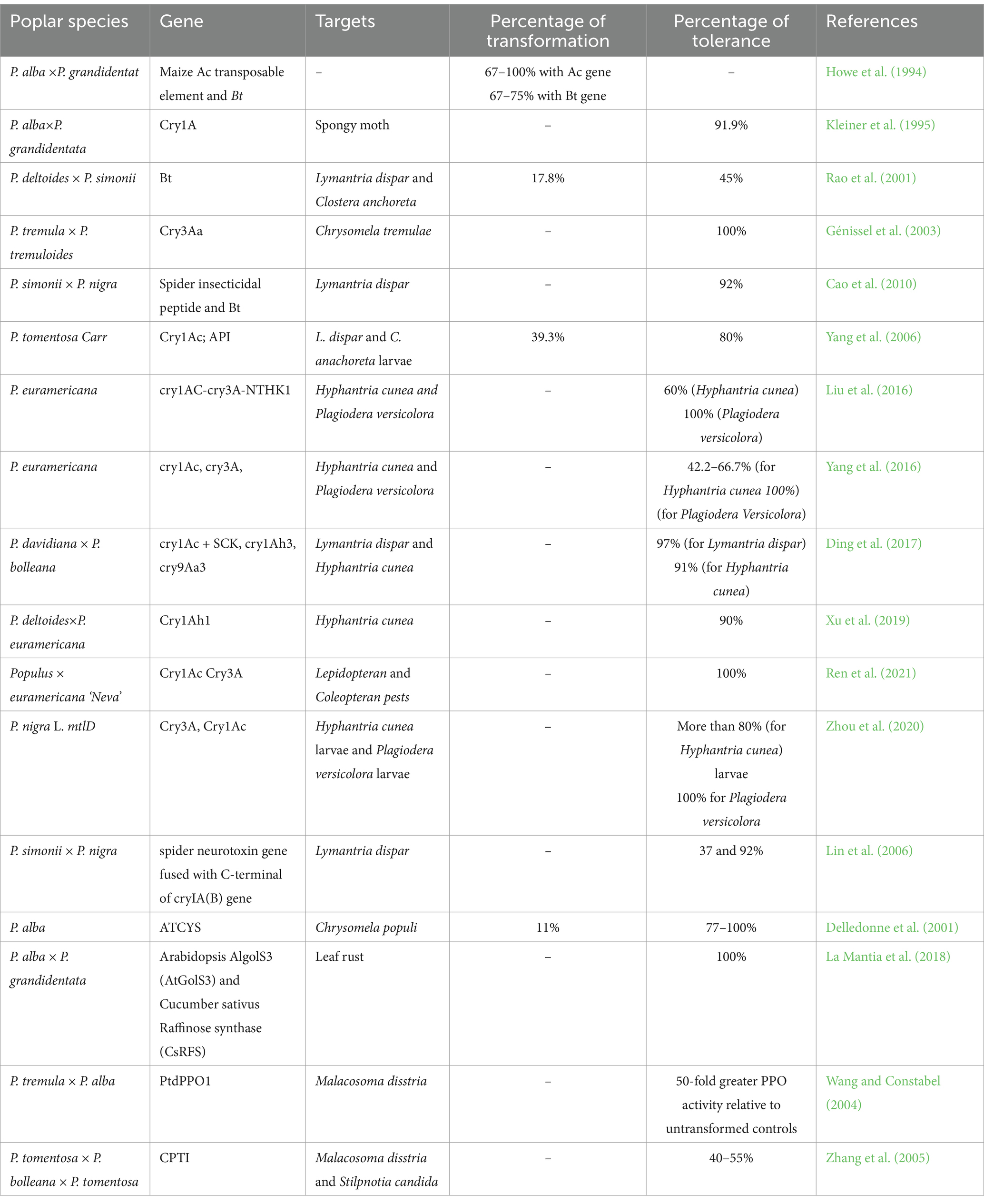
Poplars are frequently impacted by infestations of insect pests such as mites, aphids, and caterpillars, as well as bacterial, viral, and fungal infections. These pests and pathogens target all parts of the tree, damaging buds and leaves, inducing gall formation, sucking sap, altering bark structure, and boring into shoots and roots, which facilitates the transmission of plant diseases. Over time, these attacks can lead to complete defoliation, reduced tree growth, and even tree death. As a result, affected poplars become unsuitable for various uses, including furniture production, biofuel generation, and veneering (Charles et al., 2018).
2.1 Pest attacks on poplar
Insect pests are a limiting factor affecting Populus productivity worldwide (Table 2). Globally, outbreaks of pests are boosted due to climate change (Fenning, 2013). Around 525 and 300 species of insects and mites feeding on Populus have been identified as serious threats for causing economic and ecological losses in poplar plantations in Europe and North America, respectively (Charles et al., 2018). Ahmad and Faisal (2012) documented that around 133 insect species feed on poplar plantations in India. These pests hinder plant growth and increase tree mortality (Dickmann, 2001; Coyle et al., 2005). In North America, the cottonwood leaf beetle (CLB) (Chrysomela scripta) is reported as the most widespread and severe defoliator of young Populus cultivation (Coyle et al., 2005). In the Mediterranean, Saperda carcharias (large poplar borer) is reported to be one of the most damaging insects for young poplar plantations (Biselli et al., 2022). In China, major pest species destroying poplar plantations are trunk borers and defoliators such insects of the Lepidoptera (Hyphantria cunea Drury), Apocheima cinerarius Ershoff, Lymantria dispar Linnaeus, Malacosoma neustria Motschulsky, and other moth species belonging to the Notodontidae and Limacodidae and Coleoptera (Apriona germari Hope, Anoplophora glabripennis Motschulsky, and Plagiodera versicolora Laicharting). In addition, up to 40% loss of hybrid Populus plantations is reported due to poplar looper (Apochemia cinerarua) and the spongy moth (Lymantria dispar) (Hu et al., 2001; Wang et al., 2018) in China. Anaplophora glabripennis also causes massive destruction of hectares of Chinese poplar (P. simonii) plantations. Biselli et al. (2022) observed that Phloemyzus passerinii [Wooly Poplar Aphid (WPA)] causes 10% of production losses of poplar, mainly in European and American countries. Other insects, for example, Cossus cossus, Agrilus suvorovi, Megaplatypus mutatus, Paranthrene tabaniformis, Melanophila picta, and Gypsonoma aceriana, also threaten poplar farming. Recently, transcriptomic and metabolomic analyses were conducted to investigate the species-specific defense responses of Populus tremula against herbivores such as spongy moths (Lymantria dispar) and aphids (Chaitophorus populialbae). The insights gained from these studies could be valuable for developing transgenic poplar varieties with enhanced resistance to pest attacks (Pastierovič et al., 2024).
2.2 Pathogen attacks on poplar
Poplar trees are also constantly challenged by various pathogens like fungi, bacteria and viruses (Table 2). These pathogens inhibit the growth of poplar, impacting the quality and quantity of wood biomass. The different poplar culture practices and the introduction of exotic pathogens promote the widespread distribution of some specific pathogens (Newcombe et al., 1996).
2.2.1 Fungal attack on poplar
Fungi are usually considered as “primary parasites.” They infect healthy plants, which can eventually affect poplar growth and hence decrease the quality and production of wood. The diseased poplars can exhibit reduced leaf photosynthetic areas. Leaf scars created allows entry of secondary pathogens. Repeated infections and premature poplar defoliation may weaken plants, making them susceptible to insect attack, high temperatures and drought (Kebert et al., 2022). Generally, plant pathogens are categorized into three groups: (a) biotrophs (feed on living plant tissue), (b) necrotrophs (feed on dead plant tissue), and (c) hemibiotrophs (first infect living plant tissue and make them dead and then feed on dead tissues) (McCombe et al., 2023). Examples of biotrophs infecting poplar are powdery mildews by Phyllactinia spp. or Uncinula spp., leaf rust by fungus Melampsora spp., while necrotrophs including leaf blight by Septoria spp. and leaf spot by Coryneum spp. and Marssonina spp., canker (Septoria spp.) (Feau et al., 2010; Zeng et al., 2023). Among fungi, the genus Melamspora (biotrophic rust fungi), especially (Melamspora. larici-populina) is reported as the most severe and widespread fungi in poplar plantations (Polle et al., 2013). Infection with this genus is characterized by premature defoliation and reduced photosynthetic ability, resulting in loss of wood production (Polle et al., 2013). Moreover, M. larici-populina is also responsible for severe economic poplar losses in Europe and America (Duplessis et al., 2009), while Melamspora medusae caused leaf rust in P. deltoides in East-North America and the North-West USA (Newcombe et al., 1996). The other primary poplar diseases like stem canker and leaf spot in North America and Europe are caused by fungus Septoria musiva (also known as Sphaerulina musiva) (Zhao et al., 2023). Venturia spp. are found to cause shoot and leaf blight in poplar plantations in Asia, Europe and North America (Gennaro and Giorcelli, 2019). Other major fungal pathogens of Populus affecting leaf are Apioplagiostoma populi (causing bronze leaf disease) and Taphrina spp. (causing yellow blister of leaves), Entoleuca mammata (causing Hypoxylon canker), Cytospora chrysosperma, (causing canker) and Phellinus tremulae (causing aspen bracket) (Duplessis et al., 2009). The poplar blister canker disease develops upon infection with the Botryosphaeria pathogen during drought stress, commonly observed in southern China (Xing et al., 2022). Recently, black spot disease in poplar has been reported to be one of the major diseases in China affected by fungi such as Marssonina castagnei, Marssonina populi, and Marssonina brunnea (Xiong et al., 2021).
2.2.2 Bacterial diseases in poplar
A few bacteria also negatively affect the growth of poplar plantations. The attack of bacteria (Xanthomonas populi, Erwinia genus and Lonsdalea populi) causes canker, resulting in reduced wood biomass yield of poplar (Li et al., 2019). Xanthomonas populi (Ridé) Ridé and Ridé and Pseudomonas syringae Van Hall are responsible for necrosis, wilting, injury, cankers, rots and tumors in poplar vegetations (Kalinichenko et al., 2017). The fluctuating temperatures cause P. syringae growth in poplar bark (Ramstedt et al., 1994). Lonsdalea quercina caused bark canker in Populus×euramericana (Tóth et al., 2013). Recently, Pseudomonas aeruginosa (Schröter) Migula was reported to cause disease in poplar plants. It causes rot, resulting in fast wilting, with trees dying within 48 h. Agrobacterium radiobacter Beijerinck and van Delden and Agrobacterium tumefaciens cause crown gall disease upon transfer and integration of the bacterial transfer DNA (T-DNA) into the plant genome (Kwaśna et al., 2021a). Currently, the large population of the hybrid poplar Populus × euramericana in Hungary and China is severely affected by Lonsdalea populi (Zlatković et al., 2020). Bacterial wetwood of poplar (Populus alba L.) by Lelliottia nimipressuralis has been common in the territory of Ukraine since 1974. The poplar wetwood disease was also reported in Bulgaria, USA, and other countries. The primary bacterial pathogens of poplar are Xanthomonas populi, Pseudomonas syringae, Enterobacter cancerogenus in the coastal zone of Western Europe, Eastern Europe and Central Europe, respectively (Goychuk et al., 2023).
2.2.3 Viral attack in poplar
Viral pathogens such as the poplar mosaic virus, poplar decline virus, tobacco necrosis virus, tobacco mosaic virus, rhabdoviruses, cucumber mosaic virus, tobacco rattle virus, arabis mosaic virus and tomato black ring virus are also severe threats to the poplar population other than fungus and bacteria (Table 2; Wang P. et al., 2023; Wang S. et al., 2023). Poplar mosaic virus (PopMV), with a single-stranded RNA, is the most common dangerous filamentous plant virus and is widespread worldwide (UK, “former Czechoslovakia and former Yugoslavia” Holland, France, Germany, Switzerland, Denmark, Italy, Bulgaria, USA and Canada) where poplar is grown at large scale. It attacks almost all the poplar plants in the Aigeiros section, including several clones of P. x euramericana. Members of the Tacamahaca section and crosses between these species and the Aigeiros section are also affected by viruses. The symptoms of a viral attack on poplar include stunted growth, leaf discoloration, necrosis, wilting and deformities in poplar. It causes severe losses in the quantity and quality of wood (Smith et al., 2004; Naylor et al., 2005; Smith et al., 2009). The virus is generally spread by cutting diseased parts (Berg, 1964).
3 Transmission route, infection, and defense mechanism in poplar attacked by pest and pathogens and their control
The vast diversity of insect pests and pathogens poses significant challenges to forest trees, severely impacting their health and productivity. These threats are particularly serious for poplar plantations worldwide. Climate change also plays a crucial role in altering the occurrence and spread of native and invasive insect outbreaks. Insects typically target susceptible trees for feeding or establishing habitats, further exacerbating the problem. These insects attack and affect all tree parts like shoot, xylem, phloem leaves, flowers, barks, and roots (Balla et al., 2021). In addition, most insects are generally introduced into a non-native area other than their native range and spread rapidly across the country. Imported alive plants and wood materials can act as carriers for introducing many pests (Dara et al., 2019). Fungi, the most common disease agent of poplar trees, have several invasion mechanisms and an array of virulent factors. In root rot disease, rhizomorphs (clusters of intertwining fungal hyphae) and secondary metabolites play a crucial role in infection. The rhizomorphs aggregate around the tree roots, feeding on the host tissues, and can persist in the dead tissues of infected plants for extended periods. This disease is marked by root decay, premature defoliation, wilting, and the production of dwarf fruits and leaves (Balla et al., 2021). Warmer winters, due to climate change, have increased the frequency of sporulation and the rate of fungal infections. Notably, poplar’s defense mechanisms vary depending on the type of fungus involved. Rust diseases caused by the Melampsora spp. are the most common diseases in forest trees, such as poplar. Cankers are mainly caused by attacks of fungal pathogens which affect tree branches, shoots, and twigs. It has been noticed that canker-related diseases occur because of functional failure of the cambium and phloem, carbon starvation, and hydraulic failure. For instance, the fungus inoculations Botryosphaeria disease in poplar (P. alba var. pyramidalis = Populus bolleana) arrested the regeneration of callus and phloem and decreased the rate of photosynthesis and transpiration, as well as arrested the opening of the stomatal aperture and disrupted electron transport (Xing et al., 2022). Bacteria affect plants by forming colonies on their surface or within their tissues. Unlike fungi, they cannot penetrate host cells directly. Instead, they typically enter through natural openings like stomata or through wounded areas. Once inside, these bacteria secrete extracellular enzymes that break down host cells, allowing them to colonize plant tissues. Additionally, they produce polysaccharides that clog the plant’s vascular system, reducing water transport through the xylem. Beetles and leafhoppers can also act as vectors, carrying pathogens and transmitting diseases to plants. Bacterial infections often manifest through symptoms such as spots, cankers, burns, tissue rot, and hormonal imbalances, which can lead to excessive root branching and leaf epinasty (Chatterjee et al., 2008). Certain bacteria, like Agrobacterium tumefaciens and Agrobacterium rhizogenes, inject their plasmids into plant host cells through wounded areas, integrating them into the host genome. This results in tumor gall diseases and the production of hairy roots, respectively (Sharan et al., 2019). Viral pathogens are widespread in plant ecosystems, serving two roles: as agents of plant diseases and as natural enemies of pests and tree pathogens, offering indirect protection to trees. Viral infections often cause significant tissue damage and can lead to symptoms like yellowing, chlorotic lesions, necrotic spots, and ring spots on plant parts. Some stable viruses, such as tobacco mosaic virus, do not require vectors to spread, while other viruses rely on vectors, such as aphids, mites, leafhoppers, fungi, beetles and nematodes, soil, water, other plants and debris for transmission (Balla et al., 2021). Smith and Campbell (2004) reported that the poplar mosaic virus (PopMV) infection and spread depend on the poplar genotypes.
Insects are typically controlled by the release of toxic phytochemicals from plants, which either inhibit pest growth or kill the insects (Fernandez-Conradi et al., 2021). To defend against pathogen attacks, poplars utilize two types of defense mechanisms: induced and constitutive defenses. Induced defenses are activated in response to external stimuli and involve complex processes, while constitutive defenses, the first line of defense, involve non-host resistance through physical barriers and the accumulation of phytochemicals in the plant (Alkan and Fortes, 2015; Zeng et al., 2023). Induced resistance can be further classified into locally induced resistance and systemic induced resistance (SIR). SIR provides broad-spectrum, long-lasting protection against secondary infections. The exogenous application of signal molecules that trigger these defenses can enhance plant immunity and help manage pest populations (Balla et al., 2021). Some observations demonstrate that signal molecules such as salicylic acid (SA) and methyl jasmonate (MeJA) are involved in local and systemic defense responses. Recent research proved that both SA and MeJA pathways are induced in leaves of poplar upon infection with the fungus M. larici-populina proving that both hormone pathways are essential for defense response (Ullah et al., 2019; Chen et al., 2021). Upon fungus attack, the poplar trees activates constitutive defenses involving several processes such as recognition of the fungus by receptor proteins and resistance (R) proteins (PR) resulting into pattern-triggered immunity (PTI) (receptors of plant membranes recognize molecular patterns (PAMPs) of pathogens) and effector-triggered immunity (ETI) (intracellular receptors, (a nucleotide-binding leucine-rich repeat (NLR) class recognize effectors released by pests and pathogens) as well as noncoding RNA (ncRNA)-mediated defense (non-coding RNA having more than 200 nucleotides in length with having role in plant growth and development, and stress responses), initiation of hormone signaling network pathways (mitogen-activated protein kinase (MAPK) cascades and calcium-dependent protein kinase (CDPK) involved in plant growth and development and stress response), activation of defense-related genes and transcription factors (TFs) involved in controlling gene expression by binding DNA elements at 5′ non-coding regions (promoters) of desired genes and modulating transcription rate) and accumulation of phytoconstituents (De Kesel et al., 2021; Zeng et al., 2023). In addition, the pathogen-associated protein 1 (PR1) gets activated as a plant response to abiotic and biotic stresses. Total 17 PtPR1 genes were found in Populus trichocarpa (Wang P. et al., 2023; Wang S. et al., 2023). A total of 1888 lncRNAs and 52,810 mRNAs were recognized in poplar coma (Song et al., 2024). The 30 CDPK genes and 20 closely related kinase genes were identified in Populous spp. (Zuo et al., 2013), The 11 MAPKKs (PtMKKs) and 21 MAPKs (PtMPKs) were identified in the Populus trichocarpa (Hamel, 2006). A total of 104 WRKYs (TFs) have been identified in poplar (He et al., 2012). Recently, the integrated transcriptomic and transgenic analyses were applied to understand mechanisms of poplar resistance against Alternaria alternata attack (Wang W. et al., 2022; Wang Y. et al., 2022).
The most effective method for preventing leaf diseases caused by pests, fungi, bacteria, and viruses is selecting and planting pathogen-resistant poplar clones. Another approach involves using fungicides, such as copper- and carbamide-based treatments, to prevent infections. Fungal diseases can also be managed by maintaining proper spacing between poplars, reducing weed competition, and optimizing plant density, as high relative humidity contributes to disease development. Infected leaves, roots, stems, and branches should be pruned, particularly during the dormant season, to minimize pest and pathogen attacks. Additionally, poplars should be planted in appropriate soil conditions within nurseries to promote healthy growth. Additionally, the soil from infected areas must not be used and moved with equipment (Kebert et al., 2022). Proteomic and genomic technologies offer valuable tools for precisely identifying and characterizing bacterial infections by analyzing their genetic and protein markers (Zubair et al., 2022). Recent studies have shown that lactic acid bacteria (LAB) can effectively combat plant pathogens due to their high biosecurity and ability to promote plant growth (Jaffar et al., 2023). Quorum sensing (QS) molecules, such as 3-OH PAME, regulate the virulence genes in bacteria and fungi, making the identification and development of QS-quenching genes and enzymes promising for disease control (Wang P. et al., 2023; Wang S. et al., 2023). Additionally, eucalyptus oil, known for its antibacterial properties and ability to stimulate plant defense mechanisms, has been shown to reduce plant diseases and could be used to protect poplar in the future (Montesinos et al., 2023). A few genes have been reported whose expression can impart disease resistance in poplar trees. For example, the overexpression of PdbLOX2 was able to induce the resistance in P. davidiana × P. bollena against A. alternata attack, while silencing this gene increased the susceptibility of the poplar tree to A. alternata infection (Huang et al., 2022). Furthermore, the study reported that PtoMYB142 can regulate transcription of wax biosynthesis genes [fatty acid hydroxylase (CER4) and 3-ketoacyl CoA synthase (KCS6)] mediating adaption of poplars against drought conditions were highly expressed upon infection with fungal pathogens (Song et al., 2022). In addition, lignin has a vital role in protecting poplar from pest and pathogen attacks. It is a primary three-dimensional phenolic biopolymer of the secondary cell wall in vascular plants (Ma et al., 2024). It imparts strength and imperviousness to cell walls, mediating long-distance water transport in vascular tissues. In addition, it acts as a barrier to the spread of invading pathogens as it is non-degradable to pathogens, thereby preventing their penetration into the plant cell wall and the supply of water and nutrients from plant cells to pathogens. It is noticed that the gene expression of lignin increased with higher lignin content upon pathogen infection. The genes (phenylalanine ammonia lyase (PAL), HCT4-Coumarate: coenzyme A ligase (4CL), cinnamate 4-hydroxylase (C4H), cinnamoyl-CoA reductase (CCR), cinnamyl alcohol dehydrogenase (CAD) and hydroxycinnamoyl transferase) are involved in lignin biosynthesis and highly expressed during fungal infection leading to increase in lignin content (Lee et al., 2019; Zeng et al., 2023; Ma et al., 2024; Riseh et al., 2024). Hence, regulating the lignin biosynthesis pathway may be critical for improving poplar resistance against pathogen attacks (Polle et al., 2013). It is reported that the higher expression of Pto4CL1 of P. tomentosa increased the lignin content from 33.11 to 46.65%, leading to the decreased formation of cellulose, hemicellulose, and pectin (Hu et al., 2019). RNAi technology was used to down-regulate the expression of 4CL gene to modify lignin biosynthesis in P. tremula (Kovalitskaya et al., 2016). A significant 30% reduction in lignin content has been observed in poplars due to the downregulation of cinnamate 4-hydroxylase (C4H) genes (Bjurhager et al., 2010). Similarly, the downregulation of CAD genes in Populus tremula × Populus alba led to reduced lignin levels (Özparpucu et al., 2017). Dirigent (DIR) proteins have also been identified as crucial players in lignin biosynthesis. Li et al. (2022) reported that the overexpression of PtDIR11 in poplars enhanced lignin biosynthesis, thereby increasing the trees’ resistance to Septotis populiperda. Hence, gene editing can be utilized to regulate the expression of these genes to enhance lignin biosynthesis, which confers pest and pathogen resistance in poplar trees.
4 Conventional breeding in poplar
The recent global temperature, drought, humidity and climate instability render poplar plants vulnerable to pests and pathogens, severely affecting wood quality and quantity (Gullino et al., 2022). Plants have physical and physiological barriers against microbial pathogens, preventing their access to plants (Kumudini et al., 2018). Plants produce antimicrobial peptides and other molecules that cause detoxification and the inhibition of virulence factors (Silva et al., 2016). Moreover, plants also apply RNA interference (RNAi), which detects invading viruses and cleaves the RNAs of viruses (Bocos-Asenjo et al., 2022). However, these pathogens have evolved to cope with the defense systems of their plant cells by secreting cell-wall degrading enzymes, which gain access for molecules into plant cytoplasm, inhibiting host defenses and promoting susceptibility of plants toward pathogens (Kaur et al., 2022). Some viral pathogens are also reported to attack and silence the host RNAi system, promoting viral pathogenicity (Leonetti et al., 2021). The increased demand for wood and its sustainability requires approaches to improve efficient production even under environmental constraints and minimize the threats due to pests affecting wood properties for industrial purposes (Polle et al., 2013). However, controlling pests and pathogens with chemicals is increasingly considered unsafe due to their high toxicity, environmental accumulation, and harmful impacts on beneficial insects, non-target organisms, and humans (Ahmad et al., 2024). The commonly adopted traditional methods for the protection of plants from attack of poplar pests and pathogens are the use of resistant clones that have good adaptability to different soil (salinity, calcium level and pH), drought and climatic conditions coupled with the adoption of proper cultivation practices (i.e., fertilization, low plant density and irrigation) (Biselli et al., 2022). Enormous progress has been made in enhancing traits such as plant growth rate, pest and pathogen resistance, and environmental adaptations in poplar plantations by applying conventional breeding practices (Ye et al., 2011). However, these traditional methods are time-consuming because of their long-life span, costly land demand, and high labor costs. In addition, because of the heterozygosity of most Populus genotypes and inbreeding depression, it is difficult to estimate the genetic control of particular traits (Ye et al., 2011). Genetic engineering and molecular breeding methods for developing transgenic poplar plants can address the limitations of traditional breeding, such as the challenges of distant hybridisation, the complexity of cultivation, and issues with interspecific hybridisation (Begna, 2021).
These tools have enormous potential to improve two or more traits simultaneously by introducing desired exogenous genes of donor plant or non-plant origin into a particular plant genome, enabling the improvement of poplar against pests and pathogens, herbicide resistance, abiotic stress, wood properties, flowering regulation and phytoremediation (Thomas, 2022).
5 Genetic engineering and different transformation methods in poplar
Genetic transformation has been widely employed in research on various forest trees. This process involves introducing exogenous genes into tree cells, thereby altering their genetic traits (Li et al., 2023). Poplar trees were among the first forest trees used successfully for genetic engineering for gene research (Ye et al., 2011). For more than 20 years, several progress has been made in Populus transformation. The most widely used transgenic tools involve vector-mediated transformation, such as Agrobacterium tumefaciens-mediated and A. rhizogenes mediated and non-vector mediated transformation (Gene gun-mediated, pollen tube pathway, and protoplast transformation methods). The Genome editing method is an advanced approach for adding, deleting, or modifying genes within the specific genome (Li et al., 2023). Mobile genome editing techniques, such as clustered regularly interspaced short palindromic repeats (CRISPR)-associated (CRISPR-Cas) systems, RNA interference (RNAi), and nanoparticle-meditated gene transformation have been recently applied to improve poplar tree (Yin et al., 2021; Figure 1). Among these methods, Agrobacterium-mediated and gene gun-mediated transformations are the most widely used techniques for forest trees (Lv et al., 2020).
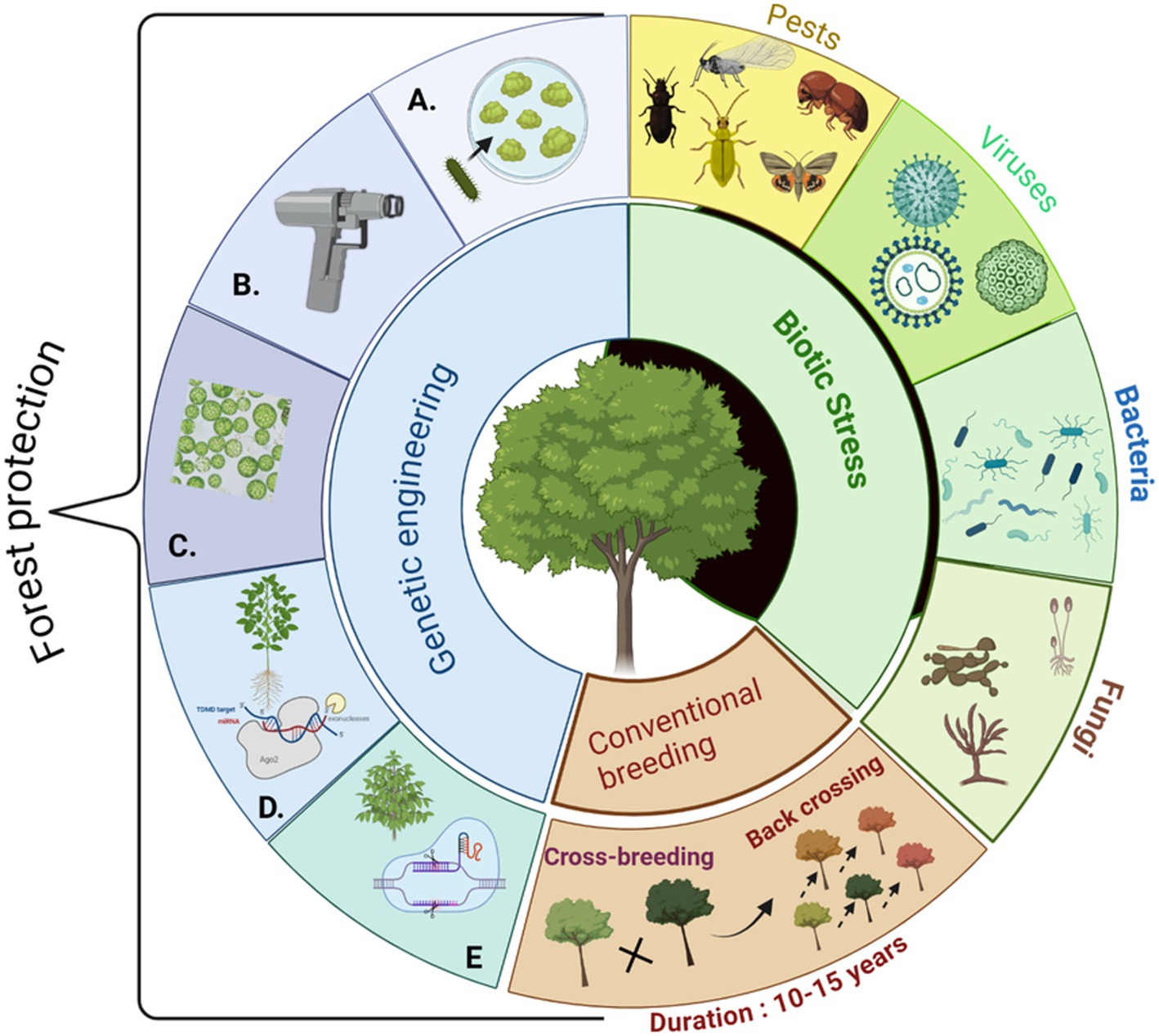
Figure 1. Summarizing forest protection using genetic engineering. Increasing biotic stressors (i.e., pests and pathogens) cause dramatic tree mortality in the forests worldwide. The conventional breeding method is time-consuming and fails to cope with the demands. Transgenic pest and disease resistance trees (i.e., Poplar) can potentially mitigate the challenges. Transgenic trees can be generated using various transformation techniques such as (A) agrobacterium-mediated transformation, (B) gene-gun mediated transformation, (C) protoplast-mediated transformation, (D) mi-RNA or RNAi-mediated transformation, and (E) CRISPR-mediated transformation techniques. (The figure is prepared using biorender.com).
5.1 Agrobacterium tumefaciens-mediated transformations: basic mechanisms
Agrobacterium tumefaciens-mediated transformation is the most preferred method for the genetic transformation of forest trees. A. tumefaciens (a gram-negative soil bacteria) infects the wounded sites in many dicotyledons, gymnosperms and a few angiosperms. It delivers its transfer DNA (T-DNA) molecules into plant cells and then integrates them into the plant genome (Chilton et al., 1980; Sekine and Shinmyo, 2020). The Agrobacterium-mediated transformation method involves removing oncogenes causing tumorigenesis, inserting exogenous genes in disarmed T-DNA, and delivering and integrating foreign genes into the plant genome (Pratiwi and Surya, 2020). The success of Agrobacterium-mediated transformation depends on different parameters such as the virulence of Agrobacterium cells, explant types and plant genotypes and regeneration of transgenic populations. Agrobacterium rhizogenes is also a relative of A. tumefaciens, which develops hairy root at the wounded site of plant cells (also known as “hairy root disease”) and can be used to transfer the T-DNA into a binary vector into developing root cells (Limpens et al., 2004; Sharan et al., 2019). Various wild-type strains of A. tumefaciens and A. rhizogenes have transformed various trees. A. tumefaciens-mediated genetic transformation system has been widely applied in various poplars, such as Populus alba × Populus glandulosa, Populus simonii × Populus nigra, and Populus tomentosa. Several attempts were made to improve A. tumefaciens-mediated transformation in poplar by optimizing several parameters such as types of explants, different strains of Agrobacterium and culture densities, incubation time and concentration of acetosyringone and sucrose (Movahedi et al., 2014, Sharan et al., 2019). Pest infestation and bacterial, fungal and viral diseases are limiting factors which affect the healthy growth of poplar trees (Li P. et al., 2024; Li Y. et al., 2024; Li Z. et al., 2024). By introducing insect and disease-resistance genes into poplar trees using A. tumefaciens, these trees can protect themselves from invading pests and diseases, enhancing their survival rate and disease-resistance capabilities. However, Agrobacterium-mediated transformation has been done in several poplars, but many other poplars remain recalcitrant to Agrobacterium-mediated transformation (Song et al., 2019).
5.2 Agrobacterium-mediated transformation for pest resistance in poplar
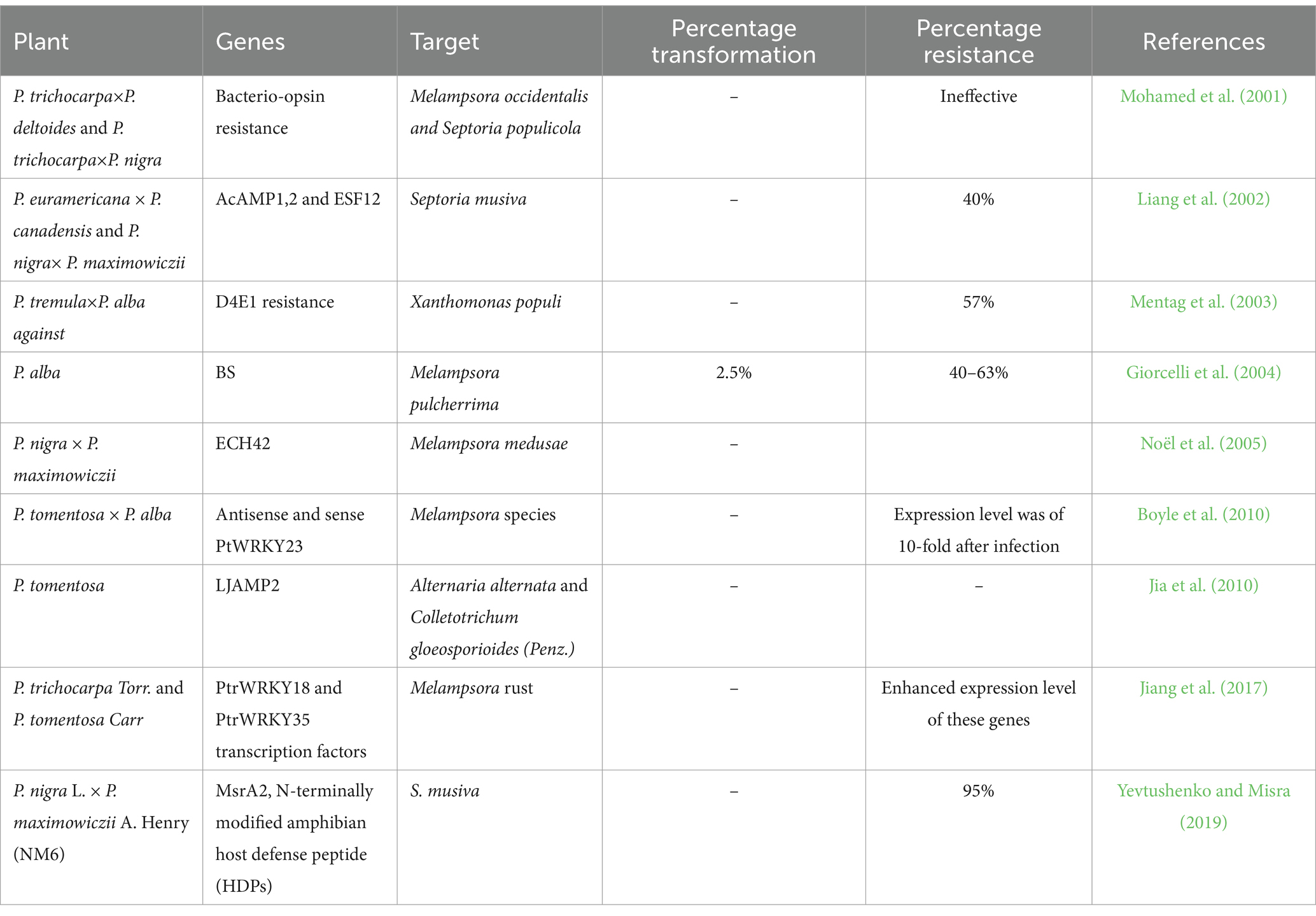
Several transgenic poplars have been developed that overexpress genes encoding different serine proteinase inhibitor proteins (Heuchelin et al., 1997; Confalonieri et al., 1998) and Bacillus thuringiensis-derived genes (Cry/Bt genes) (McCown et al., 1991; Wang et al., 1996), Androctonus australis hector insect toxin, Kunitz trypsin inhibitor (KTI) and chitinase gene for conferring pests resistance (Clemente et al., 2019; Ren et al., 2021; Table 3). However, Bt gene is the most widely used for generating pest-resistant poplar trees. The first stable transfer of Bt was reported in Populus nigra (McCown et al., 1991). Recently, the simultaneous introduction of two Bt genes into the trees’ genomes expanded the scope of insect resistance in transgenic forest trees (Dong et al., 2015; Wang et al., 2018). China has been the first nation to generate and commercialize two transgenic lepidopteran-resistant poplar lines since 2002 (Thakur et al., 2021). The plant P. alba×P. glandulosa was transformed with a Bt Gene (CRY3A) using Agrobacterium-mediated transformation method, which resulted in the development of transgenic line BGA-5 and toxic to the larvae of Anoplophora glabripennis with a growth inhibition rate of 78.6% (Zhang et al., 2006). P. × euramericana was transformed with Cry1AC and Cry3A genes to confer resistance to the poplar plants against H. cunea exhibiting mortality rate of 42.2–66.1 and 100% of Plagiodera versicolora larvae of L1 and L2 stages, respectively (Yang et al., 2016). Transgenic poplar lines ‘Shanxin’ (Populus davidiana×Populus bolleana) were developed through Agrobacterium-mediated transformation method carrying Cry1Ac + SCK, Cry1Ah3, and Cry9Aa3, respectively against fall webworm (Hyphantria cunea) and gypsy moth (Lymantria dispar) as these genes Cry1Ac + SCK, Cry1Ah3, and Cry9Aa3 were toxic to the larvae of both insects (Ding et al., 2017). Two Bt toxin genes, Cry1Ac and Cry3A, were simultaneously integrated into the genome of Populus × euramericana ‘Neva’ with the help of Agrobacterium tumefaciens, to develop transgenic poplar, which was highly resistance to Lepidopteran and Coleopteran pests (Satish et al., 2021). Other than Bt, many other genes, such as cowpea trypsin inhibitor (CPTI), cysteine proteinase inhibitor (Atcys) gene, glycine max trypsin proteinase inhibitor (KTi3 and PtdPP01 genes, etc.) were inserted into Populus species, which conferred some degree of resistance against insect pests (Table 3). La Mantia et al. (2018) observed that the overexpression of Arabidopsis AlgolS3 (AtGolS3) and Cucumber sativus Raffinose synthase (CsRFS) in Populus alba × P. grandidentata antagonizes leaf rust defense mechanism by inhibiting reactive oxygen species (ROS) and attenuating phosphatidic acid and calcium signaling pathways leading to salicylic acid (SA) defense. Lin et al. (2006) generated transgenic P. simonii×P. nigra plants by inserting the spider neurotoxin gene along with C-terminal of CryIA(B) gene resistance against Lymantria dispar. Moreover, the scorpion neurotoxin AaIT expression in hybrid Populus was responsible for developing resistance against the spongy moth (Lin et al., 2006).
5.3 Agrobacterium-mediated transformation for disease resistance in poplar
A diverse array of bacterial and fungal attacks and viral infestation causes significant losses in the poplar yield. Transgenic poplars have various antibacterial and antifungal genes encoding proteins capable of breaking down mycotoxins and inhibiting cell-wall-degrading enzymes such as rabbit defensin (NP-1), osmotin, glucanases, chitinases (CH5B), lysozyme and thaumatin were able to combat pathogens mentioned in Table 4 (Zhao et al., 1999; Juge, 2006; Karlovsky, 2011; Thakur et al., 2021). Noël et al. (2005) generated transgenic hybrid poplar plants harboring the ECH42 (Trichoderma harzianum endochitinase) gene responsible for imparting an enhanced level of resistance against Melampsora medusa, a leaf rust pathogen of poplar. Levée et al. (2009) functionally identified and characterized the transcription factor PtWRKY23 gene in P. tomentosa × P. alba whose silencing is responsible for enhanced susceptibility of transgenic poplars toward Melampsora infection. In addition, the overexpression of a transcription factor PtoWRKY60 in P. tomentosa clone 741 was noticed for conferring resistance to the fungal pathogen Dothiorella gregaria (Ye et al., 2014). Jiang et al. (2017) observed that over-expression of PtrWRKY18 and PtrWRKY35 transcription factors increased resistance in poplar transgenics against Melampsora rust. Hybrid Populus having over-expressed a wheat (Triticum aestivum) germin-like oxalate oxidase gene encoding enzyme responsible for metabolizing the oxalic acid molecules secreted by fungal pathogen Septoria musiva, showed delayed infection by the fungal pathogen (Liang et al., 2001). Interestingly, developing genetically engineered transgenic poplar resistant to bacterial pathogens is less common as bacterial damage is rare in poplar plantations. However, severe infections by Xanthomonas spp. on poplar plantations are reported (Ye et al., 2011). Mentag et al. (2003) generated transgenic P. tremula × P. alba having a gene encoding a synthetic antimicrobial peptide D4E1 imparting resistance to several fungal and bacterial pathogens. The nucleotide MsrA2 [N-terminally modified amphibian host defense peptide (HDPs) from the skin secretion of arboreal frogs] was inserted into the hybrid poplar Populus nigra L. × P. maximowiczii A through Agrobacterium-mediated transformation method. The peptide was reported to inhibit S. musiva conidia germination but is non-toxic to poplar (Yevtushenko and Misra, 2019). Certain viruses, such as the poplar decline virus, poplar mosaic virus, and arabis mosaic virus, pose significant threats to the poplar population by stunting plant growth and severely impacting wood biomass and quality (Pinon and Frey, 2005). To date, there have been no reports of developing transgenic poplar plants with improved viral resistance using the Agrobacterium tumefaciens-mediated transformation method. Therefore, this method holds potential for future use in enhancing viral resistance in poplar.
5.4 Gene gun-mediated transformations
The gene gum method (biolistic particle delivery system) has excellent potential in forest tree research. This physical method is commonly applied for genetic transformations of several plants. This method was first developed by Sanford and colleagues in 1982. The process involves the transfer of gold or tungsten microparticles (or microcarriers) coated with exogenous donor genes into receptor cells or tissues or organs with the help of accelerators like pressurized helium (He) gas and integration of genes into receptor genome and expression of the genes (Zhang et al., 2014; Cunningham et al., 2018). The efficiency of gene gun-mediated transformation depends on the factors, for example, types of receptors, culture and transformation conditions (Wang et al., 2018). In addition, this method is independent of plant genotypes compared to A. tumefaciens-mediated transformation. This method is commonly applied to generate transgenic poplars, as mentioned in Table 5 (Ozyigit and Yucebilgili Kurtoglu, 2020). The insect resistance Bt gene was co-transformed into P. nigra through gene gun mediated transformation protocol (Li et al., 2000). The Bt gene (cry3Bb) gene was successfully incorporated into the genome of poplar plastid through biolistic bombardment, generating transformed poplar with a mortality rate of 100% to Plagiodera versicolora (Xu et al., 2020). Wang et al. (2007) inserted three foreign Bacillus subtilis genes vitreoscilla hemoglobin (vgb), fructan sucrase (SacB), and bivalent stem borer resistance (BtCry3A + OC-I), and the regulatory gene (JERF3) into Populus × euramericacana ‘Guariento’ through particle bombardment method. No incorporation of pathogen and disease-resistant genes in poplar trees with the help of gene gun mediated transformation tools has been reported. The disadvantages of this gene gun-mediated transformation are low efficiency, silencing the transformed genes, inserting multiple gene copies and unstable expression of exogenous genes (Yin et al., 2021).
5.5 Protoplast transformation
The use of protoplasts for genetic transformation in plants has grown significantly in recent years. This technique involves introducing and incorporating exogenous genes into plant protoplasts, leading to the generation of transgenic plants with stable gene expression. The protoplast method has proven to be easy, fast, and efficient, with minimal or no interference from surrounding cells or the microenvironment (Yin et al., 2021; Adjei et al., 2023). Because of their versatility and efficiency, protoplast transformation systems have been optimized, established and applied to many recalcitrant non-model plants, along with the efficient delivery of several genes (Rehman et al., 2016; Naing et al., 2021; Ojuederie et al., 2022). This method is affected by several parameters such as explant types, tissue types, the composition of the digestion solutions, the pH of the digestion solution, the digestion time, the concentration of polyethene glycol (PEG) and the transformation time (Rezazadeh et al., 2011, Biswas et al., 2022). The protoplast transformation method is easy and efficient in annual herbaceous plants such as Oryza sativa, Arabidopsis thaliana and Nicotiana tabaccum (Jiang et al., 2013; Sun et al., 2018). The separation and transformation of protoplasts and regeneration from transformed protoplasts are difficult in forest trees. Advances have been made in PEG-mediated transformation method by applying liposome-mediated shock perforation and A. tumefaciens co-culture transformation method (Wu et al., 2014). The PEG-mediated method is the widely used protoplast transformation system in plants (Lenaghan and Neal Stewart, 2019). In addition, protoplasts can be transformed directly by imbibing DNA followed by PEG pre-treatment, microinjection, and electroporation. However, protoplast isolation and its transformation are complex and challenging for woody trees like poplar and have not been fully optimized and developed. Xu et al. (2020) used the leaf protoplast of poplar (P. davidiana *P. bollaena) to introduce cry3Bb genes for developing insect-resistant transgenic poplar. This method has not yet been utilized to generate transgenic poplar with pathogen-resistant genes.
5.6 Micro RNA mediated transformation
MicroRNAs (miRNAs) are endogenous, short, single-stranded, non-coding RNAs of 20–24 nucleotides, processed from hairpin RNA precursors by Dicer-like (DCL) enzymes. These are found in all eukaryotic cells and negatively regulate gene expression. After their discovery in plants, several miRNAs have been recognized with the help of high-throughput sequencing technology and bioinformatics and for there essential roles in regulating critical genes involved in plant-pathogen interactions at the transcriptional or post-transcriptional levels (Islam et al., 2022; Nizamani et al., 2023). According to the host and the specific pathogen, miRNAs can be up- or down-regulated, thereby promoting plant disease resistance by participating in hormone signaling and regulating and moderating resistance (R) genes (Yang et al., 2021). The first report of plant microRNAs was reported in Arabidopsis by Llave et al. (2002). Several studies established the pivotal roles of microRNAs in regulating biotic and abiotic stresses in several plants (Kar and Raichaudhuri, 2021). Transgenic poplar overexpressing miR159a (OX-159) showed enhanced resistance to necrotrophic fungi C. chrysosperma while enhanced susceptibility to infection by L. populi (bacterial canker) and hemi-biotrophic fungi C. gloeosporioides (Yang et al., 2023). Furthermore, in transgenic poplar (P. trichocarpa), miR472a positively regulated resistance to Colletotrichum gloeosporioides by targeting nucleotide-binding site and leucine-rich repeat domains (largest R proteins, NBS-LRR) and regulated negatively resistance to Cytospora chrysosperma (Su et al., 2018). miR156a was found to be the most stable miRNA examined as a reference gene in P. tomentosa under canker pathogen stress (Zhang et al., 2021). Several evidences proved that miRNAs can regulate and mediate biological processes during plant-insect and plant-viral interactions, ultimately conferring pest/viral resistance in plants (Zhang et al., 2022; Satish et al., 2021). To date, miRNA molecules have not yet been used to generate transgenic poplar with pest and viral resistance genes. We believe that with the growing recognition of miRNA molecules, as highlighted by the 2024 Nobel Prize, artificial mi-RNA holds the potential to be used for manipulating tree traits in the future.
5.7 RNA interference-mediated transformation
RNA interference (RNAi) is a naturally occurring cellular defense system in most eucaryotic cells. It is mediated by double-stranded RNA (dsRNA) as either a source of virus infection or because of transposon activity, both seeking need to be suppressed (Obbard et al., 2009). RNAi pathway involves the formation of several interfering molecules, i.e., small interfering RNAs (siRNAs) and microRNAs (miRNAs), generated through the activity of a dicer enzyme. These interfering molecules are then loaded on an RNA-induced silencing complex (RISC) containing argonaute protein (AGO). RISC directs the interfering molecules to their target gene, and homology-based cleavage of target mRNA occurs in the cells (Mamta and Rajam, 2017). RNAi has an important role in functional genomics research and is also a promising species-specific pest/pathogen management strategy in agroforestry (Mamta and Rajam, 2017; Joga et al., 2021; Mogilicherla et al., 2023; Sandal et al., 2023; Sellamuthu et al., 2024). RNAi tool is a sequence-based method that suppresses target gene expression for pest growth, development, and reproduction without affecting non-targeting other pest species (Whyard et al., 2009; Christiaens et al., 2020). Transgenic poplar plants harboring dsRNA targeting CYP6B53 from Lymantria dispar via A. tumefaciens-mediated transformation exhibited inhibited larval feeding and delayed growth (Sun et al., 2022). Such studies endorse the potential for using trees expressing dsRNA against target forest pests. RNAi-mediated lignin modification has also been successfully applied in poplar. The overexpression of microRNA, such as miR6443 reduces S lignin biosynthesis during shoot development in Populus tomentosa, making the plant susceptible to pathogens (Fan et al., 2020). Thus, RNAi can be utilized to modulate gene expression miR6443 to produce more lignin in poplar to confer resistance against pathogens.
5.8 Microparticle-mediated CRISPR DNA delivery for genome editing in poplar
The clustered regularly interspaced palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system is the most promising technique used for precise genetic engineering in plants, including poplar (Bewg et al., 2018; Anders et al., 2023; Sulis et al., 2023). This method is harnessed to improve sustainable production and introduce precise alterations at target sites, thereby altering plant architecture and floral development and developing biotic/abiotic resistance in trees (Borthakur et al., 2022). This method does not introduce foreign genes into the forest trees, making it safer than other genetic engineering methods. In this process, CRISPR gene-editing reagents, i.e., Cas9 protein and the guide RNA (gRNA), are generally delivered through A. tumefaciens, resulting in the stable genome integration and expression of the transfer DNA (T-DNA) in the plant genome (Hoengenaert et al., 2023). Alternative strategies other than Agrobacterium-mediated method for the delivery of gene-editing reagents into plant genomes are either through the expression of a gRNA- and Cas9-coding DNA/RNA or ribonucleoproteins (RNPs) into callus or protoplasts (Lin et al., 2018). However, this approach has drawbacks, such as inducing somaclonal variation and large genome rearrangements resulting in altered phenotype of plants (Serres et al., 1991; Fossi et al., 2019). Another commonly used method applies mechanical force like a gene gun to deliver gene-editing reagents coated with microparticles into plant tissue. Several researchers applied CRISPR-mediated gene editing for wood quality improvement and drought/pest/disease resistance in forest trees (Dort et al., 2020). In addition, microparticle-mediated DNA delivery technology has previously been used to deliver the CRISPR gene in poplar trees (Devantier et al., 1993; Nowak et al., 2004; Canto, 2016). Jang et al. (2021) and Huang et al. (2022) utilized this method for knocking out caffeoyl shikimate esterase (CSE) to improve lignocellulose biomass and root growth transcription factor PDNF-YB21 for repression of root and inducing drought resistance in transgenic poplar, respectively. However, it has not yet been applied to develop pest and pathogen resistance in poplar trees.
6 Regeneration methods used in Populus species
An efficient regeneration system is crucial for successful genetic transformation, as it enables the development of transgenic plants from a single cell carrying the desired genes. However, genetic transformation and regeneration remain significant challenges in many forest trees, including poplar. Various plant regeneration methods have been developed for poplars (Thakur et al., 2005; Li et al., 2017), which can be employed to produce transgenic trees with resistance to pests and diseases. In recent decades, significant research efforts have focused on creating transgenic poplars with enhanced resistance to abiotic stress and improved wood quantity and quality. There have been a few reports on regenerating transgenic poplar trees with biotic stress resistance. The established suspension cultures of P. alba x P. grandidentata cv. ‘Crandon’ were transformed with vectors A. tumefaciens carrying the maize Ac transposable element and an insect toxin gene isolated from Bacillus thuringiensis (Bt). These transgenic plants were regenerated by subculturing the transformed callus on the medium, supplemented with a growth regulator Thidiazuron (TDZ) of 0.11–27.0 μM (Howe et al., 1994). A. tumefaciens mediated genetic transformation and regeneration of hybrid poplar (P. alba x P. grandidentata) and transgenic quaking aspen from cuttings from young leaves were also readily achieved (Tsai et al., 1994). A. tumefaciens-mediated transformation of leaf explants of P. nigra L. was done with a Kunitz trypsin proteinase inhibitor (KTi3) gene for pest resistance, and regeneration of this transgenic leaf explants was successfully achieved (Confalonieri et al., 1998). The stems and petioles of transformed hybrid aspen (Populus tremula × P. alba) clones containing PtdPPO1 genes (conferring pest and pathogens resistant in plants) of in vitro plantlets were used for regeneration (Wang and Constabel, 2004). Further research is needed to establish a protocol for regenerating transgenic poplars with enhanced resistance to pests and pathogens from modified cells.
7 Conclusion and future perspectives
Poplars play a crucial role in supporting global ecological and socioeconomic wellbeing. The growing demand for poplar products has driven genetic engineering efforts to enhance various traits, particularly pest and disease resistance, as these trees are highly susceptible to numerous pests, fungi, and viruses. Considering the long growth cycle with low transformation tendency in forest trees, including poplar trees, it is necessary to establish a stable and efficient transformation system. Adopting pest and disease-resistant transgenic poplar plants to minimize yield loss and pesticide consumption has been successful. Many researchers have employed genetic transformation methods, including Agrobacterium tumefaciens, protoplast, gene gun, RNA interference, and miRNA-mediated transformations, to improve poplar resistance to pests and pathogens. These techniques, along with genome editing to introduce pest resistance genes and modulate lignin biosynthesis, offer promising avenues for developing transgenic poplar trees capable of withstanding pest and disease attacks, thus improving their survival rates. However, no research has been conducted on the pollen tube method, A. rhizogenes mediated and nanoparticle-mediated transformation to enhance pest/pathogen or virus resistance in Populus species. Further efforts are required to establish transgenic poplars with single/multiple genes for increasing biotic stress tolerance limits (pest/pathogens infestations) using nanoparticles, A. rhizogenes, and the pollen tube method. It will be optimal if the methods developed in poplar can be used in other forest trees to make them resistant to biotic and abiotic stresses. The use of omics technologies (i.e., genomics, transcriptomics, and proteomics), along with high-throughput screening and selection methods, accelerates the identification of successful transgenic poplar lines. Integrating big data, machine learning, and artificial intelligence (AI) into poplar breeding programs (i.e., data-driven breeding) can enhance the accuracy of predicting genetic alteration outcomes (Farooq et al., 2024), enabling more targeted and efficient transgenic strategies for trees. A key factor for success is the competence to regenerate transgenic plants from modified cells. Addressing the genotype dependency in poplar transformation is crucial for expanding the applicability of transgenic approaches. Developing “transgene-free” or non-GMO techniques, such as transient CRISPR expression, could alleviate regulatory and public concerns, facilitating the adoption of genetically improved poplar.
Author contributions
AS: Conceptualization, Formal analysis, Funding acquisition, Investigation, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing. SS: Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft. AC: Investigation, Validation, Visualization, Writing – original draft, Writing – review & editing. AR: Formal analysis, Funding acquisition, Investigation, Resources, Software, Visualization, Writing – review & editing. IS: Data curation, Formal analysis, Investigation, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. AR and AC were supported by “Excellent team grants (2023–24)” from FLD, CZU. Financial support was provided to AS by the Science and Engineering Research Board (SERB), Department of Science and Technology, New Delhi, India (ECR/2017/002478; SPG/2021/002969).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adjei, M. O., Zhao, H., Tao, X., Yang, L., Deng, S., Li, X., et al. (2023). Using a protoplast transformation system to enable functional studies in Mangifera indica L. Int. J. Mol. Sci. 24:11984. doi: 10.3390/ijms241511984
Ahmad, M. F., Ahmad, F. A., Alsayegh, A. A., Zeyaullah, M., AlShahrani, A. M., Muzammil, K., et al. (2024). Pesticides impacts on human health and the environment with their mechanisms of action and possible countermeasures. Heliyon 10:e29128. doi: 10.1016/j.heliyon.2024.e29128
Ahmad, M., and Faisal, M. (2012). Status of insect pests of poplar in India with special reference to Clostera spp. For. Bull. 12, 105–122.
Alahabadi, A., Ehrampoush, M. H., Miri, M., Aval, H. E., Yousefzadeh, S., Ghaffari, H. R., et al. (2017). A comparative study on the capability of different tree species in accumulating heavy metals from soil and ambient air. Chemosphere 172, 459–467. doi: 10.1016/j.chemosphere.2017.01.045
Alkan, N., and Fortes, A. M. (2015). Insights into molecular and metabolic events associated with fruit response to post-harvest fungal pathogens. Front. Plant Sci. 6:889. doi: 10.3389/fpls.2015.00889
Anders, C., Hoengenaert, L., and Boerjan, W. (2023). Accelerating wood domestication in forest trees through genome editing: advances and prospects. Curr. Opin. Plant Biol. 71:102329. doi: 10.1016/j.pbi.2022.102329
Andersson Gunnerås, S., Mellerowicz, E. J., Love, J., Segerman, B., Ohmiya, Y., Coutinho, P. M., et al. (2006). Biosynthesis of cellulose enriched tension wood in Populus: global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. Plant J. 45, 144–165. doi: 10.1111/j.1365-313X.2005.02584.x
Balla, A., Silini, A., Cherif-Silini, H., Chenari Bouket, A., Moser, W. K., Nowakowska, J. A., et al. (2021). The threat of pests and pathogens and the potential for biological control in forest ecosystems. Forests 12:1579. doi: 10.3390/f12111579
Begna, T. (2021). Conventional breeding methods widely used to improve self-pollinated crops. Int. J. Res. 7, 1–16. doi: 10.20431/2454-6224.0701001
Berg, T. M. (1964). Studies on poplar mosaic virus and its relation to the host. Wageningen: Veenman: Wageningen University and Research.
Bewg, W. P., Ci, D., and Tsai, C.-J. (2018). Genome editing in trees: from multiple repair pathways to long-term stability. Front. Plant Sci. 9:1732. doi: 10.3389/fpls.2018.01732
Bhalerao, R. (2003). Gene finding in Populus—the bioinformatics of an EST program (Doctoral dissertation). Stockholm, Sweden: Department of Biotechnology, Royal Institute of Technology.
Biselli, C., Vietto, L., Rosso, L., Cattivelli, L., Nervo, G., and Fricano, A. (2022). Advanced breeding for biotic stress resistance in poplar. Plan. Theory 11:2032. doi: 10.3390/plants11152032
Biswas, S., Bridgeland, A., Irum, S., Thomson, M. J., and Septiningsih, E. M. (2022). Optimization of prime editing in rice, peanut, chickpea, and cowpea protoplasts by restoration of GFP activity. Int. J. Mol. Sci. 23:9809. doi: 10.3390/ijms23179809
Bjurhager, I., Olsson, A.-M., Zhang, B., Gerber, L., Kumar, M., Berglund, L. A., et al. (2010). Ultrastructure and mechanical properties of Populus wood with reduced lignin content caused by transgenic down-regulation of cinnamate 4-hydroxylase. Biomacromolecules 11, 2359–2365. doi: 10.1021/bm100487e
Bocos-Asenjo, I. T., Niño-Sánchez, J., Ginésy, M., and Diez, J. J. (2022). New insights on the integrated management of plant diseases by RNA strategies: Mycoviruses and RNA interference. Int. J. Mol. Sci. 23:9236. doi: 10.3390/ijms23169236
Borthakur, D., Busov, V., Cao, X. H., Du, Q., Gailing, O., Isik, F., et al. (2022). Current status and trends in forest genomics. For. Res. 2:01. doi: 10.48130/FR-2022-0011
Boyle, B., Levée, V., Hamel, L., Nicole, M., and Séguin, A. (2010). Molecular and histochemical characterisation of two distinct poplar Melampsora leaf rust pathosystems. Plant Biol. 12, 364–376. doi: 10.1111/j.1438-8677.2009.00310.x
Canto, T. (2016). Transient expression systems in plants: potentialities and constraints. Adv. Technol. Protein Complex Prod. Charact., Springer. 896, 287–301. doi: 10.1007/978-3-319-27216-0_18
Cao, C.-W., Liu, G.-F., Wang, Z.-Y., Yan, S.-C., Ma, L., and Yang, C.-P. (2010). Response of the gypsy moth, Lymantria dispar to transgenic poplar, Populus simonii× P. nigra, expressing fusion protein gene of the spider insecticidal peptide and Bt-toxin C-peptide. J. Insect Sci. 10:200. doi: 10.1673/031.010.20001
Cavalcaselle, B. (1972). Ecology and ethology of some Buprestids injurious to poplar in central-southern Italy Redia. 53: 67–122.
Cerny, M., Berka, M., Dvořák, M., Milenković, I., Saiz-Fernández, I., Brzobohatý, B., et al. (2022). Defense mechanisms promoting tolerance to aggressive Phytophthora species in hybrid poplar. Front. Plant Sci. 13:1018272. doi: 10.3389/fpls.2022.1018272
Cervera, M.-T., Storme, V., Ivens, B., Gusmao, J., Liu, B. H., Hostyn, V., et al. (2001). Dense genetic linkage maps of three Populus species (Populus deltoides, P. Nigra and P. trichocarpa) based on AFLP and microsatellite markers. Genetics 158, 787–809. doi: 10.1093/genetics/158.2.787
Chanda, S., Pramanik, A., and Maiti, G. G. (2010). Taxonomic study of the section Tacamahaca Spach of the genus Populus L.(Salicaceae Mirb.) in India. Indian J. For. 33, 425–428. doi: 10.54207/bsmps1000-2010-4JF647
Charles, L. S., Dwyer, J. M., Smith, T. J., Connors, S., Marschner, P., and Mayfield, M. M. (2018). Species wood density and the location of planted seedlings drive early stage seedling survival during tropical forest restoration. J. Appl. Ecol. 55, 1009–1018. doi: 10.1111/1365-2664.13031
Charles, J. G., Nef, L., Allegro, G., Collins, C. M., Delplanque, A., Gimenez, R., et al. (2014). “Insect and other pests of poplars and willows” in Poplars and willows: trees for society and the environment (CABI Wallingford UK), 459–526.
Chatterjee, S., Newman, K. L., and Lindow, S. E. (2008). Cell-to-cell signaling in Xylella fastidiosa suppresses movement and xylem vessel colonization in grape. Mol. Plant-Microbe Interact. 21, 1309–1315. doi: 10.1094/MPMI-21-10-1309
Chen, Y., Tong, S., Jiang, Y., Ai, F., Feng, Y., Zhang, J., et al. (2021). Transcriptional landscape of highly lignified poplar stems at single-cell resolution. Genome Biol. 22, 1–22. doi: 10.1186/s13059-021-02537-2
Chilton, M.-D., Saiki, R. K., Yadav, N., Gordon, M. P., and Quetier, F. (1980). T-DNA from Agrobacterium Ti plasmid is in the nuclear DNA fraction of crown gall tumor cells. Proc. Natl. Acad. Sci. 77, 4060–4064. doi: 10.1073/pnas.77.7.4060
Christiaens, O., Whyard, S., Vélez, A. M., and Smagghe, G. (2020). Double-stranded RNA technology to control insect pests: current status and challenges. Front. Plant Sci. 11:451. doi: 10.3389/fpls.2020.00451
Clemente, M., Corigliano, M. G., Pariani, S. A., Sánchez-López, E. F., Sander, V. A., and Ramos-Duarte, V. A. (2019). Plant serine protease inhibitors: biotechnology application in agriculture and molecular farming. Int. J. Mol. Sci. 20:1345. doi: 10.3390/ijms20061345
Confalonieri, M., Allegro, G., Balestrazzi, A., Fogher, C., and Delledonne, M. (1998). Regeneration of Populus nigra transgenic plants expressing a Kunitz proteinase inhibitor (KTi 3) gene. Mol. Breed. 4, 137–145. doi: 10.1023/A:1009640204314
Coyle, D. R., Nebeker, T. E., Hart, E. R., and Mattson, W. J. (2005). Biology and management of insect pests in north American intensively managed hardwood forest systems. Annu. Rev. Entomol. 50, 1–29. doi: 10.1146/annurev.ento.50.071803.130431
Cronk, Q. C. B. (2005). Plant eco devo: the potential of poplar as a model organism. New Phytol. 166, 39–48. doi: 10.1111/j.1469-8137.2005.01369.x
Cunningham, F. J., Goh, N. S., Demirer, G. S., Matos, J. L., and Landry, M. P. (2018). Nanoparticle-mediated delivery towards advancing plant genetic engineering. Trends Biotechnol. 36, 882–897. doi: 10.1016/j.tibtech.2018.03.009
Dara, S. K., Montalva, C., and Barta, M. (2019). Microbial control of invasive forest pests with entomopathogenic fungi: a review of the current situation. Insects 10:341. doi: 10.3390/insects10100341
De Kesel, J., Conrath, U., Flors, V., Luna, E., Mageroy, M. H., Mauch-Mani, B., et al. (2021). The induced resistance lexicon: do’s and don’ts. Trends Plant Sci. 26, 685–691. doi: 10.1016/j.tplants.2021.01.001
De Tillesse, V., Nef, L., Charles, J., Hopkin, A., and Augustin, S. (2007). Damaging poplar insects. Rome: FAO.
Delledonne, M., Allegro, G., Belenghi, B., Balestrazzi, A., Picco, F., Levine, A., et al. (2001). Transformation of white poplar (Populus alba L.) with a novel Arabidopsis thaliana cysteine proteinase inhibitor and analysis of insect pest resistance. Mol. Breed. 7, 35–42. doi: 10.1023/A:1009605001253
Deng, B., Wu, L., Xiao, H., and Cheng, Q. (2023). Characterization of Pseudomonas sp. En3, an endophytic bacterium from poplar leaf Endosphere with plant growth-promoting properties. Forests 14:2203. doi: 10.3390/f14112203
Devantier, Y. A., Moffatt, B., Jones, C., and Charest, P. J. (1993). Microprojectile-mediated DNA delivery to the Salicaceae family. Can. J. Bot. 71, 1458–1466. doi: 10.1139/b93-176
Ding, L., Chen, Y., Wei, X., Ni, M., Zhang, J., Wang, H., et al. (2017). Laboratory evaluation of transgenic Populus davidiana× Populus bolleana expressing Cry1Ac+ SCK, Cry1Ah3, and Cry9Aa3 genes against gypsy moth and fall webworm. PLoS One 12:e0178754. doi: 10.1371/journal.pone.0178754
Dong, Y., Du, S., Zhang, J., Yang, M., and Wang, J. (2015). Differential expression of dual Bt genes in transgene poplar juba (Populus deltoides cv.‘juba’) transformed by two different transformation vectors. Can. J. For. Res. 45, 60–67. doi: 10.1139/cjfr-2014-0335
Dort, E. N., Tanguay, P., and Hamelin, R. C. (2020). CRISPR/Cas9 gene editing: an unexplored frontier for forest pathology. Front. Plant Sci. 11:1126. doi: 10.3389/fpls.2020.01126
Dou, C., Marcondes, W. F., Djaja, J. E., Bura, R., and Gustafson, R. (2017). Can we use short rotation coppice poplar for sugar based biorefinery feedstock? Bioconversion of 2-yearold poplar grown as short rotation coppice. Biotechnol. Biofuels 10, 1–15. doi: 10.1186/s13068-017-0829-6
Douglas, C. J. (2017). Populus as a model tree. Comparative and Evolutionary Genomics of Angiosperm Trees, Plant Genetics and Genomics: Crops and Models 61–84. doi: 10.1007/7397_2017_3
Du, W., Wang, Y., Xie, D., Li, E., Bai, Y., Shang, C., et al. (2024). Phylogenomics reveal Populus gonggaensis as a hybrid between P. Lasiocarpa and P. Cathayana (Salicaceae). Phytokeys 21, 237–161. doi: 10.3897/phytokeys.237.103012
Dunnell, K. L., and LeBoldus, J. M. (2017). The correlation between Septoria leaf spot and stem canker resistance in hybrid poplar. Plant Dis. 101, 464–469. doi: 10.1094/PDIS-06-16-0903-RE
Duplessis, S., Major, I., Martin, F., and Séguin, A. (2009). Poplar and pathogen interactions: insights from Populus genome-wide analyses of resistance and defense gene families and gene expression profiling. Crit. Rev. Plant Sci. 28, 309–334. doi: 10.1080/07352680903241063
Eckenwalder, J. E. (1996). Taxonomic signal and noise in multivariate interpopulational relationships in Populus mexicana (Salicaceae). Syst. Bot. 21, 261–271. doi: 10.2307/2419658
Eqbal, N., and Ansari, M. A. (2024). Assessment of automated systems in ICFRE institutes libraries of India: Issues and challenges. Pearl A J. Libr. Inform. Sci. 18, 92–101. doi: 10.5958/0975-6922.2024.00011.3
Fabi, A., Graziani, V., Anselmi, N., and Varvaro, L. (2008). Newly recognized bacteria involved in trunk scab and canker of white and hybrid poplars. For. Pathol. 38, 356–370. doi: 10.1111/j.1439-0329.2008.00556.x
Fan, D., Li, C., Fan, C., Hu, J., Li, J., Yao, S., et al. (2020). MicroRNA6443 mediated regulation of FERULATE 5 HYDROXYLASE gene alters lignin composition and enhances saccharification in Populus tomentosa. New Phytol. 226, 410–425. doi: 10.1111/nph.16379
Farooq, M. A., Gao, S., Hassan, M. A., Huang, Z., Rasheed, A., Hearne, S., et al. (2024). Artificial intelligence in plant breeding. Trends in Genetics, 40, 891–908.
Feau, N., Mottet, M.-J., Périnet, P., Hamelin, R. C., and Bernier, L. (2010). Recent advances related to poplar leaf spot and canker caused bySeptoria musiva. Can. J. Plant Pathol. 32, 122–134. doi: 10.1080/07060661003740009
Fenning, T. (2013). Challenges and opportunities for the World’s forests in the 21st century : Springer. (Vol. 81). Springer Science & Business Media. doi: 10.1007/978-94-007-7076-8
Fernandez-Conradi, P., Castagneyrol, B., Jactel, H., and Rasmann, S. (2021). Combining phytochemicals and multitrophic interactions to control forest insect pests. Curr. Opin. Insect Sci. 44, 101–106. doi: 10.1016/j.cois.2021.04.007
Fossi, M., Amundson, K., Kuppu, S., Britt, A., and Comai, L. (2019). Regeneration of Solanum tuberosum plants from protoplasts induces widespread genome instability. Plant Physiol. 180, 78–86. doi: 10.1104/pp.18.00906
Frewen, B. E., Chen, T. H. H., Howe, G. T., Davis, J., Rohde, A., Boerjan, W., et al. (2000). Quantitative trait loci and candidate gene mapping of bud set and bud flush in Populus. Genetics 154, 837–845. doi: 10.1093/genetics/154.2.837
Gai, Z., Zhai, J., Chen, X., Jiao, P., Zhang, S., Sun, J., et al. (2021). Phylogeography reveals geographic and environmental factors driving genetic differentiation of Populus sect. Turanga in Northwest China. Plant Sci. 12:705083. doi: 10.3389/fpls.2021.705083
Génissel, A., Leplé, J.-C., Millet, N., Augustin, S., Jouanin, L., and Pilate, G. (2003). High tolerance against Chrysomela tremulae of transgenic poplar plants expressing a synthetic cry3Aa gene from Bacillus thuringiensis ssp tenebrionis. Mol. Breed. 11, 103–110. doi: 10.1023/A:1022453220496
Gennaro, M., and Giorcelli, A. (2019). The biotic adversities of poplar in Italy: a reasoned analysis of factors determining the current state and future perspectives. Ann. Silvic. Res 43, 41–51.
Giorcelli, A., Sparvoli, F., Mattivi, F., Tava, A., Balestrazzi, A., Vrhovsek, U., et al. (2004). Expression of the stilbene synthase (StSy) gene from grapevine in transgenic white poplar results in high accumulation of the antioxidant resveratrol glucosides. Transgenic Res. 13, 203–214. doi: 10.1023/B:TRAG.0000034658.64990.7f
Goué-Mourier, M. C., Faivre-Rampant, P., Le Guerroué, B., Lefèvre, F., and Villar, M. (1996). “Molecular and genetic approaches to rust resistance (Melampsora sp.) in poplar (Populus sp.)” in Somatic cell genetics and molecular genetics of trees (Springer Netherlands), 249–254. doi: 10.1007/978-94-011-3983-0
Goychuk, A., Kulbanska, I., Shvets, M., Pasichnyk, L., Patyka, V., Kalinichenko, A., et al. (2023). Bacterial diseases of bioenergy Woody plants in Ukraine. Sustain. For. 15:4189. doi: 10.3390/su15054189
Guleria, I., Kumari, A., Lacaille-Dubois, M.-A., Saini, A. K., Kumar, V., Saini, R. V., et al. (2022). In-vitro antimicrobial, antioxidant, anti-inflammatory, and cytotoxic activities of Populus ciliata bark and leaves: a comparative study. South African J. Bot. 148, 238–250. doi: 10.1016/j.sajb.2022.04.040
Gullino, M. L., Albajes, R., Al-Jboory, I., Angelotti, F., Chakraborty, S., Garrett, K. A., et al. (2022). Climate change and pathways used by pests as challenges to plant health in agriculture and forestry. Sustain. For. 14:12421. doi: 10.3390/su141912421
Häggman, H., Sutela, S., Walter, C., and Fladung, M. (2013). “Biosafety considerations in the context of deployment of GE trees” in Challenges and opportunities for the World’s forests in the 21st century (Springer), 491–524. doi: 10.1007/978-94-007-7076-8_21
Hamel, L. P. (2006). Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 11, 192–198. doi: 10.1016/j.tplants.2006.02.007
He, H., Dong, Q., Shao, Y., Jiang, H., Zhu, S., Cheng, B., et al. (2012). Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 31, 1199–1217. doi: 10.1007/s00299-012-1241-0
Hertzberg, M., Sievertzon, M., Aspeborg, H., Nilsson, P., Sandberg, G., and Lundeberg, J. (2001). cDNA microarray analysis of small plant tissue samples using a cDNA tag target amplification protocol. Plant J. 25, 585–591. doi: 10.1046/j.1365-313x.2001.00972.x
Heuchelin, S. A., McNabb, H. S., and Klopfenstein, N. B. (1997). Agrobacterium mediated transformation of Populus× euramericana" Ogy" using the chimeric CaMV 35S-pin 2 gene fusion. Can. J. For. Res. 27, 1041–1048.
Hoengenaert, L., Van Doorsselaere, J., Vanholme, R., and Boerjan, W. (2023). Microparticle mediated CRISPR DNA delivery for genome editing in poplar. Front. Plant Sci. 14:1286663. doi: 10.3389/fpls.2023.1286663
Howe, G. T., Goldfarb, B., and Strauss, S. H. (1994). Agrobacterium-mediated transformation of hybrid poplar suspension cultures and regeneration of transformed plants. Plant Cell Tissue Organ Cult. 36, 59–71. doi: 10.1007/BF00048316
Hu, J., Qi, Q., Zhao, Y., Tian, X., Lu, H., Gai, Y., et al. (2019). Unraveling the impact of Pto4CL1 regulation on the cell wall components and wood properties of perennial transgenic Populus tomentosa. Plant Physiol. Biochem. 139, 672–680. doi: 10.1016/j.plaphy.2019.03.035
Hu, J. J., Tian, Y. C., Han, Y. F., Li, L., and Zhang, B. E. (2001). Field evaluation of insect resistant transgenic Populus nigra trees. Euphytica 121, 123–127. doi: 10.1023/A:1012015709363
Huang, Y., Ma, H., Yue, Y., Zhou, T., Zhu, Z., and Wang, C. (2022). Integrated transcriptomic and transgenic analyses reveal potential mechanisms of poplar resistance to Alternaria alternata infection. BMC Plant Biol. 22:413. doi: 10.1186/s12870-022-03793-5
ICFRE, Country report on poplars and willows period (2016). 2012 to 2015. National Poplar Commission of India, Indian Council of Forestry Research and Education. Dehradun.
Islam, W., Waheed, A., Naveed, H., and Zeng, F. (2022). MicroRNAs mediated plant responses to salt stress. Cells 11:2806. doi: 10.3390/cells11182806
Jaffar, N. S., Jawan, R., and Chong, K. P. (2023). The potential of lactic acid bacteria in mediating the control of plant diseases and plant growth stimulation in crop production a mini review. Front. Plant Sci. 13:1047945. doi: 10.3389/fpls.2022.1047945
Jang, H.-A., Bae, E.-K., Kim, M.-H., Park, S.-J., Choi, N.-Y., Pyo, S.-W., et al. (2021). CRISPR-knockout of CSE gene improves saccharification efficiency by reducing lignincontent in hybrid poplar. Int. J. Mol. Sci. 22:9750. doi: 10.3390/ijms22189750
Jansson, S., and Douglas, C. J. (2007). Populus: a model system for plant biology. Annu. Rev. Plant Biol. 58, 435–458. doi: 10.1146/annurev.arplant.58.032806.103956
Jia, Z., Sun, Y., Yuan, L., Tian, Q., and Luo, K. (2010). The chitinase gene (Bbchit1) from Beauveria bassiana enhances resistance to Cytospora chrysosperma in Populus tomentosa Carr. Biotechnol. Lett. 32, 1325–1332. doi: 10.1007/s10529-010-0297-6
Jiang, Y., Guo, L., Ma, X., Zhao, X., Jiao, B., Li, C., et al. (2017). The WRKY transcription factors PtrWRKY18 and PtrWRKY35 promote Melampsora resistance in Populus. Tree Physiol. 37, 665–675. doi: 10.1093/treephys/tpx0008
Jiang, F., Zhu, J., and Liu, H.-L. (2013). Protoplasts: a useful research system for plant cell biology, especially dedifferentiation. Protoplasma 250, 1231–1238. doi: 10.1007/s00709-013-0513-z
Joga, M. R., Mogilicherla, K., Smagghe, G., and Roy, A. (2021). RNA interference-based forest protection products (FPPs) against wood-boring coleopterans: hope or hype? Front. Plant Sci. 12:733608. doi: 10.3389/fpls.2021.733608
Johnson, A. M., Kim, H., Ralph, J., and Mansfield, S. D. (2017). Natural acetylation impacts carbohydrate recovery during deconstruction of Populus trichocarpa wood. Biotechnol. Biofuels 10, 48–12. doi: 10.1186/s13068-017-0734-z
Juge, N. (2006). Plant protein inhibitors of cell wall degrading enzymes. Trends Plant Sci. 11, 359–367. doi: 10.1016/j.tplants.2006.05.006
Kalinichenko, A., Pasichnyk, L., Osypenco, S., Patyka, V., and Usmanova, H. (2017). Bacterial diseases of energy plants. Ecol. Chem. Eng. A 24, 169–191.
Kar, M. M., and Raichaudhuri, A. (2021). Role of microRNAs in mediating biotic and abiotic stress in plants. Plant Gene 26:100277. doi: 10.1016/j.plgene.2021.100277
Karlovsky, P. (2011). Biological detoxification of the mycotoxin deoxynivalenol and its use in genetically engineered crops and feed additives. Appl. Microbiol. Biotechnol 91, 491–504. doi: 10.1007/s00253-011-3401-5
Kaur, S., Samota, M. K., Choudhary, M., Choudhary, M., Pandey, A. K., Sharma, A., et al. (2022). How do plants defend themselves against pathogens-biochemical mechanisms and genetic interventions. Physiol. Mol. Biol. Plants 28, 485–504. doi: 10.1007/s12298-022-01146-y
Kebert, M., Kostić, S., Čapelja, E., Vuksanović, V., Stojnić, S., Markić, A. G., et al. (2022). Ectomycorrhizal fungi modulate pedunculate oak’s heat stress responses through the alternation of polyamines, phenolics, and osmotica content. Plan. Theory 11:3360. doi: 10.3390/plants11233360
Kleiner, K. W., Ellis, D. D., Mccown, B. H., and Raffa, K. F. (1995). Field evaluation of transgenic poplar expressing a Bacillus thuringiensis cry1A (∞) d-endotoxin gene against forest tent caterpillar (Lepidoptera: Lasiocampidae) and gypsy moth (Lepidoptera: Lymantriidae) following winter dormancy. Environ. Entomol. 24, 1358–1364. doi: 10.1093/ee/24.5.1358
Kollert, W., Carle, J., and Rosengren, L. (2014). “Poplars and willows for rural livelihoods and sustainable development” in Poplars and willows: trees for society and the environment (Wallingford UK: CABI), 577–602. doi: 10.1079/9781780641089.0577
Kovalitskaya, Y., Dayanova, L., Azarova, A., and Shestibratov, K. (2016). RNA interference mediated down-regulation of 4-coumarate: Coenzyme a ligase in Populus tremula alters lignification and plant growth. Educ: Int. J. Environ. Sci.
Kowalski, T. (2013). Infectious forest diseases. Wallingford UK: CABI, 488–518. doi: 10.1079/9781780640402.0488
Kumudini, B. S., Jayamohan, N. S., Patil, S. V., and Govardhana, M. (2018). Primary plant metabolism during plant–pathogen interactions and its role in defense. Plant metabolites and regulation under environmental stress 215–229. doi: 10.1016/B978-0-12-812689-9.00011-X
Kwaśna, H., Szewczyk, W., Baranowska, M., and Behnke-Borowczyk, J. (2021a). Bacteria associated with vascular wilt of poplar. Arch. Microbiol. 203, 4829–4838. doi: 10.1007/s00203-021-02464-7
Kwaśna, H., Szewczyk, W., Baranowska, M., Gallas, E., Wiśniewska, M., and Behnke Borowczyk, J. (2021b). Mycobiota associated with the vascular wilt of poplar. Plan. Theory 10:892. doi: 10.3390/plants10050892
La Mantia, J., Unda, F., Douglas, C. J., Mansfield, S. D., and Hamelin, R. (2018). Overexpression of at GolS3 and Cs RFS in poplar enhances ROS tolerance and represses defense response to leaf rust disease. Tree Physiol. 38, 457–470. doi: 10.1093/treephys/tpx100
Lawrence, S. D., and Novak, N. G. (2006). Expression of poplar chitinase in tomato leads to inhibition of development in Colorado potato beetle. Biotechnol. Lett. 28, 593–599. doi: 10.1007/s10529-006-0022-7
Lee, M., Jeon, H. S., Kim, S. H., Chung, J. H., Roppolo, D., Lee, H., et al. (2019). Lignin based barrier restricts pathogens to the infection site and confers resistance in plants. EMBO J. 38:e101948. doi: 10.15252/embj.2019101948
Lenaghan, S. C., and Neal Stewart, C. (2019). An automated protoplast transformation system. Plant Genome Ed. with Cris. Syst. Methods Protoc. 1917, 355–363. doi: 10.1007/978-1-4939-8991-1_26
Leonetti, P., Stuttmann, J., and Pantaleo, V. (2021). Regulation of plant antiviral defense genes via host RNA-silencing mechanisms. Virol. J. 18, 1–10. doi: 10.1186/s12985-021-01664-3
Levée, V., Major, I., Levasseur, C., Tremblay, L., MacKay, J., and Séguin, A. (2009). Expression profiling and functional analysis of Populus WRKY23 reveals a regulatory role in defense. New Phytol. 184, 48–70. doi: 10.1111/j.1469-8137.2009.02955.x
Li, A., and He, W. (2019). Molecular aspects of an emerging poplar canker caused by Lonsdalea populi. Front. Microbiol. 10:2496. doi: 10.3389/fmicb.2019.02496
Li, P., Song, W., Zhang, Y., Yang, Y., Li, S., and Zhang, J. (2024). Drought stress enhances plastid-mediated RNA interference for efficient the willow leaf beetle management. Pestic. Biochem. Physiol. 204:106037. doi: 10.1016/j.pestbp.2024.106037
Li, L., Sun, W., Wang, P., Li, H., Rehman, S., Li, D., et al. (2022). Characterization, expression, and functional analysis of the pathogenesis-related gene PtDIR11 in transgenic poplar. Int. J. Biol. Macromol. 210, 182–195. doi: 10.1016/j.ijbiomac.2022.05.012
Li, Y., Yuan, Y., Hu, Z., Liu, S., and Zhang, X. (2023). Genetic transformation of Forest trees and its research advances in stress tolerance. Forests 15:441. doi: 10.3390/f15030441
Li, Z., Zhang, B., Fu, Y., Suo, Y., Zhang, Y., Feng, J., et al. (2024). A rapid and efficient in vivo inoculation method for introducing tree stem canker pathogens onto leaves: suitable for large-scale assessment of resistance in poplar breeding progeny. bioRxiv, 2003–2024.
Li, M., Zhang, H., Hu, J., Han, Y., and Tian, Y. (2000). Study on insect-resistant transgenic poplar plants containing both Bt and PI genes. Sci. Silvae Sin. 36, 93–97. doi: 10.3321/j.issn:1001-7488.2000.02.015
Li, S., Zhen, C., Xu, W., Wang, C., and Cheng, Y. (2017). Simple, rapid and efficient transformation of genotype Nisqually-1: a basic tool for the first sequenced model tree. Scientific reports 7:2638. doi: 10.1038/s41598-017-02651-x
Liang, H., Maynard, C. A., Allen, R. D., and Powell, W. A. (2001). Increased Septoria musiva resistance in transgenic hybrid poplar leaves expressing a wheat oxalate oxidase gene. Plant Mol. Biol 45, 619–629. doi: 10.1023/A:1010631318831
Liang, H., Catranis, C. M., Maynard, C. A., and Powell, W. A. (2002). Enhanced resistance to the poplar fungal pathogen, Septoria musiva, in hybrid poplar clones transformed with genes encoding antimicrobial peptides. Biotechnol. Lett. 24, 383–389. doi: 10.1023/A:1014552503140
Li Ling, L. L., Qi LiWang, Q. L., Han YiFan, H. Y., Wang YingChun, W. Y., and Li WenBin, L. W. (2000). A study on the introduction of male sterility of antiinsect transgenic Populus nigra by the TA29-Barnase gene. Sci Silvae Sinicae 36, 28–32.
Limpens, E., Ramos, J., Franken, C., Raz, V., Compaan, B., Franssen, H., et al. (2004). RNA interference in Agrobacterium rhizogenes transformed roots of Arabidopsis and Medicago truncatula. J. Exp. Bot. 55, 983–992. doi: 10.1093/jxb/erh122
Lin, C. S., Hsu, C. T., Yang, L. H., Lee, L. Y., Fu, J. Y., Cheng, Q. W., et al. (2018). Application of protoplast technology to CRISPR/Cas9 mutagenesis: from single-cell mutation detection to mutant plant regeneration. Plant Biotechnol. J. 16, 1295–1310. doi: 10.1111/pbi.12870
Lin, T., Wang ZhiYing, W. Z., Liu, K., Jing, T., and Zhang, C. (2006). Transformation of spider neurotoxin gene with prospective insecticidal properties into hybrid poplar Populus simonii × P. nigra. Acta Entomol. Sin. 49, 593–598. doi: 10.3321/j.issn:0454-6296.2006.04.009
Liu, X., Wang, Z., Shao, W., Ye, Z., and Zhang, J. (2017). Phylogenetic and taxonomic status analyses of the Abaso section from multiple nuclear genes and plastid fragments reveal new insights into the North America origin of Populus (Salicaceae). Front. Plant Sci. 7:2022. doi: 10.3389/fpls.2016.02022
Liu, D., Zhang, J., Dong, Y., Zhang, X., Yang, M., and Gao, B. (2016). Genetic transformation and expression of Cry1AcCry3A-NTHK1 genes in Populus× euramericana" Neva". Springer Nat. 38:177.
Llave, C., Xie, Z., Kasschau, K. D., and Carrington, J. C. (2002). Cleavage ofScarecrow-likemRNA targets directed by a class ofArabidopsismiRNA. Science 297, 2053–2056. doi: 10.1126/science.1076311
Lubrano, L. (1992). High-tech and micropropagation II. Springer Berlin Heidelberg, 151–178. doi: 10.1007/978-3-642-76422-6_8
Lv, Z., Jiang, R., Chen, J., and Chen, W. (2020). Nanoparticle mediated gene transformation strategies for plant genetic engineering. Plant J. 104, 880–891. doi: 10.1111/tpj.14973
Ma, Y.-N., Gu, Y.-L., Liu, J., Zhang, Y., Wang, X., Xia, Z., et al. (2024). Frontiers research topics February 2024. Charact. Manag. plant Pathog: Detect, 4.
Mamta, B., and Rajam, M. V. (2017). RNAi technology: a new platform for crop pest control. Physiol. Mol. Biol. Plants 23, 487–501. doi: 10.1007/s12298-017-0443-x
Martín García, J., Jactel, H., and Diez, J. J. (2011). Patterns and monitoring of Sesia apiformis infestations in poplar plantations at different spatial scales. J. Appl. Entomol. 135, 382–392. doi: 10.1111/j.1439-0418.2010.01562.x
Martínez-Arias, C., Macaya-Sanz, D., Witzell, J., and Martín, J. A. (2019). Enhancement of Populus alba tolerance to Venturia tremulae upon inoculation with endophytes showing in vitro biocontrol potential. Eur. J. Plant Pathol. 153, 1031–1042. doi: 10.1007/s10658-018-01618-6
Mazhar, A. R., and Sadeghi, S. E. (2024). Evaluation of susceptibility poplar species and clones (Populus spp.) against two poplar key pests; Monsteira unicostata and Melanophila picta in Hamadan province. J. Entomol. Soc. Iran. 44, 463–469. doi: 10.61186/jesi.44.4.8
McCombe, C. L., Catanzariti, A., Greenwood, J. R., Desai, A. M., Outram, M. A., Yu, D. S., et al. (2023). A rust fungus Nudix hydrolase effector decaps mRNA in vitro and interferes with plant immune pathways. New Phytol. 239, 222–239. doi: 10.1111/nph.18727
McCown, B. H., McCabe, D. E., Russell, D. R., Robison, D. J., Barton, K. A., and Raffa, K. F. (1991). Stable transformation of Populus and incorporation of pest resistance by electric discharge particle acceleration. Plant Cell Rep. 9, 590–594. doi: 10.1007/BF00232339
McIntyre, P. J., and Whitham, T. G. (2003). Plant genotype affects long term herbivore population dynamics and extinction: conservation implications. Ecology 84, 311–322. doi: 10.1890/0012-9658(2003)084[0311:PGALTH]2.0.CO;2
Meert, R. (2022). Sesia apiformis (Lepidoptera: Sesiidae) living in Populus root suckers. Phegea 50, 27–31. doi: 10.6084/m9.figshare.19122965
Mehri, H., Lotfollahi, P., de Lillo, E., and Azimi, S. (2020). Redescription of Aceria varia and Tegoprionus dentatus (Trombidiformes: Eriophyoidea: Eriophyidae) from Iran. Persian J. Acarol. 9, 129–139. doi: 10.22073/pja.v9i2.58457
Mentag, R., Luckevich, M., Morency, M.-J., and Seguin, A. (2003). Bacterial disease resistance of transgenic hybrid poplar expressing the synthetic antimicrobial peptide D4E1. Tree Physiol. 23, 405–411. doi: 10.1093/treephys/23.6.405
Mogilicherla, K., Chakraborty, A., Taning, N. T. C., Smagghe, G., and Roy, A. (2023). RNAi in termites (Isoptera): current status and prospects for pest management. Entomol. Gen. 43, 55–68. doi: 10.1127/entomologia/2022/1636
Mohamed, R., Meilan, R., and Strauss, S. H. (2001). Complex behavior of a copper-inducible gene expression system in transgenic poplar. For. Genet. 8, 69–72.
Montesinos, L., Baró, A., Gascón, B., and Montesinos, E. (2023). Bactericidal and plant defense elicitation activities of Eucalyptus oil decrease the severity of infections by Xylella fastidiosa on almond plants. Front. Plant Sci. 14:1122218. doi: 10.3389/fpls.2023.1122218
Movahedi, A., Zhang, J., Amirian, R., and Zhuge, Q. (2014). An efficient Agrobacterium mediated transformation system for poplar. Int. J. Mol. Sci. 15, 10780–10793. doi: 10.3390/ijms150610780
Nagaraju, D. K., Jain, R. K., Iyyanar, D., Singh, M., Kasturi, N., Prema, R. T., et al. (2023). Interception of live phratora laticollis (suffrian)(coleoptera: chrysomelidae) on poplar logs imported from Belgium and Germany. Indian J. Entomol. 85, 271–273. doi: 10.55446/IJE.2022.441
Naing, A. H., Adedeji, O. S., and Kim, C. K. (2021). Protoplast technology in ornamental plants: current progress and potential applications on genetic improvement. Sci. Hortic. 283:110043. doi: 10.1016/j.scienta.2021.110043
Naylor, M., Reeves, J., Cooper, J. I., Edwards, M.-L., and Wang, H. (2005). Construction and properties of a gene-silencing vector based on poplar mosaic virus (genus Carlavirus). J. Virol. Methods 124, 27–36. doi: 10.1016/j.jviromet.2004.10.007
Newcombe, G., Bradshaw, H. D. Jr., Chastagner, G. A., and Stettler, R. F. (1996). A major gene for resistance to Melampsora medusae f. sp. deltoidae in a hybrid poplar pedigree. Phytopathology 86, 87–94. doi: 10.1094/Phyto-86-87
Nizamani, M. M., Zhang, Q., Muhae-Ud-Din, G., and Wang, Y. (2023). High-throughput sequencing in plant disease management: a comprehensive review of benefits, challenges, and future perspectives. Phytopathol. Res. 5, 1–17. doi: 10.1186/s42483-023-00199-5
Noël, A., Levasseur, C., and Séguin, A. (2005). Enhanced resistance to fungal pathogens in forest trees by genetic transformation of black spruce and hybrid poplar with a Trichoderma harzianum endochitinase gene. Physiol. Mol. Plant Pathol. 67, 92–99. doi: 10.1016/j.pmpp.2005.09.010
Nowak, K., Luniak, N., Meyer, S., Schulze, J., Mendel, R. R., and Hänsch, R. (2004). Fluorescent proteins in poplar: a useful tool to study promoter function and protein localization. Plant Biol. 6, 65–73. doi: 10.1055/s-2004-815730
Obbard, D. J., Gordon, K. H. J., Buck, A. H., and Jiggins, F. M. (2009). The evolution of RNAi as a defence against viruses and transposable elements. Philos. Trans. R. Soc. B Biol. Sci. 364, 99–115. doi: 10.1098/rstb.2008.0168
Ojuederie, O. B., Igwe, D. O., and Popoola, J. O. (2022). Transgenic plant-mediated phytoremediation: applications, challenges, and prospects. Assist. Phytoremed. Elsevier. 102, 179–202. doi: 10.1016/B978-0-12-822893-7.00009-4
Osdaghi, E., Kakavandi, N. R., and Nejati, M. (2014). First report of Alternaria alternata causing leaf spot on Populus euphratica in Iran. Iran J Plant Pathol 50, 309–310.
Ostry, M., Ramstedt, M., Newcombe, G., and Steenackers, M. (2014). Poplars and willows: trees for society and the environment : CABI. 443–458. doi: 10.1079/9781780641089.0443
Özparpucu, M., Rüggeberg, M., Gierlinger, N., Cesarino, I., Vanholme, R., Boerjan, W., et al. (2017). Unravelling the impact of lignin on cell wall mechanics: a comprehensive study on young poplar trees downregulated for CINNAMYL ALCOHOL DEHYDROGENASE (CAD). Plant J. 91, 480–490. doi: 10.1111/tpj.13584
Ozyigit, I. I., and Yucebilgili Kurtoglu, K. (2020). Particle bombardment technology and its applications in plants. Mol. Biol. Rep. 47, 9831–9847. doi: 10.1007/s11033-020-06001-5
Park, Y. W., Baba, K., Furuta, Y., Iida, I., Sameshima, K., Arai, M., et al. (2004). Enhancement of growth and cellulose accumulation by overexpression of xyloglucanase in poplar. FEBS Lett. 564, 183–187. doi: 10.1016/S0014-5793(04)00346-1
Pastierovič, F., Mogilicherla, K., Hradecký, J., Kalyniukova, A., Dvořák, O., Roy, A., et al. (2024). Genome-wide transcriptomic and Metabolomic analyses unveiling the Defence mechanisms of Populus tremula against sucking and chewing insect herbivores. Int. J. Mol. Sci. 25:6124. doi: 10.3390/ijms25116124
Pinon, J., and Frey, P. (2005). Rust diseases of willow and poplar. Wallingford UK: CABI Publishing. 139–154. doi: 10.1079/9780851999999.0000
Polle, A., Janz, D., Teichmann, T., and Lipka, V. (2013). Poplar genetic engineering: promoting desirable wood characteristics and pest resistance. Appl. Microbiol. Biotechnol. 97, 5669–5679. doi: 10.1007/s00253-013-4940-8
Porth, I., Goessen, R., and Heinze, B. (2024). “Poplar genomics: an introduction” in The poplar genome (Springer International Publishing), 1–31. doi: 10.1007/978-3-031-50787-8_1
Pratiwi, R. A., and Surya, M. I. (2020). Agrobacterium-mediated transformation. Genet. Transform. Crop. doi: 10.5772/intechopen.91132
Ramstedt, M., ÅRström, B., and von Fircks, H. A. (1994). Dieback of poplar and willow caused by Pseudomonas syringae in combination with freezing stress. Eur. J. For. Pathol. 24, 305–315. doi: 10.1111/j.1439-0329.1994.tb00824.x
Rao, H., Wr, N., Huang, M., Fan, Y., and Wang, M. (2001). Molecular breeding of Woody plants, Proceedings of the international wood biotechnology symposium (Amsterdam, Netherlands: IWBS) Elsevier, 239–246.
Rehman, L., Su, X., Guo, H., Qi, X., and Cheng, H. (2016). Protoplast transformation as a potential platform for exploring gene function in Verticillium dahliae. BMC Biotechnol. 16, 1–9. doi: 10.1186/s12896-016-0287-4
Ren, Y., Zhou, X., Dong, Y., Zhang, J., Wang, J., and Yang, M. (2021). Exogenous gene expression and insect resistance in dual Bt toxin Populus× euramericana ‘Neva’transgenic plants. Front. Plant Sci. 12:660226. doi: 10.3389/fpls.2021.660226
Rezazadeh, R., Williams, R. R., and Harrison, D. K. (2011). Factors affecting mango (Mangifera indica L.) protoplast isolation and culture. Sci. Hortic. 130, 214–221. doi: 10.1016/j.scienta.2011.06.046
Riseh, R. S., Fathi, F., Lagzian, A., Vatankhah, M., and Kennedy, J. F. (2024). Modifying lignin: a promising strategy for plant disease control. Int. J. Biol. Macromol. 271:132696. doi: 10.1016/j.ijbiomac.2024.132696
Rottmann, W. H., Meilan, R., Sheppard, L. A., Brunner, A. M., Skinner, J. S., Ma, C., et al. (2000). Diverse effects of overexpression of LEAFY and PTLF, a poplar (Populus) homolog of LEAFY/FLORICAULA, in transgenic poplar and Arabidopsis. Plant J. 22, 235–245. doi: 10.1046/j.1365-313x.2000.00734.x
Sadeghi, S. E., Yarmand, H., Zamani, S. M., Ali, B., Zeinaly, S., Mehrabi, A., et al. (2009). “Insects associated with forest communities and poplar plantations in Iran” in Review of forests, wood products and wood biotechnology of Iran and Germany– Part III. eds. A. R. Kharazipour, C. Schopper, C. Muller, and M. Euring (Gottingen: Universitatsverlag Gottingen), 265–283.
Saint-Vincent, P. M. B., Ridout, M., Engle, N. L., Lawrence, T. J., Yeary, M. L., Tschaplinski, T. J., et al. (2020). Isolation, characterization, and pathogenicity of two Pseudomonas syringae pathovars from Populus trichocarpa seeds. Microorganisms 8:1137. doi: 10.3390/microorganisms8081137
Sandal, S., Singh, S., Bansal, G., Kaur, R., Mogilicherla, K., Pandher, S., et al. (2023). Nanoparticle-shielded dsRNA delivery for enhancing RNAi efficiency in cotton spotted bollworm Earias vittella (Lepidoptera: Nolidae). Int. J. Mol. Sci. 24:9161. doi: 10.3390/ijms24119161
Satish, D., Mukherjee, S. K., and Gupta, D. (2021). The landscape of microRNAs in plant viral infections. Plant Gene 26:100293. doi: 10.1016/j.plgene.2021.100293
Saxena, J., Pandey, V. V., and Sisodia, R. (2017). Evaluating poplar leaf extract concentration on potential plant pathogens. Int. J. Curr. Microbiol. App. Sci. 6, 2933–2942. doi: 10.20546/ijcmas.2017.609.360
Schroeder, H., and Fladung, M. (2018). Poplar clones differ in their resistance against insects feeding. Landbauforschung 68, 19–26. doi: 10.3220/LBF1534394196000
Sekine, M., and Shinmyo, A. (2020). “Agrobacterium and plant genetic engineering” in Recombinant microbes for industrial and agricultural applications, 623–639.
Sellamuthu, G., Chakraborty, A., Vetukuri, R. R., Sarath, S., and Roy, A. (2024). RNAi-biofungicides: a quantum leap for tree fungal pathogen management. Crit. Rev. Biotechnol. 1–28. doi: 10.1080/07388551.2024.2430478
Serres, R., Ostry, M., McCown, B., and Skilling, D. (1991). Somaclonal variation in Populus hybrids regenerated from protoplast culture. Woody Plant Biotechnol., 210, 59–61. doi: 10.1007/978-1-4684-7932-4_7
Seserman, D.-M. (2018). “Benefits of agroforestry systems for land equivalent ratio-case studies in Brandenburg and lower Saxony, Germany” in European agroforestry conference-agroforestry as sustainable land use. 4th ed (Federation: EURAF).
Sharan, S., Sarin, N. B., and Mukhopadhyay, K. (2019). Elicitor-mediated enhanced accumulation of ursolic acid and eugenol in hairy root cultures of Ocimum tenuiflorum L. is age, dose, and duration dependent. South Afr. J. Bot. 124, 199–210. doi: 10.1016/j.sajb.2019.05.009
Shen, Z., Sun, J., Yao, J., Wang, S., Ding, M., Zhang, H., et al. (2015). High rates of virus induced gene silencing by tobacco rattle virus in Populus. Tree Physiol. 35, 1016–1029. doi: 10.1093/treephys/tpv064
Shi, T.-L., Jia, K.-H., Bao, Y.-T., Nie, S., Tian, X.-C., Yan, X.-M., et al. (2024). High-quality genome assembly enables prediction of allele-specific gene expression in hybrid poplar. Plant Physiol. 195, 652–670. doi: 10.1093/plphys/kiae078
Silva, L. N., Zimmer, K. R., Macedo, A. J., and Trentin, D. S. (2016). Plant natural products targeting bacterial virulence factors. Chem. Rev. 116, 9162–9236. doi: 10.1021/acs.chemrev.6b00184
Smith, C. M., and Campbell, M. M. (2004). Populusgenotypes differ in infection by, and systemic spread of Poplar mosaic virus. Plant Pathol. 53, 780–787. doi: 10.1111/j.1365-3059.2004.01095.x
Smith, C. M., Rodriguez-Buey, M., Karlsson, J., and Campbell, M. M. (2004). The response of the poplar transcriptome to wounding and subsequent infection by a viral pathogen. New Phytologist, 123–136.
Smith, I. M., Dunez, J., Phillips, D. H., Lelliott, R. A., and Archer, S. A. (2009). European handbook of plant diseases. Hoboken, New Jersey, USA: John Wiley & Sons.
Song, C., Lu, L., Guo, Y., Xu, H., and Li, R. (2019). Efficient Agrobacterium-mediated transformation of the commercial hybrid poplar Populus Alba× Populus glandulosa Uyeki. Int. J. Mol. Sci. 20:2594. doi: 10.3390/ijms20102594
Song, Q., Kong, L., Yang, X., Jiao, B., Hu, J., Zhang, Z., et al. (2022). PtoMYB142, a poplar R2R3-MYB transcription factor, contributes to drought tolerance by regulating wax biosynthesis. Tree Physiol. 42, 2133–2147. doi: 10.1093/treephys/tpac060
Song, Z., Zhang, C., Song, G., Wei, H., Xu, W., Pan, H., et al. (2024). Unraveling the lncRNA-miRNA-mRNA regulatory network involved in poplar coma development through high-throughput sequencing. Int J Mol Sci 25:7403. doi: 10.3390/ijms25137403
Stanton, B. J., Neale, D. B., and Li, S. (2009). “Populus breeding: from the classical to the genomic approach” in Genetics and genomics of populus (Springer), 309–348. doi: 10.1007/978-1-4419-1541-2_14
Sterky, F., Regan, S., Karlsson, J., Hertzberg, M., Rohde, A., Holmberg, A., et al. (1998). Gene discovery in the wood-forming tissues of poplar: analysis of 5,692 expressed sequence tags. Proc. Natl. Acad. Sci. USA 95, 13330–13335. doi: 10.1073/pnas.95.22.13330
Su, Y., Li, H.-G., Wang, Y., Li, S., Wang, H.-L., Yu, L., et al. (2018). Poplar miR472a targeting NBS-LRRs is involved in effective defence against the necrotrophic fungus Cytospora chrysosperma. J. Exp. Bot. 69, 5519–5530. doi: 10.1093/jxb/ery304
Sulis, D. B., Jiang, X., Yang, C., Marques, B. M., Matthews, M. L., Miller, Z., et al. (2023). Multiplex CRISPR editing of wood for sustainable fiber production. Science 381, 216–221. doi: 10.1126/science.add4514
Sun, L., Gao, Y., Zhang, Q., Lv, Y., and Cao, C. (2022). Resistance to Lymantria dispar larvae in transgenic poplar plants expressing CYP6B53 double stranded RNA. Ann. Appl. Biol. 181, 40–47. doi: 10.1111/aab.12752
Sun, B., Zhang, F., Xiao, N., Jiang, M., Yuan, Q., Xue, S., et al. (2018). An efficient mesophyll protoplast isolation, purification and PEG-mediated transient gene expression for subcellular localization in Chinese kale. Sci. Hortic. 241, 187–193. doi: 10.1016/j.scienta.2018.07.001
Taylor, G. (2002). Populus: Arabidopsis for forestry. Do we need a model tree? Ann. Bot. 90, 681–689. doi: 10.1093/aob/mcf255
Thakur, A. K., Kumar, P., Parmar, N., Shandil, R. K., Aggarwal, G., Gaur, A., et al. (2021). Achievements and prospects of genetic engineering in poplar: a review. New For. 52, 889–920. doi: 10.1007/s11056-021-09836-3
Thakur, A. K., Sharma, S., and Srivastava, D. K. (2005). Plant regeneration and genetic transformation studies in petiole tissue of Himalayan poplar (Populus ciliata wall.). Curr. Sci. 89, 664–668.
The Plant List. (2013). Version 1.1. Published on the Internet. Available at: http://www.theplantlist.org/.
Thomas, P. (2022). Game of’Mones: Comprehending Bemisia tabaci MEAM1 nymph-based resistance and defense Phytohormone signaling in alfalfa. Riverside: University of California.
Tóth, T., Lakatos, T., and Koltay, A. (2013). Lonsdalea quercina subsp. populi subsp. nov., isolated from bark canker of poplar trees. Int. J. Syst. Evol. Microbiol. 63, 2309–2313. doi: 10.1099/ijs.0.042911-0
Trumbore, S., Brando, P., and Hartmann, H. (2015). Forest health and global change. Science 349, 814–818. doi: 10.1126/science.aac6759
Tsai, C.-J., Podila, G. K., and Chiang, V. L. (1994). Agrobacterium-mediated transformation of quaking aspen (Populus tremuloides) and regeneration of transgenic plants. Plant Cell Rep. 14, 94–97. doi: 10.1007/BF00233768
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & gray). Science 313, 1596–1604. doi: 10.1126/science.1128691
Ullah, C., Tsai, C., Unsicker, S. B., Xue, L., Reichelt, M., Gershenzon, J., et al. (2019). Salicylic acid activates poplar defense against the biotrophic rust fungus Melampsora larici Populina via increased biosynthesis of catechin and proanthocyanidins. New Phytol. 221, 960–975. doi: 10.1111/nph.15396
Uniyal, K., Chandra, G., Khan, R. U., and Singh, Y. P. (2018). Selection of potent isolates from a population of Alternaria Alternata, a leaf spot pathogen of poplar. Am. J. Appl. Math. Stat. 6, 232–238. doi: 10.12691/ajams-6-6-3
Urban, J. (2013). Biology of Byctiscus populi (L.) (Coleoptera, Attelabidae). Part II. Leafrolls, larvae and this year’s imagoes. Acta Univ. Agric. Silvic. Mendelianae Brun. 60, 155–166. doi: 10.11118/actaun201260010155
von Bargen, S., Al Kubrusli, R., Gaskin, T., Fürl, S., Hüttner, F., Blystad, D., et al. (2020). Characterisation of a novel Emaravirus identified in mosaic diseased Eurasian aspen (Populus tremula). Ann. Appl. Biol. 176, 210–222. doi: 10.1111/aab.12576
Wang, W., Bai, X.-D., Chen, K., Gu, C.-R., Yu, Q.-B., Jiang, J., et al. (2022). Role of PsnWRKY70 in regulatory network response to infection with Alternaria alternata (Fr.) keissl in Populus. Int. J. Mol. Sci. 23:7537. doi: 10.3390/ijms23147537
Wang, G., Castiglione, S., Chen Ying, C. Y., Li Ling, L. L., Han YiFan, H. Y., Tian YingChuan, T. Y., et al. (1996). Poplar (Populus nigra L.) plants transformed with a Bacillus thuringiensis toxin gene: insecticidal activity and genomic analysis.
Wang, J., and Constabel, C. P. (2004). Polyphenol oxidase overexpression in transgenic Populus enhances resistance to herbivory by forest tent caterpillar (Malacosoma disstria). Planta 220, 87–96. doi: 10.1007/s00425-004-1327-1
Wang, G., Dong, Y., Liu, X., Yao, G., Yu, X., and Yang, M. (2018). The current status and development of insect-resistant genetically engineered poplar in China. Front. Plant Sci. 9:1408. doi: 10.3389/fpls.2018.01408
Wang, Z., Du, S., Dayanandan, S., Wang, D., Zeng, Y., and Zhang, J. (2014). Phylogeny reconstruction and hybrid analysis of Populus (Salicaceae) based on nucleotide sequences of multiple single-copy nuclear genes and plastid fragments. PloS one 9:e103645. doi: 10.1371/journal.pone.0103645
Wang, S., Hu, M., Chen, H., Li, C., Xue, Y., Song, X., et al. (2023). Pseudomonas forestsoilum sp. nov. and P. Tohonis biocontrol bacterial wilt by quenching 3 hydroxypalmitic acid methyl ester. Front. Plant Sci. 14:1193297. doi: 10.3389/fpls.2023.1193297
Wang, Y., Huang, J., Li, E., Xu, S., Zhan, Z., Zhang, X., et al. (2022). Phylogenomics and biogeography of Populus based on comprehensive sampling reveal deep-level relationships and multiple intercontinental dispersals. Front. Plant Sci. 13:813177. doi: 10.3389/fpls.2022.813177
Wang, J., Su, X., Ji, L., Zhang, B., Hu, Z., Huang, R., et al. (2007). Multiple transgenes Populus xeuramericana ‘Guariento’ plants obtained by biolistic bombardment. Chin. Sci. Bull. 52, 224–230. doi: 10.1007/s11434-007-0034-2
Wang, P., Zhou, J., Sun, W., Li, H., Li, D., and Zhuge, Q. (2023). Characteristics and function of the pathogenesis-related protein 1 gene family in poplar. Plant Sci. 336:111857. doi: 10.1016/j.plantsci.2023.111857
Whyard, S., Singh, A. D., and Wong, S. (2009). Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol. 39, 824–832. doi: 10.1016/j.ibmb.2009.09.007
Wijekoon, C. P., Kalischuk, M. L., Brunelle, P., Howard, R. J., and Kawchuk, L. M. (2021). Characterization of bronze leaf disease in western Canadian aspen and poplar trees. Can. J. Plant Sci. 102, 11–19. doi: 10.1139/cjps-2021-0104
Wu, J., Zhao, C., Lin, W., Hu, R., Wang, Q., Chen, H., et al. (2014). Binding characteristics between polyethylene glycol (PEG) and proteins in aqueous solution. J. Mater. Chem. B 2, 2983–2992. doi: 10.1039/c4tb00253a
Xi, B., Clothier, B., Coleman, M., Duan, J., Hu, W., Li, D., et al. (2021). Irrigation management in poplar (Populus spp.) plantations: a review. For. Ecol. Manag. 494:119330. doi: 10.1016/j.foreco.2021.119330
Xing, J., Li, M., Li, J., Shen, W., Li, P., Zhao, J., et al. (2022). Stem canker pathogen Botryosphaeria dothidea inhibits poplar leaf photosynthesis in the early stage of inoculation. Front. Plant Sci. 13:1008834. doi: 10.3389/fpls.2022.1008834
Xiong, Q., Zhang, L., Zheng, X., Qian, Y., Zhang, Y., Zhao, L., et al. (2021). Rapid and specific detection of the poplar black spot disease caused by marssonina brunnea using loop-mediated isothermal amplification assay. Plan. Theory 10:253. doi: 10.3390/plants10020253
Xu, C., Wei, H., Wang, L., Yin, T., and Zhuge, Q. (2019). Optimization of the cry1Ah1 sequence enhances the hyper-resistance of transgenic poplars to Hyphantria cunea. Front. Plant Sci. 10:335. doi: 10.3389/fpls.2019.00335
Xu, S., Zhang, Y., Li, S., Chang, L., Wu, Y., and Zhang, J. (2020). Plastid-expressed Bacillus thuringiensis (Bt) cry3Bb confers high mortality to a leaf eating beetle in poplar. Plant Cell Rep. 39, 317–323. doi: 10.1007/s00299-019-02492-0
Yang, X., Fu, T., Yu, R., Zhang, L., Yang, Y., Xiao, D., et al. (2023). miR159a modulates poplar resistance against different fungi and bacteria. Plant Physiol. Biochem. 201:107899. doi: 10.1016/j.plaphy.2023.107899
Yang, M., Li, L., Wang, Y., Wang, J., and Liang, H. (2006). Transformation and expression of two insect-resistant genes to hybrid triploid of Chinese white poplar. Sci. Silvae Sin. 42, 61–67. doi: 10.3321/j.issn:1001-7488.2006.09.012
Yang, R. L., Wang, A. X., Zhang, J., Dong, Y., Yang, M. S., and Wang, J. M. (2016). Genetic transformation and expression of transgenic lines of Populus× euramericana with insect resistance and salt-tolerance genes. Genet. Mol. Res. 15: (10.4238). doi: 10.4238/gmr.15028635
Yang, X., Zhang, L., Yang, Y., Schmid, M., and Wang, Y. (2021). miRNA mediated regulation and interaction between plants and pathogens. Int. J. Mol. Sci. 22:2913. doi: 10.3390/ijms22062913
Ye, X., Busov, V., Zhao, N., Meilan, R., McDonnell, L. M., Coleman, H. D., et al. (2011). Transgenic Populus trees for forest products, bioenergy, and functional genomics. CRC. Crit. Rev. Plant Sci. 30, 415–434. doi: 10.1080/07352689.2011.605737
Ye, S., Jiang, Y., Duan, Y., Karim, A., Fan, D., Yang, L., et al. (2014). Constitutive expression of the poplar WRKY transcription factor PtoWRKY60 enhances resistance to Dothiorella gregaria Sacc. in transgenic plants. Tree Physiol. 34, 1118–1129. doi: 10.1093/treephys/tpu079
Yevtushenko, D. P., and Misra, S. (2019). Enhancing disease resistance in poplar through modification of its natural defense pathway. Plant Mol. Biol. 100, 481–494. doi: 10.1007/s11103-019-00874-2
Yin, Y., Wang, C., Xiao, D., Liang, Y., and Wang, Y. (2021). Advances and perspectives of transgenic technology and biotechnological application in forest trees. Front. Plant Sci. 12:786328. doi: 10.3389/fpls.2021.786328
Zegler, T. J., Moore, M. M., Fairweather, M. L., Ireland, K. B., and Fulé, P. Z. (2012). Populus tremuloides mortality near the southwestern edge of its range. For. Ecol. Manag. 282, 196–207. doi: 10.1016/j.foreco.2012.07.004
Zeng, Y., Song, H., Xia, L., Yang, L., and Zhang, S. (2023). The responses of poplars to fungal pathogens: a review of the defensive pathway. Front. Plant Sci. 14:1107583. doi: 10.3389/fpls.2023.1107583
Zhang, D., Das, D. B., and Rielly, C. D. (2014). Potential of microneedle-assisted microparticle delivery by gene guns: a review. Drug Deliv. 21, 571–587. doi: 10.3109/10717544.2013.864345
Zhang, L., Shang, C., Du, F. K., Zhao, F., Xiong, B., and Zhang, Z. (2017). Chloroplast phylogenomic analyses maternal relationships among sections in the genus Populus. Biochemical Systematics and Ecology 70, 132–140. doi: 10.1016/j.bse.2016.11.008
Zhang, B., Su, X., Li, Y., Zhang, Y., Qu, L., Wang, Y., et al. (2006). Production of Populus alba× P. glandulosa with a coleopterous insect resistant gene and analysis of insect resistance. J. Beijing For. Univ 28, 102–105. doi: 10.5555/20063075621
Zhang, Y., Waseem, M., Zeng, Z., Xu, J., Chen, C., Liu, Y., et al. (2022). MicroRNA482/2118, a miRNA superfamily essential for both disease resistance and plant development. New Phytol. 233, 2047–2057. doi: 10.1111/nph.17853
Zhang, L., Yang, X., Yin, Y., Wang, J., and Wang, Y. (2021). Identification and validation of miRNA reference genes in poplar under pathogen stress. Mol. Biol. Rep. 48, 3357–3366. doi: 10.1007/s11033-021-06369-y
Zhang, B., Zhu, W., Diao, S., Wu, X., Lu, J., Ding, C., et al. (2019). The poplar pangenome provides insights into the evolutionary history of the genus. Commun. Biol. 2:215. doi: 10.1038/s42003-019-0474-7
Zhang, G., Zou, C., and Wang, Z. (2005). Transformation system of chimeric gene for spider insecticidal peptide and Bt of Populus euramericana cv." 114/69". doi: 10.5555/20063102021
Zhao, Y., Zheng, X., Tabima, J. F., Zhu, S., Søndreli, K. L., Hundley, H., et al. (2023). Secreted effector proteins of poplar leaf spot and stem canker pathogen Sphaerulina musiva manipulate plant immunity and contribute to virulence in diverse ways. Mol. Plant-Microbe Interact. 36, 779–795. doi: 10.1094/MPMI-07-23-0091-R
Zhao, S. M., Zu, G. C., Liu, G. Q., Huang, M. R., Xu, J. X., and Sun, Y. R. (1999). Introduction of rabbit defensin NP-1 gene into poplar (P. tomentosa) by Agrobacterium mediated transformation. Yi Chuan Xue Bao 26, 711–714.
Zhou, H., Song, X., and Lu, M. Z. (2024). Growth-regulating factor 15-mediated vascular cambium differentiation positively regulates wood formation in hybrid poplar (Populus alba× P. glandulosa). Frontiers in Plant Science 15:1343312. doi: 10.3389/fpls.2024.1343312
Zhou, X., Dong, Y., Zhang, Q., Xiao, D., Yang, M., and Wang, J. (2020). Expression of multiple exogenous insect resistance and salt tolerance genes in Populus nigra L. Front. Plant Sci. 11:1123. doi: 10.3389/fpls.2020.01123
Zlatković, M., Tenorio-Baigorria, I., Lakatos, T., Tóth, T., Koltay, A., Pap, P., et al. (2020). Bacterial canker disease on Populus× euramericana caused by Lonsdalea populi in Serbia. Forests 11:1080. doi: 10.3390/f11101080
Zobrist, K., Haider, N., Stanton, B. J., and Stonex, R. (2023). Growing hybrid poplar for bioenergy in the PNW. Extension mimeo (Washington State University. Extension), vol. 123: Washington State University doi: 10.7273/000005545
Zubair, M., Wang, J., Yu, Y., Faisal, M., Qi, M., Shah, A. U., et al. (2022). Proteomics approaches: a review regarding an importance of proteome analyses in understanding the pathogens and diseases. Front. Vet. Sci. 9:1079359. doi: 10.3389/fvets.2022.1079359
Keywords: forest protection, genetic transformation, protoplast transformation, Agrobacterium-mediated transformation, salicylic acid (SA), methyl jasmonate (MeJA)
Citation: Sharan S, Chakraborty A, Roy A, Singh IK and Singh A (2024) Transgenic poplar for resistance against pest and pathogen attack in forests: an overview. Front. For. Glob. Change. 7:1490562. doi: 10.3389/ffgc.2024.1490562
Edited by:
Milica Zlatkovic, University of Novi Sad, SerbiaReviewed by:
Elena Corredoira, Spanish National Research Council (CSIC), SpainVijay Sheri, East Carolina University, United States
Alla Yemets, National Academy of Sciences of Ukraine (NAN Ukraine), Ukraine
Sivamani Elumalai, Syngenta Group, United States
Dulam Sandhya, Kakatiya University, India
Valentyna Meshkova, Ukrainian Research Institute of Forestry and Forest Melioration (URIFFM), Ukraine
Copyright © 2024 Sharan, Chakraborty, Roy, Singh and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Archana Singh, YXJjaGFuYXNpbmdoQHBtYi5kdS5hYy5pbg==; Indrakant K. Singh, aWtzaW5naEBkYi5kdS5hYy5pbg==; Amit Roy, cm95QGZsZC5jenUuY3o=
 Swati Sharan
Swati Sharan Amrita Chakraborty
Amrita Chakraborty Amit Roy
Amit Roy Indrakant K. Singh
Indrakant K. Singh Archana Singh
Archana Singh