
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. For. Glob. Change , 14 March 2024
Sec. Forest Growth
Volume 7 - 2024 | https://doi.org/10.3389/ffgc.2024.1320995
This article is part of the Research Topic Response of Plant Functional Traits to Environments and Their Roles in Ecosystem Processes View all 8 articles
 Lihui Ma1,2†
Lihui Ma1,2† Liubin Li3,4†
Liubin Li3,4† Wen Fang4
Wen Fang4 Zhi Dong4
Zhi Dong4 Yang Liu4
Yang Liu4 Chaoying Wang5
Chaoying Wang5 Yingzan Xie2
Yingzan Xie2 Mingyang Li1
Mingyang Li1 Muhammad Arif6*
Muhammad Arif6* Haiyang Wang1*
Haiyang Wang1*Genetic diversity in endangered species is of special significance in the face of escalating global climate change and alarming biodiversity declines. Rhododendron platypodum Diels, an endangered species endemic to China, is distinguished by its restricted geographical range. This study aimed to explore genetic diversity and differentiation among its populations, gathering samples from all four distribution sites: Jinfo Mountain (JFM), Zhaoyun Mountain (ZYM), Baima Mountain (BMM), and Mao’er Mountain (MEM). We employed 18 pairs of Simple Sequence Repeat (SSR) primers to ascertain the genetic diversity and structural characteristics of these samples and further utilized 19 phenotypic data points to corroborate the differentiation observed among the populations. These primers detected 52 alleles, with the average number of observed alleles (Na) being 2.89, the average number of effective alleles (Ne) being 2.12, the average observed heterozygosity (Ho) being 0.57, and the expected heterozygosity (He) being 0.50. This array of data demonstrates the efficacy of the primers in reflecting R. platypodum’s genetic diversity. SSR-based genetic analysis of the populations yielded Ho, He, and Shannon index (I) values ranging from 0.47 to 0.65, 0.36 to 0.46, and 0.53 to 0.69, respectively. Notably, the ZYM population emerged as the most genetically diverse. Further analysis, incorporating molecular variance, principal component analysis, UPGMA cluster analysis, and structure analysis, highlighted significant genetic differentiation between the Chongqing (BMM, JFM, ZYM) and Guangxi (MEM) populations. Morphological data analysis corroborated these findings. Additionally, marked genetic and morphological distinctions were evident among the three Chongqing populations (BMM, JFM, and ZYM). This suggests that, despite the observed regional differentiation, R. platypodum’s overall genetic diversity is relatively constrained compared to other species within the Rhododendron genus. Consequently, R. platypodum conservation hinges critically on preserving its genetic diversity and protecting its distinct populations.
Conserving endangered plant species is crucial to the survival of biological diversity and ecosystem stability (Torres-Díaz et al., 2021; Chen et al., 2022; Flores-Argüelles et al., 2022; Harrington et al., 2023; Singh et al., 2023). Rhododendron platypodum Diels is a rare and endangered plant species that has a limited distribution and is one of those species that requires urgent attention (Ma et al., 2021). Human activities continue to affect natural habitats (Arif et al., 2023; Boast et al., 2023) and understanding the genetic diversity and population differentiation of endangered plants such as R. platypodum becomes essential for effective conservation strategies. Scholars intended to provide an overview of the importance of genetic diversity (Planchuelo et al., 2020). Other researchers (Ma et al., 2021) have also delved into the ecological and conservation aspects of R. platypodum, with the primary objective of comprehending its genetic composition.
Adaptability, resilience, and the long-term survival of species are inextricably linked to genetic diversity within populations (Jiajia et al., 2023). Populations can withstand environmental changes, diseases, and other stressors if their gene pool is diverse. The evolution of new traits and adaptations is also facilitated by genetic variation (Ye et al., 2020). Loss of genetic diversity can lead to species being less able to adapt to changes in their environment, making them more vulnerable to extinction consequently (Johnson and Molano-Flores, 2023; Naz et al., 2024). Therefore, assessing the genetic diversity of endangered species is essential for developing effective conservation measures. R. platypodum is commonly known as the Flat Pod Rhododendron and is an endemic species that is found only in certain regions of the world (Ma et al., 2021). It has distinct ecological characteristics. Human disturbances such as habitat fragmentation, deforestation, and land development are often the cause of it thriving in disturbed habitats (He et al., 2023). It is particularly vulnerable to the impacts of these anthropogenic activities due to its limited distribution and habitat requirements (Zheng et al., 2023). It is well known for its ornamental value, but habitat loss and overexploitation have threatened the species’ survival.
It is estimated that there are approximately 850 Rhododendron species in the world, most of which grow in high-altitude subtropical areas. In China, Rhododendron forms distribution centers in Yunnan, Sichuan, and Xizang provinces (Wang et al., 2017; Yu et al., 2017). Rhododendron plants are recognized as the queen of flowers because of their large flowers, unique shapes, and vibrant colors. This makes them valuable for conservation and development (Mao and Gogoi, 2012; Shen et al., 2015). Genetic diversity can be effectively conserved for endangered plants by understanding its distribution within and between populations (Kwiecinska-Poppe et al., 2020; Zizhuo et al., 2022). The diversity of morphological traits can be used to reveal variations in phenotypic traits among populations within a distribution area. This is an important aspect of genetic diversity research (Zhao et al., 2012). The use of simple sequence repeat (SSR) markers for analyzing genetic diversity can provide more direct evidence. The genetic diversity of several Rhododendron species has been studied so far, including R. vialii (Yang et al., 2020), R. molle (Zhang et al., 2021), and R. dauricum (DePristo et al., 2011).
R. platypodum is an evergreen shrub. The plant grows on rocks or in dense forests between the elevations of 1820 and 2,130 m above sea level (Hong, 1983). This species was previously listed as vulnerable on the Red List of Rhododendrons (Gibbs et al., 2011) and the International Union for Conservation of Nature (IUCN) (Ma et al., 2021). The species R. platypodum has been classified as endangered after a reassessment of its endangerment (Zizhuo et al., 2022). R. platypodum has very limited resources, insufficient competitiveness, a narrow distribution range, and weak renewable resources (Huang et al., 2020; Zizhuo et al., 2022). At present, this species is only known to occur on Jinfo Mountain (JFM), Zhaoyun Mountain (ZYM), and Baima Mountain (BMM) in Chongqing, and Maoer Mountain (MEM) in Guangxi, China. Research has previously characterized the MEM population in terms of conservation, but the population is now uniformly classified as having R. Platypodum in it (Gibbs et al., 2011). Nevertheless, high-intensity human disturbance has adversely affected Rhododendron survival and reproduction, indicating the need for immediate protection and research.
Therefore, we selected four populations of R. platypodum in this study from JFM, ZYM, and BMM in Chongqing and MEM in Guangxi. Furthermore, morphological characteristics and SSR markers were analyzed to determine genetic diversity and population phenotypic structure. In addition, a comparative analysis of populations was carried out simultaneously. This study assesses the genetic diversity and population differentiation of R. platypodum to inform conservation strategies for its long-term survival. Specifically, the study aims to: (Chen et al., 2022) evaluate genetic diversity within and among R. platypodum populations using molecular markers; (Torres-Díaz et al., 2021) investigate the patterns of population differentiation and gene flow between different populations of R. platypodum; and (Harrington et al., 2023) identify potential genetic bottlenecks or inbreeding effects that might limit the species’ genetic diversity. Conservation strategies for endangered species such as R. platypodum require understanding genetic diversity and population differentiation. It is vitally needed to understand the genetic makeup of this species to preserve its genetic variability, enhance its resilience, and ultimately prevent its extinction. This study provides valuable insights into the phylogenetic status of R. platypodum and provides a foundation for future conservation action.
The collection of materials was systematically executed across all R. platypodum natural distribution sites, including JFM, ZYM, BMM, and MEM, from April to May 2020 (Figure 1). Each distribution site underwent meticulous sampling, adhering to a minimum distance criterion of 15 meters. This distance was determined based on the spatial distribution of individual plants within the respective sites. Due to the inherent challenges posed by the limited population of R. platypodum and the arduous accessibility associated with plants growing on cliff rock walls, a total of 61 samples were acquired from these sites, with each site ranging between 8 and 20, as delineated in Table 1. The selection process for molecular analysis necessitated the procurement of three to five leaves from healthy adult plants. These leaves were meticulously inspected to ensure pest and disease freedom. These collected leaves were subsequently preserved using silica gel after being thoroughly dried. In tandem with the molecular analysis, morphological assessment required the collection of branches and inflorescences. These botanical components were promptly sealed within zip-lock bags and stored within an icebox, ensuring their preservation and integrity for subsequent measurements.
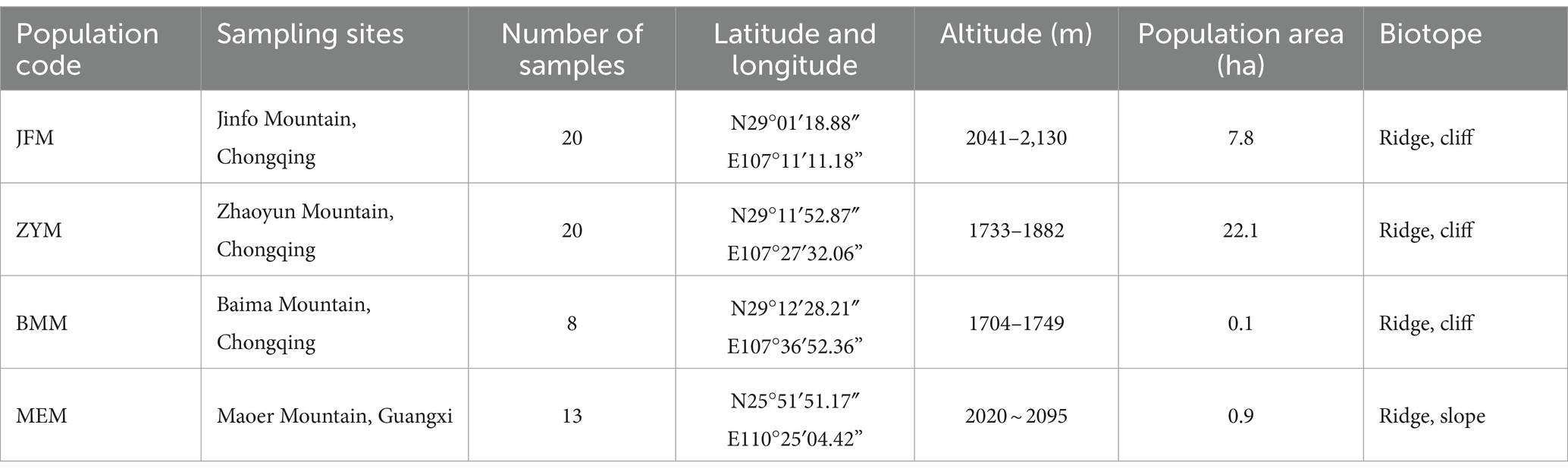
Table 1. The sampling information for this study includes the sampling site location, population number, population distribution range, and biotope.
The total DNA from each sample was extracted using the CTAB method (Jinlu et al., 2013), and the concentration of each sample was determined using Nanodrop. OD260/280 ratios were used to determine the quality of DNA.
In accordance with the published SSR study on Rhododendron sect. Ponticum G. Don (Jinlu et al., 2013; Kaiyun et al., 2020), 68 pairs of SSR primers were synthesized, and 18 pairs were selected for the experiment (Table 2). We used a reaction volume of 10 μL for the PCR amplification. As part of this system, 0.2 μL DNA template, 0.1 μL Taq enzyme, 0.2 μL dNTP mixture, and 0.5 μL of forward and reverse primers were used. We carried out the following reaction procedures: pre-denaturation at 94°C for 5 min, denaturation at 94°C for 30 s, annealing at 60°C for 30 s, extension at 72°C for 100 s for 35 cycles, and finally extension at 72°C for 10 min. Electrophoresis of PCR products was performed on an 8% polyacrylamide gel for 30–40 min at 350 V. The gel was stained with a 2% silver nitrate solution for 10–15 min before being photographed.
PCR amplification and genotyping of all samples were performed according to the steps, using SSR primers for PCR amplification and development. All samples produced effective PCR bands. Utilizing the software tools POPGENE 32 (Yeh et al., 1997) and GenAIEx ver. 6.5 (Peakall and Smouse, 2006), an array of crucial genetic parameters were computed, encompassing the count of observed alleles (Na), the tally of effective alleles (Ne), observed heterozygosity (Ho), expected heterozygosity (He), the Shannon index (I), the inbreeding coefficient (Fis), and Nei’s genetic distance. Additionally, the polymorphic information content (PIC) was ascertained using Cervus 3.0.7, and the assessment of adherence to Hardy–Weinberg equilibrium (HWE) was conducted (Slate et al., 2000). This multifaceted analysis contributes to a comprehensive exploration of the genetic dynamics under consideration. The genetic differentiation coefficient (Fst) and gene flow (Nm) were calculated using GenAIEx version 6.0 (Peakall and Smouse, 2006). Analysis of molecular variance (AMOVA), principal component analysis (PCA), and the Mantel test were also conducted using this software. The UPGMA clustering trees were drawn with MEGA using Nei’s genetic distance value (Tamura et al., 2021). We used the Bayesian clustering method in structure (Gilbert et al., 2012) to analyze R. platypodum’s genetic structure. We adopted an independent allele frequency mixing model. The number of classes (K) was set to 1–10, the length of the burn-in period and MCMC were both set to 100,000, and each K value was run ten times.
Plant phenotypic characteristics encompass the external manifestations of their underlying genetic traits, which serve as crucial indicators in the examination of genetic differentiation. In line with the morphological classification criteria pertinent to the genus Rhododendron, a total of 19 morphological indices were meticulously selected for evaluation. These encompassed 6 indices related to leaf morphology, 2 indices related to inflorescence characteristics, and 11 indices pertaining to flower morphology. To amass comprehensive leaf morphological data, each sampled individual was tasked with measuring a minimum of 15 leaves derived from 5 distinct branches. In parallel, the assessment of inflorescence morphology required the measurement of a minimum of 5 inflorescences, while flower morphology data necessitated the measurement of at least 5 flowers from more than 5 distinct inflorescences. Statistical analysis was conducted utilizing univariate covariance analysis (ANOVA) through the use of SPSS 20.0. Multiple comparisons were subsequently performed employing the Tukey method to discern significant differences among various populations. Finally, PCA was employed as an instrumental tool to ascertain phenotypic differentiation across diverse population groups.
We employed 18 SSR primer pairs to discern a total of 52 alleles within the cohort of 61 samples. Among these alleles, there was Na of 2.89, Ne of 2.12 [range = 1.07 (A67)-2.76 (A20)], Ho of 0.57 [range = 0.07 (A67)-1.00 (A20)], and He of 0.50 [range = 0.06 (A67)-0.64 (A20)]. The PIC range was 0.06 to 0.57. Overall, seven loci (A19, A20, A26, A40, A53, A58, and A64) displayed high polymorphism (PIC >0.5), while two loci (A67 and A13) showed low polymorphism (PIC <0.25). In addition, other loci displayed medium polymorphism (0.25 < PIC <0.5). Out of these 18 loci, 7 showed consistency with HWE (Table 3).
Across all populations, Na was higher than Ne. In particular, Na and Ne were maximized for the JFM and ZYM populations, respectively (Table 4). The values of Ho, He, and I for the different populations studied ranged from 0.47 to 0.65, 0.36 to 0.46, and 0.52 to 0.69, respectively. All three parameters were maximized in the ZYM population. Furthermore, it is noteworthy that the Fis value exhibited consistent negative trend across all populations under study. This observation signifies the persistence of a high level of heterozygosity within each population. This implies that the individuals comprising these populations do not stem from inbreeding processes.
Genetic variation among populations using SSR-based AMOVA was 14% and among individuals within populations was 86% (Table 5). As indicated by the results of interpopulation Fst and gene flow (Nm) (Table 6), the genetic differentiation coefficients between the four populations ranged from 0.05 to 0.19, while gene flow ranged from 1.10 to 5.13. JFM and ZYM populations displayed the lowest differentiation coefficients and the highest frequency of gene exchanges. There was the greatest differentiation coefficient among the BMM and MEM populations, with the least amount of gene exchange.
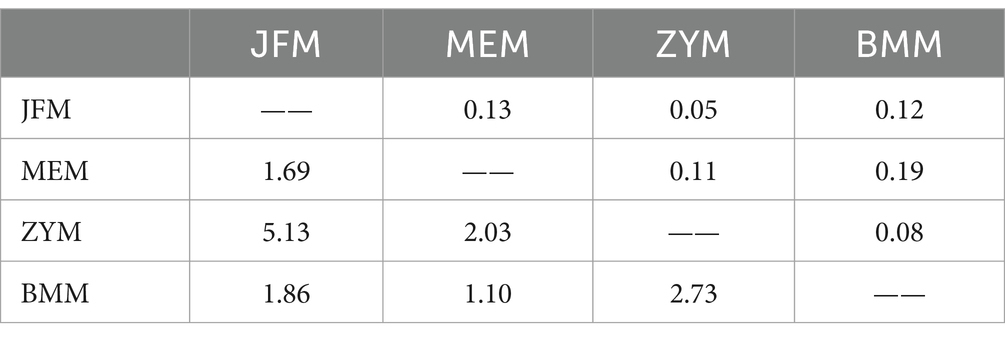
Table 6. Genetic differentiation (Fst, above diagonal) and gene flow (Nm, below diagonal) of four R. platypodum populations.
PCA results determined that the first and second PCA axes accounted for 23.54 and 14.59% of the total variation, respectively (Figure 2). On the first principal axis, the MEM population was separated from the JFM, ZYM, and BMM populations. On the second principal axis, the three populations from Chongqing appeared separated to some extent. MEM and JFM, ZYM, and BMM populations were distinguished by UPGMA clustering trees (Figure 3). The JFM population from Chongqing first gathered with the ZYM population before gathering with the BMM population. In all the populations tested by the Mantel test, genetic distance positively correlated with geographic distance (R2 = 0.606, p = 0.068) (Figure 4). This suggests that geographical isolation contributes to variation among populations.
The structure analysis revealed that if K = 2, ∆K becomes the largest, suggesting that “2” is the optimal number of clusters (Figure 5). It is evident that the populations of Chongqing and Guangxi are distinct when K = 2. When K = 3, the JFM and BMM populations are separated, and the genetic components of the ZYM population show heterozygosity with those of the JFM and BMM populations. The four populations are all separated when K = 4, despite the presence of a small amount of genetic heterozygosity between them.
Figures 6, 7 depict the examination of genetic differentiation, gene flow, and phenotypic traits within R. platypodum populations, which is further elaborated in Table 7. In the JFM population, leaf length and width were significantly wider than those in the other populations (p < 0.01). Petiole lengths in the JFM population were significantly longer than those in the ZYM population (p < 0.01) but were similar in the BMM and MEM populations. The petiole width (p < 0.01) was consistent across different populations, although it was maximized in the JFM population and minimized in the MEM population. The aspect ratio of the petioles of the MEM population was considerably greater than that of other populations (p < 0.01). MEM populations exhibited the most lateral veins, whereas ZYM populations exhibited the fewest lateral veins. In the MEM population, traits such as floral part size, flower number, corolla length, corolla lobe depth, style length, and radium of the stigma were significantly smaller than those in the other populations (p < 0.01). Compared to the other populations, the MEM population had a significantly longer pedicel, calyx, and radium of the ovary (p < 0.05).
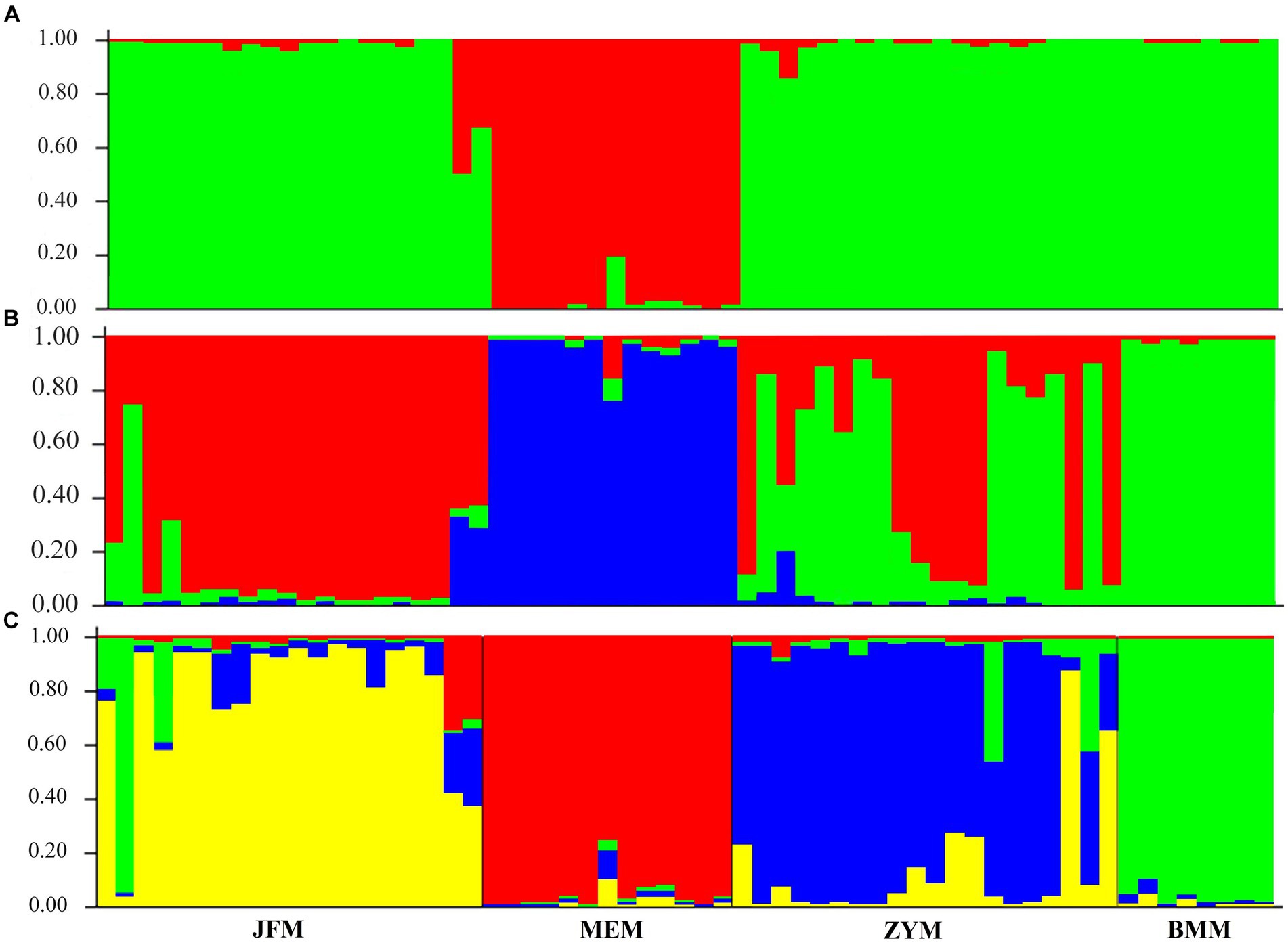
Figure 6. Genetic structure for different K values with the STRUCTURE program for 4 populations of R. platypodum. (A) K = 2, (B) K = 3, (C) K = 4.
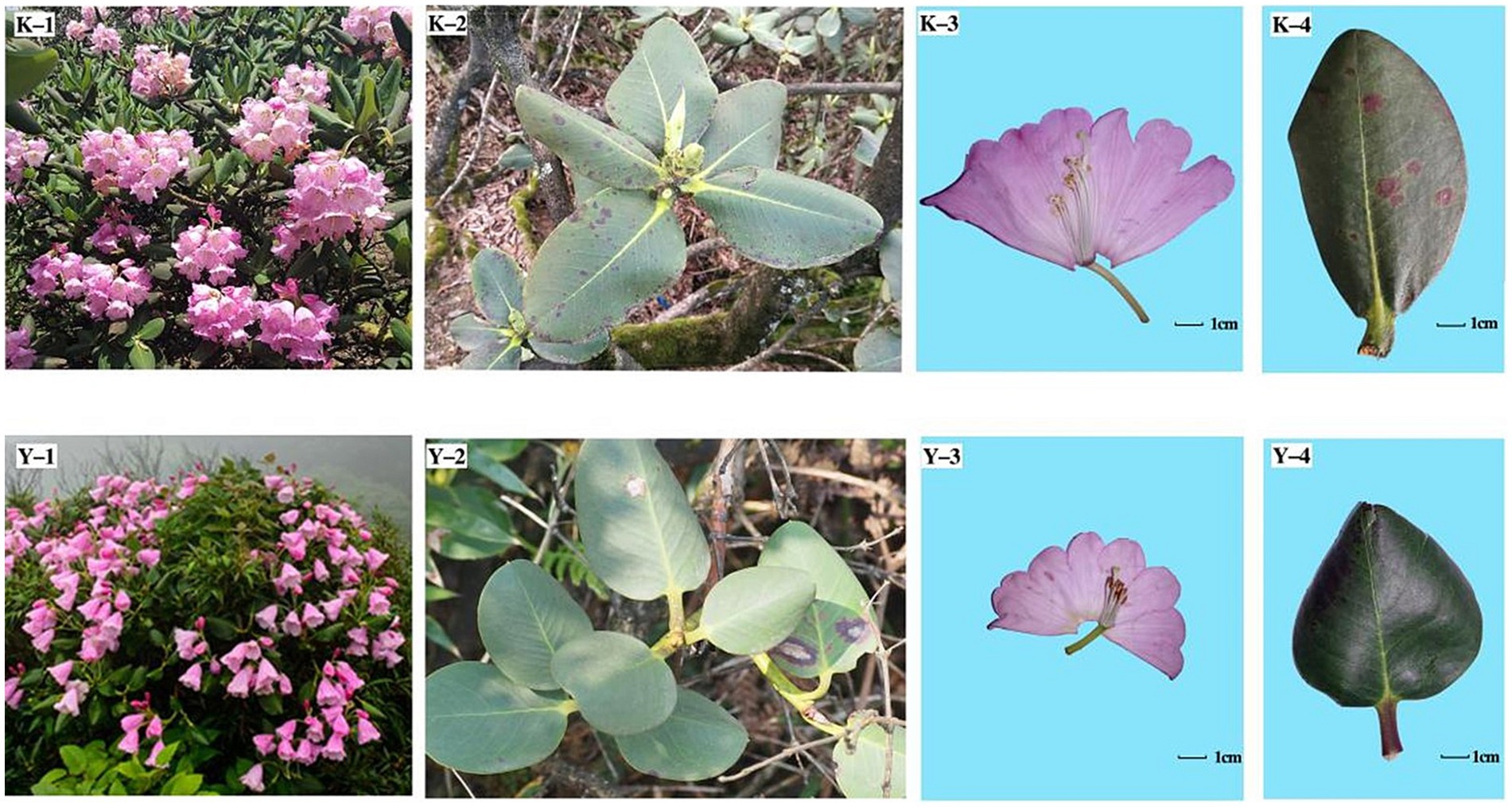
Figure 7. Phenotypes of the ZYM and MEM populations. K-1, K-2, K-3, and K-4 indicate the inflorescence, phyllo-taxy, and flower and leaf morphologies of R. platypodum (ZYM), respectively; Y-1, Y-2, Y-3, and Y-4 indicate the inflorescence, phyllo-taxy, and flower and leaf morphologies of R. platypodum (MEM), respectively.
PCA performed using the phenotypic data revealed that the JFM, ZYM, and BMM populations are highly similar, indicating that they are significantly different from the MEM population (Figure 8).
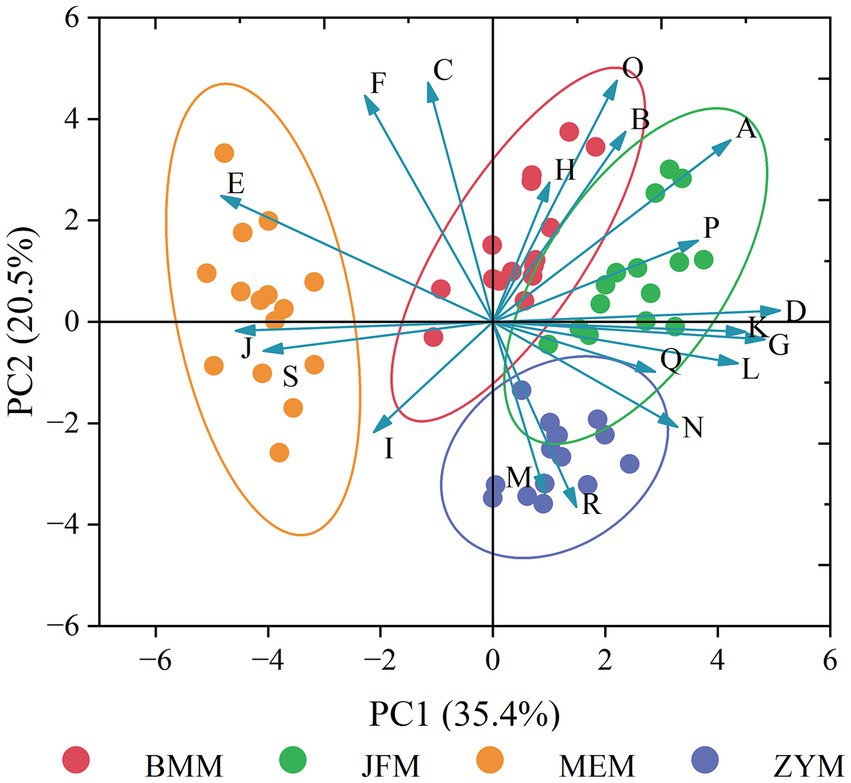
Figure 8. PCA map based on quantifiable phenotypic traits (A-S presents the differentiated quantifiable phenotypic traits: A: length of leaf; B: width of leaf; C: length of petiole; D: width of petiole; E: aspect ratio of petiole; F: number of lateral veins; G: flower number of inflorescence; H: length of inflorescence rachis; I: length of pedicel; J: length of calyx; K: length of corolla; L: depth of corolla lobe; M: number of stamen; N: length of anther; O: length of filament; P: length of style; Q: length of stigma; R: length of ovary; S: length of ovary).
In comparison with the other R. platypodum populations, the ZYM population exhibits the highest genetic diversity (Table 4). In contrast, the BMM population exhibits the lowest genetic diversity. Genetic diversity was similar between the JFM and MEM populations. ZYM plants are widely distributed and contain many plants (Table 1). This population of plants may be characterized by rich genetic diversity due to the conditions under which genetic information is exchanged between individuals. Although the BMM population has a small size and a narrow distribution, its low genetic diversity might be due to its small size and narrow distribution.
He of the population samples in this study ranged from 0.35 to 0.41 (average = 0.41). Similarly, R. platypodum had a lower Ne than R. protistum (He = 0.60) (Ma et al., 2013), R. simsii (He = 0.75) (Li et al., 2022), R. ripense (He = 0.80) (Tan et al., 2009), and R. dauricum (He = 0.57) (DePristo et al., 2011). Comparative species belong to the same genus as R. platypodum. However, the Ne of R. platypodum was significantly higher than that of R. rufohirtum (He = 0.28) (Kondo et al., 2009). The He values of cosmopolitan and stenochoric species were 0.62 and 0.42, respectively (Hsieh et al., 2013), and R. platypodum was found to be genetically similar to stenochoric species.
The Fis of all populations involved was negative, indicating no inbreeding. In other species of the genus, like Rhododendron longipedicellatum (Li et al., 2018), R. protistum (Zizhuo et al., 2022), R. rex (Li et al., 2022), R. dauricum (DePristo et al., 2011), and R. molle (Zhang et al., 2021), the situation was similar. This may be attributed to Rhododendron growth and breeding habits (Keller and Waller, 2002; Stoeckel et al., 2006). Rhododendron plants are perennial woody plants that have a long lifespan. R. platypodum exhibits mainly outbreeding and partial self-breeding (Ma et al., 2021), which suggests that the current populations are probably descended from ancestral populations with a rich genetic history.
Various levels of genetic variation have been identified within and among R. platypodum populations. The genetic diversity of populations inhabiting relatively undisturbed and less fragmented habitats may exhibit a markedly higher degree when compared to populations residing in fragmented or degraded environments. This observation aligns seamlessly with the prevailing theory positing that habitat fragmentation tends to curtail gene flow while concurrently elevating genetic isolation (Hira et al., 2023; Jiajia et al., 2023). This ultimately results in a decrease in genetic diversity within small and isolated populations. Prioritizing conservation efforts requires identifying populations with high genetic diversity, since these populations can serve as sources of genetic material for the recovery of species (Garza et al., 2020).
Plant populations exhibit a comprehensive genetic structure that reflects their evolutionary history. Therefore, it is imperative to formulate conservation strategies for endangered species based on these data. AMOVA based on SSR revealed that 86% of the total genetic variation was accounted for by intra-population genetic variation, whereas 14% was accounted for by inter-population genetic variation; these findings align with other studies (Zhao et al., 2012; Shen et al., 2015; Yu et al., 2017; Yang et al., 2020; Ma et al., 2021; Zhang et al., 2021). Moreover, our study’s K = 2 structure analysis in genetic differentiation (Figure 5) sheds light on systematic relationships in the Rhododendron genus, as previously reported by other researchers (Milne, 2004; Wilson, 2016).
Fst provides important information about population genetic structure. In this study, the three Chongqing populations and the Guangxi population exhibited Fst within a range of 0.11–0.19 (Table 6), indicating that there was significant genetic differentiation between the Chongqing population and the Guangxi population. Further confirmation of this differentiation was achieved through PCA and UPGMA cluster analysis. Structure analysis revealed significant differences between the Chongqing and Guangxi populations. Similar results were found in the morphological analysis of the data. This indicated that the Guangxi population had shorter leaves, narrower petioles, and smaller inflorescences and flowers. This is also one of the reasons for the controversy surrounding the MEM population classification.
All three Chongqing populations showed Fst values between 0.05 and 0.12, which indicates only a small to moderate degree of genetic differentiation. It was found that when K = 3, the genetic components of the ZYM population were mixed with those of the JFM and BMM populations. It is evident that they differ morphologically in terms of leaf length, leaf width, and ovary length (Table 7; Figure 7).
R. platypodum population differentiation patterns provide insight into connectivity between different populations of this species. Keeping populations healthy and preventing inbreeding requires an understanding of gene flow and barriers to gene exchange. A high degree of genetic differentiation between populations may indicate limited gene flow. This may be caused by geographical barriers, habitat fragmentation, or both (Zheng et al., 2022; Arif et al., 2024). The identification of restricted gene flow areas can assist in implementing strategies to enhance connectivity between populations, such as the creation of habitat corridors and the promotion of natural pollination vectors.
R. platypodum, with its limited resource base and confined distribution area, presents an unusual ecological context. This study reveals substantial differentiation between the Guangxi and Chongqing populations of the broad-stalked R. platypodum, along with discernible genetic and morphological variations among the three populations in Chongqing. These revelations underscore the pivotal significance of each population in preserving the genetic diversity and structural integrity of the wide-stalked R. platypodum. Presently, the JFM, BMM, and MEM populations are all situated within designated nature reserves. JFM and MEM have received particular attention as priority hotspots for safeguarding Rhododendron plant diversity in China, as evidenced by previous studies (Zhao et al., 2012; Shen et al., 2015; Yu et al., 2017; Yang et al., 2020; Ma et al., 2021; Zhang et al., 2021). Consequently, they have been subjected to increased scrutiny and more comprehensive protection measures. Notably, the ZYM population stands out with its substantial number of individuals, abundant seedlings, and extensive geographical spread. These factors collectively contribute to the highest genetic diversity observed. This population occupies a central role in the overall genetic structure of R. platypodum and is therefore endowed with the utmost conservation value. Nevertheless, it is imperative to acknowledge that, despite its ecological significance, the ZYM population currently experiences a gap in protection efforts. Therefore, it is imperative to enhance and fortify conservation measures to ensure the enduring survival and prosperity of this vital genetic reservoir.
This study has direct implications for R. platypodum conservation. The survival of the species depends on a multifaceted approach (Tsukamoto et al., 2021; Liu et al., 2022; Abbasi et al., 2023; Cao et al., 2023; Rauf et al., 2023; Naz et al., 2024): (Chen et al., 2022) Restoration efforts should aim to improve the quality and connectivity of habitats to facilitate healthy populations and gene flow (Torres-Díaz et al., 2021). R. platypodum is a valuable species in the ecosystem, and conservation efforts can be bolstered by raising awareness of its importance and role within it (Harrington et al., 2023). Protecting the habitats of the species and enforcing regulations against illegal collection and destruction of those habitats is vital (Singh et al., 2023). Genetic monitoring and research must occur continuously in order to determine the extent of changes in genetic diversity and adjust conservation strategies as necessary.
The present study employed a set of 18 SSR primers to examine 61 samples drawn from four distinct populations located in Chongqing and Guangxi. It was determined that 52 alleles were present in the comprehensive analysis, yielding an average PIC value of 0.41. R. platypodum exhibits very low genetic diversity in comparison to most Rhododendron species documented, and it closely resembles the patterns of endemic species. Corroborating the genetic analysis, the outcomes of PCA, UPGMA cluster analysis, and structure analysis consistently delineated the separation of R. platypodum into two discrete groups: the Chongqing group and the Guangxi group. Importantly, these groups exhibited substantial genetic differentiation between them. Furthermore, the phenotypic data analysis, comprising 19 distinct categories, aligned harmoniously with the genetic structure. Notably, the Chongqing and Guangxi groups displayed noteworthy disparities in various traits, encompassing leaf length, petiole width, inflorescence size, and flower size. Moreover, within the Chongqing group, significant distinctions were observed among different populations, particularly in terms of leaf length, width, and ovary size. The comprehensive findings of this study hold significant implications for R. platypodum conservation. It underscores the necessity of intensifying protection efforts, particularly in the case of the Zhaoyun Mountain population, which has the highest genetic diversity among the populations under scrutiny. This study thus serves as a valuable reference point for future conservation strategies. It reinforces the urgency of safeguarding genetic diversity within the Zhaoyun Mountain population.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
LM: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. LL: Conceptualization, Data curation, Formal analysis, Methodology, Software, Supervision, Validation, Writing – original draft, Writing – review & editing. WF: Investigation, Methodology, Visualization, Writing – original draft. ZD: Investigation, Software, Visualization, Writing – original draft. YL: Investigation, Validation, Visualization, Writing – original draft. CW: Formal Analysis, Investigation, Project administration, Writing – original draft. YX: Investigation, Project administration, Writing – original draft. ML: Validation, Visualization, Writing – review & editing. MA: Conceptualization, Data curation, Formal Analysis, Methodology, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. HW: Conceptualization, Data curation, Methodology, Resources, Supervision, Validation, Visualization, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by Chongqing Invigorating Forestry Projects through Science and Technology, grant number YLKY2018-14, and the Scientific and Technological Innovation Special Program of Social Livelihood of Chongqing, grant number CSTC2019-lykjfpxz002.
The authors would like to express their gratitude to the staff of the Senmiao Botanical Garden in Yinchuan City in China for their valuable assistance with the experimental sample.
LL was employed by China Forestry Group Co., Ltd. LL, WF, ZD, and YL were employed by Chongqing Forestry Investment Development Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbasi, U. A., Gilani, H., Hussain, K., and Ali, A. (2023). Increasing stand stature weakens the positive effects of tree richness and structural imbalance on aboveground biomass in temperate forests: the stand stature hypothesis. For. Ecol. Manag. 539:121040. doi: 10.1016/j.foreco.2023.121040
Arif, M., Jie, Z., Behzad, H. M., and Changxiao, L. (2023). Assessing the impacts of ecotourism activities on riparian health indicators along the three gorges reservoir in China. Ecol. Indic. 155:111065. doi: 10.1016/j.ecolind.2023.111065
Arif, M., Petrosillo, I., and Changxiao, L. (2024). Effects of changing riparian topography on the decline of ecological indicators along the drawdown zones of long rivers in China. Front. For. Glob. Change 7:1293330. doi: 10.3389/ffgc.2024.1293330
Boast, A. P., Wood, J. R., Bolstridge, N., Perry, G. L. W., and Wilmshurst, J. M. (2023). Ancient and modern scats record broken ecological interactions and a decline in dietary breadth of the critically endangered kākāpō parrot (Strigops habroptilus). Front. Ecol. Evol. 11:1058130. doi: 10.3389/fevo.2023.1058130
Cao, W., Muhammad, A., Cui, Z., and Li, C. (2023). Effects of different trace elements on ecophysiological characteristics of Ligustrum obtusifolium saplings. Forests 14:972. doi: 10.3390/f14050972
Chen, Y., Zhang, H.-L., Zhang, L., Nizamani, M. M., Zhou, T., Zhang, H., et al. (2022). Genetic diversity assessment of Hopea hainanensis in Hainan Island. Front. Plant Sci. 13:1075102. doi: 10.3389/fpls.2022.1075102
DePristo, M. A., Banks, E., Poplin, R., Garimella, K. V., Maguire, J. R., Hartl, C., et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498. doi: 10.1038/ng.806
Flores-Argüelles, A., Espejo-Serna, A., López-Ferrari, A. R., and Krömer, T. (2022). Diversity and vertical distribution of epiphytic angiosperms, in natural and disturbed forest on the northern coast of Jalisco, Mexico. Front. For. Glob. Change 5:828851. doi: 10.3389/ffgc.2022.828851
Garza, G., Rivera, A., Barrera, C. S. V., Martinez-Avalos, J. G., Dale, J., and Arroyo, T. P. F. (2020). Potential effects of climate change on the geographic distribution of the endangered plant species Manihot walkerae. Forests 11:689. doi: 10.3390/f11060689
Gibbs, D., Chamberlain, D., and Argent, G. The RED LIST OF rhododendrons. Botanic Gardens Conservation International, Richmond, UK, (2011).
Gilbert, K. J., Andrew, R. L., Bock, D. G., Franklin, M. T., Kane, N. C., Moore, J. S., et al. (2012). Recommendations for utilizing and reporting population genetic analyses: the reproducibility of genetic clustering using the program STRUCTURE. Mol. Ecol. 21, 4925–4930. doi: 10.1111/j.1365-294X.2012.05754.x
Harrington, C. A., Gould, P. J., and Cronn, R. (2023). Site and provenance interact to influence seasonal diameter growth of Pseudotsuga menziesii. Front. For. Glob. Change 6:1173707. doi: 10.3389/ffgc.2023.1173707
He, X., Muhammad, A., Zheng, J., Ni, X., Yuan, Z., Zhu, Q., et al. (2023). Plant diversity patterns along an elevation gradient: the relative impact of environmental and spatial variation on plant diversity and assembly in arid and semi-arid regions. Front. Environ. Sci. 11:1021157. doi: 10.3389/fenvs.2023.1021157
Hira, A., Arif, M., Zarif, N., Gul, Z., Xiangyue, L., and Yukun, C. (2023). Effects of riparian buffer and stream channel widths on ecological indicators in the upper and lower Indus River basins in Pakistan. Front. Environ. Sci. 11:1113482. doi: 10.3389/fenvs.2023.1113482
Hsieh, Y. C., Chung, J. D., Wang, C. N., Chang, C. T., Chen, C. Y., and Hwang, S. Y. (2013). Historical connectivity, contemporary isolation and local adaptation in a widespread but discontinuously distributed species endemic to Taiwan, Rhododendron oldhamii (Ericaceae). Heredity 111, 147–156. doi: 10.1038/hdy.2013.31
Huang, M., Dai, X., Yang, C., Deng, L., Yuan, C., Li, H., et al. (2020). Characteristics and diversity of typical Rhododeudrou platypodum communities in Guizhou. J. West China For. Sci. 49, 113–120. doi: 10.16473/j.cnki.xblykx1972.2020.02.017
Jiajia, L., Lijuan, L., Muhammad, A., Dongdong, D., Xin, H., and Changxiao, L. (2023). Newly formed riparian microhabitats simplify bacterial community structure and diversity. J. Soils Sediments 23, 1927–1943. doi: 10.1007/s11368-023-03454-6
Jiajia, L., Muhammad, A., Dongdong, D., Xin, H., Qianwen, G., Fan, Y., et al. (2023). The diversity of plant communities in different habitats can lead to distinct methanotrophic communities. Rhizosphere 26:100690. doi: 10.1016/j.rhisph.2023.100690
Jinlu, L., Shuo, W., Jing, Y., Ling, W., and Shiliang, Z. (2013). A modified CTAB protocol for plant DNA extraction Chinese bulletin of botany. Chin. Bull. Bot. 48, 72–78. doi: 10.3724/SP.J.1259.2013.00072
Johnson, S. A., and Molano-Flores, B. (2023). Is the endangered species act living to its full potential? The reassessment of the conservation status and recovery of Macbridea alba Chapm. As a case study. Front. Conserv. Sci. 4:1116848. doi: 10.3389/fcosc.2023.1116848
Kaiyun, C., Qiao, L., Yu, W., Donghui, L., and Jingli, Z. (2020). Genetic diversity analysis of Rhododendron vialii based on microsatellite marker. J. Yunnan Univ. Nat. Sci. 35, 818–825. doi: 10.12101/j.issn.1004-390X(n).201805029
Keller, L. F., and Waller, D. M. (2002). Inbreeding effects in wild populations. Trends Ecol. Evol. 17, 230–241. doi: 10.1016/s0169-5347(02)02489-8
Kondo, T., Nakagoshi, N., and Isagi, Y. (2009). Shaping of genetic structure along pleistocene and modern river systems in the hydrochorous riparian azalea, Rhododendron ripense (Ericaceae). Am. J. Bot. 96, 1532–1543. doi: 10.3732/ajb.0800273
Kwiecinska-Poppe, E., Haliniarz, M., Sowa, S., and Paczos-Grzeda, E. (2020). Genetic diversity and population structure of endangered plant species Anagallis foemina mill. Lysimachia foemina (mill.) U. Manns & Anderb. Physiol. Mol. Biol. Plants 26, 1675–1683. doi: 10.1007/s12298-020-00839-6
Li, T., Liu, X., Li, Z., Ma, H., Wan, Y., Liu, X., et al. (2018). Study on reproductive biology of Rhododendron longipedicellatum: a newly discovered and special threatened plant surviving in limestone habitat in Southeast Yunnan, China. Front. Plant Sci. 9:33. doi: 10.3389/fpls.2018.00033
Li, J., Yang, H., Liu, H., Li, C., and Zhou, Y. (2022). Soil nutrients and microbial community characteristics of Rhododendron rex in Central Yunnan province of China. J. Biobaased Mater. Bioenergy 16, 431–441. doi: 10.1166/jbmb.2022.2204
Liu, X., Arif, M., Wan, Z., and Zhu, Z. (2022). Dynamic evaluation of coupling and coordinating development of environments and economic development in key state-owned forests in Heilongjiang province, China. Forests 13:2069. doi: 10.3390/f13122069
Ma, Y., Chen, G., Edward, G. R., Dao, Z., Sun, W., and Guo, H. (2013). Conserving plant species with extremely small populations (PSESP) in China. Biodivers. Conserv. 22, 803–809. doi: 10.1007/s10531-013-0434-3
Ma, L. H., Zhu, H. X., Wang, C. Y., Li, M. Y., and Wang, H. Y. (2021). The complete chloroplast genome of Rhododendron platypodum (Ericaceae): an endemic and endangered species from China. Mitochondrial DNA Part B-Resour. 6, 196–197. doi: 10.1080/23802359.2020.1860714
Milne, R. I. (2004). Phylogeny and biogeography of Rhododendron subsection Pontica, a group with a tertiary relict distribution. Mol. Phylogenet. Evol. 33, 389–401. doi: 10.1016/j.ympev.2004.06.009
Naz, F., Arif, M., Xue, T., and Changxiao, L. (2024). Artificially remediated plants impact soil physiochemical properties along the riparian zones of the three gorges dam in China. Front. For. Glob. Change 7:1301086. doi: 10.3389/ffgc.2024.1301086
Peakall, R., and Smouse, P. E. (2006). GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295. doi: 10.1111/j.1471-8286.2005.01155.x
Planchuelo, G., Kowari, I., and von der Lippe, M. (2020). Endangered plants in novel urban ecosystems are filtered by strategy type and dispersal syndrome, not by spatial dependence on natural remnants. Front. Ecol. Evol. 8:18. doi: 10.3389/fevo.2020.00018
Rauf, Z., Zarif, N., Khan, A., Siddiqui, S., Fatima, S., Iqbal, W., et al. (2023). The western Himalayan fir tree ring record of soil moisture in Pakistan since 1855. Int. J. Biometeorol. 67, 1477–1492. doi: 10.1007/s00484-023-02517-0
Shen, S. K., Wu, F. Q., Yang, G. S., Wang, Y. H., and Sun, W. B. (2015). Seed germination and seedling emergence in the extremely endangered species Rhododendron protistum var. giganteum-the world's largest Rhododendron. Flora 216, 65–70. doi: 10.1016/j.flora.2015.08.006
Singh, R. A., Cheesman, A. W., Flores-Moreno, H., Preece, N. D., Crayn, D. M., and Cernusak, L. A. (2023). Temperature, nutrient availability, and species traits interact to shape elevation responses of Australian tropical trees. Front. For. Glob. Change 6:1089167. doi: 10.3389/ffgc.2023.1089167
Slate, J., Marshall, T., and Pemberton, J. (2000). A retrospective assessment of the accuracy of the paternity inference program CERVUS. Mol. Ecol. 9, 801–808. doi: 10.1046/j.1365-294x.2000.00930.x
Stoeckel, S., Grange, J., Fernandez-Manjarres, J. F., Bilger, I., Frascaria-Lacoste, N., and Mariette, S. (2006). Heterozygote excess in a self-incompatible and partially clonal forest tree species - Prunus avium L. Mol. Ecol. 15, 2109–2118. doi: 10.1111/j.1365-294X.2006.02926.x
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11 molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tan, X. X., Li, Y., and Ge, X. J. (2009). Development and characterization of eight polymorphic microsatellites for Rhododendron simsii planch (Ericaceae). Conserv. Genet. 10, 1553–1555. doi: 10.1007/s10592-008-9791-y
Torres-Díaz, C., Valladares, M. A., Acuña-Rodríguez, I. S., Ballesteros, G. I., Barrera, A., Atala, C., et al. (2021). Symbiotic interaction enhances the recovery of endangered tree species in the fragmented Maulino forest. Front. Plant Sci. 12:663017. doi: 10.3389/fpls.2021.663017
Tsukamoto, Y., Yonezawa, S., Katayama, N., and Isagi, Y. (2021). Detection of endangered aquatic plants in rapid streams using environmental DNA. Front. Ecol. Evol. 8:622291. doi: 10.3389/fevo.2020.622291
Wang, G., Deng, H., Li, Y., Wang, Q., Zong, X., Yang, X., et al. (2017). An IUCN criteria-based assessment of the endangered categories of key protected wild plants in Chongqing municipality. J. Southeast Univ. Nat. Sci. Ed. 39, 42–50. doi: 10.13718/j.cnki.xdzk.2017.09.007
Wilson, H. Systematic study of Rhododendron calophytum and its immediate allies: a systematic consideration of their relationships to the rest of subsection Fortunea within subgenus Hymenanthes, doctoral dissertation, (2016), The University of Edinburgh, South Bridge, United Kingdom
Yang, B., Zhang, G., Guo, F., Wang, M., Wang, H., and Xiao, H. (2020). A genomewide scan for genetic structure and demographic history of two closely related species, Rhododendron dauricum and R. Mucronulatum (Rhododendron, Ericaceae). Front. Plant Sci. 11:1093. doi: 10.3389/fpls.2020.01093
Ye, X. Z., Zhao, G. H., Zhang, M. Z., Cui, X. Y., Fan, H. H., and Liu, B. (2020). Distribution pattern of endangered plant Semiliquidambar cathayensis (Hamamelidaceae) in response to climate change after the last interglacial period. Forests 11:434. doi: 10.3390/f11040434
Yeh, F. C., Yang, R. C., Boyle, T. B., Ye, Z. H., and Mao, J. X. POPGENE, the user-friendly shareware for population genetic analysis, version 1.31. Molecular Biology and Biotechnology Centre, University of Alberta, Alberta. (1997).
Yu, F. Y., Skidmore, A. K., Wang, T. J., Huang, J. H., Ma, K. P., and Groen, T. A. (2017). Rhododendron diversity patterns and priority conservation areas in China. Divers. Distrib. 23, 1143–1156. doi: 10.1111/ddi.12607
Zhang, X. J., Liu, X. F., Liu, D. T., Cao, Y. R., Li, Z. H., Ma, Y. P., et al. (2021). Genetic diversity and structure of Rhododendron meddianum, a plant species with extremely small populations. Plant Divers 43, 472–479. doi: 10.1016/j.pld.2021.05.005
Zhao, B., Si, G. C., Gu, X., and Zhang, Y. L. (2012). Population variation analysis of seven Rhododendron concinnum wild populations in Qinling mountains. Bull. Bot. Res. 32, 561–566. doi: 10.7525/j.issn.1673-5102.2012.05.010
Zheng, J., Arif, M., He, X., Liu, X., and Li, C. (2023). Distinguishing the mechanisms driving multifaceted plant diversity in subtropical reservoir riparian zones. Front. Plant Sci. 14:1138368. doi: 10.3389/fpls.2023.1138368
Zheng, J., Muhammad, A., He, X., Ding, D., Zhang, S., Ni, X., et al. (2022). Plant community assembly is jointly shaped by environmental and dispersal filtering along elevation gradients in a semiarid area, China. Front. Plant Sci. 13:1041742. doi: 10.3389/fpls.2022.1041742
Keywords: biodiversity protection, plant conservation, endangered plant species, simple sequence repeat, genetic structure, phenotype differentiation
Citation: Ma L, Li L, Fang W, Dong Z, Liu Y, Wang C, Xie Y, Li M, Arif M and Wang H (2024) Genetic variability and population divergence of Rhododendron platypodum Diels in China in the context of conservation. Front. For. Glob. Change. 7:1320995. doi: 10.3389/ffgc.2024.1320995
Received: 13 October 2023; Accepted: 01 March 2024;
Published: 14 March 2024.
Edited by:
Wen Xing Long, Hainan University, ChinaReviewed by:
Jai Pal Sharma, Dr. Yashwant Singh Parmar University of Horticulture and Forestry, IndiaCopyright © 2024 Ma, Li, Fang, Dong, Liu, Wang, Xie, Li, Arif and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Arif, YXJpZmdyZWVuZXhwZXJ0QG91dGxvb2suY29t; Haiyang Wang, d2h5c3dhdUAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.