
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. For. Glob. Change , 09 March 2023
Sec. Forest Management
Volume 6 - 2023 | https://doi.org/10.3389/ffgc.2023.791766
This article is part of the Research Topic Retaining Ecosystem Legacies in Forest Management: Effects on Forest Structures and Functions View all 9 articles
Introduction: Logging impacts millions of hectares of forests globally every year, and not only affects tree cover, but also disrupts below-ground soil communities that are essential for forest ecosystems. Soil fungi are particularly vulnerable to such disturbances due to their reliance upon plant hosts as their source of carbon. Fluctuations within the major guilds of fungi important for forest function can have ramifications for plant communities and biogeochemical processes. We addressed questions about soil fungal communities in temperate forest stands with varying logging histories: (1) Do assembly patterns of soil fungal communities and functional guilds reflect historical differences in logging legacies? (2) Does sequencing of below-ground communities of fungi resemble the composition of surveys of fungal fruiting bodies? (3) How do fungal communities in the litter layer differ from those in the soil and do these assembly patterns change with logging history?
Methods: Our study took place in the H. J. Andrews Experimental Forest in western Oregon, USA. We sampled soil and litter (Oi—Oe) in three sites with different logging histories: one clear cut in 1974, one selectively logged and thinned three times between 1974 and 2001, and one unlogged. We sequenced soil fungi separately for mineral soil samples and litter samples. Additionally, we compiled fruiting-body studies from 1972 through the present to compare with our eDNA samples.
Results: We found that four decades after logging had ceased there were detectable signatures within the soil fungal communities that distinguished logged from unlogged sites, indicating a legacy that affects many generations of fungi (PERMANOVA; p < 0.001 for both soil and litter fungi). There were also significant differences between litter and mineral soil communities (PERMANOVA; p < 0.001) with higher relative abundances of pathogens within the litter layer and a greater proportion of mycorrhizal fungi in the soil.
Discussion: These results highlight the importance of including forest litter in studies, as entire guilds of fungi can be underestimated when considering a single fraction. Together, these results have repercussions for the regeneration of forests following logging, as the composition of fungal guilds important to plant functions do not fully recover even after decades of cessation.
Logging is one of the most pervasive disturbances to forests. Around 3.5 billion m3 of wood is harvested around the globe annually, affecting 29 million hectares of forest (Pepke, 2010; Crowther et al., 2015). While improvements to harvesting and processing technologies have made the logging industry more efficient, demands for timber products have increased in recent decades and that trend is predicted to continue (Zhou et al., 2005; Nepal et al., 2021). Logging not only affects tree cover, but also disrupts belowground soil communities that are essential for forest ecosystems. Fungi are especially vulnerable to logging-associated disturbances due their formation of multicellular bodies known as mycelia that ramify through their substrate, and their reliance upon plant hosts—alive or dead, depending on the species—as their source of carbon. Fungal communities play critical roles in the functioning of forest ecosystems, including structuring plant communities (Gams, 2007), driving nutrient fluxes (Delgado-Baquerizo et al., 2016), influencing forest productivity, and sequestering carbon (Frąc et al., 2018).
Three main functional groups of fungi important to forest functioning are mycorrhizal fungi, pathogens, and decomposers (Ehrenfeld et al., 2005; Baldrian, 2016). Mycorrhizal fungi take up nutrients from the soil for their host plants in exchange for photosynthetically derived carbon (Bagyaraj and Ashwin, 2017). These fungi are also able to protect their hosts from pathogenic infections and increase stress tolerance (Jayne and Quigley, 2014; Kanekar et al., 2018; Gonthier et al., 2019). There are two broad categories of mycorrhizal fungi, arbuscular mycorrhizal fungi (AMF), and ectomycorrhizal fungi (ECMF). AMF are widely distributed and associate with >80% of plant species, while ECMF associate with ∼2% of plant species, primarily large woody species that dominate temperate and some tropical forests (Aerts, 2003). Pathogenic fungi attack their host plant species, which adds nutrients and carbon to resource pools (Maron et al., 2011). Decomposers—or saprotrophs—break down organic matter by releasing extracellular enzymes, increasing access to nutrients for other organisms (Bagyaraj and Ashwin, 2017). Decomposers can also suppress pathogenic fungi, reducing mortality of vulnerable host plants (Adnan et al., 2019). Many soil fungi also produce fruiting bodies (e.g., mushrooms) to generate and release sexual spores. Prior to next generation sequencing studying fungal communities primarily consisted of collecting these reproductive structures for identification. It is useful to compare sequences found in belowground environmental DNA (eDNA) studies with macrofungi collections to discover what proportion of species are shared between different methods. Not all fungi produce fruiting bodies, therefore sequencing of DNA in soil can reveal species absent from aboveground collections. On the other hand, aboveground collections can fill in gaps resulting from biases in DNA extraction, PCR amplification, and sequencing. Studies comparing fruiting body surveys to soil sequencing surveys typically find between 10 and 20% overlap, indicating the importance of utilizing both styles of survey (Gardes and Bruns, 1996; Dahlberg et al., 1997; Baptista et al., 2015).
The Pacific Northwest of the US is dominated by ectomycorrhizal trees and continues to be one of the most active logging regions in North America. Recent estimates of logging in the area show an annual average of seven million tons of lumber are harvested in Oregon (ORFI, 2020), and nine million tons are harvested in Washington per year (Arnold, 2004). Increases in efficiency of lumber use and conservation efforts have protected some old-growth areas in these areas, but demand continues to drive increases in harvesting (Pepke, 2010). Two major logging techniques are selective logging and clearcutting. The impacts of each technique on soils are similar, but clearcutting effects tend to be far more extreme. The use of large vehicles and machines in both techniques results in compaction of soil and heavy metal pollution (Hartmann et al., 2014). Openings in the canopy allow more light to reach the soil, which decreases moisture and increases temperature. The removal of trees also eliminates hosts for symbionts and alters the chemistry of soil as litter inputs are reduced. Erosion is also far more likely, which can further alter soil chemistry and nutrient pools. Knowledge about the impacts of logging on belowground communities will not only increase understanding the role of disturbance in fungal community assembly but could also aid in active forest management and replanting efforts.
To evaluate the consequences of logging legacies on fungal composition, we sequenced fungi from soils in clearcut, selectively logged, and old-growth forest in the Pacific Northwest. We separately sampled soil (Oi—Oe) and litter because the litter layer is often ignored but contains a substantial amount of fungal biomass (Lim et al., 2010; Frey et al., 2011; Hu et al., 2019). Furthermore, the layers may differ in the composition of fungal functional guilds (McGuire et al., 2013). We also compared belowground communities to aboveground fruiting body composition. We addressed the following questions: (1) How do soil and litter fungal communities vary between forest stands with different logging histories? (2) What proportion of fungi are found both and belowground within the forest? (3) How do fungal communities in the litter layer differ from those in the soil?
Our study sites were located within The H. J. Andrews Experimental Forest (hereafter referred to as HJA; 44.2°N, 122.2°W), a mid-elevation Pacific Northwest conifer forest dominated by Pseudotsuga menziesii (Douglas-fir) and Tsuga heterophylla (Western hemlock) with stands of varying age up to 700 years. The annual mean temperature is 8.6°C, the mean annual precipitation is 2,289 mm/yr. HJA was established in 1948 by the United States Forest Service. The original focus of the research being conducted within HJA was to manipulate forest operations such as logging and forest regeneration to optimize the efficiency of the timber industry. Due to this history, the watersheds within HJA were subjected to logging of varying intensity at different times. We sampled three watersheds with different logging legacies within HJA—watershed 6 (WS6), watershed 7 (WS7), and watershed 8 (WS8) (Figure 1). WS6 and WS7 were both previously logged: WS6 was clearcut in 1974 with 9% road coverage and was replanted with P. menziesii seedlings in 1976, while WS7 had 60% of its overstory harvested in 1974 with 0% road coverage, the rest of the canopy harvested in 1984, and a final thinning in 2001 to ensure ∼4 m between each tree. WS8 was never logged, it was left as an old-growth forest control for comparison with other watersheds from the inception of HJA (Table 1). However, it did experience a fire that burned up to 70% of the standing trees in the 1850s, though this is still considered an old-growth forest within this system. The watersheds lie within the transition between western hemlock (Tsuga heterophylla) and silver fir (Abies amabilis) zones. WS6 was planted with 100% P. menziesii at a density of 383 stems/ha with a basal area of 70 ± 10 m2/ha. WS7 was planted with 100% P. menziesii at a density of 333 stems/ha with a basal area of 75 ± 4.8 m2/ha, which was reduced to 220 stems/ha in 2001. The understory of all three watersheds is comprised of vine maple (Acer circinatum), two Oregon grape species (Mahonia aquifolium and M. nervosa), whipplea (Whipplea modesta), salal (Gaultheria shallon), and rhododendron (Rhododendron macrophyllum). The overstory in WS8 is a mix of P. menziesii, T. heterophylla, A. amabilis, and western red cedar (Thuja plicata) with nearly one-third of individuals being greater than 450 years of age and the remaining around 125 years or younger (Andrews, 2022).
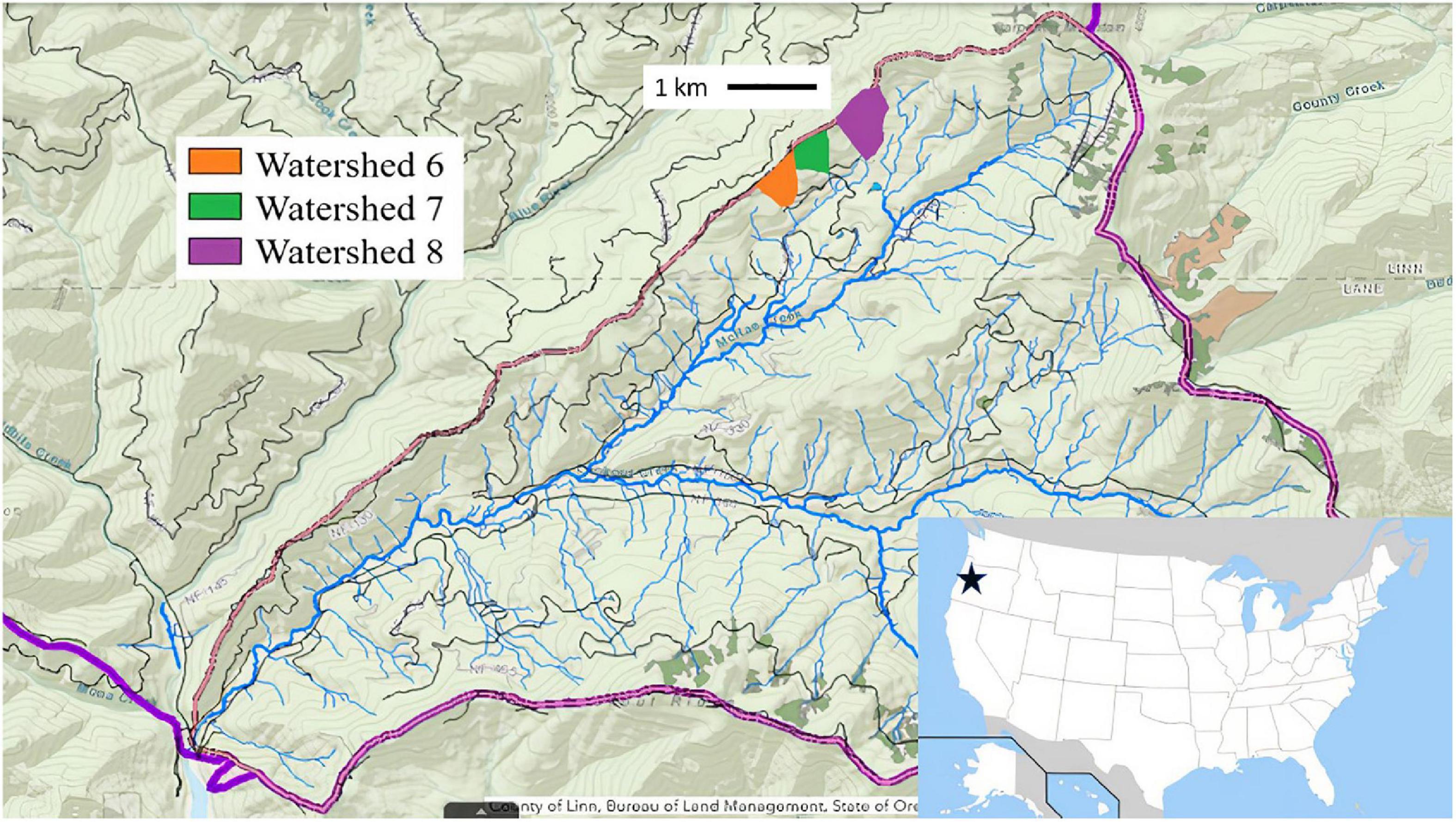
Figure 1. Map of H. J. Andrews Experimental Forest—a Long Term Ecological Research Site (LTER) in Lane and Linn County, Oregon, USA. Includes coloration of three watersheds with different management histories that were sampled for soil fungi. Watershed 6, clearcut in 1974; watershed 7, canopy harvest in 1974, 1984, and thinning in 2004; watershed 8, unlogged.
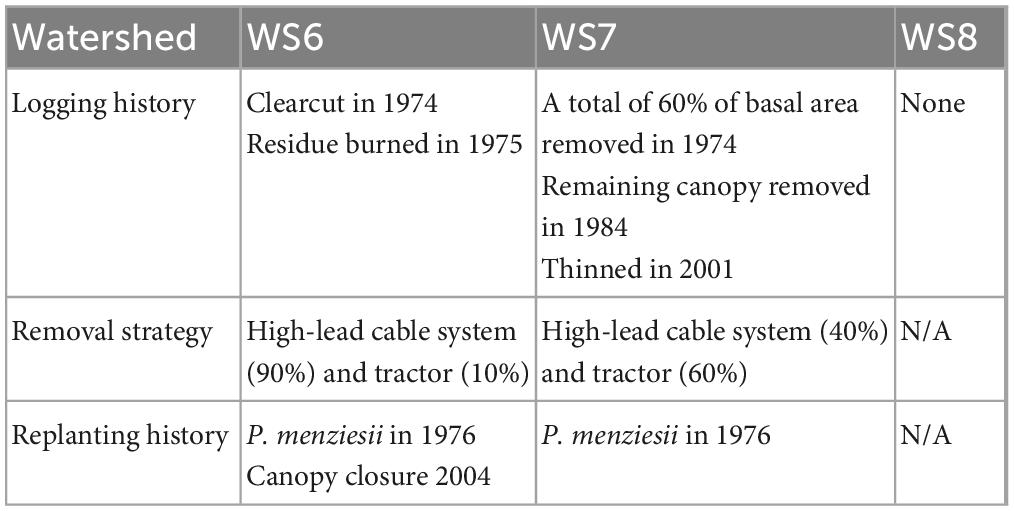
Table 1. Management history for each watershed sampled within the H. J. Andrews experimental forest.
All samples were collected on 15 November 2018. Within each watershed we sampled along three 100 m transects running east to west that were separated by 50 m north to south. Along each transect we collected five litter and five soil samples at randomized points (soil n = 45, litter n = 45). Litter samples consisted of collecting organic litter around the target point by hand while wearing EtOH-sterilized gloves to fill a sterile 500 mL bag. Soil samples consisted of 2.5 cm × 20 cm cores taken using an EtOH-sterilized corer after the litter layer was removed. Samples were stored on ice for up to 12 h before being transferred to a −20°C freezer until homogenization and DNA extraction.
Soil samples were homogenized using EtOH and UV-sterilized 2 mm aperture sieves, and litter samples were homogenized using EtOH and UV-sterilized ceramic mortar and pestles. To facilitate homogenization, liquid nitrogen was poured onto the litter samples immediately before pulverizing. For each sample, 0.25 g and 3.0 g were weighed in sterile 2 ml Eppendorf tubes for DNA extraction and moisture analysis, respectively. An additional sterile 2 ml Eppendorf tube was filled and kept as an archival sample within a −80°C freezer. All tubes were stored at −20°C before use.
The rest of each homogenized sample was air-dried at room temperature in a labeled paper bag. Air-dried soil samples were prepared for pH analysis by mixing a 1:2 ratio of sample to deionized water. For soil samples, 10 g of soil was weighed in a glass beaker and mixed with 20 ml of deionized water. For litter samples a ratio of 1:8 was used to compensate for absorption, 2.5 g of litter was weighed in a glass beaker and mixed with 20 ml of deionized water. Three readings per sample were recorded using a benchtop pH meter with a glass electrode, and the average value was calculated. Moisture was calculated for each sample by recording the weight of 3.0 g of sample in an aluminum weighing dish, drying the sample in a drying oven at 130°C for 48 h, re-weighing the sample, and subtracting the dried sample weight from the initial sample weight (Black, 1965).
High throughput sequencing was used to analyze fungal community composition for soil and litter samples. DNA was extracted from 0.25 g of each soil and litter sample using Qiagen DNeasy PowerSoil single-tube extraction kits, following the manufacturer’s protocol. The ITS1 region of the fungal rRNA gene was amplified using the ITS1-F (CTTGGTCATTTAGAGGAAGTAA) and ITS2 (GCTGCGTTCTTCATCGATGC) primer pair, the Nextra NX2 Illumina (San Diego, California) adapter, and Earth Microbiome barcoded primers. A master mix containing 12.5 μl GoTaq, 9.45 μl nuclease-free water, 0.5 μl forward (ITS1F) primer, 0.5 μl reverse (ITS2) primer, 0.5 μl Nextera (NX2) primer, and 0.05 μl BSA was added to the wells of a 96-well PCR plate. In each well, 1.0 μl of genomic DNA and 0.5 μl of a unique barcoded primer was added to the master mix for a total volume of 25 μl per well. PCR reactions were held in a thermal cycler at 94°C for 1 min, with amplification proceeding for 35 cycles at 94°C for 30 s, 52°C for 30 s, and 68°C for 30 s, and a final elongation of 7 min at 68°C.
Amplicon concentrations were quantified according to a Quant-iT PicoGreen protocol (Thermo Fisher Scientific, Waltham, MA). DNA concentrations were quantified on a Molecular Devices SpectraMax M5E multi-mode plate reader (San Jose, CA). Amplicons were pooled together according to quantification data from the PicoGreen assay to obtain an equimolar concentration of 1 ng of DNA per sample. The pooled library was purified using a Qiagen QIAquick PCR purification kit, according to the manufacturer’s protocol. The amplicon pool was sequenced using an Illumina MiSeq (San Diego, CA) instrument at the University of Oregon’s Genomics and Cell Characterization Core Facility (GC3F).
Raw sequences were demultiplexed using the Quantitative Insights into Microbial Ecology (QIIME) pipeline (Caporaso et al., 2010). We then assembled an amplicon single variant (ASV) table using the DADA2 pipeline (Callahan et al., 2016). Taxonomy was assigned using the naïve Bayesian classifier from the Ribosomal Database Project (RDP) against the UNITE database (Koljalg et al., 2005; Cole et al., 2014). We used variation-stabilizing transformation to normalize ASV counts (Anders and Huber, 2010). To assign functional guild classifications to each ASV we used the annotation tool FUNGuild set to its default parameters (Nguyen et al., 2016). Many ASVs tend to have multiple guilds assigned to them (e.g., saprotroph and pathogen). When performing our downstream analyses, we allowed for ASVs to be represented in multiple categorical subsets (i.e., the previous example would be counted once as a saprotroph and once as a pathogen).
Statistical analyses were performed in R (v3.6.3) using the vegan package (Oksanen et al., 2019). We used the Bray-Curtis dissimilarity statistic to quantify the compositional dissimilarity between our sample communities. This statistic allowed us to visualize the difference between communities within a two-dimensional plot using non-metric multidimensional scaling (NMDS). Additionally, to test for significance in our Bray-Curtis dissimilarity results we performed permutation analyses of variance (PERMANOVAs) comparing watershed, sample source (soil or litter), as well as physicochemical properties (pH, moisture). Additionally, to visualize compositional differences, we compared the relative abundances of functional guilds and the most abundant 15 genera found in each treatment. Figures were generated using ggplot2 (Wickham, 2016).
To gain an understanding of the total fungal community in the forest and because eDNA and sporocarp surveys capture different parts of the fungal community (Frøslev et al., 2019) we wanted to compare the fungal species represented by sporocarps (sexual) vs. those in the soil eDNA samples (largely asexual). We limited the lists to macrofungi [mushrooms in the general sense (Thiers and Halling, 2018), including boletes and corals as well as larger ascomycetes such as Helvella] that were identified to species. This list was then compared to a list of macrofungi collected from the LTER using existing data from the H. J. Andrews from the following publications and reports, and their associated datasets in the H. J. Andrews datafiles [Rhoades, 1972; Luoma et al., 1991 (TP109); Smith et al., 1996 (SA01401)] and iNaturalist (2022) observations through February 2021. Sporocarps in the Rhoades (1972) study were collected opportunistically between October 1970 and April 1972, those in the Smith et al. (1996) study were collected twice per year for 4 years from three forest stands, one old growth, one rotation age and one young-growth. The Luoma et al. (1991) study took place in 10 stands, with sampling occurring for about 6 weeks twice per year for 4 years. All the names in our final list (Supplementary Table 1) were verified in Index Fungorum; this process uncovered numerous typos and names changes which are enumerated in Supplementary Table 1. Many collections were identified only to genus; these were not included in our comparison with the soil eDNA because we had no way of determining which species they might match in the eDNA.
While keeping litter and mineral soil separate, the fungal communities form separate clusters by management histories (clearcut, canopy + thinned, unlogged) based on Bray-Curtis dissimilarity (Figure 2). The difference between management types was significant for both mineral soil and litter using PERMANOVA (soil: df = 2, F = 1.84, R2 = 0.0825, p < 0.001; litter: df = 2, F = 2.32, R2 = 0.104, p < 0.001). The composition of the genera that comprised each guild were distinct between management types (Figures 3–5A). Saprotrophs, pathogens, and ectomycorrhizal fungi were all significant when considered separately (Figures 3–5C and Supplementary Table 2). Similarly, if the watersheds are grouped into a binary of logged and unlogged (Figure 6) all of these categories remain significantly different (all fungi: df = 1, F = 2.55, R2 = 0.0291, p < 0.001; ECMF: df = 1, F = 2.35, R2 = 0.0275, p < 0.001; saprotroph: df = 1, F = 7.34, R2 = 0.0795, p < 0.001; pathogen: df = 1, F = 3.08, R2 = 0.125, p < 0.001).
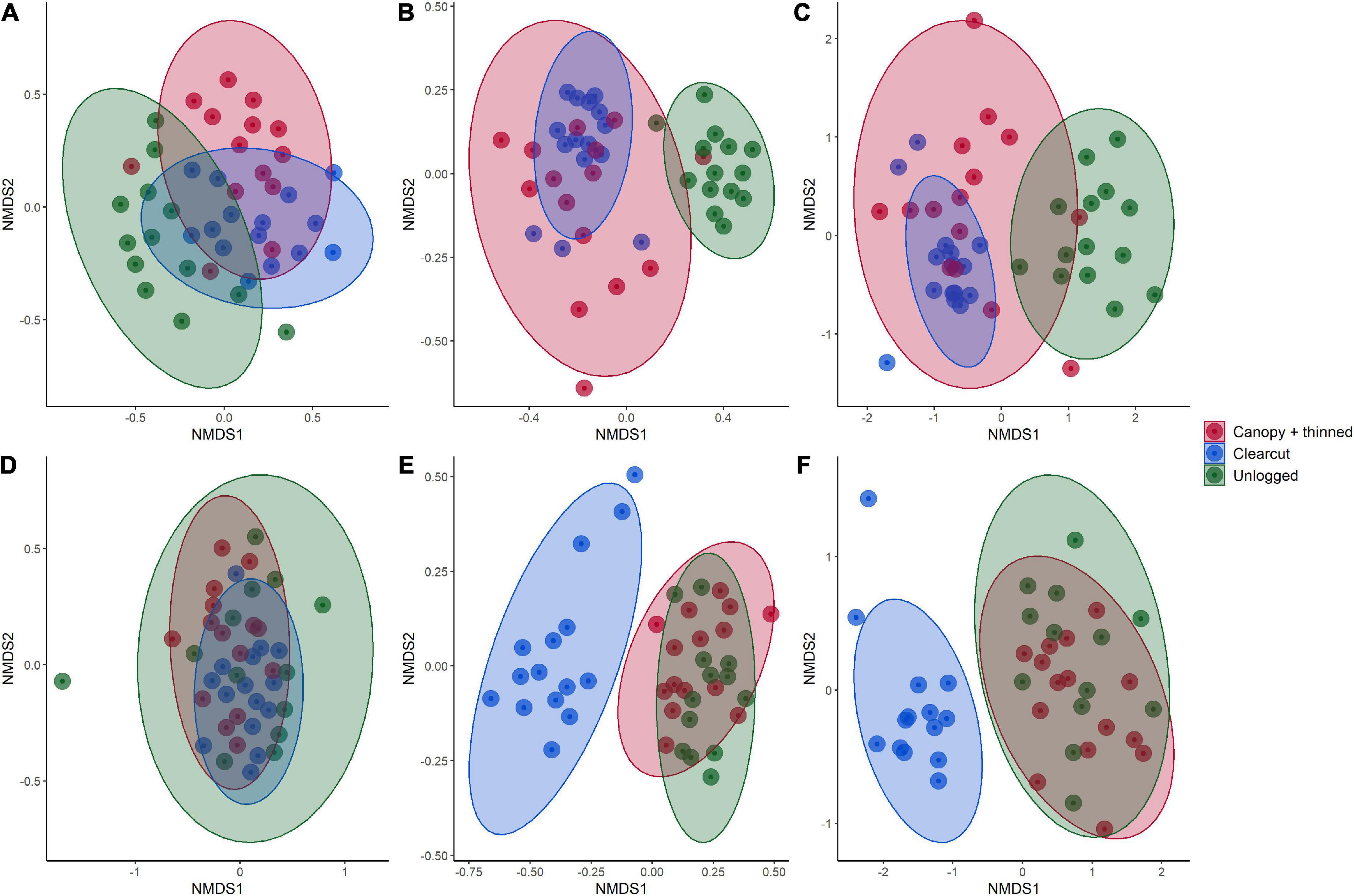
Figure 2. Non-metric multidimensional scaling ordinations based on Bray-Curtis dissimilarity between three forest stands with different logging histories sampled in H. J. Andrews Forest, Oregon. Included are ectomycorrhizal fungi, saprotrophs, and plant pathogens in soils (A–C), as well as ectomycorrhizal fungi, saprotrophs, and plant pathogens in litter (D–F). All PERMANOVA p-values are <0.001. Clearcut, watershed 6, canopy + thinned, watershed 7, unlogged, watershed 8.
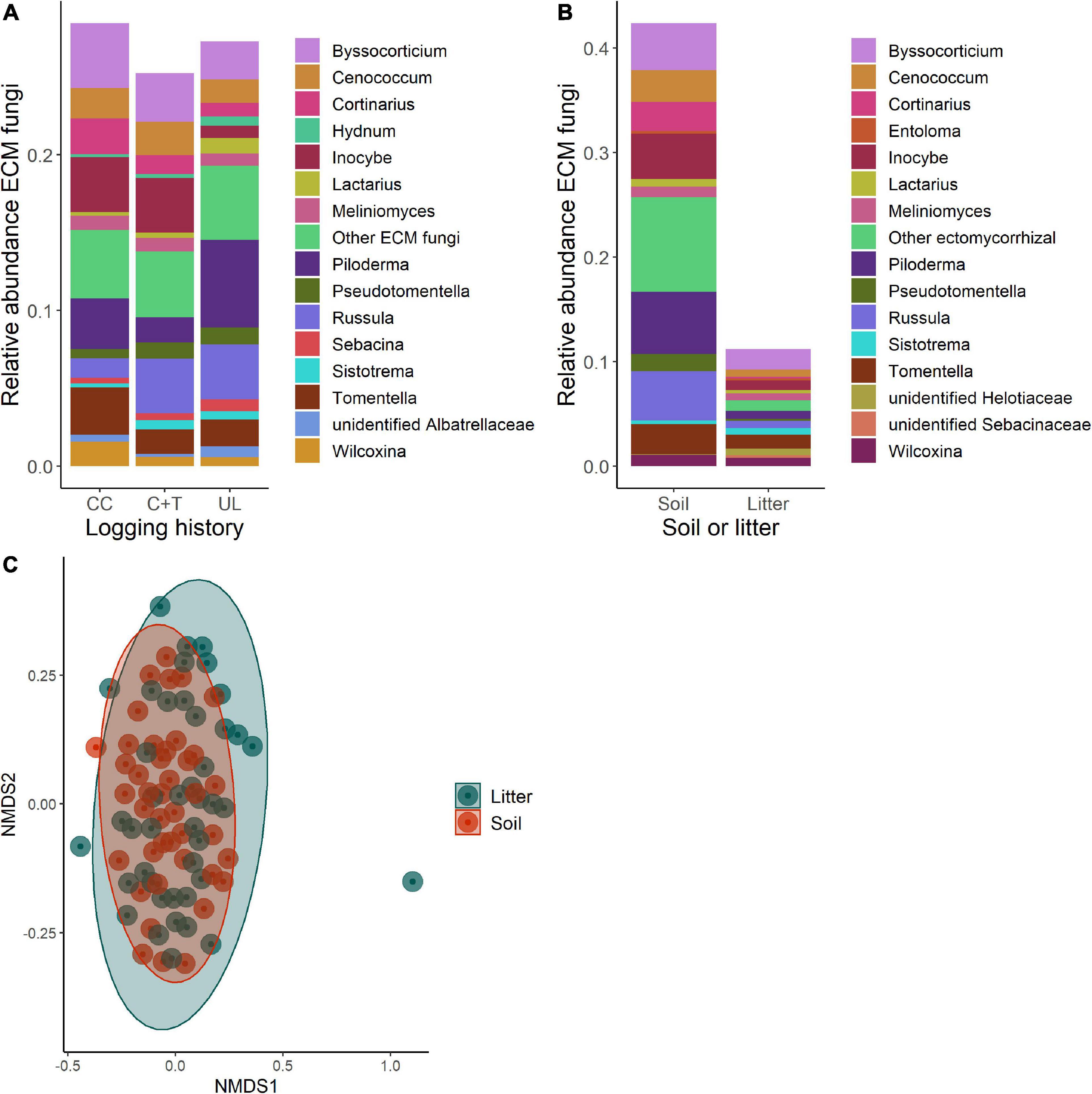
Figure 3. Comparisons of ectomycorrhizal fungal (ECM fungi) communities in soil and litter of logged and unlogged forest stands within the H. J. Andrews Forest, Oregon, USA. Clearcut (CC), watershed 6, canopy + thinned (C + T), watershed 7, unlogged (UL), watershed 8. (A) Relative abundances of ECMF genera between watersheds. (B) Relative abundances of ECMF genera between soil and litter. (C) NMDS ordination of ECMF communities based on Bray-Curtis dissimilarities between soil and litter (PERMANOVA, p = 0.578).
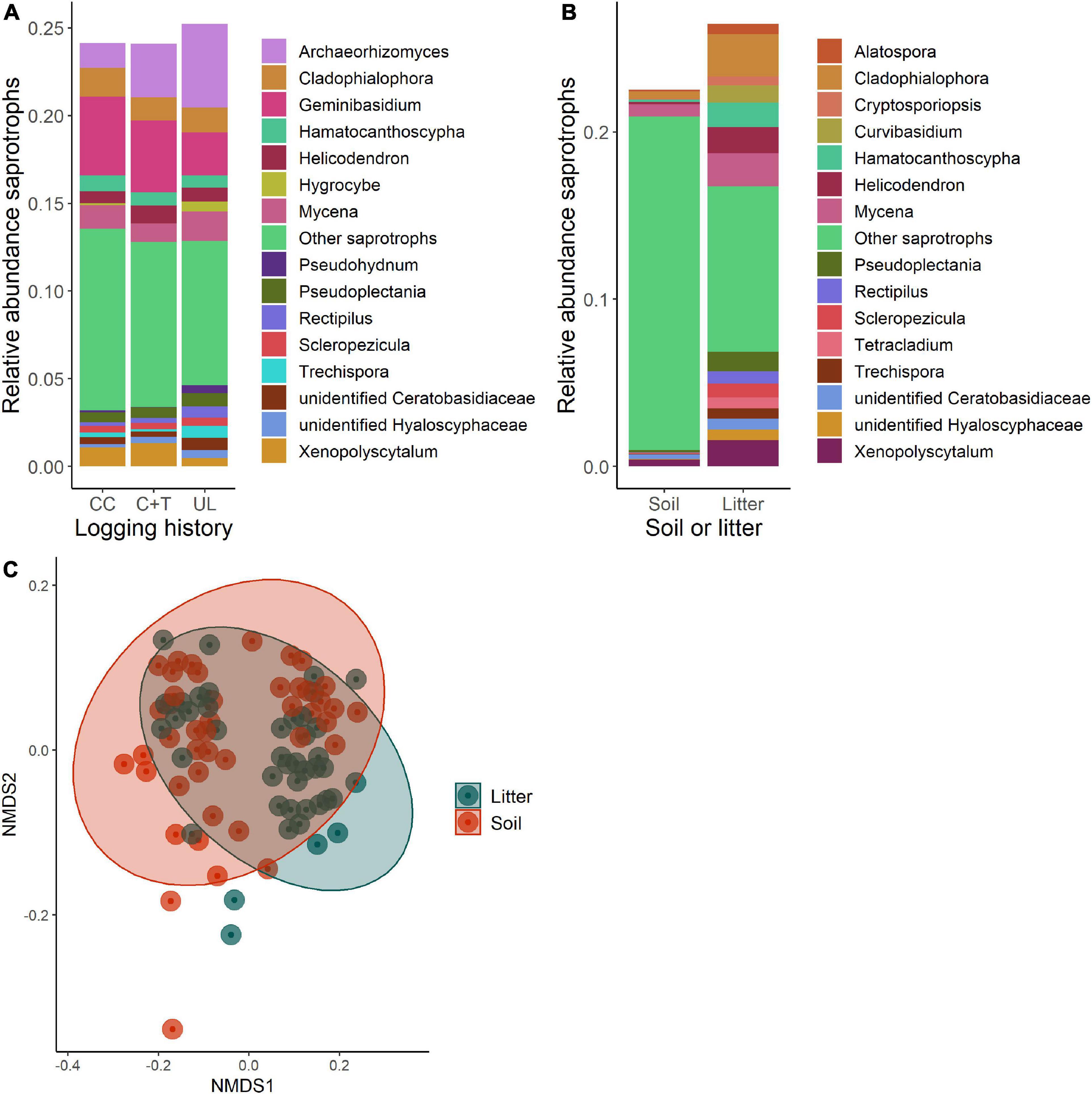
Figure 4. Comparisons of saprotroph communities in soil and litter of logged and unlogged forest stands within the H. J. Andrews Forest, Oregon, USA. Clearcut (CC), watershed 6, canopy + thinned (C + T), watershed 7, unlogged (UL), watershed 8. (A) Relative abundances of saprotroph genera between watersheds. (B) Relative abundances of saprotroph genera between soil and litter. (C) NMDS ordination of saprotroph communities based on Bray-Curtis dissimilarities between soil and litter (PERMANOVA, p = 0.028).
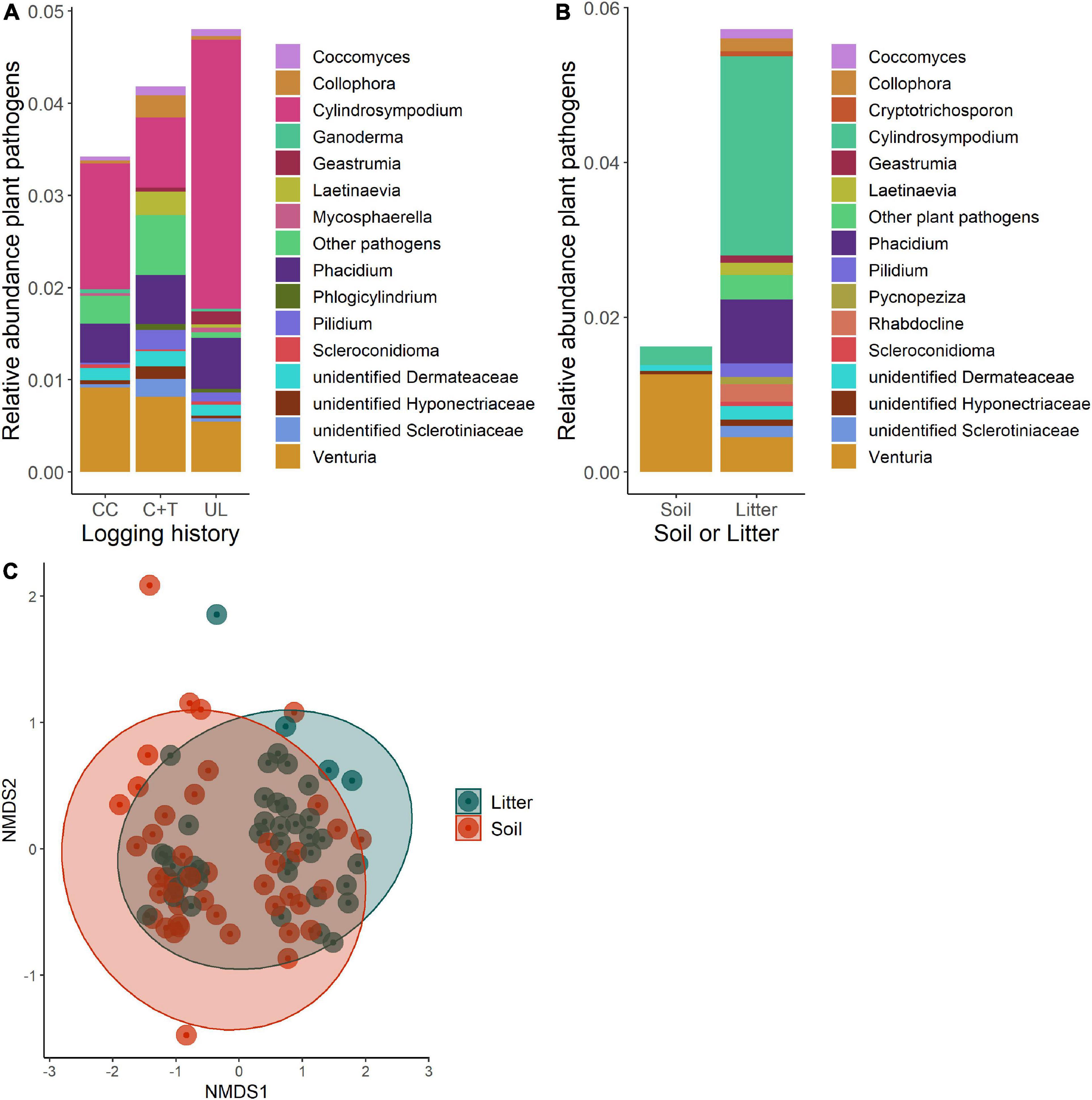
Figure 5. Comparisons of pathogen communities in soil and litter of logged and unlogged forest stands within the H. J. Andrews Forest, Oregon, USA. Clearcut (CC), watershed 6, canopy + thinned (C + T), watershed 7, unlogged (UL), watershed 8. (A) Relative abundances of pathogen genera between watersheds. (B) Relative abundances of pathogen genera between soil and litter. (C) NMDS ordination of pathogen communities based on Bray-Curtis dissimilarities between soil and litter (PERMANOVA, p = 0.005).
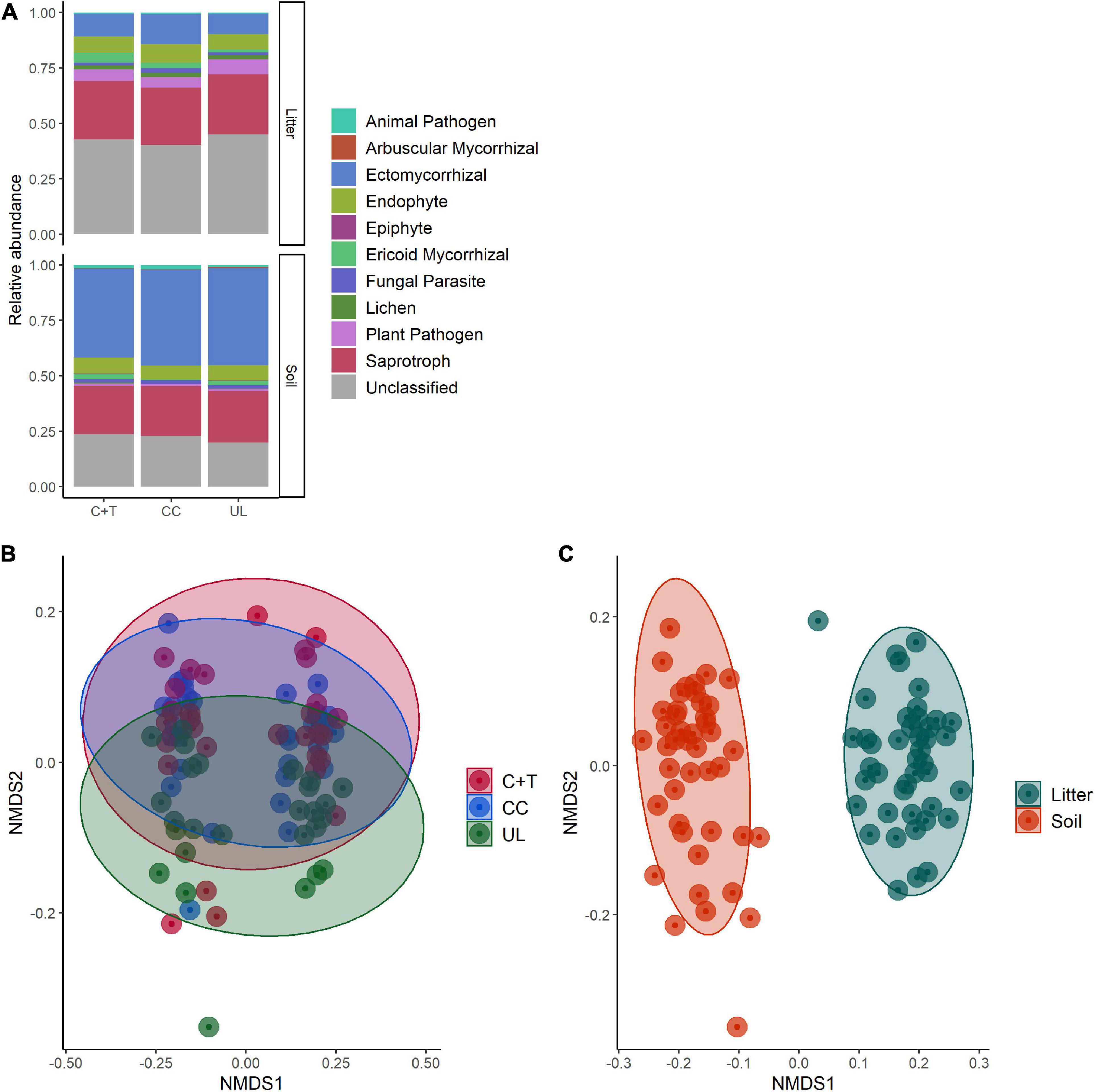
Figure 6. Comparisons of fungal communities in soil and litter of three watersheds with varying logging histories within the H. J. Andrews Forest, Oregon, USA. Clearcut (CC), watershed 6, canopy + thinned (C + T), watershed 7, unlogged (UL), watershed 8. (A) Relative abundances of functional guilds between watersheds for soil and litter. Functional guilds were assigned using FUNGuild analysis. (B) NMDS ordination of fungal communities based on Bray-Curtis dissimilarities comparing watersheds (PERMANOVA, p < 0.001). (C) NMDS ordination of fungal communities based on Bray-Curtis dissimilarities comparing soil and litter (PERMANOVA, p < 0.001).
Based on Bray-Curtis dissimilarity, the overall fungal communities separated into distinct clusters for mineral soil and litter (Oi—Oe) samples (Figure 6C). We found that the difference between these groups is highly significant using PERMANOVA (df = 1, F = 13.92, R2 = 0.141, p < 0.001). Some of these differences are apparent when comparing relative abundances of functional guilds and the genera that comprise them (Figures 3–5B). Litter communities tend to contain more pathogens, while mineral soil communities contain more ectomycorrhizal fungi (ECMF; Figure 6A). When comparing the saprotrophs and pathogens between soil and litter samples there are significant differences (df = 1, F = 2.42, R2 = 0.028, p = 0.008 and df = 1, F = 3.08, R2 = 0.035, p = 0.007, respectively). There are not significant differences between soil and litter for ectomycorrhizal fungi (df = 1, F = 0.944, R2 = 0.011, p = 0.574)—however, it was significant when accounting for an interaction with management history (df = 1, F = 1.63, R2 = 0.019, p = 0.007).
Our belowground sequences from the three sites yielded 144 macrofungi including those that form mushrooms, cups, crusts, and truffles that were identified to species. Our compilation of sporocarp data across the entire H. J. Andrews forest yielded 380 species; however, only 34 (6.49%) were shared between the sporocarp and eDNA datasets for a total of 524 species found. The entire list, indicating the kind of data (eDNA or sporocarp) and the source of the information can be found in Supplementary Table 1.
We characterized the pH and moisture content of our samples. Based on PERMANOVAs of the Bray-Curtis dissimilarities of communities, we found that pH and moisture had a significant effect on soil fungi (pH: df = 1, F = 2.63, R2 = 0.0590, p < 0.001; moisture: df = 1, F = 3.07, R2 = 0.0682, p < 0.001) while it did not have a significant effect on litter fungi (pH: df = 1, F = 1.19, R2 = 0.0282, p = 0.121; moisture: df = 1, F = 1.16, R2 = 0.0275, p = 0.128).
Pseudo replication is a potential concern with our dataset, as our logging treatments are confounded with watershed identity. This may limit the generalizability of our data to other logging systems. However, due to the close geographic proximity, shared climate, similar topography, identical understory composition, and limited physical barriers we are confident that this effect is minimal. Logging practices vary in their effects on aboveground forest structure and we found that differences in historical logging also had significant legacy effects on belowground fungal composition. We saw large differences in some taxa between logged and unlogged forests, and these differences depended on the functional groups of the fungi. We also saw differences in fungal composition between logged forests depending on whether the forest was clearcut or selectively logged. Logging causes several consistent ecological changes in forest structure, composition and function that the fungi are likely responding to. For example, the early stages of succession are lost due to replanting and since the tree ages are the same in replated forests, there are no snags, few downed logs and the canopy is open allowing more sunlight to hit the forest floor for about 30 years post cut (Franklin et al., 2002). To simplify our discussion, we only consider the genera that had very large (2× or more) differences between sampling units in our soils and litter data.
In soils, we found that ECM fungi showed strong compositional responses to logging legacies, which may have implications for forest regeneration trajectories since most of the trees in these sites depend on ECM fungi for growth and survival. We found that some fungal genera were more abundant in early to mid-successional forests whereas others were more restricted to unlogged, old growth forests. Since the dominant overstory tree hosts (P. menziesii) are the same in these sites, these results suggest that environmental filtering structures ECM fungal communities across sites due to the abiotic changes from historical logging practices. We found greater abundance (≥2×) of Piloderma and Russula in unlogged forests, which is consistent with other studies (Smith et al., 2016; Kranabetter et al., 2018) and we found more of the early successional Wilcoxina in logged forests (Dickie et al., 2013). Studies of sporocarps (fruiting bodies) have generally found that Inocybe species are largely early successional (Cripps, 1997 and refs. therein), which is consistent with our data. However, a study by Norvell and Exeter (2004) on sporocarps in the Oregon Coast range found no relationship between stand age after logging and Inocybe fruiting. The differences between the Inocybe studies could be the result of differences in methods; studies of sporocarps can only detect fruiting, not abundance of the taxon in the soil, whereas we could only detect abundance in the soil and our fruiting body survey was not concomitant—it was a compilation of more than 40 years of data. Similarly, the genus Cortinarius is considered to be later successional (Nara et al., 2003; Sun et al., 2015), but we found them to be more than 2× more abundant and more diverse in the soils of logged watersheds, with only 3/37 taxa restricted to the soils of the unlogged forest. Another study compared ECMF in logged and unlogged forests in Malaysia, finding similar trends in relative abundances of genera—nearly twice as many Russula in unlogged forests, and more Inocybe in logged forests. Together, these results suggest that some fungal clades may show similar trends due to logging regardless of location (McGuire et al., 2015).
Plant pathogens were distinctly in their abundance and composition between the logged and unlogged sites, and our data suggest that these differences are potentially linked to differences in the abundance of litter. For example, Cylindrosympodium was greater than twice as abundant in the unlogged watershed, and this genus was also greater than twice as abundant in the litter. Post clearcutting, the logged watersheds were burned, which would have significantly reduced the litter relative to the unlogged watershed which also would have had larger and older trees. We note here that FUNGuild (Nguyen et al., 2016) classified Cylindrosympodium as a pathogen, but the particular species in our data were not determinable with the exception of C. lauri, which was not common. It was originally isolated from Laurus leaves and was assumed to be plant pathogenic (Crous et al., 2007), but we found no evidence that it has been tested for pathogenicity in any study. While some species of Cylindrosympodium are thought to be plant pathogens (Crous et al., 2007), others appear to be involved in decomposition (Purahong et al., 2016; Hernandez-Restrepo et al., 2017). Deeper study of this abundant genus is warranted as its abundance in litter and in both our study and others (e.g., Jumpponen et al., 2010; Silva and Lambers, 2021) suggests it is important at the ecosystem scale. The higher abundance of pathogens could make seed recruitment difficult within the unlogged watershed but may also help maintain tree diversity as has been found in tropical systems (Song et al., 2021).
We also saw differences between the individual logging treatments. The soil fungal communities of watershed 7 (selectively logged) were similar to watershed 6 (clearcut), while the litter fungal communities were similar to watershed 8 (old-growth). This may indicate that below-ground soil fungi are more vulnerable to impacts from logging than fungi found in the litter later. Another explanation may be that the process of selectively harvesting in watershed 7, rather than clearcutting, allowed for persistence of the original above-ground fungal community. This is in contrast with the above-ground fungal community of watershed 6, which was likely wiped out when clearcut and burned with a novel assembly replacing it, perhaps representative of where the seedlings were sourced. These differences indicate that there are alternative states depending on the type of logging and regeneration strategy, and that above-ground and below-ground communities may exhibit different legacy responses (Rodriguez-Ramos et al., 2020). Even following planting, and a cessation of logging in watershed 6, 43 years before our sampling there remains a legacy effect. This is indicative of potential changes in the function of the communities between logged and unlogged watersheds.
Changes to the soil fungal community can impede or facilitate forest regeneration through their interactions with plants as they establish and mature. The seedlings of ectomycorrhizal plant species can rely upon common mycorrhizal networks (CMN) to assist in their establishment (Liang et al., 2021). Access to CMNs give seedlings a steady flow of nutrients, and—depending on the fungi involved—a source of fixed carbon from mature conspecifics in the network (Walder et al., 2012). Changes to the saprotroph community can further influence access to nutrients by altering decomposition. Thus, changes in the composition of these two guilds due to logging can influence the accessibility of nutrients and carbon for trees during early regeneration. This can ultimately slow down the process of regeneration and the storage of carbon within a forest (Allison and Treseder, 2008).
Soil and litter fungi had different compositional trends across the logging gradient due to the strong segregation of fungal functional groups across these horizons. Higher relative abundances of pathogens in the litter layer are likely due to the presence of pathogens within senesced host material that falls and constitutes the Oi layer (Wahdan et al., 2020). We saw the opposite trend with ECMF. ECMF can break down organic materials, but they are not as efficient as independent saprotrophs (Talbot et al., 2008). This explains why ECMF were detected in greater relative abundances in the mineral soil, as they forage for nutrient pools, rather than in the litter layer where the early stages of decomposition occur (Lindahl et al., 2007). This tendency may also explain why there does not seem to be a distinct difference between the ECMF found in mineral soil and litter layers—it appears that the genera in the litter samples are comprised of a subset of the genera found in the mineral soil samples. The ECMF sequences found in the litter layer may be the exploratory hyphae of the belowground mycelium searching for nutrient sources (Rosinger et al., 2018) or may be spores from fruiting bodies.
Saprotrophic fungal composition was also distinct between soil and litter, likely due to the different forms of organic material in the layers (Bödeker et al., 2016). The relative abundance of saprotrophs was also higher in litter samples. Early colonizers of senesced organic material are likely found in the litter layer, while those that are responsible for later stages of decay may be found in or near the mineral soil. ECMF and saprotrophs compete for the same organic resources, which further explains their spatial segregation (Talbot et al., 2008; Bödeker et al., 2016).
Fungal sporocarps, e.g., mushrooms, are produced when sexual reproduction has occurred (Pelkmans et al., 2016), whereas fungal DNA in soils is primarily composed of asexual hyphae and asexual spores (Camenzind et al., 2022). The two datasets combined suggest that the 16,000 acres of the HJ Andrews Forest contain at least 524 species of macrofungi. Of these only a small subset of the soil eDNA dataset shared species with the compilation of sporocarp data from HJ Andrews (6.49%), which is somewhat less than the 10% overlap typically found between sporocarp and sequencing surveys (Dahlberg et al., 1997; Richard et al., 2005; Porter et al., 2008; Baptista et al., 2015). Interestingly, there were more macrofungi in collections/observations than in the eDNA, which is unusual. However, given that we surveyed a much larger area (the whole LTER) for sporocarps than we collected soil eDNA from (several points in each of three watersheds), and the sporocarp surveys covered several years, the larger number of species in the sporocarp group is less surprising. It could be argued that we can’t compare the two data sets, even qualitatively, because they were not taken in exactly the same places and at the same time. However, studies of fruiting body formation indicate that a combination of environmental signals trigger fruiting and that these signals are species-specific (Pelkmans et al., 2016). For this reason, it can take dozens of years or more to completely sample a plot in a forest. For example, a study in Switzerland by Straatsma et al. (2001) sampled mushrooms over a 21 year period in the same plots. Only 8/408 were found in all years and many only fruited once during the 21 years. Future studies would benefit from collecting and sequencing fruiting bodies as well as soils because that is the only way to directly and accurately compare the datasets, as identification errors can occur with morphological studies, fungal diversity is poorly documented, and species identification with high-throughput sequence data can be error-prone (Thines et al., 2018; Nilsson et al., 2019).
Our data show that logging can have a legacy effect upon fungal communities lasting decades, including guilds that are important for forest ecosystem processes. Planting following logging, as was done in our sites, may minimize some of these legacy changes, as there is less of a lag between the loss of hosts for the soil fungi and their reintroduction. However, planting may introduce novel fungi into the system, which can change the trajectory of regeneration for the fungal community and add its own legacy effect. Changes to the fungal community due to legacy effects can have consequences for recovery following disturbance. Forest regeneration is an ongoing process with no real end—yet forests can reach stable states where the turnover of species is minimal. The plant communities of the forests in the Western Cascades can take decades or even more than a century following a disturbance to reach such a state depending on the severity (Franklin et al., 2002). Long term studies that track the regeneration of fungal communities in parallel to the regeneration of plants are severely lacking, limiting our understanding of how the communities influence one another. Further research is required to determine the impacts from the loss of particular fungal species on tree establishment and recruitment.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA762546.
MS performed sampling in the field, processed samples, analyzed data, and wrote the manuscript. TT aided with sample processing and helped write the lab portion of the methods. BR compiled and analyzed the fruiting-body dataset and wrote those portions of the manuscript, as well as aided with edits. LS assisted in gaining permission to sample at H. J. Andrews, consulted on sampling scheme, provided us with assistance from lab members, and helped with edits. KM helped conceive the project, aided with edits, and provided lab space as well as funding. All authors contributed to the article and approved the submitted version.
This work supported by the National Science Foundation under the LTER Grants: LTER8 DEB-2025755 (2020–2026) and LTER7 DEB-1440409 (2012–2020). Additional support was provided by the NSF Plant Biotic Interactions (PBI) 1758947. Funding was provided via the McGuire Lab start-up fund endowed by the University of Oregon.
Thanks to M. Schultz for LTER access, site suggestions, and providing access to emergency support. M. Farinacci for showing us the watersheds in person and giving US coordinates for our transects. C. J. Paulino, K. Evens, and G. Ridder for their assistance with collecting soil and litter. K. Shek for assistance with bioinformatics and data analysis. J. Jones and F. Swanson for valuable comments and suggestions. Access to sites was provided by the H. J. Andrews Experimental Forest and Long-Term Ecological Research (LTER) program, administered cooperatively by the USDA Forest Service Pacific Northwest Research Station, Oregon State University, and the Willamette National Forest.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffgc.2023.791766/full#supplementary-material
Adnan, M., Islam, W., Shabbir, A., Khan, K. A., Ghramh, H. A., Huang, Z., et al. (2019). Plant defense against fungal pathogens by antagonistic fungi with Trichoderma in focus. Microb. Pathog. 129, 7–18. doi: 10.1016/j.micpath.2019.01.042
Aerts, R. (2003). “The role of various types of mycorrhizal fungi in nutrient cycling and plant competition,” in Mycorrhizal Ecology, eds M. G. A. van der Heijden and I. R. Sanders (Berlin: Springer), 117–133. doi: 10.1093/femsec/fiz069
Allison, S. D., and Treseder, K. K. (2008). Warming and drying suppress microbial activity and carbon cycling in boreal forest soils. Glob. Chang. Biol. 14, 2898–2909. doi: 10.1111/j.1365-2486.2008.01716.x
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Andrews, H. J. (2022). Experimental watersheds and gauging stations. long term ecological research. Corvallis, OR: Oregon State University.
Arnold, J. (2004). Forest facts & figures, ed. C. Mitchell (Olympia, WA: Washington Forest Protection Association).
Bagyaraj, D. J., and Ashwin, R. (2017). Soil biodiversity: Role in sustainable horticulture. Biodivers. Horticult. Crops 5, 1–18.
Baldrian, P. (2016). Forest microbiome: Diversity, complexity and dynamics. FEMS Microbiol. Rev. 41, 109–130. doi: 10.1093/femsre/fuw040
Baptista, P., Reis, F., Pereira, E., Tavares, R. M., Santos, P. M., Richard, F., et al. (2015). Soil DNA pyrosequencing and fruitbody surveys reveal contrasting diversity for various fungal ecological guilds in chestnut orchards. Environ. Microbiol. Rep. 7, 946–954. doi: 10.1111/1758-2229.12336
Black, C. A. (1965). Methods of soil analysis: Part I physical and mineralogical properties. Madison, WI: American Society of Agronomy.
Bödeker, I. T. M., Linahl, B. D., Olson, A., and Clemmensen, K. E. (2016). Mycorrhizal and saprotrophic fungal guilds compete for the same organic substrates but affect decomposition differently. Funct. Ecol. 30, 1967–1978. doi: 10.1111/1365-2435.12677
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmed, S. P. (2016). DADA2: High resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Camenzind, T., Weimershaus, P., Lehmann, A., Aguilar-Trigueros, C., and Rillig, M. C. (2022). Soil fungi invest into asexual sporulation under resource scarcity, but trait spaces of individual isolates are unique. Environ. Microbiol. 24, 2962–2978. doi: 10.1111/1462-2920.16012
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Cole, J. R., Wang, Q., Fish, J. A., Chai, B., McGarrell, D. M., Sun, Y., et al. (2014). Ribosomal database project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, 633–642. doi: 10.1093/nar/gkt1244
Cripps, C. L. (1997). The genus Inocybe in Montana aspen stands. Mycologia 89, 670–690. doi: 10.2307/3761005
Crous, P. W., Schubert, K., Braun, U., De Hoog, G., Hocking, A., Shin, H. D., et al. (2007). Opportunistic, human-pathogenic species in the Herpotrichiellaceae are phenotypically similar to saprobic or phytopathogenic species in the Venturiaceae. Stud. Mycol. 58, 185–217. doi: 10.3114/sim.2007.58.07
Crowther, T. W., Glick, H. B., Covey, K. R., Bettigole, C., Maynard, D. S., Thomas, S. M., et al. (2015). Mapping tree density at a global scale. Nature 525, 201–205. doi: 10.1038/nature14967
Dahlberg, A., Jonsson, L., and Nylund, J. E. (1997). Species diversity and distribution of biomass above and below ground among ectomycorrhizal fungi in an old-growth Norway spruce forest in south Sweden. Can. J. Bot. 75, 1323–1335. doi: 10.1139/b97-844
Delgado-Baquerizo, M., Maestre, F. T., Reich, P. B., Jeffries, T. C., Gaitan, J. J., Encinar, D., et al. (2016). Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 7:10541. doi: 10.1038/ncomms10541
Dickie, I. A., Martinez-Garcia, L. B., Koele, N., Grelet, G. A., Tylianakis, J. M., Peltzer, D. A., et al. (2013). Mycorrhizas and mycorrhizal fungal communities throughout ecosystem development. Plant Soil 367, 11–39. doi: 10.3852/11-399
Ehrenfeld, J. G., Ravit, B., and Elgersma, K. (2005). Feedback in the plant-soil system. Annu. Rev. Environ. Resour. 30, 75–115. doi: 10.1146/annurev.energy.30.050504.144212
Frąc, M., Hannula, S. E., Belka, M., and Jędryczka, M. (2018). Fungal biodiversity and their role in soil health. Front. Microbiol. 9:707. doi: 10.3389/fmicb.2018.00707
Franklin, J. F., Spies, T. A., Van Pelt, R., Carey, A. B., Thornburgh, D. A., Berg, D. R., et al. (2002). Disturbances and structural development of natural forest ecosystems with silvicultural implications, using Douglas-fir forests as an example. For. Ecol. Manage. 155, 399–423. doi: 10.1016/S0378-1127(01)00575-8
Frey, B., Niklaus, P. A., Kremer, J., Luscher, P., and Zimmermann, S. (2011). Heavy machinery traffic impacts methane emission, abundance of methanogens and community structure in oxic forest soils. Appl. Environ. Microbiol. 77, 6060–6068. doi: 10.1128/AEM.05206-11
Frøslev, T. G., Kjøller, R., Bruun, H. H., Erjnæs, R., Hansen, A. J., Læssøe, T., et al. (2019). Man against machine: Do fungal fruitbodies and eDNA give similar biodiversity assessments across broad environmental gradients? Biol. Conserv. 233, 201–212. doi: 10.1016/j.biocon.2019.02.038
Gams, W. (2007). Biodiversity of soil-inhabiting fungi. Biodivers. Conserv. 16, 69–72. doi: 10.1007/s10531-006-9121-y
Gardes, M., and Bruns, T. D. (1996). Community structure of ectomycorrhizal fungi in a Pinus muricata forest: Above- and below-ground views. Can. J. Bot. 74, 1572–1583. doi: 10.1046/j.1365-294x.1999.00773.x
Gonthier, P., Giordano, L., Zampieri, E., Lione, G., Vizzini, A., Colpaert, J. V., et al. (2019). An ectomycorrhizal symbiosis differently affects host susceptibility to two congeneric fungal pathogens. Fungal Ecol. 39, 250–256. doi: 10.1016/j.funeco.2018.12.008
Hartmann, M., Niklaus, P. A., Zimmermann, S., Schmutz, S., Kreme, J., Abarenkov, K., et al. (2014). Resistance and resilience of the forest soil microbiome to logging-associated compaction. ISME J. 8, 226–244. doi: 10.1038/ismej.2013.141
Hernandez-Restrepo, M., Gené, J., Castañeda-Ruiz, R. F., Mena-Portales, J., Crous, P. W., and Guarro, J. (2017). Phylogeny of saprobic microfungi from Southern Europe. Stud. Mycol. 86, 53–97. doi: 10.1016/j.simyco.2017.05.002
Hu, Y., Veresoglou, S. D., Tedersoo, L., Xu, T., Ge, T., Liu, L., et al. (2019). Contrasting latitudinal diversity and co-occurrence patterns of soil fungi and plants in forest ecosystems. Soil Biol. Biochem. 131, 100–110. doi: 10.1016/j.soilbio.2019.01.001
iNaturalist (2022). H. J. Andrews experimental forest LTER. Available online at: https://www.inaturalist.org/projects/hj-andrews-experimental-forest-lter
Jayne, B., and Quigley, M. (2014). Influence of arbuscular mycorrhiza on growth and reproductive response of plants under water deficit: A meta-analysis. Mycorrhiza 24, 109–119. doi: 10.1023/a:1004891210871
Jumpponen, A., Jones, K. L., and Blair, J. (2010). Vertical distribution of fungal communities in tallgrass prairie soil. Mycologia 102, 1027–1041. doi: 10.3852/09-316
Kanekar, S. S., Cale, J. A., and Erbilgin, N. (2018). Ectomycorrhizal fungal species differentially affect the induced defensive chemistry of the lodgepole pine. Oecologia 188, 395–404. doi: 10.1007/s00442-018-4231-2
Koljalg, U., Larsson, K. H., Abarenkov, K., Nilsson, R. H., Alexander, I. J., Eberhardt, U., et al. (2005). UNITE: A database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. 166, 1063–1068. doi: 10.1111/j.1469-8137.2005.01376.x
Kranabetter, J. M., Berch, S. M., MacKinnon, J. A., Ceska, O., Dunn, D. E., and Ott, P. K. (2018). Species-area curve and distance-decay relationships indicate habitat thresholds of ectomycorrhizal fungi in an old-growth Pseudotsuga menziesii landscape. Divers. Distrib. 24, 755–764. doi: 10.1111/ddi.12720
Liang, M. X., Shi, L. Q., Burslem, D. F. R. P., Johnson, D., Fang, M., Zhang, X. Y., et al. (2021). Soil fungal networks moderate density-dependent survival and growth of seedlings. New Phytol. 230, 2061–2071. doi: 10.1111/nph.17237
Lim, Y. W., Kim, B. K., Kim, C., Jung, H. S., Kim, B.-S., Lee, J.-H., et al. (2010). Assessment of soil fungal communities using pyrosequencing. J. Microbiol. 48, 284–289. doi: 10.1007/s12275-010-9369-5
Lindahl, B. D., Ihrmark, K., Boberg, J., Trumbore, S. E., Högberg, P., Stenlid, J., et al. (2007). Spatial separation of litter decomposition and mycorrhizal nitrogen uptake in a boreal forest. New Phytol. 173, 611–620. doi: 10.1111/j.1469-8137.2006.01936.x
Luoma, D. L., Frenkel, R. E., and Trappe, J. M. (1991). Fruiting of hypogeous fungi in Oregon Douglas-fir forests: Seasonal and habitat variation. Mycologia 83, 335–353.
Maron, J. L., Marler, M., Kilronomos, J. N., and Cleveland, C. C. (2011). Soil fungal pathogens and the relationship between plant diversity and productivity. Ecol. Lett. 14, 36–41. doi: 10.1111/j.1461-0248.2010.01547.x
McGuire, K. L., Allison, S. D., Fierer, N., and Treseder, K. K. (2013). Ectomycorrhizal-dominated boreal and tropical forests have distinct fungal communities, but analogous spatial patterns across soil horizons. PLoS One 8:e68278. doi: 10.1371/journal.pone.0068278
McGuire, K. L., D’angelo, H., Brearly, F. Q., Gedallovich, S. M., Babar, N., Yang, N., et al. (2015). Responses of soil fungi to logging and oil palm agriculture in Southeast Asian tropical forests. Microb. Ecol. 69, 733–747. doi: 10.1007/s00248-014-0468-4
Nara, K., Nakaya, H., and Hogetsu, T. (2003). Ectomycorrhizal sporocarp succession and production during early primary succession on Mount Fuji. New Phytol. 158, 193–206. doi: 10.1046/j.1469-8137.2003.00724.x
Nepal, P., Johnston, C. M. T., and Ganguly, I. (2021). Effects on global forests and wood production markets of increased demand for mass timber. Sustainability 13:13943. doi: 10.3390/su132413943
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Nilsson, R. H., Anslan, S., Bahram, M., Wurzbacher, C., Baldrian, P., and Tedersoo, L. (2019). Mycobiome diversity: High-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 17, 95–109. doi: 10.1038/s41579-018-0116-y
Norvell, L. L., and Exeter, R. L. (2004). Ectomycorrhizal epigeous basidiomycete diversity in Oregon Coast range Pseudotsuga menziesii forests-preliminary observations. Memoirs N. Y. Bot. Garden 89, 159–190.
ORFI (2020). Oregon forest facts 2019-2020 edition. Portland, OR: Oregon Forest Resources Institute.
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2019). vegan: Community Ecology Package. R package version 2.5-6.
Pelkmans, J. F., Lugones, L. G., and Wösten, H. A. B. (2016). “Fruiting body formation in basidiomycetes,” in The Mycota: A comprehensive treatise on fungi as Experimental systems for basic and applied research:. I. Growth, differentiation and sexuality, 3rd Edn, ed. J. Wendland (Cham: Springer International Publishing), 387–406. doi: 10.1007/978-3-642-36821-9
Porter, T. M., Skillman, J. E., and Moncalvo, J. (2008). Fruiting body and soil rDNA sampling detects complementary assemblage of Agaricomycotina (Basidiomycota, Fungi) in a hemlock-dominated forest plot in southern Ontario. Mol. Ecol. 17, 3037–3050. doi: 10.1111/j.1365-294X.2008.03813.x
Purahong, W., Arnstadt, T., Kahl, T., Bauhus, J., Kellner, H., Hofrichter, M., et al. (2016). Are correlations between deadwood fungal community structure, wood physico-chemical properties and lignin-modifying enzymes stable across different geographical regions? Fungal Ecol. 22, 98–105. doi: 10.1016/j.funeco.2016.01.002
Rhoades, F. M. (1972). Fleshy fungi fruiting in the H. J. Andrews experimental forest: A partial list of collections from fall 1970 to spring 1972. Coniferous Forest Biome Internal Report Nr. 45. Seattle, WA: University of Washington.
Richard, F., Millot, S., Gardes, M., and Selosse, M. (2005). Diversity and specificity of ectomycorrhizal fungi retrieved from an old-growth Mediterranean forest dominated by Quercus ilex. New Phytol. 166, 1011–1023. doi: 10.1111/j.1469-8137.2005.01382.x
Rodriguez-Ramos, J. C., Cale, J. A., Cahill, J. F., Simard, S. W., Justine, K., and Erbilgin, N. (2020). Changes in soil fungal community composition depend on functional group and forest disturbance type. New Phytol. 229, 1–13. doi: 10.1111/nph.16749
Rosinger, C., Sanden, H., Matthews, B., Mayer, M., and Godbold, D. L. (2018). Patterns in ectomycorrhizal diversity, community composition, and exploration types in European beech pine, and spruce forests. Forests 9:445. doi: 10.3390/f9080445
Silva, L. C. R., and Lambers, H. (2021). Soil-plant-atmosphere interactions: Structure, function, and predictive scaling for climate change mitigation. Plant Soil 461, 5–27. doi: 10.1007/s11104-020-04427-1
Smith, J. E., Molina, R., Luoma, D. L., and Castellano, M. (1996). Ectomycorrhizal fungus diversity in Douglas-fir forests of the Oregon Cascades. Mycorrhizas in integrated systems from genes to plant development: Proceedings of the fourth European Symposium on Mycorrhizas. Brussels: European Commission.
Smith, P., House, J. I., Bustamante, M., Sobocka, J., Harper, R., Pan, G., et al. (2016). Global change pressures on soils from land use and management. Glob. Change Biol. 22, 1008–1028. doi: 10.1111/gcb.13068
Song, X., Lim, J. Y., Yang, J., and Luskin, M. S. (2021). When do Janzen-Connell effect matter? A phylogenetic meta-analysis of conspecific negative distance and density dependence experiments. Ecol. Lett. 24, 608–620. doi: 10.1111/ele.13665
Straatsma, G., François, A., and Simon, E. (2001). Species richness, abundance, and phenology of fungal fruit bodies over 21 years in a Swiss forest plot. Mycol. Res. 105, 515–523. doi: 10.1017/S0953756201004154
Sun, H., Santalahti, M., Pumpanen, J., Koster, K., Berninger, F., Raffaello, T., et al. (2015). Fungal community shifts in structure and function across a boreal forest fire chronosequence. Appl. Environ. Microbiol. 81, 7869–7880. doi: 10.1128/AEM.02063-15
Talbot, J. M., Allison, S. D., and Treseder, K. K. (2008). Decomposers in disguise: Mycorrhizal fungi as regulators of soil C dynamics in ecosystems under global change. Funct. Ecol. 22, 955–963. doi: 10.1111/j.1365-2435.2008.01402.x
Thiers, B. M., and Halling, R. E. (2018). The Macrofungi collection consortium. Appl. Plant Sci. 6:e1021. doi: 10.1002/aps3.1021
Thines, M., Crous, P. W., Aime, M. C., Aoki, T., Cai, L., Hyde, K. D., et al. (2018). Ten reasons why a sequence-based nomenclature is not useful for fungi anytime soon. IMA fungus 9, 177–183. doi: 10.5598/imafungus.2018.09.01.11
Walder, F., Niemann, H., Natarajan, M., Lehmann, M. F., Boller, T., and Wiemken, A. (2012). Mycorrhizal networks: Common goods of plants shared under unequal terms of trade. Plant Physiol. 159, 789–797. doi: 10.1104/pp.112.195727
Wahdan, S. F. M., Hossen, S., Tanunchai, B., Schädler, M., Buscot, F., and Purahong, W. (2020). Future climate significantly alters fungal plant pathogen dynamics during the early phase of wheat litter decomposition. Microorganisms 8:908. doi: 10.3390/microorganisms8060908
Keywords: fungi, logging, cascade, soil, litter, legacy, structure, forest
Citation: Spencer MW, Roy BA, Thornton TE, Silva LCR and McGuire KL (2023) Logging has legacy effects on the structure of soil fungal communities several decades after cessation in Western Cascade forest stands. Front. For. Glob. Change 6:791766. doi: 10.3389/ffgc.2023.791766
Received: 08 October 2021; Accepted: 17 February 2023;
Published: 09 March 2023.
Edited by:
Rui S. Oliveira, University of Coimbra, PortugalReviewed by:
Benoit Marçais, INRA Centre Nancy-Lorraine, FranceCopyright © 2023 Spencer, Roy, Thornton, Silva and McGuire. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Max W. Spencer, bXdzQHVvcmVnb24uZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.