
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. For. Glob. Change , 14 March 2023
Sec. Pests, Pathogens and Invasions
Volume 6 - 2023 | https://doi.org/10.3389/ffgc.2023.1124754
This article is part of the Research Topic "Forentomics": forest pest and pathogen biology, ecology, and management using omics View all 10 articles
The Eurasian spruce bark beetle (ESBB), Ips typographus, has recently caused catastrophic damage to Norway spruce (Picea abies) forests in Europe, resulting in the loss of more than 100 million cubic meters of wood. Traditional forest management strategies have failed to constrain the growing infestation rate; hence, novel measures must be deployed. A better understanding of ESBB physiology and adaptation to host allelochemicals may provide a platform for future management strategies using molecular tools such as RNA interference. To understand ESBB physiology and adaptation, the current study unraveled the gene expression dynamics of ESBB in different life stages and tissues. We obtained ESBB transcriptomes for different life stages [larvae (L1, L2, and L3), pupa, callow, and sclerotized adult] and male/female tissues (gut, fat body, and head) from callow and sclerotized adult beetles. Differential gene expression analysis (DGE) identified multiple gene families related to detoxification, digestion, resistance, and transport in different life stages and tissues of the beetle. Gene Ontology (GO) enrichment revealed 61 critical metabolic pathways enriched across all DGE comparisons. DGE analysis further pinpointed the differential expression of essential genes involved in detoxification, digestion, transport, and defense in various tissues and life stages. RT-qPCR experiments and enzymatic assays corroborated the findings further. The catalogue of differentially expressed genes identified in ESBB could aid better understanding of ESBB physiology and adaptation to hosts and serve as targets for future RNAi-based ESBB management.
Ips typographus, the Eurasian spruce bark beetle, is one of the most destructive forest pests of Norway spruce (Picea abies), causing considerable damage throughout Europe (Huang et al., 2020; Fang et al., 2021; Hlásny et al., 2021). They attack trees weakened due to drought or windthrow when the bark beetle population is in an endemic phase (Jakuš et al., 2011; Biedermann et al., 2019; Hlásny et al., 2019; Netherer and Hammerbacher, 2022). The host selection by ESBB is based on the attraction via pheromones and kairomones, viz. 2-methyl-3-buten-2-ol and 4S-(–)-cis-verbenol, and repellence due to non-host volatiles/anti-attractants such as verbenone (Faccoli et al., 2005; Zhang et al., 2012; Unelius et al., 2014; Netherer et al., 2021). Males are the pioneering sex, and after colonizing the host, they release aggregation pheromones to attract conspecifics, resulting in mass attacks. ESBBs infest healthy trees during an epidemic stage and subsequently kill them (Biedermann et al., 2019). The defense mechanism of the conifer trees can be overwhelmed by the detoxification machinery of the beetle and their resident symbionts, such as bacteria and fungi that can metabolize plant defense chemicals to semiochemicals or sequester them to less toxic and more usable carbon resource forms (Krokene and Solheim, 1998; Faldt et al., 2006; Hammerbacher et al., 2013; Davis, 2015; Cale et al., 2019; Chakraborty et al., 2020a,b). Upon encountering a suitable host, the aggressive beetles bore into the intercortical regions where they feed on the phloem tissues, lay eggs, and the emerging larvae develop by feeding on the surrounding tissues, forming characteristic galleries. After completing the development, ESBB adults leave these galleries, swarm around the forest, and attack new trees to start the cycle again (Hlásny et al., 2019).
Conifers have evolved complex constitutive and inducible defenses to protect themselves against herbivores and their associates, including anatomical barriers and secondary metabolites, such as phenolics and terpenes (mono-, sesqui-, and diterpenes) produced via phenylpropanoid and terpene synthase pathways (Franceschi et al., 2005; Biedermann et al., 2019; Celedon and Bohlmann, 2019). Herbivore insects counteract these responses by employing various coping strategies, such as avoidance, sequestration, target site mutation, and enhanced metabolism/detoxification (Heckel, 2014; Meng et al., 2015; Gaddelapati et al., 2018; Hilliou et al., 2021; Lu et al., 2021; Bras et al., 2022). During detoxification, insects produce enzymes that convert the toxic allelochemicals to less toxic, easily excretable forms by oxidation, hydroxylation, or glycosylation. Such complex biochemical reactions are carried out by enzymes categorized as phase I, phase II, and, more recently, phase III enzymes (Hilliou et al., 2021). The primary or phase I enzymes target the lipophilic groups via oxidation, dihydroxylation, or dealkylation, rendering them water-soluble forms. In contrast, the phase II or secondary enzymes attack the epoxide intermediates of the phase I molecules, which can then be excreted through body fluid. The enzymes involved in phase I detoxification include cytochrome P450s (P450), carboxyl/choline esterases (CCE), and other esterases (i.e., acetylcholinesterase-AChE, JH esterase) that carry out the oxidation, hydrolysis, and reduction of primary toxic molecules. These molecules are then conjugated with glutathione molecules by glutathione S-transferases (GSTs) or glycosylated by UDP-glucuronosyltransferases/UDP-glycosyltransferases (UGTs), which are considered phase II enzymes (Heidel-Fischer and Vogel, 2015; Krempl et al., 2016; Jin et al., 2019). Secondary enzymes also include sulfotransferase and ATP-binding cassette (ABC) transporters. However, ABC transporters have recently been reported as phase III enzymes that obtain energy from the hydrolysis of ATP to transport the substrate across the lipid membrane and facilitate the excretion of entomotoxic compounds (Jin et al., 2019; Hilliou et al., 2021).
Dendroctonus species, close relatives of ESBB, have been well studied regarding their gene expression toward host toxins. In D. armandii, sixty-four P450s were identified in different developmental stages (larvae, pupa, and adult) and validated with RT-qPCR; nineteen CYP genes were upregulated upon host feeding (Dai et al., 2015). The role of the CYP4 and CYP9 families in detoxification was also confirmed in D. rhizophagus (Cano-Ramirez et al., 2013; Sarabia et al., 2019). In D. ponderosae, three CYPs were identified as involved in monoterpene oxidation, and one CYP was implicated in aggregation. Furthermore, transcript levels of several GSTs, ABC transporters, esterases, and dehydrogenases have been reported to increase significantly in mountain pine (D. ponderosae) beetles upon host feeding (Robert et al., 2013; Chiu et al., 2019a,b). Various Ips species, like I. paraconfusus and I. pini, are reported to deploy sex-specific expression of CYPs to convert host allelochemicals to de novo produced aggregation pheromones (Huber et al., 2007; Song et al., 2013; Tittiger and Blomquist, 2017; Blomquist et al., 2021). For ESBB, the roles of most of the detoxifying enzymes and the related genes upon mass attack have yet to be identified. ESBBs are exposed to various levels of host bark chemicals throughout the life cycle; as a result, the expression or catalytic activity of beetle defense enzymes may fluctuate according to the life stage or phase of the detoxification process. Frequent ESBB outbreaks in the recent past and the projected increased frequency of such outbreaks have created a high demand for target-specific control of ESBBs, as conventional methods have proved inefficient (Hlasny et al., 2021; Joga et al., 2021). Hence, detailed knowledge of the detoxification enzyme expression dynamics in the different life-stages of the ESBB is necessary to obtain the target genes for future species-specific management. Furthermore, the recent release of the ESBB genome has opened such captivating research avenues by giving insights into the ubiquitous presence of phase I, II, and III enzymes (84 P450s, 58 GSTs, 86 UGTs, 75 ABC transporters, and 59 CCE families) in the species (Powell et al., 2021). In the present study, we performed screening of the various developmental stages from larval, callow adults, and fully emerged sclerotized beetles, and different tissues of ESBB to catalogue genes differentially expressed and putatively involved in the processes of detoxification, digestion, and transport. We captured the expression dynamics of detoxification machinery and other physiologically essential genes in ESBB. Our findings have led to a better understanding of ESBB physiology and offer valuable targets for RNAi-based control measures (Joga et al., 2021).
All ESBB samples (Figure 1 and Supplementary material 1) were taken directly from beetle-infested logs from Rouchovany in Czechia (49°04′08.0″N 16°06′15.4″ E, 360 m above sea level, a warm and drought-prone area with regular forest management by State Forest Enterprise) during the infestation season in May 2019. The average temperature was around 15°C during collection. The larvae (L1, L2, and L3), pupae, callow, and sclerotized adult beetles were snap-frozen in liquid nitrogen, and the target tissues (gut, head, and fat body) were dissected in sterile conditions and collected in RNAlater™. The sex of the collected beetles was determined by pronotum hair density, as has been described previously (Chakraborty et al., 2020a; Powell et al., 2021). The collected developmental stages and tissues were stored at −80°C until total RNA extraction, as we previously described (Powell et al., 2021). ESBBs were also reared in an insect-rearing chamber with fresh Norway spruce logs at 27 ± 1°C, 70 ± 5% humidity, and a 16:8-h light/dark (L: D) photoperiod. A new set of samples (developmental stages and tissues) was obtained from the F1 generation, and the extracted RNA and protein were used in RT-qPCR and enzymatic experiments, respectively.
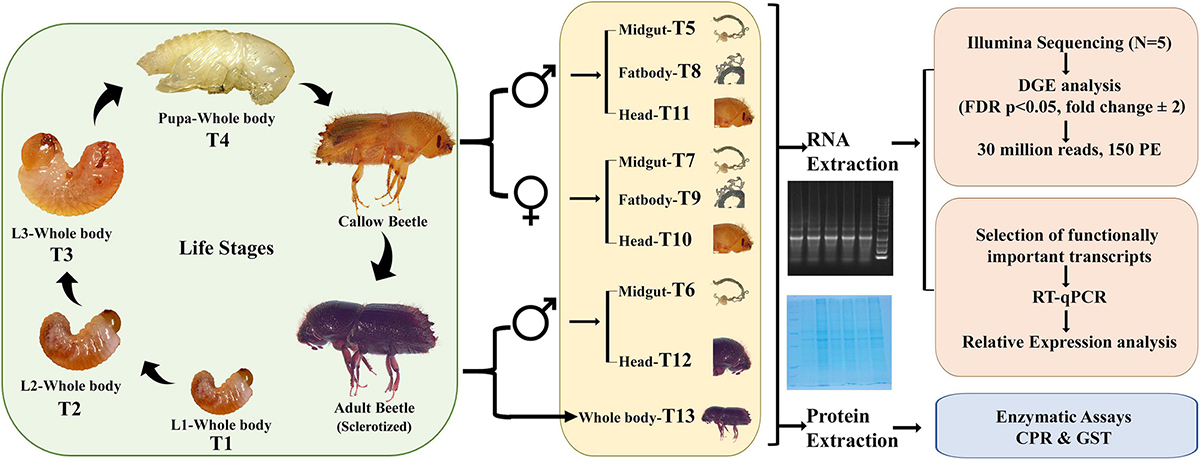
Figure 1. Study design. Larvae 1 (L1/T1), Larvae 2 (L2/T2), Larvae 3 (L3/T3), Pupa (T4), Callow male gut (T5), Sclerotized adult gut (T6), Callow female gut (T7), Callow male fat body (T8), Callow female fat body (T9), Callow female head (T10), Callow male head (T11), Sclerotized adult male head (T12), Sclerotized adult whole body (T13) (CPR, Cytochrome P450; GST, Glutathione S-transferase).
Various ESBB life stages and tissues were used to create transcriptome libraries [larval stage 1 (T1), larval stage 2 (T2), larval stage 3 (T3), pupa (T4), callow male gut (T5), sclerotized adult male gut (T6), callow female gut (T7), callow male fat body (T8), callow female fat body (T9), callow female head (T10), callow male head (T11), sclerotized adult male head (T12), and sclerotized adult whole-body (T13)] (Figure 1 and Supplementary material 1) during the Ips typographus genome study (Powell et al., 2021). Precisely, mRNA in samples was enriched using oligo (dT) beads, and cDNA libraries were prepared using NEB Next® Ultra™ RNA Library Prep Kit followed by Illumina sequencing (Illumina Novaseq6000) to generate 30 million reads (150 paired-end) for each sample. Five biological replicates were sequenced per sample (Powell et al., 2021). The raw reads were processed using CLC workbench (CLC version 21.0.5, Qiagen, Denmark). The sequence data were submitted to NCBI (PRJNA679450) and reused for the present gene expression study. For differential gene expression analysis (DGE), raw reads were mapped back to the I. typographus reference genome (Powell et al., 2021), and the read counts were obtained using CLC workbench (version 21.0.5). Gene expression quantification was obtained by employing standard pre-optimized settings and parameters, such as mapping to exon regions. The TMM-adjusted log CPM counts (similar to the EdgeR approach) were also used to correct biases in the sequence datasets and abundance, resulting in accurate estimations of relative expression levels as described by Robinson et al. (2010). A multi-factorial statistical analysis based on a negative binomial Generalized Linear Model (GLM) was used to identify differentially abundant transcripts between developmental stages and tissues in varied comparisons deploying an FDR corrected p-value < 0.05; fold change ± 2 was used as a threshold for differentially expressed transcripts (DET). Using the GLM model permitted fitting the curves to expression values without presuming that the error on the values was normally distributed. DEGs were functionally annotated using the “cloud blast” feature within the “Blast2GO Plugin” in the CLC Genomic Workbench 21.0.5 (Qiagen, Denmark). A nucleotide blast was done against the NCBI Nr database with an E-value cut-off of 1.0E-5. Both annex and GO slim were used to improve the GO term identification further by crossing the three GO categories (biological process, molecular function, and cellular component) to search for name similarities, GO term, and enzyme relationships within KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways (Roy et al., 2017; Powell et al., 2021).
Ribonucleic acid was isolated from each life stage and tissue using the PureLink™ RNA Kit from Ambion (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. For the ESBB tissue samples, 10 guts, 10 fat bodies and 5 heads were dissected and pooled. Eight L1 larvae, eight L2 larvae, five L3 larvae, five pupae, and two adults were used to obtain enough RNA from each developmental phase for downstream processing. The total extracted RNA was then treated with DNase I (TURBO DNase Kit, Ambion, Austin, TX, USA). The integrity of the purified total RNA was checked on 1.2% agarose gel. One μg of total RNA was used for cDNA synthesis using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems-Life Technologies, Waltham, MA, USA) and stored at −20°C. The cDNA samples were diluted 10-fold before being used as a template in RT-qPCR experiments. Four biological replicates from each sample were used in each RT-qPCR assay. The IDT PrimerQuest software (IDT, Belgium) was used to design the primers (Supplementary material 2). RT-qPCR was carried out for all the samples (T1-T9 and T13). The 10 μL RT-qPCR reactions contained 5.0 μL of SYBR® Green PCR Master Mix (Applied Biosystems), 1.0 μL of cDNA, 1.0 μL of 10 μM forward and reverse primers, and 3.0 μL RNase-free water (Invitrogen). The reactions were performed in an Applied Biosystems™ StepOne™ Real-Time PCR System (Applied Biosystems) with the following conditions: initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C for 1 min, and dissociation curve analysis during which temperature was increased from 60 to 95°C. The expression levels of the target genes were calculated using the 2–ΔΔCt method (Livak and Schmittgen, 2001). RPL7 and RPS7 were used as reference genes for expression normalization (Sellamuthu et al., 2022).
To consider differences in the enzyme activity levels between the ESBB developmental stages, biochemical assays for Glutathione S-transferase (GST) and Cytochrome P450 Reductase (CYP450) were performed as described previously (Habig and Jakoby, 1981; Ortego et al., 1999; Bosch-Serra et al., 2021). The developmental stages and gut samples of ESBB were collected and homogenized in sodium phosphate buffer (50 mM) solution at pH 7.0. The homogenate was centrifuged at 10,000 rpm for 10 min at 4°C. Precisely, eight L1 larvae (T1), six L2 larvae (T2), four L3 larvae (T3), three pupae (T4), two sclerotized adults, and ten guts were pooled to extract enough protein from each sample category. The extract was collected to serve as a source of the enzyme, and enzyme activities were corrected using the protein concentration as a standard correction factor. The total protein content of the enzyme solution was determined by the Bradford method using bovine serum albumin as the standard (Bradford, 1976).
The activity of glutathione S-transferase (GST) was measured using a spectrophotometric method with 5 mM reduced glutathione (GSH) and 1.5 mM 1-chloro-2, 4-dinitrobenzene (CDNB) as substrates (Habig and Jakoby, 1981; Bosch-Serra et al., 2021). One unit of glutathione S-transferase activity is the amount of enzyme-catalyzing 1 μmol of CDNB -GSH conjugate per minute at 30°C. For the GST activity, the reaction solution contained 20 μl of protein extract (Supplementary Table 2), 170 μl of 5 mM GSH in sodium phosphate buffer (pH 7.0), and 10 μL1.5 mM CDNB to make a 200 μl total reaction volume in a transparent micro-plate (96-well Thermo Scientific™ Sterilin™ Clear Microtiter™ Plates). For blanks, 20 μl of sodium phosphate buffer (50 mM pH 7.0) was used instead of protein extracts. After a 1 min incubation period at 30°C, the absorbance of the reaction product [CDNB-glutathione (GSH) conjugate] was measured at 340 nm at intervals of 1 min for up to 10 min using the microplate reader (Agilent BioTek Cytation 5). GST activity was expressed as nmol substrate conjugated⋅min-1 ⋅mg-1 protein using a CDNB molar extinction coefficient (9.6 mM–1 × cm–1). The final specific activity was expressed in μmol of CDNB-glutathione min–1 mg–1 of total protein extracted (Habig and Jakoby, 1981).
Cytochrome P450 Reductase (CYP450) activity was measured according to Ortego et al. (1999) and Bosch-Serra et al. (2021). The 200 μl reaction was prepared in a clear micro-plate (96-well Thermo Scientific™ Sterilin™ Clear Microtiter™ Plates). It comprised 20 μl of protein extract or sodium phosphate buffer (blank) (Supplementary Table 2) and 160 μl of NADPH-generating system [0.6 mM β-nicotinamide adenine dinucleotide phosphate (NADP), 2.8 mM D-glucose-6-phosphate (G6P), and 0.28 units of glucose-6-phosphate dehydrogenase (G6PD)]. The reaction was initiated by adding 20 μl of 1 mM cytochrome C in sodium phosphate buffer (pH 7.0), and the absorbance was measured at 550 nm at intervals of 1 min for up to 25 min using the microplate reader (Agilent BioTek Cytation 5). CYP450 activity was expressed as nmol substrate conjugated⋅min–1 ⋅mg–1 protein using a cytochrome C molar extinction coefficient (27.6 mM–1 × cm–1) (Margoliash and Frohwirt, 1959). The enzymatic activity was measured in cytochrome P450 equivalent unit min–1 mg–1 of the total protein extracted.
First, we conducted pairwise comparisons with Student’s t-test to consider the differences in RT-qPCR determined gene transcript levels for 19 selected genes related to detoxification. The pairwise comparisons included callow gut vs. sclerotized male gut, and callow female fat body vs. callow male fat body. For multiple comparisons in the relative gene expression of 19 genes between different developmental stages and gut tissues of ESBB, Tukey’s HSD was used. Lastly, one-way ANOVA was used to test the significance of differences in enzymatic activities between ESBB developmental stages and tissues. All these analyses were performed in RStudio (version 4.2.2). The statistical analysis deployed for DGE data analysis is described above in the “Transcriptome sequencing, library preparation, and analysis” section.
From 65 libraries (13 samples × 5 biological replicates), 5,199,150,714 reads were retrieved, and 3,728,549,638 reads were mapped back to the reference genome of I. typographus (Powell et al., 2021) to generate the count table (Supplementary Table 1). Using CLC workbench 21.0.5 (QIAGEN Aarhus, Denmark), DGE comparisons were performed among the 13 samples (T1 to T13) (Figure 1). The number of DEGs, i.e., up- or downregulated genes, were reported based on the cut-off of FDR p < 0.05 and fold change ± 2 considered as the threshold for differential expression. We performed DGE for the following perspectives: (A) life-stage comparison: T1, T2, T3, T4, and T13 (from larvae to adult); (B) tissue-specific comparison: (i) T7, T8, and T9, and (ii) T5, T9, and T11; (C) sex-specific comparison: (i) T7 vs. T5, and (ii) T9 vs. T8; and (D) comparison between gut tissues in different adults stages: T6 vs. T5 (sclerotized vs. callow male guts) (Table 1, Supplementary Figure 1, and Supplementary materials 1, 3–13).
A group-wise comparison was made between the five samples (three larval stages, pupa, and adult) to explore the differential gene expression across the developmental stages (Supplementary material 3). The principal component analysis (PCA plot) resulted in the segregation of samples from different life stages, indicating life-stage-specific gene expression in ESBB (Figure 2A). The gene expression dynamics in different life stages of ESBB were represented as a heatmap (Figure 2B). We found that detoxifying enzyme families (i.e., cytochrome P450s/CYPs, UDP-glucuronosyltransferases/UGTs, acetylcholinesterase/AchE, hydrolases, dehydrogenases, peroxidases, carboxylesterases, esterases, ABC transporters, and glutathione S-transferases/GSTs) showed differential gene expression (up- and downregulation) in larvae and adult stages (Figures 2C, D). A graph was plotted using mean CPM values (FDR corrected p < 0.05) for essential detoxification genes/enzymes (P450s, GSTs, UGTs, carboxylesterases, esterases, and ABC transporters). The expression of detoxification-related genes increased from L1 to L2, decreased in L3 and pupa, and increased in adults (Figure 2D). Some other detoxifying enzymes, such as dehydrogenases, hydrolases, and aminoacylases, also revealed a similar pattern (Supplementary Figure 2).
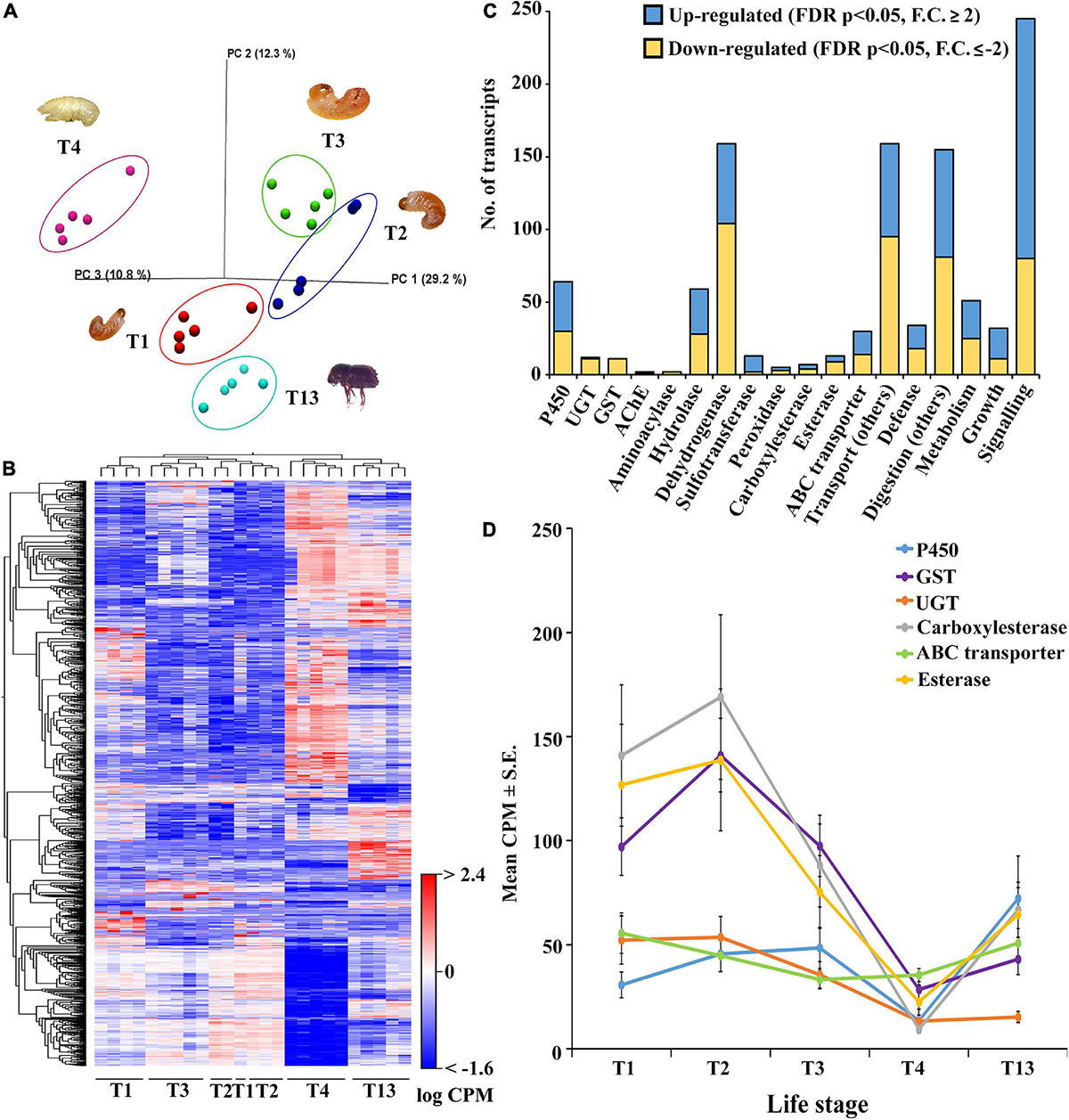
Figure 2. Group-wise comparison between different developmental life stages; larvae 1-3 (T1-T3), pupae (T4), and sclerotized adult whole body (T13) (N = 5). (A) PCA plot showing sample clustering. (B) Heat maps representing gene clustering. The color spectrum, stretching from blue to red, represents TMM-adjusted log CPM expression values obtained after DGE analysis. (C) Bar graphs showing the number of transcripts (CPM) for differentially expressed contigs of specific detoxification-related enzyme families and other essential enzymes related to defense, digestion, transport, metabolism, signaling, and growth after group-wise comparison between different developmental life stages. (D) Detoxification enzyme dynamics during different life stages. Average CPM ± standard error values of significantly expressed genes are plotted against each enzyme on the x-axis corresponding to the life stages on the y-axis. The total CPM was calculated by taking the average of all the differentially expressed contigs of each gene family. Analysis was done using CLC workbench (21.0.5, Qiagen, Denmark) with FDR p < 0.05 and a fold change ± 2 cut-off.
Most of the cytochrome and esterase family genes showed no variation in gene expression across the life stages of ESBB, indicating their importance for beetle survival under the bark (Figure 3A). The expression of cytochrome P450 genes was high in the larval and adult stages. The present study identified differentially expressed cytochrome P450 genes primarily for family 4, with a few belonging to families 6 and 9 (Figure 3). In the upregulated category, CYP4BH1, CYP6CR2, chorion peroxidase, sphingomyelin phosphodiesterase-like and esterase FE4-like gene were only over-expressed in adults, whereas CYP306A1, CYP6A1-like, cAMP and cAMP-inhibited cGMP 3′,5′-cyclic phosphodiesterase 10A-like, and palmitoleoyl-protein carboxylesterase NOTUM-like were overexpressed only in pupae (Figure 3A). In the downregulated category, most genes were downregulated in pupae compared to other ESBB life stages, with CYP410A1 showing the lowest gene expression level. In adults, the most highly upregulated gene was CYP4G27 (Figure 3A), and the lowest gene expression was recorded for GST 1 isoform D (Figure 3B). The 19 transcripts involved in pheromone biosyntheses, like ipsdienol dehydrogenase, juvenile hormone (JH) epoxide hydrolase, JH esterase, and JH acid O-methyltransferase, were also reported to be primarily overexpressed in adults (T13) and larval stage L2 (T2) and L3 (T3) and were least expressed in pupae (T4) (Supplementary material 3).
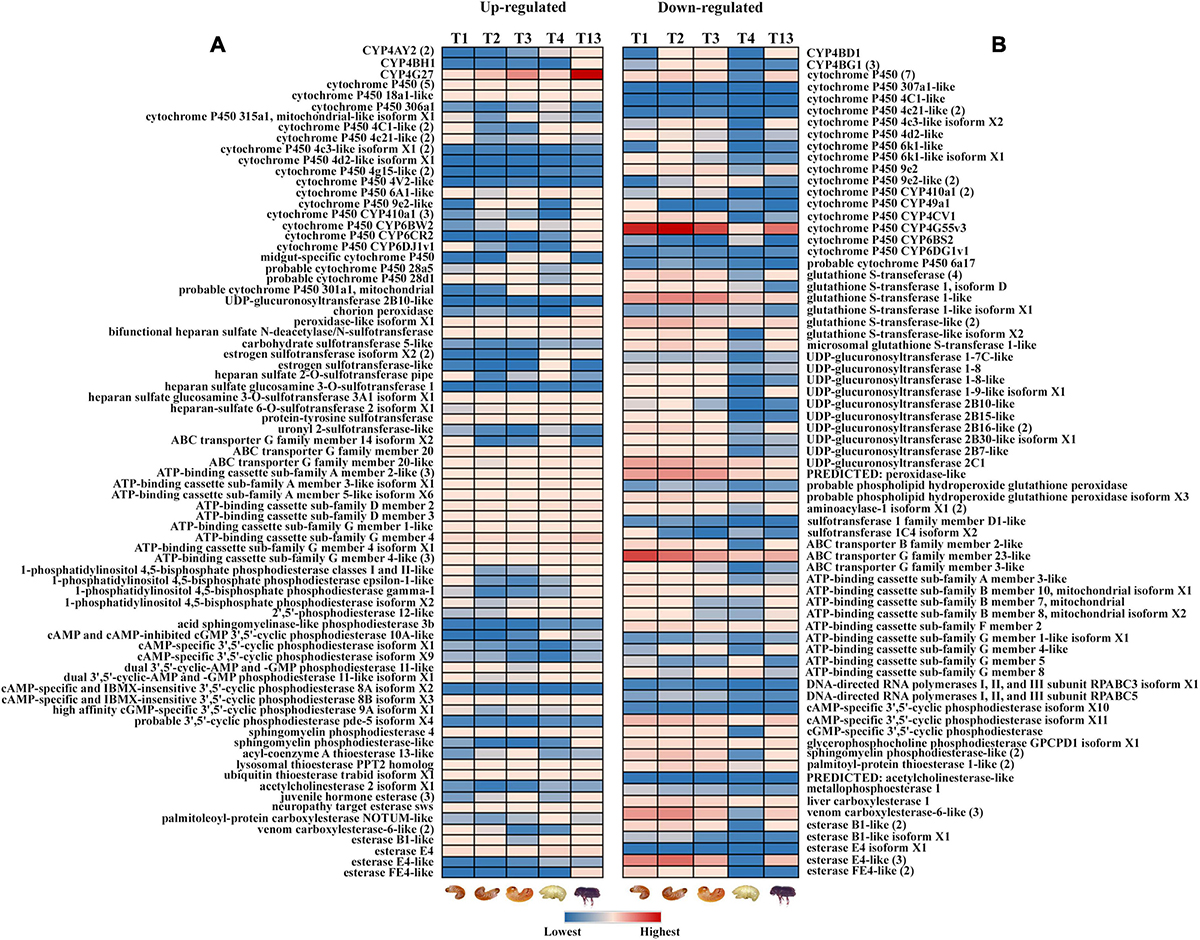
Figure 3. Expression profile of detoxification-related genes in different life stages of ESBB. (A) Upregulated genes; (B) downregulated genes. The details of the genes are given in Supplementary Excel 3. The expression level is represented in different colors indicating the TMM-adjusted log CPM expression mean values. Analysis was done with FDR p < 0.05 and fold change ± 2 cut-off in CLC workbench (21.0.5, Qiagen, Denmark).
The effect of bark feeding and encountering the host at different developmental stages was studied by comparing the feeding stages (larva and adult; T1, T2, T3, and T13) against the non-feeding stage (pupae; T4) (Supplementary materials 4–7). This revealed the complete segregation of gene expression between the feeding and non-feeding stages (Figures 4A–D). Compared to T4, the transcript levels of most of the genes of interest were significantly elevated in T1, T2, T3, and T13. The T2 vs. T4 comparison revealed the most differentially expressed gene transcripts, i.e., 8,319 (4,896 downregulated, 3,423 upregulated). We discovered that 1,798 gene transcripts were commonly upregulated, and 1,060 transcripts were commonly downregulated in all feeding stages, including larvae and adults, with the dehydrogenases and cytochromes being the most differentially expressed (Figures 5, 6 and Supplementary materials 36, 37). Two GST transcripts, namely, GST-like isoform X2 and microsomal GST 1-like, had higher expression in the feeding stages (adults and larvae) than the non-feeding pupal stage, indicating their putative involvement in digestion and the detoxification of host allelochemicals.
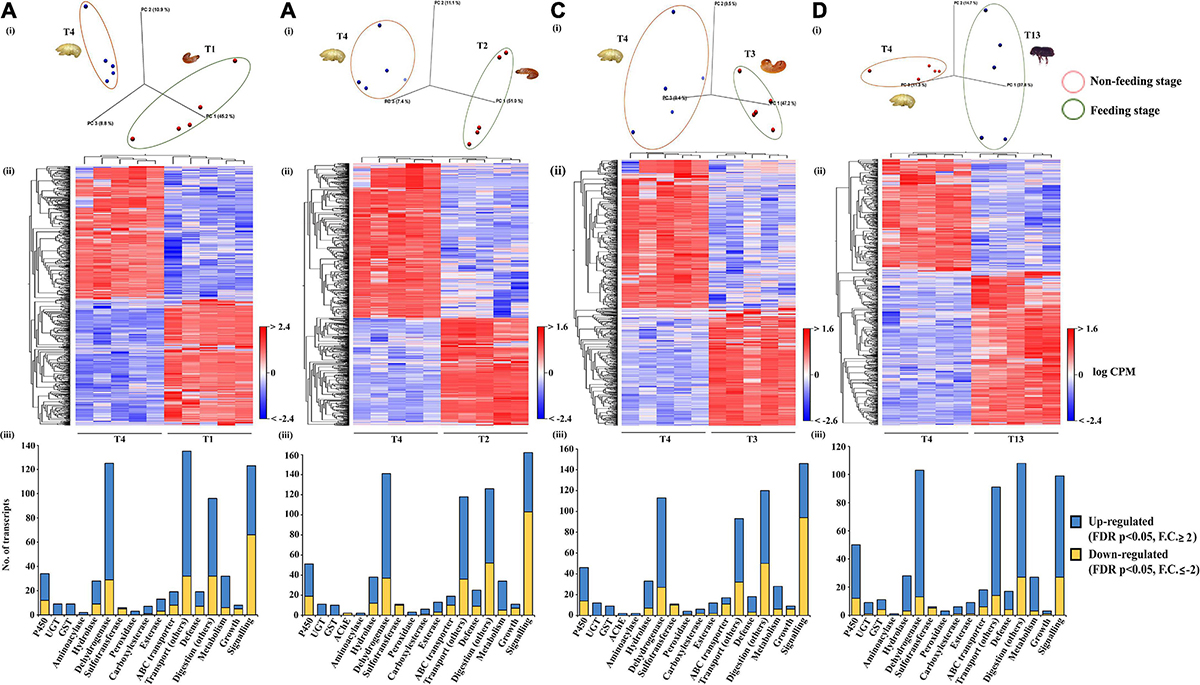
Figure 4. Feeding vs. non-feeding stage comparison. Pupa (T4, non-feeding stage) was compared with different feeding stages (T1, T2, T3, and T13) (N = 5). (A) PCA plot showing sample clustering. (B) Heat maps representing gene clustering. The color spectrum, stretching from blue to red, represents TMM-adjusted log CPM expression values obtained after DGE analysis. (C,D) Bar graphs showing differentially expressed contigs of detoxification-related enzyme families and other essential enzymes related to defense, digestion, transport, metabolism, signaling, and growth between the feeding vs. the non-feeding stage. Analysis was done using the CLC workbench using FDR p < 0.05 and fold change ± 2 as the threshold for significance.
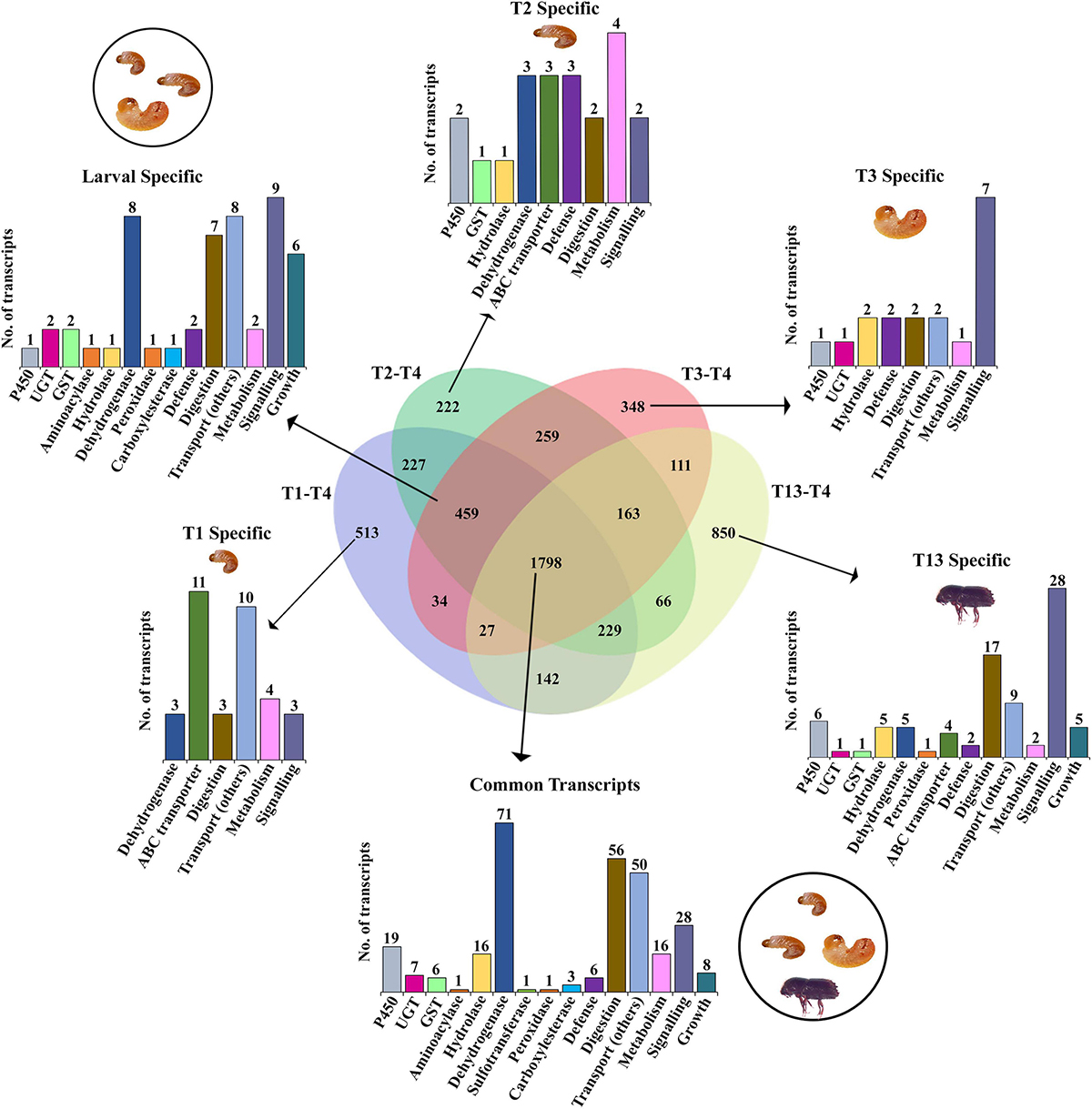
Figure 5. Venn diagram representation of feeding vs. non-feeding stage comparison for upregulated enzymes using FDR p < 0.05 and fold change ≥ 2 as the threshold for significance. Numbers within the Venn diagram represent the number of contigs found for each group. The bar graph corresponds to the number of contigs of each gene family identified from the list (Supplementary material 36).
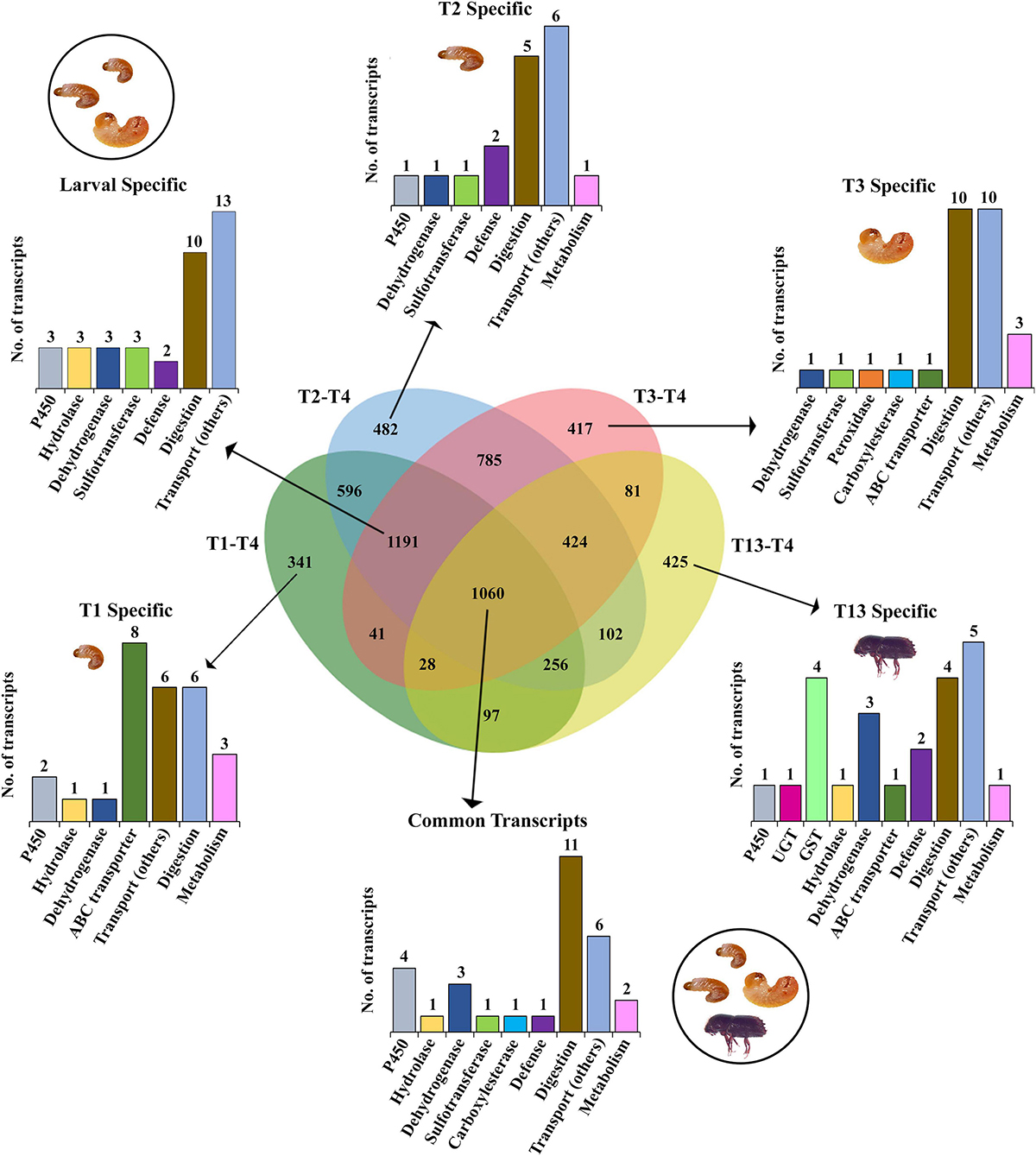
Figure 6. Venn diagram representation of feeding vs. non-feeding stage comparison for downregulated enzymes deploying FDR p < 0.05 and fold change ≤ –2 the threshold for significance. Numbers within the Venn diagram represent the number of contigs found for each group. The bar graph corresponds to the number of contigs for each gene family identified from the list (Supplementary material 37).
We compared tissues from both females and males separately to capture similarities and sex-specific differences in gene expression in different tissues, which are crucial to understand ESBB physiology (Figures 7A, B). In both males and females, the expression of genes related to detoxification, digestion, defense, and metabolism was highest in the gut, followed by in the fat body, and lowest in head tissues.
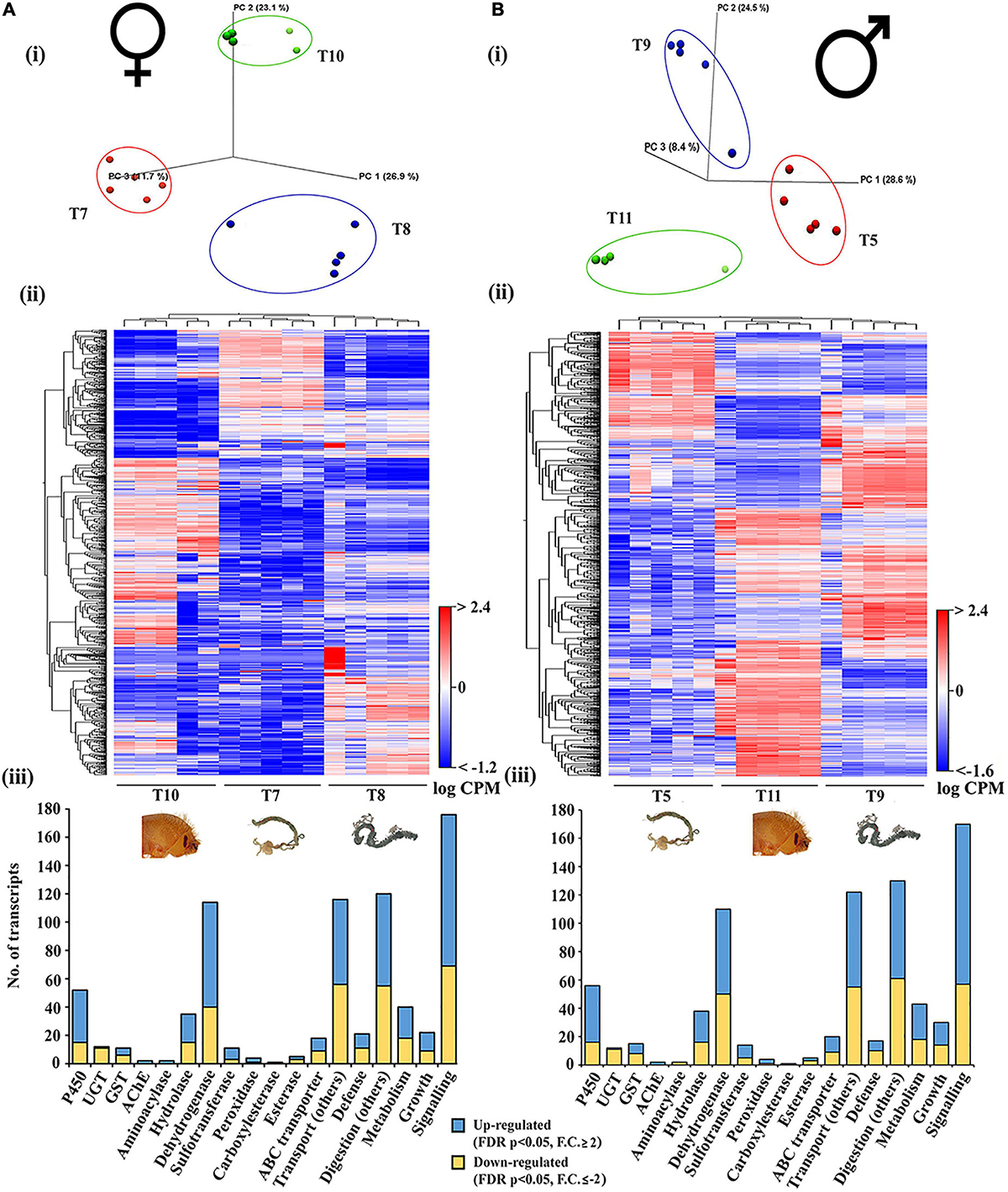
Figure 7. Tissue-specific comparison in callow male and female beetles. (A) Callow female head (T10), Callow female gut (T7), and Callow female fat body (T8) (N = 5). (B) Callow male head (T11), Callow male gut (T5), and Callow male fat body (T9) (N = 5). (i) PCA plots showing sample clustering. (ii) Heat maps representing gene clustering. The color spectrum, stretching from blue to red, represents TMM-adjusted log CPM expression values obtained after DGE analysis. (iii) Bar graphs showing differentially expressed contigs of detoxification-related enzyme families and other essential enzymes related to defense, digestion, transport, metabolism, signaling, and growth after group-wise comparison between different developmental life stages. Analysis was done using the CLC workbench using FDR p < 0.05 and fold change ± 2 as cut-offs for significance.
The callow female head (T10), fat body (T8), and gut (T7) were compared and clustered on a group-by-group basis (Figure 7A, i, ii and Supplementary material 8). There were 6,024 genes upregulated (FDR corrected p < 0.05, fold change ≥ 2), including 74 dehydrogenases, 37 CYP/P450s, 20 hydrolases, 9 ABC transporters, 8 sulfotransferases, 5 GSTs, 3 peroxidases, 2 each of the AchE, aminoacylase, and esterase sub-families, and 1 UGT and carboxylesterase (Figure 7A, iii). Among the 4,112 downregulated transcripts (FDR corrected p < 0.05, fold change ≤ −2) were 40 dehydrogenases, 15 CYP/P450s, 11 UGTs, 15 hydrolases, 9 ABC transporters, 6 GSTs, 3 of each sulfotransferase and esterase subfamilies, and 1 peroxidase. The upregulated transcripts have molecular functions that relate to defense (21 transcripts), signaling (176 transcripts), growth (22 transcripts), transport (116 transcripts, excluding ABC transporter families), digestion (120 transcripts, excluding esterase sub-families), and metabolism (40 transcripts).
The callow male head (T11), fat body (T9), and gut (T5) were compared and clustered on a group-by-group basis (Figure 7B, i, ii and Supplementary material 9). There were 10,464 transcripts substantially expressed (FDR corrected p < 0.05), with 6,178 upregulated (fold change ≥ 2) and 4,287 downregulated (fold change ≤ −2). The upregulated transcripts included 40 CYP/P450s, 1 UGT, 2 AchEs, 7 GSTs, 22 hydrolases, 60 dehydrogenases, 9 sulfotransferases, 3 peroxidases, 1 carboxylesterase, and 2 esterase subfamily genes. The downregulated transcripts included 16 CYP/P450s, 11 UGT, 8 GSTs, 2 aminoacylases, 16 hydrolases, 50 dehydrogenases, 5 sulfotransferases, 1 peroxidase, and 3 esterase subfamily genes (Figure 7B, iii). A total of 17 transcripts had molecular functions associated with defense, 130 with digestion, 30 with growth, 170 with signaling, 122 with transport (excluding ABC transporters), and digestion (excluding esterase subfamilies).
We compared the callow male and female ESBB guts (T7 vs. T5) and fat bodies (T9 vs. T8) for gene expression dynamics during maturation feeding (Supplementary Figures 3A, B).
The callow male gut had higher expression levels than the callow female gut for most of the transcripts involved in vital physiological functions, such as detoxification, defense, digestion, transport, metabolism, signaling, and growth (Supplementary Figure 3A, i, ii, iii and Supplementary material 10). Most of the transcripts were upregulated in the male gut (T5), including four cytochrome P450s (301A1, 4G15-like, 4C21-like, CYP410A1), 14 dehydrogenases, 1 ubiquinone oxidoreductase (defense), 6 serine/threonine-protein kinases and 1 phosphatase (signaling), 1 transporter protein and 1 trehalose transporter (transport), 1 serine protease, and 1 phosphodiesterase.
The callow female and male fat body (T9 vs. T8) comparison showed 7,268 differentially expressed transcripts (FDR-corrected p < 0.05); 4,501 were upregulated (fold change ≥ 2), and 2,767 were downregulated (fold change ≤ −2) (Supplementary Figure 3B,i, ii and Supplementary material 11). Most transcripts with molecular functions related to detoxification, defense, digestion, and metabolism had higher expression in the callow male fat body, which is plausibly linked to future host colonization as pioneers (Supplementary Figure 3B iii). Thirteen transcripts related to pheromone biosynthesis were differentially expressed, such as juvenile hormone esterases, epoxide hydrolase, ipsdienol dehydrogenases, juvenile hormone acid O-methyltransferase, and pheromone binding proteins between males and female ESBB fat body. Of these, nine were upregulated in male (T9) and four in female ESBB fat bodies (T8). None of these genes were found to be expressed in the gut comparison.
The current comparison dealt with the gene expression dynamics in ESBB adults during maturation feeding (callow beetles) and post-maturation feeding (sclerotized). As demonstrated by the PCA plot and heatmap (Figures 8A, B and Supplementary material 12), there was a significant difference in gene expression between sclerotized (black) and callow male gut (T6 vs. T5). A total of 7,268 gene transcripts were significantly expressed (FDR-corrected p < 0.05), with 4,501 being upregulated (fold change ≥ 2) and 2,767 being downregulated (fold change ≤ −2). The upregulated gene transcripts include 17 P450s/CYPs, 1 UGT and AchE, 19 hydrolases, 46 dehydrogenases, 6 sulfotransferases, 2 peroxidases, 2 carboxylesterases, and 7 ABC transporters. The downregulated gene transcripts include 26 P450s, 8 UGTs, 12 GSTs, 2 aminoacylases, 14 hydrolases, 62 dehydrogenases, 3 sulfotransferases, 2 peroxidases, 1 carboxylesterase, 9 esterase subfamilies, and 43 ABC transporters (Figure 8C). A total of 15 gene transcripts had molecular functions predicted to be associated with defense, 145 with signaling, 18 with growth, 103 with transport (excluding ABC transporter families), 107 with digesting (excluding esterase sub-families), and 27 with metabolism. The detoxification-related gene transcripts were plotted based on average CPM values (mean ± STD error) (Figure 8D), and the majority of these were considerably overexpressed in the callow male gut (T6), indicating that callow beetles need those enzymes in high amounts during maturation feeding under the bark before they sclerotize. When comparing the sclerotized male gut, callow male gut, and callow female gut (T6, T5, and T7) (Supplementary material 13, 34, 35), a similar propensity was observed, with the sclerotized male gut (T6) segregating from the other two (T5 and T7), thus indicating the fine-tuning of gene expression for new colony establishment in the pioneer sex (Supplementary Figure 4). The cytochrome P450s expressed substantially in the sclerotized gut (T6) belong to the known detoxification families 4 (5), family 6 (3), and family 9 (1) (Figure 9A). However, the downregulated detoxification-related genes had lower mean CPM values or transcript levels in the sclerotized male gut than in the callow one (Figure 9B).
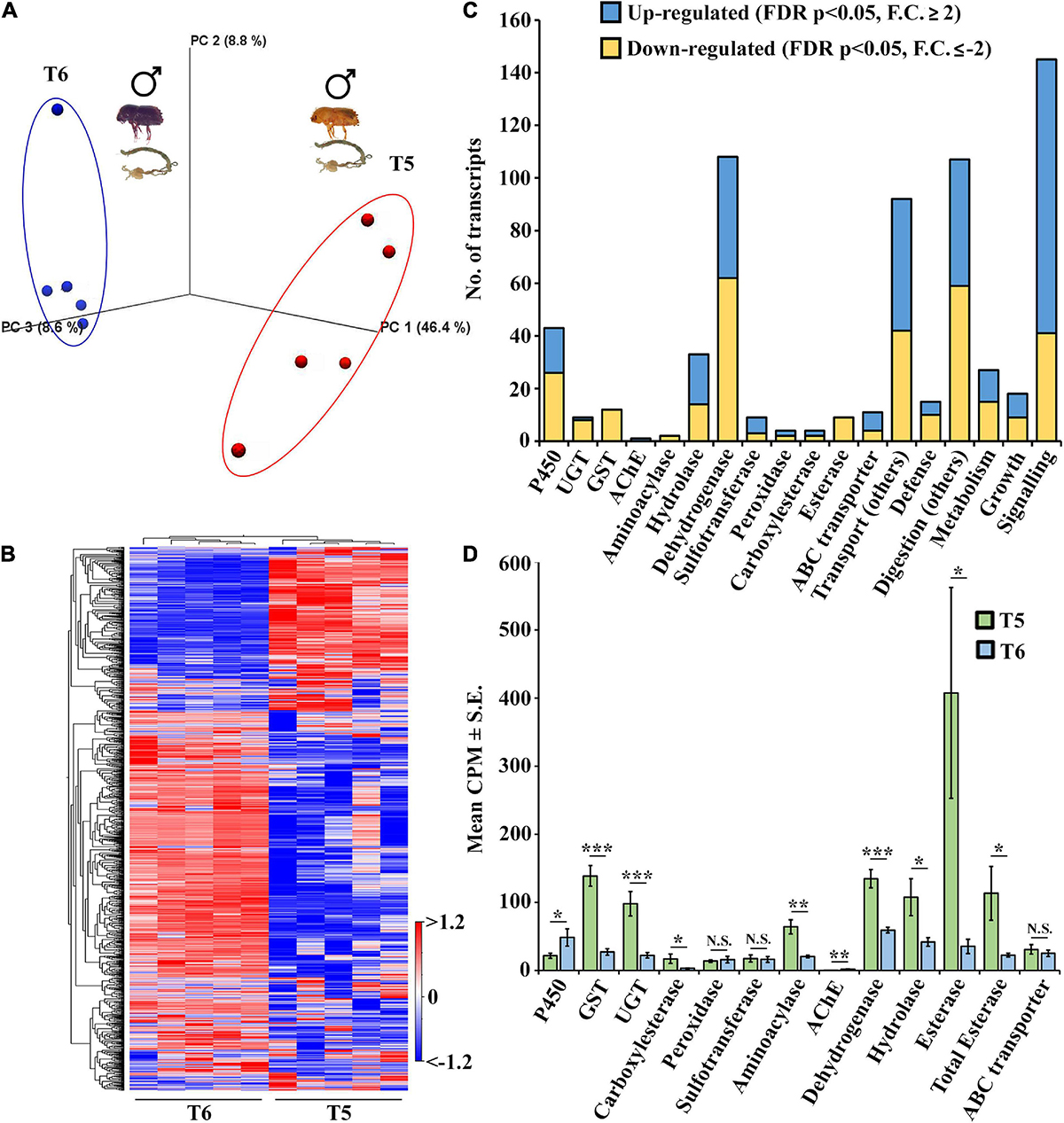
Figure 8. Comparison between sclerotized male gut (T6) vs. callow male gut (T5) N = 5. (A) PCA plots showing sample clustering. (B) Heat maps representing gene clustering. The color spectrum, stretching from blue to red, represents TMM-adjusted log CPM expression values obtained after DGE analysis. (C) Bar graphs showing the differentially expressed contigs of specific detoxification-related enzyme families and other important enzymes related to defense, digestion, transport, metabolism, signaling, and growth between sclerotized male gut (T6) vs. callow male gut (T5). (D) Detoxification genes expressed as the average of total differentially expressed genes in each family between the sclerotized male vs. the callow guts. Gene families on the x-axis plotted against their corresponding mean CPM values ± S.E. One-tailed T-test was performed to obtain the significant differences between the two stages. * represents p < 0.05, ** represents p < 0.01, *** represents p < 0.001, and NS represents non-significant difference. DGE analysis was done using CLC workbench with FDR p < 0.05 and a fold change ± 2 cut-off.
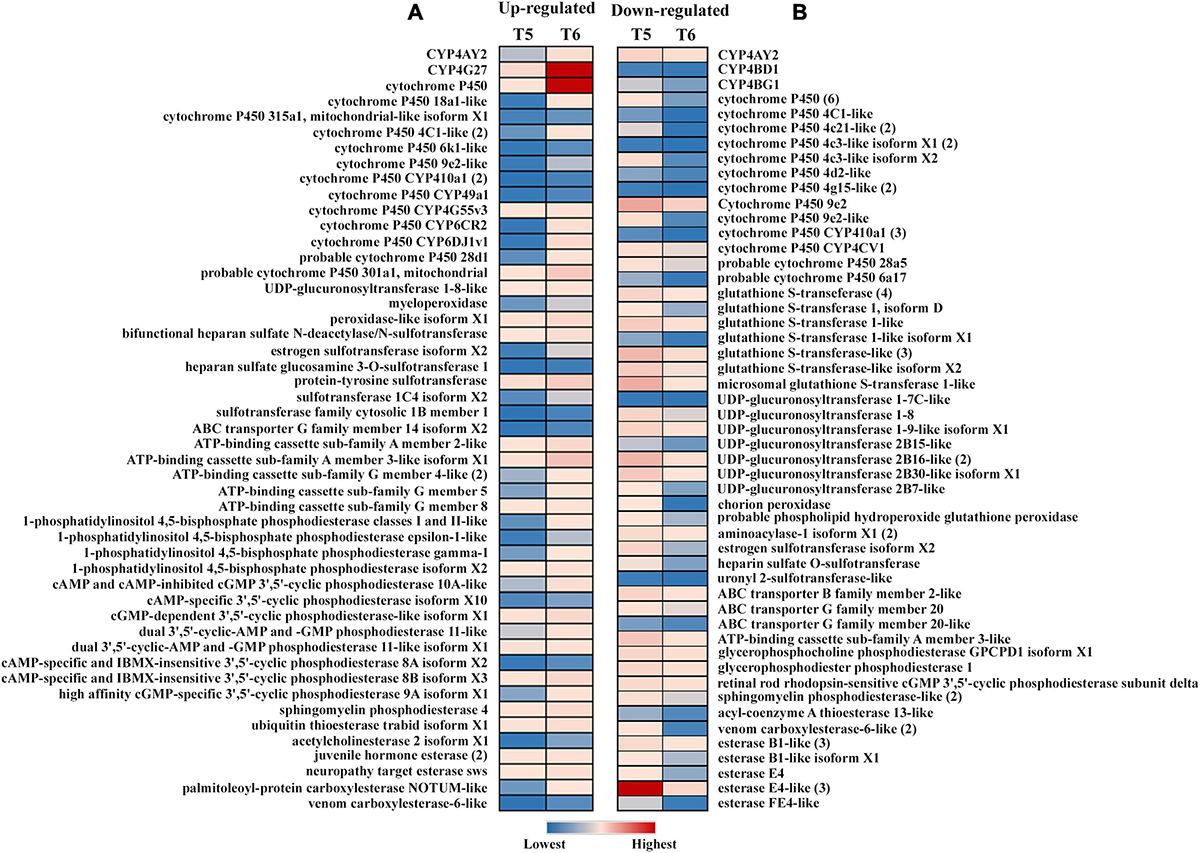
Figure 9. Expression profiles of detoxification-related genes in the sclerotized male gut (T6) vs. callow male gut (T5) (N = 5). (A) Upregulated genes FDR corrected p < 0.05 and equal or above the threshold of fold change of 2; (B) downregulated genes FDR corrected p < 0.05 and equal or below the threshold of fold change of –2. The expression level is represented in different colors indicating the TMM-adjusted log CPM expression mean values.
Gene ontology enrichment analyses for upregulated and downregulated transcripts were performed in all ten comparisons using the FDR-corrected p < 0.05 and a fold change threshold of ± 2 (Supplementary materials 14–33). According to the GO enrichment, the 62 crucial metabolic pathways enriched across seven comparisons could be involved in detoxification, digestion, and defense. Most of these were related to molecular functions (28) and biological processes (34) in the GO classification system (Figure 10). In the comparison of developmental stages (T1, T2, T3, T4, vs. T13), 37 critical pathways were enriched, including acyltransferase activity, hydrolase activity, transferase activity, immune response, stress response, and gene expression regulation. The comparison of callow and sclerotized male gut (T5 vs. T6) showed enrichment of 21 pathways in callow beetles, including catalytic activity, enzyme binding, cellular response to stress, and detection of biotic stress. Concerning specific tissues, 19, 11, 16, and 11 metabolic pathways were enriched in callow male and female fatbody (T8 vs. T9), callow male and female guts (T5 vs. T7), callow male gut, fat body, and head (T5, T9, vs. T11) and callow female gut, fat body, and head (T7, T8, vs. T10), respectively. The pheromone-related activity was only enriched in the callow male fat body vs. callow female fat body (T8 vs. T9), implying that pheromone production occurs in the ESBB fat body (Figure 10).
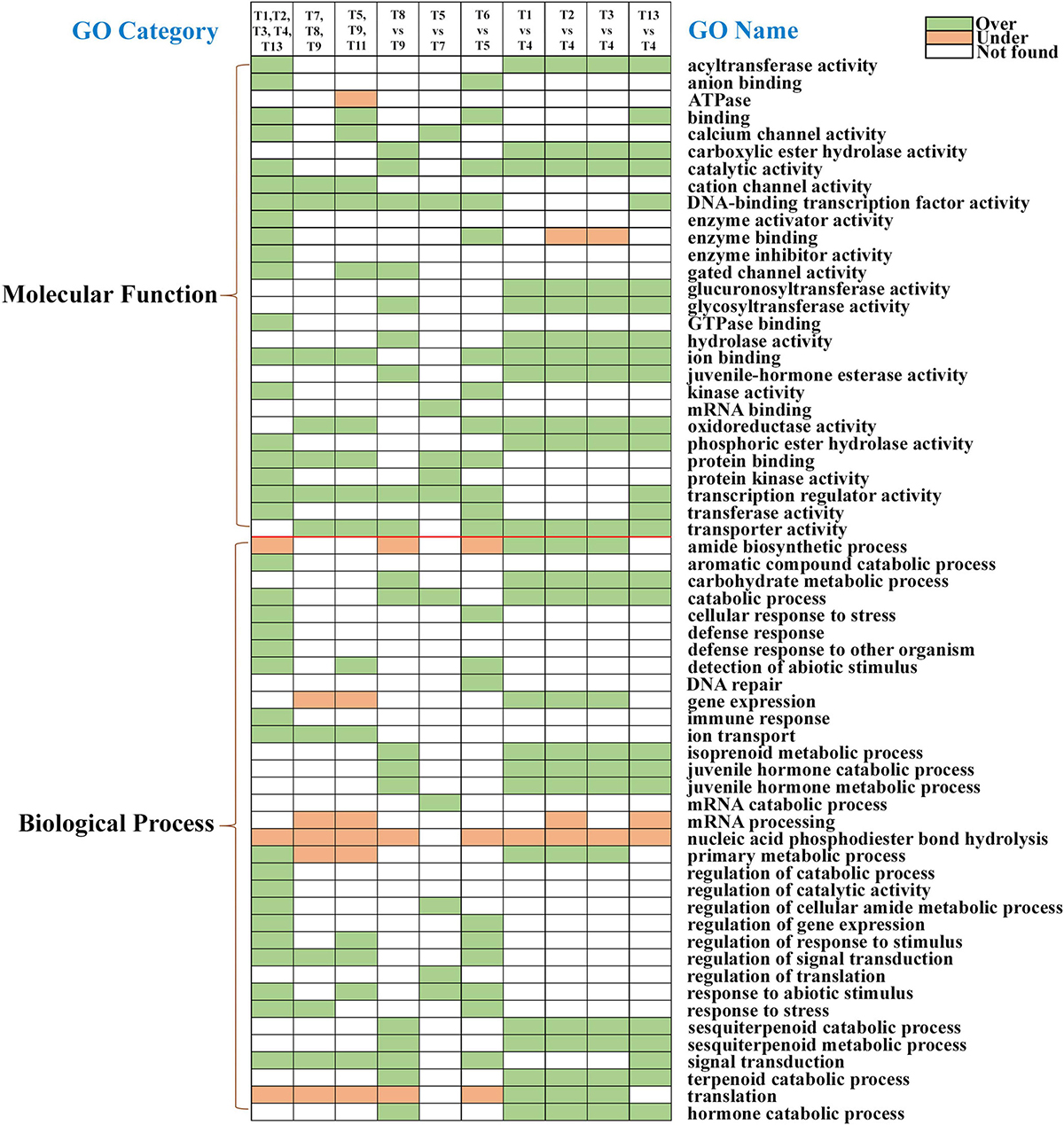
Figure 10. Critical physiological processes in different ESBB sample comparisons after GO enrichment analysis (FDR p < 0.05, fold change ≥ 2). Green boxes are over-enriched, orange boxes are under-enriched, and white boxes represent processes not occurring in a particular comparison.
The RT-qPCR was performed for 19 differentially regulated genes in different life stages, sex-specific guts, and fat body comparisons. Most of these genes revealed strong similarities in transcript levels with respect to expression patterns observed in their respective transcriptome expression (Figures 11–13).
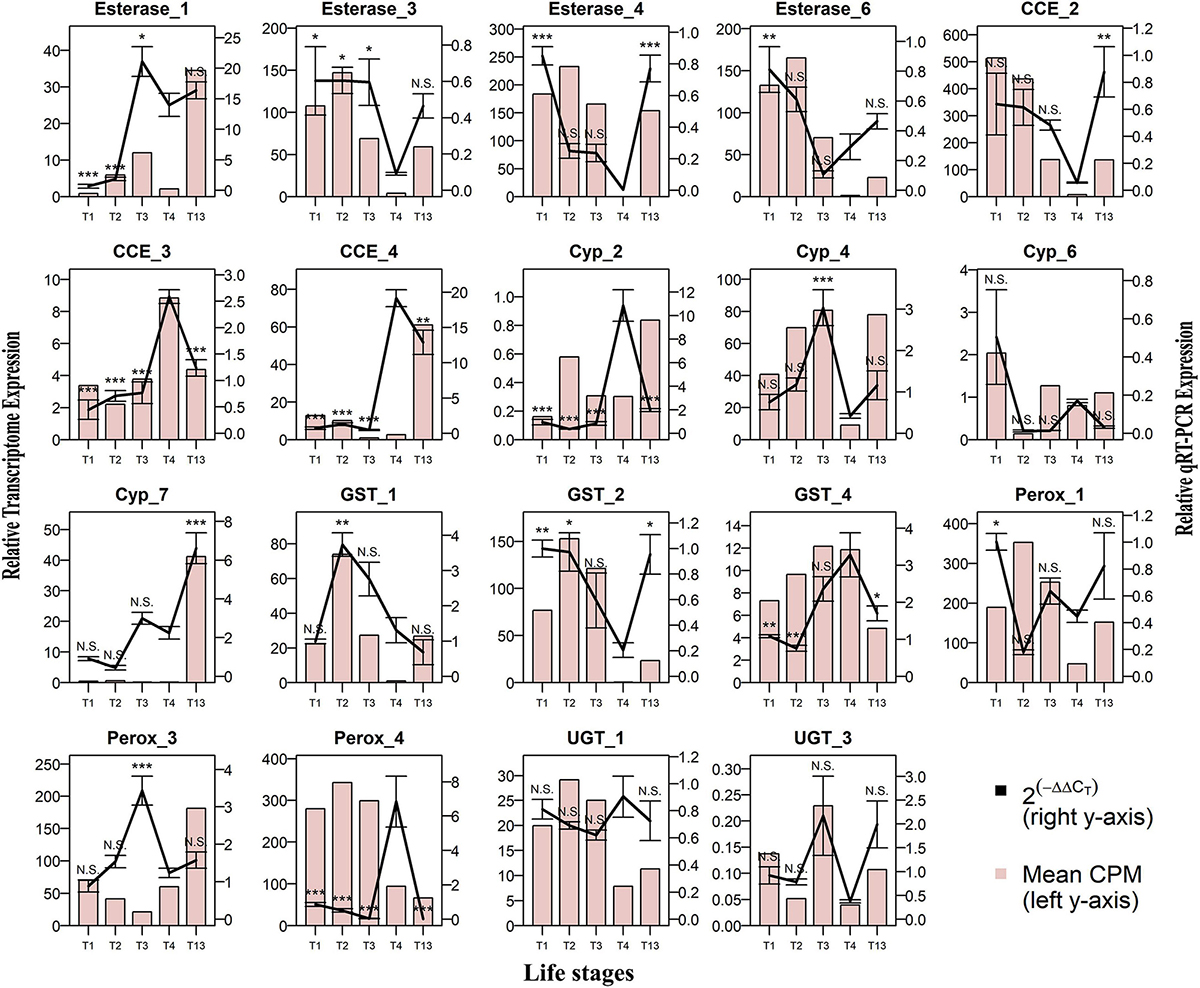
Figure 11. Comparison of RNA-seq and RT-qPCR data for the relative expression of 19 genes in developmental life stages. Life stages: 1st instar larvae (T1), 2nd instar larvae (T2), 3rd instar larvae (T3), pupae (T4), and sclerotized adult beetles (T13) on the x-axis plotted against relative expression in RNA seq (N = 5) represented as mean CPM on the left y-axis, and relative expression in RT-qPCR (N = 4) represented as mean 2^-(ΔΔCt) ± S.E. on the right y-axis. The bar graph represents CPM-based expression from RNA-seq, and the solid line represents RT-qPCR expression. Tukey’s HSD was performed with a 95% confidence level with T4 as the control group, and p-values were generated as per the fit model. * represents p < 0.05, ** represents p < 0.01, *** represents p < 0.001, and N.S. represents non-significant difference.
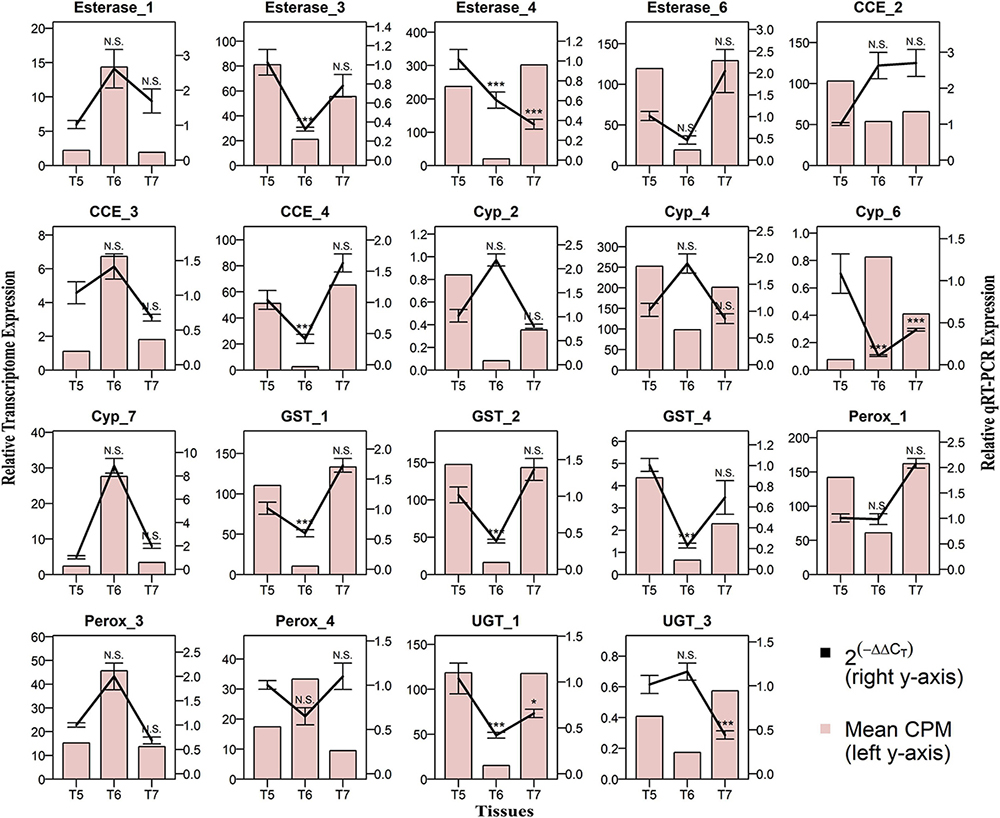
Figure 12. Comparison of RNA-seq and RT-qPCR data for the relative expression of 19 genes in gut tissues. Callow male gut (T5), sclerotized adult male gut (T6), and callow female gut (T7) on the x-axis plotted against relative expression in RNA seq (N = 5) represented as mean CPM on the left y-axis and relative expression in RT-qPCR (N = 4) represented as mean 2^-(ΔΔCt) ± S.E. on the right y-axis. The bar graph represents the mean CPM-based expression from RNA-seq transcriptome data, and the solid line represents RT-qPCR expression. Relative expressions in T6 and T7 were compared against those in T5 using Tukey’s HSD with a 95% confidence level, and p-values were generated as per the fit model. * represents p < 0.05, *** represents p < 0.001, and N.S. represents non-significant difference.

Figure 13. Comparison of RNA-seq and RT-qPCR data for the relative expression of 19 genes during RT-qPCR in the fat body tissues. Callow female fat body (T8) and callow male fat body (T9) on the x-axis plotted against relative expression in RNA seq (N = 5) represented as mean CPM on the left y-axis, and relative expression in RT-qPCR (N = 4) represented as mean 2^-(ΔΔCt) ± S.E. on the right y-axis. Light brown bars represent mean transcriptome expression, and dark green bars represent RT-qPCR expression. Student’s T-test was performed with a 95% confidence level, and p-values were generated after comparing RT-qPCR expression between T9 vs. T8. * represents p < 0.05, ** represents p < 0.01, *** represents p < 0.001, and N.S. represents the non-significant difference.
The enzyme activity assay of cytochrome P450 reductase (CYP450) and glutathione S-transferase (GST) showed stage-specific expression (Figure 14). When compared against the larval stage 1 (T1), the pupa (T4) and sclerotized adult (T13) stages had significantly higher enzyme activities of GST, while the larval stages showed constitutive expression of GST. The activity of CYP450 decreased from T1 stage to T4 and was lowest in the sclerotized adult stage (T13). Similarly, the callow male ESBB gut (T5) had a relatively low expression of CYP450 but a high expression of GST.
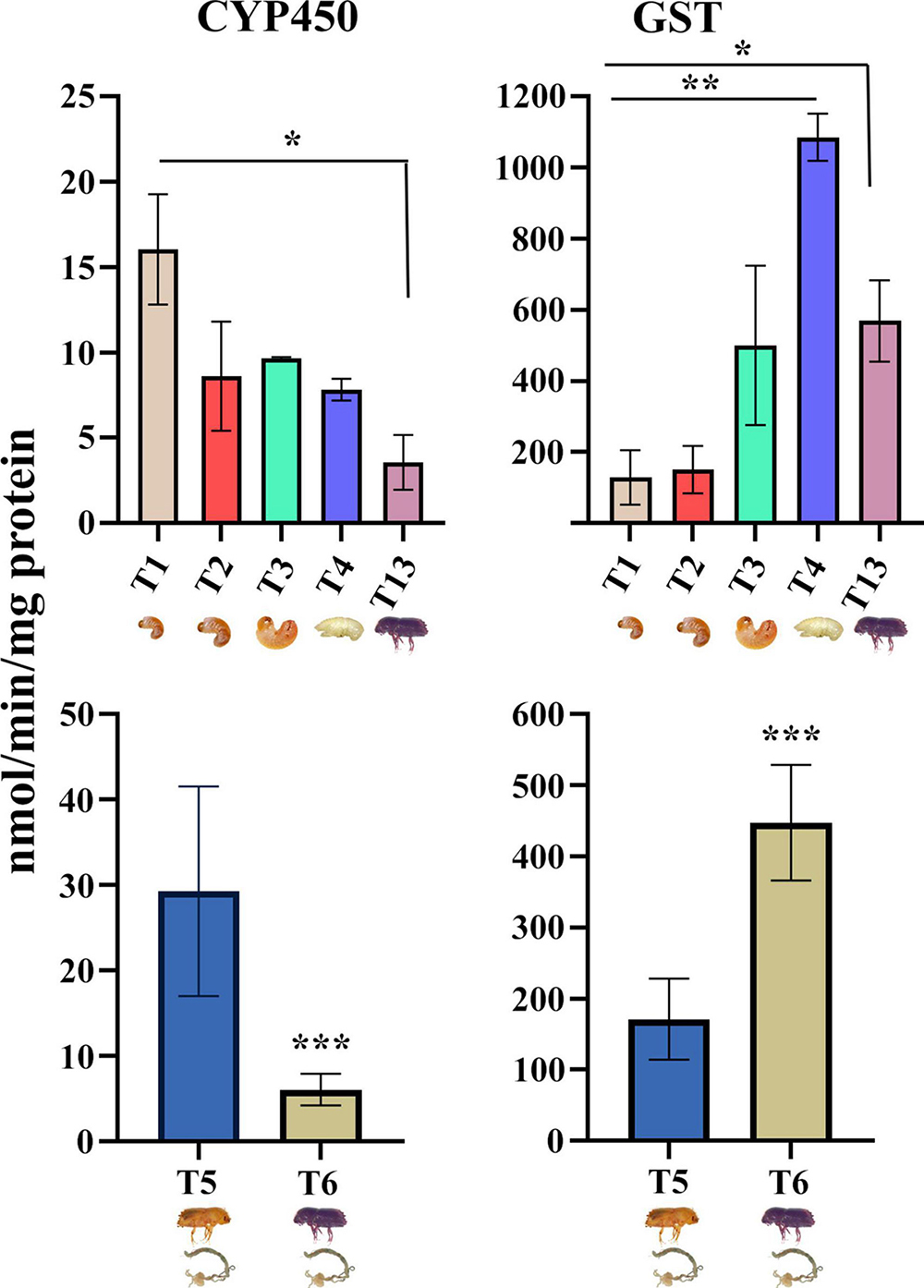
Figure 14. The activities of selected enzymes, glutathione S-transferase (GST) and cytochrome P450 (CYP450), measured in the developmental and tissue-specific stages. The bars represent mean values and whiskers standard deviations in four biological replicates. One-way ANOVA was performed between the life stages (T1, T2, T3, T4, and T13) and Student’s t-test between guts of callow and sclerotized males (T6 vs. T5). * indicates that values are significantly different from each other at * p < 0.05, ** p < 0.01, ***p < 0.001. GST, and CYP450 activity are expressed as nmol/min/mg protein.
The family Curculionidae of the order Coleoptera comprises a massive number of bark beetles that colonize tissues of a wide range of tree species. The damage caused by bark beetles has been extensive in recent years, leading to the destruction of more than 100 million m3 of spruce forest during a single year in Europe and Asia (Hlásny et al., 2019). This included 14.5 (67% forest cover) million m3 of spruce forest within Czechia in 2021 (Lubojacký et al., 2022). The revenue loss in the timber industry was so huge that the Czechia state had to support it with ca 260 million euros in 2018–2019 (Hlásny et al., 2021). The management of ESBB necessitates a thorough knowledge of its physiology and adaptation to a nutritionally limiting host. ESBB has four main phases in their life cycle: egg, larva, pupa, and adult (callow and sclerotized). The ESBB larvae (L1, L2, and L3), pupa, and adults are developed under the bark upon exposure to the host allelochemicals. When their development is complete, the sclerotized males, being the pioneer sex, attack new host trees displaying constitutive and induced defenses. The shift from callow (early) to sclerotized (black) adult is a critical phase, during which the beetles undergo various physiological and gene-transcript-level changes that prepare them to establish themselves in a new host. The mechanisms that control the growth and development of ESBBs and their resistance toward spruce allelochemicals are still unknown. RNA-seq has always been a standard method for studying complex gene expression and molecular mechanisms in non-model insects. Hence, we used RNA-seq data (PRJNA679450) from developmental stages (larvae to fully emerged sclerotized beetles) and diverse tissues of ESBB produced during the in-house ESBB genome study to evaluate the gene expression dynamics further. RT-qPCR and enzymatic assays further corroborated the transcriptome data.
All detoxifying enzyme families were downregulated in the pupae, non-feeding stage, suggesting that the expression of genes related to detoxification was costly and should be optimally expressed as per requirements, for instance, during host feeding. This phase-specific variation (Figure 2D) showed that the expression of detoxifying enzymes considerably relies on host encounters and active feeding in ESBB.
In this study, many detoxification-related cytochrome families (CYP 4, 6, and 9) were overexpressed during the adult life stage. The cytochrome gene expression of the Dendroctonus species, close relatives of I. typographus, has been extensively researched concerning the detoxification of host allelochemicals. For example, 64 P450s were identified in D. armandii at various life stages (larvae, pupae, and adults), and RT-qPCR showed upregulation of 41 of these genes in later life stages as compared to larvae. After host feeding, 19 CYP genes of these 41 genes showed overexpression as compared to the unfed beetles (Dai et al., 2015). Dendroctonus bark beetles showed a significant, non-sex-specific change in gene expression of CYP6DG1, CYP6BW5, CYP6DJ2, CYP9Z18, and CYP9Z20 during the early hours of host feeding, implying that cytochrome families 6 and 9 were also involved in the xenobiotic response in beetles as well (Sarabia et al., 2019). In D. rhizophagus, the role of CYPs from the CYP4, CYP6, and CYP9 families in detoxification was also confirmed. Recently, three CYPs involved in monoterpene oxidation and one CYP involved in aggregation were identified and characterized in D. ponderosae (Chiu et al., 2019a,b). CYP6CR2 is an epoxidase involved in exo-brevicomin biosynthesis and pheromone generation in male mountain pine beetles after they leave the brood tree, but CYP6CR2 decreases during host tree selection and mating (Song et al., 2014). In Sitophilus zeamais, terpinen-4-ol fumigation induced differential regulation of cytochrome genes related to detoxification, such as CYP4BH1 (up), CYP6BW2 (up), CYP6DJ1 (up), CYP6DG1 (up), and P450 9E2 (down) (Huang et al., 2018, 2019). Hence, cytochromes are vital components in insect adaptations, including bark beetles.
CYPs, particularly those belonging to the CYP6 gene family, are reported to play a role in detoxification, toxin resistance, and other functions such as pheromone synthesis (Nadeau et al., 2017; Ramakrishnan et al., 2022). In previous investigations, the CYP4 and CYP9 genes have also been reported to have a sex-specific expression when bark beetles are exposed to host tree allelochemicals. For instance, the sex-specific expression of four CYP4 (AY2, G27, BD1, and BG1) and cytochrome P450 9E2 genes were identified in I. paraconfusus. The CYP4AY1, CYP4BG1, and CYP9T1 genes are expressed in male I. paraconfusus, suggesting their putative involvement in synthesizing male-specific aggregation pheromones (Huber et al., 2007). Similarly, after exposing D. rhizophagus to host-tree monoterpenes, differential expression of the CYP4G27, CYP4AY1, and CYP4AY2 genes was observed in the female and male antenna and gut (Cano-Ramirez et al., 2013). In our data, the CYP4BH1 gene was highly expressed in ESBB adults, whereas the CYP4BG1 and CYP4BD1 gene expression were low. The differential expression of these genes suggested that feeding regulates their expression. While we did not determine the sex of these ESBB adults, the regulation may be sex-specific and related to pheromone biosynthesis pathways, as reported earlier for Dendroctonus ponderosae (Nadeau et al., 2017). However, this needs to be functionally validated in ESBB.
Previous research has verified that some of the identified CYP6 family cytochromes (CYP6DJ1, CYP6BW1, and CYP6BW3) in Dendroctonus spp. are involved in pheromone biosynthesis pathways during the oxidation of host terpenes (Chiu et al., 2019b). Symbiotic blue-stain fungi vectored by bark beetles synthesize bicyclic ketals, i.e., pheromones and other semiochemicals (exo-brevicomin, endo-brevicomin, and trans-conophthorin) and help the beetles to colonize healthy trees (Lee et al., 2006; Zhao et al., 2019). A recent study showed that the CYP6CR2 and CYP6DE5 genes were highly expressed under the treatment of terpenoids, whereas silencing of these genes significantly reduced the activity of P450 and increased the mortality of adults after exposure to terpenoids. CYP6CR2 expression was considerably higher in larvae and sclerotized adults, with lower expression in starving adult mountain pine beetles (Robert et al., 2013, 2016; Liu et al., 2022). A transcriptome study of the imidacloprid resistance (IR) and isogenic susceptible (IGS) strains of Aphis gossypii revealed that the resistant strain had high gene expression of cytochrome P450 4G15-like (Kim et al., 2015). We found that the CYP6CR2 gene was expressed 400-fold higher in sclerotized adult beetles than in the other stages. Cytochrome P450 4G15-like gene expression was also higher in adult ESBB, suggesting that both genes may play a role in detoxifying plant-allelochemicals.
In general, cytochrome genes are involved in many critical physiological functions in addition to detoxification. For instance, ecdysteroid 26-hydroxylase encoded by CYP18A1 is a key hormone inactivation enzyme. CYP18A1 is expressed in many of the target tissues of ecdysteroid (20E) in Drosophila larvae, and CYP18A1 inactivation slows the larval development and causes pupal mortality, while CYP18A1 overexpression causes late embryonic lethality. According to these findings, the inactivation of 20E is required for proper growth, and CYP18A1 is a crucial enzyme in this process (Guittard et al., 2011). In the current study, the CYP18A1-like gene was constitutively expressed from the initial larvae to the adult, which showed that uniform expression of this gene may aid larval development and pupal formation and adult development (Figure 3B). The enzyme Cytochrome P450 306A1 is involved in the metabolism of insect hormones, such as ecdysteroid C25-hydroxylase activity, and the breakdown of synthetic pesticides. The effective conversion of keto-diol to keto-triol in CYP306A1 transfected S2 cells via carbon 25 hydroxylation suggests that CYP306A1 works as a carbon 25 hydroxylase and plays an essential role in ecdysteroid production during insect development (Niwa et al., 2004). Cytochrome P450 306A1 gene was highly expressed in the pupal stage, suggesting that it may aid in the pupal transition into an adult. The enzyme CYP4C1 is involved in abiotic stress tolerance. For instance, silencing of CYP4C1 causes B. tabaci to have much lower heat resistance and higher cold tolerance, implying that CYP4C1 is a significant regulator in temperature adaptation and geographical distribution (Shen et al., 2021). The CYP4C1-like gene was highly expressed in the sclerotized adult ESBB, suggesting its putative involvement in winter survival, range expansion, and adaptability to different environments. Chalkbrood is the most common fungal disease in honeybees, and transcriptome investigation of the immunological defenses of A. cerana larvae to A. apis infection revealed that the fungal-infected strain had a high expression of the cytochrome P450 6A1-like gene (Guo et al., 2019). The cytochrome P450 6A1-like gene was significantly expressed in the ESBB pupal stage, suggesting that it may aid pupal resistance to fungal infection (Figure 3B). Interestingly, CYP4BD1 was expressed in all larval stages and sclerotized adults but not in the pupa (Figure 3B). CYP4BD1 expression was higher in males and did not differ significantly in females compared to unfed individuals; it might be required for male-specific physiological activities such as pheromone production and/or juvenile hormone biosynthesis (Tillman et al., 1998, 2004; Helvig et al., 2004; Huber et al., 2007).
We found UGTs and GSTs expressed in various life stages and gut tissues of ESBB (Figure 3). UGTs are endoplasmic reticulum-associated enzymes that perform glycosylation using UDP-glucose as an active sugar source. The protein structures are composed of N-terminal aglycone substrate-binding and C-terminal UDP-glycoside binding domains. About 40 UGTs in H. armigera and 44 UGTs in Bombyx mori were identified, and two UGT genes, UGT41B3 and UGT40D1, have been linked to metabolizing gossypol via glycosylation in H. armigera (Ahn et al., 2012; Krempl et al., 2016). Ecdysteroid UDP-glycosyltransferase (EGT), a baculovirus-encoded protein, activates and regulates insect molting by ecdysteroid hormones (Shen et al., 2018). UGTs were reported to aid host plant allelochemical detoxification in Spodoptera (Roy et al., 2016). The UDP-glucuronosyltransferase 2B10 (UGT2B10) is a detoxifying enzyme specializing in the N-linked glucuronidation of numerous drugs and xenobiotics. In our investigation, the UDP-glucuronosyltransferase 2B10-like gene was significantly expressed in sclerotized adults, suggesting that it may play a role in detoxification (Figure 3B). The UDP-glucuronosyltransferase 1-8 (UGT1A8) is an enzyme that participates in the glucuronidation pathway, which converts small lipophilic compounds into water-soluble, excretable metabolites (Wang et al., 2013). UDP-glucuronosyltransferase 1-8 and UDP-glucuronosyltransferase 1-8-like genes were expressed in ESBB larvae, probably facilitating food digestion and excretion.
Glutathione S-transferases are well-known for mediating insecticide resistance by reductive dehydrochlorination or conjugation reactions (Kostaropoulos et al., 2001; Enayati et al., 2005; Song et al., 2022). In S. litura, RNA-seq-based DGE studies were used to identify 8 GSTs and verify the involvement of GSTS1 in tomatine metabolism (Li et al., 2019). Previous studies have reported 16 GSTs from four different groups (delta, epsilon, sigma, and theta) in the Chinese white pine beetle, and these GSTs were expressed in all developmental stages and tissues (antennae, gut, and reproductive tissues) (Gao et al., 2020). Similarly, the present study observed GSTs expression during the different life stages and tissues of ESBB, implying its physiological importance in bark beetles (Figure 3).
The overexpression of esterases (ESTs), particularly those belonging to class I (clade A-C), aids detoxification, while class III (clade I-M), containing CCE and AChEs (clade I-M), works primarily through target site mutation-based resistance to pyrethroids, organophosphates, and carbamates, but their role in detoxification has also been documented concerning malathion detoxification (Wei et al., 2020). The NOTUM gene encodes a palmitoleoyl-protein carboxylesterase that works as a negative regulator of the Wnt signaling pathway. NOTUM knockdown lentivirus inhibits colon adenocarcinoma development in vitro and in vivo by reducing tumor proliferation, shrinking tumor size, and enhancing apoptosis (Gong et al., 2021). The palmitoleoyl-protein carboxylesterase NOTUM-like gene was significantly expressed in ESBB pupae, implying that it regulates apoptosis in the non-feeding stage. Carboxylesterase 6 has the highest expression level in the social wasp P. varia, while solitary wasps have little or no expression, suggesting that this protein may play a defensive role in social wasps (Yoon et al., 2020). The venom carboxylesterase-6-like gene was highly expressed in ESBB sclerotized adults, suggesting its putative involvement in the ESBB defense mechanism (Figure 3A). In insects, chorion peroxidase (pxt) has a role in the formation of a rigid and insoluble egg chorion by catalyzing chorion protein crosslinking through dityrosine formation and phenol oxidase-catalyzed chorion melanization (Li et al., 2004). Chorion peroxidase and peroxidase-like isoform X1 were highly expressed in sclerotized adults, suggesting that they may function in egg production and development (Figure 3A). In Myzus persicae, the overexpression of E4 and FE4 genes have a role in insecticide resistance (Field and Devonshire, 1998). In our study, the esterase FE4-like gene was expressed in all larval and sclerotized adult stages but not in the pupae, suggesting that it may influence the detoxification of host allelochemicals during feeding (Figures 3A, B). However, esterase B1-like and esterase E4 genes were expressed in pupae, suggesting that they may be involved in fat consumption for energy production during pupa-to-adult development. However, our observations need further experimental validation.
The phospholipase activity of the mitochondrial cardiolipin hydrolase gene is essential for mitochondrial fusion and fission, allowing cells to cope with the increased nucleotide demand during DNA synthesis (Huang et al., 2011). In the current study, the mitochondrial cardiolipin hydrolase-like gene was highly expressed in late larval and adult stages, suggesting its involvement in larvae-to-pupae conversion and adult development. The protein deubiquitination carried by the ubiquitin C-terminal hydrolase (UCH) and ubiquitin-specific processing protease (UBP) protein families is involved in various biological activities in animals, fungi, and plants (Wang et al., 2018). Ubiquitin carboxyl-terminal hydrolase 1-like gene was significantly expressed in pupae and adult ESBB stages compared to the larvae, suggesting its putative role in later stages of ESBB development.
ATP-binding cassette transporter proteins transport a variety of molecules across cell membranes. ATP binding cassette subfamily A member 12 (ABCA12) protein is essential for transporting fats (lipids) and enzymes in the cells and maintaining the layers of lipids within the epidermis to prevent water loss (dehydration) and to allow normal skin development (Fukuda et al., 2012; Akiyama, 2014). The expression of ABC transporters remained primarily constant throughout development, suggesting their importance throughout the life cycle of ESBB. In the larval and adult stages, the ABC transporter subfamily genes (D member 3, G member 1-like, and G member 4) exhibited higher expression than in the pupal stage (Figure 3). Interestingly, ABC transporter expression dropped dramatically during the early ESBB larval stage, but increased again in the pupal and adult stages, demonstrating its role in later developmental stages in ESBB. Interestingly, all ABC transporter subfamily A genes had higher expression in pupae, suggesting that these genes might play an essential role during the pupal stage. The ABC transporter subfamilies D and G genes expressed in other ESBB life stages may be involved in detoxification during feeding. Furthermore, ATP-binding cassette sub-family A member 2-like and ATP-binding cassette sub-family A member 3-like isoform X1 were highly expressed in ESBB pupae, suggesting that they may facilitate proper epidermis formation in ESBB. The ATP-binding cassette sub-family G member 4 (ABCG4) subunit of heterodimeric precursor transporters for eye-pigment synthesis was identified in Drosophila’s brain and eye (Oldfield et al., 2002). In the current study, the ATP-binding cassette sub-family G member 4, ATP-binding cassette sub-family G member 4-like, and ATP-binding cassette sub-family G member 4 isoform X1 genes were significantly expressed in ESBB sclerotized adults as compared to larvae and pupae, indicating that they may play a similar role in eye pigment synthesis. The ATP binding cassette subfamily G member 8 (ABCG8) and member 5 (ABCG5) genes produced sterolin-1 and sterolin-2, respectively, and combinedly formed sterolin protein (Calandra et al., 2011). Sterolin is a plant sterol transporter protein that moves substances across cell membranes and is found mainly in intestines and liver cells. Sterolin also helps cholesterol regulation; typically, about 50 percent of the cholesterol in the diet is absorbed by the body (Calandra et al., 2011). The ATP-binding cassette sub-family G member 8 and ATP-binding cassette sub-family G member 5 genes were significantly expressed in ESBB pupae, implying that they may be involved in fat absorption and accumulation to provide energy for adult conversion.
The mitochondrial glutamate dehydrogenase catalyzes the conversion of L-glutamate into alpha-ketoglutarate, an essential intermediate in the tricarboxylic acid cycle. It plays a role in insulin homeostasis and is also involved in learning and memory reactions by increasing the turnover of the excitatory neurotransmitter glutamate (Plaitakis et al., 2000, 2017). In this study, glutamate dehydrogenase mitochondrial-like protein and glutamate dehydrogenase mitochondrial-like isoform X2 genes were highly expressed from late larvae to adults, suggesting that they may be involved in energy production and ESBB development (Supplementary material 3). In the glycolytic pathway, the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a highly conserved enzyme that catalyzes the transformation of glyceraldehyde-3-phosphate to glycerate-1, 3-biphosphate, and produces NADH. In the Mortierella alpina fungus, the overexpression or knockdown of GAPDH1 and GAPDH2 genes significantly accumulated or reduced lipid content, respectively (Wang et al., 2020). In the current study, the glyceraldehyde-3-phosphate dehydrogenase-like and glyceraldehyde-3-phosphate dehydrogenase 2-like genes were highly expressed from late larvae to adults. Fungi and insects share common metabolic pathways, and our data implied that these genes may have a role in lipid accumulation to satisfy the energy requirement for ESBB while searching for a suitable host. The glucose dehydrogenase genes are expressed at all stages of Drosophila development and play an essential role in the eclosion process and cuticular remodeling (Krasney et al., 1990). Similarly, the glucose dehydrogenase (FAD, quinone)-like gene was expressed in all ESBB developmental stages, suggesting its involvement in the eclosion process and cuticular remodeling in ESBB. In Ips pini, the short-chain oxidoreductase ipsdienol dehydrogenase (IDOLDH) converts (-)-ipsdienol to ipsdienone and plays a role in determining pheromone composition (Figueroa-Teran et al., 2016). In the current study, ipsdienol dehydrogenase transcripts were expressed from initial larvae to adults, suggesting that they may have a similar role in pheromone biosynthesis and composition in ESBB (Ramakrishnan et al., 2022). The glutaryl-CoA dehydrogenase (GCDH) is an enzyme found in mitochondria and involved in the breakdown of amino acids like lysine, hydroxylysine, and tryptophan, which are building blocks of proteins (Basinger et al., 2006). The glutaryl-CoA dehydrogenase mitochondrial gene was expressed in all larval and adult stages in our study but not in the pupae, implying a function in protein formation during ESBB feeding stages. However, such observations need to be further empirically validated.
We observed tissue-specific regulation of many genes; for instance, CYP6K1-like and CYP410A1 had higher expression in the head and lower expression in the gut and fat body, respectively, implying a role in olfaction as odorant degrading enzymes (ODEs) (Baldwin et al., 2021). Similarly, juvenile hormone epoxide hydrolase 1-like (JHEH 1-like) gene transcripts were more abundant in the gut and fat body than in the head, as these two tissues are the site of production and metabolism of aggregation pheromone in ESBB (Ramakrishnan et al., 2022). Our findings (Figure 7) were consistent with earlier research showing the upregulation of detoxification-related genes and pheromone genes in the gut and fat body (Keeling et al., 2016; Li et al., 2019; Ramakrishnan et al., 2022). Cytochrome P450 4C1 (CYP4C1) transcripts are mainly engaged in the metabolism of insect hormones and the breakdown of synthetic insecticides. Whiteflies, after silencing CYP4C1, showed significantly lower heat resistance and higher cold tolerance, which indicates that CYP4C1 is a critical regulator in temperature adaptation and influences the geographical distribution and dispersal of whiteflies (Shen et al., 2021). In the present study, the cytochrome P450 4C1-like gene transcripts showed high levels in male and female fat bodies and heads of callow ESBB, suggesting a putative role in thermal adaptation during new host finding. In bark beetles, the CYP9T2 (gut-specific cytochrome P450) gene hydroxylates myrcene to ipsdienol and functions toward the end of the pheromone biosynthesis pathway (Sandstrom et al., 2006). Gut-specific cytochrome P450 transcript was expressed in both male and female callow ESBB fat body, suggesting a putative role in pheromone biosynthesis and odor degradation.
The glutathione S-transferases (GSTs) play an essential role in detoxifying xenobiotic toxins in insects, including insecticides (Balakrishnan et al., 2018). Most GSTs, such as glutathione S-transferase-like, glutathione S-transferase-like isoform X2, and microsomal glutathione S-transferase 1-like transcripts, are expressed in the callow beetle’s gut, suggesting roles in food digestion and detoxification of plant xenobiotics. UDP-glucuronosyltransferases (UGTs) are significant phase II drug metabolism and multifunctional detoxification enzymes that play an essential role in insect resistance to various plant allelochemicals and pesticides (Neumann et al., 2016; Roy et al., 2016; Cui et al., 2020). Most UGTs, such as UDP-glucuronosyltransferase 1-8, UDP-glucuronosyltransferase 1-9-like isoform X1, UDP-glucuronosyltransferase 2B10-like, UDP-glucuronosyltransferase 2B16-like, UDP-glucuronosyltransferase 2B30-like isoform X1, and UDP-glucuronosyltransferase 2C1, are expressed in the male and female callow ESBB gut, implying a role in multifunctional detoxification during maturation feeding of ESBB. Aminoacylase 1 (ACY1) was found in many tissues and organs, where it may be involved in breaking down proteins that are no longer needed. A novel aminoacylase isolated from Burkholderia sp. strain LP5_18B effectively catalyzes N-lauroyl-L-amino acids synthesis (Takakura and Asano, 2019). In the current study, aminoacylase-1 isoform X1 was expressed in both callow ESBB male and female guts, suggesting a role in dietary protein breakdown. Epoxide hydrolases (EH) are essential regulators of lipid epoxides and play a role in detoxification processes, and their inhibition can have a physiological and pathological impact (Morisseau, 2013). Epoxide hydrolase 4-like gene was expressed highly in the fat bodies of both male and female callow ESBB, suggesting that it may be required for lipid regulation and detoxification and also help in the physiological process upon conversion of immature callow to mature sclerotized (black) beetles. Glycosyl hydrolases hydrolyze the glycosidic bond between two or more carbohydrates or between a carbohydrate and a non-carbohydrate moiety. The enzymatic activity and ancestral origin suggest that glycoside hydrolase families 45 (GH45s) were likely essential for the adaptation of phytophagous beetles to feed on plants (Busch et al., 2019). GH45 transcript was highly expressed in both the gut and fat bodies of males and females of callow ESBB, suggesting roles in food digestion, detoxification, and host adaptation. The juvenile hormone epoxide hydrolase (JHEH) is an essential enzyme in the breakdown pathways of juvenile hormone (JH) in insects; it converts JH to JH-diol and hydrolyzes JH acid to JH acid-diol, and JHEH titers regulate the entire process of insect development. In the gypsy moth Lymantria dispar, the knockdown of JHEH1 slightly delayed larval development (Wen et al., 2018). The juvenile hormone epoxide hydrolase 1-like transcripts were highly expressed in the male and female guts and fat bodies of callow ESBB, suggesting that they may be involved in JH pathway regulation. The genome-wide microarray study in gut tissue of Indian silkworm Bombyx mori Sarupat race resistant against BmNPV infection of gut showed upregulation of the lactase-phlorizin hydrolase-like gene, indicating that the gene plays a role in the antiviral immune response (Lekha et al., 2015). The lactase-phlorizin hydrolase-like transcript was highly expressed in the gut of callow ESBB, probably performing a similar role by protecting the beetles from getting infected with pathogenic viruses during maturation feeding.
Sulfotransferase catalyzes the sulfate conjugation of catecholamines such as dopamine, prostaglandins, leukotriene E4, drugs, and xenobiotic substances by using 3’-phospho-5’-adenylyl sulfate (PAPS) as a sulfonate donor. Sulfonation enhances the water solubility of most chemicals and facilitates their excretion, but it can also lead to bioactivation and the formation of active metabolites (Shimada et al., 2004). In the present study, the sulfotransferase 1 family member D1-like transcript was highly expressed in the gut of callow ESBB, pointing toward similar xenobiotic catabolism. The chorion peroxidase is expressed in mature eggs of Aedes aegypti mosquitoes and is involved in forming a rigid and insoluble chorion (egg shell) by catalyzing chorion protein cross-linking (Han et al., 2000). Esterases are often present in antennae and help in odor desensitization (Guo and Smith, 2022). Esterase E4 transcripts were expressed in the head of callow ESBB, possibly performing a similar function. The ABCB subfamily contains both full-transporters and half-transporters; ABCB2 and ABCB10 are half-transporters with evolutionarily conserved roles and protect arthropods from oxidative stress (Wang et al., 2019). ABC transporter B family member 2-like and ATP-binding cassette sub-family B member 10 mitochondrial isoform X1 transcripts were expressed in both the gut and fat body of callow ESBB, suggesting that they may be involved in the transport and subsequent excretion of entomotoxic substances during maturation feeding.
Most of the upregulated genes were linked to cytochromes and peroxidases in the male gut, indicating roles in establishing an attack on the new host tree as the pioneer sex and capacity to aid digestion and detoxification of dietary material. Other key genes expressed in the gut, such as UGTs, GSTs, esterase sub-families, hydrolases, and peroxidases, were not differentially regulated, indicating conserved expression requirements. Cytochrome P450s are essential for insecticide tolerance in the endoparasitoid wasp Meteorus pulchricornis; the cytochrome P450 301A1 (mitochondrial) gene was expressed highly after insecticide exposure compared to control (Xing et al., 2021). In the current study, a probable cytochrome P450 301A1 (mitochondrial) transcript was highly expressed in a callow male gut and fat body of ESBB, suggesting a role in supporting male beetles to deal with host allelochemicals during host colonization (Supplementary Figure 3).
Apart from the detoxification-, digestion-, and defense-related genes, we observed that the genes related to pheromone biosynthesis were expressed in the fat body tissues, which are the site of pheromone production in ESBB. Our finding was coherent with the copulation and infestation mechanism where ESBB male, the pioneer sex, releases sex aggregation pheromones to attract the conspecifics during new host colonization (Ramakrishnan et al., 2022). Various Ips species, including I. paraconfusus and I. pini, have been observed to use the sex-specific expression of CYPs to convert host allelochemicals into aggregation pheromones (Huber et al., 2007; Song et al., 2013; Tittiger and Blomquist, 2017; Blomquist et al., 2021). However, the function of these genes is yet to be confirmed in I. typographus. Furthermore, additional validation of the pheromone biosynthesis pathway genes of ESBB may reveal sex-specific roles. All stages of ESBB are exposed to the chemical defenses of the trees to varying degrees. These enzymes are produced at different times in different tissues throughout life, and it is believed that variations in their expression or catalytic activity are related to the need for compound detoxification. The transcriptome of the sex pheromone gland of the sandfly, Lutzomyia longipalpis, revealed the expression of NADP + dependent farnesol dehydrogenase, which is implicated in the isoprenoid pathway (Gonzalez-Caballero et al., 2013). Furthermore, a farnesol dehydrogenase-like transcript was highly expressed in callow male ESBB fat body, implying its involvement in pheromone synthesis.
As the gene expression in the gut reflects the feeding behavior, differences in gene expression between callow and sclerotized adult guts have additional adaptive significance. Many physiologically important genes are differentially regulated in these tissues. For instance, the cytochrome P450 9E2-like gene showed higher gene expression in the sclerotized male gut of Ips typographus than in the callow male gut (Ramakrishnan et al., 2022). In the current study, the cytochrome P450 9E2-like was extensively expressed in the larval and adult stages, suggesting a role in ESBB feeding and aggregation pheromone production (Figure 9). In Aedes aegypti, transcriptome comparison between an insecticide-susceptible strain (Bora7) and insecticide-resistant strain (KhanhHoa7) showed upregulation of cytochrome P450 4C1, 4C3, 4C21, 4D1, 4D1 isoform X2, 4D2, 4D2 isoform X2, 4G15, 6A2, 6A8, 6D3, and 9E2 in the resistant strain (Lien et al., 2019). The comparative genome-wide analysis of single-base nucleotide polymorphisms between insecticide-resistant MED whitefly lines and insecticide-susceptible MED whitefly lines showed potential resistance markers (SNP) in cytochrome P450 4C21-like transcripts (Wang et al., 2022). In the current study, the cytochrome P450 4C1-like, cytochrome P450 4C21-like, cytochrome P450 4C3-like isoform X1, cytochrome P450 4C3-like isoform X2, cytochrome P450 4D2-like, cytochrome P450 4G15-like, cytochrome P450 6K1-like, Cytochrome P450 9E2, and cytochrome P450 9E2-like transcripts were expressed in the callow male gut, suggesting that they may have a role in detoxification mechanism during maturation feeding.
Furthermore, glutathione S-transferase, glutathione S-transferase 1, isoform D, glutathione S-transferase 1-like, glutathione S-transferase 1-like isoform X1, glutathione S-transferase-like, glutathione S-transferase-like isoform X2, and microsomal glutathione S-transferase 1-like transcripts were highly expressed in the callow male gut, suggesting that they may help freshly emerged callow beetles to detoxify plant allelochemicals. UDP-glucuronosyltransferase 1-8, UDP-glucuronosyltransferase 1-9-like isoform X1, UDP-glucuronosyltransferase 2B15-like, UDP-glucuronosyltransferase 2B16-like, UDP-glucuronosyltransferase 2B16-like, UDP-glucuronosyltransferase 2B30-like isoform X1, and UDP-glucuronosyltransferase 2B7-like transcripts were highly expressed in the callow male gut, suggesting roles in detoxification.
In the mosquito, Culex pipiens, the evolution of overproduced esterases is implicated in organophosphate pesticide resistance (Raymond et al., 1998). Esterase B1-like, esterase B1-like isoform X1, esterase E4, and esterase E4-like transcripts were highly expressed in the callow male gut, implying a role in ESBB digestion and detoxification. The mitochondrially encoded NADH: ubiquinone oxidoreductase core subunit 1 (MT-ND1) gene encodes a protein called NADH dehydrogenase 1. This protein is part of a large enzyme complex known as complex I, active in mitochondria that convert the energy from food to power the biochemical reactions in the cell (Valentino et al., 2004). NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9 (mitochondrial), NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 11 (mitochondrial), subunit 5 (mitochondrial), subunit 8 (mitochondrial), NADH dehydrogenase [ubiquinone] 1 subunit C2 and NADH dehydrogenase [ubiquinone] flavoprotein 1 (mitochondrial), and NADH dehydrogenase [ubiquinone] iron-sulfur protein 6 (mitochondrial) transcripts were highly expressed in the callow male gut, possibly to aid food digestion and energy metabolism.
In summary, many physiologically essential genes, such as cytochromes, GSTs, UGTs, esterases, and dehydrogenases, were differentially expressed in the callow or sclerotized ESBB gut, probably supporting different behavioral traits (i.e., such as maturation feeding, host finding, and new colony establishments). The ecological relevance of such fine-tuned gene expression demands further functional corroboration.
The enzymatic assay results drew attention toward fine-tuned protein expressions that reflect the feeding and non-feeding behavior of ESBB. The larvae are in an intense feeding and growth stage, which involves detoxification and digestion of host materials. During the pupal stage, the beetle becomes sedentary and non-feeding, and most of its energy reserves are deployed in ecdysis and metamorphosis. The pupae develop into the callow stage and again start feeding and become sclerotized, as they require energy to fly out, start a new attack, and aggregate the conspecifics (by male ESBB). Our assay results pinpointed that most GST proteins are abundant in pupae and have putative functions in body development and overwintering (for pupae). In contrast, cytochromes were expressed more in both larvae and adult ESBB and participate primarily in detoxification and digestion during host feeding. These results were in accordance with the previous studies suggesting life stage-specific expression of various genes in different insect groups (Perkin and Oppert, 2019; Oppert et al., 2020; Ernst and Westerman, 2021).
The science of insect physiology has dramatically benefited from RNA-seq-based studies. This trend will continue as omics technology becomes more accessible and less expensive. However, there are some limitations in transcriptomics studies. Regulatory proteins are more likely to influence physiological response than mRNAs with short half-lives, i.e., an increase or decrease in mRNA may not always equate to a corresponding change in protein abundance or activity. Such limitations also exist in the current study. To alleviate some of these limitations, we performed RT-qPCR to validate the most crucial DEGs with independently collected fresh samples. Furthermore, we used an enzymatic assay to cross-check the protein expression and activity of two key proteins in ESBB, namely, cytochromes and GSTs. The results of the enzymatic and RT-qPCR assays correlated positively with the transcriptome findings, with minor exceptions. The physiological functions of several crucial genes discussed in the present study need additional validation.
Overall, the current transcriptome study combined with RT-qPCR validation and enzymatic assays reveals gene expression dynamics specific to ESBB life stages, sex, and tissues. Our findings indicated that ESBB has allelochemical resistance mechanisms that are an essential precondition for the hormetic response in larval and adult ESBB growth, ultimately contributing to the widespread successful colonization of coniferous trees. The physiologically essential genes identified in this work should be functionally validated to better understand ESBB physiology and adaptation to a nutritionally challenging diet containing host defense allelochemicals. Furthermore, the RNAi-based approach can target key genes for survival and adaptation, identified in the present study, for ESBB management as described in detail by Joga et al. (2021).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/- PRJNA679450.
AR conceptualized the study and performed the RNA-seq analysis. AN and GS did the lab work. AR, AN, and KM interpreted the data. AN and AR prepared the figures. AN wrote the first draft. AR, KM, AN, and GS prepared the final draft. All authors read and approved the final draft.
This research was supported by grant no. CZ.02.1.01/0.0/0.0/15_003/0000433, “EXTEMIT–K project,” financed by the Operational Program Research, Development and Education (OP RDE), and Internal Grant Agency (A_06_22, AISHA NASEER) at the Faculty of Forestry and Wood Sciences, Czech University of Life Sciences, Prague, Czechia. AR and KM are supported by grant no. CZ.02. 1.01/0.0/0.0/16_019/0000803 financed by OP RDE.
We acknowledge the support from Faculty for Forestry and Wood Sciences (FLD), CZU, for conducting the present study. We also acknowledge Dr. Roman Modlinger and Dr. Jiri Synek, FLD, CZU, for assistance in beetle collection and maintenance. We acknowledge the Ips typographus genome manuscript team’s contribution in generating the valuable RNA-seq data. We appreciate the helpful feedback provided by the reviewers and especially the handling editor. Special thanks to Prof. Fredrik Schlyter for his constructive comments and encouragement.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffgc.2023.1124754/full#supplementary-material
Supplementary Figure 1 | Volcano plots of different comparisons were made for the study with the number of differentially expressed contigs (FDR p < 0.05, fold change ± 2). (A) Group-wise comparison between L1-L3 (T1-T3), pupa (T4), and sclerotized adult whole-body (T13); (B) T1 vs. T4; (C) T2 vs. T4; (D) T3 vs. T4; (E) T13 vs. T4; (F) tissue-specific group-wise comparison between the callow female head (T10), gut (T7) and fat body (T8); (G) tissue-specific group-wise comparison between the callow male head (T11), gut (T5) and fat body (T9); (H) sex-specific comparison between callow male gut (T5) vs. callow female gut (T7); (I) sex-specific comparison between callow female fat body (T8) vs. callow male fat body (T9); (J) comparison between sclerotized male gut (T6) vs. callow male gut (T5); and (K) group-wise comparison between all gut tissues sclerotized male gut (T6), callow male gut (T5), and callow female gut (T7). N = 5. Up-head arrows represent upregulation (fold change ≥ 2), and down-head arrows represent downregulation (fold change ≤ −2). Red dots represent significantly expressed contigs (FDR p < 0.05), and blue dots represent non-significantly expressed contigs (FDR p > 0.05). The X-axis represents the log2 fold-change plotted against −log10 (p-values).
Supplementary Figure 2 | Dynamics of other detoxification-related genes. Different life stages of ESBB on the x-axis are plotted against the average CPM values of all the contigs of a gene family on the y-axis. Different line colors represent individual gene families. The total CPM was calculated by taking the average of all the differentially expressed contigs (FDR p < 0.05, fold change ± 2) of each gene family. T1, T2, and T3 represent the first, second, and third larval stage, respectively, T4 represent pupal stage, and T13 represent the sclerotized adult stage of I. typographus.
Supplementary Figure 3 | Sex-specific comparison. (A) Callow male gut (T5) vs. Callow female gut (T7) (N = 5). (B) Callow female fat body (T8) vs. Callow male fat body (T9) (N = 5). (i) PCA plot showing sample clustering. (ii) Heatmaps representing gene clustering. The color spectrum, stretching from blue to red, represents TMM-adjusted log CPM expression values obtained after DGE analysis. (iii) Bar graphs showing differentially expressed contigs of specific detoxification-related enzyme families and other essential enzymes related to defense, digestion, transport, metabolism, signaling, and growth. Analysis was done using the CLC workbench (FDR p < 0.05 and fold change ± 2).
Supplementary Figure 4 | Group-wise comparison between sclerotized male gut (T6), callow male gut (T5), and callow female gut (T7) (N = 5). (i) PCA plot showing sample clustering. (ii) Heatmaps representing gene clustering. The color spectrum, stretching from blue to red, represents TMM-adjusted log CPM expression values obtained after DGE analysis. (iii) Bar graphs showing differentially expressed contigs of specific detoxification-related enzyme families and other essential enzymes related to defense, digestion, transport, metabolism, signaling, and growth after group-wise comparison between the three tissues. Analysis was done using CLC workbench with FDR p < 0.05 and fold change ± 2 cut off.
Supplementary Table 1 | RNA-seq statistics for ESBB life stage and tissue transcriptomes (T1-T13).
Supplementary Table 2 | Protein concentrations for four replicates used in enzymatic assays (T1-T6 and T13).
Supplementary Material 1 | Metadata file for all the life stages and tissues used for the study.
Supplementary Material 2 | Functional information about the genes used for designing primer for RT-qPCR and their functional information in NCBI BLASTn.
Supplementary Material 3 | Expression browser for group-wise comparison between all life stages (T1, T2, T3, T4, and T13) containing all the differentially expressed genes (FDR p < 0.05, fold change ± 2) and their mean CPM values for each stage (N = 5).
Supplementary Material 4 | Expression browser for all the differentially expressed genes in T1 compared against T4 as control (FDR p < 0.05, fold change ± 2) and their mean CPM values for each stage (N = 5).
Supplementary Material 5 | Expression browser for all the differentially expressed genes in T2 compared against T4 as control (FDR p < 0.05, fold change ± 2) and their mean CPM values for each stage (N = 5).
Supplementary Material 6 | Expression browser for all the differentially expressed genes in T3 compared against T4 as control (FDR p < 0.05, fold change ± 2) and their mean CPM values for each stage (N = 5).
Supplementary Material 7 | Expression browser for all the differentially expressed genes in T13 compared against T4 as control (FDR p < 0.05, fold change ± 2) and their mean CPM values for each stage (N = 5).
Supplementary Material 8 | Expression browser for group-wise comparison for tissue-specific female comparison between callow female gut (T7), callow female fat body (T8), and callow female head (T10) containing all the differentially expressed genes (FDR p < 0.05, fold change ± 2) and their mean CPM values for each stage (N = 5).
Supplementary Material 9 | Expression browser for group-wise comparison for tissue-specific male comparison between callow male gut (T5), callow male fat body (T9), and callow male head (T11) containing all the differentially expressed genes (FDR p < 0.05, fold change ± 2) and their mean CPM values for each stage (N = 5).
Supplementary Material 10 | Expression browser for all the differentially expressed genes in the callow female gut (T7) compared against callow male gut (T5) as control (FDR p < 0.05, fold change ± 2) and their mean CPM values for each stage (N = 5).
Supplementary Material 11 | Expression browser for all the differentially expressed genes in callow female fat body (T8) compared against callow male fat body (T9) as control (FDR p < 0.05, fold change ± 2) and their mean CPM values for each stage (N = 5).
Supplementary Material 12 | Expression browser for all the differentially expressed genes in the sclerotized male gut (T6) compared against callow male gut (T5) as control (FDR p < 0.05, fold change ± 2) and their mean CPM values for each stage (N = 5).
Supplementary Material 13 | Expression browser for group-wise comparison for tissue-specific male comparison between callow male gut (T5), sclerotized male gut (T6), and callow female gut (T7) containing all the differentially expressed genes (FDR p < 0.05, fold change ± 2) and their mean CPM values for each stage (N = 5).
Supplementary Material 14 | Fisher extract test result for all life stages (T1, T2, T3, T4, and T13) comparison for differentially upregulated transcript IDs (FDR p < 0.05, fold change ≥ 2).
Supplementary Material 15 | Fisher extract result for all life stage (T1, T2, T3, T4, and T13) comparison for differentially downregulated transcript IDs (FDR p < 0.05, fold change ≤ −2).
Supplementary Material 16 | Fisher extract test result for T1 vs. T4 comparison for differentially upregulated transcript IDs (FDR p < 0.05, fold change ≥ 2).
Supplementary Material 17 | Fisher extract test result for T1 vs. T4 comparison for differentially downregulated transcript IDs (FDR p < 0.05, fold change ≤ −2).
Supplementary Material 18 | Fisher extract test result for T2 vs. T4 comparison for differentially upregulated transcript IDs (FDR p < 0.05, fold change ≥ 2).
Supplementary Material 19 | Fisher extract test result for T2 vs. T4 comparison for differentially downregulated transcript IDs (FDR p < 0.05, fold change ≤ −2).
Supplementary Material 20 | Fisher extract test result for T3 vs. T4 comparison for differentially upregulated transcript IDs (FDR p < 0.05, fold change ≥ 2).
Supplementary Material 21 | Fisher extract test result for T3 vs. T4 comparison for differentially downregulated transcript IDs (FDR p < 0.05, fold change ≤ −2).
Supplementary Material 22 | Fisher extract test result for T13 vs. T4 comparison for differentially upregulated transcript IDs (FDR p < 0.05, fold change ≥ 2).
Supplementary Material 23 | Fisher extract test result for T13 vs. T4 comparison for differentially downregulated transcript IDs (FDR p < 0.05, fold change ≤ −2).
Supplementary Material 24 | Fisher extract test result for T7, T8, and T10 comparison for differentially upregulated transcript IDs (FDR p < 0.05, fold change ≥ 2).
Supplementary Material 25 | Fisher extract test result T7, T8, and T10 comparison for differentially downregulated transcript IDs (FDR p < 0.05, fold change ≤ −2).
Supplementary Material 26 | Fisher extract test result for T5, T9, and T11 comparison for differentially upregulated transcript IDs (FDR p < 0.05, fold change ≥ 2).
Supplementary Material 27 | Fisher extract test result T5, T9, and T11 comparison for differentially downregulated transcript IDs (FDR p < 0.05, fold change ≤ −2).
Supplementary Material 28 | Fisher extract test result for T7 vs. T5 comparison for differentially upregulated transcript IDs (FDR p < 0.05, fold change ≥ 2).
Supplementary Material 29 | Fisher extract test result for T7 vs. T5 comparison for differentially downregulated transcript IDs (FDR p < 0.05, fold change ≤ −2).
Supplementary Material 30 | Fisher extract test result for T8 vs. T9 comparison for differentially upregulated transcript IDs (FDR p < 0.05, fold change ≥ 2).
Supplementary Material 31 | Fisher extract test result for T8 vs. T9 comparison for differentially downregulated transcript IDs (FDR p < 0.05, fold change ≤ −2).
Supplementary Material 32 | Fisher extract test result for T6 vs. T5 comparison for differentially upregulated transcript IDs (FDR p < 0.05, fold change ≥ 2).
Supplementary Material 33 | Fisher extract test result for T6 vs. T5 comparison for differentially downregulated transcript IDs (FDR p < 0.05, fold change ≤ −2).
Supplementary Material 34 | Fisher extract test result for T5, T6, and T7 comparison for differentially upregulated transcript IDs (FDR p < 0.05, fold change ≥ 2).
Supplementary Material 35 | Fisher extract test result T5, T6, and T7 comparison for differentially downregulated transcript IDs (FDR p < 0.05, fold change ≤ −2).
Supplementary Material 36 | List of common and unique differentially upregulated (FDR p < 0.05, fold change ≥ 2) transcript IDs between life stages (T1, T2, T3, T4, and T5) described in Figure 5.
Supplementary Material 37 | List of common and unique differentially downregulated (FDR p < 0.05, fold change ≤ −2) transcript IDs between life stages (T1, T2, T3, T4, and T5) described in Figure 6.
Ahn, S. J., Vogel, H., and Heckel, D. G. (2012). Comparative analysis of the UDP-glycosyltransferase multigene family in insects. Insect Biochem. Mol. Biol. 42, 133–147. doi: 10.1016/j.ibmb.2011.11.006
Akiyama, M. (2014). The roles of ABCA12 in epidermal lipid barrier formation and keratinocyte differentiation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1841, 435–440. doi: 10.1016/j.bbalip.2013.08.009
Balakrishnan, B., Su, S., Wang, K., Tian, R. Z., and Chen, M. H. (2018). Identification, Expression, and regulation of an omega class glutathione S-transferase in Rhopalosiphum padi (L.) (Hemiptera: Aphididae) under insecticide stress. Front. Physiol. 9:427. doi: 10.3389/fphys.2018.00427
Baldwin, S. R., Mohapatra, P., Nagalla, M., Sindvani, R., Amaya, D., Dickson, H. A., et al. (2021). Identification and characterization of CYPs induced in the Drosophila antenna by exposure to a plant odorant. Sci. Rep. 11:20530. doi: 10.1038/s41598-021-99910-9
Basinger, A. A., Booker, J. K., Frazier, D. M., Koeberl, D. D., Sullivan, J. A., and Muenzer, J. (2006). Glutaric acidemia type 1 in patients of Lumbee heritage from North Carolina. Mol. Genet. Metab. 88, 90–92. doi: 10.1016/j.ymgme.2005.12.008
Biedermann, P. H. W., Muller, J., Gregoire, J. C., Gruppe, A., Hagge, J., Hammerbacher, A., et al. (2019). Bark beetle population dynamics in the Anthropocene: Challenges and solutions. Trends Ecol. Evol. 34, 914–924. doi: 10.1016/j.tree.2019.06.002
Blomquist, G. J., Tittiger, C., MacLean, M., and Keeling, C. I. (2021). Cytochromes P450: Terpene detoxification and pheromone production in bark beetles. Curr. Opin. Insect Sci. 43, 97–102. doi: 10.1016/j.cois.2020.11.010
Bosch-Serra, D., Rodríguez, M. A., Avilla, J., Sarasúa, M. J., and Miarnau, X. (2021). Esterase, glutathione s-transferase and NADPH-cytochrome P450 reductase activity evaluation in Cacopsylla pyri L.(Hemiptera: Psyllidae) individual adults. Insects 12:329. doi: 10.3390/insects12040329
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254.
Bras, A., Roy, A., Heckel, D. G., Anderson, P., and Karlsson Green, K. (2022). Pesticide resistance in arthropods: Ecology matters too. Ecol. Lett. 25, 1746–1759. doi: 10.1111/ele.14030
Busch, A., Danchin, E. G. J., and Pauchet, Y. (2019). Functional diversification of horizontally acquired glycoside hydrolase family 45 (GH45) proteins in phytophaga beetles. BMC Evol. Biol. 19:100. doi: 10.1186/s12862-019-1429-9
Calandra, S., Tarugi, P., Speedy, H. E., Dean, A. F., Bertolini, S., and Shoulders, C. C. (2011). Mechanisms and genetic determinants regulating sterol absorption, circulating LDL levels, and sterol elimination: Implications for classification and disease risk. J. Lipid Res. 52, 1885–1926. doi: 10.1194/jlr.R017855
Cale, J. A., Ding, R. S., Wang, F. A., Rajabzadeh, R., and Erbilgin, N. (2019). Ophiostomatoid fungi can emit the bark beetle pheromone verbenone and other semiochemicals in media amended with various pine chemicals and beetle-released compounds. Fungal Ecol. 39, 285–295. doi: 10.1016/j.funeco.2019.01.003
Cano-Ramirez, C., Lopez, M. F., Cesar-Ayala, A. K., Pineda-Martinez, V., Sullivan, B. T., and Zuniga, G. (2013). Isolation and expression of cytochrome P450 genes in the antennae and gut of pine beetle Dendroctonus rhizophagus (Curculionidae: Scolytinae) following exposure to host monoterpenes. Gene 520, 47–63. doi: 10.1016/j.gene.2012.11.059
Celedon, J. M., and Bohlmann, J. (2019). Oleoresin defenses in conifers: Chemical diversity, terpene synthases and limitations of oleoresin defense under climate change. New Phytol. 224, 1444–1463. doi: 10.1111/nph.15984
Chakraborty, A., Ashraf, M. Z., Modlinger, R., Synek, J., Schlyter, F., and Roy, A. (2020a). Unravelling the gut bacteriome of Ips (Coleoptera: Curculionidae: Scolytinae): Identifying core bacterial assemblage and their ecological relevance. Sci. Rep. 10:18572. doi: 10.1038/s41598-020-75203-5
Chakraborty, A., Modlinger, R., Ashraf, M. Z., Synek, J., Schlyter, F., and Roy, A. (2020b). Core mycobiome and their ecological relevance in the gut of five Ips bark beetles (Coleoptera: Curculionidae: Scolytinae). Front. Microbiol. 11:568853. doi: 10.3389/fmicb.2020.568853
Chiu, C. C., Keeling, C. I., and Bohlmann, J. (2019a). The cytochrome P450 CYP6DE1 catalyzes the conversion of α-pinene into the mountain pine beetle aggregation pheromone trans-verbenol. Sci. Rep. 9:1477. doi: 10.1038/s41598-018-38047-8
Chiu, C. C., Keeling, C. I., Henderson, H. M., and Bohlmann, J. (2019b). Functions of mountain pine beetle cytochromes P450 CYP6DJ1, CYP6BW1 and CYP6BW3 in the oxidation of pine monoterpenes and diterpene resin acids. PLoS One 14:e0216753. doi: 10.1371/journal.pone.0216753
Cui, X. P., Wang, C., Wang, X. X., Li, G. L., Liu, Z. G., Wang, H. F., et al. (2020). Molecular mechanism of the UDP-glucuronosyltransferase 2b20-like gene (AccUGT2B20-like) in pesticide resistance of Apis cerana cerana. Front. Genet. 11:592595. doi: 10.3389/fgene.2020.592595
Dai, L. L., Ma, M. Y., Wang, C. Y., Shi, Q., Zhang, R. R., and Chen, H. (2015). Cytochrome P450s from the Chinese white pine beetle, Dendroctonus armandi (Curculionidae: Scolytinae): Expression profiles of different stages and responses to host allelochemicals. Insect Biochem. Mol. Biol. 65, 35–46. doi: 10.1016/j.ibmb.2015.08.004
Davis, T. S. (2015). The ecology of yeasts in the bark beetle holobiont: A century of research revisited. Microb. Ecol. 69, 723–732. doi: 10.1007/s00248-014-0479-1
Enayati, A. A., Ranson, H., and Hemingway, J. (2005). Insect glutathione transferases and insecticide resistance. Insect Mol. Biol. 14, 3–8. doi: 10.1111/j.1365-2583.2004.00529.x
Ernst, D. A., and Westerman, E. L. (2021). Stage-and sex-specific transcriptome analyses reveal distinctive sensory gene expression patterns in a butterfly. BMC Genomics 22:584. doi: 10.1186/s12864-021-07819-4
Faccoli, M., Blazenec, M., and Schlyter, F. (2005). Feeding response to host and nonhost compounds by males and females of the spruce bark beetle Ips typographus in a tunneling microassay. J. Chem. Ecol. 31, 745–759. doi: 10.1007/s10886-005-3542-z
Faldt, J., Solheim, H., Langstrom, B., and Borg-Karlson, A. K. (2006). Influence of fungal infection and wounding on contents and enantiomeric compositions of monoterpenes in phloem of Pinus sylvestris. J. Chem. Ecol. 32, 1779–1795. doi: 10.1007/s10886-006-9109-9
Fang, J. X., Zhang, S. F., Liu, F., Cheng, B., Zhang, Z., Zhang, Q. H., et al. (2021). Functional investigation of monoterpenes for improved understanding of the relationship between hosts and bark beetles. J. Appl. Entomol. 145, 303–311. doi: 10.1111/jen.12850
Field, L. M., and Devonshire, A. L. (1998). Evidence that the E4 and FE4 esterase genes responsible for insecticide resistance in the aphid Myzus persicae (Sulzer) are part of a gene family. Biochem. J. 330, 169–173. doi: 10.1042/bj3300169
Figueroa-Teran, R., Pak, H., Blomquist, G. J., and Tittiger, C. (2016). High substrate specificity of ipsdienol dehydrogenase (IDOLDH), a short-chain dehydrogenase from Ips pini bark beetles. J. Biochem. 160, 141–151. doi: 10.1093/jb/mvw019
Franceschi, V. R., Krokene, P., Christiansen, E., and Krekling, T. (2005). Anatomical and chemical defenses of conifer bark against bark beetles and other pests. New Phytol. 167, 353–375. doi: 10.1111/j.1469-8137.2005.01436.x
Fukuda, S., Hamada, T., Ishii, N., Sakaguchi, S., Sakai, K., Akiyama, M., et al. (2012). Novel adenosine triphosphate (ATP)-binding cassette, subfamily A, member 12 (ABCA12) mutations associated with congenital ichthyosiform erythroderma. Br. J. Dermatol. 166, 218–221. doi: 10.1111/j.1365-2133.2011.10516.x
Gaddelapati, S. C., Kalsi, M., Roy, A., and Palli, S. R. (2018). Cap ‘n’ collar C regulates genes responsible for imidacloprid resistance in the Colorado potato beetle, Leptinotarsa decemlineata. Insect Biochem. Mol. Biol. 99, 54–62. doi: 10.1016/j.ibmb.2018.05.006
Gao, H. M., Dai, L. L., Fu, D. Y., Sun, Y. Y., and Chen, H. (2020). Isolation, expression profiling, and regulation via host allelochemicals of 16 glutathione S-transferases in the Chinese white pine beetle, Dendroctonus armandi. Front. Physiol. 11:546592. doi: 10.3389/fphys.2020.546592
Gong, H., Niu, Q., Zhou, Y., Wang, Y. X., Xu, X. F., and Hou, K. Z. (2021). Notum palmitoleoyl-protein carboxylesterase regulates Fas cell surface death receptor-mediated apoptosis via the Wnt signaling pathway in colon adenocarcinoma. Bioengineered 12, 5241–5252. doi: 10.1080/21655979.2021.1961657
Gonzalez-Caballero, N., Valenzuela, J. G., Mc Ribeiro, J., Cuervo, P., and Brazil, R. P. (2013). Transcriptome exploration of the sex pheromone gland of Lutzomyia longipalpis (Diptera: Psychodidae: Phlebotominae). Parasit. Vectors 6, 56. doi: 10.1186/1756-3305-6-56
Guittard, E., Blais, C., Maria, A., Parvy, J. P., Pasricha, S., Lumb, C., et al. (2011). CYP18A1, a key enzyme of Drosophila steroid hormone inactivation, is essential for metamorphosis. Dev. Biol. 349, 35–45. doi: 10.1016/j.ydbio.2010.09.023
Guo, H., and Smith, D. P. (2022). Time-Dependent Odorant Sensitivity Modulation in Insects. Insects 13:354.
Guo, R., Chen, D. F., Diao, Q. Y., Xiong, C. L., Zheng, Y. Z., and Hou, C. S. (2019). Transcriptomic investigation of immune responses of the Apis cerana cerana larval gut infected by Ascosphaera apis. J. Invertebr. Pathol. 166:107210. doi: 10.1016/j.jip.2019.107210
Habig, W. H., and Jakoby, W. B. (1981). [51] Assays for differentiation of glutathione S-transferases. Methods Enzymol. 77, 398–405.
Hammerbacher, A., Schmidt, A., Wadke, N., Wright, L. P., Schneider, B., Bohlmann, J., et al. (2013). A common fungal associate of the spruce bark beetle metabolizes the stilbene defenses of Norway spruce. Plant Physiol. 162, 1324–1336. doi: 10.1104/pp.113.218610
Han, Q., Li, G., and Li, J. (2000). Purification and characterization of chorion peroxidase from Aedes aegypti eggs. Arch. Biochem. Biophys. 378, 107–115. doi: 10.1006/abbi.2000.1821
Heckel, D. G. (2014). Insect detoxification and sequestration strategies. Annu. Plant Rev. Online 47, 77–114.
Heidel-Fischer, H. M., and Vogel, H. (2015). Molecular mechanisms of insect adaptation to plant secondary compounds. Curr. Opin. Insect Sci. 8, 8–14. doi: 10.1016/j.cois.2015.02.004
Helvig, C., Koener, J., Unnithan, G., and Feyereisen, R. (2004). CYP15A1, the cytochrome P450 that catalyzes epoxidation of methyl farnesoate to juvenile hormone III in cockroach corpora allata. Proc. Natl. Acad. Sci. U.S.A. 101, 4024–4029. doi: 10.1073/pnas.0306980101
Hilliou, F., Chertemps, T., Maibeche, M., and Le Goff, G. (2021). Resistance in the genus Spodoptera: Key insect detoxification genes. Insects 12:544. doi: 10.3390/insects12060544
Hlásny, T., Krokene, P., Liebhold, A., Montagné-Huck, C., Müller, J., Qin, H., et al. (2019). Living with bark beetles: Impacts, outlook and management options. Joensuu: European Forest Institute.
Hlasny, T., Zimova, S., and Bentz, B. (2021). Scientific response to intensifying bark beetle outbreaks in Europe and North America. For. Ecol. Manag. 499:119599. doi: 10.1016/j.foreco.2021.119599
Hlásny, T., Zimová, S., Merganičová, K., Štěpánek, P., Modlinger, R., and Turčáni, M. (2021). Devastating outbreak of bark beetles in the Czech republic: Drivers, impacts, and management implications. For. Ecol. Manag. 490:119075.
Huang, H. Y., Gao, Q., Peng, X. X., Choi, S. Y., Sarma, K., Ren, H. M., et al. (2011). piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev. Cell 20, 376–387. doi: 10.1016/j.devcel.2011.01.004
Huang, J. B., Kautz, M., Trowbridge, A. M., Hammerbacher, A., Raffa, K. F., Adams, H. D., et al. (2020). Tree defence and bark beetles in a drying world: Carbon partitioning, functioning and modelling. New Phytol. 225, 26–36. doi: 10.1111/nph.16173
Huang, Y., Liao, M., Yang, Q., Xiao, J., Hu, Z., and Cao, H. (2019). iTRAQ-based quantitative proteome revealed metabolic changes of Sitophilus zeamais in response to terpinen-4-ol fumigation. Pest Manag. Sci. 75, 444–451. doi: 10.1002/ps.5135
Huang, Y., Liao, M., Yang, Q., Xiao, J., Hu, Z., Zhou, L., et al. (2018). Transcriptome profiling reveals differential gene expression of detoxification enzymes in Sitophilus zeamais responding to terpinen-4-ol fumigation. Pestic. Biochem. Physiol. 149, 44–53. doi: 10.1016/j.pestbp.2018.05.008
Huber, D. P., Erickson, M. L., Leutenegger, C. M., Bohlmann, J., and Seybold, S. J. (2007). Isolation and extreme sex-specific expression of cytochrome P450 genes in the bark beetle, Ips paraconfusus, following feeding on the phloem of host ponderosa pine, Pinus ponderosa. Insect Mol. Biol. 16, 335–349. doi: 10.1111/j.1365-2583.2007.00731.x
Jakuš, R., Edwards-Jonášová, M., Cudlín, P., Blaženec, M., Ježík, M., Havlíèek, F., et al. (2011). Characteristics of Norway spruce trees (Picea abies) surviving a spruce bark beetle (Ips typographus L.) outbreak. Trees 25, 965–973.
Jin, M., Liao, C., Fu, X., Holdbrook, R., Wu, K., and Xiao, Y. (2019). Adaptive regulation of detoxification enzymes in Helicoverpa armigera to different host plants. Insect Mol. Biol. 28, 628–636. doi: 10.1111/imb.12578
Joga, M. R., Mogilicherla, K., Smagghe, G., and Roy, A. (2021). RNA interference-based forest protection products (FPPs) against wood-boring coleopterans: Hope or hype? Front. Plant Sci. 12:733608. doi: 10.3389/fpls.2021.733608
Keeling, C. I., Li, M., Sandhu, H. K., Henderson, H., Saint Yuen, M. M., and Bohlmann, J. (2016). Quantitative metabolome, proteome and transcriptome analysis of midgut and fat body tissues in the mountain pine beetle, Dendroctonus ponderosae Hopkins, and insights into pheromone biosynthesis. Insect Biochem. Mol. Biol. 70, 170–183. doi: 10.1016/j.ibmb.2016.01.002
Kim, J. I., Kwon, M., Kim, G.-H., Kim, S. Y., and Lee, S. H. (2015). Two mutations in nAChR beta subunit is associated with imidacloprid resistance in the Aphis gossypii. J. Asia Pac. Entomol. 18, 291–296.
Kostaropoulos, I., Papadopoulos, A. I., Metaxakis, A., Boukouvala, E., and Papadopoulou-Mourkidou, E. (2001). The role of glutathione S-transferases in the detoxification of some organophosphorus insecticides in larvae and pupae of the yellow mealworm, Tenebrio molitor (Coleoptera: Tenebrionidae). Pest Manag. Sci. 57, 501–508. doi: 10.1002/ps.323
Krasney, P. A., Carr, C., and Cavener, D. R. (1990). Evolution of the glucose-dehydrogenase gene in Drosophila. Mol. Biol. Evol. 7, 155–177.
Krempl, C., Sporer, T., Reichelt, M., Ahn, S.-J., Heidel-Fischer, H., Vogel, H., et al. (2016). Potential detoxification of gossypol by UDP-glycosyltransferases in the two Heliothine moth species Helicoverpa armigera and Heliothis virescens. Insect Biochem. Mol. Biol. 71, 49–57. doi: 10.1016/j.ibmb.2016.02.005
Krokene, P., and Solheim, H. (1998). Pathogenicity of four blue-stain fungi associated with aggressive and nonaggressive bark beetles. Phytopathology 88, 39–44. doi: 10.1094/phyto.1998.88.1.39
Lee, S., Kim, J. J., and Breuil, C. (2006). Diversity of fungi associated with the mountain pine beetle, Dendroctonus ponderosae and infested Lodgepole pines in British Columbia. Fungal Divers. 22, 91–105.
Lekha, G., Gupta, T., Awasthi, A. K., Murthy, G. N., Trivedy, K., and Ponnuvel, K. M. (2015). Genome wide microarray based expression profiles associated with BmNPV resistance and susceptibility in Indian silkworm races of Bombyx mori. Genomics 106, 393–403. doi: 10.1016/j.ygeno.2015.09.002
Li, J. S., Kim, S. R., and Li, J. Y. (2004). Molecular characterization of a novel peroxidase involved in Aedes aegypti chorion protein crosslinking. Insect Biochem. Mol. Biol. 34, 1195–1203. doi: 10.1016/j.ibmb.2004.08.001
Li, Q. L., Sun, Z. X., Shi, Q., Wang, R. M., Xu, C. C., Wang, H. H., et al. (2019). RNA-Seq analyses of midgut and fat body tissues reveal the molecular mechanism underlying Spodoptera litura resistance to tomatine. Front. Physiol. 10:8. doi: 10.3389/fphys.2019.00008
Lien, N. T. K., Ngoc, N. T. H., Lan, N. N., Hien, N. T., Tung, N. V., Ngan, N. T. T., et al. (2019). Transcriptome sequencing and analysis of changes associated with insecticide resistance in the dengue mosquito (Aedes aegypti) in Vietnam. Am. J. Trop. Med. Hyg. 100, 1240–1248. doi: 10.4269/ajtmh.18-0607
Liu, B., Fu, D., Ning, H., Tang, M., and Chen, H. (2022). Knockdown of CYP6CR2 and CYP6DE5 reduces tolerance to host plant allelochemicals in the Chinese white pine beetle Dendroctonus armandi. Pestic. Biochem. Physiol. 187:105180. doi: 10.1016/j.pestbp.2022.105180
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. Methods 25, 402–408.
Lu, K., Song, Y. Y., and Zeng, R. S. (2021). The role of cytochrome P450-mediated detoxification in insect adaptation to xenobiotics. Curr. Opin. Insect Sci. 43, 103–107. doi: 10.1016/j.cois.2020.11.004
Lubojacký, J., Lorenc, F., Samek, M., Knížek, M., and Liška, J. (2022). “Hlavní problémy v ochraně lesa v Česku v roce 2021 a prognóza na rok 2022,” in Škodliví činitelé v lesích Česka 2021/2022 – Škody zvěří. Sborník referátů z celostátního semináře s mezinárodní účastí. Průhonice, April 28, 2022, ed. F. Lorenc (Průhonice: Zpravodaj Ochrany Lesa), 17–26.
Margoliash, E., and Frohwirt, N. (1959). Spectrum of horse-heart cytochrome c. Biochem. J. 71, 570–572.
Meng, X. K., Zhang, Y. X., Bao, H. B., and Liu, Z. W. (2015). Sequence analysis of insecticide action and detoxification-related genes in the insect pest natural enemy Pardosa pseudoannulata. PLoS One 10:e0125242. doi: 10.1371/journal.pone.0125242
Morisseau, C. (2013). Role of epoxide hydrolases in lipid metabolism. Biochimie 95, 91–95. doi: 10.1016/j.biochi.2012.06.011
Nadeau, J. A., Petereit, J., Tillett, R. L., Jung, K., Fotoohi, M., MacLean, M., et al. (2017). Comparative transcriptomics of mountain pine beetle pheromone-biosynthetic tissues and functional analysis of CYP6DE3. BMC Genomics 18:311. doi: 10.1186/s12864-017-3696-4
Netherer, S., and Hammerbacher, A. (2022). “The Eurasian spruce bark beetle in a warming climate: Phenology, behavior, and biotic interactions,” in Bark beetle management, ecology, and climate change, eds R. W. Hofstetter and K. J. K. Gandhi (Amsterdam: Elsevier), 89–131.
Netherer, S., Kandasamy, D., Jirosova, A., Kalinova, B., Schebeck, M., and Schlyter, F. (2021). Interactions among Norway spruce, the bark beetle Ips typographus and its fungal symbionts in times of drought. J. Pest Sci. 94, 591–614. doi: 10.1007/s10340-021-01341-y
Neumann, E., Mehboob, H., Ramirez, J., Mirkov, S., Zhang, M., and Liu, W. Q. (2016). Age-dependent hepatic UDP-glucuronosyltransferase gene expression and activity in children. Front. Pharmacol. 7:437. doi: 10.3389/fphar.2016.00437
Niwa, R., Matsuda, T., Yoshiyama, T., Namiki, T., Mita, K., Fujimoto, Y., et al. (2004). CYP306A1, a cytochrome P450 enzyme, is essential for ecdysteroid biosynthesis in the prothoracic glands of Bombyx and Drosophila. J. Biol. Chem. 279, 35942–35949. doi: 10.1074/jbc.M404514200
Oldfield, S., Lowry, C. A., Ruddick, J., and Lightman, S. L. (2002). ABCG4: A novel human white family ABC-transporter expressed in the brain and eye. Biochim. Biophys. Acta Mol. Cell Res. 1591, 175–179. doi: 10.1016/s0167-4889(02)00269-0
Oppert, B., Perkin, L. C., Lorenzen, M., and Dossey, A. T. (2020). Transcriptome analysis of life stages of the house cricket, Acheta domesticus, to improve insect crop production. Sci. Rep. 10:3471. doi: 10.1038/s41598-020-59087-z
Ortego, F., López-Olguıìn, J., Ruız, M., and Castañera, P. (1999). Effects of toxic and deterrent terpenoids on digestive protease and detoxication enzyme activities of Colorado potato beetle larvae. Pestic. Biochem. Physiol. 63, 76–84.
Perkin, L. C., and Oppert, B. (2019). Gene expression in Tribolium castaneum life stages: Identifying a species-specific target for pest control applications. PeerJ 7:e6946.
Plaitakis, A., Kalef-Ezra, E., Kotzamani, D., Zaganas, I., and Spanaki, C. (2017). The glutamate dehydrogenase pathway and its roles in cell and tissue biology in health and disease. Biology 6:11. doi: 10.3390/biology6010011
Plaitakis, A., Metaxari, M., and Shashidharan, P. (2000). Nerve tissue-specific (GLUD2) and housekeeping (GLUD1) human glutamate dehydrogenases are regulated by distinct allosteric mechanisms: Implications for biologic function. J. Neurochem. 75, 1862–1869. doi: 10.1046/j.1471-4159.2000.0751862.x
Powell, D., Grosse-Wilde, E., Krokene, P., Roy, A., Chakraborty, A., Lofstedt, C., et al. (2021). A highly-contiguous genome assembly of the Eurasian spruce bark beetle, Ips typographus, provides insight into a major forest pest. Commun. Biol. 4:1059. doi: 10.1038/s42003-021-02602-3
Ramakrishnan, R., Hradecky, J., Roy, A., Kalinova, B., Mendezes, R. C., Synek, J., et al. (2022). Metabolomics and transcriptomics of pheromone biosynthesis in an aggressive forest pest Ips typographus. Insect Biochem. Mol. Biol. 140:103680. doi: 10.1016/j.ibmb.2021.103680
Raymond, M., Chevillon, C., Guillemaud, T., Lenormand, T., and Pasteur, N. (1998). An overview of the evolution of overproduced esterases in the mosquito Culex pipiens. Philos. Trans. R. Soc. B Biol. Sci. 353, 1707–1711. doi: 10.1098/rstb.1998.0322
Robert, J. A., Bonnett, T., Pitt, C., Spooner, L. J., Fraser, J., Yuen, M. M. S., et al. (2016). Gene expression analysis of overwintering mountain pine beetle larvae suggests multiple systems involved in overwintering stress, cold hardiness, and preparation for spring development. PeerJ 4:e2109. doi: 10.7717/peerj.2109
Robert, J. A., Pitt, C., Bonnett, T. R., Yuen, M. M. S., Keeling, C. I., Bohlmann, J., et al. (2013). Disentangling detoxification: Gene expression analysis of feeding mountain pine beetle illuminates molecular-level host chemical defense detoxification mechanisms. PLoS One 8:e77777. doi: 10.1371/journal.pone.0077777
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140.
Roy, A., George, S., and Palli, S. R. (2017). Multiple functions of CREB-binding protein during postembryonic development: Identification of target genes. BMC Genomics 18:996. doi: 10.1186/s12864-017-4373-3
Roy, A., Walker, I. W., Vogel, H., Chattington, S., Larsson, M., Anderson, P., et al. (2016). Diet dependent metabolic responses in three generalist insect herbivores Spodoptera spp. Insect Biochem. Mol. Biol. 71, 91–105. doi: 10.1016/j.ibmb.2016.02.006
Sandstrom, P., Welch, W. H., Blomquist, G. J., and Tittiger, C. (2006). Functional expression of a bark beetle cytochrome P450 that hydroxylates myrcene to ipsdienol. Insect Biochem. Mol. Biol. 36, 835–845. doi: 10.1016/j.ibmb.2006.08.004
Sarabia, L. E., Lopez, M. F., Pineda-Mendoza, R. M., Obregon-Molina, G., Gonzalez-Escobedo, R., Albores-Medina, A., et al. (2019). Time-course of CYP450 genes expression from Dendroctonus rhizophagus (Curculionidae: Scolytinae) during early hours of drilling bark and settling into the host tree. J. Insect Sci. 19:7. doi: 10.1093/jisesa/iez046
Sellamuthu, G., Bílý, J., Joga, M. R., Synek, J., and Roy, A. (2022). Identifying optimal reference genes for gene expression studies in Eurasian spruce bark beetle, Ips typographus (Coleoptera: Curculionidae: Scolytinae). Sci. Rep. 12:4671. doi: 10.1038/s41598-022-08434-3
Shen, G. W., Wu, J. X., Wang, Y., Liu, H. L., Zhang, H. Y., Ma, S. Y., et al. (2018). The expression of ecdysteroid UDP-glucosyltransferase enhances cocoon shell ratio by reducing ecdysteroid titre in last-instar larvae of silkworm, Bombyx mori. Sci. Rep. 8:17710. doi: 10.1038/s41598-018-36261-y
Shen, X. A., Liu, W. X., Wan, F. H., Lv, Z. C., and Guo, J. Y. (2021). The role of cytochrome P450 4C1 and carbonic anhydrase 3 in response to temperature stress in Bemisia tabaci. Insects 12:1071. doi: 10.3390/insects12121071
Shimada, M., Terazawa, R., Kamiyama, Y., Honma, W., Nagata, K., and Yamazoe, Y. (2004). Unique properties of a renal sulfotransferase, st1d1, in dopamine metabolism. J. Pharmacol. Exp. Ther. 310, 808–814. doi: 10.1124/jpet.104.065532
Song, M., Gorzalski, A., Nguyen, T. T., Liu, X., Jeffrey, C., Blomquist, G. J., et al. (2014). exo-Brevicomin biosynthesis in the fat body of the mountain pine beetle, Dendroctonus ponderosae. J. Chem. Ecol. 40, 181–189. doi: 10.1007/s10886-014-0381-9
Song, M. M., Kim, A. C., Gorzalski, A. J., MacLean, M., Young, S., Ginzel, M. D., et al. (2013). Functional characterization of myrcene hydroxylases from two geographically distinct Ips pini populations. Insect Biochem. Mol. Biol. 43, 336–343. doi: 10.1016/j.ibmb.2013.01.003
Song, X. W., Zhong, Q. S., Ji, Y. H., Zhang, Y. M., Tang, J., Feng, F., et al. (2022). Characterization of a sigma class GST (GSTS6) required for cellular detoxification and embryogenesis in Tribolium castaneum. Insect Sci. 29, 215–229. doi: 10.1111/1744-7917.12930
Takakura, Y., and Asano, Y. (2019). Purification, characterization, and gene cloning of a novel aminoacylase from Burkholderia sp. strain LP5_18B that efficiently catalyzes the synthesis of N-lauroyl-l-amino acids. Biosci. Biotechnol. Biochem. 83, 1964–1973. doi: 10.1080/09168451.2019.1630255
Tillman, J. A., Holbrook, G. L., Dallara, P. L., Schal, C., Wood, D. L., Blomquist, G. J., et al. (1998). Endocrine regulation of de novo aggregation pheromone biosynthesis in the pine engraver, Ips pini (Say)(Coleoptera: Scolytidae). Insect Biochem. Mol. Biol. 28, 705–715.
Tillman, J. A., Lu, F., Goddard, L. M., Donaldson, Z. R., Dwinell, S. C., Tittiger, C., et al. (2004). Juvenile hormone regulates de novo isoprenoid aggregation pheromone biosynthesis in pine bark beetles, Ips spp., through transcriptional control of HMG-CoA reductase. J. Chem. Ecol. 30, 2459–2494. doi: 10.1007/s10886-004-7945-z
Tittiger, C., and Blomquist, G. J. (2017). Pheromone biosynthesis in bark beetles. Curr. Opin. Insect Sci. 24, 68–74.
Unelius, C. R., Schiebe, C., Bohman, B., Andersson, M. N., and Schlyter, F. (2014). Non-host volatile blend optimization for forest protection against the European spruce bark beetle, Ips typographus. PLoS One 9:e85381. doi: 10.1371/journal.pone.0085381
Valentino, M. L., Barboni, P., Ghelli, A., Bucchi, L., Rengo, C., Achilli, A., et al. (2004). The ND1 gene of complex I is a mutational hot spot for Leber’s hereditary optic neuropathy. Ann. Neurol. 56, 631–641. doi: 10.1002/ana.20236
Wang, D. H., Song, W., Wei, S. W., Zheng, Y. F., Chen, Z. S., Han, J. D., et al. (2018). Characterization of the ubiquitin C-terminal hydrolase and ubiquitin-specific protease families in rice (Oryza sativa). Front. Plant Sci. 9:1636. doi: 10.3389/fpls.2018.01636
Wang, H.-L., Rao, Q., Chen, Z.-Z., and Navas-Castillo, J. (2022). Identifying potential insecticide resistance markers through genomic-level comparison of Bemisia tabaci (Gennadius) lines. Available online at: https://ssrn.com/abstract=4100303 (accessed May 4, 2022).
Wang, M., Sun, D.-F., Wang, S., Qing, Y., Chen, S., Wu, D., et al. (2013). Polymorphic expression of UDP-glucuronosyltransferase UGTlA gene in human colorectal cancer. PLoS One 8:e57045. doi: 10.1371/journal.pone.0057045
Wang, S., Chen, H., Tang, X., Zhang, H., Hao, G., Chen, W., et al. (2020). The role of glyceraldehyde-3-phosphate dehydrogenases in NADPH supply in the oleaginous filamentous fungus Mortierella alpina. Front. Microbiol. 11:818. doi: 10.3389/fmicb.2020.00818
Wang, Z. B., Tian, F. J., Cai, L. J., Zhang, J., Liu, J. L., and Zeng, X. N. (2019). Identification of candidate ATP-binding cassette transporter gene family members in Diaphorina citri (Hemiptera: Psyllidae) via adult tissues transcriptome analysis. Sci. Rep. 9:15842. doi: 10.1038/s41598-019-52402-3
Wei, D. D., He, W., Miao, Z. Q., Tu, Y. Q., Wang, L., Dou, W., et al. (2020). Characterization of esterase genes involving malathion detoxification and establishment of an RNA interference method in Liposcelis bostrychophila. Front. Physiol. 11:274. doi: 10.3389/fphys.2020.00274
Wen, R. R., Wang, B. Y., Wang, B. W., and Ma, L. (2018). Characterization and expression profiles of juvenile hormone epoxide hydrolase from Lymantria dispar (Lepidoptera: Lymantridae) and RNA interference by ingestion. J. Insect Sci. 18, 1–7. doi: 10.1093/jisesa/iey002
Xing, X., Yan, M., Pang, H., Wu, F. A., Wang, J., and Sheng, S. (2021). Cytochrome P450s are essential for insecticide tolerance in the endoparasitoid wasp Meteorus pulchricornis (Hymenoptera: Braconidae). Insects 12:651. doi: 10.3390/insects12070651
Yoon, K. A., Kim, K., Kim, W. J., Bang, W. Y., Ahn, N. H., Bae, C. H., et al. (2020). Characterization of venom components and their phylogenetic properties in some aculeate bumblebees and wasps. Toxins 12:47. doi: 10.3390/toxins12010047
Zhang, Q.-H., Schlyter, F., and Birgersson, G. (2012). 2-methyl-3-buten-2-ol: A pheromone component of conifer bark beetles found in the bark of nonhost deciduous trees. Psyche 2012:414508.
Keywords: Norway spruce, Eurasian spruce bark beetle, DGE analysis, allelochemicals, detoxification, RT-qPCR, enzyme assays, forest pest management
Citation: Naseer A, Mogilicherla K, Sellamuthu G and Roy A (2023) Age matters: Life-stage, tissue, and sex-specific gene expression dynamics in Ips typographus (Coleoptera: Curculionidae: Scolytinae). Front. For. Glob. Change 6:1124754. doi: 10.3389/ffgc.2023.1124754
Received: 15 December 2022; Accepted: 13 February 2023;
Published: 14 March 2023.
Edited by:
Ari Mikko Hietala, Norwegian Institute of Bioeconomy Research (NIBIO), NorwayReviewed by:
Melissa Hamner Mageroy, Norwegian Institute of Bioeconomy Research (NIBIO), NorwayCopyright © 2023 Naseer, Mogilicherla, Sellamuthu and Roy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amit Roy, Um95QGZsZC5jenUuY3o=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.