
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. For. Glob. Change , 08 December 2021
Sec. Pests, Pathogens and Invasions
Volume 4 - 2021 | https://doi.org/10.3389/ffgc.2021.778611
This article is part of the Research Topic Forest Resilience Against Pest and Pathogen Pandemics View all 4 articles
Eucalyptus grandis, in its native Australian range, varies in resistance to Austropuccinia psidii (syn. Puccinia psidii). The biotrophic rust fungus, A. psidii is the causal agent of myrtle rust and poses a serious threat to Australian biodiversity. The pathogen produces yellow pustules of urediniospores on young leaves and shoots, resulting in shoot tip dieback, stunted growth, and death. Dissecting the underlying mechanisms of resistance against this pathogen will contribute to improved breeding and control strategies to mitigate its devastating effects. The aim of this study was to determine the molecular dialogue between E. grandis and A. psidii, using an RNA-sequencing approach. Resistant and susceptible E. grandis seedlings grown from seed collected across its natural range were inoculated with the pandemic biotype of A. psidii. The leaf tissue was harvested at 12-h post inoculation (hpi), 1-day post inoculation (dpi), 2-dpi and 5-dpi and subjected to RNA-sequencing using Illumina 50 bp PE reads to a depth of 40 million reads per sample. Differential gene expression and gene ontology enrichment indicated that the resistant seedlings showed controlled, coordinated responses with a hypersensitive response, while the susceptible seedlings showed no systemic response against myrtle rust. Brassinosteroid signaling was apparent as an enriched term in the resistant interaction at 2-dpi, suggesting an important role of this phytohormone in defense against the pathogen. Brassinosteroid mediated signaling genes were also among the candidate genes within two major disease resistance loci (Puccinia psidii resistance), Ppr3 and Ppr5. While brassinosteroids have been tagged as positive regulators in other plant disease resistance interactions, this is the first report in the Eucalyptus – Austropuccinia psidii interaction. Furthermore, several putative resistance genes, underlying known resistance loci and implicated in the interaction have been identified and highlighted for future functional studies. This study provided further insights into the molecular interactions between E. grandis and A. psidii, contributing to our understanding of this pathosystem.
Austropuccinia psidii (G. Winter) Beenken (formerly Puccinia psidii, commonly myrtle rust) is a biotrophic fungal pathogen causing widespread devastation to the natural and commercial range of numerous plant species within the Myrtaceae family. Originating in South America, this pathogen jumped from native Myrtaceae to introduced Eucalyptus and has since spread across the world causing devastation to natural and introduced Myrtaceae species (Carnegie and Pegg, 2018). Currently, A. psidii has been discovered in South and Central America (Coutinho et al., 1998) and Florida, California and Hawaii in the United States, North America (Marlatt and Kimbrough, 1979; Rayachhetry et al., 1997; Uchida et al., 2006). The pathogen has also been reported in countries such as Japan (Kawanishi et al., 2009), Australia (Carnegie et al., 2010), China (Zhuang and Wei, 2011), South Africa (Roux et al., 2013), New Caledonia (Giblin, 2013), Indonesia (Mctaggart et al., 2016), Singapore, and New Zealand (Du Plessis et al., 2019). Myrtle rust has an unusually broad host range on various members of the Myrtaceae family, affecting approximately 480 species within 86 genera, producing yellow pustules of urediniospores on growing leaves and shoots (Coutinho et al., 1998; Soewarto et al., 2019). This results in dieback of shoot tips, stunted growth and in severe cases plant death (Carnegie et al., 2016; Pegg et al., 2017, 2020; Fensham et al., 2020). Eucalyptus spp. are amongst the most susceptible members of this family to myrtle rust, posing a risk to their natural and introduced ranges (Coutinho et al., 1998; Junghans et al., 2003a), with Eucalyptus grandis showing variable responses ranging from highly susceptible to resistant.
Plants have evolved complex and integrated defense mechanisms against pathogenic organisms, including preformed and induced defenses. Preformed defenses include factors such as cuticle waxes on the leaf surface, cell walls and secondary metabolites (Naidoo et al., 2014). Dos Santos et al. (2019) found the chemical composition of the cuticle waxes in E. grandis and E. phaeotricha contribute significantly to susceptibility against A. psidii, with various cuticular signals influencing the interaction between host and pathogen. Additionally, older leaves of E. grandis have been shown to comprise of greater amounts of waxes than younger leaves which may affect the ability of A. psidii urediniospores to adhere to or penetrate the leaf surface. This results in pre-penetration resistance in mature trees against this pathogen (Xavier et al., 2015). When a urediniospore successfully attaches to the surface of the leaf, the spore germinates and develops a germ tube. From the germ tube, an appressorium forms which gives rise to a penetration peg that infiltrates the cuticle (Coutinho et al., 1998). Following leaf infiltration, haustoria develop in the host cells (Coutinho et al., 1998; Glen et al., 2007). It is through these haustoria that the pathogen acquires nutrients from the host plant to ensure its survival (Hahn and Mendgen, 2001) and sustained release of proteins that suppress the host defense responses (Jones and Dangl, 2006). These proteins are known as effectors (or avirulence proteins) that contribute to the success of the pathogen (Jones and Dangl, 2006).
Junghans et al. (2003a) identified a locus associated with A. psidii resistance in E. grandis, termed Puccinia psidii resistance 1 (Ppr1), as per the previous classification of A. psidii. Ppr1 was shown to contribute to a large portion of resistance against A. psidii. This locus was found to be associated with numerous nucleotide-binding site leucine-rich repeat (NBS-LRR) resistance genes (R-genes) (Thumma et al., 2013), which suggests resistance against A. psidii is controlled by a combination of these genes. The discovery of this locus allowed targeted breeding programs, selecting for rust resistance to combat this pathogen. Despite the significance of Ppr1 in rust resistance, clones carrying this locus were shown to be susceptible to certain biotypes of A. psidii, indicating the locus was failing at providing resistance against A. psidii (Graça et al., 2011). Four additional loci contributing to myrtle rust resistance were discovered in E. globulus (Butler et al., 2016), termed Ppr2, Ppr3, Ppr4, and Ppr5. These quantitative trait loci (QTLs) determine if the plant shows symptoms (Ppr2 and Ppr3) and if there is a hypersensitive response (HR, Ppr4 and Ppr5) (Butler et al., 2016). The identification of additional QTLs contributing to disease resistance highlights the complexity of rust resistance. Despite the significance of these resistance loci, few studies have investigated the expression patterns of genes underlying these loci and little is known about how these genes contribute to disease resistance.
A recent study aimed to investigate the transcriptome of Melaleuca quinquenervia upon infection by A. psidii, to identify differences in responses among individuals with varying responses to the pathogen (Hsieh et al., 2018). At 5-dpi, they found that susceptible plants had higher levels of fungal transcripts than resistant plants, suggesting A. psidii could grow more successfully in susceptible leaves. Additionally, the authors found numerous differentially expressed genes (DEGs) associated with defense. These include NBS-LRR genes, WRKY transcription factors and Pathogenesis-related (PR) genes, such as PR-1, PR-2, PR-3, PR-4, and PR-5, with expression varying among resistant individuals. The authors suggest a potential role of the phenylpropanoid pathway in resistance against A. psidii, however, expression of associated genes differed among resistant samples and further studies are required to confirm this involvement. Sekiya et al. (2021), studying the interaction between E. grandis and A. psidii, found that rust resistance largely depends on the control of phenylpropanoid pathway related enzymes, corroborating the involvement of this pathway in defense against A. psidii. Tobias et al. (2018) studied the transcriptional responses of Syzygium leuhmannii challenged by A. psidii. The authors found up-regulation of receptor-like kinase genes, NBS-LRR genes as well as genes encoding enzymes involved in secondary metabolism which may contribute to disease resistance. Furthermore, they identified two TIR-NBS-LRR-type genes differentially expressed at 2-dpi with E. grandis homologs predicted within the Ppr1 locus. Santos et al. (2020) investigated the transcriptomes of resistant and susceptible E. grandis upon infection with A. psidii after 24-hpi. The authors found constitutive overexpression of defense-related genes in resistant compared to susceptible genotypes. These responses included up-regulation of salicylic acid (SA) associated genes, signal transduction and protein kinase leucine-rich receptors (PK-LRR). These studies provide some of the first insights into the transcriptional responses of Myrtaceae species challenged by A. psidii and further studies of these interactions are required to gain a deeper understanding of the molecular mechanisms governing host resistance.
The present study aimed to investigate the molecular interaction of resistant and susceptible Eucalyptus grandis challenged by A. psidii over time. In susceptible E. grandis genotypes, mycelia and haustoria have reportedly been observed between 12- and 18-hpi and in resistant genotypes, these have been observed between 18- and 1-dpi. Tobias et al. (2018) found significant transcriptome changes between resistant and susceptible samples 2-dpi whereas Hsieh et al. (2018) reported the possibility of fungal parasitic feeding stage at 5-dpi. Therefore, we hypothesize that at these and earlier time points (12-hpi, 1-, 2-, and 5-dpi) investigation of the expression profiles of resistant and susceptible seedlings would reveal molecular responses governing resistance, including changes in phytohormones, reactive oxygen species (ROS) and resistance genes. Through this, we have identified putative pathways and genes that contribute to host defense in E. grandis that can be manipulated in breeding and control programs, to mitigate the effects of myrtle rust. This is the first study to look at the transcriptome of E. grandis upon infection by A. psidii over a time course and provides further insights into the molecular interactions.
Eucalyptus grandis seedlings were grown from seed sourced from wild plants across its natural distribution in eastern Australia, ranging from Coffs Harbor in New South Wales to northern tropical regions of Queensland. Seed was germinated in glasshouse conditions, where temperatures ranged from 20 to 30°C. The seedlings, all with at least four young leaves, were inoculated to initially determine the phenotypes. Inoculation were generally carried out as described by Pegg et al. (2014). Spores of A. psidii were however collected from Syzygium jambos growing as street trees in suburbs of Brisbane, Queensland rather than using spores bulked from single spore inoculations in the laboratory. Spores were shaken into paper bags and passed through a sieve in the laboratory to remove plant debris. The collected spores were then desiccated in silica gel for 48 h and stored at −80°C. Urediniospores of the pandemic biotype were removed from −80°C storage and left at room temperature before addition of sterile distilled water with 0.05% tween 20. Spore were counted using a hemocytometer, with the concentration adjusted to 1 × 105 spore mL–1. Six-week-old E. grandis seedlings were spray inoculated with A. psidii urediniospores using a fine mist at a pressure of 2.9 kPa. Spores were applied to the upper and lower leaf surfaces, and the seedling trays were covered with a plastic sheet for 24 h in the dark at 18°C. After 24 h, the plastic covers were removed, and the seedlings were assessed for resistance and susceptibility based on their responses at 2 weeks post inoculation. Leaves were rated on a scale from 1 to 5, where rating one was considered resistant (R-interaction) and rating five was considered susceptible (S-interaction) based on the system used in Junghans et al. (2003b) (Table 1).
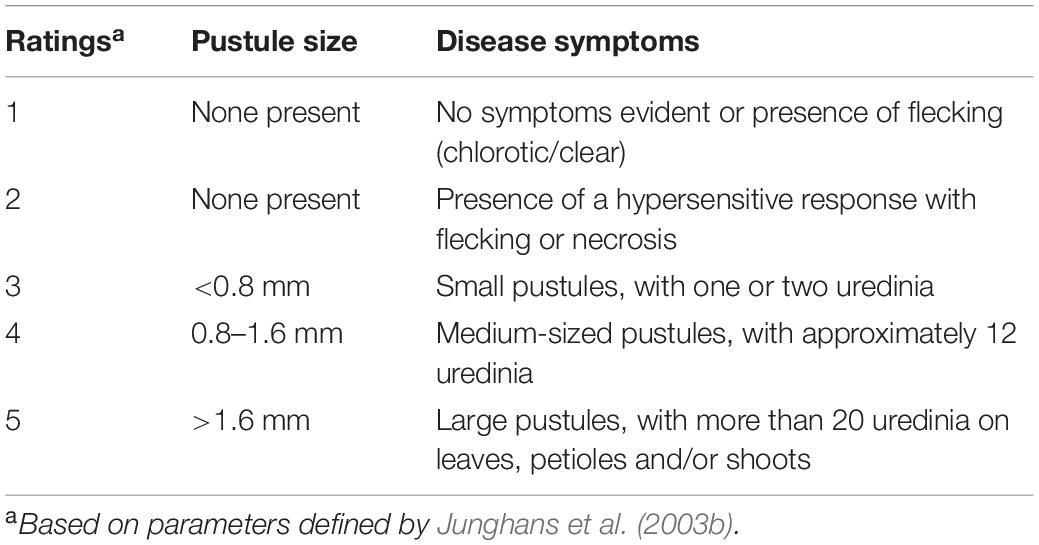
Table 1. Progression of disease symptoms observed on Eucalyptus grandis in response to Austropuccinia psidii.
Seedlings rated R and S were selected and separated out from other seedlings, and diseased tissues were removed by pruning. The seedlings were left to reshoot and after 8 weeks inoculation trials were repeated to obtain samples for RNA-seq. Seedlings were either inoculated as described above (infected), or mock-inoculated with sterile distilled water with 0.05% tween 20 (control). The youngest two sets of leaves of the infected and control plants were sampled as these are the most susceptible to A. psidii infection. Samples were collected from the seedlings at four time intervals namely, 12-hpi, 1-, 2-, and 5-dpi with every time point sampled from the same individual. Three replicates per time point were collected, comprising 14 seedlings per replicate to account for the genetic diversity within the seedlings.
Leaf samples collected from infected and control groups were subsequently frozen in liquid nitrogen. Total RNA was extracted as described by Zeng and Yang (2002) and treated with Qiagen RNase-free DNase 1 enzyme (Qiagen Inc.). Following successful RNA extraction, the samples were purified using Qiagen RNeasy Mini Kit as per the manufacturer’s instructions. To determine the concentration and quality of the extracted RNA, the samples were analyzed using Bio-Rad Experion analyzer. Purified RNA from three biological replicates were submitted to the Beijing Genomics Institute (BGI) for mRNA-sequencing using 50 bp paired-end Illumina HiSeq 2,500 with an insert size of 300 bp and a sequencing depth of 40 million reads per sample.
RNA-seq data were analyzed using the Galaxy workspace (Goecks et al., 2010) and FASTQC v0.11.3 was used to assess read quality. Reads from the R- and S-interactions at each time point (both infected and control samples) were mapped to the E. grandis v2.0 reference genome (Myburg et al., 2014) using Bowtie 2 (Langmead and Salzberg, 2012), Tophat2 v2.0.14 (Trapnell et al., 2012) and featureCounts v1.5.0-p3 (Liao et al., 2013). Pathogen transcripts were removed for the purpose of this study and will be investigated in future studies using the A. psidii reference genome (Tobias et al., 2021).
Data was normalized and variance stabilized transformation was applied in DESeq2 (Love et al., 2014) and transcripts per kilobase million (TPM) in R v3.6.0 (R Core Team, 2018). DEGs were analyzed using DESeq2 with filtering based on a two-fold change (> 1.00 or < –1.00 log2 (fold change)) and a false discovery rate of P < 0.001. We compared changes in gene expression between resistant, control relative to resistant, infected (R-interaction) and susceptible, control relative to susceptible, infected (S-interaction) to determine the effect pathogen infection would have on each interaction. Additionally, we compared susceptible, control relative to resistant, control (S-C vs. R-C) to determine the background gene expression. Heatmaps were generated using the R package ComplexHeatmap v2.0.0 (Gu et al., 2016) using log2 (fold change) values.
Identified DEGs were analyzed for Gene Ontology (GO) enrichment over-representation in the category biological processes (BP), to identify biological pathways potentially involved in the interaction. Enrichment was calculated using Fisher’s exact test and p-values were adjusted using the Benjamini and Hochberg (BH) method with a false discovery rate (FDR) of 0.05, with all calculations performed in R v3.6.0. Enrichment was determined separately for all time points as well as up- and down-regulated DEGs. Additionally, Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation enrichment was performed on DEGs to identify enriched KEGG terms using the same method.
To identify candidate genes underlying A. psidii resistance QTLs (Ppr1, Ppr2, Ppr3, Ppr4, and Ppr5), the physical positions of their flanking markers were obtained (Table 2; Junghans et al., 2003a; Mamani et al., 2010; Butler et al., 2016). Phytozome v12.11 was used to identify genes underlying these physical positions of each locus on its respective linkage group using the genome browser.
Disease progression was monitored routinely for the presence of symptoms. Disease assessments were performed on the primary and secondary foliage (youngest two expanding pairs of leaves) to avoid the influence leaf age may have on disease severity levels. Based on symptom development, seedlings were assessed 14-dpi. Different responses were observed in E. grandis seedlings following inoculations as shown in Figure 1. Symptoms were similar to those observed in previous inoculations performed by Junghans et al. (2003b). This included light chlorotic flecking on seedlings rated one. Visible HR and necrotic lesion formation on seedlings rated two. Progression of necrosis and formation of uredinia pustules were observed in ratings three and four. Severe pustules were observed in seedlings rated five, with disease commonly occurring on the petiole and stem. For the purpose of this study, plants rated one were considered resistant (R) and those rated five on the severity scale considered susceptible (S), with samples harvested from these ratings for further differential gene expression analysis.
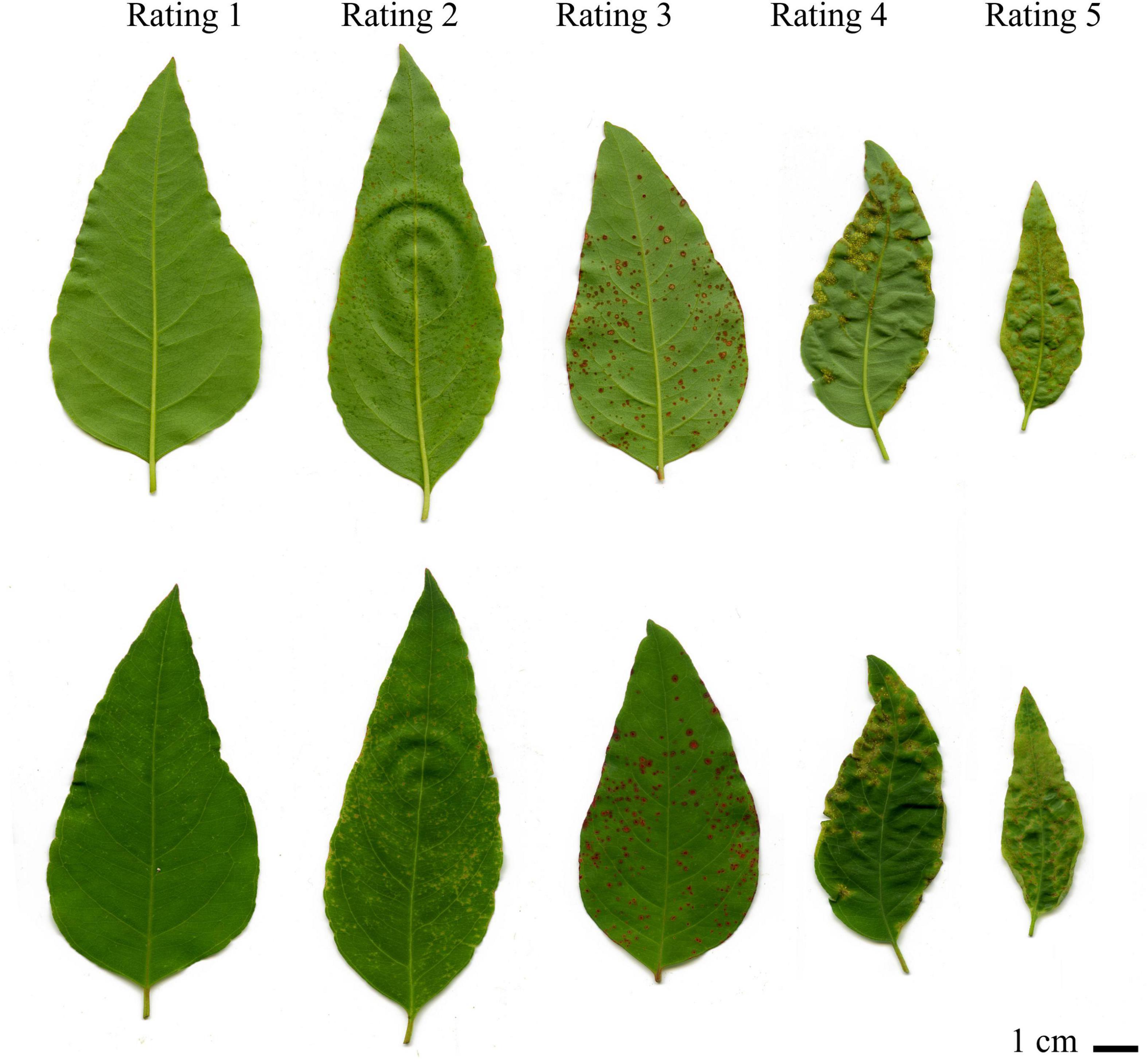
Figure 1. Disease progression of Eucayptus grandis in response to Austropuccinia psidii. The top leaves show the adaxial while the bottom shows the abaxial view of the leaves. Rating one was considered resistant while ratings two to five were considered susceptible to A. psidii. Rating one showed slight chlorotic flecking, progressing in rating two and three where the flecking became necrotic and uredinia pustules began to form. Uredinia pustules became more apparent in rating four and severe pustules began forming by rating five. Rating one and rating five were selected for further analysis.
To determine the effects pathogen infection would have on infected seedlings, we compared infected and control seedlings for both the R- and S-interactions across the time series. There were 4,681 significantly DEGs over the time course in the R-interaction, while there were 2,800 DEGs in the S-interaction over the time course (data not shown). During early infection at 12-hpi, there were 50 down- and 96 up-regulated genes in the R-interaction, while the S-interaction had 39 down- and 309 up-regulated genes (Figure 2A). The number of up-regulated genes began increasing at 1-dpi in both the interactions. Up-regulation of genes peaked in the R-interaction at 2-dpi with 2,221 genes and 5-dpi in the S-interaction, with 2,146 genes (Figure 2A). Basal gene expression in resistant and susceptible plants might contribute to disease phenotypes when a pathogen is introduced, as previously observed in the A. psidii-E. grandis pathosystem (Santos et al., 2020). To investigate this, we compared susceptible, control relative to resistant, control (S-C vs. R-C) (Figure 2B). There was a total of 209 DEGs across the time course (data not shown). These results suggest that differences in basal expression exist.
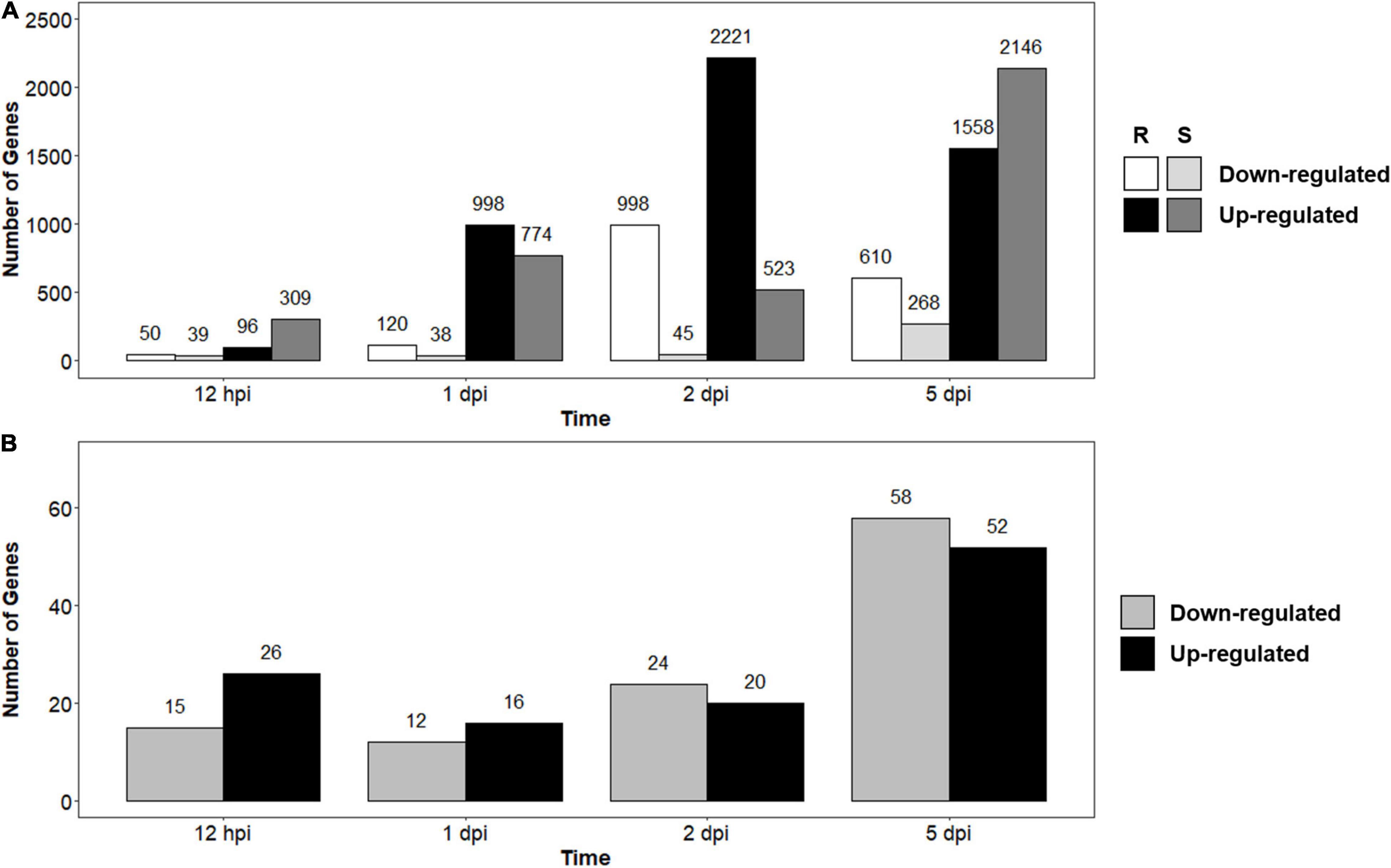
Figure 2. Number of differentially expressed genes (DEGs) identified through RNA-seq analysis over the time series in (A) Resistant (R-interaction) and susceptible (S-interaction) and (B) Susceptible, control compared to resistant, control (S-C vs. R-C) where gray bars represent genes greater expressed in the S-interaction and black bars represent genes greater expressed in the R-interaction. Genes considered down-regulated with a log2 (fold change) < –1.00 and up-regulated with log2 fold change > 1.00.
To identify pathways putatively involved in resistance and susceptibility against A. psidii, GO enrichment was performed. No over-represented GO terms were identified in S-C vs. R-C, which is expected due to the low numbers of DEGs. Figure 3 shows a heatmap of over-represented defense-related terms in both the R- and S-interaction over the time series. Defense-related terms were separated into terms associated with phytohormone signaling and responses, oxidative burst, secondary metabolism and defense responses. Additionally, up- and down-regulated terms are represented. Terms enriched in the R-interaction involve up-regulation of genes associated with brassinosteroid (BR) signaling, at 2-dpi, genes associated with a range of salicylic acid (SA) associated pathways as well as jasmonic acid (JA) and ethylene (ET) associated pathway. Enriched terms in the S-interaction involve up-regulation of genes associated with JA and abscisic acid (ABA) pathways. Interestingly, the R-interaction showed up-regulation of secondary metabolic pathways including terpenoid transport, while these responses with absent/down-regulated in the S-interaction. Moreover, defense responses such as callose deposition were up-regulated in the R-interaction, while these responses were limited in the S-interaction.
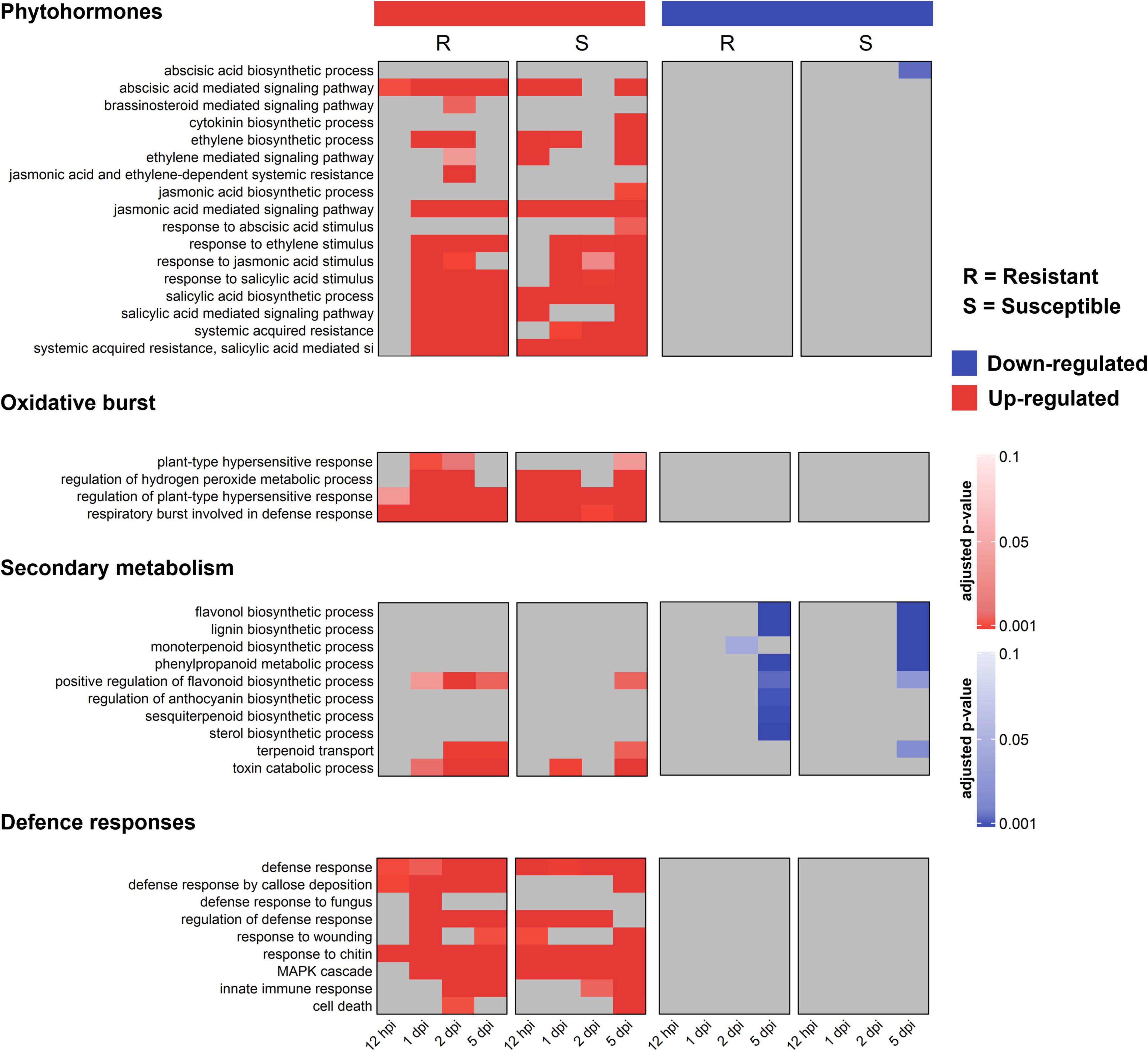
Figure 3. Gene ontology (GO) biological processes (BP) functional characterization of significantly differentially expressed genes (DEGs) in the R- and S-interactions, showing selected defense-related terms over time. Red bars represent up-regulated, and blue bars represent down-regulated GO terms. Color bars indicate significance with an adjusted p-value ≤ 0.05.
The DEGs identified in the R- and S-interaction were analyzed with KEGG to further elucidate the putative role these genes play in defense pathways. KEGG was used to determine pathways involved in defense against this pathogen. KEGG enrichment revealed up-regulation of “photosynthesis—antenna proteins” at 12-hpi, up-regulation of “terpenoid backbone biosynthesis” at 1-dpi and up-regulation of “amino sugar and nucleotide sugar metabolism” at 2-dpi unique to the R-interaction. Shared responses included plant-pathogen interaction at 1-dpi, 2-dpi, and 5-dpi. Interestingly, at 5-dpi, the S-interaction showed down-regulation of cutin, suberin and wax biosynthesis (Figure 4).
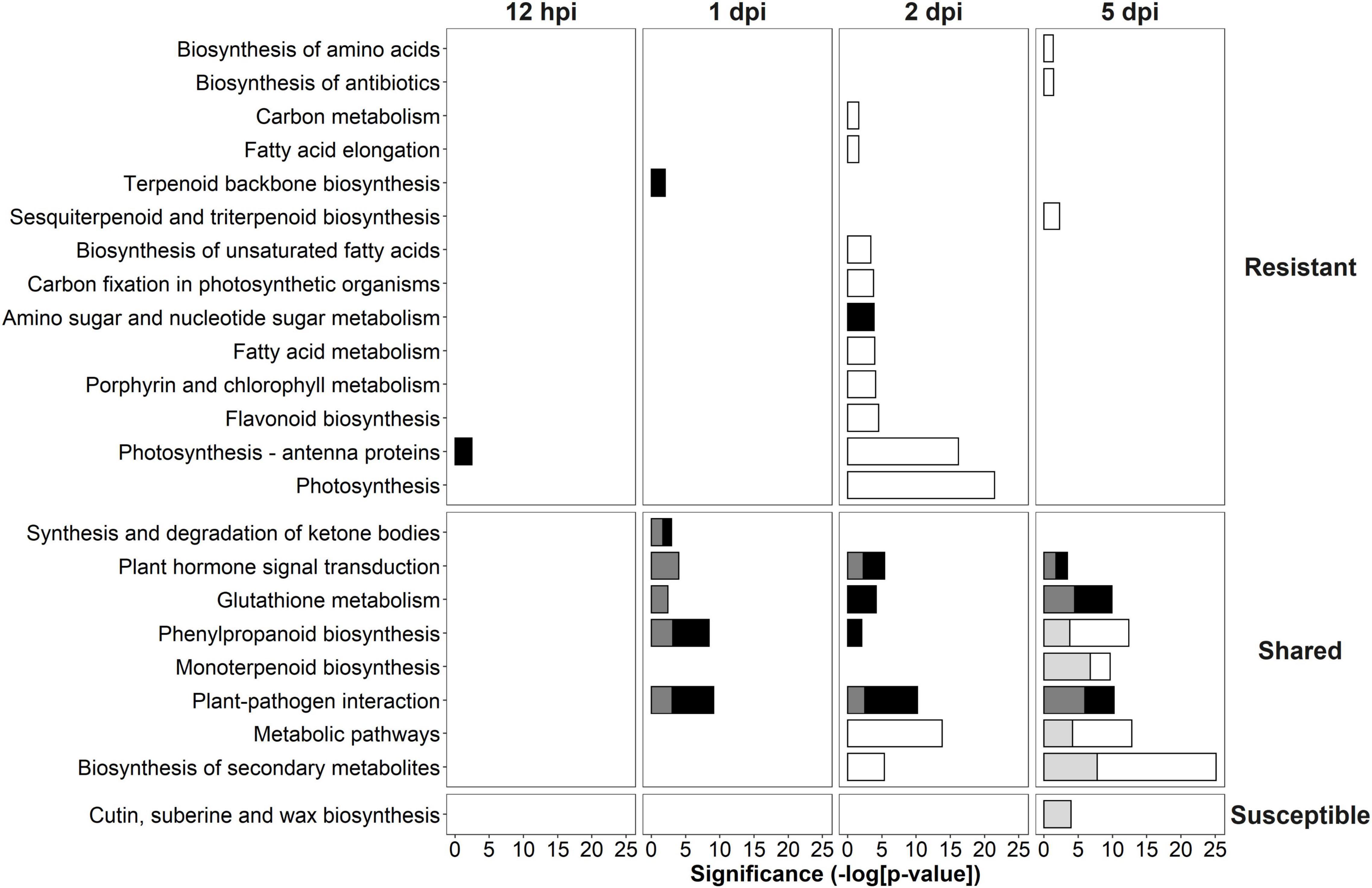
Figure 4. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of significantly differentially expressed genes (DEGs) in the R- and S-interactions over time showing the significance of each over-represented term. White bars indicate down-regulated resistant, black bars indicate up-regulated resistant, light gray bars indicate down-regulated susceptible and dark gray bars indicate up-regulated susceptible. KEGG terms were separated into those unique to the R-interaction, shared in both the R- and S-interactions and unique to the S-interaction.
In the R-interaction, BR signaling was observed at 2-dpi, with these responses absent in the S-interaction (Figure 3). Further investigations into the genes contributing to BR signaling revealed DEGs in both interactions, with the R-interaction expressing more of these genes at greater intensities. Signaling genes included BRI1-associated receptor kinase (BAK1), leucine-rich repeat protein kinase family proteins and BZR1 family proteins. These genes showed different expression patterns across the R- and S-interaction over time, with the greatest expression observed in the R-interaction at 2-dpi. Similar responses were observed at 5-dpi in the S-interaction, although these terms were not enriched in the GO analysis (Figure 5).
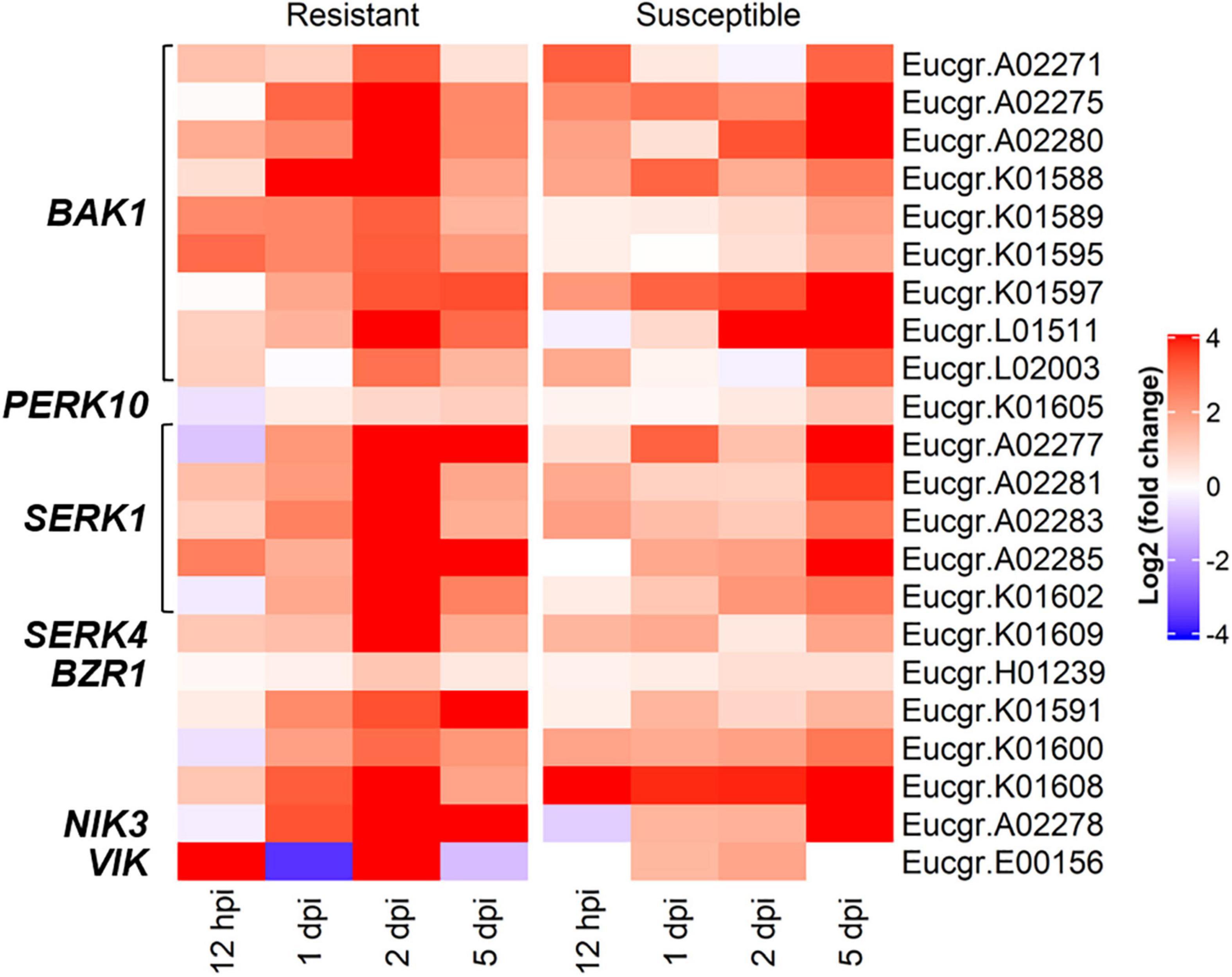
Figure 5. The brassinosteroid mediated signaling pathway associated genes showing differentially expressed genes (DEGs) in both the R- and S-interactions over time, where red and blue indicate up- and down-regulation, respectively [log2 (fold change)]. These genes were identified in GO BP enrichment.
To identify candidate genes underlying known resistance loci, the physical positions of the flanking markers were identified (Table 2) and the genes between the markers were selected for further investigation (Supplementary Figure 1). Table 3 shows the total number of genes identified within each locus, with the number of DEGs underlying these loci. Additionally, the number of R-genes and differentially expressed (DE) R-genes underlying these loci are shown (Table 3 and Supplementary Table 1). To determine the putative biological pathways that underlie the A. psidii resistance QTLs, the candidate genes identified were investigated for GO biological processes functional characterization (Supplementary Figure 1). Defense-related enriched terms included “response to salicylic acid stimulus,” “defense response,” and “salicylic acid metabolic process.” The involvement of these Ppr loci in defense-related processes highlighted throughout literature, the results of the GO BP enrichment analysis as well as the identification of numerous R-genes prompted further investigations into the expression profiles of the candidate genes underlying these QTLs.
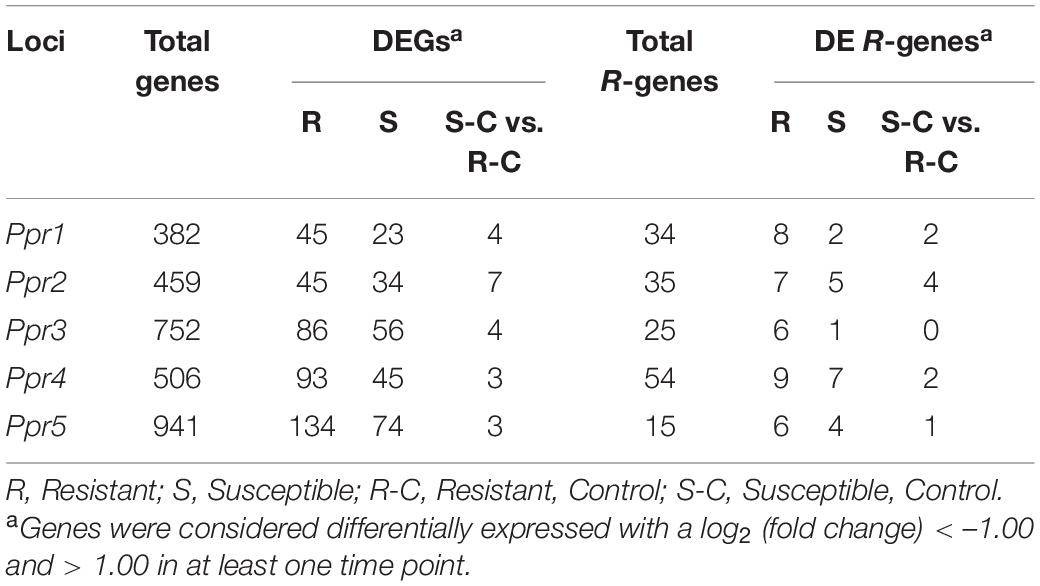
Table 3. The total number of genes identified underlying known resistance loci, showing the percentage of total and differentially expressed R-genes in both resistant and susceptible hosts.
To determine the roles the A. psidii resistance loci play in defense, DEGs underlying the QTLs were identified in the R-, S-interactions (Figure 6) and S-C vs. R-C (Figures 7A–I). Across all comparisons, there was significant differential expression of R-genes, namely NBS-LRR genes (Table 3 and Supplementary Table 2). Figure 6 is a heatmap representing log2 (fold change) values of the DE R-genes underlying these QTLs in the R- and S-interactions. Differential expression of these R-genes is greater in the R-interaction, with up-regulation of numerous genes earlier in the interaction between 1 and 2-dpi when compared to the S-interaction. The S-interaction shows similar differential expression patterns later in the time series, between 2 and 5-dpi. The DE R-genes underlying Ppr1, Ppr2, Ppr3, and Ppr5 had low TPM values, despite showing significant differential expression. Interestingly, Ppr4, a locus determining if the plant exhibits HR (Butler et al., 2016), showed increased expression of these R-genes, with expression of these genes greater than those observed at the other disease resistance QTLs. This corroborates the findings in Figure 3, where HR-associated responses were significantly enriched in both the R- and S-interactions.
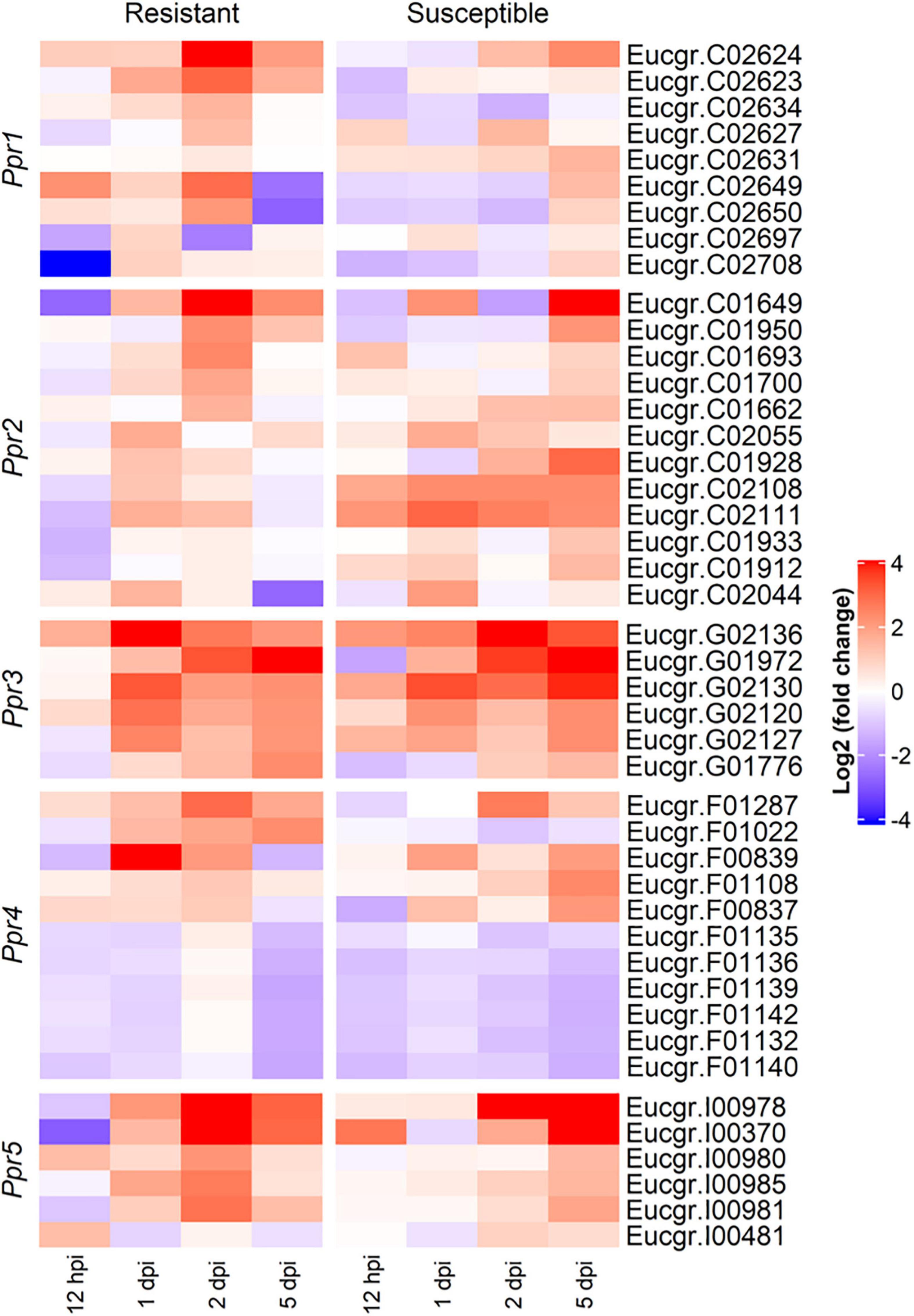
Figure 6. Heatmap showing the expression profiles of differentially expressed R (resistance) genes underlying the Austropuccinia psidii resistance loci in the R- and S-interactions over time, where red and blue represent up- and down-regulated genes, respectively. Heatmap shows log2 (fold change) with p < 0.001 representing significant DE.
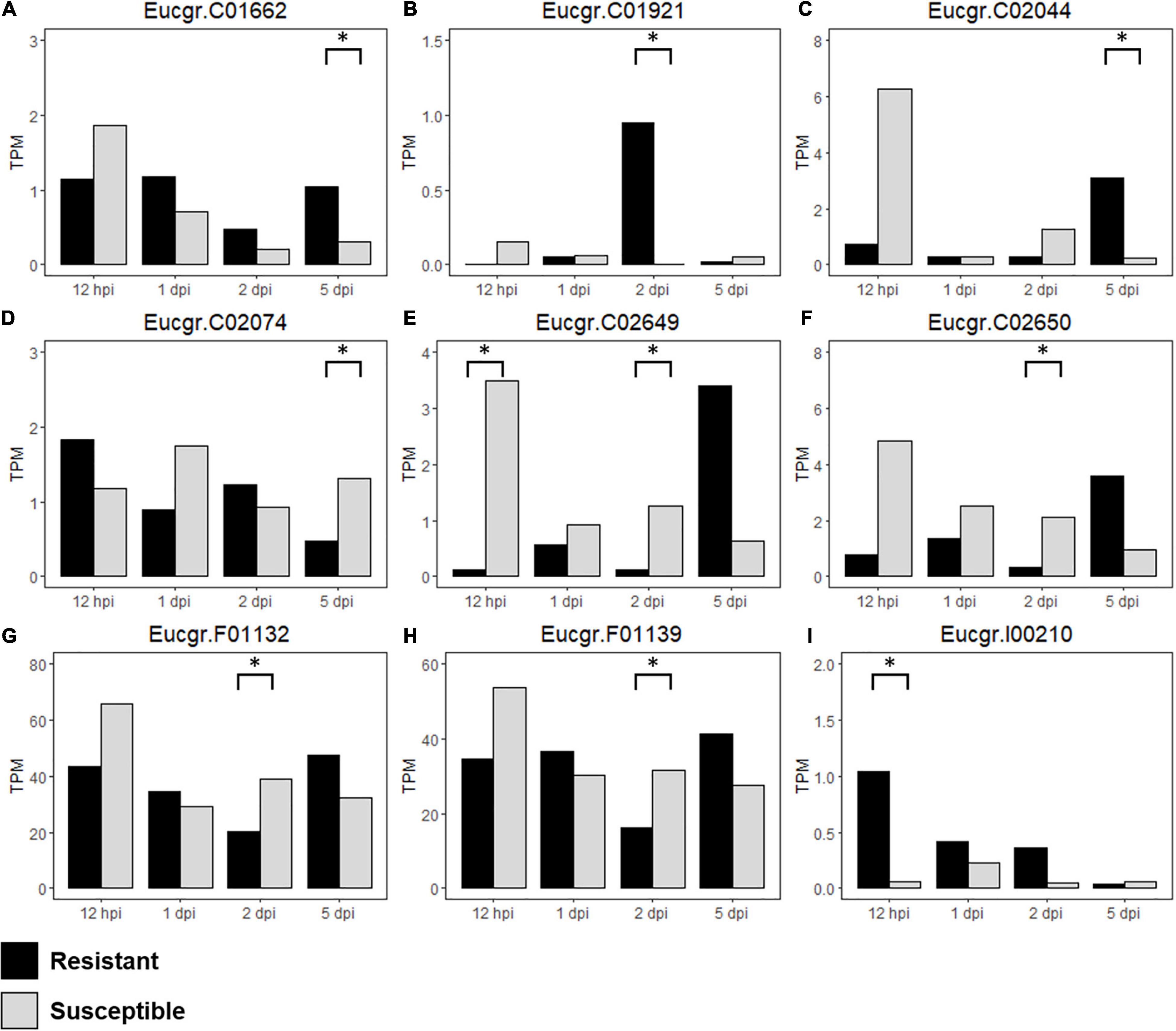
Figure 7. Differentially expressed (DE) R-genes (A–I) underlying the Austropuccinia psidii resistance loci in the susceptible, control compared to resistant, control seedlings over the time series, representing TPM values of DE R-genes in all biological replicates in the controls of the R- and S-interactions i.e., constitutive expression of the R-genes are being compared, where black represents resistant and light gray represents susceptible seedlings and asterisks represent significant DE.
To determine the differences in constitutive expression of the R-genes underlying the disease resistance loci, we analyzed S-C relative to R-C and found nine DE R-genes underlying these loci in the control samples (Table 3 and Supplementary Table 2). Figures 7A–I show the expression (TPM) of these genes over the time series. Eucgr.F01132 (Figure 7G) and Eucgr.F01139 (Figure 7H), underlying Ppr4, are highly expressed, this is similar to the findings in the R- and S-interactions, in which R-genes were highly expressed. Eucgr.I00210 (Figure 7I), underlying Ppr5, showed greater constitutive expression in R-C at 12-hpi, 1- and 2-dpi, suggesting a role in constitutive resistance against A. psidii. Interestingly, Eucgr.C01921 (Figure 7B) underlying Ppr1 showed significantly greater DE in R-C at 2-dpi than in S-C. Gene Eucgr.C02650 (Figure 7F) showed greater expression in S-C at 12-hpi, with the expression of this gene increasing R-C at 5-dpi. The timing and intensity of basal expression of these genes may contribute to the phenotypes observed when resistant and susceptible E. grandis is challenged by A. psidii. Further studies into the functional role of these genes are required to determine if they contribute to rust resistance and how these genes differ at the molecular level between resistant and susceptible seedlings.
Genes involved in BR mediated signaling was significantly over-represented in the R-interaction at 2-dpi (Figures 3, 5). To determine if the Ppr loci harbor genes associated with BR signaling, genes associated with this pathway identified through GO (GO:0009742) were compared to genes underlying the resistance loci. No BR mediated signaling genes were identified within Ppr1, Ppr2, and Ppr4. Two and four BR mediated signaling genes were identified within Ppr3 and Ppr5, respectively. These genes include a signaling kinase (BSK1), leucine-rich receptors (BRI1), protein kinases and a BR receptor (Supplementary Table 3).
The aim of this study was to compare the transcriptional responses of resistant and susceptible E. grandis with A. psidii over time. Through this, we identified pathways that may be involved in the E. grandis defense response against myrtle rust, such as the hypersensitive response, oxidative burst, and a range of phytohormones, with brassinosteroids signaling identified in resistant seedlings. Additionally, the timing of these responses varied between R- and S-seedlings, with delays and absences of responses in susceptible hosts. This suggests the importance in a coordinated, rapid response in resistance against A. psidii. Analysis of loci associated with resistance revealed these resistance loci harbor a plethora of R-genes, and we have identified candidate R-genes that are DE in the interactions, showing potential involvement in the disease outcome. Overall, we suggest a combination of these responses contribute to the resistance observed in E. grandis.
Despite the absence of physical symptoms of HR in the resistant seedlings, HR was significantly enriched in the resistant RNA-seq data, suggesting that this response is occurring at a molecular level. The plant material sourced for this study were seedlings rather than clones. This introduces genetic diversity within the each rating class, affecting the results obtained in downstream data analysis (Tobias et al., 2018). It is possible that the molecular responses observed are representative of the genetic diversity within the samples, with the HR varying among seedlings within the resistant rating class. This accounts for the variation seen in seedlings rated one, where few showed no visible infection and most showed light chlorotic flecking (Table 1). A study conducted by Wang et al. (2018) showed that HR is involved in resistance against anthracnose (Colletotrichum) in the tea plant (Camellia sinensis). Through transcriptional analysis and histochemistry, the authors found the significant involvement of the HR, where cell walls were reinforced in resistant cultivars as a result of H2O2 accumulation. Despite the involvement of HR in resistant cultivars, resistant leaves did not present with physical symptoms of disease or show signs of HR. Despite this, future studies will need to investigate the levels of HR associated compounds, such as H2O2, in resistant E. grandis to determine if this mechanism is associated with plant resistance at a molecular level, despite the lack of physical evidence of the HR.
Seedlings rated one were considered resistant while seedlings rated five were considered susceptible. This was characterized by either showing few symptoms of infection or large pustules on leaves, shoots and petioles (Table 1). Seedlings rated two can also be considered resistant, as the presence of HR is indicative of host resistance against infection. However, the presence of necrosis suggests cell death as the result of disease rather than resistance, as described by Morel and Dangl (1997). Additionally, we observed in some instances that seedlings rated two developed pustules, despite the presence of an HR. For this reason, despite an HR observed in seedlings rated two, we only selected seedlings rated one and five for RNA-seq analysis to investigate the transcriptional responses of the two extremes of host resistance and susceptibility. This is a criteria also selected by Hsieh et al. (2018) when investigating the responses of M. quinquenervia upon A. psidii infection.
Gene ontology biological processes functional characterization of DEGs in both the R- and S-interactions over time revealed interesting patterns of responses against A. psidii. In both interactions, there was significant up-regulation of defense-related terms over time, with few or no down-regulation observed. These responses appeared to begin earlier in the S-interaction, with significant over-representation of defense-related terms observed at 12-hpi. These included responses associated with SA biosynthesis and signaling, hydrogen peroxide metabolism and MAPK cascades (Figure 3). A possible explanation for the earlier increased expression of genes may be due to a lack of effective preformed defenses. A study by Hsieh et al. (2018) suggests that resistance in some individuals of M. quinquenervia against myrtle rust may be influenced by preformed defenses such as physical barriers and chemical constituents. The R-interaction may have effective preformed defenses that prevent appressorium formation at 12-hpi, while these barriers may be absent more susceptible hosts. Additionally, Xavier et al. (2001) reported that primary hyphae were observed at 12-hpi in susceptible E. grandis hosts, while only observed in the resistant hosts at 18-hpi. This is further supported by a study conducted by Dos Santos et al. (2019) in which susceptibility in E. grandis and E. phaeotricha was greatly influenced by the chemical composition of the cuticle waxes. Susceptible hosts lack effective preformed foliar compounds to prevent fungal penetration and as a result, respond to the presence of the pathogen earlier than in resistant hosts.
Despite this observation, the R-interaction shows a rapid increase in responses over 1- and 2-dpi, while similar responses are muted or absent in the S-interaction. Our study suggests that the S-interaction can induce a response against A. psidii, as evident by the changes in the transcriptome upon infection. However, it cannot implement adequate and sustained mechanisms to combat disease shown by the absence of critical defense responses by 2-dpi. A recent study showed that susceptible genotypes of S. luehmannii had no coordinated systemic response to A. psidii, with low host transcript numbers detected (Tobias et al., 2018). This is further supported by Santos et al. (2020), in which susceptible E. grandis genotypes were able to activate defense responses after 1-dpi with A. psidii, but the responses did not result in successful defense due to a lack of effective signal transduction. This suggests that susceptible E. grandis hosts and similarly of other Myrtaceae species, lack coordination in regulation of defense-related genes, which may contribute to disease susceptibility. A previous study on the interaction between A. psidii and M. quinquenervia showed susceptible plants had various up-regulated defense-related responses at 5-dpi (Hsieh et al., 2018). This is similar to the results obtained in this study, suggesting that Myrtaceae species are able to deploy defenses against A. psidii, but delays in these responses may contribute to susceptibility. Over-compensation later in the interaction at 5-dpi may be too late to prevent spread of disease.
Interesting KEGG over-represented terms included “photosynthesis—antenna proteins” up-regulated in the R-interaction at 12-hpi, with this term absent in the S-interaction over the time series. Photosynthetic processes are known to play roles in plant defense against pathogenic invasion. Santos et al. (2020) found the constitutive involvement of this pathway in the defense response in resistant E. grandis genotypes. Furthermore, “amino sugar and nucleotide sugar metabolism” was unique to the R-interaction at 2-dpi. Sugar signaling has been implicated in disease resistance, with high levels of sugars in plants contributing to defense responses. Increased plant sugars contribute to resistance by inducing expression of PR genes, interacting with the plant phytohormone biosynthesis and signaling pathways (Morkunas and Ratajczak, 2014) and increasing production of ROS (Morkunas et al., 2008). Exogenous applications of sucrose in Vitis vinifera induces the production of secondary metabolites such as phenylpropanoid, which may contribute to disease resistance modulated by sugar signaling (Ferri et al., 2011). The up-regulation of phenylpropanoid biosynthesis identified in this study (Figure 4), as well as identification of this pathway in other studies investigating interactions with myrtle rust, suggests the importance of phenylpropanoids in resistance, with sugar metabolism putatively contributing to the induction of this pathway. Moreover, sugar transport, mediated by the sugar transport protein TaHTP, was found to contribute to resistance against the biotrophic rust pathogen Puccinia triticina in wheat (Savadi et al., 2018).
Phytohormones contribute significantly to plant defense responses, with crosstalk existing between them to facilitate defense against pathogenic invasion. In this study, genes associated with phytohormones were extensively enriched in both resistant and susceptible E. grandis seedlings upon infection with A. psidii (Figure 3). These include genes relating to SA, JA, ET, ABA, and auxin. Our results suggests putative involvement of SA biosynthesis and signaling, with downstream responses such as systemic acquired resistance identified (Figure 3) in both the resistant and susceptible hosts. Santos et al. (2020) found the up-regulation of SA signaling and responsive genes when comparing mock-inoculated myrtle rust resistant and susceptible E. grandis genotypes. These findings suggest the importance of SA in E. grandis defense against A. psidii, both at a constitutive and induced level. Interestingly, GO analysis revealed the involvement of BR at 2-dpi in the R-interaction, a pathway absent in susceptible hosts.
BRs are polyhydroxylated steroidal phytohormones found in plants that have versatile roles in many plant physiological processes. These processes include those such as cell growth and development, seed germination and response to abiotic stress (Saini et al., 2015; Yu et al., 2018; Kim and Russinova, 2020). In recent years, studies have emerged implicating BRs in resistance to various pathogens, with complex molecular interactions driving host responses against pathogenic invasion (Choudhary et al., 2012). BRs have been shown to confer resistance against biotrophic bacteria (Huang et al., 2014) and exogenous application confers systemic resistance to biotrophic fungi such as Oidium sp. and Magnaporthe grisea (Nakashita et al., 2003).
Many studies on the roles BR play in modulating immune responses have focused on the leucine-rich repeat receptor-like kinase (LRR-RLK) and BRI1 (BR-insensitive 1)-associated kinase (BAK1). BAK1 is a co-receptor of BRI1 and plays a role in BR signal transduction as well as interacting with pattern recognition receptors (PRR) to induce a PTI response against invading pathogens (Li et al., 2002; Vert, 2008; De Bruyne et al., 2014). Brassinolide (BL), an active form of BR, binds to BRI1 leading to autophosphorylation and the activation of BAK1 (De Bruyne et al., 2014). This ultimately leads to downstream transcriptional changes associated with BR-signaling. BAK1 has been implicated in disease resistance against fungal pathogens including Sclerotinia sclerotiorum and Verticillium dahliae (Fradin et al., 2009; Chaparro-Garcia et al., 2011) and the identification of nine DE BAK1 genes in the present study suggests a putative role of BR and associated receptors in the defense response against A. psidii. The higher number of DE BAK1 genes in response to A. psidii in the R-interaction may contribute to disease resistance. Similarly, Santos et al. (2020) found constitutive over expression of BAK1 when comparing mock-inoculated A. psidii resistant and susceptible E. grandis at 1-dpi, corroborating the findings of the present study. Additionally, in a transcriptome study between banana and Fusarium oxysporum f. sp. cubense, up-regulation of BAK1 was identified in relatively resistant banana plants, potentially implicating this gene in the innate immune response in banana (Li et al., 2013).
In addition to BAK1, the somatic embryogenesis receptor-like protein kinase-1 (SERK1) are implicated in disease resistance in tomato against the foliar fungal pathogen, Cladosporium fulvum (Fradin et al., 2011). The SERK protein family, which includes SERK3/BAK1 has been extensively shown to contribute to disease resistance. In this study, five SERK1 genes were DE in E. grandis upon challenge by A. psidii (Figure 4). The expression of these genes is similar to that of BAK1. Further analyses of the genes involved in BR signaling revealed DE of gene BZR1 encoding brassinazole-resistant 1. This is a transcription factor responsible for repressing expression of defense-related genes (Robert-Seilaniantz et al., 2011). Further studies into the role this gene is playing during the interaction of E. grandis are required. However, up-regulation of this gene was low or absent over the time course within both interactions. This suggests defense-related responses involved in PTI may not be repressed.
Despite our results suggesting a role of BR signaling in defense against A. psidii, studies on the effector AVR2 have shown that increased BR signaling in transgenic potato plants leads to enhanced susceptibility to Phytophthora infestans. This effector manipulates the crosstalk that exists between plant growth and immunity to facilitate enhanced pathogen colonization (Turnbull et al., 2017). This highlights the complexities of BR signaling and the roles this phytohormone pathway plays in plant defense. Different pathogens employ different virulence mechanisms to elicit disease in host plants, and this may explain the different outcomes observed in BR signaling between P. infestans (oomycete) and A. psidii (fungus). Moreover, the lifestyle of the pathogen contributes to the outcome of BR in plant defense. Despite AVR2 being up-regulated during the biotrophic phase in P. infestans, this oomycete is a hemi-biotroph while A. psidii is a biotroph and studies have shown that BR increases plant defense in response to biotrophs while decreasing plant defense against hemibiotrophs and necroptrophs (De Vleesschauwer et al., 2012; Turnbull et al., 2017; Yu et al., 2018), this contributes further to the complexities of this phytohormone in plant responses. For this reason, more studies are required to understand how BRs contribute to the responses of E. grandis to A. psidii.
The identification of BR mediated signaling genes underlying A. psidii resistance loci may implicate this pathway in the resistance conferred by these loci. Ppr3 harbors two genes that are associated with BR signaling and BR receptors, while Ppr5 harbors four genes that code leucine-rich receptor proteins and protein kinases. Eucgr.G01832 (BRI1-interacting signaling kinase, BSK1) underlies Ppr3, while Eucgr.I00302 and Eucgr.I00303 (BRI1) underlies Ppr5 (Supplementary Table 3). BRs associates with BRI1, causing phosphorylation and subsequent dissociation of BRI1 from its associated inhibitors. This cascade event leads to the activation of BAK1 and the phosphorylation of BSK1 (De Bruyne et al., 2014; Yu et al., 2018). This causes activation of downstream BR signaling responses and changes in gene expression. Despite these loci harboring BR signaling genes, these were not DE in this study. However, extensive polymorphisms may exist within these genes that affect the expression as well as the final protein product, which may contribute to BR signaling and resistance against A. psidii. Further studies are required to identify single nucleotide polymorphisms (SNPs) that may exist between resistant and susceptible seedlings to determine how these genes may contribute to disease resistance.
Plants recognize pathogen effectors through resistance proteins, most commonly the nucleotide-binding domains and leucine-rich repeats (NLRs) (Jones and Dangl, 2006). Effectors are small secreted proteins (SSPs) that the pathogen deploys to facilitate the manipulation of host responses, allowing the pathogen to acquire host nutrients and reduce plant defense (Fudal et al., 2018). The recognition of these pathogenic effector proteins leads to effector-triggered immunity (ETI), resulting in oxidative burst of ROS as well as hypersensitive responses (HR) (Jones and Dangl, 2006). These responses lead to extensive modifications in defense-related gene expression and prevention of pathogen spread from the site of infection (Dodds and Rathjen, 2010).
In Eucalyptus, numerous R-gene loci conferring resistance to A. psidii have been discovered (Junghans et al., 2003a; Butler et al., 2016). Ppr1 confers resistance to myrtle rust in E. grandis while Ppr2–5 has been shown to confer resistance to myrtle rust in E. globulus, a close relative of E. grandis (Butler et al., 2016). Little is known about the changes in expression over time of genes associated with these loci and how they contribute to disease resistance. To further investigate the genes underlying these loci, we identified various DEGs in the interaction between A. psidii and E. grandis, many of which were R-genes (Figures 6, 7A–I and Supplementary Table 2). In general, there were more significantly DE R-genes in the R-interaction underlying these loci than in the S-interaction. Additionally, the R-interaction showed significant DE predominately between 1- and 2-dpi, while significant expression was only observed in the S-interaction during later stage infection. A recent study by Santos et al. (2020) identified two significantly DE R-genes underlying Ppr1 when investigating mock-inoculated resistant and susceptible E. grandis as well as susceptible infected and control E. grandis, namely Eucgr.C02650 and Eucgr.C02749. Eucgr.C02650 was down-regulated in both, while Eucgr.C02749 was up-regulated in both. In our study, we found significant DE of Eucgr.C02650 when investigating resistant, infected relative to control as well as investigations between resistant and susceptible controls.
Ppr2 and Ppr3 determine if the plant shows disease symptoms (Butler et al., 2016). In the R-interaction, these loci showed significant up-regulation at 1- and 2-dpi. In contrast, while the expression patterns are similar, the S-interaction showed fewer significant DE of these R-genes. This may contribute to the disease symptoms observed in susceptible seedlings. Ppr4 and Ppr5 determine if the plant exhibits a hypersensitive response (Butler et al., 2016). Both hosts showed down-regulation of these genes in Ppr4, suggesting the pathogen is potentially manipulating the HR that are controlled by these R-genes (Figure 6). Despite this, there was significant expression of some of these R-genes in both interactions at Ppr4 showing high TPM values. Similarly, at Ppr5, the R-interaction shows more DE than in the S-interaction, with significant up-regulation in the R-interaction at 2-dpi. This suggests an involvement of these R-genes in the HR observed in the resistant hosts. This observation correlates with those made in the GO responses in which there is a delay in HR in the S-interaction.
It is important for plant defense responses to be suppressed in the absence of plant pathogens, as constitutive expression of defense-related compounds may be detrimental to plant growth and development (Marone et al., 2013). However, constitutive expression of defense-related genes, including R-genes, has been shown to contribute to disease resistance against pathogens. For example, in Arabidopsis ADR1 codes a specific CC-NB-LRR resistance gene. Mutations in this gene led to the constitutive expression of this R-gene, conferring broad-spectrum disease resistance against biotrophic pathogens and drought tolerance (Grant et al., 2003; Chini et al., 2004). Furthermore, constitutive expression of a TIR-NB-ARC-LRR disease resistance protein conferred resistance to phytopathogenic fungi and bacteria (Wen et al., 2017). Our results suggest that the constitutive expression of R-genes underlying resistance QTLs may putatively contribute to disease resistance. These results corroborate those from a recent study investigating the transcriptome of E. grandis, which found constitutive expression of genes encoding R proteins, such as Eucgr.C02650 underlying Ppr1, that was found to be significantly constitutively DE in the present study. The authors suggest this constitutive expression of defense-related genes potentially contributes to disease resistance observed against A. psidii (Santos et al., 2020).
The identification of DE R-genes when investigating the E. grandis transcriptomes, both at the induced and constitutive levels highlights candidate genes putatively involved in the defense responses and further investigations into the molecular differences that exist within these genes between genotypes is required to elucidate their roles.
This study investigated the molecular interactions of E. grandis in response to A. psidii to investigate mechanisms underlying host resistance. We identified pathways and genes potentially involved in the interaction. This includes brassinosteroid signaling, a pathway not previously reported in the A. psidii-host interactions and various R-genes that have different expression patterns, both at the constitutive and induced levels. Moreover, our findings suggest that resistant and susceptible hosts share similar responses, but timing and intensity determines disease severity. Due to the bioinformatic nature of RNA-seq analysis, further functional studies are required to characterize the observed responses to validate the results in the present study. This includes studies that investigate the brassinosteroid levels between resistant and susceptible seedlings to confirm the involvement of this pathway in host resistance. Moreover, investigations into the identified resistance genes are required to elucidate the roles they play during the interaction. Future studies should also aim to include investigations of the pathogen responses to identify mechanisms A. psidii is employing to manipulate susceptible hosts, as it will provide insight into novel targets of pathogen control, facilitated by the use of the A. psidii reference genome (Tobias et al., 2021). In conclusion, the results from this study will contribute to improved selection and breeding of E. grandis by manipulating pathways and genes tagged as important contributors to resistance.
The data have been uploaded to the NCBI under bioproject PRJNA763498.
LSS and SN conceived the study. SN obtained funding for the study. LSS and GSP performed the experiments and collected sample material. SS and CNO analyzed the data. SS wrote the first draft of the manuscript with input from SN. SS, CNO, LSS, GSP, and SN commented and revised the manuscript. All authors approved the submitted version.
This work was supported by the South African National Research Foundation (NRF) Y-rated Grant to SN (UID105767) and post doctoral support to LSS. The authors acknowledge funding from the Technology Innovation Agency of South Africa through the Forest Molecular Genetics Cluster Program to SN, CNO, and SS. Opinions and conclusions arrived at by the author(s) are not necessarily attributed to the NRF and other funding agencies.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Simon Southerton, formerly of the CSIRO, who kindly provided the E. grandis seedlings for inoculation.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffgc.2021.778611/full#supplementary-material
Butler, J. B., Freeman, J. S., Vaillancourt, R. E., Potts, B. M., Glen, M., Lee, D. J., et al. (2016). Evidence for different QTL underlying the immune and hypersensitive responses of Eucalyptus globulus to the rust pathogen Puccinia psidii. Tree Genet. Genomes 12:39. doi: 10.1007/s11295-016-0987-x
Carnegie, A. J., Kathuria, A., Pegg, G. S., Entwistle, P., Nagel, M., and Giblin, F. R. (2016). Impact of the invasive rust Puccinia psidii (myrtle rust) on native Myrtaceae in natural ecosystems in Australia. Biol. Invasions 18, 127–144.
Carnegie, A. J., Lidbetter, J. R., Walker, J., Horwood, M. A., Tesoriero, L., Glen, M., et al. (2010). Uredo rangelii, a taxon in the guava rust complex, newly recorded on Myrtaceae in Australia. Austr. Plant Pathol. 39, 463–466. doi: 10.1071/AP10102
Carnegie, A. J., and Pegg, G. S. (2018). Lessons from the incursion of myrtle rust in Australia. Annu. Rev. Phytopathol. 56, 457–478. doi: 10.1146/annurev-phyto-080516-035256
Chaparro-Garcia, A., Wilkinson, R. C., Gimenez-Ibanez, S., Findlay, K., Coffey, M. D., Zipfel, C., et al. (2011). The receptor-like kinase SERK3/BAK1 is required for basal resistance against the late blight pathogen Phytophthora infestans in Nicotiana benthamiana. PLoS One 6:e16608. doi: 10.1371/journal.pone.0016608
Chini, A., Grant, J. J., Seki, M., Shinozaki, K., and Loake, G. J. (2004). Drought tolerance established by enhanced expression of the CC–NBS–LRR gene, ADR1, requires salicylic acid, EDS1 and ABI1. Plant J. 38, 810–822. doi: 10.1111/j.1365-313X.2004.02086.x
Choudhary, S. P., Yu, J.-Q., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L.-S. P. (2012). Benefits of brassinosteroid crosstalk. Trends Plant Sci. 17, 594–605. doi: 10.1016/j.tplants.2012.05.012
Coutinho, T. A., Wingfield, M. J., Alfenas, A. C., and Crous, P. W. (1998). Eucalyptus Rust: a disease with the potential for serious international implications. Plant Dis. 82, 819–825. doi: 10.1094/PDIS.1998.82.7.819
De Bruyne, L., Höfte, M., and De Vleesschauwer, D. (2014). Connecting growth and defense: the emerging roles of brassinosteroids and gibberellins in plant innate immunity. Mol. Plant 7, 943–959. doi: 10.1093/mp/ssu050
De Vleesschauwer, D., Van Buyten, E., Satoh, K., Balidion, J., Mauleon, R., Choi, I. R., et al. (2012). Brassinosteroids antagonize gibberellin- and salicylate-mediated root immunity in rice. Plant Physiol. 158, 1833–1846. doi: 10.1104/pp.112.193672
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genetics 11, 539–548. doi: 10.1038/nrg2812
Dos Santos, I. B., Lopes, M. D. S., Bini, A. P., Tschoeke, B. A. P., Verssani, B. A. W., Figueredo, E. F., et al. (2019). The Eucalyptus cuticular waxes contribute in preformed defense against Austropuccinia psidii. Front. Plant Sci. 9:1978. doi: 10.3389/fpls.2018.01978
Du Plessis, E., Granados, G. M., Barnes, I., Ho, W. H., Alexander, B. J. R., Roux, J., et al. (2019). The pandemic strain of Austropuccinia psidii causes myrtle rust in New Zealand and Singapore. Austr. Plant Pathol. 48, 253–256. doi: 10.1007/s13313-019-0624-x
Fensham, R. J., Carnegie, A. J., Laffineur, B., Makinson, R. O., Pegg, G. S., and Wills, J. (2020). Imminent extinction of Australian Myrtaceae by fungal disease. Trends Ecol. Evol. 35:7. doi: 10.1016/j.tree.2020.03.012
Ferri, M., Righetti, L., and Tassoni, A. (2011). Increasing sucrose concentrations promote phenylpropanoid biosynthesis in grapevine cell cultures. J. Plant Physiol. 168, 189–195. doi: 10.1016/j.jplph.2010.06.027
Fradin, E. F., Abd-El-Haliem, A., Masini, L., Van Den Berg, G. C. M., Joosten, M. H. A. J., and Thomma, B. P. H. J. (2011). Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 156, 2255–2265. doi: 10.1104/pp.111.180067
Fradin, E. F., Zhang, Z., Juarez Ayala, J. C., Castroverde, C. D. M., Nazar, R. N., Robb, J., et al. (2009). Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 150, 320–332. doi: 10.1104/pp.109.136762
Fudal, I., Balesdent, M.-H., and Rouxel, T. (2018). Effector biology in fungal pathogens of nonmodel crop plants. Trends Plant Sci. 23, 753–755. doi: 10.1016/j.tplants.2018.07.002
Giblin, F. (2013). Myrtle Rust Report: New Caledonia. Maroochydore: University of the Sunshine Coast.
Glen, M., Alfenas, A. C., Zauza, E. A. V., Wingfield, M. J., and Mohammed, C. (2007). Puccinia psidii: a threat to the Australian environment and economy - a review. Austr. Plant Pathol. 36, 1–16. doi: 10.1071/AP06088
Goecks, J., Nekrutenko, A., Taylor, J., and Team, G. (2010). Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11:R86. doi: 10.1186/gb-2010-11-8-r86
Graça, R. N., Aun, C. P., Guimarães, L. M. S., Rodrigues, B. V. A., Zauza, E. A. V., and Alfenas, A. C. (2011). A new race of Puccinia psidii defeats rust resistance in eucalypt. Austr. Plant Pathol. 40, 442–447. doi: 10.1007/s13313-011-0056-8
Grant, J. J., Chini, A., Basu, D., and Loake, G. J. (2003). Targeted activation tagging of the Arabidopsis NBS-LRR gene, ADR1, conveys resistance to virulent pathogens. Mol. Plant Microbe Interact. 16, 669–680. doi: 10.1094/MPMI.2003.16.8.669
Gu, Z., Eils, R., and Schlesner, M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. doi: 10.1093/bioinformatics/btw313
Hahn, M., and Mendgen, K. (2001). Signal and nutrient exchange at biotrophic plant-fungus interfaces. Curr. Opin. Plant Biol. 4, 322–327. doi: 10.1016/S1369-5266(00)00180-1
Hsieh, J.-F., Chuah, A., Patel, H. R., Sandhu, K. S., Foley, W. J., and KüLheim, C. (2018). Transcriptome profiling of Melaleuca quinquenervia challenged by myrtle rust reveals differences in defense responses among resistant individuals. Phytopathology 108, 495–509. doi: 10.1094/PHYTO-09-17-0307-R
Huang, P.-Y., Yeh, Y.-H., Liu, A.-C., Cheng, C.-P., and Zimmerli, L. (2014). The Arabidopsis LecRK-VI.2 associates with the pattern-recognition receptor FLS2 and primes Nicotiana benthamiana pattern-triggered immunity. Plant J. 79, 243–255. doi: 10.1111/tpj.12557
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Junghans, D. T., Alfenas, A. C., Brommonschenkel, S. H., Oda, S., Mello, E. J., and Grattapaglia, D. (2003a). Resistance to rust (Puccinia psidii Winter) in Eucalyptus: mode of inheritance and mapping of a major gene with RAPD markers. Theor. Appl. Genet. 108, 175–180. doi: 10.1007/s00122-003-1415-9
Junghans, D. T., Alfenas, A. C., and Maffia, L. A. (2003b). Escala de notas para quantificação da ferrugem em Eucalyptus. Fitopatol. Brasileira 28, 184–188.
Kawanishi, T., Uematsu, S., Kakishima, M., Kagiwada, S., Hamamoto, H., Horie, H., et al. (2009). First report of rust disease on ohia and the causal fungus, Puccinia psidii, in Japan. J. Gener. Plant Pathol. 75, 428–431. doi: 10.1007/s10327-009-0202-0
Kim, E.-J., and Russinova, E. (2020). Brassinosteroid signalling. Curr. Biol. 30, R294–R298. doi: 10.1016/j.cub.2020.02.011
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, C. Q., Shao, J. F., Wang, Y. J., Li, W. B., Guo, D. J., Yan, B., et al. (2013). Analysis of banana transcriptome and global gene expression profiles in banana roots in response to infection by race 1 and tropical race 4 of Fusarium oxysporum f. sp cubense. BMC Genomics 14:16. doi: 10.1186/1471-2164-14-851
Li, J., Wen, J., Lease, K. A., Doke, J. T., Tax, F. E., and Walker, J. C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. doi: 10.1016/s0092-8674(02)00812-7
Liao, Y., Smyth, G. K., and Shi, W. (2013). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Mamani, E. M., Bueno, N. W., Faria, D. A., Guimarães, L. M., Lau, D., Alfenas, A. C., et al. (2010). Positioning of the major locus for Puccinia psidii rust resistance (Ppr1) on the Eucalyptus reference map and its validation across unrelated pedigrees. Tree Genet. Genomes 6, 953–962. doi: 10.1007/s11295-010-0304-z
Marlatt, R. B., and Kimbrough, J. W. (1979). Puccinia psidii on Pimenta dioica in south Florida. Plant Dis. Report. 63, 510–512.
Marone, D., Russo, M. A., Laido, G., De Leonardis, A. M., and Mastrangelo, A. M. (2013). Plant nucleotide binding site-leucine-rich repeat (NBS-LRR) genes: active guardians in host defense responses. Int. J. Mol. Sci. 14, 7302–7326. doi: 10.3390/ijms14047302
Mctaggart, A. R., Roux, J., Granados, G. M., Gafur, A., Tarrigan, M., Santhakumar, P., et al. (2016). Rust (Puccinia psidii) recorded in Indonesia poses a threat to forests and forestry in South-East Asia. Austr. Plant Pathol. 45, 83–89. doi: 10.1007/s13313-015-0386-z
Morel, J.-B., and Dangl, J. L. (1997). The hypersensitive response and the induction of cell death in plants. Cell Death Diff. 4, 671–683. doi: 10.1038/sj.cdd.4400309
Morkunas, I., Bednarski, W., and Kopyra, M. (2008). Defense strategies of pea embryo axes with different levels of sucrose to Fusarium oxysporum and Ascochyta pisi. Physiol. Mol. Plant Pathol. 72, 167–178. doi: 10.1016/j.pmpp.2008.09.003
Morkunas, I., and Ratajczak, L. (2014). The role of sugar signaling in plant defense responses against fungal pathogens. Acta Physiol. Plantarum 36, 1607–1619. doi: 10.1007/s11738-014-1559-z
Myburg, A. A., Grattapaglia, D., Tuskan, G. A., Hellsten, U., Hayes, R. D., Grimwood, J., et al. (2014). The genome of Eucalyptus grandis. Nature 510:356. doi: 10.1038/nature13308
Naidoo, S., KüLheim, C., Zwart, L., Mangwanda, R., Oates, C. N., and Visser, E. A. (2014). Uncovering the defence responses of Eucalyptus to pests and pathogens in the genomics age. Tree Physiol. 34, 931–943. doi: 10.1093/treephys/tpu075
Nakashita, H., Yasuda, M., Nitta, T., Asami, T., Fujioka, S., Arai, Y., et al. (2003). Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 33, 887–898. doi: 10.1046/j.1365-313x.2003.01675.x
Pegg, G., Taylor, T., Entwistle, P., Guymer, G., Giblin, F., and Carnegie, A. (2017). Impact of Austropuccinia psidii (myrtle rust) on Myrtaceae-rich wet sclerophyll forests in south east Queensland. PLoS One 12:e0188058. doi: 10.1371/journal.pone.0188058
Pegg, G. S., Brawner, J., and Lee, D. (2014). Screening Corymbia populations for resistance to Puccinia psidii. Plant Pathol. 63, 425–436. doi: 10.1111/ppa.12097
Pegg, G. S., Entwistle, P., Giblin, F. R., and Carnegie, A. J. (2020). Fire and rust – the impact of Austropuccinia psidii (myrtle rust) on regeneration of Myrtaceae in coastal heath following wildfire. Southern For. J. For. Sci. 82, 280–291. doi: 10.2989/20702620.2020.1819154
R Core Team (2018). “R: a Language and Environment for Statistical Computing”, in: R Foundation for Statistical Computing. Vienna. Available online at: https://www.R-project.org/ (accessed January 24, 2020).
Rayachhetry, M. B., Elliott, M. L., and Van, T. K. (1997). Natural epiphytotic of the rust Puccinia psidii on Melaleuca quinquenervia in Florida. Plant Dis. 81:831. doi: 10.1094/pdis.1997.81.7.831a
Robert-Seilaniantz, A., Grant, M., and Jones, J. D. (2011). Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. doi: 10.1146/annurev-phyto-073009-114447
Roux, J., Greyling, I., Coutinho, T. A., Verleur, M., and Wingfield, M. J. (2013). The Myrtle rust pathogen, Puccinia psidii, discovered in Africa. IMA Fungus 4, 155–159. doi: 10.5598/imafungus.2013.04.01.14
Saini, S., Sharma, I., and Pati, P. K. (2015). Versatile roles of brassinosteroid in plants in the context of its homoeostasis, signaling and crosstalks. Front. Plant Sci. 6:950. doi: 10.3389/fpls.2015.00950
Santos, S. A., Vidigal, P. M. P., Guimarães, L. M. S., Mafia, R. G., Templeton, M. D., and Alfenas, A. C. (2020). Transcriptome analysis of Eucalyptus grandis genotypes reveals constitutive overexpression of genes related to rust (Austropuccinia psidii) resistance. Plant Mol. Biol. 104, 339–357. doi: 10.1007/s11103-020-01030-x
Savadi, S., Prasad, P., Bhardwaj, S. C., Kashyap, P. L., Gangwar, O. P., Khan, H., et al. (2018). Temporal transcriptional changes in SAR and sugar transport-related genes during wheat and leaf rust pathogen interactions. J. Plant Growth Regul. 37, 826–839. doi: 10.1007/s00344-017-9777-4
Sekiya, A., Marques, F. G., Leite, T. F., Cataldi, T. R., De Moraes, F. E., Pinheiro, A. L. M., et al. (2021). Network analysis combining proteomics and metabolomics reveals new insights into early responses of Eucalyptus grandis during rust infection. Front. Plant Sci. 11:604849. doi: 10.3389/fpls.2020.604849
Soewarto, J., Giblin, F., and Carnegie, A. J. (2019). Austropuccinia Psidii (myrtle rust) Global Host List. Version 4. Canberra: Australian Network for Plant Conservation.
Thumma, B., Pegg, G. S., Warburton, P., Brawner, J. T., Macdonell, P., Yang, X., et al. (2013). “Molecular tagging of rust resistance genes in eucalypts”. Final Report to Plant Health. (Canberra: CSIRO Plant Industry). 1–15.
Tobias, P. A., Guest, D. I., Külheim, C., and Park, R. F. (2018). De novo transcriptome study identifies candidate genes involved in resistance to Austropuccinia psidii (myrtle rust) in Syzygium luehmannii (riberry). Phytopathology 108, 627–640. doi: 10.1094/PHYTO-09-17-0298-R
Tobias, P. A., Schwessinger, B., Deng, C. H., Wu, C., Dong, C., Sperschneider, J., et al. (2021). Austropuccinia psidii, causing myrtle rust, has a gigabase-sized genome shaped by transposable elements. G3 11:jkaa015.
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protocols 7:562. doi: 10.1038/nprot.2012.016
Turnbull, D., Yang, L., Naqvi, S., Breen, S., Welsh, L., Stephens, J., et al. (2017). RXLR effector AVR2 up-regulates a brassinosteroid-responsive bHLH transcription factor to suppress immunity. Plant Physiol. 174, 356–369. doi: 10.1104/pp.16.01804
Uchida, J., Zhong, S., and Killgore, E. (2006). First report of a rust disease on Ohia caused by Puccinia psidii in Hawaii. Plant Dis. 90:524. doi: 10.1094/pd-90-0524c
Vert, G. (2008). Plant signaling: brassinosteroids, immunity and effectors are BAK! Curr. Biol. 18, R963–R965. doi: 10.1016/j.cub.2008.09.006
Wang, Y., Hao, X., Lu, Q., Wang, L., Qian, W., Li, N., et al. (2018). Transcriptional analysis and histochemistry reveal that hypersensitive cell death and H2O2 have crucial roles in the resistance of tea plant (Camellia sinensis (L.) O. Kuntze) to anthracnose. Hortic. Res. Engl. 5:18. doi: 10.1038/s41438-018-0025-2
Wen, Z., Yao, L., Singer, S. D., Muhammad, H., Li, Z., and Wang, X. (2017). Constitutive heterologous overexpression of a TIR-NB-ARC-LRR gene encoding a putative disease resistance protein from wild Chinese Vitis pseudoreticulata in Arabidopsis and tobacco enhances resistance to phytopathogenic fungi and bacteria. Plant Physiol. Biochem. 112, 346–361. doi: 10.1016/j.plaphy.2017.01.017
Xavier, A. A., Alfenas, A. C., Matsuoka, K., and Hodges, C. S. (2001). Infection of resistant and susceptible Eucalyptus grandis genotypes by urediniospores of Puccinia psidii. Austr. Plant Pathol. 30, 277–281. doi: 10.1071/ap01038
Xavier, A. A., Da Silva, A. C., Da Silva Guimarães, L. M., Matsuoka, K., Hodges, C. S., and Alfenas, A. C. (2015). Infection process of Puccinia psidii in Eucalyptus grandis leaves of different ages. Trop. Plant Pathol. 40, 318–325. doi: 10.1007/s40858-015-0043-7
Yu, M.-H., Zhao, Z.-Z., and He, J.-X. (2018). Brassinosteroid signaling in plant-microbe interactions. Int. J. Mol. Sci. 19:4091. doi: 10.3390/ijms19124091
Zeng, Y., and Yang, T. (2002). RNA isolation from highly viscous samples rich in polyphenols and polysaccharides. Plant Mol. Biol. Report. 20, 417–417. doi: 10.1007/BF02772130
Keywords: Austropuccinia psidii, Eucalyptus, RNA-Seq, phytohormones, resistance loci
Citation: Swanepoel S, Oates CN, Shuey LS, Pegg GS and Naidoo S (2021) Transcriptome Analysis of Eucalyptus grandis Implicates Brassinosteroid Signaling in Defense Against Myrtle Rust (Austropuccinia psidii). Front. For. Glob. Change 4:778611. doi: 10.3389/ffgc.2021.778611
Received: 17 September 2021; Accepted: 12 November 2021;
Published: 08 December 2021.
Edited by:
Mohd Faidz Mohamad Shahimin, Universiti Malaysia Perlis, MalaysiaReviewed by:
Louis Bernier, Laval University, CanadaCopyright © 2021 Swanepoel, Oates, Shuey, Pegg and Naidoo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sanushka Naidoo, c2FudXNoa2EubmFpZG9vQGZhYmkudXAuYWMuemE=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.