- 1Department of Forest and Conservation Sciences, Faculty of Forestry, The University of British Columbia, Vancouver, BC, Canada
- 2Parks Canada Agency, Mount Revelstoke and Glacier National Parks, Revelstoke, BC, Canada
Whitebark pine (Pinus albicaulis Engelm.; WBP) is an endangered subalpine tree species and requires associations with ectomycorrhizal fungi (ECMF) for survival and growth. Despite this obligate dependence, there are gaps in the identification of ECMF that associate with WBP. In addition, ECMF rarely feature in assessments of recovery actions and little is known about the relationship between ECMF and the insects and pathogens affecting WBP. We used next-generation sequencing to characterize ECMF occurring in soil and mycorrhizal root tip samples from naturally occurring mature WBP trees and seedlings as well as planted WBP seedlings in the Columbia Mountains of Interior British Columbia, Canada. ECMF data was paired with data on tree age, tree health and soil conditions. Thirty-three species and twenty-one genera of ECMF were identified with medium or high confidence from mycorrhizal root tip samples. Major groups were: generalist ascomycetes [Cenococcum, Meliniomyces (=Hyaloscypha)], Atheliales (Piloderma, Amphinema, Tylospora), non-ascomycetous generalists (e.g., Amphinema), associates of high-elevation conifers (species of Cortinarius, Russula) and Suilloids (Suillus, Rhizopogon). Differences in WBP ECMF with other, drier and southerly regions that have been studied previously, were consistent with a distinct forest type and an endemism hypothesis. Soil at the planting site and planted seedlings hosted a reduced ECMF community or were non-ectomycorrhizal, which can be explained by site factors and is expected to affect seedling survival. ECMF composition on mature trees was correlated with tree health, which may have implications for WBPs resistance to pathogens and signals that ECMF are affected by the decline of their host. Understanding the ecology of WBP ECMF and their relationship with tree performance is essential for WBP recovery efforts.
Introduction
Whitebark pine (Pinus albicaulis Engelm.; hereafter WBP) is an obligate ectomycorrhizal mutualist; that is, the formation of functioning ectomycorrhizae is essential to the health and survival of the tree (Mohatt et al., 2008). Ectomycorrhizae are a predominantly mutualistic symbiosis formed between plant roots (usually of a tree or woody shrub) and a mycorrhizal fungus that functions to exchange nutrients, photosynthates and other molecules (Smith and Read, 2008). The mycorrhiza symbiosis is formed in some 80% of land plants and is a major component of temperate forest systems; in some species, 80% of plant N and P can be taken up through ectomycorrhizal fungal partners (van der Heijden et al., 2015). Generating knowledge on the community of ectomycorrhizal fungi (ECMF) that associate with WBP forms a major contribution to WBP ecology and recovery of the tree species.
WBP is listed as endangered in Canada under the Species at Risk Act (2002) and has been proposed for listing in the United States. Its decline is attributed to four pressures: white pine blister rust (Cronartium ribicola), mountain pine beetle (Dendroctonus ponderosae), fire suppression and climate change (Environment and Climate Change Canada [ECCC], 2017). The status of the species has stimulated numerous recovery efforts coordinated by a range-wide recovery plan (Keane et al., 2012).
The WBP ECMF community has been well studied over the past 15 years. This research can be organized into three groups. The first group consists of studies aimed at documenting and describing the fungal community (Johnson et al., 1994; Mohatt, 2006; Mohatt et al., 2008; Cripps and Antibus, 2011). This group and later additions from other applications and informal reports created a working species profile of the ECMF associated with WBP; the profile consists of ∼95 associates that are summarized in Keane et al. (2012) into three ecological clusters—(1) generalists, (2) high-elevation conifer associates and (3) Suilloids (Supplementary Table 1). The Suilloid cluster associates almost exclusively with pines or sometimes a smaller subset of pine species (Mohatt et al., 2008). This character makes them particularly important. Suilloids may confer a competitive advantage to WBP over secondary successional tree species that cannot form mycorrhizal associations with Suilloid taxa. This co-dependence also means that Suilloid taxa are likely to decline concurrently with WBP (Cripps and Antibus, 2011). The species Suillus subalpinus M. M. Moser has only been documented with WBP and because of this obligate dependence on a threatened species, it has been proposed for listing on the IUCN Red List (Osmundson, 2016).
The second group of studies use the WBP ECMF community to test a range of ecological questions. These include the topics: nutrient dynamics in ECMF (Antibus et al., 2018), community assembly and environmental determinants of ECMF composition (Cripps and Antibus, 2011; Glassman et al., 2017b), the theory of island biogeography (Glassman et al., 2017a), continental biogeographic patterns in ECMF composition and function (Talbot et al., 2014; Glassman et al., 2015), ECMF soil spore banks (Glassman et al., 2015, 2016) and fire-ECMF relationships (Trusty, 2009; Trusty and Cripps, 2011; Glassman et al., 2016; Jenkins, 2017; Jenkins et al., 2018).
The third group of studies is motivated by applied recovery applications for WBP and has focused on the use of ECMF in outplanting putatively blister rust resistant seedlings (Trusty, 2009; Cripps and Grimme, 2011; Trusty and Cripps, 2011; Lonergan, 2012; Lonergan et al., 2013; Jenkins, 2017; Cripps et al., 2018; Jenkins et al., 2018). The accumulation of research on WBP ECMF has had real impacts on the practice of whitebark management. Seedling inoculation and the incorporation of criteria to support ECMF development in planting site selection are strategies being implemented in some locations where WBP is actively managed (e.g., Lonergan et al., 2013; Jenkins, 2017; Cripps et al., 2018).
There are still important gaps in our knowledge of the WBP ECMF community. The descriptive work done to create the existing species profile (Supplementary Table 1) has been concentrated in three regions of the WBP range: the Greater Yellowstone Ecosystem (MT, United States), Waterton (AB, Canada) and Yosemite (CA, United States) (Figure 1), with little research outside these areas. Yet, ECMF composition can change over large biogeographic gradients (Talbot et al., 2014), even within the mycobiome of a single host species (e.g., Defrenne et al., 2019b with Pseudotsuga menziesii). Fruiting body (mushroom) collections and sequencing of soil samples have been dominant methods for documenting the community (Mohatt et al., 2008; Trusty and Cripps, 2011). Fruiting body surveys can be poor proxies of the ECMF communities below ground (Gardes and Bruns, 1996; Durall et al., 2006; Lalli et al., 2015), though recent studies in fungal ecology (in particular in wood inhabiting fungi) demonstrate that fruiting body surveys can be comparable or complimentary to high-throughput sequencing and metabarcoding (Ovaskainen et al., 2013; Frøslev et al., 2019; Saine et al., 2020; Heine et al., 2021). ECMF composition can also follow successional patterns and change with tree age/life stage (Glassman et al., 2017b; Koizumi et al., 2018). These gaps in the species profile of the WBP ECMF community can be partially filled by sampling in other biogeographically distinct parts of the species’ range, by retrieving actual ectomycorrhizae samples and by sampling across life stages.
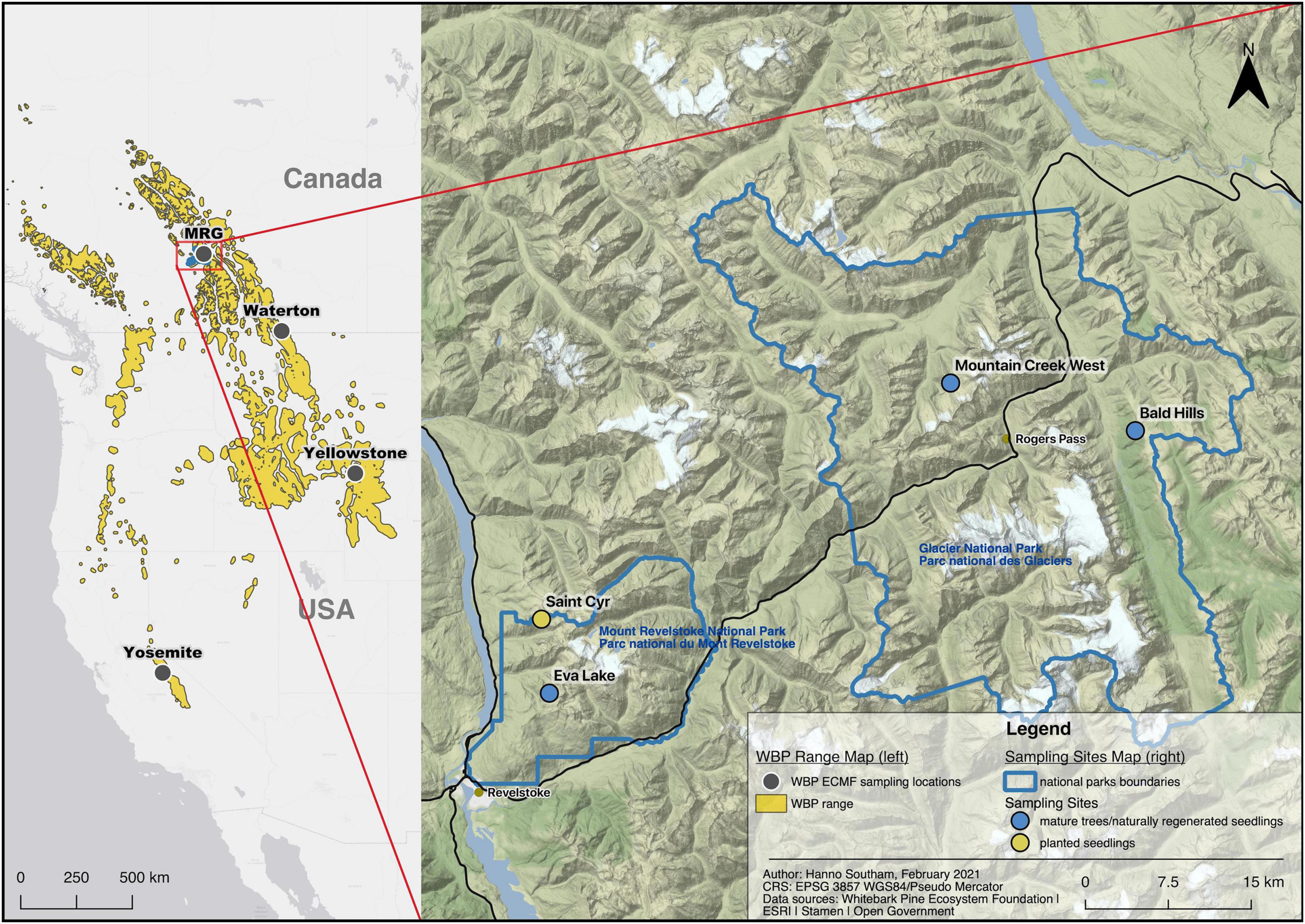
Figure 1. Locations of studies describing the ectomycorrhizal fungi associated with whitebark pine (left) and the sampling sites in this study (right). MRG: Mount revelstoke and glacier national parks. Created in QGIS v3.10.4 (QGIS.org, 2021).
In an applied ecology context, there is an ongoing research project into which, and to what extent, niche variables structure fungal and ECMF communities (van der Linde et al., 2018). These variables include host species, edaphic conditions (pH, soil nutrients, soil texture), climate conditions (temperature, precipitation) and tree/stand age. Study designs that pair sampling of ECMF with the collection of environmental/host data can contribute to this larger research project (van der Linde et al., 2018), even if it is not the primary research objective.
In the applied WBP recovery context, very little research has documented if and how planted seedlings are colonized by native ectomycorrhizal fungi and if the primary insects and forest pathogens affecting WBP (white pine blister rust and mountain pine beetle) have direct or cascading effects on its ECMF. Trusty and Cripps (2011) is the only study that has sampled ECMF from planted WBP; sampling from planted trees will indicate if the fungal component of the ectomycorrhizal mutualism is benefiting from recovery efforts. There are no studies in WBP systems examining correlations between insect or pathogen infestations and ECMF, though there is evidence from other pine systems that the death or stress of tree hosts can mediate declines or compositional shifts in ECMF (Karst et al., 2014, 2015; Treu et al., 2014). Studies that sample across varying severities of tree health will attend to the critical question of how ECMF associated with WBP are affected by insect and pathogen infestation and tree decline.
To respond to these gaps, we characterized the ECMF in ectomycorrhizal root tip and local soil samples from mature trees, naturally regenerated seedlings and planted seedlings located in Mount Revelstoke and Glacier National Parks, British Columbia, Canada (Figure 1), using next-generation sequencing. We use the terms tree community and soil community to refer to the ECMF communities from ectomycorrhizae samples and soil samples, respectively. We paired ECMF sampling with pre-existing or newly collected data on soil conditions (pH, %C, %N, C:N), tree age and tree health. This work was centered around five specific research questions: (1) Which ECMF are present on WBP and in the soils of WBP stands in this part of the species range?, (2) How do ECMF communities compare between naturally occurring stands and a planting site?, (3) Does ECMF community composition vary between seedlings and mature trees, or with tree age?, (4) Do soil properties affect the composition of ECMF communities?, and (5) Is ECMF community composition affected by tree health?
Materials and Methods
Study Site
This research was conducted in Mount Revelstoke and Glacier National parks of Canada (research permit#: GLA-2019-32862) (Figure 1). This region is characterized as an interior rainforest; lower elevations are in the Interior Cedar-Hemlock biogeoclimatic zone, which transitions to the Engelmann Spruce-Subalpine Fir zone at higher elevations. WBP occurs at subalpine elevations in mixed forests, often as a sub-dominant species to subalpine fir (Abies lasiocarpa), Engelmann spruce (Picea engelmanni) and mountain hemlock (Tsuga mertensiana).
Sampling occurred over four sites—three sites where mature trees and naturally regenerated seedlings were sampled and one site where planted seedlings were sampled (Figure 1, coordinates Supplementary Table 2). All of our sites were shared with other monitoring projects that provided data on site characteristics, tree health and transects to sample from. The mature tree and naturally regenerated seedling sites were shared with a project that is monitoring blister rust in the Canadian Rocky and Columbia Mountain regions (Smith et al., 2008, 2013; Shepherd et al., 2018). In this project, 10 m wide belt transects were established at these sites and health assessments conducted every 5 years. The planted seedling site (Saint Cyr) is also a monitoring site that tracks seedlings from putatively blister rust resistant WBP lineages. Seedlings in that project were planted in line transects and separated by > 1 m.
Climate, soil and vegetation characteristics for the sampling sites are summarized in Supplementary Table 2. The sites are centered around 2,000 m elevation and on southerly aspects. Mean annual temperature is approximately 0°C and mean annual precipitation averages 2,000 mm. The soils are Brunisols and Podzols (detailed classifications in Supplementary Table 2).
The Saint Cyr planting site was burned by a mixed severity wildfire in 2003; approximately 50% of the area burned was classified as moderate severity with the remainder equally split between low and high severity (K. Macauley, Parks Canada, pers. comms., 18 March 2020). Two groups of WBP were planted 14- (fall, 2017) and 15-years (fall, 2018) post fire, respectively. Seedlings were 2-years old when received from the nursery and were 4 and 3 years-old, respectively, at the time of sampling. Naturally occurring WBP had not been recorded at the site (pre- or post-fire). In the surrounding area (within 1 km), WBP occurs as rare sporadic individuals. The site is considered an ideal planting location because it meets the physical habitat requirements of WBP and the burn provides abundant open space for planting. However, the site’s recent history of fire and lack of naturally occurring WBP distinguish it from the naturally occurring WBP sampling sites and have significant implications for our results.
Field Sampling and Sample Processing
Thirty mature trees, 13 naturally regenerated seedlings and 10 planted seedlings were sampled in total; the distribution of samples between sites is recorded in Supplementary Table 2. Mature trees and planted seedlings were randomly selected from transects associated with other monitoring projects (see above). Equal numbers of planted seedlings were sampled from the 2017 and 2018 planting groups present at the Saint Cyr site. Naturally regenerated seedlings were randomly sampled from just outside the associated mature tree transects to avoid disturbing other monitoring projects. Mature trees sampled were a minimum of ∼5 m apart.
WBP commonly grows in multi-stemmed clumps and the outplanting at the Saint Cyr site emulated that form. In mature tree sampling, multi-stemmed trees or multiple adjacent stems whose roots could not be separated, were treated as single individuals. Occasionally, naturally regenerated seedlings germinated as a clump and could not be sampled as individual stems in the field but could be separated in processing; this added three additional individuals to our original target sample size of 10 (final sample size = 13). All naturally regenerated seedling samples were processed as individual samples but only one seedling from each of the clumps sampled was retained in comparative statistical analyses. Outplanted seedling clumps could be sampled as single stems without damaging neighboring seedlings.
Mature tree sampling followed the methods in Birch et al. (2021). Subsamples of fine roots and soil were collected from three random locations around each tree and then pooled into a single fine root sample and a single soil sample per tree. Excavated roots were followed from the bole of the tree outward until fine roots with a visible connection to the parent root could be found and the terminal fine root mass was then collected. This process was repeated at two other random locations around each tree. A soil subsample was collected adjacent to each fine root mass subsample. The L layer was removed and soil collected with a trowel to a 15 cm depth (this sample included the FH layers and surface mineral horizons). Equal volumes of the three soil subsamples were then pooled into a single sample. A tree core was also taken from the bole of each mature tree (Haglöf Increment Borer). The tree core was collected at the root collar, or at the earliest point that the stem became vertical, on the uphill side. In multi-stemmed clumps, the stem with the largest diameter was cored. Seedlings were excavated and collected in entirety. For each seedling, a single soil sample was collected adjacent to the seedling roots. To reduce possible disease transmission between trees or sample cross contamination, sampling tools were cleaned and sprayed with 10% bleach or alcohol between each tree sampled.
Fine root samples were stored at 4°C and processed within 48 h. Fine roots were rinsed free of soil and debris in water and then inspected for ECMF root tips. In the seedling samples, the entire intact root masses were scanned using WinRHIZO prior to collecting root tip samples. Root tips were examined using a 4X magnification dissecting microscope to identify ECMF colonization. Cross sections of each distinct ECMF morphotype were further examined using a 10X–40X compound microscope to confirm the absence of root hairs, presence of a fungal mantle, and presence of a Hartig net. Fifty ECMF root tips were randomly excised per sample and placed in a 1.5 ml microtube; occasionally, seedling samples contained less than fifty ECMF root tips, and all ECMF root tips that were present were thus collected. Samples were frozen and stored at −30°C. To reduce DNA cross-contamination between samples, trays and tools used for rinsing root masses were cleaned and submerged in a 10% bleach solution for 5 min between samples.
Soil samples were immediately frozen at −30°C post-field sampling and processed within 45 days. Soil samples were thawed and sieved to a final sieve pore size of 2 mm. A volume of 7.5 ml of each sieved soil sample was transferred to a Falcon tube and then re-frozen at −30°C for storage until DNA extraction and sequencing. The remainder of each sample was oven dried at 50°C and then stored in paper bags for subsequent soil chemistry analyses.
Processed fine root tip samples and 7.5 ml soil samples were shipped together on dry ice to the Integrated Microbiome Resource (Dalhousie University, Halifax, NS) for DNA extraction and subsequent next-generation sequencing.
DNA Extraction, Next-Generation Sequencing and Bioinformatics
DNA extraction and next-generation sequencing were completed for each pooled fine root tip sample and soil sample to identify the ECMF tree and soil communities, respectively. DNA was extracted using the QIAGEN PowerSoil DNA Kit, following the manufacturer’s protocol. The Internal Transcribed Spacer 2 (ITS2) region of fungi was PCR amplified and then sequenced following a 1-step amplicon library preparation protocol (Comeau et al., 2017), using modified primers ITS86(F) (Turenne et al., 1999) and ITS4(R) (White et al., 1990) that contained both the Illumina MiSeq adaptor and barcodes. Amplicon libraries were returned in the format of 1.8 Cassava demultiplexed paired-end fastq sequences.
Bioinformatics were primarily carried out in QIIME 2 (Bolyen et al., 2019). Low quality fragments were trimmed using Trimmomatic (threshold: sliding window = 5 bp, quality score = 20) (Bolger et al., 2014) and then primers were removed using CutAdapt (Martin, 2011). The ITS2 region was extracted with ITSxpress (Rivers et al., 2018). DADA2 (Callahan et al., 2016) was used for denoising, dereplication, chimera removal and production of amplicon sequence variants (ASVs). Taxonomy was assigned to ASVs using a trained Naïve Bayes classifier sourced from the Integrated Microbiome Resource (Comeau et al., 2017); the classifier was developed from the UNITE database (version 8.0, current to 2 February 2019) (Nilsson et al., 2019) with a 99% similarity threshold applied to species hypotheses and covered the entire ITS region. ASVs were filtered with three successive criteria to create the final dataset: (i) ASV frequency was > 0.1% of the dataset’s mean sample depth (corresponding to contaminant abundance between Illumina MiSeq runs) (Comeau et al., 2017), (ii) ASV was classified to the level of genus or species, and (iii) ASV was assigned to a taxon that is a known ectomycorrhizal associate. Ectomycorrhizal status was assigned to taxa with the FUNGuild database (Nguyen et al., 2016a). Taxa not described or belonging to multiple trophic guilds in FUNGuild were assigned ectomycorrhizal status manually; we relied on Tedersoo et al. (2010) as the authoritative source for ectomycorrhizal status. The main group requiring manual assignment was the Helotiales; only taxa from this group that were definitively described as ectomycorrhizal in Tedersoo et al. (2010) were retained in the dataset.
Tree Data
Tree Age
Tree cores were mounted on wooden increment core mounts, sanded and scanned at 2,400 dpi. Ring width measurements and a pith correction were done in CooRecorder (v7.8). Crossdating was completed in CDendro (v7.8) using a hybrid approach that compared cores against a reference chronology from the McMurdo site in Youngblut and Luckman (2013) and a reference chronology of cores from this project that had been successfully dated. Breaks and twists in the cores were common that are a result of the krummholz form of the trees sampled; this resulted in low cross correlations (Supplementary Table 3) and age estimates should be understood as “best estimates.”
Canopy Kill
We used existing data from the long-term health monitoring project (Smith et al., 2008, 2013; Shepherd et al., 2018) on canopy kill as a proxy for tree health; canopy kill was estimated on a discrete scale from 1 to 11 that corresponds to percentage ranges (Tomback et al., 2005). The discrete data was transformed to continuous percentages by taking the middle value of a range (e.g., 4 = 26–35% → 30.5%). In multi-stemmed clumps we averaged across stems. Dead stems were given a value of 100%.
Soil Analysis
pH
pH was measured from oven dried soil with a pH meter in a soil-water dilution following Hendershot and Lalande (2007). Because the samples contained the FH layers, a dilution ratio for organic soils was used of one-part dried soil to 10-parts deionized water. Every tenth sample was measured in duplicate for quality control.
Carbon and Nitrogen Analyses
Total C and N analyses were completed by the Stable Isotope Facility at the University of British Columbia using an Elementar vario EL cube elemental analyzer. Oven dried soil was freeze-dried, homogenized and encapsulated. Samples were measured in triplicate and the average was taken as the final %N and %C values.
Data Analyses
R (version 3.6.2) (R Core Team, 2019) was used for all data analyses. We relied primarily on the packages phyloseq (for microbiome data analyses) (McMurdie and Holmes, 2013) and ggplot (for graphics) (Wickham, 2016). Tree and soil communities were separated in analyses; within these communities, samples were typically aggregated by tree type (e.g., alpha diversity of ECMF from root tip samples (tree community) of mature trees). Prior to analyses, the raw dataset created in QIIME2 was rarefied to a library size of 1,250 reads (determined from the saturation point in number of reads vs. observed ASV curves); we did not discard samples below this library size because library size was interpreted to be an indicator of the abundance of ECMF tissue in a sample and not an artifact of inherent variability in amplification (see section “Amplification Levels”).
To describe the abundance of lineages and the composition of communities, we used the abundance metrics of number of occurrences and relative abundance; relative abundance was defined as the proportion of reads of a given ASV, species or genus in a sample. To investigate differences between tree types, we compared per-sample sequence counts, alpha-diversity and community composition. Alpha-diversity was estimated at the ASV level with Shannon’s diversity metric (; pi is the proportion of reads belonging to the ith ASV in a given sample). To test for differences in sequence count and alpha-diversity between tree types, we used ANOVA and pairwise t-tests or the non-parametric alternative Kruskal-Wallis and Dunn tests; all of these tests were housed in the rstatix package (Kassambara, 2020).
To quantify compositional differences, we used the Bray-Curtis dissimilarity metric (; Cij is the sum of the shared ASVs between two samples and Si and Sj are the total number of ASVs in the samples i and j, respectively) (Bray and Curtis, 1957) and Principal Coordinate Analysis (PCoA). To better resolve compositional differences, we agglomerated ASVs to the level of genus before running these analyses. The first two axes of PCoA were used to construct ordination plots. Genera were added to ordination plots with weighted taxa scores to identify marker genera (vegan function wascores); to reduce plot clutter, only the genera making up 90% of the relative abundance in each of the tree types were plotted. Adonis and ANOSIM tests were used to test the significance of compositional differences between tree types; estimates of beta dispersion (vegan function betadipser) or built-in diagnostic plots were used to check for location vs. dispersion effects in these tests.
To test the effects of soil (pH, %C, %N, C:N) and tree (age, canopy kill) variables on ECMF composition, we used vector fitting to the first two axes of PCoA ordination (vegan function envfit). Samples associated with planted seedlings were removed from the dataset to isolate naturally occurring patterns. Tree variables were only tested as predictors in the tree community. Ordinations were created following the method described above. For all ordination-based methods, distance matrices and PCoA analyses were done in phyloseq (McMurdie and Holmes, 2013) and statistical testing was done in vegan (Oksanen et al., 2019).
Results
Bioinformatics
Raw sequencing data contained 107 samples with a mean sample depth (sequences/sample) of 58,605. This data is available on NCBI’s Sequence Read Archive (PRJNA722685). Standard processing (isolating ITS2, denoising, dereplication, removal of chimeras) rendered 5,409 ASVs across 107 samples with a mean sample depth of 45,760. Taxonomic filtering to ECMF taxa identified at the genus or species level left 353 ASVs across 106 samples with mean sample depth of 21,242. Full summary statistics from the bioinformatic workflow broken down by sample type are in Supplementary Table 4. Representative sequences for each ASV are available on GenBank; the accession numbers for these sequences are in Supplementary Table 5.
Ectomycorrhizal Fungal Communities
Tree Community
ECMF in the tree community consisted of 224 ASVs grouping to 53 species, 26 genera and 20 families; a full list of these taxa is included as Supplementary Table 6. Of these taxa, 44 species, 24 genera and 19 families were found on mature trees. Twenty-one species, 15 genera and 12 families were confirmed on natural seedlings and 14 species, 14 genera and 13 families were confirmed on planted seedlings. Species accumulation curves indicate sampling was sufficient for mature trees and planted seedlings but not for natural seedlings (Supplementary Figure 1A).
Mature trees and naturally regenerated seedlings shared similar ECMF compositions. Dominant families/clades (large relative abundances and high percent occurrences) observed in these tree types were: Atheliaceae (genera: Piloderma, Amphinema, Tylospora), Helotiaceae [genus: Meliniomyces (=Hyaloscypha)], Cortinariaceae (genus: Cortinarius), Gloniaceae (genus: Cenococcum), Russulaceae (genera: Lactarius, Russula) and Suilloids (genera: Suillus, Rhizopogon). Major genera on planted seedlings were Meliniomyces (=Hyaloscypha), Russula, Cenococcum and Suillus with other genera occurring in trace amounts (Supplementary Table 7 and Figure 2A).
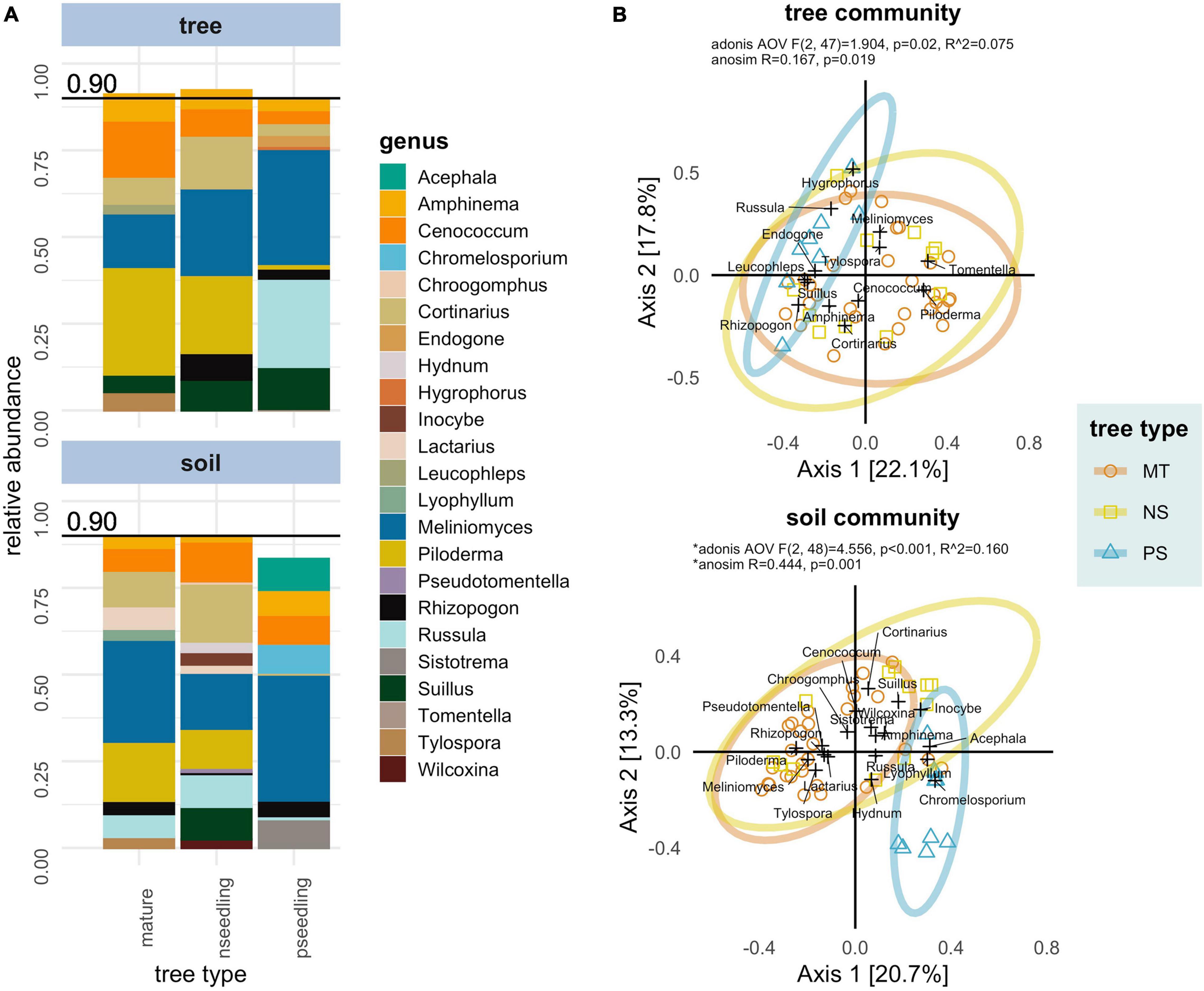
Figure 2. Compositional differences between tree types in ectomycorrhizal fungal communities associated with whitebark pine in interior British Columbia separated by sample type. Sample type: ectomycorrhizae samples (tree OR tree community) and soil samples (soil OR soil community). Tree types: mature trees (mature OR MT), naturally regenerated seedlings (nseedling OR NS) and planted seedlings (pseedling OR PS). (A) Relative abundance (proportion of reads after rarfying) of the genera making up > 0.9 of the total abundance in each tree type. (B) Principal Coordinate Analysis (PCoA) biplot ordinations based off a Bray-Curtis distance matrix. Axes show the percent of variation in community composition explained. Samples are marked as colored points; genera are added as weighted scores and marked as black crosses. Results of Adonis and ANOSIM statistics testing tree type as predictor of composition are displayed in top line. *A significant dispersion effect was detected in the soil community that could have effected these statistics.
Soil Community
ECMF in the soil community made up 283 ASVs belonging to 80 species, 33 genera, and 26 families; a complete list of the soil ECMF taxa is included as Supplementary Table 8. Sixty-eight species, 30 genera and 23 families were identified in samples associated with mature trees. Samples associated with natural seedlings confirmed 38 species, 22 genera, and 18 families and samples associated with planted seedlings contained only five species, nine genera, and nine families. Species accumulation curves were saturated for all three of the sample types (Supplementary Figure 1B).
The mature tree soil community featured Helotiaceae [genus: Meliniomyces (=Hyaloscypha)], Atheliaceae (genera: Piloderma, Amphinema, Tylospora), Cortinariaceae (genus: Cortinarius), Russulaceae (genera: Russula, Lactarius), Gloniaceae (genus: Cenococcum), Pezizales (genera: Hydnotrya, Wilcoxina), Thelephoraceae (genus: Pseudotomentella), Hydnaceae (genus: Sistotrema) and Suilloids (genera: Rhizopogon, Suillus) as the major taxa. Major taxa in the naturally regenerated seedling soil community were Helotiaceae [genus: Meliniomyces (=Hyaloscypha)], Cortinariaceae (genus: Cortinarius), Atheliaceae (genera: Piloderma, Amphinema, Tylospora), Gloniaceae (genus: Cenococcum), Russulaceae (genera: Russula, Lactarius), Inocybaceae (genus: Inocybe), Hydnaceae (genus: Hydnum), Pezizales (genera: Hydnotrya, Wilcoxina), Thelephoraceae (genera: Pseudotomentella, Tomentella) and Suilloids (genera: Suillus, Rhizopogon, Chroogomphus). The soil community associated with planted seedlings featured only one genus in more than two samples—Helotian Meliniomyces (=Hyaloscypha) (Supplementary Table 9 and Figure 2A).
Comparisons Between Tree Types
Seedling Root Structure and Colonization Levels
Root structure differed significantly between planted and naturally regenerated seedlings. Planted seedlings had greater root mass, a greater proportion of fine roots and more root tips than naturally regenerated seedlings (Figure 3).
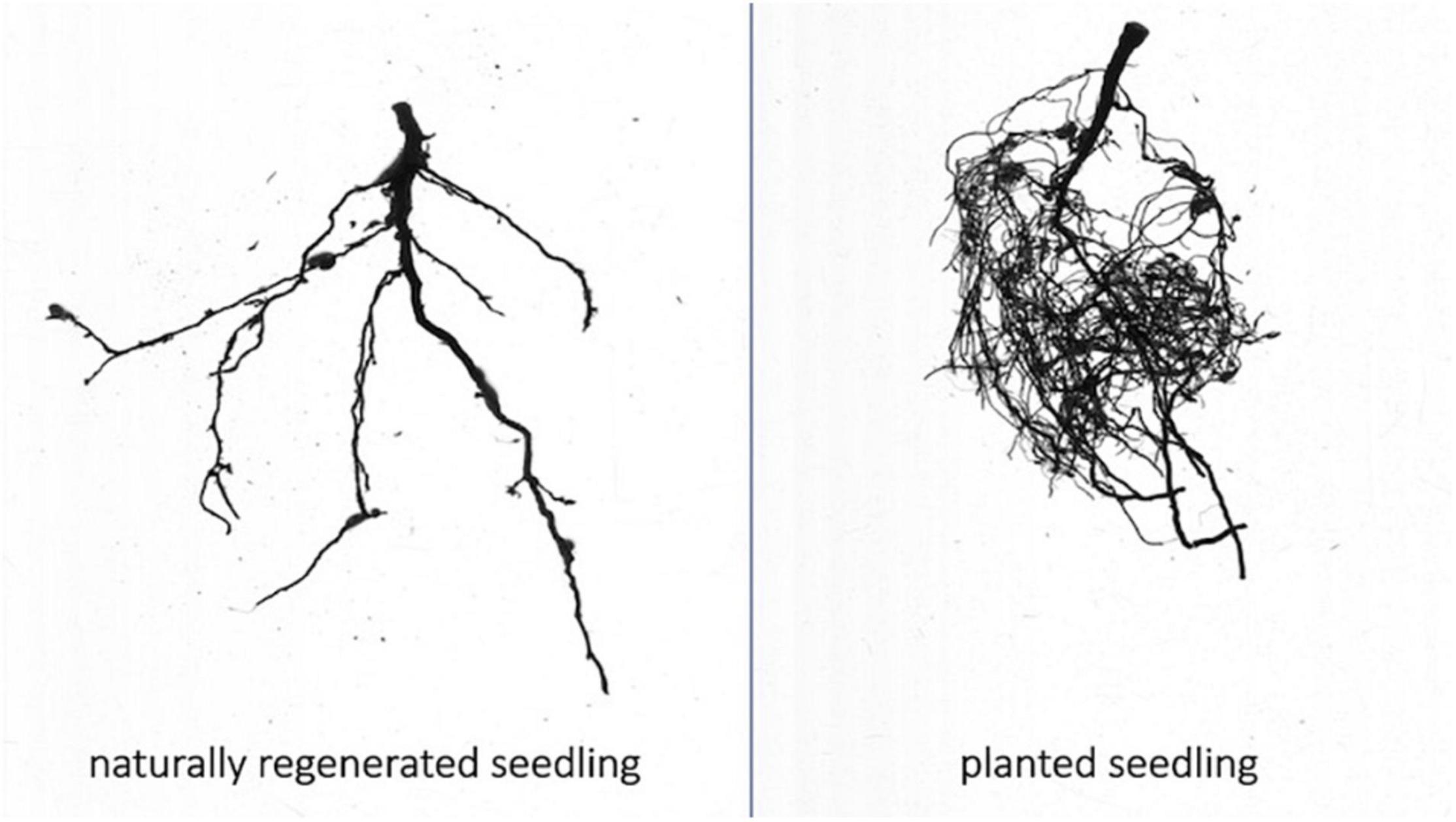
Figure 3. Representative root scans of naturally regenerated (left) and planted (right) whitebark pine seedlings from interior British Columbia. Scans taken with WinRHIZO root scanning system.
Colonization levels were consistently higher in naturally regenerated seedlings relative to planted seedlings. The mean number of colonized root tips collected in naturally regenerated seedlings was 40 and half of the seedlings met the target sample size of 50 root tips. The mean number of colonized root tips collected on planted seedlings was just five (Supplementary Table 10).
Amplification Levels
Mature trees and naturally regenerated seedlings consistently showed higher levels of amplification than planted seedlings but did not differ from each other (Figure 4 and Supplementary Table 4). Kruskal-Wallis tests showed tree type as a significant predictor of sequence count in both tree and soil communities (tree: p = 0.0022, eta2 = 0.218; soil: p = < 0.0001, eta2 = 0.505). Post-hoc Dunn tests showed that in the tree community, the difference in amplification between the mature trees and planted seedlings was significant (p = 0.001) and that between naturally regenerated and planted seedlings was nearly significant (p = 0.064). In the soil community, both comparisons were significant (mature tree vs. planted seedling: p = <0.0001, naturally regenerated seedling vs. planted seedling: p = 0.012).
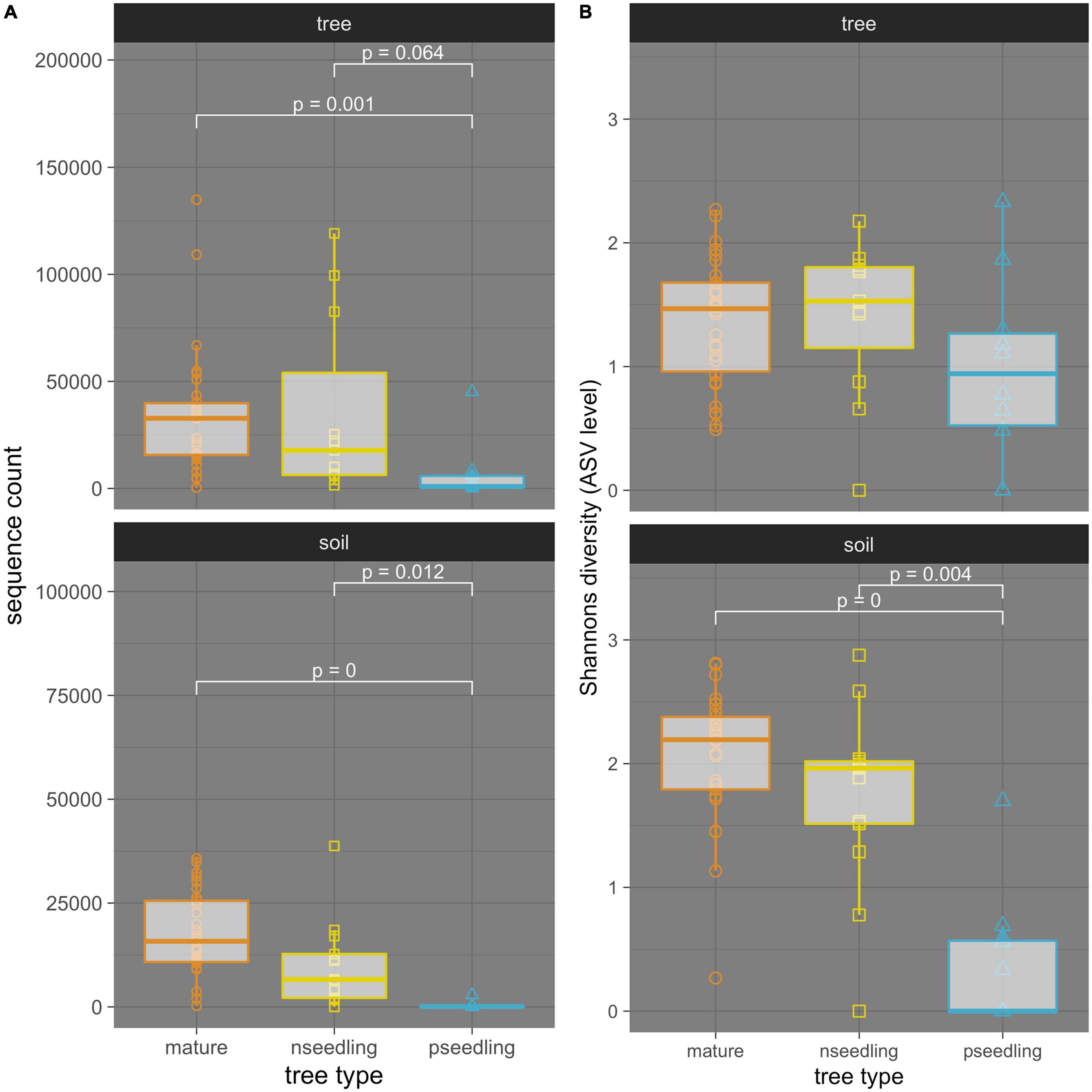
Figure 4. Comparisons between tree types of (A) amplification levels and (B) alpha-diversity of ectomycorrhizal fungi associated with whitebark pine in interior British Columbia separated by sample type. Sample type: ectomycorrhizae samples (tree OR tree community) and soil samples (soil OR soil community). Tree types: mature trees (mature), naturally regenerated seedlings (nseedling) and planted seedlings (pseedling). sequence count is the number of ITS2 sequences belonging to ectomycorrhizal fungi in each sample. Diversity calculated at the ASV level using Shannon’s diversity index. Results of Dunn tests with a Bonferroni adjustment are reported in white; only significant or near-significant pairs are shown.
Alpha Diversity
In both tree and soil communities, alpha diversity was highest in mature trees, followed by naturally regenerated seedlings, then planted seedlings (Figure 4 and Supplementary Table 11). In the tree community, mature trees, naturally regenerated seedlings and planted seedling averaged seven, six, and four ECMF species per sample; in the soil community, the mean number of ECMF species was 13, 11 and one, respectively. ANOVA/Kruskal-Wallis statistics testing the significance of tree type as a predictor of Shannon’s diversity showed this pattern was only significant in the soil community (tree: p = 0.17, = 0.07; soil: p = < 0.0001, eta2 = 0.452). Post-hoc Dunn tests in the soil community showed that the mature tree and naturally regenerated seedlings had significantly higher alpha diversity than planted seedlings but were not significantly different from each other (Figure 4).
Community Composition
Natural tree types (mature trees, naturally regenerated seedlings) were compositionally distinct from planted seedlings in both tree and soil communities. In the tree community, planted seedlings had greater relative abundance of Meliniomyces (=Hyaloscypha) and reduced relative abundances across numerous taxa (e.g., Piloderma and Cortinarius). In the soil community the compositional shift in planted seedlings was driven by a reduction in diversity. Meliniomyces (=Hyaloscypha) was the only genus to appear regularly; other genera that were frequent in soils associated with natural tree types were reduced or lost (e.g., Piloderma, Cortinarius, Suillus, Rhizopogon, Cenococcum). Two genera (Acephala, Chromelosporium) appeared at low abundances in soils associated with planted seedlings that were not present in those from natural tree types (Supplementary Table 9 and Figure 2A).
These compositional differences were confirmed graphically and with statistical tests (Figure 2B). The first two axes of Principal Coordinate Analysis (PCoA) explained 39.9 and 34.0% of the variation in community composition in the tree and soil communities, respectively; planted seedlings formed distinct groupings in both of these plots. Adonis and ANOSIM statistics were consistent with each other and showed tree type was a significant predictor of composition in both communities (tree: Adonis, p = 0.02, R2 = 0.075; ANOSIM, p = 0.019 | soil: Adonis, p = < 0.001, R2 = 0.160; ANOSIM, p = 0.001). This indicates that mature trees and naturally regenerated seedlings were compositionally distinct from planted seedlings but not from each other. In the soil community, a dispersion effect was detected that could confound these statistics; the clear separation of planted seedling samples in PCoA indicate both location and dispersion effects are present in the soil community and that the significant result for differences in composition holds.
Tree Age, Tree Health and Soil Variables as Predictors of Ectomycorrhizal Fungi Communities
Tree Variables
Tree age was not a significant predictor of ECMF community composition (envfit: p = 0.978, r2 = 0.002) but canopy kill was (envfit: p = 0.002, r2 = 0.439) (Figure 5A). Higher values of canopy kill (trees that were more diseased) typically hosted simpler communities and were typified by increased abundances of Cenococcum and Amphinema.
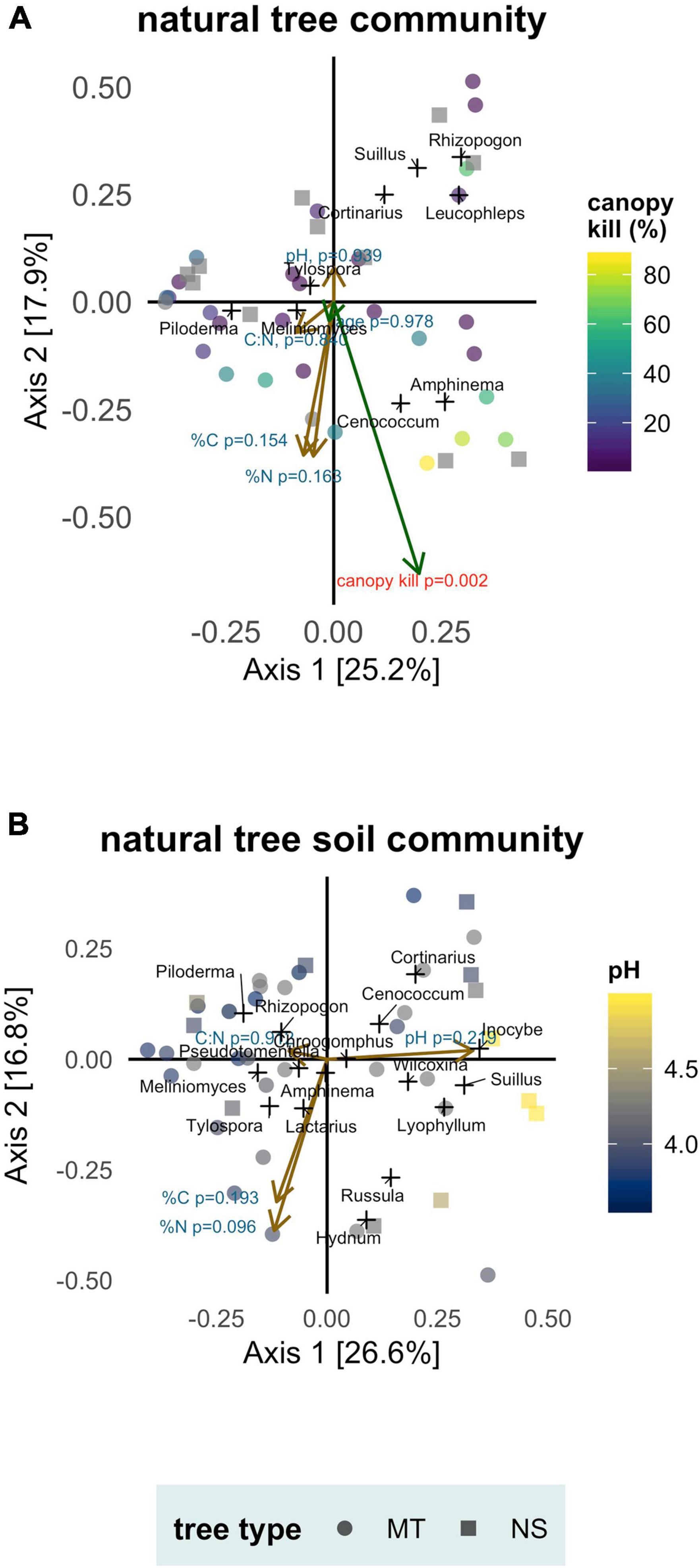
Figure 5. Soil conditions (pH, %N, %C, C:N), tree age and canopy kill as predictors of ectomycorrhizal fungal communities associated with naturally occurring whitebark pine separated by sample type: (A) mycorrhizae samples (tree community) and (B) soil samples (soil community). Tree type: mature trees (MT) and naturally regenerated seedlings (NS). Principal Coordinate Analysis biplot ordinations based off a Bray-Cutis distance matrix. Axes labels indicate percent of variation in community composition explained. Samples are marked as colored points; genera are added as weighted scores and marked as black crosses. Vectors representing soil variables are drawn in brown and vectors representing tree age and canopy kill are drawn in green. Length of the vector is proportional to its correlation with community composition; significant predictors (canopy kill) are marked with red.
Soil Variables
None of the soil variables tested (pH, %C, %N, C:N) were significant predictors of ECMF community composition in either tree or soil communities (Figure 5). In the tree community, envfit statistics from vector fitting to PCoA ordination were—pH: p = 0.939, r2 = 0.006; %C: p = 0.154, r2 = 0.132; %N: p = 0.163, r2 = 0.131; C:N: p = 0.840, r2 = 0.013. In the soil community, envfit statistics were—pH: p = 0.219, r2 = 0.109; %C: p = 0.193, r2 = 0.117; %N: p = 0.096, r2 = 0.165; C:N: p = 0.912, r2 = 0.008.
Discussion
Ectomycorrhizal Fungi of Whitebark Pine in Interior British Columbia
The set of confirmed WBP ectomycorrhizal associates reported here as the tree community is the core contribution of this research. These ECMF were identified using next-generation sequencing and barcoding from true ectomycorrhizae samples. An important caveat is that identification of sequences in “meta-barcoding” (Somervuo et al., 2017) approaches, like that applied here, can often be wrong. This is especially true at the level of species and for some groups (e.g., Cortinarius is not well resolved by ITS2). Sequence errors, intraspecific variation and errors/limited coverage in the reference database can pose problems to correct identification (Somervuo et al., 2017; Lücking et al., 2020). The taxa reported here should be understood as “first diagnoses” (Lücking et al., 2020). Still, this dataset combined with previous research on the WBP ECMF community forms a profile of the reported associates of WBP with reasonable coverage across the WBP range; this list is compiled as Supplementary Table 1.
Sixty-seven species and twenty-six genera were identified in the tree community; an additional, non-overlapping, twenty species and nine genera were identified from the paired soil sampling. We carried out a qualitative confidence assessment of the taxa in the tree community based on frequency of occurrence and whether the taxa had been reported previously as a WBP associate (Supplementary Table 12). This left thirty-three species and twenty-one genera that we confirm as WBP associates with high or medium confidence; of these, twenty-three species and three genera had not been reported as WBP associates before.
Generalist ascomycetes, Cenococcum and Meliniomyces (=Hyaloscypha), were dominant and widespread. The Atheliales (generalists: Piloderma, Amphinema, and Tylospora) were also a dominant clade. A diverse spread of taxa occurred at medium abundances and frequencies—generalists: Wilcoxina, Hydnotrya, Pseudotomentella, Tomentella, Sistotrema and Endogone (=Jimgerdemannia) and high-elevation conifer associates: species of Cortinarius, Lactarius and Russula. Suilloids (Rhizopogon and Suillus) were, as reported elsewhere, a regular and important part of the community.
Helotian taxa were abundantly co-amplified in our samples. These taxa are known as secondary root associates and they are commonly co-amplified from ectomycorrhizae samples; the nutritional modes of these taxa are, however, debated and only a few have been proven truly ectomycorrhizal (Tedersoo et al., 2009). In reviewing the ectomycorrhizal status of these taxa following Tedersoo et al. (2010), we removed all except for the lineages Acephala macrosclerotiorum and Meliniomyces bicolor (=Hyaloscypha bicolor). Even if non-ectomycorrhizal, the presence of these taxa is worth reporting because they form a significant part of the WBP rhizosphere and have been linked elsewhere with poor stand health (discussed below).
The community documented here diverges from previous descriptions of the WBP ECMF community. There is a functional shift toward generalist and high-elevation conifer associates and away from Suilloids. The dominance of the wood inhabiting Atheliales clade (Piloderma, Amphinema and Tylospora) is particularly distinctive. The shift we observed is likely the result of sampling mixed forests here vs. relatively pure WPB forests in other studies. Host plant identity is a primary determinant of ECMF composition (van der Linde et al., 2018). In our study region, WBP is subdominant to subalpine fir, Engelmann spruce and mountain hemlock, where the Atheliales have previously been shown to be a dominant ectomycorrhizal guild (Hagerman et al., 1999; Walker et al., 2014). This contrasts with the other regions where the WBP ECMF community has been sampled, where pine was the dominant host. In addition, Piloderma can access organic nitrogen pools (Heinonsalo et al., 2015) and this may be advantageous in soils, like those at our study sites, that have accumulations of recalcitrant litter.
This shift in dominant guilds could also reflect our sampling approach. Sampling was biased toward roots that were in organic soil horizons and close to the bole of the tree, potentially selecting for wood-inhabiting fungi. Talbot et al. (2014), in contrast, sampled from both organic and underlying mineral horizons in WBP forests in the Yosemite region; they reported no analogous shift, but structuring between these layers has been documented in other high-elevation pine systems (Koizumi et al., 2018). Different survey methods (fruiting body surveys vs. next-generation sequencing and metabarcoding) could also amplify this functional shift because it is driven by resupinate and hypogenous taxa, but the shift is present in comparisons with studies that employ similar methods (Talbot et al., 2014; Glassman et al., 2015, 2017a).
The second mode by which our community diverges from communities described in other parts of the species’ range is compositional. At the species level, the communities diverge, whereas they align at the genus level and higher. Of the thirty-three species and twenty-one genera identified with medium or high confidence, 70% (23/33) of the species are newly described associates of WBP but only 14% (3/21) of the genera are new. This indicates there is regional endemism, as suggested by others (Talbot et al., 2014; Rosinger et al., 2018; Defrenne et al., 2019b). Talbot et al. (2014) documented the soil ECMF community across continental North America in the Pinaceae and emphasize endemism resulting from dispersal limitation as a primary determinant of ECMF composition. Rosinger et al. (2018) and Defrenne et al. (2019b) both provide evidence for endemic structuring of ECMF within the range of single host species but also join a growing body of evidence that identifies environmental variables (e.g., climate, edaphic properties and nutrient regimes) as determinants of ECMF composition (reviewed in van der Linde et al., 2018). Our study region is climatically distinct from other regions where sampling has occurred and environmental variables, in particular mean annual precipitation (Defrenne et al., 2019b), may contribute to this compositional divergence.
Our data also raises unresolved questions in the pine-specific Suilloid group. Suillus americanus (syn. S. sibiricus), S. discolor and S. subalpinus are five-needled pine or whitebark pine specialists with broad distributions (Cripps and Antibus, 2011; Nguyen et al., 2016b). None of these species were recorded in our data. The lineage we identified as S. punctatipes (=Boletinus punctatipes) sits in the Suillus placidus/subalpinus/anomalus species complex but is molecularly (with ITS2) and/or morphologically distinct from the related species in the complex (Nguyen et al., 2016b). S. americanus, S. discolor and S. subalpinus being absent from our data is surprising because these taxa do not appear dispersal limited, and greater congruence is expected among host specialists. S. americanus has been recorded with western white pine (Pinus monticola) in our study area and with WBP in Banff National Park (C. Cripps, pers. comms., 6 May 2021), which is a good indication that processes other than dispersal limitation are restricting it from our site. S. brevipes is a second Suillus taxa recorded in our data; finding it here is surprising because it is part of the well-established/luteus clade that is near completely restricted to hard-pine hosts, whereas WBP is a soft-pine (Nguyen et al., 2016b). S. brevipes has previously been reported from soils of WBP stands in the Yosemite region (Glassman et al., 2017a) and this would represent a second rare case of “host jumping” in that clade (Nguyen et al., 2016b).
The common errors in assigning taxonomy with meta-barcoding techniques (noted above) (Somervuo et al., 2017; Lücking et al., 2020) may contribute to the questions raised in Suillus and more generally to the compositional divergence with previous studies. Re-evaluation of our data with alternative approaches to classification [e.g., phylogenetic placement of reads in reference trees or probabilistic methods (PROTAX)] may increase confidence in species and genus names assigned to sequences (Somervuo et al., 2016; Lücking et al., 2020) and aid comparison with other descriptions of the WBP ECMF community.
Assessment of Ectomycorrhizal Fungi at a Whitebark Pine Planting Site
The ECMF community on planted seedlings was clearly distinct from those on natural tree types (mature trees and naturally regenerated seedlings). Planted seedlings had fewer colonized root tips, significantly lower per-sample sequence counts, lower (though not significantly) diversity and were compositionally distinct; the relative abundance of the generalist Meliniomyces (=Hyaloscypha) was increased and numerous taxa prominent on natural tree types (e.g., Piloderma and Cortinarius) were reduced or absent. The most apparent difference, however, was observed visually when processing fine root samples. Planted seedlings had a completely distinct root structure, with far more fine-root mass than naturally regenerated seedlings. Moreover, with the exception of a single dominant morphotype that we expect is Meliniomyces (=Hyaloscypha), planted seedlings were visually non-mycorrhizal (though Russula, Suillus, and Cenococcum were identified at medium abundances through sequencing). The prolific fine root mass of planted seedlings may be an artifact of root development and fertilizer regimes in a nursery upbringing or it may be a foraging response to the lack of ectomycorrhizal partners that are the dominant source of nutrients (Defrenne et al., 2019a).
Differences in the ECMF community between the soils of the planting site and soils where naturally occurring trees were sampled were even more apparent than the differences observed from the mycorrhizal communities on tree roots. Largely, our results indicate that soils in proximity to planted seedlings did not contain ECMF tissue with DNA that could be amplified at this sequencing depth. Per-sample sequence counts and alpha diversity were significantly lower in soil from the planting site compared to soil associated with natural tree types, and Meliniomyces (=Hyaloscypha) was the single dominant genus.
Together these results reveal that the ECMF community occurring on planted seedlings did not match naturally occurring communities, nor did the soils of the planting site contain the appropriate inoculum to provide that community. This has not yet effected seedling survival (as of 2019, seedling survival was 82.4 and 94.2% for the 2017 and 2018 plantings, respectively) (Skinner et al., 2019) but we expect it will in time. High survival rates immediately after planting are common (Jenkins, 2017) and even obligately mycorrhizal tree seedlings can survive as non-mycorrhizal for a lag period (Collier and Bidartondo, 2009). Monitoring over 5–10 years will help isolate the effect of mycorrhizal status on WBP planted seedling survival (Jenkins, 2017). Note that our sample size for seedlings (natural and planted) was relatively small because of restrictions on the destructive sampling of a species protected by the Canadian Species at Risk Act and comparisons against mature trees were unbalanced. But the consistent and visibly apparent nature of the results observed at the planting site support the conclusions we make.
These differences in ECMF are largely attributable to the planting site. The planting site was located within the footprint of a 2003 mixed severity wildfire, largely lacks an ectomycorrhizal plant community and is not a naturally occurring WBP forest (presently or historically). The effect of fire on ECMF communities is an active field of research that has variable consensus. There is good evidence that fire shifts ECMF composition for long (∼50 year) timeframes and moderate evidence that fire reduces diversity and inoculum potential (measured with colonization) over decadal timeframes (∼22 and 11 years, respectively) (Karst et al., 2014; Dove and Hart, 2017; Taudière et al., 2017). We also know there is a suite of fungi, termed the “ectomycorrhizal fungal sporebank,” that is separate and compositionally distinct from the active ECMF community and exists in the soil as resistant propagules that colonize regenerating plant hosts post-disturbance (Glassman et al., 2015, 2016); in WBP forests, this community is dominated by Rhizopogon, Cenococcum, Wilcoxina and Thelephora (Glassman et al., 2015). ECMF sporebanks have been shown to persist through wildfires (Glassman et al., 2016) and it is likely this sporebank that WBP seedlings outplanted on burns are reliant on. The longevity of ECMF sporebanks is not well assessed but existing research is focused on pine associated taxa (predominantly Rhizopogon) and predicts that these sporebanks will remain viable for decades (Bruns et al., 2009; Nguyen et al., 2012); these two studies are first reports from a century long experiment that will contribute valuable knowledge to this discussion.
Paired with the influence of fire, is the importance of established trees as sources of inoculum for regenerating seedlings (Haskins and Gehring, 2005; Hubert and Gehring, 2008; Teste et al., 2009; Mueller et al., 2019; Simard et al., 2021). Established trees can provide inoculum directly [from their root system at immediate spatial scales (∼5 m)] (Teste et al., 2009) or indirectly [nearby stands that host ECMF that disperse to regenerating seedlings at local scales (∼1 km)] (Trusty and Cripps, 2011; Policelli et al., 2019). The species identity of the source tree can alter the community composition of the inoculum (Hubert and Gehring, 2008); conspecific or congeneric source trees are likely especially important for WBP because these trees will host Suilloid taxa. Even dead trees can provide a “legacy” of ECMF inoculum; prior to tree death, ECMF spore banks may form that persist to colonize successive generations (Mueller et al., 2019; Shemesh et al., 2019).
The combination of fire and lack of established ectomycorrhizal trees help explain the paucity of ECMF at our planting site. Fire would have reduced ECMF inoculum potential directly by burning live ECMF and reducing the density of the soil spore bank, and indirectly by killing ectomycorrhizal host plants. The absence of inoculation from a persistent, fire resistant soil spore bank is more puzzling. The significant lag time (14 or 15 years) between fire and planting may have led to the degeneration of the spore bank (contrary to viability estimates in Bruns et al., 2009; Nguyen et al., 2012); our knowledge of ECMF soil spore bank longevity is small and this process may well be modified in harsh high-elevation habitats. The lack of significant WBP stands at or near the planting site is another important variable; the nearest trees to the site are sporadic, isolated WBP ∼1 km away. These stands would have hosted Rhizopogon species that contribute to soil spore banks and facilitated colonization of seedlings by spore dispersal from outside the fire perimeter.
This interpretation aligns with (and builds upon) the only other study that has sampled ECMF from outplanted seedlings (Trusty and Cripps, 2011). Their results contrast with ours and document successful integration of planted seedlings with naturally occurring ECMF. They identify two key factors in this success: (1) seedlings were planted 1 year post burn and could access a viable soil spore bank, and (2) unburned WBP stands were present outside the fire perimeter that could provide Suilloid propagules (Trusty and Cripps, 2011). These same two factors likely explain the lack of integration of planted seedlings and native ECMF in our results.
Lack of Evidence for Successional or Edaphic Filtering on Ectomycorrhizal Fungi
No successional patterns were apparent in our data; we detected no compositional differences between mature trees and naturally regenerated seedlings, and tree age was not a significant predictor of ECMF composition. Similarly, none of the edaphic variables (pH, %C, %N, C:N) were significant predictors of ECMF composition. These results are surprising because these variables have been shown to be determinants of ECMF composition elsewhere (Twieg et al., 2009; Cripps and Antibus, 2011; Tedersoo et al., 2012; Glassman et al., 2017b; Koizumi et al., 2018; van der Linde et al., 2018; Defrenne et al., 2019b). In particular, Glassman et al. (2017b) and Koizumi et al. (2018) are studies of subalpine pine systems (WBP-Pinus contorta and Pinus pumila communities, respectively) at similar spatial scales that have good comparative potential. Both studies reported soil variables (dominantly pH) and tree age or life stage as strong drivers of ECMF community composition.
The continuous, mixed age nature of the forests we sampled may well explain why tree age was not significant. A shared feature of Glassman et al. (2017b) and Koizumi et al. (2018) is that individual trees were spatially separated (see Glassman et al., 2017a for an application of island biogeography to their system). Spatial separation requires seedlings to rely on soil spore bank ECMF and allows succession to occur on small spatial scales. The trees we sampled were in a continuous forest matrix where root systems of neighboring trees of different age classes overlapped and a common mycorrhizal network is likely present (Simard et al., 2012); this might promote homogeneity in ECMF communities and dampen or obscure successional processes.
Limitations in methodology may also contribute to not detecting significance in soil variables and tree age. We did not use multivariate models explicitly designed to account for the influence of multiple predictor variables simultaneously (e.g., Glassman et al., 2017b) and instead relied on simple vector fitting to ordinations. In soil sampling we did not separate organic and mineral horizons and Koizumi et al. (2018) stress that the significance of soil variables in their study was driven by stratification of ECMF in these layers. In addition, twelve soil samples were removed from analyses because of an error in sample processing, which reduced statistical power, and a second processing error allowed samples to be stored wet before air drying, which could have altered their properties.
Linking Tree Health of Ectomycorrhizal Fungi Composition
Canopy kill was a significant predictor of ECMF composition in the tree community; diseased trees hosted simpler communities with compositional differences driven by increased abundances of Cenococcum and Amphinema. Shifts in ECMF composition in response to tree stress (drought, herbivory, parasitism) are well documented in pine systems and tend to move toward ascomycete and low-carbon demanding fungi (Karst et al., 2014). This generally aligns with our observations and might suggest Amphinema has low carbon requirements.
Recent evidence linking mountain pine beetle (MPB) outbreaks to ECMF (Treu et al., 2014; Karst et al., 2015) has shaped our interpretation of these results. Treu et al. (2014) showed that ECMF composition was altered and abundance reduced in MPB-affected stands. Karst et al. (2015) showed that these changes in ECMF mediated transgenerational effects that reduced seedling survival and the capacity to generate defense compounds. Helotian taxa were identified as a dominant feature of MPB affected stands (Karst et al., 2015). These taxa were abundant in our samples, which may signal ECMF are already responding to stressors and mortality of WBP in our region.
The link between tree health and ECMF is important because it recognizes that ECMF suffer from the decline of their host (Karst et al., 2014) and that the resulting changes in ECMF can influence the success of regeneration and the capacity of the host to resist stressors and pathogens (Karst et al., 2015). There is no reason to assume that this link does not apply to WBP as well and our results are a first indication that it does. These changes in ECMF are expected to disproportionately affect pine-specific Suilloid taxa and some are at risk of being lost from the landscape altogether (Cripps and Antibus, 2011; Treu et al., 2014; Osmundson, 2016). In addition, this link highlights the importance ECMF in WBP ecology. Some ecological or organismal processes that are critical to species recovery (e.g., production of insect defense compounds) may be mediated by ECMF.
Conclusion
The major contribution of this research is the identification of the ectomycorrhizal fungal associates of WBP in the interior mountains of British Columbia, Canada. This community was clearly distinct from other regions where WBP ECMF have been sampled, which is consistent with differences in forest type and the potential for endemism. The combined list of ECMF associates of WBP compiled here can be used as a basis for management and future research. Our assessment of outplanted WBP seedlings identified a case where ECMF were mostly absent from a planting site and the planted WBP seedlings on that site. We also provide preliminary evidence that ECMF composition is correlated to WBP tree health. Both of these results have implications in WBP recovery. Planting strategy and site selection needs to incorporate ECMF colonization, ideally through direct assessments of sites, but alternatively through the consideration of site factors (fire history, source trees, ECMF spore bank) and the option for inoculation. Further work should develop the link between ECMF and tree/stand health, with the potential to develop an ECMF related indicator of WBP stand health.
WBP is part of an obligate ectomycorrhizal mutualism and as we work to restore the species, attention needs to be given to the fungal component of this mutualism. This fungal component is essential to WBP ecology and recovery actions and closely tied to the status of its host.
Data Availability Statement
Raw sequencing data is available on NCBI’s Sequence Read Archive (PRJNA722685). Representative sequences for each ASV are available on GenBank – accession numbers are in Supplementary Table 5. For other data, contact the corresponding author.
Author Contributions
SG, NS, and HS carried out the fieldwork. SG and HS preformed the sample processing and bioinformatics. HS completed the data analysis and wrote the manuscript. All authors contributed to editing and the research design.
Funding
Parks Canada (Mount Revelstoke and Glacier National Parks Field Unit) provided in-kind support to fieldwork. Funding was provided by the NSERC Discovery Grant to SS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Fieldwork for this research took place on the territories of the Syilx tmixw (Okanagan). sngaytskstx tum-xula7xw (Sinixt), Ktunaxa ɁamakɁis and Secwepemcúl’ecw (Secwépemc). Lab work and analysis occurred on the territory of the xwməθkwəy̓əm (Musqueam). We thank to the people of Mount Revelstoke and Glacier National Parks Field Unit and the Below Ground Ecosystem Group (UBC). We thank to Drs. Cathy Cripps and Joseph Birch who generously gave time to developing methods and review. We thank to a list of other people/groups who supported in various ways: Alex Newall, Heather Waterous, Allen Laroque, Spencer Bronson, and the UBC Tree Ring Lab. This manuscript was prepared in part from Southam (2020) (BSc honors thesis).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffgc.2021.750701/full#supplementary-material
Abbreviations
WBP, whitebark pine; ECMF, ectomycorrhizal fungi/fungal; ASV, amplicon sequence variant.
References
Achuff, P. L., Holland, W. D., Coen, G. M., and Van Tighem, K. (eds) (1984). Ecological Land Classification of Mount Revelstoke and Glacier National Parks, British Columbia (AIP Publication No. M-84-11). Edmonton ON: Alberta Institute of Pedology (AIP) and Environment Canada.
Ammirati, J. F., Niskanen, T., Liimatainen, K., Bojantchev, D., Peintner, U., Kuhnert-Finkernagel, R., et al. (2017). Spring and early summer species of Cortinarius, subgenus Telamonia, section Colymbadini and/Flavobasilis, in the mountains of western North America. Mycologia 109, 443–458. doi: 10.1080/00275514.2017.1349468
Antibus, R. K., Hobbie, E. A., and Cripps, C. L. (2018). Sporocarp δ15N and use of inorganic and organic nitrogen in vitro differ among host-specific suilloid fungi associated with high elevation five-needle pines. Mycoscience 59, 294–302. doi: 10.1016/j.myc.2017.11.007
Birch, J. D., Lutz, J. A., Turner, B. L., and Karst, J. (2021). Divergent, age-associated fungal communities of Pinus flexilis and Pinus longaeva. For. Ecol. Manage. 494:119277. doi: 10.1016/j.foreco.2021.119277
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bray, J. R., and Curtis, J. T. (1957). An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 27, 325–349. doi: 10.2307/1942268
Bruns, T. D., Peay, K. G., Boynton, P. J., Grubisha, L. C., Hynson, N. A., Nguyen, N. H., et al. (2009). Inoculum potential of Rhizopogon spores increases with time over the first 4 yr of a 99-yr spore burial experiment. New Phytol. 181, 463–470. doi: 10.1111/j.1469-8137.2008.02652.x
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Collier, F. A., and Bidartondo, M. I. (2009). Waiting for fungi: the ectomycorrhizal invasion of lowland heathlands. J. Ecol. 97, 950–963. doi: 10.1111/j.1365-2745.2009.01544.x
Comeau, A. M., Douglas, G. M., and Langille, M. G. I. (2017). Microbiome helper: a custom and streamlined workflow for microbiome research. MSystems 2, e00127–16. doi: 10.1128/mSystems.00127-16
Cripps, C. L., Alger, G., and Sissons, R. (2018). Designer Niches Promote Seedling Survival in Forest Restoration: a 7-Year Study of Whitebark Pine (Pinus albicaulis) Seedlings in Waterton Lakes National Park. Forests 9:477. doi: 10.3390/f9080477
Cripps, C. L., and Antibus, R. K. (2011). “Native ectomycorrhizal fungi of limber and whitebark pine: necessary for forest sustainability,” in The future of high-elevation, five-needle white pines in Western North America: proceedings of the High Five Symposium (RMRS-P-63), Missoula MT, 28-30 June 2010, eds R. E. Keane, D. F. Tomback, M. P. Murray, and C. M. Smith (Fort Collins CO: U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station), 37–44.
Cripps, C. L., and Grimme, E. (2011). “Inoculation and successful colonization of whitebark pine seedlings with native mycorrhizal fungi under greenhouse conditions,” in The future of high-elevation, five-needle white pines in Western North America: proceedings of the High Five Symposium (RMRS-P-63), Missoula MT, 28-30 June 2010, eds R. E. Keane, D. F. Tomback, M. P. Murray, and C. M. Smith (Fort Collins CO: U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station), 312–322.
Cripps, C. L., Lindgren, J. E., and Barge, E. G. (2017). Amanita alpinicola sp. nov., associated with Pinus albicaulis, a western 5-needle pine. Mycotaxon 132, 665–676. doi: 10.5248/132.665
Cripps, C. L., Smith, C., Carolin, T., and Lapp, J. (2008). Ectomycorrhizal Fungi with Whitebark Pine. Nutcracker Notes 14, 12–14.
Defrenne, C. E., McCormack, M. L., Roach, W. J., Addo-Danso, S. D., and Simard, S. W. (2019a). Intraspecific fine-root trait-environment relationships across interior douglas-fir forests of western Canada. Plants 8, 10–12. doi: 10.3390/plants8070199
Defrenne, C. E., Philpott, T. J., Guichon, S. H. A., Roach, W. J., Pickles, B. J., and Simard, S. W. (2019b). Shifts in ectomycorrhizal fungal communities and exploration types relate to the environment and fine-root traits across interior douglas-fir forests of western Canada. Front. Plant Sci. 10:643. doi: 10.3389/fpls.2019.00643
Desirò, A., Rimington, W. R., Jacob, A., Pol, N. V., Smith, M. E., Trappe, J. M., et al. (2017). Multigene phylogeny of Endogonales, an early diverging lineage of fungi associated with plants. IMA Fungus 8, 245–257. doi: 10.5598/imafungus.2017.08.02.03
Dove, N. C., and Hart, S. C. (2017). Fire Reduces Fungal Species Richness and In Situ Mycorrhizal Colonization: a Meta-Analysis. Fire Ecol. 13, 37–65. doi: 10.4996/fireecology.130237746
Durall, D. M., Gamiet, S., Simard, S. W., Kudrna, L., and Sakakibara, S. M. (2006). Effects of clearcut logging and tree species composition on the diversity and community composition of epigeous fruit bodies formed by ectomycorrhizal fungi. Can. J. Bot. 84, 966–980. doi: 10.1139/B06-045
Environment and Climate Change Canada [ECCC] (2017). Recovery Strategy for the Whitebark Pine (Pinus albicaulis) in Canada [Proposed]. Species at Risk Act Recovery Strategy Series. Ottawa: Environment and Climate Change Canada.
Frøslev, T. G., Kjøller, R., Bruun, H. H., Ejrnæs, R., Hansen, A. J., Læssøe, T., et al. (2019). Man against machine: do fungal fruitbodies and eDNA give similar biodiversity assessments across broad environmental gradients? Biol. Conserv. 233, 201–212. doi: 10.1016/j.biocon.2019.02.038
Gardes, M., and Bruns, T. D. (1996). Community structure of ectomycorrhizal fungi in a Pinus muricata forest: above- and below-ground views. Can. J. Bot. 74, 1572–1583. doi: 10.1139/b96-190
Glassman, S. I., Levine, C. R., DiRocco, A. M., Battles, J. J., and Bruns, T. D. (2016). Ectomycorrhizal fungal spore bank recovery after a severe forest fire: some like it hot. ISME J. 10, 1228–1239. doi: 10.1038/ismej.2015.182
Glassman, S. I., Lubetkin, K. C., Chung, J. A., and Bruns, T. D. (2017a). The theory of island biogeography applies to ectomycorrhizal fungi in subalpine tree “islands” at a fine scale. Ecosphere 8:e01677. doi: 10.1002/ecs2.1677
Glassman, S. I., Wang, I. J., and Bruns, T. D. (2017b). Environmental filtering by pH and soil nutrients drives community assembly in fungi at fine spatial scales. Mol. Ecol. 26, 6960–6973. doi: 10.1111/mec.14414
Glassman, S. I., Peay, K. G., Talbot, J. M., Smith, D. P., Chung, J. A., Taylor, J. W., et al. (2015). A continental view of pine-associated ectomycorrhizal fungal spore banks: a quiescent functional guild with a strong biogeographic pattern. New Phytol. 205, 1619–1631. doi: 10.1111/nph.13240
Hagerman, S. M., Jones, M. D., Bradfield, G. E., Gillespie, M., and Durall, D. M. (1999). Effects of clear-cut logging on the diversity and persistence of ectomycorrhizae at a subalpine forest. Can. J. For. Res. 29, 124–134. doi: 10.1139/cjfr-29-1-124
Haskins, K. E., and Gehring, C. A. (2005). Evidence for mutualist limitation: the impacts of conspecific density on the mycorrhizal inoculum potential of woodland soils. Oecologia 145, 123–131. doi: 10.1007/s00442-005-0115-3
Heine, P., Hausen, J., Ottermanns, R., and Roß-Nickoll, M. (2021). Comparing eDNA metabarcoding with morphological analyses: fungal species richness and community composition of differently managed stages along a forest conversion of Norway spruce towards European beech in Germany. For. Ecol. Manage. 496:119429. doi: 10.1016/j.foreco.2021.119429
Heinonsalo, J., Sun, H., Santalahti, M., Bäcklund, K., Hari, P., and Pumpanen, J. (2015). Evidences on the Ability of Mycorrhizal Genus Piloderma to Use Organic Nitrogen and Deliver It to Scots Pine. PLoS One 10:e0131561. doi: 10.1371/journal.pone.0131561
Hendershot, W. H., and Lalande, H. (2007). “Soil Reaction and Exchangeable Acidity,” in Soil Sampling and Methods of Analysis, eds M. R. Carter and E. G. Gregorich (Boca Raton, FL: CRC Press), 173–178.
Hubert, N. A., and Gehring, C. A. (2008). Neighboring trees affect ectomycorrhizal fungal community composition in a woodland-forest ecotone. Mycorrhiza 18, 363–374. doi: 10.1007/s00572-008-0185-2
Jenkins, M. L. (2017). Restoration of whitebark pine on a burn site utilizing native ectomycorrhizal suilloid fungi. Ph.D. thesis. Bozeman (MT): Montana State University.
Jenkins, M. L., Cripps, C. L., and Gains-Germain, L. (2018). Scorched Earth: suillus colonization of Pinus albicaulis seedlings planted in wildfire-impacted soil affects seedling biomass, foliar nutrient content, and isotope signatures. Plant Soil 425, 113–131.
Johnson, K., Kendall, K. C., and Coen, B. (1994). “Whitebark pine mycorrhizae: results of preliminary efforts to isolate fungi from the roots of field-collected seedlings,” in Proceedings of workshop on research and management in whitebark pine ecosystems, Glacier National Park, MT, 3 May 1994, eds K. C. Kendall and B. Coen (Fort Collins, CO: USDI, National Biological Survey, Glacier Field Station), 140.
Karst, J., Erbilgin, N., Pec, G. J., Cigan, P. W., Najar, A., Simard, S. W., et al. (2015). Ectomycorrhizal fungi mediate indirect effects of a bark beetle outbreak on secondary chemistry and establishment of pine seedlings. New Phytol. 208, 904–914. doi: 10.1111/nph.13492
Karst, J., Randall, M. J., and Gehring, C. A. (2014). Consequences for ectomycorrhizal fungi of the selective loss or gain of pine across landscapes. Botany 92, 855–865. doi: 10.1139/cjb-2014-0063
Kassambara, A. (2020). rstatix: pipe-Friendly Framework for Basic Statistical Tests. R package version 0.6.0. Available Online at: https://CRAN.R-project.org/package=rstatix. (accessed December 29, 2021)
Keane, R. E., Tomback, D. F., Aubry, C. A., Bower, A. D., Campbell, E. M., Cripps, C. L., et al. (2012). A range-wide restoration strategy for whitebark pine (Pinus albicaulis) (RMRS-GTR-279). Fort Collins, CO: U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station, doi: 10.2737/RMRS-GTR-279
Koizumi, T., Hattori, M., and Nara, K. (2018). Ectomycorrhizal fungal communities in alpine relict forests of Pinus pumila on Mt. Norikura, Japan. Mycorrhiza 28, 129–145. doi: 10.1007/s00572-017-0817-5
Lalli, G., Leonardi, M., Oddis, M., Pacioni, G., Salerni, E., Iotti, M., et al. (2015). Expanding the understanding of a forest ectomycorrhizal community by combining root tips and fruiting bodies: a case study of Tuber magnatum stands. Turk. J. Bot. 39, 527–534. doi: 10.3906/bot-1406-50
Lonergan, E. R. (2012). The Use Of Native Ectomycorrhizal Fungi In The Restoration Of Whitebark Pine. Ph.D. thesis. Bozeman (MT): Montana State University
Lonergan, E. R., Cripps, C. L., and Smith, C. M. (2013). Influence of site conditions, shelter objects, and ectomycorrhizal inoculation on the early survival of whitebark pine seedlings planted in Waterton Lakes National Park. For. Sci. 60, 603–612. doi: 10.5849/forsci.13-511
Lücking, R., Aime, M. C., Robbertse, B., Miller, A. N., Ariyawansa, H. A., Aoki, T., et al. (2020). Unambiguous identification of fungi: where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus 11:14. doi: 10.1186/s43008-020-00033-z
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Mohatt, K. R. (2006). Ectomycorrhizal fungi of whitebark pine (Pinus albicaulis) in the Northern Greater Yellowstone Ecosystem. Ph.D. thesis. Bozeman (MT): Montana State University.
Mohatt, K. R., Cripps, C. L., and Lavin, M. (2008). Ectomycorrhizal fungi of whitebark pine (a tree in peril) revealed by sporocarps and molecular analysis of mycorrhizae from treeline forests in the Greater Yellowstone Ecosystem. Botany 86, 14–25. doi: 10.1139/B07-107
Molina, R., and Trappe, J. M. (1994). Biology of the ectomycorrhizal genus, Rhizopogon: I. Host associations, host-specificity and pure culture syntheses. New Phytol. 126, 653–675. doi: 10.1111/j.1469-8137.1994.tb02961.x
Mueller, R. C., Scudder, C. M., Whitham, T. G., and Gehring, C. A. (2019). Legacy effects of tree mortality mediated by ectomycorrhizal fungal communities. New Phytol. 224, 155–165. doi: 10.1111/nph.15993
Nguyen, N. H., Hynson, N. A., and Bruns, T. D. (2012). Stayin’ alive: survival of mycorrhizal fungal propagules from 6-yr-old forest soil. Fungal Ecol. 5, 741–746. doi: 10.1016/j.funeco.2012.05.006
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016a). FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Nguyen, N. H., Vellinga, E. C., Bruns, T. D., and Kennedy, P. G. (2016b). Phylogenetic assessment of global Suillus ITS sequences supports morphologically defined species and reveals synonymous and undescribed taxa. Mycologia 108, 1216–1228. doi: 10.3852/16-106
Nilsson, R. H., Larsson, K.-H., Taylor, A. F. S., Bengtsson-Palme, J., Jeppesen, T. S., Schigel, D., et al. (2019). The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47, D259–D264. doi: 10.1093/nar/gky1022
Oksanen, J., Blanchet, G. F., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2019). vegan: community Ecology Package. R package version 2.5-6. Available Online at: https://cran.r-project.org/package=vegan. (accessed December 29, 2021)
Osmundson, T. W. (2016). Suillus subalpinus. The Global Fungal Red List Initiative. Available Online at: http://iucn.ekoo.se/iucn/species_view/442468/ (accessed June 4, 2021)
Ovaskainen, O., Schigel, D., Ali-Kovero, H., Auvinen, P., Paulin, L., Nordén, B., et al. (2013). Combining high-throughput sequencing with fruit body surveys reveals contrasting life-history strategies in fungi. ISME J. 7, 1696–1709. doi: 10.1038/ismej.2013.61
Policelli, N., Bruns, T. D., Vilgalys, R., and Nuñez, M. A. (2019). Suilloid fungi as global drivers of pine invasions. New Phytol. 222, 714–725. doi: 10.1111/nph.15660
Potvin, L. R., Richter, D. L., Jurgensen, M. F., and Dumroese, R. K. (2012). Association of Pinus banksiana Lamb. and Populus tremuloides Michx. seedling fine roots with Sistotrema brinkmannii (Bres.) J. Erikss. (Basidiomycotina). Mycorrhiza 22, 631–638. doi: 10.1007/s00572-012-0440-4
QGIS.org (2021). QGIS Geographic Information System. QGIS Association. Available Online at: http://www.qgis.org (accessed 29, December 2021)
R Core Team (2019). R: a Language And Environment For Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing.
Rivers, A. R., Weber, K. C., Gardner, T. G., Liu, S., and Armstrong, S. D. (2018). ITSxpress: software to rapidly trim internally transcribed spacer sequences with quality scores for marker gene analysis. F1000Research 7:1418. doi: 10.12688/f1000research.15704.1
Robert, V., Vu, D., Amor, A. B. H., van de Wiele, N., Brouwer, C., Jabas, B., et al. (2013). MycoBank gearing up for new horizons. IMA Fungus 4, 371–379. doi: 10.5598/imafungus.2013.04.02.16
Rosinger, C., Sandén, H., Matthews, B., Mayer, M., and Godbold, D. L. (2018). Patterns in ectomycorrhizal diversity, community composition, and exploration types in European beech, pine, and spruce forests. Forests 9:445. doi: 10.3390/f9080445
Saine, S., Ovaskainen, O., Somervuo, P., and Abrego, N. (2020). Data collected by fruit body- and DNA-based survey methods yield consistent species-to-species association networks in wood-inhabiting fungal communities. Oikos 129, 1833–1843. doi: 10.1111/oik.07502
Seitzman, B. H., Ouimette, A., Mixon, R. L., Hobbie, E. A., and Hibbett, D. S. (2011). Conservation of biotrophy in Hygrophoraceae inferred from combined stable isotope and phylogenetic analyses. Mycologia 103, 280–290. doi: 10.3852/10-195
Shemesh, H., Boaz, B. E., Millar, C. I., and Bruns, T. D. (2019). Symbiotic interactions above treeline of long-lived pines: mycorrhizal advantage of limber pine (Pinus flexilis) over Great Basin bristlecone pine (Pinus longaeva) at the seedling stage. J. Ecol. 108, 908–916. doi: 10.1111/1365-2745.13312
Shepherd, B., Jones, B., Sissons, R., Cochrane, J., Park, J., Smith, C. M., et al. (2018). Ten Years of Monitoring Illustrates a Cascade of Effects of White Pine Blister Rust and Focuses Whitebark Pine Restoration in the Canadian Rocky and Columbia Mountains. Forests 9:138. doi: 10.3390/f9030138
Simard, S. W., Beiler, K. J., Bingham, M. A., Deslippe, J. R., Philip, L. J., and Teste, F. P. (2012). Mycorrhizal networks: mechanisms, ecology and modelling. Fungal Biol. Rev. 26, 39–60. doi: 10.1016/j.fbr.2012.01.001
Simard, S. W., Roach, W. J., Beauregard, J., Burkart, J., Cook, D., Law, D., et al. (2021). Partial Retention of Legacy Trees Protect Mycorrhizal Inoculum Potential, Biodiversity, and Soil Resources While Promoting Natural Regeneration of Interior Douglas-Fir. Front. For. Glob. Chang. 3:620436. doi: 10.3389/ffgc.2020.620436
Skinner, M., Greenberg, J., Shulhof, J., Southam, H., Newall, A., and Stafl, N. (2019). Vegetation Projects 2019 Mount Revelstoke and Glacier National Parks. Revelstoke, BC: Parks Canada: Mount Revelstoke and Glacier National Parks Field Unit.
Smith, C. M., Shepherd, B., Gillies, C., and Stuart-Smith, J. (2013). Changes in blister rust infection and mortality in whitebark pine over time. Can. J. For. Res. 43, 90–96. doi: 10.1139/cjfr-2012-0127
Smith, C. M., Wilson, B., Rasheed, S., Walker, R. C., Carolin, T., and Shepherd, B. (2008). Whitebark pine and white pine blister rust in the Rocky Mountains of Canada and northern Montana. Can. J. For. Res. 38, 982–995. doi: 10.1139/X07-182
Smith, S. E., and Read, D. J. (2008). Mycorrhizal Symbiosis. San Diego: Elsevier Science & Technology.
Somervuo, P., Koskela, S., Pennanen, J., Henrik Nilsson, R., and Ovaskainen, O. (2016). Unbiased probabilistic taxonomic classification for DNA barcoding. Bioinformatics 32, 2920–2927. doi: 10.1093/bioinformatics/btw346
Somervuo, P., Yu, D. W., Xu, C. C. Y., Ji, Y., Hultman, J., Wirta, H., et al. (2017). Quantifying uncertainty of taxonomic placement in DNA barcoding and metabarcoding. Methods Ecol. Evol. 8, 398–407. doi: 10.1111/2041-210X.12721
Southam, H. (2020). Characterizing the Ectomycorrhizal Fungal Community of Whitebark Pine (Pinus albicaulis) in Mount Revelstoke and Glacier National Parks. [undergraduate research]. Vancouver (BC): University of British Columbia, doi: 10.14288/1.0398124
Talbot, J. M., Bruns, T. D., Taylor, J. W., Smith, D. P., Branco, S., Glassman, S. I., et al. (2014). Endemism and functional convergence across the North American soil mycobiome. Proc. Natl. Acad. Sci. U. S. A. 111, 6341–6346. doi: 10.1073/pnas.1402584111
Taudière, A., Richard, F., and Carcaillet, C. (2017). Review on fire effects on ectomycorrhizal symbiosis, an unachieved work for a scalding topic. For. Ecol. Manage. 391, 446–457. doi: 10.1016/j.foreco.2017.02.043
Tedersoo, L., Bahram, M., Toots, M., Diédhiou, A. G., Henkel, T. W., Kjoller, R., et al. (2012). Towards global patterns in the diversity and community structure of ectomycorrhizal fungi. Mol. Ecol. 21, 4160–4170. doi: 10.1111/j.1365-294X.2012.05602.x
Tedersoo, L., May, T. W., and Smith, M. E. (2010). Ectomycorrhizal lifestyle in fungi: global diversity, distribution, and evolution of phylogenetic lineages. Mycorrhiza 20, 217–263. doi: 10.1007/s00572-009-0274-x
Tedersoo, L., Pärtel, K., Jairus, T., Gates, G., Põldmaa, K., and Tamm, H. (2009). Ascomycetes associated with ectomycorrhizas: molecular diversity and ecology with particular reference to the Helotiales. Environ. Microbiol. 11, 3166–3178. doi: 10.1111/j.1462-2920.2009.02020.x
Teste, F. P., Simard, S. W., and Durall, D. M. (2009). Role of mycorrhizal networks and tree proximity in ectomycorrhizal colonization of planted seedlings. Fungal Ecol. 2, 21–30. doi: 10.1016/j.funeco.2008.11.003
Tomback, D. F., Keane, R. E., McCaughey, W. W., and Smith, C. (2005). Methods For Surveying And Monitoring Whitebark Pine For Blister Rust Infection And Damage. Missoula, MT: Whitebark Pine Ecosystem Foundation.
Treu, R., Karst, J., Randall, M., Pec, G. J., Cigan, P. W., Simard, S. W., et al. (2014). Decline of ectomycorrhizal fungi following a mountain pine beetle epidemic. Ecology 95, 1096–1103. doi: 10.1890/13-1233.1
Trusty, P. E. (2009). Impact Of Severe Fire On Ectomycorrhizal Fungi Of Whitebark Pine Seedlings. Ph.D. thesis. Bozeman (MT): Montana State University
Trusty, P. E., and Cripps, C. L. (2011). “fluence of Fire on Mycorrhizal Colonization of Planted and Natural Whitebark Pine Seedlings: ecology and Management Implications,” in The future of high-elevation, five-needle white pines in Western North America: proceedings of the High Five Symposium (RMRS-P-63), Missoula MT, 28-30 June 2010, eds R. E. Keane, D. F. Tomback, M. P. Murray, and C. M. Smith (Fort Collins CO: U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station), 198–202.
Turenne, C. Y., Sanche, S. E., Hoban, D. J., Karlowsky, J. A., and Kabani, A. M. (1999). Rapid Identification of Fungi by Using the ITS2 Genetic Region and an Automated Fluorescent Capillary Electrophoresis System. J. Clin. Microbiol. 37, 1846–1851. doi: 10.1128/JCM.37.6.1846-1851.1999
Twieg, B. D., Durall, D. M., Simard, S. W., and Jones, M. D. (2009). Influence of soil nutrients on ectomycorrhizal communities in a chronosequence of mixed temperate forests. Mycorrhiza 19, 305–316.
van der Heijden, M. G. A., Martin, F. M., Selosse, M., and Sanders, I. R. (2015). Mycorrhizal ecology and evolution: the past, the present, and the future. New Phytol. 205, 1406–1423. doi: 10.1111/nph.13288
van der Linde, S., Suz, L. M., Orme, C. D. L., Cox, F., Andreae, H., Asi, E., et al. (2018). Environment and host as large-scale controls of ectomycorrhizal fungi. Nature 558, 243–248. doi: 10.1038/s41586-018-0189-9
Visnovsky, S. B., Cummings, N., Guerin-Laguette, A., Wang, Y., Yamada, A., Kobayashi, H., et al. (2014). Detection of the edible ectomycorrhizal fungus Lyophyllum shimeji colonising seedlings of cultivated conifer species in New Zealand. Mycorrhiza 24, 453–463. doi: 10.1007/s00572-013-0552-5
Walker, J. K. M., Phillips, L. A., and Jones, M. D. (2014). Ectomycorrhizal fungal hyphae communities vary more along a pH and nitrogen gradient than between decayed wood and mineral soil microsites. Botany 92, 453–463. doi: 10.1139/cjb-2013-0239
Wang, T., Hamann, A., Spittlehouse, D., and Carroll, C. (2016). Locally downscaled and spatially customizable climate data for historical and future periods for North America. PLoS One 11:e0156720. doi: 10.1371/journal.pone.0156720
White, T. J., Bruns, T., Lee, S., and Taylor, J. W. (1990). ““Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: a Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (New York: Academic Press, Inc), 315–322.
Keywords: whitebark pine, Pinus albicaulis, ectomycorrhizal fungi, Suilloid fungi, Parks Canada, next-generation sequencing, endangered tree species, plant-fungal interactions
Citation: Southam H, Stafl N, Guichon SHA and Simard SW (2022) Characterizing the Ectomycorrhizal Fungal Community of Whitebark Pine in Interior British Columbia: Mature Trees, Natural Regeneration and Planted Seedlings. Front. For. Glob. Change 4:750701. doi: 10.3389/ffgc.2021.750701
Received: 31 July 2021; Accepted: 20 December 2021;
Published: 03 February 2022.
Edited by:
Verena C. Griess, ETH Zürich, SwitzerlandReviewed by:
Joanna Dames, Rhodes University, South AfricaRui S. Oliveira, University of Coimbra, Portugal
Copyright © 2022 Southam, Stafl, Guichon and Simard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hanno Southam, aGFubm9zb3V0aGFtQGdtYWlsLmNvbQ==
 Hanno Southam
Hanno Southam Natalie Stafl
Natalie Stafl Shannon H. A. Guichon
Shannon H. A. Guichon Suzanne W. Simard
Suzanne W. Simard