- 1Renewable Resources Department, University of Alberta, Edmonton, AB, Canada
- 2Univ. Lille, IMT Lille Douai, Univ. Artois, Yncrea Hauts-de-France, ULR 4515 – LGCgE, Laboratoire de Génie Civil et Géo-Environnement, Lille, France
Boreal forest soils are highly susceptible to global warming, and in the next few decades, are expected to face large increases in temperature and transformative vegetation shifts. The entire boreal biome will migrate northward, and within the main boreal forest of Western Canada, deciduous trees will replace conifers. The main objective of our research was to assess how these vegetation shifts will affect functioning of soil microbial communities and ultimately the overall persistence of boreal soil carbon. In this study, aspen and spruce forest floors from the boreal mixedwood forest of Alberta were incubated in the laboratory for 67 days without (control) and with the addition of three distinct 13C labeled substrates (glucose, aspen leaves, and aspen roots). Our first objective was to compare aspen and spruce substrate utilization efficiency (SUE) in the case of a labile C source (13C-glucose). For our second objective, addition of aspen litter to spruce forest floor mimicked future vegetation shifts, and we tested how this would alter substrate use efficiency in the spruce forest floor compared to the aspen. Tracking of carbon utilization by microbial communities was accomplished using 13C-PLFA analysis, and 13C-CO2 measurements allowed quantification of the relative contribution of each added substrate to microbial respiration. Following glucose addition, the aspen community showed a greater 13C-PLFA enrichment than the spruce throughout the 67-day incubation. The spruce community respired a greater amount of 13C glucose, and it also had a much lower glucose utilization efficiency compared to the aspen. Following addition of aspen litter, in particular aspen leaves, the aspen community originally showed greater total 13C-PLFA enrichment, although gram positive phospholipid fatty acids (PLFAs) were significantly more enriched in the spruce community. While the spruce community respired a greater amount of the added 13C-leaves, both forest floor types showed comparable substrate utilization efficiencies by Day 67. These results indicate that a shift from spruce to aspen may lead to a greater loss of the aspen litter through microbial respiration, but that incorporation into microbial biomass and eventually into the more persistent soil carbon pool may not be affected.
Introduction
The boreal forest is the largest terrestrial ecosystem and the largest terrestrial sink of carbon on Earth (Watson et al., 2000). Indeed, boreal forests account for over half of the forest carbon stocks globally (DeLuca and Boisvenue, 2012). The boreal forest forms a “green belt” between latitude 45° and 70° N, and in Canada, it stretches over 307 million ha from the Yukon to Newfoundland and Labrador. Due to the extreme climatic conditions occurring in this region, the boreal forest is characterized by a low diversity of tree species, including Abies, Larix, Picea, Pinus, and Populus (Brandt et al., 2013). In the boreal mixedwood forests of western Canada, Picea glauca (white spruce) and Populus tremuloides (trembling aspen) are the most common species (Lieffers et al., 1996). The boreal is particularly sensitive and vulnerable to global warming, and in the next few decades, is expected to face transformative changes; it is likely that the region will see a 2°C temperature increase by 2050 compared to 2000 (Price et al., 2013). Ongoing increases in atmospheric CO2 and warming have already been linked to vegetation shifts where early-successional deciduous broadleaves have been replacing late-successional conifers since the 1950s (Searle and Chen, 2017). Predictions for precipitation are not as straightforward, although frequency and intensity of extreme events, including drought, are expected to increase. Boreal tree species exhibit a wide range of responses to the changing climate, and broadleaved trees are less sensitive to moisture deficit than conifers (Hogg et al., 2017; D’Orangeville et al., 2018). In the mixedwood boreal forest of Western Canada, this will mean an accelerated shift from white spruce to trembling aspen stands.
The composition and activity of forest floor microbial communities are the result of complex interactions between biotic and abiotic factors including climate, tree species, quality and quantity of organic matter input, soil fauna, nutrient availability, and land disturbance (e.g., Myers et al., 2001; Grayston and Prescott, 2005; Quideau et al., 2013; Cleveland et al., 2014). Differences in microbial communities associated with individual plant species are hypothesized to result from variations in the quantity and quality of plant carbon inputs (Grayston and Prescott, 2005), and coniferous forest floors (i.e., surficial soil organic horizons containing litter at various stages of decomposition) typically have lower microbial biomass and activity than those under deciduous cover (Bauhus et al., 1998). Similarly, in the mixedwood boreal forest of Western Canada, forest floors from white spruce and aspen stands have distinct microbial communities and distinct properties, including greater microbial biomass and higher pH and nutrient availability in aspen forest floors (Hannam et al., 2006; Swallow and Quideau, 2013). In a systematic review comparing aspen and coniferous stands across North America, Laganière et al. (2017) concluded that forest floor carbon stocks were larger in conifer dominated stands in the majority of studies. In the mixedwood of central Alberta, Kishchuk et al. (2014) reported average soil carbon stocks of 25 and 42 Mg ha–1, for aspen and spruce forest floors, respectively. On the other hand, soil carbon under aspen appears to be more stable than under spruce (Laganière et al., 2017), and this has recently been linked to preferential retention of aspen foliage leachates compared to spruce needle leachates (Boča et al., 2020). What remains unclear, however, is how current and future vegetation shifts may be affecting carbon storage and persistence in boreal soils.
The home-field advantage (HFA) concept, which stipulates that litter decomposes faster in an area dominated by plants from which it is derived (i.e., at home) rather than under an alternative plant cover, has been proposed as a widespread phenomenon occurring in most forest ecosystems (Ayres et al., 2009). According to the HFA theory, a rapid shift in vegetation could mean a slow decomposition of the new aspen litter by the microbial community already present in the native spruce forest floor. On the other hand, when labile organic substrates are added to soil, microbial activity is commonly stimulated and a short-term change in the turnover of native organic matter [termed “priming effect” (PE)] may occur in which both added substrate and indigenous soil organic matter (SOM) are degraded (Waldrop and Firestone, 2004; Kuzyakov, 2010). A positive PE following vegetation shifts could potentially increase decomposition as fresh, more labile aspen litter is added to native spruce forest floor. Furthermore, an increasingly accepted paradigm highlights the importance of microbial anabolism, and substrate utilization efficiency (SUE), in retaining plant-derived carbon and eventually increasing the persistence of carbon in soils (Cotrufo et al., 2013; Waring et al., 2020; Neupane et al., 2021). Several interacting factors, including the composition of soil microbial communities and substrate stoichiometry, can alter SUE (Rinnan and Baath, 2009; Keiblinger et al., 2010; Kohl et al., 2021). Global carbon stocks in boreal forests are estimated to reach 1095 Pg of carbon (Bradshaw and Warkentin, 2015). As about a quarter of these stocks are stored in boreal forest floors themselves (Kurz et al., 2013), it is important to assess how aspen replacing spruce may alter the carbon balance of boreal forest floors.
Stable isotope probing (SIP) of phospholipid fatty acids (PLFAs) found in microbial membranes allows tracking of 13C-labeled substrates as they become incorporated into soil microbial communities (Waldrop and Firestone, 2004; Watzinger, 2015; Feland and Quideau, 2020). In this study, aspen and spruce forest floors from the mixedwood boreal forest of Alberta were incubated following addition of 13C-enriched glucose, aspen leaves and aspen roots that had been previously 13C-labeled, and results were compared to control forest floors (no substrate added). Our first objective was to assess if differences between aspen and spruce forest floors would foster differences in soil microbial metabolism, as indicated by estimating SUE following addition of 13C-glucose. Secondly, addition of 13C-labeled aspen litter to spruce forest floor mimicked the effects of anticipated future vegetation shifts in the boreal mixedwood forest of Western Canada. Hence, the second objective of our study was to test whether the addition of fresh aspen litter to spruce forest floor would alter microbial utilization processes, and in turn, to ascertain implications in terms of overall carbon persistence in these soils. Respired 13C-CO2 was measured to quantify the percent of CO2 coming from each added substrate, and carbon assimilation by different members of the forest floor microbial community was tracked using PLFA-SIP.
Materials and Methods
Soil Collection and Aspen Seedling Labeling
Soil samples (0–10 cm) were collected from two mature aspen and white spruce stands (>70-year-old) located in the boreal forest of Northern Alberta, Canada (Table 1). The region experiences a mean annual temperature of 0.7°C and a mean annual precipitation of 45 cm. These two stands were selected within 2 km of each other to minimize climatic variation and were established on similar fine-textured Luvisols (IUSS Working Group WRB, 2015). Both sites were selected from previous studies investigating plant and soil microbial communities in the area (Norris et al., 2013, 2016; Quideau et al., 2013). The two soil types were collected in preparation of the laboratory incubation by sampling ten locations within each stand and then composited to obtain homogenous samples. Forest floors averaged 19.5 cm for the spruce and 7 cm for the aspen stand (Table 1); while samples were contained within the forest floor layers (i.e., soil organic layers with >17% C) for the spruce stand, the aspen samples also included some underlying mineral soil. Soil samples were kept cool and transported to the laboratory at which time they were sieved to 4 mm (Kalra and Maynard, 1991), and stored in the dark until the start of the incubation experiment.

Table 1. Key properties of the aspen and spruce forest floor materials used during the 67-day laboratory incubation.
Enriched aspen leaves and roots were obtained by growing seedlings in a greenhouse as described by Norris et al. (2012). After five weeks of growth, seedlings were labeled once a week for three consecutive weeks with 99.9% 13C-CO2 (Cambridge Isotope Laboratories, Inc. Andover, MA, United States). The enriched 13CO2 gas was applied to each seedling for 30 min each week, and seedlings were harvested one week after the last pulse. Leaves and roots were separated, dried, ground and stored until the start of the incubation experiment. The abundance of 13C in the labeled plants was measured to average 228 ± 18‰ for the aspen leaves and 128 ± 35‰ for the aspen roots.
Laboratory Incubation
Prior to the start of the laboratory incubation, deionized water was added to soils to reach a moisture content equivalent to field capacity (−12 kPa), as estimated by the pressure plate method (Swallow and Quideau, 2013). Resulting moisture contents were 98 ± 3%, and 210 ± 13% for the aspen and spruce forest floors, respectively (Table 1). Sixty experimental units were constructed for each forest floor type (aspen and white spruce) and were left to equilibrate for two weeks before the labeled substrates were added. Moisture content was maintained during the preconditioning period as well as during the incubation itself by regular weighing and application of any lost moisture. Each experiment unit consisted of 2 g of equivalent dried soil in a small tube resting at the bottom of a 1 L airtight glass jar. Lids were fitted with Luer Lock valves to allow for gas sampling.
The incubation experiment began with the addition of the labelled substrates, which were either dissolved (glucose), or suspended as fine powder (leaves and roots) in deionized water. After addition to the soils, each tube was shaken to promote homogenous incorporation of the substrate. For each soil type, 15 experimental units were used as control, where no substrate was added. For the other 45 experiment units, 15 were used for incubations with a 13C labeled synthetic substrate (Sigma Aldrich, 99% D-glucose-1-13C), 15 for incubations with 13C-labeled aspen leaves, and 15 with 13C-labeled aspen roots. The quantity of substrate to add was calculated to correspond to the same amount of carbon for each substrate, which was chosen as 0.4 mg C g–1 of dry soil to optimize respiration fluxes as shown by Larionova et al. (2007). Soil samples were incubated in the dark at room temperature (20 °C) for 67 days. For the entire duration of the experiment, every 2–3 days, the jars were opened and ventilated for 30 min to maintain aerobic conditions. Before opening the jars, gas samples were taken to estimate accumulated CO2 from five randomly chosen units for the two soil types and each incubation type (control, glucose, aspen leaves, and aspen roots). In addition, the same flasks were sampled nine times for isotopic composition analysis of CO2 (δ13CO2), namely after 0.2, 2, 7, 14, 18, 25, 46, 60, and 67 days of incubation. Lastly, five experimental units for each incubation type and each soil were destructively sampled after 0.2, 14, and 67 days of incubation. These were immediately frozen at −80°C to be later freeze-dried prior to PLFA analysis.
Soil Respiration Measurements
For quantitative CO2 and δ13CO2 measurements, two aliquots were taken from the experimental unit headspaces. One aliquot was transferred to an evacuated borosilicate Labco Exetainer (Labco Limited, High Wycombe, United Kingdom), and accumulated (respired) CO2 was analyzed on a HP5890 Series II gas chromatograph and a HP3396 Series II Integrator (Hewlett-Packard, Santa Clara, CA, United States), using helium as the carrier gas. The amount of respired CO2 in each incubation unit headspace was corrected for the background CO2 concentration present in the room, and converted from ppmv to gC using the ideal gas law.
The second aliquot was transferred to a helium flushed positive pressure Labco Exetainer, and δ13CO2 of respired CO2 was measured using a CTC Combi PAL autosampler (CTC Analytics AG, Zwingen, CH) leading to a Porapak Q column on a Finnigan Gas Bench II (Thermo Electron Corporation, Waltham, MA, United States) attached to an Isotope Ratio Mass Spectrometer (IR-MS). The δ13C values of CO2 were reported against the Vienna Pee Dee Belemnite standard (isotopic ratio: RVPDB = 0.0111802) and were corrected for atmospheric contribution on each sampling date according to:
where the subscript “r,” “I,” and “a” refers to the CO2 “respired,” the total CO2 measured in the “incubation unit” and the contribution from the “atmosphere,” respectively. Atmospheric CO2 concentrations and δ13C values were measured in replicate prior to sampling of the experimental units; over the 67-day incubation period, concentration averaged 505 ± 106 ppmv, and δ13CO2 was −9.9 ± 1.2‰.
Phospholipid Fatty Acid Analysis
Phospholipid Fatty Acid analysis was conducted on 0.9 g of freeze-dried soil using a Bligh and Dyer extractant containing a 0.15M citrate buffer at the methanol:chloroform:buffer 1:1:0.9 ratio according to Quideau et al. (2016). Prior to extraction, PC(19:0/19:0) nonadecanoate surrogate standard (Avanti Polar Lipids Inc., Alabaster, AL, United States) was added to each sample to assess recovery. Extracted lipids were separated on solid phase extraction (SPE) silica columns (Agilent Technologies, Santa Clara, CA, United States), and re-dissolved in 1:1 chloroform:methanol prior to mild alkaline methanolysis and subsequent extraction with hexane to synthesize fatty acid methyl esters (FAMEs), as specified in White et al. (1997). An instrument standard of 10:0Me (methyl decanoate, Aldrich, St. Louis, MO, United States) was added prior to identification and quantification on a HP5890A Series II GC equipped with a HP7673 Injector, and an FID detector (Hewlett-Packard, Santa Clara, CA, United States). Peaks were identified using the Sherlock Microbial Identification System Version 4.5 software (MIDI, Inc., Newark, NJ, United States). PLFAs were described using the standard X:YωZ nomenclature, and their abundance in each sample was expressed as nmol PLFA g–1 of soil.
The δ13C values (‰) of individual PLFAs were measured using a 6890N Agilent GC (Agilent Technologies, Santa Clara, CA, United States) equipped with an HP-Ultra 2 column (50 m length, 0.2 mm i.d. and 0.33 μm film by J&W Scientific Columns from Agilent Technologies), and linked to a Thermo Finnigan GC Combustion III (Thermo Finnigan, Bremen, Germany) and the IRMS. Retention times for individual PLFAs were calibrated using FAME mixes GLC-30 and C4-C24 Unsaturated (Sigma Aldrich). In addition, retention times were compared to those from the GC-FID to ensure their correct assignment. Peaks with low intensity (<200 mV) were discarded due to low signal to noise ratios. Measured values were expressed in the δ-notation (‰) and corrected for the additional methanol group added during the last step of the PLFA extraction. Values were calibrated against three 20:0 isotope standards (USGS70, USGS71, and USGS72).
Microbial biomarkers were equated to PLFAs containing between 14 and 20 carbons, and included: saturated straight chained PLFAs (e.g., 15:0); saturated PLFAs with branching on the second (e.g., i15:0) or third (e.g., a15:0) carbon from the aliphatic end; saturated PLFAs with mid-chain (10-methyl) branching (e.g., 16:010Me); monounsaturated PLFAs (e.g., 16:1ω5); polyunsaturated PLFAs (18:3ω6); and cycloPLFAs (e.g., 17:0cyclo). Biomarkers were assigned to microbial groups according to the literature (e.g., Zelles, 1999; Myers et al., 2001; Frostegård et al., 2010; Quideau et al., 2016), where 10Me PLFAs are typically associated with Actinobacteria, branched saturated PLFAs to gram positive bacteria, monounsaturated PLFAs to gram negative bacteria, and polyunsaturated PLFAs to fungi. The number of selected biomarkers was 35 when using results from the MIDI software (mol%), and δ13C values (‰) were obtained for 22 of these.
Data Analysis
Data collected from the daily CO2 measurements from each individual flask were summed to estimate cumulative CO2 fluxes over the entire incubation period, and average values and standard deviations were calculated for the two forest floor types with and without substrate addition (n = 5). Statistical differences among respiration rates from different substrates and forest floor types (mgC-CO2/gCsoil) were tested for each sampling date using a one-way ANOVA followed by a Tukey’s HSD post-hoc test. A Bonferroni adjustment was used for multiple comparisons to maintain the family-wise Type I error rate at 0.05.
The percentage of respired CO2-C coming from the added substrate (%Csub) was calculated with a mixing model as in Waldrop and Firestone (2004):
where δC is the δ13C value of the respired CO2 from the control (no added substrate) soils, δT is the δ13C respired CO2 in the treated soils, and δS is the δ13C of the labeled substrate. The cumulative substrate-derived CO2 was also calculated in mgC-CO2/g soil by multiplying the amount of respired CO2 by the percentage of respired CO2-C coming from added substrate (%Csub) for each date. A Student’s t-test was conducted to detect statistical differences between aspen and spruce for each added substrate.
The difference in SOM mineralization following substrate addition, named “PE,” was calculated by comparing the amount of unlabelled CO2 production with and without substrate addition according to the equation from Blagodatsky et al. (2010):
where CO2 control is evolved CO2 from the control soil, and is the respired CO2 coming from SOM mineralisation in the amended soil according to:
where %Csub is the percentage of respired CO2-C coming from the added substrate, and [CO2]r is the total respired CO2. Data were further examined by calculating arithmetic means and standard deviations.
Microbial community structure was examined by non-metric multidimensional scaling (NMDS) ordination on the obtained PLFA values (mol %, Hellinger transformed) using the PC-ORD software version 5 (MjM Software Design, Gleneden Beach, OR, United States). Statistical differences between whole PLFA profiles were assessed using a Sorenson (Bray-Curtis) distance measurement followed by multi-response permutation procedure (MRPP; Mielke and Berry, 2001) in PC-ORD. Outputs from the MRPP test include overall significance (p); degree of separation between groups (T), where a greater value indicates a greater separation; and an indicator of within-group homogeneity (A), where homogeneity increases as values become closer to 1. Lastly, the relation between specific PLFAs and variation in the ordination space was displayed using correlation vectors when r2 > 0.5.
For individual PLFAs, the difference in 13C (εT–C in ‰) between the substrate-amended samples and the control samples was calculated using the following equation:
where δ13CPLFA(treated) is the isotopic composition of PLFA in the amended soil; and δ13CPLFA(control) is the isotopic composition of PLFA in the control (no substrate) soils.
The percent of labeled substrate-C (%) incorporated into each PLFA was estimated according to the following equation (Ziegler and Billings, 2011):
where δ13CPLFA(treated) is the isotopic composition of PLFA in the amended soil; δ13CPLFA(control) is the isotopic composition of PLFA in the control (no substrate) soil; and δ13Csubstrate is the isotopic composition of the labeled substrate. Data were also expressed in μgC/g soil to calculate the PLFA-based SUE as follows (%):
Average SUE values were computed both for total PLFA biomass, and for given groups of bacterial PLFAs (10Me, branched saturated, monounsaturated).
Results
Respiration Fluxes
Daily respiration rates decreased over time during the 67 days of the laboratory incubation (Figure 1 and Supplementary Tables 1, 2). The decrease occurred most rapidly during the first 7 days of incubation and then became progressively less. For instance, daily respiration rates from the aspen control incubation units decreased by 30% between Day 1 and Day 7 of the incubation period, but only by 9% between Day 7 and Day 16. Respiration fluxes from the aspen forest floors continued to decrease after Day 15 and reached a plateau by the end of the incubation (Figure 1). On Day 67, daily respiration rates from the aspen control units were more than five times lower than rates at the beginning of the incubation (Supplementary Table 1). Similar trends were apparent for the amended aspen incubation units (Figure 1 and Supplementary Table 1). While substrate addition boosted daily respiration rates in the first 2 days by 100–200% compared to the control (Supplementary Table 1), and resulted in significantly higher cumulative fluxes (Figure 1), all respiration rates from the aspen incubation units reached comparable low values by the end of the 2-month incubation.
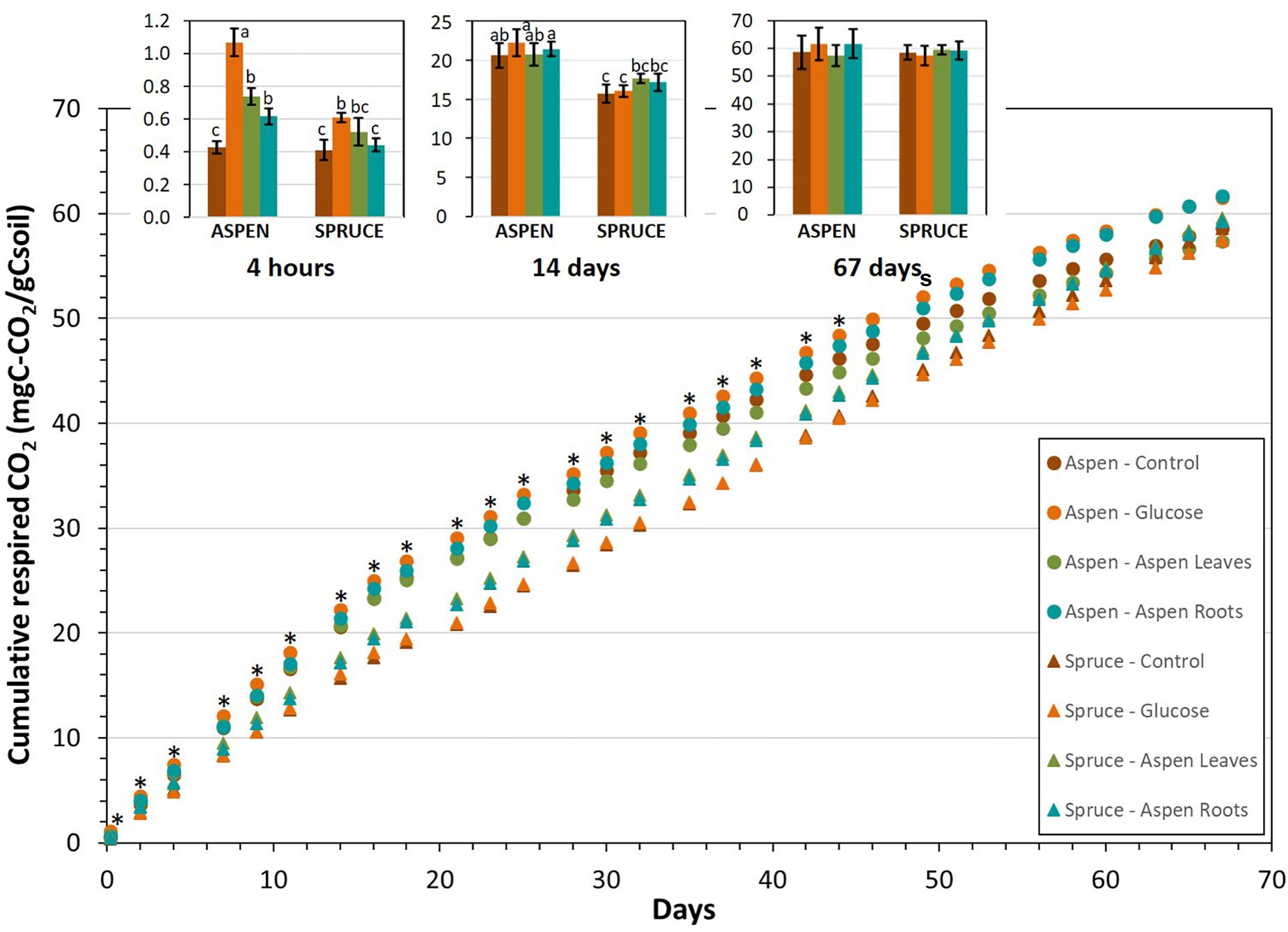
Figure 1. Average cumulative respired C-CO2 from the aspen (circles) and spruce (triangles) soil incubation units for all substrates (glucose, aspen leaves and aspen roots) during the 67-day laboratory incubation. Asterisks indicate dates where statistical differences among samples were detected using a one-way ANOVA. In addition, the inset bar graph shows average cumulative respired CO2 from the aspen and spruce soil incubation units at three dates (4 h, 14 days, and 67 days), with error bars showing standard deviations over five replicates and different letters indicating significant differences (p < 0.05).
During the first two weeks of incubation, cumulative respiration fluxes for the aspen incubation units increased faster than for the spruce, indicating that the aspen forest floor samples respired more during that period than the spruce forest floors (Figure 1). However, following this initial period, the daily rates for spruce decreased at a slower rate than for aspen, and showed no evidence to have reached a plateau by the end of the incubation (Figure 1 and Supplementary Tables 1, 2). Consequently, by Day 67, cumulative respiration rates from the control spruce incubation units (58.6 ± 2.7 mgC-CO2/gCsoil) had reached values comparable to those from aspen (58.6 ± 6.0 mgC-CO2/gCsoil). Similarly, cumulative respiration rates of the incubation units amended with the labelled substrates, which ranged from 57.4 ± 3.6 to 61.4 ± 5.3 mgC-CO2/gCsoil did not significantly differ from the control units (Figure 1).
The control incubation units (with no added substrate) for the aspen and spruce forest floors maintained respired δ13C-CO2 values around −29.1 ± 5.4 and −24.8 ± 2.3‰, respectively, throughout the incubation period (Supplementary Tables 1, 2). For both forest floors, as expected, the highest δ13C-CO2 values were observed for the incubation units amended with glucose, but these decreased rapidly within the first week of incubation. Similarly, a relatively large portion of the respiration fluxes, between 32% for spruce and 43% for aspen came from the added glucose in the first 4 h following substrate addition, but values decreased rapidly thereafter (Supplementary Figure 1). In both forest floor types, the majority of respiration fluxes coming from glucose occurred during the first week of incubation. After 2 days, glucose corresponded to about 10% of the respired CO2 fluxes for both forest floor types, and continued to decrease for the spruce incubation units to reach negligible values after one week. After 4 h of incubation in the aspen units, the percent of respired CO2 coming from the added substrates reached 23% for aspen leaves and 19% for aspen roots (Supplementary Figure 1). Corresponding values in the spruce incubation units were 15% for aspen leaves and 14% for aspen. Similarly to the glucose substrate, both aspen leaf and aspen root substrates tended to represent a higher percentage of the total respiration fluxes for the aspen forest floors compared to the spruce.
The cumulative substrate-derived CO2 fluxes (mgC-CO2/gsoil) were higher for glucose than for aspen leaves during the first 2 days of incubation, but the opposite was true for the remainder of the incubation (Figure 2). Respiration fluxes from the added glucose became significantly higher in the spruce incubation units compared to the aspen units on Day 14 of the laboratory incubation, and this remained the case for the rest of the incubation. A slightly different trend emerged following aspen leaf addition, where cumulative substrate-derived CO2 fluxes tended to be higher from the aspen units during the initial two weeks of the incubation, but then became greater from the spruce units. The spruce incubation systems showed statistically higher aspen leaf-derived CO2 fluxes after Day 46, and this remained true for the rest of the incubation; i.e., for Days 60 and 67 (Figure 2).
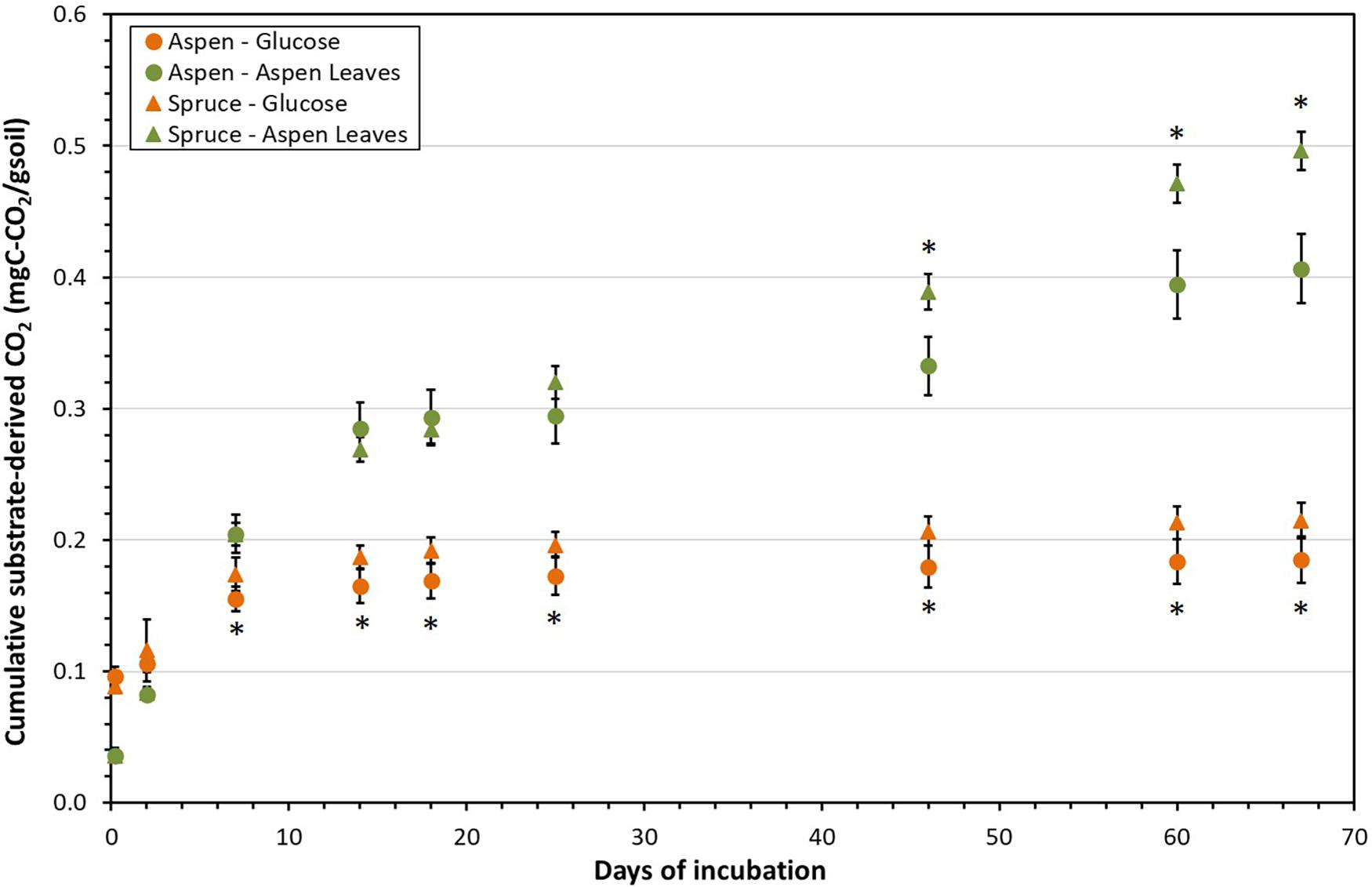
Figure 2. Cumulative substrate-derived CO2 fluxes (mgC-CO2/gsoil) from the aspen (circles) and spruce (triangles) soil incubation units following addition of glucose and aspen leaves during the 67-day laboratory incubation. Asterisks indicate dates where significant differences were detected between aspen and spruce (p < 0.05).
A positive PE was observed in most incubation systems 4 h following the addition of substrates, with the exception of the spruce forest floors following addition of aspen leaves and aspen roots (Table 2). The enhanced degradation of native organic matter following substrate addition was larger in the aspen incubation units (43, 34, and 17%, for glucose, leaves, and roots substrates, respectively; Table 2), but the PE decreased very rapidly, and was not detected past 2 days of incubation.

Table 2. Priming effect (PE%), corresponding to % increase or decrease in soil C mineralized after 4 h and 2 days of incubation following addition of glucose, aspen leaves and aspen roots.
Phospholipid Fatty Acid Profiles
For both aspen and spruce forest floor incubations, total PLFAs stayed relatively constant during all 67 days of incubation. Total PLFAs were slightly higher in the aspen forest floor incubations, with a value around 4,600 nmol g–1 of soil, compared to the spruce forest floor incubations (≈3,700 nmol g–1 of soil), but there was no significant difference between the two forest types.
The microbial soil community structure was assessed using an NMDS ordination of the PLFA data (Figure 3) and statistical patterns within the data were tested using a MRPP (Table 3). Ordination on the PLFA data yielded a two-dimensional solution with a final stress of 6.8 after 62 iterations. Testing for significance of the grouping variables (forest floor, incubation time, and treatment) identified significant differences between the two forest floor types and among the incubation times. The biggest separation was linked to forest floor type (p < 10–5) followed by incubation time (p < 10–4). As can be seen on Figure 3, points corresponding to the aspen forest floors clustered closely together for each incubation time, while the spruce data points were more spread out. However, for both forest floor types, there was a comparable shift in the composition of the microbial communities with time (Figure 3). Separation between forest floor types was also very clear, and remained for the entirety of the two months of incubation. On the other hand, there was no significant difference linked to the substrates added for either aspen or spruce (Table 3).
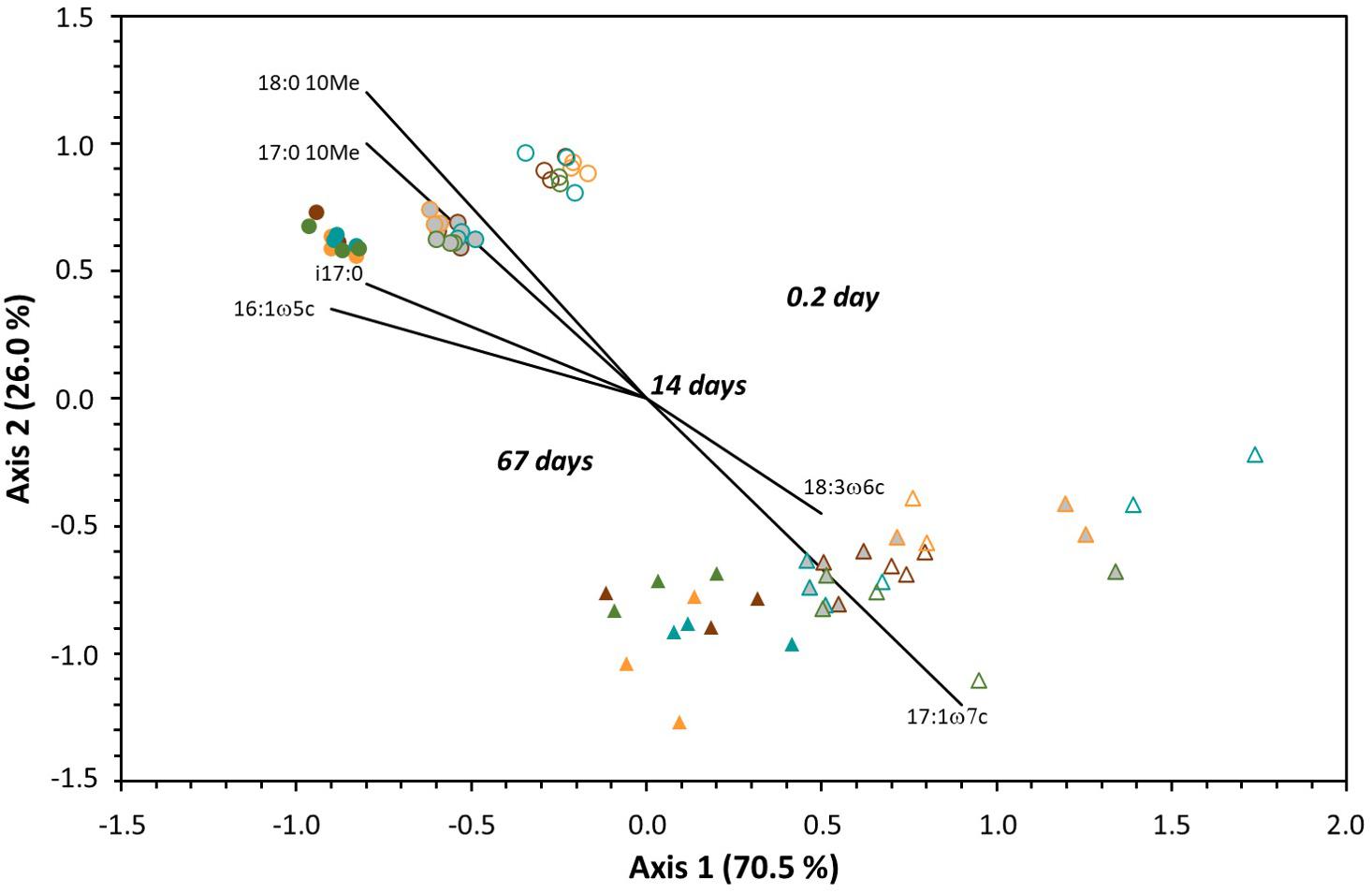
Figure 3. Non-metric multidimensional scaling ordination of PLFA profiles extracted from the aspen (circles) and spruce (triangles) soil incubation units after different incubation times, including 4 h (empty symbols), 14 days (grayed symbols), and 67 days (filled symbols). Substrates are indicated with different colors, including brown for control, orange for glucose, green for aspen leaves, and blue for aspen roots.

Table 3. Multi-response permutation procedure (MRPP) results for PLFAs extracted from the aspen and spruce soil incubation units during the 67-day laboratory incubation.
For the control forest floor incubations, δ13CPLFA values ranged from −35.1 to −24.1‰ within the aspen incubation units, and from −34.1 to −22.9‰ within the spruce forest floor incubations (Supplementary Tables 3, 4). As expected, δ13CPLFA values for the forest floors amended with glucose presented higher 13C enrichment values than the forest floors amended with other substrates (Figure 4). These enrichment values were clearly higher for aspen forest floor incubations than for spruce forest floor incubations for nearly all PLFAs, and were generally higher after 4 h than after 67 days of incubation (Figure 4A). However, different PLFA groups responded differently. Monounsaturated and straight chained saturated PLFAs became highly enriched after 4 h, while 10-methyl PLFAs proportionally became more enriched after 67 days (Figure 4A).
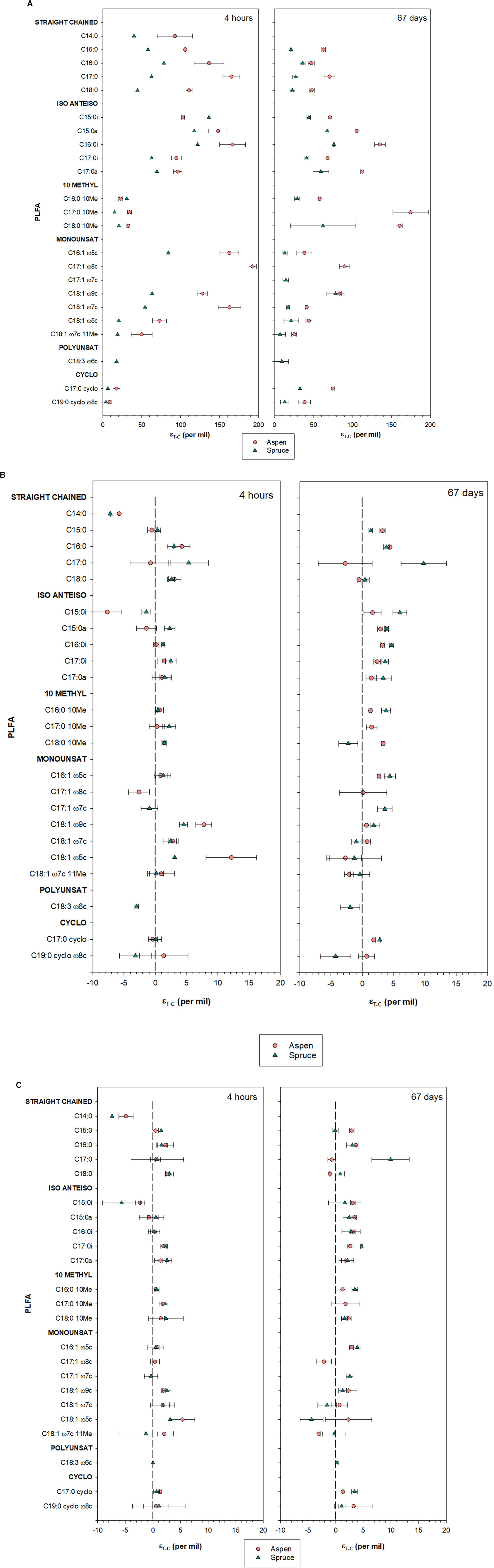
Figure 4. Differences in isotopic composition of selected PLFA biomarkers (εT–C) between substrate-amended and control incubation units after 67 days of incubation. Results are presented for both aspen and spruce soil incubation units, and added substrates included: glucose (A), aspen leaves (B), and aspen roots (C). Average values are presented with standard deviations over five experimental replicates. However, for some PLFAs, only one reliable measurement could be obtained with GC-C-IRMS analysis, and results are presented as a single point with no standard deviation.
Enrichment following addition of 13C-labeled aspen leaves and aspen roots was much smaller than was the case following glucose addition, and in several cases PLFA δ13C values were similar to those of the control incubation units, especially after only 4 h of incubation (Figures 4B,C). There was no clear difference between aspen and spruce forest floors following addition of the 13C-labeled aspen roots (Figure 4C). On the other hand, after 67 days of incubation, a slight enrichment was observed for most PLFAs in the case of the forest floors amended with aspen leaves (Figure 4B). This was particularly true of the saturated PLFAs with terminal branching, which tended to become more enriched in spruce compared to aspen forest floors (Figure 4B). This was the opposite trend to what was observed following glucose, where all microbial PLFAs were more enriched in the case of the aspen forest floor (Figure 4A).
Incorporation of substrate-C into microbial PLFAs (μgC/gsoil) showed both temporal shifts and differences linked to forest floor type (aspen and spruce), substrate added (glucose and aspen leaves), and the group of PLFAs considered (Figure 5). All PLFA groupings (10Me, branched saturated, monounsaturated, total) showed significantly higher incorporation of glucose-13C for the aspen than the spruce forest floors. As the spruce forest floors respired more of the glucose (Figure 2), the resulting PLFA-based SUE values were clearly higher for aspen than for spruce, in particular for the monounsaturated PLFAs preferentially associated with gram negative bacteria (Figure 5). Right after addition of the aspen leaves, substrate incorporation was also statistically higher for the aspen than the spruce forest floors in terms of total PLFAs, but this did not hold true for all PLFA groupings; terminally branched PLFAs (characteristic of gram positive bacteria) showed a statistically higher enrichment in the case of the spruce forest floors. After 67 days, the aspen and spruce forest floors reached comparable SUE values (Figure 5).
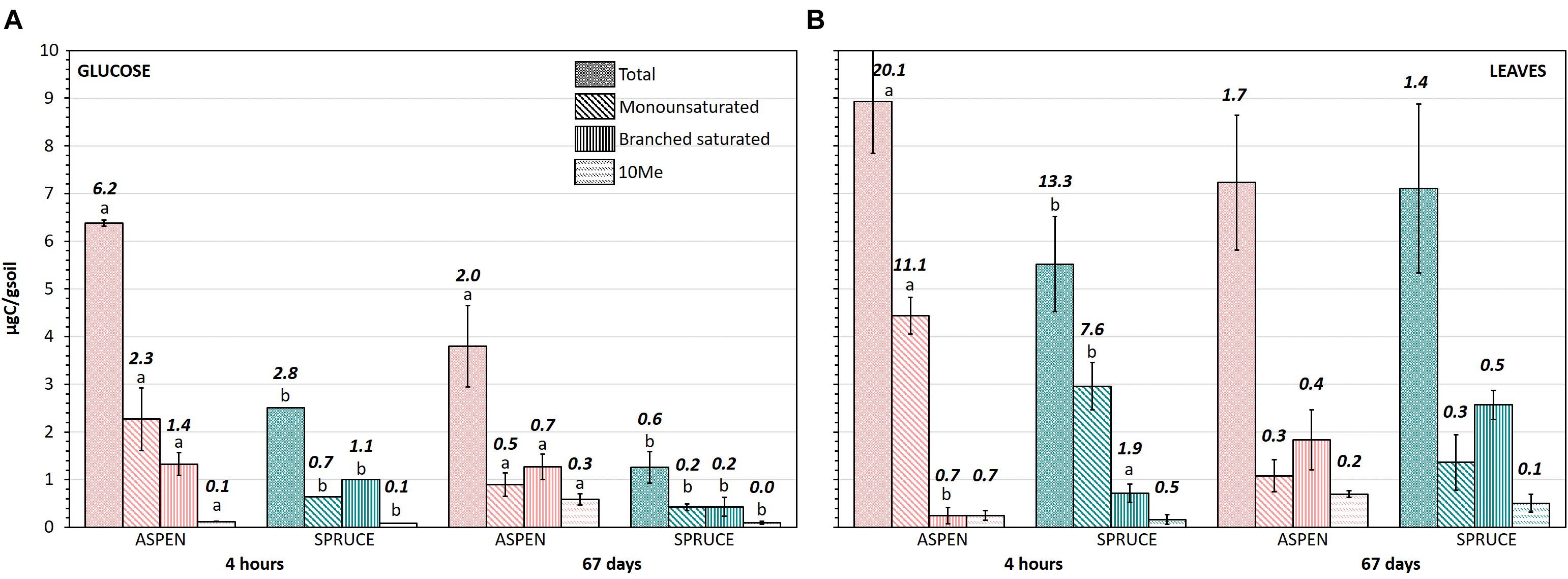
Figure 5. Incorporation of (A) glucose and (B) aspen leaves into 10Me, branched saturated, monounsaturated, and total PLFAs for both aspen and spruce soil incubation units. Error bars show standard deviations over five replicates and different letters indicate significant differences between the aspen and spruce soil incubation units for each date (4 h and 67 days) and each PLFA grouping (p < 0.05). The value indicated on top of each bar corresponds to the PLFA based-SUE value (%).
Discussion
Microbial Substrate Utilization Following Glucose Addition
Glucose addition did not significantly alter PLFA profiles in either aspen and spruce forest floors (Figure 1), and PEs, when detected, were short-lived (Table 2). Similarly, no change in PLFA profiles was detected in another study with a one-time addition of either starch or xylose to an oak woodland soil, at a level of substrate addition (400 μg C/g of soil) comparable to our study (Waldrop and Firestone, 2004), or in incubations where small quantities of carbon substrates (glucose, glutamate, oxalate or phenol) were added to a soil from a Douglas-fir/hemlock dominated site (Brant et al., 2006). Microbial communities in both aspen and spruce forest floors changed with time during the 67-day incubation (Figure 3 and Table 3), but this temporal shift was similar for the control and the labeled materials (Figure 3). In both forest floors, concentrations of saturated PLFAs, which tend to be more abundant in gram positive bacteria, generally decreased during the 67 days of incubation while monounsaturated PLFAs more typical of gram negative bacteria increased (Supplementary Tables 3, 4). The ratios corresponding to monounsaturated/saturated PLFAS increased from 1.09 (4 h) to 1.12 (14-days) to 1.24 (67 days), corresponding to a 14% increase, in the aspen incubation units; and from 0.99 to 1.04 and finally 1.07 (i.e., a smaller 8% increase) in the spruce incubation units. In addition, there were larger changes in specific PLFAs for the aspen than the spruce community (Figure 3). For instance, a shift in the iso/anteiso ratio, which has been linked to changes in bacterial membrane fluidity (Watzinger, 2015) could be detected for aspen but not for spruce; specifically, the (i15:0+i17:0)/(a15:0+a17:0) ratio increased from 1.2 (4 h) to 1.3 (14 days) to 1.4 (67 days) for aspen, while there was not any clear change for spruce.
Individual PLFAs diverged in their temporal response to 13C-glucose addition, where non-specific, straight chained PLFAs (e.g., 16:0, 17:0) as well as PLFAs with terminal branching associated with gram positive bacteria (e.g., a15:0a, i16:0i), and in particular monounsaturated gram negative PLFAs (e.g., 18:1ω7) readily picked up the label after 4 h, while 10Me PLFAs took longer to become enriched (Figure 4). This trend is similar to results from incubations with 13C-labeled glucose conducted over short time periods (0.75–48 h) where a rapid enrichment of PLFAs associated with gram positive bacteria (e.g., a15:0), and the gram negative 18:1ω7 was detected (Ziegler et al., 2005; Swallow and Quideau, 2020). The 10Me18:0 became enriched following depletion of the original glucose, which was evidence of the importance of Actinobacteria in recycling glucose originally incorporated in other bacterial biomass (Ziegler et al., 2005). Incorporation of the 13C label was increased in Actinobacteria PLFAs 5 days following 13C-glucose addition while δ13C values in 16:0 and 18:0 PLFAs were reduced (Dungait et al., 2011). Similarly, in our incubation, 10Me PLFAs maintained strong 13C enrichment even after 67 days of incubation (Figure 5).
When comparing the two forest floor types, the aspen community was clearly more efficient than the spruce at utilizing glucose for growth: all PLFAs became more 13C enriched in aspen than in spruce forest floors (Figure 4), and incorporation of 13C-glucose was greater in the aspen PLFAs, especially for gram negative at the beginning, and gram positive, including Actinobacteria, at the end of the incubation (Figure 5). These results indicate rapid and preferential glucose anabolism by gram negative bacteria in the aspen forest floor. Actinobacteria, better able to utilize more complex C substrates (Ziegler and Billings, 2011), such as those in microbial necromass, played a larger role later in the incubation. Furthermore, the aspen community respired less of the added glucose, and corresponding PLFA-based SUEs were clearly higher for the aspen (Figure 5). Together with an enhanced ability to retain labile C through mineral sorption (Boča et al., 2020), a greater microbial utilization efficiency of labile C substrates such as glucose likely plays a role in the greater and more persistent C stocks typically observed in aspen mineral horizons compared to spruce (Laganière et al., 2017; Sewell et al., 2020).
Microbial Response to Aspen Litter Addition
Distinct differences in microbial communities between the two forest floor types (Figure 3) have previously been reported in comparable Luvisolic soils from the Mixedwood forest of western Canada (e.g., Hannam et al., 2006; Swallow and Quideau, 2013; Thacker and Quideau, 2021). Similarly to these studies, differences in our study appeared to be linked to specific PLFAs rather than to broader groupings (e.g., gram positive bacteria). In the same way to what we report here where Actinobacteria PLFAs (10Me17:0, 10Me18:0) and the gram positive i17:0 were correlated to aspen (Figure 3), 10Me16:0, 10Me18:0 and i16:0 PLFAs have been associated with aspen forest floors (Swallow and Quideau, 2013). The gram negative 17:1ω7, which correlated with spruce in our study (Figure 3), has previously been recognized as an indicator PLFA for spruce forest floors (Hannam et al., 2006). Furthermore, differences between the two forest floors remained throughout the two months of incubation, which is in agreement with the work from Hannam et al. (2007), who did not detect any change in microbial community structure when forest floors of aspen and spruce were incubated in the field for 1 year following reciprocal transfer to either spruce or aspen stands. Microbial communities in aspen and spruce forest floors remained distinct 17 years following clearcutting of aspen and spruce stands where aspen was naturally regenerating in both stand types (Thacker and Quideau, 2021). In the case of ecosystems from warmer climates (e.g., Potthast et al., 2010), or more drastic vegetation and/or land management changes (e.g., Quideau et al., 2013), changes in PLFA profiles are typically detected following vegetation shifts. In the boreal forest of Western Canada, however, this does not appear to be the case, and the long-lasting legacy of the original forest floor seems to buffer microbial communities against rapid structural changes following vegetation shifts.
A greater amount of the added aspen leaves was respired by the spruce community (Figure 2). Thus, results were opposite of what would have been expected according to the HFA theory (Ayres et al., 2009), which stipulates that the microbial communities in the aspen forest floors would be better adapted to mineralize the aspen litter when compared to the communities present in the spruce forest floors. It is entirely possible that different results would be obtained if a litter decomposition study were to be conducted in situ, as divergent conditions including higher moisture and lower temperature may be slowing down decomposition processes under spruce (Sewell et al., 2020). But under laboratory conditions where abiotic limitations were lifted, there was no apparent inhibition of aspen leaf decomposition by the spruce community. Interestingly, greater aspen leaf respiration by the spruce community did not necessarily correspond to greater microbial biomass uptake, except in the case of gram positive bacteria (Figure 5). From a soil carbon balance point of view, this meant a faster loss of aspen leaf carbon from the spruce forest floors, but a comparable carbon retention in microbial biomass, as suggested by the comparable SUE in both forest floors by the end of the incubation period. Further, while our study did not detect any differences between aspen and spruce in terms of overall bacterial or fungal biomass, it is possible that species-specific differences exist, such as those observed in fungal species between spruce and beech forests (Asplund et al., 2019), which could have impacts on soil carbon persistence. Differences in soil carbon storage have been linked to distinct mycorrhizal fungal communities in boreal forest soils (Clemmensen et al., 2015). In all cases, observations from our study indicate that a transition from spruce to aspen will initially increase the loss of aspen leaf litter through microbial respiration, although incorporation into microbial biomass and eventually more persistent soil carbon may not be affected. Follow-up work is needed to test if these results hold true over a longer time period.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
EL and SQ conceived and designed the methodology. EL performed the laboratory incubation, statistical analyses, and wrote the first draft of the manuscript. Both authors discussed the results and contributed to the final version of the manuscript.
Funding
The Natural Sciences and Engineering Research Council of Canada (NSERC) provided overall financial support through a Discovery Grant (RGPIN-2014-04693) to SQ. The funding agencies had no role in the design, implementation or summation of this project.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are grateful to C. Norris for her assistance on obtaining the labeled aspen tissues, and to M. Swallow for providing guidance on the incubation and PLFA analyses. Isotopic analyses were conducted by B. Feland at the Stable Isotope Facility for Ecosystem Research (SIFER) at the University of Alberta.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffgc.2021.700751/full#supplementary-material
References
Asplund, J., Asplund, J., Kauserud, H., Ohlson, M., and Nybakken, L. (2019). Spruce and beech as local determinants of forest fungal community structure in litter, humus and mineral soil. FEMS Microbiol. Ecol. 95:fiy232. doi: 10.1093/femsec/fiy232
Ayres, E., Steltzer, H., Simmons, B. L., Simpson, R. T., Steinweg, J. M., Wallenstein, M. D., et al. (2009). Home-field advantage accelerates leaf litter decomposition in forests. Soil Biol. Biochem. 41, 606–610. doi: 10.1016/j.soilbio.2008.12.022
Bauhus, J., Paré, D., and Côté, L. (1998). Effects of tree species, stand age and soil type on microbial biomass and its activity in a southern boreal forest. Soil Biol. Biochem. 30, 1077–1089. doi: 10.1016/S0038-0717(97)00213-7
Blagodatsky, S., Blagodatskaya, E., Yuyukina, T., and Kuzyakov, Y. (2010). Model of apparent and real priming effects: linking microbial activity with soil organic matter decomposition. Soil Biol. Biochem. 42, 1275–1283. doi: 10.1016/j.soilbio.2010.04.005
Boča, A., Jacobson, A. R., and Van Miegroet, H. (2020). Aspen soils retain more dissolved organic carbon than conifer soils in sorption experiment. Front. For. Global Change 3:594473. doi: 10.3389/ffgc.2020.594473
Bradshaw, C. J. A., and Warkentin, I. G. (2015). Global estimates of boreal forest carbon stocks and flux. Glob. Planet. Change 128, 24–30. doi: 10.1016/j.gloplacha.2015.02.004
Brandt, J. P., Flannigan, M. D., Maynard, D. G., Thompson, I. D., and Volney, W. J. A. (2013). An introduction to Canada’s boreal zone: ecosystem processes, health, sustainability, and environmental issues. Environ. Rev. 21, 207–226. doi: 10.1139/er-2013-0040
Brant, J. B., Sulzman, E. W., and Myrold, D. D. (2006). Microbial community utilization of added substrates in response to long-term carbon input manipulation. Soil Biol. Biochem. 38, 2219–2232. doi: 10.1016/j.soilbio.2006.01.022
Clemmensen, K. E., Finlay, R. D., Dahlberg, A., Stenlid, J., Wardle, D. A., and Lindahl, B. D. (2015). Carbon sequestration is related to mycorrhizal fungal community shifts during long-term succession in boreal forests. New Phytol. 205, 1525–1536.
Cleveland, C. C., Reed, S. C., Keller, A. B., Nemergut, D. R., O’Neill, S. P., Ostertag, R., et al. (2014). Litter quality versus soil microbial community controls over decomposition: a quantitative analysis. Oecologia 174, 283–294. doi: 10.1007/s00442-013-2758-9
Cotrufo, M. F., Wallenstein, M. D., Boot, C. M., Denef, K., and Paul, E. (2013). The Microbial Efficiency-Matrix Stabilization (MEMS) framework integrates plant litter decomposition with soil organic matter stabilization: Do labile plant inputs form stable soil organic matter? Global Change Biol. 19, 988–995. doi: 10.1111/gcb.12113
D’Orangeville, L., Houle, D., Duchesne, L., Phillips, R. P., Bergeron, Y., and Kneeshaw, D. (2018). Beneficial effects of climate warming on boreal tree growth may be transitory. Nat. Commun. 9:3213. doi: 10.1038/s41467-018-05705-4
DeLuca, T. H., and Boisvenue, C. (2012). Boreal forest soil carbon: distribution, function and modeling. Forestry 85, 161–184. doi: 10.1093/forestry/cps003
Dungait, J. A. J., Kemmitt, S. J., Michallon, L., Guo, S., Wen, Q., Brookes, P. C., et al. (2011). Variable responses of the soil microbial biomass to trace concentrations of 13C-labelled glucose, using 13C-PLFA analysis. Eur. J. Soil Sci. 62, 117–126. doi: 10.1111/j.1365-2389.2010.01321.x
Feland, B., and Quideau, S. A. (2020). Isotope applications to soil science at the University of Alberta — an historical perspective. Can. J. Soil Sci. 100, 344–355. doi: 10.1139/cjss-2019-0153
Frostegård, Å, Tunlid, A., and Bååth, E. (2010). Use and misuse of PLFA measurements in soils. Soil Biol. Biochem. 43, 1621–1625. doi: 10.1016/j.soilbio.2010.11.021
Grayston, S. J., and Prescott, C. E. (2005). Microbial communities in forest floors under four tree species in coastal British Columbia. Soil Biol. Biochem. 37, 1157–1167. doi: 10.1016/j.soilbio.2004.11.014
Hannam, K. D., Quideau, S. A., and Kishchuk, B. E. (2006). Forest floor microbial communities in relation to stand composition and timber harvesting in northern Alberta. Soil Biol. Biochem. 38, 2565–2575. doi: 10.1016/j.soilbio.2006.03.015
Hannam, K. D., Quideau, S. A., and Kishchuk, B. E. (2007). The microbial communities of aspen and spruce forest floors are resistant to changes in litter inputs and microclimate. Appl. Soil Ecol. 35, 635–647. doi: 10.1016/j.apsoil.2006.09.005
Hogg, E. H., Michaelian, M., Hook, T. I., and Undershultz, M. E. (2017). Recent climatic drying leads to age-independent growth reductions of white spruce stands in western Canada. Global Change Biol. 23, 5297–5308. doi: 10.1111/gcb.13795
IUSS Working Group WRB (2015). World Reference Base for Soil Resources 2014 Update 2015. Rome: Food and Agriculture Organization of the United Nations.
Kalra, Y. P., and Maynard, D. C. (1991). Methods Manual for Forest Soil and Plant Analysis. Edmonton: Forestry Canada, Northwest region, Northern Forestry Center, 125.
Keiblinger, K. M., Hall, E. K., Wanek, W., Szukics, U., Hämmerle, I., Ellersdorfer, G., et al. (2010). The effect of resource quantity and resource stoichiometry on microbial carbon-use-efficiency. FEMS Microbiol. Ecol. 73, 430–440. doi: 10.1111/j.1574-6941.2010.00912.x
Kishchuk, B. E., Quideau, S. A., Wang, Y., and Prescott, C. E. (2014). Long-term soil response to variable-retention harvesting in the EMEND (Ecosystem Management Emulating Natural Disturbance) experiment, northwestern Alberta. Can. J. Soil Sci. 94, 263–279. doi: 10.4141/CJSS2013-034
Kohl, L., Myers-Pigg, A. N., Edwards, K. A., Billings, S. A., Warren, J., Podrebarac, F., et al. (2021). Microbial inputs at the litter layer translate climate into altered organic matter properties. Glob. Change Biol. 27, 435–453. doi: 10.1111/gcb.15420
Kurz, W. A., Shaw, C. H., Boisvenue, C., Stinson, G., Metsaranta, J., Leckie, D., et al. (2013). Carbon in Canada’s boreal forest – a synthesis. Environ. Rev. 21, 260–292. doi: 10.1139/er-2013-0041
Kuzyakov, Y. (2010). Priming effects: interactions between living and dead organic matter. Soil Biol. Biochem. 42, 1363–1371. doi: 10.1016/j.soilbio.2010.04.003
Laganière, J., Boča, A., Van Miegroet, H., and Paré, D. (2017). A tree species effect on soil that is consistent across the species’ range: the case of aspen and soil carbon in North America. Forests 8:113. doi: 10.3390/f8040113
Larionova, A. A., Yevdokimov, I. V., and Bykhovets, S. S. (2007). Temperature of soil respiration is dependent on concentration of readily decomposable C. Biogeosciences 4, 1073–1081. doi: 10.5194/bg-4-1073-2007
Lieffers, V. J., Macmillan, R. B., MacPherson, D., Branter, K., and Stewart, J. D. (1996). Semi-natural and intensive silvicultural systems for the boreal mixedwood forest”. For. Chron. 72, 286–292. doi: 10.5558/tfc72286-3
Mielke, P. W. Jr., and Berry, K. J. (2001). Permutation methods: A Distance Function Approach. Springer Series in Statistics. New York, NY: Springer-Verlag, 353. doi: 10.1007/978-1-4757-3449-2
Myers, R. T., Zak, D. R., White, D. C., and Peacock, A. (2001). Landscape-level patterns of microbial community composition and substrate use in upland forest ecosystems. Soil Sci. Soc. Am. J. 65, 359–367. doi: 10.2136/sssaj2001.652359x
Neupane, A., Reed, S. C., and Cusack, D. F. (2021). Warming and microbial uptake influence the fate of added soil carbon across a Hawai’ian weathering gradient. Soil Biol. Biochem 153:108080. doi: 10.1016/j.soilbio.2020.108080
Norris, C. E., Dungait, J. A. J., Joynes, A., and Quideau, S. A. (2013). Biomarkers of novel ecosystem development in boreal forest soils. Org. Geochem. 64, 9–18. doi: 10.1016/j.orggeochem.2013.08.014
Norris, C. E., Quideau, S. A., and Oh, S.-W. (2016). Microbial utilization of double-labeled aspen litter in boreal aspen and spruce soils. Soil Biol. Biochem. 100, 9–20. doi: 10.1016/j.soilbio.2016.05.013
Norris, C. E., Quideau, S. A., Landhäusser, S. M., Bernard, G. M., and Wasyleshen, R. E. (2012). Tracking stable isotope enrichment in tree seedlings with solid-state NMR spectroscopy. Sci. Rep. 2:719. doi: 10.1038/srep00719
Potthast, K., Hamer, U., and Makeschin, F. (2010). Impact of litter quality on mineralization processes in managed and abandoned pasture soils in Southern Ecuador. Soil Biol. Biochem. 42, 56–64. doi: 10.1016/j.soilbio.2009.09.025
Price, D. T., Alfaro, R. I., Brown, K. J., Flannigan, M. D., Fleming, R. A., Hogg, E. H., et al. (2013). Anticipating the consequences of climate change for Canada’s boreal forest ecosystems. Environ. Rev. 21, 322–365. doi: 10.1139/er-2013-0042
Quideau, S. A., Mcintosh, A. C., Norris, C. E., Lloret, E., Swallow, M., and Hannam, K. (2016). Extraction and analysis of microbial phospholipid fatty acids in soils. JOVE-J. Vis. Exp. 114:54360. doi: 10.3791/54360
Quideau, S. A., Swallow, M. J. B., Prescott, C. E., Graston, S. J., and Oh, S.-W. (2013). Comparing soil biogeochemical processes in novel and natural boreal forest ecosystems. Biogeosciences 10, 5651–5661. doi: 10.5194/bg-10-5651-2013
Rinnan, R., and Baath, E. (2009). Differential utilization of carbon substrates by bacteria and fungi in tundra soil. Appl. Environ. Microb. 75, 3611–3620. doi: 10.1128/AEM.02865-08
Searle, E. B., and Chen, H. Y. H. (2017). Persistent and pervasive compositional shifts of western boreal forest plots in Canada. Glob. Change Biol. 23, 857–866. doi: 10.1111/gcb.13420
Sewell, P. D., Quideau, S. A., Dyck, M., and Macdonald, E. (2020). Long-term effects of harvest on boreal forest soils in relation to a remote sensing-based soil moisture index. For. Ecol. Manage 462:117986. doi: 10.1016/j.foreco.2020.117986
Swallow, M. J., and Quideau, S. A. (2020). Rapid 13C-enrichment of microbial PLFAs in response to the application of 13C-enriched glucose and selective inhibitors. Can. J. Soil Sci. 100, 356–362. doi: 10.1139/cjss-2019-0152
Swallow, M., and Quideau, S. A. (2013). Moisture effects on microbial communities in boreal forest floors are stand-dependent. Appl. Soil Ecol. 63, 120–126. doi: 10.1016/j.apsoil.2012.09.006
Thacker, S. J., and Quideau, S. A. (2021). Rhizosphere response to predicted vegetation shifts in boreal forest floor. Soil Biol. Biochem. 154:108141. doi: 10.1016/j.soilbio.2021.108141
Waldrop, M. P., and Firestone, M. K. (2004). Microbial community utilization of recalcitrant and simple carbon compounds: impact of oak-woodland plant communities. Oecologia 138, 275–284. doi: 10.1007/s00442-003-1410-9
Waring, B., Sulman, B., Reed, S., Smith, P., Averill, C., Creamer, C., et al. (2020). From pools to flow: the PROMISE framework provides new insights on soil carbon cycling in a changing world. Glob. Change Biol 26, 6631–6643. doi: 10.1111/gcb.15365
Watson, R., Noble, I., Bolin, B., Ravindranath, N. H., Verardo, D. J., and Dokken, D. J. (2000). IPCC Special Report: Land use, Land-use Change, and Forestry. Cambridge: Cambridge University Press.
Watzinger, A. (2015). Microbial phospholipid biomarkers and stable isotope methods help reveal soil functions. Soil Biol. Biochem. 86, 98–107. doi: 10.1016/j.soilbio.2015.03.019
White, D. C., Ringelberg, D. B., MacNaughton, S. J., Alugupalli, S., and Schram, D. (1997). “Signature lipid biomarker analysis for quantitative assessment in situ of environmental microbial ecology,” in Molecular Markers in Environmental Geochemistry, Vol. 671, ed. R. P. Eganhouse (Washington, D.C: American Chemical Society), 22–34.
Zelles, L. (1999). Fatty acid patterns of phospholipids and lipopolysaccharides in the characterization of microbial communities in soil: a review. Biol. Fert. Soils 29, 111–129. doi: 10.1007/s003740050533
Ziegler, S. E., and Billings, S. A. (2011). Soil nitrogen status as a regulator of carbon substrate flows through microbial communities with elevated CO2. J. Geophys. Res. 116:G01011. doi: 10.1029/2010JG001434
Keywords: boreal, forest floor, vegetation shifts, labeled substrate, phospholipid fatty acid analysis, SIP–PLFA, microbial communities
Citation: Lloret E and Quideau S (2021) Microbial Substrate Utilization and Vegetation Shifts in Boreal Forest Floors of Western Canada. Front. For. Glob. Change 4:700751. doi: 10.3389/ffgc.2021.700751
Received: 26 April 2021; Accepted: 05 August 2021;
Published: 31 August 2021.
Edited by:
Laurent Augusto, INRA Centre Bordeaux-Aquitaine, FranceReviewed by:
Susan Ziegler, Memorial University of Newfoundland, CanadaAntra Boca, Latvia University of Agriculture, Latvia
Copyright © 2021 Lloret and Quideau. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sylvie Quideau, c3lsdmllLnF1aWRlYXVAdWFsYmVydGEuY2E=
 Emily Lloret1,2
Emily Lloret1,2 Sylvie Quideau
Sylvie Quideau