
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. For. Glob. Change, 29 June 2021
Sec. Forest Soils
Volume 4 - 2021 | https://doi.org/10.3389/ffgc.2021.696983
This article is part of the Research TopicChanges in Forest Ecosystem NutritionView all 25 articles
 Antonios Michas1
Antonios Michas1 Giovanni Pastore2
Giovanni Pastore2 Akane Chiba1,3
Akane Chiba1,3 Martin Grafe1
Martin Grafe1 Simon Clausing4
Simon Clausing4 Andrea Polle4
Andrea Polle4 Michael Schloter1,5
Michael Schloter1,5 Marie Spohn2,6
Marie Spohn2,6 Stefanie Schulz1*
Stefanie Schulz1*Phosphorus (P) solubilization is an important process for P acquisition by plants and soil microbes in most temperate forests. The abundance of inorganic P solubilizing bacteria (PSB) is affected by the P concentration in the soil and the carbon input by plants. We used a girdling approach to investigate the interplay of root-derived C and initial P content on the community composition of gcd-harboring bacteria as an example of PSB, which produce gluconic acid. We hypothesized that gcd-harboring PSB communities from P-poor sites are more vulnerable to girdling, because of their lower diversity, and that a shift in gcd-harboring PSB communities by girdling is caused by a response of few, mostly oligotrophic, taxa. We used a high-throughput metabarcoding approach targeting the gcd gene, which codes for the quinoprotein glucose dehydrogenase, an enzyme involved in the solubilization of inorganic P. We compared the diversity of gcd-harboring PSB in the mineral topsoil from two temperate beech forests with contrasting P stocks, where girdling was applied and compared our data to the respective control plots with untreated young beech trees. At both sites, gcd-harboring PSB were dominated by Proteobacteria and Acidobacteria, however, with differences in relative abundance pattern on the higher phylogenetic levels. The P-poor site was characterized by a high relative abundance of Kaistia, whereas at the P-rich site, Dongia dominated the gcd-harboring bacterial communities. Girdling induced an increase in the relative abundance of Kaistia at the P-poor site, whereas other bacterial groups of the family Rhizobiaceae were reduced. At the P-rich site, major microbial responders differed between treatments and mostly Bradyrhizobium and Burkholderia were positively affected by girdling in contrast to uncultured Acidobacteria, where reduced relative abundance was found. Overall, these effects were consistent at different time points analyzed after the introduction of girdling. Our data demonstrate that plant-derived carbon influences community structure of gcd-harboring bacteria in temperate beech forest soils.
The availability of phosphorus (P) in soil is essential for the growth of plants and the functioning of soil biomes. Soil microbes can exploit different inorganic and organic P sources via several solubilization and mineralization pathways (Sharma et al., 2013). Microbial solubilization of inorganic P in soil promotes plant nutrition (Satyaprakash et al., 2017), the mitigation of drought stress (Shintu and Jayaram, 2015) and the recovery of soil P, e.g., after ecosystem restoration processes (Liang et al., 2020), and it is accomplished by several processes, like the release of protons or secretion of organic acids and siderophores (Sashidhar and Podile, 2010). The secreted organic acids include a diverse range of acids, such as citric acid, oxalic acid, formic acid, and malic acid, among which gluconic acid is considered to be the most important (Goldstein, 1995; Rodríguez and Fraga, 1999; Alori et al., 2017). The formation of gluconic acid from glucose is the result of the expression of the bacterial gcd gene, which encodes for the quinoprotein glucose dehydrogenase and the pyrroloquinoline quinon cofactor. As Gcd is a membrane bound enzyme, the oxidation of glucose directly leads to external acidification and subsequent solubilization of mineral bound inorganic P (Goldstein et al., 1993; Goldstein, 1995; Yang et al., 2017; Rasul et al., 2019), and can be considered as an important mechanisms for the solubilization of inorganic P Gram negative bacteria.
P availability in soil affects the structure and abundance of gcd-harboring bacterial communities. Previous studies have shown that the diversity and abundance of phosphate solubilizing bacteria (PSB) increase (Pastore et al., 2020; Spohn et al., 2020) or decrease (Bergkemper et al., 2016b; Kurth et al., 2020) at forest sites with low P availability. Further gcd expression is directly controlled by phosphate concentration (Zeng et al., 2016). However, the role of the availability of other nutrients, e.g., carbon (C) or nitrogen (N), as well as the stoichiometry of those nutrients for the abundance and diversity of PSB are still poorly understood.
The input of organic material into soil, e.g., by root exudation, changes the soil nutrient content and can shift the carbon to phosphorus ratio (C:P) with pronounced effects on the structure of the plant associated microbiome and the phenotype of single microorganisms (Kuzyakov et al., 2007; Koranda et al., 2011; Meier et al., 2017; Vives-Peris et al., 2020). In addition, the release of plant exudates stimulates microbial mobilization of P in the rhizosphere (Spohn et al., 2013). A 13C-labeling experiment demonstrated that different PSB were attracted by root-derived C in P-poor (e.g., Bacillales) and P-rich (e.g., Rhizobiales) soils (Long et al., 2018). In addition, single compounds of root exudates may act as a modulator for P turnover, for example citramalic acid and salicylic acid for inorganic P solubilization (Tawaraya et al., 2006; Khorassani et al., 2011). In a recent study, Clausing et al. (2021) demonstrated that tree girdling, which reduces the release of plant photosynthates into soil (Högberg et al., 2001; Kleinsteuber et al., 2008; Zeller et al., 2008; Kaiser et al., 2010, 2015; Pena et al., 2010), significantly reduced the P uptake of plants and increased the abundance of gcd-harboring bacteria in the mineral topsoil of beech forests. Girdling significantly increased the potential for microbial inorganic P solubilization in the mineral topsoil only at the P-poor site, while other microbial driven P turnover processes like the mineralization of organic P were not significantly affected.
The results from Clausing et al. (2021) and the previously demonstrated importance of gluconic acid for inorganic P solubilization (Goldstein, 1995; Rodríguez and Fraga, 1999; Alori et al., 2017) gave us the motivation to investigate feedback loops of plant-derived C and shifts in the stoichiometry of soil C:P on the diversity of gcd-harboring PSB. For this purpose, we used a molecular barcoding approach based on the gcd gene to assess changes in diversity and community composition in response to P and C availability. We analyzed soil samples from a tree girdling experiment, which was performed at two sites with contrasting soil P stocks in temperate beech forests. Samples were taken at two time points after girdling and compared to the respective controls, where no girdling was applied. We hypothesized that (1) the gcd community from P-poor sites is more vulnerable to girdling, because of their lower diversity. (2) The increase in abundance of gcd-harboring bacteria observed by Clausing et al. (2021) is associated with an increase in the relative abundance of few taxa, rather than a uniform growth of the whole gcd community.
The experimental design has been previously described in detail by Clausing et al. (2021). Briefly, the experiment was conducted at two beech (F. sylvatica L.) forests in Germany, Bad Brückenau (BBR: 5579975 N, 3566195 E) and Luess (LUE: 5857057 N, 3585473 E), with different soil properties. Among others, soils differed in total P content; BBR topsoil contained approximately 0.9 mg Ptot g–1 of dry mass (P-rich), compared to 0.02 mg Ptot g–1 dry mass at the LUE site (P-poor). The average soil C:Porg ratios were 59 and 493 g g–1 for BBR and LUE, respectively, (Lang et al., 2017). Experimental plots (4 m2) with young beech trees (10 years) were established at both forest sites in May 2017. In June 2017, all trees in half of each plot were girdled by removal of a strip of bark around the stem. This treatment interrupts the C flux into the soil. Trees in the other half of the plot were separated by a lawn edge and left untreated. Composite samples consisting of eight soil cores per subplot were collected one (T1) and eight (T2) weeks after girdling from each replicate plot. Bulk topsoil samples from three plots per site were sieved (mesh width: 4 mm) and divided into two aliquots in the field. One aliquot was frozen in liquid nitrogen and stored at −80°C for molecular barcoding. The second aliquot was dried (40°C, 14 days) for calculating soil dry weight.
Total nucleic acids were isolated from 0.5 g of frozen soil per sample by mechanical cell disruption using a Precellys 24 homogenisator (5 m s–1; 30 s; Bertin Technologies, France), followed by organic solvent extraction according to Lueders et al. (2004). The modified protocol is described by Clausing et al. (2021). A sample without soil served as extraction blank for subsequent sequencing analysis. The amount of DNA was quantified using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Germany) and the purity was assessed based on the ratio of absorbance of 280/260 nm. Extracted samples were stored at −20°C until further processing.
The amplicon library preparation of gcd genes from the soil samples was performed in triplicates according to Bergkemper et al. (2016a) using the same primer pair as for the qPCR analysis in the study of Clausing et al. (2021), gcd-FW and gcd-RW, with the following modifications. PCR master mix contained 1× High Fidelity PCR Buffer, 1 U Platinum® Taq DNA Polymerase High Fidelity (Invitrogen, Germany), 5 pmol of each primer, 0.06% bovine serum albumin, 2 mM MgSO4, 0.2 mM dNTP, and 50 ng of DNA template in 25 μl total volume. Equal volumes of extraction blank and nuclease-free water were used as non-template controls. The amplification profile was as follows: initial denaturation 94°C for 2 min, 35 amplification cycles of 94°C (30 s), 57°C (60 s) and 72°C (45 s), and final extension at 72°C for 7 min. The presence of DNA fragments of the correct size was confirmed by agarose gel (1%) electrophoresis. A second PCR (10 cycles) was performed with 2 μl of the first PCR product and 2 μl of each of the same primers containing the overhangs for Illumina sequencing. Triplicates were combined and the products were purified using Agencourt AMPure XP beads (0.8 × sample volume; Beckman Coulter Inc., United States). The length and quantity of the fragments were assessed using a Fragment Analyzer Automated CE System (Advanced Analytical Technologies Inc., United States). Barcoded sequences and indices were incorporated by the following PCR reaction: 1 × NEBNext High Fidelity Master Mix (New England BioLabs Ltd., United Kingdom), 2.5 μl of each indexing primer (Nextera® XT Index Kit v2) and 5 ng of the purified template in a 25 μl final volume. Indexed amplicons were purified using Agencourt AMPure XP beads and analyzed by the Fragment Analyzer System as described above. Libraries were diluted to 4 nM, equimolar pooled and sequenced on a MiSeq instrument (Illumina, United States) using the MiSeq Reagent Kit v3 for 600 cycles.
The raw amplicon sequencing data was processed with a modified version of the protocol by Bergkemper et al. (2016a). Briefly, adapter sequences and low quality bases (minimum Phred score: 15) were trimmed and short reads (minimum length: 50 bp) were discarded using AdapterRemoval v2.1.7 (Schubert et al., 2016). Paired reads were merged and chimeras were removed using the qiime dada2 denoise-paired command (N-terminal trimming: 10 bp; C-terminal trimming: 235 and 170 bp for forward and reverse reads, respectively; maximum expected errors: 3) of the DADA2 R package v1.3.4 (Callahan et al., 2016) in Qiime2 v2018.8.01 (Caporaso et al., 2010). The merged reads were used to infer amplicon sequence variants (ASVs) at the same step. Open reading frames (ORFs) were predicted from the merged reads using FragGeneScan v1.19 (Rho et al., 2010) and were subsequently annotated to functions using hmmsearch (HMMER v3.1b2;2) against selected Hidden Markov Models (HMMs) obtained from the TIGRFAM v15.0 database3. Overlapping motifs were removed, results were quality filtered (maximum e-value: 1e-5) and the sequences assigned to the target gene were extracted from the original datasets. The “positive” sequences were taxonomically aligned to the non-redundant NCBI database (January 2018) using Kaiju v1.4.4 (mismatches allowed in greedy mode: 5; Menzel et al., 2016). The datasets can be found in the Sequence Read Archive4.
Diversity calculations and statistical analysis of gcd amplicon sequencing data, as well as data visualization, were performed using R v3.5.2 (R Core Team,, 2015) and RStudio v1.2.5033. Rarefaction curves were produced with the rarecurve command of the “vegan” package (Oksanen et al., 2019); the negative control contained no ASVs. The abundances of ASVs were normalized by calculating relative (%) abundances. Alpha (α−) diversity indices were calculated with the richness and diversity commands of the “microbiome” package (Lahti and Shetty, 2012-2017) and beta (β−) diversity was explored by non-metric multidimensional scaling (NMDS) analysis based on Bray-Curtis dissimilarity calculations with the metaMDS command of package “vegan.” ASVs assigned to the same genus were merged using the “phyloseq” package (McMurdie and Holmes, 2013) and the results were visualized using “phyloseq” and “ggplot2” (Wickham, 2016). Common gcd-harboring ASVs were defined as the ASVs detected in all replicate plots of each treatment and detected using the online tool of Bioinformatics and Evolutionary Genomics group, Gent, Belgium5. The impact of site and girdling treatment on Shannon diversity, was tested per sampling time point by robust 2-way ANOVAs on trimmed means using the t2way command of the “WRS2” package (Mair and Wilcox, 2019). The impact of girdling was further tested by pairwise comparisons between the girdled and the respective control plots individually per sampling point at each site by robust 1-way ANOVAs on trimmed means using the t1way command of the “WRS2” package. Additionally, the impact of site, girdling treatment and sampling time point on the taxonomic profiles of gcd gene datasets was tested by PERMANOVA with 999 permutations using the adonis command of the “vegan” package. The results are summarized in Supplementary Table 1. Differences in the relative abundances of the detected gcd-harboring genera due to site and girdling treatment, were detected by multiple comparisons (robust 1-way ANOVAs on trimmed means, as described above) followed by Benjamini-Hochberg p-value adjustment. In all tests, effects were considered significant when the p-value was <0.05.
For the analysis of the diversity of gcd-harboring PSB, we used a molecular barcoding approach based on sequencing of the gcd gene. Overall, we generated a total of 3.5 Mbp reads which could be assigned to 2,517 different ASVs. Among those, 2,515 ASVs contained ORFs and 2,496 of them had a significant hit against the gcd-specific HMM, confirming the validity of the applied method. Rarefaction analysis revealed sufficient coverage of major ASVs in all datasets (Supplementary Figure 2).
Shannon diversity analysis revealed a significantly higher diversity of gcd-harboring bacteria at the BBR site, compared to LUE, at both time points (p-values = 0.02; Figure 1A, Supplementary Table 1) and a decrease from T1 to T2. At BBR, diversity was slightly lower in the girdled plots compared to the control plots for both time points, while the opposite effect was observed at LUE. However, these effects did not reach statistical significance.
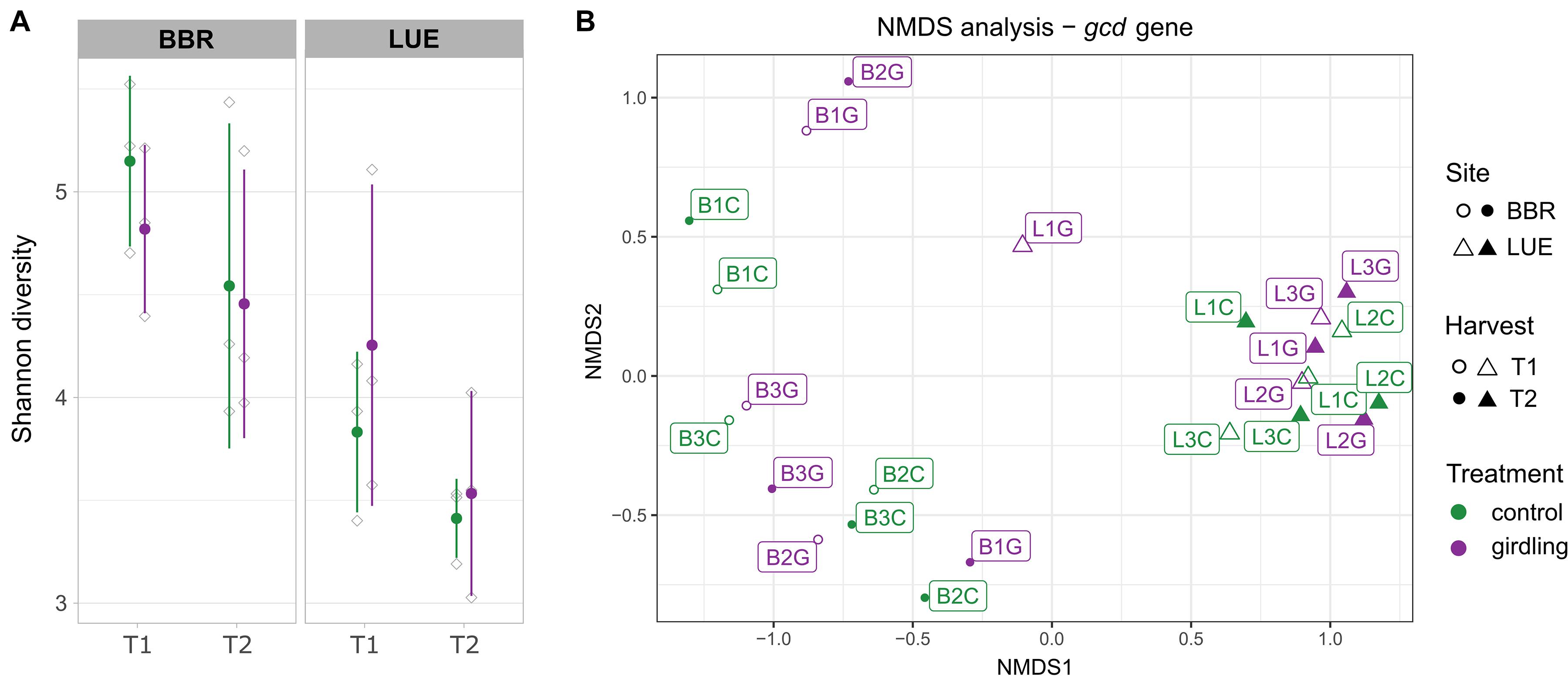
Figure 1. Diversity analysis of the amplified gcd gene based on the detected ASVs. (A) Shannon diversity. Green and purple lines include the samples obtained from the control and girdled plots, respectively. Gray shapes present the individual replicates and error bars the standard deviation between replicates. (B) Non-metric multidimensional scaling analysis (NMDS). The sampled sites, sampling time point and treatments are shown by different shape, filling and color, respectively. Labels indicate the number of the replicate sampled plot.
The comparison of β-diversity revealed that the structure of gcd-harboring bacteria differed between the two sites (p-value < 0.01). Whereas at LUE all samples clustered closely together (with one exception at T1) and neither girdling nor time point had a significant effect on the β-diversity, individual response pattern toward girdling and time point were observed at BBR for each plot. As the responses differed for each plot, an overall assessment of the shifts did not reach significance (Figure 1B and Supplementary Table 1).
At both sites, almost 90% of the obtained ASVs were annotated as putative Proteobacteria belonging to the genera Bradyrhizobium, Dongia, Kaistia, as well as other bacteria of the family Rhizobiaceae, which could not be further assigned to a genus. Moreover, a high proportion of reads linked to putative uncultured Acidobacteria was detected at both sites. However, the relative abundance of the dominant genera differed at both sites (Figure 2A). Whereas at BBR almost 50% of the reads could be assigned to uncultured Acidobacteria and Dongia, ASVs linked to Kaistia were dominating the gcd-harboring bacterial community at LUE. With the exception of Kaistia, differences in the relative abundances of the detected genera between the two sites were not statistically significant, as a result of the huge variability of β-diversity between replicates mostly at BBR (Figure 2B). Girdling influenced the relative abundance of ASVs linked to the dominant genera (Figure 2). At LUE, at both time points, an increase in relative abundance of ASVs linked to Kaistia was observed, which was complemented by a reduced relative abundance of other ASVs linked to putative members of the family Rhizobiaceae, which so far have not been further classified. Differences between the two sampling time points were only detected for ASVs, which were not part of the dominant groups listed above. For example, at the control subplots slight increases in the relative abundance of ASV grouped as Agrobacterium and Pseudomonas was visible, whereas in the subplots subjected to girdling ASVs linked to Paraburkholderia were slightly increased in relative abundance. This shift was also confirmed by a cultivation based approach (Supplementary Figure 3, Supplementary Methods).
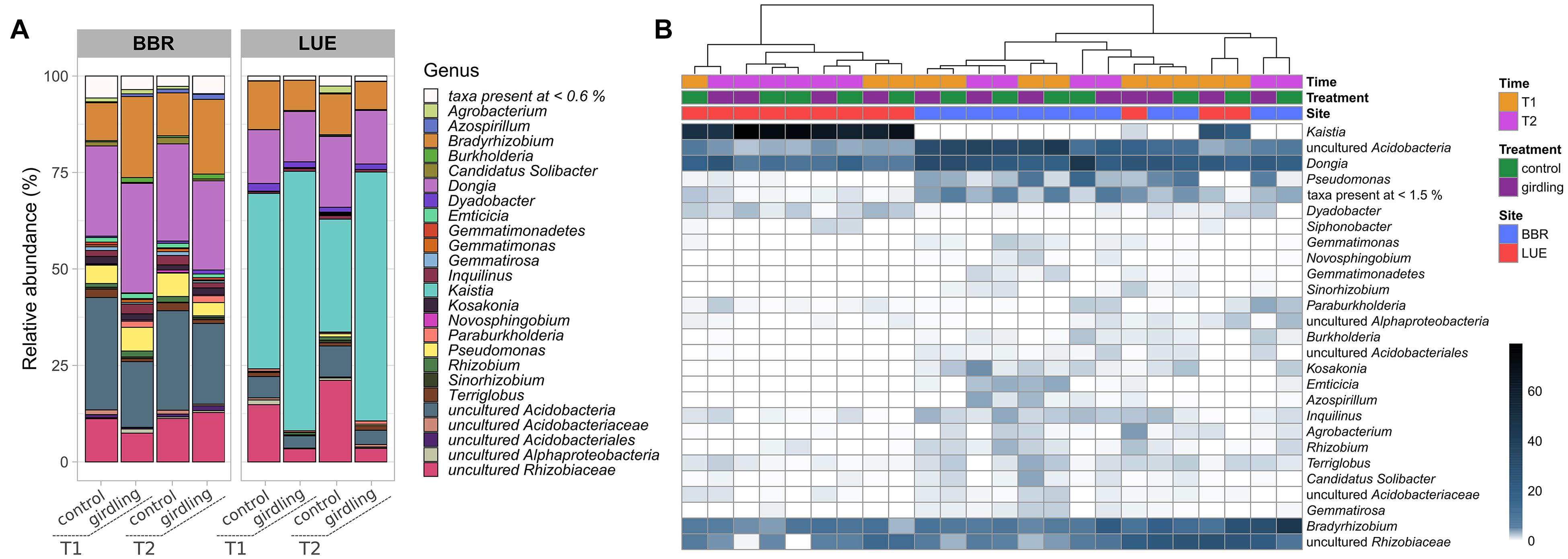
Figure 2. Taxonomic assignment at the genus level of ASVs obtained by gcd gene amplicon sequencing. (A) Stacked barplots of mean relative abundance (%) values. Only genera with >0.6% relative abundance are shown. (B) Heatmap of relative abundances (%) in individual replicate samples clustered based on their profile similarity. Only genera with >1.5% relative abundance are shown.
At BBR mostly ASVs linked to Bradyrhizobium and Burkholderia were affected by girdling, as numbers increased at both time points of sampling as a result of girdling. At the same time, there was a trend to reduced numbers of ASVs belonging to uncultured Acidobacteria. At T1 also ASVs linked to uncultured bacteria of the family Rhizobiaceae were negatively affected. In addition, differences accounting for the sampling time point were observed, which affected mostly the relative abundance of ASVs assigned to Azospirillum independent from the treatment. A time point depending response was observed for ASVs linked to Agrobacterium, which were increased in relative abundance at T2 in the control treatments compared to all other variants.
As the NMDS analysis revealed strong differences between replicate plots, we identified consistent ASVs among the replicate plots (common gcd-harboring ASVs) and compared them between the control and girdling treatments at T1 and T2 (Figure 3). The responses to girdling were more pronounced at BBR compared to LUE, as the number of unaffected ASVs was lower at BBR compared to LUE. Interestingly at both sites the overall number of common gcd-harboring ASVs for the girdled and control subplots, respectively, was lower at T2 compared to T1, indicating an increased fluctuation over time (Figure 3A). Most common gcd-harboring ASVs could be assigned to Dongia at BBR and Dongia and Kaistia at LUE, again confirming the importance of these proteobacterial groups for inorganic P solubilization at both sites. The analysis at the level of individual ASVs revealed differences also for the major bacterial genera which are shared between both sites, differences on the level of ASVs indicating differing species composition e.g., for Dongia for both sites (Figure 3B). In addition, also girdling specific effects mostly at BBR were observed for Dongia, with clear differences in the ASV composition as a result of girdling.
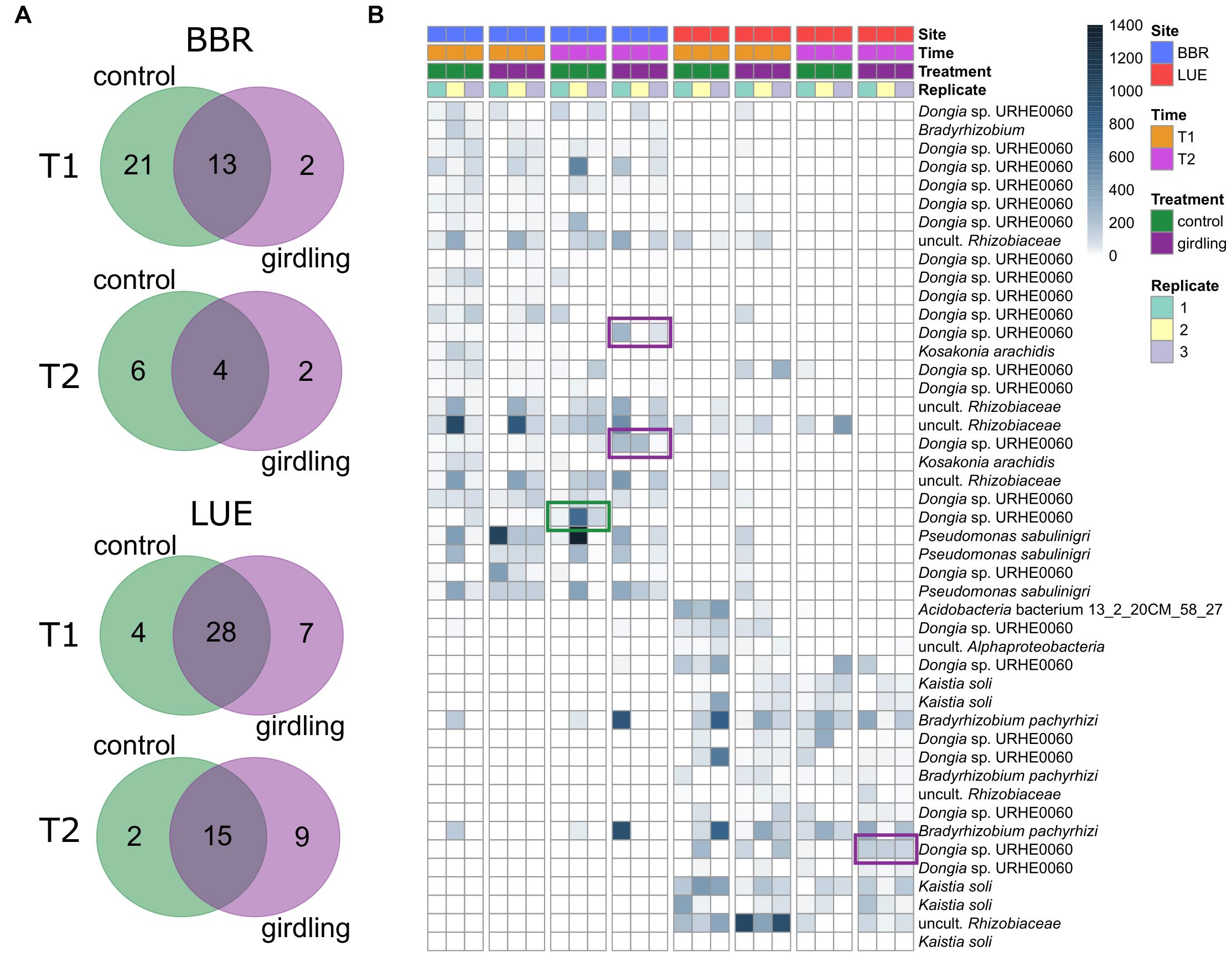
Figure 3. (A) Unique and shared common gcd-harboring ASVs between treatments. Common gcd-harboring ASVs were defined as the ASVs detected in all replicate plots of each treatment. (B) Relative abundance of unique ASVs and their taxonomic affiliation on the species level. The color bar indicates the number of reads assigned to each ASV, after normalization of the datasets. The ASVs that responded 8 weeks after girdling are marked in a green or purple frame.
Clausing et al. (2021) previously investigated the effect of girdling on the plant and soil microbial P mobilization processes at BBR (P-rich) and LUE (P-poor). Following the reduced efflux of dissolved organic C from the roots caused by girdling, plant root biomass and plant P uptake decreased especially at the P-poor site, indicating that the stimulation of gcd-harboring PSB by plant-derived C is an important factor with a strong feedback on plant performance. In this study, we investigated the diversity of gcd-harboring PSB communities as affected by girdling at two sites with contrasting P stocks.
The most abundant genera detected at both sites potentially belonged to the orders Rhizobiales, Rhodospirillales, and the phylum Acidobacteria, which have also been previously detected by both amplicon and shotgun metagenomic sequencing (Bergkemper et al., 2016a,b). Considering that these dominant genera harbor also genes encoding for phosphatases (Weon et al., 2008; Ragot et al., 2015; Kim et al., 2016; Grönemeyer et al., 2017), they can potentially exploit both inorganic and organic P sources depending on the changes in soil C:P stoichiometry. Thus, it is possible that girdling causes significant changes in the P acquisition strategy of gcd-harboring PSB, which may explain partly that the observed changes in the diversity pattern of gcd-harboring PSB were less pronounced than expected, and strong shifts might occur on the level of transcriptomes and associated phenotypes rather than the level of genomes.
However, we observed that the decrease in plant-derived C affected the diversity of gcd-harboring PSB at both sites, but response pattern of gcd-harboring PSB toward girdling were not consistent at both sites. At the P-rich site (BBR), we observed that the overall high diversity of potential gcd-harboring PSB which is present at this site, slightly decreased after the girdling of plants, while the diversity of gcd-harboring PSB at the P-poor site was favored by girdling and the subsequent low plant P uptake, which is in line with the observation of Clausing et al. (2021) who observed an increasing abundance of gcd-harboring PSB (Supplementary Figure 1).
The opposite response pattern at the two sites might be linked to differences in the stoichiometry of available C and P. In this regard, strong competition of beech roots and soil microbes in P-poor compared to P-rich conditions has been shown for LUE (Clausing and Polle, 2020). Here, the diversity and abundance of gcd-harboring PSB at LUE increased as a result of the girdling. The reduced competition of bacteria and plants for P resulted in an increase in diversity despite the reduced available C pools. Changes mostly occurred in the family of Rhizobiaceae, which opposed the seasonal fluctuation observed in the control plots. While some genera of the Rhizobiaceae were negatively impacted, Kaistia benefited from girdling. Whereas many Rhizobiaceae form tight interactions with plants (Alves et al., 2014) and may share with them the low amounts of available P, Kaistia as a mostly free living genus, might directly fight for P with plants, when plants are active in P acquisition as on the control subplots. In fact Kaistia spp. have been detected in various environments, including wastewater soils, river sediments and an anaerobic reactor (Im et al., 2004; Lee et al., 2007; Jin et al., 2012) indicating a large flexibility toward different C sources. Thus, the reduced performance by the plants as a result of girdling may have a negative impact on the plant associated bacteria of the Rhizobiaceae, but free living bacteria of the same family may benefit.
At BBR, where control subplots are characterized by a generally low soil C:P ratio, it might be argued that girdling induces a situation where C becomes the limiting factor and not all gcd-harboring PSB present in the control subplots are capable to tolerate these conditions. However, the reduced relative abundance of Acidobacteria, which can be considered as bacteria with an oligotrophic lifestyle (Kielak et al., 2016) and thus an indicator for reduced stocks of available C, is in contradiction to this postulation. In contrast, we observed an increase of ASVs assigned to Bradyrhizobium, Burkholderia, and Paraburkholderia. The latter was also confirmed by a cultivation based approach (Supplementary Figure 3). Most of these bacteria can form strong interactions with plant roots and have been often considered as part of the plant holobiont (Hassani et al., 2018). However, several Burkholderia species are known to change their lifestyle depending on plant fitness and change their mode of interaction from plant beneficial to plant pathogens (Rojas-Rojas et al., 2019). These shifts might be triggered by the availability and quality of root exudates. As plant growth promoting bacteria, Burkholderia may benefit from root exudates, whereas as plant pathogens they are capable to degrade cell wall compounds like cellulose or lignin to invade the root interior. The ability to degrade lignin has been described for a number of Burkholderia strains (Morya et al., 2019). Thus, at BBR a girdling induced shift in C quality might be more important than total C availability. Because of the high concentrations of available P, microbes, which are capable to degrade more complex organic materials like plant-derived cellulose or lignin, might become dominant members after girdling. As this flexibility in C use is restricted to some bacterial families only, the reduced diversity pattern observed may be a consequence of shifts in C quality. When T1 and T2 are compared, we expect that this is not the endpoint of community changes after girdling as, for example, the common ASVs detected in the control and girdling treatment still decreased (Figure 3). Rasche et al. (2011) observed significant girdling effects even in the second year after girdling. However, long-term girdling effects might be also a consequence of lower fine root biomass or earlier tree senescence, which interferes with the girdling effects (Kaiser et al., 2010). Differences, e.g., in relation to the increase in relative abundance of Agrobacterium at T2, were found. Agrobacterium spp. are known as degraders for bacterial cellulose (Sieger et al., 1995). Thus, their increase in relative abundance might indicate a secondary succession after girdling, where a number of bacteria which benefit from the changed C quality at T1 die off and their cell wall materials are consumed by others, again benefiting from the overall sufficiently available P.
Beyond shifts in the relative abundance of genera also shifts in ASVs belonging to one species were observed. This was mainly true for the dominant genera, including Dongia. This might indicate functional redundancy within the dominant genera, as some ASVs consistently increased in the girdled plots while being absent in the respective control and the other way around. Different preferences of genus ecotypes were also reported previously (Chase et al., 2018), but to prove this mechanism, additional isolation approaches are needed to compare the traits of responding species on whole genome level. Moreover, some bacteria might have obtained the ability to solubilize inorganic P by horizontal gene transfer, as it was demonstrated recently for some proteobacteria like Bradyrhizobium (Liang et al., 2020). The question if the identified groups received their capability for the solubilization of inorganic P by horizontal gene transfer of gcd genes or by coevolution cannot be disentangled by an amplicon sequencing approach, but needs additional isolation experiments, which allow the comparison of the phylogeny based on 16S rRNA genes and for the gcd gene for the same isolate. However, this approach is beyond the scope of this study and also comes along with all limitations related to cultivation experiments like selecting for bacteria preferring copiotrophic conditions, which especially introduced a strong bias for communities isolated from LUE, which is underlined by the low number of different PSB isolates obtained from LUE (Supplementary Figure 3).”
Short-term responses of young-beech trees toward girdling were observed at both sites with different responding taxa. As interactions of girdling and season were not significant, the different response pattern observed could be most likely linked to the different soil C:P ratios at the two sites and the factors limiting microbial performance.
The majority of observed shifts in gcd-harboring PSB community structure were consistent for the two time points. However, this does not imply that no additional long-term changes in gcd-harboring PSB community composition could be observed as a result of girdling after T2. In addition, it needs to be taken into account that the study was performed with relatively young beech trees, which may differ in their exudation pattern as well as strategies to exploit nutrients from trees of older age classes. Furthermore, tree specific effects need to be taken into account despite the fact that bulk soil was sampled, where the direct influence of the trees is limited. But phytoalexins and other compounds are highly persistent in soil and thus might strongly influence all soil microbiome members, including gcd-harboring PSB. Thus, future studies might include different tree ages or species as well as the presence and activity of other PSB that do not harbor the gcd gene, and were consequently excluded from our current analysis.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA694833 https://www.ncbi.nlm.nih.gov/genbank/, MW806976-MW807204.
AP, MSc, MSp, and SS designed the experiment. SC performed the experiment and collected samples. GP performed cultivation and sequenced isolates. MG extracted soil DNA, generated and analyzed qPCR data, prepared amplicon libraries. AC analyzed amplicon sequencing data. AM performed statistical analysis, visualized results, and wrote the manuscript. AM, MSc, MSp, and SS conceptualized the manuscript. All authors contributed to revisions and approved the final manuscript.
The work has been funded by the Deutsche Forschungsgemeinschaft in frame of the Priority Program SPP1685 “Ecosystem Nutrition” (SCHL 446/20-2, SCHU 2907/3-2, PO362/22-2, and SP1389/5-2).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer, AM, declared a shared affiliation, though no other collaboration, with the authors, SC and AP, to the handling editor.
The authors are grateful to Susanne Kublik for sequencing the amplicon libraries.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffgc.2021.696983/full#supplementary-material
Supplementary Figure 1 | Absolute counts of gcd copies g−1 of dry soil as also shown previously in Clausing et al. (2021). Green and purple bars include the samples obtained from the control and girdled plots, respectively. Errors bars present the standard deviation between replicates. Statistically significant differences between plots with girdled trees and the respective controls are presented by *.
Supplementary Figure 2 | Constructed rarefaction curves of the sequenced gcd gene libraries, showing the number of ASVs detected by means of sequencing depth.
Supplementary Figure 3 | (A) Relative number (%) of PSB cultures to total number of cultures obtained by cultivation on Pikovskaya’s agar. Green and purple bars include the samples obtained from the control and plots with girdled trees, respectively. Error bars present the standard deviation between replicates. Statistically significant differences between girdled and the respective controls are presented by *. (B) Taxonomic assignment of the isolated PSB cultures at the genus level.
Supplementary Table 1 | Statistical analysis of the copy numbers, Shannon diversity and taxonomic profiles of amplified gcd genes, as well as the relative number of PSB cultures. When applicable, the test statistics (Q and F. model), degrees of freedom (df) and the respective p-values are presented. Significant differences (p-value < 0.05) are marked with orange color.
Supplementary Methods | Description of the isolation and characterization of PSB based on cultivation on Pikovskaya agar.
Alori, E. T., Glick, B. R., and Babalola, O. O. (2017). Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 8:971. doi: 10.3389/fmicb.2017.00971
Alves, L. M. C., de Souza, J. A. M., de Mello Varani, A., and de Macedo Lemos, E. G. (2014). “The Family Rhizobiaceae,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson, (Berlin: Springer), 419–437.
Bergkemper, F., Kublik, S., Lang, F., Krüger, J., Vestergaard, G., Schloter, M., et al. (2016a). Novel oligonucleotide primers reveal a high diversity of microbes which drive phosphorus turnover in soil. J. Microbiol. Methods 125, 91–97. doi: 10.1016/j.mimet.2016.04.011
Bergkemper, F., Schöler, A., Engel, M., Lang, F., Krüger, J., Schloter, M., et al. (2016b). Phosphorus depletion in forest soils shapes bacterial communities towards phosphorus recycling systems. Environ. Microbiol. 18, 1988–2000. doi: 10.1111/1462-2920.13188
Callahan, B. J., Mcmurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Caporaso, J., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336.
Chase, A. B., Gomez-Lunar, Z., Lopez, A. E., Li, J., Allison, S. D., Martiny, A. C., et al. (2018). Emergence of soil bacterial ecotypes along a climate gradient. Environ. Microbiol. 20, 4112–4126. doi: 10.1111/1462-2920.14405
Clausing, S., Pena, R., Song, B., Müller, K., Mayer-Gruner, P., Marhan, S., et al. (2021). Carbohydrate depletion in roots impedes phosphorus nutrition in young forest trees. New Phytol. 229, 2611–2624. doi: 10.1111/nph.17058
Clausing, S., and Polle, A. (2020). Mycorrhizal Phosphorus Efficiencies and Microbial Competition Drive Root P Uptake. Front. For. Glob. Change 3:54. doi: 10.3389/ffgc.2020.00054
Grönemeyer, J. L., B nger, W., and Urek, B. (2017). Bradyrhizobium namibiense sp. nov., a symbiotic nitrogen-fixing bacterium from root nodules of lablab purpureus, hyacinth bean, in Namibia. Int. J. Syst. Evol. Microbiol. 67, 4884–4891.
Goldstein, A. H. (1995). Recent progress in understanding the molecular genetics and biochemistry of calcium phosphate solubilization by gram negative bacteria. Biol. Agric. Hortic. 12, 185–193. doi: 10.1080/01448765.1995.9754736
Goldstein, A. H., Rogers, R. D., and Mead, G. (1993). Mining by Microbe: separating phosphate from ores via bioprocessing. Nat. Biotechnol. 11, 1250–1254. doi: 10.1038/nbt1193-1250
Hassani, M. A., Durán, P., and Hacquard, S. (2018). Microbial interactions within the plant holobiont. Microbiome 6:58. doi: 10.1186/s40168-018-0445-0
Högberg, P., Nordgren, A., Buchmann, N., Taylor, A. F. S., Ekblad, A., Högberg, M. N., et al. (2001). Large-scale forest girdling shows that current photosynthesis drives soil respiration. Nature 411, 789–792. doi: 10.1038/35081058
Im, W.-T., Yokota, A., Kim, M.-K., and Lee, S.-T. (2004). Kaistia adipata gen. nov., sp. nov., a novel.ALPHA.-proteobacterium. J. Gen. Appl. Microbiol. 50, 249–254. doi: 10.2323/jgam.50.249
Jin, L., Kim, K. K., Lee, H. G., Ahn, C. Y., and Oh, H. M. (2012). Kaistia defluvii sp. nov., isolated from river sediment. Int. J. Syst. Evol. Microbiol. 62, 2878–2882. doi: 10.1099/ijs.0.038687-0
Kaiser, C., Kilburn, M. R., Clode, P. L., Fuchslueger, L., Koranda, M., Cliff, J. B., et al. (2015). Exploring the transfer of recent plant photosynthates to soil microbes: mycorrhizal pathway vs direct root exudation. New Phytol. 205, 1537–1551. doi: 10.1111/nph.13138
Kaiser, C., Koranda, M., Kitzler, B., Fuchslueger, L., Schnecker, J., Schweiger, P., et al. (2010). Belowground carbon allocation by trees drives seasonal patterns of extracellular enzyme activities by altering microbial community composition in a beech forest soil. New Phytol. 187, 843–858. doi: 10.1111/j.1469-8137.2010.03321.x
Khorassani, R., Hettwer, U., Ratzinger, A., Steingrobe, B., Karlovsky, P., and Claassen, N. (2011). Citramalic acid and salicylic acid in sugar beet root exudates solubilize soil phosphorus. BMC Plant Biol. 11:121. doi: 10.1186/1471-2229-11-121
Kielak, A. M., Barreto, C. C., Kowalchuk, G. A., van Veen, J. A., and Kuramae, E. E. (2016). The ecology of Acidobacteria: moving beyond genes and genomes. Front. Microbiol. 7:744. doi: 10.3389/fmicb.2016.00744
Kim, D. U., Lee, H., Kim, H., Kim, S. G., and Ka, J. O. (2016). Dongia soli sp. nov., isolated from soil from Dokdo. Korea. Antonie Van Leeuwenhoek 109, 1397–1402.
Kleinsteuber, S., Schleinitz, K. M., Breitfeld, J., Harms, H., Richnow, H. H., Vogt, C., et al. (2008). Molecular characterization of bacterial communities mineralizing benzene under sulfate-reducing conditions. FEMS Microbiol. Ecol. 66, 143–157. doi: 10.1111/j.1574-6941.2008.00536.x
Koranda, M., Schnecker, J., Kaiser, C., Fuchslueger, L., Kitzler, B., Stange, C. F., et al. (2011). Microbial processes and community composition in the rhizosphere of european beech - the influence of plant C exudates. Soil Biol. Biochem. 43, 551–558. doi: 10.1016/j.soilbio.2010.11.022
Kurth, J. K., Albrecht, M., Karsten, U., Glaser, K., Schloter, M., and Schulz, S. (2020). Correlation of the abundance of bacteria catalyzing phosphorus and nitrogen turnover in biological soil crusts of temperate forests of Germany. Biol. Fertil. Soils 57, 1–14.
Kuzyakov, Y., Hill, P. W., and Jones, D. L. (2007). Root exudate components change litter decomposition in a simulated rhizosphere depending on temperature. Plant Soil 290, 293–305. doi: 10.1007/s11104-006-9162-8
Lahti, L., and Shetty, S. (2012-2017). microbiome R package. Available online at: http://microbiome.github.io (accessed October 30, 2019).
Lang, F., Krüger, J., Amelung, W., Willbold, S., Frossard, E., Bünemann, E. K., et al. (2017). Soil phosphorus supply controls P nutrition strategies of beech forest ecosystems in Central Europe. Biogeochemistry 136, 5–29. doi: 10.1007/s10533-017-0375-0
Lee, H. W., Yu, H. S., Liu, Q. M., Jung, H. M., An, D. S., Im, W. T., et al. (2007). Kaistia granuli sp. nov., isolated from anaerobic granules in an upflow anaerobic sludge blanket reactor. Int. J. Syst. Evol. Microbiol. 57, 2280–2283. doi: 10.1099/ijs.0.65023-0
Liang, J. L., Liu, J., Jia, P., Yang, T. T., Zeng, Q. W., Zhang, S. C., et al. (2020). Novel phosphate-solubilizing bacteria enhance soil phosphorus cycling following ecological restoration of land degraded by mining. ISME J. 14, 1600–1613. doi: 10.1038/s41396-020-0632-4
Long, X. E., Yao, H., Huang, Y., Wei, W., and Zhu, Y. G. (2018). Phosphate levels influence the utilisation of rice rhizodeposition carbon and the phosphate-solubilising microbial community in a paddy soil. Soil Biol. Biochem. 118, 103–114. doi: 10.1016/j.soilbio.2017.12.014
Lueders, T., Manefield, M., and Friedrich, M. W. (2004). Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6, 73–78. doi: 10.1046/j.1462-2920.2003.00536.x
Mair, P., and Wilcox, R. (2019). Robust statistical methods in R using the WRS2 package. Behav. Res. Methods 52, 464–488. doi: 10.3758/s13428-019-01246-w
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Meier, I. C., Finzi, A. C., and Phillips, R. P. (2017). Root exudates increase N availability by stimulating microbial turnover of fast-cycling N pools. Soil Biol. Biochem. 106, 119–128. doi: 10.1016/j.soilbio.2016.12.004
Menzel, P., Ng, K. L., and Krogh, A. (2016). Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 7:11257.
Morya, R., Kumar, M., Singh, S. S., and Thakur, I. S. (2019). Genomic analysis of Burkholderia sp. ISTR5 for biofunneling of lignin-derived compounds. Biotechnol. Biofuels 12:277. doi: 10.1186/s13068-019-1606-5
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2019). Vegan: Community Ecology Package. Available online at: https://cran.r-project.org/package=vegan (accessed April 09, 2020).
Pastore, G., Kernchen, S., and Spohn, M. (2020). Microbial solubilization of silicon and phosphorus from bedrock in relation to abundance of phosphorus-solubilizing bacteria in temperate forest soils. Soil Biol. Biochem. 151:108050. doi: 10.1016/j.soilbio.2020.108050
Pena, R., Offermann, C., Simon, J., Naumann, P. S., Geßler, A., Holst, J., et al. (2010). Girdling affects ectomycorrhizal fungal (EMF) diversity and reveals functional differences in EMF community composition in a beech forest. Appl. Environ. Microbiol. 76, 1831–1841. doi: 10.1128/aem.01703-09
R Core Team (2015). R: A Language And Environment For Statistical Computing. Vienna: R foundation for statistical computing.
Ragot, S. A., Kertesz, M. A., and B nemann, E. K. (2015). phoD alkaline phosphatase gene diversity in soil. Appl. Environ. Microbiol. 81, 7281–7289.
Rasche, F., Knapp, D., Kaiser, C., Koranda, M., Kitzler, B., Zechmeister-Boltenstern, S., et al. (2011). Seasonality and resource availability control bacterial and archaeal communities in soils of a temperate beech forest. ISME J. 5, 389–402. doi: 10.1038/ismej.2010.138
Rasul, M., Yasmin, S., Suleman, M., Zaheer, A., Reitz, T., Tarkka, M. T., et al. (2019). Glucose dehydrogenase gene containing phosphobacteria for biofortification of Phosphorus with growth promotion of rice. Microbiol. Res. 223–225, 1–12. doi: 10.1016/j.micres.2019.03.004
Rho, M., Tang, H., and Ye, Y. (2010). FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 38:e191. doi: 10.1093/nar/gkq747
Rodríguez, H., and Fraga, R. (1999). Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 17, 319–339. doi: 10.1016/s0734-9750(99)00014-2
Rojas-Rojas, F. U., López-Sánchez, D., Meza-Radilla, G., Méndez-Canarios, A., Ibarra, J. A., and Estrada-de los Santos, P. (2019). The controversial Burkholderia cepacia complex, a group of plant growth promoting species and plant, animals and human pathogens. Rev. Argent. Microbiol. 51, 84–92.
Sashidhar, B., and Podile, A. R. (2010). Mineral phosphate solubilization by rhizosphere bacteria and scope for manipulation of the direct oxidation pathway involving glucose dehydrogenase. J. Appl. Microbiol. 109, 1–12. doi: 10.1111/j.1365-2672.2009.04654.x
Satyaprakash, M., Nikitha, T., Reddi, E. U. B., Sadhana, B., and Satya Vani, S. (2017). Phosphorous and Phosphate Solubilising Bacteria and their Role in Plant Nutrition. Int. J. Curr. Microbiol. Appl. Sci. 6, 2133–2144. doi: 10.20546/ijcmas.2017.604.251
Schubert, M., Lindgreen, S., and Orlando, L. (2016). AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res. Notes 9:88. doi: 10.1186/s13104-016-1900-2
Sharma, S. B., Sayyed, R. Z., Trivedi, M. H., and Gobi, T. A. (2013). Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus 2:587.
Shintu, P. V., and Jayaram, K. M. (2015). Phosphate solubilising bacteria (Bacillus polymyxa)-An effective approach to mitigate drought in tomato (Lycopersicon esculentum Mill.). Trop. Plant Res. 2, 17–22.
Sieger, C. H. N., Kroon, A. G. M., Batelaan, J. G., and van Ginkel, C. G. (1995). Biodegradation of carboxymethyl celluloses by Agrobacterium CM-1. Carbohydr. Polym. 27, 137–143. doi: 10.1016/0144-8617(95)00039-A
Spohn, M., Ermak, A., and Kuzyakov, Y. (2013). Microbial gross organic phosphorus mineralization can be stimulated by root exudates - A 33P isotopic dilution study. Soil Biol. Biochem. 65, 254–263. doi: 10.1016/j.soilbio.2013.05.028
Spohn, M., Zeißig, I., Brucker, E., Widdig, M., Lacher, U., and Aburto, F. (2020). Phosphorus solubilization in the rhizosphere in two saprolites with contrasting phosphorus fractions. Geoderma 366:114245. doi: 10.1016/j.geoderma.2020.114245
Tawaraya, K., Naito, M., and Wagatsuma, T. (2006). Solubilization of insoluble inorganic phosphate by hyphal exudates of arbuscular mycorrhizal fungi. J. Plant Nutr. 29, 657–665. doi: 10.1080/01904160600564428
Vives-Peris, V., de Ollas, C., Gómez-Cadenas, A., and Pérez-Clemente, R. M. (2020). Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep. 39, 3–17. doi: 10.1007/s00299-019-02447-5
Weon, H. Y., Lee, C. M., Hong, S. B., Kim, B. Y., Yoo, S. H., Kwon, S. W., et al. (2008). Kaistia soli sp. nov., isolated from a wetland in Korea. Int. J. Syst. Evol. Microbiol. 58, 1522–1524.
Yang, Y., Wang, N., Guo, X., Zhang, Y., and Ye, B. (2017). Comparative analysis of bacterial community structure in the rhizosphere of maize by high-throughput pyrosequencing. PLoS One 12:e0178425. doi: 10.1371/journal.pone.0178425
Zeller, B., Liu, J., Buchmann, N., and Richter, A. (2008). Tree girdling increases soil N mineralisation in two spruce stands. Soil Biol. Biochem. 40, 1155–1166. doi: 10.1016/j.soilbio.2007.12.009
Keywords: beech, gcd gene, girdling, P solubilizing bacteria, quinoprotein glucose dehydrogenase
Citation: Michas A, Pastore G, Chiba A, Grafe M, Clausing S, Polle A, Schloter M, Spohn M and Schulz S (2021) Phosphorus Availability Alters the Effect of Tree Girdling on the Diversity of Phosphorus Solubilizing Soil Bacterial Communities in Temperate Beech Forests. Front. For. Glob. Change 4:696983. doi: 10.3389/ffgc.2021.696983
Received: 18 April 2021; Accepted: 07 June 2021;
Published: 29 June 2021.
Edited by:
Sebastian Loeppmann, Christian-Albrechts-Universität zu Kiel, GermanyReviewed by:
Bang-Xiao Zheng, University of Helsinki, FinlandCopyright © 2021 Michas, Pastore, Chiba, Grafe, Clausing, Polle, Schloter, Spohn and Schulz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefanie Schulz, c3RlZmFuaWUuc2NodWx6QGhlbG1ob2x0ei1tdWVuY2hlbi5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.