- 1Southern Swedish Forest Research Centre, Swedish University of Agricultural Sciences, Alnarp, Sweden
- 2Department of Organismal Biology, Systematic Biology, Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden
For the last two decades, large-scale population decline of European ash (Fraxinus excelsior) has occurred in Europe because of the introduction of the alien fungal pathogen, Hymenoscyphus fraxineus, from East Asia. Since European ash is a keystone species having critical importance for biodiversity, and only a small percentage of the ash population appears to show some tolerance against the pathogen, the loss of ash trees means that other associated organisms, especially those with high or obligate associations to ash, are at risk of further species declines. In this study, we used high throughput DNA sequencing and multivariate analysis to characterize: (i) the mycobiome in aerial tissues (i.e., leaf, bark, and xylem) of ash trees showing different phenotypic response to ash dieback, (ii) the temporal variation in fungal communities across the growing season, and (iii) the similarity in fungal community structure between ash and other common trees species that may serve as an ecological niche substitute for ash microfungi. Results showed that fungal communities differed among the three tissue types, susceptibility classes, in time and between sites. Trophic analysis of functional groups using the FUNGuild tool indicated a higher presence of pathotrophic fungi in leaves than in bark and xylem. The share of pathotrophic fungi increased along a gradient of low to high disease susceptibility in both bark and xylem tissue, while the proportion of symbiotrophic fungi correspondingly decreased in both tissue types. Neighboring, alternative host trees did not share all the fungal species found in ash, however, most microfungi uniquely associated to ash in this study are generalists and not strictly host specific. The progressive disappearance of ash trees on the landscape imposes a high risk for extinction of Red-listed macrofungal species, and breeding for resistance against ash dieback should help sustain important biodiversity associated to ash. Microfungal diversity though may be less prone to such demise since most ash-associated endophytes appear to occur on a broad range of host species.
Introduction
Common ash (Fraxinus excelsior L.) is an important keystone species in European forests (Bell et al., 2008; Pautasso et al., 2013). At present, common ash populations in Europe are threatened by the alien invasive pathogen Hymenoscyphus fraxineus (T. Kowalski) Baral, Queloz, Hosoya, an ascomycete fungus and the primary causal agent of ash dieback. Hymenoscyphus fraxineus originates from Asia (Gross et al., 2014; Inoue et al., 2019) and was likely introduced to Europe through an imported nursery stock (Woodward and Boa, 2013). The pathogen spreads via wind-dispersed ascospores (Timmermann et al., 2011; Hietala et al., 2013; Chandelier et al., 2014) and infects trees primarily through the leaves and rachises (Cleary et al., 2013). Root collar infections are also common (Husson et al., 2012; Enderle et al., 2013; Chandelier et al., 2016).
In Sweden, ash has been a Red-listed species since 2010 and recently its status has worsened to become critically endangered, considered at risk of extinction in the wild (Cleary et al., 2017). Research has shown strong genotypic variation in susceptibility in natural populations of ash throughout Europe (Kjaer et al., 2011; McKinney et al., 2011; Pliura et al., 2011; Stener, 2013; Lobo et al., 2014; Stocks et al., 2017; Timmermann et al., 2017). Though the large majority of ash trees are affected (McKinney et al., 2014), a small percentage (between 1 and 5%) of the ash population shows tolerance to the pathogen (Pliura et al., 2011; Kirisits and Freinschlag, 2012). With such low proportion of ash trees projected to survive this epidemic, the fate of other associated biodiversity will likely be negatively impacted (Pautasso et al., 2013). Ecological cascade effects have occurred with the decline of other tree species affected by invasive alien pathogens (e.g., Ulmus spp. affected by Dutch elm disease) (Brasier, 1991) and can be similarly anticipated for ash ecosystems with trophic cascades on biodiversity and natural cycles (i.e., forest succession, nutrient- and carbon cycles) (Pautasso et al., 2013; Mitchell et al., 2014). Associated biodiversity, e.g., insects, bryophytes, lichens, and fungi, having high- or obligate associations with ash, or both, may suffer substantial population decline because of the declining ash tree population (Mitchell et al., 2014; Hultberg et al., 2020). An even larger threat for local extinction are Red-listed host-specific organisms such as the Tabby knot horn (Euzophera pinguis Haworth), the scarce fritillary (Euphydryas maturna L.) butterfly, the jewel beetle (Agrilus convexicollis Redtenbacher), the epiphytic bryophyte Neckera pennata (Hedw.), and macrofungi including Pyrenula nitidella (Flörke ex Schaer.) Müll. Arg. and Perenniporia fraxinea (Bull.) Ryvarden (Jönsson and Thor, 2012; Artdatabanken, 2015; Bengtsson and Stenström, 2017). Another important group of organisms are the so-called “hidden biodiversity,” including microfungi inhabiting plants as endophytes that are often known to benefit their hosts by enhancing their resistance to abiotic and biotic stresses (Faeth and Fagan, 2002; Redman et al., 2002; Arnold, 2007; Rodriguez et al., 2009; Tellenbach and Sieber, 2012). However, because microfungi cannot be detected through traditional survey methods (Halme et al., 2012), the implications of H. fraxineus on the loss of ash-associated microfungi is generally unknown.
The resource use defines the niche of the species and according to the competitive exclusion theory, two species with highly similar or identical niches cannot stably coexist in the same physical space for long periods of time if their common resources are limiting (Chase and Leibold, 2003; Mikkelson, 2005). Thus, to stably co-exist in the same host, two species must sufficiently utilize different resources to avoid competitive exclusion. The aggressiveness of H. fraxineus may therefore displace other microfungi competing for the same ecological niche. For example, it was already demonstrated that Hymenoscyphus albidus (Roberge ex Desm.) W. Phillips, an obligate, native endophyte/saprophyte of F. excelsior leaves (Kowalski and Holdenrieder, 2009) lost the niche competition race against H. fraxineus whereby local extirpation of H. albidus has occurred due to competitive exclusion (McKinney et al., 2012). Similar outcomes may be expected for other host-specific fungal species that otherwise may have an important role for the host in those ecosystems (as with H. albidus, which was a common degrader of fallen leaf rachises). For this reason, interest has surged in studying the fungal communities of ash to better understand the ecological role of Hymenoscyphus species and other endophytic fungi, their interactions and influence of the fungal communities in enhancing plant fitness and resistance.
Several studies have focused on describing fungal communities of Fraxinus species on either leaves (Scholtysik et al., 2013; Cross et al., 2017; Ibrahim et al., 2017; Trapiello et al., 2017; Schlegel et al., 2018) or twigs (Haňáčková et al., 2017; Kosawang et al., 2018), but not both. With the exception of Cross et al. (2017), the aforementioned studies focused on the culturable fraction of the endophytic fungal community. The main aim of this study was to characterize the ash mycobiome in different tissue types, across a gradient of phenotypic susceptibility to the disease, and to examine how the fungal communities are influenced over time and space. A secondary aim was to compare the similarity in fungal community composition between ash and other tree species commonly growing in the surrounding environment to determine whether they may serve as an ecological niche substitute for some microfungi associated to ash.
Materials and Methods
Study Location and Sampling Methods
Two ash seed orchards located at Snogeholm (55°32′N, 13°32′E, 50 m a.s.l.) and Trolleholm (55°57′N, 13°12′E, 100 m a.s.l.) in southern Sweden were chosen for examining fungal communities of ash phenotypes. The trials were established in 1992 (Snogeholm) and 1995 (Trolleholm) on former agricultural land, prior to the arrival of H. fraxineus in Sweden (first report in 2001). Each site comprised 106 F. excelsior clones that were phenotypically selected from plus-trees from 27 stands across southern Sweden, mainly selected for growth traits as reforestation material. Damage assessments previously conducted between 2006 and 2016 revealed large genotypic variation in susceptibility to H. fraxineus among ash clones, that damage was largely under genetic control (Stener, 2013), and there is stability in damage traits among clones over time (Stener, 2018).
The tree genotypes selected in the study were based on the previous screening in this long-term genetic trial, and represented a gradient of susceptibility from low to high based on rankings of tree vitality and shoot and crown damage in the previous assessments on a scale from 0 to 9, whereby 0 means no damage and 9 highly damaged (Stener, 2013). Up to seven trees (per individual genotype) were sampled in three susceptibility classes: resistant, intermediate and susceptible (Table 1 and Figure 1). Resistant trees had a healthy appearance of the crown with little or no evidence of crown or branch dieback and an average damage class less than 2.5. Intermediate trees exhibited damage in the form of partial crown dieback and few dead branches, and an average damage class rating between 3.5 and 4.5. Highly susceptible trees had severe crown dieback, several branches had died, and overall status moribund, with an average damage class rating higher than 5.5. Additional tree species were selected among the trees growing close to, or within, the seed orchards: silver birch (Betula pendula, Roth) and pedunculate oak (Quercus robur, L.) at Snogeholm, and European beech (Fagus sylvatica, L.), pedunculate oak, and hybrid larch (Larix x eurolepis, Mill.) at Trolleholm (Supplementary Table S1).
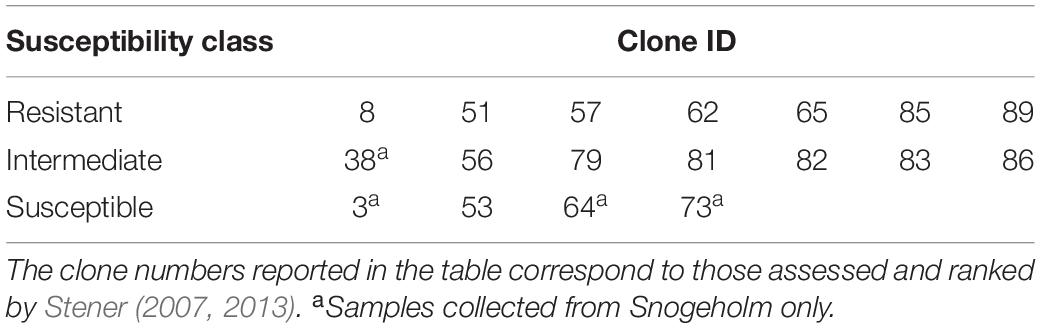
Table 1. Ash clones sampled within each susceptibility class at Snogeholm and Trolleholm seed orchards.

Figure 1. Examples of ash trees representing three susceptibility classes: Resistant (R), clone 89 at Snogeholm; Intermediate (I), clone 79 at Trolleholm; and Susceptible (S), clone 73 at Snogeholm. Photos: Marta Agostinelli.
Trees were sampled at four different time points throughout the growing season in 2015: (i) 28th of May (prior to the sporulation period of H. fraxineus), (ii) 21st of July, (iii) 13th of August (during the peak period of H. fraxineus sporulation), and (iv) 17th of September (during the presumed early infection period of H. fraxineus, prior to leaf senescence and leaf shed). At each time point, a visually healthy branch was selected from each sampled tree and two individuals of each clone were cut depending on available material. The same trees were visited at each time point. Leaves were detached from the shoot, placed in a plastic bag and labeled accordingly. Phloem (bark) and xylem tissue were separated for the shoots, and individually labeled. Both shoots and leaves were stored immediately on dry ice in the field. In total, 384 ash samples and 60 samples from neighboring tree species were collected, for a total of 444 samples (Supplementary Table S1). In the lab, samples were transferred to a −20°C freezer until they could be processed further. Then, leaflets were removed from the rachises and placed in 50 mL Falcon tubes. Samples were then lyophilized for 24 h and homogenized in a FastPrep-24 homogenizer (MP biomedicals, Santa Ana, United States). Bark and xylem were separated with a sterile scalpel and placed in two separate 25 mL Falcon tubes, then lyophilized for at least 48 h. Liquid nitrogen was used to homogenize any remaining phloem and xylem to fine powder.
DNA Extraction and Illumina MiSeq Sequencing
Genomic DNA was extracted using the E.Z.N.A. SP Plant DNA kit (Omega Bio-Tek, Inc., Norcross, United States) following manufacturer’s instructions. DNA quantity and quality were measured using NanoDrop® ND-1000 spectrophotometer (Wilmington, United States). The ITS2 region of the rDNA, a suitable marker for barcoding fungi from environmental samples (Ihrmark et al., 2012) was amplified. PCR was performed in 50 μL reactions containing 10 μL of 5X Phusion HF buffer, 0.2 mM dNTP mix, 0.3 μM of tagged primer ITS4 (White et al., 1990), 0.5 μM of tagged primer fITS7 (Ihrmark et al., 2012), 1.5 mM MgCl2, 0.02 Unit of Phusion Hot Start High-Fidelity DNA polymerase (Thermo Fisher Scientific, San Jose, United States) and 10 ng genomic DNA. Amplifications were performed in a S1000 thermal cycler (BIO-RAD, San Francisco, United States) with the following conditions: 98°C for 3 min, 31 cycles of 98°C for 15 s, 57°C for 30 s, and 72°C for 30 s, and a final extension step for 7 min at 72°C. PCR products were visualized on a 1.5% agarose gel stained with GelRed (Biotium, Hayward, CA, United States). Amplified PCR products were cleaned using Agencourt Ampure XP beads (Beckman Coulter) and pooled in an equimolar ratio to yield three sequencing libraries. After excluding some samples due to poor amplification (Supplementary Table S1), the sample set comprised 295 samples representing ash trees from different susceptibility classes and neighboring tree species across a variety of tissue types, time points, and sampling sites. Samples were sent to National Genomics Infrastructure/NGI Stockholm (Science for Life Laboratory, Stockholm, Sweden) for ThruPLEX library preparation (Rubicon Genetics, Ann Arbor, MI, United States) and sequencing on the Illumina MiSeq (Illumina, Inc., San Diego, CA, United States) on separate runs to generate paired-end reads (2 × 300 bp).
Bioinformatics and Sequence Processing
The QIIME (version 1.9.1) bioinformatics pipeline (Caporaso et al., 2010) was used to perform mycobiome analysis from raw DNA sequencing data. After checking the number of reads, read length, and sequence read orientation, quality control and quality trimming of the sequences were performed using FastQC version 0.11.5 (default settings) and Trimmomatic version 0.32 (settings: LEADING:3, TRAILING:3, SLIDINGWINDOW:4:15, MINLEN:100), respectively. Reads were then merged, barcodes extracted, and reads reoriented. Samples were then demultiplexed and reads were pooled together. Primers were then removed using the toolkit software seqtk (v1.0-r31). Chimera were detected and removed using USEARCH6.1, and global singletons were removed. Sequence reads were clustered into operational taxonomic units (OTUs) at 97% sequence identity using USEARCH as implemented in QIIME. The QIIME formatted rDNA ITS sequence UNITE database (Abarenkov et al., 2010) was used for the taxonomic assignment (release version 7.1, 2016-11-20). One representative sequence from each OTU cluster was chosen and the closest taxonomic affiliation was determined via comparison to the QIIME-implemented UNITE database that includes sequences from INSD (International Nucleotide Sequence Databases: NCBI, EMBL, DDBJ). Finally, four samples with low number of reads (i.e., < 10,000) were removed.
Statistical Analysis
Statistical analyses were performed on OTUs assigned to fungal taxa. OTUs with fewer than 10 reads globally were excluded from further analyses in order to reduce errors and artifacts introduced by the USEARCH procedure. Relative read abundance per phylum was calculated as the number of reads for phylum divided by the total number of reads in the dataset for three types of samples: all samples, ash samples only, and neighboring tree species grouped together. Rarefaction curves for host species, tissue type, and susceptibility class were generated in the software EstimateS 9.1.0 (R. K. Colwell)1 to assess sampling efficacy. The statistical analyses were conducted in R 3.2.2 (R Development Core Team, 2016).
Fungal diversity was analyzed using Hill numbers, a diversity metric for microbial sequence data from complex communities (Jost, 2006; Haegeman et al., 2013) and has previously been implemented for fungal community sequence data (Bálint et al., 2015; Nguyen D. et al., 2016). Hill numbers can be used to measure and incorporate richness (Hill 0), exponent of Shannon Index (Hill 1) and inverse Simpson (Hill 2) (Legendre and Legendre, 1998). The interpretation of the Hill numbers, as represented by Renyi diversities, is that a community is more diverse if all of the numbers are higher than in another community. Fungal diversity was assessed at different levels: (i), among tissue types (leaf, bark, xylem), (ii) among susceptibility classes (resistant, intermediate, susceptible), (iii) among individual ash clones, and (iv) among the four sampling points during the growing season. Linear models were used to explain Hill 0, Hill 1, and Hill 2, respectively, after accounting for differential sequencing depth in different samples with the square root of total read numbers per sample. The partial residuals of the Hill numbers, again after accounting for differential sequencing depth, among tissue types, susceptibility classes, clones and sampling time points, respectively, were compared with Tukey’s HSD.
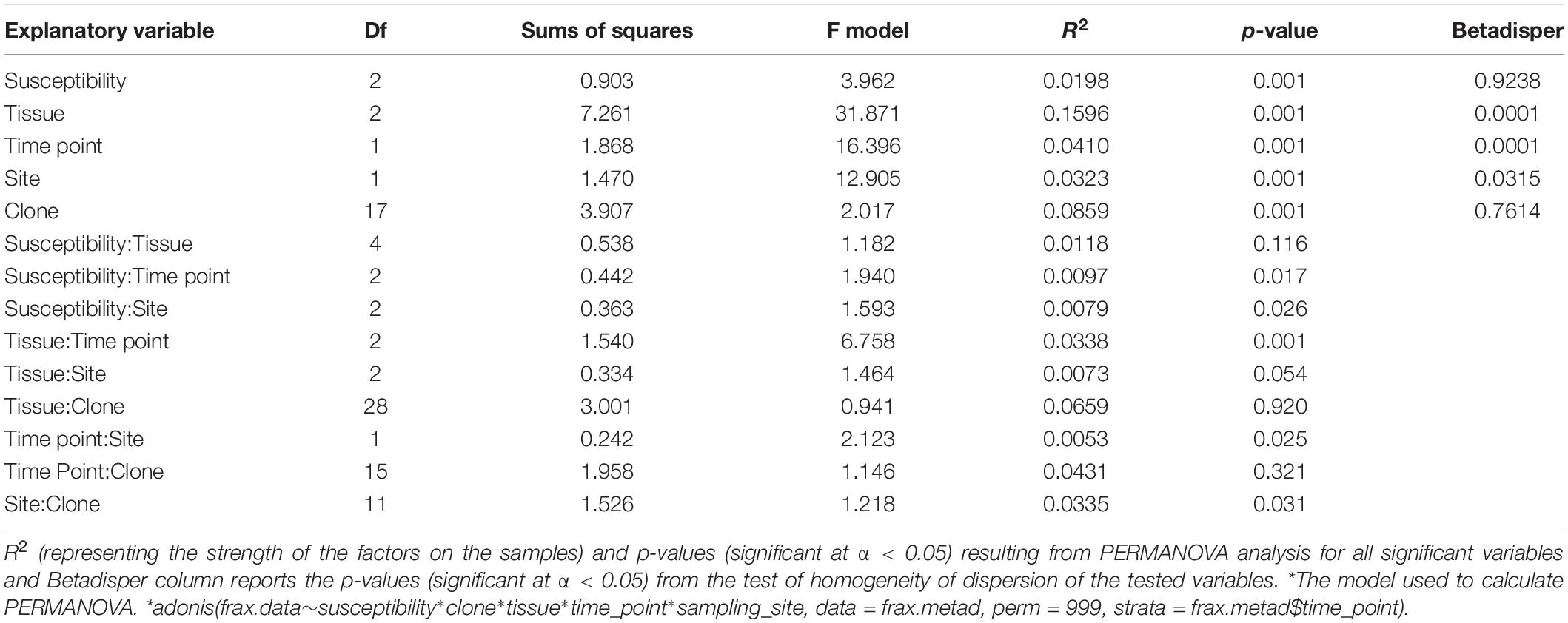
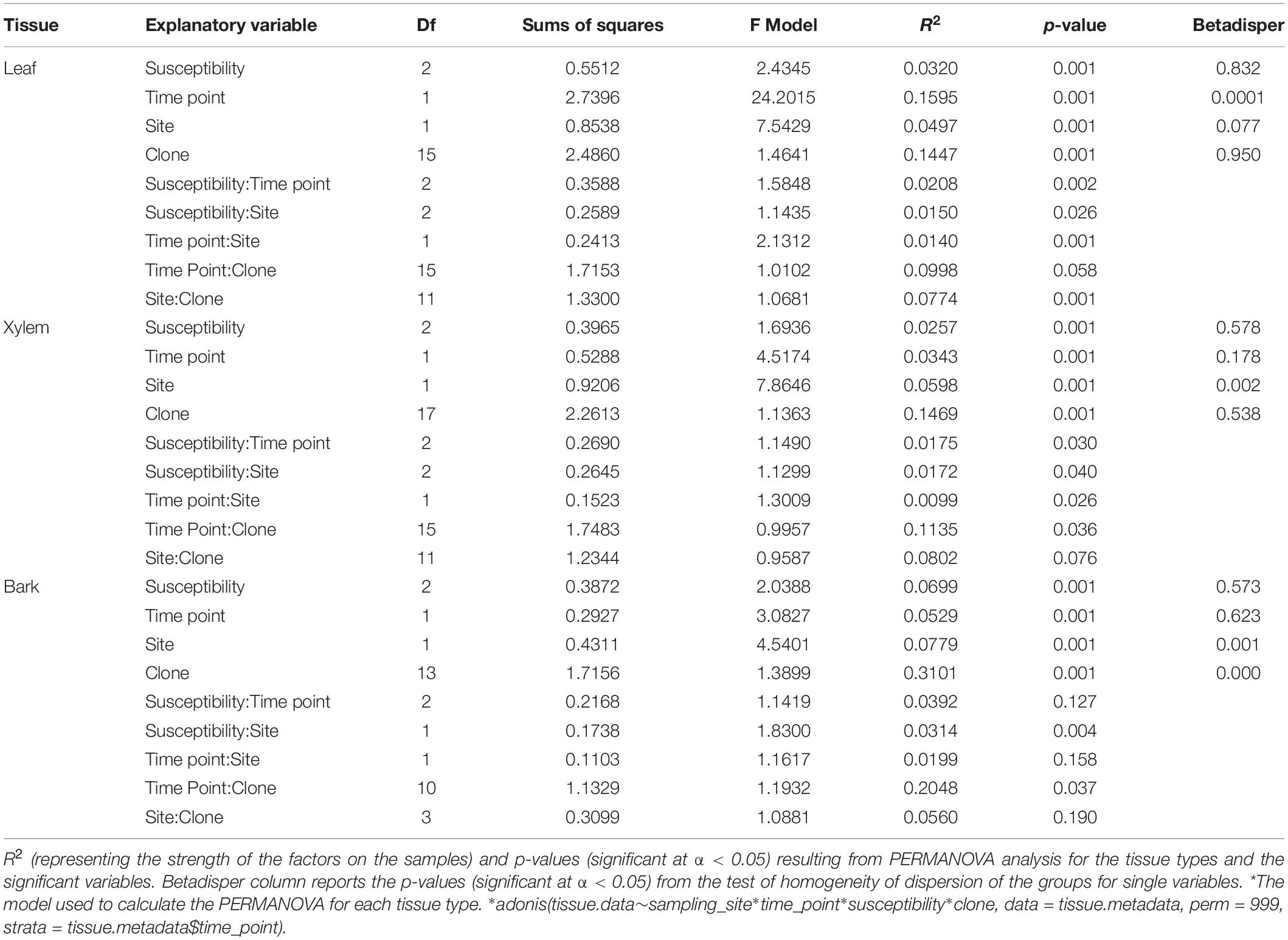
Differences in OTU richness across time, tissue types, susceptibility classes, and sites were tested with linear mixed model (lmer function in lme4 package). Variation in ash fungal community assemblages at different time points was visualized by non-metric multidimensional scaling (NMDS) using the function metaMDS in the vegan package (Oksanen et al., 2016) on a Bray-Curtis dissimilarity matrix (Oksanen et al., 2016). Variation was then estimated by a permutation analysis of variance (PERMANOVA) (Anderson, 2001) with the adonis function in the vegan package (Oksanen et al., 2016) using 999 permutations and constrained to time point. A post hoc pairwise PERMANOVA was tested for those variables whose PERMANOVA analyses was significant (p < 0.05) to assess which variables were affecting the community composition, and p-values were corrected with Bonferroni correction. For all significant variables identified in the PERMANOVA analysis, multivariate homogeneity of group dispersions analysis was performed with the function betadisper in vegan package (Oksanen et al., 2016); a p < 0.05 indicated that the criterion for homogeneous dispersion was met and the effect observed with adonis were not due to artifacts. PERMANOVA was then repeated on each tissue type to test if the variation detected for the whole communities holds at tissue level.
Venn diagrams were constructed to visualize the number of OTUs specific for each tree host and shared between host species. OTUs found to be exclusively associated to ash samples were also distinguished according to tissue type and susceptibility class.
To assess whether the fungal functional groups differed in relative abundance between host phenotypes within each tissue type, OTUs were classified into an ecological guilds using the FUNGuild database2 (Nguyen N. H. et al., 2016). Trophic levels defining the substrate utilization of the fungus (Nguyen N. H. et al., 2016) were assigned as either pathotroph (fungi damaging host cells), saprotroph (fungi degrading dead host cells), or symbiotroph (fungi exchanging resources with the host cells). Fungal taxa categorized into combined pathotroph-saprotroph or pathotroph-symbiotroph guilds include taxa that may exhibit different life styles depending on their life cycle stage and/or environmental conditions. Not all OTUs were associated with a trophic level, either due to limited taxonomic information for the OTU and/or trophic level association may be unknown for some fungal taxa. For those OTUs assigned to a trophic level, a canonical correspondence analysis (CCA) plot was constructed to visualize the degree of correspondence between the ash samples and the explanatory variables (i.e., tissue type, susceptibility class, time points, and site). CCA plots were then constructed for each of the three tissue types to visualize samples representing the highest read abundance for the two sampling sites (a total of 30 samples) and the tissue average. The trophic relative abundance composition of these samples was then visualized with pie charts.
Results
Health Condition of Trees
The seed orchards at Snogeholm and Trolleholm have been considerably affected by ash dieback. At both sites, dead trees either have been removed by the landowner, or remain standing dead or fallen, creating large gaps in both orchards. For this reason, it was not possible to obtain a similar number of replicates within each susceptibility class at each site. Many of the susceptible clones selected based on earlier assessments (Stener, 2013) were dead at the time of sample collection in 2015. Of the susceptible clones pre-selected for sampling in this study, only four were still viable at Snogeholm and only one at Trolleholm (Table 1). In the intermediate class, one of the seven clones sampled at Snogeholm was not found at Trolleholm because those trees belonging to that clone had died (Table 1). All resistant clones selected based on earlier rankings of clonal performance were still alive and had overall good vitality at both seed orchards.
Sequencing Output
From the 295 sequenced samples, 28,545,212 high quality paired-end reads were obtained (average of 96,763 reads per sample, ± 48,227 S.D) that initially clustered into 16,788 OTUs. After removal of non-fungal OTUs (8.5% belonged either to Plantae, Protista, or could not be identified due to poor or lack of BLAST hits), 14,690 OTUs remained with a total of 26,114,930 high quality reads. After removing four samples with low number of reads, 291 samples were used in the data analysis (272 belonging to ash trees and 19 to neighboring tree species). After the removal of OTUs with fewer than 10 reads globally, the final dataset included 4,430 OTUs (25,175,104 reads). Of the total number of OTUs, only 1,809 (40%) were assigned to a trophic level using FUNGuild.
Approximately 98% of reads were assigned to Ascomycota (59.3%) and Basidiomycota (38.5%), approximately 0.03% of reads was assigned to Chytridiomycota, Glomeromycota, and Zygomycota and 2.1% of reads belonged to unidentified fungi (Table 2). Similar relative abundances were found in ash samples and in neighboring tree species with the exception of Glomeromycota and Zygomycota that were more relatively abundant in neighboring tree species than in ash (Table 2). Rarefaction curves constructed for tree species (Supplementary Figure S1), tissue type (Supplementary Figure S2), and susceptibility class (Supplementary Figure S3) showed that the curves of ash samples provided a reasonably good indication of endophyte and epiphyte richness and diversity, but a broader sampling may reveal even more fungal species; similarly for neighboring tree species (Supplementary Figure S1).
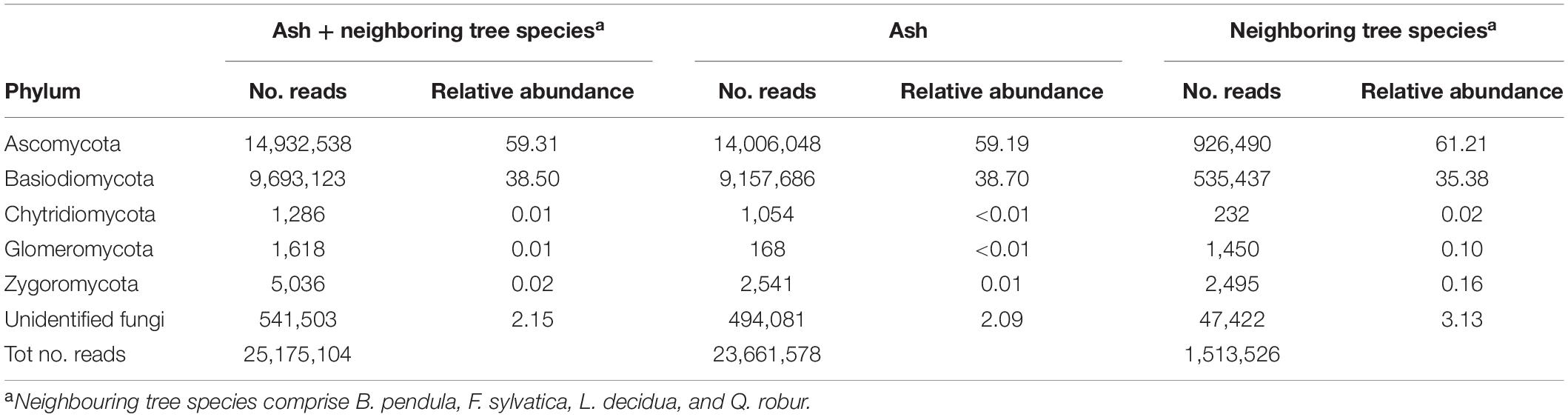
Table 2. Number of reads and relative abundance of reads for all samples, only ash trees, and neighboring tree per fungal division.
Sample and Taxonomic Diversity
A more diverse ash fungal community was detected in the earliest sampling time after leaf flush (May) when compared to later time points (e.g., August) (Tukey’s HSD, p = 0.001), though the fungal community was similarly diverse between July and September. Ash fungal community was also diverse among tissue types, with a tendency toward higher diversity in leaves compared to xylem and bark (Tukey’s HDS, p = 0.002).
OTU richness varied among the three tissue types sampled. Leaf OTU richness differed between the first time point in May and the latter time points in the season, with the trend toward increased OTU richness in all susceptibility classes especially between May and July. By August, OTU richness of both resistant and intermediate trees plateaued but decreased for susceptible trees (Supplementary Figure S4).
Bark OTU richness decreased between May and July for intermediate and susceptible but the opposite was shown for resistant trees (Supplementary Figure S5). Patterns of OTU richness in bark varied for the later time points and depended on the susceptibility class. However, no significant difference was observed in bark OTU richness over time nor across different susceptibility classes.
Xylem OTU richness differed significantly between May and later sampling dates (July and September) but no difference was observed among susceptibility classes. OTU richness appeared to decrease between May and July and then increase thereafter though the degree to which this occurred varied across trees with different phenotypic susceptibility to the disease; i.e., the increase was less pronounced in susceptible trees compared to resistant and intermediate trees. Only intermediate trees showed a decrease in OTU richness by the end of the sampling period while the OTU richness for other susceptibility phenotypes remained unchanged (Supplementary Figure S6).
The taxonomic diversity found in our study was composed of few common OTUs with high read count and many rare OTUs with low read count. Hymenoscyphus fraxineus was detected from 125 samples (42% of the total number of samples), albeit at a low level (0.03% of the total reads). Of the total number of samples in which H. fraxineus was detected, 95% of reads were from leaf samples and 90% of those were from the last sampling time point in September. Detection of H. fraxineus varied by susceptibility phenotype; approximately 83% of reads were found in the intermediate class, 9% in the high susceptibility class, and 8% in the low susceptibility class. However, site differences were evident; approximately 76% of reads were found in samples from Snogeholm and 24% from Trolleholm. The fungus was also detected on neighboring trees, albeit at low frequency (∼0.3%).
Almost 60% of the total reads were comprised by just 21 OTUs (with relative read abundance greater than 1%), with Ascomycota (n = 12) and Basidiomycota (n = 9) being the two phyla represented. Among these 21 OTUs, generalist and ubiquitous fungi were found in 99% of the samples (Table 3). With the exception of Erythrobasidium hasegawianum (Hamam., Sugiy., and Komag.), all other OTUs identified to species level were assigned to a trophic level, the majority being pathotrophs and saprotrophs. Only one OTU was assigned to the symbiotroph trophic level (Table 3).
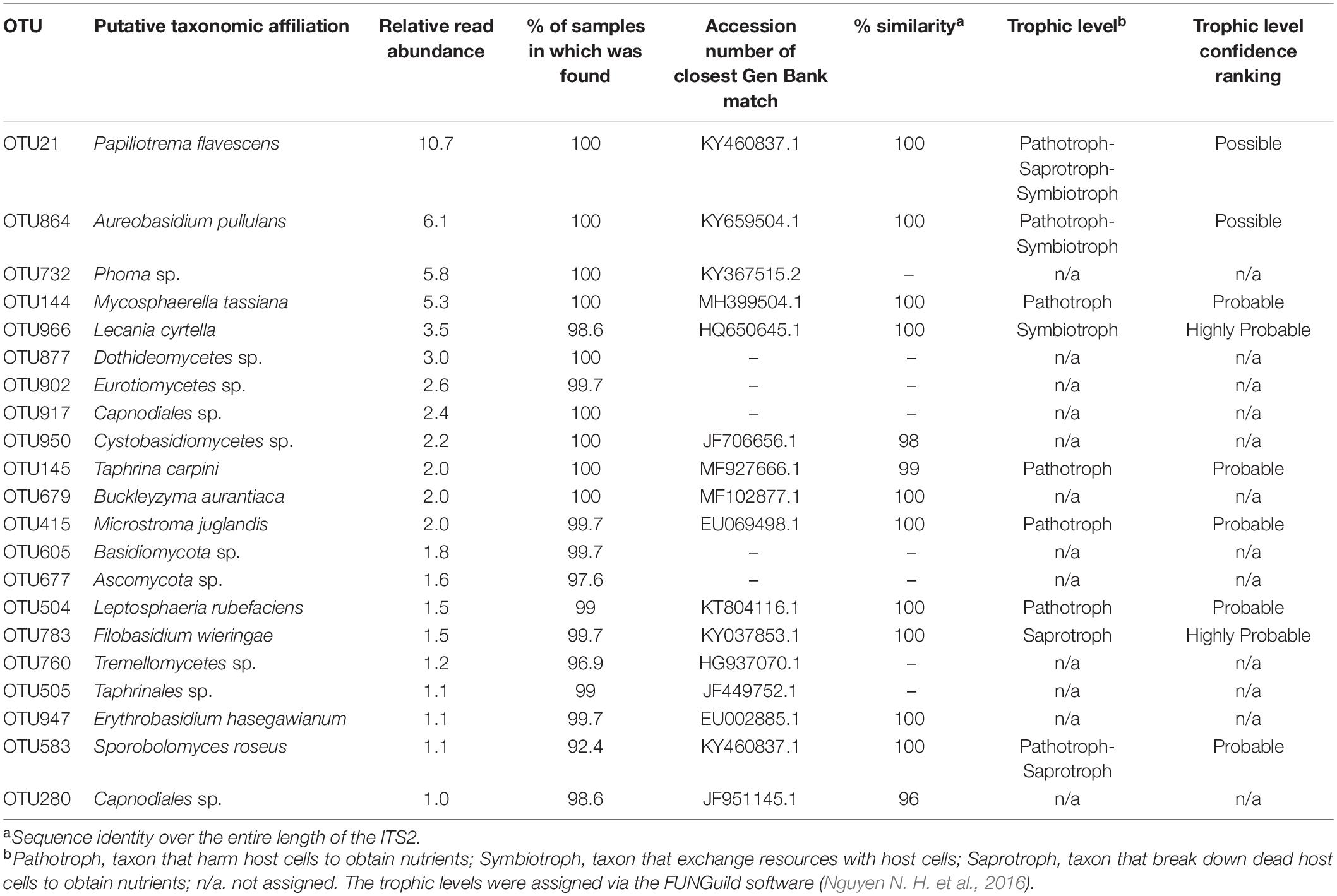
Table 3. Taxonomic and trophic level assignment of the 21 OTUs with relative read abundance greater than 1%.
Fungal Community Composition
The overall pattern of ash fungal community assemblage visualized with NMDS plots showed that fungal community composition differed among the three tissue types. Leaf fungal community differed from the communities found in bark and xylem at all sampling time points throughout the season, most notably at the first sampling in May (Figure 2). Bark and xylem had similar communities that diverged at later sampling times (Figure 2). From PERMANOVA analyses, we observed significant interactions between tissue type and time point (referring to seasonal differences), and to some degree susceptibility classes and time point (Table 4). To better understand which factors (i.e., susceptibility class, time, and site) associated with the fungal community composition, we looked at each tissue type separately (Table 5). Pairwise comparison of the fungal community in bark with phenotypic susceptibility to the disease and time points highlighted a significant difference in community composition between trees in the intermediate and susceptible classes, and among time points (except July and August) (Supplementary Table S2).
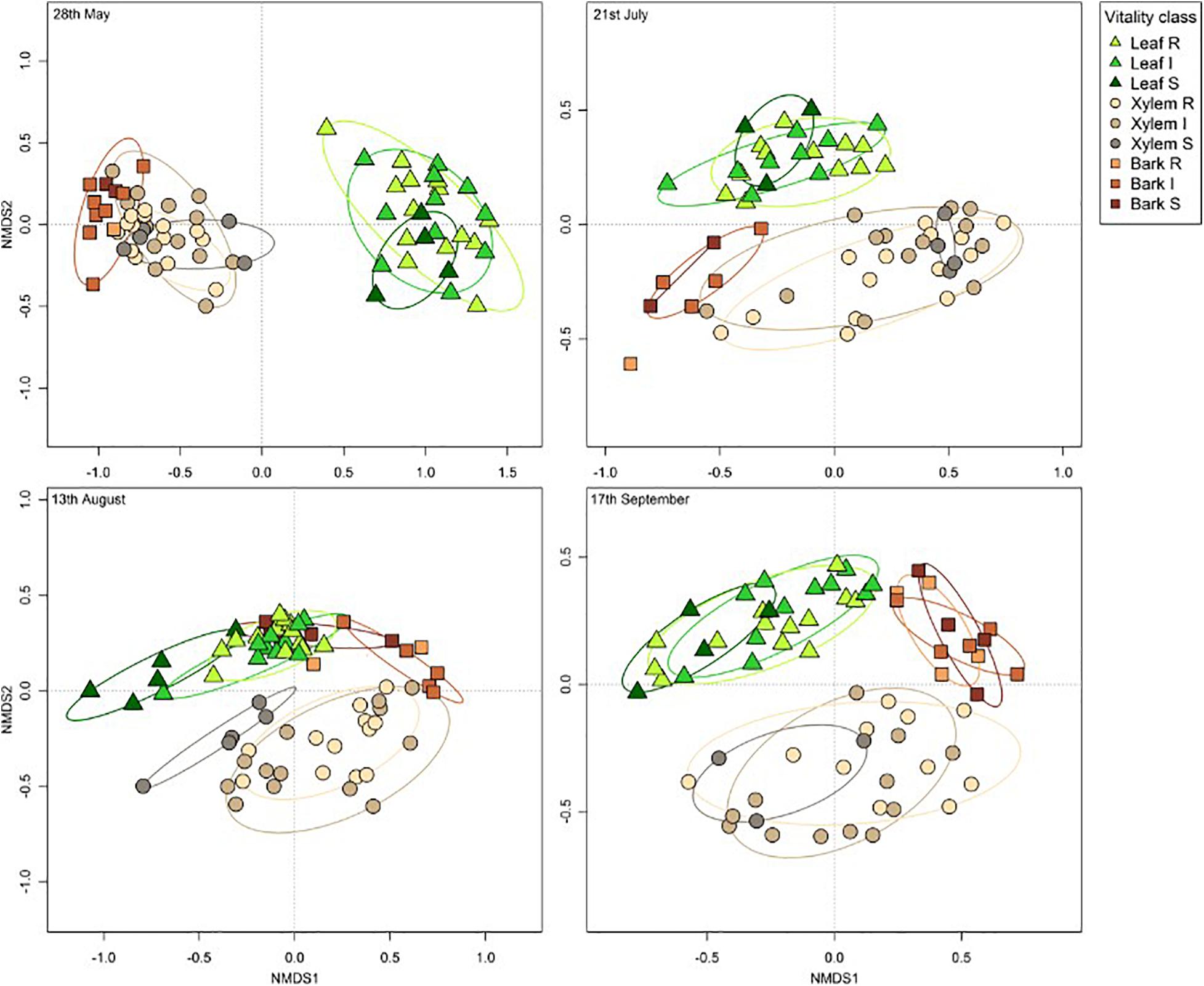
Figure 2. Non-metric multidimensional scaling (NMDS) ordination plots of ash fungal communities based on Bray-Curtis dissimilarity matrix. The plots are divided according to the four sampling time points: 28th May (stress value = 0.08), 21st July (stress value = 0.12), 13th August (stress value = 0.13), and 17th September (stress value = 0.14). Tissue types are represented by different shapes (leaf: triangle, xylem: circle, bark: square). Susceptibility classes are represented by intensity of colors for the gradient from Resistant (light) to Susceptible (dark).
Communities and Functional Groups
Of the 4,430 OTUs analyzed in this study, approximately 60% were shared between ash and the neighboring tree species, i.e., silver birch, European beech, pedunculate oak, hybrid larch (Figure 3). A small fraction of the OTUs (2%) was exclusively associated with neighboring tree species, while 37% of OTUs exclusively associated with ash. Among the Ascomycota, Dothideomycetes spp. was the most abundant class (42%), while among the Basidiomycota, Tremellomycetes spp. was most abundant (Supplementary Table S3).
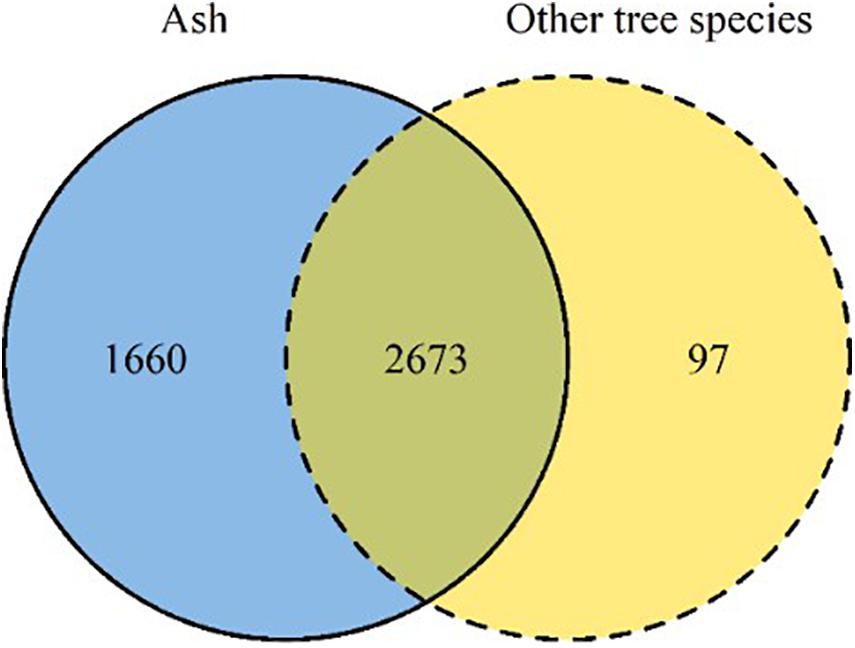
Figure 3. Venn diagram summarizing the number of OTUs detected in ash (solid line, blue) and in neighboring tree species (dashed line, yellow). The overlapping area of the two circles reports the number of OTUs shared between ash and neighboring tree species.
Among the 1,660 OTUs exclusively associated with ash, 815 OTUs were shared among all tissue types. Xylem hosted more unique OTUs relative to leaves and bark; there were twice as many unique xylem OTUs than leaves and 10 times more than bark (Figure 4A). OTUs uniquely found in xylem and bark were dominated by Ascomycota (58 and 78%, respectively) while leaf OTUs were dominated by Basidiomycota (53%). Totally, 992 OTUs were shared across all three susceptibility classes, and belong predominantly to Ascomycota (61%) and Basidiomycota (31%). Resistant and intermediate classes hosted a similar number of unique OTUs, while the high susceptibility class had much fewer (Figure 4B and Supplementary Table S4).
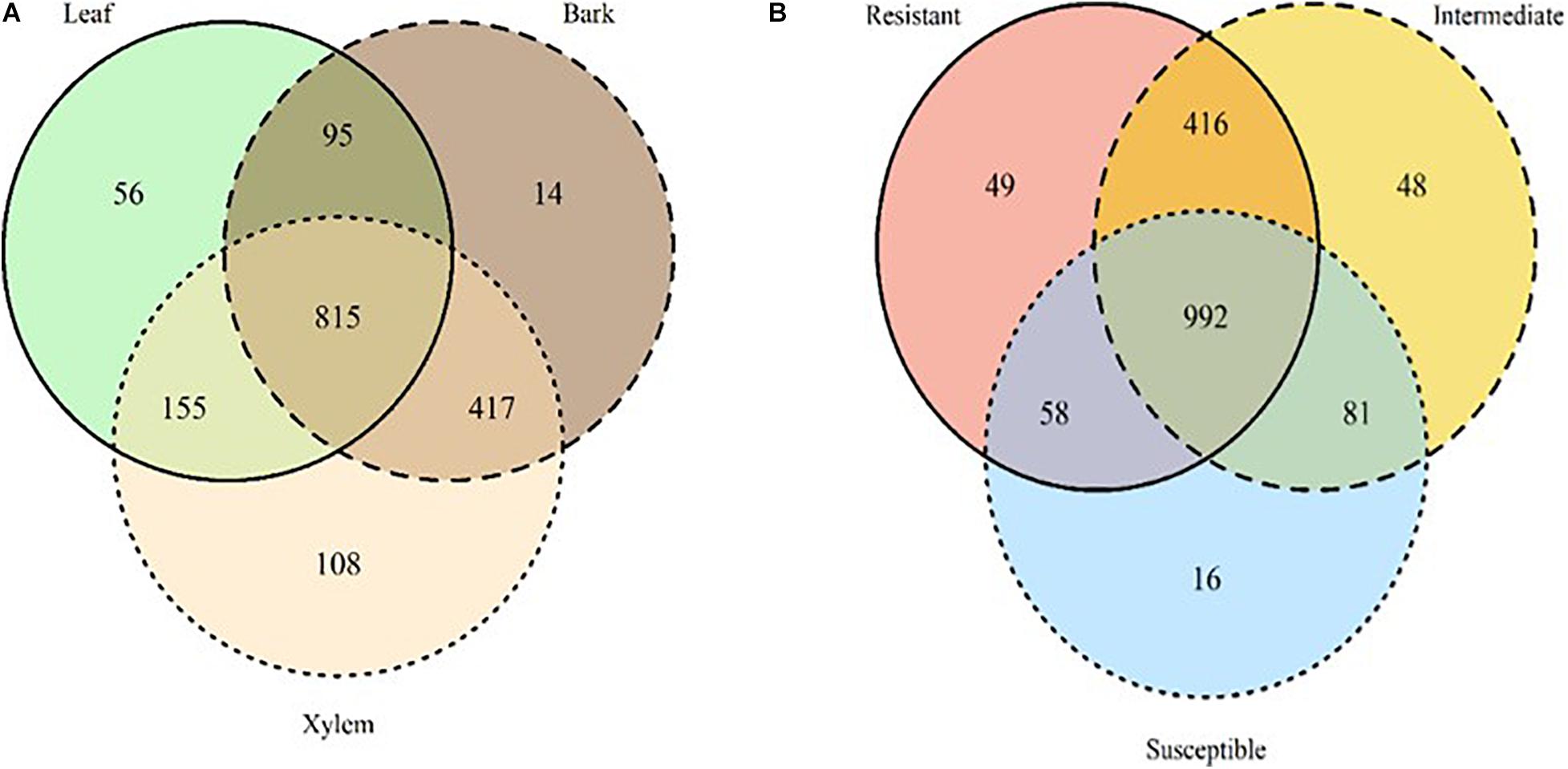
Figure 4. Venn diagrams summarizing the number of OTUs detected only in ash trees by tissue type (A) leaf: solid line, green; bark: dashed line, brown; xylem: dotted line, light beige and susceptibility classes (B) resistant: solid line, red; Intermediate: dashed line, yellow; Susceptible: dotted line, blue). The overlapping areas report the number of OTUs shared among tissues and susceptibility classes.
Among the OTUs uniquely associated with each of the phenotypes (n = 113), approximately half could not be assigned to a trophic level, either due to limitation of taxonomic assignment or lack of information about trophic affiliation (Table 6 and Supplementary Table S4). Considering all tissue types, pathotroph OTUs dominated in susceptible trees. Generally, an increase in the relative abundance of pathotrophic taxa was observed along a susceptibility gradient from low to high susceptibility in bark and xylem while the proportion of pathotrophs were similarly abundant in leaf, irrespective of phenotype (Figure 5). Saprotrophs were distributed similarly among the three tissue types and susceptibility classes while the relative abundance of the pathotroph-saprotrophic OTUs was higher in leaves compared to bark or xylem (Figure 5). Symbiotrophic OTUs were more abundant in bark and xylem than in leaf tissues. Susceptible trees had a lower abundance of symbiotrophic taxa than resistant or intermediate trees (Figure 5). The relative abundance of OTUs assigned as pathotroph-symbiotroph was marginal compared to the other more abundant trophic classes, though, it was higher in resistant and intermediate trees across all tissue types compared to susceptible trees (Figure 5). The canonical correspondence analysis (CCA) ordination plot of trophic level-assigned OTUs (Figure 6) shows the distributions of 272 ash samples in relation to tissue type, susceptibility class, time points, and site. The first and second CCA axes (eigenvalues 0.18 and 0.08, respectively) explained 4% of the variance in the data and 72% of the variance between samples and explanatory variables. A negative correlation was found between susceptibility and site, and we observed no correlation between tissue and susceptibility, site, and time. Overall, the fungal community composition appeared to separate by tissue type and site (Figure 6).
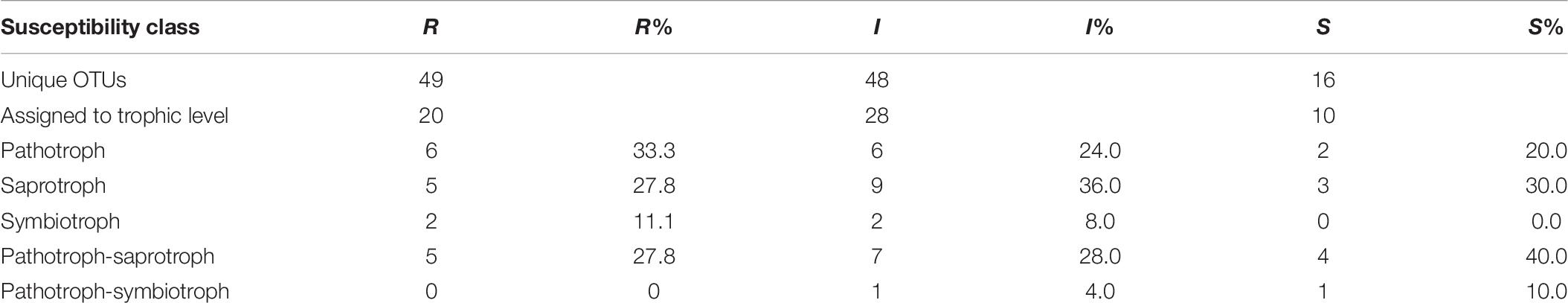
Table 6. Number of OTUs exclusively assigned to each susceptibility class (R-resistant; I-intermediate; S-susceptible) and number of OTUs assigned to a trophic level and respective relative abundance for each class.
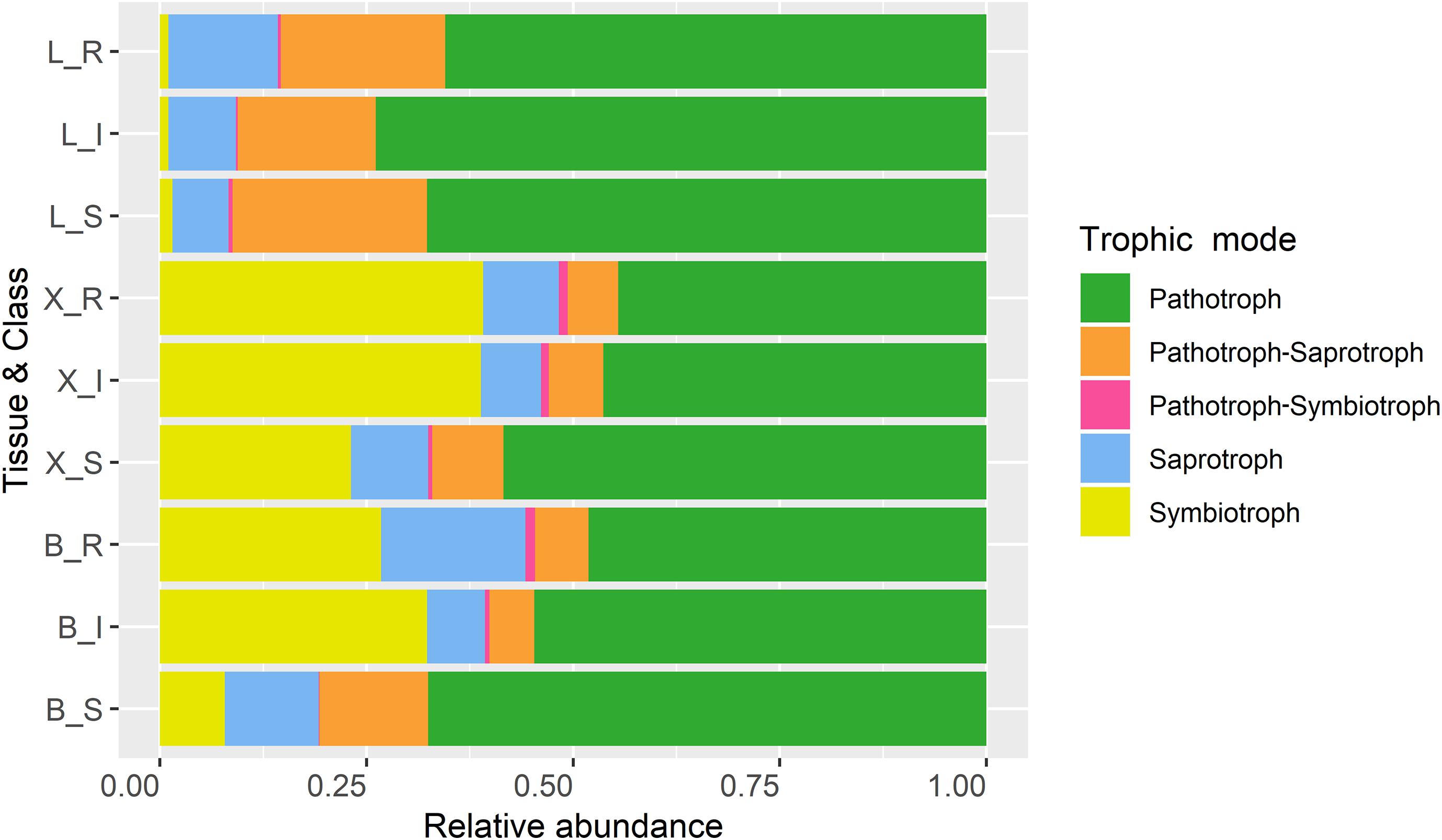
Figure 5. Trophic modes of OTUs based on their relative read abundance for ash tissue types (L, leaves; X, xylem; B, bark) and susceptibility class (R, resistant; I, intermediate; S, susceptible).
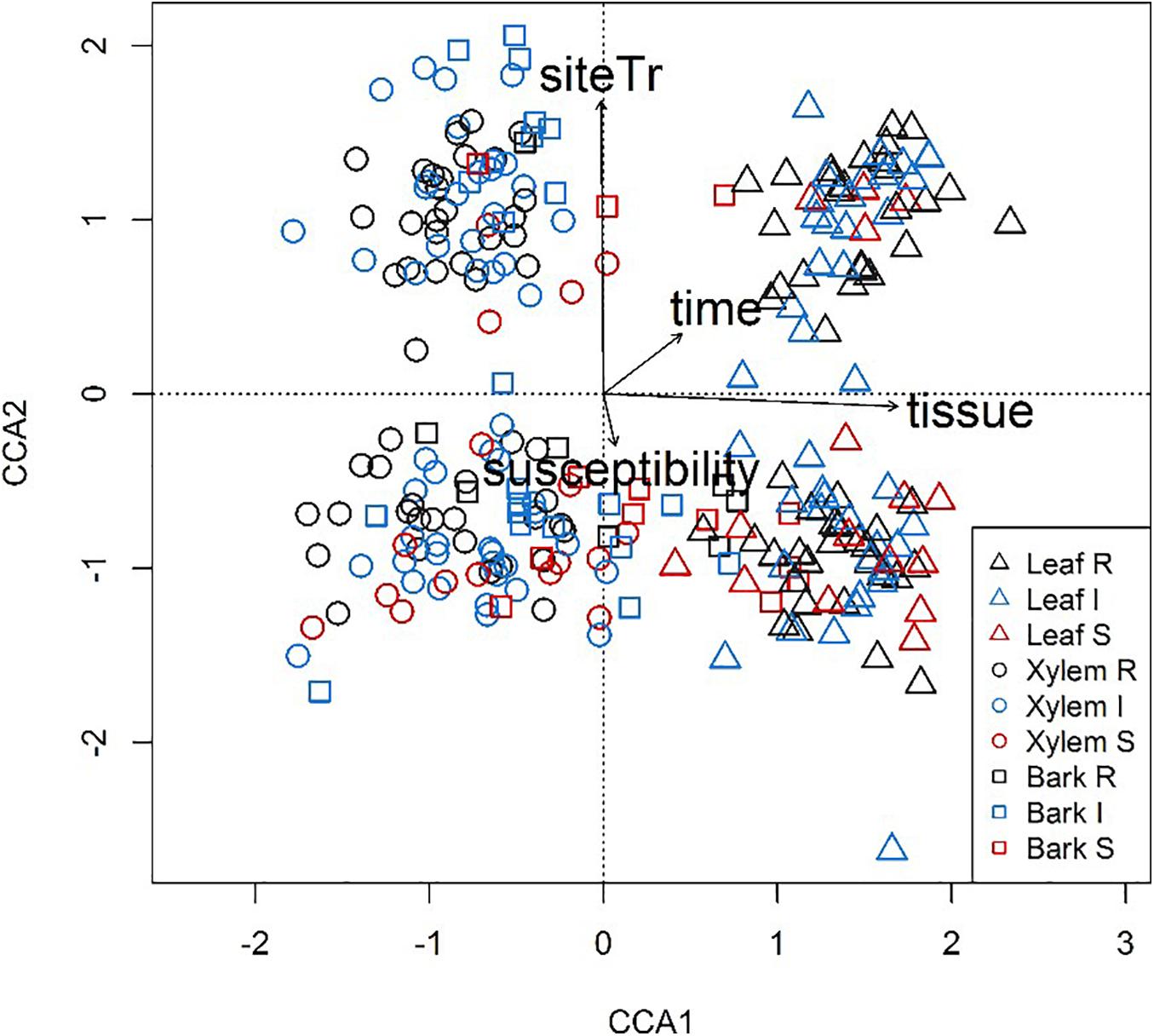
Figure 6. Variation in fungal community composition visualized by canonical correspondence analysis (CCA) plot based on trophic levels assigned by FUNGuild for all ash samples. Samples are shape- and color-coded according to tissue type (leaves, triangle; xylem, circle; bark, square) and susceptibility class (Resistant, black; Intermediate, blue; Susceptible, red).
Discussion
Prior to the introduction of the invasive alien pathogen H. fraxineus, the mycobiome of F. excelsior had rarely been the subject of studies. Now with the progressive disappearance of ash from the landscape, there is greater interest in ash fungal communities (Vasiliauskas et al., 2006; Bakys et al., 2009; Scholtysik et al., 2013) and especially the role of endophytes as enhancers of resistance in the host and against H. fraxineus (Schlegel et al., 2016; Haňáčková et al., 2017; Kosawang et al., 2018). Until recently, little focus has been directed to the impact of the ash dieback pathogen on ash fungal biodiversity. Leveraging advanced methods such as high throughput sequencing and metagenomics, and tools such as FUNGuild (Nguyen N. H. et al., 2016) and the new FungalTraits database (Põlme et al., 2021), we can now uncover the hidden fungal diversity and begin to understand the ecological role of these cryptic fungi in the ecosystems.
Our study revealed that the mycobiome of ash trees with different phenotypic susceptibility to H. fraxineus is highly diverse and varies over space and time. Previous studies have linked differences in fungal communities to the host genetic background (Bálint et al., 2013; Lamit et al., 2014; Albrectsen et al., 2018). However, limited by the study design and the death of specific ash clones, we were not able to associate specific ash genotypes to specific fungal communities. Though the genetic underpinnings of disease susceptibility are not yet worked out, we observed fungal community composition differences among trees from different susceptibility classes. The degree of decline can differentially affect fungal assemblages of trees as previously shown for declining Scots pine (Giordano et al., 2009) and pedunculate oak (Agostinelli et al., 2018) that were found to harbor a higher number of taxa and were characterized by higher colonization frequency compared to healthy-looking trees. In contrast, the pattern is inconsistent in declining ash.
Furthermore, in this study, the effect of phenotype on fungal community composition depended on tissue type. At certain time points during our sampling, tissue type influenced the composition of the fungal communities; bark hosted the highest community richness (at all sampling time points) compared to xylem and leaves, which is in line with previous work (e.g., Petrini and Fisher, 1990; Kowalski and Gajosek, 1998; Ragazzi et al., 2003). Martín et al. (2013) found a higher endophyte frequency and diversity in the xylem of Dutch elm disease susceptible elm trees than in the bark. During the late season sampling (August and September), the fungal communities in the xylem of susceptible ash trees appeared more distinct from the other phenotypes compared to communities found in leaves and bark. The marked reduction in fungal symbiotrophs and corresponding increase in pathotrophs and saprotrophs of susceptible trees versus intermediate and resistant phenotypes suggests an indirect role of tree health on the fungal assemblages in xylem tissue. In comparing fungal communities of different tissue types, some fungi were exclusively associated to different host tissue suggesting that some fungal taxa are not only host- but also tissue-specific (Kehr and Wulf, 1993; Ragazzi et al., 2003; Martín et al., 2013).
Site had a strong influence on the mycobiome of ash, but differences were probably more qualitative than quantitative (in terms of species richness). Arnold and Lutzoni (2007) and Millberg et al. (2015) showed that endophytic diversity decreased along a north-south gradient. The two sites in this study are approximately 60 km apart and are both located on previous agricultural land in the mesophytic deciduous broadleaved and coniferous-broadleaved forests (Bohn et al., 2000) which has probably limited any strong differences in species richness between the two sites. The differences in community composition may be attributable to slight differences in climatic and micro-climatic conditions between sites since Zimmerman and Vitousek (2012) found that variation in temperature and precipitation can differentially affect community. Seasonal variation in phyllosphere fungal communities have been previously observed in different hosts (Jumpponen and Jones, 2010; Unterseher et al., 2013; Cross et al., 2017; Schlegel et al., 2018).
At specific time points in the season, the fungal community was strongly influenced by tissue type with leaves exhibiting most of this seasonal variation. Newly flushed leaves have a less diverse fungal community (Faeth and Hammon, 1997; Saikkonen et al., 1998, 2004), and prolonged exposure to horizontally spreading fungal inoculum throughout the season leads to an accumulation of fungal infections in the leaves (Helander et al., 1993; Stone and Petrini, 1997; Kaneko et al., 2003; Pérez-Izquierdo et al., 2017). While a season effect was observed in all tissue types, the increase in fungal diversity and biomass was observed to a much lesser extent in both perennial tissues than in leaves. Generally, a decrease in species richness was observed in all three tissue types at the end of the season. The decrease in species richness and change in community composition in leaf samples may be related to alteration in leaf physiology as consequence of leaf senescence and the increased inoculum pressure from H. fraxineus (Cross et al., 2017). Carbohydrate metabolism in tree leaves is known to fluctuate within and between seasons, reflecting the physiological changes occurring in leaves during the phenology (Wong et al., 2009). This may lead to a changed availability of soluble carbohydrates or accumulation of potentially antifungal or fungistatic secondary metabolites, such as certain phenolics (Riipi et al., 2002), altering niche structures that may affect the fungal community composition at the end of the season, and favoring species that can utilize the available carbon sources at the end of the season.
Our results suggest that the fungal community of aerial tissues of ash trees is composed of diverse fungal species with different ecological roles. While the relative abundance of pathotrophic and saprotrophic fungi in leaves were similar among the three susceptibility classes, we speculate that there may be a trend toward increasing relative abundance with decreasing plant health status (i.e., from resistant to susceptible class). This may reflect increased colonization by new saprotrophic and pathotrophic species, or indicate that the already present species increasingly switched to these strategies as the conditions changed in declining trees. Although FUNGuild is an important tool for assigning ecological roles to fungi, the tool continues to be developed and relies on new studies that clearly link fungal taxonomy to ecology (Nguyen N. H. et al., 2016). Consequently, most of the OTUs in our study were not assigned to a trophic level due to either low resolution in identification of fungal taxa and/or lack of knowledge on their ecological role in ecosystems. Moreover, variable lifestyles of fungi on different host species creates confusion on its true trophic mode. These limitations may be improved with further studies and curation of databases with information on guilds associated to fungi (Põlme et al., 2021).
The taxonomic diversity in ash fungal communities resembled those found in other studies of broadleaved species, with Ascomycota and Basidiomycota being the two most abundant phyla (e.g., Nguyen et al., 2017). The community was composed of few species commonly found in all samples and other rare species associated with specific susceptibility classes, tissue types, and time points. Although H. fraxineus was detected at low abundance in the overall fungal community, we observed an increase in read abundance across the season, with the majority occurring in leaves collected in September. Similar observations were made by Cross et al. (2017) who showed that increased H. fraxineus biomass gave an increased read count as the season progressed. Agan et al. (2020) also showed that H. fraxineus read abundance increased vigorously toward autumn which was associated with a decline in fungal species richness. It is interesting that H. fraxineus was not detected as frequently in leaves of susceptible or resistant trees as in intermediate trees. However, the low abundance could also be explained by low efficiency of the amplification of the marker gene for H. fraxineus because as Cross et al. (2017) pointed out in their study, limited presence of H. fraxineus may be attributable to the long 3′ terminal 18S intron, which renders the species difficult to detect with ITS primers (Tedersoo et al., 2019).
With the ash population declining all over Europe, ash-associated fungal diversity might be at risk of severe decline and in some cases, possible extinction, especially for host-specific species such as Venturia fraxini (Schlegel et al., 2018). Local extirpation of Hymenoscyphus albidus, the European native, host-specific saprophyte of F. excelsior, has already occurred due to competitive exclusion by H. fraxineus (McKinney et al., 2012). Similar to Schlegel et al. (2018), our study showed that the majority of fungal species found in ash trees was not strictly host specific. Thus, endophytic diversity may be less affected by reduced host density compared to certain macrofungi and other guilds such as bryophytes, insects and lichens, which have very high- or obligate associations to ash (Mitchell et al., 2014; Hultberg et al., 2020). Breeding ash for resistance against H. fraxineus and restoration of forests with highly resistant trees can help conserve important biodiversity. More research is needed to define the critical thresholds in host density and vitality that are needed to sustain microfungal diversity in landscapes affected by ash dieback.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
MC conceived the study. MC and JW designed the study. MA and MC carried out field work. MA carried out molecular lab work. DN and MA carried out statistical analysis of sequence data set. MC, DN, and MA drafted the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank Lars-Göran Stener, Iryna Semashko, Marjan Ghasemkhani, Moritz Buck, Chloé Boldrini, and Carl Salk for their assistance with the field and lab work and data analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffgc.2021.580514/full#supplementary-material
Footnotes
References
Abarenkov, K., Henrik Nilsson, R., Larsson, K., Alexander, I. J., Eberhardt, U., Erland, S., et al. (2010). The UNITE database for molecular identification of fungi – recent updates and future perspectives. New Phytol. 186, 281–285. doi: 10.1111/j.1469-8137.2009.03160.x
Agan, A., Drenkhan, R., Adamson, K., Tedersoo, L., Solheim, H., Børja, I., et al. (2020). The relationship between fungal diversity and invasibility of a foliar niche — the case of ash dieback. J. Fungi 6:150. doi: 10.3390/jof6030150
Agostinelli, M., Cleary, M., Martín, J. A., Albrectsen, B. R., and Witzell, J. (2018). Pedunculate oaks (Quercus robur L.) differing in vitality as reservoirs for fungal biodiversity. Front. Microbiol. 9:1758. doi: 10.3389/fmicb.2018.01758
Albrectsen, B. R., Siddique, A. B., Decker, V. H. G., Unterseher, M., and Robinson, K. M. (2018). Both plant genotype and herbivory shape aspen endophyte communities. Oecologia 187, 535–545. doi: 10.1007/s00442-018-4097-3
Anderson, M. J. (2001). Permutation tests for univariate or multivariate analysis of variance and regression. Can. J. Fish. Aquat. Sci. 58, 626–639. doi: 10.1139/cjfas-58-3-626
Arnold, A. E. (2007). Understanding the diversity of foliar endophytic fungi: progress, challenges, and frontiers. Fungal Biol. Rev. 21, 51–66. doi: 10.1016/j.fbr.2007.05.003
Arnold, A. E., and Lutzoni, F. (2007). Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots? Ecology 88, 541–549. doi: 10.1890/05-1459
Bakys, R., Vasaitis, R., Barklund, P., Thomsen, I. M., and Stenlid, J. (2009). Occurrence and pathogenicity of fungi in necrotic and non-symptomatic shoots of declining common ash (Fraxinus excelsior) in Sweden. Eur. J. For. Res. 128, 51–60. doi: 10.1007/s10342-008-0238-2
Bálint, M., Bartha, L., O’Hara, R. B., Olson, M. S., Otte, J., Pfenninger, M., et al. (2015). Relocation, high-latitude warming and host genetic identity shape the foliar fungal microbiome of poplars. Mol. Ecol. 24, 235–248. doi: 10.1111/mec.13018
Bálint, M., Tiffin, P., Hallström, B., O’Hara, R. B., Olson, M. S., Fankhauser, J. D., et al. (2013). Host genotype shapes the foliar fungal microbiome of balsam poplar (Populus balsamifera). PLoS One 8:e53987. doi: 10.1371/journal.pone.0053987
Bell, S., McLoughlin, J., Reeg, T., Gavaland, A., Muehlthaler, U., Makinen, K., et al. (2008). Cultural Aspects of the Trees in Selected European Countries. FPS COST Action E42 Growing Valuable Broadleaved Tree Species. Strasbourg: COST Office, European Science Foundation, 66–78.
Bengtsson, V., and Stenström, A. (2017). “Ash Dieback – a continuing threat to veteran ash trees?,” in Dieback of European Ash (Fraxinus spp.): Consequences and Guidelines for Sustainable Management, eds R. Vasaitis and R. Enderle (Uppsala: University of Agricultural Sciences), 262–272.
Bohn, U., Gollub, G., and Hettwer, C. (2000). Map of the Natural Vegetation of Europe. Bonn: Federal Agency for Nature Conservation.
Brasier, C. M. (1991). Ophiostoma novo-ulmi sp. nov., causative agent of current Dutch elm disease pandemics. Mycopathologia 115, 151–161. doi: 10.1007/BF00462219
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chandelier, A., Gerarts, F., San Martin, G., Herman, M., Delahaye, L., and Hantula, J. (2016). Temporal evolution of collar lesions associated with ash dieback and the occurrence of Armillaria in Belgian forests. For. Pathol. 46, 289–297. doi: 10.1111/efp.12258
Chandelier, A., Helson, M., Dvorak, M., and Gischer, F. (2014). Detection and quantification of airborne inoculum of Hymenoscyphus pseudoalbidus using real-time PCR assays. Plant Pathol. 63, 1296–1305. doi: 10.1111/ppa.12218
Chase, J., and Leibold, M. (2003). Ecological Niches: Linking Classical and Contemporary Approaches. Chicago, IL: University of Chicago Press.
Cleary, M., Nguyen, D., Stener, L.-G., Stenlid, J., and Skovsgaard, J.-P. (2017). “Ash and ash dieback in Sweden: a review of disease history, current status, pathogen and host dynamics, host tolerance and management options in forests and landscapes,” in Dieback of European Ash (Fraxinus spp.) – Consequences and Guidelines for Sustainable Management, eds R. Vasaitis and R. Enderle (Uppsala: Swedish University of Agricultural Sciences), 195–208.
Cleary, M. R., Daniel, G., and Stenlid, J. (2013). Light and scanning electron microscopy studies of the early infection stages of Hymenoscyphus pseudoalbidus on Fraxinus excelsior. Plant Pathol. 62, 1294–1301. doi: 10.1111/ppa.12048
Cross, H., Sønstebø, J. H., Nagy, N. E., Timmermann, V., Solheim, H., Børja, I., et al. (2017). Fungal diversity and seasonal succession in ash leaves infected by the invasive ascomycete Hymenoscyphus fraxineus. New Phytol. 213, 1405–1417. doi: 10.1111/nph.14204
Enderle, R., Peters, F., Nakou, A., and Metzler, B. (2013). Temporal development of ash dieback symptoms and spatial distribution of collar rots in a provenance trial of Fraxinus excelsior. Eur. J. For. Res. 132, 865–876. doi: 10.1007/s10342-013-0717-y
Faeth, S. H., and Fagan, W. F. (2002). Fungal endophytes: common host plant symbionts but uncommon mutualists. Integr. Comp. Biol. 42, 360–368. doi: 10.1093/icb/42.2.360
Faeth, S. H., and Hammon, K. E. (1997). Fungal endophytes in oak trees. Long-term patterns of abundance and associations with leafminers. Ecology 78, 810–819.
Giordano, L., Gonthier, P., Varese, G. C., Miserere, L., and Nicolotti, G. (2009). Mycobiota inhabiting sapwood of healthy and declining Scots pine (Pinus sylvestris L.) trees in the Alps. Fungal Divers. 38, 69–83.
Gross, A., Holdenrieder, O., Pautasso, M., Queloz, V., and Sieber, T. N. (2014). Hymenoscyphus pseudoalbidus, the causal agent of European ash dieback. Mol. Plant Pathol. 15, 5–21. doi: 10.1111/mpp.12073
Haegeman, B., Hamelin, J., Moriarty, J., Neal, P., Dushoff, J., and Weitz, J. S. (2013). Robust estimation of microbial diversity in theory and in practice. ISME J. 7, 1092–1101. doi: 10.1038/ismej.2013.10
Halme, P., Heilmann-Clausen, J., Rämä, T., Kosonen, T., and Kunttu, P. (2012). Monitoring fungal biodiversity – towards an integrated approach. Fungal Ecol. 5, 750–758. doi: 10.1016/j.funeco.2012.05.005
Haňáčková, Z., Havrdová, L., Černý, K., Zahradník, D., and Koukol, O. (2017). Fungal endophytes in ash shoots – Diversity and inhibition of Hymenoscyphus fraxineus. Balt. For. 23, 89–106.
Helander, M. L., Neuvonen, S., Sieber, T., and Petrini, O. (1993). Simulated acid rain affects birch leaf endophyte populations. Microb. Ecol. 26, 227–234. doi: 10.1007/BF00176955
Hietala, A. M., Timmermann, V., Børja, I., and Solheim, H. (2013). The invasive ash dieback pathogen Hymenoscyphus pseudoalbidus exerts maximal infection pressure prior to the onset of host leaf senescence. Fungal Ecol. 6, 302–308. doi: 10.1016/j.funeco.2013.03.008
Hultberg, T., Sandberg, J., Felton, A., Öhman, K., Witzell, J., Rönnberg, J., et al. (2020). Ash dieback risks an extinction cascade. Biol. Conserv. 244:108516.
Husson, C., Caël, O., Grandjea, J. P., Nageleisen, L. M., and Marçais, B. (2012). Occurrence of Hymenoscyphus pseudoalbidus on infected ash logs. Plant Pathol. 61, 889–895. doi: 10.1111/j.1365-3059.2011.02578.x
Ibrahim, M., Sieber, T. N., and Schlegel, M. (2017). Communities of fungal endophytes in leaves of Fraxinus ornus are highly diverse. Fungal Ecol. 29, 10–19. doi: 10.1016/j.funeco.2017.05.001
Ihrmark, K., Bödeker, I. T. M., Cruz-Martinez, K., Friberg, H., Kubartova, A., Schenck, J., et al. (2012). New primers to amplify the fungal ITS2 region - evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 82, 666–677. doi: 10.1111/j.1574-6941.2012.01437.x
Inoue, T., Okane, I., Ishiga, Y., Degawa, Y., Hosoya, T., and Yamaoka, Y. (2019). The life cycle of Hymenoscyphus fraxineus on Manchurian ash, Fraxinus mandshurica, in Japan. Mycoscience 60, 89–94.
Jönsson, M. T., and Thor, G. (2012). Estimating coextinction risks from epidemic tree death: affiliate lichen communities among diseased host tree populations of Fraxinus excelsior. PLoS One 7:e45701. doi: 10.1371/journal.pone.0045701
Jumpponen, A., and Jones, K. L. (2010). Seasonally dynamic fungal communities in the Quercus macrocarpa phyllosphere differ between urban and nonurban environments. New Phytol. 186, 496–513. doi: 10.1111/j.1469-8137.2010.03197.x
Kaneko, R., Kakishima, M., and Tokumasu, S. (2003). The seasonal occurrence of endophytic fungus, Mycosphaerella buna, in Japanese beech, Fagus crenata. Mycoscience 44, 277–281. doi: 10.1007/S10267-003-0115-2
Kehr, R. D., and Wulf, A. (1993). Fungi associated with above-ground portions of declining oaks (Quercus robur) in Germany. For. Pathol. 23, 18–27. doi: 10.1111/j.1439-0329.1993.tb00803.x
Kirisits, T., and Freinschlag, C. (2012). Ash dieback caused by Hymenoscyphus pseudoalbidus in a seed plantation of Fraxinus excelsior in Austria. J. Agric. Ext. Rural Dev. 4, 184–191.
Kjaer, E. D., McKinney, L. V., Nielsen, L. R., Hansen, L. N., and Hansen, J. K. (2011). Adaptive potential of ash (Fraxinus excelsior) populations against the novel emerging pathogen Hymenoscyphus pseudoalbidus. Evol. Appl. 5, 219–228. doi: 10.1111/j.1752-4571.2011.00222.x
Kosawang, C., Amby, D. B., Bussaban, B., McKinney, L. V., Xu, J., Kjær, E. D., et al. (2018). Fungal communities associated with species of Fraxinus tolerant to ash dieback, and their potential for biological control. Fungal Biol. 122, 110–120. doi: 10.1016/j.funbio.2017.11.002
Kowalski, T., and Gajosek, M. (1998). Endophytic mycobiota in stems and branches of Betula pendula to a different degree affected by air pollution. Osterr. Z. Pilzk. 7, 13–24.
Kowalski, T., and Holdenrieder, O. (2009). The teleomorph of Chalara fraxinea, the causal agent of ash dieback. For. Pathol. 39, 304–308. doi: 10.1111/j.1439-0329.2008.00589.x
Lamit, L. J., Lau, M. K., Sthultz, C. M., Wooley, S. C., Whitham, T. G., and Gehring, C. A. (2014). Tree genotype and genetically based growth traits structure twig endophyte communities. Am. J. Bot. 101, 467–478. doi: 10.3732/ajb.1400034
Lobo, A., Hansen, J. K., McKinney, L. V., Nielsen, L. R., and Kjær, E. D. (2014). Genetic variation in dieback resistance: growth and survival of Fraxinus excelsior under the influence of Hymenoscyphus pseudoalbidus. Scand. J. For. Res. 29, 519–526. doi: 10.1080/02827581.2014.950603
Martín, J. A., Witzell, J., Blumenstein, K., Rozpedowska, E., Helander, M., Sieber, T. N., et al. (2013). Resistance to Dutch elm disease reduces presence of xylem endophytic fungi in elms (Ulmus spp.). PLoS One 8:e56987. doi: 10.1371/journal.pone.0056987
McKinney, L. V., Nielsen, L. R., Collinge, D. B., Thomsen, I. M., Hansen, J. K., and Kjaer, E. D. (2014). The ash dieback crisis: genetic variation in resistance can prove a long-term solution. Plant Pathol. 63, 485–499. doi: 10.1111/ppa.12196
McKinney, L. V., Nielsen, L. R., Hansen, J. K., and Kjær, E. D. (2011). Presence of natural genetic resistance in Fraxinus excelsior (Oleraceae) to Chalara fraxinea (Ascomycota): an emerging infectious disease. Heredity (Edinb) 106, 788–797. doi: 10.1038/hdy.2010.119
McKinney, L. V., Thomsen, I. M., Kjær, E. D., Bengtsson, S. B. K., and Nielsen, L. R. (2012). Rapid invasion by an aggressive pathogenic fungus (Hymenoscyphus pseudoalbidus) replaces a native decomposer (Hymenoscyphus albidus): a case of local cryptic extinction? Fungal Ecol. 5, 663–669. doi: 10.1016/j.funeco.2012.05.004
Mikkelson, G. M. (2005). Niche-based vs. neutral models of ecological communities. Biol. Philos. 20, 557–566. doi: 10.1007/s10539-005-5583-7
Millberg, J., Boberg, J., and Stenlid, J. (2015). Changes in fungal community of Scots pine (Pinus sylvestris) needles along a latitudinal gradient in Sweden. Fungal Ecol. 17, 126–139.
Mitchell, R. J., Beaton, J. K., Bellamy, P. E., Broome, A., Chetcuti, J., Eaton, S., et al. (2014). Ash dieback in the UK: a review of the ecological and conservation implications and potential management options. Biol. Conserv. 175, 95–109. doi: 10.1016/j.biocon.2014.04.019
Nguyen, D., Boberg, J., Cleary, M., Bruelheide, H., Hönig, L., Koricheva, J., et al. (2017). Foliar fungi of Betula pendula: impact of tree species mixtures and assessment methods. Sci. Rep. 7:41801. doi: 10.1038/srep41801
Nguyen, D., Boberg, J., Ihrmark, K., Stenström, E., and Stenlid, J. (2016). Do foliar fungal communities of Norway spruce shift along a tree species diversity gradient in mature European forests? Fungal Ecol. 23, 97–108. doi: 10.1016/j.funeco.2016.07.003
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P., O’Hara, R., et al. (2016). vegan: Community Ecology Package. R package version 2.4-0.
Pautasso, M., Aas, G., Queloz, V., and Holdenrieder, O. (2013). European ash (Fraxinus excelsior) dieback – A conservation biology challenge. Biol. Conserv. 158, 37–49. doi: 10.1016/j.biocon.2012.08.026
Pérez-Izquierdo, L., Zabal-Aguirre, M., Flores-Rentería, D., González-Martínez, S. C., Buée, M., and Rincón, A. (2017). Functional outcomes of fungal community shifts driven by tree genotype and spatial-temporal factors in Mediterranean pine forests. Environ. Microbiol. 13, 385–392. doi: 10.1111/1462-2920.13690
Petrini, O., and Fisher, P. J. (1990). Occurrence of fungal endophytes in twigs of Salix fragilis and Quercus robur. Mycol. Res. 94, 1077–1080. doi: 10.1016/S0953-7562(09)81336-1
Pliura, A., Lygis, V., Suchockas, V., and Bartkevičius, E. (2011). Performance of twenty four european fraxinus excelsior populations in three Lithuanian progeny trials with a special emphasis on resistance to Chalara fraxinea. Balt. For. 17, 17–34.
Põlme, S., Abarenkov, K., Nilsson, R. H., Lindahl, B., Clemmensen, K., Kauserud, H., et al. (2021). FungalTraits: a user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 105, 1–16. doi: 10.1007/s13225-020-00466-2
R Development Core Team (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Ragazzi, A., Moricca, S., Capretti, P., Dellavalle, I., and Turco, E. (2003). Differences in composition of endophytic mycobiota in twigs and leaves of healthy and declining Quercus species in Italy. For. Pathol. 33, 31–38. doi: 10.1046/j.1439-0329.2003.3062003.x
Redman, R. S., Sheehan, K. B., Stout, R. G., Rodriguez, R. J., and Henson, J. M. (2002). Thermotolerance generated by plant/fungal symbiosis. Science 298:1581. doi: 10.1126/science.1072191
Riipi, M., Ossipov, V., Lempa, K., Haukioja, E., Koricheva, J., Ossipova, S., et al. (2002). Seasonal changes in birch leaf chemistry: are there trade-offs between leaf growth and accumulation of phenolics? Oecologia 130, 380–390. doi: 10.1007/s00442-001-0826-z
Rodriguez, R. J., White, J. F., Arnold, A. E., and Redman, R. S. (2009). Fungal endophytes: diversity and functional roles. New Phytol. 182, 314–330. doi: 10.1111/j.1469-8137.2009.02773.x
Saikkonen, K., Faeth, S. H., Helander, M., and Sullivan, T. J. (1998). Fungal endophytes: a continuum of interactions with host plants. Annu. Rev. Ecol. Syst. 29, 319–343. doi: 10.1146/annurev.ecolsys.29.1.319
Saikkonen, K., Wäli, P., Helander, M., and Faeth, S. H. (2004). Evolution of endophyte-plant symbioses. Trends Plant Sci. 9, 275–280. doi: 10.1016/j.tplants.2004.04.005
Schlegel, M., Dubach, V., von Buol, L., and Sieber, T. N. (2016). Effects of endophytic fungi on the ash dieback pathogen. FEMS Microbiol. Ecol. 92:fiw142. doi: 10.1093/femsec/fiw142
Schlegel, M., Queloz, V., and Sieber, T. N. (2018). The endophytic mycobiome of European ash and Sycamore maple leave - geographic patterns, host specificity and influence of ash dieback. Front. Microbiol. 9:2345. doi: 10.3389/fmicb.2018.02345
Scholtysik, A., Unterseher, M., Otto, P., and Wirth, C. (2013). Spatio-temporal dynamics of endophyte diversity in the canopy of European ash (Fraxinus excelsior). Mycol. Prog. 12, 291–304. doi: 10.1007/s11557-012-0835-9
Stener, L.-G. (2013). Clonal differences in susceptibility to the dieback of Fraxinus excelsior in southern Sweden. Scand. J. For. Res. 28, 205–216. doi: 10.1080/02827581.2012.735699
Stener, L.-G. (2007). Studie av klonskillnader i känslighet för askskottsjuka. Arbetsrapport från Skogforsk. Nr. 648, 18.
Stener, L.-G. (2018). Genetic evaluation of damage caused by ash dieback with emphasis on selection stability over time. For. Ecol. Manage. 409, 584–592. doi: 10.1016/j.foreco.2017.11.049
Stocks, J. J., Buggs, R. J. A., and Lee, S. J. (2017). A first assessment of Fraxinus excelsior (common ash) susceptibility to Hymenoscyphus fraxineus (ash dieback) throughout the British Isles. Sci. Rep. 7:16546. doi: 10.1038/s41598-017-16706-6
Stone, J., and Petrini, O. (1997). “Endophytes of forest trees: a model for fungus-plant interactions,” in Plant Relationships Part B, eds G. C. Carroll and P. Tudzynski (Berlin: Springer Berlin Heidelberg), 129–140. doi: 10.1007/978-3-642-60647-2_8
Tedersoo, L., Drenkhan, R., Anslan, S., Morales-Rodriguez, C., and Cleary, M. (2019). High-throughput identification and diagnostics of pathogens and pests: overview and practical recommendations. Mol. Ecol. Resour. 19, 47–76. doi: 10.1111/1755-0998.12959
Tellenbach, C., and Sieber, T. N. (2012). Do colonization by dark septate endophytes and elevated temperature affect pathogenicity of oomycetes? FEMS Microbiol. Ecol. 82, 157–168. doi: 10.1111/j.1574-6941.2012.01415.x
Timmermann, V., Børja, I., Hietala, A. M., Kirisits, T., and Solheim, H. (2011). Ash dieback: pathogen spread and diurnal patterns of ascospore dispersal, with special emphasis on Norway. EPPO Bull. 41, 14–20. doi: 10.1111/j.1365-2338.2010.02429.x
Timmermann, V., Nagy, N. E., Hietala, A. M., Børja, I., and Solheim, H. (2017). Progression of ash dieback in Norway related to tree age, disease history and regional aspects. Balt. For. 23, 150–158.
Trapiello, E., Schoebel, C. N., and Rigling, D. (2017). Fungal community in symptomatic ash leaves in Spain. Balt. For. 23, 68–73.
Unterseher, M., Peršoh, D., and Schnittler, M. (2013). Leaf-inhabiting endophytic fungi of European Beech (Fagus sylvatica L.) co-occur in leaf litter but are rare on decaying wood of the same host. Fungal Divers. 60, 43–54. doi: 10.1007/s13225-013-0222-0
Vasiliauskas, R., Bakys, R., Lygis, V., Ihrmark, K., Barklund, P., and Stenlid, J. (2006). “Fungi associated with the decline of Fraxinus excelsior in the Baltic States and Sweden,” in Possible Limitations of Dieback Phenomena in Broadleaved Stands, eds T. Oszako and S. Woodward (Warsaw: Forest Research Institute), 45–53. doi: 10.5586/am.2013.031
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomial RNA genes for phyologenetics,” in PCR Protocols: A Guide to Methods and Applications, eds T. J. Innis, M. A. Gelfand, D. H. Sninsky, and J. J. White (New York, NY: Academic Press Inc.), 315–322.
Wong, B. L., Baggett, K. L., and Rye, A. H. (2009). Cold-season patterns of reserve and soluble carbohydrates in sugar maple and ice-damaged trees of two age classes following drought. Botany 87, 293–305. doi: 10.1139/B08-123
Woodward, S., and Boa, E. (2013). Ash dieback in the UK: a wake-up call. Mol. Plant Pathol. 14, 856–860. doi: 10.1111/mpp.12084
Keywords: fungal diversity, biodiversity loss, endophytic fungi, FUNGuild, Hymenoscyphus fraxineus
Citation: Agostinelli M, Nguyen D, Witzell J and Cleary M (2021) Mycobiome of Fraxinus excelsior With Different Phenotypic Susceptibility to Ash Dieback. Front. For. Glob. Change 4:580514. doi: 10.3389/ffgc.2021.580514
Received: 13 July 2020; Accepted: 01 February 2021;
Published: 08 March 2021.
Edited by:
Alberto Santini, National Research Council (CNR), ItalyReviewed by:
Antonino Malacrinò, The Ohio State University, United StatesRein Drenkhan, Estonian University of Life Sciences, Estonia
Copyright © 2021 Agostinelli, Nguyen, Witzell and Cleary. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michelle Cleary, TWljaGVsbGUuQ2xlYXJ5QHNsdS5zZQ==
 Marta Agostinelli1
Marta Agostinelli1 Diem Nguyen
Diem Nguyen Johanna Witzell
Johanna Witzell Michelle Cleary
Michelle Cleary