- 1Institute of Soil Science and Land Evaluation, University of Hohenheim, Stuttgart, Germany
- 2Genomic and Applied Microbiology & Göttingen Genomics Laboratory, Georg-August University of Göttingen, Göttingen, Germany
- 3Institute of Crop Science, University of Hohenheim, Stuttgart, Germany
- 4Forest Botany and Tree Physiology, Georg-August University of Göttingen, Göttingen, Germany
Phosphorus (P) is an essential nutrient, but European forest ecosystems are experiencing widespread declines in soil P concentrations. To clarify the roles of ectomycorrhizal (EM) and saprotrophic (SAP) fungi in P cycling in forest soils that differ in inorganic and organic P availability, we conducted an ingrowth tube (IGT) experiment over 18 months of exposure in five beech (Fagus sylvatica L.) forests in Germany. To separate the contributions of both fungal guilds in situ, two different types of IGTs were used: (i) open IGTs with micromesh windows to allow EM fungi to regrow into the tubes, and (ii) closed IGTs (without mesh windows) in which recolonization by EM hyphae was restricted. We then measured phosphomonoesterase, phosphodiesterase, peroxidase, and phenoloxidase activities in EM (open IGTs) and SAP (closed IGTs) dominated fungal assemblages. Shifts in fungal assemblages occurred over time, but the speed and extent of those shifts differed between the five forest sites. In three forest sites fungal community composition shifted toward greater dominance of EM fungi in open IGTs than in closed IGTs after a period of 18 months, but these changes were not apparent in two other sites. Although shifts in fungal community composition were noticeable between treatments, we found no effect on phosphomonoesterase activity. Since phosphomonoesterase is present in both fungal guilds, this indicates functional redundancy of fungi. Phosphodiesterase activity was slightly reduced in SAP dominated IGTs, indicating that SAP fungi contribute to phosphodiesterase activity to a lesser extent than do EM fungi. P cycling enzymes did not appear to have been influenced by the total P stocks of the forest sites but may have been affected by additional abiotic factors. Contrary to our expectation, phenoloxidase activity was unaffected by fungal community composition, and only peroxidase activity was higher in SAP dominated IGTs, indicating that the contribution of EM fungi to degradation of complex organic material is driven by the need to obtain nutrients in addition to C. Overall, the results of this study contribute to a better understanding of the roles of EM and SAP fungi in P cycling in forest soils.
Introduction
Foliar P content in European beech (Fagus sylvatica L.) forest ecosystems is declining, suggesting that these forests may encounter future P deficiency (Jonard et al., 2015; Talkner et al., 2015). Plants take up P in the form of phosphate anions HPO42– and H2PO4– (Jones and Oburger, 2011). However, inorganic P (Pinorg) is frequently adsorbed to minerals and the equilibrium concentration of orthophosphates in soil solution is low; therefore, only a minor fraction of total soil P (Ptot), which is often <1%, is plant available (Condron et al., 2005; Quiquampoix and Mousain, 2005). In forest soils, a large fraction of Ptot is bound in the organic P (Porg) pool, and this can account for 35–83% of Ptot in beech forests (Talkner et al., 2015; Lang et al., 2017; Zederer et al., 2017). Porg comprises orthophosphate monoesters (e.g., inositol phosphates), orthophosphate diesters (e.g., nucleic acids, phospholipids), phosphonates, and phosphate anhydrides (Condron et al., 2005). In contrast to the geochemical P cycle, for which exchange processes of soil P pools due to abiotic factors are well understood (Frossard et al., 2000), little is known about the importance of biological regulation mechanisms of the P cycle and the contributions of microbial groups to these processes (Tamburini et al., 2012).
Microorganisms are significantly involved in the P cycle through solubilization, immobilization, and mineralization of Porg and Pinorg (Richardson and Simpson, 2011), thus mediating the transfer of P between different pools (Plante, 2007). Ectomycorrhizal (EM) and saprotrophic (SAP) fungi are major microbial groups in forest soils and pursue functionally different lifestyles with respect to their carbon (C) acquisition strategies. EM fungi form symbioses with host trees, which supply the fungi with C through photo-assimilates such as sucrose and in return benefit from nutrients and water supplied by EM fungi (Smith and Read, 1997). Especially in nutrient poor ecosystems, mycorrhizal plants accumulate more P than non-mycorrhizal plants because the mutualistic fungi can access a larger soil volume through their branched hyphal network (Smith and Read, 1997; Lambers et al., 2008; Jansa et al., 2011). This beneficial interaction results in an increase in foliar P concentration (Wallander et al., 1997; Alvarez et al., 2009; Danielsen and Polle, 2014).
In contrast to mycorrhizal fungi associated with roots, SAP fungi degrade organic material to obtain C, thereby contributing significantly to the decomposition of organic substances in forest soils (Grinhut et al., 2007; Baldrian, 2008). Mineralization of organic compounds by SAP fungi is directly linked with the turnover of nutrients (Attiwill and Adams, 1993). The release of Pinorg is a process catalyzed by extracellular phosphatase enzymes, which are secreted into the soil by microorganisms and plants in response to P demand (Quiquampoix and Mousain, 2005). The extent of enzyme production and excretion is highly regulated by the availability of Pinorg. Low Pinorg availability stimulates phosphatase activity and high Pinorg availability inhibits it (Kandeler, 1990; Burns and Dick, 2002; Kavka and Polle, 2017). In addition to their production of specific P cycle enzymes, fungi can decompose complex organic matter due to their ability to produce extracellular per- and phenoloxidases (Baldrian, 2009; Burke et al., 2014; Bödeker et al., 2014). The cleavage of high molecular weight organic carbon compounds is necessary for fungi in order to access N and P containing compounds stored in complex biotic structures such as above- and belowground plant residues. The potential to decompose complex organic matter was previously ascribed to SAP fungi, but recent studies have demonstrated that enzymes required for organic matter degradation are present also in the genomes of EM fungi (Burke and Cairney, 2002; Talbot et al., 2013). Compared to other fungal guilds (i.e., white rot and brown rot saprotrophs), however, the abundance of genes involved in soil organic matter (SOM) decomposition is lower in EM fungi (Martino et al., 2018), indicating that EM fungi may not be able to modify SOM to the same extent as free-living SAP fungi (Zak et al., 2019). Additionally, fungi and bacteria are capable of solubilizing P from recalcitrant inorganic sources (e.g., rock material) by secretion of low molecular weight organic “acid” anions such as oxalates and gluconate (Kucey et al., 1989; Gyaneshwar et al., 2002).
Here, our goal was to disentangle the contributions of EM and SAP fungal assemblages to P mobilization in forest soils. To address this goal we used open and closed ingrowth tubes (IGTs) (Wallander et al., 2001; Wallander, 2006; Wallander and Thelin, 2008). The tubes were filled with forest soil, and by a sand barrier at the top and bottom of each tube fine root accession was avoided, while nutrient and water supply from the surrounding soil into the tubes was enabled. A lateral micromesh window in the open IGTs permitted EM fungal hyphae to recolonize the IGTs, while in closed IGTs (without micromesh windows) recolonization by EM fungi was restricted. We expected that the contribution of fungal guilds to P mobilization depended on P availability in the soil. Therefore, we selected five forests along a geosequence characterized by a gradient in soil P content (Lang et al., 2017) to study fungal abundance and community composition as well as enzyme activity.
We hypothesized: (I) In closed IGTs the fungal communities are shifted toward SAP dominated assemblage because of an EM fungal dieback resulting from a lack of available photo-assimilates. In contrast, the symbiotic EM fungi are able to recolonize the soil in the open IGTs. (II) The phosphomono-and phosphodiesterase activities depend on the proportion of SAP to EM fungi, with higher phosphatase activities in EM dominated soil, since the ability to produce high levels of phosphatases are attributed to symbiotic fungi. (III) The activities of P-cycle enzymes depend on soil P stocks and therefore acid phosphomono- and phosphodiesterase activities are site specific, having lower values in forests with high P stocks and higher values in forests with limited P stocks. (IV) If EM fungi do contribute substantially to the oxidation of organic substrates, the activities of peroxidases and phenoloxidases decrease with decreasing EM abundance in the closed IGTs.
Materials and Methods
Study Sites
The experiment was conducted in five even-aged beech (Fagus sylvatica L.) forests located across Germany (Table 1). The soils were formed from different parent materials and differed in P stocks in decreasing order as follows: Bad Brückenau (BBR), over Mitterfels (MIT), Vessertal (VES), Conventwald (CON) to Lüss (LUE) decreasing from 904 to 164 g Ptot m–2 in depth to 1 m, with ∼50–70% belonging to the Porg pool in the Ah1 horizon (Table 1). According to the IUSS Working Group WRB (2014), all investigated soils are cambisols with pH values from 3.4 to 4. For more soil properties of each site we refer to Lang et al. (2017) and http://www.ecosystem-nutrition.uni-freiburg.de/standorte.
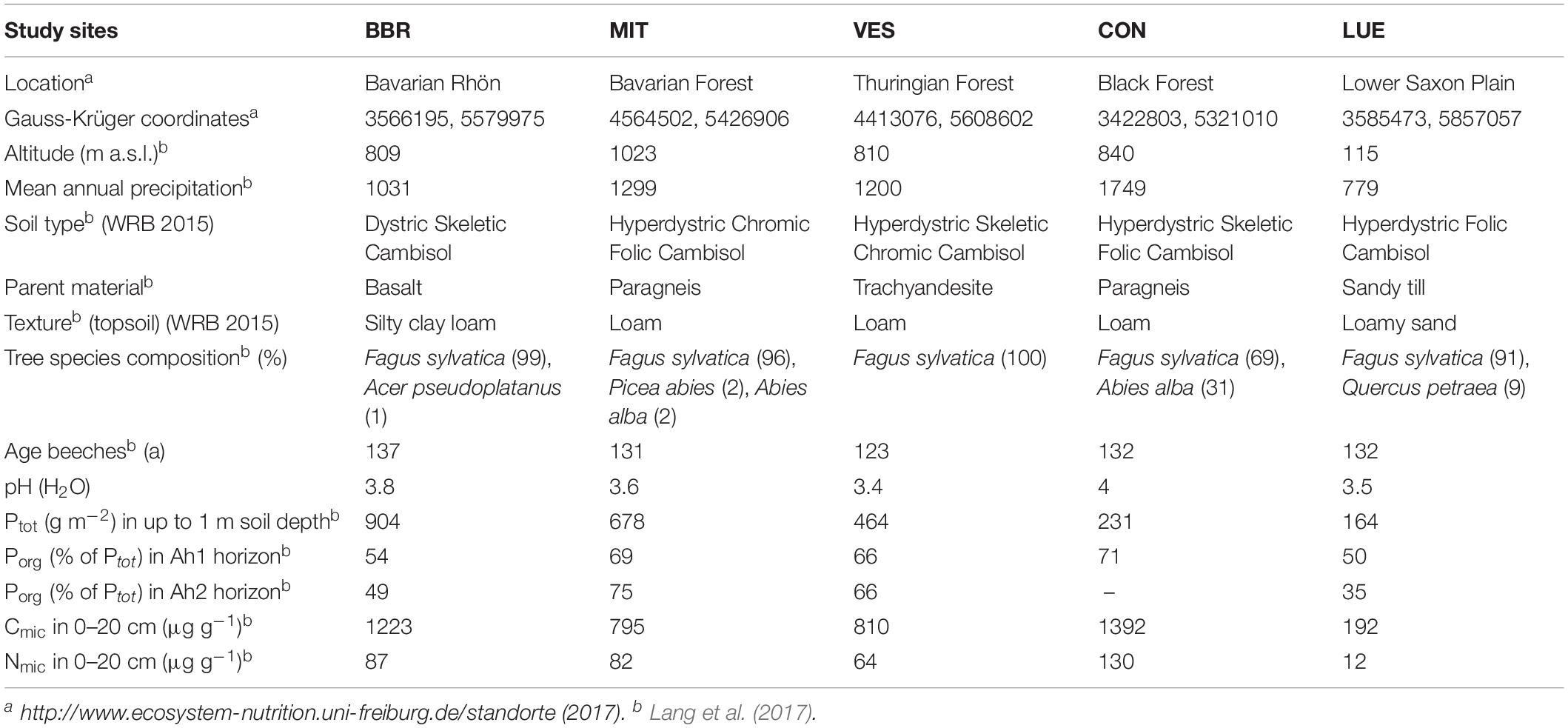
Table 1. Summary of soil characteristics at the five study sites in Bad Brückenau (BBR), Mitterfels (MIT), Vessertal (VES), Conventwald (CON), and Lüss (LUE), modified from Lang et al. (2017).
Preparation, Installation, and Sampling of Ingrowth-Tubes (IGTs)
To separate EM and SAP fungal communities, we installed in situ “open” and “closed” ingrowth tubes (IGTs, size: height of 10 cm, diameter of 5 cm) after descriptions of Johnson et al. (2001), Wallander et al. (2001), and Hendricks et al. (2006). The open IGTs were constructed with 340° micromesh windows (50 μm mesh size, SEFAR NITEX 03-50/37, SEFAR GmbH, Edling, Germany), surrounding the tubes, allowing EM derived hyphae, but not plant roots, to grow into the tubes from ambient soil (see Supplementary Figure S1). In contrast, the closed IGTs (without surrounding micromesh) did not allow any ingrowth of hyphae. All IGTs were filled with topsoil (0–20 cm) from the respective forest sites; these had been collected 1 week before tube filling and kept at 4°C. The soil was sieved (<2 mm) to minimize possible carryover of roots. The IGTs were filled to a bulk density of ∼1 g cm–3, reflecting average bulk densities of the different forest topsoils (Lang et al., 2017). Tops and bottoms of the tubes had 2 cm mesh borders (50 μm mesh size) filled with sand to avoid fine root accession and hyphal migration of saprotrophic fungi which may have been promoted by starvation of their hyphae while nutrient foraging (Lori Phillips’ and Melanie Jones’ pers. comm. in Wallander et al., 2013). The sand barriers were made of a 1:1 mixture of carbonate free mineral sand of two differing grain sizes (aquarium sand 0.4–0.8 mm, aquarium sand 1.0–2.0 mm; SCHICKER Mineral GmbH, Bad Berneck, Germany) according to Wallander et al. (2001). This sand barrier was successful as at none of the harvests were plant roots found inside of our IGTs.
In spring 2014, five 30–40 year old beech trees were selected in each of the forests and four IGT treatment sets (each consisting of one open IGT and one closed IGT) were randomly installed around each selected tree at a distance of 2.5 m from the tree. The open and closed IGT of a given set were installed at a distance of 20–30 cm from each other to expose them to comparable microhabitat conditions. The IGTs were vertically buried in the top 20 cm of the soil, in close contact to the surrounding soil. The tops of the upper sand barriers were subsequently covered with a ∼3 cm thick topsoil layer and re-mulched with litter from the surrounding forest floor. For sampling, one IGT set from each tree was removed after 3 months in summer 2014, after 6 months in autumn 2014, after 14 months in summer 2015, and after 18 months in autumn 2015. At the time of removal of each IGT set, additional soil was collected from undisturbed soil in close proximity to the IGT set. Thus, 300 samples were obtained in total (5 forest sites × 3 treatments × 5 replicates × 4 time points). The control samples were also sieved at 2 mm and all samples were frozen at −20°C until further analyses. Soil water content was determined gravimetrically by drying samples at 105°C for 24 h.
Determination of Phospholipid Fatty Acid (PLFA) Contents
For determination of microbial biomass, phospholipid fatty acid (PLFA) analysis was carried out according to Frostegård et al. (1991) and Frostegård et al. (1993). Initially, 4 g fresh soil was mixed with 18.4 ml Bligh & Dyer reagent [chloroform, methanol, and citrate buffer (pH 4), 1:2:0.8]. For lipid fractionation, the extract was pipetted onto an extraction column and subsequently diluted with chloroform, acetone, and methanol (Frostegård et al., 1991). Fractionation occurred after alkaline methylation according to Dowling et al. (1986) via methanolic potassium hydroxide, hexane chloroform solution, and acetic acid as described by Ruess et al. (2007). The resulting PLFA methyl esters were dissolved in iso-octane and measured on a gas chromatograph (GC, AutoSystem XL, PerkinElmer Inc., Norwalk, CT, United States). The GC was equipped with a flame ionization detector, an HP-5 capillary column (cross-linked 5% phenyl methyl silicone; 50 m × 0.2 mm, film thickness: 0.33 μm) and helium as carrier gas. The initial column temperature of 70°C was held for 2 min. Temperature was then increased by 30°C min–1 to 160°C, then by 3°C min–1 to 280°C and held for 15 min. Injection temperature was set at 260°C. The concentration of the fatty acid methyl ester (FAME) was calculated via an internal C24:1 FAME standard, which was added to the samples before methylation. Fungal PLFAs were represented by 18:2ω6,9, gram-positive bacterial PLFAs by i15:0, a15:0, i16:0, i17:0 and gram-negative bacterial PLFAs by cy17:0 and cy19:0. The sum of gram-positive and-negative bacterial PLFAs as well as 16:1ω7 represented the sum of bacterial PLFAs. Total microbial PFLAs were calculated as the sum of all previously mentioned PLFAs plus PLFAs 15:0 and 16:1ω5 (Frostegård and Bååth, 1996; Kandeler et al., 2006).
Molecular Analysis of Relative Fungal Abundance and Fungal Diversity (Illumina Sequencing)
Soil samples were homogenized in a ball mill Type MM400 (Retsch GmbH, Haan, Germany) in 25 ml containers fitted with a stainless steel ball (20 mm) under liquid nitrogen at 30 Hz s–1 for 2 min. DNA isolations were initiated by transferring 250 mg sample material into a bead beating tube containing glass beads (2 ml, MO BIO Laboratories Inc., Carlsbad, CA, United States). Homogenization was conducted under liquid nitrogen for 10 min in the ball mill (30 Hz s–1). DNA was extracted with the PowerSoil DNA isolation kit (MO BIO Laboratories Inc., Carlsbad, CA, United States). For DNA elution, a total of 100 μl nuclease-free water (AppliChem, Darmstadt, Germany) instead of buffer supplied by the kit was used for each sample. DNA yields were estimated with a NanoDrop ND-1000 spectrophotometer (Peqlab Biotechnologie GmbH, Erlangen, Germany).
To characterize fungal community composition, the internal transcribed spacer region ITS1 was amplified by PCR using the specific forward primer ITS1-F_KYO1 according to Toju et al. (2012) and the reverse primer ITS2 according to White et al. (1990). Both primers were labeled with Illumina MiSeq specific overhang adapters (Microsynth, Balgach, Switzerland), i.e., ITS1-F_KYO1 5′-TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG CTH GGT CAT TTA GAG GAA STA A-3′ and ITS2 5′ GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGC TGC GTT CTT CAT CGA TGC-3′. The polymerase chain reaction (PCR) mix was composed of 5 μl template DNA, 10 μl 5x Phusion GC buffer (New England Biolabs, Frankfurt a. M., Germany), 0.15 μl MgCl2 (50 mM, New England Biolabs, Frankfurt a. M., Germany), 2.5 μl DMSO (5%, New England Biolabs, Frankfurt a. M., Germany), 2.5 μl BSA (8 mg/ml, Sigma Aldrich, St. Louis, MO, United States), 1 μl of each primer (10 mmol/l, Microsynth, Wolfurt, Austria), 1 μl dNTP mix (10 mM each, Thermo Fisher Scientific, Osterode am Harz, Germany), 0.5 μl Phusion high-fidelity DNA polymerase (2 U/μl, New England Biolabs, Frankfurt a. M., Germany) and adjusted to a total volume of 50 μl with nuclease-free distilled water. PCR reactions were performed in a SensoQuest Labcycler (Göttingen, Germany). The PCR program was initiated at 98°C (hold), followed by 98°C for 30 s, 25 cycles of 10 s at 98°C (denaturation), 20 s at 47°C (annealing), 20 s at 72°C (extension) and terminated at 72°C for 5 min.
PCR products were subjected to electrophoresis in 1.2% agarose gels (Biozym LE Agarose, Biozym Scientific GmbH Hessisch Oldendorf, Germany) using 1x running buffer (recipe 5x running buffer: 89 mM tris ultra, 89 mM boric acid, 2 mM EDTA, pH 8.0–8.3) and 10 × loading buffer (600 μl glycerin (99.5%), 50 μl SDS (20%), 200 μl TBE buffer (5x), 100 μl EDTA (0.5 M, pH 8.0), 10 μl bromophenol blue, 40 μl H2O). Gel Red (10,000x) was used for staining (VWR International, Darmstadt, Germany). PCR products were observed using a Raytest Fluorescence Laser Scanner FLA-5100 and evaluated with the Aida Image Analyzer v. 4.27 (both: Raytest GmbH, Straubenhardt, Germany). All PCR reactions were performed in triplicate and pooled and purified using the innuPREP PCRpure kit (Analytik Jena, Jena, Germany). PCR products were run on an agarose gel again and cut in the range of 300–400 bp using the QIAquick Gel extraction kit (Qiagen GmbH, Hilden, Germany) as recommended by the manufacturer. Each sample was eluted in 20 μl nuclease-free water (AppliChem, Darmstadt, Germany) and a UV table (INTAS UV System Type N80M, Göttingen, Germany). Quantification of purified PCR products was done using the Quant-iT dsDNA HS assay Kit (Life Technologies GmbH, Darmstadt, Germany) and a Qubit fluorometer (Life Technologies GmbH, Darmstadt, Germany).
Amplicon libraries were normalized, pooled and sequenced on the Illumina MiSeq platform with MiSeq reagent kit v.3 as recommended by the manufacturer (Illumina, San Diego, CA, United States). Resulting paired-end ITS sequence datasets were merged with PEAR (version 0.9.10; Zhang et al., 2014), quality-filtered and primer clipped with split_libraries_fastq.py from the QIIME 1.9.0 software package (Caporaso et al., 2010). Additional primer clipping was performed with cutadapt (version 1.12) (Martin, 2011). High-quality reads were further processed with USEARCH (version 9.2.64_i86linux64; Edgar, 2010), which included, in the following order: size filter (sequences shorter 140 bp were discarded), dereplication, denoising (UNOISE), reference-based (UNITE version 7.2) chimera removal, sequence sorting by length, and OTU determination at 97% sequence identity. Finally, the high quality reads were mapped against the OTUs with USEARCH and an OTU table was generated (Edgar, 2010). Taxonomic classification of OTU sequences was inferred with parallel_assign_taxonomy_blast.py against the UNITE database (version 7.2) (Kõljalg et al., 2013). Taxonomic information was added to the OTU table with biom tools (McDonald et al., 2012). Unidentified fungal OTUs were searched with BLASTn (Altschul et al., 1990) against the nt database (version March 2017) to remove non-fungal OTUs, and only reads classified as fungi were kept. Finally, unclassified OTUs and extrinsic domain OTUs (Protista, Plantae) were removed from the table by employing filter_otu_table.py (Caporaso et al., 2010). Sample comparisons were performed at the same surveying effort, utilizing the lowest number of sequences by random selection (3,650). Fungal sequences were assigned to functional guilds using FunGuild (Nguyen et al., 2016).
The ITS1 region sequences were deposited in the National Centre for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under bioproject accession number PRJNA592056.
Easily Soluble (Psol) and Microbially Bound P (Pmic)
Easily soluble inorganically bound P (Psol) and microbially bound P fractions (Pmic) were extracted according to Hedley et al. (1982). Psol was extracted from fresh soil equivalents to 2 g air-dried soil with 30 ml ultrapure water in presence of a 2 × 3 cm anion exchange membrane (551642S, VWR-International, Darmstadt, Germany) in bicarbonate form. To extract P bound in microbial cells, a second subsample was prepared with 1 ml hexanol in addition to the soil, ultrapure water, and anion exchange membrane (PHexanol). A third subsample (PControl) included a defined P concentration of 25 μg P g–1 soil DM (spike) for later calculation correction. All subsamples were shaken on a horizontal shaker at room temperature for 18 h. Subsequently, the membranes were washed with deionized water (H2Odeion), dried and transferred to a fresh vessel. To remove the P bound onto the membranes, they were shaken in 20 ml 0.5 M HCl for 1 h. After dyeing 1 ml of each sample with 2 ml M&R reagent (Murphy and Riley, 1962), the subsamples were measured photometrically at 712 nm with a microplate reader (ELx808, Absorbance Microplate Reader, BioTek Instruments Inc., Winooski, VT, United States). Percent recovery of spiked P was expressed as:
Using R and the extraction factor kEP of 0.4 (Brookes et al., 1982) as the relative proportion of hexanol-extractable inorganic P (PHexanol) in total microbial P, Pmic was calculated as following:
Potential Enzyme Activities
The activity of acid phosphomonoesterase (EC 3.1.3.2) was determined in duplicate using the phenyl phosphate method according to Hoffmann (1968), modified by Öhlinger et al. (1996). For this, a 10 ml mixed buffer (pH 5) consisting of 1 M glacial acetic acid and 1 M sodium acetate trihydrate (1:2), as well as 5 ml 0.1 M disodium phenyl phosphate dihydrate (EC 3279-54-7) as substrate solution, were added to 5 g fresh soil. In one control sub-sample, the substrate solution was replaced by H2Odeion. All samples were incubated with constant shaking (100 rpm) in a water bath at 37°C for 3 h. Afterward, 35 ml H2Odeion was added, and samples were shaken and filtered. Next, 0.1 ml filtrate was added to 1 ml borate buffer (12.4 g boric acid, solubilized in 100 ml 1 M sodium hydroxide and H2Odeion, pH adjusted to 10 and brought to 1 L volume with H2Odeion). For measurement, 0.2 ml of 2.6-dibromchinone-4-chlorimide (EC 202-937-2) color reagent (2 mg ml–1 ethanol, 60%) was added with 5 ml H2Odeion. After 30 min and dilution with 13.9 ml H2Odeion, absorbance was measured on a microplate reader (ELx808, Absorbance Microplate Reader, BioTek Instruments Inc., Winooski, VT, United States) at 630 nm.
The activity of phosphodiesterase (EC 3.1.4.2) was measured according to Browman and Tabatabai (1978), modified by Schinner et al. (2012). In duplicate, 4 ml of 0.05 M Tris(hydroxymethyl)aminomethane [Tris(THAM)] buffer (pH 8.0) and 1 ml 5 mM bis(4-nitrophenyl) phosphate sodium salt (EC 223-739-2) as substrate solution were added to 1 g fresh soil. One of these sub-samples was prepared without substrate solution to serve as control. All samples were incubated under constant shaking (100 rpm) in a water bath at 37°C for 1 h. Subsequently, 1 ml 0.5 M calcium chloride solution and 4 ml 0.1 M Tris (THAM) buffer (pH 12.0) were pipetted into the samples. Additionally, 1 ml of the substrate solution was mixed into the control sub-samples. After filtering, all samples were measured at 405 nm using a microplate reader (ELx808, Absorbance Microplate Reader, BioTek Instruments Inc., Winooski, VT, United States).
Measurements of peroxidase and phenoloxidase activities were carried out photometrically using ABTS (2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid)) as substrate as described by Floch et al. (2007). A mixture of 50 ml H2Odeion and 0.4 g fresh soil was ultrasonically suspended (50 J s–1). Peroxidase activity was calculated by subtracting phenoloxidase activity from total enzyme activity. To this end, microplates were prepared for total enzyme activity and phenoloxidase activity analyses separately (Bach et al., 2013). For analysis of total enzymatic activity, 50 μl soil solution was pipetted into each microplate well with 140 μl modified universal buffer [MUB; 2.42 g Tris(THAM) buffer, 2.32 g maleic acid, 2.80 g citric acid monohydrate, 1.26 g boric acid≥ 99.8% solubilized in H2Odeion, adjusted to pH 3, brought to 1 L volume], 50 μl 2 mM ABTS substrate solution, and 10 μl H2O2. Total volume in each well was 250 μl.
For phenoloxidase activity, 100 μl soil solution and 100 μl MUB were mixed in each microplate well and 50 μl 2 mM ABTS substrate solution was added. A reference sample without substrate was prepared for each sample. Additionally, negative controls without soil were prepared for total enzyme and phenoloxidase activity, respectively. Microplates with sample solutions and substrate were pre-incubated at 30°C for at least 5 min (Floch et al., 2007). Measurement of the resulting stable radical cation ABTS*+ was carried out with a microplate reader (SYNERGY HTX, BioTek Instruments Inc., Winooski, VT, United States) at 30°C at wavelength 650 nm for 30 min every 3 min.
Statistical Analyses
Statistical analyses were performed with R version 3.5.2 (R Core Team, 2019). A linear mixed-effects model (nlme package version 3.1-140; Pinheiro et al., 2019) was used with treatments, harvest dates, sites, and their interaction as fixed effects; and trees (representing the replicates), position of IGT sets around the trees as cardinal direction, and sample IDs as random effects. The fit of the model was assessed based on normal distribution and heteroscedasticity of model residuals and the data were square-root or log-transformed to achieve normality of distribution when necessary. Results of the fitted models are given for the full dataset in Supplementary Table S1 and for the exposure time of 18 months in Supplementary Table S2. Tukey’s tests for comparison of means between open and closed IGTs at specific sampling dates, as well as control soils between forests, were run with the package emmeans (version 1.3.5; Lenth et al., 2019) at a significance level of 5% (see bold values in Supplementary Tables S3, S4, respectively).
Principal component analysis (PCA) was carried out on enzyme activities, PLFAs, and abundant EM and SAP genera (absolute numbers of OTUs per genus) in order to determine how enzyme activities varied as a function of fungal community differences. This was done using the “prcomp” function of the R stats package (R Core Team, 2019).
Results
Shifts in the Relative Abundances and Community Structure of Ectomycorrhizal (EM) and Saprotrophic (SAP) Fungi
At 3 and 6 months after exposure of the IGTs, no shifts in the EM:SAP ratios of the fungal communities were detected (Figures 1A,B). In the second year, 14 and 18 months after exposure, the ratio of EM:SAP fungi in the open IGTs increased in three of five forests (MIT, CON, and LUE) (Figure 1A). These shifts resulted from dynamic changes in both EM and SAP fungi (Table 2 and Supplementary Table S3). Highest recolonization of open IGTs was found in CON, with an increase in the relative abundance of EM fungi from 10 ± 7% to 54 ± 14% (Table 2 and Supplementary Table S3) and a decrease in SAP fungi from 65 ± 5% to 21 ± 10%. Similar changes occurred in the MIT and LUE soils (p = 0.002). In contrast to the strong changes in the EM and SAP fungal communities in the open IGTs, the EM:SAP fungal ratios in the closed IGTs varied only slightly over the 18 month exposure period in all the studied forests (Figure 1B).
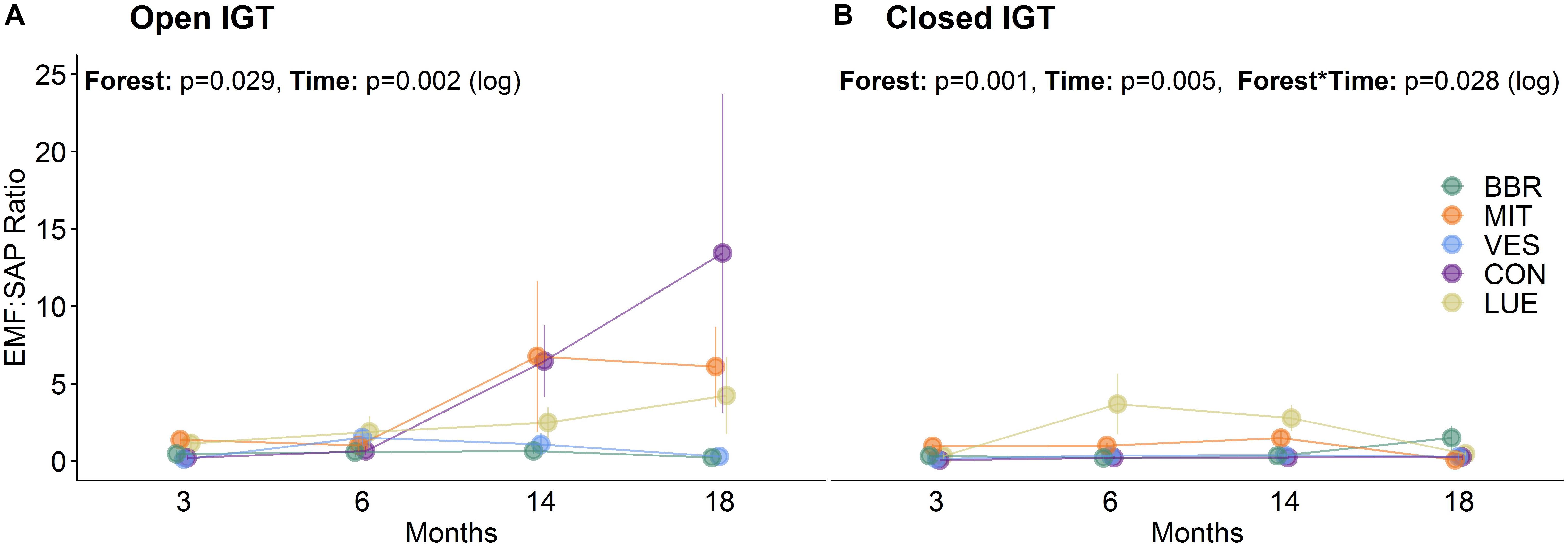
Figure 1. Ratio of ectomycorrhizal to saprotrophic fungi (EM:SAP) in (A) open ingrowth tubes (IGTs) and (B) closed IGTs based on absolute number of OTUs in Bad Brückenau (BBR), Mitterfels (MIT), Vessertal (VES), Conventwald (CON), and Lüss (LUE) over 18 months of exposure (n = 5).
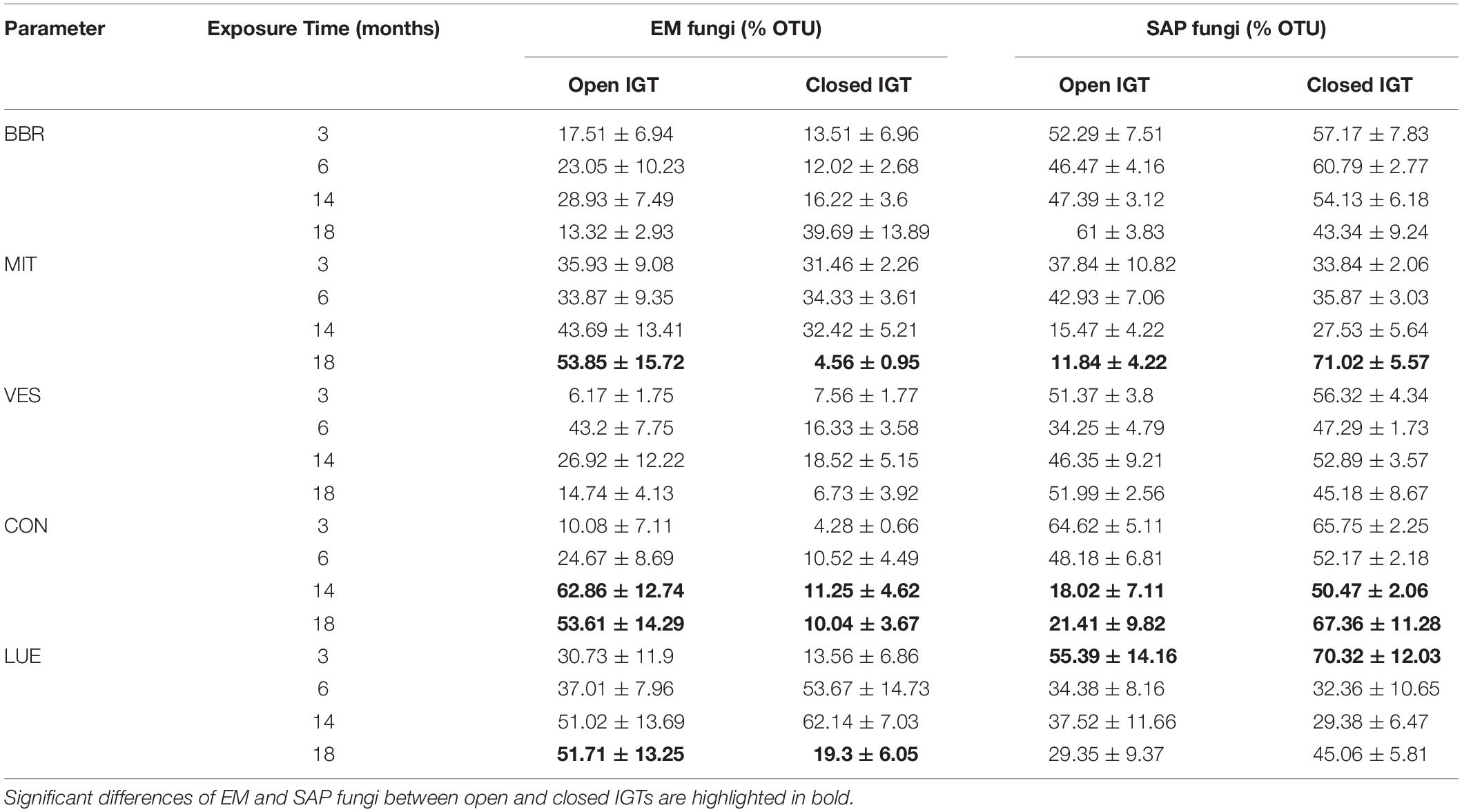
Table 2. Relative proportion of ectomycorrhizal (EM) and saprotrophic (SAP) fungi OTUs in open and closed ingrowth tubes (IGTs) based on Illumina sequencing in Bad Brückenau (BBR), Mitterfels (MIT), Vessertal (VES), Conventwald (CON), and Lüss (LUE) over 18 months of exposure (n = 5).
Community composition of EM and SAP fungi varied among the IGTs in the investigated beech forests (Figures 2A,B). Unexpectedly, we detected an increase in the abundance of EM fungal genera and a decrease in SAP fungi in the BBR forest, but in the other four forests SAP fungi increased (Figure 2B) and EM fungi decreased (Figure 2A). The decrease in relative abundance of EM fungi in the closed IGTs at MIT, VES, CON and LUE was mainly due to decreases in the genera Russula and Craterellus spp. (Figure 2A). In the closed IGTs at BBR forest, the genera Amanita, Imleria and Cenococcum spp. increased in abundance (Figure 2A). Among SAP, Solicoccozyma and Mortierella spp. were most affected by the treatments in all forests, increasing dramatically during the exposure period (Figure 2B). These results show that our experimental approach with open and closed IGTs successfully established distinct EM and SAP fungal assemblages, but other fungal groups with unclear EM status (such as Entoloma, Meliniomyces, and Sistotrema spp.) were unaffected by the treatment after 18 months of exposure (Supplementary Table S3).
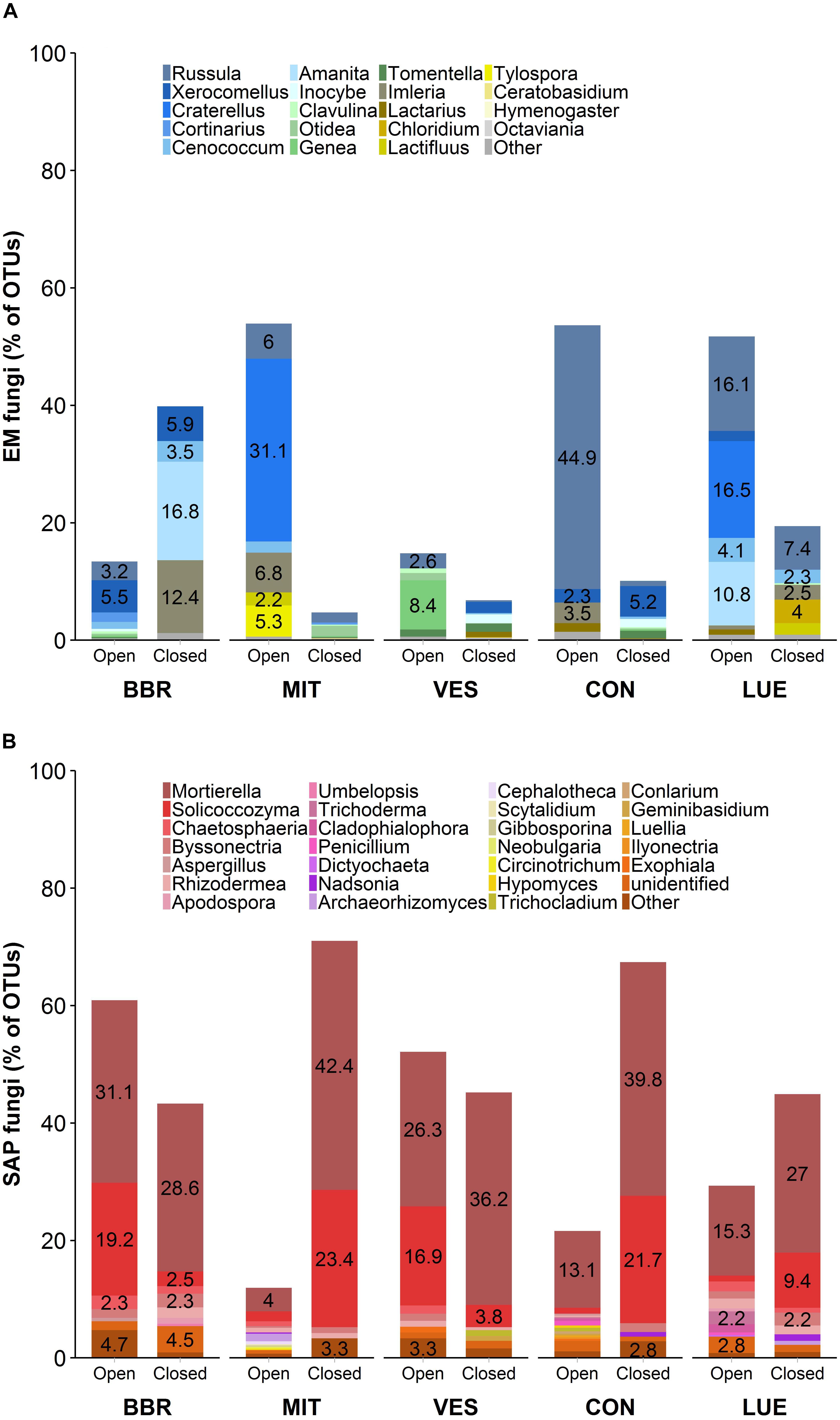
Figure 2. Fungal community composition of (A) ectomycorrhizal (EM) fungi and (B) saprotrophic (SAP) fungi in open and closed IGTs based on Illumina Sequencing data in Bad Brückenau (BBR), Mitterfels (MIT), Vessertal (VES), Conventwald (CON), and Lüss (LUE) after 18 months of exposure (n = 5).
Phospholipid Fatty Acid (PLFA) Content
Total microbial PLFA content in the closed IGTs was on average 8% lower than in the open IGTs after 18 months (Supplementary Figure S2A). Similar results were found for 18:2ω6,9 (p < 0.001), which is considered to be a fungal biomarker (Figure 3). The difference between open and closed IGTs was also mirrored in bacterial biomass, with the strongest decrease in VES (13%) (Supplementary Figure S2B). The five forests differed significantly in their fungal and bacterial PLFA contents (Figure 3 and Supplementary Figure S2): BBR and CON had the highest and LUE the lowest fungal PLFA content (p = 0.026). The fungal-to-bacterial ratios of PLFAs were unaffected by treatment after 18 months of IGT exposure.
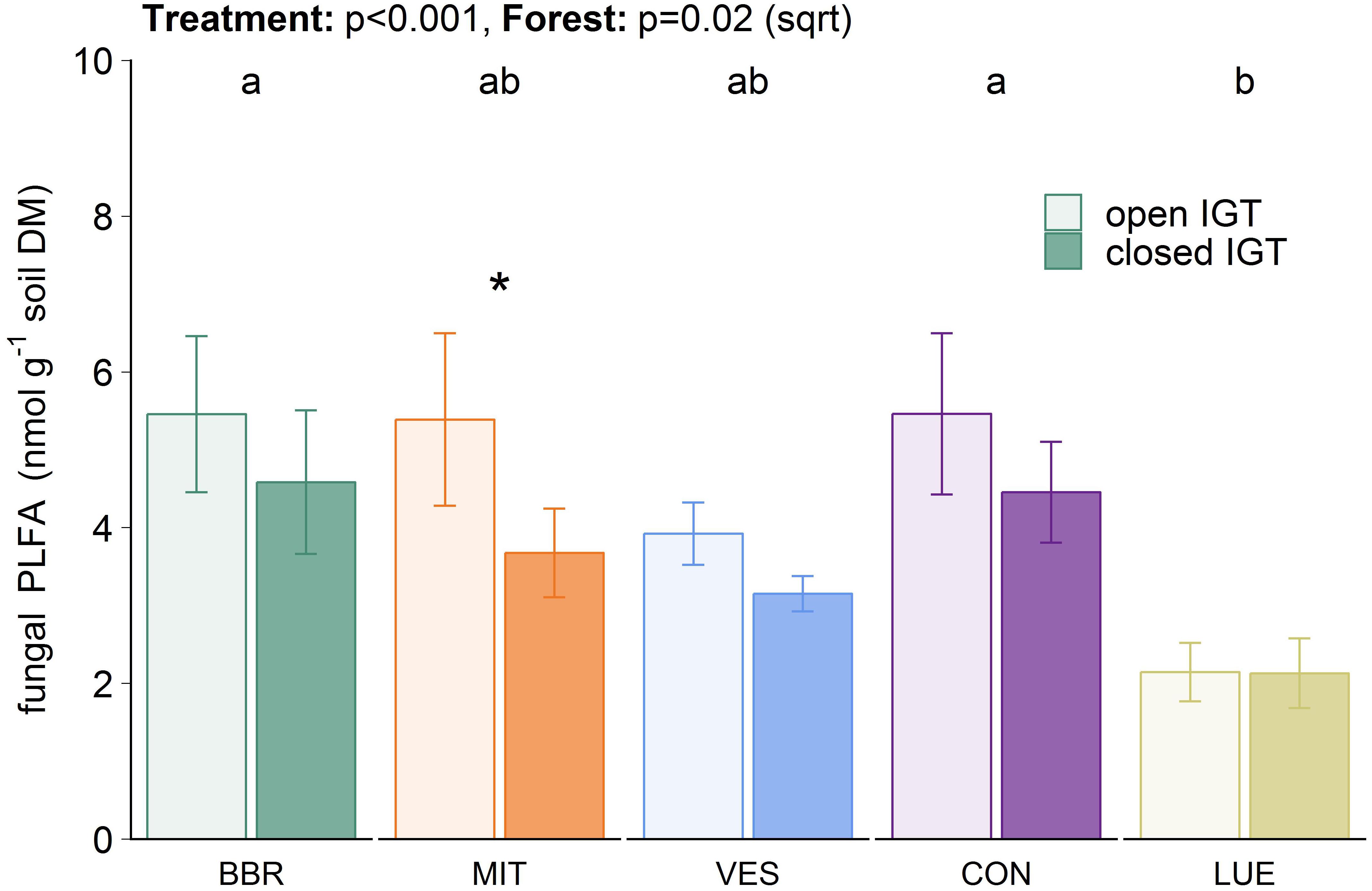
Figure 3. Fungal biomass in open and closed ingrowth tubes (IGTs) based on phospholipid fatty acid analysis in Bad Brückenau (BBR), Mitterfels (MIT), Vessertal (VES), Conventwald (CON), and Lüss (LUE) after 18 months of exposure (n = 5). Forests sharing no letter are significantly different and asterisks highlight significant differences between open and closed IGTs at the respective forest site.
Easily Soluble P Fraction (Psol) and Microbial Bound P (Pmic)
The content of water-extractable P (Psol) was not influenced by the type of IGT (Figure 4A). However, Psol content was forest-specific (p < 0.001) with much higher values in VES than in the other forests. The microbial P (Pmic) content did not differ between treatments (Figure 4B), although a trend was seen, with an increase in the Pmic content between 14 and 18 months in all forests (p = 0.083, Supplementary Table S3). The installation of the IGTs had no effect on microbially bound P values, as the Pmic values inside the cores did not change in comparison to the ambient soil (Figure 4B and Supplementary Table S4).
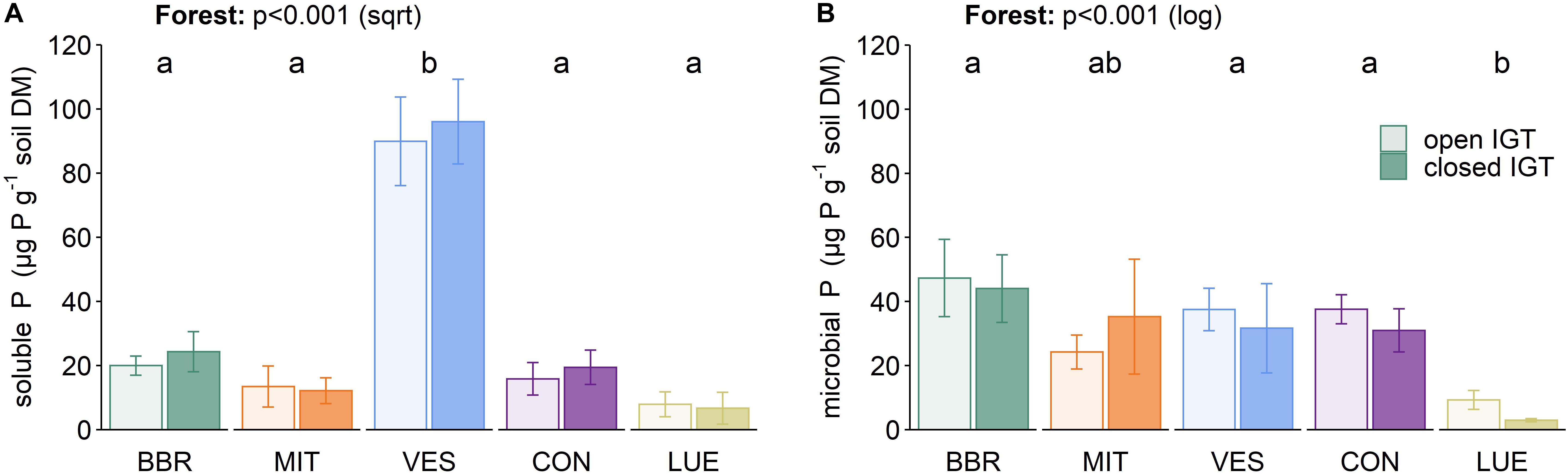
Figure 4. (A) Soluble P and microbially bound P (B) in open and closed IGTs in Bad Brückenau (BBR), Mitterfels (MIT), Vessertal (VES), Conventwald (CON), and Lüss (LUE) after 18 months exposure (n = 5). Forests sharing no letter are significantly different.
Enzyme Activities
Acid phosphomonoesterase activities (Figure 5A) were ∼40-fold higher than the activities of phosphodiesterases (Figure 5B). The acid phosphomonoesterase activity was not significantly different between open and closed IGTs after 18 months of exposure. In contrast, phosphodiesterase activity was reduced in the closed compared to the open IGTs (p < 0.001), with significant differences in all forests except BBR. Both esterase activities were forest-specific (acid phosphomonoesterase: p = 0.038, phosphodiesterase: p < 0.001).
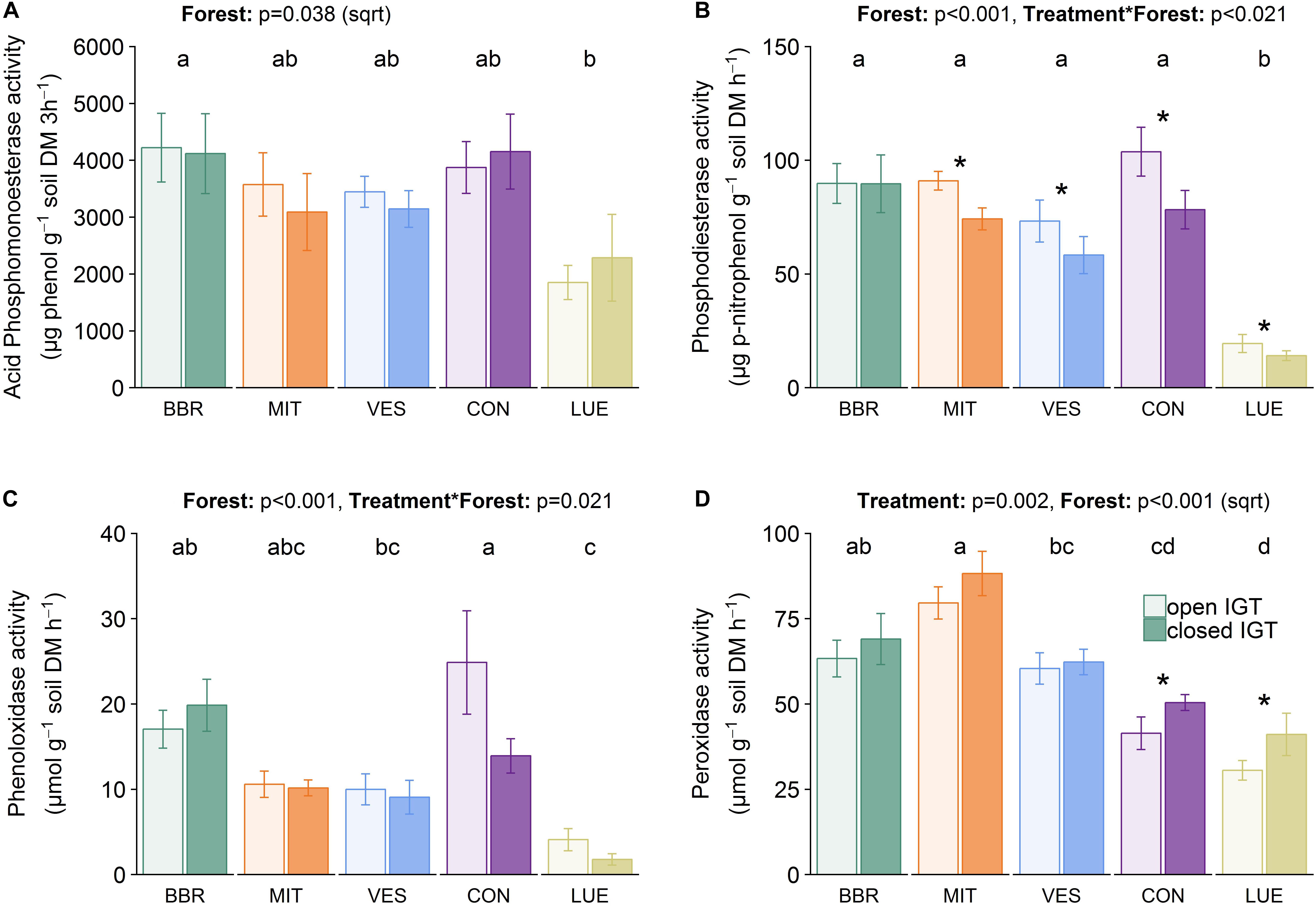
Figure 5. Enzyme activities of (A) acid phosphomonoesterase, (B) phosphodiesterase, (C) phenoloxidase, and (D) peroxidase activity in open and closed IGTs in Bad Brückenau (BBR), Mitterfels (MIT), Vessertal (VES), Conventwald (CON), and Lüss (LUE) after 18 months of exposure (n = 5). Forests sharing no letter are significantly different and asterisks highlight significant differences between open and closed IGTs at the respective forest site.
Phenol- and peroxidase activities (Figures 5C,D) varied between the forest sites (p < 0.001) after 18 months of exposure. The treatment was only significant in CON with 44% lower phenoloxidase activity in closed than in open IGTs but no treatment effects in the other forests (Figure 5C). In contrast to phenoloxidase activity, the closed IGTs were higher in peroxidase activities than the open IGTs in all forests (p = 0.002) (Figure 5D). The effect of treatment was higher in CON and LUE with increases in peroxidase activity of 21 and 34%, respectively.
Principal component analysis disclosed that the first two principal components (PCs) together accounted for 50% of the variance in enzyme activity, fungal and bacterial PLFA content, as well as abundant EM and SAP genera after 18 months of exposure (Figure 6). The fungal genera appeared to separate on axis PC2, while total fungal and bacterial PLFAs separated together with enzyme activities on axis PC1. LUE was clearly separate from the other forests. Additionally, in BBR and CON, closed and open IGTs separated from each other.
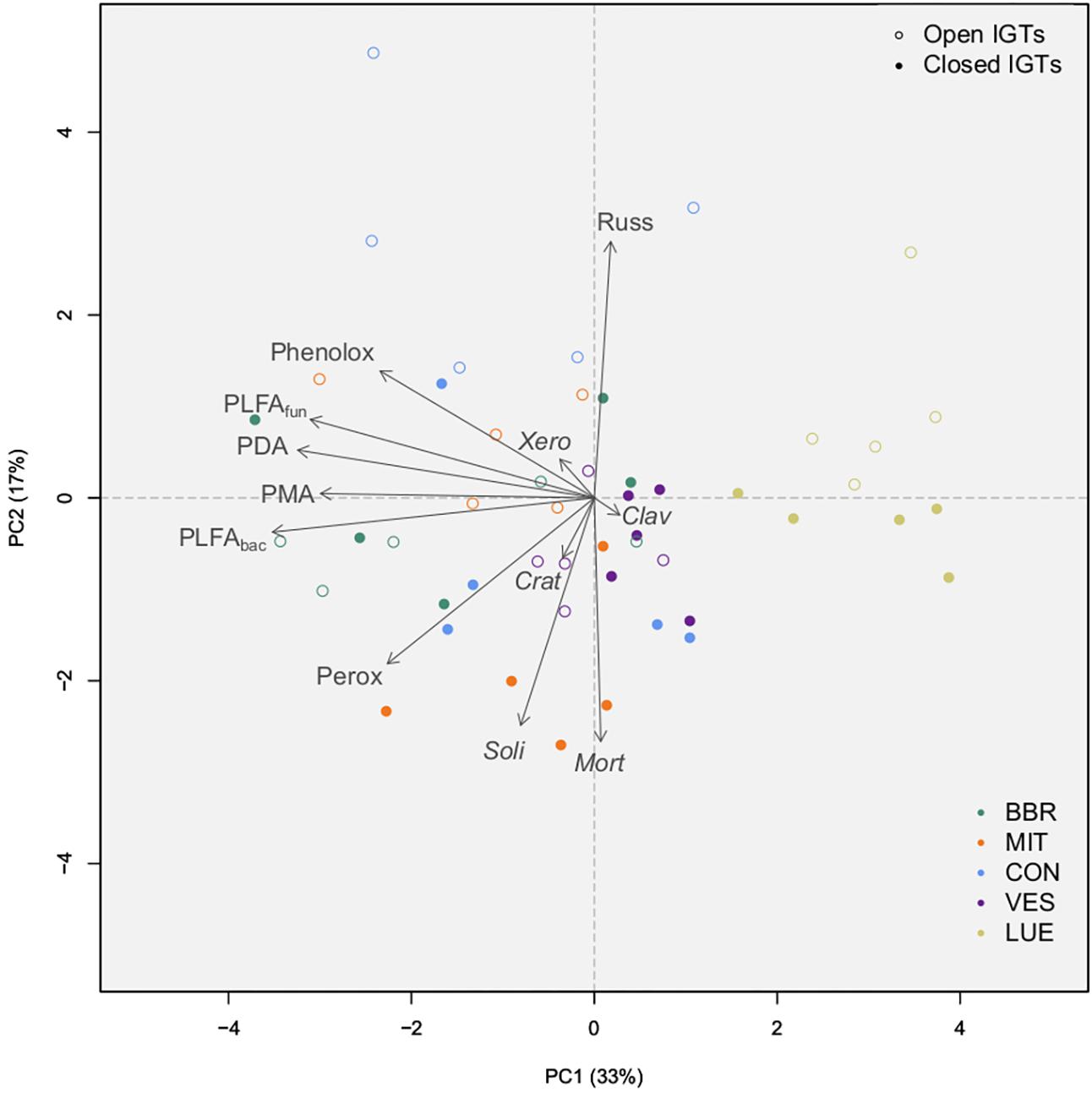
Figure 6. Results of principal component analysis of absolute amounts of PLFAs (sum of 15:0, i15:0, a15:0, i16:0, i17:0, cy17:0, cy19:0, 16:1ω5 = bacterial PLFA, 18:2ω6,9 = fungal PLFA), absolute number of OTUs from the most abundant EM and SAP fungal genera, and absolute amount of enzyme activity groups by treatment (open IGTs, closed IGTs) and forest [Bad Brückenau (BBR), Mitterfels (MIT), Vessertal (VES), Conventwald (CON) and Lüss (LUE)]. Clav, Clavulina; Crat, Craterellus; Mort, Mortierella; PDA, Phosphodieserase activity; Perox, Peroxidase activity; Phenolox, Phenoloxidase activity; PLFAbac, bacterial PLFA; PLFAfun, fungal PLFA; PMA, Phosphomonoesterase activity; Russ, Russula; Soli, Solicoccozyma; Xero, Xeromellus.
Discussion
Trenching Resulted in a Shift in the Fungal Community
An experiment with two different types of ingrowth cores was used to study the importance of fungal-fungal interactions in forest sites differing in their inorganic and organic P availability. We expected a shift in fungal communities in the closed IGTs toward SAP dominated assemblages due to the disruption in photo-assimilate availability for EM fungi. In contrast, open IGTs should have permitted the recolonization of EM fungi into the cores from the surrounding soil. Our results clearly show that shifts in fungal assemblages occurred over time, but that the speed and extent of changes differed in the five forest sites. In three forest sites (MIT, CON, and LUE), fungal community composition shifted toward a higher dominance of EM fungi in open IGTs after a period of 18 months, whereas these changes were not apparent in two other forests (BBR and VES). It could be that recolonization of the open IGTs occurred more slowly in BBR and VES than in the other forest sites, but it is unclear whether unidentified abiotic environmental factors may have precluded rapid fungal growth. Another possibility is that SAP fungal communities competed with EM fungi and thereby delayed the recolonization process. Therefore, our hypothesis was only supported by the results of three of the five studied sites, suggesting that additional factors not included in our study may have impacted fungal community composition and influenced the speed and extent of the colonization process of the IGTs.
The temporal dynamics of the fungal assemblages in the IGTs were influenced by the initial disruption of the fungal hyphal network of both EM and SAP fungi between the soil inside the IGTs and the surrounding soil. The exclusion of animal guilds, which graze specifically on EM or disturb the hyphal net such as Protura and earthworms (Yang et al., 2015; Bluhm et al., 2019) might have facilitated strong proliferation of EM compared to other fungi, which we found in some IGTS (MIT, CON, LUE). However, this suggestion is currently speculative since we do not know how soil animals vary among our forest sites. Based on low EM/SAP ratio in closed IGTs and higher EM/SAP ratio in open IGTs we were able to discuss the differences in fungal physiology in P-cycling. Our results are in accordance with data from Wallander et al. (2013). They found twice as much fungal biomass (measured as 18:2ω6,9 PLFA content) in a mesh bag in young Norway spruce (Picea abies L.) forests after 12 months of incubation than in those buried for only 2–5 months. After 18 months of exposure, we found a higher content of the fungal PLFA biomarker (18:2ω6,9) in the open IGTs than in the closed IGTs. Since EM fungi are the dominant contributors to the total 18:2ω6,9 PLFA in acidic forest soils (Wallander et al., 2001; Hagerberg and Wallander, 2002), we tentatively attribute the differences in the fungal PLFA content to recolonization by EM fungi in the open IGTs. This suggestion is supported by our amplicon data, which showed an increase in the number of EM OTUs in the open IGTs in MIT, VES, and LUE, and a stable number of EM OTUs in BBR and CON over time. In addition, SAP abundance decreased in the forest sites MIT, VES and LUE over time, whereas their abundance remained stable in BBR and CON. Overall, these changes in fungal assemblages were largely restricted to the EM and SAP fungal communities, while other fungal groups (e.g., fungi with unknown EM status or arbuscular mycorrhizal fungi) were low in OTU abundance and did not change throughout the experiment. Therefore, these groups were considered negligible in terms of P dynamics.
The EM fungal assemblages found in the open IGTs are typical for temperate beech forest sites (Lang et al., 2013). In our open IGTs, Russula, Cenococcum, and Xerocomellus spp. were the abundant groups after 18 months of exposure. These results agree with those of Pena et al. (2013), who found great interspecific variation in the temporal patterns of beech litter recolonization by EM fungi. In a litter bag experiment, Pena et al. (2013) observed an initial recolonization by EM fungi classified as contact or short-distance exploration types such as Russula cuprea, Cenococcum geophilum, Humaria spp., or Sebacina spp. (Agerer, 2001). EM fungi from the long-distance exploration types (Agerer, 2001) such as Xerocomellus pruinatus gained access to the litter inside the bags over the course of time. Additionally, we found Cortinarius spp., which have extensive SOM decomposition capacity, also in boreal forest soils (Bödeker et al., 2014). This result suggests ongoing decomposition of SOM inside the IGTs due not only to SAP fungi, but also to some EM fungi.
Fungi with the capacity to decompose complex organic material may profit from the dieback of other microorganisms by using the C and nutrients from microbial necromass in addition to plant-derived C sources. The installation of the IGTs affected the abundance and diversity of the EM fungi to a greater degree than that of the SAP fungi, as EM fungi rely on the connection with roots of their host plant. Disconnection from their host plants led to a dieback of these organisms (Högberg and Högberg, 2002; Remén et al., 2008), as seen, e.g., for Russula spp. in the closed IGTs in CON. Furthermore, disturbance of the soil by IGT installation may have brought litter and dead root material into closer contact than before to fungi and other microorganisms with saprotrophic properties. This could have provided SAP with a spatial advantage because new EM fungi had to grow into the open IGTs, whereas SAP could immediately establish their hyphal networks with the readily available resources, occupying free niches in the IGTs. Our results thus concur with those of Lindahl et al. (2010) in a Pinus sylvestris L. forest in Sweden. The authors concluded that disturbance induces rapid growth of opportunistic SAP fungi using the dying mycorrhizal mycelia. In accordance, Brabcová et al. (2016) found a specific microbial community in the vicinity of the dead mycelia of mostly non-basidiomycetous r-strategists such as Mortierella, Aspergillus, and Penicillium spp. In our study, Mortierella was the most abundant genus of SAP fungi and more abundant in the closed than in the open IGTs. In particular, in MIT 42% of the SAP OTUs belonged to this genus, in comparison to only 4% in the corresponding open IGTs. Similar to the data of Buée et al. (2009), who identified Solicoccozyma as a highly abundant genus in different forest soils in France, this genus was also abundant in our forest soils. Solicoccozyma specializes in using C from cellulose in spruce forests and thus is known to be heavily involved in the decomposition of dead plant litter (Štursová et al., 2012). We assume that the dead organic material of dying EM hyphae served as a C source in addition to the available plant litter material. This source may explain the observed increase in abundance of Solicoccozyma in the closed IGTs in three (MIT, CON, and LUE) out of five forests.
SAP and EM Fungal Assemblages Affected Enzyme Activities
We hypothesized that phosphomono-and phosphodiesterase activities depend on the proportion of SAP to EM fungi, with higher P cycling enzyme activities in soils dominated by EM fungi. All of our forest sites are characterized by low pH values (3.4–4) where fungi are the dominant microbial group and compete successfully with bacteria. As we cannot completely exclude the bacterial and animal contribution to the production of phosphatases, we focused on fungi community composition and related differences in P-cycling enzyme activity mainly to changes in fungal production of these enzymes and not to plants, bacteria and archaea (Nannipieri et al., 2011). Our results demonstrated that fungal community composition had no effect on phosphomonoesterase activity, while the exclusion of EM led to a decrease in phosphodiesterase activity of up to 38%. Although the shifts in fungal community composition were noteworthy between treatments, phosphomonoesterase activity was steady, suggesting some functional redundancy in fungal-derived P cycling enzymes (Figure 6). Therefore, the capacity to produce acid phosphomonoesterases is present in both fungal guilds to a similar extent. Our results are supported by findings of Colpaert and Laere (1996) who found, in a pot experiment with mycorrhizal and non-mycorrhizal seedlings of Pinus sylvestris L., considerable evidence that EM fungi produce extracellular phosphatases. Also, Dighton (1983, 1991) measured equal or even higher phosphatase activity production by EM fungi than by SAP basidiomycetes. Cell wall-bound and extracellular acid phosphomonoesterase activities have, indeed, been demonstrated for a wide taxonomic range of EM fungi (Pritsch and Garbaye, 2011), including genera which are abundant in our study, e.g., Clavulina, Russula, Cenococcum, Tomentella, Lactarius, and Amanita (Ho and Zak, 1979; Alvarez et al., 2005, 2006; Nygren and Rosling, 2009; Walker et al., 2014; Ning et al., 2018; Ruess et al., 2019).
In our experiment, phosphodiesterase activity was generally lower than phosphomonoesterase activity, which is in line with findings from Turner and Haygarth (2005). We found lower phosphodiesterase activities in four of our sites (exception BBR) in SAP-dominated IGTs, indicating that SAP fungi were not as involved as EM fungi in the production and secretion of phosphodiesterases. Phosphodiesterase activity has scarcely been investigated in situ (Ruess et al., 2019), and to our knowledge measurements are lacking for individual EM fungal genera. The formerly held opinion that SAP fungi are the only degraders of complex organic substances that release directly plant-available P is now being revised (Lindahl and Tunlid, 2015). EM fungi have a greater capacity to produce enzymes involved in the degradation of complex organic material than was previously thought. Genomic and field studies have demonstrated that fungi have retained many degrading enzymes and that SAP and EM fungi are more functionally similar than previously considered (Baldrian, 2009; Burke et al., 2014; Bödeker et al., 2016).
Whereas phosphomono- and phosphodiesterase directly release plant-available P, phenoloxidase and peroxidase activities may indirectly contribute to P mineralization by liberating organic P-compounds from complex organic substrates, preparing them for further cleavage. Contrary to our initial hypothesis, the phenoloxidase activity remained unchanged, whereas the peroxidase activity was higher only in SAP-dominated IGTs. This indicates that EM fungi produce phenoloxidases much as SAP fungi do under similar environmental conditions. The capacity to produce substrate-specific laccases, probably the largest class of phenoloxidases in soils, is known only for some ectomycorrhizal species, e.g., Russula delica (Matsubara and Iwasaki, 1972). Russula was one of the most abundant genera in our experiment. Luis et al. (2004, 2005) investigated the diversity of basidiomycete laccase genes in beech-oak forests in Germany and they linked 108 gene sequences to specific fungal taxa. Two-thirds belonged to EM fungi and the remaining to SAP fungi. In a follow-up experiment, Kellner et al. (2009) showed that the temporal variation in EM laccase diversity peaked with autumn litterfall, while overall phenoloxidase activity in the litter layer was similar over the seasons. This indicates the importance of EM fungi as decomposers of more complex material to gain access to organic forms of P (Talbot et al., 2013), although the presence of a gene sequence in the DNA that likely codes for phenoloxidase enzymes is not adequate proof of a particular species’ capacity to produce these enzymes in situ.
We assume from our results that peroxidase activity, at least to some extent, was derived from EM fungi. For example, Cortinarius spp. encode lignolytic class-II peroxidases in their genome, and their gene transcription correlates highly with peroxidase activity in a boreal pine forest soil (Bödeker et al., 2014). In agreement with these results, we found high peroxidase activities in BBR and MIT where Cortinarius spp. were abundant (Figure 2), but this genus was not found in VES, CON, and LUE, where peroxidase activity was lower. Additionally, Bödeker et al. (2014) found that Mn-peroxidase production is not a general feature of all Cortinarius spp. in soils. High peroxidase activity was also detected when these fungi were not abundant, indicating that the Mn-peroxidase production is not restricted to Cortinarius. Distinct peroxidase activity is also known for Lactarius spp. (Nicolás et al., 2019) and Russula spp. (Bödeker et al., 2009). All mentioned genera were present in our investigated forests. Although these genera are ecologically important, little is known about their specific functions as they comprise many members which have not been well studied. Therefore, the question remains as to what extent the EM fungal community is involved in the decomposition process to obtain nutrients other than C. Collectively, our results suggest that EM fungi are not only involved in the production of P cycle enzymes but also in per- and phenoloxidase production to decompose complex SOM to meet their own P needs. Therefore, we reject our third hypothesis that the exclusion of EM fungi in the closed IGTs led to a pronounced reduction in oxidase activity.
Forest Conditions Affected P Cycle Enzyme Activities With Implications for Pmic and Psol
P availability is acknowledged as a key driver of the production of P cycle enzymes (Quiquampoix and Mousain, 2005), so we expected that the activity of these enzymes is dependent on soil P stock, with lower values in forests with high P stocks and higher values in forests with limited P stocks. The investigated forests were selected based on a geosequence derived from total P stock values (BBR > MIT > VES > CON > LUE, see Table 1) according to Lang et al. (2017). Specific phosphomonoesterase activities (enzyme activity per microbial biomass measured as PLFAs) showed an opposite gradient from the total P stock gradient, with highest enzyme activity per biomass in LUE and lowest in BBR. For specific phosphodiesterase activity, we recorded highest values in CON, intermediate values in BBR, MIT, VES, and lowest in LUE. Therefore, we can partly confirm our hypothesis.
Some evidence indicated that in addition to P availability, soil texture may influence biotic soil components and their functions. The adsorption of enzymes to clay particles can reduce enzyme activities in situ (Leprince and Quiquampoix, 1996) and this likely depends on the soil type (Louche et al., 2010). The specific phosphodiesterase activity was lowest in LUE, although Lang et al. (2017) identified different Porg fractions by NMR spectroscopy, and found a high proportion of phosphodiesters of ∼20% of total Porg in the upper 20 cm depth. They assumed that this contrast in high P substrate availability and low phosphodiesterase activity was due to either low enzyme stability at low pH values of 3.5 and/or low microbial enzyme production, resulting in a higher proportion of phosphodiesters in comparison to phosphomonoesters in LUE.
The BBR soil was not P limited for plants, as P-fertilization did not enhance photosynthesis in contrast to LUE soil (Zavišić et al., 2018). Similarly, Heuck and Spohn (2016) concluded that the soils in BBR and MIT may not be P depleted and/or that P is not the only limiting nutrient in the soil. Apart from microbial P, N, and C supplies (Kieliszewska-Rokicka, 1992; Taniguchi et al., 2008), other abiotic factors such as moisture (Courty et al., 2007), temperature, and pH (Tibbett et al., 1998), may influence microbial activities and functions to some extent. In VES, we found much higher Psol values than in the other four forests (see Figure 4A). Surprisingly, the higher Psol values did not influence the specific enzyme activities of phosphomono- and phosphodiesterases, underscoring the possibility that microbially mediated P mobilization is also influenced by site-specific conditions.
Conclusion
Understanding the importance of fungal-fungal interactions in forest ecosystems is necessary to better disentangle the relative contributions of EM and SAP fungi in P cycling. This study investigated phosphatase and oxidase activities in EM and SAP dominated fungal assemblages in beech forest ecosystems with differing P availabilities. Taken together, our results indicate that phosphatase and oxidase activity in EM and SAP dominated assemblages was more similar than expected, with only minor differences in phosphodiesterase and peroxidase activities. They also provide new insights into biological regulation mechanisms of the P cycle and highlight the role of EM fungi in meeting P demand for the fungi themselves and for their host plant, especially in nutrient poor ecosystems.
Data Availability Statement
The datasets generated for this study can be found in the National Centre for Biotechnology Information (NCBI) Sequence Read Archive (SRA), accession number: PRJNA592056.
Author Contributions
EK and SM developed the project. KM, NK, and PN wrote the manuscript, which was read and revised by all co-authors. Fieldwork was performed by PN. Soil analyses were mainly performed by PN and PM-G. DS, RD, and AP performed the molecular biological analyses and bioinformatic analysis of the sequence data. KM performed statistics and data evaluation with support by H-PP. All authors listed have made substantial, direct, and intellectual contributions to the work, and have approved it for publication.
Funding
We are grateful to the Deutsche Forschungsgemeinschaft for supporting the Priority Program 1685 “Ecosystem Nutrition” by funding the projects Ka 1590/12-1 and Po 362/22-1.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer MG declared a shared affiliation, though no other collaboration, with several of the authors DS, RD, AP to the handling Editor.
Acknowledgments
We would like to thank H. Haslwimmer and S. Rudolph for help during soil analysis, T. Klein (University of Göttingen, Laboratory for Radio-Isotopes) for DNA extraction and preparation for the ILLUMINA analyses and K. Regan for English corrections. Furthermore, we thank the student helpers for supporting the extensive installation of soil cores and subsequent harvest campaigns in the field.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffgc.2020.00047/full#supplementary-material
FIGURE S1 | (A) Prototype of the open IGTs and (B) soil filled open and closed IGTs directly before installation.
FIGURE S2 | (A) Microbial and (B) bacterial biomass based on PLFA-analysis in the open and closed IGTs in Bad Brückenau (BBR), Mitterfels (MIT), Vessertal (VES), Conventwald (CON), and Lüss (LUE) after 18 months of exposure (n = 5). Forests sharing no letter are significantly different and asterisks highlight significant differences between open and closed IGTs at the respective forest site.
TABLE S1 | Results of the nlme-Model for the IGTs in Bad Brückenau (BBR), Mitterfels (MIT), Vessertal (VES), Conventwald (CON), and Lüss (LUE), including all sampling dates (n = 5). Level of significance: ***p < 0.001, **p < 0.01, *p < 0.05, and +p < 0.1.
TABLE S2 | Results of the nlme-Model for the IGTs in Bad Brückenau (BBR), Mitterfels (MIT), Vessertal (VES), Conventwald (CON), and Lüss (LUE), including only the last sampling date after 18 months of exposure (n = 5). Level of significance: ***p < 0.001, **p < 0.01, *p < 0.05, and +p < 0.1.
TABLE S3 | Results of open and closed IGTs in Bad Brückenau (BBR), Mitterfels (MIT), Vessertal (VES), Conventwald (CON), and Lüss (LUE) during the experiment (n = 5). Bold values represent statistically significant differences (Tukey test) between open and closed IGTs at the respective forest and sampling date.
TABLE S4 | Results of control soil in Bad Brückenau (BBR), Mitterfels (MIT), Vessertal (VES), Conventwald (CON), and Lüss (LUE) after 18 months of exposure (n = 5). Superscript letters indicate results of the Tukey test, highlighting differences between forest sites.
References
Agerer, R. (2001). Exploration types of ectomycorrhizae. Mycorrhiza 11, 107–114. doi: 10.1007/s005720100108
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Alvarez, M., Gieseke, A., Godoy, R., and Härtel, S. (2006). Surface-bound phosphatase activity in ectomycorrhizal fungi. A comparative study between a colorimetric and a microscope-based method. Biol. Fertil. Soils 42:561. doi: 10.1007/s00374-005-0053-6
Alvarez, M., Godoy, R., Heyser, W., and Härtel, S. (2005). Anatomical–physiological determination of surface bound phosphatase activity in ectomycorrhizae of Nothofagus obliqua. Soil Biol. Biochem. 37, 125–132. doi: 10.1016/j.soilbio.2004.07.028
Alvarez, M., Huygens, D., Olivares, E., Saavedra, I., Alberdi, M., and Valenzuela, E. (2009). Ectomycorrhizal fungi enhance nitrogen and phosphorus nutrition of Nothofagus dombeyi under drought conditions by regulating assimilative enzyme activities. Physiol. Plant. 136, 426–436. doi: 10.1111/j.1399-3054.2009.01237.x
Bach, C. E., Warnock, D. D., van Horn, D. J., Weintraub, M. N., Sinsabaugh, R. L., Allison, S. D., et al. (2013). Measuring phenol oxidase and peroxidase activities with pyrogallol, l-DOPA, and ABTS. Effect of assay conditions and soil type. Soil Biol. Biochem. 67, 183–191. doi: 10.1016/j.soilbio.2013.08.022
Baldrian, P. (2008). “Enzymes of saprotrophic basidiomycetes,” in British Mycological Society Symposia Series: Ecology of Saprotrophic Basidiomycetes, eds L. Boddy, J. C. Frankland, and P. van West (New York, NY: Academic Press), 19–41. doi: 10.1016/s0275-0287(08)80004-5
Baldrian, P. (2009). Ectomycorrhizal fungi and their enzymes in soils. Is there enough evidence for their role as facultative soil saprotrophs? Oecologia 161, 657–660. doi: 10.1007/s00442-009-1433-7
Bluhm, S. L., Potapov, A. M., Shrubovych, J., Ammerschubert, S., Polle, A., and Scheu, S. (2019). Protura are unique. First evidence of specialized feeding on ectomycorrhizal fungi in soil invertebrates. BMC Ecol. 19:10. doi: 10.1186/s12898-019-0227-y
Bödeker, I. T. M., Clemmensen, K. E., de Boer, W., Martin, F., Olson, Å., and Lindahl, B. D. (2014). Ectomycorrhizal Cortinarius species participate in enzymatic oxidation of humus in northern forest ecosystems. New Phytol. 203, 245–256. doi: 10.1111/nph.12791
Bödeker, I. T. M., Lindahl, B. D., Olson, Å., Clemmensen, K. E., and Treseder, K. (2016). Mycorrhizal and saprotrophic fungal guilds compete for the same organic substrates but affect decomposition differently. Funct. Ecol. 30, 1967–1978. doi: 10.1111/1365-2435.12677
Bödeker, I. T. M., Nygren, C. M. R., Taylor, A. F. S., Olson, Å., and Lindahl, B. D. (2009). ClassII peroxidase-encoding genes are present in a phylogenetically wide range of ectomycorrhizal fungi. ISME J. 3:1387. doi: 10.1038/ismej.2009.77
Brabcová, V., Nováková, M., Davidová, A., and Baldrian, P. (2016). Dead fungal mycelium in forest soil represents a decomposition hotspot and a habitat for a specific microbial community. New Phytol. 210, 1369–1381. doi: 10.1111/nph.13849
Brookes, P. C., Powlson, D. S., and Jenkinson, D. S. (1982). Measurement of microbial biomass phosphorus in soil. Soil Biol. Biochem. 14, 319–329. doi: 10.1016/0038-0717(82)90001-3
Browman, M. G., and Tabatabai, M. A. (1978). Phosphodiesterase activity of soils. Soil Sci. Soc. Am. J. 42, 284–290. doi: 10.2136/sssaj1978.03615995004200020016x
Buée, M., Reich, M., Murat, C., Morin, E., Nilsson, R. H., Uroz, S., et al. (2009). 454 Pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol. 184, 449–456. doi: 10.1111/j.1469-8137.2009.03003.x
Burke, D. J., Smemo, K. A., and Hewins, C. R. (2014). Ectomycorrhizal fungi isolated from old-growth northern hardwood forest display variability in extracellular enzyme activity in the presence of plant litter. Soil Biol. Biochem. 68, 219–222. doi: 10.1016/j.soilbio.2013.10.013
Burke, R., and Cairney, J. (2002). Laccases and other polyphenol oxidases in ecto-and ericoid mycorrhizal fungi. Mycorrhiza 12, 105–116. doi: 10.1007/s00572-002-0162-0
Burns, R. G., and Dick, R. P. (2002). Enzymes in the Environment. Activity, Ecology, and Applications. New York, NY: CRC Press.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336.
Colpaert, J. V., and Laere, A. (1996). A comparison of the extracellular enzyme activities of two ectomycorrhizal and a leaf-saprotrophic basidiomycete colonizing beech leaf litter. New Phytol. 134, 133–141. doi: 10.1111/j.1469-8137.1996.tb01153.x
Condron, L. M., Turner, B. L., and Cade-Menun, B. J. (2005). “Chemistry and dynamics of soil organic phosphorus,” in Phosphorus: Agriculture and the Environment, eds J. T. Sims and A. N. Sharpley (Madison, WI: ASA-CSSA-SSSA), 87–121. doi: 10.2134/agronmonogr46.c4
Courty, P.-E., Bréda, N., and Garbaye, J. (2007). Relation between oak tree phenology and the secretion of organic matter degrading enzymes by Lactarius quietus ectomycorrhizas before and during bud break. Soil Biol. Biochem. 39, 1655–1663. doi: 10.1016/j.soilbio.2007.01.017
Danielsen, L., and Polle, A. (2014). Poplar nutrition under drought as affected by ectomycorrhizal colonization. Environ. Exp. Bot. 108, 89–98. doi: 10.1016/j.envexpbot.2014.01.006
Dighton, J. (1983). Phosphatase production by mycorrhizal fungi. Plant Soil 71, 455–462. doi: 10.1007/978-94-009-6833-2_51
Dighton, J. (1991). Acquisition of nutrients from organic resources by mycorrhizal autotrophic plants. Experientia 47, 362–369. doi: 10.1007/bf01972078
Dowling, N. J. E., Widdel, F., and White, D. C. (1986). Phospholipid ester-linked fatty acid biomarkers of acetate-oxidizing sulphate-reducers and other sulphide-forming bacteria. Microbiology 132, 1815–1825. doi: 10.1099/00221287-132-7-1815
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Floch, C., Alarcon-Gutiérrez, E., and Criquet, S. (2007). ABTS assay of phenol oxidase activity in soil. J. Microbiol. Methods 71, 319–324. doi: 10.1016/j.mimet.2007.09.020
Frossard, E., Condron, L. M., Oberson, A., Sinaj, S., and Fardeau, J. C. (2000). Processes governing phosphorus availability in temperate soils. J. Environ. Qual. 29, 15–23. doi: 10.2134/jeq2000.00472425002900010003x
Frostegård, A., and Bååth, E. (1996). The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol. Fertil. Soils 22, 59–65. doi: 10.1007/bf00384433
Frostegård, A., Bååth, E., and Tunlio, A. (1993). Shifts in the structure of soil microbial communities in limed forests as revealed by phospholipid fatty acid analysis. Soil Biol. Biochem. 25, 723–730. doi: 10.1016/0038-0717(93)90113-p
Frostegård, A., Tunlid, A., and Bååth, E. (1991). Microbial biomass measured as total lipid phosphate in soils of different organic content. J. Microbiol. Methods 14, 151–163. doi: 10.1016/0167-7012(91)90018-l
Grinhut, T., Hadar, Y., and Chen, Y. (2007). Degradation and transformation of humic substances by Saprotrophic fungi. Processes and mechanisms. Fungal Biol. Rev. 21, 179–189. doi: 10.1016/j.fbr.2007.09.003
Gyaneshwar, P., Naresh Kumar, G., Parekh, L. J., and Poole, P. S. (2002). Role of soil microorganisms in improving P nutrition of plants. Plant Soil 245, 83–93.
Hagerberg, D., and Wallander, H. (2002). The impact of forest residue removal and wood ash amendment on the growth of the ectomycorrhizal external mycelium. FEMS Microbiol. Ecol. 39, 139–146. doi: 10.1111/j.1574-6941.2002.tb00915.x
Hedley, M. J., Stewart, J. W. B., and Chauhan, B. S. (1982). Changes in inorganic and organic soil phosphorus fractions induced by cultivation practices and by laboratory incubations. Soil Sci. Soc. Am. J. 46, 970–976. doi: 10.2136/sssaj1982.03615995004600050017x
Hendricks, J. J., Mitchell, R. J., Kuehn, K. A., Pecot, S. D., and Sims, S. E. (2006). Measuring external mycelia production of ectomycorrhizal fungi in the field. The soil matrix matters. New Phytol. 171, 179–186. doi: 10.1111/j.1469-8137.2006.01742.x
Heuck, C., and Spohn, M. (2016). Carbon, nitrogen and phosphorus net mineralization in organic horizons of temperate forests. Stoichiometry and relations to organic matter quality. Biogeochemistry 131, 229–242. doi: 10.1007/s10533-016-0276-7
Ho, I., and Zak, B. (1979). Acid phosphatase activity of six ectomycorrhizal fungi. Can. J. Bot. 57, 1203–1205. doi: 10.1139/b79-144
Hoffmann, G. (1968). Eine photometrische Methode zur Bestimmung der Phosphatase-Aktivität in Böden. Z. Pflanzenernähr. Bodenk. 118, 161–172. doi: 10.1002/jpln.19681180303
Högberg, M. N., and Högberg, P. (2002). Extramatrical ectomycorrhizal mycelium contributes one-third of microbial biomass and produces, together with associated roots, half the dissolved organic carbon in a forest soil. New Phytol. 154, 791–795. doi: 10.1046/j.1469-8137.2002.00417.x
IUSS Working Group WRB (2014). “World reference base for soil resouces 2014,” in Proceedings of the International Soil Classification SYSTEM for Naming Soils and Creating Legends for Soil Maps, Word Soil Resources Reports No. 106 (Rome: FAO).
Jansa, J., Finlay, R., Wallander, H., Smith, F. A., and Smith, S. E. (2011). “Role of mycorrhizal symbioses in phosphorus cycling,” in Phosphorus in Action, eds E. Bünemann, A. Oberson, and E. Frossard (Berlin: Springer), 137–168. doi: 10.1007/978-3-642-15271-9_6
Johnson, D., Leake, J. R., and Read, D. J. (2001). Novel in-growth core system enables functional studies of grassland mycorrhizal mycelial networks. New Phytol. 152, 555–562. doi: 10.1046/j.0028-646x.2001.00273.x
Jonard, M., Fürst, A., Verstraeten, A., Thimonier, A., Timmermann, V., Potočić, N., et al. (2015). Tree mineral nutrition is deteriorating in Europe. Glob. Chang. Biol. 21, 418–430. doi: 10.1111/gcb.12657
Jones, D. L., and Oburger, E. (2011). “Solubilization of phosphorus by soil microorganisms,” in Phosphorus in Action, eds E. Bünemann, A. Oberson, and E. Frossard (Berlin: Springer), 169–198. doi: 10.1007/978-3-642-15271-9_7
Kandeler, E. (1990). Characterization of free and adsorbed phosphatases in soils. Biol. Fertil. Soils 9, 199–202. doi: 10.1007/bf00335808
Kandeler, E., Mosier, A. R., Morgan, J. A., Milchunas, D. G., King, J. Y., Rudolph, S., et al. (2006). Response of soil microbial biomass and enzyme activities to the transient elevation of carbon dioxide in a semi-arid grassland. Soil Biol. Biochem. 38, 2448–2460. doi: 10.1016/j.soilbio.2006.02.021
Kavka, M., and Polle, A. (2017). Dissecting nutrient-related co-expression networks in phosphate starved poplars. PLoS One 12:e0171958. doi: 10.1371/journal.pone.0171958
Kellner, H., Luis, P., Schlitt, B., and Buscot, F. (2009). Temporal changes in diversity and expression patterns of fungal laccase genes within the organic horizon of a brown forest soil. Soil Biol. Biochem. 41, 1380–1389. doi: 10.1016/j.soilbio.2009.03.012
Kieliszewska-Rokicka, B. (1992). Effect of nitrogen level on acid phosphatase activity of eight isolates of ectomycorrhizal fungus Paxillus involutus cultured in vitro. Plant Soil 139, 229–238. doi: 10.1007/bf00009314
Kõljalg, U., Nilsson, R. H., Abarenkov, K., Tedersoo, L., Taylor, A. F. S., Bahram, M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277. doi: 10.1111/mec.12481
Kucey, R. M. N., Janzen, H. H., and Leggett, M. E. (1989). Microbially mediated increases in plant-available phosphorus. Adv. Agron. 42, 199–228. doi: 10.1016/s0065-2113(08)60525-8
Lambers, H., Raven, J., Shaver, G., and Smith, S. (2008). Plant nutrient-acquisition strategies change with soil age. Trends Ecol. Evol. 23, 95–103. doi: 10.1016/j.tree.2007.10.008
Lang, C., Finkeldey, R., and Polle, A. (2013). Spatial patterns of ectomycorrhizal assemblages in a monospecific forest in relation to host tree genotype. Front. Plant Sci. 4:103. doi: 10.3389/fpls.2013.00103
Lang, F., Krüger, J., Amelung, W., Willbold, S., Frossard, E., Bünemann, E. K., et al. (2017). Soil phosphorus supply controls P nutrition strategies of beech forest ecosystems in Central Europe. Biogeochemistry 136, 5–29. doi: 10.1007/s10533-017-0375-0
Lenth, R., Singmann, H., Love, J., Buerkner, P., and Herve, M. (2019). emmeans Estimated Marginal Means, aka Least-Squares Means. Available at: https://github.com/rvlenth/emmeans (accessed August 5, 2019).
Leprince, F., and Quiquampoix, H. (1996). Extracellular enzyme activity in soil. Effect of pH and ionic strength on the interaction with montmorillonite of two acid phosphatases secreted by the ectomycorrhizal fungus Hebeloma cylindrosporum. Eur. J. Soil Sci. 47, 511–522.
Lindahl, B. D., de Boer, W., and Finlay, R. D. (2010). Disruption of root carbon transport into forest humus stimulates fungal opportunists at the expense of mycorrhizal fungi. ISME J. 4, 872–881. doi: 10.1038/ismej.2010.19
Lindahl, B. D., and Tunlid, A. (2015). Ectomycorrhizal fungi-potential organic matter decomposers, yet not saprotrophs. New Phytol. 205, 1443–1447. doi: 10.1111/nph.13201
Louche, J., Ali, M. A., Cloutier-Hurteau, B., Sauvage, F.-X., Quiquampoix, H., and Plassard, C. (2010). Efficiency of acid phosphatases secreted from the ectomycorrhizal fungus Hebeloma cylindrosporum to hydrolyse organic phosphorus in podzols. FEMS Microbiol. Ecol. 73, 323–335. doi: 10.1111/j.1574-6941.2010.00899.x
Luis, P., Kellner, H., Zimdars, B., Langer, U., Martin, F., and Buscot, F. (2005). Patchiness and spatial distribution of laccase genes of ectomycorrhizal, saprotrophic, and unknown basidiomycetes in the upper horizons of a mixed forest cambisol. Microb. Ecol. 50, 570–579. doi: 10.1007/s00248-005-5047-2
Luis, P., Walther, G., Kellner, H., Martin, F., and Buscot, F. (2004). Diversity of laccase genes from basidiomycetes in a forest soil. Soil Biol. Biochem. 36, 1025–1036. doi: 10.1016/j.soilbio.2004.02.017
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12.
Martino, E., Morin, E., Grelet, G.-A., Kuo, A., Kohler, A., Daghino, S., et al. (2018). Comparative genomics and transcriptomics depict ericoid mycorrhizal fungi as versatile saprotrophs and plant mutualists. New Phytol. 217, 1213–1229. doi: 10.1111/nph.14974
Matsubara, T., and Iwasaki, H. (1972). Occurrence of laccase and tyrosinase in fungi of agaricales and comparative study of laccase from Russula delica and Russula pseudodelica. Bot. Mag. 85, 71–83. doi: 10.1007/bf02489202
McDonald, D., Clemente, J. C., Kuczynski, J., Rideout, J. R., Stombaugh, J., Wendel, D., et al. (2012). The Biological Observation Matrix (BIOM) format or How I learned to stop worrying and love the ome-ome. GigaScience 1:7.
Murphy, J., and Riley, J. P. (1962). A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 27, 31–36. doi: 10.1016/s0003-2670(00)88444-5
Nannipieri, P., Giagnoni, L., Landi, L., and Renella, G. (2011). “Role of phosphatase enzymes in soil,” in Phosphorus in Action: Biological Processes in Soil Phosphorus Cycling, eds E. Oberson, A. Bünemann, and E. Frossard (Berlin: Springer).
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild. An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Nicolás, C., Martin-Bertelsen, T., Floudas, D., Bentzer, J., Smits, M., Johansson, T., et al. (2019). The soil organic matter decomposition mechanisms in ectomycorrhizal fungi are tuned for liberating soil organic nitrogen. ISME J. 13, 977–988. doi: 10.1038/s41396-018-0331-6
Ning, C., Mueller, M. G., Egerton-Warburton, M. L., Wilson, W. A., Yan, W., and Xiang, W. (2018). Diversity and enzyme activity of ectomycorrhizal fungal communities following nitrogen fertilization in an Urban-adjacent pine plantation. Forests 9:99. doi: 10.3390/f9030099
Nygren, C. M. R., and Rosling, A. (2009). Localisation of phosphomonoesterase activity in ectomycorrhizal fungi grown on different phosphorus sources. Mycorrhiza 19, 197–204. doi: 10.1007/s00572-008-0223-0
Öhlinger, R., Margesin, R., and Kandeler, E. (1996). “Enzymes involved in phosphorus metabolism,” in Methods in Soil Biology, eds F. Schinner, R. Öhlinger, E. Kandeler, and R. Margesin (Berlin: Springer), 208–227. doi: 10.1007/978-3-642-60966-4_13
Pena, R., Tejedor, J., Zeller, B., Dannenmann, M., and Polle, A. (2013). Interspecific temporal and spatial differences in the acquisition of litter-derived nitrogen by ectomycorrhizal fungal assemblages. New Phytol. 199, 520–528. doi: 10.1111/nph.12272
Pinheiro, J., Bates, D., DebRoy, S., Sarkar, D., and R Core Team. (2019). nlme: Linear and Nonlinear Mixed Effects Models. Available at: https://svn.r-project.org/R-packages/trunk/nlme (accessed July 23, 2019).
Plante, A. F. (2007). “Soil biogeochemical cycling of inorgacnic nutrients and metals,” in Soil Microbiology, Ecology and Biochemistry, Third Edn, ed. E. A. Paul (San Diego, CA: Academic Press), 389–432. doi: 10.1016/b978-0-08-047514-1.50019-6
Pritsch, K., and Garbaye, J. (2011). Enzyme secretion by ECM fungi and exploitation of mineral nutrients from soil organic matter. Ann. For. Sci. 68, 25–32. doi: 10.1007/s13595-010-0004-8
Quiquampoix, H., and Mousain, D. (2005). “Enzymatic hydrolysis of organic phosphorus,” in Organic Phosphorus in the Environment, eds B. L. Turner, E. Frossard, and D. S. Baldwin (Wallingford: CAB International), 89–112. doi: 10.1079/9780851998220.0089
R Core Team (2019). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Remén, C., Persson, T., Finlay, R., and Ahlström, K. (2008). Responses of oribatid mites to tree girdling and nutrient addition in boreal coniferous forests. Soil Biol. Biochem. 40, 2881–2890. doi: 10.1016/j.soilbio.2008.08.006
Richardson, A. E., and Simpson, R. J. (2011). Soil microorganisms mediating phosphorus availability-update on microbial phosphorus. Plant Physiol. 156, 989–996. doi: 10.1104/pp.111.175448
Ruess, L., Schütz, K., Migge-Kleian, S., Häggblom, M. M., Kandeler, E., and Scheu, S. (2007). Lipid composition of Collembola and their food resources in deciduous forest stands-Implications for feeding strategies. Soil Biol. Biochem. 39, 1990–2000. doi: 10.1016/j.soilbio.2007.03.002
Ruess, W. R., Swanson, M. M., Kielland, K., McFarland, W. J., Olson, D. K., and Taylor, L. D. (2019). Phosphorus mobilizing enzymes of alnus-associated Ectomycorrhizal Fungi in an Alaskan Boreal Floodplain. 10:554. doi: 10.3390/f10070554
Schinner, F., Öhlinger, R., Kandeler, E., and Margesin, R. (eds) (2012). Methods in Soil Biology. Berlin: Springer.
Štursová, M., Žifèáková, L., Leigh, M. B., Burgess, R., and Baldrian, P. (2012). Cellulose utilization in forest litter and soil. Identification of bacterial and fungal decomposers. FEMS Microbiol. Ecol. 80, 735–746. doi: 10.1111/j.1574-6941.2012.01343.x
Talbot, J. M., Bruns, T. D., Smith, D. P., Branco, S., Glassman, S. I., Erlandson, S., et al. (2013). Independent roles of ectomycorrhizal and saprotrophic communities in soil organic matter decomposition. Soil Biol. Biochem. 57, 282–291. doi: 10.1016/j.soilbio.2012.10.004
Talkner, U., Meiwes, K. J., Potočić, N., Seletković, I., Cools, N., de Vos, B., et al. (2015). Phosphorus nutrition of beech (Fagus sylvatica L.) is decreasing in Europe. Ann. For. Sci. 72, 919–928. doi: 10.3389/fpls.2019.00744
Tamburini, F., Pfahler, V., Bünemann, E. K., Guelland, K., Bernasconi, S. M., and Frossard, E. (2012). Oxygen isotopes unravel the role of microorganisms in phosphate cycling in soils. Environ. Sci. Technol. 46, 5956–5962. doi: 10.1021/es300311h
Taniguchi, T., Kataoka, R., and Futai, K. (2008). Plant growth and nutrition in pine (Pinus thunbergii) seedlings and dehydrogenase and phosphatase activity of ectomycorrhizal root tips inoculated with seven individual ectomycorrhizal fungal species at high and low nitrogen conditions. Soil Biol. Biochem. 40, 1235–1243. doi: 10.1016/j.soilbio.2007.12.017
Tibbett, M., Sanders, F. E., and Cairney, J. W. G. (1998). The effect of temperature and inorganic phosphorus supply on growth and acid phosphatase production in arctic and temperate strains of ectomycorrhizal Hebeloma spp. in axenic culture. Mycol. Res. 102, 129–135. doi: 10.1017/s0953756297004681
Toju, H., Tanabe, A. S., Yamamoto, S., and Sato, H. (2012). High-Coverage ITS Primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS One 7:e40863. doi: 10.1371/journal.pone.0040863
Turner, B. L., and Haygarth, P. M. (2005). Phosphatase activity in temperate pasture soils. Potential regulation of labile organic phosphorus turnover by phosphodiesterase activity. Sci. Total Environ. 344, 27–36. doi: 10.1016/j.scitotenv.2005.02.003
Walker, J. K. M., Cohen, H., Higgins, L. M., and Kennedy, P. G. (2014). Testing the link between community structure and function for ectomycorrhizal fungi involved in a global tripartite symbiosis. New Phytol. 202, 287–296. doi: 10.1111/nph.12638
Wallander, H. (2006). External mycorrhizal mycelia – the importance of quantification in natural ecosystems. New Phytol. 171, 240–242. doi: 10.1111/j.1469-8137.2006.01803.x
Wallander, H., Ekblad, A., Godbold, D. L., Johnson, D., Bahr, A., Baldrian, P., et al. (2013). Evaluation of methods to estimate production, biomass and turnover of ectomycorrhizal mycelium in forests soils – A review. Soil Biol. Biochem. 57, 1034–1047. doi: 10.1016/j.soilbio.2012.08.027
Wallander, H., Nilsson, L. O., Hagerberg, D., and Baath, E. (2001). Estimation of the biomass and seasonal growth of external mycelium of ectomycorrhizal fungi in the field. New Phytol. 151, 753–760. doi: 10.1046/j.0028-646x.2001.00199.x
Wallander, H., and Thelin, G. (2008). The stimulating effect of apatite on ectomycorrhizal growth diminishes after PK fertilization. Soil Biol. Biochem. 40, 2517–2522. doi: 10.1016/j.soilbio.2008.06.011
Wallander, H., Wickman, T., and Jacks, G. (1997). Apatite as a P source in mycorrhizal and non-mycorrhizal Pinus sylvestris seedlings. Plant Soil 196, 123–131.
White, T. J., Bruns, T., Lee, S., and Taylor, J. L. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. 18, 315–322. doi: 10.1016/b978-0-12-372180-8.50042-1
Yang, N., Schützenmeister, K., Grubert, D., Jungkunst, H. F., Gansert, D., Scheu, S., et al. (2015). Impacts of earthworms on nitrogen acquisition from leaf litter by arbuscular mycorrhizal ash and ectomycorrhizal beech trees. Environ. Exp. Bot. 120, 1–7. doi: 10.1016/j.envexpbot.2015.06.013
Zak, D. R., Pellitier, P. T., Argiroff, W., Castillo, B., James, T. Y., Nave, L. E., et al. (2019). Exploring the role of ectomycorrhizal fungi in soil carbon dynamics. New Phytol. 223, 33–39. doi: 10.1111/nph.15679
Zavišić, A., Yang, N., Marhan, S., Kandeler, E., and Polle, A. (2018). Forest soil phosphorus resources and fertilization affect ectomycorrhizal community composition, beech P uptake efficiency, and photosynthesis. Front. Plant Sci. 9:463. doi: 10.3389/fpls.2018.00463
Zederer, D. P., Talkner, U., Spohn, M., and Joergensen, R. G. (2017). Microbial biomass phosphorus and C/N/P stoichiometry in forest floor and A horizons as affected by tree species. Soil Biol. Biochem. 111, 166–175. doi: 10.1016/j.soilbio.2017.04.009
Keywords: organic P, inorganic P, acid phosphomonoesterase, phosphodiesterase, peroxidase, phenoloxidase
Citation: Müller K, Kubsch N, Marhan S, Mayer-Gruner P, Nassal P, Schneider D, Daniel R, Piepho H-P, Polle A and Kandeler E (2020) Saprotrophic and Ectomycorrhizal Fungi Contribute Differentially to Organic P Mobilization in Beech-Dominated Forest Ecosystems. Front. For. Glob. Change 3:47. doi: 10.3389/ffgc.2020.00047
Received: 19 December 2019; Accepted: 03 April 2020;
Published: 14 May 2020.
Edited by:
Sebastian Loeppmann, Christian-Albrechts-Universität zu Kiel, GermanyReviewed by:
Martin Maier, Forstliche Versuchs-und Forschungsanstalt Baden-Württemberg (FVA), GermanyMatthias Gube, Georg August University Göttingen, Germany
Copyright © 2020 Müller, Kubsch, Marhan, Mayer-Gruner, Nassal, Schneider, Daniel, Piepho, Polle and Kandeler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karolin Müller, a2Fyb2xpbi5tdWVsbGVyQHVuaS1ob2hlbmhlaW0uZGU=
 Karolin Müller
Karolin Müller Nadine Kubsch1
Nadine Kubsch1 Sven Marhan
Sven Marhan Pascal Nassal
Pascal Nassal Dominik Schneider
Dominik Schneider Rolf Daniel
Rolf Daniel Hans-Peter Piepho
Hans-Peter Piepho Andrea Polle
Andrea Polle