
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Ecol. Evol. , 07 February 2025
Sec. Population, Community, and Ecosystem Dynamics
Volume 13 - 2025 | https://doi.org/10.3389/fevo.2025.1528335
This article is part of the Research Topic Biological Invasions in Aquatic Ecosystems: Detection, Assessment and Countermeasures View all 13 articles
 Lei Wang1,2
Lei Wang1,2 Tingting Sun1
Tingting Sun1 Huichao Jiang3,4*
Huichao Jiang3,4* Wenjing Zhang1
Wenjing Zhang1 Jianlong He3,4
Jianlong He3,4 Yuanqing Ma3,4
Yuanqing Ma3,4 Jianmin Zhao1
Jianmin Zhao1 Zhijun Dong1*
Zhijun Dong1*The frequent occurrence of Aurelia coerulea medusae in coastal waters poses a serious threat to power plants and fisheries, emphasizing the importance of early jellyfish bloom detection. Owing to the complex life cycle of jellyfish and the limitations of traditional survey methods, identifying the natural habitat of early stage jellyfish, especially polyps and ephyrae, is challenging. In this study, we aimed to identify the early habitats A. coerulea using environmental DNA (eDNA) metabarcoding technology to predict the sources of blooms. We successfully detected A. coerulea in a temperate bay (Laizhou Bay, LZB) and nearby aquaculture ponds (Dongying, DY) in March, when medusae were absent, revealing the habitats and distributions of the early stages of bloom-causing jellyfish. The relative abundance of blooming A. coerulea in the DY aquaculture ponds was significantly higher than that in the LZB. Our results suggest that coastal aquaculture ponds, as natural habitats for A. coerulea polyps and ephyrae, are an important source of A. coerulea medusa aggregates in the surrounding coastal waters. These findings suggest that jellyfish management strategies can be focused on aquaculture ponds, allowing for the source-based prevention and control of blooms before they cause damage.
Aurelia spp., the most common bloom-causing scyphozoan jellyfish, are widely distributed in harbors, lakes, and coastal waters worldwide (Lucas, 2001; Dong, 2019), disrupting ecological balance and threatening local power plants, fisheries, and aquaculture (Purcell et al., 2013; Dong, 2019). Climate change, coastal eutrophication, overfishing, and artificial construction have been proposed as important contributors to Aurelia spp. blooms (Dong et al., 2010; Duarte et al., 2012; Dong, 2019).
Aurelia spp. has a metagenetic life cycle comprising a benthic, asexual polyp and a pelagic, sexual medusa. As polyps can rapidly multiply through asexual reproduction, leading to an increased adult population, they are crucial for regulating the formation of blooms (Lucas et al., 2012). Previous studies on the population dynamics of Aurelia coerulea in Chinese coastal waters have shown that medusae appear primarily in summer and autumn (Dong et al., 2014; Wang and Sun, 2015). However, the distribution and population dynamics of polyps and ephyrae are difficult to monitor because of their cryptic morphology and habitats, which are often overlooked by traditional survey methods. This poses a considerable challenge for reliably predicting and controlling A. coerulea blooms (Ceh and Riascos, 2017), highlighting the urgent need to track the natural habitats of A. coerulea during its early life stages.
Environmental DNA (eDNA) metabarcoding is an emerging method that uses environmental samples, such as water and sediments, to investigate biodiversity and biomass (Berry et al., 2019; Ruppert et al., 2019; Skelton et al., 2022). Recently, this technique has been used for studies on invasive species detection (Roux et al., 2020; Thomas et al., 2020), pollution prediction (Li et al., 2018), dietary and trophic investigations (Yoon et al., 2017), and species distribution (Timmers et al., 2022). In addition, the high detection rate and sensitivity of eDNA detection assays demonstrate their applicability in investigations of jellyfish diversity and spatial distribution (Ames et al., 2021; Morrissey et al., 2022; Peng et al., 2023; Ye et al., 2024; Morrissey et al., 2024a, b). For example, eDNA techniques demonstrated that A. coerulea has a vertical distribution pattern in the Bohai Sea (Ye et al., 2024) and that A. coerulea aggregations were most likely to occur at the inner bottom region of Yantai Sishili Bay (Peng et al., 2023). Moreover, eDNA has been used to investigate potential polyp habitats and to detect polyps when medusae are absent (Morrissey et al., 2024a, b).
Laizhou Bay, situated in the Bohai Sea of China, is characterized by its coastal areas, which support sea cucumber Apostichopus japonicus aquaculture. A previous study showed that artificial reef structures in A. japonicus culture ponds, such as plastic sunshade nets, triangular tiles, cage substrates, and hollow bricks, provide appropriate substrates for the settlement and proliferation of A. coerulea polyps (Dong et al., 2018). Therefore, we propose that A. japonicus culture ponds may act as nursery grounds for A. coerulea, potentially representing a source of jellyfish blooms in Chinese coastal waters. In this study, we aimed to elucidate the occurrence and distribution of the bloom-causing jellyfish A. coerulea during its early life stages. Accordingly, we utilized eDNA metabarcoding based on the mitochondrial cytochrome oxidase subunit I (COI) gene to identified the presence of A. coerulea in seawater samples from aquaculture ponds and adjacent areas in Laizhou Bay during March, before blooming medusae appear. This research offers an effective method for identifying the origins of jellyfish blooms and provides a foundation for enhancing strategies to prevent and manage A. coerulea blooms.
The investigation was conducted in March 2024 at 57 sampling stations in aquaculture ponds and surrounding seawater in Laizhou Bay, including 24 stations in Laizhou Bay (LZB1–24) (Figure 1A) and 33 ponds from four aquaculture areas in Dongying (DY1-1–7, DY2-1–9, DY3-1–9, and DY4-1–7) (Figure 1B). Surface and bottom seawater samples were collected at stations with depths greater than 5 m in the LZB, whereas only surface seawater was collected at other stations and in aquaculture ponds (Figure 1A). One liter (LZB) or 0.5 liter (DY) of seawater was filtered through a 0.7 μm GF/F membrane (Whatman, Maidstone, UK) immediately after collection and then preserved in 2 mL sterile cryopreservation tubes (Beyotime, Shanghai, China). Less water was used for DY than LZB samples due to the challenges associated with filtering seawater samples collected from aquaculture ponds. The membrane samples were frozen in liquid nitrogen and stored at –80°C. Before sampling at different locations and water layers, all devices used for sample collection and filtration were sterilized with 10% bleach solution and washed at least three times with Milli-Q water. To track possible contamination during sample collection, negative controls were established by collecting 1 L/0.5 L distilled water at each station. Seawater temperature was measured in situ using an EXO2 Multiparameter Sonde (YSI, Yellow Springs, OH, USA).
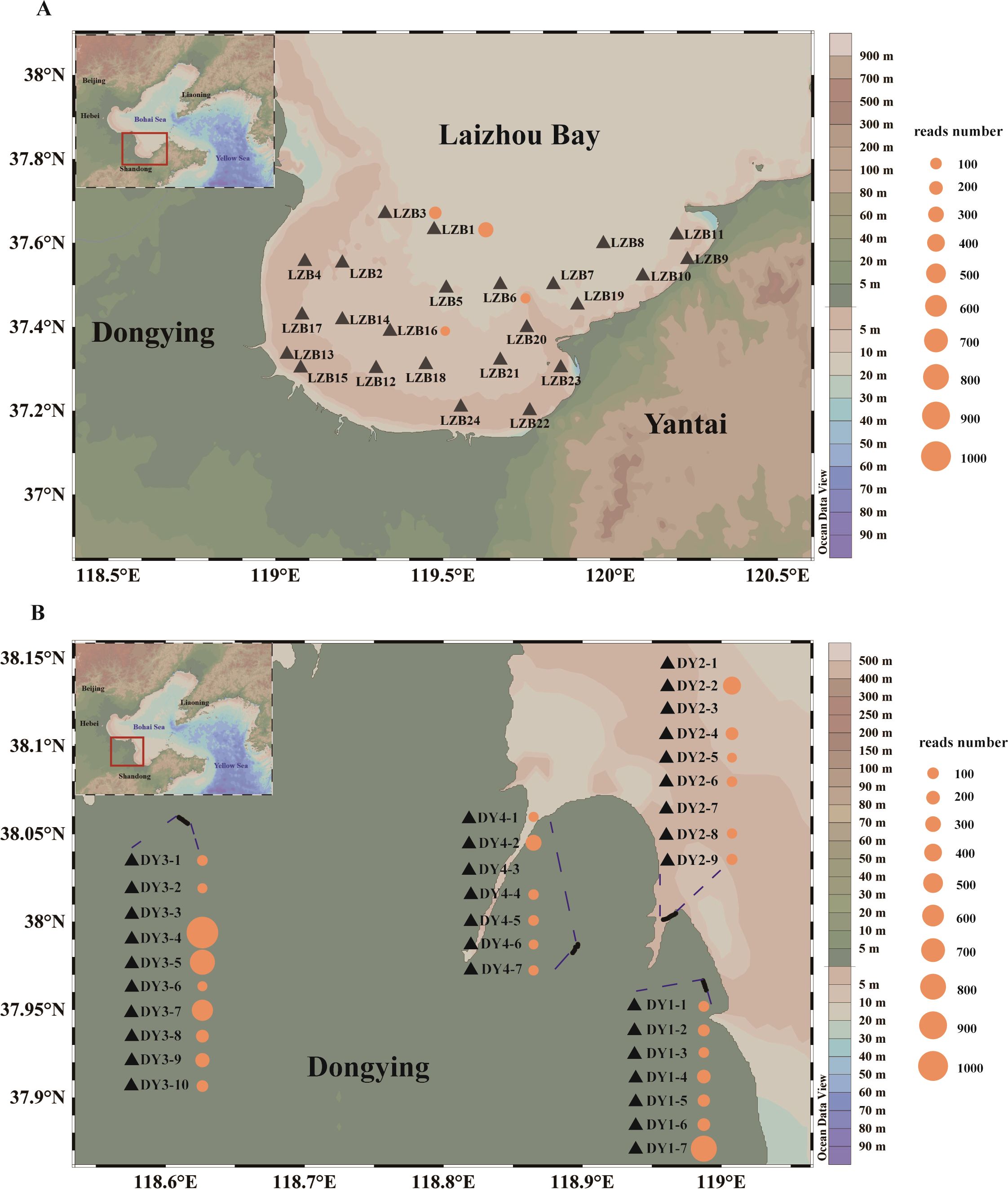
Figure 1. Sampling locations in Laizhou Bay [LZB, (A)] and Dongying aquaculture ponds [DY, (B)]. Bubbles indicate the number of reads obtained for Aurelia coelurea, where the number of reads obtained from samples collected in the DY pond is multiplied by 2 to represent equal sample sizes (1 L).
eDNA from seawater samples (n = 70) and negative controls (n = 57) was extracted following the procedure for the Qiagen DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). The concentration and purity of DNA were evaluated using the NanoDrop ND-2000C spectrophotometer (NanoDrop Technologies, USA), and the integrity was determined through 2% (w/v) agarose gel electrophoresis. After extraction, eDNA was dissolved in 80 µL of AE buffer and stored at –20°C. Polymerase Chain Reaction (PCR) was performed using the mitochondrial COI universal metazoan primers (mlCOIintF 5´-GGWACWGGWTGAACWGTWTAYCCYCC-3´ and jgHCO2198 5´-TAIACYTCIGGRTGICCRAARAAYCA-3´; Leray et al., 2013). Each PCR reaction was made up to 20 μL containing: 4 µL 5× FastPfu Buffer, 2 µL 2.5 mM dNTPs, 0.4 µL FastPfu Polymerase, 0.8× 2 µL primers (5 µM), 0.2 µL BSA, 2 µL eDNA template, and 9.8 µL double-distilled H2O. Thermocycler conditions were as follows: initial denaturation at 95°C for 3 min; 37 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 45 s; and a final extension at 72°C for 10 min. PCR products were analyzed using 2% (w/v) agarose gel electrophoresis and recovered using an AxyPrep DNA gel recovery kit (Axygen, Silicon Valley, USA). After normalization to equimolar amounts using the QuantiFluorTM-ST Blue Fluorescence Quantification System (Promega, Madison, WI, USA), paired-end sequencing (2 × 300 bp) was performed using an Illumina MiSeq platform at Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China).
The paired-end sequences were assembled using FLASH (version 1.2.11) (Magoč and Salzberg, 2011), and the merged sequences were controlled and filtered using QIIME v 1.9.1 to obtain high-quality clean reads (Caporaso et al., 2010; Bokulich et al., 2013). The quality-filtered sequences were clustered over a 97% similarity threshold into OTUs (operational taxonomic units). Chimeras were removed during clustering using UCHIME (Edgar et al., 2011). Representative sequences for each OTU were annotated using the NT database (Nucleotide Sequence Database, v20210917) in the NCBI database based on BLAST (e-value = 1e−5). All the samples were rarefied to the sequence number corresponding to the sample with the fewest sequences. Only OTUs classified as metazoans were retained (unclassified OTUs were removed). The sample featuring the fewest sequence count was employed as a reference, and subsequently, the sequence counts of all the other samples were randomly standardized to conform to this number, thereby generating normalized data.
Maps of the LZB and DY sampling stations were visualized using the Ocean Data View software (Reiner Schlitzer, Alfred Wegener Institute, Bremerhaven, Germany). Unclassified and classified Viridiplantae, fungal, bacterial, and eukaryotic reads were removed, and only OTUs annotated as metazoans were retained. To reduce falsity, more than five reads of the species detected in each sample were required. Mann–Whitney U tests were used to examine the relative abundance of A. coerulea between the LZB and DY collection stations. GraphPad Prism (version 8, San Diego, CA, USA) was used for statistical analyses, and statistical significance was set at P < 0.05.
During the sampling in March 2024, seawater temperatures ranged from 4.4°C to 10.1°C (mean ± SD = 6.6 ± 1.8°C; n = 24) at LZB stations and from 8.2°C to 11.7°C (mean ± SD = 10.1 ± 1.2°C; n = 33) in DY aquaculture ponds.
Seventy amplicon libraries were successfully constructed from seawater samples collected in the LZB (n = 37) and the surrounding DY aquaculture ponds (n = 33). No amplification of metazoan DNA was detected in the negative controls. A total of 303,289 metazoan sequences remained across all samples after quality control, which were taxonomically identified as 158 species. Twelve phyla were detected, with the top four in terms of species richness being Arthropoda (55.70%; 88 species), Cnidaria (13.92%; 22 species), Annelida (6.96%; 11 species), and Chordata (6.33%; 10 species) (Supplementary Figure S1).
A total of 17 species were classified as jellyfish, covering two classes (Hydrozoa and Scyphozoa), seven orders, 13 families, and 15 genera (Supplementary Table S1). Overall, Leptothecata (five species), Anthoathecata (four species), and Siphonophorae (three species) were the three most abundant orders in terms of species count (Supplementary Table S1). Fifteen species (88.24%) of hydrozoans and two (11.76%) of scyphozoans were identified. Among these, three species (17.65%) were shared between the two regions, three species (17.65%) were exclusively detected in the LZB samples, and 11 species (64.70%) were only detected in the DY aquaculture ponds (Supplementary Table S1). Of the jellyfish that were detected, A. coerulea had the highest read count detected in both LZB and DY samples (Supplementary Table S1).
A total of 3,641 reads of A. coerulea were obtained using COI gene amplicon sequencing. The identification percentage of the A. coerulea blasted in the NT database was 100.00%, showing reliability. Among the 3,641 reads, only 256 were detected in the eDNA samples extracted from LZB, whereas 2,885 reads were detected in the DY aquaculture ponds. Aurelia coerulea was detected in 28 of 33 aquaculture pond samples (Figure 1B) but only in 4 of the 24 LZB stations (LZB1, LZB3, LZB6, and LZB16, Figure 1A). The detection rate was 84.85% in DY aquaculture ponds, compared to 16.67% in the LZB. Moreover, the relative abundance of A. coerulea in coastal aquaculture ponds was significantly higher than that in the LZB (U = 113; P < 0.01; Figure 2).
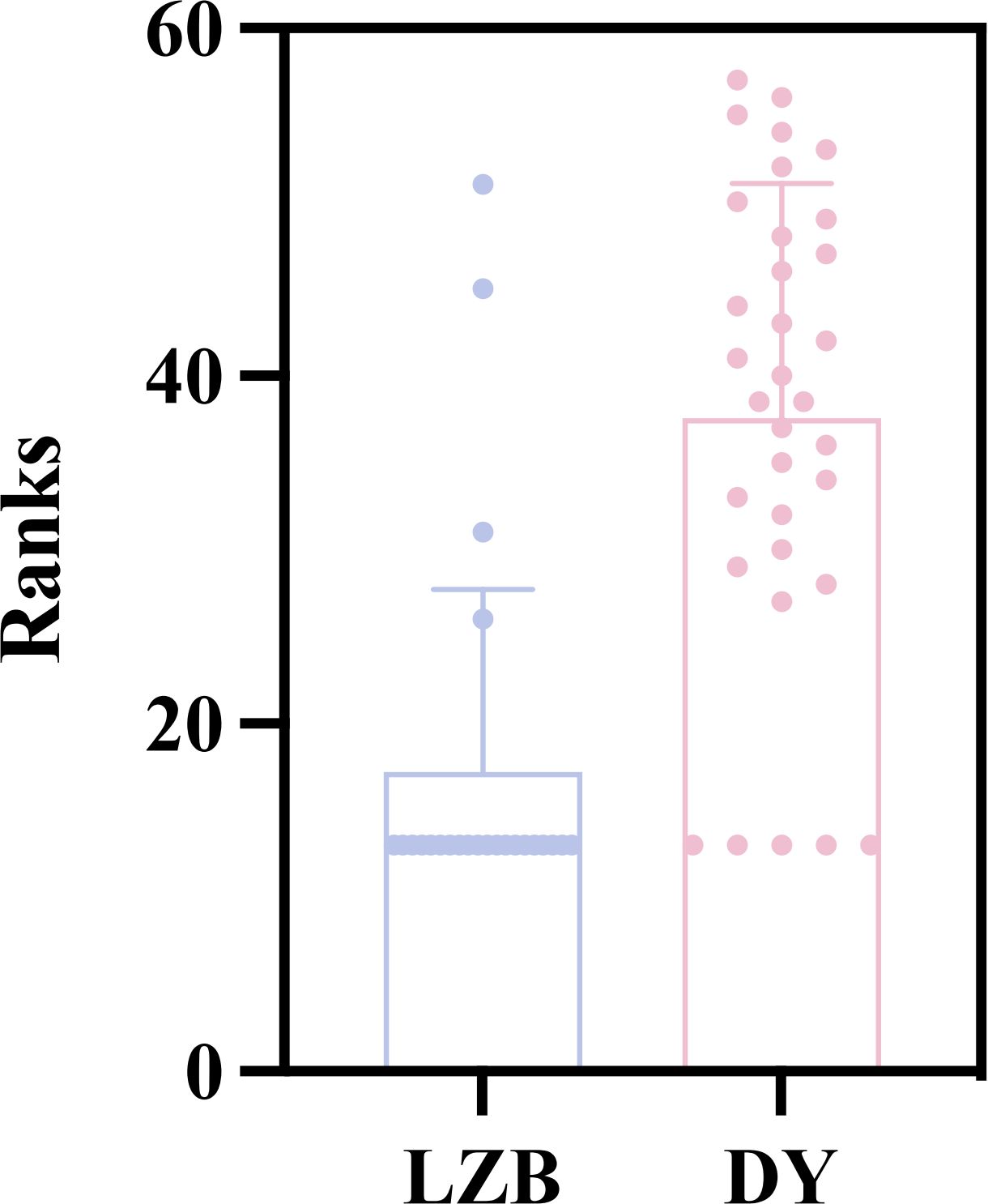
Figure 2. The relative abundance of Aurelia coerulea polyps between Laizhou Bay (LZB) and Dongying (DY) aquaculture ponds. Mann-Whitney U tests, P < 0.01; n = 24 sampling sites in the LZB, n = 33 aquaculture ponds from DY.
In the present study, we collected seawater samples from the LZB and from DY coastal aquaculture ponds in early spring (March), when A. coerulea medusae were absent (Dong et al., 2014; Feng et al., 2018). Visual surveys and trawling operations using type II zooplankton nets in LZB also failed to observe or capture A. coerulea medusae. Therefore, we revealed the habitats and distribution patterns of A. coerulea polyps and ephyrae using eDNA metabarcoding. Aurelia coerulea was detected in 84.85% of DY aquaculture ponds, with a significantly higher occurrence frequency in aquaculture ponds than in open coastal seawater. In addition, the relative abundance of A. coerulea reads in the DY aquaculture ponds was significantly higher than that in the LZB. This revealed that the early life stages of the blooming jellyfish A. coerulea were mainly distributed in the DY aquaculture ponds rather than those in the LZB coastal waters, suggesting that culture ponds are critical natural habitats for A. coerulea polyps and ephyrae.
Relatively enclosed culture ponds can provide a stable environment for the settlement of planulae and the reproduction and expansion of A. coerulea polyps (Lo et al., 2008; Purcell et al., 2013; Dong et al., 2017, 2018). Previous studies have demonstrated that A. coerulea polyps can reproduce prolifically on various artificial substrates such as marina floats and oil rig wrecks in coastal waters (van Walraven et al., 2016). Similarly, aquaculture ponds offer numerous artificial attachment sites for these polyps (Purcell et al., 2013; Dong et al., 2018). For example, the mean percent coverage of A. coerulea polyps under plastic sunshade nets and inside substrate cages was more than 40% in A. japonicus culture ponds in DY and Qingdao (Dong et al., 2018). In addition, the relatively gentle water flow in the ponds decreased the time required for A. coerulea planulae to settle (Dong et al., 2018). Furthermore, the risk of predation for polyps and ephyrae in ponds is greatly reduced compared to that in the wild. Consequently, a substantial population of A. coerulea polyps may proliferate in nearshore aquaculture environments, contributing to later blooms. This finding is further corroborated by the detection of A. coerulea in 84.85% of aquaculture pool samples through eDNA metabarcoding, underscoring the validity of this technique for elucidating the distribution of jellyfish during early life stages.
Water exchange in culture ponds and currents flowing in nearshore waters play important roles in the transport and aggregation of ephyrae, thus promoting jellyfish blooms. Water outlets and inlets create critical conditions for the exchange of A. coerulea ephyrae between aquaculture ponds and coastal waters (Dong et al., 2018). Covering these areas with nylon nets can prevent the escape of farmed organisms (sea cucumbers and shrimp) and the entry of potential predators, but does not prevent the spread of A. coerulea ephyrae or planulae. Furthermore, as ambient temperatures reach 3.9°C, strobilation is induced, and ephyrae begin to be released into the ocean at 7°C (Feng et al., 2018). Therefore, ephyrae may have already reached the LZB from the DY coastal aquaculture ponds by the time of our sampling in early spring. Since ephyrae are regarded as weak swimmers (von Montfort et al., 2023), nearshore currents play a significant role in accelerating the spread of ephyrae. Therefore, the substantial quantities of ephyrae released into offshore aquaculture ponds may serve as an important source of A. coerulea medusa aggregation in the surrounding coastal waters.
Polyps and ephyrae serve as sources of A. coerulea outbreaks; thus, tracing their origins and distribution is crucial for predicting their population dynamics and controlling blooms before they cause damage. Chemical, physical, and biological techniques have been used to control the early life stages of jellyfish. Dong et al. (2017) found that tea saponins effectively eradicate A. coerulea ephyrae and polyps in sea cucumber aquaculture ponds. In addition, high-pressure water cannons and manual eradication have been shown to be effective for polyp removal. In future research, jellyfish control strategies that focus on source management should be reinforced.
eDNA technology has proven to be a valuable tool for studying the field distribution characteristics of jellyfish (including scyphozoans and cubozoans) at different life stages (Gaynor et al., 2017; Morrissey et al., 2024b). For example, Gaynor et al. (2017) used eDNA technology to reveal the spatial and temporal disparities in the distribution of free-swimming early stage Chrysaora quinquecirrha in Barnegat Bay, on the Atlantic Coast. When medusae were absent in winter, Chironex fleckeri polyps were detected through eDNA metabarcoding technology and served as a basis for investigating potential polyp habitats (Morrissey et al., 2024b). Therefore, in this study, eDNA metabarcoding was used to compare the occurrence and distribution of the early life stages of A. coerulea in the LZB and nearshore aquaculture ponds in March 2023. The relative abundance of A. coerulea eDNA in the DY aquaculture ponds was significantly higher than that in the LZB. Therefore, coastal aquaculture ponds likely represent a notable source of A. coerulea blooms. These findings provide evidence that eDNA can serve as an effective tool for detecting the origins and distribution of jellyfish, thereby establishing a foundation for enhanced prevention and control strategies targeting the sources of A. coerulea outbreaks.
eDNA metabarcoding technology effectively demonstrated that coastal aquaculture ponds represent a notable source of A. coerulea blooms. However, due to the variability of DNA density and gene copy number in tissues, limitations exist in calculating the relative abundance of jellyfish based on read counts. Furthermore, the relationship between organismal abundance and eDNA concentrations is influenced by biotic and abiotic factors (Rourke et al., 2021; Morrissey et al., 2024b). Consequently, implementing a variety of disparate methods to comprehensively analyze the wild distribution patterns of jellyfish is required to compensate for the limitations of single analysis techniques.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
LW: Conceptualization, Investigation, Project administration, Writing – review & editing. TS: Data curation, Formal analysis, Methodology, Writing – original draft. HJ: Writing – review & editing, Investigation. WZ: Writing – original draft, Writing – review & editing. JH: Writing – review & editing. YM: Writing – review & editing, Writing – original draft. JZ: Writing – review & editing. ZD: Conceptualization, Project administration, Supervision, Writing – review & editing, Resources, Writing – original draft, Investigation.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the National Key Research and Development Program of China (2023YFC3108200), the Key Project of the NSFC-Shandong Joint Fund (U2106208), Observation and Research Station of Laizhou Bay Marine Ecosystem, MNR, and Shandong Key Laboratory of Marine Ecological Restoration (SAL202403), the Taishan Scholars Program (tsqn202211263), and Natural Science Foundation of Shandong (ZR2024QD282).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2025.1528335/full#supplementary-material
Supplementary Figure 1 | Total species from twelve metazoan phyla.
Supplementary Table 1 | Total abundance of 17 jellyfish species. LZB, Laizhou Bay; DY, Dongying.
Ames C. L., Ohdera A. H., Colston S. M., Collins A. G., Fitt W. K., Morandini A. C., et al. (2021). Fieldable environmental DNA sequencing to assess jellyfish biodiversity in nearshore waters of the Florida Keys, United States. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.640527
Berry T. E., Saunders B. J., Coghlan M. L., Stat M., Jarman S., Richardson A. J., et al. (2019). Marine environmental DNA biomonitoring reveals seasonal patterns in biodiversity and identifies ecosystem responses to anomalous climatic events. PLoS Genet. 15, e1007943. doi: 10.1371/journal.pgen.1007943
Bokulich N. A., Subramanian S., Faith J. J., Gevers D., Gordon J. I., Knight R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–U11. doi: 10.1038/nmeth.2276
Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Ceh J., Riascos J. M. (2017). Cryptic life stages in scyphozoan jellyfish: larval settlement preferences of the South American sea nettle Chrysaora plocamia. J. Exp. Mar. Biol. Ecol. 490, 52–55. doi: 10.1016/j.jembe.2017.02.007
Dong Z. J. (2019). “Blooms of the moon jellyfish Aurelia: causes, consequences and controls,” in World seas: an environmental evaluation, vol III: ecological issues and environmental impacts. Ed. Charles S. (Elsevier, Amsterdam), 163–171. doi: 10.1016/B978-0-12-805052-1.00008-5
Dong Z. J., Liu D. Y., Keesing J. K. (2010). Jellyfish blooms in China: Dominant species, causes and consequences. Mar. pollut. Bull. 60, 954–963. doi: 10.1016/j.marpolbul.2010.04.022
Dong Z. J., Liu D. Y., Keesing J. K. (2014). “Contrasting trends in populations of Rhopilema esculentum and Aurelia aurita in Chinese waters in jellyfish blooms,” in Jellyfish blooms. Eds. Pitt K. A., Lucas C. H. (Springer, Dordrecht), 207–218. doi: 10.1007/978-94-007-7015-7_9
Dong Z. J., Sun T. T., Liang L. K., Wang L. (2017). Effect of tea saponin on ephyrae and polyps of the moon jellyfish Aurelia sp.1. PLoS One 12, e0182787. doi: 10.1371/journal.pone.0182787
Dong Z. J., Wang L., Sun T. T., Liu Q. Q., Sun Y. F. (2018). Artificial reefs for sea cucumber aquaculture confirmed as settlement substrates of the moon jellyfish Aurelia coerulea. Hydrobiologia 818, 223–234. doi: 10.1007/s10750-018-3615-y
Duarte C. M., Pitt K. A., Lucas C. H., Purcell J. E., Uye S. I., Robinson K., et al. (2012). Is global ocean sprawl a cause of jellyfish blooms? Front. Ecol. Evol. 11, 91–97. doi: 10.2307/23470528
Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Feng S., Wang S. W., Sun S., Zhang F., Zhang G. T., Liu M. T., et al. (2018). Strobilation of three scyphozoans (Aurelia coelurea, Nemopilema nomurai, and Rhopilema esculentum) in the field at Jiaozhou Bay, China. Mar. Ecol. Prog. Ser. 591, 141–153. doi: 10.3354/meps12276
Gaynor J. J., Bologna P. A., Restaino D. J., Barry C. L. (2017). qPCR detection of early life history stage Chrysaora quinquecirrha (sea nettles) in Barnegat Bay, New Jersey. J. Coastal. Res. 78, 184–192. doi: 10.2112/SI78-014.1
Leray M., Yang J. Y., Meyer C. P., Mills S. C., Agudelo N., Ranwez V., et al. (2013). A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Front. Zool. 10, 34. doi: 10.1186/1742-9994-10-34
Li F. L., Peng Y., Fang W. D., Altermatt F., Xie Y. W., Yang J. H., et al. (2018). Application of environmental DNA metabarcoding for predicting anthropogenic pollution in rivers. Environ. Sci. Technol. 52, 11708–11719. doi: 10.1021/acs.est.8b03869
Lo W., Purcell J. E., Hung J., Su H., Hsu P. (2008). Enhancement of jellyfish (Aurelia aurita) populations by extensive aquaculture rafts in a coastal lagoon in Taiwan. ICES J. Mar. Sci. 65, 453–461. doi: 10.1093/icesjms/fsm185
Lucas C. H. (2001). Reproduction and life history strategies of the common jellyfish, Aurelia aurita, in relation to its ambient environment. Hydrobiologia 451, 229–246. doi: 10.1023/A:1011836326717
Lucas C. H., Graham W. M., Widmer C. (2012). Jellyfish life histories: role of polyps in forming and maintaining scyphomedusa populations. Adv. Mar. Biol. 63, 133–196. doi: 10.1016/B978-0-12-394282-1.00003-X
Magoč T., Salzberg S. L. (2011). FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Morrissey S. J., Jerry D. R., Kingsford M. J. (2022). Genetic detection and a method to study the ecology of deadly Cubozoan jellyfish. Diversity 14, 1139. doi: 10.3390/d14121139
Morrissey S. J., Jerry D. R., Kingsford M. J. (2024a). Use of eDNA to determine source locations of deadly jellyfish (Cubozoa) in an open coastal system. Coast 4, 198–212. doi: 10.3390/coasts4010011
Morrissey S. J., Jerry D. R., Kingsford M. J. (2024b). Use of eDNA to test hypotheses on the ecology of Chironex fleckeri (Cubozoa). Mar. Ecol. Prog. Ser. 728, 25–41. doi: 10.3354/meps14507
Peng S. J., Wang L., Ma Y. Q., Ye L. J., Hou C. W., Liu Y. L., et al. (2023). Application of environmental DNA metabarcoding and quantitative PCR to detect blooming jellyfish in a temperate bay of northern China. Ecol. Evol. 13, e10669. doi: 10.1002/ece3.10669
Purcell J. E., Baxter E. J., Fuentes V. (2013). “Jellyfish as products and problems for aquaculture,” in Advances in Aquaculture Hatchery Technology. Eds. Allan G., Burnell G. (Elsevier, Amsterdam), 404–430. doi: 10.1533/9780857097460.2.404
Rourke M. L., Fowler A. M., Hughes J. M., Broadhurst M. K., DiBattista J. D., Fielder S., et al. (2021). Environmental DNA (eDNA) as a tool for assessing fish biomass: a review of approaches and future considerations for resource surveys. Environ. DNA 4, 9–33. doi: 10.1002/edn3.185
Roux L. M. D., Giblot-Ducray D., Bott N. J., Wiltshire K. H., Deveney M. R., Westfall K. M., et al. (2020). Analytical validation and field testing of a specific qPCR assay for environmental DNA detection of invasive European green crab (Carcinus maenas). Environ. DNA 2, 309–320. doi: 10.1002/edn3.65
Ruppert K. M., Kline R. J., Rahman M. S. (2019). Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: a systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 17, e00547. doi: 10.1016/j.gecco.2019.e00547
Skelton J., Cauvin A., Hunter E. M. (2022). Environmental DNA metabarcoding read numbers and their variability predict species abundance, but weakly in non-dominant species. Environ. DNA 5, 1092–1104. doi: 10.1002/edn3.355
Thomas A. C., Tank S., Nguyen P. L., Ponce J., Goldberg C. S. (2020). A system for rapid eDNA detection of aquatic invasive species. Environ. DNA 2, 261–270. doi: 10.1002/edn3.25
Timmers M. A., Vicente J., Webb M., Jury C. P., Toonen R. J. (2022). Sponging up diversity: Evaluating metabarcoding performance for a taxonomically challenging phylum within a complex cryptobenthic community. Environ. DNA 4, 239–253. doi: 10.1002/edn3.163
van Walraven L., Driessen F., van Bleijswijk J., Bol A., Luttikhuizen P. C., Coolen Joop W. P., et al. (2016). Where are the polyps? Molecular identification, distribution and population differentiation of Aurelia aurita jellyfish polyps in the southern North Sea area. Mar. Biol. 163, 172. doi: 10.1007/s00227-016-2945-4
von Montfort G. M., Costello J. H., Colin S. P., Morandini A. C., Migotto A. E., Maronna M. M., et al. (2023). Ontogenetic transitions, biomechanical trade-offs and macroevolution of scyphozoan medusae swimming patterns. Sci. Rep. 13, 9760. doi: 10.1038/s41598-023-34927-w
Wang Y. T., Sun S. (2015). Population dynamics of Aurelia sp.1 ephyrae and medusa in Jiaozhou Bay, China. Hydrobiologia 754, 147–155. doi: 10.1007/s10750-014-2021-3
Ye L. J., Peng S. J., Ma Y. Q., Zhang W. J., Wang L., Sun X. Y., et al. (2024). Biodiversity and distribution patterns of blooming jellyfish in the Bohai Sea revealed by eDNA metabarcoding. BMC Ecol. Evol. 24, 37. doi: 10.1186/s12862-024-02224-3
Keywords: jellyfish blooms, Aurelia coerulea, aquaculture ponds, eDNA metabarcoding, source control
Citation: Wang L, Sun T, Jiang H, Zhang W, He J, Ma Y, Zhao J and Dong Z (2025) Coastal aquaculture ponds represent a notable source of the blooming jellyfish Aurelia coerulea. Front. Ecol. Evol. 13:1528335. doi: 10.3389/fevo.2025.1528335
Received: 14 November 2024; Accepted: 23 January 2025;
Published: 07 February 2025.
Edited by:
Wei Wang, Ocean University of China, ChinaReviewed by:
Zhenming Lv, Zhejiang Ocean University, ChinaCopyright © 2025 Wang, Sun, Jiang, Zhang, He, Ma, Zhao and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huichao Jiang, amlhbmdodWljaGFvMjAwOEAxNjMuY29t; Zhijun Dong, empkb25nQHlpYy5hYy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.