- Department of Biology, Boston University, Boston, MA, United States
Phenotypic plasticity is a fundamental property of developing organisms and is thought to play an important role in diversification. Plastic responses themselves are remarkably diverse and respond to a wide range of environmental factors. Here I focus on plasticity in response to variation in nutrition in insects since 1) nutrition is a widespread factor that impacts most organisms, and 2) insects are important models to study phenotypic plasticity. First, I provide a brief overview of our current mechanistic understanding of the regulation of nutritionally cued plasticity in insects, in both traditional as well as emerging model systems. Then, I explore evolutionary mechanisms enabling the diversification of regulation across taxa, emphasizing the role of gene duplication and gene regulatory network co-option. Furthermore, I examine nutrition-responsive phenotypes as suites of multiple traits that develop in a coordinated manner. I argue that understanding how these traits are integrated at the molecular level can shed light on the evolution of complex phenotypes. Finally, I discuss potential challenges and opportunities to further our understanding of nutrition-responsive plasticity, its regulation, and its evolution.
Introduction
Developmental plasticity is a fundamental process that allows organisms to adjust their phenotype during development in response to variable environmental conditions. The degree to which phenotypes respond to environmental factors varies across individuals, populations, and species. Traits may change as a response to an environmental factor in a continuous manner yet in extreme cases, discrete phenotypes, or polyphenisms, are produced (West-Eberhard, 2003). Various environmental factors often have an effect on development, and in many instances multiple environmental inputs act in a concerted manner to achieve a highly regulated phenotypic response (Shingleton et al., 2009; Lin et al., 2018). One of the most widespread environmental factors that can impact development is nutrition (Moczek, 1998; Karino et al., 2004; Shingleton et al., 2009). Plasticity in response to nutrition is present in diverse taxa, from insects to plants and humans (Schlichting, 1986; Moore et al., 2004; Teleman et al., 2008; Tchokponhoué et al., 2019). In some cases, nutrition is the sole environmental factor, whereas in others it acts in concert with a suite of environmental factors that elicit a specific phenotypic response (e.g. in combination with population density, social environment or temperature; Brian, 1979; Shingleton et al., 2009; Serobyan et al., 2013; Deem et al., 2024).
Insects have emerged as models to understand the regulation and evolution of plasticity, in part due to the feasibility and amenability of tools available, but also due to the enormous diversity of plastic responses they exhibit (Fjerdingstad and Crozier, 2006; Moczek, 2010; Casasa et al., 2017). Insights from Drosophila have elucidated many genetic and genomic mechanisms of plasticity, and studies of diverse insect species have continued to reveal the evolution of these responses. In this review, I summarize recent advances on the genetic and genomic mechanisms of developmental plasticity in response to nutrition across diverse insects. Then, I examine how these mechanisms have evolved, focusing on gene duplication, and gene network co-option. Finally, I emphasize the need to study the regulation of complex suites of environmentally sensitive traits, in particular how they are integrated during development.
Molecular mechanisms of nutritional plasticity across insects
The insulin/insulin-like growth factor signaling pathway (IIS) is key in regulating phenotypic changes in response to nutrition. This pathway is conserved across organisms, from yeast to vertebrates (Brogiolo et al., 2001; Barbieri et al., 2003; Vitali et al., 2018). In insects, this pathway is known to regulate body size as a function of nutrition, as well as tissue-specific relative size (Stern, 2003; Shingleton et al., 2005; Edgar, 2006; Callier and Nijhout, 2013; Casasa and Moczek, 2018). Studies on Drosophila have revealed that nutritional conditions during larval development are first detected by cells in the fat body, where the Target of Rapamycin (TOR) pathway detects amino acid levels (reviewed in Koyama and Mirth, 2018). Consequently, the fat body secretes peptide signals, which are detected by insulin-producing cells in the brain. These cells, in turn, secrete insulin-like peptides (ILPs) into the hemolymph. Across tissues, ILPs bind to the insulin receptor (InR), which activates a signal transduction cascade that promotes growth and proliferation (Brogiolo et al., 2001).
Beyond Drosophila, the IIS pathway has been implicated in mediating nutrition-responsive plasticity across a broad range of insect orders and traits (reviewed in Nijhout and McKenna, 2018; Weger and Rittschof, 2024). In the rhinoceros beetle, Trypoxylus dichotomus (Coleoptera), growth of exaggerated horns used as weapons depends on nutritional conditions during larval development. This increased sensitivity of horns to nutritional conditions is regulated by the InR, and InR knockdown results in a decrease in horn length (Emlen et al., 2012). In the butterfly Precis coenia, bombyxin, the first discovered insect ILP (discovered in Bombyx mori), regulates growth of wing imaginal discs (Nijhout and Grunert, 2002). In the clonal raider ant, Ooceraea biroi (Hymenoptera), the ILP2 is involved in the regulation of reproductive division of labor (Chandra et al., 2018). Thus, the IIS pathway is a general mechanism that mediates nutrition-responsive plasticity across insects.
In addition to the role of the IIS in mediating nutrition-responsive growth, there are multiple other pathways and insect hormones that have been identified to be involved in this process. For instance, one of the major insect hormones, juvenile hormone (JH), has been implicated in the regulation of stag beetle (Cyclommatus metallifer) mandible enlargement (Gotoh et al., 2011). Moreover, the IIS pathway regulates several downstream pathways and hormones. For example, the IIS pathway is involved in promoting biosynthesis of JH as well as ecdysone, another major insect hormone (Abrisqueta et al., 2014; Gokhale et al., 2016). At the same time, these insect hormones also have the ability to regulate the IIS pathway (Colombani et al., 2005; Mirth et al., 2014). Overall, the interplay between IIS and other mechanisms is complex, and regulation of nutrition-dependent plasticity involves crosstalk across various hormonal and signaling pathways.
Evolution of the mechanisms regulating nutrition-responsive plasticity
Developmentally plastic responses exhibit remarkable diversity across multiple levels of biological organization. Within populations, different traits display different sensitivities to nutritional levels. For instance, sexually selected traits are often extremely sensitive to variation in nutrition and result in exaggerated morphologies in high nutrition males and more moderate ones in low nutrition males (e.g. horns in rhinoceros beetles, Emlen et al., 2012). Many other traits scale proportionally to body size and are therefore moderately sensitive to nutrition (e.g. wings, legs, etc.; Shingleton et al., 2007, 2008). Finally, there are traits that are insensitive or buffered from variation in nutrition (e.g. male genitalia; Eberhard, 2009). These traits display low levels of phenotypic variation across a nutritional gradient, regardless of overall body size. Across populations and species, plastic responses are also diverse (Casasa and Moczek, 2019). For example, in the red-shouldered soapberry bug, distinct populations vary in the proportion of short- and long-wing morphs (Fawcett et al., 2018). Similarly, closely related species of onthophagine horned beetles differ in relative horn length and degree of plasticity in response to nutrition. While some species exhibit a moderate degree of horn plasticity, others display either an exaggerated degree or have entirely lost horns and plasticity (Casasa et al., 2017, 2020). Together, the evolution of plasticity, either in terms of increase or loss of plasticity, greatly contributes to diversity within and among species.
Recently, significant progress has been made in elucidating the molecular and developmental mechanisms that regulate plastic responses in many non-model insects (Emlen et al., 2012; Xu et al., 2015; Casasa and Moczek, 2018; Fawcett et al., 2018). These insights are paving the way to our understanding of how these mechanisms have evolved. Here I focus on two mechanisms that have contributed to the evolution of the underpinning of nutritionally plastic responses. First, I discuss the role of gene duplication in the evolution of nutrition-responsive plasticity. Then I review our current understanding of the role of gene co-option and gene regulatory network co-option in the evolution of plastic responses.
The duplication of key genes that regulate nutrition-responsive plasticity has contributed to the evolution of the mechanisms underlying plastic responses. The most prominent example is the duplication of the InR. Most insect taxa possess two InR copies, including ants (Lu and Pietrantonio, 2011), bees (de Azevedo and Hartfelder, 2008), brown planthoppers (Xu et al., 2015; Xu and Zhang, 2017), horned beetles (Casasa and Moczek, 2018), and red flour beetles (Sang et al., 2016). Recent phylogenetic analyses revealed that this duplication occurred around 400 million years ago within basal Insecta and a few losses occurred thereafter, including in Diptera and Lepidoptera (Kremer et al., 2018; Smykal et al., 2020). Wingless insects such as silverfish (Zygentoma), some of the most basal insects studied, have a single InR copy, whereas basal-winged insects such as mayflies (Ephemeroptera) possess two. Interestingly, other non-insect taxa, including sponges, nematodes and vertebrates, have undergone independent duplications of the InR (Smykal et al., 2020).
The extent to which InR duplication contributes to the potential origin and diversification of plastic responses is taxon-specific. In Hemiptera, there have been multiple duplication events of both the InR1 and InR2, in some cases leading to as many as four copies of InR2 (some cicadellids; Smykal et al., 2020). The two InR copies in the brown planthopper Nilaparvata lugens are the primary regulators of wing polyphenism (Xu et al., 2015), where food quality and population density are the main environmental cues that induce short- and long-winged morphologies (Lin et al., 2018). Interestingly, InR1 has a canonical role, whereas InR2 has the opposing effect and negatively regulates InR1 (Xu et al., 2015). Across other hemipteran species the regulation of wing polyphenism has diverged from that of N. lugens. For instance, in the linden bug Pyrrhocoris apterus InR1 was duplicated. Functional genetic analyses revealed that although the role of InR2 seems conserved across species, InR1a plays a similar role to InR2 in N. lugens (Smykal et al., 2020). In contrast to these studies, where InR plays a key role in the regulation of wing polyphenism, a similar phenotypic response operates differently in the red-shouldered soapberry bug J. haematoloma. In this species, knockdown of the Forkhead Box O (Foxo) transcription factor, a negative growth regulator downstream of the InR, results in a shift in frequency towards a larger proportion of short-winged individuals. Thus, Foxo is the main regulator of wing polyphenism in J. haematoloma and acts in the opposite manner compared to N. lugens (Fawcett et al., 2018). Beyond Hemiptera, InR has also duplicated in termites, cockroaches and stick insects, and expression of the three paralogs differs among termite castes of three species (Kremer et al., 2018; Smykal et al., 2020). Yet whether different InR copies have a role in termite caste determination has yet to be functionally tested. In Onthophagus horned beetles neither of the two InR copies seem to have a role in the regulation of the male horn polyphenism (Casasa and Moczek, 2018), although in the rhinoceros beetle Trypoxylus dichotomus, the single InR does play a role in regulating horn growth in response to nutrition (Emlen et al., 2012).
Additional cases of gene duplications underlying plastic responses have been found in eusocial Hymenoptera, in which caste polyphenism is also nutritionally cued. A study using the honeybee Apis mellifera found that caste-biased genes often correspond to genes that had been duplicated compared to non-duplicated ones, and expression levels were similarly higher for duplicated genes, suggesting that duplicated genes play an important role in caste determination and probably contributed to its evolution (Chau and Goodisman, 2017). Moreover, in the harvester ant Pogonomyrmex barbatus, the gene vitellogenin (Vg) has undergone duplication followed by subfunctionalization, where the two paralogs exhibit caste- and behavior-specific expression (Corona et al., 2013).
Gene duplication is by no means required for the evolution of developmental plasticity. A growing body of evidence also suggests existing developmental pathways can be redeployed in novel ways to produce diverse plastic traits, including polyphenic traits (Moczek and Nagy, 2005; Kijimoto et al., 2012; Gotoh et al., 2014; Morandin et al., 2016; Casasa and Moczek, 2018; Kapheim et al., 2020). Above, I discussed the role that key pathways, such as IIS and TOR, have in the regulation of plasticity. These pathways have an ancestral role translating a nutrition cue into a phenotypic response, yet they have been co-opted in the context of providing a nutritional sensor to novel traits or traits that ancestrally did not exhibit high nutrition responsiveness. For instance, Onthophagus beetle head horns are novel traits that respond to a nutritional gradient in a discontinuous manner (polyphenism; Moczek, 2003). One of the components of the IIS pathway, Foxo, is key in mediating this growth response (Casasa and Moczek, 2018). While Foxo’s role in negatively regulating growth is conserved, it has been redeployed in the regulation of a novel trait. Likewise, genes and gene regulatory networks that were ancestrally not directly involved in the regulation of plasticity are known to have been co-opted in the regulation and evolution of plastic traits. Examples of this include the role of doublesex (dsx) — a gene that encodes for a sex determination transcription factor — in the regulation of exaggerated, nutrition sensitive male stag beetle mandibles (Gotoh et al., 2014), and the role of Hedgehog signaling pathway — involved in anterior/posterior axis determination — in the evolution of novel and polyphenic beetle head horns (Kijimoto and Moczek, 2016). In contrast to IIS and TOR signaling pathways, which are well known to be directly involved in nutrient responsiveness (Koyama and Mirth, 2018), Dsx and Hedgehog signaling pathways are not (Ingham and McMahon, 2001; Price et al., 2015; but see Agrawal and Léopold, 2015). Instead, these examples showcase how the latter pathways display novel contributions to exaggerated nutritional responses (Gotoh et al., 2014; Kijimoto and Moczek, 2016).
Gene duplication and neofunctionalization offer an intuitive explanation for the evolution of plasticity and emergence of polyphenisms, yet the genetic mechanisms giving rise to co-option of existing pathways during the evolution of plastic traits are less clear. Generally, network co-option can occur through changes in key regulators, either in regulatory regions or protein coding sequences, that lead to recruitment of many effector genes and their regulatory elements (True and Carroll, 2002; McQueen and Rebeiz, 2020). While the precise mechanisms that led to redeployment of dsx and Hedgehog signaling pathway in the evolution of novel plastic responses have not been fully elucidated, this process was likely facilitated by pre-existing connections and these pathways’ canonical roles in insect somatic sex differentiation (i.e. sexual dimorphisms), and patterning and cell proliferation, respectively (Ingham and McMahon, 2001; Hopkins and Kopp, 2021). Moreover, in instances where the same trait has independently evolved, similar pathways have been recruited, albeit in some instances different components of the same pathway act as the primary regulators. For example, head horns in Onthophagus beetles and thoracic horns in the rhinoceros beetle T. dichotomus are thought to have evolved independently. In both systems the IIS pathway plays a key role, yet in Onthophagus beetles Foxo is the main IIS regulator, whereas in T. dichotomus InR is the key IIS horn growth regulator (Emlen et al., 2012; Casasa and Moczek, 2018). This further suggests that pre-existing connections related to the pathway’s canonical role could facilitate redeployment of the same pathways in similar contexts. Future studies delving into network architecture and rewiring of gene networks during evolution of plastic traits can shed light on the process by which gene networks are co-opted during evolution of these traits. Collectively, gene duplication, in addition to gene and gene regulatory network co-option have emerged as important processes in the evolution of nutrition-responsive plasticity, and future studies will continue to provide insights into the precise mechanisms orchestrating plastic trait evolution.
Regulation of nutritionally cued plasticity and the evolution complex phenotypes
Several nutritionally responsive phenotypes are composed of multiple traits that are well integrated to function in a coordinated manner (Hallgrímsson et al., 2002; Klingenberg, 2005; Simpson et al., 2011). This is particularly conspicuous in polyphenisms, where alternative morphologies display different suites of traits (Moran, 1992; Nijhout, 1999). Namely, morphologically variable traits are typically accompanied by behavioral differentiation. For instance, castes in Atta can be distinguished morphologically based on size, mandible and leg length, among other traits. Behaviorally, each caste is characterized by a distinct combination of tasks (e.g. fungal gardening, brood care, leaf-harvesting, etc.) at specific frequencies (Muratore et al., 2023). In Onthophagus beetles, large males display large horns accompanied by a fighter behavior; whereas small males are morphologically hornless and display a sneaker behavior (Moczek and Emlen, 2000). Thus, morphological, physiological, and behavioral traits are predicted to be coordinated during development to form complex phenotypes. Yet, in many cases, how behavioral and morphological plasticity are coordinated at the molecular level is unclear and likely involves complex regulatory mechanisms.
Understanding how trait integration is regulated is critical since this poses distinct evolutionary consequences. One possibility is that traits are regulated through separate developmental modules (i.e. behavioral trait-specific module and morphological trait-specific module) and integrated through key components that link the different modules (Figure 1A; Klingenberg, 2005; Wagner et al., 2007; Rittschof and Robinson, 2016). Modularity has been proposed to reduce the effects of pleiotropy and enhance evolvability (Wagner and Altenberg, 1996). Therefore, this scenario would allow evolutionary flexibility since traits and modules could evolve independently without disruption of one another (Klingenberg, 2005). If instead, traits are inherently integrated such that they cannot be dissociated (i.e. same module acts on both behavior and morphology; Figure 1B), then evolutionary changes on this module would result in a change in both trait types (correlated traits; Wagner et al., 2007). The first scenario could result in rapid trait coevolution: as a behavioral trait evolves, a change in morphological traits that matches this behavior could follow if advantageous. This in turn could feedback and result in further behavioral changes, reciprocally coevolving in a runaway manner. Trait decoupling across different levels (e.g. physical, molecular) has indeed been proposed to facilitate diversification (Frédérich et al., 2014; Powell et al., 2020). In addition, the environmentally responsive nature of these traits could facilitate this process since plasticity can fuel evolution (Pfennig et al., 2010; Moczek et al., 2011; Jones and Robinson, 2018). This process could theoretically occur in the second scenario as well yet constraints imposed by pleiotropy could limit the extent to which this can occur (Carroll, 2005; Williams et al., 2023).
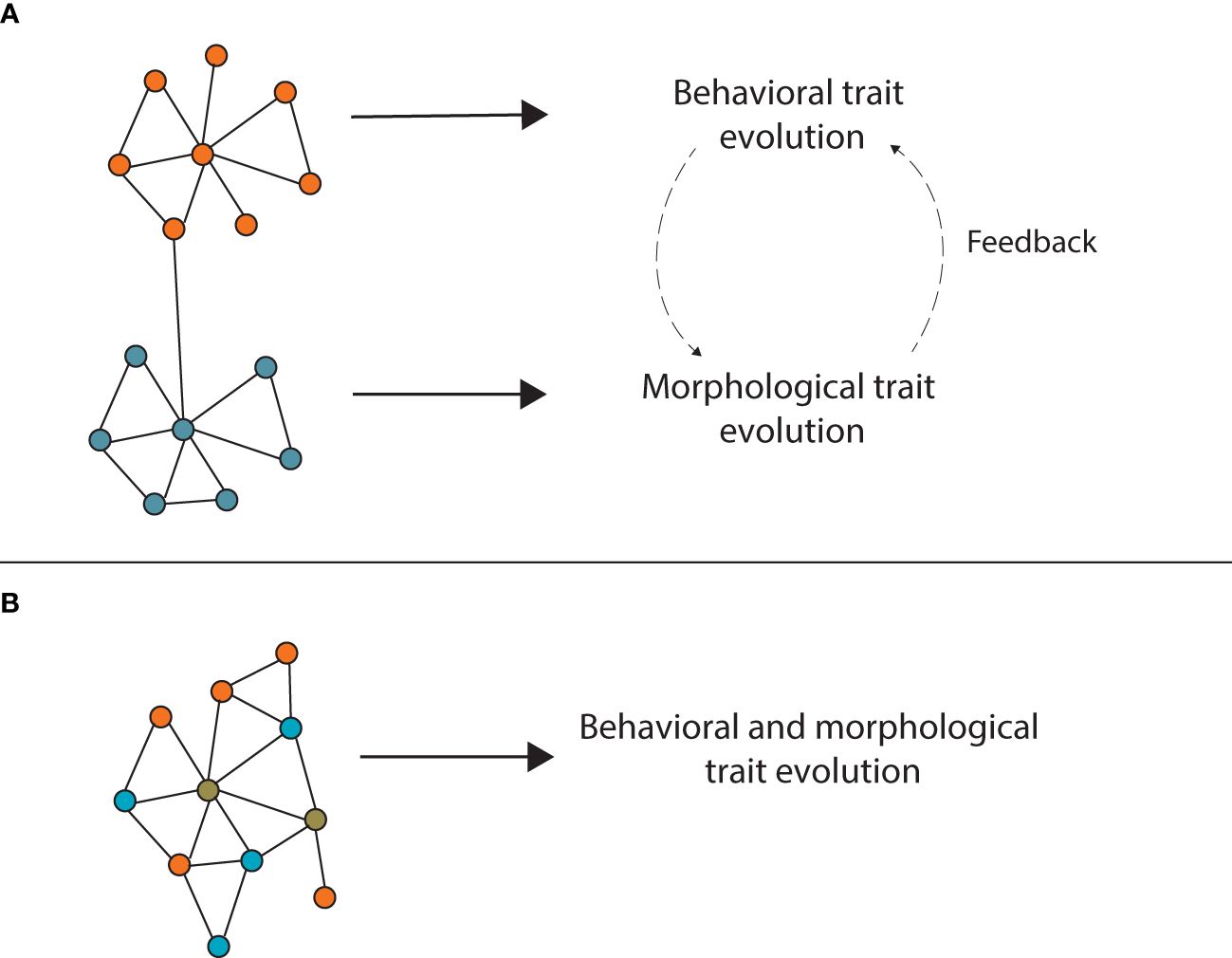
Figure 1 Regulation of trait integration under two scenarios. (A) Traits could be regulated through separate developmental modules (behavioral trait-specific, orange; or morphological-trait specific, blue) linked by key components. This scenario can result in evolutionary flexibility (see main text). (B) Alternatively, traits could be regulated by the same developmental module, where diverse components can be trait-specific (behavioral trait-specific, orange; morphological-trait specific, blue), yet are tightly interlinked among themselves and with components that contribute to both trait types (brown).
Together, providing a regulatory understanding of trait integration across nutritionally plastic phenotypes is key to further understand how complex traits evolve. As discussed above, gene duplication and gene network co-option are key processes that play a role in the evolution of plastic traits. While developmental modules can, at least in part, consist of duplicated genes or co-opted gene networks, co-option has been proposed to result in loss of tissue specificity and potentially limit evolution, particularly, the ability of traits to evolve in an independent manner (McQueen and Rebeiz, 2020). Given that several studies have implicated gene and gene network co-option in the evolution of plastic traits (Moczek and Nagy, 2005; Kijimoto et al., 2012; Gotoh et al., 2014; Morandin et al., 2016; Casasa and Moczek, 2018; Kapheim et al., 2020), identifying the gene regulatory network architecture of plastic traits, focusing on both morphological and behavioral traits, will be a critical first step. Moreover, evaluating the extent to which the modules underlying these traits overlap and the strength of module integration, can help elucidate how these complex phenotypes evolve.
Conclusions and future directions
Here I described the molecular mechanisms that underlie nutritionally cued plasticity in insects, and some of the major mechanisms by which these underpinnings have evolved. In the process, I have identified some gaps in our knowledge. Beyond these gaps, some key challenges remain. First, we need to fully understand how genomic variation contributes to the diversity of nutritionally responsive phenotypes and how this variation translates into a phenotypic response (from gene expression to behavior and morphology). Exploring the genetic variation of nutritionally cued responses from a population genomics perspective can further our understanding of how these responses evolve and allow us to identify key genomic regions involved. Second, nutritionally cued plastic responses are complex in insects and often involve thousands of genes, yet how these genes interact in a gene regulatory network remains largely unknown (but see Abouheif and Wray, 2002; Sinha et al., 2020). While this has been particularly challenging for non-model insect systems, the emergence of new technologies such as single-cell RNA sequencing and ATAC-seq (Assay for Transposase-Accessible Chromatin) now offer a more sensitive analysis of the molecular mechanisms underlying nutritionally cued plasticity and allow reconstruction of gene regulatory networks (Li et al., 2022; Traniello et al., 2023; Davidson and Moczek, 2024). Leveraging current knowledge on model insect systems and taking a comparative approach paring model with emerging model systems has proven to be particularly powerful (Li-Byarlay et al., 2014; Sheng et al., 2020). Hence, comparative studies utilizing these new technologies will further provide valuable insights on the extent of network co-option and gene regulatory network rewiring during the evolution of plastic traits. Third, our understanding of the mechanisms and evolution of nutritionally cued plasticity is incomplete without considering how organismal traits are integrated at the molecular level. Detailed functional genetic analyses in insects with complex, environmentally sensitive traits, and assessing different trait types (e.g. behavioral or morphological traits), would facilitate answering these questions and enable us to better understand how complex phenotypes evolve.
Finally, it is important to emphasize that we live in a rapidly changing world, where organisms continuously experience new nutritional environments. For instance, a shift in geographic distribution of insects as a result of climate change can impact their encounters with new nutritional environments (Janes et al., 2014). Thus, understanding how the regulation of plasticity has evolved will provide the basis to further investigate the impact of climate change on plastic responses. Doing so will enable a more accurate prediction of key aspects in the regulation of plasticity, particularly those that could pose challenges for certain species in the future. Together, insect studies have provided valuable insights into the mechanistic understanding of phenotypic plasticity. With the development of novel technologies and the increase in development of tractable insect systems, the time is ripe to expand integrative studies on the regulatory mechanisms of plasticity and their evolution.
Author contributions
SC: Conceptualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This manuscript was supported by start-up funds from Boston University.
Acknowledgments
I would like to thank the journal editors for the opportunity to contribute this work. I would also like to thank Sarah Davies, James Traniello, and two anonymous reviewers for valuable comments and feedback on the paper.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abouheif E., Wray G. A. (2002). Evolution of the gene network underlying wing polyphenism in ants. Sci. (80-.). 297, 249–252. doi: 10.1126/science.1071468
Abrisqueta M., Süren-Castillo S., Maestro J. L. (2014). Insulin receptor-mediated nutritional signalling regulates juvenile hormone biosynthesis and vitellogenin production in the German cockroach. Insect Biochem. Mol. Biol. 49, 14–23. doi: 10.1016/j.ibmb.2014.03.005
Agrawal N., Léopold P. (2015). Developmental biology: Hedgehog turns into a metabolic hormone. Curr. Biol. 25(3), R117–R119. doi: 10.1016/j.cub.2014.12.021
Barbieri M., Bonafè M., Franceschi C., Paolisso G. (2003). Insulin/IGF-I-signaling pathway: An evolutionarily conserved mechanism of longevity from yeast to humans. Am. J. Physiol. - Endocrinol. Metab. 285, 1064–1071. doi: 10.1152/ajpendo.00296.2003
Brian M. V. (1979). Caste differentiation and division of labor. Soc insects 1, 121–222. doi: 10.1016/B978-0-12-342201-9.50012-4
Brogiolo W., Stocker H., Ikeya T., Rintelen F., Fernandez R., Hafen E. (2001). An evolutionarily conserved function of the drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 11, 213–221. doi: 10.1016/S0960-9822(01)00068-9
Callier V., Nijhout H. F. (2013). Body size determination in insects: A review and synthesis of size- and brain-dependent and independent mechanisms. Biol. Rev. 88, 944–954. doi: 10.1111/brv.12033
Carroll S. B. (2005). Evolution at two levels: On genes and form. PloS Biol. 3, 1159–1166. doi: 10.1371/journal.pbio.0030245
Casasa S., Moczek A. P. (2018). Insulin signalling’s role in mediating tissue-specific nutritional plasticity and robustness in the horn-polyphenic beetle Onthophagus taurus. Proc. R. Soc B Biol. Sci. 285, 20181631. doi: 10.1098/rspb.2018.1631
Casasa S., Moczek A. P. (2019). Evolution of, and via, Developmental Plasticity: Insights through the Study of Scaling Relationships. Integr. Comp. Biol. 59, 1346–1355. doi: 10.1093/icb/icz086
Casasa S., Schwab D., Moczek A. P. (2017). Developmental regulation and evolution of scaling: novel insights through the study of Onthophagus beetles. Curr. Opin. Insect Sci. 19, 52–60. doi: 10.1016/j.cois.2016.11.004
Casasa S., Zattara E. E., Moczek A. P. (2020). Nutrition-responsive gene expression and the developmental evolution of insect polyphenism. Nat. Ecol. Evol. 4, 970–978. doi: 10.1038/s41559-020-1202-x
Chandra V., Fetter-Pruneda I., Oxley P. R., Ritger A. L., McKenzie S. K., Libbrecht R., et al. (2018). Social regulation of insulin signaling and the evolution of eusociality in ants. Sci. (80-.). 361, 398–402. doi: 10.1126/science.aar5723
Chau L. M., Goodisman M. A. D. (2017). Gene duplication and the evolution of phenotypic diversity in insect societies. Evol. (N. Y). 71, 2871–2884. doi: 10.1111/evo.2017.71.issue-12
Colombani J., Bianchini L., Layalle S., Pondeville E., Dauphin-Villemant C., Antoniewski C., et al. (2005). Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Sci. (80-.). 310, 667–670. doi: 10.1126/science.1119432
Corona M., Libbrecht R., Wurm Y., Riba-Grognuz O., Studer R. A., Keller L. (2013). Vitellogenin underwent subfunctionalization to acquire caste and behavioral specific expression in the harvester ant pogonomyrmex barbatus. PloS Genet. 9, e1003730. doi: 10.1371/journal.pgen.1003730
Davidson P. L., Moczek A. P. (2024). Genome evolution and divergence in cis-regulatory architecture is associated with condition-responsive development in horned dung beetles. PloS Genet. 20, 1–21. doi: 10.1371/journal.pgen.1011165
de Azevedo S. V., Hartfelder K. (2008). The insulin signaling pathway in honey bee (Apis mellifera) caste development - differential expression of insulin-like peptides and insulin receptors in queen and worker larvae. J. Insect Physiol. 54, 1064–1071. doi: 10.1016/j.jinsphys.2008.04.009
Deem K. D., Gregory L. E., Liu X., Ziabari O. S., Brisson J. A. (2024). Evolution and molecular mechanisms of wing plasticity in aphids. Curr. Opin. Insect Sci. 61, 101142. doi: 10.1016/j.cois.2023.101142
Eberhard W. G. (2009). Static allometry and animal genitalia. Evol. (N. Y). 63, 48–66. doi: 10.1111/evo.2009.63.issue-1
Edgar B. A. (2006). How flies get their size: genetics meets physiology. Nat. Rev. Genet. 7, 907–916. doi: 10.1038/nrg1989
Emlen D. J., Warren I. A., Johns A., Dworkin I., Lavine L. C. (2012). A mechanism of extreme growth and reliable signaling in sexually selected ornaments and weapons. Sci. (80-.). 337, 860–864. doi: 10.1126/science.1224286
Fawcett M. M., Parks M. C., Tibbetts A. E., Swart J. S., Richards E. M., Vanegas J. C., et al. (2018). Manipulation of insulin signaling phenocopies evolution of a host-associated polyphenism. Nat. Commun. 9, 1–11. doi: 10.1038/s41467-018-04102-1
Fjerdingstad E. J., Crozier R. H. (2006). The evolution of worker caste diversity in social insects. Am. Nat. 167, 390–400. doi: 10.1086/499545
Frédérich B., Olivier D., Litsios G., Alfaro M. E., Parmentier E. (2014). Trait decoupling promotes evolutionary diversification of the trophic and acoustic system of damselfishes. Proc. R. Soc B Biol. Sci. 281, 20141047. doi: 10.1098/rspb.2014.1047
Gokhale R. H., Hayashi T., Mirque C. D., Shingleton A. W. (2016). Intra-organ growth coordination in Drosophila is mediated by systemic ecdysone signaling. Dev. Biol. 418, 135–145. doi: 10.1016/j.ydbio.2016.07.016
Gotoh H., Cornette R., Koshikawa S., Okada Y., Lavine L. C., Emlen D. J., et al. (2011). Juvenile hormone regulates extreme mandible growth in male stag beetles. PloS One 6, 1–5. doi: 10.1371/journal.pone.0021139
Gotoh H., Miyakawa H., Ishikawa A., Ishikawa Y., Sugime Y., Emlen D. J., et al. (2014). Developmental Link between Sex and Nutrition; doublesex Regulates Sex-Specific Mandible Growth via Juvenile Hormone Signaling in Stag Beetles. PloS Genet. 10, 1–9. doi: 10.1371/journal.pgen.1004098
Hallgrímsson B., Willmore K., Hall B. K. (2002). Canalization, developmental stability, and morphological integration in primate limbs. Yearb. Phys. Anthropol. 45, 131–158. doi: 10.1002/(ISSN)1096-8644
Hopkins B. R., Kopp A. (2021). Evolution of sexual development and sexual dimorphism in insects. Curr. Opin. Genet. Dev. 69, 129–139. doi: 10.1016/j.gde.2021.02.011
Ingham P. W., McMahon A. P. (2001). Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 15, 3059–3087. doi: 10.1101/gad.938601
Janes J. K., Li Y., Keeling C. I., Yuen M. M. S., Boone C. K., Cooke J. E. K., et al. (2014). How the mountain pine beetle (Dendroctonus ponderosae) breached the canadian rocky mountains. Mol. Biol. Evol. 31, 1803–1815. doi: 10.1093/molbev/msu135
Jones B. M., Robinson G. E. (2018). Genetic accommodation and the role of ancestral plasticity in the evolution of insect eusociality. J. Exp. Biol. 221, 1–11. doi: 10.1242/jeb.153163
Kapheim K. M., Jones B. M., Pan H., Li C., Harpur B. A., Kent C. F. (2020). Developmental plasticity shapes social traits and selection in a facultatively eusocial bee. PNAS 117, 13615–13625. doi: 10.1073/pnas.2000344117
Karino K., Seki N., Chiba M. (2004). Larval nutritional environment determines adult size in Japanese horned beetles Allomyrina dichotoma. Ecol. Res. 19, 663–668. doi: 10.1111/j.1440-1703.2004.00681.x
Kijimoto T., Moczek A. P. (2016). Hedgehog signaling enables nutrition-responsive inhibition of an alternative morph in a polyphenic beetle. Proc. Natl. Acad. Sci. U. S. A. 113, 5982–5987. doi: 10.1073/pnas.1601505113
Kijimoto T., Moczek A. P., Andrews J. (2012). Diversification of doublesex function underlies morph-, sex-, and species-specific development of beetle horns. Proc. Natl. Acad. Sci. U. S. A. 109, 20526–20531. doi: 10.1073/pnas.1118589109
Klingenberg C. P. (2005). “Developmental constraints, modules, and evolvability,” in Variation: A central concept in biology. Eds. Hallgrimsson B., Hall B. (Elsevier, Burlington, MA), 219–247.
Koyama T., Mirth C. K. (2018). Unravelling the diversity of mechanisms through which nutrition regulates body size in insects. Curr. Opin. Insect Sci. 25, 1–8. doi: 10.1016/j.cois.2017.11.002
Kremer L. P. M., Korb J., Bornberg-Bauer E. (2018). Reconstructed evolution of insulin receptors in insects reveals duplications in early insects and cockroaches. J. Exp. Zool. Part B Mol. Dev. Evol. 330, 305–311. doi: 10.1002/jez.b.22809
Li Q., Wang M., Zhang P., Liu Y., Guo Q., Zhu Y., et al. (2022). A single-cell transcriptomic atlas tracking the neural basis of division of labour in an ant superorganism. Nat. Ecol. Evol. 6, 1191–1204. doi: 10.1038/s41559-022-01784-1
Li-Byarlay H., Rittschof C. C., Massey J. H., Pittendrigh B. R., Robinson G. E. (2014). Socially responsive effects of brain oxidative metabolism on aggression. Proc. Natl. Acad. Sci. 111, 12533–12537. doi: 10.1073/pnas.1412306111
Lin X., Xu Y., Jiang J., Lavine M., Lavine L. C. (2018). Host quality induces phenotypic plasticity in a wing polyphenic insect. Proc. Natl. Acad. Sci. U. S. A. 115, 7563–7568. doi: 10.1073/pnas.1721473115
Lu H. L., Pietrantonio P. V. (2011). Insect insulin receptors: Insights from sequence and caste expression analyses of two cloned hymenopteran insulin receptor cDNAs from the fire ant. Insect Mol. Biol. 20, 637–649. doi: 10.1111/j.1365-2583.2011.01094.x
McQueen E., Rebeiz M. (2020). On the individuality of gene regulatory networks: how does network re-use affect subsequent evolution? Curr. Top. Dev. Biol. 139, 375–405. doi: 10.1016/bs.ctdb.2020.03.002
Mirth C. K., Tang H. Y., Makohon-Moore S. C., Salhadar S., Gokhale R. H., Warner R. D., et al. (2014). Juvenile hormone regulates body size and perturbs insulin signaling in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 111, 7018–7023. doi: 10.1073/pnas.1313058111
Moczek A. P. (1998). Horn polyphenism in the beetle Onthophagus taurus: Larval diet quality and plasticity in parental investment determine adult body size and male horn morphology. Behav. Ecol. 9, 636–641. doi: 10.1093/beheco/9.6.636
Moczek A. P. (2003). The behavioral ecology of threshold evolution in a polyphenic beetle. Behav. Ecol. 14, 841–854. doi: 10.1093/beheco/arg062
Moczek A. P. (2010). Phenotypic plasticity and diversity in insects. Philos. Trans. R. Soc B Biol. Sci. 365, 593–603. doi: 10.1098/rstb.2009.0263
Moczek A. P., Emlen D. J. (2000). Male horn dimorphism in the scarab beetle, Onthophagus taurus: Do alternative reproductive tactics favour alternative phenotypes? Anim. Behav. 59, 459–466. doi: 10.1006/anbe.1999.1342
Moczek A. P., Nagy L. M. (2005). Diverse developmental mechanisms contribute to different levels of diversity in horned beetles. Evolution & Development 185, 175–185. doi: 10.1111/j.1525-142X.2005.05020.x
Moczek A. P., Sultan S., Foster S., Ledón-Rettig C., Dworkin I., Nijhout H. F., et al. (2011). The role of developmental plasticity in evolutionary innovation. Proc. R. Soc B Biol. Sci. 278, 2705–2713. doi: 10.1098/rspb.2011.0971
Moore V. M., Davies M. J., Willson K. J., Worsley A., Robinson J. S. (2004). Dietary composition of pregnant women is related to size of the baby at birth. J. Nutr. 134, 1820–1826. doi: 10.1093/jn/134.7.1820
Moran N. A. (1992). The evolutionary maintenance of alternative phenotypes. Am. Nat. 139, 971–989. doi: 10.1086/285369
Morandin C., Tin M. M. Y., Abril S., Gómez C., Pontieri L., Schiøtt M., et al. (2016). Comparative transcriptomics reveals the conserved building blocks involved in parallel evolution of diverse phenotypic traits in ants. Genome Biol. 17, 1–19. doi: 10.1186/s13059–016-0902–7
Muratore I. B., Ilieş I., Huzar A. K., Zaidi F. H., Traniello J. F. A. (2023). Morphological evolution and the behavioral organization of agricultural division of labor in the leafcutter ant Atta cephalotes. Behav. Ecol. Sociobiol. 77, 1–16. doi: 10.1007/s00265-023-03344-4
Nijhout H. F. (1999). Control mechanisms of polyphenic development in insects. Bioscience 49, 181–192. doi: 10.2307/1313508
Nijhout H. F., Grunert L. W. (2002). Bombyxin is a growth factor for wing imaginal disks in lepidoptera. Proc. Natl. Acad. Sci. U. S. A. 99, 15446–15450. doi: 10.1073/pnas.242548399
Nijhout H. F., McKenna K. Z. (2018). The distinct roles of insulin signaling in polyphenic development. Curr. Opin. Insect Sci. 25, 58–64. doi: 10.1016/j.cois.2017.11.011
Pfennig D. W., Wund M. A., Snell-Rood E. C., Cruickshank T., Schlichting C. D., Moczek A. P. (2010). Phenotypic plasticity’s impacts on diversification and speciation. Trends Ecol. Evol. 25, 459–467. doi: 10.1016/j.tree.2010.05.006
Powell S., Price S. L., Kronauer D. J. C. (2020). Trait evolution is reversible, repeatable, and decoupled in the soldier caste of turtle ants. Proc. Natl. Acad. Sci. U. S. A. 117, 6608–6615. doi: 10.1073/pnas.1913750117
Price D. C., Egizi A., Fonseca D. M. (2015). The ubiquity and ancestry of insect doublesex. Sci. Rep. 5, 1–9. doi: 10.1038/srep13068
Rittschof C. C., Robinson G. E. (2016). Behavioral genetic toolkits. Curr Topics Dev Biol. 119(1):157–204. doi: 10.1016/bs.ctdb.2016.04.001
Sang M., Li C., Wu W., Li B. (2016). Identification and evolution of two insulin receptor genes involved in Tribolium castaneum development and reproduction. Gene 585, 196–204. doi: 10.1016/j.gene.2016.02.034
Schlichting C. D. (1986). The evolution of phenotypic plasticity in plants. Annu. Rev. Ecol. Syst. 17, 667–693. doi: 10.1146/annurev.es.17.110186.003315
Serobyan V., Ragsdale E. J., Müller M. R., Sommer R. J. (2013). Feeding plasticity in the nematode pristionchus pacificus is influenced by sex and social context and is linked to developmental speed. Evol. Dev. 15, 161–170. doi: 10.1111/ede.12030
Sheng L., Shields E. J., Gospocic J., Glastad K. M., Ratchasanmuang P., Berger S. L., et al. (2020). Social reprogramming in ants induces longevity-associated glia remodeling. Sci. Adv. 6, eaba9869. doi: 10.1126/sciadv.aba9869
Shingleton A. W., Das J., Vinicius L., Stern D. L. (2005). The temporal requirements for insulin signaling during development in Drosophila. PloS Biol. 3, 1607–1617. doi: 10.1371/journal.pbio.0030289
Shingleton A. W., Estep C. M., Driscoll M. V., Dworkin I. (2009). Many ways to be small: Different environmental regulators of size generate distinct scaling relationships in Drosophila melanogaster. Proc. R. Soc B Biol. Sci. 276, 2625–2633. doi: 10.1098/rspb.2008.1796
Shingleton A. W., Frankino W. A., Flatt T., Nijhout H. F., Emlen D. J. (2007). Size and shape: The developmental regulation of static allometry in insects. BioEssays 29, 536–548. doi: 10.1002/bies.20584
Shingleton A. W., Mirth C. K., Bates P. W. (2008). Developmental model of static allometry in holometabolous insects. Proc. R. Soc B Biol. Sci. 275, 1875–1885. doi: 10.1098/rspb.2008.0227
Simpson S. J., Sword G. A., Lo N. (2011). Polyphenism in insects. Curr. Biol. 21, R738–R749. doi: 10.1016/j.cub.2011.06.006
Sinha S., Jones B. M., Traniello I. M., Bukhari S. A., Halfon M. S. (2020). Behavior-related gene regulatory networks : A new level of organization in the brain. PNAS 117, 23270–23279. doi: 10.1073/pnas.1921625117
Smykal V., Pivarci M., Provaznik J., Bazalova O., Jedlicka P., Luksan O., et al. (2020). Complex evolution of insect insulin receptors and homologous decoy receptors, and functional significance of their multiplicity. Mol. Biol. Evol. 37, 1775–1789. doi: 10.1093/molbev/msaa048
Stern D. (2003). Body-size control: how an insect knows it has grown enough. Curr. Biol. 13, 267–269. doi: 10.1016/S0960-9822(03)00197-0
Tchokponhoué D. A., N’Danikou S., Houéto J. S., Achigan-Dako E. G. (2019). Shade and nutrient-mediated phenotypic plasticity in the miracle plant Synsepalum dulcificum (Schumach. & Thonn.) Daniell. Sci. Rep. 9, 1–11. doi: 10.1038/s41598-019-41673-5
Teleman A. A., Hietakangas V., Sayadian A. C., Cohen S. M. (2008). Nutritional control of protein biosynthetic capacity by insulin via myc in drosophila. Cell Metab. 7, 21–32. doi: 10.1016/j.cmet.2007.11.010
Traniello I. M., Bukhari S. A., Dibaeinia P., Serrano G., Avalos A., Ahmed A. C., et al. (2023). Single-cell dissection of aggression in honeybee colonies. Nat. Ecol. Evol. 7, 1232–1244. doi: 10.1038/s41559-023-02090-0
True J. R., Carroll S. B. (2002). Gene co-option in physiological and morphological evolution. Annu. Rev. Cell Dev. Biol. 18, 53–80. doi: 10.1146/annurev.cellbio.18.020402.140619
Vitali V., Horn F., Catania F. (2018). Insulin-like signaling within and beyond metazoans. Biological Chemistry 399, 851–857. doi: 10.1515/hsz-2018–0135
Wagner G., Altenberg L. (1996). Complex adaptations and the evolution of evolvability. Evol. (N. Y). 50, 967–976.
Wagner G. P., Pavlicev M., Cheverud J. M. (2007). The road to modularity. Nat. Rev. Genet. 8, 921–931. doi: 10.1038/nrg2267
Weger A. A., Rittschof C. C. (2024). The diverse roles of insulin signaling in insect behavior. Front. Insect Sci. 4, 1–9. doi: 10.3389/finsc.2024.1360320
West-Eberhard M. J. (2003). Developmental plasticity and evolution (New York, NY: Oxford University Press). doi: 10.1093/oso/9780195122343.003.0008
Williams A. M., Ngo T. M., Figueroa V. E., Tate A. T. (2023). The effect of developmental pleiotropy on the evolution of insect immune genes. Genome Biol. Evol. 15, 1–16. doi: 10.1093/gbe/evad044
Xu H. J., Xue J., Lu B., Zhang X. C., Zhuo J. C., He S. F., et al. (2015). Two insulin receptors determine alternative wing morphs in planthoppers. Nature 519, 464–467. doi: 10.1038/nature14286
Keywords: developmental plasticity, nutrition, insects, evolution, regulatory mechanisms, polyphenism
Citation: Casasa S (2024) Evolution of regulatory mechanisms underlying nutrition-responsive plasticity in insects. Front. Ecol. Evol. 12:1409743. doi: 10.3389/fevo.2024.1409743
Received: 30 March 2024; Accepted: 03 June 2024;
Published: 21 June 2024.
Edited by:
Smadar Ben-Tabou De-Leon, University of Haifa, IsraelReviewed by:
Christen Kerry Mirth, Monash University, AustraliaClare C. Rittschof, University of Kentucky, United States
Copyright © 2024 Casasa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sofia Casasa, YXNjYXNhc2FAYnUuZWR1
 Sofia Casasa
Sofia Casasa