- 1Laboratorio de Ecología Molecular de Vertebrados Acuáticos (LEMVA), Departamento de Ciencias Biológicas, Universidad de Los Andes, Bogotá, Colombia
- 2Centro Colecciones y Gestión de Especies, Dirección de Conocimiento, Instituto de Investigación de Recursos Biológicos Alexander von Humboldt, Bogotá, Colombia
The development of fast, cost-effective, non-invasive, and efficient sampling alternatives, such as environmental DNA (eDNA), is crucial for understanding the changes in species biodiversity and distributions worldwide, particularly for low abundance, cryptic, and threatened species. This study utilized environmental eDNA to analyze the variety of aquatic, semi-aquatic, and terrestrial vertebrates in the Colombian Amazon and Orinoco basins. The study focused on four main subregions: Bojonawi Natural Reserve and adjacent areas (Vichada Department), Sierra de la Macarena National Park and Tillavá (Meta Department), Puerto Nariño and adjacent areas (Amazonas Department), and the Municipality of Solano (Caquetá Department). A total of 709 operational taxonomic units (OTUs) were identified across all sampling locations. The Orinoco River had the highest number of fish genera (68), while the Guayabero River had the highest number of tetrapod genera (13). New taxonomic records were found for all locations, with the highest number of previously undetected fish diversity being found in the Bita, Orinoco, and Tillavá rivers, compared to traditional surveys. Likewise, the study identified two fish species, four mammal species, and one reptile species as vulnerable. Additionally, four mammal species were identified as endangered, including the giant otter (Pteronura brasiliensis), two subspecies of the Amazon River dolphin (Inia geoffrensis geoffrensis and Inia geoffrensis humboldtiana), and the tucuxi (Sotalia fluviatilis). Standardizing the methodology and improving current DNA sequence databases for the Neotropics is essential to develop future eDNA studies and enhance our understanding of the region’s diversity.
Introduction
Biodiversity loss on a global scale is a significant environmental issue of this century (Cardinale et al., 2012; Valentini et al., 2016; Johnson et al., 2017; Crossley et al., 2020; DiBattista et al., 2020). Scientific evidence suggests that human activities, such as resource appropriation, habitat fragmentation, non-native species introduction, spread of pathogens, and direct killing of species, are predicting a sixth mass extinction (Barnosky et al., 2011; Ceballos et al., 2015, 2020; Cowie et al., 2022). Traditionally, conservation of fauna and flora has been based on observed local diversity. However, this approach has resulted in significant gaps in worldwide biological information (Pärtel et al., 2011; Delić et al., 2017). There is a relatively new ecological concept called ‘dark diversity’ (Pärtel et al., 2011; Lewis et al., 2017; Fernandes et al., 2019), which refers to the pool of species that are presumed to be present within a certain region. However, traditional sampling methods may miss these species, resulting in them being reported as absent (Moeslund et al., 2017), as diversity encompasses cryptic, rare, and endangered species that are challenging to detect but are crucial for effective ecological management and conservation programs (Rees et al., 2014; Boussarie et al., 2018). Therefore, scientists urgently require the development of rapid, cost-effective, sensitive, and non-invasive molecular tools for species monitoring and conservation (Kelly et al., 2014; Deiner et al., 2017).
Environmental DNA (eDNA) combined with high-throughput sequencing (metabarcoding) is a promising technique for detecting aquatic, semi-aquatic, and terrestrial vertebrates. It has become a powerful complement to traditional surveys, as demonstrated by several studies (Hänfling et al., 2016; Valentini et al., 2016; Sard et al., 2019; Coutant et al., 2021; Dal Pont et al., 2021; Mena et al., 2021; Pawlowski et al., 2021; Jia et al., 2022; Mas-Carrió et al., 2022). Aquatic ecosystems may be affected by various factors, including salinity, temperature, pH, solar radiation, and microorganisms. These factors can contribute to the degradation of eDNA (Thomsen et al., 2012; Barnes et al., 2014; Jo et al., 2017; Saito and Doi, 2021). Therefore, this methodology is believed to produce consistent results with the species present in the current location and time of the environmental sample (Herder et al., 2014; Rees et al., 2014; Thomsen and Willerslev, 2015; Deiner et al., 2017; Seymour, 2019).
Colombia is located in the American tropics, which is one of the most species-rich regions on Earth (Antonelli et al., 2018). It is the third most diverse country, hosting close to 10% of the planet’s biodiversity (Ministry of Environment and Sustainable Development, 2017, 2019; Sistema de Información Ambiental de Colombia, 2020; Convention on Biological Diversity, 2021). Colombia is geographically divided into five regions, two of which are the Amazon and Orinoco. According to the Geographic Institute Agustín Codazzi (2002), the Colombian Amazon region accounts for 6.4% of the total area of the Amazon basin, while the Orinoco River basin represents 35% of the Colombian territory (Correa et al., 2005; Lasso et al., 2010; Universidad Nacional de Colombia, 2013; Lasso et al., 2014; Aldana et al., 2017; Guio and Rojas, 2019; WWF, 2020). These two regions contain a variety of freshwater ecosystems and biomes, which in turn have extraordinary species diversity (Wrona and Reist, 2013). However, these ecosystems are being threatened by a multitude of anthropogenic stressors (Suring, 2020) that critically reduce freshwater biodiversity (Tickner et al., 2020; IUCN, International Union for Conservation of Nature, 2022). Not only is the loss of valuable biological information at stake, but freshwater ecosystems also provide essential goods and services to humans that are now at serious risk (Portocarrero-Aya, 2011; Faghihinia et al., 2021).
Until now, eDNA has been used in relatively few studies in Colombia. These studies include investigations on marine animals such as fishes, whales, and corals (Juhel et al., 2021; Marques et al., 2021; Stauffer et al., 2021; Polanco et al., 2021b; Mathon et al., 2022), as well as on freshwater, underground, and cave ecosystems in search of vertebrates (Martinelli-Marín et al., 2020; Lozano Mojica and Caballero, 2021; Caballero et al., 2021a, b; Polanco et al., 2021c). However, neither has implemented eDNA in the Amazon and Orinoco basins as a whole. Also, most biodiversity inventorying and monitoring studies in these two regions have relied on traditional, expensive, and time-consuming methodologies (Prieto and Arias, 2007; Lasso et al., 2010; Lasso and Morales-Betancourt, 2017; Lasso et al., 2020). On the other hand, a cost-benefit analysis of eDNA implementation has shown that, at least for the Orinoco Basin, similar or even better results have been obtained with less time, space and economic investment compared to traditional surveys (Martinelli-Marín et al., 2020). This new tool could therefore improve the detection and monitoring of freshwater species by the Colombian environmental authorities in a rapid and cost-effective manner.
To the best of our knowledge, this is the first study to use eDNA and next-generation sequencing to describe the diversity and richness of aquatic, semi-aquatic, and terrestrial vertebrates in the water bodies (rivers, streams, lakes, and lagoons) of four different geographic regions of the Colombian Amazon and Orinoco basins.
Materials and methods
Sample location
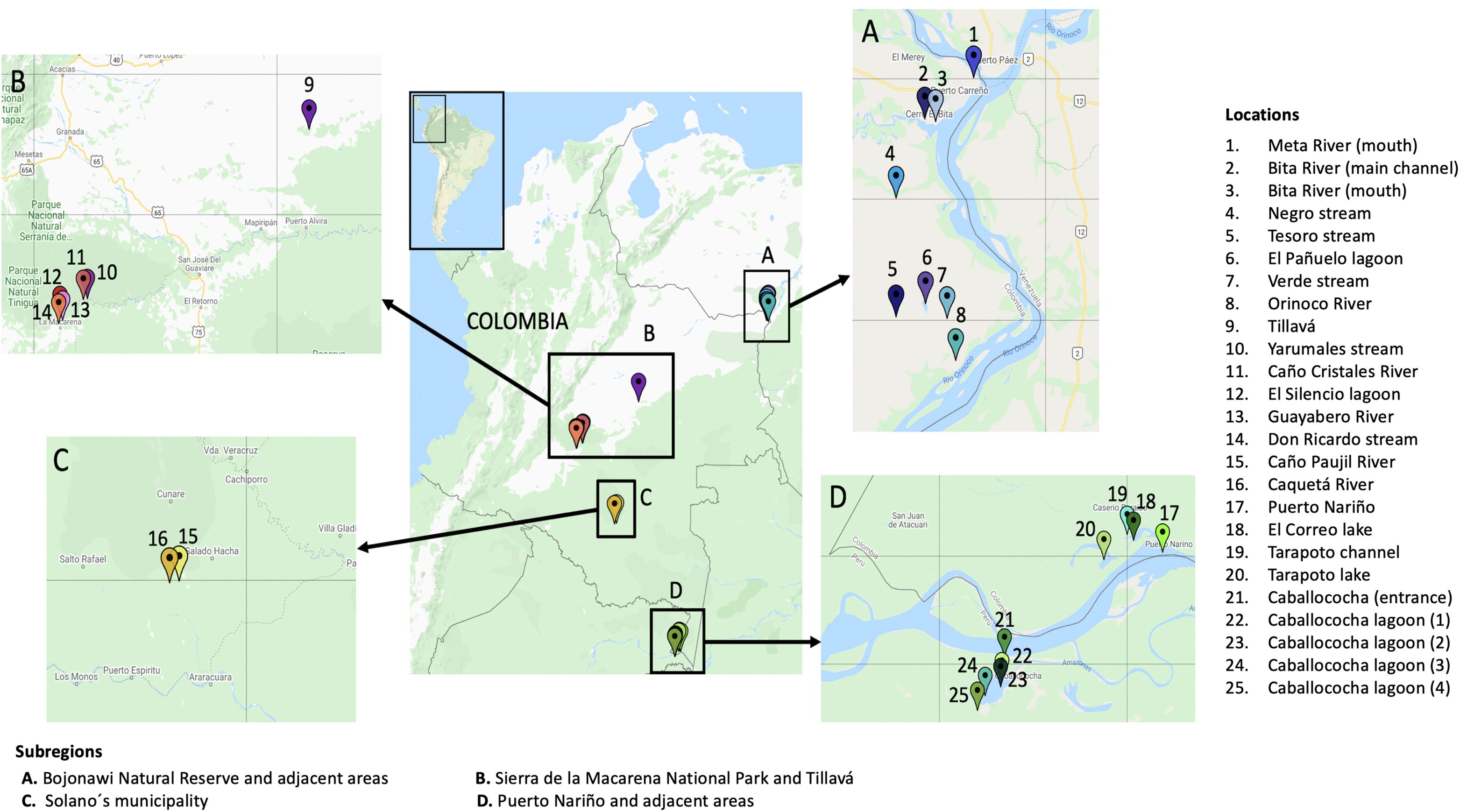
Between October 2019 and February 2020, the Aquatic Vertebrate Molecular Ecology Laboratory (LEMVA) at los Andes University, in collaboration with the Biological Resources Research Institute Alexander von Humboldt, conducted four expeditions to the Colombian Amazon and Orinoco basins. The study examined three Amazon subbasins (Caquetá, Javari, and Loretoyacu rivers) and five Orinoco subbasins (Bita, Guaviare, Meta, the Orinoco main channel, and Vichada), which were divided into four terrestrial subregions: Bojonawi Nature Reserve and adjacent areas, Puerto Nariño and adjacent areas, Sierra de la Macarena National Park, and Tillavá and Solano’s Municipality (Figure 1; Supplementary Material). All field trips were undertaken during low water seasons. During the dry season in the Amazon basin, the Orinoco basin experiences its rainy season and vice versa. As a result, the low water season in the Amazon takes place from May to December, while in the Orinoco it occurs from January to April (Guhl, 2016).
eDNA sampling
25 main locations were sampled using the methodologies employed by the NatureMetrics (2019) laboratory, as well as the studies conducted by Lozano Mojica and Caballero (2021) and Caballero et al. (2021a). At each location, up to seven water subsamples (1L each) were collected in sterile plastic bottles and then transferred to a bucket covered with a plastic bag, using sterile gloves. Prior to use, the plastic bag was decontaminated with 90% ethanol and 10% diluted bleach, and then rinsed with tap water. Water samples were collected at intervals of 10–20 metres along a linear transect using boats for rivers, canoes for lakes and lagoons, and on foot for streams. The coordinates of each sampled point were recorded using a Garmin etrex 12 channel GPS. Once all the subsamples from a location were taken, water samples were filtered using NatureMetrics eDNA collection kits. A 60 ml syringe filled with the collected water was attached to a filter disk with a 0.8 μm pore size. When the filter disk became clogged and water could no longer pass through, we detached the syringe and used a smaller one with a preserving buffer (Longmire’s solution) to protect the filter and prevent DNA degradation (Baker et al., 2018). We stored each filter in an envelope with its corresponding field information and kept them cool in a styrofoam cooler with ice packs.
eDNA extraction, amplification and sequencing
The filters were transported to Nature Metrics in Egham, Surrey, England, where DNA was extracted using the Qiagen DNeasy Blood & Tissue Kit, following the manufacturer’s instructions. The method for disc filters in buffer, with the following modifications, was based on Spens et al. (2016): proteinase K was initially added directly to the filter housing; following incubation, 1 mL of lysate was carried forward for extraction, and all DNeasy Blood and Tissue Kit (Qiagen) reagents were adjusted accordingly. The final elution was in 200 uL. The modifications aim to minimise the risk of contamination and maximise DNA yield. The DNA was purified using the DNeasy PowerClean Pro Cleanup kit to eliminate PCR inhibitors. Subsequently, DNA extracted from each filter was amplified using 12 replicates of the 12S rRNA mitochondrial gene (5´- TAGAACAGGCTCCTCTAG-3´ and 5´-TTAGATACCCCACTATGC-3´) to target fishes as part of the eDNA survey - Vertebrates pipeline (Riaz et al., 2011; Milan et al., 2020). PCR replicates were combined, and adapters were added at the 5′ end of the primers to complement Illumina Nextera index primers. The amplification mixture for each replicate consisted of 1X DreamTaq PCR Master Mix (Thermo Scientific), 0.4 μM of each of the tailed primers, 1 μL of DNA, and PCR grade water (Thermo Scientific) up to a total reaction volume of 10 μL. All PCRs were performed in the presence of both a negative control and a positive control sample (mock community with a known composition, not expected to occur in Colombia). The PCR conditions comprised an initial denaturation at 95°C for 2 minutes, followed by 10 cycles of 20 seconds at 95°C, a 30-second touchdown annealing step (-0.5°C per cycle) starting at 60°C, and 40 seconds at 72°C, 35 cycles of 20 seconds at 95°C, 30 seconds at 55°C, and 40 seconds at 72°C, and a final elongation step at 72°C for 5 minutes. Amplification success was determined by gel electrophoresis. The amplicons were purified using MagBind TotalPure NGS (Omega Biotek) magnetic beads with a bead to DNA ratio of 0.8:1 to eliminate primer dimers. The purified index PCRs were then quantified using a Qubit high sensitivity kit following the manufacturer’s protocol. Subsequently, all purified index PCRs were combined into a final library with equal concentrations. Finally, the library was sequenced using an Illumina MiSeq V2 kit at 12 pM with a 10% PhiX spike sterile in. The sequence data underwent processing through a custom bioinformatics pipeline, USEARCH v11, which included quality filtering, dereplication, and taxonomic assignment. Taxonomic assignment was considered successful when there was at least 80% agreement in the overlap. The merged sequences were processed using cutadapt 2.3 (Martin, 2011; Mathon et al., 2021) to remove forward and reverse primers. Only sequences with a trimmed length between 80–120 bp were kept. Quality filtering was performed to retain sequences with an expected error rate per base of 0.01 or lower. Dereplication was then carried out by sample, retaining only singletons. All unique reads from the samples underwent denoising in a single analysis using UNOISE (Dal Pont et al., 2021). ZOTUs (zero-radius OTUs) with a minimum abundance of 8 were retained and clustered at 99% similarity. An OTU-by-sample table was created by mapping all dereplicated reads for each sample to the OTU representative sequences with an identity threshold of 97%.
Taxonomic information was evaluated for each OTU by conducting sequence similarity searches against the NCBI nt database (GenBank) and PROTAX (Somervuo et al., 2016; Lozano Mojica and Caballero, 2021). Identifications from either source were accepted, and these were consistent at the level at which they were made. Species and genus level assignments were automatically retained if they were supported by unambiguous matches to reference sequences at ≥99% or ≥95%, respectively. Public records from GBIF were used to determine the most likely species present in Colombia when there were multiple equally good matches. This allowed for the resolution of numerous uncertain sequences to the species level. OTUs that were ≥99% similar and had similar co-occurrence patterns were combined using LULU (Frøslev et al., 2017). The OTU table was then filtered to remove low abundance OTUs from each sample (<0.05% or <10 reads). In order to eliminate any erroneous identifications, the sequences corresponding to human, food fish, and livestock were removed, as were the OTUs identified at the order level above. In addition, sequences referring to species or genera distributed in other regions of Colombia, different from those sampled, or in other countries were removed (Supplementary Material).
Before and after each step of the extraction, amplification, and sequencing process, all benches were decontaminated with CHEMGENE HLD4L wipes (STARLAB). Negative controls were not sequenced and did not produce any bands. Each step of the process had its own designated space, equipment, reagents, and consumables.
Bioinformatic analyses
Due to the large amount of ichthyological data in comparison to other vertebrates, the results were separated into two groups: fish and tetrapods. Tetrapods included amphibians, birds, mammals, and reptiles, whether they were terrestrial, aquatic, or semi-aquatic. The analyses were also divided into 18 sampling locations (Supplementary Material). Although Tillavá is not part of Sierra de la Macarena National Park, it is relatively close (296 km) compared to the other subregions. Therefore, we decided to group them. Furthermore, Tillavá belongs to the same department (Meta) and region (Orinoco) as Sierra de la Macarena National Park. Additionally, we decided to use the taxonomic level of ‘genus’ for all analyses, since it had the highest number of reads and identifications.
R studio (RStudio Team, 2020) (R Project for Statistical Computing, RRID: SCR_001905) version 3.6.0, ‘vegan’ package, was used to calculate alpha and beta diversity for all sampling sites, based on presence/absence (i.e. binary) matrices of genera. Shannon-Weiner and Simpson indices were calculated for alpha diversity (Moreno, 2001; Li et al., 2021). For the Simpson index, low (0.00–0.35), medium (0.36–0.75) and high (0.76–1.00) diversity values were used. For the Shannon index, low (0.1 - 1.5), medium (1.6 - 3.0) and high (3.1 - 4.5) diversity values were used (Magurran, 1988). Beta diversity was then assessed using the Jaccard dissimilarity index (Nakagawa et al., 2018; Wiersma, 2019; Lin et al., 2021) and principal coordinate analysis (PCoA). Similarly, the number of unique genera identified for each site and the genera shared among them were represented by a Euler diagram, both for the fish group and for the tetrapod group.
Results
High-quality vertebrate sequence data were obtained from 31 filters, resulting in a total of 709 OTUs after data curation. The fish group contained 9 orders, 35 families, 132 genera, and 249 species (Supplementary Material), while the tetrapods group contained 22 orders, 32 families, 43 genera, and 49 species (Supplementary Material). The three most predominant orders in the fish group were Siluriformes, Characiformes, and Gymnotiformes, with Characidae and Loricariidae as the leading families. Among the tetrapod group, the orders with the highest prevalence were Anura, Primates, and Rodentia, along with the Psittacidae family.
Orinoco River (RO) had the highest number of fish genera with 67, followed by Tillavá (T) (52), Meta River (RM) (45), Yarumales stream (CY) (43), Bita River (RB)(43), Puerto Nariño (PN) (37), El Silencio lagoon (LES) (33), Verde stream (CV) (31),El Pañuelo lagoon (LEP (31), Caballococha (PCC) (28), Negro stream (CN) (27), Correo-Tarapoto Redondo lakes (LCT) (25), Don Ricardo stream (CDR) (21), Guayabero River (RG) (18), Caño Cristales River (CC) (11), Caño Paujil River (CP) (11), Tesoro stream (CT) (9) and lastly Caquetá River (RC) (4). In terms of relative abundance of DNA based on percentage of reads, the most abundant genera in Puerto Nariño and adjacent areas were Prochilodus (~38%) and Cyphocharax (~38%), while for the municipality of Solano these were Acestrorhynchus (~22%) and Apteronotus (~18%) (Figure 2). In the Bojonawi Nature Reserve they were Prochilodus (~30%) and Leporinus (~21%), and in the Sierra de la Macarena National Park and Tillavá they were Cyphocharax (~20%) and Prochilodus (~13%) (Figure 2).
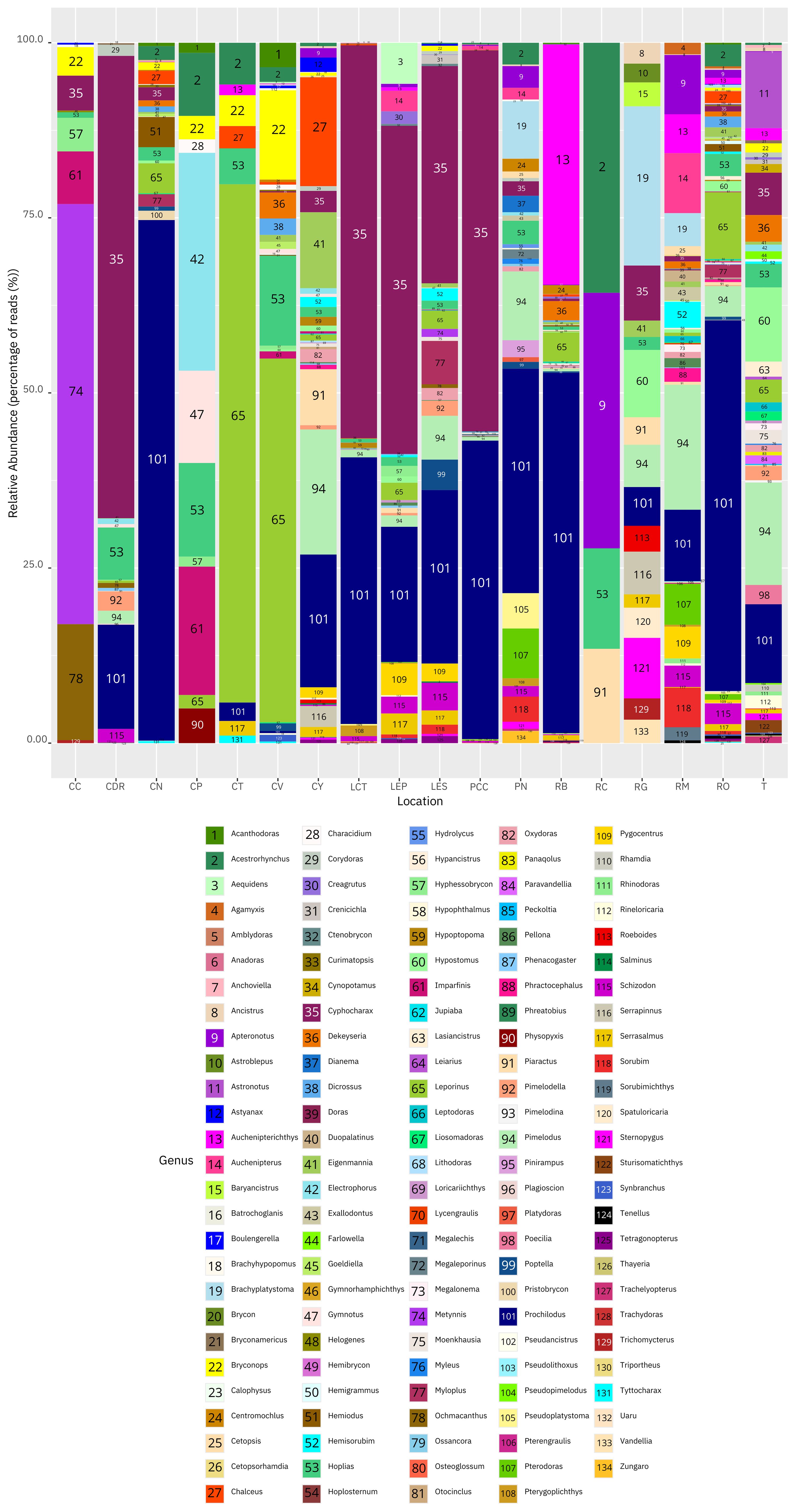
Figure 2 Histogram showing the relative abundance of DNA based on percentage of reads for fish genera in the Amazon and Orinoco basins: Caño Cristales River (CC), Don Ricardo stream (CDR), Negro stream (CN), Caño Paujil River (CP), Tesoro stream (CT), Verde stream (CV), Yarumales stream (CY), Correo and Tarapoto lakes (LCT), El Pañuelo lagoon (LEP), El Silencio lagoon (LES), Caballococha lake (PCC), Puerto Nariño (PN), Bita River (RB), Caquetá River (RC), Guayabero River (RG), Meta River (RM), Orinoco River (RO) and Tillavá (T).
For the tetrapods, the number of genera found, in order from largest to smallest, was as follows RG (13), CY (12), LES (8), RO (8), PCC (7), CP (6), RB (5), PN (5), T (5), CV (4), CDR (3), LCT (3), RM (3), CC (2), RC (2), CN (1) and LEP (1). In addition, the most abundant genera were Phalacrocorax (birds) (~65%) and Inia (mammals) (~33%) for Puerto Nariño and adjacent areas, Chelus (reptiles) (~41%) and Steatornis (birds) (~18%), in the municipality of Solano, Inia (mammals)(~41%) and Peltocephalus (reptiles) (~21%) for the Bojonawi Nature Reserve and adjacent areas, and Odocoileus (mammals) (~21%) and Bubulcus (birds) (~14%) in the Sierra de la Macarena National Park and Tillavá, respectively (Figure 3).
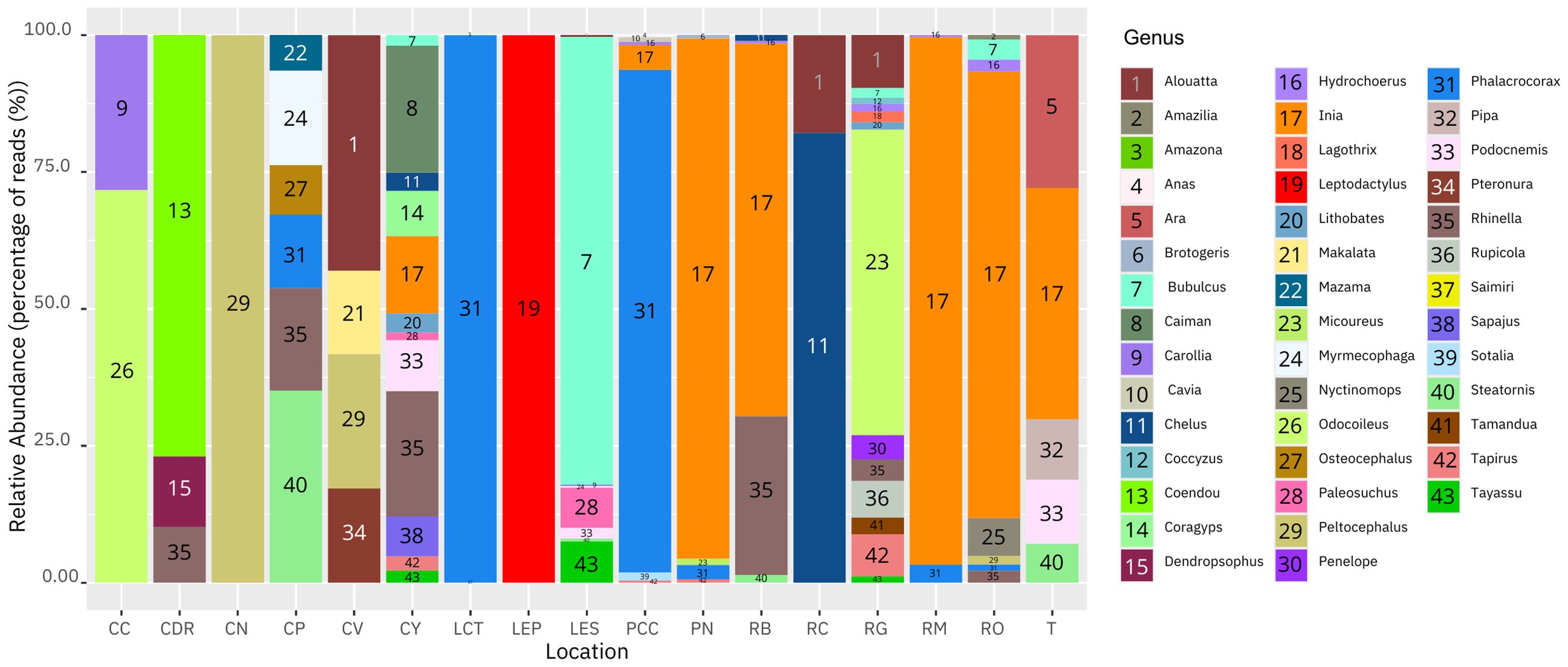
Figure 3 Histogram showing the relative abundance of DNA based on percentage of reads for tetrapods genera in the Amazon and Orinoco basins: Negro stream (CN), Verde stream (CV), El Pañuelo lagoon (LP), Bita River (RB), Meta River (RM), Orinoco River (RO), Caño Cristales River (CC), Yarumales stream (CY), El Silencio lagoon (LES), Guayabero River (RG), Don Ricardo stream (CDR), Tillavá River (T), Puerto Nariño (PN), Correo and Tarapoto lakes (LCT), Caballococha lake (PCC), Caño Paujil River (CP) and Caquetá River (RC).
Alpha diversity
In relation to the fish group, all sites in the Orinoco region showed high diversity according to Simpson’s index, including the Amazonian sites, except for RC, which showed medium diversity (Figure 4B). With regards to Shannon’s index, 10 out of the 13 Orinoco sites had relatively high diversity (CDR, CN, CV, CY, LEP, LES, RB, RM, RO and T), while the remaining three (CC, CT and RG) had medium diversity (Figure 4A). In the case of tetrapods, five locations (CY, LES, RG, RO and T) exhibited relatively high diversity values, while five other locations (CC, CDR, CV, RB and RM) showed medium diversity values, and two locations (CN and LEP) had low diversity values. Conversely, in the Amazon region, three sites (CP, PCC and PN) had high Simpson’s diversity values, while two sites (LCT and RC) had medium values (Figure 4D). The Orinoco region displayed medium (CY, LES, RG, RO and T) and low (CC, CDR, CV, RB, and RM) diversities in the Shannon’s index (Figure 4C). Additionally, CT and LEP had only one genus detected, resulting in a diversity value of H’ = 0 (Şen and Grillo, 2018). In contrast, the Amazon region exhibited medium diversity in three locations (CP, PCC and PN) and low diversity in two locations (LCT and RC) (Supplementary Material).
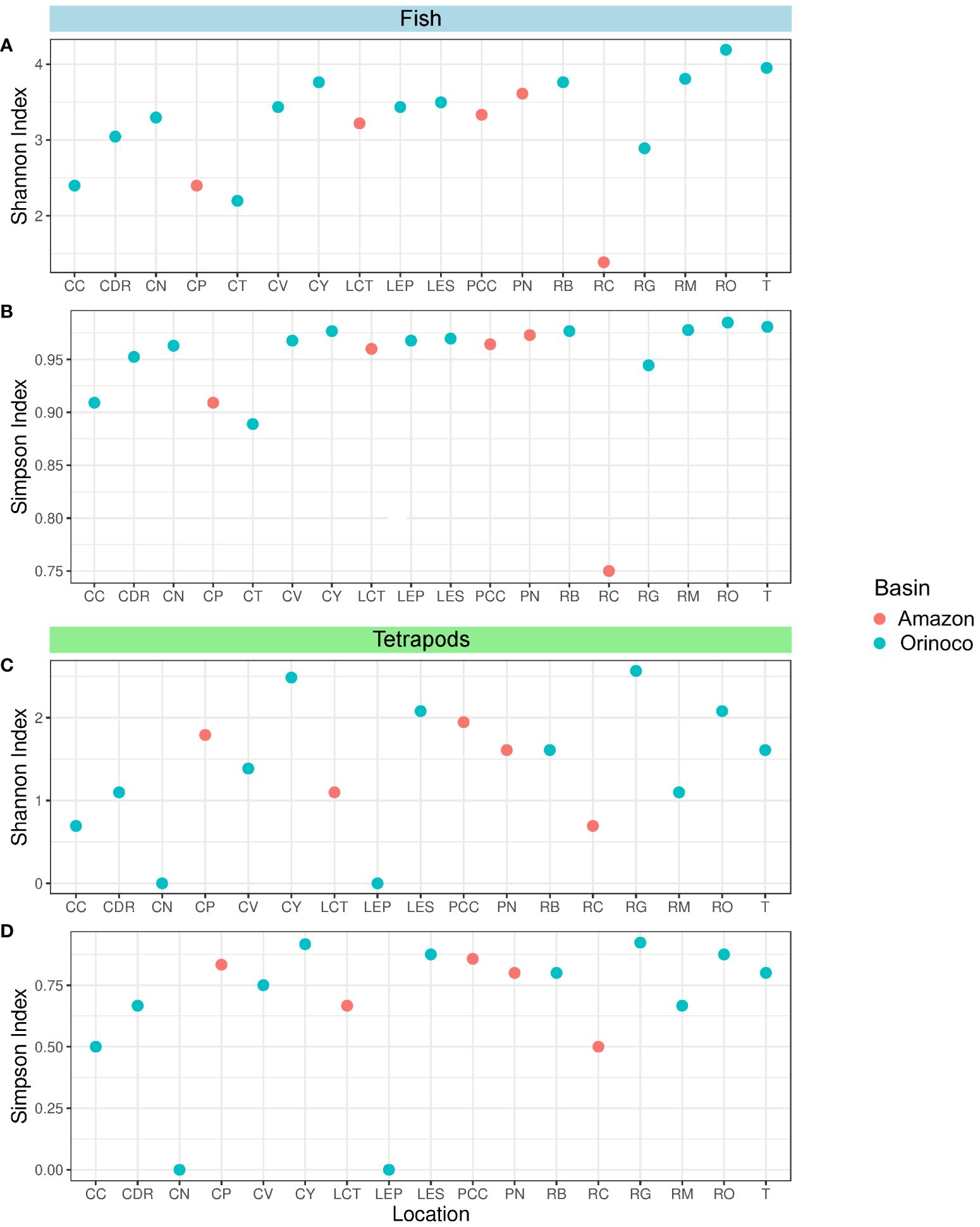
Figure 4 Dot plots showing Alpha diversity indices obtained for the fish and tetrapods groups respectively: Shannon index (A, C) and Simpson Index (B, D). Locations: Caño Cristales River (CC), Don Ricardo stream (CDR), Negro stream (CN), Caño Paujil River (CP), Verde stream (CV), Yarumales stream (CY), Correo and Tarapoto lakes (LCT), El Pañuelo lagoon (LEP), El Silencio lagoon (LES), Caballococha lake (PCC), Puerto Nariño (PN), Bita River (RB), Caquetá River (RC), Guayabero River (RG), Meta River (RM), Orinoco River (RO) and Tillavá (T).
Beta diversity
Principal coordinate analysis (PCoA) enabled us to illustrate the ecological distance based on genus diversity. As a result, the fish group was divided into four clusters (Figure 5A). The largest cluster included all locations from the Orinoco basin, except for the lowland streams of Bojonawi Nature Reserve and the Caño Cristales River (CC) for Sierra de la Macarena National Park. The second cluster was formed by Caño Negro (CN) and Caño Verde (CV), while CC formed a third cluster with the two locations of the Caquetá subbasin (Caquetá River and Caño Paujil River), in addition to the third stream of Bojonawi Nature Reserve, the Tesoro stream (CT). The fourth cluster grouped locations of the Loretocayu River and Javari River subbasins of the Amazon region (PN, LCT and PCC). In the first cluster, the Orinoco and Bita rivers showed the closest proximity as they overlapped. Additionally, these two locations were found to be closer to the Meta River, El Pañuelo lagoon, Tillavá, and Yarumales stream compared to the other locations in the group, such as Don Ricardo stream, Guayabero River, and El Silencio lagoon. The initial two locations were grouped in the upper right quadrant, while El Silencio lagoon was the only location placed in the bottom right quadrant. This indicates a significant difference in the composition of genus diversity within the group.
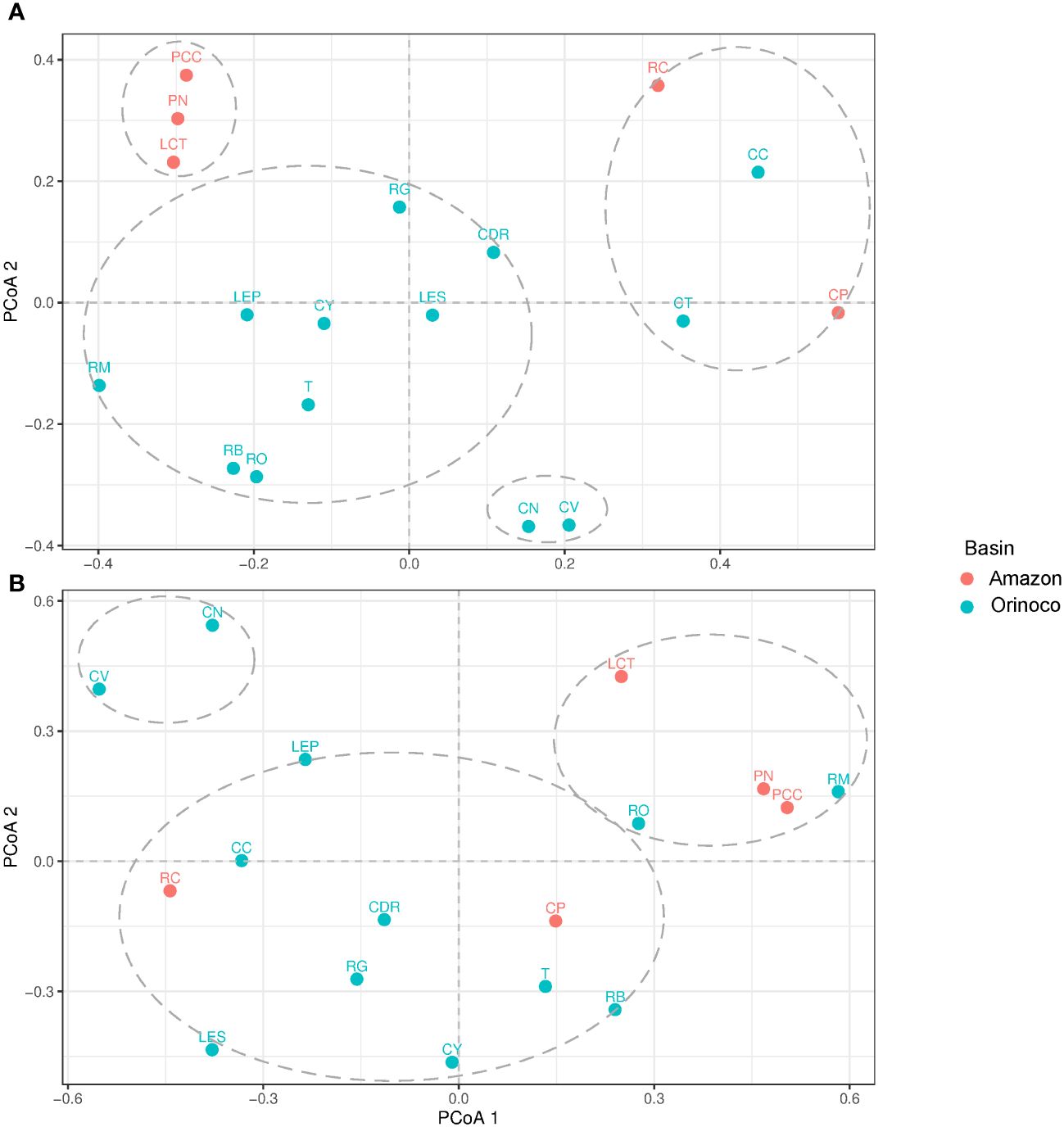
Figure 5 Principal coordinates analysis (PCoA) ordination based on the Jaccard distance of fish (A) and tetrapods (B) genera. Locations: El Silencio lagoon (LES), Caño Cristales River (CC), Guayabero River (RG), Yarumales stream (CY), Don Ricardo stream (CDR), Tillavá River (T), Verde stream (CV), Negro stream (CN), Tesoro stream (CT), Orinoco River (RO), Bita River (RB), Meta River (RM), El Pañuelo lagoon (LP), Puerto Nariño (PN), Correo and Tarapoto lakes (LCT), Caballococha (PCC), Caño Paujil River (CP) and Caquetá River (RC).
The study identified three clusters of tetrapods. The first cluster included all locations from Guaviare and Caquetá subbasins, with the Bita River (B) and El Silencio lagoon (LES) grouped together. The second cluster contained the Orinoco and Meta rivers from Bojonawi Nature Reserve, as well as samples from the Loretoyacu River and Javari River subbasins (PN, LCT and PCC). The third cluster consisted of the fish group, with Caño Negro (CN) and Caño Verde (CV) forming a separate group (Figure 5B).
Taxonomic composition by regions, subregions and locations
A total of 68 fish genera were found exclusively in one of the four sampled subregions. Specifically, 31 genera were detected in the Sierra de la Macarena National Park, 20 in Bojonawi Nature Reserve, 16 in Puerto Nariño and adjacent areas, and one in Solano’s municipality. All sampled subregions shared four genera. Sierra de la Macarena National Park and Bojonawi Nature Reserve had the most genera in common (23), followed by Sierra de la Macarena National Park, Bojonawi Nature Reserve and Puerto Nariño (21). The Amazon basin subregions did not share any genera (Figure 6A; Supplementary Material).
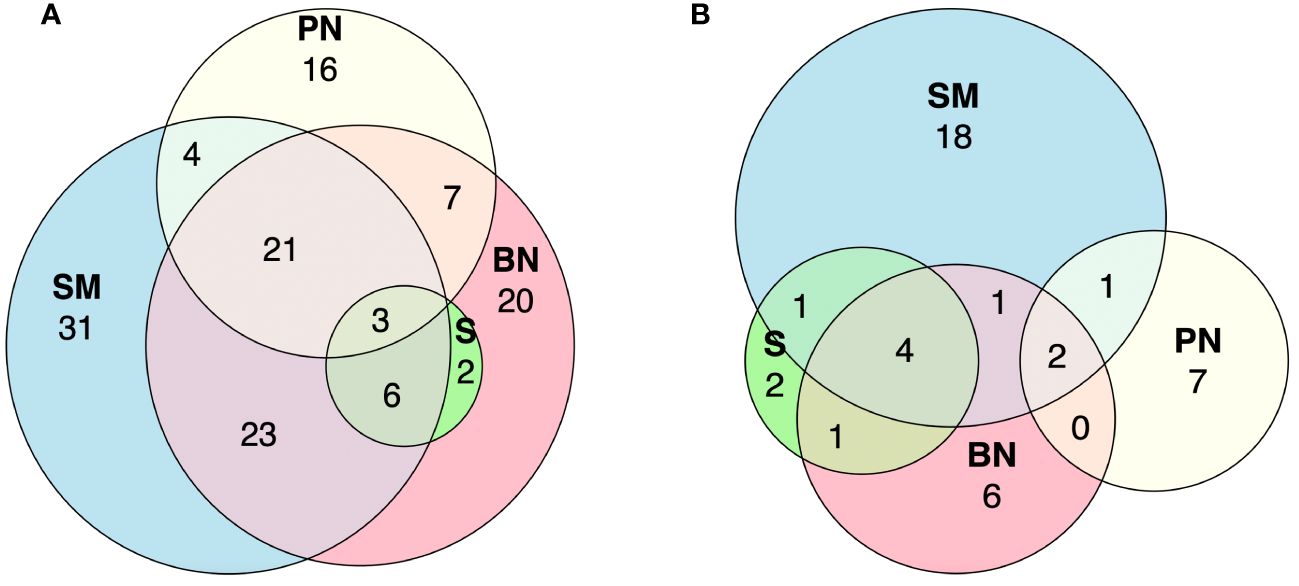
Figure 6 Euler diagrams showing the number of shared and unique genera in the four subregions sampled for fish (A) and tetrapods (B). Subregions: Bojonawi Nature Reserve and adjacent areas (BN), Sierra de la Macarena National Park and Tillavá (SM), Puerto Nariño and adjacent areas (PN) and Solano´s Municipality (S).
For the tetrapod group, 33 of the 43 genera were found in just one of the four subregions. There were 18 genera in Sierra de la Macarena and Tillavá, seven in Puerto Nariño and adjacent areas, six in the Bojonawi Nature Reserve and adjacent areas, and two in Solano’s municipality. None of the four subregions share genera between them (Figure 6B; Supplementary Material).
Discussion
This study validates the effectiveness of environmental (e)DNA sampling in evaluating the relative taxonomic richness of aquatic, semiaquatic, and terrestrial vertebrates in different macrohabitats, such as rivers, lowland streams, lakes, and lagoons in the Colombian Amazon and Orinoco hydrographic basins. We also identified organisms at the taxonomic levels of families, genera and species, and allowed the diversity of different communities to be assessed. The results obtained complement and efficiently extend traditional species inventories, although traditional methodologies require greater spatial component and field collection effort compared to eDNA (Fraija-Fernández et al., 2020; Sales et al., 2020b).
The study successfully identified cryptic genera, such as Apteronotus, Eigenmannia, Pellona, Prochilodus, and Sternopygus, which are physically similar but genetically and taxonomically distinct. Additionally, the study detected rare and low-abundance genera that are difficult to observe and not easily detectable in traditional surveys, including Anchoviella, Electrophorus, Poecilia, Synbranchus, and Phreatobius (Ota et al., 2020). Also, the analysis suggests that there are likely more species within the same genus in the Colombian Amazon and Orinoco basins than previously known, including many that are probably new to science. For instance, in the Bojonawi Nature Reserve, seven OTUs of the Prochilodus genus were reported (Supplementary Material), whereas only two species, P. mariae and P. rubrotaeniatus, are recognized in the basin (DoNascimiento et al., 2021). The high diversity found for this genus in Northern South America (Moyer et al., 2005) could be correlated with potential hybridization and introgression. This likely increases the chances of detecting particular sequences, rather than just a unique sequence for 12s.
As for the genus Electrophorus, only one species is recognised for the Orinoco Basin (E. electricus), but here we detected a second OTU, supporting the study by de Santana et al. (2019), which proposed an unexpected species diversity of electric eels. Additionally, several species of fish that were not previously reported in inventories or studies were detected using environmental DNA (eDNA), particularly in the Bojonawi Nature Reserve and Sierra de la Macarena and Tillavá. The Bita, Orinoco, and Tillavá rivers contain a high percentage of unknown fish diversity, estimated at 75% (65), 35% (113), and 52% (56), respectively (Supplementary Material). Therefore, these results support the need to associate eDNA detection and traditional capture methods to avoid biased estimates of occurrence and obstruct species conservation (Sales et al., 2020b). Additionally, they enable the rapid identification of regions with unexplored diversity to be investigated for future diversity studies in the Orinoco basin.
Furthermore, given the global decline in fisheries and the high cost, logistical demands, and invasiveness of conventional survey methods for stock assessment, eDNA has emerged as an effective complementary tool for assessing fish biomass in both freshwater and marine ecosystems (Evans and Lamberti, 2018; Gilbey et al., 2021; Jerde, 2021; Rourke et al., 2022). The study detected several commercially important fish species in the Colombian Amazon and Orinoco basins, both for consumption and ornamental use. In the Orinoco, the main orders were Acanthuriformes (Plagioscion), Clupeiformes (Pellona), Characiformes (Hoplias, Metynnis, Myleus, Myloplus, Piaractus, Prochilodus, Pygocentrus, Serrasalmus and Schizodon) and Siluriformes (Brachyplatystoma, Hemisorubim, Leiarius, Oxydoras, Pseudoplatystoma and Sorubimichtys). In the Amazon, various fish species were reported in the following genera: Chalceus, Myleus, Hoplias, Hydrolycus, Prochilodus, Serrasalmus, Pellona, Apteronotus, Osteoglossum, Brachyplatystoma, Hypostomus, Leiarius, Oxydoras, Pseudoplatystoma, Pterygoplichthys and Sorubimichthys (Salinas and Agudelo, 2000; Lasso et al., 2011). Therefore, these results suggest that eDNA could be a valuable tool for future fisheries monitoring.
Simultaneously, eDNA enables the identification, surveillance, and protection of threatened species in a more feasible manner (Thomsen et al., 2012; Laramie et al., 2015; Weltz et al., 2017; Loeza-Quintana et al., 2021; Plough et al., 2021). The current study identified two fish species as near threatened (Sorubimichthys planiceps and Sorubim lima) and two as vulnerable (Osteoglossum bicirrhosum and Zungaro zungaro) (Mojica et al., 2012). Five vulnerable species were detected in the tetrapods group: the white-lipped peccary (Tayassu pecari), the lowland tapir (Tapirus terrestris), the giant anteater (Myrmecophaga tridactyla), the common woolly monkey (Lagothrix lagotricha), and the Big-headed Amazon River Turtle (Peltocephalus dumerilianus) (Keuroghlian et al., 2013; Miranda et al., 2014; Tortoise & Freshwater Turtle Specialist Group, 1996; Varela et al., 2019; Stevenson et al., 2021). Four endangered species have been identified, including the giant otter (Pteronura brasiliensis), as well as two subspecies of the Amazon River dolphin found in Colombia (Inia geoffrensis geoffrensis and Inia geoffrensis humboldtiana), and the tucuxi (Sotalia fluviatilis) (da Silva et al., 2018, 2020; Groenendijk et al., 2023).
Furthermore, eDNA provides a clear picture of how the distribution of diversity is mainly related to environmental conditions and resources in aquatic and terrestrial systems (Mott, 2010; Wallis et al., 2021). It is expected that sampling locations in the same geographical area would be more similar to each other for biological diversity, compared to locations in different regions (Heino et al., 2015). Most of the fish and tetrapods group data were distributed according to their respective geographic basin in either the Amazon or Orinoco basins, forming two main groups (Figures 5A, B). However, for some taxa, particular combinations resulted in Amazon and Orinoco samples grouping together. A possible explanation for this date back to the late Miocene epoch (approximately 11.6 to 5.3 million years ago) when the eastern Cordillera rose, leading to the emergence of the Macarena massif and the uplift of the Vaupés swell. This event eventually caused the separation of the Orinoco and Amazon systems. These geographical events may have isolated the respective populations, both terrestrial and aquatic, leading to species dispersion among the fragmented Amazon and Orinoco basins (Correa et al., 2005; Mora et al., 2010; Renno et al., 2011; Holbourn et al., 2018; Cala-Cala, 2019; Vargas-Ramírez et al., 2020; Rizo-Fuentes et al., 2021).
It is important to highlight, that eDNA is a relatively new methodology (Peters et al., 2018), therefore limitations remain that must be considered for future studies. Primarily, there is currently no public database that includes sequences from all species described to date, nor a database with divergent sequences between closely related species (e.g., sister species) (Freeland, 2017; Cristescu and Hebert, 2018; Zaiko et al., 2018; Polanco et al., 2021a). This hinders accurate species identification, an issue that has been underlined by previous neotropical eDNA studies (Cilleros et al., 2019; Bevilaqua et al., 2020; Sales et al., 2020b). In the current study, of all the fishes detected, only 29% were assigned to the species level. This also occurred for lesser extent in the tetrapods (57%). At the same time, misidentification, which arises from error generated as a result of the workflow, could lead to either false positives or false negatives, including false haplotypes (Furlan et al., 2020). As an example, the rough-toothed dolphin (Steno bredanensis) was reported in the results for Caballococha, Amazon, but this organism is a marine species. Phylogenetic studies have identified that the Steno genetic sequence for the 12s ribosomal RNA gene is very similar to that of the Sotalia genus, which is found in the Amazon River by the Tucuxi (Sotalia fluviatilis) (Caballero et al., 2007; Cunha et al., 2011; Mosquera et al., 2015). Another example is the identification of the paujil or black guan (Penelopina nigra) in the Guayabero River, which is not found in Colombia and is found only in Central America (Pineda et al., 2008). However, when evaluating the Penelopinae subfamily phylogenetics, it was found that the Penelopina genus is indeed a sister taxon of the Penelope genus, which is reported in the Sierra de la Macarena National Nature Park with the Spix´s guan (Penelope jacquacu) (Lasso et al., 2018). Therefore, to obtain accurate biological information, especially in megadiverse freshwater systems such as the Neotropics, it is crucial to develop curated and complete molecular DNA databases, including the expansion of currently available 12s and 16s rDNA sequence databases (Bevilaqua et al., 2020; Milan et al., 2020; Jackman et al., 2021).
Furthermore, the effectiveness of organism’s detection (species/genera) between eDNA and traditional sampling varies depending on the analysis approach. As reported in Martinelli-Marín et al. (2020), for the Orinoco River, 223 species were identified by metabarcoding and 128 species by traditional sampling (Lasso et al., 2020), representing a difference of 57% between the two methods. This would also be the case if we consider the whole system (river-floodplain), represented in this case by the Orinoco River and the set of macro-habitats (channels-lagoons) of the floodplain (262 species by metabarcoding vs. 270 species by traditional methods). However, when the macrohabitats (lagoons, channels and main river) are analyzed at the individual level, there is a large difference between the two methods, with the traditional approach showing a superior efficiency of almost 40%. This pattern is consistent in the other remaining sites, except for the Solano´s municipality, Puerto Nariño and Caballococha, where there were no records for comparison (Table 1). We attribute the difference in ‘efficiency’ between the two methods to multiple aspects. First, eDNA sampling in this study was restricted to the main channel of the rivers. In contrast, the traditional methods included all macrohabitats of the basin, such as main rivers, tributaries, morichals, small ravines and others. Also, there was a significant difference in sampling time, with traditional methods taking weeks, months or even years (historic records) compared to days for eDNA. Finally, eDNA captures a specific sample at a particular time and location, whereas traditional sampling involves using various fishing gears to cover different areas in the water column at different seasons of the year which could contribute to a higher coverage of diversity.
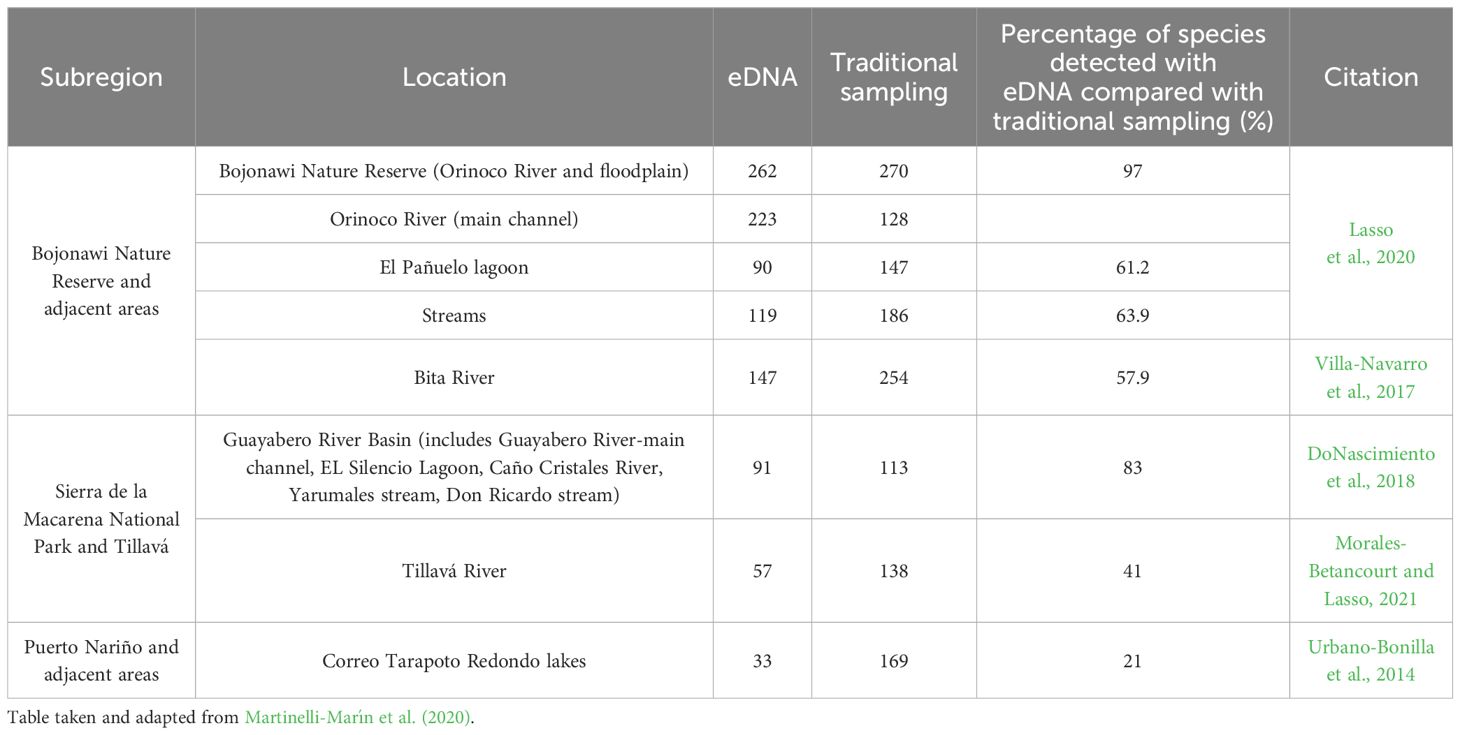
Table 1 Comparison of fish species detected by eDNA and traditional sampling in the Amazon and Orinoco basins.
In addition, the study found that the number of tetrapod genera and species was low compared to fishes, which is consistent with previous studies using traditional methods, which reported even lower records (Ruiz et al., 2007; Lasso et al., 2010; Lasso and Morales-Betancourt, 2017; Lasso et al., 2018; Méndez-López et al., 2019; Sánchéz et al., 2019; Lasso et al., 2020; Morales-Betancourt and Lasso, 2021). This may be related to the behaviour of the terrestrial animal itself, in particular its opportunistic interactions with water such as swimming, wallowing, salivation during drinking and deposition of urine or faeces (Sales et al., 2020b; Lyet et al., 2021). The results of this eDNA study are consistent with previous research showing that more aquatic, semi-aquatic and nocturnal species were detected by eDNA than by observational methods: Coendou sp., Hydrochoerus hydrochaeris, I. geoffrensis, M. tridactyla, P. brasiliensis, Tamandua tetradactyla, and T. terrestris (Sales et al., 2020a; Coutant et al., 2021). Moreover, the lack of iconic carnivores, such as the jaguar (Panthera onca), cougar (Puma concolor), and ocelot (Leopardus pardalis), in the current study may be due to their ecological characteristics, such as a relatively large home range, dietary preferences, or solitary behaviour (Gompper et al., 2006; Harper et al., 2019; Sales et al., 2020a; Broadhurst et al., 2021; Lyet et al., 2021). Alternatively, another possible reason for the low abundance of tetrapods, mainly amphibians, birds and reptiles, may be that we used universal vertebrate primers that amplify a wide range of organisms (Yang et al., 2014); some may have lower primer fidelity and less abundant reads may be too rare to detect. Hence, the identification of specific taxa depends on primers that amplify DNA exclusively from the target taxa and are sensitive enough to detect even small amounts of target DNA from low-quality samples (Macdonald and Sarre, 2017). Moreover, newer studies have employed eDNA metabarcoding with targeted or modified primers and different genetic markers to detect a wider range of amphibian, bird and reptile species (Valentini et al., 2016; Sasso et al., 2017; Wilson et al., 2017; Ushio et al., 2018; Neice and McRae, 2021; Lam et al., 2022). Also, eDNA extraction can be used to target a wide range of environments other than the aquatic ones, which favours the detection of terrestrial organisms (Ficetola et al., 2019). Alternative water sources have been used to obtain environmental DNA, for example, in Sweden, DNA samples for ungulate species identification have been obtained from browsed branches of deciduous trees (Nichols et al., 2012), while in tropical forests, salt licks have been used to detect mammals (Ishige et al., 2017).
Although eDNA is still an exploratory tool, our study demonstrates its effectiveness in the interpretation of taxon richness and diversity of aquatic, semi-aquatic and terrestrial vertebrates in different water bodies of the Colombian Amazon and Orinoco basins. With a simple water sample, we were able to detect organisms that had not been previously identified by traditional surveys which require expensive and time-consuming sampling devices and larger spatial scales. Therefore, we recommend using eDNA as an efficient tool to complement and extend biological inventories in a more rapid and non-invasive manner. However, it is necessary to develop more complete DNA sequence reference databases that include all neotropical species sequences discovered to date. Additionally, it is important to create standardized protocols based on the animal’s habitat and behavior of interest to maximize the benefits of eDNA detection.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://datadryad.org/stash, https://doi.org/10.5061/dryad.15dv41p0d.
Author contributions
DM: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. CL: Data curation, Supervision, Validation, Writing – review & editing. SC: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Funding for this project was available from a private donation to Universidad de los Andes (project “Conservación del Manatı́ Antillano (Trichechus manatus) en Colombia y el Caribe uso de nuevas tecnologı́as como apoyo efectivo en procesos de recuperación de especies amenazadas”) and from the Faculty of Sciences Research Program 2019- 2022 Initiative to SC (Programa de Investigación Facultad de Ciencias, Universidad de los Andes).
Acknowledgments
The authors would like to express their gratitude to Juan Esteban Jacome for his valuable assistance in creating the graphs. Additionally, we are thankful for the help provided by C. Romero, J. D. Lozano, L. Baldrich, M. Morales, and B. Castañeda during the field work. We would also like to thank Fundación Omacha for their hospitality at the Bojonawi Nature Reserve and Puerto Nariño.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2024.1409296/full#supplementary-material
References
Aldana J., Montes C., Martínez M., Medina N., Hahn J., Duque M. (2017). Biodiversity and ecosystem services knowledge in the Colombian Caribbean: progress and challenges. Trop. Conserv. Sci. 10, 1–41. doi: 10.1177/1940082917714229
Antonelli A., Zizka A., Carvalho F. A., Scharn R., Bacon C. D., Silvestro D., et al. (2018). Amazonia is the primary source of Neotropical biodiversity. Proc. Natl. Acad. Sci. 115, 6034–6039. doi: 10.1073/pnas.1713819115
Baker C. S., Steel D., Nieukirk S., Klinck H. (2018). Environmental DNA (eDNA) from the wake of the whales: Droplet digital PCR for detection and species identification. Front. Mar. Sci. 5, 133. doi: 10.3389/fmars.2018.00133
Barnes M. A., Turner C. R., Jerde C. L., Renshaw M. A., Chadderton W. L., Lodge D. M. (2014). Environmental conditions influence eDNA persistence in aquatic systems. Environ. Sci. Technol. 48, 1819–1827. doi: 10.1021/es404734p
Barnosky A. D., Matzke N., Tomiya S., Wogan G. O., Swartz B., Quental T. B., et al. (2011). Has the Earth’s sixth mass extinction already arrived? Nature 471, 51–57. doi: 10.1038/nature09678
Bevilaqua D. R., de Melo S. A., de Carvalho Freitas C. E., da Silva A. C. V., da Silva Batista J. (2020). First environmental DNA (eDNA) record of central Amazon in a floodplain lake: extraction method selection and validation. Braz. J. Dev. 6, 87606–87621. doi: 10.34117/bjd
Boussarie G., Bakker J., Wangensteen O. S., Mariani S., Bonnin L., Juhel J. B., et al. (2018). Environmental DNA illuminates the dark diversity of sharks. Sci. Adv. 4, 1–8. doi: 10.1126/sciadv.aap9661
Broadhurst H. A., Gregory L. M., Bleakley E. K., Perkins J. C., Lavin J. V., Bolton P., et al. (2021). Mapping differences in mammalian distributions and diversity using environmental DNA from rivers. Sci. Total Environ. 801, 1–10. doi: 10.1016/j.scitotenv.2021.149724
Caballero S., Martinelli-Marín D., Lasso Alcala C. A. (2021b). Diversidad de vertebrados en ecosistemas subterráneos de Colombia a partir de análisis de ADN ambiental. Memorias II Congreso Colombiano de espeleología 2021. Available online at: http://repository.humboldt.org.co/bitstream/handle/20.500.11761/35838/23_Caballero%20et%20al.pdf?sequence=1.
Caballero S., Ortiz M., Bohorquez L., Lozano J. M., Caicedo-Herrera D., Arévalo-González K., et al. (2021a). Mitochondrial genetic diversity, population structure and detection of antillean and amazonian manatees in Colombia: new areas and new techniques. Front. Genet. 12, 726916. doi: 10.3389/fgene.2021.726916
Caballero S., Trujillo F., Vianna J. A., Barrios H., Montiel M. G., Beltrán S., et al. (2007). Taxonomic status of the genus Sotalia: Species level ranking for “tucuxi” (Sotalia fluviatilis) and “costero” dolphins (Sotalia guianensis). Mar. Mammal Sci. 23, 358–386. doi: 10.1111/j.1748-7692.2007.00110.x
Cala-Cala P. (2019). “El escenario para la diversificación de peces neotropicales: una historia de ríos tropicales de Suramérica,” in Medio ambiente y diversidad de los peces de agua dulce de Colombia Bogotá D. C. (Colombia: Academia Colombiana de Ciencias Exactas, F sicas y Naturales), 71–113.
Cardinale B. J., Duffy J. E., Gonzalez A., Hooper D. U., Perrings C., Venail P., et al. (2012). Biodiversity loss and its impact on humanity. Nature 486, 59–67. doi: 10.1038/nature11148
Ceballos G., Ehrlich P. R., Barnosky A. D., García A., Pringle R. M., Palmer T. M. (2015). Accelerated modern human–induced species losses: Entering the sixth mass extinction. Sci. Adv. 1, 1–6. doi: 10.1126/sciadv.1400253
Ceballos G., Ehrlich P. R., Raven P. H. (2020). Vertebrates on the brink as indicators of biological annihilation and the sixth mass extinction. Proc. Natl. Acad. Sci. 117, 13596–13602. doi: 10.1073/pnas.1922686117
Cilleros K., Valentini A., Allard L., Dejean T., Etienne R., Grenouillet G., et al. (2019). Unlocking biodiversity and conservation studies in high-diversity environments using environmental DNA (eDNA): A test with Guianese freshwater fishes. Mol. Ecol. Resour. 19, 27–46. doi: 10.1111/1755-0998.12900
Convention on Biological Diversity. (2021). Colombia – Main Details. Available online at: https://www.cbd.int/countries/profile/?country=co#:~:text=Biodiversity%20Facts&text=Colombia%20is%20listed%20as%20one,butterflies%2C%20freshwater%20fishes%20and%20amphibians.
Correa H. D., Ruiz S. L., Arévalo L. M. (Eds.) (2005). Plan de acción en biodiversidad de la cuenca del Orinoco – Colombia/2005 - 2015 – Propuesta Técnica (Bogotá D.C: Corporinoquia, Cormacarena, I.A.v.H, Unitrópico, Fundación Omacha, Fundación Horizonte Verde, Universidad Javeriana, Unillanos, WWF - Colombia, GTZ – Colombia), 273.
Coutant O., Richard-Hansen C., de Thoisy B., Decotte J. B., Valentini A., Dejean T., et al. (2021). Amazonian mammal monitoring using aquatic environmental DNA. Mol. Ecol. Resour. 21, 1875–1888. doi: 10.1111/1755-0998.13393
Cowie R. H., Bouchet P., Fontaine B. (2022). The Sixth Mass Extinction: fact, fiction or speculation? Biol. Rev. 97, 640–663. doi: 10.1111/brv.12816
Cristescu M. E., Hebert P. D. (2018). Uses and misuses of environmental DNA in biodiversity science and conservation. Annu. Rev. Ecol. Evolution Systematics 49, 209–230. doi: 10.1146/annurev-ecolsys-110617-062306
Crossley M. S., Meier A. R., Baldwin E. M., Berry L. L., Crenshaw L. C., Hartman G. L., et al. (2020). No net insect abundance and diversity declines across US Long Term Ecological Research sites. Nat. Ecol. Evol. 4, 1368–1376. doi: 10.1038/s41559-020-1269-4
Cunha H. A., Moraes L. C., Medeiros B. V., Lailson C.OMMAJr. J., Da Silva V. M., Solé A. M., et al. (2011). Phylogenetic status and timescale for the diversification of Steno and Sotalia dolphins. PloS One 6, 1–7. doi: 10.1371/journal.pone.0028297
Dal Pont G., Duarte Ritter C., Agostinis A. O., Stica P. V., Horodesky A., Cozer N., et al. (2021). Monitoring fish communities through environmental DNA metabarcoding in the fish pass system of the second largest hydropower plant in the world. Sci. Rep. 11, 1–13. doi: 10.1101/2021.08.17.456687
da Silva V., Trujillo F., Martin A., Zerbini A. N., Crespo E., Aliaga-Rossel E., et al. (2018). Inia geoffrensis. The IUCN Red List of Threatened Species 2018: e.T10831A50358152. doi: 10.2305/IUCN.UK.20182.RLTS.T10831A50358152.en
da Silva V., Martin A., Fettuccia D., Bivaqua L., Trujillo F. (2020). Sotalia fluviatilis. The IUCN Red List of Threatened Species 2020: e.T190871A50386457. doi: 10.2305/IUCN.UK.2020–3.RLTS.T190871A50386457.en
Deiner K., Bik H. M., Mächler E., Seymour M., Lacoursière A., Altermatt F., et al. (2017). Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 26, 5872–5895. doi: 10.1111/mec.14350
Delić T., Trontelj P., Rendoš M., Fišer C. (2017). The importance of naming cryptic species and the conservation of endemic subterranean amphipods. Sci. Rep. 7, 1–12. doi: 10.1038/s41598-017-02938-z
de Santana C. D., Crampton W. G., Dillman C. B., Frederico R. G., Sabaj M. H., Covain R., et al. (2019). Unexpected species diversity in electric eels with a description of the strongest living bioelectricity generator. Nat. Commun. 10, 1–10. doi: 10.1038/s41467-019-11690-z
DiBattista J. D., Reimer J. D., Stat M., Masucci G. D., Biondi P., De Brauwer M., et al. (2020). Environmental DNA can act as a biodiversity barometer of anthropogenic pressures in coastal ecosystems. Sci. Rep. 10, 1–15. doi: 10.1038/s41598-020-64858-9
DoNascimiento C., Bogota J. D., Albornoz J. G., Méndez A., Villa F. A., Herrera E. E., et al. (2021). Lista de especies de peces de agua dulce de Colombia/Checklist of the freshwater fishes of Colombia. v. 2.13. Asociación Colombiana de Ictiólogos. Dataset/Checklist. doi: 10.15472/numrso
DoNascimiento C., Mesa L. M., Albornoz-Garzón J. G., Méndez-López A. L., Lasso C. A., García-Melo J. E., et al. (2018). Peces de los r os Guayabero medio, bajo Losada y bajo Duda, sierra de La Macarena, Meta, Colombia. C. In: V. Biodiversidad de la sierra de La Macarena, Meta, Colombia. Parte I. R os Guayabero medio, bajo Losada y bajo Duda. Eds. Lasso C. A., Morales-Betancourt e M. A., Escobar-Martínez I. D. Serie Fauna Silvestre Neotropical. Instituto de Investigaci n de los Recursos Biol gicos Alexander von Humboldt (IAvH). Bogot , D. C., Colombia.
Evans N. T., Lamberti G. A. (2018). Freshwater fisheries assessment using environmental DNA: A primer on the method, its potential, and shortcomings as a conservation tool. Fisheries Res. 197, 60–66. doi: 10.1016/j.fishres.2017.09.013
Faghihinia M., Xu Y., Liu D., Wu N. (2021). Freshwater biodiversity at different habitats: Research hotspots with persistent and emerging themes. Ecol. Indic. 129, 107926. doi: 10.1016/j.ecolind.2021.107926
Fernandes C. S., Batalha M. A., Bichuette M. E. (2019). Dark diversity in the dark: a new approach to subterranean conservation. Subterranean Biol. 32, 69–80. doi: 10.3897/subtbiol.32.38121
Ficetola G. F., Manenti R., Taberlet P. (2019). Environmental DNA and metabarcoding for the study of amphibians and reptiles: species distribution, the microbiome, and much more. Amphibia-Reptilia 40, 129–148. doi: 10.1163/15685381-20191194
Fraija-Fernández N., Bouquieaux M. C., Rey A., Mendibil I., Cotano U., Irigoien X., et al. (2020). Marine water environmental DNA metabarcoding provides a comprehensive fish diversity assessment and reveals spatial patterns in a large oceanic area. Ecol. Evol. 10, 7560–7584. doi: 10.1002/ece3.6482
Freeland J. R. (2017). The importance of molecular markers and primer design when characterizing biodiversity from environmental DNA. Genome 60, 358–374. doi: 10.1139/gen-2016-0100
Frøslev T. G., Kjøller R., Bruun H. H., Ejrnæs R., Brunbjerg A. K., Pietroni C., et al. (2017). Algorithm for post-clustering curation of DNA amplicon data yields reliable biodiversity estimates. Nat. Commun. 8, 1–11. doi: 10.1038/s41467-017-01312-x
Furlan E. M., Davis J., Duncan R. P. (2020). Identifying error and accurately interpreting environmental DNA metabarcoding results: A case study to detect vertebrates at arid zone waterholes. Mol. Ecol. Resour. 20, 1259–1276. doi: 10.1111/1755-0998.13170
Gilbey J., Carvalho G., Castilho R., Coscia I., Coulson M. W., Dahle G., et al. (2021). Life in a drop: Sampling environmental DNA for marine fishery management and ecosystem monitoring. Mar. Policy 124, 104331. doi: 10.1016/j.marpol.2020.104331
Gompper M. E., Kays R. W., Ray J. C., LaPoint S. D., Bogan D. A., Cryan J. R. (2006). A comparison of noninvasive techniques to survey carnivore communities in northeastern North America. Wildlife Soc. Bull. 34, 1142–1151. doi: 10.2193/0091-7648(2006)34[1142:ACONTT]2.0.CO;2
Groenendijk J., Leuchtenberger C., Marmontel M., Van Damme P. A., Wallace R., Schenck C. (2022). Pteronura brasiliensis (amended version of 2022 assessment). The IUCN Red List of Threatened Species 2023: e.T18711A244867206. doi: 10.2305/IUCN.UK.2022–2.RLTS.T18711A222719180.en
Guhl E. (2016). Colombia: bosquejo de su geografía tropical Vol. I (Bogotá D. C., Colombia: Ministerio de Cultura, Biblioteca Nacional de Colombia), 338.
Guio A. C., Rojas A. (2019). Amazonia Colombiana dinámicas territoriales. Ideas Verdes (Bogotá D. C., Colombia: Heinrich Böll Foundation), 22, 1–52. Available at: https://co.boell.org/es/2020/01/24/amazonia-colombiana-dinamicas-territoriales.
Hänfling B., Lawson Handley L., Read D. S., Hahn C., Li J., Nichols P., et al. (2016). Environmental DNA metabarcoding of lake fish communities reflects long-term data from established survey methods. Mol. Ecol. 25, 3101–3119. doi: 10.1111/mec.13660
Harper L. R., Lawson Handley L., Carpenter A. I., Ghazali M., Di Muri C., Macgregor C. J., et al. (2019). Environmental DNA (eDNA) metabarcoding of pond water as a tool to survey conservation and management priority mammals. Biol. Conserv. 238, 108225. doi: 10.1016/j.biocon.2019.108225
Heino J., Melo A. S., Siqueira T., Soininen J., Valanko S., Bini L. M. (2015). Metacommunity organisation, spatial extent and dispersal in aquatic systems: patterns, processes and prospects. Freshw. Biol. 60, 845–869. doi: 10.1111/fwb.12533
Herder J. E., Valentini A., Bellemain E., Dejean T., van Delft J. J. C. W., Thomsen P. F., et al. (2014). Environmental DNA - a review of the possible applications for the detection of (invasive) species. (Technical report N° 2013–104) (Nimega, Netherlands: RAVON).
Holbourn A. E., Kuhnt W., Clemens S. C., Kochhann K. G., Jöhnck J., Lübbers J., et al. (2018). Late Miocene climate cooling and intensification of southeast Asian winter monsoon. Nat. Commun. 9, 1–13. doi: 10.1038/s41467-018-03950-1
Instituto Geográfico Agustín Codazzi (2002). Regiones naturales. [Political Map]. Available online at: http://www2.igac.gov.co/ninos/UserFiles/Image/Mapas/regiones%20naturales.pdf.
Ishige T., Miya M., Ushio M., Sado T., Ushioda M., Maebashi K., et al. (2017). Tropical-forest mammals as detected by environmental DNA at natural saltlicks in Borneo. Biol. Conserv. 210, 281–285. doi: 10.1016/j.biocon.2017.04.023
IUCN, International Union for Conservation of Nature (2022). Freshwater biodiversity. Available online at: https://www.iucn.org/theme/species/our-work/freshwaterbiodiversity#:~:text=Almost%20one%2Dthird%20of%20freshwater,determine%20the%20threats%20they%20face.
Jackman J. M., Benvenuto C., Coscia I., Oliveira Carvalho C., Ready J. S., Boubli J. P., et al. (2021). eDNA in a bottleneck: Obstacles to fish metabarcoding studies in megadiverse freshwater systems. Environ. DNA 3, 837–849. doi: 10.1002/edn3.191
Jerde C. L. (2021). Can we manage fisheries with the inherent uncertaint from eDNA? J. fish Biol. 98, 341–353. doi: 10.1111/jfb.14218
Jia H., Zhang H., Xian W. (2022). Fish diversity monitored by environmental DNA in the Yangtze River mainstream. Fishes 7, 1–13. doi: 10.3390/fishes7010001
Jo T., Murakami H., Masuda R., Sakata M. K., Yamamoto S., Minamoto T. (2017). Rapid degradation of longer DNA fragments enables the improved estimation of distribution and biomass using environmental DNA. Mol. Ecol. Resour. 17, e25–e33. doi: 10.1111/1755-0998.12685
Johnson C. N., Balmford A., Brook B. W., Buettel J. C., Galetti M., Guangchun L., et al. (2017). Biodiversity losses and conservation responses in the Anthropocene. Science 356, 270–275. doi: 10.1126/science.aam9317
Juhel J. B., Marques V., Fernández A. P., Borrero-Pérez G. H., Martinezguerra M. M., Valentini A., et al. (2021). Detection of the elusive Dwarf sperm whale (Kogia sima) using environmental DNA at Malpelo island (Eastern Pacific, Colombia). Ecol. Evol. 11, 2956. doi: 10.1002/ece3.7057
Kelly R. P., Port J. A., Yamahara K. M., Martone R. G., Lowell N., Thomsen P. F., et al. (2014). Harnessing DNA to improve environmental management. Science 344, 1455–1456. doi: 10.1126/science.1251156
Keuroghlian A., Desbiez A., Reyna-Hurtado R., Altrichter M., Beck H., Taber A., et al. (2013). Tayassu pecari. The IUCN Red List of Threatened Species 2013: e.T41778A44051115. doi: 10.2305/IUCN.UK.2013–1.RLTS.T41778A44051115.en
Lam I. P., Sung Y. H., Fong J. J. (2022). Using eDNA techniques to find the endangered big-headed turtle (Platysternon megacephalum). PloS One 17, e0262015. doi: 10.1371/journal.pone.0262015
Laramie M. B., Pilliod D. S., Goldberg C. S. (2015). Characterizing the distribution of an endangered salmonid using environmental DNA analysis. Biol. Conserv. 183, 29–37. doi: 10.1016/j.biocon.2014.11.025
Lasso C. A., Agudelo E., Jiménez-Segura L. F., Ramírez-Gil H., Morales-Betancourt M., Ajiaco-Martínez R. E., de Paula Gutiérrez F., Usma J. S., Muñoz S. E., Sanabria A. I. (Eds.) (2011). I. Catálogo de los recursos pesqueros continentales de Colombia. Bogotá, D. C., Colombia: Serie Editorial Recursos Hidrobiológicos y Pesqueros Continentales de Colombia (Instituto de Investigación de Recursos Biológicos Alexander von Humboldt (IAvH).
Lasso C. A., Morales-Betancourt M. A. (Eds.) (2017). III. Fauna de Caño Cristales, sierra LaMacarena, Meta, Colombia. Bogotá, D. C., Colombia: Serie Editorial Fauna Silvestre Neotropical (Instituto de Investigación de Recursos Biológicos Alexander von Humboldt (IAvH).
Lasso C. A., Morales-Betancourt M. A., Cuervo A. M., Lomelín A., Amado J. C. (2018). “Aves del Área de Manejo Especial de la Macarena-AMEM, con énfasis en los ríos Guayabero medio (sector Raudal de Angosturas I), bajo Losada y bajo Duda, sierra de la Macarena, Meta, Colombia,” in V. Biodiversidad de la sierra de la Macarena, Meta, Colombia. Parte I. Ríos Guayabero medio, bajo Losada y bajo Duda (Bogotá D. C., Colombia: Instituto de Investigación de Recursos BiológicosAlexander von Humboldt, Corporaci n para el Desarrollo Sostenible del Área de Manejo Especial La MacarenaCormacarena 2018), 211–261.
Lasso C. A., Rial A., Colonnello G., MaChado A., Trujillo F. (Eds.) (2014). XI. Humedales de la Orinoquia (Colombia- Venezuela) Bogotá D. C. Colombia: Instituto de Investigación de Recursos Biológicos Alexander von Humboldt (IAvH).
Lasso C. A., Trujillo F., Morales- Betancourt M. A. (Eds.) (2020). VIII. Biodiversidad de la Reserva Natural Bojonawi, ViChada, Colombia: río Orinoco y planicie de inundación. Bogotá, D. C., Colombia: Serie Editorial Fauna Silvestre Neotropical (Bogotá D. C., Colombia: Instituto de Investigación de Recursos Biológicos Alexandervon Humboldt).
Lasso C. A., Usma J. S., Trujillo F., Rial A. (Eds.) (2010). Biodiversidad de la cuenca del Orinoco: bases científicas para la identificación de áreas prioritarias para la conservación y uso sostenible de la biodiversidad. (Bogotá, D. C., Colombia: Instituto de Investigación de Recursos Biológicos Alexander von Humboldt, WWF Colombia, Fundación Omacha, Fundación La Salle, El Instituto de Estudios de la Orinoquia (Universidad Nacional de Colombia)).
Lewis R. J., de Bello F., Bennett J. A., Fibich P., Finerty G. E., Götzenberger L., et al. (2017). Applying the dark diversity concept to nature conservation. Conserv. Biol. 31, 40–47. doi: 10.1111/cobi.12723
Li W. P., Liu Z. F., Guo T., Chen H., Xie X. (2021). Using optimal environmental DNA method to improve the fish diversity survey—From laboratory to aquatic life reserve. Water 13, 1468. doi: 10.3390/w13111468
Lin M., Simons A. L., Harrigan R. J., Curd E. E., Schneider F. D., Ruiz-Ramos D. V., et al. (2021). Landscape analyses using eDNA metabarcoding and Earth observation predict community biodiversity in California. Ecol. Appl. 31, e02379. doi: 10.1002/eap.2379
Loeza-Quintana T., Crookes S., Li P. Y., Reid D. P., Smith M., Hanner R. H. (2021). Environmental DNA detection of endangered and invasive species in Kejimkujik National Park and Historic Site. Genome 64, 172–180. doi: 10.1139/gen-2020-0042
Lozano Mojica J. D., Caballero S. (2021). Applications of eDNA metabarcoding for vertebrate diversity studies in northern Colombian water bodies. Front. Ecol. Evol. 8, 617948. doi: 10.3389/fevo.2020.617948
Lyet A., Pellissier L., Valentini A., Dejean T., Hehmeyer A., Naidoo R. (2021). eDNA sampled from stream networks correlates with camera trap detection rates of terrestrial mammals. Sci. Rep. 11, 1–14. doi: 10.1038/s41598-021-90598-5
MacDonald A. J., Sarre S. D. (2017). A framework for developing and validating taxon-specific primers for specimen identification from environmental DNA. Mol. Ecol. Resour. 17, 708–720. doi: 10.1111/1755-0998.12618
Magurran A. E. (1988). Ecological Diversity and Its Measurements (Princeton, USA: Princeton University Press). doi: 10.1007/978–94-015–7358-0
Marques V., Castagné P., Polanco A., Borrero-Pérez G. H., Hocdé R., Guérin P.É., et al. (2021). Use of environmental DNA in assessment of fish functional and phylogenetic diversity. Conserv. Biol. 35, 1944–1956. doi: 10.1111/cobi.13802
Martin M. (2011). Cutadapt removes adapter sequences from highthroughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Martinelli-Marín D., Lasso C. A., Caballero-Gaitán S. (2020). “Diversidad y riqueza de vertebrados de la Reserva Natural Bojonawi y áreas adyacentes (Orinoquia, Colombia), estimadas a partir de análisis de ADN ambiental,” in VIII. Biodiversidad de la Reserva Natural Bojonawi, ViChada, Colombia: río Orinoco y planicie de inundación. Eds. Lasso C. A., Trujillo F., Morales-Betancourt M. A. (Serie Editorial Fauna Silvestre Neotropical. Instituto de Investigación de Recursos Biológicos Alexander von Humboldt, Bogotá D.C., Colombia), 345–369.
Mas-Carrió E., Schneider J., Nasanbat B., Ravchig S., Buxton M., Nyamukondiwa C., et al. (2022). Assessing environmental DNA metabarcoding and camera trap surveys as complementary tools for biomonitoring of remote desert water bodies. Environ. DNA 4, 580–595. doi: 10.1002/edn3.274
Mathon L., Marques V., Mouillot D., Albouy C., Andrello M., Baletaud F., et al. (2022). Cross-ocean patterns and processes in fish biodiversity on coral reefs through the lens of eDNA metabarcoding. Proc. R. Soc. B 289, 20220162. doi: 10.1098/rspb.2022.0162
Mathon L., Valentini A., Guérin P. E., Normandeau E., Noel C., Lionnet C., et al. (2021). Benchmarking bioinformatic tools for fast and accurate eDNA metabarcoding species identification. Mol. Ecol. Resour. 21, 2565–2579. doi: 10.1111/1755-0998.13430
Mena L. J., Yagui H., Tejeda V., Bonifaz E., Bellemain E., Valentini A., et al. (2021). Environmental DNA metabarcoding as a useful tool for evaluating terrestrial mammal diversity in tropical forests. Ecol. Appl. 31, 1–13. doi: 10.1002/eap.2335
Méndez-López A., Sánchez-Garcés G. C., Jiménez J. C., Alfonso J., Lomelín J. C., Beltrán B., et al. (2019). “Peces,” in Parte 3. Resultados adicionales de la salida de campo caño Yarumales-río Guayabero. Informe final, Convenio N° 19–048. Eds. Lasso C. A., Morales-Betancourt M. A. (Bogotá, D. C., Colombia: Instituto de Investigación de Recursos Biológicos Alexander von Humboldt, Corporación para el Desarrollo Sostenible del Área de Manejo Especial La Macarena-Cormacarena), 20–30.
Milan D. T., Mendes I. S., Damasceno J. S., Teixeira D. F., Sales N. G., Carvalho D. C. (2020). New 12S metabarcoding primers for enhanced Neotropical freshwater fish biodiversity assessment. Sci. Rep. 10, 1–12. doi: 10.1038/s41598-020-74902-3
Ministry of Environment and Sustainable Development (2017). Biodiversity Action Plan (BAP) For the implementation of the National Policy for the Integral Management of Biodiversity and its Ecosystem Services 2016- 2030. Available online at: https://www.cbd.int/doc/world/co/co-nbsap-v3-en.pdf.
Ministry of Environment and Sustainable Development (2019). Colombia, el segundo país más biodiverso del mundo, celebra el Día Mundial de la Biodiversidad (Bogotá D.C., Colombia: Minambiente). Available at: https://www.minambiente.gov.co/index.php.
Miranda F., Bertassoni A., Abba A. M. (2014). Myrmecophaga tridactyla. The IUCN Red List of Threatened Species 2014: e.T14224A47441961. doi: 10.2305/IUCN.UK.2014–1.RLTS.T14224A47441961.en
Moeslund J. E., Brunbjerg A. K., Clausen K. K., Dalby L., Fløjgaard C., Juel A., et al. (2017). Using dark diversity and plant characteristics to guide conservation and restoration. J. Appl. Ecol. 54, 1730–1741. doi: 10.1111/1365-2664.12867
Mojica J. I., Usma J. S., Álvarez R., Lasso C. A. (Eds.) (2012). Libro rojo de peces dulceacuícolas de Colombia 2012 (Bogotá, D. C., Colombia: ARFO - Arte y Fotolito).
Mora A., Baby P., Roddaz M., Parra M., Brusset S., Hermoza W., et al. (2010). “Tectonic history of the Andes and sub-Andean zones: implications for the development of the Amazon drainage basin,” in Amazonia, landscape and species evolution: a look into the past (Oxford, UK: Blackwell Publishing), 38–60.
Morales-Betancourt M. A., Lasso C. A. (2021). Estudio de delfín de río (Inia geoffrensis), la nutria gigante (Pteronura brasiliensis) y sus interrelaciones con los otros recursos hidrobiológicos y humedales del río Tillavá, Meta. Informe final, Convenio N° 20–264 (Bogotá, D. C., Colombia: Instituto de Investigación de Recursos Biológicos Alexander von Humboldt, Corporación para el Desarrollo Sostenible del Área de Manejo Especial La Macarena-Cormacarena, Fundación Omacha).
Mosquera F., Trujillo F., Diaz M. C., Mantilla H. (2015). Estimación poblacional y densidad para Inia geoffrensis y Sotalia fluviatilis en los ecosistemas acuáticos de la Amazonia y Orinoquia Colombiana. Momentos de Ciencia 12, 93–99. Available at: https://core.ac.uk/download/288215134.pdf.
Mott C. L. (2010). Environmental constraints to the geographic expansion of plant and animal species. Nat. Educ. Knowledge 3, 72. Available at: https://www.nature.com/scitable/knowledge/library/environmental-constraints-to-the-geographic-expansion-of-13236052/.
Moyer G. R., Winemeller K. O., McPhee M. V., Turner T. F. (2005). Historical demography, selection, and coalescence of mitochondrial and nuclear genes in Prochilodus species of northern South America. Evolution 59, 599–610.
Nakagawa H., Yamamoto S., Sato Y., Sado T., Minamoto T., Miya M. (2018). Comparing local-and regional-scale estimations of the diversity of stream fish using eDNA metabarcoding and conventional observation methods. Freshw. Biol. 63, 569–580. doi: 10.1111/fwb.13094
NatureMetrics. (2019). Available at: https://www.naturemetrics.com/species-detection. [Accessed August, 2020].
Neice A. A., McRae S. B. (2021). An eDNA diagnostic test to detect a rare, secretive marsh bird. Global Ecol. Conserv. 27, e01529. doi: 10.1016/j.gecco.2021.e01529
Nichols R. V., Königsson H., Danell K., Spong G. (2012). Browsed twig environmental DNA: diagnostic PCR to identify ungulate species. Mol. Ecol. Resour. 12, 983–989. doi: 10.1111/j.1755-0998.2012.03172.x
Ota W. M., Hall C., Malloy J., Clark M. A. (2020). Environmental DNA monitoring: better tracking of endangered, rare, cryptic, and invasive species. J. Sci. Policy Governance 17, 1–8. doi: 10.38126/JSPG170117
Pärtel M., Szava-Kovats R., Zobel M. (2011). Dark diversity: shedding light on absent species. Trends Ecol. Evol. 26, 124–128. doi: 10.1016/j.tree.2010.12.004
Pawlowski J., Bonin A., Boyer F., Cordier T., Taberlet P. (2021). Environmental DNA for biomonitoring. Mol. Ecol. 30, 2931–2936. doi: 10.1111/mec.16023
Peters L., Spatharis S., Dario M. A., Dwyer T., Roca I. J., Kintner A., et al. (2018). Environmental DNA: a new low-cost monitoring tool for pathogens in salmonid aquaculture. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.03009
Pineda L., Herrera N., Ibarra R. (2008). Contribuciones a la biología de la Pava negra (Penelopina nigra) en El Salvador. Zeledonia 12, 20–24. Available at: https://dialnet.unirioja.es/servlet/articulo?codigo=4041975.
Plough L. V., Bunch A. J., Lee B. B., Fitzgerald C. L., Stence C. P., Richardson B. (2021). Development and testing of an environmental DNA (eDNA) assay for endangered Atlantic sturgeon to assess its potential as a monitoring and management tool. Environ. DNA 3, 800–814. doi: 10.1002/edn3.186
Polanco A., Marques V., Fopp F., Juhel J. B., Borrero-Pérez G. H., Cheutin M. C., et al. (2021b). Comparing environmental DNA metabarcoding and underwater visual census to monitor tropical reef fishes. Environ. DNA 3, 142–156. doi: 10.1002/edn3.140
Polanco A., Mutis M., Marques V., Villa-Navarro F., Borrero Pérez G. H., Cheutin M., et al. (2021c). Detecting aquatic and terrestrial biodiversity in a tropical estuary using environmental DNA. Biotropica 53, 1606–1619. doi: 10.1111/btp.13009
Polanco A., Richards E., Flück B., Valentini A., Altermatt F., Brosse S., et al. (2021a). Comparing the performance of 12S mitochondrial primers for fish environmental DNA across ecosystems. Environ. DNA 3, 1113–1127. doi: 10.1002/edn3.232
Portocarrero-Aya M. (2011). Conservation of freshwater biodiversity in key areas of the Colombian Amazon [Thesis]. Hull (UK): University of Hull.
Prieto A., Arias J. C. (2007). Diversidad biológica del sur de la Amazonía Colombiana (Bogotá, Colombia: Diversidad biológica y cultural del sur de la Amazonía Colombiana– Diagnóstico. Corpoamazonía, Instituto Humboldt, Instituto Sinchi, UAESPNN), 73–197.
Rees H. C., Maddison B. C., Middleditch D. J., Patmore J. R., Gough K. C. (2014). The detection of aquatic animal species using environmental DNA–a review of eDNA as a survey tool in ecology. J. Appl. Ecol. 51, 1450–1459. doi: 10.1111/1365-2664.12306
Renno J. F., Carvajal F., Pablo J., Sirvas S., Bonhomme F., Desmarais E., et al. (2011). “BIODIVERSIDAD Y EVOLUCIÓN DE LOS PECES EN AMAZONÍA”. En Biologıía de las Poblaciones de Peces Amazónicos y Piscicultura, II COLOQUIO DE LA RED DE INVESTIGACIÓN SOBRE LA ICTIOFAUNA AMAZÓNICA (RIIA). Manaus, Brasil.
Riaz T., Shehzad W., Viari A., Pompanon F., Taberlet P., Coissac E. (2011). ecoPrimers: inference of new DNA barcode markers from whole genome sequence analysis. Nucleic Acids Res. 39, e145–e145. doi: 10.1093/nar/gkr732
Rizo-Fuentes M. A., Correa C. A., Lasso C. A., Morales-Betancourt M. A., Barragán D. C., Caballero S. (2021). Phylogeography, genetic diversity and population structure of the freshwater stingray, Paratrygon aiereba (Müller & Henle 1841)(Myliobatiformes: potamotrygonidae) in the Colombian amazon and orinoco basins. Mitochondrial DNA Part A 32, 20–33. doi: 10.1080/24701394.2020.1844679
RStudio Team. (2020). RStudio: Integrated Development for R. RStudio Team. Available at: http://www.rstudio.com/.
Rourke M. L., Fowler A. M., Hughes J. M., Broadhurst M. K., DiBattista J. D., Fielder S., et al. (2022). Environmental DNA (eDNA) as a tool for assessing fish biomass: A review of approaches and future considerations for resource surveys. Environ. DNA 4, 9–33. doi: 10.1002/edn3.185
Ruiz S. L., Sánchez E., Tabares E., Prieto A., Arias J. C., Gómez R., Castellanos D., García P., Rodríguez L. (Eds.) (2007). Diversidad biológica y cultural del sur de la Amazonia Colombiana – Diagnóstico (Bogotá D. C., Colombia: Ramos López Editorial Fotomecánica Ltda).
Şen B., Grillo O. (2018). “Methods for Biodiversity Assessment: Case Study on an Area of Atlantic Forest in Southern Brazil,” in Selected Studies in Biodiversity (London, UK: IntechOpen), 45–58. doi: 10.5772/66032
Saito T., Doi H. (2021). A model and simulation of the influence of temperature and amplicon length on environmental DNA degradation rates: A meta-analysis approach. Front. Ecol. Evol. 9. doi: 10.3389/fevo.2021.623831
Sales N. G., Kaizer M. D. C., Coscia I., Perkins J. C., Highlands A., Boubli J. P., et al. (2020a). Assessing the potential of environmental DNA metabarcoding for monitoring Neotropical mammals: a case study in the Amazon and Atlantic Forest, Brazil. Mammal Rev. 50, 221–225. doi: 10.1111/mam.12183
Sales N. G., McKenzie M. B., Drake J., Harper L. R., Browett S. S., Coscia I., et al. (2020b). Fishing for mammals: Landscape-level monitoring of terrestrial and semi-aquatic communities using eDNA from riverine systems. J. Appl. Ecol. 57, 707–716.
Salinas Y., Agudelo E. (2000). Peces de importancia económica en la cuenca amazónica Colombiana Vol. 1 (Bogotá, D. C., Colombia: Instituto Amazónico de Investigaciones Cientı́fica, SINCHI. Ministerio del Medio Ambiente).
Sánchéz J. N., Bustamante Y., Losada-Prado S. (2019). “Aves,” in Parte 3. Resultados adicionales de la salida de campo caño Yarumales-río Guayabero. Informe final, Convenio N° 19- 048. Eds. Lasso C. A., Morales-Betancourt M. A. (Bogotá, D. C., Colombia: Instituto de Investigación de Recursos Biológicos Alexander von Humboldt, Corporación para el Desarrollo Sostenible del Área de Manejo Especial La Macarena-Cormacarena), 31–52.
Sard N. M., Herbst S. J., Nathan L., Uhrig G., Kanefsky J., Robinson J. D., et al. (2019). Comparison of fish detections, community diversity, and relative abundance using environmental DNA metabarcoding and traditional gears. Environ. DNA 1, 368–384. doi: 10.1002/edn3.38
Sasso T., Lopes C. M., Valentini A., Dejean T., Zamudio K. R., Haddad C. F., et al. (2017). Environmental DNA characterization of amphibian communities in the Brazilian Atlantic forest: potential application for conservation of a rich and threatened fauna. Biol. Conserv. 215, 225–232. doi: 10.1016/j.biocon.2017.09.015
Seymour M. (2019). Rapid progression and future of environmental DNA research. Commun. Biol. 2, 1–3. doi: 10.1038/s42003-019-0330-9
Sistema de Información Ambiental de Colombia-SIAC (2020). Biodiversidad en Colombia (Bogotá, CO: IDEAM). Available at: http://www.siac.gov.co/biodiversidad.
Somervuo P., Koskela S., Pennanen J., Henrik Nilsson R., Ovaskainen O. (2016). Unbiased probabilistic taxonomic classification for DNA barcoding. Bioinformatics 32, 2920–2927. doi: 10.1093/bioinformatics/btw346
Spens J., Evans A. R., Halfmaerten D., Knudsen S. W., Sengupta M. E., Mak S. S., et al. (2016). Comparison of capture and storage methods for aqueous macrobial eDNA using an optimized extraction protocol: advantage of enclosed filter. Methods Ecol. Evol. 8, 635–645. doi: 10.1111/2041-210X.12683
Stauffer S., Jucker M., Keggin T., Marques V., Andrello M., Bessudo S., et al. (2021). How many replicates to accurately estimate fish biodiversity using environmental DNA on coral reefs? Ecol. Evol. 11, 14630–14643. doi: 10.1101/2021.05.26.445742
Stevenson P. R., Defler T. R., de la Torre S., Moscoso P., Palacios E., Ravetta A. L., et al. (2021). Lagothrix lagothricha (amended version of 2020 assessment). The IUCN Red List of Threatened Species 2021: e.T160881218A192309103. doi: 10.2305/IUCN.UK.2021-1.RLTS.T160881218A192309103.en. Accessed on 20 June 2024.
Suring H. L. (2020). “Freshwater: Oasis of Life—An Overview,” in Encyclopedia of the World’s Biomes. Eds. Goldstein I. M., DellaSala A. D. (Amsterdam: Elsevier Academic Press), 1–11.
Thomsen P. F., Kielgast J., Iversen L. L., Møller P. R., Rasmussen M., Willerslev E. (2012). Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PloS One 7, 1–9. doi: 10.1371/journal.pone.0041732
Thomsen P. F., Willerslev E. (2015). Environmental DNA–An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 183, 4–18. doi: 10.1016/j.biocon.2014.11.019
Tickner D., Opperman J. J., Abell R., Acreman M., Arthington A. H., Bunn S. E., et al. (2020). Bending the curve of global freshwater biodiversity loss: an emergency recovery plan. BioScience 70, 330–342. doi: 10.1093/biosci/biaa002
Tortoise & Freshwater Turtle Specialist Group. (1996). Peltocephalus dumerilianus (errata version published in 2016). The IUCN Red List of Threatened Species 1996: e.T16511A97397017. doi: 10.2305/IUCN.UK.1996.RLTS.T16511A5972664.en
Universidad Nacional de Colombia (2013). Desmovilización y Reintegración (ODDR). Available online at: https://www.humanas.unal.edu.co/observapazyconflicto/files/4614/3144/5526/caracterizacion_de_la_orinoquia.pdf.
Urbano-Bonilla A., Mojica J. I., Agudelo-Córdoba E., Maldonado-Ocampo J. (2014). “Diversidad íctica del sistema de lagos de Tarapoto,” in Los humedales de Tarapoto: aportes al conocimiento sobre su biodiversidad y uso. Serie humedales de la Amazonia y Orinoquia. (Bogotá, D. C., Colombia: Fundación Omacha, Corpoamazonia, Universidad Nacional Sede Leticia), 156–257.
Ushio M., Murata K., Sado T., Nishiumi I., Takeshita M., Iwasaki W., et al. (2018). Demonstration of the potential of environmental DNA as a tool for the detection of avian species. Sci. Rep. 8, 1–10. doi: 10.1038/s41598-018-22817-5
Valentini A., Taberlet P., Miaud C., Civade R., Herder J., Thomsen P. F., et al. (2016). Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Mol. Ecol. 25, 929–942. doi: 10.1111/mec.13428
Varela D., Flesher K., Cartes J. L., de Bustos S., Chalukian S., Ayala G., et al. (2019). Tapirus terrestris. The IUCN Red List of Threatened Species 2019: e.T21474A45174127. doi: 10.2305/IUCN.UK.2019–1.RLTS.T21474A45174127.en
Vargas-Ramírez M., Caballero S., Morales-Betancourt M. A., Lasso C. A., Amaya L., Fritz U. (2020). Genomic analyses reveal two species of the matamata (Testudines: Chelidae: Chelus spp.) and clarify their phylogeography. Mol. Phylogenet. Evol. 148, 106823. doi: 10.1016/j.ympev.2020.106823
Villa-Navarro A. F., Lopez O. E., Albornoz G. J., Montoya D., Taphorn C. D., DoNascimiento C., et al. (2017). “Peces,” in IV. Biodiversidad del río Bita. Eds. Trujillo F., Lasso C. A. (ViChada, Colombia. Serie Editorial Fauna Silvestre Neotropical. Instituto de Investigación de Recursos Biológicos Alexander von Humboldt (IAvH, Bogotá, D. C., Colombia), 169–240.
Wallis C. I., Tiede Y. C., Beck E., Böhning-Gaese K., Brandl R., Donoso D. A., et al. (2021). Biodiversity and ecosystem functions depend on environmental conditions and resources rather than the geodiversity of a tropical biodiversity hotspot. Sci. Rep. 11, 1–14. doi: 10.1038/s41598-021-03488-1
Weltz K., Lyle J. M., Ovenden J., Morgan J. A., Moreno D. A., Semmens J. M. (2017). Application of environmental DNA to detect an endangered marine skate species in the wild. PloS One 12, e0178124. doi: 10.1371/journal.pone.0178124
Wiersma G. A. (2019). Statistical learning methods for environmental DNA (Groningen, Netherlands: University of Groningen). Available at: https://fse.studenttheses.ub.rug.nl/19753/1/mAppM_2019_WiersmaAG.pdf.
Wilson J. J., Sing K. W., Chen P. N., Zieritz A. (2017). Tracking the southern river terrapin (Batagur affinis) through environmental DNA: prospects and challenges. Mitochondrial DNA Part A 29, 862–866. doi: 10.1080/24701394.2017.1373109
Wrona F., Reist D. J. (2013). “Freshwater Ecosystems,” in Arctic Biodiversity Assesment (Denmark: Narayana Press), 442–485.
WWF Colombia. (2020). 3 cosas que tal vez no sabías sobre la Orinoquia. Available online at: https://www.wwf.org.co/?uNewsID=360852.
Yang L., Tan Z., Wang D., Xue L., Guan M. X., Huang T., et al. (2014). Species identification through mitochondrial rRNA genetic analysis. Sci. Rep. 4, 1–11. doi: 10.1038/srep04089
Keywords: eDNA metabarcoding, diversity, richness, vertebrates, Amazon basin, Orinoco basin, Colombia
Citation: Martinelli Marín D, Lasso CA and Caballero Gaitan SJ (2024) eDNA metabarcoding: an effective tool for vertebrate diversity studies in the Colombian Amazon and Orinoco basins. Front. Ecol. Evol. 12:1409296. doi: 10.3389/fevo.2024.1409296
Received: 29 March 2024; Accepted: 14 June 2024;
Published: 22 July 2024.
Edited by:
Marco Girardello, Joint Research Centre, ItalyReviewed by:
Elena Buzan, University of Primorska, SloveniaChristopher L. Jerde, University of California, Santa Barbara, United States
Copyright © 2024 Martinelli Marín, Lasso and Caballero Gaitan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniela Martinelli Marín, ZGFuaWVsYW1hcnRpbmVsbGltYXJpbkBnbWFpbC5jb20=
†Present address: Susana J. Caballero Gaitan, Department of Marine and Environmental Sciences, Halmos College of Arts and Sciences, Nova Southeastern University, Fort Lauderdale, FL, United States
 Daniela Martinelli Marín
Daniela Martinelli Marín Carlos A. Lasso2
Carlos A. Lasso2 Susana J. Caballero Gaitan
Susana J. Caballero Gaitan