
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 03 May 2024
Sec. Evolutionary Developmental Biology
Volume 12 - 2024 | https://doi.org/10.3389/fevo.2024.1385516
This article is part of the Research Topic Tunicates in Evolutionary Developmental Biology View all 12 articles
Lumen formation, as a key process of biological tube construction, is essential in various physiological processes such as nutrient and waste transporting, gas exchanging, and structural supporting. However, the mechanisms underlying tubular lumen development are still not fully understood. In the present study, we identified a matrix metalloproteinase, Nas15, which is enriched in the apical domain of the Ciona embryonic notochord. The expression level of the Nas15 gene significantly increased during notochord lumen formation and expansion. Nas15 loss-of-function resulted in abnormal notochord lumen expansion in Ciona embryos. Besides, yeast two-hybrid screening and CO-IP results indicated a Phosphatase 2 Catalytic Subunit Alpha (PPP2CA) physically interacted with Nas15. PPP2CA also involved in notochord lumen formation via localizing Nas15. Furthermore, we investigated the distribution of laminin in Nas15 disrupted embryos. In conclusion, our results revealed a mechanisms of how notochord cells regulating lumen expansion via metalloproteinase-mediated ECM localization. This findings provide insight into the mechanisms of tubular organ lumen formation and serve as a reference for research on human abnormal lumenogenesis diseases.
Tubular structures play essential roles in various organs, such as the lung, kidney, intestine, and vasculature. Due to their unique structure, biological tubes are responsible for transporting and absorbing materials (Lubarsky and Krasnow, 2003). For instance, arteries facilitate the delivery of blood throughout the body and organs, while the intestinal tube is responsible for nutrient absorption. Recent research suggests that tube structures also participate in regulating the mechanical properties of tissues (Yasuoka, 2020). In the case of the notochord, the pressure within the notochord lumen is controlled by the secretion of matrix components from notochord cells, which contributes to overall tissue stiffness regulation (Yasuoka, 2020). The vacuole of the notochord is an evolutionally conserved structure, from Amphioxus, Ciona to vertebrate (Annona et al., 2015). The proper formation of tube lumens is crucial, as any abnormalities in size or shape will lead to various diseases. For example, polycystic kidney disease (Simons and Walz, 2006) and atherosclerotic heart disease (Sarraju and Nissen, 2024) have been identified to be associated with abnormal lumen formation. Tubulogenesis is a critical process in many physiological activities, yet the underlying mechanism of lumen formation remains unclear. Multiple cellular behaviors have been identified as involved in lumen construction, including wrapping, budding, cavitation, cord vacuolation, and cell vacuolation (Lubarsky and Krasnow, 2003). Ciona notochord constructs an extracellular lumen via a complicated cell behavior, including cell intercalation, elongation, lumen initial formation/expansion, and bi-directional migration (Dong et al., 2011; Lu et al., 2019), resulting in a flattened endothelial cells covered, connected extracellular lumen, a process known as mesenchymal-epithelial transition (Dong et al., 2009; Ouyang et al., 2023).
Regarding the regulatory mechanism of lumen localization and size, numerous studies have indicated that the secretion of extracellular matrix (ECM) by Ciona notochord cells plays a crucial role in the feedback regulation of notochord morphogenesis. The ECM of Ciona notochord comprises two distinct types: the basal ECM (notochord sheath) and the apical ECM (Wei et al., 2017). The notochord sheath refers to a layer of basal lamina that covers the surface of the notochord, composed of various components such as collagen, laminin, fibronectin, and other components (Veeman et al., 2008; Segade et al., 2016; Peng et al., 2023). These ECM components are indispensable for notochord morphogenesis. Fibronectin contributes to notochord intercalation (Segade et al., 2016), whereas laminin is involved in notochord boundary formation and convergence extension (Veeman et al., 2008). The collagen surrounding the notochord forms a supracellular arch, facilitating notochord elongation and coordinated movement with multiple tissues (Peng et al., 2023). In addition to the basal notochord sheath, the apical ECM secreted into the lumen also plays a vital role in the initial construction and expansion of the lumen. Extensive research has demonstrated that vesicular trafficking is a critical factor driving lumen expansion (Dong et al., 2011; Bhattachan et al., 2020). The 14-3-3 epsilon protein mediates material transport from the basal membrane to the apical side, which is essential for lumen expansion (Mizotani et al., 2018).
Tissue morphogenesis is a highly dynamic process, necessitating constant reshaping, rebuilding, and recycling of the ECM to maintain an appropriate quantity and quality conducive to development (Bonnans et al., 2014). The proteomic analysis of Ciona notochord revealed a diverse array of vesicle transport-related proteins and ECM components (Wang et al., 2023). The DYRK1-endophilin-mediated endocytosis occurs actively in close proximity to the apical membrane of the Ciona notochord, thereby implicating the role of ECM dynamic turnover in lumen formation (Ouyang et al., 2023), which highlights the significance of the continuous remodeling of ECM in the process of lumen formation.
So, an interesting question of ECM mediated morphogenesis is how the cells regulate the ECM to feedback influence the tissue reshaping. Matrix metalloproteinase (MMPs) have been identified as key players in this process. MMPs constitute a family of calcium-dependent zinc-containing endopeptidases (Verma and Hansch, 2007) that facilitate the remodeling and degradation of various ECM components (Nagase et al., 2006). Notably, some studies have suggested the involvement of membrane-type matrix metalloproteinase in vascular lumen formation through its regulation of ECM remodeling. These findings shed light on the intricate interplay between cells and the ECM during tissue morphogenesis (Senger and Davis, 2011; Sacharidou et al., 2012).
However, the specific involvement of MMPs in apical ECM remodeling and their regulatory role in lumen formation in the Ciona notochord remains elusive. Within the astacin metalloproteinase family, nematode astacins-15 (Nas-15) emerges as a potential candidate component involved in Ciona notochord lumen formation, which has been identified in proteomic analysis of Ciona notochord (Wang et al., 2023). Previous studies have demonstrated the participation of astacin metalloproteinases in diverse developmental processes, including food digestion, early embryonic development, tissue remodeling, and differentiation (Baumann et al., 1993; Sterchi et al., 2008; Park et al., 2010; Calabria et al., 2019). By analyzing single-cell transcriptome data in the Single Cell Portal (SCP) database, it was observed that the homologous protein of Nas-15 in Ciona robusta (Cr-Nas15) exhibits high expression levels during the stages of lumen formation in the notochord of Ciona embryos (Tarhan et al., 2023). Moreover, the Ghost database revealed specific enrichment of Cr-Nas15 mRNA surrounding the apical membrane of the notochord lumen (Kusakabe et al., 2002; Satou et al., 2005). Taken together, these clues suggest that Cr-Nas15 is a promising candidate MMP involved in Ciona notochord lumen formation.
In this study, we initially identified Cr-Nas15, which features a typical ZnMc-astacin-like domain and Zinc-binding metalloprotease motif. Subsequently, using qPCR, promoter analysis, and immunofluorescence techniques, we confirmed the specific localization of Cr-Nas15 at the apical domain of notochord cells during the late tail bud stage of Ciona embryos, which overlaps spatially and temporally with notochord lumen formation. We then investigated the functional role of Cr-Nas15 in this process. Treatment with the inhibitor actinonin, overexpression of a dominant negative version of Nas-15, and tissue-specific knock-out experiments all resulted in similar lumen formation failure phenotypes, further supporting the involvement of Cr-Nas15 in Ciona notochord lumen formation. Additionally, we demonstrated that Protein Phosphatase 2 Catalytic Subunit Alpha (PPP2CA) interacts with Nas15, and the PPP2CA is also essential for lumen formation in Ciona notochord.
Adult Ciona robusta was collected from Rongcheng and Qingdao in Shandong Province, China. Following collection, the animals were temporarily housed in laboratory artificial seawater tanks, which were carefully maintained at a temperature range of 17°C to 19°C and a salinity level of 30-32‰. Sperm and eggs were obtained separately from different adult Ciona individuals. The sperm and eggs were mixed in sea water over 5 minutes for fertilization. Then, the fertilized eggs underwent a dechorionation process before proceeding with electroporation. The electroporation method for Ciona embryos was performed based on previously reported techniques (Stolfi and Christiaen, 2012). 40 µg of each plasmid was dissolved in sterile ddH2O, reaching a total volume of 80 µl. This plasmid solution was then mixed with 420 µl of electroporation buffer. The resultant mixture was transferred into a 4 mm electroporation cuvette. Subsequently, 300 µl of sea water together with dechorionated Ciona fertilized eggs were added to the cuvette. Electroporation was carried out using a pulse generator, employing a voltage of 50V and a capacitance of 1500/2000 µF. Following the electroporation step, the embryos were placed into a constant temperature incubator set at 16°C for further incubation.
In the promoter analysis experiment, the upstream 3 kb DNA sequence of Nas15 was amplified by PCR using genomic DNA from C. robusta as a template with primers Nas15 (3 k) -F and Nas15 (3k) -R. The PCR product was then inserted into the pEGFP-N1 vector to construct the Cr-Nas15 (3 kb) > GFP fusion construct.
The Ciona notochord specific expressed vector was constructed via replacing the CMV promoter of pEGFP-N1 with Ciona savignyi (Cs) brachyury upsteam 3kb promoter. Using C. robusta cDNA as a template, full-length coding sequences (CDS) of Nas15 and PPP2CA were amplified via PCR with primers Bra > Nas15-F and Bra > Nas15-R and Bra>PPP2CA-F and Bra>PPP2CA-R, respectively. The PCR products were then inserted into Brachyury (3 kb) > EGFP-N1 vector, leading to the construction of the expression plasmids Brachyury (3 kb) > Nas15-EGFP and Brachyury (3 kb) > PPP2CA-EGFP, and the construction of mutant plasmids were generated introducing mutated bases for overexpression experiments. Further, the PCR product was cloned into the linearized vectors CMV > GFP and CMV > HA, generating the expression plasmids CMV > Nas15-GFP and CMV > PPP2CA-HA for cell experiments. Additionally, the PCR product was cloned into the linearized vectors pGBKT7 and pGADT7 to construct the expression plasmids pGBKT7-Nas15 and pGADT7-PPP2CA for yeast two-hybrid assays. Finally, the CDS of Nas15 was ligated to the pET-30a vector for prokaryotic expression of Nas15 protein.
The ClonExpress II one-step cloning kit (Vazyme, Nanjing, China) was utilized to generate all constructs, and their accuracy was verified through sequencing analysis. All the PCR primers are listed in the Supplementary Table S1.
The qPCR was performed based on the previously reported techniques (Livak and Schmittgen, 2001). Nas15 qPCR primers were designed using Beacon Designer 7 software. Total RNA was extracted from various developmental stages (8, 12, 13, 15, 16, 17, 20, 22, and 24 hours post fertilization, hpf) of C. robusta embryos/larvae using RNAiso plus (TAKARA, Japan). The extracted RNA was reverse transcribed into cDNA using the HiScript II Q RT SuperMix for qPCR kit (Novogene, Nanjing, China). Quantitative PCR amplification was performed using the ChamQTM SYBR Color qPCR Master Mix kit (Novogene, Nanjing, China). Data analysis was carried out using the 2−ΔΔCt method, and graphical representation was created using Excel software. The primers are listed in the Supplementary Table S1.
23 hpf (late tailbud stage) Ciona embryos were collected and lysed in loading buffer at 100°C for 10 minutes to denature the proteins. The samples were then loaded onto an SDS-PAGE gel for electrophoresis. After electrophoresis, the protein was transferred to a polyvinylidene fluoride (PVDF) membrane. Following membrane transfer, the PVDF membrane was blocked with 5% skimmed milk at room temperature for 2 hours. Primary antibody (Nas15 1:300) was diluted in 10% goat serum and incubated with PVDF membrane for overnight at 4°C. After completion of the primary antibody incubation, the PVDF membrane was washed three times with 0.05% TBST solution for 15 minutes each time. Secondary antibody Goat Anti-Mouse IgG, HRP Conjugate (TransGen, 1:2000, HS201–01) was diluted in 10% goat serum and incubated with PVDF membrane at room temperature for 2 hours. After the secondary antibody incubation, the PVDF membrane was washed three times with 0.05% TBST solution for 15 minutes each time. Once the washing was completed, the membrane was subjected to staining and photographed.
Embryos were fixed with 4% paraformaldehyde at room temperature for 2 hours, followed by permeabilization with PBS solution containing 0.1% Triton X-100 (permeabilization was performed three times for a total of 8 hours). Blocking was carried out with 10% goat serum at room temperature for 1 hour. Incubation with the primary antibody (Nas15 1:100) was performed overnight at 4°C. Washing was performed three times with PBS solution containing 0.1% Triton X-100 (a total of 8 hours). Incubation with Alexa Fluor 488-conjugated phalloidin (Invitrogen) and the secondary antibody Goat Anti-Mouse IgG, HRP Conjugate (TransGen, 1:2000, HS201–01) was carried out overnight at 4°C. Washing was performed three times with PBS solution containing 0.1% Triton X-100 (a total of 8 hours). Finally, the embryos were mounted with DAPI for confocal microscopy observation.
14 hpf (early tailbud stage) Ciona embryos was collected and cultured into the 48-well plate (about 200 embryos per well with 2 ml filtered seawater until 16 hpf (mid tailbud stage). Then, Actinonin dissolved in DMSO (MedChemExpress) was added to a final concentration of 1.2 μm in the inhibitor treated group, and the control group was added an equal amount of DMSO. Three repeated experiments were set for each group. Then, the embryos were cultured in 18°C until 23 hpf (late tailbud stage) and collected respectively. The embryos were fixed with 4% paraformaldehyde. Following the staining with phalloidin, the embryos were mounted with DAPI for confocal microscopy observation and imaging.
The genome editing in Ciona embryos by CRISPR/Cas9 was performed based on the previously reported techniques (Stolfi et al., 2014). Using the CRISPRdi-rect (http://crispr.dbcls.jp), three Nas15 sgRNA sequences were designed and subsequently sent to Shanghai Biotechnology Company for synthesis. Then, two single strands of sgRNAs were mixed respectively and subjected to a 5-minute treatment at 100°C to ensure complete denaturing, followed by a cooling to room temperature for annealing. Subsequently, the annealed primers were ligated to the linearized Cr-U6 > sgRNA (F+E) (Addgene number: 59 986) for sgRNAs transcription in vivo. Then the constructed sgRNA expression transcription vector was then used as a template for PCR amplification of the U6>sgRNA fragment. After that, the Cr-EF1α > NLS:: Cas9::NLS::P2A-mCherry plasmid and the sgRNA PCR product was co-electroporated into Ciona embyros. Then, the embryos were cultured in 18°C until 18hpf, and the embryos with red fluorescence were sorted and collected. Genomic DNA extracted from the collected embryos was used for efficiency test using T7 endonuclease I digestion. Additionally, the Cs-Brachyury (1 kb) > NLS::Cas9::NLS::P2A::mCherry and the sgRNA PCR product was co-electroporated into Ciona embryos for phenotype observation. The sequences of sgRNAs were listed in the Supplementary Table S2.
The yeast two-hybrid experiment was performed following the protocol provided by Matchmaker GAL4-based two-hybrid system (Clontech) (Hu et al., 2021).
HEK293T cells were cultured in DMEM medium supplemented with 10% FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin. The cells were maintained at 37°C in a 5% CO2 atmosphere. After reaching 80-90% confluency, the transfection was carried out using Lipofectamine 3000 Invitrogen (Thermo Fisher) according to the manufacturer’s instructions. 48 hours post-transfection, cells were collected and lysed in a buffer containing 50 mM Tris-HCl (pH 8), 75 mM NaCl, 1 mM MgCl2, 0.05% NP-40, 100 mM sucrose, 1 mM DTT, and 1x Protease Cocktail inhibitors (Roche) at 4°C for 30 minutes. Centrifuge at 12000 rpm for 10 minutes to remove cellular debris. Afterward, the supernatant was subjected to overnight incubation with GFP-tap beads (gta 20, chromotek, Germany) on a rotator at 4°C. The immunoprecipitated samples were washed three times with lysis buffer, and then directly boiled at 95°C for 10 minutes in 2×SDS loading buffer. Subsequently, these samples were loaded into 10% SDS-PAGE gels and separated via electrophoresis.
A phylogenetic tree for Nas15 was constructed using MEGA11 software (Tamura et al., 2021). The Conserved Domains Database tool provided by the NCBI website was utilized for protein domain prediction. Multiple sequence alignment was conducted using DNAMAN software (Lynnon Biosoft Bioinformatic Solutions, San Ramon, CA, USA), and the resulting alignments were visually represented. The diameter of the lumen was measured using the software ImageJ. All Significance statistical analysis was preformed using unpaired two-tailed t test. * represents p<0.05. ** represents p<0.01. *** represents p<0.001. **** represents p<0.0001. The colocalization of Nas15::tdTomato and PPP2CA::eGFP was analyzed with ImageJ. The phosphorylation sites in Nas15 were predicted using NetPhos - 3.1 (https://services.healthtech.dtu.dk/) website. The single cell sequencing data in different tissues and stages of Cr-Nas15 were downloaded from Single Cell Portal (SCP) database (https://singlecell.broadinstitute.org/single_cell/study/SCP454/comprehensive-single-cell-transcriptome-lineages-of-a-proto-vertebrate#study-summary ) (Tarhan et al., 2023). The in situ hybridization result of Cr-Nas15 were identified on Ghost database(http://ghost.zool.kyoto-u.ac.jp/cgi-bin/photogetkh.cgi?inkey=CLSTR07818&source=kh2013) (Kusakabe et al., 2002; Satou et al., 2005).
The sequence of Cr-Nas15 was downloaded from the Ghost database. The Cr-Nas15 is a 44.2 KDa protein and the in situ hybridization data demonstrated its expression in the larval notochord of Ciona (Kusakabe et al., 2002; Satou et al., 2005; Tassy et al., 2010). Meanwhile, the single-cell transcriptome sequencing data showed that the Cr-Nas15 was recognized in larval notochord cells at the lumen formation stage (Tarhan et al., 2023). To explore the evolution of Cr-Nas15, a group of Cr-Nas15 homologous sequences in other species was identified and then a phylogenetic tree constructed using full-length sequences revealed that Cr-Nas15 clustered with proteins from vertebrates, such as Homo sapiens MeprinA and Mus musculus MeprinA, suggesting its evolutionary conservation from invertebrates to chordates (Figure 1A). We also compared the Cr-Nas15 domain structure with homologous proteins in other species using the online tool NCBI Conserved Domain Search. The results showed that they both had conserved ZnMc-astacin-like domain and Zinc-binding metalloprotease motif (Figure 1B). Because of the key function of Zinc-binding metalloprotease motif, we analyzed the primary structure of this motif. The results showed that the Cr-Nas15 together with the homologous proteins in other species had a conserved motif HEXXHXXGXXH, which was the core structure for Zinc-binding metalloprotease catalytic activity (Bond and Beynon, 1995; Trevisan-Silva et al., 2010) (Figure 1C). In summary, the Cr-Nas15 sequence structure is evolutionary conserved including a typical ZnMc-astacin-like domain and Zinc-binding metalloprotease motif, suggesting that the Cr-Nas15 may has the similar catalytic activity with that in other species.
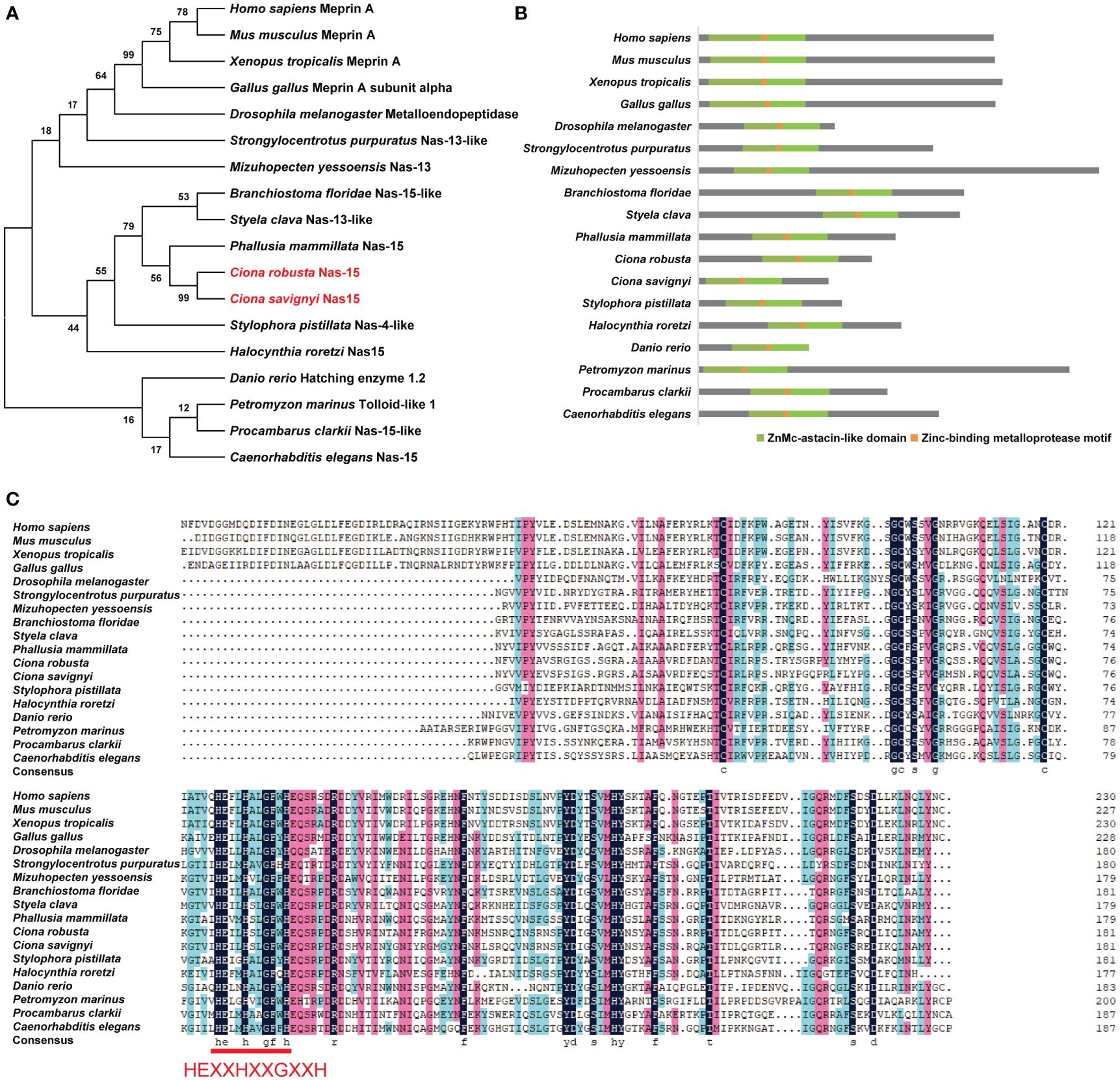
Figure 1 Phylogenetic analysis and domain composition of Nas15 in different species. (A) Phylogenetic tree analysis of Nas15 in different species. Red font indicates Ciona robusta Nas15 and C. savignyi Nas15. (B) Schematic diagram of Nas15 structure composition in diverse species. ZnMc-astacin-like domain (green) and Zinc-binding metalloprotease motif (orange) are presented in Nas15 in diverse species. (C) Protein sequence alignment of Nas15 ZnMc-astacin-like domain from diverse species. In the Zinc-binding metalloprotease motif region of Nas15 from diverse species, the amino acid sequence HEXXHXXGXXH is highly conserved.
To confirm the temporal expression of Cr-Nas15 from single-cell transcriptome sequencing dataset, we performed a qRT-polymerase chain reaction (PCR) analysis of Cr-Nas15 using different developmental-staged embryos. The result shows that the expression level is increasing from 8 hpf (mid gastrula stage) to 24 hpf (hatching larva stage), and highly expressed during 20 hpf (late tailbud stage) to 24 hpf (Figure 2A), which is consistent with the single-cell transcriptome sequencing data. To further explore the expression level of Cr-Nas15 in different tissues, we built a UMAP plot based on the single cell sequencing data of Ciona early embryos. The result indicated that Cr-Nas15 was expressed in many different tissues, such as the notochord, epidermis, muscle, and endoderm (Cao et al., 2019; Tarhan et al., 2023) (Figure 2B). Together with the qPCR (Figure 2A) and in situ hybridization data (Kusakabe et al., 2002; Satou et al., 2005), we inferred that Cr-Nas15 was expressed in notochord in lumen expansion stages. To confirm this spatial and temporal overlap between Cr-Nas15 expression and notochord lumen formation, we investigated the Cr-Nas15 expression level in notochord during entire lumen formation process by the promoter analysis. At 19 hpf (mid gastrula stage), an extracellular lumen initial formed between two adjacent notochord cells, while at 23 hpf and 24 hpf, the lumen undergoes continuous expansion and tilts (Figure 2C). The upstream 3,000 bps were cloned and fused with eGFP and electroporated into the Ciona fertilized eggs. Before the lumen formation (19 hpf), we did not observe any fluorescent signal in the notochord. But at 21 hpf (late tailbud stage) and 23 hpf, corresponding to the lumen formation and expansion phase, the eGFP signal was observed in the notochord (Figure 2D). Furthermore, to explore the subcellular localization of Cr-Nas15, we prepared a Murine polyclonal antibody with the full length of Cr-Nas15 as antigen. To validate the specificity of the antibody, the 23 hpf staged Ciona larvae were collected to preform total protein extraction. The western blot with Cr-Nas15 antibody showed a band at expected size over 40 KDa, suggesting the high efficacy of self-prepared Cr-Nas15 antibody (Figure 2E). We thus performed the Cr-Nas15 immunofluorescence experiment with Ciona larva at different developmental stages. In 19 hpf embryos before the extracellular lumen formation, the Cr-Nas15 has a low signal density in notochord, and localized at the pre-apical membrane of each notochord cells. Then, the Cr-Nas15 signal was increased and localized at the apical domain with the lumen formation in 23 hpf. In 24 hpf, in which stage the notochord cells starting a bi-directional migration, Cr-Nas15 also enriched at the apical membrane of each notochord cells (Figure 2F). The Cr-Nas15 immunofluorescence results confirmed that the Cr-Nas15 expression is increasing with the lumen formation, and indicated a special localization at the apical domain near the extracellular lumen. Together with the results of RT-qPCR, promoter analysis and immunofluorescence, we described the Cr-Nas15 spatio-temporal expression profile. The Cr-Nas15 was expressed at the notochord in lumen formation stages from 19 to 24 hpf, and localized at the apical domain, spatial closing to the extracellular lumen, suggesting a potential function of Cr-Nas15 in lumen formation.
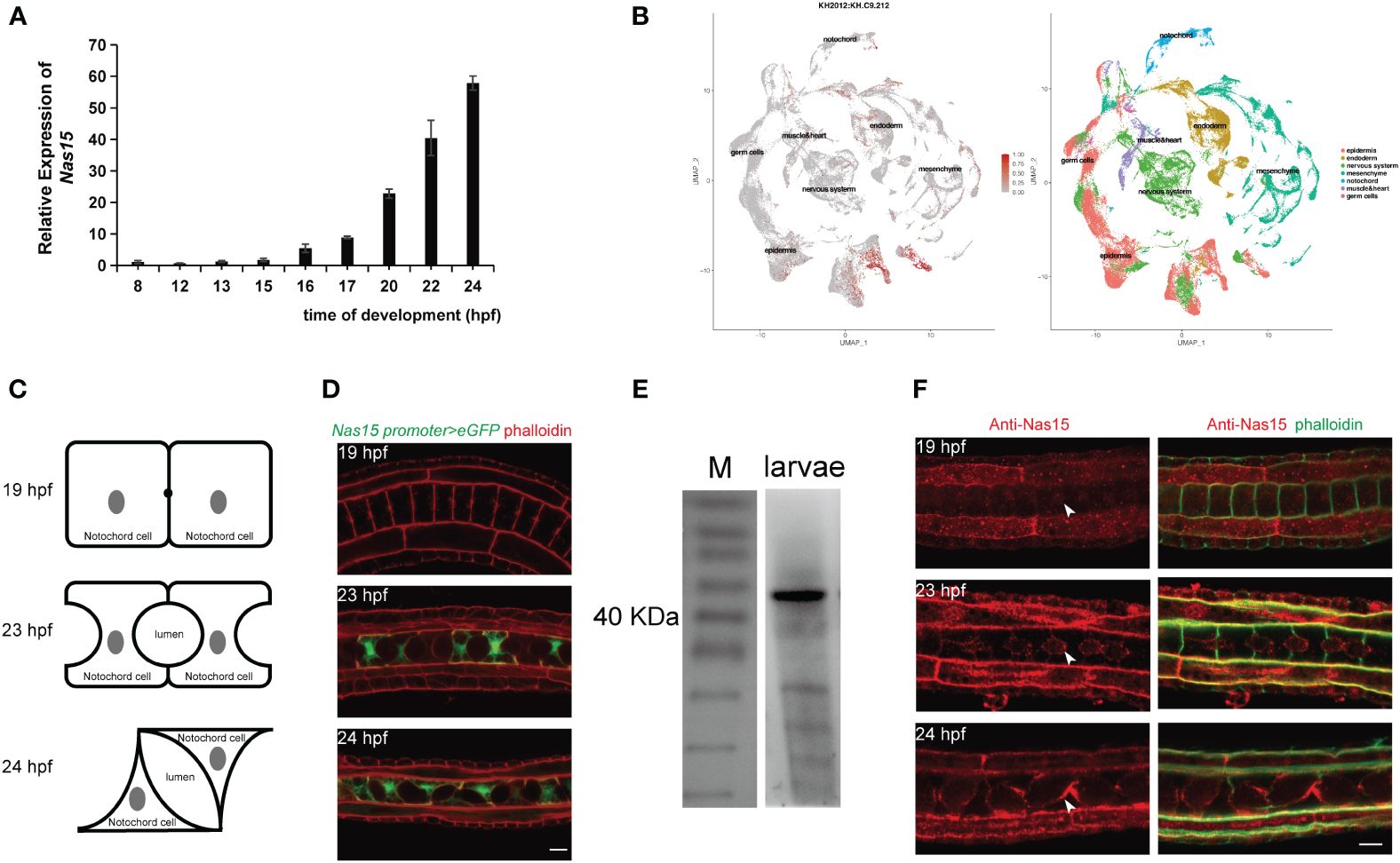
Figure 2 Expression pattern and subcellular localization of Nas15. (A) Cr-Nas15 mRNA expression levels at different developmental stages of Ciona embryo. (B) The UMAP plot displaying the expression of Cr-Nas15 in different tissues. (C) The illustration of Ciona notochord lumen in different developmental stages. (D) Expression pattern of Cr-Nas15 promotor-driven GFP in notochord tissue at diverse developmental stages of Ciona. (E) Western Blot with self-prepared Cr-Nas15 murine polyclonal antibody (F) Cr-Nas15 Immunofluorescence in Ciona embryos at different developmental stages. The white arrows represent the signal of Cr-Nas15. All scale bars represent 10 μm.
To explore the function of Cr-Nas15, actinonin, which binds at the catalytic site of astacin metalloproteinase as a competitive inhibitor (Talantikite et al., 2018). was used to block the function of all astacin metalloproteinase. In the actinonin-treated group, the embryos were cultured in sea water containing 1.2 μM DMSO dissolved actinonin from 16 hpf, while the control group was cultured in sea water with the same dose of DMSO. Then, the embryos were collected and fixed at 21 and 23 hpf stages, respectively. After dying with phalloidin to visualize the cell boundary, we observed the shape of extracellular lumen. In both 21 and 23 hpf, the lumen size in the actinonin-treated embryos obviously smaller than the control group (Figure 3A). So, we measured the diameter of the lumen in 23 hpf. Compared with the control group, the lumen diameter in actinonin-treated group was significantly smaller, both in 21hpf and 23 hpf (Figure 3B). Besides, the lumen shape was also abnormal in actinonin -treated embryos. Compared with the control group, the lumen in actinonin-treated embryos was not homogeneous growing, but expanded bialy towards one neighbor notochord cell (Figure 3A). These results indicate that the astacin metalloproteinase malfunction not only influence the Ciona notochord lumen formation, but also inhibit the lumen expansion process.
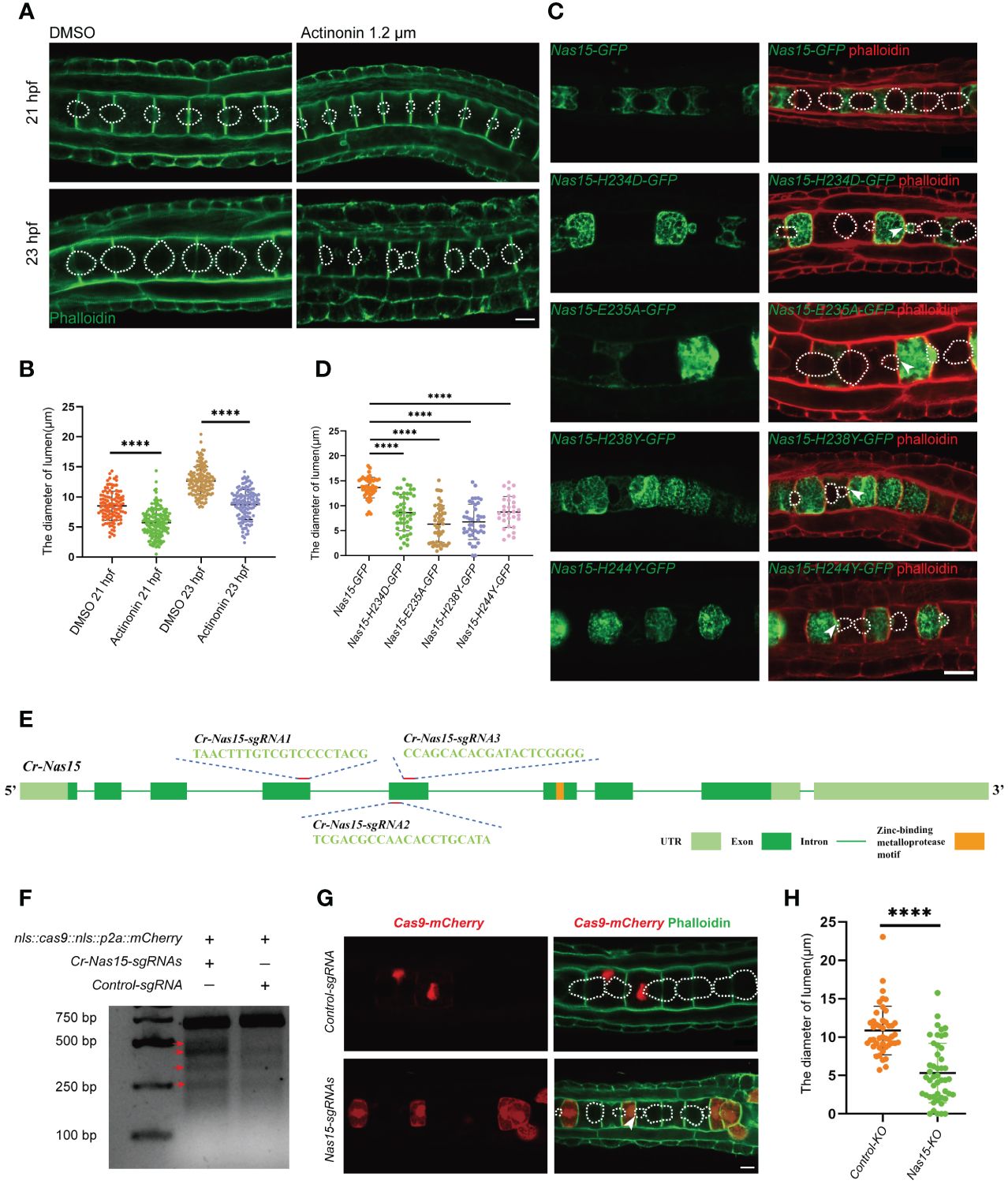
Figure 3 Cr-Nas15 loss-of-function disrupted lumen formation and expansion. (A) The phenotype of Astacin zinc metalloproteinase inhibitor Actinonin treated embryos. The dashed lines indicate the boundary of lumen. (B) Quantification of the lumen diameter of DMSO (n21 hpf = 112、n23 hpf = 136) and Actinonin (n21 hpf = 157、n23 hpf = 127) treated embryos at 21 and 23 hpf at 16 °C in A. (C) Overexpression of dominant negative Cr-Nas15 results in abnormal formation and expansion of lumen. (D) Quantification of the lumen diameter of overexpressing Nas15 CDS (nNas15-GFP = 45) and Nas15 dominant negative (nNas15-H234D-GFP=71、nNas15-E235A-GFP= 29、nNas15-H238Y-GFP = 39、nNas15-H244Y-GFP = 29) in C. (E) Cr-Nas15 gene Structure and sgRNAs design. Light green squares represent UTRs, dark green squares represent Exons, green lines represent Introns, and orange square represents Zinc-binding metalloprotease motif. (F) T7 endonuclease I test results of Cr-Nas15 knock out embryos. The red arrows show the different bands produced by the T7 endonuclease cleavage in the knockout group compared to the control group. (G) Nas15 notochord tissue specific knockout results. After the knock-out of Nas15, the formation and expansion of the notochord lumens were abnormal. (H) Quantification of the lumen diameter of Nas15-KO (n = 47) and Control-KO (n = 45) in G. All Significance statistical analysis was preformed using unpaired two-tailed t test. ****p < 0.0001. All scale bars represent 10 μm.
To further confirm the function of Cr-Nas15 involved in the notochord lumen formation, we constructed several dominant negative (DN) versions of Cr-Nas15 driven by brachyury promoter, which drives the constructs to specifically express in notochord tissue in Ciona larvae. According to previous studies, the Cr-Nas15-H234D, H238Y and H244Y lead to Nas15 loss of function by blocking the Zn binding, and the E235A lead to the Nas15 catalytic activity malfunction without influencing the Zn and substrate binding (Franco et al., 2005). The Cr-Nas15-DN and wild type (WT) expression plasmids were electroporated into the Ciona embryo, respectively. The results showed that the Cr-Nas15-WT was enriched at the notochord apical domain. However, some signals also appeared in the cytoplasm and around the contractile ring, which may be the mis-localization because of the overexpression. The notochord lumen in Cr-Nas15-WT overexpression group expanded normally, but all four DN versions led to abnormal lumen formation (Figure 3C). We measured the lumen diameter in all four DN version groups and the Cr-Nas15-WT group. The statistical results showed that the volume of lumen in each DN groups were significantly smaller than which in Cr-Nas15-WT group, phenocopying the inhibitor treatment (Figure 3D). Interestingly, the lumen shape in DN overexpression groups also different from the WT group. All four DN versions led to an asymmetrical lumen expansion. Due to the mosaic expression of Ciona, the Cr-Nas15-DN only expressed in several notochord cells. The lumen between a Cr-Nas15-DN overexpression positive cell and a negative cell always asymmetrically expanded into the negative cell (Figure 3C). These results reveal the essential function of Cr-Nas15 in Ciona notochord lumen expansion.
However, because of the side effect of dominant negative overexpression, we decided to perform a Cr-Nas15 knock out (KO) using the CRISPR/Cas9 system. Due to the important function of Zinc-binding metalloprotease motif, we designed three guiding RNA around this motif area (Figure 3E). To improve the KO efficacy, all the three guiding RNA plasmids were mixed to electroporate together with Cas9 expression plasmid. We also electroporated the control guiding RNA plasmid together with Cas9 expression plasmid as a control group. Then, the genomic DNA of KO group and control group, respectively were extracted for T7 Endonuclease I efficacy test. The knockout efficiency was determined to be approximately 16.37% by calculating the stripe grayscale values, confirming the efficiency of Cr-Nas15 gene KO (Figure 3F). So, we collected the Cr-Nas15 notochord specific KO embryos and control embryos. After the phalloidin staining, we observed the phenotype of notochord lumen formation. In Cr-Nas15 gene KO, 23 of 26 embryos have an abnormal lumen formation, while in control group, there are only 3 of 15 embryos have an abnormal lumen formation (Figure 3G). We also measured the diameter of the lumen in both Cr-Nas15 gene KO group and control group. The statistical analysis showed that the lumen in Cr-Nas15 gene KO group were significantly smaller than control group, phenocopying the inhibitor treatment and dominant negative overexpression results (Figure 3H). This result confirms that the Cr-Nas15 is an important factor regulating the Ciona notochord lumen formation.
To further find out the working mechanism of Cr-Nas15, we performed a yeast two-hybrid (Y2H) screening assay. The Protein Phosphatase 2 Catalytic Subunit Alpha (PPP2CA) was found as a candidate protein interacting with Cr-Nas15 (Figure 4A). To confirm the interaction between Cr-Nas15 and the PPP2CA, we carried out the CO-IP experiments. When the Nas15-GFP and PPP2CA-HA both existed, a band could be found in the IP group (Figure 4B). These results indicate the interaction between Cr-Nas15 and PPP2CA, suggesting the Cr-Nas15 and PPP2CA may form a complex functioning together. To confirm the Cr-Nas15 and PPP2CA formed a complex intracellular, we co-expressed the PPP2CA-eGFP and Cr-Nas15-tdTomato in Ciona notochord. The imaging data showed that the PPP2CA-eGFP and Cr-Nas15-tdTomato significantly co-localized together at the apical domain of notochord cell (Figure 4C). Together with these data, the PPP2CA and Cr-Nas15 formed a complex around the notochord apical domain. Then, we performed a co-electroporation of PPP2CA-C201R or PPP2CA WT with Cr-Nas15 fusion protein. The Cr-Nas15 showed a lower enrichment on apical membrane in PPP2CA-C201R group, suggesting the PPP2CA involved in Nas15 apical localization (Figures 4D, E). To test if the interaction between the PPP2CA and Cr-Nas15 is essential for the lumen formation, we constructed the PPP2CA-C201R as a dominant negative and overexpressed it in Ciona notochord, to observe if it can lead to same phenotype with Cr-Nas15 loss-of-function. The result showed that the lumen in PPP2CA-C201R overexpressed group is significantly smaller than the control group (Figure 4F), phenocopying the Cr-Nas15 loss-of-function. The measurement of lumen diameter also confirmed the smaller lumen phenotype of PPP2CA-C201R overexpression (Figure 4G). The identification of multiple phosphorylation sites within Nas15 implies a potential regulatory role of PPP2CA in modulating the phosphorylation levels of Nas15 (Figure 4H). In conclusion, the PPP2CA is an interaction protein of Cr-Nas15, which is essential for Ciona notochord lumen formation.
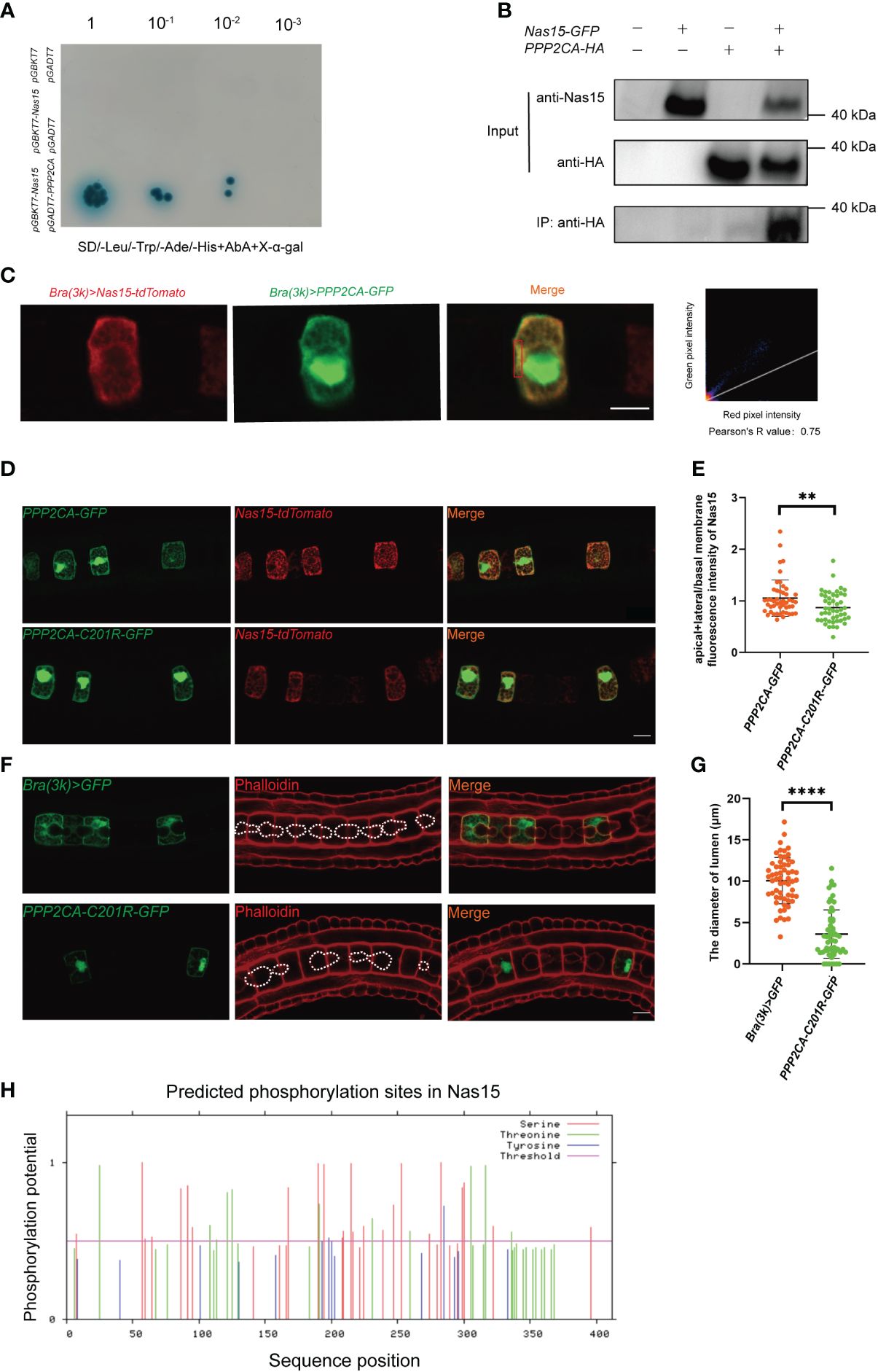
Figure 4 Cr-Nas15 interact with PPP2CA involving in lumen formation. (A) Interaction assays between Nas15 and PPP2CA by Y2H. (B) Co-IP validation of Nas15 interaction with PPP2CA in the HEK293T cell line. (C) Co-localization of Nas15 and PPP2CA in the apical domain of notochord cells. Red rectangle indicates the image area preforming colocalization test. (D) Co-electroporation of PPP2CA-C201R or PPP2CA WT with Cr-Nas15 fusion protein marker. (E) Quantification of the (apical+lateral)/basal membrane fluorescence intensity of Nas15 of overexpressing PPP2CA (nBrachyury (3k) >PPP2CA::GFP = 48) and PPP2CA dominant negative (nBrachyury (3k) > PPP2CA-C201R::GFP = 48) in D. (F) Phenotype of PPP2CA dominant negative overexpression in Ciona notochord. The dashed lines indicate the boundary of lumen. (G) Quantification of the lumen diameter of overexpressing Brachyury (3k) > GFP (nBrachyury (3k) > GFP = 59) and PPP2CA dominant negative (nBrachyury (3k) > PPP2CA-C201R::GFP = 61) in F. (H) Prediction of phosphorylation sites in Nas15. All Significance statistical analysis was preformed using unpaired two-tailed t test. **p < 0.01 and ****p < 0.0001. All scale bars represent 10 μm.
In our research, we identified a Ciona matrix metalloproteinase, Cr-Nas15. Through phylogenetic and domain analysis, we confirmed that Cr-Nas15 is conserved, including the typical ZnMc-astacin-like domain and Zinc-binding metalloprotease motif. These findings suggest that Ciona Nas15 may share a conserved function with Nas proteins in vertebrates. Additionally, employing qPCR, promoter analysis, and immunofluorescence techniques, we have demonstrated the expression of Cr-Nas15 in the larval notochord of Ciona during lumen formation stages, where it is enriched at the apical domain of Ciona notochord cells.
Subsequently, we conducted experiments involving the inhibitor actinonin treatment, dominant negative overexpression, and gene knockout using the CRISPR/Cas9 system. These experiments further confirmed the involvement of Cr-Nas15 in notochord lumen formation and expansion. Moreover, we have discovered an interaction between PPP2CA and Cr-Nas15, which stabilizes the localization of Cr-Nas15 to the notochord apical domain.
The Nas15 belongs to the astacins family, together with the Meprin, BMP1/Tolloid etc (Huxley-Jones et al., 2007), which is a large family of zinc metalloproteases in bacteria and animals (Park et al., 2010). Previous studies have demonstrated that astacin family metalloproteases play crucial roles in various biological processes, including digestion, hatching, peptide processing, morphogenesis, and pattern formation. For example, the Meprins are involved in the base membrane protein hydrolyzing, such as the collagen IV, nidogen and fibronectin, which is essential for ECM remodeling (Kruse et al., 2004). Besides, the Meprin and BMP-1/Tolloid have been found to promote the collagen maturation and assembly by digesting the procollagen (Hopkins et al., 2007; Broder et al., 2013). The HCH-1, another member of BMP-1/tolloid family, participates in the hatching of C. elegans by digesting the eggshell. Astacus embryonic astacin (AEA) is highly expressed shortly before hatching, suggesting its function in eggshell degradation (Geier and Zwilling, 1998). Importantly, the functional roles of astacin metalloproteases primarily rely on their ability to digest stable ECM complex structures, facilitating their assembly or remodeling. Interestingly, in our study, Nas15 was found to be enriched on the apical membrane near the extracellular lumen, whereas most of the stable ECM components are localized in the notochord sheath covering the basal side of notochord cells. Both the laminin, fibronectin and collagen are secreted in a polarized manner, leading to the accumulation of the basal membrane in the notochord sheath at the basal domain of the notochord (Veeman3et al., 2008; Segade et al., 2016; Peng et al., 2023). On the contrary, the ECM in the notochord lumen exhibits low density and consists of short fibers, indicating the presence of a viscous fluid composed of dissolved ECM components (Hunziker and Schenk, 1984; Dong et al., 2009; Wei et al., 2017). Therefore, it’s worth investigating why the Nas15 localizes to the apical domain instead of basal membrane. One possibility is it interacts with the dissolved ECM to contribute to lumen formation. This observation may suggest a potential novel mechanism of how matrix metalloproteases regulate morphogenesis. The astacin family proteins have been found to localize to the apical membrane in some special models. For example, Meprin is localized to the apical membranes (brush-border) of renal proximal tubules (Herzog et al., 2014). Initially, Meprin anchors to the apical domain as a transmembrane metalloprotease (TMP), and subsequently, it is released as a secreted MMP through the digestion of ADAM10 (Herzog et al., 2014). However, in our study, we didn’t find any Cr-Nas15 signal in the lumen space neither in immunofluorescence nor in fluorescence protein fusion protein marker overexpression. Cr-Nas15 specifically anchors to the apical membrane of the notochord lumen. Since the immobile Cr-Nas15 can only contact with a low-density ECM close to the apical membrane, how can the Cr-Nas15 influence the lumen formation?
One possible mechanism is keeping the apical ECM soluble. Around the notochord cells, the ECM exhibits different properties in different area. On the basal side, most of ECM component are made up by long fiber as a basement membrane. But there is no basement membrane ECM can be found but only short fiber ECM instead in the extracellular lumen (Wei et al., 2017), which suggesting a degradation mechanism may exist on the initial apical domain. During the expansion of the Ciona notochord lumen, a hydrostatic pressure is generated within the lumen space, driving its expansion (He et al., 2022). In this mechanism, the apical ECM need to be soluble. We propose that Cr-Nas15, enriched on the apical membrane, degrades the long ECM fiber to maintain a liquid lumen fluid. Therefore, Cr-Nas15 may serve as an protecting potential mechanism to prevent the accumulation of long fiber ECM in the apical domain. So, the ECM components in the initial lumen can act as a “sponge” to facilitate fluid influx (Denker and Jiang, 2012). The ECM degradation activity of Cr-Nas15 can increasing the osmotic pressure in the lumen space.
Another possible mechanism by which Cr-Nas15 influences lumen formation is by aiding in the maintenance of apical-basal (A-B) polarity. Many studies have shown that the ECM serves as a directional signal that helps cells establish A-B polarity. For example, the basement membrane interacts with integrin, providing an extracellular signal for epithelial cell A-B polarity (Manninen, 2015). In the Ciona notochord, the ECM also plays an important role in polarity maintenance (Peng et al., 2020). Thus, the specific apically localized Cr-Nas15 may influence the polarized distribution of the basement membrane ECM around notochord cells, which could in turn affect A-B polarity maintenance. In our study, loss-of-function of Cr-Nas15 delayed the initial formation of the lumen, suggesting a blocking of A-B polarity establishment. Another interesting phenotype was the asymmetrical expansion of the lumen in Cr-Nas15, indicating a decrease in the surface area of the apical membrane. Since apical membrane delivery is provided by polarized vesicle transport, we propose that the disruption of A-B polarity in Cr-Nas15 loss-of-function cells could lead to a decrease of apical domain targeting vesicles.
The interaction between cells and ECM plays a crucial role in the regulation of morphogenesis. However, it is still unclear how cells can dynamically regulate the quantity, location, and properties of the ECM to provide feedback control of tissue morphogenesis. In our study, we discovered that Cr-Nas15 has the potential ability to degrade ECM within the lumen, which can influence lumen formation. This finding provides a new and possible mechanism for how matrix metalloproteinases may be involved in tubulogenesis.
In this study, the interaction between PPP2CA and Cr-Nas15 was identified, and the dominant negative versions of PPP2CA show phenocopy with the Cr-Nas15. As PPP2CA functions as the catalytic subunit of a phosphatase, we hypothesize that the phosphorylation of Cr-Nas15 plays a crucial role in its function. While Cr-Nas15 exhibits numerous potential phosphorylation sites, the specific mechanism remains unclear. Phosphorylation is an important form of post-translational modification that can affect protein localization or activity. Therefore, we propose that PPP2CA may regulate lumen formation indirectly by modulating the function of Cr-Nas15.
The notochord is a special mid-line organ of all Chordates, acting as a trunk supporting structure and developmental signaling center (Corallo et al., 2015). As a supporting structure, the mechanical stiffness of notochord is an important factor to provide a basis of controlled and rapid movement and protection of neural tube (Sui et al., 2021). During the notochord evolution, there are two events leading to remarkable stiffness increase. The turgid rod structure appearance constructed with a ECM sheath outside and vacuolated notochord cells inside (Yasuoka, 2020), and the spine formation in which the notochord cells forming the nucleus pulposus of the intervertebral discs (Smits and Lefebvre, 2003; Raghavan et al., 2023). Before the emergence of chordates, some cells or tissues present gene expression pattern similar to that in notochord such as expression of Brachyury in the stomodeum and proctdaeumin of Polychaete or hindgut and anal pads of Drosophila (Satoh et al., 2012). A notochord-like muscle structure called axochord was reported in annelid (Lauri et al., 2014; Brunet et al., 2015), which was considered as a primitive form of notochord. These tissues be known as notochord ancestor both lack observable vacuoles. The notochord in ascidian, amphioxus and chordate both have a vacuolation which making up a turgor pressure sheath structure together with the ECM notochord sheath (Yasuoka, 2020), which structure improving the notochord stiffness by sealing a hydrostatic pressure. Interestingly, the notochord vacuolation in Ciona is special. The notochord lumen in the Amphioxus (Eakin and Westfall, 1962) and chordate such as the zebrafish, chick and mice (Choi et al., 2008; Ellis et al., 2013; Ward et al., 2018) are intercellular vacuole. However, the Ciona larval notochord construct an extracellular connected lumen (Dong et al., 2009), which is a special notochord structure in notochord evolution. Previous researches showed that the Ciona notochord expressed some evolutionarily conserved key molecule regulating notochord specific gene expression (Maguire et al., 2018; Raghavan et al., 2023), which suggests a key phylogenetic position of Ciona as a notochord origin class. Therefore, the notochord extracellular lumen may show the early form of vacuolate notochord. Compared with the intracellular vacuole inflation, the extracellular lumen formation needs some different cell behavior regulation such as the A-B polarity establishing, cell junction remodeling, apical ECM secretion and processing, etc., but the mechanism still remains unclear. Here, we found the MMP family protein Cr-Nas15 specific localized at the membrane of extracellular lumen which is essential for lumen expansion, suggesting that some pre-existing extracellular ingredient degradation such as the ECM may help the extracellular lumen formation. This study suggested that the mechanism that MMP mediated extracellular lumen opening may play as a functional replacement before intracellular notochord vacuole appeared in evolution. So, the Cr-Nas15 may play as a clue for further study of the vacuolation of notochord.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
JB: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. YG: Investigation, Visualization, Writing – review & editing. ZW: Investigation, Writing – review & editing. HP: Validation, Visualization, Writing – original draft, Writing – review & editing. BD: Funding acquisition, Methodology, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Science & Technology Innovation Project of Laoshan Laboratory (No. LSKJ202203204).
We thank members of Dong laboratory for experimental technical guidance. We are very grateful to Guo huarong’s lab for sharing the HEK293 T cell lines.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2024.1385516/full#supplementary-material
Annona G., Holland N. D., D’Aniello S. (2015). Evolution of the notochord. EvoDevo 6, 1–13. doi: 10.1186/s13227-015-0025-3
Baumann U., Wu S., Flaherty K. M., McKay D. B. (1993). Three-dimensional structure of the alkaline protease of Pseudomonas aeruginosa: a two-domain protein with a calcium binding parallel beta roll motif. EMBO J. 12, 3357–3364. doi: 10.1002/embj.1993.12.issue-9
Bhattachan P., Rae J., Yu H., Jung W., Wei J., Parton R. G., et al. (2020). Ascidian caveolin induces membrane curvature and protects tissue integrity and morphology during embryogenesis. FASEB J. 34, 1345–1361. doi: 10.1096/fj.201901281R
Bond J. S., Beynon R. J. (1995). The astacin family of metalloendopeptidases. Protein Sci. 4, 1247–1261. doi: 10.1002/pro.5560040701
Bonnans C., Chou J., Werb Z. (2014). Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 15, 786–801. doi: 10.1038/nrm3904
Broder C., Arnold P., Vadon-Le Goff S., Konerding M. A., Bahr K., Muller S., et al. (2013). Metalloproteases meprin alpha and meprin beta are C- and N-procollagen proteinases important for collagen assembly and tensile strength. Proc. Natl. Acad. Sci. U.S.A. 110, 14219–14224. doi: 10.1073/pnas.1305464110
Brunet T., Lauri A., Arendt D. (2015). Did the notochord evolve from an ancient axial muscle? The axochord hypothesis. Bioessays 37, 836–850. doi: 10.1002/bies.201500027
Calabria P. A., Shimokawa-Falcão L. H. A., Colombini M., Moura-da-Silva A. M., Barbaro K. C., Faquim-Mauro E. L., et al. (2019). Design and production of a recombinant hybrid toxin to raise protective antibodies against Loxosceles spider venom. Toxins 11, 108. doi: 10.3390/toxins11020108
Cao C., Lemaire L. A., Wang W., Yoon P. H., Choi Y. A., Parsons L. R., et al. (2019). Comprehensive single-cell transcriptome lineages of a proto-vertebrate. Nature 571, 349–354. doi: 10.1038/s41586-019-1385-y
Choi K. S., Cohn M. J., Harfe B. D. (2008). Identification of nucleus pulposus precursor cells and notochordal remnants in the mouse: implications for disk degeneration and chordoma formation. Dev. Dyn. 237, 3953–3958. doi: 10.1002/dvdy.21805
Corallo D., Trapani V., Bonaldo P. (2015). The notochord: structure and functions. Cell Mol. Life Sci. 72, 2989–3008. doi: 10.1007/s00018-015-1897-z
Denker E., Jiang D. (2012). Ciona intestinalis notochord as a new model to investigate the cellular and molecular mechanisms of tubulogenesis. Semin. Cell Dev. Biol. 23, 308–319. doi: 10.1016/j.semcdb.2012.03.004
Dong B., Deng W., Jiang D. (2011). Distinct cytoskeleton populations and extensive crosstalk control Ciona notochord tubulogenesis. Development 138, 1631–1641. doi: 10.1242/dev.057208
Dong B., Horie T., Denker E., Kusakabe T., Tsuda M., Smith W. C., et al. (2009). Tube formation by complex cellular processes in Ciona intestinalis notochord. Dev. Biol. 330, 237–249. doi: 10.1016/j.ydbio.2009.03.015
Eakin R. M., Westfall J. A. (1962). Fine structure of the notochord of amphioxus. J. Cell Biol. 12, 646–651. doi: 10.1083/jcb.12.3.646
Ellis K., Bagwell J., Bagnat M. (2013). Notochord vacuoles are lysosome-related organelles that function in axis and spine morphogenesis. J. Cell Biol. 200, 667–679. doi: 10.1083/jcb.201212095
Franco A. A., Buckwold S. L., Shin J. W., Ascon M., Sears C. L. (2005). Mutation of the Zinc-Binding Metalloprotease Motif AffectsBacteroides fragilisToxin Activity but Does Not Affect Propeptide Processing. Infection Immunity. 73, 5273–5277. doi: 10.1128/IAI.73.8.5273-5277.2005
Geier G., Zwilling R. (1998). Cloning and characterization of a cDNA coding for Astacus embryonic astacin, a member of the astacin family of metalloproteases from the crayfish Astacus astacus. Eur. J. Biochem. 253, 796–803. doi: 10.1046/j.1432-1327.1998.2530796.x
He M., Wei J., Li Y., Dong B. (2022). Nuclear factor of activated T cells-5 regulates notochord lumenogenesis in chordate larval development. Int. J. Mol. Sci. 23 (22), 14407. doi: 10.3390/ijms232214407
Herzog C., Haun R. S., Ludwig A., Shah S. V., Kaushal G. P. (2014). ADAM10 is the major sheddase responsible for the release of membrane-associated meprin A. J. Biol. Chem. 289, 13308–13322. doi: 10.1074/jbc.M114.559088
Hopkins D. R., Keles S., Greenspan D. S. (2007). The bone morphogenetic protein 1/Tolloid-like metalloproteinases. Matrix Biol. 26, 508–523. doi: 10.1016/j.matbio.2007.05.004
Hu J., Gao Y., Huang Q., Wang Y., Mo X., Wang P., et al. (2021). Flotillin-1 interacts with and sustains the surface levels of TRPV2 channel. Front. Cell Dev. Biol. 9. doi: 10.3389/fcell.2021.634160
Hunziker E. B., Schenk R. K. (1984). Cartilage ultrastructure after high pressure freezing, freeze substitution, and low temperature embedding. II. Intercellular matrix ultrastructure - preservation of proteoglycans in their native state. J. Cell Biol. 98, 277–282. doi: 10.1083/jcb.98.1.277
Huxley-Jones J., Clarke T.-K., Beck C., Toubaris G., Robertson D. L., Boot-Handford R. P. (2007). The evolution of the vertebrate metzincins; insights from Ciona intestinalis and Danio rerio. BMC Evolution Biol. 7, 1–20. doi: 10.1186/1471-2148-7-63
Kruse M. N., Becker C., Lottaz D., Kohler D., Yiallouros I., Krell H. W., et al. (2004). Human meprin alpha and beta homo-oligomers: cleavage of basement membrane proteins and sensitivity to metalloprotease inhibitors. Biochem. J. 378, 383–389. doi: 10.1042/bj20031163
Kusakabe T., Yoshida R., Kawakami I., Kusakabe R., Mochizuki Y., Yamada L., et al. (2002). Gene expression profiles in tadpole larvae of Ciona intestinalis. Dev. Biol. 242, 188–203. doi: 10.1006/dbio.2002.0538
Lauri A., Brunet T., Handberg-Thorsager M., Fischer A. H., Simakov O., Steinmetz P. R., et al. (2014). Development of the annelid axochord: insights into notochord evolution. Science 345, 1365–1368. doi: 10.1126/science.1253396
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu Q., Bhattachan P., Dong B. (2019). Ascidian notochord elongation. Dev. Biol. 448, 147–153. doi: 10.1016/j.ydbio.2018.11.009
Lubarsky B., Krasnow M. A. (2003). Tube morphogenesis: making and shaping biological tubes. Cell 112, 19–28. doi: 10.1016/S0092-8674(02)01283-7
Maguire J. E., Pandey A., Wu Y., Di Gregorio A. (2018). Investigating evolutionarily conserved molecular mechanisms controlling gene expression in the notochord. Transgenic Ascidians. p, 81–99.
Manninen A. (2015). Epithelial polarity–generating and integrating signals from the ECM with integrins. Exp. Cell Res. 334, 337–349. doi: 10.1016/j.yexcr.2015.01.003
Mizotani Y., Suzuki M., Hotta K., Watanabe H., Shiba K., Inaba K., et al. (2018). 14-3-3epsilona directs the pulsatile transport of basal factors toward the apical domain for lumen growth in tubulogenesis. Proc. Natl. Acad. Sci. U.S.A. 115, E8873–E8881. doi: 10.1073/pnas.1808756115
Nagase H., Visse R., Murphy G. (2006). Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc. Res. 69, 562–573. doi: 10.1016/j.cardiores.2005.12.002
Ouyang X., Wu B., Yu H., Dong B. (2023). DYRK1-mediated phosphorylation of endocytic components is required for extracellular lumen expansion in ascidian notochord. Biol. Res. 56, 10. doi: 10.1186/s40659-023-00422-9
Park J. O., Pan J., Mohrlen F., Schupp M. O., Johnsen R., Baillie D. L., et al. (2010). Characterization of the astacin family of metalloproteases in C. elegans. BMC Dev. Biol. . 10, 14. doi: 10.1186/1471-213X-10-14
Peng H., Qiao R., Dong B. (2020). Polarity establishment and maintenance in ascidian notochord. Front. Cell Dev. Biol. 8. doi: 10.3389/fcell.2020.597446
Peng H., Qiao J., Wang G., Shi W., Xia F., Qiao R., et al. (2023). A collagen-rich arch in the urochordate notochord coordinates cell shaping and multi-tissue elongation. Curr. Biol. 33, 5390–5403 e5393. doi: 10.1016/j.cub.2023.11.001
Raghavan R., Coppola U., Wu Y., Ihewulezi C., Negrón-Piñeiro L. J., Maguire J. E., et al. (2023). Gene expression in notochord and nuclei pulposi: a study of gene families across the chordate phylum. BMC Ecol. Evol. 23, 63. doi: 10.1186/s12862-023-02167-1
Sacharidou A., Stratman A. N., Davis G. E. (2012). Molecular mechanisms controlling vascular lumen formation in three-dimensional extracellular matrices. Cells Tissues Organs 195, 122–143. doi: 10.1159/000331410
Sarraju A., Nissen S. E. (2024). Atherosclerotic plaque stabilization and regression: a review of clinical evidence. Nat. Rev. Cardiol. 1–11. doi: 10.1038/s41569-023-00979-8
Satoh N., Tagawa K., Takahashi H. (2012). How was the notochord born? Evol. Dev. 14, 56–75. doi: 10.1111/j.1525-142X.2011.00522.x
Satou Y., Kawashima T., Shoguchi E., Nakayama A., Satoh N. (2005). An integrated database of the ascidian, Ciona intestinalis: towards functional genomics. Zoological science. 22, 837–843. doi: 10.2108/zsj.22.837
Segade F., Cota C., Famiglietti A., Cha A., Davidson B. (2016). Fibronectin contributes to notochord intercalation in the invertebrate chordate, Ciona intestinalis. Evodevo 7, 21. doi: 10.1186/s13227-016-0056-4
Senger D. R., Davis G. E. (2011). Angiogenesis. Cold Spring Harbor Perspect. Biol. 3, a005090–a005090. doi: 10.1101/cshperspect.a005090
Simons M., Walz G. (2006). Polycystic kidney disease: cell division without a c(l)ue? Kidney Int. 70, 854–864. doi: 10.1038/sj.ki.5001534
Smits P., Lefebvre V. r. (2003). Sox5 and Sox6 are required for notochord extracellular matrix sheath formation, notochord cell survival and development of the nucleus pulposus of intervertebral discs. Development 130, 1135–1148. doi: 10.1242/dev.00331
Sterchi E., Stocker W., Bond J. (2008). Meprins, membrane-bound and secreted astacin metalloproteinases. Mol. Aspects Med. 29, 309–328. doi: 10.1016/j.mam.2008.08.002
Stolfi A., Christiaen L. (2012). Genetic and genomic toolbox of the chordate Ciona intestinalis. Genetics 192, 55–66. doi: 10.1534/genetics.112.140590
Stolfi A., Gandhi S., Salek F., Christiaen L. (2014). Tissue-specific genome editing in Ciona embryos by CRISPR/Cas9. Development 141, 4115–4120. doi: 10.1242/dev.114488
Sui Z., Zhao Z., Dong B. (2021). Origin of the chordate notochord. Diversity 13 (10), 462. doi: 10.3390/d13100462
Talantikite M., Lecorche P., Beau F., Damour O., Becker-Pauly C., Ho W. B., et al. (2018). Inhibitors of BMP-1/tolloid-like proteinases: efficacy, selectivity and cellular toxicity. FEBS Open Bio. 8, 2011–2021. doi: 10.1002/2211-5463.12540
Tamura K., Stecher G., Kumar S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evolution . 38, 3022–3027. doi: 10.1093/molbev/msab120
Tarhan L., Bistline J., Chang J., Galloway B., Hanna E., Weitz E. (2023). Single Cell Portal: an interactive home for single-cell genomics data. bioRxiv. doi: 10.1101/2023.07.13.548886
Tassy O., Dauga D., Daian F., Sobral D., Robin F., Khoueiry P., et al. (2010). The ANISEED database: digital representation, formalization, and elucidation of a chordate developmental program. Genome Res. 20, 1459–1468. doi: 10.1101/gr.108175.110
Trevisan-Silva D., Gremski L. H., Chaim O. M., da Silveira R. B., Meissner G. O., Mangili O. C., et al. (2010). Astacin-like metalloproteases are a gene family of toxins present in the venom of different species of the brown spider (genus Loxosceles). Biochimie 92, 21–32. doi: 10.1016/j.biochi.2009.10.003
Veeman M. T., Nakatani Y., Hendrickson C., Ericson V., Lin C., Smith W. C. (2008). Chongmague reveals an essential role for laminin-mediated boundary formation in chordate convergence and extension movements. Development 135, 33–41. doi: 10.1242/dev.010892
Verma R. P., Hansch C. (2007). Matrix metalloproteinases (MMPs): chemical-biological functions and (Q)SARs. Bioorg Med. Chem. 15, 2223–2268. doi: 10.1016/j.bmc.2007.01.011
Wang Z., Tan Z., Bi J., Liu A., Jiang A., Dong B. (2023). Proteomic identification of intracellular vesicle trafficking and protein glycosylation requirements for lumen inflation in Ciona notochord. Proteomics, 23 (10), 2200460. doi: 10.1002/pmic.202200460
Ward L., Pang A. S. W., Evans S. E., Stern C. D. (2018). The role of the notochord in amniote vertebral column segmentation. Dev. Biol. 439, 3–18. doi: 10.1016/j.ydbio.2018.04.005
Wei J., Wang G., Li X., Ren P., Yu H., Dong B. (2017). Architectural delineation and molecular identification of extracellular matrix in ascidian embryos and larvae. Biol. Open 6, 1383–1390. doi: 10.1242/bio.026336
Keywords: Nas15, notochord, lumen expansion, ECM, ciona, tunicates
Citation: Bi J, Ge Y, Wang Z, Peng H and Dong B (2024) Matrix metalloproteinase Nas15 regulates the lumen formation and expansion in Ciona notochord. Front. Ecol. Evol. 12:1385516. doi: 10.3389/fevo.2024.1385516
Received: 13 February 2024; Accepted: 22 April 2024;
Published: 03 May 2024.
Edited by:
Paolo Sordino, Anton Dohrn Zoological Station Naples, ItalyReviewed by:
Ugo Coppola, Cincinnati Children’s Hospital Medical Center, United StatesCopyright © 2024 Bi, Ge, Wang, Peng and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongzhe Peng, cGVuZ2hvbmd6aGVAc3R1Lm91Yy5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.