
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Ecol. Evol. , 26 March 2024
Sec. Conservation and Restoration Ecology
Volume 12 - 2024 | https://doi.org/10.3389/fevo.2024.1354138
 David Thuo1,2*
David Thuo1,2* Nicholas A. Macgregor3,4
Nicholas A. Macgregor3,4 Samuel D. Merson5
Samuel D. Merson5 Dianne Scopel6
Dianne Scopel6 J. Scott Keogh2
J. Scott Keogh2 Jeremy Kenny7
Jeremy Kenny7 Jessica L. Williams3
Jessica L. Williams3 Tracey Guest6
Tracey Guest6 Shaeleigh Swan6
Shaeleigh Swan6 Steve McAlpin8
Steve McAlpin8 Leo Joseph1
Leo Joseph1Introduction: Accurately quantifying the diet of species has implications for our understanding of their ecology and conservation. Yet, determining the dietary composition of threatened and elusive species in the wild is often difficult.
Methods: This study presents the first dietary assessment of tjakura (Liopholis kintorei) using non-invasive sampling of scats and high-throughput sequencing techniques.
Results: The tjakura in Uluru consumed 48 invertebrates, 27 plants, and two vertebrate taxa. Fruit flies (Leucophenga spp.), beetles (Harpalus spp. and Omorgus spp.), mosquitos (Culicidae spp.), termites (Termitidae spp.), spiked mallow (Malvastrum americanum), bush tomatoes (Solanum centrale), and wild turnip (Brassica tournefortii) comprised the majority of the diet. Analysis of similarity revealed that food items did not differ significantly between tjakura age groups, seasons, or time since the last fire, however, adults, hot season, and fire scar of 2018 showed a relatively higher prey diversity.
Discussion: These high similarities in diet composition between age classes and fire scars indicate potential intraspecific competition when food resources are scarce. The diet diversity and potential plasticity observed in this study reflect a dietary ecology influenced by food availability rather than preference. Our study demonstrates that scat DNA metabarcoding is an important complementary tool to conventional scat analysis or indigenous knowledge as most food items we identified were previously not recorded through those methods.
Effective wildlife conservation relies on a good understanding of the relationship between populations and their environment. Animal diet is a vital component in discerning these complex interrelationships as feeding and food selection are essential in defining ecological niches, community dynamics and individual fitness (Vitt and Pianka, 2005; Romano et al., 2020; Westeen et al., 2023). Dietary data are also fundamental in identifying resource requirements (Westeen et al., 2023), ecosystem functionality (Kartzinel and Pringle, 2015) and predicting how a species will respond to environmental changes.
Anthropogenic pressure and global warming have caused extensive degradation of arid and semi-arid environments worldwide (Isbell et al., 2023). These ecosystems are normally characterised by highly diverse and rich reptile assemblages as reptiles possess an extraordinary ability for behavioural thermoregulation (Bogert, 1949; Seebacher and Franklin, 2005). However, most desert-dwelling reptile species have recently declined in abundance or completely disappeared in response to environmental pressures (McAlpin, 2001; Pimenta et al., 2005; Roll et al., 2017; Cox et al., 2022). One such species is the great desert skink (Liopholis kintorei) also known as tjakura, in the languages of central Australia’s indigenous peoples (IDA, 2022). The great desert skink, hereafter tjakura, is a large scincid lizard endemic to the arid zones of central and western Australia. Tjakura exhibits social behaviour and lives in multi-tunneled communal burrows (McAlpin, 2001; Ridley and Schlesinger, 2023). The species is currently classified as ‘Vulnerable’ due to its small population size and continuing decline in range and abundance (EPBC, 1999; Paltridge et al., 2018; IUCN, 2023). It is vulnerable to introduced predators such as cats (Felis catus) and European foxes (Vulpes vulpes) and changes in fire regimes (Moore et al., 2018). Alongside its contributions to ecological functions and regulating services (e.g. pest outbreak, zoonotic risk, and seed dispersal) (McAlpin, 2001; Valencia-Aguilar et al., 2013; de Miranda, 2017), tjakura is also a culturally important species to many Australian indigenous groups and is widely celebrated in art, dance, and indigenous lore throughout Australia’s western and central deserts.
Food availability may influence burrow occupancy and distribution of tjakura (McAlpin, 2001; Nowak et al., 2010; Ridley and Schlesinger, 2023). Active burrows of known populations have become inactive shortly after resource availability was altered by intense fire or other factors such as drought (Chapple, 2003; Moore et al., 2015; Cadenhead et al., 2016). The impact of post-fire ground cover can influence the burrow system, predation, and breeding success of tjakura. However, it is unclear how diet shifts across fire regimes and whether there is a particular post-fire age of a landscape that is optimal for the tjakura diet. The species exhibits both active and ambush foraging strategies, generally foraging away from the burrow at night or opportunistically catching prey as they pass the burrow (McAlpin, 2001). To date, dietary composition has been minimally documented but classical dietary studies have shown that tjakura predominantly consumes a wide range of invertebrates augmented with plant materials and small vertebrates (McAlpin, 2001; Chapple, 2003). However, the complete dietary profile and how it relates to fire, weather, or age of tjakura is still unknown.
The diet of tjakura is currently based on indigenous ecological knowledge and conventional, but potentially biased methods such as direct observation or morphological analysis of undigested remains in scats (McAlpin, 2001). A relatively new method, scat DNA metabarcoding has recently been shown to provide quantitatively accurate information on food items consumed by terrestrial reptiles (Mondino et al., 2022; Swinehart et al., 2023), including skinks (Tercel et al., 2022). Unlike conventional methods, DNA metabarcoding is efficient in identifying differentially digested food items. Hence it can provide a robust, non-invasive tool for the dietary characterisation of tjakura while expanding our understanding of the species’ prey choice (Browett et al., 2021; Tournayre et al., 2021; Curran et al., 2022).
This study analysed scat samples of tjakura from Uluru Kata-Tjuta National Park and the surrounding Katiti-Petermann Indigenous Protected Area in central Australia. This area has previously been considered a stronghold for tjakura as it is one of the few large populations recorded in one contigous area of sandplain habitat with good fire management (McAlpin, 2001), though the population now appear to be declining (Paltridge et al., 2018; IUCN, 2023). Comparatively, this population is genetically distinct, and there is a recommendation for it to be managed separately for conservation (Dennison et al., 2015). We aimed to broaden the knowledge of the trophic ecology of tjakura by using DNA metabarcoding assays of plants, invertebrates, and vertebrates in scat samples. We investigated whether diet composition changes between seasons, age classes and among fire scars of different years. This study provides information that could be used alongside Indigenous ecological knowledge (two-way science conservation approach) to inform conservation actions such as fire management and support the recovery of tjakura.
This study was conducted at Uluru Kata-Tjuta National Park and the adjacent Katiti-Petermann Indigenous Protected Area, the traditional land of the Anangu people, in Central Australia (−25.3445, 131.034; Figure 1). This landscape is generally flat, with undulating dunes, creek lines, and the sandstone-conglomerate Uluru, and Kata-Tjuta geological formations. The vegetation is mainly composed of one or more species of spinifex grass Triodia spp. with scattered shrubs, predominantly Acacia spp., and desert oak Allocasuarina decaisneana. The climate is arid with extreme summers and cold winters, daily temperatures from November to March regularly exceed 38°C, and the coldest month is July when the average minimum is 3°C. Frequent frosts occur from June to August, with air temperatures dropping as low as −4°C (Masters, 1996). The study area is a UNESCO World Heritage site (https://whc.unesco.org/en/list/447) recognised as a living cultural landscape where Anangu continue to practice Tjukurpa, a complex belief systems that explains the physical world, and law and moral systems, connecting Anangu to country and ancestors. For its size, the park contains one of the richest assemblages of arid zone reptiles and is an important habitat for diverse native and threatened desert-dwelling species such as tjakura, brush-tailed mulgara Dasycercus blythi and Ayers Rock trigger plant Stylidium inaequipetalum.
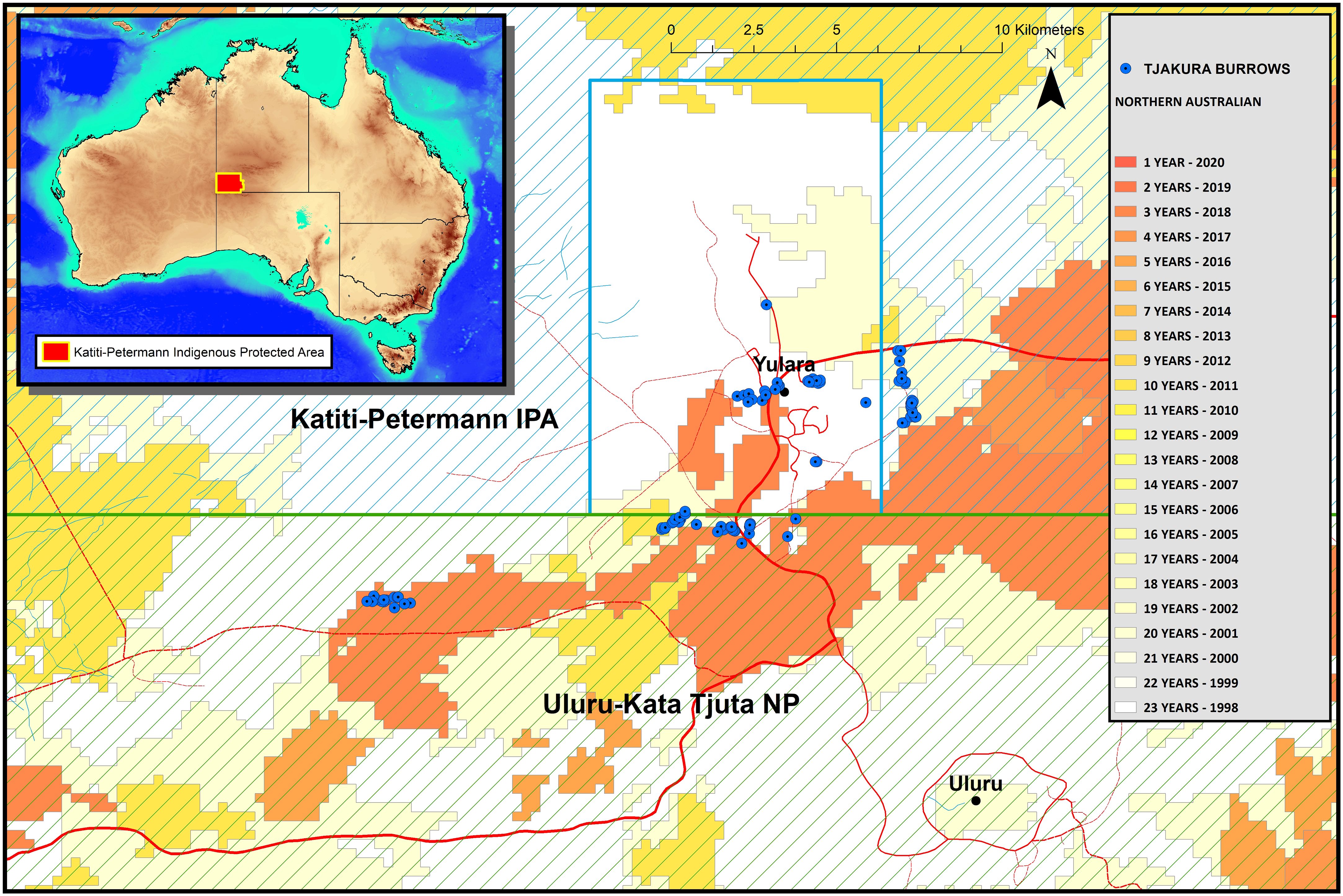
Figure 1 Map of the sampling sites within Uluru Kata-Tjuta National Park and the surrounding Katiti-Petermann Indigenous Protected Area and Yulara Conservation Area. The inset display the location of the study area in Australia, highlighted in red. The colour schemes depict time since the last fire.
Tjakura have a kin-based social structure where closely related individuals share a single multi-tunnelled burrow system (Gardner et al., 2008; McAlpin et al., 2011) and defecate in communal latrines, usually placed close to burrow entrances. Previously known and newly sighted burrows were visited between March 2022 and July 2023 (37 days of sampling in total). The sampling period spanned from kuli (hot) to wari (cold) seasons. Two to eight freshly defecated scats (based on a visual evaluation) were collected from each communal latrine and individually stored in grease-proof paper bags or 80% ethanol until DNA extraction.
Scats were sorted based on fire history (time since the burrow site was last burnt) and initially estimated age class using scat sizes (juvenile <15 mm, sub-adult 20–30 mm and adult >35 mm; IDA, 2022); however, scats of subadults could not be accurately separated from those of adults, therefore, in our analysis, subadult scats were classified as adults. We collected 97 scat samples, of which 25 were from juveniles and 72 from subadults/adults. Nine active burrows in 2022 were inactive in the subsequent sampling periods, and only 4 were sampled in both seasons. Visible beetle exoskeleton remains in the communal latrines were also collected for morphological identification (Supplementary Methods). For each sampled burrow, habitat and burrow attributes such as number of burrow entrances and estimated age of the burrow were recorded. Central Land Council, Parks Australia, and the Commonwealth Scientific and Industrial Research Organisation Animal Ethics Committee granted all relevant research and ethics permits for sample collection.
DNA was extracted from 97 scat samples using the Invitrogen ChargeSwitch® Forensic DNA Purification Kit (Invitrogen™ Life Technologies, USA) following protocols optimised by Thuo et al. (2020). Each scat was individually homogenised and approximately 40mg was used for DNA extraction. Samples were processed in batches of 24 including an extraction negative control (extraction without any scat sample). Before preparing amplicon sequencing libraries, amplification efficiency and inhibitions were first evaluated by diluting each DNA sample into three (1:1, 1:10 and 1:100) followed by a qPCR run of each dilute using the 12SV5 universal vertebrate primers (Riaz et al., 2011). We selected vertebrate assay for inhibition testing because all our DNA extract ideally contained tjakura DNA. The PCR reaction mix, and thermal cycling conditions used for the PCR efficiency test are available in Supplementary Methods 1. The DNA dilution with the highest relative proportion of the starting template (determined by Cq value) was selected for subsequent library preparations using the three assays targeting invertebrates (COI gene,ZBJ-Art; Zeale et al., 2011), vertebrates (12SV5) and plants (trnL [UAA], g and h; Taberlet et al., 2007). Three independent amplicon libraries were constructed for each sample and assay using a one-step qPCR. This step concurrently added multiplexing indices i5 and i7 and Illumina sequencing adaptors p5 and P7 at both ends of each DNA fragment. The gene-specific sequences for each primer, reaction mix, and PCR conditions are detailed in Supplementary Methods 1. The amplicon library for each assay was built by pooling 8–10 samples based on average Cq values. The pooled amplicon libraries, including all negative controls, were then purified using Agencourt™ AMPure XP Beads (Beckman Coulter Genomics, USA) following the manufacturer’s instructions. The concentration and amplicon sizes were checked using 2% agarose gel as well as a NanoDrop® ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). For each assay, pooled amplicons were combined into a single library ‘super pool’ based on equimolar concentration and agarose gel electrophoresis band sizes. The resultant three superpools were pooled into a single ‘metapool,’ purified, and sized using an Agilent 2200 TapeStation with D1000 ScreenTape assay (Agilent Technologies). The expected size of amplicons (including primers, indexes, and adaptors) was between 140 bp for trnL, 170 bp for 12SV5 and 230 bp for COI. The ‘metapool’ was sequenced using MiSeq v2 2x150 sequencing kit and PhiX control spiked at 10% at the Ramaciotti Centre for Genomics (University of New South Wales, Sydney, Australia).
The raw sequence reads were quality trimmed and sequencing adaptors were removed using TRIMMOMATIC software (Bolger et al., 2014). TRIMMOMATIC removed poor-quality bases (Q-score <30) at the end of each read and then using a sliding window of 4 bases it clipped reads when the average quality per base was below 15. The remaining high-quality sequences were analysed using the OBITOOLS program (http://metabarcoding.org/obitools; Boyer et al., 2016). The ILLUMINAPAIRED function was used to assemble forward and reverse reads to recover full amplicon sequences. Pairs that merged with a lower overlap alignment score (<0.8) were discarded before downstream analysis. Best-paired sequences were then assigned to their respective sample based on MID tags using the NGSFILTER program. For each sample, reads were collapsed into unique reads using OBIUNIQ (dereplication step) and shorter reads (shorter than 40 bp for trnL, 70 bp for 12V5 and 100 bp for COI) were removed using the OBIGREP command. OBICLEAN command was then used to remove PCR and sequencing artefacts. Lastly, human sequences were filtered from the dataset as these were likely as a result of contaminations.
Taxonomic assignments were carried out using ECOTAG. This command assigns the queried sequence to the last common ancestor when the identification is ambiguous among sibling taxa (Boyer et al., 2016). The taxonomic identification was done using respective reference databases built using the standard trnL, 12V5 and COI sequences from the EMBL (release 140; https://www.ebi.ac.uk/and NCBI (https://ftp.ncbi.nih.gov/). The ECOTAG function was set only to assign sequence to taxon if the similarity was equal or higher than 97%. Results with similarity lower than 96% were considered unclassified taxa. The final ECOTAG output files were imported in R (version 4.2.2; www.r-project.org) for downstream and statistical analysis. Some reads were assigned to species or genera that are not present in our study area; these were reassigned or discarded based on relative abundance and prior knowledge of the distribution of taxa recorded in the Atlas of Living Australia (https://www.ala.org.au/). For example, all sequences assigned to false spinifx (Triodia compacta), which occurs well to the south of our study area, were reassigned to a single genus level assignment Triodia.
Tjakura dietary description was summarised as i) percentage of relative read abundance, RRA (percentage of read count for each taxon in a sample), (ii) weighted percentage of occurrence, wPOO (percentage of occurrence for each taxon in a sample) and (iii) frequency of occurrence, FOO (proportion of samples in which a taxon was detected). All taxa read numbers were then converted to presence/absence data and patterns in dietary dissimilarity between age classes (juvenile and subadult/adult), season (summer vs. winter) and among different fire regimes (1998, 2002, 2012 and 2018) calculated based on Jaccard dissimilarity distance. Analysis of similarities (ANOSIM) was used to detect whether diet vcomposition aried between age class, seasons, and fire sites (fire scars), with 999 permutations of the test statistics. We visualized the results with hierarchical clustering analysis using NMDS (nonmetric multidimensional scaling) ordination using the metaMDS function in vegan 2.6-4 in R. Finally, we calculated Shannon diversity (Alpha diversity) by age class, seasons, and time since last fire in vegan package.
Overall, the sequencing run generated ~14 million paired-end reads from 97 scat samples, an average of 143,968 per sample. After denoising and retaining only plants, invertebrates, and vertebrates, ~10.9 million (78.3%) sequences were retained (1,603,575, 4,695,934 and 4,653,907 for plants, invertebrates, and vertebrates respectively). In total, 78 taxonomic categories across all levels of biological classifications were identified in the scats. Of these, 48 were invertebrates, 27 plants and three vertebrate taxa; including unidentified Liopholis spp. suspected to be from the individual tjakura that had produced the scats and thus excluded from the analysis. 18 plant operational taxonomic units (OTUs) were identified to species, six to genus, two to family and one to ordinal taxonomic levels. For invertebrates, we identified six species, 22 genera, 16 family/subfamily and one each to class and phylum. The three vertebrate OTUs were identified to generic level.
Among the most abundant and frequently consumed items, we detected a high proportion of plants (Malvastrum americunum (37.7% RRA and 18% FOO)), and invertebrates (Leucophenga sp. (51% RRA and 2.9% FOO), Culex sp. (6.5% RRA) and Harpalus spp. (44.3% FOO). Brassica tournefortii (22.3% RRA), Chlaenius darlingensis (14.3% RRA) and Termitidae (11.9% RRA) each contributing >10% to the diet of tjakura (Figure 2). A button quail Turnix sp. and unidentified lizard Tiliqua spp. were the only vertebrates detected and both at 0.1% RRA. Spiders (Sparassidae), butterflies (Lycaenidae), terrestrial snails (Gastropoda) and centipedes (Scutigera) were also frequently detected. We found on average four dietary taxa per scat whereas seven scat samples produced only tjakura DNA.
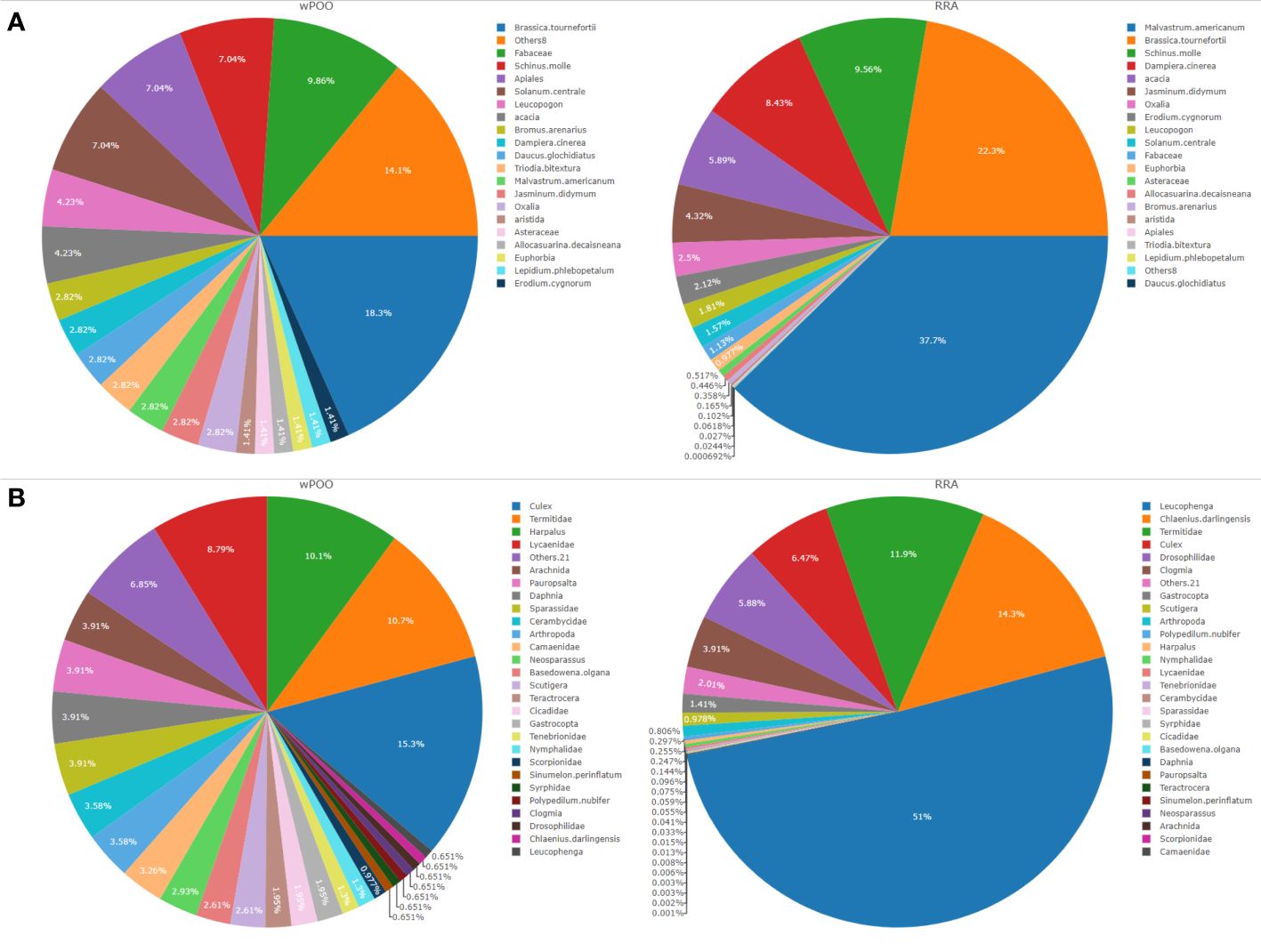
Figure 2 Dietary composition of tjakura based on (A) Weighted percentage of occurrence (wPOO) and relative read abundance (RRA) of trnL (UAA) and (B) POO and RRA of COI (ZBJ-Art) marker. All the taxa with wPOO and RRA of <1% (trnL n=8 and COI n=21) were grouped into ‘Others’.
Culex returned the highest abundance of invertebrate taxa during the two seasons followed by unidentified Termitidae and Harpalus spp. Only six plant taxa (Schinus mole, Apiales, Fabaceae, Brassica tournefortii, Triodia bitextura and Hydrocotyle trachycarpa) were detected in winter and summer seasons, with the majority of plant taxa occurring in scats collected during summer. However, ANOSIM pairwise tests revealed that these seasonal differences were not statistically significant (p = 0.7; Figure 3).
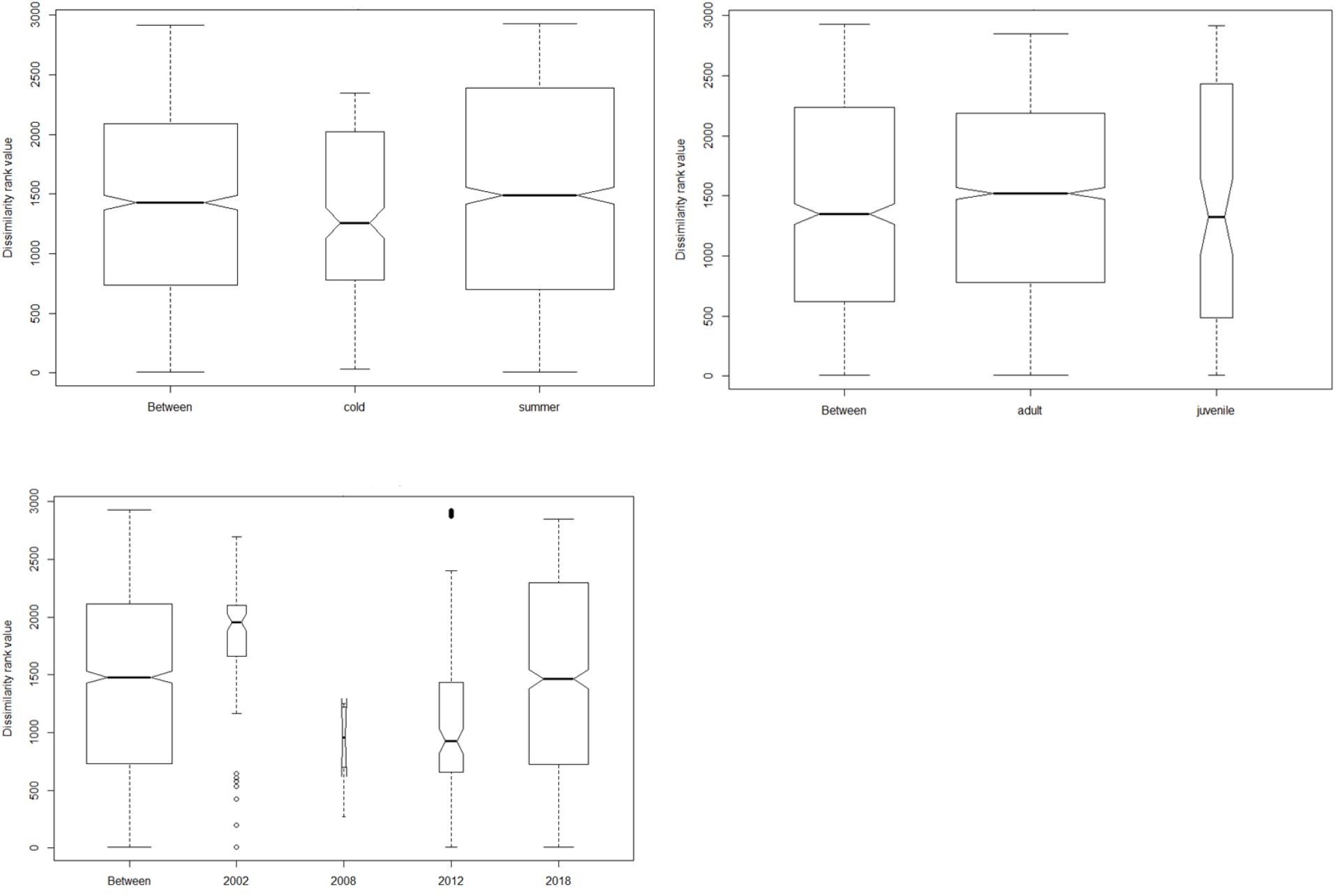
Figure 3 Analysis of similarity plots showing the differences in diet between and within seasons, age class and time since last fire. The width of the bar is directly proportional to the sample size.
We found no differences between dietary species at any taxonomic levels in comparisons among tjakura age classes and among post-fire scars as shown by the ANOSIM results in Figure 3. However, Shannon indices revealed high diversity of consumed prey in summer (H= 2.31) and in area burned in 2018 (H= 2.29). Alpha diversity values also showed that diet of subadult/adult skinks was subtly different to that of juveniles and that diet in the area burned in 2008 had the lowest diversity.
NMDS showed generally high dietary overlap between fire regimes, seasons, and tjakura age class, but considerable separation was observed between areas last burned in 2002 and 2018 and between areas burned in 2002 and 2012, and between adults and juvenile age classes (Figure 4). The kernel distribution estimation plot on NMDS1 showed that samples collected in the 2002 fire scar were the least overlapping of all fire regimes.
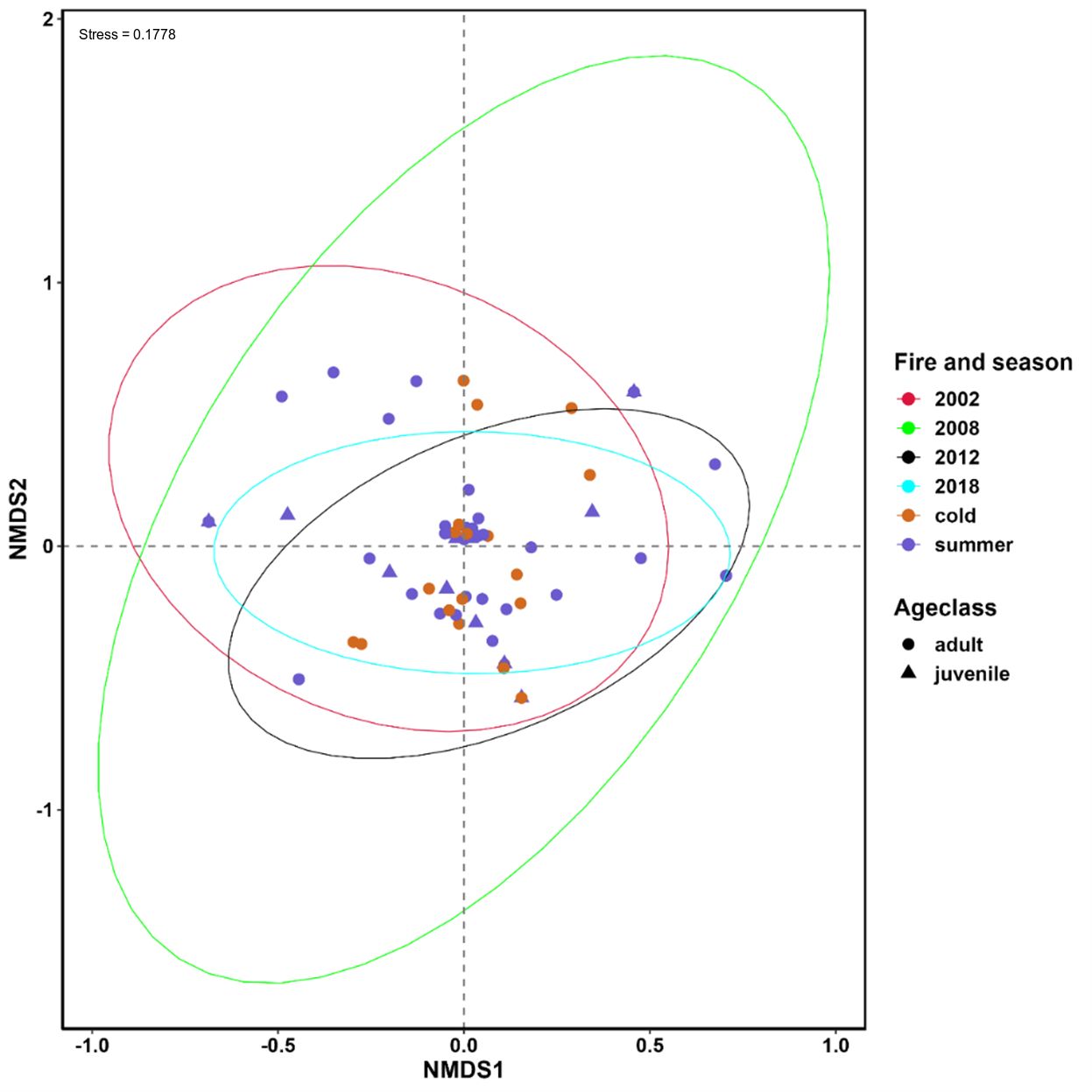
Figure 4 Nonmetric multidimensional scaling (NMDS) showing the differences in the dietary composition of tjakura between seasons, age classes and among time since the last fire. The stress level of NMDS is shown in the plot.
This study is the first application of scat DNA metabarcoding to assess the trophic ecology of tjakura. Our findings highlight the power of scat metabarcoding to assess diet and provide insights into foraging behaviour of cryptic and difficult-to-study reptile species. We found that tjakura could be considered an opportunist generalist, consuming a diverse range of invertebrates and plants supplemented at least occasionally with small vertebrate prey items. Invertebrates of the genera Leucophenga (fruit flies), Harpalus (ground beetles), Culicidae (mosquitos) and Termitidae (termites) were the most abundant and frequently consumed taxa while plants of species Malvastrum americunum, Acacia spp., Solanum centrale, Brassica tournefortii and an Apiales sp. constituted a substantial proportion of the diet (40.2% FOO). Both diurnally active prey species such as Lycaenidae and nocturnal taxa such as Lycosidae were detected which support the observation that tjakura actively forage both during the day and at night.
Termites and beetles have been described as important invertebrate taxa in the diet of a wide variety of omnivorous and insectivorous skinks (Duffield and Bull, 1998; McAlpin, 2001; Manicom and Schwarzkopf, 2011; Civantos et al., 2013). This is probably due to their high nutritive value (Razeng and Watson, 2015), abundance, and ease of capture (Civantos et al., 2013). In Australia, beetles are highly abundant and diverse, with more than 117 different families and over 23,000 described species (Lawrence and Slipinski., 2013). Our study site is characterised by termite mounds having many tjakura burrows within a few meters. This habitat structure could explain the abundance of termites in tjakura scats. In contrast to previous observations that termites (Drepanotermes spp.) are the most common prey for this species (McAlpin, 2001; Moore, 2018; IDA, 2022), our study found Culicidae (15.3% wPOO) to be the most frequently consumed insect taxon followed by termites (10.7% wPOO) and beetles (10.1% wPOO). Mosquitos are common in arid environments and often rest on vegetation (Schlein and Müller, 2012). It is possible that they are taken by tjakura purely opportunistically or accidentally while feeding on leaves or flowers. High detection of Culicidae also underscores the accuracy of molecular analysis in detecting soft-bodied prey species that leave few or no hard remains in the scats. This is highly important in that previuos traditional approaches tend to rely on morphological anaysis of prey items in scat samples. This tends to over-represent hard-bodied prey items. Other important invertebrate taxa consumed by tjakura were spiders (Sparassidae), butterflies (Lycaenidae), terrestrial snails (Gastrocopta) and centipedes (Scutigera). Leucophenga sp. and other unidentified Drosophilidae were also widely represented in the diet. Given that they are flying insects and that they accumulate in the tjakuras’ communal latrines it is possible that they were mostly consumed there. It is worth noting, however, that this could also be a result of post-defecation contamination when flies interact with the scats. The issue of contamination with non-food DNA is a common challenge in scat DNA metabarcoding studies (Pompanon et al., 2012; Ando et al., 2018; Curran et al., 2022).
Plants make up the second most consumed category and flowering and fruiting species such as Malvastrum americunum, Solanum centrale and Brassica tournefortii are the most frequently consumed taxa (Figure 2). This preference is consistent with previous observational studies that have demonstrated that tjakura and other omnivorous lizards tends to feed on easily digestible and seasonally abundant plants (McAlpin, 2001; Cooper and Vitt, 2002; Chapple, 2003). Even though the seasonal variation in the plant composition was miniscule, our study demonstrated that tjakura consumed more plant material during the summer (kuli) period when many plants are flowering (DCCEEW, 2023). During the cooler season, we only detected Schinus mole, Fabaceae, Brassica tournefortii and Triodia bitextura. This was likely an indication of decreased forage abundance and quality coupled with reduced tjakura activity as ambient temperature begin to decline. Skinks are known to reduce their feeding during cooler seasons (Wapstra and Swain, 1996). However, it should be noted that this plant diversity was conditioned by the taxonomic resolution provided by the P6 loop region of the trnL primers set.
Fire has various beneficial or detrimental influence on plant and animal communities (Pons and Clavero, 2010; Sarsha et al., 2018). Wildfire and poorly controlled fires can lead to local extinctions of threatened species such as tjakura. This can be due to reduced opportunities for dispersal and colonisation (McAlpin, 2001; Lazzari, 2019) and loss of vegetation cover which is ideal for survival of this species (Ridley et al., 2020; Ridley and Schlesinger, 2023). On the other hand, properly prescribed burning can be a driver of species richness and community composition on natural ecosystems (Cadenhead et al., 2016; Rainsford et al., 2021; Zylinski and Swan, 2022). Our study demonstrated that time since the last fire had an influence on dietary diversity though it was not statistically significant (Figure 3). Dietary diversity in the more recently burned fire scars (2018) was higher than in older fire scars. This may indicate that there is a greater variety of food items in recently burned areas, or that skinks are switching to new items in those areas if preferred resources are less available. Fire ecology studies in other animals have shown similar increases in animal diversity in recently burned areas (Zylinski and Swan, 2022). Notably, though from limited sample sizes, diet diversity in fire scars of 2008 and 2012 were lower than that in the 2002 fire scar. This could be attributed to the particularly severe Millenium Drought (1997–2009) and subsequent droughts in central Australia (Gergis et al., 2012) which may have modified the landscape at similar scale to fire. Further research is needed to disentangle possible drivers of tjakura’s dietary and habitat diversity.
Evidence of diet partitioning by age among lizards is widely recorded and juveniles consume a wider range of prey taxa than adults for energy gains (Best and Pfaffenberger, 1987; Chapple, 2003; Rato et al., 2022). In our study no significant intraspecific differences in diet were found between the two age groups. However, this could be attributed to the smaller sample size for juveniles as their scats were rarer during our surveys. The only two vertebrate taxa that we detected were consumed by adults. This is expected as juveniles are too small to take vertebrates (McAlpin, 2001) and may lack predatory skills required to handle certain types of vertebrate species. ANOSIM and diversity analysis did not support plant consumption in tjakura being related to age class as previously observed with some species of Liopholis (Chapple, 2003). However, this should be investigated further with increased sample sizes.
Our metabarcoding approach used 12S-V5 primer set designed for detection of vertebrate species (Riaz et al., 2011). Hence, our sequencing results yielded a large number of reads assigned to the host species. While this overrepresentation of host DNA can be addressed using blocking primers (Vestheim and Jarman, 2008), blocking primers may obscure the identification of ingested sloughed shed skin and filial cannibalism, which is not uncommon in lizards (Clemann et al., 2004; Huang, 2008; Manicom and Schwarzkopf, 2011; William et al., 2014; Mateo and Pleguezuelos, 2015; Labra et al., 2022). Two vertebrate taxa, representing less than 1% of the overall diet were identified from the scat samples. These were a bird and a lizard and were Little-Button quail (Turnix velox) and Tiliqua sp., respectively. Tjakura has previously been recorded feeding on small vertebrates (IDA, 2022) but to the best of our knowledge this study is the first to record a bird in their diet. Although tjakura are known to ambush passing prey, it is unlikely that they subdued a mature Turnix sp. suggesting that tjakura actively prey on eggs or chicks of ground-nesting birds such as Turnix velox (Forshaw and Knight, 2023). Two scats in our study had hair samples potentially from the readily available spinifex hopping mouse (Notomys alexis), however we did not identify any mammal species using our metabarcoding approach. In addition, beetles of family Tenebrionidae and Trogidae and weevils of family Curculionidae were only identified using morphological analysis of faecal remains. This discrepancy could be as a result of methodological differences between conventional and metabarcoding techniques. It shows the need for future studies to account for the differences in sensitivity between these methods and, if possible, combine both techniques (Kurita and Toda, 2022).
Our invertebrate metabarcoding assay also detected two parasites (mite Dermatophagoides sp. and nematode Cylicocyclus sp.) in the scats. Although we did not quantify their load on the animals this may be a potential problem for social animals such as tjakura due to high contagion probabilities. The parasites may also pose a zoonotic risk as previously observed with snake parasites (Hossain et al., 2023). The detection of vector-borne organisms underscores the ecosystem services that tjakura provides by suppressing the disease vetors (mosquitos) and shedding potential zoonotic-risk parasites.
Overall, our study showed a high overlap in dietary composition between age groups, seasons and among fire scars. This suggests potential for high intraspecific competition especially when food items are scarce. In the long term, this may strongly impact on population dynamics and other aspects of tjakura life history (Donihue et al., 2016).
The current study demonstrates that tjakura have a broad omnivorous diet dominated by invertebrates. We found at least 78 taxa of which Leucophenga, Harpalus, Culicidae, Termitidae, Malvastrum americanum, Solanum centrale, Brassica tournefortii and Apiales were some of the key consumed taxa. The detailed dietary diversity and plasticity identified in this study suggest that tjakura may be generalist feeders that are able to switch their foraging preferences in response to environmental disturbances and change in food resources and that therefore they may be resilient to some changes in food availability. However, for long-term resilience and recovery of tjakura conservation measures should ideally be implemented to support the full range of important food sources identified in this study. Our results predict potentially high intraspecific competition when food is scarce because there was no clear resource partitioning between age groups and seasons. Our data also support that recently burned areas are important for harbouring suitable and diverse food sources for tjakura but this needs to be balanced with an understanding of the minimal vegetation age post-fire that is habitable by tjakura. Another question for further study is how diet, and dietary changes, relate to fitness and survivorship. The results of this study reinforce the need for continued indigenous fire management and ecological restoration programs to promote resource availability year-round. They also demonstrate the utility of non-invasive sampling and DNA metabarcoding techniques for studying the diet of many other culturally important species. This method has provided invaluable insights that can be used alongside traditional knowledge for species conservation and in mitigation of potential human-health risks through identification of potential zoonotic spillover.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
The animal study was approved by CSIRO Wildlife, Livestock and Laboratory Animal Ethics Committee. The study was conducted in accordance with the local legislation and institutional requirements.
DT: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Project administration. NM: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. DS: Conceptualization, Funding acquisition, Resources, Writing – review & editing. SaM: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. JKeo: Funding acquisition, Resources, Writing – review & editing. JKen: Investigation, Methodology, Writing – review & editing, Resources. TG: Investigation, Methodology, Resources, Writing – review & editing, Data curation. JW: Investigation, Resources, Writing – review & editing, Data curation. SS: Investigation, Resources, Supervision, Writing – review & editing, Data curation. StM: Methodology, Resources, Supervision, Writing – review & editing, Data curation. LJ: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review & editing, Investigation.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Funding to support this study was jointly provided by the Commonwealth Scientific and Industrial Research Organisation and the Director of National Parks.
The authors would like to thank the indigenous people in central Australia, especially the Anangu people who are the traditional custodians of our study area. We are indebted to many individuals and organisations that helped with sample collection, especially Uluru Kata-Tjuta National Park Rangers, Mutitijulu Community rangers, Tjakura Rangers, Parks Australia and CSIRO. We thank the Anangu traditional owners, Director of National Parks (Permit number: R1/22), Central Land Council (Authority number: P78658), Voyages Ayers Rock Resort and CSIRO Animal Ethics Committee (permit number: AEC 2022-01) for granting us permission to conduct our study in Uluru Kata-Tjuta National Park, Katiti-Petermann IPA and adjacent land. We are grateful to Adam Slipinski and Living Li for their expertise in the morphological identification of beetle remains. We also extend our gratitude to Christopher Wilson for his assistance in preparing Figure 1 and creating our sampling maps, To Margarita Goumas, Alex Drew, Tonya Haff, Eric Doerr and Rebecca Pirzl for assistance with fieldwork and constructive comments on a draft of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2024.1354138/full#supplementary-material
Ando H., Fujii C., Kawanabe M., Ao Y., Inoue T., Takenaka A. (2018). Evaluation of plant contamination in metabarcoding diet analysis of a herbivore. Sci. Rep. 8, 2–11. doi: 10.1038/s41598-018-32845-w
Best T. L., Pfaffenberger G. S. (1987). Age and Sexual Variation in the Diet of Collared Lizards (Crotaphytus collaris) The Southwestern Naturalist, 32 (4), 415–426. Available at: https://www.jstor.org/stable/3671473.
Bogert C. M. (1949). Thermoregulation in reptiles; a factor in evolution. Evolution; Int. J. Organic Evol. 3, 195–211. doi: 10.2307/2405558
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Boyer F., Mercier C., Bonin A., Le Bras Y., Taberlet P., Coissac E. (2016). OBITOOLS: a UNIX-inspired software package for DNA metabarcoding. Molecular Ecology Resources 16 (1), 176–182. doi: 10.1111/1755-0998.12428
Browett S. S., Curran T. G., O’Meara D. B., Harrington A. P., Sales N. G., Antwis R. E., et al. (2021). Primer biases in the molecular assessment of diet in multiple insectivorous mammals. Mamm. Biol. 101, 293–304. doi: 10.1007/s42991-021-00115-4
Cadenhead N. C. R., Kearney M. R., Moore D., McAlpin S., Wintle B. A. (2016). Climate and fire scenario uncertainty dominate the evaluation of options for conserving the great desert skink. Conserv. Lett. 9, 181–190. doi: 10.1111/conl.12202
Chapple D. (2003). Ecology, life-history, and behavior in the Australian scincid genus Egernia, with comments on the evolution of complex sociality in lizards. Herpetological Monogr. 17, 145–180. doi: 10.1655/0733-1347(2003)017[0145:ELABIT]2.0.CO;2
Civantos E., Ortega J., López P., Pérez-Cembranos A., Pérez-Mellado V., Martín J. (2013). Diet selection by the threatened Chafarinas’ skink Chalcides parallelus in North Africa. Afr. J. Herpetology 62, 78–89. doi: 10.1080/21564574.2013.799101
Clemann N., Chapple D. G., Wainer J. (2004). Sexual dimorphism, diet, and reproduction in the swamp skink. Egernia coventryi. 38 (3), 461–467. doi: 10.1670/224-03A-N
Cooper W., Vitt L. J. (2002). Distribution, extent, and evolution of plant consumption by lizards. Journal of Zoology 257 (4), 487–517. doi: 10.1017/S0952836902001085
Cox N., Young B. E., Bowles P., Fernandez M., Marin J., Rapacciuolo G., et al. (2022). A global reptile assessment highlights shared conservation needs of tetrapods. Nature 605, 285–290. doi: 10.1038/s41586-022-04664-7
Curran T. G., Browett S. S., O’Neill D., O’Hanlon A., O’Reilly C., Harrington A. P., et al. (2022). One bat’s waste is another man’s treasure: a DNA metabarcoding approach for the assessment of biodiversity and ecosystem services in Ireland using bat faeces. Biodiversity Conserv. 31, 2699–2722. doi: 10.1007/s10531-022-02451-4
DCCEEW. (2023). Understanding the country, Uluru-Kata Tju t a National Park. Available at: https://www.dcceew.gov.au/sites/default/files/documents
de Miranda E. B. P. (2017). The plight of reptiles as ecological actors in the tropicss. Front. Ecol. Evol. 5. doi: 10.3389/fevo.2017.00159
Dennison S., McAlpin S., Chapple D. G., Stow A. J. (2015). Genetic divergence among regions containing the vulnerable great desert skink (Liopholis kintorei) in the Australian Arid Zone. PloS One 10, 1–14. doi: 10.1371/journal.pone.0128874
Donihue C. M., Brock K. M., Foufopoulos J., Herrel A. (2016). Feed or fight: Testing the impact of food availability and intraspecific aggression on the functional ecology of an island lizard. Funct. Ecol. 30, 566–575. doi: 10.1111/1365-2435.12550
Duffield G. A., Bull C. M. (1998). Seasonal and ontogenetic changes in the diet of the Australian skink Egernia stokesii. Herpetologica 54, 414–419.
EPBC. (1999) Environment protection and biodiversity conservation act 1999, department of agriculture, water and the environment (DAWE). Available online at: https://www.legislation.gov.au/Details/C2016C00777.
Forshaw J., Knight F. (2023). “Quail, buttonquail and plains-wanderer in Australia and New Zealand. In CSIRO Publishing. CSIRO Publishing. doi: 10.1071/9781486312603
Gardner M. G., Hugall A. F., Donnellan S. C., Hutchinson M. N., Foster R. (2008). Molecular systematics of social skinks : phylogeny and taxonomy of the Egernia group (Reptilia : Scincidae). Zool. J. Linn. Soc. 781–794. doi: 10.1111/j.1096-3642.2008.00422.x
Gergis J., Jane A., Gallant E., Braganza K., Karoly D. J., Allen K., et al. (2012). On the long-term context of the 1997 – 2009 ‘ Big Dry ‘ in South-Eastern Australia : insights from a 206-year multi-proxy rainfall reconstruction. Climatic Change 923–944. doi: 10.1007/s10584-011-0263-x
Hossain M. E., Kennedy K. J., Wilson H. L., Spratt D., Koehler A., Gasser R. B., et al. (2023). Human neural larva migrans caused by ophidascaris robertsi ascarid. Emerging Infect. Dis. 29, 1900–1903. doi: 10.3201/eid2909.230351
Huang W. (2008). Predation risk of whole-clutch filial cannibalism in a tropical skink with maternal care. doi: 10.1093/beheco/arn109
IDA A. I. D. (2022). Looking after Tjakura, Tjalapa, Mulyamiji, Warrarna, Nampu Draft National Recovery Plan for the Great Desert Skink (Liopholis kintorei). DCCEEW 2022–2032. Available at: https://www.dcceew.gov.au/sites/default/files/documents
Isbell F., Balvanera P., Mori A. S., He J. S., Bullock J. M., Regmi G. R., et al. (2023). Expert perspectives on global biodiversity loss and its drivers and impacts on people. Front. Ecol. Environ. 21, 94–103. doi: 10.1002/fee.2536
Kartzinel T. R., Pringle R. M. (2015). Molecular detection of invertebrate prey in vertebrate diets: Trophic ecology of Caribbean island lizards. Mol. Ecol. Resour. 15, 903–914. doi: 10.1111/1755-0998.12366
Kurita T., Toda M. (2022). Comparison of morphological identification and DNA metabarcoding for dietary analysis of faeces from a subtropical lizard. Wildlife Res. 50, 224–236. doi: 10.1071/WR22054
Labra A., Daigre M., Imil V. (2022). Cannibalism in microlophus lizards. J. Herpetology 56, 67–74. doi: 10.1670/20-132
Lawrence J., Slipinski A. (2013). Australian Beetles. In Australian Beetles (Vol. 1). doi: 10.1071/9780643100695
Lazzari J. (2019). Fire and fragmentation interactions : effects on reptiles and small mammals in modified semi-arid landscapes. ANU, ProQuest Dissertations Publishing, October. Available at: https://openresearch-repository.anu.edu.au/handle/1885/177008.
Manicom C., Schwarzkopf L. (2011). Diet and prey selection of sympatric tropical skinks. Austral Ecol. 36, 485–496. doi: 10.1111/aec.2011.36.issue-5
Masters P. (1996). The effects of fire-driven succession on reptiles in spinifex grasslands at Uluru national park, Northern Territory. Wildlife Res. 23, 39–48. doi: 10.1071/WR9960039
Mateo J. A., Pleguezuelos J. M. (2015). Cannibalism of an endemic island lizard (genus Gallotia). Zoologischer Anzeiger 259, 131–134. doi: 10.1016/j.jcz.2015.07.003
McAlpin S. (2001). A recovery plan for the Great Desert Skink (Egernia kintorei (Alice Springs, NT: Arid Lands Environment Centre), 29. Available at: http://155.187.2.69/biodiversity/threatened/publications/recovery/great-desert-skink/pubs/great-desert-skink.rtf.
McAlpin S., Duckett P., Stow A. (2011). Lizards cooperatively tunnel to construct a long-term home for family members. PLOS ONE 6 (5), 4–7. doi: 10.1371/journal.pone.0019041
Mondino A., Crovadore J., Lefort F., Ursenbacher S. (2022). Impact of invading species on biodiversity: Diet study of the green whip snake’s (Hierophis viridiflavus, L. 1789) in Switzerland. Global Ecol. Conserv. 38, e02239. doi: 10.1016/j.gecco.2022.e02239
Moore D. (2018). An investigation of potential threatening processes for the Vulnerable great desert skink, Liopholis kintorei. Macquarie University. Thesis. doi: 10.25949/19440302.v1
Moore D., Kearney M. R., Paltridge R., Mcalpin S., Stow A. (2015). Is fire a threatening process for a nationally listed threatened skink? Wildlife Res. 42, 207–216. doi: 10.1071/WR14227
Moore D., Kearney M. R. A. Y., Paltridge R., Mcalpin S., Stow A. (2018). Feeling the pressure at home : Predator activity at the burrow entrance of an endangered arid-zone skink. 102–109. doi: 10.1111/aec.12547
Nowak M. A., Tarnita C. E., Wilson E. O. (2010). The evolution of eusociality. Nature 466, 1057–1062. doi: 10.1038/nature09205
Paltridge R., Catt G., Cowan M., Gaikhorst G., How R., ZichyWoinarski J., et al. (2018). Liopholis kintorei — Great desert skink, tjakura, warrarna, mulyamiji. 8235, 3–6. doi: 10.2305/IUCN.UK.2018-1.RLTS.T7040A101743329.en
Pimenta B. V. S., Haddad C. F. B., Nascimento L. B., Cruz C. A. G., Pombal J. P. (2005). Comment on “Status and trends of amphibian declines and extinctions worldwide”. Science 309, 1783–1787. doi: 10.1126/science.1112996
Pompanon F., Deagle B. E., Symondson W. O. C., Brown D. S., Jarman S. N., Taberlet P. (2012). Who is eating what: Diet assessment using next generation sequencing. Mol. Ecol. 21, 1931–1950. doi: 10.1111/j.1365-294X.2011.05403.x
Pons P., Clavero M. (2010). Bird responses to fire severity and time since fire in managed mountain rangelands. Animal Conservation 13 (3), 294–305. doi: 10.1111/j.1469-1795.2009.00337.x
Rainsford F. W., Kelly L. T., Leonard S. W. J., Bennett A. F. (2021). How does prescribed fire shape bird and plant communities in a temperate dry forest ecosystem? Ecol. Appl. 31, 1–16. doi: 10.1002/eap.2308
Rato C., Dellinger T., Carretero M. A. (2022). Dietary variation is driven by landscape heterogeneity in an insular omnivorous endemic lizard, revealed by DNA metabarcoding. Diversity 14 (12), 1130. doi: 10.3390/d14121078
Razeng E., Watson D. M. (2015). Nutritional composition of the preferred prey of insectivorous birds: Popularity reflects quality. J. Avian Biol. 46, 89–96. doi: 10.1111/jav.00475
Riaz T., Shehzad W., Viari A., Pompanon F., Taberlet P., Coissac E. (2011). EcoPrimers: Inference of new DNA barcode markers from whole genome sequence analysis. Nucleic Acids Res. 39, 1–11. doi: 10.1093/nar/gkr732
Ridley J. C. H., Bull C. M., Schlesinger C. A. (2020). Location of long-term communal burrows of a threatened arid-zone lizard in relation to soil and vegetation. Austral Ecology 45 (4), 444–453. doi: 10.1111/aec.12656
Ridley J. C. H., Schlesinger C. A. (2023). Activity of tjakura (great desert skinks) at burrows in relation to plant cover and predators. Ecology and Evolution 123 (July), 1–15. doi: 10.1002/ece3.10391
Roll U., Feldman A., Novosolov M., Allison A., Bauer A. M., Bernard R., et al. (2017). The global distribution of tetrapods reveals a need for targeted reptile conservation. Nat. Ecol. Evol. 1, 1677–1682. doi: 10.1038/s41559-017-0332-2
Romano A., Séchaud R., Roulin A. (2020). Global biogeographical patterns in the diet of a cosmopolitan avian predator. J. Biogeography 47, 1467–1481. doi: 10.1111/jbi.13829
Sarsha G., Greenlees M., Shine. R. (2018). The impact of wildfire on an endangered reptile (Eulamprus leuraensis) in Australian montane swamps. International Journal OfWildland Fire 27 (7), 447–456. doi: 10.1071/WF17048
Schlein Y., Müller G. C. (2012). Diurnal resting behavior of adult Culex pipiens in an arid habitat in Israel and possible control measurements with toxic sugar baits. Acta Tropica 124, 48–53. doi: 10.1016/j.actatropica.2012.06.007
Seebacher F., Franklin C. E. (2005). Physiological mechanisms of thermoregulation in reptiles: A review. J. Comp. Physiol. B: Biochemical Systemic Environ. Physiol. 175, 533–541. doi: 10.1007/s00360-005-0007-1
Swinehart A., Partridge C., Russell A., Thacker A., Kovach J., Moore J. (2023). Diet of a threatened rattlesnake (eastern massasauga) revealed by DNA metabarcoding. Ecol. Evol. 13, 1–14. doi: 10.1002/ece3.10029
Taberlet P., Coissac E., Pompanon F., Gielly L., Miquel C., Valentini A., et al. (2007). Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 35 (3), e14–e14. doi: 10.1093/nar/gkl938
Tercel M. P. T. G., Moorhouse-Gann R. J., Cuff J. P., Drake L. E., Cole N. C., Goder M., et al. (2022). DNA metabarcoding reveals introduced species predominate in the diet of a threatened endemic omnivore, Telfair’s skink (Leiolopisma telfairii). Ecol. Evol. 12, 1–14. doi: 10.1002/ece3.8484
Thuo D., Broekhuis F., Furlan E., Bertola L. D., Kamau J., Gleeson D. M. (2020). An insight into the prey spectra and livestock predation by cheetahs in Kenya using faecal DNA metabarcoding. Zoology 143, 125853. doi: 10.1016/j.zool.2020.125853
Tournayre O., Leuchtmann M., Galan M., Trillat M., Piry S., Pinaud D., et al. (2021). eDNA metabarcoding reveals a core and secondary diets of the greater horseshoe bat with strong spatio-temporal plasticity. Environ. DNA 3, 277–296. doi: 10.1002/edn3.167
Valencia-Aguilar A., Cortés-Gómez A. M., Ruiz-Agudelo C. A. (2013). Ecosystem services provided by amphibians and reptiles in Neotropical ecosystems. Int. J. Biodiversity Science Ecosystem Serv. Manage. 9, 257–272. doi: 10.1080/21513732.2013.821168
Vestheim H., Jarman S. N. (2008). Blocking primers to enhance PCR amplification of rare sequences in mixed samples - a case study on prey DNA in Antarctic krill stomachs. Front. Zoology 5, 12. doi: 10.1186/1742-9994-5-12
Vitt L. J., Pianka E. R. (2005). Deep history impacts present-day ecology and biodiversity. Proc. Natl. Acad. Sci. United States America 102, 7877–7881. doi: 10.1073/pnas.0501104102
Wapstra E., Swain R. (1996). Feeding ecology of the tasmanian spotted skink, niveoscincus ocellatus (Squamata : scincidae). Australian Journal of Zoology 44 (2), 205–213. doi: 10.1071/ZO9960205
Westeen E. P., Martínez-Fonseca J. G., d’Orgeix C. A., Walker F. M., Sanchez D. E., Wang I. J. (2023). Dietary niche partitioning of three Sky Island Sceloporus lizards as revealed through DNA metabarcoding. Ecol. Evol. 13, 1–12. doi: 10.1002/ece3.10461
William C., Dimopoulos I., Pafili P. (2014). Sex, age, and population density affect aggressive behaviors in island lizards promoting cannibalism. Ethology 121 (Bartlett 1987), 207–314. doi: 10.1111/eth.12335
Zeale M. R. K., Butlin R. K., Barker G. L. A., Lees D. C., Jones G. (2011). Taxon-specific PCR for DNA barcoding arthropod prey in bat faeces. Mol. Ecol. Resour. 11, 236–244. doi: 10.1111/j.1755-0998.2010.02920.x
Keywords: Liopholis kintorei, scats, fire management, skink’s diet, threatened species
Citation: Thuo D, Macgregor NA, Merson SD, Scopel D, Keogh JS, Kenny J, Williams JL, Guest T, Swan S, McAlpin S and Joseph L (2024) Metabarcoding clarifies the diet of the elusive and vulnerable Australian tjakura (Great Desert Skink, Liopholis kintorei). Front. Ecol. Evol. 12:1354138. doi: 10.3389/fevo.2024.1354138
Received: 11 December 2023; Accepted: 12 March 2024;
Published: 26 March 2024.
Edited by:
David Jack Coates, Conservation and Attractions (DBCA), AustraliaReviewed by:
Guinevere O.U. Wogan, Oklahoma State University, United StatesCopyright © 2024 Thuo, Macgregor, Merson, Scopel, Keogh, Kenny, Williams, Guest, Swan, McAlpin and Joseph. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Thuo, david.thuo@csiro.au
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.