- 1School of Public Health, Lanzhou University, Lanzhou, China
- 2Institute of Archaeological Science, Fudan University, Shanghai, China
- 3Department of Radiology, The First Affiliated Hospital of Shantou University Medical College, Shantou, China
- 4Ministry of Education (MOE) Key Laboratory of Contemporary Anthropology, School of Life Sciences, Fudan University, Shanghai, China
- 5Department of History, Fudan University, Shanghai, China
- 6Criminal Justice College, China University of Political Science and Law, Beijing, China
- 7Ministry of Education (MOE) Laboratory for National Development and Intelligent Governance, Fudan University, Shanghai, China
- 8Center for the Belt and Road Archaeology and Ancient Civilizations (BRAAC), Fudan University, Shanghai, China
Introduction: The Xinjiang Mongolians, located along the Silk Road, migrated westward from Northeast Asia in the 13th and 14th centuries. Despite its significance, genetic studies on Xinjiang Mongolians have been limited compared to other Mongolian populations.
Methods: In this study, we analyzed the non-recombining region of the Y-chromosome in 165 Xinjiang Mongolian males using 108 biallelic markers and 44 STRs.
Results and discussion: Our study identified prevalent haplogroups C2a1a3-F1918 (10%), C2a1a2-M48 (8%), N1a1a-M178 (5%), and R1a1a-M17 (10%) in the Xinjiang Mongolians. Additionally, our study suggested a genetic affinity between Xinjiang Mongolians and Inner Mongolia Mongolian populations, as well as other ethnic groups from northwest China, based on the PCA analysis. The Network analysis revealed distinct branching expansion patterns in haplogroups C2a1a3a-F3796, C2a1a2-M48, and N1a1a-M178, with estimated timeframes aligning with Genghis Khan's invasion of Xinjiang in the Yuan Dynasty. Notably, our analysis of the R1a1a-M17 Network highlighted the role of Xinjiang Mongolians in the expansion of Turkic-speaking populations in Xinjiang and surrounding regions. The integration of ancient DNA data suggested that the high frequency haplogroups C2a1a3a-F3796, C2a1a2-M48, and N1a1a-M178 could be traced back to their origin in Northeast Asia. Furthermore, the estimated TMRCA of haplogroup R1a1a-M17 implied cultural and genetic influences from Turkic populations during the Qagatay Khanate period. Overall, our study provided a genetic explanation for the ethnic origin of Xinjiang Mongolians, highlighting their migration from Northeast Asia and subsequent assimilation with the local populations in Xinjiang.
1 Introduction
Xinjiang, located in northwest China, has long been a vital hub connecting ancient China to the rest of the world through the Silk Road. This region is known for its diverse ethnic makeup, with various ethnic groups migrating and settling there over the years (Tian, 2023). One such group is the Mongolian, who has a widespread presence across northern China, Mongolia, Russia, Kazakhstan, and other regions (Bai et al., 2018; Wang et al., 2021). It is estimated that there are around 10 million Mongolians worldwide (Bai et al., 2018), with approximately 6 million residing in China, mainly in Inner Mongolia. In Xinjiang, there are about 170,000 Mongolians, with the majority concentrated in Bayingol, Boltala, and Hoboksar.
Historians believe that the Mongolians can trace their roots back to the Donghu nomadic tribes (Yi, 1979; Tu, 1984). Recent genome-wide analyses of 214 ancient individuals from the Mongolian Plateau and Lake Baikal region have revealed that there are at least four ancestral origins of the Mongolians. Two of them can be traced back to their origins in northeastern and northern Mongolia before the Bronze Age, with these populations descending from hunter-gatherer communities in northeast Asia and northern Eurasia. Another ancestral source is linked to the Afanasievo culture, while the fourth source is a genetic mixture of Yamnaya pastoralists and European farmers (Jeong et al., 2020; Wang et al., 2021). A study using mitochondrial DNA has shown that Chinese Mongolians primarily carry East Asian-specific haplogroups, with a decreasing distribution from east to the west. Interestingly, some European-prevalent haplogroups have also been detected, showing an opposite distribution pattern to the East Asian-specific haplogroups (Cheng et al., 2008). Furthermore, various studies on Y-chromosomal haplogroups have identified C2*-M217 as the foundational paternal lineage among Mongolic-speaking populations. Other haplogroups such as O2*-M122, O1b*-M268/P31, N*-M231, and R-M207 have also been found in certain proportions (Wei et al., 2018; Zhang et al., 2018a; Huang et al., 2018; Wang et al., 2021). Researchers have concluded that haplogroup C2*-M217 is one of the founding paternal lineages of all Mongolic-speaking populations (Wei et al., 2018). Additionally, studies have traced the origins of specific Y-chromosome haplogroups, such as C3*-F3918 (now updated to C2a1a1-F3918), back to ancient nomadic groups like the Donghu (Zhang et al., 2018a). Detailed phylogenetic and phylogeographic analyses have suggested the northeastern Asian origin of haplogroup C2c1a1a1-M407 among Mongolic-speaking populations (Huang et al., 2018). Wang et al. have developed high-resolution Y-SNP SNaPshot panels, identifying three main ancestral sources of dominant Mongolian haplogroups: local lineage C2*-M217, incoming lineages from southern East Asia (O2*-M122, O1b*-P31, and N1*-CTS3750), and western Eurasia (R1*-M173) (Wang et al., 2021).
However, existing Y-chromosome studies of Xinjiang Mongolians primarily focused on comparing genetic variations and forensic applications across groups, leaving a gap in understanding the detailed haplogroup distribution and paternal genetic makeup of this population. For example, one study has examined the genetic diversity of Y-STR and indel markers in Xinjiang Mongolians and found their closest genetic relationship with the Gansu Dongxiang population, followed by Hulunbuir Mongols, the Daur ethnic group from Inner Mongolia, and three other ethnic minorities in Xinjiang (Liu et al., 2020). Another study has found that the genetic distance between the Mongolian ethnic group and the Uyghur and Xibe populations in Xinjiang is relatively small compared to populations from other continents (Wei et al., 2019).
Previous studies have shown that different geographic divisions and ethnic groups within the Mongolian populations have distinct genetic backgrounds (Bai et al., 2018; Zhao et al., 2020; Wang et al., 2021; He et al., 2023). However, research on the Y-chromosome analysis of Xinjiang Mongolians is lacking. In this study, our aim is to explore the paternal genetic structure of the Mongolian ethnic group in Xinjiang. We will analyze 108 Y-SNPs and 44 Y-STRs from 165 Mongolian males from Xinjiang to gain insights into the origin of Xinjiang Mongolians and their interactions with other local Xinjiang populations.
2 Materials and methods
2.1 Sample preparation
With the informed consent from the Ethics Committee of Fudan University of Life Sciences, 165 bloodstain samples were collected from unrelated and healthy Mongolian males from Xinjiang. Genomic DNA was extracted through the QIAamp DNA Blood Mini Kit (QIAGEN, Germany). In all DNA samples, 108 Y-SNP markers were hierarchically genotyped by SNaPshot (ABI SNaPshot® Multiplex Kit) as described in previous studies (Wang et al., 2014; Wen et al., 2020). The genotyping of the Xinjiang Mongolian samples was conducted utilizing the GeneAmp PCR system 9,700 (Thermo Fisher Scientific) comprising a set of 44 microsatellite markers (Zhang, 2015; Zhang et al., 2018b). The data presented in the study are deposited in the YHRD (Y-chromosomal haplotype reference database), accession number YA004579 (Xinjiang, China, [Mongolian]).
2.2 Y-SNP data analysis
Haplogroups were categorized in accordance with the ISOGG Y-DNA Haplogroup Tree 2019. We chose 167 modern Eurasian populations, with a particular focus on Central Asian/Altaic-speaking populations, for population comparisons with a consistent level of haplogroup resolution. Meanwhile, the haplogroup frequencies were determined through the direct counting method. Additionally, a principal component analysis (PCA) was conducted based on Y-SNPs using RStudio software version 2021.09.2.
2.3 Y-SNP-STR data analysis
Median-joining networks were computed using the NETWORK version 10.2.0.0 program and 15 Y-STRs data for the common haplogroups. The Time to Most Recent Common Ancestor (TMRCA) for the identified common haplogroups in both clusters was calculated using the average squared distance (ASD) estimator (Sengupta et al., 2006), with a generation time set at 25 years (Wang et al., 2014). The genealogical mutation rate OMRS in ASD method for population variation was used for the age estimates (Wang and Li, 2015). In order to ascertain TMRCA for representative genetic lineages, recent expansions within the specific population were identified. Subsequently, we examined samples within a network diagram, specifically those within a range of five mutation steps, to investigate recent historical events within the Xinjiang Mongolians (Luo et al., 2020).
3 Results
3.1 Simplified phylogenetic tree of Y-chromosomal haplogroups
According to the ISOGG Y-DNA haplogroup tree 2019-2020 and Y-SNP frequencies, we conducted a comparative analysis of haplogroup distribution among various populations in Xinjiang, including Mongolian (this study) (Supplementary Table 1), Xibe (unpublished), Hui (unpublished), Kazakh (previously reported) (Wang et al., 2023), Kyrgyz (previously reported) (Wen et al., 2022), as well as Hulunbuir, Hohhot, and Ordos Mongolian populations (previously reported) (Wang et al., 2021). To visually represent the distribution of haplogroups across these populations, we constructed a simplified phylogenetic tree (Figure 1). The figure showed that the most common haplogroups in Xinjiang Mongolians were C2a1a3-F1918 (10%), R1a1a-M17 (10%), C2a1a2-M48 (8%), and N1a1a-M178 (5%).

Figure 1 Frequencies of Y-chromosomal haplogroups and geographic locations of Xinjiang Mongolians and other reference modern populations. The figure depicts the geographical locations of the Xinjiang Mongolians and the reference population, while illustrating the investigation of Y-chromosome haplogroups in this study along with their corresponding frequencies. The branches indicate the names of the haplogroups, while the marker names are presented according to ISOGG Y-DNA Haplogroup Tree 2019.
The C2a1a3-F1918 haplogroup emerged as the predominant genetic lineage among the Xinjiang Mongolians. It was the upstream haplogroup of C2a1a3a-F3796 and had the highest prevalence among diverse groups, such as Xinjiang Kazakhs (50%), Hulunbuir Mongolians (18%), Xinjiang Xibes (14%), Ordos Mongolians (14%), and Hohhot Mongolians (12%). C2a1a3-F1918 was widely recognized as one of the main ancestral paternal lineages among Mongolic-speaking communities and spread rapidly across the Eurasian steppe during the Mongol conquests (Wei et al., 2018). Therefore, the genetic impact of the Mongol Western March on the local population in Xinjiang may be clearly evident.
C2a1a2-M48 was the third most prevalent haplogroup among Xinjiang Mongolians, with significant distribution patterns among Xinjiang Kazakhs (10%), Hohhot Mongolians (9%), and Hulunbuir Mongolians (7%). C2a1a2-M48 was the predominant paternal lineage among Tungusic-speaking populations (Liu et al., 2021).
It was worth noting that R1a1a-M17 (10%) and its downstream subhaplogroups, namely Rlalalblala-M458 (2%) and Rlalalb2-Z93 (1%), were highly prevalent among the Xinjiang Mongolians and were also widely distributed among other ethnic groups in Xinjiang. Specifically, these lineages were common among Xinjiang Kyrgyz (48%), Xinjiang Hui (17%), Xinjiang Xibe (8%), and Xinjiang Kazakh (7%). However, they were less prevalent in the Mongolian populations of Inner Mongolia, with frequencies of 5% in Ordos, 3% in Hohhot, and 1% in Hulunbuir. R1a1a-M17 was widely distributed among Indo-European-speaking populations (Koryakova and Epimakhov, 2014).
The N1a1a-M178 haplogroup emerged as the fourth most prevalent lineage among Xinjiang Mongolians, with significant distribution among the Hulunbuir (11%) and Ordos (7%) Mongolian populations. Additionally, this haplogroup had a limited presence in both the Xinjiang Hui (4%) and Xinjiang Xibe (3%) populations.
3.2 PCA analysis of Y-SNP haplotypes
At the population level, we compared the Y-SNP data of Xinjiang Mongolians with data from 167 modern Eurasian populations, including Mongolic, Tungusic, Turkic, Indo-European, Uralic, Sino-Tibetan speaking groups, and others. We conducted a principal component analysis (PCA) on these datasets to analyze the genetic relationships (Figure 2, Supplementary Table 2). The linguistic clusters were broadly distributed in the figure, with Xinjiang Mongolians falling within the overlapping confidence intervals of Han Chinese and Mongolian populations, indicating a close genetic affinity with both groups. Additionally, Xinjiang Mongolians showed significant genetic relationships with populations such as Xinjiang Hui, Xinjiang Xibe, Xinjiang Kazakh, Kyrgyzstan-Northwest Kyrgyz, Hohhot Mongolian, Hulunbuir Mongolian, Ordos Mongolian, and others.
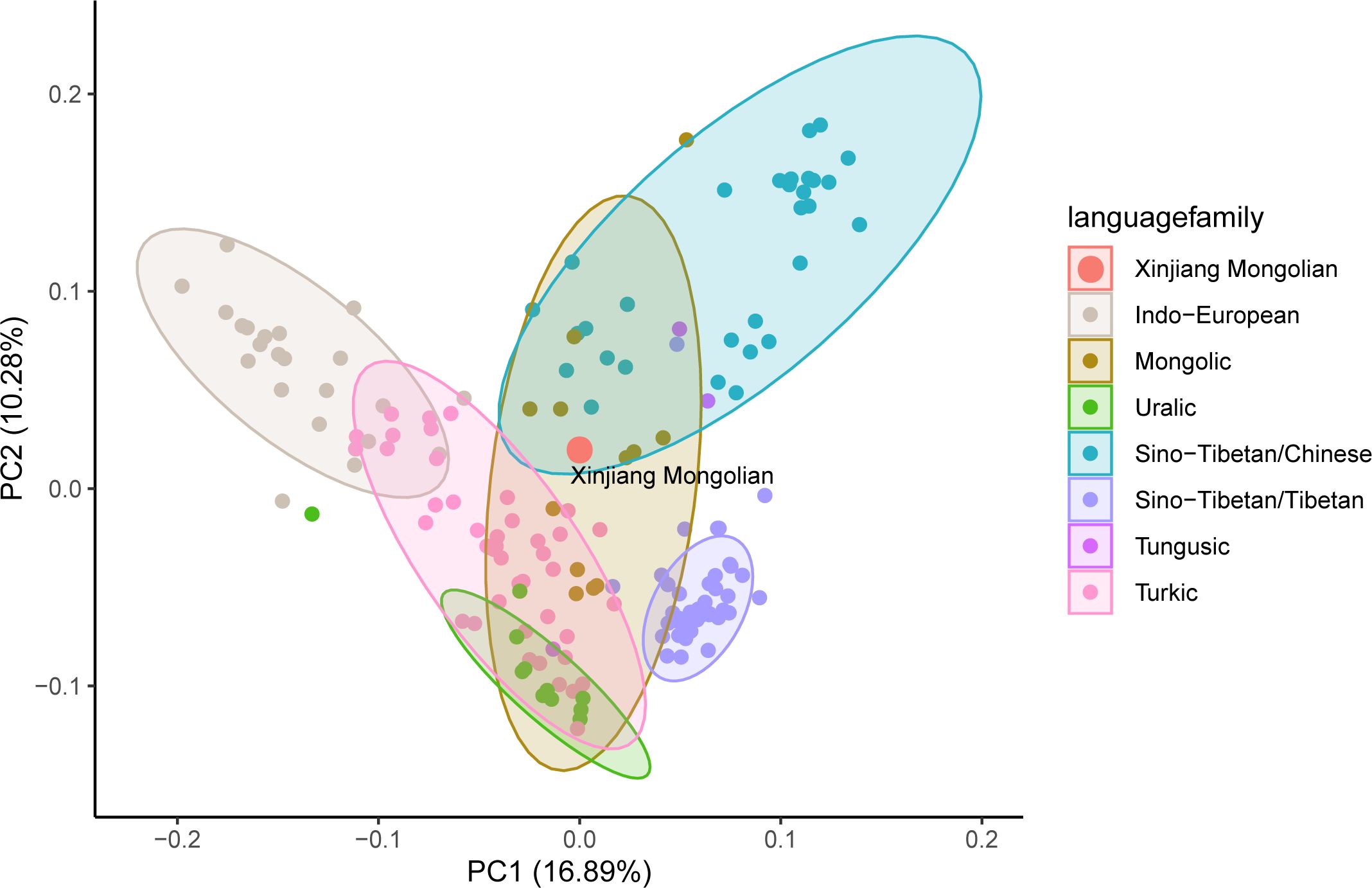
Figure 2 Genetic relationships of Xinjiang Mongolians with other reference modern populations from the principal component analysis (PCA) based on Y-chromosome haplogroups. Supplementary Table 2 provides comprehensive information regarding the populations included in this study.
3.3 Network analysis of Y-SNP-STR haplotypes
At the individual level, we conducted an analysis and investigation of four primary lineages of Xinjiang Mongolians using reduced median networks, namely C2a1a3a-F3796, R1a1a-M17, C2a1a2-M48, and N1a1a-M178. Furthermore, we categorized and cross-referenced populations based on their respective language families.
The network of haplogroup C2a1a3a-F3796 was established using 15 Y-STR profiles from 262 individuals across 16 diverse populations (Figure 3, Supplementary Table 3). The network of haplogroup C2a1a3a-F3796 displayed a characteristic branching expansion pattern, featuring a central founder haplotype and multiple sub-founders. The central haplotypes depicted in the image predominantly consist of Kazakh populations from northwest China and Central and North Asia, Xinjiang Kyrgyz populations (blue), as well as Mongolians originating from Inner and Outer Mongolia (purple). Xinjiang Mongolians (red) tended to exhibit clustering patterns with other Mongolic-speaking populations in northern China, as well as with various ethnic groups in Xinjiang, including Kyrgyz, Xibe, and Uygur populations. These clusters represented simultaneous small-scale expansions. By employing the ASD method, we estimated that the samples within the dashed ellipse had a Time to Most Recent Common Ancestor (TMRCA) of approximately 729.76 years Before Present (BP), with a standard deviation of ± 653.89 years, while excluding steps greater than five mutations.

Figure 3 A reduced median joining network was constructed for haplogroup (A) C2a1a3a-F3796, (B) C2a1a2-M48, and (C) N1a1a-M178. Haplotypes are visually represented by circles, whose sizes are proportionate to the number of individuals they represent. The connecting lines between the circles indicate mutational distance, with the shortest line representing a single mutational step.
Haplogroup C2a1a2-M48 exhibited a predominant distribution in Central and northern Asia, particularly among Tungusic-speaking and Paleo-Siberian-speaking populations. Additionally, it demonstrated a significant presence in Xinjiang Mongolians. The Xinjiang Mongolians distributed in the upper part of the figure (Figure 3, Supplementary Table 4), forming a star-shaped expansion pattern along with the Inner Mongolia Mongolian populations (purple), Xinjiang Kazakh populations (blue), and Xinjiang Uygur populations (blue), presenting an aesthetically pleasing distribution. Excluding steps greater than five mutations, we estimated TMRCA of the most recent cluster in the right clade (samples in the dashed line), yielding a result of 765.43 ± 545.09 years ago.
Haplogroup N1a1a-M178 was frequently observed in populations of northern European and northern East Asian descent. In the genetic network analysis (Figure 3, Supplementary Table 5), Xinjiang Mongolian samples exhibited proximity to other Mongolic-speaking populations (purple), while also displaying associations with a limited number of Turkic-speaking populations (blue). The upper left corner of the network revealed the involvement of Xinjiang Mongolians in the expansion of Mongolians from Inner Mongolia. Excluding steps greater than five mutations, we estimated TMRCA of this cluster (samples in the dashed line), yielding a result of 769.85 ± 457.38 years ago.
Haplogroup R1a1a-M17 was also a major haplogroup among Xinjiang Mongolians. The distribution of Xinjiang Mongolians in the network appeared to be dispersed (Figure 4, Supplementary Table 6), suggesting a diverse ancestral background among those belonging to the R1a1a-M17. Xinjiang Mongolians participated in the expansion of Turkic-speaking populations in Xinjiang (cyan) as well as in other regions (dark blue). Excluding steps greater than five mutations, we estimated TMRCA of the most recent cluster in the right clade (samples in the dashed line), yielding a result of 552.1 ± 345.13 years ago.
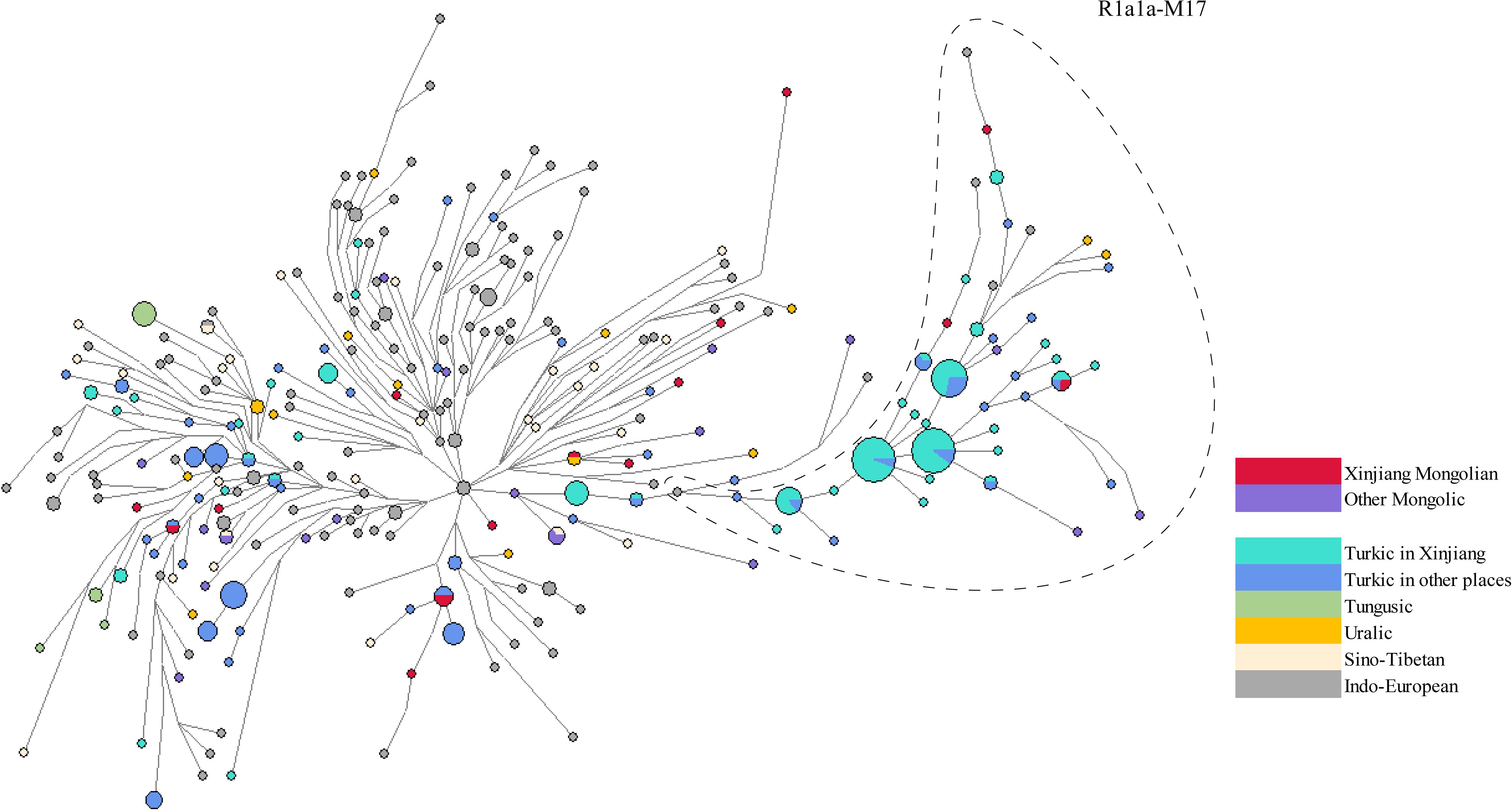
Figure 4 A Reduced Median joining network of haplogroup R1a1a-M17. Haplotypes are visually depicted as circles, where the sizes of the circles correspond to the number of individuals they represent. The lines connecting these circles indicate mutational distance, with the briefest line denoting a single mutational step.
4 Discussion
Haplogroup C2a1a3a-F3796 is renowned for being one of the ancestral paternal lineage of Mongolic-speaking populations (Wei et al., 2018). Further research has revealed that this cluster is linked to the ancient Mongolian tribe Nirun (Wei et al., 2018; Zhabagin et al., 2020). Ancient DNA evidence has shown that it can be found in Dornod, Mongolia during the Late Medieval period (Jeong et al., 2020). Wei and Li have shown that the estimated time to the most recent common ancestor (TMRCA) of C2a1a3a-F3796 is approximately 1000 ± 300 years old, with its origin in eastern Mongolia and the northern Greater Khingan Mountains before spreading westward (Wei and Li, 2022). Previous research has also uncovered a direct correlation between the expansion of the Mongolian empire after the 13th century and the dissemination of this paternal lineage (Zerjal et al., 2003). We estimated the TMRCA of approximately 729.76 ± 653.89, which could potentially provide further evidence to support this speculation. After the Mongolian expansion, it seemed that the introduction of Haplogroup C2a1a3a-F3796 into Xinjiang was observed in this group of Mongolian individuals.
Haplogroup C2a1a2-M48 represents the predominant paternal lineage in Tungusic-speaking populations (Lell et al., 2002; Pakendorf et al., 2006, 2007), which is one of the largest demographic groups in Siberia (Liu et al., 2021). C2a1a2-M48 is widely distributed throughout Central and Northern Asia, with higher frequencies among populations speaking Mongolic and Tungusic languages (Katoh et al., 2005; Malyarchuk et al., 2010; Balanovska et al., 2018; Liu et al., 2021). However, the presence of C2a1a2-M48 in these populations does not indicate a common origin but rather reflect an admixture event during the recent historical period (Liu et al., 2021). Historically, Mongolic-speaking and Tungusic-speaking populations in the outer Baikal region were closely associated, suggesting frequent genetic exchange between them (Baodungude, 1988). This haplogroup diverged approximately 15,000 years ago and underwent sub-branching around 4,000 years ago (Karmin et al., 2015). Its origin was conventionally associated with the inner regions of Central and Northern Asia (Karafet et al., 2002). Previous studies of ancient DNA have shown that haplogroup C2a1a2-M48 first appears in the Songnen Plain of China before the Last Glacial Maximum (about 20,000 years before present) and can also be found in the Songnen Plain and Russian Far East during the Neolithic Age (about 10,000 years before present) (Wang et al., 2014; Mao et al., 2021; Wang et al., 2023). Since then, this haplogroup was found among Mongolians from the Iron Age onwards (about 3,400 years before present) (Jeong et al., 2020). We undertook calculations that suggested the TMRCA of haplogroup C2a1a2-M48, thereby implying a possible link between the Xinjiang Mongolians and Genghis Khan’s expedition. The spatial and temporal distribution of haplogroup C2a1a2-M48 closely corresponded to that of haplogroup C2a1a3a-F3796. The high prevalence of haplogroup C2a1a3a-F3796 and C2a1a2-M48 in the Xinjiang Mongolians might suggest a Northeast Asian origin.
Haplogroup N1a1a-M178 is commonly found in ancient Siberian archaeological sites as well as among present-day populations of Altaic, Uralic, Russian, and Chinese descent in the Siberian region (Rootsi et al., 2007; Hu et al., 2015). Research by Rootsi et al. has suggested that the haplogroup N1c-M46 lineage, now updated to N1a1-M46, likely originates in China and experienced population bottlenecks in northern East Asia before spreading into Siberia (Rootsi et al., 2007). Ancient DNA evidence have indicated that haplogroup N1a1a-M178 emerges around 7,500 years ago in Jilin, China (Ning et al., 2020) and appeared in Siberia during the Bronze Age, approximately 5,000 years ago (Hollard et al., 2018). The emergence of Haplogroup N1a1a-M178 in Mongolians could be traced back to about 2,300 years ago (Keyser et al., 2004). Therefore, the prevalence of N1a1a-M178 in Xinjiang Mongolians might also suggest a Northeast Asian origin.
Haplogroup R1a1a-M17 is one of the most common haplogroups in Xinjiang Mongolians. It is more prevalent in Eastern Europe, South Asia, and Siberia, while less frequently observed among individuals of Middle Eastern and East Asian descent (Balanovsky et al., 2008; Koryakova and Epimakhov, 2014). Ancient DNA studies has consistently shown a correlation between the spread of the paternal lineage R1a1a-M17 and the migration of Indo-European populations, estimating that this lineage originated around 8,000 years ago during the early Holocene period (Semino et al., 2000; Keyser et al., 2009; Allentoft et al., 2015; Haak et al., 2015; Damgaard et al., 2018) Haplogroup R1a1a-M17 has been identified in the Lake Baikal region of Siberia over 7,000 years ago (Moussa et al., 2018). Moreover, R1a1a-M17 has also been detected in the Tarim Basin of Xinjiang during the Bronze Age to the Iron Age (between approximately 6,000 and 3,000 years before present), exclusively observed in male individuals from the West Eurasia, which is related to the migration of ancient people from Europe in the early Bronze Age (at least 4,000 years before present) (Li et al., 2010). Given the widespread distribution of haplogroup R1a1a-M17 across different regions and time periods, we hypothesized that individuals in the Xinjiang Mongolian population might have genetic connections to Siberia or West Eurasia. Our network and computational TMRCA analyses hinted at the possibility that Xinjiang Mongolians contributed to the population expansion of other groups, such as the Xinjiang Kyrgyz, following their westward migration. These might be attributed to the process of Turkic cultural assimilation among the Mongolians during the Chagatai Khanate period (Liu, 1994).
Haplogroup O-M175 is a significant component of the Y-chromosome gene pool in East Asians, making up approximately 75% of Chinese paternal lineages (Yan et al., 2011). This haplogroup is also prevalent in Southeast and East Asian populations (Poznik et al., 2016). Wei et al. have found that among Mongolic-speaking populations, the frequency of Haplogroup O*-M175 is 18.929% (Wei and Li, 2022). In our study, we observed a high frequency of the subhaplogroup O*-M175 in Xinjiang Mongolians, with O2*-M122 and O1b*-M268 being the most common subgroups, accounting for 17% and 8% respectively. Previous research on haplogroups O1-M119 (now updated to O1a-M119), O2-M95 (now updated to O1b1a1a-M95), and O3-M122 (now updated to O2-M122) has suggested that northern populations originate from the south and migrated northward after the initial Paleolithic settlement of East Asia around 3 million years ago (Bing et al., 2000; Wen et al., 2004; Shi et al., 2005; Wang et al., 2013). These might explain the widespread presence of O2*-M122 and O1b*-M268 among Xinjiang Mongolians.
By analyzing Y-chromosome data of the Xinjiang Mongolians and considering historical contextual information, we can make more robust inferences about their migratory history. In the 13th century, the Mongol Empire, a nomadic nation thriving on the Mongolian Plateau, founded an extensive land empire that reached into Central Asia, including present-day Xinjiang (Howorth, 1888; Man, 2014). The influence of this historical legacy can still be seen in the diverse cultural and ethnic landscape of modern Xinjiang. Genghis Khan’s conquests began in the early 1200s, and by the time of his death in 1227, he had already laid the foundations for a vast empire. His successors, particularly Kublai Khan and Hulagu Khan, continued to expand the empire. During the westward expansion, the Mongols conquered Central Asia, including the Khwarazmian Empire, Persia, Anatolia, and parts of the Caucasus (Weng, 1985; Han, 1986). They also underwent significant cultural assimilation through interactions with societies in Central Asia. Central Asia is linguistically diverse, with multiple language families present within its borders (Dalizabu, 2006). As a result, a discernible genetic component from other language families can be observed among Xinjiang Mongolians.
5 Conclusion
Based on the combined analyses of ancient DNA studies focusing on haplogroups C2a1a3a-F3796, C2a1a2-M48, and N1a1a-M178, as well as the calculated TMRCA of the haplogroups, it could be concluded that the Xinjiang Mongolians had the origin in Northeast Asia. Additionally, the analysis of haplogroup R1a1a-M17 provided further evidence of their integration with neighboring populations since their migration from Northeast Asia to Xinjiang.
There was a significant genetic similarity between the Xinjiang Mongolians and present-day Mongolians in Inner Mongolia, as well as genetic interactions with indigenous populations in Xinjiang such as Uygur, Kyrgyz, Xibe, and Kazakh. However, caution should be exercised when estimating TMRCA due to inherent errors in estimation. Overall, our study provided genetic evidence supporting the complex paternal genetic composition of Xinjiang Mongolians through admixture. To further enhance our understanding of the genetic structure and historical background of Xinjiang Mongolians, it is imperative to conduct further investigations using diverse genetic markers and incorporating genetic evidence from other geographically distinct Mongolian populations.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: FigShare, https://figshare.com/s/16d906f041f6c4489fb8.
Ethics statement
The studies involving humans were approved by Ethics Committee, School of Life Sciences, Fudan University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
YFW: Writing – original draft. LX: Writing – original draft. KW: Writing – original draft. ZJ: Writing – original draft. YF: Writing – original draft. YY: Writing – original draft. XC: Writing – original draft. HM: Writing – original draft. YX: Writing – original draft. YSW: Writing – original draft. MS: Writing – review & editing. XW: Writing – review & editing. SW: Writing – review & editing.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Natural Science Foundation of China (32111530227, 32070576, 82004468 and 41991251), National Social Science Foundation of China (19VJX074), Qian Duansheng Distinguished Scholars Program of China University of the Political Science and Law (01140065140), Cross disciplinary construction project of evidence investigation (10322308), European Research Council (ERC) grant (ERC-2019-ADG-883700-TRAM) and Lantai Young Scholars Program from Institute of Chinese History (2022LTQN602).
Acknowledgments
This is a short text to acknowledge the contributions of specific colleagues, institutions, or agencies that aided the efforts of the authors.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2024.1349231/full#supplementary-material
References
Allentoft M., Sikora M., Sjögren K., Rasmussen S., Rasmussen M., Stenderup J., et al. (2015). Population genomics of bronze age Eurasia. Nature 522, 167–172. doi: 10.1038/nature14507
Bai H., Guo X., Narisu N., Lan T., Wu Q., Xing Y., et al. (2018). Whole-genome sequencing of 175 Mongolians uncovers population-specific genetic architecture and gene flow throughout North and East Asia. Nat. Genet. 50, 1696–1704. doi: 10.1038/s41588-018-0250-5
Balanovska E., Bogunov Y., Kamenshikova E., Balaganskaya O., Agdzhoyan A., Bogunova A., et al. (2018). Demographic and genetic portraits of the Ulchi population. Russ J. Genet. 54, 1245–1253. doi: 10.1134/S1022795418100046
Balanovsky O., Rootsi S., Pshenichnov A., Kivisild T., Churnosov M., Evseeva I., et al. (2008). Two sources of the Russian patrilineal heritage in their Eurasian context. Am. J. Hum. Genet. 82, 236–250. doi: 10.1016/j.ajhg.2007.09.019
Bing S., Li J., Peter U., Jeremy M., Nilmani S., Stephen T., et al. (2000). Polynesian origins: insights from the Y chromosome. Proc. Natl. Acad. Sci. United States America 97, 8225–8228. doi: 10.1073/pnas.97.15.8225
Cheng B., Tang W., He L., Dong Y., Lu J., Lei Y., et al. (2008). Genetic imprint of the Mongol: signal from phylogeographic analysis of mitochondrial DNA. J. Hum. Genet. 53, 905–913. doi: 10.1007/s10038-008-0325-8
Dalizabu (2006). Outline of Mongolian history (Beijing: Central University for Nationalities Press).
Damgaard P., Marchi N., Rasmussen S., Peyrot M., Renaud G., Korneliussen T., et al. (2018). 137 ancient human genomes from across the Eurasian steppes. Nature 557, 369–374. doi: 10.1038/s41586-018-0094-2
Haak W., Lazaridis I., Patterson N., Rohland N., Mallick S., Llamas B., et al. (2015). Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211. doi: 10.1038/nature14317
He G., Wang M., Miao L., Chen J., Zhao J., Sun Q., et al. (2023). Multiple founding paternal lineages inferred from the newly-developed 639-plex Y-SNP panel suggested the complex admixture and migration history of Chinese people. Hum. Genomics 17, 29. doi: 10.1186/s40246-023-00476-6
Hollard C., Zvénigorosky V., Kovalev A., Kiryushin Y., Tishkin A., Lazaretov I., et al. (2018). New genetic evidence of affinities and discontinuities between bronze age Siberian populations. Am. J. Phys. Anthropol. 167, 97–107. doi: 10.1002/ajpa.23607
Howorth H. (1888). History of the Mongols, from the 9th to the 19th Century (Franklin: Burt Franklin).
Hu K., Yan S., Liu K., Ning C., Wei L., Li S., et al. (2015) The dichotomy structure of Y chromosome Haplogroup N. Available online at: http://arxiv.org/abs/1504.06463 (Accessed November 20, 2023).
Huang Y. Z., Wei L. H., Yan S., Wen S. Q., Wang C. C., Yang Y. J., et al. (2018). Whole sequence analysis indicates a recent southern origin of Mongolian Y-chromosome C2c1a1a1-M407. Mol. Genet. Genomics 293, 657–663. doi: 10.1007/s00438-017-1403-4
Jeong C., Wang K., Wilkin S., Taylor W. T., Miller B., Bemmann J. H., et al. (2020). A dynamic 6,000-year genetic history of Eurasia’s Eastern Steppe. Cell 183, 890–904. doi: 10.1016/j.cell.2020.10.015
Karafet T., Osipova L., Gubina M., Posukh O., Zegura S., Hammer M. (2002). High levels of Y-chromosome differentiation among native Siberian populations and the genetic signature of a boreal hunter-gatherer way of life. Hum. Biol. 74, 761–789. doi: 10.1353/hub.2003.0006
Karmin M., Saag L., Vicente M., Sayres M. A. W., Järve M., Talas U. G., et al. (2015). A recent bottleneck of Y chromosome diversity coincides with a global change in culture. Genome Res. 25, 459–466. doi: 10.1101/gr.186684.114
Katoh T., Munkhbat B., Tounai K., Mano S., Ando H., Oyungerel G., et al. (2005). Genetic features of Mongolian ethnic groups revealed by Y-chromosomal analysis. Gene 346, 63–70. doi: 10.1016/j.gene.2004.10.023
Keyser C., Bouakaze C., Crubézy E., Nikolaev V. G., Montagnon D., Reis T., et al. (2009). Ancient DNA provides new insights into the history of south Siberian Kurgan people. Hum. Genet. 126, 395–410. doi: 10.1007/s00439-009-0683-0
Keyser T., Blandin P., Ricaut F. X., Petkovski E., Crubézy E., Ludes B. (2004). Does the Tat polymorphism originate in northern Mongolia? Int. Congress Ser. (Elsevier), 325–327. doi: 10.1016/S0531-5131(03)01701-1
Koryakova L., Epimakhov A. V. (2014). The Urals and western Siberia in the Bronze and Iron ages (Cambridge: Cambridge university press).
Lell J., Sukernik R., Starikovskaya Y., Su B., Jin L., Schurr T., et al. (2002). The dual origin and Siberian affinities of Native American Y chromosomes. Am. J. Hum. Genet. 70, 192–206. doi: 10.1086/338457
Li C., Li H., Cui Y., Xie C., Cai D., Li W., et al. (2010). Evidence that a West-East admixed population lived in the Tarim Basin as early as the early Bronze Age. BMC Biol. 8, 15. doi: 10.1186/1741-7007-8-15
Liu Y. (1994). Study on northwest nationality history and Chagatai Khanate history (Nanjing: Nanjing University Press).
Liu B., Ma P., Wang C., Yan S., Yao H., Li Y., et al. (2021). Paternal origin of Tungusic-speaking populations: Insights from the updated phylogenetic tree of Y-chromosome haplogroup C2a-M86. Am. J. Hum. Biol. 33, e23462. doi: 10.1002/ajhb.23462
Liu Y., Yu T., Mei S., Jin X., Lan Q., Zhou Y., et al. (2020). Forensic characteristics and genetic affinity analyses of Xinjiang Mongolian group using a novel six fluorescent dye-labeled typing system including 41 Y-STRs and 3 Y-InDels. Molec. Gen. Gen. Med. 8, e1097. doi: 10.1002/mgg3.1097
Luo X. Q., Du P. X., Wang L. X., Zhou B. Y., Li Y. C., Zheng H. X., et al. (2020). Uniparental genetic analyses reveal the major origin of Fujian tanka from ancient indigenous Daic populations. Hum. Biol. 91, 257–277. doi: 10.13110/humanbiology.91.4.05
Malyarchuk B., Derenko M., Denisova G., Wozniak M., Grzybowski T., Dambueva I., et al. (2010). Phylogeography of the Y-chromosome haplogroup C in northern Eurasia. Ann. Hum. Genet. 74, 539–546. doi: 10.1111/j.1469-1809.2010.00601.x
Man J. (2014). The Mongol Empire: Genghis Khan, his heirs and the founding of modern China (New York: Random House).
Mao X., Zhang H., Qiao S., Liu Y., Chang F., Xie P., et al. (2021). The deep population history of northern East Asia from the Late Pleistocene to the Holocene. Cell 184, 3256–3266. doi: 10.1016/j.cell.2021.04.040
Moussa N., Bazaliiskii V., Goriunova O., Bamforth F., Weber A. (2018). Y-chromosomal DNA analyzed for four prehistoric cemeteries from Cis-Baikal, Siberia. J. Archaeol. Sci.: Rep. 17, 932–942. doi: 10.1016/j.jasrep.2016.11.003
Ning C., Fernandes D., Changmai P., Flegontova O., Yüncü E., Maier R., et al. (2020). The genomic formation of First American ancestors in East and Northeast Asia. BioRxiv, 336628. doi: 10.1101/2020.10.12.336628
Pakendorf B., Novgorodov I. N., Osakovskij V. L., Danilova A. P., Protod’jakonov A. P., Stoneking M. (2006). Investigating the effects of prehistoric migrations in Siberia: genetic variation and the origins of Yakuts. Hum. Genet. 120, 334–353. doi: 10.1007/s00439-006-0213-2
Pakendorf B., Novgorodov I. N., Osakovskij V. L., Stoneking M. (2007). Mating patterns amongst Siberian reindeer herders: Inferences from mtDNA and Y-chromosomal analyses. Am. J. Phys. Anthropol. 133, 1013–1027. doi: 10.1002/ajpa.20590
Poznik G. D., Xue Y., Mendez F. L., Willems T. F., Massaia A., Wilson Sayres M. A., et al. (2016). Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences. Nat. Genet. 48, 593–599. doi: 10.1038/ng.3559
Rootsi S., Zhivotovsky L. A., Baldovič M., Kayser M., Kutuev I. A., Khusainova R., et al. (2007). A counter-clockwise northern route of the Y-chromosome haplogroup N from Southeast Asia towards Europe. Eur. J. Hum. Genet. 15, 204–211. doi: 10.1038/sj.ejhg.5201748
Semino O., Passarino G., Oefner, Peter J., Lin A. A., Arbuzova S., et al. (2000). The Genetic Legacy of Paleolithic Homo sapiens sapiens in Extant Europeans: AY Chromosome Perspective. Science 290, 1155–1159. doi: 10.1126/science.290.5494.1155
Sengupta S., Zhivotovsky L. A., King R., Mehdi S. Q., Edmonds C. A., Chow C.-E. T., et al. (2006). Polarity and temporality of high-resolution Y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of central Asian pastoralists. Am. J. Hum. Genet. 78, 202–221. doi: 10.1086/499411
Shi H., Dong Y., Wen B., Xiao C., Underhill P. A., Shen P.-D., et al. (2005). Y-chromosome evidence of southern origin of the east Asian–specific haplogroup O3-M122. Am. J. Hum. Genet. 77, 408–419. doi: 10.1086/444436
Tian W. (2023). The construction of Xinjiang historical narrative under the guidance of historical materialism. west. Res., 1–17. doi: 10.16363/j.cnki.xyyj.2023.04.001
Wang M., He G., Zou X., Liu J., Ye Z., Ming T., et al. (2021). Genetic insights into the paternal admixture history of Chinese Mongolians via high-resolution customized Y-SNP SNaPshot panels. Forensic Sci. Intern.: Genet. 54, 102565. doi: 10.1016/j.fsigen.2021.102565
Wang C., Li H. (2015). Evaluating the Y chromosomal STR dating in deep-rooting pedigrees. Invest. Genet. 6, 8. doi: 10.1186/s13323-015-0025-z
Wang B., Liang J., Allen E., Chang X., Jiang Z., Yu Y., et al. (2023). Y chromosome evidence confirms northeast Asian origin of Xinjiang Kazakhs and genetic influence from 18th century expansion of Kerey clan. Front. Ecol. Evol. 11. doi: 10.3389/fevo.2023.1264718
Wang C., Wang L., Shrestha R., Zhang M., Huang X., Hu K., et al. (2014). Genetic structure of Qiangic populations residing in the western Sichuan corridor. PloS One 9, e103772. doi: 10.1371/journal.pone.0103772
Wang C., Yan S., Qin Z., Lu Y., Ding Q., Wei L., et al. (2013). Late Neolithic expansion of ancient Chinese revealed by Y chromosome haplogroup O3a1c-002611. J. System. Evol. 51, 280–286. doi: 10.1111/j.1759-6831.2012.00244.x
Wei Y., Jin X., Lan Q., Cui W., Chen C., Kong T., et al. (2019). Genetic distribution and forensic evaluation of multiplex autosomal short tandem repeats in the Chinese Xinjiang Mongolian group. J. Zhejiang Univers. Sci. B 20, 287. doi: 10.1631/jzus.B1800279
Wei L., Li H. (2022). Molecular anthropology traceability of Mongolian population (Shanghai: Shanghai Scientific and Technical Publishers).
Wei L., Yan S., Lu Y., Wen S., Huang Y., Wang L., et al. (2018). Whole-sequence analysis indicates that the Y chromosome C2*-Star Cluster traces back to ordinary Mongols, rather than Genghis Khan. Eur. J. Hum. Genet. 26, 230–237. doi: 10.1038/s41431-017-0012-3
Wen S., Du P., Sun C., Cui W., Xu Y., Meng H., et al. (2022). Dual origins of the Northwest Chinese Kyrgyz: the admixture of Bronze age Siberian and Medieval Niru’un Mongolian Y chromosomes. J. Hum. Genet. 67, 175–180. doi: 10.1038/s10038-021-00979-x
Wen S., Sun C., Song D., Huang Y., Tong X., Meng H., et al. (2020). Y-chromosome evidence confirmed the Kerei-Abakh origin of Aksay Kazakhs. J. Hum. Genet. 65, 797–803. doi: 10.1038/s10038-020-0759-1
Wen B., Xie X., Gao S., Li H., Shi H., Song X., et al. (2004). Analyses of genetic structure of Tibeto-Burman populations reveals sex-biased admixture in southern Tibeto-Burmans. Am. J. Hum. Genet. 74, 856–865. doi: 10.1086/386292
Weng D. (1985). A brief history of Mongolian (Inner Mongolia: Inner Mongolia People’s Publishing House).
Yan S., Wang C.-C., Li H., Li S.-L., Jin L.. (2011). An updated tree of Y-chromosome Haplogroup O and revised phylogenetic positions of mutations P164 and PK4. Eur. J. Hum. Genet. 19, 1013–1015. doi: 10.1038/ejhg.2011.64
Yi L. (1979). Ethnic origin of northern China and Mongolia. J. Inner Mongolia Univ. (philosophy Soc. Sci. edition), 1–23. doi: 10.13484/j.cnki.ndxbzsb.1979.z2.001
Zerjal T., Xue Y., Bertorelle G., Wells R. S., Bao W. (2003). The genetic legacy of the Mongols. Am. J. Hum. Genet. 72, 717–721. doi: 10.1086/367774
Zhabagin M., Sabitov Z., Tarlykov P., Tazhigulova I., Junissova Z., Yerezhepov D., et al. (2020). The medieval Mongolian roots of Y-chromosomal lineages from South Kazakhstan. BMC Genet. 21, 87. doi: 10.1186/s12863-020-00897-5
Zhang L. (2015). Population data for 15 autosomal STR loci in the Bouyei ethnic minority from Guizhou Province, Southwest China. Forensic Sci. Intern.: Genet. 17, 108–109. doi: 10.1016/j.fsigen.2015.04.006
Zhang J., Mo X., Zhang Y., Ding G., Wang X., Li W., et al. (2018). Genetic analysis of 26 Y-STR loci in Han population from Leshan, Southwest China. Forensic Sci. Intern.: Genet. 37, e15–e16. doi: 10.1016/j.fsigen.2018.07.022
Zhang Y., Wu X., Li J., Li H., Zhao Y., Zhou H. (2018). The Y-chromosome haplogroup C3*-F3918, likely attributed to the Mongol Empire, can be traced to a 2500-year-old nomadic group. J. Hum. Genet. 63, 231–238. doi: 10.1038/s10038-017-0357-z
Keywords: Xinjiang Mongolian, genetic diversity, Y-chromosomal lineages, Y-SNP, Y-STR
Citation: Wang Y, Xie L, Wang K, Jiang Z, Feng Y, Yu Y, Chang X, Meng H, Xu Y, Wu Y, Shi M, Wang X and Wen S (2024) Genetic origins and migration patterns of Xinjiang Mongolian group revealed through Y-chromosome analysis. Front. Ecol. Evol. 12:1349231. doi: 10.3389/fevo.2024.1349231
Received: 04 December 2023; Accepted: 13 May 2024;
Published: 25 July 2024.
Edited by:
Guanglin He, Sichuan University, ChinaReviewed by:
Levon Yepiskoposyan, Armenian National Academy of Sciences, ArmeniaWang Zhiyong, Kunming Medical University, China
Yuguo Huang, Sichuan University, China
Copyright © 2024 Wang, Xie, Wang, Jiang, Feng, Yu, Chang, Meng, Xu, Wu, Shi, Wang and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meisen Shi, c2hpbWVpc2VuMjAwMEAxNjMuY29t; Xiaoxia Wang, d2FuZ3hpYW94aWFAbHp1LmVkdS5jbg==; Shaoqing Wen, d2Vuc2hhb3FpbmdAZnVkYW4uZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Yifan Wang1,2†
Yifan Wang1,2† Lei Xie
Lei Xie Yuhang Feng
Yuhang Feng Yiran Xu
Yiran Xu Meisen Shi
Meisen Shi Xiaoxia Wang
Xiaoxia Wang Shaoqing Wen
Shaoqing Wen