- 1Agricultural Science and Technology Institute, Andong National University, Andong, Republic of Korea
- 2Institute of Environmental and Agricultural Biology (X-BIO), Tyumen State University, Tyumen, Russia
- 3Entomology Division, Nepal Agricultural Research Council, Kathmadu, Nepal
- 4Department of Zoology, Tribuvan University, Kathmandu, Nepal
- 5Department of Plant Medicine, Andong National University, Andong, Republic of Korea
- 6Department of Fishery and Beekeeping, Tashkent State Agrarian University, Tashkent, Uzbekistan
Introduction: The mites belonging to the genus Tropilaelaps are ectoparasites of honey bees, primarily infesting the larval and pupal stages. Originating from subtropical regions, these mites can cause brood malformation, bee mortality, and subsequent decline or absconding of colonies.
Material and methods: During field surveys conducted in Nepal, South Korea, and Uzbekistan to investigate honey bee pests, several populations of T. mercedesae were collected. This is the first record of T. mercedesae from Uzbekistan. The morphological characteristics of the collected populations were analyzed, and their phylogenetic relationship with other Asian populations was examined.
Results and discussion: The molecular analysis of cytochrome oxidase I gene revealed high similarity between Uzbekistan and Pakistan populations, signaling the potential invasion of subtropical honey bee parasites into the Central Asian beekeeping sectors. Phylogenetic analysis indicated the presence of four distinct lineages within the mainland-Indonesian populations of T. mercedesae. Notably, an evolutionary divergence was observed between the haplotypes from Sri Lanka and the Philippines compared to the remaining mainland Asian and Indonesian haplotypes, suggesting the possible existence of subspecies or separate species in these isolated locations. To gain a deeper understanding of this phenomenon, it is essential to continue monitoring the spread of this significant honey bee pest and conduct comprehensive morphological and molecular analyses of samples collected from these specific localities.
Introduction
Honey bees are the most important pollinator which have enormous influence on crop production and with enabling reproduction of the flowering plants, they are also important in the diversity of wild flowering plat (Klein et al., 2007). The global decline in the population of honey bees and other pollinators raised concern about the sustainability of food production in future and study the fatal factors is necessary to protect them (Potts et al., 2010). Tropilaelaps is a genus of ectoparasitic mites which are native to Asia. They are primarily parasites of wild honey bees such as A. dorsata, A. laboriosa and A. breviligula (Delfinado-Baker et al., 1985) however among Tropilaelaps species, T. mercedesae and T. clareae can infest managed honey bee colonies of A. mellifera (Kumar et al., 1993). T. mercedesae was initially described from Hanoi, Vietnam, where it was discovered within a colony of A. mellifera (Hymenoptera: Apidae) (Anderson and Morgan, 2007). Subsequently, this species expanded its distribution and is now recorded in South and Southeast Asia. The threat posed by these mites is amplified by the Western honey bee’s lack of behavioral defenses compared to the Asian Eastern honey bee, A. cerana, making them particularly vulnerable to parasitic infestations (Wongsiri et al., 1989; Delfinado-Baker, 1982). A survey conducted in 2005 in South Korea revealed that Varroa mite infestations were significantly higher than Tropilaelaps mite infestations (91% versus 25.7% infestation rate) (Lee et al., 2005), while recent studies indicate an increasing occurrence of Tropilaelaps mites in honey bees (Jung et al., 2014; Buawangpong et al., 2015; Truong et al., 2022). Additionally, dual parasitism by Varroa mites and Tropilaelaps mites can lead to more severe damage to honey bee colonies (Truong et al., 2022).
Likewise, V. destructor, these mites can only reproduce in the honey bee brood cells and exclusively feed on immature stages of honey bees due to their specific mouthpart structure (Anderson and Roberts, 2013). However, other characteristics such as having a smaller size, shorter phoretic phase, rapid locomotion and fast reproductive rate made Tropilaelaps mites an emerging threat for beekeeping industry (Anderson and Roberts, 2013; Pettis et al., 2013; Buawangpong et al., 2015; de Guzman et al., 2017). Furthermore, these ectoparasitic mites are also able to play as a vector of honey bee viral diseases and their potential in transferring Deformed Wing Virus (DWV) has been demonstrated (Forsgren et al., 2009; Wu et al., 2017). The presence of Tropilaelaps mites in honey bee colonies during the early developmental stage could potentially increase viral proliferation within the colony. This is due to the longer exposure to the virus and the increased susceptibility to viral infections caused by stress (Nazzi and Pennacchio, 2018). Recent studies by de Guzman et al. (2020) have shown that bee pupae infested with either Varroa or Tropilaelaps mites had higher levels of DWV variants compared to uninfected pupae (de Guzman et al., 2020). Additionally, Tropilaelaps mite feeding can trigger a cellular immune response in worker broods, which may be a result of mite-induced injuries, viral infections, or a combination of both factors (Khongphinitbunjong et al., 2015). Studying the genetic variation of the populations of Tropilaelaps species is necessary to understand their intraspecific genetic diversity, population structure, and evolutionary history. Furthermore, this information is crucial for accurately identifying different species through barcode gap analysis of the molecular markers. In addition, these studies not only provide invaluable information about the level of genetic divergence among the populations of this species, showing the level of the species plasticity to the adverse biotic and abiotic environmental factors, but it also provides valuable information on the movement and spread of alien species (Arca et al., 2015; Mohamadzade Namin et al., 2019). By analyzing the genetic markers of different mite populations, researchers can determine their origins, routes of transmission and patterns of dispersal. This can help in understanding the dynamics of mite infestations, predicting their potential spread to new areas, and designing effective management strategies to control their expansion (Mohamadzade Namin et al., 2019). While Tropilaelaps mites currently have their primary distribution in Asia, their potential to spread to other continents, primarily through the trade of honey bees and bee products, raises significant concerns. This pattern of potential dissemination mirrors the global spread of honey bee pests, such as Varroa destructor and Vairimorpha ceranae, which have often been unintentionally transported across borders due to the international exchange of honey bees and their related products (Higes et al., 2010; Rosenkranz et al., 2010). As a result, the risk of inadvertent introduction to new regions has become a pressing issue, given the extensive global trade and travel networks.
Our study delves into the genetic variation and relationships among T. mercedesae populations in Asia. To achieve this, we have collected samples from South Korea, conducted two expeditions to Nepal and Uzbekistan, and integrated previously archived cytochrome oxidase I sequences from GenBank. Additionally, our research aims to uncover the potential route of the recent invasion of this pest into Uzbekistan, shedding light on how it might have been introduced to this region. Furthermore, we aim to deepen our understanding of the phylogenetic relationships among the various populations of this species in Asia. This research is essential not only for academic purposes but also for practical implications in the context of global beekeeping and pollinator health. By understanding the genetic diversity and spread of Tropilaelaps mites, we can develop more effective strategies for their management and containment, as well as contribute to the broader understanding of the threat these mites pose to the health of beekeeping and pollinator populations on a global scale.
Materials and methods
Sample collection and morphological examination
The specimens of T. mercedesae were collected from the body of honey bee workers in infested A. mellifera colonies in Nepal (one locality), South Korea (four localities) and Uzbekistan (one locality) (Table 1). Specimens were cleared in lactic acid solution and mounted in Hoyer’s medium (Walter and Krantz, 2009). Photomicrographs were captured using a DP22 Olympus camera (Japan). The anatomical structures follow Evans and Till (1979). All examined specimens deposited at the acarology collection of Andong National University (South Korea). The rest of samples were stored in absolute ethanol at -20˚C prior to DNA isolation. Although several samples from one hive were available, only a single sample from each infested colony was used for molecular analysis. In total forty-eight specimens of T. mercedesae were subjected to molecular analysis.
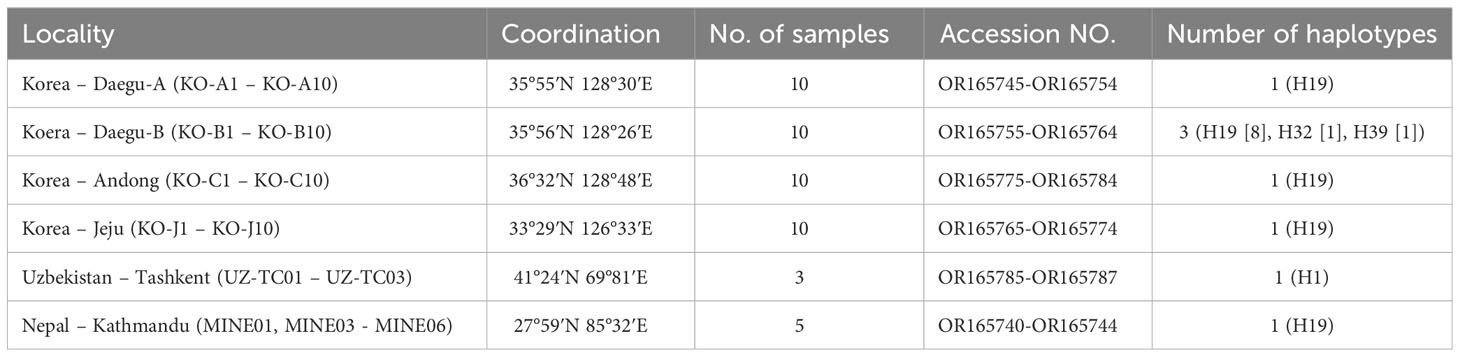
Table 1 Sampling information and the GenBank accession numbers and haplotype information of each population of T. mercedesae.
DNA extraction, primer, PCR, and sequencing
Total DNA was extracted using DNeasy Blood and Tissue kit (Qiagen, Seoul, Korea). A single specimen was crashed using sterilized pestle and used for DNA extraction. The mitochondrial COI gene was analyzed for species identification and the construction of phylogenetic evolution among T. mercedesae populations in Asia. COI has gained widespread use in evolutionary studies owing to its maternal inheritance, ease of amplification (Gupta et al., 2015), and, notably, the abundance of information available in molecular databases. This wealth of data facilitates comprehensive evolutionary analyses and enables the tracing of species movements across their distributional ranges (Mohamadzade Namin et al., 2019). The mitochondrial COI gene was amplified by the Polymerase Chain Reaction (PCR) using AccuPower PCR PreMix (Bioneer, Daejeon, Korea) with the primer set TCF1 (5’-CTATCCTCAATTATTGAAATAGGAAC-3’) and a TCR2 (5’-TAGCGGCTGTGAAATAGGCTCG-3’) (Anderson and Morgan, 2007). Amplifications were conducted with 5 min initial denaturation at 95°C followed by 35 cycles of 30 s denaturation at 95°C, 30 s annealing at 50°C, 60 s extension at 72°C, and a final extension for 5 min at 72°C. Sequencing was performed commercially by Macrogen (Deajan, South Korea). All sequences were generated in both directions. The COI sequence data obtained in this study were deposited in GenBank databases under accessions OR165740- OR165787.
Phylogenetic analysis and network construction
Sequence reads were assembled using the Bioedit v7.0.5.2 (Hall, 1999). Sixty-four previously-reported COI sequences of T. mercedesae were retrieved from NCBI database (September 2022) and incorporated in the analysis (Appendix 1). The sequences were aligned, using Clustal X version 1.8 (Thompson et al., 1997). The number of haplotypes was defined using DNAsp v5 (Librado and Rozas, 2009) and the pairwise genetic divergence among the haplotypes were estimated in MEGA7. The maximum likelihood phylogenetic relationship among haplotypes were conducted in MEGA7 (Huelsenbeck and Ronquist, 2001), using the Tamura-Nei mutation model (TN93+G+I). The jModelTest (version 2.1.3) program was used to select the best nucleotide substitution model using the default parameters (Darriba et al., 2012). Tropilaelaps clareae (EF025464) was used as an outgroup to root the tree. A haplotype network was constructed using median-joining method (Bandelt et al., 1999) in Network software version 10 to infer the relationships among haplotypes and their geographical distribution.
Result
Sequence analysis and pairwise genetic divergence
COI sequences were generated for 48 samples of T. mercedesae collected from Nepal, South Korea and Uzbekistan. The generated sequences had a consistent length of 538 base pairs. No insertions or deletions were identified in the examined sequences. Notably, BLAST comparisons between our sequence dataset and T. mercedesae sequence entries in the NCBI GenBank database revealed a high degree of similarity, ranging from 99.81% to 100%. Four distinct haplotypes were retrieved from analyzed sequences. The genetic divergence observed between the generated sequences of T. mercedesae and other congeneric species revealed significant differences. Specifically, the divergence was within the range of 11.7-11.9 when compared to T. thai, 11.9-12.27 in comparison with T. clareae, and 14.5–14.68 when contrasted with T. koenigerum.
Among Korean populations, three distinct haplotypes were found. A single haplotype (H19) was found from all populations (Andong, Daegu-A, Daegu-B, Jeju) and all three Korean haplotypes were found in Daegu-B population, although the H19 were also dominant in this population (Table 1). Only one haplotype was retrieved from the analyzed sequences from Nepal and Uzbekistan (Table 1). The genetic distance among analyzed sequences were varied from 0 to 0.37.In addition, we compared the sequences generated in this research with Sixty-four previously-reported COI sequences of T. mercedesae available in the NCBI database. A total of 112 COI sequences of T. mercedesae were analyzed, and 39 distinct haplotypes were retrieved from the analyzed sequences. Fifteen haplotypes were identified from China while the number of haplotypes from Indonesia was twelve. Three haplotypes were recognized from South Korea and Vietnam and two haplotypes form Thailand, Malaysia and Sri Lanka. Only one haplotype retrieved from the studied sequences from the rest of countries (India, Nepal, Pakistan, Papua, Philippines and Uzbekistan). The highest genetic divergence was observed between the H21 from Indonesia and H28 from Sri Lanka (0.074) (Figure 1; Appendix 2). The within country genetic divergence varied between 0.002 – 0.017 among Chinese haplotypes and 0.002 – 0.022 among Indonesian haplotypes. The highest genetic divergence in Korean and Thailand was 0.004 while in Sri Lanka, Malasia and Vietnam it was 0.002, 0.002 and 0.007 respectively (Figure 1; Appendix 2).
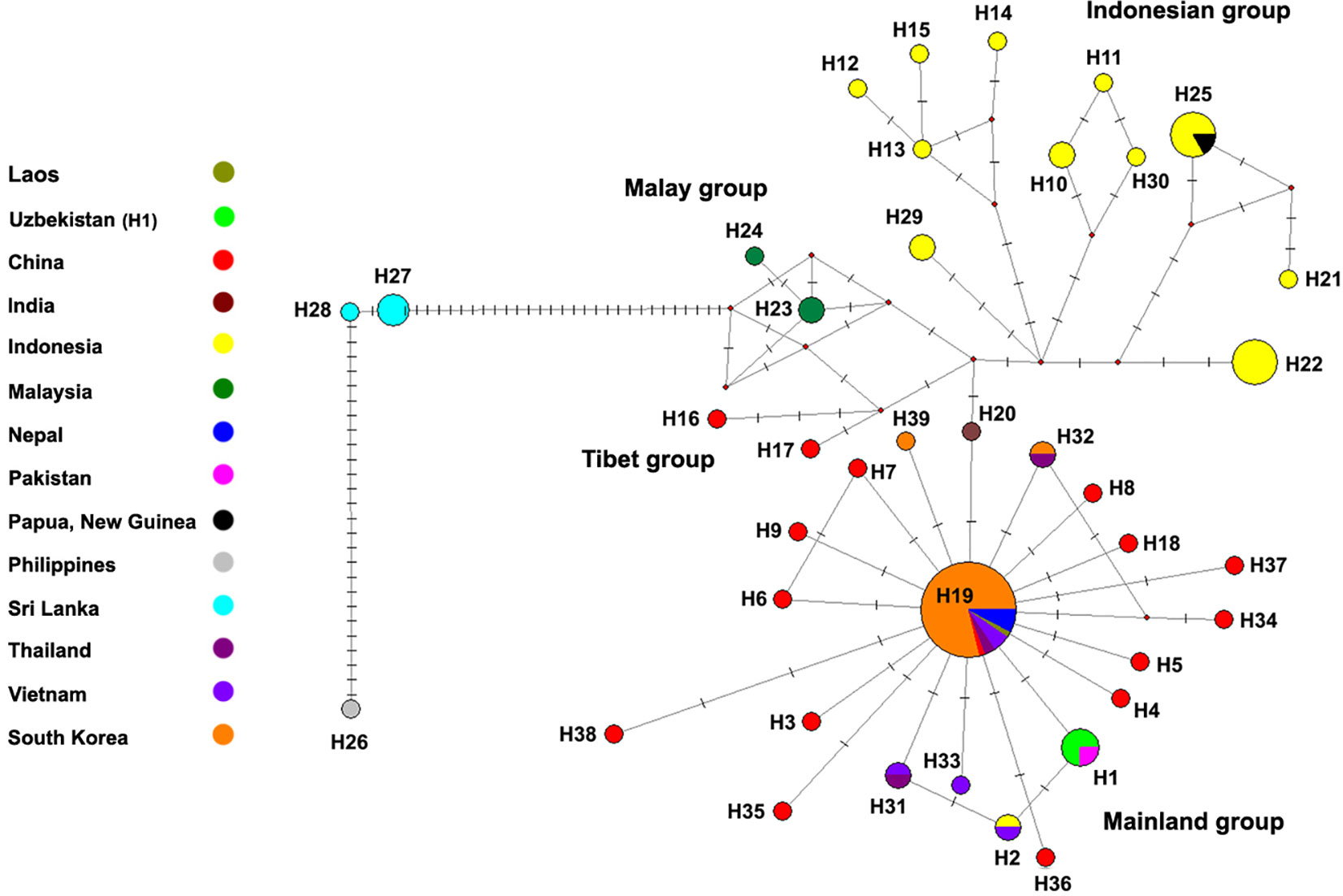
Figure 1 Median-joining haplotype network of the COI gene of the T. mercedesae. Each circle represents one haplotype. Small red dots represent median vectors and the dashes on the connectors represent the number of substitutions. The number under each circle indicates the haplotype number.
Phylogenetic analysis and network construction
Phylogenetic analysis was conducted to determine the relationships among populations of T. mercedesae based on the sequences obtained from samples collected in this study and the previously reported sequences which are available in the databases. The maximum likelihood phylogenetic tree demonstrated the occurrence of two major clades in studied populations of T. mercedesae. The genetic divergence between these two clades varied between 0.058 (between H27 from Sri Lanka and H21 from Indonesia) and 0.074 (between H28 from Sri Lanka and H23 from Malaysia). The Clade II included three haplotypes from Sri Lanka and Philippines (Palawan), however, other haplotypes from all other Asian countries are grouped in the Clade II of the phylogenetic tree. Among haplotypes of the clade I, haplotypes from Malaysia grouped in a well-supported Malay subclade (Bootstrap value [BV]=91) while other haplotypes divided into three different subclades. The Indonesian subclade (BV=50) included almost all haplotypes from Indonesia, however, H2 which is shared between Indonesia and Vietnam is grouped with other Asian mainland subclade. In this subclade, haplotypes from Borneo grouped together in a well-supported clade, while three haplotypes from Southern Indonesia islands (Java, Bali, Lombok, Sumbawa, New Guinea) are closely related to each other. Among the two haplotypes found in Sumatra, one is closely related to the haplotypes from Belitung (Figure 2). The mainland subclade (BV=74) included all haplotypes from mainland Asia except Malay and Tibet subclades. Among mainland subclade, H20 from India is distantly related with the group of other mainland haplotypes. All haplotypes which are recognized from Nepal, South Korea and Uzbekistan are also located in the mainland subclade. Among 15 Chinese haplotypes, H16 and H17 from Yunnan, are distantly related with the rest and made another Tibet subclade (BV=52).
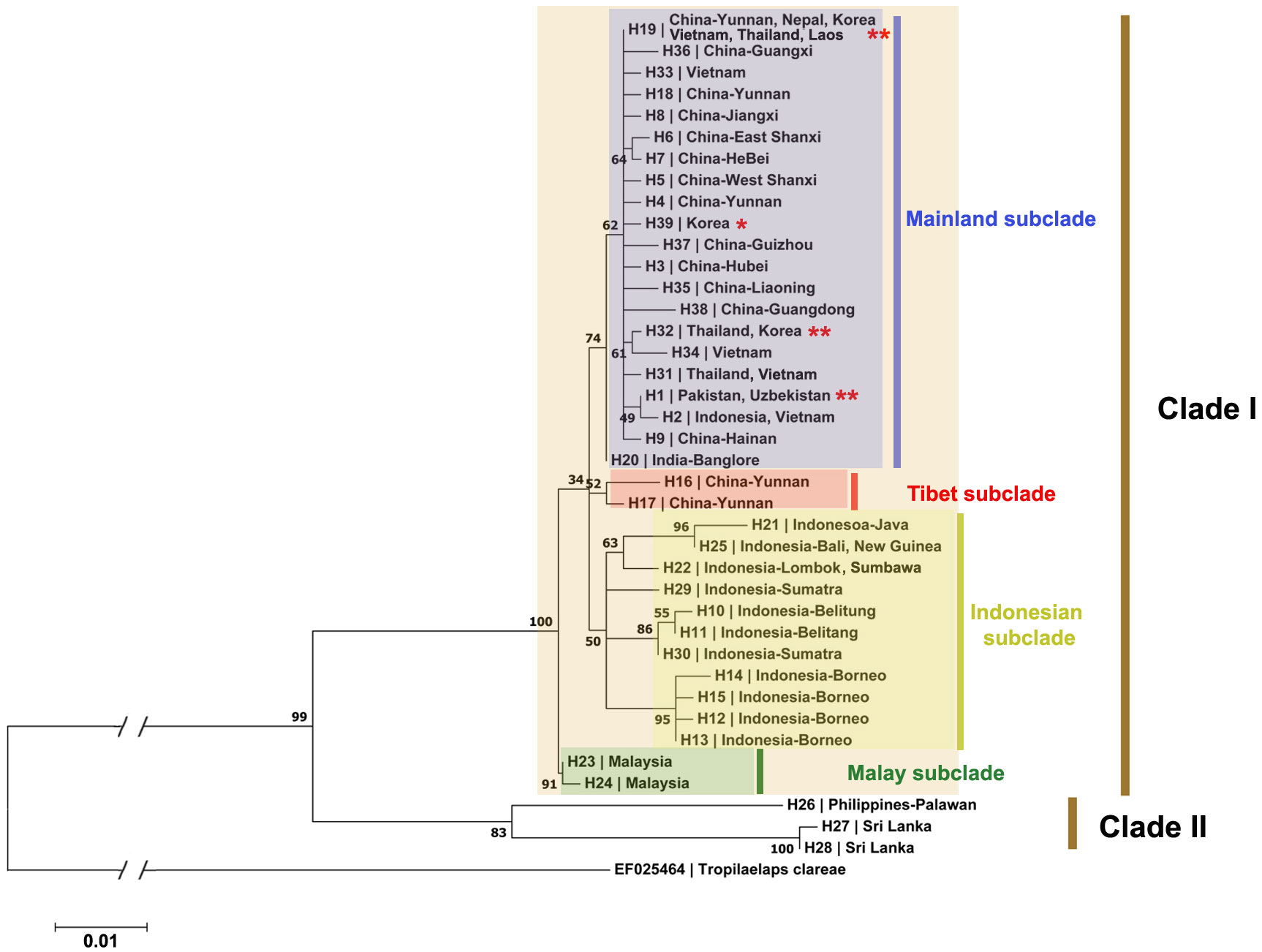
Figure 2 Maximum Likelihood phylogenetic tree of 538-bp part of COI sequences from 112 T. mercedesae sequences. The haplotypes and the localities in which the haplotypes are collected are listed in the tree. The accession numbers for the analyzed sequences are provided in Appendix 1. *: Haplotypes derived exclusively from sequences collected in this study; **: Haplotypes derived from sequences originating from both this study and GenBank.
The haplotype network also shows a large genetic distance between haplotypes from Sri Lanka and Palawan with the other recognized haplotypes of T. mercedesae in Asia. H19 which is reported from China, Nepal, South Korea, Laos and Vietnam is located in the center of the star-shaped haplotype network of mainland group and it is probably the main haplotype in East Asia, while haplotype 20 from India connected other haplotype groups to the mainland group of haplotypes (Figure 1).
Morphological variations and intraspecific differences of T. mercedesae
Following specimens were morphologically examined: South Korea: 6 females, Andong, beekeeping apiary of Andong National University, 2020.IX.15. Nepal: 2 females, Chapagaon, 2019.IX.27; 2 females, Chapagaon, 2019.IX.25. Uzbekistan: 3 females, Tashkent, Kirgiz, 2022.VIII.26. This is the first report of this species from Central Asia, Uzbekistan.
Morphological Variation: Given the absence of distinctive characteristics for distinguishing T. mercedesae from other congeneric species within the genus Tropilaelaps, a comparative analysis of specimens from Korea, Nepal, and Uzbekistan reveals significant variations across several traits. Notably, we observed variations in the degree of ventral shield sclerotization, with Korean and Nepalese populations exhibiting a higher degree of sclerotization compared to specimens from Uzbekistan. Moreover, the density of opisthogastric setae in the studied samples from Korea and Nepal was notably greater than in the Uzbekistani samples (see Figures 3A, C). Additionally, Uzbekistani specimens displayed the presence of a small ventral mucro, resembling a spin-like process, near the apex of the cheliceral movable digit, a feature absent in Korean and Nepalese specimens (see Figures 3B, D). These observed morphological distinctions may be indicative of intraspecific variations occurring in distinct geographical regions.
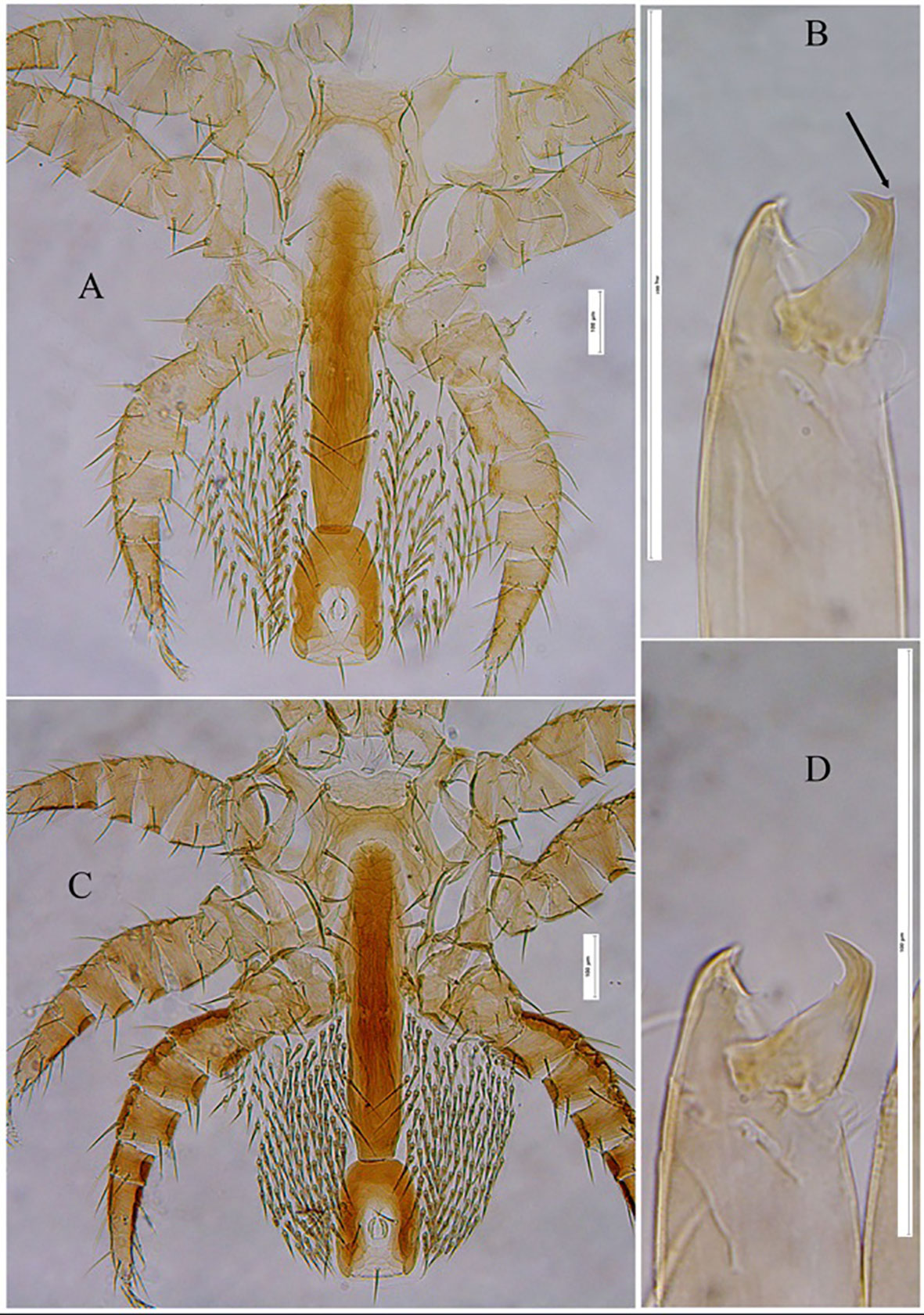
Figure 3 Tropilaelaps mercedesae female: ventral region of Idiosoma (showing different shields) and Chelicerae. (A, B) Specimen from Uzbekistan; (C, D) Specimen from Republic of Korea. The arrow indicates the spin-like process on cheliceral movable digit.
Discussion
In this study, we conducted an analysis of intraspecific genetic divergence within populations of T. mercedesae. Our research utilized sequences derived from samples collected in South Korea, Nepal, and Uzbekistan, in addition to previously archived sequences from various Asian countries available in GenBank. It’s important to note that while the samples from Nepal and Uzbekistan were obtained from single localities, those from South Korea were gathered from four different areas. Surprisingly, despite the geographical spread, samples from Daegu-A, Andong, and the isolated Jeju population all shared a common H19 haplotype. In contrast, the Daegu-B population displayed three distinct haplotypes, including the unexpected presence of H32, previously reported in Thailand, highlighting an intriguing genetic connection. All five samples from Nepal exhibited a singular haplotype, H19, which is recognized as “Mainland Asia” according to Anderson and Morgan (2007). This haplotype had previously been reported in China, Laos, Thailand, and Vietnam, making our observation of this haplotype in South Korea and Nepal noteworthy.
In the phylogenetic analysis, the relationship among Indonesian haplotypes is complex, and the identification of a main haplotype has proven challenging, likely due to the presence of isolated populations on Indonesian islands that are separated by geographical barriers. Our analysis suggests a common ancestry of Indonesian, Malay, and Tibet haplotypes, all tracing back to haplotype H20 from India (Bangalore). The distant relationship observed between Malay haplotypes and those from the mainland can be attributed to geographical barriers such as the Bilauktaung subrange and the Isthmus of Kra (Parnell, 2013). Our study also revealed that haplotypes from the Philippines and Sri Lanka are distinctly related to other Asian haplotypes. Although they fall into clade II of the phylogenetic tree, significant genetic divergence was observed between Sri Lankan haplotypes and a single Palawan haplotype (H26). This divergence suggests the potential for these mite populations to be classified as subspecies or even separate species, depending on the level of morphological differences in key characteristics.
In our investigation of the phylogenetic relationships among T. mercedesae populations in Asia, we utilized the COI gene, a widely employed mitochondrial marker with inherent advantages, including maternal inheritance and ease of amplification (Gupta et al., 2015). While COI has demonstrated its value in deciphering evolutionary relationships, it does come with the potential oversimplification of the intricate evolutionary history of T. mercedesae, particularly when considering a sequence length of approximately 500bp. To address these challenges and enhance the depth of our analysis, forthcoming studies should consider integrating both nuclear and mitochondrial genes (Fisher-Reid and Wiens, 2011) to trace the phylogenetic evolution of T. mercedesae.
The examination of detailed morphological characteristics in collected samples of T. mercedesae from South Korea, Nepal, and Uzbekistan has unveiled notable variations within their populations. Despite geographical divergence and barriers, samples from South Korea and Nepal exhibit similarities, whereas distinct morphological differences emerge when comparing Uzbekistani specimens with those from Korea and Nepal. Understanding these intraspecific morphological variations is pivotal for accurate species identification. Regrettably, the initial description of T. mercedesae by Anderson and Morgan (2007) is concise and lacks crucial details, such as the chaetotaxy of the legs, necessary for morphologically based identification. Consequently, the provided information, including both description and illustrations, proves inadequate for precise and consistent species identification. A more comprehensive and detailed morphological study of diverse T. mercedesae populations is essential. The species is closely related to the type species of the genus, T. clareae Delfinado and Baker, 1961. However, due to the absence of distinctive key characters, differentiation between these two species is impossible without knowledge of the locality and host bee of the collected sample, as highlighted by Anderson and Roberts (2013). This underscores the urgent need for a comprehensive revision to establish a robust framework for accurate identification and classification. Such a revision should encompass a thorough examination of morphological traits, including those previously overlooked, to provide a more nuanced understanding of the intricate variations within T. mercedesae populations.
Furthermore, this study also serves as the first documented presence of T. mercedesae in Uzbekistan. The samples from Uzbekistan shared a single haplotype with Pakistan, indicating this haplotype’s prevalence in Central Asia. Given the expanding global climate conditions and the potential for Tropilaelaps mites to spread, early warning systems and surveillance programs are vital to detect and monitor infestations (Pettis et al., 2013; Cont et al., 2021). Such efforts aim to enable timely interventions and control measures to protect honey bee populations. This expansion could extend beyond Asia and pose substantial threats to the apiculture industry in Europe and North America. Therefore, vigilance and preventive measures are essential. Collaborative efforts involving research institutions, beekeeping associations, and regulatory bodies are required to develop effective strategies for mitigating this threat and safeguarding honey bee populations worldwide. Raising awareness among beekeepers and promoting best hive management practices are crucial steps in this endeavor (Chantawannakul et al., 2018). Addressing the challenges posed by T. mercedesae necessitates a global, coordinated approach that combines knowledge, resources, and cooperative action.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The requirement of ethical approval was waived by Andong National University IACUC for the studies involving animals. The subjects for consideration of IACUC include all experimental spinalized animals. The bee mite is not a subject for consideration. The studies were conducted in accordance with the local legislation and institutional requirements.
Author contributions
SMN: Conceptualization, Data curation, Formal analysis, Investigation, Resources, Validation, Writing – original draft. OJ: Data curation, Investigation, Writing – review & editing. SA: Resources, Writing – review & editing. RT: Resources, Writing – review & editing. S-HK: Resources, Writing – review & editing. BK: Resources, Writing – review & editing. CJ: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The research received support from a grant to CJ via the Core research center Program of the National Research Foundation of Korea through the Ministry of Education (NRF-2018R1A6A1A03024862), RDA project (RS2023-00232847) and Uzbekistan-Collaboration Research (2022K1A3A9A05036394).
Acknowledgments
SN and CJ express their sincere appreciation to Nepal and Uzbekistan collaborators for their invaluable assistance during our expedition trips.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2024.1275995/full#supplementary-material
References
Anderson D. L., Morgan M. J. (2007). Genetic and morphological variation of bee parasitic Tropilaelaps mites (Acari: Laelapidae): New and redefined species. Exp. Appl. Acarol. 43, 1–24. doi: 10.1007/s10493-007-9103-0.
Anderson D. L., Roberts J. M. K. (2013). Standard methods for Tropilaelaps mites research. J. Apic. Res. 52, 1–16. doi: 10.3896/IBRA.1.52.4.21
Arca M., Mougel F., Guillemaud T., Dupas D., Rome Q., Perrard A., et al. (2015). Reconstructing the invasion and the demographic history of the yellow-legged hornet, Vespa velutina, in Europe. Biol. Invasions 17, 2357–2371. doi: 10.1007/s10530-015-0880-9.
Bandelt H. J., Forster P., Röhl A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036.
Buawangpong N., de Guzman L. I., Khongphinitbunjong K., Frake A. M., Burgett M., Chantawannakul P. (2015). Prevalence and reproduction of Tropilaelaps mercedesae and Varroa destructor in concurrently infested Apis mellifera colonies. Apidologie 46, 779–786. doi: 10.1007/s13592-015-0368-8.
Chantawannakul P., Ramsey S., van Engelsdorp D., Khongphinitbunjong K., Phokasem P. (2018). Tropilaelaps mite: an emerging threat to European honey bee. Curr. Opin. Insect Sci. 26, 69–75. doi: 10.1016/j.cois.2018.01.012.
Cont A. D., De Georges B., Huleux A., Duquesne V. (2021). Rapid identification of Tropilaelaps mite (Mesostigmata: Laelapidae) species using a COI barcode-HRM. J. Econ. Entomol. 114, 520–529. doi: 10.1093/jee/toaa330.
Darriba D., Taboada G., Doallo R., Posada D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772. doi: 10.1038/nmeth.2109.
de Guzman L. I., Simone-Finstrom M., Cervancia C., Tokarz P., Frake A. M. (2020). Tropilaelaps species identification and viral load evaluation of Tropilaelaps and Varroa mites and their Apis mellifera hosts in Palawan, Philippines. J. Invertebr. Pathol. 170, 107324. doi: 10.1016/j.jip.2020.107324.
de Guzman L. I., Williams G. R., Khongphinitbunjong K., Chantawannakul P. (2017). Ecology, life history, and management of tropilaelaps mites. J. Econ. Entomol. 110, 319–332. doi: 10.1093/jee/tow304.
Delfinado M. D., Baker E. W. (1961). Tropilaelaps, a new genus of mite from the Philippines (Laelapidae: Acarina). Fieldiana-Zoology 44, 53–56.
Delfinado-Baker M. (1982). New records for Tropilaelaps clareae from colonies of Apis cerana indica. Am. Bee. J. 122, 382.
Delfinado-Baker M., Underwood B. A., Baker E. W. (1985). The occurrence of Tropilaelaps mites in brood nests of Apis dorsata and A. laboriosa in Nepal, with descriptions of the nymphal stages. Am. Bee J. 125, 703–706.
Evans G. O., Till W. M. (1979). Mesostigmatid mites of Britain and Ireland (Chelicerata: Acari - Parasitiformes). An introduction to their external morphology and classification. Trans. Zool. Soc Lond. 35, 145–270. doi: 10.1111/j.1096-3642.1979.tb00059.x.
Fisher-Reid M. C., Wiens J. J. (2011). What are the consequences of combining nuclear and mitochondrial data for phylogenetic analysis? Lessons from Plethodon salamanders and 13 other vertebrate clades. BMC Evol. Biol. 11, 300. doi: 10.1186/1471-2148-11-300.
Forsgren E., de Miranda J. R., Isaksson M., Wei S., Fries I. (2009). Deformed wing virus associated with Tropilaelaps mercedesae infesting European honey bees (Apis mellifera). Exp. Appl. Acarol 47, 87–97. doi: 10.1007/s10493-008-9204-4.
Gupta A., Bhardwaj A., Sharma P., Pal Y., Kumar S. (2015). Mitochondrial DNA – a tool for phylogenetic and biodiversity search in Equines. JBES S1, 006. doi: 10.4172/2332-2543.S1-006
Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Higes M., Martin-Hernandez R., Meana A. (2010). Nosema ceranae in Europe: an emergent type C nosemosis. Apidologie 41, 375–392. doi: 10.1051/apido/2010019.
Huelsenbeck J. P., Ronquist F. (2001). MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754.
Jung C., Kim D. W., Kim J. W. (2014). Redefined species of Tropilaelaps mercedesae (Acari: Laelapidae) parasitic on Apis mellifera. Korean J. Apic. 29, 217–221. doi: 10.17519/apiculture.2014.11.29.4.217
Khongphinitbunjong K., de Guzman L. I., Tarver M. R., Rinderer T. E., Chantawannakul P. (2015). Interactions of Tropilaelaps mercedesae, honey bee viruses and immune response in Apis mellifera. J. Apic. Res. 54, 40–47. doi: 10.1080/00218839.2015.1041311.
Klein A. M., Vaissiere B. E., Cane J. H., Steffan-Dewenter I., Cunningham S. A., Kremen C., et al. (2007). Importance of pollinators in changing landscapes for world crops. P. R. Soc Lond. B. Bio. 274, 303–313. doi: 10.1098/rspb.2006.3721.
Kumar N. R., Kumar R., Mbaya J., Mwangi R. W. (1993). Tropilaelaps clareae found on Apis mellifera in Africa. Bee World 74, 101–102.
Lee M.-L., Park Y.-M., Lee M.-Y., Kim Y.-S., Kim H.-K. (2005). Density distribution of parasitic mites, Varroa destructor Anderson & Trueman and Tropilaelaps clareae Delfinado & Baker, on honeybee pupae (Apis mellifera L.) in Autumn season in Korea. Korean J. Apic 20, 103–108.
Librado P., Rozas J. (2009). DnaSP v5: software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187.
Mohamadzade Namin S., Koh Y., Osabutey A. F., Jung. C. (2019). Invasion pathway of the honeybee pest, small hive beetle, Aethina tumida (Coleoptera: Nitidulidae) in the Republic of Korea inferred by mitochondrial DNA sequence analysis. J. Asia-Pac. Entomol. 22, 963–968. doi: 10.1016/j.aspen.2019.07.008.
Nazzi F., Pennacchio F. (2018). Honey bee antiviral immune barriers as affected by multiple stress factors: A novel paradigm to interpret colony health decline and collapse. Viruses 10, 159. doi: 10.3390/v10040159.
Parnell J. (2013). The biogeography of the Isthmus of Kra region: a review. Nordic Journal of Botany 31, 1–15.
Pettis J. S., Rose R., Lichtenberg E. M., Chantawannakul P., Buawangpong N., Somana W., et al. (2013). A rapid survey technique for Tropilaelaps mite (Mesostigmata: Laelapidae) detection. J. Econ Entomol. 106, 1535–1544. doi: 10.1603/EC12339.
Potts S. G., Biesmeijer J. C., Kremen C., Neumann P., Schweiger O., Kunin W. E. (2010). Global pollinator declines; trends, impacts and drivers. Trends Ecol. Evol. 25, 345–353. doi: 10.1016/j.tree.2010.01.007.
Rosenkranz P., Aumeier P., Ziegelmann B. (2010). Biology and control of Varroa destructor. J. Invertebr. Pathol. 103, S96–S119. doi: 10.1016/j.jip.2009.07.016.
Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Truong A.-T., Yoo M.-S., Yun B.-R., Kang J. E., Noh J., Hwang T. J., et al. (2022). Prevalence and pathogen detection of Varroa and Tropilaelaps mites in Apis mellifera (Hymenoptera, Apidae) apiaries in South Korea. J. Apic. Res. 62 (1), 1–9. doi: 10.1080/00218839.2021.2013425
Walter D. E., Krantz G. W. (2009). “Collecting, rearing and preparing specimens,” in A Manual of Acarology, 3rd Edition. Eds. Krantz G. W., Walter D. E. (Texas Tech University Press, Lubbock, Texas), 83–95.
Wongsiri S., Tangkanasing P., Sylvester H. A. (1989). “The resistance behavior of Apis cerana against Tropilaelaps claraea,” in Proceedings of the first Asia-Pacific conference of entomology, Chiang Mai. 25–34.
Keywords: mtDNA, South Korea, Nepal, Uzbekistan, genetic variation, climate change, invasive species
Citation: Mohamadzade Namin S, Joharchi O, Aryal S, Thapa R, Kwon S-H, Kakhramanov BA and Jung C (2024) Exploring genetic variation and phylogenetic patterns of Tropilaelaps mercedesae (Mesostigmata: Laelapidae) populations in Asia. Front. Ecol. Evol. 12:1275995. doi: 10.3389/fevo.2024.1275995
Received: 11 August 2023; Accepted: 05 February 2024;
Published: 22 February 2024.
Edited by:
Daniele Salvi, University of L’Aquila, ItalyReviewed by:
Michael Lattorff, University of KwaZulu-Natal, South AfricaMaéva Angelique Techer, College Station, United States
Copyright © 2024 Mohamadzade Namin, Joharchi, Aryal, Thapa, Kwon, Kakhramanov and Jung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuleui Jung, Y2p1bmdAYW5kb25nLmFjLmty
 Saeed Mohamadzade Namin
Saeed Mohamadzade Namin Omid Joharchi
Omid Joharchi Sunil Aryal
Sunil Aryal Ratna Thapa4
Ratna Thapa4 Chuleui Jung
Chuleui Jung