
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
EDITORIAL article
Front. Ecol. Evol. , 14 November 2023
Sec. Biogeography and Macroecology
Volume 11 - 2023 | https://doi.org/10.3389/fevo.2023.1325165
This article is part of the Research Topic Spatial Ecology of Cells Living in Micro-Structured Environments View all 7 articles
Editorial on the Research Topic
Spatial ecology of cells living in micro-structured environments
Communities of cells make up microbiomes as well as tissues of multi-cellular organisms. These communities are spatially explicit and therefore to understand their structure and how they assemble into functional units it is crucial to understand their spatial ecology (Tilman and Kareiva, 1998). To understand ecological systems special attention should be given to how patterns observable at one scale are the product of processes operating at another (Levin, 1992). Neither tissues of metazoa, which are made of eukaryotic cells, nor microbiomes which consist of tight associations between prokaryotes and eukaryotes are an exception to this framework. For this reason, landscape/spatial ecology approaches to cancer, where metastatic tumors are seen as spatial ecologies embedded in the metazoan physiology (ecosystem), are opening new avenues of research and treatment (Pienta et al., 2008; Keymer and Marquet, 2014; Amend et al., 2016). Inspired by these studies of cancer cell communities, and to further expand the application of ecology to spatially distributed cell systems in general, the aim of this Research Topic is to put a diverse collection of studies under the same focus. Namely, how the micron-scale spatial structure of the supporting environment (habitat landscape), operating at the scale of single cells, influences community assembly and functionality at the scales of collectives of cells composing tissues and microbiomes.
At the scale of single cells, tissues composed of eukaryotic cells as well as microbial communities are all spatially explicit ecologies occurring at a given location. In these loci, the micron-scale spatial structure of their environment determines the ecological niche (Grinnell, 1917) underlying their biological functioning. At a larger scale, that of a habitat landscape, cell death and birth/differentiation and migration in the context of tissue renewal processes or meta-community colonization-extinction dynamics occurring in microbiomes are also of critical importance to understand long-term structure. As we study these systems, the spatial, temporal, and phylogenetic grain and extent (Wiens, 1989; Ladau and Eloe-Fadrosh, 2019) of our observations are of crucial consideration. In Figure 1 we show a sketch of habitat locations (lower panel; green circles) which are created (and destroyed) by existing spatial gradients in the environment (top panel) and together compose a temporally varying, spatially distributed landscape (Keymer et al., 2000).
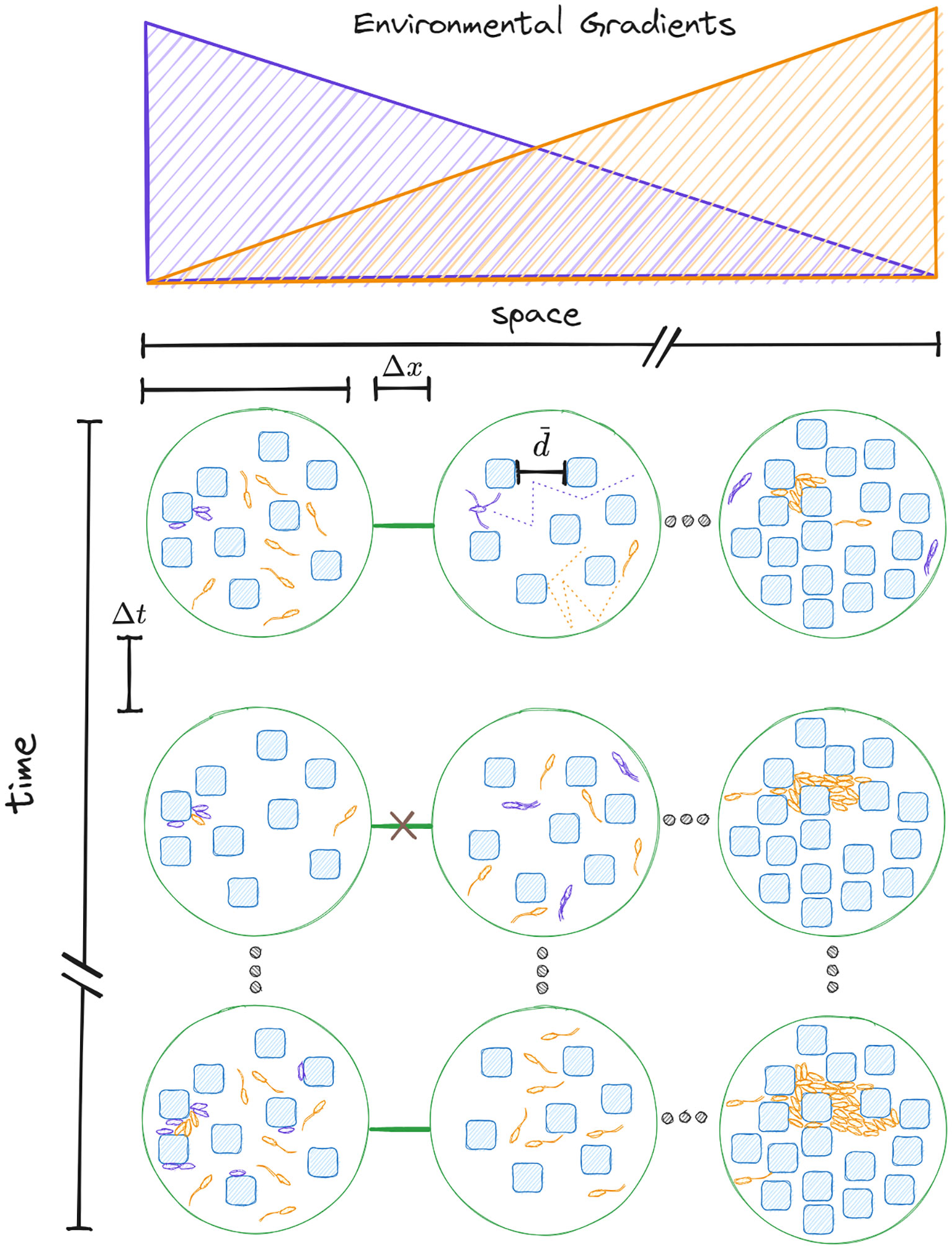
Figure 1 The spatial, temporal, as well as phylogenetic grain and extent of a cell ecology. Habitat patches (green circles) have an intra-patch characteristic porosity , as well as a scale for inter-patch adjacency relationship via ecological corridors (green line) connecting neighboring patches some distance (Δx) apart. The ability or inclination to trans-locate via ecological corridors is often a time dependent feature of the landscape, as in the case of varying nutrient gradients. Colonization-extinction processes for a generalist (Orange) and specialist (Purple) lifehistory strategy represent different scales of phylogenetic resolution co-occurring in this dynamic habitat landscape. Determining the appropriate domains of scale is dependent on the research questions in mind. For studying single cell migration, fine temporal and spatial grain is needed (See top middle patch). To study colonization-extinction dynamics of metapopulations in habitat landscapes a larger extent is necessary.
Overall the present topic addressed how micron-scale spatial structure of the environment influences the ecology of the cells inhabiting it. We present a diverse Research Topic spanning the three domains of life and encompassing differing levels of ecological organization, from the autecology of single cells to populations and communities.
In line with the perspective of the Grinnelian Niche (Grinnell, 1917), the approach of autecology considers the relationship of organisms (cells) as individuals and their biotic and abiotic environment. Investigations in this field focus primarily on the adaptations, requirements and tolerances of individuals with respect to their surroundings. In turn how do the physio-chemical characteristics of the external environment shape the physiology, morphology and behavior of individuals and secondarily, the distributions of populations in space and time (Figure 1). In the present topic two studies illustrate how this perspective is of significant value.
For instance, Kasahara et al. implement microfabrication techniques to develop and characterize a microfluidics cultivation device allowing them to precisely control environmental oxygen availability. They demonstrate the capabilities of this newly integrated on-chip environment by following the single cell physiological and growth dynamics of Escherichia coli in aerobic and anaerobic conditions. E. coli is a facultative anaerobe (Stolper et al., 2010), thus preferential growth conditions include adequate oxygen for aerobic respiration. Extending the results of this microfluidic approach may glean an insight into the role of oxygen gradients in complex spatially structured communities like biofilms (Kannan et al., 2023).
For planktonic E. coli the search for ideal oxygen conditions is also of significant importance. This is manifest in the costly investment of the signal transduction network of the chemotaxis system including oxygen specific receptors (Berg, 2004). By capturing the correct spatial and temporal grain and extent the careful observer can facilitate connections between single cell foraging behavior and population distributions. Locally, and under sufficiently long temporal extent, depletion of oxygen by present aerobic respirators will create conditions for ecological succession of redox processes (Chen et al., 2017).
Differences in physical structure can also induce shifts in the physiological and behavioral response of cells. In eukaryotic cells this feedback between cell and environment makes up the stem cell niche (Li and Xie, 2005). In the present topic, Kimmle et al. demonstrate how varying the amount of gelatin nanofibers in the microenvironment of mouse pluripotent, embryonic stem cells can impact migration, dynamic deformation and adhesion properties. In general, sparse gelatin nanofiber surfaces permit faster dispersal and the least restriction on dynamic cell deformation. Connecting these results to the longer-term biological function of stem cells including self-renewal and cell differentiation will be of great importance for characterizing single stem cell niches as well as describing tissues and organs as networks of spatially repeating stem cell niches (see Figure 1 in Keymer and Marquet, 2014).
Scaling up from the autecology of single cells to populations new organizational phenomena come to the fore. For multicellular eukaryotes, this can include morphogenesis and cancer progression. A detailed review of how inter-cellular adhesion plays a role in such processes is presented in this topic by Montel et al.
For prokaryotes such as E. coli, traveling population waves emerge as a macroscopic consequence of chemotaxis driven motility behavior of individual cells (Adler, 1969; Saragosti et al., 2011). These waves drive migration in the direction of nutrient gradients. How spatial heterogeneity influences single cell foraging (Bhattacharjee and Datta, 2019) and population level processes (Wolfe and Berg, 1989; Mattingly and Emonet, 2022) is a subject of great interest. In the present topic, Wetherington et al. consider how variance of ecological corridor properties and thus landscape connectivity shift the long-term distribution of E. coli metapopulations. Discerning the processes leading to distributional shifts as a result of landscape connectivity will require a much smaller temporal grain. Interpreting how finer-scale processes propagate to this mesoscopic (landscape-level) spatial scale will include relating individual foraging behavior to population level migration. Stepping back to see the bigger picture, macroecological results acquired with larger spatial grain and phylogenetic extent (Grilli, 2020) seem to convey more general system properties, suggesting a crossover from mesoscopic to macroscopic domains of scale.
Relevant to this discussion, an example of landscape connectivity dynamics (Keymer et al., 2000) at play is addressed in this topic by the work of Bian et al. where they study the effects of pulsed inputs of labile organic carbon (LOC) on community productivity with respect to soil moisture and available native soil organic matter (SOM). Acting as a stand-in as a connectivity parameter in a more generic sense (Stauffer and Aharony, 2018) it is not entirely surprising that moderate levels of water holding capacity leads to optimal proportions of LOC : SOM utilization. Their work adds further evidence to the importance of moisture content as a key indicator of connectivity and productivity in soil landscapes (Tecon and Or, 2017). In this same light, in the present topic, Keuschnig et al. investigate the relationship between soil aggregate size and microbial community composition. Their work uses a micro-fractionation approach which allows them to maintain and filter microbial communities by their respective soil aggregate sizes. This work pertains to much of what has been discussed above, including oxygen and other nutrient gradients, spatial structure as an environmental filter and gauge for migration capabilities.
In the current Research Topic, we collect six ostensibly disparate studies of cell communities that can be brought together under the perspective of spatial biology and landscape ecology. We hope this topic provides insights on the powerful conceptual framework ecology has to offer to understand an otherwise diverse set of biological systems from a common perspective.
MW: Writing – original draft, Writing – review & editing. PG: Writing – review & editing. JK: Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. MW acknowledges the financial support of the James S. McDonnell Foundation Postdoctoral Fellowship and NIH Grant R35-GM138354. PG acknowledges the support of the Hungarian National Research, Development and Innovation Fund under Grant Number OTKA K 116516. JK acknowledges the financial support from Shenzhen X-Institute and from ANID—Millennium Science Initiative Program—NCN19_170.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adler J. (1969). Chemoreceptors in bacteria: Studies of chemotaxis reveal systems that detect attractants independently of their metabolism. Science 166, 1588–1597. doi: 10.1126/science.166.3913.1588
Amend S. R., Roy S., Brown J. S., Pienta K. J. (2016). Ecological paradigms to understand the dynamics of metastasis. Cancer Lett. 380, 237–242. doi: 10.1016/j.canlet.2015.10.005
Bhattacharjee T., Datta S. S. (2019). Bacterial hopping and trapping in porous media. Nat. Commun. 10, 2075. doi: 10.1038/s41467-019-10115-1
Chen J., Hanke A., Tegetmeyer H. E., Kattelmann I., Sharma R., Hamann E., et al. (2017). Impacts of chemical gradients on microbial community structure. ISME J. 11, 920–931. doi: 10.1038/ismej.2016.175
Grilli J. (2020). Macroecological laws describe variation and diversity in microbial communities. Nat. Commun. 11, 4743. doi: 10.1038/s41467-020-18529-y
Grinnell J. (1917). The niche-relashionships of the california trasher. Auk 34, 427–433. doi: 10.2307/4072271
Kannan H., Sun P., Çağlar T., Yao P., Taylor B. R., Sahu K., et al. (2023). Spatiotemporal development of growth and death zones in expanding bacterial colonies driven by emergent nutrient dynamics. bioRxiv, 2023–2008. doi: 10.1101/2023.08.27.554977
Keymer J. E., Marquet P. A. (2014). The complexity of cancer ecosystems. Frontiers Ecol Evol. Complex (Copit-Arxives). doi: 10.7910/DVN/AIXLUY
Keymer J. E., Marquet P. A., Velasco-Hernández J. X., Levin S. A. (2000). Extinction thresholds and metapopulation persistence in dynamic landscapes. Am. Nat. 156, 478–494. doi: 10.1086/303407
Ladau J., Eloe-Fadrosh E. A. (2019). Spatial, temporal, and phylogenetic scales of microbial ecology. Trends Microbiol. 27, 662–669. doi: 10.1016/j.tim.2019.03.003
Levin S. A. (1992). The problem of pattern and scale in ecology: The Robert H. Macarthur award lecture. Ecology 73, 1943–1967. doi: 10.2307/1941447
Li L., Xie T. (2005). Stem cell niche: structure and function. Annu. Rev. Cell Dev. Biol. 21, 605–631. doi: 10.1146/annurev.cellbio.21.012704.131525
Mattingly H. H., Emonet T. (2022). Collective behavior and nongenetic inheritance allow bacterial populations to adapt to changing environments. Proc. Natl. Acad. Sci. 119, e2117377119. doi: 10.1073/pnas.2117377119
Pienta K. J., McGregor N., Axelrod R., Axelrod D. E. (2008). Ecological therapy for cancer:Defining tumors using an ecosystem paradigm suggests new opportunities for novel cancer treatments. Trans. Oncol. 1, 158–164. doi: 10.1593/tlo.08178
Saragosti J., Calvez V., Bournaveas N., Perthame B., Buguin A., Silberzan P. (2011). Directional persistence of chemotactic bacteria in a traveling concentration wave. Proc. Natl. Acad. Sci. 108, 16235–16240. doi: 10.1073/pnas.1101996108
Stauffer D., Aharony A. (2018). Introduction to percolation theory. (London, UK: Taylor and Francis).
Stolper D. A., Revsbech N. P., Canfield D. E. (2010). Aerobic growth at nanomolar oxygen concentrations. Proc. Natl. Acad. Sci. 107, 18755–18760. doi: 10.1073/pnas.1013435107
Tecon R., Or D. (2017). Biophysical processes supporting the diversity of microbial life in soil. FEMS Microbiol. Rev. 41, 599–623. doi: 10.1093/femsre/fux039
Tilman D., Kareiva P. (1998). Spatial Ecology: The Role of Space in Population Dynamics and Interspecific Interactions (Princeton: Princeton University Press). doi: 10.1515/9780691188362
Keywords: cell communities, spatial biology, landscape ecology, ecology, cell biology
Citation: Wetherington MT, Galajda P and Keymer JE (2023) Editorial: Spatial ecology of cells living in micro-structured environments. Front. Ecol. Evol. 11:1325165. doi: 10.3389/fevo.2023.1325165
Received: 20 October 2023; Accepted: 03 November 2023;
Published: 14 November 2023.
Edited and Reviewed by:
Peter Convey, British Antarctic Survey (BAS), United KingdomCopyright © 2023 Wetherington, Galajda and Keymer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Miles T. Wetherington, bWlsZXN3ZXRoQGdtYWlsLmNvbQ==; Juan E. Keymer, anVhbi5rZXltZXJAeC1pbnN0aXR1dGUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.